Abstract
We determined whether the multi‐kinase inhibitor sorafenib or its derivative regorafenib interacted with phosphodiesterase 5 (PDE5) inhibitors such as Viagra (sildenafil) to kill tumor cells. PDE5 and PDGFRα/β were over‐expressed in liver tumors compared to normal liver tissue. In multiple cell types in vitro sorafenib/regorafenib and PDE5 inhibitors interacted in a greater than additive fashion to cause tumor cell death, regardless of whether cells were grown in 10 or 100% human serum. Knock down of PDE5 or of PDGFRα/β recapitulated the effects of the individual drugs. The drug combination increased ROS/RNS levels that were causal in cell killing. Inhibition of CD95/FADD/caspase 8 signaling suppressed drug combination toxicity. Knock down of ULK‐1, Beclin1, or ATG5 suppressed drug combination lethality. The drug combination inactivated ERK, AKT, p70 S6K, and mTOR and activated JNK. The drug combination also reduced mTOR protein expression. Activation of ERK or AKT was modestly protective whereas re‐expression of an activated mTOR protein or inhibition of JNK signaling almost abolished drug combination toxicity. Sildenafil and sorafenib/regorafenib interacted in vivo to suppress xenograft tumor growth using liver and colon cancer cells. From multiplex assays on tumor tissue and plasma, we discovered that increased FGF levels and ERBB1 and AKT phosphorylation were biomarkers that were directly associated with lower levels of cell killing by ‘rafenib + sildenafil. Our data are now being translated into the clinic for further determination as to whether this drug combination is a useful anti‐tumor therapy for solid tumor patients. J. Cell. Physiol. 230: 2281–2298, 2015. © 2015 The Authors. Journal of Cellular Physiology Published by Wiley Periodicals, Inc.
Abbreviations
- Ad
adenovirus
- CMV
empty vector plasmid or virus
- SCR
scrambled
- si
small interfering
- SIL
sildenafil
- SOR
sorafenib
- REGO
regorafenib
- VEH
vehicle
Phosphodiesterase 5 (PDE5) inhibitors were originally developed as agents to manipulate cardio‐vascular biology that were in parallel noted to treat erectile dysfunction (Watanabe et al., 2002; Benavides et al., 2013). Inhibition of PDE5 suppresses the degradation of cyclic GMP resulting in the activation of PKG (Francis et al., 2010). cGMP/PKG, through its stimulatory actions upon the ERK, p38 MAPK, JNK, and NFκB pathways can increase the expression of inducible nitric oxide synthase (iNOS), resulting in the production of nitric oxide (NO) (Komalavilas et al., 1999; Choi et al., 2007; Das et al., 2008; Musicki et al., 2014). NO and cGMP/PKG have multiple cellular targets including (to name but a few) ion channels, receptors, phospholipases, Rho A, altered protein nitrosylation, ceramide generation, and death receptor signaling (Hayden et al., 2001; Florio et al., 2003; Choi et al., 2007; Kots et al., 2011; Russwurm et al., 2013; Musicki et al., 2014).
Prior studies from our laboratories have demonstrated that PDE5 inhibitors enhance the toxicities of multiple well established cytotoxic chemotherapies (Das et al., 2010; Booth et al., 2014; Roberts et al., 2014; Booth et al., 2015). In these studies PDE5 inhibitors, in an NOS‐dependent fashion, were show to enhance chemotherapy killing through activation of the CD95 death receptor pathway, the generation of reactive oxygen species and mitochondrial dysfunction. The mechanism(s) by which PDE5 inhibitors and chemotherapies interacted to activate CD95 were not further explored.
Sorafenib and regorafenib are multi‐kinase inhibitors approved for the treatment of liver and kidney, and colon cancers, respectively (Carr et al., 2013). Sorafenib was originally developed as an inhibitor of RAF‐1 in the ERK1/2 pathway. The steady state (7 day) Cmax for sorafenib is ~21 μM in plasma, with ~99% of the drug protein bound based on in vitro human serum binding assays; though it is known that the drug is also rapidly taken up into tissues, and in addition patient data from clinical trials would argue that a significant amount of the drug has to be bioavailable, at least in the low micro‐molar range, in a tumor based on its single agent effects by decreasing both ERK1/2 phosphorylation and reducing MCL‐1 protein expression in tumor cells that are not specifically oncogene addicted (Hotte and Hirte, 2002; Elser et al., 2007). Indeed, it has been shown that some sorafenib metabolites such as M2, M4, and M5 can have up to 10‐fold greater activity than the parent drug (Inaba et al., 2001; Li et al., 2010; Pratz et al., 2010). Our prior in vitro and in vivo data have tended to argue using several sorafenib + “drug” combinations that PDGFRβ is a major target of sorafenib for its interactions with other agents, e.g., with histone deacetylase inhibitors (Martin et al., 2009; Park et al., 2010a,2010b).
A major biological effect of sorafenib is the induction of an endoplasmic stress (ER)/unfolded protein response (UPR), with reduced expression of proteins that have short half‐lives such as MCL‐1 and BCL‐XL (e.g., Rahmani et al., 2005; Rahmani et al., 2007; Martin et al., 2009). Reduced MCL‐1 levels due to sorafenib exposure have been linked in many tumor types to increased levels of apoptosis. Studies by our group have also linked high dose single agent sorafenib exposure to an increase in the levels of autophagic markers including increased numbers of LC3‐GFP vesicles and elevated expression of Beclin1 and ATG5; however lower sorafenib concentrations only caused a modest transient alteration in autophagy flux (Martin et al., 2009; Park et al., 2010a,2010b). Other studies from our groups have shown that based on the sorafenib dose the induction of ER stress may be a “protective” or a “toxic” event in the cellular response to the drug (e.g., Rahmani et al., 2005).
The present studies determined whether the clinically relevant PDE5 inhibitors interacted with the multi‐kinase inhibitors sorafenib/regorafenib to kill tumor cells. Our data demonstrate a strong interaction between these drugs in multiple tumor cell types with killing that is due to both death receptor activation and a toxic form of autophagy. Biomarker studies reveal ERBB1/AKT signaling as a compensatory survival response.
Materials and Methods
Materials
Phospho‐/total‐antibodies were purchased from Cell Signaling Technologies (Danvers, MA) and Santa Cruz Biotechnology (Santa Cruz, CA). All drugs were purchased from Selleckchem (Houston, TX). Commercially available validated short hairpin RNA molecules to knock down RNA/protein levels were from Qiagen (Valencia, CA). Antibody reagents, other kinase inhibitors, caspase inhibitors cell culture reagents, and non‐commercial recombinant adenoviruses have been previously described (Park et al., 2008, 2010a,2010b; Martin et al., 2009; Tang et al., 2012; Cruickshanks et al., 2013). Cell lines were obtained from the ATCC (Bethesda, MD) and were not further validated beyond the validation statements of the ATCC.
Tissue microarray and immunostaining
Human HCC tissue microarrays were purchased from Imgenex Corp. Two tissue microarrays were used: one containing 40 primary HCC, 10 metastatic HCC, and 9 normal adjacent liver samples (Imgenex; IMH‐360), the other containing 46 primary HCC and 13 metastatic HCC (Imgenex; IMH‐318). Antigen retrieval was performed by the Department of Pathology, VCU (with thanks to Dr. George Alemara). Immuno‐staining was performed using anti‐PDE5 antibody (1:50) or using a mixture of anti‐PDGFRα and anti‐PDGFRβ antibodies (1:50, each). After immuno‐staining, slides were H&E stained and slides examined using a confocal microscope (×10).
Methods
Cell culture and in vitro exposure of cells to drugs
All fully established cancer lines were cultured at 37°C (5% (v/v) CO2) in vitro using RPMI supplemented with 10% (v/v) fetal calf serum and 10% (v/v) non‐essential amino acids. For short‐term cell killing assays and immunoblotting, cells were plated at a density of 3 × 103 per cm2 and 24 h after plating were treated with various drugs, as indicated. In vitro small molecule inhibitor treatments were from a 100 mM stock solution of each drug and the maximal concentration of Vehicle (DMSO) in media was 0.02% (v/v). Cells were not cultured in reduced serum media during any study.
Cell treatments, SDS‐PAGE, and Western blot analysis
Cells were treated with various drug concentrations, as indicated in the figure legends. SDS‐PAGE and immunoblotting were performed as described in references Park et al. (2008, 2010a), Martin et al. (2009), Tang et al. (2012), and Cruickshanks et al. (2013). For SDS‐PAGE and immunoblotting, cells were plated at 5 × 105 cells/cm2 and treated with drugs at the indicated concentrations and after the indicated time of treatment, lysed in whole‐cell lysis buffer (0.5 M Tris‐HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β‐mercaptoethanol, 0.02% bromophenol blue), and the samples were boiled for 30 min. The boiled samples were loaded onto 10–14% SDS‐PAGE and electrophoresis was run overnight (10–100 µg/lane based on the gel size). Proteins were electrophoretically transferred onto 0.22 µm nitrocellulose, and immunoblotted with various primary antibodies against different proteins. All immunoblots were visualized using an Odyssey infrared imager (Li‐Cor, Lincoln, NE). For presentation, immunoblots were digitally assessed using the provided Odyssey imager software. Images have their color removed and figures generated in Microsoft PowerPoint.
Recombinant adenoviral vectors; infection in vitro
We generated and purchased previously noted recombinant adenoviruses (Park et al., 2008, 2010a,2010b; Martin et al., 2009; Tang et al., 2012; Cruickshanks et al., 2013). Cells were infected with these adenoviruses at an approximate m.o.i. as indicated in the figure/legend (usually 50 m.o.i.). Cells were incubated for 24 h to ensure adequate expression of transduced gene products prior to drug exposures.
Analysis of ROS levels
ROS levels were determined in a vector 3 plate reader (PerkinElmer Life and Analytical Sciences). In brief, cancer cells were plated in 96‐well plates. Cells were loaded for 30 min with either dihydro‐DCF (10 μM) which is sensitive to oxidation by hydroxyl radicals and peroxynitrite directly and hydrogen peroxide; or 3‐amino,4‐aminomethyl‐2′,7′‐difluorescein (DAF‐FM DA, 4 μM) which is sensitive to oxidation by NO. Cells were treated with drugs and fluorescence measured 2 and 6 h afterwards. Data are presented corrected for basal fluorescence of vehicle‐treated cells at each time point.
Detection of cell death by trypan blue and live/dead assays
For trypan blue assays, floating cells were isolated along with attached cells that were harvested by trypsinization with Trypsin/EDTA for ~10 min at 37°C. For live/dead assays in 96‐well plates, plates were gently spun to sediment detached dead cells onto the plate. Cells were then incubated with green fluorescent calcein‐AM to indicate intracellular esterase activity and red fluorescent di‐ethidium bromide to detect cells with disrupted plasma membranes. Cells are visualized using a Hermes Wiscan microscope with integrated imaging software to permit total cell counting and thus, determination of the percentage dead cells (five fields per well; green = alive; yellow = dying; red = dead).
Assessment of autophagy
Cells were transfected with a plasmid to express a green fluorescent protein (GFP) and red fluorescent protein (RFP) tagged form of LC3 (ATG8). For analysis of cells transfected with the GFP‐RFP‐LC3 construct, the GFP/RFP‐positive vesicularized cells were examined under the ×40 objective of a Zeiss Axiovert fluorescent microscope.
Plasmid transfection
Plasmids
Cells were plated as described above and 24 h after plating, transfected. Plasmids (0.5 μg) expressing a specific mRNA or appropriate vector control plasmid DNA was diluted in 50 μl serum‐free and antibiotic‐free medium (one portion for each sample). Concurrently, 2 μl Lipofectamine 2,000 (Invitrogen), was diluted into 50 μl of serum‐free and antibiotic‐free medium. Diluted DNA was added to the diluted Lipofectamine 2,000 for each sample and incubated at room temperature for 30 min. This mixture was added to each well/dish of cells containing 200 μl serum‐free and antibiotic‐free medium for a total volume of 300 μl and the cells were incubated for 4 h at 37°C. An equal volume of 2× medium was then added to each well. Cells were incubated for 48 h, then treated with drugs. To assess transfection efficiency of plasmids, we used a plasmid to express GFP and defined the percentage of cells being infected as the percentage of GFP+ cells. For all cell lines the infection efficiency was >70%.
siRNA
Cells were plated in 60 mm dishes from a fresh culture growing in log phase as described above, and 24 h after plating transfected. Prior to transfection, the medium was aspirated and 1 ml serum‐free medium was added to each plate. For transfection, 10 nM of the annealed siRNA, the positive sense control doubled stranded siRNA targeting GAPDH or the negative control (a “scrambled” sequence with no significant homology to any known gene sequences from mouse, rat, or human cell lines) were used (predominantly Qiagen, Valencia, CA; occasional alternate siRNA molecules were purchased from Ambion, Inc., Austin, TX). Ten nanomoles siRNA (scrambled or experimental) was diluted in serum‐free media. Four microliters Hiperfect (Qiagen) was added to this mixture and the solution was mixed by pipetting up and down several times. This solution was incubated at room temperature for 10 min, then added drop‐wise to each dish. The medium in each dish was swirled gently to mix, then incubated at 37°C for 2 h. One milliliter of 10% (v/v) serum‐containing medium was added to each plate, and cells were incubated at 37°C for 24–48 h before re‐plating (50 × 103 cells each) onto 12‐well plates. Cells were allowed to attach overnight, then treated with drugs (0–48 h). Trypan blue exclusion assays and SDS PAGE/immunoblotting analyses were then performed at the indicated time points.
Mass spectrometry measurements
Equal numbers of cells (5.98 ± 0.02 × 106) cells were treated for 6 h and lipids were extracted. Bioactive lipid levels were quantified by liquid chromatography‐electrospray ionization‐tandem mass spectrometry (LC‐ESI‐MS/MS) with a Shimadzu LC‐20AD binary pump HPLC system (Shimadzu, Kyoto, Japan) and an Applied Biosystems 4,000 QTRAP operating in a triple quadrupole mode, as described previously (see Hait et al., 2009 for additional Methodological details) (Hait et al., 2014).
Tumor growth studies
Athymic female NCr‐nu/nu mice (National Cancer Institute) weighing 20 g were used for human tumor xenograft studies. Mice were maintained under pathogen‐free conditions in facilities approved by the American Association for Accreditation of Laboratory Animal Care and in accordance with current regulations and standards of the US Department of Agriculture, the US Department of Health and Human Services, and the National Institutes of Health. Mice were injected with 0.5 × 107 HuH7/HT29 cells unless otherwise indicated (~40 mice per separate experiment to obtain at least four usable tumors per group) in 10 µl of growth medium. HuH7/HT29 cells were in the flank. Seven days after tumor cell implantation, mice were PO administered drugs or the drug combination QD for 3 days (HuH7 tumors) or 7 days (HT29 tumors). Animals were monitored daily and tumor volume determined as indicated in the figures. When the volume of the tumor reached >1,500 mm3, animals were humanely sacrificed and the tumor and blood removed for further studies. Animal survival was plotted on a Kaplan–Meier graph and longitudinal statistical assays performed.
Multiplex assays for cytokine expression
A MAGPIX multiplex instrument with associated software was purchased from Bio‐Rad. The following Bio‐Plex assay plates were used in our assays of mouse plasma for human cytokines: Bio‐Plex Pro Human Cytokine Group I 4‐plex (Y500023JM2); Human CYTO STD GRP II 23‐PLEX (171D60001); Human CYTO HGF set (171B6008M); Human CYTO SDF‐1a set (171B6019M); Pro Human Cancer 2 18‐plex (171AC600M); BP Pro TGF‐B 3‐PLEX (171W4001M). Mouse plasma was assayed according to the instructions provided by Bio‐Rad and with Bio‐Rad technical assistance to assess human cytokine levels derived from the HT29 and HuH7 tumors.
Multiplex assays for signal transduction protein phosphorylation and expression
The following Bio‐Plex assay plates were used in our assays for tumor cell signal transduction proteins: Bio‐Plex Bio‐Plex Pro Phosphoprotein magnetic 8‐plex Assay (LQ00004IXUYDC4); Bio‐Plex Pro Phosphoprotein magnetic 15‐plex Assay (LQ000064Q3MJ1). Tumor lysates were assayed according to the instructions provided by Bio‐Rad and with Bio‐Rad technical assistance to assess human signaling changes derived from the HT29 and HuH7 tumors.
Ex vivo manipulation of tumors
Animals were euthanized by CO2 and placed in a BL2 cell culture hood on a sterile barrier mat. The bodies of the mice were soaked with 70% (v/v) EtOH and the abdominal skin around the tumor removed using small scissors, forceps, and a disposable scalpel. These implements were flame sterilized between removal of the outer and inner layers of skin. The tumor was removed and placed in a 10 cm dish containing 5 ml of RPMI cell culture media on ice. The tumor sample that had been placed in RPMI was minced with a sterile disposable scalpel into the smallest possible pieces then placed in a sterile disposable flask. The dish was rinsed with 6.5 ml of RPMI medium which was then added to the flask. A 10× solution of collagenase (Sigma, St. Louis MO; 2.5 ml, 28 U/ml final concentration) and 10× of enzyme mixture containing DNAse (Sigma; 308 U/ml final concentration) and pronase (EMD Sciences, San Diego CA; 22,500 U/ml final concentration) in a volume of 1 ml was added to the flask. The flasks were placed into an orbital shaking incubator at 37°C for 1.5 h at 150 rpm. Following digestion, the solution was passed through a 0.4 µM filter into a 50 ml conical tube. After mixing, a sample was removed for viable and total cell counting using a hemacytometer. Cells were centrifuged at 500 × g for 4 min, the supernatant removed, and fresh RPMI media containing 10% (v/v) fetal calf serum was added to give a final resuspended cell concentration of 1 × 106 cells/ml. Cells were diluted and plated in 6‐well dishes in triplicate at a concentration of 0.1–10.0 × 103 cells/well.
Data analysis
Comparison of the effects of various treatments was performed using one way analysis of variance and a two tailed Student's t‐test. Median dose effect isobologram analyses to determine synergism of drug interaction were done according to the methods of Chou and Talalay using the CalcuSyn program for Windows (Biosoft). Cells are treated with agents at a fixed concentration dose. A combination index (CI) value of <1.00 indicates synergy of interaction between the drugs, a value of 1.00 indicates additivity, and a value of >1.00 equates to antagonism of action between the agents. Statistical examination of in vivo animal survival data utilized log rank statistical analyses between the different treatment groups. Differences with a P‐value of <0.05 were considered statistically significant. Experiments shown are the means of multiple individual points from multiple experiments (±SEM).
Results
Initial studies examined the dose‐response of death receptor CD95 expressing HEP3B tumor cells to increasing concentrations of sorafenib and the PDE5 inhibitor Viagra (sildenafil). Sildenafil enhanced sorafenib toxicity in a dose‐dependent fashion (Fig. 1A). Sildenafil interacted with sorafenib to kill hepatoma cells, regardless of whether the tumor cell expressed CD95, i.e., HuH7 are CD95 null (Fig. 1B). Regorafenib is a derivative of sorafenib with greater solubility and potency in vitro and in vivo that the parent compound sorafenib. Similar data to that with sorafenib were obtained using regorafenib in combination with sildenafil (Fig. 1C). Sildenafil interacted with regorafenib to kill hepatoma cells, regardless of whether the tumor cell expressed CD95 (Fig. 1D). Similar data were obtained using other PDE5 inhibitors such as Cialis and Levitra (data not shown). Sildenafil and sorafenib also interacted with sildenafil to kill multiple other tumor cell types (Fig. 1E). Of particular note, when tumor cells were cultured in 100% heat inactivated human serum, concentrations of sorafenib/regorafenib at one quarter of their steady state Cmax values were still capable of interacting with sildenafil to rapidly kill tumor cells (Fig. 1F). This data strongly argue that the hypothesis of Houghton and Smith regarding low levels of active‐free sorafenib concentrations in patient plasma is very probably incorrect (Smith and Houghton, 2013). In colony formation assays, sildenafil and sorafenib/regorafenib interacted in a synergistic fashion to kill tumor cells, with combination index (CI) values of less than ~0.70 (Table 1).
Figure 1.
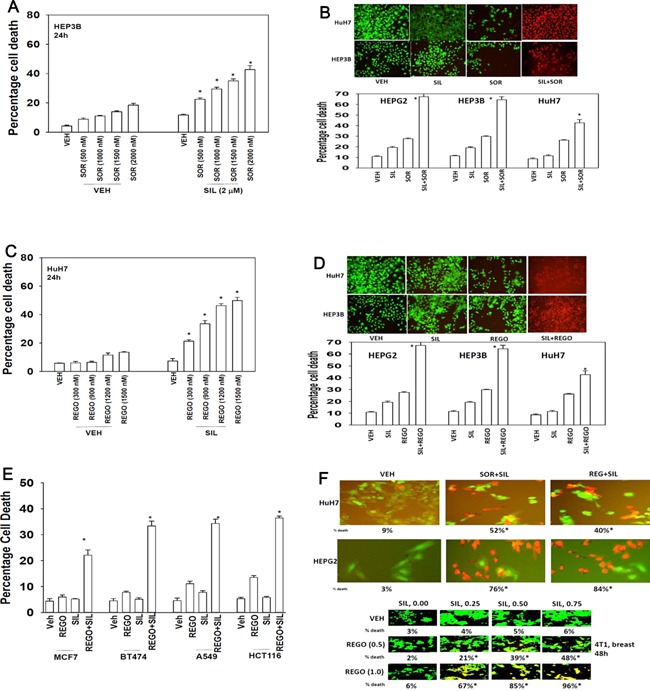
Sorafenib and PDE5 inhibitors interact to kill multiple tumor cell types. (A) HEP3B cells were treated with vehicle (DMSO), sorafenib (SOR, 500 nM–2.0 μM) and/or sildenafil (2.0 μM) as indicated. Cells were isolated 24 h after exposure and viability determined by trypan blue exclusion (n = 3, ±SEM) *P 0.05 < greater than vehicle control. (B) Hepatoma cells 24 h after plating were treated with vehicle (DMSO), sorafenib (SOR, 2.0 μM), PDE5 inhibitor (sildenafil, 2 μM); or the drugs in combination. Twenty‐four hours after treatment cells were isolated and viability determine by trypan blue (n = 3, ±SEM). *P 0.05 < greater than vehicle control. Upper images were generated using a live/dead assay using a Hermes WiScan instrument. (C) HuH7 cells were treated with vehicle (DMSO), regorafenib (REGO, 300–1,500 nM) and/or sildenafil (2.0 μM) as indicated. Cells were isolated 24 h after exposure and viability determined by trypan blue exclusion (n = 3, ±SEM) *P 0.05 < greater than vehicle control. (D) Hepatoma cells 24 h after plating were treated with vehicle (DMSO), regorafenib (REGO, 0.5 μM), PDE5 inhibitor (sildenafil, 2 μM); or the drugs in combination. Twenty‐four hours after treatment cells were isolated and viability determined by trypan blue (n = 3, ±SEM). *P 0.05 < greater than vehicle control. Upper images were generated using a live/dead assay using a Hermes WiScan instrument. (E) Tumor cells 24 h after plating were treated with vehicle (DMSO), regorafenib (REGO, 0.5 μM), sildenafil (2 μM) or the drugs in combination. Twenty‐four hours after treatment cells were isolated and viability determined by trypan blue (n = 3, ±SEM). *P 0.05 < greater than vehicle control. (F) Tumor cells were cultured in 100% heat inactivated human serum. Cells were treated with vehicle; sorafenib (5 μM) and sildenafil (2 μM); or with regorafenib (2 μM) and sildenafil (0–0.75 μM). Cells were isolated 24 h after drug exposure and viability determined using a live/dead assay using a Hermes WiScan instrument. Studies were performed in triplicate in 96‐well plates with the percentage numbers of live/dead cells being determined in three images per well (n = 3, ±SEM). *P 0.05 < greater than vehicle control.
Table 1.
Sorafenib/Regorafenib synergize with sildenafil to kill hepatoma cells
| Drug (μM) | HEPG2 | HEP3B | HuH7 | |
|---|---|---|---|---|
| REG | Sild | Cl | Cl | Cl |
| 0.5 | 1.0 | 0.66 | 0.76 | 0.79 |
| 1.0 | 2.0 | 0.53 | 0.67 | 0.59 |
| 1.5 | 3.0 | 0.49 | 0.51 | 0.46 |
| 2.0 | 4.0 | 0.39 | 0.44 | 0.39 |
| Drug (μM) | HEPG2 | HEP3B | HuH7 | |
| Sor | Sild | Cl | Cl | Cl |
| 1.5 | 2.5 | 0.56 | 0.68 | 0.63 |
| 3.0 | 5.0 | 0.49 | 0.61 | 0.39 |
| 4.5 | 7.5 | 0.45 | 0.49 | 0.43 |
| 6.0 | 10.0 | 0.52 | 0.54 | 0.38 |
Hepatoma cells were plated as single cells (250–1,500 per well) in sextuplicate and, 12 h after plating, treated with vehicle (DMSO), sorafenib (SOR, 1.5–6.0 μM), regorafenib (0.5–2.0 μM) and/or sildenafil (Sild, 1.0–10.0 μM) or with both drugs combined, as indicated at a fixed concentration ratio to perform median dose effect analyses for the determination of synergy. Twenty four hours after drug exposure, the medium was changed and cells were cultured in drug‐free medium for an additional 10 to 14 d. Cells were fixed and stained with crystal violet, and colonies of >50 cells/colony were counted. Colony formation data were entered into the CalcuSyn program and CI and fraction affected values were determined. A CI value of <0.90 to 1.00 indicates synergy, a CI value of 0.90 to 1.10 approximates to additive interactions between the drugs, and a CI value of >1.10 indicates antagonism (n = 2 independent studies).
We next determined the relative expression of PDE5 and PDGFRα/β in normal liver and in hepatoma, as well as on‐ and off‐target effects of our drugs in promoting drug combination killing. PDE5 was over‐expressed in hepatocellular carcinoma tissues but not in normal liver from the same patient (Fig. 2A; similar data were obtained in multiple other samples, Fig. 2B). Knock down of PDE5 expression enhanced regorafenib toxicity in CD95 null HuH7 cells (Fig. 2C). In prior studies, we have shown that PDGFRα/β play an important role in the biology of sorafenib in terms of its toxic combination with other therapeutic agents such as histone deacetylase inhibitors (Martin et al., 2009; Park et al., 2010; Cruickshanks et al., 2013). PDGFRα/β was over‐expressed in hepatocellular carcinoma tissues but not in normal liver tissue from the same patient (Fig. 2D). Knock down of PDGFRα/β expression enhanced the toxicity sildenafil in tumor cells (Fig. 2E).
Figure 2.
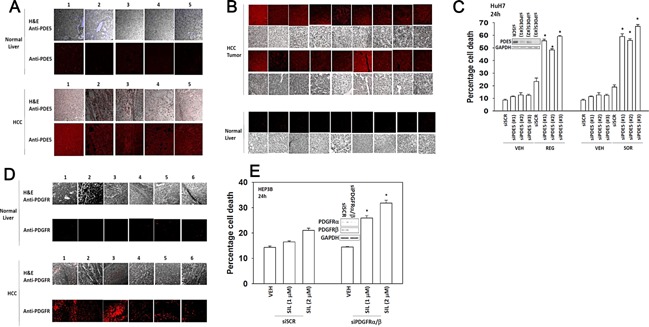
Molecular manipulation of regorafenib and sildenafil targets recapitulates the effects of drug combination treatment. (A) Tissue microarrays of normal and tumor tissue matched from the same patients were stained for expression of PDE5 followed by H&E. The panels show five matched normal liver and hepatocellular tumor sections, each from the same patient (five patients). (B) Further normal liver and liver tumor sections stained to determine the expression of PDE5. (C) HuH7 cells were transfected with a control scrambled siRNA (siSCR) or three different siRNA molecules to knock down expression of PDE5 (siPDE5 #1, #2, #3). Thirty‐six hours after transfection were treated with vehicle (DMSO) or regorafenib (REG, 0.5 μM) or with sorafenib (SOR, 2.0 μM). Twenty‐four hours after treatment cells were isolated and viability determine by trypan blue (n = 3, ±SEM). *P < 0.05 greater than corresponding value in siSCR cells. (D) Tissue microarrays were stained for expression of PDGFRα/β followed by H&E. The parts show six matched normal liver and hepatocellular tumor sections, each from the same patient (six patients). (E) HEP3B cells were transfected with a control scrambled siRNA (siSCR) or siRNA molecules to knock down expression of PDGFRα and PDGFRβ. Thirty‐six hours after transfection were treated with vehicle (DMSO) or sildenafil (SIL, 1.0–2.0 μM). Twenty‐four hours after treatment cells were isolated and viability determine by trypan blue (n = 3, ±SEM). *P < 0.05 greater than corresponding value in siSCR cells.
We next attempted to define whether biomarkers of regorafenib and sildenafil i.e., physiologic effects, could be observed in tumor cells. Sildenafil treatment increased phosphorylation of VASP‐1 (S239), which is a known cGMP dependent kinase (PKG) site (Fig. 3A–E). Identical data were obtained when cells were treated with dibutyrl‐cGMP (data not shown). Prior studies using sorafenib had shown that the drug could cause a modest endoplasmic reticulum stress response, with increased phosphorylation of eIF2α (S51) (Park et al., 2008). Regorafenib treatment increased eIF2α (S51) phosphorylation (Fig. 3A–E). Studies in future figures make use of the drug FTY720 (Gilenya, Fingolimod) which is a histone deacetylase inhibitor; an inhibitor of sphingosine‐1‐phosphate production and also down‐regulates expression of sphingosine‐1‐phosphate receptors, e.g., EDG‐1. Treatment of tumor cells for 6 h with FTY720 abolished expression of EDG‐1 (Fig. 3F). Of additional note, combined treatment of cells with regorafenib and sildenafil also abolished EDG‐1 expression within 6 h. Expression of a dominant negative eIF2α (S51A) protein abolished the ability of (regorafenib + sildenafil) treatment to reduce EDG‐1 expression within 6 h (Fig. 3G). Inhibition of the PERK‐eIF2α‐ATF4‐CHOP pathway protected cells from combination treatment lethality (Fig. 3H).
Figure 3.
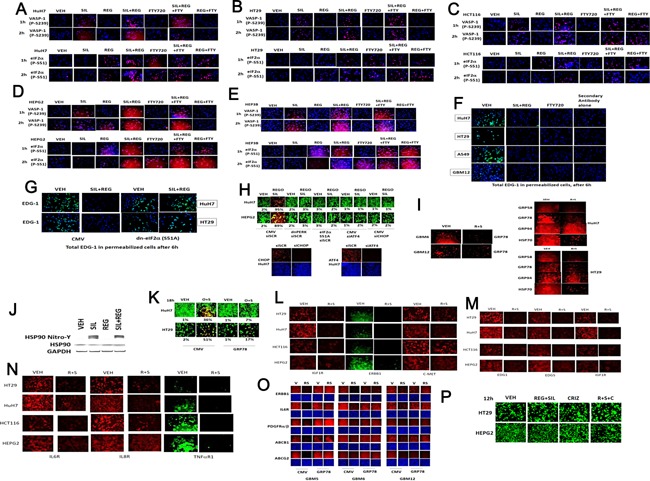
Identification of tumor cell biomarkers that respond to regorafenib and sildenafil treatment. (A–D) Cells were grown in 96‐well plates and treated for the indicated amounts of time with regorafenib (0.5 μM), sildenafil (2 μM), FTY720 (50 nM), and the drugs in combination as indicated in each part. Cells were gently fixed in situ using 2% (v/v) para‐formaldehyde with Triton ×100 for permeabilization and probed for the phosphorylation of VASP‐1 (S239); eIF2α (S51); and the expression of the sphingosine‐1‐phosphate receptor (EDG‐1). (G) HuH7 and HT29 cells were transfected with empty vector plasmid (CMV) or to express dominant negative eIF2α (S51A). Twenty‐four hours after transfection cells were treated with regorafenib (0.5 μM) and sildenafil (2 μM) for 6 h. Cells were gently fixed in situ using 2% (v/v) para‐formaldehyde with Triton ×100 for permeabilization and probed for the expression of the sphingosine‐1‐phosphate receptor (EDG‐1). (E and F) Cells were grown in 96‐well plates and treated for the indicated amounts of time with regorafenib (0.5 μM), sildenafil (2 μM), FTY720 (50 nM), and the drugs in combination as indicated in each part. Cells were gently fixed in situ using 2% (v/v) para‐formaldehyde with Triton ×100 for permeabilization and probed for the phosphorylation of VASP‐1 (S239), eIF2α (S51), and the expression of the sphingosine‐1‐phosphate receptor (EDG‐1). (G) HuH7 and HT29 cells were transfected with empty vector plasmid (CMV) or to express dominant negative eIF2α (S51A). Twenty‐four hours after transfection cells were treated with regorafenib (0.5 μM) and sildenafil (2 μM) for 6 h. Cells were gently fixed in situ using 2% (v/v) para‐formaldehyde with Triton ×100 for permeabilization and probed for the expression of the sphingosine‐1‐phosphate receptor (EDG‐1). (H) Tumor cells were transfected with scrambled siRNA (siSCR) or siRNA molecules to knock down expression of ATF4 or CHOP (siATF4, siCHOP); or tumor cells were transfected with plasmids to express dominant negative PERK or eIF2α S51A. Thirty‐six hours after transfection cells were treated with regorafenib (0.5 μM) and sildenafil (2 μM) for 6 h. Cells were gently fixed in situ using 2% (v/v) para‐formaldehyde with Triton ×100 for permeabilization and probed for the expression of the sphingosine‐1‐phosphate receptor (EDG‐1). (I) Cells were treated with vehicle, regorafenib (0.5 μM), and sildenafil (2.0 μM) for 6 h. Cells were then fixed and the expression levels of HSP70 + HSC70, GRP78, GRP94, and GRP58 determined. (J) HuH7 cells were treated with vehicle, regorafenib (0.5 μM), and sildenafil (2.0 μM) for 3 h. HSP90 was immuno‐precipitated and the levels of total and nitrosylated HSP90 in the precipitates determined. (K) Cells were transfected with empty vector CMV or with a plasmid to express GRP78. Twenty‐four hours later cells were treated with vehicle or with [regorafenib (0.5 μM) + sildenafil (2.0 μM)] for a further 24 h. Cells were isolated and viability determine by a live/dead assay (n = 3, ±SEM). #P < 0.05 lower than corresponding value in CMV transfected cells. (L) Tumor cells as indicated in each part were treated with regorafenib (0.5 μM) and sildenafil (2 μM) together for 6 h. Cells were gently fixed in situ using 2% (v/v) para‐formaldehyde with Triton ×100 for permeabilization and probed for the total expression of each of the indicated growth factor receptors. (M and N) Tumor cells as indicated in each part were treated with regorafenib (0.5 μM) and sildenafil (2 μM) together for 6 h. Cells were gently fixed in situ using 2% (v/v) para‐formaldehyde with Triton ×100 for permeabilization and probed for the total expression of each of the indicated growth factor receptors. (O) Cells were transfected with empty vector plasmid or a plasmid to express GRP78. Twenty‐four hours after transfection cells were treated with vehicle or with regorafenib (0.5 μM) and sildenafil (2 μM) together for 6 h. Cells were gently fixed in situ using 2% (v/v) para‐formaldehyde with Triton ×100 for permeabilization and probed for the total expression of each of the indicated growth factor receptors and plasma membrane pumps. (P) Cells were treated with vehicle or with [regorafenib (0.5 μM) + sildenafil (2.0 μM)] and crizotinib (1.5 μM), as indicated. Cell death was assessed 12 h later using live/dead assays.
Treatment of cells with regorafenib and sildenafil reduced the expression of HSP70, GRP78, and GRP58, but weakly modulated GRP94 levels (Fig. 3I). Sildenafil increased the nitration of HSP90, indicative of reduced HSP90 chaperone function (Fig. 3J). Over‐expression of GRP78 protected cells against regorafenib and sildenafil toxicity (Fig. 3K). Treatment of tumor cells with (regorafenib + sildenafil) treatment also rapidly reduced the expression of multiple other growth factor receptors (Fig. 3L–N). Over‐expression of the chaperone protein GRP78 reduced the ability of (regorafenib + sildenafil) treatment to reduce expression of growth factor receptors and plasma membrane transporters (Fig. 3O). Compared to changes in the expression of many other growth factor receptors, the expression of c‐MET was more modestly down‐regulated by regorafenib and sildenafil treatment; however, use of the potent clinically relevant c‐MET inhibitor crizotinib at a supra‐physiologic concentration did not noticeably enhance (regorafenib + sildenafil) lethality in liver or colon cancer cells (Fig. 3P).
PDE5 inhibitors are known to enhance the levels of reactive oxygen and reactive nitrogen species in cells (e.g., Das et al., 2010; Musicki et al., 2014). Treatment of tumor cells with sildenafil and regorafenib rapidly increased the levels of ROS and RNS (Fig. 4A). Inhibition of nitric oxide synthase enzymes using L‐NG‐Nitroarginine Methyl Ester (L‐NAME) inhibited cell killing by sildenafil and sorafenib, as did quenching of ROS production by incubation with N‐acetyl cysteine (NAC) or by expression of thioredoxin (TRX) (Fig. 4B). Knock down of inducible nitric oxide synthase (iNOS) or endothelial nitric oxide synthase (eNOS) expression suppressed killing by the drug combination (Fig. 4C). Of note, knock down of iNOS abolished the drug combination interaction but did not alter sorafenib toxicity as a single agent, whereas knock down of eNOS modestly but significantly reduced sorafenib toxicity. Sorafenib and sildenafil were still capable of interacting to kill in cells lacking eNOS expression. The combination of sildenafil and sorafenib surprisingly increased iNOS expression (Fig. 4D).
Figure 4.
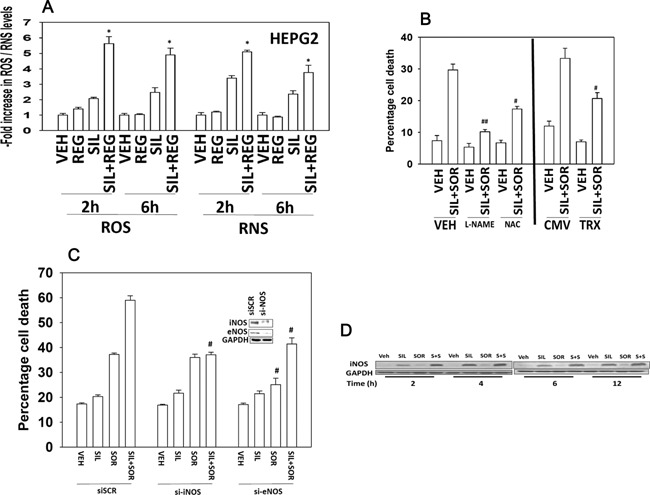
The generation of ROS/RNS following regorafenib and sildenafil treatment is a key mediator of tumor cell killing. (A) HEPG2 cells in 96‐well plates were loaded for 30 min with either dihydro‐DCF (10 μM) which is sensitive to oxidation by hydroxyl radicals and peroxynitrite directly and hydrogen peroxide (i.e., reactive oxygen species, ROS); or 3‐amino,4‐aminomethyl‐2′,7′‐difluorescein (DAF‐FM DA, 4 μM) which is sensitive to oxidation by NO (i.e., reactive nitrogen species, RNS). Cells were treated with vehicle (DMSO), regorafenib (1 μM), sildenafil (2 μM); or the drugs in combination. Cells—ROS/RNS measurements—were made in a Vector 3 plate reader at the indicated times after drug treatment (n = 3, ±SEM). *P 0.05 < greater than vehicle control. (B) Left portion of the graph: HEPG2 cells were pre‐treated with vehicle, the NOS inhibitor L‐NAME (1 μM) or the ROS quenching agent N‐acetyl cysteine (10 mM). Cells were then treated with vehicle or with sildenafil (2 μM), and sorafenib (2 μM) in combination. Twenty‐four hours after treatment cells were isolated and viability determined by trypan blue (n = 3, ±SEM). Right portion of the graph: HEPG2 cells were transfected with either an empty vector plasmid (CMV) or a plasmid to express thioredoxin (TRX). Twenty‐four hours after transfection cells were treated with vehicle or with sildenafil (2 μM), and sorafenib (2 μM) in combination. Twenty‐four hours after treatment cells were isolated and viability determined by trypan blue (n = 3, ±SEM). #P < 0.05 less than corresponding value in VEH/CMV cells; ##P < 0.05 less than corresponding value in NAC cells. (C) HEPG2 cells were transfected with a control scrambled siRNA (siSCR) or siRNA molecules to knock down expression of eNOS or iNOS. Thirty six hours after transfection cells were treated with vehicle or with sildenafil (2 μM), and sorafenib (2 μM) in combination. Twenty‐four hours after treatment cells were isolated and viability determined by trypan blue (n = 3, ±SEM). #P < 0.05 less than corresponding value in siSCR cells. (D) HEPG2 cells were treated with vehicle or with sildenafil (2 μM), and/or sorafenib (2 μM) in combination. Cells were isolated at the indicated time points and western immunoblotting performed to determine the expression of iNOS.
In prior studies, we have shown that sorafenib as a single agent can modestly promote increased numbers of autophagosomes (Park et al., 2008, 2010a,2010b; Martin et al., 2009; Cruickshanks et al., 2013). Treatment of cells with sildenafil and sorafenib increased the numbers of autophagosomes and autolysosomes in a greater than additive fashion (Fig. 5A and B). The increase in the numbers of autophagosomes and autolysosomes was suppressed by incubation of cells with either L‐NAME or NAC. Knock down of either ULK‐1, Beclin1, or ATG5 expression inhibited the drug‐induced increase in autophagosome and autolysosome levels and reduced cell killing by sildenafil and sorafenib or by sildenafil and regorafenib treatments (Fig. 5C and D).
Figure 5.
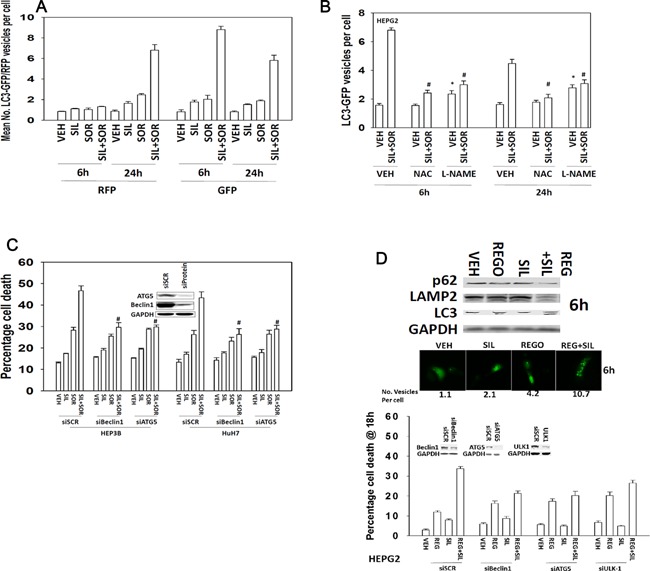
ROS/RNS regulate the induction of autophagy by drug treatment. (A and B) HEPG2 cells were transfected with a plasmid to express LC3‐GFP‐RFP. Twenty‐four hours after transfection cells were, as indicated pre‐treated with L‐NAME (1 μM) or NAC (10 mM), then treated with vehicle (DMSO), sorafenib (2 μM), sildenafil (2 μM); or the drugs in combination. The number of GFP vesicles (early autophagosomes) and RFP vesicles (late autolysosomes) were determined 6 and 24 h after drug treatment (n = 3, ±SEM). #P < 0.05 less than corresponding value in cells not treated with L‐NAME or NAC. (C) HEP3B and HuH7 cells were transfected with a control scrambled siRNA (siSCR) or siRNA molecules to knock down expression of Beclin1 or ATG5. Thirty‐six hours after transfection were treated with vehicle or with sildenafil (2 μM), and/or sorafenib (2 μM). Twenty‐four hours after treatment cells were isolated and viability determined by trypan blue (n = 3, ±SEM). #P < 0.05 less than corresponding value in siSCR cells. (D) Lower graph: HEPG2 cells were transfected with a control scrambled siRNA (siSCR) or siRNA molecules to knock down expression of Beclin1, ULK1, or ATG5. Thirty‐six hours after transfection were treated with vehicle or with sildenafil (2 μM), and/or regorafenib (0.5 μM). Twenty‐four hours after treatment cells were isolated and viability determine by trypan blue (n = 3, ±SEM); Middle images: Cells expressing LC3‐GFP were treated with vehicle or with sildenafil (2 μM), and/or regorafenib (0.5 μM) and the number of intense GFP+ vesicles counted in 40 cells and the mean number of vesicles per cell is presented (n = 3, ±SEM); Upper blots: Cells were treated with vehicle or with sildenafil (2 μM), and/or regorafenib (0.5 μM) and isolated after 6 h. Cell lysates were western blotted for the expression of the indicated proteins (n = 3).
In addition to our prior studies linking sorafenib and autophagy, we have also described how sorafenib can interact with agents that generate ROS to facilitate activation of the death receptor CD95 (Park et al., 2008, 2010a). In these studies HuH7 cells, that lack endogenous CD95 expression, were particularly resistant to the tested sorafenib drug combinations. In the present studies i.e., Figure 1, we have found that sorafenib and sildenafil interact to kill both CD95 null HuH7 cells and hepatoma cells that express CD95 (HEPG2, HEP3B), though drug‐combination killing appears to be more efficient in HEPG2 and HEP3B cells. Thus, we next determined whether inhibition of the extrinsic and/or intrinsic apoptosis pathways could reduce cell killing by sorafenib and sildenafil. In HEPG2 cells that express CD95, expression of BCL‐XL and dominant negative caspase 9 that suppress activation of the intrinsic apoptosis pathway protected cells from drug combination toxicity (Fig. 6A). Expression of the caspase 8 inhibitor c‐FLIP‐s in HEPG2 cells also was protective demonstrating that the extrinsic pathway was playing a role in cell killing. In HuH7 cells that are null for CD95, expression of BCL‐XL and dominant negative caspase 9 was protective, whereas expression of c‐FLIP‐s did not alter the cell death response (Fig. 6A).
Figure 6.
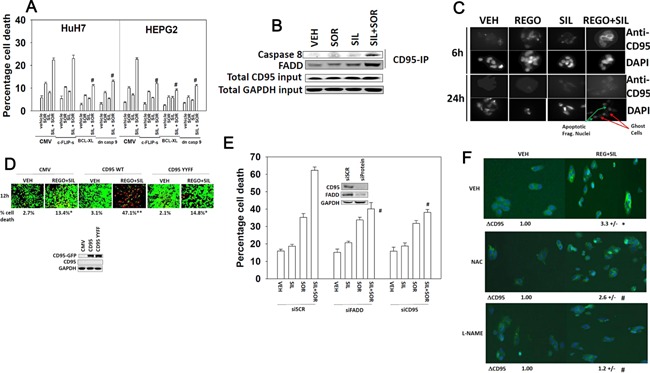
Death receptor‐dependent and independent induction of cell death by sorafenib and sildenafil treatment. (A) HuH7 and HEPG2 cells were infected with recombinant adenoviruses to express empty vector (CMV); dominant negative caspase 9, BCL‐XL, or c‐FLIP‐s. Twenty‐four hours after infection cells were then treated with sorafenib (SOR 2.0 μM) and/or sildenafil (SIL, 2.0 μM). Twenty‐four hours after drug treatment cells were isolated and viability determined by trypan blue exclusion assay (n = 3, ±SEM). #P < 0.05 less than corresponding value in CMV infected cells. (B) HEPG2 cells were treated with sorafenib (SOR 2.0 μM) and/or sildenafil (SIL, 2.0 μM). Six hours after treatment cells were lysed and prepared for immunoprecipitation of CD95. After immunoprecipitation, immunoblotting was performed to determine the levels of caspase 8 and FADD in the immunoprecipitate. Total levels of CD95 and GAPDH in the lysate are also presented. (C) HEPG2 cells grown on microscope slides were treated with vehicle (DMSO), regorafenib (0.5 μM), sildenafil (2 μM), or the drugs in combination. Cells were fixed (but not permeabilized) 6 and 24 h after drug exposure. IHC was performed to determine the plasma membrane levels of CD95 and death by DAPI staining. (D) HuH7 cells were transfected with either an empty vector plasmid (CMV); a plasmid to express CD95‐GFP or a plasmid to express CD95‐GFP‐Y232F Y291F. Twenty‐four hours after transfection cells were then treated with sorafenib (SOR 2.0 μM) and/or sildenafil (SIL, 2.0 μM). Twenty‐four hours after drug treatment cells were isolated and viability determined by using a live/dead viability assay (n = 3, ±SEM). *P < 0.05 greater than corresponding value in CMV transfected cells. (E) HEP3B cells were transfected with a control scrambled siRNA (siSCR) or siRNA molecules to knock down expression of CD95 or FADD. Thirty‐six hours after transfection were treated with vehicle or with sildenafil (2 μM), and/or sorafenib (2 μM). Twenty‐four hours after treatment cells were isolated and viability determine by trypan blue (n = 3, ±SEM). #P < 0.05 less than corresponding value in siSCR transfected cells. (F) HEP3B cells grown on microscope slides were pre‐treated with L‐NAME (1 μM) or NAC (10 mM), then treated with vehicle (DMSO), sorafenib (2 μM), sildenafil (2 μM), or the drugs in combination. Cells were fixed (but not permeabilized) 6 h after drug exposure. IHC was performed to determine the plasma membrane levels of CD95.
We further explored signaling by the extrinsic pathway in our system. Drug combination treatment caused a rapid plasma membrane localization of the death receptor CD95 that was associated with increased association of caspase 8 with CD95 (Fig. 6B and C). These data are of note because other receptors that are more highly expressed under basal conditions in the plasma membrane have their total protein expression reduced. Expression of wild type CD95 in HuH7 cells enhanced cell killing by the drug combination, but killing was not enhanced by expression of a CD95 mutant that lacked the sites of regulatory tyrosine phosphorylation (Fig. 6D). Knock down of CD95 or of FAS‐associated death domain protein (FADD) suppressed cell killing by the drug combination (Fig. 6E). As the drug combination was increasing ROS and RNS levels that were causal in cell killing, we determined whether ROS and RNS regulated CD95 activation. Incubation of cells with the NOS inhibitor L‐NAME and to a lesser extent the ROS quenching agent NAC suppressed drug‐induced CD95 activation (Fig. 6F).
Treatment of hepatoma cells in vitro increased the phosphorylation of PKR‐like endoplasmic reticulum kinase (PERK) and eIF2α (S51) as judged by western blotting (Fig. 7A). Drug combination treatment reduced the phosphorylation of MEK1 (S218/S222), AKT (T308), and of ERK1/2, and enhanced phosphorylation of JNK1/2. No obvious change was noted in MEK1 S298 phosphorylation. Similar data were obtained when cells were fixed in place and probed by immuno‐fluorescence for phosphorylation status by immuno‐fluorescence (Fig. 7B–D). The phosphorylation of the autophagy regulatory kinase mTOR was significantly reduced by drug combination treatment, which also was associated to a lesser extent with reduced expression of mTOR protein itself (Fig. 7E) i.e., both total protein and the specific activity of the mTOR protein declined. Expression of activated forms of MEK1 or AKT incompletely suppressed killing by regorafenib and sildenafil treatment (Fig. 7F). Inhibition of JNK1/2 signaling or expression of an activated form of mTOR abolished drug combination toxicity.
Figure 7.
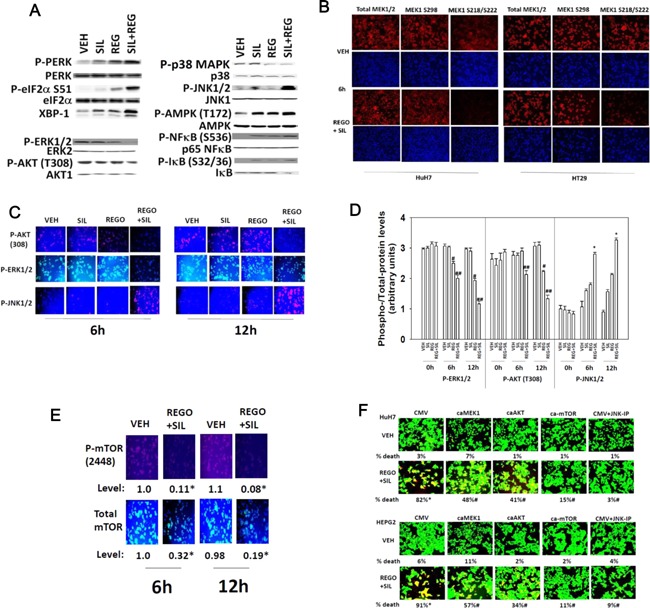
Activation of JNK and inhibition of mTOR, ERK1/2, and AKT plays key roles in regorafenib and sildenafil toxicity. (A) HuH7 cells were treated with vehicle, regorafenib (0.5 μM), sildenafil (2 μM), or the drugs in combination for 3 h. Cells were isolated and the phosphorylation/expression of the indicated proteins determined by western immunoblotting. (B and C) HuH7 and HT29 cells growing in 96‐well plates were treated with vehicle, regorafenib (0.5 μM), sildenafil (2 μM), or the drugs in combination for 6 h or for 12 h. At each time point cells were fixed in place and probed for the expression and phosphorylation of MEK1, AKT, ERK1/2, and JNK1/2 and images obtained using a Hermes WiScan system. (D) Pictorial data shown in part C was analyzed and quantified using software provided with the WiScan system. The intensity of immuno‐staining fluorescence using each Phospho‐Antibody and in parallel total expression antibody was determined and the arbitrary “specific activity” of each kinase determine in multiple wells from multiple experiments (n = 3, ±SEM). #P < 0.05 less than vehicle control value; ##P < 0.05 less than regorafenib value; *P < 0.05 greater than regorafenib value. (E) HuH7 cells growing in 96‐well plates were treated with vehicle, regorafenib (0.5 μM), sildenafil (2 μM), or the drugs in combination for 6 h or for 12 h. At each time point, cells were fixed in place and probed for the expression and phosphorylation of mTOR/mTOR (S2448) and images obtained using a Hermes WiScan system. *P < 0.05 expression/phosphorylation of mTOR lower than corresponding vehicle control. (F) HuH7 and HEPG2 cells were transfected with plasmids: CMV (empty vector); to express activated MEK1, caMEK1; to express activated AKT, caAKT; or to express activated mTOR, ca‐mTOR. Twenty‐four hours after transfection a portion of vehicle control cells were treated with the JNK inhibitory peptide (JNK‐IP, 10 μM). Cells were then treated with vehicle or with regorafenib (0.5 μM) and sildenafil (2.0 μM) and cell viability determined after 24 h using a live/dead assay using a Hermes WiScan instrument (n = 3, ±SEM). *P < 0.05 greater than vehicle control; #P < 0.05 lower than corresponding value in CMV transfected cells.
Prior reports from our group have shown that sorafenib can increase the levels of ceramide in hepatoma cells (Park et al., 2010a,2010b). Regorafenib and sildenafil treatment increased the levels of multiple ceramides and dihydro‐ceramides (Fig. 8A and B). Regorafenib, and sorafenib, both increased the levels of sphingosine‐1‐phosphate and di‐hydro sphingosine‐1‐phosphate (Fig. 8C, data not shown). The approved multiple sclerosis drug FTY720 (Gilenya, Fingolimod) inhibits the production of sphingosine‐1‐phosphate as an active site inhibitor of sphingosine kinases 1 and 2, and when phosphorylated by these kinases also down‐regulates the expression of the S1P receptor 1 (EDG‐1, see Fig. 2). Treatment of cells with low clinically relevant concentrations of FTY720 suppressed the ability of regorafenib to increase sphingosine‐1‐phosphate levels, and also increased the production of ceramide species by regorafenib (Fig. 8C). Furthermore, at early time points, prior to evident toxicity caused by (regorafenib + sildenafil), or by FTY720 treatment as a single agent, the combination of (regorafenib + sildenafil + FTY720) caused tumor cell killing (Fig. 8D and E). Similar data to that generated using FTY720 were obtained using histone deacetylase inhibitors; the well‐established chemotherapy drug all‐trans retinoic acid (ATRA); and ionizing radiation (Fig. 8F–I). The effect of vorinostat is in concordance with our pre‐clinical and clinical findings that some hepatocellular tumors are effectively inhibited in their growth and radiosensitized by sorafenib and vorinostat treatment (see our on‐going Phase I trial NCT01075113, Dent and Poklepovic, unpublished observations).
Figure 8.
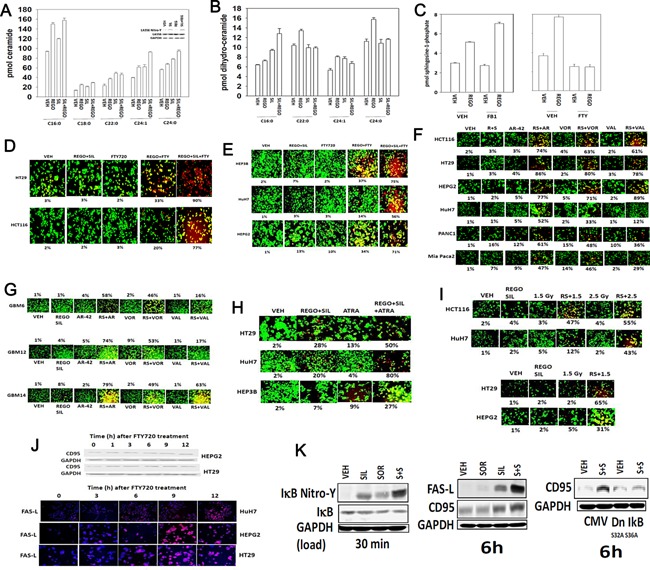
Regorafenib and sildenafil interact to increase ceramide levels. (A and B) HuH7 cells were treated with vehicle control or with regorafenib (REGO 0.5 μM) and/or sildenafil (SIL, 2.0 μM). Six hours after treatment cells were isolated and bioactive lipids extracted. Multiple bioactive lipid species were analyzed using GC/MS techniques (n = 2 in triplicate ±SEM). *P < 0.05 greater than corresponding value in VEH cells; #P < 0.05 less than corresponding value in VEH treatment; % P < 0.05 greater than value in regorafenib alone treatment. Part A, upper inset part: Treatment of cells with sildenafil (2 μM) for 3 h increases nitro‐tyrosine (Y) levels in ceramide synthase 6 (LASS6). (C) HuH7 cells were treated with vehicle control or with regorafenib (REGO 0.5 μM) and/or Fumonisin B1 (FB1, 25.0 μM) or FTY720 (50 nM). Six hours after treatment cells were isolated and bioactive lipids extracted. Multiple bioactive lipid species were analyzed using GC/MS techniques (n = 2 in triplicate ±SEM). (D and E) Tumor cells were treated with vehicle control or with regorafenib (REGO 0.5 μM) and/or sildenafil (SIL, 2.0 μM) and/or FTY720 (50 nM). Cells were examined 9 h after treatment using a live/dead assay in a Hermes WiScan instrument (n = 3, ±SEM). (F and G) Tumor cells were treated with vehicle, [regorafenib (0.5 μM) + sildenafil (2.0 μM)], the HDAC inhibitors vorinostat (500 nM), AR‐42 (250 nM), Valproate (0.1 mM), or regorafenib + sildenafil + HDAC inhibitor. Cells were examined 9 h after treatment using a live/dead assay in a Hermes WiScan instrument (n = 3, ±SEM). (H) Tumor cells were treated with vehicle control or with regorafenib (REGO 0.5 μM) and/or sildenafil (SIL, 2.0 μM) and/or ATRA (150 nM). Cells were examined 18 h after treatment using a live/dead assay in a Hermes WiScan instrument (n = 3, ±SEM). (I) Tumor cells were treated with vehicle control or with regorafenib (REGO 0.5 μM) and/or sildenafil (SIL, 2.0 μM) followed 30 min later by exposure to ionizing radiation. Cells were examined 12 h after treatment using a live/dead assay in a Hermes WiScan instrument (n = 3, ±SEM). (J) Upper blots: Tumor cells were treated with 50 nM FTY720 for the indicated times and cell lysates western blotted to determine expression of the death receptor CD95 (n = 3); Lower IHC: Tumor cells in situ in a 96‐well plate were treated with FTY720 (50 nM) for the indicated times, and cells fixed and probed for expression of FAS ligand (FAS‐L) (n = 3). (K) HEPG2 cells were treated with vehicle (DMSO), sorafenib (2 μM), sildenafil (2 μM); or the drugs in combination. Left: Cells were isolated after 30 min and immuno‐precipitation of IκB was performed in duplicate. On separate blots assessment of IκB nitro‐tyrosine and total IκB was performed. Center: Cells were isolated after 6 h and the expression of FAS‐L and CD95 determined. Right: HEPG2 cells were transfected with empty vector plasmid or plasmid to express dominant negative IκB S32A S36A. Twenty‐four hours after transfection cells were treated with vehicle (DMSO), sorafenib (2 μM), sildenafil (2 μM); or the drugs in combination. Cells were isolated after 6 h and the expression of CD95 determined.
FTY720 is an HDAC inhibitor and we discovered, similar to our prior data with vorinostat and valproate, that FTY720 treatment increased the expression of CD95 and FAS ligand (Fig. 8J). We also discovered that sildenafil treatment of cells increased the nitrosylation and inactivation of IκB and that expression of a dominant negative IκB S32A S36A protein abolished the induction of CD95 (Fig. 8K). Of note: HT29 colon cancer cells express a mutant p53, CD95, and a wild type K‐RAS; HCT116 colon cancer cells express a wild type p53, CD95, and a mutant K‐RAS; HEPG2 hepatoma cells express a wild type p53, CD95, and a mutant N‐RAS; HEP3B hepatoma cells express a mutant p53, CD95, wild type RAS proteins, and a variety of proteins associated with stable infection by Hepatitis B virus; HuH7 hepatoma cells express a mutant p53, lack CD95 expression, and have wild type RAS proteins.
FTY720, in addition to its regulatory role in S1P metabolism and biology, has also been shown to act as a histone deacetylase inhibitor (HDAC inhibitor) and in a recent manuscript was shown to increase VEGF‐D mRNA levels in an immune competent mouse (Hait et al., 2009). In prior studies using established HDAC inhibitors, e.g., valproate; vorinostat, we have observed these drugs increasing expression of CD95 and FAS‐L (Park et al., 2008, 2010a,2010b). Treatment of tumor cells with FTY720 also increased expression of CD95 and FAS‐L (Fig. 8J).
We next determined whether sorafenib/regorafenib interacted with sildenafil in vivo to kill tumor cells. Initial studies examined the ex vivo plating efficiency of tumor cells treated in vivo with regorafenib and sildenafil. Treatment of tumors with regorafenib and sildenafil in vivo caused a reduction in the ex vivo plating efficiency of isolated tumor cells following drug exposure; thus the long‐term colony forming ability of drug treated cells was reduced beyond that achieved due to the proximal anti‐tumor effects of the drugs (Fig. 9A). Based on the data in Figure 1, we next performed additional studies using pre‐formed HuH7 tumors (~150 mm3); in our prior experience HuH7 cells have a significantly greater tumorigenic potential than either HEPG2 or HEP3B cells to grow in an athymic mouse. The a priori prediction for our HuH7 tumor studies would be that due to the lack of CD95 expression, the relative amount of killing may be more modest, although data from Figure 9A argues that in vivo killing by our regorafenib and sildenafil combination occurs. Transient treatment of animals with sildenafil did not significantly alter tumor growth whereas transient treatment with sorafenib caused a non‐significant trend towards reduced growth (Fig. 9B). Combined treatment of animals with both drugs for 3 days caused a significant reduction/delay in HuH7 tumor re‐growth.
Figure 9.
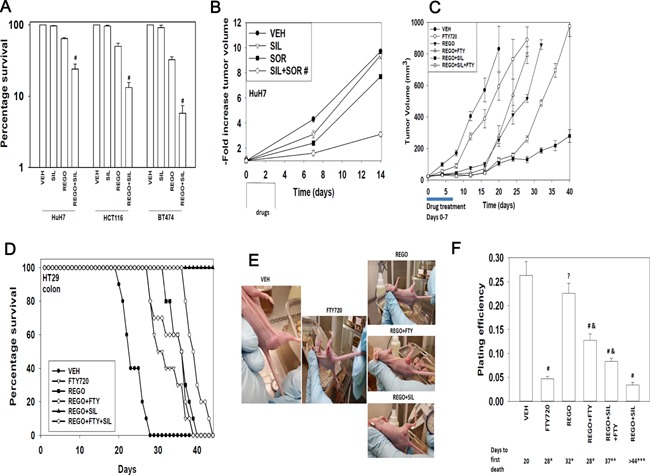
Sorafenib/regorafenib interact in vivo to suppress tumor growth. (A) HuH7 (hepatoma), HCT116 (colorectal) and BT474 (breast) tumors were formed in the flanks of athymic mice (∼150 mm3). Animals were treated PO with vehicle diluent (cremophore); sildenafil (5 mg/kg); regorafenib (25 mg/kg); or the drug combination for 24 h. Tumors were isolated and single cell suspensions of tumor cells derived. Cells were plated and colonies permitted to form for 7 days. Colonies were fixed, stained and counted, and the survival value in vehicle treated tumors defined as 100% (n = 3 × 6, ±SEM). #P < 0.05 less than survival in regorafenib treated cells. (B) HuH7 tumors were formed in the flanks of athymic mice (~150 mm3). Animals were treated PO with vehicle diluent (cremophore); sildenafil (5 mg/kg); sorafenib (25 mg/kg); or the drug combination for 3 days. Tumor volumes were measured 7 days and 14 days after the start of drug treatment (n = 2 studies, eight animals per group ± SEM). #P < 0.05 less growth than sorafenib treated cells. (C) HT29 tumors were formed in the flanks of athymic mice (∼30 mm3). Animals were treated PO with vehicle diluent (cremophore); sildenafil (5 mg/kg) and regorafenib (25 mg/kg); FTY720 (0.05 mg/kg) or the drugs in combination for 7 days. Tumor volumes were measured every 4 days after the end of drug treatment (n = 2 studies, eight animals per group ± SEM). (D) Kaplan Meier survival plot of animals treated in part C; animals were humanely sacrificed when tumor volumes exceeded 1,500 mm3. (E) Representative images of tumors from each condition, taken when tumors were of approximately the same volume (between days 15 and 30). (F) HT29 tumors, at the time of sacrifice were isolated from the animals and gently digested to obtain a single cell suspension of tumor cells. Cells were plated as single cells (100‐10,000) per well of a 6‐well plate. Colonies were permitted to form for 7–10 days after which they were fixed, stained and counted, and the plating efficiency for tumor cells ex vivo for each treatment determined (n = 8, ±SEM). (G) Athymic mice carrying pre‐formed HT29 tumors (50 mm3) were treated PO with vehicle diluent (cremophore) or sildenafil (10 mg/kg) and regorafenib (50 mg/kg) for 7 days after which animals were sacrificed, their normal tissues obtained and fixed and sealed in paraffin wax. Ten micron slices of each tissue were taken and H&E stained and examined under 10× magnification.
We then determined, based on data in Figures 1 and 8, whether the HDAC inhibitor and S1P antagonist FTY720 enhanced in vivo the anti‐tumor activity of regorafenib and of (regorafenib + sildenafil) in established HT29 colon cancer tumors. FTY720 as a single agent had a modest single agent effect on reducing tumor growth but did cause a modest significant increase in overall survival (Fig. 9C and D). To our surprise, we found that a transient 7 day treatment of tumors with regorafenib and FTY720 or with regorafenib and sildenafil and FTY720 caused the tumors exposed to FTY720 to re‐grow at a much faster rate after cessation of drug treatment than either regorafenib treatment alone or treatment with regorafenib and sildenafil (Fig. 9C). Our tumor growth data were reflected in overall animal survival (Fig. 9D). The growth of tumors treated with the double combination of (regorafenib + sildenafil) was profoundly reduced both during and for many weeks following drug exposure, which was reflected in a significant increase in animal survival (Fig. 9D).
Morphological examination of the tumors in situ in the mouse flank (pictures were chosen with tumors in each condition at the same approximate volume) demonstrated that tumors treated with FTY720 for 7 days exhibited a more vascularized appearance on their surface and were more capable of suppurating (Fig. 9E). At the time of sacrifice, portions of tumor were digested and single cells plated to determine the ex vivo plating efficiency of cells. Treatment of tumors with FTY720 as a single agent, that had a marginal effect on tumor growth and animal survival, significantly reduced ex vivo plating efficiency (Fig. 9F). Regorafenib as a single agent did not alter ex vivo colony formation that correlated with our tumor growth and survival findings and with extant clinical data where this drug has only a ~1% response rate. The reduced plating efficiency of tumors treated with regorafenib and sildenafil correlated with suppressed tumor growth and increased animal survival. Regorafenib and sildenafil treatment did not damage normal tissues as judged by H&E staining (Fig. 9G).
Based on our unexpected findings in Figure 9, we determined using a Bio‐Rad MAGPIX multiplex system the expression levels of human cytokines in mouse plasma and the expression and activity of signal transduction proteins within the established tumor itself. Regorafenib and sildenafil exposure for 7 days decreased the plasma levels of bFGF and GM‐CSF but significantly increased the expression of PDGFbb (Fig. 10A). In agreement with data in Figure 8, treatment of tumors for 7 days with FTY720 increased plasma levels of FAS‐L (Fig. 10B). For a number of cytokines, e.g., endoglin, IL6, VEGF‐A/C/D, endpoietin 2, EGF, their plasma level declined in vehicle control treated tumors over 7 days of growth which was associated with a mean of approximately sevenfold increase in tumor volume (Fig. 10B–F). In this respect, treatment of tumors with FTY720 or with regorafenib and FTY720 prevented the decline in plasma cytokine levels that were observed in mice with vehicle control treated tumors at day 7 (Fig. 10B–G). For tumors exposed to regorafenib and sildenafil, the expression of pro‐growth/pro‐angiogenic/pro‐invasion cytokines was either unchanged compared to vehicle control treated tumors or was significantly reduced (Fig. 10C–G).
Figure 10.
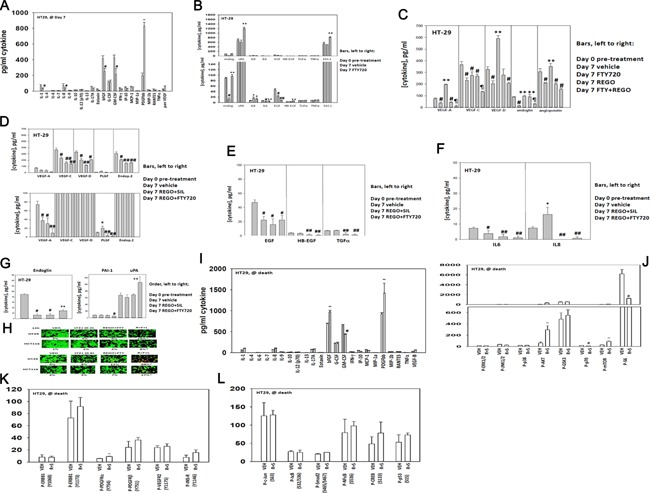
Treatment of HT29 tumors with regorafenib, sildenafil and FTY720 alters the cytokine expression levels in mouse plasma. (A–G) HT29 tumors (∼30 mm3) were formed in the flanks of athymic mice. Aliquots (∼75 μl) of mouse blood were obtained in a heparin/EDTA coated Eppendorf tube. Animals were then treated PO with vehicle diluent (cremophore); sildenafil (5 mg/kg) and regorafenib (25 mg/kg); FTY720 (0.05 mg/kg) or the drugs in combination as indicated for 7 days. Data in part A are vehicle control and [regorafenib + sildenafil] tumor data. After 7 days aliquots of mouse blood were again obtained (∼75 μl). Clarified mouse plasma free of cells was then subjected to multiplex assays in a Bio‐Rad MAGPIX system to define the expression of the noted cytokines in each part before and following treatment (n = 2 studies, eight animals per group ± SEM). *P < 0.05 greater than Day 0 pre‐treatment value; **P < 0.05 greater than Day 7 vehicle control; #P < 0.05 less than Day 0 pre‐treatment value; ##P < 0.05 less than Day 7 vehicle value; ¶P < 0.05 less than Day 7 regorafenib treatment value. (H) HT29 and HCT116 tumor cells in vitro were treated with vehicle control; [regorafenib (0.5 μM) + FTY720 (50 nM)]; the TGF β receptor inhibitor LY2157299 (0.2, 0.6 μM) or the drugs in combination as indicted for 9 h. After 9 h cell viability was determined in a Hermes WiScan instrument using a live/dead assay (n = 3, ±SEM). *P < 0.05 greater than REGO + FTY value. (I–L) HT29 tumors isolated from vehicle control treated or (regorafenib + sildenafil) treated tumors at the time of animal nadir were subjected to multiplex assays in a Bio‐Rad MAGPIX analyzer to determine the expression of cytokines in plasma and the phosphorylation of the indicated signal transduction proteins (n = 8, ±SEM). ~P < 0.05 greater than vehicle control treated tumor value; #P < 0.05 less than vehicle control treated tumor value. (M) HuH7 cells were treated with vehicle control; regorafenib (0.5 μM) and sildenafil (2 μM); BGJ398 (1 μM); Lapatinib (1 μM); MK2206 (1 μM) or in the combinations indicated in the part. Cells were examined 12 h after treatment using a live/dead assay in a Hermes WiScan instrument. Red/yellow cells = dead; green cells = alive (n = 3, ±SEM). *P < 0.05 greater than corresponding value in (regorafenib + sildenafil) treatment alone.
Of additional concern, and that correlated with the stronger tumor re‐growth response was that treatment of tumors with regorafenib and FTY720 increased expression of the pro‐growth/pro‐angiogenesis/pro‐invasion cytokine endoglin, that enhances TGF β signaling, as well as increasing uPA levels and reducing expression of the endogenous uPA inhibitor protein PAI‐1 (Fig. 10G). As treatment of tumors with FTY720 caused an increase in endoglin expression, which will enhance TGF β receptor family signaling as well as integrin signaling, we determined whether the toxicity of regorafenib and FTY720 treatment was increased in vitro using a clinically relevant TGF β inhibitor (LY2157299; Galunisertib) that is in Phase II evaluation. Inhibition of TGF β receptor 1 signaling in a dose‐dependent fashion significantly enhanced the toxicity of (regorafenib + sildenafil) treatment (Fig. 10H).
We also collected tumor and blood materials at the time of humane sacrifice of the mice from our HT29 experiments. Most notably the expression of the growth factors bFGF and of PDGFbb were both elevated and the expression of GM‐CSF reduced at the time of nadir (Fig. 10I). This was associated with increased AKT activity and reduced S6 phosphorylation (Fig. 10J). Changes in receptor phosphorylation or transcription factor phosphorylation we not significantly changed (Fig. 10K and L). Based on our data showing that drug treated tumors had stable high ERBB1 phosphorylation, expressed more FGF and had higher AKT activity we determine the impact of ERBB1/2, FGFR, and AKT inhibitors on (regorafenib + sildenafil) toxicity. Inhibition of either AKT or ERBB1/2 signaling enhanced (regorafenib + sildenafil) lethality (Fig. 10M). Collectively, the data in Figure 10 argue that the enhanced re‐growth of HT29 colorectal tumors treated with FTY720 may be due to the increased expression of multiple cytokines and that TGF β and ERBB1/2 signaling, and intracellular AKT signaling, has been validated as biomarkers for third drug combination studies.
We then examined the activities of multiple signal transduction pathways/proteins in tumors 7 days after the start of treatment. Treatment of HuH7 and HT29 tumors with (regorafenib + sildenafil) reduced plasma levels of FGF and GM‐CSF and increased the levels of PDGFbb (Figs. 10A and 11A). Regorafenib and sildenafil treatment also reduced the expression of multiple tumor growth factors in the blood of animals carrying HuH7 tumors (Fig. 11B). In both HuH7 and HT29 tumors, we observed drug combination exposure caused activation of AKT though total S6 phosphorylation was reduced in both tumor types (Fig. 11C). These findings were associated with reduced PDGFRβ phosphorylation and decreased NFκB activity (Fig. 11D and E). As observed for HT29 tumors, obvious biomarkers for third drug combinations include AKT inhibitors, FGFR inhibitors, and ERBB1/2 inhibitors. Inhibition of AKT or ERBB1/2 signaling enhanced (regorafenib + sildenafil) killing (Fig. 11F).
Figure 11.
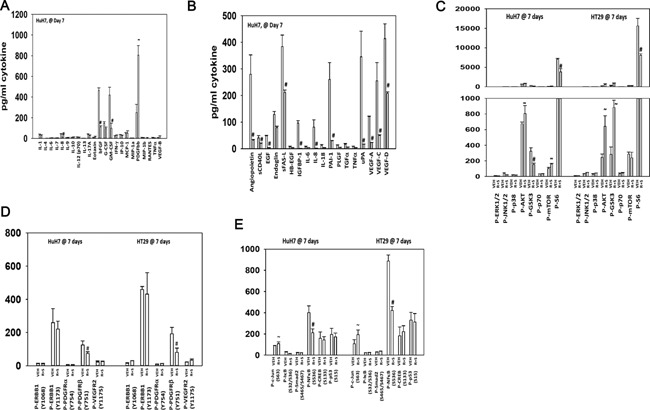
Regorafenib/sildenafil treatment alter multiple cell signaling processes in drug treated tumors at cessation of drug treatment. Parts (A–E) HuH7 and HT29 tumors, treated as described in Figures 9 and 10, were isolated 7 days after the start of drug exposure or were isolated at the time of sacrifice, when tumor volume was >1,500 mm3 (n.b. tumor mass for REGO + SIL treated tumors was only ∼500 mm3). Tumors were freeze‐thawed and lysed according to established procedures/manufacturer instructions. Tumors were subjected to multiplex assays to determine the levels of plasma cytokines/growth factors and the phosphorylation/expression of multiple membrane and intracellular signal transduction proteins (n = 8, ±SEM). #P < 0.05 lower value than vehicle control treated tumors; ~P < 0.05 greater value than in vehicle control treated tumors. (F) HuH7 cells were treated with vehicle control; regorafenib (0.5 μM) and sildenafil (2 μM); BGJ398 (1 μM); Lapatinib (1 μM); MK2206 (1 μM) or in the combinations indicated in the part. Cells were examined 12 h after treatment using a live/dead assay in a Hermes WiScan instrument. Red/yellow cells = dead; green cells = alive (n = 3, ±SEM). *P < 0.05 greater than corresponding value in (regorafenib + sildenafil) treatment alone.
Discussion
Our data demonstrated that PDE5 inhibitors and sorafenib/regorafenib interact in a greater than additive fashion to kill a genetically diverse set of hepatoma/colon cancer cells in vitro and in vivo. Knock down of PDE5 recapitulated the combinatorial effect of a PDE5 inhibitor when combined with sorafenib. And, knock down of PDGFRα/β recapitulated the combinatorial effect of sorafenib when combined with a PDE5 inhibitor. PDE5 inhibitors are known to enhance cGMP and NO levels; inhibition of NOS enzymes by L‐NAME prevented the killing interaction between sildenafil and sorafenib.
Hepatoma cells which lack CD95 expression, e.g., HuH7, are more resistant to sorafenib in combination with histone deacetylase inhibitors. In the present studies, HuH7 cells were readily killed by the combination of sorafenib and PDE5 inhibitors. In cells expressing CD95, inhibition of the extrinsic/CD95 pathway blocked cell killing whereas in HuH7 cells killing was due to activation of the mitochondrial/intrinsic pathway. Activation of CD95 was suppressed by inhibition of NOS enzymes. Studies by others have shown that nitrosylation of CD95 at its regulatory tyrosine residues inhibits CD95 signaling (Reinehr et al., 2004). Others, however, have argued that NO signaling and CD95 activation can cooperate to induce cell death (Gandelman et al., 2013; González et al., 2013).
In addition to activating the extrinsic apoptosis pathway, the drug combination also increased the numbers of autophagosomes and autolysosomes, i.e., “autophagy,” in a time dependent fashion that was inhibited by incubation of the cells with L‐NAME. Inhibition of autophagy suppressed cell killing. Nitric oxide signaling has been linked by others to the regulation of autophagy; in some primary non‐transformed cells NO can inhibit autophagy by inhibiting JNK signaling (Benavides et al., 2013; Shen et al., 2014). In some transformed cells NO signaling can promote autophagy in an mTOR‐dependent fashion which seems to play a role in tumor cell killing (Yu et al., 2012; Tripathi et al., 2013). We discovered that the drug combination inactivated mTOR, and expression of an activated form of mTOR suppressed killing, as well as autophagy as judged by GFP+ and RFP+ cells. Activation of the JNK pathway played a key role in regorafenib and sildenafil lethality. Thus nitric oxide signaling plays a role in de‐repressing a brake on toxic autophagy (mTOR) as well as promoting activation of CD95 apoptosis.
The drug combination rapidly increased the levels of both ROS and RNS. Inhibition of NOS enzymes almost abolished cell killing caused by sildenafil and regorafenib, whereas quenching of ROS levels was protective though not to the same extent. Sildenafil also increased cGMP levels, though the amount of cGMP generated by sildenafil did not appear to be strongly enhanced by sorafenib treatment, despite increased levels of VASP‐1 (S239) phosphorylation in sildenafil and regorafenib treated compared to sildenafil alone treated tumor cells (unpublished data). Knock down of iNOS expression or of PKGα/β suppressed the toxicity of sorafenib and sildenafil treatment in GI tumor cells.
One important mechanism by which NO is inactivated is by its reaction with the superoxide anion (O2 −) (Valez et al., 2013). Compared to non‐transformed cells, tumor cells generate considerably greater amounts of O2 − (Haklar et al., 2001). Indeed, it is of note that whereas sildenafil is often associated with reduced oxidative stress in non‐transformed cells it promotes oxidative stress in tumor cells (Das et al., 2010). The reaction of NO with O2 − forms the more potent oxidant peroxynitrite (ONOO−) (Hirst and Robson, 2007). Peroxynitrite causes oxidative damage and S‐nitrosylation of proteins, lipids, and DNA (Felley‐Bosco, 1998). Nitrosative stress by ONOO− has been implicated in DNA breakage, followed by poly‐ADP‐ribose polymerase (PARP) and ATM/ATR activation (Negi et al., 2010). Further studies will be needed to determine whether DNA damage is enhanced in our system and whether ATM/ATR signaling plays any role in the regulation of apoptosis/autophagy pathways and signal transduction pathways (Thompson et al., 2000; Ha et al., 2003; Sarfati et al., 2003; Deguchi et al., 2004; Zhu et al., 2005a,2005b, 2009; Black et al., 2008).
It has recently been argued that as sorafenib is ~99% protein bound based on in vitro binding assays with the drug, and thus only a very small amount of the drug is bioavailable and that in vitro studies using the drug at above ~1 μM are not valid in terms of their bench‐to‐bedside patient translatability (Smith and Houghton, 2013). These comments did not take into account that some sorafenib metabolites are at least as active, if not more active, i.e., 10‐fold, than the parent drug itself (Pratz et al., 2010). In our present studies, we noted that sorafenib concentrations as low as 500 nM or regorafenib concentrations as low as 300 nM could interact with sildenafil to kill tumor cells. However, the majority of our studies were performed using 2 μM sorafenib and 0.5 μM regorafenib. To address this issue in our present studies, we thus also performed cell killing assays in the presence of 100% human serum, and then treated cells with regorafenib or sorafenib at 25% of their respective Cmax plasma values; strong significant drug combination lethality was still observed after 24 h of treatment in hepatoma and breast cancer cells. These findings strongly argue the statements made by Houghton and Smith regarding bioactive levels of sorafenib in 100% serum/10% serum are largely incorrect.
The major plasma protein to which sorafenib binds is human serum albumin, and using radio‐labeled drug it was discovered that sorafenib is rapidly taken up into multiple tissues, particularly the liver. Although the drug is not as potently taken up by the brain, appreciable amounts can also accumulate in this tissue. The half‐life of sorafenib in plasma is ~11 h whereas its half‐life in various tissues/organs is 24–36 h (EMA, Nexavar Authorization Statement, 2006). In animals sorafenib is also known to be excreted in milk. Furthermore, we have recently noted that our pre‐clinical cell culture based studies combining pemetrexed and sorafenib (2–3 μM) have translated into multiple prolonged stable disease, prolonged partial, and complete responses in breast cancer patients in a Phase I trial (NCT01450384, Poklepovic and Dent, unpublished observations). Thus, in vitro studies in the 10–100% serum concentration range using sorafenib or regorafenib in the low micromolar 0.5–3.0 μM range are likely to have translational clinical relevance.
Based on our encouraging in vitro data, we moved our studies forward using several animal model systems in athymic mice. Short‐term exposure to sildenafil and regorafenib killed tumor cells in vivo as judged in ex vivo colony formation assays. Longer‐term exposure to sildenafil and sorafenib suppressed HuH7 tumor growth over several weeks, as was also observed using HT29 colon cancer tumors. This is of note as HuH7 cells, which lack CD95, would be predicted based on in vitro data to respond relatively poorly to the (regorafenib + sildenafil) combination. Our in vivo anti‐tumor data are likely an amalgamation of a direct anti‐tumor cell killing effect by the drug combination and increased tumor vasculature permeability caused by the actions of sildenafil on endothelial cells (Black et al., 2008). We determined whether FTY720 and other HDAC inhibitors could enhance the lethality of (regorafenib + sildenafil) treatment in our HT29 model of colon cancer n.b. regorafenib is an approved third line treatment for colorectal cancer. Combined exposure of HT29 colon cancer tumors to (regorafenib + FTY720) or to (regorafenib + sildenafil + FTY720) resulted unexpectedly in a stronger re‐growth of tumors after cessation of drug exposure than was observed in tumors treated with regorafenib or with (regorafenib + sildenafil) but not treated with FTY720. Indeed, tumor growth was reduced and animal survival was increased in tumors treated with (regorafenib + sildenafil) compared to (regorafenib + sildenafil + FTY720) (Booth et al., 2015).
We discovered that treatment of tumors with FTY720 in general caused increased expression of multiple growth/angiogenic/invasion regulatory cytokines, including FAS ligand. And, that inhibition of the CD95/FAS‐L pathway prevented the enhanced tumor re‐growth of (regorafenib + FTY720) treated tumors. As FTY720 inhibits S1P signaling, the drug has been examined for anti‐cancer properties and has recently been shown to suppress colon cancer development and growth in an inflammatory model of colitis (Liang et al., 2013). More recently, we have shown that FTY720 strongly enhances the anti‐tumor properties of (celecoxib + sildenafil) treatment in breast and brain cancer cells in vitro and in vivo (Booth et al., 2015).
In conclusion, we have demonstrated that sorafenib/regorafenib in combination with PDE5 inhibitors kills tumor cells in a greater than additive manner in a process that involves autophagy, and intrinsic and extrinsic apoptotic pathways. In vivo, regorafenib/sorafenib strongly interacted with sildenafil to suppress tumor growth. By applying multiplex technology, we were able to predict and validate various signaling biomarkers as compensatory responses of tumor cells mediating survival.
Acknowledgments
We thank Mrs. Grizzard for her support to the Dent lab and to the Betts family fund for support in the purchase of the Hermes Wiscan instrument. We also thank Dr. G. Almera for performing immuno‐fluorescence studies. We gratefully acknowledge the assistance of the VCU lipidomics/Metabolomics core in performing our ceramide and S1P analyses. PD is the holder of the Universal Inc. Chair in Signal Transduction Research.
The authors have no conflicts of interest.
Mehrad Tavallai, Hossein A. Hamed, and Jane L. Roberts share co‐first authorship.
Literature Cited
- Benavides GA, Liang Q, Dodson M, Darley‐Usmar V, Zhang J. 2013. Inhibition of autophagy and glycolysis by nitric oxide during hypoxia‐reoxygenation impairs cellular bioenergetics and promotes cell death in primary neurons. Free Radic Biol Med 65:1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black KL, Yin D, Ong JM, Hu J, Konda BM, Wang X, Ko MK, Bayan JA, Sacapano MR, Espinoza A, Irvin DK, Shu Y. 2008. PDE5 inhibitors enhance tumor permeability and efficacy of chemotherapy in a rat brain tumor model. Brain Res 1230:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Cruickshanks N, Conley A, Durrant DE, Das A, Fisher PB, Kukreja RC, Grant S, Poklepovic A, Dent P. 2014. Phosphodiesterase 5 inhibitors enhance chemotherapy killing in gastrointestinal/genitourinary cancer cells. Mol Pharmacol 85:408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Cruickshanks N, Tavallai S, Webb T, Samuel P, Conley A, Binion B, Young HF, Poklepovic A, Spiegel S, Dent P. 2015. PDE5 inhibitors enhance Celecoxib killing in multiple tumor types. J Cell Physiol 230:1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr BI, D'Alessandro R, Refolo MG, Iacovazzi PA, Lippolis C, Messa C, Cavallini A, Correale M, Di Carlo A. 2013. Effects of low concentrations of regorafenib and sorafenib on human HCC cell AFP, migration, invasion, and growth in vitro. J Cell Physiol 228:1344–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CY, Park KR, Lee JH, Jeon YJ, Liu KH, Oh S, Kim DE, Yea SS. 2007. Isoeugenol suppression of inducible nitric oxide synthase expression is mediated by down‐regulation of NF‐kappaB, ERK1/2, and p38 kinase. Eur J Pharmacol 576:151–159. [DOI] [PubMed] [Google Scholar]
- Cruickshanks N, Hamed HA, Booth L, Tavallai S, Syed J, Sajithlal GB, Grant S, Poklepovic A, Dent P. 2013. Histone deacetylase inhibitors restore toxic BH3 domain protein expression in anoikis‐resistant mammary and brain cancer stem cells, thereby enhancing the response to anti‐ERBB1/ERBB2 therapy. Cancer Biol Ther 14:982–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Xi L, Kukreja RC. 2008. Protein kinase G‐dependent cardioprotective mechanism of phosphodiesterase‐5 inhibition involves phosphorylation of ERK and GSK3beta. J Biol Chem 283:29572–29585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Durrant D, Mitchell C, Mayton E, Hoke NN, Salloum FN, Park MA, Qureshi I, Lee R, Dent P, Kukreja RC. 2010. Sildenafil increases chemotherapeutic efficacy of doxorubicin in prostate cancer and ameliorates cardiac dysfunction. Proc Natl Acad Sci USA 107:18202–18207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi A, Thompson WJ, Weinstein IB. 2004. Activation of protein kinase G is sufficient to induce apoptosis and inhibit cell migration in colon cancer cells. Cancer Res 64:3966–3973. [DOI] [PubMed] [Google Scholar]
- Elser C, Siu LL, Winquist E, Agulnik M, Pond GR, Chin SF, Francis P, Cheiken R, Elting J, McNabola A, Wilkie D, Petrenciuc O, Chen EX. 2007. Phase II trial of sorafenib in patients with recurrent or metastatic squamous cell carcinoma of the head and neck or nasopharyngeal carcinoma. J Clin Oncol 25:3766–3773. [DOI] [PubMed] [Google Scholar]
- Felley‐Bosco E. 1998. Role of nitric oxide in genotoxicity: implication for carcinogenesis. Cancer Metastasis Rev 17:25–37. [DOI] [PubMed] [Google Scholar]
- Florio T, Arena S, Pattarozzi A, Thellung S, Corsaro A, Villa V, Massa A, Diana F, Spoto G, Forcella S, Damonte G, Filocamo M, Benatti U, Schettini G. 2003. Basic fibroblast growth factor activates endothelial nitric‐oxide synthase in CHO‐K1 cells via the activation of ceramide synthesis. Mol Pharmacol 63:297–310. [DOI] [PubMed] [Google Scholar]
- Francis SH, Busch JL, Corbin JD, Sibley D. 2010. CGMP‐dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol Rev 62:525–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandelman M, Levy M, Cassina P, Barbeito L, Beckman JS. 2013. P2×7 receptor‐induced death of motor neurons by a peroxynitrite/FAS‐dependent pathway. J Neurochem 126:382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González R, Ferrín G, Aguilar‐Melero P, Ranchal I, Linares CI, Bello RI, De la Mata M, Gogvadze V, Bárcena JA, Alamo JM, Orrenius S, Padillo FJ, Zhivotovsky B, Muntané J. 2013. Targeting hepatoma using nitric oxide donor strategies. Antioxid Redox Signal 18:491–506. [DOI] [PubMed] [Google Scholar]
- Ha KS, Kim KM, Kwon YG, Bai SK, Nam WD, Yoo YM, Kim PK, Chung HT, Billiar TR, Kim YM. 2003. Nitric oxide prevents 6‐hydroxydopamine‐induced apoptosis in PC12 cells through cGMP‐dependent PI3 kinase/Akt activation. FASEB J 17:1036–1047. [DOI] [PubMed] [Google Scholar]
- Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. 2009. Regulation of histone acetylation in the nucleus by sphingosine‐1‐phosphate. Science 325:1254–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, Wise LE, Allegood JC, O'Brien M, Avni D, Reeves TM, Knapp PE, Lu J, Luo C, Miles MF, Milstien S, Lichtman AH, Spiegel S. 2014. Active, phosphorylated fingolimod inhibits histone deacetylases and facilitates fear extinction memory. Nat Neurosci 17:971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haklar G, Sayin‐Ozveri E, Yüksel M, Aktan AO, Yalçin AS. 2001. Different kinds of reactive oxygen and nitrogen species were detected in colon and breast tumors. Cancer Lett 165:219–224. [DOI] [PubMed] [Google Scholar]
- Hayden MA, Lange PA, Nakayama DK. 2001. Nitric oxide and cyclic guanosine monophosphate stimulate apoptosis via activation of the Fas‐FasL pathway. J Surg Res 101:183–189. [DOI] [PubMed] [Google Scholar]
- Hirst DG, Robson T. 2007. Nitrosative stress in cancer therapy. Front Biosci 12:3406–3418. [DOI] [PubMed] [Google Scholar]
- Hotte SJ, Hirte HW. 2002. BAY 43‐9006: Early clinical data in patients with advanced solid malignancies. Curr Pharm Des 8:2249–2253. [DOI] [PubMed] [Google Scholar]
- Inaba H, Rubnitz JE, Coustan‐Smith E, Li L, Furmanski BD, Mascara GP, Heym KM, Christensen R, Onciu M, Shurtleff SA, Pounds SB, Pui CH, Ribeiro RC, Campana D, Baker SD. 2001. Phase I pharmacokinetic and pharmacodynamics study of the multi‐kinase inhibitor sorafenib in combination with clofarabine and cytarabine in pediatric relapsed/refractory leukemia. J Clin Oncol 29:3293–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komalavilas P, Shah PK, Jo H, Lincoln TM. 1999. Activation of mitogen‐activated protein kinase pathways by cyclic GMP and cyclic GMP‐dependent protein kinase in contractile vascular smooth muscle cells. J Biol Chem 274:34301–34309. [DOI] [PubMed] [Google Scholar]
- Kots AY, Bian K, Murad F. 2011. Nitric oxide and cyclic GMP signaling pathway as a focus for drug development. Curr Med Chem 18:3299–3305. [DOI] [PubMed] [Google Scholar]
- Li L, Zhao M, Navid F, Pratz K, Smith BD, Rudek MA, Baker SD. 2010. Quantitation of sorafenib and its active metabolite sorafenib N‐oxide in human plasma by liquid chromatography‐tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 878:3033–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Nagahashi M, Kim EY, Harikumar KB, Yamada A, Huang WC, Hait NC, Allegood JC, Price MM, Avni D, Takabe K, Kordula T, Milstien S, Spiegel S. 2013. Sphingosine‐1‐phosphate links persistent STAT3 activation, chronic intestinal inflammation, and development of colitis‐associated cancer. Cancer Cell 23:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP, Park MA, Mitchell C, Walker T, Rahmani M, Thorburn A, Häussinger D, Reinehr R, Grant S, Dent P. 2009. BCL‐2 family inhibitors enhance histone deacetylase inhibitor and sorafenib lethality via autophagy and overcome blockade of the extrinsic pathway to facilitate killing. Mol Pharmacol 76:327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musicki B, Bivalacqua TJ, Champion HC, Burnett AL. 2014. Sildenafil promotes eNOS activation and inhibits NADPH oxidase in the transgenic sickle cell mouse penis. J Sex Med 11:424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi G, Kumar A, Sharma SS. 2010. Concurrent targeting of nitrosative stress‐PARP pathway corrects functional, behavioral and biochemical deficits in experimental diabetic neuropathy. Biochem Biophys Res Commun 391:102–106. [DOI] [PubMed] [Google Scholar]
- Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, Graf M, Rahmani M, Ryan K, Liu X, Spiegel S, Norris J, Fisher PB, Grant S, Dent P. 2008. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide‐dependent CD95 and PERK activation. Cancer Biol Ther 7:1648–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MA, Reinehr R, Häussinger D, Voelkel‐Johnson C, Ogretmen B, Yacoub A, Grant S, Dent P. 2010a. Sorafenib activates CD95 and promotes autophagy and cell death via Src family kinases in gastrointestinal tumor cells. Mol Cancer Ther 9:2220–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MA, Mitchell C, Zhang G, Yacoub A, Allegood J, Häussinger D, Reinehr R, Larner A, Spiegel S, Fisher PB, Voelkel‐Johnson C, Ogretmen B, Grant S, Dent P. 2010b. Vorinostat and sorafenib increase CD95 activation in gastrointestinal tumor cells through a Ca(2+)‐de novo ceramide‐PP2A‐reactive oxygen species‐dependent signaling pathway. Cancer Res 70:6313–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratz KW, Cho E, Levis MJ, Karp JE, Gore SD, McDevitt M, Stine A, Zhao M, Baker SD, Carducci MA, Wright JJ, Rudek MA, Smith BD. 2010. A pharmacodynamic study of sorafenib in patients with relapsed and refractory acute leukemias. Leukemia 24:1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani M, Davis EM, Crabtree TR, Habibi JR, Nguyen TK, Dent P, Grant S. 2007. The kinase inhibitor sorafenib induces cell death through a process involving induction of endoplasmic reticulum stress. Mol Cell Biol 27:5499–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani M, Davis EM, Bauer C, Dent P, Grant S. 2005. Apoptosis induced by the kinase inhibitor BAY 43‐9006. J Biol Chem 280:35217–35227. [DOI] [PubMed] [Google Scholar]
- Reinehr R, Görg B, Höngen A, Häussinger D. 2004. CD95‐tyrosine nitration inhibits hyperosmotic and CD95 ligand‐induced CD95 activation in rat hepatocytes. J Biol Chem 279:10364–10373. [DOI] [PubMed] [Google Scholar]
- Roberts JL, Conley A, Booth L, Cruickshanks N, Malkin M, Kukreja RC, Grant S, Poklepovic A, Dent P. 2014. PDE5 inhibitors enhance the lethality of standard of care chemotherapy in pediatric CNS tumor cells. Cancer Biol Ther 15:758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russwurm M, Russwurm C, Koesling D, Mergia E. 2013. NO/cGMP: The past, the present, and the future. Methods Mol Biol 1020:1–16. [DOI] [PubMed] [Google Scholar]
- Sarfati M, Mateo V, Baudet S, Rubio M, Fernandez C, Davi F, Binet JL, Delic J, Merle‐Beral H. 2003. Sildenafil and vardenafil, types 5 and 6 phosphodiesterase inhibitors, induce caspase‐dependent apoptosis of B‐chronic lymphocytic leukemia cells. Blood 101:265–269. [DOI] [PubMed] [Google Scholar]
- Shen C, Yan J, Erkocak OF, Zheng XF, Chen XD. 2014. Nitric oxide inhibits autophagy via suppression of JNK in meniscal cells. Rheumatology (Oxford) 53:1022–1033. [DOI] [PubMed] [Google Scholar]
- Smith MA, Houghton P. 2013. A proposal regarding reporting of in vitro testing results. Clin Cancer Res 19:2828–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Hamed HA, Cruickshanks N, Fisher PB, Grant S, Dent P. 2012. Obatoclax and lapatinib interact to induce toxic autophagy through NOXA. Mol Pharmacol 81:527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WJ, Piazza GA, Li H, Liu L, Fetter J, Zhu B, Sperl G, Ahnen D, Pamukcu R. 2000. Exisulind induction of apoptosis involves guanosine 3′,5′‐cyclic monophosphate phosphodiesterase inhibition, protein kinase G activation, and attenuated beta‐catenin. Cancer Res 60:3338–3342. [PubMed] [Google Scholar]
- Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, Wogan GN. 2013. Reactive nitrogen species regulate autophagy through ATM‐AMPK‐TSC2‐mediated suppression of mTORC1. Proc Natl Acad Sci USA 110:E2950–E2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valez V, Cassina A, Batinic‐Haberle I, Kalyanaraman B, Ferrer‐Sueta G, Radi R. 2013. Peroxynitrite formation in nitric oxide‐exposed submitochondrial particles: Detection, oxidative damage and catalytic removal by Mn‐porphyrins. Arch Biochem Biophys 529:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Ohashi K, Takeuchi K, Yamashita K, Yokoyama T, Tran QK, Satoh H, Terada H, Ohashi H, Hayashi H. 2002. Sildenafil for primary and secondary pulmonary hypertension. Clin Pharmacol Ther 71:398–402. [DOI] [PubMed] [Google Scholar]
- Yu Y, Fan SM, Yuan SJ, Tashiro S, Onodera S, Ikejima T. 2012. Nitric oxide (•NO) generation but not ROS plays a major role in silibinin‐induced autophagic and apoptotic death in human epidermoid carcinoma A431 cells. Free Radic Res 46:1346–1360. [DOI] [PubMed] [Google Scholar]
- Zhu B, Strada S, Stevens T. 2005a. Cyclic GMP‐specific phosphodiesterase 5 regulates growth and apoptosis in pulmonary endothelial cells. Am J Physiol Lung Cell Mol Physiol 289:L196–L206. [DOI] [PubMed] [Google Scholar]
- Zhu B, Vemavarapu L, Thompson WJ, Strada SJ. 2005b. Suppression of cyclic GMP‐specific phosphodiesterase 5 promotes apoptosis and inhibits growth in HT29 cells. J Cell Biochem 94:336–350. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zhang L, Alexeyev M, Alvarez DF, Strada SJ, Stevens T. 2009. Type 5 phosphodiesterase expression is a critical determinant of the endothelial cell angiogenic phenotype. Am J Physiol Lung Cell Mol Physiol 296:L220–L228. [DOI] [PMC free article] [PubMed] [Google Scholar]


