Abstract
Visualization of single RNA molecules in living cells has enabled the study of synthesis, movement, and localization of mRNAs and has provided insight into gene regulation with sub-second temporal resolution and nanometer spatial resolution. Following transcription in single cells indicates that gene activity is heterogeneous between cells and also exhibits random variability over time even within single cells. Studies of mRNAs in yeast can take advantage of the powerful genetics available in this model organism and allow mechanistic questions to be addressed. In this chapter, we describe an approach for visualizing mRNA and transcription in live yeast cells. The method is based on binding of fluorescently labeled MS2 and PP7 coat proteins to stem loops sequences that are introduced into the gene of interest. We give detailed protocols for the construction of the necessary yeast strains, for image acquisition, and for validation.
Keywords: Single-molecule, RNA, transcription, live cell, microscopy, fluorescence
INTRODUCTION
Single-molecule RNA detection in intact cells has emerged as a powerful and versatile approach for quantifying gene expression and RNA localization in yeast (Larson et al., 2011; Powrie et al., 2011; Trcek et al., 2011; Zenklusen et al., 2008). RNA visualization can be accomplished either in fixed cells, using single-molecule FISH on endogenous genes, or in live cells engineered to express high-affinity RNA-binding proteins that interact with sites on genetically-modified genes. Both approaches have advantages and disadvantages (Larson et al., 2009). Single-molecule RNA FISH protocols have recently been extensively described elsewhere (Rahman and Zenklusen, 2013; Trcek et al., 2012). The live cell approach is the only way to directly visualize RNA dynamics in single cells and is the subject of this protocol.
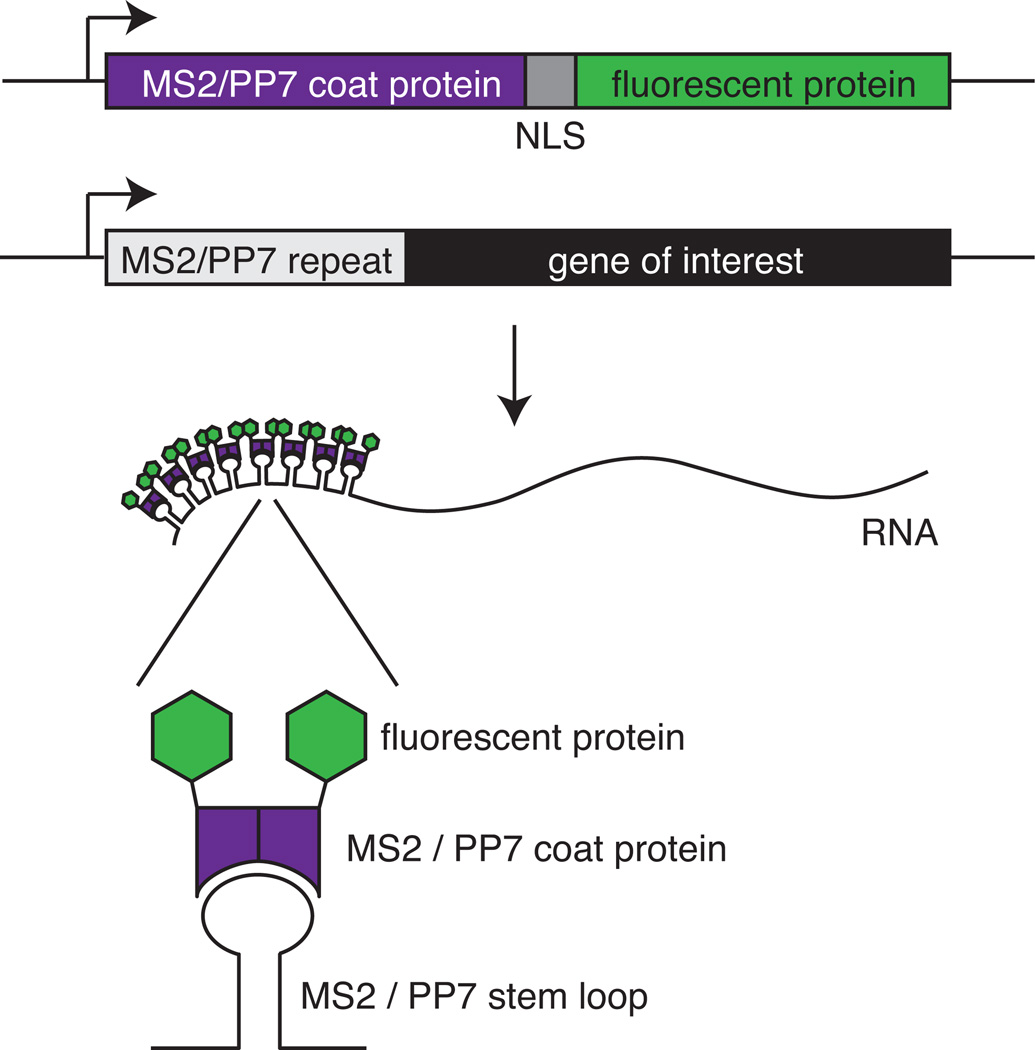
Here, we describe a method for detection of mRNA in live yeast cells using the MS2 or PP7 RNA labeling technique. The method depends on binding of RNA bacteriophage coat proteins to specific stem loop sequences in the RNA to be visualized (Fig. 1)(Bertrand et al., 1998; Chao et al., 2008; Larson et al., 2011). Each stem loop binds a dimer of a chimeric protein consisting of the phage coat protein, a nuclear localization signal (NLS), and a fluorescent protein, such as green fluorescent protein (GFP). To visualize RNAs, cells are modified in two ways: repeats of the stem loop are introduced in the genes of interest, and PP7 or MS2 coat proteins with fluorescent tags are expressed in the cell. When the genes of interest are transcribed, binding of the coat protein to the stem loop repeats in the RNA concentrates the fluorescence, resulting in a bright spot on a background of freely diffusing coat protein. Because MS2-RNA and PP7-RNA interactions are very specific, both can be used in the same cells, for example to tag two different RNAs or to tag a particular RNA in two positions (Hocine et al., 2013; Coulon et al., 2014).
Figure 1.
Schematic of the MS2 and PP7 systems. In order to visualize RNA, the gene of interest is tagged with MS2 or PP7 repeat sequences. As an example 5’ UTR tagging is shown. The cells also express the appropriate coat protein fused to a nuclear localization signal (NLS) and a fluorescent protein. When both the tagged RNA and the coat proteins are expressed, fluorescent coat protein bind to the stem loops, resulting in a punctate fluorescent spot.
The MS2/PP7 approach can been used to study the movement and location of RNAs or to measure nascent RNA formation at the transcription site (Bertrand et al., 1998; Janicki et al., 2004; Larson et al., 2011). This protocol focuses on visualizing transcription.
STRATEGIC PLANNING
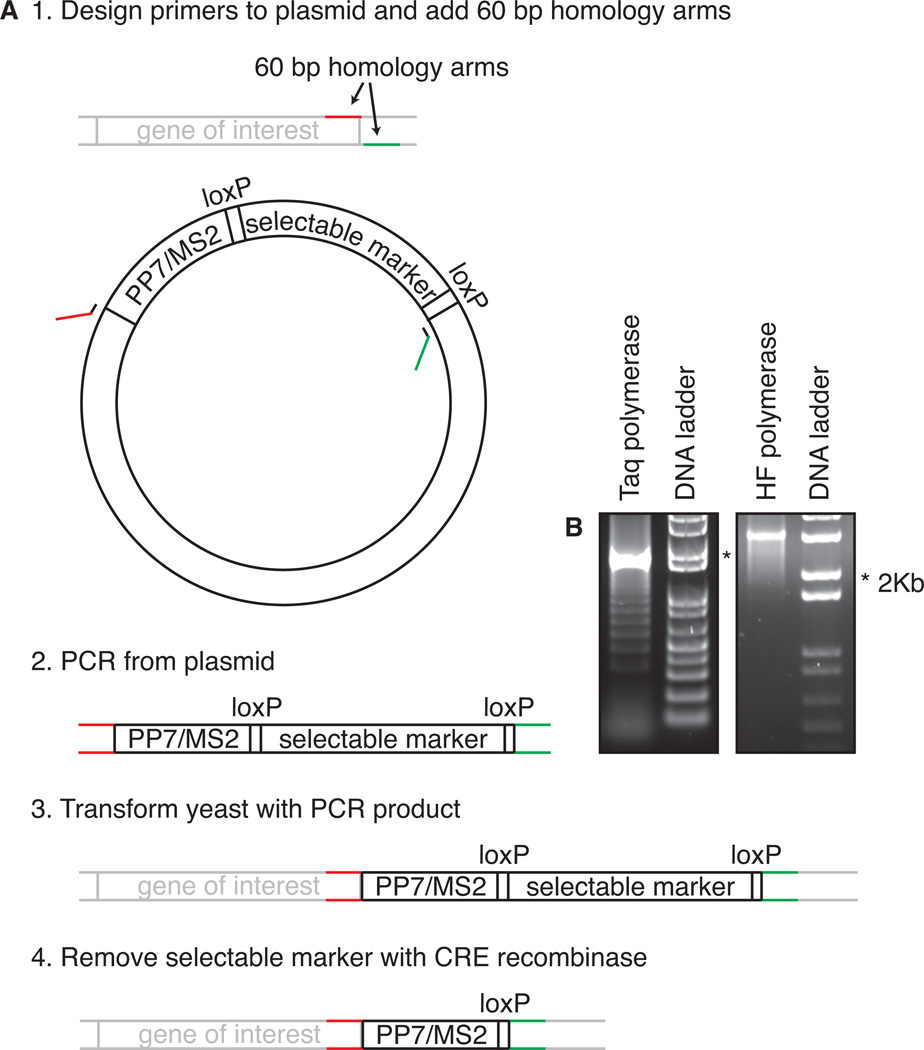
Live-cell detection of transcription and mRNAs requires a yeast strain which is engineered to carry repeats of MS2 or PP7 loops in the gene of interest and to express the corresponding phage coat protein fusion (Fig. 2). Construction of such a yeast strain is not trivial and typically requires multiple transformations carried out over weeks or months. During this construction, the stem loops are integrated in the genome, the selectable marker used to insert them is removed with CRE-lox recombination, and a plasmid with the coat protein is introduced (Haim et al., 2007). Steps are shown in Fig. 2. and described in detail in the protocol section.
Figure 2.
(A) Strategy for tagging a gene of interest with MS2 or PP7 loops. As an example, tagging in the 3’ UTR is shown, but the same applies to tagging in the 5’ UTR or in the intron. See text for details. (B) Examples of PCR reactions on a template containing PP7/MS2 repeats using Taq polymerase (left) or a high-fidelity (HF) polymerase (right).
Using such strains, live cell imaging of transcription can be accomplished with multiple imaging modalities. In this protocol we focus on wide-field or epifluorescence microscopy. Such microscopes can be optimized for sensitive detection of the low-level fluorescence in single-molecule experiments. Since yeast cells are on the order of 4–10 µm, the objective should have sufficient magnification (> 100x) to see them. It should also have a high numerical aperture (NA > 1.3) to maximize light collection from the sample.
In addition, the fluorophore should be excited in a narrow band around its excitation peak, to maximize fluorescence signal while minimizing collateral photobleaching and phototoxicity. The preferred light source is therefore monochromatic, provided by laser illumination, but a very strong light source (such as the several commercially available LED sources) in combination with a narrow band pass filter can also suffice. To repeat, the wavelength band transmitted by the excitation filter should bracket the fluorophore's excitation peak and the emission filter should capture most of the light emitted in a wavelength band bracketing its emission peak.
Moreover, the image should be recorded by a sensitive electron multiplying CCD (EM-CCD) camera. Drift in the z-axis should be corrected by an automated focusing device. Finally, cells should be maintained at a constant temperature (30°C for most yeast experiments) with a stage incubator or live-cell imaging chamber. Care must also be taken that the room itself does not fluctuate in temperature, because this temperature drift may affect the temperature inside the chamber and thus the results of the experiment.
In summary, single-molecule imaging is a demanding protocol, and small deviations from optimum in strain construction, microscope setup design, and imaging conditions during the experiment can greatly reduce the chances of success.
BASIC PROTOCOL 1
VISUALIZING GENE EXPRESSION IN LIVE YEAST USING MS2 AND PP7 LABELING
Live-cell imaging of RNA starts with construction of a yeast strain that contains a MS2 or PP7 tagged version of the gene of interest and that contains a plasmid expressing fluorescent coat protein. The first part of the protocol is focused on construction of the required strain. Standard yeast protocols (Burke et al., 2005), such as transformation protocols, are described in Chapter 7, and only steps specific for integrating stem loops will be discussed. The second part of the protocol describes how to prepare yeast for imaging and how to image transcription or diffusing RNAs.
Materials
Plasmid containing repeats of stem loop sequences and a selectable integration marker flanked by loxP sites, such as pDZ415 (24MS2SL loxP-KanMX-loxP), pDZ416 (24PP7SL loxP-KanMX-loxP), available at Addgene (www.addgene.com), or pTL030 (14MS2SL loxP-KanMX-loxP), pTL031 (14PP7SL loxP-KanMX-loxP), available from the authors.
Plasmid encoding the coat protein fused to an NLS and a fluorescent protein, such as pTL041 (pURA PADE3-PCP-NLS-2x-yeGFP) or pTL042 (pLEU PMET17-MCP-NLS-mKate2), available from the authors.
URA3 plasmid such as pSH47 encoding CRE recombinase under a GAL1 promoter.
(Selective) yeast plates and media for growth and transformation (including YPD plates, YPGAL medium, synthetic media minus uracil plates and media, 5-fluoroorotic acid plates) For recipes see: (Lundblad and Struhl, 2001; Guthrie and Fink, 2004).
PCR primers for integration of stem loop repeats.
PCR primers to check genomic integration of stem loop repeats and to check Cre-mediated excision of selectable marker.
PCR cleaning kit to purify the PCR product and remove primers, dNTPs and enzymes.
Standard materials for PCR.
Standard materials for transformation
0.7% - 1% Agarose gels.
50% glycerol.
Concanavalin A from Canavalia ensiformis (Jack bean).
Glass bottom culture dishes with a coverslip thickness which matches the objective used in the experiment (e.g. MatTek 35 mm, No. 1.5 or NEST 35 mm).
Incubator at 30°C.
- Inverted wide-field microscope (e.g. Zeiss AxioObserver) with:
- > 100 × objective, NA > 1.3 (e.g. Zeiss Plan-Apochromat 150x/1.35 Glyc DIC).
- Laser excitation source (or equivalent strong narrowband excitation)(e.g. Spectra-Physics Excelsior diode-pumped solid state lasers).
- Excitation and emission filters optimized for fluorophore (e.g. for GFP: ET470/30x excitation filter (for non-laser illumination), T495lpxr dichroic beamsplitter and ET525/50m emission filter, Chroma).
- EM-CCD (e.g. Evolve 512, Photometrics).
- Focusing device (e.g. Definite focus, Zeiss).
- Microscope stage incubator (e.g. Tokai Hit, INUB-LPS).
Integration of stem loops into yeast
To visualize mRNAs and transcription in living cells, MS2 or PP7 stem loops have to be integrated into the gene of interest.
-
1.Choose the position and number of stem loop repeats to be integrated into the gene of interest.Both the research question and technical limitations dictate the optimal position of the stem loops in the RNA. Since integration of stem loops may interfere with the normal function of the mRNA, the position and number of stem loop repeats has to be carefully considered. See ‘critical parameters’ for a detailed description.
-
2.Acquire a plasmid that encodes the desired number of stem loop repeats and a selectable marker flanked by two loxP sites.Currently, plasmids with 24 and 14 MS2/PP7 repeats are available. If different repeat numbers are desired, one has to use molecular cloning techniques to construct a plasmid with the correct number of repeats.
-
3.Design PCR primers that anneal to the plasmid containing stem loops and a selectable marker flanked by two loxP sites (Fig. 2).Design primers which anneal outside of the loop region and do not share homology with sequence within the loops to prevent the formation PCR products with fewer loops.Design each of the two primers with 60 nt 5' overhang at each end homologous to the desired integration.
-
4.PCR the loops using standard PCR protocols.Use a high-fidelity DNA polymerase enzyme. Taq DNA polymerase is not very processive, leading to partial PCR fragments that can re-anneal on themselves during the amplification, causing a ladder of PCR products with fewer loops (Fig. 2B).Use enough plasmid in your reaction (at least 30 ng in 50 µl reaction). At lower plasmid concentration, the primers are more likely to anneal on partial PCR products than on the plasmid, resulting in a ladder.
-
5.
Run a small aliquot of the PCR on a 0.7% - 1% agarose gel to check the size of product.
-
6.
"Clean" the PCR product using a commercially available PCR purification kit. Elute the cleaned PCR product in a small volume, preferably less than 10 µl.
-
7.
Transform yeast with the PCR product using standard transformation procedures and plate the transformed cells on selective plates that contain an antibiotic on which only the integrants can grow.
-
8.
After 3–4 days, re-streak several colonies on selective plates.
-
9.Check correct integration by PCR.During homologous recombination, the number of stem loops may be reduced. Loss of stem loops is more frequent for MS2 than for PP7 loops (Hocine et al., 2013). Make sure that the size of the PCR product is correct.If PCR shows that the integration was unsuccessful, repeat the transformation, but this time growing cells in the transformation mix in YPD overnight, before plating them onto selective medium.
-
10.
Make stocks of cells with correctly integrated stem loops by combining 300 µl of 50% glycerol and 700 µl of an overnight culture in a cryogenic vial. Freeze at −80°C.
Removal of the selectable marker
After integration of the stem loops into the gene of interest, the next step is to remove the selectable marker. Since the selectable marker is flanked by two loxP sites, it can be removed by expression of CRE recombinase (Fig. 2).
-
11.
Transform the cells with a URA3 plasmid encoding CRE recombinase under the control of the GAL1 promoter (e.g. pSH47) using standard transformation procedures. Plate transformed cells on plates lacking uracil.
-
12.Inoculate several colonies into YPGAL medium and grow them overnight at 30°C while shaking.For reasons not clear now, the time needed to grow different integrants in YPGAL before cells lose the selectable marker can vary. In some cases, basal gene expression of Cre from the GAL1 promoter seems sufficient to for cells to loose the marker, while other intermediate strains require culture in YPGAL for days before many undergo Cre-mediated excision.
-
13.Streak 5 µl on plates containing 5-Fluoroorotic acid (5-FOA). Incubate for 3 days at 30°C.5-FOA is converted into a toxic form when the URA3 gene is expressed. Cells that survive 5-FOA will therefore have lost the URA3 plasmid encoding the CRE recombinase.
-
14.
To test removal of the selectable marker, replica plate on YPD and selective plates.
-
15.
Pick colonies that grow on YPD plates but are unable to grow on selective plates.
-
16.
Test removal of the selective marker by PCR.
-
17.
Make stocks and freeze at −80°C.
-
18.
If dual color labeling with both MS2 and PP7 repeats is desired, repeat the above process to introduce the second stem loop cassette.
Introduction of the coat protein
-
19.
Transform the cells with a plasmid that directs the synthesis of the bacteriophage coat protein fused to an NLS and a fluorescent protein. Plate the transformed cells on selective plates. Grow for 3 days at 30°C.
-
20.
Re-streak several colonies on selective plates.
-
21.
After 3–4 days, pick colonies and make stocks.
-
22.
Use single-molecule FISH, RT-PCR or Northern blot to check normal expression levels of the tagged RNA.
Preparing yeast cells for imaging
-
23.
Four days before imaging, streak yeast cells that contain integrated stem loop repeats in the gene of interest and a plasmid encoding the coat protein onto selective plates.
-
24.The day before imaging in the morning, start a culture by inoculating one colony of yeast into selective media. Grow at 30°C while shaking.It is important that yeast cells stay in mid-log phase, since they become autofluorescent at various wavelengths in diauxic shift (Fig. 3).Grow yeast in synthetic media, lacking the amino acids for the coat protein plasmid markers, to prevent loss of plasmid and to reduce background fluorescence.Use ADE2+ strains, or supplement ade2- strains with additional adenine (20 µg/ml), to prevent accumulation of the fluorescent adenine biosynthesis intermediate phosphoribosylaminoimidazole (Rines et al., 2011).
-
25.In the evening, dilute cultures into pre-warmed medium (30°C) to a density so that the cells are around OD600 ~ 0.2–0.4 in the morning.If unsure how fast the strain grows, make a dilution series.
-
26.On the day of imaging, adjust OD600 of culture to OD600 ~ 0.2–0.4.Alternatively, re-dilute culture for imaging later in the day.If the OD600 of cultures is too low, wait until cells are ready. If culture is overgrown (OD600 > 1.0), dilute them into fresh medium and let them double at least 2–3 times before imaging.
-
27.
Pipet 300 µl of yeast culture onto concanavalin A coated glass bottom culture dishes (see recipe).
-
28.
Incubate 20–30 min at 30°C.
-
29.
Remove excess liquid.
-
30.
Wash 2 times with pre-warmed media by carefully pipetting 300 µl media onto the cells and removing the liquid.
-
31.
Add 3 ml of pre-warmed media to the cells. The cells are now ready to image.
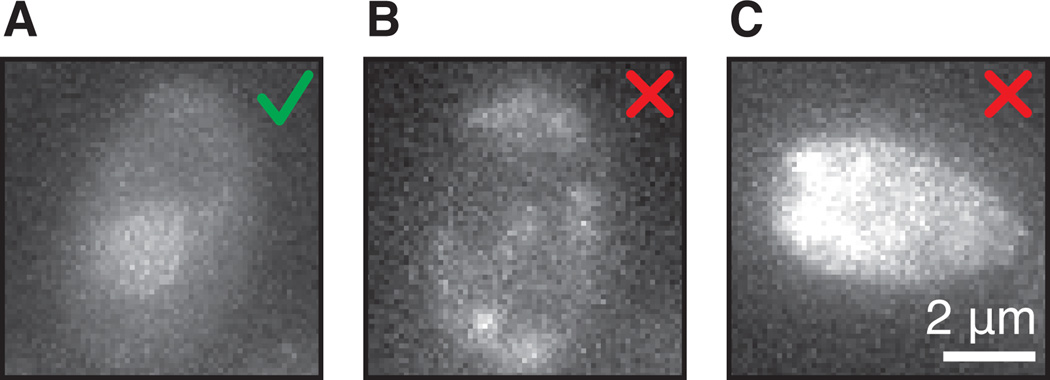
Figure 3.
(A) Example of a correct coat protein levels in the cell. There is even staining across the cell and slightly higher coat protein levels in the nucleus. (B) Example of photo toxicity. The cell shows uneven fluorescence at the cell membrane. (C) Example of auto-fluorescent cell grown into diauxic shift.
Imaging of yeast transcription
-
32.Turn on the wide-field microscope, the camera, and the lasers according to manufacturer's instructions. Let the laser equilibrate for 20 min before imaging. Make sure the stage incubator is at the correct temperature.When first setting up the stage incubator, it is important to obtain an independent measurement of the temperature in the dish, for example by using a thermocouple. Compare the dish temperature measured by the thermocouple with the desired temperature and adjust the stage temperature accordingly.
-
33.
Open the microscope software.
-
34.
Focus on the cells using transmitted light.
-
35.
Switch to laser illumination and turn on the live module of the software using 30ms-50ms exposure and low excitation power.
-
36.Find a field of view with cells that have the right levels of coat protein.The fluorescent signal from the coat protein in the cytoplasm should be at very low levels and even across the cells (Fig. 3A). Sometimes a lighter staining of the center of the cell is visible because of the vacuole. This is not a problem, but make sure there is no uneven staining at the cell membrane (Fig. 3B).The coat protein fusion contains a NLS, so the levels in the nucleus should be slightly brighter than the cytoplasm, visible as a brighter circular structure in the cell (Fig. 3A). Too much nuclear fluorescence is not preferable, because it increases the background relative to the transcription site.If no nuclear fluorescence is visible, it is possible that the excitation power is too low. Another reason why nuclear fluorescence might not be visible is that even though freely diffusing coat proteins get imported into the nucleus, coat proteins associated with the MS2/PP7-tagged RNA are also exported to the cytoplasm after its synthesis. Export of mRNA-coat protein complexes cause a decrease in the nuclear concentration of freely diffusing fluorescent coat proteins. At high expression levels of the tagged RNA, we have observed that the export of coat protein may be faster than the import of newly synthesized coat protein, resulting in a depletion of coat protein from the nucleus. In these latter conditions, one must be careful, because not all transcription events may be visible (Fig. 4 and critical parameters).Avoid highly auto-fluorescent cells (Fig 3C). Yeast cells become auto-fluorescent when they enter diauxic shift, so it is important to keep them in mid-log phase during imaging.
-
37.When imaging the strain for the first time, make sure that the strain behaves normally and the fluorescent signal in the cell does not come from aggregates. Check whether there is background fluorescence from the coat protein and transcription sites in the nucleus, to make sure that coat protein is not depleted from the nucleus (Fig. 4C and critical parameters). When using high excitation power (>10 mW) and fast exposure time (30–50 ms) diffusing RNAs in the cytoplasm can often be visible (Fig. 5), indicating proper export. After transcription shut-off, fluorescence from transcription sites should disappear and mRNAs should be degraded.Depending on the gene, basal or uninduced transcription may not be sufficient, and it may be necessary to induce gene transcription to see transcripts. For example, one can only see transcription from the GAL genes after galactose induction.
-
38.Setup the imaging conditions in the image software. For visualizing transcription, use low excitation power (for lasers, around 100–1000 µW), moderate exposure times (around 100–200 ms) and an acquisition rate with around 10–60s intervals between frames. Take 9–15 z-stacks in 0.5 µm increments, depending on the size of the cells. For longer acquisitions, use the microscopes focus feature to eliminate drift in z.Good imaging conditions are a balance between good signal to noise, low bleaching, low phototoxicity and a long enough acquisition to see fluctuations in transcription. To a first approximation, there are a ‘fixed’ number of photons that each dye emits before bleaching, and the user must decide how those photons are partitioned in the experiment (Larson, 2010; Lee et al., 2013). High excitation power will increase the signal-to-noise ratio, but will also increase bleaching and phototoxicity. Too low excitation power may prevent you from seeing the transcription site above background.Set an exposure time such that the transcription site is visible above background. Longer exposures allow for lower excitation power without losing signal, but too long exposures may result in blurring caused by movement.Both the transcription site and the entire yeast nucleus are highly mobile. For that reason, it is important to take z-stacks, to ensure that fluctuations of the transcription site are not caused by movements around the focal plane in z. Choose the number of z-stacks so that at least the nucleus and preferably the entire cell are captured. The number of z-stacks required depends on the type of cell and the growth conditions. Yeast cells range in size from 4–10 µm. Haploid cells are generally smaller than diploid cells and faster growing cells are larger than slow growing cells (Johnston et al., 1979). For example, cells in glucose are larger than cells in galactose. Mutants that affect growth rate likely also affect cell size. We recommend 9–15 z-stacks in 0.5 µm increments.The rate of transcription elongation is around 20–46 bp/s and the termination rate is ~ 70 s (Larson et al., 2011). The time a transcript is visible at the transcription site depends on the length of the gene and the position of the stem loop repeats. Choose an acquisition rate that captures the transcription event in several frames. If the event is short-lived, it is best to image with short acquisition interval, but this will shorten the total duration of acquisition. In that case, taking fewer z-stacks may help.Be aware that too much light can bleach the fluorescent proteins, but more importantly, cause phototoxic damage to the cell. Signs of phototoxicity include uneven fluorescent signal (Fig. 3B), termination of transcription and abortion of cell division.For visualizing diffusing mRNAs, use high excitation power, short exposure times (30–50ms) and fast acquisition rates. Low gain settings of the camera can reduce noise.
-
39.If necessary, induce the transcription of the gene whose transcription is to be visualized.Transcription of yeast genes can be very fast, resulting in the appearance of transcription sites within several minutes. It is therefore desired to induce the cells while on the microscope, after a field of view has been chosen and immediately before image acquisition.
-
40.
Refocus on the center of the cells using the live-mode with low laser illumination.
-
41.
Start the acquisition.
-
42.
The necessary number of cells imaged depends on the signal to noise, the number of frames per acquisition and the biological question. As a crude rule of thumb, we usually start with ~ 30 cells.
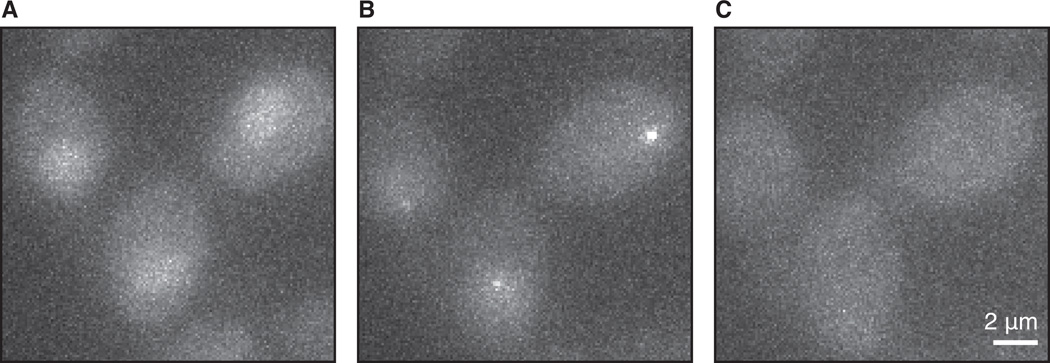
Figure 4.
Induction of transcription of GAL10, tagged with PP7 at the 5’ UTR. (A) Before induction there is no transcription site, and only background levels of coat protein are visible. (B) Upon induction with galactose, GAL10 transcription sites appear. (C) After long induction, the coat protein is depleted from the nuclear, resulting in loss of visibility of the transcription sites and nuclear staining. Shown are maximum intensity projections of 9 z-planes, 150 ms exposure.
Figure 5.
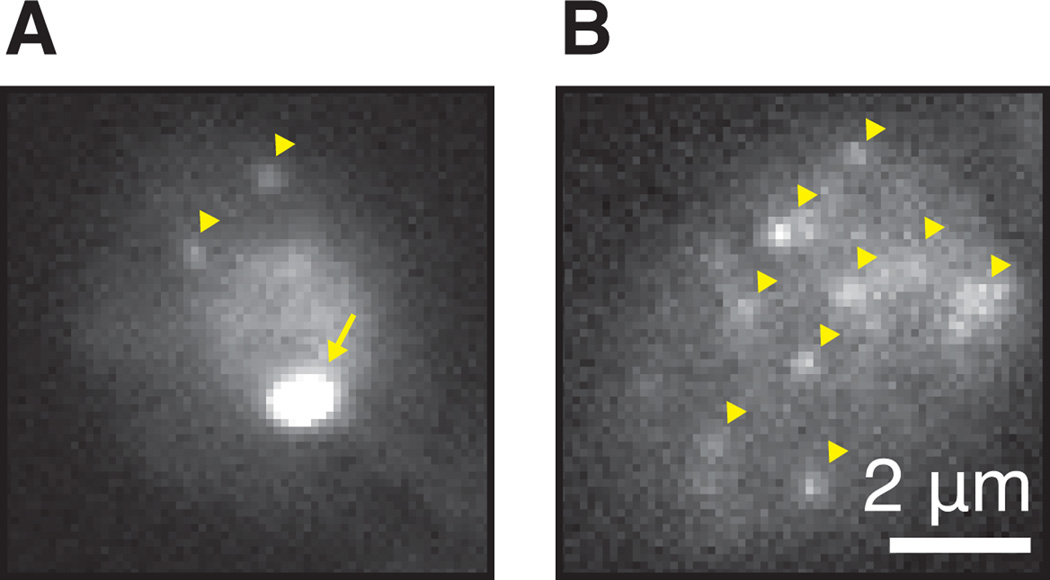
Detection of single mRNAs. (A) GAL10 PP7 tagged cell in galactose imaged with fast exposure 30ms. Note the nuclear staining, the bright transcription site (arrow) in the nucleus and two cytoplasmic RNAs (arrowheads). Frequently, the transcription site will appear saturated at light intensities where single mRNAs are visible. (B) Same as panel A with several single RNAs (arrowheads). Note that the nucleus is out of focus.
SUPPORTING PROTOCOL
Yeast grows in suspension, but in the imaging protocol described here, yeast is immobilized to a glass-bottom dish before imaging. Yeast cells are bound to the dish by letting them settle onto concanavalin A, with which the coverslip is coated. This supporting protocol describes the coating of dishes with concanavalin A.
Materials
Concanavalin A from Canavalia ensiformis (Jack bean).
PBS.
0.22 µm filter.
Glass bottom culture dishes with a coverslip thickness which matches the objective used in the experiment (e.g. MatTek 35 mm, No. 1.5 or NEST 35 mm).
Concanavalin A coating slides
Dissolve concanavalin A in PBS for a 20 mg/ml stock solution. Aliquot and store at −20°C.
Dilute stocks of concanavalin A to 1 mg/ml.
Filter sterilize.
Pipet 300 µl of dilution onto glass-bottom dish.
Wait 20 min.
Aspirate liquid.
Air dry 20 min.
COMMENTARY
Background Information
Labeling of mRNAs with the MS2 system was developed by Bertrand et al. to monitor the localization of ASH1 mRNA (Bertrand et al., 1998). The system was subsequently applied in several organisms to study the movement and distribution of mRNAs (Rook et al., 2000; Fusco et al., 2003; Forrest and Gavis, 2003; Shav-Tal et al., 2004). In an elegant study by Janicki et al., MS2 mRNA labeling was used to follow the dynamics of transcription in living cells. Using a transgene array of Lac operators and MS2 repeats, changes in chromatin structure and transcription activation were visualized and found to be correlated over time (Janicki et al., 2004). Similarly, real-time transcription measurements of reporter genes in bacteria and mammalian cells were used to quantify bursting and transcription kinetics (Golding et al., 2005; Darzacq et al., 2007). The related PP7 labeling method was used to study fluctuations in fluorescence at the transcription site of an endogenous yeast gene, from which initiation, elongation and termination kinetics could be extracted (Larson et al., 2011; Chao et al., 2008). Recently, dual tagging with MS2 and PP7 labeling was used study two alleles in the same cell and different sites on the same transcript (Coulon et al., 2014; Hocine et al., 2013).
Currently, the MS2 and PP7 labeling systems are the most sensitive methods for visualizing transcription. One limitation of them is the high background fluorescence from the coat protein. One approach to overcome this limitation has been to express in the cell two different proteins each encoding the phage coat protein but fused to different halves of a split-GFP, so that it only becomes fluorescent when the two halves are brought together by dimerization of the coat proteins on a target mRNA (Wu et al., 2014). A similar split-GFP approach can also be applied with different RNA binding proteins (Valencia-Burton et al., 2007; Ozawa et al., 2007). Although these approaches are useful for studying the localization of mRNAs, the maturation of split-GFP is too slow to study transcription dynamics. Another approach to is to use exogenously supplied fluorescent ligands that bind aptamer sequences engineered into the RNA, forming RNA-fluorophore complexes called Spinach (Paige et al., 2011) . To increase sensitivity, two different RNA-aptamer ligands are linked to the two members of a FRET pair, resulting in an increased FRET signal when the FRET pairs are brought in close proximity on the RNA (ImageTags) (Shin et al., 2014). This approach has the benefit of mitigating deterioration of signal from photobleaching, because the fluorescent ligand is continuously turning over on the RNA. Although this approach has been useful to measure transcript accumulation, the aptamer approach is presently not sensitive enough to visualize individual transcripts or the transcription site in a manner which is comparable to MS2/PP7.
Critical Parameters
Position and number of stem loop repeats
The position and number of stem loop repeats are an important consideration in the design of the experiment. Because they contain stop codons, the stem loops are usually positioned in the 5’ or 3’ untranslated regions (UTR) of a gene. For genes that are spliced, it is also possible to introduce the stem loops in the intron (Coulon et al., 2014). The optimal location depends on the research question and technical considerations.
For studies of transcription, when stem loops are positioned in the 5’ UTR, fluctuations in fluorescence signal will be caused by all transcription events including elongation, termination and release, as well as whether transcription occurs in bursts (i.e. "bursting"). When stem loops are positioned in the 3’ UTR, fluctuations will be dominated by bursting and termination/release. When positioned in an intron, changes in signal will capture transcription as well as splicing kinetics. For studies of RNA localization and distribution, 3' tagging is usually the best choice (Bertrand et al., 1998).
Apart from the particular research question, technical limitations may also affect the choice of position. Introduction of stem loops into noncoding regions can alter gene function by affecting processes including nuclear export, translation and degradation. Importantly, tagged mRNAs occasionally form brighter spots in the nucleus or cytoplasm. Nuclear spots can be a sign of incorrect nuclear export (Jensen et al., 2001). Cytoplasmic spots may be biologically functional, such as messenger ribonucleoprotein (mRNP) foci that occur during glucose starvation (Zid and O’Shea, 2014), but may possibly also be aggregates , perhaps akin to P-bodies (Sheth and Parker, 2003)
In our hands, the modification that we have found to be the least perturbative is insertion of MS2 stem loops in the 3’ UTR. The tagged mRNAs are generally properly exported, translated and degraded, and can support growth when being the sole copy under essential condition. In contrast, PP7 loops in the 3’ UTR are less well tolerated and the fluorescent signal can show spots under conditions where transcription should be off, which is perhaps caused by stabilization of the mRNA or aggregation. 5’ UTR tagging with either MS2 or PP7 loops can block translation, essentially mimicking a gene deletion. If tagging the gene makes it nonfunctional, a possible solution is to tag only one of the copies in diploid cells, so there is always a second untagged version of gene. Addition of loops to an intron can affect splicing, in the worst scenario completely abolishing it.
Both the 5’ and 3’ UTRs of a gene can contain regulatory sequences for proper processing and degradation. To minimize the effect of the stem loops on these, we recommend that the investigator positioned them close to the ORF, immediately upstream of the start codon in the 5’ UTR and/or immediately downstream of the stop codon in the 3’ UTR. A second reason to prefer this location is that yeast UTRs tend to be very short and have many isoforms (Pelechano et al., 2013). If the loops are positioned further from the ORF, transcription of the shorter isoforms may be missed. In practice, it may be necessary to test additional positions. If the stem loops are positioned in the intron, the 5’ and 3’ splice sites and the branch point should remain intact and the loops should be placed between the 5’ splice site and the branch point. To our knowledge, successful integration into S. cerevisiae introns has not been reported. In all cases, it is critical that the selectable integration marker is removed using the CRE-Lox system, otherwise the RNA contains the selectable marker and the lox sites.
The number of stem loop repeats is also a design criterion to consider. Historically, 24 loops have been used both for MS2 and PP7. However, many yeast genes do not tolerate 24 loops, causing export problems, abortion of transcription and aggregation at the transcription site or cytoplasm. Lowering the number of loops to 12–14 resolves most of these side effects. The fluorescent intensity of the RNA relative to the background decreases with decreasing number of loops, but modern microscopes are sensitive enough to still see single mRNAs above the background.
As for any modification, the effects of stem loop tagging are different per gene. For that reason it important to test the effect of the different stem loops on gene expression levels, mRNA stability, and, if relevant, splicing, using RNA FISH, RT-PCR or northern blots. In addition, when imaging transcription in live cells, make sure that the nucleus does not show a really bright fluorescent signal that does not fluctuate, suggesting aggregation. If the gene is inducible, confirm that no spots are present in un-induced conditions and only appear after induction. After removal of the transcriptional inducer, the transcription spot should disappear, and the mRNA in the cytoplasm should be degraded.
Levels of the coat protein
Every stem loop binds two coat proteins, which are each attached to fluorescent proteins. When multiple stem loops are being transcribed, the accumulation of fluorescent proteins at one site results in the formation of a punctate spot visible in the microscope. The expression level of the coat protein-XFP fusion forms the background fluorescence, above which the signal (labeled mRNAs) should be visible. The expression level of the coat protein is therefore critical for visualizing mRNA. Higher levels increase the background, thus lowering the signal-to-noise ratio. Lower levels will result in incomplete binding of coat protein to the mRNA. We get good results with low copy number plasmids that drive the synthesis of the fusion protein from low-level constitutive promoters such as the ADE3 or MET17. The genes that direct the synthesis of coat proteins can also be integrated in the genome, but in this case the expression level decreases. If chromosomally integrated constructs are used to direct synthesis of the fusion proteins, induce their transcription by growth in medium without adenine or methionine for several hours before imaging. We also note that some investigators have used coat proteins which are constitutively dimerized, which may increase sensitivity of RNA labeling under certain conditions (Wu et al., 2012). The advantage of this approach is that constitutive dimers will bind stem loops at lower effective concentrations of coat protein than would normally be required by mass action-driven binding of monomers.
The coat proteins are fused to a nuclear localization signal (NLS). After their synthesis, they are imported into the nucleus, causing an enrichment of freely diffusing coat protein in the nucleus. Despite the NLS, fluorescent coat proteins can usually also be observed in the cytoplasm.
When RNA containing stem loop repeats is being transcribed, the freely diffusing coat proteins in the nucleus bind the RNA as a dimer. During export of the tagged RNA, the associated coat proteins are also exported from the nucleus. The higher the rate of transcription, the more coat protein is exported, decreasing the amount of freely diffusing coat proteins in the nucleus that are available to bind newly synthesized RNA. Above a certain transcription rate, more RNA-coat protein complexes are exported than that newly synthesized coat proteins can be synthesized and imported, resulting in depletion of freely diffusing coat proteins in the nucleus. In that case, transcription of the target RNA can still occur, but will no longer be visible, because no fluorescent coat proteins associate with the RNA. Because each RNA is bound by many coat proteins (twice the number of stem loop repeats), the initiation rate of the tagged RNA has to be at least an order of magnitude lower than the synthesis rate of new coat protein.
If transcription of the tagged gene is inducible, we recommend starting in non-inducing conditions, where the absence of stem loops results in accumulation of the coat proteins in the nucleus. Upon induction of the tagged RNA, RNA-coat protein complexes are formed and exported, with the result that the amount coat proteins in the nucleus drops until a new steady state is reached. For many inducible genes, the steady state expression is high enough to deplete all coat proteins from the nucleus. Only the initial changes of transcription of inducible genes can likely be studied with this method (i.e. the first 30–60 minutes after induction). When imaging the RNA for the first time, check whether there is nuclear enrichment of the coat protein, which is a good indication that steady state coat protein levels are high enough. If the coat protein is depleted from the nucleus by mRNA export, the nuclear background fluorescence will no longer be visible, and the nucleus is sometimes even darker than the cytoplasm (Fig. 4C).
Choosing a fluorescent protein
The best fluorophores have high brightness, fast maturation time, low aggregation, and high photostability (Lee et al., 2013). We recommend using GFP or mCherry/Mkate2. For these fluorophores, the wavelength difference between excitation and emission peaks (stokes shift) is large enough to collect almost all of the emitted light. When using both MS2 and PP7 labeling, use fluorophores with non-overlapping emission and excitation peaks such as GFP in combination with mCherry/mKate2. Avoid fluorophores that are pH sensitive, since pH can vary within the cell.
Anticipated Results
Figure 4 shows an experiment where transcription of an inducible gene is imaged. Cells in non-inducing conditions, in which the tagged gene is not transcribed, should resemble Fig. 4A, with low background levels of coat protein in the cytoplasm and slightly higher levels in the nucleus. Upon transcriptional induction, fluorescent signal from the binding of the fusion protein to the nascent RNA should become visible in the nucleus (Fig. 4B). After long induction, the nucleus becomes depleted of coat protein and neither the transcription site nor nuclear staining are visible (Fig. 4C). When high excitation power (>10 mW) and fast exposure times (<50 ms) are used, diffusing mRNAs are visible in the cytoplasm (Fig. 5). Under optimal imaging conditions (see above), it should be possible to see single RNAs, both nascent (still being transcribed) and mature (exported to the cytoplasm). For example, in galactose-induced cells where GAL10 is tagged with 14xPP7, both cytoplasmic RNAs (Fig. 5A and Fig. 5B) and the transcription site are visible (Fig. 5A, note that the transcription site is overexposed in these imaging conditions).
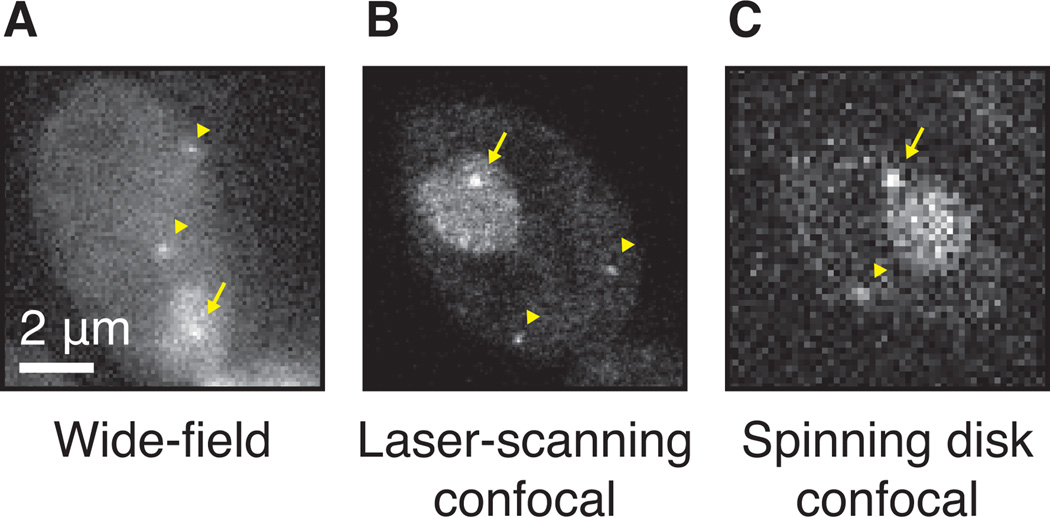
If experiments require the visualization of single RNAs, it may be worth testing confocal microscopes, such as laser-scanning or spinning disk microscopes. These microscopes are capable of optical sectioning and therefore minimize out-of-focus fluorescence originating from unbound coat protein. Figure 6 shows images of the same PP7-tagged strain, imaged with three different microscopes. Compared to a wide-field microscope, the laser-scanning microscope yields better signal to noise for the tagged RNA in the plane of focus. One disadvantage is that acquisition time is longer, resulting in slower frame rates, and the focused high-intensity laser light results in greater phototoxicity and bleaching. The spinning disk confocal microscope combines the high signal-to-noise ratio of optical sectioning with the fast acquisition rates associated with parallel point acquisition and may therefore be the best way to visualize single RNAs at high time resolution.
Figure 6.
Detection of single RNAs with different microscopes. (A) Single RNAs tagged with PP7, imaged with a wide-field or epifluorescence microscope, 50ms exposure. The transcription site is marked with an arrow, the diffusing RNAs with arrowheads. (B) Same strain as panel A, imaged with a laser-scanning microscope. (C) Same as panel A, image with a spinning disk confocal microscope, 50ms exposure.
Time Considerations
It can take several weeks to months to construct the yeast strains required for mRNA imaging. Once a strain is ready, it may take several days to test whether its expression levels are normal by Northern, RT-PCR or single-molecule FISH and to confirm that the strain is suitable for imaging (i.e. proper export, no RNA aggregation, appropriate expression levels of the tagged RNA without nuclear coat protein depletion). Several rounds of trial and error may be necessary before a good strain is obtained. After validating a good strain, optimizing the imaging conditions can take a few days. The time to collect images varies on the kinetics of the process. A typical measurement of transcription activity takes about an hour, while an acquisition of diffusing mRNAs may be less than a minute. The number of cells that are necessary to obtain robust results depends on many variables, such as the signal to noise, the number of frames per acquisition, the variation between cells and the biological question. We usually start with at least 30 cells.
Acknowledgments
Supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, the European Molecular Biology Organization and the KWF Dutch Cancer Society, grants ALTF 37–2012, KWF 2012–5394 (T.L.L.).
Footnotes
INTERNET RESOURCES
The Saccharomyces genome database (SGD) website is a great repository for information on all yeast genes.
µManager is an open source microscopy software package.
Fiji is an imaging-processing software package.
Contributor Information
Tineke L. Lenstra, Center for Cancer Research, National Cancer Institute, Building 41, Room B302, 41 Library Dr., Bethesda, MD 20814, Tineke.Lenstra@nih.gov, 301-496-4793
Daniel R. Larson, Center for Cancer Research, National Cancer Institute, Building 41, Room B201, 41 Library Dr., Bethesda, MD 20814, Dan.Larson@nih.gov, 301-496-0986
LITERATURE CITED
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Molecular Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Burke DJ, Amberg DC, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, 2005 Edition, 2005 Edition First Printing edition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Chao JA, Patskovsky Y, Almo SC, Singer RH. Structural basis for the coevolution of a viral RNA-protein complex. Nature Structural & Molecular Biology. 2008;15:103–105. doi: 10.1038/nsmb1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Ferguson ML, de Turris V, Palangat M, Chow CC, Larson DR. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife. 2014:3. doi: 10.7554/eLife.03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nature Structural & Molecular Biology. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Current biology: CB. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Current biology: CB. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular and Cell Biology. Gulf Professional Publishing; 2004. [Google Scholar]
- Haim L, Zipor G, Aronov S, Gerst JE. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nature Methods. 2007;4:409–412. doi: 10.1038/nmeth1040. [DOI] [PubMed] [Google Scholar]
- Hocine S, Raymond P, Zenklusen D, Chao JA, Singer RH. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nature methods. 2013;10:119–121. doi: 10.1038/nmeth.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Patricio K, McCarthy T, Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Molecular Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Johnston GC, Ehrhardt CW, Lorincz A, Carter BL. Regulation of cell size in the yeast Saccharomyces cerevisiae. Journal of Bacteriology. 1979;137:1–5. doi: 10.1128/jb.137.1.1-5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR. The economy of photons. Nature Methods. 2010;7:357–359. doi: 10.1038/nmeth0510-357. [DOI] [PubMed] [Google Scholar]
- Larson DR, Singer RH, Zenklusen D. A single molecule view of gene expression. Trends in Cell Biology. 2009;19:630–637. doi: 10.1016/j.tcb.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science (New York, N.Y.) 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lim WA, Thorn KS. Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PloS One. 2013;8:e67902. doi: 10.1371/journal.pone.0067902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; 2001. [Accessed November 14, 2014]. Yeast. Available at: http://onlinelibrary.wiley.com/doi/10.1002/0471142727.mb1300s82/abstract. [Google Scholar]
- Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nature Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science (New York, N.Y.) 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie EA, Zenklusen D, Singer RH. A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA in S. cerevisiae. RNA (New York, N.Y.) 2011;17:134–144. doi: 10.1261/rna.1210411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Zenklusen D. Single-molecule resolution fluorescent in situ hybridization (smFISH) in the yeast S. cerevisiae. Methods in Molecular Biology (Clifton, N.J.) 2013;1042:33–46. doi: 10.1007/978-1-62703-526-2_3. [DOI] [PubMed] [Google Scholar]
- Rines DR, Thomann D, Dorn JF, Goodwin P, Sorger PK. Live cell imaging of yeast. Cold Spring Harbor Protocols. 2011:2011. doi: 10.1101/pdb.top065482. [DOI] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIalpha 3’ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science (New York, N.Y.) 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science (New York, N.Y.) 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin I, Ray J, Gupta V, Ilgu M, Beasley J, Bendickson L, Mehanovic S, Kraus GA, Nilsen-Hamilton M. Live-cell imaging of Pol II promoter activity to monitor gene expression with RNA IMAGEtag reporters. Nucleic Acids Research. 2014;42:e90. doi: 10.1093/nar/gku297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Chao JA, Larson DR, Park HY, Zenklusen D, Shenoy SM, Singer RH. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nature Protocols. 2012;7:408–419. doi: 10.1038/nprot.2011.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Larson DR, Moldón A, Query CC, Singer RH. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell. 2011;147:1484–1497. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. RNA visualization in live bacterial cells using fluorescent protein complementation. Nature Methods. 2007;4:421–427. doi: 10.1038/nmeth1023. [DOI] [PubMed] [Google Scholar]
- Wu B, Chao JA, Singer RH. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophysical Journal. 2012;102:2936–2944. doi: 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chen J, Singer RH. Background free imaging of single mRNAs in live cells using split fluorescent proteins. Scientific Reports. 2014;4:3615. doi: 10.1038/srep03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nature Structural & Molecular Biology. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, O’Shea EK. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014;514:117–121. doi: 10.1038/nature13578. [DOI] [PMC free article] [PubMed] [Google Scholar]