Abstract
Macroautophagy (hereafter autophagy) is one of the major degradation systems in eukaryotic cells, and its dysfunction may result in diseases ranging from neurodegeneration to cancer. Although most of the autophagy-related (Atg) proteins that function in this pathway were first identified in yeast, many were subsequently shown to have homologs in higher eukaryotes including humans, and the overall mechanism of autophagy is highly conserved. The most prominent feature of autophagy is the formation of a double-membrane sequestering compartment, the phagophore; this transient organelle surrounds part of the cytoplasm and matures into an autophagosome, which subsequently fuses with the vacuole or lysosome to allow degradation of the cargo. Much attention has focused on the process involved in phagophore nucleation and expansion, but many questions remain. Here, we identified the yeast protein Icy2, which we now name Atg41, as playing a role in autophagosome formation. Atg41 interacts with the transmembrane protein Atg9, a key component involved in autophagosome biogenesis, and both proteins display a similar localization profile. Under autophagy-inducing conditions the expression level of Atg41 increases dramatically and is regulated by the transcription factor Gcn4. This work provides further insight into the mechanism of Atg9 function and the dynamics of sequestering membrane formation during autophagy.
Keywords: autophagy, lysosome, stress, vacuole, yeast
Abbreviations
- Atg
autophagy related
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
- GFP
green fluorescent protein
- IDRs
intrinsically disordered regions
- PA
protein A
- PAS
phagophore assembly site
- RT-qPCR
real-time quantitative PCR
- UTR
untranslated region
- VC
C terminus of Venus YFP
- VN
N terminus of Venus YFP
- WT
wild type
Introduction
Autophagy is involved in numerous aspects of cellular homeostasis, and autophagic dysfunction is associated with a range of diseases in humans.1 Part of the reason that autophagy is connected to a wide array of physiological pathways has to do with the unique process of sequestration involving the phagophore. In contrast to the secretory pathway, where vesicles bud off from organelles already containing their cargo, the phagophore is generated through an expansion process that provides tremendous cargo flexibility. Thus, autophagy is the primary mechanism through which cells can eliminate damaged or superfluous organelles. For this reason, the mechanism of phagophore nucleation and expansion, including the origin of the donor membranes, has been the subject of intense focus.
The biogenesis of the phagophore is thought to initiate at a nucleating site, named the phagophore assembly site (PAS). Most of the Atg proteins transiently reside at the PAS, although the ultrastructure of this site and the interactions among the Atg proteins during phagophore formation are not known. One of the key proteins that functions in formation and/or expansion of the phagophore is Atg9. This protein is predicted to contain 6 transmembrane domains, and to transit from the endoplasmic reticulum to the Golgi apparatus;2,3 Atg9 subsequently localizes to peri-mitochondrial sites that are termed Atg9 peripheral structures, Atg9 reservoirs or tubulovesicular clusters.4 As a membrane protein, Atg9 is considered to play a role in delivering or directing membrane from a donor site(s) to the PAS. In addition, Atg9 may move between these sites in successive rounds of trafficking.4,5 Anterograde movement to the PAS requires Atg11, Atg23 and Atg27, whereas retrograde movement involves Atg1-Atg13 and Atg2-Atg18.5,6 The details of Atg9 trafficking, however, are not understood.
Molecular genetic studies in Saccharomyces cerevisiae have contributed substantially to our understanding of autophagy at the molecular level.7 Genetic screens for autophagy-defective mutants have identified a series of genes whose products are involved primarily or exclusively in autophagy. These Atg proteins act at different steps of the autophagy process. Even though 40 different Atg proteins have been reported in yeast, new ones continue to be identified. Atg41 is a small protein identified in yeast as being upregulated during heat shock,8 but the structural and functional information for this protein is limited and its cellular function is not known.9-11
Here, we report that Atg41 plays a role in autophagosome formation; atg41Δ cells display a severe defect in autophagy activity. We found that ATG41 is upregulated at the transcriptional level, resulting in a substantial increase in the protein amount under autophagy-inducing conditions, and this upregulation is required for efficient autophagy. The association between Atg41 and Atg9 suggest that this protein is part of the Atg9 complex that plays a role in determining the frequency of autophagosome formation.
Results
Atg41/Icy2 is required for nonselective autophagy
We identified icy2Δ in a large-scale screen for mutants defective in mitophagy, the selective autophagic degradation of mitochondria.12 To determine whether Icy2 plays a role in nonselective autophagy, we carried out the GFP-Atg8 processing assay. Atg8 is required for autophagosome formation and the phosphatidylethanolamine (PE)-conjugated form of the protein initially lines both sides of the phagophore; Atg8–PE on the outer surface of the mature autophagosome is released, whereas the population inside the autophagosome is delivered to the vacuole and degraded.3 When fused to GFP, Atg8 is still degraded in the vacuole lumen, but the GFP moiety is relatively stable. Thus, the generation of free GFP can be used to monitor autophagic delivery to the vacuole.13 We monitored the processing of GFP-Atg8 expressed under the control of the endogenous ATG8 promoter. In growing conditions there is a low level of GFP-Atg8, and the protein is upregulated following autophagy induction (Fig. 1A). We observed a substantial decrease of free GFP in icy2Δ cells after 2 h of autophagy induced by nitrogen starvation compared to the wild type (Fig. 1A-B), suggesting that autophagy flux was reduced in the mutant.
Figure 1.
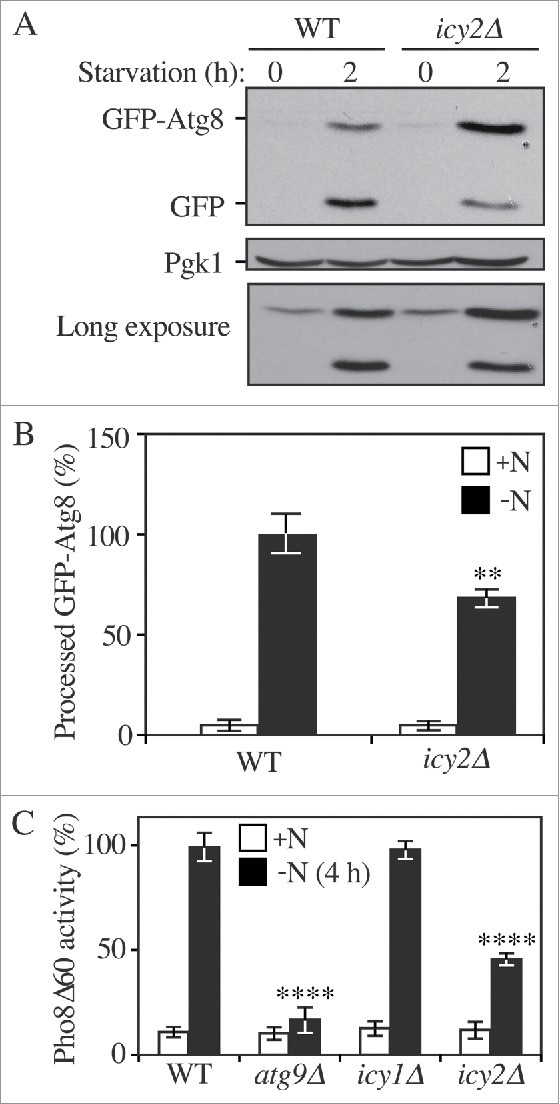
Icy2 is required for nonselective autophagy. (A) GFP-Atg8 processing assay for samples prepared from wild-type (WT; ZYY101) and icy2Δ (ZYY102) strains. Both growing samples (0 h; mid-log phase in YPD) and starvation samples (2 h; SD-N medium) were collected. Proteins were resolved by SDS-PAGE and detected by western blot with anti-YFP antibody and anti-Pgk1 (loading control) antiserum. A long exposure of a separate blot is included to show the GFP-Atg8 bands in growing conditions. (B) Quantitative analysis of processed GFP for the experiment in (A) Error bars represent the SD of 3 independent experiments. The result is examined by 2-way analysis of variance (ANOVA). p values derived from the Sidak post test are reported for the comparison between wild type and mutant. **, p<0 .01. (C) Pho8Δ60 assay for the WT (WLY176), atg9Δ (ZYY104), icy1Δ (ZYY106) and icy2Δ (ZYY105) strains. Samples were collected from growing cells (+N; mid-log phase in YPD) and after starvation (-N; SD-N medium). The Pho8Δ60 activity in the WT was set to 100% and the other samples were normalized. The error bars indicate the SD of 3 independent experiments. The result is examined by ANOVA. p values derived from the Sidak post test are reported for the comparison between wild type and mutant. ****, p < 0 .0001.
The GFP-Atg8 processing assay reflects the surface area of the inner autophagosome vesicle, which is referred to as an autophagic body once it is released into the vacuole lumen. A more quantitative measure of autophagic capacity can be achieved with the Pho8Δ60 assay, which measures the volume of the autophagosome/autophagic body. Pho8 is a vacuolar phosphatase that is synthesized as a zymogen; it is delivered to the vacuole via part of the secretory pathway and is subsequently activated in the vacuolar lumen by proteolytic removal of a C-terminal propeptide.14 Pho8Δ60 lacks the N-terminal transmembrane domain that serves as an internal, uncleaved signal sequence; the truncated protein remains in the cytosol and is only delivered to the vacuole by autophagy. Thus, autophagy activity can be measured by monitoring Pho8Δ60-dependent phosphatase activity in a strain lacking the endogenous PHO8 gene.15
The wild-type strain showed the expected increase in autophagy activity following nitrogen starvation (Fig. 1C). In contrast, an atg9Δ strain displayed essentially a complete block in Pho8Δ60-dependent phosphatase activity. Consistent with the GFP-Atg8 processing assay, the icy2Δ strain showed a significant reduction in autophagy activity compared to the wild type. Yeast express a paralog of Icy2, Icy1, and we extended the analysis by examining the requirement for this protein in autophagy. In contrast to the icy2Δ strain, however, there was no significant difference between the autophagy activity in the icy1Δ and wild-type cells. To verify that the defect seen in the icy2Δ mutant was not due to an unknown secondary mutation we transformed the null strain with a plasmid encoding the wild-type ICY2 gene under the control of its endogenous promoter. The wild-type gene fully restored the autophagy activity of the null strain suggesting that the mutant phenotype was indeed the result of deleting ICY2 (Fig. S1A).
We decided to examine whether Icy2 was sufficient to induce autophagy by overexpressing it during nutrient-rich conditions. We took advantage of the ZEO1 promoter, which allows constitutive expression of the downstream gene before and after starvation. However, overexpressing ICY2 by replacing its endogenous promoter with the ZEO1 promoter did not induce autophagy in growing conditions (Fig. S1B), and in fact caused a partial decrease in activity in starvation conditions. The inability to induce autophagy by overexpression in growing conditions likely reflects the need for the upregulation of other Atg proteins including Atg7, Atg8 and Atg9 to achieve a higher level of autophagy activity.16-18 To rule out the possibility that Icy2 affects the transcription of other ATG genes we examined the expression of the ATG7 and ATG9 genes in the icy2Δ mutant after starvation. We found that the expression of either gene was not affected by the absence of Icy2 (Fig. S1C). Similarly, the amount of the Atg9 protein was unaffected (Fig. S1D), indicating that Icy2 was not required for Atg9 translation; we do not have antibody to the endogenous Atg7 protein.
Next, to investigate whether Icy2 affects selective autophagy, we examined processing of precursor aminopeptidase I (prApe1). The precursor form of Ape1 is the major cargo of the cytoplasm-to-vacuole targeting (Cvt) pathway, a biosynthetic type of selective autophagy.19 Upon delivery to the vacuole, the propeptide of prApe1 is proteolytically cleaved and the resulting molecular weight shift can be detected by western blot. We observed a defect in prApe1 processing in the absence of Icy2 (Fig. S1E), indicating that this protein is required for an efficient Cvt pathway. Taken together, these data indicate that Icy2 is required for normal autophagy flux. Because of the autophagy-defective phenotype of the icy2Δ strain based on the GFP-Atg8 processing and Pho8Δ60 assays, and the block in the Cvt pathway, we concluded that Icy2 is involved in both selective and nonselective autophagy. We are not able to identify a previous publication describing Icy2, and therefore refer to the protein as Atg41 hereafter.
An increased Atg41 expression level during starvation is required for autophagy
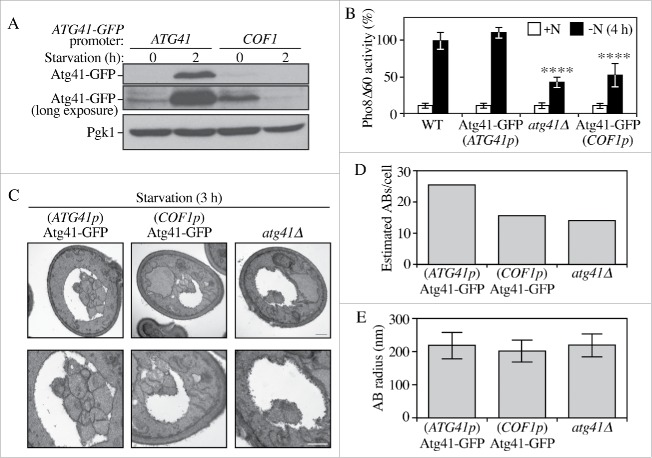
Either insufficient or excessive autophagy can be detrimental to cell physiology. Accordingly, autophagy activity is regulated at many levels, including through transcriptional control.16-18,20 To determine whether the expression level of Atg41 changed between nutrient-rich and nitrogen starvation conditions we first examined the ATG41 mRNA level by real-time quantitative PCR (RT-qPCR; Fig. 2A). A dramatic increase of ATG41 mRNA was observed after 1 h of nitrogen starvation. To determine whether the ATG41 transcriptional expression increase leads to a corresponding translational upregulation, we integrated epitope tags at the ATG41 chromosomal locus using either GFP or protein A (PA) to generate C-terminal fusions. A Pho8Δ60 assay of the ATG41-GFP strain showed a comparable autophagy activity level to the wild type, indicating that the C-terminal tagging of ATG41 did not disrupt protein function (Fig. S2A). Cells expressing tagged Atg41 were harvested from cultures in growing and nitrogen starvation conditions and protein extracts were examined by western blot (Fig. 2B-C). In agreement with the RT-qPCR results, the protein level of Atg41 detected in either of the tagged strains increased substantially during nitrogen starvation. We could not detect Atg41-GFP in growing conditions with a short-time exposure, but could observe a low level of Atg41-PA. The inability to detect the GFP-tagged protein in growing conditions probably reflects the low expression level of Atg41 during vegetative growth and the different sensitivity of the respective antibodies.
Figure 2.
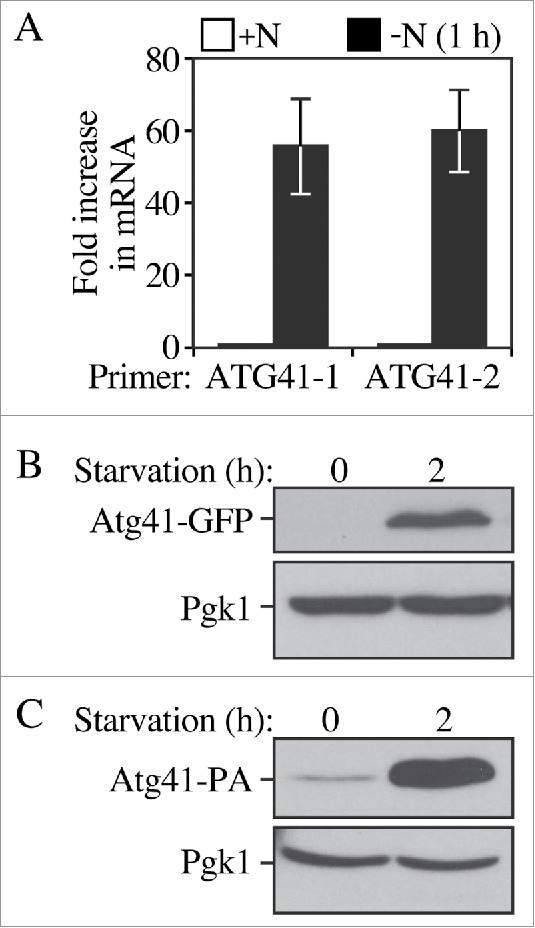
The Atg41 expression level increases after autophagy is induced. (A) The mRNA level of ATG41 was measured by RT-qPCR in the wild-type strain (WT; WLY176) before and after nitrogen starvation with 2 pairs of independent ATG41 primers. The error bars represent the SD from 3 independent experiments. (B) The protein level of Atg41-GFP was detected by western blot. Both growing (mid-log phase in YPD) and starvation (SD-N medium) samples of Atg41-GFP were collected, protein extracts resolved by SDS-PAGE and blots probed with anti-YFP antibody and anti-Pgk1 (loading control) antiserum. (C) The protein level of Atg41-PA was detected by western blot as described in B.
We took advantage of the GFP tag to examine the expression of Atg41 by fluorescence microscopy. Consistent with the western blot results, dramatically increased fluorescence intensity was observed after 2-h nitrogen starvation (Fig. S2B). To investigate the stability of the Atg41 protein, we performed the western blot equivalent of a pulse-chase experiment, relying on the low expression level observed in growing conditions (Fig. S2C). Cells expressing Atg41-GFP were first starved for nitrogen and then shifted back to rich medium conditions (YPD). Protein extracts were collected at different time points and examined by western blot. The Atg8 level was also monitored for comparison because Atg8 shows a similar dramatic increase in expression under starvation conditions. We found that the Atg41 level decreased back to the pre-starvation level within 30 min after shifting the cultures back to growing conditions, indicating that Atg41 is unstable or rapidly degraded when the cells are not under autophagy-inducing conditions. In contrast, Atg8 levels displayed a steady but slow decline consistent with the turnover of the part of the Atg8 population that was present within autophagic bodies.
The increased expression of Atg41 was of a greater magnitude than that seen for any of the characterized Atg proteins, with the exception of Atg8. In the case of Atg7, Atg8 and Atg9 the increases in protein levels are important for efficient autophagy.16-18 Accordingly, we next wanted to determine whether the increase in Atg41 was required for robust autophagy activity. Accordingly, we replaced the endogenous ATG41 promoter in the genome with the COF1 promoter. We determined empirically that the COF1 promoter drives relatively low protein expression; in addition, the expression pattern of genes under the control of the COF1 promoter showed relatively little difference between vegetative and starvation conditions (data not shown). Thus, by replacing the endogenous ATG41 promoter (ATG41p) with that of COF1 we were able to maintain a similar, low ATG41 expression level before and after nitrogen starvation (Fig. 3A). We used the C-terminal GFP-tagged strain to allow us to detect the Atg41-GFP protein level; in fact, the primary purpose of using this strain in addition to the null mutant was that it allowed us to verify the reduced level of Atg41 with a positive signal. The atg41Δ, ATG41p-ATG41-GFP and COF1p-ATG41-GFP cells were grown to mid-log phase and shifted to starvation conditions; aliquots were collected and protein extracts were analyzed by the Pho8Δ60 assay. The wild-type and ATG41p-ATG41-GFP strains displayed a robust induction of autophagy activity during nitrogen starvation (Fig. 3B). In contrast, the COF1p-ATG41-GFP cells showed a substantial reduction in autophagy activity, close to that of the atg41Δ null strain. These data suggest that the large increase in Atg41 protein seen under starvation conditions is required for normal autophagy activity.
Figure 3.
The increase in Atg41 protein level is required for autophagy. (A) The COF1 promoter was used to clamp Atg41 expression at the basal level. Both ATG41p-ATG41-GFP (ZYY107) and COF1p-ATG41-GFP (ZYY109) cells were collected before and after nitrogen starvation, and protein extracts were analyzed by western blot as in Figure 1. A long exposure is shown to visualize Atg41 in growing conditions. (B) Pho8Δ60 assay of ATG41p-ATG41-GFP (ZYY107) and COF1p-ATG41-GFP (ZYY109). The activity in the wild-type strain (WT; WLY176) was set to 100% and other values were normalized. Error bars represent the SD of 3 independent experiments. The result is examined by 2-way analysis of variance (ANOVA). p values derived from the Sidak post test are reported for the comparison between wild type and mutant. ****, p < 0 .0001. (C) Representative TEM images of ATG41p-ATG41-GFP, COF1p-ATG41-GFP and atg41Δ cells after starvation (SD-N medium). Autophagic bodies are seen as clustered vesicles in the vacuole lumen, which appears white. The lower panels provide a higher magnification of the cells in the upper panels. Scale bar: 500 nm. (D, E) Estimated average number (D) or average radius (E) of of autophagic bodies per cell for the strains in C. Estimations were based on the number (D) or the radius (E) of autophagic body cross sections observed by TEM in a sample size of more than 100 cells. The error bars indicate SD of the sample cells' radius estimated and calculated by the mathematic algorithm.
Atg41 protein levels correlate with autophagosome number
A decrease in the number and/or size of autophagosomes can result in impaired autophagy activity.21,22 Therefore, we used transmission electron microscopy to determine the effect of decreased Atg41 protein levels with regard to autophagosome formation, by measuring the number and size of autophagic bodies.23 We used a pep4Δ vps4Δ strain background for this analysis. PEP4 encodes proteinase A, which is required to activate multiple vacuolar hydrolases; deleting PEP4 causes the accumulation of intact autophagic bodies in the vacuole. VPS4 edcodes a AAA-ATPase involved in multivesicular body protein sorting, and deletion of this gene reduces the vesicular background content of the vacuole. We analyzed the atg41Δ, ATG41p-ATG41-GFP and COF1p-ATG41-GFP strains with regard to autophagic body number and size (Fig. 3C-E). Quantitative analysis of the images revealed that the number of autophagic bodies was reduced in the atg41Δ and COF1p-ATG41-GFP strains (Fig. 3D), whereas there was essentially no difference in autophagic body size (Fig. 3E). Using an algorithm to estimate total autophagic body number and size within the vacuole,23 the electron microscopy analysis indicated that the total autophagy flux in the COF1p-ATG41-GFP and atg41Δ strains was reduced by approximately 50% compared to the ATG41p-ATG41-GFP wild type (Table S1), which is consistent with the Pho8Δ60 assay results. These data further support our finding that the increased Atg41 level is required for a maximal autophagic response. Finally, because the lower autophagy activity was due to decreased autophagosome numbers, these results also suggest that the Atg41 level is important in determining the rate of autophagosome formation rather than the size/expansion, similar to Atg9.17
Atg41 localizes to Atg9 peripheral structures and associates with Atg9
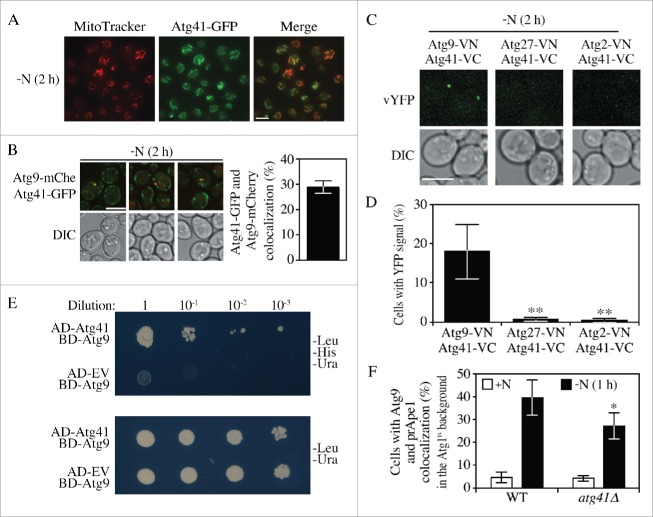
Reduced levels of Atg41 affected the number of autophagosomes formed during starvation, and we next wanted to determine the relationship of Atg41 to other components of the Atg core machinery, those proteins that are needed for autophagosome formation.24 Information on protein subcellular location can often provide useful informative with regard to protein function, and most of the Atg proteins have discrete localization patterns. Accordingly, we examined the localization of Atg41-GFP. Our analysis of Atg41 expression based on the visualization of the Atg41-GFP chimera showed a unique distribution pattern for this protein (Fig. S2B), which was reminiscent of the overall morphology of yeast mitochondria. Accordingly, we used MitoTracker Red to stain the mitochondria of cells expressing Atg41-GFP and examined them by fluorescence microscopy. We observed substantial colocalization of Atg41-GFP and MitoTracker Red (Fig. 4A). A peri-mitochondrial distribution is similar to that displayed by Atg9-GFP.4 In addition, Atg41 does not contain a mitochondrial targeting sequence or transmembrane domain, suggesting that Atg41-GFP distributes around the mitochondria rather than being directly associated with the organelle.
Figure 4.
Atg41 localizes to peripheral Atg9-containing sites. (A) Representative images for colocalization of Atg41 and MitoTracker Red. ATG41-GFP (ZYY107) cells were grown to mid-log phase and stained with MitoTracker Red in YPD medium. Cells were then washed and shifted to starvation conditions (SD-N) before fluorescence microscopy analysis; images are representative pictures from single Z-sections. Scale bar: 5 µm. (B) Representative images and the quantification for Atg41-GFP and Atg9-mCherry (ZYY125) colocalization. Cells were starved in SD-N medium prior to microscopy. Scale bar: 5 µm. (C) Representative images for a BiFC assay of strains expressing Atg41-VC and either Atg2-VN (ZYY114), Atg9-VN (ZYY113) or Atg27-VN (ZYY115). Cells were starved in SD-N medium prior to microscopy. Scale bar: 5 µm. (D) Quantitative analysis of cells showing BiFC signals described in (C). The result was examined by 2-way analysis of variance (ANOVA). p values derived from the Sidak post test are reported for the comparisons between ZYY113 and 2 other strains. **, p<0 .01. (E) Images for the yeast 2-hybrid result. The ATG41-AD and AD empty prey plasmids were cotransformed with the ATG9-BD bait plasmid. Cells were grown on SMD –Leu -Ura plates selecting for the presence of both plasmids. Cells were also grown on SMD –Leu –Ura -His plates with 10-fold serial dilution for the selection of strains having protein interactions. (F) Quantitative analysis of Atg9 and prApe1 (PAS) colocalization. WT (SKB171) and mutant (ZYY126) cells were transformed with a plasmid expressing temperature-sensitive Atg1 and cultured at the permissive temperature (24°C) with SMD medium (+N) until the mid-log phase. Samples were then shifted to SD-N and cultured at the nonpermissive temperature (37°C) for 1 h. The colocalization of Atg9–3GFP and RFP-Ape1 were counted and the frequency was determined. The result was examined by ANOVA. p values derived from the Sidak post test are reported for the comparison between wild type and mutant. *, p < 0.05.
Considering that Atg41 had a similar subcellular distribution to that of Atg9, and a decrease in either protein correlated with a reduced number of autophagic bodies, we decided to investigate the relationship between these 2 proteins. We first tagged Atg9 with mCherry, and Atg41 with GFP to see if these 2 proteins displayed a close proximity under fluorescence microscopy (Fig. 4B). The results indicated a partial colocalization of Atg9 and Atg41. To further examine whether a potential transient interaction exists, we took advantage of the bimolecular fluorescence complementation (BiFC) assay.25 In this assay, Venus YFP is split into 2 parts, the N terminus (VN) and C terminus (VC). We genomically tagged the C terminus of Atg41 with VC and the C terminus of Atg9 with VN, generating an ATG41-VC ATG9-VN strain. The YFP fluorescence signal in this strain can only be observed when VN and VC are in close proximity. For comparison, we also constructed ATG41-VC ATG2-VN and ATG41-VC ATG27-VN strains. Atg2 and Atg27 both function in Atg9 trafficking between the peripheral sites and the PAS, in retrograde and anterograde trafficking, respectively. We observed bright fluorescent dots in the cells expressing Atg41-VC Atg9-VN, but not in the other strains (Fig. 4C-D). Furthermore, we could only detect a fluorescent signal between Atg41-VC and Atg9-VN under starvation conditions; however, this may simply reflect the extremely low level of Atg41 expression during growing conditions (Fig. 2). This result suggests that Atg41 associates with (or is located in proximity to) Atg9, but not necessarily other Atg proteins involved in Atg9 trafficking.
To confirm the BiFC result, we also performed the yeast two-hybrid assay (Fig. 4E). AD-ATG41 and BD-ATG9 were cotransformed into cells, while the AD empty vector and BD-ATG9 were cotransformed as controls. The results showed that only cells with AD-ATG41 and BD-ATG9 could grow under minus histidine selection, indicating an association between Atg41 and Atg9. Since Atg9 is proposed to function in donating or directing membrane to the expanding phagophore by trafficking from donor sites to the PAS, we next investigated whether Atg41 controlled autophagy via an effect on Atg9 movement. Previous studies indicated that in the absence of Atg1 the retrograde trafficking of Atg9 was largely blocked resulting in accumulation of the protein at the PAS.5 We took advantage of this phenotype to carry out a kinetic analysis of Atg9 anterograde movement in cells expressing a temperature sensitive allele of ATG1 (Atg1ts); the strain also expressed Atg9–3GFP and RFP-Ape1 as a PAS marker. When autophagy was induced at the permissive temperature, Atg9–3GFP could shuttle between the PAS and peripheral sites, resulting in a low level of colocalization between Atg9 and prApe1 (data not shown). Similarly, the cells displayed a low level of colocalization in growing conditions (Fig. 4F). Following a shift of the Atg1ts strain to the nonpermissive temperature, Atg9–3GFP accumulated at the PAS resulting in a substantial increase in colocalization with RFP-Ape1. In the atg41Δ strain there was a significant decrease in the colocalization between these 2 proteins compared to the wild type. The Atg41 protein level in the Atg1ts strain was comparable to that in the wild-type strain after 1 h of starvation at the nonpermissive temperature (Fig. S3A), indicating that the defect in Atg9 trafficking reflected the absence of Atg41 specifically in the atg41Δ (Atg1ts) strain and not the ATG41 wild-type (Atg1ts) strain. This result indicated a kinetic defect in Atg9 anterograde trafficking in the absence of Atg41.
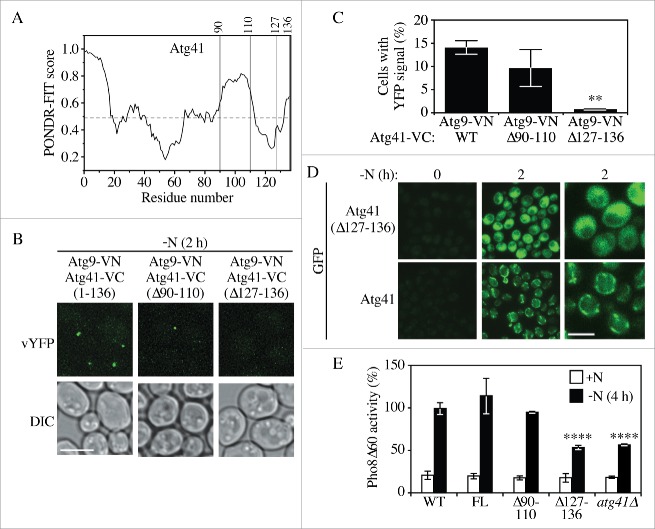
Because fluorescence microscopy, bimolecular fluorescence complementation and yeast two-hybrid data suggested that Atg41 associated with Atg9, we wanted to map the region of Atg41 that is required for this interaction, which would then allow us to examine the effect of specifically disrupting this association. We started the analysis with a computerized prediction of intrinsically disordered regions (IDRs) in Atg41. For many proteins IDRs participate in dynamic binding interactions. Furthermore, we determined that many of the yeast Atg proteins contain IDRs,26 and this appears to be conserved up to humans.27 We used PONDR-FIT, 28 an algorithm that combines several individual disorder predictors that evaluate the tendency for intrinsic disorder based on the target sequence, to identify putative IDRs in Atg41 (Fig. 5A). Based on the analysis, Atg41 contains at least 4 regions above the threshold level. Two of these potential IDRs are located at or near the N terminus. Because deletions of this part of the protein were more likely to affect folding of the remainder of the protein we focused our initial analysis on the 2 C-terminal IDRs and generated strains expressing Atg41(Δ91–110) and Atg41(Δ127–136). The C termini of these strains were tagged with VC, and Atg9 was tagged with VN to allow us to carry out the BiFC assay.
Figure 5.
The C-terminal region of Atg41 is important for its function in autophagy. (A) Tendency of intrinsic disorder and probability of an Atg41 residue being in a disordered region, predicted by PONDR-FIT. Regions of the protein above the horizontal dashed line are predicted to be disordered. The vertical lines indicate the regions chosen for deletion analysis. (B) Representative images for a BiFC assay of Atg41-VC Atg9-VN cells expressing full-length (1–136) Atg41-VC (ZYY113), and deletion mutants Atg41(Δ91–110)-VC (ZYY119) or Atg41(Δ127–136)-VC (ZYY120). Cells were starved in SD-N medium before fluorescence microscopy analysis. Scale bar: 5 µm. (C) Quantitative analysis of the BiFC assay described in (B). The result is examined by 2-way analysis of variance (ANOVA). p values derived from the Sidak post test are reported for the comparison between wild type and mutant. **, p < 0 .01. (D) Representative images showing the distribution of Atg41-GFP (ZYY107) and Atg41(Δ127–136)-GFP (ZYY121) in cells before and after starvation in SD-N medium prior to fluorescence microscopy analysis. The right column of panels corresponds to a higher magnification. Scale bar: 5 µm. (E) Pho8Δ60 assay of a wild-type (WT; WLY176) or atg41Δ (ZYY105) strain, and strains expressing full-length (FL) Atg41-VC (ZYY116) and the indicated Atg41-VC deletion mutants (ZYY117 and ZYY118). Pho8Δ60 activity was set to 100% in the WT after starvation and normalized in the other samples. Error bar represented the SD of 3 independent experiments. The result is examined by 2-way analysis of variance (ANOVA). p values derived from the Sidak post test are reported for the comparison between wild type and mutant. ****, p < 0 .0001.
Based on the bimolecular fluorescence complementation assay after autophagy induction we could still detect a fluorescent signal with Atg9-VN and Atg41(Δ91–110)-VC, similar to the results with full-length Atg41 (Fig. 5B-C). In contrast, deletion of the extreme 10 C-terminal residues in Atg41(Δ127–136)-VC abrogated the association between the 2 proteins. Because of the relatively small size of the deletion, and its location at the extreme C terminus, this mutation is unlikely to affect the stability of Atg41; indeed, analysis of Atg41(Δ127–136)-GFP by microscopy indicated that the truncation did not affect the stability of the protein relative to the wild type (Fig. 5D). Similarly, western blot indicated that Atg41(Δ127-136)-GFP was stable (Fig. S3B). Therefore, these data suggest that the extreme C-terminal region of Atg41 is important for associating with Atg9.
Having identified a region in Atg41 that was needed for its association with Atg9, we next utilized the Pho8Δ60 assay to examine the phenotypic consequence of disrupting this association (Fig. 5E). Cells expressing Atg41-VC displayed a wild-type level of autophagy activity, which was also the case for cells expressing Atg41(Δ91–110)-VC. In contrast, the Atg41(Δ127–136)-VC protein, which no longer interacted with Atg9, had a reduction of Pho8Δ60 activity similar to that seen with the atg41Δ strain. Finally, we examined the distribution pattern of the Atg41(Δ127–136) mutant. In contrast to the peri-mitochondrial localization seen with wild-type Atg41-GFP, Atg41(Δ127–136)-GFP showed a diffuse distribution pattern (Fig. 5D). Taken together, these results indicate the very C-terminal region of Atg41 is important for its association with Atg9, its correct subcellular distribution and normal autophagy activity.
The Gcn4 transcription factor regulates ATG41 expression
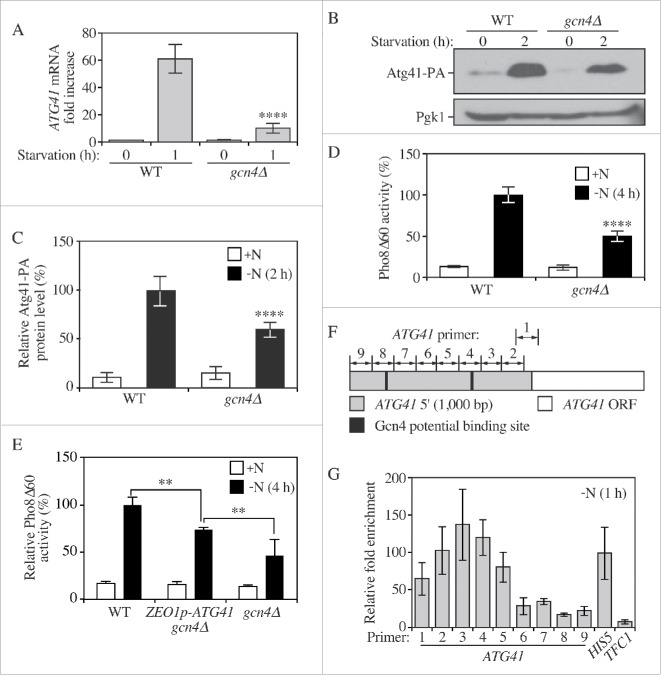
Because the Atg41 expression level changed considerably between growing and starvation conditions, we wanted to determine the mechanism involved in the transcriptional regulation of ATG41. High-throughput analysis indicates that Gcn4, the master regulator of gene expression during amino acid starvation, modulates the transcriptional level of multiple ATG genes.29 Moreover, combining the previous study and an online yeast transcriptional database, Yeastract (www.yeastract.com), indicated that the ATG41 5′ untranslated region (UTR) contains 2 putative Gcn4 binding regions.30 Therefore, we decided to investigate the possible role of Gcn4 in regulating ATG41 expression. We examined the ATG41 mRNA level by RT-qPCR. As we observed previously (Fig. 2A), there was a strong increase in ATG41 mRNA in the wild-type strain following autophagy induction by nitrogen starvation. In contrast, there was a significant reduction relative to the wild type in a gcn4Δ strain (Fig. 6A). Next, we tagged Atg41 with protein A in the gcn4Δ mutant and monitored the protein level of Atg41-PA by western blot. In agreement with the RT-qPCR results, the protein level of Atg41-PA was reduced by approximately 40% in the gcn4Δ background (Fig. 6B and C). Next, we used the Pho8Δ60 assay to determine whether the absence of Gcn4 caused an autophagy defect (Fig. 6D). We found a clear decrease of autophagy activity in the gcn4Δ background, suggesting that Gcn4-dependent regulation is necessary for optimal autophagy activity. To test whether overexpression of Atg41 in starvation conditions could rescue the gcn4Δ phenotype, we replaced the ATG41 endogenous promoter with the ZEO1 promoter in the gcn4Δ background; Gcn4 does not regulate the ZEO1 promoter. The Pho8Δ60 assay of ZEO1p-ATG41 showed a significantly increased autophagy activity compared to gcn4Δ, indicating a partial rescue of the gcn4Δ autophagy defect (Fig. 6E). The observation that the ZEO1p-ATG41 strain did not display a complete restoration of autophagy likely reflects a role for Gcn4 in regulating other genes whose products are required for full autophagy activity. Taken together, these data suggest that Gcn4 positively regulates ATG41 and that this regulation is important for autophagy.
Figure 6.
Gcn4 activates the transcription of ATG41 during nitrogen starvation. (A) The mRNA level of ATG41 in the wild-type (WT; WLY176) and gcn4Δ (ZYY122) strains was measured by RT-qPCR. Samples were collected in both growing (YPD) and starvation (SD-N) conditions. Error bars represent the SD of 3 independent experiments. The result was examined by 2-way analysis of variance (ANOVA). p values derived from the Sidak post test are reported for the comparison between wild type and mutant. ****, p < 0 .0001. (B) The Atg41-PA level of WT (ZYY108) and gcn4Δ (ZYY123) strains was analyzed by western blot. Pgk1 served as a loading control. (C) Quantitative analysis of the protein levels from the samples in (B). The protein level in the WT strain in starvation conditions was set to 100% and other samples were normalized. Error bars indicate the SD of 3 independent experiments. The result is examined by ANOVA. p values derived from the Sidak post test are reported for the comparison between wild type and mutant. ****, p < 0 .0001. (D) Pho8Δ60 assay for the WT and gcn4Δ strains. Cells were starved in SD-N. Error bars indicate the SD of 3 independent experiments. (E) Pho8Δ60 assay for the WT, ZEO1p-ATG41 gcn4Δ (ZYY127) and gcn4Δ strains. Cells were starved in SD-N. Error bars indicate the SD of 3 independent experiments. The result is examined by ANOVA. p values derived from the Sidak post test are reported for the comparison between ZYY127 and 2 other strains. **, p < 0 . 01. (F) Schematic picture showing the regions of the ATG41 5′ UTR covered by the indicated primers for analysis by ChIP; “1” corresponds to the ATG41–1 primer pair, etc. Gcn4 potential binding sites are shown as black lines. (G) RT-qPCR analysis of ChIP samples of the GCN4-PA strain (ZYY124). Numbers correspond to the primers illustrated in (F). The values were normalized to the positive control HIS5 (set to 100%). The error bars indicate the SD of 3 independent experiments.
Finally, we wanted to determine whether Gcn4 regulates ATG41 by direct binding to its promoter or indirectly by activating other transcription factors. To address this question we carried out a chromatin immunoprecipitation (ChIP) experiment using a Gcn4-PA strain. We designed 9 pairs of primers (ATG41–1 to ATG41–9) each spanning approximately 130 to 170 nucleotides of the ATG41 promoter region to ensure complete coverage including 2 potential Gcn4 binding regions predicted by computer and database analysis (Fig. 6F). In addition, we used a previously identified site in the HIS5 gene as a positive control29 and the TFC1 gene, which lacks any predicted Gcn4 binding sites, as a negative control. The RT-qPCR analysis indicated binding at the HIS5 promoter, but no binding at the TFC1 promoter as expected (Fig. 6G). For the ATG41 promoter region, we detected enrichment (at least 50% the level of the positive control) of the PCR product using primers ATG41–1 to ATG41–5, with the regions detected by primers ATG41–3 and ATG41–4 showing an even higher enrichment than the HIS5 positive control. This result is in agreement with the prediction that one of the potential Gcn4 binding sites is located in the region covered by these primers. The second predicted binding site, covered by the primer pair ATG41–8 did not show a strong enrichment. This second site is located almost 800 nucleotides away from the translational start site (compared to approximately 300 nucleotides for the enriched site), which is generally outside the region of yeast promoters, and is therefore unlikely to function in transcriptional control of ATG41. Taken together, these results indicated that Gcn4 binds directly to the promoter region of ATG41 and transcriptionally activates expression of this gene.
Discussion
In this report we identified Atg41 as a novel component of the autophagy machinery in yeast. The absence of Atg41 results in a severe defect in autophagy activity due to a decreased rate of autophagosome formation. Icy1, a paralog of Atg41/Icy2, has low sequence similarity to Atg41/Icy2, which likely explains why Icy1 does not appear to play a role in autophagy. Many of the Atg proteins are upregulated during starvation, but only Atg8 and Atg41 display a greater than 10-fold increase in protein level. Both Atg8 and Atg41 are relatively small proteins, and Atg8 functions in phagophore expansion; lower levels of Atg8 correspond to smaller autophagosomes.21 In contrast, Atg41 does not appear to play a role in determining the size of autophagosomes, but rather influences their formation rate, similar to Atg9.17 Along these lines, the localization of Atg41 shows a peri-mitochondrial distribution similar to Atg9. Furthermore, our results showed an association between Atg9 and Atg41. Considering all of these data, we propose that Atg41 is part of the Atg9 complex and is involved in the delivery of donor membrane to the expanding phagophore.
We also identified Gcn4 as a positive regulator of autophagy in agreement with previous studies.31 ATG41 mRNA levels only increased approximately 8-fold in the gcn4Δ strain compared to the nearly 60-fold increase seen in the wild type, indicating that Gcn4 plays a primary role in transcriptional regulation of ATG41. Although ATG41 transcripts increased at 6- to 7-fold higher levels in the wild type compared to the gcn4Δ strain, the protein levels only increased approximately 2-fold. This difference may reflect the role of other potential positive regulators, or translational silencing. The gcn4Δ strain displayed an approximately 50% decrease in autophagy activity, which further supports our hypothesis that a large increase in the amount of the Atg41 protein is required for normal autophagy activity. Cells do not maintain elevated levels of autophagy indefinitely, although the mechanisms involved in downregulating autophagy under inducing conditions are not fully understood. For example, at present we do not know what controls the turnover of Atg41. We have not detected this protein within the vacuole (data not shown), suggesting that it may be degraded by the proteasome. Expression from the COF1 promoter is predicted to be constant under growing and starvation conditions, yet we observed a clear decrease in Atg41-GFP when expressed at low levels (Fig. 3A), which may correspond to degradation of the protein. Future studies may provide further information on the mechanism(s) involved in regulating the expression and stability of Atg41 and in determining its role in autophagy.
Although homologs of Atg41/Icy2 are found in different fungal species, no homolog is reported in higher eukaryotes based on sequence analysis. Even though many Atg proteins are conserved from yeast to mammals, a number of Atg proteins in yeast lack the corresponding homologs in mammals; however, in many cases the potential functional counterparts of these Atg proteins in mammals exist. One example is seen with Atg17 and RB1CC1, the latter of which is considered as the mammalian counterpart. Although RB1CC1 is much larger than Atg17 (1594 amino acids compared to 417 amino acids), both proteins bind to the Atg1/ULK1 kinase and play important roles in autophagy induction.32 In yeast, Atg17, Atg29 and Atg31 form a stable complex, whereas RB1CC1 can interact with ULK1 alone. It is thus possible in this case that in mammals one protein combines the functions of several yeast proteins. We speculate that Atg41 also falls into this category. That is, it is possible that a functional counterpart exists in mammals, and this protein either functions similarly to Atg41 or combines the functions of Atg41 and at least one other yeast protein.
Materials and Methods
Yeast strains, media and growth conditions
Yeast strains used in this paper are listed in Table S2. Yeast cells are grown in rich medium (YPD: 1% [wt/vol] yeast extract [ForMedium, YEM04], 2% [wt/vol] peptone [ForMedium, PEP04], 2% [wt/vol] glucose; or SMD: 0.67% yeast nitrogen base [ForMedium, CYN0410], 2% [wt/vol] glucose, 0.5% casamino acids [ForMedium, CAS03], 1× complete amino acids and vitamins supplement (see Table S3 for details), 2% glucose). Autophagy was induced by shifting mid-log phase cells from rich medium to nitrogen starvation medium (SD-N: 0.17% yeast nitrogen base without ammonium sulfate or amino acids [ForMedium,CYN0501], 2% [wt/vol] glucose) for the indicated times.
Antibodies and antisera
Anti-YFP antibody, which detects GFP, was from Clontech (JL-8; 63281), and anti-Pgk1 was a generous gift from Dr. Jeremy Thorner (University of California, Berkeley). Antibodies to Ape1 and Atg9 were described previously.33,34 The protein A tag was detected with a nonspecific commercial antibody that is no longer available.
Plasmids
pRS405-GFP-ATG8 contains the GFP-ATG8 open reading frame with the endogenous ATG8 promoter. This plasmid was linearized and integrated into the corresponding strains listed in Table S2. pRS414-ATG41 contains 1,000 nucleotides of the ATG41 5′ UTR, the ATG41 open reading frame and 500 nucleotides of the 3′ UTR. pRS414-ATG1-ts contains the ATG1 open reading frame with the endogenous ATG1 promoter, and L88H, F112L and S158P mutations that confer a temperature-sensitive phenotype.35
Fluorescence microscopy
Yeast cells were grown to mid-log phase in rich medium and then shifted to SD-N for autophagy induction. Cells were then examined by microscopy (Olympus; Delta Vision, GE Healthcare Life Sciences, Pittsburgh, PA, USA) using a 100× objective, and a CCD camera (CoolSnap HQ; Photometrics, Tucson, AZ, USA). Twelve-image stacks were taken for each picture with a 0.3-μm distance between 2 images.
Mitochondria staining
MitoTracker Red CMXRos (Molecular Probes/ThermoFisher Scientific, M-7512) was dissolved in DMSO. The dye was added to mid-log cells growing in rich medium with a final working concentration of 200 nM and a final DMSO concentration of 0.02%. After approximately 45 min, cells were washed and grown in rich medium for another 1 h. Cells were then shifted to SD-N for autophagy induction and microscopy analysis.
Real-time quantitative PCR
Yeast cells were cultured in YPD to mid-log phase and then shifted to SD-N for autophagy induction. Cells were then collected and frozen in liquid nitrogen. Total RNA was extracted using an RNA extraction kit (NucleoSpin RNA II; Clontech, 740955.50). Reverse transcription was carried out using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems/ThermoFisher Scientific, ILT4368814). RT-qPCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems/ThermoFisher Scientific, ILT4367659). All primer information can be found in Table S4.
Chromatin immunoprecipitation
The protocol for ChIP was slightly modified from one described previously.36 After induction of autophagy, formaldehyde was added to the cells for DNA-protein cross-linking. Samples were then harvested, lysed and chromatin was isolated. DNA was then sheared by sonication and the sheared chromatin was immunoprecipitated. The protein-DNA complex was eluted from the beads and reverse cross-linking was performed. DNA was then purified and examined by quantitative PCR. All primer information can be found in Table S4.
Other methods
Western blot, Pho8Δ60, GFP-Atg8 processing, and prApe1 processing assays, and transmission electron microscopy were performed as described previously.13,15,23,37 Gene deletion, truncation or integration was performed based on a standard method.38 Genome tagging and genomic promoter exchange were performed based on a standard method.39
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Dr. Hana Popelka for assistance with the PONDR-FIT analysis.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by NIH grant GM053396 (to DJK) and a University of Michigan–Israel Partnership for Research grant (to DJK).
References
- 1.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science 2004; 306:990-5; PMID:15528435; http://dx.doi.org/ 10.1126/science.1099993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 2010; 190:1005-22; PMID:20855505; http://dx.doi.org/ 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/ 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 2005; 1:101-9.; PMID:16874040; http://dx.doi.org/ 10.4161/auto.1.2.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004; 6:79-90; PMID:14723849; http://dx.doi.org/ 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- 6.Yen WL, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell 2007; 18:581-93; PMID:17135291; http://dx.doi.org/ 10.1091/mbc.E06-07-0612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 1993; 333:169-74; PMID:8224160; http://dx.doi.org/ 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- 8.Jones DL, Petty J, Hoyle DC, Hayes A, Oliver SG, Riba-Garcia I, Gaskell SJ, Stateva L. Genome-wide analysis of the effects of heat shock on a Saccharomyces cerevisiae mutant with a constitutively activated cAMP-dependent pathway. Comp Funct Genomics 2004; 5:419-31; PMID:18629174; http://dx.doi.org/ 10.1002/cfg.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn KM, DeRisi JL, Brown PO, Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol Cell Biol 2001; 21:916-27; PMID:11154278; http://dx.doi.org/ 10.1128/MCB.21.3.916-927.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature 2003; 425:859-64; PMID:14574415; http://dx.doi.org/ 10.1038/nature02062 [DOI] [PubMed] [Google Scholar]
- 11.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 2005; 15:1456-61; PMID:16169922; http://dx.doi.org/ 10.1101/gr.3672305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, et al.. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell 2009; 20:4730-8; PMID:19793921; http://dx.doi.org/ 10.1091/mbc.E09-03-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem 2004; 279:29889-94; PMID:15138258; http://dx.doi.org/ 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. Embo J 1989; 8:2241-50; PMID:2676517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol 2008; 451:33-42; PMID:19185711; http://dx.doi.org/ 10.1016/S0076-6879(08)03203-5 [DOI] [PubMed] [Google Scholar]
- 16.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al.. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A 2012; 109:11206-10; PMID:22733735; http://dx.doi.org/ 10.1073/pnas.1200313109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol 2014; 24:1314-22; PMID:24881874; http://dx.doi.org/ 10.1016/j.cub.2014.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard A, Jin M, Gonzalez-Rodriguez P, Fullgrabe J, Delorme-Axford E, Backues SK, Joseph B, Klionsky DJ. Rph1/KDM4 Mediates Nutrient-Limitation Signaling that Leads to the Transcriptional Induction of Autophagy. Curr Biol 2015; 25(5):546-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett 2010; 584:1359-66; PMID:20146925; http://dx.doi.org/ 10.1016/j.febslet.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang YA, Sanalkumar R, O'Geen H, Linnemann AK, Chang CJ, Bouhassira EE, Farnham PJ, Keles S, Bresnick EH. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol 2012; 32:226-39; PMID:22025678; http://dx.doi.org/ 10.1128/MCB.06166-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 2008; 19:3290-8; PMID:18508918; http://dx.doi.org/ 10.1091/mbc.E07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin M, Klionsky DJ. Transcriptional regulation of ATG9 by the Pho23-Rpd3 complex modulates the frequency of autophagosome formation. Autophagy 2014; 10:1681-2; PMID:25046109; http://dx.doi.org/ 10.4161/auto.29641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Backues SK, Chen D, Ruan J, Xie Z, Klionsky DJ. Estimating the size and number of autophagic bodies by electron microscopy. Autophagy 2014; 10:155-64; PMID:24270884; http://dx.doi.org/ 10.4161/auto.26856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102-9; PMID:17909521; http://dx.doi.org/ 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- 25.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 2007; 24:767-75; PMID:17534848; http://dx.doi.org/ 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- 26.Popelka H, Uversky VN, Klionsky DJ. Identification of Atg3 as an intrinsically disordered polypeptide yields insights into the molecular dynamics of autophagy-related proteins in yeast. Autophagy 2014; 10:1093-104; PMID:24879155; http://dx.doi.org/ 10.4161/auto.28616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei Y, Su M, Soni G, Salem S, Colbert CL, Sinha SC. Intrinsically disordered regions in autophagy proteins. Proteins 2014; 82:565-78; PMID:24115198; http://dx.doi.org/ 10.1002/prot.24424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 2010; 1804:996-1010; PMID:20100603; http://dx.doi.org/ 10.1016/j.bbapap.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 2001; 21:4347-68; PMID:11390663; http://dx.doi.org/ 10.1128/MCB.21.13.4347-4368.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fedorova AV, Chan IS, Shin JA. The GCN4 bZIP can bind to noncognate gene regulatory sequences. Biochim Biophys Acta 2006; 1764:1252-9; PMID:16784907; http://dx.doi.org/ 10.1016/j.bbapap.2006.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z, Geng J, Yen WL, Wang K, Klionsky DJ. Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell 2010; 38:250-64; PMID:20417603; http://dx.doi.org/ 10.1016/j.molcel.2010.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy 2009; 5:85-7; PMID:18981720; http://dx.doi.org/ 10.4161/auto.5.1.7180 [DOI] [PubMed] [Google Scholar]
- 33.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol 1992; 119:287-99; PMID:1400574; http://dx.doi.org/ 10.1083/jcb.119.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 2000; 148:465-80; PMID:10662773; http://dx.doi.org/ 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20:5971-81; PMID:11689437; http://dx.doi.org/ 10.1093/emboj/20.21.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger SL. Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev 2010; 24:1772-86; PMID:20713519; http://dx.doi.org/ 10.1101/gad.1954910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 2002; 30:e23; PMID:11884642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953-61; PMID:9717241; http://dx.doi.org/ 10.1002/(SICI)1097-0061(199807)14:10%3c953::AID-YEA293%3e3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.