Abstract
Extracellular vesicles (EVs) are membrane surrounded structures released by cells, which have been increasingly recognized as mediators of intercellular communication. Recent reports indicate that EVs participate in important biological processes and could serve as potential source for cancer biomarkers. As an attractive EVs source with merit of non-invasiveness, human saliva is a unique medium for clinical diagnostics. Thus, we proposed a facile approach to prepare salivary extracellular vesicles (SEVs). Affinity chromatography column combined with filter system (ACCF) was developed to efficiently remove the high abundant proteins and viscous interferences of saliva. Protein profiling in the SEVs obtained by this strategy was compared with conventional centrifugation method, which demonstrated that about 70% more SEVs proteins could be revealed. To explore its utility for cancer proteomics, we analyzed the proteome of SEVs in lung cancer patients and normal controls. Shotgun proteomic analysis illustrated that 113 and 95 proteins have been identified in cancer group and control group, respectively. Among those 63 proteins that have been consistently discovered only in cancer group, 12 proteins are lung cancer related. Our results demonstrated that SEVs prepared through the developed strategy are valuable samples for proteomics and could serve as a promising liquid biopsy for cancer.
Extracellular vesicles (EVs) are defined as intact, submicron, phospholipid-rich vesicles ranging from 100 nm to 1000 nm in diameters, which shed from the surface of cells1. The functions of EVs are not completely understood yet. They are initially known as garbage cans whose job is to discard unwanted cellular substances2,3. However, recent research has revealed that these vesicles act also as important messengers for intercellular communication4. For instance, they could putatively attach or fuse with the target cell membrane, delivering surface proteins and perhaps cytoplasm to the recipient cell5,6. These properties are critical for signal transduction in the microenvironments, especially in disease pathogenesis and the tumor organotropic metastasis7,8,9. Study of gliomas tumor cells demonstrated that microvesicles from tumor cells could release to cellular surroundings and blood of tumor-bearing mice and contribute to horizontal propagation of oncogenes10. Therefore, it is of great interest to explore the proteome of EVs that originate from human body fluids, which might carry important biomarkers for the early detection of cancers11.
Human saliva is an attractive body fluid for molecular diagnostics, due to its unique composition and non-invasive sample collection. Owing to its enormous diagnostic potential, human saliva has been comprehensively explored for the detection of different oral diseases12,13 as well as systemic diseases14,15,16,17,18. Meanwhile, saliva has been recommended as a detection medium by the FDA for vulnerable populations, for instance children. Of note is that human saliva harbors plenty of extracellular vesicles (EVs), namely salivary extracellular vesicles (SEVs)19,20. SEVs studies demonstrated that tumor-secreted vesicles could enter the extracellular microenvironment and then affect and alter salivary gland in vitro21 and in vivo22, More specifically, tumor cell-specific mRNA and protein could be detected in microvesicles from saliva and blood. Unique biological information of tumor carried by EVs could also initiate the proliferation and metastasis of lung cancer23, which emphasized the association between distal tumor progression and the biomarker discovery in saliva through microvesicles.
However, one of the obstacles for SEVs preparation is the interference of high abundant amylase and other viscous proteins in saliva24. The overwhelming concentration of amylase in saliva could affect the identification and characterization of low abundant proteins as well as SEVs proteins, which are often biomarker candidates25. Meanwhile, amylase also interfere the extraction and separation of EVs by encasing and clinging globule with membrane structures and viscous proteins. Thus removal of amylase and other viscous proteins from saliva before SEVs’ extraction could benefit downstream proteomic analysis of SEVs and contribute to biomarker discovery for cancer26.
Lung cancer is one of the leading cancers for both genders worldwide and the most common causes of cancer related deaths27. The incidence of lung cancer has significantly increased in recent years, partially owing to the large smoking population and the declined air quality28. In addition, the five-year survival for lung cancer was lower than 10%. This dismal prognosis is mainly due to the fact that most patients were diagnosed at III or IV stage of disease. Therefore, early detection is the key for cure and the most effective way to reduce lung cancer deaths29. However, it’s technically challenging for medical imaging techniques and invasive biopsy to find early cancers, due to the limited resolution and specificity30. Molecular diagnostics is a promising and alternative approach for the early detection of cancers. Of note is that proteomic biomarkers have been discovered in human saliva for the detection of lung cancer31.
Since there is no standard operation procedure for pre-treatment of saliva samples as well as SEVs preparation, it is therefore necessary to establish a straightforward method as that for serum to remove high abundant amylase and viscous proteins. Starch has been previously used to specifically remove amylase from saliva, based on their strong affinity interactions32. Especially, it is easily available, economical, and feasible for practical applications. Keeping the availability and cost of starch in mind, hereby we intent to develop an affinity chromatography column combined with a filter system (ACCF) to trap amylase specifically, using starch as the stationary phase, and enrich EVs in saliva samples. In the present work, we prepared SEVs by ACCF and compared their protein profiling with that isolated by the conventional centrifugation method. The developed approach was further applied to harvest SEVs from healthy subjects and lung cancer patients, respectively. The proteome of both groups were compared through shotgun proteomics and further used for candidate biomarker discovery for the detection of lung cancer.
Results
Our human saliva contains high abundant proteins along with many viscous proteins, which interfere in the preparation of SEVs. To prepare quality SEVs for clinical applications and also minimize the interference of salivary proteins, herein we developed an affinity chromatography coupled with filter system aiming at high quality SEVs separation, which can be further used for cancer proteomics.
ACCF system development
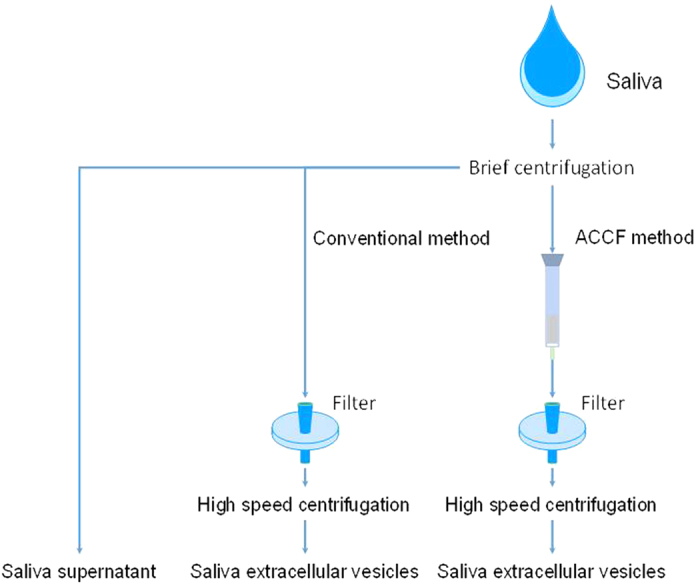
The ACCF system consisted of two parts, the affinity chromatography column (ACC) and membrane filter (F), as shown in Fig. 1. The ACC part was a column prepared in a syringe by packing 0.5 g starch (from potato, Sigma, Shanghai, China) with 3 mL phosphate buffered saline (PBS) (Shanghai Bioscience Co. Ltd), which was adequate for 300 μL saliva sample preparation. The F part was a filter with 5 μm PVDF membrane (Millipore, Billerica, MA, USA).
Figure 1. Schematic diagram for EVs isolation from human saliva.
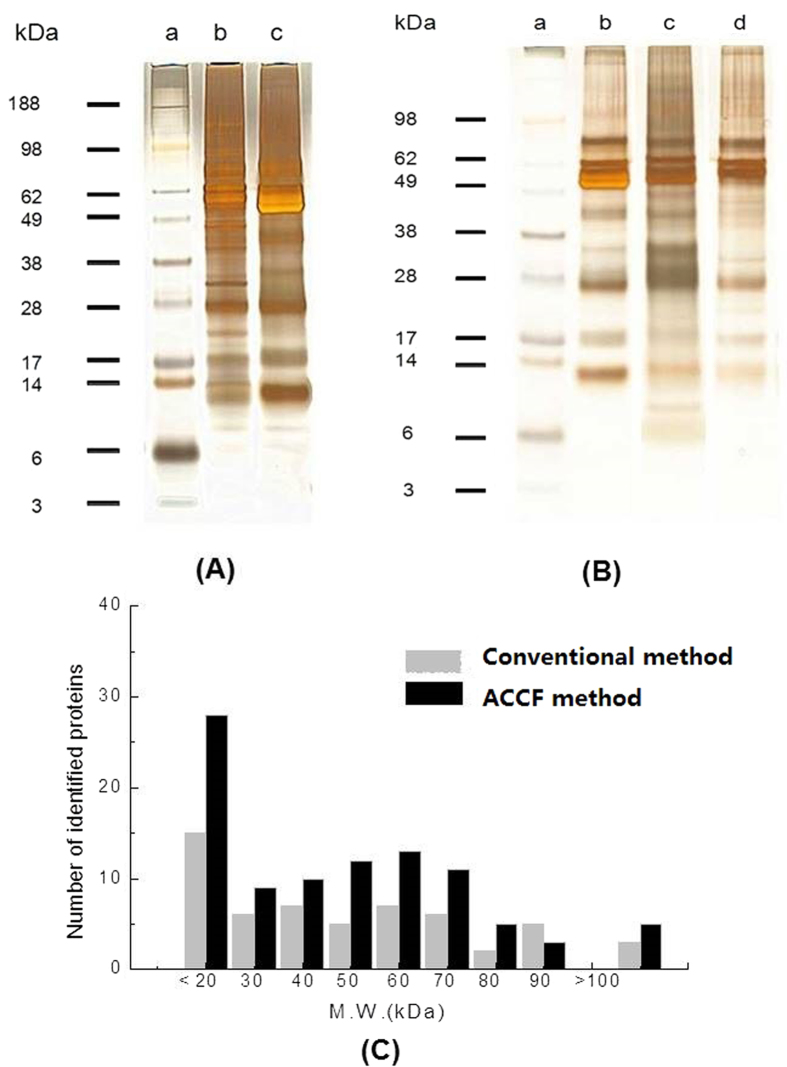
In order to evaluate purification efficiency of the ACCF system, 1D SDS-PAGE was used to separate salivary and SEVs proteins, as shown in Fig. 2A. Comparing the protein bands obtained from SEVs isolated by ACCF (lane b) and saliva (lane c), notable difference appeared between 49 kDa and 62 kDa, because this range was mostly salivary amylase33. In lane (c), amylase was a wide and strong band while other protein bands were masked in the same region. In addition, lane (b) displayed more refined protein bands than lane (c), which suggests that high content amylase create interference in the separation and detection of other low abundant proteins.
Figure 2. SDS-PAGE of salivary proteins and SEVs proteins.
(A) 1D SDS-PAGE of salivary protein. (a) Protein ladder; (b) 1.5 μg of salivary proteins prepared by ACCF method; (c) 1.5 μg of primordial salivary proteins. (B) 1D SDS-PAGE of SEVs proteins. (a) Protein ladder; (b) 1.0 μg of salivary proteins; (c) 1.0 μg of EVs’ proteins prepared by ACCF method; (d) 1.0 μg of EVs’ proteins prepared by conventional centrifugation method. (C) Molecular weight distribution of SEVs proteins prepared by conventional centrifugation method and ACCF method.
In order to compare the ACCF system with the conventional centrifugation method, the extracted SEVs proteins from both ACCF and conventional centrifugation method were separated by SDS-PAGE (Fig. 2B). The SDS-PAGE analysis revealed that proteins isolated from SEVs by using ACCF (lane c) and conventional centrifugation method (lane d) exhibited significant difference in their profiling when compared to the whole saliva protein (lane b). It was observed that the SEVs’ proteins obtained from ACCF (lane c) have some unique compositions. When lane d was compared with lane c, it was observed that there are a large number of low molecule weight proteins (≤14 kDa). We quantitatively compared the salivary proteins before and after amylase removal and found that the salivary protein concentration decreased from 1.25 μg/μL to 0.62 μg/μL. This result confirmed that amylase is the most abundant protein in saliva, which could be trapped by ACCF system. The reproducibility of these two methods was evaluated by isolating SEVs from 300 μL saliva sample. As shown in Table 1, the RSD (n = 3) for ACCF method and conventional centrifugation method was 1.85% and 2.83%, respectively.
Table 1. Reproducibility of the SEVs isolation.
| Methods | No. | SEVs protein (μg) | Average | SD | RSD (%) |
|---|---|---|---|---|---|
| ACCF method | 1 | 3.876 | 3.940 | 0.073 | 1.85 |
| 2 | 3.926 | ||||
| 3 | 4.020 | ||||
| Conventional centrifugation method | 1 | 3.324 | 3.315 | 0.094 | 2.83 |
| 2 | 3.405 | ||||
| 3 | 3.216 |
SEVs were separated from 300 μL saliva sample by ACCF and conventional centrifugation method, respectively.
Nanoparticle tracking analysis for the size distribution of SEVs
Nanoparticle tracking analysis was used to evaluate the size distribution of SEVs. The results are shown in Fig. 3. The SEVs size in primordial saliva and purified saliva sample using ACCF system was both smaller than 1,000 nm. However, for SEVs prepared from conventional centrifugation method, the particle size ranged from 109 nm to 660 nm, while for SEVs prepared through ACCF method, the diameter ranged from 99 nm to 944 nm.
Figure 3.
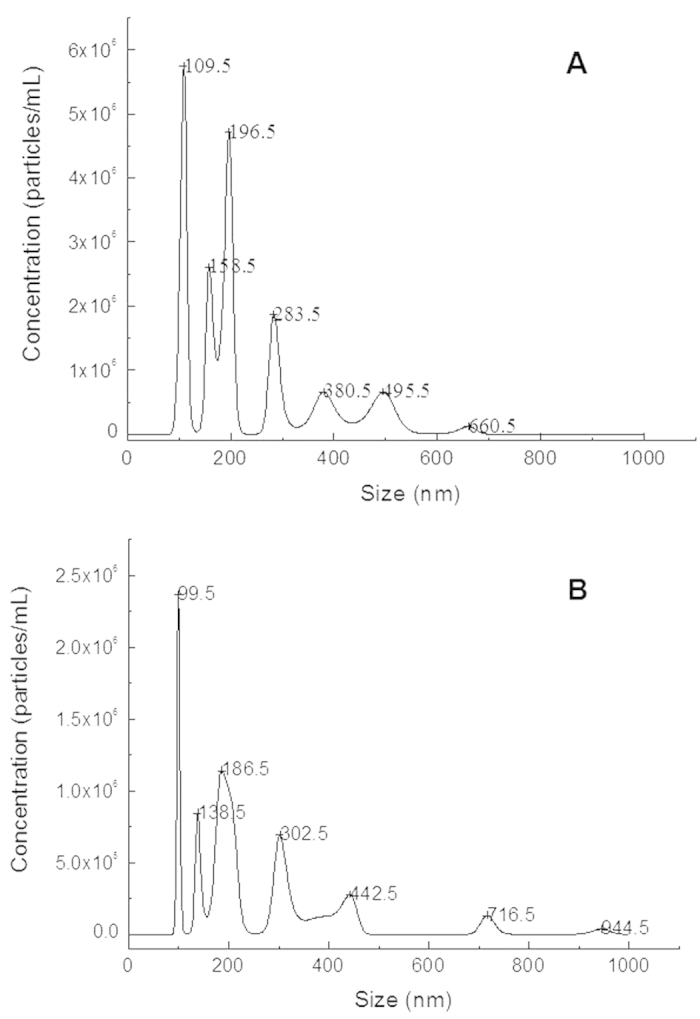
Particle size and distribution of the SEVs obtained by primordial saliva (A) and purified saliva sample using ACCF system (B).
Proteomics analysis of SEVs
LC-MS/MS based shotgun proteomic approach was applied for the proteome analysis of SEVs. All experiments (both conventional centrifugation method and ACCF method) were carried out in triplicate. For each SEVs sample obtained by ACCF method, we identified 128, 138 and 107 proteins, respectively. For each SEVs sample prepared by conventional centrifugation method, we discovered 87, 76 and 72 proteins, respectively. To extract the most reliable data, we used the overlapped results of 3 experiments. Finally, we consistently obtained 95 SEVs proteins for ACCF method and 56 SEVs proteins for conventional centrifugation method (Fig. 4). The number of identified SEVs proteins was increased more than 70% through using ACCF method (Fig. 4A). Further analysis revealed that 42 proteins were shared by both methods, which include SEVs marker protein Integrin beta-234. The ACCF method was able to discover 53 unique proteins from SEVs, while 14 proteins were unique to conventional centrifugation method. It could be observed from Fig. 2C that the use of ACCF approach for the removal of amylase led to find more number of low molecule weight proteins (<20 kDa) than conventional centrifugation method. These results were consistent with the SDS-PAGE analysis of both methods (Fig. 2B).
Figure 4. Venn diagram of identified proteins.
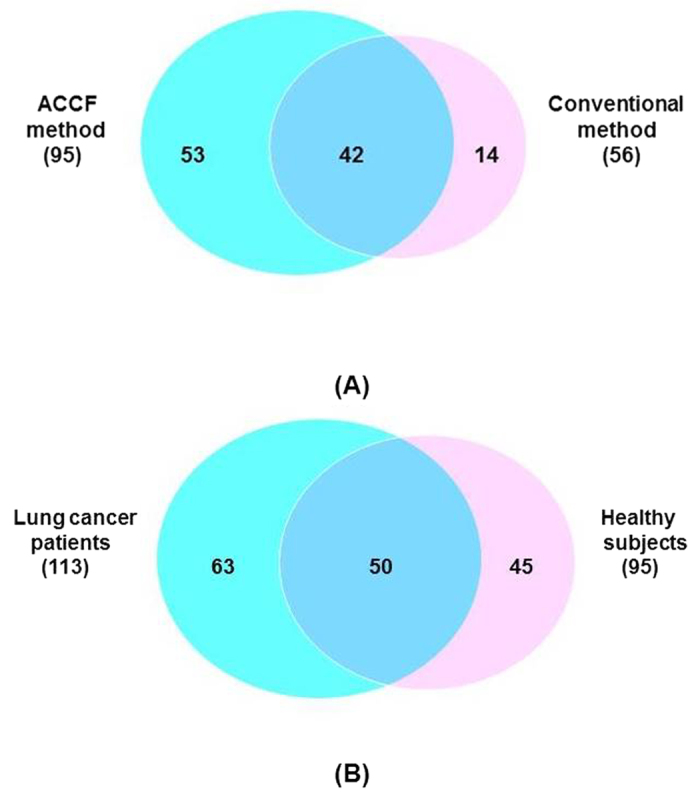
(A) Overlap of SEVs proteins prepared through ACCF method and conventional method; (B) Overlap of SEVs proteins extracted from the saliva of lung cancer patients and healthy subjects.
To further demonstrate the interference of amylase on SEVs isolation and their protein identification, the total ion chromatograms of SEVs’ peptides prepared through ACCF method and the conventional one were compared, as shown in Fig. 5. The SEVs sample obtained after removal of amylase eluted more peaks between retention time of 90 mins and 130 mins (Fig. 5B), which partially contributed to the identification of 53 unique proteins for SEVs.
Figure 5.
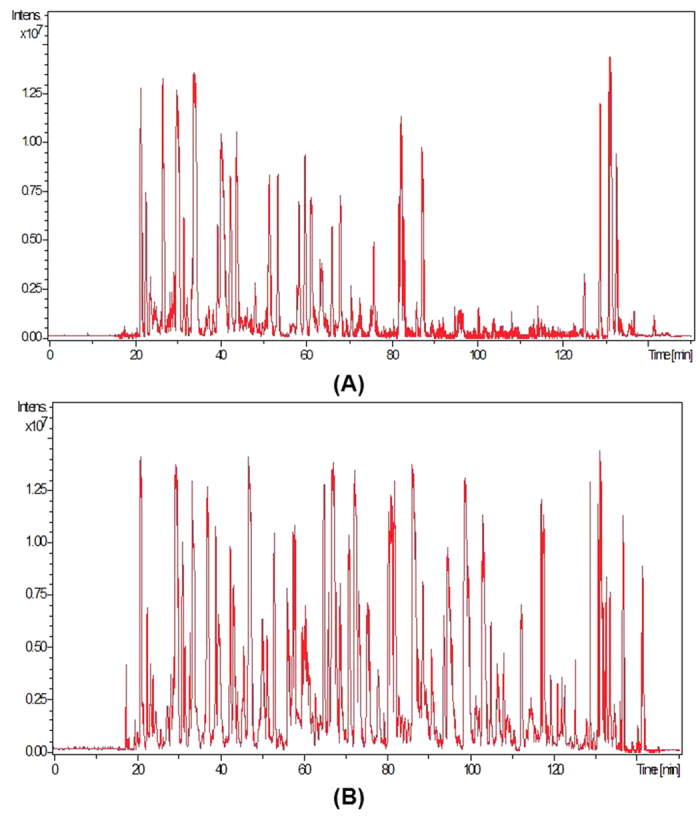
Total ion chromatogram of SEVs’ peptides from primordial saliva (A) and ACCF purified saliva (B).
Detailed information of identified SEVs proteins through conventional centrifugation method and ACCF method is shown in Tables S1 and S2, respectively. To uncover the molecular events underlying these discrepant proteins profiles, we studied the Gene Ontology information of these specific proteins, which include biological process, cellular component, molecular function, pathway, and protein class analysis by using PANTHER software (Fig. S1). The Gene Ontology pathway information of SEVs proteins obtained by ACCF approach and conventional centrifugation method was compared. We found SEVs proteins discovered through conventional centrifugation method revealed only 6 common pathways which are necessary for common cell growth and immune cell activation. Whereas, SEVs proteins obtained by ACCF method involved in 31 pathways, indicating many dynamic disease processes.
ACCF method for proteomic analysis of cancer SEVs
We further applied the ACCF system to isolate SEVs from the saliva of 3 lung cancer patients. SEVs proteins were extracted and analyzed by LC-MS/MS in parallel and technically repeated. We identified 138, 146, and 129 proteins, respectively, for the lung cancer SEVs sample prepared by ACCF method. We also used the overlapped data and 113 proteins (Table S3) were consistently discovered from the SEVs prepared from the saliva of cancer patients. The Gene Ontology data of these 113 proteins is shown in Fig. S2. When proteins were further compared with healthy subjects, 50 proteins were commonly shared by the two methods, 45 proteins presented uniquely in healthy subjects’ saliva and 63 proteins only appeared in lung cancer patients’ SEVs (Fig. 4B). The differences of SEVs’ protein between healthy subjects and lung cancer patients suggested that SEVs carried versatile biological information, which might serve as a biomarker source for lung cancer.
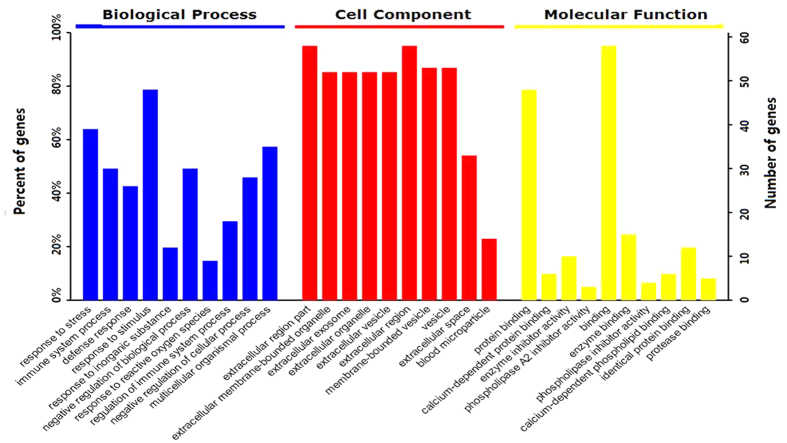
To discover potential biomarkers, we further analyzed these 63 proteins that were unique to lung cancer (Table 2). The Gene Ontology information of these 63 proteins showed that about 80% were involved in response to stimulus and 60% were related to stress response and multicellular organismal process (Fig. 6). The Gene Ontology information further revealed the extracellular nature of these proteins, they were mainly present in region part, membrane-bounded organelle, vesicle or exosome and blood microparticle. Molecular function results of these proteins showed that 90% gene referred to binding, which includes protein binding, enzyme binding, identical protein binding, and protease binding.
Table 2. List of candidate SEVs’ protein biomarkers for lung cancer.
| No. | Accession# | Protein name | Gene symbol | M.W. (kDa) | #Unique peptide | Mascot score |
|---|---|---|---|---|---|---|
| 1 | Q8WTW4 | Nitrogen permease regulator 2-like protein | NPRL2 | 43.6 | 2 | 315 |
| 2 | P01009 | Alpha-1-antitrypsin | A1AT | 46.7 | 6 | 519 |
| 3 | P01023 | Alpha-2-macroglobulin | A2M | 163.3 | 3 | 270 |
| 4 | P01011 | Alpha-1-antichymotrypsin | AACT | 47.6 | 4 | 82 |
| 5 | P12814 | Alpha-actinin-1 | ACTN1 | 103 | 2 | 269 |
| 6 | P43353 | Aldehyde dehydrogenase family 3 member B1 | AL3B1 | 51.8 | 2 | 75 |
| 7 | P50995 | Annexin A11 | ANXA11 | 54.3 | 3 | 141 |
| 8 | P04083 | Annexin A1 | ANXA1 | 38.7 | 5 | 435 |
| 9 | P07355 | Annexin A2 | ANXA2 | 38.6 | 5 | 575 |
| 10 | P12429 | Annexin A3 | ANXA3 | 36.3 | 2 | 460 |
| 11 | P08758 | Annexin A5 | ANXA5 | 35.9 | 2 | 295 |
| 12 | P08133 | Annexin A6 | ANXA6 | 75.8 | 4 | 159 |
| 13 | P84077 | ADP-ribosylation factor 1 | ARF1 | 20.6 | 2 | 264 |
| 14 | P52566 | Rho GDP-dissociation inhibitor 2 | ARHGDIB | 22.9 | 2 | 305 |
| 15 | P01024 | Complement C3 | C3 | 187.1 | 5 | 450 |
| 16 | Q5SNV9 | Uncharacterized protein C1orf167 | CA167 | 162.4 | 3 | 125 |
| 17 | P27482 | Calmodulin-like protein 3 | CALL3 | 16.8 | 2 | 93 |
| 18 | P04040 | Catalase | CATA | 59.7 | 3 | 92 |
| 19 | P07339 | Cathepsin D | CATD | 44.5 | 2 | 90 |
| 20 | P13688 | Carcinoembryonic antigen-related cell adhesion molecule 1 | CEACAM1 | 57.5 | 7 | 671 |
| 21 | P01034 | Cystatin-C | CYTC | 15.7 | 2 | 126 |
| 22 | P27487 | Dipeptidyl peptidase 4 | DPP4 | 88.2 | 3 | 85 |
| 23 | P15311 | Ezrin | EZRI | 69.4 | 2 | 43 |
| 24 | Q01469 | Fatty acid-binding protein, epidermal | FABP5 | 15.1 | 3 | 502 |
| 25 | P15328 | Folate receptor alpha | FOLR1 | 29.8 | 4 | 81 |
| 26 | P04406 | Glyceraldehyde-3-phosphate dehydrogenase | G3P | 36 | 2 | 250 |
| 27 | P28676 | Grancalcin | GRAN | 24 | 2 | 71 |
| 28 | Q58FF8 | Putative heat shock protein HSP 90-beta 2 | H90B2 | 44.4 | 2 | 33 |
| 29 | Q96A08 | Histone H2B type 1-A | HIST1H2BA | 14.1 | 11 | 1002 |
| 30 | P62805 | Histone H4 | HIST1H4A | 11.3 | 4 | 402 |
| 31 | P00738 | Haptoglobin | HPT | 45.2 | 3 | 75 |
| 32 | P02790 | Hemopexin | HPX | 51.6 | 6 | 560 |
| 33 | Q58FG1 | Putative heat shock protein HSP 90-alpha A4 | HS904 | 47.7 | 2 | 36 |
| 34 | P54652 | Heat shock-related 70 kDa protein 2 | HSP72 | 70 | 2 | 147 |
| 35 | P0DMV8 | Heat shock 70 kDa protein 1A | HSPA1A | 70 | 4 | 402 |
| 36 | P0DMV9 | Heat shock 70 kDa protein 1B | HSPA1B | 70 | 4 | 365 |
| 37 | P48741 | Putative heat shock 70 kDa protein 7 | HSPA7 | 40.2 | 3 | 43 |
| 38 | P01781 | Ig heavy chain V-III region GAL | HV320 | 12.7 | 3 | 245 |
| 39 | P30740 | Leukocyte elastase inhibitor | ILEU | 42.7 | 2 | 194 |
| 40 | P11215 | Integrin alpha-M | ITAM | 127.1 | 5 | 301 |
| 41 | P01593 | Ig kappa chain V-I region AG | KV101 | 12 | 2 | 160 |
| 42 | Q9GZZ8 | Extracellular glycoprotein lacritin | LACRT | 14.2 | 7 | 614 |
| 43 | P80188 | Neutrophil gelatinase-as | LCN2 | 22.5 | 3 | 397 |
| 44 | P00338 | L-lactate dehydrogenase A chain | LDHA | 36.6 | 4 | 115 |
| 45 | P09960 | Leukotriene A-4 hydrolase | LKHA4 | 69.2 | 2 | 227 |
| 46 | P14174 | Macrophage migration inhibitory factor | MIF | 12.4 | 6 | 86 |
| 47 | Q13421 | Mesothelin | MSLN | 68.9 | 8 | 805 |
| 48 | P26038 | Moesin | MSN | 67.8 | 9 | 713 |
| 49 | P15941 | Mucin-1 | MUC1 | 122.1 | 4 | 390 |
| 50 | P02763 | Alpha-1-acid glycoprotein 1 | ORM1 | 23.5 | 4 | 410 |
| 51 | P07237 | Protein disulfide-isomerase | PDIA1 | 57.1 | 2 | 121 |
| 52 | P05164 | Myeloperoxidase | PERM | 83.8 | 2 | 97 |
| 53 | P00558 | Phosphoglycerate kinase 1 | PGK1 | 44.6 | 4 | 217 |
| 54 | Q06830 | Peroxiredoxin-1 | PRDX1 | 22.1 | 4 | 104 |
| 55 | O43490 | Prominin-1 | PROM1 | 97.2 | 8 | 705 |
| 56 | Q6MZM9 | Proline-rich protein 27 | PRR27 | 22.7 | 2 | 46 |
| 57 | O75556 | Mammaglobin-B | SCGB2A1 | 10.8 | 4 | 330 |
| 58 | Q96QR1 | Secretoglobin family 3A member 1 | SCGB3A1 | 10.1 | 5 | 528 |
| 59 | Q687×5 | Metalloreductase STEAP4 | STEA4 | 51.9 | 2 | 80 |
| 60 | P37837 | Transaldolase | TALDO | 37.5 | 2 | 58 |
| 61 | P20061 | Transcobalamin-1 | TCO1 | 20.6 | 4 | 219 |
| 62 | P21580 | Tumor necrosis factor alpha-induced protein 3 | TNFAIP3 | 89.6 | 2 | 335 |
| 63 | P06753 | Tropomyosin alpha-3 chain | TPM3 | 32.9 | 3 | 285 |
Figure 6. Gene Ontology analysis of candidate SEVs’ protein biomarkers for lung cancer.
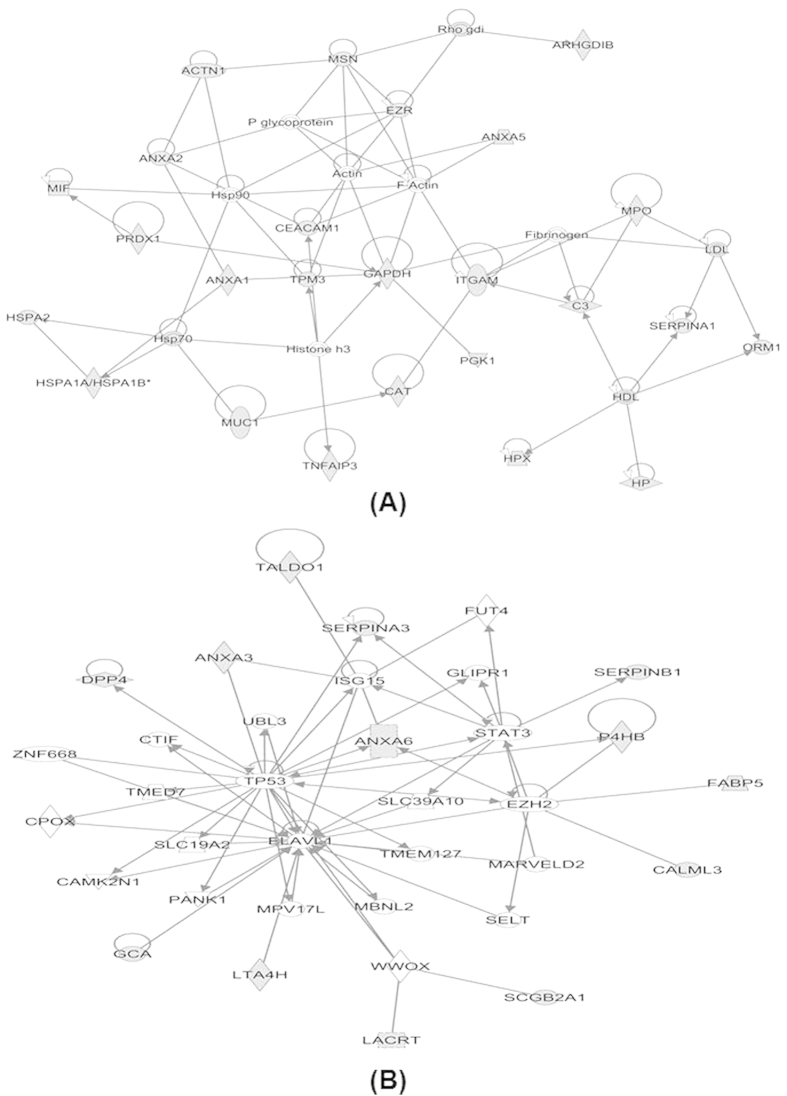
Ingenuity Pathway Analysis (IPA, Redwood City, CA, USA) analysis of these candidate SEVs’ protein biomarkers for lung cancer revealed that 25 of them (about 40%) involved in cancer network (Fig. 7A) and 13 of them (about 20%) were related with cellular movement (Fig. 7B, Table S4). Further, the literature survey showed that out of these 40% proteins, 12 proteins are lung cancer related biomarkers, which includes Annexin family members (Annexin A1, A2, A3, A5, A6, A11), Nitrogen permease regulator 2-like protein (NPRL2), Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), Mucin 1 (MUC1), Prominin-1 (PROM1), Histone H4 (HIST1H4A), and Tumor necrosis factor alpha-induced protein 3 (TNFAIP3).
Figure 7. IPA analysis of the candidate SEVs’ protein biomarkers for lung cancer.
Number of lung cancer specific proteins involve in cancer related network (A) and cellular movement related network (B). Gray symbols are SEVs proteins identified in this study.
Discussion and Conclusion
EVs have emerged as a potential biomarker source for molecular diagnostics. However, due to their small size and low abundance in body fluids, it is very challenging to efficiently separate and prepare EVs. In particular, human saliva contains high abundant proteins and viscous components that could disturb the extraction of EVs by encapsulating and overshadowing the EVs. In this case, we assumed that a portion of EVs might be attached to or gathered on proteins such as amylase and mucins hence decreasing the yield of EVs extraction. And protein aggregations with similar size of EVs may interfere in the accuracy and the quantity of proteomic analysis. To overcome these SEVs extraction issues, we developed ACCF technique in which on one hand we can remove high abundance amylase to reduce the protein interference. On the other hand, the system can filtrate out aggregated protein to release more types of EVs.
In this study, we paid much attention to the isolation of EVs and EVs proteins from saliva. We chose starch as an affinity stationary phase to specifically remove salivary amylase. After removal of amylase from saliva, we further used the processed saliva sample to prepare EVs. We observed that SEVs isolated by ACCF approach have broader diameter range (Fig. 3), which indicated that different types of SEVs were isolated. Therefore, our approach could further increase the quantity of SEVs proteins and boost their comprehensive proteomic analysis (Figs 4 and 5). Meanwhile, according to the SDS-PAGE results shown in Fig. 2, we found that low abundant proteins in saliva samples could be well resolved after amylase removal. In addition, more protein bands could be observed for SEVs proteins prepared by ACCF method than that of conventional method, especially many low molecular weight proteins as discovered by LC-MS/MS.
We further applied the developed method to prepare SEVs from lung cancer patient’ saliva as well as healthy subjects’ saliva and compared their protein profiling. We found 63 proteins that were unique to lung cancer patients (Table 2). After Gene Ontology analysis and extensive literature search we found that 12 of them were lung cancer related biomarkers, including 6 ANAX protein family members ANXA1, ANXA2, ANXA3, ANXA5, ANXA6, ANXA11. These ANAX proteins are associated with cell migration and vesicles fusion35,36. Another lung cancer related protein we found in cancer patients is NPRL2, which is a novel tumor suppressor gene and associated with cell growth and enhances sensitivity to various anticancer drugs37. CEACAM-1 and MUC1 were also presented in patients’ SEVs which mainly possess protein homodimerization activity and has been implicated in non-small-cell lung cancer development and progression38 and cancer cell signaling by promoting the synthesis and secretion of vascular endothelial growth factor (VEGF) through the AKT signaling pathway39.
Other lung cancer related proteins are PROM1, HIST1H4A and TNFAIP3. Exosome carried PROM1 has been one of the lipid raft-associated component that contributed to signal transduction and mediating intercellular communication40,41. HIST1H4A plays an important role in inducing cell death of tumor cells42. TNFAIP3 is an ubiquitin-editing enzyme, which is linked to a radioresistant phenotype of non-small cell lung cancer43.
Further EVs are known as signal transduction messengers. It has been proposed that EVs could assist tumor metastasis and diffusion after they were secreted to circulating system. Therefore, it is believed that EVs released from lung cancer tumor could carry tumor cell-specific proteins and enter saliva from blood, which has been verified in a xenografted mouse model of human lung cancer. In our study, lung cancer related proteins were identified in SEVs, which confirmed that these EVs in saliva might originate from lung cancer tumor. Therefore it is very promising to identify biomarkers from SEVs for the detection of lung cancer as well as other systemic diseases. Although further validation in a large sample set is required, the finding of candidate biomarkers in SEVs would provide a clue to the biological study of lung cancer and assist in future disease progression research. Our developed method for SEVs preparation will further expand the application of salivary diagnostics.
Materials and Methods
Saliva collection
Saliva samples were collected according to approved protocols (IRB#M15017) by Institutional Review Board (IRB) of Shanghai Jiao Tong University and all subjects provided written informed consents. The methods were carried out in accordance with the approved guidelines. All experimental protocols were approved by Bio-X Ethics Committee of Shanghai Jiao Tong University. Six healthy subjects and three lung cancer patients were recruited for this study. None of the healthy subjects had any history of malignancy, immunodeficiency, autoimmune disorders, hepatitis, and/or HIV infection. The whole saliva sample collection was performed as previously described. Briefly, saliva samples were pooled and kept on ice during the sample collection. Whole saliva sample was centrifuged at 2600 × g for 30 min at 4 °C to remove cells, bacteria, debris and food remnants, then protease inhibitor cocktail (Roche complete tablet, Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) was added to saliva supernatant to prevent protein degradation. Finally, the saliva sample was diluted with PBS at the ratio of 1:1 and ready for SEVs preparation.
SEVs preparation
To prepare SEVs by using conventional centrifugation method, the diluted saliva supernatant was filtered with 5 μm PVDF membrane and then directly centrifuged to collect EVs. In our developed method, diluted saliva supernatant was loaded into ACCF system to remove amylase. Then the clear filtrate was centrifuged at 20,000 × g for 1 h at 4 °C to collect the EVs. The pellets were washed with PBS twice and then centrifuged again at 20,000 × g for another 1 h at 4 °C to harvest the final SEVs (Fig. 1).
SEVs protein extraction
Protein extraction was the same as previously described. Briefly, the obtained SEVs were resuspended in 100 μl PBS afterwards 2 μL Triton X-100 and 5 μL protease inhibitor cocktails were added. Then the sample was placed on ice for 30 min to disrupt the membranes and centrifuged at 20,000 × g 1 h at 4 °C to remove the sediment. The soluble fraction was collected and precipitated with 10 times pre-chilled ethanol at −20 °C for 10 h. SEVs protein was further isolated by centrifugation at 15,000 × g for 30 min. Protein concentration was measured using the BCA method (BCA assay kit, Peirce, Rockford, USA).
1D SDS-PAGE and in-solution digestion
SEVs proteins were loaded into a 10% Bis-Tris Mini Gel (Life Technologies, Shanghai, China) and were run at 100 V for 60 min in MES SDS running buffer. Pre-stained protein standard (Life Technologies, Shanghai, China) was used to track protein migration. The resulting gels were stained with Fast Sliver Stain Kit (Beyotime, Beijing, China). In-solution tryptic digestion was carried out overnight at 37 °C using trypsin (Promega, Madison, WI, USA) in 50 μL 50 mM NH4HCO3.
Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA, London, United Kingdom) offers the ability to directly visualize the size and count nanoparticles in liquid suspension. SEVs’ size was measured by the nanoparticle tracking system NanoSight (NTA: LM10, London, United Kingdom) by loading SEVs prepared from 1 mL saliva.
Nano LC-MS/MS and database search
For shotgun proteomics, 10 μg proteins were taken from each cancer sample and control sample and processed in parallel. For protein identification, 1 μg protein digests from each sample were analyzed using an LC system (Nano Pump, Ultimate 3000, Dionex, Thermofisher) coupled with an ESI-Q-TOF mass spectrometer (Maxis Impact UHR Q-TOF, Impact, Bruker Daltonik, Germany). Briefly, each peptide sample was re-dissolved in 2% acetonitrile with 0.1% formic acid, and then loaded onto a peptide trap column (100 μm × 2 cm, 5 μm, Dionex, Thermofisher). Then trapped peptides were eluted to a C18 capillary column (75 μm × 15 cm, 3 μm, Dionex, Thermofisher). The peptides were eluted for 90 min with a gradient of 2–80% v/v of acetonitrile containing 0.1% v/v formic acid at flow rate of 400 nL/min. MS was performed in a positive mode using a repetitively full MS scan, followed by collision-induced dissociation of the five most dominant ions selected from the initial MS scan. The analysis of each sample was technically repeated in triplicates. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride/) with the dataset identifier PXD003269 and 10.6019/PXD003269.
Combined MS and MS/MS spectra were submitted for database search using MASCOT software (version 2.0) to identify proteins from the Human Swissprot database (548208 sequences). The peak list was directly generated from raw data using centroid algorithm with peak width set as 0.1 m/z and intensity above 100. No peak smooth or filter process was applied. The parameters for searching were enzyme of trypsin, 2 missed cleavage, fixed modifications of carbamidomethyl (C), and variable modifications of oxidation (M). A mass tolerance of 20 ppm was used for MS precursors and 0.05 Da for fragment ions. Peptide charges of +2, +3 and +4 were selected. The criteria of two peptides and C.I.% > 95 were used for protein identification, which allowed a 99% confidence level of protein identification with less than 1% false discovery rate.
Additional Information
How to cite this article: Sun, Y. et al. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci. Rep. 6, 24669; doi: 10.1038/srep24669 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 21035004, No. 21275099, No. 21305087, No. 81302005 and No. 21475086), the National Key Development of Scientific Instruments (No. 2011YQ030139) and the Key Scientific Project of Shanghai Jiao Tong University (No. YG2010ZD209, No. YG2013MS10, No. YG2014QN21 and No. YG2015MS48). H.X. is supported by the Recruitment Program of Global Youth Experts of China and National High-tech R&D Program of China (863 Program, No. 2014AA020545).
Footnotes
Author Contributions H.X. and C.-X.C. contributed to the study design. Y.S. and Z.J.X. conducted the experiments and collected the data. Y.S., Z.J.X., Z.S., K.B.S., L.-Y.F., C.-X.C. and H.X. contributed to the data analysis. X.M.N. and L.Q.Q. contributed to sample collection. The manuscript was written by Y.S. All the authors reviewed the manuscript. All aspects of the study were supervised by H.X. and C.-X.C.
References
- Heijnen H. F. G., Schiel A. E., Fijnheer R., Geuze H. J. & Sixma J. J. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 94, 3791–3799 (1999). [PubMed] [Google Scholar]
- Henry T. et al. Improved methods for producing outer membrane vesicles in gram-negative bacteria. Res. Microbiol. 155, 437–446 (2004). [DOI] [PubMed] [Google Scholar]
- Lee E. Y., Choi D. S., Kim K. P. & Gho Y. S. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom. Rev. 27, 535–555 (2008). [DOI] [PubMed] [Google Scholar]
- Barteneva N. S., Maltsev N. & Vorobjev I. A. Microvesicles and intercellular communication in the context of parasitism. Front. Cell. Infect. Mi. 3, 49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz A., Baj-Krzyworzeka M. & Baran J. Characterization and biological role of extracellular vesicles. Postepy Hig. Med. Dosw. 68, 1421–1432 (2014). [DOI] [PubMed] [Google Scholar]
- Clayton A. et al. Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J. 18, 977–979 (2004). [DOI] [PubMed] [Google Scholar]
- Baj-Krzyworzeka M. et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol. Immunother. 55, 808–818 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A. et al. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–U1209 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K. et al. Intercellular transfer of the oncogenic receptor EGFrvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 10, 619–U624 (2008). [DOI] [PubMed] [Google Scholar]
- Inal J. M. et al. Blood/plasma secretome and microvesicles. Biochim. Biophys. Acta 1834, 2317–2325 (2013). [DOI] [PubMed] [Google Scholar]
- Hu S. et al. Salivary proteomics for oral cancer biomarker discovery. Clin.. Cancer Res. 14, 6246–6252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S. et al. Salivary proteomic and genomic biomarkers for primary Sjogren’s syndrome. Arthritis Rheum. 56, 3588–3600 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLos One 5, e15573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H. et al. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol. Cell. Proteomics 11, M111.012112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell. Mol. Life Sci. 69, 3341–3350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 138, 949–U194 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H. et al. Differential proteomic analysis of human saliva using tandem mass tags quantification for gastric cancer detection. Sci. Rep. 6, 22165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winck F. V. et al. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci. Rep. 5, 16305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H. & Wong D. T. W. Proteomic analysis of microvesicles in human saliva by gel electrophoresis with liquid chromatography-mass spectrometry. Anal. Chim. Acta 723, 61–67 (2012). [DOI] [PubMed] [Google Scholar]
- Lau C. S. & Wong D. T. W. Breast cancer exosome-like microvesicles and salivary gland cells interplay alters salivary gland cell-derived exosome-like microvesicles in vitro. PLos One 7, e33037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem. 288, 26888–26897 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wei F., Schafer C. & Wong D. T. W. Detection of tumor cell-specific mRNA and protein in exosome-like microvesicles from blood and saliva. PLos One 9, e110641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado F. M. L., Vitorino R. M. P., Domingues P., Lobo M. J. C. & Duarte J. A. R. Analysis of the human saliva proteome. Expert Rev. Proteomics 2, 521–539 (2005). [DOI] [PubMed] [Google Scholar]
- Punyani S. R. & Sathawane R. S. Salivary level of interleukin-8 in oral precancer and oral squamous cell carcinoma. Clin. Oral Investig. 17, 517–524 (2013). [DOI] [PubMed] [Google Scholar]
- Deutsch O. et al. An approach to remove alpha amylase for proteomic analysis of low abundance biomarkers in human saliva. Electrophoresis 29, 4150–4157 (2008). [DOI] [PubMed] [Google Scholar]
- Herbst R. S., Heymach J. V. & Lippman S. M. Molecular origins of cancer: lung cancer. N. Engl. J. Med. 359, 1367–1380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell R. & Karachaliou N. Lung cancer in 2014 optimizing lung cancer treatment approaches. Nat. Rev. Clin. Oncol. 12, 75–76 (2015). [DOI] [PubMed] [Google Scholar]
- Hassanein M. et al. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev. Res. 5, 992–1006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M. et al. Clinical and experimental studies regarding the expression and diagnostic value of carcinoembryonic antigen-related cell adhesion molecule 1 in non-small-cell lung cancer. BMC Cancer 13, 359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C. E. et al. Blood-based lung cancer biomarkers identified through proteomic discovery in cancer tissues, cell lines and conditioned medium. Clin. Proteomics 12, 18–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz K. Historical highlights and current status of biotechnology” - from mystery to the lock and key hypothesis to the design of recombinant enzymes. Med. Chem. Res. 11, 399–421 (2002). [Google Scholar]
- Chatterton R. T. Jr., Vogelsong K. M., Lu Y.-C., Ellman A. B. & Hudgens G. A. Salivary alpha-amylase as a measure of endogenous adrenergic activity. Clin. Physiol. 16, 433–448 (1996). [DOI] [PubMed] [Google Scholar]
- Inal J. M., Fairbrother U. & Heugh S. Microvesiculation and disease. Biochem. Soc. Trans. 41, 237–240 (2013). [DOI] [PubMed] [Google Scholar]
- Madureira P. A. et al. Annexin A2 is a novel cellular redox regulatory protein involved in tumorigenesis. Oncotarget 2, 1075–1093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni G. et al. Annexin A1-containing extracellular vesicles and polymeric nanoparticles promote epithelial wound repair. J. Clin. Invest. 125, 1215–1227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senchenko V. N. et al. Simultaneous down-regulation of tumor suppressor genes RBSP3/CTDSPL, NPRL2/G21 and RASSF1A in primary non-small cell lung cancer. BMC Cancer 10, 75 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dango S. et al. Elevated expression of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM-1) is associated with increased angiogenic potential in non-small-cell lung cancer. Lung Cancer 60, 426–433 (2008). [DOI] [PubMed] [Google Scholar]
- Woo J. K. et al. Mucin 1 enhances the tumor angiogenic response by activation of the AKT signaling pathway. Oncogene 31, 2187–2198 (2012). [DOI] [PubMed] [Google Scholar]
- Zhu L. et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457, 603–U114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H. et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 13, 1672–1686 (2013). [DOI] [PubMed] [Google Scholar]
- Tao Y. F. et al. Molecular mechanism of the cell death induced by the histone deacetylase pan inhibitor LBH589 (panobinostat) in wilms tumor cells. PLos One 10, e0126566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I. et al. Regulatory roles of tumor necrosis factor alpha-induced proteins (TNFAIPs) 3 and 9 in arthritis. Clin. Immunol. 153, 73–78 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.