Summary
Chronic infection with hepatitis B virus (HBV) greatly increases the risk for liver cirrhosis and hepatocellular carcinoma (HCC). HBV isolates worldwide can be divided into 10 genotypes. Moreover, the immune clearance phase selects for mutations in different parts of the viral genome. The outcome of HBV infection is shaped by the complex interplay of the mode of transmission, host genetic factors, viral genotype and adaptive mutations, as well as environmental factors. Core promoter mutations and mutations abolishing HBeAg expression have been implicated in acute liver failure, while genotypes B, C, subgenotype A1, core promoter mutations, preS deletions, C-terminal truncation of envelope proteins, and spliced pregenomic RNA are associated with HCC development. Our efforts to treat and prevent HBV infection are hampered by the emergence of drug resistant mutants and vaccine escape mutants. This paper provides an overview of the HBV life cycle, followed by review of HBV genotypes and mutants in terms of their biological properties and clinical significance.
Introduction
About 240 million people worldwide are chronically infected with hepatitis B virus (HBV), which put them at great risk to develop liver cirrhosis and hepatocellular carcinoma (HCC) [1]. Both viral and host factors contribute to the outcome of HBV infection. In this review we will discuss the current state of knowledge regarding HBV genetic variants, including their role in establishment and maintenance of chronic infection, disease progression, pathogenesis, and treatment response. To better understand these topics, a brief overview of the HBV life cycle is essential.
1. HBV replication cycle
1.1. Genome organization and RNA transcription
HBV is the prototype of a group of hepatotropic DNA viruses infecting birds (ducks, geese, herons) and mammals (woodchucks, ground squirrels, bats) [2]. The 42-nm infectious virion has an internal capsid (core particle) of 27nm to shield a partially double stranded DNA genome of unusual structure. Upon infection of hepatocytes, the relaxed circular (RC) DNA is released and converted to covalently closed circular (ccc) DNA form [3] (Fig. 1). The cccDNA, which forms a minichromosome with high stability (see companion paper by Lucifora and Protzer), is the template for transcription of viral mRNAs utilizing host DNA-dependent RNA polymerase. As this unleashes viral protein expression and genome replication, the cccDNA is the intracellular seed for initiation of the viral life cycle.
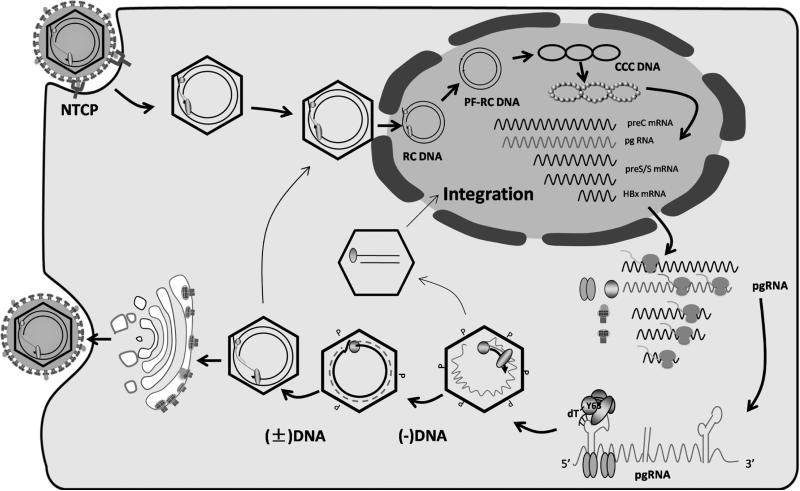
Fig. 1.
HBV life cycle. HBV enters hepatocytes through NTCP, followed by uncoating, and nuclear transport of the RC DNA. The RC DNA is converted to cccDNA, which serves as the template for transcription of the 3.5 kb preC RNA and pgRNA, 2.4- and 2.1-kb preS/S mRNAs, and 0.7-kb HBx mRNA. These RNAs are exported to cytoplasm for protein translation. pgRNA is selectively packaged inside core particles, followed by P protein-mediated (−) strand DNA synthesis (reverse transcription), pgRNA degradation, and (+) strand DNA synthesis to generate RC DNA. Such mature core particles can be enveloped for release as virions, or transported to the nucleus to generate more cccDNA. Double stranded linear DNA is an aberrant replication product of pgRNA, and the preferred template for integration into host chromosomal DNA.
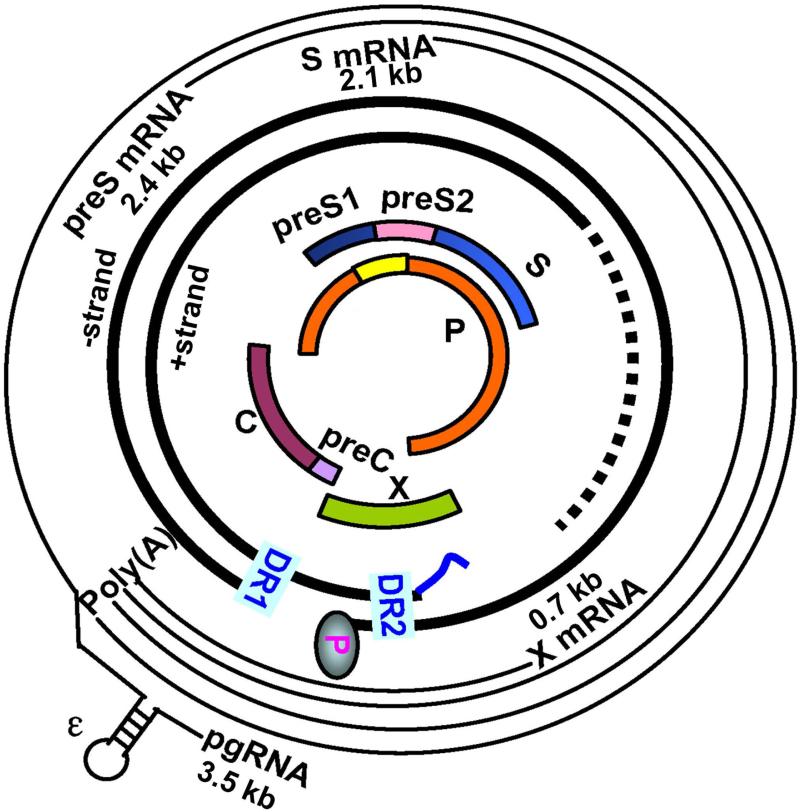
The 3.2-kb HBV genome is the smallest among DNA viruses but also one of the most compact. First, every nucleotide in the cccDNA has coding capacity. Second, the polymerase (P) gene overlaps with the 3’ end of core gene, the entire envelope gene, and the 5’ end of X gene (Fig. 2). Third, related but functionally distinct core and envelope proteins are generated by alternative translation initiation from in-frame AUG codons. Alternative use of the preS1, preS2, and S AUGs in the envelope gene generates large (preS1+preS2+S), middle (preS2+S), and small (S) envelope proteins. Similarly, expression of precore/core and core proteins depends on translation initiation from the precore AUG and core AUG, respectively. Fourth, multiple transcripts are employed to express the seven viral proteins, and the promoters and enhancers needed to generate such transcripts overlap with the coding sequences. Compactness of the HBV genome causes pleiotropic effect of many naturally occurring mutations, thus complicating our efforts to dissect molecular mechanisms of HBV pathogenesis.
Fig. 2.
Genetic organization of the HBV genome and mechanisms of viral protein translation. Shown innermost is the P (polymerase) ORF overlapping completely with the preS1/preS2/S ORF, and partially with preC/C ORF and X ORF. Next are partially double stranded DNA genome found inside virions, with the (−) strand DNA having the P protein attached to its 5’ end and the (+) strand DNA having incomplete 3’ end (dashed line). The two direct repeat (DR) sequences, DR1 and DR2, are critical for HBV DNA replication and genome circularization. The outmost are four classes of HBV RNAs transcribed from the cccDNA template: 0.7-kb X mRNA (for HBx protein), 2.1-kb S mRNA (for M and S proteins), 2.4-kb preS mRNA (for L protein), and 3.5-kb pgRNA (for core and P proteins). In addition, the 3.5-kb precore RNA (not shown) is used for HBeAg expression.
Four promoters: core, SPI, SPII, and X, drive transcriptional initiation at different positions in the HBV genome to generate the 3.5-, 2.4-, 2.1-, and 0.7-kb polyadenylated RNAs, respectively. The four classes of RNAs are co-terminal at the 3’ end due to a single polyadenylation signal in the viral genome (Fig. 2). Transcription of these RNAs is further augmented by two enhancer elements and also by HBx (X gene product), a weak transcriptional transactivator with pleiotropic effect [4]. HBx is required for initiation of HBV infection and has been implicated in hepatocarcinogenesis (see companion paper by Levrero and Zucman-Rossi).
1.2. Protein translation, genome replication, and virion release
The four classes of unspliced HBV RNAs are exported to cytoplasm for protein translation and DNA replication (Fig. 1). The 2.4- and 0.7-kb mRNAs produce L and HBx proteins, respectively, while the heterogeneous 5’ end of the 2.1-kb mRNA enables it to produce both M and S proteins, with the latter being predominant. Similarly, the longer and shorter versions of the 3.5-kb RNA, called precore RNA and pregenomic (pg) RNA, are respectively responsible for the expression of precore/core and core proteins. Cleavage of the N-terminal signal peptide as well as C-terminal arginine-rich sequence converts the precore/core protein to hepatitis B e antigen (HBeAg), a secreted protein dispensable for replication per se but critical for the establishment of chronic infection. The 5’ end of the P gene overlaps with the 3’ end of the core gene, and P protein is translated from pgRNA through ribosomal leaky scanning past the core gene AUG. The outcome is low P/core protein ratio conducive to DNA replication, which is catalyzed by probably just one copy of P protein packaged inside core particles assembled from 180 or 240 copies of core protein.
pgRNA is the only HBV transcript required for genome replication. Besides serving as bicistronic mRNA for core and P proteins, it is also the RNA molecule to be packaged inside core particle for conversion to progeny DNA [5]. Selective packaging of pgRNA but not shorter transcripts is attributed to an RNA secondary structure (ε signal) at its 5’ end. The co-packaged P protein, which contains four structural domains in the order of terminal protein-spacer -reverse transcriptase - RNase H (Fig. 3), catalyzes minus strand DNA synthesis by reverse transcription from the pgRNA template. A tyrosine residue on its terminal protein domain serves as the acceptor site for the first nucleotide of minus strand DNA. The error-prone nature of reverse transcription causes a high mutation rate, estimated at 10-3-6 substitutions per replication cycle according to various investigators [6-13]. That provides a large repertoire of genetic variants for selective outgrowth at different stages of infection. Next, the RNase H domain cleaves pgRNA, with its 5’ most remnant serving as the primer for plus strand DNA synthesis. The core particle is subsequently enveloped by the three envelope proteins and virions are budded through the multivesicular bodies [14]. Surprisingly, majority of virions contain no DNA at all, apparently because core particle assembly does not require pgRNA packaging [15]. The functional significance of such a defective but perhaps interfering form of virions in the natural course of HBV infection, if any, remains to be established.
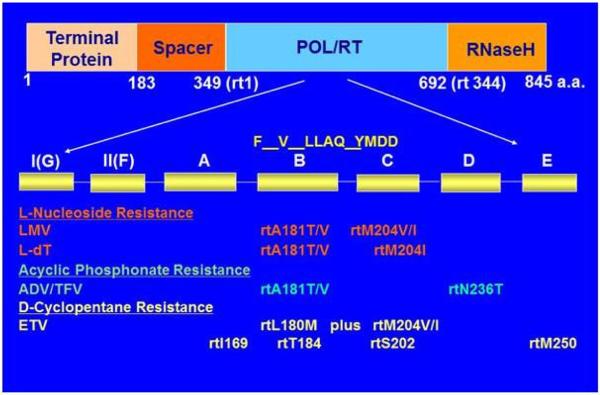
Fig. 3.
Domain structure of the P protein and mutations in its RT domain conferring resistance to NAs. The RT domain can be subdivided into seven subdomains, and mutations conferring resistance to LMV, L-dT, ADV/TFV, and ETV are shown. Adapted from Zoulim and Locarnini [155].
Alternative to their envelopment and release as infectious virions, core particles with mature viral genome can traffic back to the nucleus where the RC DNA is converted to cccDNA (Fig. 1). This intracellular route of cccDNA amplification ensures enough copies of cccDNA per cell to sustain persistent infection. Besides RC DNA, double stranded linear DNA can also be generated from pgRNA template due to the lack of template switch during DNA synthesis. Inside nucleus such linear DNA is the preferred template for integrating into host chromosomal DNA. Integration does not play any role in HBV life cycle, but may contribute to HCC formation depending on the integration site.
Besides virions, the majority of the S protein is secreted as the 22-nm subviral particles lacking internal capsids [14, 16]. Subviral particles outnumber virions by 1,000 fold or greater and are detected serologically as hepatitis B surface antigen (HBsAg). It was the serendipitous detection of HBsAg from Australia aborigines 50 years ago that led to the discovery of the etiological agent for hepatitis B [17].
2. HBV genotypes: their impact on biology and pathogenesis
2.1. Major HBV genotypes and their distributions
Liver cirrhosis and HCC do not stem from acute HBV infection. Rather, they are the consequences of repeated cycles of hepatocyte destruction and regeneration during the immune clearance phase of chronic HBV infection. Consequently, risk for such chronic liver diseases is elevated by host and viral factors that 1) predispose to the establishment of chronic infection, and 2) prolong the immune clearance phase or increase the total cycles of hepatocyte turnover during this phase. In this regard HBV isolates worldwide were initially classified into eight genotypes (A to H) and further divided into subgenotypes based on nucleotide sequence divergence of the entire genome by 8% and 4%, respectively [18-24]. While most genotypes have a genome size of 3215 nucleotides (nt), genotypes A, D, E, and G have a genome of 3221, 3182, 3212, and 3248 nt, respectively due to in-frame insertion or deletion (Table 1). Genotypes A - D are the best characterized, with genotype A most frequent in North America and Africa, genotypes B and C the dominant viruses throughout East Asia, and genotype D most common in Southern Europe and India (Table 1). Genotype E is largely restricted to Sub Saharan Africa, while genotypes F and H co-circulate in indigenous peoples of South America. Genotype G is unusual in its high infection rate in men having sex with men, co-infection with another HBV genotype and HIV, and association with increased risk for liver fibrosis [25-28]. In this regard the unique 36-nucleotide insertion in its core gene causes genotype G to overproduce core protein [29-30]. Genotypes E, G, and H have no subgenotypes, suggesting their recent origin. In contrast, genotype C has 16 subgenotypes and is probably the oldest HBV genotype [13]. Among the HBV subgenotypes, A1 is largely restricted to Africa and India [31-34], B1 to Japan, and C4 to indigenous Australians [35-36]. In Korea only the C2 subgenotype is found [37-38]. Nevertheless, geographic distribution of HBV genotypes is being increasingly altered through travel and migration, as demonstrated by the emergence of European, Asian and African genotypes in countries of low HBV endemicity such as Australia (Bannister E, personal communication).
Table 1.
Virological and clinical features of HBV genotypes A-J
| genotype | genome (nt) | Sub-genotype | geographic distribution | virological features | Clinical features |
|---|---|---|---|---|---|
| A | 32211 | A1-A4 | A1: Africa, Southern Asia; A2: Europe, North America, recently Japan |
Secretes multiple size forms of HBeAg; Rare occurrence of G1896A nonsense mutation to abolish HBeAg expression5; High prevalence of core promoter mutations6. |
Perinatal transmission of A1 in Africa associated with high HCC risk; Sexual transmission of A2 associated with greater risk for chronic infection than genotypes B and C in Japan. |
| B | 3215 | B1-B5 | B1: Japan B2: East Asia B3: Indonesia, Philippines, China |
B2-B4 are recombinants with precore/core gene derived from genotype C; In cell culture B2 has higher replication capacity but lower virion secretion than C2. |
A major cause of chronic HBV infection in East Asia through perinatal transmission. Consequently a major cause of cirrhosis and HCC; Better response to IFN therapy than genotype C. |
| C | 3215 | C1-C16 | C1-C3: Southeast Asia, East Asia C4: Australian aborigines C6-C16: Indonesia |
The oldest HBV genotype; Higher prevalence of core promoter mutations and preS deletions than genotype B; C1 rarely develops G1896A mutation5. |
A major cause of chronic HBV infection in East Asia through perinatal transmission; Delayed HBeAg seroconversion and higher risk for cirrhosis and HCC than genotype B. |
| D | 31822 | D1-D7 | D1: Middle East D2: Europe/Africa D3: worldwide D5: India D7: Africa |
Secretes less HBsAg than genotypes A-C. | A major cause of acute infection through horizontal transmission; A major cause of chronic HBV infection with anti-HBe; More likely to cause severe liver diseases than A2. |
| E | 32123 | - | Western/Central Africa | closest to genotype D | |
| F | 3215 | F1-F4 | South and Central Americas | F2 and F3 isolates rarely develop G1896A mutation5. | Genotype F infection in Alaskan natives associated with much earlier HCC development than infection with other genotypes. |
| G | 32484 | - | Mostly in men having sex with men | High core protein expression due to 36-nt insertion; unable to express HBeAg due to C1817T and G1896A. |
Co-infection with HIV, and HBV genotype A or H. |
| H | 3215 | - | Mexico, Nicarragua | Closest to genotype F; rarely develop G1896A mutation5. |
|
| I | 3215 | I1 and I2 | Southern China, Vietnam, Laos, India | A recombinant between genotypes A, C, G; A single clone characterized in cell culture and in mice showed similar biological properties as genotypes A-D. |
Higher prevalence in male than female; Associated with HCC development. |
| J | 31822 | -- | The single isolate probably originated from Borneo | the single isolate differs from other genotypes by 10.7-15.7%; a probable recombinant between gibbon HBV and HBV genotype C. |
The 88-yr old Japanese man suffered from HCC. |
A 6-nt insertion near the 3’ end of core gene.
A 33-nt deletion at the 5’ end of preS1 region.
A 3-nt deletion at the 5’ end of preS1 region.
A 36-nt insertion at the 5’ end of core gene and a 3-nt deletion at the 5’ end of preS1 region.
Due to C1858, which base pairs with G1896.
A1762T/G1764A and other mutations reduce HBeAg expression but enhance genome replication at the transcriptional level.
2.2. Co-infection by different HBV genotypes and formation of recombinant virus
The same host can be infected by different HBV genotypes, as best demonstrated by the sensitive next generation sequencing technology if one genotype dominates over the other (which is often the case) [26, 39-42]. The consequences of co-infection include genotype switch when the selection pressure changes [25-26, 43-44], and formation of recombinant virus [45-46]. Thus, the core gene of subgenotypes B2 - B5 originated from genotype C [47], while the dominant HBV found in Tibet is a recombinant between genotypes C and D [48]. Recombinants have also been identified for genotypes A/D, G/C and D/E, such as between D7 and E in Africa [49]. While most recombination breakpoints occur in the precore/core region [45], subgenotype C4 identified exclusively in Australian aborigines has a genotype C2 backbone but an S region of genotype J (see below) [36].
2.3. Minor and putative HBV genotypes: I and J
In 2000 three full-length HBV genomes from Vietnam were found to display unusual phylogenetic features. While a 2.2-kb genomic fragment was remotely related to genotype A, the remaining sequence aligned best with genotype C [50]. Further analysis revealed alignment of the preS and S regions with genotypes A and G, respectively, and the remaining sequence with subgenotype C4 [51]. Assigning such recombinant HBV to genotype I was met with resistance, because their divergence from many genotype C isolates was less than 8%. However, with the isolation of similar viruses from Laos and eastern India [52-53], the cutoff for genotypic divergence was reduced to 7.5% and genotype I was recognized [23]. This new genotype is common in Southern China bordering with Vietnam, and has been associated with HCC development [54]. Transfection experiments of a genotype I clone in cell culture and mice suggested its competence in genome replication and expression of HBsAg and HBeAg [55]. Putative genotype J was isolated from an 88-year old Japanese male who probably contracted HBV infection while stationed in Borneo, a South East Asian island. The complete genome of this single isolate (3182 nt) differs from genotypes A-I by at least 9.9%, with the least divergence from a gibbon HBV genome [56]. It is probably a recombinant between gibbon HBV and human HBV genotype C [57], or represented cross-species transmission to human. The particular genotype J isolate was infectious in mice repopulated with human hepatocytes [56].
2.4. Transmission route and viral genotype in the establishment of chronic infection
Age of transmission is the most critical host factor for the establishment of chronic infection (defined by HBsAg positivity for more than 6 months). Thus, more than 90% of perinatal transmission from HBeAg-positive mothers ends up with chronic infection, in contrast to less than 5% of transmission during adulthood. Genotypes B and C are transmitted primarily by the perinatal route leading to high prevalence of chronic HBV infection in East Asia. In contrast, genotypes A and D are usually acquired by horizontal transmission in adulthood (subgenotype A2) or adolescence (genotype D). Studies from Japan revealed higher propensity of the exotic subgenotype A2 to cause chronic infection than genotype B or C during sexual transmission [58-59]. In Europe genotype A was associated with chronic infection in contrast to a much greater contribution of genotype D to acute infection [60].
Even within the same genotype considerable differences in HBV natural history and pathogenesis can be identified, perhaps best exemplified by subgenotypes A1 and A2. A2 (European/North American) is almost always acquired in adulthood and is rarely associated with liver cancer. In stark contrast, A1 (African/Indian) is transmitted in early childhood and associated with rapid progression to liver cancer, without underlying cirrhosis, in young African males [32-33, 61]. In fact adolescents infected with subgenotype A1 are 4.5 times more likely to develop liver cancer than persons infected with other African genotypes [32-34]. Besides viral factors, contribution of confounding environmental and host factors cannot be excluded.
2.5. HBeAg and subviral particles in the establishment of chronic HBV infection
The viral factors facilitating the establishment of chronic infection are HBeAg and excess subviral particles, traits of the wild-type virus. Both antigens have been implicated in the induction of immune tolerance. HBeAg is derived from the precore/core protein by removal of the N-terminal signal peptide of 19 residues followed by furin cleavage of the C-terminal 29 residues [62]. However, genotype A has altered furin cleavage sites to produce additional larger sized forms of HBeAg [63], although the functional significance remains uncertain. Whereas the core protein activates type I T helper (Th1) cells leading to immune attack, HBeAg triggers Th2 type responses promoting immune tolerance. Importantly, it promotes a switch of the immune response to core protein from Th1 to Th2 [64-66]. HBeAg can also cross the placenta to establish immune tolerance in the developing fetus, and suppress a range of innate immune signaling pathways [67-71]. Although acute infection is observed in newborns of women infected with HBeAg-negative HBV, lack of HBeAg expression prevents the establishment of persistent infection. Similarly, genotype G is unable to express HBeAg due to double precore nonsense mutation, which may partly explain why chronic genotype G infection does not occur alone but rather in association with another HBV genotype as the source of HBeAg [25-26, 72].
3. Mutations selected at the immune clearance phase of chronic HBV infection
Immune tolerance induced by HBeAg and HBsAg does not last forever, as evidenced by two seroconversion events: loss of HBeAg followed by rise of anti-HBe antibody, and loss of HBsAg followed by rise of anti-HBs. The first seroconversion is usually accompanied by marked drop in viremia titer, while the second seroconversion coincides with clearance of viremia. The humoral and cell-mediated immunities provide the driving force for replacement of wild-type HBV with various mutants, and also greatly increase sequence heterogeneity to generate viral quasispecies [73-74]. The mutations selected help to reduce or abolish production of HBeAg or subviral particles (as they no longer promote viral persistence), to alter antigenic epitopes of the core or envelope proteins for immune escape, or to augment replication capacity to compensate for virus destruction.
3.1. Precore and core promoter mutations to suppress HBeAg expression. Genotype-dependent emergence of G1896A mutation to abolish HBeAg expression
In the Mediterranean region, patients who seroconvert from HBeAg to anti-HBe may continue to suffer from severe liver diseases, and the search for the “Mediterranean variant” led to the identification of mutations in the precore region preventing HBeAg expression [75-77]. The most common is G1896A converting TGG to TAG, a stop codon. Subsequent studies revealed presence of such mutants elsewhere. Instead of patients being infected with the G1896A mutant, the G1896A mutation arises de novo around the time of HBeAg seroconversion. Interestingly, genotypes A and D co-circulate in the Mediterranean region, but genotype A rarely develops the G1896A mutation. In this regard the precore region overlaps with the ε signal essential for pgRNA packaging, and the G-to-A substitution would improve a U1858 – G1896 base pair in genotype D but disrupt a preexisting C1858 – G1896 base pair in genotype A [78]. Genotype H, subgenotypes C1, F2, and F3 also have C1858 and are similarly hampered in developping the G1896A mutation [79-80]. As expected the G1896A mutation correlates with lower HBeAg levels in vivo [81], which are associated with improved response to interferon treatment [82]. On the other hand, presence of G1896A mutation at as low as 1% of the quasispecies pool has been shown to negatively impact HBsAg loss to tenofovir treatment in genotype A and D patients, for unknown reasons [83].
Core promoter mutations reduce HBeAg expression while enhancing genome replication
The core promoter, which drives transcription of precore RNA and pgRNA, can be divided into basic core promoter (BCP) and core upstream regulatory sequence. The A1762T/G1764A double mutation is the most common BCP mutation, which can be accompanied by additional mutations nearby [84]. The BCP mutations reduce, but do not abolish, HBeAg expression [85-86]. They provide a mechanism whereby HBV genotypes unable to develop the G1896A mutation can suppress HBeAg expression. Indeed, such mutations are more prevalent in genotype A than genotype D [87-88] and in subgenotype C1 than C2 [89-91]. Experiments performed in cell culture confirmed their down regulation of precore RNA to reduce HBeAg expression, but also revealed up regulation of pgRNA to enhance genome replication [92-93]. Whereas the A1762T/G1764A double mutation reduces HBeAg expression less than 30% while increasing genome replication 2 fold, up to 80% reduction in HBeAg expression and 20 fold increase in genome replication can be achieved when additional mutations such as T1753C, C1766T, and T1768A are present [92, 94-95]. Therefore, BCP and precore mutations have distinct functions, and they can coexist in the same genome.
Acute liver failure stems from rapid and massive virus infection of the liver accompanied by equally vigorous immune attack, and HBV isolates implicated in acute liver failure often harbor BCP mutations to augment genome replication in addition to the G1896A precore mutation to abolish HBeAg expression [96-101]. Increased pgRNA transcription will inevitably increase core protein expression to attract greater immune attack. The replication impact of BCP mutations could also explain their higher prevalence in genotype C than genotype B [90, 102-103], as genotype C isolates with wild-type BCP sequence in general displayed lower replication capacity than genotype B isolates, at least in cell culture [104]. Such a low replication capacity may extend the immune tolerance phase, while replacement of the wild-type virus by BCP mutant at the immune clearance phase could prolong survival. These features probably could explain why genotype C patients seroconvert from HBeAg to anti-HBe about a decade later than genotype B patients [105-108], and why such patients have increased lifelong risk to develop liver cirrhosis and HCC [90, 109-111]. In fact BCP mutations were strongly associated with cirrhosis and HCC development irrespective of the viral genotype [111-116]. Recent studies using ultradeep sequencing have shown that patients harboring a high proportion of BCP mutants (≥ 45%) one year after HBeAg seroconversion have a greater likelihood of progressing to cirrhosis than those with a lower mutant percentage(<10%) [116].
BCP overlaps with the X gene, with the T1753C, A1762T, G1764A, and T1768A mutations causing I127T, K130M, V131I, and F132Y mutations in HBx protein, respectively. Some of these mutations have been found to abolish the growth inhibitory effect of wild-type HBx protein, partly through down regulation of p21, a tumor suppressor [117-118]. These findings reveal an alternative mechanism whereby BCP mutations promote HCC formation.
3.2. Envelope gene mutations to alter HBsAg expression or to escape neutralization
Subviral particles are not essential for HBV life cycle, and marked reduction in the expression of envelope proteins did not impair virion secretion if a proper L/S protein ratio was maintained [119]. Furthermore, a genome defective in envelope protein expression can be rescued by envelope proteins produced by another genome in the same cell (transcomplementation). Thus, envelope protein expression can be down regulated. Since anti-preS and anti-S antibodies are neutralizing, mutating the corresponding B cell epitopes could escape neutralization to prolong survival.
Deletions in the preS domains
Large in-frame deletions have been detected in the preS regions of the envelope gene, often clustered at the 3’ end of preS1 and 5’ end of preS2 [120-122]. These deletions are mostly found in genotypes A1, B and C, probably related to their prolonged replicative phase (longest for genotype C) in association with perinatal transmission. Although preS deletions are detectable in some cirrhotic samples, their highest prevalence was found in HCC tissues [122-125]. Presence of such deletions in childhood HCC reinforces its active role in hepatocarcinogenesis [126-127]. The deletions remove C-terminal preS1 domain or N-terminal preS2 domain in the L protein, and in the latter case will prevent M protein expression. Although large deletions are introduced to P protein as well, this corresponds to the spacer region not critical for its function. Nevertheless, the junction between preS1 and preS2 domain is required for virion formation [16], therefore the deletion mutants may be impaired in transmission if unaccompanied by wild-type virus. The preS1 region also corresponds to the SPII promoter for transcription of the 2.1-kb RNA, hence such deletions could reduce S protein expression at the transcriptional level. So far limited experiments have been performed to validate the functional consequences of preS deletions. On the other hand, significant progress has been made in elucidating their mechanisms of hepatocarcinogenesis. Independent studies found that preS deletions are associated with ground glass hepatocytes [128]. In transfection experiments, these deletions caused retention of envelope proteins, triggered ER stress, oxidative stress and DNA damage, and led to cell cycle progression and transformation [129].
HBV RNA splicing removing most part of the P/envelope genes
The 3.5-kb HBV RNA can be spliced [130], although splicing is not essential for HBV DNA replication in vitro. Over a dozen splicing-derived variants have been recognized [131], and the most common are the 2.2-kb singly or doubly spliced variants. The singly spliced 2.2-kb variant has a 1.2-kb deletion between the 3’ core gene and middle of S region, which prevent the expression of full-length P protein. Instead, the N-terminal 46 residues of the P protein are fused to 47 residues encoded by a different reading frame to generate hepatitis B spliced protein (HBSP) [132]. If provided with intact P protein, this splicing variant can be packaged and replicated. In fact, virions harboring such a shortened HBV genome can circulate in blood of chronic HBV carriers, but not in those recovering from acute infection [131, 133]. In fact increased ratio of the splicing variant relative to wild-type virus in the blood correlates with liver fibrosis and HCC [134-135]. Cell culture studies suggested that HBSP promotes hepatoma cell proliferation and migration [136]. Alternatively, pathogenicity of the splicing variant could be attributed to increased expression of core and HBx proteins as well as DNA replication [133, 137], as a consequence of the large deletion which prevents the transcription of 2.4-kb and 2.1-kb RNAs.
Immune escape mutants arising at the late stage of chronic infection
The S domain contains 226 residues, with its immunodominant loop (residues 106-163) exposed on the surface of virions and subviral particles. HBV can be classified into four major serological subtypes: adr, adw, ayr, and ayw, with the “a” determinant (residues 124-147) being the major target of anti-S antibodies developed at the late immune clearance phase. Such antibodies neutralize HBV infectivity by blocking virus binding to heparan sulfate proteoglycans, the low-affinity HBV receptor [138] (see companion paper by Urban and Li). However, single amino acid substitutions such as G145R could markedly reduce the affinity of anti-S antibodies [139-140], and such immune escape mutants could survive the rising anti-HBs antibody better than wild-type virus. Such mutants could contribute to occult HBV infection, which is defined by presence of HBV DNA but no detectable HBsAg [141].
Immune escape mutants as a source of breakthrough HBV infection
The immune escape mutants selected at the late immune clearance phase could delay but not prevent virus elimination, most likely due to additional neutralization by anti-preS1 and anti-preS2 antibodies as well as limited replicative space available. On the other hand, current HBV vaccine consists of yeast-derived recombinant S protein alone [142]. At a time when hepatitis B immune globulin (HBIG) pooled from vaccine recipients was used as the only means to prevent HBV re-infection of the grafted liver, such immune escape mutants were the source of breakthrough infection [143-147]. Currently, children born to HBV infected mothers are given HBIG in addition to HBV vaccine. Unfortunately about 5-10% of such infection cannot be prevented, which could be attributed to intrauterine infection, high maternal viral load, or selection of immune escape mutants [148-149]. Experiments in cell culture suggested that G145R and other immune escape mutants such as T118K, K141E, D144G, C147R, and C149R were severely impaired in virion secretion, which nevertheless could be rescued to various extents by a new N-linked glycosylation site at residue 131 [150-152]. Although T131N/M133T double mutation is needed to create such a glycosylation site for non-A genotypes, coexistence of an escape mutation with the double mutation is not uncommon [144, 153]. Thus, M133T (or N131N/M133T) behaves as a compensatory mutation for G145R. Infection experiments using hepatitis delta virus (HDV), which employs HBV envelope proteins for entry into hepatocytes, suggested that the G145R mutant had sustained infectivity [154]. Therefore, potent immune escape, efficient rescue of virion secretion by compensatory mutation(s), and sustained infectivity could explain why G145R is the most common immune escape mutant.
3.4. Drug resistant mutants of nucleos(t)ide analogues
Current antiviral strategies against chronic HBV infection remain suboptimal [155-156]. To date five nucleos(t)ide analogues (NAs) have been approved for the treatment of HBV infection, namely lamivudine (LMV), adefovir dipivoxil (ADV), entecavir (ETV), telbivudine (LdT), and tenofovir (TDF). They act as competitive inhibitors of the reverse transcription step of HBV DNA synthesis (Fig. 1), because their incorporation into the elongating DNA strand leads to chain termination. Potent inhibitors such as ETV and TDF also target the priming step of reverse transcription and can lower viremia to undetectable level [5]. However, NAs do not affect HBeAg or HBsAg expression and are inefficient at promoting HBeAg or HBsAg seroconversion. Consequently they have to be used for prolonged period of time, which may lead to the development of drug resistance. For example, LMV resistant mutations develop in 23% of patients following 12 months of therapy, rising to 80% by five years of treatment [157]. The drug resistant mutations are located in the RT domain of the P protein (Fig. 3). For LMV, rtM204V/I confers drug resistance but also reduces replication capacity, which can be rescued by compensatory mutations such as rtL180M and/or rtV173L [158-159]. ADV resistance is conferred by rtA181T/V or rtN236T mutation. Resistance to ETV requires multiple mutations in addition to those conferring LMV resistance, meaning a higher genetic barrier. However, in the setting of prior resistance to LMV, high rates of resistance to ETV and ADV can occur because of cross-resistance. Although mutations conferring resistance to TDF seem uncommon even following 5 years of treatment [160-161], some mutations may reduce viral sensitivity to TDF therapy [162].
Due to the overlap between the P and envelope genes, drug resistant mutations in the P gene can cause amino acid substitutions in or near the “a” determinant of envelope proteins, such as F158Y, F161L, E164D, I195M, W196S. Such mutations reduced binding of anti-S antibodies suggesting immune escape [163-164]. The rtA181T mutation can convert codon 172 in the S region into a stop codon, thus truncating the envelope proteins by 55 residues. This mutant was deficient in both virion and HBsAg secretion; it had dominant-negative effect on virion secretion from the wild-type virus [165]. The deletion mutant was associated with an increased HCC risk [166], possibly due to intracellular retention of the envelope proteins leading to ER stress.
Concluding statement
Direct maternal transmission, as best exemplified by genotype C, is the most efficient mechanism whereby HBV has sustained in the human population. Induction of chronic infection is most efficient by the noncytopathic wild-type virus, and requires high doses of HBeAg and HBsAg. On the other hand, mutants replace wild-type virus during immune clearance phase because of their immune escape or higher replication. Consistent with earlier HBeAg than HBsAg seroconversion, core promoter mutations arise much earlier than preS deletions and S domain escape mutations. These mutants contribute to liver diseases including HCC through increased expression of highly immunogenic core protein (core promoter mutants), or envelope protein retention and ER stress (internal deletion of L protein or C-terminal truncation of S protein).
The clinical consequences of HBV mutants can differ between acute and chronic infection. Thus, combination of G1896A and core promoter mutations is associated with acute liver failure, whereas core promoter mutations rather than G1896A mutation is consistently associated with HCC development during chronic infection. Emergence of immune escape mutants in the setting of chronic infection will not prevent eventual HBV clearance, but transmission of such mutants to vaccine recipients may lead to breakthrough infection.
HCC formation is promoted by prolonged HBV infection, high viral load, genotypes B and C, core promoter mutations, preS deletions, truncated S protein, and DNA integration. Although genotype F in general has not been associated with HCC, in Alaska natives genotype F (subgenotype F1) is associated with much higher HCC risk than genotypes A-D [167-168]. In fact the median age of HCC diagnosis was 60 years for genotypes A, C, D combined but only 22.5 years for genotype F. Whether certain biological features unique to these particular subgenotype F1 isolates or compounding host/environmental factors are involved warrants further investigation.
Key points.
HBeAg and HBsAg (subviral particles) promote chronic HBV infection, most efficiently in infants and young children, through induction of immune tolerance. Nevertheless, HBeAg expression is dispensable for viral life cycle, and HBeAg-negative mutants with enhanced replication capacity may cause acute liver failure.
Core promoter mutations reduce HBeAg expression but enhance genome replication. They are selected at the early immune clearance phase, and associated with acute liver failure, cirrhosis, and HCC. Cirrhotic and HCC patients also have high prevalence of pre-S deletions, S region truncations, and spliced HBV RNAs. G145R mutation in the S region might pose a threat to the global vaccination program due to immune escape against anti-HBs and sustained infectivity.
Genotypes A and D have wide and overlapping geographic distributions. Genotype D is a major cause of acute adulthood infection and can persist despite HBeAg seroconversion, while subgenotype A2 is highly effective at establishing chronic infection in adults. Genotypes B and C co-circulate in East Asia and are major causes of chronic infection through perinatal transmission. Genotype C is an even greater cause of cirrhosis and HCC because of its higher prevalence of core promoter mutations and delayed HBeAg seroconversion.
Compactness of the HBV genome restricts the emergence of HBeAg-negative G1896A mutation in some HBV genotypes/subgenotypes. It also reduces biological fitness of some primary drug resistant mutants and immune escape mutants, which can be rescued by second-site mutations.
HBV can exist in quasispecies, especially at the late immune clearance phase of chronic infection. Through trans-complementation HBV mutants with defects in HBsAg or other protein expression can still persist in the host and be transmitted.
Acknowledgements
We would like to thank Dr. Yongxiang Wang and Xiaohui Bi for their help in preparation of this manuscript. PR is supported by a Melbourne Health DW Keir Medical Research Fellowship (RF-003-2015). ST has been support by NIH grants AI103648, AI113394 and AI116639.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 2.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 4.Slagle BL, Andrisani OM, Bouchard MJ, Lee CG, Ou JH, Siddiqui A. Technical standards for hepatitis B virus X protein (HBx) research. Hepatology. 2015;61:1416–1424. doi: 10.1002/hep.27360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones SA, Hu J. Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention. Emerg Microbes Infect. 2013;2:e56. doi: 10.1038/emi.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girones R, Miller RH. Mutation rate of the hepadnavirus genome. Virology. 1989;170:595–597. doi: 10.1016/0042-6822(89)90455-8. [DOI] [PubMed] [Google Scholar]
- 7.Orito E, Mizokami M, Ina Y, Moriyama EN, Kameshima N, Yamamoto M, et al. Host-independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc Natl Acad Sci USA. 1989;86:7059–7062. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Günther S, Sommer G, Von Breunig F, Iwanska A, Kalinina T, Sterneck M, et al. Amplification of full-length hepatitis B virus genomes from samples from patients with low levels of viremia: frequency and functional consequences of PCR-introduced mutations. J Clin Microbiol. 1998;36:531–538. doi: 10.1128/jcm.36.2.531-538.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fares MA, Holmes EC. A revised evolutionary history of hepatitis B virus (HBV). J Mol Evol. 2002;54:807–814. doi: 10.1007/s00239-001-0084-z. [DOI] [PubMed] [Google Scholar]
- 10.Torres C, Fernández MDB, Flichman DM, Campos RH, Mbayed VA. Influence of overlapping genes on the evolution of human hepatitis B virus. Virology. 2013;441:40–48. doi: 10.1016/j.virol.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 11.Tedder RS, Bissett SL, Myers R, Ijaz S. The ‘Red Queen’ dilemma–running to stay in the same place: reflections on the evolutionary vector of HBV in humans. Antivir Ther. 2013;18:459–496. doi: 10.3851/IMP2655. [DOI] [PubMed] [Google Scholar]
- 12.Bouckaert R, Alvarado-Mora MV, Pinho JR. Evolutionary rates and HBV: issues of rate estimation with Bayesian molecular methods. Antivir Ther. 2013;18:497–503. doi: 10.3851/IMP2656. [DOI] [PubMed] [Google Scholar]
- 13.Paraskevis D, Magiorkinis G, Magiorkinis E, Ho SY, Belshaw R, Allain JP, et al. Dating the origin and dispersal of hepatitis B virus infection in humans and primates. Hepatology. 2013;57:908–916. doi: 10.1002/hep.26079. [DOI] [PubMed] [Google Scholar]
- 14.Prange R. Host factors involved in hepatitis B virus maturation, assembly, and egress. Med Microbiol Immunol. 2012;201:449–461. doi: 10.1007/s00430-012-0267-9. [DOI] [PubMed] [Google Scholar]
- 15.Ning X, Nguyen D, Mentzer L, Adams C, Lee H, Ashley R, et al. Secretion of genome-free hepatitis B virus--single strand blocking model for virion morphogenesis of pararetrovirus. PLoS Pathog. 2011;7:e1002255. doi: 10.1371/journal.ppat.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruss V. Hepatitis B virus morphogenesis. World J Gastroenterol. 2007;13:65–73. doi: 10.3748/wjg.v13.i1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blumberg BS, Alter HJ, Visnich S. A “new” antigen in leukemia sera. JAMA. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 18.Chu CJ, Lok AS. Clinical significance of hepatitis B virus genotypes. Hepatology. 2002;35:1274–1276. doi: 10.1053/jhep.2002.33161. [DOI] [PubMed] [Google Scholar]
- 19.Norder H, Courouce AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, et al. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 20.Kramvis A, Arakawa K, Yu MC, Nogueira R, Stram DO, Kew MC. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J Med Virol. 2008;80:27–46. doi: 10.1002/jmv.21049. [DOI] [PubMed] [Google Scholar]
- 21.Kim BK, Revill PA, Ahn SH. HBV genotypes: relevance to natural history, pathogenesis and treatment of chronic hepatitis B. Antivir Ther. 2011;16:1169–1186. doi: 10.3851/IMP1982. [DOI] [PubMed] [Google Scholar]
- 22.Tong S, Li J, Wands JR, Wen YM. Hepatitis B virus genetic variants: biological properties and clinical implications. Emerg Microbes Infect. 2013;2:e10. doi: 10.1038/emi.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141–150. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- 24.Lin C-L, Kao J-H. Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med. 2015;5:a021436. doi: 10.1101/cshperspect.a021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran A, Kremsdorf D, Capel F, Housset C, Dauguet C, Petit MA, et al. Emergence of and takeover by hepatitis B virus (HBV) with rearrangements in the pre-S/S and pre-C/C genes during chronic HBV infection. J Virol. 1991;65:3566–3574. doi: 10.1128/jvi.65.7.3566-3574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato H, Orito E, Gish RG, Bzowej N, Newsom M, Sugauchi F, et al. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology. 2002;35:922–929. doi: 10.1053/jhep.2002.32096. [DOI] [PubMed] [Google Scholar]
- 27.Lacombe K, Massari V, Girard PM, Serfaty L, Gozlan J, Pialoux G, et al. Major role of hepatitis B genotypes in liver fibrosis during coinfection with HIV. AIDS. 2006;20:419–427. doi: 10.1097/01.aids.0000200537.86984.0e. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka Y, Sanchez LV, Sugiyama M, Sakamoto T, Kurbanov F, Tatematsu K, et al. Characteristics of hepatitis B virus genotype G coinfected with genotype H in chimeric mice carrying human hepatocytes. Virology. 2008;376:408–415. doi: 10.1016/j.virol.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Li K, Zoulim F, Pichoud C, Kwei K, Villet S, Wands J, et al. Critical role of the 36-nucleotide insertion in hepatitis B virus genotype G in core protein expression, genome replication, and virion secretion. J Virol. 2007;81:9202–9215. doi: 10.1128/JVI.00390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutelius D, Li J, Wands J, Tong S. Characterization of the pleiotropic effects of the genotype G-specific 36-nucleotide insertion in the context of other hepatitis B virus genotypes. J Virol. 2011;85:13278–13289. doi: 10.1128/JVI.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimbi GC, Kramvis A, Kew MC. Distinctive sequence characteristics of subgenotype A1 isolates of hepatitis B virus from South Africa. J Gen Virol. 2004;85:1211–1220. doi: 10.1099/vir.0.19749-0. [DOI] [PubMed] [Google Scholar]
- 32.Kew MC, Kramvis A, Yu MC, Arakawa K, Hodkinson J. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-saharan Africans. J Med Virol. 2005;75:513–521. doi: 10.1002/jmv.20311. [DOI] [PubMed] [Google Scholar]
- 33.Kramvis A, Kew MC. Epidemiology of hepatitis B virus in Africa, its genotypes and clinical associations of genotypes. Hepatol Res. 2007;37:S9–S19. doi: 10.1111/j.1872-034X.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- 34.Kramvis A, Kew MC. Molecular characterization of subgenotype A1 (subgroup Aa) of hepatitis B virus. Hepatol Res. 2007;37:S27–S32. doi: 10.1111/j.1872-034X.2007.00100.x. [DOI] [PubMed] [Google Scholar]
- 35.Davies J, Littlejohn M, Locarnini SA, Whiting S, Hajkowicz K, Cowie BC, et al. Molecular epidemiology of hepatitis B in the Indigenous people of northern Australia. J Gastroenterol Hepatol. 2013;28:1234–1241. doi: 10.1111/jgh.12177. [DOI] [PubMed] [Google Scholar]
- 36.Littlejohn M, Davies J, Yuen L, Edwards R, Sozzi T, Jackson K, et al. Molecular virology of hepatitis B virus, sub-genotype C4 in northern Australian Indigenous populations. J Med Virol. 2014;86:695–706. doi: 10.1002/jmv.23888. [DOI] [PubMed] [Google Scholar]
- 37.Ahn SH, Yuen L, Revill P. Clarification required for the definition of hepatitis B virus subgenotypes C1 and C2. Intervirology. 2009;52:321–322. doi: 10.1159/000237739. [DOI] [PubMed] [Google Scholar]
- 38.Ahn SH, Yuen L, Han KH, Littlejohn M, Chang HY, Damerow H, et al. Molecular and clinical characteristics of hepatitis B virus in Korea. J Med Virol. 2010;82:1126–1134. doi: 10.1002/jmv.21844. [DOI] [PubMed] [Google Scholar]
- 39.Hannoun C, Krogsgaard K, Horal P, Lindh M. Genotype mixtures of hepatitis B virus in patients treated with interferon. J Infect Dis. 2002;186:752–759. doi: 10.1086/342599. [DOI] [PubMed] [Google Scholar]
- 40.Andernach IE, Jutavijittum P, Samountry B, Yousukh A, Thammavong T, Hubschen JM, et al. A high variability of mixed infections and recent recombinations of hepatitis B virus in Laos. PloS One. 2012;7:e30245. doi: 10.1371/journal.pone.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beggel B, Neumann-Fraune M, Doring M, Lawyer G, Kaiser R, Verheyen J, et al. Genotyping hepatitis B virus dual infections using population-based sequence data. J Gen Virol. 2012;93:1899–1907. doi: 10.1099/vir.0.043042-0. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Shan X, Liang Z, Shan Y, Huang W, Zhang D, et al. Deep sequencing analysis of HBV genotype shift and correlation with antiviral efficiency during adefovir dipivoxil therapy. PloS One. 2015;10:e0131337. doi: 10.1371/journal.pone.0131337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerner PR, Friedt M, Oettinger R, Lausch E, Wirth S. The hepatitis B virus seroconversion to anti-HBe is frequently associated with HBV genotype changes and selection of preS2-defective particles in chronically infected children. Virology. 1998;245:163–172. doi: 10.1006/viro.1998.9126. [DOI] [PubMed] [Google Scholar]
- 44.Jardi R, Rodriguez-Frias F, Schaper M, Giggi E, Tabernero D, Homs M, et al. Analysis of hepatitis B genotype changes in chronic hepatitis B infection: Influence of antiviral therapy. J Hepatol. 2008;49:695–701. doi: 10.1016/j.jhep.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 45.Shi W, Carr MJ, Dunford L, Zhu C, Hall WW, Higgins DG. Identification of novel inter-genotypic recombinants of human hepatitis B viruses by large-scale phylogenetic analysis. Virology. 2012;427:51–59. doi: 10.1016/j.virol.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 46.Araujo NM. Hepatitis B virus intergenotypic recombinants worldwide: An overview. Infect Genet Evol. 2015;36:500–510. doi: 10.1016/j.meegid.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 47.Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, et al. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol. 2002;76:5985–5992. doi: 10.1128/JVI.76.12.5985-5992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui C, Shi J, Hui L, Xi H, Zhuoma Quni, et al. The dominant hepatitis B virus genotype identified in Tibet is a C/D hybrid. J Gen Virol. 2002;83:2773–2777. doi: 10.1099/0022-1317-83-11-2773. [DOI] [PubMed] [Google Scholar]
- 49.Abdou Chekaraou M, Brichler S, Mansour W, Le Gal F, Garba A, Deny P, et al. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J Gen Virol. 2010;91:1609–1620. doi: 10.1099/vir.0.018127-0. [DOI] [PubMed] [Google Scholar]
- 50.Hannoun C, Norder H, Lindh M. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J Gen Virol. 2000;81:2267–2272. doi: 10.1099/0022-1317-81-9-2267. [DOI] [PubMed] [Google Scholar]
- 51.Huy TTT, Ngoc TT, Abe K. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J Virol. 2008;82:5657–5663. doi: 10.1128/JVI.02556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olinger CM, Jutavijittum P, Hübschen JM, Yousukh A, Samountry B, Thammavong T, et al. Possible new hepatitis B virus genotype, southeast Asia. Emerg Infect Dis. 2008;14:1777. doi: 10.3201/eid1411.080437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arankalle VA, Gandhe SS, Borkakoty BJ, Walimbe AM, Biswas D, Mahanta J. A novel HBV recombinant (genotype I) similar to Vietnam/Laos in a primitive tribe in eastern India. J Viral Hepat. 2010;17:501–510. doi: 10.1111/j.1365-2893.2009.01206.x. [DOI] [PubMed] [Google Scholar]
- 54.Fang Z-L, Hué S, Sabin CA, Li G-J, Yang J-Y, Chen Q-Y, et al. A complex hepatitis B virus (X/C) recombinant is common in Long An county, Guangxi and may have originated in southern China. J Gen Virol. 2011;92:402–411. doi: 10.1099/vir.0.026666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H, Yuan Q, Ge S-X, Wang H-Y, Zhang Y-L, Chen Q-R, et al. Molecular and phylogenetic analyses suggest an additional hepatitis B virus genotype “I”. PloS One. 2010;5:e9297. doi: 10.1371/journal.pone.0009297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, et al. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83:10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Locarnini S, Littlejohn M, Aziz MN, Yuen L. Possible origins and evolution of the hepatitis B virus (HBV). Semin Cancer Biol. 2013;23:561–575. doi: 10.1016/j.semcancer.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki Y, Kobayashi M, Ikeda K, Suzuki F, Arfase Y, Akuta N, et al. Persistence of acute infection with hepatitis B virus genotype A and treatment in Japan. J Med Virol. 2005;76:33–39. doi: 10.1002/jmv.20320. [DOI] [PubMed] [Google Scholar]
- 59.Matsuura K, Tanaka Y, Hige S, Yamada G, Murawaki Y, Komatsu M, et al. Distribution of hepatitis B virus genotypes among patients with chronic infection in Japan shifting toward an increase of genotype A. J Clin Microbiol. 2009;47:1476–1483. doi: 10.1128/JCM.02081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayerat C, Mantegani A, Frei PC. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J Viral Hepat. 1999;6:299–304. doi: 10.1046/j.1365-2893.1999.00174.x. [DOI] [PubMed] [Google Scholar]
- 61.Gopalakrishnan D, Keyter M, Shenoy KT, Leena KB, Thayumanavan L, Thomas V, et al. Hepatitis B virus subgenotype A1 predominates in liver disease patients from Kerala, India. World J Gastroenterol. 2013;19:9294–9306. doi: 10.3748/wjg.v19.i48.9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Messageot F, Salhi S, Eon P, Rossignol JM. Proteolytic processing of the hepatitis B virus e antigen precursor. Cleavage at two furin consensus sequences. J Biol Chem. 2003;278:891–895. doi: 10.1074/jbc.M207634200. [DOI] [PubMed] [Google Scholar]
- 63.Ito K, Kim KH, Lok AS, Tong S. Characterization of genotype-specific carboxyl-terminal cleavage sites of hepatitis B virus e antigen precursor and identification of furin as the candidate enzyme. J Virol. 2009;83:3507–3517. doi: 10.1128/JVI.02348-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milich DR, Jones JE, Hughes JL, Price J, Raney AK, McLachlan A. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc Natl Acad Sci USA. 1990;87:6599–6603. doi: 10.1073/pnas.87.17.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Milich DR, Chen MK, Hughes JL, Jones JE. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J Immunol. 1998;160:2013–2021. [PubMed] [Google Scholar]
- 66.Chen MT, Billaud JN, Sallberg M, Guidotti LG, Chisari FV, Jones J, et al. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc Natl Acad Sci USA. 2004;101:14913–14918. doi: 10.1073/pnas.0406282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson AJ, Colledge D, Rodgers S, Wilson R, Revill P, Desmond P, et al. Stimulation of the interleukin-1 receptor and Toll-like receptor 2 inhibits hepatitis B virus replication in hepatoma cell lines in vitro. Antivir Ther. 2009;14:797–808. doi: 10.3851/IMP1294. [DOI] [PubMed] [Google Scholar]
- 68.Lang T, Lo C, Skinner N, Locarnini S, Visvanathan K, Mansell A. The hepatitis B e antigen (HBeAg) targets and suppresses activation of the toll-like receptor signaling pathway. J Hepatol. 2011;55:762–769. doi: 10.1016/j.jhep.2010.12.042. [DOI] [PubMed] [Google Scholar]
- 69.Wilson R, Warner N, Ryan K, Selleck L, Colledge D, Rodgers S, et al. The hepatitis B e antigen suppresses IL-1beta-mediated NF-kappaB activation in hepatocytes. J Viral Hepat. 2011;18:e499–507. doi: 10.1111/j.1365-2893.2011.01484.x. [DOI] [PubMed] [Google Scholar]
- 70.Jegaskanda S, Ahn SH, Skinner N, Thompson AJ, Ngyuen T, Holmes J, et al. Down-regulation of IL-18 mediated cell signalling and IFN-gamma expression by the hepatitis B virus e antigen. J Virol. 2014;88:10412–10420. doi: 10.1128/JVI.00111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Visvanathan K, Lang T, Ryan K, R W, NA S, AJV T, et al. TIR- mediated innate immune responses vary across HBV genotype and predict treatment response to Peg-IFN in HBeAg positive CHB patients. J Viral Hepat. 2015 doi: 10.1111/jvh.12477. in press. [DOI] [PubMed] [Google Scholar]
- 72.Sanchez L, Tanaka Y, Maldonado M, Mizokami M, Panduro A. Difference of hepatitis B virus genotype distribution in two groups of Mexican patients with different risk factors. Intervirology. 2007;50:9–15. doi: 10.1159/000096307. [DOI] [PubMed] [Google Scholar]
- 73.Sede M, Lopez-Ledesma M, Frider B, Pozzati M, Campos RH, Flichman D, et al. Hepatitis B virus depicts a high degree of conservation during the immune-tolerant phase in familiarly transmitted chronic hepatitis B infection: deep-sequencing and phylogenetic analysis. J Viral Hepat. 2014;21:650–661. doi: 10.1111/jvh.12196. [DOI] [PubMed] [Google Scholar]
- 74.Lin YY, Liu C, Chien WH, Wu LL, Tao Y, Wu D, et al. New insights into the evolutionary rate of hepatitis B virus at different biological scales. J Virol. 2015;89:3512–3522. doi: 10.1128/JVI.03131-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, et al. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 76.Brunetto MR, Stemler M, Bonino F, Schodel F, Oliveri F, Rizzetto M, et al. A new hepatitis B virus strain in patients with severe anti-HBe positive chronic hepatitis B. J Hepatol. 1990;10:258–261. doi: 10.1016/0168-8278(90)90062-v. [DOI] [PubMed] [Google Scholar]
- 77.Tong SP, Li JS, Vitvitski L, Trepo C. Active hepatitis B virus replication in the presence of anti-HBe is associated with viral variants containing an inactive pre-C region. Virology. 1990;176:596–603. doi: 10.1016/0042-6822(90)90030-u. [DOI] [PubMed] [Google Scholar]
- 78.Li JS, Tong SP, Wen YM, Vitvitski L, Zhang Q, Trepo C. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J Virol. 1993;67:5402–5410. doi: 10.1128/jvi.67.9.5402-5410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lok AS, Akarca U, Greene S. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc Natl Acad Sci USA. 1994;91:4077–4081. doi: 10.1073/pnas.91.9.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Norder H, Arauz-Ruiz P, Blitz L, Pujol FH, Echevarria JM, Magnius LO. The T(1858) variant predisposing to the precore stop mutation correlates with one of two major genotype F hepatitis B virus clades. J Virol. 2003;84:2083–2087. doi: 10.1099/vir.0.19034-0. [DOI] [PubMed] [Google Scholar]
- 81.Thompson AJ, Nguyen T, Iser D, Ayres A, Jackson K, Littlejohn M, et al. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology. 2010;51:1933–1944. doi: 10.1002/hep.23571. [DOI] [PubMed] [Google Scholar]
- 82.Fried MW, Piratvisuth T, Lau GK, Marcellin P, Chow WC, Cooksley G, et al. HBeAg and hepatitis B virus DNA as outcome predictors during therapy with peginterferon alfa-2a for HBeAg-positive chronic hepatitis B. Hepatology. 2008;47:428–434. doi: 10.1002/hep.22065. [DOI] [PubMed] [Google Scholar]
- 83.Bayliss J, Rosenberg G, Thompson A, Gaggar A, Kitrinos K, Subramanian M, et al. HBV variants present in treatment naïve patients can predict response to NA therapy in immune clearance disease. J Hepatol. 2015;62:S570–S571. [Google Scholar]
- 84.Okamoto H, Tsuda F, Akahane Y, Sugai Y, Yoshiba M, Moriyama K, et al. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol. 1994;68:8102–8110. doi: 10.1128/jvi.68.12.8102-8110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buckwold VE, Xu Z, Chen M, Yen TS, Ou JH. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moriyama K, Okamoto H, Tsuda F, Mayumi M. Reduced precore transcription and enhanced core-pregenome transcription of hepatitis B virus DNA after replacement of the precore-core promoter with sequences associated with e antigen-seronegative persistent infections. Virology. 1996;226:269–280. doi: 10.1006/viro.1996.0655. [DOI] [PubMed] [Google Scholar]
- 87.Tanaka Y, Hasegawa I, Kato T, Orito E, Hirashima N, Acharya SK, et al. A case-control study for differences among hepatitis B virus infections of genotypes A (subtypes Aa and Ae) and D. Hepatology. 2004;40:747–755. doi: 10.1002/hep.20365. [DOI] [PubMed] [Google Scholar]
- 88.Grabarczyk P, Garmiri P, Liszewski G, Doucet D, Sulkowska E, Brojer E, et al. Molecular and serological characterization of hepatitis B virus genotype A and D infected blood donors in Poland. J Viral hepat. 2010;17:444–452. doi: 10.1111/j.1365-2893.2009.01192.x. [DOI] [PubMed] [Google Scholar]
- 89.Chan HL, Tsui SK, Tse CH, Ng EY, Au TC, Yuen L, et al. Epidemiological and virological characteristics of 2 subgroups of hepatitis B virus genotype C. J Infect Dis. 2005;191:2022–2032. doi: 10.1086/430324. [DOI] [PubMed] [Google Scholar]
- 90.Wang Z, Tanaka Y, Huang Y, Kurbanov F, Chen J, Zeng G, et al. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J Clin Microbiol. 2007;45:1491–1496. doi: 10.1128/JCM.02157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuan J, Zhou B, Tanaka Y, Kurbanov F, Orito E, Gong Z, et al. Hepatitis B virus (HBV) genotypes/subgenotypes in China: mutations in core promoter and precore/core and their clinical implications. J Clin Virol. 2007;39:87–93. doi: 10.1016/j.jcv.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 92.Baumert TF, Rogers SA, Hasegawa K, Liang TJ. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J Clin Invest. 1996;98:2268–2276. doi: 10.1172/JCI119037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsai A, Kawai S, Kwei K, Gewaily D, Hutter A, Tong DR, et al. Chimeric constructs between two hepatitis B virus genomes confirm transcriptional impact of core promoter mutations and reveal multiple effects of core gene mutations. Virology. 2009;387:364–372. doi: 10.1016/j.virol.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parekh S, Zoulim F, Ahn SH, Tsai A, Li J, Kawai S, et al. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol. 2003;77:6601–6612. doi: 10.1128/JVI.77.12.6601-6612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jammeh S, Tavner F, Watson R, Thomas HC, Karayiannis P. Effect of basal core promoter and pre-core mutations on hepatitis B virus replication. J Gen Virol. 2008;89:901–909. doi: 10.1099/vir.0.83468-0. [DOI] [PubMed] [Google Scholar]
- 96.Carman WF, Fagan EA, Hadziyannis S, Karayiannis P, Tassopoulos NC, Williams R, et al. Association of a precore genomic variant of hepatitis B virus with fulminant hepatitis. Hepatology. 1991;14:219–222. [PubMed] [Google Scholar]
- 97.Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. New Engl J Med. 1991;324:1699–1704. doi: 10.1056/NEJM199106133242404. [DOI] [PubMed] [Google Scholar]
- 98.Liang TJ, Hasegawa K, Rimon N, Wands JR, Ben-Porath E. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. New Engl J Med. 1991;324:1705–1709. doi: 10.1056/NEJM199106133242405. [DOI] [PubMed] [Google Scholar]
- 99.Sato S, Suzuki K, Akahane Y, Akamatsu K, Akiyama K, Yunomura K, et al. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann Intern Med. 1995;122:241–248. doi: 10.7326/0003-4819-122-4-199502150-00001. [DOI] [PubMed] [Google Scholar]
- 100.Friedt M, Gerner P, Lausch E, Trubel H, Zabel B, Wirth S. Mutations in the basic core promotor and the precore region of hepatitis B virus and their selection in children with fulminant and chronic hepatitis B. Hepatology. 1999;29:1252–1258. doi: 10.1002/hep.510290418. [DOI] [PubMed] [Google Scholar]
- 101.Stuyver L, De Gendt S, Cadranel JF, Van Geyt C, Van Reybroeck G, Dorent R, et al. Three cases of severe subfulminant hepatitis in heart-transplanted patients after nosocomial transmission of a mutant hepatitis B virus. Hepatology. 1999;29:1876–1883. doi: 10.1002/hep.510290614. [DOI] [PubMed] [Google Scholar]
- 102.Lindh M, Hannoun C, Dhillon AP, Norkrans G, Horal P. Core promoter mutations and genotypes in relation to viral replication and liver damage in East Asian hepatitis B virus carriers. J Infect Dis. 1999;179:775–782. doi: 10.1086/314688. [DOI] [PubMed] [Google Scholar]
- 103.Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, et al. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218–223. doi: 10.1053/jhep.2001.20532. [DOI] [PubMed] [Google Scholar]
- 104.Qin Y, Tang X, Garcia T, Hussain M, Zhang J, Lok A, et al. Hepatitis B virus genotype C isolates with wild-type core promoter sequence replicate less efficiently than genotype B isolates but possess higher virion secretion capacity. J Virol. 2011;85:10167–10177. doi: 10.1128/JVI.00819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, et al. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology. 2001;34:590–594. doi: 10.1053/jhep.2001.27221. [DOI] [PubMed] [Google Scholar]
- 106.Chu CJ, Hussain M, Lok AS. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122:1756–1762. doi: 10.1053/gast.2002.33588. [DOI] [PubMed] [Google Scholar]
- 107.Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J Med Virol. 2004;72:363–369. doi: 10.1002/jmv.10534. [DOI] [PubMed] [Google Scholar]
- 108.Livingston SE, Simonetti JP, Bulkow LR, Homan CE, Snowball MM, Cagle HH, et al. Clearance of hepatitis B e antigen in patients with chronic hepatitis B and genotypes A, B, C, D, and F. Gastroenterology. 2007;133:1452–1457. doi: 10.1053/j.gastro.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 109.Yuen MF, Tanaka Y, Mizokami M, Yuen JC, Wong DK, Yuan HJ, et al. Role of hepatitis B virus genotypes Ba and C, core promoter and precore mutations on hepatocellular carcinoma: a case control study. Carcinogenesis. 2004;25:1593–1598. doi: 10.1093/carcin/bgh172. [DOI] [PubMed] [Google Scholar]
- 110.Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, et al. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53:1494–1498. doi: 10.1136/gut.2003.033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu S, Xie J, Yin J, Zhang H, Zhang Q, Pu R, et al. A matched case-control study of hepatitis B virus mutations in the preS and core promoter regions associated independently with hepatocellular carcinoma. J Med Virol. 2011;83:45–53. doi: 10.1002/jmv.21829. [DOI] [PubMed] [Google Scholar]
- 112.Baptista M, Kramvis A, Kew MC. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology. 1999;29:946–953. doi: 10.1002/hep.510290336. [DOI] [PubMed] [Google Scholar]
- 113.Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327–334. doi: 10.1053/gast.2003.50053. [DOI] [PubMed] [Google Scholar]
- 114.Kuang SY, Jackson PE, Wang JB, Lu PX, Munoz A, Qian GS, et al. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci USA. 2004;101:3575–3580. doi: 10.1073/pnas.0308232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Poustchi H, Mohamadkhani A, Bowden S, Montazeri G, Ayres A, Revill P, et al. Clinical significance of precore and core promoter mutations in genotype D hepatitis B-related chronic liver disease. J Viral Hepat. 2008;15:753–760. doi: 10.1111/j.1365-2893.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- 116.Tseng TC, Liu CJ, Yang HC, Chen CL, Yang WT, Tsai CS, et al. Higher proportion of viral basal core promoter mutant increases the risk of liver cirrhosis in hepatitis B carriers. Gut. 2015;64:292–302. doi: 10.1136/gutjnl-2014-306977. [DOI] [PubMed] [Google Scholar]
- 117.Kwun HJ, Jang KL. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic acids Res. 2004;32:2202–2213. doi: 10.1093/nar/gkh553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang Y, Tong S, Tai AW, Hussain M, Lok AS. Hepatitis B virus core promoter mutations contribute to hepatocarcinogenesis by deregulating SKP2 and its target, p21. Gastroenterology. 2011;141:1412–1421. doi: 10.1053/j.gastro.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Garcia T, Li J, Sureau C, Ito K, Qin Y, Wands J, et al. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J Virol. 2009;83:11152–11165. doi: 10.1128/JVI.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sugauchi F, Ohno T, Orito E, Sakugawa H, Ichida T, Komatsu M, et al. Influence of hepatitis B virus genotypes on the development of preS deletions and advanced liver disease. J Med Virol. 2003;70:537–544. doi: 10.1002/jmv.10428. [DOI] [PubMed] [Google Scholar]
- 121.Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006;130:1153–1168. doi: 10.1053/j.gastro.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 122.Chen CH, Changchien CS, Lee CM, Hung CH, Hu TH, Wang JH, et al. Combined mutations in pre-s/surface and core promoter/precore regions of hepatitis B virus increase the risk of hepatocellular carcinoma: a case-control study. J Infect Dis. 2008;198:1634–1642. doi: 10.1086/592990. [DOI] [PubMed] [Google Scholar]
- 123.Bock CT, Tillmann HL, Maschek HJ, Manns MP, Trautwein C. A preS mutation isolated from a patient with chronic hepatitis B infection leads to virus retention and misassembly. Gastroenterology. 1997;113:1976–1982. doi: 10.1016/s0016-5085(97)70018-0. [DOI] [PubMed] [Google Scholar]
- 124.Preikschat P, Gunther S, Reinhold S, Will H, Budde K, Neumayer HH, et al. Complex HBV populations with mutations in core promoter, C gene, and pre-S region are associated with development of cirrhosis in long-term renal transplant recipients. Hepatology. 2002;35:466–477. doi: 10.1053/jhep.2002.30698. [DOI] [PubMed] [Google Scholar]
- 125.Fang ZL, Sabin CA, Dong BQ, Wei SC, Chen QY, Fang KX, et al. Hepatitis B virus pre-S deletion mutations are a risk factor for hepatocellular carcinoma: a matched nested case-control study. J Gen Virol. 2008;89:2882–2890. doi: 10.1099/vir.0.2008/002824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abe K, Thung SN, Wu HC, Tran TT, Le Hoang P, Truong KD, et al. Pre-S2 deletion mutants of hepatitis B virus could have an important role in hepatocarcinogenesis in Asian children. Cancer Sci. 2009;100:2249–2254. doi: 10.1111/j.1349-7006.2009.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang HP, Hsu HY, Chen CL, Ni YH, Wang HY, Tsuei DJ, et al. Pre-S2 deletions of hepatitis B virus and hepatocellular carcinoma in children. Pediatr Res. 2010;67:90–94. doi: 10.1203/PDR.0b013e3181c1b0b7. [DOI] [PubMed] [Google Scholar]
- 128.Wang HC, Wu HC, Chen CF, Fausto N, Lei HY, Su IJ. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am J Pathol. 2003;163:2441–2449. doi: 10.1016/S0002-9440(10)63599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Su IJ, Wang LH, Hsieh WC, Wu HC, Teng CF, Tsai HW, et al. The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis. J Biomed Sci. 2014;21:98. doi: 10.1186/s12929-014-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Su T-S, Lai C-J, Huang J, Lin L-H, Yauk Y, Chang C, et al. Hepatitis B virus transcript produced by RNA splicing. J Virol. 1989;63:4011–4018. doi: 10.1128/jvi.63.9.4011-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Günther S, Sommer G, Iwanska A, Will H. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology. 1997;238:363–371. doi: 10.1006/viro.1997.8863. [DOI] [PubMed] [Google Scholar]
- 132.Soussan P, Garreau F, Zylberberg H, Ferray C, Brechot C, Kremsdorf D. In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J Clin Invest. 2000;105:55–60. doi: 10.1172/JCI8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rosmorduc O, Petit MA, Pol S, Capel F, Bortolotti F, Berthelot P, et al. In vivo and in vitro expression of defective hepatitis B virus particles generated by spliced hepatitis B virus RNA. Hepatology. 1995;22:10–19. [PubMed] [Google Scholar]
- 134.Soussan P, Pol J, Garreau F, Schneider V, Le Pendeven C, Nalpas B, et al. Expression of defective hepatitis B virus particles derived from singly spliced RNA is related to liver disease. J Infect Dis. 2008;198:218–225. doi: 10.1086/589623. [DOI] [PubMed] [Google Scholar]
- 135.Bayliss J, Lim L, Thompson AJ, Desmond P, Angus P, Locarnini S, et al. Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J Hepatol. 2013;59:1022–1028. doi: 10.1016/j.jhep.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 136.Chen W-N, Chen J-Y, Jiao B-Y, Lin W-S, Wu Y-L, Liu L-L, et al. Interaction of the hepatitis B spliced protein with cathepsin B promotes hepatoma cell migration and invasion. J Virol. 2012;86:13533–13541. doi: 10.1128/JVI.02095-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kandpal M, Samal J, Biswas B, Negi A, Mishra VC, Tyagi N, et al. Enhanced hepatitis B virus (HBV) pre-genomic RNA levels and higher transcription efficiency of defective HBV genomes. J Gen Virol. 2015;96:3109–3117. doi: 10.1099/jgv.0.000256. [DOI] [PubMed] [Google Scholar]
- 138.Sureau C, Salisse J. A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus a-determinant. Hepatology. 2013;57:985–994. doi: 10.1002/hep.26125. [DOI] [PubMed] [Google Scholar]
- 139.Waters JA, Kennedy M, Voet P, Hauser P, Petre J, Carman W, et al. Loss of the common “A” determinant of hepatitis B surface antigen by a vaccine-induced escape mutant. J Clin Invest. 1992;90:2543–2547. doi: 10.1172/JCI116148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cooreman MP, van Roosmalen MH, te Morsche R, Sunnen CM, de Ven EM, Jansen JB, et al. Characterization of the reactivity pattern of murine monoclonal antibodies against wild-type hepatitis B surface antigen to G145R and other naturally occurring “a” loop escape mutations. Hepatology. 1999;30:1287–1292. doi: 10.1002/hep.510300508. [DOI] [PubMed] [Google Scholar]
- 141.Raimondo G, Pollicino T, Cacciola I, Squadrito G. Occult hepatitis B virus infection. J Hepatol. 2007;46:160–170. doi: 10.1016/j.jhep.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 142.Gerlich WH. Prophylactic vaccination against hepatitis B: achievements, challenges and perspectives. Med Microbiol Immunol. 2015;204:39–55. doi: 10.1007/s00430-014-0373-y. [DOI] [PubMed] [Google Scholar]
- 143.McMahon G, Ehrlich PH, Moustafa ZA, McCarthy LA, Dottavio D, Tolpin MD, et al. Genetic alterations in the gene encoding the major HBsAg: DNA and immunological analysis of recurrent HBsAg derived from monoclonal antibody—treated liver transplant patients. Hepatology. 1992;15:757–766. doi: 10.1002/hep.1840150503. [DOI] [PubMed] [Google Scholar]
- 144.Carman WF, Trautwein C, van Deursen J, Colman K, Dornan E, McIntyre G, et al. Hepatitis B virus envelope variation after transplantation with and without hepatitis B immune globulin prophylaxis. Hepatology. 1996;24:489–493. doi: 10.1002/hep.510240304. [DOI] [PubMed] [Google Scholar]
- 145.Ghany MG, Ayola B, Villamil FG, Gish RG, Rojter S, Vierling JM, et al. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology. 1998;27:213–222. doi: 10.1002/hep.510270133. [DOI] [PubMed] [Google Scholar]
- 146.Protzer-Knolle U, Naumann U, Bartenschlager R, Berg T, Hopf U, Meyer zum Buschenfelde KH, et al. Hepatitis B virus with antigenically altered hepatitis B surface antigen is selected by high-dose hepatitis B immune globulin after liver transplantation. Hepatology. 1998;27:254–263. doi: 10.1002/hep.510270138. [DOI] [PubMed] [Google Scholar]
- 147.Terrault NA, Zhou S, McCory RW, Pruett TL, Lake JR, Roberts JP, et al. Incidence and clinical consequences of surface and polymerase gene mutations in liver transplant recipients on hepatitis B immunoglobulin. Hepatology. 1998;28:555–561. doi: 10.1002/hep.510280237. [DOI] [PubMed] [Google Scholar]
- 148.Carman WF, Karayiannis P, Waters J, Thomas H, Zanetti A, Manzillo G, et al. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 149.Harrison TJ, Hopes EA, Oon CJ, Zanetti AR, Zuckerman AJ. Independent emergence of a vaccine-induced escape mutant of hepatitis B virus. J Hepatol. 1991;13:S105–S107. doi: 10.1016/0168-8278(91)90037-c. [DOI] [PubMed] [Google Scholar]
- 150.Kalinina T, Iwanski A, Will H, Sterneck M. Deficiency in virion secretion and decreased stability of the hepatitis B virus immune escape mutant G145R. Hepatology. 2003;38:1274–1281. doi: 10.1053/jhep.2003.50484. [DOI] [PubMed] [Google Scholar]
- 151.Ito K, Qin Y, Guarnieri M, Garcia T, Kwei K, Mizokami M, et al. Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J Virol. 2010;84:12850–12861. doi: 10.1128/JVI.01499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kwei K, Tang X, Lok AS, Sureau C, Garcia T, Li J, et al. Impaired virion secretion by hepatitis B virus immune escape mutants and its rescue by wild-type envelope proteins or a second-site mutation. J Virol. 2013;87:2352–2357. doi: 10.1128/JVI.02701-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Song BC, Kim SH, Kim H, Ying YH, Kim HJ, Kim YJ, et al. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J Med Virol. 2005;76:194–202. doi: 10.1002/jmv.20354. [DOI] [PubMed] [Google Scholar]
- 154.Salisse J, Sureau C. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J Virol. 2009;83:9321–9328. doi: 10.1128/JVI.00678-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos (t) ide analogues. Gastroenterology. 2009;137:1593–1608. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 156.Zeisel MB, Lucifora J, Mason WS, Sureau C, Beck J, Levrero M, et al. Towards an HBV cure: state-of-the-art and unresolved questions - report of the ANRS workshop on HBV cure. Gut. 2015;64:1314–1326. doi: 10.1136/gutjnl-2014-308943. [DOI] [PubMed] [Google Scholar]
- 157.Lai CL, Dienstag J, Schiff E, Leung NW, Atkins M, Hunt C, et al. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis. 2003;36:687–696. doi: 10.1086/368083. [DOI] [PubMed] [Google Scholar]
- 158.Ono SK, Kato N, Shiratori Y, Kato J, Goto T, Schinazi RF, et al. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J Clin Invest. 2001;107:449–455. doi: 10.1172/JCI11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Delaney WE, Yang H, Westland CE, Das K, Arnold E, Gibbs CS, et al. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J Virol. 2003;77:11833–11841. doi: 10.1128/JVI.77.21.11833-11841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marcellin P, Heathcote EJ, Buti M, Gane E, de Man RA, Krastev Z, et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- 161.Marcellin P, Buti M, Krastev Z, de Man RA, Zeuzem S, Lou L, et al. Kinetics of hepatitis B surface antigen loss in patients with HBeAg-positive chronic hepatitis B treated with tenofovir disoproxil fumarate. J Hepatol. 2014;61:1228–1237. doi: 10.1016/j.jhep.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sheldon J, Camino N, Rodés B, Bartholomeusz A, Kuiper M, Tacke F, et al. Selection of hepatitis B virus polymerase mutations in HIV-coinfected patients treated with tenofovir. Antiviral Ther. 2005;10:727–734. [PubMed] [Google Scholar]
- 163.Torresi J, Earnest-Silveira L, Deliyannis G, Edgtton K, Zhuang H, Locarnini SA, et al. Reduced antigenicity of the hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology. 2002;293:305–313. doi: 10.1006/viro.2001.1246. [DOI] [PubMed] [Google Scholar]
- 164.Sloan RD, Ijaz S, Moore PL, Harrison TJ, Teo C-G, Tedder RS. Antiviral resistance mutations potentiate hepatitis B virus immune evasion through disruption of its surface antigen a determinant. Antivir Ther. 2008;13:439–447. [PubMed] [Google Scholar]
- 165.Warner N, Locarnini S. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology. 2008;48:88–98. doi: 10.1002/hep.22295. [DOI] [PubMed] [Google Scholar]
- 166.Yeh C-T, Chen T, Hsu C-W, Chen Y-C, Lai M-W, Liang K-H, et al. Emergence of the rtA181T/sW172* mutant increased the risk of hepatoma occurrence in patients with lamivudine-resistant chronic hepatitis B. BMC Cancer. 2011;11:398. doi: 10.1186/1471-2407-11-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Livingston SE, Simonetti JP, McMahon BJ, Bulkow LR, Hurlburt KJ, Homan CE, et al. Hepatitis B virus genotypes in Alaska native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5–11. doi: 10.1086/509894. [DOI] [PubMed] [Google Scholar]
- 168.Kowalec K, Minuk GY, Børresen ML, Koch A, McMahon BJ, Simons B, et al. Genetic diversity of hepatitis B virus genotypes B6, D, and F among circumpolar indigenous individuals. J Viral Hepat. 2013;20:122–130. doi: 10.1111/j.1365-2893.2012.01632.x. [DOI] [PubMed] [Google Scholar]