Abstract
Water deficit is one of the most important environmental stresses limiting plant growth and crop yield. While the identification of many key factors involved in the plant water deficit response has greatly increased our knowledge about the regulation system, the mechanisms underlying dehydration tolerance in plants are still not well understood. In our current study, we investigated the roles of the key flowering time regulator, OsGIGANTEA (OsGI), in the osmotic stress tolerance in rice. Results showed that mutation of OsGI conferred tolerance to osmotic stress generated by polyethylene glycol (PEG), increased proline and sucrose contents, and accelerated stomata movement. In addition, qRT-PCR and microarray analysis revealed that the transcript abundance of some osmotic stress response genes, such as OsDREB1E, OsAP37, OsAP59, OsLIP9, OsLEA3, OsRAB16A, and OsSalT, was significantly higher in osgi than in WT plants, suggesting that OsGI might be a negative regulator in the osmotic stress response in rice.
Keywords: GIGANTEA, osmotic stress, water deficiency, stomata, rice
Introduction
Water availability is a critical environmental factor for plant growth and development. To cope with water shortages, plants have developed multiple mechanisms to preserve cellular water homeostasis, including morphological, physiological and biochemical modulations that enhance water uptake and reduce water loss (Hincha et al., 2002; Chaves et al., 2003; Villadsen et al., 2005; Valliyodan and Nguyen, 2006; Hadiarto and Tran, 2011). A promising approach to enhance plant tolerance to dehydration stresses is the modulation of genes responsive to water deficiency (Yamaguchi-Shinozaki et al., 1995; Shinozaki et al., 1998). In recent years, several instances of crosstalk between the osmotic stress response and other signaling pathways, such as abscisic acid (ABA) signaling and flowering time regulation, have been identified (Ikegami et al., 2009; Fujita et al., 2011). For example, genes involved in flowering time regulation, such as phytochrome B (phyB) and timing of CAB expression 1 (toc1), have been shown to be negative regulators in the response to dehydration conditions (Legnaioli et al., 2009; Liu et al., 2012).
GIGANTEA (GI) is regarded as a key component of flowering time regulation in many plant species (Fowler et al., 1999; Hayama et al., 2003; Hecht et al., 2007; Higuchi et al., 2011). In Arabidopsis, GI, CONSTANS (CO), and FLOWERING LOCUS T (FT) control photoperiodic flowering responses (Fowler et al., 1999; Mouradov et al., 2002; Srikanth and Schmid, 2011). Overexpression of GI promotes early flowering while GI mutants develop a large rosette of leaves and “gigantic” size under long day conditions due to a prolonged vegetative growth phase (Koornneef et al., 1991; Fowler et al., 1999; Park et al., 1999). The rice homolog of GI, OsGI, is also a circadian gene controlling diurnal rhythms of the global transcriptome and carbohydrate metabolism (Izawa et al., 2011), and mutation of OsGI causes late flowering under short day condition (Hayama et al., 2003; Izawa et al., 2011).
Besides its role in the regulation of flowering time and circadian rhythms, GI is also involved in processes such as sucrose signaling, starch accumulation and stress tolerance (Kurepa et al., 1998; Fowler and Thomashow, 2002; Dalchau et al., 2011; Kim et al., 2013; Riboni et al., 2013; Mishra and Panigrahi, 2015). In the past decade, GI has been shown to function in the response to several abiotic stressors including cold, salt, drought and oxidative stresses (Kurepa et al., 1998; Cao et al., 2005; Kim et al., 2013; Riboni et al., 2013). Expression of Arabidopsis GI was induced 5–8 fold by cold stress (Fowler and Thomashow, 2002). GI was proposed to regulate cold acclimation through a C-repeat Binding proteins (CBFs) independent pathway (Cao et al., 2005). GI mutants displayed increased cold sensitivity because the protective role of GI in cold tolerance was reduced (Cao et al., 2005). Recent evidence suggests that GI is a negative regulator for the tolerance to salt stress (Kim et al., 2013). Under normal conditions, Arabidopsis GI interacts with salt overly sensitive 2 (SOS2) to prevent SOS2-mediated SOS1 phosphorylation and activation (Kim et al., 2013). Under salt stress condition, GI is degraded and the freed SOS2 can interact with the Ca2+-activated sensor of sodium ions, SOS3, to activate and stabilize SOS1 (Kim et al., 2013; Park et al., 2013). Therefore, the gi mutant confers enhanced salt tolerance due to the constitutive activation of SOS1 (Shi et al., 2000; Kim et al., 2013; Park et al., 2013).
In addition, GI has been shown to regulate the response to oxidative stress which increases GI abundance and promotes flowering (Qian et al., 2014). GI mutants exhibit increased activation of superoxide dismutase (SOD) and ascorbate peroxidase (APX) and tolerance to a redox cycling agent, parquat, and H2O2 (Kurepa et al., 1998; Cao et al., 2006). Furthermore, it was recently discovered that GI can lead to early flowering and drought tolerance via the abscisic acid (ABA)-dependent activation of florigens under long day condition (Riboni et al., 2013).
Despite the increasing knowledge of GI's role in Arabidopsis, little is known about the biochemical and molecular functions of its rice homolog, OsGI, in response to water deficits. In this study, we investigated the role of OsGI in osmotic stress in rice. We determined that mutation of OsGI confers tolerance to osmotic stress generated by polyethylene glycol (PEG). Mutation of OsGI results in increased proline and sucrose contents and more rapid stomata movement. In addition, transcription analysis revealed that the expression of many genes involved in drought response is altered in osgi plants.
Materials and methods
Plant materials, growth conditions, and stress treatments
The rice (Oryza sativa) osgi mutant, osgi, was obtained from the Rice Tos17 insertion mutant database at the Rice Genome Resource Center, Japan. Osgi and its wild type control, cv. Nipponbare, were used for all physiological experiments. For complementation of the osgi mutant, the OsGI coding sequence was expressed in a modified pCAMBIA1300 vector under control of the Cauliflower Mosaic virus 35S promoter (Wang et al., 2009). The construct was introduced into osgi mutants using a Agrobacterium tumefaciens-mediated transformation method (Wang et al., 2009). A modified culture medium containing 1.425 mM NH4NO3, 0.323 mM NaH2PO4, 0.513 mM K2SO4, 1.643 mM MgSO4, 0.998 mM CaCl2, 0.125 mM EDTA-Fe(II), 0.075 μM (NH4)6Mo7O24, 0.25 mM NaSiO3, 0.009 mM MnCl2, 0.019 μM H3BO3, 0.155 μM CuSO4, and 0.152 μM ZnSO4was used at pH 5.5 for hydroponic experiments(Yoshida et al., 1976; Li et al., 2014). Rice plants were grown in a growth room with a 12 h light/12 h dark cycle, a daytime temperature of 30°C and nighttime temperature of 22°C.
Rice seeds were first germinated in tap water for 2 days before being transferred into the culture media. For osmotic stress tolerance experiments, seedlings were transferred to culture media containing 21% polyethylene glycol (PEG) 8000. For gene expression analysis, plants were transferred to culture media containing 18% PEG 8000. For microarrays, leaves from 14-day-old osgi and WT seedlings grown under normal growth condition were used.
Measurement of proline and sucrose contents
Proline content was measured as previously described (Bates et al., 1973). Briefly, approximately 50 mg fresh leaves from 15-day-old rice plants grown in hydroponics were harvested for analysis. Samples were homogenized in 5 ml 3% sulfosalicylic acid, incubated at 100°C for 10 min and pelleted by centrifugation. The resulting supernatant was collected, mixed 1:1:1 (2 mL each) with glacial acetic acid and acidic ninhydrin, and incubated at 100°C for 30 min. The chromophore was toluene extracted, and absorption values for the solution were detected at 520 nm wavelength. Proline concentration was determined using a standard concentration curve and adjusted to the fresh weight of leaves.
For sucrose analysis, approximately 200 mg fresh leaves were homogenized in 2 ml deionized water and centrifuged at 4°C for 10 min. The resulting supernatant was incubated at 100°C for 3 min, and sucrose content was determined using ion chromatography (ICS-3000, DIONEX).
RNA preparation, quantitative real-time PCR
For microarray, total RNA was extracted from plant samples using RNeasy mini kits (Qiagen, USA, http://www.qiagen.com) according to the manufacturer's instructions. For quantitative reverse transcription PCR (qRT-PCR), total RNA was extracted from leaf samples using TRIzol Reagent (Invitrogen, CA, USA) according to the manufacturer's instructions.
First-strand cDNAs were synthesized from 4 μg total RNA using SuperScript II reverse transcriptase (Invitrogen), and qRT-PCR was performed using LightCycler 480 SYBR Green I Master Kit (Roche Diagnostics, USA) on a LightCycler480 thermocycler. The amplification steps and quantitative analysis of relative expression levels was performed as previously described (Li et al., 2014). RNA samples were collected from three biological replicates. Each sample was analyzed using three biological replicates and normalized to the housekeeping gene OsACTIN. All primers used for qRT-PCR analyses are listed in Supplementary Table 1.
Microarray analyses
For the microarray analysis, leaves were sampled from plants grown for 14 days after germination (DAG) under normal condition. Genes were considered significantly differentially expressed when p < 0.05 and Posterior Probability of Differential Expression (PPDE) >0.95 with a 2 fold cut-off to correct for false discovery rates (Dalchau et al., 2011). Differentially expressed genes (up- or down-regulated) between osgi and WT were compared to a previously published microarray (Jain et al., 2007). The raw microarray data files have been supplied as Supplementary File 2.
Stomata conductance (gs) and transpiration rate (Tr) measurements
Stomata conductance (gs) and transpiration rate (Tr) were recorded in osgi and WT plants inside a growth chamber on fully expanded leaves using an Li-6400 portable gas-exchange system (LI-COR) according to the manufacturer's instructions. All measurements were conducted 9 h after lights-on in saturating light (1500 μmol m−2 s−1) with 400 μmol mol−1 CO2 surrounding the leaf. Leaf temperature for all measurements was approximately 30°C (ambient temperature), and the leaf-to-air vapor pressure deficit (VPD) was kept abetween 1.5–2.5 kPa. To measure stomata movement in response to PEG treatment, osgi and WT plants were grown in hydroponics under normal condition for 5 weeks and then treated with 18% PEG 8000 at 4, 6, and 8 h after lights-on.
Statistical analysis
Statistical significance was determined using the SAS program with t-test (SAS Institute Inc., http://www.sas.com).
Results
Morphological analysis of osgi mutant rice plants
The TOS17 insertion mutant of osgi was obtained and characterized. As shown in Figure 1, the cyclic expression pattern of OsGI, which peaks at dusk in wild type (WT) plants, was completely abolished in osgi mutant plants (Figure 1A). Circadian rhythm genes, such as OsRFT1 and OsHd3a, were also suppressed in osgi plants (Supplementary Figure 1). Osgi mutants displayed a growth inhibited phenotype (Figure 1B; Itoh and Izawa, 2011). Shoot and root lengths of 30-day-old osgi plants were reduced by 35 and 15% compared to WT plants, respectively, resulting in an increased root-to-shoot ratio (Figure 1C).
Figure 1.
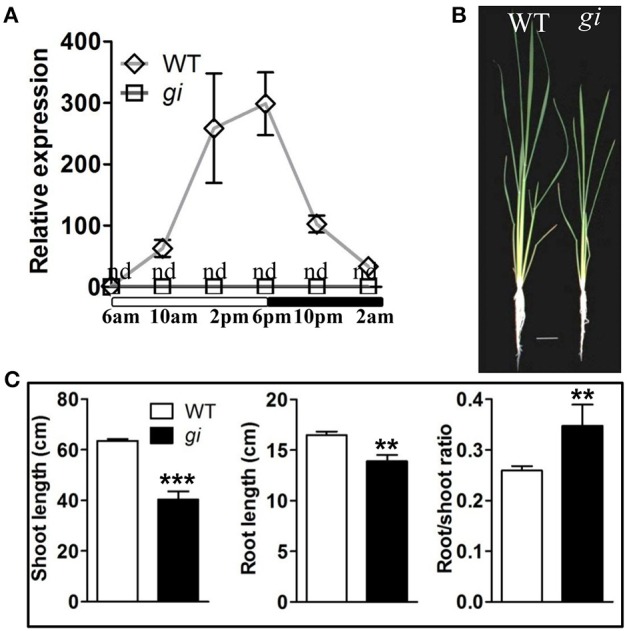
Characterization of the growth phenotype of WT and osgi plants. (A) Expression of OsGI in WT and osgi (gi) plants throughout the day. Unfilled and filled bars below the x-axis indicate times of lights-on and lights-off, respectively. Gene expression was normalized to the expression level of OsACTIN. (B) WT and osgi plants were grown hydroponically under normal growth conditions for 30 days. (C) Shoot length, root length and root/shoot ratio of WT and osgi plants. All data represent the mean of three biological replicates, with error bars indicating SD. Significant differences relative to the corresponding WT strain are indicated with asterisks (***P < 0.001; and **P < 0.01). Bars = 4 cm. nd, not detectable.
To confirm that the growth defects were caused by the mutation in OsGI, genetic complementation was carried out by introducing the OsGI coding sequence under control of the CaMV35S promoter into osgi mutants in two transgenic events, L1 and L2. As shown in Figure 2A, the overexpression of OsGI in the osgi background rescued the growth defect observed in the mutant. The relative transcript abundance of OsGI, measured by reverse-transcription PCR (RT-PCR), was significantly higher in L1 and L2 than in either WT or osgi plants (Figure 2B, Supplementary Figure 2). Compared to osgi plants, the root and shoot lengths were increased in L1 and L2 while the root-to-shoot ratio decreased (Figure 2C). Based on these results, the L1 and L2 plants were used as overexpression lines in later experiments.
Figure 2.
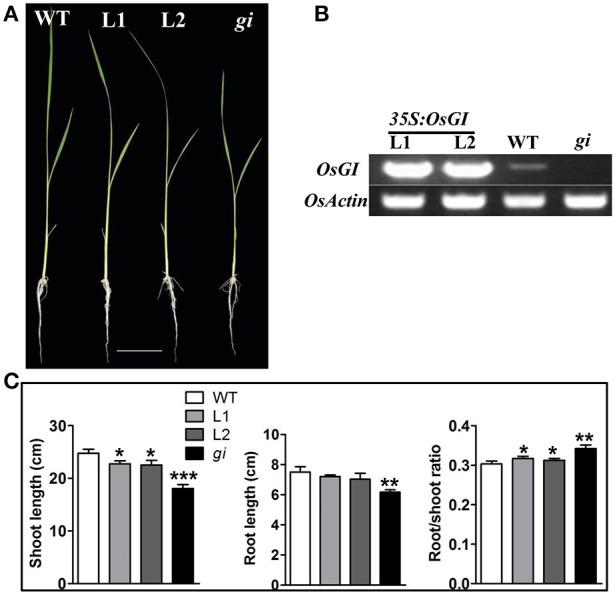
Characterization of the growth phenotype in osgi-complemented plants. (A) WT, osgi mutant (gi) and two OsGI-complemented osgi mutant lines (L1 and L2) grown hydroponically under normal conditions for 14 days. (B) OsGI expression in WT, osgi, L1 or L2 plants 4 h after lights-on measured by RT-PCR. Plants were grown under normal conditions for 14 days. (C) Comparison of shoot length, root length and root/shoot ratio of WT, L1, L2 and osgi plants. All data represent the mean of three biological replicates, with error bars indicating SD. Significant differences relative to the corresponding WT strain are indicated with asterisks (***P < 0.001; **P < 0.01; and *P < 0.05). Bars = 4 cm.
Mutation of OsGI improved plant osmotic stress tolerance
OsGI has been reported to be a molecular switch connecting flowering time regulation with multiple stress-response pathways, such as oxidative and salinity stresses (Kurepa et al., 1998; Kim et al., 2013). To investigate whether GI influences the tolerance to osmotic stress, WT and osgi plants were grown for 15 days under normal growth condition and then treated with 21% PEG 8000 for 3 days. As shown in Figure 3A, osgi mutants displayed improved osmotic stress tolerance compared to WT. After 3 days of exposure to high concentrations of PEG, WT plants were completely wilted while osgi plants remained healthy. PEG treatment decreased the fresh weight of WT and osgi plants by 55 and 30%, respectively, compared to the corresponding plants grown under normal condition (Figures 3B,C). While the water content in both WT and osgi leaves decreased during PEG treatment, osgi plants maintained a significantly higher water content than WT (Figure 3D). However, osgi plants used for the analysis of resistance to drought in soil showed the same phenotype as WT (data not shown).
Figure 3.
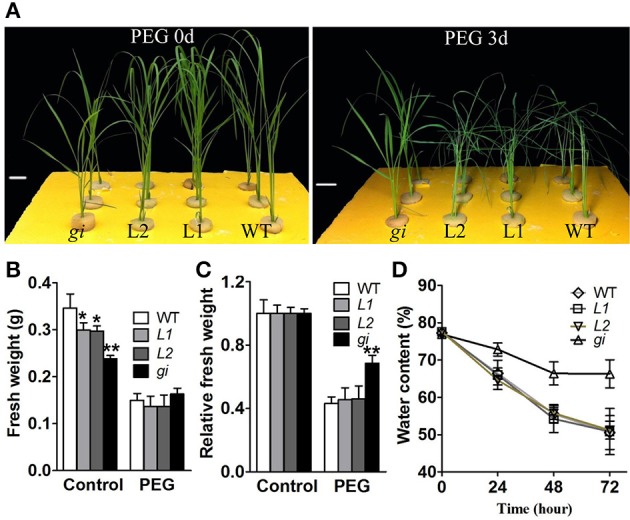
Growth characteristics of WT, L1, L2 and osgi plants under PEG-treated conditions. (A) 15-day-old WT, L1, L2 and osgi seedlings (left) were treated in 21% PEG-conditioned culture media for 3 days (right). (B) Absolute fresh weight or (C) fresh weight relative to the strain-matched control of WT, L1, L2 and osgi plants grown in control or PEG-treated culture media. (D) Water contents in WT, L1, L2 and osgi leaves were measured 24, 48, and 72 h after PEG treatment. All data represent the mean of three biological replicates, with error bars indicating SD. Significant differences relative to the corresponding WT strain are indicated with asterisks (**P < 0.01; and *P < 0.05). Bars = 4 cm.
To exclude the possibility that the improved osmotic stress tolerance in osgi plants was due to its relatively smaller size (Figures 1B, 3A), additional osmotic stress tests were performed using size-matched WT plants and osgi seedlings at 11 DAG (WT) and 14 DAG (osgi), respectively (Figures 4A,B). Results showed that osgi plants still exhibited much higher tolerance to PEG treatment than WT plants (Figure 4A).
Figure 4.
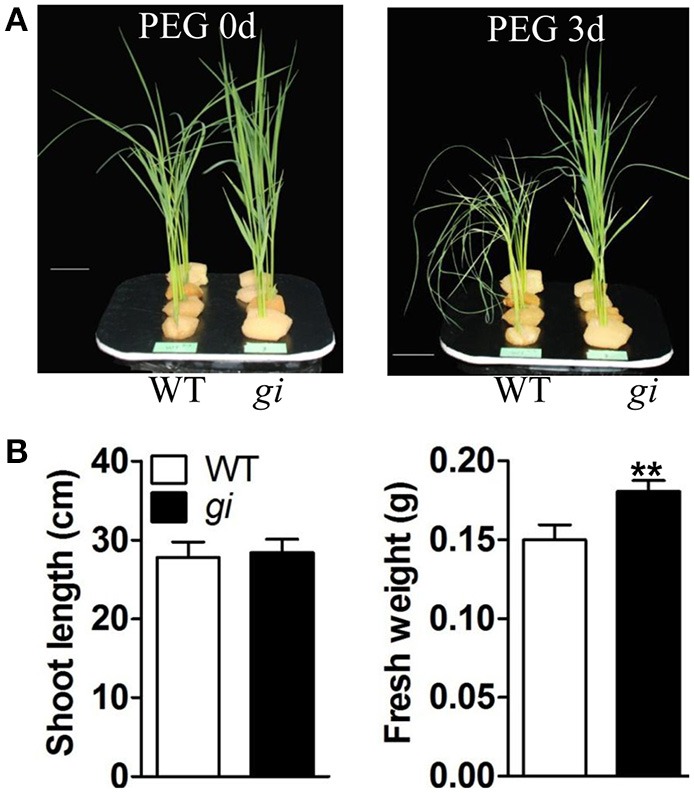
Growth characteristics of size-matched WT and osgi plants in response to PEG treatment. (A) 11-day-old WT and 14-day-old osgi seedlings, displaying the same plant height, were treated with 21% PEG condition for 3 days. (B) Shoot length and fresh weight of WT and osgi plants in (A). All data represent the mean of three biological replicates with error bars indicating SD. Significant differences relative to the corresponding WT strain are indicated with asterisks (**P < 0.01). Bars = 4 cm.
To confirm these results, the OsGI overexpression lines L1 and L2 were analyzed for their tolerance to osmotic stress. Results showed that L1 and L2 lines displayed increased sensitivity to osmotic stress under PEG exposure (Figure 3) compared to osgi mutants. However, we did not detect any differences between WT and OsGI overexpressing plants in response to PEG treatment (Figure 3). These results confirmed that OsGI functions as a negative factor in the osmotic tolerance of rice.
Mutation of OsGI increases proline and sucrose contents in rice
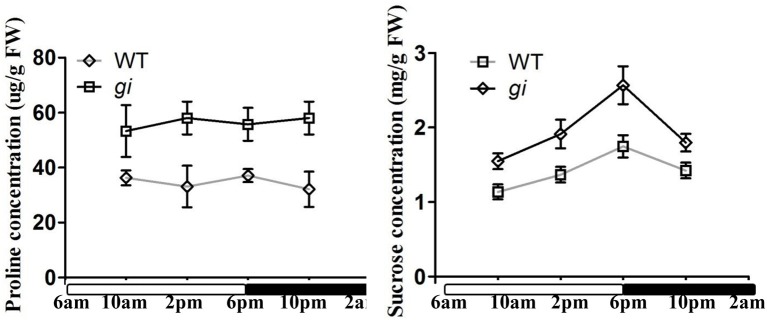
It is generally accepted that under abiotic stress conditions, plants often accumulate compatible osmolytes to maintain osmotic balance and protect their subcellular structures from damage. Several studies have shown that the accumulation of free proline is positively correlated to plant tolerance to dehydration stress (Shinozaki and Yamaguchi-Shinozaki, 2007; Xiong et al., 2012). In order to assess whether mutation of OsGI can enhance the accumulation of osmotic protectants, the free proline content in WT and osgi plants was analyzed (Figure 5). Leaves from plants grown under normal conditions were collected at four different time points at 15 DAG and the proline content measured. At all times, the proline content was higher in osgi than in WT plants. For example, 8 h after lights-on, the proline levels in osgi plants were approximately 1.7 times higher than in WT plants (Figure 5). Moreover, while sucrose levels increased throughout the day and decreased at night in both WT and osgi plants, osgi plants had higher sucrose levels than WT plants at all times (Figure 5). The sucrose content in L1 and L2 plants was not altered compared to WT plants (Supplementary Figure 3). These results suggest that osgi plants possess a much higher osmotic potential, which could be beneficial in protecting plants against osmotic stress.
Figure 5.
Analysis of proline and sucrose contents in WT and osgi plants. Proline and sucrose contents in leaves of 15-day-old WT and osgi plants grown under normal conditions. All data represent the mean of three biological replicates, with error bars indicating SD. The unfilled and filled bars below the x-axis indicate times of lights-on and lights-off, respectively.
Stomata movement in WT and osgi plants
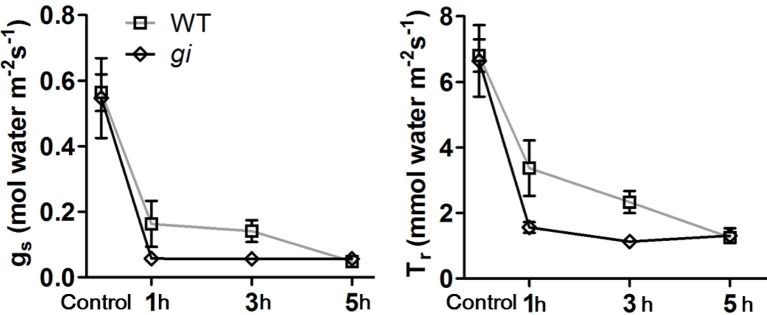
In Arabidopsis, GI has been shown to be involved in the regulation of stomata opening (Ando et al., 2013). To investigate whether OsGI participates in the regulation of stomata movement under dehydration stress conditions, the stomata conductance (gs) and transpiration rate (Tr) in WT and osgi plants were measured using LI-6400. 35-day-old WT and osgi seedlings, grown under normal conditions or treated with 18% PEG 8000 at 4, 6, and 8 h after lights-on, were used for the experiment. The gs and Tr were recorded 9 h after lights-on (1, 3, and 5 h after PEG treatment, respectively) using an Li-6400.
Under normal growth conditions, there was no difference in the gs and Trbetween WT and osgi plants (Figure 6). PEG treatment reduced the gs and Tr in both WT and osgi plants. The gs decreased by 70, 75, and 90% in WT plants after PEG treatment for 1, 3, and 5 h, respectively. In contrast, the gs in osgi seedlings decreased rapidly by about 90% after only 1 h of PEG treatment and stayed at a similarly low level after 3 and 5 h of PEG exposure. The change in Tr in response to PEG treatment followed a similar pattern as gs (Figure 6). These results indicate that under osmotic stress conditions, stomata movement changes faster in osgi mutant plants than in WT plants.
Figure 6.
Analysis of stomata conductance (gs) and transpiration rate (Tr) in WT and osgi plants. Stomata conductance (gs) and transpiration rate (Tr) in WT and osgi plants after PEG treatment for 1, 3, and 5 h. 35-day-old WT and osgi plants grown under normal conditions or treated with 18% PEG 8000 at 4, 6, and 8 h after lights-on, respectively, were used for the experiment. The gs and Tr were recorded in osgi and WT plants using an Li-6400 at 9 h after lights-on. All data represent the mean of three biological replicates, with error bars indicating SD.
Mutation of OsGI alters the expression pattern of dehydration-related genes
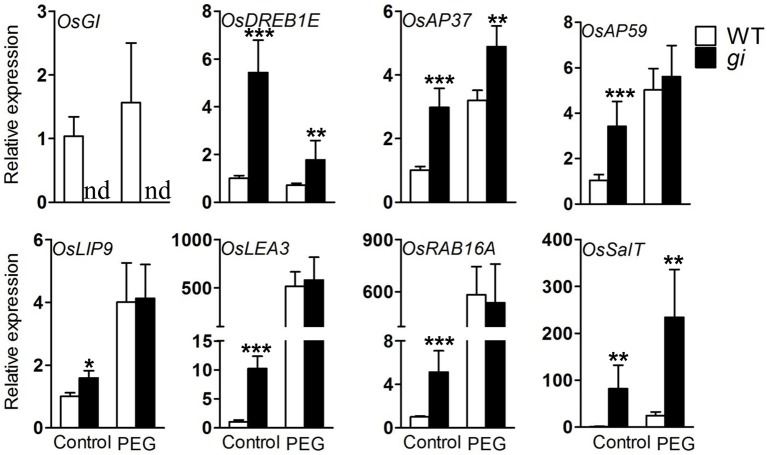
To investigate the impact of OsGI mutation on the expression of genes involved in the response to dehydration stress, qRT-PCR was carried out on leaf samples of WT and osgi plants grown under normal or PEG treatment conditions. The expression of OsGI was not affected by osmotic stress (Figure 7, Supplementary Figure 4). OsDREB1E, OsAP37, OsAP59, OsLEA3, OsRAB16A, OsLIP9, and OsSalT, which have been shown to be positively associated with osmotic stress tolerance, were selected for analysis (Chaves et al., 2003; Umezawa et al., 2006; Valliyodan and Nguyen, 2006; Shinozaki and Yamaguchi-Shinozaki, 2007; Xiao et al., 2007; Fukao et al., 2011). As expected, the expression of OsAP37, OsAP59, OsLEA3, OsRAB16A, OsLIP9, and OsSalT were induced by osmotic stress (Figure 7). Under normal conditions, the transcript abundance of all tested genes was significantly higher in osgi mutants than in the WT. Osmotic stress conditions further increased the expression of OsAP37, OsAP59, and OsSalT in osgi mutants (Figure 7). These results suggest that dehydration-responsive genes are constitutively active in osgi mutant plants.
Figure 7.
Expression of water deficiency-related genes in WT and osgi leaves. WT and osgi plants were grown under normal conditions for 15 days and then cultured in 18% PEG 8000 or normal culture media for 2 days. Leaves were sampled 8 h after lights-on. Total RNA was extracted from the leaves and used for qRT-PCR. All data represent the mean of three biological replicates, with error bars indicating SD. Expression of OsACTIN was used as the internal control. Significant differences relative to the corresponding WT strain are indicated with asterisks (***P < 0.001; **P < 0.01; and *P < 0.05).
To elucidate the molecular function of OsGI, the expression profiles in osgi mutant and WT leaves were analyzed using Affymetrix GeneChip. Microarray analysis revealed that mutation of osgi results in substantial transcriptomic reprogramming. Indeed, 1870 genes were differentially expressed in the osgi mutant background compared to WT plants (p < 0.05, PPDE > 0.95, 2-fold cut-off), of which 594 and 1267 genes were down- or up-regulated, respectively (Figure 8). Given the increased osmotic stress tolerance in osgi plants, we compared the mutant transcriptome to a dehydration-altered transcriptome of WT rice (Qian et al., 2014). In summary, 151 down-regulated and 257 up-regulated genes in osgi overlapped with the reported dehydration stress responses (Figure 8; Jain et al., 2007).
Figure 8.
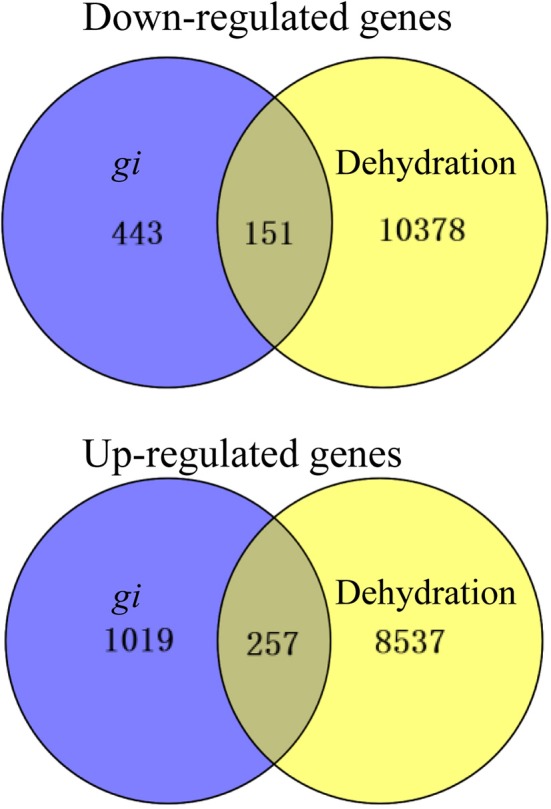
Transcriptomic analyses of osgi and WT seedlings. Venn diagram representing the number of differentially expressed genes in osgi seedlings compared to WT seedlings grown under normal conditions (top; p < 0.05, PPDE > 0.95, 2-fold cut-off) or to WT seedlings exposed to dehydration conditions (bottom; Jain et al., 2007). For the microarray, rice seedlings were grown for 14 days under normal conditions before leaves were harvested.
In order to further assess how OsGI mutation alters dehydration stress-related genes, we analyzed the expression pattern of water deficiency-related genes in leaves in our microarray. Abscisic acid (ABA) is thought to be involved in the regulation of stomata movement and plays a critical role in the response to drought (Chaves et al., 2003). Given the regulation of OsGI in stomata opening, the expression of ABA-related genes was examined in osgi plants (Table 1). A number of genes involved in ABA signaling displayed altered expression in osgi leaves, including several ABA-induced proteins and protein phosphatase 2C (PP2C) genes in subfamily A (Table 1; Schweighofer et al., 2004). Since improved antioxidant capacity is critical in order for plants to survive in water-limited conditions, the expression levels of antioxidant genes were investigated in the microarray (Mittler et al., 2004; Hu et al., 2008). Surprisingly, mutation of OsGI activated antioxidant genes including superoxide dismutase, peroxidase and thioredoxin (Table 2), indicating that osgi plants are strong Reactive Oxygen Species (ROS) scavengers. Furthermore, increased expression of chaperones which protect proteins from stress damage has been shown to improve plant tolerance to water deficits (Wang et al., 2004). Accordingly, the expression of various chaperone genes such as heat shock proteins increased in osgi plants (Table 3).
Table 1.
ABA signaling-related genes with significantly altered transcript abundance in osgi compared to WT leaves (p < 0.05).
| TIGR locus identifier | Fold change | Description |
|---|---|---|
| osgi vs. WT | ||
| ABA | ||
| Os04g52090 | 2.54 | OsAP2-39 |
| Os03g09170 | 2.63 | DRE binding factor |
| Os09g35010 | 2.11 | HvCBF4 |
| Os04g46440 | 3.23 | DREB1C |
| Os11g30500 | 3.73 | ABA induced protein |
| Os04g34600 | 1.58 | ABA induced protein |
| Os06g14370 | 4.91 | ABA induced protein |
| Os11g06720 | 1.63 | Abscisic stress ripening protein (OsASR5) |
| Os07g07050 | 3.26 | Abscisic-aldehyde oxidase |
| Os07g18120 | 1.75 | Abscisic-aldehyde oxidase |
| Os02g52780 | 1.53 | bZIP transcription factor ABI5 |
| Os05g50800 | 4.22 | ABIL1 protein |
| Os02g03960 | −2.52 | Transcription factor,OsbZIP14 |
| Os02g49860 | −11.10 | ABA induced plasma membrane protein PM 19 |
| Os02g50140 | −2.95 | ABA-induced protein |
| Os09g07380 | −1.35 | ABI3-interacting protein 2 |
| Os01g73250 | −3.29 | Abscisic stress-ripening (OsASR6) |
| PP2C | ||
| Os01g40094 | 3.98 | Protein phosphatase 2C |
| Os05g46040 | 1.47 | Protein phosphatase 2C |
| Os03g16170 | 4.06 | Protein phosphatase 2C |
| Os09g15670 | 7.29 | Protein phosphatase 2C |
| Os01g62760 | 1.55 | Protein phosphatase 2C |
| Os01g46760 | 1.52 | Protein phosphatase 2C |
TIGR locus identifiers are given for each transcript at the left. Expression ratio in osgi vs. WT leaves is shown. The annotation description of each gene is according to the TIGR and RAP general description. Positive numbers indicate increased gene expression whereas negative numbers indicate decreased gene expression.
Table 2.
Genes involved in antioxidant synthesis with significantly altered transcript abundance in osgi compared to WT leaves (p < 0.05).
| TIGR locus identifier | Fold change | Description |
|---|---|---|
| osgi vs. WT | ||
| Os07g46990 | 1.59 | Superoxide dismutase |
| Os08g44770 | 1.92 | Superoxide dismutase |
| Os06g02500 | 2.21 | Superoxide dismutase |
| Os05g25850 | 1.29 | Superoxide dismutase |
| Os01g61320 | 2.19 | Thioredoxin family protein |
| Os03g58630 | 1.24 | Thioredoxin H-type 5 (OsTrx10) |
| Os01g07376 | 1.65 | Thioredoxin H-type (OsTrx1) |
| Os02g56900 | 2.31 | Thioredoxin family protein |
| Os08g29110 | 1.44 | Thioredoxin family protein |
| Os02g53400 | 1.30 | Thioredoxin-like 5, chloroplast precursor |
| Os05g11090 | 1.45 | Thioredoxin-like 6, chloroplast precursor |
| Os04g15690 | 5.24 | DSBA-like thioredoxin domain containing protein |
| Os06g37080 | 1.55 | L-ascorbate oxidase precursor |
| Os11g42220 | 4.06 | L-ascorbate oxidase precursor |
| Os08g44340 | 2.50 | monodehydroascorbate reductase |
| Os07g49400 | 1.19 | OsAPx2—Cytosolic Ascorbate Peroxidase gene |
| Os01g19020 | 15.16 | Peroxidase 1 precursor |
| Os05g04380 | 18.38 | Peroxidase 1 precursor |
| Os01g22370 | 1.62 | Peroxidase 1 precursor |
| Os01g22230 | 2.37 | Peroxidase 1 precursor |
| Os10g39170 | 1.33 | Peroxidase 1 precursor |
| Os01g07770 | 2.62 | Peroxidase 25 precursor |
| Os06g46799 | 2.18 | Peroxidase 39 precursor |
| Os05g04500 | 9.58 | Peroxidase 63 precursor |
| Os06g13050 | 1.50 | Peroxidase family protein |
| Os06g09610 | 2.64 | Peroxiredoxin bcp |
| Os02g09940 | 5.88 | Peroxiredoxin-5, mitochondrial precursor |
| Os01g16152 | 1.77 | Peroxiredoxin-5, mitochondrial precursor |
| Os02g33450 | 1.67 | 2-cys peroxiredoxin BAS1, chloroplast precursor |
Table 3.
Genes involved in chaperone synthesis with significantly different transcript abundance in osgi vs. WT leaves (p < 0.05).
| TIGR locus identifier | Fold change | Description |
|---|---|---|
| osgi vs. WT | ||
| Os02g52150 | 2.93 | Heat shock protein (OsHSP20) |
| Os06g11610 | 3.57 | Heat shock protein |
| Os09g31486 | 2.20 | Heat shock 70 kDa protein, mitochondrial precursor |
| Os02g53420 | 1.31 | Heat shock 70 kDa protein, mitochondrial precursor |
| Os03g60620 | 1.25 | Heat shock cognate 70 kDa protein 2 |
| Os01g62290 | 2.36 | Heat shock cognate 70 kDa protein |
| Os05g38530 | 2.25 | Heat shock cognate 70 kDa protein |
| Os06g35960 | 3.03 | Heat shock factor protein |
| Os04g48030 | 2.05 | Heat shock factor protein |
| Os09g28354 | 4.39 | Heat shock factor protein |
| Os01g43590 | 2.04 | Heat shock factor protein HSF8 |
| Os08g39140 | 1.30 | Heat shock protein 81-1 |
| Os03g18200 | 1.30 | Heat shock protein DnaJ |
| Os12g31460 | 1.50 | Heat shock protein DnaJ |
| Os01g53220 | 2.13 | Heat shock factor protein (OsHsfC1b) |
| Os03g16040 | 1.84 | Heat shock protein (OsHsp17.7) |
| Os02g54140 | 2.82 | Heat shock protein |
| Os10g32550 | 1.99 | Chaperonin CPN60-1, mitochondrial precursor |
| Os03g04970 | 1.85 | Chaperonin CPN60-1, mitochondrial precursor |
| Os05g46290 | 1.69 | Chaperonin CPN60-2, mitochondrial precursor |
| Os06g09679 | 1.29 | Chaperonin, chloroplast precursor |
| Os09g26730 | 1.54 | Chaperonin, chloroplast precursor |
| Os03g25050 | 2.18 | Chaperonin |
| Os07g44740 | 1.78 | Chaperonin |
| Os03g25050 | 1.79 | Chaperonin |
Discussion
In the past decade, the identification of many key factors involved in the response to water deficits has greatly increased our understanding of molecular mechanisms participating in the osmotic response in plants. In our current study, we investigated the role of a key flowering time regulator, OsGI, in osmotic stress tolerance in rice. Our data revealed that OsGI plays a negative role in the rice response to water deficiency by regulating at both physiological and transcriptional levels. Results from our analysis suggest that in osgi mutant plants, several components of the osmotic stress response are constitutively activated.
Alteration of OsGI could have improved the plant osmotic stress response in two ways. One, the osmotic potential in the osgi mutant could have increased due to the constitutively altered morphology, metabolism and gene expression which led to the accumulation of osmoprotectants such as sucrose and proline. It has been shown that increased sucrose and proline contents improve plant tolerance to osmotic stress (Hadiarto and Tran, 2011). In osgi seedlings, concentrations of sucrose and proline increased in the leaves (Figure 5), which could have increased the plants' osmotic potential to defend against dehydration. Alteration of OsGI could also play a role in the osmotic stress response because rapid physiological adjustment has been shown to decrease water loss and improve water utilization (Hirayama and Shinozaki, 2010; Hadiarto and Tran, 2011; Xiong et al., 2012). In plants, water loss mainly occurs in transpiration via stomata opening. Stomata movement is critical for plants to reduce transpiration and to rapidly respond to osmotic stress conditions (Chaves et al., 2003; Valliyodan and Nguyen, 2006). In osgi plants, the accelerated stomata closure could have greatly improved water utilization by rapidly decreasing the transpiration rate in leaves, resulting in decreased water loss (Figures 3D, 6).
Although GI shares no known functional domains with other proteins, it has been shown to function in many pathways at the protein level (Shi et al., 2000; Kim et al., 2013; Park et al., 2013). In our study, expression of OsGI is not affected by osmotic stress at the transcriptional level (Figure 7, Supplementary Figure 4), indicating that OsGI might function purely on a protein level in the response to dehydration stress. Further study to determine the OsGI protein abundance under osmotic stress conditions will provide more insight into the molecular mechanisms by which OsGI responds to dehydration stress. Interestingly, mutation of OsGI resulted in substantial transcriptomic reprogramming, and 151 of the down- and 257 of the up-regulated genes in osgi leaves showed the same (down/up-regulated) response in WT leaves exposed to dehydration conditions (Figure 8). However, further studies are required to determine the mechanisms by which OsGI regulates the transcriptional response to osmotic pressure.
ABA is required for plants to adapt stomata movement to water-deficient conditions (Fujita et al., 2011). Because OsGI is involved in regulating stomata opening (Figure 6), ABA-related genes were examined in osgi plants. Results showed that a number of genes involved in ABA signaling were altered in osgi leaves (Table 1). Although the growth phenotype of rice osgi plants under ABA treatment was comparable to that of WT plants (data not shown), GI has been shown to function in ABA metabolism (Penfield and Hall, 2009; Riboni et al., 2013). Indeed, GI has been suggested to play a role in drought response and seed dormancy in an ABA-dependent pathway (Penfield and Hall, 2009; Riboni et al., 2013). GI serves as a core gene in the circadian rhythm of plants, and recent research revealed that around 40% of ABA-related genes are controlled by the circadian clock (Covington et al., 2008; Legnaioli et al., 2009). The circadian gene TOC1 participates in ABA signaling by regulating ABA-Related gene (ABAR) and by interacting with Abscisic Acid Insensitive 3 (ABI3) and is degraded by ZEITLUPE (ZTL) (Kurup et al., 2000; Kim et al., 2007). It is possible that GI functions in ABA signaling via TOC1 regulation by directly interacting with ZTL (Kim et al., 2007). In addition, the GI-CO-FT flowering time regulatory system is also conserved in stomata regulation (Ando et al., 2013). FT is required for maintaining H+-ATPase activation to regulate stomata opening (Kinoshita et al., 2011). GI might therefore be involved in the H+-ATPase-dependent regulation of stomata movement. Our results suggest that GI could regulate stomata movement via multiple pathways but how GI regulates stomata movement in response to osmotic stress still needs further investigation.
ROS can quickly accumulate in plants under osmotic stress and cause oxidative damage to proteins. As a result, ROS scavenging is critical for plants in order to survive under water deficient condition (Umezawa et al., 2006). In Arabidopsis, increased levels of superoxide dismutase and ascorbate peroxidase confer oxidative stress tolerance in Atgi plants (Kurepa et al., 1998; Cao et al., 2006). In rice, superoxide dismutase, peroxidase and thioredoxin are constitutively active in osgi plants to defend against oxidative damage (Table 2). Moreover, molecular chaperones are key factors in protein stabilization, and the increased expression of chaperone genes in osgi leaves is speculated to protect proteins from dehydration damage (Table 3; Wang et al., 2004).
Compared to WT, a majority of the alterations in osgi plants, such as sucrose, were observed both in the field and greenhouse (Figure 5; Izawa et al., 2011). However, transcriptome and metabolome analysis revealed that osgi plants exhibited different responses in the field and greenhouse in some pathways. While stomata conductance was higher in field-grown osgi than WT plants, there was no difference between osgi and WT plants grown under normal conditions in the greenhouse (Figure 6; Izawa et al., 2011). Conversely, osgi plants grown in the greenhouse had higher proline contents than WT plants (Figure 5) while there was no difference in the proline contents of field-grown osgi and WT plants (Izawa et al., 2011). The TCA cycle intermediate malate was decreased in field-grown but increased in greenhouse-grown osgi plants (Izawa et al., 2011). In addition, Expressions of some key genes in the phenylpropanoid pathway, a secondary metabolite pathway controlled by circadian clocks, was significantly upregulated in osgi plants in the field, but none of these genes were altered in our microarray, indicating that OsGI function is affected by the environment (Izawa et al., 2011).
The tolerance to osmotic stress prompted us to check whether osgi plants exhibit drought resistance but we did not detect any differences between WT and osgi plants grown in soil under drought condition (data not shown), nor did we find any differences between WT and osgi plants in response to salt stress. In Arabidopsis, GI-deficient plants exhibit increased sensitivity to drought but improved tolerance to salt stress (Han et al., 2013; Kim et al., 2013), suggesting divergent functions of GI in drought and salt resistance in rice and Arabidopsis. Seedlings encounter sudden dehydration stress when transferred from normal culture media to PEG-treated media, as opposed to soil-grown plants where water depletion and thus drought formation is much more gradual, commonly occurring over hours or even days. In conclusion, while osgi plants are capable of tolerating sudden osmotic stress to some degree, further study is needed to better understand the mechanisms by which OsGI is involved in drought response.
In summary, we have shown that OsGI is an essential regulator of the plant response to dehydration stress, and modification of OsGI expression might improve the osmotic tolerance of crop cultures. However, more work remains to be done to fully understand the mechanisms of action by which OsGI functions in the regulation of the plant stress response.
Author contributions
HS conceived and designed the research. SL, WY, MW, WQ, and LZ conducted the experiments and analyzed the data. SL and HS wrote the manuscript. All authors read and approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation (31172024, 31401934, 31572189). The authors thank the RGRC (Rice Genome Resource Center, Japan) for the Tos17 insertion mutant NF8540.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00465
References
- Ando E., Ohnishi M., Wang Y., Matsushita T., Watanabe A., Hayashi Y., et al. (2013). TWIN SISTER OF FT, GIGANTEA, and CONSTANS have a positive but indirect effect on blue light-induced stomatal opening in Arabidopsis. Plant Physiol. 162, 1529–1538. 10.1104/pp.113.217984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates L. S., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. 10.1007/BF00018060 [DOI] [Google Scholar]
- Cao S., Jiang S., Zhang R. (2006). The role of GIGANTEA gene in mediating the oxidative stress response and in Arabidopsis. Plant Growth Regul. 48, 261–270. 10.1007/s10725-006-0012-8 [DOI] [Google Scholar]
- Cao S., Ye M., Jiang S. (2005). Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 24, 683–690. 10.1007/s00299-005-0061-x [DOI] [PubMed] [Google Scholar]
- Chaves M. M., Maroco J. O. P., Pereira J. O. S. (2003). Understanding plant responses to drought-from genes to the whole plant. Funct. Plant Biol. 30, 239–264. 10.1071/fp02076 [DOI] [PubMed] [Google Scholar]
- Covington M. F., Maloof J. N., Straume M., Kay S. A., Harmer S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130. 10.1186/gb-2008-9-8-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalchau N., Baek S. J., Briggs H. M., Robertson F. C., Dodd A. N., Gardner M. J., et al. (2011). The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. U.S.A. 108, 5104–5109. 10.1073/pnas.1015452108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., et al. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. 10.1093/emboj/18.17.4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Thomashow M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. 10.1105/tpc.003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. 10.1007/s10265-011-0412-3 [DOI] [PubMed] [Google Scholar]
- Fukao T., Yeung E., Bailey-Serres J. (2011). The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23, 412–427. 10.1105/tpc.110.080325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadiarto T., Tran L.-S. (2011). Progress studies of drought-responsive genes in rice. Plant Cell Rep. 30, 297–310. 10.1007/s00299-010-0956-z [DOI] [PubMed] [Google Scholar]
- Han Y., Zhang X., Wang Y., Ming F. (2013). The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS ONE 8:e73541. 10.1371/journal.pone.0073541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. 10.1038/nature01549 [DOI] [PubMed] [Google Scholar]
- Hecht V., Knowles C. L., Vander Schoor J. K., Liew L. C., Jones S. E., Lambert M. J., et al. (2007). Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144, 648–661. 10.1104/pp.107.096818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Sage-Ono K., Sasaki R., Ohtsuki N., Hoshino A., Iida S., et al. (2011). Constitutive expression of the GIGANTEA ortholog affects circadian rhythms and suppresses one-shot induction of flowering in Pharbitis nil, a typical short-day plant. Plant Cell Physiol. 52, 638–650. 10.1093/pcp/pcr023 [DOI] [PubMed] [Google Scholar]
- Hincha D. K., Zuther E., Hellwege E. M., Heyer A. G. (2002). Specific effects of fructo- and gluco-oligosaccharides in the preservation of liposomes during drying. Glycobiology 12, 103–110. 10.1093/glycob/12.2.103 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. 10.1111/j.1365-313X.2010.04124.x [DOI] [PubMed] [Google Scholar]
- Hu X., Wang W., Li C., Zhang J., Lin F., Zhang A., et al. (2008). Cross-talks between Ca2+/CaM and H2O2 in abscisic acid-induced antioxidant defense in leaves of maize plants exposed to water stress. Plant Growth Regul. 55, 183–198. 10.1007/s10725-008-9272-9 [DOI] [Google Scholar]
- Ikegami K., Okamoto M., Seo M., Koshiba T. (2009). Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res. 122, 235–243. 10.1007/s10265-008-0201-9 [DOI] [PubMed] [Google Scholar]
- Itoh H., Izawa T. (2011). A study of phytohormone biosynthetic gene expression using a circadian clock-related mutant in rice. Plant Signal. Behav. 6, 1932–1936. 10.4161/psb.6.12.18207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T., Mihara M., Suzuki Y., Gupta M., Itoh H., Nagano A. J., et al. (2011). Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23, 1741–1755. 10.1105/tpc.111.083238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Arora R., Agarwal P., Ray S., Sharma P., et al. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483. 10.1104/pp.106.091900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Y., Ali Z., Park H. J., Park S. J., Cha J. Y., Perez-Hormaeche J., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 4:1352. 10.1038/ncomms2357 [DOI] [PubMed] [Google Scholar]
- Kim W. Y., Fujiwara S., Suh S. S., Kim J., Kim Y., Han L., et al. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360. 10.1038/nature06132 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Ono N., Hayashi Y., Morimoto S., Nakamura S., Soda M., et al. (2011). FLOWERING LOCUS T regulates stomatal opening. Curr. Biol. 21, 1232–1238. 10.1016/j.cub.2011.06.025 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C. J., van der Veen J. H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. 10.1007/BF00264213 [DOI] [PubMed] [Google Scholar]
- Kurepa J., Smalle J., Van Montagu M., Inze D. (1998). Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 14, 759–764. 10.1046/j.1365-313x.1998.00168.x [DOI] [PubMed] [Google Scholar]
- Kurup S., Jones H. D., Holdsworth M. J. (2000). Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21, 143–155. 10.1046/j.1365-313x.2000.00663.x [DOI] [PubMed] [Google Scholar]
- Legnaioli T., Cuevas J., Mas P. (2009). TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28, 3745–3757. 10.1038/emboj.2009.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wang C., Zhou L., Shou H. (2014). Oxygen deficit alleviates phosphate overaccumulation toxicity in OsPHR2 overexpression plants. J. Plant Res. 127, 433–440. 10.1007/s10265-014-0628-0 [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang F., Zhou J., Chen F., Wang B., Xie X. (2012). Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice. Plant Mol. Biol. 78, 289–300. 10.1007/s11103-011-9860-3 [DOI] [PubMed] [Google Scholar]
- Mishra P., Panigrahi K. C. (2015). GIGANTEA - an emerging story. Front. Plant Sci. 6:8. 10.3389/fpls.2015.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Mouradov A., Cremer F., Coupland G. (2002). Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14 (Suppl.), S111–S130. 10.1105/tpc.001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. H., Somers D. E., Kim Y. S., Choy Y. H., Lim H. K., Soh M. S., et al. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. 10.1126/science.285.5433.1579 [DOI] [PubMed] [Google Scholar]
- Park H. J., Kim W. Y., Yun D. J. (2013). A role for GIGANTEA: Keeping the balance between flowering and salinity stress tolerance. Plant Signal. Behav. 8:e24820. 10.4161/psb.24820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Hall A. (2009). A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21, 1722–1732. 10.1105/tpc.108.064022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Han X., Peng X., Lu T., Liu W., Fu Z. (2014). The circadian clock gene regulatory module enantioselectively mediates imazethapyr-induced early flowering in Arabidopsis thaliana. J. Plant Physiol. 171, 92–98. 10.1016/j.jplph.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Riboni M., Galbiati M., Tonelli C., Conti L. (2013). GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol. 162, 1706–1719. 10.1104/pp.113.217729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A., Hirt H., Meskiene I. (2004). Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9, 236–243. 10.1016/j.tplants.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C., Zhu J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. 10.1073/pnas.120170197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K., Mizoguchi T., Urao T., Katagiri T., Nakashima K., et al. (1998). Molecular responses to water stress inArabidopsis thaliana. J. Plant Res. 111, 345–351. 10.1007/BF02512195 [DOI] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Fujita M., Fujita Y., Yamaguchi-Shinozaki K., Shinozaki K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotech. 17, 113–122. 10.1016/j.copbio.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Valliyodan B., Nguyen H. T. (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Biotech. 9, 189–195. 10.1016/j.pbi.2006.01.019 [DOI] [PubMed] [Google Scholar]
- Villadsen D., Rung J. H., Nielsen T. H. (2005). Osmotic stress changes carbohydrate partitioning and fructose-2,6-bisphosphate metabolism in barley leaves. Funct. Plant Biol. 32, 1033–1043. 10.1071/FP05102 [DOI] [PubMed] [Google Scholar]
- Wang C., Ying S., Huang H., Li K., Wu P., Shou H. (2009). Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 57, 895–904. 10.1111/j.1365-313X.2008.03734.x [DOI] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. 10.1016/j.tplants.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Xiao B., Huang Y., Tang N., Xiong L. (2007). Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115, 35–46. 10.1007/s00122-007-0538-9 [DOI] [PubMed] [Google Scholar]
- Xiong J., Zhang L., Fu G., Yang Y., Zhu C., Tao L. (2012). Drought-induced proline accumulation is uninvolved with increased nitric oxide, which alleviates drought stress by decreasing transpiration in rice. J. Plant Res. 125, 155–164. 10.1007/s10265-011-0417-y [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Urao T., Shinozaki K. (1995). Regulation of genes that are induced by drought stress in Arabidopsis thaliana. J. Plant Res. 108, 127–136. 10.1007/BF02344316 [DOI] [Google Scholar]
- Yoshida S., Forno D. A., Cock J. H. K. G. (1976). Laboratory Manual for Physiological Studies of Rice, 3rd Edn. Manila: The International Rice Research Institute. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.