ABSTRACT
Macroautophagy is primarily a degradative process that cells use to break down their own components to recycle macromolecules and provide energy under stress conditions, and defects in macroautophagy lead to a wide range of diseases. Atg9, conserved from yeast to mammals, is the only identified transmembrane protein in the yeast core macroautophagy machinery required for formation of the sequestering compartment termed the autophagosome. This protein undergoes dynamic movement between the phagophore assembly site (PAS), where the autophagosome precursor is nucleated, and peripheral sites that may provide donor membrane for expansion of the phagophore. Atg9 is a phosphoprotein that is regulated by the Atg1 kinase. We used stable isotope labeling by amino acids in cell culture (SILAC) to identify phosphorylation sites on this protein and identified an Atg1-independent phosphorylation site at serine 122. A nonphosphorylatable Atg9 mutant showed decreased autophagy activity, whereas the phosphomimetic mutant enhanced activity. Electron microscopy analysis suggests that the different levels of autophagy activity reflect differences in autophagosome formation, correlating with the delivery of Atg9 to the PAS. Finally, this phosphorylation regulates Atg9 interaction with Atg23 and Atg27.
KEYWORDS: autophagy, lysosome, stress, vacuole, yeast
Introduction
Autophagy refers to a group of highly conserved cellular processes in which cytoplasmic components are degraded within the lysosome in most of the more complex eukaryotes or the vacuole in yeast and plants. The resulting macromolecular constituents are recycled and used as building blocks for anabolic pathways or to generate energy through catabolism.1 Autophagy in yeast can be divided into 2 main types, macroautophagy and microautophagy. Macroautophagy (hereafter called autophagy) is the major process in which random cytoplasm is sequestered within a double-membrane structure, the phagophore, which eventually matures into a vesicle termed the autophagosome; generation of the autophagosome is a distinguishing feature between macroautophagy and microautophagy because the latter process involves direct uptake at the vacuole limiting membrane without the use of a phagophore or autophagosome. After fusion of the autophagosome with the vacuole, and degradation in the vacuole lumen, the breakdown products are released back into the cytosol.2 Autophagy can be selective or nonselective. Nonselective autophagy is used for the turnover of bulk cytoplasm, whereas selective autophagy specifically targets damaged or superfluous organelles, including mitochondria and peroxisomes.3,4
Over the past 2 decades, 41 autophagy-related (ATG) genes have been identified in fungi.5 Among them, one subset including 18 genes is shared by both nonselective and selective autophagy and is required for autophagosome formation, and thus the corresponding gene products are termed the core machinery of autophagosome formation; these proteins can be divided into different functional groups.2,6 Although our understanding of the molecular mechanism of autophagy has increased tremendously since the discovery and initial characterization of the Atg proteins, the complex mechanisms involved in the regulation of autophagy are still unclear. Transcriptional, post-transcriptional and post-translational regulation are all used to modulate autophagy in order to adapt to different types of environmental stress,7-9 and several Atg proteins are phosphorylated.10-14
Atg9, conserved from yeast to mammals, is the only transmembrane protein identified in the yeast autophagy core machinery.15 One of the unique features of Atg9 concerns its subcellular distribution. Whereas most Atg proteins are localized to the PAS, in addition to a cytosolic pool, Atg9 has multiple punctate populations in a yeast cell. Atg9 is detected at the PAS along with most of the other Atg proteins; however, Atg9 is additionally present at peripheral sites, also termed Atg9 reservoirs and tubulovesicular clusters.16 The Atg9 peripheral sites are found adjacent to mitochondria, but are not directly associated with this organelle, and newly synthesized Atg9 is delivered to these sites through part of the secretory pathway.17 Studies using high-sensitivity microscopy have shown that most of the punctate Atg9 structures are highly mobile in the cytoplasm.18 Thus, Atg9 may move between these peripheral sites and the PAS, and earlier studies have suggested that this cycling of Atg9 is required for autophagosome formation, functioning in some manner to direct membrane to the expanding phagophore.16,19-21 In mammalian cells, ATG9 also displays dynamic cycling, moving between the trans-Golgi network and endosomes in response to nutritional changes. A subpopulation of ATG9 colocalizes with both LC3, a mammalian homolog of Atg8, and RAB7, a late endosomal protein.22
Several Atg proteins have important roles in regulating Atg9 cycling. The anterograde movement of Atg9 from peripheral sites to the PAS requires Atg23, Atg27 and Atg11 (the latter is required primarily in selective types of autophagy).20 The retrograde trafficking of Atg9 from the PAS depends on the Atg1-Atg13 complex, the Atg2-Atg18 complex and the phosphatidylinositol 3-kinase complex.16,19 Self-interaction of Atg9 is also important for its PAS localization.23
Recent studies have shown that the Atg1 kinase directly phosphorylates Atg9 in an early step of regulation.14 In this study, we identified Atg9 serine 122 (S122), a site that is not a direct target of Atg1, as being important for Atg9 function in autophagy activity. Phosphorylation of S122 regulates Atg9 anterograde trafficking by mediating its interaction with Atg23 and Atg27. These findings provide new evidence of the importance of post-translational modification of Atg9 and regulation of autophagy activity.
Results
Determination of Atg9 phosphorylation sites
Atg1 is the only protein kinase of the core autophagy machinery,24 and as such it has received considerable attention. Several of the Atg proteins are phosphorylated, but until recently it was not known if any of these proteins were Atg1 substrates. Many other protein kinases (i.e., proteins that are not part of the autophagy core machinery) also modulate autophagy activity,25-28 but in most cases the relevant targets have not been identified. Atg9 plays a key role in autophagy, and we recently demonstrated that the amount of the Atg9 protein correlates with the rate of autophagosome formation.29 The functional pool of Atg9—the part of the population actively engaged in phagophore expansion—is presumed to be that at the PAS. We carried out a SILAC analysis to identify phosphorylation sites on Atg9 that might affect its movement to the PAS, and decided to focus on modifications that were independent of direct Atg1 kinase function, because recent studies have already identified Atg1-dependent sites in this protein.14
To identify Atg1-independent phosphorylation sites we performed the SILAC analysis comparing wild-type and atg1Δ strains. Phosphorylation scores were defined as mean log2 (atg1Δ:wild type [WT]); potential Atg1 targets should have a score <-1, reflecting a decrease in phosphorylation, whereas changes in phosphorylation that were not directly dependent on Atg1 would have positive scores. From this analysis we identified 4 putative sites in Atg9 that demonstrated alterations in phosphorylation under conditions of nitrogen starvation: S864, T794, S792 and S122 (Fig. 1A). Among these sites, only S864 showed a negative enrichment score suggesting it was a direct substrate of Atg1. This site was identified in the study by Papinski et al.,14 but was relatively unaffected by Atg1 kinase activity. This previous analysis also identified S792, T794 and S122 as being phosphorylated independent of Atg1.
Figure 1.
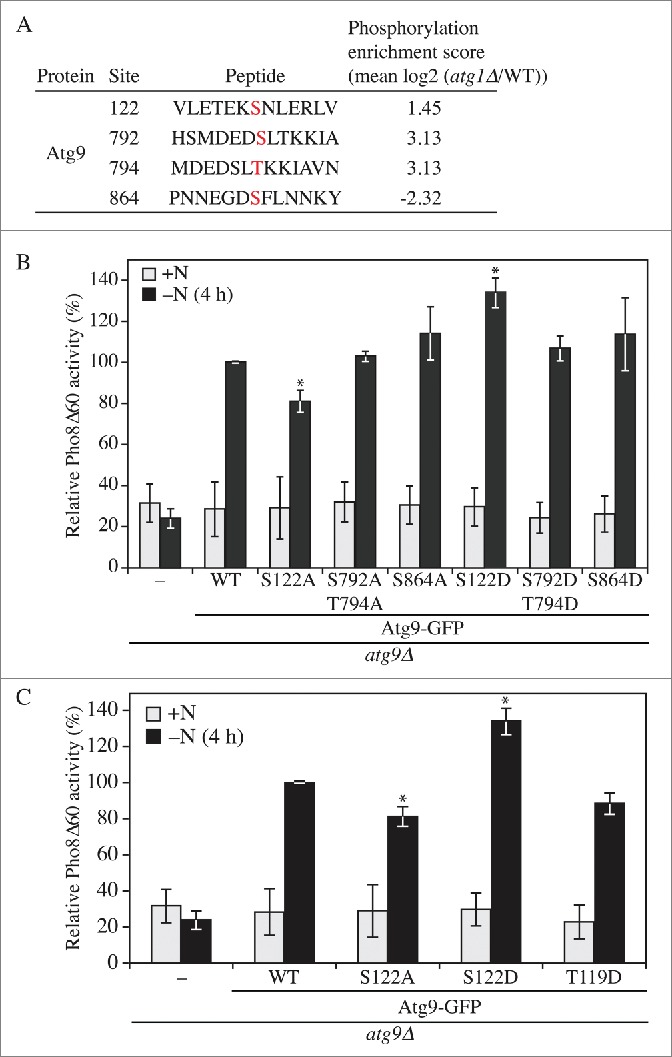
Phosphorylation of Atg9 S122 is important for nonselective autophagy. (A) Wild-type (WT; SEY6210) and atg1Δ cells were collected after 2 h nitrogen starvation and subjected to SILAC analysis. A phosphorylation enrichment score was identified for 4 sites of Atg9. (B, C) WLY176 cells were transformed with empty vector (pRS406) or a plasmid containing WT or different phosphorylation mutants of Atg9-GFP as indicated. Cells were cultured in YPD (+N) to midlog phase, and shifted to SD-N (-N) for 4 h. The Pho8Δ60 assay was performed as described in Materials and Methods. Error bars correspond to the standard deviation and were obtained from 3 independent repeats. Two-tailed t test was used for statistical significance; ∗p < 0.05.
Phosphorylation of Atg9 S122 is important for nonselective autophagy
To determine if any of these phosphorylation sites plays a role in autophagy we generated 3 nonphosphorylatable Atg9 mutants, S122A, S792A T794A, and S864A, and 3 phosphomimetic mutants, S122D, S792D T794D and S864D, through site-directed mutagenesis.30 We then transformed an atg9Δ strain with a plasmid encoding either WT Atg9 fused to the green fluorescent protein (Atg9-GFP), the Atg9 mutants or an empty vector, and monitored autophagy activity using the Pho8Δ60 assay.31 Pho8 is a cytosolic derivative of a vacuolar phosphatase. The truncated version, Pho8Δ60, lacking its N-terminal transmembrane domain, resides in the cytosol and can only be delivered to the vacuole through autophagy, where a propeptide is removed to generate the active enzyme. The atg9Δ strain with an empty vector displayed a significant block in autophagy in nitrogen starvation conditions (Fig. 1B and S1C). In contrast, the strain expressing Atg9-GFP restored the autophagy activity to ∼60% of the wild-type strain (Fig. S1A and S1E); the activity of this strain was set to 100% and used for normalization (Fig. 1B and S1C). Out of the 6 mutants tested, changes at S792, T794 and S864 did not result in significant differences relative to the WT. In contrast, S122A displayed an ∼20% decrease, and S122D an ∼30% increase, of Pho8Δ60 activity, suggesting that the phosphorylation of Atg9 S122 is important for autophagy.
Atg9 T119 was not identified as a phosphosite in the previous SILAC analysis14 or in our present study. We decided to mutate this site due to its proximity to S122, in particular to determine whether the addition of a negative charge in this part of the protein would result in a phenotype similar to that seen with the S122D mutation. In contrast to the latter, the T119D mutant did not display an increase in activity, but instead resulted in a slight decrease in Pho8Δ60 activity, suggesting the specificity of the S122D phenotype (Fig. 1C and S1D). Atg9 homologs have been previously identified in other organisms including plants and humans,32,33 and the serine at position 122 is highly conserved among fungi (Fig. S1B); similarly, human ATG9A has a serine at position 123. Thus, regulation of Atg9 through post-translational modification at this site might correspond to a conserved mechanism.
Phosphorylation of Atg9 S122 is important for selective autophagy
Atg9 is one of the key proteins in the core machinery that is shared between nonselective and selective autophagy.34 Accordingly, we next decided to examine the requirement for phosphorylation at S122 in selective types of autophagy. Aminopeptidase I (Ape1), a vacuole resident hydrolase, is initially synthesized as a cytosolic precursor (prApe1), and is delivered to the vacuole through either nonselective autophagy or the cytoplasm-to-vacuole targeting (Cvt) pathway, depending on nutrient conditions;35-38 in either case delivery is a selective process depending on a receptor protein, Atg19.5,39,40 Upon delivery to the vacuole, the propeptide of prApe1 is enzymatically removed generating the mature hydrolase. This processing event is easily detected as a change in molecular mass following SDS-PAGE. To simplify our analysis, we took advantage of the Cvt pathway phenotype of the vac8Δ background to monitor selective autophagy; in the vac8Δ strain only prApe1 accumulates in rich conditions, but there is rapid vacuolar delivery following the induction of autophagy.41 Thus, a shift from rich to starvation conditions essentially corresponds to a pulse-chase analysis, and prevents the complicating accumulation of mature Ape1 that would otherwise be present. In the atg9Δ vac8Δ strain only prApe1 could be detected even after the induction of autophagy (Fig. 2A). The block of prApe1 maturation during starvation was rescued by integration of WT ATG9 back into the chromosome. This maturation was partially blocked in cells expressing Atg9S122A after 2 h starvation. At this same time point we could not detect a difference between the strain expressing WT Atg9 and Atg9S122D, because essentially all of the protein was in the mature form. Therefore, we examined earlier time points and found accelerated maturation in the Atg9S122Dcells relative to the WT (Fig. 2B).
Figure 2.
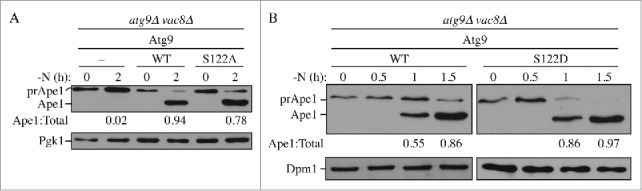
Phosphorylation of Atg9 S122 is important for selective autophagy. (A, B) Processing of prApe1 was monitored. atg9Δ vac8Δ cells, transformed with empty vector or a plasmid containing WT Atg9 and either (A) Atg9S122A or (B) Atg9S122D, were cultured in YPD to early log phase (0D600= 0.5) and shifted to SD-N for 0–2 h as indicated. Cells were collected and protein extracts were analyzed by western blot with anti-Ape1 antibody and either anti-Pgk1 or anti-Dpm1 (loading control) antiserum or antibody. The ratio of Ape1:total Ape1 (preApe1 + Ape1) was measured and normalized to that of WT cells, which was set to 100%.
Phosphorylation of Atg9 S122 regulates autophagy by mediating autophagosome formation
At a mechanistic level there are 2 fundamental ways to modulate the magnitude of autophagy: changing the number or the size of autophagosomes. In yeast, the large size of the vacuole makes it possible to accumulate autophagic bodies, which correspond to the single-membrane compartments that result from the fusion of an autophagosome with a vacuole, when their degradation is blocked. The size and number of the accumulated autophagic bodies can then be determined through morphometric analysis by transmission electron microscopy (EM). To carry out this analysis we utilized strains harboring deletions in the PEP4 gene to prevent autophagic body breakdown. The WT (pep4Δ Atg9) and mutant S122A (pep4Δ Atg9S122A) and S122D (pep4Δ Atg9S122D) strains were grown to mid-log phase and then shifted to nitrogen starvation conditions for 4 h, before being processed for imaging by EM (Fig. 3A); the size and number of autophagic bodies were quantified and estimated as described previously.21
Figure 3.
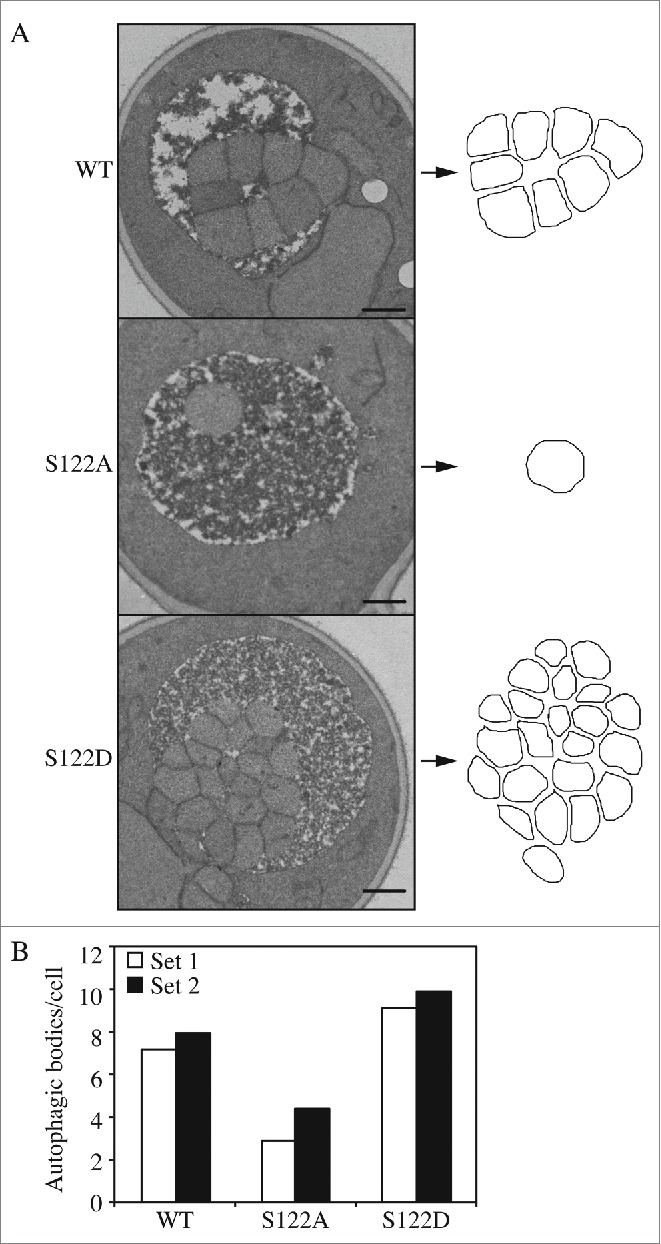
Phosphorylation of Atg9 S122 regulates the number of autophagosomes formed/formation rate. (A) Representative EM images of cells of WT and mutant Atg9 after 4 h of nitrogen starvation. Autophagic bodies are outlined on the right. Scale bar: 500 nm. (B) Estimated average number of autophagic body numbers per cell with WT or mutant Atg9 after 4 h of nitrogen starvation. Estimation was based on the number of autophagic body cross sections observed by EM in 2 independent experiments done by 2 different labs of more than 100 cells each for each strain.
The average size of the autophagic bodies in both mutants was not significantly different compared to the WT (Fig. S2). However, the Atg9S122A strain had fewer autophagic bodies per cell, whereas Atg9S122D displayed an increase (Fig. 3B). This result suggests that autophagosome formation was partially impaired in the strain expressing the S122A mutant; there were fewer autophagosomes formed, which corresponds to the lower Pho8Δ60 activity (Fig. 1C and S1D). In contrast, the higher Pho8Δ60 activity of the S122D mutant (Fig. 1C and S1D) may be explained by the observation that more autophagosomes were formed. The EM analysis thus suggests that the phosphorylation of Atg9 at S122 is important to maintain an appropriate rate of autophagosome formation, and hence the overall level of autophagy.
Phosphorylation of S122 affects autophagy through regulating Atg9 anterograde trafficking
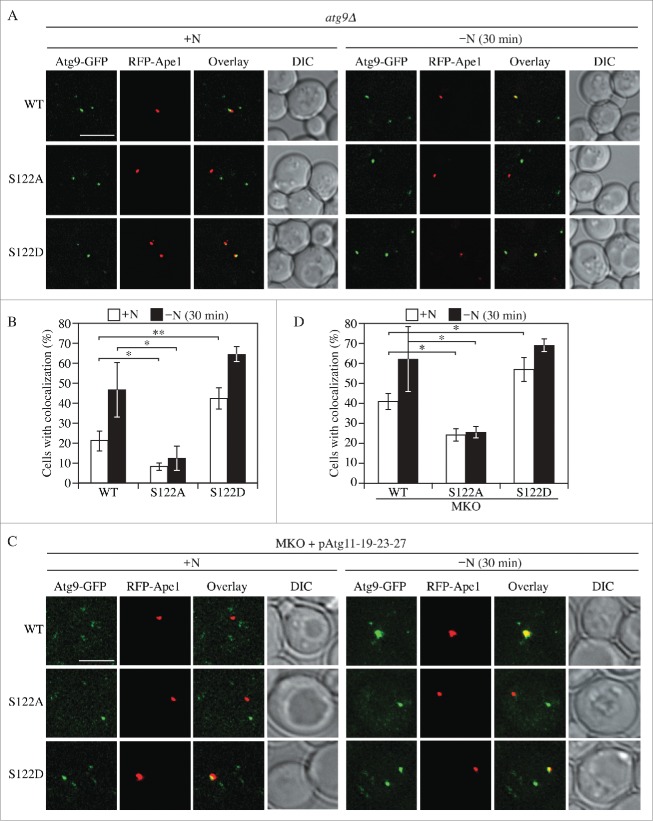
Atg9 cycles between the PAS and other cytosolic punctate structures (the peripheral sites), and has been proposed to be involved in membrane delivery to the PAS for autophagosome formation.15,42 To monitor the trafficking of Atg9, we examined cells expressing Atg9 WT, S122A or S122D under the control of the native ATG9 promoter, and tagged with GFP. We quantified the colocalization of the Atg9 chimeras with red fluorescent protein (RFP)-tagged prApe1 (RFP-Ape1), which serves as a marker for the PAS. In rich conditions (+N) Atg9 localized to multiple puncta, and the abundance of these puncta was increased after cells were shifted to starvation (-N) conditions for 30 min (Figs. 4A, B and S4A). In rich conditions, ∼21% of the WT cells displayed Atg9-GFP colocalized with RFP-Ape1, which represents the distribution of Atg9 during basal autophagy. After starvation for 30 min to induce autophagy this colocalization doubled to ∼47%. The colocalization in cells expressing Atg9S122A-GFP was less than half that seen in the WT, and was not substantially elevated after starvation (Figs. 4A, B and S4A), suggesting that the trafficking of Atg9S122A was impaired. In contrast, cells expressing Atg9S122D showed the opposite phenotype; these cells displayed ∼42% colocalization of Atg9S122D-GFP with RFP-Ape1 in rich conditions, and this increased to ∼64% after starvation (Figs. 4A, B and S4A), suggesting that the constitutive phosphorylation of Atg9 as mimicked by the phosphomimetic mutation promotes the anterograde movement of Atg9 to the PAS during autophagy. These results further indicate that Atg9 S122 phosphorylation plays an important role in regulating autophagy activity by controlling Atg9 trafficking.
Figure 4.
Phosphorylation of S122 is important for Atg9 anterograde trafficking. (A) Representative florescence microscopy images of atg9Δ cells transformed with a plasmid expressing WT or mutant Atg9-GFP. A plasmid expressing RFP-Ape1 was also used to provide a PAS marker. Cells were cultured in YPD, shifted to SD-N and collected at both 0 and 30 min nitrogen starvation. The single z-stack images are displayed at equal intensity for comparison. Scale bars: 5 μm. (B) Quantification of colocalization between Atg9-GFP and RFP-Ape1. Error bars correspond to the standard deviation and were obtained from 3 independent repeats. (C) Representative florescence microscopy images of the MKO strain expressing Atg11-Atg19-Atg23-Atg27 and RFP-Ape1, and transformed with a plasmid expressing WT or mutant Atg9-GFP. Cells were cultured, collected and imaged as in (A). (D) Quantification of colocalization between Atg9-GFP and RFP-Ape1 was performed as in (B). The ANOVA F-test was used for statistical significance. *, p < 0.05; **, p < 0.01.
Atg9 cycling between peripheral sites and the PAS can be split into 2 steps, corresponding to anterograde and retrograde trafficking.19 We wanted to determine whether phosphorylation of S122 specifically affects one of these steps. To address this point, we took advantage of a strain developed previously, the multiple-knockout (MKO) strain, which lacks 24 different ATG genes that are required for autophagosome formation.43,44 The re-addition to this strain of specific genes in various combinations allows the “in vivo reconstitution” of autophagy, and the definition of the minimal set of proteins necessary for a specific step of this process.45 Recently we showed that the MKO strain expressing Atg9, Atg11, Atg23 and Atg27 was sufficient to promote the anterograde movement of Atg9.45 We transformed this strain with the RFP-Ape1 plasmid and again examined colocalization. Similar results were observed with the MKO strain as seen as in the WT background; Atg9S122A was partially defective in PAS localization, whereas Atg9S122D had a higher percentage of localization at the PAS, in particular under nutrient-rich conditions (Figs. 4C, D and S4B). This consistent localization of Atg9 between the WT and MKO (Atg11-Atg19-Atg23-Atg27) strains, where the latter can only carry out anterograde movement, suggests that S122 phosphorylation affects Atg9 anterograde trafficking.
Phosphorylation of S122 regulates Atg9 through its interactions with Atg23 and Atg27
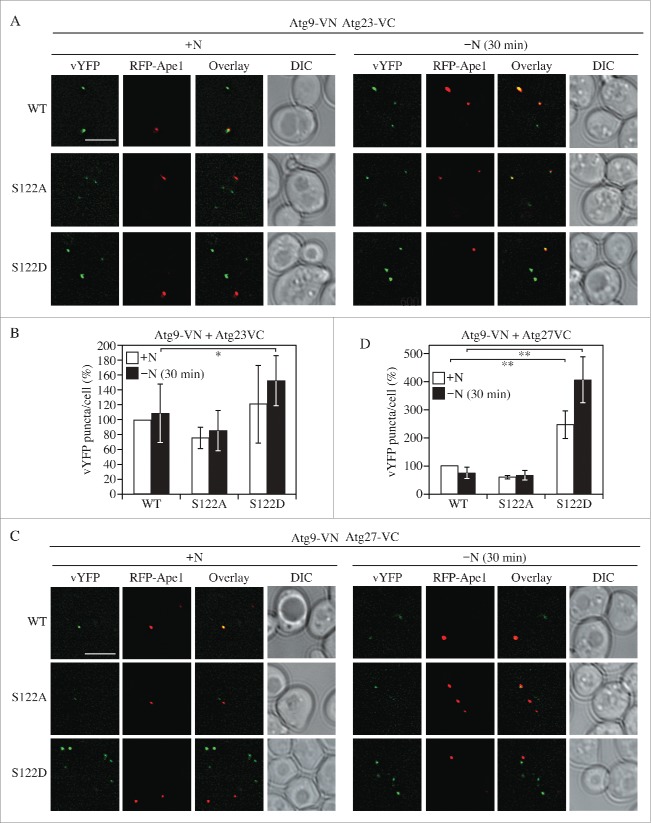
Having determined that S122 phosphorylation affects Atg9 anterograde trafficking, we next wanted to address the question of mechanism. Because of the known role of Atg11, Atg23 and Atg27 in regulating Atg9 movement,20,46 and based on our finding that the MKO strain expressing these proteins recapitulates the Atg9 localization phenotype seen with the mutant forms of Atg9, we hypothesized that the interaction of Atg9 with Atg23 and/or Atg27 might be affected in the Atg9 mutants (Atg11 is not essential for nonselective autophagy and the deletion of ATG11 has little effect on Phho8Δ60 activity47). To test this hypothesis, we used the bimolecular fluorescence complementation (BiFC) assay. Briefly, in the BiFC assay, the Venus yellow fluorescent protein (vYFP) is split into 2 fragments, VN (corresponding to the N terminus of vYFP) and VC (the C terminus of vYFP).48,49 We fused VN to Atg9 and VC to either Atg23 or Atg27 on the genome. Fluorescence from these 2 chimeras can only be detected when the 2 proteins interact and bring the 2 fragments of vYFP proximal to each other. A plasmid of RFP-Ape1 was transformed into these strains to mark the PAS.
A vYFP signal was observed both before and after N starvation (30 min), indicating that interactions between WT Atg9 and Atg23 or Atg27 exist during both basal and nonselective autophagy (Fig. 5). No vYFP fluorescence was detected in VN-Atg9, VC-Atg23 or VC-Atg27 alone (Fig. S3), which suggested that the vYFP puncta seen with combinations of these proteins (Fig. 5) represented an interaction between Atg9 and Atg23 or Atg27. vYFP signals were also observed in S122A and S122D, suggesting that these interactions were not abolished in either mutant; however, the mutants displayed different numbers of vYFP dots per cell (Fig. 5). Calculation of the number of puncta was performed automatically using CellProfiler. Upon nitrogen starvation, with Atg23, Atg9S122A cells had ∼20% fewer vYFP dots than WT, whereas Atg9S122D cells contained almost 50% more puncta (Fig. 5B and S5A). A similar, but more dramatic, trend was found with Atg27, where the Atg9S122D cells displayed almost 3 times more puncta per cell than WT upon starvation (Fig. 5D and S5B). These results suggest that phosphorylation of S122 is important for the interaction of Atg9 with Atg23 and Atg27; in particular, this interaction was partially reduced with the S122A mutant and was substantially enhanced with S122D.
Figure 5.
Phosphorylation of Atg9 S122 is important for the interaction of Atg9 with Atg23 and Atg27. (A, B) Representative fluorescence microscopy images (A) of WT or mutant Atg9-VN expressed with Atg23-VC. RFP-Ape1 was used as a PAS marker. Cells were cultured and collected as in Figure 4. Scale bars: 5 μm. (B) Quantification of vYFP puncta/cell was performed using CellProfiler. (C, D) Representative fluorescence microscopy images (C) of WT or mutant Atg9-VN expressed with Atg27-VC. (D) Quantification of vYFP puncta/cell was performed using CellProfiler. The ANOVA F-test was used for statistical significance. *, p < 0.05; **, p < 0.01.
We could barely detect the interaction of Atg9 with either Atg23 or Atg27 by co-immunoprecipitation, even with the wild-type proteins, suggesting that the binding may be weak, transient or unstable during the immunoprecipitation procedure.
Discussion
In this study, Atg9 S122 was identified through a SILAC assay as showing altered phosphorylation during autophagy induction. Among several sites of Atg9 that showed a change of phosphorylation level, S122 was the only one that showed a defect in autophagy activity when altered to a non-phosphorylatable mutant, S122A, and enhanced autophagy activity when constitutive phosphorylation was mimicked with S122D. In addition to nonselective autophagy, our data also showed that the phosphorylation of S122 is important for selective autophagy. However, based on the SILAC results, S122 displayed a 1.45-fold increase of phosphorylation level in the atg1Δ strain compared to the WT, which means that it is not a direct target of Atg1 kinase, which was previously shown to phosphorylate Atg9.14
We attempted to determine the effect of altering phosphorylation at S122, and the mechanism behind any such effect. We found that the S122A and S122D mutations had opposing effects on autophagy activity, which may be explained by changes in autophagic body (and hence autophagosome) number. That is, more Atg9 at the PAS, as seen with Atg9S122D, correlated with an increased rate of autophagosome formation. This result agrees with findings from a recent study where the absolute Atg9 levels correspond to the number, but not the size, of autophagosomes generated during autophagy.29 With the Atg9S122D mutant we altered the amount of Atg9 at the PAS, rather than the total amount of the Atg9 protein. Thus, the present results suggest that it is this pool of Atg9 in particular that accounts for autophagosome formation.
Considering that the PAS-localized pool of Atg9 is the critical one with regard to autophagy activity, how does phosphorylation affect the localization of Atg9? Using fluorescence microscopy, we found that Atg9S122A was defective in PAS localization, whereas Atg9S122D displayed a higher level of localization. Furthermore, using the MKO strain, we determined that anterograde movement was altered with the mutant version of Atg9. Finally, our data suggest that the S122D mutation results in an enhanced interaction in particular between Atg9 and Atg27, one of the components required for efficient anterograde trafficking of Atg9.21,45 We note, however, that Atg23 and Atg27 do not appear to be conserved in higher eukaryotes, so different components my take the place of these proteins if this regulatory aspect of Atg9 trafficking is conserved beyond fungi.
In summary, the phosphorylation-dependent regulation of Atg9 anterograde trafficking reveals another mechanism through which autophagy activity can be regulated. At present, the kinase or phosphatase that directly modifies Atg9 S122 remains to be identified, but these studies are currently underway.
Materials and methods
Strains, media, and growth conditions
Yeast strains used in this paper are listed in Table 1. Gene deletions or integrations were performed using a standard method.41 Cells were cultured in rich medium (YPD; 1% [w/v] yeast extract [ForMedium, YEM04], 2% [w/v] peptone [ForMedium, PEP04], and 2% [w/v] glucose) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base [ForMedium, CYN0410], 2% glucose, and auxotrophic amino acids and vitamins as needed) as indicated. Autophagy was induced through nitrogen starvation by shifting cells in mid-log phase from YPD (or SMD) to SD-N (0.17% yeast nitrogen base without ammonium sulfate or amino acids [ForMedium, CYN0501], and 2% [w/v] glucose) for the indicated times.
Table 1.
Yeast strains used in this study.
| Name | Genotype | Reference | |
|---|---|---|---|
| SEY6210 | MATα leu2–3,112 ura3–52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801 GAL | 59 | |
| SKB257 | MKO RFP-APE1 ATG9-GFP pATG11-19-23-27 | This study | |
| SKB259 | MKO RFP-APE1 ATG9-GFPS122ApATG11-19-23-27 | This study | |
| SKB260 | MKO RFP-APE1 ATG9-GFPS122DpATG11-19-23-27 | This study | |
| WLY176 | SEY6210 pho13Δ pho8Δ60 | 60 | |
| YKF001 | WLY176 atg9Δ::LEU2 ATG9-GFP::URA3 | This study | |
| YKF002 | WLY176 atg9Δ::LEU2 ATG9-GFPS122A::URA3 | This study | |
| YKF003 | WLY176 atg9Δ::LEU2 ATG9-GFPS122D::URA3 | This study | |
| YKF004 | WLY176 atg9Δ::LEU2 ATG9-GFPT119D::URA3 | This study | |
| YKF008 | SEY6210 RFP-APE1::LEU2 atg9Δ::HIS3 ATG9-GFP::URA3 | This study | |
| YKF009 | SEY6210 RFP-APE1::LEU2 atg9Δ::HIS3 ATG9-GFPS122A::URA3 | This study | |
| YKF010 | SEY6210 RFP-APE1::LEU2 atg9Δ::HIS3 ATG9-GFPS122D::URA3 | This study | |
| YKF080 | SEY6210 pep4Δ::LEU2 atg9Δ::HIS3 ATG9-GFP::URA3 | This study | |
| YKF081 | SEY6210 pep4Δ::LEU2 atg9Δ::HI3S ATG9-GFPS122A::URA3 | This study | |
| YKF082 | SEY6210 pep4Δ::LEU2 atg9Δ::HIS3 ATG9-GFPS122D::URA3 | This study | |
| YKF109 | BY4742 RFP-APE1::LEU2 ATG9-GFP::URA3 | This study | |
| YKF110 | BY4742 RFP-APE1::LEU2 ATG9-GFPS122A::URA3 | This study | |
| YKF111 | BY4742 RFP-APE1::LEU2 ATG9-GFPS122D::URA3 | This study | |
| YKF096 | SEY6210 vac8Δ::KAN atg9Δ::LEU2 | This study | |
| YKF396 | SEY6210 vac8Δ::KAN atg9Δ::LEU2 ATG9-GFP::URA3 | This study | |
| YKF397 | SEY6210 vac8Δ::KAN atg9Δ::LEU2 ATG9-GFPS122A::URA3 | This study | |
| YKF398 | SEY6210 vac8Δ::KAN atg9Δ::LEU2 ATG9-GFPS122D::URA3 | This study | |
| YKF417 | SEY6210 Atg9-VNWT::TRP1 Atg23-VC::HIS3 RFP-APE1::LEU2 | This study | |
| YKF413 | SEY6210 Atg9S122A-VN::TRP1 Atg23-VC::HIS3 RFP-APE1::LEU2 | This study | |
| YKF414 | SEY6210 Atg9S122D-VN::TRP1 Atg23-VC::HIS3 RFP-APE1::LEU2 | This study | |
| YKF415 | SEY6210 Atg9S122A-VN::TRP1 Atg27-VC::HIS3 RFP-APE1::LEU2 | This study | |
| YKF416 | SEY6210 Atg9S122D-VN::TRP1 Atg27-VC::HIS3 RFP-APE1::LEU2 | This study | |
| YKF418 | SEY6210 Atg9WT-VN:TRP1 Atg27-VC::HIS3 RFP-APE1::LEU2 | This study |
Plasmids
Integrating plasmids encoding Atg9-GFP29 and a centromeric plasmid encoding RFP-Ape1 have been published previously.50 Plasmids encoding Atg9-GFP with mutations S122A, S122D, or T119D were generated by site-directed mutagenesis51 from Atg9-GFP.
Fluorescence microscopy
For fluorescence microscopy, yeast cells were grown to OD600 ∼0.5 in SMD and shifted to SD-N for autophagy induction. Images were collected on a Deltavision Elite deconvolution microscope (GEHealthcare/Applied Precision) with a 100× objective and a CCD camera (CoolSnap HQ; Photometrics).
For quantification of Atg9-GFP colocalization with RFP-Ape1, stacks of 15 image planes were collected with a spacing of 0.2 µm to cover the entire yeast cell. Analysis was performed on an average projection of the imaging planes.
Mass spectrometry
WT and atg1Δ strains were grown in heavy and light lysine-supplemented SMD media, respectively. Upon reaching OD600 = 1.0, WT and atg1Δ cells were shifted to nitrogen starvation by culturing in SD-N media supplemented with light and heavy lysine, respectively, for 2 h. Cells were then harvested and equal numbers of cells were mixed prior to protein extraction, trypsinization, enrichment of phosphopeptides by strong cation exchange chromatography, and purification of phosphopeptides using IMAC columns.52 Desalted peptides were dissolved in 5% formic acid and analyzed on an LTQ Orbitrap Velos mass spectrometer as described previously.52 The following instrument parameters were employed: fully tryptic or LysC digestion, up to 2 missed cleavages, precursor mass tolerance of ±25 ppm, 1.0 Da product ion mass tolerance, a static modification of carbamidomethylation on cysteine (+57.0214); and dynamic modifications for phosphorylation on serine, threonine, and tyrosine (+79.9663), methionine oxidation (+15.9949), and 13C6 15N2-lysine (+8.0142). Spectra were searches using SEQUEST and matched to peptides with a 1% FDR using the target decoy approach53 and subsequently analyzed using linear discriminant analysis to derive Xcorr, ΔCn′, precursor mass error, and charge state. The Ascore method54,55 was used to assign phosphorylation sites and peptides quantified as described previously.52
Transmission electron microscopy
Two separate EM preparations were performed in 2 different labs. The first set of samples was prepared by Dr. M. Baba as described previously56 with slight modifications: In pre-fixation, the final fixative contained 0.1 M HEPES, pH 6.8, 0.1 M sorbitol, 1 mM MgCl2, 2% glutaraldehyde and 0.5% formaldehyde. 2x fixative was added directly into the starvation medium, and incubated for 5 min at room temperature (RT). Cells were collected by centrifugation and 1x fixative was added, followed by incubation at 4°C for 2.5 h. In post-fixation, 2% KMnO4was added to the cells for 5 min at RT. Cells were collected by centrifugation and the fixative solution was removed. KMnO4 (2%) was added and incubated for approximately 1 h at RT. After washing the cells, 2% low melting agarose was added followed by mixing. Subsequent steps were performed as described previously.56 The second set of samples was prepared in the Klionsky lab as described previously.21
Additional assays
Pho8Δ60 assays, western blot, prApe1 processing and EM were performed as described previously.21,49,57,58 Antibodies against Atg9, Ape1 and Pgk1 (a generous gift from Dr. Jeremy Thorner, University of California, Berkeley), and a commercial antibody that reacts with protein A (no longer available) were used as described previously.15,35 Anti-Dpm1 was from Molecular Probes/Invitrogen (A-6429).
Statistical analysis
Two-tailed Student t test, 2-tailed paired Student t test and ANOVA F-test was used to determine statistical significance.
Supplementary Material
Abbreviations
- Ape1
aminopeptidase I
- Atg
autophagy related
- BiFC
bimolecular fluorescence complementation
- Cvt
cytoplasm-to-vacuole targeting
- EM
electron microscopy
- GFP
green fluorescent protein
- MKO
multiple-knockout
- PAS
phagophore assembly site
- prApe1
precursor Ape1
- RFP
red fluorescent protein
- RT
room temperature
- SILAC
stable isotope labeling by amino acids in cell culture
- WT
wild type
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by NIH grant GM053396 to D.J.K. and GM095567 to J.W.H.
References
- [1].Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen E-L, et al.. A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy 2011; 7:1273-94; PMID:21997368; http://dx.doi.org/ 10.4161/auto.7.11.17661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/ 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Deffieu M, Bhatia-Kissova I, Salin B, Galinier A, Manon S, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. J Biol Chem 2009; 284:14828-37; PMID:19366696; http://dx.doi.org/ 10.1074/jbc.M109.005181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dunn WA Jr., Cregg JM, Kiel JAKW, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy 2005; 1:75-83; PMID:16874024; http://dx.doi.org/ 10.4161/auto.1.2.1737 [DOI] [PubMed] [Google Scholar]
- [5].Klionsky DJ, Cregg JM, Dunn WA Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al.. A unified nomenclature for yeast autophagy-related genes. Dev Cell 2003; 5:539-45; PMID:14536056; http://dx.doi.org/ 10.1016/S1534-5807(03)00296-X [DOI] [PubMed] [Google Scholar]
- [6].Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 2013; 194:341-61; PMID:23733851; http://dx.doi.org/ 10.1534/genetics.112.149013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713-20; PMID:23817233; http://dx.doi.org/ 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol 2015; 25:354-63; PMID:25759175; http://dx.doi.org/ 10.1016/j.tcb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 2015; 11:28-45; PMID:25484070; http://dx.doi.org/ 10.4161/15548627.2014.984267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, Yip CK, Klionsky DJ. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A 2013; 110:E2875-84; PMID:23858448; http://dx.doi.org/ 10.1073/pnas.1300064110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 2010; 30:1049-58; PMID:19995911; http://dx.doi.org/ 10.1128/MCB.01344-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A 2009; 106:17049-54; PMID:19805182; http://dx.doi.org/ 10.1073/pnas.0903316106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wilkinson DS, Jariwala JS, Anderson E, Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V, Dillin A, et al.. Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol Cell 2015; 57:55-68; PMID:25544559; http://dx.doi.org/ 10.1016/j.molcel.2014.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al.. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell 2014; 53:471-83; PMID:24440502; http://dx.doi.org/ 10.1016/j.molcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 2000; 148:465-80; PMID:10662773; http://dx.doi.org/ 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004; 6:79-90; PMID:14723849; http://dx.doi.org/ 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- [17].Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 2010; 190:1005-22; PMID:20855505; http://dx.doi.org/ 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 2012; 198:219-33; PMID:22826123; http://dx.doi.org/ 10.1083/jcb.201202061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 2005; 1:101-9; PMID:16874040; http://dx.doi.org/ 10.4161/auto.1.2.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yen W-L, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell 2007; 18:581-93; PMID:17135291; http://dx.doi.org/ 10.1091/mbc.E06-07-0612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Backues SK, Chen D, Ruan J, Xie Z, Klionsky DJ. Estimating the size and number of autophagic bodies by electron microscopy. Autophagy 2014; 10:155-64; PMID:24270884; http://dx.doi.org/ 10.4161/auto.26856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006; 119:3888-900; PMID:16940348; http://dx.doi.org/ 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- [23].He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell 2008; 19:5506-16; PMID:18829864; http://dx.doi.org/ 10.1091/mbc.E08-05-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 2000; 150:1507-13; PMID:10995454; http://dx.doi.org/ 10.1083/jcb.150.6.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 1998; 273:3963-6; PMID:9461583; http://dx.doi.org/ 10.1074/jbc.273.7.3963 [DOI] [PubMed] [Google Scholar]
- [26].Pfaffenwimmer T, Reiter W, Brach T, Nogellova V, Papinski D, Schuschnig M, Abert C, Ammerer G, Martens S, Kraft C. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep 2014; 15:862-70; PMID:24968893; http://dx.doi.org/ 10.15252/embr.201438932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tanaka C, Tan LJ, Mochida K, Kirisako H, Koizumi M, Asai E, Sakoh-Nakatogawa M, Ohsumi Y, Nakatogawa H. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J Cell Biol 2014; 207:91-105; PMID:25287303; http://dx.doi.org/ 10.1083/jcb.201402128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang Z, Geng J, Yen WL, Wang K, Klionsky DJ. Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell 2010; 38:250-64; PMID:20417603; http://dx.doi.org/ 10.1016/j.molcel.2010.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol 2014; 24:1314-22; PMID:24881874; http://dx.doi.org/ 10.1016/j.cub.2014.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 2008; 8:91; PMID:19055817; http://dx.doi.org/ 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Klionsky DJ. Monitoring autophagy in yeast: the Pho8Δ60 assay. Methods Mol Biol 2007; 390:363-71; PMID:17951700; http://dx.doi.org/ 10.1007/978-1-59745-466-7_24 [DOI] [PubMed] [Google Scholar]
- [32].Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 2002; 129:1181-93; PMID:12114572; http://dx.doi.org/ 10.1104/pp.011024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 2005; 280:18283-90; PMID:15755735; http://dx.doi.org/ 10.1074/jbc.M413957200 [DOI] [PubMed] [Google Scholar]
- [34].Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci U S A 1996; 93:12304-8; PMID:8901576; http://dx.doi.org/ 10.1073/pnas.93.22.12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol 1992; 119:287-99; PMID:1400574; http://dx.doi.org/ 10.1083/jcb.119.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell 2005; 16:1593-605; PMID:15659643; http://dx.doi.org/ 10.1091/mbc.E04-11-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Reggiori F, Wang C-W, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell 2004; 15:2189-204; PMID:15004240; http://dx.doi.org/ 10.1091/mbc.E03-07-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Suzuki K, Kamada Y, Ohsumi Y. Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev Cell 2002; 3:815-24; PMID:12479807; http://dx.doi.org/ 10.1016/S1534-5807(02)00359-3 [DOI] [PubMed] [Google Scholar]
- [39].Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell 2001; 7:1131-41; PMID:11430817; http://dx.doi.org/ 10.1016/S1097-2765(01)00263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Leber R, Silles E, Sandoval IV, Mazon MJ. Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole. J Biol Chem 2001; 276:29210-7; PMID:11382752; http://dx.doi.org/ 10.1074/jbc.M101438200 [DOI] [PubMed] [Google Scholar]
- [41].Scott SV, Nice DC III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem 2000; 275:25840-9; PMID:10837477; http://dx.doi.org/ 10.1074/jbc.M002813200 [DOI] [PubMed] [Google Scholar]
- [42].Legakis JE, Yen WL, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy 2007; 3:422-32; PMID:17426440; http://dx.doi.org/ 10.4161/auto.4129 [DOI] [PubMed] [Google Scholar]
- [43].Cao Y, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. A multiple ATG gene knockout strain for yeast two-hybrid analysis. Autophagy 2009; 5:699-705; PMID:19337029; http://dx.doi.org/ 10.4161/auto.5.5.8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol 2008; 182:703-13; PMID:18725539; http://dx.doi.org/ 10.1083/jcb.200801035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Backues SK, Orban DP, Bernard A, Singh K, Cao Y, Klionsky DJ. Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae. Traffic 2015; 16:172-90; PMID:25385507; http://dx.doi.org/ 10.1111/tra.12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tucker KA, Reggiori F, Dunn WA Jr., Klionsky DJ. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J Biol Chem 2003; 278:48445-52; PMID:14504273; http://dx.doi.org/ 10.1074/jbc.M309238200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA Jr., Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 2001; 153:381-96; PMID:11309418; http://dx.doi.org/ 10.1083/jcb.153.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 2007; 24:767-75; PMID:17534848; http://dx.doi.org/ 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- [49].Mao K, Liu X, Feng Y, Klionsky DJ. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 2014; 10:652-61; PMID:24451165; http://dx.doi.org/ 10.4161/auto.27852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang K, Yang Z, Liu X, Mao K, Nair U, Klionsky DJ. Phosphatidylinositol 4-kinases are required for autophagic membrane trafficking. J Biol Chem 2012; 287:37964-72; PMID:22977244; http://dx.doi.org/ 10.1074/jbc.M112.371591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 2004; 32:e115; PMID:15304544; http://dx.doi.org/ 10.1093/nar/gnh110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dephoure N, Gygi SP. A solid phase extraction-based platform for rapid phosphoproteomic analysis. Methods 2011; 54:379-86; PMID:21440633; http://dx.doi.org/ 10.1016/j.ymeth.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010; 143:1174-89; PMID:21183079; http://dx.doi.org/ 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 2007; 4:207-14; PMID:17327847; http://dx.doi.org/ 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- [55].Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 2006; 24:1285-92; PMID:16964243; http://dx.doi.org/ 10.1038/nbt1240 [DOI] [PubMed] [Google Scholar]
- [56].Wright R. Transmission electron microscopy of yeast. Microsc Res Tech 2000; 51:496-510; PMID:11169854; http://dx.doi.org/ 10.1002/1097-0029(20001215)51:6%3c496::AID-JEMT2%3e3.0.CO;2-9 [DOI] [PubMed] [Google Scholar]
- [57].Noda T, Klionsky DJ. The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol 2008; 451:33-42; PMID:19185711; http://dx.doi.org/ 10.1016/S0076-6879(08)03203-5 [DOI] [PubMed] [Google Scholar]
- [58].Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun 1995; 210:126-32; PMID:7741731; http://dx.doi.org/ 10.1006/bbrc.1995.1636 [DOI] [PubMed] [Google Scholar]
- [59].Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol 1988; 8:4936-48; PMID:3062374; http://dx.doi.org/ 10.1128/MCB.8.11.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen W-L, et al.. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell 2009; 20:4730-8; PMID:19793921; http://dx.doi.org/ 10.1091/mbc.E09-03-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.