Abstract
Background
Comparative mapping of 5S and 45S rDNA by fluorescent in situ hybridization (FISH) technique is an excellent tool to determine cytogenetic relationships among closely related species.
Results
In this study, the number and position of 5S and 45S rDNA loci in all Citrullus species and subspecies were determined. The cultivated watermelon (C. lanatus subsp. vulgaris), C. lanatus subsp. mucosospermus, C. colocynthis and C. naudinianus (or Acanthosicyos naudinianus) had two 45S rDNA loci and one 5S rDNA locus which was located syntenic to one of the 45S rDNA loci. C. ecirrhosus and C. lanatus subsp. lanatus had one 45S rDNA locus and two 5S rDNA loci, each located on a different chromosome. C. rehmii had one 5S and one 45S rDNA locus positioned on different chromosomes. The distribution of 5S and 45S rDNA in several species belonging to other genera in Benincaseae tribe was also investigated. The distribution pattern of rDNAs showed a great difference among these species.
Conclusions
The present study confirmed evolutionary closeness among cultivated watermelon (C. lanatus subsp. vulgaris), C. lanatus subsp. mucosospermus and C. colocynthis. Our result also supported that C. lanatus subsp. lanatus was not a wild form of the cultivated watermelon instead was a separate crop species. In addition, present cytogenetic analysis suggested that A. naudinianus was more closely related to Cucumis than to Citrullus or Acanthosicyos, but with a unique position and may be a link bridge between the Citrullus and the Cucumis.
Keywords: rDNA, Oligonucleotides probes, Citrullus species, Cytogenetic relationship, Fluorescence in situ hybridization
Background
The genus Citrullus belongs to the Benincaseae tribe of the Cucurbitaceae family [1]. There are four or five species in the Citrullus, which are Citrullus lanatus (Thunb.) Matsum. & Nakai, C. colocynthis (L.) Schrad., C. eccirrhosus Cogn., C. rehmii de Winter and C. naudinianus (Sond.) Hook.f. [1, 2]. C. naudinianus is so different from the other Citrullus species in gross morphology that it is placed in the genus Acanthosicyos (A. naudinianus) by Jeffrey [3]. C. lanatus includes three subspecies of C. lanatus subsp. vulgaris (cultivated watermelon), C. lanatus subsp. mucosospermus and C. lanatus subsp. lanatus. Both C. lanatus subsp. vulgaris and C. lanatus subsp. mucosospermus have been classified as C. lanatus var. lanatus. The C. lanatus subsp. lanatus is named as C. lanatus var. citroides, which are the synonyms with C. caffer and C. amarus [1, 3]. However, the latest study reveals that C. lanatus is not a biological species since C. lanatus subsp. lanatus is more closely to C. ecirrhosus than it is to subspecies of C. lanatus subsp. vulgaris and C. lanatus subsp. mucosospermus based on molecular phylogenetic study of the genus Citrullus [4]. Moreover, Chomicki and Renner [4] think that three subspecies of ‘Citrullus lanatus’ are unrelated species and there are seven species in Citrullus including C. naudinianus.
Although various analyses have been carried out aiming to establish phylogenetic relationships among different Citrullus species, the conclusions are inconsistent and the progenitor of cultivated watermelon has not yet been determined [5–11]. Cytological [12] and cross-compatibility [13] observations favored C. colocynthis as the ancestor of cultivated watermelon. Several studies established that C. lanatus var. citroides was phylogenetically sister to cultivated watermelon [3, 14–16]. However, C. ecirrhosus has been found to have closer relative relationship with cultivated watermelon based on sequencing analysis of cpDNA regions [17, 18]. Jarret and Newman [9] proposed that C. rehmii might be the ancestral to cultivated watermelon by the analysis of internal transcribed spacer sequence heterogeneity.
Mapping of 5S and 45S rDNA by fluorescent in situ hybridization (FISH) technique is an excellent tool to determine phylogenetic relationships because their map positions can reveal similarities and differences between chromosomes of related species [19–21]. To date, the position and number of rDNA loci have been determined in more than 1000 plant species with FISH [22]. These studies showed that the number and position of the 5S and 45S rDNA were usually characteristic of a given species, genus or group [23] and 5S and 45S rDNA tended to occupy similar chromosomes and positions in closely related species [24–28]. The chromosomal localization of 5S and 45S rDNA loci has been reported using the FISH technique in three C. lanatus subspecies [29]. The study [29] showed there were one 5S locus and two 45S rDNA loci on chromosomes 4 and 8 and the 5S rDNA locus was located syntenic to one of the 45S rDNA loci on chromosome 8 in C. lanatus subsp. vulgaris and C. lanatus subsp. mucosospermus. Whereas C. lanatus subsp. lanatus contained one 45S locus on chromosomes 4 and two 5S rDNA loci, one 5S rDNA locus was on chromosome 8 and the other 5S rDNA locus was on chromosome 11. The rDNA distribution has also been investigated in C. colocynthis and C. rehmii besides C. lanatus var. lanatus and C. lanatus var. citroides [30]. However, the chromosomes with rDNA sites have not been identified due to the lack of chromosome identification markers in Reddy et al.’ study [30]. Recently, we successfully labeled specific chromosomes using oligonucleotides (oligos) libraries as probes in cucumber [31]. In this study, the distribution of the 5S and 45S rDNA in all Citrullus species including three C. lanatus subspecies were investigated and the chromosomes with rDNA sites were identified using synthesized watermelon oligos probes to gain cyto-evolution at the chromosome level and resolve relationships among Citrullus species.
Methods
Plant materials and chromosome preparation
The tested Citrullus species included watermelon cultivar 97103 (C. lanatus subsp. vulgaris, 2n = 2x =22) which a draft genome has been published [29], C. lanatus subsp. mucosospermus (PI 270144, 2n = 2x =22), C. lanatus subsp. lanatus (C. amarus, PI 500335, 2n = 2x =22), C. colocynthis (PI 386021, 2n = 2x =22), C. ecirrhosus (PI 632751, 2n = 2x =22), C. rehmii (Grif 16376, 2n = 2x =22) and A. naudinianus (PI 596690, PI 671961 and Grif 14021, 2n = 2x =24) in this study. In order to learn about the rDNA distribution information in more species closely related with Citrullus species, several species belonging to other genera in Benincaseae tribe were also analyzed, including Cucumis sativus inbred line 9930 (2n = 2x =14), Cucumis metuliferus (PI 482465, 2n = 2x =24), Lagenaria siceraria (PI 379367, 2n = 2x = 22), Zehneria mariothii (PI 596649, 2n = 2x = 24), Melothria pendula (Grif 16574, 2n = 2x = 24), Benincasa fistulosa (PI 449332, 2n = 2x = 24), Coccinia sessilifolia (PI 596663, 2n = 2x = 24) and Diplocyclos palmatus (PI 618818, 2n = 2x = 24). Seeds of all PI and Grif lines were obtained from the U.S. National Plant Germplasm System (Ames, IA). The seeds of watermelon cultivar 97103 were obtained from National Engineering Research Center for Vegetables in Beijing Academy of Agriculture and Forestry Sciences. The seeds of C. sativus inbred line 9930 were obtained from Institute of Vegetables and Flowers in Chinese Academy of Agricultural Sciences. Root tips were harvested from germinated seeds, pretreated in 0.002 M 8-hydroxyquinoline at room temperature for 2 h to accumulate metaphase cells, and fixed in methanol:glacial acetic acid (3:1). After washing with water, root tips were macerated in 2 % cellulose and 1 % pectolyase at 37 °C for 2.5 h. Finally, the treated root tips were squashed and dried in flame. The slides were kept in −20 °C freezer for FISH.
rDNA probes and oligo probes
The plasmids of 5S and 45S rDNA which were cloned in the vector pUC18 were kindly provided by K. Arumuganthan, University of Nebraska. The 5S and 45S rDNA were labeled with digoxigenin-dUTP and biotin-dUTP via nick translation, respectively. Two oligos libraries were developed to identify cultivated watermelon chromosomes 4, 8 and 11 with rDNA sites reported by Guo et al. [29]. Library 1 contained 10000 oligos from watermelon chromosomes 4 and 8 at the densities of 2–3 oligos per kilobases, respectively. Library 2 contained 10000 oligos from watermelon chromosomes 8 and 11 at the densities of 2–3 oligos per kilobases, respectively. The region spanned by library 2 was located adjacently the region spanned by library 1 on chromosome 8. Thus, the chromosome with overlapping signals must be chromosome 8 and the others are either 4 or 11. Two oligos libraries were also developed to identify melon chromosomes 8, 10 and 12. Library 1 contained 10000 oligos from melon chromosomes 8 and 10 at the densities of 2–3 oligos per kilobases, respectively. Library 2 contained 10000 oligos from melon chromosomes 10 and 12 at the densities of 2–3 oligos per kilobases, respectively. The region spanned by library 2 was located adjacently the region spanned by library 1 on chromosome 10. Thus, the chromosome with overlapping signals must be chromosome 10 and the others are either 8 or 12. The oligo libraries were synthesized by MYcroarray (Ann Arbor, MI). Each synthesized oligo contained 48 bp of genomic sequence, a 5′ F primer, which included the T7 RNA polymerase promoter sequence, and a 3′ R primer. Amplification and labeling of oligos libraries was the same as the protocol developed by us [31].
Fluorescence in situ hybridization (FISH)
FISH was performed according to published protocols [32]. Firstly, slides were hybridized with oligo librariy 1 (biotin-labeled) and oligo librariy 2 (digoxigenin-labeled). After the first round of probing and image capture, coverslips were taken off carefully and wash the slides three times in 1X PBS (phosphate-buffered saline) (5 min each). The slides were then dehydrated in an ethanol series (70, 90, and 100 %, 5 min each), denatured again in 70 % formamide at 80 °C for 2 min, dehydrated in a second ethanol series, and reprobed with the 45S rDNA (biotin-labeled) and 5S rDNA (digoxigenin-labeled) sequences simultaneously. Biotin-labeled and digoxigenin-labeled probes were detected using a fluorescein isothiocyanate (FITC)-conjugated antibiotin antibody (Vector Laboratories) and a rhodamine-conjugated antidigoxigenin antibody (Roche Diagnostics), respectively. Chromosomes were counterstained with 4,6-diamidino-2-phenylindole (DAPI) in a VectaShield antifade solution (Vector Laboratories). Images were captured digitally using a CCD camera (QIMAGING, RETIGA-SRV, FAST 1394) attached to an Olympus BX63 epifluorescence microscope. Gray-scale images were captured for each color channel and then merged. Final image adjustments were done with Adobe Photoshop (Adobe Systems).
Results
Chromosome identification using watermelon oligos probes in Citrullus species and A. naudinianus
It is difficult to distinguish each chromosome in watermelon because the chromosomes are relatively small and are morphologically similar. Two oligos probes were developed to identify watermelon chromosomes 4, 8 and 11. As expected, two oligo probes produced bright FISH signals on cultivated watermelon chromosomes 4, 8 and 11 (Fig. 1a1, a3). We also performed FISH on metaphase chromosomes in all Citrullus species including three C. lanatus subspecies and A. naudinianus in order to investigate the potential of oligo probes for cross-species hybridization. Two probes generated FISH signals with similar intensity on three pairs of chromosomes in C. lanatus subsp. mucosospermus (Fig. 1b1, b3), C. colocynthis (Fig. 1c1, c3), C. lanatus subsp. lanatus (Fig. 1d1, d3), C. ecirrhosus (Fig. 1e1, e3), C. rehmii (Fig. 1f1, f3), and on two pairs of chromosomes in A. naudinianus (Fig. 1g1, g3). The counterparts of cultivated watermelon chromosomes 4, 8 and 11 in other Citrullus species or subspecies and A. naudinianus were also designated as chromosomes 4, 8 and 11 based on the homeologous relationship with the corresponding chromosome of watermelon.
Fig. 1.
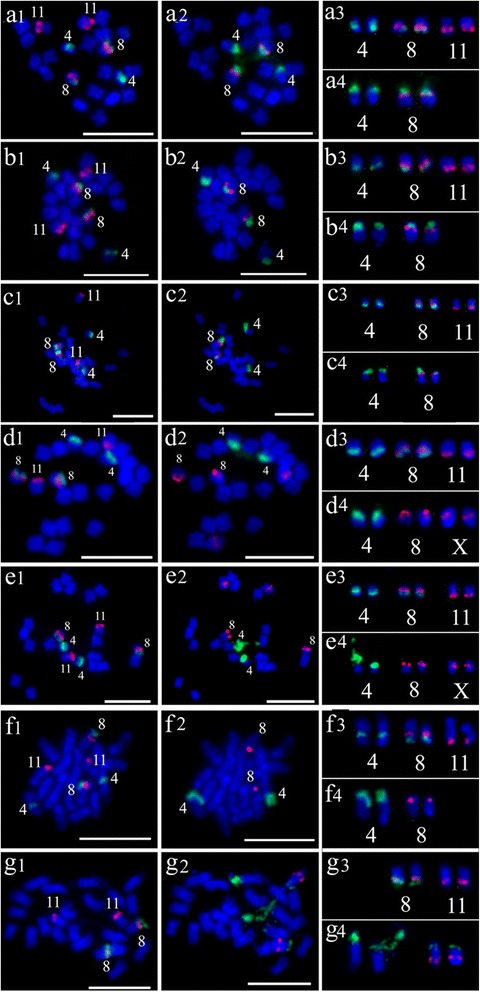
FISH of watermelon oligos probes, 5S and 45S rDNA in Citrullus species and Acanthosicyos naudinianus. a1-g1 FISH oligos probes Library 1 (green) and Library 2 (red) on metaphase chromosomes of C. lanatus subsp. vulgaris (a1), C. lanatus subsp. mucosospermus (b1), C. colocynthis (c1), C. lanatus subsp. lanatus (d1), C. ecirrhosus (e1), C. rehmii (f1) and A. naudinianus (g1), respectively. a2-g2 The same cells in A1-G1 were reprobed with 5S rDNA (red) and 45S rDNA (green) probes. a3-g3 The chromosomes with the signals of the oligos probes Library 1 (green) and Library 2 (red) in a1-g1. a4-g4 The chromosomes with the signals of 5S rDNA (red) and 45S rDNA (green) probes in a2-g2. Bars, 5 mm
Chromosomal distribution of 5S and 45S rDNA in Citrullus species and A. naudinianus
To determine the distribution of 5S and 45S rDNA in Citrullus species and A. naudinianus, following the FISH analysis of using oligos probes, the slides were washed and reprobed with 5S and 45S rDNA sequences simultaneously. The results were summarized Table 1. Consist with previous results [29], the genome of cultivated watermelon contained one 5S locus (red) and two 45S rDNA loci (green) on the chromosomes 4 and 8 (Fig. 1a2, a4). The 5S rDNA locus (red) was colocalized with the 45S rDNA loci (green) on the chromosome 8 (Fig. 1a2, a4). The number and location of 5S (red) and 45S (green) rDNA in the genomes of C. lanatus subsp. mucosospermus (Fig. 1b2, b4) and C. colocynthis (Fig. 1c2, c4) were identical to those in the genome of cultivated watermelon (Fig. 1a2, a4).
Table 1.
The number of 5S and 45S rDNA loci in different species
| Species | 2n | No. of 5S rDNA loci (Chromosome(s) bearing 5S rDNA) | No. of 45S rDNA loci (Chromosome(s) bearing 45S rDNA) |
|---|---|---|---|
| Citrullus lanatus subsp. vulgaris | 22 | 1 (8) | 2 (4, 8) |
| C. lanatus subsp. mucosospermus | 22 | 1 (8) | 2 (4, 8) |
| C. colocynthis | 22 | 1 (8) | 2 (4, 8) |
| C. lanatus subsp. lanatus | 22 | 2 (8, ?) | 1 (4) |
| C. ecirrhosus | 22 | 2 (8, ?) | 1 (4) |
| C. rehmii | 22 | 1 (8) | 1 (4) |
| Acanthosicyos naudinianus | 24 | 1 (?) | 2 (?, ?) |
| Lagenaria siceraria | 22 | 1 | 2 |
| Zehneria mariothii | 24 | 1 | 2 |
| Melothria pendula | 24 | 1 | 2 |
| Cucumis metuliferus | 24 | 1 | 2 |
| Benincasa fistulosa | 24 | 2 | 2 |
| Coccinia sessilifolia | 24 | 1 | 3 |
| Diplocyclos palmatus | 24 | 1 | 3 |
| Cucumis sativus | 14 | 1 | 5 |
The genomes of the C. lanatus subsp. lanatus (Fig. 1d2, d4) and C. ecirrhosus (Fig. 1e2, e4) contained two 5S loci (red) and one 45S rDNA locus (green) on the chromosomes 4. One 5S rDNA locus (with strong signals) located at the terminal end of chromosome 8, while the other 5S rDNA site (with weak signals) located intercalary near to the centromere on an unidentified chromosome x (Fig. 1d4, e4) instead chromosome 11 as that previously reported [29]. The genome of C. rehmii contained one 5S rDNA locus (red) on the chromosomes 8 and one 45S rDNA locus (green) on the chromosomes 4 (Fig. 1f2, f4). Like the cultivated watermelon, the genome of A. naudinianus also contained one 5S locus (red) and two 45S rDNA loci (green) (Fig. 1g2, g4). The 5S rDNA locus was located syntenic to one of the 45S rDNA loci. However, the 5S and 45S rDNA were far apart. Moreover, both 5S and 45S rDNA loci in A. naudinianus weren’t located on chromosomes 4 and 8 but on other chromosomes. In addition, A. naudinianus had 24 chromosomes while other Citrullus species had 22 chromosomes. We analyzed three A. naudinianus accessions (PI 596690, PI 671961 and Grif 14021) and the results were same.
Distribution of 5S and 45S rDNA in several genera of Benincaseae tribe
The distribution of 5S and 45S rDNA were also investigated in eight species from other genera in Benincaseae tribe, including Lagenaria siceraria (Fig. 2a), Zehneria mariothii (Fig. 2b), Melothria pendula (Fig. 2c), Cucumis metuliferus (Fig. 2d), Benincasa fistulosa (Fig. 2e), Coccinia sessilifolia (Fig. 2f), Diplocyclos palmatus (Fig. 2g) and Cucumis sativus (Fig. 2h) (Table 1). There was only one 5S locus (red) in these species except for B. fistulosa with two 5S loci (Fig. 2e). However, the number of 45S rDNA loci (green) showed a great difference. There were two 45S rDNA loci in L. siceraria (Fig. 2a), Z. mariothii (Fig. 2b), M. pendula (Fig. 2c), C. metuliferus (Fig. 2d) and B. fistulosa (Fig. 2e), three 45S rDNA loci in C. sessilifolia (Fig. 2f) and D. palmatus (Fig. 2g), and five 45S rDNA loci in C. sativas (Fig. 2h), respectively. The distribution pattern of 5S and 45S rDNA and chromosome number were exactly same between L. siceraria (Fig. 2a) and cultivated watermelon (Fig. 1a2, a4), also between C. sessilifolia (Fig. 2f) and D. palmatus (Fig. 2g).
Fig. 2.
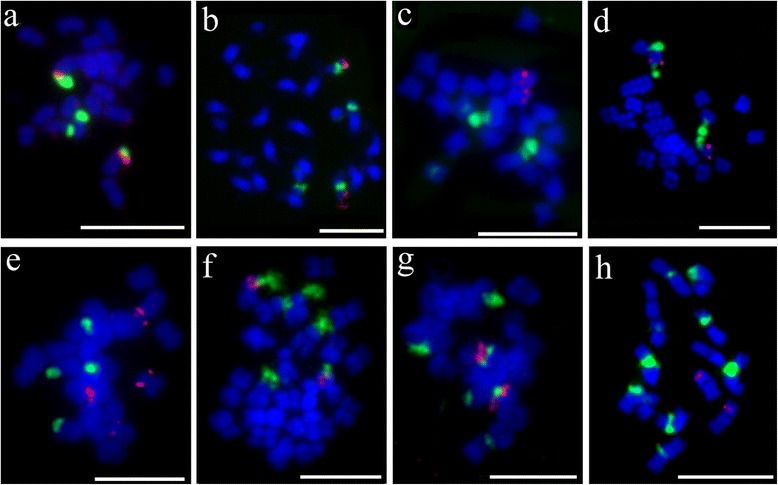
FISH of 5S rDNA (red) and 45S rDNA (green) probes in eight species from other genera in Benincaseae tribe. a Lagenaria siceraria. b Zehneria mariothii. c Melothria pendula. d Cucumis metuliferus. e Benincasa fistulosa. f Coccinia sessilifolia. g Diplocyclos palmatus. h Cucumis sativus. Bars, 5 mm
The 5S rDNA locus was located syntenic to one of the 45S rDNA loci in L. siceraria (Fig. 2a), Z. mariothii (Fig. 2b), C. metuliferus (Fig. 2d), C. sessilifolia (Fig. 2f) and D. palmatus (Fig. 2g). However, only in C. metuliferus, the 5S and 45S rDNA were far apart (Fig. 2d). We found that C. metuliferus (Fig. 2d) had same number and location of 5S and 45S rDNA loci as A. naudinianus (Fig. 1g2, g4). To further confirm the close relationship between C. metuliferus and A. naudinianus, we performed FISH on the chromosomes of C. metuliferus and A. naudinianus using two melon oligos probes which were used to identify chromosomes 8, 10 and 12 with 5S and 45S rDNA loci in Cucumis species (unpublished data). Two melon probes generated distinct FISH signals on three pairs of chromosomes in C. metuliferus (Fig. 3a1, a3) and A. naudinianus (Fig. 3b1, b3). Like melon, the chromosomes with same oligo probes signals were also designated as chromosomes 8, 10 and 12 in C. metuliferus and A. naudinianus. Following the FISH analysis of using oligos probes, the slides were washed and reprobed with 5S and 45S rDNA sequences simultaneously. The FISH results showed that two 45S rDNA loci (green) were on the chromosomes 8 and 10 in C. metuliferus (Fig. 3a2, a4) and A. naudinianus (Fig. 3b2, b4). The 5S rDNA locus (red) was located syntenic to the 45S rDNA loci on chromosome 10 (Fig. 3a4, b4). We also tested melon oligos probes on chromosomes of other Citrullus species and watermelon oligos probes on chromosomes of Cucumis species and L. siceraria which was closely related to watermelon. We did not detect unambiguous signals in these species using any of the four probes (data not shown). This was consistent with our previous study that unambiguous signals weren’t detected on chromosomes of watermelon and Cucurbita pepo (2n = 4x = 40) using any of three cucumber oligo probes [31]. Therefore, cross-genus hybridization was not feasible using oligo probes.
Fig. 3.
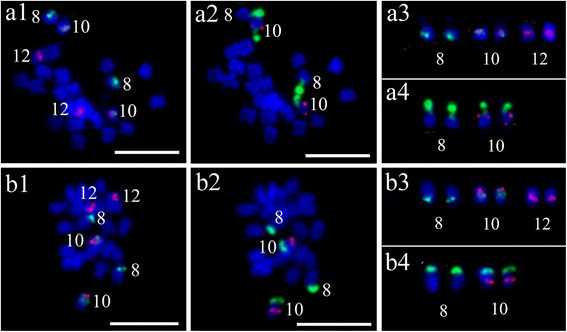
FISH of melon oligos probes, 5S and 45S rDNA in Cucumis metuliferus and Acanthosicyos naudinianus. a1-b1 FISH oligos probes Library 1 (green) and Library 2 (red) on metaphase chromosomes of C. metuliferus (a1) and A. naudinianus (b1). a2-b2 The same cells in a1-b1 were reprobed with 5S rDNA (red) and 45S rDNA (green) probes. a3-b3 The chromosomes with the signals of the oligos probes Library 1 (green) and Library 2 (red) in a1-b1. a4-b4 The chromosomes with the signals of 5S rDNA (red) and 45S rDNA (green) probes in a2-b2. Bars, 5 mm
Discussion
The genus Citrullus belongs to the Benincaseae tribe of the Cucurbitaceae family [1]. The Benincaseae is a relatively large tribe with 204–214 species in 24 genera [33]. The phylogenetic relationships among species in this tribe were assessed by morphological [34, 35], geographical [4, 36], biochemical [37] or nucleotide data [4, 36, 38, 39]. In this study, we investigated the distribution of 5S and 45S rDNA in several genera located on the different branches of the phylogenetic tree of the tribe Benincaseae [36]. We found L. siceraria and cultivated watermelon had exactly same rDNA distribution. Similarly, C. sessilifolia had identical rDNA distribution pattern as D. palmatus. This was in accordance with the results based on the molecular phylogenetic studies, which revealed a close relationship between the genera Citrullus and Lagenaria, and between the genera Coccinia and Diplocyclos [4, 36].
Consist with previous studies [29, 30], the present FISH analysis demonstrated that the rDNA gene loci showed wide differences among the Citrullus species. C. lanatus subsp. vulgaris (cultivated watermelon) had same number and location of 5S and 45S rDNA loci to those in C. lanatus subsp. mucosospermus and C. colocynthis, but differed from those in C. lanatus subsp. lanatus. Our results are concordant with assumptions that the progenitor of the cultivated watermelon might be C. colocynthis [12, 13] and C. lanatus subsp. mucosospermus is the recent ancestor of C. lanatus subsp. vulgaris [29]. Several reports based on nuclear and plastid data of the genus Citrullus also confirmed evolutionary closeness among cultivated watermelon, C. lanatus subsp. mucosospermus and C. colocynthis. For example, the analyses of the sequence variation at cpDNA regions also confirmed the relationship because some of the C. lanatus var. lanatus accessions shared a unique substitution at the trnE-trnT region with all C. colocynthis accessions [40]. Schaefer et al. [36] also confirmed that the watermelon and C. colocynthis evolved from a common ancestor. Our results also supported Chomicki and Renner’ findings [4], which revealed that C. lanatus subsp. lanatus was not a wild form or progenitor of the cultivated watermelon but instead was a separate crop species, domesticated independently. In addition, present cytogenetic analysis showed that C. lanatus subsp. lanatus and C. ecirrhosus were more closely related to each other than they are to other Citrullus species. Based on the current FISH analysis and previous molecular phylogenetics [4] and cytogenetic studies [29, 30], a scheme about phylogenetic relationships among the Citrullus species was given (Fig. 4).
Fig. 4.
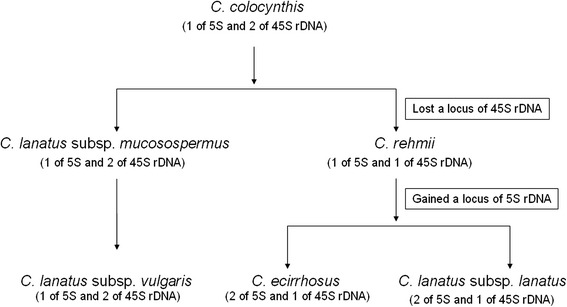
A scheme showing phylogenetic relationships among the Citrullus species
One of the most striking observations from the present results was that the number, size and location of 5S and 45S rDNA signals were exactly identical between A. naudinianus and C. metuliferus in genus Cucumis. Moreover, two melon oligos libraries generated distinct FISH signals in A. naudinianus. The number and location of rDNA and oligos probes signals were completely identical between A. naudinianus and C. metuliferus. However, unambiguous signals weren’t detected when melon oligos probes hybridized to chromosomes of Citrullus species and watermelon oligos probes hybridized to chromosomes of Cucumis species. In addition, A. naudinianus had 24 chromosomes while other Citrullus species had 22 chromosomes. In the genus Cucumis, C. sativus is the only species with chromosome number 14, whereas the rest of the Cucumis species have a basic number 24 [41]. Our previous study also confirmed that the A. naudinianus had closer genetic affinity with the genus Cucumis than with Citrullus by comparative genomic in situ hybridization [42]. All above results suggest that A. naudinianus is more closely related to Cucumis than to Citrullus or Acanthosicyos. Interestingly, both watermelon and melon oligos probes generated distinct FISH signals on A. naudinianus chromosomes although cross-genus hybridization was not feasible using oligo probes in other species. Therefore, A. naudinianus could be a link bridge between the Citrullus and the Cucumis.
Consistent identification of individual chromosomes in a species is the foundation for successful cytogenetic research. FISH signals are reliable markers for chromosome identification. Most common FISH probes used in chromosome identification have been repetitive DNA elements or large genomic DNA clones [43–47]. Large genomic clones often contain dispersed repetitive sequences that will cause high background signal in FISH and cannot be used as chromosome-specific FISH probes [48]. The watermelon chromosomes are relatively small in size and are morphologically similar. Repetitive DNA elements were unavailable for differentiating all watermelon chromosomes. Recently, eleven BAC clones were used to differentiate eleven watermelon chromosomes and assign linkage groups to their corresponding chromosomes [49]. Furthermore, using the eleven BAC clones, 5S and 45S rDNA loci were assigned to chromosomes 4 and 8 in the genomes of cultivated watermelon and C. lanatus subsp. mucosospermus, and chromosomes 4, 8 and 11 in C. lanatus subsp. lanatus [29]. However, in present study we found that one of 5S rDNA sites was on an unidentified chromosome rather than on chromosome 11 by using oligos probes to identify chromosome 11 in C. lanatus subsp. lanatus.
Conclusions
The present study confirmed evolutionary closeness among cultivated watermelon, C. lanatus subsp. mucosospermus and C. colocynthis. Our result also supported that C. lanatus subsp. lanatus was not a wild form of the watermelon instead was a separate crop species. In addition, present cytogenetic analysis suggested that A. naudinianus was more closely related to Cucumis than to Citrullus or Acanthosicyos, but with a unique position and may be a link bridge between the Citrullus and the Cucumis. In future, we will develope chromosome-specific oligos probes for identifying each watermelon chromosome. Furthermore, bulked oligo probes will be designed to cover an entire watermelon chromosome. Thus, interchromosomal rearrangements and karyotype evolution among related Citrullus species can be revealed in more detail through cross-species chromosome painting. The information will be useful for collection and utilization of genetic resources for cultivated watermelon improvement.
Ethics
Not applicable.
Consent to publish
Not applicable.
Availability of data and materials
All data are contained within the manuscript.
Acknowledgments
The authors are grateful to Kassandra Semrau from MYcroarray (http://mycroarray.com) for designing oligo probes and Dr. H. Miao from Institute of Vegetables and Flowers in Chinese Academy of Agricultural Sciences for supplying the seeds of C. sativus inbred line 9930. We also thank the anonymous reviewers for their helpful comments.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 31271350) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Abbreviations
- FISH
fluorescent in situ hybridization
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YHH designed the study, performed the analysis and wrote the paper. YX, JMW and ZYL participated in the design of the study, analyzed the data and critically revised the manuscript. KPL, YXW, HZ, YW and XML carried out the experiments and acquisition of original data. KPL analyzed the data and participated in writing. All authors read and approved the final manuscript.
Contributor Information
Kun-Peng Li, Email: 896492496@qq.com.
Yun-Xiang Wu, Email: 463727355@qq.com.
Hong Zhao, Email: zhaohong@nercv.org.
Yan Wang, Email: 1024489848@qq.com.
Xing-Ming Lü, Email: 2856423758@qq.com.
Ji-Ming Wang, Email: pomology@163.com.
Yong Xu, Email: xuyong@nercv.org.
Zong-Yun Li, Email: zongyunli@jsnu.edu.cn.
Yong-Hua Han, Email: hanyonghua@jsnu.edu.cn.
References
- 1.Robinson RW, Decker-Walters DS. Cucurbits. New York: CAB International; 1997. [Google Scholar]
- 2.De Winter B. A new species of Citrullus (Benincaseae) from the Namib Desert, Namibia. Bothalia. 1990;20:209–11. doi: 10.4102/abc.v20i1.900. [DOI] [Google Scholar]
- 3.Jeffrey C. Cucurbitaceae. In: Hanelt P, editor. Mansfeld’s encyclopedia of agricultural and horticultural crops. 3. Berlin: Springer; 2001. pp. 1510–57. [Google Scholar]
- 4.Chomicki G, Renner SS. Watermelon origin solved with molecular phylogenetics including Linnaean material: another example of museomics. New Phytol. 2015;205:526–32. doi: 10.1111/nph.13163. [DOI] [PubMed] [Google Scholar]
- 5.Khoshoo TN, Vij P. Biosystematics of Citrullus vulgaris var. fistulosus. Caryologia. 1963;16:541–52. [Google Scholar]
- 6.Zamir D, Navot N, Rudich J. Enzyme polymorphism in Citrullus lanatus and C. colocynthis in Israel and Sinai. Plant Syst Evol. 1984;146:163–70. doi: 10.1007/BF00989543. [DOI] [Google Scholar]
- 7.Navot N, Zamir D. Isozyme and seed protein phylogeny of the genus Citrullus (Cucurbitaceae) Plant Syst Evol. 1987;156:61–7. doi: 10.1007/BF00937202. [DOI] [Google Scholar]
- 8.Jarret RL, Merrick LC, Holms T, Evans J, Aradhya MK. Simple sequence repeats in watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai] Genome. 1997;40:433–41. doi: 10.1139/g97-058. [DOI] [PubMed] [Google Scholar]
- 9.Jarret RL, Newman M. Phylogenetic relationships among species of Citrullus and the placement of C. rehmii De Winter as determined by Internal Transcribed Spacer (ITS) sequence heterogeneity. Genet Resour Crop Evol. 2000;47:215–22. doi: 10.1023/A:1008754719102. [DOI] [Google Scholar]
- 10.Levi A, Thomas CE, Keinath AP, Wehner TC. Genetic diversity among watermelon (Citrullus lanatus and Citrullus colocynthis) accessions. Genet Resour Crop Evol. 2001;48:559–66. doi: 10.1023/A:1013888418442. [DOI] [Google Scholar]
- 11.Wasylikowa K, van der Veen M. An archaeobotanical contribution to the history of watermelon, Citrullus lanatus (Thunb.) Matsum. & Nakai (syn. C. vulgaris Schrad.) Veget Hist Archaeobot. 2004;13:213–7. doi: 10.1007/s00334-004-0039-6. [DOI] [Google Scholar]
- 12.Whitaker TW. Cytogenetical and phylogenetic studies in the Cucurbitaceae. Bot Gaz. 1933;93:780–90. doi: 10.1086/334347. [DOI] [Google Scholar]
- 13.Shimotsuma M. Cytogenetical studies in the genus Citrullus. IV. Intra and inter-specific hybrids between C. colocynthis Schr. and C. vulgaris Schrad. Jpn J Gen. 1960;35:303–12. doi: 10.1266/jjg.35.303. [DOI] [Google Scholar]
- 14.Zeven AC, Zhukovsky PM. Dictionary of cultivated plants and their centres of diversity. Wageningen: Centre for Agriculture Publishing and Documentation; 1975. [Google Scholar]
- 15.Maynard DN. An introduction to the watermelon. Alexandria: ASHS Press; 2001. [Google Scholar]
- 16.Singh NP, Matta NK. Levels of seed proteins in Citrullus and Praecitrullus accessions. Plant Syst Evol. 2010;290:47–56. doi: 10.1007/s00606-010-0347-5. [DOI] [Google Scholar]
- 17.Dane F, Lang P. Sequence variation at cpDNA regions of watermelon and related wild species: implications for the evolution of Citrullus haplotypes. Am J Bot. 2004;91:1922–9. doi: 10.3732/ajb.91.11.1922. [DOI] [PubMed] [Google Scholar]
- 18.Dane F, Lang P, Bakhtiyarova R. Comparative analysis of chloroplast DNA variability in wild and cultivated Citrullus species. Theor Appl Genet. 2004;108:958–66. doi: 10.1007/s00122-003-1512-9. [DOI] [PubMed] [Google Scholar]
- 19.Cerbah M, Coulaud J, Siljak-Yakovlev S. rDNA organization and evolutionary relationships in the genus Hypochaeris (Asteraceae) J Hered. 1998;89:312–8. doi: 10.1093/jhered/89.4.312. [DOI] [Google Scholar]
- 20.Shan F, Yan G, Plummer A. Cyto-evolution of Boronia genomes revealed by fluorescence in situ hybridization with rDNA probes. Genome. 2003;46:507–13. doi: 10.1139/g03-009. [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Gill BS. Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome. 1994;37:717–25. doi: 10.1139/g94-102. [DOI] [PubMed] [Google Scholar]
- 22.Garcia S, Garnatje T, Kovařík A. Plant rDNA database: ribosomal DNA loci information goes online. Chromosoma. 2012;121:389–94. doi: 10.1007/s00412-012-0368-7. [DOI] [PubMed] [Google Scholar]
- 23.Garcia S, Garnatje T, McArthur ED, Siljak-Yakovlev S, Vallès J. Ribosomal DNA, heterochromatin, and correlation with genome size in diploid and polyploid North American endemic sagebrushes (Artemisia, Asteraceae) Genome. 2009;52:1012–24. doi: 10.1139/G09-077. [DOI] [PubMed] [Google Scholar]
- 24.Maluszynska J, Heslop-Harrison JS. Physical mapping of rDNA loci in Brassica species. Genome. 1993;36:774–81. doi: 10.1139/g93-102. [DOI] [PubMed] [Google Scholar]
- 25.Seijo JG, Lavia GI, Fernandez A, Krapovickas A, Ducasse D, Moscone EA. Physical mapping of the 5S and 18S-25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipanensis are the wild diploid progenitors of A. hypogaea (Leguminosae) Am J Bot. 2004;91:1294–303. doi: 10.3732/ajb.91.9.1294. [DOI] [PubMed] [Google Scholar]
- 26.Cai Q, Zhang D, Lui ZL, Wang XR. Chromosomal localization of 5S and 18S rDNA in five species of subgenes strobus and their implications for genome evolution of pinus. Ann Bot. 2006;97:715–22. doi: 10.1093/aob/mcl030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung MC, Lee YI, Cheng YY, Chou YJ, Lu CF. Chromosomal polymorphism of ribosomal genes in the genus Oryza. Theor Appl Genet. 2008;116:745–53. doi: 10.1007/s00122-007-0705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berjano R, Roa F, Talavera S, Guerra M. Cytotaxonomy of diploid and polyploid Aristolochia (Aristolochiaceae) species based on the distribution of CMA/DAPI bands and 5S and 45S rDNA sites. Plant Syst Evol. 2009;280:219–27. doi: 10.1007/s00606-009-0184-6. [DOI] [Google Scholar]
- 29.Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 2013;45:51–8. doi: 10.1038/ng.2470. [DOI] [PubMed] [Google Scholar]
- 30.Reddy UK, Aryal N, Islam-Faridi N, Tomason YR, Levi A, Nimmakayala P. Cytomolecular characterization of rDNA distribution in various Citrullus species using fluorescent in situ hybridization. Genet Resour Crop Evol. 2013;60(7):2091. doi: 10.1007/s10722-013-9976-1. [DOI] [Google Scholar]
- 31.Han Y, Zhang T, Thammapichai P, Weng Y, Jiang J. Chromosome-specific painting in Cucumis species ysing bulked oligonucleotides. Genetics. 2015;200(3):771–9. doi: 10.1534/genetics.115.177642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang JM, Gill BS, Wang GL, Ronald PC, Ward DC. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci U S A. 1995;92:4487–91. doi: 10.1073/pnas.92.10.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer H, Renner SS. Phylogenetic relationships in the order Cucurbitales and a new classification of the gourd family (Cucurbitaceae) Taxon. 2011;60:122–38. [Google Scholar]
- 34.Jeffrey C. Notes on Cucurbitaceae, including a proposed new classification of the family. Kew Bull. 1962;15:337–71. doi: 10.2307/4115586. [DOI] [Google Scholar]
- 35.Jeffrey C. A note on pollen morphology in Cucurbitaceae. Kew Bull. 1964;17:473–6. doi: 10.2307/4113823. [DOI] [Google Scholar]
- 36.Schaefer H, Heibl C, Renner SS. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc R Soc B. 2009;276:843–51. doi: 10.1098/rspb.2008.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunnill PM, Fowden L. The amino acids of seeds of the Cucurbitaceae. Phytochemistry. 1965;4:933–44. doi: 10.1016/S0031-9422(00)86271-8. [DOI] [Google Scholar]
- 38.Chung SM, Decker-Walters DS, Staub JE. Genetic relationships within the Cucurbitaceae as assessed by consensus chloroplast simple sequence repeats (ccSSR) marker and sequence analyses. Can J Bot. 2003;81:814–32. doi: 10.1139/b03-074. [DOI] [Google Scholar]
- 39.Decker-Walters DS, Chung SM, Staub JE. Plastid sequence evolution: a new pattern of nucleotide substitutions in the Cucurbitaceae. J Mol Evol. 2004;58:606–14. doi: 10.1007/s00239-004-2585-z. [DOI] [PubMed] [Google Scholar]
- 40.Dane F, Liu J. Diversity and origin of cultivated and citron type watermelon (Citrullus lanatus) Genet Resour Crop Evol. 2007;54:1255–65. doi: 10.1007/s10722-006-9107-3. [DOI] [Google Scholar]
- 41.Dane F, Tsuchiya T. Chromosome studies in the genus Cucumis. Euphytica. 1976;25:367–74. doi: 10.1007/BF00041569. [DOI] [Google Scholar]
- 42.Chang KW, Shen JJ, Li ZY, Han YH. Genomic homology between Citrullus lanatus and its close relatives revealed by comparative genomic in situ hybridization. Plant Sci J. 2012;30:402–6. doi: 10.3724/SP.J.1142.2012.40402. [DOI] [Google Scholar]
- 43.Mukai Y, Nakahara Y, Yamamoto M. Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome. 1993;36:489–94. doi: 10.1139/g93-067. [DOI] [PubMed] [Google Scholar]
- 44.Kato A, Lamb JC, Birchler JA. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci U S A. 2004;101:13554–9. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng ZK, Buell CR, Wing RA, Gu MH, Jiang JM. Toward a cytological characterization of the rice genome. Genome Res. 2001;11:2133–41. doi: 10.1101/gr.194601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Findley SD, Cannon S, Varala K, Du J, Ma J, Hudson ME, et al. A fluorescence in situ hybridization system for karyotyping soybean. Genetics. 2010;185:727–44. doi: 10.1534/genetics.109.113753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong ZY, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci U S A. 2011;108:7908–13. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki G, Ogaki Y, Hokimoto N, Xiao L, Kikuchi-Taura A, Harada C, et al. Random BAC FISH of monocot plants reveals differential distribution of repetitive DNA elements in small and large chromosome species. Plant Cell Rep. 2012;31:621–8. doi: 10.1007/s00299-011-1178-8. [DOI] [PubMed] [Google Scholar]
- 49.Ren Y, Zhao H, Kou Q, Jiang J, Guo S, Zhang H, et al. A high resolution genetic map anchoring scaffolds of the sequenced watermelon genome. PLoS One. 2012;7:e29453. doi: 10.1371/journal.pone.0029453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.


