Abstract
Introduction: We describe the clinical and microbiologic courses of two patients with ventricular assist device infections secondary to Corynebacterium striatum treated with daptomycin. In both cases, the pathogen was initially susceptible to daptomycin (minimum inhibitory concentration [MIC] <0.125 mg/L) but became resistant (MIC >256 mg/L) during therapy. Methods: The clonal nature of the isolates was determined by pulse-field gel electrophoresis (PFGE). Daptomycin binding was assessed by fluorescence microscopy using daptomycin–boron-dipyrromethene (bodipy). Induction and stability of daptomycin resistance were assessed by culturing strains in the presence of low concentrations of daptomycin or passage of resistant strains on daptomycin-free medium and repeat MIC testing, respectively. Results: PFGE revealed that resistant clinical isolates were genetically indistinguishable from their parent strains, but the two pairs were unrelated to each other. The resistant strains had 7.5–15 times lower binding of daptomycin-bodipy compared to the related susceptible strains (p ≤ 0.0002). High-level daptomycin resistance (MIC >256 mg/L) was generated in vitro for both susceptible parent strains after overnight culture in the presence of daptomycin. One of the resistant strains maintained a high-level resistance phenotype up to 5 days of passage on daptomycin-free medium, whereas the other strain reverted back to a susceptible phenotype (MIC = 0.38 mg/L) after one passage on daptomycin-free medium, with a concomitant increase in daptomycin binding. Conclusions: High-level daptomycin resistance in C. striatum was readily generated in vitro and during the course of therapy in these patients. This resistance appears to be mediated by reduced daptomycin binding. Providers should be cautious about using long-term daptomycin monotherapy for C. striatum infections.
Introduction
Corynebacterium striatum is a gram-positive bacillus that is a common constituent of human skin flora. In recent years, C. striatum has been recognized as an important opportunistic pathogen that has been implicated in a variety of invasive infections.1,4,10,13 These infections frequently involve the presence of medical devices, such as pacemakers, prosthetic heart valves, or left ventricular assist devices (LVAD).3,4 In addition, clinical isolates of C. striatum are frequently resistant to a variety of antimicrobials, limiting treatment options.5,8,13
Invasive infections secondary to multidrug-resistant C. striatum have been successfully managed with daptomycin-based therapy.1,11 Two recent reports, however, have described the emergence of high-level daptomycin resistance in C. striatum strains isolated from endovascular infections.6,13 These reports suggest that high-level daptomycin resistance can be selected rapidly in vitro, but little is known about the mechanism of resistance. The objectives of this study were, therefore, to describe the clinical and microbiological courses of two cases of LVAD infections in which initially daptomycin-susceptible strains of C. striatum developed high-level resistance during daptomycin therapy and to characterize the resistance phenotype of these strains in vitro.
Case Reports
Patient No. 1
A 67-year-old man with ischemic cardiomyopathy had an LVAD (HeartMate II; Thoratec) placed 3 years earlier as destination therapy.12 He developed 2 days of erythema and swelling in his epigastrium, which progressed despite oral amoxicillin–clavulanate therapy. On presentation, his temperature was 37.8°C and he had brown drainage from his driveline exit site. Ultrasound revealed a complex fluid collection in the epigastrium that tracked to the driveline.
Due to a history of a severe vancomycin reaction, the patient was started on daptomycin at a dose of 6 mg/kg/day. Blood cultures were negative for growth after 5 days of incubation. After 5 days of daptomycin, he had incision and drainage of an abscess at the inferior portion of the sternal wound. Intraoperative cultures were positive for the growth of a presumptive Corynebacterium species that was definitively identified as C. striatum by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MALDI Biotyper, software version 3.1; Bruker Daltonics). This isolate was susceptible to daptomycin with a minimum inhibitory concentration (MIC) of 0.0625 mg/L, as determined by Etest®.
After 16 days of daptomycin therapy, he developed fever and leukocytosis. A single blood culture yielded C. striatum. This isolate was now resistant to daptomycin with an MIC >256 mg/L (Table 1). Therapy was switched to linezolid, and subsequent blood cultures were sterile. The patient continued suppressive treatment with linezolid for 4 months before transitioning to hospice care.
Table 1.
MICs of Daptomycin-Susceptible Parent Strains (T48082 and W40308) and Daptomycin-Resistant Derivatives (S32472 and W49297)
| MIC (mg/L) | ||||
|---|---|---|---|---|
| Patient No. 1 | Patient No. 2 | |||
| T48082 | S32472 | W40308 | W49297 | |
| Ampicillin | 2 | 2 | 32 | 32 |
| Ceftriaxone | 8 | 8 | >32 | >32 |
| Ciprofloxacin | >32 | >32 | >32 | >32 |
| Clindamycin | >256 | >256 | >256 | >256 |
| Daptomycin | 0.064 | >256 | 0.125 | >256 |
| Erythromycin | 16 | 8 | >256 | 32 |
| Gentamicin | 4 | 4 | 0.064 | 0.5 |
| Linezolid | 0.25 | 0.5 | — | 0.25 |
| Penicillin | 1 | 0.5 | >32 | 16 |
| Tetracycline | 64 | 64 | 64 | 32 |
| Vancomycin | 0.5 | 0.5 | 0.5 | 1 |
MICs were determined by Etest.® Daptomycin MICs were confirmed by macrobroth dilution.
MIC, minimum inhibitory concentration.
Patient No. 2
A 67-year-old man with nonischemic cardiomyopathy had an LVAD (HeartMate II) placed as destination therapy. In the subsequent months, he had bacteremia and driveline exit site infection with both methicillin-susceptible Staphylococcus aureus and a coagulase-negative Staphylococcus spp. After a 6-week course of therapy with vancomycin followed by nafcillin, he was maintained on oral doxycycline for suppression. Five months post-LVAD placement, he developed a subxyphoid abscess overlying the driveline and two of two blood cultures and intraoperative debridement cultures grew C. striatum that was susceptible to vancomycin (MIC 0.5 mg/L) and daptomycin (MIC 0.125 mg/L) but resistant to tetracycline (MIC ≥64 mg/L) (Table 1).
The patient was maintained on vancomycin for 7 months as suppressive therapy. He then developed a recurrent subxyphoid abscess, and cultures from an operative debridement grew multiple strains of methicillin-susceptible S. aureus, one of which had a vancomycin MIC of 4 mg/L. Cefazolin was added to the vancomycin therapy. After subsequent bacteremia with Pseudomonas aeruginosa, antibiotics were changed to daptomycin at 6 mg/kg/day and oral levofloxacin. Three months after the initiation of daptomycin, he had recurrent bacteremia with C. striatum and P. aeruginosa. This C. striatum isolate was now resistant to daptomycin with an MIC >256 mg/L. He developed recurrent infection with P. aeruginosa and progressive multiple organ dysfunction and was transitioned to hospice care.
Materials and Methods
Pulse-field gel electrophoresis and susceptibility testing
Strain clonality was assessed by pulse-field gel electrophoresis (PFGE), as described previously, with slight modifications following restriction digest with swaI (performed at ARUP Laboratories).9 Band patterns were assessed visually for differences. Strains were considered indistinguishable if no differences in banding were visible and closely related, possibly related, or different if 2–3, 4–6, or >7 differences in band pattern were detected, respectively. Susceptibility testing was performed by Etest (bioMérieux) according to the manufacturer's instructions using Mueller–Hinton agar containing 5% sheep blood (Remel) that was incubated at 35°C under ambient air conditions. In addition to Etest, daptomycin resistance was confirmed by macrobroth dilution using cation-adjusted Mueller–Hinton broth (Remel) with 5% lysed horse blood supplemented with calcium to a final concentration of 50 mg/L.
Daptomycin–boron-dipyrromethene binding
Each strain was prepared as previously described14 before imaging with a Zeiss LSM 510 META inverted fluorescence microscope (Keck Imaging Centre, University of Washington). Images from each sample were collected using identical camera exposures. ImageJ software was used to subtract background fluorescence and measure the fluorescence intensities of individual cells and cell clusters. Daptomycin–boron-dipyrromethene (bodipy) binding to each resistant derivative was compared to that of the respective parent strain. In addition, strain W40308 and its daptomycin-resistant derivative W49297 were grown in the presence of subinhibitory concentrations of ampicillin (0.5× MIC) overnight before binding experiments to evaluate the impact of ampicillin exposure in daptomycin binding. At least 50 cells were used to quantitatively assess the binding for each strain. To normalize the binding signal to the quantity of cells per measurement frame, binding was reported as the ratio of the daptomycin–bodipy to 4′,6-diamidino-2-phenylindole (DAPI) signal intensity. Values are reported as mean ± standard error. Statistical significance was assessed using Student's two-tailed t test.
In vitro resistance selection and phenotype stability
The daptomycin-susceptible parent strains from each pair (W40308 and T48082) were grown in the presence of daptomycin to assess the proclivity toward resistance selection. A few colonies from each strain were inoculated into 5 ml of calcium supplemented Mueller–Hinton broth (Difco) to create a 1 McFarland suspension. Daptomycin was added to achieve a concentration of 1 mg/L, and the cells were incubated with gentle shaking at 37°C. After 24 hours, 100 μl of the cell suspension was inoculated into calcium-supplemented brain heart infusion agar (BHIA; Difco) containing 1, 2, or 4 mg/L of daptomycin and 50 mg/L of elemental calcium and incubated at 37°C overnight. The daptomycin MICs of surviving colonies were reevaluated by Etest the following day. Conversely, resistant strains (W49297 and S32472) were serially passed on BHIA in the absence of daptomycin and the Etest MICs checked daily for up to 5 days to assess the stability of the resistance phenotype.
Results
PFGE and susceptibility testing
PFGE analysis revealed that strains S32472 and W49297 were isogenic derivatives of T48082 and W40308, respectively (Fig. 1). MICs of the two daptomycin-susceptible parent strains (T48082 and W40308) and the daptomycin-resistant derivatives (S32472 and W49297) are shown in Table 1.
FIG. 1.
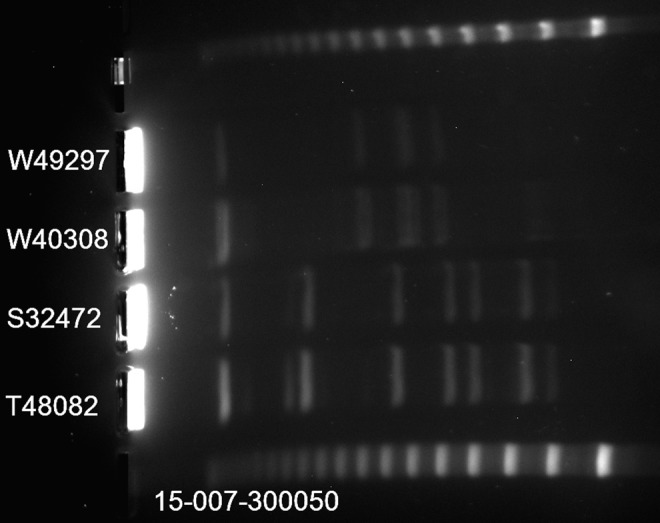
Pulse-field gel electrophoresis banding pattern after restriction digest with swaI. Top and bottom lanes of the lambda phage size marker ladder (48.5 kb concatemers).
Daptomycin-bodipy binding
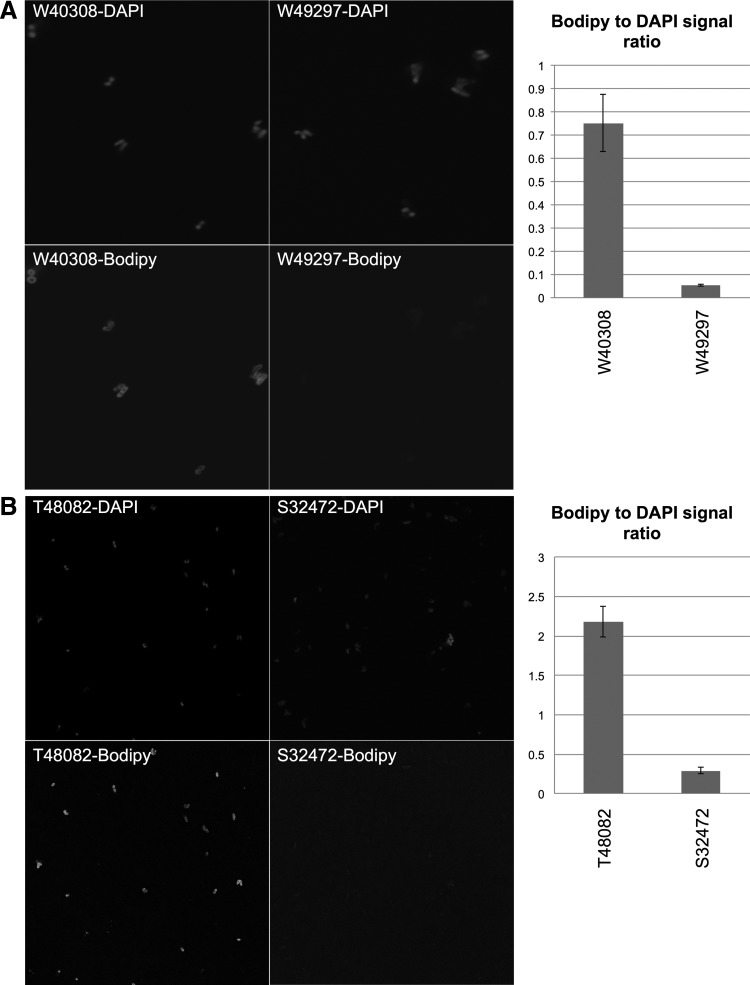
For patient No. 1, daptomycin binding was 7.5-fold lower to the resistant derivative (S32472) than to the daptomycin-susceptible parent strain (T48082) (ratio of daptomycin-bodipy to DAPI signal 0.29 ± 0.03 [mean ± standard error] and 2.18 ± 0.19, respectively, p = 0.0001 by Student's two-tailed t-test) (Fig. 2). For patient No. 2, daptomycin binding was 15-fold lower to the resistant derivative (W49297) than to the daptomycin-susceptible parent strain (W40308) (ratio of daptomycin-bodipy to DAPI signal 0.05 ± 0.003 and 0.75 ± 0.12, respectively, p = 0.0002) (Fig. 2). The addition of ampicillin had no significant effect on daptomycin binding to the resistant strain (W49297; 0.05 vs. 0.07, without and with ampicillin, respectively, p = 0.164) but nearly doubled daptomycin binding to the susceptible strain (W40308; 0.75 vs. 1.45, p = 0.0005).
FIG. 2.
4′,6-Diamidino-2-phenylindole (DAPI) and daptomycin–boron-dipyrromethene (bodipy) binding to (A) daptomycin-susceptible parent strain W40308 and its daptomycin-resistant derivative W49297 from patient No. 2 and (B) daptomycin-susceptible parent strain T48082 and its daptomycin-resistant derivative S32472 from patient No. 1. Representative fields are shown along with the bodipy:DAPI signal ratio taken from at least 50 organisms.
In vitro resistance selection and phenotype stability
After overnight exposure to 1 mg/L of daptomycin, both susceptible parent strains were able to grow on BHIA containing 4 mg/L of daptomycin. Colonies isolated from these plates demonstrated a daptomycin MIC >256 mg/L by Etest. When these experiments were repeated in the presence of ampicillin at 0.5× MIC in addition to 1 mg/L of daptomycin, daptomycin resistance was delayed by 48 hours in W40308 and 96 hours in T48082. When resistance did emerge, the daptomycin MIC was >256 mg/L by Etest. After subculture on daptomycin-free medium, the resistant isolate S32472 lost its resistance phenotype and the daptomycin MIC fell to 0.38 mg/L. Daptomycin-bodipy binding to this now susceptible isolate also increased to a level similar to that seen with the susceptible parent strain (data not shown). Resistant strain W49297, in contrast to strain S32472, maintained an MIC >256 mg/L after subculture on daptomycin-free BHIA for all 5 days tested.
Discussion
In this study, we observed that daptomycin resistance in C. striatum can develop rapidly in vitro and confirmed that this resistance phenotype can be unstable. The lack of stability seen here differs from a prior study, in which all daptomycin-resistant isolates maintained their resistant phenotype after multiple rounds of subculture in the absence of daptomycin.6 This discrepancy may be related, in part, to different experimental conditions, but the variable stability of daptomycin resistance may suggest that daptomycin resistance in C. striatum does not always emerge by homogeneous mechanisms.
Regarding the mechanism(s) involved in this resistance, a prior study found that resistance was associated with the loss of daptomycin-induced membrane depolarization. In that study, resistance occurred without changes in cell surface charge, which is a common phenotypic feature documented in many daptomycin-nonsusceptible staphylococci and enterococci.13 We demonstrated that daptomycin resistance in C. striatum was associated with a dramatic reduction in daptomycin binding and that loss of the resistance phenotype was concomitant with an increase in daptomycin binding, suggesting that antibiotic binding was key to resistance development. The genetic features of daptomycin-resistant C. striatum remain unknown, but the rapid development of in vitro resistance and the >12 log2 dilution difference in susceptible and resistant phenotypes in our C. striatum isolates are different from the general pattern of the development of resistance for either S. aureus or Enterococcus spp., suggesting that potential membrane alterations may be regulated differently in C. striatum. Recently, high-level daptomycin resistance in Streptococcus mitis has been linked to both a reduction in and redistribution of cardiolipin.7 We were unable to evaluate changes in phospholipid content in our strains, however, this may be an area for future investigation.
C. striatum is an emerging pathogen associated with infections of medical devices.4 In this study, daptomycin-resistant C. striatum developed in two patients with LVAD infections and prolonged exposure to daptomycin. With our report, three of the four instances of daptomycin-resistant C. striatum reported in the literature have been isolated from patients with LVAD infections.6 Due to inherent problems with device removal, the management of LVAD infections is often limited to antimicrobial therapy for sustained periods against biofilm-embedded organisms.3 Consequently, LVAD infections are an ideal setting for the emergence of antimicrobial resistance.
Daptomycin is an attractive option for the treatment of medical device infections. It is bactericidal against a wide array of gram-positive bacteria, including multiply-resistant gram-positive pathogens such as methicillin-resistant S. aureus, coagulase-negative Staphylococcus spp., and other common pathogens involved in medical device infections. In addition, daptomycin is well tolerated and administered once daily, making it convenient for outpatient therapy.2 However, the rapid development of daptomycin resistance in vitro and the clinical failure of daptomycin monotherapy in our patients with C. striatum infections should raise some concerns when considering this agent as monotherapy for the treatment of this pathogen.6,13
Both of our patients had been receiving daptomycin at a dose of 6 mg/kg/day. There are no data to guide the optimal dosing of daptomycin for C. striatum infections. Of the two reports describing successful daptomycin therapy for C. striatum endocarditis, one patient received 10 mg/kg/day1 and the other received 6 mg/kg/day in combination with rifampin.11 In the patients reported to develop resistance on daptomycin therapy, one occurred when the patient was receiving 6 mg/kg/day13 and the other when the patient was receiving 8 mg/kg/day.6 It is not known if higher doses of daptomycin would have any benefit in preventing the development of resistance in C. striatum; however, given the concentration-dependent pharmacodynamics of daptomycin and its excellent safety profile, it would be reasonable to consider. Alternatively, combination therapy may help prevent the emergence of daptomycin resistance. In this study, we observed that subinhibitory concentrations of ampicillin nearly doubled daptomycin binding to susceptible C. striatum and delayed the selection of resistance under the conditions tested. The addition of ampicillin also appeared to reduce the prevalence of daptomycin-resistant clones because only two colonies (∼20 CFU/ml) were isolated from resistance screening plates versus nearly confluent growth, which was observed in the absence of ampicillin. Synergy between beta-lactams and daptomycin has been well documented in other gram-positive pathogens. However, unlike many strains of daptomycin-nonsusceptible S. aureus and enterococci, the addition of ampicillin did not appear to improve daptomycin binding to daptomycin-resistant C. striatum.
In conclusion, high-level daptomycin resistance in C. striatum develops in patients during the course of therapy with significant clinical consequences and is readily reproduced in vitro. We demonstrate for the first time that this resistance is due, in part, to reduced daptomycin binding. We recommend caution in the long-term use of daptomycin to treat C. striatum infections, particularly in the setting of an indwelling device such as an LVAD.
Acknowledgments
We thank Cubist Pharmaceuticals, a wholly owned subsidiary of Merck, for providing daptomycin-bodipy powder used in the drug-binding experiments. This work was funded internally by the University of Washington. W.O.H. is supported, in part, by an institutional training grant (T32 AI007044) awarded through the National Institute of Allergy and Infectious Diseases. B.J.W. has received research funding from Merck and Actavis.
Disclosure Statement
No competing financial interests exist.
References
- 1.Fernandez Guerrero M.L., Molins A., Rey M., Romero J., and Gadea I. 2012. Multidrug-resistant Corynebacterium striatum endocarditis successfully treated with daptomycin. Int. J. Antimicrob. Agents 40:373–374 [DOI] [PubMed] [Google Scholar]
- 2.He W., Zhang Y., Chen H., Zhao C., and Wang H. 2014. Efficacy and safety of daptomycin for the treatment of infectious disease: a meta-analysis based on randomized controlled trials. J. Antimicrob. Chemother. 69:3181–3189 [DOI] [PubMed] [Google Scholar]
- 3.Koval C.E., and Rakita R. 2013. Ventricular assist device related infections and solid organ transplantation. Am. J. Transplant. 13(Suppl 4):348–354 [DOI] [PubMed] [Google Scholar]
- 4.Lee P.P., Ferguson D.A., Jr., and Sarubbi F.A. 2005. Corynebacterium striatum: an underappreciated community and nosocomial pathogen. J. Infect. 50:338–343 [DOI] [PubMed] [Google Scholar]
- 5.Mashavi M., Soifer E., Harpaz D., and Beigel Y. 2006. First report of prosthetic mitral valve endocarditis due to Corynebacterium striatum: successful medical treatment. Case report and literature review. J. Infect. 52:e139–e141 [DOI] [PubMed] [Google Scholar]
- 6.McElvania TeKippe E., Thomas B.S., Ewald G.A., Lawrence S.J., and Burnham C.A. 2014. Rapid emergence of daptomycin resistance in clinical isolates of Corynebacterium striatum… a cautionary tale. Eur. J. Clin. Microbiol. Infect. Dis. 33:2199–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra N.N., Alvarez D.N., Seeprsaud R., Tran T.T., Garcia-de-la-Maria C., Miro J.M., Rybak M.J., Arias C.A., Sullam P.M., and Bayer A.S. 2015. Phenotypic and Genotypic Mechanisms of Daptomycin-resistance (DAP-R) in Streptococcus mitis. The 55th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, CA [Google Scholar]
- 8.Oliva A., Belvisi V., Iannetta M., Andreoni C., Mascellino M.T., Lichtner M., Vullo V., and Mastroianni C.M. 2010. Pacemaker lead endocarditis due to multidrug-resistant Corynebacterium striatum detected with sonication of the device. J. Clin. Microbiol. 48:4669–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otsuka Y., Ohkusu K., Kawamura Y., Baba S., Ezaki T., and Kimura S. 2006. Emergence of multidrug-resistant Corynebacterium striatum as a nosocomial pathogen in long-term hospitalized patients with underlying diseases. Diagn. Microbiol. Infect. Dis. 54:109–114 [DOI] [PubMed] [Google Scholar]
- 10.Rufael D.W., and Cohn S.E. 1994. Native valve endocarditis due to Corynebacterium striatum: case report and review. Clin. Infect. Dis. 19:1054–1061 [DOI] [PubMed] [Google Scholar]
- 11.Shah M., and Murillo J.L. 2005. Successful treatment of Corynebacterium striatum endocarditis with daptomycin plus rifampin. Ann. Pharmacother. 39:1741–1744 [DOI] [PubMed] [Google Scholar]
- 12.Slaughter M.S., Rogers J.G., Milano C.A., Russell S.D., Conte J.V., Feldman D., Sun B., Tatooles A.J., Delgado R.M., 3rd., Long J.W., et al. 2009. Advanced heart failure treated with continuous-flow left ventricular assist device. N. Engl. J. Med. 361:2241–2251 [DOI] [PubMed] [Google Scholar]
- 13.Tran T.T., Jaijakul S., Lewis C.T., Diaz L., Panesso D., Kaplan H.B., Murray B.E., Wanger A., and Arias C.A. 2012. Native valve endocarditis caused by Corynebacterium striatum with heterogeneous high-level daptomycin resistance: collateral damage from daptomycin therapy?. Antimicrob. Agents Chemother. 56:3461–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werth B.J., Steed M.E., Ireland C.E., Tran T.T., Nonejuie P., Murray B.E., Rose W.E., Sakoulas G., Pogliano J., Arias C.A., et al. . 2014. Defining daptomycin resistance prevention exposures in vancomycin-resistant Enterococcus faecium and E. faecalis. Antimicrob. Agents Chemother. 58:5253–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]