Abstract
The Atg8/LC3/GABARAP family of proteins, a group that has structural homology with ubiquitin, connects with a large set of binding partners to function in macroautophagy (hereafter autophagy). This interaction occurs primarily via a conserved motif termed the LC3-interacting region (LIR), or the Atg8-interacting motif (AIM). The consensus sequence for this motif, [W/F/Y]xx[L/I/V], can be found in many proteins, but only some of them are physiological partners containing a functional LIR/AIM. Because the structure of many full-length partners has not been, or cannot be, solved, the structural context of the LIR/AIM within the native protein conformation is not obvious. Here we suggest that the functional LIR/AIM is a short linear motif (SLiM) protein-binding module, arising from an intrinsically disordered region. This finding enables the rapid elimination of some false Atg8/LC3/GABARAP-binding proteins, and connects the exponentially growing knowledge on disordered SLiMs with autophagy.
Keywords: AIM motif, Atg8, GABARAP, intrinsic disorder, LC3, LIR motif, SLiM
Abbreviations
- AIM
Atg8-interacting motif
- IDPR
intrinsically disordered protein region
- LIR
LC3-interacting region
- SLiM
short linear motif
Introduction
Both specific and nonspecific types of autophagy rely on the sequestration of cargo within the double-membrane phagophore, which subsequently matures into an autophagosome. This process is mediated by cargo receptors that interact with the Atg8/LC3/GABARAP family of proteins and thereby link the cargo with the autophagy machinery.1 The cargo receptors mediate this specific interaction via the LC3-interacting region, a term typically used to refer to the sequence in higher eukaryotes, or an Atg8-interacting motif, which is the term used in yeast. The LIR/AIM has the consensus sequence [W/F/Y]xx[L/I/V], where “x” stands for any amino acid, and is often flanked by diverse sequences containing Ser, Thr and/or the negatively charged residues Glu and/or Asp.2-4 This consensus sequence is also utilized by some autophagy proteins that are not autophagy receptors (e.g., Atg1, Atg3, ULK1), but function in autophagy via interaction with the Atg8/LC3/GABARAP family. The known exceptions from the consensus sequence are the CALCOCO2/NDP52 and TAX1BP1 proteins that have the atypical LIR motif sequence LVV,3 and Atg7 with Phe, Ile and Ala in an atypical AIM-like sequence spanning 12 amino acids.5
The general mechanism of binding via the LIR/AIM motif is that an aromatic residue (W/Y/F) and a hydrophobic residue (L/I/V) bind in 2 hydrophobic pockets formed by the ubiquitin-like fold of the Atg8/LC3/GABARAP proteins. Thus, the LIR/AIM motif ensures the specificity of the interaction between Atg8/LC3/GABAPAP and their physiological binding partners. The consensus sequence [W/F/Y]xx[L/I/V] can be found in many proteins, even in those that do not interact with the Atg8/LC3/GABARAP family in the cell. For example, in the yeast Saccharomyces cerevisiae, Atg2 has 22 putative AIM motifs predicted by the ELM algorithm (http://elm.eu.org),6 and Atg5, Atg31 and Atg20 have 6, 2, and 5 putative AIM motifs, respectively. Therefore, a simple match of the [W/F/Y]xx[L/I/V] pattern in the protein sequence by the ELM algorithm does not necessarily indicate the presence of a functional LIR/AIM motif; although the ELM algorithm deserves credit for the incorporation of filters that include taxonomy, cellular compartment, evolutionary conservation and structural features, the latter filters out SMART/PFAM globular domains. This leads to the question of whether the functional LIR/AIM motif has a certain structural association in the native conformation of the physiological binding partner, and can thereby be identified using an additional criterion. The structural context of the functional LIR/AIM is often hidden in the unsolved structure of the full-length protein. Analysis presented in this work reveals a structural feature common to all peptides that bear a functional LIR/AIM motif, and we discuss useful aspects of this finding.
Results and Discussion
The significance of the interaction between the Atg8/LC3/GABARAP family proteins and their physiological binding partners in autophagy has encouraged a number of studies to solve the crystal structure of the corresponding complexes. The structures where Atg19 and Atg32 interact with Atg8 show that the AIM motif of the receptor provides a β strand and forms an intermolecular β sheet with the β2 sheet of Atg8 (Fig. 1). In comparison, the structure of the Atg3–Atg8 complex from Plasmodium falciparum reveals that the Atg3-AIM binds to Atg8 in an isolated β-bridge (single pair β sheet hydrogen bond formation) conformation (Fig. 1). Analysis of the order/disorder propensity of amino acid sequences corresponding to these 3 Atg8-binding partners by the PONDR-FIT algorithm7,8 shows a high flexibility of the AIM-bearing peptides. This indicates that the AIM along with its flanking sequence in each of these Atg8-binding proteins is located within an intrinsically disordered protein region (IDPR) (Fig. 1, right column) that exhibits a characteristic PONDR-FIT profile where stretches of 30 or more amino acid residues have high scores (near or above 0.5). Along these lines, it is important to note that a PONDR-FIT score of a particular amino acid residue needs to be evaluated in the context of its surrounding sequence. This means that a lower-score residue flanking or in a high-score region is still part of an IDPR, and, vice versa, a residue with a score near 0.5 in a low-score region is part of a folded domain. To facilitate the interpretation of the PONDR-FIT results, we present a schematic representation using dark-gray round rectangles and bold lines above each profile.
Figure 1.
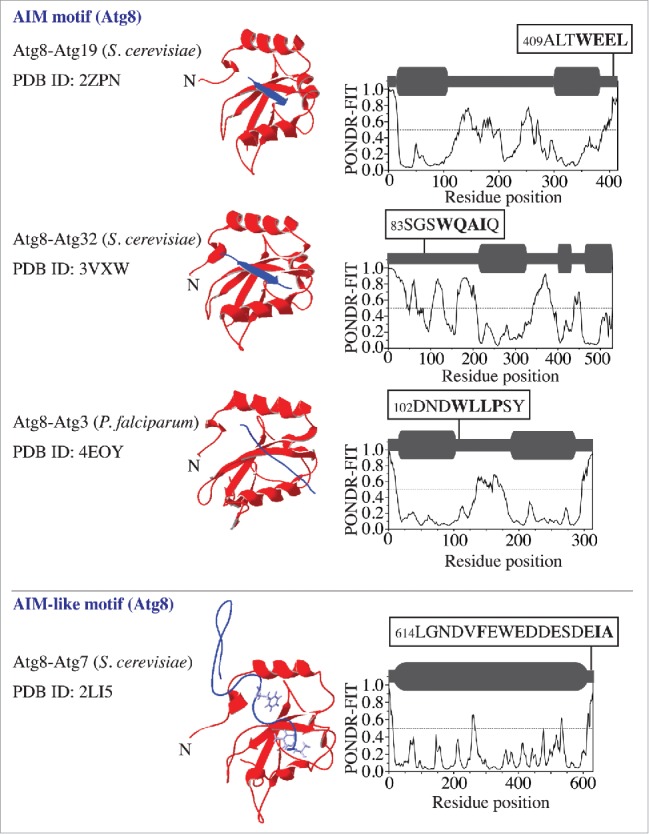
Atg8 in a complex with its binding proteins (Atg19, Atg32, and Atg3) that contain an AIM motif, and with Atg7, which contains an AIM-like motif. The left column shows the name of the complex, the organism in parentheses, and the Protein Data Bank identification (PDB ID) number. The middle column shows 3D structures, where the Atg8 ribbon structure is depicted in red and the peptide containing the AIM or AIM-like motif in the binding partner is in purple. The right column shows the PONDR-FIT score of the corresponding full-length partner protein. The schematic representation of each PONDR-FIT profile is shown in dark gray above the graph; structurally stable domains are represented by round rectangles and IDPRs by bold lines. The AIM/AIM-like motif (marked in bold) along with its flanking sequence is highlighted in black boxes at the corresponding position in the amino acid sequence.
Consistent with the finding for the AIM motif (Fig. 1), the AIM-like motif at the Atg7 C terminus, which is required for interaction with, and activation of, Atg85 is also located within the primary intrinsically disordered region (Fig. 1). In this case, Phe619 and Ile629 in Atg7, which bind in the W and L pockets of Atg8, map onto the sequence spanning more than 10 residues. This sequence does not form an intermolecular β sheet that is seen in a true AIM, but stays in an extended conformation (Fig. 1).
In contrast to Atg3, Atg7, Atg19, and Atg32, there are proteins in the autophagy core machinery that have a structure and/or autophagy function irreconcilable with Atg8 interaction, and that contain false AIM motifs predicted by the ELM algorithm. To demonstrate a few examples and to illustrate how PONDR-FIT eliminates false putative AIM motifs, we show the PONDR-FIT results for Atg12, Atg17 and Atg27 along with the crystal structures of Atg12 and Atg17 (Fig. 2). Atg12, a component of the Atg12–Atg5-Atg16 complex, has 2 false AIM motifs in the α helix and β sheet of the ubiquitin-like folded domain. These 2 motifs have a very low PONDR-FIT score. Atg17, the scaffold protein in the Atg17-Atg31-Atg29 trimer involved in autophagy initiation, from S. cerevisiae carries 4 false AIM motifs. The first 3 of these motifs have homologs in L. thermotolerans that are located in α helical rods of the structured crescent. Again, all 4 motifs in S. cerevisiae Atg17 have a very low PONDR-FIT score. Finally, Atg27, the integral membrane protein required for the movement of Atg9 to the phagophore assembly site, has a single false AIM motif, with a low PONDR-FIT score, located in the structured N-terminal domain.
Figure 2.
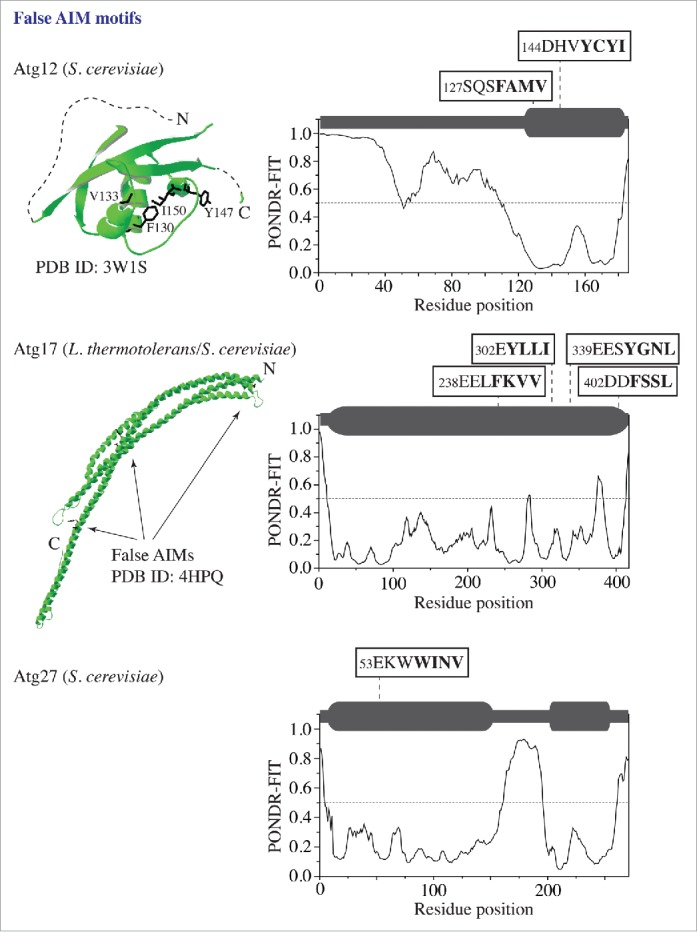
False putative AIM motifs in proteins (Atg12, Atg17, and Atg27) that are not Atg8-interacting partners. The 3D structure (in green) of the ubiquitin-like domain in S. cerevisiae Atg12 and the L. thermotolerans Atg17 crescent were obtained from the PDB database. The PONDR-FIT result for each protein containing the false AIM motif(s) is shown in the right column. For other details, please refer to the Figure 1 legend.
To probe the native conformation of the LIR motif, we analyzed the amino acid sequences of the LIR-containing proteins that have been crystalized in a complex with LC3 or GABARAP. The result is very similar to that observed for the AIM; the LIR motif maps onto a protein domain with a high PONDR-FIT score, which corresponds to an IDPR that forms an intermolecular β sheet upon interaction with LC3 or GABARAP (Fig. 3). For human CALR (calreticulin), the crystal structure of the individual protein (PDB ID:3POW) confirms the position of 3 LIR-motif residues (Trp200, Asp201 and Phe202) in a disordered region. Leu203 is missing in this structure along with the downstream IDPR consisting of 100 residues (Fig. S1, dashed line). This would indicate that the LIR/AIM-bearing peptides localize to IDPRs. In most cases, these flexible regions undergo disorder-to-order transition into a β sheet, which is induced by interaction with Atg8/LC3/GABARAP. To further verify this finding, we tested the sequences of other Atg8/LC3/GABARAP-binding proteins3,9 that were not crystalized in a complex with their ubiquitin-like partner, but for which the amino acid sequences are available in the UniProt database (http://www.uniprot.org/uniprot). The outcome was again the same, the LIR/AIM motif of Atg1, Atg3, Atg30, ATG4B, TBC1D5, BNIP3, FUNDC1, TP53INP1, STBD1, or RB1CC1/FIP200 overlaps with an IDPR detected by a high PONDR-FIT score (Fig. 4). The results in Figures 1, 3 and 4 are in agreement with a recent review showing that the LIR motif of human ULK1 (357FVMV) is located in its disordered serine- and proline-rich domain, and the AIM motif of S. cerevisiae Atg34 and Atg36 is at the disordered C or N terminus, respectively.10 Similarly, these results are also consistent with the recently disclosed AIM motifs in S. cerevisiae Atg39 and Atg40,11 and the LIR motif in human JIP112 (Fig. S2). Taken together, our analysis reveals that the native conformation of the LIR/AIM motif in autophagy proteins is located within an intrinsically disordered protein region.
Figure 3.
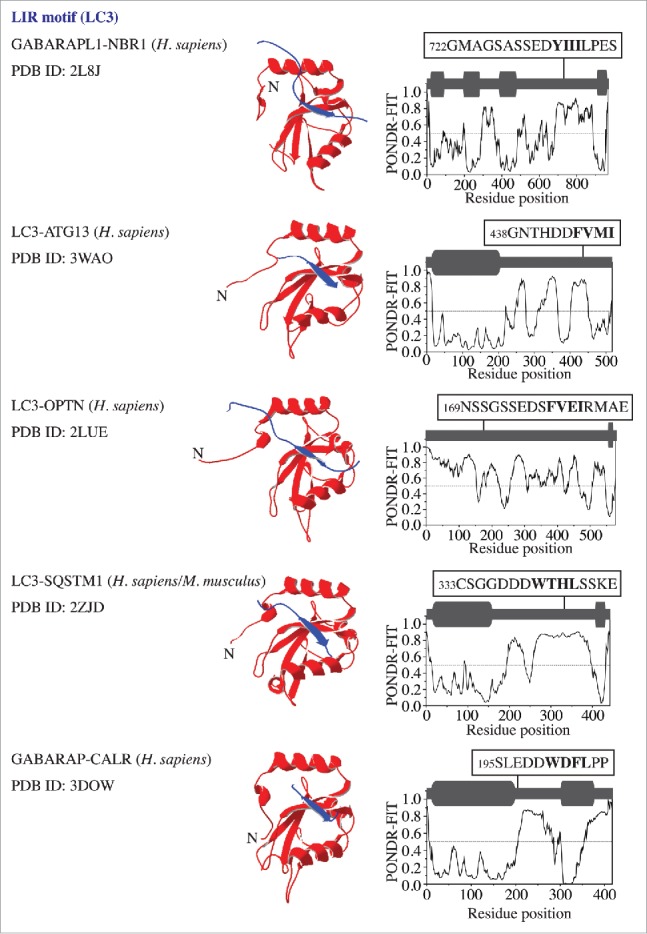
LC3/GABARAP proteins in a complex with binding partners (NBR1, ATG13, OPTN/optineurin, SQSTM1/p62, and CALR) that contain a LIR motif. For more details, please refer to the Figure 1 legend.
Figure 4.
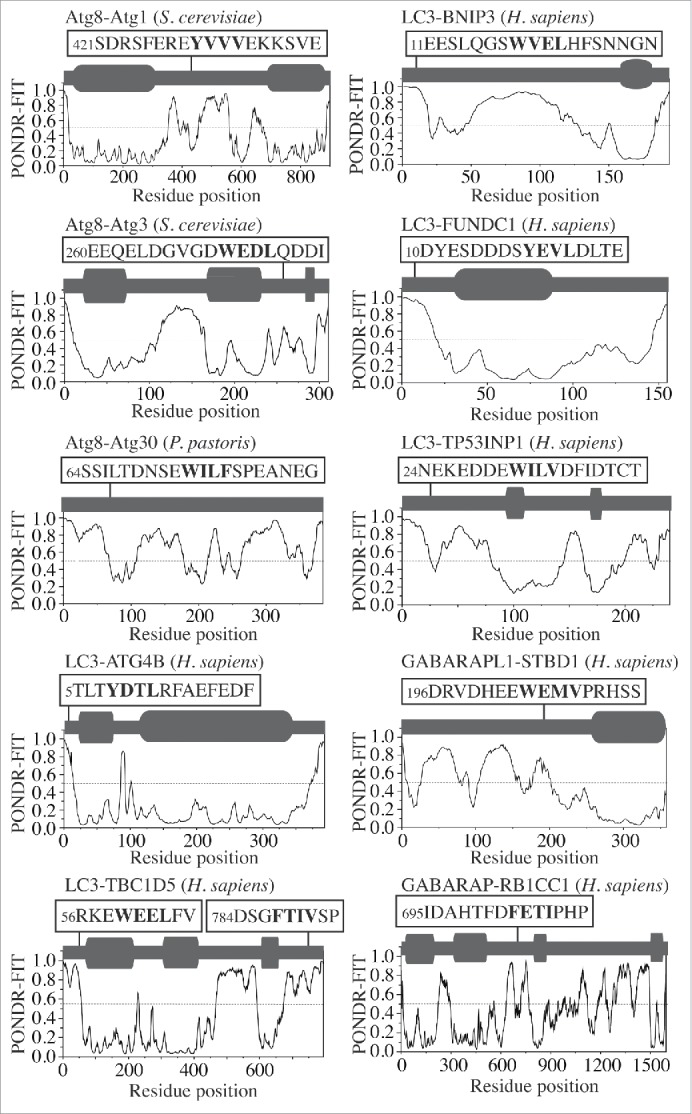
PONDR-FIT profiles of autophagy proteins (Atg1, Atg3, Atg30, ATG4B, TBC1D5, BNIP3, FUNDC1, TP53INP1, STBD1 and RB1CC1/FIP200) that contain an AIM/LIR motif. 3D structures are not available for these complexes. For more details, please refer to the Figure 1 legend.
The LIR/AIM is only 4 amino acids and its association with an IDPR identifies it as a structurally flexible protein interaction module often known as a short linear motif (SLiM). SLiMs are compact (3–10 contiguous amino acids) binding modules of proteins associated with their flexible domains. The majority of SLiMs is found in IDPRs, but they can also be present in flexible loops of globular domains. SLiMs engage in specific, but transient interactions. Because they regulate low-affinity interactions due to their limited binding surface, they function as rapid molecular switches. SLiM function, evolution, regulation, disease association, and therapeutic potential are comprehensively summarized in a recent review.13
Several additional lines of evidence support the identification of a functional LIR/AIM as a typical flexible SLiM. The SLiM or its flanking sequence is often posttranslationally modified.14 Indeed, posttranslational modification of the LIR/AIM motif or its flanking sequence was reported for several Atg8/LC3/GABARAP-binding proteins. For example, phosphorylation of BNIP3 on Ser17 and Ser24 flanking the LIR motif directly promotes its binding to LC3B and GABARAPL2.15 Phosphorylation of OPTN/optineurin by TBK1 (TANK-binding kinase 1) on Ser177, directly upstream of the LIR motif, enhances its binding to LC3 and thereby autophagy.16 Similarly, phosphorylation of Atg36 on Ser31 and phosphorylation of Atg32 on Ser81, Ser83 and Ser85 upstream of the AIM motif facilitates their functional binding to Atg8.17 Conversely, phosphorylation of FUNDC1 negatively regulates LIR binding; Tyr18 in the LIR motif is phosphorylated by SRC kinase when the interaction between FUNDC1 and LC3 needs to be inhibited under physiological (nonhypoxia) conditions.18 Intrinsic disorder is in general important for protein phosphorylation,19 because it allows for easy access of a kinase to its substrate. Thus, the LIR/AIM motif and its flanking sequence apparently take advantage of the plasticity of the disordered region that harbors them.
SLiMs have a specific amino acid composition, which determines their characteristics. For example, they are enriched in certain hydrophobic (Tyr, Phe, Trp, Leu) and charged (Asp, Arg) residues.20 Indeed, these residues form a significant portion of the LIR/AIM motif consensus sequence and its flanking sequence (Figs. 1, 3, and 4).
Structural plasticity enables SLiMs to precisely fit their binding partner, which is often achieved by binding, coupled with folding. SLiMs often undergo a disorder-to-order transition and form α helices or β sheets upon binding;13 a feature similar to what was observed in the solved 3D structures of LIR/AIM-containing complexes (Figs. 1 and 3).
SLiMs and the disordered regions that surround them primarily bind onto the surface of globular domains.13 The LIR/AIM binds on the surface of the ubiquitin-like fold of Atg8/LC3/GABARAP. Identification of the LIR/AIM as a disordered SLiM explains how so many diverse amino acid sequences of various Atg8/LC3/GABARP-binding partners can all accommodate 2 specific (aromatic and hydrophobic) pockets on one globular surface. This type of interaction is recognized as a many-to-one binding scenario, where many IDPRs interact with the same rigid partner, but not at the same time.20
The LIR/AIM as a disordered SLiM represents a fraction from thousands of other experimentally validated SLiMs.13 Another autophagy-related example of this binding mechanism is the well-known SLiM in the binding partners of YWHA/14-3-3 proteins. The YWHA/14-3-3 proteins interact with many different, mostly phosphorylated, protein partners that have diverse cellular functions. An earlier report showed that almost all YWHA/14-3-3 binding sites are inside intrinsically disordered regions.21 Recent autophagy studies link the YWHA/14–3-3 proteins with the autophagy machinery when they revealed that the YWHA/14-3-3 docking sites are created by phosphorylation on Ser234 and Ser295 in human BECN122 and on Ser761 near the C terminus of mammalian ATG9.23 These docking sites are also associated with structurally plastic protein domains (Fig. S3).10
The revelation that the native conformation of a functional LIR/AIM motif in autophagy proteins is within an IDPR, and hence outside of a stably folded protein domain, has useful consequences. One such consequence is that it allows better identification of the functional LIR/AIM motif, and filtering out of false positive matches of the [W/F/Y]xx[L/I/V] pattern recognized by the ELM algorithm. Complementation of the prediction of a LIR/AIM (e.g., by the ELM algorithm) with a prediction of disorder (e.g., by PONDR-FIT) from protein amino acid sequence analysis yields a method for rapid assessment of a protein's putative ability to bind an Atg8/LC3/GABARAP protein. In particular, if this method maps a putative LIR/AIM motif onto an IDPR detected by PONDR-FIT, then the motif is a promising candidate suitable for further experimental validation. Conversely, a putative LIR/AIM mapped on a protein region with a very low PONDR-FIT score for intrinsic disorder yields an unlikely candidate for a functional motif, due to the rigid nature of that region.
The method just described can provide fast preliminary insight into the Atg8/LC3/GABARAP-binding ability of any new autophagy protein that emerges from future studies. This method represents an alternative to the iLIR web source24 that attempts to incorporate a possible disorder of the LIR motif by including the ANCHOR predictor.25,26 The important caveat regarding iLIR is that ANCHOR is not a general disorder prediction method. More importantly, ANCHOR is not designed to find linear motifs; rather, ANCHOR finds disordered binding regions, which are longer than short linear motifs and which bind on the principle of gaining stabilizing energy via folding upon binding to a globular protein. In contrast, short linear motifs utilize per residue binding strength, and only approximately 60% of SLiMs increase affinity of binding by disorder-to-order transition.13 To quote the authors who designed ANCHOR: “…disordered binding sites and sequence specific binding motifs capture different aspects of certain binding regions.”25 As a consequence, ANCHOR in iLIR does not yield a suitable output with respect to intrinsic disorder. Illustrative examples are Atg32, for which iLIR predicts one of 3 extended LIR motifs within a structured intermembrane space region (which is obviously unlikely to bind cytosolic Atg8), or Atg20 that has a putative extended LIR in its globular PX domain that binds phosphatidylinositol-3-phosphate. For Atg19, for example, iLIR finds the experimentally verified functional AIM motif, but generates a negative result from ANCHOR. This is correct with respect to the ANCHOR algorithm in itself (as it is not designed to find short linear motifs), but not with respect to iLIR that uses ANCHOR to find functional LIR/AIM motifs that undergo disorder-to-order transition; Atg19 clearly does undergo this transition (Fig. 1; compare the IDPR in the PONDR-FIT result [right column] with the intermolecular β sheet in the crystal structure [the middle column]).
Another useful aspect of a functional LIR/AIM recognized as an IDPR-based SLiM is that it connects the entire knowledge on this type of structurally plastic binding modules with the field of autophagy. SLiM-associated mutations are linked to various human diseases.13 It is plausible that mutation in a LIR/AIM can have a severe consequence when a specific cargo fails to connect with the autophagy machinery. In analogy with currently known therapeutics affecting SLiM-mediated interactions,13 therapeutics (e.g., inhibitory peptides) that recognize and mimic the LIR/AIM motif of a specific protein or modify its nearby phosphorylable residue might become successful negative regulators of autophagy, for example in cancer cells that utilize autophagy for their own survival.27
Materials and Methods
Modeling
3D structures of autophagy proteins in a complex with Atg8/LC3/GABARAP were created using DeepView/Swiss-PDBViewer v. 4.0.4. available at http://www.expasy.org/spdbv/.28 The published pdb files were downloaded from the PDB database (http://www.rcsb.org/pdb).
Amino acid sequence analysis
Amino acid sequences of autophagy proteins were downloaded from the UniProt database (http://www.uniprot.org/uniprot). The disorder propensity of each sequence was determined by the PONDR-FIT algorithm (http://www.disprot.org/pondr-fit.php)8 available in the DisProt database (http://www.disprot.org/index.php).7
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by National Institutes of Health grant GM053396 to D.J.K. and the Protein Folding Diseases FastForward Initiative, University of Michigan.
References
- 1.Mijaljica D, Nazarko TY, Brumell JH, Huang WP, Komatsu M, Prescott M, Simonsen A, Yamamoto A, Zhang H, Klionsky DJ, et al.. Receptor protein complexes are in control of autophagy. Autophagy 2012; 8:1701-5; PMID:22874568; http://dx.doi.org/ 10.4161/auto.21332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci 2013; 126:3237-47; PMID:23908376; http://dx.doi.org/ 10.1242/jcs.126128 [DOI] [PubMed] [Google Scholar]
- 3.Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 2014; 53:167-78; PMID: 24462201; http://dx.doi.org/ 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 4.Hurley JH, Schulman BA. Atomistic autophagy: The structures of cellular self-digestion. Cell 2014; 157:300-11; PMID:24725401; http://dx.doi.org/ 10.1016/j.cell.2014.01.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noda NN, Satoo K, Fujioka Y, Kumeta H, Ogura K, Nakatogawa H, Ohsumi Y, Inagaki F. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell 2011; 44:462-75; PMID:22055191; http://dx.doi.org/ 10.1016/j.molcel.2011.08.035 [DOI] [PubMed] [Google Scholar]
- 6.Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Kruger D, Grebnev G, Kuban M, et al.. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res 2014; 42:D259-266; PMID:24214962; http://dx.doi.org/ 10.1093/nar/gkt1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, et al.. Disprot: The database of disordered proteins. Nucleic Acids Res 2007; 35:D786-93; PMID:17145717; http://dx.doi.org/ 10.1093/nar/gkl893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. Pondr-fit: A meta-predictor of intrinsically disordered amino acids. BBA-Proteins Proteom 2010; 1804:996-1010; http://dx.doi.org/ 10.1016/j.bbapap.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Overvatn A, Kirkin V, Johansen T. Atg8 family proteins act as scaffolds for assembly of the ULK complex: Sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem 2012; 287:39275-90; PMID:23043107; http://dx.doi.org/ 10.1074/jbc.M112.378109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popelka H, Klionsky DJ. Posttranslationally-modified protein structures in the autophagy machinery: An integrative perspective. FEBS J 2015; 282:3474-88; PMID:26108642; http://dx.doi.org/ 10.1111/febs.13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015; 522:359-62; PMID:26040717; http://dx.doi.org/ 10.1038/nature14506 [DOI] [PubMed] [Google Scholar]
- 12.Fu MM, Nirschl JJ, Holzbaur ELF. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev Cell 2014; 29:577-90; PMID:24914561; http://dx.doi.org/ 10.1016/j.devcel.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Roey K, Uyar B, Weatheritt RJ, Dinkel H, Seiler M, Budd A, Gibson TJ, Davey NE. Short linear motifs: Ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem Rev 2014; 114:6733-78; PMID:24926813; http://dx.doi.org/ 10.1021/cr400585q [DOI] [PubMed] [Google Scholar]
- 14.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al.. Classification of intrinsically disordered regions and proteins. Chem Rev 2014; 114:6589-631; PMID:24773235; http://dx.doi.org/ 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu YY, Massen S, Terenzio M, Lang V, Chen-Lindner S, Eils R, Novak I, Dikic I, Hamacher-Brady A, Brady NR. Modulation of serines 17 and 24 in the LC3-interacting region of BNIP3 determines pro-survival mitophagy versus apoptosis. J Biol Chem 2013; 288:1099-113; PMID:23209295; http://dx.doi.org/ 10.1074/jbc.M112.399345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al.. Phosphorylation of the autophagy receptor optineurin restricts salmonella growth. Science 2011; 333:228-33; PMID:21617041; http://dx.doi.org/ 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep 2013; 14:441-9; PMID:23559066; http://dx.doi.org/ 10.1038/embor.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al.. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 2012; 14:177-85; PMID:22267086; http://dx.doi.org/ 10.1038/ncb2422 [DOI] [PubMed] [Google Scholar]
- 19.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 2004; 32:1037-49; PMID:14960716; http://dx.doi.org/ 10.1093/nar/gkh253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uversky VN. Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Design 2013; 19:4191-213; PMID:23170892; http://dx.doi.org/ 10.2174/1381612811319230005 [DOI] [PubMed] [Google Scholar]
- 21.Bustos DM, Iglesias AA. Intrinsic disorder is a key characteristic in partners that bind 14-3-3 proteins. Proteins 2006; 63:35-42; PMID:16444738; http://dx.doi.org/ 10.1002/prot.20888 [DOI] [PubMed] [Google Scholar]
- 22.Wang RC, Wei YJ, An ZY, Zou ZJ, Xiao GH, Bhagat G, White M, Reichelt J, Levine B. AKT-mediated regulation of autophagy and tumorigenesis through BECLIN1 phosphorylation. Science 2012; 338:956-9; PMID: 23112296; http://dx.doi.org/ 10.1126/science.1225967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weerasekara VK, Panek DJ, Broadbent DG, Mortenson JB, Mathis AD, Logan GN, Prince JT, Thomson DM, Thompson JW, Andersen JL. Metabolic-stress-induced rearrangement of the 14-3-3 zeta interactome promotes autophagy via a ULK1-and AMPK-regulated 14-3-3 zeta interaction with phosphorylated ATG9. Mol Cell Biol 2014; 34:4379-88; PMID:25266655; http://dx.doi.org/ 10.1128/MCB.00740-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalvari I, Tsompanis S, Mulakkal NC, Osgood R, Johansen T, Nezis IP, Promponas VJ. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy 2014; 10:913-25; PMID:24589857; http://dx.doi.org/ 10.4161/auto.28260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meszaros B, Simon I, Dosztanyi Z. Prediction of protein binding regions in disordered proteins. Plos Comput Biol 2009; 5:e1000376; PMID: 19412530; http://dx.doi.org/ 10.1371/journal.pcbi.1000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dosztanyi Z, Meszaros B, Simon I. Anchor: Web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009; 25:2745-6; PMID:19717576; http://dx.doi.org/ 10.1093/bioinformatics/btp518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov 2007; 6:304-12; PMID:17396135; http://dx.doi.org/ 10.1038/nrd2272 [DOI] [PubMed] [Google Scholar]
- 28.Guex N, Peitsch MC. Swiss-model and the swiss-pdbviewer: An environment for comparative protein modeling. Electrophoresis 1997; 18:2714-23; PMID:9504803; http://dx.doi.org/ 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


