Abstract
The metastasis-associated lung adenocarcinoma transcription 1 (MALAT1) is a highly conserved long non-coding RNA (lncRNA) gene. However, little is known about the pathological role of lncRNA MALAT1 in glioma. In the present study, we explored the expression level of lncRNA MALAT1 in primary glioma tissues as well as in U87 and U251 glioma cell lines. Using qRT-PCR, we found that the expression of lncRNA MALAT1 was significantly increased in glioma tissues compared with that of paracancerous tissues. Meanwhile, the expression of MALAT1 was highly expressed in U98 and U251 cells. In order to explore the function of MALAT1, the expression of MALAT1 was greatly reduced in U87 and U251 cells transfected with siRNA specifically targeting MALAT1. Consequently, cell viability of U87 and U251 cells were drastically decreased after the knockdown of MALAT1. Concomitantly, the apoptosis rate of the two cell lines was dramatically increased. Furthermore, the expression levels of some tumor markers were reduced after the knockdown of MALAT1, such as CCND1 and MYC. In summary, the current study indicated a promoting role of MALAT1 in the development of glioma cell.
Keywords: RNA, Long Noncoding; MALAT1 Long Non-coding RNA, Human; Glioma; Apoptosis
INTRODUCTION
Glioma represents the most common form of primary brain tumors. According to the WHO (World Health Organization) classification in 2007 (1), gliomas are generally categorized into astrocytomas with well differentiation (WHO grade I–II), anaplastic astrocytomas (WHO grade III), and the most aggressive glioblastoma multiforme (GBM, WHO grade IV). Although great progress has been made in surgery, radiotherapy as well as chemotherapy for the treatment of GBM, yet its prognosis is still very poor with a median survival of only about 15 months (2,3). Therefore, identification of novel molecules is an urgent need for the diagnosis, treatment of gliomas, especially the GBM, where lncRNAs are promising candidates.
Long non-coding RNAs (lncRNAs) are defined as genomic transcripts longer than 200 nucleotides (nt) without protein coding function due to a lack of an open reading frame of significant length (shorter than 100 amino acids) (4,5). Mounting evidence has demonstrated that lncRNAs are involved in a range of genetic phenomena, including X-chromosome inactivation, DNA methylation, as well as other transcriptional, post-transcriptional and epigenetic regulations (6,7,8,9). The establishment of new bioinformatical strategies has allowed the identification of great amount of novel lncRNA transcripts. Meanwhile, more and more evidence is being provided concerning the ways by which lncRNAs function. It is clear now that lncRNAs could regulate gene expressions possibly through their interactions with DNAs, RNAs, or chromatin remodeling proteins (10). It has also been generally accepted that lncRNAs may be key regulators in diverse brain disorders, including gliomas.
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear enriched abundant transcript 2 (NEAT2), is a lncRNA with a length of about 8000-nt, which is ubiquitously expressed in human tissues and highly phylogenetic conserved in mammals (11). Previous studies have revealed that MALAT1 was usually overexpressed and functioned in a variety of cancers, such as esophageal squamous cell carcinoma, gastric cancer, hepatocellular cancer, pancreatic cancer, colorectal cancer, cervical cancer and prostate cancer (12,13,14,15,16,17,18). Experiment showed that lncRNA MALAT1 was involved in regulation of hyperproliferation, epithelial-mesenchymal transition (EMT) and metastasis of cancer cells through modulation of oncogenic transcription factors such as p53 and c-Myc, favoring cancer progression (19,20). MALAT1 might also function in the tumorigenesis and progression of glioma. Multivariate analysis by Ma and colleagues has suggested that increased level of lncRNA MALAT1 was an independent predictor for poor prognosis in glioma patients. However, the functionality of lncRNA MALAT1 in glioma cells, especially in GBM, remains elusive.
In this study, we detected the expression profile of MALAT1 in glioma patients and cell lines via quantitative reverse transcription PCR (qRT-PCR) assay, and explored the function of MALAT1 on cell growth in U87 and U251 glioma cell lines with specific small interfering RNAs for MALAT1. Furthermore, the involvement of factors displaying important functions in glioma tumorigenesis and development was also determined in this process.
MATERIALS AND METHODS
Patients information
Thirty seven glioma tissue samples and the corresponding adjacent normal brain tissues with a distance 1 cm away from the cancerous tissues were selected. The samples were collected between 2011 and 2015 from the Yishui Hospital, and the pathological information was retrieved. None of the patients had ever received chemotherapy or radiotherapy prior to surgery.
Cell culture
Normal glia cell line NHA and the human malignant glioma cell lines U87 and U251 were acquired from the Beijing Zhongyuan Company (Beijing, China). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 µg/mL of streptomycin and 100 units/mL of penicillin at 37℃ in a humidified atmosphere containing 5% CO2.
Quantitative real-time PCR (qRT-PCR)
The first strand of cDNA was synthesized from 1 μg purified totalRNA by reverse transcription with the Superscript III First-Strand Synthesis System kit (Invitrogen, Waltham, MA, USA). Briefly, 10 μL reaction system was firstly prepared with 1 μg RNA, 1 μL RT-primer, 1 μL 10 mM dNTP and DEPC-treated water and then maintained at 65℃ for 5 minutes. Secondly, the 10 μL reaction system containing 2 μL 10 × RT buffer solution, 2 μL 0.1 MDTT, 4 μL 25 mM MgCl2, 1 μL RNaseOUT and 1 μL SuperScript III RT was added and mixed evenly to prepare a total 20 μL reaction system and the reaction was carried out at 50℃ for 50 minutes and at 85℃ for 5 minutes sequentially. Finally, 1 μL RNaseH was added to react at 37℃ for 20 minutes and then the reverse transcription product was collected. With the obtaining cDNA as a template and using SYBR Premix Ex Taq II (TaKaRa Bio, Otsu, Japan), qPCR was performed on ABI7300 (Invitrogen). The reaction condition was as follows: melting at 95℃ for 10 seconds, annealing at 95℃ for 5 seconds, and extension at 60℃ for 20 seconds, for a total of 45 cycles. The relative expression levels of lncRNAs or coding genes were quantified using 2-ΔΔCT method. GAPDH was used as internal control. The primer sequences were as follows: MALAT1: sense, TGATAGCCAAATTGAGACAA; antisense, TTCAGGGTGAGGAAGTAAAA; CCND1: sense, GCTGCGAAGTGGAAACCATC, antisense, CCTCCTTCTGCACACATTTGAA; MYC: sense, GGCTCCTGGCAAAAGGTCA, antisense, CTGCGTAGTTGTGCTGATGT; GAPDH, sense, AATGGACAACTGGTCGTGGAC, antisense, CCCTCCAGGGGATCTGTTTG.
Knockdown of MALAT1 in glioma cells
The siRNA specifically targeting MALAT1 was constructed by GeneChem Company (Shanghai, China), the sequence was as follows: sense, GGGCUUCUCUUAACAUUUAUU, antisense, UAAAUGUUAAGAGAAGCCCUU. U87 and U251 with high level of MALAT1 cells were transfected with siRNA targeting MALAT1 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells transfected with Scramble was used as the negative control, the Scramble siRNA sequence was as follows: sense, AACAUCUCUUGGGCUUUUAUU, antisense, GAGAAUUAAGUAAAUGCCCUU. Cells were collected after transfection for RNA extraction, western blot, MTT cell growth assay, and apoptosis assay.
Determination of glioma cell growth
A total of 2 × 103 cells were seeded into 96-well plates and cultured in medium with 10% FBS. A total of 5 groups were set: 0 hours, 24 hours, 48 hours, 72 hours and 96 hours, with three replicates for each time point. At each time point, cells in one well were treated with MTT solution (5 mg/mL in PBS) 20 μL, and incubated for 4 hours, the supernatant medium was then discarded followed by the addition of 150 μL DMSO (dimethylsulfoxide). After vibrated for 10 minutes, the values were measured at 570 nm wavelength.
Determination of glioma cell invasion
The invasion assays were performed using a Transwell chamber (Corning, NY, USA). A total of 5 × 104 cells in serum-free medium were placed into the upper chamber coated with Matrigel (BD Biosciences, San Jose, CA, USA). Then, medium containing 10% FBS was added to the lower chamber. After 24 hours of incubation, the cells invaded through the membrane were fixed and stained with methanol and 0.1% crystal violet. The cells were imaged using an IX70 inverted microscope (Olympus, Tokyo, Japan).
Determination of cell apoptosis
Flow cytometric analyses with fluorescein isothiocyanate (FITC)-labelled annexin V and propidium iodide were performed according to the manufacturer’s protocol (BD Pharmingen, BD Biosciences). Briefly, cells were seeded in 6 well plates at a density of 50,000 cells per well and grow to a confluency of 70% before treatment with nucleosides. Following exposure to the test nucleosides, the cells were harvested and analysed by flow cytometry on a BD LSR II flow cytometer. The data was plotted as a scattergraph with FITC-Annexin V (green fluorescence) represented on the X-axis versus PI (red fluorescence) on the Y-axis.
Western blot analysis
U87 and U251 cells (1 × 106 cells/well) were seeded into 100 mL culture dishes for 24 hours and then transfected with Scramble and siRNA targeting MALAT1. After incubation for 24 hours, the cells were harvested and prepared in NP-40 lysis buffer (Beyotime Biotechnology Company, Shanghai, China) for 30 minutes on ice. Protein concentrations were determined using a BCA protein assay kit (Beijing ComWin Biotech Company, Beijing, China), and resolved on 12%–15% SDS-PAGE gels, then transferred to PVDF (polyvinylidene difluoride) membranes (Roche Applied Science, Basel, Switzerland) and blocked with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) overnight at 4°C. After washing with TBST, the membranes were incubated with primary antibodies against CCND1 (sc-249, dilutions of 1:2,000, Santa Cruz Biotech Company, CA, USA), MYC (sc-40, dilutions of 1:1,000, Santa Cruz Biotech Company), and GAPDH (sc-32233, dilutions of 1:5,000, Santa Cruz Biotech Company) overnight at 4°C. Subsequently, the membranes were incubated with secondary antibodies (Zhongshanjinqiao Biotech Company, Beijing, China) at room temperature for 2 hours at a dilution of 1:5,000, respectively. Antibody-bound proteins were detected, normalized to GAPDH and quantified in a ChemiDoc™ XRS system (Universal Hood II, Bio-Rad, Hercules, CA, USA) using an Immun-Star™ HRP Chemiluminescence Kit (Bio-Rad).
Statistical analysis
The statistical analyses were performed using SPSS 17.0 (SPSS, Chicago, IL, USA). The significance of the difference between various groups was analyzed using Student’s t-test. P value < 0.05 was considered statistically significant.
Ethics statement
This study was approved by the institutional review board of Qingdao University and Yishui Hospital, Shandong, China (IRB No. 2013-11-012). Informed consent was submitted from the patients for use of tissues in this study.
RESULTS
MALAT1 was highly expressed in glioma patients and cell lines
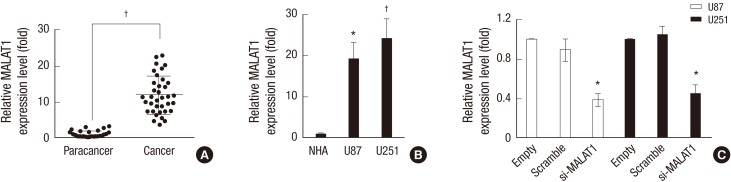
The relative expression level of MALAT1 in glioma patients were examined with qRT-PCR assay, and the level of MALAT1 in glioma samples were compared to that in adjacent normal brain tissues. The result showed that the expression of MALAT1 was significantly increased in cancerous tissues compared with that in paracancerous tissues (Fig. 1A). Meanwhile, the expression of MALAT1 in U87 and U251 cells were also verified via qRT-PCR assay, and the expression of MALAT1 was also greatly up-regulated in these two malignant glioma cell lines when compared with that in the normal human astrocytes (NHA) cell line (Fig. 1B). To further explore the function of MALAT1 in glioma growth, U87 and U251 cells were transfected with siRNA targeting MALAT1. The endogenous expression of MALAT1 was effectively knocked down (Fig. 1C).
Fig. 1.
MALAT1 was highly expressed in glioma patients and cell lines. (A) 37 glioma tissue samples and adjacent normal brain samples were selected, and then qRT-PCR assay was employed to detect the relative expression level of MALAT1 in glioma patients. The adjacent normal brain tissues (paracancer group) were used as the negative control. (B) Human malignant glioma cell lines U87 and U251 cells were cultured, and then qRT-PCR assay was employed to detect the relative expression level of MALAT1 in both cells. The normal glia cell line NHA was used as the negative control. (C) U87 and U251 cells were transfected with siRNA targeting MALAT1, QRT-PCR assay was employed to detect the endogenous expression of MALAT1 to confirm the knockdown efficiency. The glioma cells transfected with Scramble was used as the negative control.
*P < 0.05 vs. the control; † P < 0.01 vs. the control.
Knockdown of MALAT1 decreased the growth of glioma cells
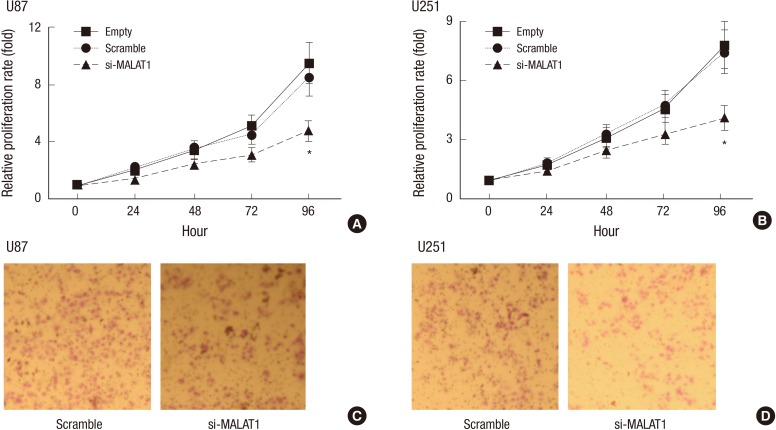
As indicated that MALAT1 could be effectively knocked down, cell growth was then detected via MTT assay. Glioma cells generally manifest powerful growth ability, which, however, was greatly attenuated with knockdown of MALAT1 in a time dependent manner, especially at 72 hours and 96 hours post transfection of MALAT1 siRNA (Fig. 2A and 2B). Next, we studied if MALAT1 knockdown would also affect cell invasion in glioma cell lines. We found that both U87 and U251 cells displayed reduced cell mobility after MALAT1 knockdown (Fig. 2C and 2D).
Fig. 2.
Knockdown of MALAT1 decreased the growth of glioma cells. (A, B) U87 and U251 cells were seeded into 96-well plates and cultured for 0 hours, 24 hours, 48 hours, 72 hours and 96 hours respectively, MTT assay was employed to detect the effect of MALAT1 knockdown on cell growth at different time points in both cells. The glioma cells transfected with Scramble was used as the negative control. *P < 0.05 vs. the control. (C, D) U87 and U251 cells in serum-free medium were placed into the upper transwell chamber, the invasion assays in vitro were carried out to detect the effect of MALAT1 knockdown on cell invasion after 24 hours of incubation. The glioma cells transfected with Scramble was used as the negative control.
*P < 0.05 vs. the control.
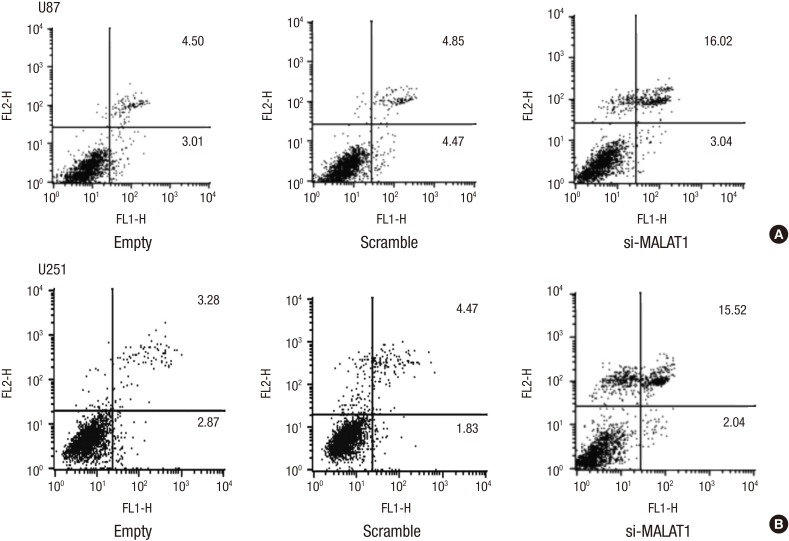
Knockdown of MALAT1 increased the apoptosis rate of glioma cells
We further examined the effect of MALAT1 knockdown on the apoptosis rate of glioma cells, which was determined by Annexin V-FITC/PI double staining assay. The results indicated that U87 and U251 cells transfected with siRNA targeting MALAT1 showed a much higher apoptosis rate than the cells with scramble siRNA (Fig. 3A and 3B).
Fig. 3.
Knockdown of MALAT1 increased the apoptosis rate of glioma cells. (A, B) U87 and U251 cells were seeded in 6 well plates and grow to a confluency of 70%, then both cells were transfected with si-MALAT1 or Scramble respectively. Annexin V-FITC/PI double staining assay was employed to detect the effect of MALAT1 knockdown on the apoptosis rate in U87 (A) and U251 (B) cells. The glioma cells transfected with Scramble was used as the negative control. FL2-H, FL2-Height.
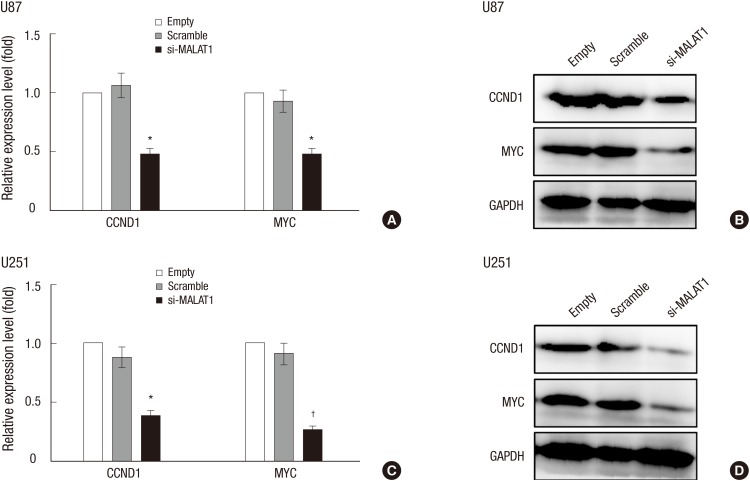
CCND1 and MYC was down-regulated with MALAT1 knockdown
It was reported that oncogenic genes CCND1 (21) and MYC (22) was expressed at high levels in glioma cells. We thus examined the change of expression levels of CCND1 and MYC in response to MALAT1 knockdown via qRT-PCR and western blot analysis. The results showed that the expression of these two proteins were remarkably inhibited in both U87 (Fig. 4A and 4B) and U251 (Fig. 4C and 4D) glioma cells transfected with siRNA targeting MALAT1.
Fig. 4.
CCND1 and MYC was down-regulated with MALAT1 knockdown. U87 and U251 cells were seeded in 60 mm plates and grow to a confluency of 70%, then both cells were transfected with si-MALAT1 or Scramble respectively. QRT-PCR and western blot analysis were employed to verify the change of expression levels of CCND1 and MYC in response to MALAT1 knockdown in U87 cells (A, B) and U251 cells (C, D). The cells transfected with Scramble was used as the negative control.
*P < 0.05 vs. the control; † P < 0.01 vs. the control.
DISCUSSION
LncRNAs have been suggested to play important roles in both normal brain development and different pathologic processes including gliomagenesis. Certain types of lncRNAs like TSLC1-AS1, NEAT1 and HOTAIRM1, are closely involved in the initiation, progression, invasion, recurrence and stem-like characteristics of glioma, and might therefore be exploited for the purposes of sub-classification, early diagnosis and prognosis prediction of glioma. Qin et al. (23) suggested that lncRNA TSLC1-AS1, the antisense transcript of tumor suppressor gene TSLC1, serves as a tumor suppressor in the tumorigenesis of glioma through modulating the expression of TSLC1. Resultantly, lncRNA TSLC1-AS1 may serve as a biomarker and novel therapeutic target for glioma. Zhen et al. (24) demonstrated that lncRNA NEAT1 promotes the development of glioma by regulating the miR-449b-5p/c-Met axis. Zhang et al. (25) have suggest the potential roles of lncRNA in gliomagenesis, and may help to understand the pathogenesis of gliomas associated with Isocitrate Dehydrogenase 1 (IDH1) mutation. Ke et al. (26) provided a comprehensive demonstration of the HOTAIR-miR-326-FGF1 axis in human glioma.
Despite the widely reported involvement in a range of cancers and the aberrant expression profile in glioma patients, the functionality and regulation of lncRNA MALAT1 in glioma are yet largely unknown. The current study, for the first time as we know, explored the possible role and associated regulatory machinery of MALTA1 in glioma cells. The results showed that the expression level of MALAT1 was much higher in glioma tissues than in para-cancerous tissues, which was supported by the results from the comparison between glioma cell lines U87, U251 and normal glia cell line NHA, with the cancer cell lines exhibiting much higher level of MALAT1. Successful knockdown of MALAT1 expression with specific siRNA in both U87 and U251 cell line resulted in the retardation of cell growth and the promotion of cell apoptosis rate, accompanied with the down regulated expression of CCND1 and MYC, suggesting a proto-oncogenic role of MALAT1 in glioma.
MALAT1 has been suggested to participate in cancer cell growth and metastasis of a list of cancer cells and we added glioma onto the list. In gastric cancer cells, with recruitment of SF2/ASF, MALAT1 could facilitate cell cycle progression and promote cancer cell growth (27). Meanwhile, silencing of MALAT1 with siRNA in esophageal squamous cell carcinoma cells drastically suppressed the cell proliferation through G2/M arrest (28). In cervical cancer cells, suppressing MALAT1 expression decreased the expression of cyclinD1, cyclinE, and CDK6, favoring cell cycle retardation (29). We also found that inhibition of endogenous MALAT1 retarded the cell growth, and decreased the expression of CCND1 and MYC, two important regulators of cell cycle. Taken together, we might propose that knockdown of MALAT1 expression in glioma cells induced cell cycle retardation, followed with induction of cell apoptosis, thus inhibit the growth of glioma. As MALAT1 has been proposed to function generally through two mechanisms, one is the control of alternative splicing and the other is the transcriptional regulation. How MALAT1 exert its function on cell proliferation and apoptosis in glioma cells remains further investigation. Furthermore, due to the involvement of MALAT1 in the invasion and metastasis of MALAT1 in cancer cells, especially in nonsmall cell lung cancer (NSCLC) and the general invasive growth of glioma, it is tempting to detect the role of MALAT1 in the invasion of glioma cells.
Clinical studies have also suggested that lncRNA MALAT1 overexpression could be an independent indicator for poor prognosis in several types of cancers, such as lung (30), liver (31) and bladder cancers (32). Ma et al. also (33) showed that the expression level of MALAT1 conversely correlated with the overall survival (OS) in glioma patients and through multivariate analysis, they pointed out that MALAT1 overexpression was an independent factor for unfavorable prognosis. Together with our results, MALAT1 probably serves as a proto-oncogenic role in glioma. Nonetheless, comprehensive studies are needed to determine in which step MALAT1 could affect glioma, the initiation step or the development process.
Footnotes
DISCLOSURE: The authors have no potential conflicts of interest to disclose.
AUTHOR CONTRIBUTION: Conception and design of study: Xiang J, Guo S, Xiang J. Data collection and analysis: Xiang J, Jiang S, Xu Y. Writing the manuscript: Xiang J, Li J, Li L, Xiang J. Discussion and manuscript revision: Xiang J, Xu Y, Li J, Xiang J. Approval of the final version of the manuscript: all authors.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 3.Tate MC, Aghi MK. Biology of angiogenesis and invasion in glioma. Neurotherapeutics. 2009;6:447–457. doi: 10.1016/j.nurt.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XQ, Leung GK. Long non-coding RNAs in glioma: functional roles and clinical perspectives. Neurochem Int. 2014;77:78–85. doi: 10.1016/j.neuint.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu WT, Lu X, Tang GH, Ren JJ, Liao WJ, Ge PL, Huang JF. LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J Gastroenterol. 2014;20:6314–6321. doi: 10.3748/wjg.v20.i20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36:2403–2407. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 16.Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie P, Zhou G, Li G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett. 2014;7:2135–2141. doi: 10.3892/ol.2014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sowalsky AG, Xia Z, Wang L, Zhao H, Chen S, Bubley GJ, Balk SP, Li W. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Cancer Res. 2015;13:98–106. doi: 10.1158/1541-7786.MCR-14-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 20.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, Zeng X, Tian D, Xu H, Cai Q, Wang J, Chen Q. MicroRNA-383 inhibits anchorage-independent growth and induces cell cycle arrest of glioma cells by targeting CCND1. Biochem Biophys Res Commun. 2014;453:833–838. doi: 10.1016/j.bbrc.2014.10.047. [DOI] [PubMed] [Google Scholar]
- 22.Annibali D, Whitfield JR, Favuzzi E, Jauset T, Serrano E, Cuartas I, Redondo-Campos S, Folch G, Gonzàlez-Juncà A, Sodir NM, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin X, Yao J, Geng P, Fu X, Xue J, Zhang Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int J Clin Exp Pathol. 2014;7:3065–3072. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 2015 doi: 10.1007/s13277-015-3843-y. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XQ, Kiang KM, Wang YC, Pu JK, Ho A, Cheng SY, Lee D, Zhang PD, Chen JJ, Lui WM, et al. IDH1 mutation-associated long non-coding RNA expression profile changes in glioma. J Neurooncol. 2015;125:253–263. doi: 10.1007/s11060-015-1916-9. [DOI] [PubMed] [Google Scholar]
- 26.Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, Li Z, Liu XB, Li ZQ, Wang ZH, et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6:21934–21949. doi: 10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557–564. doi: 10.1016/j.biopha.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu H, He Y, Lin L, Qi Z, Ma L, Li L, Su Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2015 doi: 10.1007/s13277-015-3946-5. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 30.Guo F, Yu F, Wang J, Li Y, Li Y, Li Z, Zhou Q. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed Rep. 2015;3:309–312. doi: 10.3892/br.2015.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8:5395–5402. [PMC free article] [PubMed] [Google Scholar]
- 32.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 33.Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36:3355–3359. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]