Abstract
Bacillus fortis IAGS162 has been previously shown to induce systemic resistance in tomato plants against Fusarium wilt disease. In the first phase of current study, the ISR determinant was isolated from extracellular metabolites of this bacterium. ISR bioassays combined with solvent extraction, column chromatography and GC/MS analysis proved that phenylacetic acid (PAA) was the potential ISR determinant that significantly ameliorated Fusarium wilt disease of tomato at concentrations of 0.1 and 1 mM. In the second phase, the biochemical basis of the induced systemic resistance (ISR) under influence of PAA was elucidated by performing non-targeted whole metabolomics through GC/MS analysis. Tomato plants were treated with PAA and fungal pathogen in various combinations. Exposure to PAA and subsequent pathogen challenge extensively re-modulated tomato metabolic networks along with defense related pathways. In addition, various phenylpropanoid precursors were significantly up-regulated in treatments receiving PAA. This work suggests that ISR elicitor released from B. fortis IAGS162 contributes to resistance against fungal pathogens through dynamic reprogramming of plant pathways that are functionally correlated with defense responses.
Keywords: induced systemic resistance (ISR), Bacillus, tomato, fusarium wilt, phenylacetic acid
Introduction
During the long history of coevolution between host plants and pathogens, plants have developed their own strategies to combat with pathogens. Plant interactions with other organisms lead to cross-talk between signaling pathways that helps to activate different defense responses against pathogens (Beckers and Spoel, 2006). These interactions can eventually cross the border between the aerial parts of plants and the roots (Garcia-Brugger et al., 2006; Pieterse et al., 2009; van Dam, 2009). In case of plant-pathogen interactions, plants can produce immune signals from the infection sites (Ausubel, 2005; Jones and Dangl, 2006). These immune signals activate batteries of defense responses (Chisholm et al., 2006; Dodds and Rathjen, 2010).
Induced systemic resistance (ISR) is a great regulatory potential of plants that activates appropriate cellular defense responses before or upon pathogen attack. ISR activates plant immunity in a similar way as induced by pathogens or insects that are specifically directed against invaders in incompatible interactions (Conrath et al., 2002). ISR is accompanied with augmented expression of defense related genes, increased accumulation of secondary metabolites, and defense associated proteins (van Loon et al., 1998; Conrath, 2006; van Hulten et al., 2006; Zamioudis and Pieterse, 2012). These induced defense mechanisms are dependent on jasmonic acid (JA) and ethylene (ET) signaling in the plants (van Loon et al., 1998; Conrath et al., 2006). In addition, these altered traits directly and indirectly influence defense related mechanisms of host plants and their growth attributes. Finally, plants defend themselves against invading pathogens through a combination of induced and constitutive defenses that negatively affect pathogen performance. Another type of resistance mechanism in plants is termed as “Systemic acquired resistance” (SAR). SAR is associated with the perception of elicitors from avirulent pathogens (Thakur and Sohal, 2013). SAR is phenotypically similar to ISR, which is also effective against diverse pathogens. SAR is dependent on salicylic acid (SA) signaling pathway (Park et al., 2008) while ISR typically relies on the JA and ET signaling pathways (Pieterse et al., 2002).
Plant responses to infectious agents are mediated by recognition of microbial signaling molecules (Gómez-Vásquez et al., 2004; Garcia-Brugger et al., 2006; Conrath, 2011). These microbial signaling molecules associated with some beneficial microbes, are termed as microbe-associated molecular patterns (MAMPs) (Boller and Felix, 2009; Mishra et al., 2009). These MAMPs upon recognition initiate basal defense responses throughout the plant body (Montesano et al., 2003; Ryan and Pearce, 2003; Gómez-Vásquez et al., 2004). MAMPs mediated ISR is not related with direct activation of plant defense related genes, rather it implies quicker and stronger activation of some basal plant defense responses upon attack of a pathogen (Zipfel et al., 2004, 2006; Mishra et al., 2009). This MAMPs based resistance is effective enough to hinder infection and pathogen establishment inside plant body (Katagiri and Tsuda, 2010). Plant growth-promoting rhizobacteria can elicit ISR in plants by secreting MAMPs, which are also termed as ISR elicitors or determinants (van Loon et al., 1998; Bakker et al., 2003; Persello-Cartieaux et al., 2003; Lugtenberg and Kamilova, 2009). Along with MAMPs, recently some phytohormones have been found capable to elicit induced resistance in some plants. Some studies have revealed new insights into the role of auxins in plant defense (Kazan and Manners, 2009). Similar to the application of the defense-eliciting hormones, SA and JA, exogenous application of auxins can positively affect resistance against some pathogens (Sharaf and Farrag, 2004).
Several studies concerning the screening of bacterial strains capable to elicit ISR in plants have been conducted, yet relatively few studies have focused on the recognition of MAMPs produced by these bacteria and dynamic changes of metabolic responses in plants under influence of these MAMPs. Some secondary metabolites produced by bacteria have been recognized as ISR elicitors. These include as 4-aminocarbonyl phenylacetate secreted by Klebsiella oxytoca C1036 (Park et al., 2009), N-alkylated benzylamine derivative produced by Pseudomonas putida BTP1 (Ongena et al., 2005) and dimethyl disulfide produced extracellularly by Bacillus cereus C1L (Huang et al., 2012). Some bacteria produce volatile organic compounds as 2-3-butanediol and acetoin, which can trigger ISR in host plants upon recognition (Ryu et al., 2004).
Metabolomics is a promising analytical technology that has been used to unravel the metabolic fluctuations of plant molecules and the re-programing associated with various plant pathways (De Vos et al., 2007; Zhao et al., 2015). This technique is considered one of the most rapidly growing areas of modern science. The transcriptional or protein profile cannot be directly linked with metabolic changes (Zhao et al., 2015). We previously proved that B. fortis IAGS162 is a beneficial rhizospheric bacterium capable of managing fusarium wilt disease of tomato by ISR phenomenon in tomato plants (Akram et al., 2013). This study reports phenylacetic acid (PAA) as an ISR elicitor that is secreted extracellularly by this beneficial bacterial strain. It also describes significant metabolic rerouting in the plant pathways under influence of this ISR elicitor. Moreover the compatible host pathogen interactions have been characterized by a lower level of certain defense-related mechanisms compared with host pathogen interactions in the presence of ISR elicitor that leads to a more dynamic metabolic response over the course of colonization. We have also discussed the potential relevance of these changes in host defense responses.
Methodologies
Microbial Treatments Preparation
The tomato wilt pathogen “F. oxysporum f.sp. lycopersici” was provided by Fungal biotechnology lab, Institute of Agricultural sciences, University of the Punjab, Lahore. This fungus was grown on potato dextrose agar (PDA Difco) for 1 week. Conidia were harvested by scraping and conidial suspension was prepared in sterilized water at a concentration of 105 conidia/ml. ISR capable bacterial strain Bacillus fortis IAGS162 (Akram et al., 2013) was cultured in Luria Bertani (LB) broth media overnight at 35°C. Cell free culture filtrate (CFCF) was obtained by centrifugation at 4000 g for 20 min and used for ISR assay. Bacterial pallet was re-suspended in sterilized distilled water to a final concentration of 104 cfu/ml for application. Intracellular metabolites were extracted by sonicating the bacterial cell suspension at resonance amplitude for 30 s to obtain intra-cellular components.
Preliminary Screening of ISR Determinants from B. fortis IAGS162
This experiment was performed for preliminary screening of ISR determinant/s either present in intracellular metabolites or CFCF of B. fortis IAGS162 against fusarium wilt, as previously described by Akram et al. (2015). Briefly, tomato plants of fusarium susceptible variety were raised from sterilized seeds, inside plastic pots containing sterilized commercial potting mix. Fourteen days after emergence, tomato plants were treated with 50 ml of intra-cellular metabolites and CFCF of B. fortis IAGS162 separately. Here plants in positive control were provided with 50 ml of a water based formulation of B. fortis IAGS162, while the water treated control plants got 50 ml of distilled water. Three days after treatments with abovementioned substances, the wilt pathogen was provided by adding 50 ml of the F. oxysporum conidial suspension. The pots were incubated in a greenhouse for 15 days under natural day light conditions. Each treatment had five replicate plants, and experiment was performed twice. To determine the DI, wilting was scored based on the criteria developed by Epp (1987). The equation described by Cachinero et al. (2002) was used to calculate the DI.
| (1) |
where, ni = the number of diseased plants, si = value of the disease score, N = the total number of plants observed, and S = maximum rank of disease score.
Isolation of ISR Determinants from CFCF of B. fortis IAGS162
Cell free culture filtrate of B. fortis IAGS162 was prepared as described previously and extracted twice with series of organic solvents (Figure 1). All the organic extracts were thoroughly dried, then dissolved in 10% dimethyl sulfoxide (DMSO). These were amended in Murashige and Skoog (MS) medium at a concentration of 0.1%, and ISR bioassay was performed as described below. The extract with the positive ISR activity was partitioned into sub-fractions by performing silica gel column chromatography by stepwise elution method by using methanol and ethyl acetate. These sub-fractions were dissolved in 10% DMSO after drying, and 10 μl of each DMSO based formulation was again used in ISR bioassay.
FIGURE 1.
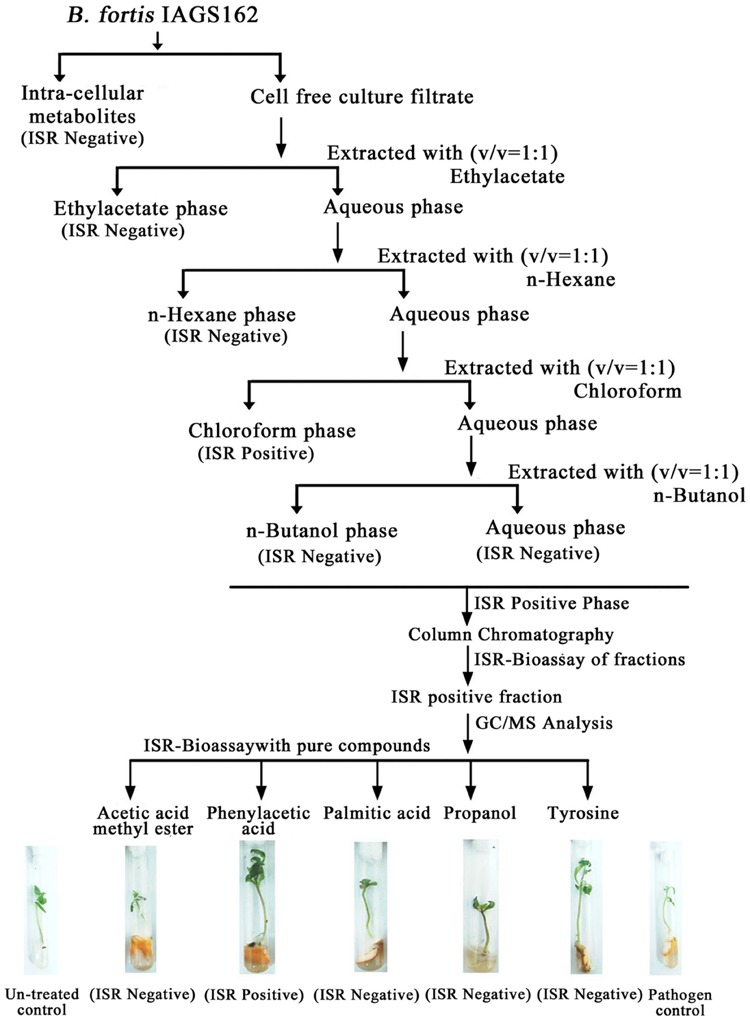
Extraction procedure and ISR activity of crude and purified metabolites from B. fortis IAGS162.
The sub-fraction showing ISR activity was subjected to GC/MS analysis as described by Akram et al. (2015). The compounds identified by GC/MS analysis, present in the ISR-active sub-fraction were purchased from Sigma–Aldrich. ISR bioassay was again performed by adding these pure compounds in MS media at three varying concentrations viz: 0.01, 0.1, and 1 mM.
ISR Bioassay
For ISR bioassays and metabolomic analysis, tomato plants of fusarium susceptible variety were grown in MS medium inside glass culture tubes in a growth chamber for seedling development (25°C, 16 h light). At the age of 5 days, growth media was added with different elicitor preparations that were to be tested for ISR activity. Two days after elicitor application, tomato plants were treated by adding 10 μL of the pathogen inoculum in the form of an aqueous spore suspension at a concentration of 105 conidia/ml. Tomato plants were incubated under same conditions as indicated above. DI was recorded after 1 week of incubation, as described earlier. The experiment included 10 replicates of each treatment and was repeated twice.
Tomato Metabolome Analysis for Elucidation of PAA Mediated ISR Mechanism
Another independent test tube bioassay was performed to elucidate PAA mediate ISR process in tomato plants against fusarium wilt disease as described above. This experiment included four treatments: untreated control plants, pathogen challenged plants, PAA (0.01 mM) treated plants, PAA (0.01 mM) + pathogen treated plants. After 1 week of treatment applications, shoot samples of plants were taken for GC/MS analysis. The metabolite extraction process was carried out by using a single-phase solvent (chloroform/methanol/water). This solvent system has been developed to recover a wide range of metabolites (Catchpole et al., 2005; Beckmann et al., 2007). Three replicate shoot samples, obtained from five plants of two independent experiments, were used in this GC-MS analysis.
Metabolomic Analysis
Methodology proposed by Lisec et al. (2006) was used to perform tomato metabolome analysis. Metabolites were identified by comparing their spectral similarity in the NIST library. Metabolite levels were determined using the Mzmine software package1. Obtained values were log10-transformed (Steinfath et al., 2008) and normalized to show identical medium peak sizes per sample group. Statistical analyses and graphical representations were performed using DSAASTAT and the ClustVis: a web based multivariate data analysis tool. The PCA analysis was performed using the ‘bpca’ algorithm in ClustVis online tool2. The metabolite data were summarized using the heatmap function in ClustVis tool with row wise scaling and correlation-based clustering.
Statistical Analysis
The data were analyzed by performing analysis of variance (ANOVA), and the significance of treatment was determined by Duncan’s new multiple range test (DNMRT) at P ≤ 0.05 with the software DSAASTAT (Onofri Italy).
Results
Preliminary Screening of ISR Determinants from B. fortis IAGS162
In this study, we tested mainly the control efficacy of intracellular metabolites and CFCF of B. fortis IAGS162 against fusarium wilt of tomato in greenhouse. Data regarding disease index showed a significant response of different treatments on plant survival. Both CFCF and alive cells of B. fortis IAGS162 (used as a positive control) displayed efficacy in control of fusarium wilt disease (Table 1). Indeed, treatment with CFCF was as effective as the alive bacterial cells and resulted in approximately 70% reduction in disease index as compared to the pathogen control. In contrast the plants treated with intracellular metabolites exhibited no considerable protection against fusarium wilt disease (Table 1). Thus, these results suggested that CFCF from B. fortis IAGS162 could be mainly responsible for the suppression of fusarium wilt disease and carry the potential ISR determinant/s.
Table 1.
Potential of B. fortis IAGS162 and its components to induce systemic resistance in tomato against fusarium wilt.
| Treatments | Disease index (DI) |
|---|---|
| Alive cells | 22.18 ± 03.82D |
| Heat killed cells | 81.06 ± 11.26AB |
| Intra-cellular components | 78.92 ± 06.91B |
| CFCF | 34.38 ± 02.08C |
| Pathogen control | 86.53 ± 11.47A |
| Non-treated Control | – |
CFCF, cell free culture filtrates. Numbers with ± represent Standard error between different replicates of same treatments. Different letters shows levels of significance as governed by ANOVA and DNMRT at P ≤ 0.05.
Isolation of ISR Determinants from CFCF of B. fortis IAGS162
In search for the ISR determinant/s, CFCF of B. fortis IAGS162 was extracted by using organic solvents system and ISR bioassays were performed by using extracts obtained. Compared to the rest of the treatments, chloroform fraction elicited ISR in tomato plants against fusarium wilt disease (Figure 1). Biochemcials present in this fraction were further sub-divided by performing silica gel chromatography, and the sub-fractions were again used in ISR bioassays. GC/MS analysis identified five compounds in ISR active sub-fraction including, acetic acid methyl ester, PAA, palmitic acid, propanol, and tyrosine (Figure 2).
FIGURE 2.
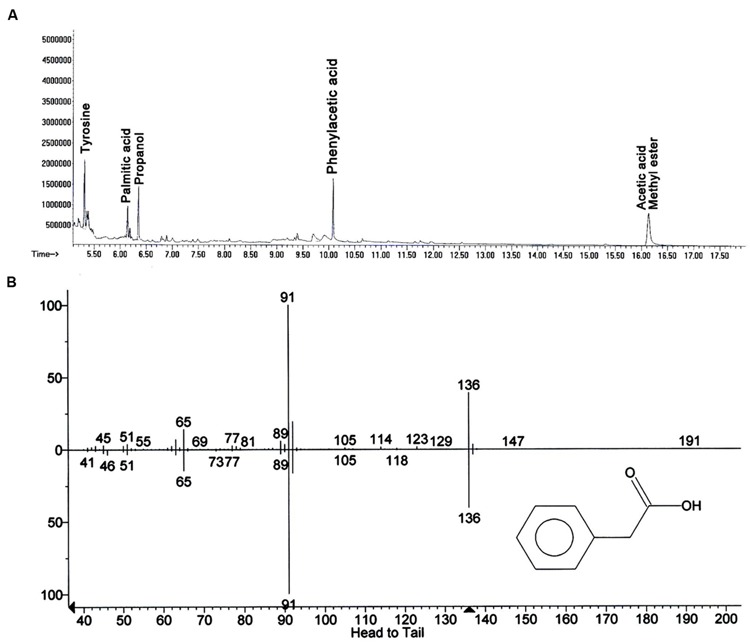
Elucidation of biochemicals presents in ISR active sub-fraction in cell free culture filtrates of B. fortis IAGS162. (A) Chromatogram of Gas Chromatography and Mass Spectrometry analysis (GC/MS) for identification of the ISR determinant/s of B. fortis IAGS162 present in the ISR-active sub-fraction. (B) The mass spectrum analysis obtained by electrospray ionization of Phenylacetic acid.
To finally confirm the potential of ISR determinant/s, use of pure biochemicals as present in ISR active sub-fraction was focused upon. In these ISR bioassays, only PAA was found to suppress fusarium wilt (Figures 1 and 3). Thus the results indicated that 0.1 and 1.0 mM PAA significantly (P ≤ 0.05) reduced disease index up to 58.4 and 76.7%, respectively, as compared to that in the pathogen control (Figure 3). The other compounds failed to elicit significant (P ≤ 0.05) reduction in DI (Figures 1 and 3). Conspicuously, tomato plants receiving PAA provided better growth as compared to the rest of the treatments in this ISR bioassay that can be attributed toward plant growth promoting capability of PAA (Figure 1).
FIGURE 3.
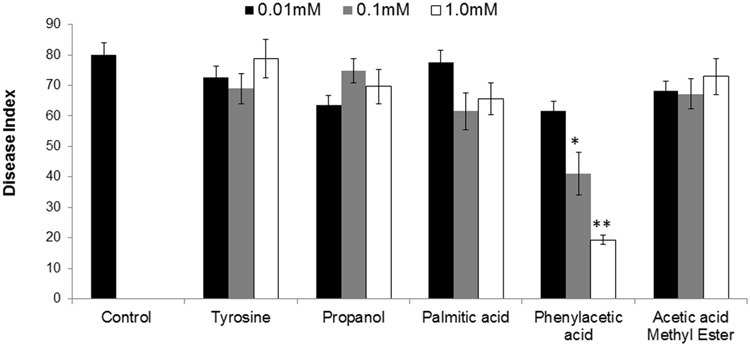
Influence of root treatment with chemicals present in ISR active sub-fraction on the disease development on tomato plants after inoculation with fusarium wilt pathogen. Sterile distilled water was used as positive control. ISR eliciting sub-fraction was subjected to GC/MS analysis and chemicals present were purchased and subjected to ISR bioassay. Vertical bar represents standard errors. Asterisks indicate statistically significant reduction in disease index as compared to pathogen control as governed by ANOVA at P ≤ 0.05.
GC/MS Analysis of Tomato Metabolome
Metabolites are considered as signaling molecules as they are associated with physiological processes. To elucidate ISR process that may be involved in PAA-mediated resistance to fusarium wilt disease, we analyzed the whole metabolome of tomato plants inoculated with the pathogen and PAA in either combination. Central metabolites changed in response to PAA + F. oxysporum, PAA, and F. oxysporum alone were normalized to respective control and were expressed in fold change (Figure 4). Here we identified more than 60 metabolites whose levels were seemed to be altered in response to different treatments (Figure 7).
FIGURE 4.
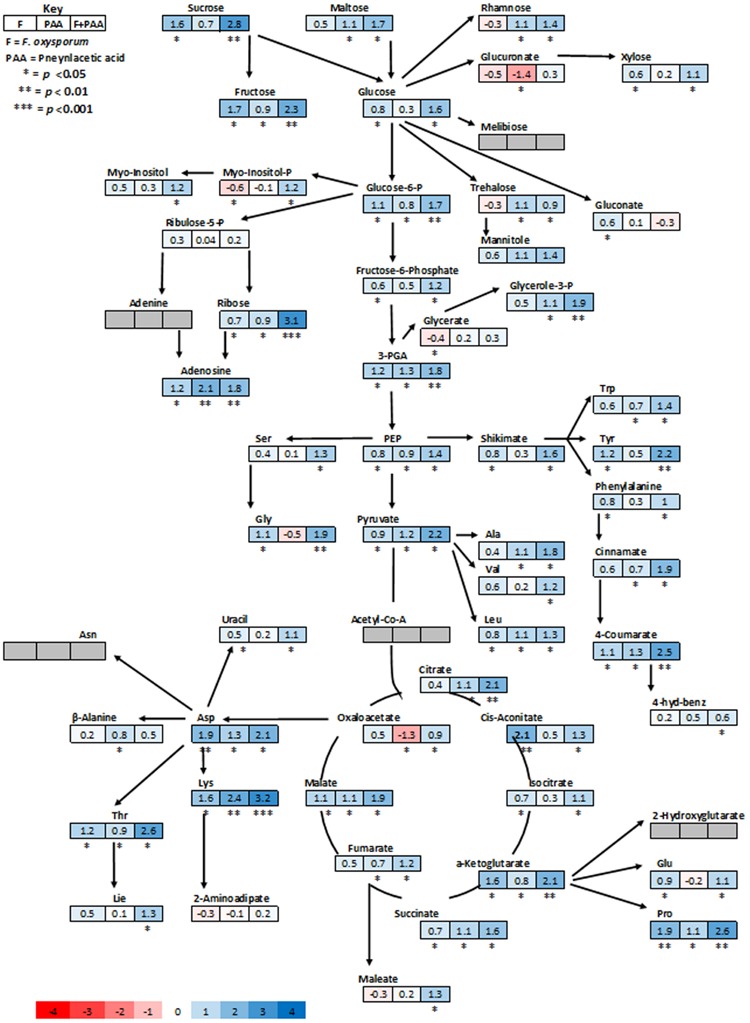
Schematic representations of tomato metabolites in fold change as compared to untreated control plants. Different colors represent levels of metabolite fold change where blue is increasing and red is decreasing. Mean values of two independent experiments are presented here.
FIGURE 7.
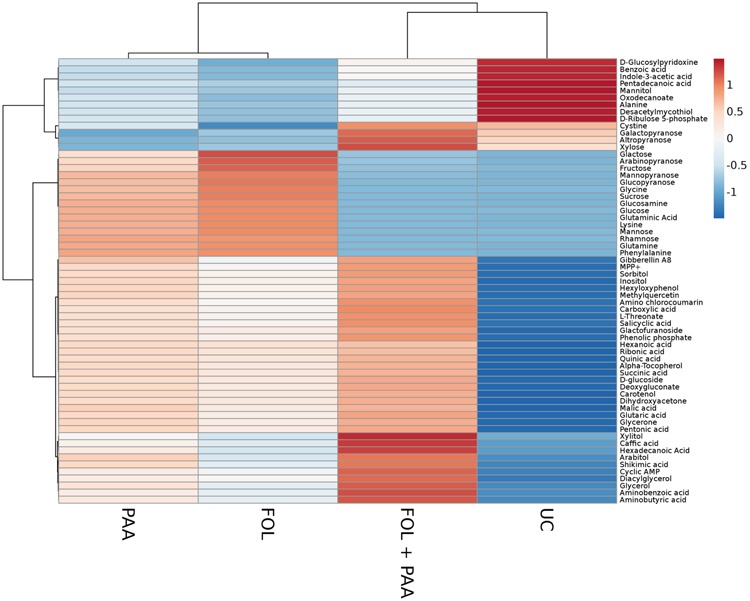
Heat map illustrating the different metabolite levels in tomato plants under different treatments. Each row represents differentially expressed metabolites while each column represents a treatment. Mean values of two independent experiments are presented here. Metabolites are clustered using average linkage hierarchical clustering. The colors in the heat map represent the intensity of the log2-fold change in metabolite levels. FOL, F. oxysporum f.sp. lycopersici. PAA, phenylacetic acid. UC, non-treated control.
As expected, the most represented categories included metabolites involved in defense pathways, metabolites involved in cell communication and signaling, and metabolites implicated in primary and secondary metabolism. The PCA scores plot revealed a clear separation of the all different treatments and demonstrating the significant and differential effect of the treatments on the metabolic level (Figure 5). Quantities of more than 40 metabolites appeared significantly increased or repressed in treatments receiving PAA in either combinations as compared to rest of the treatments.
FIGURE 5.
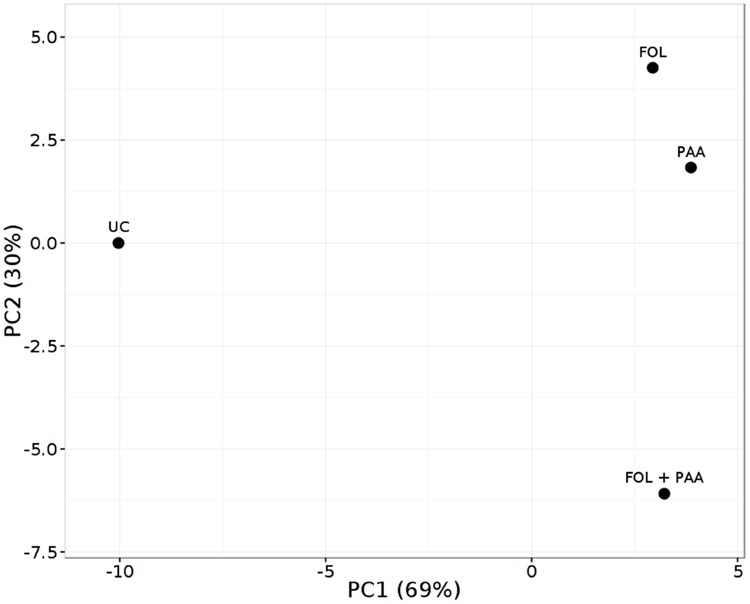
Principal component analysis (PCA) score plot of metabolite finger printing of tomato shoots 7 days after treatment applications. FOL, F. oxysporum f.sp. lycopersici. PAA, phenylacetic acid. UC, non-treated control.
In general, analysis of tomato plants challenged by PAA and F. oxysporum showed that the primary metabolism was significantly reprogrammed in both cases but with different consequences (Figure 4). Number of metabolites involved in the shikimate and the phenylpropanoid pathways were up-regulated under influence of either pathogen alone on in combination to PAA as compared to the control plants (Figures 4 and 6). However, some metabolites specifically precursors of phenylpropanoid pathway were not significantly up-regulated by pathogen alone viz: (tryptophan, cinnamic acid, 4-hydroxybenzene) although they were induced by PAA in combination to the pathogen (Figures 4 and 6).
FIGURE 6.
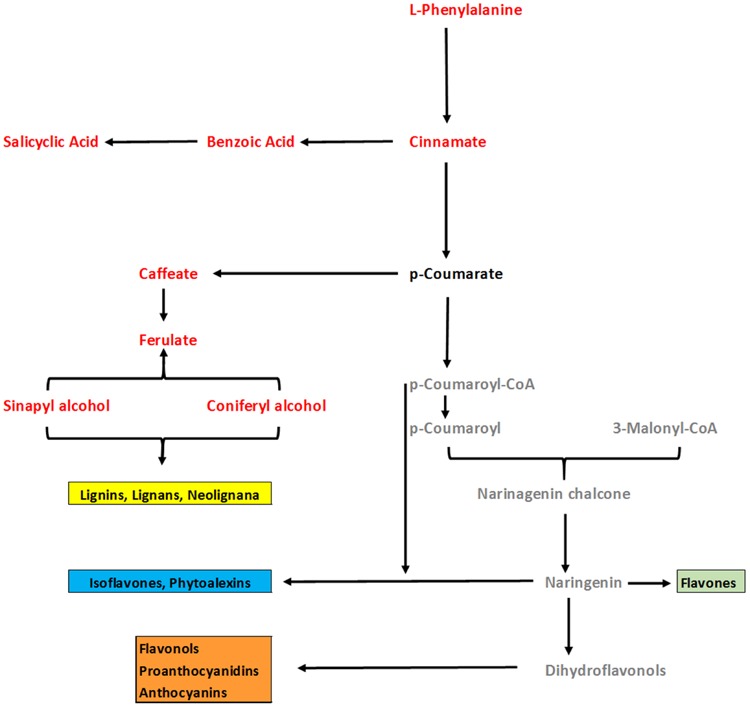
An overview of changes in phenylpropenoid pathway of tomato plants under influence of PAA and fusarium wilt pathogen. Comparison was made between two treatments viz: 1 = Plants receiving PAA and F. oxysporum, 2 = plants receiving F. oxysporum alone (considered as control). Metabolites in red font color show significant increase over control as governed by ANOVA at P ≤ 0.05. Black font represent no significant change as compared to control. Gray color represent metabolites not detected.
In accordance with these observations, plants treated with pathogen alone showed a significant down-regulation of some glycolytic, amino acid metabolism and TCA pathway intermediates viz: (Glucoronate, Myo-inositol, treahlose, 2-aminoadipitate and malic acid) but an increase was observed under synergistic effect of PAA and F. oxysporym (Figure 4). In the same way, in tomato plants primed with PAA in either combination, the glycolytic and TCA pathways were up-regulated, and their precursors were also significantly increased. This may be related to the different elicitation and signaling progression between the two elicitors (PAA and F. oxysporum) (Figure 4). Along with that, some resemblance was also observed between PAA mediated ISR and pathogen triggered SAR responses. It gives us clue that PAA-mediated activation and up-regulations of metabolites may contribute to PAA-induced resistance against fusarium wilt disease.
Discussion
Searching ISR Determinant/s from B. fortis IAGS162
The potential importance of a beneficial rhizospheric bacterial strain was first reported when author observed ISR eliciting capability of B. fortis IAGS162 against fusarium wilt of tomato (Akram et al., 2013). Beside a limited number of researches screening the potential MAMPs from these beneficial symbiotic microbes, a lot of studies to date appears to describe screening the beneficial microbes. However, there do not exist follow up studies that elucidate detailed mechanisms behind MAMPs mediated ISR in crop plants against diseases. Initial studies were conducted to search the ISR determinant/s responsible for elicitation of ISR in tomato against fusarium wilt disease. In preliminary experiments, CFCF significantly reduced the disease effect, clued that ISR determinant/s resides in CFCF of B. fortis IAGS162. The largest significant reduction in disease index was obtained by CFCF treatment (Table 1). The ISR elicitors from beneficial microbes can be either produced in intracellular matrix or secreted extracellularly in the rhizosphere. In some studies, it was shown that ISR determinant/s were retained in CFCF of some microbes (Van Peer and Schippers, 1992; Leeman et al., 1996; Gomez-Gomez and Boller, 2002). Here chloroform phase of culture filtrates of B. fortis IAGS162, was containing ISR elicitor as directed by ISR bioassays. The metabolites present in this phase were further separated into sub-fractions and subjected to GC/MS analysis. Further ISR bioassays guided that among different compounds present in ISR sub-fractions, only PAA effectively switched on systemic resistance in tomato plants against fusarium wilt disease (Figure 4).
Auxins have long been recognized as regulators of plant growth, but some recent studies have discovered the role of auxins in plant defense mechanisms. Some phytohormones including auxins have recently emerged as re-modulators of plant defense against pathogens (Robert-Seilaniantz et al., 2011). Recent advances in plant immunity suggest that auxins and JA act synergistically and their signaling pathways share many commonalities (Kazan and Manners, 2009). In Arabidopsis, jasmonate (JA) signaling repressor has been shown to be stimulated by auxins (Grunewald et al., 2009). Auxins and some defense related phytoalexins are synthesized by the same tryptophan pathway (Tan et al., 2007). PAA, also known as benzeneacetic acid, is an aromatic carboxylic acid having a phenyl functional group. It is also a naturally occurring auxin that is present in many plant species (Abe et al., 1974; Schneider and Wightman, 1986) and produced by some bacteria (Sarwar and Franckenberger, 1995; Hwang et al., 2001; Kim et al., 2004: Somers et al., 2005; Sumayo et al., 2013). In this study, PAA effectively elicited ISR in tomato plants when applied at concentrations of 0.1 and 1 mM. In another study, this kind of ISR was elicited by PAA along with 1-hexadecene and linolic acid secreted by Ochrobactrum lupini KUDC1013 (Sumayo et al., 2013). In the same way, rhizospheric application of an auxin (IAA) trigerred ISR in tomato plants against fusarium wilt of tomato along with positive effect on growth and fruit yield (Sharaf and Farrag, 2004).
Elucidation of Mechanism behind PAA Induced ISR in Tomato
Metabolites are the signaling molecules, involved in both constitutive and inducible plant defense responses that interfere negatively with pathogen performance. GC/MS is an analytical technique in metabolomics for detection, identification, and analysis of small molecules (Budak et al., 2015). Plants accumulate some metabolites to protect them from both biotic and abiotic stress for their survival. First phase of this study suggested that PAA is an ISR determinant produced by B. fortis IAGS162. However, a conclusive demonstration that PAA mediated ISR elicitation activity leads to significant increase in levels of plant metabolites during infection process was still lacking. Hence it was required to elucidate its role in basal resistance to pathogens. For that purpose, we carried out a complete metabolome analysis of tomato plants treated with PAA and infected with F. oxysporum to determine what responses activated by PAA either alone or in combination to pathogen, may be involved in tomato defense against the fungal pathogen F. oxysporum. The metabolites induced by both PAA and F. oxysporum infection not only encode defense-related biochemicals (phytoalexins), but also other biochemicals involved in primary and secondary metabolisms.
In our study, PAA application in either combination caused significant increase in some sugar contents viz: sucrose, maltose, glucose and fructose. Carbohydrates are considered the major source of energy for the metabolic changes that occur in plants. Their availability is of major relevance to the plant growth and might be associated with the induction of defense responses in plants. This increase in sugar contents might lead to increased biosynthesis of numerous precursors involved in other secondary metabolic pathways. Furthermore, increased sugars production has been speculated to provide the phosphate sugars that are used for antioxidant pathway activity and phenolic synthesis (Shetty and Wahlqvist, 2004). We also observed dynamic changes in polyols such as myo-inositol, under influence of PAA in tomato plants. Accumulation of polyols is reported to be associated with stress tolerance in plants (Stoop et al., 1996).
Phenylpropanoids are secondary metabolites that play a basic role in signaling and plant defense responses against abiotic or biotic constraints (Nugroho, 2002; Reuben et al., 2013). GC/MS analysis confirmed that some precursor compounds of phenylpropenoid pathway like L-phenylalanine, cinnamic acid, benzoic acid, caffeic acid, and salicyclic acid, were distinctively increased in tomato plants receiving PAA. L-phenylalanine is a precursor of a wide range of natural products. Enzymatic deamination of L-phenylalanine directs the energy flow to the various branches of the general phenypropanoid metabolism (Vogt, 2010). Along with that, inoculation of tomato plants with PAA led to increased synthesis of benzoic acid, a precursor of salicyclic acid biosynthesis. Salicyclic acid is a key signaling compound triggering local and systemic resistance in plants against both biotic and abiotic stresses (Shah and Klessig, 1999). Caffeic acid is mainly involved in lignification of plant cell walls and may have other physiological functions (Rastogi and Dwivedi, 2008). These results suggest that up-regulation of these compounds are likely to re-enforce plant defense responses against invading pathogen.
Phenylpropanoid-based polymers in plants, such as lignin, suberin, or condensed tannins, contribute to the physical stability and robustness toward environmental damages from drought or wounding (Conrath et al., 2001; Anterola and Lewis, 2002; Reuben et al., 2013). In accordance with the results of GC/MS analysis, an overview of PAA-modulated changes in quantities of precursors involved in the phenylpropanoid pathway showed that this pathway is clearly involved in ISR, leading to the production of phytoalexins such as lignin, flavonoids and anthocyanins. Tomato plants treated with PAA in combination with the pathogen challenge, provided a maximum increase in the levels of defense-related biochemicals, suggesting a positive synergistic effect of PAA and pathogen. Furthermore, some metabolites triggered by PAA treatment displayed a similar behavior after F. oxysporum infection, suggesting that at least a part of the plant defense responses elicited by PAA mediate same signaling pathways as by this pathogenic fungus. In line with the observation that PAA-induced ISR resembles pathogen-triggered immunity, it can be concluded that PAA primed ISR can be distinguished by the much more pronounced induction in the phenylpropanoid pathway.
Conclusion
Based on our findings, colonization of B. fortis IAGS162 can play a role in suppression of fusarium wilt disease by secreting PAA in plant rhizosphere. In our attempts to elucidate the mechanism underlying ISR induced by PAA, we showed that its application extensively re-modulates whole metabolome in tomato plants. This study also provides an insight that phenylpropenoid pathway might be the main factor in this defense process against a fungal pathogen. Undoubtedly, future research on auxins measurement, or the study of auxin-responsive or auxin polar transport genes should be performed to draw a more complete story.
Author Contributions
WA and TA designed the study. BA drafted the manuscript and critically revised the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Citrus Education and Research Centre, University of Florida, USA, for providing us assistance to Perform GCMS analysis. We are also thankful to Fungal biotechnology lab, Institute of Agricultural sciences, University of the Punjab, Lahore providing Microbial Strains.
Footnotes
References
- Abe U., Uchiyama M., Sato R. (1974). Isolation of phenylacetic acid and its p-hydroxy derivative as auxin like substances from Undaria pinnatifida. Agric. Biol. Chem. 38 897–898. 10.1271/bbb1961.38.897 [DOI] [Google Scholar]
- Akram W., Anjum T., Ali B. (2015). Searching ISR determinant/s from Bacillus subtilis IAGS174 against Fusarium wilt of tomato. Biocontrology 60 271–280. 10.1007/s10526-014-9636-1 [DOI] [Google Scholar]
- Akram W., Anjum T., Ali B., Ahmad A. (2013). Screening of native bacillus strains to induce systemic resistance in tomato plants against fusarium wilt in split root system and its field applications. Int. J. Agric. Biol. 15 1289–1294. [Google Scholar]
- Anterola A. M., Lewis N. G. (2002). Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61 221–294. 10.1016/S0031-9422(02)00211-X [DOI] [PubMed] [Google Scholar]
- Ausubel F. M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6 973–979. 10.1038/ni1253 [DOI] [PubMed] [Google Scholar]
- Bakker P., Ran L. X., Pieterse C. M. J., Van Loon L. C. (2003). Understanding the involvement of rhizobacteria-mediated induction of systemic resistance in biocontrol of plant diseases. Can. J. Plant Pathol. Rev. Can. Phytopathol. 25 5–9. 10.1080/07060660309507043 [DOI] [Google Scholar]
- Beckers G. J. M., Spoel S. H. (2006). Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol. 8 1–10. 10.1055/s-2005-872705 [DOI] [PubMed] [Google Scholar]
- Beckmann M., Enot D. P., Overy D. P., Draper J. (2007). Representation, comparison, and interpretation of metabolome fingerprint data for total composition analysis and quality trait investigation in potato cultivars. J. Agric. Food Chem. 55 3444–3451. 10.1021/jf0701842 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger-signals by pattern recognition receptors. Annu. Rev. Plant Biol. 60 379–406. 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Budak H., Hussain B., Khan Z., Ozturk N. Z., Ullah N. (2015). From genetics to functional genomics: improvement in drought signaling and tolerance in wheat. Front. Plant Sci. 6:1012 10.3389/fpls.2015.01012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachinero J. M., Hervas A., Jimenez-Diaz R. M., Tena M. (2002). Plant defence reactions against Fusarium wilt in chickpea induced by incompatible race 0 of Fusarium oxysporum f.sp. ciceris and non-host isolates of F. oxysporum. Plant Pathol. 51 765–776. 10.1046/j.1365-3059.2002.00760.x [DOI] [Google Scholar]
- Catchpole G. S., Beckmann M., Enot D. P., Mondhe M., Zywicki B., Taylor J., et al. (2005). Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc. Natl. Acad. Sci. U.S.A. 102 14458–14462. 10.1073/pnas.0503955102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S. T., Coaker G., Day B., Staskawicz B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814. 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Conrath U. (2006). Systemic acquired resistance. Plant Signal. Behav. 1 179–184. 10.4161/psb.1.4.3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U. (2011). Molecular aspects of defence priming. Trends Plant Sci. 16 524–531. 10.1016/j.tplants.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Conrath U., Beckers G. J. M., Flors V., García-Agustín P., Jakab G. (2006). Priming: getting ready for battle. Mol. Plant Microbe. Interact. 19 1062–1071. 10.1094/MPMI-19-1062 [DOI] [PubMed] [Google Scholar]
- Conrath U., Pieterse C. M., Mauch-Mani B. (2002). Priming in plant-pathogen interactions. Trends Plant Sci. 7 210–216. 10.1016/S1360-1385(02)02244-6 [DOI] [PubMed] [Google Scholar]
- Conrath U., Thulke O., Katz V., Schwindling S., Kohler A. (2001). Priming as a mechanism in induced systemic resistance of plants. Eur. J. Plant Pathol. 107 113–119. 10.1023/A:1008768516313 [DOI] [Google Scholar]
- De Vos R. C., Moco S., Lommen A., Keurentjes J. J., Bino R. J., Hall R. D. (2007). Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2 778–791. 10.1038/nprot.2007.95 [DOI] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11 539–548. 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- Epp D. (1987). “Somaclonal variation in banana: a case study with fusarium wilt,” in Banana and Plantain Breeding Strategies, eds Persley G. J., De Langhe E. A. (Canberra: ACIAR Publication; ). [Google Scholar]
- Garcia-Brugger A., Lamotte O., Vandelle E., Bourque S., Lecourieux D., Poinssot B., et al. (2006). Early signaling events induced by elicitors of plant defenses. Mol. Plant Microbe. Interact. 19 711–724. 10.1094/MPMI-19-0711 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L., Boller T. (2002). Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7 251–256. 10.1016/S1360-1385(02)02261-6 [DOI] [PubMed] [Google Scholar]
- Gómez-Vásquez R., Day R., Buschmann H., Randles S., Beeching J. R., Cooper R. M. (2004). Phenylpropanoids, phenylalanine ammonia lyase and peroxidases in elicitor-challenged cassava (Manihot esculenta) suspension cells and leaves. Ann. Bot. 94 87–97. 10.1093/aob/mch107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Cannoot B., Friml J., Gheysen G. (2009). Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 5:e1000266 10.1371/journal.ppat.1000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. J., Tsay J. F., Chang S. Y., Yang H. P., Wu W. S., Chen C. Y. (2012). Dimethyl disulfide is an induced systemic resistance elicitor produced by Bacillus cereus C1L. Pest. Manag. Sci. 68 1306–1310. 10.1002/ps.3301 [DOI] [PubMed] [Google Scholar]
- Hwang B. K., Lim S. W., Kim B. S., Lee J. Y., Moon S. S. (2001). Isolation and in vivo and in vitro antifungal activity of phenylacetic acid and sodium phenylacetate from Streptomyces humidus. Appl. Environ. Microbiol. 67 3739–3745. 10.1128/AEM.67.8.3739-3745.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Katagiri F., Tsuda K. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant. Biol. 13 459–465. 10.1016/j.pbi.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2009). Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci. 14 373–382. 10.1016/j.tplants.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Kim Y., Cho J. Y., Kuk J. H., Moon J. H., Cho J. I., Kim Y. C., et al. (2004). Identification and antimicrobial activity of phenylacetic acid produced by Bacillus licheniformis isolated from fermented soybean, chungkook-jang. Curr. Microbiol. 48 312–317. 10.1007/s00284-003-4193-3 [DOI] [PubMed] [Google Scholar]
- Leeman M., den Ouden F. M., van Pelt J. A., Dirkx F. P. M., Steijl H., Bakker P. A. H. M., et al. (1996). Iron availability affects induction of systemic resistance to Fusarium wilt of radish by Pseudomonas fluorescens. Phytopathology 86 149–155. 10.1094/Phyto-86-149 [DOI] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzerm L., Fernie A. R. (2006). Gas chromatography mass spectrometry based metabolite profiling in plants. Nat. Protocols 1 387–396. 10.1038/nprot.2006.59 [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63 541–556. 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Mishra A. K., Sharma K., Misra R. S. (2009). Purification and characterization of elicitor protein from Phytophthora colocasiae and basic resistance in Colocasia esculenta. Microbiol. Res. 164 688–693. 10.1016/j.micres.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Montesano M., Brader G., Palva E. T. (2003). Pathogen derived elicitors: searching for receptors in plants. Mol. Plant Pathol. 4 73–79. 10.1046/j.1364-3703.2003.00150.x [DOI] [PubMed] [Google Scholar]
- Nugroho L. (2002). Activities of enzymes involved in the phenylpropanoid pathway in constitutively salicylic acid-producing tobacco plants. Plant Physiol. Biochem. 40 755–760. 10.1016/S0981-9428(02)01437-7 [DOI] [Google Scholar]
- Ongena M., Jourdan E., Schäfer M., Kech C., Budzikiewicz H., Luxen A., et al. (2005). Isolation of an N-alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in bean. Mol. Plant Microbe Interact. 18 562–569. 10.1094/MPMI-18-0562 [DOI] [PubMed] [Google Scholar]
- Park M. R., Kim Y. C., Lee S., Kim I. S. (2009). Identification of an ISR-related metabolite produced by rhizobacterium Klebsiella oxytoca C1036 active against soft-rot disease pathogen in tobacco. Pest Manag. Sci. 65 1114–1117. 10.1002/ps.1800 [DOI] [PubMed] [Google Scholar]
- Park S. W., Vlot A. C., Klessig D. F. (2008). Systemic acquired resistance: the elusive signal(s). Curr. Opin. Plant Biol. 11 436–442. 10.1016/j.pbi.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Persello-Cartieaux F., Nussaume L., Robaglia C. (2003). Tales from the underground: molecular plant-rhizobacteria interactions. Plant Cell Environ. 26 189–199. 10.1046/j.1365-3040.2003.00956.x [DOI] [Google Scholar]
- Pieterse C. M. J., Leon-Reyes A., Van der Ent S., Van Wees S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5 308–316. 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Van Wees S. C. M., Ton J., Van Pelt J. A., Van Loon L. C. (2002). Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol. 4 535–544. 10.1111/j.1438-8677.2011.00549.x [DOI] [Google Scholar]
- Rastogi S., Dwivedi U. N. (2008). Manipulation of lignin in plants with special reference to O-methyltransferase. Plant Sci. 174 264–277. 10.1016/j.plantsci.2007.11.014 [DOI] [Google Scholar]
- Reuben S., Rai A., Bhinu V. S., Rodrigues A., Swarup S. (2013). A bacterial quercetin oxidoreductase QuoA-mediated perturbation in the phenylpropanoid metabolic network increases lignification with a concomitant decrease in phenolamides in Arabidopsis. J. Exp. Bot. 64 5183–5194. 10.1093/jxb/ert310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J. D. G. (2011). Hormone crosstalk in plant disease and defense: more than just JASMONATE SALICYLATE antagonism. Annu. Rev. Phytopathol. 2011. 49 317–343. 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- Ryan C. A., Pearce G. (2003). Systemins: a functionally defined family of peptide signal that regulate defensive genes in Solanaceae species. Proc. Natl. Acad. Sci. U.S.A. 100 14577–14580 10.1073/pnas.1934788100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C. M., Farag M. A., Hu C. H., Reddy M. S., Kloepper J. W., Pare P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134 1017–1026. 10.1104/pp.103.026583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwar M., Franckenberger W. T. J. (1995). Fate of lphenylalanine in soil and its effect on plant growth. Soil Sci. Soc. Am. J. 59 1625–1630. 10.2136/sssaj1995.03615995005900060017x [DOI] [Google Scholar]
- Schneider E. A., Wightman F. (1986). Auxins on non-flowering plants. I. Occurrence of 3 indoleacetic acid and phenylacetic acid in vegetative and fertile fronds of the ostrich fern (Matteucia struthiopteris). Physiol. Plant 68 396–402. 10.1111/j.1399-3054.1986.tb03372.x [DOI] [Google Scholar]
- Shah Y., Klessig D. F. (1999). “Salicylic acid: signal perception and transduction,” in Biochemistry and Molecular Biology of Plant Hormones, eds Hooykaas P. J. J., Hall M. A., Libbenga K. R. (Amsterdam: Elsevier Science; ), 513–541. [Google Scholar]
- Sharaf E. F., Farrag A. A. (2004). Induced resistance in tomato plants by IAA against Fusarium oxysporum lycopersici. Pol. J. microbiol. 53 111–116. [PubMed] [Google Scholar]
- Shetty K., Wahlqvist M. L. (2004). A model for the role of the praline-linked pentosephosphate pathway in phenolic phytochemical biosynthesis and mechanism of action for human health and environmental application. Asia Pac. J. Clin. Nutr. 13 1–24. [PubMed] [Google Scholar]
- Somers E., Ptacek D., Gysegom P., Srinivasan M., Vanderleyden J. (2005). Azospirillum brasilense produces the auxinlike phenylacetic acid by using the key enzyme for indole-3- acetic acid biosynthesis. Appl. Environ. Microbiol. 71 1803–1810. 10.1128/AEM.71.4.1803-1810.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfath M., Groth D., Lisec J., Selbig J. (2008). Metabolite profile analysis: from raw data to regression and classification. Physiol. Plant. 132 150–161. 10.1111/j.1399-3054.2007.01006.x [DOI] [PubMed] [Google Scholar]
- Stoop J. H. M., Williamson J. D., Pharr D. M. (1996). Mannitol metabolism in plants: a method for coping with stress. Trends Plant Sci. 1 139–144. 10.1016/S1360-1385(96)80048-3 [DOI] [Google Scholar]
- Sumayo M., Hahm M. S., Ghim Y. (2013). Determinants of Plant Growth-promoting Ochrobactrum lupini KUDC1013 Involved in Induction of Systemic Resistance against Pectobacterium carotovorum subsp. carotovorum in Tobacco Leaves. Plant Pathol. J. 29 174–181. 10.5423/PPJ.SI.09.2012.0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645. 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Thakur M., Sohal B. S. (2013). Role of elicitors in inducing resistance in plants against pathogen infection: a review. ISRN Biochem. 10:762412 10.1155/2013/762412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam N. M. (2009). How plants cope with biotic interactions. Plant Biol. 11 1–5. 10.1111/j.1438-8677.2008.00179.x [DOI] [PubMed] [Google Scholar]
- van Hulten M., Pelser M., van Loon L. C., Pieterse C. M., Ton J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103 5602–5607. 10.1073/pnas.0510213103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon L. C., Bakker P. A., Pieterse C. M. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36 453–483. 10.1146/annurev.phyto.36.1.453 [DOI] [PubMed] [Google Scholar]
- Van Peer R., Schippers B. (1992). Lipopolysaccharides of plant-growth promoting Pseudomonas sp. strain WCS417R induce resistance in carnation to Fusarium wilt. Neth. J. Plant Pathol. 98 129–139. 10.1007/BF01996325 [DOI] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid biosynthesis. Mol. Plant. 3 2–20. 10.1093/mp/ssp106 [DOI] [PubMed] [Google Scholar]
- Zamioudis C., Pieterse C. M. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant Microbe. Interact. 25 139–150. 10.1094/MPMI-06-11-0179 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhao J., Zhao C., Zhou H., Li Y., Zhang J., et al. (2015). A metabolomics study delineating geographical location-associated primary metabolic changes in the leaves of growing tobacco plants by GC-MS and CE-MS. Sci. Rep. 5:16346 10.1038/srep16346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D. G., Boller T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D. G., Felix G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]


