Summary
Ribosome profiling is a powerful method for globally assessing the activity of ribosomes in a cell. Despite its application in many organisms, ribosome profiling studies in bacteria have struggled to obtain the resolution necessary to precisely define translational pauses. Here we report improvements that yield much higher resolution in E. coli profiling data, enabling us to more accurately assess ribosome pausing and refine earlier studies of the impact of polyproline motifs on elongation. We comprehensively characterize pausing at proline-rich motifs in the absence of elongation factor EFP. We find that only a small fraction of genes with strong pausing motifs have reduced ribosome density downstream and identify features that explain this phenomenon. These features allow us to predict which proteins likely have reduced output in the efp knockout strain.
Introduction
Ribosome profiling is a powerful approach for analyzing the mechanism of protein synthesis in living cells (Ingolia et al., 2009). In this approach, ribosome footprints are determined globally by isolating and deep sequencing protected mRNA fragments. Ideally, ribosome profiling reveals the position of the ribosome at single-nucleotide resolution, as is routinely observed in yeast (Ingolia et al., 2009). Sites of high ribosome occupancy within ORFs correspond to translational pauses (Artieri and Fraser, 2014; Ingolia et al., 2011) that can arise from the interaction of the nascent polypeptide with the ribosome (Dimitrova et al., 2009; Nakatogawa and Ito, 2002) or from mRNA elements such as high secondary structure or stretches of rare codons (Doma and Parker, 2006; Roche and Sauer, 1999). Variations in elongation rates have been broadly implicated in gene regulation (Ito and Chiba, 2013), codon bias (Plotkin and Kudla, 2011), mRNA decay (Doma and Parker, 2006), and protein folding (Zhang et al., 2009). Ribosome profiling may provide a means to further flesh out these promising links between changes in elongation rate and important biological outcomes.
There has been considerable focus recently on pausing at consecutive proline codons due to proline’s tendency to act as both a poor peptidyl donor at the end of the nascent chain and as a poor peptidyl acceptor on the incoming aminoacyl-tRNA (Pavlov et al., 2009; Wohlgemuth et al., 2008). In all forms of life, a specific factor facilitates elongation through consecutive Pro codons. EFP in bacteria and (e/a)IF5A in eukarya and archaea enter paused ribosomes where they enhance peptidyl-transfer rates through direct engagement of the peptidyl-transferase center or the peptidyl-tRNA itself (Doerfel et al., 2013; Gutierrez et al., 2013; Ude et al., 2013). In E. coli, the activity of EFP depends on an extension of the Lys34 side chain with a β-lysine moiety by two EFP-modifying enzymes, EpmA and EpmB (Navarre et al., 2010; Yanagisawa et al., 2010). A third enzyme, EpmC, adds a hydroxyl group to this modified Lys residue, although the biological significance of this additional modification is unclear (Peil et al., 2012).
Since EFP is critical for rapid translation of polyproline stretches (Doerfel et al., 2013; Ude et al., 2013) and many such motifs occur in essential genes in E. coli, it is surprising that loss of EFP causes only a modest reduction in growth rate and a few relatively minor phenotypes (Zou et al., 2012). Indeed, mass spectrometry studies have found that the levels of most PPP-containing proteins are not substantially altered by deletion of efp (Hersch et al., 2013; Peil et al., 2013). Given this discrepancy between the behavior of the knockout strains and data obtained in biochemical studies, it is clear that we do not yet fully understand the determinants of polyproline pausing in vivo and how such pauses in turn affect the level of protein output.
Here we report analyses of translational pausing based on ribosome profiling data from E. coli cells lacking EFP. Despite its high resolution in yeast, ribosome profiling has suffered from poor resolution in other systems, including E. coli. Through a few changes in the profiling method, we obtained profiling data at much higher resolution, allowing us to characterize pausing in bacteria with unprecedented clarity. Through our analysis, we define the characteristics of genes with reduced protein output in the efp mutant and propose a model for how these determinants are integrated at the molecular level.
Results and Discussion
Preparation of bacterial profiling samples
We generated fifteen ribosome profiling libraries from E. coli K-12 MG1655 and its mutants in which genes encoding EFP or its modifying enzymes were deleted (Table S1). We obtained biological replicates of the wild-type and efp mutant libraries in Luria broth and a complete MOPS medium. In isolating and cloning ribosome footprints, we followed the published bacterial protocol (Li et al., 2012; Oh et al., 2011) with a few modifications as described in the Experimental Procedures. Our improved resolution does not arise from changes in the library preparation procedure but from changes in the computational methods.
Assigning density to the 3′-end of reads reveals the ribosome’s position precisely
The poor resolution in bacterial ribosome profiling data contrasts sharply with the precision obtained in experiments in yeast, where RNase I is used to generate ribosome footprints. Since RNase I reliably degrades mRNA to the edge of the ribosome, assigning ribosome occupancy to the 5′-end of the reads conveys precise information about the position of the ribosome on the mRNA and the reading frame of translation (Ingolia et al., 2009). This is especially true for mRNA fragments 28 nt in length that are fully trimmed by the nuclease. In contrast, because RNase I is inhibited by bacterial ribosomes (Datta and Burma, 1972), the bacterial protocol uses micrococcal nuclease (MNase), an enzyme known to cleave RNA in a sequence selective manner (Dingwall et al., 1981). MNase yields a broad distribution of fragment lengths, none of which is superior at conveying information about position or reading frame. Given the apparent variation at both ends of the RNA fragments, in early bacterial profiling studies, ribosome occupancy was broadly distributed across the center of the reads and not assigned specifically to the 5′- or 3′-end (Li et al., 2012; Oh et al., 2011). This has the undesirable effect of blurring the signal.
Building on the finding that most of the variation occurs at the 5′-end of the reads (O’Connor et al., 2013), we found that assigning ribosome density to the 3′-end yields precise information about ribosome position. In plots of average density derived from genes aligned at their start codons (Figure 1A), the peak corresponding to initiating ribosomes is about 20 nt wide using the center-assignment method. 5′-assignment also yields a distribution more than 10 nt wide, now shifted upstream of the start codon, as anticipated. In contrast, 3′-assignment reveals a strong peak downstream of the start codon and its width is only 1 – 2 nt. As shown in Figure 1B, the average density at stop codons follows the same pattern: center- and 5′-assignment blur the position of terminating ribosomes that is sharply defined when reads are assigned to their 3′-ends.
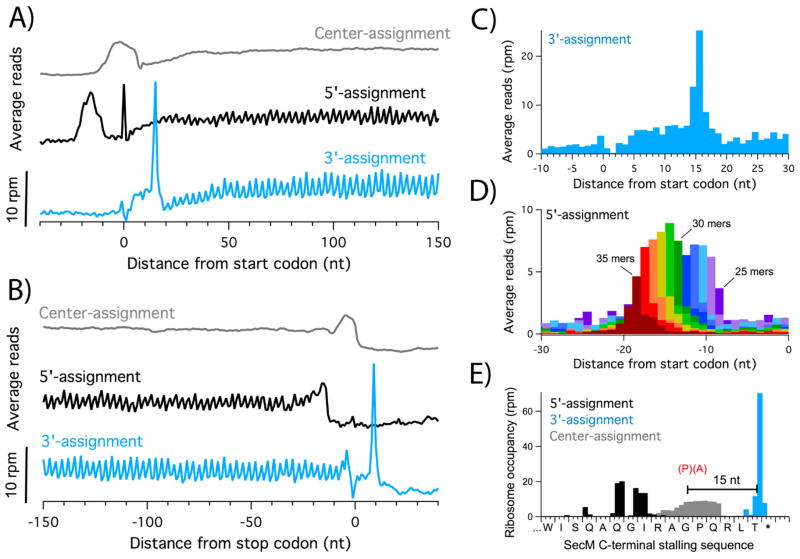
Figure 1.
Three strategies for assigning ribosome occupancy. A) Average ribosome occupancy aligned at the start codon. Reads from the Δefp3 dataset were assigned using the center-assignment strategy (grey) or the 5′ or 3′-ends of the reads (black and blue, respectively). B) Average ribosome occupancy aligned at the stop codon. C) Expanded view at the start codon using the 3′-assignment method. D) Average density aligned at start codons using the 5′-assignment strategy. Each plot represents reads of a single length; eleven plots are shown in different colors for 25 – 35 nt reads. E) Comparison of the three assignment strategies at the stalling motif in secM, where the ribosome stalls with the Gly codon in the P site.
Not only is 3′-assignment more precise, it is also more accurate than the alternative methods. A more detailed view of the average density aligned at start codons reveals that the initiation peak is 15 nt downstream of the first nucleotide of the start codon (Figure 1C); this corresponds to the distance from the P-site codon to the 3′-boundary of the ribosome as established by structural and biochemical experiments (Hartz et al., 1988; Yusupova et al., 2001). The fact that these distances match existing biochemistry and structure so closely suggests that MNase degrades mRNA fragments right up to the 3′-boundary protected by the ribosome. As such, the observed variation in read length must occur primarily at the 5′-end. We verified this by assigning ribosome density to the 5′-end using reads of a single length and found that, indeed, the initiation peak differs by a single nucleotide as a function of increasing read length (Figure 1D). This heterogeneity limits the accuracy of 5′- and center-assignment methods.
As a clear illustration of these assignment methods, ribosome density on the translational pause site in the secM gene is shown in Figure 1E. Although it would be difficult to determine the exact site of stalling using either the center- or 5′-assignment methods because of the width of the ribosome density, the 3′-assigned reads map to a single nucleotide 15 nt downstream of the glycine codon in the sequence RAGP. This peak correlates with the site of stalling determined using a variety of biochemical and structural approaches (Bhushan et al., 2011; Muto et al., 2006). Taken together, these findings establish that the 3′-end of reads in bacterial ribosome profiling data contain precise and accurate information about the position of the ribosome that is discarded by assigning occupancy to the center of reads. This analysis holds true for other published bacterial ribosome profiling data as well. Indeed, we are not alone in making this observation: a recent study of the function of LepA in E. coli employed the 3′-assignment method to analyze pausing at stop codons and Gly codons with high precision (Balakrishnan et al., 2014).
Variability in pausing at Pro-Pro motifs in bacteria lacking EFP
Our ability to assign ribosome footprints at higher resolution yields dramatic results in the analysis of pauses at polyproline motifs. Because the signal is no longer blurred, the signal to noise ratio is higher, substantially raising the pause scores and making it easier to identify pauses with confidence. For example, in the absence of EFP, the ribosome density is strongly enriched at the RPPP sequence in recG with a pause score of 381 in the Δefp1 dataset and 475 in the Δefp2 dataset (Figure 2A). The density is highest at the first nucleotide of the second Pro codon (Figure 2A, inset), consistent with previous observations that stalling occurs with the second Pro codon in the P site and the third in the A site (Doerfel et al., 2013; Peil et al., 2013; Tanner et al., 2009; Woolstenhulme et al., 2013).
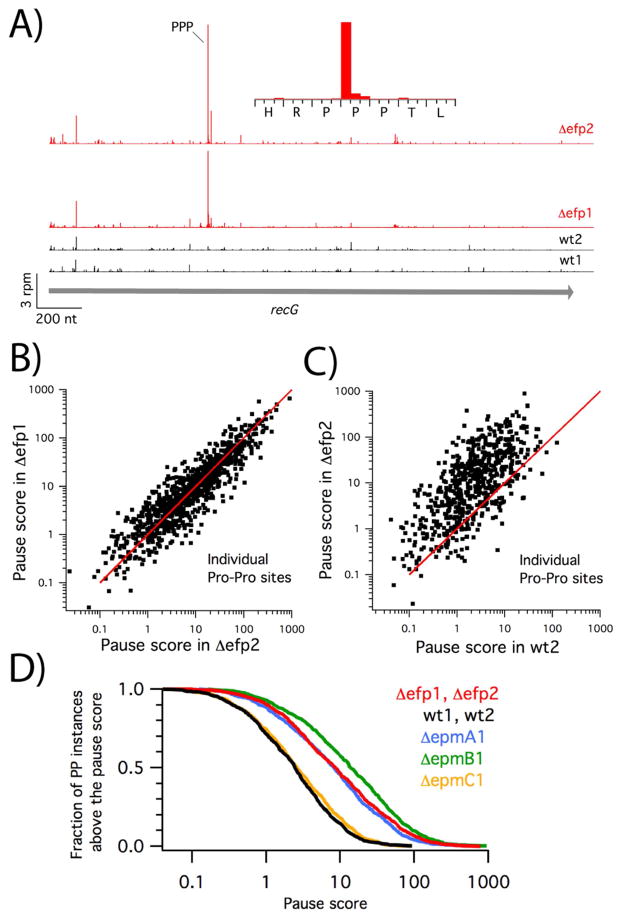
Figure 2.
Ribosome pausing at Pro-Pro motifs. A) Ribosome density in the recG gene for two biological replicates of wild-type (black) and efp knockout cells (red). Inset: expanded view of the strong pause site in the Δefp2 data. B) Correlation of pause scores at 1537 individual instances of Pro-Pro motifs in two biological replicates of the efp knockout strain. The red line represents y = x. C) Pause scores at individual instances of Pro-Pro motifs in the wt2 and Δefp2 data. D) Fraction of Pro-Pro instances with a pause score above a given strength, calculated from the average of the Δefp1 and Δefp2 data (red), wt1 and wt2 data (black), or from the ΔepmA1 (blue), ΔepmB1 (green), or ΔepmC1 data (yellow).
Analyzing pausing at Pro-Pro motifs across the transcriptome in cells lacking EFP, we observe pause scores with a range of over 1,000-fold (Figure 2B). The fact that these scores are reproducible in two biological replicates, Δefp1 and Δefp2, suggests that random error is not a main source of this variation. It seems likely that biases in library preparation may partially explain the variation: as observed in other profiling data, we see a large range of peak intensities across a given gene, as high as 1,000-fold on highly translated genes. From RNAseq experiments performed in parallel, we know that certain sequences are enriched while others are suppressed during library cloning. Given these concerns, in characterizing ribosome pausing, we are careful to use approaches that minimize noise, calculating averages rather than focusing on individual pausing sites.
EFP affects pause strength at a vast majority of proline-rich motifs
We find that loss of EFP amplifies pausing at the vast majority of instances of Pro-Pro in the genome. In a plot comparing the pause score at a given instance of Pro-Pro in the Δefp2 dataset with the score in the wt2 dataset, the score is almost always markedly higher when EFP is lacking (Figure 2C). Taking the ratio of the pause score at single sites in the mutant versus the wild-type dataset, we calculate that, on average, pausing is about 10-fold higher at Pro-Pro motifs in the absence of EFP.
A recent profiling study came to the conclusion that only a small fraction (2.8%) of Pro-Pro motifs induced significant pauses in an efp knockout strain (Elgamal et al., 2014). In this study, the authors defined significant pauses as greater than 10-fold above the gene average. In our analyses, pausing signals are generally higher because the 3′-assignment strategy gives us higher resolution. In our data, a cumulative distribution plot of the fraction of Pro-Pro sites higher than a given pause score reveals that about 47% of Pro-Pro instances have a pause score above 10 in the efp mutant (Figure 2D).
We find a similar enrichment in global Pro-Pro pausing in bacteria lacking the EFP-modifying enzymes EpmA and EpmB (also known as PoxA and YjeK respectively), but not in bacteria lacking EpmC (YfcM). These data are consistent with earlier studies showing that modification of EFP with the β-lysyl moiety is essential for its stall-relieving function, whereas hydroxylation of the modified Lys34 side chain is not (Doerfel et al., 2013; Navarre et al., 2010; Peil et al., 2013; Ude et al., 2013).
The sequence context determines pause strength at Pro codons
Our data allow us to discern rules that govern pausing at polyproline motifs in vivo. In earlier work, we and others established that a major determinant of pausing at Pro-Pro motifs is the identity of the aminoacyl-tRNA reacting at the A site when two Pro residues have been incorporated into the nascent peptide (Doerfel et al., 2013; Peil et al., 2013; Tanner et al., 2009; Woolstenhulme et al., 2013). In our analysis of pausing at PP(X), where the parentheses denote that the X codon is positioned in the A site, we observe very strong pauses at PPW, PPN, PPD, and PPP (Figure 3A); for example, the average pause at PPW is 90-fold above the gene average, suggesting a substantial inhibition of elongation at this motif. No enrichment of density at these sequences is seen in the corresponding RNAseq data, confirming that the high pause scores are not due to cloning bias (Figure S1). PPW, PPN, PPD, and PPP also induce the strongest pauses in toeprinting experiments (Woolstenhulme et al., 2013) and reduce protein expression when placed in front of a lacZ reporter gene (Peil et al., 2013). The weakest pauses are observed where the aminoacyl-tRNA recruited to the A site has a hydrophobic or aromatic side chain: PPI, PPL, and PPM are weak (~ 6.8, 5.6, and 3.0 respectively) and PPY and PPF have minimal pause scores of 1.6.
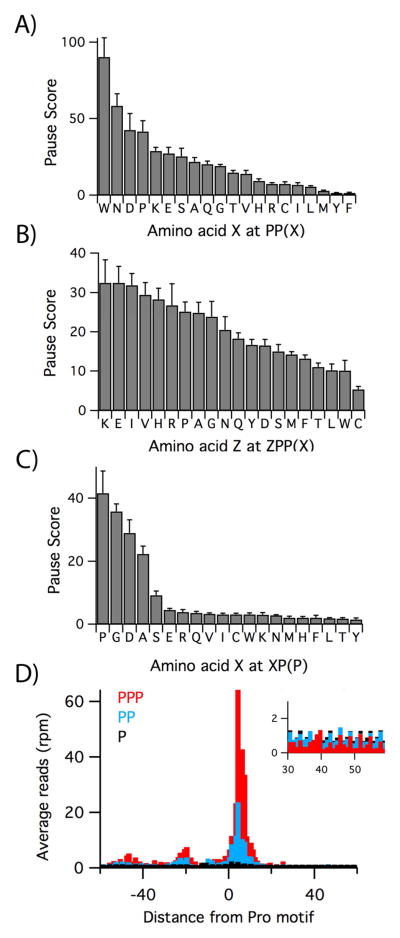
Figure 3.
Effect of protein context on proline pauses. A) Pause scores for PP(X) where X is the codon in the ribosomal A site. These numbers represent the average of the scores computed for each of the four efp knockout datasets, with standard error of the mean. B) Effect of the −3 residue (Z) in the nascent polypeptide on pausing at ZPP(X). C) Effect of the −2 residue in the nascent peptide on pausing at Pro-Pro, with the second Pro codon in the A site. D) Average ribosome density at one, two, or three Pro codons in the combined Δefp3 and Δefp4 data. Inset: close-up of the density 30 – 60 nt downstream of the pause site. See also Figure S1 and Table S2.
The sequence of the peptide upstream of the stalling motif also affects the intensity of pausing. As a further refinement of our analysis of pausing at PP(X) motifs, we found that the residue just upstream, Z in ZPP(X), has a small effect on the average pause score (Figure 3B). In general, motifs associated with high pausing scores have Lys, Glu, Ile, Val, His, Arg, and Pro at the Z (−3) position and were observed to reduce lacZ expression in a reporter system (Starosta et al., 2014a). We note that the presence of Arg at the −3 position enhances stalling in the RXP(P) motif characterized in our previous study (Woolstenhulme et al., 2013) and at the RPPP pause in recG in Figure 2A. In contrast, three amino acids that yield lower pause scores when placed at the −3 position, Cys, Leu, and Thr, suppress stalling on the PPP motif and partially restore lacZ expression (Starosta et al., 2014a).
Robust stalling can also occur at consecutive Pro codons in the ribosomal P and A sites. We calculated the average pause score for the XP(P) motifs and found that in addition to PPP, seen above, strong pauses occur at GPP, DPP, and APP and a weaker pause at SPP (Figure 3C). GPP, DPP, and APP have the strongest effect of any XP(P) sequence on lacZ reporter expression (Peil et al., 2013). These data suggest that the penultimate residue in the nascent chain can inhibit the reactivity of Pro-tRNAPro bound in the A site. Taken together, these data help refine our understanding of how protein context affects pausing at Pro-Pro motifs.
The consequences of pausing on upstream and downstream ribosome occupancy
Comparison of the ribosome occupancy averaged at one, two, or three consecutive Pro codons in the Δefp3 and Δefp4 datasets reveals that pausing can have broad consequences for protein synthesis (Figure 3D). Pro has a relatively high pause score in the P site, even in the wild-type strain, suggesting that EFP is not fully efficient in relieving pausing (see Table S2). Pausing is ten-fold higher (24) with two consecutive Pro codons and higher still (64) for three. Upstream of these large pauses, two additional peaks appear in the PP and PPP plots; the spacing of these peaks, roughly 25 nt apart, is consistent with two ribosomes stacked behind the Pro-stalled ribosome. We also note that there is a reduction of density downstream of the PP and PPP stalling motifs. For a single Pro codon, the average density 20 – 60 nt downstream is 0.98, close to the expected value of 1.0. With two Pro codons, however, the downstream density drops slightly to 0.90, and with three, it drops to 0.52 (see Figure 3D, inset). These data are consistent with a model in which pausing at the Pro motif leads to ribosome queuing; downstream ribosomes continue translation, leaving an exposed segment of mRNA where ribosome occupancy is diminished.
Pausing reduces downstream ribosome density in a fraction of genes
In certain genes, ribosome density sharply declines downstream of a polyproline motif in the efp knockout strain. For example, in the cysQ gene, a pause is observed at the PPG motif, with 11-fold higher ribosome density upstream of this motif than downstream (Figure 4A, top); we define this ratio as the asymmetry score (AS). This asymmetry is the result of the loss of EFP since in the wild-type strain, strong ribosome density is found distributed throughout the gene. In contrast to these dramatic changes in cysQ, however, the large majority of genes do not show a reduction in ribosome density downstream of polyproline motifs in the absence of EFP. Ribosome density is the same before and after the PPD motif in the katE gene, for example, despite the existence of a strong pause (Figure 4A, bottom).
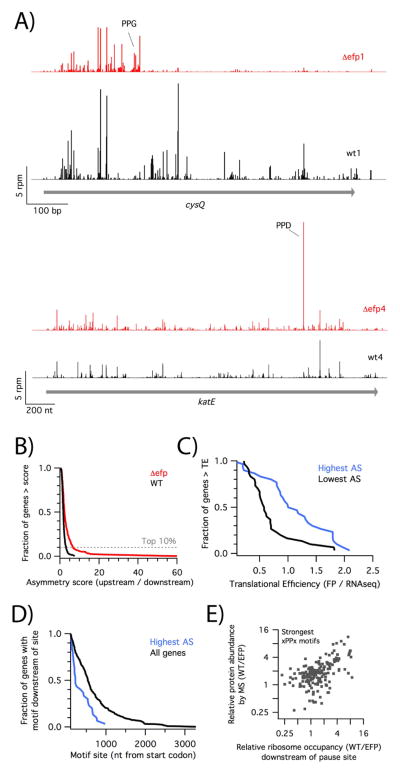
Figure 4.
Reduced ribosome density downstream of pause sites. A) Ribosome occupancy on the cysQ and katE genes in wild-type (black) and efp mutant data (red). B) For a subset of 300 genes containing strong pausing motifs, we calculated an asymmetry score (AS), the density upstream of the pause divided by the density downstream. The fraction of genes above a given AS is plotted for the combined four wild-type (black) or efp mutant datasets (red). C) For 30 genes with the highest or lowest AS in the efp knockout, we plotted the fraction of genes with more than the given translational efficiency. D) For 30 genes with the highest AS in the efp knockout, the fraction of genes with the pausing motif after a specific position was plotted. E) Correlation of the relative protein abundance in MS data vs the relative ribosome occupancy downstream of the pause site for genes with strong pausing motifs. See also Figure S2 and Table S3.
To better understand why the loss of EFP results in asymmetric ribosome density on some genes but not others, we analyzed the profiles of 300 genes containing the strongest Pro-containing stalling motifs (PPW, PPN, PPD, PPP, PPG and GPP, DPP, and APP). We focused on these because we observed that strong pauses occur commonly in genes where ribosome density is reduced downstream of the motif (Figure S2) but are not sufficient in and of themselves to explain this change. To help identify other contributing factors, we ranked these 300 genes carrying a strong pausing motif by their asymmetry score. About 10% of these genes have high asymmetry scores in the efp knockout strain (AS > 6.5, Figure 4B).
We identify three factors that explain why some genes have higher asymmetry scores. First, genes with reduced ribosome density downstream of pausing motifs (i.e. high AS scores) tend to be highly translated. The defects that we see in elongation may lower the rate from the normal 20 codons per second into the range of values modeled for initiation (0.004 – 4 s−1) (Subramaniam et al., 2014). Genes with high initiation rates may be particularly vulnerable, as elongation is more likely to become rate limiting for the overall translation cycle. For our 300 genes with strong pausing motifs, we calculated the translational efficiency in the wild-type strain by dividing ribosome density by RNAseq density. We found that genes displaying the highest asymmetry tend to be translated more efficiently than those with the lowest asymmetry (Figure 4C).
These data are consistent with the recent observation by Navarre and co-workers that increasing the strength of the Shine-Dalgarno sequence in the atpA gene increases its dependence on EFP (Hersch et al., 2014). They propose that ribosomes form a queue that eventually extends as far back as the start codon, preventing subsequent 30S ribosomes from loading on the mRNA. These traffic jams are intensified by high rates of initiation. Our data expand on their findings, showing that it is generally true that genes in which ribosome density is depleted downstream of strong pauses tend to have higher TE and presumably higher rates of initiation.
Second, genes with high asymmetry scores tend to have strong pause sites early in the ORF (Figure 4D). In 14 of the top 30 genes, the motif is found less than 200 nt downstream from the start codon. Because the AS is computed using density upstream of the pause site, we cannot reliably calculate scores for motifs within 100 nt of the start codon. However, by inspection we found that seven of 34 genes with pausing motifs within 100 nt of the start codon showed very strong pauses and pronounced depletion of ribosome density compared to the wild-type. These observations about the polarity of pause site location are consistent with a model in which pausing leads to a queue of ribosomes that sterically blocks initiation. Given that queues of only 2 – 3 stacked ribosomes are observed in our data and elsewhere (Subramaniam et al., 2014), the likelihood of initiation being blocked is higher when the pause site is close to the start codon.
Lastly, five of the ten genes with top asymmetry scores have compound motifs (made up of two or more consecutive strong motifs). For example, the ribosome could stall either at PPP or PPN in the extended PPPN motif in the valS gene, at PPP or PPW in the PPPW motif in gntX, or at GPP, PPP or PPG in the GPPPG motif in gpr. In these compound motifs, the ribosome is expected to pause at consecutive codons. This result underscores the importance of the strength of the stalling motif in reducing downstream ribosome density. If stacking of ribosomes behind the paused ribosome is an important mechanism for blocking initiation, then increasing the length of the pause at a single site early in the ORF might have more of an effect on gene expression than adding strong pausing motifs further downstream.
These three factors may rationalize why pausing reduces downstream ribosome density so strongly on the cysQ transcript (Figure 4A). Ribosomes pause at the compound motif DPPG in this protein, composed of two strong pausing motifs. This motif is positioned early in the gene, 199 nt downstream from the start codon, and cysQ is relatively well translated (in the top 10th percentile, with a translational efficiency of 2.1). The strong asymmetry score for cysQ likely has consequences for its expression. Indeed, we observe that there is seven-fold more density downstream of the pausing motif in the wild-type strain than in the efp mutant. For genes where there is differential downstream density, our data predict that protein output will be reduced by proline-stimulated pausing in the absence of EFP.
Proteins whose level is sensitive to the loss of EFP
To further define the effects of pausing on protein output, we compared our profiling results with quantitative mass spectrometry data obtained by Wilson and co-workers from strains nearly identical to ours (Peil et al., 2013). We wondered whether relative ribosome occupancy (RRO) downstream of a pause site (comparing the wild type vs. mutant strain) would correlate with effects on protein output as evaluated by mass spectrometry. This metric was used instead of the asymmetry score because it explicitly compares ribosome footprints between the wild-type and efp mutant cells and is thus inherently sensitive to differences in mRNA concentration between the two strains. For the 178 proteins with strong pausing motifs for which MS and profiling data are both sufficiently deep, we observe a good correlation of r = 0.63 (Figure 4E), suggesting that both methods broadly identify the same set of genes as sensitive to the loss of EFP.
We next asked whether our data can predict additional proteins whose output is likely to be affected by depletion of EFP. Indeed, the depth of our datasets allows us to compute RRO and asymmetry scores for 801 of the 1424 genes encoding proteins with PP motifs. In contrast, the output ratios for only 482 of these proteins could be determined in the SILAC MS study. As presented in Table S3, we document an RRO score of two or more on 126 genes, 42 of which were not detected by MS; we would predict that many of these would exhibit diminished output in the efp knockout strain. For example, we find that the CadC protein has an RRO score of 11 and the highest asymmetry score (84), making it a likely candidate for reduced expression in the EFP knockout strain. Although it was not observed in the MS study, given the importance of this protein in the discovery of the function of EFP and its modifying enzymes (Ude et al., 2013), it certainly makes sense that CadC is one of the most EFP-dependent proteins in the cell. We also note that valS, encoding the valyl-tRNA synthetase, has an RRO score of 5 and an asymmetry score of 29. The PPP motif in the ValS protein is essential for its function and is highly conserved (Starosta et al., 2014b). It seems likely that stalling at the PPP motif in the absence of EFP lowers the levels of ValS and charged Val-tRNAVal. This is consistent with our observation of increased ribosome density with Val codons in the A site: in the LB data, we observe pause scores of 2.7 in the efp knockout and 1.2 in the wild-type strain. We anticipate that the increased depth and resolution of our analysis may help in deciphering phenotypes associated with loss of EFP in bacteria.
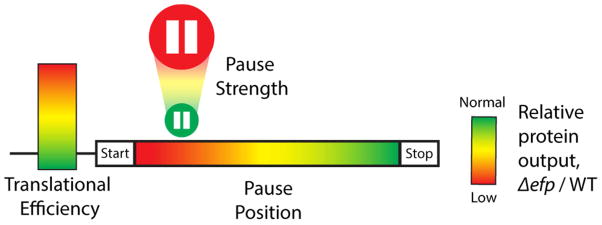
A model for how pausing during elongation affects protein output
Our improved computational approaches allow us to use ribosome profiling data in E. coli to define molecular features associated with pausing that contribute to overall reduced protein output. The strength of the pause, the location (5′ polarity) of the pause, and the translational efficiency of the gene all correlate with an impact on protein output (Figure 5). Our findings can be explained by a model in which strong pauses lead to ribosome queuing, which when proximal to the start site, in the context of a rapidly initiating ORF, leads to steric occlusion and thus to an overall reduction of the rate of initiation. This model expands the conclusions of an earlier study highlighting the importance of initiation rates in determining the effects of pausing (Hersch et al., 2014). There is no necessary link between pausing and reduced protein output, since elongation is not normally rate-limiting. We note that others who have correlated pause strength and protein output have used strongly expressed reporters with the pausing motifs near the 5′-end (Peil et al., 2013; Starosta et al., 2014a), an ideal set-up for favoring diminished protein output. With these new insights, future profiling studies should be more easily parsed to identify significant effects on the biology of the cell.
Figure 5.
Three factors that determine how pausing affects protein output. Translational efficiency is represented by a scale of low to high initiation levels; pause strength by the size of the pause button; and 5′-polarity by the position along the gene. Red represents reduced protein levels and green represents normal levels in the efp mutant.
Experimental Procedures
Bacterial strains and growth conditions
The ORFs encoding EFP and its modifying enzymes were deleted from E. coli K-12 MG1655 by Lambda Red recombination (Datsenko and Wanner, 2000). Cultures were grown to an OD600 of 0.4 at 37 °C in LB media or in MOPS media supplemented with 0.2% glucose, all 20 amino acids, and other nutrients (Teknova). Pauses with Ser codons in the A site were observed in the LB data, as were pauses at Ile codons in the MOPS data, presumably due to depletion of these amino acids during growth or other issues with media formulation.
Ribosome profiling
Sequencing data are available from the Gene Expression Omnibus, accession number GSE64488. Libraries were generated as described previously (Oh et al., 2011) except that ribosome protected fragments 20 – 40 nt in length were selected by polyacrylamide gel electrophoresis. Pause scores were calculated by taking the ribosome density at the first nucleotide of the codon of interest, dividing by the mean of the ribosome density for the entire gene, then averaging the score for all instances of the motif. Asymmetry scores were computed by taking the ratio of upstream / downstream density at strong pausing motifs. Details of library preparation, calculations, and other aspects of data processing are described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The 3′-end of footprints best defines occupancy in bacterial ribosome profiling
Though pausing at Pro-Pro motifs is widespread, context determines the intensity
Strong pauses near the 5′-end of genes reduce downstream ribosome density
Acknowledgments
The authors thank Edward Wallace, Rasi Subramaniam, Eric Mills, and Gene-Wei Li for helpful discussions and feedback. This work was supported by National Institutes of Health Grant GM110113 to ARB. CJW was supported by the Protein Translation Research Network funded by the National Institutes of Health Grant GM105816.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artieri CG, Fraser HB. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014;24:2011–2021. doi: 10.1101/gr.175893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan R, Oman K, Shoji S, Bundschuh R, Fredrick K. The conserved GTPase LepA contributes mainly to translation initiation in Escherichia coli. Nucleic Acids Res. 2014;42:13370–13383. doi: 10.1093/nar/gku1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, Berninghausen O, Wilson DN, Beckmann R. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta AK, Burma DP. Association of ribonuclease I with ribosomes and their subunits. J Biol Chem. 1972;247:6795–6801. [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Lomonossoff GP, Laskey RA. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981;9:2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgamal S, Katz A, Hersch SJ, Newsom D, White P, Navarre WW, Ibba M. EF-P dependent pauses integrate proximal and distal signals during translation. PLoS Genet. 2014;10:e1004553. doi: 10.1371/journal.pgen.1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Hersch SJ, Elgamal S, Katz A, Ibba M, Navarre WW. Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J Biol Chem. 2014;289:28160–28171. doi: 10.1074/jbc.M114.593277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SJ, Wang M, Zou SB, Moon KM, Foster LJ, Ibba M, Navarre WW. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. mBio. 2013;4:e00180–00113. doi: 10.1128/mBio.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Chiba S. Arrest Peptides: Cis-Acting Modulators of Translation. Annual Review of Biochemistry. 2013;82 doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, Edvokimova E, Prost LR, Kumar R, Ibba M, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor PB, Li GW, Weissman JS, Atkins JF, Baranov PV. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 2013;29:1488–1491. doi: 10.1093/bioinformatics/btt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci U S A. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Lassak J, Atkinson GC, Virumae K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci U S A. 2013;110:15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Virumae K, Atkinson GC, Tenson T, Remme J, Wilson DN. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat Chem Biol. 2012;8:695–697. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. Embo J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta AL, Lassak J, Peil L, Atkinson GC, Virumae K, Tenson T, Remme J, Jung K, Wilson DN. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res. 2014a;42:10711–10719. doi: 10.1093/nar/gku768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta AL, Lassak J, Peil L, Atkinson GC, Woolstenhulme CJ, Virumae K, Buskirk A, Tenson T, Remme J, Jung K, et al. A Conserved Proline Triplet in Val-tRNA Synthetase and the Origin of Elongation Factor P. Cell reports. 2014b;9:476–483. doi: 10.1016/j.celrep.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, O’Shea EK. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell. 2014;159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem. 2009;284:34809–34818. doi: 10.1074/jbc.M109.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci U S A. 2013;110:E878–887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nature structural & molecular biology. 2010;17:1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nature structural & molecular biology. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- Zou SB, Hersch SJ, Roy H, Wiggers JB, Leung AS, Buranyi S, Xie JL, Dare K, Ibba M, Navarre WW. Loss of elongation factor P disrupts bacterial outer membrane integrity. J Bacteriol. 2012;194:413–425. doi: 10.1128/JB.05864-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.