Abstract
We evaluated cellular metabolism profiles of HIV-1 and HIV-2 infected primary human monocyte-derived macrophages (MDMs). First, HIV-2 GL-AN displays faster production kinetics and greater amounts of virus as compared to HIV-1s: YU-2, 89.6 and JR-CSF. Second, quantitative LC–MS/MS metabolomics analysis demonstrates very similar metabolic profiles in glycolysis and TCA cycle metabolic intermediates between HIV-1 and HIV-2 infected macrophages, with a few notable exceptions. The most striking metabolic change in MDMs infected with HIV-2 relative to HIV-1-infected MDMs was the increased levels of quinolinate, a metabolite in the tryptophan catabolism pathway that has been linked to HIV/AIDS pathogenesis. Third, both HIV-1 and HIV-2 infected MDMs showed elevated levels of ribose-5-phosphate, a key metabolic component in nucleotide biosynthesis. Finally, HIV-2 infected MDMs display increased dNTP concentrations as predicted by Vpx-mediated SAMHD1 degradation. Collectively, these data show differential metabolic changes during HIV-1 and HIV-2 infection of macrophages.
Keywords: HIV-1, HIV-2, Metabolites, TCA, Glycolysis, Macrophages, LC–MS/MS
Introduction
Human Immunodeficiency Virus type 1 (HIV-1) is the predominant lentivirus infecting the global population, with most patients progressing to acquired immune deficiency syndrome (AIDS) unless treated with complete antiretroviral therapy (cART) (Kanki et al., 1997; Nyamweya et al., 2013; Rowland-Jones, 2006). HIV type 2 (HIV-2) infection also contributes to the disease burden, but is more localized to West Africa. Importantly, only 30% of the HIV-2 infected individuals progress to AIDS, indicating that HIV-2 is significantly less pathogenic than HIV-1 (De Cock et al., 2012; Gilbert et al., 2003). However, the mechanisms responsible for the discrepancy in disease progression between these two viruses remain unclear.
Cellular tropism is an important factor that differentiates HIV-1 and HIV-2 pathogenesis (Blaak et al., 2008). HIV infects both activated CD4+T cells and macrophages (Aggarwal et al., 2013; Cory et al., 2013; Cosenza et al., 2004; Koppensteiner et al., 2012; McElrath et al., 1989; Rong and Perelson, 2009). However, the ability of HIV-1 and HIV-2 to infect human primary macrophages varies (Marchant et al., 2006) and suggests that viral envelope and co-receptor usage are important determinants for their cellular tropism (Bhattacharya et al., 2003). For example, HIV-2 capsid is more sensitive human TRIM5α than HIV-1 capsid (Takeuchi et al., 2013). Host Lv2 protein has HIV-2 specific envelope restriction function (McKnight et al., 2001). Recently Lv4 was reported to be another capsid specific host restriction factor inhibiting SIVmac and HIV-2 infection (Pizzato et al., 2015). Several studies have reported that host sterile alpha motif (SAM) and histidine/aspartic acid (HD) domain protein 1 (SAMHD1) acts as a myeloid specific host anti-HIV/SIV restriction factor (Hrecka et al., 2011; Laguette et al., 2011). Moreover, the viral accessary protein, viral protein x (Vpx), which is specifically encoded by HIV-2 and many SIV strains, directs the DCAF1 E3 ubiquitin ligase to degrade SAMHD1 (Ahn et al., 2012; Hrecka et al., 2011), thereby counter-acting SAMHD1's anti-viral effect in non-dividing myeloid cells. SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase (dNTPase) (Goldstone et al., 2011; Powell et al., 2011), whose antiviral role is to deplete cellular dNTPs to kinetically suppress the viral reverse transcription step in various non-dividing cell types including macrophages, dendritic cells (Pauls et al., 2013; Planelles, 2011) and resting human primary CD4+ T cells (Baldauf et al., 2012; Hrecka et al., 2011; Laguette et al., 2011; Lahouassa Contents lists available at ScienceDirect et al., 2012; Gelais et al., 2012). The Vpx-mediated SAMHD1 depletion led to elevation of cellular dNTP levels in non-dividing cells, which in turn promoted acceleration of proviral DNA synthesis for HIV-1 infection in primary human monocyte-derived macrophages (MDMs) (Kim et al., 2012).
Our previous metabolomics study compared metabolic changes in human primary activated CD4+ T cells and PMA-treated U1 cells, a chronically infected HIV-1 cell to the U937 parental control cells (Hollenbaugh et al., 2011). We showed that U1 cells had reduced glucose uptake and glycolysis intermediates as compared to HIV-1 infected activated CD4+ T cells (Hollenbaugh et al., 2011). In this study, we compared the metabolic profiles of primary human MDMs infected with HIV-1 and HIV-2 strains. We also investigate the effect of Vpx on the metabolic changes of macrophages by employing HIV-2 GL-AN and GL-ANΔVpx strains during acute infection. The data presented in this study reports several key differential metabolic changes during HIV-1 and HIV-2 infection of macrophages.
Results
Robust replication kinetics of HIV-2 GL-AN in MDMs
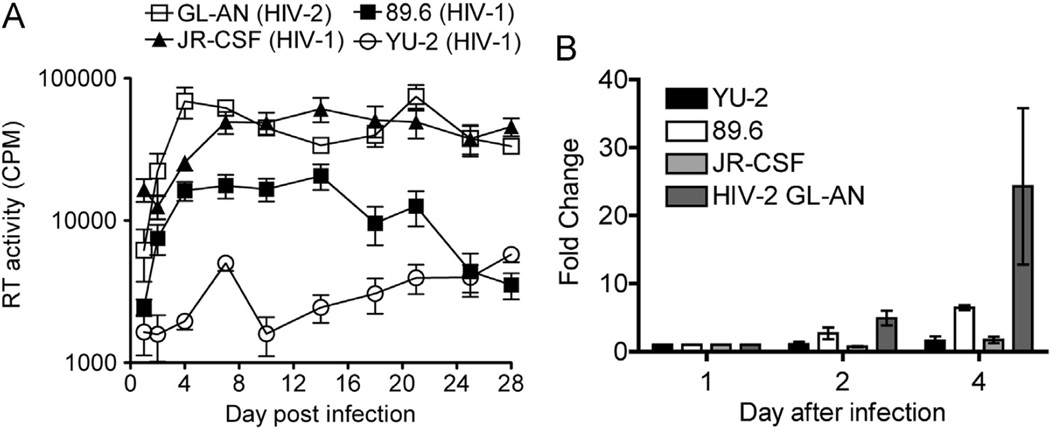
The replication and production kinetics of HIV-2 (GL-AN) and HIV-1 strains (YU-2, 89.6 and JR-CSF) were evaluated in human primary terminally-differentiated/non-dividing monocyte-derived macrophages (MDMs) from three healthy donors. In this study, infectious viruses containing both their own Env and VSV-G proteins on the virion surface were used to enhance their initial infectivity of macrophages, removing a potential entry restriction (Bhattacharya et al., 2003). The viruses were incubated with MDMs for five hours before removal and washing the cells with culture medium to remove free viruses. Cell-free sample aliquots were harvested at days 1, 2, 4, 7, 10, 14, 18, 21, 25 and 28 after infection and analyzed for HIV RT activity (Fig. 1A). HIV-2 GL-AN replicates much more rapidly (24.2-fold higher as compared to day 1 post infection) than the three HIV-1 strains: 89.6 (6.5-fold), JRCSF (1.7-fold) and YU-2 (1.6-fold) over the first 4 days of viral culture (Fig. 1B). HIV-2 GL-AN reached the maximum production rate at day 4 after infection, while the HIV-1 strains required closer to 8 days for maximum viral production in MDMs (Fig. 1A). Interestingly, HIV-1 JR-CSF displayed a higher maximal viral production, determined by RT activity, than HIV-1 YU-2 and 89.6 strains in MDMs.
Fig. 1.
HIV reverse transcription (RT) activity. (A) The time course for days 1–28 are plotted for virus generation. MDMs were infected GL-AN, JR-CSF, 89.6 and YU-2 viruses (triplicated wells). (B) Analysis of days 1–4 of the time course. Day 1 was set at 1 for all the donors and fold increases of RT-activity are plotted for each virus. Each sample was analyzed in triplicate wells for HIV RT activity to determine counts per minute (CPM) at each time point. Data was plotted as mean and S.E.M. for three independent human primary MDM donors.
Phenotyping the MDMs used for metabolomic analysis
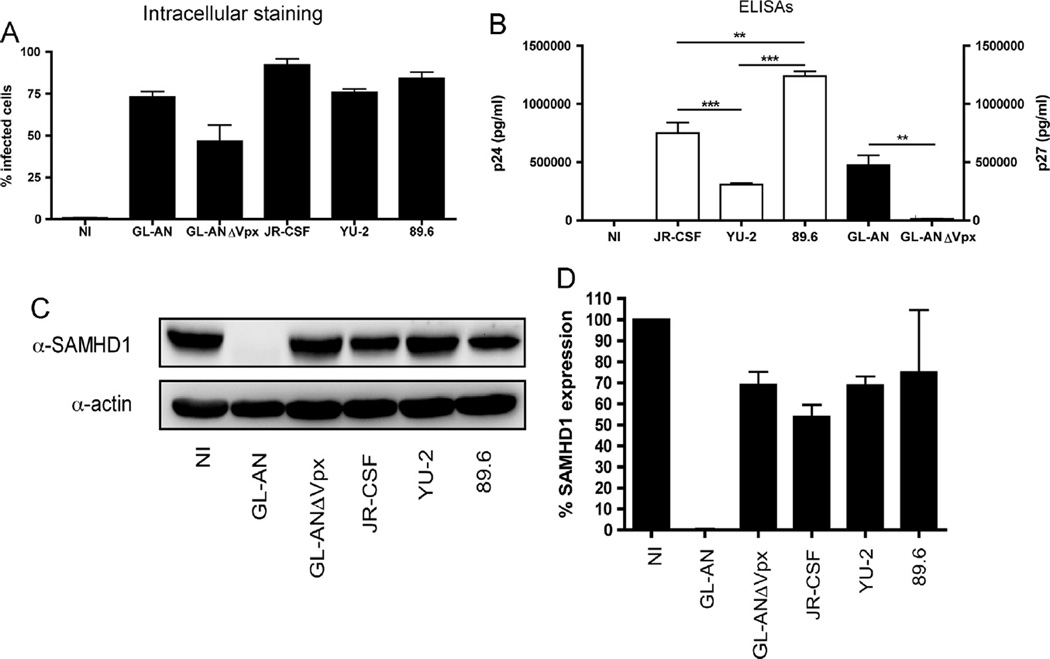
Since GL-AN had the highest viral production in MDMs, we next wanted to examine whether HIV-1 and HIV-2, with and without Vpx, differentially impacted the cellular metabolism of MDMs. Due to the issues of restricted ability to infect primary macrophages and the requirement of high biomass for HPLC–MS/ MS evaluation, we infected MDMs with the maximum amount of the viruses possible for 24 h before replacing the culture medium. Cells, cellular supernatants, cellular lysates and cellular metabolites were collected at day 4 after infection for multiple assay analyses. Formaldehyde fixed MDMs were intracellular stained using an antibody that recognizes p24/p27 (Fig. 2A) to determine the percentage of cells infected by each individual virus. Infection with the GL-AN, JR-CSF, YU-2 or the 89.6 strain resulted in 75% or greater infection as detected by FACS analysis, while GL-ANΔVpx infection resulted in 50% infectivity, based on positive FACS staining of cells from three independent donors. The non-infected (NI) control MDMs showed less than 1% positive staining for all donors, indicating a low level of background staining.
Fig. 2.
Characterization of MDMs used for metabolomic analysis. Three independent MDM donors were infected with VSV-G pseduotyped GL-AN, GL-ANÄVpx, JR-CSF, 89.6 and YU-2 viruses or not infected (NI). At day 4 after infection, MDMs and supernatant were collected for multiple analyses. (A) MDMs were intracellular stained for capsid protein and analyzed by FACS. The percentage of infected MDMs were graphed. (B) Cell-free supernatants were analyzed by ELISA for p24 antigen (HIV-1) or p27 antigen (HIV-2) levels. Significant differences (Student T test) between the different viruses are displayed. (C) Immunoblot analysis for SAMHD1 and actin protein levels was done for the different MDM samples. (D) SAMHD1 expression levels were quantitated and displayed for the three independent donors.
Next, we determined viral production by ELISA with the culture supernatant collected (Fig. 2B). For the HIV-1 viruses, infection with the 89.6 strain produced significantly higher amounts of p24 antigen relative to infection with the JR-CSF (**; p < 0.01; T test) or YU-2 strains (***; p < 0.001) (Fig. 2B). Moreover, the JR-CSF strain produced significantly higher levels (***; p < 0.001) of p24 antigen than did the YU-2 strain. We detected a significant increase (**; p < 0.01) in p27 antigen levels for GL-AN (Vpx+) as compared to GL-ANΔVpx, which exhibited a very low level of detectable p27 in the culture medium.
SAMHD1 is a known host cellular restriction factor for retroviruses (Baldauf et al., 2012; Berger et al., 2011a; Hrecka et al., 2011; Laguette et al., 2011; Lahouassa et al., 2012; Gelais et al., 2012; White et al., 2012). Our previous studies showed no augmentation of SAMHD1 protein levels in MDMs transduced by single-round D3HIV-1 vector or treatment with virus-like particles not containing Vpx (Hollenbaugh et al., 2014; Kim et al., 2012). Immunoblot analysis (Fig. 2C) for SAMHD1 validated complete SAMHD1 degradation (below the level of detection) during GL-AN infection (Fig. 2C). SAMHD1 level remained high in MDMs infected with all of the replicative competent HIV-1 strains and GL-ANΔVpx, suggesting that infection with replicative competent viruses do not promote SAMHD1 degradation by day four after infection (Figs. 2C and D).
Evaluation of glycolysis, TCA cycle and UDP-sugars metabolic intermediates
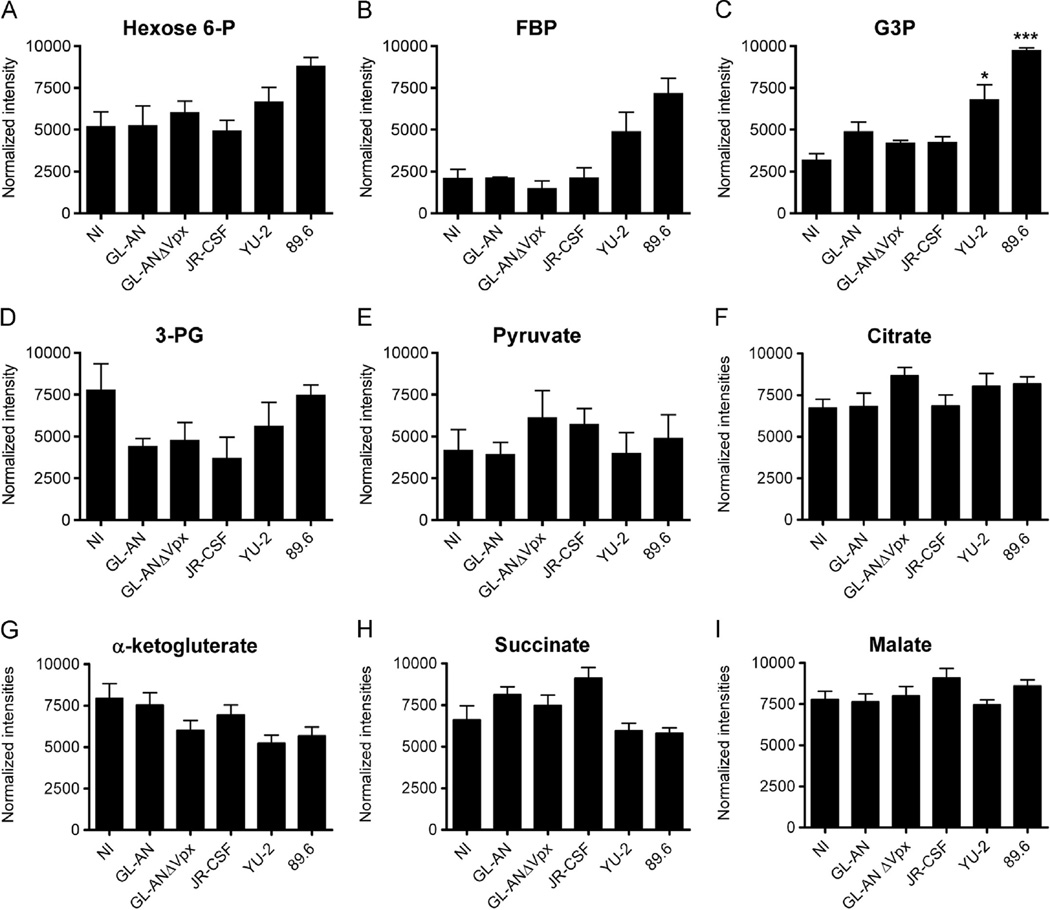
We previously used liquid chromatograph tandem mass spectrum (LC–MS/MS) analysis to evaluate metabolic intermediates in HIV-1 infected human primary activated CD4+ T cells as well as myeloid cell lines: U937 and U1 (Hollenbaugh et al., 2011). In this study, cellular metabolites were isolated from non-infected (NI) or HIV-infected human primary MDMs infected after 4 days, and analyzed using LC–MS/MS. Selected normalized cellular metabolites intensities are plotted for glycolysis and the TCA cycle intermediates in Fig. 3. For glycolysis, the levels of hexose 6-phosphate (Fig. 3A) or fructose 1,6-bisphosphate (FBP: Fig. 3B) did not appreciably change, although we did observe a trend of higher FBP levels for YU-2 and 89.6 infected MDMs. For glyceraldehyde 3-phosphate (G3P), levels were significantly higher relative to the NI group for YU-2 (*, p < 0.05) and 89.6 (***, p < 0.001), whereas GL-AN, GL-ANΔVpx and JR-CSF infected MDMs were not significantly different (Fig. 2C). No significant differences were observed for 3-phosphoglycerate (3-PG; Fig. 3D) or pyruvate (Fig. 3E). Similarly, there was little change in pool sizes for TCA cycle intermediates, including citrate (Fig. 3F), α-ketoglutarate (Fig. 3G), succinate (Fig. 3H) or malate (Fig. 3I), and were not significantly different relative to NI MDMs. Collectively, these data indicate that substantial shifts in glycolytic or TCA cycle pool sizes do not occur at day 4 after infection of MDMs.
Fig. 3.
Evaluation of cellular intermediates. HPLC–MS/MS analysis was used to determine changes in glycolysis intermediates for (A) hexose 6-P, (B) fructose 1,6 bisphosphate (FBP), (C) glyceraldehyde 3-phosphate (G3P), (D) 3-phosphoglycerate (3-PG), and (E) pyruvate. Significant increases are denoted by * (p < 0.05) and *** (p < 0.001). TCA cycle intermediates for (F) citrate, (G) á-ketoglutarate, (H) succinate and (I) malate are plotted. Normalized intensities are graphed as means and S.E.M. for three independent donors.
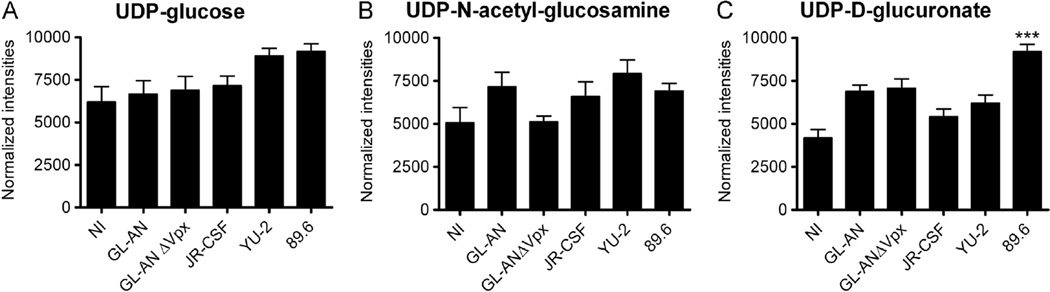
The HIV envelope protein is highly glycosylated (Raska et al., 2010). Therefore, we analyzed UDP-sugars pools (Fig. 4), which are both used for gluconeogenesis and to glycosylate proteins. No significant changes were detected for UPD-glucose (Fig. 4A) or UDP-N-acetyl-glucosamine (Fig. 4B). In contrast, UDP-Dglucuronate pool size in 89.6 infected MDMs were significantly (***, p < 0.001) increased relative to NI MDMs (Fig. 4C).
Fig. 4.
Evaluation of UDP-sugars. We monitored changes in (A) UDP-glucose, (B) UDP-N-acetyl-glucosamine and (C) UDP-D-glucuronate. Data are graphed as means and S.E. M. are plotted for three independent donors. Significant increase is denoted by *** (p < 0.001).
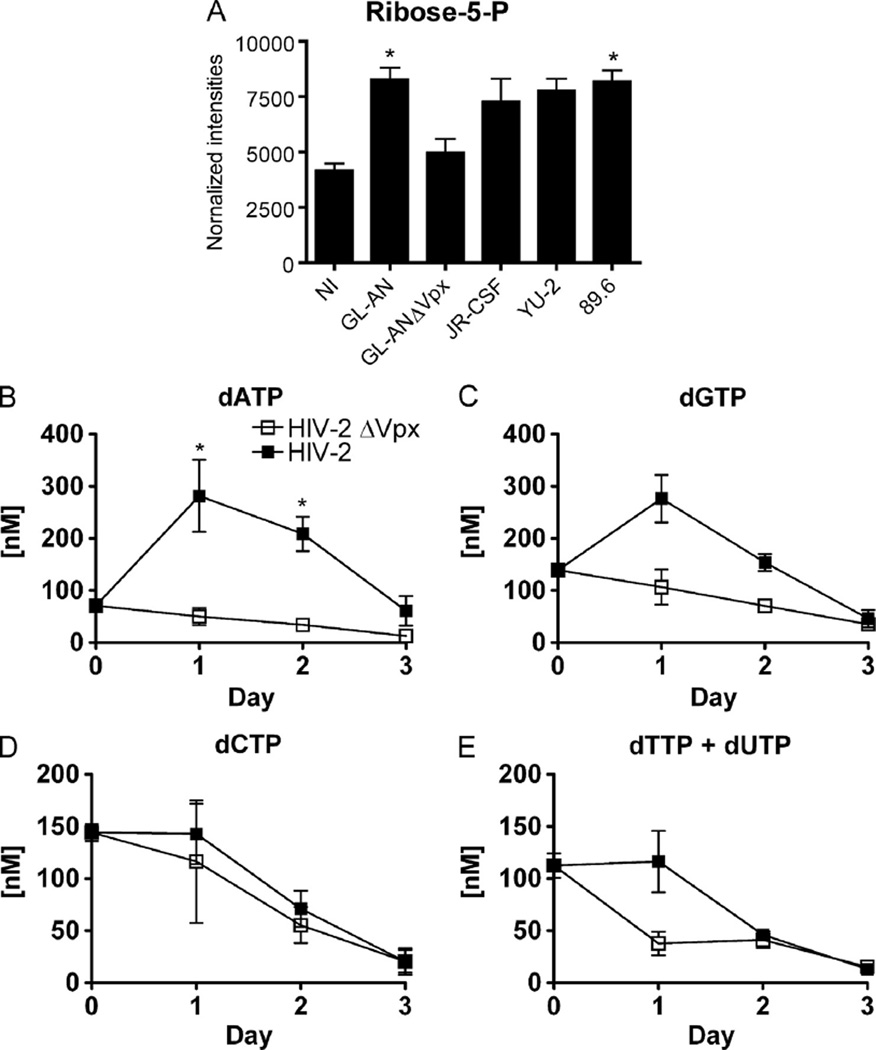
Effect of Vpx on ribose pool size and dNTP levels
Ribonucleotide reductase and the salvage pathway for deoxynucleosides, in conjunction with dNTPase activity of SAMHD1, collectively modulate cellular dNTP levels. When cells are exposed to Vpx from HIV-2 strains or virus-like particles (VLPs), cellular dNTP levels increased due to the elimination of SAMHD1 protein in non-dividing cells and resting CD4+ T cells (Baldauf et al., 2012; Hrecka et al., 2011; Kim et al., 2012; Laguette et al., 2011; Lahouassa et al., 2012). The effect of Vpx on the pentose phosphate pathway intermediate ribose-5P was evaluated, since it is involved in de novo ribonucleoside synthesis. Indeed, as shown in Fig. 5A, MDMs infected with wild type GL-AN (Vpx+) displayed a significant increase (*, p < 0.05) in ribose-5-P, whereas GL-ANΔVpx did not affect ribose-5-P levels in MDMs. Interestingly, increases in ribose-5-P were also observed in HIV-1 infected MDMs (Fig. 5A). Collectively, these data suggest that HIV infection of MDMs generally lead to an increase in ribose-5-P pool size, likely necessary to generate the ribonucleoside triphosphates used for new viral genomes.
Fig. 5.
Ribose and dNTP concentrations. The normalized levels of (A) ribose-5-Phosphate is shown for the different viral infected MDMs. Means and S.E.M. are plotted for three independent donors. Significant increase is denoted by * (p < 0.05) when compared to NI MDMs. Determining dNTP concentrations in HIV-2 infected MDMs. Seven day maturated MDMs were infected with either GL-AN (filled squares) or GL-ANÄVpx (open squares). The dNTP levels were determined for (B) dATP, (C) dGTP, (D) dCTP and (E) dTTPþdUTP at days 0–3 after infection for four independent MDM donors. Significant difference* (p < 0.05; Student T test) are indicated. The data are graphed as means and S.E.M. for three independent donors.
Modulation of dNTP levels by Vpx in MDMs have been well characterized using VLPs. Previously reports have shown that VLPs containing Vpx or HIV-1 engineered with Vpx promote SAMHD1 degradation and led to a rapid increase in dNTPs by 24 h (Berger et al., 2011b; Hollenbaugh et al., 2014; Kim et al., 2012; Sunseri et al., 2011). HIV-1 strains do not contain Vpx; therefore, HIV-1 does not promote rapid degradation of SAMHD1, which would led to the rapid increase dNTP concentration during the first 12 h of infection (Kim et al., 2012). Therefore, we focused on changes in dNTP levels for GL-AN and GL-ANΔVpx infected MDMs. The dATP concentration was significantly increased (*, p < 0.05) at days 1 and 2 after infection (Fig. 5B). The concentration of dGTP also increased at day 1, but not significantly (Fig. 5C). The concentrations of dCTP (Fig. 5D) and dTTP+dUTP (Fig. 5E) remained relatively comparable without increasing from day 0. As expected, these data showed that Vpx in GL-AN virus containing Vpx promoted the increase in dNTPs, but this increase was much more transient in duration as compared to VLP treated MDMs (Hollenbaugh et al., 2014). Collectively, the importance of elevated dNTP concentration is most marked early after infection, when the majority of reverse transcription is thought to take place.
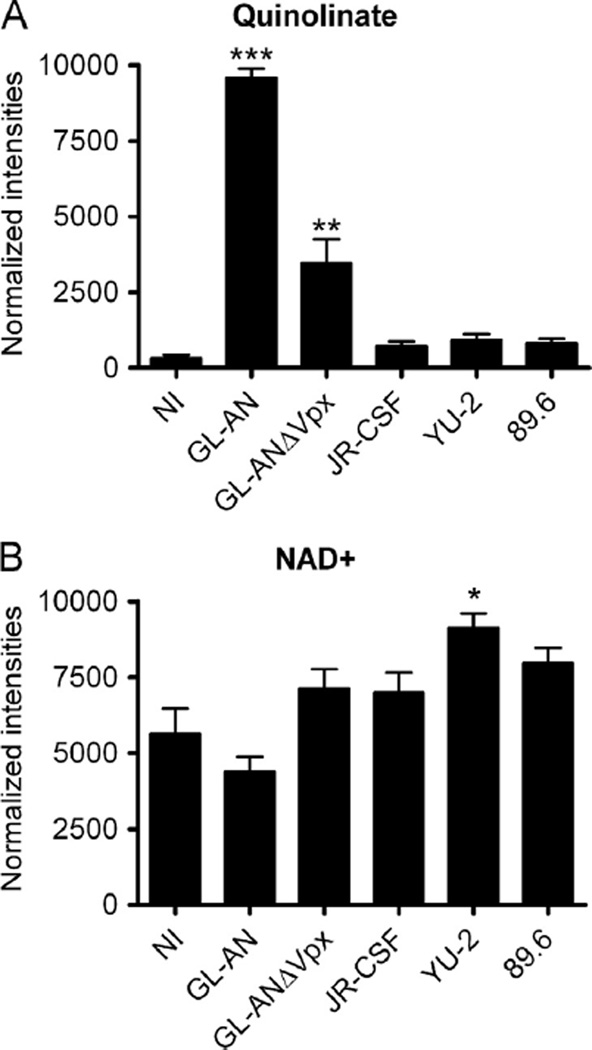
Quinlinate accumulates in HIV-2 infected MDMs
The most striking difference detected with LC–MS/MS analysis was the induction of quinolinate (2,3-pyridine-dicarboxylic acid) levels, which is part of the kynurenine pathway leading to nicotinamide adenine dinucleotide (NAD+) generation. Quinolinate pool size was significantly increased for GL-AN (***; p < 0.001) and GL-ANΔVpx (**; p < 0.01) as compared to NI MDMs (Fig. 6A). Moreover, quinolinate pool sizes were slightly, but not significantly, elevated for all of the HIV-1 infected MDMs as compared to NI MDMs. Quinolinate is further metabolized into NAD+, which is an essential oxidizing reagent, accepting electrons from molecules in TCA cycle. We detected a significant increase (**; p < 0.05) in NAD+ for YU-2 infected MDMs (Fig. 6B), while a reduced level of NAD+ was detected in GL-AN infected MDMs. Collectively, GL-AN and GL-ANΔVpx infected MDMs have increased quinolinate pool sizes, but this does not significantly diminish NAD+ levels in infected MDMs. Additional studies are required to validate the role of this metabolic activation for HIV infection and to examine the mechanisms of strain specific changes in quinolinate levels.
Fig. 6.
Evaluating metabolites in the kynurenine pathway. We evaluated the pool sizes for (A) quinolinate and (B) NADþ metabolites. Significant increase is denoted by * (p < 0.05) and *** (p < 0.001). Data are graphed as means and S.E.M. are plotted for three independent MDM donors.
Discussion
We evaluated whether cellular metabolic pathways might influence the difference between HIV-1 and HIV-2 in infected MDMs. Using an in vitro model, we observed that GL-AN (HIV-2) had higher production of virus as compared to the HIV-1 strains: YU-2, JR-CSF and 89.6 (Fig. 1). When we compared GL-AN and GL-ANΔVpx from an infection of MDMs standpoint, it was not surprisingly to find that the GL-ANΔVpx strain was attenuated relative to the GL-AN strain (Fig. 2). These data are further supported by SIV models, in which Vpx promoted high levels of virus production, but Vpx was not essential in order for the animals to develop AIDS (Shingai et al., 2015).
The LC–MS/MS methodology is a very useful way of evaluating metabolite pool sizes in cells, without directly measuring all enzyme activities associated with the complex pathways. Indeed, this method provides a broad picture of the cellular metabolite environment. For this study, we detected an enhancement in early glycolysis intermediates: FBP and G3P for YU-2 and 89.6 viruses (Fig. 3), yet no significant changes were detected in any of the TCA cycle intermediates. These data are in contrast to our finding using the chronically HIV-1 infected U1 cell line (Hollenbaugh et al., 2011), but fit more in line with HIV-1 infection of activated CD4+ T cells (Hegedus et al., 2014; Hollenbaugh et al., 2011). This discrepancy may be linked to 1) using a cell line that has been selected over time that can produce low levels of HIV-1 as compared to acutely infected human primary MDMs, and 2) U1 cells do not have a functional Tat protein (Folks et al., 1987), which modulates Akt activity (Chugh et al., 2007), and it in turn influences many intracellular signaling pathways of cellular metabolism (Rathmell et al., 2003). UDP-sugar pool sizes were also evaluated (Fig. 4). The UDP-glucose pool was not significantly changed, but UDP-glucuronate level was significantly increased for the 89.6 strain (Fig. 4C). UDP-glucuronate participates in the biosynthesis of glycosaminoglycans such as hyaluronan, chondroitin sulfate, and heparan sulfate found in the extracellular matrix (Yang et al., 2013).
Ribose-5-P is a building block for ribonucleosides and an intermediate in the pentose phosphate pathway. It was significantly increased for GL-AN and 89.6 strains, and to a lesser extent in the GL-ANΔVpx, JR-CSF and YU-2 strains (Fig. 5A). We previously reported that U1 cells showed a decrease in ribose-5-P pool size as compared to U937 cell (uninfected control cells) (Hollenbaugh et al., 2011). The discrepancy between our recent and published finding may be due to 1) analysis of human primary MDMs vs. transformed cells, for the U1 cells have been clonally selected over time, 2) timing of the analysis may influence results. Our preliminary data using virus-like particles with and without Vpx (SIVmac251) showed no significant differences in ribose-5-P pool size (data not shown). The concentrations of dNTPs were monitored in GL-AN and GL-ANΔVpx infected MDMs, in order to evaluate the contribution of Vpx. A significant increase in dATP (Fig. 6B) was detected in GL-AN infected MDMs, and an increase in dGTP concentration (Fig. 6B). However, these dNTP increases were much lower in magnitude and shorter in duration as compared to our data using VLPs (Hollenbaugh et al., 2014; Kim et al., 2012). Collectively, HIV-2 infected MDMs do not maintain high levels of dNTPs, since they are only needed during the reverse transcription and gap repair during viral DNA integration (Amie et al., 2013; Van Cor-Hosmer et al., 2013), which are very early events in the viral life cycle.
The most impressive change, in any of the metabolites, monitored occurred with quinolinate, which exhibited a >12-fold increase in HIV-2 infected MDMs, while the GL-ANΔVpx strain had 2-fold higher pool size as compared to 89.6, YU-2 and JR-CSF strains. The accumulation of the quinolinate metabolite did not correlate to a significant reduction in NAD+, a downstream metabolite. The kynurenine pathway starts with the degradation of tryptophan, an essential amino acid. Various diseases including HIV/AIDS, neurodegenerative disease, lipid metabolism, cancer, aging and chronic inflammation have been associated with the dysfunction of this pathway (Huengsberg et al., 1998; Maddison and Giorgini, 2015; Suzuki et al., 2010; van der Goot and Nollen, 2013). Routy et al. have recently reviewed the interplay between tryptophan metabolism and immune function (Routy et al., 2015). The depletion of tryptophan at the site of infection can be a protective immune mechanism for microorganisms, however prolonged depletion of tryptophan can lead to immunosuppression by T cell exhaustion. The quinolinic acid/tryptophan ratio was reported to be a biomarker of neurological disease in SIV-infected macaques (Drewes et al., 2015). In HIV-1 infected individuals, cART was shown to normalize the tryptophan metabolism (Jenabian et al., 2015). Tryptophan catabolism is associated with an increase in immune activation and HIV-mediated inflammation (Jenabian et al., 2015). We can speculate that HIV-2 infection may promote a higher immune activation state, which in turn leads to better control of the virus. The change in tryptophan metabolism may contribute to the multifactorial nature as to why HIV-1 and HIV-2 infected individuals have different disease onsets.
Materials and methods
Ethics statement
These experiments used primary human primary monocytes obtained from human buffy coats (New York Blood Services, Long Island, NY). These are pre-existing materials that are publicly available, and there is no subject-identifying information associated with the cells. As such, the use of these samples does not represent human subjects research because: 1) materials were not collected specifically for this study, and 2) we are not able to identify the subjects.
Cells
Primary human monocytes were isolated from the peripheral blood buffy coats by positive selection using MACS CD14+ beads as previously described (Hollenbaugh et al., 2014). Monocytes were maturated into monocyte-derived macrophages (MDMs) in the presence of 5 ng/ml human GM-CSF (Miltenyl Biotec) treated at days 0 and 2 of maturation. MDMs were used at day 7 or afterwards for experiments. For the HIV-1 RT activity assay, 250,000 monocytes were maturated in 24-well plates. For the metabolomics analysis, 2 million monocytes were maturated in 3 cm dishes (Hollenbaugh et al., 2011). To determine dNTP levels using the HIV-1 RT-based assay (Diamond et al., 2004), 1 million monocytes were maturated in 6-well plates.
Generation of viruses
The p89.6, pYU-2 and pJR-CSF were obtained through NIH AIDS Reagent Program. GL-AN and GL-AN-2ΔVpx strains were a kind gift from Dr. Florence Margottin-Goguet (Université of Paris Descartes, Paris, France). To generate viruses, 293 T cells were grown to 80% confluency in T225 flasks before transfection with with 40 µg of HIV plasmid DNA and 20 µg of pVSV-G DNA using 180 µl/ µg PEI (1 mg/ml) in 30 ml of DMEM complete medium/flask. The day 1 medium was discarded and replaced with 30 ml of fresh DMEM medium. At days 2 and 3, medium was harvested, centrifuged at 400 g for 7 min to remove cellular debris, and then stored in a T75 flask at 4 °C until experimental use.
HIV-1 RT activity assay
Tubes containing 300 µl of samples were centrifuged at 11 K RPM (9740 g) at 4 °C for 2 h to pellet the virus (Jouan MR23i, Br4i, Sigma 3k30). The pellet was solubilized by vortexing in the presence of 30 µl virus solubilization buffer (0.5% Triton X-100, 0.8 M NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 20% glycerol, and 0.05 M Tris HCl). Then 10 µl of each sample were added to 75 µl RT reaction mixture (0.06 M Tris HCl, pH 7.8, 0.012 M MgCl2, 6 mM dithiothreitol, 6 µg/ml poly rAoligo dT(12–18) [The Midland Certified Reagent Co. Inc. Midland, TX]), 96 µg/ml dATP (Sigma-Aldrich, St. Louis, MO), and 1 µM of 0.08 mCi/mL 3H-thymidine-5′-triphosphate 87.0 Ci/mmol (Moravek Biochemicals, Brea, CA) and incubated at 37 °C for 3 h. The reaction was stopped and the nucleic acid was precipitated overnight at 4 °C in the presence of 10% trichloroacetic acid (100 µl) containing 0.05% sodium pyrophosphate. The acid insoluble product was harvested onto filter paper (cat # 6005422, Perkin Elmer, Waltham, MA) using a Perkin Elmer Harvester (FilterMate Harvester). The radioactive activity was read on a Perkin Elmer TopCount NXT (Microplate Scintillation & Luminescence Counter). The percent of control was calculated. The method by Belen’kii and Schinazi was used to determine the median effective concentrations (EC50) (Belen'kii and Schinazi, 1994).
ELISA kits and intracellular staining
Cell culture supernatants were collected and centrifuged for 1 min at 15,000 rpm in a microcentrifuge to remove cellular debris. Samples were stored at −80 °C before evaluation of either p24 or p27 antigen. For the ELISA, HIV-1 p24 Antigen Capture Assay (Advanced Bioscience Laboratories, Cat# 5421) and SIV p27 Antigen ELISA (ZeptoMetrix Corporation, Cat# 0801201) were used. Intracellular staining was carried out as follows. At day for post infection, medium was removed and cell washed once with PBS. One milliliter of 10× Trypsin was added to the 3 cm dish for 5 minutes. Cells were gentle removed using a cell scraper and collected in a 1.5 eppendorf tube containing 0.5 ml of medium. The tube was centrifuged at 5000 rpm in a microcentrifuge for 30 sec. Supernatant was carefully removed and 1 ml of 4% formaldehyde was added to the tube. Cells pellet was resuspended and the tube was allowed to side at room temperature for 20 min to fix the cells and inactive the HIV. The formaldehyde was removed after fixing the cells by centrifugation at 5000 rpm for 30 s. Cell pellet was resuspended in 50 µl of 1% saponin solution. Ten microliters of FBS was added to the tube and then allowed to sit for 10 min. The primary antibody (Clone 39/6.14; ZeptoMetrix Corporation, Cat# 0801081) can recognize both p24 and p27 antigens and was added at 1:500 dilution in 1% saponin solution. Cells were incubated at room temperature for 20 min before washing three times with PBS. The secondary anti-mouse-PE antibody (Biolegend: Cat # 653203) was added at 1:500 dilution in 1% saponin solution and then allowed to incubate at room temperature for 20 min. Cells were washed thrice with PBS before being collected by flow analysis using a BD Accuri C6 flow cytometer. Data were analyzed using FlowJo software and data plotted using Prism software.
Western blot analysis
Samples were processed in radioimmune precipitation buffer containing 1 µm DTT, 10 µm PMSF, 10 µl/ml phosphatase inhibitor (Sigma), and 10 µl/ml protease inhibitor (Sigma). The cells were sonicated with three 5-s pulses to ensure compete lysis. Cellular debris was removed by 15,000 rpm centrifugation for 10 min. Supernatants were stored at −80 °C before use. Cell lysates (20 µg) were resolved on 8% SDS-PAGE. Proteins were transferred to nitrocellulose membrane. The membrane was blocked with 2% nonfat milk in TBST for 1 h followed by the addition of primary SAMHD1 antibody (Abcam; catalog # ab119751) and incubation overnight at 4 °C. The next day, the membrane was washed (3X, 20 min with TBST) and stained with goat anti-mouse HRP (GE Healthcare; Catalog # NA931V) for 1 h at room temperature. Membrane was washed (3X with TBST) and developed using the SuperSignal West Femto Kit (Thermo Scientific). Images were captured using the Bio-Rad ChemiDoc Imager. ImageLab Analysis software (Bio-Rad) was used to analyze the data sets. Data were normalized using actin as a loading control.
Primer extension assay
This is a HIV-1 RT-based assay protocol, which was followed as previously described (Kim et al., 2012). MDMs were removed from the 6-well plate using 200 µl of 60% cold methanol and scraped. Methanol was transferred to a 1.5 ml eppendorf tube. The well was washed with an addition 200 µl of 60% methanol and then combined in the tube. The tubes were heated at 95 °C for 3 min then placed on ice for 1 min. Tubes were vortexed for 30 s, followed by clearing the cellular debris with a 14,000 rpm centrifugation for 3 min. Supernatant was transferred to a new tube and dried using a SpeedVac (Thermo Scientific). The dNTP pellets were resuspended in 20 µl water. Two microliters of sample were used in the primer extension assay. 5′ 32P-end-labeled primer (5′-GTCCCTCTTCGGGCGCCA-3′) was individually annealed to one of four different templates (3′-CAGGGAGAAGCCCGCGGTN-5′). The template:primer complex was extended by HIV-1 reverse transcriptase, generating one additional nucleotide extension product for one of four dNTPs contained in the sample. In this assay, the molar amount of product is equal to that of each dNTP contained in the extracted samples, which allows us to calculate and compare the dNTP concentrations for the different treatments (Diamond et al., 2004).
Liquid chromatography-tandem mass spectrometry (LC–MS/MS): Culture medium was replaced with serum-free DMEM media lacking sodium pyruvate. After 1 h at 37 °C, the medium was aspirated and dry ice cold 80% methanol was added to the tissue culture plate. Samples were stored at −80 °C for 5 min followed by processing with a rubber policeman to remove attached cells. Supernatant was transferred to a 15 ml conical tube and vortexed for 1 min. Samples were centrifuged at 3500 RPM for 5 min. The supernatant was transferred to a new tube. Pellets were washed with 1 ml of dry ice cold 80% methanol. Samples were centrifuged and supernatants consolidated for each sample. Samples were dried under nitrogen gas. Once dried, samples were stored at −80 °C until analyzed.
Samples were removed from the −80 °C and allowed to equilibrate to room temperature. The cell pellet was resuspended in 70 µl 50% methanol and liquid transferred to 1.5 ml centrifuge tube. Cellular debris was removed by 15 K RPM centrifugation for 10 min and 65 µl of supernatant was transferred to a LC–MS/MS vial and capped. Ten microliters of sample were autoloaded into LC-20 AD HPLC system (Shimadzu) for metabolite separation. The LC was coupled to a mass spectrometer running in negative mode with an ESI source using reversed phase chromatography with an amine-based ion pairing agent. A Synergi Hydro-RP column (150 by 2 mm with a 5 µm particle size; Phenomenex) was used with the following LC parameters: autosampler temperature, 4 °C; column temperature, 40 °C; flow rate, 200 µl/min. The LC solvents were as follows: solvent A, 100% methanol; solvent B, 10 mM tributylamine and 15 mM acetic acid in 97:3 water–methanol. The HPLC gradients were as follows: for t = 0, 100% B; for t = 5, 100% B; for t = 10, 80% B; for t = 20, 80% B; for t = 35, 35% B; for t = 38, 5% B; for t = 42, 5% B; for t = 43, 100% B; and for t = 50, 100% B. Mass spectrometric analyses were performed with a TSQ Quantum Ultra triple-quadrupole mass spectrometer (Thermo Fisher Scientific) with mass spectrometry parameters as per (Bajad et al., 2006).
Data analysis
Data was graphed and Statistical analysis done using Prism software. Student T-test and ANOVA analysis, Kruskal-Wallis test followed by Dunns comparison, were used to evaluate statistical significance.
Acknowledgments
We would like to thank Xenia Schafer and Dr. Cody Spencer for the technical assistance with the LC–MS/MS instrument. This study was supported by NIH, USA AI049781 (B.K.), GM104198 (B. K.), AI081773 (J.M.), and MH100999 (R.F.S.).
References
- Aggarwal A, McAllery S, Turville SG. Revising the role of myeloid cells in HIV pathogenesis. Curr. HIV/AIDS Rep. 2013;10:3–11. doi: 10.1007/s11904-012-0149-1. [DOI] [PubMed] [Google Scholar]
- Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, Skowronski J. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 2012;287:12550–12558. doi: 10.1074/jbc.M112.340711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amie SM, Noble E, Kim B. Intracellular nucleotide levels and the control of retroviral infections. Virology. 2013;436:247–254. doi: 10.1016/j.virol.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 2012;18:1682–1689. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belen'kii MS, Schinazi RF. Multiple drug effect analysis with confidence interval. Antivir. Res. 1994;25:1–11. doi: 10.1016/0166-3542(94)90089-2. [DOI] [PubMed] [Google Scholar]
- Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV–1 infection. PLoS Pathog. 2011a;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger G, Durand S, Goujon C, Nguyen XN, Cordeil S, Darlix JL, Cimarelli A. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV–1-derived lentiviral vectors. Nat. Protoc. 2011b;6:806–816. doi: 10.1038/nprot.2011.327. [DOI] [PubMed] [Google Scholar]
- Bhattacharya J, Peters PJ, Clapham PR. CD4-independent infection of HIV and SIV: implications for envelope conformation and cell tropism in vivo. Aids. 2003;17(4):S35–S43. doi: 10.1097/00002030-200317004-00004. [DOI] [PubMed] [Google Scholar]
- Blaak H, Boers PH, van der Ende ME, Schuitemaker H, Osterhaus AD. CCR5-restricted HIV type 2 variants from long-term aviremic individuals are less sensitive to inhibition by beta-chemokines than low pathogenic HIV type 1 variants. AIDS Res. Human Retrovir. 2008;24:473–484. doi: 10.1089/aid.2007.0001. [DOI] [PubMed] [Google Scholar]
- Chugh P, Fan S, Planelles V, Maggirwar SB, Dewhurst S, Kim B. Infection of human immunodeficiency virus and intracellular viral Tat protein exert a pro-survival effect in a human microglial cell line. J. Mol. Biol. 2007;366:67–81. doi: 10.1016/j.jmb.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory TJ, Schacker TW, Stevenson M, Fletcher CV. Overcoming pharmacologic sanctuaries. Curr. Opin. HIV AIDS. 2013;8:190–195. doi: 10.1097/COH.0b013e32835fc68a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosenza MA, Zhao ML, Lee SC. HIV-1 expression protects macrophages and microglia from apoptotic death. Neuropathol. Appl. Neurobiol. 2004;30:478–490. doi: 10.1111/j.1365-2990.2004.00563.x. [DOI] [PubMed] [Google Scholar]
- De Cock KM, Jaffe HW, Curran JW. The evolving epidemiology of HIV/AIDS. Aids. 2012;26:1205–1213. doi: 10.1097/QAD.0b013e328354622a. [DOI] [PubMed] [Google Scholar]
- Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes JL, Meulendyke KA, Liao Z, Witwer KW, Gama L, Ubaida-Mohien C, Li M, Notarangelo FM, Tarwater PM, Schwarcz R, Graham DR, Zink MC. Quinolinic acid/tryptophan ratios predict neurological disease in SIV-infected macaques and remain elevated in the brain under cART. J. Neurovirol. 2015;21:449–463. doi: 10.1007/s13365-015-0334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- Gilbert PB, McKeague IW, Eisen G, Mullins C, Gueye NA, Mboup S, Kanki PJ. Comparison of HIV-1 and HIV–2 infectivity from a prospective cohort study in Senegal. Stat. Med. 2003;22:573–593. doi: 10.1002/sim.1342. [DOI] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV–1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Hegedus A, Kavanagh Williamson M, Huthoff H. HIV–1 pathogenicity and virion production are dependent on the metabolic phenotype of activated CD4+ T cells. Retrovirology. 2014;11:98. doi: 10.1186/s12977-014-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbaugh JA, Munger J, Kim B. Metabolite profiles of human immunodeficiency virus infected CD4+ T cells and macrophages using LC–MS/MS analysis. Virology. 2011;415:153–159. doi: 10.1016/j.virol.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbaugh JA, Tao S, Lenzi GM, Ryu S, Kim DH, Diaz-Griffero F, Schinazi RF, Kim B. dNTP pool modulation dynamics by SAMHD1 protein in monocyte-derived macrophages. Retrovirology. 2014;11:63. doi: 10.1186/s12977-014-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huengsberg M, Winer JB, Gompels M, Round R, Ross J, Shahmanesh M. Serum kynurenine-to-tryptophan ratio increases with progressive disease in HIV-infected patients. Clin. Chem. 1998;44:858–862. [PubMed] [Google Scholar]
- Jenabian MA, El-Far M, Vyboh K, Kema I, Costiniuk CT, Thomas R, Baril JG, LeBlanc R, Kanagaratham C, Radzioch D, Allam O, Ahmad A, Lebouche B, Tremblay C, Ancuta P, Routy JP Montreal Primary Infection and Slow Progressor Study Groups. Immunosuppressive tryptophan catabolism and gut mucosal dysfunction following early HIV infection. J. Infect. Dis. 2015;212:355–366. doi: 10.1093/infdis/jiv037. [DOI] [PubMed] [Google Scholar]
- Kanki PJ, Peeters M, Gueye-Ndiaye A. Virology of HIV-1 and HIV-2: implications for Africa. Aids. 1997;11(Suppl B):S33–S42. [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 2012;287:21570–21574. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppensteiner H, Brack-Werner R, Schindler M. Macrophages and their relevance in human immunodeficiency virus Type I infection. Retrovirology. 2012;9:82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DC, Giorgini F. The kynurenine pathway and neurodegenerative disease. Semin. Cell Dev. Biol. 2015;40:134–141. doi: 10.1016/j.semcdb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Marchant D, Neil SJ, McKnight A. Human immunodeficiency virus types 1 and 2 have different replication kinetics in human primary macrophage culture. J. Gen. Virol. 2006;87:411–418. doi: 10.1099/vir.0.81391-0. [DOI] [PubMed] [Google Scholar]
- McElrath MJ, Pruett JE, Cohn ZA. Mononuclear phagocytes of blood and bone marrow: comparative roles as viral reservoirs in human immunodeficiency virus type 1 infections. Proc. Natl. Acad. Sci. USA. 1989;86:675–679. doi: 10.1073/pnas.86.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight A, Griffiths DJ, Dittmar M, Clapham P, Thomas E. Characterization of a late entry event in the replication cycle of human immunodeficiency virus type 2. J. Virol. 2001;75:6914–6922. doi: 10.1128/JVI.75.15.6914-6922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyamweya S, Hegedus A, Jaye A, Rowland-Jones S, Flanagan KL, Macallan DC. Comparing HIV-1 and HIV-2 infection: lessons for viral immunopathogenesis. Rev. Med. Virol. 2013;23:221–240. doi: 10.1002/rmv.1739. [DOI] [PubMed] [Google Scholar]
- Pauls E, Ballana E, Este JA. Nucleotide embargo by SAMHD1: a strategy to block retroviral infection. Antivir. Res. 2013;97:180–182. doi: 10.1016/j.antiviral.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Pizzato M, McCauley SM, Neagu MR, Pertel T, Firrito C, Ziglio S, Dauphin A, Zufferey M, Berthoux L, Luban J. Lv4 is a capsid-specific antiviral activity in human blood cells that restricts viruses of the SIVMAC/SIVSM/HIV-2 lineage prior to integration. PLoS Pathog. 2015;11:e1005050. doi: 10.1371/journal.ppat.1005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planelles V. Restricted access to myeloid cells explained. Viruses. 2011;3:1624–1633. doi: 10.3390/v3091624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell RD, Holland PJ, Hollis T, Perrino FW. The Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 2011;286:43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska M, Takahashi K, Czernekova L, Zachova K, Hall S, Moldoveanu Z, Elliott MC, Wilson L, Brown R, Jancova D, Barnes S, Vrbkova J, Tomana M, Smith PD, Mestecky J, Renfrow MB, Novak J. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J. Biol. Chem. 2010;285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol. Cell. Biol. 2003;23:7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L, Perelson AS. Modeling HIV persistence, the latent reservoir, and viral blips. J. Theor. Biol. 2009;260:308–331. doi: 10.1016/j.jtbi.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy JP, Mehraj V, Vyboh K, Cao W, Kema I, Jenabian MA. Clinical relevance of kynurenine pathway in HIV/AIDS: an immune checkpoint at the crossroads of metabolism and inflammation. AIDS Rev. 2015;17:96–106. [PubMed] [Google Scholar]
- Rowland-Jones S. Protective immunity against HIV infection: lessons from HIV-2 infection. Futur. Microbiol. 2006;1:427–433. doi: 10.2217/17460913.1.4.427. [DOI] [PubMed] [Google Scholar]
- Shingai M, Welbourn S, Brenchley JM, Acharya P, Miyagi E, Plishka RJ, Buckler-White A, Kwong PD, Nishimura Y, Strebel K, Martin MA. The expression of functional Vpx during pathogenic SIVmac infections of rhesus macaques suppresses SAMHD1 in CD4+ memory T cells. PLoS Pathog. 2015;11:e1004928. doi: 10.1371/journal.ppat.1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. SAMHD1 restricts HIV–1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology. 2012;9:105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunseri N, O'Brien M, Bhardwaj N, Landau NR. Human immunodeficiency virus type 1 modified to package Simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J. Virol. 2011;85:6263–6274. doi: 10.1128/JVI.00346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Suda T, Furuhashi K, Suzuki M, Fujie M, Hahimoto D, Nakamura Y, Inui N, Nakamura H, Chida K. Increased serum kynurenine/tryptophan ratio correlates with disease progression in lung cancer. Lung Cancer. 2010;67:361–365. doi: 10.1016/j.lungcan.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Takeuchi JS, Perche B, Migraine J, Mercier-Delarue S, Ponscarme D, Simon F, Clavel F, Labrosse B. High level of susceptibility to human TRIM5alpha conferred by HIV-2 capsid sequences. Retrovirology. 2013;10:50. doi: 10.1186/1742-4690-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cor-Hosmer SK, Kim DH, Daly MB, Daddacha W, Kim B. Restricted 5'-end gap repair of HIV-1 integration due to limited cellular dNTP concentrations in human primary macrophages. J. Biol. Chem. 2013;288:33253–33262. doi: 10.1074/jbc.M113.486787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot AT, Nollen EA. Tryptophan metabolism: entering the field of aging and age-related pathologies. Trends Mol. Med. 2013;19:336–344. doi: 10.1016/j.molmed.2013.02.007. [DOI] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Carlos Valle-Casuso J, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology. 2012;436:81–90. doi: 10.1016/j.virol.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yang Z, Lv H, Lou Y, Wang J, Wu N. Bridging HIV-1 cellular latency and clinical long-term non-progressor: an interactomic view. PLoS One. 2013;8:e55791. doi: 10.1371/journal.pone.0055791. [DOI] [PMC free article] [PubMed] [Google Scholar]