Summary
Diverse intercropping system has been used to control disease and improve productivity in the field. In this research, the bacterial communities in salt–alkali soils of monoculture and intercropping mulberry and soybean were studied using 454‐pyrosequencing of the 16S rDNA gene. The dominant taxonomic groups were Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, Bacteroidetes, Planctomycetes and Gemmatimonadetes and these were present across all samples. However, the diversity and composition of bacterial communities varied between monoculture and intercropping samples. The estimated bacterial diversity (H') was higher with intercropping soybean than in monoculture soybean, whereas H' showed an opposite pattern in monoculture and intercropping mulberry. Populations of Actinobacteria, Acidobacteria, and Proteobacteria were variable, depending on growth of plants as monoculture or intercropped. Most of Actinobacteria and Chloroflexi were found in intercropping samples, while Acidobacteria and Proteobacteria were present at a higher percentage in monoculture samples. The plant diversity of aboveground and microbial diversity of belowground was linked and soil pH seemed to influence the bacterial community. Finally, the specific plant species was the major factor that determined the bacterial community in the salt–alkali soils.
Introduction
Alkalization and salinization of soils the two largest factors limiting global agricultural productivity (Liu et al., 2010). Alkalization and salinization are especially problematic in the Songneng Plain in China which has 3.2 × 106 ha salt‐affected land and is one of the three largest soda saline‐alkali areas in the world. Every year, about 20 × 103 ha land becomes salinized/alkalized (Wang et al., 2009). Saline soils negatively impact plant growth due to osmotic inhibition of water absorption (Munns, 2002) and an excess of sodium ions that affects critical biochemical processes. The pernicious effects of alkalinity on plant growth are related to high pH, which disrupts ion balance (Shi and Zhao, 1997) and poor micronutrient availability in soil (Alam et al., 1999). Therefore, salt or alkali soils can tremendously negatively affect productivity of both natural grasslands and crop plants grown on cultivated lands. The productive capacity of alkaline soils could be improved by growing plants adapted to sodic soils (Gupta et al., 1990). In addition, soil organic matter content and availability of soil inorganic nitrogen can be improved via reclamation agroforestry (Singh et al., 1997).
Intercropping has been used for many years to grow two or more crop species together simultaneously (Willey, 1979). Compared with monoculture, intercropping often leads to greater stability of yields (Wiley, 1979), resilience to perturbations (Trenbath, 1999) and reduced N‐leaching (Hauggaard‐Nielsen et al., 2003). Use of a tree/N‐fixing crop intercropping system can be an especially sustainable and beneficial agricultural practice, as the N‐fixing crop provides natural N fertilizer for growing trees. Nitrogen‐fixing legumes may even be improved by intercropping (Neumann et al., 2007), because the intercropped cereals or trees can be more competitive for nitrogen in soil, forcing the legume crop to fix more atmospheric N2 (Hauggaard‐Nielsen et al., 2003). In addition, legume/cereal mixtures, grown in pots, together achieved greater P uptake from soils (El Dessougi et al., 2003) than can either species could take up by itself. In the field, increased P uptake by intercropped maize/faba bean also was observed (Li et al., 2003).
In saline and alkaline soils, excessive amounts of salts have an adverse effect on biological activity including soil enzyme activity (Rao and Pathak, 1996), nitrogen mineralization (McClung and Frankenberger, 1985) and soil microbial biomass (Tripathi et al., 2006). Soil microbial communities are important in managed field systems, as they are involved in most nutrient transformations in soil and thereby influence the aboveground plant productivity (Van Der Heijden et al., 2008). Soil microbial communities are greatly influenced by salinity (Rietz and Haynes, 2003), as well as other abiotic and biotic environmental variables (Alexander, 1977). In alkaline soils, the biological activity of microbial communities improves, when a crop, grass or tree cover is used (Rao and Ghai, 1985).
Soil is a complex and dynamic habitat for microorganisms, which are involved in many key process required for ecosystem functionality (Lim et al., 2010). The diversity of microorganisms in soils can often exceed that, found in other environments (Rosselló‐Mora and Amann, 2001) and is higher by orders of magnitude than the biodiversity of plants and animals. In order to better understand the changes of bacterial communities in monoculture versus intercropped plants (mulberry and soybean) in salt–alkali soils, a 16S rDNA gene‐based pyrosequencing approach was employed using field‐grown mulberry/soybean intercropped under salt–alkali conditions. To our knowledge, this is the first report on the changes of bacterial communities of mulberry/soybean intercropping under salt–alkali stress conditions.
Results
Pyrosequencing analysis of the V1‐V3 region of the 16S rDNA genes resulted in 71 660 high‐quality sequence reads with a read length of ≥ 400 bp across all four samples. The average read length was 482 bp. The read numbers were uneven per sample, ranging from 17 193 to 18 819, with an average of 17 915 (Table S1). We were able to classify 65 926 (92%) of the quality sequences below the domain level. As fewer sequences result in lower numbers for predicted operational taxonomic units (OTUs) and species richness (Will et al., 2010), all samples were randomly reduced to the same size using MOTHUR, based on the sample with the smallest number of reads.
Bacterial diversity and richness
To determine rarefaction curves and other measures of diversity, OTUs were identified at genetic distances of 3%, 5% and 10%. Rarefaction curves indicated consistent differences in all four samples (Fig. 1). At 10% genetic distances, almost all rarefaction curves reached saturation, indicating that the surveying effort covered almost the full extent of taxonomic diversity at this genetic distance. At 3% and 5% genetic distances, rarefaction curves suggested that the sequencing effort was not large enough to capture the complete diversity of these communities, as the curves did not level off with increasing sample size. The comparison of mean Chao 1 richness estimates of mulberry rhizosphere soils and with soybean rhizosphere soils showed different values at genetic distances of 3% (6609 OTUs and 8296 OTUs respectively), 5% (4980 OTUs and 6083 OTUs respectively) and 10% (2552 OTUs and 2990 OTUs respectively), and the richness was higher in soybean rhizosphere soils (P < 0.05) (Table 1). Analysis of differences of ways of planting by employing Student's t‐test at genetic distances of 3%, 5% and 10% showed that the intercropping patterns did not vary significantly in the predicted number of OTUs (P > 0.05). The same conclusion was seen using the Ace richness index (Table 1). Moreover, comparison of the mean Shannon diversity index of all samples revealed that the highest bacterial diversity at all analysed genetic distances was found in SM, followed by IS, IM and MM. The predicted richness and diversity in the soybean monoculture rhizosphere soils exceeded that of the corresponding mulberry monoculture soils. Thus, an influence of plant species on bacterial richness and diversity was observed. Intercropping impacted overall bacterial diversity.
Figure 1.
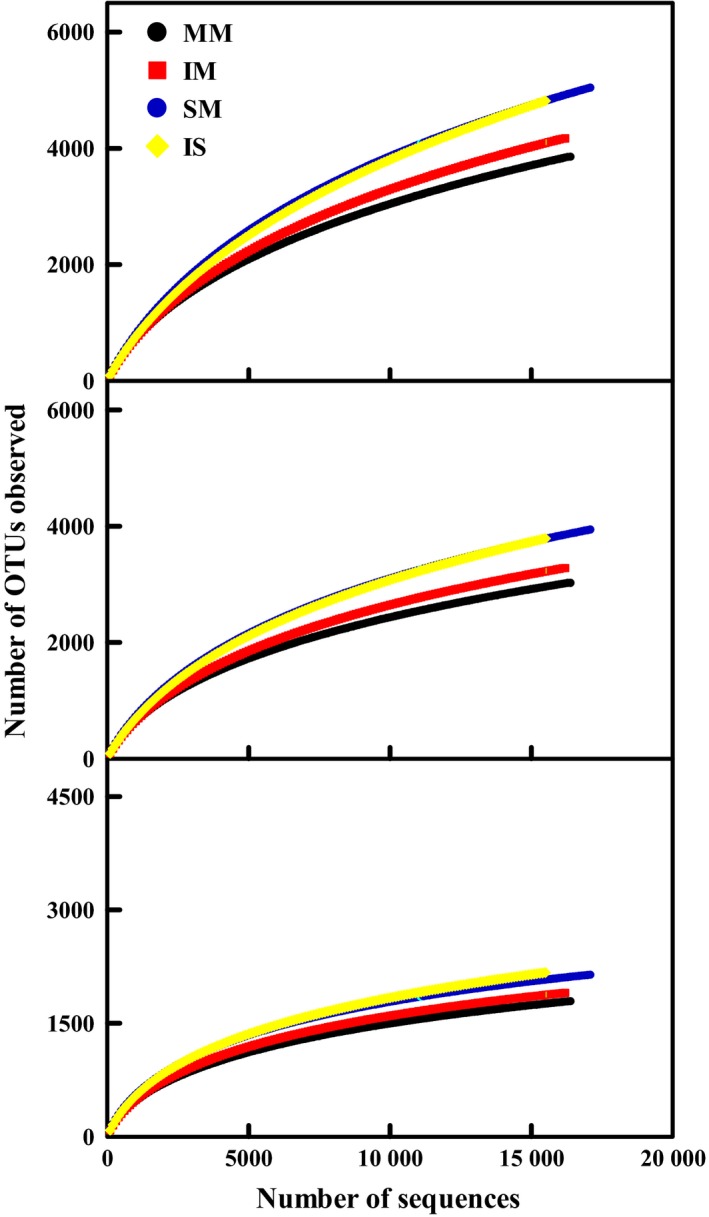
Rarefaction curves indicating the observed number of OTUs at genetic distances of 3%, 5% and 10% in all soil samples. MM, mulberry monoculture; IM, intercropping mulberry; SM, soybean monoculture; IS, intercropping soybean.
Table 1.
Bacterial diversity as assessed by Shannon index (H′) and species richness estimation in all soils
| Genetic distance (%) | Sample | H′ | Chao1 | ACE |
|---|---|---|---|---|
| No. operational taxonomic units | ||||
| 3 | MM | 7.40 ± 0.016a | 6407 ± 266.36a | 7739 ± 213.15a |
| IM | 7.71 ± 0.021b | 6810 ± 260.62a | 8156 ± 214.78a | |
| SM | 7.88 ± 0.016a | 8070 ± 270.39a | 9694 ± 229.47a | |
| IS | 7.75 ± 0.021a | 8522 ± 328.80a | 10735 ± 276.82a | |
| 5 | MM | 7.10 ± 0.016a | 4883 ± 226.78a | 5545 ± 167.02a |
| IM | 7.38 ± 0.016b | 5076 ± 207.96a | 5000 ± 157.25a | |
| SM | 7.51 ± 0.021a | 5942 ± 212.02a | 5911 ± 165.00a | |
| IS | 7.39 ± 0.021a | 6223 ± 255.32a | 7236 ± 196.01a | |
| 10 | MM | 6.23 ± 0.021a | 2526 ± 126.78a | 2441 ± 88.28a |
| IM | 6.53 ± 0.021b | 2578 ± 112.48a | 2560 ± 86.23a | |
| SM | 6.66 ± 0.016a | 2847 ± 115.32a | 2774 ± 81.74a | |
| IS | 6.59 ± 0.021a | 3124 ± 145.14a | 3049 ± 105.02a | |
Different letters following the mean values within each column indicates significant differences at P < 0.05.
MM, mulberry monoculture; IM, intercropping mulberry; SM, soybean monoculture; IS, intercropping soybean.
Distribution of taxa and phylotypes across all samples
The 65 926 classifiable sequences were affiliated with 11 bacterial phyla (Fig. 2). The groups accounted for 96% of all sequences, and a few sequences (< 1%) could not be shown (e.g. Spirochaetes). The dominant phyla across all samples were Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, Bacteroidetes, Planctomycetes and Gemmatimonadetes, representing 24.1%, 18.9%, 17.6%, 13.3%, 10.0%, 4.6% and 3.6%, respectively, of all sequences that were classified below the domain level. These dominant bacterial phyla were found in all samples (Fig. 3). Other sequences belonged to Verrucomicrobia, Nitrospirae, Armatimonadetes and unclassified bacteria of ‘Candidatedivision TM7’, and they were always found in very low proportions (< 2%).
Figure 2.
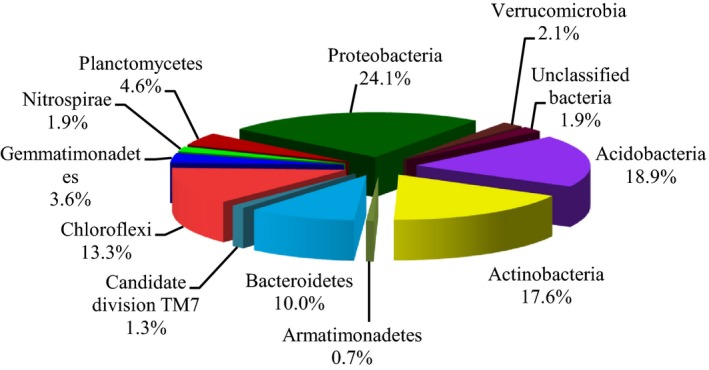
Proportional distribution of different phyla. MM, mulberry monoculture; IM, intercropping mulberry; SM, soybean monoculture; IS, intercropping soybean.
Figure 3.
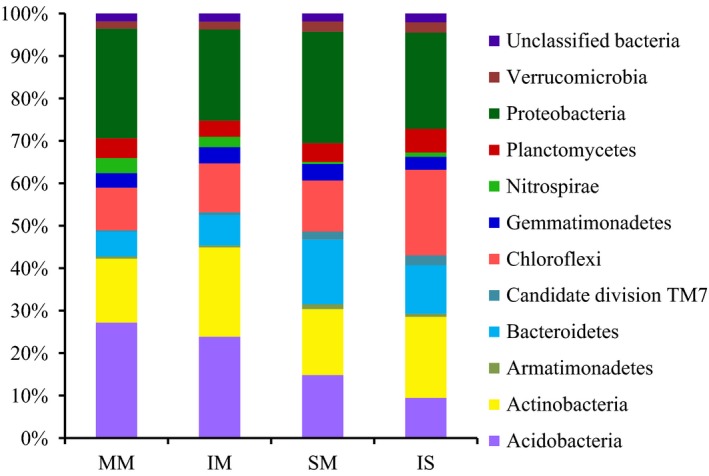
Relative abundance of bacterial phyla for each soil library. MM, mulberry monoculture; IM, intercropping mulberry; SM, soybean monoculture; IS, intercropping soybean.
Comparative analysis of the four soil samples revealed a distinct distribution of the bacterial phyla (Fig. 3). On average, Chloroflexi and Bacteroidetes showed a higher relative abundance in soybean rhizosphere soils than in mulberry rhizosphere soils, whereas Acidobacteria and Nitrospirae showed the opposite pattern. The phyla of Actinobacteria, Acidobacteria, Proteobacteria and Chloroflexi were found in variable proportions, depending on the use of monoculture or intercropping; most of Actinobacteria and Chloroflexi were found in intercropping samples, whereas Acidobacteria and Proteobacteria were present at higher percentages in monoculture samples.
At the classes' level, the bacterial community composition also revealed distinction between different soils (Table 2). Within the Acidobacteria, all samples were dominated by Acidobacteria and Holophagae, and significant differences were observed between monoculture (MM and SM) and intercropping (IM and IS) soils (P < 0.05). Furthermore, members of Chloroflexi phylum showed a opposite pattern to the one observed for Anaerolineae, which Anaerolineae shows a higher abundance in intercropping libraries than monoculture libraries. In comparison to MM libraries, the IM libraries showed a decrease in the OTUs number of Deltaproteobacteria, while other classes of Proteobacteria showed no significant differences. Within in Bacteroidetes, all samples were dominated by Sphingobacteria, and SM libraries had more OTUs number than IS libraries (P < 0.05). In constrast, the Planctomycetes, Actinobacteria and Gemmatimonadetes were present in similar proportions in monoculture and intercropping samples.
Table 2.
Taxonomic classification of bacterial reads retrieved from four samples at phylum and class levels from 16S rDNA gene pyrosequencing

Shared bacterial OTUs
Venn diagrams revealed that the sum of total observed OTUs in the four soil samples was 12 174 (Fig. 4), and 417 OTUs were common all of the soil samples. Moreover, the distribution of sequences demonstrated once again that each plant rhizosphere had its own microbial population.
Figure 4.
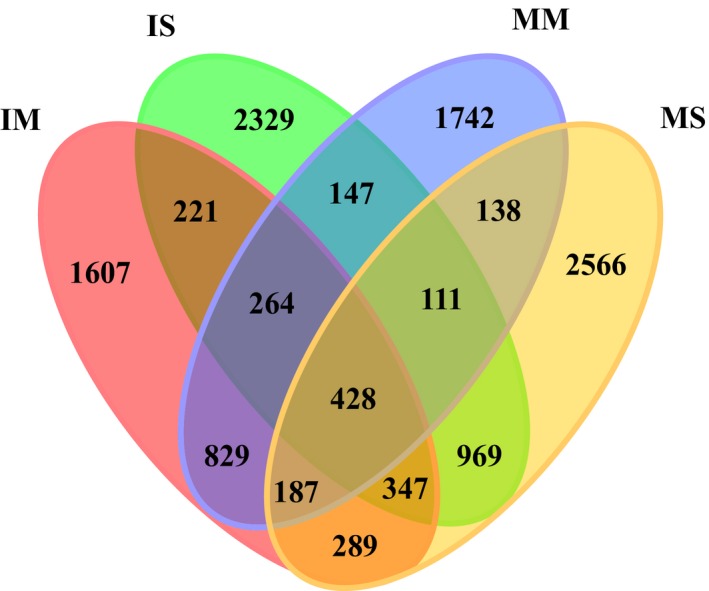
Venn diagram showing the shared bacterial OTUs (at a distance of 0.03) in all soil samples. MM, mulberry monoculture; IM, intercropping mulberry; SM, soybean monoculture; IS, intercropping soybean.
Hierarchically clustered heatmap analysis, based on the microbial community profiles at the genus level, was used to identify the different composition of these four microbial community structures (Fig. 5). The MM and IM groups were separated from SM and IS groups, suggesting clear distinctions of microbial community structure between mulberry and soybean groups. This was supported by the principal component analysis (PCA) with the weighted Unifrac distance (Fig. 6). Overall, the two PCA axes explained 77.36% of the variation between the different communities. The PCA score plot revealed that the mulberry and soybean rhizosphere soils harboured characteristic bacterial communities. Mulberry samples (MM and IM) were clustered together and were well separated from that soybean samples (SM and IS), whereas there was a little distinction between SM and IM samples. These results suggested that plant species had the greatest affect on the bacterial communities in the soil used to support those plants.
Figure 5.
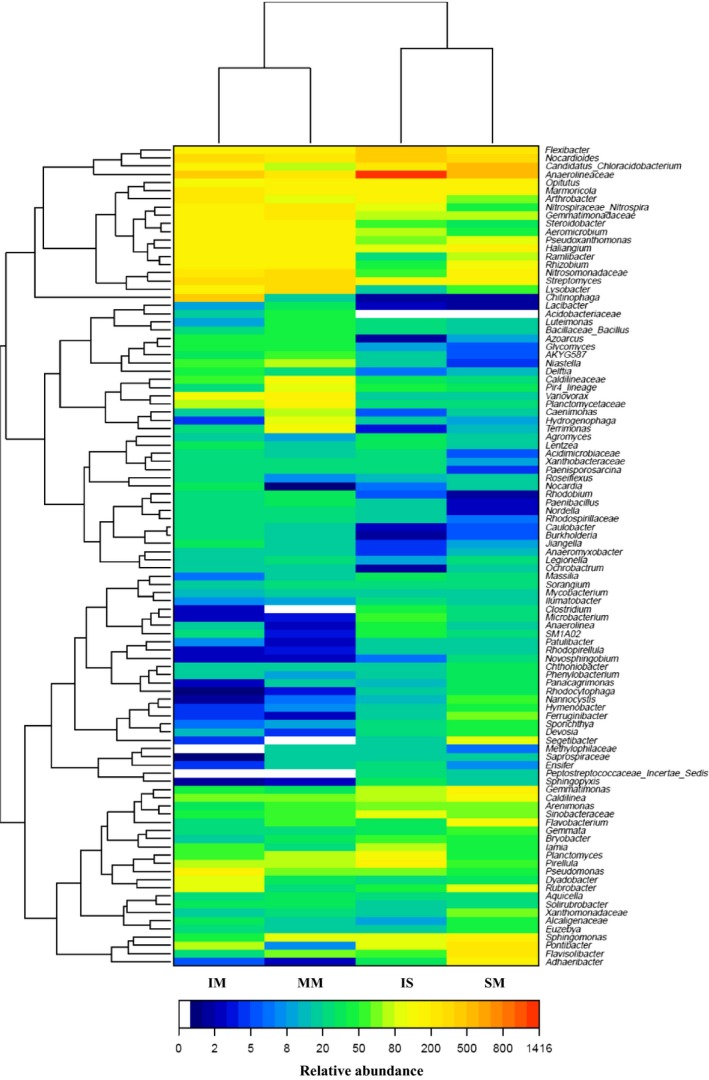
Hierarchical cluster analysis of 100 predominant bacterial communities among the four samples. The Y‐axis is the clustering of the most abundant OTUs (5% distance) in reads. The OTUs were ordered by genus. Sample communities were clustered based on complete linkage method. The colour intensity of scale indicates relative abundance of each OTU read. Relative abundance was defined as the number of sequences affiliated with that OTU divided by the total number of sequences per sample.
Figure 6.
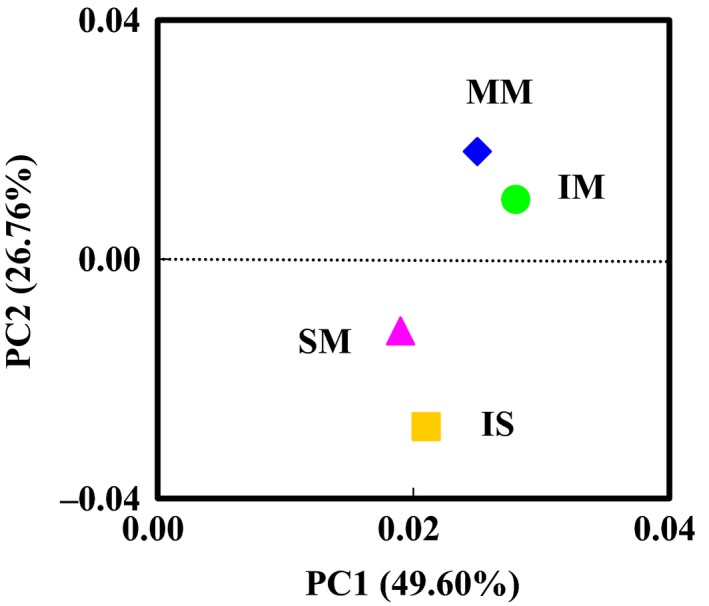
Principal component analysis (PCA) of bacterial communities from mulberry monoculture (MM), intercropping mulberry (IM), soybean monoculture (SM), and intercropping soybean (IS) based on pyrosequencing of the 16S rDNA gene. PCA were generated using the presence of each OUT (at a distance level of 3%) found in each clone library. Principal components (PCs) 1 and 2 explained 49.60% and 26.76% of the variance respectively.
At the genus level, comparison of the relative abundances revealed significant differences between monoculture and intercropping soil bacterial communities (Fig. 5). Anaerolinea, Nocardioides, Candidatus_Chloracidobacterium, Flexibacter, Streptomyces and Nitrosomonadaceae were the most abundance genera across all soil samples, representing 3.44%, 2.71%, 3.03%, 1.81%, 1.59% and 1.81% of all classified sequences in monoculture soils and 8.75%, 3.22%, 1.87%, 2.71%, 1.93% and 1.19% in intercropping soils. This indicated these genera might be indigenous in the salinized meadow soil sampled. The distribution of the dominant genera varied significantly between monoculture and intercropping soils. Anaerolineaceae (2.95%), Nocardioides (2.15%), Candidatus_Chloracidobacterium (1.05%), Arthrobacter (1.43%), Pontibacter (0.52%), Chitinophaga (3.02%), Pseudomonas (1.02%), Rubrobacter (0.64%) and Nocardia (0.62%) showed a higher relative abundance in mulberry intercropping soils than in corresponding monoculture soils, whereas Pir4_lineage (0.54%), Terrimonas (0.56%), Hydrogenophaga (0.49%), Caenimonas (0.48%), Acidobacteriaceae (0.27%) and Lacibacter (0.24%) showed the opposite pattern (Fig. 5). Candidatus_Chloracidobacterium (5.36%), Nitrosomonadaceae (1.09%), Flavisolibacter (2.70%), Pontibacter (2.00%), Ramlibacter (0.74%), Adhaeribacter (1.53%), Flavobacterium (0.85%), Xanthomonadaceae (0.60%), Segetibacter (0.79%), Alcaligenaceae (0.38%) and Ferruginibacter (0.58%) were present in higher proportions in soybean monoculture soils compared with corresponding intercropping soils (Fig. 5).
Correlations of environmental data and bacterial communities
To investigate relationships between soil microbial community composition and measured environmental variables, different bacterial phyla and proteobacterial classes were analysed using canonical correspondence analysis (CCA) (Fig. 7). The influence of the environmental parameter on the CCA bioplot is indicated by arrows in which lengths are proportional to their importance (Liu et al., 2009). pH had the longest arrow, indicating that it was most important in influencing the bacterial community. Conductivity was also significantly linked to bacterial phyla variance in CCA. With the exception of Nitrospirae, the relative abundances of all of bacterial phyla and classes positively correlated with soil pH and conductivity. Water content (WC) was also negatively correlated with Chloroflexi, Candidatedivision TM7 and Planctomycetes.
Figure 7.
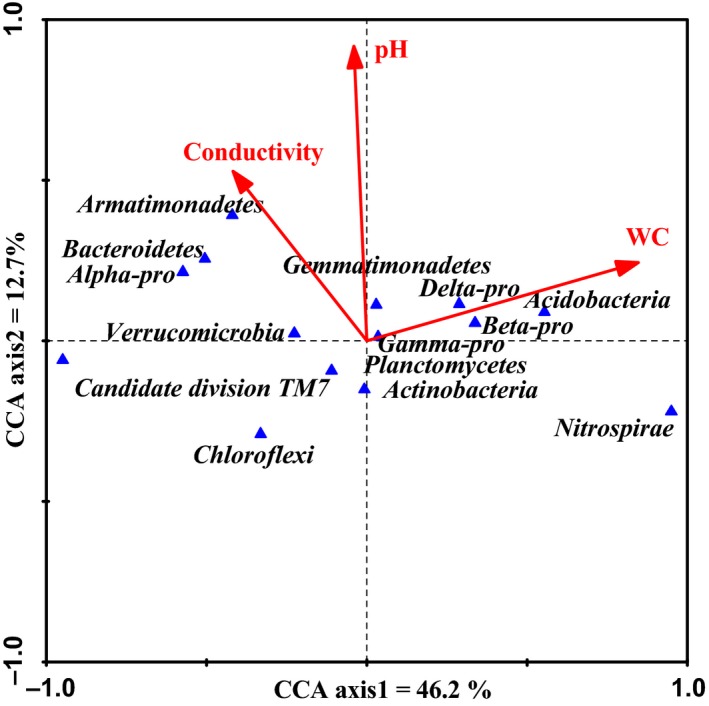
Canonical correspondence analysis (CCA) for soil bacterial community and soil environmental variables. Arrows indicated the direction and magnitude of measurable variables associated with bacterial community structures. Each triangle represents a different bacterial phyla or proteobacterial class.
Discussion
Our previous studied have indicated, with an intercropped soybean/mulberry crop, the aboveground biomass of intercropped mulberry increased 65.7% compared with monoculture mulberry during the co‐growing stage (Zheng et al., 2011). The beneficial effect may be attributed to the improvement of soil nutrient element and the reduction in pH (Li et al., 2007), as well as microbial activity and community composition in intercropping systems (Bastida et al., 2008).
Using 454‐pyrsequencing, we assessed bacterial communities' composition and diversity in salt–alkali soils of monoculture and intercropping mulberry and soybean collected from Songnen Plain of China. In this study, for microbial analysis of soil, the dominant taxonomic groups were Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, Bacteroidetes and Planctomycetes (Fig. 2). These phyla have been described as common inhabitants of non‐saline–alkali soils (Fierer et al., 2012; Kuramae et al., 2012). Shannon diversity analyses revealed a richer bacterial community in intercropping mulberry soil than that of monoculture soil (Table 1). This observation may be supported by previous studies which demonstrate that the bacterial communities of intercropping mulberry have higher diverse than monoculture. The presence of soybean plants contributed to attenuate eventual bacterial community variations occurring in intercropping mulberry. Indeed, tree‐based intercropping systems present a more heterogeneous vegetation cover, a patchier distribution of plant litter and rooting patterns that can affect soil properties and microbial communities (Reynolds et al., 2007; Lacombe et al., 2009).
The bacterial community structure in soybean rhizosphere was influenced from that of the communities in mulberry rhizosphere (Fig. 3). Several studies indicated that the interactions among plant species, growth stages and soil types could influence microbial communities in rhizosphere soil (Marschner et al., 2001; Wieland et al., 2001; Song et al., 2007). In the rhizosphere of soybean, the bacterial community structure in rhizosphere soils of monoculture and intercropping were different. Most of Actinobacteria and Chloroflexi were found in intercropping samples, whereas Acidobacteria and Proteobacteria were present at higher percentages in monoculture samples. The high similarities were found in monoculture and intercropping mulberry, indicating that an important integrated role of intercropping in shaping the soil microbial communities structure. Actinobacteria have a critical role in decomposition of soil organic materials, such as cellulose and chitin (Sykes and Skinner, 1973). Actinobacteria of intercropping soils were higher than that of monocropped soils, which may be due to the presence of more organic matter used by plants in intercropping soils. Comparing to control soil (pH 9.14), the pH of soil of monoculture and intercropping soybean was significantly decreased to 7.30 and 7.60 (Table S2), respectively, which may be due to legume as P‐mobilizing plants species can release protons in alkaline soils (Li et al., 2014). The abundance of the phylum Acidobacteria correlates with the soil pH (Hartman et al., 2008; Jones et al., 2009) and acidobacterial subgroups 1 and 2 decreased with the increase in pH (Lauber et al., 2009), which is in contrast to our study. In our study, pH of intercropping mulberry (pH 7.32) were lower than that of monoculture soils (pH 7.73) (Table S2). The pH reduction in intercropping mulberry may be due to intercropped with legume. However, monoculture mulberry soils contained more Acidobacteria than the corresponding intercropped soils. Acidobacteria are capable of degradation of plant litter in soils (Eichorst et al., 2011), the presence of mulberry debris in monoculture samples and soybean root exudates and litter in intercropping samples may have contributed to the observed differences.
To investigate relationships between soil microbial community composition and measured environmental variables, different bacterial phyla and proteobacterial classes were analysed using CCA (Fig. 7). The pH and conductivity seemed to affect soil bacterial communities. Soil property such as pH and conductivity are expected to control microbial community composition (Drenovsky et al., 2004). Hence, a greater spatial heterogeneity of microbial communities was also expected in intercropping systems of salt–alkali soils. Fierer and Jackson (2006) demonstrated that pH influenced the overall diversity and composition of microbial communities in a range of terrestrial and aquatic environments. Salinity could also affect bacterial composition and diversity across a variety of habitats (Wakelin et al., 2012).
The recent studies of overyielding in agriculture intercropping systems found a important mechanism underlying such facilitation is the ability of some crop species to chemically mobilize otherwise‐unavailable forms of one or more limiting soil nutrients such as phosphorus (Li et al., 2014). Plants do not take up organic P directly; rather, organic P is first hydrolysed by microbial or root‐related phosphatases (Li et al., 2014). Therefore, phosphate‐solubilizing bacteria have an important role in soils with low concentration of available phosphorus. Phosphate‐solubilizing bacteria were present in different proportions in monoculture and intercropping soils in our study. Burkholderia, Pseudomonas and Arthrobacter showed more abundance in intercropping soils than that monoculture soils. These bacteria can improve solubilization of fixed soil phosphorus and applied phosphates resulting in higher crop yields (Nautiyal, 1999).
Legumes and non‐legumes can ‘complement’ each other in the use of N sources as both the legume and non‐legume utilize soil inorganic N sources, but nodule in leguminous plants can also fix atmospheric N2 in symbiosis with Rhizobium (Jensen, 1996). IS libraries had low Rhizobium than SM libraries (Fig. 5). However, most studies showed a opposite results that N2 fixation of legumes may be improved by intercropping when the non‐legume is a strong competitor for soil inorganic N (Giller et al., 1991; Karpenstein‐Machan and Stuelpnagel, 2000; Hauggaard‐Nielsen et al., 2001). The integration of trees with crops on moderately alkali soils improved soil total nitrogen and carbon status (Kaur et al., 2000), due to organic matter inputs in the form of litter fall and fine roots from trees, which showed plant‐soil feedback process are also important.
Plants release enormous chemicals through their roots, at a significant carbon cost, to combat pathogenic microorganisms and attract beneficial ones (Badri et al., 2009). The activity and effects of beneficial rhizosphere microorganisms on plant growth and health are well documented for bacteria like by Pseudomonas and Burkholderia (Badri et al., 2009). Pseudomonas showed a higher relative abundance in intercropping mulberry and soybean respectively. Similar as Pseudomonales, Burkholderia was present at higher relative abundance in intercropping mulberry. These bacteria protect several major agricultural crops against disease phenomenon that is likely to be also important in natural ecosystems (Van Der Heijden et al., 2008).
Principal component analysis clearly demonstrated that the bacterial communities were separated into mulberry and soybean groups, suggesting that plant species was the major factor in determining the bacterial community in the salt–alkali soils (Fig. 5). The distribution of sequences demonstrated that each plant rhizosphere had its own microbial population (Fig. 4), which indicated the appearance of some new microbial population were stimulated by plant diversity. Previous studies also observed that soil carry‐over influences of plant diversity are mediated by a general stimulated of soil microorganisms (Bartelt‐Ryser et al., 2005). Changes in relative abundance of different OTUs also demonstrated the important effects of intercropping.
In conclusion, in intercropped plants growing in salt–alkali soils, the diversity and composition of bacterial communities varied between monoculture and intercropping samples and some of beneficial plant rhizobacteria (phosphate‐solubilizing bacteria) are more abundant in intercropping samples. Soil pH under intercropping was significantly diminished in relation to control soil. Therefore, mulberry–soybean intercropping in the perspective of biology is good strategy to improve salt–alkali soils.
Materials and Methods
Field plots
The field site used in this research was located at the Yuejin village, Zhaozhou County of Heilongjiang province of Songnen Plain (latitude, 45° 70′ N; longitude, 125° 27′ E, northeast China). The average annual temperature in the region is 3.37°C and the mean temperature in July is 22.4°C. The mean annual precipitation is 418.9 mm and nearly 73.1% of total rainfall is received by northwest monsoons from July to September. The mean annual elevation is 1597.1 mm, annual sunshine hours are 3014.4 h, the active accumulated temperature (≥ 10°C) is 2921.3°C per year, and a frost‐free period of 137 days. The soil is classified as a salinized meadow soil, with a pH of 9.04, a salinity content of 0.37%. Selected soil properties were as follows: pH 9.14; salinity content 0.37%; Cl−, 0.23 mol kg−1; CO3 2−, 0.09 mol kg−1; HCO3 −, 0.47 mol kg−1; SO4 2−, 0.16 mol kg−1; Na+, 0.11 mol kg−1; K+, 2.15 mol kg−1; Ca2+, 0.48 mol kg−1 and Mg2+, 0.15 mol kg−1.
The experimental design consisted of three blocks. Each block was divided into three plots representing the three plantation systems. Each plot unit comprised 12 rows that were 5 m long and 0.66 m wide, each 39.6 m2 in size. Plots and blocks were separated from each other by 1 m walkways. Treatments levels included (1) soybean monoculture, (2) mulberry monoculture and (3) soybean intercropped with mulberry. For the intercropped treatment, two soybean rows were intercropped with two rows of mulberry. The single cropping plots consisted of 12 rows of one plant species. Edges of each plot were sown with a mix of soybean and mulberry to minimize edge effects but these plants were not included in the harvest.
The grafted mulberry (root stocks of ‘Qiuyu’ cultivar with the scion of ‘Tieba’ cultivar) was planted on 10 May 2011 using approximately 30 cm seedlings at the spacing of 0.20 m within the rows. The soybeans (Glycine max cv. Heinong 34) were sown manually, and seedlings in each row were thinned after emergence to leave a density of 30 plants m−2. Prior to sowing soybeans, fertilizer in the form of organic manure (30 000 kg hm−2) and (NH4)2HPO4 (150 kg hm−2) were applied and the soil was disked to a depth of 10 cm. A conventional herbicide treatment was applied to the soybean at the 2–3 trifoliate leaf stages.
Soil samples were collected from all plots on 5 August 2011, when the soybean was showing early pod formation (R3 stage). Rhizosphere soils, adhering to the roots (Nazih et al., 2001), were collected by shaking the soil off the roots. Part of the sample was used for DNA extraction for determination of microbial populations while part of the sample was extracted for soil property analyses. For DNA extraction, soil samples were immediately placed into sterile centrifuge tubes under ice, transferred to laboratory within 24 h and kept frozen at −80°C until DNA extraction. For soil property analyses, soil samples were taken to the laboratory, and vegetation, roots and stones were removed prior to soil sieving (< 2 mm). Soil pH (soil/H2O ratio of 1:2) was measured using a pH meter with a glass electrode. Salinity was measured by determining the electrical conductivity (EC, dS m−1) of soil water extract (water to soil ratio, 2.5:1, vol:wt) using a conductivity meter. The soil WC was measured by weighting fresh soil samples. The samples were then dried in the oven at 105°C to a constant mass. Oven dry weight was then determined. Gravimetric soil WC was calculated as (wet soil weight‐dry soil weight)/dry soil weight.
DNA extraction, amplification of 16S rDNA genes and pyrosequencing
Total microbial community DNA was isolated from 1 g of soil per sample using an Omega Bio‐Tek E.Z.N.A. Soil DNA extraction kit (Omega Bio‐Tek, Atlanta, GA, USA) according to the manufacturer's instructions. The extracted DNA was examined following electrophoresis in 1% agarose gel and the DNA was normalized to the same concentration prior to amplification. A ~455 bp region of the 16S rDNA gene, covering the V1‐V3 region was selected to construct the community library through tag pyrosequencing. The V1‐V3 region was amplified with universal primers 8F (5′‐AGAGTTTGATCCTGGCTCAG‐3′) and 533R (5′‐TTACCGCGGCTGCTGGCAC‐3′) containing the A and B sequencing adaptors (454 Life Sciences, Shanghai, China). For each sample, three independent amplification reactions were performed. The PCR mixture (final volume, 50 μl) contained 5 μM of each primer, ~5 ng of template DNA, 5× FastPfu PCR buffer and 2.5 U of FastPfu DNA Polymerase (MBI Fermentas, USA). The amplification conditions consisted of an initial denaturation at 95°C for 2 min and 25 cycles of denaturation at 95°C at 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, followed by a final extension period at 72°C for 5 min. During amplification, negative control reactions lacking template DNA were also performed to check for experimental contamination. The amplicons were then purified once by gel electrophoresis/isolation and an additional two times using the Wizard SV Geland PCR Clean‐Up System (Promega, Madison, WI, USA). Pyrosequencing was then carried on a Roche massively parallel 454 GS‐FlX sequencer according to standard protocols.
Sequence analysis
Pyrosequencing flowgrams were converted to sequence reads using MOTHUR software (http://www.mothur.org) and analysed. The acquired sequences were filtered by evaluating data quality and removing primers and barcodes. Sequence were filtered by: (1) selecting sequences which contained barcode and forward primer and eliminate sequences with even a single base pair, (2) removing sequences shorter than 150 bp, with ambiguous base pairs, or with more than two wrong matches in primer and (3) eliminating barcodes and forward primers. After filtering, the effective sequences were clustered to OTUs based on phylum, class and genus levels using MOTHUR (Sun et al., 2009).
Microbial community analysis
In order to divide OTUs, optimized sequences were reduced to 350 bp length and compared using silva108. Rarefaction curves based on identified OTU, species richness estimator Chao 1 were generated with MOTHUR for each sample (Chao, A. 1984). Taxonomic assignment of sequences was done using the Ribosomal Database Project classifier (minimum confidence of 80%). After phylogenetic allocation of the sequences to the phylum and genus levels, the relative abundance of a given phylogenetic group was set as the number of sequences per sample. Venn diagrams of unique and OTUs (0.03 cut‐off value) were drawn in order to highlight similarities and shared sequences between the different analysed samples. Hierarchical cluster (Heatmap) analyses were generated in MOTHUR using the gplots package of R. UniFrac, which described the similarity and difference between the four samples.
Statistical analysis
All experimental data were processed by Microsoft EXCEL 2010. Data were analysed using one‐way analysis of variance ANOVA. Values shown are the means of three replicates ± SD.
Supporting information
Table S1. Number of 16S rDNA gene sequences derived from all soil samples. Mulberry monoculture (MM), intercropping mulberry (IM), soybean monoculture (SM) and intercropping soybean (IS).
Table S2. Soil property from all soil samples. Mulberry monoculture (MM), intercropping mulberry (IM), soybean monoculture (SM) and intercropping soybean (IS). Different letters following the mean values within each column indicates significant differences at P < 0.05.
Acknowledgements
This research was supported by the “Twelfth Five‐Year” National Science and Technology Support Program of China (2011BAD08B02‐3), the Major Project for Heilongjiang Province Science and Technology Program (GZ13B004) and the Fundamental Research Funds for the Central Universities (2572014AA16). The authors are so appreciate the great helpful suggestions and construction development from Prof. Sun during the early course of this research and appreciate the helpful suggestions from Prof. Finer and Drs. Xiuli Zhang for their critical reading of this manuscript.
Microbial Biotechnology (2016) 9(3), 293–304
References
- Alam, S.M. , Naqvi, S.S.M. and Ansari, R . (1999) Impact of soil pH on nutrient uptake by crop plants In Handbook of Plant and Crop Stress. Mohammad P.(ed.). Marcel Dekker, pp. 51–60. [Google Scholar]
- Alexander, M .(1977) Introduction to Soil Microbiology. John Wiley & Sons, pp. 223–225. [Google Scholar]
- Badri, D.V. , Weir, T.L. , van der Lelie, D. , and Vivanco, J.M. (2009) Rhizosphere chemical dialogues: plant–microbe interactions. Curr Opin Biotechnol, 20: 642–650. [DOI] [PubMed] [Google Scholar]
- Bartelt‐Ryser, J. , Joshi, J. , Schmid, B. , Brandl, H. , and Balser, T. (2005) Soil feedbacks of plant diversity on soil microbial communities and subsequent plant growth. Perspect Plant Ecol, 7: 27–49. [Google Scholar]
- Bastida, F. , Kandeler, E. , Moreno, J. , Ros, M. , García, C. , and Hernández, T. (2008) Application of fresh and composted organic wastes modifies structure, size and activity of soil microbial community under semiarid climate. Appl Soil Ecol, 40: 318–329. [Google Scholar]
- Drenovsky, R. , Vo, D. , Graham, K. , and Scow, K. (2004) Soil water content and organic carbon availability are major determinants of soil microbial community composition. Microbial Ecol, 48: 424–430. [DOI] [PubMed] [Google Scholar]
- Eichorst, S.A. , Kuske, C.R. , and Schmidt, T.M. (2011) Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria . Appl Environ Microbiol, 77: 586–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Dessougi, H. , zu Dreele, A. , and Claassen, N. (2003) Growth and phosphorus uptake of maize cultivated alone, in mixed culture with other crops or after incorporation of their residues. J Plant Nutr Soil Sci, 166: 254–261. [Google Scholar]
- Fierer, N. , and Jackson, R.B. (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci, 103: 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer, N. , Leff, J.W. , Adams, B.J. , Nielsen, U.N. , Bates, S.T. , Lauber, C.L. , et al (2012) Cross‐biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA, 109: 21390–21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giller, K.E. , Ormesher, J. , and Awah, F.M. (1991) Nitrogen transfer from Phaseolus bean to intercropped maize measured using 15 N‐enrichment and 15 N‐isotope dilution methods. Soil Biol Biochem, 23: 339–346. [Google Scholar]
- Gupta, S. , Sinha, A. , and Rana, R. (1990) Biomass dynamics and nutrient cycling in a sodic grassland. Int J Ecol Environ Sci, 16: 57–70. [Google Scholar]
- Hartman, W.H. , Richardson, C.J. , Vilgalys, R. , and Bruland, G.L. (2008) Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc Natl Acad Sci USA, 105: 17842–17847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauggaard‐Nielsen, H. , Ambus, P. , and Jensen, E.S. (2001) Interspecific competition, N use and interference with weeds in pea‐barley intercropping. Field Crop Res, 70: 101–109. [Google Scholar]
- Hauggaard‐Nielsen, H. , Ambus, P. , and Jensen, E.S. (2003) The comparison of nitrogen use and leaching in sole cropped versus intercropped pea and barley. Nurt Cycl Agroecosys, 65: 289–300. [Google Scholar]
- Jensen, E.S. (1996) Grain yield, symbiotic N2 fixation and interspecific competition for inorganic N in pea‐barley intercrops. Plant Soil, 182: 25–38. [Google Scholar]
- Jones, R.T. , Robeson, M.S. , Lauber, C.L. , Hamady, M. , Knight, R. , and Fierer, N. (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J, 3: 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpenstein‐Machan, M. , and Stuelpnagel, R. (2000) Biomass yield and nitrogen fixation of legumes monocropped and intercropped with rye and rotation effects on a subsequent maize crop. Plant Soil, 218: 215–232. [Google Scholar]
- Kaur, B. , Gupta, S. , and Singh, G. (2000) Soil carbon, microbial activity and nitrogen availability in agroforestry systems on moderately alkaline soils in northern India. Appl Soil Ecol, 15: 283–294. [Google Scholar]
- Kuramae, E.E. , Yergeau, E. , Wong, L.C. , Pijl, A.S. , van Veen, J.A. , and Kowalchuk, G.A. (2012) Soil characteristics more strongly influence soil bacterial communities than land‐use type. FEMS Microbiol Ecol, 79: 12–24. [DOI] [PubMed] [Google Scholar]
- Lacombe, S. , Bradley, R.L. , Hamel, C. , and Beaulieu, C. (2009) Do tree‐based intercropping systems increase the diversity and stability of soil microbial communities? Agr Ecosyst Environ, 131: 25–31. [Google Scholar]
- Lauber, C.L. , Hamady, M. , Knight, R. , and Fierer, N. (2009) Pyrosequencing‐based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol, 75: 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Zhang, F. , Li, X. , Christie, P. , Sun, J. , Yang, S. , et al (2003) Interspecific facilitation of nutrient uptake by intercropped maize and faba bean. Nutr Cycl Agroecosys, 65: 61–71. [Google Scholar]
- Li, L. , Li, S.M. , Sun, J.H. , Zhou, L.L. , Bao, X.G. , Zhang, H.G. , et al (2007) Diversity enhances agricultural productivity via rhizosphere phosphorus facilitation on phosphorus‐deficient soils. Proc Natl Acad Sci USA, 104: 11192–11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Tilman, D. , Lambers, H. , and Zhang, F.S. (2014) Plant diversity and overyielding: insights from belowground facilitation of intercropping in agriculture. New Phytol, 203: 63–69. [DOI] [PubMed] [Google Scholar]
- Lim, Y.W. , Kim, B.K. , Kim, C. , Jung, H.S. , Kim, B.S. , Lee, J.H. , et al (2010) Assessment of soil fungal communities using pyrosequencing. J Microbiol, 48: 284–289. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Luo, Y. , Teng, Y. , Li, Z. , and Christie, P. (2009) Prepared bed bioremediation of oily sludge in an oilfield in northern China. J Hazard Mater, 161: 479–484. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Guo, W. , and Shi, D. (2010) Seed germination, seedling survival, and physiological response of sunflowers under saline and alkaline conditions. Photosynthetica, 48: 278–286. [Google Scholar]
- Marschner, P. , Yang, C.H. , Lieberei, R. , and Crowley, D.E. (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem, 33: 1437–1445. [Google Scholar]
- McClung, G. , and Frankenberger, W. (1985) Soil nitrogen transformations as affected by salinity. Soil Sci, 139: 405–411. [Google Scholar]
- Munns, R. (2002) Comparative physiology of salt and water stress. Plant, Cell Environ, 25: 239–250. [DOI] [PubMed] [Google Scholar]
- Nautiyal, C.S. (1999) An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett, 170: 265–270. [DOI] [PubMed] [Google Scholar]
- Nazih, N. , Finlay‐Moore, O. , Hartel, P. , and Fuhrmann, J. (2001) Whole soil fatty acid methyl ester (FAME) profiles of early soybean rhizosphere as affected by temperature and matric water potential. Soil Biol Biochem, 33: 693–696. [Google Scholar]
- Neumann, A. , Schmidtke, K. , and Rauber, R. (2007) Effects of crop density and tillage system on grain yield and N uptake from soil and atmosphere of sole and intercropped pea and oat. Field Crops Res, 100: 285–293. [Google Scholar]
- Rao, D. , and Ghai, S. (1985) Urease and dehydrogenase activity of alkali and reclaimed soils. Soil Res, 23: 661–665. [Google Scholar]
- Rao, D. , and Pathak, H. (1996) Ameliorative influence of organic matter on biological activity of salt‐affected soils. Arid Land Res Manag, 10: 311–319. [Google Scholar]
- Reynolds, P.E. , Simpson, J.A. , Thevathasan, N.V. , and Gordon, A.M. (2007) Effects of tree competition on corn and soybean photosynthesis, growth, and yield in a temperate tree‐based agroforestry intercropping system in southern Ontario, Canada. Ecol Eng, 29: 362–371. [Google Scholar]
- Rietz, D. , and Haynes, R. (2003) Effects of irrigation‐induced salinity and sodicity on soil microbial activity. Soil Biol Biochem, 35: 845–854. [Google Scholar]
- Rosselló‐Mora, R. , and Amann, R. (2001) The species concept for prokaryotes. FEMS Microbiol Rev, 25: 39–67. [DOI] [PubMed] [Google Scholar]
- Shi, D. , and Zhao, K. (1997) Effects of NaCl and Na2CO3 on growth of Puccinellia tenuiflora and on present state of mineral elements in nutrient solution. Acta Pratacu Sin, 6: 51–61. [Google Scholar]
- Singh, G. , Singh, N. , Dagar, J. , Singh, H. , and Sharma, V. (1997) An evaluation of agriculture, forestry and agroforestry practices in a moderately alkali soil in northwestern India. Agroforest Syst, 37: 279–295. [Google Scholar]
- Song, Y. , Zhang, F. , Marschner, P. , Fan, F. , Gao, H. , Bao, X. , et al (2007) Effect of intercropping on crop yield and chemical and microbiological properties in rhizosphere of wheat (Triticum aestivum L.), maize (Zea mays L.), and faba bean (Vicia faba L.). Biol Fert Soils, 43: 565–574. [Google Scholar]
- Sun, Y.M. , Zhang, N.N. , Wang, E.T. , Yuan, H.L. , Yang, J.S. , and Chen, W.X. (2009) Influence of intercropping and intercropping plus rhizobial inoculation on microbial activity and community composition in rhizosphere of alfalfa (Medicago sativa L.) and Siberian wild rye (Elymus sibiricus L.). FEMS Microbiol Ecol, 70: 218–226. [DOI] [PubMed] [Google Scholar]
- Sykes, G. and Skinner, F.A . (1973) Actinomycetales: characteristics and practical importance In Society for Applied Bacteriology Symposium, Sykes G. and Skinner F.A. (eds.) Academic Press, pp. 191‐192. [Google Scholar]
- Trenbath, B. (1999) Multispecies cropping systems in India: predictions of their productivity, stability, resilience and ecological sustainability. Agroforest Syst, 45: 81–107. [Google Scholar]
- Tripathi, S. , Kumari, S. , Chakraborty, A. , Gupta, A. , Chakrabarti, K. , and Bandyapadhyay, B.K. (2006) Microbial biomass and its activities in salt‐affected coastal soils. Biol Fert Soils, 42: 273–277. [Google Scholar]
- Van Der Heijden, M.G. , Bardgett, R.D. , and Van Straalen, N.M. (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett, 11: 296–310. [DOI] [PubMed] [Google Scholar]
- Wakelin, S.A. , Anand, R.R. , Reith, F. , Gregg, A.L. , Noble, R.R. , Goldfarb, K.C. , et al (2012) Bacterial communities associated with a mineral weathering profile at a sulphidic mine tailings dump in arid Western Australia. FEMS Microbiol Ecol, 79: 298–311. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Seki, K. , Miyazaki, T. , and Ishihama, Y. (2009) The causes of soil alkalinization in the Songnen Plain of Northeast China. Paddy Water Environ,, 7: 259–270. [Google Scholar]
- Wieland, G. , Neumann, R. , and Backhaus, H. (2001) Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl Environ Microbiol, 67: 5849–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley, R. (1979) Intercropping‐its importance and research needs. Part, 1: 1–10. [Google Scholar]
- Will, C. , Thürmer, A. , Wollherr, A. , Nacke, H. , Herold, N. , Schrumpf, M. , et al (2010) Horizon‐specific bacterial community composition of German grassland soils, as revealed by pyrosequencing‐based analysis of 16S rRNA genes. Appl Environ Microbiol, 76: 6751–6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey, R . (1979) Intercropping: its Importance and Research Needs. Part 2, Agronomy and Research Approaches. Commonwealth Agricultural Bureaux, pp. 150–151. [Google Scholar]
- Zheng, X.Y. , Zhao, L. , Xu, N. , Zhang, H.H. , and Sun, G.Y. (2011) Interspecific interaction of below‐ground and above‐ground indices in mulberry‐soybean intercropping system. Soils, 43: 493–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Number of 16S rDNA gene sequences derived from all soil samples. Mulberry monoculture (MM), intercropping mulberry (IM), soybean monoculture (SM) and intercropping soybean (IS).
Table S2. Soil property from all soil samples. Mulberry monoculture (MM), intercropping mulberry (IM), soybean monoculture (SM) and intercropping soybean (IS). Different letters following the mean values within each column indicates significant differences at P < 0.05.


