Abstract
We examined the interaction between OSU‐03012 (also called AR‐12) with phosphodiesterase 5 (PDE5) inhibitors to determine the role of the chaperone glucose‐regulated protein (GRP78)/BiP/HSPA5 in the cellular response. Sildenafil (Viagra) interacted in a greater than additive fashion with OSU‐03012 to kill stem‐like GBM cells. Treatment of cells with OSU‐03012/sildenafil: abolished the expression of multiple oncogenic growth factor receptors and plasma membrane drug efflux pumps and caused a rapid degradation of GRP78 and other HSP70 and HSP90 family chaperone proteins. Decreased expression of plasma membrane receptors and drug efflux pumps was dependent upon enhanced PERK‐eIF2α‐ATF4‐CHOP signaling and was blocked by GRP78 over‐expression. In vivo OSU‐03012/sildenafil was more efficacious than treatment with celecoxib and sildenafil at killing tumor cells without damaging normal tissues and in parallel reduced expression of ABCB1 and ABCG2 in the normal brain. The combination of OSU‐03012/sildenafil synergized with low concentrations of sorafenib to kill tumor cells, and with lapatinib to kill ERBB1 over‐expressing tumor cells. In multiplex assays on plasma and human tumor tissue from an OSU‐03012/sildenafil treated mouse, we noted a profound reduction in uPA signaling and identified FGF and JAK1/2 as response biomarkers for potentially suppressing the killing response. Inhibition of FGFR signaling and to a lesser extent JAK1/2 signaling profoundly enhanced OSU‐03012/sildenafil lethality. J. Cell. Physiol. 230: 1982–1998, 2015. © 2015 The Authors. Journal of Cellular Physiology Published by Wiley Periodicals, Inc.
Abbreviations
- PDGF
platelet‐derived growth factor
- EGF
epidermal growth factor
- CEL
celecoxib also called Celebrex
- OSU
OSU‐03012 also called AR‐12
- SIL
sildenafil also called Viagra
- VAR
vardenafil also called Levitra
- COX
cyclooxygenase
- P
phospho‐
- ca
constitutively active
- WT
wild type
- PERK
PKR like endoplasmic reticulum kinase
- HSP
heat shock protein
- GRP
glucose‐regulated protein
OSU‐03012, is a derivative of the drug celecoxib (Celebrex), and lacks cyclooxygenase (COX2) inhibitory activity (Zhu et al., 2004; Johnson et al., 2005). COX2 is over‐expressed in several tumor types and drugs that inhibit COX2, that is, celecoxib have been shown to cause tumor cell‐specific increases in cell death, and that are also associated with a lower rate of growth (Koehne and Dubois, 2004; Cui et al., 2005; Kang et al., 2006; Klenke et al., 2006). Prolonged treatment with COX2 inhibitors can reduce the incidence of developing cancer, which, in addition, argues that COX2 inhibitors have cancer preventative effects (Kashfi and Rigas, 2005; Narayanan et al., 2006). Expression levels of COX2 do not simplistically correlate with tumor cell sensitivity to COX2 inhibitors (Kulp et al., 2004; Patel et al., 2005). Thus, COX2 inhibitors must have additional cellular targets to explain their anti‐tumor biological actions.
Compared to the parent drug celecoxib, OSU‐03012 (developed by Dr. Ching‐Shih Chen at Ohio State University in 2004 and also known as AR‐12, under licence from Ohio State University to Arno Therapeutics, NJ) has a greater level of bio‐availability in pre‐clinical large animal models to the parent compound and has an order of magnitude greater efficacy at killing tumor cells (Yacoub et al., 2006; Park et al., 2008; Booth et al., 2012a). Based on encouraging pre‐clinical data OSU‐03012 underwent Phase I evaluation in patients with solid and liquid tumors. Studies from the initial Phase I trial noted that the “C max after single dose was dose‐proportional but high PK variability was observed, likely due to inadequate disintegration and dissolution of the formulation in the stomach” (ASCO 2013 meeting. http://meetinglibrary.asco.org/content/115148‐132) The C max of OSU‐03012 in plasma after 1 day at the MTD of 800 mg BID was ∼1 to 2 μM. After 28 days of treatment, the C max was ∼2 to 3 μM with the peak C max in some patients being ∼8 μM. Thus, even considering the problems associated with differential OSU‐03012 drug absorption in different patients, our use of OSU‐03012 in prior in vitro studies and in the present manuscript of ∼1.0 to 8.0 μM of the drug is clinically relevant.
Initially, the tumoricidal effects of OSU‐03012 in transformed cells were argued to be via direct inhibition of the enzyme PDK‐1, within the PI3K pathway (Zhu et al., 2004). And, in the low micro‐molar range in cells, it has been shown that OSU‐03012 can lower AKT phosphorylation, presumably by PDK‐1 inhibition. In our previous studies, inhibition of either ERK1/2 or phosphatidyl‐inositol 3 kinase signaling enhanced the toxicity of OSU‐03012 (Yacoub et al., 2006; Park et al., 2008; Booth et al., 2012a,2012b). However, our data has also strongly argued that OSU‐03012 toxicity, and in addition its radiosensitizing effects, could not simplistically be attributed to suppression of AKT signaling (Yacoub et al., 2006; Park et al., 2008; Booth et al., 2012a,2012b). Specifically, our prior studies have argued that OSU‐03012 killed tumor cells through mechanisms, which involved enhanced endoplasmic reticulum (ER) stress signaling through activation of PKR‐like endoplasmic reticulum kinase (PERK), down‐regulation/reduced half‐life of the ER and plasma membrane localized HSP70 family chaperone GRP78/BiP/HSPA5, and a caspase‐independent, cathepsin‐dependent autophagy‐dependent form of tumor cell death (Yacoub et al., 2006; Park et al., 2008; Booth et al., 2012a,2012b). One of the hallmarks of any potentially useful anti‐cancer drug is that it is found to be relatively non‐toxic to “normal” cells/tissues and we previously noted that OSU‐03012, alone or in combination with other modalities had an excellent therapeutic window comparing toxicity in normal non‐transformed cells to tumor cells both in vitro and in vivo.
ER stress signaling is mediated by three proximal sensors, PERK, the IRE1 (inositol‐requiring protein 1α)/X‐box binding protein 1 (XBP1) system and activating transcription factor 6 (ATF6). GRP78 plays a key role in regulating the ER stress response; under resting conditions the majority of GRP78 is associated with PERK and IRE1 and keeps these proteins in an inactive state (Rao et al., 2012; Gorbatyuk and Gorbatyuk, 2013; Roller and Maddalo, 2013). GRP78, as a chaperone, also plays an important role in the protein folding processes that occur in the ER including during cancer, liver disease, and virus replication (Roux, 1990; Earl et al., 1991; Anderson et al., 1992; Hogue and Nayak, 1992; Carleton and Brown, 1997; Xu et al., 1997, 1998; Bolt, 2001; Shen et al., 2002; Dimcheff et al., 2004; He, 2006; Spurgers et al., 2010; Moreno and Tiffany‐Castiglioni, 2014; Reid et al., 2014). When high levels of unfolded protein are present in the ER, for example, in a tumor cell or in a virally infected cell, GRP78 disassociates from PERK and IRE1 resulting in their activation, and GRP78 binds to the unfolded proteins in the ER as a chaperone (Lee, 2007; Luo and Lee, 2013; Chen et al., 2014, and references therein). Activation of PERK‐eIF2α signaling acts to prevent the majority of cellular proteins from being synthesized and IRE1 signaling enhances the expression of additional GRP78 protein. Virus infection can cause a profound ER stress response, which could by cyto‐toxic or prevent virus protein synthesis, thus some viruses make targeting proteins, similar to mammalian GADD34 and Nck1, that relocate protein phosphatase 1 with eIF2α thereby preventing eIF2α phosphorylation and high levels of toxic ER stress signaling (Rathore et al., 2013). As GRP78 chaperones the unfolded protein(s), free GRP78 eventually becomes available to re‐associate with PERK and IRE1 thereby shutting off the signaling system (Lee, 2007; Luo and Lee, 2013; Chen et al., 2014). Of note, however, is that prolonged ER stress signaling downstream of PERK and IRE1 can facilitate transformed cell killing, though in our laboratory not primary cell killing (see above), arguing that a prolonged reduction of GRP78 in transformed cells reduces cell viability (Booth et al., 2014a,2014b).
We have published several manuscripts showing that knock down/inhibition of PERK signaling suppressed OSU‐03012 toxicity but that this effect appeared to be only partially eiF2α dependent as the ability of OSU‐03012 to cause high PERK phosphorylation was not reflected in the more modest phosphorylation increase of eIF2α. The induction of PERK activity by OSU‐03012 was associated with reduced GRP78 and HSP90 expression and increased HSP70 expression. More recently, however, we have noted that the ability of sildenafil to strongly enhance OSU‐03012 or celecoxib lethality was dependent on an even greater increase in PERK activity, which was now associated with very high levels of eIF2α phosphorylation.
Tumors of the brain are difficult to control. Untreated adult glioblastoma (GBM) patients have a mean survival of several months that is only prolonged up to 12–16 months by aggressive therapeutic intervention. New therapeutic approaches that could translate to the clinic for this malignancy are urgently required. In two recently published studies, we demonstrated that phosphodiesterase 5 (PDE5) inhibitors enhanced the toxicity of standard of care chemotherapies in bladder and pediatric CNS tumors (Booth et al., 2014c,2014d; Roberts et al., 2014). In two additional even more recent studies, we then noted that celecoxib or OSU‐03012 lethality is also enhanced by PDE5 inhibitors (Booth et al., 2014a,2014b). As celecoxib, OSU‐03012 and PDE5 inhibitors all readily cross the blood–brain barrier, the present studies have attempted to further understand how celecoxib and OSU‐03012 lethality is regulated by PDE5 inhibitors in malignant brain tumor cells, with specific attention to the regulation of GRP78 expression and activity of other HSP90 and HSP70 family chaperones.
Materials and Methods
Materials
Phospho‐/total‐antibodies were purchased from Cell Signaling Technologies (Danvers, MA) and Santa Cruz Biotech. (Santa Cruz, CA). All drugs, including OSU‐03012, were purchased from Selleckchem (Houston, TX). Commercially available validated short hairpin RNA molecules to knock down RNA/protein levels were from Qiagen (Valencia, CA). At least two different validated siRNA molecules were independently used to confirm the effects observed were not due to non‐specific effects. Antibody reagents, other kinase inhibitors, caspase inhibitors cell culture reagents, and non‐commercial recombinant adenoviruses have been previously described (Booth et al., 2014a,2014b,2014c,2014d). The plasmid to express GRP78/BiP/HSPA5 was kindly provided to the Dent Laboratory by Dr. A.S. Lee (University of Southern California, Los Angeles, CA). Previously characterized semi‐established GBM5/GBM6/GBM12/GBM14 GBM cells were supplied by Dr. C.D. James (University of California, San Francisco) and Dr. J.N. Sarkaria (Mayo Clinic, Rochester MN) and were not further characterized by ourselves (Giannini et al., 2005).
Methods
Mammalian cell culture and in vitro exposure of cells to drugs
All fully established cancer lines were cultured at 37°C (5% (v/v CO2) in vitro using RPMI supplemented with 10% (v/v) fetal calf serum and 10% (v/v) non‐essential amino acids. All primary human GBM cells were cultured at 37°C (5% (v/v) CO2) in vitro using RPMI supplemented with 2% (v/v) fetal calf serum and 10% (v/v) non‐essential amino acids at 37°C (5% (v/v CO2). GBM5/6/12/14 stem cells were cultured in StemCell Technologies NeuroCult NS‐A Basal Medium supplemented with 20 μg/ml bFGF, 20 μg/ml epidermal growth factor (EGF), and 2 mM heparin. CD133+ glioma cells from this population were isolated by fluorescence‐activated cell sorting analysis. Cells, for example, GBM12, grew as neurospheres and were characterized for multiple stem cell markers, including CD44, SOX2, CD133, CD15, CD36, Integrin B6, and MAP2. Neurosphere GBM cells had an approximate 10‐fold greater tumorigenicity in vivo than parental wild type (WT) GBM cells (data not shown). For short‐term cell killing assays and immunoblotting, cells were plated at a density of 3 × 103 per cm2 and 24 h after plating were treated with various drugs, as indicated. In vitro small molecule inhibitor treatments were from a 100 mM stock solution of each drug and the maximal concentration of vehicle (DMSO) in media was 0.02% (v/v). Cells were not cultured in growth factor free media during any study.
Cell treatments, SDS–PAGE, and Western blot analysis
Cells were treated with various drug concentrations, as indicated in the figure legends. Samples were isolated at the indicated times and SDS–PAGE and immunoblotting was performed as described in Booth et al. (2014a,2014b,2014c,2014d). Immunoblots were observed by using an Odyssey IR imaging system (LI‐COR Biosciences, Lincoln, NE).
Recombinant adenoviral and other virus vectors; infection in vitro
We generated and purchased previously noted recombinant adenoviruses as per Booth et al. (2014a,2014b,2014c,2014d). Cells were infected with these adenoviruses at an approximate m.o.i. as indicated in the figure/legend (usually an m.o.i. of 0.1–50). Cells were incubated for 24 h to ensure adequate expression of transduced gene products prior to drug exposures. Viruses were purchased from Research BioLabs (Philadelphia, PA).
Detection of cell death by trypan blue assay
Cells were harvested by trypsinization with Trypsin/EDTA for ∼10 min at 37°C. Harvested cells were combined with the culture media containing unattached cells and the mixture centrifuged (800 rpm, 5 min). Cell pellets were resuspended in PBS and mixed with trypan blue agent. Viability was determined microscopically using a hemocytometer (2014a,2014b,2014c,2014d). Five hundred cells from randomly chosen fields were counted and the number of dead cells was counted and expressed as a percentage of the total number of cells counted. Cell killing was confirmed using the Sceptor instrument (Millipore, Billerica, MA), which measured tumor cell size/sub G1 DNA as an indication of tumor cell viability.
Cell death measurements by live/dead assay
Cells were grown in 96‐well plates with each well containing ∼10,000 cells in 200 ml of media. Cells were treated with the indicated concentrations of drugs for the indicated amounts of time in each panel. Plates were then centrifuged (500 rpm, 5 min) to re‐adhere floating dead cells to the base of each well. The media was removed and live/dead assay reagent added and cells incubated for 10 min before the reagent was removed. Cells were imaged in a Hermes WiScan instrument under 10× magnification. Green cells = viable; yellow/red cells = dieing/dead. The numbers of viable and dead cells were counted manually from several images taken from one well together with images from another two wells.
Plasmid transfection
Plasmids
Cells were plated as described above and 24 h after plating, transfected. Plasmids (0.5 μg) expressing a specific mRNA or appropriate vector control plasmid DNA was diluted in 50 μl serum‐free and antibiotic‐free medium (1 portion for each sample). Concurrently, 2 μl Lipofectamine 2000 (Invitrogen), was diluted into 50 μl of serum‐free and antibiotic‐free medium. Diluted DNA was added to the diluted Lipofectamine 2000 for each sample and incubated at room temperature for 30 min. This mixture was added to each well/dish of cells containing 200 μl serum‐free and antibiotic‐free medium for a total volume of 300 μl and the cells were incubated for 4 h at 37°C. An equal volume of 2× medium was then added to each well. Cells were incubated for 48 h, then treated with drugs. To assess transfection efficiency of plasmids, we used a plasmid to express GFP and defined the percentage of cells being infected as the percentage of GFP+ cells. For all cell lines, the infection efficiency was >70%.
siRNA
Cells were plated in 60 mm dishes from a fresh culture growing in log phase as described above, and 24 h after plating transfected. Prior to transfection, the medium was aspirated and 1 ml serum‐free medium was added to each plate. For transfection, 10 nM of the annealed siRNA, the positive sense control doubled stranded siRNA targeting GAPDH or the negative control (a “scrambled” sequence with no significant homology to any known gene sequences from mouse, rat, or human cell lines) were used (predominantly Qiagen; occasional alternate siRNA molecules were purchased from Ambion, Inc., Austin, TX). At least two different validated siRNA molecules were independently used to confirm the effects observed were not due to non‐specific effects. Ten nanomolar of siRNA (scrambled or experimental) was diluted in serum‐free media. Four microliters Hiperfect (Qiagen) was added to this mixture and the solution was mixed by pipetting up and down several times. This solution was incubated at room temperature for 10 min, then added drop‐wise to each dish. The medium in each dish was swirled gently to mix, then incubated at 37°C for 2 h. One milliliter of 10% (v/v) serum‐containing medium was added to each plate, and cells were incubated at 37°C for 24–48 h before re‐plating (50 × 103 cells each) onto 12‐well plates. Cells were allowed to attach overnight, then treated with drugs (0–48 h). Trypan blue exclusion assays and SDS–PAGE/immunoblotting analyses were then performed at the indicated time points.
Animal studies
Athymic mice were treated by oral gavage with vehicle (cremophore); celecoxib (50 mg/kg) + sildenafil (10 mg/kg); OSU‐03012 (50 mg/kg) + sildenafil (10 mg/kg) for 14 days QD after which animals were humanely sacrificed and normal tissue/organs obtained. Organs were fixed and 10 μm sections taken, de‐parafinized and H&E stained (special thanks to Dr. Hope Richards, VCU Department of Pathology). Color images were taken at 10× magnification (special thanks to Dr. Steven Grant, VCU, Department of Hematology/Oncology). Athymic nude mice (∼20 g) were injected with 1 × 107 BT474 cells into their fourth mammary fat pad. Seven days after injection, animals with ∼50 mm3 tumors were treated by oral gavage with vehicle (cremophore); celecoxib (10 mg/kg) + sildenafil (5 mg/kg); OSU‐03012 (10 mg/kg) + sildenafil (5 mg/kg) for 5 days after which normal tissues were taken for IHC (GRP78, ABCB1, and ABCG2) and the tumors and blood isolated for multiplex assays.
Data analysis
Comparison of the effects between various in vitro drug treatments was performed after analysis of variance using the Student's t‐test. Differences with a P‐value of <0.05 were considered statistically significant. Experiments shown are the means of multiple individual points from multiple studies (± SEM).
Results
GBM stem cells compared to parental non‐selected GBM cells exhibited higher basal levels of phosphorylated eIF2α, but stem cells nevertheless also expressed more GRP78/BiP/HSPA5 protein (Fig. 1A). Stem cell‐like GBM cells expressed considerably more MCL‐1 and sphingosine‐1‐phosphate receptor 1 (EDG‐1) than parental WT cells. Of note, the intensity of each image was normalized to the most intense staining image for each antibody probe; WT cells did express MCL‐1, EDG‐1, and GRP78 albeit at lower levels than stem cells (Fig. 1A, lower images). In GBM cells, it was noted that stem cell selected cells were modestly but significantly more sensitive to OSU + sildenafil also called Viagra (SIL) treatment than parental WT cells. In the majority of GBM cell lines, treatment using 1 μM OSU‐03012 caused a greater toxic response when combined with sildenafil than did treatment with the parental compound 5 μM celecoxib and sildenafil. Regardless of whether breast cancer cells were parental or generated to be anoikis resistant, the relative ability of celecoxib also called Celebrex (CEL) + SIL treatment to kill parental or anoikis cells preferentially appeared to be more unpredictable than in GBM cells and stochastic in nature (data not shown). Prolonged high concentration dosing of mice with [OSU‐03012 + sildenafil] or [celecoxib + sildenafil] did not cause obvious frank damage to normal tissues from an athymic mouse (Fig. 1C). In contrast, treatment of animals with lower doses of [OSU‐03012 + sildenafil] or [celecoxib + sildenafil] significantly reduced tumor volumes after 5 days of treatment and reduced the long‐term viability of in vivo treated cells, as judged using ex vivo colony formation assays (Fig. 1D, data not shown).
Figure 1.
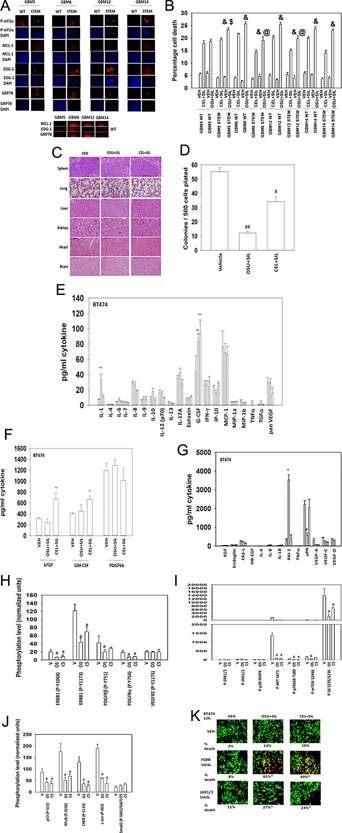
OSU‐03012 and sildenafil interact to kill tumor cells regardless of stem‐like or anoikis‐like properties, and that the tumor response to this combination provides biomarkers for the addition of a third agent. A: GBM5/6/12/14 cells or stem‐like selected variants of these cells were plated in 96‐well plates and 24 h after plating cells were cyto‐centrifuged to sediment cells to the plate, and cells were fixed with 2% paraformaldehyde containing Triton X100 for permeabilization. Immuno‐fluorescence was performed to determine the expression of GRP78/MCL‐1/P‐eIF2α, EDG‐1, and counter‐staining was with DAPI and detected using a Hermes Wiscan machine. The brightness/contrast for each IF image was set to show the most intense staining and all other images were presented with the identical same brightness/contrast setting. B: GBM cells or stem‐like variants of these cells were treated with vehicle; celecoxib (5 μM) + sildenafil (2 μM); or OSU‐03012 (1 μM) + sildenafil and the viability of cells determined by trypan blue exclusion assay after 24 h. (n = 3, ±SEM) and P < 0.05 greater than corresponding value in celecoxib treated cells; $ P < 0.05 greater than corresponding value in wild type cells; @ P < 0.05 less than corresponding value in wild type cells. C: Athymic mice were treated with vehicle (Cremophore); OSU‐03012 (50 mg/kg) + sildenafil (10 mg/kg); celecoxib (50 mg/kg + sildenafil (10 mg/kg) for 14 days (nota bene: below in section (D) that anti‐tumor effects were observed using half the doses of these drugs and for half as long). Animals were sacrificed, the normal tissue organs collected and fixed. Sections (10 μm) were taken and H&E stained by The Department of Pathology, VCU). D: Athymic nude mice were injected with 1 × 107 BT474 cells into their fourth mammary fat pad. Seven days after injection, animals with ∼50 mm3 tumors were treated by oral gavage with vehicle (cremophore); celecoxib (10 mg/kg) + sildenafil (5 mg/kg); OSU‐03012 (10 mg/kg) + sildenafil (5 mg/kg) for 5 days after which tumors were isolated, and plated as single cells to determine the ex vivo colony formation ability of in vivo treated tumors (n = 3 ± SEM). # P < 0.05 less than vehicle control; ## P < 0.05 less than [CEL + SIL] value. E–J: Tumors treated for 5 days in section (D) were isolated as was mouse blood and plasma collected. Multiplex assays were performed on tumor lysates and on clarified mouse plasma to detect changes in signal transduction parameters and on plasma cytokine levels (n = 4 tumors ± SEM). The treatments are presented in groups of three bars, with bars on the left being vehicle control; bars in the center being [OSU + SIL]; and bars on the right being [CEL + SIL]. ∼ P < 0.05 greater than vehicle control value; # P < 0.05 less than vehicle control value. K: BT474 cells were treated with vehicle control; OSU‐03012 (1 μM) and and sildenafil (2 μM); the FGFR inhibitor BGJ398 (1 μM); the JAK1/2 inhibitor ruxolitinib (1 μM) as indicated for 12 h. Cells viability was determined using a live/dead assay in a Hermes WiScan machine where red/yellow cells = dead; green cells = alive (n = 3 ± SEM). *P < 0.05 greater than corresponding value in vehicle control treated cells.
Based on these findings, we obtained normal brain and liver and obtained tumor material and plasma from [OSU‐03012 + sildenafil] and [celecoxib + sildenafil] treated animals carrying human BT474 mammary tumors that had been treated for 5 days with these drug combinations. We discovered that both drug combination treatments increased the levels of G‐CSF, and for [CEL + SIL] bFGF and GM‐CSF (Fig. 1E,F). Treatment with [OSU + SIL] strongly enhanced expression of the inhibitor PAI‐1 and reduced expression of its cognate cytokine uPA (Fig. 1G). Both drug combinations reduced the phosphorylation of ERBB1 and of the platelet‐derived growth factor (PDGF)Rα/β, but not VEGFR2 (Fig. 1H). The reduced activity of ERBB1 and PDGFRα/β correlated with reduced AKT, p70 S6K, and mTOR activity in drug treated tumors (Fig. 1I). The reduced activity of signal transduction pathways correlated with reduced activity of the transcription factors p53; NFkB; CREB; and c‐Jun, but not Smad2 (Fig. 1J). Based on our biomarker data, we then determined whether inhibition of specific additional pathways could enhance [OSU + SIL] lethality. Based on data examining G‐CSF, bFGF, and GM‐CSF levels we determined whether the clinically relevant FGFR pan inhibitor BGJ398 or the FDA approved JAK1/2 inhibitor ruxolitinib enhanced [OSU + SIL] lethality. Inhibition of FGFR signaling profoundly enhanced [OSU + SIL] lethality (Fig. 1K). Inhibition of JAK1/2 signaling more modestly, though significantly, also enhanced [OSU + SIL] lethality.
Expression of ERBB1, ERBB2, and of ERBB3 and ERBB4 in BT474 breast cancer cells was reduced by OSU + SIL treatment (Fig. 2A). Treatment of GBM cells with CEL + SIL and with OSU + SIL rapidly reduced total expression of ERBB1 (GBM12) and ERBB1 vIII (GBM6) within 6 h, whereas expression of c‐MET was reduced to a much lesser extent and in a cell‐dependent fashion (Fig. 2B). Drug combination exposures rapidly reduced expression of S1PR1 (EDG1) and S1PR5 (EDG5) as they did in a cell type dependent fashion for the IL6R and the IL8R (Fig. 2C,D). Expression of the IGF1R and of TNFαR1 were also reduced by the drug combination (Fig. 2E). Drug combination treatment also reduced the total expression of multiple plasma membrane associated drug efflux pumps; pumps that are generally over‐expressed in recurrent patient tumors and are partly responsible for chemotherapy resistance as well as for a functional blood–brain barrier (Fig. 2F). Identical data were obtained in vivo examining normal brain and liver tissues (Fig. 2G,H). Similar data showing knock down of ABCB1 and ABCG2 expression were also obtained in stem cell selected variants of GBM5/6/12/14 (data not shown). In previous studies, we have shown that expression of the death receptor CD95 (FAS) does not alter after particular these drug exposures and in fact becomes plasma membrane localized and clustered, observations indicative of CD95 activation (Booth et al., 2014a,2014b).
Figure 2.
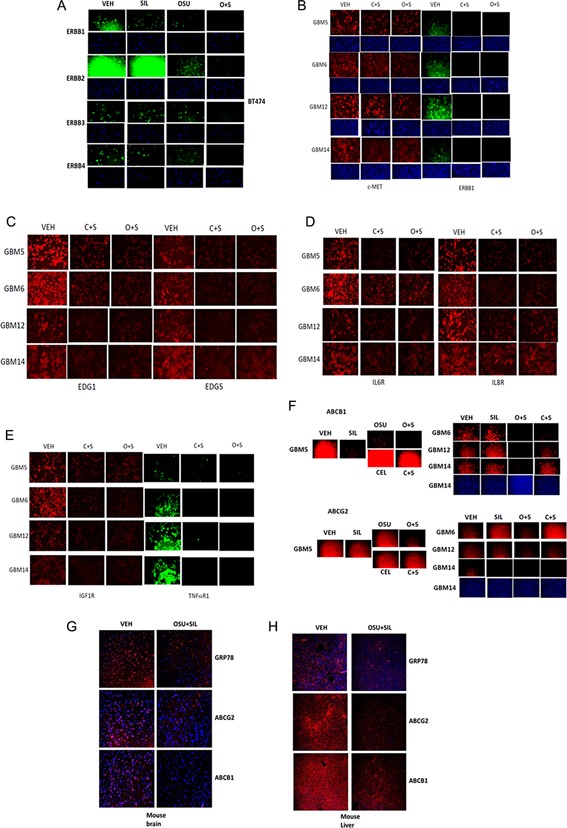
Treatment of tumor cells with [OSU‐03012 + sildenafil] or [celecoxib + sildenafil] rapidly reduces the expression of multiple growth factor receptors. A: BT474 mammary carcinoma cells known to strongly over‐express ERBB2 and: B–F: GBM5/6/12/14 cells; in 96‐well plates were treated with vehicle; celecoxib (5 μM) + sildenafil (2 μM); or OSU‐03012 (1 μM) + sildenafil and after 6 h treatment were fixed with 2% (v/v) paraformaldehyde containing Triton X100 to permeabilized cells. Immuno‐staining of the indicated growth factor receptors and plasma membrane drug transporters was performed using standard techniques and IF images detected using a Hermes Wiscan machine. G,H: Normal tissues from mice treated in Figure 1 section (D), brain and liver, were fixed, sectioned and immune‐stained to determine the total expression of GRP78, ABCB1, and ABCG2. Images were examined under 10× magnification using a Hermes WiScan instrument.
As treatment of cells with CEL + SIL or with OSU + SIL weakly reduced expression of c‐MET compared to other growth factor receptors, we determined whether a clinically relevant inhibitor of c‐MET (crizotinib) could enhance the lethality of CEL + SIL or with OSU + SIL. In a cell type dependent fashion, that did not correlate with basal expression or down‐regulation of c‐MET protein, crizotinib enhanced CEL + SIL or OSU + SIL lethality (Fig. 3A). The lethality of CEL + SIL was potently enhanced in all GBM cells by the multi‐kinase inhibitor sorafenib (Fig. 3B,C). Unlike [OSU + SIL] treatment in Fig. 2, the JAK1/2 inhibitor ruxolitinib did not significantly enhance CEL + SIL toxicity and the IGF1R inhibitor OSI‐906 only enhanced CEL + SIL toxicity in GBM6 and GBM14 cells. In GBM cells that expressed ERBB1 and ERBB1 vIII, the ERBB1/2 inhibitor lapatinib strongly enhanced CEL + SIL lethality. In a previous manuscript, we demonstrated that the histone deacetylase inhibitor and blocker of sphingosine‐1‐phosphate receptor signaling, FTY720 (multiple sclerosis drug: Gilenya), enhanced CEL + SIL lethality. The more potent HDAC inhibitors AR‐42, vorinostat enhanced CEL + SIL lethality in GBM cells with AR‐42 being most potent on a molar basis (Fig. 3D). Similar cell killing data were obtained in multiple GI tumor cell types (Fig. 3E). The lethality of the OSU + SIL drug combination was also enhanced by HDAC inhibitors (Fig. 3F,G). The multiple sclerosis drug FT720 as well as another multiple sclerosis drug (mono‐methyl fumarate [MMF]) enhanced OSU + SIL lethality (Fig. 3H).
Figure 3.
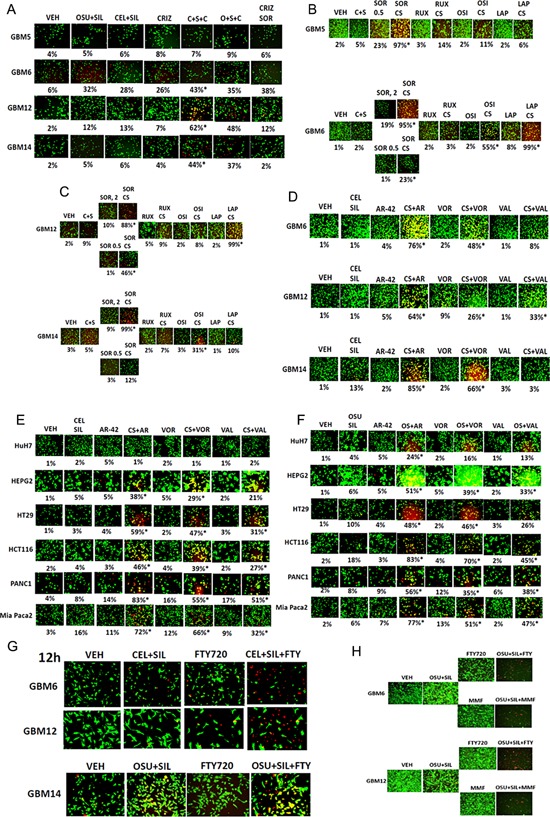
Third drug combinations with [OSU‐03012 + sildenafil] and with [celecoxib + sildenafil] to further enhance tumor cell killing. A: GBM5/6/12/14 cells in 96‐well plates were treated with vehicle; celecoxib (5 μM) + sildenafil (2 μM); or OSU‐03012 (1 μM) + sildenafil, with or without the drug crizotinib (1 μM) and after 12 h of treatment were subjected to live/dead assays where the percentage cell death is noted below each image (n = 3 ± SEM). *P < 0.05 greater than death value in two drug treated cells. B,C: GBM5/6/12/14 cells in 96‐well plates were treated with vehicle; celecoxib (5 μM) + sildenafil (2 μM), with or without the drugs: sorafenib (0.5, 2.0 μM); ruxolitinib (0.5 μM); OSI‐994 (1 μM); lapatinib (1 μM), as indicated. After 12 h of treatment cells were subjected to live/dead assays where the percentage cell death is noted below each image (n = 3 ± SEM). *P < 0.05 greater than death value in two drug treated cells. D–F: GBM5/6/12/14 cells or [Huh7, HEPG2 liver], [HT29, HCT116 colon], [PANC1, Mia Paca2 pancreatic] cells in 96‐well plates were treated with vehicle; celecoxib (5 μM) + sildenafil (2 μM) or OSU‐03012 (1 μM) + sildenafil with or without the drugs: AR‐42 (250 nM); vorinostat (500 nM); valproate (0.1 mM), as indicated. After 12 h of treatment, cells were subjected to live/dead assays where the percentage cell death is noted below each image (n = 3 ± SEM). *P < 0.05 greater than death value in two drug treated cells. G,H: GBM cells were treated with vehicle; celecoxib (5 μM) + sildenafil (2 μM) or OSU‐03012 (1 μM) + sildenafil with or without FTY720 (50 nM) with or without MMF (5 μM). After 12 h of treatment, cells were subjected to live/dead assays.
HDAC inhibitors increase the acetylation of HSP90, which reduces its chaperone function. The acetylation of heat shock protein (HSP) 90 was increased and the expression of the HSP90 client protein c‐FLIP‐s reduced by AR‐42 treatment (Fig. 4A, left blots; in agreement with Panner et al., 2007). This collectively argues that the combination of OSU + SIL + AR‐42 is inhibiting the functions of both GRP78 and HSP90, which may explain the strong killing interaction between the drugs. Furthermore, over‐expression of HSP90 did not significantly protect cells from OSU‐03012 toxicity as a single agent, though HSP90 over‐expression did modestly reduce the enhancement of cell death caused by IRE1 knock down; this effect was abolished by AR‐42 (Fig. 4A, right graph). Sildenafil and OSU‐03012 interacted to decrease total protein expression of HSP90α/β in multiple GBM cell isolates (Fig. 4A, upper IF images). Thus OSU‐03012 inhibits HSP90 function as a single agent and combined with sildenafil also considerably reduces HSP90 protein levels. Based on our findings showing the effect of sildenafil and OSU‐03012 on HSP90 function, we determined whether a clinically relevant HSP90 inhibitor (SNX2112/SNX5422, 250 nM in vitro; patient C max @ 100 mg/m2 is ∼3 μM) could enhance the lethality of OSU‐03012 and of [OSU + SIL] in GBM cells. SNX2112 at its IC90 concentration for HSP90 inhibition enhanced the toxicity of [OSU + SIL] (Fig. 4B).
Figure 4.
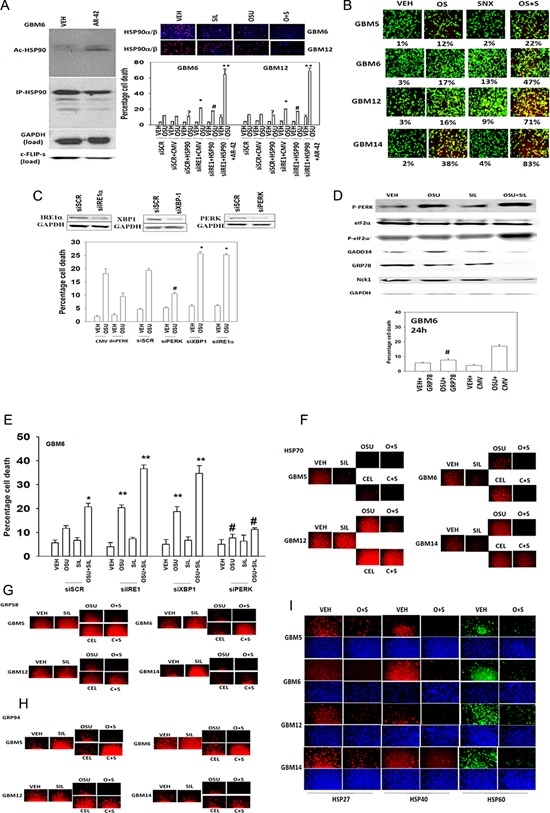
The regulation of chaperone function and expression by OSU‐03012, sildenafil and HDAC inhibitors. A: (Left blots) GBM6 cells were treated with AR‐42 (250 nM) for 90 min, after which HSP90 was immunoprecipitated and probed for total HSP90 expression and HSP90 acetylation. In parallel tumor, cell lysate was immunoblotted for the total expression of GAPDH and of c‐FLIP‐s. Right graph: GBM6 and GBM12 cells were infected with an empty vector virus or a virus to express HSP90, and in parallel transfected with either a scrambled siRNA (siSCR) or an siRNA to knock down IRE1 expression. Twenty‐four hours after infection, cells were treated with vehicle or OSU‐03012 (1 μM). Cells were isolated after 24 h and viability determined by trypan blue exclusion assay (n = 3 ± SEM). *P < 0.05 greater than siSCR + CMV value; **P < 0.05 greater than siIRE1 + CMV value; # P < 0.05 less than siIRE1 + CMV value; ? P > 0.90 between siSCR + CMV and siSCR + HSP90 values. IF images above graph: GBM6/12 cells were treated with vehicle; OSU‐03012 (1 μM); sildenafil (2 μM), or the drugs in combination and after 6 h treatment were fixed with 2% (v/v) paraformaldehyde containing Triton X100 to permeabilized cells. Immuno‐staining of HSP90α/β was performed using standard techniques and IF images detected using a Hermes Wiscan machine. B: GBM cells were treated for 12 h with vehicle, [OSU‐03012 (1 μM) + sildenafil (2 μM)], SNX2112 (250 nM), or the three drugs combined. Cell viability under each condition was determined by live/dead assays (n = 3 ± SEM). C: GBM6 cells were transfected with siSCR or siRNA molecules to knock down expression of IRE1, XBP1, and PERK. In parallel sets of plates, cells were transfected with empty vector plasmid (CMV) or to express dominant negative PERK. Twenty‐four hours after transfection, cells were treated with vehicle or with OSU‐03012 (2 μM). Twenty‐four hours after drug treatment, cell viability was determined by trypan blue exclusion assay (n = 3 ± SEM). # P < 0.05 lower than corresponding value in siSCR cells; *P < 0.05 greater than corresponding value in siSCR cells. D: (Upper blots) GBM6 cells were treated with OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for 6 h after which cells were lysed. Immunoblotting and phospho‐immunoblotting was performed to detect the expression/phosphorylation of the indicated proteins. Lower Graphs: GBM6 and GBM12 cells were transfected with empty vector (CMV) or to express GRP78. Twenty‐four hours after transfection, cells were treated with 2 μM OSU‐03012 and 24 h later viability determined via trypan blue exclusion assay (n = 3 ± SEM). # P < 0.05 less than corresponding value in CMV transfected cells. E: GBM6 cells were transfected with scrambled control (siSCR) or siRNA molecules to knock down IRE1, XBP1, or PERK. Twenty‐four hours after transfection, cells were treated with OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for 24 h. Cell viability was determined by trypan blue exclusion assay (n = 3 ± SEM). *P < 0.05 greater than OSU‐03012 single agent treatment in siSCR cells; **P < 0.05 greater than corresponding values in siSCR cells; # P < 0.05 less than corresponding values in siSCR cells. F–I: GBM cells were treated with vehicle; celecoxib (5 μM), sildenafil (2 μM), OSU‐03012 (1 μM) as single agents or in the indicated combinations for 6 h. Cells were then fixed with 2% (v/v) paraformaldehyde containing Triton X100 to permeabilize cells. Immuno‐staining of total HSP70 and HSC70 expression combined (part F) or expression of GRP94 and GRP58 (parts G and H) or expression of HSP27/HSP40/HSP60 (part I) was performed using standard techniques and IF images detected using a Hermes Wiscan machine.
As GRP78 function appeared to be playing a central role in our studies, we further explored the impact of altered ER stress signaling on the lethality of our CEL + SIL and OSU + SIL drug combinations. Knock down of PERK expression or expression of a dominant negative (dn) PERK protein suppressed single agent OSU lethality (Fig. 4C). Over‐expression of GRP78 protected cells from single agent OSU killing (Fig. 4D, lower graphs). Treatment of cells with OSU + SIL enhanced the phosphorylation of PERK to a greater extent than OSU alone, and profoundly enhanced eIF2α phosphorylation, which correlated with significantly reduced expression of GADD34, Nck1, and GRP78 (Fig. 4D, upper blots). Of note, OSU‐03012 as a single agent modestly increased the expression of GADD34 that was associated with enhanced PERK phosphorylation but not with enhanced eIF2α phosphorylation. Thus it is probable that GADD34 and Nck1, via the localized actions of protein phosphatase 1, attenuates the ER stress signal from PERK by facilitating dephosphorylation of eIF2α and that this inhibitory effect on eIF2α is overcome by the addition of sildenafil which reduces, below basal levels, the expression of GADD34 and Nck1, and de facto thus also eIF2α‐associated PP1 activity. To test this hypothesis further, cells were treated with OSU + SIL for 12 h and total eIF2α protein immunoprecipitated; in cells treated with OSU + SIL, the amount of PP1 and PP1 activity co‐precipitating with eIF2α was reduced (data not shown).
Knock down of IRE1 and XBP‐1 enhanced OSU + SIL lethality whereas knock down of ATF6 had no effect (Fig. 4E). Previously, we have shown that OSU‐03012 and celecoxib both modestly enhance expression of the HSP70 chaperone protein (Park et al., 2008). OSU‐03012 treatment modestly enhanced HSP70 chaperone expression in a cell type dependent fashion, an effect that was abolished by co‐exposure of cells with sildenafil resulting in a profound suppression of HSP70 levels (Fig. 4F). Treatment of cells with OSU‐03012 and sildenafil or celecoxib and sildenafil reduced expression of the ER‐localized chaperone proteins GRP58 and GRP94 (Fig. 4G,H). Treatment of cells with OSU‐03012 and sildenafil reduced expression of the ER‐localized chaperone proteins HSP27, HSP40, and HSP60 (Fig. 4I).
Treatment of GBM cells with OSU + SIL resulted in a large increase in eIF2α phosphorylation compared to individual drug treatments (Fig. 5A,B). The reduction in EDG‐1 expression (total and cell surface) caused by OSU + SIL was blocked by expression of a dn eIF2α S51A protein (Fig. 5C–G). Treatment of cells with OSU + SIL increased expression of ATF4 in an eIF2α‐dependent fashion (Fig. 6A,B). The reduction in EDG‐1 expression caused by OSU + SIL treatment was prevented by knock down of ATF4 or of CHOP (GADD153) (Fig. 6C–F). The OSU + SIL‐induced increase in CHOP expression was prevented by expression of dn eIF2α and the down‐regulation of EDG‐1 levels was blocked by over‐expression of GRP78 (Fig. 7A–D). Expression of dn eIF2α also prevented the drug‐induced reduction in MCL‐1 and BCL‐XL expression (Fig. 7E, data not shown). Over‐expression of GRP78 prevented the drug‐induced increase in eIF2α phosphorylation arguing that the kinase that phosphorylates eIF2α is probably inhibited by GRP78 (Fig. 7F). Over‐expression of GRP78 also prevented the drug combination‐induced rapid declines in the expression of EDG‐1, ERBB1, IL6R, IGF1R, and the PDGFRα/β, as well as drug efflux/blood–brain‐barrier ABC transporters, suggesting that the reduction in receptor expression and transporter expression is also a result of reduced GRP78 levels and enhanced ER stress signaling (Fig. 7G,H). Knock down of CHOP expression suppressed CEL + SIL and OSU + SIL lethality (Fig. 7I).
Figure 5.
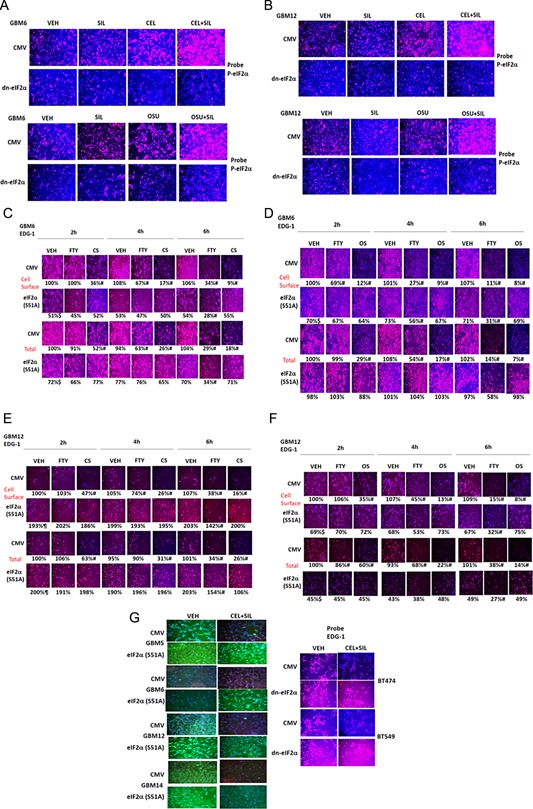
Celecoxib and sildenafil interact to increase eIF2α serine 51 phosphorylation which facilitates reduced expression of S1PR1 in GBM cells. A,B: GBM6/12 cells were plated in 96‐well plates and transfected with either empty vector or with a plasmid to express eIF2α S51A. Twenty‐four hours after transfection, cells were treated with vehicle, OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for 6 h. Cells were fixed and permeabilized and immuno‐fluorescence performed to determine the levels of phospho‐S51 eIF2α. C–G: GBM6/12 cells were plated in 96‐well plates and transfected with either empty vector or with a plasmid to express eIF2α S51A. Twenty‐four hours after transfection, cells were treated with vehicle, celecoxib (5 μM)/OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination, or with FTY720 (50 nM), for 2, 4, and 6 h, as indicated. Cells were fixed and permeabilized or not permeabilized as indicated and immuno‐fluorescence performed to determine the levels of S1PR1 (EDG‐1) in the plasma membrane and in the whole permeabilized cell (n = 3 ± SEM). # P < 0.05 less than vehicle treated value at that time point; $ P < 0.05 less than corresponding value in CMV transfected cells.
Figure 6.
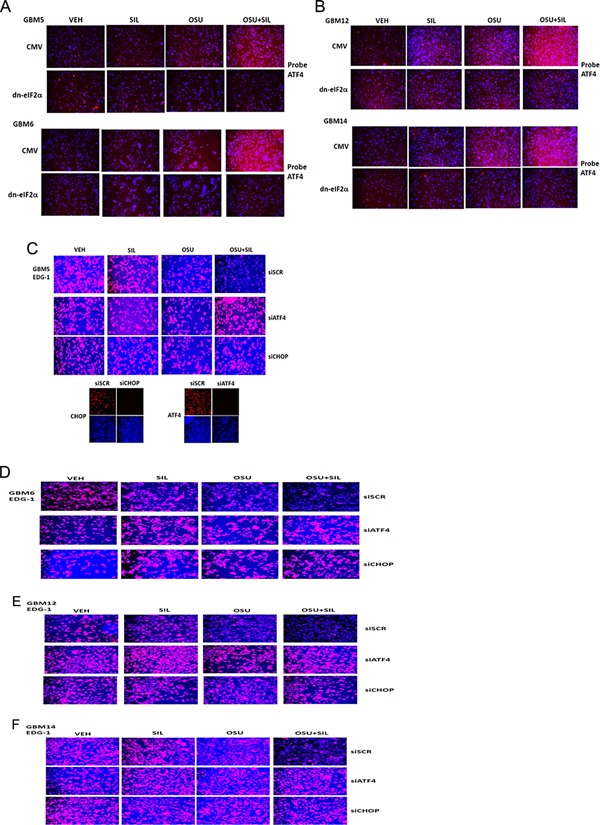
Increased expression of ATF4 after [OSU‐03012 + sildenafil] treatment is eIF2α dependent and the drug‐induced suppression of EDG‐1 expression requires ATF4/CHOP signaling. A,B: GBM cells were transfected with empty vector CMV or to express dominant negative eIF2α. Twenty‐four hours after transfection, cells were treated with vehicle, OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for 6 h. Cells were fixed and permeabilized and immuno‐fluorescence performed to determine the levels of ATF4. C–F: GBM cells were transfected with a scrambled siRNA (siSCR) or siRNA molecules to knock down the expression of ATF4 or CHOP. Twenty‐four hours after transfection, cells were treated with vehicle, OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for 6 h. Cells were fixed and permeabilized and immuno‐fluorescence performed to determine the total cellular levels of S1PR1 (EDG‐1).
Figure 7.
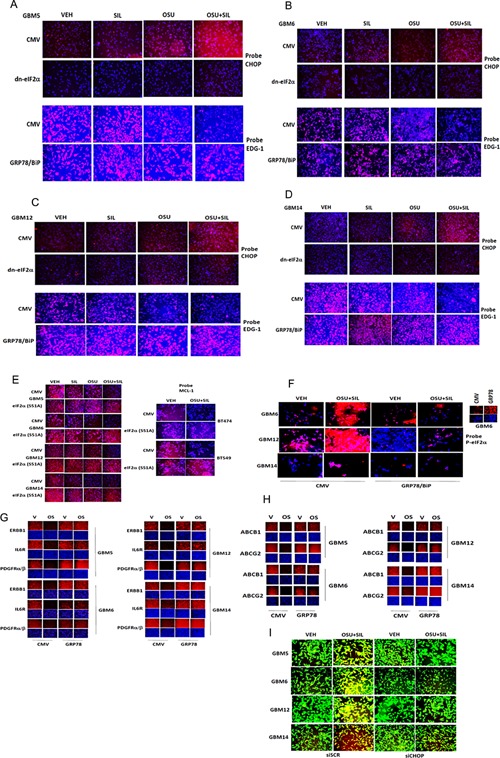
Activation of eIF2α signaling promotes expression of CHOP and down‐regulation of EDG‐1 that is blocked by over‐expression of GRP78. A–D: GBM cells as indicated were transfected with empty vector (CMV), with a plasmid to express GRP78, or with a plasmid to express dominant negative eIF2α. Twenty‐four hours after transfection, cells were treated with vehicle, OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for 6 h. Cells were fixed and permeabilized and immuno‐fluorescence performed to determine the total cellular levels of S1PR1 (EDG‐1), and CHOP (GADD153). E: Tumor cells were transfected with empty vector CMV or to express dominant negative eIF2α. Twenty‐four hours after transfection, cells were treated with vehicle, OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for 6 h. Cells were fixed and permeabilized and immuno‐fluorescence performed to determine the levels of MCL‐1. F: Tumor cells were transfected with empty vector CMV or to express GRP78. Twenty‐four hours after transfection, cells were treated with vehicle, OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for 6 h. Cells were fixed and permeabilized and immuno‐fluorescence performed to determine the levels of eIF2α serine 51 phosphorylation. G,H: Tumor cells were transfected with empty vector CMV or to express GRP78. Twenty‐four hours after transfection, cells were treated with vehicle or with OSU‐03012 (1 μM) and sildenafil (2 μM) for 6 h. Cells were fixed and permeabilized and immuno‐fluorescence performed to determine the total cellular levels of the indicated growth factor receptors and drug efflux pump proteins. I: GBM cells were transfected with a scrambled siRNA (siSCR) or an siRNA molecule to knock down the expression of CHOP. Twenty‐four hours after transfection, cells were treated with vehicle or with OSU‐03012 (1 μM) and sildenafil (2 μM). After 24 h of treatment, cells were subjected to live/dead assays where green cells are alive and yellow/red cells are dead.
In prior studies, we have shown that OSU‐03012 reduces the expression of GRP78 through protein destabilization/lower half‐life without significantly altering GRP78 promoter activity and that celecoxib increases GRP78 expression, with both drugs causing similar amounts of PERK phosphorylation. This implies that both drugs, that are very similar in structure, are probably binding to GRP78 with OSU‐0312 destabilizing the protein and celecoxib likely inhibiting the chaperone function of the protein, which both collectively result in increased PERK activity. Treatment of GBM cells with celecoxib and sildenafil increased GRP78 expression, that is, sildenafil did not alter the ability of celecoxib to increase GRP78 levels (Fig. 8A). Nevertheless, this increase in GRP78 expression was associated with increased eIF2α phosphorylation. Treatment of GBM cells with OSU‐03012 and sildenafil reduced expression of GRP78 and strongly enhanced eIF2α phosphorylation. Treatment of GBM cells with OSU‐03012 rapidly reduced cell surface and total expression of GRP78, an effect that was further enhanced by sildenafil (Fig. 8B–E). In dose–response studies, we discovered that OSU‐03012 + sildenafil treatment more rapidly and completely reduced the expression of GRP78, HSP70, and HSP90 than OSU‐03012 treatment alone (Fig. 8F). Similar data were obtained in primary mouse hepatocytes, and of note, as judged by DAPI staining no cell killing was observed (Fig. 8G). There are at least four kinases that phosphorylate eIF2α: PERK; PKR; GCN2; HRI. The nitric oxide synthase inhibitor L‐NAME abolished the ability of OSU‐03012 to reduce GRP78 levels (Fig. 8H). However, the cGMP‐dependent kinase inhibitor reduced the ability of both OSU‐03012 and [OSU‐03012 + sildenafil] to lower GRP78 levels arguing that OSU‐03012 may mediate some of its biologic effects through modulation of cGMP signaling. Knock down of PERK uniformly reduced the ability of OSU + SIL to increase eIF2α phosphorylation in all lines tested (Fig. 8I). However, in a cell isolate dependent fashion, we also found that PKR and HRI could contribute to the drug‐induced eIF2α phosphorylation (Fig. 8I,J).
Figure 8.
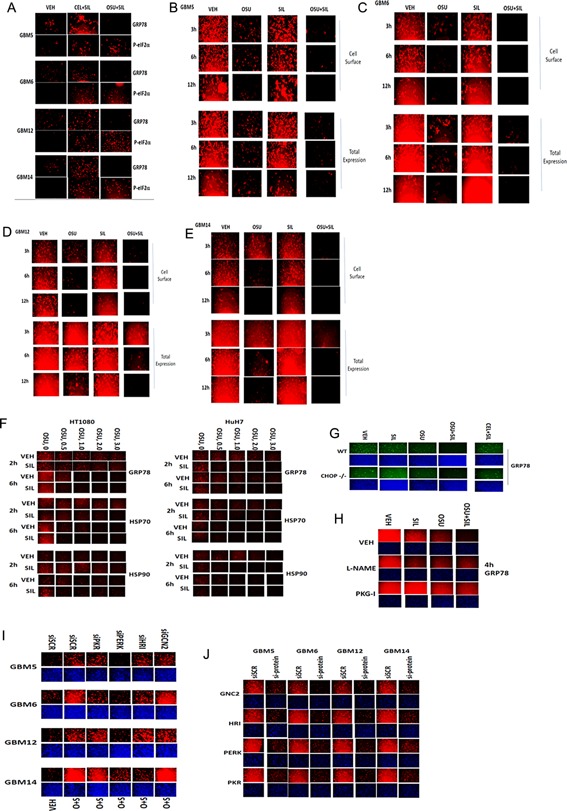
Sildenafil facilitates a rapid further reduction in GRP78 protein expression levels in cells treated with OSU‐03012. A: GBM cells were treated with vehicle, celecoxib (5 μM)/OSU‐03012 (1 μM), sildenafil (2 μM) or the drugs in combination for 6 h. Cells were fixed and permeabilized and immuno‐fluorescence performed to determine the total cellular levels of GRP78. B–E: GBM cells were treated with vehicle, OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination for the indicated amounts of time. At each time point, cells were fixed with paraformaldehyde and portions of fixed cells also being permeabilized with Triton X100. F: HuH7 (hepatoma) and HT1080 (sarcoma) were treated with vehicle, OSU‐03012 (0–3 μM), sildenafil (2 μM), or the drugs in combination. Cells after 2 and 6 h drug exposure were fixed in place and permeabilized. Immuno‐fluorescence was performed to determine the total cellular levels of GRP78; HSP90; HSP70. G: Primary mouse hepatocytes (wild type C57 black or CHOP −/− C57 black) were plated and 12 h after plating treated with vehicle, OSU‐03012 (1 μM), sildenafil (2 μM), or the drugs in combination. Six hours after drug exposure cells were fixed in place and permeabilized. Immuno‐fluorescence was performed to determine the total cellular levels of GRP78. H,I: GBM cells were transfected with scrambled siRNA or siRNA molecules to knock down the expression of PERK, PKR, GCN2, or HRI. Twenty‐four hours after transfection, cells were treated with vehicle or OSU‐03012 (1 μM) + sildenafil and the cells fixed and permeabilized 6 h later. Immuno‐fluorescence antibody staining was performed to determine the total levels of phospho‐eIF2α serine 51 under each condition.
Discussion
The present studies were initiated to determine the role of the ER chaperone protein GRP78/BiP/HSPA5 in the cell death response of tumor cells exposed to the drugs celecoxib or OSU‐03012 when combined with PDE5 inhibitors. We discovered that sildenafil (and other PDE5 inhibitors, unpublished results) enhanced the ability of OSU‐03012 to suppress GRP78 expression and blocked the ability of celecoxib to increase GRP78 expression. This was associated with increased expression of ATF4 and CHOP and elevated cell killing; with decreased expression of multiple growth factor receptors as well as plasma membrane drug transporters, effects that were blocked by GRP78 over‐expression. Killing was evident in GBM, breast, and GI tumor cells, and in cells selected for stem‐ness and for anoikis‐resistance.
In a prior study, we demonstrated that the HDAC inhibitor and S1P signaling inhibitor FTY720 enhanced the toxicity of CEL + SIL in vitro and in vivo (Booth et al., 2014b). The toxicity of CEL + SIL and OSU + SIL treatments were enhanced to a greater extent than FTY720 by high affinity HDAC inhibitors such as the clinically relevant drugs AR‐42 and vorinostat. Vorinostat is an FDA approved HDAC inhibitor for lymphoma and AR‐42 will in 2015 be re‐entering the clinic combined with the multi‐kinase inhibitor pazopanib for a phase I trial in sarcoma and renal carcinoma at The Medical College of Virginia Hospitals, Richmond, Virginia (Poklepovic and Dent, unpublished observations). HDAC inhibitors were also shown to cause inactivating acetylation of the HSP90 chaperone, implying that the combination of OSU + SIL + AR‐42 will probably reduce the chaperone functions of both GRP78, HSP70, HSC70, and HSP90, resulting in a tumor cell predisposed to the stimulation of cell death processes.
For GBM cells, CEL + SIL treatment only modestly down‐regulated c‐MET expression in GBM5 and GBM12 cells and in two of our four GBM isoaltes, GBM12 and GBM14, the c‐MET inhibitor crizotinib enhanced CEL + SIL and OSU + SIL lethality. However, c‐MET expression was effectively down‐regulated in GBM14 and not down‐regulated in GBM5 arguing that simplistic changes in receptor expression are not a valid biomarkers, and that the phosphorylation level of c‐MET may be a much better predictor of whether a tumor cell has any survival addition to signals from this receptor. As the diabetes drug metformin has also been shown to reduce GRP78 expression, we believe this agent could also interact with OSU‐03012 and sildenafil to profoundly enhance OSU + SIL lethality.
Our prior studies have linked OSU‐03012‐induced ER stress signaling by PERK as being causal in the toxicity of OSU‐03012. The present studies demonstrated that the drug combination of CEL + SIL or OSU + SIL strongly activated both PERK and eIF2α that was associated with decreased GADD34 and Nck1 and GRP78 expression. Reduced GADD34/Nck1 expression will facilitate eIF2α signaling by reducing co‐localization of the Ser/Thr phosphatase PP1 with eIF2α. Knock down of CHOP prevented drug combination down‐regulation of growth factor receptors and suppressed drug combination lethality, demonstrating that enhanced ER stress signaling distal to PERK and eIF2α was essential for drug combination toxicity. As published previously, we noted that the IRE1 arm of the ER stress response was protective against OSU‐03012 and OSU + SIL treatment, and that the ATF6 arm appeared not to significantly alter the response. Knock down of IRE1/XBP‐1 also enhanced OSU‐03012 and sildenafil combination toxicity. Further studies will need to understand how and why IRE1 signaling in our system is protective, for example, modulation of RIDD or XBP‐1 function (Tong et al., 2011; Donnelly et al., 2013; Sano et al., 2013; Maurel et al., 2014). Preliminary studies suggest the drug combination regulates viability downstream of IRE1 via altered EDEM expression (unpublished observations).
We have previously shown that OSU‐03012 as a single agent increases the levels of autophagosomes and autolysosomes and that this effect is significantly enhanced by sildenafil; enhanced autophagy in this system is cytotoxic as judged by the ability of ATG5 or Beclin1 knock down to prevent formation of autophagic vesicles and to prevent tumor cell killing. We also demonstrated that this elevated level of autophagy was suppressed by over‐expression of GRP78, and that GRP78 over‐expression protected tumor cells. In the present studies, we noted that the drug combination reduced the expression of both MCL‐1 and BCL‐XL, and as we have previously shown that this down‐regulation can lead to increased levels of unbound Beclin1, which stimulate autophagy, the ER stress signaling pathway will likely lead to increased toxic tumor cell autophagy as well as to increased mitochondrial instability. Thus cell death pathways through autophagy and caspase activation will become engaged following OSU + SIL treatment, suggestive that tumor cell resistance mechanisms will be subverted and become ineffective (Fig. 9).
Figure 9.
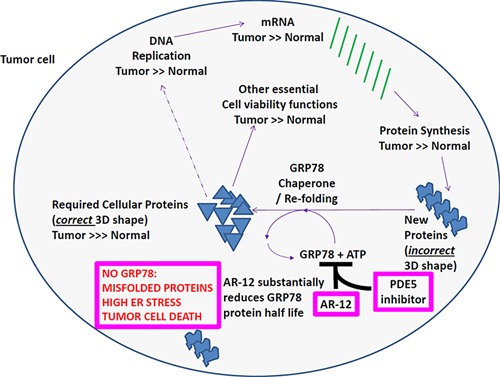
Possible molecular mechanisms by which OSU‐03012 (AR‐12) and PDE inhibitors could kill tumor cells. The ability of OSU‐03012 (AR‐12) alone, or enhanced by a PDE5 inhibitor, to reduce GRP78 expression and the function of multiple other chaperones will prevent the correct folding of essential tumor cell proteins resulting in dead tumor cells. The combination of OSU‐03012 and a PDE5 inhibitor will also reduce the expression of plasma membrane signaling receptors whose expression is essential for tumor cell growth and invasion. The loss of GRP78 and other chaperones in tumor cells will result in much lower expression levels of key oncogenic kinases, including PI3K‐AKT signaling (Liu et al., 2013, 2015; Tsai et al., 2015). Collectively our data strongly argue that it is a strong possibility that our drug combination will abolish the blood–brain barrier and have profound anti‐tumor capabilities.
Sildenafil was developed as an inhibitor of PDE5 with cardio‐protective effects, and serendipitously became an approved thereapeutic for erectile dysfunction (Zusman et al., 1999). PDE5 expression is not confined to the corpus cavernosum in the human penis and is expressed in the wider vasculature, myocardium, and tumor cells (Karami‐Tehrani et al., 2012; Shan et al., 2012). We have independently validated that PDE5 is over‐expressed in liver, colorectal, and lung cancer cells compared to normal tissues, and that viral infection increases PDE5 expression (unpublished observations). PDE5 catalyzes the degradation of cyclic GMP; that is, thus PDE5 inhibitors increase cGMP levels (Zhang et al., 2012). Nitric oxide (NO) induces smooth muscle relaxation via the actions of cGMP (Kato et al., 2012). NO at nanomolar levels binds tightly to a heme group in NO‐guanylyl cyclase (GC), also known as soluble guanylyl cyclase, and causes a ∼150‐fold activation of the enzyme (Potter, 2011). Activation of NO‐GC elevates cGMP levels, which initiate the cGMP signaling pathway, in part through activation of cGMP‐dependent protein kinase (PKG) (Friebe and Koesling, 2009). It is known in non‐tumor cells that cGMP/PKG, through its stimulatory actions upon the ERK, p38 MAPK, JNK, and NFκB pathways can increase the expression of inducible nitric oxide synthase (iNOS) (Komalavilas et al., 1999; Choi et al., 2007; Das et al., 2008). Thus, it is possible in our system that increased levels of NO activate GC and increase cGMP levels, that activates signaling pathways, which increase iNOS levels; and increased iNOS levels lead to further increases in cellular NO.
Elevation of cGMP or overexpression of constitutively active PKG can result in phosphorylation and activation of the JNK pathway and promote apoptosis (Thompson et al., 2000; Deguchi et al., 2004; Zhu et al., 2005a, 2009; Zhu and Strada, 2007). Our prior findings showed that OSU + SIL interacted to cause toxic JNK activation (Booth et al., 2012b). High concentrations of sildenafil and vardenafil induce caspase‐dependent apoptosis of B‐chronic lymphocytic leukemia cells but not in normal B cells, suggesting a tumor selective toxicity of PDE5 inhibitors (Sarfati et al., 2003). PDE5 inhibitors enhance tumor/vasculature permeability and efficacy of chemotherapy in a rat brain tumor model (Black et al., 2008). When transiently expressed in HT29 colon cancer cells, constitutively activated mutants of PKG beta, inhibit colony formation and induce apoptosis (Zhu et al., 2005b). In PC12 cells cGMP signaling, via activation of the AKT pathway prevents apoptosis (Ha et al., 2003). Others have argued that cGMP and NO kills cells through activation of the CD95/FAS‐L pathway (Hayden et al., 2001). These latter findings are similar to the data in our most recent manuscript examining this drug where OSU‐03012 and sildenafil interacted to kill through CD95 activation.
In conclusion, treatment of cells with OSU + SIL rapidly reduces the expression of the key chaperone protein GRP78, as well as reducing the chaperone functions of multiple other survival factors. The drug combination also largely abolishes the blood–brain barrier.
Acknowledgments
Support for the present study was funded in part by philanthropic monies supplied by Massey Cancer Center (to P.D.). Support for the present study was funded from PHS grants from the National Institutes of Health [R01‐CA141704, R01‐CA150214, R01‐DK52825, and R01‐CA61774]. Thanks to Mrs. Grizzard for her support to the Dent lab and to Dr. H. F. Young and the Betts family fund for support in the purchase of the Hermes Wiscan instrument. P.D. is the holder of the Universal, Inc. Chair in Signal Transduction Research. P.D. wishes to thank Dr. Hylemon (VCU) for supplying primary mouse hepatocytes.
The authors have no conflicts of interest to report.
Literature Cited
- Anderson K, Stott EJ, Wertz GW. 1992. Intracellular processing of the human respiratory syncytial virus fusion glycoprotein: Amino acid substitutions affecting folding, transport and cleavage. J Gen Virol 73:1177–1188. [DOI] [PubMed] [Google Scholar]
- Black KL, Yin D, Ong JM, Hu J, Konda BM, Wang X, Ko MK, Bayan JA, Sacapano MR, Espinoza A, Irvin DK, Shu Y. 2008. PDE5 inhibitors enhance tumor permeability and efficacy of chemotherapy in a rat brain tumor model. Brain Res 1230:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt G. 2001. The measles virus (MV) glycoproteins interact with cellular chaperones in the endoplasmic reticulum and MV infection upregulates chaperone expression. Arch Virol 146:2055–2068. [DOI] [PubMed] [Google Scholar]
- Booth L, Cazanave SC, Hamed HA, Yacoub A, Ogretmen B, Chen CS, Grant S, Dent P. 2012b. OSU‐03012 suppresses GRP78/BiP expression that causes PERK‐dependent increases in tumor cell killing. Cancer Biol Ther 13:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Cruickshanks N, Ridder T, Chen CS, Grant S, Dent P. 2012a. OSU‐03012 interacts with lapatinib to kill brain cancer cells. Cancer Biol Ther 13:1501–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Conley A, Cruickshanks N, Ridder T, Grant S, Poklepovic A, Dent P. 2014d. HDAC inhibitors enhance the lethality of low dose salinomycin in parental and stem‐like GBM cells. Cancer Biol Ther 15:305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Cruickshanks N, Conley A, Durrant DE, Das A, Fisher PB, Kukreja RC, Grant S, Poklepovic A, Dent P. 2014c. Phosphodiesterase 5 inhibitors enhance chemotherapy killing in gastrointestinal/genitourinary cancer cells. Mol Pharmacol 85:408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Cruickshanks N, Grant S, Poklepovic A, Dent P. 2014a. Regulation of OSU‐03012 toxicity by ER stress proteins and ER stress‐inducing drugs. Mol Cancer Ther 13:2384–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Cruickshanks N, Tavallai S, Webb T, Samuel P, Conley A, Binion B, Young HF, Poklepovic A, Spiegel S, Dent P. 2014b. PDE5 inhibitors enhance celecoxib killing in multiple tumor types. J Cell Physiol 230:1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton M, Brown DT. 1997. The formation of intramolecular disulfide bridges is required for induction of the Sindbis virus mutant ts23 phenotype. J Virol 71:7696–7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WT, Zhu G, Pfaffenbach K, Kanel G, Stiles B, Lee AS. 2014. GRP78 as a regulator of liver steatosis and cancer progression mediated by loss of the tumor suppressor PTEN. Oncogene 33:4997–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CY, Park KR, Lee JH, Jeon YJ, Liu KH, Oh S, Kim DE, Yea SS. 2007. Isoeugenol suppression of inducible nitric oxide synthase expression is mediated by down‐regulation of NF‐kappaB, ERK1/2, and p38 kinase. Eur J Pharmacol 576:151–159. [DOI] [PubMed] [Google Scholar]
- Cui W. Yu CH Hu KQ. 2005. In vitro and in vivo effects and mechanisms of celecoxib‐induced growth inhibition of human hepatocellular carcinoma cells. Clin Cancer Res 11:8213–8221. [DOI] [PubMed] [Google Scholar]
- Das A, Xi L, Kukreja RC. 2008. Protein kinase G‐dependent cardioprotective mechanism of phosphodiesterase‐5 inhibition involves phosphorylation of ERK and GSK3beta. J Biol Chem 283:29572–29585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi A, Thompson WJ, Weinstein IB. 2004. Activation of protein kinase G is sufficient to induce apoptosis and inhibit cell migration in colon cancer cells. Cancer Res 64:3966–3973. [DOI] [PubMed] [Google Scholar]
- Dimcheff DE, Faasse MA, McAtee FJ, Portis JL. 2004. Endoplasmic reticulum (ER) stress induced by a neurovirulent mouse retrovirus is associated with prolonged BiP binding and retention of a viral protein in the ER. J Biol Chem 279:33782–33790. [DOI] [PubMed] [Google Scholar]
- Donnelly N, Gorman AM, Gupta S, Samali A. 2013. The eIF2α kinases: Their structures and functions. Cell Mol Life Sci 70:3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Moss B, Doms RW. 1991. Folding, interaction with GRP78‐BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol 65:2047–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe A, Koesling D. 2009. The function of NO‐sensitive guanylyl cyclase: What we can learn from genetic mouse models. Nitric Oxide 21:149–156. [DOI] [PubMed] [Google Scholar]
- Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, Schroeder MA, James CD. 2005. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol 7:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Gorbatyuk OS. 2013. The molecular chaperone GRP78/BiP as a therapeutic target for neurodegenerative disorders: A mini review. J Genet Syndr Gene Ther 4: pii: 128. PMCID: 3674464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KS, Kim KM, Kwon YG, Bai SK, Nam WD, Yoo YM, Kim PK, Chung HT, Billiar TR, Kim YM. 2003. Nitric oxide prevents 6‐hydroxydopamine‐induced apoptosis in PC12 cells through cGMP‐dependent PI3 kinase/Akt activation. FASEB J 17:1036–1047. [DOI] [PubMed] [Google Scholar]
- Hayden MA, Lange PA, Nakayama DK. 2001. Nitric oxide and cyclic guanosine monophosphate stimulate apoptosis via activation of the Fas‐FasL pathway. J Surg Res 101:183–189. [DOI] [PubMed] [Google Scholar]
- He B. 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ 13:393–403. [DOI] [PubMed] [Google Scholar]
- Hogue BG, Nayak DP. 1992. Synthesis and processing of the influenza virus neuraminidase, a type II transmembrane glycoprotein. Virology 188:510–517. [DOI] [PubMed] [Google Scholar]
- Johnson AJ, Smith LL, Zhu J, Heerema NA, Jefferson S, Mone A, Grever M, Chen CS, Byrd JC. 2005. A novel celecoxib derivative, OSU‐03012, induces cytotoxicity in primary CLL cells and transformed B‐cell lymphoma cell line via a caspase‐and Bcl‐2‐independent mechanism. Blood 105:2504–2509. [DOI] [PubMed] [Google Scholar]
- Kang SG, Kim JS, Park K, Kim JS, Groves MD, Nam DH. 2006. Combination celecoxib and temozolomide in C6 rat glioma orthotopic model. Oncol Rep 15:7–13. [PubMed] [Google Scholar]
- Karami‐Tehrani F, Moeinifard M, Aghaei M, Atri M. 2012. Evaluation of PDE5 and PDE9 expression in benign and malignant breast tumors. Arch Med Res 43:470–475. [DOI] [PubMed] [Google Scholar]
- Kashfi K, Rigas B. 2005. Is COX‐2 a ‘collateral' target in cancer prevention? Biochem Soc Trans 33:724–727. [DOI] [PubMed] [Google Scholar]
- Kato M, Blanton R, Wang GR, Judson TJ, Abe Y, Myoishi M, Karas RH, Mendelsohn ME. 2012. Direct binding and regulation of RhoA by cyclic GMP‐dependent protein kinase Iα. J Biol Chem 287:41342–41351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenke FM, Gebhard MM, Ewerbeck V, Abdollahi A, Huber PE, Sckell A. 2006. The selective Cox‐2 inhibitor celecoxib suppresses angiogenesis and growth of secondary bone tumors: An intravital microscopy study in mice. BMC Cancer 12:6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehne CH Dubois RN. 2004. COX‐2 inhibition and colorectal cancer. Semin Oncol 31:12–21. [DOI] [PubMed] [Google Scholar]
- Komalavilas P, Shah PK, Jo H, Lincoln TM. 1999. Activation of mitogen‐activated protein kinase pathways by cyclic GMP and cyclic GMP‐dependent protein kinase in contractile vascular smooth muscle cells. J Biol Chem 274:34301–34309. [DOI] [PubMed] [Google Scholar]
- Kulp SK, Yang YT, Hung CC, Chen KF, Lai JP, Tseng PH, Fowble JW, Ward PJ, Chen CS. 2004. 3‐Phosphoinositide‐dependent protein kinase‐1/Akt signaling represents a major cyclooxygenase‐2‐independent target for celecoxib in prostate cancer cells. Cancer Res 64:1444–1451. [DOI] [PubMed] [Google Scholar]
- Lee AS. 2007. GRP78 induction in cancer: Therapeutic and prognostic implications. Cancer Res 67:3496–3499. [DOI] [PubMed] [Google Scholar]
- Lin YG, Shen J, Yoo E, Liu R, Yen HY, Mehta A, Rajaei A, Yang W, Mhawech‐Fauceglia P, DeMayo FJ, Lydon J, Gill P, Lee AS. 2015. Targeting the glucose‐regulated protein‐78 abrogates Pten‐null driven AKT activation and endometrioid tumorigenesis. Oncogene, DOI: 10.1038/onc.2015.4 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Li X, Gao W, Zhou Y, Wey S, Mitra SK, Krasnoperov V, Dong D, Liu S, Li D, Zhu G, Louie S, Conti PS, Li Z, Lee AS, Gill PS. 2013. Monoclonal antibody against cell surface GRP78 as a novel agent in suppressing PI3K/AKT signaling, tumor growth, and metastasis. Clin Cancer Res 19:6802–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Lee AS. 2013. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 32:805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel M, Chevet E, Tavernier J, Gerlo S. 2014. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci 39:245–254. [DOI] [PubMed] [Google Scholar]
- Moreno JA, Tiffany‐Castiglioni E. 2014. The chaperone Grp78 in protein folding disorders of the nervous system. Neurochem Res 40:329–335. [DOI] [PubMed] [Google Scholar]
- Narayanan BA, Narayanan NK, Pittman B, Reddy BS. 2006. Adenocarcina of the mouse prostate growth inhibition by celecoxib: Downregulation of transcription factors involved in COX‐2 inhibition. Prostate 66:257–265. [DOI] [PubMed] [Google Scholar]
- Panner A, Murray JC, Berger MS, Pieper RO. 2007. Heat shock protein 90 alpha recruits FLIPS to the death‐inducing signaling complex and contributes to TRAIL resistance in human glioma. Cancer Res 67:9482–9489. [DOI] [PubMed] [Google Scholar]
- Park MA, Yacoub A, Rahmani M, Zhang G, Hart L, Hagan MP, Calderwood SK, Sherman MY, Koumenis C, Spiegel S, Chen CS, Graf M, Curiel DT, Fisher PB, Grant S, Dent P. 2008. OSU‐03012 stimulates PKR‐like endoplasmic reticulum‐dependent increases in 70‐kDa heat shock protein expression, attenuating its lethal actions in transformed cells. Mol Pharmacol 73:1168–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MI, Subbaramaiah K, Du B, Chang M, Newman RA, Cordon‐Cardo C, Thaler HT, Dannenberg AJ. 2005. Celecoxib inhibits prostate cancer growth: Evidence of a cyclooxygenase‐2‐independent mechanism. Clin Cancer Res 11:1999–2007. [DOI] [PubMed] [Google Scholar]
- Potter LR. 2011. Guanylyl cyclase structure, function and regulation. Cell Signal 23:1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R, Fiskus W, Ganguly S, Kambhampati S, Bhalla KN. 2012. HDAC inhibitors and chaperone function. Adv Cancer Res 116:239–262. [DOI] [PubMed] [Google Scholar]
- Rathore APS, Ng ML, Vasudevan SG. 2013. Differential unfolded protein response during Chikungunya and Sindbis virus infection: CHIKV nsP4 suppresses eIF2α phosphorylation. Virol J 10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Shurtleff AC, Costantino JA, Tritsch SR, Retterer C, Spurgers KB, Bavari S. 2014. HSPA5 is an essential host factor for Ebola virus infection. Antiviral Res 109:171–174. [DOI] [PubMed] [Google Scholar]
- Roberts JL, Booth L, Conley A, Cruickshanks N, Malkin M, Kukreja RC, Grant S, Poklepovic A, Dent P. 2014. PDE5 inhibitors enhance the lethality of standard of care chemotherapy in pediatric CNS tumor cells. Cancer Biol Ther 15:758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller C, Maddalo D. 2013. The molecular chaperone GRP78/BiP in the development of chemoresistance: Mechanism and possible treatment. Front Pharmacol 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux L. 1990. Selective and transient association of Sendai virus HN glycoprotein with BiP. Virology 175:161–166. [DOI] [PubMed] [Google Scholar]
- Sano R, Reed JC. 2013. ER stress‐induced cell death mechanisms. Biochim Biophys Acta 1833:3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfati M, Mateo V, Baudet S, Rubio M, Fernandez C, Davi F, Binet JL, Delic J, Merle‐Beral H. 2003. Sildenafil and vardenafil, types 5 and 6 phosphodiesterase inhibitors, induce caspase‐dependent apoptosis of B‐chronic lymphocytic leukemia cells. Blood 101:265–269. [DOI] [PubMed] [Google Scholar]
- Shan X, Quaile MP, Monk JK, French B, Cappola TP, Margulies KB. 2012. Differential expression of PDE5 in failing and nonfailing human myocardium. Circ Heart Fail 5:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Meunier L, Hendershot LM. 2002. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J Biol Chem 277:15947–15956. [DOI] [PubMed] [Google Scholar]
- Spurgers KB, Alefantis T, Peyser BD, Ruthel GT, Bergeron AA, Costantino JA, Enterlein S, Kota KP, Boltz RC, Aman MJ, Delvecchio VG, Bavari S. 2010. Identification of essential filovirion‐associated host factors by serial proteomic analysis and RNAi screen. Mol Cell Proteomics 9:2690–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WJ, Piazza GA, Li H, Liu L, Fetter J, Zhu B, Sperl G, Ahnen D, Pamukcu R. 2000. Exisulind induction of apoptosis involves guanosine 3′,5′‐cyclic monophosphate phosphodiesterase inhibition, protein kinase G activation, and attenuated beta‐catenin. Cancer Res 60:3338–3342. [PubMed] [Google Scholar]
- Tong L, Heim RA, Wu S. 2011. Nitric oxide: A regulator of eukaryotic initiation factor 2 kinases. Free Radic Biol Med 50:1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YL, Zhang Y, Tseng CC, Stanciauskas R, Pinaud F, Lee AS. 2015. Characterization and mechanism of stress‐induced translocation of 78‐kilodalton glucose regulated protein (GRP78) to the cell surface. J Biol Chem pii: jbc.M114.618736 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A, Bellamy AR, Taylor JA. 1998. BiP (GRP78) and endoplasmin (GRP94) are induced following rotavirus infection and bind transiently to an endoplasmic reticulum‐localized virion component. J Virol 72:9865–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Jensen G, Yen TS. 1997. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol 71:7387–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub A, Park MA, Hanna D, Hong Y, Mitchell C, Pandya AP, Harada H, Powis G, Chen CS, Koumenis C, Grant S, Dent P. 2006. OSU‐03012 promotes caspase‐independent but PERK‐, cathepsin B‐, BID‐, and AIF‐dependent killing of transformed cells. Mol Pharmacol 70:589–603. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yan G, Ji J, Wu J, Sun X, Shen J, Jiang H, Wang H. 2012. PDE5 inhibitor promotes melanin synthesis through the PKG pathway in B16 melanoma cells. J Cell Biochem 113:2738–2743. [DOI] [PubMed] [Google Scholar]
- Zhu J, Huang JW, Tseng PH, Yang YT, Fowble JW, Shiau CW, et al. 2004. From the cyclooxygenase‐2 inhibitor celecoxib to a novel class of 3‐phosphoinositide‐dependent protein kinase‐1 inhibitors. Cancer Res 64:4309–4318. [DOI] [PubMed] [Google Scholar]
- Zhu B, Strada SJ. 2007. The novel functions of cGMP‐specific phosphodiesterase 5 and its inhibitors in carcinoma cells and pulmonary/cardiovascular vessels. Curr Top Med Chem 7:437–454. [DOI] [PubMed] [Google Scholar]
- Zhu B, Strada S, Stevens T. 2005a. Cyclic GMP‐specific phosphodiesterase 5 regulates growth and apoptosis in pulmonary endothelial cells. Am J Physiol Lung Cell Mol Physiol 289:L196–L206. [DOI] [PubMed] [Google Scholar]
- Zhu B, Vemavarapu L, Thompson WJ, Strada SJ. 2005b. Suppression of cyclic GMP‐specific phosphodiesterase 5 promotes apoptosis and inhibits growth in HT29 cells. J Cell Biochem 94:336–350. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zhang L, Alexeyev M, Alvarez DF, Strada SJ, Stevens T. 2009. Type 5 phosphodiesterase expression is a critical determinant of the endothelial cell angiogenic phenotype. Am J Physiol Lung Cell Mol Physiol 296:L220–L228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman RM, Morales A, Glasser DB, Osterloh IH. 1999. Overall cardiovascular profile of sildenafil citrate. Am J Cardiol 83:35C–44C. [DOI] [PubMed] [Google Scholar]


