Abstract
Through genome-wide association analysis and an independent replication study using a total of 1131 bladder cancer cases and 12 558 non-cancer controls of Japanese populations, we identified a susceptibility locus on chromosome 15q24. SNP rs11543198 was associated with bladder cancer risk with odds ratio (OR) of 1.41 and P-value of 4.03 × 10−9. Subgroup analysis revealed rs11543198 to have a stronger effect in male smokers with OR of 1.66. SNP rs8041357, which is in complete linkage disequilibrium (r2 = 1) with rs11543198, was also associated with bladder cancer risk in Europeans (P = 0.045 for an additive and P = 0.025 for a recessive model), despite much lower minor allele frequency in Europeans (3.7%) compared with the Japanese (22.2%). Imputational analysis in this region suggested CYP1A2, which metabolizes tobacco-derived carcinogen, as a causative candidate gene. We also confirmed the association of previously reported loci, namely SLC14A1, APOBEC3A, PSCA and MYC, with bladder cancer. Our finding implies the crucial roles of genetic variations on the chemically associated development of bladder cancer.
INTRODUCTION
Bladder cancer is one of the most frequent cancers, which causes ∼150 000 death per year in the world (1). Both environmental and genetic factors are involved in the development of bladder cancer, and tobacco smoking is known to be the most important factor to increase the risk of bladder cancer; current or former smokers have a 2- to 6-fold higher risk than never-smokers (2,3). In addition, occupational exposures to industrial chemicals (4–6), arsenic contamination in drinking water (7) and infectious diseases (8) also increase the bladder cancer risk. Bladder cancer incidence in males is nearly 3-fold higher than that in females (3), probably due to the higher prevalence of tobacco smoking and occupational exposure in males. On the other hand, familial aggregation of bladder cancer has also been reported (9,10), suggesting the importance of genetic factors in bladder cancer development.
NAT2 and GSTM1 are involved in the detoxification of carcinogen (11), and an NAT2 slow-metabolizer genotype and a GSTM1 null genotype were indicated their association with an increased risk of bladder cancer (12,13). In addition, recent genome-wide association studies (GWASs) in European populations have identified multiple genetic factors associated with bladder cancer (14–18). However, no GWAS has yet been conducted in Asian populations. So, to identify genetic factors associated with the risk of bladder cancer, we conducted a genome-wide association study in the Japanese population.
RESULTS
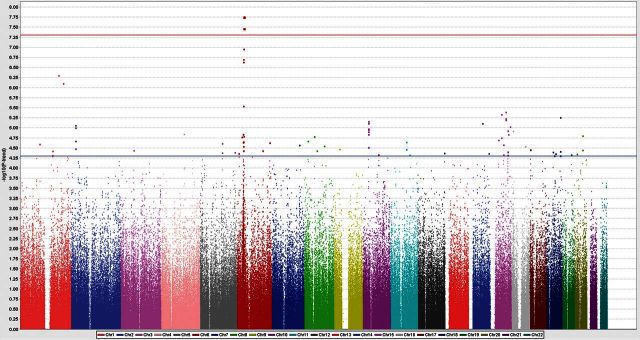
In this study, we performed a two-stage GWAS approach. A study design and the information of subjects used in this analysis are shown in Supplementary Material, Fig. S1 and Table S1. Case samples in the screening and replication stages were recruited from a collaborative network consisting of 11 university-affiliated hospitals in Japan. The genotyping information for non-cancer controls was obtained from the BioBank Japan project. In the screening stage, a total of 539 bladder cancer cases were genotyped by using Human OmniExpress Exome chip. After an initial standard quality control procedure, we obtained genotyping results of 554 389 SNP loci in 531 cases and compared them with those of 5581 controls. Principle component analysis indicated that all subjects used in GWAS were of the Asian ancestry (Supplementary Material, Fig. S2). We performed statistical analysis using a Cochran–Armitage trend test and obtained the genomic inflation factor lambda to be 1.0493, indicating the low possibility of population stratification (Supplementary Material, Fig. S3). From the association analysis, 82 SNPs in 45 distinct genomic regions were indicated suggestive associations (P < 5 × 10−5). Among them, three SNPs on chromosome 6p21 passed the genome-wide significance threshold (P = 1.76–3.39 × 10−8, Fig. 1 and Supplementary Material, Table S2).
Figure 1.
Manhattan plot showing the genome-wide P-values of association. The P-values were calculated by Cochran–Armitage trend test. The y-axis represents the −log10 P-values of 554 389 SNPs, and x-axis shows their chromosomal positions. The horizontal blue line shows the threshold of P ≤ 5 × 10−5 for selecting 84 SNPs for replication analysis.
To validate these associations, we selected 64 SNPs through linkage disequilibrium analysis with the criteria of pair-wise r2 of <0.8. In the replication analysis, we attempted genotyping of a new set of 592 cases and 6964 controls for these 64 SNPs and successfully obtained the genotype information at 59 SNP loci. Among these 59 SNPs, the only one SNP, rs11543198, on chromosome 15q24 revealed significant association with a P-value of 1.22 × 10−4 (P < 8.47 × 10−4 = 0.05/59, Table 1 and Supplementary Material, Table S3). The combined analysis of the GWAS and replication data indicated SNP rs11543198 to have the genome-wide significance of the association (Pmeta = 4.03 × 10−9, odds ratio (OR) = 1.41, Table 1).
Table 1.
Summary of GWAS and replication analyses
| SNP | Case |
Control |
Ptrenda | ORb | (95% CI) | Phetc | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | |||||
| rs11543198 | ||||||||||
| GWAS | 16 | 140 | 375 | 283 | 1913 | 3385 | 6.22 × 10−6 | 1.48 | (1.25–1.75) | 0.48 |
| Replication | 21 | 158 | 413 | 316 | 2382 | 4265 | 1.22 × 10−4 | 1.36 | (1.16–1.59) | |
| Combinedd | 4.03 × 10−9 | 1.41 | (1.26–1.59) | |||||||
A total of 1131 bladder cancer cases and 12 545 controls were analyzed.
P-value obtained from Cochrane–Armitage trend test.
Odds ratios (OR) and confidence interval (CI) are calculated using the non-susceptible allele (A) as reference.
The P-values of heterogeneities across two stages were examined by using the Breslow–Day test.
Meta-analysis of two stage was conducted by using a Cochran–Mantel–Haenszel test.
For validation, we examined the association of this locus with bladder cancer risk in 3508 bladder cancer cases and 5101 controls of European ancestry (17). As rs11543198 was not genotyped in the European GWAS, we used SNP rs8041357, which is in complete linkage disequilibrium (D’ = 1, r2 = 1) with rs11543198 in Asians and Europeans based on the 1000 Genomes Project data (19). In agreement with the 1000 Genomes data, the frequency of the minor allele G was 22.2% in the Japanese controls, compared with 3.7% in Europeans (Supplementary Material, Table S4). Owing to much lower allele frequency, the association in Europeans was weaker but the direction and magnitude of association were similar in both populations.
As gender and smoking status are known to be important risk factors for bladder cancer, we conducted subgroup analysis. As a result, SNP rs11543198 showed a stronger effect among males and smokers (OR of 1.58 and 1.65, respectively) (Table 2). As males are more likely to use tobacco, we also conducted subgroup analysis stratified by both gender and smoking status. SNP rs11543198 exhibited stronger effect among males (OR of 1.66 for smoker and 1.49 for never-smoker) than females (OR of 1.29 for smoker and 1.12 for never-smoker), irrespective of smoking status (Table 2). Although we did not observe significant heterogeneity among each group, our findings suggested possible functional interaction of this genetic factor with tobacco smoking, occupational exposure to carcinogen or some hormonal factors.
Table 2.
Subgroup analysis stratified by gender and smoking status
| SNP | Case |
Control |
Ptrenda | OR (95% CI) b | Phetc | ||||
|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | ||||
| rs11543198 | |||||||||
| Male | 16 | 148 | 427 | 309 | 2220 | 3876 | 3.01 × 10−8 | 1.58 (1.34–1.87) | 0.07 |
| Female | 6 | 45 | 97 | 229 | 1697 | 3049 | 0.32 | 1.16 (0.86–1.55) | |
| Smoker | 11 | 95 | 294 | 295 | 2101 | 3714 | 8.63 × 10−7 | 1.65 (1.34–1.87) | 0.10 |
| Nonsmoker | 11 | 98 | 230 | 243 | 1816 | 3211 | 0.01 | 1.30 (0.86–1.55) | |
| Male/smoker | 10 | 90 | 281 | 218 | 1579 | 2816 | 1.63 × 10−6 | 1.66 (1.35–2.04) | 0.15 |
| Male/nonsmoker | 6 | 58 | 146 | 91 | 641 | 1060 | 3.24 × 10−3 | 1.49 (1.14–1.95) | |
| Female/smoker | 1 | 5 | 13 | 77 | 522 | 898 | 0.54 | 1.29 (0.57–2.95) | |
| Female/nonsmoker | 5 | 40 | 84 | 152 | 1175 | 2151 | 0.47 | 1.12 (0.82–1.54) | |
A total of 739 bladder cancer cases and 11 380 controls were analyzed.
P-value obtained from Cochrane–Armitage trend test.
ORs and CI are calculated using the non-susceptible allele (A) as reference.
The P-value of heterogeneities across four subgroups was examined by using the Breslow–Day test.
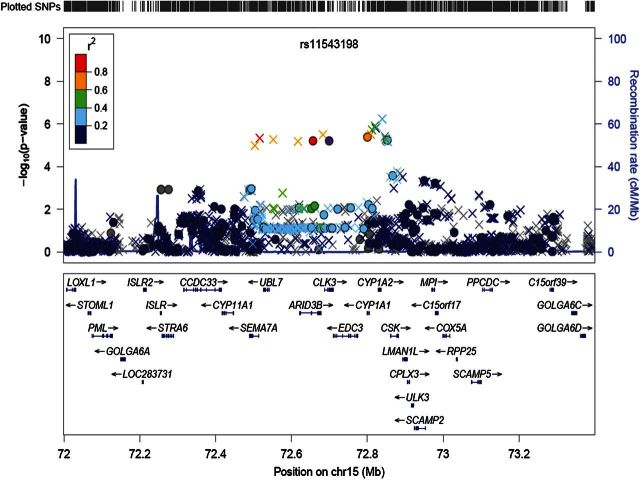
We also conducted imputation analysis around this SNP region of chromosome 15q24 in our GWAS cohort and found that 29 SNPs within a 341-kb region including SEMA7A-UBL7-ARID3B-CLK3-EDC3-CYP1A1-CYP1A2-CSK genes showed similar association as rs11543198 (P < 1 × 10−5, Fig. 2 and Supplementary Material, Table S5). Among these 29 SNPs, rs1350194 in the 3′ flanking region of CYP1A2 revealed the strongest association (P = 5.80 × 10−7 with OR of 1.55, Supplementary Material, Table S5). Moreover, an A allele of SNP rs2069514 (CYP1A2*1C), which is known to have the lower CYP1A2 activity (20), was indicated to reduce the disease risk (P = 1.42 × 10−6 with OR = 0.66, Supplementary Material, Table S5). These two SNPs (rs1350194 and rs2069514) were in modest linkage disequilibrium with rs11543198 (r2 = 0.215 and 0.468, respectively). As CYP1A2 metabolizes some polycyclic aromatic hydrocarbons (PAHs) to carcinogenic intermediates (21,22), the higher CYP1A2 activity generates more carcinogenic metabolites and then is likely to enhance the risk of tobacco-related cancers. Therefore, our result suggests that CYP1A2 on chromosome 15q24 may be a causative gene candidate to increase the risk of bladder cancer.
Figure 2.
Regional association plots around rs11543198 on 15q24 (1.4 Mb). Upper panel: P-values of genotyped SNPs (circle) and imputed SNPs (cross) are plotted (as −log10 P-value) against their physical position on chromosome 15 (NCBI Build 36). SNP rs11543198 is represented by purple circle. The genetic recombination rates estimated from 1000 Genomes samples (JPT + CHB) are shown with a blue line. SNP's color indicates LD with rs11543198 according to a scale from r2 = 0 to 1 based on pair-wise r2-values from HapMap JPT. Lower panel; gene annotations from the University of California Santa Cruz genome browser.
In addition, we also examined previously reported susceptible loci in our GWAS cohort (14–18). We successfully genotyped 10 SNPs as shown in Table 3 and confirmed the significant association of the loci in the SLC14A1, APOBEC3A, PSCA and MYC genes with P-values of <0.05 (Table 3). Two SNPs on TACC3 and UGT1A loci revealed weak trends, but the remaining four loci were not replicated. These results may imply the genetic heterogeneity between European and Asian populations.
Table 3.
Result of previously reported SNPs
| SNP | Chr. | Chr. location | Gene | Relative Pos. | Case |
Control |
Ptrenda | ORb | RAFb | Alleleb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | 11 | 12 | 22 | Case | Control | 1 | 2 | |||||||
| rs17674580 | 18 | 43309911 | SLC14A1 | 0 | 5 | 86 | 439 | 28 | 579 | 4619 | 1.8 × 10−4 | 1.54 | 0.091 | 0.061 | A | G |
| rs1014971 | 22 | 39332623 | APOBEC3A | −20904 | 117 | 256 | 157 | 920 | 2545 | 1761 | 0.0074 | 1.19 | 0.462 | 0.420 | T | C |
| rs2294008 | 8 | 143761931 | PSCA | 0 | 241 | 228 | 61 | 2079 | 2416 | 730 | 0.0092 | 1.20 | 0.670 | 0.629 | T | C |
| rs9642880 | 8 | 128718068 | MYC | −30247 | 63 | 226 | 241 | 491 | 2165 | 2569 | 0.039 | 1.15 | 0.332 | 0.301 | T | G |
| rs798766 | 4 | 1734239 | TACC3 | 0 | 15 | 174 | 341 | 154 | 1461 | 3611 | 0.056 | 1.17 | 0.192 | 0.169 | A | G |
| rs11892031 | 2 | 234565283 | UGT1A8 | 0 | 526 | 4 | 0 | 5143 | 83 | 0 | 0.13 | 2.11 | 0.996 | 0.992 | T | G |
| rs1495741 | 8 | 18272881 | NAT2 | 14158 | 55 | 232 | 244 | 559 | 2394 | 2628 | 0.62 | 1.03 | 0.322 | 0.315 | A | G |
| rs401681 | 5 | 1322087 | CLPTM1L | 0 | 229 | 245 | 56 | 2359 | 2288 | 579 | 0.64 | 0.97 | 0.663 | 0.670 | G | A |
| rs710521 | 3 | 189645933 | LEPREL1 | 28584 | 298 | 203 | 29 | 2948 | 1996 | 282 | 0.92 | 0.99 | 0.754 | 0.755 | T | C |
| rs8102137 | 19 | 30296853 | CCNE1 | −6048 | 2 | 89 | 439 | 37 | 834 | 4355 | 0.92 | 0.99 | 0.912 | 0.913 | G | A |
We analyzed 539 bladder cancer cases and 5581 controls at GWAS stage. Chr., chromosome; Pos., position in the NCBI Build 36.3.
P-values were obtained from Cochrane-Armitage trend test.
Allele 1 is a risk allele in the previous GWAS studies. OR was calculated using allele 2 as a reference.
DISCUSSION
Here, we reported the result of GWAS analysis using a total of 1131 Japanese bladder cancer cases and 12 558 controls. Our data indicated that SNP rs11543198 on chromosome 15q24 was significantly associated with bladder cancer risk. SNP rs11543198 is located within the CLK3 gene, which encodes a serine/threonine protein kinase. CLK3 regulates localization of SR family of splicing factors; however, its association with human carcinogenesis was not reported so far. Although SNP rs11543198 alters amino acid sequence of minor isoforms of CLK3 (X1, X2, X5 and X6), this SNP does not affect amino acid sequence of catalytic domain. Therefore, SNP rs11543198 is not likely to be a causative variation.
Imputation analysis of 29 SNPs in the 341-kb region including this SNP locus suggested an SNP near the CYP1A2 gene to be a causative candidate because of its stronger association than the marker SNP rs11543198 as well as its biological relevance. Interestingly, CYP1A2 belongs to the cytochrome P450 superfamily, members of which encode monooxygenases involved in metabolism of various substrates including drugs and play essential roles in the synthesis of cholesterol, steroids and other lipids (23,24). CYP1A2, which is highly expressed in the liver, is activated by the exposure to PAHs (25,26) and metabolizes them. It is well know that CYP1A2 metabolizes heterocyclic aromatic amines contained in tobacco smoke (27,28) and generates carcinogenic intermediates (21,22). In addition, genetic variations in the CYP1A2 locus are known to affect its enzymatic activity (29,30). CYP1A2 activity exhibits a significant degree of inter-individual diversity owing to both environmental and genetic factors. Hence, associations of CYP1A2 variations with various cancers including bladder, breast, colorectal and lung cancers have been repeatedly investigated, but the results are controversial (31–34). Our first GWAS in the Asian population revealed that CYP1A2 locus is significantly associated with bladder cancer. Moreover, SNP rs11543198 showed the stronger effect on smokers compared with never-smokers in both males and females. Considering the role of CYP1A2 in the metabolism of tobacco-derived carcinogen, our finding suggested the interesting gene-environmental interaction on the development of bladder cancer.
Among genes associated with bladder cancer in previously reported GWAS, our analyses revealed that SNPs in the SLC14A1, APOBEC3A-CBX6, PSCA and MYC loci were associated with bladder cancer risk in the Japanese population (P < 0.05). Association of these four SNPs with bladder cancer was also reported in the Chinese population (35,36). Thus, these variations are common bladder cancer loci among the European and Asian populations.
SLC14A1 functions as a urea transporter and regulates urine concentration and body-fluid balance. Although the function of SNP rs17674580 in SLC14A1 was not fully elucidated, our previous analysis revealed that SLC14A1-SLC14A2 locus was associated with blood urea nitrogen level (37). Thus, this genetic variation would be associated with the kidney function and urine concentration and subsequently affect on the bladder cancer risk. SNP rs1014971 on chromosome 22q13.1 is located ∼25 kb centromeric to APOBEC3A. As APOBEC3A was expressed only in peripheral blood leukocytes and spleen (38), the role of this variation in the carcinogenesis of bladder cancer is not yet clarified.
SNPs rs2294008 and rs9642880 are located on chromosome 8q24, and these loci are not in the same linkage disequilibrium (LD) block. SNP rs9642880, which is located in the LD block adjacent to MYC, a well-known oncogene, was shown to be associated with MYC mRNA and protein expression (39). SNP rs2294008 is associated with cell surface localization and higher expression level of PSCA (40,41). PSCA, which is up-regulated in various tumors including bladder cancer, was shown to be involved in cell renewal and proliferation (42). Therefore, the association of these SNPs with bladder cancer risk can be explained by the growth promoting effect of MYC and cell surface PSCA.
Taken together, we here demonstrated the important roles of genetic factors on the development of bladder cancer. Particularly, the stronger effect of rs11543198 on smokers is very important because it implies a possibility that a small change in the life style, quitting or avoiding smoking, may contribute to the improvement of individuals carrying the risk genotype. Although further prospective analysis is necessary, we hope that our finding would further emphasize the significance of tobacco control.
MATERIALS AND METHODS
Study population
Characteristics of each cohort are shown in Supplementary Material, Table S1. In this study, we conducted GWAS and replication analyses using a total of 1131 bladder cancer cases and 12 558 controls. Case samples in GWAS and replication were obtained from a collaboration network consisting of Iwate Medical University, Okayama University, Kochi Medical School, Kyoto Prefectural University of Medicine, Kanazawa University, Yamagata University, University of Tsukuba, Nagoya City University, Gifu University, Kagoshima University and Ehime University. Control samples in GWAS and replication consisted of healthy volunteers (n = 1919) and subjects with other diseases (n = 10 639, cerebral aneurysm, chronic obstructive pulmonary disease, glaucoma, nephrolithiasis, nephrotic syndrome, epilepsy, atopic dermatitis and Grave's disease) obtained from Biobank Japan Project supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan. In the BioBank Japan Project, DNA and serum of patients were collected through a collaborating network of 66 hospitals throughout Japan. More than 200 000 individuals with 47 common diseases, irrespective of prior treatment, were enrolled in this project from 2003. A list of participating hospitals is provided at the BioBank Japan website (http://www.biobankjp.org/english/index.html). Subjects with a history of any cancers were excluded from controls. Smoking information of both cases and controls was obtained by oral interview. This project was approved by the ethical committees at each institute.
SNP genotyping
Genomic DNA was extracted from peripheral blood leukocytes or normal tissue using a standard method. In GWAS, we genotyped 539 bladder cancer cases and 5769 non-cancer controls (cerebral aneurysm, primary sclerosing cholangitis, chronic obstructive pulmonary disease, glaucoma and healthy volunteers) using Human OmniExpress and HumanExome (Supplementary Material, Fig. S1). A total of 951 117 SNPs including 925 436 autosomal SNPs were genotyped by both platforms. Among the 925 436 autosomal SNPs, 223 273 SNPs were monomorphic in our case–controls sample set. We excluded the following samples from analysis: closely related samples, gender mismatch and subjects whose ancestries were estimated to be distinct from East-Asian populations using principle component analysis. Then, we applied SNP quality control as follows: call rate ≥0.99 in case and control samples, minor allele frequency (MAF) of <0.01 and P-value of Hardy–Weinberg Equilibrium in control group ≥1 × 10−6. Consequently, 554 389 SNPs on autosomal chromosomes passed the quality control filters. Among 82 SNPs showing P < 5 × 10−5, we selected 64 SNPs by linkage disequilibrium analysis with the criteria of pair-wise r2 of >0.8. In the replication analysis, we genotyped 64 SNPs in 592 bladder cancer cases by using multiplex PCR-based Invader assay (Third Wave Technologies). A total of 6964 non-cancer controls (nephrolithiasis, epilepsy, atrophic dermatitis and Grave's disease) from Biobank Japan were genotyped by Human OmniExpress exome beadchip.
Statistical analysis
The association of SNPs with bladder cancer risk was tested by Cochran–Armitage trend test. To characterize population structure in the GWAS cohort, we performed principal component analysis (43). In the GWAS, the genetic inflation factor λ was derived by P-values obtained by Cochran–Armitage trend test for all the tested SNPs. The quantile–quantile plot was drawn using R program. The ORs were calculated using the non-susceptible allele as references, unless it was stated otherwise. The combined analysis of GWAS and replication stage was verified by conducting the Mantel–Haenszel method. Heterogeneity across two stages was examined by using the Breslow–Day test (44). We considered P = 5 × 10−8 (GWAS and meta-analysis) and 8.33 × 10−4 (0.05/59, replication analysis) as the significant threshold after Bonferroni correction for multiple testing.
Replication analysis in Europeans
Genotypes of rs8041357 in 3508 bladder cancer cases and 5101 controls of European ancestry were extracted from the previous GWAS data set (17). Each participating study obtained informed consent from study participants and approval from its respective Institutional Review Board for this study. Genome-wide genotyping was conducted using HumanHap 1M or HumanHap610-Quad BeadCHIP (Supplementary Material, Table S1). Genotypes of rs8041357 were in Hardy–Weinberg equilibrium in controls (P > 0.05). Association was evaluated for an additive model adjusting for age, sex, smoking status (ever/never), study sites and significant EVs as in the original GWAS (17). Owing to low allele frequency for allele G (3.7%), we also analyzed the results using a recessive genetic model, evaluating the effect in risk homozygotes (AA) versus a combined group of rare homozygotes (GG) and heterozygotes (AG), adjusting for the same covariates.
Imputation analysis
Imputation of ungenotyped SNPs was conducted by MACH (45) and minimac (46) using data of JPT/CHS/CHD subjects from 1000 genome project phase 1 (release 16 March 2012) as a reference. We excluded SNPs that met the following criteria: MAF < 0.01, Hardy–Weinberg Equilibrium P-value < 1 × 10−6 or large allele frequency difference between reference panel and GWAS (>0.16).
Software
For general statistical analysis, we employed R statistical environment version 2.9.1 (cran.r-project.org). The Haploview software version 4.2 (47) was used to draw Manhattan plot. Primer3-webv0.3.0 (http://frodo.wi.mit.edu) web tool was used to design primers. MACH (45) (http://www.sph.umich.edu/csg/abecasis/MACH/), minimac (46) (http://genome.sph.umich.edu/wiki/Minimac) and mach2dat (http://genome.sph.umich.edu/wiki/Mach2dat:_Association_with_MACH_output) were used for imputation analysis.
SUPPLEMENTARY MATERIAL
FUNDING
This work was conducted as a part of the BioBank Japan Project that was supported by the Ministry of Education, Culture, Sports, Science and Technology of the Japanese government. The work was supported in part by the Intramural Research Program (IRP) of the National Cancer Institute of the US National Institutes of Health.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the patients and the members of the Rotary Club of Osaka-Midosuji District 2660 Rotary International in Japan, who donated their DNA for this work. We also thank Satomi Takahashi (the University of Tokyo) and the technical staff of the Laboratory for Genotyping Development, Center for Integrative Medical Science, RIKEN for their technical support.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Freedman N.D., Silverman D.T., Hollenbeck A.R., Schatzkin A., Abnet C.C. Association between smoking and risk of bladder cancer among men and women. JAMA. 2011;306:737–745. doi: 10.1001/jama.2011.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castelao J.E., Yuan J.M., Skipper P.L., Tannenbaum S.R., Gago-Dominguez M., Crowder J.S., Ross R.K., Yu M.C. Gender- and smoking-related bladder cancer risk. J. Natl Cancer Inst. 2001;93:538–545. doi: 10.1093/jnci/93.7.538. [DOI] [PubMed] [Google Scholar]
- 4.Colt J.S., Baris D., Stewart P., Schned A.R., Heaney J.A., Mott L.A., Silverman D., Karagas M. Occupation and bladder cancer risk in a population-based case-control study in New Hampshire. Cancer Causes Control. 2004;15:759–769. doi: 10.1023/B:CACO.0000043426.28741.a2. [DOI] [PubMed] [Google Scholar]
- 5.Kogevinas M., t Mannetje A., Cordier S., Ranft U., Gonzalez C.A., Vineis P., Chang-Claude J., Lynge E., Wahrendorf J., Tzonou A., et al. Occupation and bladder cancer among men in Western Europe. Cancer Causes Control. 2003;14:907–914. doi: 10.1023/b:caco.0000007962.19066.9c. [DOI] [PubMed] [Google Scholar]
- 6.Goebell P.J., Villanueva C.M., Rettenmeier A.W., Rubben H., Kogevinas M. Environmental exposure, chlorinated drinking water, and bladder cancer. World J. Urol. 2004;21:424–432. doi: 10.1007/s00345-003-0389-1. [DOI] [PubMed] [Google Scholar]
- 7.Chen C.J., Kuo T.L., Wu M.M. Arsenic and cancers. Lancet. 1988;1:414–415. doi: 10.1016/s0140-6736(88)91207-x. [DOI] [PubMed] [Google Scholar]
- 8.Badawi A.F., Mostafa M.H., Probert A., O'Connor P.J. Role of schistosomiasis in human bladder cancer: evidence of association, aetiological factors, and basic mechanisms of carcinogenesis. Eur. J. Cancer Prev. 1995;4:45–59. doi: 10.1097/00008469-199502000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Aben K.K., Witjes J.A., Schoenberg M.P., Hulsbergen-van de Kaa C., Verbeek A.L., Kiemeney L.A. Familial aggregation of urothelial cell carcinoma. Int. J. Cancer. 2002;98:274–278. doi: 10.1002/ijc.10191. [DOI] [PubMed] [Google Scholar]
- 10.Murta-Nascimento C., Silverman D.T., Kogevinas M., García-Closas M., Rothman N., Tardón A., García-Closas R., Serra C., Carrato A., Villanueva C. Risk of bladder cancer associated with family history of cancer: do low-penetrance polymorphisms account for the increase in risk? Cancer Epidemiol. Biomarkers Prev. 2007;16:1595–1600. doi: 10.1158/1055-9965.EPI-06-0743. [DOI] [PubMed] [Google Scholar]
- 11.Hein D.W. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat. Res. 2002;506:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 12.Risch A., Wallace D.M., Bathers S., Sim E. Slow N-acetylation genotype is a susceptibility factor in occupational and smoking related bladder cancer. Hum. Mol. Genet. 1995;4:231–236. doi: 10.1093/hmg/4.2.231. [DOI] [PubMed] [Google Scholar]
- 13.García-Closas M., Malats N., Silverman D., Dosemeci M., Kogevinas M., Hein D.W., Tardón A., Serra C., Carrato A., García-Closas R. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueroa J.D., Ye Y., Siddiq A., Garcia-Closas M., Chatterjee N., Prokunina-Olsson L., Cortessis V.K., Kooperberg C., Cussenot O., Benhamou S., et al. Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum. Mol. Genet. 2014;23:1387–1398. doi: 10.1093/hmg/ddt519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Closas M., Ye Y., Rothman N., Figueroa J.D., Malats N., Dinney C.P., Chatterjee N., Prokunina-Olsson L., Wang Z., Lin J., et al. A genome-wide association study of bladder cancer identifies a new susceptibility locus within SLC14A1, a urea transporter gene on chromosome 18q12.3. Hum. Mol. Genet. 2011;20:4282–4289. doi: 10.1093/hmg/ddr342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rafnar T., Vermeulen S.H., Sulem P., Thorleifsson G., Aben K.K., Witjes J.A., Grotenhuis A.J., Verhaegh G.W., Hulsbergen-van de Kaa C.A., Besenbacher S., et al. European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum. Mol. Genet. 2011;20:4268–4281. doi: 10.1093/hmg/ddr303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothman N., Garcia-Closas M., Chatterjee N., Malats N., Wu X., Figueroa J.D., Real F.X., Van Den Berg D., Matullo G., Baris D., et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 2010;42:978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X., Ye Y., Kiemeney L.A., Sulem P., Rafnar T., Matullo G., Seminara D., Yoshida T., Saeki N., Andrew A.S., et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium G.P. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakajima M., Yokoi T., Mizutani M., Kinoshita M., Funayama M., Kamataki T. Genetic polymorphism in the 5′-flanking region of human CYP1A2 gene: effect on the CYP1A2 inducibility in humans. J. Biochem. 1999;125:803–808. doi: 10.1093/oxfordjournals.jbchem.a022352. [DOI] [PubMed] [Google Scholar]
- 21.Lewtas J., Walsh D., Williams R., Dobiáš L. Air pollution exposure–DNA adduct dosimetry in humans and rodents: evidence for non-linearity at high doses. Mutat. Res. 1997;378:51–63. doi: 10.1016/s0027-5107(97)00097-3. [DOI] [PubMed] [Google Scholar]
- 22.Boobis A.R., Lynch A.M., Murray S., de la Torre R., Solans A., Farré M., Segura J., Gooderham N.J., Davies D.S. CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res. 1994;54:89–94. [PubMed] [Google Scholar]
- 23.Guengerich F.P. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 24.Guengerich F.P. Catalytic selectivity of human cytochrome P450 enzymes: relevance to drug metabolism and toxicity. Toxicol. Lett. 1994;70:133–138. doi: 10.1016/0378-4274(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 25.DeCaprio A.P. Biomarkers: coming of age for environmental health and risk assessment. Environ. Sci. Technol. 1997;31:1837–1848. [Google Scholar]
- 26.Elovaara E., Mikkola J., Stockmann-Juvala H., Luukkanen L., Keski-Hynnilä H., Kostiainen R., Pasanen M., Pelkonen O., Vainio H. Polycyclic aromatic hydrocarbon (PAH) metabolizing enzyme activities in human lung, and their inducibility by exposure to naphthalene, phenanthrene, pyrene, chrysene, and benzo (a) pyrene as shown in the rat lung and liver. Arch. Toxicol. 2007;81:169–182. doi: 10.1007/s00204-006-0135-8. [DOI] [PubMed] [Google Scholar]
- 27.Manabe S., Tohyama K., Wada O., Aramaki T. Detection of a carcinogen, 2-amino-1-methyl-6-phenylimidazo [4, 5-b] pyridine (PhIP), in cigarette smoke condensate. Carcinogenesis. 1991;12:1945–1947. doi: 10.1093/carcin/12.10.1945. [DOI] [PubMed] [Google Scholar]
- 28.Wogan G.N., Hecht S.S., Felton J.S., Conney A.H., Loeb L.A. Seminars in Cancer Biology. Vol. 14. Elsevier; 2004. Environmental and chemical carcinogenesis; pp. 473–486. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima M., Yokoi T., Mizutani M., Shin S., Kadlubar F.F., Kamataki T. Phenotyping of CYP1A2 in Japanese population by analysis of caffeine urinary metabolites: absence of mutation prescribing the phenotype in the CYP1A2 gene. Cancer Epidemiol. Biomarkers Prev. 1994;3:413–421. [PubMed] [Google Scholar]
- 30.Chida M., Yokoi T., Fukui T., Kinoshita M., Yokota J., Kamataki T. Detection of three genetic polymorphisms in the 5′-flanking region and intron 1 of human CYP1A2 in the Japanese population. Jpn. J. Cancer Res. 1999;90:899–902. doi: 10.1111/j.1349-7006.1999.tb00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Z., Li Y.L., Zhao L., Zhang C.L. Role of CYP1A2 1F polymorphism in cancer risk: evidence from a meta-analysis of 46 case-control studies. Gene. 2013;524:168–174. doi: 10.1016/j.gene.2013.04.038. [DOI] [PubMed] [Google Scholar]
- 32.Zhenzhen L., Xianghua L., Ning S., Zhan G., Chuanchuan R., Jie L. Current evidence on the relationship between three polymorphisms in the CYP1A2 gene and the risk of cancer. Eur. J. Cancer Prev. 2013;22:607–619. doi: 10.1097/CEJ.0b013e32835f3bd2. [DOI] [PubMed] [Google Scholar]
- 33.Deng S.Q., Zeng X.T., Wang Y., Ke Q., Xu Q.L. Meta-analysis of the CYP1A2-163C>A polymorphism and lung cancer risk. Asian Pac. J. Cancer Prev. 2013;14:3155–3158. doi: 10.7314/apjcp.2013.14.5.3155. [DOI] [PubMed] [Google Scholar]
- 34.Ma Z., Guo W., Gong T., Niu H.J., Wang R.W., Jiang Y.G. CYP1A2 rs762551 polymorphism contributes to risk of lung cancer: a meta-analysis. Tumour Biol. 2014;35:2253–2257. doi: 10.1007/s13277-013-1298-6. [DOI] [PubMed] [Google Scholar]
- 35.Wang P., Ye D., Guo J., Liu F., Jiang H., Gong J., Gu C., Shao Q., Sun J., Zheng S.L., et al. Genetic score of multiple risk-associated single nucleotide polymorphisms is a marker for genetic susceptibility to bladder cancer. Genes Chromosomes Cancer. 2014;53:98–105. doi: 10.1002/gcc.22121. [DOI] [PubMed] [Google Scholar]
- 36.Wang M., Chu H., Lv Q., Wang L., Yuan L., Fu G., Tong N., Qin C., Yin C., Zhang Z., et al. Cumulative effect of genome-wide association study-identified genetic variants for bladder cancer. Int. J. Cancer. 2014;135:2653–2660. doi: 10.1002/ijc.28898. [DOI] [PubMed] [Google Scholar]
- 37.Kamatani Y., Matsuda K., Okada Y., Kubo M., Hosono N., Daigo Y., Nakamura Y., Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 38.Bogerd H.P., Wiegand H.L., Hulme A.E., Garcia-Perez J.L., O'Shea K.S., Moran J.V., Cullen B.R. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M., Zhang W., Yuan L., Fu G., Wei Q., Zhang Z. Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis. 2009;30:991–996. doi: 10.1093/carcin/bgp091. [DOI] [PubMed] [Google Scholar]
- 40.Fu Y.P., Kohaar I., Rothman N., Earl J., Figueroa J.D., Ye Y., Malats N., Tang W., Liu L., Garcia-Closas M., et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc. Natl Acad. Sci. USA. 2012;109:4974–4979. doi: 10.1073/pnas.1202189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanikawa C., Urabe Y., Matsuo K., Kubo M., Takahashi A., Ito H., Tajima K., Kamatani N., Nakamura Y., Matsuda K. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat. Genet., 2012;44:430–434. doi: 10.1038/ng.1109. S431–432. [DOI] [PubMed] [Google Scholar]
- 42.Gu Z., Thomas G., Yamashiro J., Shintaku I., Dorey F., Raitano A., Witte O., Said J., Loda M., Reiter R. Prostate stem cell antigen (PSCA) expression increases with high Gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19:1288–1296. doi: 10.1038/sj.onc.1203426. [DOI] [PubMed] [Google Scholar]
- 43.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 44.Breslow N.E., Day N.E. Statistical methods in cancer research. Volume II – the design and analysis of cohort studies. IARC Sci. Publ. 1987;82:1–406. [PubMed] [Google Scholar]
- 45.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrett J., Fry B., Maller J., Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.