Abstract
Mitochondrial biogenesis, the generation of new mitochondrial DNA and proteins, has been linked to osteoclast (OC) differentiation and function. In this study we used mice with mutations in key alternative NF-κB pathway proteins, RelB and NIK, to dissect the complex relationship between mitochondrial biogenesis and osteoclastogenesis. OC precursors lacking either NIK or RelB, RANKL were unable to increase mitochondrial DNA or OxPhos protein expression, associated with lower oxygen consumption rates. Transgenic OC precursors expressing constitutively active NIK showed normal RANKL-induced mitochondrial biogenesis (OxPhos expression and mitochondria copy number) compared to controls, but larger mitochondrial dimensions and increased oxygen consumption rates, suggesting increased mitochondrial function. To deduce the mechanism for mitochondrial biogenesis defects in NIK- and RelB-deficient precursors, we examined expression of genes known to control this process. PGC-1β (Ppargc1b) expression, but not PGC-1α, PPRC1 or ERRα, was significantly reduced in RelB−/− and NIK−/− OCs. Because PGC-1β has been reported to positively regulate both mitochondrial biogenesis and differentiation in OCs, we retrovirally overexpressed PGC-1β in RelB−/− cells, but surprisingly found that it did not affect differentiation, nor restore RANKL-induced mitochondrial biogenesis. To determine whether the blockade in osteoclastogenesis in RelB-deficient cells precludes mitochondrial biogenesis, we rescued RelB−/− differentiation via overexpression of NFATc1. Mitochondrial parameters in neither WT nor RelB-deficient cultures were affected by NFATc1 overexpression, and bone resorption in RelB −/− was not restored. Furthermore, NFATc1 co-overexpression with PGC-1β, while allowing OC differentiation, did not rescue mitochondrial biogenesis or bone resorption in RelB−/− OCs, by CTX-I levels. Thus, our results indicate that the alternative NF-κB pathway plays dual, but distinct roles in controlling the independent processes of OC differentiation and OC mitochondrial biogenesis. Furthermore, the inability of PGC-1β to drive mitochondrial biogenesis in OCs without RelB indicates a cell-type specificity in mitochondria regulation.
Keywords: osteoclasts, transcription factors, molecular pathways-remodeling
INTRODUCTION
Recently, it has been shown that the alternative NF-κB pathway is important in regulating mitochondria content and oxidative phosphorylation function within skeletal muscle (1). Mice lacking alternative pathway proteins showed defects in muscle fiber composition, contractile force, and mitochondria size. Mitochondrial biogenesis is the orchestrated synthesis of nuclear and mitochondrial-encoded DNA, proteins, and mitochondria membranes, and biogenesis of these network of organelles are vital for their inherent functions of fatty acid oxidation, oxidative phosphorylation (OxPhos), mitochondrial ROS (mROS) production, among many other metabolic roles. Although the stimulus for activating the pathway in skeletal muscle cells was not identified in the study, the downstream transcriptional target was PPARγ co-factor 1β (Ppargc1b or PGC-1β), a known regulator of mitochondrial biogenesis in this cell type. Interestingly, like skeletal myocytes, osteoclasts (OCs) are very mitochondria-rich, and several studies have outlined the importance of regulating mitochondrial biogenesis in these cells (2–7). In particular, global or conditional deletion of PGC-1β in OC precursors resulted in defects in both differentiation and mitochondrial biogenesis (2,3), leading to a working model in which PGC-1β is a direct regulator of both processes(8). However, previous experiments have not resolved whether mitochondrial biogenesis is a prerequisite for full OC differentiation or for bone resorption. In fact, one study suggested that reduced mitochondrial function is associated with increased bone resorption (4).
Receptor Activator of NF-κB ligand (RANKL) is a TNF superfamily member ligand that is, in most circumstances, required to propagate signals for differentiation and function of the OC(9). Engagement of its receptor RANK leads to the activation of several downstream signaling pathways, including both classical and alternative NF-κB. Activation of the alternative pathway allows de novo NF-κB Inducing Kinase (NIK) to escape TRAF3-mediated degradation and phosphorylate IKKα to allow the key transcriptional subunit in the pathway, RelB, to translocate into the nucleus (10,11). This pathway, along with many other signals that are activated upon RANK engagement, allows mono-nuclear myeloid precursor cells to fuse and differentiate into mature, multinucleated tartrate resistant acid phosphatase (TRAP) positive OCs (10). The specialized role of a mature OC is to polarize onto bone, acidify its resorptive lacunae, and secrete enzymes to degrade the inorganic and organic bone matrix (9,12).
RelB and NIK support osteoclastogenesis in vitro; in the absence of either protein there is a dramatic reduction in the number of mature OCs, which can be rescued by overexpressing RelB, but not classical NF-κB subunit p65(13). Conversely, transgenic expression of a constitutively activated form of NIK (lacking the TRAF3 interaction domain) leads to enhanced osteoclastogenesis at low doses of RANKL, as well as significantly increased resorptive activity of each OC(14). This indicates that RANKL-dependent activation of the alternative pathway is important in controlling not only differentiation, but also resorptive functions of OCs (10). However, RelB target genes that regulate cellular processes controlling OC resorptive function have not been established.
In this study, using multiple genetic models in which alternative NF-κB pathway proteins are altered (via ablation of RelB and NIK, or constitutive activation of NIK), we investigate the relationship between mitochondrial biogenesis and OC differentiation. Here we establish that the alternative NF-κB pathway is an important regulator of the mitochondrial biogenesis that occurs during OC differentiation. However, these processes can be uncoupled; osteoclastogenesis can be restored in RelB-deficient cultures without rescuing the defect in mitochondrial biogenesis. Furthermore, although PGC-1β expression is modulated by alternative NF-κB, this factor does not mediate RANKL-induced mitochondrial biogenesis in the absence of RelB.
MATERIALS AND METHODS
Reagents and mice
CatK.NT3 transgenic mice were generated by mating Cathepsin K-cre mice to homozygous NT3 mice (which have the NIKΔT3 transgene knocked into the ROSA26 locus)(14). Controls were Cre-negative littermates. RelB−/− and NIK −/− mice were generated from respective RelB+/− and NIK +/− parents(13,15). All mice are on a C57Bl/6 background and age and sex-matched pairs of mice were used for in vitro experiments. Mice were between the ages of 6 and 10 weeks old, averaging 20–24 g, housed in specific pathogen-free barrier facility, with 12 hour light and dark cycles, without treatment of any drugs or compounds or subject to any surgical procedures. All cages, components, bedding, and water are autoclaved and cages changed once a week, with room monitoring by sentinel mice tested for pathogens quarterly by Washington University’s Department of Comparative Medicine (DCM) Diagnostic Laboratory. Daily health checks of each cage is monitored by DCM staff, supervisor, and veterinarian. For each experimental and control pair of mice, the basal health of the animals were not different. All protocols were approved by the Institutional Animal Studies Committee at Washington University School of Medicine (permits 20110067 and 20140058).
Supernatant from the CMG 14–12 cell line was used as a source of M-CSF and glutathione-S-transferase RANKL (GST-RANKL) was purified as previously described(16,17).
The following antibodies were used for western immuno-blotting: Anti-Oxphos cocktail antibody was obtained from Abcam (ab110413). Anti-β-actin AC-74 was obtained from Sigma (A2228). Anti-NFATc1 7A6 antibody was obtained from Santa-Cruz (sc-7294). Anti-β3 integrin antibody was obtained from Cell Signaling Technology (Cell Signaling 4702).
Bone marrow macrophage isolation and osteoclast differentiation
Primary bone marrow macrophage (BMM) isolation was as previously described(18). Bone marrow was extracted from long bones of 6–10 week old mice with αMEM (Sigma) in 10% FBS (Gibco) containing 100 IU/ml penicillin and streptomycin, and BMMs expanded in 1:10 dilution of CMG 14-12 cell supernatant (containing M-CSF, equivalent to 100ng/mL) for 4 days. For osteoclast differentiation, cells were cultured at the following densities: 9x103 cells per well in 96-well tissue-culture plastic (TPP), 4.2x104 in 48-well plates or 5x105 in 6-well plates. 30 ng/ml GST-RANKL and CMG 14-12 supernatant supplying an equivalent 20 ng/ml M-CSF were added daily with media change until TRAP+ multinucleated cells became confluent. For TRAP staining, cells were fixed in 4% ultrapure formaldehyde (Polysciences) and 0.1% Triton X-100 in PBS for 5 min, and stained for TRAP according to the commercial kit protocol (Sigma).
Retroviral overexpression
pMX retroviral vectors containing NFATc1 or PGC-1β and a blasticidin resistance selectable marker, were transfected into separate platE cells by calcium phosphate-precipitation method(19). The virus-containing supernatant was harvested 48hrs after transfection and mixed with equal volumes αMEM, along with 100 ng/ml MCSF equivalent, 4 μg/mL hexadimethrine bromide (Sigma), then added to BMMs previously cultured for 2 days in 100 ng/ml M-CSF equivalent. After 24hr viral incubation, BMMs were selected for 2 days with 1 μg/ml blasticidin (Life Technologies), before culturing for osteoclastogenesis. The same empty vector-transduced cultures served as the negative control for NFATc1 or PGC-1β overexpression, since experiments were performed in parallel. For co-transduction of NFATc1 and PGC-1β, pMX.puro.NFATc1 (containing a puromycine resistance gene) and pMX.bsr.PGC-1β (containing a blasticidin resistance gene) constructs were transfected into separate cultures of platE cells, and the virus-containing supernatant was harvested as above and added at 1:1 ratios to RelB −/− BMMs for 24 hrs; both pMX.puro.NFATc1 and pMX.bsr.PGC-1β viral supernatant were used to infect the BMMs simultaneously, then selected for 2 days with 1 μg/ml blasticidin (Life Technologies) and 2 μg/ml puromycin (Life Technologies) before culturing for osteoclastogenesis. Empty vector pMX.puro and pMX.bsr viral supernatants were used for the controls and to complement the pMX.puro.NFATc1 or pMX.bsr.PGC-1β as appropriate such that all cultures were doubly transduced and selected.
Bone resorption assays
Retrovirally transduced and selected BMMs were cultured on bovine bone slices for 6 days in 1:50 dilution of CMG 14-12 cell supernatant and 30ng/ml GST-RANKL, replaced with fresh media and cytokines every 24 hrs. For pit assay, bone slices were incubated in 0.5N NaOH for 30 sec and cells scraped off using a cotton swab, then incubated with 20 μg/ml peroxidase-conjugated wheat germ agglutinin in PBS (Sigma) for 30 min, washed with PBS 3 times, and exposed to 3,3′-diaminobenzidine (Sigma) for 15 min before washing and drying. BioQuant OSTEO 2010 (BioQuant Image Analysis Corporation) was used to quantify pit area. For assessment of CTX release in culture, media was collected for the final 24 h of culture, between days 6 and 7 and analyzed using the CrossLaps for Culture CTX-I ELISA kit (Immunodiagnosticsystems AC-07F1) per manufacturer instructions.
Quantitative real-time PCR
Cells were scraped in TRIzol (Life Technologies), and the aqueous layer was extracted using phenol:chloroform. Equal volume of 70% ethanol was added to the aqueous fraction and the rest of the RNA isolation followed that of the NucleoSpin RNA II kit (Clontech). 1 μg of RNA was reverse transcribed into cDNA using RNA to cDNA Ecodry premix kit (Clontech). qRT-PCR was performed on an ABI7300 Real-Time PCR system (Applied Biosystems) using Clontech SYBR Advantage Premix. For each target, the amplification reaction was as follows: 95°C for 5 seconds and 60°C for 31 seconds for 40 cycles, followed by melt curve analysis to determine primer specificity. Relative expression of target gene was calculated as follows: 1000*2−(Ct target gene-Ct Cyclophilin). Primer sequences are as follows for murine target genes: ATP5b (For CGTGAGGGCAATGATTTATACCAT, Rev TCCTGGTCTCTGAAGTATTCAGCAA), Cyclophilin A (For AGCATACAGGTCCTGGCATC, Rev TTCACCTTCCCAAAGACCAC), Cytochrome C (For GGAGGCAAGCATAAGACTGG, Rev TCCATCAGGGTATCCTCTCC), ERRα (For GGAGTACGTCCTGCTGAAAGCT, Rev CACAGCCTCAGCATCTTCAATG), NFATc1 (For GGTAACTCTGTCTTTCTAACCTTAAGCTC, Rev GTGATGACCCCAGCATGCACCAGTCACAG), PGC-1α (For CGGAAATCATATCCAACCAG, Rev TGAGAACCGCTAGCAAGTTTG), PGC-1β (For CTCCAGGCAGGTTCAACCC, Rev GGGCCAGAAGTTCCCTTAGG), PPARγ (For ACTCATACATAAAGTCCTTCCCGCT, Rev ATGGTGATTTGTCCGTTGTCTTTCC)
Mitochondria copy number quantification
Total DNA was extract as previously described(20). After quantitation by Nanodrop (Thermo), 15 ng of total DNA was used per well (replicated in technical triplicates), and the relative amplification of a mitochondria genome marker NADH Dehydrogenase subunit 1 (ND1) to a genomic DNA marker Lipoprotein lipase (LPL) was determined by q-PCR on an ABI7300 Real-Time PCR system (Applied Biosystems) using Clontech SYBR Advantage Premix. For each target, the amplification reaction was as follows: 95°C for 5 seconds and 60°C for 31 seconds for 40 cycles, followed by melt curve analysis to determine primer specificity. Relative mitochondria copy number was calculated as follows: 2∧−(Ct ND1-Ct LPL). Primer sequences are as follows for murine target genes: LPL (For GGATGGACGGTAAGAGTGATTC, Rev ATCCAAGGGTAGCAGACAGGT), ND1 (For CCCATTCGCGTTATTCTT, Rev AAGTTGATCGTAACGGAAGC)
Western immunoblotting analysis
Cells were washed with PBS and lysed with RIPA buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1mM β-glycerophosphate, 1 mM Na3VO4, 1 mM NaF) and protease inhibitor cocktail (Thermo). After incubation on ice and pelleting cellular debris at 16k x g rcf, the protein was quantitated using a BCA kit (Biorad) and 20–40 μg of protein was loaded into a 10% or 12% SDS-PA gel (only with OxPhos protein cocktail) and subjected to gel electrophoresis. Protein was subsequently transferred to a PVDF membrane via semi-dry transfer apparatus (Biorad) and blocked for 1 hr in 5% milk in TBS 0.1% tween. Primary antibody was incubated at 4° C overnight, followed by a secondary antibody incubation for 1 hr before adding WesternBright Quantum HRP substrate (Advansta) and imaged using a chemiluminescence imager (Syngene).
Transmission electron microscopy
BMMs were cultured on bovine bone slices similar to the protocol for bone resorption, and then fixed with 5% glutaraldehyde in 0.16 M collidine buffer (pH 7.4) overnight. Bone slices were decalcified in 5% EDTA/0.1% glutaraldehyde for 7 days and then fixed in 2% OsO4/3% K-ferrocyanide for 2 hr, dehydrated in ethanol and embedded in Epon LX 112. Sections were examined using a JEOL 100SX transmission electron microscope.
Mitochondria cross-sectional area measurement
Transmission electron microscopy images were taken at 25000x magnification with a 500nm scale bar attached to each image. The scale function in ImageJ software was used to set a pixel:nm ratio, and the freehand selection tool was used to outline the perimeter of each mitochondrion. ImageJ’s integrated measurements outputs a cross-sectional area based on the shapes drawn on the picture.
Oxygen consumption measurement
To measure oxygen consumption rate (OCR), BMMs and OCs were analyzed in an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience). BMMs or OCs were cultured in XF96 cell culture microplates (4k cells/well) in GST-RANKL and CMG supernatant (as described above for osteoclast differentiation) for 3–4 days. Cells were cultured in technical quadruplicate wells and 3 basal OCR readings were taken preceding the addition of mitochondrial drugs. 1.5 μM FCCP (fluoro-carbonyl cyanide phenylhydrazone), which uncouples ATP synthesis from oxygen consumption, was injected into the culture wells 35 min post basal OCR readings. To maintain the proton gradient after disruption by FCCP, cells will increase oxygen consumption to provide a maximum OCR reading (21).
Statistics
Measurements are expressed as mean “±” SD. p-values were obtained using multiple comparison in a one-way ANOVA test for multiple comparisons, with unpaired non-parametric t-test when only 2 groups are present, using GraphPad Prism v6 built-in statistical analysis. p-values less than 0.05 are considered significant. All qRT-PCR and qPCR data is expressed as a mean from at least 3 independent biological experiments (starting with different mice), each experiment with at least 3 technical replicates. All oxygen consumption results were conducted in technical triplicates or quadruplicates, and a representative experiment shown. Data from pit assay is representative from at least 3 independent experiments, averaged from at least independent 4 bone slices. For measurement of cell culture CTX-I from bone resorption assay, at least 3 independent bone slices were used per condition.
RESULTS
Alternative NF-κB signaling controls mitochondrial biogenesis and respiratory function
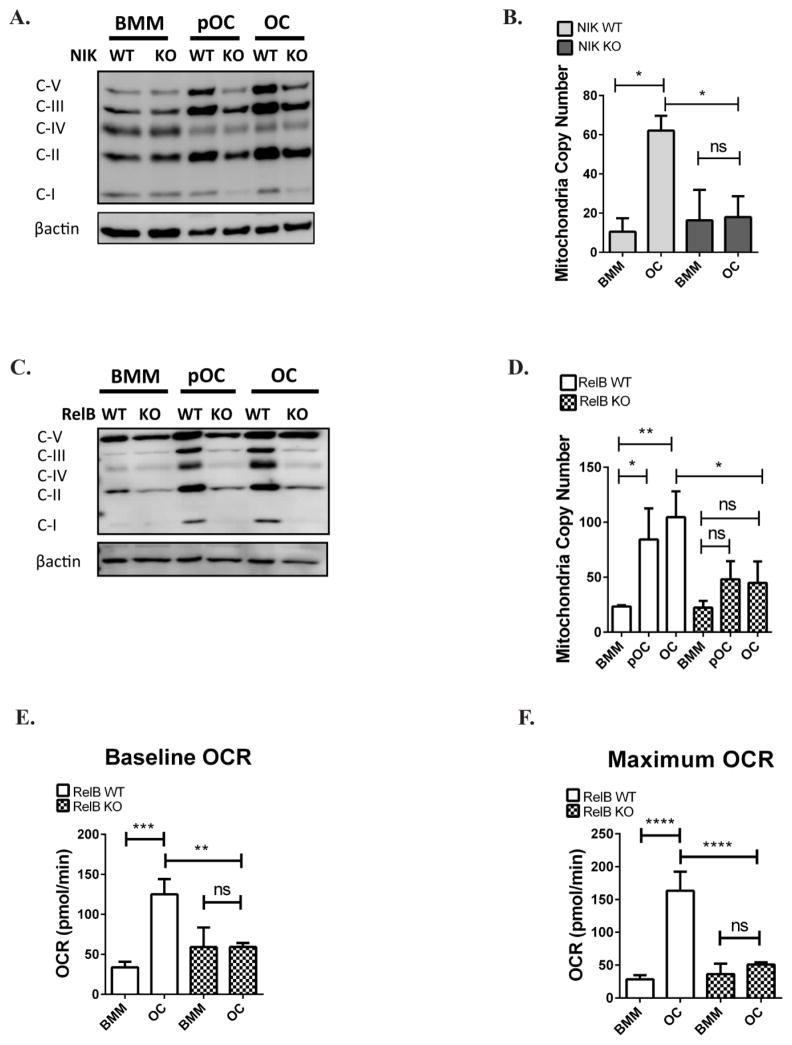
Oxidative phosphorylation (OxPhos) is important for generation of intracellular ATP by mitochondria, and expression of its components is controlled by the alternative NF-κB pathway in skeletal muscle(1). NIK and RelB are signaling components of the alternative NF-κB pathway, representing the upstream kinase and obligate transcription factor subunit, respectively. We therefore assayed whether ablation of these alternative NF-κB signaling proteins affected OxPhos subunit expression during RANKL-induced OC differentiation. Since NIK is more upstream in the pathway, we compared OxPhos protein expression between NIK−/− and NIK+/+ cells and found that upregulation of OxPhos proteins following RANKL stimulation was significantly reduced in NIK−/− cells (Fig 1A). Transcript levels of Cytochrome C and ATP5b were also blunted in NIK −/− cells (Fig S1A,B). Because increased expression of OxPhos components is usually associated with mitochondrial biogenesis, ie. an increase in total mitochondria content, we also examined mitochondrial DNA copy number (normalized to genomic DNA content) in NIK deficient cultures. Although RANKL upregulates mitochondrial copy number (mtCN) in WT cultures during osteoclastogenesis, NIK −/− cultures failed to fully upregulate mtCN in mature OCs (Fig 1B). Because RelB is downstream and in the same pathway as NIK, we assessed the same parameters in RelB−/− BMMs, preOCs, and OCs(13). We observed that OxPhos expression and mtCN upregulation also are blunted in the absence of RelB (Fig 1C,D, S1C,D). We next assayed whether respiration of oxygen (which acts as the final electron acceptor on the electron transport chain) is also changed during osteoclastogenesis. We observed that compared to control OCs, RelB−/− cells maintained lower basal and maximum oxygen consumption rates (Fig 1 E,F). Therefore the defects in both RelB−/− and NIK−/− cells suggest that they are both important components of the alternative NF- κB pathway regulating mitochondrial biogenesis. These differences are specific to alternative NF-κB, as conditional deletion of the classical NF-κB subunit p65 in OCs does not alter these mitochondrial parameters (Fig S2).
Figure 1. Mitochondrial biogenesis and respiratory function is dependent on alternative NF-κB.
RelB KO or NIK KO bone marrow macrophages (BMMs) and WT controls were cultured in M-CSF and RANKL for 2 days to generate pre-osteoclasts (pOCs) or 4 days to generate mature osteoclasts (OCs). (A,C) BMM, pOC, and OC lysates were immuno-blotted for oxidative phosphorylation (OxPhos) subunits - Complex I (NDUFB8), Complex II (SDHB), Complex III (UQCRC2), Complex IV (MTCO1), and Complex V (ATP5A). Results are representative of 3–4 independent experiments. (B,D) Mitochondrial DNA copy number (mtCN), normalized to genomic DNA, was determined by qPCR. n=3. (E,F) Baseline oxygen consumption rates (OCR) and maximum OCR (after stimulation with mitochondrial drug FCCP) were measured in BMM and OC using the Seahorse Flux Analyzer. Technical triplicates from an experiment shown. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant. Light gray and dark gray bars indicate NIK WT and KO, respectively. Solid bars and checkered bars indicate RelB WT and KO, respectively.
Constitutively activated NIK alters mitochondria morphology and size, and enhances respiratory capacity
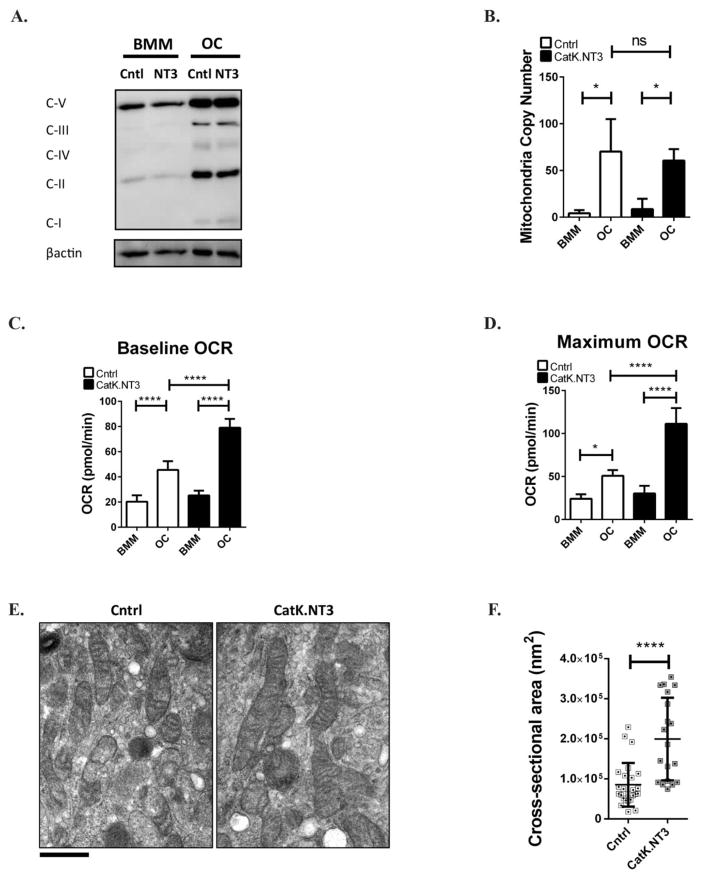
Having demonstrated that decreased alternative NF-κB signaling blocks upregulation of mitochondria in response to RANKL, we next examined the effects of activation of this pathway using CatK.NT3 mice. In this model, expression of a mutant NIK transgene is activated by Cre recombinase under the control of the Cathepsin K promoter, resulting in constitutive activation of the alternative NF-κB pathway and increased sensitivity to RANKL(14). When CatK.NT3 cells are grown in a saturating dose of RANKL they differentiate slightly faster than control cells, but both control and CatK.NT3 cultures ultimately achieve the same numbers of mature OCs. Compared to controls, CatK.NT3 OCs have similar levels of OxPhos protein expression and mtCN (Fig 2 A,B). However, when respiratory capacity was assessed, CatK.NT3 OCs had increased basal and maximum oxygen consumption rate (OCR) at day 3 of in vitro culture (Fig 2 C,D). Mature CatK.NT3 OCs cultured on bovine bone were analyzed for mitochondria morphology by transmission electron microscopy (Fig 2E). Compared to control OCs, CatK.NT3 OCs had increased cross-sectional area of mitochondria (Fig 2F), and appeared to contain denser cristae and increased inner mitochondrial membranes. These data suggest that although constitutive activation of alternative NF-κB does not further increase mitochondria content or total OxPhos protein expression, mitochondria function and morphology as a whole is altered to respire more oxygen.
Figure 2. Mitochondria morphology and respiratory capacity is enhanced when alternative NF-κB is constitutively activated.
(A) BMM and OC (4 days RANKL) lysates from Cntrl or CatK.NT3 cultures were immuno-blotted for oxidative phosphorylation (OxPhos) as in Fig 1. (B) Total DNA was extracted to calculate mtCN. n=3 (C,D) Cntrl or CatK.NT3 BMMs were cultured in M-CSF and RANKL for 3 days, and baseline oxygen consumption rates (OCR) and maximum OCR (after stimulation with mitochondrial drug FCCP) were measured in BMM and OC using the Seahorse Flux Analyzer. Technical triplicates from an experiment shown. (E) Cntrl or CatK.NT3 BMMs were cultured in M-CSF and RANKL for 6 days on bovine bone slices and analyzed by transmission electron microscopy. Scale bar: 500nm. Mitochondria cross-sectional area was quantitated in 9 cells (20–25 total mitochondria) using ImageJ software analysis. *p<0.05, ****p<0.0001; ns, not significant. White and black bars indicate Cntrl and CatK.NT3, respectively.
PGC-1β overexpression does not restore mitochondria defects in alternative NF-κB pathway deficient osteoclasts
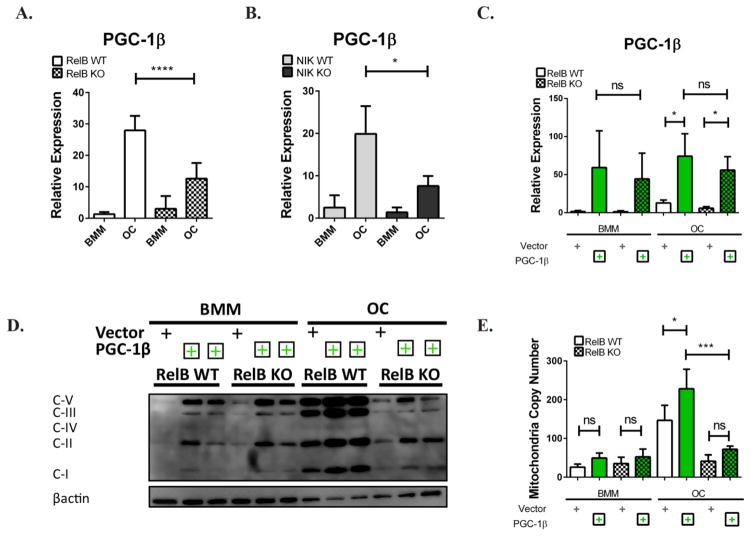
Next we investigated how alternative NF-κB modulates mitochondrial biogenesis in OCs. Regulation of mitochondrial biogenesis is thought to be controlled by nuclear co-factors of the PGC1 family (which includes PGC-1α, PGC-1β, and PRC1). Recently, it has been shown that PGC-1β expression in myotubes is controlled by alternative NF-κB activation, mediating differentiation-associated mitochondrial biogenesis(1). PGC-1β is also reported to be downstream of RANKL, acting to modulate both OC differentiation (through cFOS and NFATc1) and mitochondrial biogenesis to facilitate bone resorption (3). To determine whether PGC-1β might be the link between RelB and mitochondria in OCs, we examined PGC-1β expression during osteoclastogenesis in vitro. In both RelB−/− and NIK−/− cells, PGC-1β transcriptional upregulation was blunted relative to WT controls during differentiation. (Fig 3 A,B). We checked other nuclear hormone receptor family members (PPRC1, PGC-1α, and ERRα) but did not detect their upregulation during osteoclastogenesis in WT control, nor differences in expression in RelB−/− cells (Fig S3). ChIP analysis also demonstrated RelB binding to a conserved intronic NF-κB site within PGC-1β, enriched after RANKL stimulation, suggesting that PGC-1β is likely a direct RelB transcriptional target in OCs (Fig S4).
Figure 3. PGC-1β does not rescue mitochondria defects in alternative NF-κB deficient osteoclasts.
(A,B) RelB −/− or NIK −/− BMMs (and their respective WT controls) were cultured in M-CSF and RANKL for 4 days, and qRT-PCR was used to assess PGC-1β expression. n=3. (C–E) Plat-E cells were transfected with retroviral constructs containing PGC-1β or empty vector, and virus was used to transduce RelB −/− or RelB +/+ BMMs. BMMs were cultured in M-CSF and RANKL for 4 days to generate OCs. (C) Level of PGC-1β mRNA was assessed via qRT-PCR n=3. (D) Cell lysates were immuno-blotted for OxPhos subunits as in Fig 1. Two independent clones of PGC-1β retrovirus were used for overexpression. (E) qPCR to assess mtCN. n=4. *p<0.05, ***p<0.001, ****p<0.0001; ns, not significant. Light gray and dark gray bars indicate NIK WT and KO, respectively. Solid bars and checkered bars indicate RelB WT and KO, respectively. Green color indicates overexpression with PGC-1β.
Complete ablation of PGC-1β reduces both mitochondrial biogenesis and osteoclastogenesis(2). Therefore, we retrovirally overexpressed PGC-1β in RelB −/− OC precursors to determine whether PGC-1β deficiency contributed to defects in both of these processes. qRT-PCR demonstrated that the constructs overexpressed PGC-1β to a similar extent in RelB +/+ and RelB−/− BMM and OC cultures (Fig 3C). Mitochondria OxPhos subunit expression increased in both RelB +/+ and −/− BMMs with PGC-1β overexpression, as predicted, using two independent clones of constructs containing PGC-1β (Fig 3D, left half). However, while RelB +/+ OCs were able to further increase their OxPhos subunit expression with PGC-1β over-expression, RelB −/− OCs maintained the same levels of subunit expression as BMMs (Fig 3C, right half), and in fact never reached levels comparable to WT OCs transduced with empty vector. mtCN follows the same trend as OxPhos expression; although the increase in mtCN with PGC-1β does not reach statistical significance in BMMs, overexpression of PGC-1β significantly increases mtCN in WT OCs, compared to empty vector controls, but not in RelB-deficient OCs (Fig 3E). Although PGC-1β can drive biogenesis in BMMs, independent from RelB, there is a requirement for RelB to further upregulate mitochondria parameters in RANKL-stimulated cells. Thus, the molecular mechanism for increased mitochondrial biogenesis during differentiation appears to be different in OCs than skeletal myocytes.
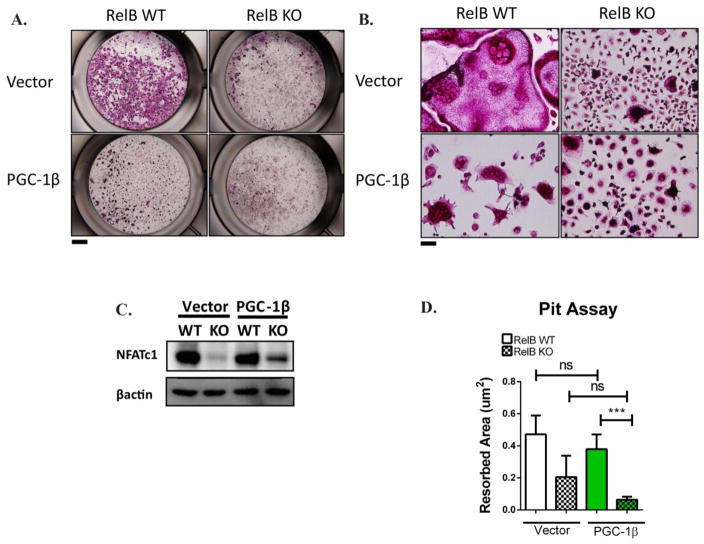
PGC-1β overexpression does not restore differentiation defects in RelB−/− cells
Since the proposed mechanism for PGC-1β to promote differentiation (ie. via cFOS) is distinct from that of mitochondrial biogenesis, we also examined the effect of PGC-1β overexpression on osteoclastogenesis. Interestingly, PGC-1β did not rescue OC differentiation in RelB−/− cells, although it clearly altered the shape of WT OCs (Fig 4A,B). WT OCs overexpressing PGC-1β were TRAP positive and multinucleated, but were poorly spread with clumped nuclei and smaller cytoplasmic area compared to empty vector transduced OCs. Despite the change in OC morphology on plastic, PGC-1β did not alter bone resorption in WT OCs (Fig 4D). Supporting our observation that PGC-1β over-expression did not change osteoclastogenesis per se, we observed no differences in NFATc1 expression (a master transcription regulator of osteoclastogenesis) in either RelB +/+ or RelB−/− cells overexpressing PGC-1β (Fig 4C, Fig S5A). Not surprisingly given its inability to upregulate either differentiation or mitochondria in RelB-deficient OCs, there also was no change in resorption in these cultures (Fig S5B).
Figure 4. PGC-1β does not restore differentiation defects in RelB KO cells.
RelB −/− or RelB +/+ BMMs were transduced with retroviral constructs containing PGC-1β or empty vector, which were then cultured in M-CSF and RANKL for 4 days. (A) Representative wells stained with TRAP to assess OC differentiation, with magnified image (B) to evaluate multi-nucleation. Scale bar for (A) is 1mm, scale bar for (B) is 10mm. In (A) representative TRAP image of 3 independent experiments, where the same empty vector served as negative control for NFATc1 and PGC-1β overexpression (C) Cell lysates were immuno-blotted to assess NFATc1 and β3 integrin protein expression. (D) OCs were grown from transduced BMMs on bovine bone slices for 6 days in M-CSF and RANKL, and area resorbed was quantified by pit assay. Data from bone resorption averaged from at least 4 bone slices (representative of 3 independent experiments). ***p<0.001; ns, not significant. Solid bars and checkered bars indicate RelB WT and KO, respectively. Green color indicates overexpression with PGC-1β.
Alternative NF-κB regulates mitochondria independent from OC differentiation to regulate bone resorption
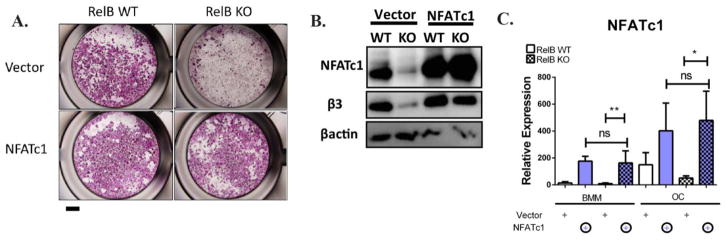
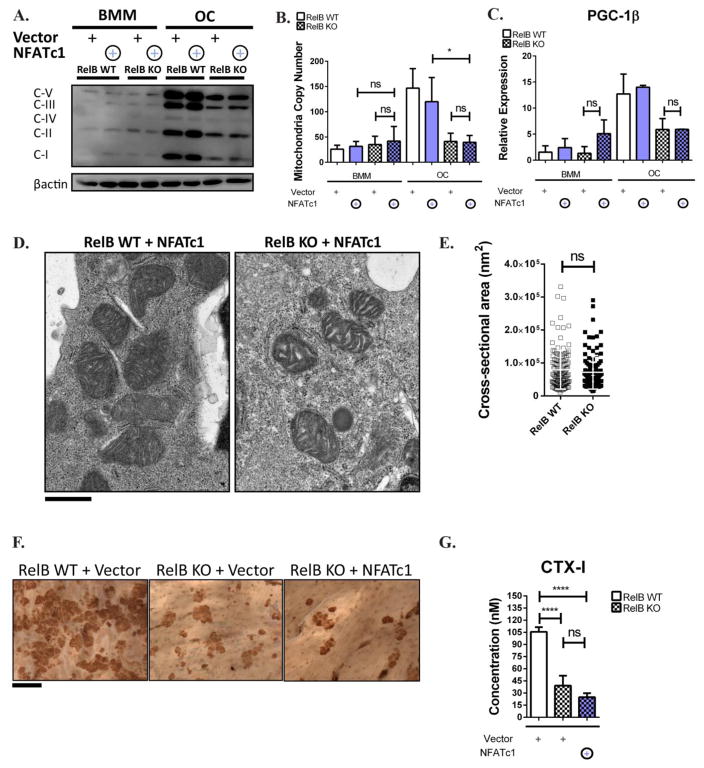
RelB−/− bone marrow derived precursors have severe OC differentiation defects, so it is possible that the failure in PGC-1β-driven mitochondrial biogenesis is secondary to the failure in differentiation. In vitro, RelB+/+ BMMs upregulate NFATc1, a master transcription regulator of osteoclastogenesis, during culture with RANKL, but RelB−/− do not, suggesting that RelB is upstream of NFATc1. Other studies have successfully used NFATc1 to rescue differentiation defects in OCs to study the effect of molecules independent from differentiation, (15–21). Therefore to understand the dependence of mitochondrial biogenesis on differentiation, we tried to rescue RelB−/− differentiation in vitro by retroviral overexpression of NFATc1. TRAP staining showed rescue of differentiation (Fig 5A), accompanied by an increase in β3 integrin expression (a marker of OC differentiation) (Fig 5B), while NFATc1 was equally over-expressed in RelB +/+ and −/− cells (Fig 5B,C). Despite the restored OC differentiation in RelB−/− cultures, the defects in mitochondria parameters persisted: RelB−/− OCs with NFATc1 overexpression maintained reduced OxPhos protein expression and mtCN compared to RelB+/+ controls (Fig 6A,B). Furthermore, NFATc1 overexpression in RelB−/− OCs did not alter PGC-1β levels (Fig 6C). Interestingly, mirroring the effects of alternative NF-κB activation, TEM analysis showed that mitochondrial structure in NFATc1-rescued RelB−/− OCs was altered, with reduced inner membrane folds separated by irregular gaps (Fig 6D), while average cross-sectional area was not significantly different (Fig 6E). Importantly, RelB−/− OCs with rescued differentiation did not resorb bone any more than those transduced with empty vector (Fig 6F,G).
Figure 5. Alternative NF-κB regulates OC differentiation through NFATc1.
RelB −/− or RelB +/+ BMMs were transduced with retroviral constructs containing NFATc1 or empty vector.. (A) Representative wells stained with TRAP to assess OC differentiation after 4 days culture in RANKL. Scale bar is 1mm. Representative TRAP image of 3 independent experiments, where the same empty vector served as negative control for NFATc1 and PGC-1β overexpression (B) Western immunoblotting on BMM and OC (3 days after RANKL stimulation) lysates to assess NFATc1 and β3 integrin protein expression. (C) qRT-PCR to assess NFATc1 transcript expression. n=4 *p<0.05, **p<01; ns, not significant.. Solid bars and checkered bars indicate RelB WT and KO, respectively. Purple color indicates overexpression with NFATc1.
Figure 6. Alternative NF-κB Controls OC differentiation through NFATc1, but independent from mitochondria biogenesis and bone resorption.
BMMs were transduced with retroviral constructs containing NFATc1 or empty vector, as in Fig 5. (A) BMMs and day 4 OC lysates blotted with OxPhos antibody cocktail as in Fig 1. (B) qPCR to assess mtCN. n=4. (C) qRT-PCR to assess PGC-1β transcript expression. n=3. (D) OCs were grown for 6 days on bovine bone slices, fixed, dehydrated, and analyzed by transmission electron microscopy. Scale bar: 500nm. (E) Mitochondria cross-sectional area was quantitated in 12 cells (between 115 and 130 total mitochondria) using ImageJ software analysis. Plat-E cells were transfected with retroviral constructs containing NFATc1 or empty vector, as in Fig 5. (F) OCs were grown on bovine bone slices to visualize resorption lacunae using peroxidase-conjugated wheat germ agglutinin, with area resorbed visualized. Scale bar 100 um. (G) Media was collected from bone slices in D, and levels of collagen degradation product CTX-I was measured by ELISA, averaged from 3 bone slices. *p<0.05, ****p<0.0001; ns, not significant. Solid bars and checkered bars indicate RelB WT and KO, respectively. Purple color indicates overexpression with NFATc1.
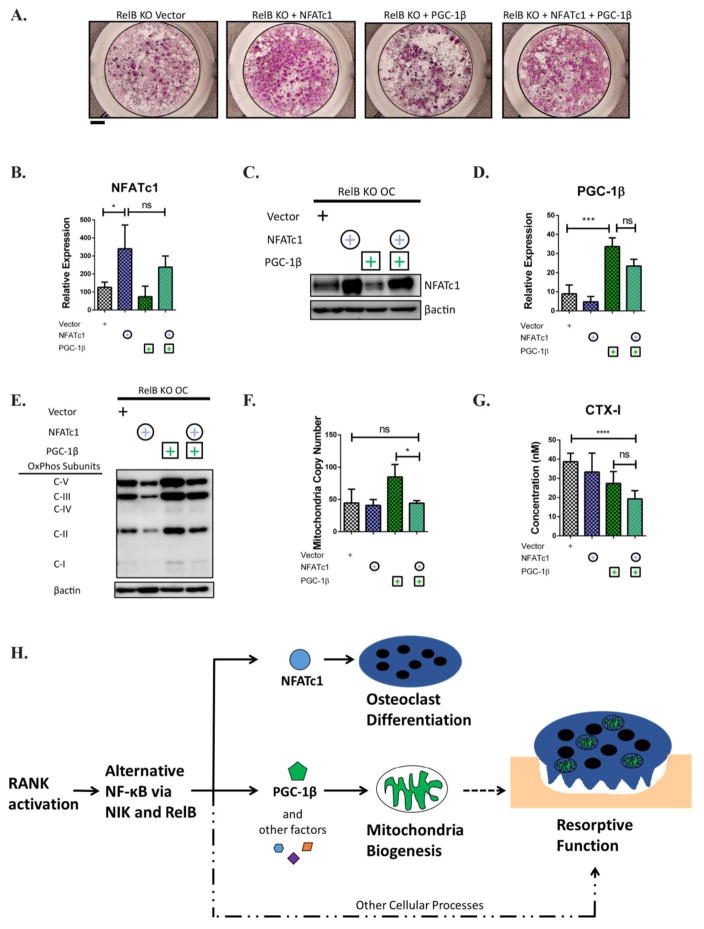
To rule out the possibility that PGC-1β requires upregulation of NFATc1 to enhance mitochondrial biogenesis, we co-overexpressed NFATc1 and PGC-1β in RelB −/− cells. The addition of NFATc1 restored OC differentiation as assessed by TRAP staining (Fig 7A). NFATc1 and PGC-1β over-expression in the double overexpressing cultures was not significantly different from single overexpressing conditions at the mRNA or protein levels (Fig 7B–D). However, addition of NFATc1 to PGC-1β overexpression did not increase OxPhos expression or mitochondria copy number above PGC-1β overexpression alone, and in fact levels were slightly lower in the NFATc1/PGC-1β overexpressing OCs. Most importantly bone resorption was not increased with the addition of NFATc1 (Fig 7E–G). Taken together this supports the model that RelB controls OC resorptive capacity by two distinct and independent pathways: differentiation via NFATc1, and mitochondrial biogenesis through PGC-1β and other factors (Fig 7H).
Figure 7. Co-expression of NFATc1 and PGC-1β fails to rescue mitrochondrial biogenesis or bone resorption in RelB-deficient OCs.
RelB −/− BMMs were transduced with retroviral constructs containing either NFATc1 or PGC-1β, or co-transduced with both, and cultured in M-CSF and RANKL for 4 days. (A) Representative wells stained with TRAP to assess OC differentiation. Scale bar is 1mm. (B) qRT-PCR to assess NFATc1 transcript expression in OCs. n=3. (C) Western immunoblotting on OC lysates to assess NFATc1 protein expression. n=3. (D) qRT-PCR to assess PGC-1β transcript expression in OCs. n=3. (E) OC lysates were immuno-blotted for OxPhos subunits as in Fig 1. Image is representative of 3. (F) qPCR to assess mtCN. n=3. (G) OCs were grown on bovine bone slices for 6 days and media was collected to assess levels of collagen degradation product CTX-I as measured by ELISA, averaged from at least 5 bone slices. *p<0.05, ***p<0.001, ****p<0.0001; ns, not significant. Checkered bars indicate RelB KO. Green and purple colors indicate overexpression with PGC-1β or NFATc1, respectively. (H) Proposed Model. The RANK-activated alternative NF-κB pathway, via NIK and RelB, controls OC differentiation and mitochondrial biogenesis through independent pathways. See text for details.
DISCUSSION
Bone resorption by OCs must be tightly regulated to maintain homeostasis. This may occur at the level of OC differentiation, and thus cell number, or at the level of cellular activation, controlling the amount resorbed per cell(9). Because OCs and their precursors interact with other cells in the bone microenvironment in important ways, it is of interest to understand how various signals affect differentiation and/or function. In this study, we have discovered that the alternative NF-κB plays two distinct roles in regulating bone resorption in the metabolically active OCs; OC differentiation and OC mitochondrial biogenesis are independently regulated by the pathway.
Mitochondrial content (as measured by abundance of OxPhos mRNA and protein expression and by mtDNA copy number) significantly increases as OC precursor cells differentiate into mature OCs. The process of mitochondrial biogenesis has been extensively studied in many cellular contexts, but relatively little has been explored in the OC (22–24). Some studies focusing on the transcriptional co-activator PGC-1β have suggested that OC differentiation is directly linked to mitochondrial biogenesis (2,3). Mice in which PGC-1β is completely ablated in OC lineage cells show defects in mitochondrial biogenesis (2), involving ROS signaling and the pCREB pathway, as well as in OC differentiation. Therefore, it is not clear whether mitochondrial biogenesis itself plays a role in OC maturation, or whether PGC-1β has distinct roles in these two processes. Similar to the experiments with PGC-1β-deficient precursor cells, we found that cells lacking alternative NF-κB pathway activation (via ablation of either NIK or RelB) showed defects in mitochondrial biogenesis, with poor induction of PGC-1β expression, as well as OC differentiation. These experiments establish alternative NF-κB as an upstream regulator of PGC-1β, but do not resolve the question of dependence of differentiation on mitochondrial biogenesis, or vice versa. To begin to address this, we used retrovirus-mediated overexpression to restore PGC-1β levels in RelB-deficient cultures. Interestingly, although PGC-1β increased mitochondrial content in WT and RelB−/− BMMs to a similar extent, PGC-1β overexpression further increased mitochondria only in WT cells stimulated to differentiate with RANKL. Additionally, PGC-1β overexpression did not alter NFATc1 expression or rescue osteoclastogenesis in RelB−/− cultures. Although some level of PGC-1β may be required for osteoclastogenesis, as suggested by the PGC-1β−/− studies, it does not appear that upregulation of PGC-1β beyond a threshold level significantly promotes differentiation.
In order to address the role of OC differentiation in mitochondrial biogenesis we utilized NFATc1, a critical transcription factor that has been shown to restore osteoclastogenesis in a variety of mouse models(15–21). Retroviral expression of NFATc1 did not alter mitochondrial parameters in either WT or RelB-deficient OC cultures, although it did rescue the differentiation defect in the latter. To assess whether PGC-1β requires differentiation signals controlled by NFATc1 to promote mitochondrial biogenesis in the OC lineage, we co-overexpressed both factors in RelB −/− cells. Despite restoring OC formation, the addition of NFATc1 in RelB −/− cells overexpressing PGC-1β did not increase mitochondria copy number nor OxPhos expression beyond PGC-1β alone, indicating that mitochondrial biogenesis in differentiating OCs occurs independent of NFATc1. The additional factors downstream of alternative NF-κB that regulate mitochondria remain to be elucidated.
Further evidence supporting a role for alternative NF-κB in controlling mitochondria independent of differentiation comes from our study of OCs derived from CatK.NT3 mice, which express a constitutively active NIK under the control of the Cathepsin K promoter. At RANKL doses that generate similar numbers of OCs between control and CatK.NT3 cultures, the latter have normal numbers of mitochondria but increased basal and maximal oxygen consumption rates. More interestingly, transmission electron micrographs show that CatK.NT3 OCs grown on bone contain mitochondria with increased inner mitochondrial membrane folds, and an increase in average mitochondria cross-sectional area. Conversely, mature RelB-deficient OCs (generated by NFATc1 overexpression) grown on bone have morphologically abnormal mitochondria, with less folding of the inner mitochondrial membrane and gaps in the cristae. Thus, alternative NF-κB appears to control the quality of mitochondria in addition to their quantity.
Our studies point to a model in which RANKL-activated alternative NF-κB regulates bone resorption in multiple ways (Fig 7H). Since overexpression of NFATc1 in RelB-deficient precursors fully restores differentiation, NFATc1 is the primary mediator of osteocolastogenesis downstream of alternative NF-κB. However, NFATc1-rescued RelB−/− OCs resorb far less bone than their WT counterparts, indicating that RelB controls resorption independent of differentiation. We also demonstrate that RelB controls mitochondrial biogenesis in response to RANKL, but this event is not exclusively controlled by PGC-1β. As in skeletal muscle, RelB regulates PGC-1β upregulation during OC differentiation. However, since overexpression of PGC-1β in RelB-deficient cells, even with overexpression of NFATc1, is not sufficient to support RANKL-induced mitochondrial biogenesis, there appear to be other RelB-dependent factors involved in this process. A recent study showed that an E26 transformation-specific family protein ASXL2 interacts with PPARγ to regulate mitochondria biogenesis and OC differentiation, via two distinctive pathways(34). Although cFOS retroviral expression was able to rescue ASXL2 −/− OC differentiation defects, it was unable to rescue mitochondria biogenesis defects. PGC-1β was also unable to restore mitochondria biogenesis in ASXL2 −/− OC lineage cells, similar to our observations in RelB −/− OC precursors. Thus in both studies, mitochondrial biogenesis was controlled independent of OC differentiation, and does not depend on PGC-1β alone.
The remaining question is the link between mitochondrial biogenesis and bone resorption. Acidification of the resorptive lacuna requires the activity of a V-ATPase and thus a ready supply of intracellular ATP, providing a teleological argument for expansion of mitochondria to support bone resorption by mature OCs. We find that RelB −/− OCs rescued with NFATc1 have lower mitochondrial content and poor resorption, while CatK.NT3 OCs have increased respiration and enhanced resorption. However, the existence of these correlations does not prove that the changes in mitochondria cause alterations in resorption in these models. Interestingly PGC-1β overexpression in WT cells does not further increase bone resorption even though OxPhos expression and mtCN increases, possibly because past a certain threshold of biogenesis, there is no additional effect on bone resorption. Further studies to directly manipulate mitochondria without affecting OC differentiation will likely clarify the role of mitochondria in OC resorptive function.
Supplementary Material
Acknowledgments
This work has been supported by National Institute of Health grants T32AR060719 (R.Z.), RO1AR053628 (R.F.), and RO1AR052705 (D.V.N), as well as Shriners Hospitals for Children (R.F.). We would like to thank Dr. Manolis Pasparakis (University of Cologne) for providing the p65fl/fl mice, Dr. Edward J. Pearce (Washington University) for help with oxygen consumption measurements, Dr. Wandy L. Beatty (Molecular Microbiology Imaging Facility, Washington University) for electron microscopy, Dr. Wei Zou (Washington University) for help with retroviral overexpression assays, and Dr. Gabriel Mbalaviele (Washington University) for critical review of the manuscript.
Footnotes
Authors’ contributions: R.Z., R.F., and D.V.N contributed to experimental design, data interpretation, data analysis and edited the manuscript. R.Z. conducted the experiments.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Bakkar N, Ladner K, Canan BD, et al. IKKalpha and alternative NF-kappaB regulate PGC-1beta to promote oxidative muscle metabolism. J Cell Biol. 2012;196(4):497–511. doi: 10.1083/jcb.201108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishii KA, Fumoto T, Iwai K, et al. Coordination of PGC-1beta and iron uptake in mitochondrial biogenesis and osteoclast activation. Nature medicine. 2009;15(3):259–66. doi: 10.1038/nm.1910. [DOI] [PubMed] [Google Scholar]
- 3.Wei W, Wang X, Yang M, et al. PGC1beta mediates PPARgamma activation of osteoclastogenesis and rosiglitazone-induced bone loss. Cell Metab. 2010;11(6):503–16. doi: 10.1016/j.cmet.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazaki T, Iwasawa M, Nakashima T, et al. Intracellular and extracellular ATP coordinately regulate the inverse correlation between osteoclast survival and bone resorption. J Biol Chem. 2012;287(45):37808–23. doi: 10.1074/jbc.M112.385369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J, Ye S, Fujiwara T, Manolagas SC, Zhao H. Steap4 plays a critical role in osteoclastogenesis in vitro by regulating cellular iron/reactive oxygen species (ROS) levels and cAMP response element-binding protein (CREB) activation. J Biol Chem. 2013;288(42):30064–74. doi: 10.1074/jbc.M113.478750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Z, Wei W, Yang M, Du Y, Wan Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell metabolism. 2014;20(3):483–98. doi: 10.1016/j.cmet.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nature medicine. 2007;13(12):1496–503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- 8.Wan Y. PPARgamma in bone homeostasis. Trends Endocrinol Metab. 2010;21(12):722–8. doi: 10.1016/j.tem.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annu Rev Pathol. 2008;3:457–84. doi: 10.1146/annurev.pathmechdis.3.121806.151431. [DOI] [PubMed] [Google Scholar]
- 10.Novack DV. Role of NF-kappaB in the skeleton. Cell Res. 2011;21(1):169–82. doi: 10.1038/cr.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun SC. The noncanonical NF-kappaB pathway. Immunological reviews. 2012;246(1):125–40. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kular J, Tickner J, Chim SM, Xu J. An overview of the regulation of bone remodelling at the cellular level. Clin Biochem. 2012;45(12):863–73. doi: 10.1016/j.clinbiochem.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Vaira S, Johnson T, Hirbe AC, et al. RelB is the NF-kappaB subunit downstream of NIK responsible for osteoclast differentiation. Proc Natl Acad Sci U S A. 2008;105(10):3897–902. doi: 10.1073/pnas.0708576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, McCoy K, Davis JL, et al. NIK stabilization in osteoclasts results in osteoporosis and enhanced inflammatory osteolysis. PLoS One. 2010;5(11):e15383. doi: 10.1371/journal.pone.0015383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aya K, Alhawagri M, Hagen-Stapleton A, Kitaura H, Kanagawa O, Novack DV. NF-(kappa)B-inducing kinase controls lymphocyte and osteoclast activities in inflammatory arthritis. The Journal of clinical investigation. 2005;115(7):1848–54. doi: 10.1172/JCI23763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2000;15(8):1477–88. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- 17.McHugh KP, Hodivala-Dilke K, Zheng MH, et al. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. The Journal of clinical investigation. 2000;105(4):433–40. doi: 10.1172/JCI8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novack DV, Yin L, Hagen-Stapleton A, et al. The IkappaB function of NF-kappaB2 p100 controls stimulated osteoclastogenesis. J Exp Med. 2003;198(5):771–81. doi: 10.1084/jem.20030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou W, Deselm CJ, Broekelmann TJ, et al. Paxillin contracts the osteoclast cytoskeleton. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2012;27(12):2490–500. doi: 10.1002/jbmr.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra R, Nogee DP, Zechner JF, et al. The transcriptional coactivators, PGC-1alpha and beta, cooperate to maintain cardiac mitochondrial function during the early stages of insulin resistance. Journal of molecular and cellular cardiology. 2012;52(3):701–10. doi: 10.1016/j.yjmcc.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang SC, Everts B, Ivanova Y, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nature immunology. 2014;15(9):846–55. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epple H, Cremasco V, Zhang K, Mao D, Longmore GD, Faccio R. Phospholipase Cgamma2 modulates integrin signaling in the osteoclast by affecting the localization and activation of Src kinase. Mol Cell Biol. 2008;28(11):3610–22. doi: 10.1128/MCB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takayanagi H, Kim S, Koga T, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3(6):889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 24.Matsuo K, Galson DL, Zhao C, et al. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J Biol Chem. 2004;279(25):26475–80. doi: 10.1074/jbc.M313973200. [DOI] [PubMed] [Google Scholar]
- 25.Kim K, Kim JH, Youn BU, Jin HM, Kim N. Pim-1 regulates RANKL-induced osteoclastogenesis via NF-kappaB activation and NFATc1 induction. Journal of immunology. 2010;185(12):7460–6. doi: 10.4049/jimmunol.1000885. [DOI] [PubMed] [Google Scholar]
- 26.Song I, Kim JH, Kim K, Jin HM, Youn BU, Kim N. Regulatory mechanism of NFATc1 in RANKL-induced osteoclast activation. FEBS letters. 2009;583(14):2435–40. doi: 10.1016/j.febslet.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 27.Choi SW, Son YJ, Yun JM, Kim SH. Fisetin Inhibits Osteoclast Differentiation via Downregulation of p38 and c-Fos-NFATc1 Signaling Pathways. Evidence-based complementary and alternative medicine : eCAM. 2012;2012:810563. doi: 10.1155/2012/810563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita T, Yao Z, Li F, et al. NF-kappaB p50 and p52 regulate receptor activator of NF-kappaB ligand (RANKL) and tumor necrosis factor-induced osteoclast precursor differentiation by activating c-Fos and NFATc1. J Biol Chem. 2007;282(25):18245–53. doi: 10.1074/jbc.M610701200. [DOI] [PubMed] [Google Scholar]
- 29.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23(9):459–66. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilkerson R, Bravo L, Garcia I, et al. The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb Perspect Biol. 2013;5(5):a011080. doi: 10.1101/cshperspect.a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luedde T, Heinrichsdorff J, de Lorenzi R, De Vos R, Roskams T, Pasparakis M. IKK1 and IKK2 cooperate to maintain bile duct integrity in the liver. Proc Natl Acad Sci U S A. 2008;105(28):9733–8. doi: 10.1073/pnas.0800198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeda K, Kobayashi Y, Udagawa N, et al. Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nature medicine. 2012;18(3):405–12. doi: 10.1038/nm.2653. [DOI] [PubMed] [Google Scholar]
- 33.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic research. 1999;8(4):265–77. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 34.Izawa T, Rohatgi N, Fukunaga T, Wang Q, Silva MJ, Gardner MJ, McDaniel ML, Abumrad NA, Semenkovich CF, Teitelbaum SL, Zou W. ASXL2 regulates glucose, lipid and skeletal homeostasis. [Accepted 2015];Cell Reports. doi: 10.1016/j.celrep.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.