Abstract
Copy number variations associate with different developmental phenotypes and represent a major cause of congenital anomalies of the kidney and urinary tract (CAKUT). Because rare pathogenic copy number variations are often large and contain multiple genes, identification of the underlying genetic drivers has proven to be difficult. Here we studied the role of rare copy number variations in 80 patients from the KIMONO-study cohort for which pathogenic mutations in three genes commonly implicated in CAKUT were excluded. In total, 13 known or novel genomic imbalances in 11 of 80 patients were absent or extremely rare in 23,362 population controls. To identify the most likely genetic drivers for the CAKUT phenotype underlying these rare copy number variations, we used a systematic in silico approach based on frequency in a large dataset of controls, annotation with publicly available databases for developmental diseases, tolerance and haploinsufficiency scores, and gene expression profile in the developing kidney and urinary tract. Five novel candidate genes for CAKUT were identified that showed specific expression in the human and mouse developing urinary tract. Among these genes, DLG1 and KIF12 are likely novel susceptibility genes for CAKUT in humans. Thus, there is a significant role of genomic imbalance in the determination of kidney developmental phenotypes. Additionally, we defined a systematic strategy to identify genetic drivers underlying rare copy number variations.
Keywords: Child, Congenital anomalies of the kidney and urinary tract, Copy number variation, Genetics, Renal hypodysplasia
INTRODUCTION
Congenital anomalies of the kidney and urinary tract (CAKUT) are the leading cause of end-stage renal disease in childhood.1 A solitary functioning kidney represents a frequent phenotype among the spectrum of CAKUT, and may profoundly impact long-term clinical outcome.2,3 Phenotypes underlying a solitary functioning kidney of congenital origin include conditions causing the primary absence of normally developed unilateral renal parenchyma,4 such as unilateral renal agenesis (URA), multicystic dysplastic kidney (MCDK), and renal hypodysplasia (RHD), or conditions leading to recurrent infections or severe hydronephrosis that require nephrectomy in childhood, such as vesicoureteral reflux (VUR), ureteropelvic junction obstruction (UPJO) and congenital megaureter.5
Genetic factors play a major role in the pathogenesis of CAKUT in mammals.6–9 Familial aggregation is identified in approximately 14% of cases,10 with different modes of inheritance.11–14 Both point mutations and structural variants are implicated in disease determination.7,15,16 Approximately 6–20% of isolated CAKUT-phenotypes underlying a solitary functioning kidney are caused by single-gene defects12,17–19 with pathogenic mutations in HNF1B (MIM189907),20 PAX2 (MIM167409),21 and DSTYK (MIM612666)22 among the most frequently implicated. Copy number variations (CNVs), defined as gain or loss of germ line DNA of the size ranging from 1 kilobase (Kb) to several megabases (Mb),23 have been associated with multiple human phenotypes, including neurodevelopmental diseases (intellectual disability, autism, schizophrenia, epilepsy), cardiac defects, lung disease, craniofacial malformations, and others.24–28 We recently demonstrated that rare genic CNVs represent a major molecular determinant of kidney malformations, accounting for up to 17% of patients with RHD.15 The identification of rare recurrent gene-disrupting deletions or duplications can establish a genetic diagnosis, therefore improving patient care and genetic counseling. Nevertheless, given that CNVs usually affect the dosage of multiple genes at the same time, the identification of the major genetic drivers underlying such events has proven to be difficult.29
In this study, we investigated the role of rare CNVs in patients with a solitary functioning kidney from the KIMONO (KIdney of MONofunctional Origin)-study. Here, patients carrying pathogenic point mutations in HNF1B, PAX2, and DSTYK were excluded before analysis of structural variants. CNV detection was performed using high-density single nucleotide polymorphisms microarrays in 80 patients and compared to over 23,000 population controls. To identify the most likely drivers for the CAKUT phenotype in carriers of pathogenic CNVs, we performed a systematic in silico approach using bioinformatic resources and expression profiling in the developing human and mouse kidney. By using this approach, we identified five high-priority genetic drivers and we propose DLG1 and KIF12 as novel candidate susceptibility genes for human kidney and urinary tract malformations. This study presents an analytical pipeline to help interpret the functional consequence of CNVs and provides a list of novel candidate CAKUT susceptibility genes for follow-up validation and functional studies.
RESULTS
Patient characteristics
The KIMONO-GENE cohort included 80 patients (Supplementary Table 1), of whom 54 (68%) patients were males. The vast majority of subjects were White. Primary kidney parenchyma defects (URA, MCDK, and RHD) were present in 66 (83%) patients. Thirty-seven (46%) patients had a solitary functioning kidney with additional CAKUT such as VUR, UPJO or a megaureter. A non-isolated CAKUT phenotype was identified in 26 (30%) patients, in whom neurocognitive (n=7, 9%), cardiac (n=4, 5%), and gastrointestinal defects (n=4, 5%) were most frequently noted. Additional clinical parameters are presented in Supplementary Table 2. DNA of family members was not available for genetic analysis.
Exclusion of mutations in HNF1B, PAX2, and DSTYK
Prior to microarray analysis for CNVs, individuals were screened for point mutations in the coding regions of HNF1B, PAX2 and DSTYK by Sanger sequencing.18,22 No pathogenic mutations were identified. An overview of all single nucleotide variants identified in this cohort is presented in Supplementary Table 3.
Genomic disorders are frequent in the KIMONO-GENE cohort
CNV analysis in the 80 patients from the KIMONO-GENE cohort identified 118 large CNVs (defined as CNV size >100 Kb; 1.48 CNVs per patient; Supplementary Table 4). Median large CNV-size was 174,888 bp [IQR 136,364–263,382 bp]. The majority of these large CNVs were duplications (n=68, 58% versus deletions: n=50, 42%). Annotation of large (≥100 Kb) rare (<1:1,000) CNVs identified 5/80 patients (6%), compared to 0.5% of our 23,362 controls (OR = 16.1 [95% CI 6.9 – 37.8]; Fisher’s exact test: P = 4.0 × 10−6).30 These patients carried 6 genomic imbalances with significant overlap with known genomic disorders (Table 1). As expected, a substantial proportion (50%) of these patients had a non-isolated CAKUT phenotype. Among the pathogenic CNVs, one patient harbored the 3q29 microdeletion syndrome that has been associated with CAKUT in animal studies,31,32 while another patient carried a 570 Kb duplication at the PKD1 locus on chromosome 16p13.3. Importantly, the 16p13.11 duplication overlaps with a duplication identified by our previous study in a different patient affected by RHD, thus representing a recurrent genomic disorder.15 Interestingly, none of the patients had a deletion affecting the HNF1B locus on chromosome 17q12.
Table 1.
Known diagnostic genomic disorders identified in the KIMONO-GENE cohort
| Chr | CNV type |
Start (Mb) |
End (Mb) |
Size (Mb) |
Syndrome | N. of genes |
KIMONO cohort (n=80) |
Controls (n=23,362) |
Sample | Pheno- type |
Family history for CAKUT |
Additio- nal CAKUT |
Non- isolated CAKUT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2q11 | del | 96.96 | 97.24 | 0.28 | 2q11.2 deletion* | 4 | 1 | 0 | KIM_1¶ | RHD | N | N | Y |
| 3q29 | del | 197.22 | 198.83 | 1.61 | 3q29 microdeletion | 34 | 1 | 0 | KIM_2 | MCDK | Y | Y | Y |
| 15q13.3 | del | 28.52 | 28.76 | 0.24 | 15q13.3 microdeletion* | 15 | 1 | 0 | KIM_1¶ | RHD | Y | N | Y |
| 16p13.3 | dup | 1.94 | 2.51 | 0.57 | 16p13.3 polycystic kidney disease* | 42 | 1 | 0 | KIM_3 | MCDK | Y | Y | N |
| 16p13.11 | dup | 15.00 | 15.16 | 0.16 | 16p13.11 duplication | 7 | 1 | 9 | KIM_4 | URA | N | Y | N |
| 16p12.2 | del | 21.75 | 22.32 | 0.57 | 16p12.1 distal deletion | 17 | 1 | 16 | KIM_5 | UPJO | N | N | N |
CNV start and end positions are based on UCSC genome build hg18. The identified CNVs showed at least 70% overlap to known genomic disorders, CNV-size >100 Kb and frequency in controls <1:1,000.
These CNVs show ≥10% overlap with a known genomic disorder.
Two rare known CNVs were identified in patient KIM_1.
CAKUT, congenital anomalies of the kidney and urinary tract; CNV, copy number variation; del, deletion; dup, duplication; MCDK, multicystic dysplastic kidney; UPJO, ureteropelvic junction obstruction; RHD, renal hypodysplasia and URA, unilateral renal agenesis.
We next restricted our search for pathogenic CNVs to very rare events (frequency <1:4,000) and identified seven independent novel rare CNVs in 7/80 patients (9%; Table 2), including three single gene deletions on chromosome 10q21.1, 13q21.32, and Xq12. The Xq12 deletion was found in a male patient, indicating a complete loss-of-function mechanism. Consistent with prior reports,15,33 2/80 patients (2.5%) were found to carry more than one large rare CNV. Both patients were affected by a non-isolated CAKUT phenotype.
Table 2.
Novel potentially pathogenic CNVs identified in the KIMONO-GENE cohort
| Chr | CNV type |
Start (Mb) |
End (Mb) |
Size (Mb) |
N. of genes |
KIMONO cohort (n=80) |
Controls (n=23,362) |
Sample | Phenotype | Family history for CAKUT |
Additio- nal CAKUT |
Non- isolated CAKUT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1q44 | del | 246.41 | 246.71 | 0.30 | 12 | 1 | 5 | KIM_6 | URA | N | N | Y |
| 3p26 | dup | 1.13 | 1.64 | 0.50 | 2 | 1 | 5 | KIM_2* | MCDK | Y | Y | Y |
| 4q25 | del | 113.14 | 113.54 | 0.40 | 6 | 1 | 0 | KIM_7 | URA | N | N | Y |
| 9q32 | dup | 115.88 | 116.21 | 0.33 | 9 | 1 | 0 | KIM_8 | Complex CAKUT | N | Y | Y |
| 10q21.1 | del | 57.04 | 57.32 | 0.28 | 1 | 1 | 5 | KIM_9 | RHD | N | N | Y |
| 13q21.32 | del | 66.10 | 66.41 | 0.32 | 1 | 1 | 1 | KIM_10 | MCDK | Y | N | N |
| Xq12 | del | 65.68 | 65.93 | 0.26 | 1 | 1 | 3 | KIM_11 | VUR | N | N | N |
CNV start and end positions are based on UCSC genome build hg18. The identified novel rare CNVs have a size >250 Kb and a frequency in controls <1:4,000.
Multiple CNVs were detected in subject KIM_2 (see Table 1).
CAKUT, congenital anomalies of the kidney and urinary tract; CNV, copy number variation; del, deletion; dup, duplication; MCDK, multicystic dysplastic kidney; RHD, renal hypodysplasia; URA, unilateral renal agenesis and VUR, vesicoureteral reflux.
Analysis of the non-coding portion of the genome identified two large intergenic CNVs that were absent in our 23,362 controls, flank additional potential candidate genes for CAKUT, and affect regions with high conservation across mammals (Supplementary Table 5, Supplementary Figure 2).
In total, we identified 13 known or novel genomic disorders in 11 patients (14%), and two unique non-coding CNVs of unknown functional significance.
Systematic in silico approach identifies potential genetic drivers for 6 CNV phenotypes
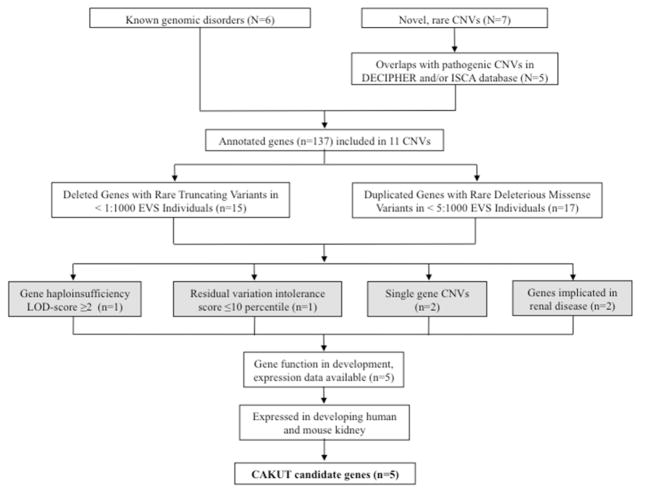
To define the candidate genetic drivers of the CNV phenotype for the 13 known or novel genomic disorders, we established a systematic in silico approach by using publicly available databases and bioinformatic resources (Figure 1, methods, and supplementary methods). The 13 identified large, rare, genic CNVs included a total of 151 genes. All genes underlying the CNVs overlapping with known genomic disorders were retained (n=119). Novel rare CNVs were annotated against the International Standards for Cytogenomic Arrays Consortium (ISCA) database and Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources Consortium (DECIPHER) to select CNVs that overlapped with variants of likely pathogenic significance (Supplementary Table 6 and Supplemental Figure 1). After alignment, we discarded the deletion on chromosome 1q44 and the duplication on chromosome 3p26 from our gene prioritization pipeline, as both CNVs showed significant overlap with benign ISCA variants (Supplementary Figure 1). In total, 137 genes were assessed for their potential pathogenic role. We cross-annotated our genes with the Exome Variant Server (EVS) and included all deleted genes that carry truncating mutations in <1:1,000 individuals and all duplicated genes that carry deleterious missense variants in <5:1,000 individuals (Supplementary Table 7 and 8, respectively). We chose these criteria to eliminate genes that harbor an excessive burden of rare deleterious variants. The resulting 32 genes were then interrogated for the haploinsufficiency logarithm of the odds (HI-LOD) score34 (only the genes underlying deletions) and the residual variation intolerance (RVI)-score35 (Supplementary Tables 9 [whole deletions] and 10–11 [prioritized genes], respectively). We defined a HI-LOD score ≥2 or the 10th percentile of the calculated RVI-score as threshold values for genes that are more likely to result in a phenotype when mutated. Using these criteria we identified two genes. We next included all single-gene CNVs in our systematic approach, as these variants may directly point to the genetic defect (n=2). Finally, we included all genes that are implicated in renal disease and genes for which mutations in murine orthologs lead to abnormal kidney and urinary tract development (n=2, from which 1 overlapped [DLG1] with our previous criteria). The resulting list included five high-priority candidate genes: DLG1 (MIM601014), EDA2R (MIM300276), KIF12 (MIM611278), PCDH9 (MIM605514), and TRAF7 (MIM606692) (Table 3). Consistent with the results from our filtering pipeline, truncating variants are extremely rare in these genes according to the EVS database (Supplementary Table 12), suggesting that deleterious mutations have been eliminated by purifying selection.
Figure 1. From CNVs to candidate genes for CAKUT.
All identified CNVs (N) were included in the analysis. For all novel, rare CNVs, deletions and duplications that showed significant overlap to pathogenic or uncertain pathogenic CNVs in public databases were included (see Supplementary Figure 1 and Supplementary Table 6). After annotation of gene content (n), genes that displayed rare truncating variants (deletions) and rare missense variants (duplications) in the Exome Variant Sever Database (http://evs.gs.washington.edu/EVS) were selected. We then assessed haploinsufficiency (HI-)LOD-scores and residual variation intolerance (RVI) scores for the prioritized genes (threshold values: HI LOD ≥2 and/or RVI-score <10th percentile) and included the prioritized genes within single gene CNVs as well as those genes that are implicated in renal disease. One gene met >1 threshold value for inclusion (DLG1). Gene expression profiles in the developing mouse kidney for all high-priority genes were evaluated by using GUDMAP (http://www.gudmap.org) and Genepaint (http://www.genepaint.org/) databases. Finally, we performed immunofluorescence studies in an E14.5 mouse kidney. By using this systematic bioinformatic approach, we prioritized 5 candidate genes for CAKUT.
CAKUT, congenital anomalies of the kidney and urinary tract; CNV, copy number variation and LOD, logarithm of the odds. Web-resources: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources Consortium (DECIPHER; http://decipher.sanger.ac.uk/); International Standards For Cytogenomic Arrays Consortium (ISCA; https://www.iscaconsortium.org/).
Table 3.
Identified candidate genes for CAKUT by using an in silico systematic approach
| Gene | MIM | Corresponding CNV | HI-LOD score* | RVI-score** | RVI-percentile** | Genes included in CNV | Implicated in renal disease | Expression data in developing mouse kidney |
|---|---|---|---|---|---|---|---|---|
| DLG1 | *601014 | 3q29 microdeletion | 4.66 | 0.40 | 76.41 | 34 | CAKUT (mice) | Yes |
| EDA2R | *300276 | Xq12 deletion | −3.91 | 0.48 | 79.25 | 1 | - | Yes |
| KIF12 | *611278 | 9q32 duplication | NA | −0.13 | 43.98 | 9 | Polycystic kidney disease (mice) | Yes |
| PCDH9 | *603581 | 13q21.32 deletion | −1.09 | −0.13 | 44.09 | 1 | - | Yes |
| TRAF7 | *606692 | 16p13.3 duplication | NA | −1.51 | 3.50 | 42 | - | Yes |
Based on 34 and
based on 35.
Genes were selected on the basis of at least one of the following criteria: individual gene HI-LOD-score ≥2, RVI-score ≤10 percentile, gene included in single gene CNV and/or gene implicated in renal disease (in mice or human models). For all candidates, expression data in the developing mouse kidney was evaluated using GUDMAP (http://www.gudmap.org) and Genepaint (www.genepaint.org/).
CAKUT, congenital anomalies of the kidney and urinary tract; CNV, copy number variation; HI-LOD, haploinsufficiency logarithm of the odds; OMIM, Online Mendelian Inheritance in Man and RVI, residual variation intolerance score.
Expression profiles of candidate genes for CAKUT
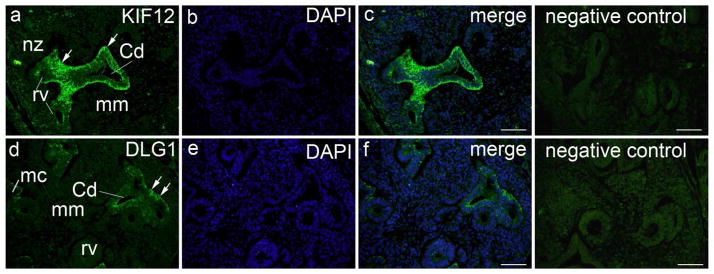
We evaluated gene expression profiles for all high-priority genes in the developing mouse kidney by using GUDMAP (https://www.gudmap.org/) and GenePaint (www.genepaint.org/) databases. According to these databases, all five genes were expressed in the developing mouse kidney and urinary tract. We subsequently performed localization studies using immunofluorescence antibody staining in the developing embryonic human and mouse kidney at the 6th gestational week and at embryonic day E14.5, respectively. As predicted from data implicating Dlg1 in urinary tract malformations in mice,31 this gene was specifically expressed in the human ureteric bud (UB) (Figure 2) and showed moderate expression in the mouse UB and metanephric mesenchyme (MM) structures (Supplementary Figure 3D). Figure 2A demonstrates strong and specific expression of KIF12 in the human UB stalk at 6th week of gestation and, to a lesser extent, in the mouse MM and developing nephrons at E14.5 (Supplementary Figure 3D). EDA2R, PCDH9, and TRAF7 showed also specific expression in human UB compartment at the 6th week of gestation (Supplementary Figure 3E, G and I) and, to a lesser extent, in the E14.5 mouse embryonic kidney (Supplementary Figure 3F, H and J).
Figure 2. Expression of KIF12 and DLG1 in the developing human kidney.
Transversal section through lumbosacral part of human embryo (6th week of development): a) within the nephrogenic zone (nz), KIF12 is weakly expressed in the developing nephron (renal vesicle – rv) and negative in the metanephric mesenchyme (mm). KIF12 is strongly expressed (arrows) in the UB stalk and UB-derived structures, such as the epithelium of collecting ducts (Cd), while the surrounding mesenchyme is negative; b) DAPI nuclear staining; c) merge of a and b; negative isotype control. d) DLG1 is weakly or not expressed in the developing nephron (renal vesicle – rv, metanephric cup - mc) and negative in the metanephric mesenchyme (mm), while it is moderately expressed (arrows) in the epithelium of collecting ducts (Cd); e) DAPI nuclear staining; f) merge of d and f; negative isotype control. Immunostaining of Kif12 and Dlg1, magnification ×40, scale bar 25Xm.
DISCUSSION
CAKUT has an overall prevalence of about 1%36,37 and accounts for 40–50% of pediatric cases of chronic kidney disease.38,39 Establishing an early molecular diagnosis in such patients may therefore significantly affect therapeutic trajectories, risk stratification for complications that develop later in life, and clinical outcome. In this study we identified rare known or new genomic imbalances in 14% of patients affected by solitary kidney but without point mutations in common CAKUT susceptibility genes. Although our study cohort includes patients with different developmental causes of solitary kidney, our results, together with recent data published by our group and others15,40 highlights that, overall, human kidney and urinary tract development is particularly sensitive to gene dosage.
Our understanding on the role of structural variations in the determination of both Mendelian and complex diseases has tremendously improved in the recent decade. In fact, genomic disorders have been implicated in a wide variety of developmental phenotypes such as schizophrenia, autism, epilepsy, intellectual disability, cardiac malformations, craniofacial malformations, CAKUT, and others.15,24–28 While these studies are fundamental in helping to explain a large fraction of the heritability for these traits, in formulating accurate diagnoses, in improving counseling, and in personalizing therapeutic strategies, CNVs offer major hurdles in their functional interpretation and in the identification of the underlying molecular defect that drives the phenotype(s) observed in patients. When a CNV harbors multiple genes, different simplified scenarios can present: a) only one gene is the major driver for the phenotype, with the others having no or negligible effect; b) one gene is the main driver for the phenotype but the other genes act in an interactive manner to modify its penetrance and expressivity; finally, c) multiple genes may act as drivers with reciprocal interaction effects, making CNV dissection extremely difficult.29 The identification of genetic drivers for CNVs requires identification of independent mutations and functional proof of causation in animal models. When the single genetic driver model holds true, identification of independent coding point mutations and validation in genetically engineered mice can be cumbersome but relatively straightforward. However, as soon as the genetic architecture underlying the CNV becomes more complex,41 this approach is likely to become inadequate. Investigators leading this field have optimized assays to model loss- and gain-of-function mutations in a time- and cost-effective manner using zebrafish mutants,41 which also allow testing for interaction. By using this approach, KCTD13 has been identified as the major driver for the neuroanatomical phenotypes of loss or gain in copy number on chromosome 16p11.2.41 More recently, the same group was able to solve the molecular defects underlying the chromosome 8q24.3 duplication syndrome and to demonstrate interaction between SCRIB and PUFA.42
We recently reported on the potential for functional interpretation of genes underlying large pathogenic CNVs using quantitative-PCR validation, extensive literature and database searches to improve the diagnostic workup of a patient with a complex developmental phenotype comprising CAKUT, intellectual disability, limb defects, and other anomalies.15,43 Here we propose an original pipeline to facilitate identification of high-priority genetic drivers for CNVs. This approach may be useful to narrow down the list of genes for follow-up resequencing studies and functional modeling in genetically engineered animal systems. These in silico analyses and expression studies in embryonic human and mouse kidney identified five potential candidate genes for CAKUT from a total of 151 transcriptional units underlying 13 rare CNVs.
Our findings strongly support DLG1 as the main genetic driver of the renal phenotype for the 3q29 microdeletion syndrome and as a novel susceptibility gene for CAKUT, and provide the rationale for interpreting results from our analytical approach. First, the 3q29 microdeletion syndrome was not found in >23,000 population controls and it has been previously described in another individual with a horseshoe kidney.44 Second, DLG1 shows a very high haploinsufficiency LOD score (4.66), indicating that heterozygous deletions of this gene are unlikely to be tolerated. 34 Third, DLG1 is hypothesized to have a role in cellular polarity establishment, cell-cell adhesion and cellular proliferation31 and Dlg1 knock-out mice strikingly mirror the phenotypes observed in our patient, showing various abnormalities of the kidney and urinary tract, including MCDK, bilateral megaureter, duplex kidney and genital malformations,31 and craniofacial malformations including cleft palate.31,32 Consistently with these data, DLG1 was expressed in the human and mouse developing urinary tract (Figure 2 and Supplementary Figure 3). Altogether, these data suggest that our strategy can identify high-priority genetic drivers and CAKUT candidate genes from rare CNV data.
Following this approach, we found a novel duplication of chromosome 9q32 that was absent in >23,000 population controls in a patient with a congenital megabladder, RHD, and congenital VUR, which spans the KIF12 locus. KIF12 (Kinesin family member 12) plays an important role in intracellular transport and spindle formation during mitosis, and has been identified as a modifier gene for renal disease severity, defined by kidney weight, length and volume, in autosomal recessive polycystic kidney disease in mice.45 Moreover, the transcription of Kif12 in mice is regulated by Hnf1b,46 which is the most commonly implicated gene in isolated CAKUT.17,18 We showed that KIF12 is highly and specifically expressed in the UB-derived structures during human kidney development (Figure 2) and in UB- and MM- derived structures in E14.5 embryonic mouse kidneys (Supplementary Figure 3). The gain of copy number at this locus suggests increased gene dosage as a molecular mechanism, although loss-of-function cannot be excluded since the CNV proximal breakpoint is located only 16 Kb from KIF12 translation starting site. Another potentially relevant structural variant was a deletion on the chromosome Xq12 locus in found in a male patient affected by isolated congenital VUR. This deletion disrupts EDA2R (Ectodysplasia A2 receptor), which has been proposed to cause hypohydrotic ectodermal dysplasia, a syndrome phenotypically characterized by hyperthermia, deficiency of sweat glands, hyperpigmentation around the eyes, sparse hair, eyebrows and eyelashes, and hypodontia with irregularly shaped teeth.47 Although our patient did not show such a phenotype, it is notable that a similar deletion on chromosome X is reported in the ISCA database in one patient with congenital hydronephrosis and intrauterine growth retardation.
Finally, the chromosome 16p13.3 duplication, found in a girl with a prenatally involuted MCDK and an ureterocele, was absent in >23,000 population controls and contains PKD1, TRAF7 and TSC2. Loss-of-function mutations in PKD1 and TSC2 have both been implicated in renal disease (autosomal dominant polycystic kidney disease and tuberous sclerosis, respectively),48–50 representing the most obvious candidates, but surprisingly our prioritization pipeline predicted TRAF7 (Tumor Necrosis Factor Receptor Associated Family 7) as the most likely genetic driver. The function of TRAF7 is largely unknown and limited to the regulation of cell survival in meningiomas.51 As this finding appears counterintuitive to our previous understanding on the role of the 16p-locus in renal disease, additional studies will be required for a precise dissection of this CNV. The remaining high-priority genetic driver identified here, PCDH9, is a member of the protocadherin family of genes and has predominantly been described within the central nervous system, where it is associated with a poor survival in glioma patients.52
In addition to these five candidate genes, we identified two intergenic CNVs that were absent in over 23,000 population controls. These rare intergenic CNVs flank additional potential CAKUT candidate genes and possibly affect conserved regulatory elements that might be important in kidney and urinary tract development.
Our approach has several limitations and is designed to explore only the simplistic model in which the phenotype associated to a CNV is mostly driven by the effect of a single gene. Moreover, the DNA from family members in the KIMONO-Study was not available for CNV analysis, thus limiting the possibility to assess for inheritance and, therefore, to strengthen our inference of genetic causation. Finally, in this study we limited our search to deletions and duplications of 100 Kb or larger in order minimize false positives and to harmonize data from platforms with different coverage. Therefore, it is possible that a fraction of patients, unknown at the present moment, might harbor pathogenic CNVs of smaller size, below the detection threshold provided by our approach. Great promises are currently held by whole-genome sequencing, which can provide assessment of structural variants (including inversions, gene fusions, and translocations) of a wide range of sizes, from few base pairs to several megabases.
In summary, we identified known or novel genomic disorders in 14% of patients with a solitary functioning kidney of congenital origin, confirming the importance of CNV analysis in improving diagnosis, risk stratification of developmental diseases that manifest later in life, and individualization of care. By using a novel prioritization strategy based on publicly available bioinformatics tools, coupled with expression profiling in the developing human and mouse urinary tract, we identified five potential genetic drivers underlying these CNVs, and propose DLG1 and KIF12 as high-priority novel candidate genes for CAKUT. The KIMONO-study thereby proposes a first approach to identify the genetic drivers of CNVs implicated in CAKUT, in order to devise comprehensive genetic and functional follow-up studies.
CONCISE METHODS
Additional Methods are reported in the Supplementary Material.
Participants
The KIMONO-study, a large cohort study of over 400 patients with a solitary functioning kidney has been previously described.3 Individuals were enrolled from April 2012 until May 2013. The institutional review boards of the VU University Medical Center and Columbia University Medical Center approved the study protocol.
Genetic analyses
Sanger sequencing for HNF1B, PAX2, and DSTYK
Genomic DNA was purified from peripheral blood samples using standard methods. Specific primers were used to direct PCR at all exons and exon-intron boundaries of HNF1B, PAX2, and DSTYK as previously described (Supplementary Table 13).18,22
CNV analysis
Genome-wide genotyping was performed in all subjects using the Illumina OmniExpress platform (730,525 markers; Illumina Inc, San Diego CA). Genotype calls and quality controls were performed in GenomeStudio (v2011.1; Illumina Inc, San Diego CA) and PLINK.53 Single nucleotide polymorphisms with missingness greater than 5% and significant deviation from Hardy-Weinberg were removed, and all samples with a genotype call rate below 99% were excluded from structural variants analysis. The CNV calls were determined with generalized genotyping methods implemented in the PennCNV program.54 Additional quality control and validation for CNVs were performed as previously described.15,30 Briefly: samples with a logR ratio (LRR) standard deviation greater than 0.35 and with a weaviness factor greater than 0.05 were excluded; based on experimental validation by quantitative PCR on over 100 structural variants,15 only CNVs with a confidence score ≥30 (threshold used to define high-quality CNVs) were included in the analyses; finally, all resulting CNVs were manually inspected in GenomeStudio software (Genome Viewer) to eliminate possible artifacts and somatic events. Annotation to the human reference genome and case-control association were performed using original Perl scripts generated in the lab.15 We compared CNV data from cases to data derived from 23,362 anonymized adult and pediatric population controls selected from eleven cohorts after stringent quality control. These controls include more than 13,000 individuals used in our previous CNV study on kidney malformations15 plus additional ~10,000 individuals from dbGap studies [Verbitsky et al.30, and unpublished data]. All controls had been genotyped on high-density Illumina platforms (Human- Hap-550, 610-Quad, 660W, 1M, 1M-Duo and Omni1) as part of case-control or longitudinal studies of complex traits. The race/ethnicity distribution for the controls was: White (85%), Black/African American (10%), Asian (4%), and Others (1%). CNV frequencies were calculated on the basis of the entire control data set of 23,362 individuals. To minimize the different detectance rate for small structural variants between genotyping platforms with different coverage density, we restricted our analyses to CNVs ≥100 Kb. We annotated CNVs with a frequency of ≤1:1,000 in controls with significant overlap (>10% in size) with known genomic disorders and subsequently used more strict frequency filters (<1:4,000) to identify potential novel genomic disorders. Finally, we annotated all large rare intergenic CNVs that were absent in controls.
Systematic in silico analysis to identify genetic drivers for CAKUT-associated CNVs
To prioritize candidate susceptibility genes for CAKUT underlying rare CNVs, we developed an automated in silico approach that relies on publicly available bioinformatics tools (Figure 1 and Supplementary Methods).
Immunofluorescence staining
High-priority candidate genes for CAKUT from our in silico prioritization study were then tested for expression in the developing human and mouse kidney and urinary tract. Immunofluorescence staining on paraffin-embedded tissues sections was performed on embryonic human kidney (6th week of development) and C57BL/6 mouse kidney at day E14.5 mice to investigate expression levels of all candidate gene products (except PKD1 and TSC2 as their role in human renal disease is well established).49,50 A normal human embryo, without any sign of abnormality or maceration was collected after tubal pregnancy from the Department of Pathology, University Hospital of Split, Croatia. The embryonic tissue was treated as postmortem material with permission of the Ethics and Drug Committee of the Clinical Hospital of Split in accordance with the 1964 Helsinki Declaration. The following rabbit polyclonal primary antibodies were used: anti-Dlg1 (55085-1-AP, also called SAP97), anti-Kif12 (12035-1-AP), anti-Traf7 (11780-1-AP; all three provided by Proteintech Group Inc., Chicago IL, USA), anti-Eda2r (NBP1-76710) and anti-Pcdh9 (NBP1-86073) (both provided by Novus Biologicals LLC, Littleton CO, USA). As negative controls, the primary or the secondary antibody was omitted from the staining procedure and additionally irrelevant IgG (MA5-16384, Thermo Fisher Scientific) was used instead of the primary antibodies. No staining was observed in either negative control.
Statistics
Continuous variables are presented as mean (standard deviation; SD) or median [interquartile range; IQR] for variables with Gaussian and non-Gaussian distribution, respectively. For blood pressure and anthropometric data, z-scores were calculated using standardized pediatric reference values based on age and gender.55,56 Qualitative variables are shown as counts (proportion). The difference in CNV frequency between cases and controls was tested using Fisher’s Exact test and nominal P-values are reported.
Supplementary Material
Acknowledgments
We thank all patients and their family members for participating. We are grateful to Cathy L. Mendelsohn for reviewing the immunohistochemistry results. This study was supported by the American Heart Association Grant in Aid 13GRNT14680075, the NIDDK 1R01DK103184 and 1R21DK098531 grants, the Columbia University Irving Institute/Clinical Trials Office pilot grant (all to SSC), and the Joint Italian Ministry of Health and NIH Young Investigators Finalized Research (to SSC and GMG). SSC is supported by the Paul Marks Scholar Award. RW is funded by a Ter Meulen Fund stipend from the Royal Netherlands Academy for Arts and Sciences (2012/225) and a grant from Fonds NutsOhra, Zorgsubsidies, Amsterdam, The Netherlands (project-number: 1101-058). KV is supported by the American Association of University Women (AAUW) International Fellowship.
Footnotes
Disclosure
All authors declare to have no relationships with companies that may have a financial interest in the information contained in the manuscript. We declare to have no conflict of interest.
- 1000 genomes project: www.1000genomes.org/
- DECIPHER: http://decipher.sanger.ac.uk/
- Exome Variant Server: http://evs.gs.washington.edu/EVS
- International Standards For Cytogenomic Arrays Consortium (ISCA): https://www.iscaconsortium.org/
- Genepaint: www.genepaint.org/
- GenitoUrinary Development Molecular Anatomy Project (GUDMAP): http://www.gudmap.org
- National Center for Biotechnology Information - Single Nucleotide Polymorphism Database (dbSNP): http://www.ncbi.nlm.nih.gov/SNP/
- Polymorphisms Phenotyping, version 2 (Polyphen-2): http://genetics.bwh.harvard.edu/pph2
- UCSC Genome Bioinformatics: http://genome.ucsc.edu/index.html
References
- 1.Wuhl E, van Stralen KJ, Verrina E, et al. Timing and outcome of renal replacement therapy in patients with congenital malformations of the kidney and urinary tract. Clin J Am Soc Nephrol. 2013;8:67–74. doi: 10.2215/CJN.03310412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanna-Cherchi S, Ravani P, Corbani V, et al. Renal outcome in patients with congenital anomalies of the kidney and urinary tract. Kidney Int. 2009;76:528–33. doi: 10.1038/ki.2009.220. [DOI] [PubMed] [Google Scholar]
- 3.Westland R, Kurvers RA, van Wijk JA, Schreuder MF. Risk factors for renal injury in children with a solitary functioning kidney. Pediatrics. 2013;131:e478–85. doi: 10.1542/peds.2012-2088. [DOI] [PubMed] [Google Scholar]
- 4.Westland R, Schreuder MF, van Goudoever JB, Sanna-Cherchi S, van Wijk JA. Clinical Implications of the Solitary Functioning Kidney. Clin J Am Soc Nephrol. 2013 doi: 10.2215/CJN.08900813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abou Jaoude P, Dubourg L, Bacchetta J, Berthiller J, Ranchin B, Cochat P. Congenital versus acquired solitary kidney: is the difference relevant? Nephrol Dial Transplant. 2011;26:2188–94. doi: 10.1093/ndt/gfq659. [DOI] [PubMed] [Google Scholar]
- 6.Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genet. 2007;8:791–802. doi: 10.1038/nrg2205. [DOI] [PubMed] [Google Scholar]
- 7.Vivante A, Kohl S, Hwang DY, Dworschak GC, Hildebrandt F. Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr Nephrol. 2014 doi: 10.1007/s00467-013-2684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanna-Cherchi S, Caridi G, Weng PL, et al. Genetic approaches to human renal agenesis/hypoplasia and dysplasia. Pediatr Nephrol. 2007;22:1675–84. doi: 10.1007/s00467-007-0479-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vainio S, Lin Y. Coordinating early kidney development: lessons from gene targeting. Nat Rev Genet. 2002;3:533–43. doi: 10.1038/nrg842. [DOI] [PubMed] [Google Scholar]
- 10.McPherson E. Renal anomalies in families of individuals with congenital solitary kidney. Genet Med. 2007;9:298–302. doi: 10.1097/gim.0b013e3180544516. [DOI] [PubMed] [Google Scholar]
- 11.Ashraf S, Hoskins BE, Chaib H, et al. Mapping of a new locus for congenital anomalies of the kidney and urinary tract on chromosome 8q24. Nephrol Dial Transplant. 2010;25:1496–501. doi: 10.1093/ndt/gfp650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saisawat P, Kohl S, Hilger AC, et al. Whole-exome resequencing reveals recessive mutations in TRAP1 in individuals with CAKUT and VACTERL association. Kidney Int. 2013 doi: 10.1038/ki.2013.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanna-Cherchi S, Reese A, Hensle T, et al. Familial vesicoureteral reflux: testing replication of linkage in seven new multigenerational kindreds. J Am Soc Nephrol. 2005;16:1781–7. doi: 10.1681/ASN.2004121034. [DOI] [PubMed] [Google Scholar]
- 14.Weng PL, Sanna-Cherchi S, Hensle T, et al. A recessive gene for primary vesicoureteral reflux maps to chromosome 12p11–q13. J Am Soc Nephrol. 2009;20:1633–40. doi: 10.1681/ASN.2008111199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanna-Cherchi S, Kiryluk K, Burgess KE, et al. Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet. 2012;91:987–97. doi: 10.1016/j.ajhg.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mefford HC, Clauin S, Sharp AJ, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–69. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber S, Moriniere V, Knuppel T, et al. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol. 2006;17:2864–70. doi: 10.1681/ASN.2006030277. [DOI] [PubMed] [Google Scholar]
- 18.Thomas R, Sanna-Cherchi S, Warady BA, Furth SL, Kaskel FJ, Gharavi AG. HNF1B and PAX2 mutations are a common cause of renal hypodysplasia in the CKiD cohort. Pediatr Nephrol. 2011;26:897–903. doi: 10.1007/s00467-011-1826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang DY, Dworschak GC, Kohl S, et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 2014 doi: 10.1038/ki.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bingham C, Bulman MP, Ellard S, et al. Mutations in the hepatocyte nuclear factor-1beta gene are associated with familial hypoplastic glomerulocystic kidney disease. Am J Hum Genet. 2001;68:219–24. doi: 10.1086/316945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favor J, Sandulache R, Neuhauser-Klaus A, et al. The mouse Pax2(1Neu) mutation is identical to a human PAX2 mutation in a family with renal-coloboma syndrome and results in developmental defects of the brain, ear, eye, and kidney. Proc Natl Acad Sci U S A. 1996;93:13870–5. doi: 10.1073/pnas.93.24.13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanna-Cherchi S, Sampogna RV, Papeta N, et al. Mutations in DSTYK and dominant urinary tract malformations. N Engl J Med. 2013;369:621–9. doi: 10.1056/NEJMoa1214479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–76. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–71. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osoegawa K, Vessere GM, Utami KH, et al. Identification of novel candidate genes associated with cleft lip and palate using array comparative genomic hybridisation. J Med Genet. 2008;45:81–6. doi: 10.1136/jmg.2007.052191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–5. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golzio C, Katsanis N. Genetic architecture of reciprocal CNVs. Current opinion in genetics & development. 2013;23:240–8. doi: 10.1016/j.gde.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verbitsky M, Sanna-Cherchi S, Fasel DA, et al. Genomic imbalances in pediatric patients with chronic kidney disease. J Clin Invest. 2015;125:2171–8. doi: 10.1172/JCI80877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iizuka-Kogo A, Ishidao T, Akiyama T, Senda T. Abnormal development of urogenital organs in Dlgh1-deficient mice. Development. 2007;134:1799–807. doi: 10.1242/dev.02830. [DOI] [PubMed] [Google Scholar]
- 32.Ahn SY, Kim Y, Kim ST, Swat W, Miner JH. Scaffolding proteins DLG1 and CASK cooperate to maintain the nephron progenitor population during kidney development. J Am Soc Nephrol. 2013;24:1127–38. doi: 10.1681/ASN.2012111074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girirajan S, Rosenfeld JA, Coe BP, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–31. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birth Defects Monitoring Program (BDMP)/Commission on Professional and Hospital Activities (CPHA) surveillance data, 1988–1991. Teratology. 1993;48:658–75. doi: 10.1002/tera.1420480608. [DOI] [PubMed] [Google Scholar]
- 37.Metropolitan Atlanta Congenital Defects Program surveillance data, 1988–1991. Teratology. 1993;48:695–709. doi: 10.1002/tera.1420480610. [DOI] [PubMed] [Google Scholar]
- 38.Centers for Disease C, Prevention. State-specific trends in chronic kidney failure--United States, 1990–2001. MMWR Morb Mortal Wkly Rep. 2004;53:918–20. [PubMed] [Google Scholar]
- 39.Harambat J, van Stralen KJ, Kim JJ, Tizard EJ. Epidemiology of chronic kidney disease in children. Pediatr Nephrol. 2012;27:363–73. doi: 10.1007/s00467-011-1939-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caruana G, Wong MN, Walker A, et al. Copy-number variation associated with congenital anomalies of the kidney and urinary tract. Pediatr Nephrol. 2015;30:487–95. doi: 10.1007/s00467-014-2962-9. [DOI] [PubMed] [Google Scholar]
- 41.Golzio C, Willer J, Talkowski ME, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–7. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dauber A, Golzio C, Guenot C, et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am J Hum Genet. 2013;93:798–811. doi: 10.1016/j.ajhg.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Materna-Kiryluk A, Kiryluk K, Burgess KE, et al. The emerging role of genomics in the diagnosis and workup of congenital urinary tract defects: a novel deletion syndrome on chromosome 3q13.31-22.1. Pediatr Nephrol. 2014;29:257–67. doi: 10.1007/s00467-013-2625-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi E, Piccini F, Zollino M, et al. Cryptic telomeric rearrangements in subjects with mental retardation associated with dysmorphism and congenital malformations. J Med Genet. 2001;38:417–20. doi: 10.1136/jmg.38.6.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mrug M, Li R, Cui X, Schoeb TR, Churchill GA, Guay-Woodford LM. Kinesin family member 12 is a candidate polycystic kidney disease modifier in the cpk mouse. J Am Soc Nephrol. 2005;16:905–16. doi: 10.1681/ASN.2004121083. [DOI] [PubMed] [Google Scholar]
- 46.Gong Y, Ma Z, Patel V, et al. HNF-1beta regulates transcription of the PKD modifier gene Kif12. J Am Soc Nephrol. 2009;20:41–7. doi: 10.1681/ASN.2008020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wisniewski SA, Trzeciak WH. A rare heterozygous TRAF6 variant is associated with hypohidrotic ectodermal dysplasia. Br J Dermatol. 2012;166:1353–6. doi: 10.1111/j.1365-2133.2012.10871.x. [DOI] [PubMed] [Google Scholar]
- 48.Brook-Carter PT, Peral B, Ward CJ, et al. Deletion of the TSC2 and PKD1 genes associated with severe infantile polycystic kidney disease--a contiguous gene syndrome. Nat Genet. 1994;8:328–32. doi: 10.1038/ng1294-328. [DOI] [PubMed] [Google Scholar]
- 49.Fedeles SV, Tian X, Gallagher AR, et al. A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat Genet. 2011;43:639–47. doi: 10.1038/ng.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rakowski SK, Winterkorn EB, Paul E, Steele DJ, Halpern EF, Thiele EA. Renal manifestations of tuberous sclerosis complex: Incidence, prognosis, and predictive factors. Kidney Int. 2006;70:1777–82. doi: 10.1038/sj.ki.5001853. [DOI] [PubMed] [Google Scholar]
- 51.Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125:351–8. doi: 10.1007/s00401-013-1093-x. [DOI] [PubMed] [Google Scholar]
- 52.Wang C, Yu G, Liu J, et al. Downregulation of PCDH9 predicts prognosis for patients with glioma. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2012;19:541–5. doi: 10.1016/j.jocn.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National High Blood Pressure Education Program Working Group on High Blood Pressure in C Adolescents. The fourth report on the diagnosis, evaluation, and treatment of high blood pressure in children and adolescents. Pediatrics. 2004;114:555–76. [PubMed] [Google Scholar]
- 56.Schonbeck Y, Talma H, van Dommelen P, et al. Increase in prevalence of overweight in Dutch children and adolescents: a comparison of nationwide growth studies in 1980, 1997 and 2009. PloS one. 2011;6:e27608. doi: 10.1371/journal.pone.0027608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.