Abstract
The major histocompatibility complex (MHC) has been known to play a critical role in immune recognition since the 1950s. It was a surprise, then, in the 1970s when the first report appeared indicating MHC might also function in social signaling and in mate choice. Since this seminal discovery, MHC signaling has been found throughout vertebrates and its known functions have expanded beyond mate choice to include a suite of behaviors from kin-biased cooperation, parent-progeny recognition to pregnancy block. The widespread occurrence of MHC in social signaling has revealed conserved behavioral-genetic mechanisms that span vertebrates and includes humans. The identity of the signal’s chemical constituents and the receptors responsible for the perception of the signal have remained elusive, but recent advances have enabled the identification of the key components of the behavioral circuit. In this chapter we organize recent findings from the literature and discuss them in relation to four non-mutually exclusive models wherein MHC functions as a signal of (i) individuality, (ii) relatedness, (iii) genetic compatibility and (iv) quality. We also synthesize current mechanistic studies, showing how knowledge about the molecular basis of MHC signaling can lead to elegant and informative experimental manipulations. Finally, we discuss current evidence relating to the primordial functions of the MHC, including the possibility that its role in social signaling may be ancestral to its central role in adaptive immunity.
MHC-based social signaling
MHC (also known as HLA in humans and H-2 in mice) signaling mediates both immune recognition during the adaptive immune response (discussed in the previous chapter), and social signaling that enhances both the recognition of optimal mates and kin-biased behaviors1. Social signaling meditated by the MHC was first discovered in regards to mate preferences in laboratory mice (Mus musculus)2, a full three decades after the histocompatibility functions were described by George Snell3. Thirty years later, social signaling via MHC has been described throughout vertebrates including mammals, birds, reptiles, amphibians, and teleost fish (see Table 1). MHC social signaling has been identified in over 20 species of vertebrates and is likely the basis for a vertebrate-wide chemosensory communication system. The original observation of MHC disassortative mating preferences seems to be common, but not omnipresent in vertebrates4; it by no means is the only behavior facilitated by MHC, nor is it the only type of observed MHC-based mate preference. In addition to MHC-mediated mating preferences, MHC signaling also facilitates cooperative behavior with kin, parent-progeny recognition and pregnancy block. In the following sections we will present the current evidence for MHC as a signal of relatedness, individuality, genetic compatibility and quality. MHC-mediated behaviors are diverse, and though general patterns exist within vertebrates, the exact function of MHC-based social signaling will be species specific and highly context-dependant.
Table 1.
Summary of studies investigating MHC-genotype signaling in social communication.
| Species | MHC-based mate preference |
MHC-Mediated Cooperative behavior |
Phenotype matching system |
Sources |
|---|---|---|---|---|
| Mammals | ||||
| House Mice (Mus musculus) |
MHC disassortative |
Female Communal Nesting |
Familial imprinting |
Yamazaki et al. 19762, 198820, 200721; Manning et al. 199222; |
| Bank voles (Clethrionomys glareolus) |
MHC disassortative |
Unknown | Unknown | Radwan et al. 200823 |
| Malagasy giant jumping rat (Hypogeomys antimena) |
MHC assortative |
Unknown | Unknown | Sommer 200524 |
| Humans (Homo sapiens) |
MHC disassortative |
Unknown | Unknown | Wedekind et al. 199525; Havlicek & Roberts 200826 |
| Mandrill (Mandrillus sphinx) |
MHC disassortative |
Unknown | Unknown | Setchell et al. 200927 |
| Fat-tailed dwarf lemur (Cheirogaleus medius) |
MHC supertype- disassortative and maximal diversity |
Unknown | Unknown | Schwensow et al. 200828 |
| Grey mouse lemur (Microcebus murinus) |
MHC disassortative (cryptic) |
Unknown | Unknown | Schwensow et al. 200829 |
| Domestic sheep (Ovis aries) |
No MHC preference |
Paterson & Pemberton 199730 | ||
| Birds | ||||
| Savannah sparrows (Passerculus sandwichensis) |
MHC disassortative |
Unknown | Unknown | Freeman-Galant et al. 200331 |
| House Sparrow (Passer domesticus) |
MHC assortative and optimal diversity |
Unknown | Unknown | Bonneaud et al. 200632 |
| Seychelles warbler (Acrocephalus sechellensis) |
MCH maximal diversity |
Unknown | Richardson et al. 200533 | |
| Great reed warbler (Acrosephalus arundinaceus) |
No MHC preference |
H. Westerdahl 200434 | ||
| Red jungle Fowl (Gallus gallus) |
MHC disassortative (cryptic) |
Unknown | Unknown | Gillingham et al. 200935 |
| Peafowel (Pavo cristatus) |
MHC maximal diversity (cryptic) |
Unknown | Hale et al. 200936 | |
| Reptiles | ||||
| Sand lizards (Lacerta agilis) |
MHC disassortative |
Unknown | Unknown | Olsson et al. 200337 |
| Tuatara (Sphenodon punctatus) |
MHC disassortative |
Kin avoidance during territory acquisition |
Unknown | Miller et al. 200938 |
| Amphibians | ||||
| Afriacn clawed Frog (Xenopus laevis) |
Unknown | Tadpole schooling |
Self-referent | Villinger & Waldman 200839 |
| Tiger Salamanders (Ambystoma tigrinum) |
MHC assortative |
Unknown | Unknown | Bos et al. 200940 |
| Fish | ||||
| Zebrafish (Danio rerio) |
Unknown | Unknown but kin groups grow faster than non- kin groups |
Familial imprinting |
Gerlach et al. 200741 & 200842 |
| Three-spined stickle back (Gasterosteus aculeatus) |
Optimal MHC diversity |
Unknown | Self-referent | Aeschlimann et al. 200343; Reusch et al. 200144; Milinski 200645 |
| Atlantic Salmon (Salmo salar) |
MHC disassortative |
Schooling with kin |
Self-referent | Rajakaruna et al. 200646; Consuegra & de Leaniz 200847 |
| Chinook salmon (Oncorhynchus tshawytscha) |
MHC disassortative |
Unknown | Unknown | Neff et al. 200848 |
| Arctic Char (Salvelinus alpinus) |
Unknown | Schooling with kin |
Self-referent | Olsen et al. 200249 |
| Brown Trout (Salmo trutta L) |
Optimal MHC diversity |
Unknown | Self-referent | Forsberg et al. 200750 |
| Brook Trout (Salvelinus fontinalis) |
Unknown | Schooling with kin |
Self-referent | Rajakaruna et al. 200646 |
| Whitefish (Coregonus sp.) |
No MHC preference (cryptic) |
Wedekind et al. 200451 |
Signaling of MHC-genotype: molecular mechanisms
For three decades after the discovery of MHC-mediated social singling in laboratory mice2, the actual mechanism of how MHC genotype was ascertained in conspecifics remained a mystery. Early on it was discovered that MHC genotype could be discriminated by chemical cues detected by the olfactory system. These studies showed that mice could discriminate MHC odortypes either through training5 or in the absence of training6. However, the nature of the signaling odorants remained elusive. This mystery was at least partially solved by the discovery that peptides known to bind MHC molecules also bound receptors in the vomeronasal organ (VNO)7. It was later shown that a similar process was working in the olfactory epithelium8.
The critical role of MHC-presented peptides during adaptive immune recognition is well established9. MHC-bound peptides are presented at the cell surface for interrogation by T lymphocytes; when the peptides are of foreign origin (e.g. from a pathogen) an immune response is initiated. The majority of MHC alleles encode unique structural aspects of the peptide binding region of the molecule, and these variants provide great specificity in the peptides they present. Because there is physical correspondence between MHC allelic variants and the amino acid sequence of their bound peptides, it was hypothesized that MHC peptides could serve as ligands for odorant receptors that had similar binding specificity, thus allowing information about MHC genotype to be conveyed. Physiological recordings from vomeronasal sensory neurons stimulated with synthetic peptides proved this to be the case7.
Detection of peptides in the olfactory system
The olfactory system of mammals is anatomically divided into two regions: the main olfactory epithelia (MOE) and the vomeronasal organ (VNO). Traditionally these two organs were viewed as functioning in largely non-overlapping modalities with the VNO being specialized for detection of nonvolatile small molecules, and proteins that typically signaled the sexual and social status of conspecifics (pheromones), while the MOE was thought to specialize as a general detection system for volatile substances.
The initial experiments to determine if the olfactory system was capable of detecting peptides was conducted in the VNO of mice. Leinders-Zufall and coworkers (2004) tested the hypothesis that dissociated MHC class I peptides (rather than MHC-peptide complexes, MHC molecules, or their volatile metabolites) could be detected in the VNO Two peptides known to be presented either by the H-2Db haplotype (AAPDNRETF) or H-2Kd haplotype (SYFPEITHI) were synthesized. These peptides were applied individually to ex-vivo preparations of mouse VNO. Both peptides activated a relatively specific subset of V2R–positive neurons in the basal zone of the VNO as revealed by extracellular field potential recordings and fluorescence imaging. The vomeronasal sensory neurons (VSNs) responded with high sensitivity at concentrations down to 10−12M.
As predicted by the hypothesis that peptides can signal MHC genotype, the peptide binding by the VSNs responded in an MHC allele-specific manner. Not only was the VSN response specific to the amino acid sequence of each peptide, but the pattern of specificity mimicked the binding properties of MHC molecules. Amino acid substitutions (underlined) at non-anchor positions (e.g. SYIPSAEKI) usually continued to stimulate the same neurons. In contrast, substitution of peptide anchor residues (underlined) with alanine (e.g. AAPDARETA or SAFPEITHA) abolished stimulation of these neurons. These VSN binding properties provide a neurophysiological basis for identifying the MHC genotype of individuals, because peptides are reverse-image “molds” of the antigen-binding site of MHC molecules. Thus, sensory receptors that detect peptides in an MHC-like fashion could in principle function as an MHC genotyping system10. These results point to the structural importance of peptide anchor residues in binding VSN receptors and, given the similar binding properties of MHC molecules, reveal the convergent ligand-binding properties of these unrelated molecules.
The same lab group applied the same hypotheses to the MOE sensory neurons, traditionally viewed as generalist receptors of volatile chemosignals8. Contrary to conventional wisdom, they discovered that non-volatile, fluorescent tagged MHC peptides gain access to the MOE without direct nasal contact to the peptide containing fluid. Most importantly, these peptides activate neurons at subnanomolar concentrations in an allele specific fashion, similar to the patterns found in the VNO. There were, however, some important physiological differences in peptide detection between the two olfactory organs11. First, a different transduction mechanism is used in the MOE during recognition of peptides12. Second, when anchor residues are substituted with alanine (eg. AAPDARETA and SAFPEITHA), olfactory sensory neurons (OSNs) cease firing at normal stimulation concentrations, but firing resumes at higher concentrations. Third, MOE-dependent peptide recognition does not induce pregnancy block13, despite normal MHC odor (mating) preferences. These experiments show that discrimination of MHC genotype by the two olfactory systems is achieved with separate neurological, physiological and behavioral response pathways.
If peptides are the odorants whereby MHC genotype is discriminated, then experimental manipulation of peptides should alter behavioral responses in an MHC-like fashion. The above series of elegant experiments shows that both the VNO and MOE can detect peptides in an MHC-like fashion, providing a mechanistic basis for the discrimination of MHC odortypes. These findings stimulated research confirming that both mice and stickleback fish (Gasterosteus aculeatus) behavior is manipulated by the addition of peptides. The experimental addition of peptides to an MHC similar odor source causes animals to respond as if it were an MHC dissimilar odor source for both mating- and odor-preferences8, 14 and pregnancy block7.
Signaling of MHC-genotype without peptides
Due to the general non-volatility of peptides15,16, the question has remained whether peptides can explain all of the observed MHC-mediated behavioral patterns. This question was recently addressed by experimentally removing all of the peptide components from the urine of two MHC-congenic strains of mice. Mice that had been trained to discriminate between the urine odors of these two strains could continue to discriminate using the peptide-free urine17. These results suggest that non-peptide volatile odorants provide signals conveying MHC genotype information. However, odor-training experiments can introduce confounding behavioral artifacts18 and this result should be confirmed in a paradigm that does not use training. If these results are confirmed, making yet a third independent mechanism for identifying MHC genotype, it underscores the functional importance of this olfactory ability and the importance of the associated behavioral responses.
Though it has been shown that peptides signal MHC genotype in mammals (mice) and fish (sticklebacks), the utilization of peptides in other vertebrates is undocumented. It has been questioned weather olfaction can explain MHC-mediated behavior in birds whose olfactory prowess has long been questioned19. No other mechanisms have been as thoroughly tested as peptide and volatile olfaction signaling of MHC genotype, and more work is needed to test whether these mechanisms drive MHC mediated behavior in other taxa.
Sources are limited to first reports and reviews. Blank boxes indicate no finding would be expected given the observed result.
MHC as a signal in individual recognition
Individual recognition is an important component of social behavior. Traits that specifically signal individual identity are predicted to be genetically determined, highly variable, cheap to produce (i.e. not condition-dependent), and signal variants are expected to have equal fitness at equilibrium (reviewed in Tibbetts and Dale 200752). MHC is an ideal candidate gene for understanding the mechanistic bases of individual recognition because it is a genetically determined trait associated with social behavior and is extremely variable (there are 109 MHC phenotypes in mice53). MHC was hypothesized to contribute to individual recognition as early as 197554. Since then, the concept of individual recognition has been invoked in many studies addressing MHC-associated cues in social signaling (e.g. Ref.17). However, many authors tacitly use different definitions of this term, and do not distinguish between individual recognition in the strict sense52, and other forms of social recognition, which can include discrimination of familiar vs. unfamiliar conspecifics, kin vs. non-kin, same-genotype vs. different-genotype and potential mates. We define individual recognition as being characterized by individual specificity in three elements of social communication: signaling; signal perception and template matching by the signal receiver; and a functional response by the receiver55. This definition includes any case where receivers have a template of a specific individual based on a learned signal, and differs from kin-recognition where the template is based on phenotype matching. Here, we review studies that have sought to characterize MHC haplotype-specific odortypes, which have mainly focused on MHC-correlated volatile profiles, and their relation to pregnancy block and scent marking.
MHC congenic strains of mice, which share the same background genome, but have unique MHC haplotypes, are a model system with which to understand behavioral responses to individuals of same- or different-haplotype at a single locus. One extrapolation from studies demonstrating MHC haplotype-dependent behavior17 in congenic strains is the possibility that, in outbred populations where MHC allelic polymorphism is likely to be very high, MHC phenotypes would be key mediators of individual recognition. For example, it has long been understood that MHC congenic strains have unique volatile organic compound signatures that are used in chemical communication15. More recently, several groups have identified suites of volatile organic compounds that are regulated by MHC odortypes15,16,56. As predicted by a model of individual recognition, some of these suites are unaffected by environmental variation57; furthermore, volatile profiles from MHC congenic mice activate overlapping but distinct subsets of neurons in the mouse main olfactory bulb58. The authors of such studies in congenic strains often conclude that the physiological machinery is in place for volatile profiles to mediate individual-specific behaviors (e.g. Ref. 57). However, counter-part experiments using outbred wild mice in a more ecologically realistic setting are lacking. Given that some genotypes will inevitably be shared between individuals, more naturalistic work is needed to understand how these volatile signatures function as signals of individuality (as defined above) or as signals of relatedness or genotype.
Pregnancy Block
Pregnancy block, also known as the Bruce effect, occurs when recently mated female laboratory mice are exposed to the odors of an unfamiliar male59. Upon exposure to an unfamiliar male odor, prolactin release from the anterior pituitary in the mated female is suppressed, resulting in pregnancy failure, reabsorption of the fetus, and the onset of estrus60. The signal responsible for pregnancy block is considered to be individual specific because the unfamiliar male and the mate both express odors capable of inducing pregnancy block. Thus, females have to learn the identity of their mate (i.e. form a memory) in order to suppress pregnancy block upon perception of the mate’s odors.
Pregnancy block can be induced by the presence of an unfamiliar male or simply his soiled bedding or urine, and direct physical contact with the odorant seems necessary60. However, in at least one case volatiles alone (i.e. no direct contact) can induce pregnancy block61. The memory developed during pregnancy block is dependent on activation of sensory neurons in the VNO; however, the specific chemical constituents that bind receptors in these neurons have proven difficult to find. Three different classes of molecules associated with individual odors have recently been investigated: MHC and MHC peptides, major urinary proteins (MUPs), and volatiles. Peele and colleagues recently investigated the relative roles of MUPs and volatiles62. They found that low molecular weight fractionations (which excludes MUPs) from urine were more effective in blocking pregnancy than those of high molecular weight, suggesting a role of volatile compounds in the odor. However, the low molecular weight fraction from the unfamiliar male resulted in only 50% pregnancy block, as opposed to 90% pregnancy block via unfamiliar male whole urine. Moreover, a recent study called these findings into doubt by showing that, contrary to Hilda Bruce’s original finding, urine from castrated or juvenile males was sufficient to induce pregnancy block. These results suggest that, although volatiles can contribute to the occurrence of pregnancy block, they are not necessary to induce it63.
MHC-associated odors have also been shown to be sufficient to induce pregnancy block in several studies, implicating it’s involvement during individual recognition. MHC was originally identified when it was found that unfamiliar males differing only at the MHC could induce pregnancy block61. Since then, searches for an MHC-odortype mechanism have targeted MHC molecules themselves, MHC peptides, and possible associated volatiles. MHC peptides were the first specific odorant to be identified that induces pregnancy block7 (see above).
The finding that sensory neurons in the VNO respond selectively to MHC peptides was biologically validated by investigating the role of peptides in producing pregnancy block7. As predicted, it was found that pregnancy block upon exposure to MHC peptides from an unfamiliar, MHC-dissimilar male was equally effective as exposure to whole urine from an unfamiliar, MHC-dissimilar male. In this case, the peptides had to be delivered on a urinary background (regardless of whether the urine was from a familiar or unfamiliar male). A more recent study, however, found that peptides alone (administered more frequently than in Ref.7) were sufficient to induce pregnancy block63. These studies show that the suite of peptides presented by an individual’s MHC molecules can, when excreted in urine, be used as odorants in chemical signaling. Because of the large diversity of MHC haplotypes in a population, there is potential for individual specific odortypes simply in excreted MHC peptides. Such odortypes are detectable by vomeronasal sensory neurons that have binding specificity for these peptides similar to that of MHC molecules7. Where these peptide signals originate, however, remains to be found. Surprisingly, there is disagreement about whether peptides can be found in mouse urine7,60,64. Peptides have not been reported in other mediums of chemical communication such as saliva, tears, or skin excretions but, we are not aware of any directed searches for peptides in these secretions.
Although MHC peptides are clearly sufficient to induce pregnancy block in inbred mice, it should be noted that the experiments described above do not demonstrate individual recognition in a strict sense. Because peptides from an unfamiliar male with the same MHC genotype as the female’s mate would not be expected to induce pregnancy block, MHC peptides in the context of pregnancy block might be more likely to signal the presence of an unfamiliar male. If individuality is perceived during pregnancy block, it would likely be conveyed via coupling with sensory neurons activated by the urinary background, and neurons in the VNO have been found to be capable of individual mice of the same laboratory strain64. Finally, while pregnancy block provides an attractive system in which to test hypotheses concerning social signaling and behavior, the system is ultimately hindered by the fact that the adaptive significance of pregnancy block, which is only observed in certain laboratory strains of mice, has not been determined for natural populations. It has been suggested that the Bruce effect functions to prevent infanticide from males who have recently displaced the dominant, territorial male4,65.
Scent-marking
In addition to the MHC, growing evidence indicates that major urinary proteins (MUPs) are another chemical signal critical to social communication and individual recognition in mice. MUPs are protein pheromones encoded by a polymorphic, multi-gene family. In a series of experiments, the laboratory of Jane Hurst has tested the relative roles of MUPs and MHC in individual recognition in mice using the scent-marking behavioral paradigm. First, it was shown that wild-derived males presented with a scent mark from another male expressing a different MUP-type will investigate and counter-mark the marks significantly more than the control66. Second, it was shown that scent-marks associated with MHC haplotype (in MHC-congenic strains) were not necessary or sufficient to influence investigation time of male mice of congenic MHC strains. Rather, investigation time was increased only when the stimulus odor differed from the genomic background of the test animal67. A third experiment tested whether wild female mice could discriminate between scent marks from congenic males whose MHC and MUP genotype were controlled. Results showed that females could discriminate between individual males only when the males differed with respect to MUP haplotype; females could not discriminate between individual males that had the same MUP haplotype, and could not discriminate between males that had different MHC haplotypes68. These three experiments indicate that, in the context of scent-marking and countermarking, MUP genotype, and not MHC genotype, is the greatest determinant of individuality in urinary odors. However, it should be noted that in light of previous research, it is anomalous that the mice in these experiments did not discriminate between urinary odors that differed with respect to MHC genotype67,68. Previous studies have documented the ability of either MHC-congenics (e.g. Ref. 5) or wild-derived mice69 to distinguish urinary odors that only differed genetically at the MHC.
Because MUPs are likely to be polygenic, polymorphic signals only in a few rodent species it is unlikely that the functions discovered in Mus will have generality across vertebrates. The results from the Hurst group studies suggest that there are key differences in signals that are conserved across taxa (e.g. MHC) and signals that are species-specific (e.g. MUPs) for the identification of individual conspecifics68. They also reveal the curious but apparent finding that signals of individuality are limited to specific behavioral interactions.
Taken together, the individual recognition studies reviewed above show that MHC may play an important role in individual recognition in certain instances (for example in pregnancy block), but also indicate that they may not be specific enough for individual recognition in the strict sense. Many of the studies focusing on individual recognition and the MHC have utilized congenic strains of mice, which provide a unique opportunity to study the role of a single locus or haplotype in chemical communication. However, the use of inbred stains of animals may limit our broader understanding of behavior and ecology, as 60 years of domestication has modified behaviors70. So, more studies will be needed to determine the role of MHC in individual recognition in outbred populations; we know of no such examples except for the aforementioned examples from the Hurst lab using the scent-marking behavioral paradigm.
MHC as a signal in kin recognition
Kin recognition using polymorphic genetic systems allows individuals to engage in behaviors specific to kin or non-kin. An individual’s fitness is a product of both its own reproductive success (i.e. direct fitness) and the reproduction of close relatives (i.e. indirect fitness); thus, proper identification of kin facilitates cooperation (or at least decreased antagonism) with relatives and promotes behaviors that increase fitness71. Additionally, recognition of kin allows for the prevention of inbreeding, which reduces the homozygous expression of deleterious recessive alleles. In order for a genetic system to be used accurately to recognize kin, it must contain enough allelic polymorphism to allow discrimination between related and unrelated individuals. Kin recognition systems that can discriminate among a range of different-degree relatives have been reported72. MHC is the most polymorphic genetic system in vertebrates4 and has long been considered to play a role in kin recognition by mediating cooperation1, parent-offspring identification73, and mating preferences that prevent inbreeding74.
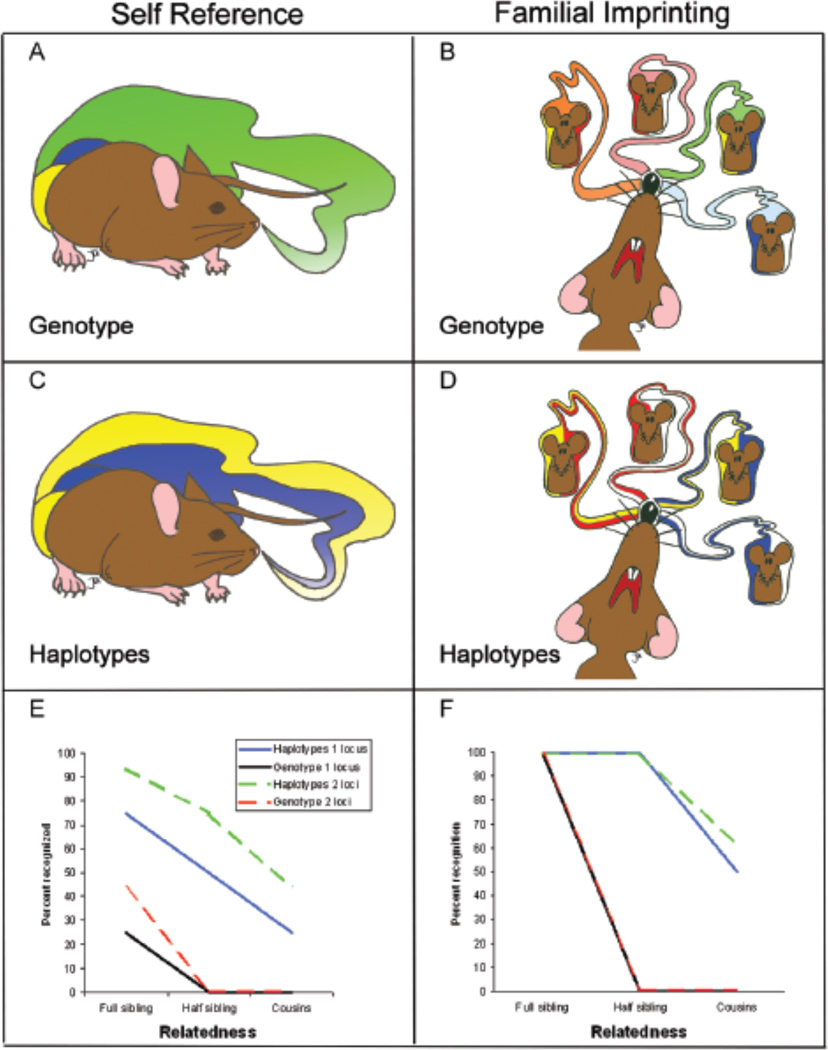
Two major phenotype matching mechanisms exist for MHC-based kin recognition within vertebrates (Figure 1). The first is a self-referent system in which individuals use their own MHC odortype as a template to recognize other individuals as kin39,43,46,49,50.The second is familial imprinting where individuals imprint upon the MHC odortypes of kin early in development and afterwards extrapolate the learned MHC signals of kin to other unfamiliar individuals20,42,75. The degree to which familial imprinting and self-referent systems identify kin differ remarkably (Figure 1) and only familial imprinting systems can identify kin that do not share odortypes with a focal individual, however the ability to recognize kin that do not share odortypes also allows for the false recognition of unrelated individuals due to imprinting on unrelated odortypes that could occur in mixed litters where odortypes produced by half siblings could be based on haplotypes from an unrelated individual. Both of these systems can be used to identify kin through odortypes based on either specific MHC haplotypes (both haplotypes providing a specific odor) or by odortypes based on a blended odor of both haplotypes. For example self-referent systems recognize either 25 or 75% of full siblings depending on whether specific haplotypes are recognized or only the genotypic odor of the combination of haplotypes76 (Figure 1). Currently few studies have been conducted to determine the specifics of phenotype matching systems used in nature and more research is needed to determine the relative prevalence of familial imprinting vs. self-referent systems and the nature of the odortypes (individual haplotypic or blended odors) used. Interestingly, the two systems most described in nature are familial imprinting on haplotypes and self reference based on exact genotype (blended odortype) which are the best and worst of the theorized kin recognition systems respectively (Figure 1). Regardless of the phenotype matching system used, kin recognition is likely one of the major functions of MHC-mediated signaling and the very existence of familial imprinting is strong evidence supporting this hypothesis because kin recognition is the only function that is enhanced by familial imprinting; self reference will be superior for functions involving genetic compatibility, individuality, or quality65.
Figure 1.
Possible phenotype matching systems using MHC-based odors and their effectiveness for the recognition of kin. Two kin recognition mechanisms that exist in nature are self reference (A, B, E) and familial imprinting (C,D,F). Phenotype matching can be based on haplotypes (e.g. allele specific odors) or on genotypes (e.g. combined haplotype odor). Self-referent phenotype matching can be based on odors associated with genotype (A) or both haplotypes (B). Familial imprinting can be based on odors associated with the genotypes (C) or haplotypes (D) present in the nest (e.g. parents or siblings). The prevalence of these phenotype matching systems in nature is largely untested; current evidence suggests that the primary phenotype matching system in mice is haplotype-based familial imprinting. The effectiveness of each phenotype matching system for recognizing three classes of kin are plotted for one or two unlinked polymorphic loci (E & F). MHC haplotypes contain multiple loci that are inherited in a linked “one locus” system or as multiple unlinked regions “multiple loci” depending on taxa. Models assume that all individuals are heterozygous, that no alleles are shared between unrelated individuals and that all combinations of parental genotypes are found within litters. Haplotype signaling is always superior to genotype signaling and two-locus systems provides an advantage in self-referent systems compared to single-locus systems. (Illustrations by J.L.K; graphic design by Linda Morrison).
Phenotype matching systems can identify more kin if multiple polymorphic unlinked loci are used, presuming a match at any locus is a signal for relatedness77. Though the impact of multiple unlinked loci has minimal impact on familial imprinting systems it has profound consequences on self-referent systems, were multiple loci dramatically improve kin recognition (Figure 1). Within both teleost fishes and amphibians, taxa where self-referent systems are common, the MHC is not inherited as a single unit but rather as two or four separate unlinked loci78,79. Whether this is coincidence or represents the inefficiency of self-referent systems favoring translocations that breakup the MHC linkage group will await more phylogentic data. Within both teleost fishes and amphibians it has been shown that MHC Class II genes are sufficient, but not necessary, for kin recognition. It has been proposed that other unlinked MHC genes provide additional information used in kin recognition46,49. Likewise, in house mice it has been observed that when MHC signals of relatedness are controlled for, signals from a different polymorphic locus (MUPs, see below) can also be used as signals of relatedness. In nature, it is highly likely that both MUPs and MHC are utilized for kin recognition in tandem80.
Cooperative behavior
Proper identification of kin can result in cooperative behaviors between relatives; MHC mediated signaling has been shown to both promote cooperation and deter antagonism between individuals (Table 1). Schooling is an important cooperative behavior in fish and tadpoles that results in enhanced foraging and predator avoidance. Several salmonid species along with the African clawed frog (Xenopus laevis) have been shown to preferentially form schools with relatives that share MHC haplotypes39,46,49 and it has been previously shown that kin-based schools have higher survival rates and larger territories81. Another MHC-mediated cooperative behavior has been documented in house mice; female mice communally nest and nurse offspring and it has been demonstrated that females preferentially nest with familiar sisters. When no familiar sisters are available, they preferentially nest with MHC-similar females1. Competition over territories is fierce in many species of vertebrates and can result in serious injury; evidence suggesting that MHC signaling prevents territorial competition between kin has recently been demonstrated in tuataras (Sphenodon punctatus)38. Scores of other kin-based cooperative behaviors have been documented within vertebrates and it is quite probable that we have only just begun to document those that are mediated by MHC signaling; however, it is not our intent to imply that all cooperative behaviors will be MHC-mediated. In fact, the precision of kin recognition systems will be enhanced as more polymorphic systems are used in signaling.
Parent-progeny recognition
Parent-progeny recognition prevents the expense associated with parental investment into unrelated individuals. In species that provide parental care individuals are at risk of providing for unrelated individuals, especially under conditions of communal living or in systems that involve extra-pair matings. Under these circumstances an identification system that could ensure parental care was only provided to legitimate genetic offspring would be highly adaptive, and many such systems have been documented82. Female house mice nest communally and are therefore at risk of providing parental care to unrelated pups. Yamazaki and others73 showed that female house mice can identify pups with which they share an MHC haplotype from congenic pups (genetically identical individuals with the exception of MHC type). This same study has shown that pups at the age of 15–21 days are also capable of recognizing and preferring their parents bedding to that of a MHC dissimilar congenic individual. This preference was reversible by cross-fostering, again showing the role of familial imprinting within MHC singling in house mice. Currently this study offers the only evidence that MHC-mediated signaling is involved in parent-progeny recognition, and though it was conducted with inbred strains of mouse, it reveals the potential of MHC signaling that may be important in nature.
Inbreeding avoidance
Degradation of fitness due to inbreeding is a result of increased homozygosity of deleterious recessive alleles that are identical by descent. These recessive alleles combine more frequently when related individuals reproduce compared to outbred matings. Direct assessment of the fitness costs of full-sibling level inbreeding within vertebrates has been conducted and early studies showed a 10% decline in litter size83,84. However these experiments only measured litter size reductions, and they failed to assess the fitness consequences of the inbred progeny in their natural context. In an experiment where the fitness impacts of a single generation of full-sibling inbreeding were assessed under seminatural conditions, it was found that outbred males had fivefold higher fitness than inbred males, with the consequences effectively approaching lethality for inbred sons. Daughters suffered an additional 20% reduction in fitness compared to previous assessments85. Likewise cousin-level inbreeding was shown to reduce male fitness by 34% and when the infectious agent Salmonella was present in the populations, the fitness decline in males was 57%86. Disassortative MHC-based mating preferences function as a mechanism of inbreeding avoidance due to their highly polymorphic nature. Only closely related individuals are likely to share MHC haplotypes; thus a mating preference for MHC-dissimilar individuals will decrease the likelihood of inbreeding. The extent to which inbreeding can be avoided is dictated by whether a self-referent or familial imprinting mechanism is utilized by a particular species76, both of which are found within vertebrates.
An ironic piece of evidence supporting haplotype based familial imprinting in regards to MHC-based inbreeding avoidance behavior within house mice has come from a study by Sherborne and colleagues80. This experiment attempted to elucidate the relative importance of MHC and MUPs in mediating inbreeding avoidance behavior, and its conclusion was that MHC is not involved in inbreeding avoidance behavior. House mice were released into seminatural enclosures with only full-sibling and half-sibling counterparts; inbreeding avoidance was assessed by the proportion of full-sibling vs. half-sibling matings, and genetic analysis was used to determine if there was either an MHC or MUP-based signal mediating inbreeding avoidance The data showed that in fact no full-sibling inbreeding avoidance occurred, despite the fact that mice sharing exact MUP genotypes avoided mating with each other. This led the authors to conclude that MUPs are exclusively responsible for inbreeding avoidance in house mice and that MHC plays no role. However, this conclusion is unwarranted due to a flaw in the experimental design. All experimental individuals had been caged with individuals from birth that contained haplotypes that were found in all experimental animals (half-siblings and full-siblings) within the seminatural enclosures. This design allowed MHC familial imprinting to occur on all of the tested haplotypes; thus, animals upon entering the enclosures found themselves surrounded by individuals that would all be recognized as relatives by MHC-based systems. This situation forced the mice to make mate choices based on other non-MHC cues and they utilized MUPs, preferring to mate with individuals that did not share exact genotypes. These results suggest MUPs are utilized in mate choice, but contrary to the conclusions of the paper, the design does not allow for the exclusion of a role for MHC. Furthermore, MUP-based mating preferences are self-referent and not imprinted80, thus they do not offer the protection against inbreeding that familial imprinted MHC preferences do.
MHC as a signal of genetic compatibility in mate choice
Genetic compatibility, broadly defined, refers to the degree to which the genes, within and between haploid genomes, that make up an organism interact to increase or decrease fitness. Levels of genetic compatibility range from inviable offspring (e.g. between species matings), severely reduced fitness (e.g. inbreeding) or modifications to fitness as different alleles interact within genotypes. The fitness consequences of genetic compatibility might be so severe that finding a mate with the “right genes” to compliments one’s own genome provides more indirect benefits than finding the “best genes” within high quality individuals87. In order to make MHC-based mate choice (or gamete fusion88,89 decisions in regards to genetic compatibility, individuals must possess the means to assess there own MHC types (see section on phenotype matching systems above). MHC-mediated odors readily signal information about the genetic compatibility between mates and MHC-disassortative mating preferences (Table 1) lead to the production of offspring with compatible genotypes both at the MHC and throughout the genome76. The mechanisms of MHC-mediated genetic compatibility described below are MHC heterozygote advantage, offspring harboring different MHC genotypes than their parents (moving target), and the avoidance of inbreeding.
Heterozygote Advantage/Superiority
MHC-disassortative mate preferences, by their very nature produce MHC heterozygous offspring, which are hypothesized to have superior immunocompetence90,91. Multiple lines of evidence now support the fitness-enhancing role of MHC-heterozygosity92–98. It was initially argued that MHC heterozygotes would have an advantage (overdominance) because they could present a wider variety of peptide antigens to the immune system making them more likely than MHC-homozygotes to recognize, and mount an immune response against, disease-causing agents. However, this mechanistic hypothesis has largely been rejected since experimental infections with single pathogens reveal that heterozygotes do not generally have an advantage over both homozygotes99. An alternative mechanism postulated that heterozygote advantage emerges over multiple infections because resistance is generally dominant and heterozygotes will benefit from the resistance profile of each allele, which masks some of the susceptibilities of each allele. This hypothesis was experimentally confirmed by laboratory-based experiments using coinfections with parasites having opposite MHC resistance/susceptibility profiles, which demonstrated that heterozygotes are more fit than either homozygote99. Recent studies on wild salmon100 and vole101 populations demonstrate that MHC heterozygotes have increased fitness under natural conditions of multi-parasite infection as well. The fitness enhancing nature of MHC heterozygote advantage in laboratory and natural settings is an example of the adaptive significance of MHC mediated signaling.
Moving Target
In addition to heterozygote advantage, selection could also favor MHC-disassortative mate preferences if the offspring genotype provided a moving target against pathogen adaptation causing pathogens adapted to either parent to be at a disadvantage in progeny that are MHC-dissimilar to both parents102. This hypothesis predicts that pathogens evolve to partially escape MHC-mediated immune recognition and that MHC-dissimilar offspring are more fit than their parents when challenged with parent-adapted pathogens. Like heterozygote advantage, mate choice decisions driven by moving target processes function to maximize genetic compatibility and are thus most effectively achieved using an MHC, self-referent phenotype matching system.
Numerous examples highlight the capacity of pathogens to rapidly adapt to escape MHC-mediated immune recognition103–110. There has been one experimental study designed to test the other prediction of the moving target hypothesis – that MHC-dissimilar offspring will be more fit than their parents when challenged with parent-adapted pathogens. MHC did influence the trajectory of adaptation by this fungal pathogen (Cryptococcus neoformans), but the large virulence increase in post-passage pathogen lines showed no specificity for the host MHC genotype of passage111. The most likely explanation is that this pathogen is a generalist that infects most birds and mammals. The passages in mice selected for adaptations to “mouseness”, which likely swamped any adaptations to MHC. Future passage studies should use pathogens specialized on the host of passage.
There is anecdotal evidence from human studies demonstrating the importance of offspring genetic diversity in reducing the probability of mother-to-child-transmission of chronic infectious disease agents (e.g. HIV-1112,113) suggest that there would be a significant selective advantage to mate choice decision making that promoted genetic diversity in offspring. There is also evidence linking increased HLA dissimilarity between mother and offspring with significantly reduced chances of vertical transmission of HIV-1114,115). The extent of pathogen adaptation during chronic infection of the parent and its’ impact on mother-to-child transmission dynamics was not addressed in the above studies. Despite this, they do support the possible role of MHC- disassortative mate preferences in producing offspring of higher quality that are more resistant to infection by chronic parasites of their parents.
Optimal MHC heterozygosity
MHC-disassortative mate choice may carry a cost if maximal MHC diversity in offspring is not optimal in regards to genetic compatibility. For instance, during the process of negative selection in the thymus, T cells with high affinity for MHC-peptide complexes are instructed to terminate themselves via apoptosis9. It follows then that MHC diversity may have an upper limit beyond which the fitness benefit of having multiple ways to present peptides from foreign invaders is offset by the cost of an increasingly limited T cell repertoire, which could ultimately reduce abilities to recognize these invaders116. If such a fitness cost exists, then it will have important implications on the evolution of MHC-disassortative mating preferences. Indeed, it has been observed that individuals with intermediate versus maximal MHC diversity harbor lower parasite burdens in experimental infections117. Additionally, it was recently shown that intermediate and not maximal levels of MHC diversity lead to significantly higher lifetime reproductive success in stickleback offspring118. Thus, it seems that maximum MHC diversity can be a costly trait.
If intermediate rather than maximal MHC diversity is optimal then an MHC-typing system could allow individuals to “optimize” the MHC diversity within their offspring so long as individuals had the capacity to obtain “quantitative” information regarding MHC diversity in their potential mate. Studies with sticklebacks have shown that females are in fact capable of such quantitative estimates of MHC diversity (also known as allele counting)119. Additionally, by estimating the extent of intra-individual MHC class IIB allele diversity within a population, it was also demonstrated that individuals with intermediate rather than maximal MHC diversity were most frequent, indicating selection for intermediate levels of MHC diversity. Subsequent experimental findings in sticklebacks43 and brown trout50 suggest that much of the selection for individuals with intermediate MHC diversity derives from female preference for MHC-dissimilar mates. Together, these studies indicate that maximal MHC diversity is not always optimal and that female preference for MHC-dissimilar mates is a primary driving force behind selection for the production of individuals with intermediate rather than maximal MHC diversity.
Inbreeding avoidance
Though inbreeding avoidance has already been covered within the kin recognition section it is important to stress that it also falls under the umbrella of MHC as a signal of genetic compatibility. In fact, inbreeding avoidance may be the single most adaptive result of MHC-disassortative mating preferences in many species of vertebrates, as both sibling and cousin level inbreeding have been found to have devastating effects on the fitness of offspring85,86. In addition, as covered in the evolution of MHC section below, growing evidence suggests that MHC mediated kin recognition to avoid inbreeding may have been the ancestral function of MHC molecules, which were later co-opted for use in the adaptive immune system120.
MHC and signals of quality in mate choice
In contrast to MHC-mediated signals that directly convey MHC genotype information (relatedness, compatibility or individuality), the disease resistance functions of MHC can also influence mate choice by modulating the expression of secondary sexual characters. Only high-quality, disease-resistant individuals should be able to invest in costly, sexually selected advertisements121, thus creating a correlation between MHC genotype and these condition-dependent traits (Table 2). By endowing an individual with genetic resistance to parasites, MHC genotype can indirectly influence signals of quality by allowing more physiological resources to be devoted to signaling rather than to the immune response122. von Schantz and colleagues123 were the first to report an association between MHC and a sexually selected trait; they found that spur length in male pheasants (Phasianus colchicus) was correlated with fitness and dependent on MHC genotype. In a study on great snipes (Gallinago media), females preferred males carrying specific MHC allelic lineages. Males with these genotypes were also larger, and females of this species are generally known to favor larger males124. A study in white-tailed deer (Odocoileus virginianus) found that MHC divergent heterozygous males had larger antlers and body size, which was correlated with lower abundance of abomasal nematodes122. Finally, a study on a canonical sexually selected trait, trains in male peacocks (Pavo cristatus), showed that the train length reflects genetic diversity at the MHC36. The above examples show that MHC-genotype can influence the expression of secondary sexual traits that are used as signals of quality. However, MHC-genotype itself is not necessarily used in the signal.
Table 2.
MHC correlations with secondary sexual traits and mating preferences.
| Species | MHC correlation with mate preference |
MHC correlation with traits of quality |
Sources |
|---|---|---|---|
| Great snipe (Gallinago media) |
MHC allele-specific preference |
Body size | Ekblom et al. 2004124 |
| Peafowl (Pavo cristatus) |
MHC heterozygosity | Train length | Hale et al. 200936 |
| Pheasants (Phasianus colchicus) |
MHC genotype | Spur length | von Schantz et al. 1996123 |
| White-tailed deer (Odocoileus virginianus) |
MHC divergent heterozygotes |
Antler and body size; reduced parasitism |
Ditchkoff et al. 2001122 |
An alternative way that MHC-genotype can indirectly influence the expression of secondary sexual characteristics is if MHC social signals themselves are costly to produce, and therefore only high quality individuals will be able to invest more in the production of this signal. This hypothesis has recently been tested by the laboratory of Manfred Milinski, which identified the first example of condition-dependent MHC signaling125. They had previously shown that female three-spined sticklebacks prefer males with optimal, rather than maximal, MHC allelic differences (relative to her own genotype), and that this mate choice is mediated by excreted MHC peptides (discussed below)14,44. Now, they show that females do not send this signal at all and that, remarkably, males only send this signal when they are in the reproductive state. These data suggest that MHC signaling is not simply a byproduct of MHC-peptide presentation, but that it is actively regulated in a fashion consistent with it being a costly signal. The authors suggest that shedding MHC-peptide complexes will create localized deficiencies of this critical immunological component, and therefore represents a trade off between immune defense and social signaling125.
MHC-mediated signals of quality may allow an individual to gain either direct benefits for themselves or indirect genetic benefits for their offspring. Avoidance of parasitism is perhaps the most likely direct benefit of MHC-mediated mate choice. Social behaviors are an opportunity for parasites to transmit to new hosts; in turn, hosts will gradually develop behavioral mechanisms to avoid parasites126. Individuals of a particular MHC-genotype may be resistant to local parasites at any given time, and choosing such an individual as a mate would provide a direct benefit of reduced risk of parasitism. Although there are several examples of mate choice for parasite-free individuals127–129, there are surprisingly few examples of studies that link MHC-dependent resistance to pathogens and subsequent mate choice117.
MHC evolution: what are the primordial functions?
Since the immune recognition function of MHC genes in adaptive immunity was discovered far earlier than MHC-mediated behaviors, and since it was so central to the complex system of vertebrate adaptive immunity, it was initially assumed that MHC-mediated behaviors were a derived function. However, Brown argued that since kin-selected behaviors (inbreeding avoidance and kin-biased cooperation) are present in the ancestral lineages leading to vertebrates and that adaptive immunity is a derived character in vertebrates, it is most parsimonious to hypothesize that MHC-mediated kin recognition functions were primordial74. The controversy continues until this day.
Boehm has recently written a tour-de-force, synthetic review that evaluates self and non-self recognition systems that exist across plants, fungi and animals, with a special emphasis on how quality recognition is maintained in the face of the rapid diversification of these highly polymorphic systems120. Quality control (the ability to accurately discriminate between self and non-self) is of particular importance in immune recognition systems that must achieve self tolerance to protect against auto-immune disease120,130.
Jawless fish are the one lineage of vertebrates that appear to have a non-MHC based adaptive immune recognition system131,132. A high diversity of lymphocyte receptors in this group are created by combinatorial assembly of receptor modules, but the critical difference from other vertebrates is that there is no junctional diversity created by mutagenic joining mechanisms133. Thus, the lymphocyte receptor repertoire for jawless fish is predictable and self tolerance could be achieved by Darwinian selection for self-compatible receptor modules120. In contrast, jawed vertebrates achieve higher lymphocyte receptor diversity by the mutagenic VDJ combinatorial joining process, which creates the problem of unpredictable receptor specificities that can lead to auto-aggressive T-cells. These potentially harmful receptors are eliminated during the evaluation of lymphocyte (T-cell) receptors in the thymus of jawed vertebrates. Boehm argues that it seems unlikely that an MHC-peptide presentation system could emerge de-novo to create the modern jawed vertebrate immune recognition system, which allows self-tolerance in the face of somatic generation of unpredictable lymphocyte receptors. It would be far more likely that a pre-existing MHC-peptide kin recognition system could be co-opted for immune recognition120. Discovery of the MHC homologues and their function in jawless fish offers one of the most promising approaches for discriminating between these two hypotheses and identifying the primordial function of MHC genes. Tunicates have a highly polymorphic histocompatibility-type (fusion) locus that functions both in allo-recognition to control colony fusion and gamete fusion134, at least in some species135. It was thought that identifying the nature of this locus might clarify the early history of MHC genes. After a two-decade search the locus was identified to be a member of the immunoglobulin super family, but it appears to not have homology to MHC genes136–138. These findings further focus the search for primordial MHC functions towards jawless fish.
The facts that within vertebrates there are completely different mechanisms controlling adaptive immune recognition and that in close relatives to vertebrates (tunicates) histocompatibility functions are controlled by genes unrelated to vertebrate histocompatibility genes, highlights the evolutionary flexibility of how similar functions can be achieved through different genetic systems. It is currently difficult to discriminate between the different proposed primordial function of MHC genes. However, the initial assumption that immune recognition must be the primordial function of MHC genes, should no longer be the default assumption.
Conclusions
In this chapter we have demonstrated the significance of MHC signaling in regards to four aspects of social communication. First, studies in mice show that MHC peptides, and to a lesser extent MHC-mediated urinary odors, signal individuality in the context of pregnancy block. MHC does not signal individuality during scent-marking, rather, a species specific signal (MUP) is used. Second, MHC as a signal of relatedness is found across vertebrate taxa and plays a role in cooperation, parent-offspring identification, and inbreeding avoidance via two different phenotype matching mechanisms: self-reference or familial imprinting. Third, MHC signals are used to determine the genetic compatibility of a potential mate, and can result in the production of heterozygous offspring. Phenotype matching is likely to be the primary means by which compatibility mating decisions are made. In some animals, mate choice for MHC compatibility is so finely tuned that they can optimize the degree of MHC heterozygosity in their offspring. Fourth, information regarding MHC genotype can be signaled indirectly through correlated characters (Table 2) and a recent study demonstrated that, at least within one species, MHC signaling itself may be condition-dependent and therefore a signal of individual quality. Taken together, these studies suggest that MHC-mediated signaling is conserved across vertebrates, but takes on unique functions depending on the life-history of the organism. Since both kin recognition and genetic compatibility mate choice are common, phenotype matching is likely to be the most common mechanism driving MHC social signaling.
Appreciating the distinction between both modes of phenotype matching (self-reference and familial imprinting) is paramount in understanding the role MHC-mediated signaling plays in social communication. Though substantial overlap in functionality exists between these phenotype matching systems, there are tradeoffs. Self-referent systems facilitate mating preferences that generate offspring with an immunological advantage by allowing the assessment of genetic compatibility. Familial imprinting systems of phenotype matching facilitate the identification of siblings, half-siblings, and cousins; in species where either cooperative behavior or avoiding inbreeding is important (e.g. communal nesting species or species that live in high-density populations), a familial imprinting system provides an advantage over a self-referent system because self/non-self discrimination is not required to increase indirect fitness. That these two systems are differentially utilized by different groups of vertebrates highlights the highly context-dependent nature of social signaling. It is important to note, however, that phenotype matching mechanisms have been described in a relatively small number of species (Table 1), and more studies are needed.
The remarkable fact that a single genetic system controls major components of both immune recognition and social recognition begs the question of which recognition system constituted the primordial function of MHC genes. The convergent evolution of similar peptide binding properties of MHC, VSN and OSN receptor molecules provides the molecular basis by which MHC genotype influences both immune and social recognition, and implies that these distinct receptor families have responded to selective pressures that required information regarding MHC genotype (bound peptides) be associated with discriminatory sensory systems. Finally, the ubiquitous presence of various modes of self versus non-self discrimination across all three domains of life coupled with the derived nature of the adaptive immune system in vertebrates further suggests that MHC-mediated social signaling evolved for the purpose of discrimination between conspecifics and consequently represents the ancestral state. Tracing the function of MHC molecules across vertebrate evolution holds the greatest promise of resolving the relative importance of immune versus social communication in MHC evolution.
References
- 1.Manning CJ, Wakeland EK, Potts WK. Communal nesting patterns in mice implicate MHC genes in kin recognition. Nature. 1992;360:581–583. doi: 10.1038/360581a0. [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki K, Boyse EA, Mike V, et al. Control of mating preferences in mice by genes in the major histocompatibility complex. J. Exp. Med. 1976;144:1324–1335. doi: 10.1084/jem.144.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snell GD. Methods for the study of histocompatibility genes. J Genet. 1948;49:87–108. doi: 10.1007/BF02986826. [DOI] [PubMed] [Google Scholar]
- 4.Piertney SB, Oliver MK. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi M, Yamazaki K, Beauchamp GK, Bard J, Thomas L, Boyse EA. Distinctive urinary odors governed by the major histocompatibility locus of the mouse. Proc. Natl. Acad. Sci. 1981;78:5817–5820. doi: 10.1073/pnas.78.9.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh PB, Brown RE, Roser B. MHC antigens in urine as olfactory recognition cues. Nature. 1987;327:161–164. doi: 10.1038/327161a0. [DOI] [PubMed] [Google Scholar]
- 7.Leinders-Zufall T, Brennan P, Widmayer P, et al. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- 8.Spehr M, Kelliher KR, Li XH, Boehm T, Leinders-Zufall T, Zufall F. Essential role of the main olfactory system in social recognition of major histocompatibility complex peptide ligands. J Neurosci. 2006;26:1961–1970. doi: 10.1523/JNEUROSCI.4939-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy KP, Travers P, Janeway C, Walport M. Immunobiology. N.Y: Garland Science; 2008. [Google Scholar]
- 10.Boehm T, Zufall F. MHC peptides and the sensory evaluation of genotype. Trends Neurosci. 2006;29:100–107. doi: 10.1016/j.tins.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Spehr M, Spehr J, Ukhanov K, Kelliher KR, Leinders-Zufall T, Zufall F. Parallel processing of social signals by the mammalian main and accessory olfactory systems. Cell Mol Life Sci. 2006;63:1476–1484. doi: 10.1007/s00018-006-6109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelliher KR, Spehr M, Li XH, Zufall F, Leinders-Zufall T. Pheromonal recognition memory induced by TRPC2-independent vomeronasal sensing. Euro J NeuroScience. 2006;23:3385–3390. doi: 10.1111/j.1460-9568.2006.04866.x. [DOI] [PubMed] [Google Scholar]
- 13.Kelliher KR, Spehr M, Li XH, Zufall F, Leinders-Zufall T. Relative roles of the main and accessory olfactory systems in behavioral responses to MHC class I peptides: Bruce effect. Chem Senses. 2005;30:A18. [Google Scholar]
- 14.Milinski M, Griffiths S, Wegner KM, Reusch TB, Haas-Assenbaum A, Boehm T. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc Natl Acad Sci U S A. 2005;102:4414–4418. doi: 10.1073/pnas.0408264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer AG, Beauchamp GK, Yamazaki K. Volatile signals of the major histocompatibility complex in male mouse urine. Proc. Natl. Acad. Sci. USA. 1997;94:2210–2214. doi: 10.1073/pnas.94.6.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willse A, Belcher AM, Preti G, et al. Identification of major histocompatibility complex-regulated body odorants by statistical analysis of a comparative gas chromatography/mass spectrometry experiment. Anal Chem. 2005;77:2348–2361. doi: 10.1021/ac048711t. [DOI] [PubMed] [Google Scholar]
- 17.Kwak J, Opiekun MC, Matsumura K, Preti G, Yamazaki K, Beauchamp GK. Major histocompatibility complex-regulated odortypes: peptide-free urinary volatile signals. Physiol Behav. 2009;96:184–188. doi: 10.1016/j.physbeh.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Carroll LS, Penn DJ, Potts WK. Discrimination of MHC-derived odors by untrained mice is consistent with divergence in peptide-binding region residues. Proc Natl Acad Sci U S A. 2002;99:2187–2192. doi: 10.1073/pnas.042244899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelano B, Edwards SV. An Mhc component to kin recognition and mate choice in birds: Predictions, progress, and prospects. American Naturalist. 2002;160:S225–S237. doi: 10.1086/342897. [DOI] [PubMed] [Google Scholar]
- 20.Yamazaki K, Beauchamp GK, Kupniewski J, Bard J, Thomas L, Boyse EA. Familial imprinting determines H-2 selective mating preferences. Science. 1988;240:1331–1332. doi: 10.1126/science.3375818. [DOI] [PubMed] [Google Scholar]
- 21.Yamazaki K, Beauchamp GK, Daisuke Y. Advances in Genetics. Vol. 59. Academic Press; 2007. Genetic Basis for MHC-Dependent Mate Choice; pp. 129–145. [DOI] [PubMed] [Google Scholar]
- 22.Manning CJ, Wakeland EK, Potts WK. Communal nesting patterns in mice implicate MHC genes in kin recognition. Nature. 1992b;360:581–583. doi: 10.1038/360581a0. [DOI] [PubMed] [Google Scholar]
- 23.Jacek R, Aleksandra T, Agnieszka K. MHC and Preferences for Male Odour in the Bank Vole. Ethology. 2008;114:827–833. [Google Scholar]
- 24.Sommer S. Major histocompatibility complex and mate choice in a monogamous rodent. Behavioral Ecology and Sociobiology. 2005;58:181–189. [Google Scholar]
- 25.Wedekind C, Seebeck T, Bettens F, Paepke A. MHC-dependent mate preferences in humans. Proc. R. Soc. Lond. B. 1995;260:245–249. doi: 10.1098/rspb.1995.0087. [DOI] [PubMed] [Google Scholar]
- 26.Havlicek J, Roberts SC. MHC-correlated mate choice in humans: A review. Psychoneuroendocrinology. 2009;34:497–512. doi: 10.1016/j.psyneuen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Setchell JM, Charpentier MJ, Abbott KM, Wickings EJ, Knapp LA. Opposites attract: MHC-associated mate choice in a polygynous primate. J Evol Biol. 2009 doi: 10.1111/j.1420-9101.2009.01880.x. [DOI] [PubMed] [Google Scholar]
- 28.Schwensow N, Fietz J, Dausmann K, Sommer S. MHC-associated mating strategies and the importance of overall genetic diversity in an obligate pair-living primate. Evolutionary Ecology. 2008;22:617–636. [Google Scholar]
- 29.Schwensow N, Eberle M, Sommer S. Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proceedings of the Royal Society B-Biological Sciences. 2008;275:555–564. doi: 10.1098/rspb.2007.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson S, Pemberton JM. No evidence for major histocompatibility complex-dependent mating patterns in a free-living ruminant population. Proc. R. Soc. Lond. B. 1997;264:1813–1819. doi: 10.1098/rspb.1997.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman-Gallant CR, Meguerdichian M, Wheelwright NT, Sollecito SV. Social pairing and female mating fidelity predicted by restriction fragment length polymorphism similarity at the major histocompatibility complex in a songbird. Molecular Ecology. 2003;12:3077–3083. doi: 10.1046/j.1365-294x.2003.01968.x. [DOI] [PubMed] [Google Scholar]
- 32.Bonneaud C, Chastel O, Federici P, Westerdahl H, Sorci G. Complex Mhc-based mate choice in a wild passerine. Proc Biol Sci. 2006;273:1111–1116. doi: 10.1098/rspb.2005.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson DS, Komdeur J, Burke T, von Schantz T. MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc Biol Sci. 2005;272:759–767. doi: 10.1098/rspb.2004.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westerdahl H. No evidence of an MHC-based female mating preference in great reed warblers. Molecular Ecology. 2004;13:2465–2470. doi: 10.1111/j.1365-294X.2004.02238.x. [DOI] [PubMed] [Google Scholar]
- 35.Gillingham MAF, Richardson DS, Lovlie H, Moynihan A, Worley K, Pizzari T. Cryptic preference for MHC-dissimilar females in male red junglefowl, Gallus gallus. Proceedings of the Royal Society B-Biological Sciences. 2009;276:1083–1092. doi: 10.1098/rspb.2008.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hale ML, Verduijn MH, Moller AP, Wolff K, Petrie M. Is the peacock’s train an honest signal of genetic quality at the major histocompatibility complex? J Evol Biol. 2009;22:1284–1294. doi: 10.1111/j.1420-9101.2009.01746.x. [DOI] [PubMed] [Google Scholar]
- 37.Olsson M, Madsen T, Nordby J, Wapstra E, Ujvari B, Wittsell H. Major histocompatibility complex and mate choice in sand lizards. Proceedings of the Royal Society of London Series B-Biological Sciences. 2003;270:S254–S256. doi: 10.1098/rsbl.2003.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller HC, Moore JA, Nelson NJ, Daugherty CH. Influence of major histocompatibility complex genotype on mating success in a free-ranging reptile population. Proc Biol Sci. 2009;276:1695–1704. doi: 10.1098/rspb.2008.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villinger J, Waldman B. Self-referent MHC type matching in frog tadpoles. Proc Biol Sci. 2008;275:1225–1230. doi: 10.1098/rspb.2008.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bos DH, Williams RN, Gopurenko D, Bulut Z, Dewoody J. Condition-dependent mate choice and a reproductive disadvantage for MHC-divergent male tiger salamanders. Molecular Ecology. 2009;18:3307–3315. doi: 10.1111/j.1365-294X.2009.04242.x. [DOI] [PubMed] [Google Scholar]
- 41.Gerlach G, Hodgins-Davis A, MacDonald B, Hannah R. Benefits of kin association: related and familiar zebrafish larvae (Danio rerio) show improved growth. Behavioral Ecology and Sociobiology. 2007;61:1765–1770. [Google Scholar]
- 42.Gerlach G, Hodgins-Davis A, Avolio C, Schunter C. Kin recognition in zebrafish: a 24-hour window for olfactory imprinting. Proc Biol Sci. 2008;275:2165–2170. doi: 10.1098/rspb.2008.0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aeschlimann PB, Haeberli MA, Reusch TBH, Boehm T, Milinski M. Female sticklebacks Gasterosteus aculeatus use self-reference to optimize MHC allele number during mate selection. 2003 [Google Scholar]
- 44.Reusch T, Haeberli MA, Aeschlimann PB, Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001;414:300–302. doi: 10.1038/35104547. [DOI] [PubMed] [Google Scholar]
- 45.Milinski M. The major histocompatibility complex, sexual selection, and mate choice. Annual Review of Ecology Evolution and Systematics. 2006;37:159–186. [Google Scholar]
- 46.Rajakaruna RS, Brown JA, Kaukinen KH, Miller KM. Major histocompatibility complex and kin discrimination in Atlantic salmon and brook trout. Mol Ecol. 2006;15:4569–4575. doi: 10.1111/j.1365-294X.2006.03113.x. [DOI] [PubMed] [Google Scholar]
- 47.Consuegra S, Garcia de Leaniz C. MHC-mediated mate choice increases parasite resistance in salmon. Proc Biol Sci. 2008;275:1397–1403. doi: 10.1098/rspb.2008.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neff BD, Garner SR, Heath JW, Heath DD. The MHC and non-random mating in a captive population of Chinook salmon. Heredity. 2008;101:175–185. doi: 10.1038/hdy.2008.43. [DOI] [PubMed] [Google Scholar]
- 49.Olsen KH, Grahn M, Lohm J. Influence of MHC on sibling discrimination in Arctic char, Salvelinus alpinus (L.) J Chem Ecol. 2002;28:783–795. doi: 10.1023/a:1015240810676. [DOI] [PubMed] [Google Scholar]
- 50.Forsberg LA, Dannewitz J, Petersson E, Grahn M. Influence of genetic dissimilarity in the reproductive success and mate choice of brown trout -females fishing for optimal MHC dissimilarity. J Evol Biol. 2007;20:1859–1869. doi: 10.1111/j.1420-9101.2007.01380.x. [DOI] [PubMed] [Google Scholar]
- 51.Wedekind C, Walker M, Portmann J, Cenni B, Müller R, Binz T. MHC-linked susceptibility to a bacterial infection, but no MHC-linked cryptic female choice in whitefish. Journal of Evolutionary Biology. 2004;17:11–18. doi: 10.1046/j.1420-9101.2004.00669.x. [DOI] [PubMed] [Google Scholar]
- 52.Tibbetts EA, Dale J. Individual recognition: it is good to be different. Trends in Ecology & Evolution. 2007;22:529–537. doi: 10.1016/j.tree.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Singh PB. Chemosensation and genetic individuality. Reproduction. 2001;121:529–539. doi: 10.1530/rep.0.1210529. [DOI] [PubMed] [Google Scholar]
- 54.Thomas L. Symbiosis as an immunologic problem. The immune system and infectious diseases. In: Neter E, Milgrom F, editors. Fourth International Convocation of Immunology. Basel: S. Karger; 1975. pp. 2–11. [Google Scholar]
- 55.Tibbetts EA, Sheehan MJ, Dale J. A testable definition of individual recognition. Trends in Ecology & Evolution. 2008;23:356–356. doi: 10.1016/j.tree.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 56.Novotny MV, Soini HA, Koyama S, Wiesler D, Bruce KE, Penn DJ. Chemical identification of MHC-influenced volatile compounds in mouse urine I: Quantitative proportions of major chemosignals. Journal of Chemical Ecology. 2007;33:417–434. doi: 10.1007/s10886-006-9230-9. [DOI] [PubMed] [Google Scholar]
- 57.Kwak J, Willse A, Matsumura K, et al. Genetically-based olfactory signatures persist despite dietary variation. PLoS One. 2008;3:e3591. doi: 10.1371/journal.pone.0003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schaefer ML, Young DA, Restrepo D. Olfactory fingerprints for major histocompatibility complex-determined body odors. J. Neurosci. 2001;21:2481–2487. doi: 10.1523/JNEUROSCI.21-07-02481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruce HM. An exteroceptive block to pregnancy in the mouse. Nature. 1959;164:105. doi: 10.1038/184105a0. [DOI] [PubMed] [Google Scholar]
- 60.Brennan PA. Outstanding issues surrounding vomeronasal mechanisms of pregnancy block and individual recognition in mice. Behav Brain Res. 2009;200:287–294. doi: 10.1016/j.bbr.2008.10.045. [DOI] [PubMed] [Google Scholar]
- 61.Yamazaki K, Beauchamp GK, Wysocki CJ, Bard J, Thomas L, Boyse EA. Recognition of H-2 types in relation to the blocking of pregnancy in mice. Science. 1983b;221:186–188. doi: 10.1126/science.6857281. [DOI] [PubMed] [Google Scholar]
- 62.Peele P, Salazar I, Mimmack M, Keverne EB, Brennan PA. Low molecular weight constituents of male mouse urine mediate the pregnancy block effect and convey information about the identity of the mating male. Eur J Neurosci. 2003;18:622–628. doi: 10.1046/j.1460-9568.2003.02790.x. [DOI] [PubMed] [Google Scholar]
- 63.Thompson RN, McMillon R, Napier A, Wekesa KS. Pregnancy block by MHC class I peptides is mediated via the production of inositol 1,4,5-trisphosphate in the mouse vomeronasal organ. J Exp Biol. 2007;210:1406–1412. doi: 10.1242/jeb.02753. [DOI] [PubMed] [Google Scholar]
- 64.He J, Ma L, Kim S, Nakai J, Yu CR. Encoding gender and individual information in the mouse vomeronasal organ. Science. 2008;320:535–538. doi: 10.1126/science.1154476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slev PR, Nelson AC, Potts WK. Sensory neurons with MHC-like peptide binding properties: disease consequences. Curr Opin Immunol. 2006;18:608–616. doi: 10.1016/j.coi.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 66.Hurst JL, Payne CE, Nevison CM, et al. Individual recognition in mice mediated by major urinary proteins. Nature. 2001;414:631–634. doi: 10.1038/414631a. [DOI] [PubMed] [Google Scholar]
- 67.Hurst JL, Thom MD, Nevison CM, Humphries RE, Beynon RJ. MHC odours are not required or sufficient for recognition of individual scent owners. Proc Biol Sci. 2005;272:715–724. doi: 10.1098/rspb.2004.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheetham SA, Thom MD, Jury F, Ollier WE, Beynon RJ, Hurst JL. The genetic basis of individual-recognition signals in the mouse. Curr Biol. 2007;17:1771–1777. doi: 10.1016/j.cub.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 69.Penn D, Potts W. Untrained mice distinguish MHC-determined odors. Physiol. Behav. 1998;64:235–243. doi: 10.1016/s0031-9384(98)00052-3. [DOI] [PubMed] [Google Scholar]
- 70.Manning CJ, Potts WK, Wakeland EK, Dewsbury DA. What’s wrong with MHC mate choice experiments. In: Doty RL, Müller-Schwarze D, editors. Chemical Signals in Vertebrates. VI. New York, N.Y: Plenum; 1992. pp. 229–235. [Google Scholar]
- 71.Hamilton WD. The genetical evolution of social behaviour I. II. J Theor Biol. 1964;7:1–52. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 72.Mateo JM. Kin-recognition abilities and nepotism as a function of sociality. Proc Biol Sci. 2002;269:721–727. doi: 10.1098/rspb.2001.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamazaki K, Beauchamp GK, Curran M, Bard J, Boyse EA. Parent-progeny recognition as a function of MHC odortype identity. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10500–10502. doi: 10.1073/pnas.180320997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown JL. Some paradoxical goals of cells and organisms: the role of the MHC. In: Pfaff DW, editor. Ethical Questions in Brain and Behavior. New York: Springer Verlag; 1983. pp. 111–124. [Google Scholar]
- 75.Penn D, Potts W. MHC-disassortative mating preferences reversed by cross-fostering. Proc. R. Soc. London. B. 1998d;265:1299–1306. doi: 10.1098/rspb.1998.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Potts WK, Wakeland EK. Evolution of MHC genetic diversity: a tale of incest, pestilence and sexual preference. Trends Genet. 1993;9:408–412. doi: 10.1016/0168-9525(93)90103-o. [DOI] [PubMed] [Google Scholar]
- 77.Paterson S, Hurst JL. How effective is recognition of siblings on the basis of genotype. J Evol Biol. 2009;22:1875–1881. doi: 10.1111/j.1420-9101.2009.01796.x. [DOI] [PubMed] [Google Scholar]
- 78.Sato A, Figueroa F, Murray BW, et al. Nonlinkage of major histocompatibility complex class I and class II loci in bony fishes. Immunogenetics. 2000;51:108–116. doi: 10.1007/s002510050019. [DOI] [PubMed] [Google Scholar]
- 79.Liu Y, Kasahara M, Rumfelt LL, Flajnik MF. Xenopus class II A genes: studies of genetics, polymorphism, and expression. Dev Comp Immunol. 2002;26:735–750. doi: 10.1016/s0145-305x(02)00034-4. [DOI] [PubMed] [Google Scholar]
- 80.Sherborne AL, Thom MD, Paterson S, et al. The genetic basis of inbreeding avoidance in house mice. Curr Biol. 2007;17:2061–2066. doi: 10.1016/j.cub.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown GE, Brown JA. Do kin always make better neighbours? The effects of territory quality. Behavioral Ecology & Sociobiology. 1993;33:225–231. [Google Scholar]
- 82.Johnston RE, Muller-Schwartz D, Sorensen PW. Advances in Chemical Signals in Vertebrates. New York: Klewer/Plenum; 1999. [Google Scholar]
- 83.Lynch CB. Inbreeding effects upon animals derived from a wild population of Mus musculus. Evolution. 1977;31:526–537. doi: 10.1111/j.1558-5646.1977.tb01042.x. [DOI] [PubMed] [Google Scholar]
- 84.Connor JL, Belucci MJ. Natural selection resisting inbreeding depression in captive wild house mice (Mus musculus) Evolution. 1979;33:929–940. doi: 10.1111/j.1558-5646.1979.tb04747.x. [DOI] [PubMed] [Google Scholar]
- 85.Meagher S, Penn DJ, Potts WK. Male-male competition magnifies inbreeding depression in wild house mice. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3324–3329. doi: 10.1073/pnas.060284797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ilmonen P, Penn DJ, Damjanovich K, et al. Experimental infection magnifies inbreeding depression in house mice. Journal of Evolutionary Biology. 2008;21:834–841. doi: 10.1111/j.1420-9101.2008.01510.x. [DOI] [PubMed] [Google Scholar]
- 87.Neff BD, Pitcher TE. Genetic quality and sexual selection: an integrated framework for good genes and compatible genes. Mol Ecol. 2005;14:19–38. doi: 10.1111/j.1365-294X.2004.02395.x. [DOI] [PubMed] [Google Scholar]
- 88.Wedekind C, Chapuisat M, Macas E, Rülicke T. Non-random fertilization in mice correlates with MHC and something else. Heredity. 1996;77:400–409. doi: 10.1038/hdy.1996.160. [DOI] [PubMed] [Google Scholar]
- 89.Rülicke T, Chapuisat M, Homberger FR, Macas E, Wedekind C. MHC-genotype of progeny influenced by parental infection. Proc. R. Soc. Lond. B. 1998;265:711–716. doi: 10.1098/rspb.1998.0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- 91.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibity complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 92.Hill AV, Allsopp CE, Kwiatkowski D, et al. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 93.Thursz MR, Thomas HC, Greenwood BM, Hill AV. Heterozygote advantage for HLA class-II type in hepatitis B virus infection. Nat. Genet. 1997;17:11–12. doi: 10.1038/ng0997-11. [DOI] [PubMed] [Google Scholar]
- 94.Carrington M, Nelson GW, Martin MP, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 95.Arkush KD, Giese AR, Mendonca HL, McBride AM, Marty GD, Hedrick PW. Resistance to three pathogens in the endangered winter-run chinook salmon (Oncorhynchus tshawytscha): effects of inbreeding and major histocompatibility complex genotypes. Canadian Journal of Fisheries and Aquatic Science. 2002;59:966–975. [Google Scholar]
- 96.Penn DJ, Damjanovich K, Potts WK. MHC heterozygosity confers a selective advantage against multiple- strain infections. Proc Natl Acad Sci U S A. 2002;99:11260–11264. doi: 10.1073/pnas.162006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Froeschke G, Sommer S. MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the Southern Kalahari. Molecular Biology and Evolution. 2005;22:1254–1259. doi: 10.1093/molbev/msi112. [DOI] [PubMed] [Google Scholar]
- 98.Hraber P, Kuiken C, Yusim K. Evidence for human leukocyte antigen heterozygote advantage against hepatitis C virus infection. Hepatology. 2007;46:1713–1721. doi: 10.1002/hep.21889. [DOI] [PubMed] [Google Scholar]
- 99.McClelland EE, Penn DJ, Potts WK. Major Histocompatibility Complex Heterozygote Superiority during Coinfection. Infect Immun. 2003;71:2079–2086. doi: 10.1128/IAI.71.4.2079-2086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kekalainen J, Vallunen JA, Primmer CR, Rattya J, Taskinen J. Signals of major histocompatibility complex overdominance in a wild salmonid population. Proc Biol Sci. 2009;276:3133–3140. doi: 10.1098/rspb.2009.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oliver MK, Telfer S, Piertney SB. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris) Proc Biol Sci. 2009;276:1119–1128. doi: 10.1098/rspb.2008.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Penn D, Potts W. The evolution of mating preferences and major histocompatibility genes. Am. Nat. 1999;153:145–164. doi: 10.1086/303166. [DOI] [PubMed] [Google Scholar]
- 103.Bertoletti A, Sette A, Chisari FV, et al. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells [see comments] Nature. 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 104.Moskophidis D, Zinkernagel RM. Immunobiology of cytotoxic T-cell escape mutants of lymphocytic choriiomeningitis virus. J. Viriol. 1995;69:2187–2193. doi: 10.1128/jvi.69.4.2187-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McMichael AJ, Phillips RE. Escape of human immunodeficiency virus from immune control. Annu Rev Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- 106.Jeffery KJ, Usuku K, Hall SE, et al. HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-I-associated myelopathy. Proc Natl Acad Sci U S A. 1999;96:3848–3853. doi: 10.1073/pnas.96.7.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Allen TM, O’Connor DH, Jing P, et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 108.Bowen DG, Walker CM. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J Exp Med. 2005;201:1709–1714. doi: 10.1084/jem.20050808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Furukawa Y, Tara M, Izumo S, Arimura K, Osame M. HTLV-I viral escape and host genetic changes in the development of adult T cell leukemia. Int J Cancer. 2006;118:381–387. doi: 10.1002/ijc.21328. [DOI] [PubMed] [Google Scholar]
- 110.Goulder PJ, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 111.McClelland EE, Granger DL, Potts WK. MHC-dependent susceptibility to Cryptococcus neoformans in mice. Infection and Immunity. 2003;71:4815–4817. doi: 10.1128/IAI.71.8.4815-4817.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Buseyne F, Janvier G, Teglas JP, et al. Impact of heterozygosity for the chemokine receptor CCR5 32-bp-deleted allele on plasma virus load and CD4 T lymphocytes in perinatally human immunodeficiency virus-infected children at 8 years of age. J Infect Dis. 1998;178:1019–1023. doi: 10.1086/515660. [DOI] [PubMed] [Google Scholar]
- 113.Misrahi M, Teglas JP, N’Go N, et al. CCR5 chemokine receptor variant in HIV-1 mother-to-child transmission, disease progression in children French Pediatric HIV Infection Study Group. Jama. 1998;279:277–280. doi: 10.1001/jama.279.4.277. [DOI] [PubMed] [Google Scholar]
- 114.MacDonald KS, Embree J, Njenga S, et al. Mother-child class I HLA concordance increases perinatal human immunodeficiency virus type 1 transmission. J Infect Dis. 1998;177:551–556. doi: 10.1086/514243. [DOI] [PubMed] [Google Scholar]
- 115.Polycarpou A, Ntais C, Korber BT, et al. Association between maternal and infant class I and II HLA alleles and of their concordance with the risk of perinatal HIV type 1 transmission. AIDS Res Hum Retroviruses. 2002;18:741–746. doi: 10.1089/08892220260139477. [DOI] [PubMed] [Google Scholar]
- 116.Woelfing B, Traulsen A, Milinski M, Boehm T. Does intra-individual major histocompatibility complex diversity keep a golden mean. Philos Trans R Soc Lond B Biol Sci. 2009;364:117–128. doi: 10.1098/rstb.2008.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wegner KM, Kalbe M, Kurtz J, Reusch TB, Milinski M. Parasite selection for immunogenetic optimality. Science. 2003;301:1343. doi: 10.1126/science.1088293. [DOI] [PubMed] [Google Scholar]
- 118.Kalbe M, Eizaguirre C, Dankert I, et al. Lifetime reproductive success is maximized with optimal major histocompatibility complex diversity. Proc Biol Sci. 2009;276:925–934. doi: 10.1098/rspb.2008.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reusch TB, Haberli MA, Aeschlimann PB, Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001;414:300–302. doi: 10.1038/35104547. [DOI] [PubMed] [Google Scholar]
- 120.Boehm T. Quality control in self/nonself discrimination. Cell. 2006;125:845–858. doi: 10.1016/j.cell.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 121.Zahavi A. Mate selection-a selection for a handicap. J. theor. Biol. 1975;53:205–214. doi: 10.1016/0022-5193(75)90111-3. [DOI] [PubMed] [Google Scholar]
- 122.Ditchkoff SS, Lochmiller RL, Masters RE, Hoofer SR, Van Den Bussche RA. Major-histocompatibility-complex-associated variation in secondary sexual traits of white-tailed deer (Odocoileus virginianus): evidence for good-genes advertisement. Evolution. 2001;55:616–625. doi: 10.1554/0014-3820(2001)055[0616:mhcavi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 123.Von-Schantz THWGGMGKP. MHC genotype and male ornamentation: genetic evidence for the Hamilton-Zuk model. Proc R Soc Lond B. 1996;263:265–271. doi: 10.1098/rspb.1996.0041. [DOI] [PubMed] [Google Scholar]
- 124.Ekblom R, Saether SA, Grahn M, Fiske P, Kalas JA, Hoglund J. Major histocompatibility complex variation and mate choice in a lekking bird, the great snipe (Gallinago media) Mol Ecol. 2004;13:3821–3828. doi: 10.1111/j.1365-294X.2004.02361.x. [DOI] [PubMed] [Google Scholar]
- 125.Milinski M, Griffiths SW, Reusch TB, Boehm T. Costly major histocompatibility complex signals produced only by reproductively active males, but not females, must be validated by a ‘maleness signal’ in three-spined sticklebacks. Proc Biol Sci. 2009 doi: 10.1098/rspb.2009.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zuk M. The role of parasites in sexual selection: current evidence and future directions. Adv.Study Behav. 1992;21:39–68. [Google Scholar]
- 127.Penn D, Potts WK. Chemical signals and parasite-mediated sexual selection. Trends Ecol. Evol. 1998;13:391–396. doi: 10.1016/s0169-5347(98)01473-6. [DOI] [PubMed] [Google Scholar]
- 128.Kavaliers M, Choleris E, Agmo A, Pfaff DW. Olfactory-mediated parasite recognition and avoidance: linking genes to behavior. Hormones and Behavior. 2004;46:272–283. doi: 10.1016/j.yhbeh.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 129.Moller AP, Saino N. Parasites, immunology of hosts, and host sexual selection. J Parasitol. 1994;80:850–858. [PubMed] [Google Scholar]
- 130.Boehm T. Co-evolution of a primordial peptide-presentation system and cellular immunity. Nat Rev Immunol. 2006;6:79–84. doi: 10.1038/nri1749. [DOI] [PubMed] [Google Scholar]
- 131.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 133.Guo P, Hirano M, Herrin BR, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scofield VL, Schlumpberger JM, West LA, Weissman IL. Protochordate allorecognition is controlled by a Mhc-like gene system. Nature. 1982;295:499–502. doi: 10.1038/295499a0. [DOI] [PubMed] [Google Scholar]
- 135.Grosberg RK, Hart MW. Mate selection and the evolution of highly polymorphic self/nonself recognition genes. Science. 2000;289:2111–2114. doi: 10.1126/science.289.5487.2111. [DOI] [PubMed] [Google Scholar]
- 136.De Tomaso AW, Nyholm SV, Palmeri KJ, et al. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.De Tomaso AW, Weissman IL. Evolution of a protochordate allorecognition locus. Science. 2004;303:977. doi: 10.1126/science.1094952. [DOI] [PubMed] [Google Scholar]
- 138.Khalturin K, Bosch TC. Self/nonself discrimination at the basis of chordate evolution: limits on molecular conservation. Curr Opin Immunol. 2007;19:4–9. doi: 10.1016/j.coi.2006.11.001. [DOI] [PubMed] [Google Scholar]