Abstract
DNA methylation in gene promoters leads to gene silencing and is the therapeutic target of methylation inhibitors such as 5-Aza-2′-deoxycytidine (5-Aza-CdR). By analyzing the time series RNA-seq data (days 5, 9, 13, 17) obtained from human bladder cells exposed to 5-Aza-CdR with 0.1 uM concentration, we showed that 5-Aza-CdR can affect isoform switching and differential exon usage (i.e., exon-skipping), in addition to its effects on gene expression. We identified more than 2,000 genes with significant expression changes after 5-Aza-CdR treatment. Interestingly, 29 exon-skipping events induced by treatment were identified and validated experimentally. Particularly, exon-skipping event in Enhancer of Zeste Homologue 2 (EZH2) along with expression changes showed significant down regulation on Day 5 and Day 9 but returned to normal level on Day 13 and Day 17. EZH2 is a component of the multi-subunit polycomb repressive complex PRC2, and the down-regulation of exon-skipping event may lead to the regain of functional EZH2 which was consistent with our previous finding that demethylation may cause regain of PRC2 in demethylated regions. In summary, our study identified pervasive transcriptome changes of bladder cancer cells after treatment with 5-Aza-CdR, and provided valuable insights into the therapeutic effects of 5-Aza-CdR in current clinical trials.
DNA methylation and histone modification play crucial roles in regulation of gene expression in mammalian developments as well as human diseases, such as cancer1,2. During tumorigenesis, the promoter regions of tumor suppressor genes could undergo abnormal hypermethylation, which lead to the silencing of these genes3,4,5. Moreover, transient exposure to low doses of DNA-demethylation agents can trigger durable antitumor effects in tumors6,7. Recently, clinical trials have been focused on investigating the possible utility of methylation inhibitors in solid tumors, either alone or in combination with other demethylation drugs8,9. Thus, reactivation of tumor suppressor genes by demethylation agents has become a possible and promising approach for cancer therapy.
Alternative splicing is closely associated with differentiation and development, and is a major source for protein diversity10. It enables cells to generate proteins of different coding sequences and functions from a single gene. Genome-wide approaches have revealed that tumorigenesis often involved large-scale alterations in alternative splicing11. Researchers also found that demethylation drugs could target transcribed regions, which suggest that the effects of demethylation drugs are not limited to the reactivation of promoters of silenced genes, but are prone to change exon recognition6,12,13. The demonstration that intragenic DNA methylation could affect elongation efficiency indicated that DNA methylation may facilitate exon inclusion14. A recent study further proved that intragenic DNA methylation modulated exon recognition, thus it is necessary to investigate the relationship between demethylation treatment and alternative splicing, which was generally overlooked in previous studies15.
DNA methyltransferases (DNMT) inhibitors, such as 5-azacytidine (5-Aza-CR) and 5-Aza-2′-deoxycytidine (5-Aza-CdR), were approved by the FDA for the treatment of myelodysplastic syndrome16,17. Therefore, a comprehensive understanding of how these demethylation drugs affect gene reactivation and alternative splicing is necessary for understanding their therapeutic effects and exploring new cancer therapies. In this study, we treated human bladder cell line UM-UC-3 with 5-AZA-CdR for 24 hours, then monitored expression changes at 5, 9, 13 and 17 days after treatment and employed deep RNA sequencing to analyze alterations in gene expression and alternative splicing. Additionally, we measured whole-genome methylation levels by the Illumina 450K methylation array at 5 and 17 days, to correlate with gene expression changes.
Results
Isoform expression changes induced by 5-Aza-CdR treatment
To explore the potential regulatory effects of 5-Aza-CdR, UM-UC-3 cells were treated with 0.1 uM 5-Aza-CdR for 24 hours, then collected at 5, 9, 13 and 17 days after treatment. Cells at the four time points together with untreated UM-UC-3 cells were then sequenced using paired-end Illumina RNA-Seq protocol, and two replicate experiments were performed for each sample. Approximately 20 Gb RNA-seq raw data for each replicate was generated after barcode removal and filtering of low-quality reads.
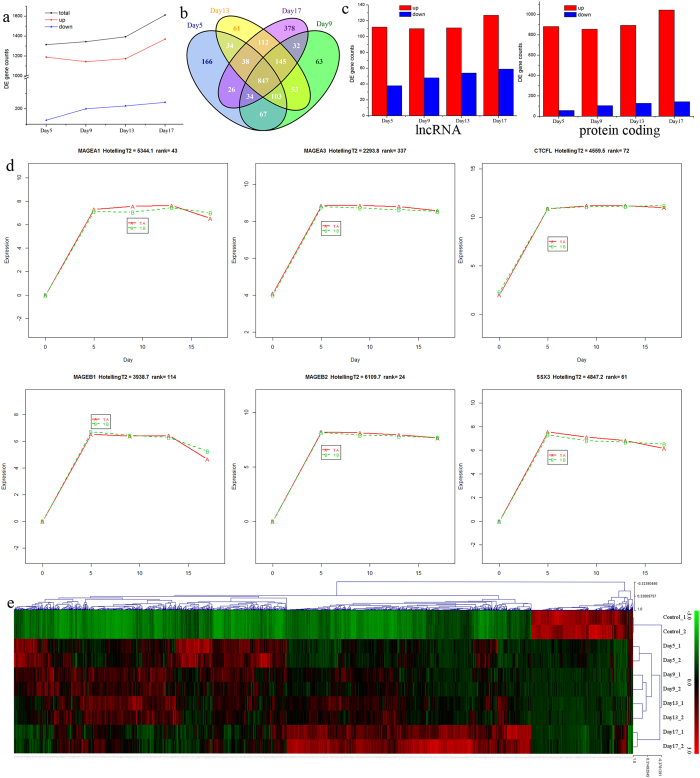
RNA-seq data generated from untreated UM-UC-3 cells (control) and cells collected from four time points (Day 5, Day 9, Day 13 and Day 17) were aligned to human genome using Tophat218 with GENCODE annotation (GRCh37.p13, GENCODE release 19). For all samples, we obtained more than 92% mapping ratio, which indicated high quality and reliability of the sequencing data. Differentially expressed (DE) genes were identified by comparing RNA-seq data obtained from each treatment with untreated cells. In total, 1315, 1344, 1393 and 1612 DE genes were found on Day 5, Day 9, Day 13 and Day 17, respectively (Fig. 1a). Among those DE genes, 847 of them were shared in all four time points (Fig. 1b). About 85% (Day 5: 90%, Day 9: 85%, Day 13: 84%, Day 17: 85%) DE genes were up regulated after 5-Aza-CdR treatment. Furthermore, the numbers of up and down regulated DE genes were positively correlated with the treatment time of 5-Aza-CdR and the most abundant DE genes were always found after 17 days treatment (Fig. 1a).
Figure 1. Dynamic transcriptome changes induced by 5-Aza-CdR treatment.
(a) The number of differentially expressed genes across different time points. (b) Venn diagram showing overlapped differentially expressed genes found in each time point. (c) The number of up and down regulated genes found in the two RNA types: protein coding RNAs and lncRNAs. (d) Tumor suppressor gene expression pattern. (Y axis indicates normalized expression, X axis indicates different 5-Aza-CdR exposure time, A and B indicated two replicates) (e) Hierarchical clustering of differentially expressed genes.
Based on RNA type annotation in GENCODE, about 72% and 12% of the DE genes are annotated as protein coding RNAs and lncRNAs, respectively (Fig. 1c, Supplementary Table S1). The numbers of protein coding RNAs and lncRNAs shared similar distributions as previously described DE genes across different time points (Fig. 1a,c). Here, we found that the expression of HOTAIR as well as several other tumor-suppressor lncRNAs such as H19 and MEG3 were reactivated after 5-Aza-CdR treatment for 5 days and maintained sustainable growth till Day 1719,20.
Previous studies on bladder cancer cells exposed to the 5-Aza-CdR for 8 days revealed that around 120 genes showed considerable changes21,22. Similarly, around 40 of previously reported DE genes were also identified in this study, while 30 of them were tumor suppressor genes (Fig. 1d, Supplementary Fig. S1), such as MAGEA1, MAGEA3, MAGEA12, MAGEB1, MAGEB2, SSX1, SSX3 and CTCFL. We later examined the expression patterns of all DE genes and found that the expression for most genes increased with longer treatment time (Fig. 1e). Therefore, the effects of demethylation treatment on bladder cancer cells can be maintained for a long period of time.
Detection of differentially expressed exons
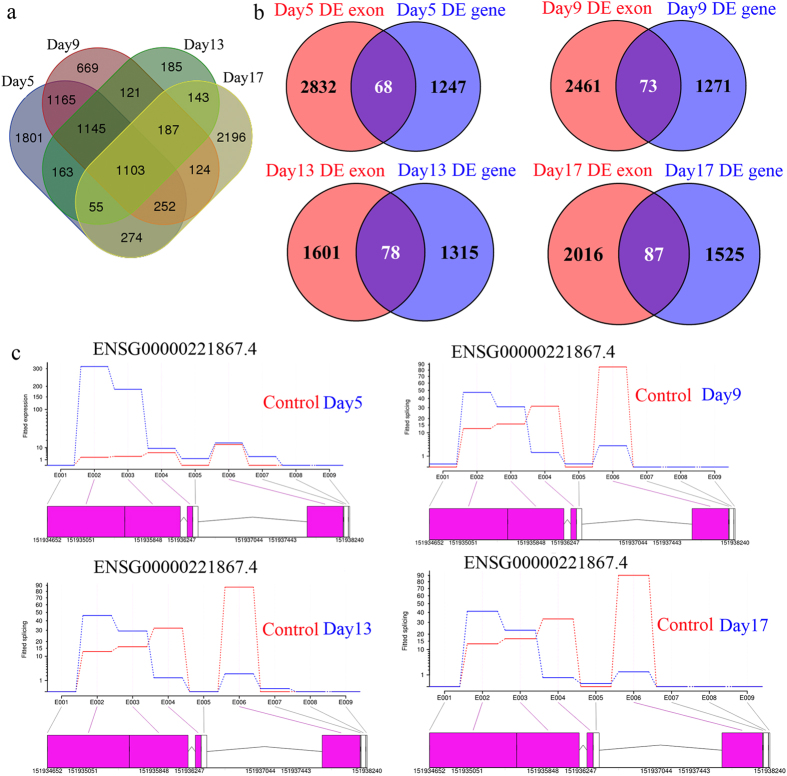
Identification of DE exons can inform us on how the demethylation treatment affects exons recognition, and it can shed lights on the potential regulatory role of DNA methylation on alternative splicing. In total, 5958 (up: 3362, down: 2596), 4766 (up: 2364, down: 2402), 3102 (up: 1940, down: 1162) and 4334 (up: 2557, down: 1777) DE exons were identified on Day 5, Day 9, Day 13 and Day 17, respectively (Padj < 0.05). Among them, 1103 exons were observed with significant changes across all time points (Fig. 2a). Unlike DE genes, the most abundant DE exons were identified on Day 5, followed with steady decrease on Day 9 and Day 13 and increased again on Day 17 (Fig. 2a). We also tried to measure the overlap between DE exons and DE genes. We found that 68, 73, 78 and 87 DE genes from Day 5, Day 9, Day 13 and Day 17 contained at least one DE exon (Fig. 2b). For instance, significant changes were found on exon 2, 3, 4 and 6 of tumor antigen MAGEA3 across all time points, while MAGEA3 itself was also known as a DE gene throughout 5-Aza-CdR treatment (Figs 2c and 1e). Interestingly, we also note that most DE genes do not contain any DE exon regardless of the exposure time (Fig. 2b). Thus, most of the genes may not be subject to isoform switching, despite that the overall expressions changed considerably. Functional analysis revealed that genes with DE exons were mainly associated with biological processes such as translational elongation, biopolymer methylation and DNA methylation. In summary, 5-Aza-CdR can not only induce changes in gene expression, but also exon-level changes for a small subset of DE genes.
Figure 2. Summary of differentially expressed exons after 5-Aza-CdR treatment.
(a) Venn diagram showing overlapped differentially expressed exons found in each time point. (b) Venn diagram showing overlapped genes identified as both differentially expressed genes and genes containing differentially expressed exons. (c) Visualization of differentially expressed exons found in MAGEA3 (ENSG00000221867.4) after 5, 9, 13, and 17 days treatment (differentially expressed exons were highlighted in red).
Identification of exon-skipping events
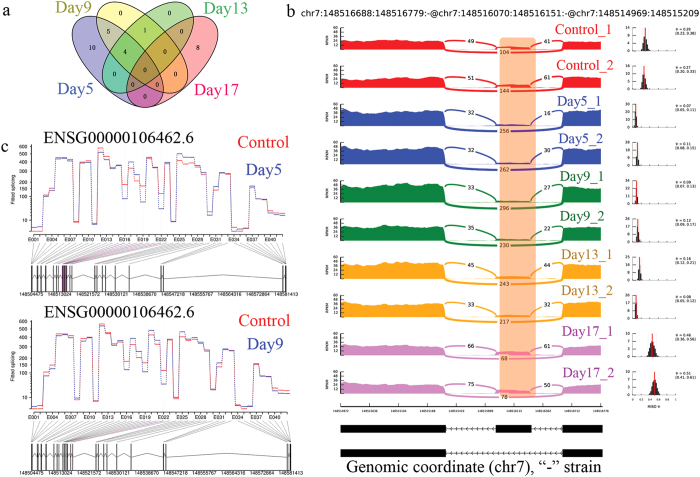
We next tried to investigate whether demethylation treatment would induce alternative splicing in bladder cancer cells, since it has been reported in other cell lines14. Alternative splicing events can be classified into different types: skipped exon, mutually exclusive exon, alternative 5′ splice site, alternative 3′ splice site23. Here, we focused on exon-skipping events, since they are likely to be affected by methylation. DE exon-skipping were identified by comparing RNA-seq data from each time point with control using MISO24. Initially, we found 43, 26, 14 and 25 exon-skipping events, which showed considerable changes on Day 5, Day 9, Day 13 and Day 17 (Fig. 3a). To further validate our findings, we compared our DE exon-skipping results with previous DE exons. Intuitively, the DE exon-skipping events should involve some DE exons. We identified 28 (up:8 down:11), 10 (up:4 down:6), 5 (up:3 down:2) and 8 (up:5 down:3) overlapped DE exon-skipping events while 19 of them altered its coding sequence (Table 1). All five DE exon-skipping events found on Day 13 were included on Day 9 while four of them were also found on Day 5 except RRBP1 (Table 1). In addition, nine exon-skipping events on Day 9 were also found on Day 5, again except RRBP1. However, eight exon-skipping events found on Day 17 together with ten events on Day 5 could not be observed at any other time points. Consequently, it seemed that exon-skipping events on Day 5 continued to show similar exon-skipping behavior on Day 9 and Day 13. After that, 17 days treatment resulted in eight other new DE exon-skipping events. One particular down regulated DE exon-skipping event was found in EZH2 gene on Day 5 and Day 9 which was located in one of the EZH2 transcripts annotated as being subject to nonsense-mediated decay by Ensemble (ENST00000483012.1) (Fig. 3b,c). EZH2 belongs to PRC2/EED-EZH2 complex which catalyzes ‘Lys-27′ methylation of histone H3 and leads to transcriptional repression of the target genes. Therefore, this exon-skipping event is expected to cause the regain of functional EZH2 gene.
Figure 3. Exon recognition changes induced by 5-Aza-CdR treatment.
(a) Venn diagram showing overlapped differentially expressed skipped exon events found in each time point. (b) Sashimi plot showing the expression changes of EZH2 exon skipping (skipped exon was highlighted with orange rectangle) events at each time point. (ΔΨ > 0.15) (c) Visualization of differentially expressed exons found in EZH2 after 5, 9, 13, and 17 days treatment (differentially expressed exons were highlighted in red).
Table 1. List of exon-skipping events identified by MISO and DEXSeq.
| Gene name | Changes induced by 5-Aza-CdR |
Genomic coordinate (skipped exon) | Alter protein sequence | Gene body demethylation | |||
|---|---|---|---|---|---|---|---|
| Day5 | Day9 | Day13 | Day17 | ||||
| YDJC | down | N/A | N/A | N/A | chr22:21983299-21983476 | no | yes |
| DLGAP5 | down | down | N/A | N/A | chr14:55615312-55615402 | yes | yes |
| LAS1L | down | down | N/A | N/A | chrX:64744444-64744494 | yes | no |
| EZH2 | down | down | N/A | N/A | chr7:148516070-148516151 | yes | yes |
| TARBP2 | down | N/A | N/A | N/A | chr12:53898919-53899046 | yes | yes |
| UAP1 | down | down | N/A | N/A | chr1:162562525-162562572 | yes | no |
| NUMA1 | down | N/A | N/A | N/A | chr11:71723447-71723488 | yes | yes |
| CSNK1G3 | down | N/A | N/A | N/A | chr5:122941033-122941056 | yes | yes |
| KRT8 | down | down | down | N/A | chr12:53343240-53343362 | no | yes |
| DPH7 | down | N/A | N/A | N/A | chr9:140470761-140470854 | no | no |
| MCM3 | down | down | down | N/A | chr6:52130033-52130183 | yes | yes |
| INSIG1 | up | N/A | N/A | N/A | chr7:155095535-155095605 | yes | yes |
| ACYP1 | up | N/A | N/A | N/A | chr14:75528387-75528465 | yes | yes |
| ORMDL1 | up | up | N/A | N/A | chr2:190647740-190647849 | no | no |
| FAM13B | up | N/A | N/A | N/A | chr5:137354644-137354835 | yes | no |
| PHKA1 | up | up | up | N/A | chrX:71840575-71840751 | yes | no |
| IKBIP | up | up | up | N/A | chr12:99028074-99028191 | yes | yes |
| ASPM | up | N/A | N/A | N/A | chr1:197069561-197074315 | yes | yes |
| NUMB | up | N/A | N/A | N/A | chr14:73745989-73746132 | yes | yes |
| RRBP1 | N/A | up | up | N/A | chr20:17660644-17660720 | no | yes |
| DIS3 | N/A | N/A | N/A | up | chr13:73355427-73355494 | no | no |
| HRAS | N/A | N/A | N/A | up | chr11:533277-533358 | yes | yes |
| SRP9 | N/A | N/A | N/A | up | chr1:225974564-225974687 | yes | yes |
| PPHLN1 | N/A | N/A | N/A | up | chr12:42768440-42768535 | no | yes |
| TIAL1 | N/A | N/A | N/A | up | chr10:121336359-121336417 | no | yes |
| SMIM7 | N/A | N/A | N/A | down | chr19:16764846-16764936 | no | no |
| SCARB1 | N/A | N/A | N/A | down | chr12:125267229-125267357 | yes | yes |
| CTC-429P9.4 | N/A | N/A | N/A | down | chr19:16764846-16764936 | no | no |
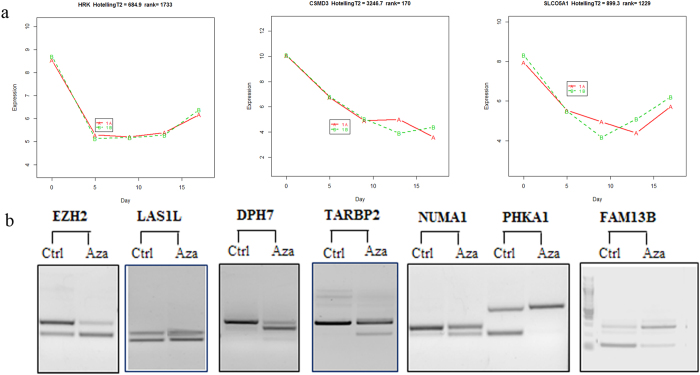
Finally, seven DE exon-skipping events were further validated by experimental verification. PCR results showed that bands with larger fragment sizes (fragment contained skipped exon) of EZH2, LAS1L, DPH7, TARBP2 and NUMA1 became lighter after 5-Aza-CdR treatment while bands with smaller fragment sizes (fragment without skipped exon) became darker, which indicated the down regulation of skipped exons. Among them, only DPH7 was not identified with protein sequence change. Opposite observations can be found in another two up-regulated skipped exons of PHKA1 and FAM13B, where expressions of fragment containing skipped exons were found to be increased after treatment (Fig. 4b, Table 1). Both of these two events resulted in protein sequence changes. In summary, 5-Aza-CdR treatments were also capable of inducing dynamic changes on exon-skipping events and most of the changes were consistent from Day 7 till Day 13. Some of these exon-skipping events may result in gene functional changes, such as truncated protein in EZH2.
Figure 4. PcG (EZH2) mediated gene expression alteration and validation of exon-skipping events.
(a) Differentially expressed PcG (EZH2) targeted genes induced by exon-skipping changes in EZH2. Aberrant exon recognition changes in EZH2 found in Day 5 and Day 9 resulted in corresponding inhibition of targeted genes. (b) Experimental validations of seven differentially expressed exon-skipping events after 5-Aza-CdR treatment.
Comparison of DNA methylation and gene expression after 5-Aza-CdR treatment
To evaluate whether DNA methylation is responsible for our findings above, we assayed the UM-UC-3 cells treated with 5-Aza-CdR for 5 and 17 days, by the Illumina 450K methylation array. In total, 18,906 and 18,740 genes were demethylated after 5-Aza-CdR treatment on Day 5 and Day 7, respectively. Meanwhile, previous results indicated that 1,315 and 1,612 DE genes were identified at the same time points. Among them, over 60% DE genes (mostly protein-coding genes) showed demethylation after drug treatment, while the rest were mostly non-coding RNAs such as lncRNAs and pseudogenes (Supplementary Fig. S2). Furthermore, among the 28 genes with exon-skipping events, 22 were demethylated after drug treatment and 19 of them showed demethylation within their gene bodies (Table 1). The observation is consistent with the hypothesis that methylation in the gene body may have an impact on splicing25. Altogether, these results provided concrete evidence that 5-Aza-CdR treatment can cause isoform switching and exon skipping events.
Discussion
Low dosage (0.1 uM) of 5-Aza-CdR treatment was widely used in current clinical trials because of its high demethylation efficacy and low toxicity. In addition, it has been shown that demethylation is not the main driving force in phenotypic changes by high dosage of 5-Aza-CdR26,27, which is why our study on 5-Aza-CdR only focused on low dose treatment. The efficiency of 5-Aza-CdR in regulating DNA methylation changes indicated that drug-induced demethylation may be an effective therapeutic intervention in cancer, since some tumor suppressor genes were reactivated as reported above (Fig. 1e). Furthermore, the number of DE genes continued to increase on Day 13 and Day 17 and there were 847 DE genes which are shared through all time points (Fig. 1a,b). Among all DE genes, we observed expression changes of a small set of lncRNAs. LncRNAs are known to be involved in cancer progression, and received more and more attention recently because of their potential as biomarkers and novel therapeutic targets for cancer28. For instance, an oncogenic lncRNA HOTAIR interacts with PRC2 complex to repress the HOXD locus and a new study indicates that HOTAIR is over-expressed in breast tumors28,29. All the results suggested that the gene reactivation induced by 5-Aza-CdR appeared to be progressive and long-lasting, maintaining for at least 17 days, which also supported by recent studies6,7,30.
Previous studies showed that exons are more highly methylated than introns and the degree of methylation differs at exon–intron boundaries31. Most recent research further showed that DNA methylation had an effect in regulating exon skipping15.Consequently, DNA methylation can possibly mediate RNA splicing32,33. In this research, we also found a small set of exon-skipping events induced by 5-Aza-CdR (Table 1). Combined with our previous results, it seemed that the longest exposure time to 5-Aza-CdR always produced more DE genes, DE exons and exon-skipping events, which may lead to alteration of gene function. However, due to tissue-specific DNA methylation pattern, we expect that DE genes (and isoform switching events) may be varied between different cell types25.
Unlike other inhibitors targeted to inhibit the overall expression of a polycomb group (PcG) protein EZH234,35,36,37, we found one skipped-exon event (chr7:148516070-148516151) from EZH2, which showed significant down-regulation after 5 and 9 days post 5-Aza-CdR treatment (Fig. 3b,c). This down-regulated exon was known as a poison exon which could lead to the nonsense-mediated decay of EZH2 by introducing premature termination codon38. Thus, the exclusion of this poison exon could cause the regain of functional EZH2. EZH2 concerts with other proteins (EED, SUZ12 and RBBP4) to form the PRC2, which can initiate polycomb-mediated gene repression39. PcG marks genes that are prone to cancer-specific DNA hypermethylation and the remaining of PcGs may lead to the exclusion of DNA methylation for certain gene40,41,42. On the other hand, demethylation may cause regain of PRC2 in demethylated regions43. Interestingly, we found that a small set of PRC2 targeted genes (HRK, CSMD3 and SLCOSA1) were inhibited on Day 5 and Day 9 (Fig. 4a)44. These particular genes were validated to be unable to gain chromatin accessibility despite the promoter demethylation in our previous findings43. Perhaps the down-regulation of EZH2 exon-skipping event on Day 5 and Day 9 could lead to the regain of PRC2 while the recovery of exon-skipping in Day 13 and Day 17 could cause the loss of PRC245. In general, permanent gene silencing require DNA methylation coupled with PRC2, such as MYT1 and CNR146. However, our results demonstrated that DNA methylation may play a key role to silence CNR1 in UM-UC-3 cell line, since after demethylation treatment the expression of CNR1 was activated regardless of the exon change in EZH2. In addition, another 9 genes marked with PcG and reported to be hypermethylated in cancer showed significant changes throughout the demethylation treatment (Supplementary Fig. S3)40. Thus, reactivation of expression for those genes may be caused by demethylation treatment by 5-Aza-CdR. Other exon-skipping events were found to be associated with tumor necrosis factor-mediated signaling pathway (KRT8) and neuroblast proliferation (ASPM and NUMB)47,48. Based on our results and previous studies, the methylation that occurred in transcribed regions may contribute to nucleosome destabilization and reduced efficiency of splicing, while inhibition of DNA methylation led to aberrant splicing6,15, since all these genes with DE exon-skipping events did not show significant changes on transcript level (Fig. 4b).
Take together, our study demonstrated that DNMT inhibitor 5-Aza-CdR can alter expression patterns of many genes on both the isoform and exon level. More importantly, we showed that DNA methylation was associated with alternative splicing and 5-Aza-CdR was able to change the exon-skipping in EZH2. This study provides valuable information on how demethylation drugs affect bladder cancer cells, thus shedding light on ongoing and future clinical trials that evaluate demethylation drugs.
Materials and Methods
Tissue Culture and 5-Aza-CdR Treatment
UM-UC-3 cells were procured from American Type Culture Collection (ATCC), and were used for all in vitro studies. No human subjects were used in the study. The methods were carried out in accordance with approved guidelines, and the experimental protocols were approved by USC.
UM-UC-3 cells were maintained in MEM medium, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. UM-UC-3 cells were treated with 0.1 uM of 5-Aza-CdR (Sigma-Aldrich). The medium was changed 24 hr later. RNA was harvested 5, 9, 13 and 17 days after drug treatment and was extracted using Direct-zol™ RNA MiniPrep (Zymo).
Library preparation and Illumina sequencing
The RNA-seq library was generated using Illumina TruSeq RNA Sample Preparation kit and was sequenced using 100 bp paired-end model at the University of Southern California Epigenome Center according to the manufacturer’s specifications. Generally, 20 Gb raw RNA-seq data for each time point was obtained and two replicates for each time point were sequenced.
Bioinformatics analyses
Raw RNA-seq data were first assessed by Fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Adapters were then removed by cutadapt package (https://code.google.com/p/cutadapt/). After that, filtered reads were aligned to the human genome by TopHat2 using Gencode annotation (GRCh37.p13, GENCODE release 19)18. Normalized read count for all genes were obtained using HTseq and subsequent DE genes were identified by DESeq49,50 (padj < 0.01,fold change > 10). Individual gene expression was analyzed using “timecourse” package from R (http://www.bioconductor.org/packages/release/bioc/html/timecourse.html). Hierarchical clustering was performed by MEV package (http://www.tm4.org/mev.html). All DE genes were then imported into the DAVID website for functional and pathway enrichment analysis51. DE exons were identified by R package DEXSeq which focused on finding differential exon usage using RNA-seq exon counts between samples with different experimental designs (p-value < 0.05)52. DE exons can be visualized using built in DEXSeq plot function. MISO served as the main tool for exploring exon-skipping events53. Pre-build GFF3 annotation files were downloaded from MISO website (http://miso.readthedocs.org/en/fastmiso/annotation.html) and customized Perl scripts were used to screen and located the genes containing exon-skipping events. Differentially expressed exon-skipping events were detected with “compare_miso–compare-samples” option and further filtered with “–num-sum-inc-exc 10–delta-psi 0.15–bayes-factor 10” parameters according to MISO documentation (http://miso.readthedocs.org/en/). All exon-skipping events were visualized by sashimi_plot (http://miso.readthedocs.org/en/fastmiso/sashimi.html).
Illumina Infinium HM450 DNA methylation assay
The Infinium DNA methylation assay was performed at the University of Southern California Epigenome Center according to the manufacturer’s specifications. The HM450 BeadChip examines the DNA methylation status of 482,421 CpG sites, covering 99% of RefSeq genes and intergenic regions. The DNA methylation level is reported as a beta value, ranging from 0 (not methylated) to 1 (fully methylated). The delta beta value 0.2 was selected as cut-off to identify genes with demethylation after drug treatment.
Experimental validation of identified skipped-exon events
RT-PCR primers were designed to amplify upstream and downstream exons if the potential skipped exons were less than 200 bp. If the potential skipped exon is larger than 200 bp, which would introduce bias selection for PCR amplification, a forward primer was designed at the potentially skipped exon with similar amplification efficiency. Primer sequences are all available upon request.
Additional Information
Accession codes: All RNA-Seq sequences were deposited in the NCBI SRA database with accession number: SRP063667.
How to cite this article: Ding, X.-L. et al. Isoform switching and exon skipping induced by the DNA methylation inhibitor 5-aza-2'-deoxycytidine. Sci. Rep. 6, 24545; doi: 10.1038/srep24545 (2016).
Supplementary Material
Acknowledgments
This work was supported by NIH grant (5R01CA138794, GL) NIH grant (HG006465, KW), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Doctorate Fellowship Foundation of Nanjing Forestry University (2011YB019).
Footnotes
Author Contributions X.-L.D. and X.Y. carried out experiments and analyses, and wrote the manuscript. K.W. and G.L. designed the study and revised the manuscript.
References
- You J. S. & Jones P. A. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell 22, 9–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S. B. & Jones P. A. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 11, 726–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. & Baylin S. B. The fundamental role of epigenetic events in cancer. Nat Rev Genet 3, 415–428 (2002). [DOI] [PubMed] [Google Scholar]
- Jones P. A. & Baylin S. B. The epigenomics of cancer. Cell 128, 683–692 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Molecular origins of cancer: Epigenetics in cancer. New Engl J Med 358, 1148–1159 (2008). [DOI] [PubMed] [Google Scholar]
- Yang X. J. et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26, 577–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. C. et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21, 430–446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens R. A. et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 1, 598–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangle J. et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget 4, 2067–2079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco R. F., Allo M., Schor I. E., Kornblihtt A. R. & Misteli T. Epigenetics in alternative pre-mrna splicing. Cell 144, 16–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. J. & Manley J. L. Alternative pre-mrna splicing regulation in cancer: Pathways and programs unhinged. Genes Dev 24, 2343–2364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton A. M. et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res 21, 1074–1086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea A. K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz M. C., Dickerson D. R., Schmitt M. & Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol 11, 1068–1075 (2004). [DOI] [PubMed] [Google Scholar]
- Maunakea A. K., Chepelev I., Cui K. & Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res 23, 1256–1269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. J., Lay F., Han H. & Jones P. A. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci 31, 536–546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaux P. et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase iii study. Lancet Oncol 10, 223–232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gejman R. et al. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocr Metab 93, 4119–4125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A., Ripoche M. A., Yoshimizu T. & Dandolo L. The H19 gene: Regulation and function of a non-coding rna. Cytogenet Genome Res 113, 188–193 (2006). [DOI] [PubMed] [Google Scholar]
- Liang G., Gonzales F. A., Jones P. A., Orntoft T. F. & Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidine. Cancer Res 62, 961–966 (2002). [PubMed] [Google Scholar]
- Qiu X. N. et al. Equitoxic doses of 5-azacytidine and 5-aza-2′-deoxycytidine induce diverse immediate and overlapping heritable changes in the transcriptome. Plos One 5, e12994, 10.1371/journal.pone.0012994 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A. J., Clark F. & Smith C. W. J. Understanding alternative splicing: Towards a cellular code. Nat Rev Mol Cell Bio 6, 386–398 (2005). [DOI] [PubMed] [Google Scholar]
- Katz Y., Wang E. T., Airoldi E. M. & Burge C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7, 1009–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews. Genetics 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A. & Jones P. A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 (2004). [DOI] [PubMed] [Google Scholar]
- Kelly T. K., De Carvalho D. D. & Jones P. A. Epigenetic modifications as therapeutic targets. Nat Biotechnol 28, 1069–1078 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M. & Rinn J. L. Large non-coding RNAs: Missing links in cancer? Hum Mol Genet 19, R152–R161 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nana-Sinkam S. P. & Croce C. M. Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Mol Oncol 5, 483–491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- Laurent L. et al. Dynamic changes in the human methylome during differentiation. Genome Res 20, 320–331 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda T. B. et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther 8, 1579–1588 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W. et al. Selective inhibition of EZH2 by a small molecule inhibitor blocks tumor cells proliferation. P Natl Acad Sci USA 109, 21360–21365 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson S. K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 8, 890–896 (2012). [DOI] [PubMed] [Google Scholar]
- McCabe M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012). [DOI] [PubMed] [Google Scholar]
- Kim E. et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27, 617–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund K., Adams P. D. & Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia 28, 44–49 (2014). [DOI] [PubMed] [Google Scholar]
- Schlesinger Y. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39, 232–236 (2007). [DOI] [PubMed] [Google Scholar]
- Ohm J. E. et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39, 237–242 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam E. N. et al. Frequent switching of polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. P Natl Acad Sci USA 105, 12979–12984 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiyan K. et al. Functional DNA demethylation is accompanied by chromatin accessibility. Nucleic Acids Res 41, 3973–3985 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I. et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40, 499–507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A. P., Dietrich N., Pasini D., Hansen K. H. & Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Gene Dev 20, 1123–1136 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E. et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature 446, 824–824 (2007). [DOI] [PubMed] [Google Scholar]
- Mahmoud A. A. & Siddiqui I. A. Abnormal spindle-like microcephaly gene detection in an autosomal recessive microcephalic saudi patient with attention deficit hyperactivity disorder and mental retardation. Neurosciences 18, 278–280 (2013). [PubMed] [Google Scholar]
- Sedo C. A., Schatten H., Combelles C. M. & Rawe V. Y. The nuclear mitotic apparatus (NUMA) protein: Localization and dynamics in human oocytes, fertilization and early embryos. Mol Hum Reprod 17, 392–398 (2011). [DOI] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. & Huber W. Htseq–a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G. Jr. et al. David: Database for annotation, visualization, and integrated discovery. Genome Biol 4, P3 (2003). [PubMed] [Google Scholar]
- Anders S., Reyes A. & Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res 22, 2008–2017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E. T., Airoldi E. M. & Burge C. B. Analysis and design of rna sequencing experiments for identifying isoform regulation. Nat Methods 7, 1009–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.