Abstract
The rate of protein synthesis varies according to the mRNA sequence in ways that affect gene expression. Global analysis of translational pausing is now possible with ribosome profiling. Here, we revisit an earlier report that Shine-Dalgarno sequences are the major determinant of translational pausing in bacteria. Using refinements in the profiling method as well as biochemical assays, we find that SD motifs have little (if any) effect on elongation rates. We argue that earlier evidence of pausing arose from two factors. First, in previous analyses, pauses at Gly codons were difficult to distinguish from pauses at SD motifs. Second, and more importantly, the initial study preferentially isolated long ribosome-protected mRNA fragments that are enriched in SD motifs. These findings clarify the landscape of translational pausing in bacteria as observed by ribosome profiling.
Introduction
The ribosome profiling method developed by Ingolia and Weissman is a powerful tool for obtaining global information about protein synthesis (Ingolia et al., 2009). In this approach, the positions of ribosomes on mRNAs are determined by sequencing ribosome-protected mRNA fragments. Perhaps the most common use of this method is to compare the number of ribosomes per gene under different conditions to monitor changes in gene expression. But the ribosome profiling method is capable of providing more detailed mechanistic insights as well: in the few short years since its development, profiling studies have explored the interaction of the nascent chain with chaperones (Liu et al., 2013; Oh et al., 2011) and observed non-canonical events like frameshifting (Michel et al., 2012), stop-codon readthrough (Dunn et al., 2013), and termination/recycling defects (Guydosh and Green, 2014; Young et al., 2015).
Because it has high resolution, ribosome profiling has the potential to reveal the location and strength of translational pauses throughout the genome. Increased levels of ribosome occupancy at specific sites provide evidence for slower elongation rates (Ingolia et al., 2011). Ribosome pausing plays a critical role in the regulation of gene expression in bacteria (Ito and Chiba, 2013) and in mRNA surveillance pathways in eukaryotes (Doma and Parker, 2006). In addition, many studies argue that elongation rates may be optimized to promote protein folding (Kim et al., 2015; Zhang and Ignatova, 2011). Ribosome profiling will continue to shed light on these important areas of research by providing a clearer picture of translational pauses in living cells.
In a pioneering study applying the ribosome profiling method to bacteria, ribosome occupancy was enriched at Shine-Dalgarno (SD) sequences (Li et al., 2012). While SD sequences upstream of the start codon have a well-characterized role in initiation, these data suggested that elongation is retarded by transient base-pairing between SD motifs within open reading frames and the anti-Shine Dalgarno sequence (aSD) in 16S rRNA. SD-associated pauses were reported to account for > 70% of strong pauses genome-wide, leading the authors to conclude that pausing at SD motifs was the primary determinant of translational pausing in bacteria (Li et al., 2012).
Here we revisit these observations using refinements in the method developed in our work on ribosome pausing in bacteria lacking EFP (Woolstenhulme et al., 2015). These refinements improved the resolution significantly. For technical reasons, the bacterial protocol produces ribosome footprints that vary in length. Earlier studies distributed information about the position of the ribosome over multiple nucleotides at the center of reads, blurring the signal (Li et al., 2012; Oh et al., 2011). We and others found that by assigning ribosome occupancy to the 3’-end of the reads, we obtain a more precise measurement of ribosome position (Balakrishnan et al., 2014; Nakahigashi et al., 2014; Woolstenhulme et al., 2015). With this higher resolution, we see that the previously observed enrichment of ribosome occupancy at SD motifs can explained by pauses at Gly codons and by failure to isolate the entire population of ribosome-protected mRNA fragments. We conclude that SD motifs probably account for a small fraction of translational pauses in vivo.
Results
Two signals and two distinct phenomena
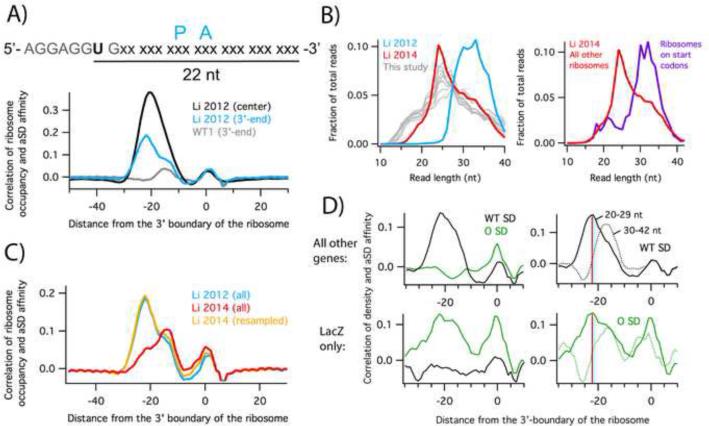
We previously established that assigning ribosome occupancy to the 3’-end of ribosome profiling reads gives higher resolution (Woolstenhulme et al., 2015). To determine if these refinements might shed light on pauses at SD motifs, we re-analyzed the data of Li et al. 2012 with both the center- and 3’-assignment strategies, observing the extent to which ribosome occupancy correlates with affinity of the mRNA to the aSD in the 16S rRNA. We employed a cross-correlation function to determine the optimal displacement between maps of aSD affinity and ribosome occupancy (Figure 1A). The small peak at zero reflects cloning bias (Figure S1E) and can be ignored. In the center-assigned data (black), a single broad peak was observed, as reported earlier. In the 3’-assigned data (blue), however, the single peak resolves into two peaks; one at −15 and another at −22. The peak at −22 corresponds to high ribosome density when the SD motif is 22 nt upstream of the 3’-end of the reads (Figure 1A). In this position, the SD is 10 nt upstream of the A-site codon as previously reported (Li et al., 2012), consistent with known optimal spacing of the SD for participating in initiation (Chen et al., 1994). The peak at −15, on the other hand, is not caused by SD:aSD pairing, as will be discussed below. These correlation plots show that the center-assignment method conflates two signals, one associated with SD pausing (−22), and one that is not (−15).
Figure 1.
High ribosome occupancy at Shine-Dalgarno motifs is due to the isolation of long mRNA fragments. A) The cross-correlation of aSD affinity and ribosome occupancy reveals the position of the SD motif that is optimal for pausing the ribosome. Ribosome occupancy was assigned to the center or 3’-end of the reads. B) Distribution of mapped read lengths. C) Cross-correlation plots calculated with the entire libraries of Li et al. 2012 (blue) and 2014 (red) or only the longer reads from the 2014 library (orange), resampled to match the 2012 read length distribution. D) Cross-correlation plots from cells expressing orthogonal ribosomes and a lacZ reporter with a complementary SD sequence (Li et al. 2012). Top panels include all endogenous genes; bottom panels only the lacZ reporter. Correlations were computed using the affinity for the wild-type (black) or orthogonal aSD sequence (green). The right two panels were computed with either long (30 – 42 nt) or short (20 – 29 nt) reads; the red line indicates the peak at –22 associated with apparent SD pausing. See also Figure S1.
Apparent SD pauses arise from the preferential selection of long mRNA fragments
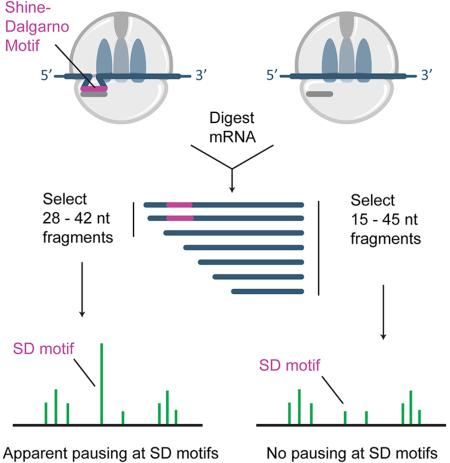
In our preparation of 19 ribosome profiling libraries from the same E. coli strain grown under similar conditions, we observed little or no correlation between ribosome occupancy and SD affinity at the −22 position, suggesting that SD pauses are absent in our data (Figure 1A, grey). A thorough discussion of quality control for these libraries is given in Figures S2 and S3. Systematically varying steps in the procedure that might affect pausing, we tested the effect of antibiotics in the media, differences in methods for harvesting and lysing cells, and treatments of the lysate intended to stabilize ribosome complexes. The single factor that affected the correlation between SD motifs and ribosome occupancy is the isolation of RNA fragments. While the initial bacterial studies by Oh et al. 2011 and Li et al. 2012 selected 28 – 42 nt ribosome-protected fragments by PAGE, we cut more broadly and isolated 15 – 45 nt fragments. This difference is clearly reflected in the distribution of read lengths in the sequenced libraries (Figure 1B, left). A later study by Li et al. (2014) cut more broadly and the size distribution is similar to our studies (Figure 1B, left).
Differences in ribosome footprint lengths are relevant because RNA fragments containing SD motifs are longer than those without them (O'Connor et al., 2013). This phenomenon can be clearly seen in footprints from ribosomes with start codons in the P site. These 70S ribosomes have completed initiation but have not yet begun elongating; presumably strong SD-aSD pairing remains intact. These footprints are significantly longer (30 – 40 nt) than footprints elsewhere in coding sequences (Figure 1B, right). We speculate that SD-containing reads are longer at the 5’-end because the interaction between the mRNA and the aSD protects the fragment from nuclease digestion.
We wondered whether by isolating fragments from the upper end of the length distribution, the earlier study may have inadvertently enriched for SD-containing mRNA fragments. To test this idea, we compared the data from the original study, where 28 – 42 nt fragments were isolated, with data from the same lab in which 15 – 45 nt fragments were isolated (Li et al., 2014). With the newer data, the cross-correlation plots contain a peak of similar intensity at −15 but a marked reduction in the peak at −22 that reflects SD pausing (red and blue traces, Figure 1C). These data suggest that the intensity of the −15 peak is independent of RNA fragment length, but that the relative proportion of ribosomes found at SD motifs is reduced when a broader selection of mRNA fragments is sequenced. Moreover, when we computationally remove shorter reads from the Li et al. 2014 library so that it has the same read length distribution as the earlier library, the cross-correlation plots are nearly identical (yellow and blue traces, Figure 1C). Taken together, these data indicate that the initial study over-estimated the strength of SD pauses because the protocol failed to isolate the full range of ribosome-protected footprints.
The ribosome protects RNA fragments that pair with the aSD
One of the most compelling experiments in the initial report of pausing at SD motifs involved mutant ribosomes in which the sequence of the aSD had been altered. These orthogonal ribosomes translate only a single mRNA species in the cell, a lacZ reporter containing the complementary SD sequence. Within the lacZ coding sequence translated by orthogonal ribosomes, ribosome density was enriched at mutant SD motifs but not at wild-type SD motifs (Figure 1D, bottom left); conversely, ribosome density across all other genes was enriched at wild-type SD motifs but not mutant SD motifs (Figure 1D, top left). The observation that enrichment occurs at the type of SD motif in the coding sequence that was used to initiate translation provides strong evidence that the increased density arises from elongating ribosomes and not from initiation events within coding sequences.
Although Li et al. (2012) interpreted the high ribosome density near SD motifs as evidence of translational pausing, our findings suggest it arises from preferential selection of long mRNA fragments that are protected against nuclease digestion by the SD-aSD interaction. To test this hypothesis using the orthogonal ribosome data, we calculated the cross-correlation between aSD affinity and ribosome occupancy using either short reads (20 – 29 nt) or long reads (30 – 42 nt). As expected, long reads from cellular mRNAs translated by normal ribosomes have a strong correlation at the –22 position with the wild-type SD motif. In contrast, shorter reads had a much lower correlation at –22 but had a strong peak at –15 (Figure 1D, top right); this peak is inconsistent with SD pausing and arises from another source as detailed below. The same pattern was seen for the orthogonal ribosomes translating lacZ: a strong correlation with the mutant aSD sequence was observed at –22 for the long reads but not the short reads (Figure 1D, bottom right). These data show that longer RNA fragments are enriched in SD motifs that pair with rRNA, indicating that this enrichment arises from base-pairing of mRNA with the ribosome and not from the SD sequence itself. These analyses support our hypothesis that the SD-aSD interaction protects the 5’-end of RNA fragments from digestion.
SD motifs make a minimal contribution to global translational pausing
To compare the enrichment of ribosome density near SD motifs in different profiling libraries, we used a different metric, calculating the average ribosome occupancy and aSD affinity for all RNA hexamers. Plots of the Li et al. data and of our own data are shown in Figure 2A with their linear fits. As expected, hexamers with high affinity for the aSD sequence have high ribosome occupancy in the Li et al. 2012 data, as much as 3-fold higher than hexamers with low affinity. The slope of the linear fit (0.28) reflects this strong dependence of ribosome occupancy on aSD affinity. In contrast, there is a much weaker dependence in the Li et al. 2014 data (slope = 0.07), and hexamers with the highest affinity have only about 1.5-fold more occupancy than those with low affinity. In our data, there is essentially no dependence at all, with a slope of −0.01 and no obvious enrichment of ribosomes on high affinity hexamers. These findings are similar to those reported with the cross-correlation analysis in Figure 1 and are independent of the method of assigning ribosome density (using the 3’-end, Figure 2A, or center of reads, Figure S1).
Figure 2.
A) Linear fits of the average ribosome density and aSD affinity for all RNA hexamers. B) Read length distributions and cross-correlation plots for two ribosome profiling datasets with the strongest or weakest correlation between ribosome density and aSD affinity. See also Table S1.
Using the same metric, our analysis of 20 E. coli ribosome profiling datasets from several labs reveals wide variations in levels of SD pausing (Table S1). For the datasets with the highest and lowest SD correlations, differences in the isolation of RNA fragments account for the observed outcomes. Balakrishnan et al. (2014) report isolating fragments 20 – 30 nt in length (Figure 2B, left), effectively discarding long reads that contain SD motifs. In a cross-correlation analysis of these data, we see an anti-correlation between aSD affinity and ribosome occupancy as evidenced by the dip at – 22 in cross-correlation plots (Figure 2B, right) and the negative slope (–0.10, Table S1). In contrast, Haft et al. (2014) enriched for SD-containing motifs by isolating long fragments (Figure 2B, left). Their data exhibit a robust peak at –22 in the cross-correlation plots and slopes of 0.21 – 0.25, nearly as high as Li et al. 2012. These findings further support the conclusion that the correlation between aSD affinity and ribosome density strongly depends on the length of the mRNA fragments isolated.
Differences in the isolation of RNA fragments do not explain all the variability we observe, however. With the exception of Balakrishnan et al. 2014, all of the studies reported isolating 28 – 42 nt fragments following the original protocol and thus might be expected to show higher correlations of aSD affinity and ribosome density. In some cases, this discrepancy can be explained by the fact that the actual length distribution is substantially different than the reported range of isolated RNA fragments. Other steps in the protocol may also have an effect. In the data of Oh et al. (2011), for example, cultures treated with chloramphenicol and centrifuged showed lower levels of SD pausing (0.08) than untreated cells that were filtered and flash frozen (0.15, Table S1).
Isolating a broad distribution of RNA fragments (15 – 45 nt), we observe an absence of a correlation between aSD affinity and ribosome density that is highly reproducible. We systematically varied steps in the procedure, generating 19 libraries that all have essentially no SD pausing, with slopes near zero (Table S1). We conclude that differences in the isolation of RNA fragments have the greatest impact on enrichment of reads containing SD motifs.
Pausing at SD motifs is not observed in vitro
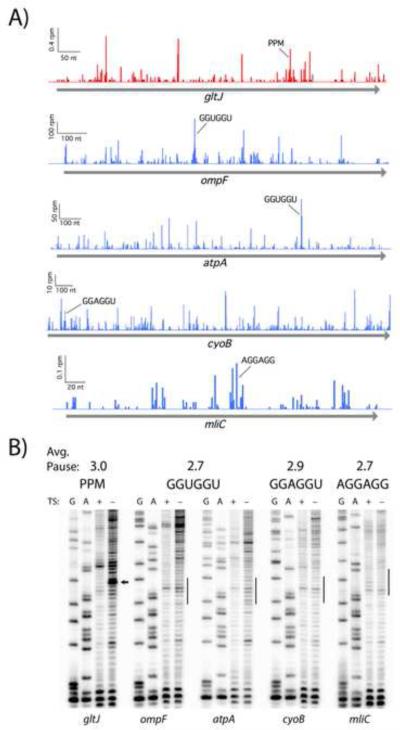
Given the questions raised by our analyses of the profiling data, we set out to determine the extent to which SD motifs impact elongation in vitro using a biochemical assay. We selected three hexamers, GGUGGU, GGAGGU, and AGGAGG, based on their high affinity for the aSD as well as their high pause scores in the original paper (Li et al., 2012). We define pause scores as the ribosome occupancy at the motif of interest divided by the mean occupancy for the gene, averaged over all instances of that motif (Woolstenhulme et al., 2015). For these three hexamers, each of which has a pause score of 2.7 or higher, we identified instances in endogenous genes with high occupancy (Figure 3A). For comparison, we evaluated pausing at Pro-Pro-Met; this tripeptide motif has an pause score of 3.0 in bacteria lacking EFP (Woolstenhulme et al., 2015), roughly the same strength as the three SD hexamers of interest in the Li et al. 2012 dataset.
Figure 3.
Pauses at Shine-Dalgarno motifs are not detected in an in vitro translation assay. A) SD pauses appear in profiling data from MG1655 (Li et al. 2012, blue). Likewise, pauses appear at Pro-Pro-Met (PPM) in a mutant lacking EFP (Woolstenhulme et al. 2015, red). B) Toeprinting analysis of four strong SD motifs and a Pro-Pro-Met control with roughly equivalent pause scores in ribosome profiling data. Expected pausing sites are indicated with an arrow or line. Thiostrepton (TS) traps the ribosome at start codons: bands seen in both treated and untreated lanes are reverse transcriptase artifacts whereas true toeprints appear in only the untreated lane.
To determine if these motifs induce translational pauses, we employed conventional toeprinting assays that have been widely used for decades to assess pause strength (Hartz et al., 1989; Sachs et al., 2002; Vazquez-Laslop et al., 2008; Woolstenhulme et al., 2013). mRNA constructs encoding the motifs within their endogenous sequence context were translated in a reconstituted translation system and a radiolabeled primer was annealed to the 3’-end of the transcripts and extended by reverse transcriptase. When reverse transcriptase encounters a paused ribosome, it arrests 15 – 16 nt downstream of the first nucleotide in the P-site codon. Strong pauses elicit strong cDNA bands on a PAGE gel. In control lanes, the general elongation inhibitor thiostrepton is added to the reaction; primer extension products that appear both with and without thiostrepton are ignored as they represent truncated cDNAs generated by reverse transcriptase even in the absence of translation, perhaps due to sequence or secondary structural elements that impede polymerization by RT.
The toeprinting data reveal a robust pause at Pro-Pro-Met (since there is no EFP present in the translation reaction) but provide no evidence of pausing at SD motifs (Figure 3B). The thiostrepton-sensitive band for the Pro-Pro-Met-containing gene gltJ corresponds to pausing where the second Pro codon is positioned in the P site, consistent with earlier biochemical data (Doerfel et al., 2013; Ude et al., 2013) and our previous ribosome profiling study (Woolstenhulme et al., 2015). This shows that the toeprinting assay is sensitive enough to detect pauses with an average pause score of 3. In contrast, there are no thiostrepton-sensitive bands at the relevant positions with the four SD-containing constructs that we assayed: ompF and atpA with GGUGGU, cyoB with GGAGGU, and mliC with AGGAGG. The lack of observable pausing in this in vitro experiment is consistent with our inability to observe pausing on such motifs in our genomic analysis. Together, these data suggest that SD motifs are not a major source of translational pausing in bacteria.
Gly codons appear to pause ribosomes when bound in the E site
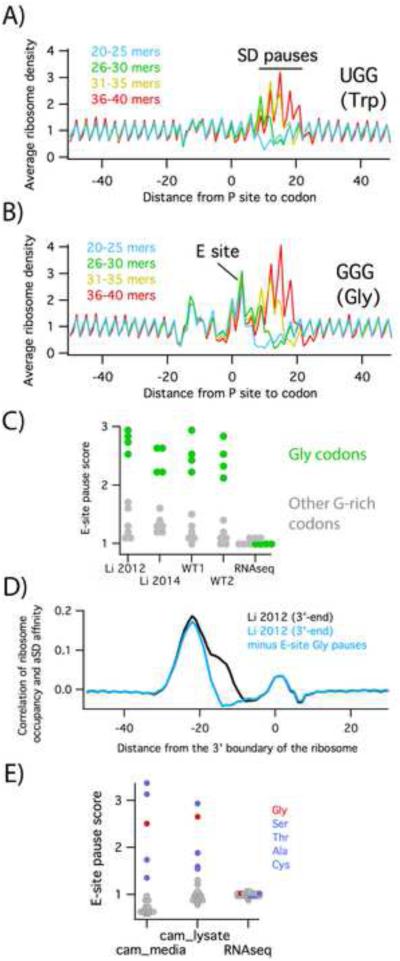
As noted above, when we look at the cross-correlation using the 3’-assignment method (Figure 1A), we observe two peaks. Initially, it was unclear why ribosome occupancy and aSD affinity should be correlated at the −15 position, about 7 nt downstream from the optimal distance for SD-aSD interactions. To explore the origins of the −15 peak, we used the Li et al. 2014 data to calculate the average ribosome density on G-rich codons, all of which have high affinity for the aSD. Plots of average density at these codons display a signature typical of SD pauses. (Note that these plots appear to be flipped compared to the cross-correlation plots; the codon starts at 0 and the signal represents ribosome density shifted to line up with the P site). For example, ribosome density is enhanced when the P site is 10 – 20 nt downstream of the UGG codon (Figure 4A) because UGG can interact strongly with the aSD. The intensity of the peak depends on the length of the reads used in the calculation; the strongest signal is seen with reads 36 – 40 nt in length. The peaks are weaker when 31 – 35 or 26 – 30 nt reads are used and are not detectable with 20 – 25 nt reads. This length dependence is consistent with what we observed above (Figure 1C and 2B); SD motifs are enriched in long reads. Pauses at AGG and CGG follow a similar pattern in their position and read length dependence (Figure S4). In each of these cases, the pausing signatures likely reflect interaction of SD motifs with the aSD.
Figure 4.
Pauses at Gly codons. A) The average ribosome density at UGG codons was calculated for subsets of the Li et al. 2014 library containing reads of various lengths. B) In the same dataset, ribosome density surrounding GGG codons shows an additional peak corresponding to ribosome pausing with Gly codons in the E site. Plots for the other eight codons containing two guanosines are shown in Figure S4. C) E-site pause scores for all ten G-rich codons. Gly codons are highlighted in green. D) Cross-correlation plots using data from Li et al. 2012, before (black) or after (blue) subtracting pauses due to Gly codons in the E site. E) E-site pause scores for all twenty amino acids from samples treated with chloramphenicol in the media or the lysate.
In contrast, pausing at GGN codons has a more complex pattern that provides a clue to the origins of the −15 peak seen in Figure 1A. In plots of the average ribosome density on GGG codons, for example, there is a strong enhancement of ribosome density when the GGG codon is positioned in the ribosomal E site (Figure 4B). These pauses are significantly stronger for the four Gly codons (GGN) than other G-rich codons (Figure S4, quantified in Figure 4C). Gly pauses differ from SD pauses in that they are not read-length dependent; strong pauses are seen for all read lengths when Gly codons are positioned in the ribosomal E site. Gly pauses are also observed in our own data (Figure 4C), consistent with the fact that our libraries exhibit a robust −15 peak in the cross correlation analysis but not the −22 peak associated with SD pausing (Figure 1A). No pausing is evident in the RNAseq samples, indicating that the observed pauses are not the result of cloning or sequencing artifacts. As a final evidence of its origin, the −15 peak disappears when the pauses associated with in-frame Gly codons in the E site are computationally subtracted (Figure 4D). Taken together, these data indicate that the −15 peak arises from pausing on Gly codons and not from SD-aSD interactions.
Although the profiling data show that ribosomes pause with Gly codons in the E site, the biochemical significance of this observation is less clear. Presumably having a Gly residue at the −2 position in the nascent polypeptide inhibits ribosome function in some way. However, we have been unable to detect pausing at Gly codons in toeprinting assays, despite the fact that the pauses in the profiling data are roughly the same strength as the Pro-Pro-Met control in Figure 3B. Indeed, the absence of toeprints at the atpA, cyoB, and ompF SD motifs in Figure 3 argues against pausing at Gly codons, since these motifs are translated as Gly-Gly. What might account for this discrepancy?
It may be that methods of arresting translation after cell lysis generate pauses that do not reflect the in vivo translational landscape. Gly codons are not the only ones that cause pausing when positioned in the ribosomal E site: pauses at Ser, Thr, Ala, and Cys are observed as well (Figure 4E). These pauses are strikingly similar to those observed when chloramphenicol is added to a culture to arrest translation prior to harvesting the cells (Figure 4E). As shown previously by Mori and co-workers, chloramphenicol arrests ribosomes in a sequence-specific manner, pausing ribosomes when the same five amino acids are encoded in the E site (Nakahigashi et al., 2014). This sequence specificity was also observed by Mankin and co-workers, who detected pauses with Gly, Ser, Ala, and Thr codons in the E site using toeprinting assays, but only in the presence of chloramphenicol (Orelle et al., 2013). Given that the activity of chloramphenicol depends on the sequence being translated, and that the lysates are translationally active (the method of preparing lysates resembles methods for preparing extracts for in vitro translation), it makes sense that adding chloramphenicol to arrest translation leads to pausing artifacts in ribosome profiling.
Discussion
Our findings raise questions about whether Shine-Dalgarno motifs are a major determinant of translational pausing in bacteria. The earlier report of strong pauses (Li et al., 2012) conflated two signals, one from true SD motifs and another from Gly codons. With the higher resolution provided by 3’-assignment, we were able to resolve these two signals. In retrospect, using either 3’-assignment or center-assignment (Figure S1), we clearly see that selection for longer RNA fragments in the initial paper artificially enriched SD-containing reads in the library (O'Connor et al., 2013). These two factors together explain the initial claims of SD pausing in the bacterial system, though they do not explain it completely. In our own data, we fail to observe even modest enrichment in ribosome occupancy at SD motifs. We have systematically varied every step of the library preparation protocol but have not been able to reproduce the small enrichment at SD-motifs that remains in Li et al. 2014.
We provide evidence for an absence of SD-pausing using standard toeprinting assays (Figure 3). Pauses were not detected on SD motifs even though pauses of equivalent strength at polyproline motifs were readily detected by this approach. Given that the toeprinting assay is widely used to detect pausing during elongation, there is every reason to expect that this method would similarly detect pauses induced by SD-motifs with equivalent pause scores. Although there may be differences between ribosome activity in vitro and in vivo, taken together, the lack of SD pausing in our profiling data and the lack of observable pausing in vitro suggest that SD motifs are not a major source of translational pausing in bacteria.
We note that two single-molecule studies indicate that internal SD motifs can promote pausing during elongation. In the first (Wen et al., 2008), in an optical tweezers experiment, ribosomes arrest at two internal SD motifs. We argue that the interpretation of this observation is not straightforward: in the optical tweezers setup, ribosomes are continually unwinding a very strong hairpin, only a small fraction complete the synthesis of the 80-mer product, and the rate of translation is quite slow (0.5 codons/s). A second single-molecule study using fluorescence approaches is potentially more convincing: in their analysis, Puglisi and co-workers found that a strong SD motif was able to inhibit the ribosome’s ability to exit the pre-translocation (hybrid) state by 3 – 4 fold (Chen et al., 2014). Here again, however, translation is at least 100-fold slower than observed in vivo. These caveats raise doubts about the relevance of these studies in understanding pausing in vivo where processivity and translation rates are much higher.
A more compelling biochemical argument is put forward by Borg and Ehrenberg (2015) who revisit the question in bulk translocation assays under in vivo-like conditions. Over the years, Ehrenberg and co-workers have developed an in vitro translation system in which the buffer and factor concentrations are carefully fine-tuned to achieve rates like those observed in vivo (~20 codons/s). In this study, they examined three SD motifs of varying affinities and found that they had no effects on the rate of translocation (Borg and Ehrenberg, 2015). Noting the discrepancy with Puglisi’s single-molecule study, they remark that their time scales are more than 100-fold shorter than those in the single-molecule work. Perhaps SD motifs induce pausing if translation is sufficiently slow or otherwise limited by the in vitro system.
Although our findings argue that SD motifs are not the primary source of translational pausing in E. coli, they certainly do not rule out the possibility that SD motifs may affect elongating ribosomes under specific circumstances that are biologically important. SD motifs have well-characterized roles in frameshifting in bacteria: in the dnaX gene, an internal SD motif contributes to −1 frameshifting at a slippery sequence followed by an mRNA hairpin (Larsen et al., 1994). In vitro studies on this system have shown that the downstream hairpin blocks translocation (Caliskan et al., 2014; Chen et al., 2014), resulting in a kinetic pause; this in turn allows different codons and reading frames to be sampled on the slippery sequence (Caliskan et al., 2015; Yan et al., 2015). In the metastable state where the ribosome slips on the message, the SD motif stabilizes the interaction with the mRNA in a new position.
Another well-characterized programmed frameshift in E. coli occurs in the prfB gene where +1 frameshifting promotes synthesis of full-length RF2 protein by avoiding termination at an in-frame stop codon. In this elegant genetic circuit, low levels of RF2 increase ribosome pausing on the UGA stop codon, triggering frameshifting. High levels of frameshifting depend on a conserved SD motif positioned upstream of the UGA codon (Weiss et al., 1988). Here again, the primary pausing event (i.e. the kinetic pause) is the slow rate of peptide release due to the limiting amounts of RF2, and the SD motif probably promotes mRNA movement on the slippery sequence.
Our improved methods allow us to detect pauses when Gly codons are positioned in the E site. These pauses are not dependent on read length (unlike the SD-motif pauses) and are observed with all four Gly codons (GGN). These observations suggest that these pauses result from features related to the amino acid and not from interaction with the mRNA. We have been unable to detect pauses at Gly codons in toeprinting assays, suggesting that protein synthesis is different in the ribosome profiling workup and the in vitro translation system we use for toeprinting. We note that using toeprinting assays, others have reported pauses with codons for Gly and other small amino acids in the E site when chloramphenicol is included in the reaction (Orelle et al., 2013). These pauses match those observed when chloramphenicol is added to the culture prior to harvesting cells (Nakahigashi et al., 2014). Chloramphenicol binds the peptidyl-transferase center and has variable effects depending on the peptide and aminoacyl-tRNA sequence (Wilson, 2009); presumably it arrests ribosomes more effectively with Gly, Ala, Ser, Cys, or Thr in the second to last position of the nascent peptide. We are currently working to understand how antibiotics and ongoing translation in the cell lysate affect the pausing landscape in ribosome profiling data.
In conclusion, by analyzing ribosome profiling data at higher resolution, we have obtained a more accurate view of translational pausing in bacteria. Although ribosome profiling is a powerful tool for observing pauses at a global level, not all the potential pitfalls are understood. It is difficult to know when the method is accurately portraying what is happening in living cells given our uncertainty of how the pausing landscape ought to look. We anticipate that as findings from profiling studies are corroborated by genetic and biochemical methods, a more complete picture of ribosome pausing will emerge.
Experimental Procedures
Ribosome profiling
Libraries were prepared as described (Woolstenhulme et al., 2015) with a few modifications: an overnight culture grown in MOPS media supplemented with 1% glucose and other nutrients (Teknova) was diluted 1:100 into 400 mL fresh media and grown at 37 °C to an OD600 of 0.25. Cell pellets were cryogenically pulverized using a Spex 6870 freezer mill with 5 cycles of 1 min grinding at 5 Hz and 1 min cooling. Ribosome footprints 15 – 45 nt were gel purified, cloned, and sequenced. RNAseq libraries were created by mild alkaline hydrolysis of total RNA; fragments between 20 – 40 nt were cloned and sequenced.
Analyses of profiling data were performed with python scripts. Only genes with an average of one or more reads per codon were included. To determine the cross-correlation of ribosome occupancy and aSD affinity, we created an aSD-affinity profile by scanning overlapping eight nt windows across all coding sequences. The free energy of hybridization of the aSD sequence (CACCUCCU) and each octamer was predicted using RNAsubopt in the Vienna RNA package (Lorenz et al., 2011). The affinity for each octamer was assigned to its seventh position. The aSD-affinity profile and ribosome profile were cross-correlated using the numpy correlate function as described (Li et al., 2012). To quantify the relationship between aSD affinity and ribosome occupancy, we first computed the lowest energy of hybridization of each RNA hexamer to the aSD sequence. We then calculated the average ribosome occupancy 23 – 28 nt downstream of the first nt in the hexamer.
Toeprinting analyses
The in vitro translation constructs contain a constant region followed by a 33 nt sequence from an E. coli gene containing an SD motif. Toeprinting assays were performed using the PURExpress system (New England Biolabs) as described in detail in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
The authors thank Gene-Wei Li and Jonathan Weissman for stimulating discussions and advice on the profiling protocol. This work was funded by NIH grant GM110113 to AB. FM and CW were supported by NIH training grant GM007445 and the Protein Translation Research Network (NIH grant GM105816), respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The GEO accession number for the sequencing data reported in this paper is GSE72899.
Author Contributions
Conceptualization, F.M., R.G., and A.B.; Methodology, F.M., C.W., and A.B.; Investigation, F.M., C.W., and A.B.; Writing – Original Draft, A.B.; Writing – Review & Editing, F.M., R.G., and A.B; Supervision, A.B.
References
- Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42:13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg A, Ehrenberg M. Determinants of the rate of mRNA translocation in bacterial protein synthesis. J Mol Biol. 2015;427:1835–1847. doi: 10.1016/j.jmb.2014.10.027. [DOI] [PubMed] [Google Scholar]
- Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV. Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014;157:1619–1631. doi: 10.1016/j.cell.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan N, Peske F, Rodnina MV. Changed in translation: mRNA recoding by −1 programmed ribosomal frameshifting. Trends Biochem Sci. 2015;40:265–274. doi: 10.1016/j.tibs.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Johansson M, Tsai A, O'Leary SE, Puglisi JD. Dynamic pathways of −1 translational frameshifting. Nature. 2014;512:328–332. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3' untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes & development. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Chiba S. Arrest Peptides: Cis-Acting Modulators of Translation. Annual Review of Biochemistry. 2013;82 doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, Barral JM, Skach WR. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348:444–448. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- Larsen B, Wills NM, Gesteland RF, Atkins JF. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Han Y, Qian SB. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms for molecular biology : AMB. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012;22:2219–2229. doi: 10.1101/gr.133249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi K, Takai Y, Shiwa Y, Wada M, Honma M, Yoshikawa H, Tomita M, Kanai A, Mori H. Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC genomics. 2014;15:1115. doi: 10.1186/1471-2164-15-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor PB, Li GW, Weissman JS, Atkins JF, Baranov PV. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 2013;29:1488–1491. doi: 10.1093/bioinformatics/btt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orelle C, Carlson S, Kaushal B, Almutairi MM, Liu H, Ochabowicz A, Quan S, Pham VC, Squires CL, Murphy BT, et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob Agents Chemother. 2013;57:5994–6004. doi: 10.1128/AAC.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MS, Wang Z, Gaba A, Fang P, Belk J, Ganesan R, Amrani N, Jacobson A. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods (San Diego, Calif. 2002;26:105–114. doi: 10.1016/S1046-2023(02)00013-0. [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. The A-Z of bacterial translation inhibitors. Critical reviews in biochemistry and molecular biology. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell reports. 2015;11:13–21. doi: 10.1016/j.celrep.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci U S A. 2013;110:E878–887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wen JD, Bustamante C, Tinoco I., Jr. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3'UTRs In Vivo. Cell. 2015;162:872–884. doi: 10.1016/j.cell.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr Opin Struct Biol. 2011;21:25–31. doi: 10.1016/j.sbi.2010.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.