Abstract
RNA editing, which adds sequence information to RNAs posttranscriptionally, is a widespread phenomenon throughout eukaryotes. The most complex form of this process is the uridine (U) insertion/deletion editing that occurs in the mitochondria of kinetoplastid protists. RNA editing in these flagellates is specified by trans acting guide RNAs and entails the insertion of hundreds and deletion of dozens of U residues from mitochondrial RNAs to produce mature, translatable mRNAs. An emerging model indicates that the machinery required for trypanosome RNA editing is much more complicated than previously appreciated. A family of RNA editing core complexes (RECCs), which contain the required enzymes and several structural proteins, catalyze cycles of U insertion and deletion. A second, dynamic multi-protein complex, the Mitochondrial RNA Binding 1 (MRB1) complex, has recently come to light as another essential component of the trypanosome RNA editing machinery. MRB1 likely serves as the platform for kinetoplastid RNA editing, and plays critical roles in RNA utilization and editing processivity. MRB1 also appears to act as a hub for coordination of RNA editing with additional mitochondrial RNA processing events. This review highlights the current knowledge regarding the complex molecular machinery involved in trypanosome RNA editing.
INTRODUCTION
The term “RNA editing” describes site-specific posttranscriptional changes in an RNA sequence other than pre-mRNA splicing and 3’-polyadenylation1. RNA editing was first described in trypanosomatids, when it was found that four non-encoded uridine (U) residues were added to the mitochondrial (mt) mRNA encoding cytochrome c oxidase 2 (cox2), thus repairing a frameshift in the gene2. This process, whose discovery was quite surprising, stood out because information not encoded within a gene unexpectedly appears in its transcript. A year after this report, a cytidine (C) base in the mRNA encoding apolipoprotein B (apoB) expressed in the mammalian small intestine was shown to be converted into a U3. This “C-to-U” type editing occurs through a simple deamination of the C and results in the creation of an internal stop codon and a truncated, functionally altered apoB protein.
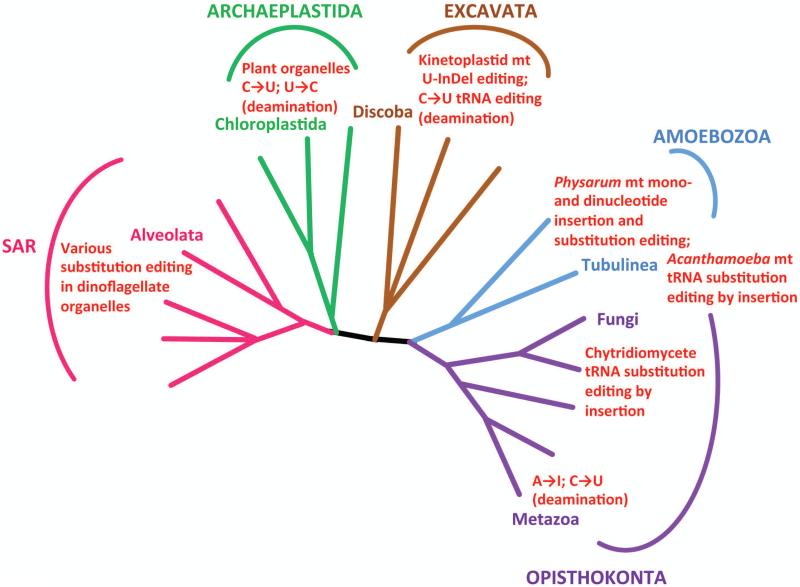
Later studies revealed that the RNA editing phenomenon is a widespread occurrence throughout eukaryotes (Fig. 1). C-to-U conversion editing is a common processing event for transcripts of plant chloroplasts and mitochondria, and a few U-to-C substitutions also occur in the latter organelle4. Another common type of RNA editing involving deamination is adenosine (A) to inosine (I) conversion. A-to-I editing represents the most common form of editing in metazoans5, 6. Insertion/deletion editing, as seen in trypanosomes, is not as widely distributed throughout eukaryotes as nucleotide substitution editing, although insertion/deletion editing coexists with substitution-type editing in mtRNAs of the slime mold Physarum polycephalum, albeit through a different mechanism not involving trans-acting RNAs7. RNA editing is not restricted to mRNAs, as tRNA8 and rRNA7, 9 also have been reported to undergo this process.
Figure 1. Distribution of multiple types of RNA editing across eukaryotes.
Phylogenetic tree based on Adl, et al.85. Only branches with clades that have a demonstrated type of RNA editing are labeled. Adjacent red text summarizes the type of editing.
When these phenomena were discovered, it was not clear how information could be added to or changed in an RNA molecule after it has been transcribed from a gene. Decades of research into the various types of RNA editing have begun to reveal their respective underlying mechanisms10, 11. Despite the diversity of RNA editing systems, two recurrent features of the molecular machinery ensure the site-specificity of modifications. The first is the utilization of double stranded RNAs to designate the RNA editing site. This can occur either in trans, by hybridization of two separate transcripts, or in the form of intramolecular hairpin loops. The second is the interplay of the substrate RNAs with protein complexes to recognize editing sites and facilitate their modification. The most complicated of these systems, in terms of the numbers of RNAs and proteins involved, remains trypanosome U insertion/deletion editing.
In this review, we summarize the current state of knowledge about the machinery underlying the bewilderingly complex process of RNA editing in trypanosomes. In addition to the well-characterized catalytic machinery, termed the editosome or RNA editing core complex (RECC)12, 13, numerous additional dynamically interacting ribonucleoprotein complexes that are essential for RNA editing and further maturation of edited RNAs have been recently uncovered. This review will focus on work done primarily in Trypanosoma brucei, the causative agent of human African trypanosomiasis, which has become a model trypanosomatid thanks to its possessing a robust RNAi pathway, a feature lacking in most easily cultured trypanosomes and related flagellates14.
A. URIDINE INSERTION/DELETION RNA EDITING AND THE RNA EDITING CORE COMPLEX
Basic mechanism of uridine insertion/deletion RNA editing
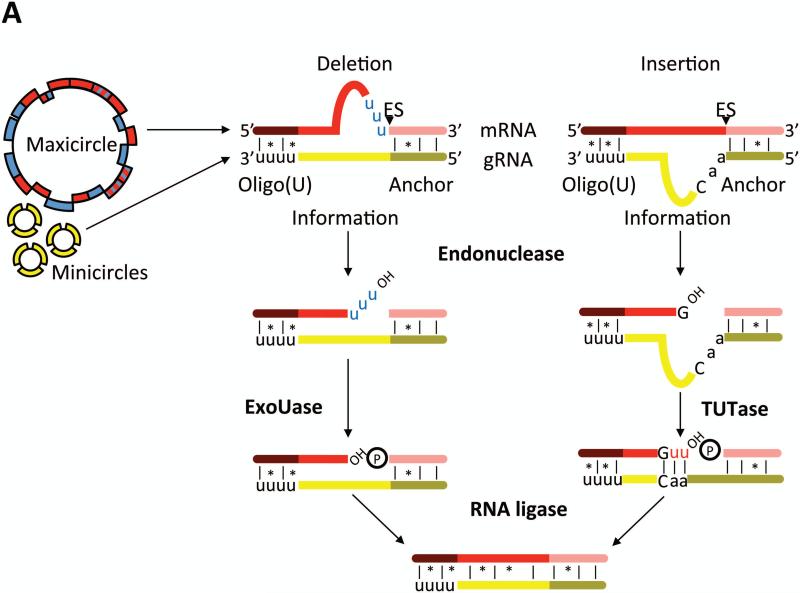
Mt DNA of T. brucei and related trypanosomatid flagellates, also known as kinetoplast DNA, is composed of two types of mutually interlocked circular DNA molecules: about a dozen identical maxicircles encode all 18 protein-coding genes and two truncated mt rRNAs, while thousands of heterogenous minicircles carry most guide (g) RNA genes15. Although six maxicircle transcripts exist as standard open reading frames, 12 require U-insertion/deletion RNA editing to generate translatable RNAs. Nine of these transcripts undergo an extreme form of the process called pan-editing. For example, cox3 RNA is remodeled by the insertion of 547 Us and deletion of 41 Us, and thus editing essentially creates the open reading frame of the RNA16. The remaining three edited RNAs are each modified in small domains, and thus called minimally edited RNAs. The sequence information for editing is provided by small trans-acting gRNAs that act as templates for U-insertion/deletion17, 18 (Fig. 2A). The sole exception is the cox2 RNA, which contains in its own 3’ UTR the template that guides insertion of four U's in its protein coding sequence19. Deep sequencing of the gRNA population of the insect vector stage (procyclic form; PF) T. brucei identified 642 major classes of gRNAs, accounting for complete sets of gRNAs for the majority of edited RNAs20. This study revealed significant redundancy in the gRNA population, which is possible because guiding interactions comprise both Watson-Crick and G-U basepairing, and very substantial variation in the abundances of different gRNA classes.
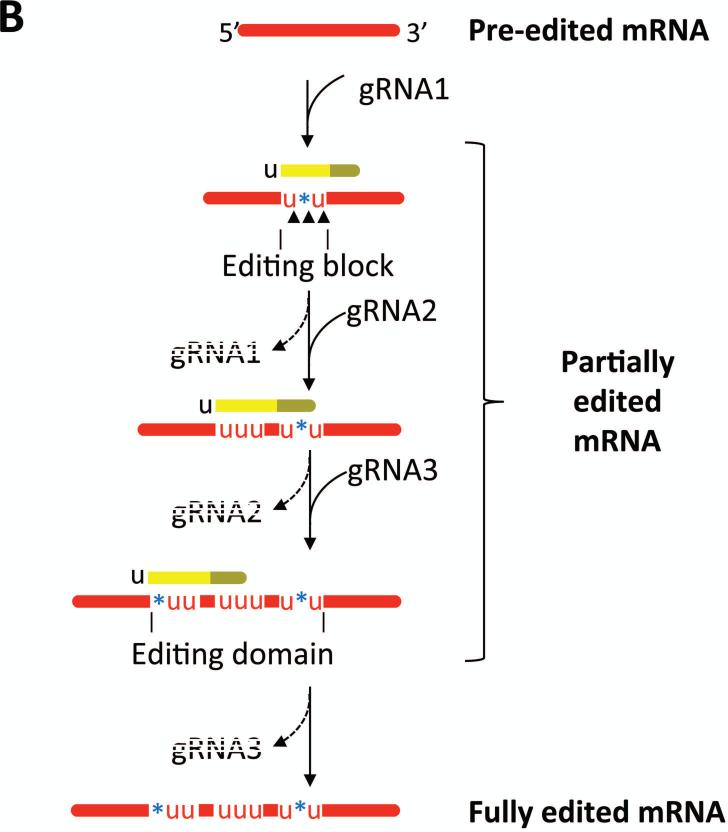
Figure 2. Mechanism of U insertion/deletion editing.
A) Pre-edited mRNAs are transcribed from mitochondrial maxicircles, while the majority of gRNAs are transcribed from the minicircle component of mitochondrial DNA. The gRNA 5’ anchor region (olive) basepairs with the mRNA and the gRNA information region (yellow) directs the number of U's inserted or deleted. The gRNA 3’ oligo(U) tail stabilizes the gRNA/mRNA interaction. Enzymes contained within the RNA editing core complex (RECC) catalyze mRNA endonucleolytic cleavage at an editing sites, U insertion by a 3’ TUTase, and U deletion by a U-specific exoribonuclease as directed by the sequences of gRNAs. Cleaved mRNAs are resealed by RNA ligases. B) Multi-round editing entails sequential utilization of multiple gRNAs. Because the anchor region of a given gRNA basepairs with edited mRNA sequence specified by the prior gRNA, editing progresses in a 3’ to 5’ direction along an mRNA. Multiple black arrowheads symbolize multiple editing sites within an editing block, as defined by the hybridized gRNA. Dashed gRNA labels indicate that they are turned over during/after an editing block has been processed. An editing domain is a stretch of mRNA sequence that requires the gRNA cascade for its processing.
Editing ensues when the 5’ anchor domain of a gRNA forms a primarily Watson-Crick duplex with its cognate mRNA just downstream of the regions whose editing it will direct (Fig. 2A). Apart from the anchor region, the remainder of the gRNA has only sporadic complementarity to the mRNA. This includes the 3’-oligo(U) tail, which has been postulated to basepair with the mRNA downstream of the region to be edited, thereby strengthening the gRNA-mRNA interaction 21. The enzymes that catalyze editing are contained within RECC22-32, and editing proceeds via an “enzyme cascade”. Following gRNA-mRNA anchor duplex formation, an editing endonuclease recognizes a mismatch between the gRNA and mRNA and cleaves the mRNA at this editing site (Fig. 2A). Depending on the sequence of the gRNA, U's will then either be added to the 3’ end of the 5’ cleavage product by a terminal uridylyltransferase (TUTase) or deleted by a U-specific exoribonuclease. At this point, the 5’ and 3’ mRNA fragments are bridged by the gRNA, and the two fragments are subsequently resealed by an RNA ligase.
One gRNA specifies the edited sequence at multiple adjacent editing sites, and following the utilization of a given gRNA, it is complementary to the edited RNA. Pan-edited mRNAs require the sequential utilization of dozens of gRNAs for their complete editing (Fig. 2B). For example, editing of cox3 and ATP synthase subunit 6 (A6) mRNAs requires 40 and 32 different gRNAs, respectively 20. The first gRNA-mRNA anchor is formed between the gRNA anchor domain and the small 3’ never edited sequence present in all pan-edited mRNAs. Subsequent gRNAs form anchor duplexes with the edited mRNA sequences directed by the 3’-proximal gRNA, dictating that editing progress in the 3’ to 5’ direction along the mRNA (Fig. 2B). This scenario necessitates the removal of fully base paired gRNAs to free up the single stranded sequence for binding the next gRNA. The REH1 helicase appears to function in this capacity to some extent33 (see Section E). However, the precise mechanism of gRNA removal and recruitment remain pressing questions.
Sequential gRNA utilization and resultant 3’ to 5’ editing progression produce a highly complex steady state RNA population containing large numbers of partially edited RNAs that are processed to different extents at their 3’-ends and unedited at their 5’-ends34 (Fig. 2B). In addition, partially edited RNAs typically contain “junction regions” between the 5’ unedited and 3’ fully edited sequences. Junctions contain edited RNA sequence that matches neither the unedited nor the fully edited sequence, and are believed to be regions of active editing that are ultimately corrected to the proper sequence, although their precise origins are unknown. It has been postulated that junctions arise from inaccurate pairing between cognate mRNA and gRNA or utilization of non-cognate gRNAs34. Experiments aimed at defining the origin and role of junction sequences, using next generation sequencing of editing intermediates in cells depleted of specific components of the editing machinery, will provide important mechanistic insights into the process.
The RNA editing core complex (RECC)
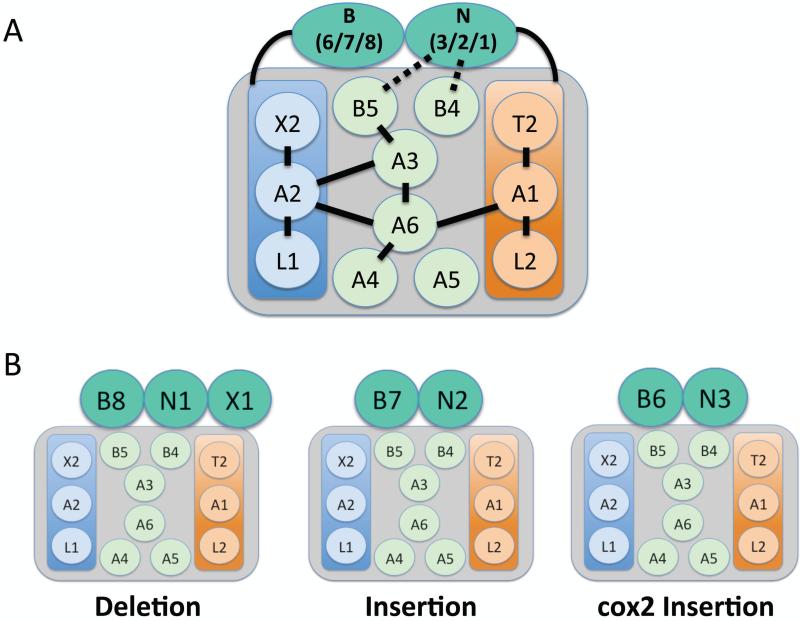
RECC contains the enzymes that catalyze an editing cycle22-32. Extensive biochemical and genetic experiments over the past 15 years have revealed three distinct types of RECC (Fig. 3), which share a common set of twelve proteins, including four enzymes: RNA editing ligases 1 and 2 (KREL1 and KREL2), 3’-TUTase (KRET2), and a U-specific exoribonuclease (KREX2). Also present are six interaction proteins containing predicted OB-folds (KREPA1-6) and two proteins with degenerate RNase III motifs (KREPB4 and KREPB5) (Fig. 3). Two heterotrimeric subcomplexes with stable protein-protein interactions exist within the common RECC proteins. These subcomplexes are comprised of 1) KRET2-KREPA1-KREL2, which catalyzes gRNA-directed U-insertion in vitro, and 2) KREX2-KREPA2-KREL1, which catalyzes gRNA-directed U deletion in vitro35. Within these subcomplexes, KREPA1 and KREPA2 bind and stimulate the activities of their partner proteins28, 35-38. KREPA1 and KREPA2 also bind directly to the KREPA3 and KREPA6 interactions proteins to bridge the insertion and deletion subcomplexes within RECC39 (Fig. 3A).
Figure 3. RNA editing core complex (RECC or editosome).
A) RECC contains 12 common proteins, of which three (KREX2, KREPA2, and KREL1) comprise a deletion subcomplex and three (KRET2, KREPA1, and KREL2) comprise an insertion subcomplex. OB-fold containing proteins KREPA3, KREPA6, KREPA4, and KREPA5 maintain protein-protein interactions. Zinc-finger containing KREPB4 and KREPB5 are thought to interact with endonucleases KREN1, KREN2, and KREN3 and their respective partners KREPB8, KREPB7, and KREPB6. KRENs interact with the insertion subcomplex and their KRPEB partners interact with the deletion subcomplex. B) There are three distinct classes of RECC, which differ in the associated KREN endonuclease and KREP partner proteins. KREN1/KREPB6 also associate with exoribonuclease KREX1. KREN1 containing RECCs catalyze U deletion, KREN2 containing RECCs catalyze U insertion, and KREN3 containing RECCs catalyze insertion specifically into cox2 mRNA.
A major advance in our understanding of RECC architecture was the discovery that the complex comprising the twelve common RECC proteins interacts with three RNase III family endonucleases (KREN1, KREN2, KREN3) and their partner proteins (KREPB8, KREPB7, and KREPB6, respectively) to form three RECC variants with differing functions25, 40 (Fig. 3B). KREN1/KREPB8 are also joined by a U-specific exoribonuclease, KREX1. Remarkably, both in vitro and in vivo studies show that KREN1/KREPB8/KREX1 RECC functions in U deletion, KREN2/KREPB7 RECC functions in U insertion, and KREN3/KREPB6 RECC is specific for the cis-editing of cox2 mRNA25, 41. Interestingly, KREX1, present only in KREN1/KREPB8/KREX1 RECC variants, is apparently the primary exoribonuclease in RNA editing. Although KREX2 is a component of the heterotrimeric deletion subcomplex and is active in vitro, this enzyme is dispensable in both PF and mammalian bloodstream form (BF) T. brucei42, and harbors a deletion that renders it catalytically inactive in Leishmania43. With regard to RECC endonuclease activities, several lines of evidence support a model in which KREN1-3 form heterodimers with the KREPB4 and KREPB5 proteins that possess degenerate RNase III motifs44. Interestingly, RNase III family endoribonucleases typically act as a homodimers cleaving double-stranded RNA45. Thus, the model in which catalytically active KREN1-3 molecules form heterodimers with inactive KREPB4 and/or KREPB5 is especially satisfying because it explains why only the mRNA strand duplexed to gRNA is cleaved during the editing cycle.
In vitro editing reactions with purified RECC result in precise, gRNA-directed U-insertion or deletion into a single editing site18. However, neither site-to-site progression nor gRNA exchange takes place in these reactions (with the exception of one report of two-site editing46), thereby implicating additional factors in the editing process in vivo. Research over the past several years has uncovered another multi-protein complex that is essential for RNA editing, termed the mt RNA binding complex 147 (MRB1; a.k.a. RNA editing substrate binding complex (RESC)48). Evidence to date suggests that MRB1 is the key to understanding RNA recruitment to RECC and the mechanism of editing progression47-50. Below we describe the discovery of MRB1 and the current model of its function in kinetoplastid RNA editing. MRB1 is thought to serve as the platform for kinetoplastid RNA editing and, additionally, as a hub for coordination of editing with other mt RNA processing machineries47, 48, 50-52.
B. DISCOVERY OF THE MRB1 COMPLEX AND ITS CONNECTION TO RNA EDITING
Concurrent discovery of the MRB1 complex
The MRB1 complex was discovered in 2008 in three independent studies, each of which used different purification techniques, thus leading to differing proposed compositions51, 53, 54. Panigrahi and colleagues53 used a monoclonal antibody (McAb) generated against a 20S fraction of T. brucei mitochondria to immunoprecipitate a complex of 16 proteins, all of unknown function. Within this set of proteins was the target of the McAb, Tb927.7.2570 (later termed GAP2 or GRBC1; Table 1), and a related protein with 31% sequence identity (GAP1; a.k.a. GRBC2), both of which contained no recognizable motifs. TAP tagging of GAP2 and two other proteins from the original purification identified 14 common proteins. Of these, several suggested the ability to interact with RNA, including a DEAD box RNA helicase (REH2; Tb927.4.1500), a C2H2 zinc finger protein (Tb927.6.1680), and a protein with an RRM RNA binding domain and an RGG box (TbRGG2; Tb927.10.10830). The majority of the other proteins lacked recognizable motifs or homologs outside the kinetoplastid flagellates. Based on the presence of several putative RNA binding proteins, the complex was named put-MRB1 (putative Mt RNA Binding complex 1).
Table 1.
MRB1 complex subunits
| Subcomplex | Name | Alias | Domains | TriTrypDB # | References* |
|---|---|---|---|---|---|
| Core | GAP1 | GRBC2 | Tb927.2.3800 | 48,51-54,60,61 | |
| Core | GAP2 | GRBC1 | Tb927.7.2570 | 48,51-54,60,61 | |
| Core | MRB3010 | GRBC6 | RPS2 | Tb927.5.3010 | 48,50,52,59,68,71 |
| Core | MRB5390 | GRBC4 | Tb11.02.5390 | 48,52,58 | |
| Core | MRB8620 | GRBC3 | Tb927.11.16860 | 48,52,70 | |
| Core | MRB11870 | GRBC5 | pentein | Tb927.10.11870 | 48,52,71 |
| Core | MRB0880 | GRBC7 | Tb927.11.9140 | 48,52 | |
| Core** | RBP30 | none | RRM | Tb927.5.2100 | na |
| Core** | none | none | Tb927.9.1420 | na | |
| Core** | none | none | Tb927.10.10120 | na | |
| TbRGG2 | TbRGG2 | none | RRM, RGG | Tb927.10.10830 | 48,49,52,58,63,74 |
| TbRGG2 | MRB1860 | REMC2 | Tb927.2.1860 | 48,52 | |
| TbRGG2 | MRB4160§ | REMC5 | Tb927.4.4160 | 48,52,69 | |
| TbRGG2 | MRB800 | REMC3 | Tb927.7.800 | 48,52 | |
| TbRGG2 | MRB8170§ | REMC5A | Tb927.8.8170 | 48,52,69 | |
| TbRGG2 | MRB8180¶ | REMC4 | Tb927.8.8180 | 48,52 | |
| TbRGG2 | PhyH | none | PhyH | Tb927.9.7260 | na |
| Unknown | MRB10130 | REMC1 | ARM/HEAT | Tb927.10.10130 | 48,52,53 |
Listed are publications in which a given protein was either analyzed by RNAi-silencing, used as bait in a protein-protein interaction study, or in which recombinant protein was characterized.
Designated as core components in Reference 48, but not found in any other MRB1 purifications.
MRB4160 and MRB8170 are paralogues arising from a gene duplication that share ~85% amino acid identity.
MRB8180 is paralogous with Tb927.4.4150. These paralogues share ~99% amino acid identity, and thus are designated as a single protein here.
Hashimi and co-workers54 identified GAP1 and GAP2 in a TAP purification of TbRGG1, a mt protein that binds synthetic oligo(U) ribonucleotide in vitro55. Reciprocal purifications of GAP1 and GAP2 returned a set of 14 proteins, of which 10 overlapped with those in the Panigrahi et al. study53. This 14-protein complex was again termed put-MRB1. Finally, Weng and colleagues51 purified a related complex from the model kinetoplastid, Leishmania tarentolae. These authors originally isolated the L. tarentolae homologs of T. brucei GAP1/2 in immunoprecipitates of the MRP1/2 RNA binding proteins that impact cytochrome b (cyB) mRNA editing56, 57. Reciprocal TAP purifications of L. tarentolae GAP1 and GAP2 revealed 12 proteins interacting in an RNA-independent manner, of which six or seven were also present in purifications of put-MRB1 from other laboratories. Weng and colleagues51 referred to the co-purified proteins resembling put-MRB1 as the GRBC (guide RNA binding complex) for reasons discussed below. Thus, three separate studies identified similar mt complexes, and the tantalizing presence of several proteins with predicted RNA binding domains piqued great interest in its function. However, the disparate lists of proteins from each laboratory left the composition of the complex unresolved, and suggested that association of some of the components might be sub-stoichiometric or transient. This ill-defined complex is now commonly referred to as the MRB1 complex47, 58, 59, and its composition has been refined and its function probed. Although the MRB1 complex was originally identified by association with the TbRGG154 or MRP1/251 proteins, these interactions were shown to be strictly RNA dependent and these proteins have not often been identified with the MRB1 complex in subsequent studies. Thus, TbRGG1 and MRP1/2 will not be further discussed here.
Evidence linking the MRB1 complex to RNA editing
The MRB1 complex was initially linked to U-insertion/deletion RNA editing by functional analyses of the GAP1 and GAP2 proteins. Both proteins are essential for growth in PF and BF T. brucei 51, 60. Strikingly, RNAi-mediated depletion of either protein causes massive destabilization of the entire gRNA population and consequent inhibition of editing of all RNAs that require trans-acting gRNAs (i.e., all RNAs except cox2)51, 60. This finding accounts for the naming of these proteins GAP1/2 (guide RNA associated proteins)60 or GRBC2/1 (guide RNA binding complex)51, and hereafter they will be referred to by their more common name, GAP1/247, 59. GAP1 and GAP2 are mutually dependent for their stability60, form a stable complex, and are resolved in native PAGE as an ~200 kDa particle, the predicted size of a tetramer51. In vitro transcription/translation studies showed that, while GAP1 can form a dimer, both GAP1 and GAP2 are required for optimal formation of the tetramer51. Moreover, GAP1/2 co-expressed in E. coli form a ~200 kDa complex, consistent with a heterotetramer of α2/β2 configuration61. In accordance with their ability to stabilize gRNAs, GAP1/2 interact with RNA in vivo. In cells lacking organellar RNA due to depletion of the mt RNA polymerase, the sedimentation co-efficient of GAP1/2 on glycerol gradients is significantly reduced60. Endogenous gRNAs are associated with TAP-purified GAP1/251. Finally, in vitro, the recombinant GAP1/2 heterotetramer binds synthetic gRNAs with a kD of about 200 nM61. Together, these studies identified a gRNA binding and stabilizing complex in the mitochondria of kinetoplastid protozoa, with the GAP1/2 heterotetramer as the likely gRNA binding component. The definition of the MRB1 complex remained in flux, however, pending more in depth analysis of its composition as described below.
Interestingly, the MRB1 complex is evolutionarily ancestral as it is apparently present already in the earliest-branching kinetoplastid Perkinsela, an aflagellar endosymbiont of another protist Paramoeba62. All six protein-coding transcripts of Perkinsela undergo editing, and the level of conservation between RECC and the MRB1 complex of T. brucei on one side and Perkinsela on the other is about the same, since in the latter flagellate we were able to identify homologues of more than half of the subunits of both complexes (David et al., submitted).
C. ARCHITECTURE OF THE MRB1 COMPLEX
Global studies reveal overall MRB1 architecture
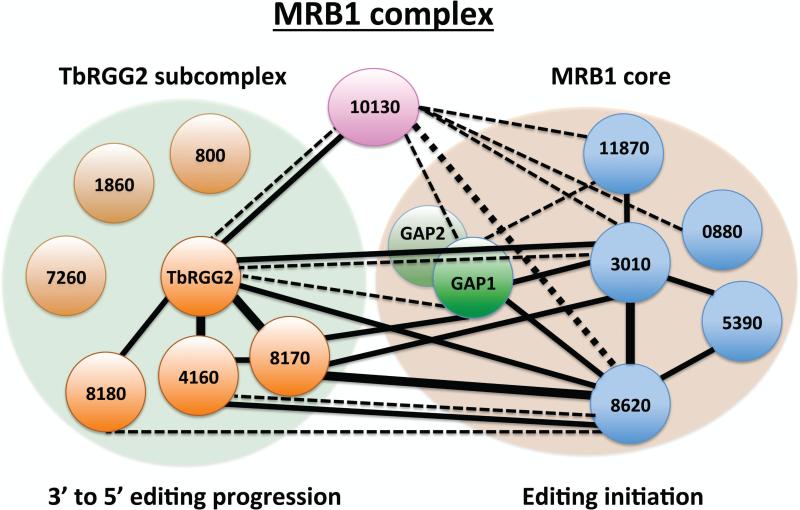
The physical and functional association of GAP1/2 with gRNAs, plus their interaction with other potential RNA binding proteins within the MRB1 complex, spurred a flurry of studies into the organization and function of the complex. Of the 31 potential MRB1 components, only six proteins were in common in all three original studies51, 53, 54. Thus, to understand the function of the MRB1 complex, it first became imperative to elucidate its composition, including the identification of stable sub-complexes, RNA dependent and independent contacts, and transiently associating proteins.
To this end, Ammerman and co-workers52 embarked on a comprehensive yeast two-hybrid screen to define direct protein-protein contacts. Each protein was given the designation MRBXXX, where XXX denotes the last 3-5 digits of the gene identifier in TriTrypDB (http://tritrypdb.org), unless it was known by a previously published name (Table 1). In all, 961 protein-protein pairs were tested in both bait and prey directions and screened for weak and strong interactions. In this screen, 19 of the 31 proteins exhibited interactions in at least one direction, with a total of 61 interactions. Remarkably, a small number of proteins accounted for the majority of interactions. GAP1, MRB3010, MRB8620, MRB8170, MRB4160, and TbRGG2 were involved in 30 of 31 strong interactions and 24 of 31 weak interactions, while MRB10130 engaged in a large number of weak interactions52. To confirm yeast two-hybrid results, the genes of MRB1 subunits that exhibited interactions were in situ tagged and resultant proteins tandem affinity purified. Purifications were performed in the presence or absence of RNases to determine interactions involving RNA. Co-purifying proteins identified by western blot analysis or mass spectrometry validated numerous yeast two-hybrid contacts, and importantly, revealed RNase-resistant interactions between some MRB1 components that did not exhibit direct interactions in the yeast two-hybrid screen. The combined yeast two-hybrid and in vivo studies thus defined an RNA-independent particle containing GAP1/2 and at least four other proteins, termed the MRB1 core (Table 1 and Fig. 4).
Figure 4. MRB1 complex organization.
MRB1 is comprised of a core that facilitates editing initiation, a TbRGG2 subcomplex that functions in 3’ to 5’ editing progression, and the MRB10130 protein that may bridge and organize the two subcomplexes. MRB1 core contains the GAP1/2 heterotetramer (bright green) that binds and stabilizes gRNAs and which appears to be dissociable and engages in additional interactions. Light green circle (TbRGG2 subcomplex) and light brown circle (MRB1 core) represent largely RNA-independent interactions determined in studies by Ammerman and colleagues52 and Aphasizheva and colleagues48. Black lines indicate direct interactions by yeast two-hybrid screen52. Solid lines, strong interactions; dotted lines, weak interactions; thin lines, interaction in one direction; thick lines, interaction in both directions. The MRB1 complex has also been referred to as RESC (RNA editing substrate binding complex), MRB1 core as GRBC (gRNA binding complex), and TbRGG2 subscomplex as REMC (RNA editing mediator complex)48.
The same study identified a second MRB1 sub-complex containing TbRGG2, an RNA binding protein previously reported to significantly impact RNA editing63 (Table 1 and Fig. 4). In contrast to the MRB1 core, TbRGG2 exhibited interactions with GAP1/2 and other core components that were partially sensitive to RNase treatment, and thus termed “RNA-enhanced”. Consistent with the observation that a small amount of TbRGG2 remains in association with the core after RNase treatment, TbRGG2 exhibited yeast two-hybrid interactions with MRB1 core components, MRB3010 and MRB8620. TbRGG2 also displayed strong yeast two-hybrid interactions with MRB4160 and MRB8170, paralogues arising from a chromosome-region duplication in T. brucei64, and with MRB8180. Like TbRGG2, the interaction between MRB8170 and the MRB1 core was strongly RNA-enhanced in pulldown experiments. In addition, co-purifications of tagged and endogenous TbRGG2 and MRB8170 were somewhat RNase sensitive, suggesting that multiple TbRGG2 subcomplexes are present on a given mRNA, and/or interactions between TbRGG2 subcomplex components are not as stable as those between MRB1 core proteins. Together, these data suggested that a TbRGG2 subcomplex(es), comprising at least TbRGG2, MRB4160, MRB8170, and MRB8180 associates with the MRB1 core in a manner that is significantly strengthened by RNA (Fig. 4).
One protein that could not be confidently placed in either the MRB1 core or TbRGG2 subcomplex is MRB10130, a protein comprised almost entirely of α-helical repeats resembling Armadillo (ARM) or HEAT repeats. MRB10130 emerged from the MRB1 yeast two-hybrid screen as a protein exhibiting one of the highest numbers of interactions52. It displayed weak interactions with numerous components of the MRB1 core, a strong interaction with TbRGG2, and weak interactions with a few proteins that were reportedly components of the KPAP1 polyadenylation and MERS1 RNA decay complexes51. In vivo pulldowns of MRB10130 indicated relatively RNase-sensitive interactions with GAP1/2 and MRB8170, and somewhat less RNase-sensitive interactions with TbRGG2 and the MRB1 core protein, MRB3010. The wide array of weak MRB10130 protein contacts is noteworthy in light of known functions of several ARM/HEAT family proteins, which often act as protein complex organizers65. It will be of interest to determine whether MRB10130 plays a similar role in organizing MRB1 function. Finally, several proteins consistently co-purified with MRB1 complex components in pulldown experiments from Ammerman and colleagues, yet could not be confidently placed in either the core or TbRGG2 subcomplexes because they failed to interact in the yeast-two hybrid screen and specific antibodies to these proteins were not available. These include MRB800, MRB1860, MRB0880, and Tb927.9.7260 (a.k.a. Tb09.160.5320).
More recently, Aphasizheva and co-workers48 reported an extensive study combining in vivo tagging and mass spectrometry of MRB1 complex proteins. Label-free quantitative mass spectrometry of both untreated and RNase-treated samples was used to place proteins within each complex and to quantify their interactions. Importantly, this approach validated the model put forth by the Ammerman study52 of the overall architecture of the MRB1 complex, again revealing the presence of an MRB1 core mediated by direct protein-protein interactions that interacts with the TbRGG2 subcomplex through RNA-enhanced contacts (Table 1 and Fig. 4). Assignment of specific proteins to each complex, which was also in agreement with the earlier study, was followed by inclusion of additional proteins in the subcomplexes. Specifically, MRB0880 makes numerous contacts with MRB1 core components, while MRB800, MRB1860, and Tb927.9.7260 cluster with TbRGG2 subcomplex proteins (Fig. 4). Also labeled as MRB1 core components were three additional proteins (Tb927.10.10120, Tb927.5.2100, and Tb927.9.1420) that were not identified in any other MRB1 study. These proteins appear to have relatively few and weak contacts, and so are not designated here as bone fide MRB1 core proteins. This group also places MRB10130 as part of the TbRGG2 subcomplex. However, their mass spectrometry results are entirely consistent with the aforementioned yeast two-hybrid study, showing MRB10130 making numerous contacts with several components of both the MRB1 core and TbRGG2 subcomplexes. Thus, we consider the role of MRB10130 in the architecture of the MRB1 complex to be unresolved and more consistent with a role as a complex organizer (Fig. 4). We note that the Aphasizheva study48 introduces new nomenclature for all MRB1 complexes and proteins based on their putative functions (Table 1). However, we prefer to stay with the more common MRB designations until the functions of these exciting but poorly understood complexes and proteins are better defined.
Some proteins that were reported in the original MRB1 purifications were not present in either of the more in-depth studies of MRB1 architecture. MRB6070, MRB1590, and TbRGG3 (previously MRB1820) were shown to engage in entirely RNA-dependent interactions with MRB1, and thus do not represent bone fide components of the complex52, 66, 67. The REH2 RNA helicase also comprises a complex separate from MRB150, and its essential role in RNA editing is reviewed in Section E. Collectively, the two global studies48, 52 provide very strong evidence for the existence of an MRB1 complex comprised of the core of about seven proteins that interact in an RNA-independent manner and a TbRGG2 subcomplex, which itself contains seven proteins that associate with the MRB1 core in an RNA-enhanced manner (Fig. 4).
Roles of specific proteins in maintaining MRB1 complex architecture
MRB1 subcomplex size and integrity have been commonly investigated by analysis of their sedimentation within a density gradient upon ultracentrifugation. While this approach is subject to variability, it is especially useful in examining the impact of RNAi-silencing of specific components on the sedimentation, and thus integrity, of an examined subcomplex. Typically, components of both the MRB1 core and TbRGG2 subcomplex tend to sediment between 20-30S, and this distribution markedly shifts towards lighter fractions upon RNase treatment, likely reflecting both the contribution of RNA to the complex's sedimentation value and the RNA-enhanced interaction between the MRB1 core and TbRGG2 subcomplex48, 52, 68, 69. Analysis of GAP1/2 sedimentation patterns is somewhat complicated, however, by their extraordinarily heterodisperse pattern on glycerol density gradients, which reveals a distribution that does not entirely coincide with that of other MRB1 complex components48, 52, 58, 60, 68, 69. Indeed, in addition to their roles as integral MRB1 core components, GAP1/2 are present in association with the REH2 RNA helicase50, 59 and, separately, the TbRGG3 RNA binding protein66 in the absence of some MRB1 core components, suggesting additional roles for GAP1/2 in mt RNA metabolism in addition to their functions in the MRB1 core. Furthermore, GAP1/2 downregulation does not appear to affect the interaction of the other MRB1 core subunits with each other70. The incorporation of GAP1/2 into higher molecular weight complexes, like other MRB1 components, requires mt RNA58, 60.
Initial studies implicate MRB3010 and MRB11870 as important structural components of the MRB1 core. Knockdown of MRB3010 causes both GAP1 and GAP2 to shift from a broad distribution between 10-40S to 10-20S fractions on glycerol gradients68. Moreover, the MRB3010-GAP1 interaction requires MRB11870, since MRB3010 immunoprecipitates contain substantially less GAP1 when MRB11870 is depleted71. In contrast, knockdown of either MRB5390 or MRB0880 failed to greatly affect the sedimentation of GAP1/2 on glycerol gradients, suggesting that these may be peripheral components of the MRB1 core48, 58.
The core proteins MRB0880 and MRB11870 likely play integral roles in the association between the MRB1 core and TbRGG2 subcomplex. Upon MRB0880 depletion, TbRGG2 markedly shifts to lighter gradient fractions and its overall abundance is modestly decreased48. Additionally, co-immunoprecipitations demonstrated that the association between the core protein MRB3010 and the TbRGG2 subcomplex is significantly decreased when MRB11870 is depleted71. Several components of the TbRGG2 subcomplex have also been implicated in bridging its interaction with the MRB1 core. For example, when either TbRGG2 or MRB8180 is downregulated, sedimentation of GAP1/2 shifts towards the top of the gradient48, 58. Likewise, MRB8180 depletion causes TbRGG2 to shift to lighter gradient fractions48, likely reflecting a disruption of the complete MRB1 complex. Kafková and colleagues69 showed that knockdown of MRB8170 causes both GAP1/2 and MRB3010 to redistribute to lighter gradient fractions, and co-depletion of the paralogous MRB8170 and MRB4160 lead to an even more pronounced shift in these core proteins. Thus, within the TbRGG2 subcomplex, TbRGG2, MRB8180, MRB4160, and MRB8170 all appear to be important in maintaining the interaction between the MRB1 core and the TbRGG2 subcomplex. Because TbRGG2, MRB4160, and MRB8170 all bind RNA in vitro63, 69, it cannot be distinguished whether the impact of these proteins involves direct protein-protein contacts or whether it is secondary via the loss of RNAs that are required for the enhanced interaction between the core and TbRGG2 subcomplexes. Future experiments with defined mutants defective in binding either RNA or specific proteins will be important in answering this question.
It is difficult to disentangle the roles of specific proteins in the maintenance of the TbRGG2 subcomplex, as its structure and composition are somewhat elusive, and evidence suggests the existence of distinct variants. For example, MRB4160 and MRB8170 share 77% amino acid identity, with major differences primarily located in their N-termini69. Using antibodies specific to MRB8170, Kafková, et al.69 showed that MRB8170 is entirely absent from immunoprecipitations of tagged MRB4160, indicating that these two proteins are present in mutually exclusive variants of the TbRGG2 subcomplex. Additional evidence suggests that the TbRGG2 subcomplex is dynamic. TbRGG2, MRB8170, and MRB4160, but not other TbRGG2 subcomplex proteins, were identified in association with the mRPN1 endoribonuclease that may function in gRNA processing in PF T. brucei72, 73. Furthermore, MRB8170 does not entirely co-sediment with TbRGG252, consistent with additional interactions of this protein. As with the MRB1 core, RNA clearly contributes to incorporation of the TbRGG2 subcomplex into higher order complexes, as RNase-treatment of mt extracts causes both TbRGG2 and MRB8170 to dramatically redistribute to lighter gradient fractions63, 69. Potential heterogeneity notwithstanding, the impacts of both MRB818048 and MBB8170/416069 knockdowns on TbRGG2 gradient sedimentation have been investigated. Interestingly, neither has a very dramatic impact on TbRGG2-containing complexes. In fact, depletion of MRB8170/4160 actually shifts the peak of TbRGG2 towards slightly heavier fractions of the gradient, hinting at complicated dynamics between components of the MRB1 complex. Moreover, the observation that MRB8170/4160 knockdown has only modest effects on TbRGG2 sedimentation, but relatively large effects on the sedimentation of MRB1 core components, is somewhat paradoxical given that it is not considered a bona fide subunit of the core.
As described above, MRB10130 exhibits numerous contacts with both MRB1 core and TbRGG2 subcomplex proteins48, 52. Consistent with a potential role in complex organization, MRB10130 depletion causes redistribution of GAP1/2 to heavier glycerol gradient fractions48, suggesting that GAP1/2 fail to properly interact with MRB1 and become free to engage in non-MRB1 interactions. The abundance of TbRGG2 as visualized in glycerol gradient fractions is significantly reduced upon MRB10130 RNAi48. Additional studies showed that depletion of the core MRB11870 subunit caused disruption of core-MRB10130 binding, as well as core-TbRGG2 subcomplex interactions71. In light of the direct MRB10130-MRB11870 interaction identified by yeast two-hybrid analysis52, these observations suggest that loss of TbRGG2 subcomplex components upon MRB11870 disruption is secondary to disruption of MRB10130 interactions with the MRB1 core. These data are consistent with a model in which MRB10130 bridges the MRB1 core and TbRGG2 subcomplex, and/or facilitates their interaction (Fig. 4).
MRB1 complex interaction with RECC
The MRB1 complex clearly interacts with gRNAs51 and mRNAs48, 50, and knockdown studies reveal its important role in RNA editing (see Section D). Nevertheless, its interactions with the RECC complex are highly transitory. MRB1 complex proteins have been observed in RECC purifications and vice versa23, 51, 63, but this is not common and entire complexes are not represented. Furthermore, sedimentation of 20S RECC is essentially unperturbed by depletion of MRB1 complex proteins, including those of both the MRB1 core and TbRGG2 subcomplex48, 51, 58, 63, 69. Nevertheless, recent evidence indicates that RECC does interact with at least GAP1/2. RECC present in 30-40S regions of glycerol gradients co-precipitates with GAP1/2 in an RNase-sensitive manner and is quantitatively super-shifted with anti-GAP1/2 antibodies48. It will be of interest to define the precise nature of complex(es) associated with RECC. More precisely, do GAP1/2 interact with RECC by themselves, or in the context of the MRB1 core particle, the entire MRB1 complex, or some other particle?
D. MRB1 COMPLEX FUNCTION
Emerging evidence points to the MRB1 complex as the platform for RNA editing47, 48, 50. While early studies assumed that RECC was the complex on which editing occurred, it was always paradoxical that purified 20S RECC contains little or no mRNA or gRNA13, 22, 48. Furthermore, in in vitro RNA editing assays, purified RECC performs very poorly and is essentially non-processive. In contrast to RECC, affinity purified MRB1 complex contains readily detectable mRNA and gRNA48, 50, 51, 59. The presence of these RNA species in association with MRB1, together with functional studies on complex components as described next, are consistent with the MRB1 complex acting as the site of kinetoplastid RNA editing.
MRB1 core
As previously stated, the GAP1/2 component of the MRB1 core binds and stabilizes gRNAs. The functions of other MRB1 core components have been examined in T. brucei engineered for inducible RNAi-silencing of individual proteins48, 58, 68, 71. When gRNA abundance is examined in these cells, it is clear that all other MRB1 core proteins are dispensable for gRNA stabilization. Thus, the GAP1/2 heterotetramer is the gRNA-binding and stabilizing component of the MRB1 core.
To determine whether depletion of other MRB1 core proteins impacts RNA editing, a commonly used assay is qRT-PCR with two primer sets: one that targets pre-edited mRNA sequences near their 3’ ends and another targeting edited mRNA sequences near their 5’ ends. Because editing progresses from 3’ to 5’ along an mRNA, the latter primers assess the generation of nearly fully edited mRNAs. Using this assay, cells depleted of MRB3010 or MRB11870 typically exhibit an 80-90% decrease in edited mRNAs and 2 to 4-fold increases in pre-edited RNAs, especially for pan-edited transcripts48, 68, 71. Minimally edited RNAs sometimes show lesser, but still obvious, defects. Remarkably, editing of cox2 mRNA is also substantially affected in MRB3010 or MRB11870 knockdown, despite the reliance of this mRNA only on a cis-acting gRNA19. Because cox2 mRNA editing is independent of trans-acting gRNAs, and thus GAP1/2, this finding indicates that the MRB1 core has a function in RNA editing that extends beyond simply presenting GAP1/2-bound gRNAs to RECC.
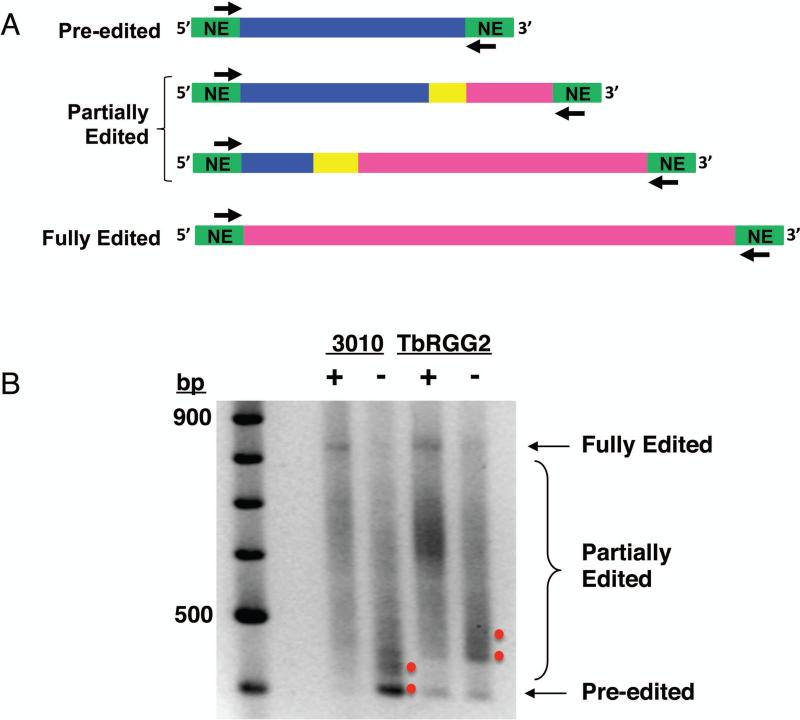
To assess in more detail the relative impacts of MRB1 core proteins on editing initiation as opposed to the 3’ to 5’ progression of editing, Ammerman and colleagues49, 68, 71 employed a “full gene PCR” assay. Here, end point PCR is performed with primers annealing to 5’ and 3’ never-edited regions present on all edited RNAs (Fig. 5). Because U-insertion is ten times more common than U-deletion, gel-resolution of the amplicons reveals pre-edited, partially edited, and fully edited RNA populations and indicates approximate points at which editing is compromised upon RNAi-silencing (Fig. 5). Full gene PCRs for MRB3010- and MRB11870-depleted cells reveal defects at 3’-most editing sites, indicating a defect in editing initiation, consistent with significantly increased pre-edited RNAs in MRB3010 and MRB11870 knockdowns68, 71. Deep sequencing of mt mRNA populations currently underway will provide more detailed information regarding the precise editing intermediates that accumulate in cells depleted of specific MRB1 components.
Figure 5. Full gene PCR assay used to analyze the step of editing in which a given MRB1 protein functions.
A) End point PCR is performed with primers targeting the 5’ and 3’ never edited regions present at the ends of all pan-edited RNAs to amplify all versions of a given RNA (pre-edited, partially edited, fully edited). Green, never edited; pink, fully edited; yellow, junction region; blue, pre-edited. B) Agarose gel analysis of full gene PCR amplicons of ATPase 6 RNA in cells either expressing (+) or depleted of (-) MRB3010 or TbRGG2. Red dots indicate products that accumulate in the absence of each protein. Note that pre-edited RNA accumulates upon MRB3010 depletion but not upon TbRGG2 depletion, signifying a role in editing initiation for the former, but not the latter.
The RNAi-silencing phenotypes of two other MRB1 core proteins, MRB0880 and MRB5390, appear somewhat less dramatic and more mRNA-specific than those following the depletion of MRB3010 or MRB1187048, 58. Results regarding MRB5390 are inconsistent, as one qRT-PCR study reports modest decreases in only edited cox3, ribosomal protein subunit 12 (RPS12), and maxicircle unknown reading frame 2 (MURF2) mRNAs, while at the same time observing increases in almost all pre-edited RNAs, suggesting possible effects on pre-edited mRNA stabilization58. Another study reported substantial decreases in many edited mRNAs by qRT-PCR, while concurrent Northern blot analysis of RPS12 mRNA revealed little increase in pre-edited or partially edited mRNA48. These differences may reflect different RNAi-efficiencies in the two studies. The role of another MRB1 core component, MRB8620 has been somewhat paradoxical based on one study. In this report48, edited RNAs are either unaffected or increased 1.5 to 2-fold when MRB8620 is knocked down. Mass spectrometry studies by these authors indicate that MRB8620 maintains numerous contacts with the large ribosomal subunit; thus, it may play a more substantial role in post-editing events than other MRB1 core components. However, a more recent study has shown that RNAi-mediated repression of MRB8620 indeed results in an inhibition of editing due to the comprised integrity of the MRB1 core, albeit to a lesser degree than when the other studied core MRB1 subunits are down-regulated70. In this background, transcripts requiring RNA editing accumulated on the GAP1/2 heterotetramer, indicating a disruption of RNA trafficking during the RNA editing process when the MRB1 core is disrupted. The MRB1 core components are essential for growth of PF48, 51, 58, 60, 68, 71 and, when examined, BF60, 68, 71 T. brucei. Only under culturing conditions that induces the PF to rely entirely on the mitochondrion for its energy metabolism did MRB8620-downregulation affect flagellate fitness, most likely due to its subtle RNAi phenotype70. Interestingly, depletion of most of these proteins also results in a slight increase in gRNA abundance48, 70, 71, suggesting that gRNAs are consumed in the editing process (Fig. 2B) and, thus, build up when editing is inhibited by MRB1 core protein knockdown. Finally, all of the studies described above analyzed the effects of MRB1 core protein depletion on never-edited mt mRNAs and rRNAs, with little or no apparent effects on these RNA populations. Thus, the MRB1 core contains GAP1/2 as its gRNA-binding component, impacts early events in the editing process apart from gRNA delivery, and possesses a function restricted to RNA editing.
TbRGG2 subcomplex
This subcomplex, which interacts with the MRB1 core in an RNA-enhanced manner48, 52 (and see above), is named for the first of its components to be studied: TbRGG2. This protein was initially identified by mass spectrometry of immunoprecipitated RECC complexes23, and it is one of the few MRB1 components with recognizable domains63. It contains a G-rich N-terminus comprising GWG and RG/RGG repeats and a canonical RRM type RNA binding domain at its C-terminus. Recombinant TbRGG2 binds synthetic pre-edited mRNA, edited mRNA, and gRNA in vitro, and exhibits an almost 10-fold higher affinity for pre-edited mRNA (Kd ~4 nM) than for the latter two transcripts 63, 74. Surprisingly, high affinity RNA binding maps to the G-rich N-terminus, and requires both GWG and RGG repeats. The RRM domain displays lower affinity RNA binding (Kds = 80-150 nM), with a relative preference for gRNA compared to the full-length protein or N-terminus. Because of the requirement for iterative mRNA-gRNA annealing and unwinding events during RNA editing, Ammerman and colleagues49 also tested whether recombinant TbRGG2 possesses these activities. Indeed, TbRGG2 displays robust mRNA-gRNA annealing activity in vitro, and exhibits RNA unwinding activity in an E. coli reporter assay. Subsequent deletion studies mapped annealing activity to the protein's G-rich N-terminus and unwinding to the RRM-containing C-terminus74. Thus, in vitro studies show that TbRGG2 binds pre-edited mRNA with high affinity and possesses the ability to modulate RNA-RNA interactions.
To address the role of TbRGG2 in RNA editing, three different groups analyzed mRNA levels in TbRGG2 knockdowns by qRT-PCR with essentially the same results48, 58, 63. Editing of pan-edited RNAs is dramatically compromised in TbRGG2-depleted cells, whereas editing of the three minimally edited RNAs remains unaffected. Furthermore, when measured, large decreases in the abundance of edited mRNAs were often not accompanied by increases in corresponding pre-edited mRNAs58, 63. These findings suggested a role for TbRGG2 in the 3’ to 5’ progression of editing with minimal effects on editing initiation. Utilizing full gene PCRs and primer extension assays with primers targeted to different parts of the RPS12 and A6 editing domains, Ammerman and co-workers49 confirmed that TbRGG2 depletion has minimal effects on editing initiation, modest effects at mRNA 3’ editing sites, and much more dramatic effects on 5’ editing sites, consistent with an impact on the 3’ to 5’ progression of editing (see also Fig. 5). Conventional sequence analysis of ~100 cDNAs derived from RPS12 full gene PCRs in TbRGG2 replete vs. depleted cells revealed two regions of the RPS12 mRNA where editing was prone to stalling. Referencing cDNA sequences with a then available gRNA database75 suggested that sites of stalling corresponded both to the 3’ end of one gRNA and to internal regions of another gRNA. Thus, it was not possible from this small cDNA dataset to distinguish whether TbRGG2 functions in gRNA exchange or in utilization of adjacent editing sites within a single mRNA-gRNA duplex. Deep sequencing analyses of edited mRNAs from TbRGG2 replete and depleted cells are currently underway to resolve this question. In addition to revealing stalling at specific points during editing progression, the sequences of the cDNA clones from the TbRGG2-silenced cells displayed significant decreases in the lengths of the junction regions that lie between 3’ fully edited and 5’ unedited regions. Junction regions are thought to be sites of active editing at the time when RNA is harvested from the cells, and their reduction upon ablation of TbRGG2 is consistent with impaired gRNA utilization. In vivo complementation studies of these RNAi knockdowns with TbRGG2 deletion and point mutants showed that the RRM domain-containing C-terminus is completely incapable of rescuing the knockdown, while the N-terminus harboring high affinity RNA-binding and annealing activities can partially complement cell growth and RNA editing74. A point mutant lacking RNA unwinding activity provided full complementation, suggesting that the RNA annealing function of TbRGG2 is more important in editing than its in vitro RNA unwinding activity. Collectively, in vivo studies implicate TbRGG2 in facilitating gRNA utilization critical for active editing and its resulting 3’ to 5’ progression.
The only other TbRGG2 subcomplex proteins that have been studied in detail are the paralogous MRB4160 and MRB8170 proteins69, which are devoid of any motifs or homology to known proteins. Nevertheless, in vitro UV cross-linking assays with synthetic RNAs demonstrated that both recombinant proteins possess RNA-binding activity. Like TbRGG2, MRB8170 displayed five to six-fold higher affinity for mRNA than gRNA in filter binding assays, although in contrast to TbRGG2 the Kds of MRB8170 for pre-edited and edited mRNA are similar (15-20 nM). In vivo, only the MRB4160/8170 double knockdown exhibits a growth defect, and the two proteins are partially redundant with respect to mt mRNA levels. No defects in mRNA levels were observed in MRB4160 knockdowns, whereas the ablation of MRB8170 resulted in modest defects in MURF2 and A6 mRNA editing and cox3 and NADH dehydrogenase subunit 7 (ND7) pre-edited mRNA stability. The double MRB4160/8170 knockdown exhibited widespread editing defects including both pan-edited mRNAs, and to a lesser extent minimally edited RNAs, and also exhibited the likely defect in the stability of pre-edited cox3 mRNA that was observed in the MRB8170 single knockdown. Similar to TbRGG2 knockdowns, pre-edited mRNA levels were not increased for pan-edited RNAs whose editing was affected by concurrent MRB4160/8170 knockdown69. This suggests MRB4160/8170 may play a role in editing progression similar to TbRGG2, although this hypothesis has not been directly tested. Analysis of mRNA levels by qRTPCR in cells depleted of other TbRGG2 subcomplex components, namely MRB8180, MRB800, and MRB1860, has been reported, with the caveat that the latter two proteins were only depleted to 60% of wild type levels and pre-edited mRNA levels were not reported48. MRB1860 depletion impacted only edited ND7 mRNA, while MRB8180 and MRB800 had distinct transcript-specific effects on edited mRNA levels. Importantly, depletion of TbRGG2 subcomplex components does not affect gRNA levels. Thus, the effects of TbRGG2 subcomplex on RNA editing likely reflect roles in RNA trafficking and utilization.
MRB10130
One published study has touched on the impact of depleting the ARM/HEAT repeat protein, MRB1013048. qRT-PCR analyses revealed a dramatic decrease in almost all edited mRNAs. While pre-edited mRNAs were not tested this way, Northern blot analysis of RPS12 mRNA showed substantial accumulation of pre-edited mRNA. In addition, the levels of several gRNAs were increased. These authors also reported modest decreases in some never-edited RNAs upon MRB10130 depletion, although this may be due to the analysis being performed at a time point after the dramatic growth defect had already ensued. In general, the phenotype of MRB10130 is more reminiscent of MRB1 core proteins than TbRGG2 subcomplex proteins, consistent with this protein's numerous physical interactions with core components.
Current model for MRB1 complex function
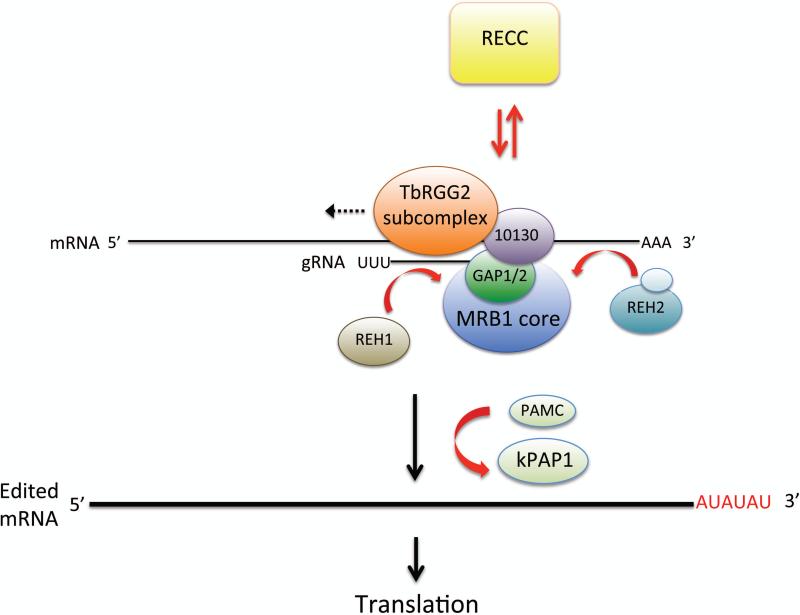
Even though we are still far from understanding the mechanisms by which the MRB1 complex manifests its essential role(s) in RNA editing, a model is emerging based on the above data47, 48, 50 (Figs. 4 and 6). The MRB1 core appears to be critical for the initiation of editing and very early editing events. One function of the core is likely to present GAP1/2-bound gRNAs to RECC. However, the effects of MRB1 component knockdowns on cox2 mRNA editing indicate that it has additional key functions in initial editing events. The TbRGG2 subcomplex apparently functions in gRNA utilization, thus facilitating 3’ to 5’ editing progression. The RNA binding and annealing capacity of TbRGG2 almost certainly contributes to this function. The roles of other TbRGG2 subcomplex components may be more RNA-related, potentially facilitating specific mRNA-gRNA interactions via their own RNA-binding activities (e.g., MRB8170 and MRB4160). Numerous outstanding questions remain, including the precise nature of the RNA-enhanced interaction between the MRB1 core and TbRGG2 subcomplex and the potential role of MRB10130 in its coordination. Other key unanswered questions are the route of gRNA and mRNA trafficking, the precise involvement of distinct proteins in this process, and the nature of the interaction between MRB1 and RECC.
Figure 6. Current model for the coordination of numerous complexes in mitochondrial U insertion/deletion RNA editing and mRNA processing.
The MRB1 complex (comprised of MRB1 core [including GAP1/2], TbRGG2 subcomplex, and MRB10130) serves as a platform for the editing reaction. RECC, containing the catalytic enzymes, associates transiently with MRB1-associated RNAs. TbRGG2 subcomplex facilitates editing 3’ go 5’ progression (dotted arrow). RNA helicases REH1 and REH2 appear to promote RNA association with MRB1. Following the completion of editing, kPAP1, potentially with assistance from PAMC, catalyzes a long A/U tail on the 3’ end of fully edited mRNA, rendering it competent for translation.
E. OTHER COMPLEXES ASSOCIATED WITH MRB1
In addition to its role as a platform for the RNA editing reaction, the MRB1 complex appears to act as a hub for numerous mt RNA processing reactions47 (Fig. 6). Several proteins interact with MRB1 transiently and/or sub-stoichiometrically, and below we describe the evidence for the physical and functional interaction of these complexes with MRB1.
RNA Helicases
Soon after the discovery that the site-specificity of trypanosome RNA editing is mediated by the hybridization of gRNAs to their cognate mRNAs, the participation of RNA helicases in the process was hypothesized. Helicase activity is likely required to unwind tight duplexes comprised of gRNA-edited mRNA, so that subsequent gRNAs can anchor, thereby permitting editing progression 76, 77 (Fig. 2B). RNA helicases have also been proposed to be involved in the remodeling of the ribonucleoprotein complexes that interact with transcripts during the course of RNA editing. So far, two mt RNA editing helicases have been characterized.
RNA editing helicase 1 (REH1), originally called mHel61p, is among the first proteins involved in RNA editing to be cloned from T. brucei76. This DExD/H box family protein exhibits in vitro RNA unwinding activity, consistent with a potential role in melting duplexed gRNA-mRNA to enable subsequent editing by the next gRNA33 (Fig. 2B). Further support for this role is that ablation of REH1 results in a decrease of some edited mRNAs, with the 3’ to 5’ progression being compromised33, 76. However, while REH1 appears to exhibit some RNA-dependent interaction with RECC33, it does not associate with the bulk of mt RNA helicase activity76, 77.
REH2 is a much larger mt DExD/H box helicase (~240 kDa as compared to the ~60 kDa REH1 in T. brucei) that associates with the majority of mt RNA unwinding activity78. This finding agrees with REH2 being discovered due to its association with MRB1 and RECC. These interactions appear to be via RNA linkers59 as was also demonstrated for REH133, 48. REH2 interacts with gRNAs in a manner dependent on its double stranded RNA binding domain and DExD/H box helicase motifs. Ablation of REH2 leads to a decrease in edited RNAs. gRNA stability may also be compromised60, 78, although this has not been observed in all studies50. Interestingly, RNA immunoprecipitation/RNAseq studies showed that REH2 appears to associate with a cohort of gRNAs that direct later editing blocks, compared to MRB3010, which associates with gRNAs biased towards those directing early editing blocks50. Moreover, REH2 depletion leads to a decrease in pre-edited and partially edited RNAs associated with MRB3010, suggesting that REH2 acts in trans to facilitate mRNA recruitment to the MRB1 core59. In sum, emerging evidence suggests that REH1 and REH2 helicases play important roles in RNA editing, although the mechanistic details await further investigation.
Nudix Hydrolase
A protein dubbed mt edited mRNA stability factor 1 (MERS1) was initially found, along with GAP1/2, associated substoichiometrically with the MRP1/2 complex in Leishmania57. This protein was later shown to be associated in an RNA-dependent manner with MRB1 in isolations of GAP1 and GAP251, 54. MERS1 bears a motif that places it in the Nudix Hydrolase superfamily of proteins. Such proteins are found throughout prokaryotes, eukaryotes and even viruses, and are mainly pyrophosphohydrolases that cleave nucleotide di- and triphosphates linked to another “X” moiety (hence the name NUDIX), i.e. NDP-X to NMP and P-X, and thus are involved in many metabolic pathways involving sugars79. In terms of participation in RNA metabolism, a Nudix hydrolase conserved among opisthokonts called Dcp2 is involved in the mRNA decapping. To date, little is known about the role of MERS1 in shaping the mt transcriptome of trypanosomes. RNAi-silencing of the protein in T. brucei leads to a destabilization of edited mRNAs51, although further studies will be required to establish the mechanistic connections between MERS1 and other mt RNA processing events, including RNA editing.
kPAP
Mt RNAs possess non-encoded 3’ tails, and the presence of two RNA populations with short (~20 nt) and longer (~120 nt) tails was reported two decades ago80. More recent studies have revealed that these tails are comprised of A, U, or A/U, with the longer tails generally having a higher percentage of U, and tail composition apparently being quite RNA-specific81-83. Short A-tails stabilize edited RNAs, but not pre-edited RNAs, both in vitro and in vivo, and only a small amount of editing is required for A-tail mediated stabilization83, 84. Long tails, present primarily on never-edited and fully edited RNAs, appear to facilitate mRNA association with the small ribosomal subunit, and thus translation82.
Kinetoplast poly(A) polymerase 1 (kPAP1) functions in both long and short 3’-tail synthesis, and it resides in the kPAP complex with a heterodimer of two pentatricopeptide (PPR) proteins, termed kPAF1 and kPAF282, 83. In vitro assays demonstrated that kPAF1/2 promotes long 3’ tail synthesis82. kPAF1/2 does not stimulate long A-tail synthesis, but rather requires interspersed U's added by RET1 to generate long tails. The kPAP1 complex engages in transient and RNA-mediated interactions with RECC and MRB148, 51, 83. kPAP1 also apparently makes several weak contacts with the mt ribosome, assisting the connection between poly A/U tail synthesis and translation48. Numerous questions remain regarding the interactions between the kPAP complex and others involved in mitochondrial RNA processing. At what point and with which proteins does kPAP1 add short tails to mt RNAs? How does the kPAP complex recognize only translatable (fully edited and never-edited) RNAs for long A/U tail synthesis?
PAMC
The so-called polyadenylation mediator complex (PAMC) exhibited protein-mediated interactions with the MRB1 core in one recent study48, although its components had not been previous identified in association with MRB1. While this complex does not strongly interact with the kPAP complex, ablation of one of its subunits (but not others) suggested a role in both short and long 3’ tail synthesis, thereby leading to its name. The role of the PAMC complex in mt RNA polyadenylation and processing await further investigation.
CONCLUSION
The discovery of MRB1 provided important clues to longstanding questions in the kinetoplastid RNA editing field, such as why RECC appeared to lack stable RNA association and why it failed to catalyze processive RNA editing in vitro. Evidence to date suggests that MRB1 is the platform for editing, and that it facilitates RNA recruitment to RECC and 3’ to 5’ progression of editing. However, these studies are in their infancy. Future work is needed to address the degree of heterogeneity and dynamism within the MRB1 complex, the biochemical functions of MRB1 complex components, and the precise protein-RNA interactions that facilitate the byzantine process that is U-insertion/deletion RNA editing. Interactions between MRB1 and other RNA processing machineries provide a further layer of complexity in kinetoplastid mt gene expression.
Acknowledgements
This work was supported by National Institutes of Health RO1 AI061580 to L.K.R. and Czech Grant Agency 15-21974S, RNPnet FP7 program (289007) and Praemium Academiae award to J.L.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 2.Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 3.Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 4.Maier RM, Zeltz P, Kossel H, Bonnard G, Gualberto JM, Grienenberger JM. RNA editing in plant mitochondria and chloroplasts. Plant Mol Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 5.Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003;278:1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 6.Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gott JM, editor. Mechanisms and Functions of RNA editing in Physarum polycephalum. Caister Academic Press; Norfolk, U.K.: 2013. pp. 17–40. [Google Scholar]
- 8.Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Rev RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valach M, Moreira S, Kiethega GN, Burger G. Trans-splicing and RNA editing of LSU rRNA in Diplonema mitochondria. Nucleic Acids Res. 2014;42:2660–2672. doi: 10.1093/nar/gkt1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maris C, Masse J, Chester A, Navaratnam N, Allain FH. NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor. RNA. 2005;11:173–186. doi: 10.1261/rna.7190705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Barnes RL, Shi H, Kolev NG, Tschudi C, Ullu E. Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery. PLoS Pathog. 2012;8:e1002678. doi: 10.1371/journal.ppat.1002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen RE, Englund PT. Network news: the replication of kinetoplast DNA. Annu Rev Microbiol. 2012;66:473–491. doi: 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- 16.Feagin JE, Abraham JM, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 17.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 18.Seiwert SD, Stuart K. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- 19.Golden DE, Hajduk SL. The 3'-untranslated region of cytochrome oxidase II mRNA functions in RNA editing of African trypanosomes exclusively as a cis guide RNA. RNA. 2005;11:29–37. doi: 10.1261/rna.7170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koslowsky D, Sun Y, Hindenach J, Theisen T, Lucas J. The insect-phase gRNA transcriptome in Trypanosoma brucei. Nucleic Acids Res. 2014;42:1873–1886. doi: 10.1093/nar/gkt973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koslowsky DJ, Reifur L, Yu LE, Chen W. Evidence for U-tail stabilization of gRNA/mRNA interactions in kinetoplastid RNA editing. RNA Biol. 2004;1:28–34. doi: 10.4161/rna.1.1.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J Am Soc Mass Spectrom. 2003;14:728–735. doi: 10.1016/S1044-0305(03)00126-0. [DOI] [PubMed] [Google Scholar]
- 24.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 28.Ernst NL, Panicucci B, Igo RP, Jr., Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3' terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 29.McManus MT, Shimamura M, Grams J, Hajduk SL. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusche LN, Huang CE, Piller KJ, Hemann M, Wirtz E, Sollner-Webb B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol Cell Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruz-Reyes J, Zhelonkina AG, Huang CE, Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol Cell Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, Herrera J, Zhou S, Maslov DA, Simpson L. Trypanosome REH1 is an RNA helicase involved with the 3'-5' polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc Natl Acad Sci U S A. 2011;108:3542–3547. doi: 10.1073/pnas.1014152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koslowsky DJ, Bhat GJ, Read LK, Stuart K. Cycles of progressive realignment of gRNA with mRNA in RNA editing. Cell. 1991;67:537–546. doi: 10.1016/0092-8674(91)90528-7. [DOI] [PubMed] [Google Scholar]
- 35.Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 36.Ernst NL, Panicucci B, Carnes J, Stuart K. Differential functions of two editosome exoUases in Trypanosoma brucei. RNA. 2009;15:947–957. doi: 10.1261/rna.1373009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park YJ, Budiarto T, Wu M, Pardon E, Steyaert J, Hol WG. The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2. Nucleic Acids Res. 2012;40:6966–6977. doi: 10.1093/nar/gks369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao G, Rogers K, Li F, Guo Q, Osato D, Zhou SX, Falick AM, Simpson L. Uridine insertion/deletion RNA editing in Trypanosomatids: specific stimulation in vitro of Leishmania tarentolae REL1 RNA ligase activity by the MP63 zinc finger protein. Protist. 2010;161:489–496. doi: 10.1016/j.protis.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnaufer A, Wu M, Park YJ, Nakai T, Deng J, Proff R, Hol WG, Stuart KD. A protein-protein interaction map of trypanosome ~20S editosomes. J Biol Chem. 2010;285:5282–5295. doi: 10.1074/jbc.M109.059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carnes J, Zelaya Soares C, Wickham C, Stuart K. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J Biol Chem. 2011 doi: 10.1074/jbc.M111.228965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carnes J, Lewis Ernst N, Wickham C, Panicucci B, Stuart K. KREX2 is not essential for either procyclic or bloodstream form Trypanosoma brucei. PLoS One. 2012;7:e33405. doi: 10.1371/journal.pone.0033405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers K, Gao G, Simpson L. Uridylate-specific 3' 5'-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J Biol Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- 44.Guo X, Carnes J, Ernst NL, Winkler M, Stuart K. KREPB6, KREPB7, and KREPB8 are important for editing endonuclease function in Trypanosoma brucei. RNA. 2012;18:308–320. doi: 10.1261/rna.029314.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Alatortsev VS, Cruz-Reyes J, Zhelonkina AG, Sollner-Webb B. Trypanosoma brucei RNA editing: coupled cycles of U deletion reveal processive activity of the editing complex. Mol Cell Biol. 2008;28:2437–2445. doi: 10.1128/MCB.01886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashimi H, Zimmer SL, Ammerman ML, Read LK, Lukes J. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol. 2013;29:91–99. doi: 10.1016/j.pt.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aphasizheva I, Zhang L, Wang X, Kaake RM, Huang L, Monti S, Aphasizhev R. RNA binding and core complexes constitute the U-insertion/deletion editosome. Mol Cell Biol. 2014;34:4329–4342. doi: 10.1128/MCB.01075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression through intrinsic pause sites. RNA. 2010;16:2239–2251. doi: 10.1261/rna.2285510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madina BR, Kumar V, Metz R, Mooers BH, Bundschuh R, Cruz-Reyes J. Native mitochondrial RNA-binding complexes in kinetoplastid RNA editing differ in guide RNA composition. RNA. 2014;20:1142–1152. doi: 10.1261/rna.044495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ammerman ML, Downey KM, Hashimi H, Fisk JC, Tomasello DL, Faktorova D, Kafkova L, King T, Lukes J, Read LK. Architecture of the trypanosome RNA editing accessory complex, MRB1. Nucleic Acids Res. 2012;40:5637–5650. doi: 10.1093/nar/gks211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol Cell Proteomics. 2008;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA. 2008;14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaili N, Goringer U, Benne R, et al. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J Biol Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- 56.Vondruskova E, van den Burg J, Zikova A, Ernst NL, Stuart K, Benne R, Lukes J. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- 57.Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Acestor N, Panigrahi AK, Carnes J, Zikova A, Stuart KD. The MRB1 complex functions in kinetoplastid RNA processing. RNA. 2009;15:277–286. doi: 10.1261/rna.1353209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madina BR, Kumar V, Mooers BH, Cruz-Reyes J. Native variants of the MRB1 complex exhibit specialized functions in kinetoplastid RNA editing. PLoS One. 2015;10:e0123441. doi: 10.1371/journal.pone.0123441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hashimi H, Cicova Z, Novotna L, Wen YZ, Lukes J. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aphasizheva I, Aphasizhev R. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol Cell Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanifuji G, Kim E, Onodera NT, Gibeault R, Dlutek M, Cawthorn RJ, Fiala I, Lukes J, Greenwood SJ, Archibald JM. Genomic characterization of Neoparamoeba pemaquidensis (Amoebozoa) and its kinetoplastid endosymbiont. Eukaryot Cell. 2011;10:1143–1146. doi: 10.1128/EC.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fisk JC, Ammerman ML, Presnyak V, Read LK. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem. 2008;283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jackson AP. Evolutionary consequences of a large duplication event in Trypanosoma brucei: chromosomes 4 and 8 are partial duplicons. BMC Genomics. 2007;8:432. doi: 10.1186/1471-2164-8-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu W, Kimelman D. Mechanistic insights from structural studies of beta-catenin and its binding partners. J Cell Sci. 2007;120:3337–3344. doi: 10.1242/jcs.013771. [DOI] [PubMed] [Google Scholar]
- 66.McAdams NM, Ammerman ML, Nanduri J, Lott K, Fisk JC, Read LK. An arginine-glycine-rich RNA binding protein impacts the abundance of specific mRNAs in the mitochondria of Trypanosoma brucei. Eukaryot Cell. 2015;14:149–157. doi: 10.1128/EC.00232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shaw PL, McAdams NM, Hast MA, Ammerman ML, Read LK, Schumacher MA. Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ammerman ML, Hashimi H, Novotna L, Cicova Z, McEvoy SM, Lukes J, Read LK. MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA. 2011;17:865–877. doi: 10.1261/rna.2446311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kafkova L, Ammerman ML, Faktorova D, Fisk JC, Zimmer SL, Sobotka R, Read LK, Lukes J, Hashimi H. Functional characterization of two paralogs that are novel RNA binding proteins influencing mitochondrial transcripts of Trypanosoma brucei. RNA. 2012;18:1846–1861. doi: 10.1261/rna.033852.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang Z, Faktorova D, Krizova A, Kafkova L, Read LK, Lukes J, Hashimi H. Integrity of the core mitochondrial RNA binidng complex 1 is vital for trypanosome RNA editing. RNA. 2015 doi: 10.1261/rna.052340.115. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ammerman ML, Tomasello DL, Faktorova D, Kafkova L, Hashimi H, Lukes J, Read LK. A core MRB1 complex component is indispensable for RNA editing in insect and human infective stages of Trypanosoma brucei. PLoS One. 2013;8:e78015. doi: 10.1371/journal.pone.0078015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carnes J, Lerch M, Kurtz I, Stuart K. Bloodstream form Trypanosoma brucei do not require mRPN1 for gRNA processing. RNA. 2015;21:28–35. doi: 10.1261/rna.045708.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madina BR, Kuppan G, Vashisht AA, Liang YH, Downey KM, Wohlschlegel JA, Ji X, Sze SH, Sacchettini JC, Read LK, et al. Guide RNA biogenesis involves a novel RNase III family endoribonuclease in Trypanosoma brucei. RNA. 2011;17:1821–1830. doi: 10.1261/rna.2815911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foda BM, Downey KM, Fisk JC, Read LK. Multifunctional G-rich and RRM-containing domains of TbRGG2 perform separate yet essential functions in trypanosome RNA editing. Eukaryot Cell. 2012;11:1119–1131. doi: 10.1128/EC.00175-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ochsenreiter T, Cipriano M, Hajduk SL. KISS: the kinetoplastid RNA editing sequence search tool. RNA. 2007;13:1–4. doi: 10.1261/rna.232907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Missel A, Souza AE, Norskau G, Goringer HU. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol Cell Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Missel A, Goringer HU. Trypanosoma brucei mitochondria contain RNA helicase activity. Nucleic Acids Res. 1994;22:4050–4056. doi: 10.1093/nar/22.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hernandez A, Madina BR, Ro K, Wohlschlegel JA, Willard B, Kinter MT, Cruz-Reyes J. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem. 2010;285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhat GJ, Souza AE, Feagin JE, Stuart K. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 1992;52:231–240. doi: 10.1016/0166-6851(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 81.Zimmer SL, McEvoy SM, Menon S, Read LK. Additive and transcript-specific effects of KPAP1 and TbRND activities on 3' non-encoded tail characteristics and mRNA stability in Trypanosoma brucei. PLoS One. 2012;7:e37639. doi: 10.1371/journal.pone.0037639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell. 2011;42:106–117. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3' adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol Cell Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]