Novel serological tests allowed for the detection of otherwise unrecognized cases of Middle East respiratory syndrome coronavirus infection among contacts in a hospital-associated respiratory illness outbreak in Jordan in April 2012, resulting in a total of 9 test-positive cases.
Keywords: MERS-CoV, Middle East respiratory syndrome, novel coronavirus, Jordan, seroepidemiology
Abstract
Background. In April 2012, the Jordan Ministry of Health investigated an outbreak of lower respiratory illnesses at a hospital in Jordan; 2 fatal cases were retrospectively confirmed by real-time reverse transcription polymerase chain reaction (rRT-PCR) to be the first detected cases of Middle East respiratory syndrome (MERS-CoV).
Methods. Epidemiologic and clinical characteristics of selected potential cases were assessed through serum blood specimens, medical record reviews, and interviews with surviving outbreak members, household contacts, and healthcare personnel. Cases of MERS-CoV infection were identified using 3 US Centers for Disease Control and Prevention serologic tests for detection of anti–MERS-CoV antibodies.
Results. Specimens and interviews were obtained from 124 subjects. Seven previously unconfirmed individuals tested positive for anti–MERS-CoV antibodies by at least 2 of 3 serologic tests, in addition to 2 fatal cases identified by rRT-PCR. The case-fatality rate among the 9 total cases was 22%. Six subjects were healthcare workers at the outbreak hospital, yielding an attack rate of 10% among potentially exposed outbreak hospital personnel. There was no evidence of MERS-CoV transmission at 2 transfer hospitals having acceptable infection control practices.
Conclusions. Novel serologic tests allowed for the detection of otherwise unrecognized cases of MERS-CoV infection among contacts in a Jordanian hospital-associated respiratory illness outbreak in April 2012, resulting in a total of 9 test-positive cases. Serologic results suggest that further spread of this outbreak to transfer hospitals did not occur. Most subjects had no major, underlying medical conditions; none were on hemodialysis. Our observed case-fatality rate was lower than has been reported from outbreaks elsewhere.
(See the Editorial Commentary by Lucey on pages 1234–6.)
In April 2012, the Jordan Ministry of Health (JMoH) investigated a cluster of 13 suspected pneumonia cases among healthcare personnel, of which 2 were fatal, at a hospital in the city of Zarqa [1]. Despite testing for multiple potential pathogens, the investigation did not identify a known etiology for these infections. Following the discovery of Middle East respiratory syndrome coronavirus (MERS-CoV) in September 2012 [2], specimens from the 2 fatal cases in Jordan were retrospectively tested and both yielded positive results for MERS-CoV by real-time reverse transcription polymerase chain reaction (rRT-PCR), and were reported to the World Health Organization (WHO). These were the first confirmed human cases of infection with this emergent virus, which continues to appear as sporadic cases and clusters internationally, and which is now the focus of worldwide public health investigation and response [3, 4].
Using newly developed serologic assays to determine MERS-CoV antibody responses among case contacts in this outbreak, epidemiologists from the JMoH, US Centers for Disease Control and Prevention (CDC), and regional partners conducted a retrospective seroepidemiologic investigation to (1) confirm whether surviving outbreak members had presence of antibodies to MERS-CoV, (2) ascertain whether viral transmission occurred among household contacts or to other healthcare personnel, and (3) describe the clinical features of all detected MERS-CoV infections in Jordan.
METHODS
Epidemiologic Investigation Methods
We interviewed and collected serum specimens from available members of the initial outbreak (who were admitted to the focal outbreak hospital during the period from 15 March to 30 April 2012 with fever and dry cough, and with radiological evidence of pneumonia), their household contacts (who reported usually sleeping under the same roof as a defined outbreak member during February–April 2012), a sample of healthcare personnel from 3 medical institutions that admitted outbreak subjects (nonsystematic enrollment, with preference toward those reporting close contact with outbreak members), and field investigators from the JMoH. Hospitalized subjects meeting the initial outbreak case definition were subsequently transferred from the focal outbreak hospital to 2 other hospitals in Amman. Participating healthcare personnel were employed at one of these hospitals or at JMoH during February–April 2012.
Epidemiologic data were obtained through medical record reviews and personal interviews during our May 2013 investigation. Interviews were conducted in Arabic, and documented contact history (with outbreak members, household members, visiting travelers, and animals) and occupational exposures. We conducted medical record reviews and key informant interviews with clinicians who provided medical care to patients with suspected infection and heads of infection control units at each medical institution and at the JMoH. Informed consent was obtained prior to serum collection and interviews. As a public health response to a disease outbreak, this investigation did not require institutional review board review.
Laboratory Investigation Methods
All work with live MERS-CoV was done in CDC Biosafety Level 3 (BSL-3) containment facilities in Atlanta, Georgia. Serum samples were inactivated using 2 × 106 rads γ-irradiation and stored at −80°C until use.
To maximize specificity, we defined MERS-CoV antibody positivity as subjects having correlated, positive laboratory results from the HKU5.2N screening enzyme-linked immunosorbent assay (ELISA), as well as confirmed positive results by either the MERS-CoV immunofluorescence assay (IFA) or the MERS-CoV microneutralization assay (MNT) (Supplementary Table 1). An initial indeterminate test result was recorded for those subjects having only a single, uncorrelated positive test result.
Antibody Detection by HKU5.2 Nucleocapsid ELISA and MERS-CoV IFA and MNT
Genetic sequencing data indicate that MERS-CoV is a β-coronavirus (subgroup 2c) similar to the bat CoVs HKU4 and HKU5. The recombinant btHKU5.2 nucleocapsid protein–based ELISA was developed by the CDC to detect the presence of antibodies that cross-react with the HKU5.2 N protein in serum samples from possible MERS cases. If cross-reactive antibodies were detected in serum samples, then confirmation of MERS-specific antibodies was determined by either MERS-CoV MNT or IFA. Pi-BatCoV HKU5.2 nucleocapsid (N) gene in pET-28b (+) plasmid was provided by Dr Susanna Lau, University of Hong Kong. His-tagged recombinant protein was expressed in Escherichia coli and purified by metal affinity chromatography. Recombinant HKU5.2N protein indirect ELISA was developed using a modified version of the severe acute respiratory syndrome (SARS) CoV N ELISA described by Haynes et al [5]. Sera were considered positive when the optical density (OD) values were at or above the 0.43 cutoff value (mean absorbance at 405 nm of sera from US blood donors plus 3 standard deviations). The overall specificity of the assay was determined after screening 545 serum samples from donors in the United States and the Middle East and persons with other non-MERS respiratory infections (eg, human coronavirus [hCoV] OC43, hCoV-229E, SARS-CoV, hCoV-NL63, rhinovirus, human metapneumovirus, H1N1). The assay specificity was 96.7% (527/545). Serum from HKU1 human serum was not available for evaluation; however, HKU1 mouse hyperimmune serum did not cross-react with the HKU5.2 N protein. At a screening dilution of 1:400, sera with OD values at or near the cutoff were titered with serial 4-fold dilutions (1:100–1:6400) and further evaluated using MERS-CoV (Hu/Jordan-N3/2012) (GenBank KC776174.1) IFA and MNT.
MERS-CoV Immunofluorescence
Indirect immunofluorescence was performed by screening sera at a dilution of 1:50 or 1:100 on paraformaldehyde-fixed, acetone-methanol–permeabilized, MERS-CoV–infected or –uninfected control Vero cells. The source of the positive control for this assay was a serum sample from a patient infected with MERS-CoV Hu/England-N1/2012 (provided by M. Zambon, Public Health England). Antihuman immunoglobulin (Ig) G, IgM, and IgA fluorescein isothiocyanate conjugate was used and specific fluorescence was detected under an immunofluorescence microscope. A positive result was scored when fluorescent intensity equaled or was higher than that of the positive control. A weakly positive result was scored when fluorescent intensity was lower than that of the positive control.
MERS-CoV Microneutralization
Serum samples were tested for the presence of neutralizing antibodies to MERS-CoV using a modified MNT method described for SARS-CoV [6]. The neutralization titer was measured as the reciprocal of the highest serum dilution that completely inhibited Vero cell monolayer lysis in at least 1 of the 3 triplicate wells. Controls were included for each MNT assay performed, including the input virus back titration and mock-infected cells. All assay results were confirmed in 3 separate assays, and representative data are presented.
Statistical Methods
Tests of statistical significance were performed between the MERS-CoV antibody–positive and –negative subjects, including Fisher exact test and χ2 tests for categorical variables using SAS software version 9.3 (SAS Institute, Cary, North Carolina).
RESULTS
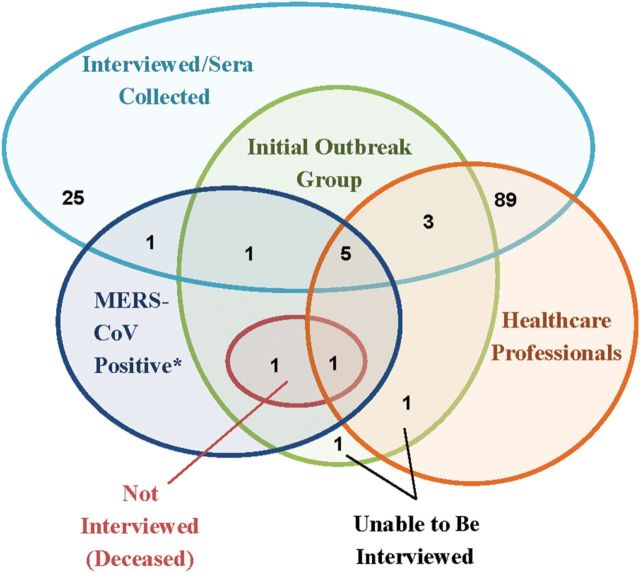
Serologic specimens and interviews were obtained from 124 subjects. We obtained serologic specimens and data from 9 of the 11 (82%) surviving members meeting the initial outbreak case definition; the remaining 2 subjects were unable to be interviewed (1 member was lost to follow-up and 1 did not consent) (Figure 1). We also enrolled 26 household contacts and 89 subjects who did not meet the initial outbreak case definition who worked in healthcare and allied professions. Among the healthcare personnel interviewed, 58% were nurses, 21% were physicians, and the remaining were allied health professionals; approximately half were employed at the focal outbreak hospital.
Figure 1.
Venn diagram of numbers of subjects in the Middle East respiratory syndrome coronavirus (MERS-CoV) investigation. *Tested positive by serologic antibody and/or real-time reverse transcription polymerase chain reaction.
Seven of the 124 subjects tested positive for anti-MERS-CoV antibodies by both HKU5.2 ELISA and IFA (Table 1 and Supplementary Figure 1), and all but 1 also had detectable neutralizing antibody titers as determined by MNT. The subject who did not have detectable neutralizing antibodies was test-positive both by HKU5.2N ELISA and by a confirmative IFA. Demographic and epidemiologic comparisons of seropositive and seronegative subjects are provided in Supplementary Table 2.
Table 1.
Serological Data for Positive and Indeterminate Specimensa in Jordan Investigation Subjects
| Subject Number | HKU5.2N ELISA Titerb | MERS-CoV_Jordan IFAc | MERS-CoV_Jordan MNTd | Initial Interpretatione | Final Interpretationf |
|---|---|---|---|---|---|
| Outbreak member 01g | 1600 | Positive | 80 | Positive | Positive |
| Outbreak member 02 | >6400 | Positive | 160 | Positive | Positive |
| Outbreak member 03 | 400 | Positive | 20 | Positive | Positive |
| Outbreak member 04 | >6400 | Positive | 80 | Positive | Positive |
| Outbreak member 06 | 1600 | Positive | 20 | Positive | Positive |
| Outbreak member 09 | 400 | Positive | 40 | Positive | Positive |
| Outbreak member 11 | 1600 | Positive | <20 | Positive | Positive |
| Household member 303 | 1600 | Positive | 80 | Positive | Positive |
| Outbreakh member 05 | <100 | Weakly positive/negativei | <20 | Indeterminate | Negative |
| Outbreakh member 07 | <100 | Weakly positive/negativei | <20 | Indeterminate | Negative |
| Outbreakh member 10 | <100 | Weakly positive/negativei | <20 | Indeterminate | Negative |
| Outbreakh member 12g,j | <100 | Negative | <20 | Negative | Negative |
| Healthcare personnel 308 | 400 | Weakly positive/negativei | <20 | Positive | Negative |
| Healthcare personnel 330 | 400 | Negative | <20 | Indeterminate | Negative |
| Healthcare personnel 361 | 400 | Negative | <20 | Indeterminate | Negative |
| Healthcare personnel 390 | 400 | Negative | <20 | Indeterminate | Negative |
| Household contact 299 | 1600 | Negative | <20 | Indeterminate | Negative |
Abbreviations: ELISA, enzyme-linked immunosorbent assay; IFA, immunofluorescence assay; MERS-CoV, Middle East respiratory syndrome coronavirus; MNT, microneutralization titer.
a Outbreak member 08 was lost to follow-up, and outbreak member 13 did not consent. Outbreak members 01 and 12 were previously laboratory-confirmed positive by real-time reverse transcription polymerase chain reaction (rRT-PCR) and died. Serum samples from outbreak members 01 and 12 were collected prior to death and stored.
b Serum specimens with optical density (OD) values ≥0.43 at a 1:400 dilution against HKU5.2N ELISA were considered to be positive. Specimens were further titered against HKU5.2N at 1:100, 1:400, 1:1600, and 1:6400 dilutions. The antibody titer was taken to be the highest antibody dilution above the cutoff OD that yielded a ratio of the absorbance of the positive serum and negative serum (P/N) > 3. The value is the reciprocal of the dilution.
c Serum specimens that were positive by HKU5.2N ELISA were screened at either 1:50 or 1:100 by indirect IFA using MERS-CoV_Jordan–infected Vero cells.
d MNT is the highest serum sample dilution that protects at least 1 of 3 wells from complete lysis of the cell monolayers. A sample with a <20 titer was considered negative for the presence of MERS-CoV neutralizing antibodies. Sera from MERS cases were used as positive controls and had neutralizing titers ranging from 1:20 to 1:160.
e Initial interpretation: 2 criteria constitute a positive test result: HKU5.2N ELISA must be positive and the MERS-CoV_Jordan indirect IFA assay must be positive. An indeterminate test result was recorded when only 1 of the criteria was achieved.
f Final interpretation: All positive and indeterminant specimens were screened by MERS-CoV_Jordan MNT and/or rescreened by MERS-CoV IFA. Two criteria constitute a positive result: HKU5.2N ELISA must be positive and the MERS-CoV_Jordan IFA or MNT must be positive.
g Fatal MERS-CoV cases.
h Outbreak members conformed to the original outbreak definition; however, some were retrospectively determined to be MERS-CoV test negative. They were part of the original, defined outbreak that our investigation used to trace a priori contacts and exposures, so this descriptive title is retained.
i HKU5.2 N ELISA OD values for serum specimens from outbreak members 05, 07, and 10 and from healthcare personnel 308 were near the assay cutoff OD value and rescreened by serial dilution. These serum samples were initially weakly positive by IFA and considered initially indeterminate. Upon rescreen by IFA, the samples were determined to be negative for the presence of MERS-CoV antibodies.
j Although outbreak member 12 was positive for MERS-CoV by rRT-PCR, his sera were antibody negative. Presumably, this subject died before an antibody response was detectable. This case is considered to be confirmed by current WHO MERS-CoV diagnostic guidelines.
Sera from the 2 fatal cases (designated outbreak subjects 01 and 12) having positive rRT-PCR tests were also tested by the 3 described serology tests. A serum sample from outbreak subject 01 (taken 16 days after onset of respiratory symptoms) was positive by HKU5.2N ELISA and IFA and had detectable MERS-CoV neutralizing antibodies. Two serum specimens from outbreak subject 12 (collected 26 and 32 days after onset) were negative for anti–MERS-CoV antibodies.
Of the 7 subjects found to be positive for anti–MERS-CoV antibodies during this investigation, 6 were surviving members of the initial outbreak group and 1 was previously unrecognized. Thus, including the 2 fatal cases previously detected and reported, a total of 9 individuals in this outbreak had evidence of MERS-CoV infections by acute rRT-PCR tests (n = 2) or convalescent antibody tests (n = 7). The case-fatality rate among all test-positive subjects was 22% (2 of 9). We documented that each serologic test–positive subject had unprotected MERS-CoV exposure(s) to at least 1 rRT-PCR test-positive subject. An additional 8 subjects had single positive test results by either HKU5.2N ELISA or IFA, but their MERS-CoV antibody status was considered indeterminate because both tests were not positive (Table 1).
Healthcare Personnel
We obtained specimens and data from a total of 97 healthcare personnel who worked during February–April, 2012, representing a majority of intensive care (intensive care unit [ICU] and coronary care unit [CCU]) personnel at the outbreak hospital as well as other personnel having close contact with initial outbreak investigation members (Figure 1). These included 8 surviving outbreak members who were healthcare personnel at the focal outbreak hospital and were not lost to follow-up, 49 other personnel at the focal outbreak hospital, 16 personnel at transfer hospital A, 20 personnel at transfer hospital B, and JMoH's 4 outbreak investigators. Of the 57 healthcare personnel at the focal outbreak hospital who survived and the 1 who died, 6 (10%) had cases of MERS-CoV. Our investigation provided no evidence of MERS-CoV infections or transmission events among personnel at the 2 receiving transfer hospitals, even though some patients were transferred temporally close to their symptom onset dates. Interviews with surviving subjects and family members revealed that transmission opportunities among healthcare personnel were not restricted to the workplace.
Household Contacts
We obtained serologic specimens from members of 11 households, including those from the initial outbreak group and another 26 subjects who had resided in those outbreak member households during the outbreak period. One household was lost to follow-up, and 1 did not consent for participation. From one of these households was the symptomatic wife of an initial outbreak investigation member who tested positive for MERS-CoV antibodies. Twelve household subjects were children <18 years old, all of whom were serologically test negative.
Summary of Underlying Conditions, Symptoms, and Clinical Findings
A summary of underlying conditions for test-positive subjects, including the 2 fatal cases initially identified by rRT-PCR (outbreak members 01 and 12), is presented in Table 2. Of the 9 test-positive subjects, 66% were male, with a median age of 40 years (range, 25–60 years) at illness onset. We found no evidence of underlying immunodeficiency or immunosuppressant medications/therapies among any of these subjects. One subject had an atrial septal defect, 2 had a history of hypertension, 2 were smokers at the time of illness, and 1 reported a pregnancy of 5 months’ gestation. Although diabetes mellitus has been observed as a potential risk factor for MERS-CoV [7], none of the subjects reported here had a prior diagnosis of diabetes mellitus and, based on serum glucose values taken during their hospitalizations, none had indications of undiagnosed diabetes mellitus.
Table 2.
Underlying Conditions and Presenting Symptomatologies for Test-Positive Middle East Respiratory Syndrome-Coronavirus Subjectsa
| Characteristic | Test-Positive MERS-CoV Subjects |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 01 | 02 | 03 | 04 | 06 | 09 | 11 | 12 | HHM-303 | |
| Age at symptom onset, y | 40 | 31 | 60 | 35 | 46 | 45 | 41 | 25 | 39 |
| Sex | Female | Male | Male | Male | Male | Male | Female | Male | Female |
| Underlying conditions | None | Atrial septal defect | Hypertension | Hypertension | None | None | None | None | Pregnancy |
| Smoking status | Current | Past | ND | ND | ND | Current | ND | ND | ND |
| Presenting symptoms | Fever Cough Dyspnea |
Fever Malaise Chest pain Cough Dyspnea |
Cough Malaise |
Fever Cough Chest pain |
Fever Cough Dyspnea Chest pain Sore throat |
Fever Cough Dyspnea |
Fever Chills Malaise Chest pain |
Fever Cough Dyspnea |
Fever Rhinorrhea Headache Cough |
Abbreviations: HHM, household member; MERS-CoV, Middle East respiratory syndrome coronavirus; ND, not documented.
a All information for HHM-303, who did not present to medical care, and the ages for all subjects except outbreak member 12 were obtained from patient or informant interview. All other information was gathered from medical records.
The most common presenting symptoms, as documented in medical charts, included fever (89%), cough (89%), dyspnea (56%), chest pain (44%), and malaise (33%). Eight subjects presented for hospital care a median of 5 days after symptom onset (range, 1–14 days). Of these patients, 7 (88%) had cough, 7 (88%) had documented fever (temperature (≥38.0°C), 6 (75%) had dyspnea, 5 (63%) had chest pain, and 5 (63%) had malaise at some point during their disease course. Less common symptoms included chills (38%), wheezing (25%), and diarrhea, vomiting, sore throat, palpitations, and confusion (13% each).
Seven subjects had abnormal chest radiographic findings reported within 3 days of presentation, and 3 of those 7 had bilateral findings. Of the remaining 4 subjects with initial unilateral findings, 3 went on to develop bilateral infiltrates later in their hospitalization, documented either by chest radiography or computed tomography (CT). One subject (outbreak member 12) received an initial diagnosis of pericarditis, and a CT scan with abnormal pulmonary findings was reported 4 days later (Table 3).
Table 3.
Documented Clinical Findings Among Test-Positive Middle East Respiratory Syndrome-Coronavirus Subjects at the Time of Initial Presentation for Medical Care
| Finding | Test-Positive MERS-CoV Subjects |
|||||||
|---|---|---|---|---|---|---|---|---|
| 01 | 02 | 03 | 04 | 06 | 09 | 11 | 12 | |
| Clinical course | ||||||||
| Days ill before hospital presentation | 7 | 10 | 9 | 2 | 1 | 3 | 2 | 14 |
| Clinical interventions | ||||||||
| Intensive care (CCU or ICU admission) | Yes | Yes | No | Yes | No | No | No | Yes |
| Respiratory support | MV | Maska | ND | Maska | Maska | Maska | ND | MV |
| Pressor support (dopamine or norepinephrine) | Yes | ND | ND | ND | ND | ND | ND | ND |
| Complicationsb | Yes | ND | ND | ND | ND | ND | ND | Yes |
| Days hospitalized | 11 | 16 | 0 | 8 | 6 | 10 | 4 | 22 |
| Disposition | Death | Discharge | Refused admission | Discharge | Discharge | Discharge | Discharge | Death |
| Laboratory measurementsc | ||||||||
| Initial (range) | ||||||||
| WBC, ×109/L | 8.1 (4.9–17.6) | ND (5.8–9.2) | 4.0 (4.0) | 7.1 (5.2–7.1) | 6.6 (3.9–8.3) | 2.8 (2.8–4.9) | 5.8 (5.8) | 19.2 (5.3–35) |
| Neutrophils, ×109/L | ND (6.9–16.5) | ND (6.6) | ND | 5.5 (3.2–5.5) | ND (1.6) | 1.6 (1.6–3.2) | 4.2 (4.2) | ND (1.0–31.92) |
| Lymphocytes, ×109/L | ND (0.3–1.1) | ND (0.5–1.7) | ND | 1.4 (1.3–1.4) | ND (2.0) | 0.9 (0.9–1.0) | 1.0 (1.0) | ND (0.7–2.1) |
| Hemoglobin, g/dL | 11.1 (8.9–10.8) | ND (12–14.4) | 15.0 (15.0) | 12.3 (10.5–12.3) | 14.6 (14.3–14.6) | 12.1 (12.1–14.4) | 9.5 (9.5) | 11.1 (9.5–12.2) |
| Platelets, ×109/L | 277 (120–302) | ND (184–216) | 222 (222) | 260 (215–260) | 295 (191–295) | 172 (134–183) | 229 (229) | 419 (122–605) |
| ALT, U/L | ND (40–62) | ND | ND | ND (26) | ND (59) | 29 | 8 (8) | 24 (24–353) |
| AST, U/L | ND (93–96) | ND | ND | ND | ND | ND | ND | 23 (23–62) |
| Blood urea, mmol/L | 6.8 (6.1–37.5) | ND (9.3) | 12.9 (12.9) | ND (3.7–9.3) | 8.2 (4.6–11.1) | 9.6 (3.7–9.6) | 5 (5) | 7.5 (7.5–20.3) |
| Serum creatinine, μmol/L | 71 (35–133) | ND (62) | ND | ND (82–115) | 97 (90–150) | 106 (103–106) | 62 (62) | 72 (40–178) |
| PT, sec | 13 (13–21.5) | ND | 14 (14) | 14 (13.2–14) | ND (13.2–15) | 16 (13.6–16) | ND | ND (12.7–34.5) |
| PTT, sec | 26 (26–38.1) | ND | 30 (30) | 25 (25–27.3) | ND (29.3–35) | 37 (23.2–37) | ND | ND (29.9–50.8) |
| INR | 1.0 (1.0–1.82) | ND | 1.07 (1.07) | 1.07 (1.02–1.07) | ND (1.02–1.2) | 1.35 (1.06–1.35) | ND | ND (0.97–3.52) |
| Chest radiography | ||||||||
| Findings within 3 d of presentation | Right side opacity | Right lobar pneumonia | Bilateral consolidation; pneumonia | Bilateral lobar pneumonia | Right side bronchial congestions with bronchovascular markings | Left side consolidation, right side infiltrate | ND | Elevated diaphragmatic level, right side; cardiomegaly |
Subject Household Member (HHM)-303 had no medical chart as she did not present to medical care.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; CCU, coronary care unit; ICU, intensive care unit; INR, international normalized ratio; MERS-CoV, Middle East respiratory syndrome coronavirus; MV, mechanical ventilation; ND, not documented; PT, prothrombin time; PTT, partial thromboplastin time; WBC, white blood cell.
a Received supplemental oxygen by mask without mechanical ventilation.
b Subject 01 had hyperkalemia, ventricular tachycardia, disseminated intravascular coagulation cardiac arrest. Subject 12 had pericarditis, pleural and pericardial effusion, and supraventricular tachycardia.
c Laboratory measurements at hospital presentation and the range over the course of illness are shown.
Seven of the 8 subjects (88%) who presented to medical care were admitted; 1 refused admission. Six subjects (75%) required respiratory support with at least supplemental oxygen, and 4 subjects (50%) received intensive care (in CCU or ICU), but only the 2 (25%) patients who died required mechanical ventilation, of which 1 patient also required pressor support (dopamine and norepinephrine) for cardiorespiratory failure. Complications among hospitalized subjects were also limited to the 2 patients who died, 1 of whom had hyperkalemia with associated ventricular tachycardia, disseminated intravascular coagulation, and eventual cardiac arrest; the other had pericarditis, pericardial and pleural effusions, and supraventricular tachycardia late in the course of illness.
Although leukopenia (<4.0 × 109/L) was observed in 2 subjects, lymphopenia (<1.5 × 109/L) was observed in 6 of the 7 subjects who had documented complete blood counts with differentials (86%). Elevated leukocyte counts (>11 × 109/L) were observed during the course for 2 subjects (25%), both of whom died. These 2 subjects also had laboratory abnormalities consistent with multiorgan system failure late in the course of disease. These included evidence of elevated alanine aminotransferase and aspartate aminotransferase (>40 U/L) and significant coagulopathy with an international normalized ratio of >1.5, as well as thrombocytopenia (<140 × 109/L). In addition, the 2 subjects who died had elevated serum creatinine measurements (≥133 µmol/L) on the day of their deaths. A third case had an isolated elevated creatinine measurement, but had a subsequent normal value the following day. No patient received hemodialysis (Table 3).
Outbreak member 01 died 17 days after onset of symptoms (on day 11 of hospitalization) and outbreak member 12 died 35 days after onset of symptoms (on day 22 of hospitalization). The remaining 7 subjects survived, and the 5 who were hospitalized were discharged following a median of 8 days (range, 4–16 days). Despite having respiratory symptoms, the pregnant household subject did not seek medical care due to concerns regarding receiving chest radiography and medications. This pregnancy resulted in stillbirth during the course of her illness [8].
Surviving subjects and the family members of deceased patients reported that contact with animals was rare in this urbanized area, and no contact with camels was identified among subjects having early symptom onsets. Furthermore, none of the subjects had traveled to, or had received visitors from, the Arabian Peninsula shortly prior to symptom onset.
Infection Control
At the focal outbreak hospital, there were no physical barriers between CCU and ICU beds, spaced approximately 3 meters, with the exception of cloth drapes in the CCU. Isolation or negative-pressure rooms were not present, and infection control compliance issues were reported during the outbreak. Infection control insufficiencies were not noted at the 2 receiving transfer hospitals.
DISCUSSION
We used novel serologic assays to determine antibody responses of subjects from a MERS-CoV outbreak investigation in Jordan, including the earliest cases of this emerging virus yet discovered. In addition to 2 fatal cases confirmed by rRT-PCR and reported to WHO, we discovered 7 previously unconfirmed and unreported MERS-CoV infections. Detection of these 7 additional antibody-positive subjects, including healthcare personnel from the focal outbreak hospital and a family contact of 1 antibody-positive subject, and the establishment of contacts with MERS-CoV infected subjects when potentially infectious, suggests that human-to-human transmission of MERS-CoV occurred. Although community exposures were possible, healthcare-associated transmission was a plausible explanation for healthcare personnel infections. MERS-CoV infections were not detected among healthcare personnel at a transfer hospital having better adherence to infection control measures.
Compared with published descriptions of Saudi Arabian and French cases [9–12], among the 9 total Jordanian cases identified through our collaborative investigation, subjects were younger and had fewer underlying medical conditions, and there was a lower case-fatality rate. Although all subjects with MERS-CoV infection in our investigation had acute respiratory illnesses during the outbreak period, 78% of those who were infected survived. Most subjects had no underlying medical conditions and none were on hemodialysis or had indications of diabetes mellitus. One newly detected subject, who was a household contact, did not seek medical care. Our data support the probability that, in outbreak settings, infections may remain undetected among subjects who have mild symptoms, lack predisposing conditions, or have barriers to accessing appropriate diagnostic care. Therefore, the true MERS-CoV case-fatality rate may be lower than that based on symptomatic, hospitalized cases alone.
The presenting symptoms we observed were largely consistent with those of previously described MERS-CoV cases [13–16] and included fever with respiratory symptoms such as cough and dyspnea, and associated infiltrates on chest radiography. On initial presentation, many subjects did not have evidence of bilateral pneumonia. Although gastrointestinal symptoms such as vomiting and diarrhea were documented for 2 subjects, we did not observe these as presenting symptoms, as they were in Saudi Arabian and French cases. Once hospitalized, lymphopenia, a prominent laboratory feature among previously described cases, was observed in the majority of our subjects. However, other laboratory abnormalities observed in previous reports, such as thrombocytopenia, were limited mostly to the 2 fatal cases late in the course of illness, consistent with multiorgan system failure. Also, unlike previously reported cases, renal failure was not a prominent clinical feature among our subjects, as renal dysfunction was observed only in the 2 fatal cases on the day of death.
Rapid isolation of patients with suspected MERS-CoV and rigorous infection control practices at the receiving transfer hospitals may have been important in preventing transmission at these locations. Hospitals should have established policies and procedures for the rapid identification of suspected or known MERS-CoV cases and implementation of appropriate infection prevention measures. The CDC recommends standard, contact, and airborne precautions for the management of hospitalized patients with known or suspected MERS-CoV infection [17].
One Jordanian patient was initially hospitalized with pericarditis, a manifestation similar to 1 MERS-CoV case occurring in the Kingdom of Saudi Arabia [9]. Although this Jordanian patient's serologic specimens tested negative for MERS-CoV antibodies at periods throughout his hospital stay, 1 acute specimen collected several days before death was confirmed positive for the virus by rRT-PCR. These laboratory findings and the patient's exposure in the CCU, where he was situated in the bed directly next to another patient with rRT-PCR–confirmed MERS-CoV, collectively suggest the likelihood that the patient was nosocomially infected with MERS-CoV and died before an antibody response was detectable.
Based on the knowledge of SARS-CoV antibody responses, IgG and neutralizing antibodies to SARS-CoV peaked 4 months following a patient's recovery from acute infection [18]. Antibody levels did decline over time, but detectable SARS-CoV neutralizing antibodies persisted up to 2 years after onset of SARS-CoV symptoms [19, 20]. Approximately 13 months had passed between our May 2013 investigation and the April 2012 outbreak. Although this was sufficient time for infected subjects to produce an antibody response to MERS-CoV, the role of waning immunity on the antibody response [21] and whether persistence of these antibodies is important for protection from reinfection remain unclear.
We implemented a rigorous case definition based on an ELISA-positive result plus at least 1 correlating assay result to maximize specificity. Infections with SARS-CoV triggered humoral and cellular immune responses in all studied humans [22], and high titers of neutralizing antibodies were observed in response to SARS-CoV infections, but such characteristics of the MERS-CoV immunologic response remain unknown. As for those indeterminate laboratory findings among subjects with documented MERS-CoV exposure(s) but having only an ELISA-positive result and mild or absent respiratory symptoms, it is possible that the viral exposure to these subjects did not trigger a long-lasting IFA- or MNT-recognizable immune response.
Because obtaining appropriate lower respiratory specimens from subjects having mild or asymptomatic infections is challenging, the use of serologic assays to identify otherwise undetected cases of MERS-CoV has been demonstrated to be a useful tool. Serological surveys have been conducted in retrospective case investigations around instances of MERS-CoV importations in Europe [23], as well as for establishing estimates of MERS-CoV seroprevalence among populations at risk [24]. Further validation of serologic assays and assessments of how they complement rRT-PCR testing is needed.
Our investigation was unable to find evidence of any exposure (either zoonotic contacts, human contacts from the Arabian Peninsula, or among hospitalized contacts preceding the earliest symptomatic cases) that might explain the origin of the virus. The precise route(s) of MERS-CoV transmission remains unclear overall, but several MERS-CoV sequences have been identified in dromedary camel nasal secretions, including one that is indistinguishable from that found in infected humans [25].
In conclusion, the Jordan respiratory illness outbreak in April 2012 resulted in a total of 9 test-positive MERS-CoV subjects. The source of the virus in these earliest known MERS-CoV cases remains unknown. Compared with other reports, the improved survivability we observed is perhaps related to the youth and relative lack of underlying illnesses among the subjects we investigated. Infection control practices at both transfer receiving hospitals may have been important in preventing MERS-CoV transmission in those facilities. Since the discovery of the MERS-CoV, enhanced surveillance for severe acute respiratory illnesses in Jordan has been implemented. International severe acute respiratory infection surveillance, collaborative investigations, and vigilance among healthcare providers are necessary components for addressing and preventing the further spread of MERS-CoV worldwide.
Supplementary Material
Notes
Members of the Jordan MERS-CoV Investigation Team. Dr Nabil Sabri, Dr Mohammad Al Azhari, Dr Hala Khazali, Dr Mohammad Al Maayah, Dr Adel Bilbeisi, Dr Naim Dawood, Dr Bilal Al Zubi (Jordan Ministry of Health); Dr Jawad Meflih (Eastern Mediterranean Public Health Network); Dr Tony Mounds, Dr Julia Fitzner, Dr Akram Eltom, Dr Ali Mafi, (World Health Organization); Congrong Miao, Dr Hayat Caidi, Suvang Trivedi, Shifaq Kamili, Dr Aron J. Hall, Aaron Curns, Jessica Moore, Huong Pham, Dr Chris Zimmerman (National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention [CDC]); Dr Eileen Farnon, Dr Genessa Giorgi, and Dr Russell Gerber (Center for Global Health, CDC).
Acknowledgments. The authors thank the directors and staff of the 3 hospitals in this investigation; Dr Ray R. Arthur, Sudhir Bunga, Catherine C. Chow, Kira A. Christian, Serena Fuller, C. J. McKnight, Myron G. Schultz (Global Disease Detection Operations Center, Division of Global Health Protection, Center for Global Health, CDC) for outbreak investigation logistical support; Dr Maha Talaat and Dr Emad Mohareb (Naval Medical Research Unit 3) for consultation; and Dr Maria Zambon (Public Health England), Dr Thorsten Wolff (the Robert Koch Institute), Dr Susanna Lau (University of Hong Kong), Dr Ella Mendelson (Israel Ministry of Health), and Dr Emad M. Elassal for providing serum and isolates. Positive control sera from MERS-CoV–infected patients were provided by Public Health England and by the Robert Koch Institute. A clinical isolate of MERS-CoV (Hu/Jordan-N3/2012) was provided by the Jordan Ministry of Health and Naval Medical Research Unit 3 (Cairo, Egypt).
Author contributions. D. C. P. had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Financial support. This work was supported by the US Global Disease Detection Operations Center Outbreak Response Contingency Fund.
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hijawi B, Abdallat M, Sayaydeh A, et al. Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. East Mediterr Health J. 2013;19:S12–8. [PubMed] [Google Scholar]
- 2.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Update: severe respiratory illness associated with Middle East respiratory syndrome coronavirus (MERS-CoV)—worldwide, 2012-2013. MMWR Morb Mortal Wkly Rep. 2013;62:1–4. [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. Global Alert and Response (GAR): novel coronavirus infection—update (Middle East respiratory syndrome coronavirus); ,. Geneva, Switzerland: WHO; Available at: http://www.who.int/csr/disease/coronavirus_infections/en . Accessed 2 May 2014. [Google Scholar]
- 5.Haynes LM, Miao C, Harcourt JL. Recombinant protein-based assays for detection of antibodies to severe acute respiratory syndrome coronavirus spike and nucleocapsid proteins. Clin Vaccine Immunol. 2007;14:331–3. doi: 10.1128/CVI.00351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sui J, Li W, Murakami A, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A. 2004;101:2536–41. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Tawfiq JA, Hinedi K, Ghandour J, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): a case-controlled study of hospitalized patients. Clin Infect Dis. 2014;59:160–5. doi: 10.1093/cid/ciu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payne DC, Iblan I, Alqasrawi S, et al. Stillbirth during infection with Middle East respiratory syndrome coronavirus (MERS-CoV) [Epub ahead of print] J Infect Dis. 2014 doi: 10.1093/infdis/jiu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arabi YM, Arifi AA, Balkhy HH, et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med. 2014;160:389–97. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 10.Guery B, Poissy J, el Mansouf L, et al. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–72. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. Epidemiological, demographic and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet. 2013;13:752–61. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assiri A, McGreer A, Perl TM, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:886. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guberina H, Witzke O, Timm J, et al. A patient with severe respiratory failure caused by novel human coronavirus. Infection. 2014;42:203–6. doi: 10.1007/s15010-013-0509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosten C, Seilmaier M, Corman VM, et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–51. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bermingham A, Chand MA, Brown CS, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17 pii:20290 Available at: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20290 . Accessed 2 May 2014. [PubMed] [Google Scholar]
- 16.Memish ZA, Zumla AI, Al-Hakeem RF, et al. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;13:1–8. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 17.US Centers for Disease Control and Prevention. Interim infection and prevention control recommendations for hospitalized patients with Middle Eastern respiratory syndrome coronavirus (MERS-CoV) Available at: http://www.cdc.gov/coronavirus/mers/infection-prevention-control.html. Accessed 2 May 2014.
- 18.Cao W, Liu W. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357:1162–3. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 19.Lie W, Fontanet A, Zhang PH, et al. Two year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis. 2006;193:792–5. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo H, Zeng G, Ren X, et al. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology. 2006;11:49–53. doi: 10.1111/j.1440-1843.2006.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie Y, Wang G, Shi X, et al. Neutralizing antibodies in patients with severe acute respiratory syndrome associated coronavirus infection. J Infect Dis. 2004;190:1119–26. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu M. SARS immunity and vaccination. Cell Mol Immunol. 2004;3:193–8. [PubMed] [Google Scholar]
- 23.Reuss A, Litterst A, Drosten C, et al. Contact investigation for imported case of Middle East respiratory syndrome, Germany. Emerg Infect Dis. 2014;20:620–5. doi: 10.3201/eid2004.131375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gierer S, Hofmann-Winkler H, Albuali WH, et al. Lack of MERS coronavirus neutralizing antibodies in humans, Eastern Province, Saudi Arabia. Emerg Infect Dis. 2013;19:20134–6. doi: 10.3201/eid1912.130701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briese T, Mishra N, Jain K, et al. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. mBio. 2014;5:e01146–14. doi: 10.1128/mBio.01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.