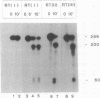
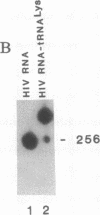
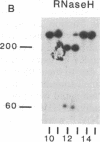
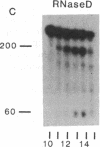
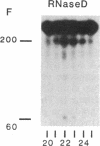
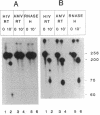
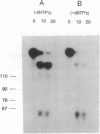
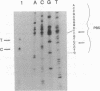
Abstract
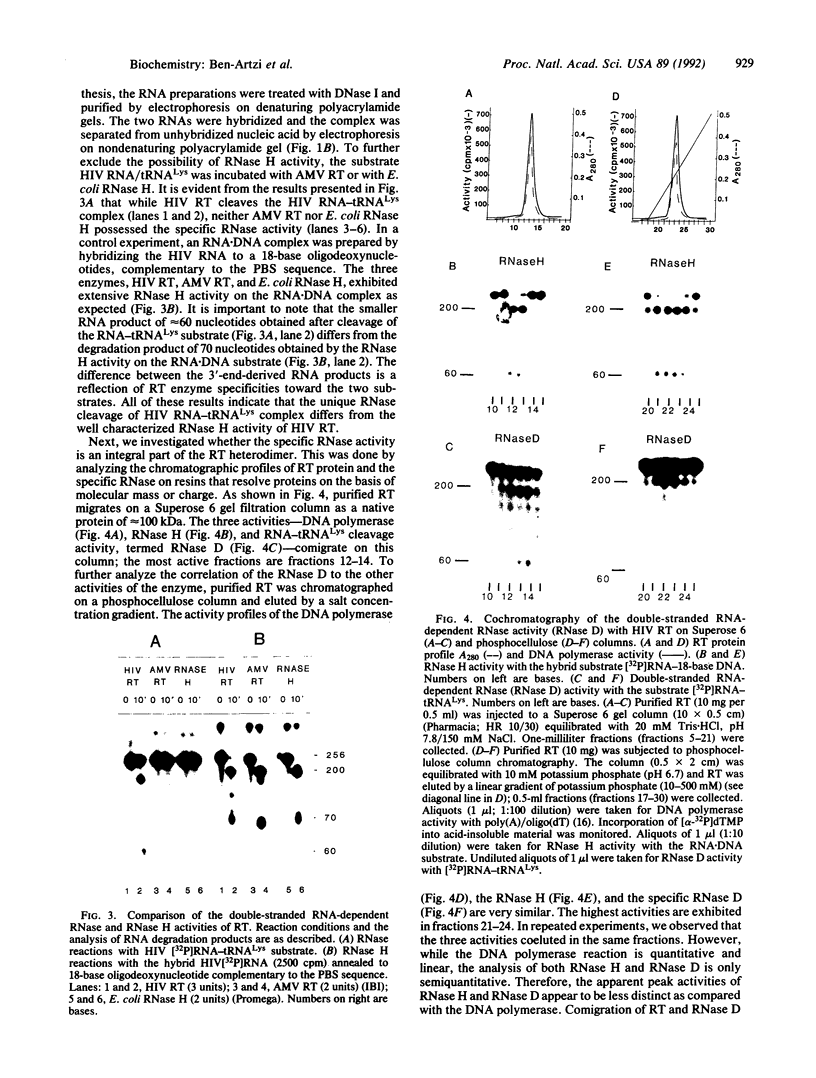
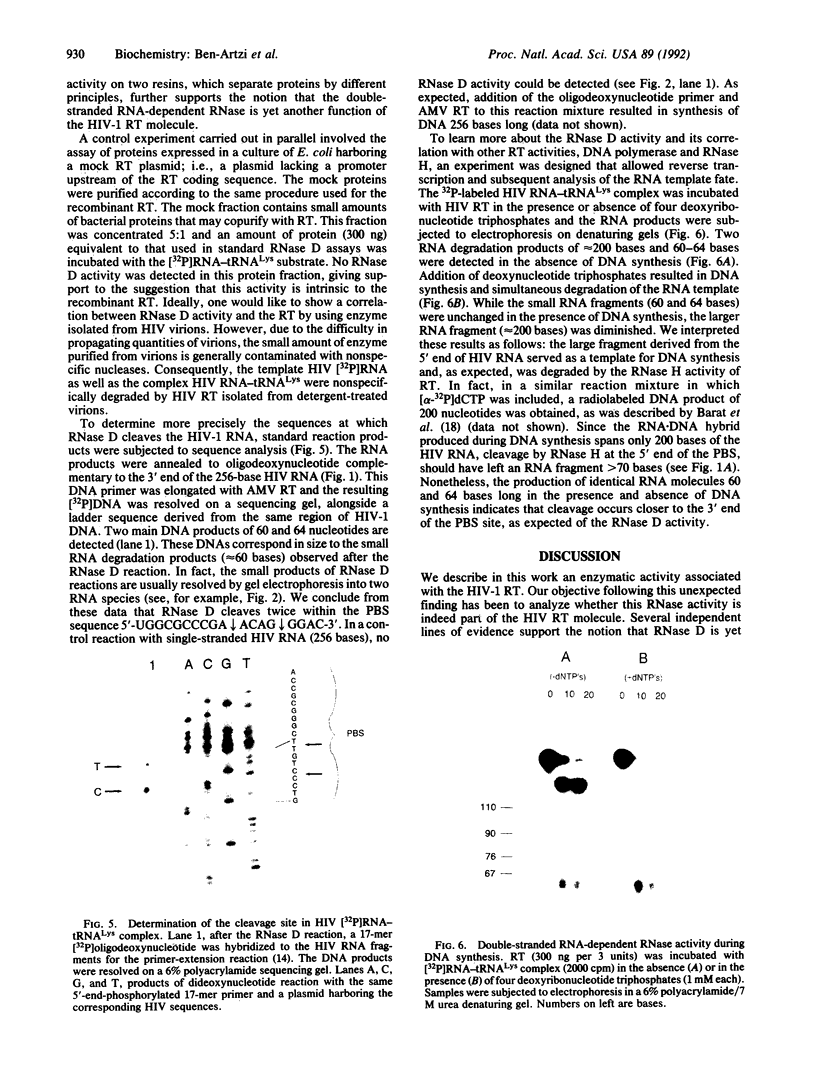
Early events in the retroviral replication cycle include the conversion of viral genomic RNA into linear double-stranded DNA. This process is mediated by the reverse transcriptase (RT), a multifunctional enzyme that possesses RNA-dependent DNA polymerase, DNA-dependent DNA polymerase, and RNase H activities. In the course of studies of a recombinant RT of human immunodeficiency virus type 1 (HIV-1), we observed an additional, unexpected activity of the enzyme. The purified RT catalyzes a specific cleavage in HIV-1 RNA hybridized to tRNALys, the primer for HIV-1 reverse transcription. The cleavage at the primer binding site (PBS) of HIV RNA is dependent on the double-stranded structure of the HIV RNA-tRNALys complex. This RNase activity appears to be distinct from the RNase H activity of HIV-1 RT, as the substrate specificity and the products of the two activities are different. Moreover, Escherichia coli RNase H and avian myeloblastosis virus RT are unable to cleave the HIV RNA-tRNALys complex. We refer to this unusual activity as RNase D. Two lines of evidence indicate that the specific RNase D activity is an integral part of recombinant HIV RT. The specific RNase D activity comigrates with the other RT activities, DNA polymerase, and RNase H upon filtration on a Superose 6 gel column or chromatography on a phosphocellulose column. Moreover, three recombinant HIV-1 RT preparations expressed and purified in different laboratories by various procedures exhibit RNase D activity. Sequence analysis indicated that RNase D activity cleaves the substrate HIV-1 RNA-tRNALys at two distinct sites within the PBS sequence 5'-UGGCGCCCGA decreases ACAG decreases GGAC-3'. The sequence specificity of RNase D activity suggests that it might be involved in two stages during the reverse transcription process: displacement of the PBS to enable copying of tRNALys sequences into plus-strand DNA or to facilitate the second template switch, which was postulated to occur at the PBS sequence.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970 Jun 27;226(5252):1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- Barat C., Le Grice S. F., Darlix J. L. Interaction of HIV-1 reverse transcriptase with a synthetic form of its replication primer, tRNA(Lys,3). Nucleic Acids Res. 1991 Feb 25;19(4):751–757. doi: 10.1093/nar/19.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman H. E., Phelps W., Feigenbaum L., Ostrove J. M., Adachi A., Howley P. M., Khoury G., Ginsberg H. S., Martin M. A. Trans-activation of the human immunodeficiency virus long terminal repeat sequence by DNA viruses. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9759–9763. doi: 10.1073/pnas.83.24.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S. P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquir Immune Defic Syndr. 1990;3(8):817–831. [PubMed] [Google Scholar]
- Hizi A., McGill C., Hughes S. H. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug M. S., Berger S. L. Ribonuclease H activities associated with viral reverse transcriptases are endonucleases. Proc Natl Acad Sci U S A. 1989 May;86(10):3539–3543. doi: 10.1073/pnas.86.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Sharmeen L., Taylor J. Specificities involved in the initiation of retroviral plus-strand DNA. J Virol. 1990 Feb;64(2):592–597. doi: 10.1128/jvi.64.2.592-597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J Virol. 1990 Sep;64(9):4321–4328. doi: 10.1128/jvi.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March P. E., Gonzalez M. A. Characterization of the biochemical properties of recombinant ribonuclease III. Nucleic Acids Res. 1990 Jun 11;18(11):3293–3298. doi: 10.1093/nar/18.11.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi V., Lazarus G. M., Miles L. M., Meyers C. A., Debouck C. Recombinant HIV-1 reverse transcriptase: purification, primary structure, and polymerase/ribonuclease H activities. Arch Biochem Biophys. 1989 Sep;273(2):347–358. doi: 10.1016/0003-9861(89)90493-1. [DOI] [PubMed] [Google Scholar]
- Mölling K., Bolognesi D. P., Bauer H., Büsen W., Plassmann H. W., Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat New Biol. 1971 Dec 22;234(51):240–243. doi: 10.1038/newbio234240a0. [DOI] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Panet A., Haseltine W. A., Baltimore D., Peters G., Harada F., Dahlberg J. E. Specific binding of tryptophan transfer RNA to avian myeloblastosis virus RNA-dependent DNA polymerase (reverse transcriptase). Proc Natl Acad Sci U S A. 1975 Jul;72(7):2535–2539. doi: 10.1073/pnas.72.7.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Schatz O., Mous J., Le Grice S. F. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5' exonuclease activity. EMBO J. 1990 Apr;9(4):1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M., Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970 Jun 27;226(5252):1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- Wöhrl B. M., Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990 Nov 6;29(44):10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]
- di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986 Mar 14;231(4743):1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]