Abstract
The heteroduplex mobility assay (HMA) is widely used to characterize strain variants of human viruses. To determine whether it can detect small sequence differences in homologous templates, we constructed a series of deletion constructs (1–10 bp deletions) in the multiple cloning site (MCS) of pBluescript II. After PCR amplification of the MCS using a mixture of wild-type and one of the deletion constructs, the resulting PCR amplicons were electrophoresed using 15% polyacrylamide gels. Two types of heteroduplexes exhibited retarded electrophoretic migration compared with individual homoduplexes. Therefore, we applied this HMA to detect transcription activator-like effector nucleases (TALEN)-induced insertion and/or deletion (indel) mutations at an endogenous locus. We found that TALEN in vivo activity was easily estimated by the degree of multiple HMA profiles derived from TALEN-injected F0 embryos. Furthermore, TALEN-injected F0 founder fish produced several unique HMA profiles in F1 embryos. Sequence analysis confirmed that the different HMA profiles contained distinct indel mutations. Thus, HMA is a rapid and sensitive analytical method for the detection of the TALEN-mediated genome modifications.
Introduction
Artificial site-specific nucleases such as transcription activator-like (TAL) effector nucleases (TALENs) and zinc finger nucleases (ZFNs) offer powerful tools for genome editing by enabling gene disruption (Bogdanove & Voytas 2011; Carroll 2011). The TAL effector, which was originally identified from the plant pathogenic Xanthomonas, performs nucleotide recognition using some simple rules. Site-specific genome sequences are recognized by their central TAL effector repeats, which have 34 identical amino acids repeats except for two highly variable residues at positions 12 and 13 (RVDs: repeat variable diresidues). An individual TAL effector repeat recognizes one nucleotide using the following RVD code (NI=A, HD=C, NG=T, NN=G or A) (Deng et al. 2012). Using these repeats along with their FokI nuclease catalytic domain, TALENs can induce targeted DNA double-strand breaks (DSBs) in a genomic locus. The DSBs are repaired through nonhomologous end joining (NHEJ), which directly connects the ends of the broken strands to efficiently introduce insertion and/or deletion (indel) mutations. Thus, TALEN technology can be used to generate targeted gene disruptions in various types of model organisms including zebrafish.
It is important to evaluate TALEN activity when disrupting a targeted gene at an endogenous locus. In zebrafish embryos, efficient cleavage at the target site by a TALEN could result in a large number of indel mutations in somatic cells during early embryogenesis. The frequency of an indel mutation can be estimated by the degree of Cel-1 or T7 endonuclease cleavage (Mussolino et al. 2011; Tesson et al. 2011), because these two endonucleases digest at the mismatched sites of heteroduplexes. Therefore, the resulting cleaved fragments and homoduplex products are detected using agarose gel electrophoresis. More recently, high resolution melt analysis (HRMA) was applied to detect TALEN-induced genome modifications (Dahlem et al. 2012) by taking advantage of the different melting temperatures between heteroduplex and homoduplex complexes and distinct HRMA profiles. More recently, we have developed a simple modified lacZ assay to detect TALEN-induced frameshifts in an endogenous target loci (Hisano et al. 2013). However, as effective as these techniques may be, they do not provide reliable information on variations that originated from individual mutant alleles.
The heteroduplex mobility assay (HMA) is widely used to characterize strains of various human viruses with slight sequence divergence (Kostrikis et al. 1995; Zou 1997). HMA is based on the denaturing and annealing of PCR-amplified nucleotide strands that are not fully complementary and therefore generate homoduplex and heteroduplexes. The heteroduplexes could be separated from the homoduplexes by polyacrylamide gel electrophoresis because the former migrate more slowly due to an opened single-strand configuration surrounding the mismatched region.
Here, we demonstrate the use of HMA to detect TALEN-induced genome modifications. We used PCR amplification at the targeted genomic region to take advantage of TALENs producing various indel mutations at endogenous loci to form heteroduplexes. We could therefore evaluate whether HMA distinguishes between sequence differences induced by TALENs at a target genomic site in zebrafish.
Results
HMA to detect heteroduplexes
Transcription activator-like effector nucleases predominantly induce small deletions at an endogenous locus (Sander et al. 2011). Because HMA is not well characterized, we examined whether HMA can detect small sequence differences using a series of deletion constructs in the multiple cloning site (MCS) of pBluescript II (Fig. 1A and Fig. S1 in Supporting Information). After PCR amplification of the MCS from a one-to-one mixture of pBluescript wild-type (wt) and one of pBluescript delta (Δ1–Δ10b) mutations, the resulting PCR amplicons were separated on either a 15% polyacrylamide gel (HMA) or a 2% agarose gel. The pBluescript wt/Δ2-wt/Δ10b lanes showed slow migration bands in the polyacrylamide gel (Fig. 1B), but only a single sharp or broad band in the 2% agarose gel (Fig. 1C). We concluded that the HMA profiles derived from a mixture of wild-type and one of the deletion constructs were composed of two fast-migrating homoduplex bands and two slow-migrating heteroduplex bands (Fig. 2A). We reached this conclusion because the two retarded bands (heteroduplexes) were selectively digested at the mismatched sites by T7 endonuclease and had expected band sizes, whereas no digestion was observed in the homoduplexes derived from the pBluescript wt or Δ9. Furthermore, direct sequencing of individual bands in pBluescript wt/Δ9 showed that the higher and lower bands approximately 170 bp corresponded to the homoduplexes derived from pBluescript wt and Δ9, respectively, and the upper two bands corresponded to the heteroduplexes derived from pBluescript wt/Δ9 (Fig. S2 in Supporting Information). The mobility distance between homoduplexes and heteroduplexes became larger as the number of bp deletions increased from 2 to 7. Individual heteroduplexes showed unique HMA profiles that did not fully depend on the length of the mismatched single strand, as the pBluescript Δ10a and Δ10b had 10-bp gaps in different regions, but displayed completely distinct HMA profiles (Fig. 1A,B). When we carried out HMA with a mixture of wild-type and all deletion constructs (Δ1–Δ10a), we observed multiple HMA profiles above the homoduplex band (Fig. 2B), whereas a similar band pattern was observed in a 2% agarose gel (Fig. 2C). Thus, HMA is a very simple and sensitive method for detecting a small sequence differences in homologous templates.
Figure 1.
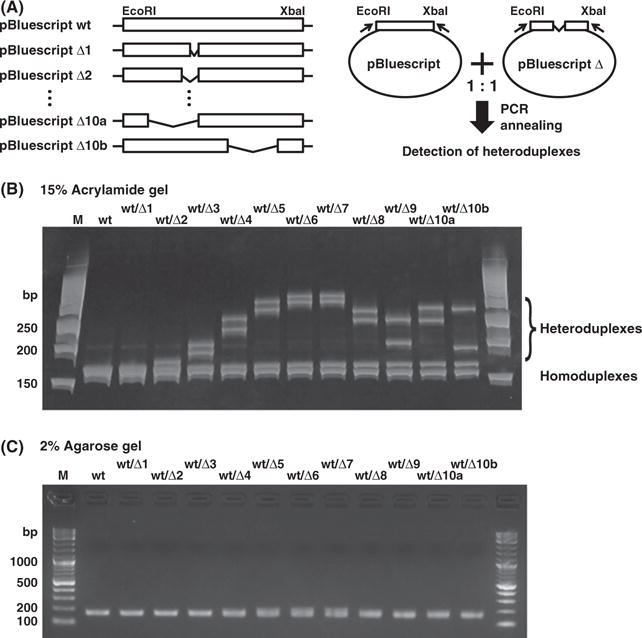
Heteroduplex mobility assay detection of small sequence differences in pBluescript-derived constructs. (A) Schematic representation of the pBluescript-derived constructs (left) and HMA (right). A series of the pBluescript deletion constructs (1–10 bp nucleotide deletions) was generated between EcoRI and XbaI sites. HMA was used to detect heteroduplexes. The MCS was amplified from a mixture of pBluescript wild type (wt) and a pBluescript Δ (delta: Δ1–Δ10b) by PCR. Both pBluescript Δ10a and Δ10b have 10-bp deletion in different region. (B) PCR amplicons for the MCS of the pBluescript-derived constructs were analyzed by electrophoresis on a 15% polyacrylamide gel. PCR amplicons from the mixtures were separated into homoduplexes (lower bands) and heteroduplexes (upper bands). Distinct HMA profiles can be seen for all mixtures except for pBluescript wt and wt/Δ1. (C) A single sharp or broad band was observed in all PCR amplicons in a 2% agarose gel. PCR products were visualized by staining with ethidium bromide. (HMA, heteroduplex mobility assay; M, DNA molecular weight marker; MCS, multiple cloning site).
Figure 2.
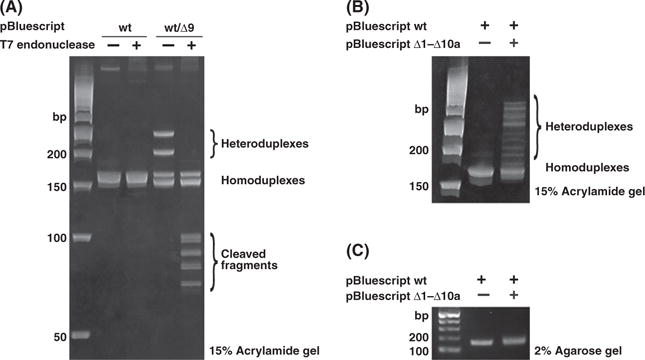
Selective cleavages of heteroduplexes by T7 endonuclease. (A) PCR amplicons from pBluescript wt and pBluescript wt/Δ9 were treated with T7 endonuclease. The two heteroduplex bands (slow mobility) in pBluescript wt/Δ9 sample were selectively cleaved, whereas no homoduplex bands were. (B) HMA of a mixture of pBluescript-derived constructs (wild type and Δ1–Δ10a) in a 15% polyacrylamide gel. PCR products from the mixtures showed multiple HMA profiles. (C) Single broad band was observed in a 2% agarose gel electrophoresis. (HMA, heteroduplex mobility assay).
Evaluation of TALEN in vivo activity by HMA in F0 zebrafish embryos
Mutant alleles induced by a TALEN can be monitored at several stages by (1) evaluating the TALEN in vivo activity in F0 embryos, (2) isolating potential F0 founders producing the mutant alleles and (3) genotyping F1 fish possessing the mutant alleles (Fig. 3A). We have applied HMA to detect TALEN-mediated genome modifications at all of these steps. To investigate the developmental function of the lipid mediator sphingosine-1-phosphate (S1P) (Kawahara et al. 2009; Hisano et al. 2012), we constructed TALENs for several S1P receptors (S1PR1, S1PR5a and S1PR5b) using the pCS2-TAL3DD/RR vector system (Table S1 in Supporting Information) (Dahlem et al. 2012). Forward- and reverse-TALEN mRNAs (400 pg each) were injected into zebrafish embryos at the 1- to 2-cell stage, and genomic DNA was prepared from these embryos at 1 day postfertilization (dpf). Most of the indel mutations occur in somatic cells of F0 embryos during embryogenesis. PCR amplicons for the TALEN target site were analyzed with HMA. We found that the intensity of homoduplexes in the indicated TALEN-injected embryos at the expected molecular sizes was weaker than those in uninjected embryos (Fig. 3B). Additionally, multiple bands of slow migration were detected in S1PR1- and S1PR5a-TALEN-injected embryos, whereas only a few such bands were detected in S1PR5b-TALEN-injected embryos. To investigate the genome modifications induced by TALENs, a wide target genome region (approximately 400–600 bp) was subcloned into a pGEM-T Easy vector. Random sequencing analysis indicated that TALENs predominantly induce small indel mutations (Fig. 3D). The efficacy of S1PR1-, S1PR5a- and S1PR5b-TALEN-induced indel mutations was approximately 25%, 40% and 20%, respectively, suggesting that the degree of multiple HMA profiles is correlated well with the TALEN activity for endogenous loci. These results showed that HMA is a very simple assay for estimating TALEN in vivo activity in F0 zebrafish embryos.
Figure 3.

Application of HMA for detecting TALEN-mediated genome modifications in zebrafish. (A) TALENs were injected into zebrafish embryos, and genomic DNA was isolated from TALEN-injected embryos at 1 dpf. TALEN in vivo activity for an endogenous locus was estimated by HMA profiles of the PCR amplicons. Potential founders were mated with wild-type fish (WT). F1 fish with indel mutant alleles was identified by HMA. (B) TALEN target regions were amplified from the genomic DNA of uninjected and the TALEN-injected embryos (S1PR1, S1PR5a and S1PR5b). PCR amplicons derived from the TALEN-injected embryos provided multiple heteroduplex bands. (C) The intensity of PCR amplicons from TALEN-injected embryos was weaker than those of uninjected embryos on a 2% agarose gel. (D) PCR amplicons were subcloned into a pGEM-T Easy vector, and individual inserts were randomly sequenced. Deleted and inserted nucleotides in the DNA sequences are indicated by red dashes and red letters, respectively. The wild-type sequence is shown at the top. The number of nucleotides deleted (−) and inserted (+) is indicated to the right with the detection number. Blue: TALEN target sequences. Green: spacer sequences. (HMA, heteroduplex mobility assay; TALEN, transcription activator-like effector nucleases).
Identification of potential F0 founders by HMA
We also examined whether HMA is suitable for identifying potential founders responsible for the mutant alleles. TALEN-injected F0 embryos raised to adulthood were mated with wild-type fish. Genomic DNA from individual F1 embryos was isolated at 1 dpf. PCR amplicons from F1 embryos derived from an S1PR5a-TALEN-injected F0 founder exhibited four unique migration patterns of heteroduplexes (Fig. 4A). The two slow-migrating bands were confirmed as heteroduplexes, because the bands (in d) were selectively cleaved by T7 endonuclease (Fig. S3 in Supporting Information). Sequencing analysis of plasmid DNAs containing individual four bands confirmed that the two migrated bands were composed of the wild-type homoduplex (upper) and the deletion mutant homoduplex (lower) and that the two slower bands corresponded to the heteroduplex of the wild-type and deletion mutant (Fig. S4 in Supporting Information). We then amplified the target genomic region from all F1 embryos by PCR, and the resulting fragments were subcloned into a pGEM-T Easy vector. DNA sequencing determined that the F1 embryos a, m and o had a 2-bp insertion plus 6-bp deletion mutant allele and a wild-type allele; F1 embryos b and f contained a 5-bp deletion mutant allele and a wild-type allele; F1 embryos d and i contained a 6-bp deletion allele and a wild-type allele; and the F1 embryo l had an 11-bp deletion allele and a wild-type allele (Fig. 4C). We did not observe indel mutations in F1 embryos c, e, g, h, j, k, n and p (Fig. 4C). Thus, the same HMA profile exhibits identical indel mutations. Furthermore, we could estimate the germ-line transmission rate from the HMA pattern as 50% (8/16) in the S1PR5a-injected F0 founder. Similar results were obtained from an S1PR1-TALEN-injected F0 founder (Fig. S5 in Supporting Information). In the case of S1PR5b-TALEN, we observed similar HMA profiles for the PCR amplicons (a and d in Fig. 4D) and confirmed that these F1 embryos possessed identical indel mutations (Fig. 4F). These differences were not observed when using a 2% agarose gel (Fig. 4B,E). The germ-line transmission rates of the S1PR5b-TALEN-derived F0 founders were 13% and 6%, which are lower than those of either the S1PR5a-TALEN founders (50% and 31%) (Fig. 4C and Fig. S6 in Supporting Information) or the S1PR1-TALEN founders (38% and 44%) (Fig. S5 in Supporting Information). Moreover, the estimated frequencies of the germ-line transmission in the TALEN-injected F0 founders were correlated with the degree of multiple HMA profiles in TALEN-derived F0 embryos. Therefore, HMA can identify potential F0 founders.
Figure 4.
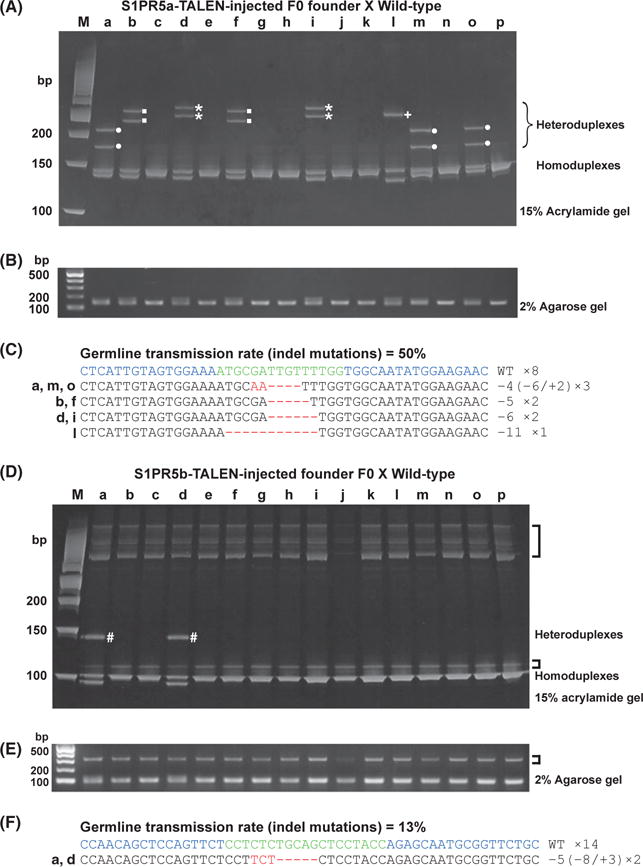
Detection of S1PR5a and S1PR5b mutant alleles by HMA in F1 embryos. (A–C) S1PR5a mutant alleles. (D–F) S1PR5b mutant alleles. (A, D) PCR amplicons from individual F1 genomic DNAs were electrophoresed on a 15% polyacrylamide gel. For S1PR5a, four distinct HMA profiles were detected (●, ■, ⋆, +); for S1PR5b, only one unique HMA profile was detected (#). (B, E) Band patterns of PCR fragments from S1PR5a (B) and S1PR5b (E) target regions were similar in all lanes of a 2% agarose gel. Nonspecific bands amplified by S1PR5b primer sets were observed in all samples (brackets). (C, F) PCR amplicons were subcloned into a pGEM-T Easy vector, and inserts from individual F1 embryos were sequenced. Deleted and inserted nucleotides in the DNA sequences are indicated by red dashes and red letters, respectively. The wild-type sequence is shown at the top. The number of nucleotides deleted (−) and inserted (+) is indicated to the right with the detection number. The germ-line transmission rate is indicated at the top. Blue: TALEN target sequences. Green: spacer sequences. (HMA, heteroduplex mobility assay; TALEN, transcription activator-like effector nucleases).
Genotyping of F1 fish containing mutant alleles by HMA
Finally, we examined whether HMA can genotype adult F1 fish. Genomic DNA was prepared from the fin clips of individual F1 fish, and PCR amplicons for the TALEN target site were analyzed by HMA on a 15% polyacrylamide gel. We observed five unique HMA profiles in F1 fish derived from the S1PR5a-TALEN-injected F0 founder (Fig. 5A). The HMA profiles in F1 fish d, g, h and k (Fig. 5A) are very similar to some HMA profiles in F1 embryos (Fig. 4A). Sequencing analysis showed that the similar HMA profiles contained identical indel mutations, suggesting that similar HMA profiles can be used to estimate the indel mutation before precise DNA sequencing analysis. We also found additional novel HMA profiles (a and j in Fig. 5A). These F1 fish contain new mutant alleles that were confirmed by DNA sequencing (Fig. 5A,B). From these results, we propose that HMA is a rapid and powerful method for genotyping mutant fish.
Figure 5.
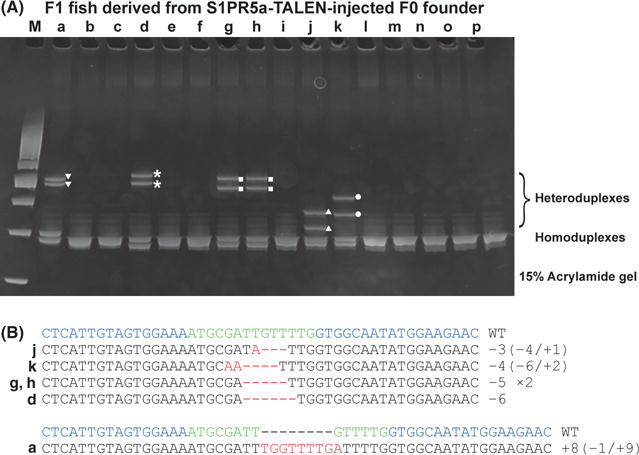
Genotyping of S1PR5a mutant alleles by HMA from fin clips of F1 fish. (A) Genomic DNA was prepared from the fin clips of individual F1 fish. PCR amplicons from individual F1 genomic DNA were electrophoresed on a 15% polyacrylamide gel. Five HMA profiles were detected (▼,⋆, ■, ▲ and ●). (B) Deleted and inserted nucleotides of DNA sequences are indicated by red dashes and red letters, respectively. The wild-type sequence is shown at the top. The number of nucleotides deleted (−) and inserted (+) is indicated to the right with the detection number. Blue: TALEN target sequences. Green: spacer sequences. (HMA, heteroduplex mobility assay; TALEN, transcription activator-like effector nucleases).
Discussion
Transcription activator-like effector nucleases can achieve targeted gene disruption in various types of model organisms including zebrafish (Perez-Pinera et al. 2012). To detect TALEN activity for an endogenous locus, several useful methods such as the T7 endonuclease assay, high resolution melt analysis (HRMA) and a modified lacZ assay were developed (Mussolino et al. 2011; Dahlem et al. 2012; Hisano et al. 2013). Because mutant alleles mediated by TALENs should be monitored at several stages in the generation of knockout fish, a more simple and economical method for screening large numbers of genomic DNA samples is desired. In this study, we demonstrated a novel use of HMA to detect the TALEN-mediated genome modifications.
We found that HMA is a rapid and reproducible method that can detect heteroduplexes derived from PCR amplicons for a TALEN target site. Heteroduplex structures exhibited significantly reduced electrophoretic mobility compared to that of homoduplexes (Figs 1B and 2A): that finding was confirmed by the direct sequencing of four bands in the HMA (Fig. S2 in Supporting Information). This retardation could be explained by the single-stranded DNA found in the heteroduplexes. Because the migration of PCR amplicons in polyacrylamide gels is dependent on the molecular size and configuration, the electrophoresis of heteroduplex structures offers significant sensitivity for the detection of TALEN-induced indel mutations.
Injection of forward- and reverse-TALEN mRNAs into zebrafish embryos resulted in various indel mutations during early embryogenesis. We were able to visualize the different indel mutations as multiple HMA profiles. HMA was a reliable analytical method for the estimation of TALEN activity at an endogenous locus based on the degree of multiple HMA profiles correlating well with TALEN in vivo activity (Fig. 3B,D). Quantification of variation was not easily detected by the T7 endonuclease assay and HRMA. We also applied the HMA to identify potential F0 founders that produced useful mutant alleles. When such potential founders were mated with wild-type fish, very unique HMA profiles from the PCR-amplified genomic region of the F1 embryos were observed. Additionally, we were able to estimate the transmission rates from the different HMA patterns. Thus, HMA can be used for rapid screening of a large number of F0 founders containing potential mutant alleles. We did not need any special equipment to perform HMA, and HMA is very economical compared with other methods. Finally, after genomic DNA was prepared from the fin clips of F1 fish, we conducted HMA to genotype individual F1 fish finding several HMA patterns. Some of the HMA profiles of the F1 fish were very similar to those of F1 embryos. In these cases, DNA sequencing confirmed identical indel mutations, suggesting that HMA is very suitable for genotyping. During the preparation of this manuscript, Chen et al. have reported that HMA is acceptable for the detection of the ZFN-mediated genome modification (Chen et al. 2012). Both their and our results show that HMA is useful for the detection of small genome alterations in various model organisms. Moreover, we show that HMA has several advantages when establishing knockout fish by TALEN: (i) the evaluation of TALEN activity for an endogenous target in TALEN-injected F0 embryos, (ii) the detection and estimation of germ-line transmission of TALEN-mediated indel mutations in F1 embryos; and (iii) the genotyping of TALEN-mediated mutations from the fin clips of adult fish. Our characterization of HMA in genome editing can be applied to the identification of designer nuclease-induced genome modifications not only in targeted gene disruptions of various model organisms but also in regenerative medicine using stem cell therapy.
Experimental procedures
Construction of pBluescript-derived constructs and TALEN plasmids
To construct pBluescript deletion series plasmids, complementary oligonucleotides with deletions of 1–10 bp were designed between the EcoRI site and XbaI sites of pBluescript II (Table S1 in Supporting Information). To begin, 15 μL of individual sense and antisense oligonucleotides (1 μg/μL) was mixed and denatured at 98 °C for 3 min. Then, the samples were incubated at 65 °C for 10 min and cooled down to room temperature. The annealed oligonucleotides were ligated into the EcoRI/XbaI site of pBluescript II.
Transcription activator-like effector nucleases plasmids were constructed as previously described with some modifications (Cermak et al. 2011). Six TAL effector repeat modules were assembled into pFUS vectors (intermediate array vectors) (Sakuma et al. 2013). The intermediate arrays and last repeat were then assembled into pCS2TAL3DD to generate a forward TALEN or into pCS2TAL3RR to generate a reverse TALEN (Dahlem et al. 2012).
Injection of TALEN mRNA
Transcription activator-like effector nucleases mRNAs were synthesized from pCS2-derived TALEN constructs (Table S1 in Supporting Information), which include appropriate TAL effector repeats by SP6 RNA polymerase using the mMESSAGE mMACHINE SP6 kit (Ambion) according to the manufacturer’s protocol. Forward- and reverse-TALEN mRNAs (400 pg each) were injected together into the blastomeres at the 1–2 cell stage of zebrafish embryos.
Preparation of genomic DNA
To detect TALEN activity at an endogenous locus, genomic DNA was isolated from TALEN-injected embryos at 1 day postfertilization (dpf) using the Gentra Puregene Tissue kit (Qiagen) according to the manufacturer’s protocol. To genotype F1 growing fish, their tail fins were amputated and genomic DNA was isolated also using the Gentra Puregene Tissue kit. To identify founder fish, potential founder zebra-fish were out-crossed with wild-type zebrafish, and individual embryos (1 dpf stage) were incubated in 50 μL of DNA extraction buffer (10 mM Tris-HCl [pH 7.5], 10 mM EDTA and 200 μg/mL proteinase K), for 3 h to overnight at 55 °C, and heated for 3 min at 95 °C to inactivate the proteinase K.
Heteroduplex mobility assay
T3 and T7 primers were used for HMA of the pBluescript-derived construct (Table S1 in Supporting Information). To detect TALEN-induced mutations by HMA, a short fragment (70–150 bp) that included the target site was amplified from genomic DNA. The primers used are listed in Table S1 in Supporting Information. Three-step PCR was carried out: 30 cycles of 98 °C for 10 s, 55 °C for 30 s and 72 °C for 30 s. Two-step PCR was also carried out for HMA: 94 °C for 2 min, and 30 cycles of 98 °C for 10 s and 68 °C for 30 s. PCR amplicons were electrophoresed on either 15% polyacrylamide gels or 2% agarose gels. After HMA, the target site was amplified from the genomic DNA of TALEN mRNA–-injected embryos or out-crossed F1 embryos using different primer pairs (the amplicon was 400–600 bp). Subsequently, the PCR fragment was cloned into a pGEM-T Easy vector (Promega). After plasmid purification, sequence analysis was carried out using the M13 forward primer (Table S2 in Supporting Information).
T7 endonuclease assay
The MCS of pBluescript wt/wt and pBluescript wt/Δ9 was amplified by PCR using T3 and T7 primers. The S1PR5a-TALEN target region was amplified from S1PR5a WT/WT and S1PR5a WT/-6 using S1PR5a-HMA-F and S1PR5a-HMA-R primers. PCR fragments were divided into two groups, and 3U of T7 endonuclease (New England Biolabs) was added to one group and incubated at 37 °C for 15 min. The samples were subjected to electrophoresis on a 15% polyacrylamide gel and were visualized by staining with ethidium bromide.
Supplementary Material
Figure S1 The formation of a heteroduplex.
Figure S2 Sequencing of pBluescript constructs (0/D9).
Figure S3 T7 endonuclease assay of S1PR5a-TALEN-injected F1 embryos (c and d).
Figure S4 Sequencing of S1PR5a-TALEN-injected F1 embryos.
Figure S5 Detection of S1PR1 mutant alleles by HMA in F1 embryos.
Figure S6 Detection of S1PR5a and S1PR5b mutant alleles by HMA in F1 embryos.
Table S1 Amino acid sequences of TALENs used in this study
Table S2 Primers used in this study
Acknowledgments
The authors thank R. Fukuoka, M. Komeno, M. Hayashi and S. Ohara for zebrafish maintenance; T. Sakuma and T. Yamamoto for pFUS-A2A, pFUS-A2B; D.F. Voytas for the Golden Gate TALEN Kit; and P. Karagiannis for valuable comments. This work was supported in part by the Funding Program for Next Generation World-Leading Researchers (NEXT Program) and by the Japan Society for the Promotion of Science.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web site:
References
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang X, Wang T, Li Z, Guan G, Hong Y. Efficient detection, quantification and enrichment of subtle allelic alterations. DNA Res. 2012;19:423–433. doi: 10.1093/dnares/dss023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano Y, Nishi T, Kawahara A. The functional roles of S1P in immunity. J Biochem. 2012;152:305–311. doi: 10.1093/jb/mvs090. [DOI] [PubMed] [Google Scholar]
- Hisano Y, Ota S, Arakawa K, Muraki M, Kono N, Oshita K, Sakuma T, Tomita M, Yamamoto T, Okada Y, Kawahara A. Quantitative assay for TALEN activity at endogenous genomic loci. Biology Open. 2013 doi: 10.1242/bio.20133871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara A, Nishi T, Hisano Y, Fukui H, Yamaguchi A, Mochizuki N. The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science. 2009;323:524–527. doi: 10.1126/science.1167449. [DOI] [PubMed] [Google Scholar]
- Kostrikis LG, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho DD. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Gersbach CA. Advances in targeted genome editing. Curr Opin Chem Biol. 2012;16:268–277. doi: 10.1016/j.cbpa.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Hosoi S, Woltjen K, Suzuki K, Kashiwagi K, Wada H, Ochiai H, Miyamoto T, Kawai N, Sasakura Y, Matsuura S, Okada Y, Kawahara A, Hayashi S, Yamamoto T. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells. 2013;18:315–326. doi: 10.1111/gtc.12037. [DOI] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Zou S. A practical approach to genetic screening for influenza virus variants. J Clin Microbiol. 1997;35:2623–2627. doi: 10.1128/jcm.35.10.2623-2627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The formation of a heteroduplex.
Figure S2 Sequencing of pBluescript constructs (0/D9).
Figure S3 T7 endonuclease assay of S1PR5a-TALEN-injected F1 embryos (c and d).
Figure S4 Sequencing of S1PR5a-TALEN-injected F1 embryos.
Figure S5 Detection of S1PR1 mutant alleles by HMA in F1 embryos.
Figure S6 Detection of S1PR5a and S1PR5b mutant alleles by HMA in F1 embryos.
Table S1 Amino acid sequences of TALENs used in this study
Table S2 Primers used in this study


