Abstract
In the present study a major protein has been purified from the venom of Indian Daboia russelii russelii using gel filtration, ion exchange and Rp-HPLC techniques. The purified protein, named daboxin P accounts for ~24% of the total protein of the crude venom and has a molecular mass of 13.597 kDa. It exhibits strong anticoagulant and phospholipase A2 activity but is devoid of any cytotoxic effect on the tested normal or cancerous cell lines. Its primary structure was deduced by N-terminal sequencing and chemical cleavage using Edman degradation and tandem mass spectrometry. It is composed of 121 amino acids with 14 cysteine residues and catalytically active His48 -Asp49 pair. The secondary structure of daboxin P constitutes 42.73% of α-helix and 12.36% of β-sheet. It is found to be stable at acidic (pH 3.0) and neutral pH (pH 7.0) and has a Tm value of 71.59 ± 0.46°C. Daboxin P exhibits anticoagulant effect under in-vitro and in-vivo conditions. It does not inhibit the catalytic activity of the serine proteases but inhibits the activation of factor X to factor Xa by the tenase complexes both in the presence and absence of phospholipids. It also inhibits the tenase complexes when active site residue (His48) was alkylated suggesting its non-enzymatic mode of anticoagulant activity. Moreover, it also inhibits prothrombinase complex when pre-incubated with factor Xa prior to factor Va addition. Fluorescence emission spectroscopy and affinity chromatography suggest the probable interaction of daboxin P with factor X and factor Xa. Molecular docking analysis reveals the interaction of the Ca+2 binding loop; helix C; anticoagulant region and C-terminal region of daboxin P with the heavy chain of factor Xa. This is the first report of a phospholipase A2 enzyme from Indian viper venom which targets both factor X and factor Xa for its anticoagulant activity.
Introduction
Haemostasis, one of the most important physiological processes of vertebrates, involves four crucial steps for sustaining equilibrium, namely, (i) vasoconstriction, to reduce blood flow from the site of injury (ii) platelet activation, aggregation and adherence leading to the platelet plug formation at the injured site (iii) initiation of coagulation cascade involving the extrinsic, intrinsic and the common pathway, forming fibrin mesh on the platelet plug and (iv) fibrinolysis leading to the dissolution of the clot formed, in order to restore normal blood flow [1]. Any malfunction in this vital process leads to two major pathophysiological conditions, haemorrhage or thrombosis, at large. Interestingly, the components of the haemostatic system of the prey/victim are one of the most vulnerable targets in snake envenomation. Owing to this, there has been a quest amongst the venom researchers to unfold the underlying mechanism and explore the therapeutic potentiality of these venom proteins since the last few decades. Recent trend of research reveals a pursuit for direct inhibitors of factor Xa (FXa) and thrombin from the venom of snakes and saliva of hematophagous animals as one of the most thrived components for anticoagulant and antithrombotic drug discovery [2–7].
Daboia russelii russelii (Daboia r. russelii), a venomous snake of the Indian subcontinent, contains an intricate blend of various biologically active protein families. Consumptive coagulopathy is one of the most manifested systemic pathological condition observed in viper envenomated victims [8]. The procoagulant venom proteins like RVV-X and RVV-V activators accelerate the blood coagulation cascade and the simultaneous down regulation of activated protein C causes severe depletion of the coagulation factors thereby depriving the victim of the serine proteases shortly [8]. This scenario is further worsened by the anticoagulant and haemorrhagic components like phospholipase A2 (PLA2) enzymes, L-amino acid oxidase (LAAO) and metalloproteases leading to hypovolemic shock and even death [9].
PLA2 enzymes are one of the major protein families in Russell’s viper proteome with various isoforms [10,11]. Although the members of this family share a highly conserved structural and catalytic scaffold, plethora of pharmacological activities are reported among the isoforms apart from prey digestion [12–15]. The subtle differences in their activities is mainly attributed to the presence of various pharmacological sites (responsible for non-enzymatic activity via protein-protein interaction) which are distinct from the catalytic site (responsible for enzymatic activity via protein-phospholipid interaction) [16]. The presence of minor amino acid variations in the exposed regions of these isoforms one of the vital causes of diverse pharmacological sites [3]. The ability of the snake venom PLA2 enzymes to inhibit blood coagulation was first reported by Boffa & Boffa in Vipera berus venom [17]. Henceforth, many anticoagulant PLA2 enzymes have been reported from snake venom by several researchers. These enzymes are classified into strong, weak and non-anticoagulant based on the concentration required to delay clot formation and amino acid residues in the predicted anticoagulant region (54th to 77th residues) [16,18]. These enzymes act either by hydrolyzing the procoagulant phospholipids or target the coagulation factors for its activity. CM-I and CM-II from Naja nigricollis and Vipoxin from Vipera ammodtes ammodytes hydrolyze the phospholipids required for the formation of the extrinsic and intrinsic tenase complex [19–21]. CM-IV from Naja nigricollis, AtxA from Vipera ammodytes ammodytes, CBc from Crotalus durissus terrificus and MtxII from Bothrops asper, target FXa of the prothrombinase complex and impede the conversion of prothrombin to thrombin [6,22]. While Nn-PLA2 from Naja naja and Nk-PLA2- β from Naja kaouthia target thrombin directly to exhibit anticoagulant effect [23,24]. On the other hand, VRV-PL-IIIb from Daboia russelii, CHA-E6b & CHA-E6a from Crotalus horridus and Tj-PLA2 from Trimeresurus jerdoni inhibit ADP and collagen induced platelet aggregation [25–27].
In the present study we have isolated and characterized a strong anticoagulant PLA2 enzyme, from the venom of Indian Daboia russelii russelii. Enzymatically it hydrolyses the phospholipids required for the formation of the tenase complexes and non-enzymatically it targets factor X (FX) and activated factor X (FXa) for exhibiting its anticoagulant activity. Hence, this purified anticoagulant PLA2 enzyme was named as daboxin P (Daboia russelii russelii FX inhibitor PLA2 enzyme).
Materials and Methods
Crude venom and chemicals/reagents
Crude venom of Daboia r. russelii (1 gm) was purchased from Irula Snake Catchers Society, Tamil Nadu, India. Few individuals (~3–4 individuals) of each snake species are caught from the forest and captivated for nearly 3–4 weeks before venom extraction. During this period venom is milked from the snakes for 4–5 times before releasing back to the forest. None of the captivated snakes are harmed or killed. Secretory phospholipase A2 (sPLA2) assay kit was obtained from Cayman Chemical Company (MI, USA). 4-vinyl pyridine, hydroxylamine hydrochloride, Cyanogen bromide activated Sepharose® 4B and bovine plasma fibrinogen were purchased from Sigma-Aldrich (MO, USA). BNPS-skatole [2-(2-Nitrophenylsulfenyl)-3-methyl-3-bromoindolenine)] was purchased from Bioworld (OH, USA). The Edman sequencing reagents were purchased from Applied Biosystems chemicals (Foster city, CA). The rest of the reagents and chemicals were of analytical grade and obtained from Merck Millipore (MA, USA) or Sigma (MO, USA).
Enzymes and chromogenic substrates
Serine proteases factor XIa (FXIa), factor IXa (FIXa), factor VIIa (FVIIa), factor X (FX), factor Xa (FXa) and Russell’s viper venom-X activator (RVV-X) were obtained from Haematologic Technologies Inc. (Vermont, USA), factor XIIa (FXIIa) was procured from Merck Calbiochem (Darmstadt, Germany), factor VIII (FVIII) from Creative Biomart (NY, USA), phospholipid blend from Avanti Polar Lipids Inc. (Alabama, USA) and tissue factor Innovin from Siemens (Murburg, Germany). The chromogenic substrates namely spectrozyme (MeSO2-D-CHG-Gly-Arg-pNA.AcOH) was purchased from Sekisui Diagnostics (MA, USA), rest of the substrates like S-2366 (pyroGlu-Pro-Arg-pNA•HCl), S-2302 (H-D-Pro-Phe-Arg-pNA•2HCl), S-2222 (Bz-IIe-Glu(γ-OR)-Gly-Arg-pNA•HCl), S-2765 (Z-D-Arg-Gly-Arg-pNA•2HCl)S-2238 (H-D-Phe-Pip-Arg-pNA•2HCl) and S-2288 (H-D-Ile-Pro-Arg-pNA•2HCl) were procured from Chromogenix (NJ,USA).
Cell culture
Human embryonic kidney (HEK)-293 and Michigan Cancer Foundation (MCF) -7 cell lines were purchased from National Centre for Cell Science (Pune, India). Dulbecco modified Eagle’s media (DMEM) was obtained from Himedia (Mumbai, India), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and trypan blue from Sigma Aldrich (Mo, USA), fetal bovine serum (FBS), streptomycin-penicillin (Strep-Pen), trypsin/EDTA solution and dimethyl sulfoxide (DMSO) were procured from Thermo Fischer (MA, USA).
Columns
HiloadTM 16/600 Superdex 75 prep grade and Hiprep CM FF 16/10 columns were purchased from GE Healthcare life Sciences (Bucks, UK). Jupiter C18 column was procured from Phenomenex (CA, USA) and Hypersil Gold C18 column was obtained from Thermo Scientific (MA, USA).
Purification of the major anticoagulant protein
Crude Daboia r. russelii venom was fractionated on HiloadTM 16/600 Superdex 75 prep grade column (1x120 ml) pre-equilibrated with 50 mM of Tris-Cl pH 7.4 using Äkta Purifier HPLC system, GE Healthcare (Uppsala, Sweden). Fractionation was carried out at a flow rate of 1 ml/min under isocratic conditions with the same buffer. Each eluted fraction was assayed for PLA2 and anticoagulant activity. P6 with highest anticoagulant and PLA2 activity was further subjected to cation exchange chromatography on Hiprep CM FF 16/10 (1.6x10 cm) using Äkta Purifier HPLC system. Elution was carried out at a flow rate of 2.25 ml/min with a linear gradient of 50 mM Tris-Cl, pH 7.4 containing 0.8 M NaCl. Ion exchange fraction, CM-II with highest PLA2 and anticoagulant activity was subjected to Rp-HPLC using Jupiter C18 column (3 μ, 4.6 x 250 mm, 300 Å). The column was pre-equilibrated with milli q containing 0.1% trifluoro acetic acid (TFA) and fractionated using 80% acetonitrile (MeCN) containing 0.1%TFA on Äkta Purifier HPLC system. The homogeneity of the purified protein (5 μg of the protein treated with 0.45 μl of β-mercaptoethanol) was validated on SDS-PAGE and stained using PierceTM silver staining kit, Life technologies, Thermo Fischer Scientific (MA, USA) [28]. This Rp-HPLC purified protein, named as daboxin P was used for characterization in the following experiments.
Biophysical characterization
Determination of molecular mass
Molecular mass of daboxin P was determined by electrospray ionization mass spectrometry (ESI-MS) using LCQ fleet Ion Trap, Thermo Scientific (MA, USA). Ion spray voltage was set at 4.4 kV. For nebulization nitrogen gas was used. Solvent (50% MeCN containing 0.1% formic acid) was delivered into the system at a flow rate of 50 μl/min by Accela 600 pump. Spectra were recorded in positive mode with mass to charge (m/z) ranging from 800 to 2000. Analysis and deconvolution of the spectra was performed by Promas for Xcaliber.
Primary structure determination
N-terminal sequencing: N-terminal sequencing of the native purified protein was performed by automated Edman degradation process using Protein sequencer PPSQ 31A Shimadzu Asia Pacific, (Singapore). Briefly, 89 μg (6545.56 pmol) of the purified protein (dissolved in 100 μl of milli q water) was dried on PVDF membrane and loaded onto the sequencer. Sequencing was performed for 30 cycles or till the chromatogram was readable.
Pyridylethylation: Prior to chemical cleavage, daboxin P was reduced and alkylated according to the method developed by Joseph and colleagues [29,30]. Briefly, 1 mg of daboxin P was dissolved in 920 μl of denaturing buffer (6 M guanidine hydrochloride and 0.1 M Tris-Cl pH 8.5) and treated with β-mercaptoethanol (30 μl). N2 gas was purged over the reaction mixture to expel any dissolved oxygen and incubated at 37°C for 150 min. 50 μl of 4-vinyl pyridine (containing 100 ppm hydroquinone) was added and further incubated at 37°C for 150 min. The pyridylethylated sample was desalted by Rp-HPLC on Jupiter C18 column (4.6 x 250 mm, 3 μm, 300 Å) with a linear gradient of 80% MeCN containing 0.1% TFA at a flow rate of 0.8 ml/min. Elution was monitored at 215 and 280 nm.
Chemical cleavage: The chemical cleavage of pyridylethylated protein by BNPS-skatole and hydroxylamine hydrochloride were carried out according to the protocol developed by Crimmins and co-workers and Milner and colleagues respectively with minor modifications [31,32]. For cleavage with BNPS-skatole, 900 μg of the pyridylethylated protein was dissolved in 0.1% of TFA containing 6 M guanidine hydrochloride (pH 5.0). To this 1.5 mg/ml of BNPS skatole (dissolved in 100% acetic acid) was added and incubated at 37°C for 24 h. The reaction was stopped by addition of equal volume of milli q water. Centrifugation was carried out at 12,000 rpm for 20 min and supernatant was collected.
For cleavage with hydroxylamine hydrochloride, 1 mg of pyridylethylated protein was dissolved in denaturing buffer (6 M guanidine hydrochloride, 0.1 M Tris-Cl, pH 9.0, 2 M hydroxylamine hydrochloride and 0.2 M K2CO3) and incubated at 45°C for 4 h. After the cleavage experiments, the samples were subjected to Jupiter C18 column using a linear gradient of 5–50% of 80% MeCN containing 0.1% TFA. The mass of the fractions obtained were checked using ESI-MS (Accela LCQ fleet ion trap, Thermo scientific, U.S.A) for the cleaved and uncleaved peptide fragments. The cleaved fractions were lyophilized and reconstituted in 100 μl of milli q water and loaded onto PVDF membrane for Edman degradation using Automated Protein sequencer PPSQ 31A, Shimadzu.
Enzymatic cleavage/ Tandem mass spectrometry: Daboxin P (50 μg) was treated with 50 mM of ammonium bicarbonate, 1% Protease Max and 0.5 M dithiothreitol at 56°C for 20 min. For alkylation, 550 mM of iodoacetamide was added and incubated in dark for 15 min. Following this, trypsin (1 μg/μl in 50 mM acetic acid) in the presence of 1% Protease Max was added to the reaction and incubated at 37°C for 3 h. Reaction was stopped by 100%TFA. Centrifugation was carried out at 12,000 rpm for 10 min and the supernatant was subjected to Tandem mass spectrometry as described previously [11]. The MS/MS spectra obtained were analyzed using Proteome Discoverer 3.1 with Sequest program.
Sequence alignment and phylogenetic analysis
PLA2 enzymes having maximum sequence similarity with daboxin P were retrieved by blastp analysis from the NCBI database and aligned using multiple sequence alignment software DNAMAN 4.1.5.1 (Lynnon BioSoft). The phylogenetic relationship among the PLA2 enzymes exhibiting anticoagulant activity were determined by the Mega 5.05 software using neighbor joining method with a bootstrap of 1000 replicates [33]. The evolutionary distance among the sequences was calculated with p-distance.
Secondary structure determination
The secondary structure was analyzed by circular dichroism (CD) spectroscopy using Jasco Spectropolarimeter J-810 (Tokyo, Japan). Briefly, 200 μl of 0.4 mg/ml of daboxin P (dissolved in milli q water) was subjected to the spectropolarimeter. The spectrum of the native protein was recorded at far UV scan from 260–190 nm at a speed of 50 nm/min in quartz cuvette (0.1cm path length) at room temperature.
The effect of different pH (phosphate buffered saline at pH 3.0, 7.4 & 12) and temperature (from 25°C to 100°C) on structural conformation of daboxin P (0.4 mg/ml) was determined under the same experimental conditions as described above. Three scans were recorded for each experimental condition and subtracted from blank to obtain the final spectra of the protein.
Biochemical and biological characterization
PLA2 activity
The PLA2 activity of daboxin P was assayed using the sPLA2 assay kit (Cayman, MI, USA) according to the manufacturer’s protocol using diheptanoylthio-phosphatidylcholine as substrate. The specific activity was expressed in micromoles of phosphatidylcholine hydrolyzed per min per mg of enzyme. Bee venom PLA2 enzyme (0.001 μg/μl) was considered as the positive control for the experiment. The results are mean ± standard deviation (SD) of three independent experiments.
Alkylation
The alkylation of His48 of daboxin P was performed as described by Maduwage and co-workers [34]. Briefly, 150 μg of daboxin P was treated with 15 μl of p-Bromophenacyl Bromide (pBpB) (100 μg dissolved in 30 μl of 100% ethanol). Equal amount of the protein was treated with 15 μl of ethanol as the vehicle control. The reaction mixtures were incubated at room temperature for 20 h and subsequently dialyzed in 20 mM of Tris-Cl, pH 7.4 at 4°C for 3 times. The dialyzed sample was lyophilized in a pre-conditioned lyophilizer (112N-G, Hahntech, South Korea) at –80°C. The lyophilized sample was reconstituted in appropriate buffer to carry out the desired assays. The extent of alkylation was verified by analyzing it using ESI-MS.
Cytotoxicity study
Cell culture: The HEK-293 and MCF-7 cell lines were grown in DMEM media enriched with 10% FBS and 1% antibiotic (Strep-Pen). The cells were grown on 96 well plate to a confluency of 70–80% at 37°C for 2–3 days in CO2 (5%) incubator (Eppendorf, Hamburg, Germany). Cell viability was assessed by trypan blue stain and the cells were quantified on a haemocytometer.
The cytotoxic activity of the crude venom of Daboia r. russelii and daboxin P were determined by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) based colorimetric method [35]. Briefly, in two independent set of experiments, the HEK-293 and MCF-7 cells were plated on 96 well plate at a concentration of 1x105 cells/well and incubated for 48 h at 37°C in CO2 (5%) incubator. Subsequently, the cells were incubated with different concentration of crude Daboia r. russelii venom and daboxin P for 24 h at 37°C in CO2 (5%) incubator. Cells treated with 0.9% NaCl was considered as the vehicle control. After incubation, the changes in morphological pattern (if any) of the treated cells were observed in an Inverted microscope (Axio Vert A1, Zeiss, Jena, Germany) at 10X magnification. 20 μl of MTT (5 mg/ml) was added and incubated at 37°C for 2 h. Following this, the formazan crystals formed by the viable cells were dissolved by 150 μl of MTT solution. The amount of formazan formed by the viable cells was quantified at 590 nm using MultiSkan Go spectrophotometer (Thermo Scientific, MA, USA). The percentage of cell viability was calculated by considering the cells without the venom treatment as 100% viable. The results are mean ± SD of three independent experiments.
In vitro-anticoagulant activities
The anticoagulant experiments were carried out with platelet poor human plasma (PPP) (whole blood centrifuged at 3000 rpm for 20 min at 16°C) and clot formation was monitored using Tulip Coastat-1 coaguloanalyser (Alto Santa Cruz, Goa, India). For each assay, clotting time of plasma with Tris-Cl buffer (20 mM, pH 7.4) was taken as the normal clotting time. The results are mean ± SD of three independent experiments.
Recalcification time (RT): Different concentrations of daboxin P (0.001, 0.01, 0.1, 1 μM) were pre-incubated with 150 μl of plasma at 37°C for 2 min [36]. Formation of plasma clot was initiated by addition of 100 μl of 50 mM CaCl2.
Activated partial thromboplastin time (APTT): APTT was determined using Liquecelin (APTT reagent) according to the manufacturer’s instructions. Briefly, different concentrations of the purified protein (0.01, 0.1, 1 μM) were pre-incubated with human plasma (50 μl) and APTT reagent (50 μl) for 3 min at 37°C. Plasma clot was formed by addition of 25 mM CaCl2 (50 μl).
Prothrombin time (PT): PT was measured using Uniplastin (PT reagent) according to the instructions of manufacturer. In brief, different concentrations of daboxin P were pre-incubated with 50 μl of human plasma at 37°C for 2 min. PT reagent (50 μl) was added to initiate the clot formation.
Stypven time: Briefly, in a reaction volume of 300 μl different concentrations of daboxin P were pre-incubated with human plasma (75 μl) for 3 min at 37°C [36]. To this 75 μl of RVV-X (10 ng/ml) was added and incubated for 2 min at the same temperature. Clotting was initiated by addition of 25 mM of CaCl2.
Thrombin time: To determine the thrombin time, different amounts of the protein (1, 3, 5 μg) were incubated with human plasma (50 μl) for 3 min at 37°C [36]. To this 50 μl of thrombin (10 u/ml) was added to initiate clot formation.
Fibrinogenolytic activity: Fibrinogenolytic activity was determined according to the method developed by Ouyang C and Teng CM [37]. In brief, different amounts of daboxin P were pre-incubated with 2 mg/ml fibrinogen (dissolved in 50 mM Tris-Cl, pH 7.5, 0.15 M NaCl) for 24 h at 37°C. For positive control, equal amount of fibrinogen was incubated with thrombin (3 μl of 10 units/ml) while for negative control fibrinogen with 50 mM Tris-Cl, pH 7.5 was incubated under the same experimental conditions. The formation of fibrinogen degradation products were analyzed on 12.5% glycine SDS-PAGE stained with Coomassie Brilliant Blue R-250.
In-vivo anticoagulant activity
The in-vivo anticoagulant activity of daboxin P was determined by the FeCl3-induced carotid artery thrombosis model with minor modifications [38–40]. Briefly, C57BL/6 male mice (9–11 weeks old, 24–28 g) were anesthetized with ketamine (75 mg/kg) and medetomidine (1 mg/kg) by intraperitoneal injection (i.p.). Daboxin P (10 mg/kg) was injected into the mice (n = 6) via its tail vein. Following this, the right carotid artery was exposed using blunt dissection and a doppler flow probe (Model MA0.5VB, Transonic System Inc., Ithaca, NY, USA) connected to a perivascular flow module (TS420, Transonic System Inc., Ithaca, NY, USA) was then attached to the carotid artery to monitor blood flow. A 2x2 mm piece of filter paper (soaked in 10% FeCl3 solution) was placed on the surface of the carotid artery for 3 min to initiate the thrombus formation. Saline treated mice (n = 5) were considered as the negative control for the experiment. Time-to-occlusion (TTO) is defined as the time taken for the blood flow to reach zero after the application of FeCl3. Maximum measurement time was considered for 60 min after the application of FeCl3. TTO was recorded as 60 min, if no occlusion occurred by this time. The animal experiments were approved by Institutional Animal Care and Use Committee, National University of Singapore
Serine protease specificity
The effect of daboxin P on the amidolytic activity of serine proteases were determined using specific chromogenic substrates. The activated serine proteases involved in the extrinsic and intrinsic tenase complex namely, FXIIa (60 nM), FXIa (0.125 nM), FIXa (333 nM), FXa (0.43 nM), FVIIa (10 nM), sTF (30 nM) were reconstituted in the activation buffer containing 50 mM Tris-Cl pH 7.4, 1 mg/ml of BSA, 140 mM NaCl and 5 mM CaCl2 [41]. The respective chromogenic substrates, S-2302 (1 mM), S-2366 (1 mM), spectrozyme (0.4 mM), S-2765 (0.65 mM) and S-2288 (500 μM) were reconstituted in milli q water to the desired working concentration. Briefly, to a reaction volume of 200 μl, each of the serine proteases was pre-incubated with different concentration of daboxin P (0.01, 0.1 and 1 μM) for 5 min. Subsequently, respective chromogenic substrates were added to trigger the amidolytic reaction. For each assay, the rate of p-nitroaniline formation upon hydrolysis of the substrate by the specific enzyme was quantified at 405 nm using UV-Vis MultiSkan GO spectrophotometer. The hydrolysis of substrate in the absence of daboxin P was considered as 100%.
Reconstitution of the tenase complexes
Extrinsic tenase complex (ETC) with and without phospholipids: The extrinsic tenase complex was reconstituted under in-vitro conditions in the presence of FVIIa (5 nM) and Innovin (10X) (tissue factor, thromboplastin and Ca+2 ions) with 67 μM of phospholipid vesicles (DOPC:DOPS 7:3) in the activation buffer. The reaction mixture was incubated at 37°C for 15 min [20]. Similarly, the complex was formed without phospholipids with 20 nM of FVIIa and soluble tissue factor, sTF (60 nM) and incubated for 15 min at 37°C.To both the experiments, different concentrations of daboxin P (0.01, 0.1, 1 and 3 μM) were added and incubated for 15 min. Following this, the reactions were incubated with FX (30 nM) for 15 min. Both the experiments were stopped by addition of quenching buffer (50 mM Tris-Cl pH 7.4, 1 mg/ml of BSA, 140 mM NaCl and 50 mM EDTA). The amount of FXa generated was determined by addition of 500 μM of S-2222 and the rate of hydrolysis was quantified at 405 nm using UV-Vis MultiSkan GO spectrophotometer.
Intrinsic tenase complex with and without phospholipids (ITC): Reconstitution of the intrinsic tenase complex under in-vitro conditions was carried out in the presence of phospholipids (67 μM) in activation buffer with FVIIIa, FIXa, FX and Ca+2 ions at 37°C [42,43]. Briefly, 3 nM of FVIII was incubated with 500 pM of thrombin for 10 min to activate FVIII. Thrombin was then deactivated by 10 nM of variegin, a thombin inhibitor from Amblyomma variegatum [44]. After incubation of the reaction mixture with 1 nM of FIXa for 10 min different concentrations of daboxin P (0.01, 0.1, 1 and 3 μM) were added and incubated for 15 min. Following this, 25 nM of FX was added and incubated for 15 min. Similarly, the complex was formed devoid of the phospholipid blend [42] with FVIII (40 nM) and thrombin (4 nM) and incubated for 10 min. Thrombin was deactivated with 40 nM of variegin. To this 10 nM of FIXa was added and incubated for 10 min. Then, different concentrations of daboxin P (0.01, 0.1, 1 and 3 μM) were incubated for 15 min. Finally, 1 μM of FX was added and incubated for 15 min. The reactions in both the experiments were terminated by addition of the quenching buffer. The amount of FXa generated was quantified by the hydrolysis of the chromogenic substrate S-2222 (500 μM) at 405 nm using UV-Vis MultiSkan GO spectrophotometer.
For determining the inhibitory concentration (IC50) of daboxin P on extrinsic and intrinsic tenase complex, different concentration of daboxin P were pre-incubated with each of the reconstituted complexes described above in the presence of phospholipids in the activation buffer.
Prothrombinase complex: Prothrombinase complex was reconstituted in the presence of phospholipid (67 μM) with FXa, FVa, and Ca+2 ions under in-vitro conditions at 37°C [19]. Briefly, 10 pM of FXa was pre-incubated with 1 nM of FVa for 15 min. To this, different concentrations of daboxin P (0.01, 0.1, 1 and 3 μM) were added and incubated for 15 min. Thereafter, prothrombin (12.5 nM) was added and the reaction mixture was incubated for another 15 min. In another set of experiment, FXa (10 pM) was pre-incubated with different concentrations of daboxin P followed by incubation with FVa (1 nM) and prothrombin (12.5 nM) for 15 min each. For both the experiments, the reactions were stopped with quenching buffer. The amount of thrombin generated by the complex was determined by the rate of hydrolysis of the chromogenic substrate S-2238 (250 μM) at 405 nm using UV-Vis MultiSkan GO spectrophotometer.
Fluorescence emission spectroscopy
Interaction of daboxin P with the coagulation factor FX and FXa were analyzed using fluorescence emission spectrophotometer (LS 55, Perkin Elmer, Palo Alto, CA). In brief, 1 μM of daboxin P (dissolved in 20 mM Tris-Cl, pH 7.4) was incubated with either 0.05 μM of FX or 0.1 μM of FXa (reconstituted in the same buffer) for different time intervals (10 min & 20 min) at room temperature in the presence and absence of 5 mM CaCl2. The change in the fluorescence emission spectra of the individual components and the mixtures were measured over a wavelength of 200 to 500 nm with an excitation wavelength of 280 nm at room temperature using a quartz cuvette (1 cm path length).
Affinity chromatography
The association of daboxin P with FX and FXa were evaluated by affinity column chromatography using CNBr activated sepharose® 4B (Sigma, Aldrich). Briefly, 40 μg of daboxin P was covalently linked to 0.1 g of CNBr activated matrix (swollen overnight in ice cold 1 mM HCl) in the presence of coupling buffer containing 0.1 M NaHCO3 and 0.5 M NaCl, pH 8.3. Subsequently, the unbound protein molecules of daboxin P were washed away with the same buffer. To avoid non-specific binding, the immobilized matrix was treated with 0.2 M glycine for 2 h with gentle shaking at room temperature. The blocking solution was removed by an alternative wash with acidic (0.1 M acetate buffer, pH 4.0) and basic buffer (coupling buffer) for 5 times. FX or FXa (8 μg) were added to the daboxin P immobilized matrix and incubated for 1 h at room temperature with mild shaking. The unbound FX or FXa molecules were washed away with the same buffer. Elution of FX or FXa was carried out with 20 mM HCl containing increasing molar concentrations of NaCl (0.5, 1, 1.5 and 3 M). The flow through and eluents were analyzed on 12.5% glycine SDS-PAGE under non-reducing condition and visualized after staining with PierceTM silver staining kit.
Modelling and molecular docking studies
Three dimensional (3D) molecular modelling: For the in-silico 3D modelling of daboxin P, online server I-TASSER was employed [45–47]. This server predicts the secondary conformation of a given protein by profile-profile alignment (PPA) threading techniques. The model with maximum C-score and TM score was selected for generation of ribbon model using DS ViewerPro 5.0 (Accelrys Inc.) software. Superimposition of the predicted model with the crystal structure of ammodytoxin A, AtxA (Pdb 3G8G), was carried out using DS ViewerPro 5.0 software.
Molecular docking: The equilibrated model of daboxin P was docked with FXa (PDB: 2BOH) using the online web server, PatchDock [48,49]. This server uses the geometry-based docking algorithm to find optimum candidate solutions and the RMSD (root mean square deviation) clustering to remove redundant solutions [49]. Each solution was given score based on the geometric fit as well as atomic desolvation energy [50]. In the present analysis, a default RMSD value of 4 Å was considered for clustering solutions. The ribbon model of the docked complex of daboxin P and FXa was generated using DS ViewerPro 5.0 (Accelrys Inc.).
Identification of interface residues and hot spot residues: The daboxin P-FXa docked model with maximum surface contact area and minimum free energy generated by PatchDock server was selected for evaluating the interface residues using PDBsum server [51,52]. The interface residues of a protein are those residues whose contact distances from the interacting protein partner are less than 6 Å [53].
Contact map analysis: The interaction between the chains of FXa and daboxin P were also analyzed using CMA (Contact Map Analysis), one of the servers of the online software SPACE (Structure Prediction and Analysis based on Complementarity with Environment) [54]. The server analyses the contact area between the residues of two protein chains or within a single chain of a given PDB file based on shape and chemical complementarity. It displays residue to residue contacts for a pair of amino acid residues involved in the interaction in the form of contact map. Contact maps provide more concrete demonstration of protein structure than its 3D atomic coordinates. For investigating the residue-residue contacts between daboxin P and FXa, a contact area threshold above 8Å2 has been considered for the present analysis.
Results
Purification of daboxin P, an anticoagulant PLA2 enzyme
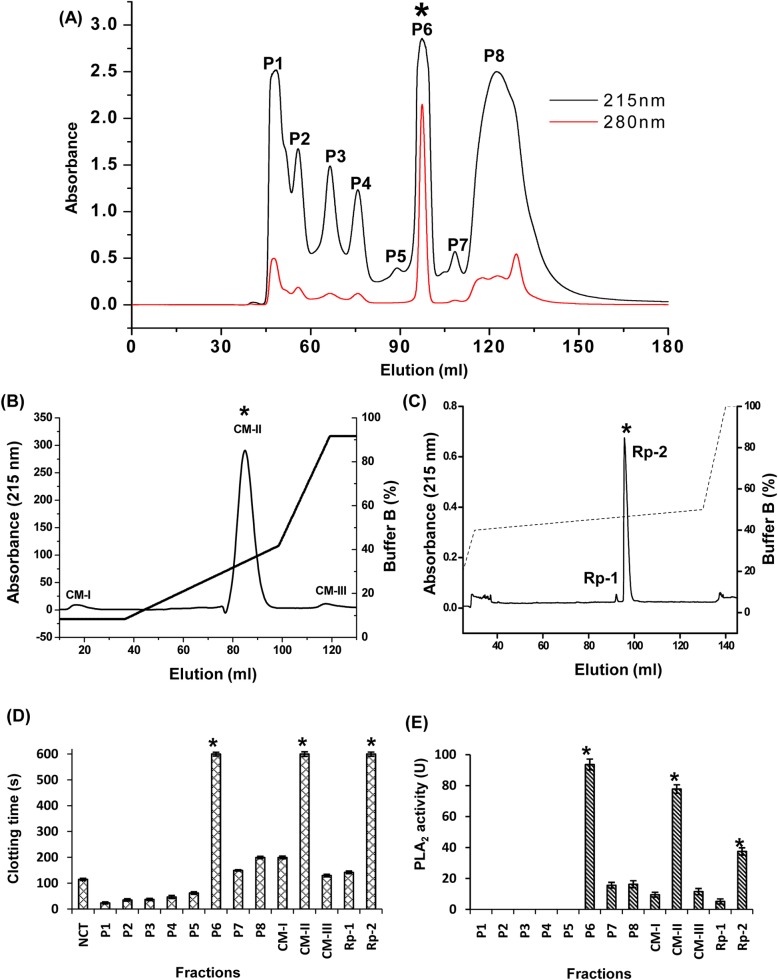
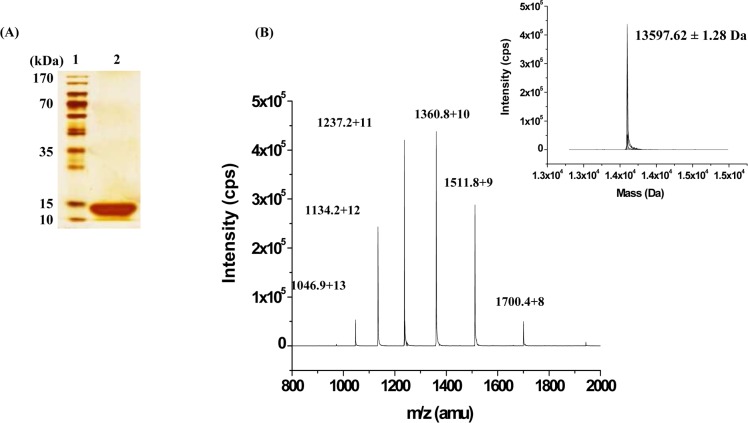
Fractionation of crude venom of Daboia r. russelii on size exclusion chromatography has resolved it into 8 prominent peaks (P1 to P8) (Fig 1A). Each individual peaks were screened for PLA2 and anticoagulant activity. Peaks, P1 to P5 were found to be devoid of PLA2 activity but exhibited procoagulant effect on human plasma while peaks P6 to P8 showed PLA2 activity and anticoagulant effect on human plasma (Fig 1D & 1E). Peak P6 which exhibited highest anticoagulant (>600 s) and PLA2 activity (93.69 μmol/min) was subjected to cation exchange chromatography. It resolved P6 into one major (CM-II) and two minor peaks (CM-I & CM-III) (Fig 1B). The major peak CM-II which showed highest anticoagulant (>600 s) and PLA2 activity (77.82 μmol/min) (Fig 1D & 1E), was further fractionated using Rp-HPLC, which resolved into one minor (Rp-1) and one major (Rp-2) peaks (Fig 1C). The homogeneity of the major peak, Rp-2 with highest PLA2 activity and anticoagulant activity was assessed by SDS-PAGE under reduced state and named as daboxin P (Fig 2A). The molecular mass of the protein was determined by ESI-MS which showed the presence of 6 mass to charge (m/z) ratio peaks ranging from +8 to +13 charges corresponding to a deconvoluted molecular mass of 13597.62 ± 1.28 Da (Fig 2B).
Fig 1. Purification of the major protein from the crude venom of Indian Daboia r. russelii.
(A): Size exclusion chromatography of crude D. r. russelii venom. 20 mg of crude venom was dissolved in 50 mM of Tris–Cl pH 7.4 and fractionated on Hiload 16/600 superdex 75 preparative grade column pre-equilibrated with the same buffer. Fractions were eluted at a flow rate of 1 ml/min and monitored at 215 & 280 nm. For each, 1 ml fractions were collected and peaks were pooled (P1 to P8). (B): Ion exchange chromatography profile of P6: The gel filtration peak, P6 was loaded onto CM FF 16/10, a weak cation exchanger column pre-equilibrated with 50 mM of Tris-Cl pH 7.4. Fractionation was carried out at a flow rate of 2.25 ml/min and eluted with a linear gradient of the same buffer containing 0.8 M NaCl and monitored at 215 nm. (C): Rp-HPLC profile of CM-II: Ion exchange fraction CM-II was loaded on Jupiter C18 column pre-equilibrated with buffer A (0.1% TFA). Fractionation was carried out at a flow rate of 0.8 ml/min with a linear gradient of buffer B (80% MeCN+0.1% TFA) and monitored at 215 nm. (D): Recalcification time of the fractions obtained from the chromatographic steps. Clotting time of plasma in presence of Tris-Cl buffer (20 mM, pH 7.4) was considered as normal clotting time (NCT). 1 μg of each fraction was incubated with plasma for 2 min followed by addition of 50 mM CaCl2 to initiate clot formation which was monitored using Tulip Coastat-1 coagulo analyser. (E): PLA2 activity of the chromatographic fractions using sPLA2 assay kit. 0.01 μg of each fraction was used for screening PLA2 activity using diheptanoylthio-phosphatidylcholine as the substrate. The amount of substrate hydrolyzed was quantified at 414 nm for 10 min at room temperature. * indicates the peak of interest in each purification step.
Fig 2. Homogeneity of the purified protein, daboxin P.
(A) 12.5% glycine SDS-PAGE profile of the purified protein after silver staining. Lane 1: PageRulerTM pre-stained protein marker (170–10 kDa). Lane 2: daboxin P after treatment with β-mercaptoethanol. (B): ESI-MS spectra. The spectra show a series of multiple charged ions corresponding to a homogenous peptide. Inset: Reconstructed mass of daboxin P (cps: counts per second, amu: atomic mass unit).
Biophysical characterization
Sequencing
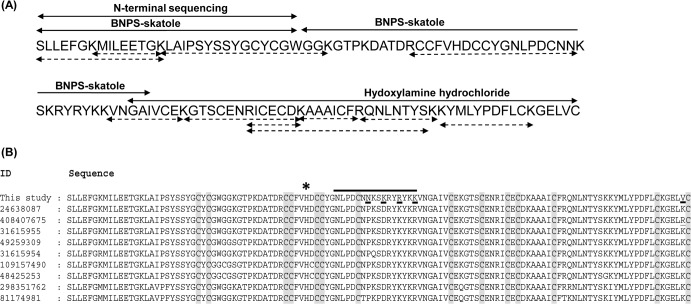
The first 30 residues were determined by N-terminal sequencing (Fig 3A). The cleavage of pyridylethylated daboxin P with BNPS-skatole yielded two peptide fragments of molecular mass 11.7 kDa and 3.6 kDa (data not shown) while cleavage with hydroxylamine hydrochloride resulted into two peptide fragments of molecular mass 6.6 kDa and 8.6 kDa (data not shown). Sequencing of these peptides by Automated Edman degradation revealed the rest of the amino acid residues (Fig 3A). On the other hand from tandem mass spectrometry, eleven peptide fragments of daboxin P were obtained (Table A in S1 File). These peptide fragments obtained by chemical and enzymatic cleavage were assembled and overlapped to decipher the complete sequence of daboxin P (Fig 3A). The homology of the protein sequence was analyzed by multiple sequence alignment with PLA2 enzymes using online blastp algorithm as represented in Fig 3B.
Fig 3. Primary structure of daboxin P.
(A): Amino acid sequence was deciphered by Edman degradation sequencing and ESI-LC MS/MS. The peptide sequences obtained from N-terminal sequencing and chemical cleavage by BNPS-skatole and hydroxylamine hydrochloride are shown with two headed solid arrows whereas peptide sequences obtained after ESI-LC MS/MS of the tryptic digested fragments are indicated with two headed doted arrows. (B): Multiple sequence alignment of daboxin P with the PLA2 enzymes from different subspecies of Daboia russelii (24638087: D. r. russelii, 408407675: D. r. siamensis, 31615955: D. r. pulchella, 49259309: D. r. russelii, 31615954: D r.pulchella, 109157490: D. r. pulchella, 48425253: D. r. pulchella, 298351762: D. r. russelii, 81174981: D. r. russelii). The conserved cys residues are highlighted in grey and the amino acid substitutions in daboxin P are underlined. * indicates the His residue at the active site. The predicted anticoagulant region is highlighted with a solid black line.
Sequence analysis
The calculated molecular mass of daboxin P was found to be 13596.70 Da which is in agreement with the observed mass of 13597.62 ± 1.28 Da from ESI-MS. It has 121 amino acid with 14 cysteine residues which should correspond to 7 disulphide bonds, a characteristic feature of group IIA viper PLA2 enzymes [55,56]. It has a theoretical pI of 8.5, calculated by online software ExPASy ProtParam [57]. At the active site, histidine residue was observed followed by an aspartate residue, a common trait for Ca+2 dependent catalytic PLA2 enzymes [58]. At the predicted anticoagulant region (53NLPDCNNKSKRYRYKK68) positively charged amino acid residues (Lys and Arg) were observed which propose it to be a strong anticoagulant PLA2 enzyme.
Homology search by blastp shows its similarity to VRV-PL-VIIIa (96%) isolated from Daboia r. russelii at position 59th (Asn), 62nd (Lys), 65th (Arg), 68th (lys) and 120th (Val) (Fig 3B) [59].
Phylogenetic relationship
Daboxin P showed close phylogenetic relation with the reported anticoagulant PLA2 enzymes which are basic, like vurtoxin which is a strong anticoagulant enzyme reported from Vipera renardi and FXa binding enzymes like DPLA2 from Daboia russelii pulchella, AtxA and AtxC from V. a. ammodytes with a bootstrap value of 99 [59–61] (Figure A in S1 File).
Secondary structure determination
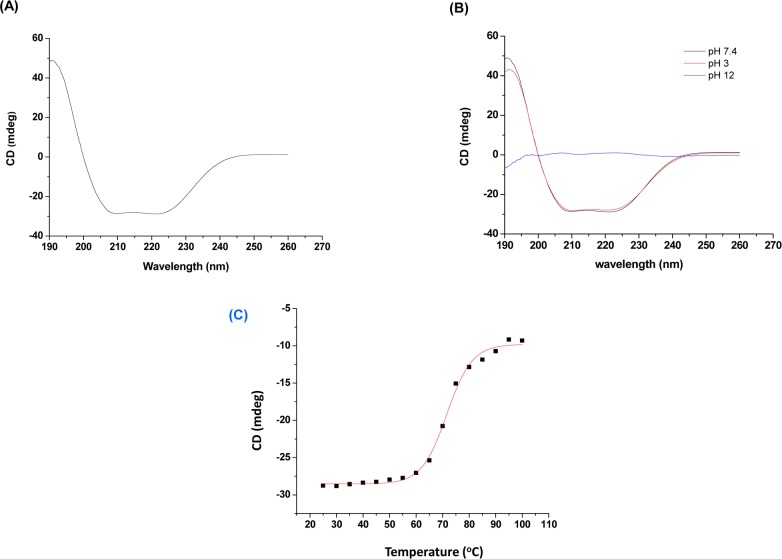
The far UV CD spectrum has shown negative minima at 222 nm and 208 nm and positive maxima at 190 nm, typical for α-helical pattern of PLA2 enzymes (Fig 4A). The percentage of secondary structure was determined by online software K2D3 which calculated 42.73% of α-helix and 12.36% of β-sheet in daboxin P [62,63].
Fig 4. Far-UV circular dichroism (CD) spectra of daboxin P (0.4 mg/ml).
(A): in milli q water at 25°C, (B): at different pH (3.0, 7.4 & 12) at 25°C. (C) Melting curve of daboxin P (dissolved in milli q water) at 222 nm considering temperature as a function. The curve was plotted using sigmoidal curve fit and Tm value was determined by Boltzman equation using Origin (OriginLab).
The CD spectrum at acidic pH (3.0) and neutral pH (7.4) showed stable structural conformation. However, the structure was distorted completely at alkaline pH (12.0) with residual α-helix of 5.51% & β-sheet of 23.75% (Fig 4B). On the other hand, its secondary structure showed a Tm (melting temperature) value of 71.59 ± 0.4°C when scanned over a range of temperature (Fig 4C).
Functional characterization
Catalytic activity
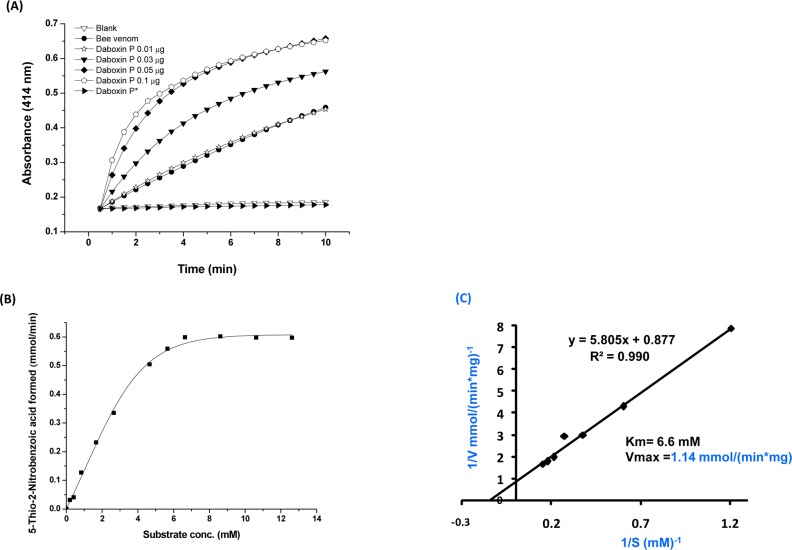
Daboxin P exhibited catalytic activity on diheptanoylthio-phosphatidylcholine (PC) in a dose dependent manner (Fig 5A). The Michaelis Menten plot with different substrate concentration showed a hyperbolic curve for enzyme activity and the Km (6.6 mM) and Vmax (1.14 mmol/(min*mg)) were determined using Lineweaver Burk-plot (Fig 5B & 5C). Alkylation with pBpB has led to the complete loss of its enzymatic activity signifying the crucial role of histidine residue in catalysis (Fig 5A).
Fig 5. Phospholipase A2 activity of daboxin P.
(A) Progress curve of diheptanoyl thio-PC cleavage by daboxin P, bee venom PLA2 enzyme and histidine modified daboxin P* at 414 nm. (B) Michaelis-Menten’s curve for sPLA2 assay. (C) The Lineweaver-Burk plot of daboxin P for determination of Km and Vmax.
Cytotoxicity
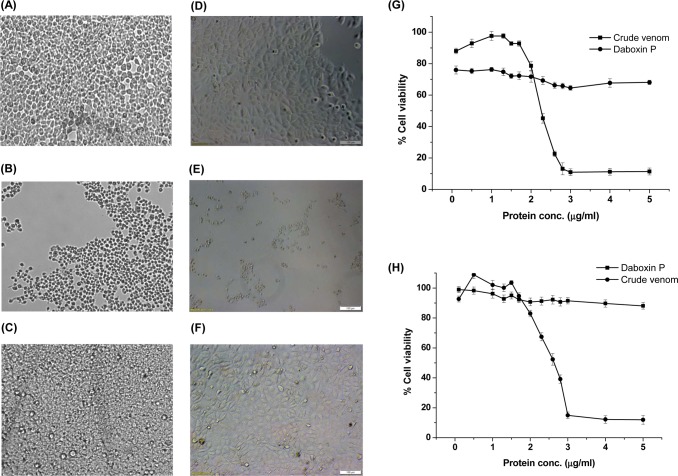
The crude venom of Daboia r. russelii has shown prominent cytotoxic effect on HEK-293 and MCF-7 cells in a dose dependent manner (Fig 6). At a concentration of 2.8 and 3 μg/ml of crude venom, only 13.09% of HEK-293and 14.97% of MCF-7 cells respectively (Fig 6B, 6E, 6G and 6H) remained viable while the same concentrations of daboxin P did not show any cytotoxic effect, suggesting that the cytotoxic effect of the crude venom is not due to this protein (Fig 6C and 6F–6H).
Fig 6. Cytotoxic effect of crude Daboia r. russelii venom and daboxin P.
Microscopic images were photographed at 10X magnification under Inverted microscope (Axio Vert A1., Zeiss) after treatment with venom samples for 24 h (A): HEK-293 cells treated with 0.9% NaCl were considered as negative control (B): HEK-293 cells treated with crude Daboia r. russelii venom (5 μg/ml) (C): HEK-293 cells treated with daboxin P (5 μg/ml) (D): MCF-7 cells treated with 0.9% NaCl were considered as negative control (E): MCF-7 cells treated with crude Daboia r. russelii venom (5 μg/ml) (F): MCF-7 cells treated with daboxin P (5 μg/ml). Percentage cell viability (G): HEK-293; (H): MCF-7 after treatment with crude venom and daboxin P using MTT based colorimetric assay. Percentage cell viability was calculated by considering the cells without venom treatment as 100% viable. The results are mean ± SD of three independent experiments.
In-vitro anticoagulant activity
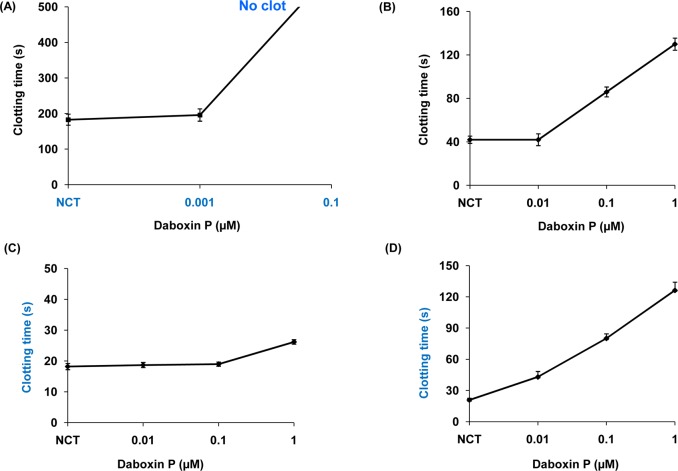
Daboxin P exhibited anticoagulant effect on the various coagulation assays in a dose dependent manner (Fig 7). At a concentration of 0.01 μM, it prolonged the recalcification time beyond 600 s indicating it to be a strong anticoagulant enzyme (Fig 7A). 1 μM of the protein delayed activated partial thromboplastin time up to 130 s but did not exhibit prominent effect on prothrombin time (Fig 7B & 7C). Daboxin P exhibited anticoagulant effect on stypven time in a dose dependent manner displaying its inhibitory effect on FX (Fig 7D). Nevertheless, it did not show any inhibitory effect on thrombin time and was devoid of fibrinogenolytic activity (Figure B i & ii in S1 File). This suggests that daboxin P might target a component/coagulation factor upstream of the common pathway.
Fig 7. Anticoagulant activities of daboxin P on platelet poor human plasma (PPP).
(A): Recalcification time, different concentrations of daboxin P (0.001, 0.01, 0.1) were pre-incubated with 150 μl of plasma at 37°C for 2 min. 100 μl of 50 mM CaCl2 was added to initiate clot formation. (B): Activated partial thromboplastin time, daboxin P (0.01, 0.1 & 1 μM) was pre-incubated with 50 μl of plasma and 50 μl APTT reagent (Liquecelin) for 3 min at 37°C. 50 μl of 25 mM CaCl2 was added to form clot. (C): Prothrombin time, different concentrations of daboxin P (0.01, 0.1 & 1 μM) were pre-incubated with 50 μl of plasma at 37°C for 2 min. 50 μl of PT reagent (Uniplastin) was added to initiate the clot formation. (D): Stypven time, daboxin P (0.01, 0.1 & 1 μM) was pre-incubated with 75 μl plasma for 3 min at 37°C. 75 μl RVV-X (10 ng/ml) was added and incubated for 2 min. 25 mM of CaCl2 was added to initiate clot formation. For all the experiments, the clot formation was monitored using Tulip Coastat-1 coagulo analyser and the time taken for clot formation in the presence of Tris-Cl buffer (20 mM, pH 7.4) was considered as normal clotting time (NCT). The results are mean ± SD of three independent experiments.
In-vivo anticoagulant activity
Daboxin P exhibited anticoagulant effect in the carotid artery of mouse treated with FeCl3. Mice administered with saline had a time-to-occlusion within 10 min. However, daboxin P administration resulted in ~4-fold increase (35.18 ± 21.58 min) in the time-to-occlusion in comparison to the saline treated mice (8.29 ± 2.61 min) (Fig 8). This result was in accordance with the in-vitro anticoagulant assays on human plasma and highlighted the anticoagulant property of daboxin P under in-vivo condition.
Fig 8. Effect of daboxin P on the time to occlusion (TTO) in FeCl3 induced carotid artery thrombosis in mice.
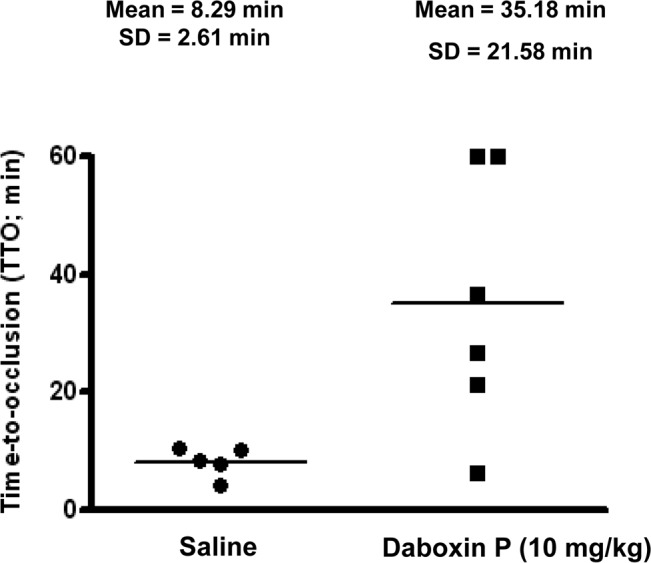
C57BL/6 male mice anesthetized with ketamine (75 mg/kg) and medetomidine (1 mg/kg) (i.p) were injected (i.p.) with daboxin P (10 mg/kg) in tail vein. Saline treated mice were considered as negative control. Each data-point represents the time-to-occlusion (TTO) of a single mouse. Maximum experimental time was considered for 60 min after FeCl3 induction.
Serine protease specificity
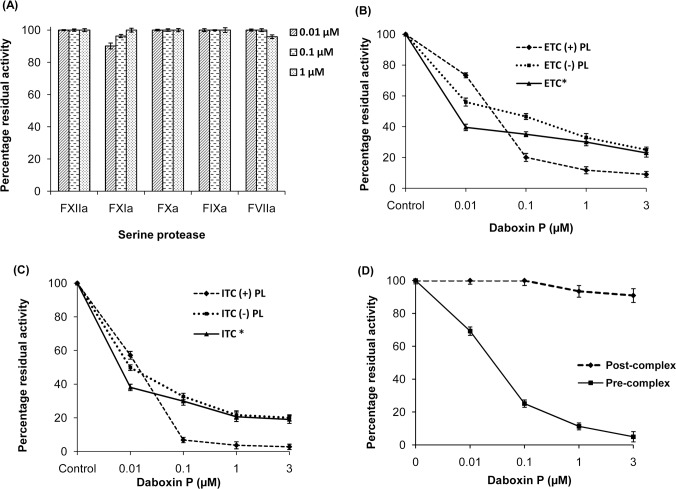
Screening for inhibitory effect of daboxin P on the amidolytic activity of various serine proteases involved in the extrinsic (FVIIa), intrinsic (FXIIa, FXIa, FIXa) and common (FXa) pathway revealed that it did not inhibit any of these serine proteases when assayed using respective synthetic chromogenic substrates (Fig 9A). However, it exerted inhibitory effect on both the extrinsic (ETC) (IC50 = 0.43 nM) and intrinsic (ITC) (IC50 = 39.20 nM) tenase complexes causing hindrance in the activation of FX to FXa (Fig 10). The residual activity of ETC and ITC was observed to be 9.05% ± 1.9 and 2.8% ± 1.6 respectively upon treatment with 3 μM of daboxin P (Fig 9B & 9C). Further, when the tenase complexes were reconstituted without phospholipid, the residual activity of ETC and ITC was found to be 25% ± 2.7 and 20.05% ± 2.3 respectively. Moreover, when the tenase complexes were treated with alkylated daboxin P (3 μM), only 23% ± 1.8 (ETC) and 19.03% ± 1.8 (ITC) of residual activity remained which is comparable to the residual activity of both the complexes reconstituted without phospholipids (Fig 9B & 9C). Interestingly, the thrombin formation by the prothrombinase complex was unaffected when daboxin P was added after the formation of the FXa-FVa complex. However, only 11.28% of residual activity remained when it was pre-incubated with FXa followed by addition of FVa for the formation of prothrombinase complex (Fig 9D).
Fig 9. Percentage residual amidolytic activity of various serine proteases and complexes pre-incubated with daboxin P.
(A): Residual activity of FXIIa, FXIa, FXa, FIXa and FVIIa; (B): Activity of extrinsic tenase complex (ETC) (C): intrinsic tenase complex (ITC), in the presence or absence of phospholipid and alkylated daboxin P (indicated by *). (D): Residual activity of prothrombinase complex. Daboxin P was either pre-incubated with FXa followed by addition of FVa (pre-complex) or after reconstitution of FXa-FVa complex (post-complex). The rate of hydrolysis of respective chromogenic substrates for all the assays was measured at 405 nm using Multiskan Go spectrophotometer. Activity of the serine protease/complex without daboxin P was considered as 100%. The results are mean ± SD of three independent experiments.
Fig 10. Inhibition curve (IC50) of daboxin P for the extrinsic and intrinsic tenase complex.
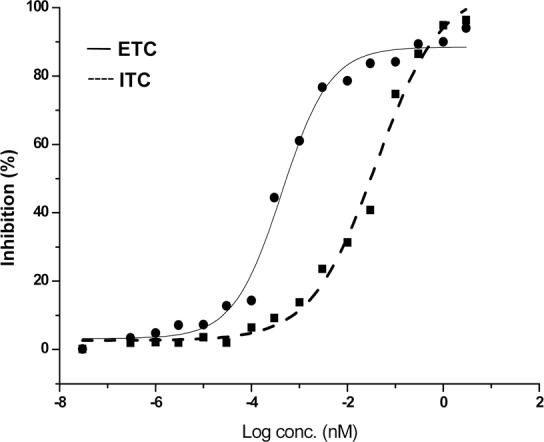
Different concentrations of daboxin P (1x10-6, 1x10-5, 1x10-4, 1x10-3, 1x10-2, 1x10-1, 1, 3x10-6, 3x10-5, 3x10-4, 3x10-3, 3x10-2, 3x10-1 and 3 μM) were pre-incubated with reconstituted tenase complexes as described above. The IC50 was calculated by fitting the points by non-linear curve fit using Origin (OriginLab, Northampton, MA).
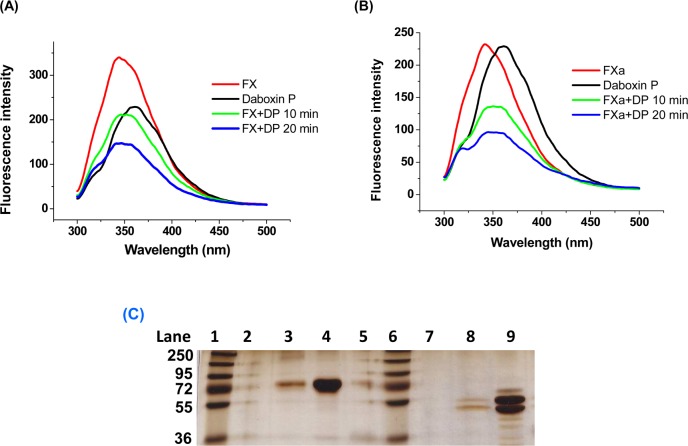
Fluorescence emission spectroscopy
The fluorescence emission spectrum of FX and FXa was quenched by daboxin P with increasing incubation time (Fig 11A & 11B). The presence or absence of Ca+2 ions did not show any effect on the quenching spectra for both FX and FXa when treated with daboxin P (data not shown).
Fig 11. Interaction of daboxin P with FX and FXa.
(A): Fluorescence emission spectra of daboxin P, FX, and the complex (daboxin P + FX). (B) Fluorescence emission spectra of daboxin P, FXa, and the complex (daboxin P + FXa). 1 μM of daboxin P (DP) was pre-incubated with either 0.05 μM of FX or 0.1 μM of FXa for different time intervals (10 min & 20 min) at room temperature. The emission spectra of the individual proteins and the complexes were measured from 200 to 500 nm with an excitation wavelength of 280 nm using quartz cuvette (1 cm path length). (C): Electrophoretic profile of the flow through and elute obtained after affinity column chromatography. Lane 1: PageRulerTM Plus pre-stained protein ladder (250–10 kDa), Lane 2: flow through (FX) Lane 3: FX after elution with 1.0 M NaCl, Lane 4: control (FX); Lane 5: blank; Lane 6: PageRulerTM Plus pre-stained protein ladder (250–10 kDa), Lane 7: flow through (FXa); Lane 8: FXa after elution with 1.0 M NaCl, Lane 9: control (FXa).
Affinity column chromatography
The electrophoretic profile of the flow through for both FX and FXa did not show any protein bands suggesting the binding of these proteins to the daboxin P immobilized CNBr matrix. Nevertheless, the eluent at 1.0 M NaCl showed prominent protein bands for both the serine proteases suggesting the disturbance of the protein-protein interaction at high salt concentration (Fig 11C).
Molecular modelling and protein-protein docking
The predicted 3D model of daboxin P has shown structural similarity to the crystal structure of AtxA (PDB: 3G8G) with a RMSD value of 0.9 Å which validates the predicted model (Figure C i in S1 File). The conserved critical regions and amino acid residues in daboxin P are shown in Figure C ii in S1 File.
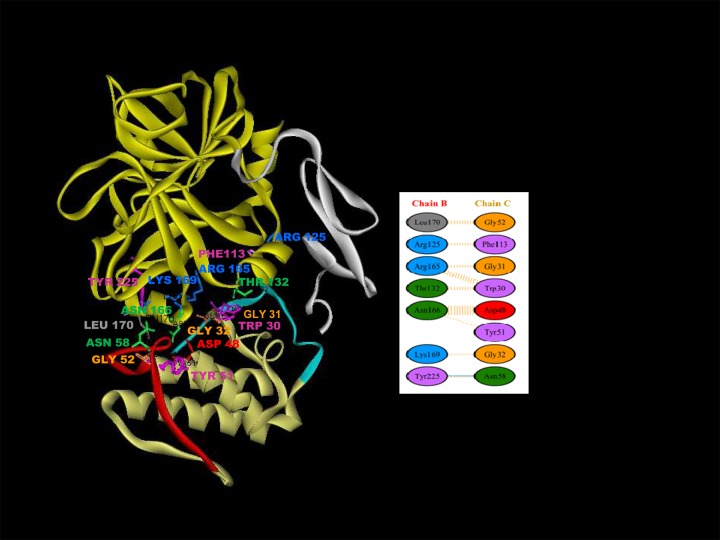
The best protein-protein docked model of daboxin P and FXa chosen in terms of minimum free energy and maximum surface contact area, was obtained from PatchDock web server (Fig 12). Daboxin P interacts with the heavy chain (B-Chain) of FXa with a geometric shape complementarity score of 12234. Geometric scoring refers to good molecular shape complementarity between the docked chains due to optimal fit with wide interface areas and lesser steric clashes [49]. The approximate interface area of the complex was found to be 1837 Å2 with atomic contact energy (ACE) of 472.62 kcal/mol.
Fig 12. 3D ribbon model of the docked complex of FXa and daboxin P.
The interface surface residues involved in the interaction were predicted by PDBsum. The Ca+2 binding loop (Trp30, Gly31, Gly32); helix C (Asp48, Tyr51, Gly52); anticoagulant region (Asn58) and C-terminal region (Phe113) of daboxin P interact with the heavy chain of FXa (Thr132, Arg165, Lys169, Asn166, Leu170, Tyr225 and Arg125) are represented in scaled ball and stick. Inset: Diagram illustrating the interaction of the seven residues of chain B (heavy chain of FXa) with eight residues of chain C (daboxin P) as predicted by PDBsum server. Orange line denotes non-bonded contacts and blue line denote hydrogen bond.
The interface and possible interacting residues across the interface of the complex were predicted by PDBsum. The total number of interface residues in protein-protein complex was found to be 31 and the interface area for each chain involved in the interaction was observed to be more than ~500 Å2. The docked complex was stabilized by molecular interactions like hydrogen bonding and non- bonded contacts. The interface amino acid residues of FXa (chain B) and daboxin P (chain C) involved in the interaction are shown in Fig 12 inset and the interface plot statistics is summarized in Table B in S1 File.
The docked complex shows interaction of daboxin P with the heavy chain (Chain B) of FXa at eight critical regions some of which are reported to be crucial for FVa binding [64]. The daboxin P residues Trp30, Gly31, Gly32 of the Ca+2 binding loop; Asp48, Tyr51, Gly52 of helix C; Asn58 of the anticoagulant region and Phe113 of C-terminal region interact with residues Thr132, Arg165, Lys169, Asn166, Leu170, Tyr225 and Arg125 of heavy chain of FXa (Table 1).
Table 1. Critical residues of daboxin P and FXa involved in interaction based on PDBsum analysis.
| Daboxin P | FXa |
|---|---|
| Trp30, Gly31, Gly32 (Ca+2 binding loop) | Thr132, Arg165, Lys169 (Heavy chain) |
| Asp48, Tyr51, Gly52 (Helix C) | Asn166,Lys169,Leu170 (Heavy chain) |
| Asn58 (Anticoagulant region) | Tyr225(Heavy chain) |
| Phe113 (C-terminal region) | Arg125(Heavy chain) |
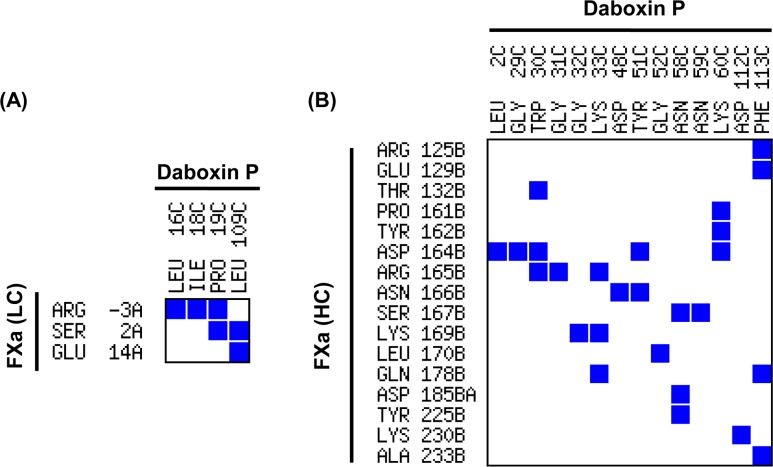
Contact map analysis shows fourteen residues of daboxin P share a contact area greater than 8 Å2 with sixteen residues of the heavy chain of FXa while only four residues of daboxin P are found to share a contact area greater than the threshold limit (8 Å2) with three residues of the light chain of FXa. This suggests the interaction of daboxin P with the light and heavy chain of FXa (Fig 13) (Table C in S1 File).
Fig 13. Contact map of daboxin P-FXa complex generated using online server Contact Map Analysis.
(A): residue to residue contact of light chain of FXa and daboxin P (B): residue to residue contact of heavy chain of FXa and daboxin P. The residue to residue contact area of the interacting amino acid residues for the chains has been considered above 8 Å2 for the design of the contact map.
Discussion
Snake venom is a plethora of pharmacologically active components with plausible therapeutic relevance in the treatment of coagulopathic abnormalities, hypotension and as diagnostic tools. Anticoagulant proteins are one such components of the venom mostly sought for the treatment of thrombosis and thromboembolism. Daboxin P, a non-toxic PLA2 enzyme purified from the venom of Indian Daboia r. russelii exhibits strong anticoagulant activity under in-vitro and in-vivo conditions. Dissection of the coagulation cascade reveals its effect upstream of the common pathway. However, it did not exhibit inhibitory effect on the amidolytic activity of the tested serine proteases. Nonetheless, daboxin P inhibited the activation of FX to FXa by both the tenase complexes in the presence and absence of phospholipids suggesting its enzymatic and non-enzymatic mode of action. Apart from phospholipids and calcium ions, FX is common in the tenase complexes hence daboxin P might be interacting with FX to exhibit its non-enzymatic mechanism of anticoagulant activity. Fluorescence emission spectroscopy supports this hypothesis where we observed the quenching of emission spectra of FX upon incubation with daboxin P for different time intervals. Apart from inhibiting the tenase complexes, daboxin P also inhibited the thrombin formation by the prothrombinase complex when pre-incubated with FXa in presence of phospholipids and Ca+2 ions followed by addition of FVa. Thus, daboxin P might interact with FXa to a region other than the active site or inhibit the complex formation by creating a steric hindrance. The quenched fluorescence emission spectra of FXa upon incubation with daboxin P suggest the probable interaction of daboxin P with FXa for exhibiting its anticoagulant activity non-enzymatically. This interaction is further supported by the affinity chromatography studies where FX and FXa were coupled to daboxin P immobilized CNBr resin and eluted at 1 M NaCl.
CM-IV, a PLA2 enzyme isolated from the venom of Naja nigricollis exhibits anticoagulant activity by inhibiting both the extrinsic tenase complex and prothrombinase complex [6,15]. It interacts with FVIIa and FXa through its anticoagulant region which shares partial sequence similarity to a region of tissue factor and partly to the light chain of FVa respectively [6,15]. However, the anticoagulant region of daboxin P does not share any such sequence similarity with FVa or TF, suggesting a different mechanism of action for daboxin P to exhibit its anticoagulant effect. Ammodytoxin A (AtxA), a PLA2 enzyme from the venom of Vipera ammodytes ammodytes inhibits prothrombinase complex by binding to FVa binding site on FXa [22]. Sequence alignment of daboxin P with AtxA shows ~76% sequence similarity with minor substitutions at helix A, helix B, Ca+2 binding loop, β-wing, helix D and C-terminal region (Figure D in S1 File). Such variations in the amino acid sequence of the PLA2 enzymes are well documented in literature which are known to confer wide range of pharmacological specificity to these proteins towards various physiological targets [65].
Based on molecular docking analysis, Faure and co-workers reported the interaction of the heavy chain of FXa with the Ca+2 binding loop, helix C, β-wing and the C-terminal region of AtxA, while light chain of FXa is involved in interaction with helix A, B and the β-wing [3,22]. On the other hand, the present in-silico molecular docking analysis shows the interaction of the Ca+2 binding loop (Trp30, Gly31, Gly32); helix C (Asp48, Tyr 51 and Gly52), anticoagulant region (Asn58) and C-terminal region (Phe113) of daboxin P with the heavy chain of FXa (Thr132, Arg165, Lys169, Asn166, Leu170, Tyr225 and Arg125 of FXa) but not with its light chain. The residues of helix A (Leu3, Met7, Leu10), helix B (Asn16, Pro17, Leu18, Thr19) and the anticoagulant region (Arg68) of AtxA are reported to be involved in the interaction with the light chain of FXa [22]. However, most of these crucial residues were found to be substituted in daboxin P (Lys7, Leu16, Ala17, Ile18, Pro19, Lys68) due to which daboxin P might not have shown any interaction with the light chain of FXa in the docking study. Nevertheless, contact map analysis shows interaction of the light chain of FXa with helix B (Leu16, Ile18, Pro19) and C-terminal region (Leu109) of daboxin P while the heavy chain of FXa (Arg125B, Glu129B, Thr132B, Pro161B, Tyr162B, Asp164B, Arg165B, Asn166B, Ser167B, Lys169B, Leu170B, Gln178B, Asp185B, Tyr225B, Lys230B, Ala233B) is found to interact with helix A (Leu2), Ca+2 binding loop (Gly29, Trp30, Gly31, Gly32, Lys33), helix C (Asp48, Tyr51, Gly52), anticoagulant region (Asn58, Asn59, Lys60), and the C-terminal region (Asp112, Phe113) of daboxin P (Fig 13).
It has been reported that Arg165, Lys169 and Lys230 of FXa form the core for FVa binding site while Arg125 and Glu129 are crucial for the complex formation [64]. Hence, binding of daboxin P to FXa might hinder the association of FXa and FVa for the formation of the prothrombinase complex in presence or absence of other co-factors. Norledge B.V. and colleagues reported the involvement of Asp4 and Asp7 of EGF-II of light chain and Asp185a, Lys186 and Lys134 of heavy chain of FXa for binding to TF/FVIIa complex based on site directed mutagenesis and molecular docking [66]. With docking analysis it has been observed that daboxin P binds to FXa (discussed above) in close vicinity to FVIIa-TF binding region on FX. Thus, interaction of daboxin P with those proximal regions might create a steric hindrance in light chain and heavy chain regions of FX and inhibit the binding of FVIIa-TF or FIXa-FVIIIa. The residues involved in this interaction could not be confirmed by docking analysis as the crystal structure of FX is not available. Based on the biochemical, biophysical and docking analysis, it is proposed that daboxin P might be exhibiting its anticoagulant activity by interacting with FX and FXa. However, site-directed mutagenesis of the critical amino acids residues needs to be carried out to confirm the interaction of daboxin P with FX and FXa.
Thus, present study evaluates daboxin P as a probable natural inhibitor of FX and FXa from snake venom. FXa is one of the most pivotal components of the physiological system which plays critical role not only in haemostasis but also in intracellular signal transduction leading to pathophysiological events like fibrosis, cancer and tissue modelling restoration [67]. Hence, thriving for natural FX/FXa inhibitors will eventually lead to manipulation/management of various pathophysiological aspects of human system.
Supporting Information
Table A. Peptide fragments of daboxin P obtained from tandem mass spectrometry. MH+ stands for mass/charge (m/z) of the peptide (protonated molecular ions), Z represents the number of charges a peptide carries after ionization, Score implies the sum of all peptide cross correlation (Xcorr) values. Figure A. The Phylogenetic relationship of daboxin P with the reported anticoagulant PLA2 enzymes from snake venom. The phylogenetic tree was constructed by neighbour joining (NJ) tree using Mega 5 software with a bootstrap value of 1000. Anticoagulant PLA2 enzymes with complete sequence were obtained from Pubmed Protein Database (http://www.ncbi.nlm.nih.gov/pubmed) and aligned using ClustalW. The sequences used in the study were CBc (Crotalus durissus terrificus), CB crotoxin (Crotalus durissus collilineatus), CBb (Crotalus durissus terrificus), PLA2 F17 (Crotalus durissus terrificus), CBa2 (Crotalus durissus terrificus), Cdc-9 (Crotalus durissus cumanensis), Cdc-10 (Crotalus durissus cumanensis), bID-PLA2 (Bothrops leucurus), Bothropstoxin (Bothrops jararacussu), DPLA2 (Daboia russellii russellii), vurtoxin (Vipera renardi), AtxA (Vipera ammodytes ammodytes), AtxC (Vipera ammodytes ammodytes), bAhp (Gloydius halys), BP III (Protobothrops flavoviridis), II-BP (Protobothrops flavoviridis), Ts-K49b (Trimeresurus stejnegeri), MTxII (Bothrops asper), PLA2 homology (Bothrops atrox), BthA-I-PLA2 (Bothrops jararacussu), Vur PL2B (Vipera renardi), EC-I-PLA2 (Echis carinatus), PLa2Vb (Vipera berus berus), CbII (Pseudocerastes fieldi), HDP-1P (Vipera nikolskii), HD-2P (Vipera nikolskii), Caudoxin (Bitis caudalis), APLA2-2 (Ophiophagus hannah); CM-III/CM-IV (Naja nigricollis); CM-III (Naja mossambica); CM-II (Naja mossambica) and CM-I (Naja mossambica).Figure B. Effect of daboxin P on thrombin time and fibrinogen. (i): Different amount of daboxin P (1, 3, 5 μg) were pre-incubated with 50 μl platelet poor plasma for 3 min at 37°C. 50 μl of thrombin (10 u/ml) was added to initiate clot formation and monitored on Tulip Coastat-1 coagulo analyser. Clotting time in presence of Tris buffer (20 mM, pH 7.4) was taken as normal clotting time (NCT). The results are mean ± SD of three independent experiments. (ii): Assessment of fibrinogenolytic activity on SDS-PAGE, different amount of daboxin P (0.01, 0.1, 1, 5 μg) were pre-incubated with 300 μl of 2 mg/ml fibrinogen for 24 h at 37°C. Fibrinogen with thrombin (3 μl of 10 units/ml) was considered as the positive control and fibrinogen with buffer (50 mM Tris-Cl, pH 7.5) was taken as the negative control. Fibrinogen degradation products were analyzed on 12.5% glycine SDS-PAGE, stained with CBB R-250. (C: Fibrinogen, Thr: Thrombin, Rvv: crude Russell’s viper venom). Figure C. Three dimensional (3D) molecular modelling (i): The 3D ribbon structure of daboxin P (in red, blue, green & white) was superimposed with X-ray crystallographic structure of AtxA (PDB 3G8G) (in yellow), (ii): 3D ribbon model of daboxin P as predicted by I-TASSER server. The Ca+2 binding loop (YGCYCGCGG in torquise blue), anticoagulant region (53NLPDCNNKSKRYRYKK68) (in yellow) and the active site His residue (in stick) are highlighted. Table B. Summary of interface plot statistics of the docked model of daboxin P and FXa. Table C. List of displayed residue to residue contact area of the light (A) and heavy chain (B) of FXa with daboxin P (C). Figure D. Sequence alignment of daboxin P with ammodytoxin A. The amino acid residues varying in daboxin P with respect to ammodytoxin A are underlined. The predicted anticoagulant region in both the PLA2 enzymes is highlighted in red.
(DOCX)
Acknowledgments
RD acknowledges DBT, Govt. of India for the twinning project (BT/43/NE/TBP/2010) and also DBT, DST, and UGC for various projects to the department. MS is a recipient of DST Inspire fellowship and thanks India Research Initiative, NUS for the short term financial support to carry out part of this work in Protein Science Laboratory, Department of Biological Sciences, NUS. The authors also acknowledge Prof. R. M. Kini for his critical comments.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work was supported by a grant to RD by DBT, Govt. of India under the twinning programme to NER region (BT/43/NE/TBP/2010), and also DBT, DST, and UGC for various projects to the department. MS is a recipient of DST Inspire fellowship and thanks India Research Initiative, NUS for the short term financial support to carry out part of this work in Protein Science Laboratory, Department of Biological Sciences, NUS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sajevic T, Leonardi A, Krizaj I. Haemostatically active proteins in snake venoms. Toxicon 2011; 57(5):627–645. 10.1016/j.toxicon.2011.01.006 [DOI] [PubMed] [Google Scholar]
- 2.Atoda H, Ishikawa M, Mizuno H, Morita T. Coagulation factor X-binding protein from Deinagkistrodon acutus venom is a Gla domain-binding protein. Biochemistry 1998; 37(50):17361–17370. [DOI] [PubMed] [Google Scholar]
- 3.Faure G, Saul F. Structural and Functional Characterization of Anticoagulant, FXa-binding Viperidae Snake Venom Phospholipases A2. Acta Chim Slov 2011; 58(4):671–677. [PubMed] [Google Scholar]
- 4.Ledizet M, Harrison LM, Koskia RA, Cappello M. Discovery and pre-clinical development of antithrombotics from hematophagous invertebrates. Curr Med Chem Cardiovasc Hematol Agents 2005; 3(1):1–10. [DOI] [PubMed] [Google Scholar]
- 5.Saikia D, Thakur R, Mukherjee AK. An acidic phospholipase A(2) (RVVA-PLA(2)-I) purified from Daboia russelli venom exerts its anticoagulant activity by enzymatic hydrolysis of plasma phospholipids and by non-enzymatic inhibition of factor Xa in a phospholipids/Ca(2+) independent manner. Toxicon 2011; 57(6):841–850. 10.1016/j.toxicon.2011.02.018 [DOI] [PubMed] [Google Scholar]
- 6.Stefansson S, Kini RM, Evans HJ. The basic phospholipase A2 from Naja nigricollis venom inhibits the prothrombinase complex by a novel nonenzymatic mechanism. Biochemistry 1990; 29(33):7742–7746. [DOI] [PubMed] [Google Scholar]
- 7.Thakur R, Kumar A, Bose B, Panda D, Saikia D, Chattopadhyay P, et al. A new peptide (Ruviprase) purified from the venom of Daboia russelii russelii shows potent anticoagulant activity via non-enzymatic inhibition of thrombin and factor Xa. Biochimie 2014; 105:149–158. 10.1016/j.biochi.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 8.Cheng AC, Wu HL, Shi GY, Tsai IH. A novel heparin-dependent inhibitor of activated protein C that potentiates consumptive coagulopathy in Russell's viper envenomation. J Biol Chem 2012; 287(19):15739–15748. 10.1074/jbc.M111.323063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maduwage K, Isbister GK. Current treatment for venom-induced consumption coagulopathy resulting from snakebite. PLoS Negl Trop Dis 2014; 8(10):e3220 10.1371/journal.pntd.0003220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faure G, Bon C. Several isoforms of crotoxin are present in individual venoms from the South American rattlesnake Crotalus durissus terrificus. Toxicon 1987; 25(2):229–234. [DOI] [PubMed] [Google Scholar]
- 11.Sharma M, Das D, Iyer JK, Kini RM, Doley R. Unveiling the complexities of Daboia russelii venom, a medically important snake of India, by tandem mass spectrometry. Toxicon 2015; 107(Pt B):266–281. 10.1016/j.toxicon.2015.06.027 [DOI] [PubMed] [Google Scholar]
- 12.Faure G, Bon C. Crotoxin, a phospholipase A2 neurotoxin from the South American rattlesnake Crotalus durissus terrificus: purification of several isoforms and comparison of their molecular structure and of their biological activities. Biochemistry 1988; 27(2):730–738. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez JM, Lomonte B. Phospholipase A2 myotoxins from Bothrops snake venoms. Toxicon 1995; 33(11):1405–1424. [DOI] [PubMed] [Google Scholar]
- 14.Kini RM. Phospholipase A2 A Complex Multifunctional Protein Puzzle In: Kini RM, editor. Venom Phospholipase A2 Enzymes: Structure, Function and Mechanism. Chichester, England: John Wiley & Sons, 1997: 1–28. [Google Scholar]
- 15.Kini RM. Structure-function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon 2005; 45(8):1147–1161. [DOI] [PubMed] [Google Scholar]
- 16.Kini RM, Evans HJ. Structure-function relationships of phospholipases. The anticoagulant region of phospholipases A2. J Biol Chem 1987; 262(30):14402–14407. [PubMed] [Google Scholar]
- 17.Boffa MC, Boffa GA. A phospholipase A2 with anticoagulant activity. II. Inhibition of the phospholipid activity in coagulation. Biochim Biophys Acta 1976; 429(3):839–852. [DOI] [PubMed] [Google Scholar]
- 18.Verheij HM, Boffa MC, Rothen C, Bryckaert MC, Verger R, de Haas GH. Correlation of enzymatic activity and anticoagulant properties of phospholipase A2. Eur J Biochem 1980; 112(1):25–32. [DOI] [PubMed] [Google Scholar]
- 19.Stefansson S, Kini RM, Evans HJ. The inhibition of clotting complexes of the extrinsic coagulation cascade by the phospholipase A2 isoenzymes from Naja nigricollis venom. Thromb Res 1989; 55(4):481–491. [DOI] [PubMed] [Google Scholar]
- 20.Kini RM, Evans HJ. The role of enzymatic activity in inhibition of the extrinsic tenase complex by phospholipase A2 isoenzymes from Naja nigricollis venom. Toxicon 1995; 33(12):1585–1590. [DOI] [PubMed] [Google Scholar]
- 21.Atanasov VN, Danchev D, Mitewa M, Petrova S. Hemolytic and anticoagulant study of the neurotoxin vipoxin and its components—basic phospholipase A2 and an acidic inhibitor. Biochemistry (Mosc) 2009; 74(3):276–280. [DOI] [PubMed] [Google Scholar]
- 22.Faure G, Gowda VT, Maroun RC. Characterization of a human coagulation factor Xa-binding site on Viperidae snake venom phospholipases A2 by affinity binding studies and molecular bioinformatics. BMC Struct Biol 2007; 7:82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta S, Gogoi D, Mukherjee AK. Anticoagulant mechanism and platelet deaggregation property of a non-cytotoxic, acidic phospholipase A2 purified from Indian cobra (Naja naja) venom: inhibition of anticoagulant activity by low molecular weight heparin. Biochimie 2015; 110:93–106. 10.1016/j.biochi.2014.12.020 [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee AK, Kalita B, Thakur R. Two acidic, anticoagulant PLA2 isoenzymes purified from the venom of monocled cobra Naja kaouthia exhibit different potency to inhibit thrombin and factor Xa via phospholipids independent, non-enzymatic mechanism. PLOS ONE 2014; 9(8):e101334 10.1371/journal.pone.0101334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasad BN, Kemparaju K, Bhatt KG, Gowda TV. A platelet aggregation inhibitor phospholipase A2 from Russell's viper (Vipera russelli) venom: isolation and characterization. Toxicon 1996; 34(10):1173–1185. [DOI] [PubMed] [Google Scholar]
- 26.Wang YM, Parmelee J, Guo YW, Tsai IH. Absence of phospholipase A(2) in most Crotalus horridus venom due to translation blockage: comparison with Crotalus horridus atricaudatus venom. Toxicon 2010; 56(1):93–100. 10.1016/j.toxicon.2010.03.015 [DOI] [PubMed] [Google Scholar]
- 27.Lu QM, Jin Y, Wei JF, Wang WY, Xiong YL. Biochemical and biological properties of Trimeresurus jerdonii venom and characterization of a platelet aggregation-inhibiting acidic phospholipase A2. J Nat Toxins 2002; 11(1):25–33. [PubMed] [Google Scholar]
- 28.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227(5259):680–685. [DOI] [PubMed] [Google Scholar]
- 29.Joseph JS, Chung MC, Jeyaseelan K, Kini RM. Amino acid sequence of trocarin, a prothrombin activator from Tropidechis carinatus venom: its structural similarity to coagulation factor Xa. Blood 1999; 94(2):621–631. [PubMed] [Google Scholar]
- 30.Ward M. Pyridylethylation of Cysteine Residues In: Walker John M., editor. The Protein Protocols Handbook. New York: Humana Press, 2002: 461–463. [Google Scholar]
- 31.Crimmins DL, Mische SM, Denslow ND. Chemical cleavage of proteins in solution In: Coligan John E, Dunn Ben M, Speicher David W, and Wingfield Paul T, editors. Current Protocols in Protein Science. New Jersey: Wiley Online Library, 2005: 1–11. [DOI] [PubMed] [Google Scholar]
- 32.Milner SJ, Thomas SM, Ballard FJ, Francis GL. Optimization of the hydroxylamine cleavage of an expressed fusion protein to produce recombinant human insulin-like growth factor (IGF)-I. Biotechnol Bioeng 1996; 50(3):265–272. [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28(10):2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maduwage K, Hodgson WC, Konstantakopoulos N, O'Leary MA, Gawarammana I, Isbister GK. The in vitro toxicity of venoms from South Asian hump-nosed pit vipers (Viperidae: Hypnale). J Venom Res 2011; 2:17–23. [PMC free article] [PubMed] [Google Scholar]
- 35.van de Loosdrecht AA, Beelen RH, Ossenkoppele GJ, Broekhoven MG, Langenhuijsen MM. A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J Immunol Methods 1994; 174(1–2):311–320. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee Y, Mizuguchi J, Iwanaga S, Kini RM. Hemextin AB complex, a unique anticoagulant protein complex from Hemachatus haemachatus (African Ringhals cobra) venom that inhibits clot initiation and factor VIIa activity. J Biol Chem 2005; 280(52):42601–42611. [DOI] [PubMed] [Google Scholar]
- 37.Ouyang C, Teng CM. Fibrinogenolytic enzymes of Trimeresurus mucrosquamatus venom. Biochim Biophys Acta 1976; 420(2):298–308. [DOI] [PubMed] [Google Scholar]
- 38.Eckly A, Hechler B, Freund M, Zerr M, Cazenave JP, Lanza F, et al. Mechanisms underlying FeCl3-induced arterial thrombosis. J Thromb Haemost 2011; 9(4):779–789. 10.1111/j.1538-7836.2011.04218.x [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Cheng Q, Xu L, Feuerstein GZ, Hsu MY, Smith PL, et al. Effects of factor IX or factor XI deficiency on ferric chloride-induced carotid artery occlusion in mice. J Thromb Haemost 2005; 3(4):695–702. [DOI] [PubMed] [Google Scholar]
- 40.Wu W, Li H, Navaneetham D, Reichenbach ZW, Tuma RF, Walsh PN. The kunitz protease inhibitor domain of protease nexin-2 inhibits factor XIa and murine carotid artery and middle cerebral artery thrombosis. Blood 2012; 120(3):671–677. 10.1182/blood-2012-03-419523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girish V. Exactin-A specific inhibitor of FX activation by extrinsic tenase complex isolated from Hemachatus haemachatus venom National University of Singapore, 2012. [Google Scholar]
- 42.Zhang Y, Ribeiro JM, Guimaraes JA, Walsh PN. Nitrophorin-2: a novel mixed-type reversible specific inhibitor of the intrinsic factor-X activating complex. Biochemistry 1998; 37(30):10681–10690. [DOI] [PubMed] [Google Scholar]
- 43.Mathur A, Bajaj SP. Protease and EGF1 domains of factor IXa play distinct roles in binding to factor VIIIa. Importance of helix 330 (helix 162 in chymotrypsin) of protease domain of factor IXa in its interaction with factor VIIIa. J Biol Chem 1999; 274(26):18477–18486. [DOI] [PubMed] [Google Scholar]
- 44.Koh CY, Kazimirova M, Trimnell A, Takac P, Labuda M, Nuttall PA, et al. Variegin, a novel fast and tight binding thrombin inhibitor from the tropical bont tick. J Biol Chem 2007; 282(40):29101–29113. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008; 9(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 2010; 5(4):725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods 2015; 12(1):7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 2005; 33(Web Server issue):W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duhovny D, Nussinov N, Wolfson HJ. Efficient Unbound Docking of Rigid Molecules. In: Gusfield In et al. , editor. Berlin: Springer Verlag, 2002: 185–200. [Google Scholar]
- 50.Zhang C, Vasmatzis G, Cornette JL, DeLisi C. Determination of atomic desolvation energies from the structures of crystallized proteins. J Mol Biol 1997; 267(3):707–726. [DOI] [PubMed] [Google Scholar]
- 51.Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res 2001; 29(1):221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res 2005; 33(Database issue):D266–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruger DM, Gohlke H. DrugScorePPI webserver: fast and accurate in silico alanine scanning for scoring protein-protein interactions. Nucleic Acids Res 2010; 38(Web Server issue):W480–W486. 10.1093/nar/gkq471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sobolev V, Eyal E, Gerzon S, Potapov V, Babor M, Prilusky J, et al. SPACE: a suite of tools for protein structure prediction and analysis based on complementarity and environment. Nucl Acids Res 2005; 33:39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davidson FF, Dennis EA. Evolutionary relationships and implications for the regulation of phospholipase A2 from snake venom to human secreted forms. J Mol Evol 1990; 31(3):228–238. [DOI] [PubMed] [Google Scholar]
- 56.Heinrikson RL, Krueger ET, Keim PS. Amino acid sequence of phospholipase A2-alpha from the venom of Crotalus adamanteus. A new classification of phospholipases A2 based upon structural determinants. J Biol Chem 1977; 252(14):4913–4921. [PubMed] [Google Scholar]
- 57.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein Identification and Analysis Tools on the ExPASy Server In: Walker John M., editor. The Proteomics Protocols Handbook. New York: Humana Press, 2005: 571–607. [Google Scholar]
- 58.Fleer EA, Verheij HM, de Haas GH. Modification of carboxylate groups in bovine pancreatic phospholipase A2. Identification of aspartate-49 as Ca2+-binding ligand. Eur J Biochem 1981; 113(2):283–288. [DOI] [PubMed] [Google Scholar]
- 59.Gowda VT, Schmidt J, Middlebrook JL. Primary sequence determination of the most basic myonecrotic phospholipase A2 from the venom of Vipera russelli. Toxicon 1994; 32(6):665–673. [DOI] [PubMed] [Google Scholar]
- 60.Saul FA, Prijatelj-Znidarsic P, Vulliez-le Normand B, Villette B, Raynal B, Pungercar J, et al. Comparative structural studies of two natural isoforms of ammodytoxin, phospholipases A2 from Vipera ammodytes ammodytes which differ in neurotoxicity and anticoagulant activity. J Struct Biol 2010; 169(3):360–369. 10.1016/j.jsb.2009.10.010 [DOI] [PubMed] [Google Scholar]
- 61.Tsai IH, Wang YM, Cheng AC, Starkov V, Osipov A, Nikitin I, et al. cDNA cloning, structural, and functional analyses of venom phospholipases A(2) and a Kunitz-type protease inhibitor from steppe viper Vipera ursinii renardi. Toxicon 2011; 57(2):332–341. 10.1016/j.toxicon.2010.12.012 [DOI] [PubMed] [Google Scholar]
- 62.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta 2005; 1751(2):119–139. [DOI] [PubMed] [Google Scholar]
- 63.Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins Structure, Function, and Bioinformatics 2011; 80 (2). [DOI] [PubMed] [Google Scholar]
- 64.Rudolph AE, Porche-Sorbet R, Miletich JP. Definition of a factor Va binding site in factor Xa. J Biol Chem 2001; 276(7):5123–5128. [DOI] [PubMed] [Google Scholar]
- 65.Kini RM, Chan YM. Accelerated evolution and molecular surface of venom phospholipase A2 enzymes. J Mol Evol 1999; 48(2):125–132. [DOI] [PubMed] [Google Scholar]
- 66.Norledge BV, Petrovan RJ, Ruf W, Olson AJ. The tissue factor/factor VIIa/factor Xa complex: a model built by docking and site-directed mutagenesis. Proteins 2003; 53(3):640–648. [DOI] [PubMed] [Google Scholar]
- 67.Borensztajn K, Peppelenbosch MP, Spek CA. Factor Xa: at the crossroads between coagulation and signaling in physiology and disease. Trends Mol Med 2008; 14(10):429–440. 10.1016/j.molmed.2008.08.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Peptide fragments of daboxin P obtained from tandem mass spectrometry. MH+ stands for mass/charge (m/z) of the peptide (protonated molecular ions), Z represents the number of charges a peptide carries after ionization, Score implies the sum of all peptide cross correlation (Xcorr) values. Figure A. The Phylogenetic relationship of daboxin P with the reported anticoagulant PLA2 enzymes from snake venom. The phylogenetic tree was constructed by neighbour joining (NJ) tree using Mega 5 software with a bootstrap value of 1000. Anticoagulant PLA2 enzymes with complete sequence were obtained from Pubmed Protein Database (http://www.ncbi.nlm.nih.gov/pubmed) and aligned using ClustalW. The sequences used in the study were CBc (Crotalus durissus terrificus), CB crotoxin (Crotalus durissus collilineatus), CBb (Crotalus durissus terrificus), PLA2 F17 (Crotalus durissus terrificus), CBa2 (Crotalus durissus terrificus), Cdc-9 (Crotalus durissus cumanensis), Cdc-10 (Crotalus durissus cumanensis), bID-PLA2 (Bothrops leucurus), Bothropstoxin (Bothrops jararacussu), DPLA2 (Daboia russellii russellii), vurtoxin (Vipera renardi), AtxA (Vipera ammodytes ammodytes), AtxC (Vipera ammodytes ammodytes), bAhp (Gloydius halys), BP III (Protobothrops flavoviridis), II-BP (Protobothrops flavoviridis), Ts-K49b (Trimeresurus stejnegeri), MTxII (Bothrops asper), PLA2 homology (Bothrops atrox), BthA-I-PLA2 (Bothrops jararacussu), Vur PL2B (Vipera renardi), EC-I-PLA2 (Echis carinatus), PLa2Vb (Vipera berus berus), CbII (Pseudocerastes fieldi), HDP-1P (Vipera nikolskii), HD-2P (Vipera nikolskii), Caudoxin (Bitis caudalis), APLA2-2 (Ophiophagus hannah); CM-III/CM-IV (Naja nigricollis); CM-III (Naja mossambica); CM-II (Naja mossambica) and CM-I (Naja mossambica).Figure B. Effect of daboxin P on thrombin time and fibrinogen. (i): Different amount of daboxin P (1, 3, 5 μg) were pre-incubated with 50 μl platelet poor plasma for 3 min at 37°C. 50 μl of thrombin (10 u/ml) was added to initiate clot formation and monitored on Tulip Coastat-1 coagulo analyser. Clotting time in presence of Tris buffer (20 mM, pH 7.4) was taken as normal clotting time (NCT). The results are mean ± SD of three independent experiments. (ii): Assessment of fibrinogenolytic activity on SDS-PAGE, different amount of daboxin P (0.01, 0.1, 1, 5 μg) were pre-incubated with 300 μl of 2 mg/ml fibrinogen for 24 h at 37°C. Fibrinogen with thrombin (3 μl of 10 units/ml) was considered as the positive control and fibrinogen with buffer (50 mM Tris-Cl, pH 7.5) was taken as the negative control. Fibrinogen degradation products were analyzed on 12.5% glycine SDS-PAGE, stained with CBB R-250. (C: Fibrinogen, Thr: Thrombin, Rvv: crude Russell’s viper venom). Figure C. Three dimensional (3D) molecular modelling (i): The 3D ribbon structure of daboxin P (in red, blue, green & white) was superimposed with X-ray crystallographic structure of AtxA (PDB 3G8G) (in yellow), (ii): 3D ribbon model of daboxin P as predicted by I-TASSER server. The Ca+2 binding loop (YGCYCGCGG in torquise blue), anticoagulant region (53NLPDCNNKSKRYRYKK68) (in yellow) and the active site His residue (in stick) are highlighted. Table B. Summary of interface plot statistics of the docked model of daboxin P and FXa. Table C. List of displayed residue to residue contact area of the light (A) and heavy chain (B) of FXa with daboxin P (C). Figure D. Sequence alignment of daboxin P with ammodytoxin A. The amino acid residues varying in daboxin P with respect to ammodytoxin A are underlined. The predicted anticoagulant region in both the PLA2 enzymes is highlighted in red.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.