Abstract
Coinfections with common bacterial respiratory pathogens and influenza viruses are well-known causes of disease, often via synergistic interactions between the influenza virus, the bacteria, and the human host. However, relatively little is known about interactions between atypical bacteria and influenza viruses. A recent report by Reinton et al. explored this issue by analyzing data from 3,661 patients seeking medical assistance for the presence of Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Bordetella pertussis, as well as influenza A or B virus in nasal swab specimens. The report, however, did not accurately assess the epidemiologic interactions of these pathogens. We aimed to describe the interactions between these bacterial species and influenza infections. Strong and highly statistically significant antagonistic interspecies interactions were detected between C. pneumoniae and influenza virus [odds ratio (OR): 0.09; p<0.0001) and M. pneumoniae and influenza virus infections (OR: 0.29; p=0.003). No association was detected between B. pertussis and influenza infection (p=0.34), contrary to the initial report, and coinfection was not detected at a higher-than-by-chance frequency within the population. Further support of these results is supplied by the analysis of two earlier investigations reporting data on influenza virus and these atypical bacteria. Our results supplement the large body of literature regarding interactions between influenza virus and typical respiratory pathogens, providing a fuller picture of the spectrum of interactions between influenza viruses and respiratory bacteria. Further, we demonstrate the importance of choosing the most appropriate reference populations for the analysis being performed and describe the pitfalls that may occur when care is not taken in this regard.
Introduction
The traditional view of infectious diseases focuses on single infections. However, simultaneous infection with multiple pathogen species is increasingly recognized as both common and important for disease manifestation, as well as treatment. In the upper respiratory tract, where exposure to microbial species is common, interactions between pathogens, often mediated by the host, are particularly relevant. Owing in large part to the devastating role of bacteria–influenza coinfections during the 1918 influenza pandemic, the most well-known, and perhaps well-studied, example of such interspecies interactions in the respiratory tract is that between bacterial pathogens—including Streptococcus pneumoniae, Streptococcus pyogenes, Staphylococcus aureus, and Haemophilus influenzae—and influenza viruses [1, 2]. These coinfections remain important today, highlighted by the excess mortality associated with bacterial infections during recent influenza epidemics and pandemics [3, 4]. While coinfections between influenza viruses and the above-mentioned bacteria are particularly salient, numerous investigations have aimed to quantify the prevalence and describe the clinical importance of coinfections between influenza and atypical bacteria, such as Klebsiella pneumoniae, Enterococcus, Mycobacterium tuberculosis, Mycoplasma and Chlamydophila pneumoniae, and Bordetella pertussis, among others [5–8].
Recently, Reinton et al. [9] set out to characterize the prevalence of coinfection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, or Bordetella pertussis with influenza A or B viruses to determine whether any of these atypical bacterial infections have antagonistic or synergistic effects on influenza virus infection, or vice versa. The authors used polymerase chain reaction (PCR) to detect bacterial DNA and influenza RNA in nasal swab specimens collected from patients seen for respiratory illness by primary care physicians in Oslo, Norway, during a 2011 M. pneumoniae epidemic. Out of an original sample of 26,039 patients tested for M. pneumoniae, C. pneumoniae, and B. pertussis, 3,661 were also tested for influenza A or B. The authors found that the odds of detection of influenza virus RNA in patients positive for B. pertussis was significantly greater than the odds of detecting influenza virus in patients positive for M. pneumoniae [odds ratio (OR): 2.6, 95 % confidence interval (CI) [1.5–4.9]; p<0.005] and concluded that the “presence of B. pertussis DNA and influenza virus RNA [were detected together] at a higher-than-by-chance frequency in [the] population.”
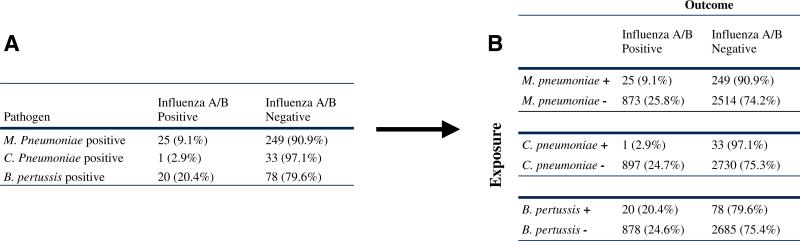
However, Reinton et al.'s original analyses were performed only in patients positive for each bacterial species (see Fig. 1a) and did not include certain necessary reference groups (i.e., bacteria-uninfected, influenza-negative individuals). These negative–negative reference groups are required in order to complete the relevant 2 × 2 contingency tables for this question (Fig. 1b), and without them, it is impossible to assess the effect of the presence of each bacterium on the detection of influenza, or to determine whether bacteria and influenza were detected together at a higher- or lower-than-by-chance frequency in the population.
Fig. 1.
Transformation of the contingency table for bacteria–influenza interactions. a Contingency table reported by Reinton et al. [9]. b Updated 2 × 2 contingency table (based on data from Reinton et al. [9]) to assess associations between atypical bacteria and influenza virus
When assessing whether an exposure (e.g., bacteria) affects the frequency of an outcome (e.g., influenza) in a population, we aim to answer the question “what difference would we have seen in the outcome if, instead of being exposed, the population had been unexposed?” Therefore, it is critically important to correctly define the unexposed, or reference, population. To conclude, for example, that B. pertussis DNA and influenza virus RNA are detected together at an increased frequency in a population implies an analysis against a B. pertussis-uninfected reference group. Here, we aimed to properly determine whether influenza virus RNAwas detected together with bacterial DNA from any of the three pathogens B. pertussis, M. pneumoniae, or C. pneumoniae at a higher- or lower-than-by-chance in the population. Put differently, we aimed to determine if the carriage of any of these atypical bacterial species affects the likelihood of detection of viral RNA.
Methods
Data for the primary analyses were initially identified and retrieved from a previously published report by Reinton et al. [9]. To find supporting data, we searched the literature for studies that collected presence/absence data on any of the three atypical bacterial pathogens M. pneumoniae, C. pneumoniae, or B. pertussis and influenza virus, and selected reports with sufficient power to detect interactions. Two studies were identified for which these data were collected with moderate to sufficient sample sizes: an analysis by Lieberman et al. from 1998 which included all three of the bacteria detected by Reinton et al., but is slightly underpowered for the statistical assessment of interspecies interactions, and a study by Renner et al. from 1983 that included only M. pneumoniae and influenza infections, but was well powered for an analysis of pathogen interaction.
Data were placed into respective 2 × 2 configurations as demonstrated in Fig. 1b and Fisher's exact test was used to calculate ORs, 95 % CIs, and probabilities of type I error. All statistical analyses were performed within the open-source R computing environment [10].
Results
Utilizing the full population for which data on the presence/absence of each bacterium and influenza virus were available (n=3,661), we re-analyzed the data from Reinton et al. with complete 2 × 2 contingency tables (Fig. 1b) and calculated, for each of the three bacterial species studied (B. pertussis, M. pneumoniae, C. pneumoniae), an OR for the effect of the presence of each bacterium on the detection of influenza RNA (Table 1). Contrary to claims made in the initial report, B. pertussis was not associated with the detection of influenza RNA (OR: 0.78, 95 % CI [0.48–1.29]; p=0.34), demonstrated clearly by the fact that there was no significant difference in the prevalence of influenza in B. pertussis-positive versus B. pertussis-negative individuals (20.4 % vs. 24.6 %; p= 0.34; Fig. 1b) in this population. On the other hand, the prevalence of influenza infection was highly significantly decreased in the presence (vs. absence) of infection with either M. pneumoniae or C. pneumoniae. Specifically, the prevalence of influenza in those infected with M. pneumoniae was only 9.1 %, much less than half the prevalence of influenza in M. pneumoniae-uninfected individuals (25.8 %; OR: 0.29, 95 % CI [0.19–0.44]; p<0.0001), and the prevalence of influenza among those infected with C. pneumoniae was only 2.9 %, compared to a nearly ten-fold higher prevalence of influenza among those without C. pneumoniae infection (24.7 %; OR: 0.09, 95 % CI [0.01–0.68]; p=0.003). Thus, while current infection with B. pertussis was not significantly associated with the detection of influenza virus RNA, current infection with M. or C. pneumoniae did significantly reduce the risk of detection of viral RNA in individuals from Reinton et al.'s Norwegian population by 64.7 % and 88.2 %, respectively.
Table 1.
Detection of influenza in bacteria-positive vs. bacteria-negative samples
| Odds ratio [95 % CI] | p-Value | Study supplying data | |
|---|---|---|---|
| M. pneumoniae (pos. vs. neg.) | 0.29 [0.19-0.44] | <0.0001 | Reinton et al. |
| 0.30 [0.03-1.47] | 0.143 | Lieberman et al. | |
| 0.07 [0.002-0.41] | <0.0001 | Renner et al. | |
| C. pneumoniae (pos. vs. neg.) | 0.09 [0.01-0.68] | 0.003 | Reinton et al. |
| 0.60 [0.16-1.89] | 0.45 | Lieberman et al. | |
| B. pertussis (pos. vs. neg.) | 0.78 [0.48-1.29] | 0.34 | Reinton et al. |
| 0.00 [0.00-1.06] | 0.056 | Lieberman et al. |
To further investigate these interspecies interactions, we analyzed previous reports from the literature that described and were of sufficient power to assess interactions between any of these three atypical bacterial pathogens and influenza virus (we identified only two papers that fit these criteria; see the Methods section and Supplementary Table 1 for the data analyzed). In a study by Lieberman et al., we found a >50 % reduction in the detection of influenza virus in M. pneumoniae-infected versus uninfected individuals (13 % vs. 34 %, respectively; OR: 0.30), which is in strong agreement with our analyses of Reinton et al.'s data (OR: 0.29); however, because the study was not powered for this particular analysis, the effect did not reach statistical significance (95 % CI [0.03–1.47]; p=0.143). Analysis of the data from Lieberman et al. also suggested a trend towards fewer influenza virus infections in C. pneumoniae-infected versus uninfected individuals, again in agreement with our analysis of Reinton et al.'s data, although the study was even less well powered for this analysis (p=0.45). Interestingly, also embedded in Lieberman et al.'s data was a strong trend towards reduced influenza infections among individuals infected (n=9) as compared to those uninfected (n=113) with B. pertussis (0 % vs. 33 %, respectively; 95 % CI [0.00–1.06]; p=0.056), further supporting our assessment that B. pertussis and influenza coinfection did not occur at a higher-than-expected frequency in the Norwegian population sampled by Reinton et al. A third study, published by Renner et al. [11], did provide data on M. pneumoniae and influenza, and was sufficiently powered to detect significant differences in the influenza infection status in M. pneumoniae-infected versus uninfected individuals; re-analysis of this data also demonstrated that influenza infection was highly significantly decreased in M. pneumoniae-infected as compared to M. pneumoniae-infected individuals (0.8 % vs. 10.2 %; OR 0.07, 95 % CI [0.002–0.405]; p<0.0001).
Discussion
Of primary interest here was a re-analysis of data from a recent report by Reinton et al. documenting the infection status of three important atypical bacterial respiratory pathogens, M. pneumoniae, C. pneumoniae, and B. pertussis, and influenza virus in 3,661 individuals during a recent M. pneumoniae epidemic in Norway in 2011. Contrary to the initial report, which, as described, employed inappropriate reference groups for the particular research question at hand, we found highly statistically significant evidence of strong antagonistic interactions between influenza virus infection status and the presence of M. pneumoniae (OR: 0.29) or C. pneumoniae (OR: 0.09) and, also in contrast to the conclusions made in the initial report by Reinton et al., no evidence of interaction between B. pertussis and influenza virus.
While a number of other studies have evaluated the prevalence of coinfection between influenza virus and these atypical bacteria, most report only the numbers of coinfections in a subset of the sample population (e.g., bacterial coinfections only in influenza-infected patients) [12–15]. Such studies are important to address the question of the prevalence of coinfection given a certain disease state, but often do not provide the necessary data (i.e., double-negative reference populations) to assess the association between the bacterial infections and the detection of influenza virus. Some studies that do report groups sufficient to complete a 2 × 2 table for exposure (i.e., atypical bacterial infection status) and outcome (i.e., influenza status) seem to be in agreement with our analysis of Reinton et al.'s data reported herein; however, because these studies did not have coinfection as a primary concern, they were largely underpowered to detect statistically significant differences [13, 14]. For example, our re-analyses of Lieberman et al.'s and Renner et al.'s data were in agreement with our re-analysis of Reinton et al.'s data in demonstrating antagonistic interactions between both M. pneumoniae or C. pneumoniae and influenza virus detection.
The mechanisms underlying the observed reductions described here of influenza virus detection in M. pneumoniae-infected and C. pneumoniae-infected patients, as compared to their bacteria-uninfected counterparts, have not been directly investigated. However, the antagonistic relationship between M. or C. pneumoniae and influenza virus infection (and the lack of association of B. pertussis with influenza virus infection) could be explained by the innate immune pathways activated by the detection of each of these pathogens, in particular those triggered by Toll-like receptor (TLR)-mediated recognition. Specifically, while the innate immune response to B. pertussis is primarily TLR-4-mediated [16], the responses to both M. pneumoniae [17, 18] and C. pneumoniae [19] are driven in large part by TLR-2-mediated pathways. This distinction is important in the context of our analysis, as proper clearance of influenza is also known to rely heavily on TLR-2 recognition [20, 21]. These commonalities in the innate immune recognition of influenza virus and M. and C. pneumoniae could abrogate coinfections with these pathogens and explain the antagonistic effects described here. Similarly, the divergent pathways utilized for the detection of B. pertussis and influenza virus could explain the lack of association found between B. pertussis status and influenza infection.
We hope that this report serves two purposes. Firstly, using data from Reinton et al., we describe an important and generally unrecognized antagonism between two atypical bacterial pathogens (i.e., M. pneumoniae and C. pneumoniae) and influenza virus infection in a notably large sample population. In light of the often synergistic interactions between influenza virus and more common bacterial pathogens, such as S. pneumoniae and S. aureus, the antagonistic relationships described herein elucidate a fuller understanding of the potential spectrum of interactions between influenza viruses and respiratory bacterial pathogens. We also clearly demonstrate that no association exists between infection with B. pertussis and influenza virus.
Secondly, we hope to highlight the importance of choosing the most appropriate analytic approach for the research question at hand. In this particular case, by utilizing the full data set provided by Reinton et al., including 3,661 cases with complete bacterial and influenza virus statuses, rather than only the cases positive for bacteria as selected by Reinton et al., we were able to calculate ORs to address the association between the presence of each bacterium and influenza virus (which may reflect a causal effect). While Reinton et al. described the relative frequency of the detection of influenza virus in B. pertussis-infected versus M. pneumoniae-infected patients, our re-analysis with complete 2 × 2 tables provides important information on whether influenza virus RNA was detected together with each bacterium at a frequency greater-than, less-than, or no-different-than that which would be expected in the population given no association between the pathogens.
As the threat of infectious disease pandemics becomes increasingly relevant, and our understanding of multispecies interactions grows, along with our capacity to detect and analyze such interactions, it is critical to ensure that we utilize data to their full potential, including selecting the most appropriate and informative analytic methods for the research questions posed.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10096-014-2120-0) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflicts of interest.
Contributor Information
M. J. Mina, Department of Ecology and Evolutionary Biology, Princeton University, Princeton, NJ 08544, USA Department of Global Health, Emory University Rollins School of Public Health, 1518 Clifton Road, Atlanta, GA 30322, USA.
R. M. Burke, Department of Epidemiology, Emory University Rollins School of Public Health, 1518 Clifton Road, Atlanta, GA 30322, USA
K. P. Klugman, Department of Global Health, Emory University Rollins School of Public Health, 1518 Clifton Road, Atlanta, GA 30322, USA Department of Epidemiology, Emory University Rollins School of Public Health, 1518 Clifton Road, Atlanta, GA 30322, USA.
References
- 1.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198(7):962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brundage JF, Shanks GD. Deaths from bacterial pneumonia during 1918–19 influenza pandemic. Emerg Infect Dis. 2008;14(8):1193–1199. doi: 10.3201/eid1408.071313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1)—United States, May–August 2009. MMWR Morb Mortal Wkly Rep. 2009;58(38):1071–1074. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC) Severe coinfection with seasonal influenza A (H3N2) virus and Staphylococcus aureus—Maryland, February–March 2012. MMWR Morb Mortal Wkly Rep. 2012;61(16):289–291. [PubMed] [Google Scholar]
- 5.Redford PS, Mayer-Barber KD, McNab FW, Stavropoulos E, Wack A, Sher A, O'Garra A. Influenza A virus impairs control of Mycobacterium tuberculosis coinfection through a type I interferon receptor-dependent pathway. J Infect Dis. 2014;209(2):270–274. doi: 10.1093/infdis/jit424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waki N, Yajima N, Suganuma H, Buddle BM, Luo D, Heiser A, Zheng T. Oral administration of Lactobacillus brevis KB290 to mice alleviates clinical symptoms following influenza virus infection. Lett Appl Microbiol. 2014;58(1):87–93. doi: 10.1111/lam.12160. [DOI] [PubMed] [Google Scholar]
- 7.Falsey AR, Becker KL, Swinburne AJ, Nylen ES, Formica MA, Hennessey PA, Criddle MM, Peterson DR, Baran A, Walsh EE. Bacterial complications of respiratory tract viral illness: a comprehensive evaluation. J Infect Dis. 2013;208(3):432–441. doi: 10.1093/infdis/jit190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Kaabi N, Solh Z, Pacheco S, Murray L, Gaboury I, Le Saux N. A comparison of group A Streptococcus versus Streptococcus pneumoniae pneumonia. Pediatr Infect Dis J. 2006;25(11):1008–1012. doi: 10.1097/01.inf.0000243198.63255.c1. [DOI] [PubMed] [Google Scholar]
- 9.Reinton N, Manley L, Tjade T, Moghaddam A. Respiratory tract infections during the 2011 Mycoplasma pneumoniae epidemic. Eur J Clin Microbiol Infect Dis. 2013;32(6):835–840. doi: 10.1007/s10096-013-1818-8. [DOI] [PubMed] [Google Scholar]
- 10.R Development Core Team . R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2012. [Google Scholar]
- 11.Renner ED, Helms CM, Johnson W, Tseng CH. Coinfections of Mycoplasma pneumoniae and Legionella pneumophila with influenza A virus. J Clin Microbiol. 1983;17(1):146–148. doi: 10.1128/jcm.17.1.146-148.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bezerra PG, Britto MC, Correia JB, Duarte Mdo C, Fonceca AM, Rose K, Hopkins MJ, Cuevas LE, McNamara PS. Viral and atypical bacterial detection in acute respiratory infection in children under five years. PLoS One. 2011;6(4):e18928. doi: 10.1371/journal.pone.0018928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pientong C, Ekalaksananan T, Teeratakulpisarn J, Tanuwattanachai S, Kongyingyoes B, Limwattananon C. Atypical bacterial pathogen infection in children with acute bronchiolitis in northeast Thailand. J Microbiol Immunol Infect. 2011;44(2):95–100. doi: 10.1016/j.jmii.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Lieberman D, Shvartzman P, Lieberman D, Ben-Yaakov M, Lazarovich Z, Hoffman S, Mosckovitz R, Ohana B, Leinonen M, Luffy D, Boldur I. Etiology of respiratory tract infection in adults in a general practice setting. Eur J Clin Microbiol Infect Dis. 1998;17(10):685–689. doi: 10.1007/s100960050161. [DOI] [PubMed] [Google Scholar]
- 15.Freymuth F, Vabret A, Brouard J, Toutain F, Verdon R, Petitjean J, Gouarin S, Duhamel JF, Guillois B. Detection of viral, Chlamydia pneumoniae and Mycoplasma pneumoniae infections in exacerbations of asthma in children. J Clin Virol. 1999;13(3):131–139. doi: 10.1016/S1386-6532(99)00030-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fransen F, Stenger RM, Poelen MC, van Dijken HH, Kuipers B, Boog CJ, van Putten JP, van Els CA, van der Ley P. Differential effect of TLR2 and TLR4 on the immune response after immunization with a vaccine against Neisseria meningitidis or Bordetella pertussis. PLoS One. 2010;5(12):e15692. doi: 10.1371/journal.pone.0015692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love W, Dobbs N, Tabor L, Simecka JW. Toll-like receptor 2 (TLR2) plays a major role in innate resistance in the lung against murine Mycoplasma. PLoS One. 2010;5(5):e10739. doi: 10.1371/journal.pone.0010739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu HW, Jeyaseelan S, Rino JG, Voelker DR, Wexler RB, Campbell K, Harbeck RJ, Martin RJ. TLR2 signaling is critical for Mycoplasma pneumoniae-induced airway mucin expression. J Immunol. 2005;174(9):5713–5719. doi: 10.4049/jimmunol.174.9.5713. [DOI] [PubMed] [Google Scholar]
- 19.Prebeck S, Kirschning C, Dürr S, da Costa C, Donath B, Brand K, Redecke V, Wagner H, Miethke T. Predominant role of toll-like receptor 2 versus 4 in Chlamydia pneumoniae-induced activation of dendritic cells. J Immunol. 2001;167(6):3316–3323. doi: 10.4049/jimmunol.167.6.3316. [DOI] [PubMed] [Google Scholar]
- 20.Tuvim MJ, Gilbert BE, Dickey BF, Evans SE. Synergistic TLR2/6 and TLR9 activation protects mice against lethal influenza pneumonia. PLoS One. 2012;7(1):e30596. doi: 10.1371/journal.pone.0030596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan AC, Mifsud EJ, Zeng W, Edenborough K, McVernon J, Brown LE, Jackson DC. Intranasal administration of the TLR2 agonist Pam2Cys provides rapid protection against influenza in mice. Mol Pharm. 2012;9(9):2710–2718. doi: 10.1021/mp300257x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.