Abstract
Ribonucleotides incorporated during nuclear DNA replication are removed by RNase H2-dependent ribonucleotide excision repair (RER). When RER is defective, topoisomerase 1 (Top1) incises the DNA at unrepaired ribonucleotides, initiating their removal but accompanied by slow growth, replicative stress and a strongly increased rate of short deletions. Here we show that these phenotypes are incurred by a high level of ribonucleotides incorporated by a variant of the major leading strand replicase, DNA polymerase ε (Pol ε), but not by orthologous variants of the lagging strand replicases, Pols α or δ. Moreover, in the absence of RER, increased ribonucleotide incorporation by Pol ε, but not by Pols α or δ, is lethal in combination with loss of RNase H1. Several explanations for this asymmetry are considered, including the idea that Top1 incision at ribonucleotides relieves torsional stress in the nascent leading strand but not in the discontinuous nascent lagging strand, where preexisting nicks prevent the accumulation of superhelical tension.
In DNA synthesis reactions containing cellular concentrations of deoxyribonucleoside and ribonucleoside triphosphates (dNTPs and rNTPs), the replicative DNA polymerases of Saccharomyces cerevisiae, DNA polymerases α (Pol α), δ (Pol δ) and ε (Pol ε), frequently incorporate ribonucleotides into DNA1. This fact motivated studies to determine if ribonucleotides are incorporated by DNA polymerases during nuclear replication in vivo, and if so, whether ribonucleotides in the genome have biological effects. To focus on ribonucleotide incorporation by a replicative DNA polymerase (replicase) rather than by the RNA primase that initiates lagging strand Okazaki fragments, our first studies examined ribonucleotide incorporation by Pol ε, the primary leading strand replicase2,3 encoded by the S. cerevisiae POL2 gene. We used a variant of Pol ε in which a glycine replaced a conserved methionine (M644) adjacent to the “steric gate” tyrosine (Y645) in the polymerase active site that prevents ribonucleotide incorporation by B family polymerases4,5. Purified M644G Pol ε incorporates about 10 times more ribonucleotides than does wild type Pol ε6. In a pol2-M644G yeast strain, deleting the RNH201 gene (rnh201Δ) encoding the catalytic subunit of RNase H2 results in increased incorporation of ribonucleotides into genomic DNA in excess over those detected in the pol2-M644G or rnh201Δ single mutant strains6. This observation demonstrates that M644G Pol ε incorporates ribonucleotides into DNA in vivo, and that RNase H2-dependent repair removes them, as had been predicted earlier7,8. This repair has been reconstituted in vitro9 and designated Ribonucleotide Excision Repair (RER).
We have also examined ribonucleotides incorporated by the major lagging strand replicase Pol δ, using a variant in which a conserved leucine adjacent to the steric gate tyrosine was replaced with methionine. Like its Pol ε counterpart, purified L612M Pol δ also incorporates about 10 times more ribonucleotides than its wild type parent10. In yeast encoding the pol3-L612M variant, deleting RNH201 also resulted in the presence of ribonucleotides in genomic DNA in excess over those detected in the pol3-L612M or rnh201Δ single mutant strains. Moreover, strand-specific probing of genomic DNA demonstrated that equivalent Pol ε variants in Schizosaccharomyces pombe11 and S. cerevisiae10,12,13 preferentially incorporate ribonucleotides into the nascent leading strand DNA, whereas the S. cerevisiae pol3-L612M variant preferentially incorporates ribonucleotides into the nascent lagging strand10. These preferences strongly support the interpretation, based on mutational signatures2,3, that Pol ε is primarily a leading strand replicase and that Pol δ is primarily a lagging strand replicase. They further demonstrate that RER removes ribonucleotides from both the leading and lagging strands.
In our original study of ribonucleotide incorporation in vitro6, we speculated that the transient presence of ribonucleotides in DNA might serve positive signaling functions, such as directing mismatch repair (MMR) to correct replication errors in the nascent leading strand. Subsequent genetic and biochemical studies have supported this hypothesis10,14. Such a signaling role, and evidence that a ribonucleotide imprint in S. pombe signals for mating type switching15, are examples of beneficial effects of ribonucleotides in DNA, with other possibilities discussed elsewhere1,16–18. Nonetheless, ribonucleotides are non-canonical constituents in DNA (i.e., lesions), and as such can elicit damage that requires repair via an RNA-DNA Damage Response (RDDR). For example, when RNase H2 cleavage at a ribonucleotide generates a nick in DNA, an attempt to ligate this nick can generate an adenylated 5´-RNA-DNA junction that can elicit genome instability if not deadenylated by aprataxin19. Moreover, in the absence of RER, topoisomerase 1 (Top1) can cleave the DNA backbone where ribonucleotides are present20,21, thereby initiating removal of ribonucleotides13. In doing so, Top1 generates DNA ends with a 5´-OH and a 2´–3´-cyclic phosphate that must be processed to allow eventual ligation. These atypical “dirty” DNA ends are non-ligatable and are problematic for the cell. For example, compared to pol2-M644G or rnh201Δ single mutant strains, the pol2-M644G rnh201Δ double mutant strain that contains large numbers of unrepaired ribonucleotides in the genome grows slowly, is sensitive to the replication inhibitor hydroxyurea (HU), and has a strongly elevated rate of 2–5 base pair deletions in repetitive DNA sequences6,21–23. These negative consequences are largely suppressed by deleting the TOP1 gene encoding Top113,21. In addition, the pol2-M644G rnh201Δ strain does not survive the additional loss of RNase H124, which can incise DNA substrates containing four or more consecutive ribonucleotides25,26.
A particularly interesting feature of the 2–5 base pair deletion mutagenesis observed in the pol2-M644G rnh201Δ double mutant strain is its asymmetry. For example, the rate of deleting a CA dinucleotide from a CACA repeat in the URA3 reporter gene is 160-fold higher when URA3 is in one orientation relative to a nearby replication origin as compared to the other orientation6. This bias suggests that the mutagenesis results from incorporation of ribonucleotides during Pol ε-mediated leading strand replication of the 5´-CACA-containing template21,22. This led us to question whether unrepaired ribonucleotides incorporated by the lagging strand replicases Pols α and δ may also contribute to replicative stress or genome instability. Here we investigate this using variants of Pols α and δ. We first show that like M644G Pol ε6 and L612M Pol δ10, the L868M Pol α variant is also much more likely than its wild type parent enzyme to incorporate ribonucleotides during DNA synthesis in vitro and also during replication in vivo. We further show that like L612M Pol δ, L868M Pol α also preferentially incorporates ribonucleotides into the nascent lagging strand in vivo, and that these ribonucleotides are repaired by RER. Surprisingly, however, Top1-dependent removal of ribonucleotides, and the resulting RNA-DNA damage, are only observed for ribonucleotides incorporated by Pol ε, not Pols α or δ. This implies that Top1-dependent ribonucleotide removal and its adverse consequences preferentially result from unrepaired ribonucleotides in the nascent leading strand. Three models to explain this asymmetry are considered, including a possible difference in the need to relieve torsional stress in the replicated duplexes behind the fork during DNA synthesis.
RESULTS
Ribonucleotide incorporation by L868M Pol α and L612M Pol δ
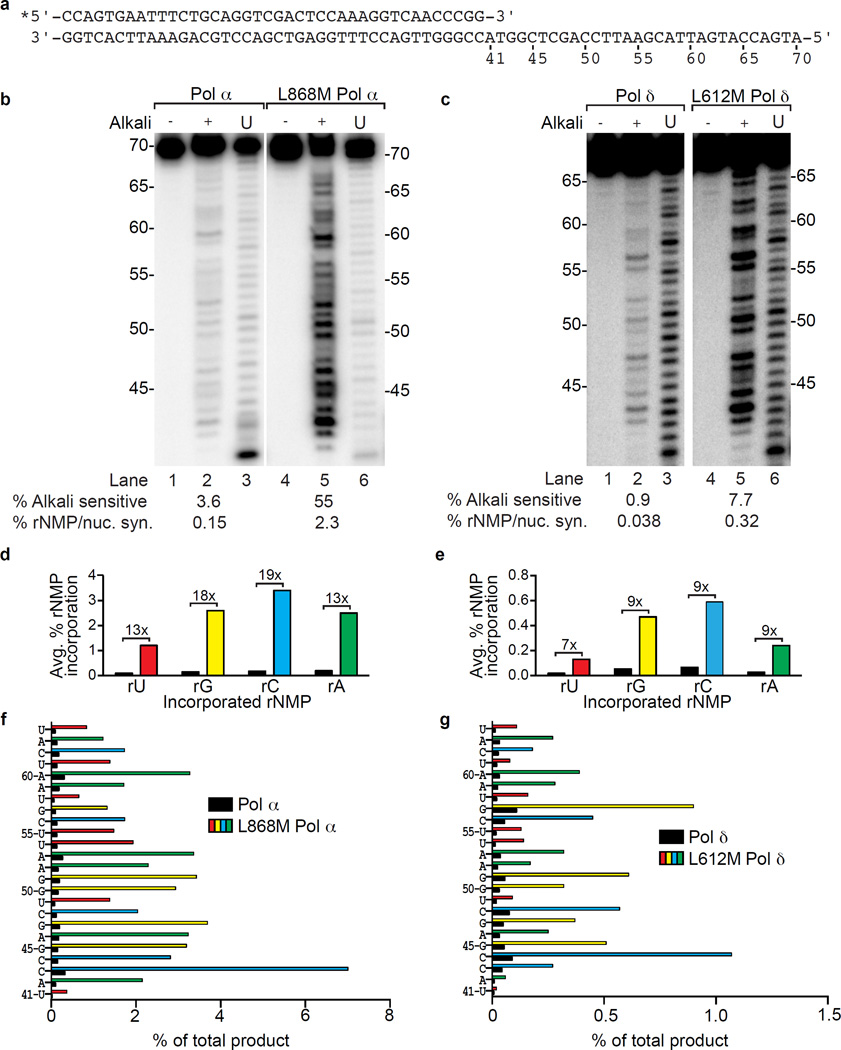
We previously showed that substituting glycine for a conserved methionine (Met644) in the Pol ε active site, or substituting methionine for Leu612 at the equivalent location in Pol δ, increases ribonucleotide incorporation during DNA synthesis in vitro6,10. Here we examined a Pol α variant in which the equivalent conserved leucine (Leu868) was changed to methionine27. We measured stable incorporation of ribonucleotides (rNMPs) during extension of a 40-mer primer hybridized to a 70-mer template (Fig. 1a). Polymerization reactions contained all four dNTPs and rNTPs at concentrations estimated to be present in yeast1. Full-length reaction products were isolated and treated with 0.3M KOH to hydrolyze the DNA backbone at positions where rNMP are present. Band intensities were used to quantify the percentage of rNMPs incorporated at 24 positions. To avoid rNMP incorporation by the RNA primase of the 4-subunit Pol α-primase complex, we used the Pol α catalytic subunit only. In order to directly compare ribonucleotide incorporation by both lagging strand replicases, we extended our initial study of 3-subunit L612M Pol δ10. The results (Fig. 1) for wild type Pols α and δ are similar to those reported in our initial study1. In comparison, average rNMP incorporation by L868M Pol α and L612M Pol δ is 15- and 8-fold higher, respectively (values below lanes in Fig. 1b,c), corresponding to an average of about one rNMP incorporated per 40 dNMPs by L868M Pol α and about one rNMP incorporated per 300 dNMPs by L612M Pol δ. As for the wild type enzymes, ribonucleotide incorporation by L868M Pol α and L612M Pol δ varies among the four rNMPs (rC > rG > rA > rU, Fig. 1d,e) and at 24 individual nucleotide positions (Fig. 1f,g). The fact that rNMP incorporation probability in vitro varies by polymerase, by the identity of the base attached to the ribose and by the surrounding sequence may be relevant to any negative and/or positive biological consequences of ribonucleotides in the genome. This possibility is supported by studies showing hotspots for ribonucleotide-dependent deletion mutations6,21, by strand-specific effects on MMR efficiency10, and by the results below.
Figure 1.
Ribonucleotide incorporation in vitro by variants of Pols α and δ. (a) Sequence of primer-template used for reactions in panels b and c. (b) Stable rNMP incorporation into DNA. The lane marked U depicts the product generated by Pol α or L868M Pol α prior to gel purification, as described in1. Lanes marked with (−) and (+) depict gel-purified products treated with 0.3 M KCl (−) or KOH (+). The percentages of alkali-sensitive products and rNMP incorporated per nucleotide synthesized are shown below each lane. The mean and range for duplicate measurements was 3.6 ± 0.3 for Pol α and 55 ± 0.5 for L868M Pol α. (c) As in panel b, but for DNA products made by Pol δ (0.9 ± 0.2) and L612M Pol δ (7.7 ± 1.5). (d) Average frequency of ribonucleotide incorporation for rU, rA, rC and rG calculated from panel b. The relative difference in ribonucleotide incorporation between Pol α and L868M Pol α is shown above each base. (e) As in panel d, but for Pol δ and L612M Pol δ using data from panel c. (f) Percentage of rNMP incorporation by Pol α or L868M Pol α at each of 24 template positions. The position and identity of each incorporated ribonucleotide is displayed on the Y-axis. (g) As in panel f, but for ribonucleotide incorporation by Pol δ or L612M Pol δ.
RER removes ribonucleotides incorporated by all three replicases and operates on both strands
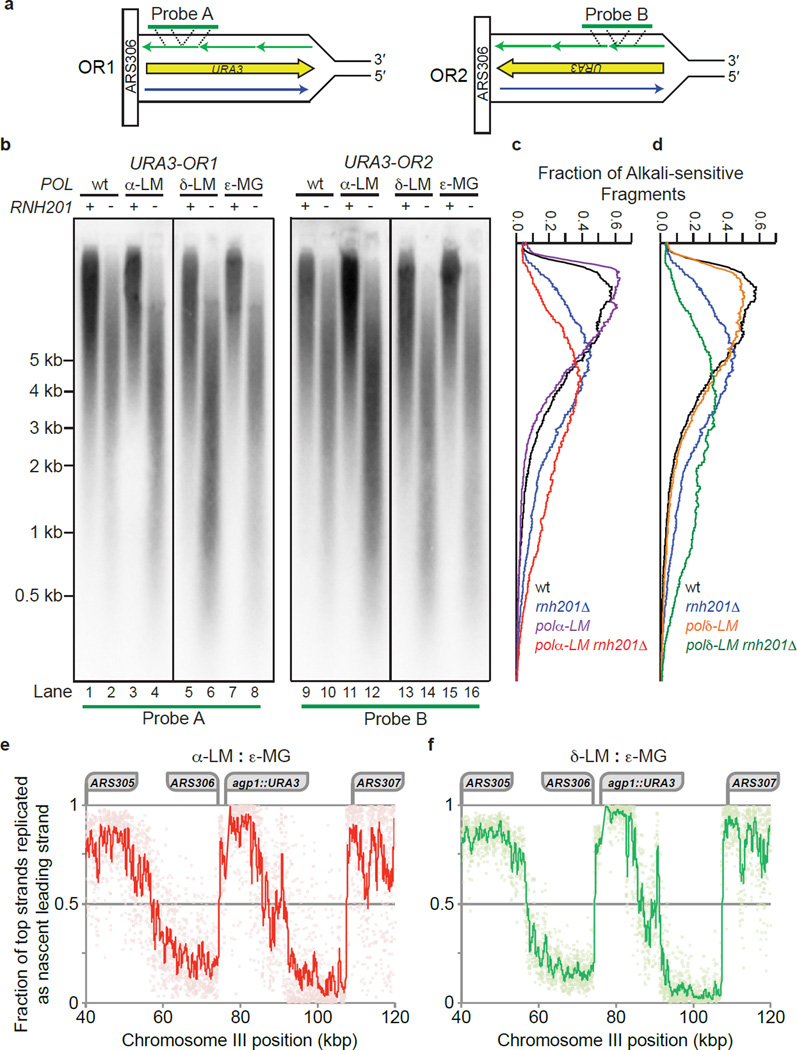
To test for strand-specific incorporation of ribonucleotides and their removal by RER, genomic DNA was isolated from 16 yeast strains (Supplementary Table 1) that differ in the genes encoding the replicases, the catalytic subunit of RNase H2 (Rnh201) and/or Top1. The DNA samples were treated with KOH to hydrolyze molecules containing ribonucleotides. Fragments were separated by electrophoresis in an alkaline-agarose gel, transferred to a nylon filter and probed with radiolabeled DNA complementary to either strand of the URA3 gene located next to the ARS306 replication origin (Fig. 2a and Supplementary Fig. 1a). The results (Fig. 2b and Supplementary Fig. 1b) indicate that, compared to all RNH201+ genomes, alkaline hydrolysis of the all rnh201Δ genomes yields shorter DNA fragments, demonstrating that RER is occurring and that in its absence unrepaired ribonucleotides are present in the genome. As expected based on earlier studies using a leading strand-specific probe11–13, nascent leading strand DNA fragments in the pol2-M644G rnh201Δ strain (Supplementary Fig. 1b, lanes 8 and 16) are shorter than leading strand fragments in the single mutant rnh201Δ strain (Supplementary Fig. 1b, lanes 2 and 10) indicating that M644G Pol ε preferentially incorporates ribonucleotides into the nascent leading strand and that they are removed by RER. Similarly, lagging strand DNA fragments in the pol3-L612M rnh201Δ strain (Fig. 2b, lanes 6 and 14, and green scan in panel d) are shorter than lagging strand fragments in the rnh201Δ strain encoding wild type replicases (Fig. 2b, lanes 2 and 10, and blue scan in panel d). This indicates that RER also removes ribonucleotides incorporated into the nascent lagging strand by L612M Pol δ. The results for L612M Pol δ (Fig. 2b) and M644G Pol ε (Supplementary Fig. 1b) serve as positive controls for the new analysis of ribonucleotides incorporated into the nascent lagging strand by DNA polymerase α. Importantly, lagging strand DNA fragments in the pol1-L868M rnh201Δ strain (Fig. 2b, lanes 4 and 12 and red scan in panel c) are shorter than lagging strand fragments in the rnh201Δ strain encoding wild type replicases (Fig. 2b, lanes 2 and 10, and blue scan in panel c), indicating that ribonucleotides are incorporated into the nascent lagging strand by Pol α and that these are subject to RER.
Figure 2.
Strand-specific probing for ribonucleotides in nascent lagging strand genomic DNA. (a) Depicted is the URA3 reporter gene on chromosome III placed adjacent to the ARS306 replication origin in each of the two orientations. Template strands are in black, the nascent leading strand (top strand) synthesized by Pol ε is blue and the nascent lagging strand (bottom strand) synthesized by Pols α and δ is purple. The annealing location of strand-specific radiolabeled probes A and B are indicated with dotted lines. (b) Detection of alkali-sensitive sites in nascent lagging strand yeast genomic DNA was performed as described13, with smaller fragments indicating the accumulation of unrepaired ribonucleotides in the absence of RNH201. Strains harbor one of four versions of the replicative DNA polymerases: wt (wild type for all enzymes), α-LM (pol1-L868M), δ-LM (pol3-L612M) or ε-MG (pol2-M644G). We note that Okazaki fragment-sized DNA (e.g. 150–300 base pair) fragments were not observed among the products of the alkaline hydrolysis of genomic DNA from the α-LM rnh201Δ or δ-LM rnh201Δ strains. Smaller DNA fragments observed for the ε-MG rnh201Δ mutant when using probes that anneal to the nascent lagging strand (lanes 8 and 16) may be related to the close proximity of URA3 to ARS306 (1.6 kb). These alkali-sensitive sites may arise during ribonucleotide incorporation by Pol ε into the nascent leading strand during bidirectional synthesis proceeding from this origin in the opposite direction (to the left of the origin in panel a). In addition, the small fragments that hybridize to the ‘lagging strand’ probe may be generated by synthesis performed by M644G Pol ε as it replicates from the adjacent ARS307 origin. (c) and (d) The data presented in panel b (for both Probe A and B) were quantified to determine the average fraction of total alkali-sensitive fragments at each position along the membrane as described in13. The vertical axis corresponds to the DNA marker positions in panel b. Curves are derived by plotting the average values from two independent experiments. (e) This panel depicts the fraction of replication events in which the bottom strand is replicated as the nascent lagging strand, as estimated from pol1-L868M rnh201Δ HydEn-seq data (N=1). Noise was reduced via comparison with pol2-M644G rnh201Δ (N=4; see Supplementary Methods for calculations). Pale diamonds represent data for 20 bp bins. The trend line represents a 20-bin (400 bp) moving average. Loci of interest are labeled above. (f) As per panel e, but calculated from pol3-L612M rnh201Δ (N=2) rather than pol1-L868M rnh201Δ data.
To confirm and extend the observation of strand-specificity of ribonucleotide incorporation during replication in vivo seen in the Southern blots, we mapped ribonucleotides by a new procedure, HydEn-seq28 (Hydrolytic 5' DNA End-sequencing). In this procedure, DNA ends are generated by alkaline hydrolysis of ribonucleotides in genomic DNA isolated from RNase H2-deficient strains (rnh201Δ), fragment libraries are prepared, and the locations of unrepaired ribonucleotides (5' read ends) are mapped by high throughput sequencing. HydEn-seq results at ARS306 and the adjacent URA3 locus are shown in Figures 2e,f. These panels depict the fraction of replication events in which the bottom strand is replicated as the nascent lagging strand, as estimated by comparing the HydEn-seq data for the pol2-M644G rnh201Δ and pol1-L868M rnh201Δ strains (Fig. 2e) and the HydEn-seq data for the pol2-M644G rnh201Δ and pol3-L612M rnh201Δ strains (Fig. 2f). The nascent bottom strand will be the nascent lagging strand immediately to the right of any given bi-directional origin. The abrupt switches in strand-specificity clearly identify and correspond to ARS306 and its neighboring replication origins, ARS305 and ARS307. The bottom strand is replicated as the nascent lagging strand during at least 80% of replication events from ARS306, through the agp1::URA3 locus, and for more than 8 kb toward ARS307 (approximately 97.5% of events at URA3; Fig. 2e,f). The high fraction of HydEn-seq ends detected to the right of each origin (see schematic in Fig. 2a) indicate that L868M Pol α and L612M Pol δ synthesize, and insert ribonucleotides into, the nascent lagging strand.
As expected of variations in replication origin usage and timing in a cell population, the strand preferences are not absolute (note the scatter in the data points, each depicting 20-nucleotide bins). These data leave open the possibility that forks emanating from ARS307 may replicate the URA3 locus adjacent to ARS306 during perhaps as many as 2.5% of replication events. If ribonucleotide incorporation was a uniform process, then given that Pol α initiates Okazaki fragment synthesis and the frequency with which L868M Pol α incorporates ribonucleotides, it would be reasonable to expect Okazaki fragment-sized alkaline hydrolysis products from pol1-L868M rnh201Δ genomic DNA. However, HydEn-seq data reveal that ribonucleotide incorporation is non-uniform across the genome, with no 5´-DNA ends detected at about 80% of base pairs in these genomes, compared to 5´-DNA end read counts of 10 or even greater than 100 at other base pairs28). This non-uniform distribution means that most ribonucleotides will be concentrated in relatively few Okazaki fragments, which greatly reduces the likelihood that DNA fragments corresponding to Okazaki fragments of about 200 bases will be observed in Southern blots performed using genomic DNA from the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains. Indeed, DNA fragments of this size are not abundant in the Southern blots that probe the URA3 locus (Fig. 2b), or following visualization of SYBR® Gold-stained genomic DNA isolated from the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains that has been subjected to alkaline hydrolysis and agarose gel electrophoresis (Supplementary Fig. 2).
Top1-dependent removal is specific to ribonucleotides incorporated by Pol ε
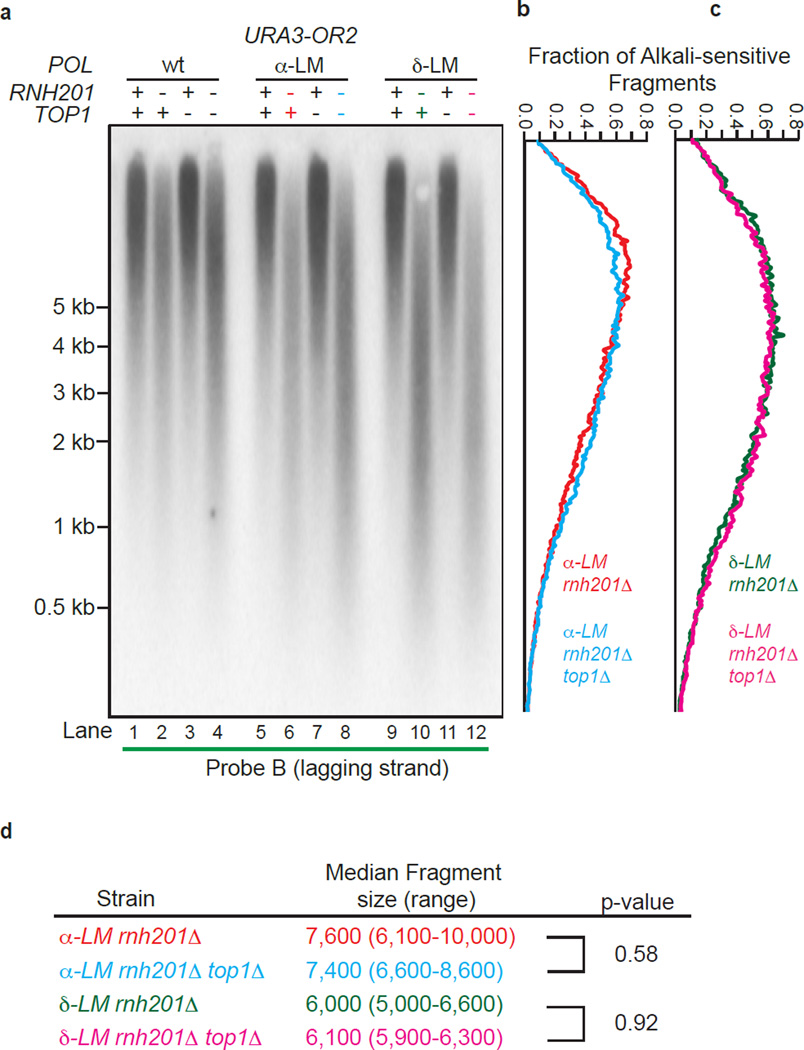
A recent study suggests that yeast Okazaki fragments are about 180 nucleotides long29. Pol α is thought to synthesize about 20 nucleotides, with the remainder synthesized by Pol δ30–32. Thus, the ability of purified L868M Pol α and L612M Pol δ to incorporate approximately one rNMP per 40 and 300 dNMPs, respectively (Fig. 1) predicts that if RER is the only process that removes them, approximately 50% of Okazaki fragments in pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains might contain an alkali-sensitive site that would result in an Okazaki-size fragment. While a trace amount of fragments of this size are observed in the scans of genomic DNA from these strains (Fig. 2b), the vast majority of fragments are larger. In addition to the possible explanation for this fact mentioned above, it could be that ribonucleotide removal is initiated by Top1 cleavage, which has previously been shown to remove ribonucleotides from the nascent leading strand in the pol2-M644G rnh201Δ strain13. Top1-dependent ribonucleotide removal from the nascent leading strand is evident from the shorter DNA fragments in the pol2-M644G rnh201Δ top1Δ triple mutant strain as compared to the pol2-M644G rnh201Δ double mutant strain (see Fig. 1 in13). In contrast, little difference in fragment sizes is observed in the equivalent triple versus double mutant strains encoding the pol1-L868M and pol3-L612M variants of the lagging strand replicases (Fig. 3b, compare lanes 6 to 8 and lanes 10 to 12, the scans in Fig. 3b,c and the quantitation of mean DNA fragment sizes in Fig. 3d). Thus Top1 has little or no role in removing ribonucleotides from the nascent lagging strand in the RER-defective pol1-L6868M rnh201Δ and pol3-L612M rnh201Δ strains.
Figure 3.
Lack of Top1-initiated ribonucleotide removal in pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains. (a) Detection of alkali-sensitive sites in nascent lagging strand DNA was performed using strains that were proficient or deficient in Top1 using the same approach as in Figure 2. All strains contain the URA3 reporter gene in orientation 2 (see Fig. 2a). (b) and (c) The data in panel a were quantified to determine the fraction of total alkali-sensitive fragments at each position along the membrane. The curves are derived using data from three independent experiments. (d) Median DNA fragment sizes (and range) in bases were determined as in13 using quantitation of the alkali-sensitivity data from five (α-LM rnh201Δ and δ-LM rnh201Δ strains) or three (α-LM rnh201Δ top1Δ and δ-LM rnh201Δ top1Δ strains) independent experiments. P values were calculated using a two-tailed Welch’s t-test.
Lack of Top1-dependent RNA-DNA damage in pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains
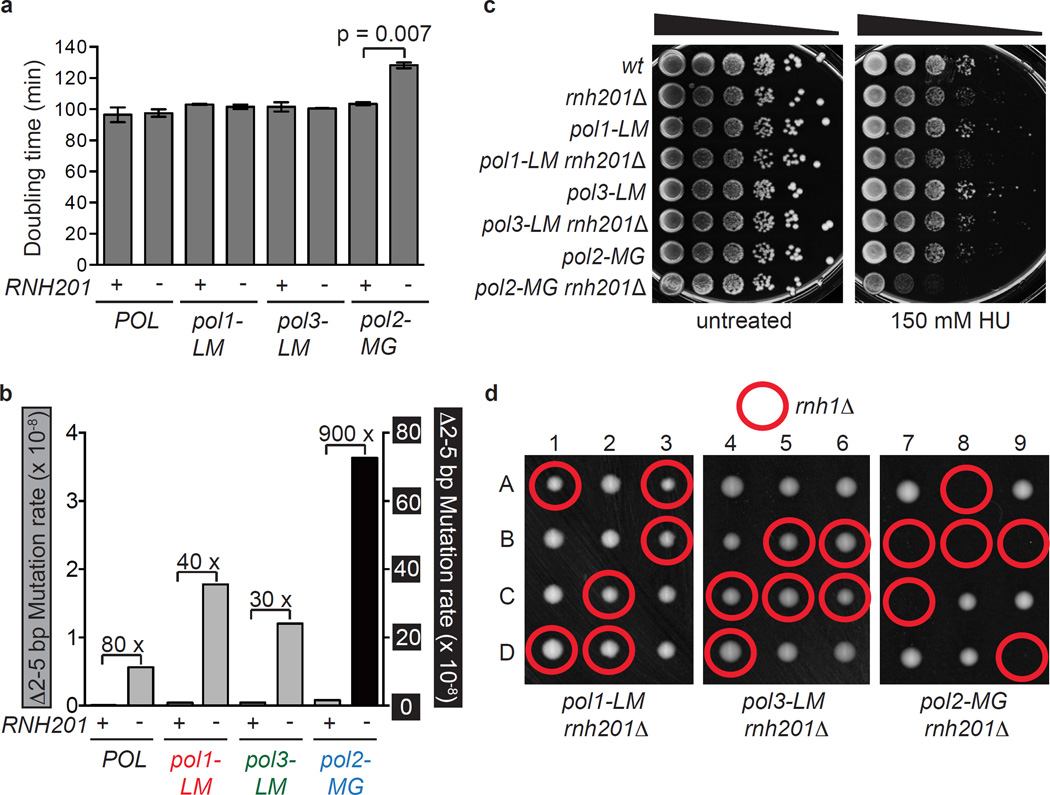
In pol2-M644G rnh201Δ strains defective in RER, Top1-dependent removal of ribonucleotides incorporated by M644G Pol ε is associated with several indicators of RNA-DNA damage including slow growth, sensitivity to treatment with the replication inhibitor hydroxyurea (HU), and genome instability in the form of 2–5 base pair deletions in repetitive sequences6,13. The data indicating that Top1 has little or no role in removing ribonucleotides from the nascent lagging strand (Fig. 3) suggests that these Top1-dependent RNA-DNA damage phenotypes may not be observed in the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains. This is indeed the case. Compared to earlier results with the pol2-M644G rnh201Δ strain6,13, and compared to the single mutant controls, the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains grow on solid YPDA (rich) medium with normal colony sizes (not shown), and they have doubling times that are equivalent to that of an RNH201 strain (Fig. 4a). This is in contrast to the pol2-M644G rnh201Δ mutant that has an increased doubling time (Fig. 4a). Also unlike the pol2-M644G rnh201Δ mutant, the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains progress normally through the cell cycle (Supplementary Fig. 3a). Furthermore, dNTP pools in the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains are not elevated compared to an rnh201Δ strain with wild-type replicases (POL) (Supplementary Fig. 3b). This is distinct from the increased cellular dNTP abundance measured in the pol2-M644G rnh201Δ mutant compared to an rnh201Δ mutant6. In the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains, the rate of 2–5 base pair deletions characteristic of Top1-dependent mutagenesis in RER-defective yeast6,21,22 is within two to three-fold of the rate in the rnh201Δ single mutant strain encoding wild type replicases (Fig. 4b and Supplementary Tables 2 and 3), as compared to the 130-fold increase in the pol2-M644G rnh201Δ mutant. In addition, the distribution of 2–5 base pair deletions within the URA3 mutational reporter gene is remarkably similar in the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains as compared to their distribution in the rnh201Δ single mutant strain encoding wild type replicases (Supplementary Fig. 4 and see Fig. 1 in22). Moreover, unlike the pol2-M644G rnh201Δ strain, the pol1-L868M rnh201Δ and pol3-L612M rnh201Δ strains are not sensitive to HU (Fig. 4c). Thus, despite the fact that L868M Pol α, L612M Pol δ and M644G Pol ε all incorporate an increased number of ribonucleotides into the nuclear genome, only the pol2-M644G variant causes an increase in Top1-dependent RNA-DNA damage in an RER-defective strain.
Figure 4.
RNase H2 is dispensable for maintaining genome integrity in strains with increased capacity to incorporate ribonucleotides into lagging strand DNA. (a) Deletion of RNH201 does not affect growth rate in pol1-L868M or pol3-L612M mutator strains, but does cause an increase in doubling time (Dt) in the leading strand mutator variant, pol2-M644G. P = 0.007 (two-tailed Student’s t test). (b) 2–5 base pair deletion mutation rates were calculated from the data in Supplementary Table 2 and Supplementary Figure 3 for the URA3-OR2 reporter. The relative fold difference in rate between RNH201 and rnh201Δ is shown above each pair of bars. The 2–5 base pair deletion rates corresponding to those displayed are presented in Supplementary Table 3. (c) Loss of RNH201 does not confer sensitivity to 150 mM HU in the pol1-LM or pol3-LM strains. The experiment was performed in triplicate and the data displayed is a representative example of n=3 independent biological replicates of this test. (d) Tetrad analysis of RNH1/rnh1Δ diploids in the pol1-LM rnh201Δ, pol3-LM rnh201Δ and pol2-MG rnh201Δ backgrounds. Columns1–9 are tetrad dissections and A–D are haploid spore colonies. Plates were photographed after 3 days growth at 30°C on rich medium.
Asymmetric synthetic lethality upon deletion of RNH1
Tetrad dissection of a diploid pol2-M644G rnh201Δ strain previously demonstrated that deleting the gene encoding RNase H1 (RNH1) results in lethality24. That result is recapitulated here (Fig. 4d, right panel, and see micro-colonies in Supplementary Fig. 5). This synthetic lethality contrasts with the survival and normal haploid spore colony sizes of triple mutant pol1-L868M rnh201Δ rnh1Δ and pol3-L612M rnh201Δ rnh1Δ strains (Fig. 4d). Thus, RNase H1 is essential only when the ribonucleotide burden is increased in the nascent leading strand but not when the ribonucleotide burden is increased in the nascent lagging strand.
DISCUSSION
Among many known types of DNA repair, two pathways are renowned for their strand specificity, transcription-coupled nucleotide excision repair (TC-NER) and MMR. As its name implies, TC-NER is coupled to transcription rather than replication, and TC-NER selectively removes DNA lesions from the transcribed DNA strand (reviewed in33). MMR is also strand-specific, but differs from TC-NER in that it removes replication errors from the newly replicated DNA strand while leaving the parental strand intact, and importantly, MMR efficiently corrects replication errors present in both the nascent leading and nascent lagging DNA strands34. The current study adds a third type of DNA repair whose strand-specificity is distinct from both TC-NER and MMR, Top1-dependent repair removes a different type of replication error / lesion that is much more abundant than single base mismatches13, and unlike MMR, it preferentially operates on only one of the two the nascent DNA strands. To our knowledge, this type of strand-specific repair is unique in the DNA repair field.
The consequences of increased ribonucleotide incorporation into DNA by the three nuclear replicases are summarized in Table 1. The results allow several novel conclusions. The observation of increased ribonucleotides in the genome of the pol1-L868M rnh201Δ strain demonstrate that L868M Pol α incorporates ribonucleotides as it extends RNA primers to initiate DNA synthesis of Okazaki fragments. This result demonstrates that just as for base-base mismatches made by L868M Pol α27,35, some ribonucleotides incorporated by Pol α survive the Okazaki fragment maturation process catalyzed by Pol δ-dependent strand displacement36. Here, we show that just as for ribonucleotides incorporated into the nascent leading strand by Pol ε and into the nascent lagging strand by Pol δ, ribonucleotides incorporated into the nascent lagging strand by Pol α are also subject to RER. Collectively, our results demonstrate that ribonucleotides are incorporated by all three replicases in a strand-specific manner, thereby supporting the current model for the primary roles of the polymerases in leading and lagging strand replication. This study clearly shows that, like MMR, RER also operates on both nascent DNA strands, and further reveals that the phenotypic consequences of ribonucleotides incorporated by the three variant replicases are different (Table 1). Relevant to this concept of asymmetric genome instability, unrepaired ribonucleotides in nascent leading strand DNA may be recombinogenic. This possibility is supported by previous observations of increased recombination in the absence of RNase H2 activity37,38.
Table 1.
Summary of phenotypes in polymerase variant yeast strains.
| Phenotype | L868M Pol α | L612M Pol δ | M644G Pol ε |
|---|---|---|---|
| RNase H2-Dependent Repair | Yes | Yes | Yes |
| Top1-Dependent Removal | No | No | Yes |
| Phenotypes of rnh201Δ Derivatives | |||
| Doubling time | Normal | Normal | Increased |
| Rate Δ2–5 bp | Wild-type | Wild-type | Elevated |
| HU-Sensitivity | Normal | Normal | Sensitive |
| Viability in rnh1Δ | Viable | Viable | Inviable |
We suggest three testable ideas for the observed asymmetries in Top1-dependent RNA-DNA damage by ribonucleotides incorporated into the nascent leading and lagging strands and note that these three ideas are not mutually exclusive. The first idea stems from the observation that in the RER-defective replicase variant strains examined here, the density of ribonucleotides is higher in the nascent leading strand (Supplementary Fig. 1b) than in the nascent lagging strand (Fig. 2b,c,d). Thus, one possibility (see model in Fig. 5a) is that at least some Top1-dependent RNA-DNA damage is only observable when ribonucleotide density exceeds a certain threshold. This idea is consistent with the fact that compared to a pol2-M644G rnh201Δ strain, an rnh201Δ strain encoding all wild type replicases exhibits relatively normal growth and resistance to HU (Fig. 4a,c and6,13,39), and by the fact that a wild type strain survives loss of HNT3 (Aprataxin), whereas deletion of HNT3 in a pol2-M644G strain with elevated ribonucleotide incorporation into the nascent leading strand confers slow growth and genome instability, phenotypes that are substantially improved upon loss of RNase H2 activity19.
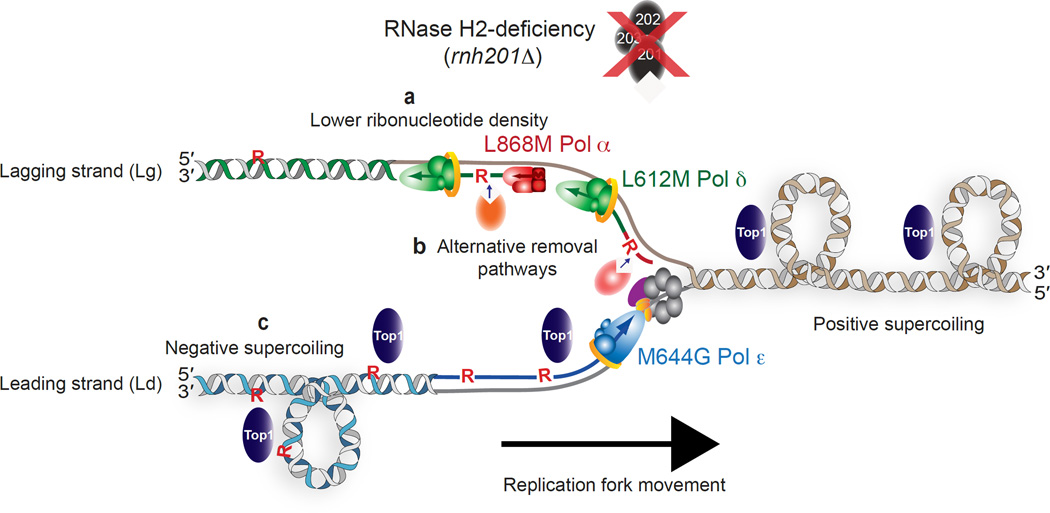
Figure 5.
A model depicting three possibilities for strand-specific consequences of unrepaired ribonucleotides in the genomes of RER-defective yeast strains. (a) Failure to observe a Top1-dependent effect in the L868M Pol α or L612M Pol δ strains may be related to the fact that these enzymes incorporate fewer ribonucleotides into DNA than does the M644G Pol ε variant. This possibility is supported by the demonstration that deletion of RNH1 is lethal in the pol2-M644G rnh201Δ mutant but not in the pol1-L868M or pol3-L612M strains lacking RNH201 (Fig. 4d), a result that may be directly related to ribonucleotide density. Generation of alternative variants of Pol α and Pol δ that elevate ribonucleotide incorporation into nascent lagging strand DNA is an approach that could be taken to test this idea. (b) A second possibility is the existence of alternative ribonucleotide-repair pathways available on the lagging strand, perhaps involving enzymes involved in Okazaki fragment maturation, such as the Fen1 or Exo1 nucleases or the Dna2 helicase. (c) A third idea involves the possibility that negative supercoils accumulate in leading strand DNA in the wake of the replication fork. This type of superhelical tension may be related to the continuous nature of the leading strand, in contrast to the discontinuity of the lagging strand in the form of preexisting DNA nicks that may allow for DNA rotation and negate the need for relief of such supercoiling by the action of Top1. As a consequence, Top1 incision at ribonucleotides in the nascent leading strand then initiates the RNA-DNA damage observed in a pol2-M644G rnh201Δ mutant.
A second possibility (Fig. 5b) is that ribonucleotides are removed from the nascent lagging strand by a mechanism that is less available to the leading strand. For example, replication errors generated by Pol α are removed by MMR27,35, and some ribonucleotides may be removed during Okazaki fragment maturation36 by processes that are independent of RNase H2 or Top1. An additional repair process, and/or the distinctly non-uniform distribution of newly incorporated ribonucleotides mentioned above, may explain why fragments the size of Okazaki fragments are not observed in pol1-L868M and pol3-L612M strains lacking RNase H2 and/or Top1, despite the biochemical potential of L868M Pol α and L612M Pol δ to incorporate a rNMP into one of every two Okazaki fragments (Fig. 1).
A third possibility is suggested by the fact that several of the phenotypes that reflect RNA-DNA damage in the pol2-M644G rnh201Δ strain, e.g., slow growth, HU-sensitivity and deletion-mutagenesis, result from Top1-dependent incision at ribonucleotides in the nascent leading strand13. These observed asymmetries in RNA-DNA damage might result from preferential Top1 incision at ribonucleotides in the nascent leading strand (Fig. 5c). These incisions could relieve torsional stress that may accumulate in the nascent leading strand behind the replication fork (see Fig. 3 in40). Indeed, the possibility of replication-induced negative supercoiling providing a structural change in DNA that regulates different transactions was discussed in a recent review41. Theoretically, nicking at rare lesions such as deoxyuracil incorporated during replication might also be important for relief of torsional stress that accumulates behind DNA replication forks, although published data42 suggests that deoxyuracil is present in DNA at less than 1% of the frequency of incorporated ribonucleotides. In principle, Top1 incision at ribonucleotides would not be needed to relieve torsional stress in the discontinuously replicated nascent lagging strand, because frequent DNA ends would prevent torsional stress from accumulating. In this scenario, only the nascent leading strand would contain the “dirty”, unligatable DNA ends generated by Top1 cleavage at ribonucleotides that initiate genome instability. Further support for this model of Top1-dependent effects being leading strand-specific is the demonstration that Top1 localizes to the replisome43 through an interaction with the CMG complex44, a helicase that selects Pol ε on the leading strand45. In this way, Top1 is well positioned to incise at ribonucleotides incorporated into the nascent leading strand by Pol ε.
METHODS
Methods and any associated references are available in the online version of the paper at xxx.
Supplementary Material
Acknowledgments
We thank Kasia Bebenek, Clinton Orebaugh and Scott Williams for helpful comments on the manuscript and all Kunkel lab members for thoughtful discussions. We acknowledge the NIEHS Molecular Genetics Core Facility for sequence analysis of 5-FOA-resistant mutants, and the NIEHS Flow Cytometry Center for FACS analysis. This work was supported by Project Z01 ES065070 to T.A.K. from the Division of Intramural Research of the NIH, NIEHS, by the Swedish Cancer Society to A.C. and by NIH grant GM032431 to P.M.B.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors have no competing financial interests to declare.
REFERENCES
- 1.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, et al. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 5.Pavlov YI, Shcherbakova PV, Kunkel TA. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics. 2001;159:47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 8.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci U S A. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparks JL, et al. RNase H2-Initiated Ribonucleotide Excision Repair. Molecular Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lujan SA, et al. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JS, et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghodgaonkar MM, et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalgaard JZ. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 2012;28:592–597. doi: 10.1016/j.tig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Williams JS, Kunkel TA. Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair (Amst) 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldecott KW. Molecular biology. Ribose--an internal threat to DNA. Science. 2014;343:260–261. doi: 10.1126/science.1248234. [DOI] [PubMed] [Google Scholar]
- 19.Tumbale P, Williams JS, Schellenberg MJ, Kunkel TA, Williams RS. Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity. Nature. 2014;506:111–115. doi: 10.1038/nature12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 21.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst) 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho JE, Kim N, Li YC, Jinks-Robertson S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Repair (Amst) 2013;12:205–211. doi: 10.1016/j.dnarep.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazzaro F, et al. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chon H, et al. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41:3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci U S A. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clausen AR, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.2957. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 34.Lujan SA, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niimi A, et al. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potenski CJ, Niu H, Sung P, Klein HL. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams JS, et al. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase epsilon. DNA Repair (Amst) 2012;11:649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 41.Yu H, Droge P. Replication-induced supercoiling: a neglected DNA transaction regulator? Trends Biochem Sci. 2014;39:219–220. doi: 10.1016/j.tibs.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Nilsen H, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 43.Bermejo R, et al. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 45.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.