Abstract
Recent genomewide association studies confirm that HLA genes have the strongest associations with several autoimmune diseases, including type 1 diabetes (T1D), providing an impetus to reduce this genetic association to practice through an HLA-based disease predictive model. However, conventional model-building methods tend to be suboptimal when predictors are highly polymorphic with many rare alleles combined with complex patterns of sequence homology within and between genes. To circumvent this challenge, we describe an alternative methodology; treating complex genotypes of HLA genes as “objects” or “exemplars”, one focuses on systemic associations of disease phenotype with “objects” via similarity measurements. Conceptually, this approach assigns disease risks base on complex genotype profiles instead of specific disease-associated genotypes or alleles. Effectively, it transforms large, discrete and sparse HLA genotypes into a matrix of similarity-based covariates. By the Kernel representative theorem and machine learning techniques, it uses a penalized likelihood method to select disease-associated exemplars in building predictive models. To illustrate this methodology, we apply it to a T1D study with eight HLA genes (HLA-DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1 and –DPB1), to build a predictive model. The resulted predictive model has an area under curve of 0.92 in the training set, and 0.89 in the validating set indicating that this methodology is useful to build predictive models with complex HLA genotypes.
Keywords: Generalized linear model, kernel machine, multi-allelic genotypes, penalized regression, prediction, similarity regression, statistical learning
Introduction
Following the successes of next generation sequencing technologies, a goal of future biotechnology innovation is to produce fully phased diploids of human genomes, i.e., a pair of phased haplotypes with multiple single nucleotide polymorphisms (SNPs) [Tewhey, et al. 2011; Yang, et al. 2011]. Within a functional gene, multiple phased SNP alleles, together with all monomorphic nucleotides, represent fully phased sequences that are useful to decipher functional transcript or protein sequences. Indeed, it is of great interest to turn multiple phased SNPs into multi-allelic polymorphisms. Probably, one of best example genes is the Human Leukocyte Antigen (HLA) genes, located between 6p22.1 and 6p21.3 on chromosome 6. For example, the HLA-DRB1 gene consists of a pair of alleles in each individual, and each allele corresponds to a phased sequence [Marsh 2000]. By the most recent counting statistics (http://www.ebi.ac.uk/ipd/imgt/hla/), HLA-DRB1 has over 1,868 alleles, which code 1,364 proteins. The exceptional polymorphisms and presumed multi-functionality have presented challenges to studying its associations with diseases, most prominently autoimmune disorders such as type 1 diabetes (T1D) [Noble 2015]. Further, the high polymorphism also hinders the translation of positive discoveries from bench to bedside, because of limited sample sizes associated with many less common alleles and multiple testing with numerous alleles.
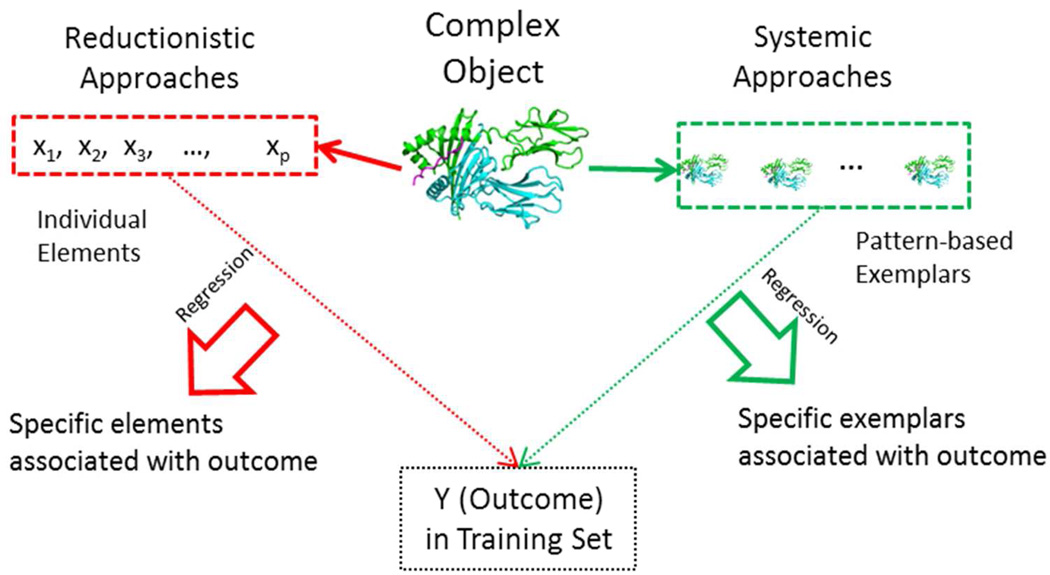
There is a need for a new analytic approach to overcoming this challenge. In genetic epidemiology, as perhaps in most scientific endeavors, mostly commonly used data analysis tools are regression-based methods focusing on individual covariates or elements. For example, genetic analysis tends to focus on individual alleles and/or genotypes in candidate gene studies or individual alleles of SNPs in genomewide association studies. In other words, our reductionistic approach is to discover “needles in the haystack” (see Figure 1 for illustration). Inevitably, the covariate-specific regression approach encounters challenges in dealing with too many covariates, as is the case with too many alleles in HLA genes. In recent years of omics research, the scientific community has paid an increasing attention to a “systems biology” approach, focusing on profiles of multiple genes and their joint associations with phenotype, i.e., “systemic” or “holistic” associations. Rather than reducing to specific elements, a systemic approach tends to address if a variation at the system level is functional. If two systems have similar variation profiles, they likely have a similar phenotype, and similarity patterns are often visually displayed [Cardinal-Fernandez, et al. 2014; Smith, et al. 2014]. Extending the idea of systemic analysis to analyze complex HLA genotypes, our interest is to develop an analytic framework to quantify systemic associations of a disease phenotype with genotype profiles, rather than individual alleles or genotypes. Towards this goal, we treat genotype profiles of multiple genes as “objects” or “exemplars”, measure similarity of subjects’ genotypes with exemplars’, and assess systemic associations of the phenotype with similarity measurements via the kernel machine [Hastie, et al. 2015]. Conceptually, this approach focuses on “objects” in the kernel machine, rather than individual elements in “objects”, leading to object-oriented regression (OOR). Hereafter, “genotype profiles”, “objects” or “exemplars” are used interchangeably. To illustrate OOR in Figure 1, we show that the systemic approach identifies a set of exemplars, representing systemic variations, and assesses which exemplars associate with phenotype.
Figure 1.
An illustration of typical reductionistic and holistic approaches: the conventional reductionistic approach reduces complex object into an array of individual elements (genes, SNPs, or alleles), and discovers specific elements associated with outcome. The systemic approach identifies a set of commonly observed data patterns (arrays of individual elements, combination of genotypes, haplotypes, or interesting objects) as exemplars (or systems), and discovers specific exemplars associated with outcome.
While being independently motivated, OOR has a close connection with three recent applications of the kernel machine. Recently, Wu and colleagues (2010 and 2011) have published two high impact papers, introducing kernel-based methods for testing gene-set associations and for assessing rare variants in case-control studies [Kwee, et al. 2008; Wu, et al. 2010; Wu, et al. 2011]. The key idea is to encapsulate genetic associations of main effects and interactions by modeling their kernel function. Extending the same idea, Minnier and colleagues have described a method for risk classification with multiple independent gene sets, in which they model gene-specific kernel functions via single value decomposition [Minnier, et al. 2015].
In term of the analytic objective, OOR has a closer connection with Zhu and Hastie’s kernel logistic regression and support vector machine[Zhu and Hastie 2005]. Without modeling the kernel function, the kernel logistic regression proposes to use “import points” to reduce the kernel space. Taking their idea of “import points” further, OOR introduces exemplars that can be derived internally or externally. More importantly, we assume that coefficients associated with many exemplars are approximately zero, so that OOR uses the penalized likelihood to de-select those uninformative exemplars that are not associated with the disease phenotype, leaving a set of “informative exemplars” for the predictive model.
In the remainder of the manuscript, the first section on Methodology provides statistical motivations for OOR, lays out the OOR framework, identifies approaches for choosing exemplars and builds up predictive models. Further, the Methodology section describes a general flow from converting complex genotypes to similarity measures and to building predictive models. Besides detailing choices of exemplars and selection of predictors, the Methodology section describes how to assess the stability of choosing the penalty parameter and how to assess the concordance of informative exemplars through bootstrap. To illustrate OOR, the Application section describes the T1D study and illustrates the utility of OOR for exploring disease associations with HLA genes and for building a T1D predictive model. The Results section describes associations of HLA-DRB1 with T1D and a T1D predictive model with six HLA genes. The manuscript ends with conclusions and discussions on OOR and related results.
Methodology
Motivation
Consider genotypes arising from studies of highly polymorphic genes. To be specific, the motivating study is a case-control study of T1D and eight class II HLA genes (HLA-DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1, and -DPB1) [Delli, et al. 2010; Delli, et al. 2012]. Because of their structural polymorphisms, only one of HLA-DRB3, -DRB4 and -DRB5 alleles appears on any single chromosome, and hence HLA-DRB345 is used to denote genotypes of all three genes hereafter. Each gene consists of two alleles, and each allele represents a fully phased nucleotide sequence. If the jth gene has, say mj, possible sequence variations, a genotype with a pair of alleles can take one of mj(mj + 1)/2 possible genotypic polymorphisms under the Hardy-Weinberg equilibrium (HE), i.e., statistically independent within a locus. An array of genotypes at multiple gene loci is referred to as a genotype profile. If these genes were in linkage-equilibrium (LE), i.e., statistical independence between loci, the total number of genotype profiles could theoretically be as large as their cross-products ∏jmj(mj + 1)/2. The total number of genotype profiles could easily exceed the typical sample sizes of most population-based studies. In practice, however, the observed number of genotype profiles is much smaller than the theoretical total, due to biological constraints: 1) HLA genetic polymorphisms are highly selected within populations; 2) Paired HLA gene alleles within loci tend to deviate from HWE; 3) Genotype profiles of multiple HLA genes tend to deviate from LE because of physical adjacency or gene-gene interactions; and 4) the genetic region covering HLA genes is known to have relatively lower recombination rate than the rest of the genome, despite presence of “recombination hot spots” in the region [Cullen, et al. 1997; Jeffreys and May 2004; Marsh 2000]. These complexities lead to typical observations that some genotype profiles are over-represented and that many others are completely absent. Such phenomenon presents challenges to HLA association analysis, if one aims to examine disease associations with individual alleles/genotypes within a single gene, to investigate one genetic association after stratifying over genotypes of another gene, or to carry out haplotype analysis with two or more genes.
Instead of focusing on individual alleles or genotypes, a complementary approach is a systemic approach, i.e., focusing on genotype profiles and examining their overall associations with outcome [Bell and Koithan 2006; Fang and Casadevall 2011]. In other words, treating observed genotype profiles as exemplars, one computes similarities of subjects’ genotypes with exemplars, and assesses if similarity to exemplars associates the disease phenotype. Given the sample size n in a case-control study, the total number of possible exemplars, if derived internally from the study, is at most n. As noted above, the actual number of unique genotype patterns formed by eight HLA genes is less than the sample size n. Treating all unique genotype profiles as exemplars, one can directly assess T1D association with subjects’ similarity measures of all these exemplars. Formalization of these observations motivates the proposal of OOR.
Consider a profile of m genotypes denoted by g̰i = (gi1, gi2,…,gim) observed on the ith subject (i=1,2,…,n). Over all subjects, unique genotype profiles are identified and are denoted as the kth exemplar (k=1,2,…,q). By observed genotypes, one measures the subject’s similarity with the kth exemplar via a similarity function, denoted as , which is referred to as a kernel function [Cristianini and Shawe-Taylor 2000; Hastie, et al. 2009; Minnier, et al. 2015; Zhu and Hastie 2005]. The analytic objective of OOR is to assess genetic association with the disease phenotype, denoted as (yi = 0 for control and yi = 1 for case). Without imposing any parametric assumptions, one can use the Representer’s theorem[Kimeldor and Wahba 1971] to capture this genetic association by the following representation
| [1] |
where the summation is over all observed samples, the kernel function K(g̰i,g̰k) measures similarity between the ith and kth genotype profiles, θk is a vector of kernel parameters, and α is an intercept so that the summation is centered around zero. Wu et al (2011) assuming a Gaussian distribution to model kernel parameters, as a way to testing rare variant associations [Wu, et al. 2011] and to examine SNP-set association [Wu, et al. 2010]. Recently, Minnier et al (2015) uses single value decomposition to simplify the similarity matrix K(g̰i,g̰k) as a way to approximate the above representation. Zhu and Hastie (2005) suggest reducing the above summation over all observed samples to a summation over a set of “import points” with fewer kernel parameters to be estimated.
In the current context, we note that the number of unique genotype profiles tends to be smaller than the sample size. Let denote all q unique genotype profiles, where k = 1,2,…,q. Many terms, sharing identical unique genotype profiles, in the above representation can be merged, and a more compact representation of the equation [1] may be written as
| [2] |
where the internal summation is over kernel parameters with the same unique , and the summation of parameters can be reparametrized into a new parameter βk. This observation leads us to propose the following logistic regression model:
| [3] |
where regression coefficient βk quantifies the disease association with the kth similarity measure of g̰i with . Let denote an exemplar. If βk does not equal zero (βk ≠ 0) it implies that subjects similar to the kth exemplar are at either increased or decreased risk. Similarly, (βk = 0) suggests that being similar to the kth exemplar is inconsequential to their disease risk. Rather than modeling this kernel parameter βk, we assume that many βk are probably equal to zero. Identifying those non-zero βk and estimating their values are primary analytic objectives for OOR. By focusing on estimation of the regression coefficients βk, the interpretation of βk hinges directly on the similarity of genotype profiles with exemplars’, rather than specific alleles or genotypes in traditional covariate-specific regression analysis.
An Object-Oriented Regression Framework
The motivation for OOR is straightforward, and its presentation in equation [3] is deceptively simple. In reality, to use OOR, we have to address three distinct methodological issues: 1) choice of a similarity measure, 2) choice of exemplars, and 3) selection of informative exemplars with non-zero βk coefficients. Different choices of these three methodological components lead to various versions of methods in the OOR framework.
Similarity Measures
Purely from a theoretical consideration, choice of the similarity measure needs to ensure that the kernel function is symmetric and semi-positive definite [Kimeldor and Wahba 1971; Zhu and Hastie 2005]. In practice, many similarity measures are suitable but are context dependent. Here we use a similarity measure suitable for genetic analysis. Suppose the exemplar for HLA gene loci, where a pair of alleles denotes the genotype at the jth gene locus. When measuring the similarity with the exemplar, we consider the following function
| [4] |
where I (.) is an indicator function and each is the identity-by-state measure commonly used in genetic analysis[Bishop and Williamson 1990]. The above similarity measure takes value between 0 and 1, ranging from no similarity to identity, respectively. However, the current measure has not accommodated potentially different functional significances of individual genes or individual alleles. One way to generalize the above similarity measure is to introduce gene-specific or allele-specific weights in the calculation.
Choice of Exemplars
In the derivation of OOR formulation[3], we have used observed but unique genotype profile as exemplar. However, from the application perspective, one can choose exemplars either externally or internally, depending on research questions of interest. For example, one may choose exemplars externally from literature. In contrast, one can choose exemplars internally, the focus of this manuscript. When selecting exemplars internally, one may choose all unique genotype profiles as exemplars, as described above. When dealing a large number of alleles or sequence variations, like HLA, one may consider clustering analysis via a n*n similarity matrix K =|K(g̰i,g̰k)|n*n, with pairwise measurement of similarity., and choose those “centroids” of clusters as exemplars.
Selections of Informative Exemplars
Following the identification of exemplars , the next analytic objective of OOR is to select those exemplars whose similarity measures significantly associate with the disease phenotype of interest. As noted above, one expects that many regression coefficients βk equal zero, and should be de-selected from the logistic regression model [3], leaving only those informative exemplars whose similarity measures associate with the disease phenotype. Since the number of exemplars can still be relatively large, we consider penalized likelihood methods to select informative exemplars, to avoid over-fitting. Using the same notation as above, the penalized log likelihood function may be written as
| [5] |
where λ is a tuning parameter to determine the penalty level, |β|1 and |β|2 are l1 -norm and l2-norm, respectively. By choosing η to take value 0, or 1, or 0.5, the above penalized likelihood method corresponds to LASSO, ridge regression and elastic net, respectively. The tuning parameter λ is estimated to have a minimum prediction error, and is chosen by the cross-validation.
In contrast, the best-known traditional strategy of selecting variables is a hybrid of forward and backward stepwise selection of predictors based upon information criterion (IC) measures such as Akaike’s IC (AIC). Given the extensive literature on the likelihood-based estimation, it suffices to note that under the logistic regression model [3], one can use a log-likelihood function similar to [5], except replacing the penalty component with λ by 2(1+q).
Penalty Parameter and Section of Informative Exemplars
It is known in the literature that the turning parameter in penalized likelihood methods imposes a penalty on parameter estimations, trading biases in estimated regression coefficients with estimated variances [Cox and O'Sullivan 1989; Friedman, et al. 2010; Sun, et al. 2013; Tibshirani, et al. 2012]. Cross-validation is commonly recommended to estimate the penalty parameter. However, the cross-validation procedure is a random process, resulting in a random estimate. The randomness may affect selection of exemplars. Here, we recommend to repeat cross-validation process multiple times, and to estimate its empirical distribution, based on which we will then evaluate the stability of variable selections with fixed penalty parameter (see discussion below). Computationally, we estimate penalty parameter with 10 fold cross-validation (a default recommendation in cv.glmnet, an R implementation of GLMNET), and repeat the computation, say 100 times. All empirically estimated parameters are used to construct an empirical distribution.
Assessing Stability of Exemplar Selection with Fixed Penalty Parameter (λ)
A major challenge facing, practically, all variable selection procedures, dealing with complex or high dimensional data, is the stability of selected variables [Meinshausen and Buhlmann 2010]. Selection of informative exemplars is not an exception. Upon assessing empirical distribution of penalty parameter estimates above, we are interested in stability of selecting informative exemplars. To address this issue, we use the bootstrap method. Briefly, we randomly draw sample observations from the study population with replacement, while keeping the same sample size. On each bootstrap sample, we perform a penalized likelihood analysis with fixed penalty parameters. Then, we compute Kappa statistics to measure if selected exemplars are consistently selected [Agresti 1988; Cohen 1968].
T1D Case-control study
A case-control study of T1D and HLA genes motivates the OOR development, and is reported elsewhere [Zhao, et al. 2016]. Briefly, this study identified 970 T1D patients whose ages range from year 1 to 18, from geographically diverse clinics in Sweden, and treated them as cases. From comparable regions, the study identified 448 controls who were free from T1D. From all study subjects, the study collected blood samples, following Human Subjects review and approval, and extracted their DNA. Focusing on HLA genes, this study used the next generation sequencing technologies to assess high-resolution genotypes of HLA genes (HLA-DRB1, -DRB345, -DQA1, -DQB1, -DPA1 and DPB1). The analytic objective of this study is to investigate T1D associations with HLA genes, and to build a predictive model of T1D status with these HLA genotypes. To be inclusive, we randomly chose 479 cases and 226 controls as a training set and the remainder as validation set (222 controls and 483 cases). Allelic frequencies of all genes among controls and cases are largely comparable between training and validating sets (for illustration, the supplementary Table S1 includes allelic frequencies of HLA-DRB1 in among controls and cases from the training and validation set).
Results
Application to HLA-DRB1
To illustrate OOR in dealing with complex HLA data, we first consider T1D association with the HLA-DRB1 gene alone. Table 1 lists genotypic distribution of HLA-DRB1 among, respectively, controls and cases above and below the diagonal line, respectively. For those homozygous genotypes falling diagonal line, genotypic frequencies among controls and cases are denoted in numerator and denominator (#/#), respectively. The first impression from reading this genotype frequency table is that the genotypic distribution with only 44 alleles is sparse and has only 159 unique genotypes, much smaller than the theoretically possible number of genotypes, i.e., 990 (=44×45/2) computed under HWE. Second, certain genotypes exhibit substantially different frequencies between cases and controls, implying their associations with T1D. For example, the homozygote DRB1*04:01:01/04:01:01 has frequencies of 0.4/9.3 among controls and cases, respectively, implying a rate ratio of 23.25. At the other extreme, the heterozygote DRB1*15:01:01/07:01:01 has frequencies of 4.4 among controls and 0 among cases, implying that this heterozygote appears to be protective against T1D. For those common genotypes, direct assessment of T1D associations is practical with the current sample size, and has often been reported. Even in such a case, the analysis has to deal with zero frequency, such as DRBA*15:01:01/07:01:01, with special statistical treatment, like Firth-correction[Firth 1993]. For many less common genotypes, the rigorous assessment is difficult, because of the sparseness, small sample sizes, and large number of comparisons. Given the desire to examine T1D associations with the gene as whole, we are motivated to seek an alternative analytic approach.
Table 1.
Estimated genotypic frequencies of HLA-DRB1 in the training set among controls (below the diagonal line) and among cases (above the diagonal line). Genotypic frequencies of homozygous genotypes among controls and cases are denoted in numerator/denominator, respectively.
| HLA- DRB1 |
*01:01:01 | *01:02:01 | *01:03 | *03:01:01 | *03:05:01 | *04:01:01 | *04:02:01 | *04:03:01 | *04:04:01 | *04:05:01 | *04:05:04 | *04:06:01 | *04:07:01 | *04:08:01 | *04:10:01 | *04:13 | *07:01:01 | *08:01:01 | *08:02:01 | *08:03:02 | *08:04:01 | *09:01:02 | *10:01:01 | *11:01:01 | *11:01:02 | *11:02:01 | *11:03 | *11:04:01 | *12:01:01 | *13:01:01 | *13:02:01 | *13:03:01 | *13:05:01 | *14:01:01 | *14:02 | *14:04 | *14:12:01 | *14:54:01 | *15:01:01 | *15:02:01 | *15:03:01 | *16:01:01 | *16:02:01 | *16:09 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| *01:01:01 | 0.9/0.2 | . | . | 2.7 | . | 11.3 | . | . | 0.4 | 0.2 | . | . | . | . | . | . | 0.6 | 0.8 | . | . | . | . | . | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . |
| *01:02:01 | 0.4 | . | . | 0.6 | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . | . | . | . | . | . | . | 0.2 | 0.2 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *01:03 | . | . | . | 0.2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *03:01:01 | 3.5 | 0.4 | . | 0.4/5.6 | . | 22.1 | 1 | 0.2 | 6.3 | 1.3 | . | . | 0.2 | . | . | . | 0.8 | 1 | . | . | . | 0.4 | . | 0.4 | 0.2 | . | . | . | . | . | 1.9 | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . |
| *03:05:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:01:01 | 0.9 | . | . | 1.3 | . | 0.4/9.3 | 0.2 | . | 2.3 | 0.2 | . | . | 0.2 | 0.4 | . | . | 2.3 | 4 | 0.2 | . | . | 1.7 | 0.2 | 1 | . | . | . | 0.2 | 0.4 | 3.1 | 3.8 | . | . | . | . | 0.2 | . | . | 0.2 | 0.2 | . | 0.4 | 0.2 | . |
| *04:02:01 | . | 0.4 | . | . | . | . | 0/0.2 | . | . | . | . | . | . | 0.2 | . | . | 0.2 | 0.2 | . | . | . | . | . | . | . | . | . | . | 0.2 | . | 0.2 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:03:01 | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:04:01 | 1.3 | . | . | 1.8 | . | 0.4 | . | . | 0.4/0.4 | 0.2 | . | . | . | . | . | . | 0.2 | 1 | . | . | . | . | . | . | . | . | . | . | . | 0.6 | 1 | 0.2 | . | . | . | . | . | . | 0.2 | . | . | . | . | . |
| *04:05:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.2 | . | . | . | 0.2 | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:05:04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:06:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:07:01 | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:08:01 | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.2 | . | . | 0.2 | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:10:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *07:01:01 | 1.3 | . | 0.4 | 0.4 | . | 3.1 | . | . | 1.8 | . | 0.4 | . | 0.4 | . | . | . | 0.9/0 | 0.4 | . | . | . | 0.4 | . | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *08:01:01 | 0.4 | . | . | 0.4 | . | 2.2 | . | 0.4 | 0.4 | . | . | . | . | . | . | . | 0.9 | . | . | . | . | 0.2 | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *08:02:01 | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *08:03:02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *08:04:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *09:01:02 | 0.4 | . | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *10:01:01 | 0.4 | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:01:01 | 1.3 | . | . | 0.9 | . | 1.3 | . | . | 0.4 | . | . | . | . | . | . | . | 0.4 | 0.4 | . | . | . | . | . | 0.4/0 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . |
| *11:01:02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:02:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:04:01 | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . | . |
| *12:01:01 | . | 0.4 | 0.4 | . | . | 0.4 | . | . | 0.4 | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.2 | . | . | . | . | . |
| *13:01:01 | 2.7 | . | . | 1.8 | . | 2.2 | . | . | 0.9 | . | . | . | 0.9 | 0.4 | . | . | 1.3 | 0.4 | . | . | . | 0.4 | . | 0.4 | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *13:02:01 | 0.4 | . | . | . | . | 2.2 | . | . | 0.9 | . | . | . | . | . | . | . | 1.8 | 0.4 | . | . | . | . | . | 0.9 | . | . | . | 0.9 | 0.4 | . | 0/0.2 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *13:03:01 | 0.4 | . | . | 0.4 | . | 0.4 | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *13:05:01 | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:01:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:02 | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:12:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:54:01 | 0.4 | . | . | 0.4 | . | . | 0.4 | . | 0.4 | . | . | . | . | . | . | . | . | 0.9 | . | . | . | . | . | 0.4 | . | . | . | . | . | . | 0.4 | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . |
| *15:01:01 | 1.8 | . | 0.9 | 2.7 | . | 4.9 | . | 0.4 | 0.9 | . | . | . | . | . | . | . | 4.4 | 0.9 | 0.4 | . | . | 0.4 | . | 1.3 | . | . | . | 0.9 | 0.4 | 2.2 | 1.3 | . | . | . | 0.4 | . | . | . | 2.2/0 | . | . | . | . | . |
| *15:02:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . |
| *15:03:01 | . | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *16:01:01 | . | . | . | 0.4 | . | 0.4 | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . | . | . | . | . | . | . | . | 0.4 | 0.9 | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . |
| *16:02:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *16:09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0.4 | . | . | . | . | . |
Consider OOR model for capturing T1D association with HLA-DRB1 via the equation[3], without invoking any assumptions. Because of varying allelic frequencies and deviations from HWE for some alleles, many theoretically possible genotypes are absent, i.e., they have zero frequency in both cases and controls (Table 1). Consequently, by treating all unique genotypes as exemplars, we have a total of q=159 exemplars in the equation [3]. Among these 159 regression coefficients we expect that most equal zero, leaving a few informative exemplars.
In this application, the element in the similarity matrix takes a value of 1 for identity, 0.5 for sharing one allele, and 0 for sharing no alleles, between a pair of subjects. The similarity matrix among 705 subjects, in which a pair of subjects shares both alleles (red), share one allele (black) and share no alleles (green) is shown as a heat map (Figure 2). From a HLA-DRB1 perspective, it is possible to identify a group of subjects who are identical (red squares falling on the diagonal line), and another group of subjects who share only one allele (black rectangles). Note that for genotypes with extreme frequencies in cases and controls, such as the exemplar DRBA*15:01:01/07:01:01, many controls share one allele with this exemplar, even though no control is identical to this exemplar.
Figure 2.
The computed similarity matrix (705 by 705) among 705 subjects in the training set, and each element takes value 0 (green), 0.5 (black) and 1 (red) to indicate that the paired subject shared zero allele, one allele and both alleles, respectively.
Following the above OOR formulation, we perform a univariate association analysis by regressing the disease outcome on one exemplar-specific similarity measurement a time, to gain insight into its marginal association. Detailed results from univariate analysis include estimated log odds ratios, standard errors, Z-scores and p-values, which are listed in the supplementary table (Table S2), together with exemplars and associated genotypes. For more intuitive interpretation, rounded integers of Z-scores are shown in a matrix format (Table 2), where absolute values of 2 or greater, corresponding to a significance level of 0.05 or better (without correcting multiple comparisons) are presented for simplicity. Results from these univariate analyses reveal that HLA-DRB1*03:01:01 and *04:01:01 are positively associated with T1D, which are colored in red stripes. On the other hand, six alleles HLA-DRB1* 07:01:01, *11:01:01, *11:04:01, 12:01:01, 13:01:01 and 15:01:01:01 are protective against T1D, which are colored in green stripes. It is interesting to note that heterozygous genotype with one risk and one protective allele tends to have a positive association with T1D.
Table 2.
Estimated Z-scores (rounded to their integers and equal or above 2) are extracted from marginal association analysis by OOR. Two major alleles (HLA-DRB1*03:01:01 and *04:01:01) assert strong risk associations (red strips). Six alleles (HLA-DRB1*07:01:01, *11:01:01, *11:01:01, *11:04:01, *12:01:01, *13:01:01 and *15:01:01) assert strong protective associations with T1D.
| *01:01:01 | *01:02:01 | *01:03 | *03:01:01 | *03:05:01 | *04:01:01 | *04:02:01 | *04:03:01 | *04:04:01 | *04:05:01 | *04:05:04 | *04:06:01 | *04:07:01 | *04:08:01 | *04:10:01 | *04:13 | *07:01:01 | *08:01:01 | *08:02:01 | *08:03:02 | *08:04:01 | *09:01:02 | *10:01:01 | *11:01:01 | *11:01:02 | *11:02:01 | *11:03 | *11:04:01 | *12:01:01 | *13:01:01 | *13:02:01 | *13:03:01 | *13:05:01 | *14:01:01 | *14:02 | *14:04 | *14:12:01 | *14:54:01 | *15:01:01 | *15:02:01 | *15:03:01 | *16:01:01 | *16:02:01 | *16:09 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| *01:01:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *01:02:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *01:03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *03:01:01 | 6 | 7 | 7 | 7 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *03:05:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:01:01 | 8 | 10 | . | 12 | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:02:01 | . | . | . | 7 | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:03:01 | . | . | . | 7 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:04:01 | . | . | . | 6 | . | 11 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:05:01 | . | . | . | 7 | . | 11 | . | . | 2 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:05:04 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:06:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:07:01 | . | . | . | 7 | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:08:01 | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:10:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *04:13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *07:01:01 | −4 | . | −6 | 3 | . | 6 | −5 | . | −4 | . | −6 | . | −6 | . | . | . | −6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *08:01:01 | . | . | . | 7 | . | 9 | . | . | . | . | . | . | . | . | . | . | −5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *08:02:01 | . | . | . | 7 | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *08:03:02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *08:04:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *09:01:02 | . | . | . | 7 | . | 10 | . | . | . | 2 | . | . | . | . | . | . | −5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *10:01:01 | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | . | −6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:01:01 | −2 | . | . | 5 | . | 8 | . | . | . | . | . | . | . | . | . | . | −7 | −3 | . | . | . | . | . | −4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:01:02 | . | . | . | 7 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:02:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:03 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *11:04:01 | . | . | . | 6 | . | 9 | . | . | . | . | . | . | . | . | . | . | −7 | . | . | . | . | . | . | −5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *12:01:01 | . | −2 | −3 | . | . | 9 | . | . | . | . | . | . | . | −2 | . | . | −6 | . | . | . | . | . | −3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *13:01:01 | −3 | −4 | . | 4 | . | 7 | . | . | −2 | . | . | . | −5 | −4 | . | . | −8 | −3 | . | . | . | −4 | . | −6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *13:02:01 | . | . | . | 6 | . | 9 | . | . | . | . | . | . | . | . | . | . | −5 | . | . | . | . | . | . | −4 | . | . | . | −3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *13:03:01 | . | . | . | 7 | . | 10 | . | . | . | . | . | . | . | −2 | . | . | −6 | . | . | . | . | . | . | . | . | . | . | −4 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *13:05:01 | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:01:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | −3 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:04 | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:12:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *14:54:01 | . | . | . | 6 | . | . | −2 | . | . | . | . | . | . | . | . | . | . | −2 | . | . | . | . | . | −5 | . | . | . | −4 | . | . | −3 | −4 | . | . | . | . | . | . | . | . | . | . | . | . |
| *15:01:01 | −7 | . | −8 | . | . | 4 | . | −8 | −7 | . | . | . | . | . | . | . | −10 | −8 | −8 | . | . | −8 | . | −9 | . | . | . | −8 | . | −10 | −8 | . | . | . | −8 | . | . | . | −7 | . | . | . | . | . |
| *15:02:01 | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | −3 | . | . | . | . | . | . |
| *15:03:01 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | −2 | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *16:01:01 | . | . | . | 6 | . | 10 | . | . | . | . | . | . | . | . | . | . | −6 | . | . | . | . | . | . | . | . | . | . | . | . | −5 | −2 | . | . | . | . | . | . | . | −8 | . | . | . | . | . |
| *16:02:01 | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| *16:09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | −8 | . | . | . | . | . |
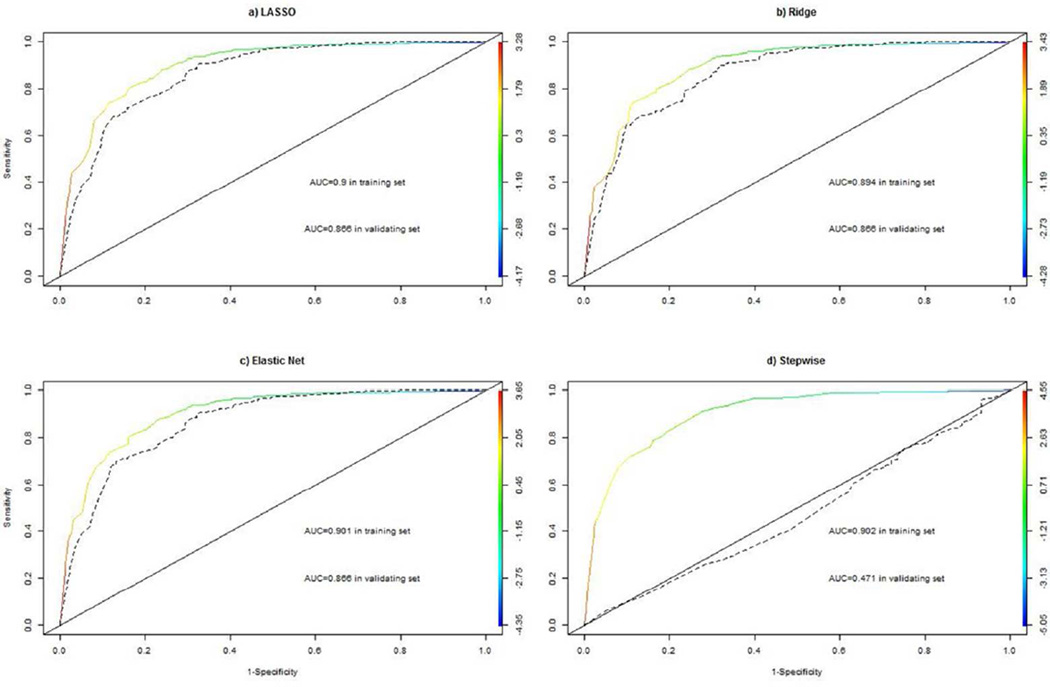
The next step of OOR is to select informative exemplars. For an empirical comparison purpose, we use four different estimation methods: LASSO, Ridge, Elastic Net and stepwise methods, discussed above. All estimated regression coefficients are listed the supplementary Table (Table S3). LASSO selects 18 predictors, from 159 exemplars, with estimated coefficients, i.e., log odds ratios. Interestingly, positive coefficients tend to associate with those exemplars from patients, while negative associations tend to associate with exemplars from controls.
In contrast, the Ridge regression produced estimated coefficients for all of exemplars, without de-selecting any exemplar. For interpretation, all exemplars in Table S3 are sorted by corresponding coefficients. Unlike LASSO estimates, estimated coefficients by Ridge regression take modest values around zero. As expected, directionalities of estimated coefficients tend to be consistent with case/control sources of all exemplars. Further, for those exemplars selected by LASSO, ridge estimates are consistent, in directionality, to those obtained by LASSO. The third column of Table S3 shows coefficients estimated by the Elastic Net, which selected 39 exemplars. Majority of selected exemplars overlap with those selected by LASSO. Quantitatively, estimated coefficients are highly correlated between the Elastic Net and LASSO (not shown). Lastly, the stepwise regression selected 14 exemplars, and ten of them overlap with those selected LASSO. Despite this seemingly high concordance, many estimated coefficients tend to be large, in comparison with their counterparts obtained by LASSO.
To gain insights into performances of predictive models with selected exemplars by these four procedures, we carried out receiver’s operating characteristics (ROC) curve analysis; evaluating sensitivity, specificity and area under curve (AUC) for all four predictive models (Figure 3). ROC curves and associated AUC values in the training set and in the validating set are shown for LASSO (Fig 3a), Ridge (Fig 3b), Elastic Net (Fig 3c) and Stepwise (Fig 3d). In the training set, estimated ROC curves and AUC values are around 0.9, and are largely comparable across four different methods. In the validation set, estimated AUC values slightly reduced to 0.866, as expected. Interestingly, differences among AUC values by three methods are less than 0.001. The comparability of the ROC curve analysis, across these three methods, demonstrates that there are probably many predictive models, with different exemplars, with comparable predictive performances.
Figure 3.
Estimated sensitivity, 1-specificity, and area under curve (AUC) for predictive models with selected exemplars by LASSO, Ridge regression, the Elastic Net, and Stepwise regression in the training set (colored solid line) and also in the validating set (dashed black line). The colored bar indicates corresponding values of risk scores under respective models.
In contrast with the result from the stepwise regression analysis, however, estimated AUC fell down to below 0.5, the null value. This result suggests that the stepwise procedure probably over-fits the training data set, with excessively large regression coefficient estimates.
Application to all Class II HLA Genes
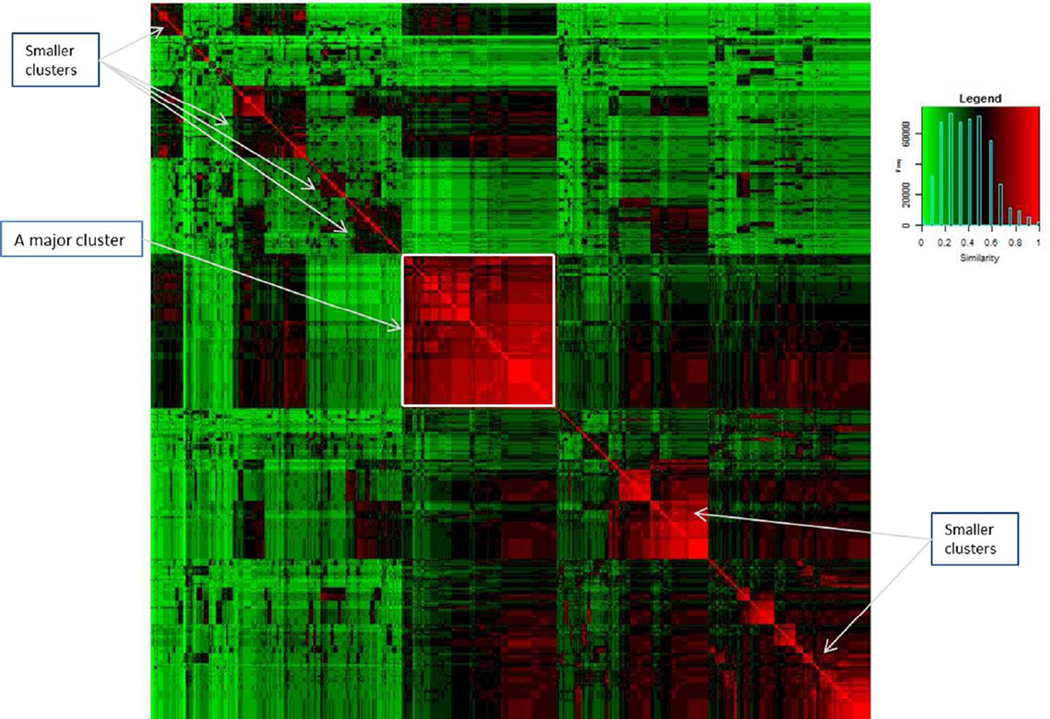
The next task is to build a T1D predictive model applying OOR to all eight HLA-DRB1, -DRB3, -DRB4, - DRB5, -DQA1, -DQB1, -DPA1, and -DPB1 class II HLA genes. We use the same training set to explore exemplars and to build a predictive model, and then validate the predictive model in the validation set. With respect to the similarity measure, we use the unweighted mean similarity measure defined in equation [4], denoted by , where n=705 and each element takes a value between 0 and 1. For easy visualization, we use hierarchical clustering algorithm to organize this similarity matrix, and present its heatmap (Figure 4). The central diagonal cluster (red square, highlighted by an annotation arrow) indicates the presence of many subjects who are either identical or highly similarly to each other. Additionally, there are multiple smaller clusters of highly similar subjects, indicated by annotation arrows. Clustering patterns indicate that subjects in the lower right corner tend to carry more common genotype profiles, because more individuals carry common genotype profiles and their pairwise similarity measures tend to be high. On the other hand, those in the upper left corner tend to have smaller clusters of individuals with relatively similarity measures, probably because their genotype profiles have relatively low frequencies and relatively smaller groups of individuals carry similar genotype profiles. Interestingly, there are subjects in the upper right corner with relatively low similarity measures, probably because individuals with common genotype profiles tend to segregate from those with less common genotype profiles. Patterns of clusters are intuitive and are useful to guide data explorations, even though it is difficult to quantify patterns with any definitive conclusions.
Figure 4.
Estimated similarity matrix with each element measuring the unweighted identity-by-state at HLA-DRB1, -DRB3,-DRB4, -DRB5, -DQA1, -DQB1, -DPA1 and -DPB1. Color range from green, to dark and to red correspond to low, medium and high similarity, denoted in the legend.
The visual display of the similarity matrix indicates that there are duplicated genotype profiles and multiple clusters. Without relying on clustering result in this application, we have chosen all unique genotype profiles in the training set as exemplars. In total, there are 499 exemplars in the training set. As part of the descriptive analysis, we apply OOR to perform univariate association analysis of T1D with all exemplars; estimated coefficients, standard errors, Z-scores and their p-values are listed along with HLA genotypes (Table S4). Exemplars are sorted by Z-scores, directionalities for which are concordant with case and control status.
Now the next task is to select informative exemplar and to build a predictive model. We have chosen not to use the stepwise procedure, because of the over-fitting problem. The Ridge regression is not considered, because it tends to retain most of exemplars and is not consistent with the prior assumption that there are only a small number of informative exemplars. Finally, we have not included the Elastic Net, since it has a comparable performance to LASSO. To focus our analytic exploration, we have chosen LASSO to build a T1D predictive model with estimated regression coefficients (Table 3). There are 26 informative exemplars selected by LASSO. Estimated coefficients are positive for those exemplars derived from cases, while those are negative for those exemplars derived from controls. For example, a subject who is highly similar to exemplars, such as D1612, is at relatively high risk for T1D. On the other extreme, a subject who is similar to, such as N000982, would have relatively low risk.
Table 3.
Estimated coefficients for informative exemplars for predicting type 1 diabetes with class II HLA genes (HLA-DRB1, -DRb345, -DQA1, -DQB1, -DPA1 and -DPB1) using LASSO.
| ID | Exemplars | DRB1 | DRB345 | DQA1 | DQB1 | DPA1 | DPB1 | Coef | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D1612 | *04:01:01 | *04:01:01 | DRB4*01:03:01 | DRB4*01:03:01 | *03:01:01 | *03:01:01 | *03:02:01 | *03:02:01 | *01:03:01 | *01:03:01 | *02:01:02 | *04:01:01 | 3.05 |
| 2 | D1868 | *07:01:01 | *04:01:01 | DRB4*01:01:01 | DRB4*01:03:01 | *02:01 | *03:02 | *02:02:01 | *03:02:01 | *01:03:01 | *02:01:01 | *02:01:02 | *03:01:01 | 2.69 |
| 3 | D1214 | *04:01:01 | *13:02:01 | DRB3*03:01:01 | DRB4*01:03:01 | *03:01:01 | *01:02:01 | *03:02:01 | *06:09 | *01:03:01 | *01:03:01 | *02:01:02 | *03:01:01 | 2.54 |
| 4 | D1209 | *04:05:01 | *01:01:01 | DRB4*01:01:01 | Non-amplified | *03:02 | *01:01:01 | *03:02:01 | *05:01:01 | *01:03:01 | *02:01:01 | *02:01:02 | *09:01:01 | 1.80 |
| 5 | D405 | *03:01:01 | *03:01:01 | DRB3*02:02:01 | DRB3*02:02:01 | *05:01:01 | *05:01:01 | *02:01:01 | *02:01:01 | *01:03:01 | *01:03:01 | *04:01:01 | *30:01 | 1.74 |
| 6 | D1344 | *03:01:01 | *03:01:01 | DRB3*02:02:01 | DRB3*02:02:01 | *05:01:01 | *05:01:01 | *02:01:01 | *02:01:01 | *01:03:01 | *01:03:01 | *02:01:02 | *03:01:01 | 1.52 |
| 7 | D1569 | *03:01:01 | *03:01:01 | DRB3*01:01:02 | DRB3*01:01:02 | *05:01:01 | *05:01:01 | *02:01:01 | *02:01:08 | *01:03:01 | *02:01:02 | *01:01:01 | *04:01:01 | 1.29 |
| 8 | D2102 | *04:05:01 | *09:01:02 | DRB4*01:03:01 | DRB4*01:03:01 | *03:02 | *03:02 | *03:02:01 | *03:03:02 | *01:03:01 | *02:01:01 | *04:01:01 | *13:01:01 | 1.04 |
| 9 | D1437 | *04:01:01 | *04:01:01 | DRB4*01:03:01 | DRB4*01:03:01 | *03:01:01 | *03:01:01 | *03:02:01 | *03:02:01 | *01:03:01 | *01:03:01 | *03:01:01 | *04:01:01 | 0.89 |
| 10 | D1596 | *03:01:01 | *03:01:01 | DRB3*01:01:02 | DRB3*01:01:02 | *05:01:01 | *05:01:01 | *02:01:01 | *02:01:08 | *01:03:01 | *01:03:01 | *04:01:01 | *04:01:01 | 0.83 |
| 11 | D704 | *04:01:01 | *09:01:02 | DRB4*01:03:01 | DRB4*01:03:01 | *03:01:01 | *03:02 | *03:02:01 | *03:03:02 | *01:03:01 | *02:02:02 | *04:01:01 | *05:01:01 | 0.77 |
| 12 | D459 | *04:01:01 | *04:01:01 | DRB4*01:03:01 | DRB4*01:03:01 | *03:01:01 | *03:01:01 | *03:02:01 | *03:02:01 | *01:03:01 | *01:03:01 | *04:01:01 | *04:01:01 | 0.75 |
| 13 | D624 | *03:01:01 | *03:01:01 | DRB3*01:01:02 | DRB3*01:01:02 | *05:01:01 | *05:01:01 | *02:01:01 | *02:01:01 | *01:03:01 | *02:01:04 | *02:01:02 | *13:01:01 | 0.68 |
| 14 | D2055 | *03:01:01 | *04:01:01 | DRB3*01:01:02 | DRB4*01:03:01 | *05:01:01 | *03:02 | *02:01:01 | *03:02:01 | *01:03:01 | *02:01:02 | *01:01:01 | *04:01:01 | 0.57 |
| 15 | D1499 | *04:01:01 | *01:01:01 | DRB4*01:03:01 | Non-amplified | *03:02 | *01:01:01 | *03:02:01 | *05:01:01 | *01:03:01 | *02:02:01 | *04:02:01 | *19:01 | 0.51 |
| 16 | N005872 | *07:01:01 | *07:01:01 | DRB4*01:01:01 | DRB4*01:01:01 | *02:01 | *02:01 | *02:02:01 | *02:02:01 | *02:01:01 | *02:01:01 | *10:01 | *11:01:01 | −0.37 |
| 17 | N001991 | *15:01:01 | *13:01:01 | DRB3*02:02:01 | DRB5*01:01:01 | *01:02:01 | *01:03:01 | *06:02:01 | *06:03:01 | *01:03:01 | *02:02:01 | *02:01:02 | *19:01:01 | −0.52 |
| 18 | N002842 | *03:01:01 | *16:01:01 | DRB3*02:02:01 | DRB5*02:02 | *05:01:01 | *01:02:02 | *02:01:01 | *05:02:01 | *01:03:01 | *02:01:01 | *04:02:01 | *14:01 | −0.65 |
| 19 | N005182 | *07:01:01 | *15:01:01 | DRB4*01:03:01 | DRB5*01:01:01 | *02:01 | *01:02:01 | *03:03:02 | *06:02:01 | *01:03:01 | *02:01:01 | *04:01:01 | *13:01:01 | −0.70 |
| 20 | N003698 | *12:01:01 | *15:01:01 | DRB3*02:02:01 | DRB5*01:01:01 | *05:05:01 | *01:02:01 | *03:01:01 | *06:02:01 | *01:03:01 | *02:02:01 | *04:02:01 | *19:01:01 | −0.97 |
| 21 | N001707 | *04:04:01 | *14:54:01 | DRB3*02:02:01 | DRB4*01:03:01 | *03:01:01 | *01:01:01 | *03:02:01 | *05:03:01 | *01:03:01 | *01:04 | *04:01:01 | *15:01 | −1.41 |
| 22 | N002460 | *04:07:01 | *13:01:01 | DRB3*01:01:02 | DRB4*01:03:01 | *03:02 | *01:03:01 | *03:01:01 | *06:03:01 | *01:03:01 | *01:03:01 | *04:01:01 | *04:02:01 | −1.91 |
| 23 | N002709 | *07:01:01 | *15:01:01 | DRB4*01:03:01 | DRB5*01:01:01 | *02:01 | *01:02:01 | *02:02:01 | *06:02:01 | *01:03:01 | *01:03:01 | *04:01:01 | *04:02:01 | −2.96 |
| 24 | N004319 | *04:07:01 | *07:01:01 | DRB4*01:03:01 | DRB4*01:03:01 | *03:01:01 | *02:01 | *03:01:01 | *03:03:02 | *01:03:01 | *01:03:01 | *02:01:02 | *04:01:01 | −3.61 |
| 25 | N004385 | *14:02 | *15:01:01 | DRB3*01:01:02 | DRB5*01:01:01 | *05:03 | *01:02:01 | *03:01:01 | *06:02:01 | *01:03:01 | *01:03:01 | *03:01:01 | *06:01 | −3.95 |
| 26 | N000982 | *14:54:01 | *15:02:01 | DRB3*02:02:01 | DRB5*01:02 | *01:01:01 | *01:03:01 | *05:03:01 | *06:01:01 | *01:03:01 | *02:01:01 | *02:01:02 | *14:01 | −4.18 |
With estimated coefficients as weights from the training set, we now construct a risk score as weighted sum
| [5] |
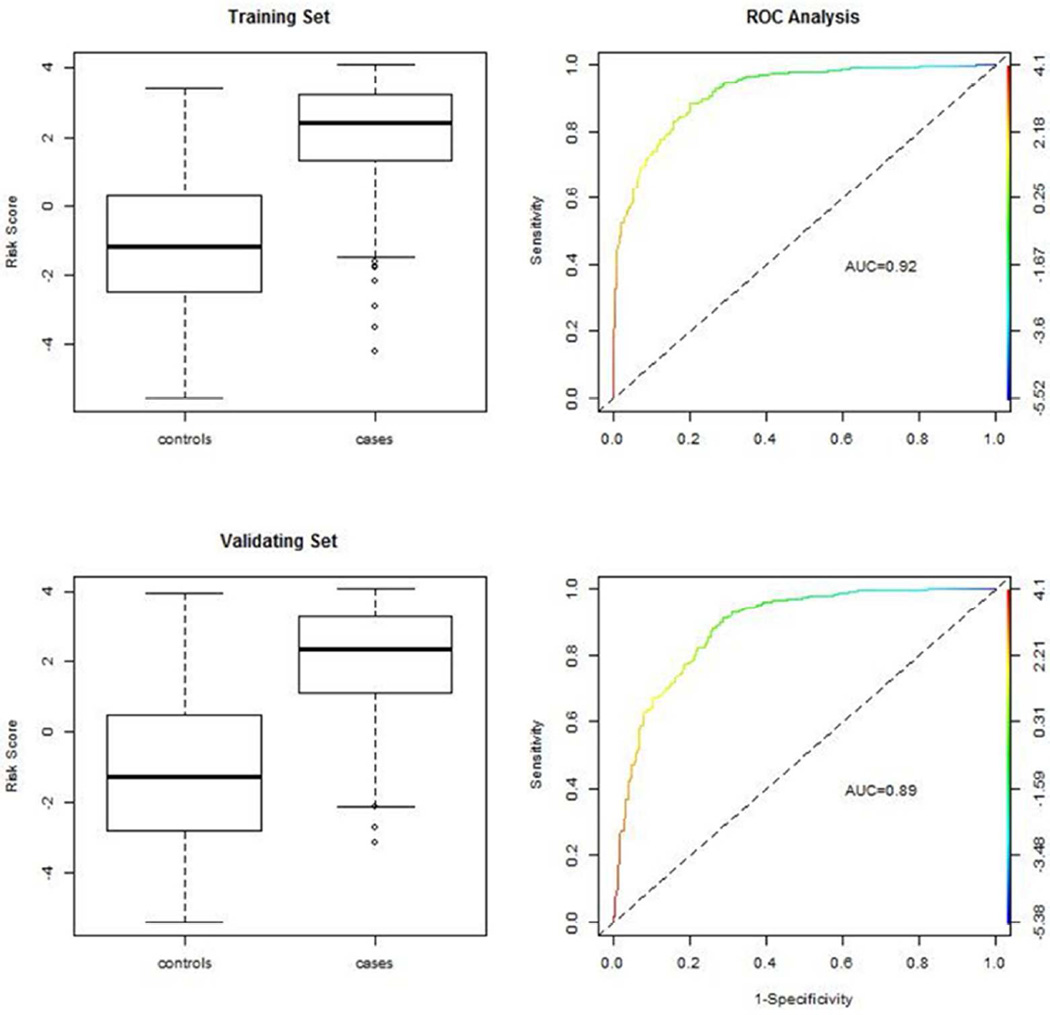
where the summation is over all those 26 selected exemplars and β̂k is the estimated coefficient listed in Table 3. To evaluate empirical distributions of risk scores, we draw boxplots for risk scores among controls and cases in the training set (Figure 5). Clearly, risk scores among cases are generally greater than those among controls in the training set, and their difference is statistically significant (p-value< 0.001, not shown). Risk scores among controls have a symmetric distribution, while those among patients are somewhat skewed. With risk scores ranging from −5.52 to 4.1, computed sensitivity (y-axis of ROC curve) and 1-specificity (x-axis) form an ROC curve with AUC=0.92 in the training set.
Figure 5.
Evaluation of T1D predictive model with class II HLA genes (HLA-DRB1, -DRB345, -DQA1, -DQB1, -DPA1 and –DPB1) in the training set (top panels) and in the validating set (bottom panels). The box plots show distributions of risk scores in training and validating set. ROC curves are shown on the left hand panels.
To validate the above predictive model, we computed risk scores for all samples in the validating set with fixed exemplars and associated weights in the model above [5]. On the lower left panel, the boxplot shows the distribution of risk scores among controls and cases (Figure 4). Clearly, empirical distributions of risk scores in the validating set are largely comparable to those in the training set. Furthermore, ROC analysis on the validating set reveals a comparable sensitivity-specificity curve with AUC=0.89 (Figure 5).
Stability in Selecting Exemplars
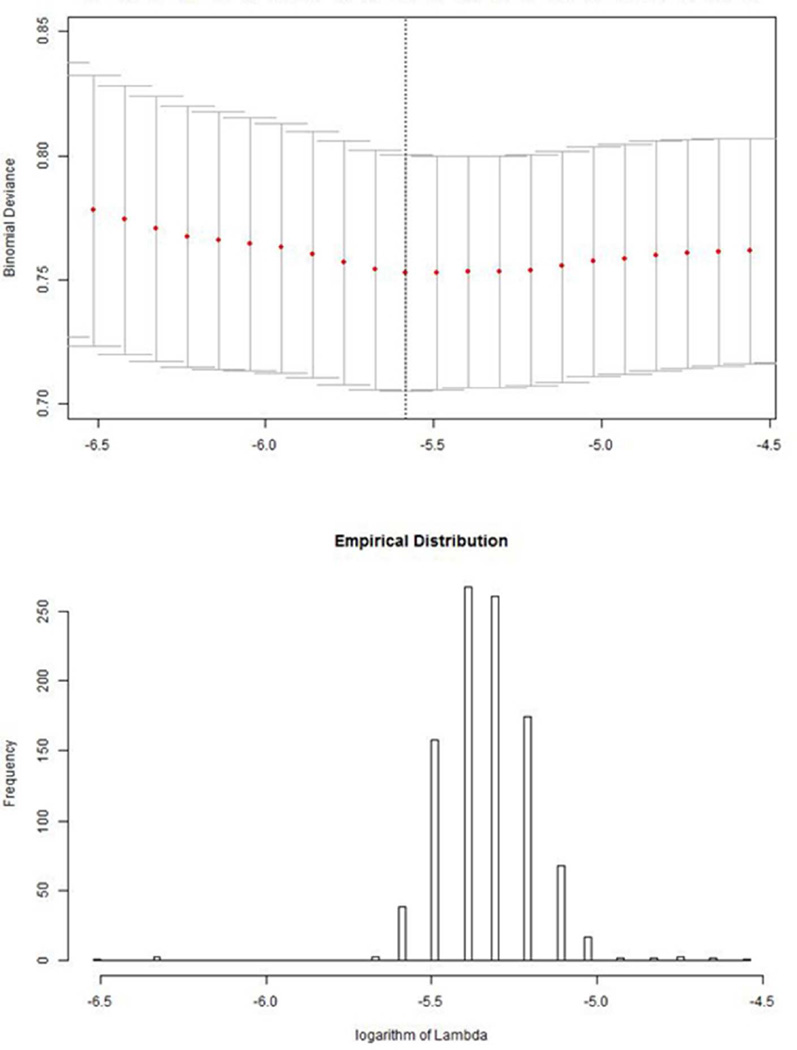
It is known that the choice of the penalty parameter (λ) has a direct and profound impact on the selection of informative exemplars [Friedman, et al. 2010; Tibshirani, et al. 2012]. Conventional cross-validation is used to determine the penalty value that achieves the minimum deviance (misclassification error or AUC). The top panel of Figure 6 shows an XY plot of deviance versus different penalty parameter values (on log scale). It achieves the minimum with the logarithmic value of the estimated penalty parameter somewhere between −6.0 to −5.5. The flatness of this function implies that cross-validation likely has a major influence on estimated penalty parameters. To assess its influence, we repeated the estimation of the penalty parameter 1000 times, and estimated corresponding values. The lower panel of Figure 6 shows the empirical distribution of estimated penalty parameter. Interestingly, estimated penalty values in the training set are discrete with a total of 15 unique values, probably because of discreteness in the similarity matrix.
Figure 6.
Empirical distribution of estimated penalty parameter (λ) from repeated cross-validation estimate (top panel) with the profile deviance function with varying penalty parameter.
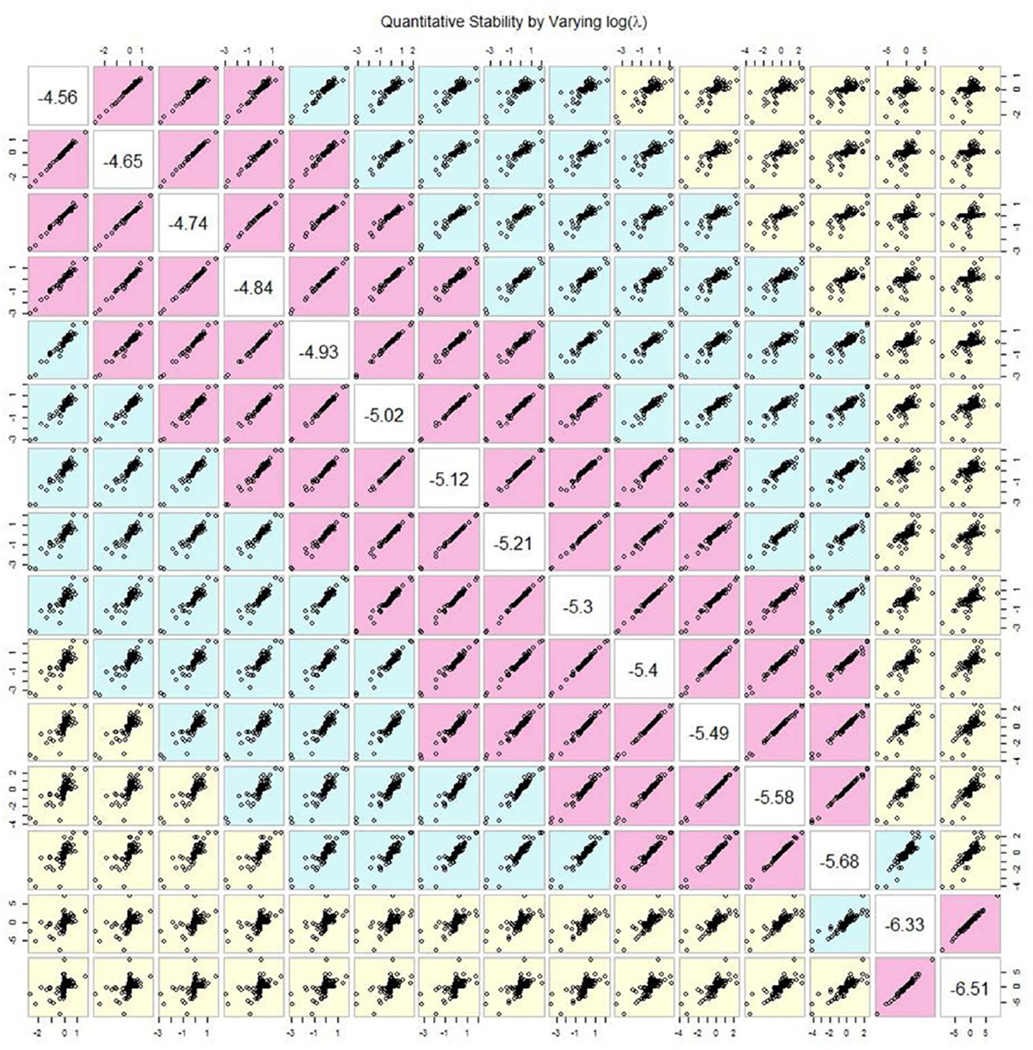
Given that the value of penalty parameter affects variable selection, it is of interest, first, whether the selected variables are stable with different penalty parameter values, and, second, whether the selection is stable even with a fixed penalty parameter. To address this question, we performed a bootstrap analysis with these fifteen different penalty parameter values. On each of 1000 bootstrap samples, we performed LASSO with respectively fixed lambda values, and selected informative exemplars by the penalized likelihood. For qualitative comparisons, we choose to use Kappa statistics to measure overlaps of selected exemplars [Agresti 1988; Cohen 1968]; the large Kappa value corresponds to the greater overlap of selected exemplars by two LASSO estimations with different penalty parameter values. Averaged Kappa values, along with their standard deviations, are computed across all Bootstrap samples (Table 4, Kappa values in the upper triangle, and standard deviations in the lower triangle). The results indicate that concordance among these fifteen penalty values is around 80% for adjacent penalty values. Concordances degrade as the differences of penalty parameter values increases, as expected. To gain further insight into quantitative concordance of estimated coefficients with different penalty values, we compute averaged coefficient estimates over all bootstraps, and plot a pairwise XY plot for mean coefficients with different penalty values (indicated in the diagonal boxes) (Figure 7). It is obvious that estimated coefficient averages are highly correlated with each other, if their penalty values of the pair are close. Otherwise, estimated coefficients, with different penalty values, could differ.
Table 4.
Averaged Kappa values over 1,000 bootstrap samples, to measure concordance of selected informative exemplars across LASSO estimations with different penalty parameter values (on the logarithmic scale). Averaged Kappa values are shown above the diagonal and are highlighted yellow if the value is greater than 0.8. Standard deviations for estimated Kappa values are recorded below the diagonal line.
| −log(lamda) | −6.512 | −6.326 | −5.675 | −5.582 | −5.489 | −5.396 | −5.303 | −5.210 | −5.117 | −5.024 | −4.931 | −4.838 | −4.745 | −4.652 | −4.559 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −6.512 | 0.87 | 0.54 | 0.50 | 0.46 | 0.43 | 0.39 | 0.37 | 0.34 | 0.31 | 0.29 | 0.27 | 0.25 | 0.23 | 0.21 | |
| −6.326 | 0.04 | 0.61 | 0.56 | 0.53 | 0.49 | 0.45 | 0.42 | 0.39 | 0.36 | 0.34 | 0.31 | 0.29 | 0.27 | 0.25 | |
| −5.675 | 0.06 | 0.06 | 0.92 | 0.86 | 0.80 | 0.75 | 0.70 | 0.65 | 0.60 | 0.56 | 0.52 | 0.48 | 0.45 | 0.42 | |
| −5.582 | 0.07 | 0.06 | 0.03 | 0.92 | 0.86 | 0.80 | 0.75 | 0.70 | 0.65 | 0.60 | 0.56 | 0.52 | 0.48 | 0.45 | |
| −5.489 | 0.07 | 0.07 | 0.05 | 0.04 | 0.93 | 0.86 | 0.80 | 0.75 | 0.70 | 0.65 | 0.60 | 0.56 | 0.52 | 0.49 | |
| −5.396 | 0.07 | 0.07 | 0.05 | 0.05 | 0.03 | 0.93 | 0.86 | 0.80 | 0.75 | 0.70 | 0.65 | 0.60 | 0.56 | 0.53 | |
| −5.303 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | 0.03 | 0.93 | 0.86 | 0.80 | 0.75 | 0.70 | 0.65 | 0.61 | 0.56 | |
| −5.210 | 0.07 | 0.07 | 0.06 | 0.06 | 0.06 | 0.05 | 0.04 | 0.93 | 0.86 | 0.80 | 0.74 | 0.69 | 0.65 | 0.61 | |
| −5.117 | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | 0.05 | 0.04 | 0.93 | 0.86 | 0.80 | 0.75 | 0.70 | 0.65 | |
| −5.024 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | 0.04 | 0.93 | 0.86 | 0.80 | 0.75 | 0.70 | |
| −4.931 | 0.06 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | 0.04 | 0.93 | 0.86 | 0.81 | 0.75 | |
| −4.838 | 0.06 | 0.06 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | 0.04 | 0.93 | 0.87 | 0.81 | |
| −4.745 | 0.06 | 0.06 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | 0.04 | 0.93 | 0.87 | |
| −4.652 | 0.06 | 0.06 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | 0.04 | 0.93 | |
| −4.559 | 0.06 | 0.06 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.06 | 0.05 | 0.04 |
Figure 7.
Pairwise XY plots for averaged coefficient estimates over 1000 bootstrap samples with one penalty value (X-axis) vs another penalty value (Y-axis). Logarithmic values of penalty parameters are shown on the diagonal line.
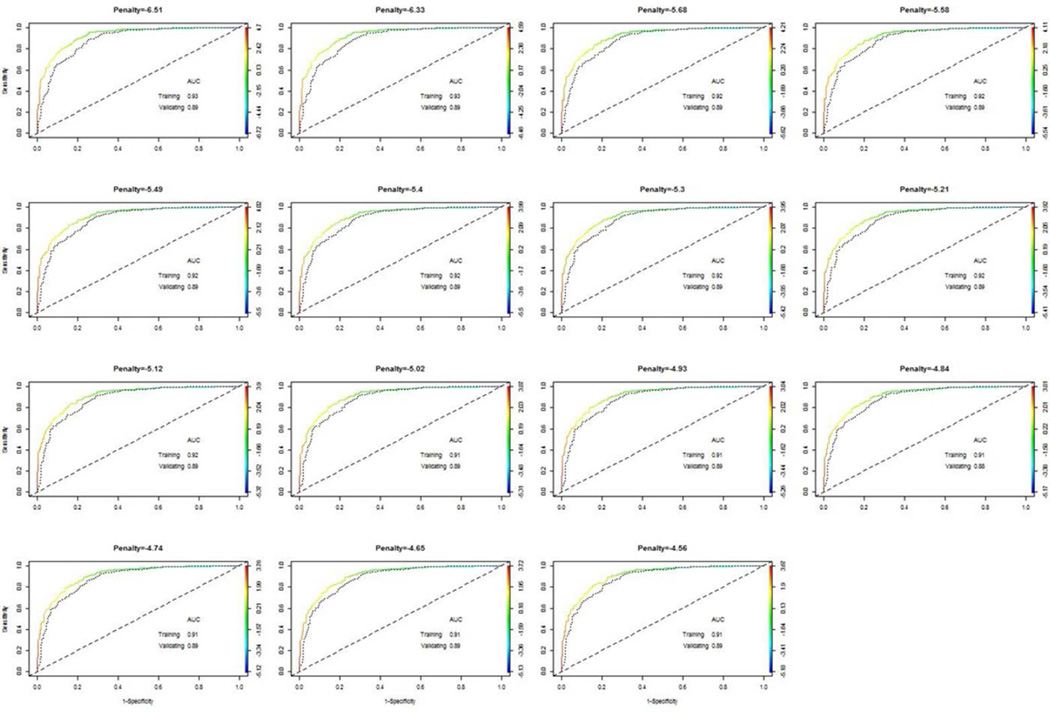
As noted earlier, it appears that multiple predictive models on a single training set have comparable performance. The question is if predictive models, with different penalty parameter values, have similar performances, even though selected informative exemplars and associated coefficients vary. For this purpose, we use LASSO, with fixed penalty parameter value, to select informative exemplars and to construct corresponding predictive models. On each predictive model, we perform ROC analysis on the training set as well as on the validating set (Figure 8). Fifteen ROC curve analysis results with estimated AUC values suggested that the ROC curves are largely comparable. AUC values in the training set vary from 0.91 to 0.93, while these values in the validation set are around 0.89.
Figure 8.
Results from ROC analyses on all predictive models with selected exemplars by LASSO, when penalty parameters are fixed to one of fifteen unique values on the logarithmic scale. AUC values are computed in the training set (colored curve) and also in the validating set (dotted black curve)
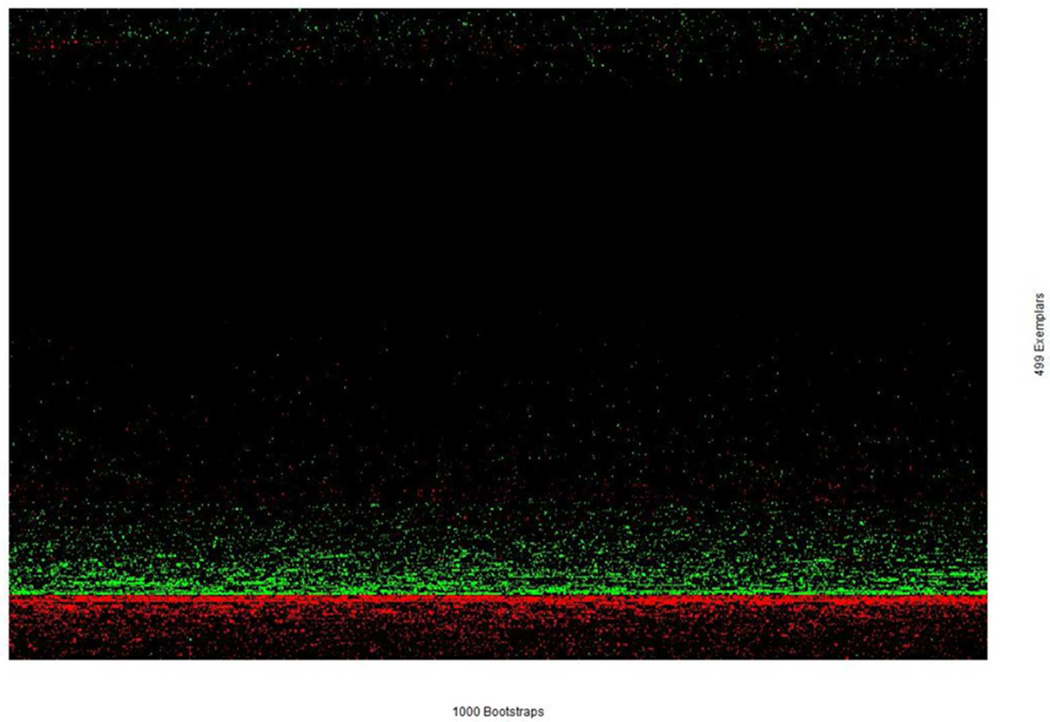
In light of comparable performance and also high concordances of selected exemplars across different penalty parameter values, we chose the medium penalty parameter value (log(λ)= −5.21) to evaluate the stability of individual coefficient estimates across 1000 bootstrap samples. The estimated coefficients for all 499 exemplars across 1000 bootstrap samples, after performing two-way clustering analysis are shown in Figure 9. All values are truncated at −2 and 2, for easy visualization. Interestingly, estimated coefficients, with a fixed penalty value, are remarkably consistent across all 1000 bootstrap samples, despites some subtle variations.
Figure 9.
Estimated LASSO coefficients across 1000 bootstrap samples with the fixed penalty parameter value at log(λ)=−5.21. While the color intensity corresponds to the magnitudes of coefficients, green denotes positive values, while red denotes negative values
Discussion
In this paper, we describe a new approach, termed objective-oriented regression (OOR), to explore disease associations or to build predictive models with highly polymorphic genes. To circumvent the challenge with complex genotypes, OOR transforms the usual genotype-specific (or allele-specific) regression approach to a regression problem with similarity measurements of subjects with exemplars. By using this new “metric”, one regresses disease phenotype on similarities of all subjects with exemplars to explore disease associations with exemplars. Through applying LASSO, OOR allows one to construct a parsimonious predictive model with informative exemplars.
While closely connecting with well-established kernel machine methods, there are differences worth noting. First, exemplars of OOR can be derived internally or externally. Second, OOR actually focuses on disease associations with exemplar-specific similarity measures and interprets association results accordingly, which is different from earlier applications of the kernel machine. Third, by LASSO, OOR builds a parsimonious predictive model with informative exemplars. Fourth, it is quite convenient to apply exemplar-based predictive models to large databases, provided that similarity with exemplars can be measured. Fifth, OOR analyzes on genotype profiles, alleviating the need for haplotypes of multiple HLA genes.
To illustrate the construction of predictive models by OOR, we built a predictive model with all HLA genes (DRB1, DRB345, DQA1, DQB1, DPA1 and DPB1), followed with assessing its performance and the stability of the selected predictors with varying penalty parameter values. On the training set, OOR selected 26 informative exemplars as predictors, and the predictive model had an admirable sensitivity and specificity profile with the AUC of 0.92. Fixing exemplars and regression coefficients, we applied our predictive model to an independently selected validating set, and ROC analysis revealed comparable sensitivity and specificity to those in the training data set, and AUC of 0.89. If further validated by external data sets, this predictive model is ready to be useful for screening T1D in the general population. Note that our analysis has not adjusted for population stratification, since there are two major ethnic populations in Sweden (Swedish and Finish). Given genetic closeness between Swedish and Finish populations, this confounding effect, if any, is not likely to alter such a strong predictive association.
An important property worthy commenting is that OOR results are complementary to allele-specific or genotype-specific results from conventional regression analyses. Recall that genotype-specific regression analysis of HLA genes is typically confined only to those common genotypes, such as HLA-DRB1*03:01:01/03:01:01 or *04:01:01/04:01:01, for which numbers of observations are sufficiently large for meaningful statistical analysis. For genotypes of less frequencies or uneven distributions between cases and controls, the analysis becomes more difficult or infeasible without special handling. In contrast, OOR assesses disease association with the similarity of subject’s genotypes with exemplars’ genotypes, bypassing the limitation noted above. For example, DRB1*15:01:01/07:01:01 has frequencies of 4.4 and 0.0 in controls and cases, respectively (Table 1). When examining phenotype association with similarity to this genotype, many cases and controls have variable similarities, ranging from 0, 0.5, and 1 and yielding robust result with Z=−10 (Table 2).
The idea of using similarity measures has numerous connections with methods developed and used in statistical genetics [Khoury, et al. 1993; Vogel 1997]. While tracing all of these connections is not intended here, it suffices to note that classical and modern genetics aim to discover outcome-associated susceptibility genes through exploiting correlatedness of subjects within families, because shared disease genes, prior to their discovery, likely lead to increasing similarities among related individuals. In the early day of genetics, segregation and linkage methods were developed to characterize and discover genes through familial aggregations of cases [Khoury, et al. 1993; Thompson 1986a; Thompson 1986b]. More recently, some research groups have proposed to assess similarity of genetic markers in cases and controls, and to use similarity regression as a way to discover disease genes [Wessel and Schork 2006]. Further, Tzeng and colleagues have described an association analysis idea of regressing “similarity of traits” on “similarity of genotypes” [Tzeng, et al. 2009]. Beyond modeling on response, similarity of genotypes has been used in assessing associations of variance components on haplotypes [Tzeng and Zhang 2007]. While bearing the same interest in similarities, OOR has a different analytic goal from discovering disease-associated SNPs.
With respect to the data mining literature in computer sciences, OOR has a close connection with a class of approaches known as the k-nearest neighbor methods (kNN)[Biau, et al. 2012; Houle, et al. 2010]. The key idea underlying kNN is that objects, in “close neighborhood” defined by certain characteristics, tend to have similar outcome. In essence, one can use the kNN to make predictions, without any modeling assumption. Such approaches are known sometimes as nonparametric predictive methods. However, kNN is less efficient, partly because it has not taken advantage of the fact that “distant neighbors” may also inform the disease associations and can thus be combined to improve prediction accuracy. In comparison, OOR utilizes all of neighboring information with multiple exemplars. At the conceptual level, OOR could be thought of as an extension to nearest neighbor regression estimates [Devroye, et al. 1994].
Another closely related method is the Grade of Membership technique, known as GoM [Kovtun, et al. 2004; Manton, et al. 1986; Pomarol-Clotet, et al. 2010]. Conceptually, GoM aims to model the joint distribution of outcome and covariates through introducing a set of latent membership variables that create clusters of subjects. Under sensible distributional assumption on these latent membership variables, one can derive a marginalized likelihood after integrating over all GoM latent membership variables, for estimation and inference. At the end of the GoM analysis, one can interpret parameters as properties associated with individuals, rather than specific marginal interpretation of individual covariates. In this regard, OOR, just like GoM, utilizes similarity information to achieve the analytic goal, but differs on modeling assumptions and related implementations. The primary advantage of OOR is that it requires no distributional assumptions on latent membership and makes inferences purely based on empirical evidence.
OOR has potential for two major extensions. First, OOR is constructed for binary disease phenotype under the logistic regression model [3]. Naturally, by extending the logistic regression to the generalized linear model [McCullagh and Nelder 1989], one can generalize OOR to studies with other types of phenotypes, such as continuous, categorical or censored phenotypes, with appropriate choice of the link function. The second extension is to consider other covariate types, such as text strings (e.g. from web searches), electronic signals or images. Further, covariates can be high dimensional data, where the number of dimension is far greater than the sample size (Zhao, et al. 2015, submitted). For these diverse applications, the key is to choose context-dependent similarity measure to define “similarity metrics” between subjects with respect to their covariate profiles.
Supplementary Material
Acknowledgments
Authors would like to thank Dr. Chad He for discussions on variable selection techniques, and to thank Dr. Neil Risch for bringing GoM to our attention. Authors would also like to thank two anonymous reviewers whose comments have substantially improved the presentation of this manuscript. This work is supported in part by European Fund for Research on Diabetes (EFSD), the Swedish Child Diabetes Foundation (Barndiabetesfonden), the National Institutes of Health (DK63861, DK26190), the Swedish Research Council including a Linne grant to Lund University Diabetes Centre, an equipment grant from the KA Wallenberg Foundation, the Skane County Council for Research and Development as well as the Swedish Association of Local Authorities and Regions (SKL), and also by the institutional developmental fund at Fred Hutchinson Cancer Research Center (LPZ).
Footnotes
Conflict of Interest
Authors declare that there is no conflict of interest with this work.
References
- Agresti A. A model for agreement between ratings on an ordinal scale. Biometrics. 1988;44(539–548):539–548. [Google Scholar]
- Bell IR, Koithan M. Models for the study of whole systems. Integr Cancer Ther. 2006;5(4):293–307. doi: 10.1177/1534735406295293. [DOI] [PubMed] [Google Scholar]
- Biau G, Devroye L, Dujmovic V, Krzyzak A. An affine invariant k-nearest neighbor regression estimate. Journal of Multivariate Analysis. 2012;112:24–34. [Google Scholar]
- Bishop DT, Williamson JA. The power of identity-by-state methods for linkage analysis. Am J Hum Gnet. 1990;46:254–265. [PMC free article] [PubMed] [Google Scholar]
- Cardinal-Fernandez P, Nin N, Ruiz-Cabello J, Lorente JA. Systems medicine: a new approach to clinical practice. Arch Bronconeumol. 2014;50(10):444–451. doi: 10.1016/j.arbres.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Cohen J. Weighted Kappa: Nominal scale agreement with provision for scaled disagreement or partial credit. Psychological Bulletin. 1968;70(4):213–220. doi: 10.1037/h0026256. [DOI] [PubMed] [Google Scholar]
- Cox DD, O'Sullivan F. Generalized Nonparametric Regression via Penalized Likelihood. AMS. 1989:1–31. [Google Scholar]
- Cristianini N, Shawe-Taylor J. An introduction to support vector machines: and other kernel-based learning methods. Cambridge; New York: Cambridge University Press; 2000. [Google Scholar]
- Cullen M, Noble J, Erlich H, Thorpe K, Beck S, Klitz W, Trowsdale J, Carrington M. Characterization of recombination in the HLA class II region. Am J Hum Genet. 1997;60(2):397–407. [PMC free article] [PubMed] [Google Scholar]
- Delli AJ, Lindblad B, Carlsson A, Forsander G, Ivarsson SA, Ludvigsson J, Marcus C, Lernmark A Better Diabetes Diagnosis Study G. Type 1 diabetes patients born to immigrants to Sweden increase their native diabetes risk and differ from Swedish patients in HLA types and islet autoantibodies. Pediatr Diabetes. 2010;11(8):513–520. doi: 10.1111/j.1399-5448.2010.00637.x. [DOI] [PubMed] [Google Scholar]
- Delli AJ, Vaziri-Sani F, Lindblad B, Elding-Larsson H, Carlsson A, Forsander G, Ivarsson SA, Ludvigsson J, Kockum I, Marcus C, et al. Zinc transporter 8 autoantibodies and their association with SLC30A8 and HLA-DQ genes differ between immigrant and Swedish patients with newly diagnosed type 1 diabetes in the Better Diabetes Diagnosis study. Diabetes. 2012;61(10):2556–2564. doi: 10.2337/db11-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devroye L, Gyorfi L, Krzyzak A, Lugosi G. On the Strong Universal Consistency of Nearest-Neighbor Regression Function Estimates. Annals of Statistics. 1994;22(3):1371–1385. [Google Scholar]
- Fang FC, Casadevall A. Reductionistic and holistic science. Infect Immun. 2011;79(4):1401–1404. doi: 10.1128/IAI.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth D. Bias Reduction of Maximum Likelihood Estimates. Biometrika. 1993;80(1):27–38. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. Springer series in statistics,. Second edition, corrected 7th printing ed. New York: Springer; 2009. The elements of statistical learning : data mining, inference, and prediction; p. xxii.p. 745. p 1 online resource. [Google Scholar]
- Hastie T, Tibshirani R, Wainwright M. Statistical learning with sparsity : the lasso and generalizations. Boca Raton: CRC Press, Taylor & Francis Group; 2015. [Google Scholar]
- Houle ME, Kriegel HP, Kroger P, Schubert E, Zimek A. Can Shared-Neighbor Distances Defeat the Curse of Dimensionality? Scientific and Statistical Database Management. 2010;6187:482–500. [Google Scholar]
- Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36(2):151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Beaty TH, Cohen BH. Fundamentals of genetic epidemiology. New York: Oxford University Press; 1993. [Google Scholar]
- Kimeldor G, Wahba G. Some Results on Tchebycheffian Spline Functions. Journal of Mathematical Analysis and Applications. 1971;33(1):82-&. [Google Scholar]
- Kovtun M, IAkushevich I, Manton KG, Tolley HD. Grade of Membership Analysis: One Possible Appraoch to Foundations. Cornell University Library. 2004 [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. American Journal of Human Genetics. 2008;82(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manton KG, Stallard E, Woodbury MA, Yashin AI. Applications of the Grade of Membership Technique to Event History Analysis - Extensions to Multivariate Unobserved Heterogeneity. Mathematical Modelling. 1986;7(9–12):1375–1391. [Google Scholar]
- Marsh SGEPPBLD. The HLA FactsBook. San Diego: Academic Press; 2000. [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Model. New York: Chapman and Hall; 1989. [Google Scholar]
- Meinshausen N, Buhlmann P. Stability selection. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2010;72:417–473. [Google Scholar]
- Minnier J, Yuan M, Liu JS, Cai T. Risk Classification with an Adaptive Naive Bayes Kernel Machine Model. J Am Stat Assoc. 2015;110(509):393–404. doi: 10.1080/01621459.2014.908778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble JA. Immunogenetics of type 1 diabetes: A comprehensive review. J Autoimmun. 2015 doi: 10.1016/j.jaut.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Pomarol-Clotet E, Salvador R, Murray G, Tandon S, McKenna PJ. Are There Valid Subtypes of Schizophrenia? A Grade of Membership Analysis. Psychopathology. 2010;43(1):53–62. doi: 10.1159/000260044. [DOI] [PubMed] [Google Scholar]
- Smith SE, Slaughter BD, Unruh JR. Imaging methodologies for systems biology. Cell Adh Migr. 2014;8(5):468–477. doi: 10.4161/cam.29152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Wang JH, Fang YX. Consistent Selection of Tuning Parameters via Variable Selection Stability. Journal of Machine Learning Research. 2013;14:3419–3440. [Google Scholar]
- Tewhey R, Bansal V, Torkamani A, Topol EJ, Schork NJ. The importance of phase information for human genomics. Nat Rev Genet. 2011;12(3):215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EA. Genetic epidemiology: a review of the statistical basis. [Review] Statistics in Medicine. 1986a;5(4):291–302. doi: 10.1002/sim.4780050402. [DOI] [PubMed] [Google Scholar]
- Thompson EA. Pedigree analysis in human genetics. Baltimore: The Johns Hopkins University Press; 1986b. [Google Scholar]
- Tibshirani R, Bien J, Friedman J, Hastie T, Simon N, Taylor J, Tibshirani RJ. Strong rules for discarding predictors in lasso-type problems. J R Stat Soc Series B Stat Methodol. 2012;74(2):245–266. doi: 10.1111/j.1467-9868.2011.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng JY, Zhang D. Haplotype-based association analysis via variance-components score test. Am J Hum Genet. 2007;81(5):927–938. doi: 10.1086/521558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng JY, Zhang D, Chang SM, Thomas DC, Davidian M. Gene-trait similarity regression for multimarker-based association analysis. Biometrics. 2009;65(3):822–832. doi: 10.1111/j.1541-0420.2008.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel FMAG. Human Genetics(third edition) New York: Springer-Verlag; 1997. [Google Scholar]
- Wessel J, Schork NJ. Generalized genomic distance-based regression methodology for multilocus association analysis. Am J Hum Genet. 2006;79(5):792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. American Journal of Human Genetics. 2010;86(6):929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chen X, Wong WH. Completely phased genome sequencing through chromosome sorting. Proc Natl Acad Sci U S A. 2011;108(1):12–17. doi: 10.1073/pnas.1016725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LP, Alshiekh S, Carlsson A, Elding-Larsson H, Forsander G, Ivarsson SA, Ludvigsson J, Kockum I, Marcus C, Persson M, et al. Next generation sequencing reveals that HLA-DRB3, - DRB4 and -DRB5 may be associated with islet autoantibodies and risk for childhood type 1 diabetes. Diabetes. 2016 doi: 10.2337/db15-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hastie T. Kernel logistic regression and the import vector machine. Journal of Computational and Graphical Statistics. 2005;14(1):185–205. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.