Abstract
This rapid report focuses on the pharmacodynamic mechanism of the carboplatin/paclitaxel combination and correlates it with its cytotoxicity. Consistent with the synergistic to additive antitumor activity (the combination index ranging from 0.53 to 0.94), cells exposed to this combination had significantly increased carboplatin-DNA adduct formation when compared to that of carboplatin alone (450 ± 30 versus 320 ± 120 adducts per 108 nucleotides at 2 h, p = 0.004). Removal of paclitaxel increased the repair of carboplatin-DNA adducts: 39.4 versus 33.1 adducts per 108 nucleotides per hour in carboplatin alone (p = 0.021). This rapid report provides the first pharmacodynamics data to support the use of carboplatin/paclitaxel combination in the clinic.
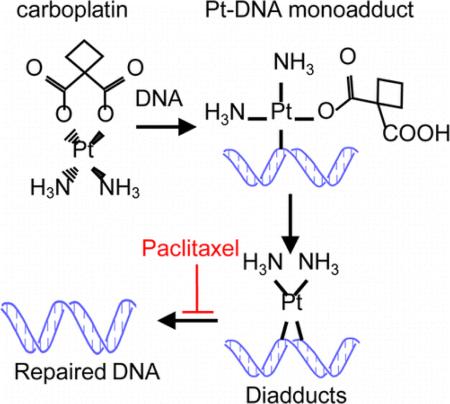
The platinum/taxane combination is one of the most commonly prescribed regimens in the clinic to treat lung, ovarian, bladder, and many other cancer types, and forms the backbone of combination therapy in clinical practice and clinical trials. However, preclinical studies show a wide range of drug–drug interactions and antitumor activities of this combination, ranging from synergism1 and additivity2 to antagonism.3-5 This regimen was designed based on the different antineoplasm mechanisms in which carboplatin exerts cytotoxicity mainly through the induction of carboplatin-DNA adducts while paclitaxel relies on the antidepolymerization of microtubules and cell cycle arrest at the G2/M phase irrespective of p53 status.6-8 It has been proposed that cell cycle arrest induced by paclitaxel hinders the repair of carboplatin-DNA adducts and enhances the antitumor activity. However, this hypothesis has yet to be validated.
Our group has reported the use of accelerator mass spectrometry (AMS) to detect cellular carboplatin-DNA monoadducts, the precursors of all forms of carboplatin-DNA damage. AMS measures 14C at the attomole (10−18-21) level or less in milligram-sized specimens with a few percent precision during repeated measurements.9 This is equivalent to less than one 14C-labeled drug molecule per cell in 105 cells. After treatment with 14C-labeled carboplatin, AMS can measure 14C bound to genomic DNA and allow the calculation of carboplatin-DNA adducts.10 Therefore, the AMS-based ap-proach allows precise measurement of carboplatin-DNA adduct formation and repair to study drug activity and mechanisms.
In this study, we aimed to determine the feasibility of using the pharmacodynamic end point of carboplatin-DNA adduct level modulation by paclitaxel to justify the use of this regimen in the clinic. Furthermore, the compatibility of AMS with translational research (low drug doses, low radiation exposures, and ease of use) can be applied to study many other chemotherapeutic agents or combinations and can have broad clinical applications.
We used a p53 mutant human bladder urothelial carcinoma cell line 5637 (ATCC, Manassas, VA, USA) to show the proof of principle. These cells were maintained with the recom-mended medium. The MTS assay was performed to determine the growth inhibition IC50 (the concentration required for 50%inhibition of cell growth) as described in the manufacturer's instructions (Promega, Madison, WI, USA). In brief, after overnight culture, 5637cells were treated with carboplatin and/or paclitaxel for 4 h to mimic the in vivo half-life of carboplatin and paclitaxel of 1.3–6h.11-13 After treatment, the cells were washed and cultured with medium at 37 °C for 68 h. After treatment with MTS, the absorption was measured at 490 nm using a SpectraMax M3 microplate reader (Molecular Devices, Sunnyvale, CA, USA). The median effect method, proposed by Chou and Talalay, was used to determine the nature (synergism, additivity, and antagonism) of drug and drug interaction.14,15 This method, using the combination index (CI) equation, allowed quantitative determination of drug interactions at increasing levels of cytotoxicity: CI < 0.9 indicates synergism; CI 0.9-1.1 indicates additivity; and CI > 1.1 indicates antagonism. Dm is the antilog of the x-axis intercept, meaning the concentration of carboplatin, paclitaxel, or combination needed to induce 50% of cell killing. Fa is the fraction of cell death induced by drug treatment and ranges from 0 to 1, with 0 meaning no cell killing, and 1 representing 100% of cell killing. The r values of 0.95 or above indicate good conformity of the dose–effect data with respect to the median-effect principle.
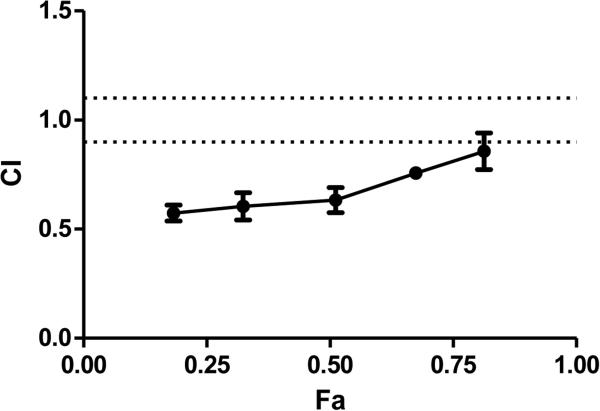
The cytotoxic activities of carboplatin and paclitaxel were first determined individually on 5637 cells. There was a dose– dependent reduction in cell viability with increasing dose for both drugs. IC50 values were 290 μM for carboplatin and 0.08 μM for paclitaxel. The cytotoxicity of carboplatin/paclitaxel combination on 5637 cells was then evaluated.15 With the carboplatin/paclitaxel combination, the Dm value was 130 μM (Table 1), less than the calculated IC50 of the combination at 150 μM [(290 + 0.08) ÷ 2]. This corresponds to the CI value of 0.71, indicating moderate synergism (Figure 1). A similar synergistic effect was observed at IC25 and IC30.AtIC75, the CI value was 0.94, falling into the additivity category. The r values for all these experiments were 0.95 or higher, indicating outstanding conformity of the dose–effect data.
Table 1.
CI Values and Anti-tumor Activity*
| Carboplatin/PTX | IC25 | IC30 | IC50 | IC75 |
|---|---|---|---|---|
| CI Value | 0.62 | 0.53 | 0.71 | 0.94 |
| Drug activity | Syn | Syn | Syn | Add |
Abbreviations: PTX, paclitaxel; Syn, synergism; Ant, antagonism; Add, additivity.
Figure 1.
Median effect method to determine the antitumor activity of carboplatin/paclitaxel combination. At the IC50 concentration, the CI value was 0.71, indicating moderate synergism. At the IC75 concentration, the CI value was 0.94, indicating additivity. Each experiment was repeated at least three times.
We next determined whether the additive to synergistic effect of this combination was associated with paclitaxel-mediated changes in the formation of carboplatin-DNA adducts. Cells were dosed and incubated with 290 μM carboplatin or 290 μM carboplatin combined with 0.08 μM paclitaxel for 4 h, each including 0.43 μM[14C]carboplatin (106 dpm/ml). After washing, cells were maintained with 14C carboplatin-free culture media. DNA was extracted from cells and converted to graphite and measured by AMS for 14C quantification as previously described.16
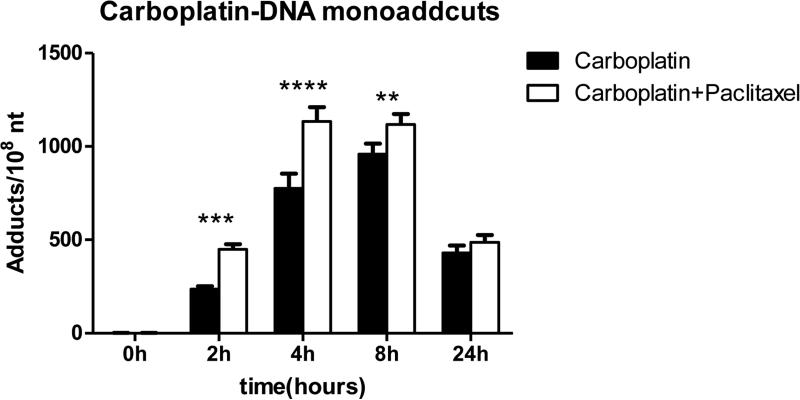
Compared to cells treated with carboplatin alone, cells treated with the carboplatin/paclitaxel combination had consistently higher carboplatin-DNA adducts: the carboplatin-DNA adduct levels were 320 ± 120 and 780 ± 80 adducts per 108 nucleotides for carboplatin alone at 2 and 4 h, respectively, compared to 450 ± 30 (p = 0.004) and 1130 ± 80 (p = 0.002) in the combination. To mimic the in vivo drug metabolism and clearance, carboplatin and carboplatin/paclitaxel were removed from the culture medium by washing 4 h after incubation. After the removal, cells treated with carboplatin alone continued to form DNA adducts during the next 4 h, from 780 ± 80 at 4 h (when carboplatin was washed) to 960 ± 60 adducts/108 nucleotides at 8 h (p = 0.068). However, in cells treated with the carboplatin/paclitaxel combination the DNA adduct levels slightly decrease from 1130 ± 80 to 1120 ± 60 (p = 0.79). At 8 h, the DNA adduct levels were slightly higher and nearly statistically significant in the combination group than the carboplatin alone group (1120 ± 60 versus 960 ± 60, p = 0.054). From 8 to 24 h, the carboplatin-DNA adducts in the combination group decreased from 1120 ± 60 to 490 ± 40 adducts per 108 nucleotides or 39.4 adducts per 108 nucleotides per hour, while the adduct levels in the carboplatin group decreased from 960 ± 60 to 430 ± 40 or 33.1 adducts per 108 nucleotides per hour (p = 0.021). Because of this increased DNA repair in the combination arm after the removal of drugs, the DNA adduct levels between these two groups were not statistically significant at 24 h: 490 ± 40 for the combination versus 430 ± 40 in the carboplatin group (p = 0.21).
This is the first mechanistic study to determine how paclitaxel affects the formation and repair of carboplatin-DNA adducts. Our findings suggest that the addition of paclitaxel increased carboplatin-DNA adduct levels possibly through interference of DNA repair from cell cycle arrest at the G2/M phase by paclitaxel. Our findings support the synergistic effect of carboplatin and paclitaxel. Here, we used ultrasensitive AMS that is able to quantify one carboplatin-DNA adduct per 100 million nucleotides or less from a small plate of cells. Hence, this technology allowed the measurement of DNA adducts under physiologically relevant drug concentrations. As shown in Figure 2, addition of paclitaxel in culture significantly increased the carboplatin-DNA adduct levels at both 2 and 4 h after incubation. One plausible explanation is that G2/M arrest decreases nucleotide excision repair of carboplatin-DNA damage since this repair activity is greatest during G1 of the cell cycle.17 Although this hypothesis requires additional experiments for validation and correlation with in vivo studies, it is supported by the observed increase in DNA repair of carboplatin-DNA monoadducts after the cells were washed and allowed to recover in drug-free cell culture medium. Our results are contradictory to another report showing that paclitaxel and docetaxel decreased cisplatin-DNA adduct formation.18 In that report, white blood cells were collected 3 h after taxane infusion and incubated with cisplatin continuously in vitro. This is not clinically and physiologically relevant as clinically there is no waiting period between infusions of two drugs. Infusion of taxane followed by a 3 h waiting period could possibly induce cell cycle arrest and decrease adduct formation. In our case, cells were treated with carboplatin/paclitaxel concurrently followed by washing. This mimics the clinical situation, in which these two drugs are administered together with similar plasma half-lives of ~4h.11,12
Figure 2.
Paclitaxel increased carboplatin-DNA adduct formation. **, p = 0.068; ***, p = 0.004; ****, p = 0.0015. Each experiment was repeated at least three times.
In conclusion, carboplatin and paclitaxel achieved synergistic antitumor activity, possibly through the inhibition of DNA repair and accumulation of carboplatin-DNA adducts. Furthermore, the AMS method described in this study can be used for other mechanistic studies involving DNA damage.
Acknowledgments
Funding
This study was supported by the VA Career Development Award-2 (PI: Pan), VA Merit (PI: Pan; grant # 1I01BX001784), the NCI Cancer Center Support Grant (PI: de Vere White; P30 CA 093373), and the Research Resource for Biomedical AMS at Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344 and supported by the National Institute of General Medical Sciences (PI; Turteltaub; P41 GM103483-15).
Abbreviations
- AMS
accelerator mass spectrometry
- MTS
3-(4,5-dimethylth-iazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4- sulfophenyl)-2H-tetrazolium
Footnotes
The work reported here does not represent the views or opinions of the Department of Veterans Affairs or the United States Government.
The authors declare no competing financial interest.
References
- 1.Konecny G, Untch M, Slamon D, Beryt M, Kahlert S, Felber M, Langer E, Lude S, Hepp H, Pegram M. Drug interactions and cytotoxic effects of paclitaxel in combination with carboplatin, epirubicin, gemcitabine or vinorelbine in breast cancer cell lines and tumor samples. Breast Cancer Res. Treat. 2001;67:223–233. doi: 10.1023/a:1017980411398. [DOI] [PubMed] [Google Scholar]
- 2.Huang GC, Liu SY, Lin MH, Kuo YY, Liu YC. The synergistic cytotoxicity of cisplatin and taxol in killing oral squamous cell carcinoma. Jpn. J. Clin. Oncol. 2004;34:499–504. doi: 10.1093/jjco/hyh091. [DOI] [PubMed] [Google Scholar]
- 3.Stamelos VA, Robinson E, Redman CW, Richardson A. Navitoclax augments the activity of carboplatin and paclitaxel combinations in ovarian cancer cells. Gynecol. Oncol. 2013;128:377–382. doi: 10.1016/j.ygyno.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Guminski AD, Harnett PR, deFazio A. Carboplatin and paclitaxel interact antagonistically in a megakaryoblast cell line–a potential mechanism for paclitaxel-mediated sparing of carboplatin-induced thrombocytopenia. Cancer Chemother. Pharmacol. 2001;48:229–234. doi: 10.1007/s002800100279. [DOI] [PubMed] [Google Scholar]
- 5.Cabrespine A, Bay JO, Barthomeuf C, Cure H, Chollet P, Debiton E. In vitro assessment of cytotoxic agent combinations for hormone-refractory prostate cancer treatment. Anti-Cancer Drugs. 2005;16:417–422. doi: 10.1097/00001813-200504000-00008. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg B. Platinum Complexes for the Treatment of Cancer: Why the Research Goes On. Vol. 3. Verlag Helvetica Chimica; Zurich, Switzerland: 1999. [Google Scholar]
- 7.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 8.Das GC, Holiday D, Gallardo R, Haas C. Taxol-induced cell cycle arrest and apoptosis: dose-response relationship in lung cancer cells of different wild-type p53 status and under isogenic condition. Cancer Lett. 2001;165:147–153. doi: 10.1016/s0304-3835(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 9.Turteltaub KW, Dingley KH. Application of accelerated mass spectrometry (AMS) in DNA adduct quantification and identification. Toxicol. Lett. 102. 1998;103:435–439. doi: 10.1016/s0378-4274(98)00344-0. [DOI] [PubMed] [Google Scholar]
- 10.Henderson PT, Pan CX. Human microdosing for the prediction of patient response. Bioanalysis. 2010;2:373–376. doi: 10.4155/bio.10.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara K, Yamauchi H, Suzuki S, Ishikawa H, Tanaka Y, Fujiwara M, Kohno I. The platelet-sparing effect of paclitaxel is not related to changes in the pharmacokinetics of carboplatin. Cancer Chemother. Pharmacol. 2001;47:22–26. doi: 10.1007/s002800000212. [DOI] [PubMed] [Google Scholar]
- 12.Sharma H, Thatcher N, Baer J, Zaki A, Smith A, McAucliffe CA, Crowther D, Owens S, Fox BW. Blood clearance of radioactively labelled cis-diammine 1,1-cyclobutane dicarboxylate platinum (II) (CBDCA) in cancer patients. Cancer Chemother. Pharmacol. 1983;11:5–7. doi: 10.1007/BF00257407. [DOI] [PubMed] [Google Scholar]
- 13.Walle T, Walle UK, Kumar GN, Bhalla KN. Taxol metabolism and disposition in cancer patients. Drug Metab. Dispos. 1995;23:506–512. [PubMed] [Google Scholar]
- 14.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 15.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 16.Ognibene TJ, Bench G, Vogel JS, Peaslee GF, Murov S. A high-throughput method for the conversion of CO2 obtained from biochemical samples to graphite in septa-sealed vials for quantification of 14C via accelerator mass spectrometry. Anal. Chem. 2003;75:2192–2196. doi: 10.1021/ac026334j. [DOI] [PubMed] [Google Scholar]
- 17.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 18.Ma J, Verweij J, Planting AS, Kolker HJ, Loos WJ, de Boer-Dennert M, van der Burg ME, Stoter G, Schellens JH. Docetaxel and paclitaxel inhibit DNA-adduct formation and intracellular accumulation of cisplatin in human leukocytes. Cancer Chemother. Pharmacol. 1996;37:382–384. doi: 10.1007/s002800050401. [DOI] [PubMed] [Google Scholar]