Abbreviations
- 3-MA
3-methyladenine
- ABC
avidin-biotin peroxidase complex
- AIM
Atg8-family interacting motif
- ALIS
aggresome-like induced structures
- Ape1
aminopeptidase I
- ARN
autophagy regulatory network
- ASFV
African swine fever virus
- Atg
autophagy-related
- AV
autophagic vacuole
- BDI
bright detail intensity
- CASA
chaperone-assisted selective autophagy
- CLEAR
coordinated lysosomal enhancement and regulation
- CLEM
correlative light and electron microscopy
- CMA
chaperone-mediated autophagy
- cryo-SXT
cryo-soft X-ray tomography
- Cvt
cytoplasm-to-vacuole targeting
- DAMP
danger/damage-associated molecular pattern
- DQ-BSA
dequenched bovine serum albumin
- e-MI
endosomal microautophagy
- EBSS
Earle's balanced salt solution
- EM
electron microscopy
- ER
endoplasmic reticulum
- FACS
fluorescence-activated cell sorter
- FRET
fluorescence resonance energy transfer
- GAP
GTPase activating protein
- GBP
guanylate binding protein
- GFP
green fluorescent protein
- HIV-1
human immunodeficiency virus type 1
- HKP
housekeeping protein
- HSV-1
herpes simplex virus type 1
- Hyp-PDT
hypericin-based photodynamic therapy
- ICD
immunogenic cell death
- IHC
immunohistochemistry
- IMP
intramembrane particle
- LAMP2
lysosomal-associated membrane protein 2
- LAP
LC3-associated phagocytosis
- LIR
LC3-interacting region
- LN
late nucleophagy
- MAP1LC3/LC3, microtubule associated protein 1 light chain 3; MDC
monodansylcadaverine
- MEC
mammary epithelial cell
- mRFP
monomeric red fluorescent protein
- mtDNA
mitochondrial DNA
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- MVB
multivesicular body
- NASH
nonalcoholic steatohepatitis
- ncRNA
noncoding RNA
- NET
neutrophil extracellular trap
- NVJ
nucleus-vacuole junction
- PAMP
pathogen-associated molecular pattern
- PAS
phagophore assembly site
- PDT
photodynamic therapy
- PE
phosphatidylethanolamine
- PI3K
phosphoinositide 3-kinase
- PMN
piecemeal microautophagy of the nucleus
- PMSF
phenylmethylsulfonylfluoride
- POF
postovulatory follicle
- PSSM
position-specific scoring matrix
- PtdIns3K
phosphatidylinositol 3-kinase
- PtdIns3P
phosphatidylinositol 3-phosphate
- PTM
posttranslational modification
- PVM
parasitophorus vacuole membrane
- qRT-PCR
quantitative real-time reverse transcription polymerase chain reaction
- RBC
red blood cell
- RCBs
Rubisco-containing bodies
- Rluc
Renilla reniformis luciferase
- ROS
reactive oxygen species
- SD
standard deviation
- SKL
serine-lysine-leucine (a peroxisome targeting signal)
- SOD
superoxide dismutase
- TEM
transmission electron microscopy
- tfLC3
tandem fluorescent LC3
- TORC1
TOR complex I
- TR-FRET
time-resolved fluorescence resonance energy transfer
- TVA
tubulovesicular autophagosome
- UPR
unfolded protein response
- UPS
ubiquitin-proteasome system
- V-ATPase
vacuolar-type H+-ATPase
- xLIR
extended LIR-motif
In 2008 we published the first set of guidelines for standardizing research in autophagy. Since then, research on this topic has continued to accelerate, and many new scientists have entered the field. Our knowledge base and relevant new technologies have also been expanding. Accordingly, it is important to update these guidelines for monitoring autophagy in different organisms. Various reviews have described the range of assays that have been used for this purpose. Nevertheless, there continues to be confusion regarding acceptable methods to measure autophagy, especially in multicellular eukaryotes.
For example, a key point that needs to be emphasized is that there is a difference between measurements that monitor the numbers or volume of autophagic elements (e.g., autophagosomes or autolysosomes) at any stage of the autophagic process versus those that measure flux through the autophagy pathway (i.e., the complete process including the amount and rate of cargo sequestered and degraded). In particular, a block in macroautophagy that results in autophagosome accumulation must be differentiated from stimuli that increase autophagic activity, defined as increased autophagy induction coupled with increased delivery to, and degradation within, lysosomes (in most higher eukaryotes and some protists such as Dictyostelium) or the vacuole (in plants and fungi). In other words, it is especially important that investigators new to the field understand that the appearance of more autophagosomes does not necessarily equate with more autophagy. In fact, in many cases, autophagosomes accumulate because of a block in trafficking to lysosomes without a concomitant change in autophagosome biogenesis, whereas an increase in autolysosomes may reflect a reduction in degradative activity. It is worth emphasizing here that lysosomal digestion is a stage of autophagy and evaluating its competence is a crucial part of the evaluation of autophagic flux, or complete autophagy.
Here, we present a set of guidelines for the selection and interpretation of methods for use by investigators who aim to examine macroautophagy and related processes, as well as for reviewers who need to provide realistic and reasonable critiques of papers that are focused on these processes. These guidelines are not meant to be a formulaic set of rules, because the appropriate assays depend in part on the question being asked and the system being used. In addition, we emphasize that no individual assay is guaranteed to be the most appropriate one in every situation, and we strongly recommend the use of multiple assays to monitor autophagy. Along these lines, because of the potential for pleiotropic effects due to blocking autophagy through genetic manipulation, it is imperative to target by gene knockout or RNA interference more than one autophagy-related protein. In addition, some individual Atg proteins, or groups of proteins, are involved in other cellular pathways implying that not all Atg proteins can be used as a specific marker for an autophagic process. In these guidelines, we consider these various methods of assessing autophagy and what information can, or cannot, be obtained from them. Finally, by discussing the merits and limits of particular assays, we hope to encourage technical innovation in the field.
Introduction
Many researchers, especially those new to the field, need to determine which criteria are essential for demonstrating autophagy, either for the purposes of their own research, or in the capacity of a manuscript or grant review.1 Acceptable standards are an important issue, particularly considering that each of us may have his/her own opinion regarding the answer. Unfortunately, the answer is in part a “moving target” as the field evolves.2 This can be extremely frustrating for researchers who may think they have met those criteria, only to find out that the reviewers of their papers have different ideas. Conversely, as a reviewer, it is tiresome to raise the same objections repeatedly, wondering why researchers have not fulfilled some of the basic requirements for establishing the occurrence of an autophagic process. In addition, drugs that potentially modulate autophagy are increasingly being used in clinical trials, and screens are being carried out for new drugs that can modulate autophagy for therapeutic purposes. Clearly it is important to determine whether these drugs are truly affecting autophagy, and which step(s) of the process is affected, based on a set of accepted criteria. Accordingly, we describe here a basic set of contemporary guidelines that can be used by researchers to plan and interpret their experiments, by clinicians to evaluate the literature with regard to autophagy-modulating therapies, and by both authors and reviewers to justify or criticize an experimental approach.
Several fundamental points must be kept in mind as we establish guidelines for the selection of appropriate methods to monitor autophagy.2 Importantly, there are no absolute criteria for determining autophagic status that are applicable in every biological or experimental context. This is because some assays are inappropriate, problematic or may not work at all in particular cells, tissues or organisms.3-6 For example, autophagic responses to drugs may be different in transformed versus nontransformed cells, and in confluent versus nonconfluent cells, or in cells grown with or without glucose.4 In addition, these guidelines are likely to evolve as new methodologies are developed and current assays are superseded. Nonetheless, it is useful to establish guidelines for acceptable assays that can reliably monitor autophagy in many experimental systems. It is important to note that in this set of guidelines the term “autophagy” generally refers to macroautophagy; other autophagy-related processes are specifically designated when appropriate.
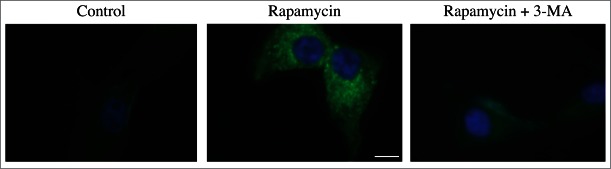
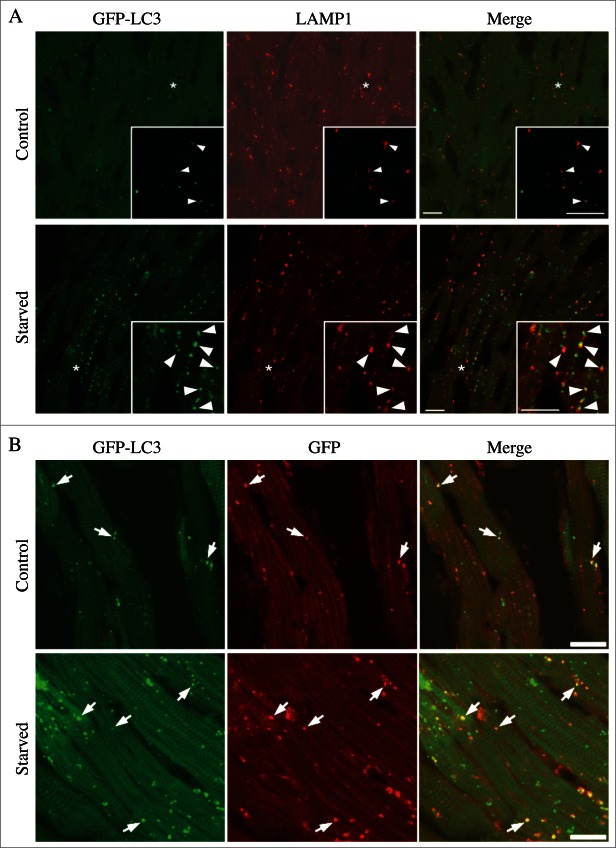
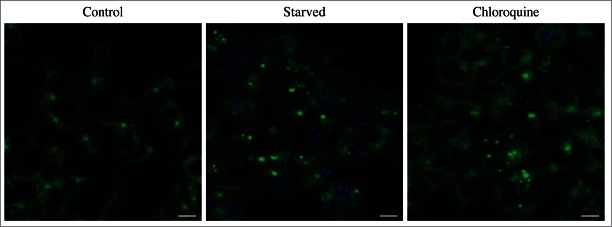
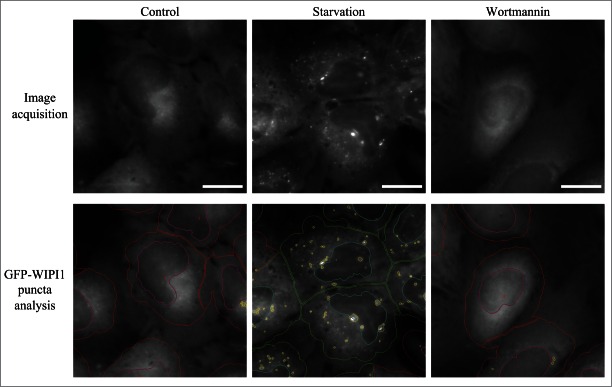
For the purposes of this review, the autophagic compartments (Fig. 1) are referred to as the sequestering (pre-autophagosomal) phagophore (PG; previously called the isolation or sequestration membrane5,6),7 the autophagosome (AP),8 the amphisome (AM; generated by the fusion of autophagosomes with endosomes),9 the lysosome, the autolysosome (AL; generated by fusion of autophagosomes or amphisomes with a lysosome), and the autophagic body (AB; generated by fusion and release of the internal autophagosomal compartment into the vacuole in fungi and plants). Except for cases of highly stimulated autophagic sequestration (Fig. 2), autophagic bodies are not seen in animal cells, because lysosomes/autolysosomes are typically smaller than autophagosomes.6,8,10 One critical point is that autophagy is a highly dynamic, multi-step process. Like other cellular pathways, it can be modulated at several steps, both positively and negatively. An accumulation of autophagosomes (measured by transmission electron microscopy [TEM] image analysis,11 as green fluorescent protein [GFP]-MAP1LC3 [GFP-LC3] puncta, or as changes in the amount of lipidated LC3 [LC3-II] on a western blot), could, for example, reflect a reduction in autophagosome turnover,12-14 or the inability of turnover to keep pace with increased autophagosome formation (Fig. 1B).15 For example, inefficient fusion with endosomes and/or lysosomes, or perturbation of the transport machinery,16 would inhibit autophagosome maturation to amphisomes or autolysosomes (Fig. 1C), whereas decreased flux could also be due to inefficient degradation of the cargo once fusion has occurred.17 Moreover, GFP-LC3 puncta and LC3 lipidation can reflect the induction of a different/modified pathway such as LC3-associated phagocytosis (LAP),18 and the noncanonical destruction pathway of the paternal mitochondria after fertilization.19,20
Figure 1.
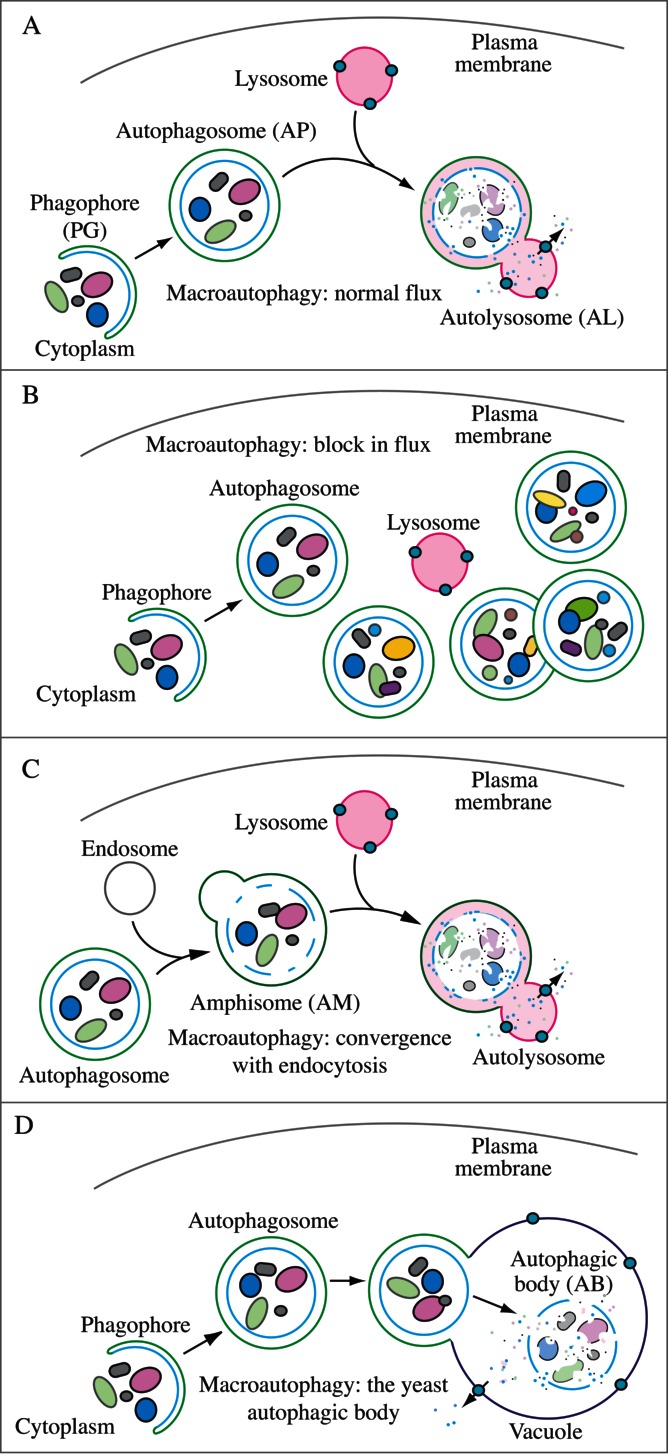
Schematic model demonstrating the induction of autophagosome formation when turnover is blocked versus normal autophagic flux, and illustrating the morphological intermediates of macroautophagy. (A) The initiation of autophagy includes the formation of the phagophore, the initial sequestering compartment, which expands into an autophagosome. Completion of the autophagosome is followed by fusion with lysosomes and degradation of the contents, allowing complete flux, or flow, through the entire pathway. This is a different outcome than the situation shown in (B) where induction results in the initiation of autophagy, but a defect in autophagosome turnover due, for example, to a block in fusion with lysosomes or disruption of lysosomal functions will result in an increased number of autophagosomes. In this scenario, autophagy has been induced, but there is no or limited autophagic flux. (C) An autophagosome can fuse with an endosome to generate an amphisome, prior to fusion with the lysosome. (D) Schematic drawing showing the formation of an autophagic body in fungi. The large size of the fungal vacuole relative to autophagosomes allows the release of the single-membrane autophagic body within the vacuole lumen. In cells that lack vacuolar hydrolase activity, or in the presence of inhibitors that block hydrolase activity, intact autophagic bodies accumulate within the vacuole lumen and can be detected by light microscopy. The lysosome of most higher eukaryotes is too small to allow the release of an autophagic body.
Figure 2.
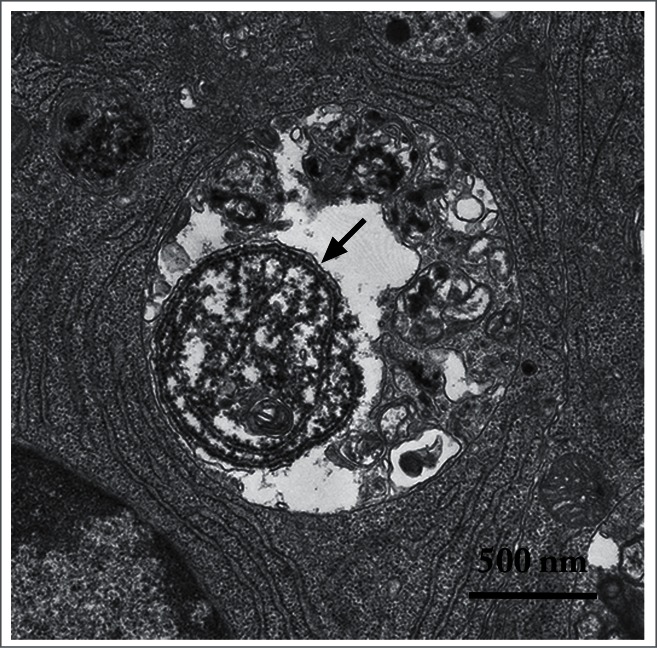
An autophagic body in a large lysosome of a mouse epithelial cell from a seminal vesicle in vitro. The arrow shows the single limiting membrane covering the sequestered rough ER. Image provided by A.L. Kovács.
Accordingly, the use of autophagy markers such as LC3-II must be complemented by assays to estimate overall autophagic flux, or flow, to permit a correct interpretation of the results. That is, autophagic activity includes not just the increased synthesis or lipidation of Atg8/LC3 (LC3 is the mammalian homolog of yeast Atg8), or an increase in the formation of autophagosomes, but, most importantly, flux through the entire system, including lysosomes or the vacuole, and the subsequent release of the breakdown products. Therefore, autophagic substrates need to be monitored dynamically over time to verify that they have reached the lysosome/vacuole, and whether or not they are degraded. By responding to perturbations in the extracellular environment, cells tune the autophagic flux to meet intracellular metabolic demands. The impact of autophagic flux on cell death and human pathologies therefore demands accurate tools to measure not only the current flux of the system, but also its capacity,21 and its response time, when exposed to a defined stress.22
One approach to evaluate autophagic flux is to measure the rate of general protein breakdown by autophagy.6,23 It is possible to arrest the autophagic flux at a given point, and then record the time-dependent accumulation of an organelle, an organelle marker, a cargo marker, or the entire cargo at the point of blockage; however, this approach, sometimes incorrectly referred to as autophagic flux, does not assess complete autophagy because the experimental block is usually induced (at least in part) by inhibiting lysosomal proteolysis, which precludes the evaluation of lysosomal functions. In addition, the latter assumes there is no feedback of the accumulating structure on its own rate of formation.24 In an alternative approach, one can follow the time-dependent decrease of an autophagy-degradable marker (with the caveat that the potential contribution of other proteolytic systems and of new protein synthesis need to be experimentally addressed). In theory, these nonautophagic processes can be assessed by blocking autophagic sequestration at specific steps of the pathway (e.g., blocking further induction or nucleation of new phagophores) and by measuring the decrease of markers distal to the block point.12,14,25 The key issue is to differentiate between the often transient accumulation of autophagosomes due to increased induction, and their accumulation due to inefficient clearance of sequestered cargos by both measuring the levels of autophagosomes at static time points and by measuring changes in the rates of autophagic degradation of cellular components.17 Both processes have been used to estimate “autophagy,” but unless the experiments can relate changes in autophagosome quantity to a direct or indirect measurement for autophagic flux, the results may be difficult to interpret.26 A general caution regarding the use of the term “steady state” is warranted at this point. It should not be assumed that an autophagic system is at steady state in the strict biochemical meaning of this term, as this implies that the level of autophagosomes does not change with time, and the flux through the system is constant. In these guidelines, we use steady state to refer to the baseline range of autophagic flux in a system that is not subjected to specific perturbations that increase or decrease that flux.
Autophagic flux refers to the entire process of autophagy, which encompasses the inclusion (or exclusion) of cargo within the autophagosome, the delivery of cargo to lysosomes (via fusion of the latter with autophagosomes or amphisomes) and its subsequent breakdown and release of the resulting macromolecules back into the cytosol (this may be referred to as productive or complete autophagy). Thus, increases in the level of phosphatidylethanolamine (PE)-modified Atg8/LC3 (Atg8–PE/LC3-II), or even the appearance of autophagosomes, are not measures of autophagic flux per se, but can reflect the induction of autophagic sequestration and/or inhibition of autophagosome or amphisome clearance. Also, it is important to realize that while formation of Atg8–PE/LC3-II appears to correlate with the induction of autophagy, we do not know, at present, the actual mechanistic relationship between Atg8–PE/LC3-II formation and the rest of the autophagic process; indeed, it may be possible to execute “self-eating” in the absence of LC3-II.27
As a final note, we also recommend that researchers refrain from the use of the expression “percent autophagy” when describing experimental results, as in “The cells displayed a 25% increase in autophagy.” Instead, it is appropriate to indicate that the average number of GFP-Atg8/LC3 puncta per cell is increased or a certain percentage of cells displayed punctate GFP-Atg8/LC3 that exceeds a particular threshold (and this threshold should be clearly defined in the Methods section), or that there is a particular increase or decrease in the rate of cargo sequestration or the degradation of long-lived proteins, when these are the actual measurements being quantified.
In a previous version of these guidelines,2 the methods were separated into 2 main sections—steady state and flux. In some instances, a lack of clear distinction between the actual methodologies and their potential uses made such a separation somewhat artificial. For example, fluorescence microscopy was initially listed as a steady-state method, although this approach can clearly be used to monitor flux as described in this article, especially when considering the increasing availability of new technologies such as microfluidic chambers. Furthermore, the use of multiple time points and/or lysosomal fusion/degradation inhibitors can turn even a typically static method such as TEM into one that monitors flux. Therefore, although we maintain the importance of monitoring autophagic flux and not just induction, this revised set of guidelines does not separate the methods based on this criterion. Readers should be aware that this article is not meant to present protocols, but rather guidelines, including information that is typically not presented in protocol papers. For detailed information on experimental procedures we refer readers to various protocols that have been published elsewhere.28-43,44 Finally, throughout the guidelines we provide specific cautionary notes, and these are important to consider when planning experiments and interpreting data; however, these cautions are not meant to be a deterrent to undertaking any of these experiments or a hindrance to data interpretation.
Collectively, we propose the following guidelines for measuring various aspects of selective and nonselective autophagy in eukaryotes.
A. Methods for monitoring autophagy
1. Transmission electron microscopy
Autophagy was first detected by TEM in the 1950s (reviewed in ref. 6). It was originally observed as focal degradation of cytoplasmic areas performed by lysosomes, which remains the hallmark of this process. Later analyses revealed that it starts with the sequestration of portions of the cytoplasm by a special double-membrane structure (now termed the phagophore), which matures into the autophagosome, still bordered by a double membrane. Subsequent fusion events expose the cargo to the lysosome (or the vacuole in fungi or plants) for enzymatic breakdown.
The importance of TEM in autophagy research lies in several qualities. It is the only tool that reveals the morphology of autophagic structures at a resolution in the nm range; shows these structures in their natural environment and position among all other cellular components; allows their exact identification; and, in addition, it can support quantitative studies if the rules of proper sampling are followed.11
Autophagy can be both selective and nonselective, and TEM can be used to monitor both. In the case of selective autophagy, the cargo is the specific substrate being targeted for sequestration—bulk cytoplasm is essentially excluded. In contrast, during nonselective autophagy, the various cytoplasmic constituents are sequestered randomly, resulting in autophagosomes in the size range of normal mitochondria. Sequestration of larger structures (such as big lipid droplets, extremely elongated or branching mitochondria or the entire Golgi complex) is rare, indicating an apparent upper size limit for individual autophagosomes. However, it has been observed that under special circumstances the potential exists for the formation of huge autophagosomes, which can even engulf a complete nucleus.25 Cellular components that form large confluent areas excluding bulk cytoplasm, such as organized, functional myofibrillar structures, do not seem to be sequestered by macroautophagy. The situation is less clear with regard to glycogen.45-47
After sequestration, the content of the autophagosome and its bordering double membrane remain morphologically unchanged, and clearly recognizable for a considerable time, which can be measured for at least many minutes. During this period, the membranes of the sequestered organelles (for example, the ER or mitochondria) remain intact, and the density of ribosomes is conserved at normal levels. Degradation of the sequestered material and the corresponding deterioration of ultrastructure commences and runs to completion within the amphisome and the autolysosome after fusion with a late endosome and lysosome (the vacuole in fungi and plants), respectively (Fig. 1).48 The sequential morphological changes during the autophagic process can be followed by TEM. The maturation from the phagophore through the autolysosome is a dynamic and continuous process,49 and, thus, the classification of compartments into discrete morphological subsets can be problematic; therefore, some basic guidelines are offered below.
In the preceding sections the “autophagosome,” the “amphisome” and the “autolysosome” were terms used to describe or indicate 3 basic stages and compartments of autophagy. It is important to make it clear that for instances (which may be many) when we cannot or do not want to differentiate among the autophagosomal, amphisomal and autolysosomal stage we use the general term “autophagic vacuole”. In the yeast autophagy field the term “autophagic vesicle” is used to avoid confusion with the primary vacuole, and by now the 2 terms are used in parallel and can be considered synonyms. It is strongly recommended, however, to use only the term “autophagic vacuole” when referring to macroautophagy in higher eukaryotic cells. Autophagosomes, also referred to as initial autophagic vacuoles (AVi), typically have a double membrane. This structure is usually distinctly visible by EM as 2 parallel membrane layers (bilayers) separated by a relatively narrower or wider electron-translucent cleft, even when applying the simplest routine EM fixation procedure (Fig. 3A).50,51 This electron-translucent cleft, however, is less visible or not visible in freeze-fixed samples, suggesting it is an artifact of sample preparation (see ref. 25, 68 and Fig. S3 in ref. 52). In the case of nonselective autophagy, autophagosomes contain cytosol and/or organelles appearing morphologically intact as also described above.48,53 Amphisomes54 can sometimes be identified by the presence of small intralumenal vesicles.55 These intralumenal vesicles are delivered into the lumen by fusion of the autophagosome/autophagic vacuole (AV) limiting membrane with multivesicular endosomes, and care should therefore be taken in the identification of the organelles, especially in cells that produce large numbers of multivesicular body (MVB)-derived exosomes (such as tumor or stem cells).56 Late/degradative autophagic vacuoles/autolysosomes (AVd or AL) typically have only one limiting membrane; frequently they contain electron dense cytoplasmic material and/or organelles at various stages of degradation (Fig. 3A and B);48,53 although late in the digestion process, they may contain only a few membrane fragments and be difficult to distinguish from lysosomes, endosomes, or tubular smooth ER cut in cross-section. Unequivocal identification of these structures and of lysosomes devoid of visible content requires immuno-EM detection of a cathepsin or other lysosomal hydrolase (e.g., ACP2 [acid phosphatase 2, lysosomal]57,58) that is detected on the limiting membrane of the lysosome.59 Smaller, often electron dense, lysosomes may predominate in some cells and exhibit hydrolase immunoreactivity within the lumen and on the limiting membrane.60
Figure 3.
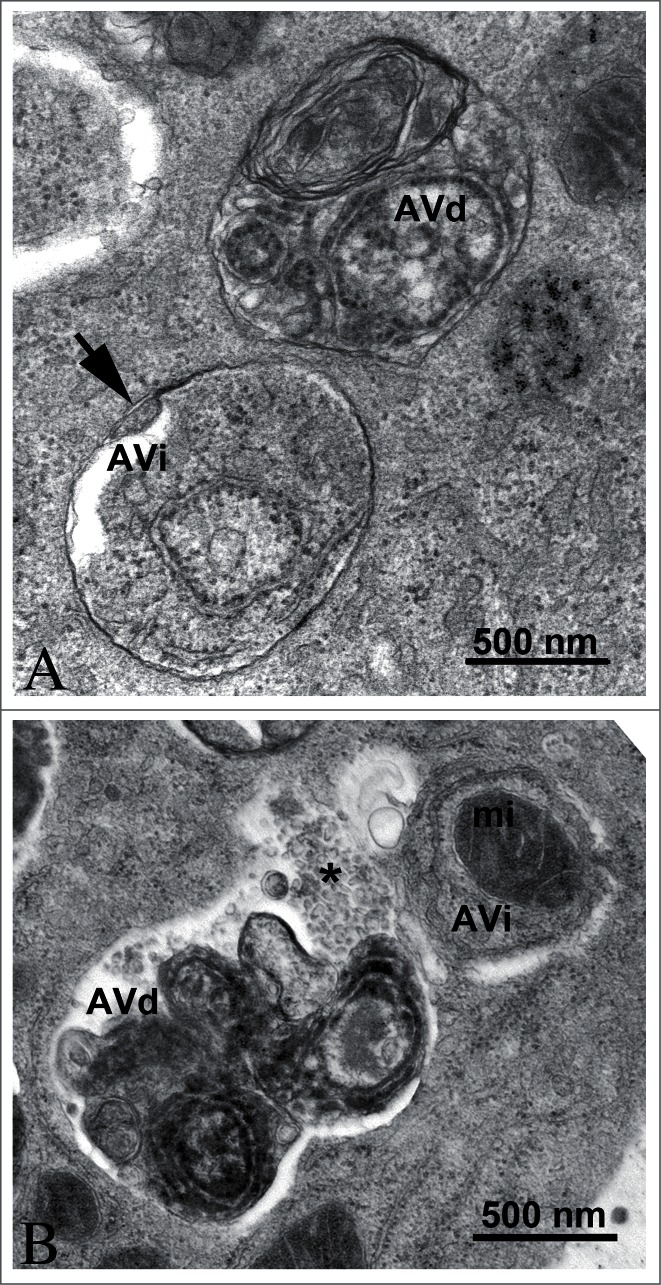
TEM images of autophagic vacuoles in isolated mouse hepatocytes. (A) One autophagosome or early initial autophagic vacuole (AVi) and one degradative autophagic vacuole (AVd) are shown. The AVi can be identified by its contents (morphologically intact cytoplasm, including ribosomes, and rough ER), and the limiting membrane that is partially visible as 2 bilayers separated by a narrow electron-lucent cleft, i.e., as a double membrane (arrow). The AVd can be identified by its contents, partially degraded, electron-dense rough ER. The vesicle next to the AVd is an endosomal/lysosomal structure containing 5-nm gold particles that were added to the culture medium to trace the endocytic pathway. (B) One AVi, containing rough ER and a mitochondrion, and one AVd, containing partially degraded rough ER, are shown. Note that the limiting membrane of the AVi is not clearly visible, possibly because it is tangentially sectioned. However, the electron-lucent cleft between the 2 limiting membranes is visible and helps in the identification of the AVi. The AVd contains a region filled by small internal vesicles (asterisk), indicating that the AVd has fused with a multivesicular endosome. mi, mitochondrion. Image provided by E.-L. Eskelinen.
In addition, structural proteins of the lysosome/late endosome, such as LAMP1 and LAMP2 or SCARB2/LIMP-2, can be used for confirmation. No single protein marker, however, has been effective in discriminating autolysosomes from the compartments mentioned above, in part due to the dynamic fusion and “kiss-and-run” events that promote interchange of components that can occur between these organelle subtypes. Rigorous further discrimination of these compartments from each other and other vesicles ultimately requires demonstrating the colocalization of a second marker indicating the presence of an autophagic substrate (e.g., LC3-CTSD [cathepsin D] colocalization) or the acidification of the compartment (e.g., mRFP/mCherry-GFP-LC3 probes [see Tandem mRFP/mCherry-GFP fluorescence microscopy], or Bodipy-pepstatin A detection of CTSD in an activated form within an acidic compartment), and, when appropriate, by excluding markers of other vesicular components.57,61,62
The sequential deterioration of cytoplasmic structures being digested can be used for identifying autolysosomes by TEM. Even when the partially digested and destroyed structure cannot be recognized in itself, it can be traced back to earlier forms by identifying preceding stages of sequential morphological deterioration. Degradation usually leads first to increased density of still recognizable organelles, then to vacuoles with heterogenous density, which become more homogenous and amorphous, mostly electron dense, but sometimes light (i.e., electron translucent). It should be noted that, in pathological states, it is not uncommon that active autophagy of autolysosomes and damaged lysosomes (“lysosophagy”) may yield populations of double-membrane limited autophagosomes containing partially digested amorphous substrates in the lumen. These structures, which are enriched in hydrolases, are seen in swollen dystrophic neurites in some neurodegenerative diseases,60 and in cerebellar slices cultured in vitro and infected with prions.63
It must be emphasized that in addition to the autophagic input, other processes (e.g., endosomal, phagosomal, chaperone-mediated) also carry cargo to the lysosomes,64,65 in some cases through the intermediate step of direct endosome fusion with an autophagosome to form an amphisome. This process is exceptionally common in the axons of neurons.66 Therefore, strictly speaking, we can only have a lytic compartment containing cargos arriving from several possible sources; however, we still may use the term “autolysosome” if the content appears to be overwhelmingly autophagic. Note that the engulfment of apoptotic cells via phagocytosis also produces lysosomes that contain cytoplasmic structures, but in this case it originates from the dying cell; hence the possibility of an extracellular origin for such content must be considered when monitoring autophagy in settings where apoptotic cell death may be reasonably expected or anticipated.
For many physiological and pathological situations, examination of both early and late autophagic vacuoles yields valuable data regarding the overall autophagy status in the cells.15 Along these lines, it is possible to use immunocytochemistry to follow particular cytosolic proteins such as SOD1/CuZn superoxide dismutase and CA/carbonic anhydrase to determine the stage of autophagy; the former is much more resistant to lysosomal degradation.67
In some autophagy-inducing conditions it is possible to observe multi-lamellar membrane structures in addition to the conventional double-membrane autophagosomes, although the nature of these structures is not fully understood. These multi-lamellar structures may indeed be multiple double layers of phagophores68 and positive for LC3,69 they could be autolysosomes,70 or they may form artifactually during fixation.68
Special features of the autophagic process may be clarified by immuno-TEM with gold-labeling,71,72 using antibodies, for example, to cargo proteins of cytoplasmic origin and to LC3 to verify the autophagic nature of the compartment. LC3 immunogold labeling also makes it possible to detect novel degradative organelles within autophagy compartments. This is the case with the autophagoproteasome73 where costaining for LC3 and ubiquitin-proteasome system (UPS) antigens occurs. The autophagoproteasome consists of single-, double-, or multiple-membrane LC3-positive autophagosomes costaining for specific components of the UPS. It may be that a rich multi-enzymatic (both autophagic and UPS) activity takes place within these organelles instead of being segregated within different cell domains.
Although labeling of LC3 can be difficult, an increasing number of commercial antibodies are becoming available, among them good ones to visualize the GFP moiety of GFP-LC3 reporter constructs.74 It is important to keep in mind that LC3 can be associated with nonautophagic structures (see Xenophagy, and Noncanonical use of autophagy-related proteins). LC3 is involved in specialized forms of endocytosis like LC3-associated phagocytosis. In addition, LC3 can decorate vesicles dedicated to exocytosis in nonconventional secretion systems (reviewed in ref. 75,76). Antibodies against an abundant cytosolic protein will result in high labeling all over the cytoplasm; however, organelle markers work well. Because there are very few characterized proteins that remain associated with the completed autophagosome, the choices for confirmation of its autophagic nature are limited. Furthermore, autophagosome-associated proteins may be cell type-specific. At any rate, the success of this methodology depends on the quality of the antibodies and also on the TEM preparation and fixation procedures utilized. With immuno-TEM, authors should provide controls showing that labeling is specific. This may require quantitative comparison of labeling over different cellular compartments not expected to contain antigen and those containing the antigen of interest.
In clinical situations it is difficult to demonstrate autophagy clearly in tissues of formalin-fixed and paraffin-embedded biopsy samples retrospectively, because (1) tissues fixed in formalin have low or no LC3 detectable by routine immunostaining, because phospholipids melt together with paraffin during the sample preparation, and (2) immunogold electron microscopy of many tissues not optimally fixed for this purpose (e.g., using rapid fixation) produces low-quality images. Combining antigen retrieval with the avidin-biotin peroxidase complex (ABC) method may be quite useful for these situations. For example, immunohistochemistry can be performed using an antigen retrieval method, then tissues are stained by the ABC technique using a labeled anti-human LC3 antibody. After imaging by light microscopy, the same prepared slides can be remade into sections for TEM examination, which can reveal peroxidase reaction deposits in vacuoles within the region that is LC3-immunopositive by light microscopy.77 In addition, statistical information should be provided due to the necessity of showing only a selective number of sections in publications.
We note here again that for quantitative data it is necessary to use proper volumetric analysis rather than just counting numbers of sectioned objects. On the one hand, it must be kept in mind that even volumetric morphometry/stereology only shows either steady state levels, or a snapshot in a changing dynamic process. Such data by themselves are not informative regarding autophagic flux, unless carried out over multiple time points. Alternatively, investigation in the presence and absence of flux inhibitors can reveal the dynamic changes in various stages of the autophagic process.12,21,78,79,42 On the other hand, if the turnover of autolysosomes is very rapid, a low number/volume will not necessarily be an accurate reflection of low autophagic activity. However, quantitative analyses indicate that autophagosome volume in many cases does correlate with the rates of protein degradation.80-82 One potential compromise is to perform whole cell quantification of autophagosomes using fluorescence methods, with qualitative verification by TEM,83 to show that the changes in fluorescent puncta reflect corresponding changes in autophagic structures.
One additional caveat with TEM, and to some extent with confocal fluorescence microscopy, is that the analysis of a single plane within a cell can be misleading and may make the identification of autophagic structures difficult. Confocal microscopy and fluorescence microscopy with deconvolution software (or with much more work, 3-dimensional TEM) can be used to generate multiple/serial sections of the same cell to reduce this concern; however, in many cases where there is sufficient structural resolution, analysis of a single plane in a relatively large cell population can suffice given practical limitations. Newer EM technologies, including focused ion beam dual-beam EM, should make it much easier to apply three-dimensional analyses. An additional methodology to assess autophagosome accumulation is correlative light and electron microscopy (CLEM), which is helpful in confirming that fluorescent structures are autophagosomes.84-86 Along these lines, it is important to note that even though GFP fluorescence will be quenched in the acidic environment of the autolysosome, some of the GFP puncta detected by light microscopy may correspond to early autolysosomes prior to GFP quenching. The mini Singlet Oxygen Generator (miniSOG) fluorescent flavoprotein, which is less than half the size of GFP, provides an additional means to genetically tag proteins for CLEM analysis under conditions that are particularly suited to subsequent TEM analysis.87 Combinatorial assays using tandem monomeric red fluorescent protein (mRFP)-GFP-LC3 (see Tandem mRFP/mCherry-GFP fluorescence microscopy) along with static TEM images should help in the analysis of flux and the visualization of cargo structures.88
Another technique that has proven quite useful for analyzing the complex membrane structures that participate in autophagy is 3-dimensional electron tomography,89,90 and cryoelectron microscopy (Fig. 4). More sophisticated, cryo-soft X-ray tomography (cryo-SXT) is an emerging imaging technique used to visualize autophagosomes.91 Cryo-SXT extracts ultrastructural information from whole, unstained mammalian cells as close to the “near-native” fully-hydrated (living) state as possible. Correlative studies combining cryo-fluorescence and cryo-SXT workflow (cryo-CLXM) have been applied to capture early autophagosomes.
Figure 4.
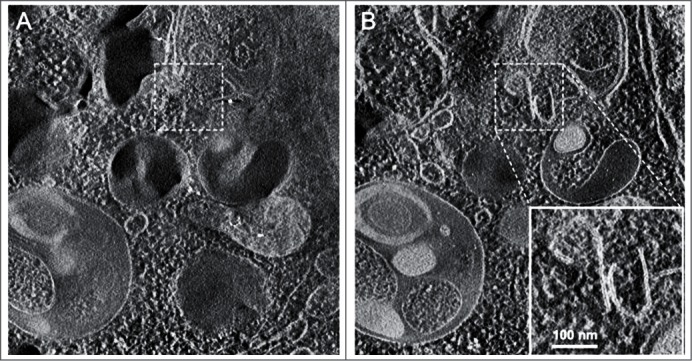
Cryoelectron microscopy can be used as a three-dimensional approach to monitor the autophagic process. Two computed sections of an electron tomogram of the autophagic vacuole-rich cytoplasm in a hemophagocyte of a semi-thin section after high-pressure freezing preparation. The dashed area is membrane-free (A) but tomography reveals newly formed or degrading membranes with a parallel stretch (B). Image published previously2185 and provided by M. Schneider and P. Walter.
Finally, although only as an indirect measurement, the comparison of the ratio of autophagosomes to autolysosomes by TEM can support alterations in autophagy identified by other procedures.92 In this case it is important to always compare samples to the control of the same cell type and in the same growth phase, and to acquire data at different time points, as the autophagosome/autolysosome ratio varies in time in a cell context-dependent fashion, depending on their clearance activity. It may also be necessary to distinguish autolysosomes from telolysosomes/late secondary lysosomes (the former are actively engaged in degradation, whereas the latter have reached an end point in the breakdown of lumenal contents) because the lysosome number generally increases when autophagy is induced. An additional category of lysosomal compartments, especially common in disease states and aged postmitotic cells such as neurons, is the residual body. This category includes ceroid and lipofuscin, lobulated vesicular compartments of varying size composed of highly indigestible complexes of protein and lipid and abundant, mostly inactive, acid hydrolases. Reflecting end-stage unsuccessful incomplete autolysosomal digestion, lipofuscin is fairly easily distinguished from AVs and lysosomes by TEM but can be easily confused with autolysosomes in immunocytochemistry studies at the light microscopy level.57
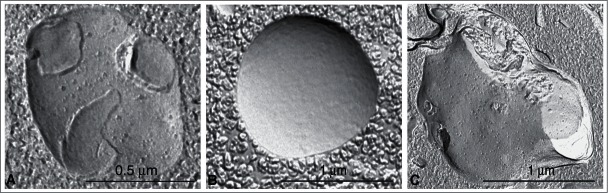
TEM observations of platinum-carbon replicas obtained by the freeze fracture technique can also supply useful ultrastructural information on the autophagic process. In quickly frozen and fractured cells the fracture runs preferentially along the hydrophobic plane of the membranes, allowing characterization of the limiting membranes of the different types of autophagic vacuoles and visualization of their limited protein intramembrane particles (IMPs, or integral membrane proteins). Several studies have been carried out using this technique on yeast,93 as well as on mammalian cells or tissues; first on mouse exocrine pancreas,94 then on mouse and rat liver,95,96 mouse seminal vesicle epithelium,25,68 rat tumor and heart,97 or cancer cell lines (e.g., breast cancer MDA-MB-231)98 to investigate the various phases of autophagosome maturation, and to reveal useful details about the origin and evolution of their limiting membranes.6,99-102
The phagophore and the limiting membranes of autophagosomes contain few, or no detectable, IMPs (Fig. 5A, B), when compared to other cellular membranes and to the membranes of lysosomes. In subsequent stages of the autophagic process the fusion of the autophagosome with an endosome and a lysosome results in increased density of IMPs in the membrane of the formed autophagic compartments (amphisomes, autolysosomes; Fig. 5C).6,25,93-96,103,104 Autolysosomes are delimited by a single membrane because, in addition to the engulfed material, the inner membrane is also degraded by the lytic enzymes. Similarly, the limiting membrane of autophagic bodies in yeast (and presumably plants) is also quickly broken down under normal conditions. Autophagic bodies can be stabilized, however, by the addition of phenylmethylsulfonylfluoride (PMSF) or genetically by the deletion of the yeast PEP4 gene (see The Cvt pathway, mitophagy, pexophagy, piecemeal microautophagy of the nucleus and late nucleophagy in yeast and filamentous fungi.). Thus, another method to consider for monitoring autophagy in yeast (and potentially in plants) is to count autophagic bodies by TEM using at least 2 time points.105 The advantage of this approach is that it can provide accurate information on flux even when the autophagosomes are abnormally small.106,107 Thus, although a high frequency of “abnormal” structures presents a challenge, TEM is still very helpful in analyzing autophagy.
Figure 5.
Different autophagic vacuoles observed after freeze fracturing in cultured osteosarcoma cells after treatment with the autophagy inducer voacamine.101 (A) Early autophagosome delimited by a double membrane. (B) Inner monolayer of an autophagosome membrane deprived of protein particles. (C) Autolysosome delimited by a single membrane rich in protein particles. In the cross-fractured portion (on the right) the profile of the single membrane and the inner digested material are easily visible. Images provided by S. Meschini, M. Condello and A. Giuseppe.
Cautionary notes: Despite the introduction of many new methods TEM maintains its special role in autophagy research. There are, however, difficulties in utilizing TEM. It is relatively time consuming, and needs technical expertise to ensure proper handling of samples in all stages of preparation from fixation to sectioning and staining (contrasting). After all these criteria are met, we face the most important problem of proper identification of autophagic structures. This is crucial for both qualitative and quantitative characterization, and needs considerable experience, even in the case of one cell type. The difficulty lies in the fact that many subcellular components may be mistaken for autophagic structures. For example, some authors (or reviewers of manuscripts) assume that almost all cytoplasmic structures that, in the section plane, are surrounded by 2 (more or less) parallel membranes are autophagosomes. Structures appearing to be limited by a double membrane, however, may include swollen mitochondria, plastids in plant cells, cellular interdigitations, endocytosed apoptotic bodies, circular structures of lamellar smooth endoplasmic reticulum (ER), and even areas surrounded by rough ER. Endosomes, phagosomes and secretory vacuoles may have heterogenous content that makes it possible to confuse them with autolysosomes. Additional identification problems may arise from damage caused by improper sample taking or fixation artifacts.50,51,108,109
Whereas fixation of in vitro samples is relatively straightforward, fixation of excised tissues requires care to avoid sampling a nonrepresentative, uninformative, or damaged part of the tissue. For instance, if 95% of a tumor is necrotic, TEM analysis of the necrotic core may not be informative, and if the sampling is from the viable rim, this needs to be specified when reported. Clearly this introduces the potential for subjectivity because reviewers of a paper cannot request multiple images with a careful statistical analysis with these types of samples. In addition, ex vivo samples are not typically randomized during processing, further complicating the possibility of valid statistical analyses. Ex vivo tissue should be fixed immediately and systematically across samples to avoid changes in autophagy that may occur simply due to the elapsed time ex vivo. It is recommended that for tissue samples, perfusion fixation should be used when possible. For yeast, rapid freezing techniques such as high pressure freezing followed by freeze substitution (i.e., dehydration at low temperature) may be particularly useful.
Quantification of autophagy by TEM morphometry has been rather controversial, and unreliable procedures still continue to be used. For the principles of reliable quantification and to avoid misleading results, excellent reviews are available.11,110-112 In line with the basic principles of morphometry we find it necessary to emphasize here some common problems with regard to quantification. Counting autophagic vacuole profiles in sections of cells (i.e., number of autophagic profiles per cell profile) may give unreliable results, partly because both cell areas and profile areas are variable and also because the frequency of section profiles depends on the size of the vacuoles. However, estimation of the number of autophagic profiles per cell area is more reliable and correlates well with the volume fraction mentioned below.53 There are morphometric procedures to measure or estimate the size range and the number of spherical objects by profiles in sections;111 however, such methods have been used in autophagy research only a few times.32,107,113,114
Proper morphometry described in the cited reviews will give us data expressed in µm3 autophagic vacuole/µm3 cytoplasm for relative volume (also called volume fraction or volume density), or µm2 autophagic vacuole surface/µm3 cytoplasm for relative surface (surface density). Examples of actual morphometric measurements for the characterization of autophagic processes can be found in several articles.21,108,111,115,116 It is appropriate to note here that a change in the volume fraction of the autophagic compartment may come from 2 sources; from the real growth of its size in a given cytoplasmic volume, or from the decrease of the cytoplasmic volume itself. To avoid this so-called “reference trap,” the reference space volume can be determined by different methods.112,117 If different magnifications are used for measuring the autophagic vacuoles and the cytoplasm (which may be practical when autophagy is less intense) correction factors should always be used.
In some cases, it may be prudent to employ tomographic reconstructions of the TEM images to confirm that the autophagic compartments are spherical and are not being confused with interdigitations observed between neighboring cells, endomembrane cisternae or damaged mitochondria with similar appearance in thin-sections (e.g., see ref. 118), but this is obviously a time-consuming approach requiring sophisticated equipment. In addition, interpretation of tomographic images can be problematic. For example, starvation-induced autophagosomes should contain cytoplasm (i.e., cytosol and possibly organelles), but autophagosome-related structures involved in specific types of autophagy should show the selective cytoplasmic target, but may be relatively devoid of bulk cytoplasm. Such processes include selective peroxisome or mitochondria degradation (pexophagy or mitophagy, respectively),119,120 targeted degradation of pathogenic microbes (xenophagy),121-126 a combination of xenophagy and stress-induced mitophagy,127 as well as the yeast biosynthetic cytoplasm-to-vacuole targeting (Cvt) pathway.128 Furthermore, some pathogenic microbes express membrane-disrupting factors during infection (e.g., phospholipases) that disrupt the normal double-membrane architecture of autophagosomes.129 It is not even clear if the sequestering compartments used for specific organelle degradation or xenophagy should be termed autophagosomes or if alternate terms such as pexophagosome,130 mitophagosome and xenophagosome should be used, even though the membrane and mechanisms involved in their formation may be identical to those for starvation-induced autophagosomes; for example, the double-membrane vesicle of the Cvt pathway is referred to as a Cvt vesicle.131
The confusion of heterophagic structures with autophagic ones is a major source of misinterpretation. A prominent example of this is related to apoptosis. Apoptotic bodies from neighboring cells are readily phagocytosed by surviving cells of the same tissue.132,133 Immediately after phagocytic uptake of apoptotic bodies, phagosomes may have double limiting membranes. The inner one is the plasma membrane of the apoptotic body and the outer one is that of the phagocytizing cell. The early heterophagic vacuole formed in this way may appear similar to an autophagosome or, in a later stage, an early autolysosome in that it contains recognizable or identifiable cytoplasmic material. A major difference, however, is that the surrounding membranes are the thicker plasma membrane type, rather than the thinner sequestration membrane type (9–10 nm, versus 7–8 nm, respectively).109 A good feature to distinguish between autophagosomes and double plasma membrane-bound structures is the lack of the distended empty space (characteristic for the sequestration membranes of autophagosomes) between the 2 membranes of the phagocytic vacuoles. In addition, engulfed apoptotic bodies usually have a larger average size than autophagosomes.134,135 The problem of heterophagic elements interfering with the identification of autophagic ones is most prominent in cell types with particularly intense heterophagic activity (such as macrophages, and amoeboid or ciliate protists). Special attention has to be paid to this problem in cell cultures or in vivo treatments (e.g., with toxic or chemotherapeutic agents) causing extensive apoptosis.
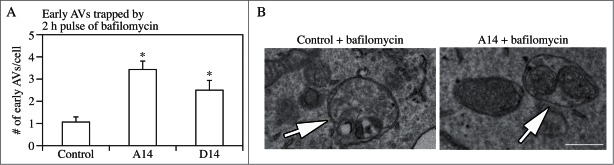
The most common organelles confused with autophagic vacuoles are mitochondria, ER, endosomes, and also (depending on their structure) plastids in plants. Due to the cisternal structure of the ER, double-membrane-like structures surrounding mitochondria or other organelles are often observed after sectioning,136 but these can also correspond to cisternae of the ER coming into and out of the section plane.50 If there are ribosomes associated with these membranes they can help in distinguishing them from the ribosome-free double-membrane of the phagophore and autophagosome. Observation of a mixture of early and late autophagic vacuoles that is modulated by the time point of collection and/or brief pulses of bafilomycin A1 (a vacuolar-type H+-ATPase [V-ATPase] inhibitor) to trap the cargo in a recognizable early state42 increases the confidence that an autophagic process is being observed. In these cases, however, the possibility that feedback activation of sequestration gets involved in the autophagic process has to be carefully considered. To minimize the impact of errors, exact categorization of autophagic elements should be applied. Efforts should be made to clarify the nature of questionable structures by extensive preliminary comparison in many test areas. Elements that still remain questionable should be categorized into special groups and measured separately. Should their later identification become possible, they can be added to the proper category or, if not, kept separate.
For nonspecialists it can be particularly difficult to distinguish among amphisomes, autolysosomes and lysosomes, which are all single-membrane compartments containing more or less degraded material. Therefore, we suggest in general to measure autophagosomes as a separate category for a start, and to compile another category of degradative compartments (including amphisomes, autolysosomes and lysosomes). All of these compartments increase in quantity upon true autophagy induction; however, in pathological states, it may be informative to discriminate among these different forms of degradative compartments, which may be differentially affected by disease factors.
In yeast, it is convenient to identify autophagic bodies that reside within the vacuole lumen, and to quantify them as an alternative to the direct examination of autophagosomes. However, it is important to keep in mind that it may not be possible to distinguish between autophagic bodies that are derived from the fusion of autophagosomes with the vacuole, and the single-membrane vesicles that are generated during microautophagy-like processes such as micropexophagy and micromitophagy.
Conclusion: EM is an extremely informative and powerful method for monitoring autophagy and remains the only technique that shows autophagy in its complex cellular environment with subcellular resolution. The cornerstone of successfully using TEM is the proper identification of autophagic structures, which is also the prerequisite to get reliable quantitative results by EM morphometry. EM is best used in combination with other methods to ensure the complex and holistic approach that is becoming increasingly necessary for further progress in autophagy research.
2. Atg8/LC3 detection and quantification
Atg8/LC3 is the most widely monitored autophagy-related protein. In this section we describe multiple assays that utilize this protein, separating the descriptions into several subsections for ease of discussion.
a. Western blotting and ubiquitin-like protein conjugation systems
The Atg8/LC3 protein is a ubiquitin-like protein that can be conjugated to PE (and possibly to phosphatidylserine137). In yeast and several other organisms, the conjugated form is referred to as Atg8–PE. The mammalian homologs of Atg8 constitute a family of proteins subdivided in 2 major subfamilies: MAP1LC3/LC3 and GABARAP. The former consists of LC3A, B, B2 and C, whereas the latter family includes GABARAP, GABARAPL1 and GABARAPL2/GATE-16.138 After cleavage of the precursor protein mostly by the cysteine protease ATG4B,139,140 the nonlipidated and lipidated forms are usually referred to respectively as LC3-I and LC3-II, or GABARAP and GABARAP–PE, etc. The PE-conjugated form of Atg8/LC3, although larger in mass, shows faster electrophoretic mobility in SDS-PAGE gels, probably as a consequence of increased hydrophobicity. The positions of both Atg8/LC3-I (approximately 16–18 kDa) and Atg8–PE/LC3-II (approximately 14–16 kDa) should be indicated on western blots whenever both are detectable. The differences among the LC3 proteins with regard to function and tissue-specific expression are not known. Therefore, it is important to indicate the isoform being analyzed just as it is for the GABARAP subfamily.
The mammalian Atg8 homologs share from 29% to 94% sequence identity with the yeast protein and have all, apart from GABARAPL3, been demonstrated to be involved in autophagosome biogenesis.141 The LC3 proteins are involved in phagophore formation, with participation of GABARAP subfamily members in later stages of autophagosome formation, in particular phagophore elongation and closure.142 Some evidence, however, suggests that at least in certain cell types the LC3 subfamily may be dispensable for bulk autophagic sequestration of cytosolic proteins, whereas the GABARAP subfamily is absolutely required.143 Due to unique features in their molecular surface charge distribution,144 emerging evidence indicates that LC3 and GABARAP proteins may be involved in recognizing distinct sets of cargoes for selective autophagy.145-147 Nevertheless, in most published studies, LC3 has been the primary Atg8 homolog examined in mammalian cells and the one that is typically characterized as an autophagosome marker per se. Note that although this protein is referred to as “Atg8” in many other systems, we primarily refer to it here as LC3 to distinguish it from the yeast protein and from the GABARAP subfamily. LC3, like the other Atg8 homologs, is initially synthesized in an unprocessed form, proLC3, which is converted into a proteolytically processed form lacking amino acids from the C terminus, LC3-I, and is finally modified into the PE-conjugated form, LC3-II (Fig. 6). Atg8–PE/LC3-II is the only protein marker that is reliably associated with completed autophagosomes, but is also localized to phagophores. In yeast, Atg8 amounts increase at least 10-fold when autophagy is induced.148 In mammalian cells, however, the total levels of LC3 do not necessarily change in a predictable manner, as there may be increases in the conversion of LC3-I to LC3-II, or a decrease in LC3-II relative to LC3-I if degradation of LC3-II via lysosomal turnover is particularly rapid (this can also be a concern in yeast with regard to vacuolar turnover of Atg8–PE). Both of these events can be seen sequentially in several cell types as a response to total nutrient and serum starvation. In cells of neuronal origin a high ratio of LC3-I to LC3-II is a common finding.149 For instance, SH-SY5Y neuroblastoma cell lines display only a slight increase of LC3-II after nutrient deprivation, whereas LC3-I is clearly reduced. This is likely related to a high basal autophagic flux, as suggested by the higher increase in LC3-II when cells are treated with NH4Cl,150,151 although cell-specific differences in transcriptional regulation of LC3 may also play a role. In fact stimuli or stress that inhibit transcription or translation of LC3 might actually be misinterpreted as inhibition of autophagy. Importantly, in brain tissue, LC3-I is much more abundant than LC3-II and the latter form is most easily discernable in enriched fractions of autophagosomes, autolysosomes and ER, and may be more difficult to detect in crude homogenate or cytosol.152 Indeed, when brain crude homogenate is run in parallel to a crude liver fraction, both LC3-I and LC3-II are observed in the liver, but only LC3-I may be discernible in brain homogenate (L. Toker and G. Agam, personal communication), depending on the LC3 antibody used.153 In studies of the brain, immunoblot analysis of the membrane and cytosol fraction from a cell lysate, upon appropriate loading of samples to achieve quantifiable and comparative signals, can be useful to measure LC3 isoforms.
Figure 6.
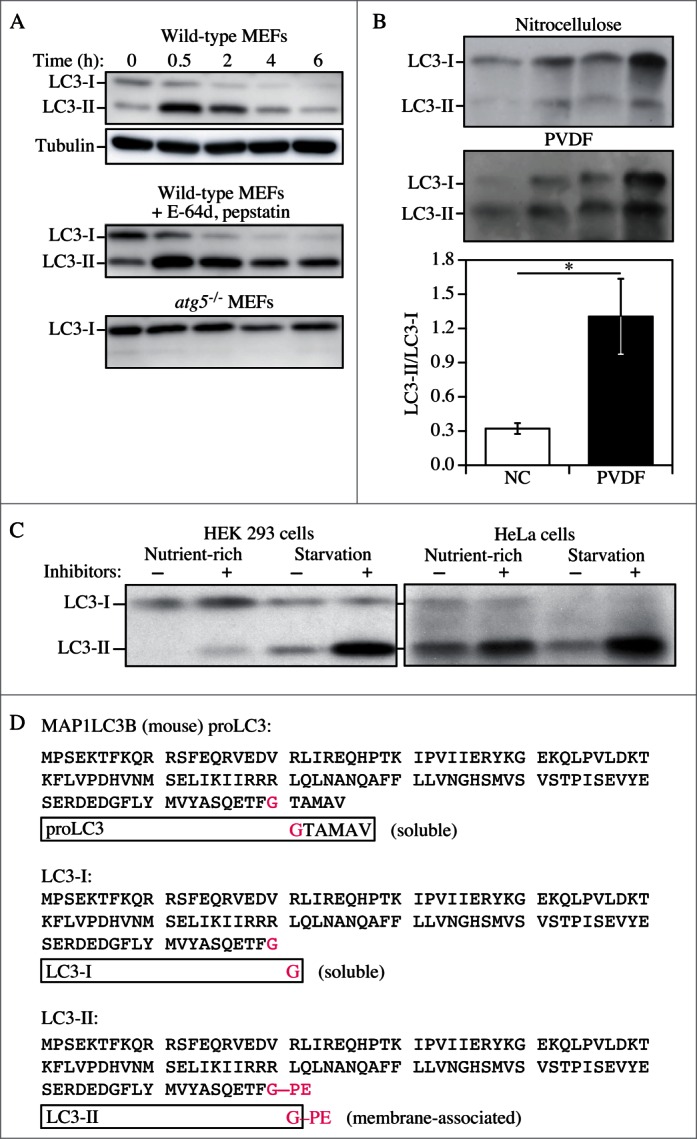
LC3-I conversion and LC3-II turnover. (A) Expression levels of LC3-I and LC3-II during starvation. Atg5+/+ (wild-type) and atg5-/- MEFs were cultured in DMEM without amino acids and serum for the indicated times, and then subjected to immunoblot analysis using anti-LC3 antibody and anti-tubulin antibody. E-64d (10 μg/ml) and pepstatin A (10 μg/ml) were added to the medium where indicated. Positions of LC3-I and LC3-II are marked. The inclusion of lysosomal protease inhibitors reveals that the apparent decrease in LC3-II is due to lysosomal degradation as easily seen by comparing samples with and without inhibitors at the same time points (the overall decrease seen in the presence of inhibitors may reflect decreasing effectiveness of the inhibitors over time). Monitoring autophagy by following steady state amounts of LC3-II without including inhibitors in the analysis can result in an incorrect interpretation that autophagy is not taking place (due to the apparent absence of LC3-II). Conversely, if there are high levels of LC3-II but there is no change in the presence of inhibitors, this may indicate that induction has occurred but that the final steps of autophagy are blocked, resulting in stabilization of this protein. This figure was modified from data previously published in ref. 26, and is reproduced by permission of Landes Bioscience, copyright 2007. (B) Lysates of 4 human adipose tissue biopsies were resolved on 2-12% polyacrylamide gels, as described previously.217 Proteins were transferred in parallel to either a PVDF or a nitrocellulose membrane, and blotted with anti-LC3 antibody, and then identified by reacting the membranes with an HRP-conjugated anti-rabbit IgG antibody, followed by ECL. The LC3-II/LC3-I ratio was calculated based on densitometry analysis of both bands. *, P < 0.05. (C) HEK 293 and HeLa cells were cultured in nutrient-rich medium (DMEM containing 10% fetal calf serum) or incubated for 4 h in starvation conditions (Krebs-Ringer medium) in the absence (−) or presence (+) of E-64d and pepstatin at 10 μg/ml each (Inhibitors). Cells were then lysed and the proteins resolved by SDS-PAGE. Endogenous LC3 was detected by immunoblotting. Positions of LC3-I and LC3-II are indicated. In the absence of lysosomal protease inhibitors, starvation results in a modest increase (HEK 293 cells) or even a decrease (HeLa cells) in the amount of LC3-II. The use of inhibitors reveals that this apparent decrease is due to lysosome-dependent degradation. This figure was modified from data previously published in ref. 174, and is reproduced by permission of Landes Bioscience, copyright 2005. (D) Sequence and schematic representation of the different forms of LC3B. The sequence for the nascent (proLC3) from mouse is shown. The glycine at position 120 indicates the cleavage site for ATG4. After this cleavage, the truncated LC3 is referred to as LC3-I, which is still a soluble form of the protein. Conjugation to PE generates the membrane-associated LC3-II form (equivalent to Atg8–PE).
The pattern of LC3-I to LC3-II conversion seems not only to be cell specific, but also related to the kind of stress to which cells are subjected. For example, SH-SY5Y cells display a strong increase of LC3-II when treated with the mitochondrial uncoupler CCCP, a well-known inducer of mitophagy (although it has also been reported that CCCP may actually inhibit mitophagy154). Thus, neither assessment of LC3-I consumption nor the evaluation of LC3-II levels would necessarily reveal a slight induction of autophagy (e.g., by rapamycin). Also, there is not always a clear precursor/product relationship between LC3-I and LC3-II, because the conversion of the former to the latter is cell type-specific and dependent on the treatment used to induce autophagy. Accumulation of LC3-II can be obtained by interrupting the autophagosome-lysosome fusion step (e.g., by depolymerizing acetylated microtubules with vinblastine), by inhibiting the ATP2A/SERCA Ca2+ pump, by specifically inhibiting the V-ATPase with bafilomycin A1155-157 or by raising the lysosomal pH by the addition of chloroquine,158,159 although some of these treatments may increase autophagosome numbers by disrupting the lysosome-dependent activation of MTOR (mechanistic target of rapamycin [serine/threonine kinase] complex 1 [MTORC1; note that the original term “mTOR” was named to distinguish the “mammalian” target of rapamycin from the yeast proteins160], a major suppressor of autophagy induction),161,162 or by inhibiting lysosome-mediated proteolysis (e.g., with a cysteine protease inhibitor such as E-64d, the aspartic protease inhibitor pepstatin A, the cysteine, serine and threonine protease inhibitor leupeptin or treatment with bafilomycin A1, NH4Cl or chloroquine158,163,164). Western blotting can be used to monitor changes in LC3 amounts (Fig. 6);26,165 however, even if the total amount of LC3 does increase, the magnitude of the response is generally less than that documented in yeast. It is worth noting that since the conjugated forms of the GABARAP subfamily members are usually undetectable without induction of autophagy in mammalian and other vertebrate cells,166,167 these proteins might be more suitable than LC3 to study and quantify subtle changes in autophagy induction.
In most organisms, Atg8/LC3 is initially synthesized with a C-terminal extension that is removed by the Atg4 protease. Accordingly, it is possible to use this processing event to monitor Atg4 activity. For example, when GFP is fused at the C terminus of Atg8 (Atg8-GFP), the GFP moiety is removed in the cytosol to generate free Atg8 and GFP. This processing can be easily monitored by western blot.168 It is also possible to use assays with an artificial fluorogenic substrate, or a fusion of LC3B to phospholipase A2 that allows the release of the active phospholipase for a subsequent fluorogenic assay,169 and there is a fluorescence resonance energy transfer (FRET)-based assay utilizing CFP and YFP tagged versions of LC3B and GABA-RAPL2/GATE-16 that can be used for high-throughput screening.170 Another method to monitor ATG4 activity in vivo uses the release of Gaussia luciferase from the C terminus of LC3 that is tethered to actin.171 Note that there are 4 Atg4 homologs in mammals, and they have different activities with regard to the Atg8 subfamilies of proteins.172 ATG4A is able to cleave the GABARAP subfamily, but has very limited activity toward the LC3 subfamily, whereas ATG4B is apparently active against most or all of these proteins.139,140 The ATG4C and ATG4D isoforms have minimal activity for any of the Atg8 homologs. In particular because a C-terminal fusion will be cleaved immediately by Atg4, researchers should be careful to specify whether they are using GFP-Atg8/LC3 (an N-terminal fusion, which can be used to monitor various steps of autophagy) or Atg8/LC3-GFP (a C-terminal fusion, which can only be used to monitor Atg4 activity).173
Cautionary notes: There are several important caveats to using Atg8/LC3-II or GABARAP-II to visualize fluctuations in autophagy. First, changes in LC3-II amounts are tissue- and cell context-dependent.153,174 Indeed, in some cases, autophagosome accumulation detected by TEM does not correlate well with the amount of LC3-II (Z. Tallóczy, R.L.A. de Vries, and D. Sulzer, unpublished results; E.-L. Eskelinen, unpublished results). This is particularly evident in those cells that show low levels of LC3-II (based on western blotting) because of an intense autophagic flux that consumes this protein,175 or in cell lines having high levels of LC3-II that are tumor-derived, such as MDA-MB-231.174 Conversely, without careful quantification the detectable formation of LC3-II is not sufficient evidence for autophagy. For example, homozygous deletion of Becn1 does not prevent the formation of LC3-II in embryonic stem cells even though autophagy is substantially reduced, whereas deletion of Atg5 results in the complete absence of LC3-II (see Fig. 5A and supplemental data in ref. 176). The same is true for the generation of Atg8–PE in yeast in the absence of VPS30/ATG6 (see Fig. 7 in ref. 177). Thus, it is important to remember that not all of the autophagy-related proteins are required for Atg8/LC3 processing, including lipidation.177 Vagaries in the detection and amounts of LC3-I versus LC3-II present technical problems. For example, LC3-I is very abundant in brain tissue, and the intensity of the LC3-I band may obscure detection of LC3-II, unless the polyacrylamide crosslinking density is optimized, or the membrane fraction of LC3 is first separated from the cytosolic fraction.44 Conversely, certain cell lines have much less visible LC3-I compared to LC3-II. In addition, tissues may have asynchronous and heterogeneous cell populations, and this variability may present challenges when analyzing LC3 by western blotting.
Second, LC3-II also associates with the membranes of nonautophagic structures. For example, some members of the PCDHGC/γ-protocadherin family undergo clustering to form intracellular tubules that emanate from lysosomes.178 LC3-II is recruited to these tubules, where it appears to promote or stabilize membrane expansion. Furthermore, LC3 can be recruited directly to apoptotic cell-containing phagosome membranes,179,180 macropinosomes,179 the parasitophorous vacuole of Toxoplasma gondii,181 and single-membrane entotic vacuoles,179 as well as to bacteria-containing phagosome membranes under certain immune activating conditions, for example, toll-like receptor (TLR)-mediated stimulation in LC3-associated phagocytosis.182,183 Importantly, LC3 is involved in secretory trafficking as it has been associated with secretory granules in mast cells184 and PC12 hormone-secreting cells.185 LC3 is also detected on secretory lysosomes in osteoblasts186 and in amphisome-like structures involved in mucin secretion by goblet cells.187 Therefore, in studies of infection of mammalian cells by bacterial pathogens, the identity of the LC3-II labeled compartment as an autophagosome should be confirmed by a second method, such as TEM. It is also worth noting that autophagy induced in response to bacterial infection is not directed solely against the bacteria but can also be a response to remnants of the phagocytic membrane.188 Similar cautions apply with regard to viral infection. For example, coronaviruses induce autophagosomes during infection through the expression of nsp6; however, coronaviruses also induce the formation of double-membrane vesicles that are coated with LC3-I, the nonlipidated form of LC3 that plays an autophagy-independent role in viral replication.189,190 Similarly, nonlipidated LC3 marks replication complexes in flavivirus (Japanese encephalitis virus)-infected cells and is essential for viral replication.191 Along these lines, during herpes simplex virus type 1 (HSV-1) infection, an LC3+ autophagosome-like organelle that is derived from nuclear membranes and that contains viral proteins is observed,192 whereas influenza A virus directs LC3 to the plasma membrane via a LC3-interacting region (LIR) motif in its M2 protein.193 Moreover, in vivo studies have shown that coxsackievirus (an enterovirus) induces formation of autophagy-like vesicles in pancreatic acinar cells, together with extremely large autophagy-related compartments that have been termed megaphagosomes;194 the absence of ATG5 disrupts viral replication and prevents the formation of these structures.195
Third, caution must be exercised in general when evaluating LC3 by western blotting, and appropriate standardization controls are necessary. For example, LC3-I may be less sensitive to detection by certain anti-LC3 antibodies. Moreover, LC3-I is more labile than LC3-II, being more sensitive to freezing-thawing and to degradation in SDS sample buffer. Therefore, fresh samples should be boiled and assessed as soon as possible and should not be subjected to repeated freeze-thaw cycles. Alternatively, trichloroacetic acid precipitation of protein from fresh cell homogenates can be used to protect against degradation of LC3 by proteases that may be present in the sample. A general point to consider when examining transfected cells concerns the efficiency of transfection. A western blot will detect LC3 in the entire cell population, including those that are not transfected. Thus, if transfection efficiency is too low, it may be necessary to use methods, such as fluorescence microscopy, that allow autophagy to be monitored in single cells. The critical point is that the analysis of the gel shift of transfected LC3 or GFP-LC3 can be employed to follow LC3 lipidation only in highly transfectable cells.196
When dealing with animal tissues, western blotting of LC3 should be performed on frozen biopsy samples homogenized in the presence of general protease inhibitors (C. Isidoro, personal communication; see also Human).197 Caveats regarding detection of LC3 by western blotting have been covered in a review.26 For example, PVDF membranes may result in a stronger LC3-II retention than nitrocellulose membranes, possibly due to a higher affinity for hydrophobic proteins (Fig. 6B; J. Kovsan and A. Rudich, personal communication), and Triton X-100 may not efficiently solubilize LC3-II in some systems.198 Heating in the presence of 1% SDS, or analysis of membrane fractions,44 may assist in the detection of the lipidated form of this protein. This observation is particularly relevant for cells with a high nucleocytoplasmic ratio, such as lymphocytes. Under these constraints, direct lysis in Laemmli loading buffer, containing SDS, just before heating, greatly improves LC3 detection on PVDF membranes, especially when working with a small number of cells (F. Gros, unpublished observations).199 Analysis of a membrane fraction is particularly useful for brain where levels of soluble LC3-I greatly exceed the level of LC3-II.
One of the most important issues is the quantification of changes in LC3-II, because this assay is one of the most widely used in the field and is often prone to misinterpretation. Levels of LC3-II should be compared to actin (e.g., ACTB), but not to LC3-I (see the caveat in the next paragraph), and, ideally, to more than one “housekeeping” protein (HKP). Actin and other HKPs are usually abundant and can easily be overloaded on the gel200 such that they are not detected within a linear range. Moreover, actin levels may decrease when autophagy is induced in many organisms from yeast to mammals. For any proteins used as “loading controls” (including actin, tubulin and GAPDH) multiple exposures of the western blot are generally necessary to ensure that the signals are detected in the linear range. An alternative approach is to stain for total cellular proteins with Coomassie Brilliant Blue and Ponceau Red,201 but these methods are generally less sensitive and may not reveal small differences in protein loading. Stain-Free gels, which also stain for total cellular proteins, have been shown to be an excellent alternative to HKPs.202
It is important to realize that ignoring the level of LC3-I in favor of LC3-II normalized to HKPs may not provide the full picture of the cellular autophagic response.153,203 For example, in aging skeletal muscle the increase in LC3-I is at least as important as that for LC3-II.204,205 Quantification of both isoforms is therefore informative, but requires adequate conditions of electrophoretic separation. This is particularly important for samples where the amount of LC3-I is high relative to LC3-II (as in brain tissues, where the LC3-I signal can be overwhelming). Under such a scenario, it may be helpful to use gradient gels to increase the separation of LC3-I from LC3-II and/or cut away the part of the blot with LC3-I prior to the detection of LC3-II. Furthermore, since the dynamic range of LC3 immunoblots is generally quite limited, it is imperative that other assays be used in parallel in order to draw valid conclusions about changes in autophagy activity.
Fourth, in mammalian cells LC3 is expressed as multiple isoforms (LC3A, LC3B, LC3B2 and LC3C206,207), which exhibit different tissue distributions and whose functions are still poorly understood. A point of caution along these lines is that the increase in LC3A-II versus LC3B-II levels may not display equivalent changes in all organisms under autophagy-inducing conditions, and it should not be assumed that LC3B is the optimal protein to monitor.208 A key technical consideration is that the isoforms may exhibit different specificities for antisera or antibodies. Thus, it is highly recommended that investigators report exactly the source and catalog number of the antibodies used to detect LC3 as this might help avoid discrepancies between studies. The commercialized anti-LC3B antibodies also recognize LC3A, but do not recognize LC3C, which shares less sequence homology. It is important to note that LC3C possesses in its primary amino acid sequence the DYKD motif that is recognized with a high affinity by anti-FLAG antibodies. Thus, the standard anti-FLAG M2 antibody can detect and immunoprecipitate overexpressed LC3C, and caution has to be taken in experiments using FLAG-tagged proteins (M. Biard-Piechaczyk and L. Espert, personal communication). Note that according to Ensembl there is no LC3C in mouse or rat.
In addition, it is important to keep in mind the other subfamily of Atg8 proteins, the GABARAP subfamily (see above).141,209 Certain types of mitophagy induced by BNIP3L/NIX are highly dependent on GABARAP and less dependent on LC3 proteins.210,211 Furthermore, commercial antibodies for GABARAPL1 also recognize GABARAP,138,143 which might lead to misinterpretation of experiments, in particular those using immunohistochemical techniques. Sometimes the problem with cross-reactivity of the anti-GABARAPL1 antibody can be overcome when analyzing these proteins by western blot because the isoforms can be resolved during SDS-PAGE using high concentration (15%) gels, as GABARAP migrates faster than GABARAPL1 (M. Boyer-Guittaut, personal communication; also see Fig. S4 in ref. 143). Because GABARAP and GABARAPL1 can both be proteolytically processed and lipidated, generating GABARAP-I or GABARAPL1-I and GABARAP-II or GABARAPL1-II, respectively, this may lead to a misassignment of the different bands. As soon as highly specific antibodies that are able to discriminate between GABARAP and GABARAPL1 become available, we strongly advise their use; until then, we advise caution in interpreting results based on the detection of these proteins by western blot. Antibody specificity can be assessed after complete inhibition of GABARAP (or any other Atg8 family protein) expression by RNA interference.143,167 In general, we advise caution in choosing antibodies for western blotting and immunofluorescence experiments and in interpreting results based on stated affinities of antibodies unless these have been clearly determined. As with any western blot, proper methods of quantification must be used, which are, unfortunately, often not well disseminated; readers are referred to an excellent paper on this subject (see ref. 212). Unlike the other members of the GABARAP family, almost no information is available on GABARAPL3, perhaps because it is not yet possible to differentiate between GABA-RAPL1 and GABARAPL3 proteins, which have 94% identity. As stated by the laboratory that described the cloning of the human GABARAPL1 and GABARAPL3 genes,209 their expression patterns are apparently identical. It is worth noting that GABARAPL3 is the only gene of the GABARAP subfamily that seems to lack an ortholog in mice.209 GABARAPL3 might therefore be considered as a pseudogene without an intron that is derived from GABARAPL1. Hence, until new data are published, GABARAPL3 should not be considered as the fourth member of the GABARAP family.
Fifth, in non-mammalian species, the discrimination of Atg8–PE from the nonlipidated form can be complicated by their nearly identical SDS-PAGE mobilities and the presence of multiple isoforms (e.g., there are 9 in Arabidopsis). In yeast, it is possible to resolve Atg8 (the nonlipidated form) from Atg8–PE by including 6 M urea in the SDS-PAGE separating gel,213 or by using a 15% resolving gel without urea (F. Reggiori, personal communication). Similarly, urea combined with prior treatment of the samples with (or without) phospholipase D (that will remove the PE moiety) can often resolve the ATG8 species in plants.214,215 It is also possible to label cells with radioactive ethanolamine, followed by autoradiography to identify Atg8–PE, and a C-terminal peptide can be analyzed by mass spectrometry to identify the lipid modification at the terminal glycine residue. Special treatments are not needed for the separation of mammalian LC3-I from LC3-II.
Sixth, it is important to keep in mind that ATG8, and to a lesser extent LC3, undergoes substantial transcriptional and posttranscriptional regulation. Accordingly, to obtain an accurate interpretation of Atg8/LC3 protein levels it is also necessary to monitor the mRNA levels. Without analyzing the corresponding mRNA it is not possible to discriminate between changes that are strictly reflected in altered amounts of protein versus those that are due to changes in transcription (e.g., the rate of transcription, or the stability of the message). For example, in cells treated with the calcium ionophore A23187 or the ER calcium pump blocker thapsigargin, an obvious correlation is found between the time-dependent increases in LC3B-I and LC3B-II protein levels, as well as with the observed increase in LC3B mRNA levels.216 Clinically, in human adipose tissue, protein and mRNA levels of LC3 in omental fat are similarly elevated in obese compared to lean individuals.217
Seventh, LC3-I can be fully degraded by the 20S proteasome or, more problematically, processed to a form appearing equal in size to LC3-II on a western blot (LC3-T); LC3-T was identified in HeLa cells and is devoid of the ubiquitin conjugation domain, thus lacking its adaptor function for autophagy.218
Eighth, a general issue when working with cell lines is that we recommend that validation be performed to verify the cell line(s) being used, and to verify the presence of genetic alterations as appropriate. Depending on the goal (e.g., to indicate general applicability of a particular treatment) it may be important to use more than one cell line to confirm the results. It is also critical to test for mycoplasma because the presence of this contaminant can significantly alter the results with regard to any autophagic response. For these reasons, we also recommend the use of low passage numbers for nonprimary cells or cell lines (no more than 40 passages or 6 months after thawing).
Finally, we would like to point out that one general issue with regard to any assay is that experimental manipulation could introduce some type of stress—for example, mechanical stress due to lysis, temperature stress due to heating or cooling a sample, or oxidative stress on a microscope slide, which could lead to potential artifacts including the induction of autophagy—even maintaining cells in higher than physiologically normal oxygen levels can be a stress condition.219 Special care should be taken with cells in suspension, as the stress resulting from centrifugation can induce autophagy. This point is not intended to limit the use of any specific methodology, but rather to note that there are no perfect assays. Therefore, it is important to verify that the positive (e.g., treatment with rapamycin, torin1 or other inducers) and negative (e.g., inhibitor treatment) controls behave as expected in any assays being utilized. Similarly, plasmid transfection or nucleofection can result in the potent induction of autophagy (based on increases in LC3-II or SQSTM1/p62 degradation). In some cell types, the amount of autophagy induced by transfection of a control empty vector may be so high that it is virtually impossible to examine the effect of enforced gene expression on autophagy (B. Levine, personal communication). It is thus advisable to perform time course experiments to determine when the transfection effect returns to acceptably low levels and to use appropriate time-matched transfection controls (see also the discussion in GFP-Atg8/LC3 fluorescence microscopy). This effect is generally not observed with siRNA transfection; however, it is an issue for plasmid expression constructs including those for shRNA and for viral delivery systems. The use of endotoxin-free DNA reduces, but does not eliminate, this problem. In many cells the cationic polymers used for DNA transfection, such as liposomes and polyplex, induce large tubulovesicular autophagosomes (TVAs) in the absence of DNA.220 These structures accumulate SQSTM1 and fuse slowly with lysosomes. Interestingly, these TVAs appear to reduce gene delivery, which increases 8–10 fold in cells that are unable to make TVAs due to the absence of ATG5. Finally, the precise composition of media components and the density of cells in culture can have profound effects on basal autophagy levels and may need to be modified empirically depending on the cell lines being used. Along these lines various types of media, in particular those with different serum levels (ranging from 0–15%), may have profound effects with regard to how cells (or organs) perceive a fed versus starved state. For example, normal serum contains significant levels of cytokines and hormones that likely regulate the basal levels of autophagy; thus, the use of dialyzed serum might be an alternative for these studies. In addition, the amino acid composition of the medium/assay buffer may have profound effects on initiation or progression of autophagy. For example, in the protozoan parasite Trypanosoma brucei starvation-induced autophagy can be prevented by addition of histidine to the incubation buffer.221 For these reasons, the cell culture conditions should be fully described. It is also important to specify duration of autophagy stimulation, as long-term autophagy can modify signal transduction pathways of importance in cell survival.222
Conclusion: Atg8/LC3 is often an excellent marker for autophagic structures; however, it must be kept in mind that there are multiple LC3 isoforms, there is a second family of mammalian Atg8-like proteins (GABARAPs), and antibody affinity (for LC3-I versus LC3-II) and specificity (for example, for LC3A versus LC3B) must be considered and/or determined. Moreover, LC3 levels on their own do not address issues of autophagic flux. Finally, even when flux assays are carried out, there is a problem with the limited dynamic range of LC3 immunoblots; accordingly, this method should not be used by itself to analyze changes in autophagy.
b. Turnover of LC3-II/Atg8–PE
Autophagic flux is often inferred on the basis of LC3-II turnover, measured by western blot (Fig. 6C)174 in the presence and absence of lysosomal, or vacuolar degradation. However, it should be cautioned that such LC3 assays are merely indicative of autophagic “carrier flux”, not of actual autophagic cargo/substrate flux. It has, in fact, been observed that in rat hepatocytes, an autophagic-lysosomal flux of LC3-II can take place in the absence of an accompanying flux of cytosolic bulk cargo.223 The relevant parameter in LC3 assays is the difference in the amount of LC3-II in the presence and absence of saturating levels of inhibitors, which can be used to examine the transit of LC3-II through the autophagic pathway; if flux is occurring, the amount of LC3-II will be higher in the presence of the inhibitor.174 Lysosomal degradation can be prevented through the use of protease inhibitors (e.g., pepstatin A, leupeptin and E-64d), compounds that neutralize the lysosomal pH such as bafilomycin A1, chloroquine or NH4Cl,16,149,158,164,224,225 or by treatment with agents that block the fusion of autophagosomes with lysosomes (note that bafilomycin A1 will ultimately cause a fusion block as well as neutralize the pH,156 but the inhibition of fusion may be due to a block in ATP2A/SERCA activity226).155-157,227 Alternatively, knocking down or knocking out LAMP2 (lysosomal-associated membrane protein 2) represents a genetic approach to block the fusion of autophagosomes and lysosomes (for example, inhibiting LAMP2 in myeloid leukemic cells results in a marked increase of GFP-LC3 dots and endogenous LC3-II protein compared to control cells upon autophagy induction during myeloid differentiation [M.P. Tschan, unpublished data]).228 This approach, however, is only valid when the knockdown of LAMP2 is directed against the mRNA region specific for the LAMP2B spliced variant, as targeting the region common to the 3 variants would also inhibit chaperone-mediated autophagy, which may result in the compensatory upregulation of macroautophagy.92,229,230
Increased levels of LC3-II in the presence of lysosomal inhibition or interfering with autophagosome-lysosome fusion alone (e.g., with bafilomycin A1) may be indicative of autophagic carrier flux (to the stage of cargo reaching the lysosome), but to assess whether a particular treatment alters complete autophagic flux through substrate digestion, the treatment plus bafilomycin A1 must be compared with results obtained with treatment alone as well as with bafilomycin A1 alone. An additive or supra-additive effect in LC3-II levels may indicate that the treatment enhances autophagic flux (Fig. 6C). Moreover, higher LC3-II levels with treatment plus bafilomycin A1 compared to bafilomycin A1 alone may indicate that the treatment increases the synthesis of autophagy-related membranes. If the treatment by itself increases LC3-II levels, but the treatment plus bafilomycin A1 does not increase LC3-II levels compared to bafilomycin A1 alone, this may indicate that the treatment induced a partial block in autophagic flux. Thus, a treatment condition increasing LC3-II on its own that has no difference in LC3-II in the presence of bafilomycin A1 compared to treatment alone may suggest a complete block in autophagy at the terminal stages.231 This procedure has been validated with several autophagy modulators.232 With each of these techniques, it is essential to avoid assay saturation. The duration of the bafilomycin A1 treatment (or any other inhibitor of autophagic flux such as chloroquine) needs to be relatively short (1–4 h)233 to allow comparisons of the amount of LC3 that is lysosomally degraded over a given time frame under one treatment condition to another treatment condition. A dose-curve and time-course standardization for the use of autophagic flux inhibitors is required for the initial optimization of the conditions to detect LC3-II accumulation and avoid nonspecific or secondary effects. By using a rapid screening approach, such as a colorimetric-based platform method,234 it is possible to monitor a long time frame for autolysosome accumulation, which closely associates with autophagy efficiency.235 Positive control experiments using treatment with known autophagy inducers, along with bafilomycin A1 versus vehicle, are important to demonstrate the utility of this approach in each experimental context. The same type of assay monitoring the turnover of Atg8–PE can be used to monitor flux in yeast, by comparing the amount of Atg8 present in a wild-type versus a pep4Δ strain following autophagy induction;236 however, it is important to be aware that the PEP4 knockout can influence yeast cell physiology. PMSF, which inhibits the activity of Prb1, can also be used to block Atg8–PE turnover.
An additional methodology for monitoring autophagy relies on the observation that in some cell types a subpopulation of LC3-II exists in a cytosolic form (LC3-IIs).237-239 The amount of cytosolic LC3-IIs and the ratio between LC3-I and LC3-IIs appears to correlate with changes in autophagy and may provide a more accurate measure of autophagic flux than ratios based on the total level of LC3-II.239 The validity of this method has been demonstrated by comparing autophagic proteolytic flux in rat hepatocytes, hepatoma cells and myoblasts. One advantage of this approach is that it does not require the presence of autophagic or lysosomal inhibitors to block the degradation of LC3-II.
Due to the advances in time-lapse fluorescence microscopy and the development of photoswitchable fluorescent proteins, autophagic flux can also be monitored by assessing the half-life of the LC3 protein240 post-photoactivation or by quantitatively measuring the autophagosomal pool size and its transition time.241 These approaches deliver invaluable information on the kinetics of the system and the time required to clear a complete autophagosomal pool. Nonetheless, care must be taken for this type of analysis as changes in translational/transcriptional regulation of LC3 might also affect the readout.
Finally, autophagic flux can be monitored based on the turnover of LC3-II, by utilizing a luminescence-based assay. For example, a reporter assay based on the degradation of Renilla reniformis luciferase (Rluc)-LC3 fusion proteins is well suited for screening compounds affecting autophagic flux.242 In this assay, Rluc is fused N-terminally to either wild-type LC3 (LC3WT) or a lipidation-deficient mutant of LC3 (G120A). Since Rluc-LC3WT, in contrast to Rluc-LC3G120A, specifically associates with the autophagosomal membranes, Rluc-LC3WT is more sensitive to autophagic degradation. A change in autophagy-dependent LC3 turnover can thus be estimated by monitoring the change in the ratio of luciferase activities between the 2 cell populations expressing either Rluc-LC3WT or Rluc-LC3G120A. In its simplest form, the Rluc-LC3-assay can be used to estimate autophagic flux at a single time point by defining the luciferase activities in cell extracts. Moreover, the use of a live cell luciferase substrate makes it possible to monitor changes in autophagic activity in live cells in real time. This method has been successfully used to identify positive and negative regulators of autophagy from cells treated with microRNA, siRNA and small molecule libraries.242-248
Cautionary notes: The main caveat regarding the measurement of LC3-IIs/LC3-I is that this method has only been tested in isolated rat hepatocytes and H4-II-E cells. Thus, it is not yet known whether it is generally applicable to other cell types. Indeed, a soluble form of LC3-II (i.e., LC3-IIs) is not observed in many standard cell types including HeLa, HEK 293 and PC12. In addition, the same concerns apply regarding detection of LC3-I by western blotting. It should be noted that the LC3-IIs/LC3-I ratio must be analyzed using the cytosolic fractions rather than the total homogenates. Furthermore, the same caveats mentioned above regarding the use of LC3 for qualitatively monitoring autophagy also apply to the use of this marker for evaluating flux.
The use of a radioactive pulse-chase analysis, which assesses complete autophagic flux, provides an alternative to lysosomal protease inhibitors,148 although such inhibitors should still be used to verify that degradation is lysosome-dependent. In addition, drugs must be used at concentrations and for time spans that are effective in inhibiting fusion or degradation, but that do not provoke cell death. Thus, these techniques may not be practical in all cell types or in tissues from whole organisms where the use of protease inhibitors is problematic, and where pulse labeling requires artificial short-term culture conditions that may induce autophagy. Another concern when monitoring flux via LC3-II turnover may be seen in the case of a partial autophagy block; in this situation, agents that disrupt autophagy (e.g., bafilomycin A1) will still result in an increase in LC3-II. Thus, care is needed in interpretation. For characterizing new autophagy modulators, it is ideal to test autophagic flux at early (e.g., 4 h) and late (e.g., 24 h) time-points, since in certain instances, such as with calcium phosphate precipitates, a compound may increase or decrease flux at these 2 time points, respectively.233 Moreover, it is important to consider assaying autophagy modulators in a long-term response in order to further understand their effects. Finally, many of the chemicals used to inhibit autophagy, such as bafilomycin A1, NH4Cl (see Autophagy inhibitors and inducers) or chloroquine, also directly inhibit the endocytosis/uncoating of viruses (D.R. Smith, personal communication), and other endocytic events requiring low pH, as well as exit from the Golgi (S. Tooze, personal communication). As such, agents that neutralize endosomal compartments should be used only with extreme caution in studies investigating autophagy-virus interactions.
One additional consideration is that it may not be absolutely necessary to follow LC3-II turnover if other substrates are being monitored simultaneously. For example, an increase in LC3-II levels in combination with the lysosomal (or ideally autophagy-specific) removal of an autophagic substrate (such as an organelle249,250) that is not a good proteasomal substrate provides an independent assessment of autophagic flux. However, it is probably prudent to monitor both turnover of LC3-II and an autophagosome substrate in parallel, due to the fact that LC3 might be coupled to endosomal membranes and not just autophagosomes, and the levels of well-characterized autophagosome substrates such as SQSTM1 can also be affected by proteasome inhibitors.251
Another issue relates to the use of protease inhibitors (see Autophagy inhibitors and inducers). When using lysosomal protease inhibitors, it is of fundamental importance to assess proper conditions of inhibitor concentration and time of pre-incubation to ensure full inhibition of lysosomal cathepsins. In this respect, 1 h of pre-incubation with 10 µg/ml E-64d is sufficient in most cases, since this inhibitor is membrane permeable and rapidly accumulates within lysosomes, but another frequently used inhibitor, leupeptin, requires at least 6 h pre-incubation.59 Moreover, pepstatin A is membrane impermeable (ethanol or preferably DMSO must be employed as a vehicle) and requires a prolonged incubation (>8 h) and a relatively high concentration (>50 µg/ml) to fully inhibit lysosomal CTSD (Fig. 7). An incubation of this duration, however, can be problematic due to indirect effects (see GFP-Atg8/LC3 lysosomal delivery and proteolysis). At least in neurons, pepstatin alone is a less effective lysosomal proteolytic block, and combining a cysteine protease inhibitor with it is most effective.59 Also, note that the relative amount of lysosomal CTSB (cathepsin B) and CTSD is cell-specific and changes with culture conditions. A possible alternative to pepstatin A is the pepstatin A, BODIPY® FL conjugate,252,253 which is transported to lysosomes via endocytosis. In contrast to the protease inhibitors, chloroquine (10–40 µM) or bafilomycin A1 (1–100 nM) can be added to cells immediately prior to autophagy induction. Because cysteine protease inhibitors upregulate CTSD and have potential inhibitory activity toward calpains and other cysteine proteases, whereas bafilomycin A1 can have potential significant cytotoxicity, especially in cultured neurons and pathological states, the use of both methods may be important in some experiments to exclude off-target effects of a single method.
Figure 7.
Effect of different inhibitors on LC3-II accumulation. SH-SY5Y human neuroblastoma cells were plated and allowed to adhere for a minimum of 24 h, then treated in fresh medium. Treatments were as follows: rapamycin (Rap), (A) 1 µM, 4 h or (B) 10 µM, 4 h; E-64d, final concentration 10 µg/ml from a 1 mg/ml stock in ethanol (EtOH); NH4Cl (NH4+), final concentration 10 mM from a 1 M stock in water; pepstatin A (Pst), final concentration 10 µg/ml from a 1 mg/ml stock in ethanol, or 68.6 µg/ml from a 6.86 mg/ml stock in DMSO; ethanol or DMSO, final concentration 1%. Pre-incubations in (B) were for 1 or 4 h as indicated. 10 mM NH4Cl (or 30 µM chloroquine, not shown) were the most effective compounds for demonstrating the accumulation of LC3-II. E-64d was also effective in preventing the degradation of LC3-II, with or without a preincubation, but ammomium chloride (or chloroquine) may be more effective. Pepstatin A at 10 µg/ml with a 1 h pre-incubation was not effective at blocking degradation, whereas a 100 µM concentration with 4 h pre-incubation had a partial effect. Thus, alkalinizing compounds are more effective in blocking LC3-II degradation, and pepstatin A must be used at saturating conditions to have any noticeable effect. Images provided by C. Isidoro. Note that the band running just below LC3-I at approximately 17.5 kDa may be a processing intermediate of LC3-I; it is detectable in freshly prepared homogenates, but is less visible after the sample is subjected to a freeze-thaw cycle.
Conclusion: It is important to be aware of the difference between monitoring the steady-state level of Atg8/LC3 and autophagic flux. The latter may be assessed by following Atg8/LC3 in the absence and presence of autophagy inhibitors, and by examining the autophagy-dependent degradation of appropriate substrates. In particular, if there is any evidence of an increase in LC3-II (or autophagosomes), it is essential to determine whether this represents increased flux, or a block in fusion or degradation through the use of inhibitors such as chloroquine or bafilomycin A1. In the case of a suspected impaired degradation, assessment of lysosomal function is then required to validate the conclusion and to establish the basis.
c. GFP-Atg8/LC3 lysosomal delivery and partial proteolysis
GFP-LC3B (hereafter referred to as GFP-LC3) has also been used to follow flux. It should be cautioned that, as with endogenous LC3, an assessment of autophagic GFP-LC3 flux is a carrier flux that cannot be equated with, and is not necessarily representative of, an autophagic cargo flux. When GFP-Atg8 or GFP-LC3 is delivered to a lysosome/vacuole, the Atg8/LC3 part of the chimera is sensitive to degradation, whereas the GFP protein is relatively resistant to hydrolysis (note, however, that GFP fluorescence is quenched by low pH; see GFP-Atg8/LC3 fluorescence microscopy and Tandem mRFP/mCherry-GFP fluorescence microscopy). Therefore, the appearance of free GFP on western blots can be used to monitor lysis of the inner autophagosome membrane and breakdown of the cargo in metazoans (Fig. 8A),236,254,255 or the delivery of autophagosomes to, and the breakdown of autophagic bodies within, the fungal and plant vacuole.214,215,236,256 Reports on Dictyostelium and mammalian cells highlight the importance of lysosomal pH as a critical factor in the detection of free GFP that results from the degradation of fused proteins. In these cell types, free GFP fragments are only detectable in the presence of nonsaturating levels of lysosomotropic compounds (NH4Cl or choroquine) or under conditions that attenuate lysosomal acidity; otherwise, the autophagic/degradative machinery appears to be too efficient to allow the accumulation of the proteolytic fragment (Fig. 8B,C).37,257 Hence, a reduction in the intensity of the free GFP band may indicate reduced flux, but it may also be due to efficient turnover. Using a range of concentrations and treatment times of compounds that inhibit autophagy can be useful in distinguishing between these possibilities.258 Since the pH in the yeast vacuole is higher than that in mammalian or Dictyostelium lysosomes, the levels of free GFP fragments are detectable in yeast even in the absence of lysosomotropic compounds.30 Additionally, in yeast the diffuse fluorescent haze from the released GFP moiety within the vacuole lumen can be observed by fluorescence microscopy.
Figure 8.
GFP-LC3 processing can be used to monitor delivery of autophagosomal membranes. (A) atg5-/- MEFs engineered to express Atg5 under the control of the Tet-off promoter were grown in the presence of doxycyline (Dox; 10 ng/ml) for one week to suppress autophagy. Cells were then cultured in the absence of drug for the indicated times, with or without a final 2 h starvation. Protein lysates were analyzed by western blot using anti-LC3 and anti-GFP antibodies. The positions of untagged and GFP-tagged LC3-I and LC3-II, and free GFP are indicated. This figure was modified from data previously published in ref. 255, FEBS Letters, 580, Hosokawa N, Hara Y, Mizushima N, Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size, pp. 2623–2629, copyright 2006, with permission from Elsevier. (B) Differential role of unsaturating and saturating concentrations of lysosomal inhibitors on GFP-LC3 cleavage. HeLa cells stably transfected with GFP-LC3 were treated with various concentrations of chloroquine (CQ) for 6 h. Total lysates were prepared and subjected to immunoblot analysis. (C) CQ-induced free GFP fragments require classical autophagy machinery. Wild-type and atg5-/- MEFs were first infected with adenovirus GFP-LC3 (100 viral particles per cell) for 24 h. The cells were then either cultured in regular culture medium with or without CQ (10 µM), or subjected to starvation in EBSS in the absence or presence of CQ for 6 h. Total lysates were prepared and subjected to immunoblot analysis. Panel (B) and (C) are modified from the data previously published in ref. 257.
The dynamic movement to lysosomes of GFP-LC3, or of its associated cargo, also can be monitored by time-lapse fluorescence microscopy, although, as mentioned above, the GFP fluorescent signal is more sensitive to acidic pH than other fluorophores (see GFP-Atg8/LC3 fluorescence microscopy). A time-course evaluation of the cell population showing GFP-LC3 puncta can serve to monitor the autophagic flux, since a constant increase in the number of cells accumulating GFP-LC3 puncta is suggestive of defective fusion of autophagosomes with lysosomes. Conversely, a decline implies that GFP-LC3 is delivered to properly acidified lysosomes and may, in addition, reflect proteolytic elimination within them, although the latter needs to be independently established. In either case, it can be problematic to use GFP fluorescence to follow flux, as new GFP-LC3 is continuously being synthesized. A potential solution to this problem is to follow the fluorescence of a photoactivatable version of the fluorescent protein,259 which allows this assay to be performed essentially as a pulse-chase analysis. Another alternative to follow flux is to monitor GFP-LC3 fluorescence by adding lysosomal protease or fusion inhibitors to cells expressing GFP-LC3 and monitoring changes in the number of puncta. In this case, the presence of lysosomal inhibitors should increase the number of GFP-LC3-positive structures, and the absence of an effect on the total number of GFP-LC3 puncta or on the percentage of cells displaying numerous puncta is indicative of a defect(s) in autophagic flux.260 The combination of protease inhibitors (to prevent the degradation of GFP) or compounds that modify lysosomal pH such as NH4Cl or chloroquine, or compounds that block fusion of autophagosomes with lysosomes such as bafilomycin A1 or others (e.g., vinblastine) may be most effective in preventing lysosome-dependent decreases in GFP-LC3 puncta. However, because the stability of GFP is affected by lysosomal pH, researchers may also consider the use of protease inhibitors whether or not lysosomotropic compounds or fusion inhibitors are included.
Cautionary notes: The GFP-Atg8 processing assay is used routinely to monitor autophagy in yeast. One caveat, however, is that this assay is not always carried out in a quantitative manner. For example, western blot exposures need to be in the linear range. Accordingly, an enzymatic assay such as the Pho8Δ60 assay may be preferred (see Autophagic protein degradation),261,262 especially when the differences in autophagic activity need to be determined precisely (note that an equivalent assay has not been developed for higher eukaryotic cells); however, as with any enzyme assay, appropriate caution must be used regarding, for example, substrate concentrations and linearity. The Pho8Δ60 assay also requires a control to verify equal Pho8Δ60 expression in the different genetic backgrounds or conditions to be tested;261 differences in Pho8Δ60 expression potentially affect its activity and may thus cause misinterpretation of results. Another issue to keep in mind is that GFP-Atg8 processing correlates with the surface area of the inner sphere of the autophagosome, and thus provides a smaller signal than assays that measure the volume of the autophagosome. Therefore, Pgk1-GFP processing30 or the Pho8Δ60 assay are generally more sensitive assays.
The main limitation of the GFP-LC3 processing assay in mammalian cells is that it seems to depend on cell type and culture conditions (N. Hosokawa and N. Mizushima, unpublished data). Apparently, GFP is more sensitive to mammalian lysosomal hydrolases than to the degradative milieu of the yeast vacuole or the lysosomes in Drosophila. Alternatively, the lower pH of mammalian lysosomes relative to that of the yeast vacuole may contribute to differences in detecting free GFP. Under certain conditions (such as Earle's balanced salt solution [EBSS]-induced starvation) in some cell lines, when the lysosomal pH becomes particularly low, free GFP is undetectable because both the LC3-II and free GFP fragments are quickly degraded.257 Therefore, if this method is used it should be accompanied by immunoblotting and include controls to address the stability of nonlysosomal GFP such as GFP-LC3-I. It should also be noted that free GFP can be detected when cells are treated with nonsaturating doses of inhibitors such as chloroquine, E-64d and bafilomycin A1. The saturating concentrations of these lysosomal inhibitors vary in different cell lines, and it would be better to use a saturating concentration of lysosomal inhibitors when performing an autophagic flux assay.257 Therefore, caution must be exercised in interpreting the data using this assay; it would be helpful to combine an analysis of GFP-LC3 processing with other assays, such as the monitoring of endogenous LC3-II by western blot.
Along these lines, a caution concerning the use of the EGFP fluorescent protein for microscopy is that this fluorophore has a relatively neutral pH optimum for fluorescence,263 and its signal diminishes quickly during live cell imaging due to the acidic environment of the lysosome. It is possible to circumvent this latter problem by imaging paraformaldehyde-fixed cultures that are maintained in a neutral pH buffer, which retains EGFP fluorescence (M. Kleinman and J.J. Reiners, personal communication). Alternatively, it may be preferable to use a different fluorophore such as mRFP or mCherry, which retain fluorescence even at acidic pH.264 On the one hand, a putative advantage of mCherry over mRFP is its enhanced photostability and intensity, which are an order of magnitude higher (and comparable to GFP), enabling acquisition of images at similar exposure settings as are used for GFP, thus minimizing potential bias in interpretation.265 On the other hand, caution is required when evaluating the localization of mCherry fusion proteins during autophagy due to the persistence of the mCherry signal in acidic environments; all tagged proteins are prone to show enrichment in lysosomes during nonselective autophagy of the cytoplasm, especially at higher expression levels. In addition, red fluorescent proteins (even the monomeric forms) can be toxic due to oligomer formation.266 Dendra2 is an improved version of the green-to-red photoswitchable fluorescent protein Dendra, which is derived from the octocoral Dendronephthya sp.267 Dendra2 is capable of irreversible photoconversion from a green to a red fluorescent form, but can be used also as a normal GFP or RFP vector. This modified version of the fluorophore has certain properties including a monomeric state, low phototoxic activation and efficient chromophore maturation, which make it suitable for real-time tracking of LC3 and SQSTM1 (Fig. 9; K. Kaarniranta, personal communication). Another alternative to mRFP or mCherry is to use the Venus variant of YFP, which is brighter than mRFP and less sensitive to pH than GFP.268
Figure 9.
Movement of activated pDendra2-hp62 (SQSTM1; orange) from the nucleus (middle) to an aggregate in ARPE-19 cells, revealed by confocal microscopy. Cells were exposed to 5 µM MG132 for 24 h to induce the formation of perinuclear aggregates.2186 The cells were then exposed to a UV pulse (the UV-induced area is shown by red lines that are inside of the nucleus) that converts Dendra2 from green to red, and the time shown after the pulse is indicated. SQSTM1 is present in a small nuclear aggregrate, and is shuttled from the nucleus to a perinuclear large protein aggregate (detected as red). Scale bar: 5 µm. Image provided by K. Kaarniranta.
The pH optimum of EGFP is important to consider when using GFP-LC3 constructs, as the original GFP-LC3 marker269 uses the EGFP variant, which may result in a reduced signal upon the formation of amphisomes or autolysosomes. An additional caveat when using the photoactivatable construct PA-GFP263 is that the process of activation by photons may induce DNA damage, which could, in turn, induce autophagy. Also, GFP is relatively resistant to denaturation, and boiling for 5 min may be needed to prevent the folded protein from being trapped in the stacking gel during SDS-PAGE.
As noted above (see Western blotting and ubiquitin-like protein conjugation systems), Atg4/ATG4 cleaves the residue(s) that follow the C-terminal glycine of Atg8/LC3 that will be conjugated to PE. Accordingly, it is critical that any chimeras be constructed with the fluorescent tag at the amino terminus of Atg8/LC3 (unless the goal is to monitor Atg4/ATG4 activity).
Finally, lysosomal inhibition needs to be carefully controlled. Prolonged inhibition of lysosomal hydrolases (>6 h) is likely to induce a secondary autophagic response triggered by the accumulated undigested autophagy cargo. This secondary autophagic response can complicate the analysis of the autophagic flux, making it appear more vigorous than it would in the absence of the lysosomal inhibitors.
Conclusion: The GFP-Atg8/LC3 processing assay, which monitors free GFP generated within the vacuole/lysosome, is a convenient way to follow autophagy, but it does not work in all cell types, and is not as easy to quantify as enzyme-based assays. Furthermore, the assay measures the flux of an autophagic carrier, which may not necessarily be equivalent to autophagic cargo flux.
d. GFP-Atg8/LC3 fluorescence microscopy
LC3B, or the protein tagged at its N terminus with a fluorescent protein such as GFP (GFP-LC3), has been used to monitor autophagy through indirect immunofluorescence or direct fluorescence microscopy (Fig. 10), measured as an increase in punctate LC3 or GFP-LC3.269,270 The detection of GFP-LC3/Atg8 is also useful for in vivo studies using transgenic organisms such as Caenorhabditis elegans,271 Dictyostelium discoideum,272 filamentous ascomycetes,273-277 Ciona intestinalis,278 Drosophila melanogaster,279-281 Arabidopsis thaliana,282 Zea mays,283 Trypanosoma brucei,221,284,285 Leishmania major286-288 and mice.153 It is also possible to use anti-LC3/Atg8 antibodies for immunocytochemistry or immunohistochemistry (IHC),197,289-294 procedures that have the advantages of detecting the endogenous protein, obviating the need for transfection and/or the generation of a transgenic organism, as well as avoiding potential artifacts resulting from overexpression. For example, high levels of overexpressed GFP-LC3 can result in its nuclear localization, although the protein can still relocate to the cytosol upon starvation. The use of imaging cytometry allows rapid and quantitative measures of the number of LC3 puncta and their relative number in individual or mixed cell types, using computerized assessment, enumeration, and data display (e.g., see refs. 44, 295). In this respect, the alternative use of an automated counting system may be helpful for obtaining an objective number of puncta per cell. For this purpose, the WatershedCounting3D plug-in for ImageJ may be useful.296,297 Changes in the number of GFP-Atg8 puncta can also be monitored using flow cytometry (see Autophagic flux determination using flow and multispectral imaging cytometry).221
Figure 10.
Changes in the detection and localization of GFP-LC3 upon the induction of autophagy. U87 cells stably expressing GFP-LC3 were treated with PBS, rapamycin (200 nM), or rapamycin in combination with 3-MA (2 mM) for 24 h. Representative fluorescence images of cells counterstained with DAPI (nuclei) are shown. Scale bar: 10 µm. This figure was modified from Figure 6 published in ref. 270, Badr et al. Lanatoside C sensitizes glioblastoma cells to tumor necrosis factor–related apoptosis-inducing ligand and induces an alternative cell death pathway. Neuro-Oncology, 13:1213–24, 2011, by permission of Oxford University Press.
Monitoring the endogenous Atg8/LC3 protein obviously depends on the ability to detect it in the system of interest, which is not always possible. If the endogenous amount is below the level of detection, the use of an exogenous construct is warranted. In this case, it is important to consider the use of stable transformants versus transient transfections. On the one hand, stable transformants may have reduced background resulting from the lower gene expression, and artifacts resulting from recent exposure to transfection reagents (see below) are eliminated. Furthermore, with stable transformants more cells can be easily analyzed because nearly 100% of the population will express tagged LC3. On the other hand, a disadvantage of stable transfectants is that the integration sites cannot always be predicted, and expression levels may not be optimal. Therefore, it is worth considering the use of stable episomal plasmids that avoid the problem of unsuitable integration.264 An important advantage of transient transfection is that this approach is better for examining the immediate effects of the transfected protein on autophagy; however, the transient transfection approach restricts the length of time that the analysis can be performed, and consideration must be given to the induction of autophagy resulting from exposure to the transfection reagents (see below). One word of caution is that optimizing the time of transient expression of GFP-LC3 is necessary, as some cell types (e.g., HeLa cells) may require 1 day for achieving optimal expression to visualize GFP-LC3 puncta, whereas neuronal cell lines such as SH-SY5Y cells typically need at least 48 h of expression prior to performing GFP-LC3 puncta analyses. In addition, a double transfection can be used (e.g., with GFP-LC3 and the protein of interest) to visually tag the cells that express the protein being examined.
A disadvantage of transfecting GFP-LC3 with liposomes is that frequently it leads to an unstable efficiency of transfection, causing a reduction in the number of cells effectively expressing GFP-LC3, and degradation of the plasmid, thus decreasing the numbers of GFP-LC3 puncta. Stable cell lines expressing GFP-LC3 can be generated using lentiviral systems and efficiently selected through antibiotic resistance leading to uniform and prolonged expression levels. These stable cell lines are sensitive to autophagy inducers as measured by the LC3-II/LC3-I ratio by western blot, and also show increased numbers of cytoplasmic GFP-LC3 puncta upon autophagic stimuli (R. Muñoz-Moreno, R. I. Galindo, L. Barrado-Gil and C. Alonso, unpublished results).
In conclusion, there is no simple rule for the use of stable versus transient transfections. When stable transfections are utilized through a nonlentiviral system, it is worthwhile screening for stable clones that give the best signal-to-noise ratio; when transient transfections are used, it is worthwhile optimizing the GFP-LC3 DNA concentration to give the best signal-to-noise ratio. In clones, the uniformity of expression of GFP-LC3 facilitates “thresholding” when scoring puncta-positive cells (see below). However, there is also a need to be aware that a single cell clone may not be representative of the overall pool. Using a pool of multiple selected clones may reduce artifacts that can arise from the selection and propagation of individual clones from a single transfected cell (although the use of a pool is also problematic as its composition will change over time). Another possibility is using fluorescence-activated cell sorter (FACS) sorting to select a mixed stable population with uniform GFP-LC3 expression levels.298 Optimization, together with including the appropriate controls (e.g., transfecting GFP-LC3G120A as a negative control), will help overcome the effects of the inherent variability in these analyses. For accurate interpretations, it is also important to assess the level of overexpression of the GFP-LC3 constructs relative to endogenous LC3 by western blot.
An additional use of GFP-LC3 is to monitor colocalization with a target during autophagy-related processes such as organelle degradation or the sequestration of pathogenic microbes.299-302 Preincubation of cells stably expressing GFP-LC3 with leupeptin can help stabilize the GFP-LC3 signal during fluorescence microscopy, especially under conditions of induced autophagic flux. Leupeptin is an inhibitor of lysosomal cysteine and serine proteases and will therefore inhibit degradation of membrane-conjugated GFP-LC3 that is present within autolysosomes.
Cautionary notes: Quantification of autophagy by measuring GFP-LC3 puncta (or LC3 by immunofluorescence) can, depending on the method used, be more tedious than monitoring LC3-II by western blot; however, the former may be more sensitive and quantitative. Ideally, it is preferable to include both assays and to compare the 2 sets of results. In addition, if GFP-LC3 is being quantified, it is better to determine the number of puncta corresponding to GFP-LC3 on a per cell basis (or per cell area basis) rather than simply the total number (or percentage) of cells displaying puncta. This latter point is critical because, even in nutrient-rich conditions, cells display some basal level of GFP-LC3 puncta. There are, however, practical issues with counting puncta manually and reliably, especially if there are large numbers per cell. Nevertheless, manual scoring may be more accurate than relying on a software program, in which case it is important to ensure that only appropriate puncta are being counted (applicable programs include ImageJ, Imaris, and the open-source software CellProfiler303). Moreover, when autophagosome-lysosome fusion is blocked, larger autophagosomes are detected, possibly due to autophagosome-autophagosome fusion, or to an inability to resolve individual autophagosomes when they are present in large numbers. Although it is possible to detect changes in the size of GFP-Atg8/LC3 puncta by fluorescence microscopy, it is not possible to correlate size with autophagy activity without additional assay methods. Size determinations can be problematic by fluorescence microscopy unless careful standardization is carried out,304 and size estimation on its own without considering puncta number per cell is not recommended as a method for monitoring autophagy; however, it is possible to quantify the fluorescence intensity of GFP-Atg8/LC3 at specific puncta, which does provide a valid measure of protein recruitment.305
In addition to autophagosome size, the number of puncta visible to the eye will also be influenced by both the level of expression of GFP-LC3 in a given cell (an issue that can be avoided by analyzing endogenous LC3 by immunofluorescence) and by the exposure time of the microscope, if using widefield microscopy. Another way to account for differential GFP-LC3 expression levels and/or exposure is to normalize the intensity of GFP-LC3 present in the puncta to the total GFP-LC3 intensity in the cell. This can be done either on the population level306 or individual cell level.298 In many cell types it may be possible to establish a threshold value for the number of puncta per cell in conditions of “low” and “high” autophagy.307 This can be tested empirically by exposing cells to autophagy-inducing and -blocking agents. Thus, cell populations showing significantly greater proportions of cells with autophagosome numbers higher than the threshold in perturbation conditions compared to the control cells could provide quantitative evidence of altered autophagy. It is then possible to score the population as the percentage of cells displaying numerous autophagosomes. This approach will only be feasible if the background number of puncta is relatively low. For this method, it is particularly important to count a large number of cells and multiple representative sections of the sample. Typically, it is appropriate to score on the order of 50 or more cells, preferably in at least 3 different trials, depending on the particular system and experiment, but the critical point is that this determination should be based on statistical power analysis. Accordingly, high-content imaging analysis methods enable quantification of GFP-LC3 puncta (or overall fluorescence intensity) in thousands of cells per sample (e.g., see refs. 243, 258, 308). When using automated analysis methods, care must be taken to manually evaluate parameters used to establish background threshold values for different treatment conditions and cell types, particularly as many systems image at lower magnifications that may be insufficient to resolve individual puncta. Another note of caution is that treatments affecting cell morphology, leading to the “rounding up” of cells, for example, can result in apparent changes in the number of GFP-LC3 puncta per cell. To avoid misinterpretation of results due to such potential artifacts, manual review of cell images is highly recommended. If cells are rounding up due to apoptosis or mitosis, it is easy to automatically remove them from analysis based on nuclear morphology (using DAPI or Hoechst staining) or cell roundness. If levels of autophagy in the rounded up cells are of particular interest, images can be acquired as z-stacks and either analyzed as a z-series or processed to generate maximum projection or extended depth-of-field images and than analyzed.309
To allow comparisons by other researchers attempting to repeat these experiments, it is critical that the authors also specify the baseline number of puncta that are used to define “normal” or “low” autophagy. Furthermore, the cells should be counted using unbiased procedures (e.g., using a random start point followed by inclusion of all cells at regular intervals), and statistical information should be provided for both baseline and altered conditions, as these assays can be highly variable. One possible method to obtain unbiased counting of GFP-LC3 puncta in a large number of cells is to perform multispectral imaging flow cytometry (see Autophagic flux determination using flow and multispectral imaging cytometry).310 Multispectral imaging flow cytometry allows characterization of single cells within a population by assessing a combination of morphology and immunofluorescence patterns, thereby providing statistically meaningful data.311 This method can also be used for endogenous LC3, and, therefore, is useful for nontransfected primary cells.312 For adherent cell cultures, one caution for flow cytometry is that the techniques necessary to produce single cell suspensions can cause significant injury to the cells, leading to secondary changes in autophagy. Therefore, staining for plasma membrane permeabilization (e.g., cell death) before versus after isolation is an important control, and allowing a period of recovery between harvesting the culture and staining is also advisable.313
An important caveat in the use of GFP-LC3 is that this chimera can associate with aggregates, especially when expressed at high levels in the presence of aggregate-prone proteins, which can lead to a misinterpretation of the results.314 Of note, GFP-LC3 can associate with ubiquitinated protein aggregates;315 however, this does not occur if the GFP-LC3 is expressed at low levels (D.C. Rubinsztein, unpublished observations). These aggregates have been described in many systems and are also referred to as aggresome-like induced structures (ALIS),315-317 dendritic cell ALIS,318 SQSTM1/p62 bodies/sequestosomes319 and inclusions. Indeed, many pathogen-associated molecular patterns (PAMPs) described to induce the formation of autophagosomes in fact trigger massive formation of SQSTM1 bodies (L.H. Travassos, unpublished observations). Inhibition of autophagy in vitro and in vivo leads to the accumulation of these aggregates, suggesting a role for autophagy in mediating their clearance.315,316,320-322 One way to control for background levels of puncta is to determine fluorescence from untagged GFP.
The receptor protein SQSTM1 is required for the formation of ubiquitinated protein aggregates in vitro (see SQSTM1 and related LC3 binding protein turnover assays).319 In this case, the interaction of SQSTM1 with both ubiquitinated proteins and LC3 is thought to mediate delivery of these aggregates to the autophagy system.323,324 Many cellular stresses can induce the formation of aggregates, including transfection reagents,315 or foreign DNA (especially if the DNA is not extracted endotoxin free). SQSTM1-positive aggregates are also formed by proteasome inhibition or puromycin treatment and can be found in cells exposed to rapamycin for extended periods where the rates of autophagy are elevated.325 Calcium phosphate transfection of COS7 cells or lipofectamine transfection of MEFs (R. Pinkas-Kramarski, personal communication), primary neurons (A.R. La Spada, personal communication) or neuronal cells (C.T. Chu, personal communication) transiently increases basal levels of GFP-LC3 puncta and/or the amount of LC3-II. One solution to this artifact is to examine GFP-LC3 puncta in cells stably expressing GFP-LC3; however, as transfection-induced increases in GFP-LC3 puncta and LC3-II are often transient, another approach is to use cells transfected with GFP, with cells subjected to a mock time-matched transfection as the background (negative) control. A lipidation-defective LC3 mutant where glycine 120 is mutated to alanine is targeted to these aggregates independently of autophagy (likely via its interaction with SQSTM1, see above); as a result, this mutant can serve as another specificity control.315 When carrying out transfections it may be necessary to alter the protocol depending on the level of background fluorescence. For example, changing the medium and waiting 24 to 48 h after the transfection can help to reduce the background level of GFP-LC3 puncta that is due to the transfection reagent (M. I. Colombo, personal communication). Similarly, when using an mCherry-GFP-SQSTM1 double tag (see Tandem mRFP/mCherry-GFP fluorescence microscopy) in transient transfections it is best to wait 48 h after transfection to reduce the level of aggregate formation and potential inhibition of autophagy (T. Johansen, personal communication). An additional consideration is that, in addition to transfection, viral infection can activate stress pathways in some cells and possibly induce autophagy, again emphasizing the importance of appropriate controls, such as control viruses expressing GFP.326
Ubiquitinated protein aggregate formation and clearance appear to represent a cellular recycling process. Aggregate formation can occur when autophagy is either inhibited or when its capacity for degradation is exceeded by the formation of proteins delivered to the aggregates. In principle, formation of GFP-LC3-positive aggregates represents a component of the autophagy process. However, the formation of GFP-LC3-positive ubiquitinated protein aggregates does not directly reflect either the induction of autophagy (or autophagosome formation) or flux through the system. Indeed, formation of ubiquitinated protein aggregates that are GFP-LC3 positive can occur in autophagy-deficient cells.315 Therefore, it should be remembered that GFP-LC3 puncta likely represent a mix of ubiquitinated protein aggregates in the cytosol, ubiquitinated protein aggregates within autophagosomes and/or more “conventional” phagophores and autophagosomes bearing other cytoplasmic cargo (this is one example where CLEM could help in resolving this question84). In Dictyostelium, inhibition of autophagy leads to huge ubiquitinated protein aggregates containing SQSTM1 and GFP-Atg8, when the latter is co-expressed; the large size of the aggregates makes them easily distinguishable from autophagosomes. Saponin treatment has been used to reduce background fluorescence under conditions where no aggregation of GFP-LC3 is detected in hepatocytes, GFP-LC3 stably-transfected HEK 293326 and human osteosarcoma cells, and in nontransfected cells;327 however, because treatment with saponin and other detergents can provoke artifactual GFP-LC3 puncta formation,328 specificity controls need to be included in such experiments. In general, it is preferable to include additional assays that measure autophagy rather than relying solely on monitoring GFP-LC3. In addition, we recommend that researchers validate their assays by demonstrating the absence or reversal of GFP-LC3 puncta formation in cells treated with pharmacological or RNA interference-based autophagy inhibitors (Table 1). For example, 3-MA is commonly used to inhibit starvation- or rapamycin-induced autophagy,329 but it has no effect on BECN1-independent forms of autophagy,83,151 and some data indicate that this compound can also have stimulatory effects on autophagy (see Autophagy inhibitors and inducers).330
Table 1.
Genetic and pharmacological regulation of autophagy.1
| Method | Comments | |
|---|---|---|
| 1. | 3-methyladenine | A PtdIns3K inhibitor that effectively blocks an early stage of autophagy by inhibiting the class III PtdIns3K, but not a specific autophagy inhibitor. 3-MA also inhibits the class I PI3K and can thus, at suboptimal concentrations in long-term experiments, promote autophagy in some systems, as well as affect cell survival through AKT and other kinases. 3-MA does not inhibit BECN1-independent autophagy. |
| 2. | 10-NCP | 10-(4′-N-diethylamino)butyl)-2-chlorophenoxazine; an AKT inhibitor that induces autophagy in neurons.1200 |
| 3. | 17-AAG | An inhibitor of the HSP90-CDC37 chaperone complex; induces autophagy in certain systems (e.g., neurons), but impairs starvation-induced autophagy and mitophagy in others by promoting the turnover of ULK1.458 |
| 4. | Akti-1/2 | An allosteric inhibitor of AKT1 and AKT2 that promotes autophagy in B-cell lymphoma.1495 |
| 5. | AR7 | AR7 was developed as a highly potent and selective enhancer of CMA through antagonizing RARA/RARα; AR7 is the first small molecule developed to selectively stimulate CMA without affecting macroautophagy.1496 |
| 6. | ARN5187 | Lysosomotropic compound with a dual inhibitory activity against the circadian regulator NR1D2/REV-ERBβ and autophagy.1497 |
| 7. | ATG4C74A | An active site mutant of ATG4 that is defective for autophagy.1498 |
| 8. | Bafilomycin A1 | A V-ATPase inhibitor that causes an increase in lysosomal/vacuolar pH, and, ultimately, blocks fusion of autophagosomes with the vacuole; the latter may result from inhibition of ATP2A/SERCA.226 |
| 9. | Betulinic acid | A pentacyclic triterpenoid that promotes paralell damage in mitochondrial and lysosomal compartments, and, ultimately, jeopardizes lysosomal degradative capacity.235 |
| 10. | Calcium | An autophagy activator that can be released from ER or lysosomal stores under stress conditions; however, calcium can also inhibit autophagy.216,1245 |
| 11. | Chloroquine, NH4Cl | Lysosomotropic compounds that elevate/neutralize the lysosomal/vacuolar pH.163 |
| 12. | DFMO | α-difluoromethylornithine, an irreversible inhibitor of ODC1 (ornithine decarboxylase 1) that blocks spermidine synthesis and ATG gene expression.1499 |
| 13. | E-64d | A membrane-permeable cysteine protease inhibitor that can block the activity of a subset of lysosomal hydrolases; should be used in combination with pepstatin A to inhibit lysosomal protein degradation. |
| 14. | ESC8 | A cationic estradiol derivative that induces autophagy and apoptosis simultaneously by downregulating the MTOR kinase pathway in breast cancer cells. |
| 15. | Everolimus | An inhibitor of MTORC1 that induces both autophagy and apoptosis in B-cell lymphoma primary cultures.1495 |
| 16. | Fumonisin B1 | An inhibitor of ceramide synthesis that interferes with macroautophagy. |
| 17. | Gene deletion | This method provides the most direct evidence for the role of an autophagic component; however, more than one gene involved in autophagy should be targeted to avoid indirect effects. |
| 18. | HMOX1 induction | Mitophagy and the formation of iron-containing cytoplasmic inclusions and corpora amylacea are accelerated in HMOX1-transfected rat astroglia and astrocytes of GFAP-HMOX1 transgenic mice. Heme derived ferrous iron and carbon monoxide, products of the HMOX1 reaction, promote macroautophagy in these cells.1500-1502 |
| 19. | Knockdown | This method (including miRNA, RNAi, shRNA and siRNA) can be used to inhibit gene expression and provides relatively direct evidence for the role of an autophagic component. However, the efficiency of knockdown varies, as does the stability of the targeted protein. In addition, more than one gene involved in autophagy should be targeted to avoid misinterpreting indirect effects. |
| 20. | KU-0063794 | An MTOR inhibitor that binds the catalytic site and activates autophagy.341,1503 |
| 21. | Leupeptin | An inhibitor of cysteine, serine and threonine proteases that can be used in combination with pepstatin A and/or E-64d to block lysosomal protein degradation. Leupeptin is not membrane permeable, so its effect on cathepsins may depend on endocytic activity. |
| 22. | microRNA | Can be used to reduce the levels of target mRNA(s) or block translation. |
| 23. | MLN4924 | A small molecule inhibitor of NAE (NEDD8 activating enzyme);1504 induces autophagy by blockage of MTOR signals via DEPTOR and the HIF1A-DDIT4/REDD1-TSC1/2 axis as a result of inactivation of CUL/cullin-RING ligases.1505-1507 |
| 24. | NAADP-AM | Activates the lysosomal TPCN/two-pore channel and induces autophagy.1225 |
| 25. | NED-19 | Inhibits the lysosomal TPCN and NAADP-induced autophagy.1225 |
| 26. | NVP-BEZ235 | A dual inhibitor of PIK3CA/p110 and the MTOR catalytic site that activates autophagy.1508,1509 |
| 27. | Pathogen-derived | Virally-encoded autophagy inhibitors including HSV-1 ICP34.5, Kaposi sarcoma-associated herpesvirus vBCL2, γ-herpesvirus 68 M11, ASFV vBCL2, HIV-1 Nef and influenza A virus M2.566,892,896,897,902 |
| 28. | Pepstatin A | An aspartyl protease inhibitor that can be used to partially block lysosomal degradation; should be used in combination with other inhibitors such as E-64d. Pepstatin A is not membrane permeable. |
| 29. | Protease inhibitors | These chemicals inhibit the degradation of autophagic substrates within the lysosome/vacuole lumen. A combination of inhibitors (e.g., leupeptin, pepstatin A and E-64d) is needed for complete blockage of degradation. |
| 30. | PMI | p62 (SQSTM1)-mediated mitophagy inducer is a pharmacological activator of autophagic selection of mitochondria that operates without collapsing the mitochondrial membrane potential (ΔΨm) and hence by exploiting the autophagic component of the process.713 |
| 31. | Rapamycin | Binds to FKBP1A/FKBP12 and inhibits MTORC1; the complex binds to the FRB domain of MTOR and limits its interaction with RPTOR, thus inducing autophagy, but only providing partial MTORC1 inhibition. Rapamycin also inhibits yeast TOR. |
| 32. | Resveratrol | A natural polyphenol that affects many proteins1510 and induces autophagy via activation of AMPK.1511,1512 |
| 33. | RNAi | Can be used to inhibit gene expression. |
| 34. | RSVAs | Synthetic small-molecule analogs of resveratrol that potently activate AMPK and induce autophagy.1513 |
| 35. | Saikosaponin-d | A natural small-molecule inhibitor of ATP2A/SERCA that induces autophagy and autophagy-dependent cell death in apoptosis-resistant cells.1514 |
| 36. | Tat-Beclin 1 | A cell penetrating peptide that potently induces macroautophagy.1080,1226 |
| 37. | Thapsigargin | An inhibitor of ATP2A/SERCA that inhibits autophagic sequestration through the depletion of intracellular Ca2+ stores;216,1515 however, thapsigargin may also block fusion of autophagosomes with endosomes by interfering with recruitment of RAB7, resulting in autophagosome accumulation.1516 |
| 38 | TMS | Trans-3,5,4-trimethoxystilbene upregulates the expression of TRPC4, resulting in MTOR inhibition.1517 |
| 39. | Torin1 | A catalytic MTOR inhibitor that induces autophagy and provides more complete inhibition than rapamycin (it inhibits all forms of MTOR).1193 |
| 40. | Trehalose | An inducer of autophagy that may be relevant for the treatment of different neurodegenerative diseases.1241,1518,1519 |
| 41. | Tunicamycin | A glycosylation inhibitor that induces autophagy due to ER stress.1520 |
| 42. | Vacuolin-1 | A RAB5A activator that reversibly blocks autophagosome-lysosome fusion.1521 |
| 43. | Vinblastine | A depolymerizer of both normal and acetylated microtubules that interferes with autophagosome-lysosome fusion.227 |
| 44. | Wortmannin | An inhibitor of PI3K and PtdIns3K that blocks autophagy, but not a specific inhibitor (see 3-MA above). |
This table is not meant to be complete, as there are many compounds and genetic methods that regulate autophagy, and new ones are being discovered routinely.
Another general limitation of the GFP-LC3 assay is that it requires a system amenable to the introduction of an exogenous gene. Accordingly, the use of GFP-LC3 in primary nontransgenic cells is more challenging. Here again, controls need to be included to verify that the transfection protocol itself does not artifactually induce GFP-LC3 puncta or cause LC3 aggregation. Furthermore, transfection should be performed with low levels of constructs, and the transfected cells should be followed to determine 1) when sufficient expression for detection is achieved, and 2) that, during the time frame of the assay, basal GFP-LC3 puncta remain appropriately low. In addition, the demonstration of a reduction in the number of induced GFP-LC3 puncta under conditions of autophagy inhibition is helpful. For some primary cells, delivering GFP-LC3 to precursor cells by infection with recombinant lentivirus, retrovirus or adenovirus,331 and subsequent differentiation into the cell type of interest, is a powerful alternative to transfection of the already differentiated cell type.74
To implement the scoring of autophagy via fluorescence microscopy, one option is to measure pixel intensity. Since the expression of GFP-LC3 may not be the same in all cells—as discussed above—it is possible to use specific imaging software to calculate the standard deviation (SD) of pixel intensity within the fluorescence image and divide this by the mean intensity of the pixels within the area of analysis. This will provide a ratio useful for establishing differences in the degree of autophagy between cells. Cells with increased levels of autophagic activity, and hence a greater number of autophagosomes in their cytosol, are associated with a greater variability in pixel intensity (i.e., a high SD). Conversely, in cells where autophagy is not occurring, GFP-LC3 is uniformly distributed throughout the cytosol and a variation in pixel intensity is not observed (i.e., a low SD; M. Campanella, personal communication).
Although LC3-II is primarily membrane-associated, it is not necessarily associated with autophagosomes as is often assumed; the protein is also found on phagophores, the precursors to autophagosomes, as well as on amphisomes and phagosomes (see Western blotting and ubiquitin-like protein conjugation systems).183,332,333 Along these lines, yeast Atg8 can associate with the vacuole membrane independent of lipidation, so that a punctate pattern does not necessarily correspond to autophagic compartments.334 Thus, the use of additional markers is necessary to specify the identity of an LC3-positive structure; for example, ATG12–ATG5-ATG16L1 would be present on a phagophore, but not an autophagosome, and thus colocalization of LC3 with any of these proteins would indicate the former structure. In addition, the site(s) of LC3 conjugation to PE is not definitively known, and levels of Atg8–PE/LC3-II can increase even in autophagy mutants that cannot form autophagosomes.335 One method that can be used to examine LC3-II membrane association is differential extraction in Triton X-114, which can be used with mammalian cells,331 or western blot analysis of total membrane fractions following solubilization with Triton X-100, which is helpful in plants.214,215 Importantly, we stress again that numbers of GFP-LC3 puncta, similar to steady state LC3-II levels, reflect only a snapshot of the numbers of autophagy-related structures (e.g., autophagosomes) in a cell at one time, not autophagic flux.
Finally, we offer a general note of caution with regard to using GFP. First, the GFP tag is large, in particular relative to the size of LC3; therefore, it is possible that a chimera may behave differently from the native protein in some respects. Second, GFP is not native to most systems, and as such it may be recognized as an aberrant protein and targeted for degradation, which has obvious implications when studying autophagy. Third, some forms of GFP tend to oligomerize, which may interfere with protein function and/or localization. Fourth, EGFP inhibits polyubiquitination336 and may cause defects in other cellular processes. Fifth, not all LC3 puncta represent LC3-II and correspond to autophagosomes.190,191,337,338 Accordingly it would be prudent to complement any assays that rely on GFP fusions (to Atg8/LC3 or any protein) with additional methods that avoid the use of this fluorophore. Similarly, with the emergence of “super-resolution” microscopy methods such as photoactivated localization microscopy (PALM), new tags are being used (e.g., the EosFP green to red photoconvertible fluorescent protein, or the Dronpa GFP-like protein) that will need to be tested and validated.339
Conclusion: GFP-LC3 provides a marker that is relatively easy to use for monitoring autophagy induction (based on the appearance of puncta), or colocalization with cargo; however, monitoring this chimera does not determine flux unless utilized in conjunction with inhibitors of lysosomal fusion and/or degradation. In addition, it is recommended that results obtained by GFP-LC3 fluorescence microscopy be verified by additional assays.
e. Tandem mRFP/mCherry-GFP fluorescence microscopy
A fluorescence assay that is designed to monitor flux relies on the use of a tandem monomeric RFP-GFP-tagged LC3 (tfLC3; Fig. 11).264 The GFP signal is sensitive to the acidic and/or proteolytic conditions of the lysosome lumen, whereas mRFP is more stable. Therefore, colocalization of both GFP and mRFP fluorescence indicates a compartment that has not fused with a lysosome, such as the phagophore or an autophagosome. In contrast, a mRFP signal without GFP corresponds to an amphisome or autolysosome. Other fluorophores such as mCherry are also suitable instead of mRFP,319 and an image-recognition algorithm has been developed to quantify flux of the reporter to acidified compartments.340-342 One of the major advantages of the tandem mRFP/mCherry-GFP reporter method is that it enables simultaneous estimation of both the induction of autophagy and flux through autophagic compartments without requiring the use of any lysosomal inhibitors. The competence of lysosomal digestion of the substrate requires additional analysis using methods described above. The use of more than one time point allows visualization of increased early autophagosomes followed by increases in late autophagosomes as an additional assurance that flux has been maintained.343 In addition, this method can be used to monitor autophagy in high-throughput drug screening studies.341 The quantification of “yellow only” (where the yellow signal results from merging the red and green channels) and “red only” dots in a stable tandem-fluorescent LC3-reporter cell line can be automated by a Cellomics microscope that can be used to assess a huge population of cells (1,000 or more) over a large number of random fields of view.233,344 Notably, organelle-specific variations of the tandem mRFP/mCherry-GFP reporter system have successfully been used to analyze selective types of autophagy, such as pexophagy345 and mitophagy346,347 in mammalian cells.
Figure 11.
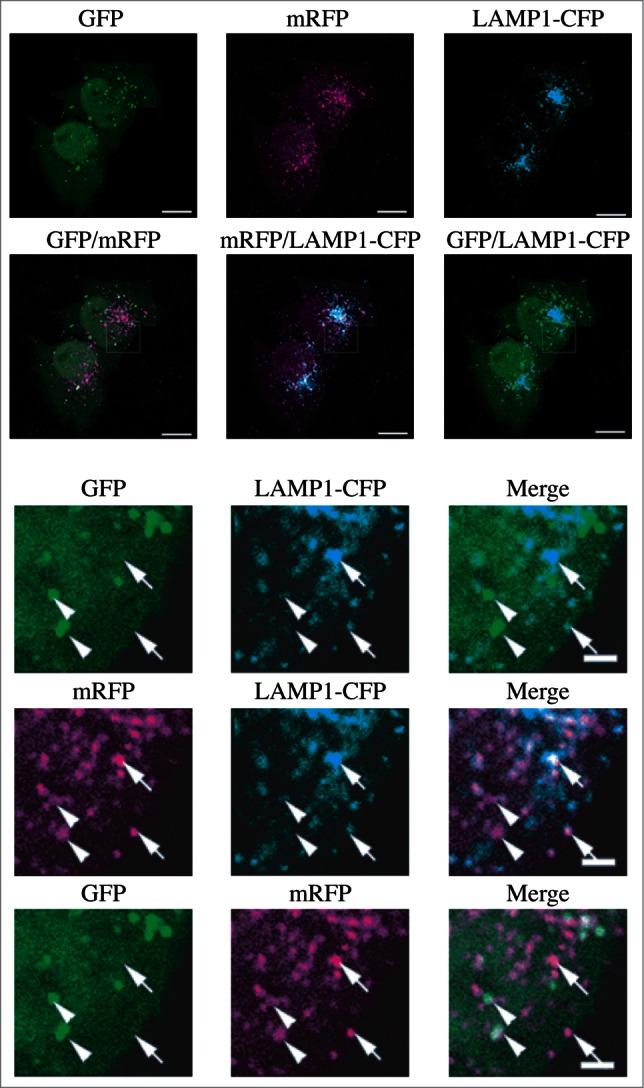
The GFP and mRFP signals of tandem fluorescent LC3 (tfLC3, mRFP-GFP-LC3) show different localization patterns. HeLa cells were cotransfected with plasmids expressing either tfLC3 or LAMP1-CFP. Twenty-four h after transfection, the cells were starved in Hanks balanced salt solution for 2 h, fixed and analyzed by microscopy. The lower panels are a higher magnification of the upper panels. Bar: 10 µm in the upper panels and 2 µm in the lower panels. Arrows in the lower panels point to (or mark the location of) typical examples of colocalized signals of mRFP and LAMP1. Arrowheads point to (or mark the location of) typical examples of colocalized particles of GFP and mRFP signals. This figure was previously published in ref. 264, and is reproduced by permission of Landes Bioscience, copyright 2007.
An alternative dual fluorescence assay involves the Rosella pH biosensor. This assay monitors the uptake of material to the lysosome/vacuole and complements the use of the tandem mRFP/mCherry-GFP reporter. The assay is based upon the genetically encoded dual color-emission biosensor Rosella, a fusion between a relatively pH-stable fast-maturing RFP variant, and a pH-sensitive GFP variant. When targeted to specific cellular compartments or fused to an individual protein, the Rosella biosensor provides information about the identity of the cellular component being delivered to the lysosome/vacuole for degradation. Importantly, the pH-sensitive dual color fluorescence emission provides information about the environment of the biosensor during autophagy of various cellular components. In yeast, Rosella has been successfully used to monitor autophagy of cytosol, mitochondria (mitophagy) and the nucleus (nucleophagy).348-350 Furthermore, the Rosella biosensor can be used as a reporter under various conditions including nitrogen depletion-dependent induction of autophagy.348,349 The Rosella biosensor can also be expressed in mammalian cells to follow either nonselective autophagy (cytoplasmic turnover), or mitophagy.349
Cautionary notes: The use of tandem mRFP/mCherry-GFP-LC3/Atg8 reporters in live imaging experiments can be complicated by the motion of LC3/Atg8 puncta. As a consequence, conventional confocal microscopy may not allow visualization of colocalized mRFP/mCherry-GFP puncta. In this case, GFP colocalized puncta represent newly formed autophagic structures whereas mRFP/mCherry-only puncta are ambiguous. Spinning disk confocal microscopy or rapid acquisition times may be required for imaging tandem mRFP/mCherry-GFP proteins, although these techniques require a brighter fluorescent signal associated with what may be undesirably higher levels of transgene expression. One solution is to use the mTagRFP-mWasabi-LC3 chimera,351 as mTagRFP is brighter than mRFP1 and mCherry, and mWasabi is brighter than EGFP.352 Another possibility is to use fixed cells; however, this presents an additional concern: The use of tandem mRFP/mCherry-GFP relies on the quenching of the GFP signal in the acidic autolysosome; however, fixation solutions are often neutral or weak bases, which will increase the pH of the entire cell. Accordingly, the GFP signal may be restored after fixation (Fig. 12), which would cause an underestimation of the amount of signal that corresponds only to RFP (i.e., in the autolysosome). Thus, the tissue or cell samples must be properly processed to avoid losing the acidic environment of the autolysosomes. In addition, there may be weak fluorescence of EGFP even in an acidic environment (pH between 4 and 5).263,331 Therefore, it may be desirable to choose a monomeric green fluorescent protein that is more acid sensitive than EGFP for assaying autophagic flux.
Figure 12.
GFP fluorescence in the autolysosome can be recovered upon neutralization of the pH. (A) GFP-LC3 emits green fluorescence in the autolysosomes of post-mortem processed heart sections. Cryosections of 3.8% paraformaldehyde-fixed ventricular myocardium from 3-wk-old GFP-LC3 transgenic mice at the baseline (control) or starved for 24 h (starved) were processed for immunostaining using a standard protocol (buffered at pH 7.4). Most of the GFP-LC3 puncta are positive for LAMP1, suggesting that the autolysosomes had recovered GFP fluorescence. (B) Colocalization between GFP-LC3 direct fluorescence (green) and indirect immunostaining for GFP (red). Sections processed as in (A) were immunostained for GFP using a red fluorescence-tagged secondary antibody, and the colocalization with GFP fluorescence was examined by confocal microscopy. Almost all of the red puncta emit green fluorescence. Image provided by Xuejun Wang.
Another caution in the interpretation of the tandem fluorescent marker is that colocalization of GFP and mRFP/mCherry might also be seen in the case of impaired proteolytic degradation within autolysosomes or altered lysosomal pH. Finally, expression of tandem mRFP-GFP-LC3 is toxic to some cancer cell lines relative to GFP-LC3 or RFP-LC3 (K.S. Choi, personal communication). The cytotoxicity of DsRed and its variants such as mRFP1 is associated with downregulation of BCL2L1/Bcl-xL.353 In contrast to mRFP-GFP-LC3, overexpression of mTagRFP-mWasabi-LC3 does not appear to be toxic to HeLa cells (J. Lin, personal communication).
The Rosella assay has not been tested in a wide range of mammalian cell types. Accordingly, the sensitivity and the specificity of the assay must be verified independently until this method has been tested more extensively and used more widely.
Finally, it may be desirable to capture the dynamic behavior of autophagy in real time, to generate data revealing the rate of formation and clearance of autophagosomes over time, rather than single data points. For example, by acquiring signals from 2 fluorescent constructs in real time, the rate of change in colocalization signal as a measure of the fusion rate and recycling rate between autophagosomes and lysosomes can be assessed.354 Importantly, due to the integral dynamic relationship of autophagic flux with the onset of apoptosis and necrosis, it is advantageous to monitor cell death and autophagic flux parameters concomitantly over time, which FRET-based reporter constructs make possible.355
In addition, as the metabolic control of autophagy is becoming increasingly clear, highlighting a tight network between the autophagy machinery, energy sensing pathways and the cell's metabolic circuits,356,357 mitochondrial parameters such as fission and fusion rate as well as the cell's ATP demand should be monitored and correlated with autophagic flux data. This will provide a better understanding of the variability of autophagy and cell death susceptibility.
Tandem fluorescent markers show real-time changes in autophagosome fusion with lysosomes, due to entry into an acidic environment; however, fusion is not definitive evidence of substrate or carrier degradation. Lysosomes may be able to fuse, but be unable to degrade newly delivered cargo, as occurs in some lysosomal storage diseases. Best practice would be to perform an autophagic flux assay in parallel with quantification of tandem fluorescent markers to confirm completion of carrier flux.
Conclusion: The use of tandem fluorescent constructs, which display different emission signals depending on the environment (in particular, GFP fluorescence is sensitive to an acidic pH), provides a convenient way to monitor autophagic flux in many cell types.
f. Autophagic flux determination using flow and multispectral imaging cytometry
Whereas fluorescence microscopy, in combination with novel autophagy probes, has permitted single-cell analysis of autophagic flux, automation for allowing medium- to high-throughput analysis has been challenging. A number of methods have been developed that allow the determination of autophagic flux using flow cytometry,225,311,327,358-361 and commercial kits are now available for monitoring autophagy by flow cytometry. These approaches make it possible to capture data or, in specialized instruments, high-content, multiparametric images of cells in flow (at rates of up to 1,000 cells/sec for imaging, and higher in nonimaging flow cytometers), and are particularly useful for cells that grow in suspension. Optimization of image analysis permits the study of cells with heterogeneous LC3 puncta, thus making it possible to quantify autophagic flux accurately in situations that might perturb normal processes (e.g., microbial infection).360,362 Since EGFP-LC3 is a substrate for autophagic degradation, total fluorescence intensity of EGFP-LC3 can be used to indicate levels of autophagy in living mammalian cells.358 When autophagy is induced, the decrease in total cellular fluorescence can be precisely quantified in large numbers of cells to obtain robust data. In another approach, soluble EGFP-LC3-I can be depleted from the cell by a brief saponin extraction so that the total fluorescence of EGFP-LC3 then represents that of EGFP-LC3-II alone (Fig. 13A).326,327 Since EGFP-LC3 transfection typically results in high relative levels of EGFP-LC3-I, this treatment significantly reduces the background fluorescence due to nonautophagosome-associated reporter protein. By comparing treatments in the presence or absence of lysosomal degradation inhibitors, subtle changes in the flux rate of the GFP-LC3 reporter construct can be detected. If it is not desirable to treat cells with lysosomal inhibitors to determine rates of autophagic flux, a tandem mRFP/mCherry-EGFP-LC3 (or similar) construct can also be used for autophagic flux measurements in flow cytometry experiments (see Tandem mRFP/mCherry-GFP fluorescence microscopy).359
Figure 13.
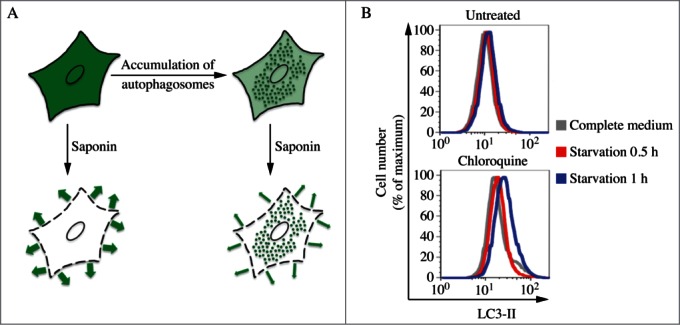
Saponin extraction allows quantification of LC3-II fluorescence by FACS. (A) Schematic diagram of the effects of the saponin wash. Due to the reorganization of the EGFP-LC3 reporter protein, induction of autophagosome formation does not change the total levels of fluorescence in EGFP-LC3-transfected cells. However, extraction of EGFP-LC3-I with saponin results in a higher level of fluorescence in cells with proportionally higher levels of EGFP-LC3-II-containing autophagosomes. This figure was previously published in ref. 327. (B) Saponin extraction can also be used to measure flux of endogenous LC3 protein. Human osteosarcoma cells were starved of amino acids and serum by incubation in EBSS, for the indicated times in the presence or absence of a 1 h chloroquine (50 µM) treatment. Cells were then washed with PBS containing 0.05% saponin and processed for FACS analysis for endogenous LC3. Image provided by K.E. Eng and G.M. McInerney.
These methods, however, require the cells of interest to be transfected with reporter constructs. Since the saponin extraction method can also be combined with intracellular staining for endogenous LC3 protein, subtle changes in autophagic flux can be measured without the need for reporter transfections (Fig. 13B).
Cautionary notes: Care must be taken when applying flow cytometry measurements to adherent cells, particularly neurons and other cells with interdigitated processes, as the preparation of single cell suspensions entails significant levels of plasma membrane disruption and injury that can secondarily induce autophagy.
Users of the saponin extraction method should carefully titrate saponin concentrations and times of treatment to ensure specific extraction of LC3-I in their systems. Also, it has been observed in some cell types that saponin treatment can lead to nonautophagic aggregation of LC3,328 which should be controlled for in these assays (see GFP-Atg8/LC3 fluorescence microscopy).
Cell membrane permeabilization with digitonin and extraction of the nonmembrane-bound form of LC3 allows combined staining of membrane-associated LC3-II protein and any markers for detection of autophagy in relation to other cellular events/processes. Based on this approach, a method for monitoring autophagy in different stages of the cell cycle was developed.363 Thus, the presence of basal or starvation-induced autophagy is detected in G1, S, and G2/M phases of the cell cycle in MEFs with doxycycline-regulated ATG5 expression. In these experiments cells were gated based on their DNA content and the relative intensity of GFP-LC3-II and LC3-II expression. This approach might also be used for the detection of autophagic flux in different stages of the cell cycle or subG1 apoptotic cell population by measuring accumulation of LC3-II in the presence or absence of lysosomal inhibitors.
Although GFP-LC3 can be used as a reporter for flow cytometry, it is more stable (which is not necessarily ideal for flux measurements) than GFP-SQSTM1 or GFP-NBR1 (NBR1 is a selective autophagic substrate with structural similarity to SQSTM1364). GFP-SQSTM1 displays the largest magnitude change following the induction of autophagy by amino acid deprivation or rapamycin treatment, and may thus be a better marker for following autophagic flux by this method (confirmed in SH-SY5Y neuronal cell lines stably expressing GFP-SQSTM1; E.M. Valente, personal communication).365
Conclusion: Medium- to high-throughput analysis of autophagy is possible using flow and multispectral imaging cytometry (Fig. 14). The advantage of this approach is that larger numbers of cells can be analyzed with regard to GFP-LC3 puncta, cell morphology and/or autophagic flux, and concomitant detection of surface markers can be included, potentially providing more robust data than is achieved with other methods. A major disadvantage, however, is that flow cytometry only measures changes in total GFP-LC3 levels, which can be subject to modification by changes in transcription or translation, or by pH, and this approach cannot accurately evaluate localization (e.g., to autophagosomes) or lipidation (generation of LC3-II) without further permeabilization of the cell.
Figure 14.
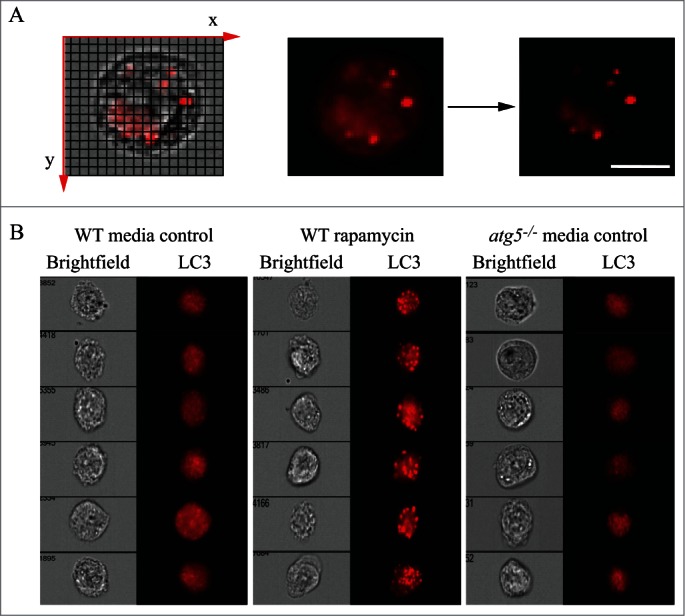
Assessing autophagy with multispectral imaging cytometry. (A) Bright Detail Intensity (BDI) measures the foreground intensity of bright puncta (that are 3 pixels or less) within the cell image. For each cell, the local background around the spots is removed before intensity calculation. Thus, autophagic cells with puncta have higher BDI values. (B) Media control (untreated wild type), rapamycin-treated wild-type and atg5-/- MEFs were gated based on BDI. Representative images of cells with high or low BDI values. Scale bar: 10 µm. Images provided by M.L. Albert.
g. Immunohistochemistry
Immunodetection of ATG proteins (particularly LC3 and BECN1) has been reported as a prognostic factor in various human carcinomas, including lymphoma,197,366 breast carcinoma,367 endometrial adenocarcinoma,368,369 head and neck squamous cell carcinoma,370-372 hepatocellular carcinoma,373,374 gliomas,375 non-small cell lung carcinomas,376 pancreatic377 and colon adenocarcinomas,378-380 as well as in cutaneous and uveal melanomas.381,382 Unfortunately, the reported changes often reflect overall diffuse staining intensity rather than appropriately compartmentalized puncta. Therefore, the observation of increased levels of diffuse LC3 staining (which may reflect a decrease in autophagy) should not be used to draw conclusions that autophagy is increased in cancer or other tissue samples. Importantly, this kind of assay should be performed as recommended by the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK).383 As we identify new drugs for modulating autophagy in clinical applications, this type of information may prove useful in the identification of subgroups of patients for targeted therapy.384-386
In mouse and rat tissues, endogenous LC3, ATG4B, and ATG9A have been detected by immnohistochemical analyses using both paraffin sections and cryosections.293,387-389 When autophagosomes are absent, the localization pattern of LC3 in the cells of various tissues is diffuse and cytosolic. Moreover, intense fibrillary staining of LC3 is detectable along dendrites of intact neurons, whereas granular staining for LC3 appears mainly in the perikarya of neurons in CTSD- or CTSB- and CTSL (cathepsin L)-deficient mouse brains.293 LC3 puncta are also observed in mice in the peripheral nerves, specifically in Schwann cells after neurodegeneration,390 and Paneth cells of the small intestine from human Crohn disease patients and mouse models of intestinal inflammation driven by ER-stress exhibit strong LC3 puncta staining.391,392 In various neurodegenerative states, LC3 puncta may be numerous in neurites, especially within dystrophic swellings and, in many cases, these vacuoles are amphisomes or autolysosomes, reflecting the delayed or inhibited degradation of LC3 despite the presence of abundant hydrolase activity.57,66 In developing inner ear and retinal tissue in chicken, BECN1 is detected by immunofluorescence; in chick retina AMBRA1 is also detected.393-395 Finally, in non-mammalian vertebrates, BECN1 was detected during follicular atresia in the ovary of 3 fish species using paraffin sections; a punctate immunostaining for BECN1 is scattered throughout the cytoplasm of the follicular cells when they are in intense phagocytic activity for yolk removal.396
Cautionary notes: One problem with LC3 IHC is that in some tissues this protein can be localized in structures other than autophagosomes. For example, in murine hepatocytes and cardiomyocytes under starved conditions, endogenous LC3 is detected not only in autophagosomes but also on lipid droplets.397 In neurons in ATG7-deficient mice, LC3 accumulates in ubiquitin- and SQSTM1-positive aggregates.398 In neurons in aging or neurodegenerative disease states, LC3 is commonly present in autolysosomes and may be abundant in lipofuscin and other lysosomal residual bodies.57 Thus, immunodetection of LC3 in cytoplasmic granules is not sufficient to monitor autophagy in vivo. To evaluate autophagy by the methods of immunohistochemistry, it is necessary to identify the autophagosomes directly using the ABC technique for TEM observation (see Transmission electron microscopy).77
Conclusion: It has not been clearly demonstrated that IHC of ATG proteins in tissues corresponds to autophagy activity, and this area of research needs to be further explored before we can make specific recommendations.
3. SQSTM1 and related LC3 binding protein turnover assays
In addition to LC3, SQSTM1/p62 or other receptors such as NBR1, can also be used as protein markers, at least in certain settings.26,399 For example, SQSTM1 can be detected as puncta by IHC in cancer cells, similar to LC3.372 The SQSTM1 protein serves as a link between LC3 and ubiquitinated substrates.84 SQSTM1 and SQSTM1-bound polyubiquitinated proteins become incorporated into the completed autophagosome and are degraded in autolysosomes, thus serving as an index of autophagic degradation (Fig. 15). Inhibition of autophagy correlates with increased levels of SQSTM1 in mammals and Drosophila, suggesting that steady state levels of this protein reflect the autophagic status.61,389,400-404 Similarly, decreased SQSTM1 levels are associated with autophagy activation. The phosphorylation of SQSTM1 at Ser403 appears to regulate its role in the autophagic clearance of ubiquitinated proteins, and anti-phospho-SQSTM1 antibodies can be used to detect the modified form of the protein.324
Figure 15.
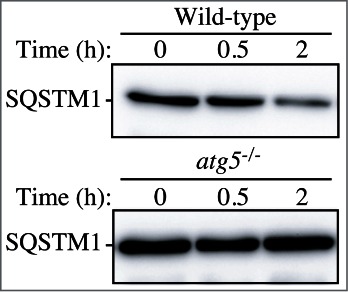
Regulation of the SQSTM1 protein during autophagy. The level of SQSTM1 during starvation. Atg5+/+ and atg5-/- MEFs were cultured in DMEM without amino acids and serum for the indicated times, and then subjected to immunoblot analysis using anti-SQSTM1 antibody (Progen Biotechnik, GP62). This figure was previously published in ref. 26, and is reproduced by permission of Landes Bioscience, copyright 2007.
Cautionary notes: SQSTM1 changes can be cell type and context specific. In some cell types, there is no change in the overall amount of SQSTM1 despite strong levels of autophagy induction, verified by the tandem mRFP/mCherry-GFP-LC3 reporter as well as ATG7- and lysosome-dependent turnover of cargo proteins (C.T. Chu, personal observation). In other contexts, a robust loss of SQSTM1 does not correlate with increased autophagic flux as assessed by a luciferase-based measure of flux;245 a decrease of SQSTM1 can even relate to a blockage of autophagy due to cleavage of the protein, together with other autophagy proteins, by caspases or calpains.405 SQSTM1 may be transcriptionally upregulated under certain conditions,317,406-409 further complicating the interpretation of results. For example, SQSTM1 upregulation, and at least transient increases in the amount of SQSTM1, is seen in some situations where there is an increase in autophagic flux.410-412 One such case is seen during retinoic acid-induced differentiation of AML cells where SQSTM1 is upregulated407 with concomitant increased autophagic flux.413 Activation of a signaling pathway, e.g., RAF1/Raf-MAP2K/MEK-MAPK/ERK, can also upregulate SQSTM1 transcription.414 SQSTM1 mRNA is also upregulated following prolonged starvation, which can restore the SQSTM1 protein level to that before starvation.415,416 In the same way, physical exercise, especially when performed during starvation, increases the SQSTM1 mRNA level in skeletal muscle, and can lead to an incorrect interpretation of autophagic flux if only the protein level is measured.417,418 Another instance when both mRNA and protein levels of SQSTM1 are elevated even though autophagic flux is not impaired is observed in aneuploid human and murine cells that are generated by introduction of 1 or 2 extra chromosomes.419,420 Thus, appropriate positive and negative controls are needed prior to the use of SQSTM1 as a flux indicator in a particular cellular context, and we recommend monitoring the SQSTM1 mRNA level as part of a complete analysis, or determining the SQSTM1 protein level in the presence of actinomycin D.
Of interest, SQSTM1 hyperexpression at both gene and protein levels can be observed in muscle atrophy induced by cancer, though not by glucocorticoids, suggesting that the stimulus inducing autophagy may also be relevant to the differential regulation of autophagy-related proteins.421 One solution to problems relating to variations in SQSTM1 expression levels is to use a HaloTag®-p62 (SQSTM1) chimera.422 The chimeric protein can be covalently labeled with HaloTag® ligands, and the loss of signal can then be monitored without interference by subsequent changes in protein synthesis. Similarly, a stable cell line expressing EGFP-tagged SQSTM1 under the control of an inducible promoter can be used to assess the rates of SQSTM1 degradation, taking into account the limitations outlined above (see Autophagic flux determination using flow and multispectral imaging cytometry).365 A similar system exists in Drosophila in which a GFP-tagged SQSTM1 can be expressed using the UAS-GAL4 system.423 It is worth noting that tetracycline can reduce autophagy levels; therefore, the appropriate control of only tetracycline addition has to be included if using an inducible promoter that responds to this drug.424 Yet another solution is to employ a radioactive pulse-chase assay to measure the rates of SQSTM1 degradation.425
SQSTM1 contains a LIR motif as well as a ubiquitin binding domain, and appears to act by linking ubiquitinated substrates with the autophagic machinery. Nonetheless, it would be prudent to keep in mind that SQSTM1 contains domains that interact with several signaling molecules,426 and SQSTM1 may be part of MTORC1.427 Thus, it may have additional functions that need to be considered with regard to its role in autophagy. In the context of autophagy as a stress response, the complexity of using SQSTM1 as an autophagy marker protein is underscored by its capacity to modulate the NFE2L2/NRF2 anti-oxidant response pathway through a KEAP1 binding domain.428,429 In fact, SQSTM1 may, itself, be transcriptionally induced by NFE2L2.430 Furthermore, it is preferable to examine endogenous SQSTM1 because overexpression of this protein leads to the formation of protein inclusions. In fact, even endogenous SQSTM1 becomes Triton X-100-insoluble in the presence of protein aggregates and when autophagic degradation is inhibited; thus, results with this protein are often context-dependent. Indeed, there is a reciprocal crosstalk between the UPS and autophagy, with SQSTM1 being a key link between them.431 First, SQSTM1 participates in proteasomal degradation, and its level may also increase when the proteasome is inhibited.432 Accordingly, the SQSTM1 degradation rate should be analyzed in the presence of an inhibitor such as epoxomicin or lactacystin to determine the contribution from the proteasome (see Autophagy inhibitors and inducers for potential problems with MG132).433 Second, the accumulation of SQSTM1 due to autophagy inhibition can impair UPS function by competitively binding ubiquitinated proteins, preventing their delivery to, and degradation by, the proteasome.434 Accordingly, it may be advisable to measure the UPS flux by using UbG76V-GFP, a ubiquitin-proteasome activity reporter, when SQSTM1 accumulation is observed. Thus, it is very important to determine whether autophagy alone or in conjunction with the UPS accounts for substrate degradation induced by a particular biological change. A number of stressors that impair the UPS induce the aggregation/dimerization of SQSTM1, and this can be seen by the detection of a high molecular mass (˜150 kDa) protein complex by western blot, which is recognized by SQSTM1 antibodies (R. Franco, personal communication).435,436 Although the accumulation of this protein complex can be related to the accumulation of ubiquitinated SQSTM1-bound proteins, or the dimerization/inactivation of SQSTM1 (R. Franco, personal communication),437 evaluation of the ratio between SQSTM1 (aggregates/dimers) and SQSTM1 monomers is likely a better measurement of changes in SQSTM1 dynamics linked to autophagy or the UPS.
SQSTM1 is also a substrate for CASP6/caspase 6 and CASP8 (as well as CAPN1/calpain 1), which may confound its use in examining cell death and autophagy.438 This is one reason why SQSTM1 degradation should also be analyzed in the presence of a pan-caspase inhibitor such as Q-VD-OPh before concluding that autophagy is activated based on a decrease of this protein.405 Another issue is that some phosphatidylinositol 3-kinase (PtdIns3K) inhibitors such as LY294002, and to a lesser extent wortmannin (but apparently not 3-MA),329 can inhibit protein synthesis;439 this might in turn affect the turnover of SQSTM1 and LC3, which could influence conclusions that are drawn from the status of these proteins regarding autophagic flux or ALIS formation. Accordingly, it may be advisable to measure protein synthesis and proteasome activity along with autophagy under inhibitory or activating conditions. With regard to protein synthesis, it is worth noting that this can be monitored through a nonradioactive method.440
Western blot analysis of cell lysates prepared using NP40- or Triton X-100-containing lysis buffers in autophagic conditions typically shows a reduction in SQSTM1 levels. However, this does not necessarily indicate that SQSTM1 is degraded, because SQSTM1 aggregates are insoluble in these detergent lysis conditions.317,441 Moreover, in some instances SQSTM1 levels do not change in the soluble fractions despite autophagic degradation, a finding that might be explained by simultaneous transcriptional induction of the gene encoding SQSTM1, since the soluble fraction accounts only for the diffuse or free form of SQSTM1. Accumulation of SQSTM1 in the Triton X-100-insoluble fraction can be observed when autophagy-mediated degradation is inhibited. Under conditions of higher autophagic flux, accumulation of SQSTM1 in Triton X-100-insoluble fractions may not be observed and SQSTM1 levels may be reduced or maintained. The simplest approach to circumvent many of these problems is using lysis buffer that allows identification of the entire cellular pool of SQSTM1 (e.g., containing 1% SDS); however, additional assessment of both Triton X-100-soluble and -insoluble fractions will provide further information regarding the extent of SQSTM1 oligomerization.398 Note, when performing a western blot using an SQSTM1 antibody, it is always a good idea to include a positive control in which SQSTM1 accumulates, such as an atg8a mutant (e.g., see Fig. S3 in ref. 442).
To conclusively establish SQSTM1 degradation by autophagy, SQSTM1 levels in both Triton X-100-soluble and -insoluble fractions need to be determined upon treatment with autophagy inducers in combination with autophagy inhibitors, such as those that inhibit the autolysosomal degradation steps (e.g., protease inhibitors, chloroquine or bafilomycin A1). Additionally, an alteration in the level of SQSTM1 may not be immediately evident with changes observed in autophagic flux upon certain chemical perturbations (S. Sarkar, personal communication). Whereas LC3 changes may be rapid, clearance of autophagy substrates may require a longer time. Therefore, if LC3 changes are assessed at 6 h or 24 h after a drug treatment, SQSTM1 levels can be tested not only at the same time points, but also at later time points (24 h or 48 h) to determine the maximal impact on substrate clearance. An alternative method is immunostaining, with and without autophagy inhibitors, for SQSTM1, which will appear as either a diffuse or punctate pattern. Experiments with autophagy inducers and inhibitors, in combination with western blot and immunostaining analyses, best establish autophagic degradation based on SQSTM1 turnover. A final point, however, is that empirical evidence suggests that the species specificity of antibodies for detecting SQSTM1 must be taken into account. For example, some commercial antibodies recognize both human and mouse SQSTM1, whereas others detect the human, but not the mouse protein.443 Another issue with detecting SQSTM1 in the context of human diseases is that it can be mutated (e.g., in Paget disease of bone).444 Thus, care should be taken to ensure that potential mutations are not affecting the epitopes that are recognized by anti-SQSTM1 antibodies when using western blotting to detect this protein.
As an alternative, the SQSTM1:BECN1 protein level ratio can be used as a readout of autophagy.445 Since both decreased SQSTM1 levels and increased BECN1 levels correlate with enhanced autophagy (as noted in the present article), a decreased SQSTM1:BECN1 protein level ratio (when derived from the same protein extract) may, cautiously, be interpreted as augmented autophagy, keeping in mind that SQSTM1 gene expression varies significantly under different conditions and may obscure the meaning of a change in the amount of SQSTM1 protein. As a general note, using ratios of the levels of proteins changing in opposite directions, rather than the protein levels themselves, could be beneficial since it overcomes the loading normalization issue. The often-used alternative approach of housekeeping proteins to normalize for loading biases among samples is sometimes problematic as levels of the HKPs change under various physiological, pathological and pharmacological conditions.446-450
Finally, a novel protein family of autophagy receptors, named CUET (from Cue5/Tollip), was identified, which in contrast to SQSTM1 and NBR1 has members that are present in all eukaryotes.451 The CUET proteins also possess a ubiquitin-binding CUE-domain and an Atg8-family interacting motif (AIM)/LIR sequence that interacts with Atg8/LC3. In their absence, cells are more vulnerable to the toxicity resulting from aggregation-prone proteins showing that CUET proteins, and more generally autophagy, play a critical evolutionarily conserved role in the clearance of cytotoxic protein aggregates.451 Experiments in yeast have shown that Cue5 and the cytoplasmic proteins that require this autophagy receptor for rapid degradation under starvation conditions could be potentially good marker proteins for measuring autophagic flux.
Special caution must be taken when evaluating SQSTM1 levels in models of protein aggregation. Small protoaggregates often stain positively for SQSTM1 and may be similar in size to autophagic puncta. Similarly, GFP-u/GFP-degron reporters (designed as an unstable variant that undergoes proteasome-dependent degradation) will mark SQSTM1-positive protein inclusions. Last, some types of aggregates and inclusions will release soluble SQSTM1 or GFP-u/GFP-degron under cell lysis or denaturing conditions, which can skew the interpretation of soluble SQSTM1 and/or proteasomal function, accordingly.
Conclusion: There is not always a clear correlation between increases in LC3-II and decreases in SQSTM1. Thus, although analysis of SQSTM1 can assist in assessing the impairment of autophagy or autophagic flux, we recommend using SQSTM1 only in combination with other methods detailed in these guidelines to monitor flux. See also the discussion in Autophagic flux determination using flow and multispectral imaging cytometry.
4. TOR/MTOR, AMPK and Atg1/ULK1
Atg1/ULK1 are central components in autophagy that likely act at more than one stage of the process. There are multiple ULK isoforms in mammalian cells including ULK1, ULK2, ULK3, ULK4 and STK36.452 ULK3 is a positive regulator of the Hedgehog signaling pathway,453 and its overexpression induces both autophagy and senescence.454 Along these lines, ectopic ULK3 displays a punctate pattern upon starvation-induced autophagy induction.454 ULK3, ULK4 and STK36, however, lack the domains present on ULK1 and ULK2 that bind ATG13 and RB1CC1/FIP200.455 Thus, ULK3 may play a role that is restricted to senescence and that is independent of the core autophagy machinery. ULK2 has a higher degree of identity with ULK1 than any of the other homologs, and they may have similar functions that are tissue specific. However, ULK1 may be the predominant isoform involved in autophagy, as knockdown of ULK2 does not affect movement of ATG9.456 Similarly, pharmacological inhibition of ULK1 and ULK2, with the compound MRT68921, blocks macroautophagy and expression of a drug-resistant ULK1 mutant is sufficient to rescue this block.457 The stability and activation of ULK1, but not ULK2, is dependent on its interaction with the HSP90-CDC37 chaperone complex. Pharmacological or genetic inhibition of the chaperone complex increases proteasome-mediated turnover of ULK1, impairing its kinase activity and ability to promote both starvation-induced autophagy and mitophagy.458
AMP-activated protein kinase (AMPK) is a multimeric serine/threonine protein kinase comprised of PRKAA1/AMPKα1 or PRKAA2/AMPKα2 (α, catalytic), the PRKAB1/AMPKβ1 or PRKAB2/AMPKβ2 (β, scaffold), and the PRKAG1/AMPKγ1, PRKAG2/AMPKγ2 or PRKAG3/AMPKγ3 (γ, regulatory) subunits. The enzyme activity of AMPK is dependent on phosphorylation of the PRKAA/α-subunit on Thr172,459,460 and, therefore, can be conveniently monitored by western blotting with a phosphospecific antibody against this site. In some cells, Thr172 is phosphorylated by CAMKK2/CaMKKβ, whereas in others it is a substrate of the STK11/LKB1 kinase. Regulation of AMPK activity is mediated primarily by Thr172-dephosphorylating protein phosphatases such as PPP1/PP1 (protein phosphatase 1) and PPP2/PP2A (protein phosphatase 2).461 Thr172 dephosphorylation is modulated by adenine nucleotides that bind competitively to regulatory sites in the PRKAG/γ-subunit. AMP and ADP inhibit dephosphorylation and promote AMPK activity, whereas Mg2+-ATP has the opposite effect.460 Thus, AMPK acts as a fine-tuned sensor of the overall cellular energy charge that regulates cellular metabolism to maintain energy homeostasis. Overexpression of a dominant negative mutant (R531G) of PRKAG2, the γ-subunit isoform 2 of AMPK that is unable to bind AMP, makes it possible to analyze the relationship between AMP modulation (or alteration of energetic metabolism) and AMPK activity.462,463 Activation of AMPK is also associated with the phosphorylation of downstream enzymes involved in ATP-consuming processes, such as fatty acid (ACAC [acetyl-CoA carboxylase]) and cholesterol (HMGCR [3-hydroxy-3-methylglutaryl-CoA reductase]) biosynthesis.
The role of AMPK in autophagy is complex and highly dependent on both cell type and metabolic conditions. Furthermore, as noted above, there are 2 isoforms of the catalytic subunit, PRKAA1/AMPKα1 and PRKAA2/AMPKα2, and these may have distinct effects with regard to autophagy (C. Koumenis, personal communication). In liver cells, AMPK suppresses autophagy at the level of cargo sequestration, as indicated by the rapid sequestration-inhibitory effects of a variety of AMPK activators, whereas it appears to stimulate autophagy in many other cell types, including fibroblasts, colon carcinoma cells and skeletal muscle.464-473 Autophagy-promoting effects of AMPK are most evident in cells cultured in a complete medium with serum and amino acids, where cargo sequestration is otherwise largely suppressed.470 Presumably, AMPK antagonizes the autophagy-inhibitory effect of amino acids (at the level of phagophore assembly) by phosphorylating proteins involved in MTORC1 signaling, such as TSC2474 and RPTOR475 as well the MTORC1 target ULK1 (see below).476-478
Compound C is an effective and widely used inhibitor of activated (phosphorylated) AMPK.479,480 However, being a nonspecific inhibitor of oxidative phosphorylation,481,482 this drug has been observed to inhibit autophagy under conditions where AMPK is already inactive or knocked out,483 and it has even been shown to stimulate autophagy by an AMP-independent mechanism.482,484 Compound C thus cannot be used as a stand-alone indicator of AMPK involvement, but can be used along with shRNA-mediated inhibition of AMPK.
TORC1 is an autophagy-suppressive regulator that integrates growth factor, nutrient and energy signals. In most systems, inhibition of MTOR leads to induction of autophagy, and AMPK activity is generally antagonistic toward MTOR function. MTORC1 mediates the autophagy-inhibitory effect of amino acids, which stimulate the MTOR protein kinase through a RRAG GTPase dimer. INS/insulin and growth factors activate MTORC1 through upstream kinases including AKT/protein kinase B and MAPK1/ERK2-MAPK3/ERK1 when the energy supply is sufficient, whereas energy depletion may induce AMPK-mediated MTORC1 inhibition and autophagy stimulation, for example, during glucose starvation. In contrast, amino acid starvation can strongly induce autophagy even in cells completely lacking AMPK catalytic activity.485
AMPK and MTORC1 regulate autophagy through coordinated phosphorylation of ULK1. Under glucose starvation, AMPK promotes autophagy by directly activating ULK1 through phosphorylation, although the exact AMPK-mediated ULK1 phosphorylation site(s) remains unclear (Table 2).473,476-478 Under conditions of nutrient sufficiency, high MTORC1 activity prevents ULK1 activation by phosphorylating alternate ULK1 residues and disrupting the interaction between ULK1 and AMPK. There are commercially available phospho-specific antibodies that recognize different forms of ULK1. For example, phosphorylation at Ser555, an AMPK site, is indicative of increased autophagy in response to nutrient stress, whereas Ser757 is targeted by MTOR to inhibit autophagy. Even the autophagy-suppressive effects of AMPK could, conceivably, be mediated through ULK1 phosphorylation, for example, at the inhibitory site Ser638.486 AMPK inhibits MTOR by phosphorylating and activating TSC2.487 Therefore, AMPK is involved in processes that synergize to activate autophagy, by directly activating ULK1, and indirectly impairing MTOR-dependent inhibition of ULK1. The identification of ULK1 as a direct target of MTORC1 and AMPK represents a significant step toward the definition of new tools to monitor the induction of autophagy. However, further studies directed at identifying physiological substrates of ULK1 will be essential to understand how ULK1 activation results in initiation of the autophagy program. Along these lines, ULK1 phosphorylates AMBRA1,488 and the MLCK-like protein Sqa,489 as well as ATG13, ATG9 and RB1CC1/FIP200.423,490-493 Furthermore, following amino acid starvation or MTOR inhibition, the activated ULK1 phosphorylates BECN1 on Ser14, enhancing the activity of the complexes containing ATG14 and PIK3C3/VPS34. This BECN1 phosphorylation by ULK1 is required for full autophagic induction.494 In addition, ULK1 binds to, and phosphorylates, RPTOR, leading to inhibition of MTORC1.495 Furthermore, ULK1 itself appears to be able to mediate inhibitory AMPK phosphorylation to generate a negative feedback loop.496 Note that caution should be taken to use appropriate inhibitors of phosphatases (e.g., sodium fluoride, and beta-glycerophosphate) in cell lysis buffer before analyzing the phosphorylation of AMPK and ULK1 at serine and threonine sites.
Table 2.
Phosphorylation targets of AKT, AMPK, GSK3B, MTORC1, PKA and Atg1/ULK1.
| Protein and phosphorylation site | Main kinase | Function | Ref |
|---|---|---|---|
| AMBRA1 S52 | TORC1 | Inhibits AMBRA1-dependent activation of ULK1 | 501 |
| Atg1 | TORC1 | Inhibits Atg1 kinase activity | 504 |
| Atg1 | PKA | Regulation of kinase activity | 1522 |
| Atg9 | Atg1 | Recruitment of Atg protein to the PAS | 493 |
| Atg13 | TORC1 | Interaction with Atg1, assembly of Atg1 kinase complex | 504,1523 |
| Atg13 | PKA | Regulates localization to the PAS | 1524 |
| BECN1 S14 | ULK1 | Increases the activity of the PtdIns3K | 494 |
| BECN1 S90 | MAPKAPK2-MAPKAPK3 | Stimulates macroautophagy | 1525 |
| BECN1 S91, S94 (S93, S96 in human) | AMPK | Required for glucose starvation-induced macroautophagy | 1526 |
| BECN1 Y229, Y233 | EGFR | Inhibits macroautophagy | 523 |
| BECN1 S234, S295 | AKT | Suppresses macroautophagy | 522 |
| LC3 S12 | PKA | Inhibits macroautophagy by reducing recruitment to phagophores | 343 |
| MTOR S2448 | AKT | Correlates with the activity of MTORC1 | 1527 |
| MTOR S2481 | Autophosphorylation | Necessary for MTORC1 formation and kinase activity | 1528 |
| NBR1 T586 | GSK3A/B | Modulates protein aggregation | 1529 |
| RPS6KB T389 | MTORC1 (apparently indirect, through reduction of dephosphorylation) | Necessary for protein activity | 1530 |
| RPS6KB S371 | GSK3B | Necessary for T389 phosphorylation and the activity of RPS6KB | 1531 |
| RPTOR S792 | AMPK | Suppresses MTORC1 | 475 |
| SQSTM1 S403 | ULK1 (also TBK1, CSNK, CDK1) | Promotes autophagic degradation of SQSTM1 and its substrates | 1532 |
| ULK1 S555 | AMPK (direct) | Necessary for ATG13-ULK1 interaction and for autophagy mediated by ULK complex | 477 |
| ULK1 S317, S467, S555, S574, S777 | AMPK (direct) | Necessary for the kinase activity of ULK1 | 477,478 |
| ULK1 S757 | MTORC1 | Prevents ULK1 interaction with AMPK | 478 |
| ULK1 S758 | MTORC1 | Facilitates ULK1 interaction with AMPK | 478,512 |
| ULK1 S637 | MTORC1, AMPK | Facilitates ULK1 interaction with AMPK | 477,512 |
| ULK1 (uncertain site between 278 and 351) | Autophosphorylation | Modulates the conformation of the C-terminal tail and prevents its interaction with ATG13 | 492,1533 |
TORC1 activity can be monitored by following the phosphorylation of its substrates, such as EIF4EBP1/4E-BP1/PHAS-I and RPS6KB/p70S6 kinase or the latter's downstream target, RPS6/S6, for which good commercial antibodies are available.497-499 In mammalian cells, the analysis should focus on the phosphorylation of RPS6KB1/S6K1 at Thr389, and EIF4EBP1 at Thr37 and Thr46, which are directly phosphorylated by MTORC1.500 The MTORC1-dependent phosphorylation of EIF4EBP1 can be detected as a molecular mass shift by western blot.499 Examining the phosphorylation status of RPS6KB and EIF4EBP1 may be a better method for monitoring MTORC1 activity than following the phosphorylation of proteins such as RPS6, because the latter is not a direct substrate of MTORC1 (although RPS6 phosphorylation is a good readout for RPS6KB1/2 activities, which are directly dependent on MTOR), and it can also be phosphorylated by other kinases such as RPS6KA/RSK. Furthermore, the mechanisms that determine the selectivity as well as the sensitivity of MTORC1 for its substrates seem to be dependent on the integrity and configuration of MTORC1. For example, rapamycin strongly reduces RPS6KB1 phosphorylation, whereas its effect on EIF4EBP1 is more variable. In the case of rapamycin treatment, EIF4EBP1 can be phosphorylated by MTORC1 until rapamycin disrupts MTORC1 dimerization and its integrity, whereas RPS6KB1 phosphorylation is quickly reduced when rapamycin simply interacts with MTOR in MTORC1 (see Autophagy inhibitors and inducers for information on catalytic MTOR inhibitors such as torin1).500 Since it is likely that other inhibitors, stress, and stimuli may also affect the integrity of MTORC1, a decrease or increase in the phosphorylation status of one MTORC1 substrate does not necessarily correlate with changes in others, including ULK1. Therefore, reliable anti-phospho-ULK1 antibodies should be used to directly examine the phosphorylation state of ULK1, along with additional experimental approaches to analyze the role of the MTOR complex in regulating autophagy. The MTORC1-mediated phosphorylation of AMBRA1 on Ser52 has also been described as relevant to ULK1 regulation and autophagy induction.488,501 In line with what is described for ULK1, the anti-phospho-AMBRA1 antibody, which is commercially available, could be used to indirectly measure MTORC1 activity.501
Activation/assembly of the Atg1 complex in yeast (composed of at least Atg1-Atg13-Atg17-Atg31-Atg29) or the ULK1 complex in mammals (ULK1-RB1CC1/FIP200-ATG13-ATG101) is one of the first steps of autophagy induction. Therefore, activation of this complex can be assessed to monitor autophagy induction. In yeast, dephosphorylation of Atg13 is associated with activation/assembly of the core complex that reflects the reduction of TORC1 and PKA activities. Therefore, assessing the phosphorylation levels of this protein by immunoprecipitation or western blotting502-505 can be used not only to follow the early steps of autophagy but also to monitor the activity of some of the upstream nutrient-sensing kinases. Because this protein is not easily detected when cells are lysed using conventional procedures, a detailed protocol has been described.506 In addition, the autophosphorylation of Atg1 at Thr226 is required for its kinase activity and for autophagy induction; this can be detected using phospho-specific antibodies, by immunoprecipitation or western blotting (Fig. 16).507,508 In Drosophila, TORC1-dependent phosphorylation of Atg1 and Atg1-dependent phosphorylation of Atg13 can be indirectly determined by monitoring phosphorylation-induced electromobility retardation (gel shift) of protein bands in immunoblot images.423,509,510 Nutritional starvation suppresses TORC1-mediated Atg1 phosphorylation,423,509 while stimulating Atg1-mediated Atg13 phosphorylation.423,509,510 In mammalian cells, the phosphorylation status of ULK1 at the activating sites (Ser317, 777, 467, 555, 637, or Thr574) or dephosphorylation at inactivating sites (Ser637, 757) can be determined by western blot using phospho-specific antibodies.477,478,480,486,511,512 In general, the core complex is stable in mammalian cells, although, as noted above, upstream inhibitors (MTOR) or activators (AMPK) may interact dynamically with it, thereby determining the status of autophagy.
Figure 16.
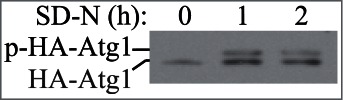
S. cerevisae cells transformed with a plasmid encoding HA-Atg1 were cultured to mid-log phase and shifted to SD-N (minimal medium lacking nitrogen that induces a starvation response). Immunoblotting was done with anti-HA antibody. The upper band corresponds to autophosphorylation of Atg1. This figure was modified from data previously published in ref. 508, and is reproduced by permission of the American Society for Cell Biology, copyright 2011.
One additional topic that bears on ULK1 concerns the process of LC3-associated phagocytosis (see Noncanonical use of autophagy-related proteins). LAP is a type of phagocytosis in macrophages that involves the conjugation of LC3 to single-membrane pathogen-containing phagosomes, a process that promotes phagosome acidification and fusion with lysosomes.182 Although ULK1 is not required for LAP, in this context it is important to note that UNC-51 (the Atg1 homolog in C. elegans) is required for apoptotic cell corpse clearance (a process corresponding to LAP) during embryonic development in worms,513 whereas this process is mediated by LAP in mammals,180 and does not require UNC-51 in C. elegans Q cell neuroblasts.514 In human macrophages infected with Mycobacterium tuberculosis, MORN2 is recruited at the phagosome membrane containing M. tuberculosis to induce the recruitment of LC3, and subsequent maturation into phagolysosomes. In addition, MORN2 drives trafficking of M. tuberculosis to a single-membrane compartment. Thus, in certain conditions MORN2 can be used to help to make the distinction between autophagy and LAP.515
Cautionary notes: A decrease in TORC1 activity is a good measure for autophagy induction; however, TORC1 activity does not necessarily preclude autophagy induction because there are TOR-independent mechanisms that induce autophagy both in mammals and yeast.516-520 Along these lines, whereas in most systems inhibition of MTOR leads to the induction of autophagy, there are instances in commonly used cancer cell lines in which MTOR appears to be a positive effector.521 Also, MTOR suppression does not always induce autophagy, such as when BECN1 undergoes inhibitory phosphorylation by the growth factor signaling molecules EGFR and AKT.522,523 Note that the effect of everolimus in EGFR-transgenic mice is not mainly attributable to autophagy although it suppresses MTOR and induces autophagy in EGFR-driven lung cancer cell lines.524 In adult skeletal muscle, active MTORC1 phosphorylates ULK1 at Ser757 to inhibit the induction of autophagosome formation. Thus, induction of autophagy requires inhibition of MTORC1 and not of MTORC2.525,526 There is also evidence that inhibition of MTORC1 is not sufficient to maintain autophagic flux, but requires additional activation of FOXO transcription factors for the upregulation of autophagy gene expression.468 In addition, MTORC1 is downstream of AKT; however, oxidative stress inhibits MTOR, thus allowing autophagy induction, despite the concomitant activation of AKT.150 Also, persistent MTORC1 inhibition can cause downregulation of negative feedback loops on IRS-MTORC2-AKT that results in the reactivation of MTORC2 under conditions of ongoing starvation.222,415,527 Along these lines, both TORC1 and autophagy can be active in specific cell subpopulations of yeast colonies.520 Thus, it is necessary to be cautious in deciding how to monitor the TOR/MTOR pathway, and to verify that the pathway being analyzed displays TOR/MTOR-dependent inhibition.
In addition, the regulation of autophagy by MTOR can be ULK1-independent. During mycobacterial infection of macrophages, MTOR induces the expression of MIR155 and MIR31 to sustain the activation of the WNT5A and SHH/sonic hedgehog pathways. Together, these pathways contribute to the expression of lipoxygenases and downregulation of IFNG-induced autophagy.528 Signaling pathways can be monitored by western blotting, and TaqMan miRNA assays are available to detect these miRNAs.
One problem in monitoring assembly of the ULK1 complex is the low abundance of endogenous ULK1 in many systems, which makes it difficult to detect phospho-ULK1 by western blot analysis. In addition, Atg1/ULK1 is phosphorylated by multiple kinases, and the amount of phosphorylation at different sites can increase or decrease during autophagy induction. Thus, although there is an increase in phosphorylation at the activating sites upon induction, the overall phosphorylation states of ULK1 and ATG13 are decreased under conditions that lead to induction of autophagy; therefore, monitoring changes in phosphorylation by following molecular mass shifts upon SDS-PAGE may not be informative. In addition, such phosphorylation/dephosphorylation events are expected to occur relatively early (1–2 h) in the signaling cascade of autophagy. Therefore, it is necessary to optimize treatment time conditions. Finally, in Arabidopsis and possibly other eukaryotes, the ATG1 and ATG13 proteins are targets of autophagy, which means that their levels may drop substantially under conditions that induce autophagic turnover.256
At present, the use of Atg1/ULK1 kinase activity as a tool to monitor autophagy is limited because only a few physiological substrates have been identified, and the importance of the Atg1/ULK1-dependent phosphorylation has not always been determined. Nonetheless, Atg1/ULK1 kinase activity appears to increase when autophagy is induced, irrespective of the pathway leading to induction. As additional physiological substrates of Atg1/ULK1 are identified, it will be possible to follow their phosphorylation in vivo as is done with analyses for MTOR. Nonetheless, it must be kept in mind that monitoring changes in the activity of Atg1/ULK1 is not a direct assay for autophagy, although such changes may correlate with autophagy activity. Furthermore, in some cells ULK1 has functions in addition to autophagy, such as in axonal transport and outgrowth, and its activity state may thus reflect its role in these processes.529-534 Accordingly, other methods as described throughout these guidelines should also be used to follow autophagy directly.
Finally, there is not a complete consensus on the specific residues of ULK1 that are targeted by AMPK or MTOR. Similarly, apparently contradictory data have been published regarding the association of AMPK and MTOR with the ULK1 kinase complex under different conditions. Therefore, caution should be used in monitoring ULK1 phosphorylation or the status of ULK1 association with AMPK until these issues are resolved.
Conclusion: Assays for Atg1/ULK1 can provide detailed insight into the induction of autophagy, but they are not a direct measurement of the process. Similarly, since MTOR substrates such as RPS6KB1 and EIF4EBP1 are not recommended readouts for autophagy, their analysis needs to be combined with other assays that directly monitor autophagy activity.
5. Additional autophagy-related protein markers
Although Atg8/LC3 has been the most extensively used protein for monitoring autophagy, other proteins can also be used for this purpose. Here, we discuss some of the more commonly used or better-characterized possibilities.
a. Atg9
Atg9 is the only integral membrane Atg protein that is essential for autophagosome formation in all eukaryotes. Mammalian ATG9 displays partial colocalization with GFP-LC3.535 Perhaps the most unique feature of Atg9, however, is that it localizes to multiple discrete puncta, whereas most Atg proteins are detected primarily in a single punctum or diffusely within the cytosol. Yeast Atg9 may cycle between the phagophore assembly site (PAS) and peripheral reservoirs;536 the latter correspond to tubulovesicular clusters that are precursors to the phagophore.537 Anterograde movement to the PAS is dependent on Atg11, Atg23, Atg27 and actin. Retrograde movement requires Atg1-Atg13, Atg2-Atg18 and the PtdIns3K complex I.538 Mutants such as atg1Δ accumulate Atg9 primarily at the PAS, and this phenotype forms the basis of the “transport of Atg9 after knocking out ATG1” (TAKA) assay.106 In brief, this is an epistasis analysis in which a double-mutant strain is constructed (one of the mutations being atg1Δ) that expresses Atg9-GFP. If the second mutated gene encodes a protein that is needed for Atg9 anterograde transport, the double mutant will display multiple Atg9-GFP puncta. In contrast, if the protein acts along with or after Atg1, all of the Atg9-GFP will be confined to the PAS. Monitoring the localization of ATG9 has not been used extensively in higher eukaryotes, but this protein displays the same type of dependence on Atg1/ULK1 and PtdIns3P for cycling as seen in yeast,535,538 suggesting that it is possible to follow this ATG9 as an indication of ULK1 and ATG13 function.492
b. Atg12–Atg5
ATG5, ATG12 and ATG16L1 associate with the phagophore and have been detected by fluorescence or immunofluorescence (Fig. 17).539,540 The endogenous proteins form puncta that can be followed to monitor autophagy upregulation. Under physiological conditions, these proteins are predominantly diffusely distributed throughout the cytoplasm. Upon induction of autophagy, for example during starvation, there is a marked increase in the proportion of cells with punctate ATG5, ATG12 and ATG16L1. Furthermore, upstream inhibitors of autophagosome formation result in a block in this starvation-induced puncta formation, and this assay is very robust in some mammalian cells. Conversely, downstream inhibition of autophagy at the level of autophagosome elongation, such as with inhibition of LC3/GABARAP expression, results in an accumulation of the phagophore-associated ATG5, ATG12 and ATG16L1 immunofluorescent puncta.541
Figure 17.
Confocal microscopy image of HCT116 cells immunostained with antibody specific to human ATG12. Cells were starved for 8 h or treated with chloroquine (50 µM) for 3 h. Scale bar: 10 µm. Image provided by M. Llanos Valero, M.A. de la Cruz and R. Sanchez-Prieto.
ATG12–ATG5 conjugation has been used in some studies to measure autophagy. In Arabidopsis and some mammalian cells it appears that essentially all of the ATG5 and ATG12 proteins exist in the conjugated form and the expression levels do not change, at least during short-term starvation.214,539,540,542 Therefore, monitoring ATG12–ATG5 conjugation per se may not be a useful method for following the induction of autophagy. It is worth noting, however, that in some cell lines free ATG5 can be detected,543 suggesting that the amount of free ATG5 may be cell line-dependent; free ATG5 levels also vary in response to stress such as DNA damage.544 One final parameter that may be considered is that the total amount of the ATG12–ATG5 conjugate may increase following prolonged starvation as has been observed in hepatocytes and both mouse and human fibroblasts (A.M. Cuervo, personal communication; S. Sarkar, personal communication).
c. ATG14
Yeast Atg14 is the autophagy-specific subunit of the Vps34 complex I,545 and a human homolog, named ATG14/ATG14L/BARKOR, has been identified.546-549 ATG14 localizes primarily to phagophores. The C-terminal fragment of the protein, named the BATS domain, is able to direct GFP and BECN1 to autophagosomes in the context of a chimeric protein.550 ATG14-GFP or BATS-GFP detected by fluorescence microscopy or TEM can be used as a phagophore marker protein; however, ATG14 is not localized exclusively to phagophores, as it can also be detected on mature autophagosomes as well as the ER.550,551 Accordingly, detection of ATG14 should be carried out in combination with other phagophore and autophagosome markers. A good antibody that can be used to detect endogenous ATG14 is now available commercially (D.-H. Kim, personal communication).
d. ATG16L1
ATG16L1 has been used to monitor the movement of plasma membrane as a donor for autophagy, and thus an early step in the process. Indeed, ATG16L1 is located on phagophores, but not on completed autophagosomes.344,552 ATG16L1 can be detected by immuno-TEM, by immunostaining of Flag epitope-tagged ATG16L1, and/or by the use of GFP-tagged ATG16L1.
e. Atg18/WIPI family
Yeast Atg18553,554 and Atg21335 (or the mammalian WIPI homologs555) are required for both macroautophagy (i.e., nonselective sequestration of cytoplasm) and autophagy-related processes (e.g., the Cvt pathway,556,557 specific organelle degradation,119 and autophagic elimination of invasive microbes122,123,125,126,553,558). These proteins bind phosphatidylinositol 3-phosphate (PtdIns3P) that is present at the phagophore and autophagosome559,560 and also PtdIns(3,5)P2. Human WIPI1 and WIPI2 function downstream of the class III phosphatidylinositol 3-kinase complex I (PIK3C3/VPS34, BECN1, PIK3R4/VPS15, ATG14) and upstream of both the ATG12 and LC3 ubiquitin-like conjugation systems.559,561,562 Upon the initiation of the autophagic pathway, WIPI1 and WIPI2 bind PtdIns3P and accumulate at limiting membranes, such as those of the ER, where they participate in the formation of omegasomes and/or autophagosomes. On the basis of quantitative fluorescence microscopy, this specific WIPI protein localization has been used as an assay to monitor autophagy in human cells.560 Using either endogenous WIPI1 or WIPI2, detected by indirect fluorescence microscopy or EM, or transiently or stably expressed tagged fusions of GFP to WIPI1 or WIPI2, basal autophagy can be detected in cells that display WIPI puncta at autophagosomal membranes. In circumstances of increased autophagic activity, such as nutrient starvation or rapamycin administration, the induction of autophagy is reflected by the elevated number of cells that display WIPI puncta when compared to the control setting. Also, in circumstances of reduced autophagic activity such as wortmannin treatment, the reduced number of WIPI puncta-positive cells reflects the inhibition of autophagy. Basal, induced and inhibited formation of WIPI puncta closely correlates with both the protein level of LC3-II and the formation of GFP-LC3 puncta.560,562 Accordingly, WIPI puncta can be assessed as an alternative to LC3. Automated imaging and analysis of fluorescent WIPI1 (Fig. 18) or WIPI2 puncta represent an efficient and reliable opportunity to combine the detection of WIPI proteins with other parameters. It should be noted that there are 2 isoforms of WIPI2 (2B and 2D),562 and in C. elegans WIPI4 (EPG-6) has been identified as the WIPI homolog required for autophagy.563 Thus, these proteins, along with the currently uncharacterized WDR45B/WIPI3, provide additional possibilities for monitoring phagophore and autophagosome formation.
Figure 18.
Automated WIPI1 puncta image acquisition and analysis monitors the induction and inhibition of autophagy. Stable U2OS clones expressing GFP-WIPI1 were selected using 0.6 μg/ml G418 and then cultured in 96-well plates. Cells were treated for 3 h with nutrient-rich medium (control), nutrient-free medium (EBSS), or with 233 nM wortmannin. Cells were fixed in 3.7% paraformaldehyde and stained with DAPI (5 μg/ml in PBS). An automated imaging and analysis platform was used to determine the number of both GFP-WIPI1 puncta-positive cells and the number of GFP-WIPI1 puncta per individual cell.470 Cells without GFP-WIPI1 puncta are highlighted in red (cell detection) and purple (nuclei detection), whereas GFP-WIPI1 puncta-positive cells are highlighted in yellow (GFP-WIPI1 puncta detection), green (cell detection) and blue (nuclei detection). Scale bars: 20 µm. Images provided by S. Pfisterer and T. Proikas-Cezanne.
Cautionary notes: With regard to detection of the WIPI proteins, endogenous WIPI1 puncta cannot be detected in many cell types,559 and the level of transiently expressed GFP-WIPI1 puncta is cell context-dependent;559,560 however, this approach has been used in human and mouse cell systems470,560 and mCherry-Atg18 also works well for monitoring autophagy in transgenic Drosophila,135 although one caution with regard to the latter is that GFP-Atg18 expression enhances Atg8 lipidation in the fat body of fed larvae. GFP-WIPI1 and GFP-WIPI2 have been detected on the completed (mature) autophagosome by freeze-fracture analysis,102 but endogenous WIPI2 has not been detected on mRFP-LC3- or LAMP2-positive autophagosomes or autolysosomes using immunolabeling.559 Accordingly, it may be possible to follow the formation and subsequent disappearance of WIPI puncta to monitor autophagy induction and flux using specific techniques. As with GFP-LC3, overexpression of WIPI1 or WIPI2 can lead to the formation of aggregates, which are stable in the presence of PtdIns3K inhibitors.
f. BECN1/Vps30/Atg6
BECN1 (yeast Vps30/Atg6) and PIK3C3/VPS34 are essential partners in the autophagy interactome that signals the onset of autophagy,545,564,565 and many researchers use this protein as a way to monitor autophagy. BECN1 is inhibited by its binding to the anti-apoptotic protein BCL2.566 Autophagy is induced by the release of BECN1 from BCL2 by pro-apoptotic BH3 proteins, phosphorylation of BECN1 by DAPK1 (at Thr119, located in the BH3 domain),567 or phosphorylation of BCL2 by MAPK8/JNK1 (at Thr69, Ser70 and Ser87).568,569 The relationship between BECN1 and BCL2 is more complex in developing cerebellar neurons, as it appears that the cellular levels of BCL2 are, in turn, post-translationally regulated by an autophagic mechanism linked to a switch from immaturity to maturity.570,571 It is important to be aware, however, that certain forms of macroautophagy are induced in a BECN1-independent manner and are not blocked by PtdIns3K inhibitors.83,572 Interestingly, caspase-mediated cleavage of BECN1 inactivates BECN1-induced autophagy and enhances apoptosis in several cell types,573 emphasizing that the crosstalk between apoptosis and autophagy is complex.
Although a population of BECN1 may localize in proximity to the trans-Golgi network,574 it is also present at the ER and mitochondria.566 In keeping with these observations, in cerebellar organotypic cultures BECN1 co-immunoprecipitates with BCL2 that is primarily localized at the mitochondria and ER; and in a mouse model of neurodegeneration, autophagic vacuoles in Purkinje neurons contain partially digested organelles that are immunoreactive for BCL2.571,575 In addition, BECN1 and PIK3C3/VPS34 can be present in multiple complexes, so caution must be exercised when monitoring localization. On induction of autophagy by various stimuli the presence of BECN1- and PIK3C3/VPS34-positive macroaggregates can be detected in the region of the Golgi complex by immunofluorescence.150,576 Thus, BECN1-GFP puncta detected by fluorescence microscopy or TEM may serve as an additional marker for autophagy induction;577 however, it should be noted that caspase cleavage of BECN1 can be detected in normal culture conditions (S. Luo, personal communication), and cleaved BECN1 is translocated into the nucleus,578 thus care needs to be taken with these assays under stress conditions in which more pronounced BECN1 cleavage occurs. In addition, as with any GFP chimeras there is a concern that the GFP moiety interferes with correct localization of BECN1. To demonstrate that BECN1 or PtdIns3K macroaggregates are an indirect indication of ongoing autophagy, it is mandatory to show their specific association with the process by including appropriate controls with inhibitors (e.g., 3-MA) or autophagy gene silencing. When a BECN1-independent autophagy pathway is induced, such aggregates are not formed regardless of the fact that the cell expresses BECN1 (e.g., as assessed by western blotting; C. Isidoro, personal communication). As BECN1-associated PtdIns3K activity is crucial in autophagosome formation in BECN1-dependent autophagy, the measurement of PtdInsk3K in vitro lipid kinase activity in BECN1 immunoprecipitates can be a useful technique to monitor the functional activity of this complex during autophagy modulation.522,523,579
g. DRAM1
DRAM1 is a gene induced by activated TP53 in response to different types of cellular stress, including DNA damage.580,581 DRAM1 is a small hydrophobic protein with 6 transmembrane domains. It is detected as a subpopulation in the Golgi and cis-Golgi, colocalizing with GOLGB1/giantin and GOLGA2/GM130, and also in early and late endosomes and lysosomes, colocalizing with EEA1 and LAMP2.581 The elimination of DRAM1 by siRNA blocks autophagy,581,582 as effectively as elimination of BECN1, indicating it is an essential component for this process, although its mechanism of action is not known. The time course of autophagy as a consequence of DRAM1 activation can be monitored by immunoblot by following the disappearance of the VRK1 protein, a direct target of this process.581 Detection of DRAM1 RNA is very easy by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) during autophagy; 580,581 however, detection of the DRAM1 protein is very difficult because of its small size and hydrophobicity, features that complicate the generation of specific antibodies, which in general have very low sensitivity. A commercial DRAM1 antibody may allow the detection of this protein in rat skeletal muscle (D.W. Russ, personal communication).
h. ZFYVE1/DFCP1
ZFYVE1 binds PtdIns3P that localizes to the ER and Golgi. Starvation induces the translocation of ZFYVE1 to punctate structures on the ER; the ER population of ZFYVE1 marks the site of omegasome formation.583 ZFYVE1 partially colocalizes with WIPI1 upon nutrient starvation562 and also with WIPI2.559
i. STX17
STX17 is a SNARE protein that is recruited to completely sealed autophagosomes, but not to phagophores.584,585 As little STX17 is present on autolysosomes, STX17 is enriched on completed autophagosomes among autophagy-related structures. However, STX17 as a competence factor may be recruited just prior to fusion of autophagosomes with lysosomes, and not all autophagosomes are positive for this protein. Moreover, it is also present at the ER and mitochondria.
j. TECPR1
TECPR1 binds ATG5 through an AFIM (ATG5 [five] interacting motif). TECPR1 competes with ATG16L1 for binding to ATG5, suggesting that there is a transition from the ATG5-ATG16L1 complex that is involved in phagophore expansion to an ATG5-TECPR1 complex that plays a role in autophagosome-lysosome fusion. TECPR1 thus marks lysosomes and autolysosomes.586
Conclusion: Proteins other than Atg8/LC3 can be monitored to follow autophagy, and these can be important tools to define specific steps of the process. For example, WIPI puncta formation can be used to monitor autophagy, but, similar to Atg8/LC3, should be examined in the presence and absence of lysosomal inhibitors. Analysis of WIPI puncta should be combined with other assays because individual members of the WIPI family might also participate in additional, uncharacterized functions apart from their role in autophagy. At present, we caution against the use of changes in BECN1 localization as a marker of autophagy induction. It is also worth considering the use of different markers depending on the specific autophagic stimuli.
6. Sphingolipids
Sphingolipids are ubiquitous membrane lipids that can be produced in a de novo manner from the ER or by cleavage of sphingomyelin by phosphodiesterases (sphingomyelinases). The multiple different metabolites of the sphingolipid pathway, which are distinct by even a single double bond, carbon chain length of the fatty acid, or presence of a phosphate group, can have quite varied cellular functions. Sphingolipids were first recognized for their role in the architecture of membrane bilayers affecting parameters such as bilayer stiffness, neighboring lipid order parameter and microdomain/raft formation. They also act as second messengers in vital cellular signaling pathways and as key determinants of cellular homostasis in what is called a sphingolipid rheostat.587 Sphingolipids participate in the formation of different membrane structures and subcellular organelles, such as mitochondria and ER, and are also involved in the fusion and biophysical properties of cell membranes.588
Ceramides, positioned at the core of sphingolipid metabolism, play several roles that affect multiple steps of macroautophagy, by inhibition of nutrient transporters,589 by modulation of BCL2-BECN1 association at the level of AKT signaling,590 and by regulation of mitophagy.591 The latter function is regulated by a particular ceramide species, steroyl (C18:0)-ceramide, a sphingolipid generated by CERS1 (ceramide synthase 1). C18-ceramide, in association with LC3-II, targets damaged mitochondria for autophagic sequestration in response to ceramide stress, leading to tumor suppression.591-593 The binding of ceramide to LC3-II can be detected using anti-ceramide and anti-LC3 antibodies by immunofluorescence and confocal microscopy, co-immunoprecipitation using anti-LC3 antibody followed by liquid chromatography-tandem mass spectrometry, using appropriate standards (targeted lipidomics), or labeling cells with biotin-sphingosine to generate biotin-ceramide, and immunoprecipitation using avidin-columns followed by western blotting to detect LC3-II. It should be noted that inhibitors of ceramide generation, and mutants of LC3 with altered ceramide binding (F52A or I35A), and/or that are conjugation defective (e.g., G120A), should be used as negative controls.
Other sphingolipids are also involved in autophagy. For example, accumulation of endogenous sphingosine-1-phosphate, a pro-survival downstream metabolite from ceramide triggers ER-stress associated macroautophagy, by activation of AKT.594 In addition, gangliosides, have been implicated in autolysosome morphogenesis.595 To analyze the role of gangliosides in autophagy, 2 main technical approaches can be used: co-immunoprecipitation and fluorescence resonance energy transfer. For the first method, lysates from untreated or autophagy-induced cells have to be immunoprecipitated with an anti-LC3 polyclonal antibody (a rabbit IgG isotypic control should be used as a negative control). The obtained immunoprecipitates are subjected to ganglioside extraction, and the extracts run on an HPTLC aluminum-backed silica gel and analyzed for the presence of specific gangliosides by using monoclonal antibodies. Alternatively, the use of FRET by flow cytometry appears to be highly sensitive to small changes in distance between 2 molecules and is thus suitable to study molecular interactions, for example, between ganglioside and LC3. Furthermore, FRET requires ∼10 times less biological material than immunoprecipitation.
Conclusion: Sphingolipids are bioactive molecules that play key roles in the regulation of autophagy at various stages, including upstream signal transduction pathways to regulate autophagy via transcriptional and/or translational mechanisms, autolysosome morphogenesis, and/or targeting phagophores to mitochondria for degradation via sphingolipid-LC3 association.204,593,596
7. Transcriptional, translational and posttranslational regulation
The induction of autophagy in certain scenarios is accompanied by an increase in the mRNA levels of certain autophagy genes, such as ATG7,597,598 ATG8/Lc3,599,600 ATG9,601 Atg12,602 and Atg14,603 and an autophagy-dedicated microarray was developed as a high-throughput tool to simultaneously monitor the transcriptional regulation of all genes involved in, and related to, autophagy.604 The mammalian gene that shows the greatest transcriptional regulation in the liver (in response to starvation and circadian signals) is Ulk1, but others also show more limited changes in mRNA levels including Gabarapl1, Bnip3 and, to a minor extent, Lc3b (J.D. Lin, personal communication). In several mouse and human cancer cell lines, ER stress and hypoxia increase the transcription of Lc3/LC3, Atg5/ATG5 and Atg12/ATG12 by a mechanism involving the unfolded protein response (UPR). Similarly, a stimulus-dependent increase in LC3B expression is detected in neural stem cells undergoing autophagy induction.605 Increased expression of Atg5 in vivo after optic nerve axotomy in mice606 and increased expression of Atg7, Becn1 and Lc3a during neurogenesis at different embryonic stages in the mouse olfactory bulb are also seen.607 LC3 and ATG5 are not required for the initiation of autophagy, but mediate phagophore expansion and autophagosome formation. In this regard, the transcriptional induction of LC3 may be necessary to replenish the LC3 protein that is turned over during extensive ER stress- and hypoxia-induced autophagy.602,608 In the clinical setting, tissue expression of ATG5, LC3A and LC3B and their respective proteins accompanies elevated autophagy flux in human adipose tissue in obesity.217,609 Thus, assessing the mRNA levels of LC3 and other autophagy-related genes by northern blot or qRT-PCR may provide correlative data relating to the induction of autophagy. Downregulation of autophagy-related mRNAs has been observed in human islets under conditions of lipotoxicity409 that impair autophagic flux.610 It is not clear if these changes are sufficient to regulate autophagy, however, and therefore these are not direct measurements.
Several transcription factors of the nuclear receptor superfamily modulate gene expression of autophagy genes. For instance, NR1D1/Rev-erbα represses Ulk1, Bnip3, Atg5, Park2/parkin and Becn1 gene expression in mouse skeletal muscle by directly binding to regulatory regions in their DNA sequences. Consistently, nr1d1-/- mice display an increased LC3-II/LC3-I ratio, as well as PARK2 and BNIP3 protein levels, elevated autophagic flux as measured upon different inhibitor (3-MA, NH4Cl, bafilomycin A1 and chloroquine) treatment and autophagosomes detected by EM of skeletal muscle sections.611 The nuclear receptors PPARA (peroxisome proliferator-activated receptor alpha) and NR1H4/FXR (nuclear receptor subfamily 1, group H, member 4) also regulate hepatic autophagy in mice. Indeed, PPARA and NR1H4 compete for the control of lipophagy in response to fasting and feeding nutritional cues, respectively.612 NR1H4 may also inhibit autophagy via inhibition of CREB-CRTC2 complex assembly.613 Consistent with in vitro studies utilizing human cancer cell lines,614,615 in human adipose tissue explants, E2F1 binds the LC3B promoter, in association with increased expression of several autophagy genes and elevated adipose tissue autophagic flux.217,609 In this instance, classical promoter analysis studies, including chromatin immunoprecipitation and ATG promoter-luciferase constructs provide insights on the putative transcriptional regulation of autophagy genes by demonstrating promoter binding in situ, and promoter activity in vitro.609
Of note, large changes in Atg gene transcription just prior to Drosophila salivary gland cell death (that is accompanied by an increase in autophagy) are detected for Atg2, Atg4, Atg5 and Atg7, whereas there is no significant change in Atg8a or Atg8b mRNA.616,617 Autophagy is critical for Drosophila midgut cell death, which is accompanied by transcriptional upregulation of all of the Atg genes tested, including Atg8a (Fig. 19).281,618 Similarly, in the silkworm (Bombyx mori) larval midgut619 and fat body,620 the occurrence of autophagy is accompanied by an upregulation of the mRNA levels of several Atg genes. Transcriptional upregulation of Drosophila Atg8a and Atg8b is also observed in the fat body following induction of autophagy at the end of larval development,621 and these genes as well as Atg2, Atg9 and Atg18 show a more than 10-fold induction during starvation.622 Atg5, Atg6, Atg8a and Atg18 are upregulated in the ovary of starved flies,623 and an increase in Drosophila Atg8b is observed in cultured Drosophila l(2)mbn cells following starvation (S. Gorski, personal communication). An upregulation of plant ATG8 may be needed during the adaptation to reproductive growth; a T-DNA inserted mutation of rice ATG8b blocked the change from vegetative growth to reproductive growth in both homozygous and heterozygous plant lines (M.-Y. Zhang, unpublished results).
Figure 19.
pGFP-Atg8a can be used to monitor autophagy in Drosophila melanogaster. The autophagosome marker pGFP-Atg8a, results in expression of Atg8a fused to GFP from the endogenous Atg8a promoter.281 Live imaging of gastric caeca from Drosophila melanogaster midgut pGFP-Atg8a puncta (green) and Hoechst 33342 (blue). Midgut from early third instar larvae prior to the onset of cell death (top) and from dying midgut at 2 h after puparium formation (bottom). Bar: 25 µm. Image provided by D. Denton and S. Kumar.
Similarly, the upregulation of autophagy-related genes (Lc3, Gabarapl1, Bnip3, Atg4b, Atg12l) has been documented at the transcriptional and translational level in several other species (e.g., C. elegans,624 mouse, rat, human,625 trout, Arabidopsis and maize) under conditions of ER stress,602 and diverse types of prolonged (several days) catabolic situations including cancer cachexia, diabetes mellitus, uremia and fasting.215,468,626-628 Along these lines, ATG9 and ATG16L1 are transcriptionally upregulated upon influenza virus infection (H. Khalil, personal communication), and in C. elegans, the FOXA transcription factor PHA-4 and the TFEB ortholog HLH-30 regulate the expression of several autophagy-related genes (see Methods and challenges of specialized topics/model systems. C. elegans).624,629,1704 Such prolonged induction of the expression of ATG genes has been thought to allow the replenishment of critical proteins (e.g., LC3 and GABARAP) that are destroyed during autophagosome fusion with the lysosome.630 The polyamine spermidine increases life span and induces autophagy in cultured yeast and mammalian cells, as well as in nematodes and flies. In aging yeast, spermidine treatment triggers epigenetic deacetylation of histone H3 through inhibition of histone acetyltransferases, leading to significant upregulation of various autophagy-related transcripts.631
In addition to the ATG genes, transcriptional upregulation of VMP1 (a protein that is involved in autophagy regulation and that remains associated with the completed autophagosome) can be detected in mammalian cells subjected to rapamycin treatment or starvation, and in tissues undergoing disease-induced autophagy such as cancer.632 VMP1 is an essential autophagy gene that is conserved from Dictyostelium to mammals,322,633 and the VMP1 protein regulates early steps of the autophagic pathway.561 VMP1 is poorly expressed in mammalian cells under nutrient-normal conditions, but is highly upregulated in cells undergoing autophagy, and the expression of VMP1 induces autophagosome formation. The GLI3 transcription factor is an effector of KRAS that regulates the expression and promoter activity of VMP1, using the histone acetyltransferase EP300/p300 as a co-activator.634
A gene regulatory network, named CLEAR (coordinated lysosomal expression and regulation) that controls both lysosome and autophagosome biogenesis was identified using a systems-biology approach.625,635,636 The basic helix-loop-helix transcription factor TFEB acts as a master gene of the CLEAR network and positively regulates the expression of both lysosomal and autophagy genes, thus linking the biogenesis of 2 distinct types of cellular compartments that cooperate in the autophagic pathway. TFEB activity is regulated by starvation and is controlled by both MAPK1/ERK2- and MTOR-mediated phosphorylation at specific serine residues;625,637,638 thus, it can serve as a new tool for monitoring transcriptional regulation connected with autophagy. TFEB is phosphorylated by MTORC1 on the lysosomal surface, preventing its nuclear translocation. A lysosome-to-nucleus signaling mechanism transcriptionally regulates autophagy and lysosomal biogenesis via MTOR and TFEB.638 A very useful readout of endogenous TFEB activity is the evaluation of TFEB subcellular localization, as activation of TFEB correlates with its translocation from the cytoplasm to the nucleus. This shift can be monitored by immunofluorescence using antibodies against TFEB. TFEB localization may also be studied to monitor MTOR activity, as in most cases TFEB nuclear localization correlates with inhibition of MTOR. However, due to the low expression levels of TFEB in most cells and tissues, it may be difficult to visualize the endogenous protein. Thus a TFEB nuclear translocation assay was developed in a HeLa cell line stably transfected with TFEB-GFP. This fluorescence assay can be used to identify the conditions and factors that promote TFEB activation.638 TFE3 and MITF, 2 other members of the MiT/TFE family of transcription factors, in some cases can compensate for TFEB and are regulated in a similar manner.639,640
Similar to TFEB, the erythroid transcription factor GATA1 and its coregulator ZFPM1/FOG1 induce the transcription of multiple genes encoding autophagy components. This developmentally regulated transcriptional response is coupled to increases in autophagosome number as well as the percent of cells that contain autophagosomes.641 FOXO transcription factors, especially FOXO1 and FOXO3, also play critical roles in the regulation of autophagy gene expression.468,603,642 A zinc finger family DNA-binding protein, ZKSCAN3 is a master transcriptional repressor of autophagy and lysosome biogenesis; starvation and MTOR inhibition with torin1 induce nucleus-to-cytoplasm translocation of ZKSCAN3.643 Finally, CEBPB/C/EBPβ is a transcription factor that regulates autophagy in response to the circadian cycle.644
Although less work has been done on post-transcriptional regulation, several studies implicate microRNAs in controlling the expression of proteins associated with autophagy.243,247,248,645-647
Cautionary notes: Most of the ATG genes do not show significant changes in mRNA levels when autophagy is induced. Even increases in LC3 mRNA can be quite modest and are cell type- and organism-dependent.648 In addition, it is generally better to follow protein levels, which, ultimately, are the significant parameter with regard to the initiation and completion of autophagy. However, ATG protein amounts do not always change significantly and the extent of increase is again cell type- and tissue-dependent. Finally, changes in autophagy protein levels are not sufficient evidence of autophagy induction and must be accompanied by additional assays as described herein. Thus, monitoring changes in mRNA levels for either ATG genes or autophagy regulators may provide some evidence supporting upregulation of the potential to undergo autophagy, but should be used along with other methods.
Another general caution pertains to the fact that in any cell culture system mixed populations of cells (for example, those undergoing autophagy or not) exist simultaneously. Therefore, only an average level of protein or mRNA expression can be evaluated with most methods. This means that the results regarding specific changes in autophagic cells could be hidden due to the background of the average data. Along these lines, experiments using single-cell real-time PCR to examine gene expression in individual cardiomyocytes with and without signs of autophagy revealed that the transcription of MTOR markedly and significantly increases in autophagic cells in intact cultures (spontaneously undergoing autophagy) as well as in cultures treated with proteasome inhibitors to induce autophagy (V. Dosenko, personal communication). Finally, researchers need to realize that mammalian cell lines may have mutations that alter autophagy signaling or execution; this problem can be avoided by using primary cells.
Conclusion: Although there are changes in ATG gene expression that coincide with, and may be needed for, autophagy, this has not been carefully studied experimentally. Therefore, at the present time we do not recommend the monitoring of ATG gene transcription as a general readout for autophagy unless there is clear documentation that the change(s) correlates with autophagy activity.
8. Posttranslational modification of ATG proteins
Autophagy is controlled by posttranslational modification (PTM) of ATG proteins such as phosphorylation, ubiquitination, acetylation, oxidation and cleavage, which can be monitored to analyze the status of the process.343,438,519,523,649-652 The global deacetylation of proteins, which often accompanies autophagy, can be conveniently measured by quantitative immunofluorescence with antibodies specifically recognizing acetylated lysine residues.653 Indeed, depletion of the nutrient supply causes autophagy in yeast or mammalian cells by reducing the nucleo-cytosolic pool of acetyl-coenzyme A, which provides acetyl groups to acetyltransferases, thus reducing the acetylation level of hundreds of cytoplasmic and nuclear proteins.654 A global deacetylation of cellular proteins is also observed in response to so-called “caloric restriction mimetics”, that is, a class of pharmacological agents that deplete the nucleo-cytosolic pool of acetyl-coenzyme A, inhibit acetyltransferases (such as EP300) or activate deacetylases (such as SIRT1). All these agents reduce protein acetylation levels in cells as they induce autophagy.655 One prominent ATG protein that is subjected to pro-autophagic deacetylation is LC3.656,657
9. Autophagic protein degradation
Protein degradation assays represent a well-established methodology for measuring autophagic flux, and they allow good quantification. The general strategy is first to label cellular proteins by incorporation of a radioactive amino acid (e.g., [14C]- or [3H]-leucine, [14C]-valine or [35S]-methionine; although valine may be preferred over leucine due to the strong inhibitory effects of the latter on autophagy), preferably for a period sufficient to achieve labeling of the long-lived proteins that best represent autophagic substrates, and then to follow this with a long cold-chase so that the assay starts well after labeled short-lived proteins are degraded (which occurs predominantly via the proteasome). Next, the time-dependent release of acid-soluble radioactivity from the labeled protein in intact cells or perfused organs is measured.3,658,659 Note that the inclusion of the appropriate unlabeled amino acid (i.e., valine, leucine or methionine) in the starvation medium at a concentration equivalent to that of other amino acids in the chase medium is necessary; otherwise, the released [14C]-amino acid is effectively re-incorporated into cellular proteins, which results in a significant underestimation of protein degradation. A newer method of quantifying autophagic protein degradation is based on L-azidohomoalanine (AHA) labeling.660 When added to cultured cells, L-azidohomoalanine is incorporated into proteins during active protein synthesis. After a click reaction between an azide and an alkyne, the azide-containing proteins can be detected with an alkyne-tagged fluorescent dye, coupled with flow cytometry. The turnover of specific proteins can also be measured in a pulse-chase regimen using the Tet-ON/OFF or GeneSwitch systems and subsequent western blot analysis.661-663
In this type of assay a considerable fraction of the measured degradation will be nonautophagic, and thus it is important to also measure, in parallel, cell samples treated with autophagy-suppressive concentrations of 3-MA or amino acids, or obtained from mutants missing central ATG components (however, it is important to note that these controls are only appropriate assuming that nonautophagic proteolytic activity remains unchanged, which is unlikely); these values are then subtracted from the total readouts. The complementary approach of using compounds that block other degradative pathways, such as proteasome inhibitors, may cause unexpected results and should be interpreted with caution due to crosstalk among the degradative systems. For example, blocking proteasome function may activate autophagy.664-667 Thus, when using inhibitors it is critical to know whether the inhibitors being used alter autophagy in the particular cell type and context being examined. In addition, because 3-MA could have some autophagy-independent effects in particular settings it is advisable to verify that the 3-MA-sensitive degradation is also sensitive to general lysosomal inhibitors (such as NH4Cl or leupeptin).
The use of stable isotopes, such as 13C and 15N, in quantitative mass spectrometry-based proteomics allows the recording of degradation rates of thousands of proteins simultaneously. These assays may be applied to autophagy-related questions enabling researchers to investigate differential effects in global protein or even organelle degradation studies.668,669 Stable isotope labeling with amino acids in cell culture (SILAC) can also provide comparative information between different treatment conditions, or between a wild type and mutant.
Another assay that could be considered relies on the limited proteolysis of a BHMT (betaine–homocysteine S-methyltransferase) fusion protein. The 44-kDa full-length BHMT protein is cleaved in hepatocyte amphisomes in the presence of leupeptin to generate 32-kDa and 10-kDa fragments.670-673 Accumulation of these fragments is time dependent and is blocked by treatment with autophagy inhibitors. A modified version of this marker, GST-BHMT, can be expressed in other cell lines where it behaves similar to the wild-type protein.674 Additional substrates may be considered for similar types of assays. For example, the neomycin phosphotransferase II-GFP (NeoR-GFP) fusion protein is a target of autophagy.675 Transfection of lymphoblastoid cells with a plasmid encoding NeoR-GFP followed by incubation in the presence of 3-MA leads to an accumulation of the NeoR-GFP protein as measured by flow cytometry.676
A similar western blot assay is based on the degradation of a cytosolic protein fused to GFP. This method has been used in yeast and Dictyostelium cells using GFP-Pgk1 and GFP-Tkt-1 (phosphoglycerate kinase and transketolase, respectively). In this case the relative amount of free GFP versus the complete fusion protein is the relevant parameter for quantification; although it may not be possible to detect clear changes in the amount of the full-length chimera, especially under conditions of limited flux.30,37 As described above for the marker GFP-Atg8/LC3, nonsaturating levels of lysosomal inhibitors are also needed in Dictyostelium cells to slow down the autophagic degradation, allowing the accumulation and detection of free GFP. It should be noted that this method monitors bulk autophagy since it relies on the passive transit of a cytoplasmic marker to the lysosome. Consequently, it is important to determine that the marker is distributed homogeneously in the cytoplasm.
One of the most useful methods for monitoring autophagy in Saccharomyces cerevisiae is the Pho8Δ60 assay. PHO8 encodes a vacuolar phosphatase, which is synthesized as a zymogen before finally being transported to and activated in the vacuole.677 A molecular genetic modification that eliminates the first 60 amino acids prevents the mutant (Pho8Δ60) from entering the ER, leaving the zymogen in the cytosol. When autophagy is induced, the mutant zymogen is delivered to the vacuole nonselectively inside autophagosomes along with other cytoplasmic material. The resulting activation of the zymogen can be easily measured by enzymatic assays for phosphatase activity.261 To minimize background activity, it is preferable to have the gene encoding the cytosolic phosphatase (PHO13) additionally deleted (although this is not necessary when assaying certain substrates).
Cautionary notes: Measuring the degradation of long-lived proteins requires prior radiolabeling of the cells, and subsequent separation of acid-soluble from acid-insoluble radioactivity. The labeling can be done with relative ease both in cultured cells and in live animals.3 In cells, it is also possible to measure the release of an unlabeled amino acid by chromatographic methods, thereby obviating the need for prelabeling;678 however, it is important to keep in mind that amino acid release is also regulated by protein synthesis, which in turn is modulated by many different factors. In either case, one potential problem is that the released amino acid may be further metabolized. For example, branched chain amino acids are good indicators of proteolysis in hepatocytes, but not in muscle cells where they are further oxidized (A.J. Meijer, personal communication). In addition, the amino acid can be reincorporated into protein; for this reason, such experiments can be carried out in the presence of cycloheximide, but this raises additional concerns (see Turnover of autophagic compartments). In the case of labeled amino acids, a nonlabeled chase is added where the tracer amino acid is present in excess (being cautious to avoid using an amino acid that inhibits autophagy), or by use of single pass perfused organs or superfused cells.679,680 The perfused organ system also allows for testing the reversibility of effects on proteolysis and the use of autophagy-specific inhibitors in the same experimental preparation, which are crucial controls for proper assessment.
If the autophagic protein degradation is low (as it will be in cells in replete medium), it may be difficult to measure it reliably above the relatively high background of nonautophagic degradation. It should also be noted that the usual practice of incubating the cells under “degradation conditions,” that is, in a saline buffer, indicates the potential autophagic capacity (maximal attainable activity) of the cells rather than the autophagic activity that prevails in vivo or under rich culture conditions. Finally, inhibition of a particular degradative pathway is typically accompanied by an increase in a separate pathway as the cell attempts to compensate for the loss of degradative capacity.229,666 This compensation might interfere with control measurements under conditions that attempt to inhibit macroautophagy; however, as the latter is the major degradative pathway, the contributions of other types of degradation over the course of this type of experiment are most often negligible. Another issue of concern, however, is that most pharmacological protease inhibitors have “off target” effects that complicate the interpretation of the data.
The Pho8Δ60 assay requires standard positive and negative controls (such as an atg1Δ strain), and care must be taken to ensure the efficiency of cell lysis. Glass beads lysis works well in general, provided that the agitation speed of the instrument is adequate. Instruments designed for liquid mixing with lower speeds should be avoided. We also recommend against holding individual sample tubes on a vortex, as it is difficult to maintain reproducibility; devices or attachments are available to allow multiple tubes to be agitated simultaneously. Finally, it is also important to realize that the deletion of PHO8 can affect yeast cell physiology, especially depending on the growth conditions, and this may in turn have consequences for the cell wall; cells under starvation stress generate thicker cell walls that can be difficult to degrade enzymatically.
Conclusion: Measuring the turnover of long-lived proteins is a standard method for determining autophagic flux. Newer proteomic techniques that compare protein levels in autophagy-deficient animals relative to wild-type animals are promising,681 but the current ratiometric methods are affected by both protein synthesis and degradation, and thus analyze protein turnover, not just degradation.
10. Selective types of autophagy
Although autophagy can be nonselective, in particular during starvation, there are many examples of selective types of autophagy.
a. The Cvt pathway, mitophagy, pexophagy, piecemeal microautophagy of the nucleus and late nucleophagy in yeast and filamentous fungi
The precursor form of aminopeptidase I (prApe1) is the major cargo of the Cvt pathway in yeast, a biosynthetic autophagy-related pathway.128 The propeptide of prApe1 is proteolytically cleaved upon vacuolar delivery, and the resulting shift in molecular mass can be monitored by western blot. Under starvation conditions, prApe1 can enter the vacuole through nonselective autophagy, and thus has been used as a marker for both the Cvt pathway and autophagy. The yeast Cvt pathway is unique in that it is a biosynthetic route that utilizes the autophagy-related protein machinery, whereas other types of selective autophagy are degradative. The latter include pexophagy, mitophagy, reticulophagy, ribophagy and xenophagy, and each process has its own marker proteins, although these are typically variations of other assays used to monitor the Cvt pathway or autophagy. One common type of assay involves the processing of a GFP chimera similar to the GFP-Atg8/LC3 processing assay (see GFP-Atg8/LC3 lysosomal delivery and proteolysis). For example, yeast pexophagy utilizes the processing of Pex14-GFP and Pot1/Fox3/thiolase-GFP,682,683 whereas mitophagy can be monitored by the generation of free GFP from Om45-GFP, Idh1-GFP, Idp1-GFP or mito-DHFR-GFP.684,685-688 Localization of these mitochondrially targeted proteins (or specific MitoTracker dyes) or similar organelle markers such as those for the peroxisome (e.g., GFP-SKL with Ser-Lys-Leu at the C terminus that acts as a peroxisomal targeting signal, acyl-CoA oxidase 3 [Aox3-EYFP] that allows simultaneous observation of peroxisome-vacuole dynamics with the single FITC filter set, or GFP-catalase) can also be followed by fluorescence microscopy.553,683,689-691 In addition, yeast mitophagy requires both the Slt2 and Hog1 signaling pathways; the activation and phosphorylation of Slt2 and Hog1 can be monitored with commercially available phospho-specific antibodies (Fig. 20).508 It is also possible to monitor pexophagy in yeasts by the disappearance of activities of specific peroxisome markers such as catalase, alcohol oxidase or amine oxidase in cell-free extracts,692 or permeabilized cell suspensions. Catalase activity, however, is a useful marker only when peroxisomal catalases are the only such enzymes present or when activities of different catalases can be distinguished. In S. cerevisiae there are 2 genes, CTT1 and CTA1, encoding catalase activity, and only one of these gene products, Cta1, is localized in peroxisomes. Activities of both catalases can be distinguished using an in-gel activity assay after PAGE under nondenaturing conditions by staining with diaminobenzidine.693,694 Plate assays for monitoring the activity of peroxisomal oxidases in yeast colonies are also available.689,695 The decrease in the level of endogenous proteins such as alcohol oxidase, Pex14 or Pot1 can be followed by western blotting,553,696-699 TEM,700 fluorescence microscopy 553,701,702 or laser confocal scanning microscopy of GFP-labeled peroxisomes.703,704
Figure 20.
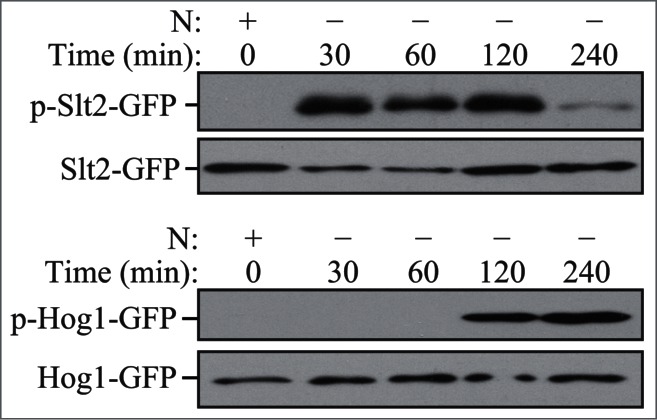
S. cerevisae cells were cultured to mid-log phase and shifted to SD-N for the indicated times. Samples were taken before (+) and at the indicated times after (–) nitrogen starvation. Immunoblotting was done with anti-phospho-Slt2 and anti-phospho-Hog1 antibody. This figure was modified from data previously published in ref. 508, and is reproduced by permission of the American Society for Cell Biology, copyright 2011.
Bimolecular fluorescence complementation (BiFC) may be useful to study protein-protein interactions in the autophagic pathway.705-707 In this assay, a protein of interest is cloned into a vector containing one half of a fluorescent reporter (e.g., YFP), while a second protein is cloned into a different vector containing the other half of the reporter. Constructs are cotransfected into cells. If the 2 proteins of interest interact, the 2 halves of the reporter are brought into close proximity and a fluorescent signal is reconstituted, which can be monitored by confocal microscopy. This assay can be used to determine protein interactions without prior knowledge of the location or structural nature of the interaction interface. Moreover, it is applicable to living cells, and relatively low concentrations of recombinant protein are required to generate a detectable signal.
In yeast, nonselective autophagy can be induced by nitrogen starvation conditions, whereas degradative types of selective autophagy generally require a carbon source change or ER stress for efficient induction. For example, in S. cerevisiae, to induce a substantial level of mitophagy, cells need to be precultured in a nonfermentable carbon source such as lactate or glycerol to stimulate the proliferation of mitochondria (although this is not the case in Pichia pastoris). After sufficient mitochondria proliferation, shifting the cells back to a fermentable carbon source such as glucose will cause the autophagic degradation of superfluous mitochondria.685 It should be noted that in addition to carbon source change, simultaneous nitrogen starvation is also required for efficient mitophagy induction. This is possibly because excessive mitochondria can be segregated into daughter cells by cell division if growth continues.685 A similar carbon source change from oleic acid or methanol to ethanol or glucose (with or without nitrogen starvation) can be used to assay for pexophagy.708 Mitophagy can also be induced by treatment with ROS, to induce mitochondria damage.709 In addition, mitophagy can be induced by culturing the cells in a nonfermentable carbon source to post-log phase. In this case, mitophagy may be induced because the energy demand is lower at post-log phase and the mitochondrial mass exceeds the cell's needs.120,710,711 It has been suggested that this type of mitophagy, also known as “stationary phase mitophagy,” reflects a quality-control function that culls defective mitochondria that accumulate in nondividing, respiring cells.712 The recently developed tool PMI that pharmacologically induces mitophagy without disrupting mitochondrial respiration713 should provide further insight as it circumvents the acute, chemically induced, blockade of mitochondrial respiration hitherto adopted to dissect the process. Similarly, pexophagy can be induced by culturing the cells in a peroxisome proliferation medium to post-log phase (J.-C. Farré, unpublished results). Along these lines, it should also be realized that selective types of autophagy continuously occur at a low level under noninducing conditions. Thus, organelles such as peroxisomes have a finite life span and are turned over at a slow rate by autophagy-related pathways.714
Piecemeal microautophagy of the nucleus (PMN, also micronucleophagy) is another selective autophagic subtype, which targets portions of the nucleus for degradation.715-717 In S. cerevisiae, the nuclear outer membrane, which is continuous with the nuclear ER, forms contact sites with the vacuolar membrane. These nucleus-vacuole junctions (NVJs) are generated by interaction of the outer nuclear membrane protein Nvj1 with the vacuolar protein Vac8.718 Nvj1 further recruits the ER-membrane protein Tsc13, which is involved in the synthesis of very-long-chain fatty acids (VLCFAs) and Swh1/Osh1, a member of a family of oxysterol-binding proteins. Upon starvation the NVJs bulge into the vacuole and subsequently a PMN-vesicle pinches off into the vacuole. PMN vesicles thus contain nuclear material and are limited by 3 membranes with the outermost derived from the vacuole, and the 2 inner ones from the nuclear ER. It is not clear which nuclear components are removed by PMN, but since PMN is not a cell death mechanism per se, most likely superfluous material is recycled. During PMN the NVJs are selectively incorporated into the PMN vesicles and degraded. Accordingly, PMN can be monitored using the proteins that are associated with the NVJs as markers. To quantitatively follow PMN, an assay analogous to the above-described GFP-Atg8/LC3 processing assay has been established using either GFP-Swh1/Osh1 or Nvj1-GFP. These GFP chimeras are, together with the PMN-vesicles, degraded in the vacuole. Thus, the formation of the relatively proteolysis-resistant GFP detected in western blots correlates with the PMN rate. In fluorescence microscopy, PMN can be visualized with the same constructs, and a chimera of mCherry fused to a nuclear localization signal (NLS-mCherry) can also be used. To assure that the measured PMN rate is indeed due to selective micronucleophagy, appropriate controls such as cells lacking Nvj1 or Vac8 should be included. Detailed protocols for the described assays are provided in ref. 719.
Late nucleophagy (LN) is another type of selective degradation of the nucleus, which specifically targets bulk nucleoplasm for degradation after prolonged periods (20–24 h) of nitrogen starvation.720 LN induction occurs in the absence of the essential PMN proteins Nvj1 and Vac8 and, therefore, the formation of NVJs. Although some components of the core Atg machinery are required for LN, Atg11 and the Vps34-containing PtdIns3K complex I are not needed. LN can be monitored by employing a nuclear-targeted version of the Rosella biosensor (n-Rosella) and following either its accumulation (by confocal microscopy), or degradation (by immunoblotting), within the vacuole.720 Dual labeling of cells with Nvj1-EYFP, a nuclear membrane reporter of PMN, and the nucleoplasm-targeted NAB35-DsRed.T3 (NAB35 is a target sequence for the Nab2 RNA-binding protein, and DsRed.T3 is the pH-stable, red fluorescent component of n-Rosella) allows detection of PMN soon after the commencement of nitrogen starvation, whereas delivery to the vacuole of the nucleoplasm reporter, indicative of LN, is observed only after prolonged periods of nitrogen starvation. Few cells show simultaneous accumulation of both reporters in the vacuole indicating PMN and LN are temporally and spatially separated.720
In contrast to unicellular yeasts, filamentous fungi form an interconnected mycelium of multinucleate hyphae containing up to 100 nuclei in a single hyphal compartment. A mycelial colony grows by tip extension with actively growing hyphae at the colony margin surrounded by an older, inner hyphal network that recycles nutrients to fuel the hyphal tips. By labeling organelle markers with GFP it is possible to show in Aspergillus oryzae that macroautophagy mediates degradation of basal hyphal organelles such as peroxisomes, mitochondria and entire nuclei.721 In contrast to yeast, PMN has not been observed in filamentous ascomycetes.723 In Magnaporthe oryzae, germination of the condiospore and formation of the appressorium are accompanied by nuclear degeneration in the spore.275 The degradation of nuclei in spores requires the nonselective autophagy machinery, whereas conserved components of the PMN pathway such as Vac8 and Tsc13 are dispensable for nuclear breakdown during plant infection.723 Nuclei are proposed to function in storage of growth-limiting nutrients such as phosphate and nitrogen.724,725 Similar to nuclei, mitochondria and peroxisomes are also preferentially degraded in the basal hyphae of filamentous ascomycetes.275,721,723-726
Cautionary notes: The Cvt pathway has been demonstrated to occur only in yeast. In addition, the sequestration of prApe1 is specific, even under starvation conditions, as it involves the recognition of the propeptide by a receptor, Atg19, which in turn interacts with the scaffold protein Atg11.727,728 Thus, unless the propeptide is removed or ATG19 is deleted, prApe1 is recognized as a selective substrate. Overexpression of prApe1 saturates import by the Cvt pathway, and the precursor form accumulates, but is rapidly matured upon autophagy induction.305 In addition, mutants such as vac8Δ and tlg2Δ accumulate prApe1 under rich conditions, but not during autophagy.505,729 Accordingly, it is possible to monitor the processing of prApe1 when overexpressed, or in certain mutant strains to follow autophagy induction. However, under the latter conditions it must be kept in mind that the sequestering vesicles are substantially smaller than typical autophagosomes generated during nonselective autophagy; the Cvt complex (prApe1 bound to Atg19) is smaller than typical peroxisomes or mitochondrial fragments that are subject to autophagic degradation. Accordingly, particular mutants may display complete maturation of prApe1 under autophagy-inducing conditions, but may still have a defect in other types of selective autophagy, as well as being unable to induce a normal level of nonselective autophagy.106 For this reason, it is good practice to evaluate autophagosome size and number by TEM. Actually, it is much simpler to monitor autophagic bodies (rather than autophagosomes) in yeast. First, the vacuole is easily identified, making the identification of autophagic bodies much simpler. Second, autophagic bodies can be accumulated within the vacuole, allowing for an increased sample size. It is best to use a strain background that is pep4Δ vps4Δ to prevent the breakdown of the autophagic bodies, and to eliminate confounding vesicles from the multivesicular body pathway. One caveat to the detection of autophagic bodies, however, is that they may coalesce in the vacuole lumen, making it difficult to obtain an accurate quantification. Finally, it is important to account for biases in sample sectioning to obtain an accurate estimate of autophagic body number or size.105
In general, when working with yeast it is preferable to use strains that have the marker proteins integrated into the chromosome rather than relying on plasmid-based expression, because plasmid numbers can vary from cell to cell. The GFP-Atg8, or similar, processing assay is easy to perform and is suitable for analysis by microscopy as well as western blotting; however, particular care is needed to obtain quantitative data for GFP-Atg8, Pex14-GFP or Om45-GFP, etc. processing assays (see cautionary notes for GFP-Atg8/LC3 lysosomal delivery and proteolysis). An alternative is an organelle-targeted Pho8Δ60 assay. For example, mitoPho8Δ60 can be used to quantitatively measure mitophagy.686 In addition, for the GFP-Atg8 processing assay, 2 h of starvation is generally sufficient to detect a significant level of free (i.e., vacuolar) GFP by western blotting as a measure of nonselective autophagy. For selective types of autophagy, the length of induction needed for a clearly detectable free GFP band will vary depending on the rate of cargo delivery/degradation. Usually 6 h of mitophagy induction is needed to be able to detect free GFP (e.g., from Om45-GFP) by western blot under starvation conditions, whereas stationary phase mitophagy typically requires 3 days before a free GFP band is observed. However, as with animal systems (see Animal mitophagy and pexophagy), it would be prudent to follow more than one GFP-tagged protein, as the kinetics, and even the occurrence of mitophagic trafficking, seems to be protein species-dependent, even within the mitochondrial matrix.730
Care should be taken when choosing antibodies to assess the degree of mitochondrial protein removal by autophagy; the quality and clarity of the result may vary depending on the specifics of the antibody. In testing the efficiency of mitophagy clearer results may be obtained by using antibodies against mtDNA-encoded proteins. This experimental precaution may prove critical to uncover subtle differences that could be missed when evaluating the process with antibodies against nuclear encoded, mitochondrially imported proteins (M. Campanella, personal communication).
b. Aggrephagy
Aggrephagy is the selective removal of aggregates by macroautophagy.731 This process can be followed in vitro (in cell culture) and in vivo (in mice) by monitoring the levels of an aggregate-prone protein such as an expanded polyglutamine (polyQ)-containing protein or mutant SNCA/α-synuclein (synuclein, alpha [non A4 component of amyloid precursor]). Levels are quantified by immunofluorescence, immunogold labeling or traditional immunoblot. In yeast, degradation of SNCA aggregates can be followed by promoter shut-off assays. Espression of the inducible GAL1 promoter of GFP-tagged SNCA is stopped by glucose repression. The removal of aggregates is thus monitored with fluorescence microscopy. The contribution of autophagy to SNCA aggregate clearance can be studied by the use of different autophagy mutants or by pharmacological treatment with the proteinase B inhibitor PMSF.732,733 Similarly, fluorescently tagged aggregated proteins such as polyQ80-CFP can be monitored via immunoblot and immunofluorescence. In addition to fluorescence methods, aggregates formed by a splice variant of CCND2 (cyclin D2) can also be monitored in electron-dense lysosomes and autophagosomes by immunogold labeling and TEM techniques.734 A polyQ80-luciferase reporter, which forms aggregates, can also be used to follow aggrephagy.735 A nonaggregating polyQ19-luciferase or untagged full-length luciferase serves as a control. The ratio of luciferase activity from these 2 constructs can be calculated to determine autophagic flux.
Autophagic degradation of endogenous aggregates such as lipofuscin can be monitored in some cell types by fluorescence microscopy, utilizing the autofluorescence of lipofuscin particles. Although under normal conditions almost 99% of the lipofuscin particles are located in the autophagosomes/lysosomes, an impairment of macroautophagy leads to free lipofuscin in the cytosol.736,737 The amount of lipofuscin in primary human adipocytes can be reduced by activation of macroautophagy, and the amount of lipofuscin is dramatically reduced in adipocytes from patients with type 2 diabetes and chronically enhanced macroautophagy.294
Cautionary notes: Caution must be used when performing immunoblots of aggregated proteins, as many protein aggregates fail to enter the resolving gel and are retained in the stacking gel. In addition, the polyQ80-luciferase in the aggregated state lacks luciferase activity whereas soluble polyQ80-luciferase retains activity. Therefore, caution must be used when interpreting results with these vectors, as treatments that increase aggrephagy or enhance protein aggregation can lead to a decrease in luciferase activity.738 Finally, soluble polyQ reporters can be degraded by the proteasome; thus, changes in the ratio of polyQ19-luciferase:polyQ80-luciferase may also reflect proteasomal effects and not just changes in autophagic flux.
c. Allophagy
In C. elegans, mitochondria, and hence mitochondrial DNA, from sperm are eliminated by an autophagic process. This process of allogeneic (nonself) organelle autophagy is termed “allophagy.”739,740 During allophagy in C. elegans, both paternal mitochondria and membranous organelles (a sperm-specific membrane compartment) are eliminated by the 16-cell stage (100–120 min post-fertilization).741,742 The degradation process can be monitored in living embryos with GFP::ubiquitin, which appears in the vicinity of the sperm chromatin (labeled for example with mCherry-histone H2B) on the membranous organelles within 3 min after fertilization. GFP fusions and antibodies specific for LGG-1 and LGG-2 (Atg8/LC3 homologs), which appear next to the sperm DNA, membranous organelles and mitochondria (labeled with CMXRos or mitochondria-targeted GFP) within 15 to 30 min post-fertilization, can be used to verify the autophagic nature of the degradation. TEM can also be utilized to demonstrate the presence of mitochondria within autophagosomes in the early embryo.
Conclusion: There are many assays that can be used to monitor selective types of autophagy, but caution must be used in choosing an appropriate marker(s). The potential role of other degradative pathways for any individual organelle or cargo marker should be considered, and it is advisable to use more than one marker or technique.
d. Animal mitophagy and pexophagy
There is no consensus at the present time with regard to the best method for monitoring mitophagy in animals. As with any organelle-specific form of autophagy, it is necessary to demonstrate: i) increased levels of autophagosomes containing mitochondria, ii) maturation of these autophagosomes that culminates with mitochondrial degradation, which can be blocked by specific inhibitors of autophagy or of lysosomal degradation, and iii) whether the changes are due to selective mitophagy or increased mitochondrial degradation during nonselective autophagy. Techniques to address each of these points have been reviewed.42,743
Antibodies against phosphorylated ubiquitin (p-S65-Ub) have very recently been described as novel tools to detect the activation of PINK1-PARK2-mediated mitophagy.744 p-S65-Ub is formed by the kinase PINK1 specifically upon mitochondrial stress, and is amplified in the presence of the E3 Ub ligase PARK2 (reviewed in ref. 745).746 p-S65-Ub antibodies have been used to demonstrate stress-induced activation of PINK1 in various cells including primary human fibroblasts (Fig. 21). Phosphorylated poly-ubiquitin chains specifically accumulate on damaged mitochondria, and staining with p-S65-Ub antibodies can be used, in addition to translocation of PARK2, to monitor the initiation of mitophagy. Given the complete conservation of the epitopes across species, mitochondrial p-S65-Ub could also be detected in mouse primary neurons upon mitochondrial depolarization. Furthermore, the p-S65-Ub signal partially colocalizes with mitochondrial, lysosomal, and total ubiquitin markers in cytoplasmic granules that appear to increase with age and disease in human postmortem brain samples.744 Along with the excellent performance of p-S65-Ub antibodies in a range of applications, these findings highlight the potential for future biomarker development.
Figure 21.
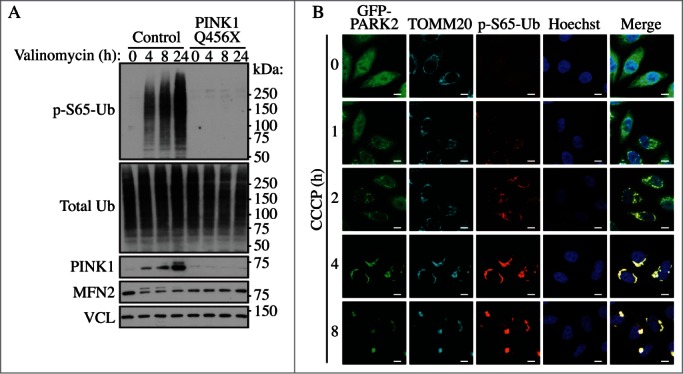
PINK1-dependent phosphorylation of ubiquitin (p-S65-Ub) upon mitophagic stress. (A) Human dermal fibroblasts from healthy controls or Parkinson disease patients carrying a PINK1 loss-of-function mutation (Q456X) were treated with valinomycin for the indicated times and lysates were analyzed by western blot. The p-S65-Ub signal is almost undetectable under nonstress conditions in controls, but is strongly induced in a PINK1 kinase-dependent manner during its stabilization on the outer mitochondrial membrane. MFN2 serves as a control substrate and VCL (vinculin) as a loading control. (B) HeLa cells stably expressing GFP-PARK2 (wild type) were treated with CCCP for the indicated times, fixed and stained with p-S65-Ub (red) and GFP-PARK2 (green) as well as mitochondrial (TOMM20, cyan) and nuclear (Hoechst, blue) markers. The p-S65-Ub staining is almost undetectable in nonstressed cells, but rapidly accumulates on damaged mitochondria where it functions to activate PARK2. On mitochondria, PINK1 and PARK2 together amplify the p-S65-Ub signal. Scale bar: 10 µm. Image provided by F.C. Fiesel and W. Springer.
Ultrastructural analysis at early time points can be used to establish selective mitophagy, although a maturation inhibitor may be needed to trap early autophagosomes with recognizable cargo (Fig. 22). Depending on the use of specific imaging techniques, dyes for living cells or antibodies for fixed cells have to be chosen. In any case, transfection of the phagophore and autophagosome marker GFP-LC3 to monitor the initiation of mitophagy, or RFP-LC3 to assess mitophagy progression, and visualization of mitochondria (independent of their mitochondrial membrane potential) makes it possible to determine the association of these 2 cellular components. Qualitatively, this may appear as fluorescence colocalization or as rings of GFP-LC3 surrounding mitochondria in higher resolution images.747,748 For live cell imaging microscopy, mitochondria should be labeled by a matrix-targeted fluorescent protein transfection or by mitochondria-specific dies. When using matrix-targeted fluorophores for certain cell lines (e.g., SH-SY5Y), it is important to allow at least 48 h of transient expression for sufficient targeting/import of mitochondrial GFP/RFP prior to analyzing mitophagy. MitoTracker probes are lipophilic cations that include a chloromethyl group and a fluorescent moiety. They concentrate in mitochondria due to their negative charge and react with the reduced thiols present in mitochondrial matrix proteins.749-751 After this reaction the probe can be fixed and remains in the mitochondria independent of altered mitochondrial function or mitochondrial membrane potential.750,752,753 This method can thus be used when cells remain healthy as the dye will remain in the mitochondria and is retained after fixation, although, as stated above, accumulation is dependent on the membrane potential. In addition, some of the MitoTracker probes, including MitoTracker Green FM and MitoTracker Red FM, are not well retained after fixation. Antibodies that specifically recognize mitochondrial proteins such as VDAC, TOMM20 or COX4I1 (cytochrome c oxidase subunit IV isoform I) may be used to visualize mitochondria in immunohistochemical experimental procedures.754,755 In neuronal cells, stabilized PINK1 on the mitochondrial outer membrane that accumulates in response to certain forms of acute mitochondrial damage is also a useful marker because it differentiates between healthy mitochondria and those that have lost their membrane potential. Redistribution of cardiolipin to the outer mitochondrial membrane acts as an elimination signal for mitophagy in mammalian cells, including primary neurons, and an ANXA5 (annexin A5) binding assay for externalized cardiolipin can also be considered a good marker for damaged mitochondria and early mitophagy.145 Colocalization analyses of mitochondria and autophagosomes provide an indication of the degree of autophagic sequestration. TEM can be used to demonstrate the presence of mitochondria within autophagosomes (referred to as mitophagosomes during mitophagy), and this can be coupled with bafilomycin A1 treatment to prevent fusion with the lysosome.42 To quantify early mitophagy, the percentage of LC3 puncta (endogenous, RFP- or GFP-LC3 puncta) that colocalize with mitochondria and the number of colocalizing LC3 puncta per cell—as assessed by either confocal microscopy or high-throughput imaging—in response to mitophagic stimuli can be employed as well.756 In addition, the percentage of lysosomes that colocalize with mitochondria can be used to quantify macroautophagy-mediated delivery of mitochondria. Overall, it is important to quantify mitophagy at various stages (initiation, progression, and late mitophagy) to identify stimuli that elicit this process.757,758
Figure 22.
Autophagosomes with recognizable cargo are rare in cells. (A) To assess relative rates of autophagosome formation, the fusion inhibitor bafilomycin A1 (10 nM) was applied for 2 h prior to fixation with 2% glutaraldehyde in order to trap newly formed autophagosomes. Two different PINK1 shRNA lines (A14 and D14) exhibit increased AV formation over 2 h compared to the control shRNA line. *, p < 0.05 vs. Control. (B) Autophagosomes in bafilomycin A1-treated control cells contain a variety of cytoplasmic structures (left, arrow), while mitochondria comprise a prominent component of autophagosomes in bafilomycin A1-treated (PINK1 shRNA) cells (right, arrow). Scale bar: 500 nm. These data indicate induction of selective mitophagy in PINK1-deficient cells. This figure was modified from Figure 2 published in ref. 1951, Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Human Molecular Genetics 2010; 19:R28-R37.
The fusion process of mitophagosomes with hydrolase-containing lysosomes represents the next step in the degradation process. To monitor the amount of fused organelles via live cell imaging microscopy, MitoTracker® Green FM and LysoTracker® Red DND-99 may be used to visualize the fusion process (Fig. 23). Independent of the cell-type specific concentration used for both dyes, we recommend exchanging MitoTracker® Green FM with normal medium (preferably phenol-free and CO2 independent to reduce unwanted autofluorescence) after incubation with the dye, whereas it is best to maintain the LysoTracker® Red stain in the incubation medium during the acquisition of images. Given that these fluorescent dyes are extremely sensitive to photobleaching, it is critical to perform live cell mitophagy experiments via confocal microscopy, preferably by using a spinning disc confocal microscope for long-term imaging experiments. For immunocytochemical experiments, antibodies specific for mitochondrial proteins and an antibody against LAMP1 (lysosomal-associated membrane protein 1) can be used. Overlapping signals appear as a merged color and can be used as indicators for successful fusion of autophagosomes that contain mitochondria with lysosomal structures.759 To measure the correlation between 2 variables by imaging techniques, such as the colocalization of 2 different stainings, we recommend some form of correlation analysis to assess the value correlating with the strength of the association. This may use, for example, ImageJ software or other colocalization scores that can be derived from consideration not only of pixel colocalization, but also from a determination that the structures have the appropriate shape. During live-cell imaging, the 2 structures (autophagosomes and mitochondria) should move together in more than one frame. Mitophagy can also be quantitatively monitored using a mitochondria-targeted version of the pH-dependent Keima protein.760 The peak of the excitation spectrum of the protein shifts from 440 nm to 586 nm when mitochondria are delivered to acidic lysosomes, which allows easy quantification of mitophagy (Fig. 24). However, it should be noted that long exposure time of the specimen to intense laser light lead to a similar spectral change. Finally, a mitochondrially-targeted version of the tandem mCherry-GFP fluorescent reporter (see Tandem mRFP/mCherry-GFP fluorescence microscopy) using a targeting sequence from the mitochondrial membrane protein FIS1346,347 can be used to monitor mitophagic flux.347
Figure 23.
Human fibroblasts showing colocalization of mitochondria with lysosomes. The degree of colocalization of mitochondria with lysosomes in human fibroblasts was measured via live cell imaging microscopy at 37°C and 5% CO2 atmosphere using the ApoTome® technique. LysoTracker® Red DND-99 staining was applied to mark lysosomal structures (red), and MitoTracker® Green FM to visualize mitochondria (green). Hoechst 33342 dye was used to stain nuclei (blue). A positive colocalization is indicated by yellow signals (merge) due to the overlap of LysoTracker® Red and MitoTracker® Green staining (white arrows). Scale bars: 10 µm. Statistical evaluation is performed by calculating the Pearson's coefficient for colocalizing pixels. Image provided by L. Burbulla and R. Krüger.
Figure 24.
Detection of mitophagy in primary cortical neurons using mitochondria-targeted Keima. Neurons transfected with mito-Keima were visualized using 458-nm (green, mitochondria at neutral pH) and 561-nm (red, mitochondria in acidic pH) laser lines and 575-nm band pass filter. Compared with the control (A) wild-type PINK1 overexpression (B) increases the number of the mitochondria exposed to acidic conditions. Scale bar: 2 µm. (C) Quantification of red dots suggests increased mitophagy in wild-type PINK1 but not in the kinase dead (kd) PINK1K219M-overexpressing neurons. Image provided by V. Choubey and A. Kaasik.
The third and last step of the degradation process is the monitoring of the amount of remaining mitochondria by analyzing the mitochondrial mass. This final step provides the opportunity to determine the efficiency of degradation of dysfunctional, aged or impaired mitochondria. Mitochondrial mass can be measured by a flow cytometry technique using MitoTracker® Green FM or MitoTracker Deep Red FM,750 on a single cell basis, by either live cell imaging or immunocytochemistry (using antibodies specifically raised against different mitochondrial proteins). Alternatively, mitochondrial content in response to mitophagic stimuli (in the presence and absence of autophagy inhibitors to assess the contribution of mitophagy) in live or fixed cells can be quantified at the single-cell level as the percentage of cytosol occupied by mitochondrial-specific fluorescent pixels using NIH ImageJ.758 Immunoblot analysis of the levels of mitochondrial proteins from different mitochondrial subcompartments is valuable for validating the data from flow cytometry or microscopy studies, and it should be noted that outer mitochondrial membrane proteins in particular can be degraded by the proteasome, especially in the context of mitochondrial depolarization.761,762 EM can also be used to verify loss of entire mitochondria, and PCR (or fluorescence microscopy) to quantify mitochondrial DNA (mtDNA). A reliable estimation of mtDNA can be performed by real-time PCR of the MT-ND2 (mitochondrially encoded NADH dehydrogenase 2) gene expressed as a ratio of mtDNA:nuclear DNA by normalizing to that of TERT (telomerase reverse transcriptase) genomic DNA.763 The spectrophotometric measurement of the activity of CS (citrate synthase), a mitochondrial matrix enzyme of the TCA cycle, which remains highly constant in these organelles and is considered a reliable marker of their intracellular content, can also be used to estimate the mitochondrial mass.763
In addition to monitoring the steady state levels of different steps of mitophagy—whether by single-cell analyses of LC3 mitochondrial colocalization or by immunoblotting for mitochondrial markers—investigation of the mitophagic flux is needed to determine whether mitophagy is impaired or activated in response to stimuli, and at which steps. Therefore, appropriate treatment (pharmacological inhibition and/or siRNA-mediated knockdown of ATG genes) may be applied to prevent mitochondrial degradation at distinct steps of the process. A recent method using flow cytometry in combination with autophagy and mitophagy inhibitors has been developed to determine mitophagic flux using MitoTracker probes.750
Certain cellular models require stress conditions to measure the mitochondrial degradation capacity, as basal levels are too low to reliably assess organelle clearance. However, one exception has been identified in Drosophila where large numbers of mitochondria are cleared by mitophagy during developmentally triggered autophagy.764 Hence, in many cases, it may be useful to pretreat the cells with uncoupling agents, such as CCCP, that stimulate mitochondrial degradation and allow measurements of mitophagic activity; however, it should be kept in mind that, although helpful to stimulate mitochondrial degradation, this treatment is not physiological and promotes the rapid degradation of outer membrane-localized mitochondrial proteins. In part for this reason a milder mitophagy stimulus has been developed that relies on a combination of antimycin A and oligomycin, inhibitors of the electron transport chain and ATP synthase, respectively;765 this treatment is less toxic, and the resulting damage is time dependent. Another method to induce mitophagy is by expressing and activating a mitochondrially localized fluorescent protein photosensitizer such as Killer Red.766 The excitation of Killer Red results in an acute increase of superoxide, due to phototoxicity, that causes mitochondrial damage resulting in mitophagy.767 The advantage of using a genetically encoded photosensitizer is that it allows for both spatial and temporal control in inducing mitophagy. Finally, the forced targeting of AMBRA1 to the external mitochondrial membrane is sufficient to induce massive mitophagy.768
A new classification suggests that mitophagy can be divided into 3 types.769 Type 1 mitophagy, involves the formation of a phagophore, and typically also requires mitochondrial fission; the PtdIns3K containing BECN1 mediates this process. In contrast, type 2 mitophagy is independent of BECN1 and takes place when mitochondria have been damaged, resulting in depolarization; sequestration involves the coalescence of GFP-LC3 membranes around the mitochondria rather than through fission and engulfment within a phagophore. In type 3 mitophagy, mitochondrial fragments or vesicles from damaged organelles are sequestered through a microautophagy-like process that is independent of ATG5 and LC3, but requires PINK1 and PARK2.
Although the process of pexophagy is prominent and well described in yeast cells,696,770 relatively little work has been done in the area of selective mammalian peroxisome degradation by autophagy (for a review see ref. 771). Typically, peroxisomes are induced by treatment with hypolipidemic drugs such as clofibrate or dioctyl phthalate, which bind to a subfamily of nuclear receptors, referred to as peroxisome proliferator-activated receptors.772 Degradation of excess organelles is induced by drug withdrawal, although starvation without prior proliferation can also be used. EPAS1 activation in liver-specific vhl-/- and vhl-/- hif1a-/- mice reduces peroxisome abundance by pexophagy, whereas ER and mitochondrial protein levels are not affected.773 Pexophagy can also be induced by the expression of a nondegradable active EPAS1 variant.773 Induction of pexophagy in response to endogenous and exogenous reactive oxygen species (ROS) and reactive nitrogen species has been observed in mammalian cells. In this setting, pexophagy is induced via ROS/reactive nitrogen species-mediated activation of ATM,774,775 repression of MTORC1 and phosphorylation of PEX5 by ATM;776,777 ATM phosphorylation of PEX5 at S141 triggers PEX5 ubiquitination and binding of SQSTM1 to peroxisomes targeted for pexophagy.777 Loss of peroxisomes can be followed enzymatically or by immunoblot, monitoring enzymes such as ACOX/fatty acyl-CoA oxidase (note that this enzyme is sometimes abbreviated “AOX,” but should not be confused with the enzyme alcohol oxidase that is frequently used in assays for yeast pexophagy) or CAT/catalase, and also by EM, cytochemistry or immunocytochemistry.778-781 Finally, a HaloTag®-PTS1 marker that is targeted to peroxisomes has been used to fluorescently label the organelle.782 An alternative approach uses a peroxisome-specific tandem fluorochrome assay (RFP-EGFP localizing to peroxisomes by the C-terminal addition of the tripeptide SKL, or a peroxisomal membrane protein tagged with mCherry-mGFP), which has been used to demonstrate the involvement of ACBD5/ATG37, NBR1 and SQSTM1 in mammalian pexophagy.345,783
Cautionary notes: There are many assays that can be used to monitor specific types of autophagy, but caution must be used in choosing an appropriate marker(s). To follow mitophagy it is best to monitor more than one protein and to include an inner membrane or matrix component in the analysis. In particular, it is not sufficient to follow a single mitochondrial outer membrane protein because these can be degraded independently of mitophagy. Although the localization of PARK2 to mitochondria as monitored by fluorescence microscopy is associated with the early stages of protonophore uncoupler (CCCP)-driven mitochondria degradation,250 this by itself cannot be used as a marker for mitophagy, as these events can be dissociated.784 Moreover, mitophagy elicited in a number of disease models does not involve mitochondrial PARK2 translocation.145,347,785 Along these lines, recent studies implicate an essential role for TRAF2, an E3 ubiquitin ligase, as a mitophagy effector in concert with PARK2 in cardiac myocytes; whereby mitochondrial proteins accumulate differentially with deficiency of either, indicating nonredundant roles for these E3 ubiquitin ligases in mitophagy.786 This finding necessitates an integrated approach to assess mitophagy based on a broad evaluation of multiple mitochondrial effectors and proteins.
PARK2 translocates to damaged mitochondria and ubiquitinates a wide range of outer membrane proteins including VDAC1, MFN1/2 and TOMM20/TOM20.755,761,762,787 This results in the preferential degradation of mitochondrial outer membrane proteins by the proteasome, while inner membrane proteins and mitochondrial DNA788 remain intact. Monitoring loss of a single protein such as TOMM20 by western blot or fluorescence microscopy to follow mitophagy may thus be misleading, as noted above.787 MitoTracker dyes are widely used to stain mitochondria and, when colocalized with GFP-LC3, they can function as a marker for mitophagy. However, staining with MitoTracker dyes depends on mitochondrial membrane potential (although MitoTracker Green FM is less sensitive to loss of membrane potential), so that damaged, or sequestered nonfunctional mitochondria may not be stained. In vitro this can be avoided by labeling the cells with MitoTracker before the induction by the mitophagic stimuli.750,789 One additional point is that MitoTracker dyes might influence mitochondrial motility in axons (D. Ebrahimi-Fakhari, personal communication).
Although it is widely assumed that macroautophagy is the major mechanism for degradation of entire organelles, there are multiple mechanisms that may account for the disappearance of mitochondrial markers. These include proteasomal degradation of outer membrane proteins and/or proteins that fail to correctly translocate into the mitochondria, degradation due to proteases within the mitochondria, and reduced biosynthesis or import of mitochondrial proteins. PINK1 and PARK2 also participate in an ATG gene-independent pathway for lysosomal degradation of small mitochondria-derived vesicles.790 Furthermore, the PINK1-PARK2 mitophagy pathway is also transcriptionally upregulated in response to starvation-triggered generalized autophagy, and is intertwined with the lipogenesis pathway.791-794 In addition to mitophagy, mitochondria can be eliminated by extrusion from the cell (mitoptosis).795,759,755,742 Transcellular degradation of mitochondria, or transmitophagy, also occurs in the nervous system when astrocytes degrade axon-derived mitochondria.796 Thus, it is advisable to use a variety of complementary methods to monitor mitochondria loss including TEM, single cell analysis of LC3 fluorescent puncta that colocalize with mitochondria, and western blot, in conjunction with flux inhibitors and specific inhibitors of autophagy induction compared with inhibitors of the other major degradation systems (see cautions in Autophagy inhibitors and inducers). To monitor and/or rule out changes in cellular capacity to undergo mitochondrial biogenesis, a process that is tightly coordinated with mitophagy and can dictate the outcome following mitophagy-inducing insults especially in primary neurons and other mitochondria-dependent cells, colocalization analysis after double staining for the mitochondrial marker TOMM20 and BrdU (for visualization of newly synthesized mtDNA) can be performed (Fig. 25).
Figure 25.
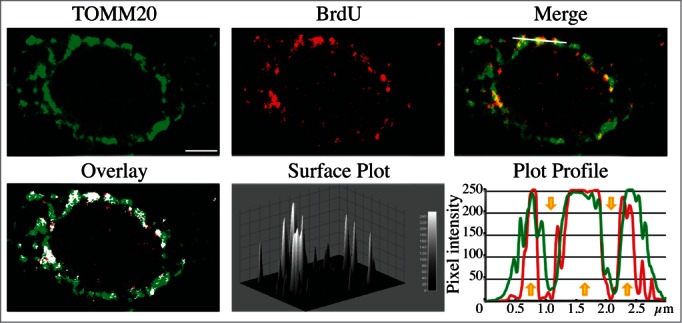
Confocal microscopy deconvolved (AutoQuant X3) images and colocalization image analysis (ImageJ 1.47; Imaris 7.6) through a local approach showing perinuclear mitochondrial biogenesis in hippocampal neuronal cultures. The upper channels show TOMM20 (green channel), BrdU (for visualization of newly synthesized mitochondrial DNA, red channel), and merged fluorescence channels. Overlay, corresponds to the spatial pattern of software thresholded colocalized structures (white spots) layered on the merged fluorescence channels. Surface Plot, or luminance intensity height, is proportional to the colocalization strength of the colocalized structures (white spots). Plot Profile, corresponds to the spatial intensity profiles of the fluorescence channels of the white line positioned in the Merge image. Yellow arrows indicate a qualitative evaluation of the spatial association trends for the fluorescence intensities. Arrows pointing up indicate an increase in the colocalization, while arrows pointing down show a decrease. Scale bar: 2 μm. This figure was modified from previously published data2187 and provided by F. Florenzano.
Likewise, although the mechanism(s) of peroxisomal protein degradation in mammals awaits further elucidation, it can occur by both autophagic and proteasome-dependent mechanisms.797 Thus, controls are needed to determine the extent of degradation that is due to the proteasome. Moreover, 2 additional degradation mechanisms have been suggested: the action of the peroxisome-specific LONP2/Lon (lon peptidase 2, peroxisomal) protease and the membrane disruption effect of 15-lipoxygenase.798
e. Chlorophagy
Besides functioning as the primary energy suppliers for plants, chloroplasts represent a major source of fixed carbon and nitrogen to be remobilized from senescing leaves to storage organs and newly developing tissues. As such, the turnover of these organelles has long been considered to occur via an autophagy-type mechanism. However, while the detection of chloroplasts within autophagic body-like vesicles or within vacuole-like compartments has been observed for decades, only recently has a direct connection between chloroplast turnover and autophagy been made through the analysis of atg mutants combined with the use of fluorescent ATG8 reporters.799,800 In fact, it is now clear that chlorophagy, the selective degradation of chloroplasts by macroautophagy, can occur via several routes, including the encapsulation of whole chloroplasts, or the budding of chloroplast material into small distinct autophagic vesicles called Rubisco-containing bodies (RCBs) and ATI1 plastid-associated bodies (ATI-PS), which then transport chloroplast cargo to the vacuole.799,801 Chloroplasts produce long tubes called stromules that project out from the organelle outer membrane. Recent studies suggest that stromules are part of the chlorophagy process, by which the stromule tips presumably containing unwanted or damaged chloroplast material are engulfed by autophagic membranes using ESCRTII endocytic machinery that depends on ATG8.802 The appearance of RCBs is tightly linked with leaf carbon status, indicating that chlorophagy through RCBs represents an important route for recycling plant nutrients provided in plastid stores.
f. Chromatophagy
Autophagy has been known for its pro-survival role in cells under metabolic stress and other conditions. However, excessively induced autophagy may be cytotoxic and may lead to cell death. Chromatophagy (chromatin-specific autophagy) comes into view as one of the autophagic responses that can contribute to cell death.803 Chromatophagy can be seen in cells during nutrient depletion, such as arginine starvation, and its phenotype consists of giant-autophagosome formation, nucleus membrane rupture and histone-associated-chromatin/DNA leakage that is captured by autophagosomes.803 Arginine starvation can be achieved by adding purified arginine deiminase to remove arginine from the culture medium, or by using arginine-dropout medium. The degradation of leaked nuclear DNA/chromatin can be observed by fluorescence microscopy; with GFP-LC3 or anti-LC3 antibody, and LysoTracker Red or anti-LAMP1, multiple giant autophagosomes or autolysosomes containing leaked nuclear DNA can be detected. In addition, the chromatophagy-related autophagosomes also contain parts of the nuclear outer-membrane, including NUP98 (nucleoporin 98kDa), indicating that the process involves a fusion event.803
g. Ferritinophagy
Ferritinophagy is a selective form of autophagy that functions in intracellular iron processing.804 Iron is recruited to ferritin for storage and to prevent the generation of free radical iron.805,806 To release iron from ferritin, the iron-bound form is sequestered within an autophagosome.807 Fusion with a lysosome leads to breakdown of ferritin and release of iron. Furthermore, iron can be acidified in the lysosome, converting it from an inactive state of Fe3+ to Fe2+.808,809 Iron can be detected in the autolysosome via TEM.808 Colocalization of iron with autolysosomes may also be determined utilizing calcein AM to tag iron.808,810 NCOA4 is a cargo receptor that recruits ferritin to the autophagosome.804
h. Intraplastidial autophagy
Intraplastidial autophagy is a process whereby plastids of some cell types adopt autophagic functions, engulfing and digesting portions of the cytoplasm. These plastids are characterized by formation of invaginations in their double-membrane envelopes that eventually generate a cytoplasmic compartment within the plastidial stroma, isolated from the outer cytoplasm. W. Nagl coined the term “plastolysome” to define this special plastid type.811 Initially, the engulfed cytoplasm is identical to the outer cytoplasm, containing ribosomes, vesicles and even larger organelles. Lytic activity was demonstrated in these plastids, in both the cytoplasmic compartment and the stroma. Therefore, it was suggested that plastolysomes digest themselves together with their cytoplasmic cargo, and transform into lytic vacuoles. Intraplastidial autophagy has been reported in plastids of suspensor cells of Phaseolus coccineus811 and Phaseolus vulgaris,812 where plastids transformed into autophagic vacuoles during the senescence of the suspensor. This process was also demonstrated in petal cells of Dendrobium813 and in Brassica napus microspores experimentally induced towards embryogenesis.814 All these reports established a clear link between these plastid transformations and their engagement in autophagy. At present, descriptions of this process are limited to a few, specialized plant cell types. However, pictures of cytoplasm-containing plastids in other plant cell types have been occasionally published, although the authors did not make any mention of this special plastid type. For example, this has been seen in pictures of fertile and Ogu-INRA male sterile tetrads of Brassica napus,815 and Phaseolus vulgaris root cells.816 Possibly, this process is not as rare as initially thought, but authors have only paid attention to it in those cell types where it is particularly frequent.
i. Lipophagy
The specific macroautophagic degradation of lipid droplets represents another type of selective autophagy.817 Lipophagy requires the core autophagic machinery and can be monitored by following triglyceride content, or total lipid levels using BODIPY 493/503 or HCS LipidTOX neutral lipid stains with fluorescence microscopy, cell staining with Oil Red O, the cholesterol dye filipin III,818 or ideally label-free techniques such as CARS or SRS microscopy. BODIPY 493/503 should be used with caution, however, when performing costains (especially in the green and red spectra) because this commonly used fluorescent marker of neutral lipids is highly susceptible to bleed-through into the other fluorescence channels (hence often yielding false positives), unlike the LipidTOX stain that has a narrow emission spectrum.819 In addition, BODIPY 493/503 cannot be used to monitor lipophagy in C. elegans because it stains both lipid droplets and the lysosome.820 TEM can also be used to monitor lipid droplet size and number, as well as lipid droplet-associated double-membrane structures, which correspond to autophagosomes.817,821,822 The transcription factor TFEB positively regulates lipophagy,624 and promotes fatty acid β-oxidation,823 thus providing a regulatory link between different lipid degradation pathways.824 Accordingly, TFEB overexpression rescues fat accumulation and metabolic syndrome in a diet-induced model of obesity.823,825 The regulation of expression of lipid droplet regulators (such as the PLIN/perilipin family) and of autophagy adaptors (such as the TBC1D1 family) during starvation and disease is one of several areas in this topic that deserves further exploration.826-828
Cautionary notes: With regard to changes in the cellular neutral lipid content, the presence and potential activation of cytoplasmic lipases that are unrelated to lysosomal degradation must be considered.
j. Lysophagy
Lysophagy is a selective macroautophagy process that participates in cellular quality control through lysosome turnover. By eliminating ruptured lysosomes, lysophagy prevents the subsequent activation of the inflammasome complex and innate immune response.829,830
k. Oxiapoptophagy
There are now several lines of evidence indicating that autophagy is an essential process in vascular functions. Autophagy can be considered as atheroprotective in the early stages of atherosclerosis and detrimental in advanced atherosclerotic plaques.831 Currently, little is known about the molecules that promote autophagy on the cells of the vascular wall. As increased levels of cholesterol oxidation products (also named oxysterols) are found in atherosclerotic lesions,832 the part taken by these molecules has been investigated, and several studies support the idea that some of them could contribute to the induction of autophagy.833,834 It is now suggested that oxysterols, especially 7-ketocholesterol, which can be increased under various stress conditions in numerous age-related diseases not only including vascular diseases but also neurodegenerative diseases,835 could trigger a particular type of autophagy termed oxiapoptophagy (OXIdation + APOPTOsis + autoPHAGY)836 characterized by the simultaneous induction of oxidative stress associated with apoptosis and autophagic criteria in different cell types from different species.837,838 As oxiapoptophagy has also been observed with 7β-hydroxycholesterol and 24S-hydroxycholesterol, which are potent inducers of cell death, it is suggested that oxiapoptophagy could characterize the effect of cytotoxic oxysterols.837
l. Reticulophagy
Starvation in yeast induces a type of selective macroautophagy of the ER, which depends on the autophagy receptors Atg39 and Atg40.839 ER stress also triggers an autophagic response,840 which includes the formation of multi-lamellar ER whorls and their degradation by a microautophagic mechanism.841 ER-selective autophagy has been termed ER-phagy or reticulophagy.842,843 Selective autophagy of the ER has also been observed in mammalian cells,844 and FAM134B has been identified as an ER-specific macroautophagy receptor that appears to be functionally homologous to Atg40.845 Since reticulophagy is selective, it should be able to act in ER quality control,846 sequester parts of the ER that are damaged, and eliminate protein aggregates that cannot be removed in other ways. It may also serve to limit stress-induced ER expansion,841 for example by reducing the ER to a normal level after a particular stress condition has ended.
m. Ribophagy
Autophagy is also used for the selective removal of ribosomes, particularly upon nitrogen starvation.847 This process can be monitored by western blot, following the generation of free GFP from Rpl25-GFP or Rpl5-GFP,848 or the disappearance of ribosomal subunits such as Rps3. Vacuolar localization of Rpl25-GFP or Rpl5-GFP can also be seen by fluorescence microscopy. The Rkr1/Ltn1 ubiquitin ligase acts as an inhibitor of 60S ribosomal subunit ribophagy via, at least, Rpl25 as a target, and is antagonized by the deubiquitinase Ubp3-Bre5 complex.847,848 Rkr1/Ltn1 and Ubp3-Bre5 likely contribute to adapt ribophagy activity to both nutrient supply and protein translation.
n. RNA-silencing components
Several components of the RNA-silencing machinery are selectively degraded by autophagy in different organisms. This was first shown for the plant AGO1/ARGONAUTE1 protein, a key component of the Arabidopsis RNA-induced silencing complex (RISC) that, after ubiquitination by a virus encoded F-box protein, is targeted to the vacuole.849 AGO1 colocalizes with Arabidopsis ATG8a-positive bodies and its degradation is impaired by various drugs such as 3-MA and E64d, or in Arabidopsis mutants in which autophagy is compromised such as the TOR-overexpressing mutant line G548 or the atg7-2 mutant allele (P. Genschik, unpublished data). Moreover, this pathway also degrades AGO1 in a nonviral context, especially when the production of miRNAs is impaired. In mammalian cells, not only the main miRNA effector AGO2, but also the miRNA-processing enzyme DICER1, is degraded as a miRNA-free entity by selective autophagy.850 Chemical inhibitors of autophagy (bafilomycin A1 and chloroquine) and, in HeLa cells, depletion of key autophagy components ATG5, ATG6 or ATG7 using short interfering RNAs, blocks the degradation of both proteins. Electron microscopy shows that DICER1 is associated with membrane-bound structures having the hallmarks of autophagosomes. Moreover, the selectivity of DICER1 and AGO2 degradation might depend on the autophagy receptor CALCOCO2/NDP52, at least in these cell types. Finally, in C. elegans, AIN-1, a homolog of mammalian TNRC6A/GW182 that interacts with AGO and mediates silencing, is also degraded by autophagy.851 AIN-1 colocalizes with SQST-1 that acts as a receptor for autophagic degradation of ubiquitinated protein aggregates and also directly interacts with LGG-1 (Atg8/LC3) contributing to cargo specificity.
o. Vacuole import and degradation pathway
In yeast, gluconeogenic enzymes such as fructose-1,6-bisphosphatase (Fbp1/FBPase), malate dehydrogenase (Mdh2), isocitrate lyase (Icl1) and phosphoenolpyruvate carboxykinase (Pck1) constitute the cargo of the vacuole import and degradation (Vid) pathway.852 These enzymes are induced when yeast cells are glucose starved (grown in a medium containing 0.5% glucose and potassium acetate). Upon replenishing these cells with fresh glucose (a medium containing 2% glucose), these enzymes are degraded in either the proteasome853-855 or the vacuole852,856 depending on the duration of starvation. Following glucose replenishment after 3 d glucose starvation, the gluconeogenic enzymes are delivered to the vacuole for degradation.857 These enzymes are sequestered in specialized 30- to 50-nm Vid vesicles.858 Vid vesicles can be purified by fractionation and gradient centrifugation; western blotting analysis using antibodies against organelle markers and Fbp1, and the subsequent verification of fractions by EM facilitate their identification.858 Furthermore, the amount of marker proteins in the cytosol compared to the Vid vesicles can be examined by differential centrifugation. In this case, yeast cells are lysed and subjected to differential centrifugation. The Vid vesicle-enriched pellet fraction and the cytosolic supernatant fraction are examined with antibodies against Vid24, Vid30, Sec28 and Fbp1.859-861
The distribution of Vid vesicles containing cargo destined for endosomes, and finally for the vacuole, can be examined using FM 4–64, a lipophilic dye that primarily stains endocytic compartments and the vacuole limiting membrane.862 In these experiments, starved yeast cells are replenished with fresh glucose and FM 4–64, and cells are collected at appropriate time points for examination by fluorescence microscopy.860 The site of degradation of the cargo in the vacuole can be determined by studying the distribution of Fbp1-GFP, or other Vid cargo markers in wild-type and pep4Δ cells.863 Cells can also be examined for the distribution of Fbp1 at the ultrastructural level by immuno-TEM.864
As actin patch polymerization is required for the delivery of cargo to the vacuole in the Vid pathway, distribution of Vid vesicles containing cargo and actin patches can be examined by actin staining (with phalloidin conjugated to rhodamine) using fluorescence microscopy.864 The distribution of GFP tagged protein and actin is examined by fluorescence microscopy. GFP-Vid24, Vid30-GFP and Sec28-GFP colocalize with actin during prolonged glucose starvation and for up to 30 min following glucose replenishment in wild-type cells; however, colocalization is less obvious by the 60-min time point.859,864
p. Xenophagy
The macroautophagy pathway has emerged as an important cellular factor in both innate and adaptive immunity. Many in vitro and in vivo studies have demonstrated that genes encoding macroautophagy components are required for host defense against infection by bacteria, parasites and viruses. Xenophagy is often used as a term to describe autophagy of microbial pathogens, mediating their capture and delivery to lysosomes for degradation. Since xenophagy presents an immune defense, it is not surprising that microbial pathogens have evolved strategies to overcome it. The interactions of such pathogens with the autophagy system of host cells are complex and have been the subject of several excellent reviews.121-126,865-871 Here we will make note of a few key considerations when studying interactions of microbial pathogens with the autophagy system. Importantly, autophagy should no longer be considered as strictly antibacterial, and several studies have described the fact that autophagy may serve to either restrict or promote bacterial replication both in vivo872 and in vitro (reviewed in refs. 874, 875).
LC3 is commonly used as a marker of macroautophagy. However, studies have established that LC3 can promote phagosome maturation independently of macroautophagy through LC3-associated phagocytosis (see cautionary notes in Atg8/LC3 detection and quantification, and Noncanonical use of autophagy-related proteins). Other studies show that macroautophagy of Salmonella enterica serovar Typhimurium (S. typhimurium) is dependent on ATG9, an essential macroautophagy protein, whereas LC3 recruitment to bacteria does not require ATG9.875 In contrast, macroautophagy of these bacteria requires either glycan-dependent binding of LGALS8/galectin-8 (lectin, galactoside-binding, soluble, 8) to damaged membranes and subsequent recruitment of the cargo receptor CALCOCO2/NDP52876 or ubiquitination of target proteins (not yet identified) and recruitment of 4 different ubiquitin-binding receptor proteins, SQSTM1,877 CALCOCO2/NDP52,878 TAX1BP1/CALCOCO3879 and OPTN.880 Therefore, the currently available criteria to differentiate LAP from macroautophagy include: i) LAP involves LC3 recruitment to bacteria in a manner that requires ROS production by an NADPH oxidase. It should be noted that most cells express at least one member of the NADPH oxidase family. Targeting expression of the common CYBA/p22phox subunit is an effective way to disrupt the NADPH oxidases. Scavenging of ROS by antioxidants such as resveratrol and alpha-tocopherol is also an effective way to inhibit LAP. In contrast, N-acetylcysteine, which raises cellular glutathione levels, does not inhibit LAP.881 ii) Macroautophagy of bacteria requires ATG9, whereas LAP apparently does not.875 iii) LAP involves single-membrane structures. For LAP, CLEM (with LC3 as a marker) is expected to show single-membrane structures that are LC3+ with LAP.182 In contrast, macroautophagy is expected to generate double-membrane structures surrounding cargo (which may include single membrane phagosomes, giving rise to triple-membrane structures875). It is anticipated that more specific markers of LAP will be identified as these phagosomes are further characterized.
Nonmotile Listeria monocytogenes can be targeted to double-membrane autophagosomes upon antibiotic treatment,882 which indicates that macroautophagy serves as a cellular defense to microbes in the cytosol. However, subsequent studies have revealed that macroautophagy can also target pathogens within phagosomes, damaged phagosomes or the cytosol. Therefore, when studying microbial interactions by EM, many structures can be visualized, with any number of membranes encompassing microbes, all of which may be LC3+.883,884 As discussed above, single-membrane structures that are LC3+ may arise through LAP, and we cannot rule out the possibility that both LAP and macroautophagy may operate at the same time to target the same phagosome. Indeed macroautophagy may facilitate phagocytosis and subsequent bacterial clearance (X. Li and M. Wu, submitted). Macroautophagy is not only induced by intracellular bacteria, but also can be activated by extracellular bacteria such as Pseudomonas aeruginosa and Klebsiella pneumoniae, which may involve complex mechanisms.885-887 Furthermore, macroautophagy can be induced by all intracellular and extracellular Gram-negative bacteria via a common mechanism involving naturally produced bacterial outer membrane vesicles;888,889 these vesicles enter human epithelial cells, resulting in autophagosome formation and inflammatory responses mediated via the host pathogen recognition receptor NOD1.888
Viruses can also be targeted by autophagy, and in turn can act to inhibit autophagy. For example, infection of a cell by influenza and dengue viruses890 or enforced expression of the hepatitis B virus C protein891 have profound consequences for autophagy, as viral proteins such as NS4A stimulate autophagy and protect the infected cell against apoptosis, thus extending the time in which the virus can replicate. Conversely, the HSV-1 ICP34.5 protein inhibits autophagy by targeting BECN1.892 While the impact of ICP34.5's targeting of BECN1 on viral replication in cultured permissive cells is minimal, it has a significant impact upon pathogenesis in vivo, most likely through interfering with activation of CD4+ T cells,893,894 and through cell-intrinsic antiviral effects in neurons.895 Also, viral BCL2 proteins, encoded by large DNA viruses, are able to inhibit autophagy by interacting with BECN1565 through their BH3 homology domain. An example of these include γ-herpesvirus68,896 Kaposi sarcoma-associated herpesvirus566 and African swine fever virus (ASFV) vBCL2 homologs.897 ASFV encodes a protein homologous to HSV-1 ICP34.5, which, similar to its herpesvirus counterpart, inhibits the ER stress response activating PPP1/protein phosphatase 1; however, in contrast to HSV-1 ICP34.5 it does not interact with BECN1. ASFV vBCL2 strongly inhibits both autophagy (reviewed in ref. 898) and apoptosis.899
HIV-1 utilizes the initial, nondegradative stages of autophagy to promote its replication in macrophages. In addition, the HIV-1 protein Nef acts as an anti-autophagic maturation factor protecting the virus from degradation by physically blocking BECN1.900-902 Autophagy contributes to limiting viral pathogenesis in HIV-1 nonprogressor-infected patients by targeting viral components for degradation.903
Care must be taken in determining the role of autophagy in viral replication, as some viruses such as vaccinia virus use double-membrane structures that form independently of the autophagy machinery.904 Similarly, dengue virus replication, which appears to involve a double-membrane compartment, requires the ER rather than autophagosomes,905 whereas coronaviruses and Japanese encephalitis virus use a nonlipidated version of LC3 (see Atg8/LC3 detection and quantification).190,191 Yet another type of variation is seen with hepatitis C virus, which requires BECN1, ATG4B, ATG5 and ATG12 for initiating replication, but does not require these proteins once an infection is established.906
Finally, it is important to realize that there may be other macroautophagy-like pathways that have yet to be characterized. For example, in response to cytotoxic stress (treatment with etoposide), autophagosomes are formed in an ATG5- and ATG7-independent manner (see Noncanonical use of autophagy-related proteins).27 While this does not rule out involvement of other macroautophagy regulators/components in the formation of these autophagosomes, it does establish that the canonical macroautophagy pathway involving LC3 conjugation is not involved. In contrast, RAB9 is required for this alternative pathway, potentially providing a useful marker for analysis of these structures. Returning to xenophagy, M. tuberculosis can be targeted to autophagosomes in an ATG5-independent manner.907 Furthermore, up to 25% of intracellular S. typhimurium are observed in multi-lamellar membrane structures resembling autophagosomes in atg5-/- MEFs.877 These findings indicate that an alternate macroautophagy pathway is relevant to host-pathogen interactions. Moreover, differences are observed that depend on the cell type being studied. Yersinia pseudotuberculosis is targeted to autophagosomes where they can replicate in bone marrow-derived macrophages,908 whereas in RAW264.7 and J774 cells, bacteria are targeted both to autophagosomes, and LC3-negative, single-membrane vacuoles (F. Lafont, personal communication).
One key consideration has recently emerged in studying xenophagy. Whereas the basal autophagic flux in most cells is essential for their survival, infecting pathogens can selectively modulate antibacterial autophagy (i.e., xenophagy) without influencing basal autophagy. This may help pathogens ensure prolonged cellular (i.e., host) survival. Thus, in the case of xenophagy it would be prudent to monitor substrate (pathogen)-specific autophagic flux to understand the true nature of the perturbation of infecting pathogens on autophagy (D. Kumar, personal communication). Furthermore, this consideration particularly limits the sensitivity of LC3 western blots for use in monitoring autophagy regulation.
q. Zymophagy
Zymophagy was originally defined as a specific mechanism that eliminates pancreatitis-activated zymogen granules in the pancreatic acinar cells and, thus, prevents deleterious effects of prematurely activated and intracellularly released proteolytic enzymes, when impairment of secretory function occurs.909 Therefore, zymophagy is primarily considered to be a protective mechanism implemented to sustain secretory homeostasis and to mitigate pancreatitis. The presence of zymogen granules, however, is not only attributed to pancreatic acinar cells. Thus, zymophagy was also reported in activated secretory Paneth cells of the crypts of Lieberkuhn in the small intestine.910 Note that one of the major functions of Paneth cells is to prevent translocation of intestinal bacteria by secreting hydrolytic enzymes and antibacterial peptides to the crypt lumens. The similarity in mechanisms of degradation of secretory granules in these 2 different types of secretory cells sustains the concept of the protective role of autophagy when “self-inflicted” damage may occur due to overreaction and/or secretory malfunction in specialized cells.
Zymophagy can be monitored by TEM, identifying autophagosomes containing secretory granules, by following SQSTM1 degradation by western blot, and by examining the subcellular localization of VMP1-EGFP, which relocates to granular areas of the cell upon zymophagy induction. Colocalization of PRSS1/trypsinogen (which is packaged within zymogen granules) and LC3, or of GFP-ubiquitin (which is recruited to the activated granules) with RFP-LC3 can also be observed by indirect or direct immunofluorescence microscopy, respectively. Active trypsin is also detectable in zymophagosomes and participates in the early onset of acute pancreatitis (F. Fortunato et al., unpublished data).
11. Autophagic sequestration assays
Although it is useful to employ autophagic markers such as LC3 in studies of autophagy, LC3-II levels or LC3 dots cannot quantify actual autophagic activity, since LC3-II is not involved in all cargo sequestration events, and LC3-II can be found on phagophores and nonautophagosomal membranes in addition to autophagosomes. Thus, quantification of autophagic markers such as LC3 does not tell how much cargo material has actually been sequestered inside autophagosomes. Moreover, LC3 and several other autophagic markers cannot be used to monitor noncanonical autophagy. Autophagic sequestration assays constitute marker-independent methods to measure the sequestration of autophagic cargo into autophagosomal compartments, and are among the few functional autophagy assays described to date. Macroautophagic cargo sequestration activity can be monitored using either an (electro)injected, inert cytosolic marker such as [3H]-raffinose911 or an endogenous cytosolic protein such as LDH/lactate dehydrogenase,912 in the latter case along with treatment with a protease inhibitor (e.g., leupeptin) or other inhibitors of lysosomal activity (e.g., bafilomycin A1)216 to prevent intralysosomal degradation of the protein marker. The assay simply measures the transfer of cargo from the soluble (cytosol) to the insoluble (sedimentable) cell fraction (which includes autophagic compartments), with no need for a sophisticated subcellular fractionation. Electrodisruption of the plasma membrane followed by centrifugation through a density cushion was originally used to separate cytosol from sedimentable cell fractions in primary hepatocytes.913 This method has also been used in various human cancer cell lines and mouse embryonic fibroblasts, where the LDH sequestration assay has been validated with pharmacological agents as well as genetic silencing or knockout of key factors of the autophagic machinery (N. Engedal, unpublished results).143,216,914 Moreover, a downscaling and simplification of the method that avoids the density cushion has been introduced.914 Homogenization and sonication techniques have also been successfully used for the LDH sequestration assay.658,915 The endogenous LDH cargo marker can be quantified by an enzymatic assay, or by western blotting. In principle, any intracellular component can be used as a cargo marker, but cytosolic enzymes having low sedimentable backgrounds are preferable. Membrane-associated markers are less suitable, and proteins such as LC3, which are part of the sequestering system itself, will have a much more complex relationship to the autophagic flux than a pure cargo marker such as LDH.
In yeast, sequestration assays are typically done by monitoring protease protection of an autophagosome marker or a cargo protein. For example, prApe1, and GFP-Atg8 have been used to follow completion of the autophagosome.916 The relative resistance or sensitivity to an exogenous protease in the absence of detergent is an indication of whether the autophagosome (or other sequestering vesicle) is complete or incomplete, respectively. Thus, this method also distinguishes between a block in autophagosome formation versus fusion with the vacuole. The critical issues to keep in mind involve the use of appropriate control strains and/or proteins, and deciding on the correct reporter protein. In addition to protease protection assays, sequestration can be monitored by fluorescence microscopy during pexophagy of methanol-induced peroxisomes, using GFP-Atg8 as a pexophagosome marker and BFP-SKL to label the peroxisomes. The vacuolar sequestration process during micropexophagy can also be monitored by formation of the vacuolar sequestering membrane stained with FM 4–64.689,697
Sequestration assays can be designed to measure flux through individual steps of the autophagy pathway. For example, intralysosomally degraded sequestration probes such as [14C]-lactate or LDH will mark prelysosomal compartments in the absence of degradation inhibitors. Hence, their accumulation in such compartments can be observed when fusion with lysosomes is suppressed, for example, by a microtubule inhibitor such as vinblastine.917 Furthermore, lactate hydrolysis can be used to monitor the overall autophagic pathway (autophagic lactolysis).918 One caveat, however, is that inhibitors may affect sequestration indirectly, for example, by modifying the uptake and metabolism (including protein synthesis) of autophagy-suppressive amino acids (see Autophagy inhibitors and inducers). Under some conditions, such as amino acid starvation, sequestered LDH en route through the autophagosome-lysosome pathway can also be detected in the absence of inhibitors.216
A variation of this approach applicable to mammalian cells includes live cell imaging. Autophagy induction is monitored as the movement of cargo, such as mitochondria, to GFP-LC3-colocalizing compartments, and then fusion/flux is measured by delivery of cargo to lysosomal compartments.331,919 In addition, sequestration of fluorescently tagged cytosolic proteins into membranous compartments can be measured, as fluorescent puncta become resistant to the detergent digitonin.920 Use of multiple time points and monitoring colocalization of a particular cargo with GFP-LC3 and lysosomes can also be used to assess sequestration of cargo with autophagosomes as well as delivery to lysosomes.758
In the Drosophila fat body, the localization of free cytosolic mCherry changes from a diffuse to a punctate pattern in an Atg gene-dependent manner, and these mCherry dots colocalize with the lysosomal marker Lamp1-GFP during starvation (G. Juhasz, unpublished data). Thus, the redistribution of free cytosolic mCherry may be used to follow bulk, nonselective autophagy due to its stability and accumulation in autolysosomes.
Cautionary notes: The electro-injection of radiolabeled probes is technically demanding, but the use of an endogenous cytosolic protein probe is very simple and requires no pretreatment of the cells other than with a protease inhibitor. Another concern with electro-injection is that it can affect cellular physiology, so it is necessary to verify that the cells behave properly under control situations such as amino acid deprivation. An alternate approach for incorporating exogenous proteins into mammalian cell cytosol is to use “scrape-loading,” a method that works for cells that are adherent to tissue culture plates.921 Finally, these assays work well with hepatocytes but may be problematic with other cell types, and it can be difficult to load the cell while retaining the integrity of the compartments in the post-nuclear supernatant (S. Tooze, unpublished results). General points of caution to be addressed with regard to live cell imaging relate to photobleaching of the fluorophore, cell injury due to repetitive imaging, autofluorescence in tissues containing lipofuscin, and the pH sensitivity of the fluorophore.
There are several issues to keep in mind when monitoring sequestration by the protease protection assay in yeast.916 First, as discussed in Selective types of autophagy, prApe1 is not an accurate marker for nonselective autophagy; import of prApe1 utilizes a receptor (Atg19) and a scaffold (Atg11) that make the process specific. In addition, vesicles that are substantially smaller than autophagosomes can effectively sequester the Cvt complex. Another problem is that prApe1 cannot be used as an autophagy reporter for mutants that are not defective in the Cvt pathway, although this can be bypassed by using a vac8Δ background.922 At present, the prApe1 assay cannot be used in any system other than yeast. The GFP-Atg8 protease protection assay avoids these problems, but the signal-to-noise ratio is typically substantially lower. In theory, it should be possible to use this assay in other cell types, and protease protection of GFP-LC3 and GFP-SQSTM1 has been analyzed in HeLa cells.923 Finally, tendencies of GFP-LC3 and particularly GFP-SQSTM1 to aggregate may make LC3 and SQSTM1 inaccessible to proteases.
Conclusion: Sequestration assays represent the most direct method for monitoring autophagy, and in particular for discriminating between conditions where the autophagosome is complete (but not fused with the lysosome/vacuole) or open (that is, a phagophore). These assays can also be modified to measure autophagic flux.
12. Turnover of autophagic compartments
Inhibitors of autophagic sequestration (e.g., amino acids, 3-MA or wortmannin) can be used to monitor the disappearance of autophagic elements (phagophores, autophagosomes, autolysosomes) to estimate their half-life by TEM morphometry/stereology. The turnover of the autophagosome or the autolysosome will be differentially affected if fusion or intralysosomal degradation is inhibited.12,14,25,924 The duration of such experiments is usually only a few hours; therefore, long-term side effects or declining effectiveness of the inhibitors can be avoided. It should be noted that fluorescence microscopy has also been used to monitor the half-life of autophagosomes, monitoring GFP-LC3 in the presence and absence of bafilomycin A1 or following GFP-LC3 after starvation and recovery in amino acid-rich medium (see Atg8/LC3 detection and quantification).16,925
Cautionary notes: The inhibitory effect must be strong and the efficiency of the inhibitor needs to be tested under the experimental conditions to be employed. Cycloheximide is sometimes used as an autophagy inhibitor, but its use in long-term experiments is problematic because of the many potential indirect effects. Cycloheximide inhibits translational elongation, and therefore protein synthesis. In addition, it decreases the efficiency of protein degradation in several cell types (A.M. Cuervo, personal communication) including hematopoietic cells (A. Edinger, personal communication). Treatment with cycloheximide causes a potent increase in MTORC1 activity, which can decrease autophagy in part as a result of the increase in the amino acid pool resulting from suppressed protein synthesis (H.-M. Shen, personal communication; I. Topisirovic, personal communication).926,927 In addition, at high concentrations (in the millimolar range) cycloheximide inhibits complex I of the mitochondrial respiratory chain,928,929 but this is not a problem, at least in hepatocytes, at low concentrations (10–20 µM) that are sufficient to prevent protein synthesis (A.J. Meijer, personal communication).
Conclusion: The turnover of autophagic compartments is a valid method for monitoring autophagic-lysosomal flux, but cycloheximide must be used with caution in long-term experiments.
13. Autophagosome-lysosome colocalization and dequenching assay
Another method to demonstrate the convergence of the autophagic pathway with a functional degradative compartment is to incubate cells with the bovine serum albumin derivative dequenched (DQ)-BSA that has been labeled with the red-fluorescent BODIPY TR-X dye; this conjugate will accumulate in lysosomes. The labeling of DQ-BSA is so extensive that the fluorophore is self-quenched. Proteolysis of this compound results in dequenching and the release of brightly fluorescent fragments. Thus, DQ-BSA is useful for detecting intracellular proteolytic activity as a measure of a functional lysosome.930
Furthermore, DQ-BSA labeling can be combined with GFP-LC3 to monitor colocalization, and thus visualize the convergence, of amphisomes with a functional degradative compartment (DQ-BSA is internalized by endocytosis). This method can also be used to visualize fusion events in real-time experiments by confocal microscopy (live cell imaging). Along similar lines, other approaches for monitoring convergence are to follow the colocalization of RFP-LC3 and LysoSensor Green (M. Bains and K.A. Heidenreich, personal communication), mCherry-LC3 and LysoSensor Blue,332 or tagged versions of LC3 and LAMP1 (K. Macleod, personal communication) or CD63331 as a measure of the fusion of autophagosomes with lysosomes. It is also possible to trace autophagic events by visualizing the pH-dependent excitation changes of the coral protein Keima.760 This quantitative technique is capable of monitoring the fusion of autophagosomes with lysosomes, that is, the formation of an autolysosome, and the assay does not depend on the analysis of LC3.
Cautionary notes: Some experiments require the use of inhibitors (e.g., 3-MA or wortmannin) or overexpression of proteins (e.g., RAB7 dominant negative mutants) that may also affect the endocytic pathway or the delivery of DQ-BSA to lysosomes (e.g., wortmannin causes the swelling of late endosomes931). In this case, the lysosomal compartment can be labeled with DQ-BSA overnight before treating the cells with the drugs, or prior to the transfection.
Conclusion: DQ-BSA provides a relatively convenient means for monitoring lysosomal protease function and can also be used to follow the fusion of amphisomes with the lysosome. Colocalization of autophagosomes (fluorescently tagged LC3) with lysosomal proteins or dyes can also be monitored.
14. Tissue fractionation
The study of autophagy in the organs of larger animals, in large numbers of organisms with very similar characteristics, or in tissue culture cells provides an opportunity to use tissue fractionation techniques as has been possible with autophagy in rat liver.35,54,932-937 Because of their sizes (smaller than nuclei but larger than membrane fragments [microsomes]), differential centrifugation can be used to obtain a subcellular fraction enriched in mitochondria and organelles of the autophagy-lysosomal system, which can then be subjected to density gradient centrifugation to enrich autophagosomes, amphisomes, autolysosomes and lysosomes.35,54,937-941 Any part of such a fraction can be considered to be a representative sample of tissue constituents and used in quantitative biochemical, centrifugational and morphological studies of autophagic particle populations.
The simplest studies of the autophagic process take advantage of sequestered marker enzymes, changes in location of these enzymes, differences in particle/compartment size and differential sensitivity of particles of different sizes to mechanical and osmotic stress (e.g., acid hydrolases are found primarily in membrane-bound compartments and their latent activities cannot be measured unless these membranes are lysed). Such a change in enzyme accessibility can be used to follow the time course of an exogenously induced, or naturally occurring, autophagic process.932,934,936
Quantitative localization of enzymatic activity (or any other marker) to specific cytoplasmic particle populations and changes in the location of such markers during autophagy can be assessed by using rate sedimentation ultracentrifugation.938 Similar results can be obtained with isopycnic centrifugation where particles enter a density gradient (sometimes made with sucrose but iso-osmotic media such as iodixanol, metrizamide and Nycodenz may be preferred as discussed below under Cautionary notes) and are centrifuged until they reach locations in the gradient where their densities are equal to those of the gradient.938
The fractionation of organelles can also be evaluated by protein-correlation-profiling, a quantitative mass spectrometry-based proteomics approach. Similar to the biochemical assays described above, gradient profiles of marker proteins can be recorded and compared to proteins of interest.362 Compared to classical biochemical approaches, protein-correlation-profiling allows the proteome-wide recording of protein gradient profiles.
Particle populations in subcellular fractions evaluated with quantitative biochemical and centrifugational approaches can also be studied with quantitative morphological methods. Detailed morphological study of the particle populations involved in the autophagic process usually requires the use of EM. The thin sections required for such studies pose major sampling problems in both intact cells942 and subcellular fractions.938 With the latter, 2,000,000 sections can be obtained from each 0.1 ml of pellet volume, so any practical sample size is an infinitesimally small subsample of the total sample.938 However, through homogenization and resuspension, complex and heterogeneous components of subcellular fractions become randomly distributed throughout the fraction volume. Therefore, any aliquot of that volume can be considered a random sample of the whole volume. What is necessary is to conserve this property of subcellular fractions in the generation of a specimen that can be examined with the electron microscope. This can be done with the use of a pressure filtration procedure.942,938 Because of the thinness of the sections, multiple sections of individual particles are possible so morphometric/stereological methods942 must be used to determine the volume occupied by a given class of particles, as well as the size distribution and average size of the particle class. From this information the number of particles in a specific particle class can be calculated.944 Examination of individual profiles gives information on the contents of different types of particles and their degree of degradation, as well as their enclosing membranes.932,934
Cautionary notes: When isolating organelles from tissues and cells in culture it is essential to use disruption methods that do not alter the membrane of lysosomes and autophagosomes, compartments that are particularly sensitive to some of those procedures. For example teflon/glass motor homogenization is suitable for tissues with abundant connective tissue, such as liver, but for circulating cells or cells in culture, disruption by nitrogen cavitation is a good method to preserve lysosomal membrane stability;945 however, this method is not suitable for small samples and may not be readily available. Other methods, including “Balch” or “Dounce” homogenizers also work well.946,947 During the isolation procedure it is essential to always use iso-osmotic solutions to avoid hypotonic or hypertonic disruption of the organelles. In that respect, because lysosomes are able to take up sucrose if it is present at high concentrations, the use of sucrose gradients for the isolation of intact lysosome-related organelles is strongly discouraged. It should also be noted that several commercially available kits for subcellular fractionation contain reducing compounds such as dithiothreitol, which may affect the redox status of any prepared fractions. Since numerous proteins involved in autophagy are redox sensitive (an area requiring much additional experimentation), there exists the potential for redox-active compounds in kits to interfere with results. As such, it is suggested to make solutions for fractionation within the laboratory, whenever possible.
As with the isolation of any other intracellular organelle, it is essential to assess the purity of each preparation, as there is often considerable variability from experiment to experiment due to the many steps involved in the process. Correction for purity can be done through calculation of recovery (percentage of the total activity present in the homogenate) and enrichment (dividing by the specific activity in the homogenate) of enzymes or protein markers for those compartments (e.g., HEX/β-hexosaminidase is routinely used to assess lysosomal purity, but enzymes such as CTSB may also be used and may provide more accurate readouts).945 Because of the time-consuming nature of quantitative morphological studies, such studies should not be carried out until simpler biochemical procedures have established the circumstances most likely to give meaningful morphometric/stereological results.
Finally, it is worthwhile noting that not all lysosomes are alike. For example, there are differences among primary lysosomes, autolysosomes and telolysosomes. Furthermore, what we refer to as “lysosomes” are actually a very heterogeneous pool of organelles that simply fulfill 5 classical criteria, having a pH <5.6, mature cathepsins, the presence of LAMP proteins, a single membrane, and the absence of endosomal and recycling compartment markers (e.g., M6PR/mannose-6-phosphate receptor or RAB5). But even applying those criteria we can separate lysosomes with clear differences in their proteome and other properties, and these distinct populations of lysosomes are likely to participate in different functions in the cell (see Chaperone-mediated autophagy).948
Conclusion: Considering the limited methods available for in vivo analysis of autophagy, tissue fractionation is a valid, although relatively laborious, method for monitoring autophagy. Care must be taken to ensure that sample analysis is representative.
15. Analyses in vivo
Monitoring autophagic flux in vivo or in organs is one of the least developed areas at present, and ideal methods relative to the techniques possible with cell culture may not exist. Importantly, the level of basal autophagy, time course of autophagic induction, and the bioavailability of autophagy-stimulating and -inhibiting drugs is likely tissue specific. Moreover, basal autophagy or sensitivity to autophagic induction may vary with animal age, sex or strain background. Therefore methods may need to be optimized for the tissue of interest. One method for in vivo studies is the analysis of GFP-LC3/Atg8 (see GFP-Atg8/LC3 fluorescence microscopy). Autophagy can be monitored in tissue (e.g., skeletal muscle, liver, brain and retina) in vivo in transgenic mice systemically expressing GFP-LC3,153,606,949,950 or in other models by transfection with GFP-LC3 plasmids or in transgenic strains that possess either mCherry- or GFP-LC3/Atg8 under control of either inducible or LC3/Atg8 promoter sequences.281,468,764 It should be noted that tissues such as white adipose tissue, ovary, and testes and some brain regions such as the hypothalamus do not appear to express the Actb promoter-driven GFP-Lc3 transgene strongly enough to allow detection of the fluorescent protein.153 In addition, tissue-specific GFP-LC3 mice have been generated for monitoring cardiac myocytes.951,952 In these settings, GFP fluorescent puncta are indicative of autophagic structures; however, the use of a lysosomal fusion or protease inhibitor would be needed to assess flux. Cleavage of GFP-LC3 to generate free GFP can be evaluated as one method to monitor the completion of autophagy. This has been successfully performed in mouse liver,257,747 suggesting the GFP-LC3 cleavage assay may also be applied to in vivo studies. Note that the accumulation of free GFP in the mouse brain is minimal after autophagy is induced with rapamycin (autophagy induction based on GFP-LC3 imaging and SQSTM1 IHC; M. Lipinski, personal communication), but significant when autophagic flux is partially blocked after traumatic brain injury.950 Thus, caution needs to be taken when interpreting results of these assays in different tissues. We also recommend including a control under conditions known to induce autophagic flux such as starvation. A simple methodology to measure autophagic flux in the brain was described.953 This strategy combines the generation of adeno-associated virus and the use of the dynamic fluorescent reporter mCherry-GFP-LC3, that allows an extended transduction and stable expression of mCherry-GFP-LC3 after intracerebroventricular injection in newborn animals. With this approach, a widespread transduction level is achieved along neurons at the central nervous system when newborn pups are injected, including pyramidal cortical and hippocampal neurons, Purkinje cells, and motor neurons in the spinal cord and also, to a lesser extent, in oligodendrocytes.953 The use of different serotypes of adeno-associated virus could be used to transduce other cell types at the CNS.953,954 This methodology allows a reproducible and sensitive mCherry-GFP-LC3 detection, and a strong LC3 flux when animals are treated with autophagy inducers including rapamycin and trehalose.955 Therefore, these combined strategies can be applied to monitor autophagy activity in mice and also determine autophagy alterations in animal models of diseases affecting the nervous system.953,954 Alternatively, confocal laser scanning microscopy, which makes it possible to obtain numerous sections and substantial data about spatial localization features, can be a suitable system for studying autophagic structures (especially for whole mount embryo in vivo analysis).956 In addition, this method can be used to obtain quantitative data through densitometric analysis of fluorescent signals.957
Another possibility is immunohistochemical staining, an important procedure that may be applicable to human studies as well considering the role of autophagy in neurodegeneration, myopathies and cardiac disease where samples may be limited to biopsy/autopsy tissue. Immunodetection of LC3 as definite puncta is possible in paraffin-embedded tissue sections and fresh frozen tissue, by either IHC or immunofluorescence;197,958-964 however, this methodology has not received extensive evaluation, and does not lend itself well to dynamic assays. Other autophagic substrates can be evaluated via IHC and include SQSTM1, NBR1, ubiquitinated inclusions and protein aggregates. Similarly, autophagy can be evaluated by measuring levels of these autophagic substrates via traditional immunoblot; however, their presence or absence needs to be cautiously interpreted as some of these substrates can accumulate with either an increase or a decrease in autophagic flux (see SQSTM1 and related LC3 binding protein turnover assays). Bone marrow transfer has been used to document in vivo the role of autophagy in the reverse cholesterol transport pathway from peripheral tissues or cells (e.g., macrophages) to the liver for secretion in bile and for excretion,965 and a study shows that TGM2 (transglutaminase 2) protein levels decrease in mouse liver in vivo upon starvation in an autophagy-dependent manner (and in human cell lines in vitro in response to various stimuli; M. Piacentini, personal communication), presenting additional possible methods for following autophagy activity. In that respect, it is noteworthy to mention that TGM2 can negatively affect autophagy by modifying ITPR1 (inositol 1,4,5-trisphosphate receptor, type 1) and suppressing its Ca2+-release activity.966
It is also possible to analyze tissues ex vivo, and these studies can be particularly helpful in assessing autophagic flux as they avoid the risks of toxicity and bioavailability of compounds such as bafilomycin A1 or other autophagy inhibitors. Along these lines, autophagic flux can be determined by western blot in retinas placed in culture for 4 h with protease inhibitors.967,968 This method could be used in tissues that can remain “alive” for several hours in culture such as the retina,967,968 brain slices,950,969 and spinal cord slices.970
Several studies have demonstrated the feasibility of monitoring autophagic flux in vivo in skeletal muscle. Starvation is one of the easiest and most rapid methods for stimulating the autophagic machinery in skeletal muscles. 12 h of fasting in mice may be sufficient to trigger autophagy in muscle,971,972 but the appropriate time should be determined empirically. It is also important to consider that the expression of autophagy-related factors, as well as the autophagic response to various stimuli and disease states, can differ between muscles of different fiber type, metabolic, and contractile properties.153,974-976 Thus, which muscle(s) or portion of muscle(s) used for analysis should be carefully considered and clearly outlined. Although food deprivation does not induce detectable macroautophagy in the brain it induces macroautophagy in the retina, and by the use of in vivo injection of leupeptin autophagic flux can be evaluated with LC3 lipidation by western blot.968 Although difficult to standardize and multifactorial, exercise may be a particularly appropriate stimulus to use for assessing macroautophagy in skeletal muscle.950,977 Data about the autophagic flux can be obtained by treating mice with, for example, chloroquine,972 leupeptin969,978 or colchicine224 and then monitoring the change in accumulation of LC3 (see cautionary notes). This type of analysis can also be done with liver, by comparing the LC3-II level in untreated liver (obtained by a partial hepatectomy) to that following subsequent exposure to chloroquine (V. Skop, Z. Papackova and M. Cahová, personal communication). Additional reporter assays to monitor autophagic flux in vivo need to be developed, including tandem fluorescent-LC3 transgenic mice, or viral vectors to express this construct in vivo in localized areas. One of the challenges of studying autophagic flux in intact animals is the demonstration of cargo clearance, but studies of fly intestines that combine sophisticated mosaic mutant cell genetics with imaging of mitochondrial clearance reveal that such analyses are possible.764
Another organ particularly amenable to ex vivo analysis is the heart, with rodent hearts easily subjected to perfusion by the methods of Langendorff established in 1895 (for review see ref. 978). Autophagy has been monitored in perfused hearts,979 where it is thought to be an important process in several modes of cardioprotection against ischemic injury.980 It should be noted that baseline autophagy levels (as indicated by LC3-II) appear relatively high in the perfused heart, although this may be due to perceived starvation by the ex vivo organ, highlighting the need to ensure adequate delivery of metabolic substrates in perfusion media, which may include the addition of INS/insulin. Another concern is that the high partial pressure of oxygen of the perfusate (e.g., buffer perfused with 95%/5% [O2/CO2]) used in the Langendorff method makes this preparation problematic for the study of autophagy because of the high levels of oxidation (redox disturbances) resulting from the preparation. Therefore, great caution should be exercised in interpretation of these results.
Human placenta also represents an organ suitable for ex vivo studies, such as to investigate pregnancy outcome abnormalities. Autophagy has been evaluated in placentas from normal pregnancies981-983 identifying a baseline autophagy level (as indicated by LC3-II) in uneventful gestation. In cases with abnormal pregnancy outcome, LC3-II is increased in placentas complicated by intrauterine growth restriction in cases both from singleton pregnancies984 and from monochorionic twins pregnancies.985 Moreover, placentas from pregnancies complicated by preeclampsia show a higher level of LC3-II than normal pregnancies.986 Finally, placentas from acidotic newborns developing neonatal encephalopathy exhibit a higher IHC LC3 expression than placentas from newborn without neonatal encephalopathy.987 For this reported association, further investigations are needed to assess if autophagy protein expression in placentas with severe neonatal acidosis could be a potential marker for poor neurological outcome.
The retina is a very suitable organ for ex vivo as well as in vivo autophagy determination. The retina is a part of the central nervous system, is readily accessible and can be maintained in organotypic cultures for some time allowing treatment with protease and autophagy inhibitors. This allows determination of autophagic flux ex vivo in adult and embryonic retinas by western blot394,967 as well as by flow cytometry and microscopy analysis.968 Moreover, only 4 h of leupeptin injection in fasted mice allows for autophagic flux assessment in the retina968 indicating 2 things: first, food deprivation induces autophagy in selected areas of the central nervous system; and second, leupeptin can cross the blood-retinal barrier.
In vivo analysis of the autophagic flux in the brain tissue of neonatal rats can also be performed. These studies use the intraperitoneal administration of the acidotropic dye monodansylcadaverine (MDC) to pup rats 1 h before sacrifice, followed by the analysis of tissue labeling through fluorescence or confocal laser scanning microscopy (365/525-nm excitation/emission filter). This method was adapted to study autophagy in the central nervous system after its validation in cardiac tissue.988 MDC labels acidic endosomes, lysosomes, and late-stage autophagosomes, and its labeling is upregulated under conditions that increase autophagy.989 In a neonatal model of hypoxic-ischemic brain injury, where autophagy activation is a direct consequence of the insult,990 MDC labeling is detectable only in the ischemic tissue, and colocalizes with LC3-II.991 The number of MDC- and LC3-II-positive structures changes when autophagy is pharmacologically up- or downregulated.991,992 Whether this method can also be used in adult animals needs to be determined. Furthermore, it should be kept in mind that staining with MDC is not, by itself, a sufficient method for monitoring autophagy (see Acidotropic dyes).
Another approach that can be used in vivo in brain tissue is to stain for lysosomal enzymes. In situations where an increase in autophagosomes has been shown (e.g., by immunostaining for LC3 and immunoblotting for LC3-II), it is important to show whether this is due to a shutdown of the lysosomal system, causing an accumulation of autophagosomes, or whether this is due to a true increase in autophagic flux. The standard methods described above for in vitro research, such as the study of clearance of a substrate, are difficult to use in vivo, but if it can be demonstrated that the increase in autophagosomes is accompanied by an increase in lysosomes, this makes it very likely that there has been a true increase in autophagic flux. Lysosomal enzymes can be detected by IHC (e.g., for LAMP1 or CTSD) or by classical histochemistry to reveal their activity (e.g., ACP/acid phosphatase or HEX/β-hexosaminidase).993-995
Some biochemical assays may be used to at least provide indirect correlative data relating to autophagy, in particular when examining the role of autophagy in cell death. For example, cellular viability is related to high CTSB activity and low CTSD activities.996 Therefore, the appearance of the opposite levels of activities may be one indication of the initiation of autophagy (lysosome)-dependent cell death. The question of “high” versus “low” activities can be determined by comparison to the same tissue under control conditions, or to a different tissue in the same organism, depending on the specific question.
Cautionary notes: The major hurdle with in vivo analyses is the identification of autophagy-specific substrates and the ability to “block” autophagosome degradation with a compound such as bafilomycin A1. Regardless, it is still essential to adapt the same rigors for measuring autophagic flux in vitro to measurements made with in vivo systems. Moreover, as with cell culture, to substantiate a change in autophagic flux it is not adequate to rely solely on the analysis of static levels or changes in LC3-II protein levels on western blot using tissue samples. To truly measure in vivo autophagic flux using LC3-II as a biomarker, it is necessary to block lysosomal degradation of the protein. Several studies have successfully done this in select tissues in vivo. Certain general principles need to be kept in mind: (a) Any autophagic blocker, whether leupeptin, bafilomycin A1, chloroquine or microtubule depolarizing agents such as colchicine or vinblastine, must significantly increase basal LC3-II levels. The turnover of LC3-II or rate of basal autophagic flux is not known for tissues in vivo, and therefore short treatments (e.g., 4 h) may not be as effective as blocking for longer times (e.g., 12 to 24 h). (b) The toxicity of the blocking agent needs to be considered (e.g., treating animals with bafilomycin A1 for 2 h can be quite toxic), and food intake must be monitored. If long-term treatment is needed to see a change in LC3-II levels, then confirmation that the animals have not lost weight may be needed. Mice may lose a substantial portion of their body weight when deprived of food for 24 h, and starvation is a potent stimulus for the activation of autophagy. (c) The bioavailability of the agent needs to be considered. For example, many inhibitors such as bafilomycin A1 or chloroquine have relatively poor bioavailability to the central nervous system. To overcome this problem, intracerebroventricular injection can be performed.
A dramatic increase of intracellular free poly-unsaturated fatty acid levels can be observed by proton nuclear magnetic resonance spectroscopy in living pancreatic cancer cells within 4 h of autophagy inhibition by omeprazole, which interacts with the V-ATPase and probably inhibits autophagosome-lysosome fusion. Omeprazole is one of the most frequently prescribed drugs worldwide and shows only minor side effects even in higher doses. Proton nuclear magnetic resonance spectroscopy is a noninvasive method that can be also applied as localized spectroscopy in magnetic resonance tomography and therefore opens the possibility of a noninvasive, clinically applicable autophagy monitoring method, although technical issues still have to be solved.997
When analyzing autophagic flux in vivo, one major limitation is the variability between animals. Different animals do not always activate autophagy at the same time. To improve the statistical relevance and avoid unclear results, these experiments should be repeated more than once, with each experiment including several animals. Induction of autophagy in a time-dependent manner by fasting mice for different times requires appropriate caution. Mice are nocturnal animals, so they preferentially move and eat during the night, while they mostly rest during daylight. Therefore, in such experiments it is better to start food deprivation early in the morning, to avoid the possibility that the animals have already been fasting for several hours. The use of chloroquine for flux analysis is technically easier, since it only needs one intraperitoneal injection per day, but the main concern is that chloroquine has some toxicity. Chloroquine suppresses the immunological response in a manner that is not due to its pH-dependent lysosomotropic accumulation (chloroquine interferes with lipopolysaccharide-induced Tnf/Tnf-α gene expression by a nonlysosmotropic mechanism),998 as well as through its pH-dependent inhibition of antigen presentation.999 Therefore, chloroquine treatment should be used for short times and at doses that do not induce severe collateral effects, which may invalidate the measurement of the autophagic flux, and care must be exercised in using chloroquine for studies on autophagy that involve immunological aspects. It is also important to have time-matched controls for in vivo analyses. That is, having only a zero hour time point control is not sufficient because there may be substantial diurnal changes in basal autophagy.644 For example, variations in basal flux in the liver associated with circadian rhythm may be several fold,1000 which can equal or exceed the changes due to starvation. Along these lines, to allow comparisons of a single time-point it is important to specify what time of day the measurement is taken and the lighting conditions under which the animals are housed. It is also important that the replicate experiments are conducted at the same time of day. Controlling for circadian effects can greatly reduce the mouse-to-mouse variability in autophagy markers and flux (J.A. Haspel and A.M.K. Choi, personal communication).
When analyzing the basal autophagic level in vivo using GFP-LC3 transgenic mice,153 one pitfall is that GFP-LC3 expression is driven by the Cmv/cytomegalovirus enhancer and Actb/β-actin (CAG) promoter, so that the intensity of the GFP signal may not always represent the actual autophagic activity, but rather the CAG promoter activity in individual cells. For example, GFP-LC3 transgenic mice exhibit prominent fluorescence in podocytes, but rarely in tubular epithelial cells in the kidney,153 but a similar GFP pattern is observed in transgenic mice carrying CAG promoter-driven non-tagged GFP.1001 Furthermore, proximal tubule-specific ATG5-deficient mice1002 display a degeneration phenotype earlier than podocyte-specific ATG5-deficient mice,1003 suggesting that autophagy, and hence LC3 levels, might actually be more prominent in the former.
One caution in using approaches that monitor ubiquitinated aggregates is that the accumulation of ubiquitin may indicate a block in autophagy or inhibition of proteasomal degradation, or it may correspond to structural changes in the substrate proteins that hinder their degradation. In addition, only cytosolic and not nuclear ubiquitin is subject to autophagic degradation. It is helpful to analyze aggregate degradation in an autophagy-deficient control strain, such as an autophagy mutant mouse, whenever possible to determine whether an aggregate is being degraded by an autophagic mechanism. This type of control will be impractical for some tissues such as those of the central nervous system because the absence of autophagy leads to rapid degeneration. Accordingly, the use of Atg16l1 hypomorphs or Becn1 heterozygotes may help circumvent this problem.
Conclusion: Although the techniques for analyzing autophagy in vivo are not as advanced as those for cell culture, it is still possible to follow this process (including flux) by monitoring, for example, GFP-LC3 by fluorescence microscopy, and SQSTM1 and NBR1 by IHC and/or western blotting.
16. Clinical setting
Altered autophagy is clearly relevant in neurodegenerative disease, as demonstrated by the accumulation of protein aggregates, for example in Alzheimer disease,1004,1005 Parkinson disease,1006 polyglutamine diseases,1007 muscle diseases,1008 and amyotrophic lateral sclerosis.1009 Further evidence comes from the observations that the crucial mitophagy regulators PINK1 and PARK2 show loss-of-function mutations in autosomal recessive juvenile parkinsonism,1010 and that the putative ribophagy regulator VCP/p97 (an ortholog of yeast Cdc48) as well as the autophagy receptor OPTN are mutated in motor neuron disease.1011,1012 In addition to neurodegenerative diseases, alterations in autophagy have also been implicated in other neurological diseases including some epilepsies, neurometabolic and neurodevelopmental disorders.969,1013-1015 A very useful nonspecific indicator of deficient aggrephagy in autopsy brain or biopsy tissue is SQSTM1 IHC.1016,1017 For clinical attempts to monitor autophagy alterations in peripheral tissues such as blood, it is important to know that eating behavior may be altered as a consequence of the disease,1018 resulting in a need to control feeding-fasting conditions during the analyses. Recently, altered autophagy was also implicated in schizophrenia, with BECN1 transcript levels decreasing in the postmortem hippocampus in comparison to appropriate controls.1019 In the same hippocampal postmortem samples, the correlation between the RNA transcript content for ADNP (activity-dependent neuroprotective homeobox) and its sister protein ADNP2 is deregulated,1020 and ADNP as well as ADNP2 RNA levels increase in peripheral lymphocytes from schizophrenia patients compared to matched healthy controls, suggesting a potential biomarker.1019
Similarly, autophagy inhibition plays a key role in the pathogenesis of inherited autophagic vacuolar myopathies (including Danon disease, X-linked myopathy with excessive autophagy, and infantile autophagic vacuolar myopathy), all of which are characterized by lysosomal defects and an accumulation of autophagic vacuoles.1021 Autophagic vacuolar myopathies and cardiomyopathies can also be secondary to treatment with autophagy-inhibiting drugs (chloroquine, hydroxychloroquine and colchicine), which are used experimentally to interrogate autophagic flux and clinically to treat malaria, rheumatological diseases, and gout.964 Autophagy impairment has also been implicated in the pathogenesis of inclusion body myositis, an age-associated inflammatory myopathy that is currently refractory to any form of treatment,1022-1024 along with other muscular dystrophies such as tibial muscular dystrophy.1025 In all these striated muscle disorders, definitive tissue diagnosis used to require ultrastructural demonstration of accumulated autophagic vacuoles; more recently, it has been shown that IHC for LC3 and/or SQSTM1 can be used instead.962-964,1026
In addition, altered basal autophagy levels are seen in rheumatoid arthritis,1027,1028 and osteoarthritis.1029 Other aspects of the immune response associated with dysfunctional autophagy are seen in neutrophils from patients with familial Mediterranean fever1030 and in monocytes from patients with TNF receptor-associated periodic syndrome,1031 2 autoinflammatory disorders. Moreover, auto-phagy regulates an important neutrophil function, the generation of neutrophil extracellular traps (NETs).1024,1032 The important role of autophagy in the induction of NET formation has been studied in several neutrophil-associated disorders such as gout,1033 sepsis,1034 and lung fibrosis.1035 Furthermore, there is an intersection between autophagy and the secretory pathway in mammalian macrophages for the release of IL1B,1036 demonstrating a possible alternative role of autophagy for protein trafficking. This role has also been implied in neutrophils through exposure of protein epitopes on NETs by acidified LC3-positive vacuoles in sepsis1034 and anti-neutrophil cytoplasmic antibody associated vasculitis.1037 Patients with chronic kidney disease also have impaired autophagy activation in leukocytes, which is closely related to their cardiac abnormalities. There is also evidence for altered autophagy in pancreatic beta cells,1038,1039 and in adipocytes 217,1040,1041 of patients with type 2 diabetes.1042 However, autophagy also plays an important role in the development in vitro of giant phagocytes, a long-lived neutrophil subpopulation, derived from neutrophils of healthy individuals.1043,1044
Photodynamic therapy (PDT), an FDA-approved anticancer therapy, has high selectivity for tumor cell elimination by eliciting efficient apoptosis and autophagy induction and fulfills the need to merge a direct cytotoxic action on tumor cells with potent immunostimulatory effects (i.e., immunogenic cell death, ICD).1045 A few photosensitizers, such as Photofrin, Hypericin, Foscan, 5-ALA and Rose Bengal acetate, are associated with danger/damage-associated molecular pattern (DAMP) exposure and/or release that is a requisite to elicit ICD. Rose Bengal acetate PDT is the first treatment to induce autophagic HeLa cells to express and release DAMPS, thus suggesting a possible role of the autophagic cells in ICD induction.1046 Similarly, the photosensitizer Hypocrellin B-acetate is able to induce macroautophagy at very low concentrations.1047
A crucial role for therapy-induced autophagy in cancer cells has recently emerged, in modulating the interface of cancer cells and the immune system;1048 primarily, by affecting the nature of danger signaling (i.e., the signaling cascade that facilitates the exposure and/or release of danger signals) associated with ICD.1045,1048-1051 This is an important point considering the recent clinical surge in the success of cancer immunotherapy in patients, and the emerging clinical relevance of ICD for positive patient prognosis. Several notorious autophagy-inducing anticancer therapies induce ICD including mitoxantrone, doxorubicin, oxaliplatin, radiotherapy, certain oncolytic viruses and hypericin-based photodynamic therapy (Hyp-PDT).1051-1054 In fact, in the setting of Hyp-PDT, ER stress-induced autophagy in human cancer cells suppresses CALR (calreticulin) surface exposure (a danger signal crucial for ICD) thereby leading to suppression of human dendritic cell maturation and human CD4+ and CD8+ T cell stimulation.1053 Conversely, chemotherapy (mitoxantrone or oxaliplatin)-induced autophagy facilitates ATP secretion (another crucial ICD-associated danger signal) thereby facilitating ICD and anti-tumor immunity in the murine system, the first documented instance of autophagy-based ICD modulation.1055 The role of ATP as a DAMP becomes clear when the extracellular concentration of ATP becomes high and elicits activation of the purinergic receptor P2RX7. P2RX7 is involved in several pathways, including the sterile immune response, and its activation induces cancer cell death through PI3K, AKT and MTOR.1056,1057 In addition, cells lacking the essential chaperone-mediated autophagy (CMA) gene LAMP2A fail to expose surface CALR after treatment with both Hyp-PDT and mitoxantrone.1058 These observations have highlighted the important, context-dependent role of therapy-induced autophagy, in modulating the cancer cell-immune cell interface by regulating the emission of ICD-associated danger signals.1059 Recent studies also have implicated insufficient autophagy in the pathogenesis of nonresolving vital organ failure and muscle weakness during critical illness, 2 leading causes of death in prolonged critically ill patients.1060,1061 Finally, a block of autophagy with consequent accumulation of autophagy substrates is detected in liver fibrosis,1062,1063 and lysosomal storage diseases.1064
It is important to note that disease-associated autophagy defects are not restricted to macroautophagy but also concern other forms of autophagy. CMA impairment, for instance, is associated with several disease conditions, including neurodegenerative disorders,229,1065 lysosomal storage diseases,1066,1067, nephropathies1068 and diabetes.1069 In addition, it is very important to keep in mind that although human disease is mostly associated with inhibited autophagy, enhanced autophagy has also been proposed to participate in, and even contribute to, the pathogenesis of human diseases, such as chronic obstructive pulmonary disease,1070 and adipocyte/adipose tissue dysfunction in obesity.217,1040 Along these lines, chloroquine decreases diabetes risk in patients treated with the drug for rheumatoid arthritis.1071
A set of recommendations regarding the design of clinical trials modulating autophagy can be found in ref. 1072.
Cautionary notes: To establish a role for autophagy in modulating the interface with the immune system, specific tests need to be performed where genes encoding autophagy-relevant components (e.g., ATG5, ATG7 or BECN1) have been knocked down through RNA silencing or other protein- or gene-specific targeting technologies.1053,1055,1058 Usage of chemical inhibitors such as bafilomycin A1, 3-MA or chloroquine can create problems owing to their off-target effects, especially on immune cells, and thus their use should be subjected to due caution, and relevant controls are critical to account for any off-target effects. In the context of ICD, consideration should be given to the observations that autophagy can play a context-dependent role in modulating danger signaling;1053,1055,1058 and thus, all the relevant danger signals (e.g., surface exposed CALR or secreted ATP) should be (re-)tested for new agents/therapies in the presence of targeted ablation of autophagy-relevant proteins/genes, accompanied by relevant immunological assays (e.g., in vivo rodent vaccination/anti-tumor immunity studies or ex vivo immune cell stimulation assays), in order to implicate autophagy in regulating ICD or general immune responses.
17. Cell death
In several cases, autophagy has been established as the cause of cell death;83,281,354,764,1073-1081 although opposite results have been reported using analogous experimental settings.1082 Furthermore, many of the papers claiming a causative role of autophagy in cell death fail to provide adequate evidence.1083 Other papers suffer from ambiguous use of the term “autophagic cell death,” which was coined in the 1970s1084 in a purely morphological context to refer to cell death with autophagic features (especially the presence of numerous secondary lysosomes); this was sometimes taken to suggest a role of autophagy in the cell death mechanism, but death-mediation was not part of the definition.1085 Unfortunately, the term “autophagic cell death” is now used in at least 3 different ways: (a) Autophagy-associated cell death (the original meaning). (b) Autophagy-mediated cell death (which could involve a standard mechanism of cell death such as apoptosis, but triggered by autophagy). (c) A distinct mechanism of cell death, independent of apoptosis or necrosis. Clearly claim (b) is stronger than claim (a), and needs to be justified by proof that inhibiting autophagy, through either genetic or chemical means, prevents cell death.1086 Claim (c) is still stronger, because, even if the cell death is blocked by autophagy inhibition, proof needs to be provided that the cell death mechanism is not apoptosis or necrosis.1087 In view of the current confusion it may be preferable to replace the term “autophagic cell death” by other terms such as “autophagy-associated cell death” or “autophagy-mediated cell death,” unless the criteria in claim (c) above have been satisfied. Along these lines, it is preferable to use the term “autophagy-dependent cell death” instead of “autophagy-mediated cell death” when it is proven that autophagy is a pre-requisite for the occurrence of cell death, but it is not proven that autophagy mechanistically mediates the switch to cell death. It is important to note that a stress/stimulus can in many circumstances induce different cell death pathways at the same time, which might lead to a “type” of cell death with mixed phenotypes.1088,1089 Furthermore, inhibition of one cell death pathway (e.g., apoptosis) can either induce the compensatory activation of a secondary mechanism (e.g., necrosis),1090,1091 or attenuate a primary mechanism (e.g., liponecrosis).1088
The role of autophagy in the death of plant cells is less ambiguous, because plants are devoid of the apoptotic machinery and use lytic vacuoles to disassemble dying cells from inside.1092 This mode of cell death governs many plant developmental processes and was named “vacuolar cell death”.1093 Recent studies have revealed a key role of autophagy in the execution of vacuolar cell death, where autophagy sustains the growth of lytic vacuoles.1094,1095 Besides being an executioner of vacuolar cell death, autophagy can also play an upstream, initiator role in immunity-associated cell death related to the pathogen-triggered hypersensitive response.1092,1096
Upon induction by starvation during multicellular development in the protist Dictyostelium, autophagy (or at least Atg1) is required to protect against starvation-induced cell death, allowing vacuolar developmental cell death to take place instead.1097,1098 Autophagy may be involved not only in allowing this death to occur, but also, as during vacuolar cell death in plants, in the vacuolization process itself.1099
Recently, a novel form of autophagy-dependent cell death has been described, autosis, which not only meets the criteria in claim (c) (i.e., blocked by autophagy inhibition, independent of apoptosis or necrosis), but also demonstrates unique morphological features and a unique ability to be suppressed by pharmacological or genetic inhibition of the Na+,K+-ATPase.1080 In addition, the demonstration that autophagy is required for cell death during Drosophila development where caspases and necrosis do not appear to be involved may be the best known physiologically relevant model of cell death that involves autophagy.281,764
Cautionary notes: In brief, rigorous criteria must be met in order to establish a death-mediating role of autophagy, as this process typically promotes cell survival. These include a clear demonstration of autophagic flux as described in this article, as well as verification that inhibition of autophagy prevents cell death (claim [b] above; if using a knockdown approach, at least 2 ATG genes should be targeted), and that other mechanisms of cell death are not responsible (claim [c] above). As part of this analysis, it is necessary to examine the effect of the specific treatment, conditions or mutation on cell viability using several methods.1090 In the case of postmitotic cells such as neurons or retinal cells, cell death—and cell rescue by autophagy inhibition—can usually be established in vivo by morphological analysis,1100 and in culture by cell counts and/or measurement of the release of an enzyme such as LDH into the medium at early and late time points; however, a substantial amount of neuronal cell death occurs during neurogenesis, making it problematic to carry out a correct analysis in vivo or ex vivo.1101,1102 In populations of rapidly dividing cells, the problems may be greater. A commonly used method is the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay or a related assay using a similar, or a water-soluble, tetrazolium salt. The main concern with the MTT assay is that it measures mitochondrial activity, but does not allow a precise determination of cellular viability or cell death, whereas methods that show cell death directly (e.g., trypan blue exclusion, or LDH release assay) fail to establish the viability of the remaining cell population.1103 Accordingly, a preferred alternative is to accurately quantify cell death by appropriate cytofluorometric or microscopy assays.1090 Moreover, long-term clonogenic assays should be employed when possible to measure the effective functional survival of cells.
Conclusion: In most systems, ascribing death to autophagy based solely on morphological criteria is insufficient; autophagic cell death can only be demonstrated as death that is suppressed by the inhibition of autophagy, through either genetic or chemical means.1086 In addition, more than one assay should be used to measure cell death. In this regard, it is important to mention that neither changes in mitochondrial activity/potential, nor caspase activation or externalization of phosphatidylserine can be accurately used to determine cell death as all these phenomena have been reported to be reversible. Only the determination of cellular viability (ratio between dead/live cells) can be used to accurately determine cell death progression.
18. Chaperone-mediated autophagy
The primary characteristic that makes CMA different from the other autophagic variants described in these guidelines is that it does not require formation of intermediate vesicular compartments (autophagosomes or microvesicles) for the import of cargo into lysosomes.1104,1105 Instead, the CMA substrates are translocated across the lysosomal membrane through the action of HSPA8/HSC70 (heat shock 70kDa protein 8) located in the cytosol and lysosome lumen, and the lysosome membrane protein LAMP2A. To date, CMA has only been identified in mammalian cells, and accordingly this section refers only to studies in mammals.
The following section discusses methods commonly utilized to determine if a protein is a CMA substrate (see ref. 1106 for experimental details):
Analysis of the amino acid sequence of the protein to identify the presence of a KFERQ-related motif that is an absolute requirement for all CMA substrates.1107
Colocalization studies with lysosomal markers (typically LAMP2A and/or LysoTracker) to identify a fraction of the protein associated with lysosomes. The increase in association of the putative substrate under conditions that upregulate CMA (such as prolonged starvation) or upon blockage of lysosomal proteases (to prevent the degradation of the protein) helps support the hypothesis that the protein of interest is a CMA substrate. However, association with lysosomes is necessary but not sufficient to consider a protein an authentic CMA substrate, because proteins delivered by other pathways to lysosomes will also behave in a similar manner. A higher degree of confidence can be attained if the association is preferentially with the subset of lysosomes active for CMA (i.e., those containing HSPA8 in their lumen), which can be separated from other lysosomes following published procedures.948
Co-immunoprecipitation of the protein of interest with cytosolic HSPA8. Due to the large number of proteins that interact with this chaperone, it is usually better to perform affinity isolation with the protein of interest and then analyze the isolated proteins for the presence of HSPA8 rather than vice versa.
Co-immunoprecipitation of the protein of interest with LAMP2A.1108 Due to the fact that the only antibodies specific for the LAMP2A variant (the only 1 of the 3 LAMP2 variants involved in CMA92,1109) are generated against the cytosolic tail of LAMP2A, where the substrate also binds, it is necessary to affinity isolate the protein of interest and then analyze for the presence of LAMP2A. Immunoblot for LAMP2A in the precipitate can only be done with the antibodies specific for LAMP2A and not just those that recognize the lumenal portion of the protein that is identical in the other LAMP2 variants. If the protein of interest is abundant inside cells, co-immunoprecipitations with LAMP2A can be done in total cellular lysates, but for low abundance cellular proteins, preparation of a membrane fraction (enriched in lysosomes) by differential centrifugation may facilitate the detection of the population of the protein bound to LAMP2A.
Selective upregulation and blockage of CMA to demonstrate that degradation of the protein of interest changes with these manipulations. Selective chemical inhibitors for CMA are not currently available. Note that general inhibitors of lysosomal proteases (e.g., bafilomycin A1, NH4Cl, leupeptin) also block the degradation of proteins delivered to lysosomes by other autophagic and endosomal pathways. The most selective way to block CMA is by knockdown of LAMP2A, which causes this protein to become a limiting factor.92 The other components involved in CMA, including HSPA8, HSP90AA1, GFAP, and EEF1A/eF1α, are all multifunctional cellular proteins, making it difficult to interpret the effects of knockdowns. Overexpression of LAMP2A1108 is also a better approach to upregulate CMA than the use of chemical modulators. The 2 compounds demonstrated to affect degradation of long-lived proteins in lysosomes,1110 6-aminonicotinamide and geldanamycin, lack selectivity, as they affect many other cellular processes. In addition, in the case of geldanamycin, the effect on CMA can be the opposite (inhibition rather than stimulation) depending on the cell type (this is due to the fact that the observed stimulation of CMA is actually a compensatory response to the blockage of HSP90AA1 in lysosomes, and different cells activate different compensatory responses).1111
The most conclusive way to prove that a protein is a CMA substrate is by reconstituting its direct translocation into lysosomes using a cell-free system.1106 This method is only possible when the protein of interest can be purified, and it requires the isolation of the population of lysosomes active for CMA. Internalization of the protein of interest inside lysosomes upon incubation with the isolated organelle can be monitored using protease protection assays (in which addition of an exogenous protease removes the protein bound to the cytosolic side of lysosomes, whereas it is inaccessible to the protein that has reached the lysosomal lumen; note that pre-incubation of lysosomes with lysosomal protease inhibitors before adding the substrate is required to prevent the degradation of the translocated substrate inside lysosomes).1112 The use of exogenous protease requires numerous controls (see ref. 1106) to guarantee that the amount of protease is sufficient to remove all the substrate outside lysosomes, but will not penetrate inside the lysosomal lumen upon breaking the lysosomal membrane.
The difficulties in the adjustment of the amount of protease have led to the development of a second method that is more suitable for laboratories that have no previous experience with these procedures. In this case, the substrate is incubated with lysosomes untreated or previously incubated with inhibitors of lysosomal proteases, and then uptake is determined as the difference of protein associated with lysosomes not incubated with inhibitors (in which the only remaining protein will be the one associated with the cytosolic side of the lysosomal membrane) and those incubated with the protease inhibitors (which contain both the protein bound to the membrane and that translocated into the lumen).1113
Confidence that the lysosomal internalization is by CMA increases if the uptake of the substrate can be competed with proteins previously identified as substrates for CMA (e.g., GAPDH/glyceraldehyde-3-phosphate dehydrogenase or RNASE1/ribonuclease A, both commercially available as purified proteins), but is not affected by the presence of similar amounts of nonsubstrate proteins (such as SERPINB/ovalbumin or PPIA/cyclophilin A). Blockage of uptake by pre-incubation of the lysosomes with antibodies against the cytosolic tail of LAMP2A also reinforces the hypothesis that the protein is a CMA substrate. It should be noted that several commercially available kits for lysosome isolation separate a mixture of lysosomal populations and do not enrich in the subgroup of lysosomes active for CMA, which limits their use for CMA uptake assays.
In other instances, rather than determining if a particular protein is a CMA substrate, the interest may be to analyze possible changes in CMA activity under different conditions or in response to different modifications. We enumerate here the methods, from lower to higher complexity, that can be utilized to measure CMA in cultured cells and in tissues (see ref. 1106 for detailed experimental procedures).
Measurement of changes in the intracellular rates of degradation of long-lived proteins, when combined with inhibitors of other autophagic pathways, can provide a first demonstration in support of changes that are due to CMA. For example, CMA is defined as lysosomal degradation upregulated in response to serum removal but insensitive to PtdIns3K inhibitors.
Measurement of levels of CMA components is insufficient to conclude changes in CMA because this does not provide functional information, and changes in CMA components can also occur under other conditions. However, analysis of the levels of LAMP2A can be used to support changes in CMA detected by other procedures. Cytosolic levels of HSPA8 remain constant and are not limiting for CMA, thus providing no information about this pathway. Likewise, changes in total cellular levels of LAMP2A do not have an impact on this pathway unless they also affect their lysosomal levels (i.e., conditions in which LAMP2A is massively overexpressed lead to its targeting to the plasma membrane where it cannot function in CMA). It is advisable that changes in the levels of these 2 CMA components are confirmed to occur in lysosomes, either by colocalization with lysosomal markers when using image-based procedures or by performing immunoblot of a lysosomal enriched fraction (purification of this fraction does not require the large amounts of cells/tissue necessary for the isolation of the subset of lysosomes active for CMA).
Tracking changes in the subset of lysosomes active for CMA. This group of lysosomes is defined as those containing HSPA8 in their lumen (note that LAMP2A is present in both lysosomes that are active and inactive for CMA, and it is the presence of HSPA8 that confers CMA capability). Immunogold or immunofluorescence against these 2 proteins (LAMP2A and HSPA8) makes it possible to quantify changes in the levels of these lysosomes present at a given time, which correlates well with CMA activity.948
Analysis of lysosomal association of fluorescent artificial CMA substrates. Two different fluorescent probes have been generated to track changes in CMA activity in cultured cells using immunofluorescence or flow cytometry analysis.948 These probes contain the KFERQ and context sequences in frame with photoswitchable or photoactivated fluorescent proteins. Activation of CMA results in the mobilization of a fraction of the cytosolic probe to lysosomes and the subsequent change from a diffuse to a punctate pattern. CMA activity can be quantified as the number of fluorescent puncta per cell or as the decay in fluorescence activity over time because of degradation of the artificial substrate. Because the assay does not allow measuring accumulation of the substrate (which must unfold for translocation), it is advisable to perform a time-course analysis to determine gradual changes in CMA activity. Antibodies against the fluorescent protein in combination with inhibitors of lysosomal proteases can be used to monitor accumulation of the probe in lysosomes over a period of time, but both the photoswitchable and the unmodified probe will be detected by this procedure.1114 As for any other fluorescence probe based on analysis of intracellular “puncta” it is essential to include controls to confirm that the puncta are indeed lysosomes (colocalization with LysoTracker or LAMPs and lack of colocalization with markers of cytosolic aggregation such as ubiquitin) and do not reach the lysosomes through other autophagic pathways (insensitivity to PtdIns3K inhibitors and sensitivity to LAMP2A knockdown are good controls in this respect).
Direct measurement of CMA using in vitro cell free assays. Although the introduction of the fluorescent probes should facilitate measurement of CMA in many instances, they are not applicable for tissue samples. In addition, because the probes measure binding of substrate to lysosomal membranes it is important to confirm that enhanced binding does not result from defective translocation. Last, the in vitro uptake assays are also the most efficient way to determine primary changes in CMA independently of changes in other proteolytic systems in the cells. These in vitro assays are the same ones described in the previous section on the identification of proteins as substrates of CMA, but are performed in this case with purified proteins previously characterized to be substrates for CMA. In this case the substrate protein is always the same, and what changes is the source of lysosomes (from the different tissues or cells that are to be compared). As described in the previous section, binding and uptake can be analyzed separately using lysosomes previously treated or not with protease inhibitors. The analysis of the purity of the lysosomal fractions prior to performing functional analysis is essential to conclude that changes in the efficiency to take up the substrates results from changes in CMA rather than from different levels of lysosomes in the isolated fractions. Control of the integrity of the lysosomal membrane and sufficiency of the proteases are also essential to discard the possibility that degradation is occurring outside lysosomes because of leakage, or that accumulation of substrates inside lysosomes is due to enhanced uptake rather than to decreased degradation.
Cautionary notes: The discovery of another selective form of protein degradation in mammals named endosomal microautophagy (e-MI)1115 has made it necessary to reconsider some of the criteria that applied in the past for the definition of a protein as a CMA substrate. The KFERQ-like motif, previously considered to be exclusive for CMA, is also used to mediate selective targeting of cytosolic proteins to the surface of late endosomes. Once there, substrates can be internalized in microvesicles that form from the surface of these organelles in an ESCRT-dependent manner. HSPA8 has been identified as the chaperone that binds this subset of substrates and directly interacts with lipids in the late endosomal membrane, acting thus as a receptor for cytosolic substrates in this compartment. At a practical level, to determine if a KFERQ-containing protein is being degraded by CMA or e-MI the following criteria can be applied: (a) Inhibition of lysosomal proteolysis (for example with NH4Cl and leupeptin) blocks degradation by both pathways. (b) Knockdown of LAMP2A inhibits CMA but not e-MI. (c) Knockdown of components of ESCRTI and II (e.g., VPS4 and TSG101) inhibits e-MI but not CMA. (d) Interfering with the capability to unfold the substrate protein blocks its degradation by CMA, but does not affect e-MI of the protein. In this respect, soluble proteins, oligomers and protein aggregates can undergo e-MI, but only soluble proteins can be CMA substrates. (e) In vitro uptake of e-MI substrates can be reconstituted using isolated late endosomes whereas in vitro uptake of CMA substrates can only be reconstituted using lysosomes.
Another pathway that needs to be considered relative to CMA is chaperone-assisted selective autophagy (CASA).1116 CASA is dependent on HSPA8 and LAMP2 (although it is not yet known if it is dependent solely on the LAMP2A isoform). Thus, a requirement for these 2 proteins is not sufficient to conclude that a protein is degraded by CMA. It should also be noted that LAMP1 and LAMP2 share common function as revealed by the embryonic lethal phenotype of lamp1-/- lamp2y/- double-deficient mice.1117 In addition to CMA, LAMP2 is involved in the fusion of late endosomes and autophagosomes or phagosomes.1118,1119 LAMP2C, one of the LAMP2 isoforms, can also function as an RNA/DNA receptor in RNautophagy and DNautophagy pathways, where RNA or DNA is taken up directly by lysosomes in an ATP-dependent manner.1120-1123 LAMP1 and LAMP2 deficiency does not necessarily affect protein degradation under conditions when CMA is active,1117 and the expression levels of neuronal CMA substrates does not change upon loss of LAMP2.1120,1124,1125
Conclusion: One of the key issues with the analysis of CMA is verifying that the protein of interest is an authentic substrate. Methods for monitoring CMA that utilize fluorescent probes are available that eliminate the need for the isolation of CMA-competent lysosomes, one of the most difficult aspects of assaying this process.
19. Chaperone-assisted selective autophagy
CASA is a recently identified, specialized form of autophagy whereby substrate proteins are ubiquitinated and targeted for lysosomal degradation by chaperone and co-chaperone proteins.1116 The substrate protein does not require a KFERQ motif, which differentiates CASA from CMA. In CASA the substrate protein is recognized by the chaperone HSPA8, the small heat shock proteins HSPB6 and HSPB8, the ubiquitin ligase STUB1/CHIP, which forms a multidomain complex with the co-chaperone BAG3, and the receptor proteins SYNPO2/myopodin (synaptopodin 2) and SQSTM1. Following ubiquitination the substrate protein is loaded onto the CASA machinery. SYNPO2 and SQSTM1 then bind to core components of the autophagosome (VPS18 and LC3, respectively) resulting in engulfment of the substrate protein and associated multidomain complex into the autophagosome, and subsequent lysosomal degradation.1116,1126
To date, CASA has only been reported in muscle with the FLN (filamin) family of proteins being the most studied target. However, CASA may also be capable of targeting nonmuscle proteins for degradation as demonstrated by an in vitro study on BAG3-mediated degradation of mutant HTT.1127,1128
Conclusion: Given that the autophagy machinery involved in CASA is very similar to that in other forms of autophagy there are currently no specific markers or inhibitors available to study this process specifically, but the involvement of BAG3 and ubiquitination of client proteins is highly suggestive of CASA activity.
B. Comments on additional methods
1. Acidotropic dyes
Among the older methods for following autophagy is staining with acidotropic dyes such as monodansylcadaverine,1129 acridine orange,1130 Neutral Red,956 LysoSensor Blue1131 and LysoTracker Red.280,1132 It should be emphasized that, whereas these dyes are useful to identify acidified vesicular compartments, they should not be relied upon to compare differences in endosomal or lysosomal pH between cells due to variables that can alter the intensity of the signal. For example, excessive incubation time and/or concentrations of LysoTracker Red can oversaturate labeling of the cell and mask differences in signal intensity that reflect different degrees of acidification within populations of compartments.1133 Use of these dyes to detect, size, and quantify numbers of acidic compartments must involve careful standardization of the conditions of labeling and ideally should be confirmed by ancillary TEM and/or immunoblot analysis. Reliable measurements of vesicle pH require ratiometric measurements of 2 dyes with different peaks of optimal fluorescence (e.g., LysoSensor Blue and LysoSensor Yellow) to exclude variables related to uptake.62,1133
Cautionary notes: Although MDC was first described as a specific marker of autophagic vacuoles1134 subsequent studies have suggested that this, and other acidotropic dyes, are not specific markers for early autophagosomes,331 but rather label later stages in the degradation process. For example, autophagosomes are not acidic, and MDC staining can be seen in autophagy-defective mutants540 and in the absence of autophagy activation.1135 MDC may also show confounding levels of background labeling unless narrow bandpass filters are used. However, in the presence of vinblastine, which blocks fusion with lysosomes, MDC labeling increases, suggesting that under these conditions MDC can label late-stage autophagosomes.989 Along these lines, cells that overexpress a dominant negative version of RAB7 (the T22N mutant) show colocalization of this protein with MDC; in this case fusion with lysosomes is also blocked1136 indicating that MDC does not just label lysosomes. Nevertheless, MDC labeling could be considered to be an indicator of autophagy when the increased labeling of cellular compartments by this dye is prevented by treatment with specific autophagy inhibitors.
Overall, staining with MDC or its derivative monodansylamylamine (MDH)1129 is not, by itself, a sufficient method for monitoring autophagy. Similarly, LysoTracker Red, Neutral Red and acridine orange are not ideal markers for autophagy because they primarily detect lysosomes and an increase in lysosome size or number could reflect an increase in nonprofessional phagocytosis (often seen in embryonic tissues1137) rather than autophagy. These markers are, however, useful for monitoring selective autophagy when used in conjunction with protein markers or other dyes. For example, increased colocalization of mitochondria with both GFP-LC3 and LysoTracker Red can be used as evidence of autophagic cargo delivery to lysosomes. Moreover, LysoTracker Red has been used to provide correlative data on autophagy in D. melanogaster fat body cells (Fig. 26).279,280 However, additional assays, such as GFP-Atg8/LC3 fluorescence and EM, should be used to substantiate results obtained with acidotropic dyes whenever possible to rule out the possibility that LAP is involved (see Noncanonical use of autophagy-related proteins). Finally, one important caution when co-imaging with LysoTracker Red and a green-fluorescing marker (e.g., GFP-LC3 or MitoTracker Green) is that it is necessary to control for rapid red-to-green photoconversion of the LysoTracker, which can otherwise result in an incorrect interpretation of colocalization.1138
Figure 26.
LysoTracker Red stains lysosomes and can be used to monitor autophagy in Drosophila. Live fat body tissues from Drosophila were stained with LysoTracker Red (red) and Hoechst 33342 (blue) to stain the nucleus. Tissues were isolated from fed (left) or 3-h starved (right) animals. Scale bar: 25 µm. This figure was modified from data presented in ref. 280, Developmental Cell, 7, Scott RC, Schuldiner O, Neufeld TP, Role and regulation of starvation-induced autophagy in the Drosophila fat body, pp. 167–78, copyright 2004, with permission from Elsevier.
Some of the confusion regarding the interpretation of results with these dyes stems in part from the nomenclature in this field. Indeed, the discussion of acidotropic dyes points out why it is advisable to differentiate between the terms “autophagosome” and “autophagic vacuole,” although they are occasionally, and incorrectly, used interchangeably. The autophagosome is the sequestering compartment generated by the phagophore. The fusion of an autophagosome with an endosome or a lysosome generates an amphisome or an autolysosome, respectively.884 The early autophagosome is not an acidic compartment, whereas amphisomes and autolysosomes are acidic. As noted in the section Transmission electron microscopy, earlier names for these compartments are “initial autophagic vacuole (AVi),” “intermediate or intermediate/degradative autophagic vacuole (AVi/d)” and “degradative autophagic vacuole (AVd),” respectively. Thus, acidotropic dyes can stain late autophagic vacuoles (in particular autolysosomes), but not the initial autophagic vacuole, the early autophagosome.
A recently developed dye for monitoring autophagy, Cyto-ID, stains vesicular structures shortly after amino acid deprivation, which extensively colocalize with RFP-LC3-positive structures, while colocalizing partially with lysosomal probes.1139 Moreover, unlike MDC, Cyto-ID does not show background fluorescence under control conditions and the 2 dyes colocalize only marginally. Furthermore, the Cyto-ID signal responds to well-known autophagy modulators. Therefore, this amphiphilic dye, which partitions in hydrophobic environments, may prove more selective for autophagic vacuoles than the previously discussed lysosomotropic dyes.
With the above caveats in mind, the combined use of early and late markers of autophagy is highly encouraged, and when quantifying mammalian lysosomes, it is important to keep in mind that increases in both lysosome size and number are frequently observed. Finally, to avoid confusion with the plant and fungal vacuole, the equivalent organelle to the lysosome, we recommend the use of the term “autophagosome” instead of “autophagic vacuole” when possible, that is, when the specific nature of the structure is known.
Conclusion: Given the development of better techniques that are indicators of autophagy, the use of acidotropic dyes to study this process is discouraged, and relying entirely on such dyes is not acceptable.
2. Autophagy inhibitors and inducers
In many situations it is important to demonstrate an effect resulting from inhibition or stimulation of autophagy (see ref. 1140 for a partial listing of regulatory compounds), and a few words of caution are worthwhile in this regard. Most chemical inhibitors of autophagy are not entirely specific, and it is important to consider possible dose- and time-dependent effects. Accordingly, it is generally preferable to analyze specific loss-of-function Atg mutants. However, it must be kept in mind that some apparently specific Atg gene products may have autophagy-independent roles (e.g., ATG5 in cell death, and the PIK3C3/VPS34-containing complexes—including BECN1—in apoptosis, endosomal function and protein trafficking), or may be dispensable for autophagy (see Noncanonical use of autophagy-related proteins).27,543,573,1141-1144 Therefore, the experimental conditions of inhibitor application and their side effects must be carefully considered. In addition, it must be emphasized once again that autophagy, as a multistep process, can be inhibited at different stages. Sequestration inhibitors, including 3-MA, LY294002 and wortmannin, inhibit class I phosphoinositide 3-kinases (PI3Ks) as well as class III PtdIns3Ks.132,330,1145 The class I enzymes generate products such as PtdIns(3,4,5)P3 that inhibit autophagic sequestration, whereas the class III product (PtdIns3P) generally stimulates autophagic sequestration. The overall effect of these inhibitors is typically to block autophagy because the class III enzymes that are required to activate autophagy act downstream of the negative regulatory class I enzymes, although cell death may ensue in cell types that are dependent upon high levels of AKT for survival. The effect of 3-MA (but not that of wortmannin) is further complicated by the fact that it has different temporal patterns of inhibition, causing a long-term suppression of the class I PI3K, but only a transient inhibition of the class III enzyme. In cells incubated in a complete medium for extended periods of time, 3-MA may, therefore (particularly at suboptimal concentrations), promote autophagy by inhibition of the class I enzyme.330 Thus, wortmannin may be considered as an alternative to 3-MA for autophagy inhibition.330 However, wortmannin can induce the formation of vacuoles that may have the appearance of autophagosomes, although they are swollen late endocytic compartments.931 Furthermore, studies have demonstrated that inhibition of autophagy with 3-MA or wortmannin can have effects on cytokine transcription, processing and secretion, particularly of IL1 family members,1146-1148 but 3-MA and wortmannin also inhibit the secretion of some cytokines and chemokines (e.g., TNF, IL6, CCL2/MCP-1) in an autophagy-independent manner (J. Harris, unpublished observations).1146,1149 Thus, in studies where the effect of autophagy inhibition on specific cellular processes is being investigated, it is important to confirm results using other methods, such as RNA silencing. Due to these issues, it is of great interest that inhibitors with specificity for the class III PtdIns3Ks, and their consequent effects on autophagy, have been described.244,1150,1151
A mutant mouse line carrying a floxed allele of Pik3c3 has been created.1152 This provides a useful genetic tool that will help in defining the physiological role of the class III PtdIns3K with bona fide specificity by deleting the class III kinase in a cell type-specific manner in a whole animal using the Cre-LoxP strategy. For example, the phenotype resulting from a knockout of Pik3c3 specifically in the kidney glomerular podocytes (Pik3c3pdKO) indicates that there is no compensation by other classes of PtdIns3Ks or related Atg genes, thus highlighting the functional specificity and physiological importance of class III PtdIns3K in these cells.
Cycloheximide, a commonly used protein synthesis inhibitor in mammals, is also an inhibitor of sequestration in vivo,12-14,78,924,1153-1157 and in various cell types in vitro,466,1158 and it has been utilized to investigate the dynamic nature of the regression of various autophagic elements.12-14,25,78,1154,1155 The mechanism of action of cycloheximide in short-term experiments is not clear, but it has no direct relation to the inhibition of protein synthesis.466 This latter activity, however, may complicate certain types of analysis when using this drug.
A significant challenge for a more detailed analysis of the dynamic role of autophagy in physiological and pathophysiological processes, for instance with regard to cancer and cancer therapy, is to find more specific inhibitors of autophagy signaling which do not affect other signaling cascades. For example, in the context of cellular radiation responses it is well known that PI3Ks, in addition to signaling through the PI3K-AKT pathway, have a major role in the regulation of DNA-damage repair.1159 However, 3-MA, which is a nonspecific inhibitor of these lipid kinases, can alter the function of other classes of this enzyme, which are involved in the DNA-damage repair response. This is of particular importance for investigations into the role of radiation-induced autophagy in cellular radiation sensitivity or resistance.1160,1161
Most other inhibitory drugs act at post-sequestration steps. These types of agents have been used in many experiments to both inhibit endogenous protein degradation and to increase the number of autophagic compartments. They cause the accumulation of sequestered material in either autophagosomes or autolysosomes, or both, because they allow autophagic sequestration to proceed. The main categories of these types of inhibitors include the vinca alkaloids (e.g., vinblastine) and other microtubule poisons that inhibit fusion, inhibitors of lysosomal enzymes (e.g., leupeptin, pepstatin A and E-64d), and compounds that elevate lysosomal pH (e.g., inhibitors of V-ATPases such as bafilomycin A1 and concanamycin A [another V-ATPase inhibitor], and weak base amines including methyl- or propylamine, chloroquine, and Neutral Red, some of which slow down fusion). Ammonia is a very useful agent for the elevation of lysosomal pH in short-term experiments, but it has been reported to cause a stimulation of autophagy during long-term incubation of cells in a full medium,1162 under which conditions a good alternative might be methylamine or propylamine.1163 Along these lines, it should be noted that the half-life of glutamine in cell culture media is approximately 2 weeks due to chemical decomposition, which results in media with lowered glutamine and elevated ammonia concentrations that can affect the autophagic flux (either inhibiting or stimulating autophagy, depending on the concentration1164). Thus, to help reduce experimental variation, the use of freshly prepared cell culture media with glutamine is advised. A special note of caution is also warranted in regard to chloroquine. Although this chemical is commonly used as an autophagy inhibitor, chloroquine may initially stimulate autophagy (F.C. Dorsey, personal communication; R. Franco, personal communication). In addition, culture conditions requiring acidic media preclude the use of chloroquine because intracellular accumulation of the chemical is dramatically reduced by low pH.1165 To overcome this issue, it is possible to use acid compounds that modulate autophagy, such as betulinic acid and its derivatives.235,1166-1168 Betulinic acid damages lysosomal function differing from traditional inbibitors (e.g., chloroquine, NH4Cl or bafilomycin A1) that raise the lysosomal pH; betulinic acid interacts with pure phospholipid membranes,235,1169 and is capable of changing membrane permeability.235,1170,1171 The lysosomal damage mediated by betulinic acid is capable of compromising autophagy without any incremental damage when lysosomal function is altered by lysosomal inhibitors (e.g., chloroquine or bafilomycin A1);235 however, betulinic acid is not lysosome specific, and will affect other organelles such as mitochondria.
Some data suggest that particular nanomaterials may also be novel inhibitors of autophagy, by as yet unidentified mechanisms.1172
It is worth noting that lysosomal proteases fall into 3 general groups, cysteine, aspartic acid and serine proteases. Therefore, the fact that leupeptin, a serine and cysteine protease inhibitor, has little or no effect does not necessarily indicate that lysosomal degradation is not taking place; a combination of leupeptin, pepstatin A and E-64d may be a more effective treatment. However, it should also be pointed out that these protease inhibitors can exert inhibitory effects not only on lysosomal proteases, but also on cytosolic proteases; that is, degradation of proteins might be blocked through inhibition of cytosolic instead of lysosomal proteases. Conversely, it should be noted that MG132 (Z-leu-leu-leu-al) and its related peptide aldehydes are commonly used as proteasomal inhibitors, but they can also inhibit certain lysosomal hydrolases such as cathepsins and calpains.1173 Thus, any positive results using MG132 do not rule out the possibility of involvement of the autophagy-lysosomal system. Therefore, even if MG132 is effective in inhibiting autophagy, it is important to confirm the result using more specific proteasomal inhibitors such as lactacystin or epoxomicin. Finally, there are significant differences in cell permeability among protease inhibitors. For example, E-64d is membrane permeable, whereas leupeptin and pepstatin A are not (although there are derivatives that display greater permeability such as pepstatin A methyl ester).1174 Thus, when analyzing whether a protein is an autophagy substrate, caution should be taken in utilizing these protease inhibitors to block autophagy.
As with the PtdIns3K inhibitors, many autophagy-suppressive compounds are not specific. For example, okadaic acid1175 is a powerful general inhibitor of both type 1 (PPP1) and type 2A (PPP2) protein phosphatases.1176 Bafilomycin A1 and other compounds that raise the lysosomal pH may have indirect effects on any acidified compartments. Moreover, treatment with bafilomycin A1 for extended periods (18 h) can cause significant disruption of the mitochondrial network in cultured cells (M.E. Gegg, personal communication), and either bafilomycin A1 or concanamycin A cause swelling of the Golgi in plants,1177 and increase cell death by apoptosis in cancer cells (V.A. Rao, personal communication). Furthermore, bafilomycin A1 may have off-target effects on the cell, particularly on MTORC1.487,527,1178 Bafilomycin A1 is often used at a final concentration of 100 nM, but much lower concentrations such as 1 nM may be sufficient to inhibit autophagic-lysosomal degradation and are less likely to cause indirect effects.157,225,1179 For example, in pulmonary A549 epithelial cells bafilomycin A1 exhibits concentration-dependent effects on cellular morphology and on protein expression; at concentrations of 10 and 100 nM the cells become more rounded accompanied by increased expression of VIM (vimentin) and a decrease in CDH1/E-cadherin (B. Yeganeh, M. Post and S. Ghavami, unpublished observations). Thus, appropriate inhibitory concentrations should be empirically determined for each cell type.231
Although these various agents can inhibit different steps of the autophagic pathway, their potential side effects must be considered in interpretation of the secondary consequences of autophagy inhibition, especially in long-term studies. For example, lysosomotropic compounds can increase the rate of autophagosome formation by inhibiting MTORC1, as activation of lysosomally localized MTORC1 depends on an active V-ATPase (as well as RRAG GTPases162).487,1180 Along these lines, chloroquine treatment may cause an apparent increase in the formation of autophagosomes possibly by blocking fusion with the lysosome (F.C. Dorsey and J.L. Cleveland, personal communication). This conclusion is supported by the finding that chloroquine reduces the colocalization of LC3 and LysoTracker despite the presence of autophagosomes and lysosomes (A.K. Simon, personal communication). This mechanism might be cell-type specific, as other studies report that chloroquine prevents autolysosome clearance and degradation of cargo content, but not autophagosome-lysosome fusion.1181-1184 Concanamycin A blocks sorting of vacuolar proteins in plant cells in addition to inhibiting vacuolar acidification.1185 Furthermore, in addition to causing the accumulation of autophagic compartments, many of these drugs seem to stimulate sequestration in many cell types, especially in vivo.79,326,924,1154,1158,1186-1190 Although it is clear why these drugs cause the accumulation of autophagic compartments, it is not known why they stimulate sequestration. One possibility, at least for hepatocytes, is that the inhibition of protein degradation reduces the intracellular amino acid pool, which in turn upregulates sequestration. A time-course study of the changes in both the intra- and extracellular fractions may provide accurate information regarding amino acid metabolism. For these various reasons, it is important to include appropriate controls; along these lines, MTOR inhibitors such as rapamycin or amino acid deprivation can be utilized as positive controls for inducing autophagy. In many cell types, however, the induction of autophagy by rapamycin is relatively slow, or transient, allowing more time for indirect effects.
Several small molecule inhibitors, including torin1, PP242, KU-0063794, PI-103 and NVP-BEZ235, have been developed that target the catalytic domain of MTOR in an ATP-competitive manner.225,1191-1195 In comparison to rapamycin, these catalytic MTOR inhibitors are more potent, and hence are stronger autophagy agonists in most cell lines.341,1193,1196 The use of these second-generation MTOR inhibitors may reveal that some reports of MTOR-independent autophagy may actually reflect the use of the relatively weak inhibitor rapamycin. Furthermore, the use of these compounds has revealed a role for MTORC1 and MTORC2 as independent regulators of autophagy.1197
Neurons, however, seem to be a particular case in regard to their response to MTOR inhibitors. Rapamycin may fail to activate autophagy in cultured primary neurons, despite its potent stimulation of autophagy in some cancer cell lines,75,544,1198 Interestingly, both rapamycin and catalytic MTOR inhibitors do not induce a robust autophagy in either cultured primary mouse neurons or human neuroblastoma SH-SY5Y cells, which can differentiate into neuron-like cells, whereas the drugs do elicit a potent autophagic response in cultured astrocytes (J. Diaz-Nido and R. Gargini, personal communication). This suggests a differential regulation of autophagy in neurons. It has been suggested that control of neuronal autophagy may reflect the particular physiological adaptations and metabolic requirements of neurons, which are very different from most peripheral cell types.1199 For example, acute starvation in transgenic mice expressing GFP-LC3 leads to a potent induction of autophagy in the liver, muscle and heart but not in the brain.153 Along these lines, glucose depletion may be much more efficient at inducing autophagy than rapamycin or amino acid starvation in neurons in culture (M. Germain and R. Slack, personal communication). Indeed treatment of cultured primary mouse neurons and human neuroblastoma SH-SY5Y cells with 2-deoxy-glucose, which hampers glucose metabolism and leads to activation of AMPK, results in robust autophagy induction (J. Diaz-Nido and R. Gargini, personal communication). Interestingly, a number of compounds can also be quite efficient autophagy inducers in neurons including the CAPN/calpain inhibitor calpeptin.1200-1202 Thus, it has been suggested that autophagy induction in neurons may be achieved by molecular mechanisms relying on AMPK or increases in intracellular calcium concentration.1199 An example where changes in cytosolic calcium levels, due to the incapacity of the mitochondria to buffer Ca2+ release, result in an increase in autophagy is seen in a cellular model of the neurodegenerative disease Friedreich ataxia, based on FXN/frataxin silencing in SH-SY5Y human neuroblastoma cells.1203
Finally, a specialized class of compounds with α,β-unsaturated ketone structure tends to induce autophagic cell death, accompanied by changes in mitochondrial morphology. Since the cytotoxic action of these compounds is efficiently blocked by N-acetyl-L-cysteine, the β-position in the structure may interact with an SH group of the targeted molecules.1204 Due to the potential pleiotropic effects of various drug treatments, it is incumbent upon the researcher to demonstrate that autophagy is indeed inhibited, by using the methodologies described herein. Accordingly, it is critical to verify the effect of a particular biochemical treatment with regard to its effects on autophagy induction or inhibition when using a cell line that was previously uncharacterized for the chemical being used. Similarly, cytotoxicity of the relevant chemical should be assessed.
The use of gene deletions/inactivations (e.g., in primary or immortalized atg-/- MEFs,540 plant T-DNA or transposon insertion mutants,282,1205 or in vivo using transgenic knockout models1206,1207 including Cre-lox based “conditional” knockouts320,321) or functional knockdowns (e.g., with RNAi against ATG genes) is the preferred approach when possible because these methods allow a more direct assessment of the resulting phenotype; however, different floxed genes are deleted with varying efficiency, and the proportion deleted must be carefully quantified.1208 Studies also suggest that microRNAs may be used for blocking gene expression.243,645,646,1209,246-248 In most contexts, it is advisable when using a knockout or knockdown approach to examine multiple autophagy-related genes to exclude the possibility that the phenotype observed is due to effects on a nonautophagic function(s) of the corresponding protein, especially when examining the possibility of autophagic cell death. This is particularly the case in evaluating BECN1, which interacts with anti-apoptotic BCL2 family proteins,566 or when low levels of a target protein are sufficient for maintaining autophagy as is the case with ATG5.255 With regard to ATG5, a better approach may be to use a dominant negative (K130R) version.1144,1198,1210 Also noteworthy is the role of ATG5 in mitotic catastrophe544 and several other nonautophagic roles of ATG proteins (see Noncanonical use of autophagy-related proteins).75 Along these lines, and as stated above for the use of inhibitors, when employing a knockout or especially a knockdown approach, it is again incumbent upon the researcher to demonstrate that autophagy is actually inhibited, by using the methodologies described herein.
Finally, we note that the long-term secondary consequences of gene knockouts or knockdowns are likely much more complex than the immediate effects of the actual autophagy inhibition. To overcome this concern, inducible knockout systems might be useful.255,404 One additional caveat to knockdown experiments is that PAMP recognition pathways can be triggered by double-stranded RNAs (dsRNA), like siRNA probes, or the viral vector systems that deliver shRNA.1211 Some of these, like TLR-mediated RNA recognition,1212 can influence autophagy by either masking any inhibitory effect or compromising autophagy independent of the knockdown probe. Therefore, nontargeting (scrambled) siRNA or shRNA controls should be used with the respective transfection or transduction methods in the experiments that employ ATG knockdown. Another strategy to specifically interfere with autophagy is to use dominant negative inhibitors. Delivery of these agents by transient transfection, adenovirus, or TAT-mediated protein transduction offers the possibility of their use in cell culture or in vivo.1210 However, since autophagy is an essential metabolic process for many cell types and tissues, loss of viability due to autophagy inhibition always has to be a concern when analyzing cell death-unrelated questions. In this respect it is noteworthy that some cell-types of the immune system such as dendritic cells333 seem to tolerate loss of autophagy fairly well, whereas others such as T and B cells are compromised in their development and function after autophagy inhibition.1213,1214
In addition to pharmacological inhibition, RNA silencing, gene knockout and dominant negative RAB and ATG protein expression, pathogen-derived autophagy inhibitors can also be considered to manipulate autophagy. Along these lines ICP34.5, viral BCL2 homologs and viral FLIP of herpesviruses block autophagosome formation,566,892,1215 whereas M2 of influenza virus and HIV-1 Nef block autophagosome degradation.362,902 However, as with other tools discussed in this section, transfection or transduction of viral autophagy inhibitors should be used in parallel with other means of autophagy manipulation, because these proteins are used for the regulation of usually more than one cellular pathway by the respective pathogens.
There are fewer compounds that act as inducers of autophagy, but the initial characterization of this process was due in large part to the inducing effects of glucagon, which appears to act through indirect inhibition of MTOR via the activation of STK11/LKB1-AMPK.935,936,1216 Currently, the most commonly used inducer of autophagy is rapamycin, an allosteric inhibitor of MTORC1 (although as mentioned above, catalytic inhibitors such as torin1 are increasingly being used). Nevertheless, one caution is that MTOR is a major regulatory protein that is part of several signaling pathways, including for example those that respond to INS/insulin, EGF/epidermal growth factor and amino acids, and it thus controls processes other than autophagy, so rapamycin will ultimately affect many metabolic pathways.504,1217-1219 In particular, the strong effects of MTOR on protein synthesis may be a confounding factor when analyzing the effects of rapamycin. MTOR-independent regulation can be achieved through lithium, sodium valproate and carbamazepine, compounds that lower the myo-inositol 1,4,5-triphosphate levels,1220 as well as FDA-approved compounds such as verapamil, trifluoperazine and clonidine.1221,1222 In vivo treatment of embryos with cadmium results in an increase in autophagy, probably to counter the stress, allowing cell survival through the elimination/recycling of damaged structures.956 Autophagy may also be regulated by the release of calcium from the ER under stress conditions;297,1175,1223,1224 however, additional calcium signals from other stores such as the mitochondria and lysosomes could also play an important role in autophagy induction. The activation of the lysosomal TPCN/two-pore channel (two pore segment channel), by nicotinic acid adenine dinucleotide phosphate (NAADP) induces autophagy, which can selectively be inhibited by the TPCN blocker NED-19, or by pre-incubation with BAPTA, showing that lysosomal calcium also modulates autophagy.1225 Cell penetrating autophagy-inducing peptides, such as Tat-vFLIP or Tat-Beclin 1 (Tat-BECN1), are also potent inducers of autophagy in cultured cells as well as in mice.1216,1227 Other cell penetrating peptides, such as Tat-wtBH3D or Tat-dsBH3D, designed to disrupt very specific regulatory interactions such as the BCL2-BECN1 interaction, are potent, yet very specific, inducers of autophagy in cultured cells.1227
In contrast to other PtdIns3K inhibitors, caffeine induces macroautophagy in the food spoilage yeast Zygosaccharomyces bailii,1228 mouse embryonic fibroblasts,1229 and S. cerevisiae1230 at millimolar concentrations. In higher eukaryotes this is accompanied by inhibition of the MTOR pathway. Similarly, in budding yeast caffeine is a potent TORC1 inhibitor suggesting that this drug induces autophagy via inhibition of the TORC1 signalling pathway; however, as with other PtdIns3K inhibitors caffeine targets other proteins, notably Mec1/ATR and Tel1/ATM, and affects the cellular response to DNA damage.
Another autophagy inducer is the histone deacetylase inhibitor valproic acid.1231,1232 The mechanism by which valproic acid stimulates autophagy is not entirely clear but may occur due to inhibition of the histone deacetylase Rpd3, which negatively regulates the transcription of ATG genes (most notably ATG81233) and, via deacetylation of Atg3, controls Atg8 lipidation.1234
It is also possible, depending on the organism or cell system, to modulate autophagy through transcriptional control. For example, this can be achieved either through overexpression or post-translational activation of the gene encoding TFEB (see Transcriptional and translational regulation), a transcriptional regulator of the biogenesis of both lysosomes and autophagosomes.625,635 Similarly, adenoviral-mediated expression of the transcription factor CEBPB induces autophagy in hepatocytes.644 Recently, it has been shown that either the genetic ablation or the knockdown of the nucleolar transcription factor RRN3/TIF-IA, a crucial regulator of the recruitment of POLR1/RNA polymerase I to ribosomal DNA promoters, induces autophagy in neurons and in MCF-7 cancer cells, respectively, linking ribosomal DNA transcription to autophagy.1235,1236
Relatively little is known about direct regulation via the ATG proteins, but there is some indication that tamoxifen acts to induce autophagy by increasing the expression of BECN1 in MCF7 cells.1237 However, BECN1 does not appear to be upregulated in U87MG cells treated with tamoxifen, whereas the levels of LC3-II and SQSTM1 are increased, while LAMP2B is downregulated and CTSD and CTSL activities are almost completely blocked (K.S. Choi, personal communication). Thus, the effect of tamoxifen may differ depending on the cell type. Other data suggest that tamoxifen acts by blocking cholesterol biosynthesis, and that the sterol balance may determine whether autophagy acts in a protective versus cytotoxic manner.1238,1239 Finally, screens have identified small molecules that induce autophagy independently of rapamycin and allow the removal of misfolded or aggregate-prone proteins,1222,1240 suggesting that they may prove useful in therapeutic applications. However, caution should be taken because of the crosstalk between autophagy and the proteasomal system. For example, trehalose, an MTOR-independent autophagy inducer,1241 can compromise proteasomal activity in cultured primary neurons.1242
Because gangliosides are implicated in autophagosome morphogenesis, pharmacological or genetic impairment of gangliosidic compartment integrity and function can provide useful information in the analysis of autophagy. To deplete cells of gangliosides, an inhibitor of CERS/ceramide synthase, such as a fungal metabolite produced by Fusarium moniliforme (fumonisin B1), or, alternatively, siRNA to CERS or ST8SIA1, can be used.595
Finally, in addition to genetic and chemical compounds, it was recently reported that electromagnetic fields can induce autophagy in mammalian cells. Studies of biological effects of novel therapeutic approaches for cancer therapy based on the use of noninvasive radiofrequency fields reveals that autophagy, but not apoptosis, is induced in cancer cells in response to this treatment, which leads to cell death.1244 This effect is tumor specific and different from traditional ionizing radiation therapy that induces apoptosis in cells.
Conclusion: Considering that pharmacological inhibitors or activators of autophagy have an impact on many other cellular pathways, the use of more than one methodology, including molecular methods, is desirable. Rapamycin is less effective at inhibiting MTOR and inducing autophagy than catalytic inhibitors; however, it must be kept in mind that catalytic inhibitors also affect MTORC2. The main concern with pharmacological manipulations is pleiotropic effects of the compound being used. Accordingly, genetic confirmation is preferred whenever possible.
3. Basal autophagy
Basal levels of LC3-II or GFP-LC3 puncta may change according to the time after addition of fresh medium to cells, and this can lead to misinterpretations of what basal autophagy means. This is particularly important when comparing the levels of basal autophagy between different cell populations (such as knockout versus wild-type clones). If cells are very sensitive to nutrient supply and display a high variability of basal autophagy, the best experimental condition is to monitor the levels of basal autophagy at different times after the addition of fresh medium. One example is the chicken lymphoma DT40 cells (see Chicken B-lymphoid DT40 cells) and their knockout variant for all 3 ITPR isoforms.1244-1246 In these cells, no differences in basal levels of LC3-II can be observed up to 4 h after addition of fresh medium, but differences can be observed after longer times (J.M. Vicencio and G. Szabadkai, personal communication). This concept should also be applied to experiments in which the effect of a drug upon autophagy is the subject of study. If the drugs are added after a time in which basal autophagy is already high, then the effects of the drug can be masked by the cell's basal autophagy, and wrong conclusions may be drawn. To avoid this, fresh medium should be added first (followed by incubation for 2–4 h) in order to reduce and equilibrate basal autophagy in cells under all conditions, and then the drugs can be added. The basal autophagy levels of the cell under study must be identified beforehand to know the time needed to reduce basal autophagy.
A similar caution must be exercised with regard to cell culture density and hypoxia. When cells are grown in normoxic conditions at high cell density, HIF1A/HIF-1α is stabilized at levels similar to that obtained with low-density cultures under hypoxic conditions.1247 This results in the induction of BNIP3 and BNIP3L and “hypoxia”-induced autophagy, even though the conditions are theoretically normoxic.1248 Therefore, researchers need to be careful about cell density to avoid accidental induction of autophagy.
It should be realized that in yeast species, medium changes can trigger a higher “basal” level of autophagy in the cells. In the methylotrophic yeast species P. pastoris and Hansenula polymorpha a shift of cells grown in batch from glucose to methanol results in stimulation of autophagy.1249,1250 A shift to a new medium can be considered a stress situation. Thus, it appears to be essential to cultivate the yeast cells for a number of hours to stabilize the level of basal autophagy before performing experiments intended to study levels of (selective) autophagy (e.g., pexophagy). Finally, plant root tips cultured in nutrient-sufficient medium display constitutive autophagic flux (i.e., a basal level), which is enhanced in nutrient-deprived medium.1132,1251,1252
Conclusion: The levels of basal autophagy can vary substantially and can mask the effects of the experimental parameters being tested. Changes in media and growth conditions need to be examined empirically to determine the effects on basal autophagy and the appropriate times for subsequent manipulations.
4. Experimental systems
Throughout these guidelines we have noted that it is not possible to state explicit rules that can be applied to all experimental systems. For example, some techniques may not work in particular cell types or organisms. In each case, efficacy of autophagy promoters, inhibitors and measurement techniques must be empirically determined, which is why it is important to include appropriate controls. Differences may also be seen between in vivo or perfused organ studies and cell culture analyses. For example, INS/insulin has no effect on proteolysis in suspended rat hepatocytes, in contrast to the result with perfused rat liver. The INS/insulin effect reappears, however, when isolated hepatocytes are incubated in stationary dishes1253,1254 or are allowed to settle down on the matrix (D. Häussinger, personal communication). The reason for this might be that autophagy regulation by insulin and some amino acids requires volume sensing via integrin-matrix interactions and also intact microtubules.1255-1257 Along these lines, the use of whole embryos makes it possible to investigate autophagy in multipotent cells, which interact among themselves in their natural environment, bypassing the disadvantages of isolated cells that are deprived of their normal network of interactions.956 In general, it is important to keep in mind that results from one particular system may not be generally applicable to others.
Conclusion: Although autophagy is conserved from yeast to human, there may be tremendous differences in the specific details among systems. Thus, results based on one system should not be assumed to be applicable to another.
5. Nomenclature
To minimize confusion regarding nomenclature, we make the following notes: In general, we follow the conventions established by the nomenclature committees for each model organism whenever appropriate guidelines are available, and briefly summarize the information here using “ATG1” as an example for yeast and mammals. The standard nomenclature of autophagy-related wild-type genes, mutants and proteins for yeast is ATG1, atg1 (or atg1Δ in the case of deletions) and Atg1, respectively, according to the guidelines adopted by the Saccharomyces Genome Database (http://www.yeastgenome.org/gene_guidelines.shtml). For mammals we follow the recommendations of the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/mgihome/nomen/), which dictates the designations Atg1, atg1 and ATG1 (for all rodents), respectively, and the guidelines for human genes established by the HUGO Nomenclature Committee (http://www.genenames.org/guidelines.html), which states that human gene symbols are in the form ATG1 and recommends that proteins use the same designation without italics, as with ATG1; mutants are written for example as ATG1-/-.1258
C. Methods and challenges of specialized topics/model systems
There are now a large number of model systems being used to study autophagy. These guidelines cannot cover every detail, and as stated in the Introduction this article is not meant to provide detailed protocols. Nonetheless, we think it is useful to briefly discuss what techniques can be used in these systems and to highlight some of the specific concerns and/or challenges. We also refer readers to the 3 volumes of Methods in Enzymology that provide additional information for “nonstandard” model systems.39-41
1. C. elegans
C. elegans has a single ortholog of most yeast Atg proteins; however, 2 nematode homologs exist for Atg4, Atg8 and Atg16.1260-1262 Multiple studies have established C. elegans as a useful multicellular genetic model to delineate the autophagy pathway and associated functions (see for example refs. 271, 633, 742, 743, 1263). The LGG-1/Atg8/LC3 reporter is the most commonly used tool to detect autophagy in C. elegans. Similar to Atg8, which is incorporated into the double membrane of autophagic vacuoles during autophagy,148,269,600 the C. elegans LGG-1 localizes into cytoplasmic puncta under conditions known to induce autophagy. Fluorescent reporter fusions of LGG-1/Atg8 with GFP, DsRED or mCherry have been used to monitor autophagosome formation in vivo, in the nematode. These reporters can be expressed either in specific cells and tissues or throughout the animal.271,742,1263,1264 Caution should be taken, however, when using protein markers fused to mCherry in worms. mCherry aggregates in autophagy-inducing conditions, such as fasting, even if not fused to LGG-1 or other autophagy markers (E. O'Rourke, personal communication); therefore mCherry puncta may not be a good readout to monitor autophagy in C. elegans. LGG-2 is the second LC3 homolog and is also a convenient marker for autophagy either using specific antibodies741 or fused to GFP,1265 especially when expressed from an integrated transgene to prevent its germline silencing.741 The exact function of LGG-1 versus LGG-2 remains to be addressed.1266
For observing autophagy by GFP-LGG-1/2 (LC3) fluorescence in C. elegans, it is best to use integrated versions of the marker741,742,1267 (GFP::LGG-1 and GFP::LGG-2; Fig. 27) rather than extrachromosomal transgenic strains271,1265 because the latter show variable expression among different animals or mosaic expression (C. Kang, personal communication; V. Galy, personal communication). Nevertheless, evaluation of GFP::LGG-1 puncta is mostly restricted to seam cells, which is tedious because of a small number of puncta/cell even in autophagy-inducing conditions (<5/cell), error prone due to high background levels in the GFP channel, and extremely difficult to visualize in the adult. To increase signal to noise, it is also possible to carry out indirect immunofluorescence microscopy using antibodies against endogenous LGG-1, 633,742 or LGG-2;741 however, anti-LGG-1 and anti-LGG-2 antibodies are not commercially available. In addition, with the integrated version, or with antibodies directed against endogenous LGG-1, it is possible to perform a western blot analysis for lipidation, at least in embryos (LGG-1-I is the nonlipidated soluble form and LGG-1-II/LGG-1–PE is the lipidated form).1267,742,633
Figure 27.
GFP::LGG-1 and GFP::LGG-2 are autophagy markers in C. elegans. (A–F) Animals were generated that carry an integrated transgene expressing a GFP-tagged version of lgg-1, the C. elegans ortholog of mammalian MAP1LC3. Representative green fluorescence images in the pharyngeal muscles of (A) control RNAi animals without starvation, (B) control RNAi animals after 9 d of starvation, (C) atg-7 RNAi animals after 9 d of starvation, (D) starvation-hypersensitive gpb-2 mutants without leucine after 3 d of starvation, and (E) gpb-2 mutants with leucine after 3 d of starvation. The arrows show representative GFP::LGG-1-positive punctate areas that label pre-autophagosomal and autophagosomal structures. (F) The relative levels of PE-conjugated and unconjugated GFP::LGG-1 were determined by western blotting. These figures were modified from data previously published in ref. 1267, Kang, C., Y.J. You, and L. Avery. 2007. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes & Development. 21:2161–2171, Copyright © 2007, Genes & Development by Cold Spring Harbor Laboratory Press and Kang, C., and L. Avery. 2009. Systemic regulation of starvation response in Caenorhabditis elegans. Genes & Development. 23:12–17, Copyright © 2011, Genes & Development by Cold Spring Harbor Laboratory Press, www.genesdev.org. (G–H) GFP:LGG-2 serves as a marker for autophagosomes in early C. elegans embryos. (G) GFP::LGG-2 expressed in the germline from an integrated transgene reveals the formation of autophagosomes (green) around sperm-inherited membranous organelles (red). DNA of the 2 pronuclei is stained (blue). (H) Later during development, GFP::LGG-2-positive structures are present in all cells of the embryo. Scale bar: 10 µm. Images provided by V. Galy.
The LGG-1 precursor accumulates in the atg-4.1 mutant, but is undetectable in wild-type embryos.1260 Moreover, the banding pattern of LGG-1 or LGG-1 fused to fluorescent proteins in western blots may not be easy to interpret in larvae or the adult C. elegans because enrichment for a fast running band (the lipidated form) is not observed in some autophagy-inducing conditions, such as fasting. In the embryos of some autophagy mutants, including epg-3, epg-4, epg-5, and epg-6 mutants, levels of LGG-1-I and LGG-1-II are elevated.563,633,1268,1269 In an immunostaining assay, endogenous LGG-1 forms distinct punctate structures, mostly at the ∼64- to 100-cell embryonic stage. LGG-1 puncta are absent in atg-3, atg-7, atg-5 and atg-10 mutant embryos,633,1261 but dramatically accumulate in other autophagy mutants.563,633 The widely used GFP::LGG-1 reporter forms aggregates in atg-3 and atg-7 mutant embryos, in which endogenous LGG-1 puncta are absent, indicating that GFP::LGG-1 could be incorporated into protein aggregates during embryogenesis. Immunostaining for endogenous VPS-34 is also a useful marker of autophagy induction in C. elegans embryos.1270
A variety of protein aggregates, including PGL granules (PGL-1-PGL-3-SEPA-1) and the C. elegans SQSTM1 homolog SQST-1, are selectively degraded by autophagy during embryogenesis; impaired autophagy activity results in their accumulation and the generation of numerous aggregates.633,1262 Thus, degradation of these autophagy substrates can also be used to monitor autophagy activity, with similar cautionary notes to those described in section A3 (see SQSTM1 and related LC3 binding protein turnover assays) for the SQST-1 turnover assay. Similar to mammalian cells, the total amount of GFP::LGG-1 along with SQST-1::GFP transcriptional expression coupled with its posttranscriptional accumulation can be informative with regard to autophagic flux in the embryo and in adult animals (again with the same cautionary notes described in section A3).629,1271
As with its mammalian counterpart, loss of the C. elegans TP53 ortholog, cep-1, increases autophagosome accumulation1272 and extends the animal's life span.1273 bec-1- and cep-1-regulated autophagy is also required for optimal life-span extension and to reduce lipid accumulation in response to silencing FRH-1/frataxin, a protein involved in mitochondrial respiratory chain functionality.1274 FRH-1 silencing also induces mitophagy in an evolutionarily conserved manner.1271 Moreover, the products of C. elegans mitophagy regulatory gene homologs (PDR-1/PARK2, PINK-1/PINK1, DCT-1/BNIP3, and SQST-1/SQSTM1) are required for induction of mitophagy (monitored through the Rosella biosensor1275) and life-span extension following FRH-1 silencing and iron deprivation.1271 The TFEB ortholog HLH-30 transcriptionally regulates macroautophagy and promotes lipid degradation,624,824 and worm life-span analyses uncovered a direct role for HLH-30/TFEB in life-span regulation in C. elegans, and likely in mammals.624,629,823
For a more complete review of methods for monitoring autophagy in C. elegans see ref. 1276. Note that most of these approaches have been optimized to monitor autophagy in embryos or early larval stages, and that autophagy markers in the adult C. elegans are currently rather poorly characterized or lacking.
2. Chicken B-lymphoid DT40 cells, retina and inner ear
The chicken B-lymphoid DT40 cell line represents a suitable tool for the analysis of autophagic processes in a nonmammalian vertebrate system. In DT40 cells, foreign DNA integrates with a very high frequency by homologous recombination compared to random integration. This makes the cell line a valuable tool for the generation of cellular gene knockouts. Generally, the complete knockout of genes encoding autophagy-regulatory proteins is preferable compared to RNAi-mediated knockdown, because in some cases these proteins function normally when expressed at reduced levels.255 Different Atg-deficient DT40 cell lines already exist, including atg13-/-, ulk1-/-, ulk2-/-, ulk1/2-/-,1278 becn1-/-, and rb1cc1/fip200-/- (B. Stork, personal communication). Many additional non-autophagy-related gene knockout DT40 cell lines have been generated and are commercially available.1278
DT40 cells are highly proliferative (the generation time is approximately 10 h), and knockout cells can be easily reconstituted with cDNAs by retroviral gene transfer for the mutational analysis of signaling pathways. DT40 cells mount an autophagic response upon starvation in EBSS,1277 and autophagy can be analyzed by a variety of assays in this cell line. Steady state methods that can be used include TEM, LC3 western blotting and fluorescence microscopy; flux measurements include monitoring LC3-II turnover and tandem mRFP/mCherry-GFP-LC3 fluorescence microscopy. Using atg13-/- and ulk1/2-/- DT40 cells, it was shown that ATG13 and its binding capacity for RB1CC1/FIP200 are mandatory for both basal and starvation-induced autophagy, whereas ULK1/2 and in vitro-mapped ULK1-dependent phosphorylation sites of ATG13 appear to be dispensable for these processes.1277
Another useful system is chick retina, which can be used for monitoring autophagy at different stages of development. For example, lipidation of LC3 is observed during starvation, and can be blocked with a short-term incubation with 3-MA.393,394 LEP-100 antibody is commercially available for the detection of this lysosomal protein. In the developing chicken inner ear, LC3 flux can be detected in otic vesicles cultured in a serum-free medium exposed to either 3-MA or chloroquine.395
One of the salient features of chicken cells, including primary cells such as chicken embryo fibroblasts, is the capacity of obtaining rapid, efficient and sustained transcript/protein downregulation with replication-competent retrovirus for shRNA expression.1279 In chicken embryo fibroblasts, nearly complete and general (i.e., in nearly all cells) protein downregulation can be observed within a few days after transfection of the shRNA retroviral vector.167
Cautionary notes: Since the DT40 cell line derives from a chicken bursal lymphoma, not all ATG proteins and autophagy-regulatory proteins are detected by the commercially available antibodies produced against their mammalian orthologs; however, commercially available antibodies for mammalian LC3 and GABARAP have been reported to detect the chicken counterparts in western blots.167 The chicken genome is almost completely assembled, which facilitates the design of targeting constructs. However, in the May 2006 chicken (Gallus gallus) v2.1 assembly, 5% of the sequence has not been anchored to specific chromosomes, and this might also include genes encoding autophagy-regulatory proteins. It is possible that there is some divergence within the signaling pathways between mammalian and nonmammalian model systems. One example might be the role of ULK1/2 in starvation-induced autophagy described above. Additionally, neither rapamycin nor torin1 seem to be potent inducers of autophagy in DT40 cells, although MTOR activity is completely repressed as detected by the level of phosphorylated RPS6KB via western blotting.1277 Finally, DT40 cells represent a transformed cell line, being derived from an avian leukosis virus-induced bursal lymphoma. Thus, DT40 cells release avian leukosis virus into the medium, and the 3′-long terminal repeat has integrated upstream of the MYC gene, leading to increased MYC expression.1280 Both circumstances might influence basal and starvation-induced autophagy.
3. Chlamydomonas
The unicellular green alga Chlamydomonas reinhardtii is an excellent model system to investigate autophagy in photosynthetic eukaryotes. Most of the ATG genes that constitute the autophagy core machinery including the ATG8 and ATG12 ubiquitin-like systems are conserved as single-copy genes in the nuclear genome of this model alga. Autophagy can be monitored in Chlamydomonas by western blotting through the detection of Atg8 lipidation as well as an increase in the abundance of this protein in response to autophagy activation.292 Localization of Atg8 by immunofluorescence microscopy can also be used to study autophagy in Chlamydomonas since the cellular distribution of this protein changes drastically upon autophagy induction. The Atg8 signal is weak and usually detected as a single spot in nonstressed cells, whereas autophagy activation results in the localization of Atg8 in multiple spots with a very intense signal.292,1281,1282 Finally, enhanced expression of ATG8 and other ATG genes has also been reported in stressed Chlamydomonas cells.1281 These methodological approaches have been used to investigate the activation of autophagy in Chlamydomonas under different stress conditions including nutrient (nitrogen or carbon) limitation, rapamycin treatment, ER stress, oxidative stress, photo-oxidative damage or high light stress.292,1281,1282
4. Drosophila
Drosophila provides an excellent system for in vivo analysis of autophagy, partly because the problem of animal-to-animal variability can be circumvented by the use of clonal mutant cell analysis, a major advantage of this model system. In this scenario, somatic clones of cells are induced that either overexpress the gene of interest, or silence the gene through expression of a transgenic RNA interference construct, or homozygous mutant cells are generated. These gain- or loss-of-function clones are surrounded by wild-type cells, which serve as an internal control for autophagy induction. In such an analysis, autophagy in these genetically distinct cells is always compared to neighboring cells of the same tissue, thus eliminating most of the variability and also ruling out potential non-cell-autonomous effects that may arise in mutant animals. Along these lines, clonal analysis should be an integral part of in vivo Drosophila studies when possible. Multiple steps of the autophagic pathway can now be monitored in Drosophila due to the recent development of useful markers, corresponding to every step of the process. Interested readers may find further information in 2 reviews with a detailed discussion of the currently available assays and reagents for the study of autophagy in Drosophila.135,1283
A commercial rabbit monoclonal anti-GABARAP (anti-Atg8) antibody can be used to detect endogenous levels of Drosophila Atg8a in both immunostaining and immunoblotting experiments.1284 Western blotting and fluorescence microscopy have been used successfully in Drosophila by monitoring flies expressing human GFP-LC3,88,279 GFP-Atg8a1285 or using any of several antibodies directed against the endogenous Atg8 protein.510,623,1286 In addition, cultured Drosophila (S2) cells can be stably transfected with GFP fused to Drosophila Atg8a, which generates easily resolvable GFP-Atg8a and GFP-Atg8a–PE forms that respond to autophagic stimuli (S. Wilkinson, personal communication); stable S2 cells with GFP-Atg8a under the control of a 2-kb Atg8a 5′ UTR are also available.1287 Similarly, cultured Drosophila cells (l[2]mbn or S2) stably transfected with EGFP-HsLC3B respond to autophagy stimuli (nutrient deprivation) and inhibitors (3-MA, bafilomycin A1) as expected, and can be used to quantify GFP-LC3 puncta, which works best using fixed cells with the aid of an anti-GFP antibody.1288 However, in the Drosophila eye, overexpression of GFP-Atg8 results in a significant increase in Atg8–PE by western blot, and this occurs even in control flies in which punctate GFP-Atg8 is not detected by immunofluorescence (M. Fanto, unpublished results), and in transfected Drosophila Kc167 cells, uninducible but persistent GFP-Atg8 puncta are detected (A. Kiger, unpublished results). In contrast, expression of GFP-LC3 under the control of the ninaE/rh1 promoter in wild-type flies does not result in the formation of LC3-II detectable by western blot, nor the formation of punctate staining; however, increased GFP-LC3 puncta by immunofluorescence or LC3-II by western blot are observed upon activation of autophagy.442 Autophagy can also be monitored with mCherry-Atg18, which is displayed in punctate patterns that are very similar to mCherry-Atg8a.135 Tandem fluorescence reporters have been established in Drosophila in vivo, where GFP-mCherry-Atg8a can be expressed in the nurse cells of the developing egg chamber or in other cell types.135,1077 A Drosophila transgenic line (UAS-Ref[2]P-GFP) and different specific antibodies against Ref(2)P, the Drosophila SQSTM1 homolog, are available to follow Ref(2)P expression and localization.402,423,1289 The advantage of UAS-Ref(2)P-GFP over the antibody against endogenous Ref(2)P is that its accumulation is independent of Ref(2)P promoter regulation and unambiguously reflects autophagy impairment (M. Robin and B. Mollereau, unpublished results). Finally, it is worth noting that Atg5 antibody can be used in the Drosophila eye and the staining is similar to GFP-LC3.1290 In addition, Atg5-GFP and Atg6-GFP constructs are available in Drosophila.1291
5. Erythroid cells
The unique morphology of red blood cells (RBCs) is instrumental to their function. These cells have a bi-concave shape provided by a highly flexible membrane and a cytoplasm deficient in organelles. This architecture allows unimpeded circulation of the RBC even through the thinnest blood vessels, thereby delivering O2 to all the tissues of the body. Erythroid cells acquire this unique morphology upon terminal erythroid maturation, which commences in the bone marrow and is completed in the circulation. This process involves extrusion of the pycnotic nucleus through a specialized form of asymmetric division, and degradation of the ribosome and mitochondria machinery via a specialized form of autophagy (Fig. 28). In the context of RBC biogenesis, autophagy exerts a unique function to sculpt the cytoplasm, with the mature autophagic vacuoles engulfing and degrading organelles, such as mitochondria and ribosomes, whose presence would impair the flexibility of the cells.
Figure 28.
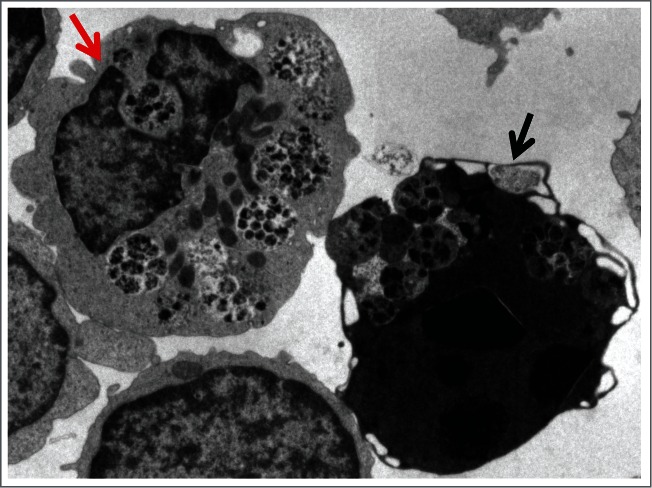
Transmission electron micrograph of erythroblasts obtained from the blood of regular donors after 10 days of culture in the presence of KITLG/SCF, IL3, EPO and dexamethasone. Original magnification 3000X. This figure shows 2 erythroblasts containing autophagic vacuoles. One erythroblast (red arrow) has the morphology of a live cell with several autophagic vacuoles that have engulfed cytoplasmic organelles. The other erythroblast (black arrow) has the electron-dense cytoplasm characteristic of a dead cell and is in the process of shedding its autolysosomes from the cytoplasm to the extracellular space. Image provided by A.R. Migliaccio and M. Zingariello.
Another unique feature of erythropoiesis is that expression of genes required for autophagosome assembly/function, such as LC3B, does not appear to be regulated by nutrient deprivation, but rather is upregulated by the erythroid-specific transcription factor GATA1.641 FOXO3, a transcription factor that modulates RBC production based on the levels of O2 present in the tissues,1292 amplifies GATA1-mediated activation of autophagy genes641 and additional genes required for erythroid maturation.1293 Furthermore, lipidation of the cytosolic form of LC3B into the lipidated LC3-II form is controlled by EPO (erythropoietin), the erythroid-specific growth factor that ensures survival of the maturing erythroid cells. The fact that the genes encoding the autophagic machinery are controlled by the same factors that regulate expression of genes encoding important red cell constituents (such as red blood cell antigens and cytoskeletal components, globin, and proteins mediating heme biosynthesis),1294-1296 ensures that the process of terminal maturation progresses in a highly ordered fashion.
The importance of autophagy for RBC production has been established through the use of mutant mouse strains lacking genes encoding proteins of the autophagy machinery (BNIP3L, ULK1, ATG7).1297-1300 These mutant mice exhibit erythroid cells blocked at various stages of terminal erythroid maturation and are anemic. Abnormalities of the autophagic machinery are also linked to anemia observed in certain human diseases, especially those categorized as ribosomopathies. As in other cell types, in erythroid cells TP53 activation may influence the functional consequences of autophagy—to determine cell death rather than maturation. TP53, through MDM2, is the gatekeeper to ensure normal ribosome biosynthesis by inducing death of cells lacking sufficient levels of ribosomal proteins. Diseases associated with congenic or acquired loss-of-function mutations of genes encoding ribosomal proteins, such as Diamond-Blackfan anemia or myelodysplastic syndrome, are characterized by activated TP53 and abnormally high levels of autophagic death of erythroid cells and anemia. Conversely, the anemia of at least certain Diamond-Blackfan anemia patients may be treated with glucocorticoids that inhibit TP53 activity.
6. Filamentous fungi
As in yeast, autophagy is involved in nutrient recycling during starvation.275,276,1301-1306 In addition, macroautophagy seems to be involved in many normal developmental processes such as sexual and asexual reproduction, where there is a need for reallocation of nutrients from one part of the mycelium to another to supply the developing spores and spore-bearing structures.276,726,1301,1302,1304,1307-1309 Similarly, autophagy also affects conidial germination under nitrogen-limiting conditions.276 In Podospora anserina, autophagy has been studied in relation to incompatibility reactions between mating strains where it seems to play a prosurvival role.274,1307 During aging of this long-standing aging model, autophagy is increased (numbers of GFP-Atg8 puncta and increased autophagy-dependent degradation of a GFP reporter protein) and acts as a prosurvival pathway.1310 Of special interest to many researchers of autophagy in filamentous fungi has been the possible involvement of autophagy in plant and insect pathogen infection and growth inside the host.275,709,1301,1302,1311-1314 Autophagy also appears to be necessary for the development of aerial hyphae,276,1302,1307,1312 and for appresorium function in M. oryzae, Colletotrichum orbiculare and Metarhizium robertsii.275,1311,1312,1314 Some of these effects could be caused by the absence of autophagic processing of storage lipids (lipophagy) to generate glycerol for increasing turgor and recycling the contents of spores into the incipient appressorium, as a prerequisite to infection.1301,1312,1313
Methods for functional analysis of autophagy have been covered in a review article (see ref. 1315). Most studies on autophagy in filamentous fungi have involved deleting some of the key genes necessary for autophagy, followed by an investigation of what effects this has on the biology of the fungus. Most commonly, ATG1, ATG4 and/or ATG8 have been deleted.275,1301,1302,1304,1305,1307,1312,1314,1316,1317 To confirm that the deletion(s) affects autophagy, the formation of autophagic bodies in the wild type and the mutant can be compared. In filamentous fungi the presence of autophagic bodies can be detected using MDC staining,275,1301 TEM275,1302 or fluorescence microscopy to monitor Atg8 tagged with a fluorescent protein.276,1304,1307 This type of analysis is most effective after increasing the number of autophagic bodies by starvation or alternatively by adding the autophagy-inducing drug rapamycin,276,1301 in combination with decreasing the degradation of the autophagic bodies through the use of the protease inhibitor PMSF.275,1302,1304,1307 In filamentous fungi it might also be possible to detect the accumulation of autophagic bodies in the vacuoles using differential interference contrast microscopy, especially following PMSF treatment.1304,1307 Additional information regarding the timing of autophagy induction can be gained by monitoring transcript accumulation of ATG1 and/or ATG8 using qRT-PCR.1302
Autophagy has been investigated intensively in Aspergilli, and in particular in the genetically amenable species Aspergillus nidulans, which is well suited to investigate intracellular traffic.1318 In A. oryzae, autophagy has been monitored by the rapamycin-induced and Atg8-dependent delivery of DsRed2, which is normally cytosolic, to the vacuoles.276 In A. nidulans, autophagy has been monitored by the more “canonical” GFP-Atg8 proteolysis assays, by monitoring the delivery of GFP-Atg8 to the vacuole (by time-lapse microscopy), and by directly following the biogenesis of GFP-Atg8-labeled phagophores and autophagosomes, which can be tracked in large numbers using kymographs traced across the hyphal axis. In these kymographs, the autophagosome cycle starting from a PAS “draws” a cone whose apex and base correspond to the “parental” PAS punctum and to the diameter of the “final” autophagosome, respectively.1319 Genetic analyses revealed that autophagosomes normally fuse with the vacuole in a Rab7-dependent manner. However, should Rab7 fusogenic activity be mutationally inactivated, autophagosomes can traffic to the endosomes in a RabB/Rab5- and CORVET-dependent manner.1319 An important finding was that RabO/Rab1 plays a key role in A. nidulans autophagy (and actually can be observed on the phagophore membranes). This finding agrees with previous work in S. cerevisiae demonstrating that Ypt1 (the homolog of RAB1) is activated by the Trs85-containing version of TRAPP, TRAPPIII, for autophagy.1320,1321 This crucial involvement of RabO/Ypt1 points at the ER as one source of membrane for autophagosomes. The suitability of A. nidulans for in vivo microscopy has been exploited to demonstrate that nascent phagophores are cradled by ER-associated structures resembling mammalian omegasomes.1319 The macroautophagic degradation of whole nuclei that has been observed in A. oryzae721 might be considered as a specialized version of reticulophagy. Finally, autophagosome biogenesis has also been observed using a PtdIns3P-binding GFP-tagged FYVE domain probe in mutant cells lacking RabB/Rab5. Under these genetic conditions Vps34 cannot be recruited to endosomes and is entirely at the disposition of autophagy,1320 such that PtdIns3P is only present in autophagic membranes.
Mitophagy has been studied in M. oryzae, by detecting the endogenous level of porin (a mitochondrial outer membrane protein) by western blot, and by microscopy observation of vacuolar accumulation of mito-GFP.709 Mitophagy is involved in regulating the dynamics of mitochondrial morphology and/or mitochondrial quality control, during asexual development and invasive growth in M. oryzae. Pexophagy has also been studied in rice-blast fungus and it serves no obvious biological function, but is naturally induced during appressorial development, likely for clearance of excessive peroxisomes prior to cell death.1322 Methods to monitor pexophagy in M. oryzae include microscopy observation of the vacuolar accumulation of GFP-SRL (peroxisome-localized GFP), and detection of the endogenous thiolase,1323 or Pex14 levels.
7. Food biotechnology
Required for yeast cell survival under a variety of stress conditions, autophagy has the potential to contribute to the outcome of many food fermentation processes. For example, autophagy induction is observed during the primary fermentation of synthetic grape must1323 and during sparkling wine production (secondary fermentation).1324 A number of genome-wide studies have identified vacuolar functions and autophagy as relevant processes during primary wine fermentation or for ethanol tolerance, based on gene expression data or cell viability of knockout yeast strains.1323,1325-1329 However, determining the relevance of autophagy to yeast-driven food fermentation processes requires experimentation using some of the methods available for S. cerevisiae as described in these guidelines.
Autophagy is a target for some widespread food preservatives used to prevent yeast-dependent spoilage. For example, the effect of benzoic acid is exacerbated when concurrent with nitrogen starvation.1330 This observation opened the way to devise strategies to improve the usefulness of sorbic and benzoic acid, taking advantage of their combination with stress conditions that would require functional autophagy for yeast cell survival.1228 Practical application of these findings would also require extending this research to other relevant food spoilage yeast species, which would be of obvious practical interest.
In the food/health interface, the effect of some food bioactive compounds on autophagy in different human cell types has already attracted some attention.1331,1332 Interpreting the results of this type of research, however, warrants 2 cautionary notes.1333 First, the relationship between health status and autophagic activity is obviously far from being direct. Second, experimental design in this field must take into account the actual levels of these molecules in the target organs after ingestion, as well as exposure time and their transformations in the human body. In addition, attention must be paid to the fact that several mechanisms might contribute to the observed biological effects. Thus, relevant conclusions about the actual involvement of autophagy on the health-related effect of food bioactive compounds would only be possible by assaying the correct molecules in the appropriate concentrations.
8. Honeybee
The reproductive system of bees, or insects whose ovaries exhibit a meroistic polytrophic developmental cycle can be a useful tool to analyze and monitor physiological autophagy. Both queen and worker ovaries of Africanized A. mellifera display time-regulated features of cell death that are, however, linked to external stimuli.1334 Features of apoptosis and autophagy are frequently associated with the degeneration process in bee organs, but only more recently has the role of autophagy been highlighted in degenerating bee tissues. The primary method currently being used to monitor autophagy is following the formation of autophagosomes and autolysosomes by TEM. This technique can be combined with cytochemical and immunohistochemical detection of acid phosphatase as a marker for autolysosomes.1335,1336 Acidotropic dyes can also be used to follow autophagy in bee organs, as long as the cautions noted in this article are followed. The honeybee genome has been sequenced, and differential gene expression has been used to monitor Atg18 in bees parasitized by Varroa destructor.1337
9. Human
Considering that much of the research conducted today is directed at understanding the functioning of the human body, in both normal and disease states, it is pertinent to include humans and primary human tissues and cells as important models for the investigation of autophagy. Although clinical studies are not readily amenable to these types of analyses, it should be kept in mind that the MTORC1 inhibitor rapamycin, the lysosomal inhibitors chloroquine and hydroxychloroquine, and the microtubule depolymerizing agent colchicine are all available as clinically approved drugs. However, these drugs have serious side effects, which often impede their clinical use to study autophagy (e.g., severe immunosuppressive effects of rapamycin; gastrointestinal complaints, bone marrow depression, neuropathy and acute renal failure induced by colchicine; gastrointestinal complaints, neuropathy and convulsions, and retinopathy induced by [hydroxy]chloroquine). Theses side effects may in part be exacerbated by potential inhibition of macroautophagy in itself by these drugs.1338 In cancer treatment, for example, autophagy-inhibiting drugs are used in combination with other anticancer drugs to increase their potency. Conversely, normal tissues such as kidney induce macroautophagy in response to anticancer drugs to resist their toxicity;1339 additional blockade of autophagy could worsen normal tissue toxicity and cause serious side effects. Therefore, the potential for serious adverse effects and toxicity of these drugs warrants caution, especially when studying a role of autophagy in high-risk patients, such as the critically ill. Fortunately, it is possible to obtain fresh biopsies of some human tissues. Blood, in particular, as well as samples of adipose and muscle tissues, can be obtained from needle biopsies or from elective surgery. For example, in a large study, adipocytes were isolated from pieces of adipose tissue (obtained during surgery) and examined for INS/insulin signaling and autophagy. It was demonstrated that autophagy was strongly upregulated (based on LC3 flux, EM, and lipofuscin degradation) in adipocytes obtained from obese patients with type 2 diabetes compared with nondiabetic subjects.294 In another study utilizing human adipose tissue biopsies and explants, elevated autophagic flux in obesity was associated with increased expression of several autophagy genes.217,609
The study of autophagy in the blood has revealed that SNCA may represent a further marker to evaluate the autophagy level in T lymphocytes isolated from peripheral blood.1340 In these cells it has been shown that (a) knocking down the SNCA gene results in increased macroautophagy, (b) autophagy induction by energy deprivation is associated with a significant decrease of SNCA levels, (c) macroautophagy inhibition (e.g., with 3-MA or knocking down ATG5) leads to a significant increase of SNCA levels, and d) SNCA levels negatively correlate with LC3-II levels. Thus, SNCA, and in particular the 14-kDa monomeric form, can be detected by western blot as a useful tool for the evaluation of macroautophagy in primary T lymphocytes. In contrast, the analysis of SQSTM1 or NBR1 in freshly isolated T lymphocytes fails to reveal any correlation with either LC3-II or SNCA, suggesting that these markers cannot be used to evaluate basal macroautophagy in these primary cells. Conversely, LC3-II upregulation is correlated with SQSTM1 degradation in neutrophils, as demonstrated in a human sepsis model.1034
A major caveat of the work concerning autophagy on human tissue is the problem of postmortem times, agonal state, premortem clinical history (medication, diet, etc.) and tissue fixation. Time to fixation is typically longer in autopsy material than when biopsies are obtained. For tumors, careful sampling to avoid necrosis, hemorrhagic areas and non-neoplastic tissue is required. The problem of fixation is that it can diminish the antibody binding capability; in addition, especially in autopsies, material is not obtained immediately after death.1341,1342 The possibilities of postmortem autolysis and fixation artifacts must always be taken into consideration when interpreting changes attributed to autophagy.1343 Analyses of these types of samples require not only special antigen retrieval techniques, but also histopathological experience to interpret autophagy studies by IHC, immunofluorescence or TEM. Nonetheless, at least one recent study demonstrated that LC3 and SQSTM1 accumulation can be readily detected in autopsy-derived cardiac tissue from patients with chloroquine- and hydroxychloroquine-induced autophagic vacuolar cardiomyopathy.962 Despite significant postmortem intervals, sections of a few millimeters thickness cut from fresh autopsy brain and fixed in appropriate glutaraldehyde-formalin fixative for EM, can yield TEM images of sufficient ultrastructural morphology to discriminate different autophagic vacuole subtypes and their relative regional abundance in some cases (R. Nixon, personal communication).
The situation is, however, typically problematic with TEM, where postmortem delays can cause vacuolization. Researchers experienced in the analysis of TEM images corresponding to autophagy should be able to identify these potential artifacts because autophagic vacuoles should contain cytoplasm. While brain biopsies may be usable for high quality TEM (Fig. 29, 30), this depends upon proper handling at the intraoperative consultation stage, and such biopsies are performed infrequently except for brain tumor diagnostic studies. Conversely, biopsies of organs such as the digestive tract, the liver, muscle and the skin are routinely performed and thus nearly always yield high-quality TEM images. When possible, nonsurgical biopsies are preferable since surgery is usually performed in anesthetized and fasting patients, 2 conditions possibly affecting autophagy. Moreover, certain surgical procedures require tissue ischemia-reperfusion strategies that can also affect autophagy level.1344 An analysis that examined liver and skeletal muscle from critically ill patients utilized tissue biopsies that were taken within 30 ± 20 min after death and were flash-frozen in liquid nitrogen followed by storage at -80°C.1061 Samples could subsequently be used for EM and western blot analysis.
Figure 29.
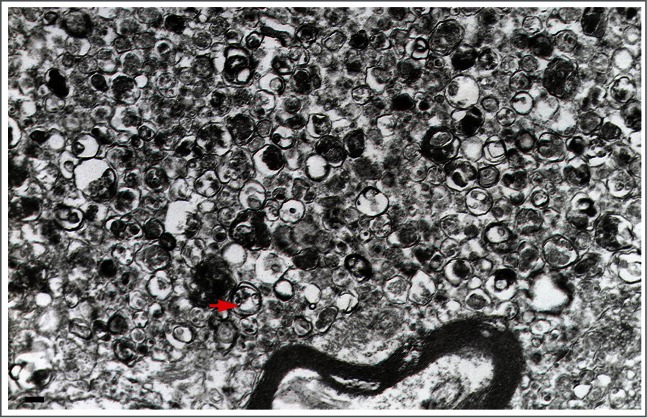
A large dystrophic neurite from a brain biopsy of a patient with Gerstmann-Sträussler-Scheinker disease not unlike those reported for Alzheimer disease.60 This structure is filled with innumerable autophagic vacuoles, some of which are covered by a double membrane. Electron dense lysosomal-like structures are also visible. The red arrow points to a double-membrane autophagic vacuole. Scale bar: 200 nm. Image provided by P. Liberski.
Figure 30.
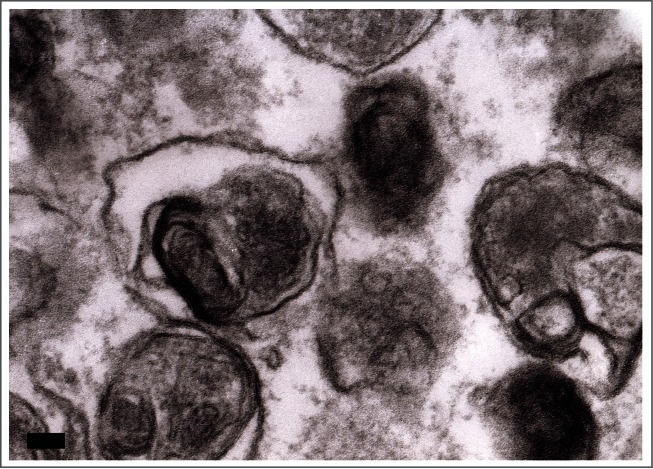
A high-power electron micrograph from a brain biopsy showing autophagic vacuoles in a case of ganglioglioma. Scale bar: 200 nm. Image provided by P. Liberski.
A major limitation of studying patient biopsies is that only static measurements can be performed. This limitation does not apply, however, for dynamic experiments on tissue biopsies or cells derived from biopsies, as described above.294 Multiple measurements over time, especially when deep (vital) organs are involved, are impossible and ethically not justifiable. Hence, quantitative flux measurements are virtually impossible in patients. To overcome these problems to the extent possible and to gain a more robust picture of the autophagic status, observational studies need to include 2 different aspects. First, a static marker for phagophore or autophagosome formation needs to be measured. This can be done by assessing ultrastructural changes with TEM and/or on the molecular level by measuring LC3-II protein levels. Second, accumulation of autophagy substrates, such as SQSTM1 and (poly)ubiquitinated proteins, can provide information on the overall efficacy of the pathway and can be a surrogate marker of the consequences of altered autophagic flux, especially when autophagy is insufficient, although these changes can also be affected by the ubiquitin-proteasome system as mentioned above. In addition, and even more so when problems with specific pathways are suspected (e.g., mitophagy), specific substrates of these pathways should be determined. Again, none of these measurements on its own provides enough information on (the efficacy of) autophagy, because other processes may confound every single parameter. However, the combination of multiple analyses should be informative. Of note, there has been recent interest in assessing markers of autophagy and autophagic flux in right atrial biopsy samples obtained from patients undergoing cardiac surgery.1345,1346 Evidence to date suggests that cardiac surgery may be associated with an increase in autophagic flux, and that this response may protect the heart from perioperative cardiac ischemia-reperfusion injury.1345 Although still in its infancy with regard to autophagy, it is worth pointing out that mathematical modeling has the power to bridge whole body in vivo data with in vitro data from tissues and cells. The usefulness of so-called hierarchical or multilevel modeling has thus been demonstrated when examining the relevance of INS/insulin signaling to glucose uptake in primary human adipocytes compared with whole-body glucose homeostasis.1347
Lipophagy is an important pathway of lipid droplet clearance in hepatocytes, and the extent of lipophagy modulates the lipid content in these cells. Hepatocytes break down lipid droplets through lipophagy as a pathway of endogenous lipid clearance in response to hormones or daily rhythms of nutrient supply.1062 LC3-II colocalizes with lipid droplets, indicating a role for autophagy in the mobilization of free fatty acids.817 Little is known regarding the changes of lipophagy under pathological conditions, such as drug toxicity, alcoholic steatohepatitis or nonalcoholic steatohepatitis (NASH). The accumulation of lipid droplets in hepatocytes activates ATG5 in the droplets, and initiates a lipophagy process; in addition, increased influx of fatty acids in hepatocytes results in oxidant stress, ER stress and autophagy,1348,1349 as indicated by the fact that there is enhanced staining of LC3-II in NASH tissue.1348,1350 However, autophagic flux is impaired in liver specimens of NASH patients as indicated by increased levels of SQSTM1.1351 Therefore, the value of using LC3-II staining in tissue as an indication of autophagy or lipophagy is in question.
A stepwise process can be proposed for linking changes in the autophagic pathway to changes in disease outcome. First, in an observational study, the changes in the autophagic pathway (see above) should be quantified and linked to changes in disease outcome. To prove causality, a subsequent autophagy-modifying intervention should be tested in a randomized study. Before an intervention study is performed in human patients, the phenotype of (in)active autophagy contributing to poor outcome should be established in a validated animal model of the disease. For the validation of the hypothesis in an animal model, a similar 2-step process is suggested, with the assessment of the phenotype in a first stage, followed by a proof-of-concept intervention study (see Large animals).
10. Hydra
Hydra is a freshwater cnidarian animal that provides a unique model system to test autophagy. The process can be analyzed either in the context of nutrient deprivation, as these animals easily survive several weeks of starvation,1352,1353 or in the context of regeneration, because in the absence of protease inhibitors, bisection of the animals leads to an uncontrolled wave of autophagy. In the latter case, an excess of autophagy in the regenerating tip immediately after amputation is deleterious.1354-1356 Most components of the autophagy and MTOR pathways are evolutionarily conserved in Hydra.1353 For steady-state measurements, autophagy can be monitored by western blot for ATG8/LC3, by immunofluorescence (using antibodies to ATG8/LC3, lysobisphosphatidic acid or RPS6KA/RSK), or with dyes such as MitoFluor Red 589 and LysoTracker Red. Flux measurements can be made by following ATG8/LC3 turnover using lysosomal protease inhibitors (leupeptin and pepstatin A) or in vivo labeling using LysoTracker Red. It is also possible to monitor MTOR activity with phosphospecific antibodies to RPS6KB and EIF4EBP1 or to examine gene expression by semiquantitative RT-PCR, using primers that are designed for Hydra. Autophagy can be induced by RNAi-mediated knockdown of Kazal1,1354,1355 or with rapamycin treatment, and can be inhibited with wortmannin or bafilomycin A1.1352,1353
11. Large animals
This section refers in particular to mammals other than humans. Assessment of autophagy (and, in particular, autophagic flux) in clinically relevant large animal models is critical in establishing its (patho)physiological role in multiple disease states. For example, evidence obtained in swine suggests that upregulation of autophagy may protect the heart against damage caused by acute myocardial infarction/heart attack.1357 Ovine models of placental insufficiency leading to intrauterine growth restriction have shown that there is no change in the expression of markers of autophagy in the fetus in late gestation1358 or in the lamb at 21 d after birth.1359 Furthermore, there is an increase in markers of autophagy in the placenta of human intrauterine growth restriction pregnancies.1360 Studies in rabbits suggest a protective role of upregulated autophagy against critical illness-induced multiple organ failure and muscle weakness,1361,1362 which is corroborated by human studies.1060,1061 Conversely, autophagy may contribute to the pathogenesis of some types of tissue injury, at least in the lung.1363,1364
Autophagy also plays an important role in the development and remodeling of the bovine mammary gland. In vitro studies with the use of a 3-dimensional culture model of bovine mammary epithelial cells (MECs) have shown that this process is involved in the formation of fully developed alveoli-like structures.1365 Earlier studies show that intensified autophagy is observed in bovine MECs at the end of lactation and during the dry period, when there is a decrease in the levels of lactogenic hormones, increased expression of auto/paracrine apoptogenic peptides, increased influence of sex steroids and enhanced competition between the intensively developing fetus and the mother organism for nutritional and bioactive compounds.1366,1367 These studies were based on some of the methods described elsewhere in these guidelines, including GFP-Atg8/LC3 fluorescence microscopy, TEM, and western blotting of LC3 and BECN1. Creation of a specific GFP-LC3 construct by insertion of cDNA encoding bovine LC3 into the pEGFP-C1 vector makes it possible to observe induction of autophagy in bovine MECs in a more specific manner than can be achieved by immunofluorescence techniques, in which the antibodies do not show specific reactivity to bovine cells and tissues.1365,1367 However, it is important to remember that definitive confirmation of cause-and-effect is challenging for studies on large animals, given the lack or poor availability of specific antibodies and other molecular tools, the frequent inability to utilize genetic approaches, and the often prohibitive costs of administering pharmacological inhibitors in these translational preparations.
In contrast with cell culture experiments, precise monitoring of autophagic flux is practically impossible in vivo in large animals. Theoretically, repetitive analyses of small tissue biopsies should be performed to study ultrastructural and molecular alterations over time in the presence or absence of an autophagy inhibitor (e.g., chloroquine). However, several practical problems impede applicability of this approach. First, repetitive sampling of small needle biopsies in the same animal (a major challenge by itself) could be assumed to induce artifacts following repetitive tissue destruction, especially when deep (vital) organs are involved. In addition, chemical inhibitors of autophagy have considerable side effects and toxicity, hampering their usage. Also, the general physical condition of an animal may confound results obtained with administration of a certain compound, for instance altered uptake of the compound when perfusion is worse.
Therefore, in contrast to cells, where it is more practical to accurately document autophagic flux, we suggest the use of a stepwise approach in animal models to provide a proof of concept with an initial evaluation of sequellae of (in)active autophagy and the relation to the outcome of interest.
First, prior to an intervention, the static ultrastructural and molecular changes in the autophagic pathway should be documented and linked to the outcome of interest (organ function, muscle mass or strength, survival, etc.). These changes can be evaluated by light microscopy, EM and/or by molecular markers such as LC3-II. In addition, the cellular content of specific substrates normally cleared by autophagy should be quantified, as, despite its static nature, such measurement could provide a clue about the results of altered autophagic flux in vivo. These autophagic substrates can include SQSTM1 and (poly)ubiquitinated substrates or aggregates, but also specific substrates such as damaged mitochondria. As noted above, measurement of these autophagic substrates is mainly informative when autophagic flux is prohibited/insufficient, and, individually, all have specific limitations for interpretation. As mentioned several times in these guidelines, no single measurement provides enough information on its own to reliably assess autophagy, and all measurements should be interpreted in view of the whole picture. In every case, both static measurements reflecting the number of autophagosomes (ultrastructural and/or molecular) and measurements of autophagic substrates as surrogate markers of autophagic flux need to be combined. Depending on the study hypothesis, essential molecular markers can further be studied to pinpoint at which stage of the process autophagy may be disrupted.
Second, after having identified a potential role of autophagy in mediating an outcome in a clinically relevant large animal model, an autophagy-modifying intervention should be tested. For this purpose, an adequately designed, randomized controlled study of sufficient size on the effect of a certain intervention on the phenotype and outcome can be performed in a large animal model. Alternatively, the effect of a genetic intervention can be studied in a small animal model with clinical relevance to the studied disease.
As mentioned above, exact assessment of autophagic flux requires multiple time points, which cannot be done in the same animal. Alternatively, different animals can be studied for different periods of time. Due to the high variability between animals, however, it is important to include an appropriate control group and a sufficiently high number of animals per time point as corroborated by statistical power analyses. This requirement limits feasibility and the number of time points that can be investigated. The right approach to studying autophagy in large animals likely differs depending on the question that is being addressed. Several shortcomings regarding the methodology, inherent to working with large animals, can be overcome by an adequate study design. As for every study question, the use of an appropriate control group with a sufficient number of animals is crucial in this regard.
12. Lepidoptera
Some of the earliest work in the autophagy field was carried out in the area of insect metamorphosis.1084 Microscopy and biochemical research revealed autophagy during the metamorphosis of American silkmoths and the tobacco hornworm, Manduca sexta, and included studies of the intersegmental muscles, but they did not include molecular analysis of autophagy. Overall, these tissues cannot be easily maintained in culture, and antibodies against mammalian proteins often do not work. Accordingly, these studies were confined to biochemical measurements and electron micrographs. During metamorphosis, the bulk of the larval tissue is removed by autophagy and other forms of proteolysis.1368 Bombyx mori is now used as a representative model among Lepidoptera, for studying not only the regulation of autophagy in a developmental setting, but also the relations between autophagy and apoptosis. The advantages of this model are the large amount of information gathered on its developmental biology, physiology and endocrinology, the availability of numerous genetic and molecular biology tools, and a completely sequenced genome.1369 The basic studies of B. mori autophagy have been carried out in 4 main larval systems: the silk gland, the fat body, the midgut and the ovary.
The techniques used for these studies are comparatively similar, starting from EM, which is the most widely used method to follow the changes of various autophagic structures and other features of the cytosol and organelles that are degraded during autophagy.619,1370-1373 Immuno-TEM also can be used, when specific antibodies for autophagic markers are available. As in other model systems the use of Atg8 antibodies has been reported in Lepidoptera. In B. mori midgut619 and fat body,620 as well as in various larval tissues of Galleria mellonella1374 and Helicoverpa armigera,1375 the use of custom antibodies makes it possible to monitor Atg8 conversion to Atg8–PE by western blotting. Moreover transfection of GFP-Atg8 or mCherry-GFP-Atg8 has been used to study autophagy in several lepidopteran cell lines.1375 Activation of TOR can be monitored with a phosphospecific antibody against EIF4EBP1.620 Acidotropic dyes such as MDC and LysoTracker Red staining have been used as markers for autophagy in silkmoth egg chambers combined always with additional assays.1370,1371 Acid phosphatase also can be used as a marker for autolysosomal participation in these tissues.619,1372,1376 Systematic cloning and analysis revealed that homologs of most of the Atg genes identified in other insect species such as Drosophila are present in B. mori, and 14 Atg genes have now been identified in the silkworm genome, as well as other genes involved in the TOR signal transduction pathway.1377-1379 Variations in the expression of several of these genes have been monitored not only in silkworm larval organs, where autophagy is associated with development,619,1377,1378,1380 but also in the fat body of larvae undergoing starvation.1377,1381
In the IPLB-LdFB cell line, derived from the fat body of the caterpillar of the gypsy moth Lymantria dispar, indirect immunofluorescence experiments have demonstrated an increased number of Atg8-positive dots in cells with increased autophagic activity; however, western blotting did not reveal the conversion of Atg8 into Atg8–PE. Instead, a single band with an approximate molecular mass of 42 kDa was observed that was independent of the percentage of cells displaying punctate Atg8 (D. Malagoli, unpublished results). In contrast, with B. mori midgut, the use of an antibody specific for BmAtg8 makes it possible to monitor BmAtg8 processing to BmAtg8–PE by western blotting.619 Thus, the utility of monitoring Atg8 in insects may depend on the particular organism and antibody.
13. Marine invertebrates
The invaluable diversity of biological properties in marine invertebrates offers a unique opportunity to explore the different facets of autophagy at various levels from cell to tissue, and throughout development and evolution. For example, work on the tunicate Ciona intestinalis has highlighted the key role of autophagy during the late phases of development in lecithotrophic organisms (larvae during metamorphosis feed exclusively from the egg yolk resources).278,1382 This work has also helped in pinpointing the coexistence of autophagy and apoptosis in cells as well as the beneficial value of combining complementary experimental data such as LC3 immunolabeling and TUNEL detection. This type of approach could shed a new light on the close relationship between autophagy and apoptosis and provide valuable information about how molecular mechanisms control the existing continuum between these 2 forms of programmed cell death. Autophagy plays a key role in the resistance to nutritional stress as is known to be the case in many Mediterranean bivalve molluscs in the winter. For example, the European clam Ruditapes decussatus is able to withstand strict fasting for 2 mo, and this resistant characteristic is accompanied by massive macroautophagy in the digestive gland (Fig. 31). This phenomenon, observed by TEM, demonstrates once again the advantage of using this classical ultrastructural method to study autophagy in unconventional biological models for which molecular tools may not be operational. Finally, autophagy also appears to play a role in the cell renewal process observed during the regeneration of the carnivorous sponge Asbestopluma hypogea.1383 The presence of the autophagic machinery in this sister group of Eumetazoans should incite interest into considering the study of the molecular networks that regulate autophagy within an evolutionary framework.
Figure 31.
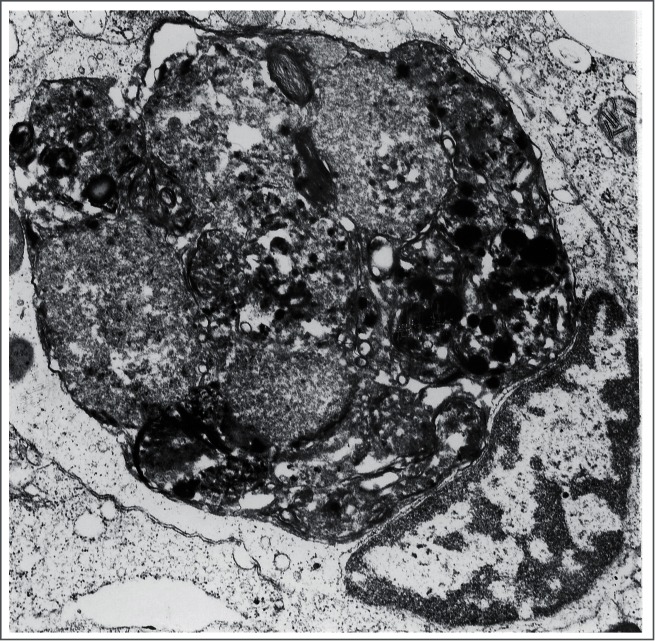
Macroautophagy in the digestive gland of Ruditapes decussatus (Mollusca, Bivalvia) subjected to a strict starvation of 2 months. Image provided by S. Baghdiguian.
14. Neotropical teleosts
In tropical environments, fish have developed different reproductive strategies, and many species have the potential for use as a biological model in cell and molecular biology, especially for studying the mechanisms that regulate gametogenesis and embryo development. In these fish, the ovary is a suitable experimental model system for studying autophagy and its interplay with cell death programs due to the presence of post-ovulatory follicles (POFs) and atretic follicles, which follow different routes during ovarian remodeling after spawning.1384 In the fish reproductive biology, POFs are excellent morphological indicators of spawning, whereas atretic follicles are relevant biomarkers of environmental stress. In addition, many freshwater teleosts of commercial value do not spawn spontaneously in captivity, providing a suitable model for studying the mechanisms of follicular atresia under controlled conditions.1385 When these species are subjected to induced spawning, the final oocyte maturation (resumption of meiosis) occurs, and POFs are formed and quickly reabsorbed in ovaries after spawning.1386 Assessment of autophagy in fish has been primarily made using TEM at different times of ovarian regression.1387 Due to the difficulty of obtaining antibodies specific for each fish species, immunodetection of ATG-proteins (mainly LC3 and BECN1) by IHC associated with analyses by western blotting can be performed using antibodies that are commercially available for other vertebrates.396 Such studies suggest dual roles for autophagy in follicular cells;1384 however, evaluation of the autophagic flux in different conditions is critical for establishing its physiological role during follicular regression and ovarian remodeling after spawning. Given the ease of obtaining samples and monitoring them during development, embryos of these fish are also suitable models for studying autophagy that is activated in response to different environmental stressors, particularly in studies in vivo.
15. Odontoblasts
Odontoblasts are long-lived dentin-forming postmitotic cells, which evolved from neural crest cells early during vertebrate evolution. These cells are aligned at the periphery of the dental pulp and are maintained during the entire healthy life of a tooth. As opposed to other permanent postmitotic cells such as cardiac myocytes or central nervous system neurons, odontoblasts are significantly less protected from environmental insult such as dental caries and trauma. Mature odontoblasts develop a well-characterized autophagy-lysosomal system, including a conspicuous autophagic vacuole that ensures turnover and degradation of cell components. Immunocytochemical and TEM studies make it possible to monitor age-related changes in autophagic activity in human odontoblasts.1388
16. Planarians
Because planarians are one of the favorite model systems in which to study regeneration and stem cell biology, these flatworms represent a unique model where it is possible to investigate autophagy in the context of regeneration, stem cells and growth. Currently the method used to detect autophagy is TEM. A detailed protocol adapted to planarians has been described.1389,1390 However, complementary methods to detect autophagy are also needed, since TEM cannot easily distinguish between activation and blockage of autophagy, which would both be observed as an accumulation of autophagosomes. Other methods to detect autophagy are being developed (C. González-Estévez, personal communication), including IHC and western blotting approaches for the planarian homolog of LC3. Several commercial antibodies against human LC3 have been tried for cross-reactivity without success, and 3 planarian-specific antibodies have been generated. Some preliminary results show that LysoTracker Red can be a useful reagent to analyze whole-mount planarians. Most of the components of the autophagy and MTOR signaling machinery are evolutionarily conserved in planarians. Whether autophagy genes vary at the mRNA level during starvation and after depletion of MTOR signaling components is still to be determined.
17. Plants
As stated above with regard to other organisms, staining with MDC or derivatives (such as monodansylamylamine) is not sufficient for detection of autophagy, as these stains also detect vacuoles. The same is true for the use of LysoTracker Red, Neutral Red or acridine orange. The fluorophore of the red fluorescent protein shows a relatively high stability under acidic pH conditions. Thus, chimeric RFP fusion proteins that are sequestered within autophagosomes and delivered to the plant vacuole can be easily detected by fluorescence microscopy. Furthermore, fusion proteins with some versions of RFP tend to form intracellular aggregates, allowing the development of a visible autophagic assay for plant cells.1391 For example, fusion of cytochrome b5 and the original (tetrameric) RFP generate an aggregated cargo protein that displays cytosolic puncta of red fluorescence and, following vacuolar delivery, diffuse staining throughout the vacuolar lumen. However, it is not certain whether these puncta represent autophagosomes or small vacuoles, and therefore these data should be combined with immuno-TEM or with conventional TEM using high-pressure frozen and freeze-substituted samples.1392
In plant studies, GFP-Atg8 fluorescence is typically assumed to correspond to autophagosomes; however, as with other systems, caution needs to be exercised because it cannot be ruled out that Atg8 is involved in processes other than autophagy. Immunolabeled GFP-Atg8 can be detected both on the inner and outer membrane of an autophagosome in an Arabidopsis root cell, using chemical fixation (see Fig. 6b in ref. 1393), suggesting that it will be a useful marker to monitor autophagy. Arabidopsis cells can be stably transfected with GFP fused to plant ATG8, and the lipidated and nonlipidated forms can be separated by SDS-PAGE.214 Furthermore, the GFP-ATG8 processing assay is particularly robust in Arabidopsis and can be observed by western blotting.215,256 Two kinds of GFP-ATG8 transgenic seeds are currently available from the Arabidopsis Biological Resource Center, each expressing similar GFP-ATG8a transgenes but having different promoter strength. One transgene is under the control of the stronger Cauliflower mosaic virus 35S promoter,542 while the other uses a promoter of the Arabidopsis ubiquitin10 gene.1394 In the GFP-ATG8 processing assay, the former has a higher ratio of GFP-ATG8a band intensity to that of free GFP than does the latter.1394 Since free GFP level reflects vacuolar delivery of GFP-ATG8, the ubiquitin promoter line may be useful when studying an inhibitory effect of a drug/mutation on autophagic delivery. Likewise, the 35S promoter line may be used for testing potential autophagy inducers.
Thus, as with other systems, autophagosome formation in plants can be monitored through the combined use of fluorescent protein fusions to ATG8, immunolabeling and TEM (Fig. 32). A tandem fluorescence reporter system is also available in Arabidopsis.1395 The number of fluorescent Atg8-labeled vesicles can be increased by pretreatment with concanamycin A, which inhibits vacuolar acidification;1095,1393 however, this may interfere with the detection of MDC and LysoTracker Red. It is also possible to use plant homologs of SQSTM1 and NBR1 in Arabidopsis1395 (the NBR1 homolog is called JOKA2 in tobacco1396) as markers for selective autophagy when constructed as fluorescent chimeras. In addition, detection of the NBR1 protein level by western blot, preferably accompanied by qPCR analysis of its transcript level, provides reliable semi-quantitative data about autophagic flux in plant cells.1397
Figure 32.
Detection of macroautophagy in tobacco BY-2 cells. (A) Induction of autophagosomes in tobacco BY-2 cells expressing YFP-NtAtg8 (shown in green for ease of visualization) under conditions of nitrogen limitation (Induced). Arrowheads indicate autophagosomes that can be seen as a bright green dot. No such structure was found in cells grown in normal culture medium (Control). Scale bar: 10 µm. N, nucleus; V, vacuole. (B) Ultrastructure of an autophagosome in a tobacco BY-2 cell cultured for 24 h without a nitrogen source. Scale bar: 200 µm. AP, autophagosome; CW, cell wall; ER, endoplasmic reticulum; P, plastid. Image provided by K. Toyooka.
It has been assumed that, just as in yeast, autophagic bodies are found in the vacuoles of plant cells, since both microautophagy and macroautophagy are detected in plant cells.1398 The data supporting this conclusion are mainly based on EM studies showing vesicles filled with material in the vacuole of the epidermis cells of Arabidopsis roots; these vesicles are absent in ATG4a and ATG4b mutant plants.282 However, it cannot be excluded that these vacuolar vesicles are in fact cytoplasmic/protoplasmic strands, or that they arrived at the vacuole independent of macroautophagy; although the amount of such strands would not be expected to increase following treatment with concanamycin. Immunolabeling with an antibody to detect ATG8 could clarify this issue.
Other methods described throughout these guidelines can also be used in plants.1399 For example, in tobacco cells cultured in sucrose starvation medium, the net degradation of cellular proteins can be measured by a standard protein assay; this degradation is inhibited by 3-MA and E-64c (an analog of E-64d), and is thus presumed to be due to autophagy.1400-1402
Cautionary notes: Although the detection of vacuolar RFP can be applied to both plant cell lines and to intact plants, it is not practical to measure RFP fluorescence in intact plant leaves, due to the very high red autofluorescence of chlorophyll in the chloroplasts. Furthermore, different autophagic induction conditions cause differences in protein synthesis rates; thus, special care should be taken to monitor the efficiency of autophagy by quantifying the intact and processed cargo proteins.
18. Protists
An essential role of autophagy during the differentiation of parasitic protists (formerly called protozoa) is clearly emerging. Only a few of the known ATG genes are present in these organisms, which raises the question about the minimal system that is necessary for the normal functioning of autophagy. The reduced complexity of the autophagic machinery in many protists provides a simplified model to investigate the core mechanisms of autophagosome formation necessary for selective proteolysis; accordingly, protist models have the potential to open a completely new area in autophagy research. Some of the standard techniques used in other systems can be applied to protists including indirect immunofluorescence using antibodies generated against ATG8 and the generation of stable lines expressing mCherry- or GFP-fused ATG8 for live microscopy and immuno-TEM analyses. Extrachromosomal constructs of GFP-ATG8 also work well with lower eukaryotes,287,288,1403 as do other fluorescently tagged ATG proteins including ATG5 and ATG12.
The unicellular amoeba Dictyostelium discoideum provides another useful system for monitoring autophagy.1404 The primary advantage of Dictyostelium is that it has a unique life cycle that involves a transition from a unicellular to a multicellular form. Upon starvation, up to 100,000 single cells aggregate by chemotaxis and form a multicellular structure that undergoes morphogenesis and cell-type differentiation. Development proceeds via the mound stage, the tipped aggregate and a motile slug, and culminates with the formation of a fruiting body that is composed of a ball of spores supported by a thin, long stalk made of vacuolized dead cells. Development is dependent on autophagy and, at present, all of the generated mutants in Dictyostelium autophagy genes display developmental phenotypes of varying severity.1404,1405 D. discoideum is also a versatile model to study infection with human pathogens and the role of autophagy in the infection process. The susceptibility of D. discoideum to microbial infection and its strategies to counteract pathogens are similar to those in higher eukaryotes.1406 Along these lines, Dictyostelium utilizes some of the proteins involved in autophagy that are not present in S. cerevisiae including ATG101 and VMP1, in addition to the core Atg proteins. The classical markers GFP-ATG8 and GFP-ATG18 can be used to detect autophagosomes by fluorescence microscopy. Flux assays based on the proteolytic cleavage of cytoplasmic substrates are also available.37,322
One cautionary note with regard to the use of GFP-ATG8 in protists is that these organisms display some “nonclassical” variations in their ATG proteins (see LC3-associated apicoplast) and possibly a wide phylogenetic variation since they constitute a paraphyletic taxon.1407 For example, Leishmania contains many apparent ATG8-like proteins (the number varying per species; e.g., up to 25 in L. major) grouped in 4 families, but only one labels true autophagosomes even though the others form puncta,287 and ATG12 requires truncation to provide the C-terminal glycine before it functions in the canonical way. Unusual variants in protein structures also exist in other protists, including apicomplexan parasites, for example, the malaria parasite Plasmodium spp. or Toxoplasma gondii, which express ATG8 with a terminal glycine not requiring cleavage to be membrane associated.1408 Thus, in each case care needs to be applied and the use of the protein to monitor autophagy validated. In addition, due to possible divergence in the upstream signaling kinases, classical inhibitors such as 3-MA, or inducers such as rapamycin, which are not as potent for trypanosomes1409 or apicomplexan parasites as in mammalian cells or yeast, must be used with caution (I. Coppens, personal communication);1403 however, RNAi knockdown of TORC1 (e.g., TOR1 or RPTOR) is effective in inducing autophagy in trypanosomes. In addition, small molecule inhibitors of the protein-protein interaction of ATG8 and ATG3 in Plasmodium falciparum have been discovered that are potent in cell-based assays and useable at 1–10 µM final concentration.1410,1411 Note that although the lysosomal protease inhibitors E64 and pepstatin block lysosomal degradative activity in Plasmodium, these inhibitors do not affect ATG8 levels and associated structures, suggesting a need for alternate methodologies to investigate autophagy in this model system.1412
In conventional autophagy, the final destination of autophagosomes is their fusion with lysosomes for intracellular degradation. However, T. gondii and certain stages of Plasmodium (insect and hepatic) lack degradative lysosomes, which makes questionable the presence of canonical autophagosomes and a process of autophagy in these parasites. Nevertheless, if protists employ their autophagic machineries in unconventional manners, studies of their core machinery of autophagy will provide information as to how autophagy has changed and adapted through evolution.
The scuticociliate Philasterides dicentrarchi has proven to be a good experimental organism for identifying autophagy-inducing drugs or for autophagy initiation by starvation-like conditions, since this process can be easily induced and visualized in this ciliate.1413 In scuticociliates, the presence of autophagic vacuoles can be detected by TEM, fluorescence microscopy or confocal laser scanning microscopy by using dyes such as MitoTracker Deep Red FM and MDC.
Finally, a novel autophagy event has been found in Tetrahymena thermophila, which is a free-living ciliated protist. A remarkable, virtually unique feature of the ciliates is that they maintain spatially differentiated germline and somatic nuclear genomes within a single cell. The germline genome is housed in the micronucleus, while the somatic genome is housed in the macronucleus. These nuclei are produced during sexual reproduction (conjugation), which involves not only meiosis and mitosis of the micronucleus and its products, but also degradation of some of these nuclei as well as the parental old macronucleus. Hence, there should be a mechanism governing the degradation of these nuclei. The inhibition of PtdIns3Ks with wortmannin or LY294002 results in the accumulation of additional nuclei during conjugation.1414 During degradation of the parental old macronucleus, the envelope of the nucleus becomes MDC- and LysoTracker Red-stainable without sequestration of the nucleus by a double membrane and with the exposure of certain sugars and phosphatidylserine on the envelope.1415 Subsequently, lysosomes fuse only to the old parental macronucleus, but other co-existing nuclei such as developing new macro- and micronuclei are unaffected.1415 Using gene technology it has been shown that ATG8 and VPS34 play critical roles in nuclear degradation.1416,1417 Knockout mutations of the corresponding genes result in a block in nuclear acidification, suggesting that these proteins function in lysosome-nucleus fusion. In addition, the envelope of the nucleus in the VPS34 knockout mutant does not become stainable with MDC. This evidence suggests that selective autophagy may be involved in the degradation of the parental macronucleus and implies a link between VPS34 and ATG8 in controlling this event.
19. Rainbow trout
Salmonids (e.g., salmon, rainbow trout) experience long periods of fasting often associated with seasonal reductions in water temperature and prey availability or spawning migrations. As such, they represent an interesting model system for studying and monitoring the long-term induction of autophagy. Moreover, the rainbow trout (Oncorhynchus mykiss) displays unusual metabolic features that may allow us to gain a better understanding of the nutritional regulation of this degradative system (i.e., a high dietary protein requirement, an important use of amino acids as energy sources, and an apparent inability to metabolize dietary carbohydrates). It is also probably one of the most deeply studied fish species with a long history of research carried out in physiology, nutrition, ecology, genetics, pathology, carcinogenesis and toxicology.1418 Its relatively large size compared to model fish, such as zebrafish or medaka, makes rainbow trout a particularly well-suited alternative model to carry out biochemical and molecular studies on specific tissues or cells that are impossible to decipher in small fish models. The genomic resources in rainbow trout are now being extensively developed; a high-throughput DNA sequencing program of EST has been initiated associated with numerous transcriptomics studies,1419-1422 and the full genome sequence is now available.
Most components of the autophagy and associated signaling pathways (AKT, TOR, AMPK, FOXO) are evolutionarily conserved in rainbow trout;628,1423-1425 however, not all ATG proteins and autophagy-regulatory proteins are detected by the commercially available antibodies produced against their mammalian orthologs. Nonetheless, the expressed sequence transcript databases facilitate the design of targeting constructs. For steady-state measurement, autophagy can be monitored by western blot or by immunofluorescence using antibodies to ATG8/LC3.1425 Flux measurements can be made in a trout cell culture model (for example, in primary culture of trout myocytes) by following ATG8/LC3 turnover in the absence and presence of bafilomycin A1. It is also possible to monitor the mRNA levels of ATG genes by real-time PCR using primer sequences chosen from trout sequences available in the above-mentioned expressed sequence transcript database. A major challenge in the near future will be to develop for this model the use of RNAi-mediated gene silencing to analyze the role of some signaling proteins in the control of autophagy, and also the function of autophagy-related proteins in this species.
20. Sea urchin
Sea urchin embryo is an appropriate model system for studying and monitoring autophagy and other defense mechanisms activated during physiological development and in response to stress.956 This experimental model offers the possibility of detecting LC3 through both western blot and immunofluorescence in situ analysis. Furthermore, in vivo staining of autolysosomes with acidotropic dyes can also be carried out. Studies on whole embryos make it possible to obtain qualitative and quantitative data for autophagy and also to get information about spatial localization aspects in cells that interact among themselves in their natural environment. Furthermore, since embryogenesis of this model system occurs simply in a culture of sea water, it is very easy to study the effects of inducers or inhibitors of autophagy by adding these substances directly into the culture. Exploiting this potential, it has recently been possible to understand the functional relationship between autophagy and apoptosis induced by cadmium stress during sea urchin development. In fact, inhibition of autophagy by 3-MA results in a concurrent reduction of apoptosis; however, using a substrate for ATP production, methyl pyruvate, apoptosis (assessed by TUNEL assay and cleaved CASP3 immunocytochemistry) is substantially induced in cadmium-treated embryos where autophagy is inhibited. Therefore, autophagy could play a crucial role in the stress response of this organism since it could energetically contribute to apoptotic execution through its catabolic role.1426 Cautionary notes include the standard recommendation that it is always preferable to combine molecular and morphological parameters to validate the data.
21. Ticks
In the hard tick Haemaphysalis longicornis, endogenous autophagy-related proteins (Atg6 and Atg12) can be detected by western blotting and/or by immunohistochemical analysis of midgut sections.1427,1428 It is also possible to detect endogenous Atg3 and Atg8 by western blotting using antibodies produced against the H. longicornis proteins (R. Umemiya-Shirafuji, unpublished results). Commercial antibodies against mammalian ATG orthologs (ATG3, ATG5, and BECN1) can also be used for western blotting. However, when the tick samples include blood of a host animal, the animal species immunized with autophagy-related proteins should be checked before use to avoid nonspecific background cross-reactivity. In addition to these methods, TEM is recommended to detect autophagosomes and autolysosomes. Although acidotropic dyes can be useful as a marker for autolysosomes in some animals, careful attention should be taken when using the dyes in ticks. Since the midgut epithelial cells contain acidic organelles (e.g., lysosomes) that are related to blood digestion during blood feeding, this method may cause confusion. It is difficult to distinguish between autophagy (autolysosomes) and blood digestion (lysosomes) with acidotropic dyes. Another available monitoring method is to assess the mRNA levels of tick ATG genes by real-time PCR.1429,1430 However, this method should be used along with other approaches such as western blotting, immunostaining, and TEM as described in this article. Unlike model insects, such as Drosophila, powerful genetic tools to assess autophagy are still not established in ticks. However, RNAi-mediated gene silencing is now well established in ticks,1431 and is currently being developed to analyze the function of autophagy-related genes in ticks during nonfeeding periods (R. Umemiya-Shirafuji, unpublished results) and in response to pathogen infection. Recently, “omics” technologies such as transcriptomics and proteomics have been applied to the study of apoptosis pathways in Ixodes scapularis ticks in response to infection with Anaplasma phagocytophilum.1432 I. scapularis, the vector of Lyme disease and human granulocytic anaplasmosis, is the only tick species for which genome sequence information is available (assembly JCVI_ISG_i3_1.0; http://www.ncbi.nlm.nih.gov/nuccore/NZ_ABJB000000000). For related tick species such as I. ricinus, mapping to the I. scapularis genome sequence is possible,1433 but for other tick species more sequence information is needed for these analyses.
22. Zebrafish
Zebrafish (Danio rerio) have many characteristics that make them a valuable vertebrate model organism for the analysis of autophagy. For example, taking advantage of the transparency of embryos, autophagosome formation can be visualized in vivo during development using transgenic GFP-Lc3 and GFP-Gabarap fish.36,1434,1435 Visualization of later-stage embryos is enhanced when medium is supplemented with 1-phenyl-2-thiourea, which inhibits melanogenesis, or through the use of strains with mutations affecting pigment production. Lysosomes can also be readily detected in vivo by the addition of LysoTracker Red to fish media prior to visualization. Additionally, protocols have been developed to monitor Lc3 protein levels and conjugation to PE by western blot analysis using commercially available Lc3 antibodies.36,1436
Because of their translucent character and external fertilization and development, zebrafish have proven to be an exceptional choice for developmental research. In situ hybridization of whole embryos can be performed to determine expression patterns. Knockdown of gene function is performed by treatment with morpholinos; the core autophagy machinery protein Gabarap,1437 and regulatory proteins such as the phosphoinositide phosphatase Mtmr14,1438 Raptor and Mtor,1439 have all been successfully knocked down by morpholino treatment. The CRISPR/Cas system is now being used for efficient targeted gene deletions.
Zebrafish are ideal organisms for in vivo drug discovery and/or verification because of their relatively small size and aqueous habitat, and several chemicals have been identified that modulate zebrafish autophagy activity.1436 Many chemicals can be added to the media and are absorbed directly through the skin. Because of simple drug delivery and rapid embryonic development, zebrafish are a promising organism for the study of autophagy's role in disease including Huntington disease,1201 Alzheimer disease,1440 and myofibrillar myopathy.1441,1442 In the case of infection, studies in zebrafish have made important contributions to understanding the role of bacterial autophagy in vivo.1443,1444 Zebrafish studies have also contributed to understanding the role of autophagy in different aspects of development, including cardiac morphogenesis, caudal fin regeneration,1445 and muscle and brain development.1434,1446,1447
D. Noncanonical use of autophagy-related proteins
1. LC3-associated phagocytosis
Although the lipidation of LC3 to form LC3-II is a commonly used marker of macroautophagy, studies have established that LC3-II can also be targeted to phagosomes to promote maturation independently of traditional autophagy, in a noncanonical autophagic process termed LC3-associated phagocytosis.1,26,1448 LAP occurs upon engulfment of particles that engage a receptor-mediated signaling pathway, resulting in the recruitment of some but not all of the autophagic machinery to the phagosome. These autophagic components facilitate rapid phagosome maturation and degradation of engulfed cargo, and play roles in the generation of signaling molecules and regulation of immune responses.179,180,1449 LAP thus represents a unique process that marries the ancient pathways of phagocytosis and autophagy.
Despite overlap in molecular machinery, there currently exist several criteria by which to differentiate LAP from macroautophagy: (a) Whereas LC3-decorated autophagosomes can take hours to form, LC3 can be detected on LAP-engaged phagosomes as early as 10 min after phagocytosis, and PtdIns3P can also be seen at LAP-engaged phagosomes minutes after phagocytosis.180,182,1449 (b) EM analysis reveals that LAP involves single-membrane structures.182 In contrast, macroautophagy is expected to generate double-membrane structures surrounding cargo. (c) Whereas most of the core autophagy components are required for LAP, the 2 processes can be distinguished by the involvement of the pre-initiation complex. RB1CC1, ATG13, and ULK1 are dispensable for LAP, which provides a convenient means for distinguishing between the 2 processes.180,1449 (d) LAP involves LC3 recruitment in a manner that requires ROS production by the NADPH oxidase family, notably CYBB/NOX2/gp91phox. It should be noted that most cells express at least one member of the NADPH oxidase family. Silencing of the common subunits, CYBB or CYBA/p22phox, is an effective way to disrupt NADPH oxidase activity and therefore LAP. It is anticipated that more specific markers of LAP will be identified as this process is further characterized.
Finally, an ATG5- and CTSL-dependent cell death process has been reported that can be activated by the small molecule NID-1; this process depends on PtdIns3K signaling, generates LC3B puncta and single-membrane vacuoles, and results in the clearance of SQSTM1. Thus, LAP and/or related processes can be co-opted to cause cell death in some cases.1450
2. LC3-associated apicoplast
In the Apicomplexa parasitic protists (e.g., T. gondii and Plasmodium spp.), the single ATG8 homolog localizes to an endosymbiotic nonphotosynthetic plastid, called the apicoplast.1408,1451-1454 This organelle is the product of a secondary endosymbiotic event, in which a red alga was endocytosed by an auxotrophic eukaryote (ancestor of an apicomplexan parasite); the apicoplast is the main remnant of this red alga. This organelle is approximately 300 nm in diameter, and is composed of 4 membranes that trace their ancestry to 3 different organisms. The outermost membranes of the apicoplast are derived from the plasma membrane of the auxotrophic eukaryote and the plasma membrane of the internalized alga. ATG8 is located in the outermost membranes that are enriched in PtdIns3P, which marks autophagic structures in mammalian cells; at that location it plays a role in the centrosome-mediated inheritance of the organelle in daughter cells during parasite division (M. Lévêque and S. Besteiro, unpublished results). Consequently, caution must be taken when identifying stress-induced autophagosomes by electron microscopy or by fluorescence microscopy with ATG8 labeling in these parasites.
3. LC3 conjugation system for IFNG-mediated pathogen control
Similar to LAP, LC3 localizes on the parasitophorus vacuole membrane (PVM) of T. gondii.181 The parasitophorus vacuole is a vesicle-like structure formed from host plasma membrane during the invasion of T. gondii, and it sequesters and protects the invasive T. gondii from the hostile host cytoplasm. The cell-autonomous immune system uses IFNG-induced effectors, such as immunity related GTPases and guanylate binding proteins (GBPs), to attack and disrupt this type of membrane structure; consequently, naked T. gondii in the cytoplasm are killed by a currently unknown mechanism. Intriguingly, proper targeting of these effectors onto the PVM of T. gondii requires the autophagic ubiquitin-like conjugation system, including ATG7, ATG3, and the ATG12–ATG5-ATG16L1 complex, although the necessity of LC3-conjugation itself for the targeting is not yet clear. In contrast, up- or downregulation of canonical autophagy using rapamycin, wortmannin, or starvation do not significantly affect the IFNG-mediated control of T. gondii. Furthermore, the degradative function or other components of the autophagy pathway, such as ULK1/2 and ATG14, are dispensable. Many groups have confirmed the essential nature of the LC3-conjugation system for the control of T. gondii,1455-1457 and the same or a similar mechanism also functions against other pathogens such as murine norovirus and Chlamydia trachomatis.1208,1455 Although topologically and mechanistically similar to LAP, the one notable difference is that the parasitophorous vacuole of T. gondii is actively made by the pathogen itself using host membrane, and the LC3-conjugation system-dependent targeting happens even in nonphagocytic cells. GBP-mediated lysis of pathogen-containing vacuoles is important for the activation of noncanonical inflammasomes,1458 but the targeting mechanism of GBPs to the vacuoles is unknown. Considering the necessity of the LC3-conjugation system to target GBPs to the PVM of T. gondii, this system may play crucial roles in the general guidance of various effector molecules to target membranes as well as in selective autophagosome-dependent sequestration, phagophore membrane expansion and autophagosome maturation.
4. Intracellular trafficking of bacterial pathogens
Some ATG proteins are involved in the intracellular trafficking and cell-to-cell spread of bacterial pathogens by noncanonical autophagic pathways. For example, ATG9 and WIPI1, but not ULK1, BECN1, ATG5, ATG7 or LC3B are required for the establishment of an endoplasmic reticulum-derived replicative niche after cell invasion with Brucella abortus.1459 In addition, the cell-to-cell transmission of B. abortus seems to be dependent on ULK1, ATG14 and PIK3C3/VPS34, but independent of ATG5, ATG7, ATG4B and ATG16L1.1460
5. Other processes
ATG proteins are involved in various other nonautophagic processes, particularly apoptosis and noncanonical protein secretion, as discussed in various papers.27,75,76,544,572,1449,1461-1465,1466
E. Interpretation of in silico assays for monitoring autophagy
The increasing availability of complete (or near complete) genomes for key species spanning the eukaryotic domain provides a unique opportunity for delineating the spread of autophagic machinery components in the eukaryotic world.1467,1468 Fast and sensitive sequence similarity search procedures are already available; an increasing number of experimental biologists are now comfortable “BLASTing” their favorite sequences against the ever-increasing sequence databases for identifying putative homologs in different species.1469 Nevertheless, several limiting factors and potential pitfalls need to be taken into account.
In addition to sequence comparison approaches, a number of computational tools and resources related to autophagy have become available online. All the aforementioned methods and approaches may be collectively considered as “in silico assays” for monitoring autophagy, in the sense that they can be used to identify the presence of autophagy components in different species and provide information on their known or predicted associations.
In the following sections we briefly present relevant in silico approaches, highlighting their strengths while underscoring some inherent limitations, with the hope that this information will provide guidelines for the most appropriate usage of these resources.
1. Sequence comparison and comparative genomics approaches
Apart from the generic shortcomings when performing sequence comparisons (discussed in ref. 1470), there are some important issues that need to be taken into account, especially for autophagy-related proteins. Since autophagy components seem to be conserved throughout the eukaryotic domain of life, the deep divergent relations of key subunits may reside in the so called “midnight zone” of sequence similarity: i.e., genuine orthologs may share even less than 10% sequence identity at the amino acid sequence level.1471 This is the case with autophagy subunits in protists1472,1473 and with other universally conserved eukaryotic systems, as for example the nuclear pore complex.1474 This low sequence identity is especially pronounced in proteins that contain large intrinsically disordered regions.1475 In such cases, sophisticated (manual) iterative database search protocols, including proper handling of compositionally biased subsequences and considering domain architecture may assist in eliminating spurious similarities or in the identification of homologs that share low sequence identity with the search molecule.1473-1475
Genome-aware comparative genomics methods1476 can also provide invaluable information on yet unidentified components of autophagy. However, care should be taken to avoid possible Next Generation Sequencing artifacts (usually incorrect genome assemblies): these may directly (via a similarity to a protein encoded in an incorrectly assembled genomic region) or indirectly (via propagating erroneous annotations in databases) give misleading homolog assignments (V.J. Promponas, I. Iliopoulos and C.A. Ouzounis, submitted). In addition, taking into account other types of high-throughput data available in publicly accessible repositories (e.g., EST/RNAseq data, expression data) can provide orthogonal evidence for validation purposes when sequence similarities are marginal.1474
2. Web-based resources related to autophagy
A number of autophagy-related resources are now available online, providing access to diverse data types ranging from gene lists and sequences to comprehensive catalogs of physical and indirect interactions. In the following we do not attempt to review all functionalities offered by the different servers, but to highlight those that (a) offer possibilities for identifying novel autophagy-related proteins or (b) characterize features that may link specific proteins to autophagic processes. Two comments regarding biological databases in general also apply to autophagy-related resources as well: (a) the need for regular updates, and (b) data and annotation quality. Nevertheless, these issues are not discussed further herein.
a. The THANATOS database
THANATOS (THe Apoptosis, Necrosis, AuTophagy OrchestratorS) is a resource being developed by the CUCKOO Workgroup at the Huazhong University of Science and Technology (Wuhan, Hubei,China). THANATOS is still under development (Y. Xue, personal communication) and it is focused on the integration of sequence data related to the main mechanisms leading to programmed cell death in eukaryotes. A simple web interface assists in data retrieval, using keyword searches, browsing by species and cell death type, performing BLAST searches with user-defined sequences, and by requesting the display of orthologs among predefined species. A Java application is also available to download for standalone usage of the THANATOS resource. The THANATOS database is publicly available online at the URL http://thanatos.biocuckoo.org/.
b. The Human Autophagy Database (HADb)
The human autophagy database, developed in the Laboratory of Experimental Hemato-Oncology (Luxembourg), lists over 200 human genes/proteins related to autophagy.604 These entries have been manually collected from the biomedical literature and other online resources604 and there is currently no information that the initially published list has been further updated. For each gene there exists information on its sequence, transcripts and isoforms (including exon boundaries) as well as links to external resources. HADb provides basic search and browsing functionalities and is publicly available online at the URL http://autophagy.lu/.
c. The Autophagy Database
The Autophagy Database is a multifaceted online resource providing information for proteins related to autophagy and their homologs across several eukaryotic species, with a focus on functional and structural data.1477 It is developed by the National Institute of Genetics (Japan) under the Targeted Proteins Research Program of the Ministry of Education, Culture, Sports, Science and Technology (http://www.tanpaku.org/). This resource is regularly updated and as of August 2014 contained information regarding 312 reviewed protein entries; when additional data regarding orthologous/homologous proteins from more than 50 eukaryotes is considered, the total number of entries reaches approximately 9,000. In addition to the browse functionalities offered under the “Protein List” and the “Homologs” menus, an instance of the NCBI-BLAST software facilitates sequence-based queries against the database entries. Moreover, interested users may download the gene list or the autophagy dump files licensed under a Creative Commons Attribution-ShareAlike 2.1 Japan License. The Autophagy Database is publicly available online at the URL http://www.tanpaku.org/autophagy/index.html.
d. The Autophagy Regulatory Network (ARN)
The most recent addition to the web-based resources relevant to autophagy research is the Autophagy Regulatory Network (ARN), developed at the Eötvös Loránd University and Semmelweis University (Budapest, Hungary) in collaboration with the Institute of Food Research and The Genome Analysis Centre (Norfolk, UK). Maintanence and hosting the ARN resource is secured at The Genome Analysis Centre until at least 2019. ARN is an integrated systems-level resource aiming to collect and provide an interactive user interface enabling access to validated or predicted protein-protein, transcription factor-gene and miRNA-mRNA interactions related to autophagy in human.1479 ARN contains data from 26 resources, including an in-house extensive manual curation, the dataset of the ChIP-MS study of Behrends et al.,464 ADB and ELM. As of June 2015, a total of more than 14,000 proteins and 386 miRNAs are present in ARN, including 38 core autophagy proteins and 113 predicted regulators. Importantly, all autophagy-related proteins are linked to major signaling pathways. A flexible—in terms of both content and format—download functionality enables users to locally use the ARN data under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The autophagy regulatory network resource is publicly available online at the URL http://autophagy-regulation.org.
e. Prediction of Atg8-family interacting proteins
Being central components of the autophagic core machinery, Atg8-family members (e.g., LC3 and GABARAP in mammals) and their interactome have attracted substantial interest.464,1479,1480 During the last decade, a number of proteins have been shown to interact with Atg8 homologs via a short linear peptide; depending on context, different research groups have described this peptide as the LIR,319 the LC3 recognition sequence (LRS),661 or the AIM.1481 Recently, 2 independent efforts resulted in the first online available tools for identification of these motifs (LIR-motifs for brevity) in combination with other sequence features, which may signify interesting targets for further validation (see below).
f. The iLIR server
The iLIR server is a specialized web server that scans an input sequence for the presence of a degenerate version of LIR, the extended LIR-motif (xLIR).1482 Currently, the server also reports additional matches to the “canonical” LIR motif (WxxL), described by the simple regular expression x(2)-[WFY]-x(2)-[LIV]. Kalvari and colleagues have also compiled a position-specific scoring matrix (PSSM) based on validated instances of the LIR motif, demonstrating that many of the false positive hits (i.e., spurious matches to the xLIR motif) are eliminated when a PSSM score >15 is sought. In addition, iLIR also overlays the aforementioned results to segments that reside in or are adjacent to disordered regions and are likely to form stabilizing interactions upon binding to another globular protein as predicted by the ANCHOR package.1483 A combination of an xLIR match with a high PSSM score (>13) and/or an overlap with an ANCHOR segment is shown to give reliable predictions.1482 It is worth mentioning that, intentionally, iLIR does not provide explicit predictions of functional LIR-motifs but rather displays all the above information accompanied by a graphical depiction of query matches to known protein domains and motifs; it is up to the user to interpret the iLIR output. As mentioned in the original iLIR publication, a limitation of this tool is that it does not handle any noncanonical LIR motifs at present. The iLIR server was jointly developed by the University of Warwick and University of Cyprus and is freely available online at the URL http://repeat.biol.ucy.ac.cy/iLIR.
g. The Eukaryotic Linear Motif resource (ELM)
The Eukaryotic Linear Motif resource1484 is a generic resource for examining functional sites in proteins in the form of short linear motifs, which have been manually curated from the literature. Sophisticated filters based on known (or predicted) query features (such as taxonomy, subcellular localization, structural context) are used to narrow down the results lists, which can be very long lists of potential matches due to the short lengths of ELMs. This resource has incorporated 4 entries related to the LIR-motif (since May 2014; http://elm.eu.org/infos/news.html), while another 3 are being evaluated as candidate ELM additions (Table 3). Again, the ELM resource displays matches to any motifs and users are left with the decision as to which of them are worth studying further. ELM is developed/maintained by a consortium of European groups coordinated by the European Molecular Biology Laboratory and is freely available online at the URL http://elm.eu.org.
Table 3.
Eukaryotic linear motif entries related to the LIR-motif (obtained from http://elm.eu.org/).
| ELM identifier | ELM | Description | Status |
|---|---|---|---|
| LIG_LIR_Gen_1 | [EDST].{0,2}[WFY]..[ILV] | Canonical LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | ELM |
| LIG_LIR_Apic_2 | [EDST].{0,2}[WFY]..P | Apicomplexa-specific variant of the canonical LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | ELM |
| LIG_LIR_Nem_3 | [EDST].{0,2}[WFY]..[ILVFY] | Nematode-specific variant of the canonical LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | ELM |
| LIG_LIR_LC3C_4 | [EDST].{0,2}LVV | Noncanonical variant of the LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | ELM |
| LIG_AIM | [WY]..[ILV] | Atg8-family interacting motif found in Atg19, SQSTM1/p62, ATG4B and CALR/calreticulin, involved in autophagy-related processes. | Candidate |
| LIG_LIR | WxxL or [WYF]xx[LIV] | The LIR might link ubiquitinated substrates that should be degraded to the autophagy-related proteins in the phagophore membrane. | Candidate |
| LIG_GABARAP | W.FL | GABAA receptor binding to clathrin and CALR; possibly linked to trafficking. | Candidate |
h. The ncRNA-associated cell death database (ncRDeathDB)
The noncoding RNA (ncRNA)-associated cell death database (ncRDeathDB),1485 most recently developed at the Harbin Medical University (Harbin, China) and Shantou University Medical College (Shantou, China), documents a total of more than 4,600 ncRNA-mediated programmed cell death entries. Compared to previous versions of the miRDeathDB,1486-1488 the ncRDeathDB further collected a large amount of published data describing the roles of diverse ncRNAs (including microRNA, long noncoding RNA/lncRNA and small nucleolar RNA/snoRNA) in programmed cell death for the purpose of archiving comprehensive ncRNA-associated cell death interactions. The current version of ncRDeathDB provides an all-inclusive bioinformatics resource on information detailing the ncRNA-mediated cell death system and documents 4,615 ncRNA-mediated programmed cell death entries (including 1,817 predicted entries) involving 12 species, as well as 2,403 apoptosis-associated entries, 2,205 autophagy-associated entries and 7 necrosis-associated entries. The ncRDeathDB also integrates a variety of useful tools for analyzing RNA-RNA and RNA-protein binding sites and for network visualization. This resource will help researchers to visualize and navigate current knowledge of the noncoding RNA component of cell death and autophagy, to uncover the generic organizing principles of ncRNA-associated cell death systems, and to generate valuable biological hypotheses. The ncRNA-associated cell death interactions resource is publicly available online at the URL http://www.rna-society.org/ncrdeathdb.
3. Dynamic and mathematical models of autophagy
Mathematical modeling methods and approaches can be used as in silico models to study autophagy. For example, a systems pharmacology approach has been used to build an integrative dynamic model of interaction between macroautophagy and apoptosis in mammalian cells.1489 This model is a general predictive in silico model of macroautophagy, and the model has translated the signaling networks that control cell fate concerning the crosstalk of macroautophagy and apoptosis to a set of ordinary differential equations.1489,1490 The model can be adapted for any type of cells including cancer cell lines and drug interventions by adjusting the numerical parameters based on experimental data.1490 Another example is seen with an agent-based mathematical model of autophagy that focuses on the dynamic process of autophagosome formation and degradation in cells,1491 and there is a mathematical model of macroautophagy that can be used to interpret the formation of autophagosomes in single cells.1492 As this aspect of the field progresses we will likely start to see these models used to help predict and understand autophagic responses to new therapeutic treatments.
Conclusions and future perspectives
There is no question that research on the topic of autophagy has expanded dramatically since the publication of the first set of guidelines.2 To help keep track of the field we have published a glossary of autophagy-related molecules and processes,1493,1494 and now include the glossary as part of these guidelines.
With this continued influx of new researchers we think it is critical to try to define standards for the field. Accordingly, we have highlighted the uses and caveats of an expanding set of recommended methods for monitoring macroautophagy in a wide range of systems (Table 4). Importantly, investigators need to determine whether they are evaluating levels of early or late autophagic compartments, or autophagic flux. If the question being asked is whether a particular condition changes autophagic flux (i.e., the rate of delivery of autophagy substrates to lysosomes or the vacuole, followed by degradation and efflux), then assessment of steady state levels of autophagosomes (e.g., by counting GFP-LC3 puncta, monitoring the amount of LC3-II without examining turnover, or by single time point electron micrographs) is not sufficient as an isolated approach. In this case it is also necessary to directly measure the flux of autophagosomes and/or autophagy cargo (e.g., in wild-type cells compared to autophagy-deficient cells, the latter generated by treatment with an autophagy inhibitor or resulting from ATG gene knockdowns). Collectively, we strongly recommend the use of multiple assays whenever possible, rather than relying on the results from a single method.
Table 4.
Recommended methods for monitoring autophagy.
| Method | Description | |
|---|---|---|
| 1. | Electron microscopy | Quantitative electron microscopy, immuno-TEM; monitor autophagosome number, volume, and content/cargo. |
| 2. | Atg8/LC3 western blotting | Western blot. The analysis is carried out in the absence and presence of lysosomal protease or fusion inhibitors to monitor flux; an increase in the LC3-II amount in the presence of the inhibitor is usually indicative of flux. |
| 3. | GFP-Atg8/LC3 lysosomal delivery and proteolysis | Western blot +/− lysosomal fusion or degradation inhibitors; the generation of free GFP indicates lysosomal/vacuolar delivery. |
| 4. | GFP-Atg8/LC3 fluorescence microscopy | Fluorescence microscopy, flow cytometry to monitor vacuolar/lysosomal localization. Also, increase in punctate GFP-Atg8/LC3 or Atg18/WIPI, and live time-lapse fluorescence microscopy to track the dynamics of GFP-Atg8/LC3-positive structures. |
| 5. | Tandem mRFP/mCherry-GFP fluorescence microscopy, Rosella | Flux can be monitored as a decrease in green/red (yellow) fluorescence (phagophores, autophagosomes) and an increase in red fluorescence (autolysosomes). |
| 6. | Autophagosome quantification | FACS/flow cytometry. |
| 7. | SQSTM1 and related LC3 binding protein turnover | The amount of SQSTM1 increases when autophagy is inhibited and decreases when autophagy is induced, but the potential impact of transcriptional/translational regulation or the formation of insoluble aggregates should be addressed in individual experimental systems. |
| 8. | MTOR, AMPK and Atg1/ULK1 kinase activity | Western blot, immunoprecipitation or kinase assays. |
| 9. | WIPI fluorescence microscopy | Quantitative fluorescence analysis using endogenous WIPI proteins, or GFP- or MYC-tagged versions. Suitable for high-throughput imaging procedures. |
| 10. | Bimolecular fluorescence complementation | Can be used to monitor protein-protein interaction in vivo. |
| 11. | FRET | Interaction of LC3 with gangliosides to monitor autophagosome formation. |
| 12. | Transcriptional and translational regulation | Northern blot, or qRT-PCR, autophagy-dedicated microarray. |
| 13. | Autophagic protein degradation | Turnover of long-lived proteins to monitor flux. |
| 14. | Pex14-GFP, GFP-Atg8, Om45-GFP, mitoPho8Δ60 | A range of assays can be used to monitor selective types of autophagy. These typically involve proteolytic maturation of a resident enzyme or degradation of a chimera, which can be followed enzymatically or by western blot. |
| 15. | Autophagic sequestration assays | Accumulation of cargo in autophagic compartments in the presence of lysosomal protease or fusion inhibitors by biochemical or multilabel fluorescence techniques. |
| 16. | Turnover of autophagic compartments | Electron microscopy with morphometry/stereology at different time points. |
| 17. | Autophagosome-lysosome colocalization and dequenching assay | Fluorescence microscopy. |
| 18. | Sequestration and processing assays in plants | Chimeric RFP fluorescence and processing, and light and electron microscopy. |
| 19. | Tissue fractionation | Centrifugation, western blot and electron microscopy. |
| 20. | Degradation of endogenous lipofuscin | Fluorescence microscopy. |
As a final reminder, we stated at the beginning of this article that this set of guidelines is not meant to be a formulaic compilation of rules, because the appropriate assays depend in part on the question being asked and the system being used. Rather, these guidelines are presented primarily to emphasize key issues that need to be addressed such as the difference between measuring autophagy components, and flux or substrate clearance; they are not meant to constrain imaginative approaches to monitoring autophagy. Indeed, it is hoped that new methods for monitoring autophagy will continue to be developed, and new findings may alter our view of the current assays. Similar to the process of autophagy, this is a dynamic field, and we need to remain flexible in the standards we apply.
Acknowledgments
In a rapidly expanding and highly dynamic field such as autophagy, it is possible that some authors who should have been included on this article have been missed. D.J.K. extends his apologies to researchers in the field of autophagy who, due to oversight or any other reason, could not be included on this article. I also note that two of our colleagues on this manuscript have passed away: Arlette Darfeuille-Michaud and Wouter van Doorn.
Funding
This work was supported in part by the National Institutes of Health, including Public Health Service grant GM053396 to D.J.K. Due to space and other limitations, it is not possible to include all other sources of financial support.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of Veterans Affairs, the U.S. Food and Drug Administration or the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Glossary
- 3-MA (3-methyladenine):
An inhibitor of class I PI3K and class III PtdIns3K, which results in macroautophagy inhibition due to suppression of class III PtdIns3K,329 but may under some conditions show the opposite effect.330 At concentrations >10 mM 3-MA inhibits other kinases such as AKT (Ser473), MAPK/p38 (Thr180/Tyr182) and MAPK/JNK (Thr183/Tyr185).1534
- 11′-deoxyverticillin A (C42):
An epipolythiodioxopiperazine fungal secondary metabolite that is used as an anticancer drug; it triggers apoptotic and necrotic cell death, and enhances macroautophagy through the action of PARP1 and RIPK1.1535
- 12-ylation:
The modification of substrates by covalent conjugation to ATG12, first used to describe the autocatalytic conjugation of ATG12 to ATG3.1536
- 14-3-3ζ:
See YWHAZ.
- ABT737:
A BH3 mimetic that competitively disrupts the interaction between BECN1 and BCL2 or BCL2L1, thus inducing macroautophagy.1537 It should be noted, however, that by its inhibitory action on the anti-apoptotic BCL2 family members, ABT737 also leads to apoptosis.1538
- ACBD5 (acyl-CoA binding domain containing 5):
ACBD5 is the human ortholog of fungal Atg37; it is a peroxisomal protein that is required for pexophagy.345,1539 See also Atg37.
- Acetyl-coenzyme A:
A central energy metabolite that represses macroautophagy if present in the cytosol.1540,1541
- Acinus:
A protein that in Drosophila regulates both endocytosis and macroautophagy; the acn mutant is defective in autophagosome maturation, whereas stabilization of endogenous Acn by mutation of its caspase cleavage site,1542 or overexpression of Acn leads to excessive macroautophagy.1543 Note that Acn can also induce DNA condensation or fragmentation after its activation by CASP3 in apoptotic cells.
- ActA:
A L. monocytogenes protein that recruits the Arp2/3 complex and other actin-associated components to the cell surface to evade recognition by xenophagy; this effect is independent of bacterial motility.1544
- Adaptophagy:
Selective degradation of signaling adaptors downstream of TLRs or similar types of receptor families.1545
- ADNP (activity-dependent neuroprotective homeobox):
A protein that interacts with LC3B and shows an increased expression in lymphocytes from schizophrenia patients.1019
- AEG-1:
See MTDH.
- AEN/ISG20L1 (apoptosis-enhancing nuclease):
A protein that localizes to nucleolar and perinucleolar regions of the nucleus, which regulates macroautophagy associated with genotoxic stress; transcription of AEN is regulated by TP53 family members.1546
- AGER/RAGE (advanced glycosylation end product-specific receptor):
A member of the immunoglobulin gene superfamily that binds the HMGB1 (high mobility group box 1) chromatin binding protein.1547 AGER overexpression enhances macroautophagy and reduces apoptosis. This can occur in response to ROS, resulting in the upregulation of macroautophagy and the concomitant downregulation of apoptosis, favoring tumor cell survival in response to anticancer treatments that increase ROS production.1548 See also HMGB1.
- Aggrephagy:
The selective removal of aggregates by a macroautophagy-like process.731
- AGS3:
See GPSM1.
- Aggresome:
An aggregation of misfolded proteins formed by a highly regulated process mediated by HDAC6 or BAG3.1549,1550 This process requires protein transport by a dynein motor and microtubule integrity. Aggresomes form at the microtubule-organizing center and are surrounded by a cage of the intermediate filament protein VIM/vimentin. Note that not all proteins that aggregate and form filaments like HTT or MAPT form aggregsomes.
- AHA (L-azidohomoalanine):
An amino acid analog used for labeling newly synthesized protein and monitoring autophagic protein degradation.660
- AICAR (aminoimidazole-4-carboxamide riboside):
Cell permeable nucleotide analog that is an activator of AMPK; inhibits macroautophagy472 through mechanisms that are not related to its effect on AMPK.483,1551
- AIM (Atg8-family interacting motif):
A short peptide motif that allows interaction with Atg8.1481 See WXXL and LIR/LRS.
- AKT/PKB (v-akt murine thymoma viral oncogene homolog 1):
A serine/threonine kinase that negatively regulates macroautophagy in some cellular systems.
- Alfy:
See WDFY3.
- ALIS (aggresome-like induced structures):
These structures may function as protein storage compartments and are cleared by macroautophagy.315 SQSTM1 may regulate their formation and macroautophagic degradation.317 See also DALIS.
- Allophagy:
The selective degradation of sperm components by macroautophagy; this process occurs in C. elegans.739
- ALOX5 (arachidonate 5-lipoxygenase):
See lipoxygenases.
- ALOX15 (arachidonate 15-lipoxygenase):
See lipoxygenases.
- ALR:
See autophagic lysosome reformation.
- ALS2/alsin (amyotrophic lateral sclerosis 2 [juvenile]):
A guanine nucleotide exchange factor for the small GTPase RAB5 that regulates endosome and autophagosome fusion and trafficking; loss of ALS2 accounts for juvenile recessive amyotrophic lateral sclerosis, juvenile primary lateral sclerosis, and infantile-onset ascending hereditary spastic paralysis.1552,1553
- ALSFTD:
See C9orf72.
- AMBRA1 (autophagy/beclin-1 regulator 1):
A positive regulator of macroautophagy. AMBRA1 interacts with both BECN1 and ULK1, modulating their activity.488,501,1206 Also, a role in both PARK2-dependent and -independent mitophagy has been described for AMBRA1.768 AMBRA1 activity is regulated by dynamic interactions with DDB1 and TCEB2/Elongin B, the adaptor proteins of the E3 ubiquitin ligase complexes containing CUL4/Cullin 4 and CUL5, respectively.1554 Finally, AMBRA1 is the macroautophagy adaptor linking this process to cell proliferation, by negatively regulating the oncogenic protein MYC through the latter's phosphorylation status.1555
- AMFR/gp78 (autocrine motility factor receptor, E3 ubiquitin protein ligase):
An ER-associated E3 ubiquitin ligase that degrades the MFN/mitofusin mitochondrial fusion proteins and induces mitophagy.1556
- Amiodarone:
An FDA-approved antiarrhythmic drug that induces macroautophagic flux via AMPK- and AKT-mediated MTOR inhibition.1557,1558
- Amphisome (AM):
Intermediate compartment formed by the fusion of an autophagosome with an endosome (this compartment can be considered a type of autophagic vacuole and may be equivalent to a late autophagosome, and as such has a single limiting membrane); the amphisome has not yet fused with a lysosome.1559 Amphisomes can also fuse with the plasma membrane to release the macroautophagic cargo (exosomal pathway). See also exophagy.
- AMPK (AMP-activated protein kinase):
A sensor of energy level that is activated by an increase in the AMP/ATP ratio via the STK11/LKB1 kinase. Phosphorylates the MTORC1 subunit RPTOR to cause induction of macroautophagy. AMPK also activates the TSC1/2 complex (thus inhibiting RHEB), and binds and directly phosphorylates (and activates) ULK1 as part of the ULK1 kinase complex, which includes ATG13, ATG101 and RB1CC1.477,478 The yeast homolog of AMPK is Snf1.472,1560 Conversely, ULK1 can phosphorylate AMPK through a negative feedback loop.496 AMPK is a heterotrimeric enzyme composed of the PRKAA1/AMPKα1 or PRKAA2/AMPKα2 subunit, the PRKAB1/AMPKβ1 or PRKAB2/AMPKβ2 subunit and the PRKAG1/AMPKγ1, PRKAG2/AMPKγ2 or PRKAG3/AMPKγ subunits.
- Ams1/α-mannosidase:
A cargo of the Cvt pathway; Ams1 forms an oligomer in the cytosol similar to prApe1.
- AMSH1/3:
Two Arabidopsis deubiquitinating enzymes that have been linked to plant macroautophagy.1561,1562
- APC (activated protein C):
APC (PROC that has been activated by thrombin) modulates cardiac metabolism and augments macroautophagy in the ischemic heart by inducing the activation of AMPK in a mouse model of ischemia/reperfusion injury.1563
- Ape1 (aminopeptidase I):
A resident vacuolar hydrolase that can be delivered in its precursor form (prApe1) to the vacuole through either the cytoplasm-to-vacuole targeting (Cvt) pathway or macroautophagy, in vegetative or starvation conditions, respectively.128 The propeptide of prApe1 is removed upon vacuolar delivery, providing a convenient way to monitor localization of the protein and the functioning of these pathways, although it must be noted that delivery involves a receptor and scaffold so that its transit involves a type of selective macroautophagy even in starvation conditions. See also Atg11, Atg19 and cytoplasm-to-vacuole targeting pathway.
- Ape1 complex/prApe1 complex:
A large protein complex comprised of multiple prApe1 dodecamers localized in the cytosol.131
- Ape4:
An aspartyl aminopeptidase that binds the Atg19 receptor and is transported to the vacuole through the Cvt pathway.1564
- APMA (autophagic macrophage activation):
A collection of macroautophagy-related processes in cells of the reticulo-endothelial system. APMA includes (1) convergence of phagocytosis and the macroautophagic machinery, (2) enhanced microbicidal properties of autolysosomes in comparison to standard phagolysosomes, (3) macroautophagic modulation of pathogen recognition receptor signaling, (4) cooperation between immunity-related GTPases and ATG proteins in attacking parasitophorus vacuoles, and (5) enhanced antigen presentation. APMA is thus recognized as a complex outcome of macroautophagy stimulation in macrophages, representing a unique composite process that brings about a heightened state of immunological activation.1565
- Appressorium:
A specialized infection structure produced by pathogenic fungi to rupture the outer layer of their host and gain entry to host cells. In plant pathogenic fungi, such as the rice blast fungus M. oryzae, formation of appressoria follows macroautophagy in conidia and recycling of the spore contents to the developing infection cell.275,1316
- ARD1:
See NAA10.
- Are1:
See Ayr1.
- Are2:
See Ayr1.
- ARRB1/β-arrestin-1 (arrestin, beta 1):
Members of the arrestin/beta-arrestin protein family are thought to participate in agonist-mediated desensitization of G-protein-coupled receptors and cause specific dampening of cellular responses to stimuli such as hormones, neurotransmitters, or sensory signals. ARRB1 is a cytosolic protein and acts as a cofactor in the ADRBK/BARK (adrenergic, beta, receptor kinase)-mediated desensitization of beta-adrenergic receptors. Besides the central nervous system, it is expressed at high levels in peripheral blood leukocytes, and thus the ADRBK/beta-arrestin system is thought to play a major role in regulating receptor-mediated immune functions. This protein plays a neuroprotective role in the context of cerebral ischemia through regulating BECN1-dependent autophagosome formation.1566
- ARHI:
See DIRAS3.
- ARN5187:
Lysosomotropic compound with dual inhibitory activity against the circadian regulator NR1D2/REV-ERBβ and autophagy. Although ARN5187 and chloroquine have similar lysosomotropic potency and are equivocal with regard to autophagy inhibition, ARN5187 has a significantly improved in vitro anticancer activity.1497
- ASB10 (ankyrin repeat and SOCS box containing 10):
The ASB family of proteins mediate ubiquitination of protein substrates via their SOCS box and as such have been implicated as negative regulators of cell signaling. ASB10 colocalizes with aggresome biomarkers and pre-autophagic structures and may form ALIS.1567
- ATF4 (activating transcription factor 4):
A transcription factor that is induced by hypoxia, amino acid starvation and ER stress, and is involved in the unfolded protein response, playing a critical role in stress adaptation; ATF4 binds to a cAMP response element binding site in the LC3B promoter, resulting in upregulation of LC3B,1568 and also directs a macroautophagy gene transcriptional program in response to amino acid depletion and ER stress.408
- ATF5 (activating transcription factor 5):
A transcription factor that is upregulated by the BCR-ABL protein tyrosine kinase, a macroautophagy repressor, through the PI3K-AKT pathway that inhibits FOXO4, a repressor of ATF5 transcription; one of the targets of ATF5 is MTOR.1569
- Atg (autophagy-related):
Abbreviation used for most of the components of the protein machinery that are involved in selective and nonselective macroautophagy and in selective microautophagy.1570
- ATG-11/EPG-7: A scaffold protein mediating the macroautophagic degradation of the C. elegans SQSTM1 homolog SQST-1.1585 ATG-11/EPG-7 interacts with SQST-1 and also with multiple ATG proteins. ATG-11/EPG-7 itself is degraded by macroautophagy. ATG-13/EPG-1: The highly divergent homolog of Atg13 in C. elegans. ATG-13/EPG-1 directly interacts with the C. elegans Atg1 homolog UNC-51.1733 See also Atg13.Atg1:
A serine/threonine protein kinase that functions in recruitment and release of other Atg proteins from the PAS.1571 The functional homologs in higher eukaryotes are ULK1 and ULK2, and in C. elegans UNC-51.
- Atg2:
A protein that interacts with Atg18; in atg2Δ mutant cells Atg9 accumulates primarily at the PAS.1572,1573
- Atg3:
A ubiquitin-conjugating enzyme (E2) analog that conjugates Atg8/LC3 to phosphatidylethanolamine (PE) after activation of the C-terminal residue by Atg7.1574,1575 ATG3 can also be conjugated to ATG12 in higher eukaryotes.1536 See also 12-ylation.
- Atg4:
A cysteine protease that processes Atg8/LC3 by removing the amino acid residue(s) that are located on the C-terminal side of what will become the ultimate glycine. Atg4 also removes PE from Atg8/LC3 in a step referred to as “deconjugation”.213 Mammals have 4 ATG4 proteins (ATG4A to ATG4D), but ATG4B appears to be the most relevant for macroautophagy and has the broadest range of activity for all of the Atg8 homologs.172,1576 See also deconjugation.
- Atg5:
A protein containing ubiquitin folds that is part of the Atg12–Atg5-Atg16 complex, which acts in part as an E3 ligase for Atg8/LC3–PE conjugation.1577
- Atg6:
See Vps30.
- Atg7:
A ubiquitin activating (E1) enzyme homolog that activates both Atg8/LC3 and Atg12 in an ATP-dependent process.1578,1579
- Atg8:
- A ubiquitin-like protein that is conjugated to PE; involved in cargo recruitment into, and biogenesis of, autophagosomes. Autophagosomal size is regulated by the amount of Atg8.107 Since Atg8 is selectively enclosed into autophagosomes, its breakdown allows measurement of the rate of macroautophagy. Mammals have several Atg8 homologs that are members of the LC3 and GABARAP subfamilies, which are also involved in autophagosome formation.142,148,600 The C. elegans homologs are LGG-1 and LGG-2.
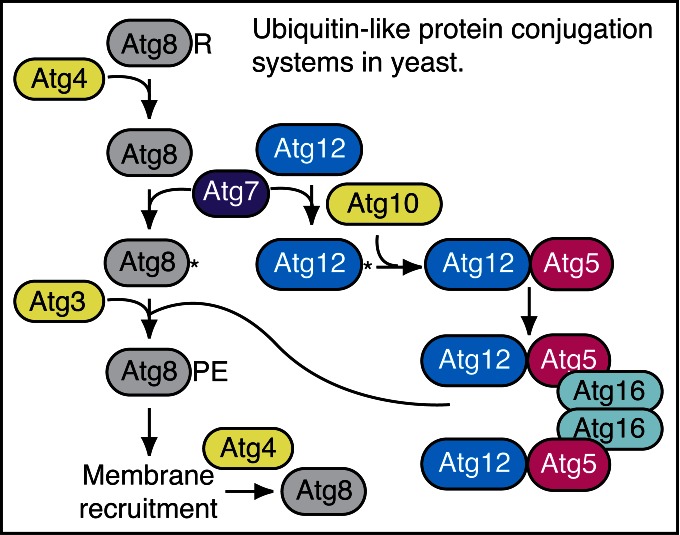
- Atg9:
A transmembrane protein that may act as a lipid carrier for expansion of the phagophore. In mammalian cells, ATG9A localizes to the trans-Golgi network and endosomes, whereas in fungi this protein localizes in part to peripheral sites (termed Atg9 reservoirs or tubulovesicular clusters) that are localized near the mitochondria, and to the PAS.536,1580 Whereas mammalian ATG9A is ubiquitously expressed, ATG9B is almost exclusively expressed in the placenta and pituitary gland.1581
- Atg9 peripheral sites/structures:
In yeast, these are peri-mitochondrial sites where Atg9 localizes, which are distinct from the phagophore assembly site.536,537 The Atg9 peripheral sites may be the precursors of the phagophore.
- Atg10:
A ubiquitin conjugating (E2) enzyme analog that conjugates Atg12 to Atg5.1582
- Atg11:
- A scaffold protein that acts in selective types of macroautophagy including the Cvt pathway, mitophagy and pexophagy. Atg11 binds Atg19, Pichia pastoris Atg30 (PpAtg30) and Atg32 as part of its role in specific cargo recognition. It also binds Atg9 and is needed for its movement to the PAS.1583 Atg11 in conjunction with receptor-bound targets may activate Atg1 kinase activity during selective macroautophagy.1584 Homologs of Atg11 include RB1CC1 in mammals (although RB1CC1 does not appear to function as an Atg11 ortholog), ATG-11/EPG-7 in C. elegans,1585 and ATG11 in Arabidopsis.1586
- Atg12:
A ubiquitin-like protein that modifies an internal lysine of Atg5 by covalently binding via its C-terminal glycine.1577 In mouse and human cells, ATG12 also forms a covalent bond with ATG3, and this conjugation event plays a role in mitochondrial homeostasis.1536 The C. elegans homolog is LGG-3.
- Atg13:
A component of the Atg1 complex that is needed for Atg1 kinase activity. Atg13 is highly phosphorylated in a PKA- and TOR-dependent manner in rich medium conditions. During starvation-induced macroautophagy in yeast, Atg13 is partially dephosphorylated. In mammalian cells, at least MTOR and ULK1 phosphorylate ATG13. The decreased phosphorylation of Atg13/ATG13 that results from TOR/MTOR inhibition is partly offset in terms of the change in molecular mass by the ULK1-dependent phosphorylation that occurs upon ULK1 activation.505,1587 The C. elegans ortholog is ATG-13/EPG-1.
- Atg14:
A component of the class III PtdIns3K complex that is necessary for the complex to function in macroautophagy.1588 Also known as ATG14/ATG14L/BARKOR in mammals,548 or EPG-8 in C. elegans.1269
- Atg15:
A yeast vacuolar protein that contains a lipase/esterase active site motif and is needed for the breakdown of autophagic and Cvt bodies within the vacuole lumen (as well as MVB-derived and other subvacuolar vesicles) and the turnover of lipid droplets.1589-1591
- Atg16:
A component of the Atg12-Atg5-Atg16 complex. Atg16 dimerizes to form a large complex.1592 There are 2 mammalian homologs, ATG16L1 and ATG16L2; mutations in either of the corresponding genes correspond to risk alleles associated with Crohn disease.1593,1594
- Atg17:
- A yeast protein that is part of the Atg1 kinase complex. Atg17 is not essential for macroautophagy, but modulates the magnitude of the response; smaller autophagosomes are formed in the absence of Atg17.106,503 In yeast, Atg17 exists as part of a stable ternary complex that includes Atg31 and Atg29; this complex functions as a dimer.1595-1597 The functional counterpart of this complex in mammalian cells may be RB1CC1.
- Atg18:
A yeast protein that binds to PtdIns3P (and PtdIns[3,5]P2) via its WD40 β-propeller domain. Atg18 interacts with Atg2, and in atg18Δ cells Atg9 accumulates primarily at the PAS. Atg18 has additional nonautophagic functions, such as in retrograde transport from the vacuole to the Golgi complex, and in the regulation of PtdIns(3,5)P2 synthesis; the latter function affects the vacuole's role in osmoregulation.553 See also WIPI.
- Atg19:
A receptor for the Cvt pathway that binds Atg11, Atg8 and the propeptide of precursor aminopeptidase I. Atg19 is also a receptor for Ams1/α-mannosidase, another Cvt pathway cargo.1598,1599
- Atg20/Snx42:
A yeast PtdIns3P-binding sorting nexin that is part of the Atg1 kinase complex and associates with Snx4/Atg24.1600 Atg20 is a PX-BAR domain-containing protein involved in pexophagy. M. oryzae Snx41 (MoSnx41) is homologous to both yeast Atg20 and Snx41, and carries out functions in both pexophagy and nonautophagy vesicular trafficking.1601
- Atg21:
A yeast PtdIns3P binding protein that is a homolog of, and partially redundant with, Atg18.335 See also WIPI.
- Atg22:
A yeast vacuolar amino acid permease that is required for efflux after autophagic breakdown of proteins.1602,1603
- Atg23:
A yeast peripheral membrane protein that associates and transits with Atg9.538,1604,1605
- Atg24:
See Snx4.
- Atg25:
A coiled-coil protein required for macropexophagy in H. polymorpha.1606
- Atg26:
A sterol glucosyltransferase that is required for micro- and macropexophagy in P. pastoris, but not in S. cerevisiae.1607,1608
- Atg27:
A yeast integral membrane protein that is required for the movement of Atg9 to the PAS; the absence of Atg27 results in a reduced number of autophagosomes under autophagy-inducing conditions.1609
- Atg28:
A coiled-coil protein involved in micro- and macropexophagy in P. pastoris.1610
- Atg29:
A yeast protein required for efficient nonselective macroautophagy in fungi. Part of the yeast Atg17-Atg31-Atg29 complex that functions at the PAS for protein recruitment and initiation of phagophore formation.1595-1597,1611
- Atg30:
A protein required for the recognition of peroxisomes during micro- and macropexophagy in P. pastoris. It binds the peroxin PpPex14 and the selective autophagy receptor protein PpAtg11.708
- Atg31:
A yeast protein required for nonselective macroautophagy in fungi. Part of the yeast Atg17-Atg31-Atg29 complex that functions at the PAS for protein recruitment and initiation of phagophore formation.1595-1597,1612
- Atg32:
A mitochondrial outer membrane protein that is required for mitophagy in yeast. Atg32 binds Atg8 and Atg11 preferentially during mitophagy-inducing conditions.687,688 See also BCL2L13.
- Atg33:
A mitochondrial outer membrane protein that is required for mitophagy in yeast.686
- Atg34:
A protein that functions as a receptor for import of Ams1/α-mannosidase during macroautophagy (i.e., under starvation conditions) in yeast.1613 This protein was initially referred to as Atg19-B based on predictions from in silico studies.1614
- Atg35:
The Atg35 protein relocates to the peri-nuclear structure and specifically regulates MIPA formation during micropexophagy; the atg35Δ mutant is able to form pexophagosomes during macropexophagy.1615
- Atg36:
Atg36 is a pexophagy receptor, which localizes to the membrane of peroxisomes in S. cerevisiae. Atg36 binds Atg8 and the scaffold protein Atg11 that links receptors for selective types of autophagy to the core autophagy machinery.1616
- Atg37:
Atg37 is a conserved acyl-CoA-binding protein that is required specifically for pexophagy in P. pastoris at the stage of phagophore formation.345 See also ACBD5.
- Atg38:
Atg38 physically interacts with Atg14 and Vps34 via its N terminus. Atg38 is required for macroautophagy as an integral component of the PtdIns3K complex I in yeast, and Atg38 functions as a linker connecting the Vps15-Vps34 and Vps30/Atg6-Atg14 subcomplexes to facilitate complex I formation.1617
- Atg39:
A receptor for selective macroautophagic degradation of nuclear membrane in yeast.839
- Atg40:
A receptor that functions in yeast reticulophagy.839 See also FAM134B.
- Atg41:
A protein that interacts with Atg9 and is needed for efficient Atg9 movement to the PAS in yeast.1955
- ATG101:
An ATG13-binding protein conserved in various eukaryotes but not in S. cerevisiae. Forms a stable complex with ULK1/2-ATG13-RB1CC1 (i.e., not nutrient-dependent) required for macroautophagy and localizes to the phagophore.1618,1619 The C. elegans homolog is EPG-9.
- ATI1/2 (ATG8-interacting protein 1/2):
Two closely related ATG8-binding proteins in Arabidopsis, which are unique to plants and define a stress-induced and ER-associated compartment that may function in a direct, Golgi-independent, ER-to-vacuole trafficking pathway.1620 ATI1 is also found in plastids following abiotic stress where it interacts with both ATG8 and plastid-localized proteins to act in their delivery to the central vacuole in an ATG5-dependent manner.801
- ATM (ATM serine/threonine kinase):
A protein kinase that activates TSC2 via the STK11/LKB1-AMPK cascade in response to elevated ROS, resulting in inhibition of MTOR and activation of macroautophagy.774
- ATP13A2 (ATPase type 13A2):
A transmembrane lysosomal type 5 P-type ATPase that is mutated in recessive familial atypical parkinsonism, with effects on lysosomal function.1621 Loss of ATP13A2 function inhibits the clearance of dysfunctional mitochondria.1622
- ats-1 (Anaplasmatranslocated substrate-1):
A type IV secretion effector of the obligatory intracellular bacterium Anaplasma phagocytophilum that binds BECN1 and induces autophagosome formation; the autophagosomes traffic to, and fuse with, A. phagocytophilum-containing vacuoles, delivering macroautophagic cargoes into the vacuole, which can serve as nutrients for bacterial growth.1623,1624
- ATRA (all-transretinoic acid):
A signaling molecule derived from vitamin A that actives macroautophagy and cell differentiation as demonstrated in leukemia cells.413,1625,1626
- AtTSPO (Arabidopsis thaliana TSPO-related):
An ER- and Golgi-localized polytopic membrane protein transiently induced by abiotic stresses. AtTSPO binds ATG8 and heme in vivo and may be involved in scavenging of cytosolic porphyrins through selective macroautophagy.1627
- AUTEN-67 (autophagy enhancer-67):
An inhibitor of MTMR14, which enhances macroautophagy.1628
- Autophagic lysosome reformation (ALR):
A self-regulating tubulation process in which the macroautophagic generation of nutrients reactivates MTOR, suppresses macroautophagy and allows for the regeneration of lysosomes that were consumed as autolysosomes.527 See also autolysosome.
- Autolysosome (AL):
A degradative compartment formed by the fusion of an autophagosome (or initial autophagic vacuole/AVi) or amphisome with a lysosome (also called degradative autophagic vacuole/AVd). Upon completion of degradation the autolysosome can become a residual body,1559,1629 or the autolysosomal membrane can be recycled to generate mature lysosomes during macroautophagic flux. This regenerative process, referred to as autophagic lysosome reformation, relies on the scission of extruded autolysosomal membrane tubules by the mechanoenzyme DNM2 (dynamin 2).527,1630
- Autophagic body (AB):
The inner membrane-bound structure of the autophagosome that is released into the vacuolar lumen following fusion of the autophagosome with the vacuole limiting membrane. In S. cerevisiae, autophagic bodies can be stabilized by the addition of the proteinase B inhibitor PMSF to the medium or by the deletion of the PEP4 or ATG15 genes. Visualization of the accumulating autophagic bodies by differential interference contrast using light microscopy is a convenient, but not easily quantified, method to follow macroautophagy.93
- Autophagic cell death:
A historically ambiguous term describing cell death with morphological features of increased autophagic vacuoles. This term is best reserved for cell death contexts in which specific molecular methods, rather than only pharmacological or correlative methods, are used to demonstrate increased cell survival following inhibition of macroautophagy.
- Autophagic stress:
A pathological situation in which induction of autophagy exceeds the cellular capacity to complete lysosomal degradation and recycling of constituents; may involve a combination of bioenergetics, acidification and microtubule-dependent trafficking deficits, to which neurons may be particularly vulnerable.15
- Autophagic vacuole:
A term typically used for mammalian cells that collectively refers to autophagic structures at all stages of maturation. We recommend using this term when the specific identity of autophagosomes, amphisomes and autolysosomes are not distinguished.
- AutophagamiR:
A term to describe miRNAs that function in the regulation of macroautophagy.1631
- Autophagist:
A researcher working in the field of autophagy.
- Autophagolysosome (APL):
A degradative compartment formed by the fusion of an LC3-containing phagosome (see LAP) or an autophagosome that has sequestered a partial or complete phagosome with a lysosome. In contrast to a phagolysosome, formation of the autophagolysosome involves components of the macroautophagic machinery. Note that this term is not interchangeable with “autophagosome” or “autolysosome”.884
- Autophagoproteasome (APP):
A cytosolic membrane-bound compartment denoted by a limiting single, double or multiple membrane, which contains both LC3 and UPS antigens. The autophagoproteasome may be derived from the inclusion of ubiquitin-proteasome structures within either early or late autophagosomes containing cytoplasmic material at various stages of degradation.73
- Autophagosome (AP):
A cytosolic membrane-bound compartment denoted by a limiting double membrane (also referred to as initial autophagic vacuole, AVi, or early autophagosome). The early autophagosome contains cytoplasmic inclusions and organelles that are morphologically unchanged because the compartment has not fused with a lysosome and lacks proteolytic enzymes. Notably, the double-membrane structure may not be apparent with certain types of fixatives. Although in most cases the term autophagosome refers to a double-membrane compartment, the late autophagosome may also appear to have a single membrane (also referred to as an intermediate or intermediate/degradative autophagic vacuole, AVi/d).1559,1629
- Autophagy:
This term summarizes all processes in which intracellular material is degraded within the lysosome/vacuole and where the macromolecular constituents are recycled.
- Autophagy:
A journal devoted to research in the field of autophagy (http://www.tandfonline.com/toc/kaup20/current#.VdzKoHjN5xu).
- Autophagy adaptor:
A LIR-containing protein that is not itself a cargo for macroautophagy.
- Autophagy receptor:
A LIR/AIM-containing protein that targets specific cargo for degradation and itself becomes degraded by macroautophagy (e.g., SQSTM1, NBR1, OPTN, Atg19).1632
- Autophagy-like vesicles (ALVs):
Double-membraned vesicles (70–400 nm) that accumulate in cells infected by a number of different viruses. These vesicles also have been referred to as compound membrane vesicles (CMVs) or as double-membraned vesicles (DMVs).
- Autosis:
A form of macroautophagy-dependent cell death that requires Na+,K+-ATPase activity (in addition to the macroautophagy machinery).1080 Morphologically, autosis has increased numbers of autophagosomes and autolysosomes, and nuclear convolution during its early stages, followed by focal swelling of the perinuclear space. Autosis occurs in response to various types of stress including starvation and hypoxia-ischemia.
- Ayr1:
A triacylglycerol lipase involved in macroautophagy in yeast.1633 Enzymes that participate in the metabolism of lipid droplets including Dga1 and Lro1 (acyltransferases involved in triacylglycerol synthesis) and Are1/2 (Acyl-CoA:sterol acyltransferases) that generate the major components of lipid droplets, triacylglycerols and steryl esters, are required for efficient macroautophagy. Deletion of the genes encoding Yeh1 (a steryl ester hydrolase), Ayr1 or Ldh1 (an enzyme with esterase and triacylglycerol lipase activities) also partially blocks macroautophagy. Finally, Ice2 and Ldb16, integral membrane proteins that participate in formation of ER-lipid droplet contact sites that may be involved in lipid transfer between these sites are also needed for efficient macroautophagy.
- AZD8055:
A novel ATP-competitive inhibitor of MTOR kinase activity. AZD8055 shows excellent selectivity against all class I PI3K isoforms and other members of the PI3K-like kinase family. Treatment with AZD8055 inhibits MTORC1 and MTORC2 and prevents feedback to AKT.1195
- Bafilomycin A1(BAFA1/BAF):
An inhibitor of the V-type ATPase as well as certain P-type ATPases that prevents acidification and alters the membrane potential of certain compartments; treatment with bafilomycin A1 ultimately results in a block in fusion of autophagosomes with lysosomes, thus preventing the maturation of autophagosomes into autolysosomes.156,157,226 Note that the abbreviation for bafilomycin A1 is not “BFA,” as the latter is the standard abbreviation for brefeldin A; nor should BAF be confused with the abbreviation for the caspase inhibitor boc-asp(o-methyl)fluoremethylketone.
- BAG3 (BCL2-associated athanogene 3):
A stress-induced co-chaperone that utilizes the specificity of HSP70 molecular chaperones toward non-native proteins as the basis for targeted, ubiquitin-independent macroautophagic degradation in mammalian cells (“BAG3-mediated selective macroautophagy”); BAG3 is induced by stress and during cell aging, and interacts with HSP70 and dynein to target misfolded protein substrates to aggresomes, leading to their selective degradation.1559,1634 BAG3 also interacts with HSPB6 and HSPB8 to target substrates for chaperone-assisted selective autophagy via a ubiquitin-dependent mechanism.1116
- BAG6/BAT3 (BCL2-associated athanogene 6):
BAG6 tightly controls macroautophagy by modulating EP300 intracellular localization, affecting the accessibility of EP300 to its substrates, TP53 and ATG7. In the absence of BAG6 or when this protein is located exclusively in the cytosol, macroautophagy is abrogated, ATG7 is hyperacetylated, TP53 acetylation is abolished, and EP300 accumulates in the cytosol, indicating that BAG6 regulates the nuclear localization of EP300.1635
- BARA (β-α repeated, autophagy-specific):
A domain at the C terminus of Vps30/Atg6 that is required for targeting PtdIns3K complex I to the PAS.1636 The BARA domain is also found at the C terminus of BECN1 and in UVRAG.
- Barkor:
See ATG14.
- Basal autophagy:
Constitutive autophagic degradation that proceeds in the absence of any overt stress or stimulus. Basal autophagy is important for the clearance of damaged proteins and organelles in normal cells (especially fully differentiated, nondividing cells).
- BATS (Barkor/Atg14[L] autophagosome targeting sequence) domain:
A protein domain within ATG14 that is required for the recruitment of the class III PtdIns3K to LC3-containing puncta during macroautophagy induction; the predicted structure of the BATS domain suggests that it senses membrane curvature.550
- Bck1:
A MAPKKK downstream of Pkc1 and upstream of Mkk1/2 and Slt2 that controls cell integrity in response to cell wall stress; Bck1 is required for pexophagy683 and mitophagy.508 See also Slt2 and Hog1.
- BCL2 family of proteins:
There are 3 general classes of BCL2 proteins; anti-apoptotic proteins include BCL2, BCL2L1/Bcl-XL, BCL2L2/BCL-W and MCL1 that inhibit macroautophagy, the pro-apoptotic BH3-only proteins include BNIP3, BAD, BIK, PMAIP1/NOXA, BBC3/PUMA and BCL2L11/Bim/BimEL that induce macroautophagy, and the pro-apoptotic effector proteins BAX and BAK1. Interaction of BCL2 with BECN1 prevents the association of the latter with the class III PtdIns3K; however, anti-apoptotic BCL2 proteins require BAX and BAK1 to modulate macroautophagy.1637
- BCL2L13/BCL-RAMBO (BCL2-like 13 [apoptosis facilitator]):
BCL2L13 is a mammalian holomog of Atg32, which is located in the mitochondrial outer membrane and has an LC3-interacting region. BCL2L13 induces mitochondrial fission and mitophagy.1638 See also Atg32.
- BCL10 (B-cell CLL/lymphoma 10):
The adaptor protein BCL10 is a critically important mediator of T cell receptor (TCR)-to-NFKB signaling. After association with the receptor SQSTM1, BCL10 is degraded upon TCR engagement. Selective macroautophagy of BCL10 is a pathway-intrinsic homeostatic mechanism that modulates TCR signaling to NFKB in effector T cells.1639
- BEC-1:
The C. elegans ortholog of BECN1.
- Beclin 1:
See BECN1.
- BECN1/Beclin 1 (beclin 1, autophagy related):
A mammalian homolog of yeast Vps30/Atg6 that forms part of the class III PtdIns3K complex involved in activating macroautophagy.1640 BECN1 interacts with many proteins including BCL2, VMP1, ATG14, UVRAG, PIK3C3 and RUBCN/Rubicon through its BH3, coiled-coil and BARA domains, the latter including the evolutionarily conserved domain (ECD).1641 The C. elegans ortholog is BEC-1.
- BECN1s (BECN1 short isoform):
A splice variant of BECN1 that lacks the sequence corresponding to exons 10 and 11; BECN1s associates with the mitochondrial outer membrane and is required for mitophagy.1642 BECN1s can bind ATG14 and activate PIK3C3/VPS34, but does not bind UVRAG.
- BECN2/Beclin 2 (beclin 2):
A mammalian-specific homolog of yeast Vps30/Atg6 that forms part of the class III PtdIns 3K complex involved in activating macroautophagy and that also functions in the endolysosomal degradation of G protein-coupled receptors (independently of the class III PtdIns3K complex).1643
- Betulinic acid:
Betulinic acid and its derivatives activate macroautophagy as a rescue mechanism to deal with damaged mitochondria;235,1167,1168,1644 however, betulinic acid impairs lysosomal integrity and converts macroautophagy into a detrimental process, leading to accumulation of nonfunctional autolysosomes that can be detected over a long time frame.235
- BH domain:
BCL2 homology domain. There are 4 domains of homology, consisting of BH1, BH2, BH3 and BH4.
- BH3 domain:
A BCL2 homology (BH) domain that is found in all BCL2 family proteins, whether they are pro-apoptotic or anti-apoptotic. A BH3 domain is also present in BECN1 and mediates the interaction with anti-apoptotic proteins possessing a BH3 receptor domain (i.e., BCL2, BCL2L1/bcl-xL, BCL2L2/BCL-W and MCL1).
- BH3-only proteins:
A series of proteins that contain a BH3 domain (but not any other BCL2 homology domains). Several BH3-only proteins (BNIP3, BAD, BIK, PMAIP1/NOXA, BBC3/PUMA and BCL2L11/Bim/BimEL) can competitively disrupt the inhibitory interaction between BCL2 and BECN1 to allow the latter to act as an allosteric activator of PtdIns3K and to activate macroautophagy.
- Bif-1:
See SH3GLB1.
- BIPASS (BAG-instructed proteasomal to autophagosomal switch and sorting):
Upon proteasomal impairment, cells switch to autophagy to ensure proper clearance of substrates (the proteasome-to-autophagy switch). Following this proteasome impairment, increasing the BAG3:BAG1 ratio ensures the initiation of BIPASS.1645
- BNIP3 (BCL2/adenovirus E1B 19kDa interacting protein 3):
- Identified in a yeast two-hybrid screen as interacting through its amino terminal 40 amino acids with BCL2 and adenovirus E1B.1646 Originally classified as a pro-apoptotic protein, BNIP3 promotes mitophagy through direct interaction with LC3B-II mediated by a conserved LIR motif that overlaps with its BCL2 interacting region.1647,1648 BNIP3 also modulates mitochondrial fusion through inhibitory interactions with OPA1 via its carboxy terminal 10 amino acids.1649 BNIP3 is transcriptionally regulated by HIF1A,1650 E2Fs,1651 FOXO3,468 TP531652 and NFKB1653 and is most highly expressed in adult heart and liver.1654,1655

- BNIP3L/NIX (BCL2/adenovirus E1B 19kDa interacting protein 3-like):
Identified as a BNIP3 homolog, BNIP3L is required for mitophagy in red blood cells.1299,1300 Like BNIP3, BNIP3L is hypoxia-inducible and also interacts with LC3B-II and GABARAP through a conserved LIR motif in its amino terminus.210 BNIP3L also interacts with RHEB at the mitochondria and the LC3-BNIP3L-RHEB complex promotes mitochondrial turnover and efficient mitochondrial function.1656
- Bre5:
A cofactor for the deubiquitinase Ubp3. See also Ubp3.
- C/EBPβ:
See CEBPB.
- C9orf72/ALSFTD:
C9orf72 plays an important role in the regulation of endosomal trafficking, and interacts with RAB proteins involved in macroautophagy and endocytic transport. C9orf72 contains a DENN (differentially expressed in normal and neoplasia)-like domain, suggesting that it may function as a GDP-GTP exchange factor for a RAB GTPase, similar to other DENN proteins. The normal function of C9orf72 remains unknown but it is highly conserved and expressed in many tissues, including the cerebellum and cortex. Hexanucleotide (GGGGCC) repeat expansions in a noncoding region of the C9orf72 gene are the major cause of familial ALS and frontotemporal dementia.1657
- C12orf5:
See TIGAR.
- C12orf44:
See ATG101
- Ca-P60A/dSERCA:
The Drosophila ER Ca2+-translocating ATPase. Inhibition of Ca-P60A with bafilomycin A1 blocks autophagosome-lysosome fusion.226
- Cad96Ca/Stit/Stitcher (Cadherin 96Ca):
A Drosophila receptor tyrosine kinase that is orthologous to the human proto-oncogene RET. Cad96Ca suppresses macroautophagy in epithelial tissues through Akt1-TORC1 signaling in parallel to InR (Insulin-like receptor). This endows epithelial tissues with starvation resistance and anabolic development during nutritional stress.1658
- Caf4:
A component of the mitochondrial fission complex that is recruited to degrading mitochondria to facilitate mitophagy-specific fission.705
- CAL-101:
A small molecule inhibitor of the PIK3CD/p110δ subunit of class 1A phosphoinositide 3-kinase; treatment of multiple myeloma cells results in macroautophagy induction.1659
- Calcineurin:
See PPP3R1.
- CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2):
A receptor that binds to the bacterial ubiquitin coat and Atg8/LC3 to target invasive bacteria, including S. typhimurium and Streptococcus pyogenes for autophagosomal sequestration.878
- Calpains:
A class of calcium-dependent, non-lysosomal cysteine proteases that cleaves and inactivates ATG5 and the ATG12–ATG5 conjugate, hence establishing a link between reduced Ca2+ concentrations and induction of macroautophagy.1660
- CALR (calreticulin):
A chaperone that is mainly associated with the ER lumen, where it performs important functions such as Ca2+ buffering, and participates in protein folding and maturation of, as well as antigen loading on, MHC molecules.1661 An extracellular role for CALR has emerged where it acts as an “eat me” signal on the surface of cancer cells.1662 Importantly, in the context of Hyp-PDT, macroautophagy suppresses CALR surface exposure by reducing ER-associated proteotoxicity.1053,1058,1663 Disruption of LAMP2A also affects CALR surface exposure.1058
- CaMKKβ:
See CAMKK2.
- CAMKK2 (calcium/calmodulin-dependent protein kinase kinase 2, beta):
Activates AMPK in response to an increase in the cytosolic calcium concentration,1664 resulting in the induction of macroautophagy.1223
- CAPNS1 (calpain, small subunit 1):
The regulatory subunit of micro- and millicalpain; CAPNS1-deficient cells are macroautophagy defective and display a substantial increase in apoptotic cell death.1665
- CASA (chaperone-assisted selective autophagy):
A degradative process that utilizes the Drosophila co-chaperone Starvin or its mammalian homolog BAG3 to direct the degradation of aggregated substrates through the action of HSPA8, HSPB8, the STUB1/CHIP ubiquitin ligase and SQSTM1.1116 The requirement for ubiquitination of the substrates (and the absence of a requirement for the KFERQ motif) along with the involvement of the ATG proteins differentiate this process from CMA, which also uses chaperones for lysosome-dependent degradation.
- Caspases (cysteine-dependent aspartate-directed proteases):
A class of proteases that play essential roles in apoptosis (formerly called programmed cell death type I) and inflammation. Several pro-apoptotic caspases cleave essential macroautophagy proteins, resulting in the inhibition of macroautophagy.438 For example, CASP3 and CASP8 cleave BECN1 and inhibit macroautophagy.1666,1667
- CCCP (carbonyl cyanide m-chlorophenylhydrazone):
Protonophore and uncoupler of oxidative phosphorylation in mitochondria; stimulates mitochondrial degradation inducing mitophagic activity.250
- CCDC88A/GIV (coiled-coil domain containing 88A):
A guanine nucleotide exchange factor for GNAI3 that acts to downregulate macroautophagy.1668 CCDC88A disrupts the GPSM1-GNAI3 complex in response to growth factors, releasing the G protein from the phagophore or autophagosome membrane; GNAI3-GTP also activates the class I PI3K, thus inhibiting macroautophagy. See also GNAI3.
- CCI-779 (temsirolimus):
A water-soluble rapamycin ester that induces macroautophagy.
- Cdc48:
Yeast homolog of VCP that is a type II AAA+-ATPase that extracts ubiquitinated proteins from the membrane as part of the ER-associated protein degradation pathway and during ER homeotypic fusion,1669 but is also required for nonselective macroautophagy.1670 See also Shp1 and VCP.
- CD46:
A cell-surface glycoprotein that interacts with the scaffold protein GOPC to mediate an immune response to invasive pathogens including Neisseria and Group A Streptococcus. Interaction of pathogens via the Cyt1 cytosolic tail induces macroautophagy, which involves GOPC binding to BECN1. CD46 is also used as a cellular receptor by several pathogens.1671
- CDKN1A/p21 (cyclin-dependent kinase inhibitor 1A [p21, Cip1]):
A cyclin-dependent kinase inhibitor that is associated with the induction of macroautophagy in melanoma cells upon exposure to a telomeric G-quadruplex stabilizing agent.1672
- CDKN1B/p27 (cyclin-dependent kinase inhibitor 1B [p27, Kip1]):
A cyclin-dependent kinase inhibitor that is phosphorylated and stabilized by an AMPK-dependent process and stimulates macroautophagy.1673
- CDKN2A (cyclin-dependent kinase inhibitor 2A):
The CDKN2A locus encodes 2 overlapping tumor suppressors that do not share reading frame: p16INK4a and p14ARF. The p14ARF tumor suppressor protein (p19ARF in mouse) can localize to mitochondria and induce macroautophagy. Tumor-derived mutant forms of p14ARF that do not affect the p16INK4a coding region are impaired for macroautophagy induction, thus implicating this activity in tumor suppression by this commonly mutated locus.1674 This gene also encodes a smaller molecular weight variant called smARF. See also smARF.
- CEBPB/C/EBPβ (CCAAT/enhancer binding protein [C/EBP], beta):
A transcription factor that regulates several autophagy genes; CEBPB is induced in response to starvation, and the protein levels display a diurnal rhythm.1000
- Cell differentiation:
This is a process through which a cell commits to becoming a more specialized cell type having a distinct form and a specific function(s). Autophagy is activated during the differentiation of various normal and cancerous cells, as revealed, for example, in adipocytes, erythrocytes, lymphocytes and leukemia cells.452
- CEP-1 (C. elegans P-53-like protein):
See TP53.
- Ceramide:
Ceramide is a bioactive sphingolipid, which plays a mitochondrial receptor role to recruit LC3-II-associated phagophores to mitochondria for degradation in response to ceramide stress and DNM1L-mediated mitochondrial fission; the direct binding between ceramide and LC3-II involves F52 and I35 residues of LC3B.591
- Chaperone-mediated autophagy (CMA):
An autophagic process in mammalian cells by which proteins containing a particular pentapeptide motif related to KFERQ are transported across the lysosomal membrane and degraded.1675,1676 The translocation process requires the action of the integral membrane protein LAMP2A and both cytosolic and lumenal HSPA8.1677,1678
- CHKB (choline kinase beta):
A kinase involved in phosphatidylcholine synthesis; mutations in CHKB cause mitochondrial dysfunction leading to mitophagy and megaconial congenital muscular dystrophy.1679
- Chloroquine (CQ):
Chloroquine and its derivatives (such as 3-hydroxychloroquine) raise the lysosomal pH and ultimately inhibit the fusion between autophagosomes and lysosomes, thus preventing the maturation of autophagosomes into autolysosomes, and blocking a late step of macroautophagy.1680
- CHMP1A (charged multivesicular body protein 1A):
CHMP1A is a member of the CHMP family of proteins that are involved in multivesicular body sorting of proteins to the interiors of lysosomes. CHMP1A regulates the macroautophagic turnover of plastid constituents in Arabidopsis thaliana.802
- Chromatophagy:
A form of macroautophagy that involves nuclear chromatin/DNA leakage captured by autophagosomes or autolysosomes.803
- Ciliophagy:
Degradation by macroautophagy of proteins involved in the process of ciliogenesis (formation of primary cilia). Ciliophagy can modulate ciliogenesis positively or negatively depending on whether the subset of proteins degraded in autophagosomes are activators or inhibitors of the formation of primary cilia.
- CISD2/NAF-1 (CDGSH iron sulfur domain 2):
An integral membrane component that associates with the ITPR complex; CISD2 binds BCL2 at the ER, and is required for BCL2 to bind BECN1, resulting in the inhibition of macroautophagy.1681 CISD2 was reported to be associated with the ER, but the majority of the protein is localized at mitochondria, and mutations in CISD2 are associated with Wolfram syndrome 2; accelerated macroautophagy in cisd2-/- mice may cause mitochondrial degradation, leading to neuron and muscle degeneration.1682
- CLEAR (coordinated lysosomal expression and regulation) gene network:
A regulatory pathway involving TFEB, which regulates the biogenesis and function of the lysosome and associated pathways including macroautophagy.636 See also PPP3R1 and TFEB.
- CLEC16A (C-type lectin domain family 16, member A):
See Ema.
- Clg1:
A yeast cyclin-like protein that interacts with Pho85 to induce macroautophagy by inhibiting Sic1.1683
- CLN3 (ceroid-lipofuscinosis, neuronal 3):
An endosomal/lysosomal protein whose deficiency causes inefficient autolysosome clearance and accumulation of autofluorescent lysosomal storage material and ATP5G/subunit c (ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C [subunit 9]).1684,1685 In human, recessive CLN3 mutations cause juvenile neuronal ceroid lipofuscinosis (JNCL; Batten disease). Recessive CLN3 mutations have also been reported in cases of autophagic vacuolar myopathy and non-syndromic retinal disease.1686,1687
- COG (conserved oligomeric Golgi) complex:
A cytosolic tethering complex that functions in the fusion of vesicles within the Golgi complex, but also participates in macroautophagy and facilitates the delivery of Atg8 and Atg9 to the PAS.1688
- Connexins:
See gap junction protein.
- CORM (CO-releasing molecule):
Carbon monoxide, partly through activation of macroautophagy, exerts cardioprotective effects in a mouse model of metabolic syndrome-induced myocardial dysfunction.1689
- Corynoxine/Cory:
An oxindole alkaloid isolated from Uncaria rhynchophylla (Miq.) Jacks (Gouteng in Chinese) that is a Chinese herb that acts as a MTOR-dependent macroautophagy inducer.1690
- Corynoxine B/Cory B:
An isomer of corynoxine, also isolated from the Chinese herb Uncaria rhynchophylla (Miq.) Jacks that acts as a BECN1-dependent macroautophagy inducer.1691
- Crinophagy:
Selective degradation of secretory granules by fusion with the lysosome, independent of macroautophagy.1692 See also zymophagy.
- Cryptides:
Peptides with a cryptic biological function that are released from cytoplasmic proteins by partial degradation or processing through macroautophagy (e.g., neoantimocrobial peptide released from ribosomal protein FAU/RPS30).1693
- CSNK2 (casein kinase 2):
A serine/threonine protein kinase that disrupts the BECN1-BCL2 complex to induce macroautophagy.1694 CSNK2 also phosphorylates ATG16L1, in particular on Ser139, to positively regulate macroautophagy. See also PPP1.
- Ctl1:
A multi-transmembrane protein in the fission yeast Schizosaccharomyces pombe that binds to Atg9 and is required for autophagosome formation.1695
- Cue5:
A yeast receptor similar to mammalian SQSTM1 that binds ubiquitin through its CUE domain and Atg8 via its C-terminal AIM.451 Some Cue5-dependent substrates are ubiquitinated by Rsp5. See also CUET.
- CUET (Cue5/TOLLIP):
A family of macroautophagy receptor proteins containing a CUE domain that are involved in macroautophagic clearance of protein aggregates. See also Cue5.451
- CUP-5 (coelomocyte uptake defective mutant-5):
The ortholog of human MCOLN1 (mucolipin 1), in C. elegans CUP-5 localizes to lysosomes, and is required for endo-lysosomal transport, lysosomal degradation,1696-1698 and proteolytic degradation in autolysosomes.1699
- CUPS (compartment for unconventional protein secretion):
A compartment located near ER exit sites that is involved in the secretion of Acb1; Grh1 is localized to the CUPS membrane, and Atg8 and Atg9 are subsequently recruited under starvation conditions.1700 Atg8 and Atg9 function in Acb1 secretion, but rapamycin-induced macroautophagy does not result in CUPS formation.
- Cvt body:
The single-membrane vesicle present inside the vacuole lumen that results from the fusion of a Cvt vesicle with the vacuole.131
- Cvt complex:
A cytosolic protein complex consisting primarily of prApe1 dodecamers in the form of an Ape1 complex that are bound to the Atg19 reeptor. This complex may also contain Ams1 and Ape4, but prApe1 is the predominant component.131
- Cvt vesicle:
The double-membrane sequestering vesicle of the Cvt pathway.131
- Cysmethynil:
A small-molecule inhibitor of ICMT (isoprenylcysteine carboxyl methyltransferase); treatment of PC3 cells causes an increase in LC3-II and cell death with macroautophagic features.1701
- Cytoplasm-to-vacuole targeting (Cvt) pathway:
A constitutive, biosynthetic pathway in yeast that transports resident hydrolases to the vacuole through a selective macroautophagy-like process.1702 See also Ams1, Ape1, Ape4 and Atg19.
- DAF-2 (abnormal dauer formation):
Encodes the C. elegans insulin/IGF1-like receptor homolog that acts through a conserved PI3K pathway to negatively regulate the activity of DAF-16/FOXO and limit life span. DAF-2 inhibits macroautophagy by a mechanism that remains to be elucidated.271,1703,1704
- DAF-16:
A C. elegans FOXO transcription factor ortholog.
- DALIS (dendritic cell aggresome-like induced structures):
Large poly-ubiquitinated protein aggregates formed in dendritic cells. These are similar to aggresomes, but they do not localize to the microtubule-organizing center. DALIS are transient in nature and small DALIS have the ability to move and form larger aggregates; they require proteasome activity to clear them.318 See also ALIS.
- DAMP (danger/damage-associated molecular pattern):
DAMPs are recognized by receptors (DDX58/RIG-I-like receptors [RLRs] or TLRs) of the innate surveillance response system. DAMPs include “non-self” molecules such as viral RNA, or products of necroptosis such as HMGB1.295 Response includes activation of macroautophagy to clear the DAMP molecule(s).1705
- DAP (death-associated protein):
A conserved phosphoprotein that is a substrate of MTOR and inhibits macroautophagy; inhibition of MTOR results in dephosphorylation of DAP and inhibition of macroautophagy, thus limiting the magnitude of the autophagic response.1706
- DAPK1 (death-associated protein kinase 1):
A kinase that phosphorylates Thr119 of BECN1 to activate it by causing dissociation from BCL2L1/Bcl-xL and BCL2, thus activating macroautophagy.1707
- DAPK3 (death-associated protein kinase 3):
See Sqa.
- DCN (decorin):
An archetypical member of the small leucine rich proteoglycans that functions as a soluble pro-autophagic and pro-mitophagic signal. DCN acts as a partial agonist for KDR/VEGFR2 and MET for endothelial cell macroautophagy and tumor cell mitophagy, respectively. DCN elicits these processes in a PEG3-dependent manner to induce endothelial cell macroautophagy, and in a TCHP/mitostatin-dependent manner for tumor cell mitophagy. It is postulated that induction of these fundamental cellular programs underlies the oncostatic and angiostatic properties of DCN.1708
- Dcp-1 (death caspase-1):
A Drosophila caspase that localizes to mitochondria and positively regulates macroautophagic flux.1709
- Dcp2/DCP2 (decapping mRNA 2):
A decapping enzyme involved in the downregulation of ATG transcripts.1710 See also Dhh1.
- DCT-1:
The C. elegans homolog of BNIP3 and BNIP3L, which functions downstream of PINK-1 and PDR-1 to regulate mitophagy under conditions of oxidative stress.1275
- DDIT4/DIG2/RTP801/REDD1 (DNA-damage-inducible transcript 4):
The DDIT4 protein is notably synthesized in response to glucocorticoids or hypoxia and inhibits MTOR, resulting in the induction of macroautophagy and enhanced cell survival.1711
- Deconjugation:
- The Atg4/ATG4-dependent cleavage of Atg8–PE/LC3-II that releases the protein from PE (illustrated for the nascent yeast protein that contains a C-terminal arginine). The liberated Atg8/LC3 can subsequently go through another round of conjugation. Atg8*, activated Atg8.
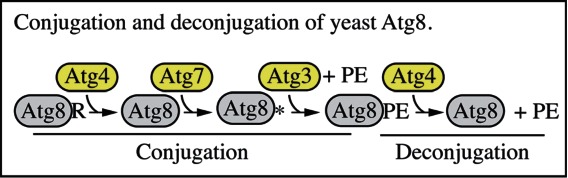
- Decorin:
See DCN.
- Decoupled signaling:
When limited for an auxotrophic requirement, yeast cells fail to induce the expression of autophagy genes even when growing slowly, which contributes to decreased cell viability.1712
- Desat1:
A Drosophila lipid desaturase that localizes to autophagosomes under starvation conditions; the Desat mutant is defective in macroautophagy induction.1713
- DFCP1:
See ZFYVE1.
- Dga1:
See Ayr1.
- Dhh1:
An RCK member of the RNA-binding DExD/H-box proteins involved in mRNA decapping; Dhh1 in S. cerevisiae and Vad1 in Cryptococcus neoformans bind certain ATG transcripts, leading to the recruitment of the Dcp2 decapping enzyme and mRNA degradation.1710 See also Dcp2.
- Diacylglycerol:
A lipid second messenger that contributes to macroautophagic targeting of Salmonella-containing vacuoles.1714
- DIG2:
See DDIT4.
- DIRAS3 (DIRAS family, GTP-binding RAS-like 3):
A protein that interacts with BECN1, displacing BCL2 and blocking BECN1 dimer formation, thus promoting the interaction of BECN1 with PIK3C3 and ATG14, resulting in macroautophagy induction.1715
- Dnm1:
A dynamin-related GTPase that is required for both mitochondrial and peroxisomal fission. Dnm1 is recruited to degrading mitochondria by Atg11, or to degrading peroxisomes by both Atg11 and Atg36 (or PpAtg30), to mediate mitophagy- or pexophagy-specific fission.705,1716 See also DNM1L.
- DNM1L/Drp1 (dynamin 1-like):
The mammalian homolog of yeast Dnm1. PRKA-mediated phosphorylation of rat DNM1L on Ser656 (Ser637 in humans) prevents both mitochondrial fission and some forms of mitophagy in neurons.1717 See also Dnm1.
- DNM2 (dynamin 2):
DNM2 is recruited to extruded autolysosomal membranes during the process of autophagic lysosome reformation and catalyzes their scission, promoting the regeneration of nascent protolysosomes during macroautophagic flux.1630 See also autophagic lysosome reformation.
- dom (domino):
A Drosophila SWI2/SNF2 chromatin remodeling protein. A loss-of-function mutation at the dom locus synergizes with genotypes depressed in macroautophagy pathway activity.1718
- Dopamine:
A neurotransmitter whose accumulation outside vesicles induces macroautophagy and cell degeneration.1719
- DOR:
See TP53INP2.
- DRAM1 (damage-regulated autophagy modulator 1):
DRAM1 gene expression is induced by TP53 in response to DNA damage that results in cell death by macroautophagy.580 DRAM1 is an endosomal-lysosomal membrane protein that is required for the induction of macroautophagy. The knockdown of DRAM1 causes downregulation of VRK1 by macroautophagy, similar to the effect of knocking down BECN1.
- Draper:
A Drosophila homolog of the Caenorhabditis elegans engulfment receptor CED-1 that is required for macroautophagy associated with cell death during salivary gland degradation, but not for starvation-induced macroautophagy in the fat body.1720
- Drs:
See SRPX.
- E2F1:
A mammalian transcription factor that upregulates the expression of BNIP3, LC3, ULK1 and DRAM1 directly, and ATG5 indirectly.614 E2F1 plays a role during DNA damage- and hypoxia-induced macroautophagy.
- EAT (early autophagy targeting/tethering) domain:
The C-terminal domain of Atg1, which is able to tether vesicles.1721 This part of the protein also contains the binding site for Atg13.
- EAT-2 (eating abnormal):
A ligand-gated ion channel subunit closely related to the non-alpha subunit of nicotinic acetylcholine receptors, which functions to regulate the rate of pharyngeal pumping. eat-2 loss-of-function mutants are dietary restricted and require macroautophagy for the extension of life span.1703,1722,1723
- EDTP:
See MTMR14.
- EEA1 (early endosome antigen 1):
A RAB5 effector used as a common marker for early endosome vesicles.
- EEF1A1/EF1A/eF1α (eukaryotic translation elongation factor 1 alpha 1):
Multifunctional member of the family of G-proteins with different cellular variants. The lysosomal variant of this protein acts coordinately with GFAP at the lysosomal membrane to modulate the stability of the CMA translocation complex. Release of membrane bound EEF1A1 in a GTP-dependent manner promotes disassembly of the translocation complex and consequently reduces CMA activity.1724
- eF1α:
See EEF1A1.
- EGFR (epidermal growth factor receptor):
A tyrosine kinase receptor that negatively regulates macroautophagy through PI3K, AKT, and MTOR modulation.523
- EGO complex:
The Ego1, Ego3 and Gtr2 proteins form a complex that positively regulates yeast microautophagy.1725
- eIF2α kinase:
See EIF2S1 kinase.
- EIF2AK2/PKR (eukaryotic translation initiation factor 2-alpha kinase 2):
A mammalian EIF2S1/EIF2 alpha kinase that induces macroautophagy in response to viral infection.558
- EIF2AK3/PERK (eukaryotic translation initiation factor 2-alpha kinase 3):
A mammalian EIF2S1/EIF2 alpha kinase that may induce macroautophagy in response to ER stress.602
- EIF2S1 (eukaryotic translation initiation factor 2, subunit 1, alpha, 35kDa):
An initiation factor that is involved in stress-induced translational regulation of macroautophagy.
- EIF2S1/eIF2α kinase:
There are 4 mammalian EIF2S1/EIF2 alpha kinases that respond to different types of stress. EIF2AK2 and EIF2AK3 induce macroautophagy in response to virus infection and ER stress, respectively.602,1726 See also Gcn2, EIF2AK2 and EIF2AK3.
- Elaiophylin:
A natural compound late-stage macroautophagy inhibitor that results in lysosomal membrane permeabilization and decreased cell viability.1727 See also LMP.
- Ema (endosomal maturation defective):
Ema is required for phagophore expansion and for efficient mitophagy in Drosophila fat body cells. It is a transmembrane protein that relocalizes from the Golgi to phagophores following starvation.1728 The vertebrate ortholog CLEC16A regulates mitophagy and is a susceptibility locus for many autoimmune disorders.1729,1730
- Embryoid bodies/EBs:
Three-dimensional aggregates of pluripotent stem cells including embryonic stem cells and induced pluripotent stem cells.
- EMC6/TMEM93 (ER membrane protein complex subunit 6):
A novel ER-localized transmembrane protein, which interacts with both RAB5A and BECN1 and colocalizes with the omegasome marker ZFYVE1/DFCP1.1731 EMC6 enhances autophagosome formation when overexpressed.
- Endorepellin:
The anti-angiogenic C-terminal cleavage product of HSPG2/perlecan. Endorepellin engages KDR/VEGFR2 and ITGA2/α2β1 integrin in a novel mechanism termed dual receptor antagonism for achieving endothelial cell specificity and function. Endorepellin evokes endothelial cell macroautophagy downstream of KDR and in a PEG3-dependent manner.1732
- Endosomal microautophagy (e-MI):
A form of autophagy in which cytosolic proteins are sequestered into late endosomes/MVBs through a microautophagy-like process. Sequestration can be nonselective or can occur in a selective manner mediated by HSPA8. This process differs from chaperone-mediated autophagy as it does not require substrate unfolding and it is independent of the CMA receptor LAMP2A.1115 This process occurs during MVB formation and requires the ESCRT-I and ESCRT-III protein machinery. See also endosome and multivesicular body.
- Endosome:
The endosomal compartments receive molecules engulfed from the extracellular space and are also in communication with the Golgi apparatus. The endosomal system can be viewed as a series of compartments starting with the early endosome. From there, cargos can be recycled back to the plasma membrane; however, more typically, internalized cargo is transported to the late endosome/MVB. These latter compartments can fuse with lysosomes. Ensosomal maturation from early endosomes is a dynamic process that involves a progressive reduction in lumenal pH. In mammalian cells, early and/or multivesicular endosomes fuse with autophagosomes to generate amphisomes.
- EP300/p300 (E1A binding protein p300):
An acetyltransferase that inhibits macroautophagy by acetylating ATG5, ATG7, ATG12 and/or LC3.656 EP300 is also involved in the GLI3-dependent transcriptional activation of VMP1 in cancer cells.634 See also GLI3.
- EPAS1/HIF2A/Hif-2α (endothelial PAS domain protein 1):
Part of a dimeric transcription factor in which the α subunit is regulated by oxygen; the hydroxylated protein is degraded by the proteasome. EPAS1 activation in mouse liver augments peroxisome turnover by pexophagy, and the ensuing deficiency in peroxisomal function encompass major changes in the lipid profile that are reminiscent of peroxisomal disorders.773
- epg(ectopic PGL granules) mutants:
C. elegans mutants that are defective in the macroautophagic degradation of PGL-1, SEPA-1 and/or SQST-1.633 The EPG-3, ATG-11/EPG-7, EPG-8 and EPG-9 proteins are homologs of VMP1, Atg11/RB1CC1, ATG14 and ATG101, respectively, whereas ATG-13/EPG-1 may be a homolog of ATG13.1733
- EPG-1:
See ATG-13.
- EPG-2:
A nematode-specific coiled-coil protein that functions as a scaffold protein mediating the macroautophagic degradation of PGL granule in C. elegans. EPG-2 directly interacts with SEPA-1 and LGG-1. EPG-2 itself is also degraded by macroautophagy.633
- EPG-3:
A metazoan-specific macroautophagy protein that is the homolog of human VMP1. EPG-3/VMP1 are involved in an early step of autophagosome formation.633
- EPG-4:
An ER-localized transmembrane protein that is the homolog of human EI24/PIG8. EPG-4 is conserved in multicellular organisms, but not in yeast. EPG-4 functions in the progression of omegasomes to autophagosomes.633
- EPG-5:
A novel macroautophagy protein that is conserved in multicellular organisms. EPG-5 regulates lysosome degradative capacity and thus could be involved in other pathways that terminate at this organelle.633 Mutations in the human EPG5 gene lead to Vici syndrome.1734
- EPG-6:
A WD40 repeat PtdIns3P-binding protein that directly interacts with ATG-2.563 EPG-6 is the C. elegans functional homolog of yeast Atg18 and probably of mammalian WDR45/WIPI4. EPG-6 is required for the progression of omegasomes to autophagosomes. See also Atg18.
- EPG-7:
See ATG-11.
- EPG-8:
An essential macroautophagy protein that functions as the homolog of yeast Atg14 in C. elegans.1269 EPG-8 is a coiled-coil protein and directly interacts with the C. elegans BECN1 homolog BEC-1. See also Atg14.
- EPG-9:
A protein with significant homology to mammalian ATG101 in C. elegans.1268 EPG-9 directly interacts with ATG-13/EPG-1. See also ATG101.
- EPG-11:
See PRMT-1.
- EPM2A/laforin (epilepsy, progressive myoclonus type 2A, Lafora disease [laforin]):
A member of the dual specificity protein phosphatase family that acts as a positive regulator of macroautophagy probably by inhibiting MTOR, as EPM2A deficiency causes increased MTOR activity.1736 Mutations in the genes encoding EPM2A or the putative E3-ubiquitin ligase NHLRC1/malin, which form a complex, are associated with the majority of defects causing Lafora disease, a type of progressive neurodegeneration. See also NHLRC1.
- ER-phagy:
See reticulophagy.
- ERK1:
See MAPK3.
- ERK2:
See MAPK1.
- ERMES (ER-mitochondria encounter structure):
A complex connecting the endoplasmic reticulum and the mitochondrial outer membrane in yeast. The core components of ERMES are the mitochondrial outer membrane proteins Mdm10 and Mdm34, the ER membrane protein Mmm1, and the peripheral membrane protein Mdm12. ERMES plays an important role in yeast mitophagy presumably by supporting the membrane lipid supply for the growing phagophore membrane.1737
- Everolimus (RAD-001):
An MTOR inhibitor similar to rapamycin that induces macroautophagy.
- ESC8:
A macroautophagy inducer that bears a cationic estradiol moiety and causes downregulation of p-MTOR and its downstream effectors including p-RPS6KB.1738
- EVA1A/FAM176A/TMEM166 (eva-1 homolog A [C. elegans]):
An integral membrane protein that induces macroautophagy and cell death when overexpressed.1739,1740 See also TMEM166.
- EXOC2/SEC5L1 (exocyst complex component 2):
A component of the exocyst complex; EXOC2 binds RALB, BECN1, MTORC1, ULK1 and PIK3C3 under nutrient-rich conditions and prevents these components from interacting with EXOC8/EXO84, thus inhibiting macroautophagy.1741 See also RALB and EXOC8.
- EXOC8/EXO84 (exocyst complex component 8):
A component of the exocyst complex, and an effector of RALB that is involved in nucleation and/or expansion of the phagophore; EXOC8 binds RALB under nutrient-poor conditions, and stimulates the formation of a complex that includes ULK1 and the class III PtdIns3K.1741 See also RALB and EXOC2.
- Exophagy:
A process in yeast and mammalian cells that is used for protein secretion that is independent of the secretory pathway (i.e., unconventional secretion), and dependent on Atg proteins and the Golgi protein Grh1; Acb1 (acyl-coenzyme A-binding protein) uses this route for delivery to the cell surface.1742-1744 See also secretory autophagy.
- FAM48A:
See SUPT20H.
- FAM134B (family with sequence similarity 134, member B):
ER-resident receptors that function in reticulophagy through interaction with LC3 and GABARAP.845
- FAM176A:
See EVA1A.
- Fasudil:
A ROCK (Rho-associated, coiled-coil containing protein kinase) inhibitor that enhances macroautophagy.1745
- Far11:
A MAP kinase target that is involved in the dephosphosphorylation of Atg13 and the induction of macroautophagy.1746 Far11 interacts with Pph21, Pph22 and Pph3 and may coordinate different cellular stress responses by regulating phosphatase activity.
- Ferritinophagy:
The selective degradation of ferritin through a macroautophagy-like process.804 This process involves a specificity receptor, NCOA4.
- FEZ1 (fasciculation and elongation protein zeta 1 [zygin I]):
FEZ1 interacts with ULK1 or with UVRAG, and forms a trimeric complex with either component by also binding SCOC.1747 FEZ1 appears to be a negative regulator of macroautophagy when it is bound only to ULK1, and this inhibition is relieved upon formation of the trimeric complex containing SCOC. Similarly, the SCOC-FEZ1-UVRAG complex is inhibitory; dissociation of UVRAG under starvation conditions allows the activation of the class III PtdIns3K complex. See also SCOC.
- FIP200:
See RB1CC1.
- FIG4 (FIG4 phosphoinositide 5-phosphatase):
A phospholipid phosphatase that controls the generation and turnover of the PtdIns(3,5)P2 phosphoinositide. Loss of FIG4 causes a decrease of PtdIns(3,5)P2 levels, enlargement of late endosomes and lysosomes and cytosolic vacuolization.1748 In human, recessive mutations in FIG4 are responsible for the neurodegenerative Yunis-Varón syndrome, familial epilepsy with polymicrogyria, and Charcot-Marie-Tooth type 4J neuropathy. Haploinsufficiency of FIG4 may also be a risk factor for amyotrophic lateral sclerosis.
- Fis1:
A component of the mitochondrial fission complex. Fis1 also plays a role in peroxisomal fission by recruiting Dnm1 to peroxisomes; it interacts with Atg11 to facilitate mitophagy- and pexophagy-specific fission.705,1716 See also Dnm1.
- FKBP1A (FK506 binding protein 1A, 12kDa):
An immunophilin that forms a complex with rapamycin and inhibits MTOR.
- FKBP5/FKBP51 (FK506 binding protein 5):
An immunophilin that forms a complex with FK506 and rapamycin; FKBP5 promotes macroautophagy in irradiated melanoma cells, thus enhancing resistance to radiation therapy.1749 FKBP5 also associates with BECN1 and shows synergistic effects with antidepressants on macroautophagy in cells, mice and humans, possibly explaining its requirement in antidepressant action.1750
- FKBP12:
See FKBP1A.
- FKBP51:
See FKBP5.
- FLCN (folliculin):
A tumor suppressor mutated in Birt-Hogg-Dubé syndrome.1751 FLCN interacts with GABARAP and this association is modulated by the presence of either FNIP1 (folliculin interacting protein 1) or FNIP2. ULK1 can induce FLCN phosphorylation, which modulates the FLCN-FNIP-GABARAP interaction.1752 FLCN is also linked to MTOR modulation through its interaction with the RRAG GTPases on lysosomes.1753,1754
- FM 4–64:
A lipophilic dye that primarily stains endocytic compartments and the yeast vacuole limiting membrane.
- FNBP1L (formin binding protein 1-like):
An F-BAR-containing protein that interacts with ATG3 and is required for the macroautophagy-dependent clearance of S. typhimurium, but not other types of autophagy.1755
- FNIP1 (folliculin interacting protein 1):
An interactor with the tumor suppressor FLCN. FNIP1464 and its homolog FNIP21752 can also interact with GABARAP.
- FOXO1 (forkhead box O1):
A mammalian transcription factor that regulates macroautophagy independent of transcriptional control; the cytosolic form of FOXO1 is acetylated after dissociation from SIRT2, and binds ATG7 to allow induction of macroautophagy in response to oxidative stress or starvation.1756 FOXO1 can also be deacetylated by SIRT1, which leads to upregulation of RAB7 and increased autophagic flux.1757 The C. elegans ortholog is DAF-16. See also SIRT1.
- FOXO3 (forkhead box O3):
A transcription factor that stimulates macroautophagy through transcriptional control of autophagy-related genes.642,1758 The C. elegans ortholog is DAF-16.
- Frataxin:
See FXN.
- Fsc1:
A type I transmembrane protein localizing to the vacuole membrane in the fission yeast S. pombe; required for the fusion of autophagosomes with vacuoles.1695
- FUNDC1 (FUN14 domain containing 1):
A mitochondrial outer membrane protein that functions as a receptor for hypoxia-induced mitophagy.1759 FUNDC1 contains a LIR and binds LC3.
- FUS (FUS RNA binding protein):
A DNA/RNA binding protein involved in DNA repair, gene transcription, and RNA splicing. FUS has also been implicated in tumorigenesis and RNA metabolism, and multiple missense and nonsense mutations in FUS are associated with amyotrophic lateral sclerosis. Macroautophagy reduces FUS-positive stress granules.1760
- FXN (frataxin):
A nuclear-encoded protein involved in iron-sulfur cluster protein biogenesis. Reduced expression of the C. elegans homolog, FRH-1, activates autophagy in an evolutionarily conserved manner.1274
- FYCO1 (FYVE and coiled-coil domain containing 1):
A protein that interacts with LC3, PtdIns3P and RAB7 to move autophagosomes toward the lysosome through microtubule plus end-directed transport.1761
- Gαi3:
See GNAI3.
- GABA (γ-aminobutyric acid):
GABA inhibits the selective autophagy pathways mitophagy and pexophagy through Sch9, leading to oxidative stress, which can be mitigated by the Tor1 inhibitor rapamycin.1762
- GNAI3 (guanine nucleotide binding protein [G protein], alpha inhibiting activity polypeptide 3):
A heterotrimeric G protein that activates macroautophagy in the GDP-bound (inactive) form, and inhibits it when bound to GTP (active state).1763,1764 See also GPSM1, RGS19, MAPK1/3 and CCDC88A.
- GABARAP [GABA(A) receptor-associated protein]:
A homolog of LC3.534,1765 The GABARAP family includes GABARAP, GABARAPL1/Atg8L/GEC1, and GABARAPL2/GATE-16/GEF2. The GABARAP proteins are involved in autophagosome formation and cargo recruitment.142
- GADD34:
See PPP1R15A.
- GAIP:
See RGS19.
- Gap junction proteins/connexins:
Multispan membrane proteins that mediate intercellular communication through the formation of hemi-channels or gap junctions at the plasma membrane. These proteins act as endogenous inhibitors of autophagosome formation by directly interacting and sequestering at the plasma membrane essential ATG proteins required for autophagosome biogenesis.
- GATA1:
A hematopoietic GATA transcription factor, expressed in erythroid precursors, megakaryocytes, eosinophils, and mast cells, that provides the differentiating cells with the requisite macroautophagy machinery and lysosomal components to ensure high-fidelity generation of erythrocytes.641 See also ZFPM1/FOG1.
- GATE-16:
See GABARAP.
- Gaucher disease (GD):
Caused by mutations in the gene encoding GBA/glucocerebrosidase (glucosidase, beta, acid), Gaucher disease is the most common of the lysosomal storage disorders and can increase susceptibility to Parkinson disease.1766-1768
- GBA/glucocerbrosidase (glucosidase, beta acid):
A lysosomal enzyme that breaks down glucosylceramide to glucose and ceramide. Mutations cause Gaucher disease and are associated with increased risk of Parkinson disease. Loss of GBA is also associated with impaired autophagy and failure to clear dysfunctional mitochondria, which accumulate in the cell.1769
- Gcn2:
A mammalian and yeast EIF2S1/eIF2α serine/threonine kinase that causes the activation of Gcn4 in response to amino acid depletion, thus positively regulating macroautophagy.1726
- Gcn4:
A yeast transcriptional activator that controls the synthesis of amino acid biosynthetic genes and positively regulates macroautophagy in response to amino acid depletion.1726
- GCN5L1:
A component of the mitochondrial acetyltransferase activity that modulates mitophagy and mitochondrial biogenesis.1770
- GEEC (GPI-enriched endocytic compartments) pathway:
A form of clathrin-independent endocytosis that contributes membrane for phagophore expansion.1771
- GFAP (glial fibrilary acid protein):
intermediate filament protein ubiquitously distributed in all cell types that bears functions beyond filament formation. Monomeric and dimeric forms of this protein associate with the cytosolic side of the lysosomal membrane and contribute to modulating the stability of the CMA translocation complex in a GTP-dependent manner coordinated with EEF1A/eF1α also at the lysosomal membrane.1724
- GFER/ERV1 (growth factor, augmenter of liver regeneration):
A flavin adenine dinucleotide-dependent sulfhydryl oxidase that is part of a disulfide redox system in the mitochondrial intermembrane space, and is also present in the cysosol and nucleus. Downregulation of GFER results in elevated levels of the mitochondrial fission GTPase DNM1L/DRP1, and decreased mitophagy.1772
- GILT:
See IFI30.
- GIV/Girdin:
See CCDC88A.
- GLI3 (GLI family zinc finger 3):
A C2H2 type of zinc finger transcription factor that plays a role in the transcriptional activation of VMP1 during the induction of autophagy by the oncogene KRAS.634 See also EP300.
- Glycophagy (glycogen autophagy):
The selective sequestration of glycogen and its subsequent vacuolar hydrolysis to produce glucose; this can occur by a micro- or macroautophagic process and has been reported in mammalian newborns and adult cardiac tissues as well as filamentous fungi.46,1308,1309,1773-1775
- GOPC/PIST/FIG/CAL (Golgi-associated PDZ and coiled-coil motif-containing protein):
Interacts with BECN1, and the SNARE protein STX6 (syntaxin 6). GOPC can induce autophagy via a CD46-Cyt-1 domain-dependent pathway following pathogen invasion.1671
- Gp78:
See AMFR.
- GPNMB (glycoprotein [transmembrane] nmb):
A protein involved in kidney repair that controls the degradation of phagosomes through macroautophagy.1776
- GPSM1/AGS3 (G-protein signaling modulator 1):
A guanine nucleotide dissociation inhibitor for GNAI3 that promotes macroautophagy by keeping GNAI3 in an inactive state.1668 GPSM1 directly binds LC3 and recruits GNAI3 to phagophores or autophagosomes under starvation conditions to promote autophagosome biogenesis and/or maturation. See also GNAI3.
- Granulophagy:
The process of bulk autophagic degradation of mRNP granules. The process has been characterized in S. cerevisiae and mammalian cells and is dependent on Cdc48/VCP in addition to the core autophagic machinery. The process is partially impaired by disease-causing mutations in VCP.1777
- GSK3B/GSK-3β (glycogen synthase kinase 3 beta):
A regulator of macroautophagy. GSK3B may act positively by inhibiting MTOR through the activation of TSC1/2 and by activating ULK1 through KAT5.1778 GSK3B modulates protein aggregation through the phosphorylation of the macroautophagy receptor NBR1.1529 GSK3B, however, it is also reported to be a negative regulator of macroautophagy. See also KAT5.
- HDAC6 (histone deacetylase 6):
A microtubule-associated deacetylase that interacts with ubiquitinated proteins. HDAC6 stimulates autophagosome-lysosome fusion by promoting the remodeling of F actin, and the quality control function of macroautophagy.665,666,1779 HDAC is also a biomarker of aggresomes.1780
- HIF1A/HIF-1α (hypoxia-inducible factor 1, alpha subunit [basic helix-loop-helix transcription factor]):
A dimeric transcription factor in which the α subunit is regulated by oxygen; the hydroxylated protein is degraded by the proteasome. HIF1A-mediated expression of BNIP3 results in the disruption of the BCL2-BECN1 interaction, thus inducing macroautophagy.1781,1782 HIF1A also regulates xenophagic degradation of intracellular E. coli.1783
- HK2 (hexokinase 2):
The enzyme responsible for phosphorylation of glucose at the beginning of glycolysis; during glucose starvation, HK2 switches from a glycolytic role and directly binds to and inhibits MTORC1 to induce macroautophagy.1784
- HLH-30:
C. elegans ortholog of the helix-loop-helix transcription factor TFEB.
- HMGB1 (high mobility group box 1):
A chromatin-associated nuclear protein that translocates out of the nucleus in response to stress such as ROS; HMGB1 binds to BECN1, displacing BCL2, thus promoting macroautophagy and inhibiting apoptosis.295 In addition, macroautophagy promotes the release of HMGB1 from the nucleus and the cell, and extracellular HMGB1 can further induce macroautophagy through binding AGER.1785,1786 See also AGER.
- Hog1:
A yeast MAPK involved in hyperosmotic stress, which is a homolog of mammalian MAPK/p38; Hog1 is required for mitophagy, but not other types of selective autophagy or nonselective autophagy.1787 See also Pbs2, Slt2 and MAPK.
- Hrr25:
A casein kinase δ/ϵ homologous protein kinase regulating diverse cellular processes such as DNA repair and vesicular trafficking. Hrr25 phosphorylates the C terminus of Atg19, which is essential for Atg19 binding to Atg11 and subsequent Cvt vesicle formation.1788 Hrr25 also phosphorylates Atg36, and this phosphorylation is required for the interaction of Atg36 with Atg11 and subsequent pexophagy.1789
- HSC70: See HSPA8. HSP70 (heat shock protein 70): The major cytosolic heat shock-inducible member of the HSP70 family. This form accumulates in the lysosomal lumen in cancer cells. HSP70 is also a biomarker of aggresomes.1794 See also HSPA1A. HSP90: See HSP90AA1. HSP90AA1/HSP90/HSPC1 (heat shock protein 90kDa alpha [cytosolic], class A member 1): A cytosolic chaperone that is also located in the lysosome lumen. The cytosolic form helps to stabilize BECN1, and promotes macroautophagy.1795 The lysosomal form of HSP90AA1 contributes to the stabilization of LAMP2A during its lateral mobility in the lysosomal membrane.1796HSPA1A (heat shock protein family A [Hsp70] member 1A):
The major cytosolic stress-inducible version of the HSP70 family. This protein localizes to the lysosomal lumen in cancer cells, and pharmacological inhibition leads to lysosome dysfunction and inhibition of autophagy.1790
- HSPA5/GRP78/BiP (heat shock protein 5 family A [Hsp70] member 5):
A master regulator of the UPR. This chaperone, maintaining ER structure and homeostasis, can also facilitate macroautophagy.1791
- HSPA8/HSC70 (heat shock protein family A [Hsp70] member 8):
This multifunctional cytosolic chaperone is the constitutive member of the HSP70 family of chaperones and participates in targeting of cytosolic proteins to lysosomes for their degradation via chaperone-mediated autophagy.1792 The cytosolic form of the protein also regulates the dynamics of the CMA receptor, whereas the lumenal form (lys-HSPA8) is required for substrate translocation across the membrane.1793 This chaperone plays a role in the targeting of aggregated proteins (in a KFERQ-independent manner) for degradation through chaperone-assisted selective autophagy,1116 and in KFERQ-dependent targeting of cytosolic proteins to late endosomes for microautophagy.1115 See also chaperone-assisted selective autophagy, chaperone-mediated autophagy, and endosomal microautophagy.
- HSPC1:
See HSP90AA1.
- HTRA2/Omi (HtrA serine peptidase 2):
A nuclear-encoded mitochondrial serine protease that was reported to degrade HAX1, a BCL2 family-related protein, to allow macroautophagy induction.1797 In this study, knockdown of HTRA2, or the presence of a protease-defective mutant form, results in decreased basal macroautophagy that may lead to neurodegeneration. Separate studies, however, indicate that mitochondrial HTRA2 plays a role in mitochondrial quality control; in this case loss of the protein leads to increased macroautophagy and in particular mitophagy.1798-1800
- Hypersensitive response:
A rapid and locally restricted form of programmed cell death as part of the plant immune response to pathogen attack. The hypersensitive response is activated by different immune receptors upon recognition of pathogen-derived effector proteins, and can be positively regulated by macroautophagy.1092,1096,1801
- IAPP (islet amyloid polypeptide):
A 37 amino acid polypeptide derived from processing of an 89 amino acid precursor, which is coexpressed with INS/insulin by pancreatic β-cells. IAPP aggregation is implicated in the pathogenesis of type 2 diabetes. Macroautophagy regulates IAPP levels through SQSTM1-dependent lysosomal degradation.1802-1804
- iC-MA (immune cell-mediated autophagy):
IL2-activated natural killer cell- and T cell-induced macroautophagy.1805
- Ice2:
See Ayr1.
- ICP34.5:
A neurovirulence gene product encoded by the herpes simplex virus type 1 (nns) that blocks EIF2S1-EIF2AK2 induction of autophagy.1726 ICP34.5-dependent inhibition of autophagy depends upon its ability to bind to BECN1.892
- IDP (Intrinsically disordered protein):
A protein that does not possess unique structure and exists as a highly dynamic ensemble of interconverting conformations.1806-1809 IDPs are very common in nature1810 and have numerous biological functions that complement the functional repertoire of ordered proteins.1811-1814 Many proteins involved in autophagy are IDPs.1815,1816
- IDPR (intrinsically disordered protein region):
A protein region without unique structure that may be biologically important. IDPRs are considered as a source of functional novelty,1817 and they are common sites of protein-protein interactions1818 and posttranslational modifications.1819
- IFI30/GILT (interferon, gamma-inducible protein 30):
A thiol reductase that controls ROS levels; in the absence of IFI30 there is an increase in oxidative stress that results in the upregulation of macroautophagy.1820
- IKK (IκB kinase):
An activator of the classical NFKB pathway composed of 3 subunits (CHUK/IKKα/IKK1, IKBKB/IKKβ/IKK2, IKBKG/IKKγ/NEMO) that are required for optimal induction of macroautophagy in human and mouse cells.1821
- iLIR:
A web resource for prediction of Atg8 family interacting proteins (http://repeat.biol.ucy.ac.cy/iLIR).1482
- Iml1 complex:
A protein complex containing Iml1, Npr2 and Npr3 that regulates non-nitrogen-starvation-induced autophagosome formation; the complex partially localizes to the PAS.1822 See also non-nitrogen-starvation (NNS)-induced autophagy.
- Immunoamphisomes:
An organelle derived from the fusion of endosomes/phagosomes with autophagosomes that regulate dendritic cell-mediated innate and adaptive immune responses.1823
- Immunophagy:
A sum of diverse immunological functions of autophagy.1824
- InlK:
An internalin family protein on the surface of L. monocytogenes that recruits vault ribonucleoprotein particles to escape xenophagy.1825
- Innate immune surveillance:
Recognition and response system for the sensing of DAMPs, including pathogens and products of somatically mutated genes. Innate surveillance responses include activation of macroautophagy to degrade DAMPs.1705 See also DAMP.
- IMPA/inositol monophosphatase:
An enzyme that regulates the level of inositol 1,4,5-triphosphate (IP3) levels. Inhibition of IMPA stimulates macroautophagy independent of MTOR.1220
- IP3R:
See ITPR.
- IRGM (immunity-related GTPase family, M):
Involved in the macroautophagic control of intracellular pathogens.1826 In mouse, this protein is named IRGM1.
- Irs4:
Irs4 and Tax4 localize to the PAS under autophagy-inducing conditions in yeast and play a role in the recruitment of Atg17.1827 These proteins have partially overlapping functions and are required for efficient nonselective macroautophagy and pexophagy.
- Isolation membrane:
See phagophore.
- ITM2A (integral membrane protein 2A):
A target of PRKA/PKA-CREB that interacts with the V-ATPase and interferes with macroautophagic flux.1828
- ITPR1/2/3 (inositol 1,4,5-trisphosphate receptor, type 1/2/3):
A large tetrameric intracellular Ca2+-release channel present in the ER that is responsible for the initiation/propagation of intracellular Ca2+ signals that can target the cytosol and/or organelles. The ITPR is activated by inositol 1,4,5-trisphosphate produced in response to extracellular agonists. Many proteins regulate the ITPR including anti-apoptotic BCL2-family proteins and BECN1. The ITPR can inhibit autophagy by scaffolding BECN1 as well as by driving Ca2+-dependent ATP production,1220,1244,1246 whereas BECN1-dependent sensitization of ITPR-mediated Ca2+ release (e.g., in response to starvation) can promote macroautophagic flux.297
- JNK1:
See MAPK8.
- Jumpy:
See MTMR14.
- JUN/c-Jun/JunB (jun proto-oncogene):
A mammalian transcription factor that inhibits starvation-induced macroautophagy.1829
- KAT5/TIP60 (K[lysine] acetyltransferase 5):
In response to growth factor deprivation, KAT5 is phosphorylated and activated by GSK3 and then acetylates and activates ULK1.1778
- Kcs1:
A yeast inositol hexakisphosphate/heptakisposphate kinase; the kcs1Δ strain has a decrease in macroautophagy that may be associated with an incorrect localization of the PAS.1830
- KDM4A (lysine [K]-specific demethylase 4A):
A mammalian demethylase that regulates the expression of a subset of ATG genes.597,598 See also Rph1.
- KEAP1 (kelch-like ECH-associated protein 1):
An E3 ubiquitin ligase responsible for the degradation of transcription factor NFE2L2/NRF2 and the NFKB activator IKBKB/IKKβ. KEAP1 is a substrate for SQSTM1-dependent sequestration. SQSTM1 influences oxidative stress-related gene transcription and regulates the NFKB pathway via its interaction with KEAP1.428,1831,1832
- KIAA0226:
See RUBCN.
- KIAA1524/CIP2A/cancerous inhibitor of protein phosphatase 2A:
KIAA1524/CIP2A suppresses MTORC1-associated PPP2/PP2A activity in an allosteric manner thereby stabilizing the phosphorylation of MTORC1 substrates and inhibiting autophagy. KIAA1524/CIP2A can be degraded by autophagy in an SQSTM1-dependent manner.1833
- KillerRed:
A red fluorescent protein that produces a high amount of superoxide upon excitation. The construct with a mitochondria targeting sequence (mitoKillerRed) can be used to induce mitochondria damage and subsequent mitophagy.766,767
- Knockdown:
An experimental technique to reduce protein expression without altering the endogenous gene encoding that protein, through the means of short DNA or RNA oligonucleotides (miRNA, RNAi, shRNA, siRNA) that are complementary to the corresponding mRNA transcript.
- Knockout:
Targeted inactivation of an endogenous genetic locus (or multiple loci) via homologous recombination or gene targeting technology.
- Ku-0063794:
A catalytic MTOR inhibitor that increases macroautophagic flux to a greater level than allosteric inhibitors such as rapamycin; short-term treatment with Ku-0063794 can inhibit both MTORC1 and MTORC2, but the effects on flux are due to the former.341 See also WYE-354.
- KU55933:
An inhibitor of the class III PtdIns3K, which inhibits autophagosome formation at concentrations not affecting the class I PI3K.244 Also inhibits ATM.
- LACRT (lacritin):
A prosecretory mitogen primarily in tears and saliva that transiently accelerates autophagic flux in stressed cells.1834 Lacritin targets heparanase-deglycanated SDC1 (syndecan 1) on the cell surface,1835 and accelerates flux by stimulating the acetylation of FOXO3 as a novel ligand for ATG101 and by promoting the coupling of stress acetylated FOXO1 with ATG7.1836
- Laforin:
See EPM2A.
- LAMP2 (lysosomal-associated membrane protein 2):
A widely expressed and abundant single-span lysosomal membrane protein. Three spliced variants of the LAMP2 gene have been described. Knockout of the entire gene results in altered intracellular vesicular trafficking, defective lysosomal biogenesis, inefficient autophagosome clearance and alterations in intracellular cholesterol metabolism.1837-1839 In human, deficiency of LAMP2 causes a cardioskeletal autophagic vacuolar myopathy, called Danon disease.1840
- LAMP2A (lysosomal-associated membrane protein 2A):
One of the spliced variants of the LAMP2 gene that functions as a lysosomal membrane receptor for chaperone-mediated autophagy.1108 LAMP2A forms multimeric complexes that allow translocation of substrates across the lysosome membrane.1796 Regulation of LAMP2A is partly achieved by dynamic movement into and out of lipid microdomains in the lysosomal membrane.1793
- Late nucleophagy:
A process in which bulk nucleoplasm is delivered to the vacuole after prolonged periods of nitrogen starvation and subsequently degraded within the vacuole lumen.720
- LC3:
See MAP1LC3.
- LC3-associated phagocytosis (LAP):
Phagocytosis in macrophages that involves the conjugation of LC3 to single-membrane phagosomes, a process that promotes phagosome acidification and fusion with lysosomes.182 TLR signaling is required for LAP and leads to the recruitment of the BECN1 complex to phagosomes. See also NADPH oxidase.
- Ldb16:
See Ayr1.
- Ldh1:
See Ayr1.
- LGG-1:
A C. elegans homolog of Atg8.
- LGG-2:
A C. elegans homolog of Atg8.
- LGG-3:
A C. elegans homolog of Atg12.
- Lipophagy:
Selective degradation of lipid droplets by lysosomes contributing to lipolysis (breakdown of triglycerides into free fatty acids). In mammals, this selective degradation has been described to occur via macroautophagy (macrolipophagy),817 whereas in yeast, microlipophagy of cellular lipid stores has also been described. This process is distinct from the PNPLA5-dependent mobilization of lipid droplets as contributors of lipid precursors to phagophore membranes.
- Lipoxygenases:
Mycobacterial infection-responsive expression of these proteins, such as ALOX5 and ALOX15, inhibits IFNG-induced macroautophagy in macrophages.528
- LIR/LRS (LC3-interacting region):
This term refers to the WXXL-like sequences (consensus sequence [W/F/Y]-X-X-[I/L/V]) found in proteins that bind to the Atg8/LC3/GABARAP family of proteins (see also AIM and WXXL-motif).364 The core LIR residues interact with 2 hydrophobic pockets of the ubiquitin-like domain of the Atg8 homologs.
- LITAF (lipopolysaccharide-induced TNF factor):
An activator of inflammatory cytokine secretion in monocytes that has other functions in different cell types; LITAF is a positive regulator of macroautophagy in B cells.1841 LITAF associates with autophagosomes, and controls the expression of MAP1LC3B.
- LKB1:
See STK11.
- LMP (lysosome membrane permeabilization):
The process by which lysosomal membranes become disrupted through the action of lysosomotropic agents, detergents or toxins.1842 LMP blocks lysosomal activity and thus autophagy and induces the release of lysosomal content to the cytoplasm including cathepsins that can induce cell death.1843,1844
- LON2 (LON protease 2):
A protease localized to the peroxisome matrix that impedes pexophagy in Arabidopsis.1845
- Long-lived protein degradation (LLPD):
Macroautophagy is a primary mechanism used by cells to degrade long-lived proteins, and a corresponding assay can be used to monitor autophagic flux;3 a useful abbreviation is LLPD.486
- Lro1:
See Ayr1.
- Lucanthone:
An anti-schistosome compound that inhibits a late stage of macroautophagy; treatment results in deacidification of lysosomes and the accumulation of autophagosomes.1846
- LRPPRC (leucine-rich pentatricopeptide repeat containing):
A mitochondrion-associated protein that binds BCL2 and PARK2 to control the initiation of general autophagy and mitophagy.1847,1848
- LRRK2 (leucine-rich repeat kinase 2):
A large multidomain, membrane-associated kinase and GTPase whose Parkinson disease-associated mutations affect the regulation of macroautophagy.196,1849
- LRS (LC3 recognition sequence):
See LIR/LRS.
- LRSAM1 (leucine rich repeat and sterile alpha motif containing 1):
A human leucine-rich repeat protein that potentially interacts with GABARAPL2; knockdown of LRSAM1 results in a defect in anti-Salmonella autophagy.1850
- Ltn1:
See Rkr1.
- LY294002:
An inhibitor of phosphoinositide 3-kinases and PtdIns3K; it inhibits macroautophagy.1851
- LYNUS (lysosomal nutrient sensing):
A complex including MTORC1 and the V-ATPase located on the lysosomal surface that senses nutrient conditions.825 The LYNUS complex regulates TFEB activity.
- Lys05:
A dimeric chloroquine derivative that accumulates in the lysosome and inhibits macroautophagy.1852,1853
- Lysophagy:
The macroautophagic removal of damaged lysosomes.829,830
- Lysosome:
A degradative organelle in higher eukaryotes that compartmentalizes a range of hydrolytic enzymes and maintains a highly acidic pH. A primary lysosome is a relatively small compartment that has not yet participated in a degradation process, whereas secondary lysosomes are sites of present or past digestive activity. The secondary lysosomes include autolysosomes and telolysosomes. Autolysosomes/early secondary lysosomes are larger compartments actively engaged in digestion, whereas telolysosomes/late secondary lysosomes do not have significant digestive activity and contain residues of previous digestions. Both may contain material of either autophagic or heterophagic origin.
- Macroautophagy:
The largely nonselective autophagic sequestration of cytoplasm into a double- or multiple-membrane-delimited compartment (an autophagosome) of non-lysosomal/vacuolar origin and its subsequent degradation by the lysosomal/vacuolar system. Note that certain proteins and organelles may be selectively degraded via a macroautophagy-related process, and, conversely, some cytosolic components such as cytoskeletal elements are selectively excluded.
- MAGEA3 (melanoma antigen family A3):
MAGEA3 and MAGEA6 form a complex with the E3 ligase TRIM28, resulting in the degradation of AMPK and the subsequent increase in MTOR activity, which in turn causes a downregulation of macroautophagy.1854 See also TRIM28.
- MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3):
A homolog of yeast Atg8, which is frequently used as a phagophore or autophagosome marker. Cytosolic LC3-I is conjugated to phosphatidylethanolamine to become phagophore- or autophagosome-associated LC3-II.269 The LC3 family includes LC3A, LC3B, LC3B2 and LC3C. These proteins are involved in the biogenesis of autophagosomes, and in cargo recruitment.142 Vertebrate LC3 is regulated by phosphorylation of the N-terminal helical region by PRKA/PKA.343
- MAP1S (microtubule-associated protein 1S):
A ubiquitiously distributed homolog of the neuron-specifc MAP1A and MAP1B with which LC3 was originally copurified. It is required for autophagosome trafficking along microtubular tracks.1855,1856
- MAP3K7/MEKK7/TAK1 (mitogen-activated protein kinase kinase kinase 7):
Required for TNFSF10/TRAIL-induced activation of AMPK and for optimal macroautophagy induction by multiple stimuli.1857
- MAPK1 (mitogen-activated protein kinase 1):
A kinase that along with MAPK3 phosphorylates and stimulate RGS19/Gα-interacting protein/GAIP, which is a GTPase activating protein (GAP) for the trimeric GNAI3 protein that activates macroautophagy,1858 and which may be involved in BECN1-independent autophagy.83 Constitutively active MAPK1/3 also traffics to mitochondria to activate mitophagy.758
- MAPK3:
See MAPK1.
- MAPK8/JNK1:
A stress-activated kinase that phosphorylates BCL2 at Thr69, Ser70 and Ser87, causing its dissociation from BECN1, thus inducing macroautophagy.569
- MAPK8IP1/JIP1 (mitogen-activated protein kinase 8 interacting protein 1):
A LIR-containing LC3-binding protein that mediates the retrograde movement of RAB7-positive autophagosomes in axons.1859 Movement toward the proximal axon involves activation of dynein, whereas binding of LC3 to MAPK8IP1 prevents activation of kinesin. The DUSP1/MKP1 phosphatase may dephosphorylate Ser421, promoting binding to dynein.
- MAPK9/JNK2:
A stress-activated kinase that prevents the accumulation of acidic compartments in cells undergoing macroautophagic flux, thus keeping stressed cells alive.1860
- MAPK14 (mitogen-activated protein kinase 14):
A signaling component that negatively regulates the interaction of ATG9 and SUPT20H/FAM48A, and thus inhibits macroautophagy. In addition, MAPK14-mediated phosphorylation of ATG5 at T75 negatively regulates autophagosome formation.1861 The widely used pyridinyl imidazole class inhibitors of MAPK14 including SB202190 interfere with macroautophagy in a MAPK/p38-independent manner and should not be used to monitor the role of this signaling pathway in macroautophagy.1862,1863 The yeast homolog is Hog1. See also Hog1.
- MAPK15/ERK7/ERK8 (mitogen activated protein kinase 15):
MAPK15 is a LIR-containing protein that interacts with LC3B, GABARAP and GABARAPL1.1864 This kinase is localized in the cytoplasm and can be recruited to macroautophagic membranes through its binding to ATG8-like proteins. MAPK15 responds to starvation stimuli by self-activating through phosphorylation on its T-E-Y motif, and its activation contributes to the regulation of macroautophagy.
- MAPKAPK2 (mitogen-activated protein kinase-activated protein kinase 2):
MAPKAPK2 is a Ser/Thr protein kinase downstream of MAPK/p38. Its activation contributes to starvation-induced macroautophagy by phosphorylating BECN1.1525 See also BECN1.
- MAPKAPK3 (mitogen-activated protein kinase-activated protein kinase 3):
MAPKAPK3 shares a similar function with MAPKAPK2 in macroautophagy.1525 See also MAPKAPK2 and BECN1.
- Matrine:
A natural compound extract from traditional Chinese medicine that inhibits autophagy by elevating lysosomal pH and interfering with the maturation of lysosomal proteases.1865
- MB21D1/cGAS (Mab-21 domain containing 1):
A cytosolic sensor that produces cGAMP to initiate IFN production via TMEM173/STING upon binding microbial DNA.1866 MB21D1 also binds to BECN1, releasing RUBCN, resulting in the induction of macroautophagy to eliminate cytosolic pathogens and cytosolic DNA; the latter serves to downregulate the immune response to prevent overactivation.
- MDC (monodansylcadaverine):
A lysosomotropic autofluorescent compound that accumulates in acidic compartments such as autolysosomes, and also labels (but is not specific for) autophagosomes.1,1134
- MDK-ALK axis:
MDK (midkine [neurite growth-promoting factor 2]) is a growth factor for which increased levels are associated with a poor prognosis in malignant tumors. MDK promotes resistance to cannabinoid-evoked autophagy-mediated cell death via stimulation of ALK (anaplastic lymphoma receptor tyrosine kinase). Targeting of the MDK-ALK axis could help to improve the efficacy of antitumoral therapies based on the stimulation of macroautophagy-mediated cancer cell death.1867,1868
- Mdm10:
A component of the ERMES complex in yeast that is required for mitophagy. See also ERMES.1737
- Mdm12:
A component of the ERMES complex in yeast. Mdm12 colocalizes with Atg32-Atg11 and is required for mitophagy. See also Atg11, Atg32, and ERMES.705,1737
- Mdm34:
A component of the ERMES complex in yeast. Mdm34 colocalizes with Atg32-Atg11 and is required for mitophagy. See also Atg11, Atg32, and ERMES.705,1737
- Mdv1:
A component of the mitochondrial fission complex. It plays a role in mediating mitophagy-specific fission.705 See also Dnm1.
- MEFV/TRIM20/pyrin (Mediterannean fever):
The gene encoding MEFV is a site of polymorphisms associated with familial Mediterranean fever; MEFV/TRIM20 acts as a receptor for selective macroautophagy of several inflammasome components.1869
- Mega-autophagy:
The final lytic process during developmental programmed cell death in plants that involves tonoplast permeabilization and rupture, resulting in the release of hydrolases from the vacuole, followed by rapid disintegration of the protoplast at the time of cell death.1398,1870,1871 This term has also been used to refer to the rupture of the yeast vacuole during sporulation, which results in the destruction of cellular material, including nuclei that are not used to form spores.1872
- Megaphagosomes:
Very large (5–10 μm) double-membraned, autophagy-related vesicles that accumulate in cells infected by coxsackievirus and, possibly, influenza virus.194
- MGEA5/NCOAT/O-GlcNAcase/oga-1(meningioma expressed antigen 5 [hyaluronidase]):
MGEA5 removes the O-GlcNAc modification and regulates the macroautophagy machinery by countering the action of OGT.1873 See also OGT.
- Microautophagy:
An autophagic process involving direct uptake of cytosol, inclusions (e.g., glycogen) and organelles (e.g., ribosomes, peroxisomes) at the lysosome/vacuole by protrusion, invagination or septation of the sequestering organelle membrane.
- MIPA (micropexophagic apparatus):
A curved double-membrane structure formed by the PAS that may serve as a scaffold for completion of the sequestration of peroxisomes during micropexophagy; fusion with the vacuolar sequestering membranes encloses the organelles within an intralumenal vesicle.1874 See also vacuolar sequestering membranes.
- Mitochondrial spheroid:
A mitochondrial structure formed in PARK2-deficient cells treated with a mitochondrial uncoupler (such as CCCP).1875,1876 Under this condition, mitophagy fails to occur and a damaged mitochondrion can transform into a spheroid containing cytosolic components in the newly formed lumen.
- MIR21(microRNA 21):
A miRNA that is overexpressed in almost all types of solid tumors and is involved in cancer chemoresistance. MIR21 modulates macroautophagy and the sensitivity of tumor cells toward drugs that induce macroautophagy.1877
- Mir31(microRNA 31):
A mouse miRNA that targets PPP2/PP2A to inhibit IFNG-induced macroautophagy in macrophages during mycobacterial infection.528 See also Mir155.
- MIR95:
A human miRNA that inhibits macroautophagy and blocks lysosome function via repression of SUMF1.247
- MIR101:
A human miRNA that inhibits macroautophagy and the expression of STMN1, RAB5A and ATG4D.243
- Mir155:
A mouse miRNA that targets PPP2/PP2A to inhibit IFNG-induced macroautophagy in macrophages during mycobacterial infection.528 See also Mir31.
- MIR205:
A microRNA precursor that impairs the autophagic flux in castration-resistant prostate cancer cells by downregulating the lysosome-associated proteins RAB27A and LAMP3.1878
- MITF (microphthalmia-associated transcription factor):
A transcription factor belonging to the microphthalmia/transcription factor E (MiT/TFE) family, along with TFEB and TFE3; MITF binds to symmetrical DNA sequences (E-boxes; 5′-CACGTG-3′), and regulates lysosomal biogenesis and macroautophagy (including the genes BCL2, UVRAG, ATG16L1, ATG9B, GABARAPL1, and WIPI1). MITF shares a common mechanism of regulation with TFEB and TFE3; MITF can partially compensate when TFEB is lost upon specific stimuli or in specific cell types.639,1879 See also TFEB.
- Mitophagic body:
The single-membrane vesicle present inside the vacuole lumen following the fusion of a mitophagosome with a vacuole.
- Mitophagosome:
An autophagosome containing mitochondria and no more than a small amount of other cytoplasmic components, as observed during selective macromitophagy.42,748
- Mitophagy:
The selective autophagic sequestration and degradation of mitochondria; can occur by a micro- or macroautophagic process.1880
- Mitostatin:
See TCHP.
- Mkk1/2:
A MAPKK downstream of Bck1 that is required for mitophagy and pexophagy in yeast.1787 See also Bck1 and Slt2.
- MLN4924:
An inhibitor of NAE1 (NEDD8-activating enzyme E1 subunit 1) that is required for CUL/CULLIN-RING E3 ligase activation; treatment with MLN4924 induces macroautophagy through the accumulation of the MTOR inhibitory protein DEPTOR.1505
- Mmm1:
A component of the ERMES complex in yeast that is required for mitophagy. See also ERMES.1737
- MORN2 (MORN repeat containing 2):
MORN2 is a membrane occupation and recognition nexus (MORN)-motif protein that was identified in mouse testis. The gene localizes on chromosome 17E3, spanning approximately 7 kb; Morn2 contains 669 nucleotides of open reading frame, and encodes 79 amino acids.1881 MORN domains have the sequence GKYQGQWQ. MORN2 promotes the recruitment of LC3 in LAP, and MORN2 co-immunopreciptates with LC3.515
- MREG (melanoregulin):
A cargo sorting protein that associates with MAP1LC3 in LC3-associated phagocytosis.1882,1883
- MTDH/AEG-1 (metadherin):
An oncogenic protein that induces noncanonical (BECN1- and class III PtdIns3K-independent) macroautophagy as a cytoprotective mechanism.1884
- MTM-3:
A C. elegans myotubularin lipid phosphatase that is an ortholog of human MTMR3 and MTMR4; MTM-3 acts upsteam of EPG-5 to catalyze the turnover of PtdIns3P and promote autophagosome maturation.1885
- MTM1 (myotubularin 1):
A PtdIns3P and PtdIns(3,5)P2 3-phosphatase.1886 Mutations affecting MTM1 lead to myotubular myopathy and alteration of macroautophagy.
- MTMR3 (myotubularin related protein 3):
This protein localizes to the phagophore and negatively regulates macroautophagy. See also MTMR14.1887
- MTMR6 (myotubularin related protein 6):
A PtdIns3P-phosphatase; knockdown of MTMR6 increases the level of LC3-II.1888
- MTMR7 (myotubularin related protein 7):
A PtdIns3P-phosphatase; knockdown of MTMR7 increases the level of LC3-II.1888
- MTMR8 (myotubularin related protein 8):
A phosphoinositide phosphatase with activity toward PtdIns3P and PtdIns(3,5)P2; MTMR8 in a complex with MTMR9 inhibits macroautophagy based on the formation of WIPI1 puncta.1889
- MTMR9 (myotubularin related protein 9):
A catalytically inactive myotubularin that increases the activity of other members of the MTMR family and controls their substrate specificity; MTMR8-MTMR9 preferentially dephosphorylates PtdIns3P and thus inhibits macroautophagy.1889
- MTMR13:
See SBF2.
- MTMR14/Jumpy (myotubularin related protein 14):
A member of the myotubularin family that is a PtdIns3P-phosphatase; knockdown increases macroautophagic activity.1888,1890 MTMR14 regulates the interaction of WIPI1 with the phagophore. The Drosophila homolog is EDTP.
- MTOR (mechanistic target of rapamycin [serine/threonine kinase]):
The mammalian ortholog of TOR. Together with its binding partners it forms either MTOR complex 1 (MTORC1) or MTOR complex 2 (MTORC2). See also TORC1 and TORC2.
- MTORC1/2 (MTOR complex 1/2):
See TORC1 and TORC2.
- Multivesicular body (MVB)/multivesicular endosome:
An endosome containing multiple 50- to 80-nm vesicles that are derived from invagination of the limiting membrane. Under some conditions the MVB contains hydrolytic enzymes in which case it may be considered to be a lysosome or autolysosome with ongoing microautophagy.
- Multivesicular body sorting pathway:
A process in which proteins are sequestered into vesicles within the endosome through the invagination of the limiting membrane. This process is usually, but not always, dependent upon ubiquitin tags on the cargo and serves as one means of delivering integral membrane proteins destined for degradation into the vacuole/lysosome lumen. ESCRT (endosomal sorting complex required for transport) complexes are required for the formation of MVBs and for autophagosome maturation.1891
- MYO1C (myosin IC):
A class I myosin that functions as an actin motor protein essential for the trafficking of cholesterol-rich lipid rafts from intracellular storage compartments to the plasma membrane; MYO1C is important for efficient autophagosome-lysosome fusion.1892
- MYO6 (myosin VI):
A unique, minus-end directed actin motor protein required for autophagosome maturation and fusion with a lysosome via delivery of early endosomes to autophagosomes; mediated by the interaction of MYO6 with the alternative ESCRT-0 protein TOM1.879,1893
- NAA10/ARD1 (N[alpha]-acetyltransferase 10, NatA catalytic subunit):
A protein that interacts with and stabilizes TSC2 by acetylation, resulting in repression of MTOR and induction of macroautophagy.1894
- NACC1/NAC1 (nucleus accumbens associated 1, BEN and BTB [POZ] domain containing):
A transcription factor that increases the expression and cytosolic levels of HMGB1 in response to stress, thereby increasing macroautophagy activity.1895
- NADPH oxidases:
These enzymes contribute to macroautophagic targeting of Salmonella in leukocytes and epithelial cells through the generation of reactive oxygen species.881 The CYBB/NOX2 NADPH oxidase in macrophages is required for LC3-associated phagocytosis.
- NAF-1:
See CISD2.
- NAMPT/visfatin (nicotinamide phosphoribosyltransferase):
NAMPT is a protein that catalyzes the condensation of nicotinamide with 5-phosphoribosyl-1-pyrophosphate to yield nicotinamide mononucleotide, one step in the biosynthesis of nicotinamide adenine dinucleotide. The protein belongs to the nicotinic acid phosphoribosyltransferase (NAPRTase) family and is thought to be involved in many important biological processes, including metabolism, stress response and aging. NAMPT promotes neuronal survival through inducing macroautophagy via regulating the TSC2-MTOR-RPS6KB1 signaling pathway in a SIRT1-dependent manner during cerebral ischemia.1896
- NAPA/αSNAP (N-ethylmaleimide-sensitive factor attachment protein, alpha):
A key regulator of SNARE-mediated vesicle fusion. Loss of NAPA promotes noncanonical macroautophagy in human epithelial cells by interrupting ER-Golgi vesicle trafficking and triggering Golgi fragmentation.1897
- NBR1 (neighbor of BRCA1 gene 1):
A selective substrate of macroautophagy with structural similarity to SQSTM1. Functions as a receptor that binds ubiquitinated proteins and LC3 to allow the degradation of the former by a macroautophagy-like process.364 NBR1 shows specificity for substrates including peroxisomes783 and ubiquitinated aggregates.364 Phosphorylation of NBR1 by GSK3A/B prevents the aggregation of ubiquitinated proteins.1529
- NCOA4 (nuclear receptor coactivator 4):
A selective cargo receptor that is involved in iron homeostasis through the recycling of ferritin by macroautophagy.804 See also ferritinophagy.
- NDP52:
See CALCOCO2.
- Necroptosis:
A form of programmed necrotic cell death;1898 induction of macroautophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance.1899
- NFKB/NF-κB (nuclear factor of kappa light polypeptide gene enhancer in B-cells):
NFKB activates MTOR to inhibit macroautophagy.1900
- NH4Cl (ammonium chloride):
A weak base that is protonated in acidic compartments and neutralizes them; inhibits the clearance of autophagosomes and amphisomes.
- NHLRC1/EPM2B/malin (NHL repeat containing E3 ubiquitin protein ligase 1):
A putative E3-ubiquitin ligase, which forms a complex with EPM2A/laforin. Recessive mutations in the genes EPM2A, or NHLRC1/EMP2B are found in the majority of cases of Lafora disease, a very rare type of progressive neurodegeneration associated with impaired macroautophagy.1901
- Nitric oxide:
A gas and a messenger that has complex regulatory roles in macroautophagy, depending on its concentration and the cell type.344,1902-1904
- NID-1 (novel inducer of cell death 1):
A small molecule that induces activation of an ATG5- and CTSL-dependent cell death process reminiscent of macroautophagy.1450
- NIX:
See BNIP3L.
- NOD (nucleotide-binding oligomerization domain):
An intracellular peptidoglycan (or pattern recognition) receptor that senses bacteria and induces macroautophagy, involving ATG16L1 recruitment to the plasma membrane during bacterial cell invasion.1905
- Non-nitrogen-starvation (NNS)-induced autophagy:
A type of macroautophagy that is induced when yeast cells are shifted from rich to minimal medium; this process is controlled in part by the Iml1, Npr2 and Npr3 proteins.1822
- Noncanonical autophagy:
A functional macroautophagy pathway that only uses a subset of the characterized ATG proteins to generate an autophagosome. BECN1-independent,83,1463 and ATG5-ATG7-independent27 forms of macroautophagy have been reported.
- NPY (neuropeptide Y):
An endogenous neuropeptide produced mainly by the hypothalamus that mediates caloric restriction-induced macroautophagy.1906
- NR1D1/Rev-erba (nuclear receptor subfamily 1, group D, member 1):
A nuclear receptor that represses macroautophagy in mouse skeletal muscle. nr1d1-/- mice display increased autophagy gene expression along with consistent changes in autophagy protein levels and macroautophagic flux.611
- NRBF2 (nuclear receptor binding factor 2):
NRBF2 is the mammalian homolog of yeast Atg38, and is a binding partner of the BECN1-PIK3C3 complex; NRBF2 is required for the assembly of the ATG14-BECN1-PIK3C3/VPS34-PIK3R4/VPS15 complex and regulates macroautophagy.1907,1908 Nrbf2 knockout mice display impaired ATG14-linked PIK3C3 lipid kinase activity and impaired macroautophagy.
- NSP2:
A nonstructural protein of Chikungunya virus that interacts with human CALCOCO2 (but not the mouse ortholog) to promote viral replication. In contrast, binding of SQSTM1 to ubiquitinated capsid leads to viral degradation through macroautophagy.1909
- Nucleophagy:
The selective autophagic degradation of the nucleus or parts of the nucleus.
- Nucleus-vacuole junction (NVJ):
Junction formed by the interaction between Nvj1, a membrane protein of the outer nuclear membrane, and Vac8 of the vacuole membrane, that are necessary for micronucleophagy.718 See also piecemeal microautophagy of the nucleus.
- NUPR1/p8 (nuclear protein, transcriptional regulator, 1):
A transcriptional regulator that controls macroautophagy by repressing the transcriptional activity of FOXO3.1910
- NVP-BGT226 (8-[6-methoxy-pyridin-3-yl]-3-methyl-1-[4- piperazin-1-yl-3-trifl uoromethyl-phenyl]-1,3-dihydroimidazo[4,5-c ]quinolin-2-one maleate):
A class I PI3K and MTOR dual inhibitor that induces macroautophagy.1911
- NVT (Nbr1-mediated vacuolar targeting):
A pathway used for the delivery of cytosolic hydrolases (Lap2 and Ape2) into the vacuole in S. pombe that involves interaction with Nbr1 and relies on the ESCRT machinery.1912
- OATL1:
See TBC1D25.
- OGT/ogt-1 (O-linked N-acetylglucosamine [GlcNAc] transferase):
OGT is a nutrient-dependent signaling transferase that regulates the autophagy machinery by adding the O-GlcNAc modification. Similar to phosphorylation, this modification is involved in signaling.1873
- Omegasome:
ZFYVE1-containing structure located at the ER that is involved in autophagosome formation during amino acid starvation.583
- Omi:
See HTRA2.
- Oncophagy:
A general term describing cancer-related autophagy.1913
- OPTN (optineurin):
An autophagy receptor that functions in the elimination of Salmonella; OPTN has a LIR and a ubiquitin-binding domain, allowing it to link tagged bacteria to the autophagy machinery.880 Phosphorylation of OPTN by TBK1 increases its affinity for LC3. OPTN may function together with CALCOCO2/NDP52 and TAX1BP1/CALCOCO3. See also CALCOCO2, TAX1BP1 and TBK1.
- Organellophagy:
General terminology for autophagic processes selective for organelles such as peroxisomes, mitochondria, the nucleus, and ER.704,1914
- Oxiapoptophagy:
A type of cell death induced by oxysterols that involves OXIdation + APOPTOsis + autoPHAGY.837,838
- Oxidized phospholipids:
Oxidized phospholipids induce macroautophagy, and in ATG7-deficient keratinocytes and melanocytes the levels of phospholipid oxidation are elevated.1915,1916
- Oxysterols:
Oxysterols are cholesterol oxide derivatives obtained either from auto-oxidation or by enzymatic oxidation of cholesterol (http://lipidlibrary.aocs.org/Primer/content.cfm?ItemNumber=39304). Some of them (7-ketocholesterol, 7β-hydroxycholesterol, 24[S]-hydroxycholesterol) can induce a complex type of cell death named oxiapoptophagy.836-838 See also oxiapoptophagy.
- P0:
A plant virus-encoded F-box protein that targets AGO1/ARGONAUTE1 to macroautophagy in order to suppress RNA silencing.849
- p8:
See NUPR1.
- p14ARF:
See CDKN2A.
- p27/p27Kip1:
See CDKN1B.
- p38α:
See MAPK14.
- p38IP:
See SUPT20H.
- p53:
See TP53.
- p62:
see SQSTM1.
- p97:
See VCP.
- PARK2/parkin (parkin RBR E3 ubiquitin protein ligase):
An E3 ubiquitin ligase (mutated in autosomal recessive forms of Parkinson disease) that is recruited from the cytosol to mitochondria following mitochondrial depolarization, mitochondrial import blockade or accumulation of unfolded proteins in the mitochondrial matrix, or ablation of the rhomboid protease PARL, to promote their clearance by mitophagy.250,1917-1920 PINK1-dependent phosphorylation of Ser65 in the ubiquitin-like domain of PARK2 and in ubiquitin itself (see phosphorylated ubiquitin/p-S65-Ub) promotes activation and recruitment of PARK2 to mitochondria (reviewed in ref. 745),1921 and USP8 deubiquitination of K6-linked ubiquitin on PARK2 to promote its efficient recruitment.1922
- PARK7/DJ-1 (parkinson protein 7):
An oncogene product whose loss of function is associated with Parkinson disease; overexpression suppresses macroautophagy through the MAPK8/JNK pathway.1923
- Parkin:
See PARK2.
- PARL (presenilin associated, rhomboid-like):
The mammalian ortholog of Drosophila rhomboid-7, a mitochondrial intramembrane protease; regulates the stability and localization of PINK1.1920,1924,1925 A missense mutation in the N terminus has been identified in some patients with Parkinson disease.1926 See also PINK1.
- PARP1 (poly [ADP-ribose] polymerase 1):
A nuclear enzyme involved in DNA damage repair; doxorubicin-induced DNA damage elicits a macroautophagic response that is dependent on PARP1.1927 In conditions of oxidative stress, PARP1 promotes macroautophagy through the STK11/LKB1-AMPK-MTOR pathway.1928
- PAS:
See phagophore assembly site.
- PAWR/par-4 (PRKC, apoptosis, WT1, regulator):
A cancer selective apoptosis-inducing tumor suppressor protein that functions as a positive regulator of macroautophagy when overexpressed.1929,1930
- PBPE:
- A selective and high affinity ligand of the microsomal antiestrogen-binding site (AEBS). PBPE induces protective macroautophagy in cancer cells through an AEBS-mediated accumulation of zymostenol (5α-cholest-8-en-3β-ol).1239,1931
- Pbs2:
A yeast MAPKK upstream of Hog1 that is required for mitophagy.1787
- Pcl1:
A yeast cyclin that activates Pho85 to stimulate macroautophagy by inhibiting Sic1.1683
- Pcl5:
A yeast cyclin that activates Pho85 to inhibit macroautophagy through degradation of Gcn4.1683
- PDPK1/PDK1 (3-phosphoinositide dependent protein kinase 1):
An activator of AKT. Recruited to the plasma membrane and activated by PtdIns(3,4,5)P3 which is generated by the class I phosphoinositide 3-kinase.
- PEA15/PED (phosphoprotein enriched in astrocytes 15):
A death effector domain-containing protein that modulates MAPK8 in glioma cells to promote macroautophagy.1932
- PDCD6IP (programmed cell death 6 interacting protein):
PDCD6IP is an ESCRT-associated protein that interacts with the ATG12–ATG3 conjugate to promote basal macroautophagy.1933 See also 12-ylation.
- PEG3 (paternally expressed 3):
A DCN (decorin)- and endorepellin-induced, genomically imprinted tumor suppressor gene that is required for macroautophagy in endothelial cells.1708 PEG3 colocalizes with and phyiscally binds to canonical macroautophagic markers such as BECN1 and LC3. Moreover, loss of PEG3 ablates the DCN- or endorepellin-mediated induction of BECN1 or MAP1LC3A; basal expression of BECN1 mRNA and BECN1 protein requires PEG3. See also DCN and endorepellin.
- Peripheral structures:
See Atg9 peripheral structures.
- PERK:
See EIF2AK3.
- PES/pifithrin-µ (2-phenylethynesulfonamide):
A small molecule inhibitor of HSPA1A/HSP70–1/HSP72; PES interferes with lysosomal function, causing a defect in macroautophagy and chaperone-mediated autophagy.1934
- peup(peroxisome unusual positioning):
Mutants isolated in Arabidopsis thaliana that accumulate aggregated peroxisomes.1935 The peup1, peup2 and peup4 mutants correspond to mutations in ATG3, ATG18a and ATG7.
- Pexophagic body:
The single-membrane vesicle present inside the vacuole lumen following the fusion of a pexophagosome with a vacuole.
- Pexophagosome:
An autophagosome containing peroxisomes, but largely excluding other cytoplasmic components; a pexophagosome forms during macropexophagy.1936
- Pexophagy:
A selective type of autophagy involving the sequestration and degradation of peroxisomes; it can occur by a micro- or macroautophagy-like process (micro- or macropexophagy).130
- PGRP (peptidoglycan-recogntion protein):
A cytosolic Drosophila protein that induces autophagy in response to invasive L. monocytogenes.1937
- Phagolysosome:
The product of a single-membrane phagosome fusing directly with a lysosome in a process that does not involve macroautophagy (we include this definition here simply for clarification relative to autolysosome, autophagosome and autophagolysosome).884
- Phagophore (PG):
- Phagophore assembly site (PAS):
A perivacuolar compartment or location that is involved in the formation of Cvt vesicles, autophagosomes and other sequestering compartments used in macroautophagy and related processes in fungi. The PAS may supply membranes during the formation of the sequestering vesicles or may be an organizing center where most of the autophagic machinery resides, at least transiently. The PAS or its equivalent is yet to be defined in mammalian cells.177,1939
- Pho8:
A yeast vacuolar phosphatase that acts upon 3′ nucleotides generated by Rny1 to generate nucleosides.1940 A modified form of Pho8, Pho8Δ60, is used in an enzymatic assay for monitoring macroautophagy in yeast. See also Rny1 and Pho8Δ60 assay.
- Pho23:
A component of the yeast Rpd3L histone deacetylase complex that negatively regulates the expression of ATG9 and other ATG genes.601
- Pho80:
A yeast cyclin that activates Pho85 to inhibit macroautophagy in response to high phosphate levels.1683
- Pho8Δ60 assay:
An enzymatic assay used to monitor macroautophagy in yeast. Deletion of the N-terminal cytosolic tail and transmembrane domain of Pho8 prevents the protein from entering the secretory pathway; the cytosolic mutant form is delivered to the vacuole via macroautophagy, where proteolytic removal of the C-terminal propeptide by Prb1 generates the active enzyme.261,262,677
- Pho85:
A multifunctional cyclin-dependent kinase that interacts with at least 10 different cyclins or cyclin-like proteins to regulate the cell cycle and responses to nutrient levels. Pho85 acts to negatively and positively regulate macroautophagy, depending on its binding to specific cyclins.1683 See also Clg1, Pcl1, Pcl5, Pho80 and Sic1.
- Phosphatidylinositol 3-kinase (PtdIns3K):
- A family of enzymes that add a phosphate group to the 3′ hydroxyl on the inositol ring of phosphatidylinositol. The 3′ phosphorylating lipid kinase isoforms are subdivided into 3 classes (I-III) and the class I enzymes are further subdivided into class IA and IB. The class III phosphatidylinositol 3-kinases (see PIK3C3 and Vps34) are stimulatory for macroautophagy, whereas class I enzymes (referred to as phosphoinositide 3-kinases) are inhibitory.1941 The class II PtdIns3K substantially contributes to PtdIns3P generation and autophagy in Pik3c3 knockout MEFs, also functioning as a positive factor for macroautophagy induction.1942 In yeast, Vps34 is the catalytic subunit of the PtdIns3K complex. There are 2 yeast PtdIns3K complexes, both of which contain Vps34, Vps15 (a regulatory kinase), and Vps30/Atg6. Complex I includes Atg14 and Atg38 and is involved in autophagy, whereas complex II contains Vps38 and is involved in the vacuolar protein sorting (Vps) pathway. See also phosphoinositide 3-kinase.
- Phosphatidylinositol 3-phosphate (PtdIns3P):
The product of the PtdIns3K. PtdIns3P is present at the PAS, and is involved in the recruitment of components of the macroautophagic machinery. It is important to note that PtdIns3P is also generated at the endosome (e.g., by the yeast PtdIns3K complex II). Additionally, FYVE-domain probes block PtdIns3P-dependent signaling, presumably by sequestering the molecule away from either interactions with downstream effectors or preventing its interconversion by additional kinases.1943 Thus, general PtdIns3P probes such as GFP-tagged FYVE and PX domains are generally not good markers for the macroautophagy-specific pool of this phosphoinositide.
- Phosphatidylinositol 3,5-bisphosphate (PtdIns[3,5]P2):
This molecule is generated by PIKFYVE (phosphoinositide kinase, FYVE finger containing) and is abundant at the membrane of the late endosome. Its function is relevant for the replication of intracellular pathogens such as the bacteria Salmonella,1944 and ASFV.1945 PtdIns(3,5)P2 also plays a role in regulating macroautophagy.1946
- Phosphoinositide 3-kinase/PI3K:
The class I family of enzymes that add a phosphate group to the 3′ hydroxyl on the inositol ring of phosphoinositides. PI3K activity results in the activation of MTOR and the inhibition of macroautophagy.
- Phosphoinositides (PI) or inositol phosphates:
These are membrane phospholipids that control vesicular traffic and physiology. There are several different phosphoinositides generated by quick interconversions by phosphorylation/dephosphorylation at different positions of their inositol ring by a number of kinases and phosphatases. The presence of a particular PI participates in conferring membrane identity to an organelle.
- Phosphorylated ubiquitin/p-S65-Ub:
Phosphorylated ubiquitin is essential for PINK1-PARK2-mediated mitophagy and plays a dual role in the intial activation and recruitment of PARK2 to damaged mitochondria (reviewed in ref. 745) Specific antibodies can be used to faithfully detect PINK1-PARK2-dependent mitophagy at early steps;744 however, the exact functions of p-S65-Ub during the different phases of mitophagy remain unclear.
- Piecemeal microautophagy of the nucleus (PMN)/micronucleophagy:
A process in which portions of the yeast nuclear membrane and nucleoplasm are invaginated into the vacuole, scissioned off from the remaining nuclear envelope and degraded within the vacuole lumen.715,716
- PI4K2A/PI4KIIα (phosphatidylinositol 4-kinase type 2 alpha):
A lipid kinase that generates PtdIns4P, which plays a role in autophagosome-lysosome fusion.1947 PI4K2A is recruited to autophagosomes through an interaction with GABARAP or GABARAPL2 (but the protein does not bind LC3).
- PIK3C3 (phosphatidylinositol 3-kinase, catalytic subunit type 3):
The mammalian homolog of yeast Vps34, a class III PtdIns3K that generates PtdIns3P, which is required for macroautophagy.1941 In mammalian cells there are at least 3 PtdIns3K complexes that include PIK3C3/VPS34, PIK3R4/VPS15 and BECN1, and combinations of ATG14, UVRAG, AMBRA1, SH3GLB1 and/or RUBCN. See also phosphatidylinositol 3-kinase)
- PIK3CB/p110β (phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta):
A catalytic subunit of the class IA phosphoinositide 3-kinase; this subunit plays a positive role in macroautophagy induction that is independent of MTOR or AKT, and instead acts through the generation of PtdIns3P, possibly by acting as a scaffold for the recruitment of phosphatases that act on PtdIns(3,4,5)P3 or by recruiting and activating PIK3C3.1948
- PIK3R4/p150/VPS15 (phosphoinositide-3-kinase, regulatory subunit 4):
The mammalian homolog of yeast Vps15, PIK3R4 is a core component of all complexes containing PIK3C3 and is required for macroautophagy.1949 PIK3R4 interacts with the kinase domain of PIK3C3, to regulate its activity and also functions as a scaffold for binding to NRBF2 and ATG14.1907,1908 While PIK3R4 is classified as a protein serine/threonine kinase, it possesses an atypical catalytic domain and lacks catalytic activity, at least in vitro (J. Murray, personal communication). PIK3R4 also interacts with RAB GTPases, including RAB51950 that may be responsible for recruitment of PIK3C3-PIK3R4-complexes to sites of autophagosome formation.
- PINK1/PARK6 (PTEN induced putative kinase 1):
A mitochondrial protein kinase (mutated in autosomal recessive forms of Parkinson disease) that is normally degraded in a membrane potential-dependent manner to maintain mitochondrial structure and function,1924,1951 suppressing the need for mitophagy.757 Upon mitochondrial depolarization, mitochondrial import blockade, accumulation of unfolded proteins in the mitochondrial matrix or ablation of the inner membrane protease PARL, PINK1 is stabilized and activated, phosphorylating ubiquitin (see phosphorylated ubiquitin/p-S65-Ub) and PARK2 for full activation and recruitment of PARK2 (reviewed in ref. 745) to facilitate mitophagy.1917-1921,1952 See also PARL.
- PKA (protein kinase A):
A serine/threonine kinase that negatively regulates macroautophagy in yeast;1953 composed of the Tpk1/2/3 catalytic and Bcy1 regulatory (inhibitory) subunits. The mammalian PKA homolog, PRKA, directly phosphorylates LC3.343 Bacterial toxins that activate mammalian PRKA can also inhibit autophagy.1954 In addition, cAMP inducers, such as β2-adrenergic agonists (D.A.P. Gonçalves, personal communication), CALC/calcitonin gene-related peptide (J. Machado, personal communication) and forskolin plus isobutilmethylxantine (W.A. Silveira, personal communication), block the conversion of LC3-I to LC3-II in C2C12 myotubes and adult skeletal muscles. Phosphorylation of the fission modulator DNM1L by mitochondrially-localized PRKA blocks mitochondrial fragmentation and autophagy induced by loss of endogenous PINK1 or by exposure to a neurotoxin in neuronal cell cultures.1717 See also DNM1L.
- PKB:
See AKT.
- Pkc1:
A yeast serine/threonine kinase involved in the cell wall integrity pathway upstream of Bck1; required for pexophagy and mitophagy.1787 See also Bck1 and Slt2.
- PKCδ:
See PRKCD.
- PKR:
See EIF2AK2.
- Plastolysome:
A plant plastid that transforms into a lytic compartment, with acid phosphatase activity, engulfing and digesting cytoplasmic regions in particular cell types and under particular developmental processes.811-814
- PLEKHM1:
An autophagic adaptor protein that contains a LIR motif, which directs binding to all of the LC3/GABARAP proteins. PLEKHM1 also interacts with GTP-bound RAB7 and the HOPS (homotypic fusion and protein sorting) complex. PLEKHM1 is present on the cytosolic face of late endosomes, autophagosomes, amphisomes and lysosomes, and serves to coordinate endocytic and macroautophagic pathway convergence at, and fusion with, the lysosome.1956
- PMT7:
A phloroglucinol derivative used as a chemotherapeutic drug to target glycolytic cancer cells.1957
- PND (programmed nuclear destruction):
A yeast cell death-related process that occurs during gametogenesis involving a noncanonical type of vacuole-dependent degradation.1872
- PNPLA5 (patatin-like phospholipase domain containing 5):
A lipase that mobilizes neutral lipid stores (e.g., triglycerides in lipid droplets) to enhance macroautophagic capacity of the cell by contributing lipid precursors for membrane biogenesis (thus enhancing macroautophagic capacity) and signaling.1958 This process should not be confused with the process of lipophagy, which is uptake of lipid droplets for triglyceride degradation in autolysosomes.
- PNS (peri-nuclear structure):
A punctate structure in P. pastoris marked by Atg35, which requires Atg17 for recruitment and is involved in micropexophagy; the PNS may be identical to the PAS.1615
- Polyphenol:
A class of plant phytochemicals that have been described as autophagy regulators in diferent disease models, such as neurodegenerative disease (reviewed in ref. 1959) including Parkinson disease,1960 and cancer (reviewed in ref. 1961).
- PP242:
A pharmacological catalytic kinase inhibitor of TOR; inhibits TORC1 and TORC2.
- PPARs (peroxisome proliferator-activated receptors):
Ligand-activated transcription factors, members of the nuclear receptor superfamily, consisting of 3 isotypes: PPARA/PPARα/NR1C1 (peroxisome proliferator-activated receptor alpha), PPARD/PPARδ/NR1C2, and PPARG/PPARγ/NR1C3.772 PPAR-mediated signaling pathways regulate, or are regulated by, molecules involved in macroautophagy.1962,1963
- PPI (protein-protein interaction):
Proper biological activity of many proteins depends on physical interactions with other proteins. Specific PPI has a functional objective. Therefore, complete understanding of protein function requires consideration of proteins in the context of their binding partners.1964,1965 Often, interactions beween proteins and protein complexes are presented in a form of large densely connected networks (PPI networks). Such network-based representation of PPIs provide the means for a more complete understanding of physiological and pathogenic mechanisms.1966
- PPM1D/Wip1 (protein phosphatase, Mg2+/Mn2+ dependent, 1D):
A protein phosphatase that negatively regulates ATM and macroautophagy.1967
- PPP1 (protein phosphatase 1):
A serine/threonine protein phosphatase that regulates ATG16L1 by dephosphorylation of CSNK2-modified Ser139 to inhibit macroautophagy. See also CSNK2.1694
- PPP1R15A/GADD34 (protein phosphatase 1, regulatory subunit 15A):
A protein that is upregulated by growth arrest and DNA damage; PPP1R15A binds to and dephosphorylates TSC2, leading to MTOR suppression and macroautophagy induction.1968
- PPP2 (protein phosphatase 2):
A serine/threonine protein phosphatase that positively regulates macroautophagy via BECN1.1969
- PPP2R5A (protein phosphatase 2, regulatory subunit B′, alpha):
B56 subunit of PPP2/PP2A, a phosphatase that binds to and dephosphorylates GSK3B at Ser9 to make it active and thus activate macroautophagy.528
- PPP3R1 (protein phosphatase 3, regulatory subunit B, alpha):
A regulatory subunit of the calcium-dependent phosphatase PPP3/calcineurin. In response to a calcium pulse via the lysosomal calcium channel MCOLN1, PPP3 dephosphorylates Ser142 and Ser211 of TFEB, leading to nuclear localization and upregulation of the CLEAR network.1970 See also CLEAR and TFEB.
- prApe1 (precursor Ape1):
See Ape1.
- Pre-autophagosomal structure (PAS):
See phagophore assembly site.
- PRKA (protein kinase, cAMP-dependent):
The mammalian homolog of yeast PKA. See also PKA.
- PRKCD/PKCδ (protein kinase C, delta):
PRKCD regulates MAPK8 activation. PRKCD also activates NADPH oxidases, which are required for antibacterial macroautophagy.1714 See also NADPH oxidases.
- PRKD1 (protein kinase D1):
A serine/threonine kinase that activates PIK3C3/VPS34 by phosphorylation; recruited to phagophore membranes.1971
- PRMT-1/EPG-11: An arginine methyltransferase in C. elegans that is the homolog of PRMT1.1735 PRMT-1/EPG-11 regulates the association of PGL granules with EPG-2 and LGG-1 puncta. PRMT-1/EPG-11 directly methylates arginine residues in the RGG domain of PGL-1 and PGL-3.Programmed cell death (PCD):
Regulated self-destruction of a cell. Type I is associated with apoptosis and is marked by cytoskeletal breakdown and condensation of cytoplasm and chromatin followed by fragmentation. Type II is associated with macroautophagy and is characterized by the presence of autophagic vacuoles (autophagosomes) that sequester organelles. Type III is marked by the absence of nuclear condensation, and the presence of a necrotic morphology with swelling of cytoplasmic organelles (oncosis). These categories of cell death are based on morphological criteria, and the Nomenclature Committee on Cell Death now recommends the use of terms that are more precise and refer to different types of regulated cell death (RCD).1091
- PROPPINs (β-propellers that bind phosphoinositides):
A WD40-protein family conserved from yeast to human.1972 These proteins fold as 7-bladed β-propellers, and each blade contains 4 antiparallel β-strands. With 2 lipid binding sites at the circumference of their propeller they bind PtdIns3P and PtdIns(3,5)P2.1973-1975 The S. cerevisiae PROPPINs are Atg18, Atg21 and Hsv2, and the mammalian counterparts are termed WIPIs.
- Proteaphagy:
The selective macroautophagic degradation of the 26S proteasome.1976 Proteaphagy is stimulated by either starvation or proteasome activation.
- Proto-lysosomes:
Vesicles derived from autolysosomes that mature into lysosomes during autophagic lysosome reformation.527 See also autophagic lysosome reformation.
- Protophagy:
Autophagy-like processes in microbial populations. The term summarizes all self-destructing patterns in prokaryotic colonies including bacterial cannibalism, autolysis, programmed cell death, and other processes, in which a part of the colony is lysed and consumed by neighboring prokaryotic cells to recycle matter and energy.1977
- PSEN (presenilin):
A protease that is part of the γ-secretase complex. Mutations in PSEN1 result in the accumulation of autophagosomes resulting at least in part from a defect in lysosomal acidification; one of the V-ATPase subunits does not target properly to the lysosome.61,1978
- PTEN (phosphatase and tensin homolog):
A 3′ phosphoinositide phosphatase that dephosphorylates PtdIns(3,4,5)P3, thereby inhibiting PDPK1/PDK1 and AKT activity.
- PTM (posttranslational modification):
After biosynthesis, many proteins undergo covalent modifications that are often catalyzed by special enzymes that recognize specific target sequences in particular proteins. PTMs provide dramatic extension of the structures, properties, and physico-chemical diversity of amino acids, thereby diversifying structures and functions of proteins.1979 There are more than 300 physyological PTMs.1980 Some PTMs (e.g., phosphorylation, acetylation, glycosylation, etc.) are reversible by the action of specific deconjugating enzymes. The interplay between modifying and demodifying enzymes allows for rapid and economical control of protein function.1979 PTMs clearly play a role in regulating the macroautophagy machinery.651,1981
- PTP4A3 (protein tyrosine phosphatase type IVA, member 3):
A plasma membrane- and endosome-localized prenylated protein phosphatase that stimulates macroautophagy; PTP4A3 is also an autophagic substrate.1982
- PTPRS/PTPσ (protein tyrosine phosphatase, receptor type, S):
A dual domain protein tyrosine phosphatase that antagonizes the action of the class III PtdIns3K; loss of PTPRS results in hyperactivation of basal and induced macroautophagy.1983
- PULKA (p-ULK1 assay):
This acronym describes the analysis of Ser317 phosphorylated (activated) ULK1 puncta by fluorescence microscopy.1984
- RAB1:
See Ypt1.
- RAB4A:
This small GTPase was previously called HRES-1/Rab4, as it is encoded by the antisense strand of the HRES-1 human endogenous retroviral locus in region q42 of human chromosome 1.1985 It has been recently designated as RAB4A to distinguish it from RAB4B on human chromosome 19. RAB4A regulates the endocytic recycling of surface proteins, such as CD4, CD247/CD3ζ, CD2AP, and TFRC/CD71, which control signal transduction through the immunological synapse in human T lymphocytes.1985,1986 Among these proteins, CD4 and CD247 are targeted by RAB4A for lysosomal degradation via macroautophagy.1985-1987 Beyond T lymphocytes, RAB4A generally promotes the formation of LC3+ autophagosomes and the accumulation of mitochondria during macroautophagy.1988 During accelerated macroautophagy, RAB4A also promotes the lysosomal degradation of intracellular proteins, such as DNM1L/Drp1 that initiates the fission and turnover of mitochondria.971,1989 Thus, RAB4A-mediated depletion of DNM1L selectively inhibits mitophagy and causes the accumulation of mitochondria in patients and mice with lupus.1987 The formation of interconnected mitochondrial tubular networks is enhanced by constitutively active RAB4AQ72L upon starvation, which may contribute to the retention of mitochondria during macroautophagy.1988
- RAB7:
A small GTPase of the RAS oncogene family functioning in transport from early to late endosomes and from late endosomes to lysosomes.1990 RAB7 is also needed for the clearance of autophagic compartments, most likely for the fusion of amphisomes with lysosomes.1136,1991 The yeast homolog is Ypt7.
- RAB8:
A small GTPase of the RAS oncogene family. RAB8A functions in secretory autophagy,1036 whereas RAB8B plays a role in degradative autophagy.1992
- RAB11:
A small GTPase that is required for autophagosome formation; ULK1 and ATG9 localize in part to RAB11-positive recycling endosomes.1993 See also TBC1D14.
- RAB12:
A small GTPase that controls degradation of the amino acid transporter SLC36A4/PAT4 (solute carrier family 36 [proton/amino acid symporter], member 4) and indirectly regulates MTORC1 activity and macroautophagy.1994
- RAB21:
A small GTPase that is required for autophagosome-lysosome fusion. Starvation induces RAB21 activity that promotes VAMP8 trafficking to the lysosome, where VAMP8 is needed to mediate fusion. See also SBF2.1995
- RAB24:
A small GTPase with unusual characteristics that associates with autophagic vacuoles and is needed for the clearance of autolysosomes under basal conditions.1996,1997
- RAB32:
A small GTPase that localizes to the ER, and enhances autophagosome formation under basal conditions.1998
- RAB33B:
A small GTPase of the medial Golgi complex that binds ATG16L1 and plays a role in autophagosome maturation by regulating fusion with lysosomes.1999 RAB33B is a target of TBC1D25/OATL1, which functions as a GAP.2000
- RABG3b:
A RAB GTPase that functions in the differentiation of tracheary elements of the Arabidopsis xylem through its role in macroautophagy; this protein is a homolog of RAB7/Ypt7.1094
- RAD001 (Everolimus):
An orally administered derivative of rapamycin.
- RAG:
See RRAG.
- RAGE:
See AGER.
- RAL:
A RRAS-like subfamily in the RAS family, RAL small GTPases typically function downstream of the RRAS effector RALGDS/RalGEF and are inhibited by RALGAP, a heterodimeric GAP structurally analogous to TSC1/2 that functions as a GAP for RHEB.2001,2002 The RAL subfamily includes mammalian RALA and RALB, Drosophila Rala, and C. elegans RAL-1. Mammalian RALB regulates exocytosis, the immune response and an anabolic/catabolic switch. In nutrient-rich conditions RALB-GTP binds EXOC2/Sec5 and EXOC8/Exo84, and through the latter associates with MTORC1 to promote anabolic metabolism.2003 Under starvation conditions RALB-GTP nucleates phagophore formation through assembly of a ULK1-BECN1-PIK3C3 complex, also via interaction with the EXOC8/Exo84 protein.1741 Although RALB direct activation and indirect inactivation (through MTORC1) of macroautophagy appears contradictory, RALB may function as a critical anabolic/catabolic switch in response to global and local nutrient contexts. RALB may be an analog of yeast Sec4.2004 See also EXOC2, Sec4/RAB40B and EXOC8.
- RALGAP:
A heterodimeric complex consisting of catalytic alpha and regulatory beta subunits, RALGAP inactivates RAL small GTPases. RALGAP is structurally analogous to the TSC1/2 GAP, and like TSC1/2 is phosphorylated and inhibited by AKT.2001,2005 An additional partner of the RALGAP complex, NKIRAS1/kappaB-Ras, also inhibits RAL function.2006 See also RAL.
- RANS (required for autophagy induced under non-nitrogen-starvation conditions) domain:
Also referred to as domain of unknown function 3608 (DUF3608; PFAM: PF12257, http://pfam.xfam.org/family/PF12257), this sequence in Iml1 is required for non-nitrogen starvation-induced autophagy.1822 This domain is spread throughout the eukaryotes (see for example, http://pfam.xfam.org/family/PF12257#tabview=tab7) and frequently reported in combination with a DEP (Dishevelled, Egl-10, and Pleckstrin) domain (PFAM: PF00610), which is also the case with Iml1.1822 See also non-nitrogen starvation (NNS)-induced autophagy.
- Rapamycin:
Allosteric TOR (in particular, TOR complex 1) inhibitor, which induces autophagy. TOR complex 2 is much less sensitive to inhibition by rapamycin.
- RAPTOR:
See RPTOR.
- Ras:
See RRAS.
- RB1-E2F1 (Retinoblastoma 1-E2 transcription factor 1):
RB1 is a tumor suppressor that promotes growth arrest, and protects against apoptosis. E2F1 regulates the transition from the G1 to the S phase in the cell cycle, and is a pro-apoptotic member of the E2F transcription family. In addition to controlling the cell cycle and apoptosis, the interaction between RB1 and E2F1 regulates macroautophagy; RB1 and E2F1 downregulate and upregulate BCL2, respectively, resulting in the induction of macroautophagy or apoptosis.615
- RB1CC1/FIP200 (RB1-inducible coiled-coil 1):
A putative mammalian functional counterpart of yeast Atg17. RB1CC1 is a component of the ULK1 complex.1533 In addition, RB1CC1 interacts with other proteins in several signaling pathways, suggesting the possibility of macroautophagy-independent functions, and a potential role in linking other cellular functions and signaling pathways to macroautophagy.
- Reactive oxygen species (ROS):
Chemically-reactive molecules that contain oxygen, including hydrogen peroxide, the hydroxyl radical ·OH, and the superoxide radical ·O2-. Hydrogen peroxide transiently inhibits delipidation of LC3 by ATG4, which is permissive for starvation-induced autophagy.519 Superoxide is essential for triggering injury-induced mitochondrial fission and mitophagy.757
- Ref(2)P:
The Drosophila homolog of SQSTM1.
- Residual body:
A lysosome that contains indigestible material such as lipofuscin.2007
- Resveratrol:
- Reticulophagy:
The selective degradation of ER by a macroautophagy-like process.843 Macroautophagy counterbalances ER expansion during the unfolded protein response. Activation of the UPR in yeast induces reticulophagy.
- RGS19/GAIP (regulator of G-protein signaling 19):
A GTPase activating protein that inactivates GNAI3 (converting it to the GDP-bound form) and stimulates macroautophagy.2009 See also GNAI3.
- RHEB (Ras homolog enriched in brain):
A small GTP-binding protein that activates MTOR when it is in the GTP-bound form.280
- Ribophagy:
The selective sequestration and degradation of ribosomes by a macroautophagy-like process.847
- Rim15:
A yeast kinase that regulates transcription factors in response to nutrients. Rim15 positively regulates macroautophagy and is negatively regulated by several upstream kinases including TOR, PKA, Sch9 and Pho85.1683,2010
- RIPK1 (receptor [TNFRSF]-interacting serine-threonine kinase 1):
RIPK1 inhibits basal macroautophagy independent of its kinase function, through activation of MAPK1/3 and inhibition of TFEB.2011
- Rkr1:
A yeast ubiquitin ligase that antagonizes ribophagy.848
- RNASET2/RNS2 (ribonuclease T2):
A conserved class II RNase of the T2 family that localizes to the lumen of the ER (or an ER-related structure) and vacuole in Arabidopsis, and to lysosomes in zebrafish; RNASET2 is involved in rRNA turnover, and rns2 mutants display constitutive macroautophagy, likely due to a defect in cellular homeostasis.2012,2013
- RNF216 (ring finger protein 216):
An E3 ubiquitin ligase that mediates the ubiquitination and the subsequent degradation of BECN1, thus acting as a negative regulator of macroautophagy.2014
- Rny1:
A yeast vacuolar RNase that hydrolyzes RNA that has been delivered to the vacuole via macroautophagy into 3′ nucleotides.1940 See also Pho8.
- Rpd3:
A yeast histone deacetylase that negatively regulates the expression of ATG8.1233 See also Sin3/SIN3 and Ume6.
- Rph1:
A histone demethylase that negatively regulates the expression of ATG7; demethylase activity is not required for transcriptional repression.597,598
- RPN10:
A component of the 26S proteasome lid. RPN10 acts as a receptor that binds ATG8 during proteaphagy in Arabidopsis.1976
- RPS6KB1/p70S6 kinase/S6K1 (ribosomal protein S6 kinase, 70kDa, polypeptide 1):
A substrate of MTORC1, in mammalian cells RPS6KB1/2 inhibits INSR (insulin receptor), which in turn causes a reduction in the activity of the class I PI3K and subsequently MTORC1; this may represent a feedback loop to help maintain basal levels of macroautophagy.1145,1218 Conversely, under conditions of long-term starvation RPS6KB1/2 levels may fall sufficiently to allow reactivation of MTORC1 to prevent excessive macroautophagy. In Drosophila, the RPS6KB1/2 ortholog S6k may act in a more direct manner to positively regulate macroautophagy.280
- RPS6KB2:
See RPS6KB1.
- RPTOR/raptor (regulatory associated protein of MTOR, complex 1):
A component of MTORC1. RPTOR interacts with ULK1, allowing MTORC1 to phosphorylate both ULK1 and ATG13, and thus repress ULK1 kinase activity and autophagy.490,491,2015 This interaction also permits a negative feedback loop to operate, whereby ULK1 phosphorylates RPTOR to inhibit MTORC1 activity.495,2016
- RRAG (Ras-related GTP binding):
A GTPase that activates MTORC1 in response to amino acids.2017 There are RRAGA, B, C and D isoforms.
- RRAS/RAS (related RAS viral [r-ras] oncogene homolog):
The small GTPase RRAS is an oncogene involved in the regulation of several cellular signaling pathways. RRAS can upregulate or downregulate autophagy through distinct signaling pathways that depend on the cellular contexts.2018
- Rsp5:
A yeast E3 ubiquitin ligase that is responsible for the autophagic clearance of certain cytosolic proteins via Cue5.451 See also Cue5.
- RUBCN/Rubicon/KIAA0226 (RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein):
RUBCN is part of a PtdIns3K complex (RUBCN-UVRAG-BECN1-PIK3C3-PIK3R4) that localizes to the late endosome/lysosome and inhibits macroautophagy.546,547
- SAHA/vorinostat (suberoylanilide hydroxamic acid):
An HDAC inhibitor that induces macroautophagy;2019 however, SAHA/vorinostat treatment has also been reported to suppress macroautophagy (e.g., see ref. 2020), suggesting context dependency.
- Saikosaponin d:
An ATP2A/SERCA inhibitor that induces macroautophagy and macroautophagy-dependent cell death in apoptosis-defective cells.1514
- SBF2/MTMR13 (SET binding factor 2):
A catalytically inactive myotubularin that is also a RAB21 guanine nucleotide exchange factor (GEF) required with RAB21 for autophagosome-lysosome fusion. Starvation induces SBF2 RAB21 GEF activity that promotes VAMP8 trafficking to the lysosome, where VAMP8 is needed to mediate fusion. See also RAB21.1995 The Drosophila homolog is Sbf.
- Sch9:
A yeast kinase that functions in parallel with PKA to negatively regulate macroautophagy. Sch9 appears to function in parallel with TOR, but is also downstream of the TOR kinase.2010
- SCOC (short coiled-coil protein):
A protein in the Golgi that interacts with FEZ1 in a complex with either ULK1 or UVRAG; the ternary complex with ULK1 promotes macroautophagy, whereas the complex with UVRAG has a negative effect by sequestering the latter from the BECN1-containing PtdIns3K complex.1747 See also FEZ1.
- SEA (Seh1-associated) protein complex:
A complex found in yeast that includes the Seh1 nucleoporin and the COPII component Sec13 (also a nucleoporin), in addition to Npr2 and Npr3, and 4 other relatively uncharacterized proteins; the SEA complex associates with the vacuole, potentially acting as a membrane coat and is involved in protein trafficking, amino acid biogenesis, and the starvation response including macroautophagy.2021
- Sec1:
Functions with the plasma membrane SNAREs Sso1/Sso2 and Sec9 to form the site for vesicle-mediated exocytosis; as with Sso1/Sso2 and Sec9, temperature sensitive sec1 mutations also abrogate macroautophagic delivery of GFP-Atg8.2022 See also Sso1/Sso2.
- Sec2:
A guanine nucleotide exchange factor for Sec4 that normally functions in exocytosis. Upon the induction of macroautophagy, Sec2 function is diverted to promote membrane delivery to the PAS.2004
- Sec4:
A Rab family GTPase that normally functions in exocytosis; under macroautophagy-inducing conditions yeast Sec4 is needed for the anterograde movement of Atg9 to the PAS.2004 The mammalian homolog is RAB40B.
- SEC5L1:
See EXOC2.
- Sec9:
Plasma membrane SNARE light chain that forms a complex with Sso1/Sso2 to generate the target complex of vesicle exocytosis; as with Sso1/Sso2, loss of Sec9 function blocks macroautophagy at an early stage by disrupting targeting of Atg9 to the Atg9 peripheral sites and PAS.2023 See also Sso1/Sso2, and Atg9 peripheral sites/structures.
- Sec18:
Homolog of mammalian NSF, an ATPase globally responsible for SNARE disassembly. Loss of function inhibits SNARE-dependent early and late events of macroautophagy (that is, vesicular delivery of Atg9 to the Atg9 peripheral sites and PAS2023 and fusion of autophagosomes with the vacuole2024). See also Atg9 peripheral sites/structures.
- Sec22:
A vesicle SNARE involved in ER and Golgi transport; mutations in Sec22 also block Atg9 trafficking to the Atg9 peripheral sites and PAS. Crosslinking experiments suggest Sec22 may be the v-SNARE responsible for the macroautophagy functions of the ordinarily plasma membrane Sso1/Sso2-Sec9 t-SNARE complex.2023 See also Sso1/Sso2, and Atg9 peripheral sites/structures.
- Secretory autophagy:
A biosynthetic mode of autophagy that occurs in mammalian cells.1036,2025 Secretory autophagy depends on the ATG proteins, RAB8A and the Golgi protein GORASP2/GRASP55, and is used for the extracellular delivery (via unconventional secretion) of proteins such as the cytokines IL1B and IL18, and HMGB1. See also exophagy.
- SEPA-1 (suppressor of ectopic P granule in autophagy mutants-1):
A C. elegans protein that is involved in the selective degradation of P granules through a macroautophagy-like process.1262 SEPA-1 self-oligomerizes and functions as the receptor for the accumulation of PGL-1 and PGL-3 aggregates. SEPA-1 directly binds PGL-3 and LGG-1.
- Septin cages:
Septins are GTP-binding proteins that assemble into nonpolar filaments (characterized as unconventional cytoskeleton), often acting as scaffolds for the recruitment of other proteins. Septin cages form in response to infection by Shigella; the cages surround the bacteria, preventing intercellular spread, and serve to recruit autophagy components such as SQSTM1 and LC3.2026
- SERPINA1/A1AT (serpin peptidase inhibitor, clade A [alpha-1 antiproteinase, antitrypsin], member 1):
SERPINA1 is the must abundant circulating protease inhibitor and is synthesized in the liver. A point mutation in the SERPINA1 gene alters protein folding of the gene product, making it aggregation prone; the proteasomal and macroautophagic pathways mediate degradation of mutant SERPINA1.2027
- sesB (stress-sensitive B):
A Drosophila mitochondrial adenine nucleotide translocase that negatively regulates autophagic flux, possibly by increasing cytosolic ATP levels.1709 See also Dcp-1.
- SESN2 (sestrin 2):
A stress-inducible protein that reduces oxidative stress, inhibits MTORC1 and induces macroautophagy, also acting as an AMPK activator.2028 SESN2 physically associates with ULK1 and SQSTM1, promotes ULK1-dependent phosphorylation of SQSTM1, and facilitates autophagic degradation of SQSTM1 targets such as KEAP1.1532,2029 SESN2 suppresses MTORC1 in response to diverse stresses including DNA damage,2030 ER stress,2031 nutritional stress,822,2029 or energetic stress.2032
- SH3GLB1/Bif-1 (SH3-domain GRB2-like endophilin B1):
A protein that interacts with BECN1 via UVRAG and is required for macroautophagy. SH3GLB1 has a BAR domain that may be involved in deforming the membrane as part of autophagosome biogenesis.2033 SH3GLB1 activity is regulated by phosphorylation at residue T145, which in starved neurons occurs via CDK5.2034 SH3GLB1 regulates autophagic degradation of EGFR,2035 NTRK1,2034 and CHRNA1.2036 Turnover of CHRNA1 is coregulated by TRIM63.2036
- SHH (sonic hedgehog):
A ligand of the sonic hedgehog pathway. Activation of this pathway suppresses IFNG-induced macroautophagy in macrophages during mycobacterial infection.528
- Shp1/Ubx1:
A yeast Ubx (ubiquitin regulatory x)-domain protein that is needed for the formation of autophagosomes during nonselective macroautophagy; Shp1 binds Cdc48 and Atg8–PE, and may be involved in extracting the latter during phagophore expansion.1670
- Sic1:
A yeast cyclin-dependent kinase inhibitor that blocks the activity of Cdc28-Clb kinase complexes to control entry into the S phase of the cell cycle. Sic1 is a negative regulator of macroautophagy that inhibits Rim15.1683
- Signalphagy:
A type of macroautophagy that degrades active signaling proteins.2037
- Sin3/SIN3 (SIN3 transcription regulator family member):
Part of the Rpd3L regulatory complex including Rpd3 and Ume6 in yeast, which downregulates transcription of ATG8 in growing conditions.1233 In mammalian cells knockdown of both SIN3A and SIN3B is needed to allow increased expression of LC3. See also Rpd3 and Ume6.
- Sirolimus:
An immunosuppressant also referred to as rapamycin.
- SIRT1 (sirtuin 1):
A NAD+-dependent protein deacetylase that is activated by caloric restriction or glucose deprivation; SIRT1 can induce macroautophagy through the deacetylation of autophagy-related proteins and/or FOXO transcription factors.2038 Deacetylation of K49 and K51 of nuclear LC3 leads to localization in the cytosol and association with phagophores.657 See also SIRT2.
- SIRT2 (sirtuin 2):
A NAD+-dependent protein deacetylase sharing homology with SIRT1 that is involved in neurodegeneration and might play a role in macroautophagy activation through regulation of the acetylation state of FOXO1.1756 Under prolonged stress the SIRT2-dependent regulation of FOXO1 acetylation is impaired, and acetylated FOXO1 can bind to ATG7 in the cytoplasm and directly affect macroautophagy.
- SIRT3 (sirtuin 3):
A mitochondrial NAD+-dependent protein deacetylase sharing homology with SIRT1, which is responsible for deacetylation of mitochondrial proteins and modulation of mitophagy.2039,2040
- SIRT5:
A mitochondrial SIRT1 homolog with NAD+-dependent protein desuccinylase/demalonylase activity; SIRT5 modulates ammonia-induced macroautophagy.2041
- SIRT6:
A member of the sirtuin family with nuclear localization, that is associated with chromatin and promotes the repair of DNA. The involvement of SIRT6 in senescence has been proposed, possibly by the modulation of IGF-AKT signaling; a role for SIRT6 in macroautophagy linked to senescence has been determined.2042
- SIRT7:
A member of the sirtuin family that is highly expressed in the nucleus/nucleolus where it interacts with POLR1/RNA polymerase I as well as with histones. Many lines of evidence point to a role for SIRT7 in oncogenic transformation and tumor growth. The involvement of SIRT7 in macroautophagy was recently suggested in a model of acute cardiovascular injury, where loss of SIRT7 activates autophagy in cardiac fibroblasts.2043
- SLAPs (spacious Listeria-containing phagosomes):
SLAPs can be formed by L. monocytogenes during infection of macrophages or fibroblasts if bacteria are not able to escape into the cytosol.2044 SLAPs are thought to be immature autophagosomes in that they bear LC3 but are not acidic and do not contain lysosomal degradative enzymes. The pore-forming toxin listeriolysin O is essential for SLAPs formation and is thought to create small pores in the SLAP membrane that prevent acidification by the v-ATPase. SLAP-like structures have been observed in a model of chronic L. monocytogenes infection,2045 suggesting that autophagy may contribute to the establishment/maintenance of chronic infection.
- SLC1A5 (solute carrier family 1 [neutral amino acid transporter], member 5):
A high affinity, Na+-dependent transporter for L-glutamine; a block of transport activity leads to inhibition of MTORC1 signaling and the subsequent activation of macroautophagy.340 See also SLC7A5.
- SLC7A5 (solute carrier family 7 [amino acid transporter light chain, L system], member 5):
A bidirectional transporter that allows the simultaneous efflux of L-glutamine and influx of L-leucine; this transporter works in conjunction with SLC1A5 to regulate MTORC1.340
- SLC9A3R1 (solute carrier family 9, subfamily A [NHE3, cation proton antiporter 3], member 3 regulator 1):
A scaffold protein that competes with BCL2 for binding to BECN1, thus promoting macroautophagy.2046
- SLC25A1 (solute carrier family 25 [mitochondrial carrier; citrate transporter], member 1):
This protein maintains mitochondrial activity and promotes the movement of citrate from the mitochondria to the cytoplasm, providing cytosolic acetyl-coenzyme A. Inhibition of SLC25A1 results in the activation of macroautophagy and mitophagy.2047
- SLC38A9 (solute carrier family 38, member 9):
A multi-spanning membrane protein that localizes to the lysosome as part of the RRAG-Ragulator complex. SLC38A9 functions as a transceptor (transporter-receptor) to link amino acid status with MTORC1 activity.2048-2050
- Slg1/Wsc1:
A yeast cell surface sensor in the Slt2 MAPK pathway that is required for mitophagy.508 See also Slt2.
- SLR (sequestosome 1/p62-like receptor):
A protein that acts as a macroautophagy receptor, and in proinflammatory or other types of signaling.2051
- Slt2:
A yeast MAPK that is required for pexophagy and mitophagy.508 See also Pkc1, Bck1 and Mkk1/2.
- smARF (short mitochondrial ARF):
A small isoform of CDKN2A/p19ARF that results from the use of an alternate translation initiation site, which localizes to mitochondria and disrupts the membrane potential, leading to a massive increase in macroautophagy and cell death.2052
- SNAP29 (synaptosomal-associated protein, 29kDa):
A SNARE protein required for fusion of the completed autophagosome with a lysosome in metazoans.584,585,2053
- SNAPIN (SNAP-associated protein):
An adaptor protein involved in dynein-mediated late endocytic transport; SNAPIN is needed for the delivery of endosomes from distal processes to lysosomes in the neuronal soma, allowing maturation of autolysosomes.149
- SNCA/α-synuclein:
A presynaptic protein relevant for Parkinson disease pathogenesis because of its toxicity resulting from aggregation. SNCA degradation in neuronal cells involves the autophagy-lysosomal pathway via macroautophagy and chaperone-mediated autophagy.2054 Conversely, SNCA accumulation over time might impair autophagy function, and an inhibitory interaction of SNCA with HMGB1 has been reported.2055 This interaction can be reversed by the natural autophagy inducer corynoxine B. Similarly, in human T lymphocytes the aggregated form of SNCA, once generated, can be degraded by macroautophagy, whereas interfering with this pathway can result in the abnormal accumulation of SNCA. Hence, SNCA can be considered as an autophagy-related marker of peripheral blood lymphocytes.1340
- Snx4/Atg24:
A yeast PtdIns3P-binding sorting nexin that is part of the Atg1 kinase complex and binds Atg20.1600 Snx4/Atg24 is also involved in recycling from early endosomes. In the filamentous fungus M. oryzae, Atg24 is required for mitophagy.709
- SNX18:
A PX-BAR domain-containing protein involved in phagophore elongation.2056
- SpeB:
A cysteine protease secreted by Streptococcus pyogenes that degrades macroautophagy components at the bacterial surface, leading to autophagy escape.2057 The lack of SpeB allows capture and killing of cytoplasmic S. pyogenes by the macroautophagy system.126,2057
- Spautin-1 (specific and potent autophagy inhibitor-1):
An inhibitor of USP10 and USP13, identified in a screen for inhibitors of macroautophagy, which promotes the degradation of the PIK3C3/VSP34-BECN1 complex.2058
- Spermidine:
A natural polyamine that induces macroautophagy through the inhibition of histone acetylases such as EP300.631,2059
- Sphingolipids:
Sphingolipids are a major class of lipids. Some metabolites including ceramide, sphingosine and sphingosine 1-phosphate are bioactive signaling molecules. Ceramide and sphingosine 1-phosphate are positive regulators of macroautophagy.2060,2061
- SPNS/spinster:
A putative lysosomal efflux permease required for autophagic lysosome reformation.2062
- Sqa (spaghetti-squash activator):
A myosin light chain kinase-like protein that is a substrate of Atg1 in Drosophila; required for starvation-induced autophagosome formation, and the mammalian homolog DAPK3 is also involved in ATG9 trafficking.489
- SQST-1:
The C. elegans homolog of SQSTM1.
- SQSTM1/p62 (sequestosome 1):
An autophagy receptor that links ubiquitinated proteins to LC3. SQSTM1 accumulates in cells when macroautophagy is inhibited. SQSTM1 interaction with LC3 requires a WXXL or a LIR motif analogous to the interaction of Atg8 with Atg19.84 SQSTM1 also interacts with HDAC6 to regulate microtubule acetylation and autophagosome turnover.2063 See also HDAC6 and LIR/LRS.
- SRPX/Drs (sushi-repeat-containing protein, x-linked):
An apoptosis-inducing tumor suppressor that is involved in the maturation of autophagosomes.2064
- SseL:
A Salmonella deubiquitinase secreted by a type III secretion system; deubiquitination of aggregates and ALIS decreases host macrophage macroautophagic flux and results in an environment more favorable to bacterial replication.2065
- Ssk1:
A yeast component of the Hog1 signaling cascade that is required for mitophagy.508 See also Hog1.
- Sso1/Sso2:
Highly homologous plasma membrane syntaxins (SNAREs) of S. cerevisiae involved in exocytosis; the Sso1/Sso2 proteins also control the movement of Atg9 to the Atg9 peripheral sites and PAS during macroautophagy and the Cvt pathway.2023
- STAT3 (signal transducer and activator of transcription 3 [acute-phase response factor]):
A transcription factor that also functions in the cytosol as a suppressor of macroautophagy.2066 STAT3 binds EIF2AK2/PKR and inhibits the phosphorylation of EIF2S1.
- Stationary phase lipophagy:
A type of lipophagy that occurs in yeast cells entering quiescence.2067,2068
- STK3 (serine/threonine kinase 3):
The mammalian homolog of the Hippo/Ste20 kinase, which can phosphorylate LC3 on Thr50; this modification is needed for the fusion of autophagosomes with lysosomes.2069
- STK4/MST1 (serine/threonine kinase 4):
As with STK3, STK4 can phosphorylate LC3.2069 STK4 also phosphorylates Thr108 of BECN1, promoting the interaction of BECN1 with BCL2 or BCL2L1, inhibiting macroautophagy.2070
- STK11/LKB1 (serine/threonine kinase 11):
A kinase that is upstream of, and activates, AMPK.1673
- STX5 (syntaxin 5):
A Golgi-localized SNARE protein involved in vesicular transport of lysosomal hydrolases, a process that is critical for lysosome biogenesis; STX5 is needed for the later stages of autophagy.2071
- STX12/STX13/STX14 (syntaxin 12):
A genetic modifier of mutant CHMP2B in frontotemporal dementia that is required for autophagosome maturation; STX12 interacts with VTI1A.2072
- STX17 (syntaxin 17):
An autophagosomal SNARE protein required for fusion of the completed autophagosome with an endosome or lysosome in metazoans.584,585 STX17 is also required for recruitment of ATG14 to the ER-mitochondria contact sites.2073
- Sui2:
The yeast homolog of EIF2S1/eIF2α.
- SUPT20H/FAM48A (suppressor of Ty 20 homolog [S. cerevisiae]):
A protein that interacts with the C-terminal domain of ATG9; this interaction is negatively regulated by MAPK14.2074
- Sunitinib:
An autofluorescent multitarget tyrosine kinase inhibitor with lysosomotropic properties; sunitinib interferes with autophagic flux by blocking trafficking to lysosomes.2075
- Symbiophagy:
A process in which invertebrates such as the coralline demosponge Astrosclera willeyana degrade part of their symbiotic bacterial community, as part of a biomineralization pathway that generates the sponge skeleton.2076
- Syx13 (Syntaxin 13):
The Drosophila homolog of human STX12 that is required for autophagosome maturation.2072
- TAB2 (TGF-beta activated kinase 1/MAP3K7 binding protein 2):
MAP3K7-binding protein that consitutively interacts with TAB3 and inhibits macroautophagy; upon macroautophagy induction these proteins dissociate from BECN1 and bind MAP3K7.2077,2078
- TAB3 (TGF-beta activated kinase 1/MAP3K7 binding protein 3):
See TAB2.
- TAK1:
See MAP3K7.
- TAKA (transport of Atg9 after knocking outATG1) assay:
An epistasis analysis that examines the localization of Atg9-GFP in a double mutant, where one of the mutations is a deletion of ATG1.106 In atg1Δ mutants, Atg9-GFP is restricted primarily to the PAS; if the second mutation results in a multiple puncta phenotype, the corresponding protein is presumably required for anterograde transport of Atg9 to the PAS.728 This analysis can be combined with localization of RFP-Ape1 to determine if any of the Atg9-GFP puncta reach the PAS, in which case that punctum would colocalize with the RFP-Ape1 PAS marker.
- Tamoxifen:
- A triphenylethylenic compound widely used for the management of estrogen receptor-positive breast cancers. This drug is a dual modulator of ESR (estrogen receptor) and a high affinity ligand of the microsomal antiestrogen binding site (AEBS). Tamoxifen induces protective macroautophagy in cancer cells through an AEBS-mediated accumulation of zymostenol (5α-cholest-8-en-3β-ol).1239,1931,2079
- TARDBP/TDP-43 (TAR DNA binding protein):
A DNA/RNA binding protein that stabilizes Atg7 mRNA.2080
- TASCC (TOR-autophagy spatial coupling compartment):
A compartment located at the trans Golgi where autolysosomes and MTOR accumulate during RRAS-induced senescence to provide spatial coupling of protein secretion (anabolism) with degradation (catabolism); for example, amino acids generated from autophagy would quickly reactivate MTOR, whereas autophagy would be rapidly induced via MTOR inhibition when nutrients are again depleted.2081
- TAX1BP1/CALCOCO3 (Tax1 [human T-cell leukemia virus type I] binding protein 1):
An autophagy receptor that contains a LIR motif and a double zinc-finger ubiquitin binding domain. TAX1BP1 interacts with ubiquitinated substrates, such as S. typhimurium, and recruits LC3-positive autophagosomal membrane.879,1893,2082
- Tax4:
See Irs4.1827
- TBC1D7 (TBC1 domain family, member 7):
This protein is the third functional subunit of the TSC1-TSC2 complex upstream of MTORC1. Loss of function of TBC1D7 results in an increase of MTORC1 signaling, delayed induction of macroautophagy and enhancement of cell growth under poor growth conditions.2083 Mutations in TBC1D7 have been associated with intellectual disability, macrocrania, and delayed autophagy.2084,2085
- TBC1D14 (TBC1 domain family, member 14):
TBC1D14 colocalizes and interacts with ULK1 and upon overexpression causes tubulation of ULK1-positive endosomes, inhibiting autophagosome formation.1993 TBC1D14 binds activated RAB11, but does not function as a GAP. TBC1D14 localizes to the Golgi complex during amino acid starvation. See also RAB11.
- TBC1D25/OATL1 (TBC1 domain family, member 25):
A Tre2-Bub2-Cdc16 (TBC) domain-containing GAP for RAB33B; TBC1D25 is recruited to phagophores and autophagosomes via direct interaction with the Atg8 family proteins (via a LIR/LRS-like sequence), and it regulates the interaction of autophagosomes with lysosomes by inactivating RAB33B.2000 Overexpression of TBC1D25 inhibits autophagosome maturation at a step prior to fusion, suggesting that it might interfere with a tethering/docking function of RAB33B. See also RAB33B and LIR/LRS.
- TBK1 (TANK-binding kinase 1):
A serine/threonine protein kinase that is similar to IKK involved in the activation of NFKB.2086 TBK1 binds and directly phosphorylates OPTN at Ser177 (in humans) within the LIR, increasing the affinity of the latter for LC3.880
- TCHP/mitostatin (trichoplein, keratin filament binding):
A DCN (decorin)-inducible tumor suppressor gene that functions in, and is required for, tumor cell mitophagy. TCHP/mitostatin responds to DCN as well as canonical cues (e.g., nutrient deprivation and rapamycin) for mitophagic induction. DCN regulates mitostatin in a PPARGC1A/PGC-1α-dependent manner. Moreover, DCN-induced mitophagy is entirely dependent on TCHP for angiogenic inhibition.2087
- TECPR1 (tectonin beta-propeller repeat containing 1):
A protein that interacts with ATG5 and WIPI2, and localizes to the phagophore (localization is dependent on WIPI2); TECPR1 is needed for phagophore formation during macroautophagic elimination of Shigella, but not for starvation-induced autophagy.2088 TECPR1 also localizes to autophagosomes that target other pathogenic microbes such as group A Streptococcus, to depolarized mitochondria and to protein aggregates, suggesting a general role in selective macroautophagy. TECPR1 also plays a role in fusion of the autophagosome with the lysosome by competing with ATG16L1 to bind ATG5 and PtdIns3P, recruiting ATG5 to the lysosome membrane.2089
- TECPR2:
A WD repeat- and TECPR domain-containing protein that plays a role in macroautophagy; mutation of TECPR2 results in a form of monogenic hereditary spastic paraparesis.2090,2091
- TFE3 (transcription factor binding to IGHM enhancer 3):
A transcription factor belonging to the microphthalmia/transcription factor E (MiT/TFE) family, along with TFEB and MITF.639,1879 See also TFEB and MITF.
- TFEB (transcription factor EB):
A transcription factor that positively regulates the expression of genes involved in lysosomal biogenesis (those in the CLEAR network636), and also several of those involved in macroautophagy (including UVRAG, WIPI, MAP1LC3B and ATG9B); the use of a common transcription factor allows the coordinated expression of genes whose products are involved in the turnover of cytoplasm.625 See also CLEAR and PPP3R1.
- TGFB1/TGF-β (transforming growth factor, beta 1):
A cytokine that activates macroautophagy through the SMAD and MAPK8 pathways. TGFB1 induces the expression of several ATG genes including BECN1.
- TGM2/TG2/TGase 2 (transglutaminase 2):
An enzyme that catalyzes the formation of an isopeptide bond between a free amine group (e.g., protein- or peptide-bound lysine) and the acyl group at the end of the side chain of protein- or peptide-bound glutamine (protein crosslinking); TGM2 interacts with SQSTM1 and is involved in the macroautophagic clearance of ubiquitinated proteins.780,2092
- THC (Δ9-tetrahydrocannabinol):
The main active component of the hemp plant Cannabis sativa. The anticancer activity of THC in several animal models of cancer relies on its ability to stimulate autophagy-mediated cancer cell death. This effect occurs via THC binding to cannabinoid receptors, and the subsequent triggering of an ER stress-related response, which leads in turn to the inhibition of the AKT-MTORC1 axis.2093-2095
- TIGAR/C12orf5 (TP53 induced glycolysis regulatory phosphatase):
A protein that modulates glycolysis, causing an increase in NADPH, which results in a lower ROS level; this reduces the sensitivity to oxidative stress and apoptosis, but also has the effect of lowering the level of macroautophagy.2096
- Timosaponin A-III:
A medicinal saponin that induces a type of macroautophagy with some features that are distinct from rapamycin-induced macroautophagy.2097
- Tlg2:
A yeast endocytic SNARE light chain involved in early stages of the Cvt pathway729 and in autophagosome membrane formation.2023 Deletion of TLG2 results in a modest impairment in Atg9 delivery to the PAS.
- TLR (toll-like receptor):
A family of receptors that induces macroautophagy following binding to a corresponding PAMP.
- TM9SF1 (transmembrane 9 superfamily member 1):
A protein with 9 transmembrane domains that induces macroautophagy when overexpressed.2098
- TMEM59 (transmembrane protein 59):
A type-I transmembrane protein able to induce an unconventional autophagic process involving LC3 labeling of single-membrane endosomes through direct interaction with ATG16L1.2099
- TMEM74 (transmembrane protein 74):
An integral membrane protein that induces macroautophagy when overexpressed.1739,1740
- TMEM166:
See EVA1A.
- TNFAIP3/A20 (tumor necrosis factor, alpha-induced protein 3):
An E3 ubiquitin ligase that also functions as a deubiquitinating enzyme that removes K63-linked ubiquitin from BECN1, thus limiting macroautophagy induction in response to TLR signaling.2100 In contrast, TNFAIP3 restricts MTOR signaling, acting as a positive factor to promote macroautophagy in CD4 T cells.2101
- TNFSF10/TRAIL (tumor necrosis factor superfamily, member 10):
Induces macroautophagy by activating AMPK, thus inhibiting MTORC1 during lumen formation.
- TOLLIP (toll interacting protein):
A mammalian ubiquitin-binding receptor protein similar to yeast Cue5 that contains a CUE domain and plays a role in the macroautophagic removal of protein aggregates.451 See also Cue5 and CUET.
- TOR (target of rapamycin):
A serine/threonine protein kinase that negatively regulates yeast macroautophagy. Present in 2 complexes, TORC1 and TORC2. TORC1 is particularly sensitive to inhibition by rapamycin. TORC1 regulates macroautophagy in part through Tap42-protein phosphatase 2A, and also by phosphorylating Atg13 and Atg1.
- TORC1 (TOR complex I):
A rapamycin-sensitive protein complex of TOR that includes at least Tor1 or Tor2 (MTOR), Kog1 (RPTOR), Lst8 (MLST8), and Tco89.2102 MTORC1 also includes DEPTOR and AKT1S1/PRAS40.2103 In mammalian cells, sensitivity to rapamycin is conferred by RPTOR. TORC1 directly regulates macroautophagy.
- TORC2 (TOR complex II):
A relatively rapamycin-insensitive protein complex of TOR that includes at least Tor2 (MTOR), Avo1 (MAPKAP1/SIN1), Avo2, Avo3 (RICTOR), Bit61, Lst8 (MLST8) and Tsc11; MTORC2 also includes FKBP8/FKBP38, and PRR5/Protor-1.2102-2104 A critical difference in terms of components relative to TORC1 is the replacement of RPTOR by RICTOR. TORC2 is primarily involved with regulation of the cytoskeleton, but this complex functions to positively regulate macroautophagy during amino acid starvation.2105 Finally, studies also support the idea that TORC2 activity is required to sustain autophagosome biogenesis,2106 whereas it exerts an inhibitory effect on CMA,2107 suggesting that a switch in TORC2 substrates may contribute to coordinating the activity of these 2 types of autophagy.
- Torin1:
A selective catalytic ATP-competitive MTOR inhibitor that directly inhibits both TORC1 and TORC2.1193
- TP53/p53 (tumor protein 53):
A tumor suppressor. Nuclear TP53 activates macroautophagy, at least in part, by stimulating AMPK and DRAM1, whereas cytoplasmic TP53 inhibits macroautophagy.1273 Note that the official name for this protein in rodents is TRP53. The TP53 C. elegans ortholog, cep-1, also regulates macroautophagy.1272,1274
- TP53INP1 (tumor protein p53 inducible nuclear protein 1):
A stress-response protein that promotes TP53 transcriptional activity; cells lacking TP53INP1 display reduced basal and stress-induced autophagy,2108 whereas its overexpression enhances autophagic flux.2109 TP53INP1 interacts directly with LC3 via a functional LIR and stimulates autophagosome formation.2110 Cells lacking TP53INP1 display reduced mitophagy; TP53INP1 interacts with PARK2 and PINK1, and thus could be a recognition molecule involved in mitophagy.2111
- TP53INP2/DOR (tumor protein p53 inducible nuclear protein 2):
A mammalian and Drosophila regulatory protein that shuttles between the nucleus and the cytosol; the nuclear protein interacts with deacetylated LC3657 and GABARAPL2 and stimulates autophagosome formation.2112 TP53INP2 also interacts with GABARAP and VMP1 and is needed for the recruitment of BECN1 and LC3 to autophagosomes. TP53INP2 translocates from the nucleus to phagophores during macroautophagy induction and binds VMP1 and LC3 directly.2113 In addition, TP53INP2 modulates muscle mass in mice through the regulation of macroautophagy.2114
- TPCN/two-pore channel (two pore segment channel):
TPCNs are endolysosomal cation channels that maintain the proton gradient and membrane potential of endosomal and lysosomal membranes. TPCN2 physically interacts with MTOR and regulates MTOR reactivation and macroautophagic flux.2115,2116
- TPR (translocated promoter region, nuclear basket protein):
TPR is a component of the nuclear pore complex that presumably localizes at intranuclear filaments or nuclear baskets. Nuclear pore complex components, including TPR, are jointly referred to as nucleoporins. TPR was originally identified as the oncogenic activator of the MET and NTRK1/trk proto-oncogenes. Knockdown of TPR facilitates macroautophagy. TPR depletion is not only responsible for TP53 nuclear accumulation, which also activates the TP53-induced macroautophagy modulator DRAM, but also contributes to HSF1 and HSP70 mRNA trafficking, and transcriptional regulation of ATG7 and ATG12.2117
- TRAF2 (TNF receptor-associated factor 2):
An E3 ubiquitin ligase that plays an essential role in mitophagy in unstressed cardiac myocytes, as well as those treated with TNF or CCCP.786
- TRAF6 (TNF receptor-associated factor 6, E3 ubiquitin protein ligase):
An E3 ubiquitin ligase that ubiquitinates BECN1 to induce TLR4-triggered macroautophagy in macrophages.2100
- TRAIL:
See TNFSF10.
- Transgenic:
Harboring genetic material of another species/organism or extra copies of an endogenous gene, usually gained through transfer by genetic engineering.
- Transmitophagy/transcellular mitophagy:
A process in which axonal mitochondria are degraded in a cell-nonautonomous mechanism within neighboring cells.796
- TRAPPII (transport protein particle II):
A guanine nucleotide exchange factor for Ypt1 and perhaps Ypt31/32 that functions in macroautophagy in yeast.2118 TRAPPII is composed of Bet3, Bet5, Trs20, Trs23, Trs31, Trs33 and the unique subunits Trs65, Trs120 and Trs130.
- TRAPPIII (transport protein particle III):
A guanine nucleotide exchange factor for Ypt1 that functions in macroautophagy in yeast.1321 TRAPPIII is composed of Bet3, Bet5, Trs20, Trs23, Trs31, Trs33 and a unique subunit, Trs85.
- TRIB3 (tribbles pseudokinase 3):
A pseudokinase that plays a crucial role in the mechanism by which various anticancer agents (and specifically cannabinoids, the active components of marijuana and their derived products) activate macroautophagy in cancer cells. Cannabinoids elicit an ER stress-related response that leads to the upregulation of TRIB3 whose interaction with AKT impedes the activation of this kinase, thus leading to a decreased phosphorylation of TSC2 and AKT1S1/PRAS40. These events trigger the inhibition of MTORC1 and the induction of macroautophagy.2094 Conversely, TRIB3 binding to SQSTM1 via its UBA and LIR motifs interferes with autophagic flux, in particular of ubiquitinated proteins, and also reduces the efficiency of the UPS, promoting tumor progression due to the accumulation of tumor-promoting factors.2093,2119,2120
- Trichostatin A:
An inhibitor of class I and class II HDACs that induces autophagy.2121
- TRIM5/TRIM5α (tripartite motif containing 5):
A selective macroautophagy receptor for xenophagy; TRIM5 binds the HIV-1 capsid.1984
- TRIM20:
See MEFV.
- TRIM21:
An antigen in autoimmune diseases such as systemic lupus erythematosus, and Sjögren syndrome, TRIM21 is a receptor for selective autophagy of IRF3 dimers, a key transcriptional activator of type I interferon responses.1869
- TRIM28 (tripartite motif containing 28):
TRIM28 is an E3 ligase that is part of a ubiquitin ligase complex that targets PRKAA1, leading to ubiquitination and proteasomal degradation in part through the upregulation of MTOR activity.1854 See also MAGEA3.
- TRIM50 (tripartite motif containing 50):
TRIM50 is a cytoplasmic E3-ubiquitin ligase,2122 which interacts and colocalizes with SQSTM1 and promotes the formation and clearance of aggresome-associated polyubiquitinated proteins through HDAC6-mediated interaction and acetylation.2123,2124
- TRIM63/MURF-1 (tripartite motif containing 63, E3 ubiquitin protein ligase):
Muscle-specific atrophy-related E3 ubiquitin ligase2125,2126 that cooperates with SH3GLB1 to regulate autophagic degradation of CHRNA1 in skeletal muscle, particularly upon muscle-atrophy induction.2036
- TRPC4 (transient receptor potential cation channel, subfamily C, member 4):
A cation channel in human umbilical vascular endothelial cells; upregulation of TRPC4 increases the intracellular Ca2+ concentration resulting in activation of CAMKK2, which leads to MTOR inhibition and the induction of macroautophagy.1517
- Trs85:
A component of the TRAPPIII complex that is required specifically for macroautophagy.699
- Trs130:
A component of the TRAPPII complex that is required for the transport of Atg8 and Atg9 to the PAS.2118
- TSC1/2 (tuberous sclerosis 1/2):
A stable heterodimer (composed of TSC1/hamartin and TSC2/tuberin) inhibited by AKT and MAPK1/3 (phosphorylation causes dissociation of the dimer), and activated by AMPK. TSC1/2 acts as a GAP for RHEB, thus inhibiting MTOR.
- TSPO (translocator protein [18kDa]):
TSPO is a mitochondrial protein that interacts with VDAC1 to modulate the efficiency of mitophagy.2127
- Tubulovesicular autophagosome (TVA):
Cationic lipoplex and polyplex carriers used for nonviral gene delivery enter mammalian cells by endocytosis and fuse with autophagosomes, generating large tubulovesicular structures (tubulovesicular autophagosomes) that immunostain for LC3; these structures do not fuse efficiently with lysosomes and interfere with gene expression.220
- Tubulovesicular cluster (TVC):
A structure identified morphologically in yeast that corresponds to the Atg9 peripheral sites.537 See also Atg9 peripheral sites/structures.
- UBE2N (ubiquitin-conjugating enzyme E2N):
A ubiquitin-conjugating enzyme involved in PARK2-mediated mitophagy.2128,2129 UBE2N activity may be only partly redundant with that of UBE2L3, UBE2D2 and UBE2D3, as it is also involved during later steps of mitophagy.
- Ubiquitin:
A 76-amino acid protein that is conjugated to lysine residues. Ubiquitin is traditionally considered part of the ubiquitin-proteasome system and tags proteins for degradation; however, ubiquitin is also linked to various types of autophagy including aggrephagy (see SQSTM1 and NBR1). Lysine linkage-specific monoclonal antibodies, which are commercially available, can be used to investigate the degradation pathway usage.2130 Proteins covalently tagged with polyubiquitin chains via K48 are destined for proteasomal degradation, whereas proteins tagged with K63-linked ubiquitin are degraded via the macroautophagy pathway. In addition, phosphorylated forms of ubiquitin have been identified including p-S65-Ub, which is specifically generated during PINK1-PARK2-mediated mitophagy. Potentially, several PTMs of the modifier ubiquitin may turn out to be highly relevant and specific for distinct forms of selective autophagy (reviewed in ref. 745). See also p-S65-Ub.
- Ubp3:
A yeast deubiquitinase that forms a complex with Bre5 and is required for ribophagy.847 Conversely, the Ubp3-Bre5 complex inhibits mitophagy.2131
- UBQLN/Ubiquilins:
Receptor proteins that deliver ubiquitinated substrates to the proteasome. Ubiquilins may aid in the incorporation of protein aggregates into autophagosomes, and also promote the maturation of autophagosomes at the stage of fusion with lysosomes.2132
- ULK family (unc-51 like autophagy activating kinase):
- Ume6:
A component of the Rpd3L complex that binds to the URS1 sequence in the ATG8 promoter and downregulates transcription in growing conditions.1233 See also Rpd3 and Sin3/SIN3.
- UNC-51:
The C. elegans Atg1/ULK1/ULK2 homolog. See also Atg1.
- UPR (unfolded protein response):
A coordinated process to adapt to ER stress, providing a mechanism to buffer fluctuations in the unfolded protein load. The activation of this pathway is often related with macroautophagy.
- USP8 (ubiquitin specific peptidase 8):
A deubiquitinase that removes K6-linked ubiquitin chains from PARK2 to promote PARK2 recruitment to depolarized mitochondria and mitophagy.1922
- USP15 (ubiquitin specific peptidase 15):
A deubiquitinating enzyme that antagonizes PARK2-mediated mitophagy.2136 See also USP30.
- USP30:
A deubiquitinating enzyme that antagonizes PARK2-mediated mitophagy.2137 USP30 is also a substrate of PARK2 and is subject to proteasome-mediated degradation. See also USP15.
- USP36:
A deubiquitinating enzyme that negatively regulates selective macroautophagy in Drosophila and human cells.2138
- UVRAG (UV radiation resistance associated):
- A Vps38 homolog that can be part of the class III PtdIns3K complex. UVRAG functions in several ways to regulate macroautophagy: 1) It disrupts BECN1 dimer formation and forms a heterodimer that activates macroautophagy. 2) It binds to SH3GLB1 to allow activation of class III PtdIns3K to stimulate macroautophagy. 3) It interacts with the class C Vps/HOPS proteins involved in fusion of autophagosomes or amphisomes with the lysosome. 4) It competes with ATG14 for binding to BECN1, thus directing the class III PtdIns3K to function in the maturation step of macroautophagy.2139 MTORC1 phosphorylates UVRAG to inhibit macroautophagy.2140 In contrast, MTORC1 can also phosphorylate UVRAG to stimulate PIK3C3 activity and autophagic lysosome reformation.2141 UVRAG also has an autophagy-independent function, interacting with membrane fusion machinery to facilitate the cellular entry of enveloped viruses.2142
- Vacuolar cell death:
One of the 2 major types of cell death in plants (another type is necrosis), wherein the content of the dying cell is gradually engulfed by growing lytic vacuoles without loss of protoplast turgor, and culminates in vacuolar collapse.1093 Vacuolar cell death is commonly observed during plant development, for example in the embryo-suspensor and xylem elements, and critically depends on macroautophagy.1095 A similar type of macroautophagy-dependent vacuolar cell death is required for Dictyostelium development.2143
- Vacuolar-type H+-ATPase (V-ATPase):
A ubiquitously expressed proton pump that is responsible for acidifying lysosomes and the yeast or plant vacuole, and therefore is important for the normal progression of autophagy. Inhibitors of the V-ATPase (e.g., bafilomycin A1) are efficient macroautophagy inhibitors.156,157
- Vacuolar sequestering membranes (VSM):
Extensions/protrusions of the vacuole limiting membrane along the surface of peroxisomes that occurs during micropexophagy.2144
- Vacuole:
The fungal and plant equivalent of the lysosome; this organelle also carries out storage and osmoregulatory functions.2145 The bona fide plant equivalent of the lysosome is the lytic vacuole.
- Vacuole import and degradation (Vid):
A degradative pathway in yeast in which a specific protein(s) is sequestered into small (30- to 50-nm) single-membrane cytosolic vesicles that fuse with the vacuole allowing the contents to be degraded in the lumen. This process has been characterized for the catabolite-induced degradation of the gluconeogenic enzyme Fbp1/fructose-1,6-bisphosphatase in the presence of glucose, and sequestration is thought to involve translocation into the completed vesicle. An alternate pathway for degradation of Fbp1 by the ubiquitin-proteasome system has also been described.2146
- Vacuolin-1:
A small chemical that potently and reversibly inhibits the fusion between autophagosomes or endosomes with lysosomes by activating RAB5A.1521
- Valinomycin:
A K+ ionophore that destroys the electrochemical gradient across the mitochondrial membane and is widely used as a stimulator of mitophagy, similar to CCCP.2147
- Vam3:
A yeast syntaxin homolog needed for the fusion of autophagosomes with the vacuole.2148
- VAMP3 (vesicle-associated membrane protein 3):
A SNARE protein that facilitates the fusion of MVBs with autophagosomes to generate amphisomes.2149
- VAMP7 (vesicle-associated membrane protein 7):
VAMP7 is a SNARE protein that colocalizes with ATG16L1 vesicles and phagophores, and is required, along with STX7 (syntaxin 7), STX8 and VTI1B, for autophagosome formation.2150 VAMP7 is also involved in the maturation of autophagosomes by facilitating fusion with a lysosome.2149
- VAMP8 (vesicle-associated membrane protein 8):
A SNARE protein that, in conjunction with VTI1B, is needed for the fusion of autophagosomes with lysosomes.2151
- VCP/p97 (valosin-containing protein):
A type II AAA+-ATPase that is a protein segregase required for autophagosome maturation under basal conditions or when the proteasomal system is impaired; mutations of VCP result in the accumulation of immature, acidified autophagic vacuoles that contain ubiquitinated substrates.2152,2153 See also Cdc48.
- Verteporfin:
An FDA-approved drug; used in photodynamic therapy, but it inhibits the formation of autophagosomes in vivo without light activation.2154
- VHL (von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase):
VHL serves as the substrate recognition subunit of a ubiquitin ligase that targets the α subunit of the heterodimeric transcription factor HIF1 for degradation. This interaction requires the hydroxylation of HIF1A on one or both of 2 conserved prolyl residues by members of the EGLN family of prolyl hydroxylases.2155
- VirG:
A Shigella protein that is required for intracellular actin-based motility; VirG binds ATG5, which induces xenophagy; IcsB, a protein secreted by the type III secretion system, competitively blocks this interaction.2156
- VMP1 (vacuole membrane protein 1):
A multispanning membrane protein that is required for macroautophagy.632,2157 VMP1 regulates the levels of PtdIns3P,2158 binding of the ATG12–ATG5-ATG16L1 complex, and lipidation of LC3.2159
- Vps1:
A dynamin-like GTPase required for peroxisomal fission. It interacts with Atg11 and Atg36 on peroxisomes that are being targeted for degradation by pexophagy.1716 See also Dnm1.
- Vps11:
A member of the core subunit of the homotypic fusion and protein sorting (HOPS) and class C core vacuole/endosome tethering (CORVET) complexes, originally found in yeast but also conserved in higher eukaryotes.2160,2161 These complexes are important for correct endolysosomal trafficking, as well as the trafficking of black pigment cell organelles, melanosomes; zebrafish Vps11 is involved in maintaining melanosome integrity, possibly through an autophagy-dependent mechanism.2162
- Vps30/Atg6:
A component of the class III PtdIns3K complex. Vps30/Atg6 forms part of 2 distinct yeast complexes (I and II) that are required for the Atg and Vps pathways, respectively. See also BECN1 and phosphatidylinositol 3-kinase.1588
- Vps34:
The yeast phosphatidylinositol 3-kinase; the lipid kinase catalytic component of the PtdIns3K complex I and II.1941 See also PIK3C3 and phosphatidylinositol 3-kinase.
- Vps38:
A yeast component of the class III PtdIns3K complex II, which directs it to function in the vacuolar protein sorting pathway.
- VTC (vacuolar transporter chaperone):
A complex composed of Vtc1, Vtc2, Vtc3 and Vtc4 that is required for microautophagy in yeast.2163
- Vti1:
A yeast soluble SNARE that, together with Sec18/NSF, is needed for the fusion of autophagosomes with the vacuole.2024 In mammalian cells, the SNARE proteins VAMP8 and VTI1B mediate the fusion of antimicrobial and canonical autophagosomes with lysosomes.2151
- WAC (WW domain containing adaptor with coiled-coil):
A positive regulator of macroautophagy that interacts with BECN1, WAC also negatively regulates the UPS.1747
- WDFY3/ALFY (WD repeat and FYVE domain containing 3):
A scaffold protein that targets cytosolic protein aggregates for autophagic degradation.2164 WDFY3 interacts directly with ATG5,2165 GABARAP proteins,146 and SQSTM1.2166
- WDR45/WIPI4 (WD repeat domain 45):
See WIPI.
- WHAMM:
A nucleation-promoting factor that directs the activity of the Arp2/3 complex to function in autophagosome formation.2167 WHAMM colocalizes with LC3, ZFYVE1 and SQSTM1 and acts in autophagosome biogenesis through a mechanism dependent on actin comet tail formation.
- WIPI (WD repeat domain, phosphoinositide interacting):
The WIPI proteins are putative mammalian homologs of yeast Atg18 and Atg21. There are 4 WIPI proteins in mammalian cells. WIPI1/WIPI49 and WIPI2 localize with LC3 and bind PtdIns3P.555 WIPI2 is required for starvation-induced macroautophagy.559 WDR45/WIPI4 is also involved in macroautophagy. In humans, WDR45 is localized on the X-chromosome and so far only de novo loss-of-function mutations are described. Heterozygous and somatic mutations cause neurodegeneration with brain iron accumulation,2168 while hemizygous mutations result in early-onset epileptic encephalopathy.2169 Impaired autophagy has been shown in lymphoblastoid cell lines derived from affected patients, showing abnormal colocalization of LC3-II and ATG9A. Furthermore, lymphoblastoid cell lines from affected subjects, show increased levels of LC3-II, even under normal conditions.2170 Surprisingly, complete Wdr45 knockout mice develop normally, but show neurodegeneration, as of 9 months of age, thereby indicating overlapping activity of the 4 WIPI proteins in mammals.2171 WDR45/WIPI4 appears to be the member of the mammalian WIPI protein family that binds ATG2.464,563
- WNT (wingless-type MMTV integration site family):
Cysteine-rich glycosylated secreted proteins that determine multiple cellular functions such as neuronal development, angiogenesis, tumor growth, and stem cell proliferation. Signaling pathways of WNT such as those that involve CTNNB1/beta-catenin can suppress macroautophagy.2172,2173
- WNT5A:
A ligand of the WNT signaling pathway. Activation of the WNT5A-CTNNB1 pathway suppresses IFNG-induced autophagy in macrophages during mycobacterial infection.528
- Wortmannin (WM):
An inhibitor of PI3K and PtdIns3K; it inhibits macroautophagy due to the downstream effect on PtdIns3K.1851
- WXXL motif:
An amino acid sequence present in proteins that allows an interaction with Atg8/LC3/GABARAP proteins; the consensus is [W/F/Y]-X-X-[I/L/V]. Also see AIM and LIR/LRS.1481
- WYE-354:
A catalytic MTOR inhibitor that increases macroautophagic flux to a greater level than allosteric inhibitors such as rapamycin (and may be used to induce macroautophagy in cell lines that are resistant to rapamycin and its derivatives); short-term treatment with WYE-354 can inhibit both MTORC1 and MTORC2, but the effects on flux are due to the former.341 See also Ku-0063794.
- XBP1 (X-box binding protein 1):
A component of the ER stress response that activates macroautophagy. The XBP1 yeast ortholog is Hac1.2174
- Xenophagy:
Cell-autonomous innate immunity defense, whereby cells eliminate intracellular microbes (e.g., bacteria, fungi, parasites and/or viruses) by sequestration into autophagosomes with subsequent delivery to the lysosome.2175
- Xestospongin B:
An antagonist of the ITPR that dissociates the inhibitory interaction between ITPR and BECN1 and induces macroautophagy.2176
- Yeh1:
See Ayr1.
- Ykt6:
A prenylated vesicle SNARE involved in Golgi transport and fusion with the vacuole (including Cvt vesicle delivery to the vacuole2177); temperature sensitive ykt6 mutations also prevent closure of the phagophore.2023
- Ymr1:
A yeast PtdIns3P-specific phosphatase involved in autophagosome maturation.2178,2179
- Ypk1:
A downstream effector of TORC2 that stimulates macroautophagy under conditions of amino acid depletion.2105 TORC2 activation of Ypk1 results in inhibition of the PPP3/calcineurin-Cmd1/calmodulin phosphatase, which otherwise dephosphorylates and inhibits Gcn2, a positive regulator of macroautophagy. See also Gcn2.
- Ypt1:
- Ypt7:
A yeast homolog of mammalian RAB7, needed for the fusion of autophagosomes with the vacuole.
- YWHAZ/14-3-3/(tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta):
A member of the 14-3-3 family of proteins that inhibits macroautophagy; direct interaction with PIK3C3 negatively regulates kinase activity, and this interaction is disrupted by starvation or C2-ceramide.2182
- ZFPM1/FOG1 (zinc finger protein, FOG family member 1):
A cofactor of GATA1, a positive regulator of macroautophagy gene transcription.641 See also GATA1.
- ZFYVE1/DFCP1 (zinc finger, FYVE domain containing 1):
A PtdIns3P-binding protein that localizes to the omegasome.583 Knockdown of ZFYVE1 does not result in a macroautophagy-defective phenotype.
- ZFYVE26/spastizin/SPG15 (zinc finger, FYVE domain containing 26):
A protein involved in a complicated form of hereditary spastic paraparesis; it interacts with the macroautophagy complex BECN1-UVRAG-RUBCN and is required for autosphagosome maturation.2183
- ZIPK:
See Sqa.
- ZKSCAN3/ZNF306 (zinc finger with KRAB and SCAN domains 3):
A zinc finger family transcription factor harboring Kruppel-associated box and SCAN domains that functions as a master transcriptional repressor of autophagy and lysosome biogenesis. ZKSCAN3 represses the transcription of more than 60 genes integral to, or regulatory for, autophagy and lysosome biogenesis and/or function and a subset of these genes, including MAP1LC3B and WIPI2, are its direct targets. Starvation and torin1 treatment induce translocation of ZKSCAN3 from the nucleus to the cytoplasm.643
- Zoledronic acid:
A bisphosphonate that induces macroautophagy and may result in autophagic cell death in prostate and breast cancer cells.2184
- Zymophagy:
The selective degradation of activated zymogen granules by a macroautophagy-like process that is dependent on VMP1, SQSTM1 and the ubiquitin protease USP9X.909 See also crinophagy.
Quick guide
1.Whenever possible, use more than one assay to monitor autophagy.
2.Whenever possible, include flux measurements for autophagy (e.g., using tandem fluorochrome assays such as RFP-EGFP-LC3 or, preferably, cargo-specific variations thereof).
3.Whenever possible, use genetic inhibition of autophagy to complement studies with nonspecific pharmacological inhibitors such as 3-MA.
4.For analysis of genetic inhibition, a minimum of 2 ATG genes (including for example BECN1, ATG7 or ULK1) should be targeted to help ensure the phenotype is due to inhibition of autophagy.
5.When monitoring GFP-LC3 puncta formation, provide quantification, ideally in the form of number of puncta per cell.
6.For the interpretation of decreased SQSTM1 levels, use a pan-caspase inhibitor to ensure that the reduced SQSTM1 amount is not due to a caspase-induced cleavage of the protein.
7.Whenever possible, monitor autophagic responses using both short-term and long-term assays.
Index
A
Acridine Orange 97, 113
Alzheimer disease 92, 109, 117 Ape1 187, 194, 195, 209, 215 Apicoplast 115, 117 Atg8–PE conjugation 182
Atg9 peripheral sites 182, 206, 208, 211
Atg12–Atg5 conjugation 67
ATG16L1 67, 68, 70, 71, 92, 118
Atg18 68, 69, 71, 106, 108, 115, 121, 183, 190, 200, 203, 213
Autophagic body 33, 34, 76, 82, 190
Autophagosome 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 46, 47, 48, 49, 50, 52, 53, 54, 55, 56, 57, 59, 60, 61, 66, 67, 68, 69, 70, 71, 72, 76, 77, 78, 79, 82, 83, 84, 85, 86, 87, 88, 91, 94, 97, 98, 99, 100, 101, 102, 104, 105, 107, 108, 110, 111, 113, 114, 115, 116, 117, 118, 120, 121, 186, 187, 188, 189, 190, 191, 192, 193, 194, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 207, 208, 209, 210, 212, 213, 214, 215, 216, 217, 218, 219, 220
B
Bafilomycin A1 40, 42, 43, 47, 48, 50, 55, 63, 71, 78, 79, 84, 86, 87, 90, 91, 93, 95, 99, 100, 106, 111, 116, 191, 192 BHMT 73
C
C. elegans 66, 69, 71, 72, 77, 83, 84, 104, 105, 180, 181, 182, 183, 185, 187, 188, 189, 190, 191, 192, 193, 195, 198, 204, 206, 208, 210, 212
Calcium 46, 48, 53, 55, 101, 102, 186, 203
Cell death 34, 37, 48, 51, 53, 55, 57, 62, 71, 75, 82, 83, 91, 93, 94, 99, 100, 101, 102, 107, 108, 112, 113, 117, 119, 120, 184, 186, 187, 188, 189, 191, 193, 195, 197, 198, 199, 200, 202, 203, 205, 207, 210, 212, 214
Chicken DT40 cells 103, 105
Chlamydomonas 106
Chloroquine 43, 44, 47, 48, 49, 50, 55, 59, 63, 67, 71, 84, 90, 91, 92, 93, 99, 100, 105, 109, 111, 181, 187, 196
Class III PtdIns3K 55, 99, 186, 189, 191, 192, 197, 198, 201, 204, 208, 210, 218, 219 Correlative light and electron microscopy (CLEM) 30, 37, 38, 54, 85
Cvt pathway 39, 40, 68, 74, 76, 87, 181, 182, 183, 188, 210
D
Dictyostelium 32, 48, 51, 54, 72, 73, 94, 115, 212
DQ-BSA 30, 87, 88
Drosophila 50, 51, 61, 62, 65, 69, 71, 80, 87, 94, 98, 106, 112, 116, 186, 192, 193, 195, 196, 204, 206, 207, 210, 211, 212, 214, 216, 218
E
Electron microscopy 30, 34, 35, 61, 84, 117, 121
Endosomal microautophagy 96, 196, 200
EPG 69, 105, 189, 190, 197, 204, 212, 213, 214, 216, 217, 219, 220
Epithelial cell 34, 85, 92, 100, 111, 116, 204, 205
Erythroid cell 106, 107
F
Flow cytometry 51, 53, 59, 60, 70, 73, 79, 80, 90, 96, 121
Fluorescence microscopy 35, 36, 37, 38, 44, 46, 47, 48, 49, 51, 52, 53, 54, 59, 68, 69, 75, 76, 77, 79, 80, 81, 82, 83, 84, 86, 87, 89, 92, 105, 106, 111, 113, 114, 117, 121, 203
Flux 32, 33, 34, 35, 37, 38, 39, 41, 43, 46, 47, 48, 49, 50, 51, 52, 53, 54, 57, 59, 60, 61, 62, 63, 66, 69, 71, 73, 74, 77, 79, 80, 81, 85, 86, 87, 89, 90, 91, 92, 94, 99, 103, 105, 109, 110, 111, 113, 114, 115, 116, 121, 187, 190, 195, 196, 198, 201, 202, 203, 205, 212, 214, 216, 217, 219, 220
G
GFP-Atg8 35, 43, 46, 48, 49, 50, 51, 52, 54, 59, 71, 73, 74, 76, 86, 87, 89, 98, 106, 107, 108, 112, 114, 115, 121, 126, 148, 212
GFP-LC3 34, 36, 38, 44, 47, 48, 49, 50, 51, 52, 53, 54, 56, 57, 58, 59, 60, 61, 67, 68, 69, 78, 79, 81, 82, 87, 88, 89, 92, 97, 98, 101, 103, 105, 106, 111, 117, 121, 220
H
Honeybee 108
Hydra 110, 111
I
ICP34.5 55, 85, 101, 194
Inhibitors 33, 35, 37, 42, 45, 47, 48, 49, 50, 51, 54, 55, 57, 59, 62, 63, 64, 65, 66, 67, 69, 70, 72, 73, 74, 77 80, 81, 84, 86, 87, 88, 90, 91, 93, 95, 96, 97, 98, 99, 100, 101, 102, 103, 106, 109, 110, 111, 115, 116, 121, 187, 192, 195, 196, 208, 212, 213, 214
K
Keima 79, 80, 88
L
Large animals 110, 111, 112
Late nucleophagy 30, 39, 74, 76, 195
LC3-II 34, 35, 41, 43, 44, 45, 46, 47, 48, 49, 50, 52, 53, 54, 59, 60, 63, 68, 70, 71, 86, 90, 91, 102, 103, 105, 106, 107, 109, 110, 111, 117, 121, 193, 194, 195, 202, 204, 208, 219, 220
LC3-associated phagocytosis (LAP) 30, 34, 37, 43, 66, 84, 117, 195, 197, 198
Lepidoptera 112
Lipofuscin 38, 61, 77, 87, 109, 121, 205
Lipophagy 71, 83, 107, 110, 201, 209, 214
LIR motif 44, 62, 119, 120, 192, 205, 214, 215, 217, 220
LysoTracker Red 82, 97, 98, 110, 111, 112, 113, 114, 117
M
3-MA 30, 51, 54, 55, 62, 69, 71, 73, 84, 87, 88, 93, 99, 105, 106, 109, 114, 115, 116, 186
MDC 30, 91, 97, 196
Mitophagy 39, 40, 43, 45, 55, 57, 64, 70, 74, 75, 76, 77, 78, 79, 80, 81, 92, 105, 108, 110, 180, 182, 183, 185, 186, 187, 189, 190, 191, 192, 193, 194, 195, 196, 197, 200, 201, 202, 204, 205, 207, 208, 209, 210, 211, 212
MitoTracker 74, 78, 79, 80, 81, 95, 115
MTOR 47, 55, 62, 63, 64, 65, 66, 67, 72, 81, 87, 93, 100, 101, 102, 105, 109, 110, 111, 113, 117, 121, 187, 188, 189, 190, 191, 194, 195, 196, 197, 198, 199, 200, 201, 202, 204, 205, 206, 208, 209, 210, 211, 212, 213, 215, 216, 217, 218, 219
N
Neotropical teleosts 112
Nucleophagy 39, 57, 74, 75, 76, 201, 205
O
Odontoblasts 113
P
Parkinson disease 75, 92, 199, 202, 206, 208, 209, 214
Pexophagy 39, 40, 57, 74, 75, 77, 80, 81, 86, 103, 108, 180, 182, 183, 185, 189, 190, 191, 192, 193, 194, 195, 197, 200, 202, 207, 213
Pho8Δ60 assay 50, 74, 76, 121, 194, 207
Photodynamic therapy 30, 93, 213
Planarians 113
Plants 32, 34, 35, 39, 40, 46, 54, 81, 94, 100, 113, 114, 121, 183, 197, 212
PolyQ protein turnover 77
prApe1 74, 76, 86, 87, 187, 194, 195, 209
Protists 32, 40, 114, 115, 117, 118
PtdIns3P 67, 68, 69, 99, 108, 117, 189, 197, 198, 204, 207, 208, 209, 214, 215, 219, 220
R
Rainbow trout 115, 116
Reticulophagy 74, 83, 108, 183, 191, 205
Rosella 57, 76, 105, 121
S
Saponin 54, 59, 210
Sea Urchin 116
Sequestration assays 86, 87, 121
SQSTM1 46, 81, 84, 86, 87, 89, 90, 92, 97, 102, 105, 106, 109, 110, 111, 117, 120, 121, 187, 191, 192, 193, 194, 197, 200, 201, 205, 206, 211, 212, 214, 216, 217, 219
T
TAKA assay 67
Tandem mRFP/mCherry-GFP-LC3 61, 105
Ticks 116
Torin1 46, 55, 65, 72, 100, 102, 105, 216, 220
Trehalose 55, 89, 102
V
V-ATPase 40, 41, 55, 91, 99, 100, 201, 202, 209, 213, 218
Viral BCL2 85, 101
Viral FLIP 101
W
Western blot 34, 42, 43, 44, 45, 46, 48, 49, 50, 51, 52, 54, 62, 63, 64, 65, 66, 69, 70, 73, 75, 76, 78, 81, 83, 84, 86, 90, 91, 92, 104, 105, 106, 108, 109, 110, 111, 112, 113, 114, 116, 117, 121
WIPI1 68, 69, 118, 197, 198, 213
WIPI2 68, 69, 209, 213, 214
Wortmannin 55, 62, 68, 87, 88, 99, 111, 115, 118, 213
X
Xenophagy 32, 40, 74, 84, 85, 180, 194, 211, 213, 214
Y
Yeast 34, 35, 38, 39, 40, 41, 42, 43, 45, 47, 49, 50, 54, 55, 57, 63, 65, 66, 67, 68, 69, 72, 73, 74, 75, 76, 77, 80, 81, 83, 84, 86, 87, 92, 102, 103, 104, 107, 108, 114, 115, 187, 188, 189, 190, 191, 192, 193, 196, 197, 198, 199, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 216, 217, 218, 219
Z
Zebrafish 116, 117, 205, 213
Zymophagy 86, 188, 214
References
- 1.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; http://dx.doi.org/ 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4:151-75; http://dx.doi.org/ 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy 2007; 3:181-206; http://dx.doi.org/ 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 4.Xia HG, Najafov A, Geng J, Galan-Acosta L, Han X, Guo Y, Shan B, Zhang Y, Norberg E, Zhang T, et al.. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol 2015; 210:705-16; http://dx.doi.org/ 10.1083/jcb.201503044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klionsky DJ. The autophagosome is overrated! Autophagy 2011; 7:353-4; http://dx.doi.org/ 10.4161/auto.7.4.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eskelinen E-L, Reggiori F, Baba M, Kovacs AL, Seglen PO. Seeing is believing: The impact of electron microscopy on autophagy research. Autophagy 2011; 7:935-56; http://dx.doi.org/ 10.4161/auto.7.9.15760. [DOI] [PubMed] [Google Scholar]
- 7.Seglen PO. Regulation of autophagic protein degradation in isolated liver cells In: Glaumann H and Ballard FJ, eds. Lysosomes: Their Role in Protein Breakdown. London: Academic Press, 1987:369-414. [Google Scholar]
- 8.de Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol 1966; 28:435-92; http://dx.doi.org/ 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 9.Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun 1988; 151:40-7; http://dx.doi.org/ 10.1016/0006-291X(88)90556-6. [DOI] [PubMed] [Google Scholar]
- 10.Dice JF, Klionsky DJ. Artophagy, the art of autophagy–macroautophagy. Autophagy 2010; 6. [DOI] [PubMed] [Google Scholar]
- 11.Lucocq JM, Hacker C. Cutting a fine figure: On the use of thin sections in electron microscopy to quantify autophagy. Autophagy 2013; 9:1443-8; http://dx.doi.org/ 10.4161/auto.25570. [DOI] [PubMed] [Google Scholar]
- 12.Kovács J, Fellinger E, Karpati AP, Kovács AL, Laszlo L, Réz G. Morphometric evaluation of the turnover of autophagic vacuoles after treatment with Triton X-100 and vinblastine in murine pancreatic acinar and seminal vesicle epithelial cells. Virchows Arch B Cell Pathol Incl Mol Pathol 1987; 53:183-90; http://dx.doi.org/ 10.1007/BF02890242. [DOI] [PubMed] [Google Scholar]
- 13.Kovács J, Fellinger E, Karpati PA, Kovács AL, Laszlo L. The turnover of autophagic vacuoles: evaluation by quantitative electron microscopy. Biomed Biochim Acta 1986; 45:1543-7. [PubMed] [Google Scholar]
- 14.Kovács J, Laszlo L, Kovács AL. Regression of autophagic vacuoles in pancreatic acinar, seminal vesicle epithelial, and liver parenchymal cells: a comparative morphometric study of the effect of vinblastine and leupeptin followed by cycloheximide treatment. Exp Cell Res 1988; 174:244-51; http://dx.doi.org/ 10.1016/0014-4827(88)90158-9. [DOI] [PubMed] [Google Scholar]
- 15.CT Chu. Autophagic stress in neuronal injury and disease. J Neuropathol Exp Neurol 2006; 65:423-32; http://dx.doi.org/ 10.1097/01.jnen.0000229233.75253.be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 2006; 281:36303-16; http://dx.doi.org/ 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- 17.Kovács AL, Reith A, Seglen PO. Accumulation of autophagosomes after inhibition of hepatocytic protein degradation by vinblastine, leupeptin or a lysosomotropic amine. Exp Cell Res 1982; 137:191-201; http://dx.doi.org/ 10.1016/0014-4827(82)90020-9. [DOI] [PubMed] [Google Scholar]
- 18.Bestebroer J, V'Kovski P, Mauthe M, Reggiori F. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic 2013; 14:1029-41; http://dx.doi.org/ 10.1111/tra.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo SM, Ge ZJ, Wang ZW, Jiang ZZ, Wang ZB, Ouyang YC, Hou Y, Schatten H, Sun QY. Unique insights into maternal mitochondrial inheritance in mice. Proc Natl Acad Sci USA 2013; 110:13038-43; http://dx.doi.org/ 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Politi Y, Gal L, Kalifa Y, Ravid L, Elazar Z, Arama E. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev Cell 2014; 29:305-20; http://dx.doi.org/ 10.1016/j.devcel.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Toth S, Nagy K, Palfia Z, Rez G. Cellular autophagic capacity changes during azaserine-induced tumour progression in the rat pancreas. Up-regulation in all premalignant stages and down-regulation with loss of cycloheximide sensitivity of segregation along with malignant transformation. Cell Tissue Res 2002; 309:409-16; http://dx.doi.org/ 10.1007/s00441-001-0506-7. [DOI] [PubMed] [Google Scholar]
- 22.Loos B, Engelbrecht AM. Cell death: a dynamic response concept. Autophagy 2009; 5:590-603; http://dx.doi.org/ 10.4161/auto.5.5.8479. [DOI] [PubMed] [Google Scholar]
- 23.Seglen PO, Gordon PB, Grinde B, Solheim A, Kovacs AL, Poli A. Inhibitors and pathways of hepatocytic protein degradation. Acta Biol Med Ger 1981; 40:1587-98. [PubMed] [Google Scholar]
- 24.Ktistakis NT, Andrews S, Long J. What is the advantage of a transient precursor in autophagosome biogenesis? Autophagy 2011; 7:118-22; http://dx.doi.org/ 10.4161/auto.7.1.13697. [DOI] [PubMed] [Google Scholar]
- 25.Kovács AL, Réz G, Pálfia Z, Kovács J. Autophagy in the epithelial cells of murine seminal vesicle in vitro. Formation of large sheets of nascent isolation membranes, sequestration of the nucleus and inhibition by wortmannin and 3-ethyladenine. Cell Tissue Res 2000; 302:253-61; http://dx.doi.org/ 10.1007/s004410000275. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007; 3:542-5; http://dx.doi.org/ 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 27.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009; 461:654-8; http://dx.doi.org/ 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 28.Kanki T, Kang D, Klionsky DJ. Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy 2009; 5:1186-9; http://dx.doi.org/ 10.4161/auto.5.8.9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grander D, Kharaziha P, Laane E, Pokrovskaja K, Panaretakis T. Autophagy as the main means of cytotoxicity by glucocorticoids in hematological malignancies. Autophagy 2009; 5:1198-200; http://dx.doi.org/ 10.4161/auto.5.8.10122. [DOI] [PubMed] [Google Scholar]
- 30.Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy 2010; 6:794-7; http://dx.doi.org/ 10.4161/auto.6.6.12348. [DOI] [PubMed] [Google Scholar]
- 31.Raju D, Jones NL. Methods to monitor autophagy in H. pylori vacuolating cytotoxin A (VacA)-treated cells. Autophagy 2010; 6:138-43; http://dx.doi.org/ 10.4161/auto.6.1.10222. [DOI] [PubMed] [Google Scholar]
- 32.Geng J, Klionsky DJ. Determining Atg protein stoichiometry at the phagophore assembly site by fluorescence microscopy. Autophagy 2010; 6:144-7; http://dx.doi.org/ 10.4161/auto.6.1.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanlund JM, Kregel KC, Oberley TD. Investigating autophagy: quantitative morphometric analysis using electron microscopy. Autophagy 2010; 6:270-7; http://dx.doi.org/ 10.4161/auto.6.2.10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Ney PA. Reticulocyte mitophagy: monitoring mitochondrial clearance in a mammalian model. Autophagy 2010; 6:405-8; http://dx.doi.org/ 10.4161/auto.6.3.11245. [DOI] [PubMed] [Google Scholar]
- 35.Seglen PO, Brinchmann MF. Purification of autophagosomes from rat hepatocytes. Autophagy 2010; 6:542-7; http://dx.doi.org/ 10.4161/auto.6.4.11272. [DOI] [PubMed] [Google Scholar]
- 36.He C, Klionsky DJ. Analyzing autophagy in zebrafish. Autophagy 2010; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calvo-Garrido J, Carilla-Latorre S, Mesquita A, Escalante R. A proteolytic cleavage assay to monitor autophagy in Dictyostelium discoideum. Autophagy 2011; 7:1063-8; http://dx.doi.org/ 10.4161/auto.7.9.16629. [DOI] [PubMed] [Google Scholar]
- 38.Xu F, Liu XH, Zhuang FL, Zhu J, Lin FC. Analyzing autophagy in Magnaporthe oryzae. Autophagy 2011; 7:525-30; http://dx.doi.org/ 10.4161/auto.7.5.15020. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky DJ. Autophagy: Lower Eukaryotes and Non-Mammalian Systems, Part A. Amsterdam: Academic Press/Elsevier, 2008. [Google Scholar]
- 40.Klionsky DJ. Autophagy in Disease and Clinical Applications, Part C. Amsterdam: Academic Press/Elsevier, 2008. [Google Scholar]
- 41.Klionsky DJ. Autophagy in Mammalian Systems, Part B. Amsterdam: Academic Press/Elsevier, 2008. [Google Scholar]
- 42.Zhu J, Dagda RK, Chu CT. Monitoring mitophagy in neuronal cell cultures. Methods Mol Biol 2011; 793:325-39; http://dx.doi.org/ 10.1007/978-1-61779-328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky DJ. Protocol URL. [Google Scholar]
- 44.Chu CT, Plowey ED, Dagda RK, Hickey RW, Cherra SJ 3rd, Clark RS. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol 2009; 453:217-49; http://dx.doi.org/ 10.1016/S0076-6879(08)04011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh PK, Singh S. Changing shapes of glycogen-autophagy nexus in neurons: perspective from a rare epilepsy. Front Neurol 2015; 6:14; http://dx.doi.org/ 10.3389/fneur.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy. Microscopy Res Tech 2004; 64:10-20; http://dx.doi.org/ 10.1002/jemt.20046. [DOI] [PubMed] [Google Scholar]
- 47.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract 2006; 202:631-8; http://dx.doi.org/ 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. Monitoring autophagy by electron microscopy in mammalian cells. Methods Enzymol 2009; 452:143-64; http://dx.doi.org/ 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- 49.Eskelinen E-L. Maturation of autophagic vacuoles in mammalian cells. Autophagy 2005; 1:1-10; http://dx.doi.org/ 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 50.Eskelinen E-L. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy 2008; 4:257-60; http://dx.doi.org/ 10.4161/auto.5179. [DOI] [PubMed] [Google Scholar]
- 51.Eskelinen E-L, Kovacs AL. Double membranes vs. lipid bilayers, and their significance for correct identification of macroautophagic structures. Autophagy 2011; 7:931-2; http://dx.doi.org/ 10.4161/auto.7.9.16679. [DOI] [PubMed] [Google Scholar]
- 52.Biazik J, Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy 2015; 11:439-51; http://dx.doi.org/ 10.1080/15548627.2015.1017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eskelinen E-L. Fine structure of the autophagosome In: Deretic V, ed. Autophagosome and Phagosome. Totowa, NJ: Humana Press, 2008:11-28; http://dx.doi.org/ 10.1007/978-1-59745-157-4. [DOI] [Google Scholar]
- 54.Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 1998; 273:21883-92; http://dx.doi.org/ 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 55.Eskelinen E-L. Macroautophagy in mammalian cells In: Saftig P, ed. Lysosomes. Georgetown, TX: LandesBioscience/Eurekah.com, 2005. [Google Scholar]
- 56.Turturici G, Tinnirello R, Sconzo G, Geraci F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am J Physiol Cell Phys 2014; 306:C621-33; http://dx.doi.org/ 10.1152/ajpcell.00228.2013. [DOI] [PubMed] [Google Scholar]
- 57.Yang DS, Lee JH, Nixon RA. Monitoring autophagy in Alzheimer's disease and related neurodegenerative diseases. Methods Enzymol 2009; 453:111-44; http://dx.doi.org/ 10.1016/S0076-6879(08)04006-8. [DOI] [PubMed] [Google Scholar]
- 58.Yokota S, Himeno M, Kato K. Immunocytochemical localization of acid phosphatase in rat liver. Cell Struct Funct 1989; 14:163-71; http://dx.doi.org/ 10.1247/csf.14.163. [DOI] [PubMed] [Google Scholar]
- 59.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci 2008; 28:6926-37; http://dx.doi.org/ 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropath Exp Neuro 2005; 64:113-22. [DOI] [PubMed] [Google Scholar]
- 61.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, et al.. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 2010; 141:1146-58; http://dx.doi.org/ 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JH, McBrayer MK, Wolfe DM, Haslett LJ, Kumar A, Sato Y, Lie PP, Mohan P, Coffey EE, Kompella U, et al.. Presenilin 1 maintains lysosomal Ca2+ homeostasis via TRPML1 by regulating vATPase-mediated lysosome acidification. Cell Rep 2015; 12:1430-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonati T, Reimann RR, Falsig J, Baral PK, O'Connor T, Hornemann S, Yaganoglu S, Li B, Herrmann US, Wieland B, et al.. The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature 2013; 501:102-6; http://dx.doi.org/ 10.1038/nature12402. [DOI] [PubMed] [Google Scholar]
- 64.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nature Rev Mol Cell Biol 2007; 8:622-32; http://dx.doi.org/ 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 65.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 66.Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J Neurosci 2011; 31:7817-30; http://dx.doi.org/ 10.1523/JNEUROSCI.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rabouille C, Strous GJ, Crapo JD, Geuze HJ, Slot JW. The differential degradation of two cytosolic proteins as a tool to monitor autophagy in hepatocytes by immunocytochemistry. J Cell Biol 1993; 120:897-908; http://dx.doi.org/ 10.1083/jcb.120.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kovács AL, Pálfia Z, Réz G, Vellai T, Kovács J. Sequestration revisited: integrating traditional electron microscopy, de novo assembly and new results. Autophagy 2007; 3:655-62; http://dx.doi.org/ 10.4161/auto.4590. [DOI] [PubMed] [Google Scholar]
- 69.Gao W, Kang JH, Liao Y, Ding WX, Gambotto AA, Watkins SC, Liu YJ, Stolz DB, Yin XM. Biochemical isolation and characterization of the tubulovesicular LC3-positive autophagosomal compartment. J Biol Chem 2010; 285:1371-83; http://dx.doi.org/ 10.1074/jbc.M109.054197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lajoie P, Guay G, Dennis JW, Nabi IR. The lipid composition of autophagic vacuoles regulates expression of multilamellar bodies. J Cell Sci 2005; 118:1991-2003; http://dx.doi.org/ 10.1242/jcs.02324. [DOI] [PubMed] [Google Scholar]
- 71.Mayhew TM. Quantitative immunoelectron microscopy: alternative ways of assessing subcellular patterns of gold labeling. Methods Mol Biol 2007; 369:309-29; http://dx.doi.org/ 10.1007/978-1-59745-294-6. [DOI] [PubMed] [Google Scholar]
- 72.Mayhew TM, Lucocq JM, Griffiths G. Relative labelling index: a novel stereological approach to test for non-random immunogold labelling of organelles and membranes on transmission electron microscopy thin sections. J Microsc 2002; 205:153-64; http://dx.doi.org/ 10.1046/j.0022-2720.2001.00977.x. [DOI] [PubMed] [Google Scholar]
- 73.Isidoro C, Biagioni F, Giorgi FS, Fulceri F, Paparelli A, Fornai F. The role of autophagy on the survival of dopamine neurons. Curr Top Med Chem 2009; 9:869-79. [PubMed] [Google Scholar]
- 74.Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 2007; 26:79-92; http://dx.doi.org/ 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep 2013; 14:143-51; http://dx.doi.org/ 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V. Secretory autophagy. Curr Opin Cell Biol 2015; 35:106-16; http://dx.doi.org/ 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saito T, Asai K, Sato S, Takano H, Adach A, Sasaki Y, Namimatsu S, Mizuno K. Proof of myocardial autophagy by combining antigen retrieval and the avidin-biotin peroxidase complex method. Int J Cardiol 2013; 168:4843-4; http://dx.doi.org/ 10.1016/j.ijcard.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 78.Kovács J. Regression of autophagic vacuoles in seminal vesicle cells following cycloheximide treatment. Exp Cell Res 1983; 144:231-4; http://dx.doi.org/ 10.1016/0014-4827(83)90460-3. [DOI] [PubMed] [Google Scholar]
- 79.Réz G, Csak J, Fellinger E, Laszlo L, Kovács AL, Oliva O, Kovács J. Time course of vinblastine-induced autophagocytosis and changes in the endoplasmic reticulum in murine pancreatic acinar cells: a morphometric and biochemical study. Eur J Cell Biol 1996; 71:341-50. [PubMed] [Google Scholar]
- 80.Kovács AL, Grinde B, Seglen PO. Inhibition of autophagic vacuole formation and protein degradation by amino acids in isolated hepatocytes. Exp Cell Res 1981; 133:431-6; http://dx.doi.org/ 10.1016/0014-4827(81)90336-0. [DOI] [PubMed] [Google Scholar]
- 81.Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci USA 1983; 80:2179-83; http://dx.doi.org/ 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mortimore GE, Lardeux BR, Adams CE. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J Biol Chem 1988; 263:2506-12. [PubMed] [Google Scholar]
- 83.Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Amer J Pathol 2007; 170:75-86; http://dx.doi.org/ 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603-14; http://dx.doi.org/ 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Orvedahl A, Sumpter R Jr., Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al.. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 2011; 480:113-7; http://dx.doi.org/ 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Razi M, Tooze SA. Correlative light and electron microscopy. Methods Enzymol 2009; 452:261-75; http://dx.doi.org/ 10.1016/S0076-6879(08)03617-3. [DOI] [PubMed] [Google Scholar]
- 87.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol 2011; 9:e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Castillo K, Rojas-Rivera D, Lisbona F, Caballero B, Nassif M, Court F, Schuck S, Ibar C, Walter P, Sierralta J, et al.. BAX inhibitor-1 regulates autophagy by controlling the IRE1α branch of the unfolded protein response. EMBO J 2011; 30:4465-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009; 5:1180-5; http://dx.doi.org/ 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 90.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Auto-phagy 2010; 6:301-3; http://dx.doi.org/ 10.4161/auto.6.2.11134. [DOI] [PubMed] [Google Scholar]
- 91.Duke EM, Razi M, Weston A, Guttmann P, Werner S, Henzler K, Schneider G, Tooze SA, Collinson LM. Imaging endosomes and autophagosomes in whole mammalian cells using correlative cryo-fluorescence and cryo-soft X-ray microscopy (cryo-CLXM). Ultramicroscopy 2014; 143:77-87; http://dx.doi.org/ 10.1016/j.ultramic.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA 2006; 103:5805-10; http://dx.doi.org/ 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct 1995; 20:465-71; http://dx.doi.org/ 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- 94.Rez G, Meldolesi J. Freeze-fracture of drug-induced autophagocytosis in the mouse exocrine pancreas. Lab Investig 1980; 43:269-77. [PubMed] [Google Scholar]
- 95.Punnonen E-L, Pihakaski K, Mattila K, Lounatmaa K, Hirsimaki P. Intramembrane particles and filipin labelling on the membranes of autophagic vacuoles and lysosomes in mouse liver. Cell Tissue Res 1989; 258:269-76; http://dx.doi.org/ 10.1007/BF00239447. [DOI] [PubMed] [Google Scholar]
- 96.Fengsrud M, Erichsen ES, Berg TO, Raiborg C, Seglen PO. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur J Cell Biol 2000; 79:871-82; http://dx.doi.org/ 10.1078/0171-9335-00125. [DOI] [PubMed] [Google Scholar]
- 97.Dickey JS, Gonzalez Y, Aryal B, Mog S, Nakamura AJ, Redon CE, Baxa U, Rosen E, Cheng G, Zielonka J, et al.. Mito-tempol and dexrazoxane exhibit cardioprotective and chemotherapeutic effects through specific protein oxidation and autophagy in a syngeneic breast tumor preclinical model. PloS One 2013; 8:e70575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rao VA, Klein SR, Bonar SJ, Zielonka J, Mizuno N, Dickey JS, Keller PW, Joseph J, Kalyanaraman B, Shacter E. The antioxidant transcription factor Nrf2 negatively regulates autophagy and growth arrest induced by the anticancer redox agent mitoquinone. J Biol Chem 2010; 285:34447-59; http://dx.doi.org/ 10.1074/jbc.M110.133579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nature Rev Mol Cell Biol 2007; 8:931-7; http://dx.doi.org/ 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 100.Krick R, M(u)he Y, Prick T, Bredschneider M, Bremer S, Wenzel D, Eskelinen E-L, Thumm M. Piecemeal microautophagy of the nucleus: genetic and morphological traits. Autophagy 2009; 5:270-2; http://dx.doi.org/ 10.4161/auto.5.2.7639. [DOI] [PubMed] [Google Scholar]
- 101.Meschini S, Condello M, Calcabrini A, Marra M, Formisano G, Lista P, De Milito A, Federici E, Arancia G. The plant alkaloid voacamine induces apoptosis-independent autophagic cell death on both sensitive and multidrug resistant human osteosarcoma cells. Autophagy 2008; 4:1020-33; http://dx.doi.org/ 10.4161/auto.6952. [DOI] [PubMed] [Google Scholar]
- 102.Proikas-Cezanne T, Robenek H. Freeze-fracture replica immunolabelling reveals human WIPI-1 and WIPI-2 as membrane proteins of autophagosomes. J Cell Mol Med 2011; 15:2007-10; http://dx.doi.org/ 10.1111/j.1582-4934.2011.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kovacs J, Rez G, Kovacs AL, Csak J, Zboray G. Autophagocytosis: freeze-fracture morphology, effects of vinblastine and influence of transcriptional and translational inhibitors. Acta Biol Med Germanica 1982; 41:131-5. [PubMed] [Google Scholar]
- 104.Hirsimaki Y, Hirsimaki P, Lounatmaa K. Vinblastine-induced autophagic vacuoles in mouse liver and Ehrlich ascites tumor cells as assessed by freeze-fracture electron microscopy. Eur J Cell Biol 1982; 27:298-301. [PubMed] [Google Scholar]
- 105.Backues SK, Chen D, Ruan J, Xie Z, Klionsky DJ. Estimating the size and number of autophagic bodies by electron microscopy. Autophagy 2014; 10:155-64; http://dx.doi.org/ 10.4161/auto.26856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell 2005; 16:3438-53; http://dx.doi.org/ 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 2008; 19:3290-8; http://dx.doi.org/ 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sigmond T, Feher J, Baksa A, Pasti G, Palfia Z, Takacs-Vellai K, Kovacs J, Vellai T, Kovacs AL. Qualitative and quantitative characterization of autophagy in Caenorhabditis elegans by electron microscopy. Methods Enzymol 2008; 451:467-91; http://dx.doi.org/ 10.1016/S0076-6879(08)03228-X. [DOI] [PubMed] [Google Scholar]
- 109.Kovács AL, Vellai T, Müller F. Autophagy in Caenorhabditis elegans In: Klionsky DJ, ed. Autophagy. Georgetown, Texas: Landes Bioscience, 2004:217-23. [Google Scholar]
- 110.Weibel ER. Practical Methods for Biological Morphometry. Academic Press, New York, 1979. [Google Scholar]
- 111.Williams MA. Quantitative methods in biology: Practical methods in electron microscopy. Amsterdam, New York, Oxford: North-Holland Publishing Company, 1977. [Google Scholar]
- 112.Howard V, Reed MG. Unbiased stereology; three dimensional measurement in microscopy. U Bios Scientific Publishers, 1998. [Google Scholar]
- 113.AL Kovacs. A simple method to estimate the number of autophagic elements by electron microscopic morphometry in real cellular dimensions. BioMed Res Intl 2014; 2014:578698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xie Z, Nair U, Geng J, Szefler MB, Rothman ED, Klionsky DJ. Indirect estimation of the area density of Atg8 on the phagophore. Autophagy 2009; 5:217-20; http://dx.doi.org/ 10.4161/auto.5.2.7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Punnonen EL, Reunanen H. Effects of vinblastine, leucine, and histidine, and 3-methyladenine on autophagy in Ehrlich ascites cells. Exp Mol Pathol 1990; 52:87-97; http://dx.doi.org/ 10.1016/0014-4800(90)90061-H. [DOI] [PubMed] [Google Scholar]
- 116.Kovacs AL, Laszlo L, Fellinger E, Jakab A, Orosz A, Rez G, Kovacs J. Combined effects of fasting and vinblastine treatment on serum insulin level, the size of autophagic-lysosomal compartment, protein content and lysosomal enzyme activities of liver and exocrine pancreatic cells of the mouse. Comp Biochem Phys B Comp Biochem 1989; 94:505-10; http://dx.doi.org/ 10.1016/0305-0491(89)90189-2. [DOI] [PubMed] [Google Scholar]
- 117.Griffiths G. Fine structure immunocytochemistry Heidelberg, Germany: Springer-Verlag, 1993; http://dx.doi.org/ 10.1007/978-3-642-77095-1. [DOI] [Google Scholar]
- 118.Reyes FC, Chung T, Holding D, Jung R, Vierstra R, Otegui MS. Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell 2011; 23:769-84; http://dx.doi.org/ 10.1105/tpc.110.082156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dunn WA Jr., Cregg JM, Kiel JAKW, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy 2005; 1:75-83; http://dx.doi.org/ 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 120.Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy 2011; 7:297-300; http://dx.doi.org/ 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Belanger M, Rodrigues PH, Dunn WA Jr., Progulske-Fox A. Autophagy: a highway for Porphyromonas gingivalis in endothelial cells. Autophagy 2006; 2:165-70; http://dx.doi.org/ 10.4161/auto.2828. [DOI] [PubMed] [Google Scholar]
- 122.Birmingham CL, Brumell JH. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy 2006; 2:156-8; http://dx.doi.org/ 10.4161/auto.2825. [DOI] [PubMed] [Google Scholar]
- 123.Colombo MI, Gutierrez MG, Romano PS. The two faces of autophagy: Coxiella and Mycobacterium. Autophagy 2006; 2:162-4; http://dx.doi.org/ 10.4161/auto.2827. [DOI] [PubMed] [Google Scholar]
- 124.Ogawa M, Sasakawa C. Shigella and autophagy. Autophagy 2006; 2:171-4; http://dx.doi.org/ 10.4161/auto.2829. [DOI] [PubMed] [Google Scholar]
- 125.Vergne I, Singh S, Roberts E, Kyei G, Master S, Harris J, de Haro S, Naylor J, Davis A, Delgado M, et al.. Autophagy in immune defense against Mycobacterium tuberculosis. Autophagy 2006; 2:175-8; http://dx.doi.org/ 10.4161/auto.2830. [DOI] [PubMed] [Google Scholar]
- 126.Yoshimori T. Autophagy vs. Group A Streptococcus. Autophagy 2006; 2:154-5; http://dx.doi.org/ 10.4161/auto.2822. [DOI] [PubMed] [Google Scholar]
- 127.Gorbunov NV, McDaniel DP, Zhai M, Liao PJ, Garrison BR, Kiang JG. Autophagy and mitochondrial remodelling in mouse mesenchymal stromal cells challenged with Staphylococcus epidermidis. J Cell Mol Med 2015; 19:1133-50; http://dx.doi.org/ 10.1111/jcmm.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett 2010; 584:1359-66; http://dx.doi.org/ 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, Higgins DE, Brumell JH. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy 2007; 3:442-51; http://dx.doi.org/ 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- 130.Klionsky DJ. Protein transport from the cytoplasm into the vacuole. J Membr Biol 1997; 157:105-15; http://dx.doi.org/ 10.1007/s002329900220. [DOI] [PubMed] [Google Scholar]
- 131.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol 1997; 139:1687-95; http://dx.doi.org/ 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dini L, Pagliara P, Carla EC. Phagocytosis of apoptotic cells by liver: a morphological study. Micros Res Tech 2002; 57:530-40; http://dx.doi.org/ 10.1002/jemt.10107. [DOI] [PubMed] [Google Scholar]
- 133.Kroemer G, El-Deiry WS, Golstein P, Peter ME, Vaux D, Vandenabeele P, Zhivotovsky B, Blagosklonny MV, Malorni W, Knight RA, et al.. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ 2005; 12:1463-7; http://dx.doi.org/ 10.1038/sj.cdd.4401724. [DOI] [PubMed] [Google Scholar]
- 134.Rez G, Palfia Z, Fellinger E. Occurrence and inhibition by cycloheximide of apoptosis in vinblastine-treated murine pancreas. A role for autophagy? Acta Biol Hungarica 1991; 42:133-40. [PubMed] [Google Scholar]
- 135.Nagy P, Varga A, Kovács AL, Takáts S, Juhász G. How and why to study autophagy in Drosophila: It's more than just a garbage chute. Methods 2015; 75:151-61; http://dx.doi.org/ 10.1016/j.ymeth.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Giammarioli AM, Gambardella L, Barbati C, Pietraforte D, Tinari A, Alberton M, Gnessi L, Griffin RJ, Minetti M, Malorni W. Differential effects of the glycolysis inhibitor 2-deoxy-D-glucose on the activity of pro-apoptotic agents in metastatic melanoma cells, and induction of a cytoprotective autophagic response. Intl J Cancer 2012; 131:E337-47. [DOI] [PubMed] [Google Scholar]
- 137.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem 2006; 281:3017-24; http://dx.doi.org/ 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 138.Le Grand JN, Chakrama FZ, Seguin-Py S, Fraichard A, Delage-Mourroux R, Jouvenot M, Boyer-Guittaut M. GABARAPL1 (GEC1): Original or copycat? Autophagy 2011; 7:1098-107; http://dx.doi.org/ 10.4161/auto.7.10.15904. [DOI] [PubMed] [Google Scholar]
- 139.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem 2003; 278:51841-50; http://dx.doi.org/ 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 140.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem 2004; 279:36268-76; http://dx.doi.org/ 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 141.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004; 117:2805-12; http://dx.doi.org/ 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 142.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 2010; 29:1792-802; http://dx.doi.org/ 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Szalai P, Hagen LK, Saetre F, Luhr M, Sponheim M, Overbye A, Mills IG, Seglen PO, Engedal N. Autophagic bulk sequestration of cytosolic cargo is independent of LC3, but requires GABARAPs. Exp Cell Res 2015; 333:21-38; http://dx.doi.org/ 10.1016/j.yexcr.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 144.Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 2004; 9:611-8; http://dx.doi.org/ 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- 145.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, et al.. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol 2013; 15:1197-205; http://dx.doi.org/ 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lystad AH, Ichimura Y, Takagi K, Yang Y, Pankiv S, Kanegae Y, Kageyama S, Suzuki M, Saito I, Mizushima T, et al.. Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep 2014; 15:557-65; http://dx.doi.org/ 10.1002/embr.201338003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell 2012; 48:329-42; http://dx.doi.org/ 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem 2000; 275:5845-51; http://dx.doi.org/ 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 149.Cai Q, Lu L, Tian J-H, Zhu Y-B, Qiao H, Sheng Z-H. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron 2010; 68:73-86; http://dx.doi.org/ 10.1016/j.neuron.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Castino R, Fiorentino I, Cagnin M, Giovia A, Isidoro C. Chelation of lysosomal iron protects dopaminergic SH-SY5Y neuroblastoma cells from hydrogen peroxide toxicity by precluding autophagy and Akt dephosphorylation. Toxicol Sci 2011:523-41; http://dx.doi.org/ 10.1093/toxsci/kfr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, Nerini-Molteni S, Sale P, Vago R, Arena G, et al.. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ 2010; 17:962-74; http://dx.doi.org/ 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 152.Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, Ohno M, Schmidt SD, Wesson D, Bandyopadhyay U, Jiang Y, et al.. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer's disease ameliorates amyloid pathologies and memory deficits. Brain 2011; 134:258-77; http://dx.doi.org/ 10.1093/brain/awq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15:1101-11; http://dx.doi.org/ 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Padman BS, Bach M, Lucarelli G, Prescott M, Ramm G. The protonophore CCCP interferes with lysosomal degradation of autophagic cargo in yeast and mammalian cells. Autophagy 2013; 9:1862-75; http://dx.doi.org/ 10.4161/auto.26557. [DOI] [PubMed] [Google Scholar]
- 155.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic 2008; 9:574-87; http://dx.doi.org/ 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 2008; 4:849-950; http://dx.doi.org/ 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 157.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 1998; 23:33-42; http://dx.doi.org/ 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 158.Ahlberg J, Berkenstam A, Henell F, Glaumann H. Degradation of short and long lived proteins in isolated rat liver lysosomes. Effects of pH, temperature, and proteolytic inhibitors. J Biol Chem 1985; 260:5847-54. [PubMed] [Google Scholar]
- 159.Yoon YH, Cho KS, Hwang JJ, Lee SJ, Choi JA, Koh JY. Induction of lysosomal dilatation, arrested autophagy, and cell death by chloroquine in cultured ARPE-19 cells. Invest Ophthalmol Vis Sci 2010; 51:6030-7; http://dx.doi.org/ 10.1167/iovs.10-5278. [DOI] [PubMed] [Google Scholar]
- 160.Thomas G, Hall MN. TOR signalling and control of cell growth. Curr Opin Cell Biol 1997; 9:782-7; http://dx.doi.org/ 10.1016/S0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 161.G Juhasz. Interpretation of bafilomycin, pH neutralizing or protease inhibitor treatments in autophagic flux experiments: novel considerations. Autophagy 2012; 8:1875-6; http://dx.doi.org/ 10.4161/auto.21544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li M, Khambu B, Zhang H, Kang JH, Chen X, Chen D, Vollmer L, Liu PQ, Vogt A, Yin XM. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J Biol Chem 2013; 288:35769-80; http://dx.doi.org/ 10.1074/jbc.M113.511212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Seglen PO, Grinde B, Solheim AE. Inhibition of the lysosomal pathway of protein degradation in isolated rat hepatocytes by ammonia, methylamine, chloroquine and leupeptin. Eur J Biochem 1979; 95:215-25; http://dx.doi.org/ 10.1111/j.1432-1033.1979.tb12956.x. [DOI] [PubMed] [Google Scholar]
- 164.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 1991; 266:17707-12. [PubMed] [Google Scholar]
- 165.McLeland CB, Rodriguez J, Stern ST. Autophagy monitoring assay: qualitative analysis of MAP LC3-I to II conversion by immunoblot. Methods Mol Biol 2011; 697:199-206; http://dx.doi.org/ 10.1007/978-1-60327-198-1. [DOI] [PubMed] [Google Scholar]
- 166.Chakrama FZ, Seguin-Py S, Le Grand JN, Fraichard A, Delage-Mourroux R, Despouy G, Perez V, Jouvenot M, Boyer-Guittaut M. GABARAPL1 (GEC1) associates with autophagic vesicles. Auto-phagy 2010; 6:495-505; http://dx.doi.org/ 10.4161/auto.6.4.11819. [DOI] [PubMed] [Google Scholar]
- 167.Maynard S, Ghosh R, Wu Y, Yan S, Miyake T, Gagliardi M, Rethoret K, Bedard PA. GABARAP is a determinant of apoptosis in growth-arrested chicken embryo fibroblasts. J Cell Physiol 2015; 230:1475-88; http://dx.doi.org/ 10.1002/jcp.24889. [DOI] [PubMed] [Google Scholar]
- 168.Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol 2001; 152:51-64; http://dx.doi.org/ 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shu CW, Drag M, Bekes M, Zhai D, Salvesen GS, Reed JC. Synthetic substrates for measuring activity of autophagy proteases: autophagins (Atg4). Autophagy 2010; 6:936-47; http://dx.doi.org/ 10.4161/auto.6.7.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li M, Chen X, Ye Q-Z, Vogt A, Yin X-M. A High-throughput FRET-based Assay for Determination of Atg4 Activity. Autophagy 2012; 8:401-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ketteler R, Seed B. Quantitation of autophagy by luciferase release assay. Autophagy 2008; 4:801-6; http://dx.doi.org/ 10.4161/auto.6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin X-M. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem 2011; 286:7327-38; http://dx.doi.org/ 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Klionsky DJ. For the last time, it is GFP-Atg8, not Atg8-GFP (and the same goes for LC3). Autophagy 2011; 7:1093-4; http://dx.doi.org/ 10.4161/auto.7.10.15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 2005; 1:84-91; http://dx.doi.org/ 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 175.Castino R, Lazzeri G, Lenzi P, Bellio N, Follo C, Ferrucci M, Fornai F, Isidoro C. Suppression of autophagy precipitates neuronal cell death following low doses of methamphetamine. J Neurochem 2008; 106:1426-39; http://dx.doi.org/ 10.1111/j.1471-4159.2008.05488.x. [DOI] [PubMed] [Google Scholar]
- 176.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res 2007; 100:914-22; http://dx.doi.org/ 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 177.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20:5971-81; http://dx.doi.org/ 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hanson HH, Kang S, Fernandez-Monreal M, Oung T, Yildirim M, Lee R, Suyama K, Hazan RB, Phillips GR. LC3-dependent intracellular membrane tubules induced by gamma-protocadherins A3 and B2: a role for intraluminal interactions. J Biol Chem 2010; 285:20982-92; http://dx.doi.org/ 10.1074/jbc.M109.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 2011; 13:1335-43; http://dx.doi.org/ 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA 2011; 108:17396-401; http://dx.doi.org/ 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 181.Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, Fujita N, Saitoh T, Akira S, Yoshimori T, et al.. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 2014; 40:924-35; http://dx.doi.org/ 10.1016/j.immuni.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al.. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007; 450:1253-7; http://dx.doi.org/ 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 183.Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunol Rev 2009; 227:203-20; http://dx.doi.org/ 10.1111/j.1600-065X.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- 184.Ushio H, Ueno T, Kojima Y, Komatsu M, Tanaka S, Yamamoto A, Ichimura Y, Ezaki J, Nishida K, Komazawa-Sakon S, et al.. Crucial role for autophagy in degranulation of mast cells. J Allergy Clin Immunol 2011; 127:1267-76 e6; http://dx.doi.org/ 10.1016/j.jaci.2010.12.1078. [DOI] [PubMed] [Google Scholar]
- 185.Ishibashi K, Uemura T, Waguri S, Fukuda M. Atg16L1, an essential factor for canonical autophagy, participates in hormone secretion from PC12 cells independently of autophagic activity. Mol Biol Cell 2012; 23:3193-202; http://dx.doi.org/ 10.1091/mbc.E12-01-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, Virgin HW. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell 2011; 21:966-74; http://dx.doi.org/ 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan JL, Saitoh T, Akira S, Seglen PO, et al.. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J 2013; 32:3130-44; http://dx.doi.org/ 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 2009; 6:137-49; http://dx.doi.org/ 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 189.Cottam EM, Maier HJ, Manifava M, Vaux LC, Chandra-Schoenfelder P, Gerner W, Britton P, Ktistakis NT, Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 2011; 7:1335-47; http://dx.doi.org/ 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 2010; 7:500-8; http://dx.doi.org/ 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sharma M, Bhattacharyya S, Nain M, Kaur M, Sood V, Gupta V, Khasa R, Abdin MZ, Vrati S, Kalia M. Japanese encephalitis virus replication is negatively regulated by autophagy and occurs on LC3-I- and EDEM1-containing membranes. Autophagy 2014; 10:1637-51; http://dx.doi.org/ 10.4161/auto.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, et al.. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 2009; 10:480-7; http://dx.doi.org/ 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 2014; 15:239-47; http://dx.doi.org/ 10.1016/j.chom.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol 2010; 84:12110-24; http://dx.doi.org/ 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Alirezaei M, Flynn CT, Wood MR, Whitton JL. Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe 2012; 11:298-305; http://dx.doi.org/ 10.1016/j.chom.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Plowey ED, Cherra SJ 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem 2008; 105:1048-56; http://dx.doi.org/ 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nicotra G, Mercalli F, Peracchio C, Castino R, Follo C, Valente G, Isidoro C. Autophagy-active beclin-1 correlates with favourable clinical outcome in non-Hodgkin lymphomas. Modern pathology: Pathol 2010; 23:937-50; http://dx.doi.org/ 10.1038/modpathol.2010.80. [DOI] [PubMed] [Google Scholar]
- 198.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol 2008; 445:77-88; http://dx.doi.org/ 10.1007/978-1-59745-157-4. [DOI] [PubMed] [Google Scholar]
- 199.Gros F, Arnold J, Page N, Decossas M, Korganow AS, Martin T, Muller S. Macroautophagy is deregulated in murine and human lupus T lymphocytes. Autophagy 2012; 8:1113-23; http://dx.doi.org/ 10.4161/auto.20275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Welinder C, Ekblad L. Coomassie staining as loading control in Western blot analysis. J Proteome Res 2011; 10:1416-9; http://dx.doi.org/ 10.1021/pr1011476. [DOI] [PubMed] [Google Scholar]
- 201.Colella AD, Chegenii N, Tea MN, Gibbins IL, Williams KA, Chataway TK. Comparison of Stain-Free gels with traditional immunoblot loading control methodology. Anal Biochem 2012; 430:108-10; http://dx.doi.org/ 10.1016/j.ab.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 202.Ghosh R, Gilda JE, Gomes AV. The necessity of and strategies for improving confidence in the accuracy of western blots. Expert Rev Proteomics 2014; 11:549-60; http://dx.doi.org/ 10.1586/14789450.2014.939635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci USA 2005; 102:13807-12; http://dx.doi.org/ 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Russ DW, Boyd IM, McCoy KM, McCorkle KW. Muscle-specificity of age-related changes in markers of autophagy and sphingolipid metabolism. Biogerontology 2015; 16:747-59. [DOI] [PubMed] [Google Scholar]
- 205.Russ DW, Krause J, Wills A, Arreguin R. “SR stress” in mixed hindlimb muscles of aging male rats. Biogerontology 2012; 13:547-55; http://dx.doi.org/ 10.1007/s10522-012-9399-y. [DOI] [PubMed] [Google Scholar]
- 206.He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, et al.. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem 2003; 278:29278-87; http://dx.doi.org/ 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 207.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 2011; 12:226; http://dx.doi.org/ 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zois CE, Koukourakis MI. Radiation-induced autophagy in normal and cancer cells: towards novel cytoprotection and radio-sensitization policies? Autophagy 2009; 5:442-50; http://dx.doi.org/ 10.4161/auto.5.4.7667. [DOI] [PubMed] [Google Scholar]
- 209.Xin Y, Yu L, Chen Z, Zheng L, Fu Q, Jiang J, Zhang P, Gong R, Zhao S. Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics 2001; 74:408-13; http://dx.doi.org/ 10.1006/geno.2001.6555. [DOI] [PubMed] [Google Scholar]
- 210.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, et al.. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 2010; 11:45-51; http://dx.doi.org/ 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielmann Y, Stangler T, Hersch N, Hoffmann B, Merkel R, Willbold D. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy 2009; 5:690-8; http://dx.doi.org/ 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 212.Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: pitfalls of densitometry. Electrophoresis 2009; 30:1845-55; http://dx.doi.org/ 10.1002/elps.200800720. [DOI] [PubMed] [Google Scholar]
- 213.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000; 151:263-76; http://dx.doi.org/ 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 2010; 62:483-93; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04166.x. [DOI] [PubMed] [Google Scholar]
- 215.Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 2009; 149:220-34; http://dx.doi.org/ 10.1104/pp.108.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Engedal N, Torgersen ML, Guldvik IJ, Barfeld SJ, Bakula D, Saetre F, Hagen LK, Patterson JB, Proikas-Cezanne T, Seglen PO, et al.. Modulation of intracellular calcium homeostasis blocks autophagosome formation. Autophagy 2013; 9:1475-90; http://dx.doi.org/ 10.4161/auto.25900. [DOI] [PubMed] [Google Scholar]
- 217.Kovsan J, Bluher M, Tarnovscki T, Kloting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schon MR, et al.. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab 2011; 96:E268-77; http://dx.doi.org/ 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- 218.Gao Z, Gammoh N, Wong PM, Erdjument-Bromage H, Tempst P, Jiang X. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy 2010; 6:126-37; http://dx.doi.org/ 10.4161/auto.6.1.10928. [DOI] [PubMed] [Google Scholar]
- 219.King JS, Veltman DM, Insall RH. The induction of autophagy by mechanical stress. Autophagy 2011; 7:1490-9; http://dx.doi.org/ 10.4161/auto.7.12.17924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Roberts R, Al-Jamal WT, Whelband M, Thomas P, Jefferson M, van den Bossche J, Powell PP, Kostarelos K, Wileman T. Autophagy and formation of tubulovesicular autophagosomes provide a barrier against nonviral gene delivery. Autophagy 2013; 9:667-82; http://dx.doi.org/ 10.4161/auto.23877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Schmidt RS, Butikofer P. Autophagy in Trypanosoma brucei: amino acid requirement and regulation during different growth phases. PloS One 2014; 9:e93875; http://dx.doi.org/ 10.1371/journal.pone.0093875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bernard M, Dieude M, Yang B, Hamelin K, Underwood K, Hebert MJ. Autophagy fosters myofibroblast differentiation through MTORC2 activation and downstream upregulation of CTGF. Autophagy 2014; 10:2193-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Saetre F, Hagen LK, Engedal N, Seglen PO. Novel steps in the autophagic-lysosomal pathway. FEBS J 2015; 282:2202-14; http://dx.doi.org/ 10.1111/febs.13268. [DOI] [PubMed] [Google Scholar]
- 224.Ju JS, Varadhachary AS, Miller SE, Weihl CC. Quantitation of “autophagic flux” in mature skeletal muscle. Autophagy 2010; 6:929-35; http://dx.doi.org/ 10.4161/auto.6.7.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, et al.. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol 2008; 183:101-16; http://dx.doi.org/ 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mauvezin C, Nagy P, Juhasz G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun 2015; 6:7007; http://dx.doi.org/ 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol 2010; 11:89; http://dx.doi.org/ 10.1186/1471-2121-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gonzalez-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquere S, Eskelinen EL, Pierron G, Saftig P, Kroemer G. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci 2005; 118: 3091-102; http://dx.doi.org/ 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- 229.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant {alpha}-synuclein by chaperone-mediated autophagy. Science 2004; 305:1292-5; http://dx.doi.org/ 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 230.Trincheri NF, Follo C, Nicotra G, Peracchio C, Castino R, Isidoro C. Resveratrol-induced apoptosis depends on the lipid kinase activity of Vps34 and on the formation of autophagolysosomes. Carcinogenesis 2008; 29:381-9; http://dx.doi.org/ 10.1093/carcin/bgm271. [DOI] [PubMed] [Google Scholar]
- 231.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, Klionsky DJ. In search of an “autophagomometer”. Autophagy 2009; 5:585-9; http://dx.doi.org/ 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 232.Sarkar S, Ravikumar B, Rubinsztein DC. Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods Enzymol 2009; 453:83-110; http://dx.doi.org/ 10.1016/S0076-6879(08)04005-6. [DOI] [PubMed] [Google Scholar]
- 233.Sarkar S, Korolchuk V, Renna M, Winslow A, Rubinsztein DC. Methodological considerations for assessing autophagy modulators: a study with calcium phosphate precipitates. Autophagy 2009; 5:307-13; http://dx.doi.org/ 10.4161/auto.5.3.7664. [DOI] [PubMed] [Google Scholar]
- 234.Martins WK, Severino D, Souza C, Stolf BS, Baptista MS. Rapid screening of potential autophagic inductor agents using mammalian cell lines. Biotechnol J 2013; 8:730-7; http://dx.doi.org/ 10.1002/biot.201200306. [DOI] [PubMed] [Google Scholar]
- 235.Martins WK, Costa ET, Cruz MC, Stolf BS, Miotto R, Cordeiro RM, Baptista MS. Parallel damage in mitochondrial and lysosomal compartments promotes efficient cell death with autophagy: The case of the pentacyclic triterpenoids. Sci Rep 2015; 5:12425; http://dx.doi.org/ 10.1038/srep12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem 2004; 279:29889-94; http://dx.doi.org/ 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Karim MR, Kanazawa T, Daigaku Y, Fujimura S, Miotto G, Kadowaki M. Cytosolic LC3 ratio as a sensitive index of macroautophagy in isolated rat hepatocytes and H4-II-E cells. Autophagy 2007; 3:553-60; http://dx.doi.org/ 10.4161/auto.4615. [DOI] [PubMed] [Google Scholar]
- 238.Kim CH, Kim KH, Yoo YM. Melatonin protects against apoptotic and autophagic cell death in C2C12 murine myoblast cells. J Pineal Res 2011; 50:241-9; http://dx.doi.org/ 10.1111/j.1600-079X.2010.00833.x. [DOI] [PubMed] [Google Scholar]
- 239.Karim MR, Kanazawa T, Daigaku Y, Fujimura S, Miotto G, Kadowaki M. Cytosolic LC3 ratio as a sensitive index of macroautophagy in isolated rat hepatocytes and H4-II-E cells. Autophagy 2007; 3:553-60. [DOI] [PubMed] [Google Scholar]
- 240.Tsvetkov AS, Arrasate M, Barmada S, Ando DM, Sharma P, Shaby BA, Finkbeiner S. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol 2013; 9:586-92; http://dx.doi.org/ 10.1038/nchembio.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Loos B, du Toit A, Hofmeyr JH. Defining and measuring autophagosome flux-concept and reality. Autophagy 2014:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Farkas T, Hoyer-Hansen M, Jaattela M. Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux. Autophagy 2009; 5:1018-25; http://dx.doi.org/ 10.4161/auto.5.7.9443. [DOI] [PubMed] [Google Scholar]
- 243.Frankel LB, Wen J, Lees M, H(o)yer-Hansen M, Farkas T, Krogh A, Jaattela M, Lund AH. microRNA-101 is a potent inhibitor of autophagy. EMBO J 2011:4628-41; http://dx.doi.org/ 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Farkas T, Daugaard M, Jaattela M. Identification of small molecule inhibitors of phosphatidylinositol 3-kinase and autophagy. J Biol Chem 2011; 286:38904-12; http://dx.doi.org/ 10.1074/jbc.M111.269134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Szyniarowski P, Corcelle-Termeau E, Farkas T, Hoyer-Hansen M, Nylandsted J, Kallunki T, Jaattela M. A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy 2011; 7:892-903; http://dx.doi.org/ 10.4161/auto.7.8.15770. [DOI] [PubMed] [Google Scholar]
- 246.Nguyen HT, Dalmasso G, Muller S, Carriere J, Seibold F, Darfeuille-Michaud A. Crohn's disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology 2014; 146:508-19; http://dx.doi.org/ 10.1053/j.gastro.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 247.Frankel LB, Di Malta C, Wen J, Eskelinen EL, Ballabio A, Lund AH. A non-conserved miRNA regulates lysosomal function and impacts on a human lysosomal storage disorder. Nat Commun 2014; 5:5840; http://dx.doi.org/ 10.1038/ncomms6840. [DOI] [PubMed] [Google Scholar]
- 248.Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis 2012; 33:2018-25; http://dx.doi.org/ 10.1093/carcin/bgs266. [DOI] [PubMed] [Google Scholar]
- 249.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem 2006; 281:4035-41; http://dx.doi.org/ 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 250.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008; 183:795-803; http://dx.doi.org/ 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Nogalska A, Terracciano C, D'Agostino C, King Engel W, Askanas V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol 2009; 118:407-13; http://dx.doi.org/ 10.1007/s00401-009-0564-6. [DOI] [PubMed] [Google Scholar]
- 252.Chahory S, Keller N, Martin E, Omri B, Crisanti P, Torriglia A. Light induced retinal degeneration activates a caspase-independent pathway involving cathepsin D. Neurochem Int 2010; 57:278-87; http://dx.doi.org/ 10.1016/j.neuint.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 253.Padron-Barthe L, Courta J, Lepretre C, Nagbou A, Torriglia A. Leukocyte Elastase Inhibitor, the precursor of L-DNase II, inhibits apoptosis by interfering with caspase-8 activation. Biochim Biophys Acta 2008; 1783:1755-66; http://dx.doi.org/ 10.1016/j.bbamcr.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 254.Gutierrez MG, Saka HA, Chinen I, Zoppino FC, Yoshimori T, Bocco JL, Colombo MI. Protective role of autophagy against Vibrio cholerae cytolysin, a pore-forming toxin from V. cholerae. Proc Natl Acad Sci USA 2007; 104:1829-34; http://dx.doi.org/ 10.1073/pnas.0601437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett 2006; 580:2623-9; http://dx.doi.org/ 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 256.Suttangkakul A, Li F, Chung T, Vierstra RD. The ATG1/13 protein kinase complex is both a regulator and a substrate of autophagic recycling in Arabidopsis. Plant Cell 2011; 23: 3761-79; http://dx.doi.org/ 10.1105/tpc.111.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy 2011; 7:188-204; http://dx.doi.org/ 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PloS One 2009; 4:e7124; http://dx.doi.org/ 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Patterson GH, Lippincott-Schwartz J. Selective photolabeling of proteins using photoactivatable GFP. Methods 2004; 32:445-50; http://dx.doi.org/ 10.1016/j.ymeth.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 260.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem 2006; 281:29776-87; http://dx.doi.org/ 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 261.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol 2008; 451:33-42; http://dx.doi.org/ 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 262.DJ Klionsky. Monitoring autophagy in yeast: the Pho8Delta60 assay. Methods Mol Biol 2007; 390:363-71; http://dx.doi.org/ 10.1007/978-1-59745-466-7. [DOI] [PubMed] [Google Scholar]
- 263.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J 1997; 73:2782-90; http://dx.doi.org/ 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3:452-60; http://dx.doi.org/ 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 265.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 2004; 22:1567-72; http://dx.doi.org/ 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 266.Strack RL, Keenan RJ, Glick BS. Noncytotoxic DsRed derivatives for whole-cell labeling. Methods Mol Biol 2011; 699:355-70; http://dx.doi.org/ 10.1007/978-1-61737-950-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 2006; 24:461-5; http://dx.doi.org/ 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 268.Rekas A, Alattia JR, Nagai T, Miyawaki A, Ikura M. Crystal structure of venus, a yellow fluorescent protein with improved maturation and reduced environmental sensitivity. J Biol Chem 2002; 277:50573-8; http://dx.doi.org/ 10.1074/jbc.M209524200. [DOI] [PubMed] [Google Scholar]
- 269.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; http://dx.doi.org/ 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Badr CE, Wurdinger T, Nilsson J, Niers JM, Whalen M, Degterev A, Tannous BA. Lanatoside C sensitizes glioblastoma cells to tumor necrosis factor-related apoptosis-inducing ligand and induces an alternative cell death pathway. Neuro-oncology 2011; 13:1213-24; http://dx.doi.org/ 10.1093/neuonc/nor067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Meléndez A, Tallóczy Z, Seaman M, Eskelinen E-L, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003; 301:1387-91; http://dx.doi.org/ 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 272.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem 2003; 278:17636-45; http://dx.doi.org/ 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 273.Liu XH, Liu TB, Lin FC. Monitoring autophagy in Magnaporthe oryzae. Methods Enzymol 2008; 451:271-94; http://dx.doi.org/ 10.1016/S0076-6879(08)03219-9. [DOI] [PubMed] [Google Scholar]
- 274.Pinan-Lucarre B, Paoletti M, Dementhon K, Coulary-Salin B, Clave C. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol Microbiol 2003; 47:321-33; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03208.x. [DOI] [PubMed] [Google Scholar]
- 275.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006; 312:580-3; http://dx.doi.org/ 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 276.Kikuma T, Ohneda M, Arioka M, Kitamoto K. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot Cell 2006; 5:1328-36; http://dx.doi.org/ 10.1128/EC.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Nolting N, Bernhards Y, Poggeler S. SmATG7 is required for viability in the homothallic ascomycete Sordaria macrospora. Fungal Genet Biol 2009; 46:531-42; http://dx.doi.org/ 10.1016/j.fgb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 278.Baghdiguian S, Martinand-Mari C, Mangeat P. Using Ciona to study developmental programmed cell death. Semin Cancer Biol 2007; 17:147-53; http://dx.doi.org/ 10.1016/j.semcancer.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 279.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell 2004; 7:179-92; http://dx.doi.org/ 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 280.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7:167-78; http://dx.doi.org/ 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 281.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 2009; 19:1741-6; http://dx.doi.org/ 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004; 16:2967-83; http://dx.doi.org/ 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Li F, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 2015; 27:1389-408; http://dx.doi.org/ 10.1105/tpc.15.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Brennand A, Rico E, Rigden DJ, Van Der Smissen P, Courtoy PJ, Michels PA. ATG24 Represses Autophagy and Differentiation and Is Essential for Homeostasy of the Flagellar Pocket in Trypanosoma brucei. PloS One 2015; 10:e0130365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Li FJ, Shen Q, Wang C, Sun Y, Yuan AY, He CY. A role of autophagy in Trypanosoma brucei cell death. Cell Microbiol 2012; 14:1242-56; http://dx.doi.org/ 10.1111/j.1462-5822.2012.01795.x. [DOI] [PubMed] [Google Scholar]
- 286.Besteiro S, Williams RA, Morrison LS, Coombs GH, Mottram JC. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J Biol Chem 2006; 281:11384-96; http://dx.doi.org/ 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- 287.Williams RA, Tetley L, Mottram JC, Coombs GH. Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol Microbiol 2006; 61:655-74; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05274.x. [DOI] [PubMed] [Google Scholar]
- 288.Williams RA, Woods KL, Juliano L, Mottram JC, Coombs GH Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy 2009; 5:159-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Elsasser A, Vogt AM, Nef H, Kostin S, Mollmann H, Skwara W, Bode C, Hamm C, Schaper J. Human hibernating myocardium is jeopardized by apoptotic and autophagic cell death. J Am Coll Cardiol 2004; 43:2191-9; http://dx.doi.org/ 10.1016/j.jacc.2004.02.053. [DOI] [PubMed] [Google Scholar]
- 290.Knaapen MW, Davies MJ, De Bie M, Haven AJ, Martinet W, Kockx MM. Apoptotic versus autophagic cell death in heart failure. Cardiovasc Res 2001; 51:304-12; http://dx.doi.org/ 10.1016/S0008-6363(01)00290-5. [DOI] [PubMed] [Google Scholar]
- 291.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, et al.. Myocytes die by multiple mechanisms in failing human hearts. Circ Res 2003; 92:715-24; http://dx.doi.org/ 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 292.Perez-Perez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 2010; 152:1874-88; http://dx.doi.org/ 10.1104/pp.109.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, Gotow T, Peters C, von Figura K, Mizushima N, et al.. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Amer J Pathol 2005; 167:1713-28; http://dx.doi.org/ 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.O]st A, Svensson K, Ruishalme I, Brannmark C, Franck N, Krook H, Sandstrom P, Kjolhede P, Stralfors P. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med 2010; 16:235-46; http://dx.doi.org/ 10.1007/s00894-009-0539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ 3rd, et al.. Endogenous HMGB1 regulates autophagy. J Cell Biol 2010; 190:881-92; http://dx.doi.org/ 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gniadek TJ, Warren G. WatershedCounting3D: a new method for segmenting and counting punctate structures from confocal image data. Traffic 2007; 8:339-46; http://dx.doi.org/ 10.1111/j.1600-0854.2007.00538.x. [DOI] [PubMed] [Google Scholar]
- 297.Decuypere J-P, Welkenhuyzen K, Luyten Y, Ponsaerts R, Dewaele M, Molgó J, Agostinis P, Missiaen L, De Smedt H, Parys JB, et al.. IP3 receptor-mediated Ca2+ signaling and autophagy induction are interrelated. Autophagy 2011; 7:1472-89; http://dx.doi.org/ 10.4161/auto.7.12.17909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Xu Y, Yuan J, Lipinski MM. Live imaging and single-cell analysis reveal differential dynamics of autophagy and apoptosis. Autophagy 2013; 9:1418-30; http://dx.doi.org/ 10.4161/auto.25080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Amer AO, Swanson MS. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell Microbiol 2005; 7:765-78; http://dx.doi.org/ 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753-66; http://dx.doi.org/ 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 301.Ogawa M, Sasakawa C. Intracellular survival of Shigella. Cell Microbiol 2006; 8:177-84; http://dx.doi.org/ 10.1111/j.1462-5822.2005.00652.x. [DOI] [PubMed] [Google Scholar]
- 302.Reyes L, Eiler-McManis E, Rodrigues PH, Chadda AS, Wallet SM, Belanger M, Barrett AG, Alvarez S, Akin D, Dunn WA Jr., et al.. Deletion of lipoprotein PG0717 in Porphyromonas gingivalis W83 reduces gingipain activity and alters trafficking in and response by host cells. PloS One 2013; 8:e74230; http://dx.doi.org/ 10.1371/journal.pone.0074230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 2011; 27:1179-80; http://dx.doi.org/ 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science 2005; 310:310-4; http://dx.doi.org/ 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 305.Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol 2008; 182:129-40; http://dx.doi.org/ 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, et al.. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell 2010; 18:1041-52; http://dx.doi.org/ 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Brady NR, Hamacher-Brady A, Yuan H, Gottlieb RA The autophagic response to nutrient deprivation in the HL-1 cardiac myocyte is modulated by Bcl-2 and sarco/endoplasmic reticulum calcium stores. FEBS J 2007; 274:3184-97; http://dx.doi.org/ 10.1111/j.1742-4658.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 308.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Tr 2008; 112:389-403; http://dx.doi.org/ 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 309.Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T, Shen Y, Rameh L, Yankner B, Tsai LH, et al.. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell 2010; 38:500-11; http://dx.doi.org/ 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Dolloff NG, Ma X, Dicker DT, Humphreys RC, Li LZ, El-Deiry WS. Spectral imaging-based methods for quantifying autophagy and apoptosis. Cancer Biol Ther 2011; 12:349-56; http://dx.doi.org/ 10.4161/cbt.12.4.17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007; 315:1398-401; http://dx.doi.org/ 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 312.Phadwal K, Alegre-Abarrategui J, Watson AS, Pike L, Anbalagan S, Hammond EM, Wade-Martins R, McMichael A, Klenerman P, Simon AK. A novel method for autophagy detection in primary cells: Impaired levels of macroautophagy in immunosenescent T cells. Autophagy 2012; 8:677-89; http://dx.doi.org/ 10.4161/auto.18935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Davey HM, Hexley P. Red but not dead? Membranes of stressed Saccharomyces cerevisiae are permeable to propidium iodide. Environ Microbiol 2011; 13:163-71; http://dx.doi.org/ 10.1111/j.1462-2920.2010.02317.x. [DOI] [PubMed] [Google Scholar]
- 314.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 2007; 3:323-8; http://dx.doi.org/ 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 315.Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, Bazett-Jones DP, Brumell JH. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2006; 2:189-99; http://dx.doi.org/ 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 316.Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH. Ubiquitinated-protein aggregates form in pancreatic [beta]-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes 2007; 56:930-9. [DOI] [PubMed] [Google Scholar]
- 317.Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci USA 2011; 108:1427-32; http://dx.doi.org/ 10.1073/pnas.1014156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Pierre P. Dendritic cells, DRiPs, and DALIS in the control of antigen processing. Immunol Rev 2005; 207:184-90; http://dx.doi.org/ 10.1111/j.0105-2896.2005.00300.x. [DOI] [PubMed] [Google Scholar]
- 319.Pankiv S, Høyvarde Clausen T, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; http://dx.doi.org/ 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 320.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al.. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885-9; http://dx.doi.org/ 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 321.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al.. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880-4; http://dx.doi.org/ 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 322.Calvo-Garrido J, Escalante R. Autophagy dysfunction and ubiquitin-positive protein aggregates in Dictyostelium cells lacking Vmp1. Autophagy 2010; 6:100-9; http://dx.doi.org/ 10.4161/auto.6.1.10697. [DOI] [PubMed] [Google Scholar]
- 323.Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Auto-phagy 2006; 2:138-9; http://dx.doi.org/ 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 324.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 Phosphorylation of p62/SQSTM1 Regulates Selective Autophagic Clearance of Ubiquitinated Proteins. Mol Cell 2011; 44:279-89; http://dx.doi.org/ 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 325.Lerner C, Bitto A, Pulliam D, Nacarelli T, Konigsberg M, Van Remmen H, Torres C, Sell C. Reduced mammalian target of rapamycin activity facilitates mitochondrial retrograde signaling and increases life span in normal human fibroblasts. Aging Cell 2013; 12:966-77; http://dx.doi.org/ 10.1111/acel.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Köchl R, Hu XW, EYW Chan, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006; 7:129-45; http://dx.doi.org/ 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 327.Eng KE, Panas MD, Karlsson Hedestam GB, McInerney GM. A novel quantitative flow cytometry-based assay for autophagy. Autophagy 2010; 6:634-41; http://dx.doi.org/ 10.4161/auto.6.5.12112. [DOI] [PubMed] [Google Scholar]
- 328.Ciechomska IA, Tolkovsky AM. Non-autophagic GFP-LC3 puncta induced by saponin and other detergents. Autophagy 2007; 3:586-90; http://dx.doi.org/ 10.4161/auto.4843. [DOI] [PubMed] [Google Scholar]
- 329.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA 1982; 79:1889-92; http://dx.doi.org/ 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen H-M. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem 2010; 285:10850-61; http://dx.doi.org/ 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 2005; 1:23-36; http://dx.doi.org/ 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- 332.Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E, Calvo TG, Larribere L, Megias D, Mulero F, et al.. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell 2009; 16:103-14; http://dx.doi.org/ 10.1016/j.ccr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 2010; 32:227-39; http://dx.doi.org/ 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Tamura N, Oku M, Sakai Y. Atg8 regulates vacuolar membrane dynamics in a lipidation-independent manner in Pichia pastoris. J Cell Sci 2010; 123:4107-16; http://dx.doi.org/ 10.1242/jcs.070045. [DOI] [PubMed] [Google Scholar]
- 335.Stromhaug PE, Reggiori F, Guan J, Wang C-W, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell 2004; 15:3553-66; http://dx.doi.org/ 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Baens M, Noels H, Broeckx V, Hagens S, Fevery S, Billiau AD, Vankelecom H, Marynen P. The dark side of EGFP: defective polyubiquitination. PloS One 2006; 1:e54; http://dx.doi.org/ 10.1371/journal.pone.0000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Cali T, Galli C, Olivari S, Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem Biophys Res Commun 2008; 371:405-10; http://dx.doi.org/ 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- 338.Al-Younes HM, Al-Zeer MA, Khalil H, Gussmann J, Karlas A, Machuy N, Brinkmann V, Braun PR, Meyer TF. Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy 2011; 7:814-28; http://dx.doi.org/ 10.4161/auto.7.8.15597. [DOI] [PubMed] [Google Scholar]
- 339.Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, Betzig E. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci USA 2007; 104:20308-13; http://dx.doi.org/ 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al.. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009; 136:521-34; http://dx.doi.org/ 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Nyfeler B, Bergman P, Triantafellow E, Wilson CJ, Zhu Y, Radetich B, Finan PM, Klionsky DJ, Murphy LO. Relieving autophagy and 4EBP1 from rapamycin resistance. Mol Cell Biol 2011; 31:2867-76; http://dx.doi.org/ 10.1128/MCB.05430-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Singh K, Sharma A, Mir MC, Drazba JA, Heston WD, Magi-Galluzzi C, Hansel D, Rubin BP, Klein EA, Almasan A. Autophagic flux determines cell death and survival in response to Apo2L/TRAIL (dulanermin). Mol Cancer 2014; 13:70; http://dx.doi.org/ 10.1186/1476-4598-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Cherra SJ III, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol 2010; 190:533-9; http://dx.doi.org/ 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Sarkar S, Korolchuk VI, Renna M, Imarisio S, Fleming A, Williams A, Garcia-Arencibia M, Rose C, Luo S, Underwood BR, et al.. Complex inhibitory effects of nitric oxide on autophagy. Mol Cell 2011; 43:19-32; http://dx.doi.org/ 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Nazarko TY, Ozeki K, Till A, Ramakrishnan G, Lotfi P, Yan M, Subramani S. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol 2014; 204:541-57; http://dx.doi.org/ 10.1083/jcb.201307050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG, Siddiqui A. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci USA 2014; 111:6413-8; http://dx.doi.org/ 10.1073/pnas.1321114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep 2013; 14:1127-35; http://dx.doi.org/ 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Rosado CJ, Mijaljica D, Hatzinisiriou I, Prescott M, Devenish RJ. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 2008; 4:205-13; http://dx.doi.org/ 10.4161/auto.5331. [DOI] [PubMed] [Google Scholar]
- 349.Mijaljica D, Rosado CJ, Devenish RJ, Prescott M. Biosensors for monitoring autophagy In: Serra PA, ed. Biosensors-Emerging Materials and Applications Croatia: InTech, 2011:383-400. [Google Scholar]
- 350.Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ 2007; 14:1647-56; http://dx.doi.org/ 10.1038/sj.cdd.4402167. [DOI] [PubMed] [Google Scholar]
- 351.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev 2010; 90:1103-63; http://dx.doi.org/ 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 352.Zhou C, Zhong W, Zhou J, Sheng F, Fang Z, Wei Y, Chen Y, Deng X, Xia B, Lin J. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy 2012; 8:1215-26; http://dx.doi.org/ 10.4161/auto.20284. [DOI] [PubMed] [Google Scholar]
- 353.Zhou J, Lin J, Zhou C, Deng X, Xia B. Cytotoxicity of red fluorescent protein DsRed is associated with the suppression of Bcl-xL translation. FEBS Lett 2011; 585:821-7; http://dx.doi.org/ 10.1016/j.febslet.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 354.Wen Y, Zand B, Ozpolat B, Szczepanski MJ, Lu C, Yuca E, Carroll AR, Alpay N, Bartholomeusz C, Tekedereli I, et al.. Antagonism of tumoral prolactin receptor promotes autophagy-related cell death. Cell Rep 2014; 7:488-500; http://dx.doi.org/ 10.1016/j.celrep.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Loos B, Genade S, Ellis B, Lochner A, Engelbrecht AM. At the core of survival: autophagy delays the onset of both apoptotic and necrotic cell death in a model of ischemic cell injury. Exp Cell Res 2011; 317:1437-53; http://dx.doi.org/ 10.1016/j.yexcr.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 356.Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic Control of Autophagy. Cell 2014; 159:1263-76; http://dx.doi.org/ 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Loos B, Engelbrecht AM, Lockshin RA, Klionsky DJ, Zakeri Z. The variability of autophagy and cell death susceptibility: Unanswered questions. Autophagy 2013; 9:1270-85; http://dx.doi.org/ 10.4161/auto.25560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy 2008; 4:621-8; http://dx.doi.org/ 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 359.Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol 2011; 9:38; http://dx.doi.org/ 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.de la Calle C, Joubert PE, Law HK, Hasan M, Albert ML. Simultaneous assessment of autophagy and apoptosis using multispectral imaging cytometry. Autophagy 2011; 7:1045-51; http://dx.doi.org/ 10.4161/auto.7.9.16252. [DOI] [PubMed] [Google Scholar]
- 361.Degtyarev M, Reichelt M, Lin K. Novel quantitative autophagy analysis by organelle flow cytometry after cell sonication. PloS One 2014; 9:e87707; http://dx.doi.org/ 10.1371/journal.pone.0087707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, et al.. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 2009; 6:367-80; http://dx.doi.org/ 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Kaminskyy V, Abdi A, Zhivotovsky B. A quantitative assay for the monitoring of autophagosome accumulation in different phases of the cell cycle. Autophagy 2011; 7:83-90; http://dx.doi.org/ 10.4161/auto.7.1.13893. [DOI] [PubMed] [Google Scholar]
- 364.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al.. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505-16; http://dx.doi.org/ 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 365.Larsen KB, Lamark T, Overvatn A, Harneshaug I, Johansen T, Bjorkoy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy 2010; 6:784-93; http://dx.doi.org/ 10.4161/auto.6.6.12510. [DOI] [PubMed] [Google Scholar]
- 366.Huang JJ, Li HR, Huang Y, Jiang WQ, Xu RH, Huang HQ, Lv Y, Xia ZJ, Zhu XF, Lin TY, et al.. Beclin 1 expression: a predictor of prognosis in patients with extranodal natural killer T-cell lymphoma, nasal type. Autophagy 2010; 6:777-83; http://dx.doi.org/ 10.4161/auto.6.6.12784. [DOI] [PubMed] [Google Scholar]
- 367.Sivridis E, Koukourakis MI, Zois CE, Ledaki I, Ferguson DJ, Harris AL, Gatter KC, Giatromanolaki A. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Amer J Pathol 2010; 176:2477-89; http://dx.doi.org/ 10.2353/ajpath.2010.090049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Sivridis E, Giatromanolaki A, Liberis V, Koukourakis MI. Auto-phagy in endometrial carcinomas and prognostic relevance of ‘stone-like’ structures (SLS): what is destined for the atypical endometrial hyperplasia? Autophagy 2011; 7:74-82; http://dx.doi.org/ 10.4161/auto.7.1.13947. [DOI] [PubMed] [Google Scholar]
- 369.Giatromanolaki A, Koukourakis MI, Koutsopoulos A, Chloropoulou P, Liberis V, Sivridis E. High Beclin 1 expression defines a poor prognosis in endometrial adenocarcinomas. Gynecol Oncol 2011; 123:147-51; http://dx.doi.org/ 10.1016/j.ygyno.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 370.Chen Y, Lu Y, Lu C, Zhang L. Beclin-1 expression is a predictor of clinical outcome in patients with esophageal squamous cell carcinoma and correlated to hypoxia-inducible factor (HIF)-1alpha expression. Pathol Oncol Res 2009; 15:487-93; http://dx.doi.org/ 10.1007/s12253-008-9143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wan XB, Fan XJ, Chen MY, Xiang J, Huang PY, Guo L, Wu XY, Xu J, Long ZJ, Zhao Y, et al.. Elevated Beclin 1 expression is correlated with HIF-1[a] in predicting poor prognosis of nasopharyngeal carcinoma. Autophagy 2010; 6:395-404; http://dx.doi.org/ 10.4161/auto.6.3.11303. [DOI] [PubMed] [Google Scholar]
- 372.Sakakura K, Takahashi H, Kaira K, Toyoda M, Oyama T, Chikamatsu K. Immunological significance of the accumulation of autophagy components in oral squamous cell carcinoma. Cancer Sci 2015; 106:1-8; http://dx.doi.org/ 10.1111/cas.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Shi YH, Ding ZB, Zhou J, Qiu SJ, Fan J. Prognostic significance of Beclin 1-dependent apoptotic activity in hepatocellular carcinoma. Autophagy 2009; 5:380-2; http://dx.doi.org/ 10.4161/auto.5.3.7658. [DOI] [PubMed] [Google Scholar]
- 374.Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, Shi GM, Wang XY, Ke AW, Wu B, et al.. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res 2008; 68:9167-75; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 375.Pirtoli L, Cevenini G, Tini P, Vannini M, Oliveri G, Marsili S, Mourmouras V, Rubino G, Miracco C. The prognostic role of Beclin 1 protein expression in high-grade gliomas. Autophagy 2009; 5:930-6; http://dx.doi.org/ 10.4161/auto.5.7.9227. [DOI] [PubMed] [Google Scholar]
- 376.Karpathiou G, Sivridis E, Koukourakis MI, Mikroulis D, Bouros D, Froudarakis ME, Giatromanolaki A. Light-chain 3A autophagic activity and prognostic significance in non-small cell lung carcinomas. Chest 2011; 140:127-34; http://dx.doi.org/ 10.1378/chest.10-1831. [DOI] [PubMed] [Google Scholar]
- 377.Fujii S, Mitsunaga S, Yamazaki M, Hasebe T, Ishii G, Kojima M, Kinoshita T, Ueno T, Esumi H, Ochiai A. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci 2008; 99:1813-9; http://dx.doi.org/ 10.1111/j.1349-7006.2008.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Li BX, Li CY, Peng RQ, Wu XJ, Wang HY, Wan DS, Zhu XF, Zhang XS. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy 2009; 5:303-6; http://dx.doi.org/ 10.4161/auto.5.3.7491. [DOI] [PubMed] [Google Scholar]
- 379.Koukourakis MI, Giatromanolaki A, Sivridis E, Pitiakoudis M, Gatter KC, Harris AL. Beclin 1 over- and underexpression in colorectal cancer: distinct patterns relate to prognosis and tumour hypoxia. Brit J Cancer 2010; 103:1209-14; http://dx.doi.org/ 10.1038/sj.bjc.6605904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Giatromanolaki A, Koukourakis MI, Harris AL, Polychronidis A, Gatter KC, Sivridis E. Prognostic relevance of light chain 3 (LC3A) autophagy patterns in colorectal adenocarcinomas. J Clin Pathol 2010; 63:867-72; http://dx.doi.org/ 10.1136/jcp.2010.079525. [DOI] [PubMed] [Google Scholar]
- 381.Sivridis E, Koukourakis MI, Mendrinos SE, Karpouzis A, Fiska A, Kouskoukis C, Giatromanolaki A. Beclin-1 and LC3A expression in cutaneous malignant melanomas: a biphasic survival pattern for beclin-1. Melanoma Res 2011; 21:188-95; http://dx.doi.org/ 10.1097/CMR.0b013e328346612c. [DOI] [PubMed] [Google Scholar]
- 382.Giatromanolaki AN, Charitoudis G St, Bechrakis NE, Kozobolis VP, Koukourakis MI, Foerster MH, Sivridis EL. Autophagy patterns and prognosis in uveal melanomas. Modern Pathol 2011; 24:1036-45; http://dx.doi.org/ 10.1038/modpathol.2011.63. [DOI] [PubMed] [Google Scholar]
- 383.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005; 97:1180-4; http://dx.doi.org/ 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 384.Kuwahara Y, Oikawa T, Ochiai Y, Roudkenar MH, Fukumoto M, Shimura T, Ohtake Y, Ohkubo Y, Mori S, Uchiyama Y. Enhancement of autophagy is a potential modality for tumors refractory to radiotherapy. Cell Death Dis 2011; 2:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Hou YJ, Dong LW, Tan YX, Yang GZ, Pan YF, Li Z, Tang L, Wang M, Wang Q, Wang HY. Inhibition of active autophagy induces apoptosis and increases chemosensitivity in cholangiocarcinoma. Lab Invest 2011; 91:1146-57; http://dx.doi.org/ 10.1038/labinvest.2011.97. [DOI] [PubMed] [Google Scholar]
- 386.O'Donovan TR, O'Sullivan GC, McKenna SL. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy 2011; 7:509-24; http://dx.doi.org/ 10.4161/auto.7.5.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Yoshimura K, Shibata M, Koike M, Gotoh K, Fukaya M, Watanabe M, Uchiyama Y. Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy 2006; 2:200-8; http://dx.doi.org/ 10.4161/auto.2744. [DOI] [PubMed] [Google Scholar]
- 388.Tamura H, Shibata M, Koike M, Sasaki M, Uchiyama Y. Atg9A protein, an autophagy-related membrane protein, is localized in the neurons of mouse brains. J Histochem Cytochem 2010; 58:443-53; http://dx.doi.org/ 10.1369/jhc.2010.955690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Cui J, Bai XY, Shi S, Cui S, Hong Q, Cai G, Chen X. Age-related changes in the function of autophagy in rat kidneys. Age 2011; 10.1007/s11357-011-9237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Marinelli S, Nazio F, Tinari A, Ciarlo L, D'Amelio M, Pieroni L, Vacca V, Urbani A, Cecconi F, Malorni W, et al.. Schwann cell autophagy counteracts the onset and chronification of neuropathic pain. Pain 2014; 155:93-107; http://dx.doi.org/ 10.1016/j.pain.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 391.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, et al.. Paneth cells as a site of origin for intestinal inflammation. Nature 2013; 503:272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Thachil E, Hugot JP, Arbeille B, Paris R, Grodet A, Peuchmaur M, Codogno P, Barreau F, Ogier-Denis E, Berrebi D, et al.. Abnormal activation of autophagy-induced crinophagy in Paneth cells from patients with Crohn's disease. Gastroenterology 2012; 142:1097-9 e4; http://dx.doi.org/ 10.1053/j.gastro.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 393.Mellén MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ 2008; 15:1279-90; http://dx.doi.org/ 10.1038/cdd.2008.40. [DOI] [PubMed] [Google Scholar]
- 394.Mellén MA, de la Rosa EJ, Boya P. Autophagy is not universally required for phosphatidyl-serine exposure and apoptotic cell engulfment during neural development. Autophagy 2009; 5:964-72; http://dx.doi.org/ 10.4161/auto.5.7.9292. [DOI] [PubMed] [Google Scholar]
- 395.Aburto MR, Sanchez-Calderon H, Hurle JM, Varela-Nieto I, Magarinos M. Early otic development depends on autophagy for apoptotic cell clearance and neural differentiation. Cell Death Dis 2012; 3:e394; http://dx.doi.org/ 10.1038/cddis.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Morais RD, Thome RG, Lemos FS, Bazzoli N, Rizzo E. Autophagy and apoptosis interplay during follicular atresia in fish ovary: a morphological and immunocytochemical study. Cell Tissue Res 2012; 347:467-78; http://dx.doi.org/ 10.1007/s00441-012-1327-6. [DOI] [PubMed] [Google Scholar]
- 397.Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, Arai H, Tanaka K, Kominami E, Uchiyama Y. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun 2009; 382:419-23; http://dx.doi.org/ 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 398.Komatsu M, Waguri S, Koike M, Sou Y-S, Ueno T, Hara T, Mizushima N, Iwata J-I Ezaki J, Murata S, et al.. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007; 131:1149-63; http://dx.doi.org/ 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 399.Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS, Opferman JT, Slack RS. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J 2011; 30:395-407; http://dx.doi.org/ 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Komatsu M, Wang QJ, Holstein GR, Friedrich VL Jr., Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA 2007; 104:14489-94; http://dx.doi.org/ 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, Rout MP, Chait BT, Zhong Y, Heintz N, Yue Z. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci 2006; 26:8057-68; http://dx.doi.org/ 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol 2008; 180:1065-71; http://dx.doi.org/ 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Bartlett BJ, Isakson P, Lewerenz J, Sanchez H, Kotzebue RW, Cumming RC, Harris GL, Nezis IP, Schubert DR, Simonsen A, et al.. p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 2011; 7:572-83; http://dx.doi.org/ 10.4161/auto.7.6.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab 2009; 10:507-15; http://dx.doi.org/ 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 405.El-Khoury V, Pierson S, Szwarcbart E, Brons NH, Roland O, Cherrier-De Wilde S, Plawny L, Van Dyck E, Berchem G. Disruption of autophagy by the histone deacetylase inhibitor MGCD0103 and its therapeutic implication in B-cell chronic lymphocytic leukemia. Leukemia 2014; 28:1636-46; http://dx.doi.org/ 10.1038/leu.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Nakaso K, Yoshimoto Y, Nakano T, Takeshima T, Fukuhara Y, Yasui K, Araga S, Yanagawa T, Ishii T, Nakashima K. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson's disease. Brain Res 2004; 1012:42-51; http://dx.doi.org/ 10.1016/j.brainres.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 407.Trocoli A, Bensadoun P, Richard E, Labrunie G, Merhi F, Schlafli AM, Brigger D, Souquere S, Pierron G, Pasquet JM, et al.. p62/SQSTM1 upregulation constitutes a survival mechanism that occurs during granulocytic differentiation of acute myeloid leukemia cells. Cell Death Differ 2014; 21:1852-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 2013; 41:7683-99; http://dx.doi.org/ 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Cnop M, Abdulkarim B, Bottu G, Cunha DA, Igoillo-Esteve M, Masini M, Turatsinze JV, Griebel T, Villate O, Santin I, et al.. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes 2014; 63:1978-93; http://dx.doi.org/ 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- 410.Colosetti P, Puissant A, Robert G, Luciano F, Jacquel A, Gounon P, Cassuto JP, Auberger P. Autophagy is an important event for megakaryocytic differentiation of the chronic myelogenous leukemia K562 cell line. Autophagy 2009; 5:1092-8; http://dx.doi.org/ 10.4161/auto.5.8.9889. [DOI] [PubMed] [Google Scholar]
- 411.Toepfer N, Childress C, Parikh A, Rukstalis D, Yang W. Atorvastatin induces autophagy in prostate cancer PC3 cells through activation of LC3 transcription. Cancer Biol Ther 2011; 12:691-9; http://dx.doi.org/ 10.4161/cbt.12.8.15978. [DOI] [PubMed] [Google Scholar]
- 412.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res 2011; 109:296-308; http://dx.doi.org/ 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Trocoli A, Mathieu J, Priault M, Reiffers J, Souquere S, Pierron G, Besancon F, Djavaheri-Mergny M. ATRA-induced upregulation of Beclin 1 prolongs the life span of differentiated acute promyelocytic leukemia cells. Autophagy 2011; 7:1108-14; http://dx.doi.org/ 10.4161/auto.7.10.16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Kim JH, Hong SK, Wu PK, Richards AL, Jackson WT, Park JI. Raf/MEK/ERK can regulate cellular levels of LC3B and SQSTM1/p62 at expression levels. Exp Cell Res 2014; 327:340-52; http://dx.doi.org/ 10.1016/j.yexcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 2014; 10:431-41; http://dx.doi.org/ 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.B'Chir W, Chaveroux C, Carraro V, Averous J, Maurin AC, Jousse C, Muranishi Y, Parry L, Fafournoux P, Bruhat A. Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation. Cell Signal 2014; 26:1385-91; http://dx.doi.org/ 10.1016/j.cellsig.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 417.Jamart C, Naslain D, Gilson H, Francaux M. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am J Physiol Endocrinol Metab 2013; 305:E964-74; http://dx.doi.org/ 10.1152/ajpendo.00270.2013. [DOI] [PubMed] [Google Scholar]
- 418.Sanchez AM, Bernardi H, Py G, Candau RB. Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am J Physiol Regul Integr Comp Physiol 2014; 307:R956-69; http://dx.doi.org/ 10.1152/ajpregu.00187.2014. [DOI] [PubMed] [Google Scholar]
- 419.Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 2012; 8:608; http://dx.doi.org/ 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell 2011; 144:499-512; http://dx.doi.org/ 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Penna F, Costamagna D, Pin F, Camperi A, Fanzani A, Chiarpotto EM, Cavallini G, Bonelli G, Baccino FM, Costelli P. Autophagic degradation contributes to muscle wasting in cancer cachexia. Amer J Pathol 2013; 182:1367-78; http://dx.doi.org/ 10.1016/j.ajpath.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 422.BenYounes A, Tajeddine N, Tailler M, Malik SA, Shen S, Metivier D, Kepp O, Vitale I, Maiuri MC, Kroemer G. A fluorescence-microscopic and cytofluorometric system for monitoring the turnover of the autophagic substrate p62/SQSTM1. Autophagy 2011; 7:883-91; http://dx.doi.org/ 10.4161/auto.7.8.15538. [DOI] [PubMed] [Google Scholar]
- 423.Chang Y-Y, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 2009; 20:2004-14; http://dx.doi.org/ 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Jiang Y, Zhu J, Wu L, Xu G, Dai J, Liu X. Tetracycline inhibits local inflammation induced by cerebral ischemia via modulating autophagy. PloS One 2012; 7:e48672; http://dx.doi.org/ 10.1371/journal.pone.0048672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 2009; 452:181-97; http://dx.doi.org/ 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 426.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009; 137:1001-4; http://dx.doi.org/ 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell 2011; 44:134-46; http://dx.doi.org/ 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al.. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 2010; 12:213-23. [DOI] [PubMed] [Google Scholar]
- 429.Gonzalez Y, Aryal B, Chehab L, Rao VA. Atg7- and Keap1-dependent autophagy protects breast cancer cell lines against mitoquinone-induced oxidative stress. Oncotarget 2014; 5:1526-37; http://dx.doi.org/ 10.18632/oncotarget.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 2010; 285:22576-91; http://dx.doi.org/ 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 2010; 584:1393-8; http://dx.doi.org/ 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 432.Bardag-Gorce F, Francis T, Nan L, Li J, He Lue Y, French BA, French SW. Modifications in p62 occur due to proteasome inhibition in alcoholic liver disease. Life Sci 2005; 77:2594-602; http://dx.doi.org/ 10.1016/j.lfs.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 433.Myeku N, Figueiredo-Pereira ME. Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: association with sequestosome 1/p62. J Biol Chem 2011; 286:22426-40; http://dx.doi.org/ 10.1074/jbc.M110.149252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell 2009; 33:517-27; http://dx.doi.org/ 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Monick MM, Powers LS, Walters K, Lovan N, Zhang M, Gerke A, Hansdottir S, Hunninghake GW. Identification of an autophagy defect in smokers' alveolar macrophages. J Immunol 2010; 185:5425-35; http://dx.doi.org/ 10.4049/jimmunol.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Vallelian F, Deuel JW, Opitz L, Schaer CA, Puglia M, Lonn M, Engelsberger W, Schauer S, Karnaukhova E, Spahn DR, et al.. Proteasome inhibition and oxidative reactions disrupt cellular homeostasis during heme stress. Cell Death Differ 2015; 22:597-611; http://dx.doi.org/ 10.1038/cdd.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Long J, Garner TP, Pandya MJ, Craven CJ, Chen P, Shaw B, Williamson MP, Layfield R, Searle MS. Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NF-kappaB signalling. J Mol Biol 2010; 396:178-94; http://dx.doi.org/ 10.1016/j.jmb.2009.11.032. [DOI] [PubMed] [Google Scholar]
- 438.Norman JM, Cohen GM, Bampton ET. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy 2010; 6:1042-56; http://dx.doi.org/ 10.4161/auto.6.8.13337. [DOI] [PubMed] [Google Scholar]
- 439.Lelouard H, Schmidt EK, Camosseto V, Clavarino G, Ceppi M, Hsu HT, Pierre P. Regulation of translation is required for dendritic cell function and survival during activation. J Cell Biol 2007; 179:1427-39; http://dx.doi.org/ 10.1083/jcb.200707166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 2009; 6:275-7; http://dx.doi.org/ 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 441.Lim J, Kim HW, Youdim MB, Rhyu IJ, Choe KM, Oh YJ. Binding preference of p62 towards LC3-ll during dopaminergic neurotoxin-induced impairment of autophagic flux. Autophagy 2011; 7:51-60; http://dx.doi.org/ 10.4161/auto.7.1.13909. [DOI] [PubMed] [Google Scholar]
- 442.Fouillet A, Levet C, Virgone A, Robin M, Dourlen P, Rieusset J, Belaidi E, Ovize M, Touret M, Nataf S, et al.. ER stress inhibits neuronal death by promoting autophagy. Autophagy 2012; 8:915-26; http://dx.doi.org/ 10.4161/auto.19716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Waguri S, Komatsu M. Biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. Methods Enzymol 2009; 453:181-96; http://dx.doi.org/ 10.1016/S0076-6879(08)04009-3. [DOI] [PubMed] [Google Scholar]
- 444.Hocking LJ, Lucas GJ, Daroszewska A, Mangion J, Olavesen M, Cundy T, Nicholson GC, Ward L, Bennett ST, Wuyts W, et al.. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget's disease. Hum Mol Genet 2002; 11:2735-9; http://dx.doi.org/ 10.1093/hmg/11.22.2735. [DOI] [PubMed] [Google Scholar]
- 445.Kara NZ, Toker L, Agam G, Anderson GW, Belmaker RH, Einat H. Trehalose induced antidepressant-like effects and autophagy enhancement in mice. Psychopharmacology 2013; 229:367-75; http://dx.doi.org/ 10.1007/s00213-013-3119-4. [DOI] [PubMed] [Google Scholar]
- 446.Beasley CL, Pennington K, Behan A, Wait R, Dunn MJ, Cotter D. Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: Evidence for disease-associated changes. Proteomics 2006; 6:3414-25; http://dx.doi.org/ 10.1002/pmic.200500069. [DOI] [PubMed] [Google Scholar]
- 447.Behan AT, Byrne C, Dunn MJ, Cagney G, Cotter DR. Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol Psychiatr 2009; 14:601-13; http://dx.doi.org/ 10.1038/mp.2008.7. [DOI] [PubMed] [Google Scholar]
- 448.Chetcuti A, Adams LJ, Mitchell PB, Schofield PR. Microarray gene expression profiling of mouse brain mRNA in a model of lithium treatment. Psychiat Genet 2008; 18:64-72; http://dx.doi.org/ 10.1097/YPG.0b013e3282fb0051. [DOI] [PubMed] [Google Scholar]
- 449.Focking M, Dicker P, English JA, Schubert KO, Dunn MJ, Cotter DR. Common proteomic changes in the hippocampus in schizophrenia and bipolar disorder and particular evidence for involvement of cornu ammonis regions 2 and 3. Arch Gen Psychiat 2011; 68:477-88; http://dx.doi.org/ 10.1001/archgenpsychiatry.2011.43. [DOI] [PubMed] [Google Scholar]
- 450.Nielsen J, Hoffert JD, Knepper MA, Agre P, Nielsen S, Fenton RA. Proteomic analysis of lithium-induced nephrogenic diabetes insipidus: mechanisms for aquaporin 2 down-regulation and cellular proliferation. Proc Natl Acad Sci USA 2008; 105:3634-9; http://dx.doi.org/ 10.1073/pnas.0800001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Lu K, Psakhye I, Jentsch S. Autophagic Clearance of PolyQ Proteins Mediated by Ubiquitin-Atg8 Adaptors of the Conserved CUET Protein Family. Cell 2014; 158:549-63; http://dx.doi.org/ 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 452.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12:823-30; http://dx.doi.org/ 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Maloverjan A, Piirsoo M, Michelson P, Kogerman P, Osterlund T. Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway. Exp Cell Res 2010; 316:627-37; http://dx.doi.org/ 10.1016/j.yexcr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 454.Young ARJ, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavaré S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev 2009; 23:798-803; http://dx.doi.org/ 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Chan EY, Tooze SA. Evolution of Atg1 function and regulation. Autophagy 2009; 5:758-65; http://dx.doi.org/ 10.4161/auto.8709. [DOI] [PubMed] [Google Scholar]
- 456.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 2007; 282:25464-74; http://dx.doi.org/ 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 457.Petherick KJ, Conway OJ, Mpamhanga C, Osborne SA, Kamal A, Saxty B, Ganley IG. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem 2015; 290:11376-83; http://dx.doi.org/ 10.1074/jbc.C114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R, Jung CH, Suen DF, et al.. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell 2011; 43:572-85; http://dx.doi.org/ 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev 2011; 25:1895-908; http://dx.doi.org/ 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Carling D, Mayer FV, Sanders MJ, Gamblin SJ. AMP-activated protein kinase: nature's energy sensor. Nat Chem Biol 2011; 7:512-8; http://dx.doi.org/ 10.1038/nchembio.610. [DOI] [PubMed] [Google Scholar]
- 461.Samari HR, Moller MT, Holden L, Asmyhr T, Seglen PO. Stimulation of hepatocytic AMP-activated protein kinase by okadaic acid and other autophagy-suppressive toxins. Biochem J 2005; 386:237-44; http://dx.doi.org/ 10.1042/BJ20040609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Dando I, Donadelli M, Costanzo C, Dalla Pozza E, D'Alessandro A, Zolla L, Palmieri M. Cannabinoids inhibit energetic metabolism and induce AMPK-dependent autophagy in pancreatic cancer cells. Cell Death Dis 2013; 4:e664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Hawley SA, Ross FA, Chevtzoff C, Green KA, Evans A, Fogarty S, Towler MC, Brown LJ, Ogunbayo OA, Evans AM, et al.. Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab 2010; 11:554-65; http://dx.doi.org/ 10.1016/j.cmet.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; http://dx.doi.org/ 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Chiacchiera F, Matrone A, Ferrari E, Ingravallo G, Lo Sasso G, Murzilli S, Petruzzelli M, Salvatore L, Moschetta A, Simone C. p38alpha blockade inhibits colorectal cancer growth in vivo by inducing a switch from HIF1alpha- to FoxO-dependent transcription. Cell Death Differ 2009; 16:1203-14; http://dx.doi.org/ 10.1038/cdd.2009.36. [DOI] [PubMed] [Google Scholar]
- 466.Kovács AL, Seglen PO. Inhibition of hepatocytic protein degradation by methylaminopurines and inhibitors of protein synthesis. Biochim Biophys Acta 1981; 676:213-20; http://dx.doi.org/ 10.1016/0304-4165(81)90189-6. [DOI] [PubMed] [Google Scholar]
- 467.Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem 2009; 284:31484-92; http://dx.doi.org/ 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al.. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007; 6:458-71; http://dx.doi.org/ 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 469.Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 2011; 145:607-21; http://dx.doi.org/ 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Pfisterer SG, Mauthe M, Codogno P, Proikas-Cezanne T. Ca2+/calmodulin-dependent kinase (CaMK) signaling via CaMKI and AMP-activated protein kinase contributes to the regulation of WIPI-1 at the onset of autophagy. Mol Pharmacol 2011; 80: 1066-75; http://dx.doi.org/ 10.1124/mol.111.071761. [DOI] [PubMed] [Google Scholar]
- 471.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett 2008; 582:46-53; http://dx.doi.org/ 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Samari HR, Seglen PO. Inhibition of hepatocytic autophagy by adenosine, aminoimidazole-4-carboxamide riboside, and N6-mercaptopurine riboside. Evidence for involvement of amp-activated protein kinase. J Biol Chem 1998; 273:23758-63; http://dx.doi.org/ 10.1074/jbc.273.37.23758. [DOI] [PubMed] [Google Scholar]
- 473.Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, Candau R. AMPK promotes skeletal muscle autophagy through activation of Forkhead FoxO3a and interaction with Ulk1. J Cell Biochem 2011. [DOI] [PubMed] [Google Scholar]
- 474.Inoki K, Zhu T, Guan K-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003; 115:577-90; http://dx.doi.org/ 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 475.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008; 30:214-26; http://dx.doi.org/ 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Egan D, Kim J, Shaw RJ, Guan K-L. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy 2011; 7:643-4; http://dx.doi.org/ 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al.. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456-61; http://dx.doi.org/ 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; http://dx.doi.org/ 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al.. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 2001; 108:1167-74; http://dx.doi.org/ 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Sharma A, Singh K, Mazumder S, Hill BT, Kalaycio M, Almasan A. BECN1 and BIM interactions with MCL-1 determine fludarabine resistance in leukemic B cells. Cell Death Dis 2013; 4:e628; http://dx.doi.org/ 10.1038/cddis.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Emerling BM, Viollet B, Tormos KV, Chandel NS. Compound C inhibits hypoxic activation of HIF-1 independent of AMPK. FEBS Lett 2007; 581:5727-31; http://dx.doi.org/ 10.1016/j.febslet.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Vucicevic L, Misirkic M, Janjetovic K, Vilimanovich U, Sudar E, Isenovic E, Prica M, Harhaji-Trajkovic L, Kravic-Stevovic T, Bumbasirevic V, et al.. Compound C induces protective autophagy in cancer cells through AMPK inhibition-independent blockade of Akt/mTOR pathway. Autophagy 2011; 7:40-50; http://dx.doi.org/ 10.4161/auto.7.1.13883. [DOI] [PubMed] [Google Scholar]
- 483.Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem 2006; 281:34870-9; http://dx.doi.org/ 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 484.Grotemeier A, Alers S, Pfisterer SG, Paasch F, Daubrawa M, Dieterle A, Viollet B, Wesselborg S, Proikas-Cezanne T, Stork B. AMPK-independent induction of autophagy by cytosolic Ca2+ increase. Cell Signal 2010; 22:914-25; http://dx.doi.org/ 10.1016/j.cellsig.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 485.Williams T, Forsberg LJ, Viollet B, Brenman JE. Basal autophagy induction without AMP-activated protein kinase under low glucose conditions. Autophagy 2009; 5:1155-65; http://dx.doi.org/ 10.4161/auto.5.8.10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci USA 2011; 108:4788-93; http://dx.doi.org/ 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H-ATPase. Science 2011; 334:678-83; http://dx.doi.org/ 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al.. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 2010; 191:155-68; http://dx.doi.org/ 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J 2011; 30:636-51; http://dx.doi.org/ 10.1038/emboj.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Jung CH, Jun CB, Ro S-H, Kim Y-M, Otto NM, Cao J, Kundu M, Kim D-H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20:1992-2003; http://dx.doi.org/ 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al.. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20:1981-91; http://dx.doi.org/ 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Chan EYW, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol 2009; 29:157-71; http://dx.doi.org/ 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al.. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell 2014; 53:471-83; http://dx.doi.org/ 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15:741-50; http://dx.doi.org/ 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Jung CH, Seo M, Otto NM, Kim DH. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy 2011; 7:1212-21; http://dx.doi.org/ 10.4161/auto.7.10.16660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Loffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M, Campbell DG, Wesselborg S, Alessi DR, Stork B. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 2011; 7:696-706; http://dx.doi.org/ 10.4161/auto.7.7.15451. [DOI] [PubMed] [Google Scholar]
- 497.Erlich S, Alexandrovich A, Shohami E, Pinkas-Kramarski R. Rapamycin is a neuroprotective treatment for traumatic brain injury. Neuobiol Dis 2007; 26:86-93; http://dx.doi.org/ 10.1016/j.nbd.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 498.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem 2006; 281:8518-27; http://dx.doi.org/ 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 499.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC Jr., Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 1997; 277:99-101; http://dx.doi.org/ 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 500.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell 2010; 38:768-74; http://dx.doi.org/ 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al.. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15:406-16; http://dx.doi.org/ 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 502.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2008; 19:668-81; http://dx.doi.org/ 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell 2005; 16:2544-53; http://dx.doi.org/ 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 2000; 150:1507-13; http://dx.doi.org/ 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Scott SV, Nice DC III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem 2000; 275:25840-9; http://dx.doi.org/ 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 506.Miller-Fleming L, Cheong H, Antas P, Klionsky DJ. Detection of Saccharomyces cerevisiae Atg13 by western blot. Autophagy 2014; 10:514-7; http://dx.doi.org/ 10.4161/auto.27707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics 2010; 185:871-82; http://dx.doi.org/ 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol 2011; 193:755-67; http://dx.doi.org/ 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Kim M, Park HL, Park HW, Ro SH, Nam SG, Reed JM, Guan JL, Lee JH. Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy 2013; 9:1201-13; http://dx.doi.org/ 10.4161/auto.24811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Nagy P, Karpati M, Varga A, Pircs K, Venkei Z, Takats S, Varga K, Erdi B, Hegedus K, Juhasz G. Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy 2014; 10:453-67; http://dx.doi.org/ 10.4161/auto.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Singh K, Matsuyama S, Drazba JA, Almasan A. Autophagy-dependent senescence in response to DNA damage and chronic apoptotic stress. Autophagy 2012; 8:236-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Shang L, Wang X. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy 2011; 7:924-6; http://dx.doi.org/ 10.4161/auto.7.8.15860. [DOI] [PubMed] [Google Scholar]
- 513.Ruck A, Attonito J, Garces KT, Nunez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD, et al.. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 2011; 7:386-400; http://dx.doi.org/ 10.4161/auto.7.4.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Li W, Zou W, Yang Y, Chai Y, Chen B, Cheng S, Tian D, Wang X, Vale RD, Ou G. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J Cell Biol 2012; 197:27-35; http://dx.doi.org/ 10.1083/jcb.201111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Abnave P, Mottola G, Gimenez G, Boucherit N, Trouplin V, Torre C, Conti F, Ben Amara A, Lepolard C, Djian B, et al.. Screening in planarians identifies MORN2 as a key component in LC3-associated phagocytosis and resistance to bacterial infection. Cell Host Microbe 2014; 16:338-50; http://dx.doi.org/ 10.1016/j.chom.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 516.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S, Pierron G, Codogno P. NF- [kappa]B activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem 2006; 281:30373-82. [DOI] [PubMed] [Google Scholar]
- 517.Liu Z, Lenardo MJ. Reactive oxygen species regulate autophagy through redox-sensitive proteases. Dev Cell 2007; 12:484-5; http://dx.doi.org/ 10.1016/j.devcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 518.Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, Ghidoni R, Codogno P. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem 2004; 279:18384-91; http://dx.doi.org/ 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 519.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007; 26:1749-60; http://dx.doi.org/ 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Cap M, Stepanek L, Harant K, Vachova L, Palkova Z. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell 2012; 46:436-48; http://dx.doi.org/ 10.1016/j.molcel.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 521.Zeng X, Kinsella TJ. Mammalian target of rapamycin and S6 kinase 1 positively regulate 6-thioguanine-induced autophagy. Cancer Res 2008; 68:2384-90; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-6163. [DOI] [PubMed] [Google Scholar]
- 522.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 2012; 338:956-9; http://dx.doi.org/ 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al.. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013; 154:1269-84; http://dx.doi.org/ 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Yasugi M, Takigawa N, Ochi N, Ohashi K, Harada D, Ninomiya T, Murakami T, Honda Y, Ichihara E, Tanimoto M, et al.. Everolimus prolonged survival in transgenic mice with EGFR-driven lung tumors. Exp Cell Res 2014; 326:201-9; http://dx.doi.org/ 10.1016/j.yexcr.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 525.Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M, Frank S, Tintignac LA, Sinnreich M, Ruegg MA. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab 2013; 17:731-44; http://dx.doi.org/ 10.1016/j.cmet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 526.Castets P, Ruegg MA. MTORC1 determines autophagy through ULK1 regulation in skeletal muscle. Autophagy 2013; 9:1435-7; http://dx.doi.org/ 10.4161/auto.25722. [DOI] [PubMed] [Google Scholar]
- 527.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al.. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010; 465:942-6; http://dx.doi.org/ 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Holla S, Kurowska-Stolarska M, Bayry J, Balaji KN. Selective inhibition of IFNG-induced autophagy by Mir155- and Mir31-responsive WNT5A and SHH signaling. Autophagy 2014; 10:311-30; http://dx.doi.org/ 10.4161/auto.27225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Mochizuki H, Toda H, Ando M, Kurusu M, Tomoda T, Furukubo-Tokunaga K. Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PloS One 2011; 6:e19632; http://dx.doi.org/ 10.1371/journal.pone.0019632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Wairkar YP, Toda H, Mochizuki H, Furukubo-Tokunaga K, Tomoda T, Diantonio A. Unc-51 controls active zone density and protein composition by downregulating ERK signaling. J Neurosci 2009; 29:517-28; http://dx.doi.org/ 10.1523/JNEUROSCI.3848-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Loh SH, Francescut L, Lingor P, Bahr M, Nicotera P. Identification of new kinase clusters required for neurite outgrowth and retraction by a loss-of-function RNA interference screen. Cell Death Differ 2008; 15:283-98; http://dx.doi.org/ 10.1038/sj.cdd.4402258. [DOI] [PubMed] [Google Scholar]
- 532.Zhou X, Babu JR, da Silva S, Shu Q, Graef IA, Oliver T, Tomoda T, Tani T, Wooten MW, Wang F. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci USA 2007; 104:5842-7; http://dx.doi.org/ 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Tomoda T, Kim JH, Zhan C, Hatten ME. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev 2004; 18:541-58; http://dx.doi.org/ 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Mol Brain Res 2000; 85:1-12; http://dx.doi.org/ 10.1016/S0169-328X(00)00218-7. [DOI] [PubMed] [Google Scholar]
- 535.Young ARJ, EYW Chan, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006; 119:3888-900; http://dx.doi.org/ 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 536.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 2005; 1:101-9; http://dx.doi.org/ 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 2010; 190:1005-22; http://dx.doi.org/ 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004; 6:79-90; http://dx.doi.org/ 10.1016/S1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 539.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 2003; 116:1679-88; http://dx.doi.org/ 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 540.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 2001; 152:657-68; http://dx.doi.org/ 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, et al.. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell 2012; 21:532-46; http://dx.doi.org/ 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Phys 2005; 138:2097-110; http://dx.doi.org/ 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006; 8:1124-32; http://dx.doi.org/ 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 544.Maskey D, Yousefi S, Schmid I, Zlobec I, Perren A, Friis R, Simon HU. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun 2013; 4:2130; http://dx.doi.org/ 10.1038/ncomms3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 2001; 152:519-30; http://dx.doi.org/ 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al.. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11:385-96; http://dx.doi.org/ 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 547.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009; 11:468-76; http://dx.doi.org/ 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 2008; 105:19211-6; http://dx.doi.org/ 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360-72; http://dx.doi.org/ 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci USA 2011; 108:7769-74; http://dx.doi.org/ 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 2010; 190:511-21; http://dx.doi.org/ 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010; 12:747-57; http://dx.doi.org/ 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA Jr., Klionsky DJ. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell 2001; 12:3821-38; http://dx.doi.org/ 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Barth H, Meiling-Wesse K, Epple UD, Thumm M. Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett 2001; 508:23-8; http://dx.doi.org/ 10.1016/S0014-5793(01)03016-2. [DOI] [PubMed] [Google Scholar]
- 555.Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 2004; 23:9314-25; http://dx.doi.org/ 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 556.Monastyrska I, Klionsky DJ. Autophagy in organelle homeostasis: peroxisome turnover. Mol Aspects Med 2006; 27:483-94; http://dx.doi.org/ 10.1016/j.mam.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem 2005; 280:41785-8; http://dx.doi.org/ 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- 558.Tallóczy Z, Virgin HW IV, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2006; 2:24-9; http://dx.doi.org/ 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 559.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010; 6:506-22; http://dx.doi.org/ 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 560.Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, Nordheim A. Human WIPI-1 puncta-formation: A novel assay to assess mammalian autophagy. FEBS Lett 2007; 581:3396-404; http://dx.doi.org/ 10.1016/j.febslet.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 561.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010; 6:764-76; http://dx.doi.org/ 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Mauthe M, Jacob A, Freiberger S, Hentschel K, Stierhof YD, Codogno P, Proikas-Cezanne T. Resveratrol-mediated autophagy requires WIPI-1 regulated LC3 lipidation in the absence of induced phagophore formation. Autophagy 2011; 7:1448-61; http://dx.doi.org/ 10.4161/auto.7.12.17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovacs AL, Yu L, Zhang H. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell 2011; 21:343-57; http://dx.doi.org/ 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 564.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22:124-31; http://dx.doi.org/ 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res 2007; 17: 839-49; http://dx.doi.org/ 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 566.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927-39; http://dx.doi.org/ 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 567.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 2009; 10:285-92; http://dx.doi.org/ 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Wei Y, Sinha S, Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 2008; 4:949-51; http://dx.doi.org/ 10.4161/auto.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008; 30:678-88; http://dx.doi.org/ 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Lossi L, Gambino G, Ferrini F, Alasia S, Merighi A. Posttranslational regulation of BCL2 levels in cerebellar granule cells: A mechanism of neuronal survival. Dev Neurobiol 2009; 69:855-70; http://dx.doi.org/ 10.1002/dneu.20744. [DOI] [PubMed] [Google Scholar]
- 571.Lossi L, Gambino G, Salio C, Merighi A. Autophagy regulates the post-translational cleavage of BCL-2 and promotes neuronal survival. Sci World J 2010; 10:924-9; http://dx.doi.org/ 10.1100/tsw.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ 2008; 15:1318-29; http://dx.doi.org/ 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 573.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18:571-80; http://dx.doi.org/ 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2001; 2:330-5; http://dx.doi.org/ 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Amritraj A, Peake K, Kodam A, Salio C, Merighi A, Vance JE, Kar S. Increased activity and altered subcellular distribution of lysosomal enzymes determine neuronal vulnerability in Niemann-Pick type C1-deficient mice. Am J Pathol 2009; 175:2540-56; http://dx.doi.org/ 10.2353/ajpath.2009.081096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Castino R, Bellio N, Follo C, Murphy D, Isidoro C. Inhibition of PI3k class III-dependent autophagy prevents apoptosis and necrosis by oxidative stress in dopaminergic neuroblastoma cells. Toxicol Sci 2010; 117:152-62; http://dx.doi.org/ 10.1093/toxsci/kfq170. [DOI] [PubMed] [Google Scholar]
- 577.Yue Z, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N. A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron 2002; 35:921-33; http://dx.doi.org/ 10.1016/S0896-6273(02)00861-9. [DOI] [PubMed] [Google Scholar]
- 578.Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ 2010; 17:268-77; http://dx.doi.org/ 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 2005; 1:46-52; http://dx.doi.org/ 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 580.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126:121-34; http://dx.doi.org/ 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 581.Valbuena A, Castro-Obregon S, Lazo PA. Downregulation of VRK1 by p53 in response to DNA damage is mediated by the autophagic pathway. PloS One 2011; 6:e17320; http://dx.doi.org/ 10.1371/journal.pone.0017320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Lorin S, Pierron G, Ryan KM, Codogno P, Djavaheri-Mergny M. Evidence for the interplay between JNK and p53-DRAM signalling pathways in the regulation of autophagy. Autophagy 2010; 6:153-4; http://dx.doi.org/ 10.4161/auto.6.1.10537. [DOI] [PubMed] [Google Scholar]
- 583.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685-701; http://dx.doi.org/ 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012; 151:1256-69; http://dx.doi.org/ 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 585.Takats S, Nagy P, Varga A, Pircs K, Karpati M, Varga K, Kovacs AL, Hegedus K, Juhasz G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol 2013; 201:531-9; http://dx.doi.org/ 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Chen D, Zhong Q. A tethering coherent protein in autophagosome maturation. Autophagy 2012; 8:985-6; http://dx.doi.org/ 10.4161/auto.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Taniguchi M, Kitatani K, Kondo T, Hashimoto-Nishimura M, Asano S, Hayashi A, Mitsutake S, Igarashi Y, Umehara H, Takeya H, et al.. Regulation of autophagy and its associated cell death by “sphingolipid rheostat”: reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. J Biol Chem 2012; 287:39898-910; http://dx.doi.org/ 10.1074/jbc.M112.416552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Justice MJ, Petrusca DN, Rogozea AL, Williams JA, Schweitzer KS, Petrache I, Wassall SR, Petrache HI. Effects of lipid interactions on model vesicle engulfment by alveolar macrophages. Biophys J 2014; 106:598-609; http://dx.doi.org/ 10.1016/j.bpj.2013.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc Natl Acad Sci USA 2008; 105:17402-7; http://dx.doi.org/ 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Pattingre S, Bauvy C, Levade T, Levine B, Codogno P. Ceramide-induced autophagy: to junk or to protect cells? Autophagy 2009; 5:558-60; http://dx.doi.org/ 10.4161/auto.5.4.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, et al.. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol 2012; 8:831-8; http://dx.doi.org/ 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Jiang W, Ogretmen B. Ceramide stress in survival versus lethal autophagy paradox: ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Autophagy 2013; 9:258-9; http://dx.doi.org/ 10.4161/auto.22739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Jiang W, Ogretmen B. Autophagy paradox and ceramide. Biochim Biophys Acta 2014; 1841:783-92; http://dx.doi.org/ 10.1016/j.bbalip.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Lepine S, Allegood JC, Park M, Dent P, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ 2011; 18:350-61; http://dx.doi.org/ 10.1038/cdd.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Matarrese P, Garofalo T, Manganelli V, Gambardella L, Marconi M, Grasso M, Tinari A, Misasi R, Malorni W, Sorice M. Evidence for the involvement of GD3 ganglioside in autophagosome formation and maturation. Autophagy 2014; 10:750-65; http://dx.doi.org/ 10.4161/auto.27959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Russ DW, Wills AM, Boyd IM, Krause J. Weakness, SR function and stress in gastrocnemius muscles of aged male rats. Exp Gastroenterol 2014; 50:40-4; http://dx.doi.org/ 10.1016/j.exger.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 597.Bernard A, Jin M, Gonzalez-Rodriguez P, Fullgrabe J, Delorme-Axford E, Backues SK, Joseph B, Klionsky DJ. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr Biol 2015; 25:546-55; http://dx.doi.org/ 10.1016/j.cub.2014.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Bernard A, Klionsky DJ. Rph1 mediates the nutrient-limitation signaling pathway leading to transcriptional activation of autophagy. Autophagy 2015; 11:718-9; http://dx.doi.org/ 10.1080/15548627.2015.1018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Nara A, Mizushima N, Yamamoto A, Kabeya Y, Ohsumi Y, Yoshimori T. SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct Funct 2002; 27:29-37; http://dx.doi.org/ 10.1247/csf.27.29. [DOI] [PubMed] [Google Scholar]
- 600.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147:435-46; http://dx.doi.org/ 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol 2014; 24:1314-22; http://dx.doi.org/ 10.1016/j.cub.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2[alpha] phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 2007; 14:230-9; http://dx.doi.org/ 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 603.Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem 2012; 287:39107-14; http://dx.doi.org/ 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Moussay E, Kaoma T, Baginska J, Muller A, Van Moer K, Nicot N, Nazarov PV, Vallar L, Chouaib S, Berchem G, et al.. The acquisition of resistance to TNFalpha in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy 2011; 7:760-70; http://dx.doi.org/ 10.4161/auto.7.7.15454. [DOI] [PubMed] [Google Scholar]
- 605.Mitroulis I, Kourtzelis I, Kambas K, Rafail S, Chrysanthopoulou A, Speletas M, Ritis K. Regulation of the autophagic machinery in human neutrophils. Eur J Immunol 2010; 40:1461-72; http://dx.doi.org/ 10.1002/eji.200940025. [DOI] [PubMed] [Google Scholar]
- 606.Rodriguez-Muela N, Germain F, Marino G, Fitze PS, Boya P. Autophagy promotes survival of retinal ganglion cells after optic nerve axotomy in mice. Cell Death Differ 2012; 19:162-9; http://dx.doi.org/ 10.1038/cdd.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Vázquez P, Arroba AI, Cecconi F, de la Rosa EJ, Boya P, De Pablo F. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy 2012; 8:187-99. [DOI] [PubMed] [Google Scholar]
- 608.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al.. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 2010; 120:127-41; http://dx.doi.org/ 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Haim Y, Blüher M, Slutsky N, Goldstein N, Klöting N, Harman-Boehm I, Kirshtein B, Ginsberg D, Gericke M, Jurado EG, et al.. Elevated autophagy gene expression in adipose tissue of obese humans: A potential non-cell-cycle-dependent function of E2F1. Autophagy 2015; 11:2074-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Las G, Serada SB, Wikstrom JD, Twig G, Shirihai OS. Fatty acids suppress autophagic turnover in beta-cells. J Biol Chem 2011; 286:42534-44; http://dx.doi.org/ 10.1074/jbc.M111.242412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, Hesselink MK, Paquet C, Delhaye S, Shin Y, et al.. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med 2013; 19:1039-46; http://dx.doi.org/ 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 2014; 516:112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, et al.. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 2014; 516:108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 2008; 27:4860-4; http://dx.doi.org/ 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 615.Jiang H, Martin V, Gomez-Manzano C, Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N, Fueyo J. The RB-E2F1 pathway regulates autophagy. Cancer Res 2010; 70:7882-93; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, Coughlin SM, Zuyderduyn SD, Jones SJ, Marra MA. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol 2003; 13:358-63; http://dx.doi.org/ 10.1016/S0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- 617.Lee C-Y, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol 2003; 13:350-7; http://dx.doi.org/ 10.1016/S0960-9822(03)00085-X. [DOI] [PubMed] [Google Scholar]
- 618.Denton D, Shravage B, Simin R, Baehrecke EH, Kumar S. Larval midgut destruction in Drosophila: not dependent on caspases but suppressed by the loss of autophagy. Autophagy 2010; 6:163-5; http://dx.doi.org/ 10.4161/auto.6.1.10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Franzetti E, Huang ZJ, Shi YX, Xie K, Deng XJ, Li JP, Li QR, Yang WY, Zeng WN, Casartelli M, et al.. Autophagy precedes apoptosis during the remodeling of silkworm larval midgut. Apoptosis 2012; 17:305-24; http://dx.doi.org/ 10.1007/s10495-011-0675-0. [DOI] [PubMed] [Google Scholar]
- 620.Tian L, Ma L, Guo E, Deng X, Ma S, Xia Q, Cao Y, Li S. 20-Hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy 2013; 9:1172-87; http://dx.doi.org/ 10.4161/auto.24731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Juhasz G, Puskas LG, Komonyi O, Erdi B, Maroy P, Neufeld TP, Sass M. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ 2007; 14:1181-90; http://dx.doi.org/ 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Erdi B, Nagy P, Zvara A, Varga A, Pircs K, Menesi D, Puskas LG, Juhasz G. Loss of the starvation-induced gene Rack1 leads to glycogen deficiency and impaired autophagic responses in Drosophila. Autophagy 2012; 8:1124-35; http://dx.doi.org/ 10.4161/auto.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Barth JM, Szabad J, Hafen E, Kohler K. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ 2011; 18:915-24; http://dx.doi.org/ 10.1038/cdd.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 2013; 15:668-76; http://dx.doi.org/ 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; http://dx.doi.org/ 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. The FASEB J 2004; 18:39-51; http://dx.doi.org/ 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 627.Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008; 178:1339-53; http://dx.doi.org/ 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Seiliez I, Gutierrez J, Salmeron C, Skiba-Cassy S, Chauvin C, Dias K, Kaushik S, Tesseraud S, Panserat S. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Comp Biochem Phys B 2010; 157:258-66; http://dx.doi.org/ 10.1016/j.cbpb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 629.Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al.. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 2013; 4:2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.M Sandri. Autophagy in health and disease. 3. Involvement of autophagy in muscle atrophy. Am J Physiol Cell Physiol 2010; 298:C1291-7; http://dx.doi.org/ 10.1152/ajpcell.00531.2009. [DOI] [PubMed] [Google Scholar]
- 631.Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al.. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 2009; 11:1305-14; http://dx.doi.org/ 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 632.Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, Seux M, Nowak J, Gonzalez CD, Iovanna JL, et al.. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem 2007; 282:37124-33; http://dx.doi.org/ 10.1074/jbc.M706956200. [DOI] [PubMed] [Google Scholar]
- 633.Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al.. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 2010; 141:1042-55; http://dx.doi.org/ 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 634.Lo Re AE, Fernandez-Barrena MG, Almada LL, Mills LD, Elsawa SF, Lund G, Ropolo A, Molejon MI, Vaccaro MI, Fernandez-Zapico ME. Novel AKT1-GLI3-VMP1 pathway mediates KRAS oncogene-induced autophagy in cancer cells. J Biol Chem 2012; 287:25325-34; http://dx.doi.org/ 10.1074/jbc.M112.370809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Sardiello M, Palmieri M, Di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al.. A gene network regulating lysosomal biogenesis and function. Science 2009; 325:473-7. [DOI] [PubMed] [Google Scholar]
- 636.Palmieri M, Impey S, Kang H, Di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 2011; 20:3852-66; http://dx.doi.org/ 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 637.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012; 8:903-14; http://dx.doi.org/ 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al.. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095-108; http://dx.doi.org/ 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Nezich CL, Wang C, Fogel AI, Youle RJ. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. J Cell Biol 2015; 210:435-50; http://dx.doi.org/ 10.1083/jcb.201501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al.. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 2015; 524:361-5; http://dx.doi.org/ 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Kang YA, Sanalkumar R, O'Geen H, Linnemann AK, Chang CJ, Bouhassira EE, Farnham PJ, Keles S, Bresnick EH. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol 2012; 32:226-39; http://dx.doi.org/ 10.1128/MCB.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007; 6:472-83; http://dx.doi.org/ 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 643.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell 2013; 50:16-28; http://dx.doi.org/ 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J 2011; 30:4642-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, et al.. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet 2011; 43:242-5; http://dx.doi.org/ 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 646.Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru AH, van Adrichem AJ, et al.. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood 2011; 118:916-25; http://dx.doi.org/ 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Roccaro AM, Sacco A, Jia X, Azab AK, Maiso P, Ngo HT, Azab F, Runnels J, Quang P, Ghobrial IM. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood 2010; 116:1506-14; http://dx.doi.org/ 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Martinet W, De Meyer GR, Andries L, Herman AG, Kockx MM. In situ detection of starvation-induced autophagy. J Histochem Cytochem 2006; 54:85-96; http://dx.doi.org/ 10.1369/jhc.5A6743.2005. [DOI] [PubMed] [Google Scholar]
- 649.Banreti A, Sass M, Graba Y. The emerging role of acetylation in the regulation of autophagy. Autophagy 2013; 9:819-29; http://dx.doi.org/ 10.4161/auto.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Jin M, Klionsky DJ. Regulation of autophagy: Modulation of the size and number of autophagosomes. FEBS Lett 2014; 588:2457-63; http://dx.doi.org/ 10.1016/j.febslet.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biology 2015; 25:354-63; http://dx.doi.org/ 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 2015; 11:28-45; http://dx.doi.org/ 10.4161/15548627.2014.984267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Pietrocola F, Marino G, Lissa D, Vacchelli E, Malik SA, Niso-Santano M, Zamzami N, Galluzzi L, Maiuri MC, Kroemer G. Pro-autophagic polyphenols reduce the acetylation of cytoplasmic proteins. Cell Cycle 2012; 11:3851-60; http://dx.doi.org/ 10.4161/cc.22027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Mariño G, Pietrocola F, Madeo F, Kroemer G. Caloric restriction mimetics: natural/physiological pharmacological autophagy inducers. Autophagy. 2014;10:1879-82. doi: 10.4161/auto.36413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Madeo F, Pietrocola F, Eisenberg T, Kroemer G. Caloric restriction mimetics: towards a molecular definition. Nat Rev Drug Discov 2014; 13:727-40; http://dx.doi.org/ 10.1038/nrd4391. [DOI] [PubMed] [Google Scholar]
- 656.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem 2009; 284:6322-8; http://dx.doi.org/ 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, et al.. Deacetylation of Nuclear LC3 Drives Autophagy Initiation under Starvation. Mol Cell 2015. [DOI] [PubMed] [Google Scholar]
- 658.Pattingre S, Petiot A, Codogno P. Analyses of G[a]-interacting protein and activator of G-protein-signaling-3 functions in macroautophagy. Methods Enzymol 2004; 390:17-31; http://dx.doi.org/ 10.1016/S0076-6879(04)90002-X. [DOI] [PubMed] [Google Scholar]
- 659.Bauvy C, Meijer AJ, Codogno P. Assaying of autophagic protein degradation. Methods Enzymol 2009; 452:47-61; http://dx.doi.org/ 10.1016/S0076-6879(08)03604-5. [DOI] [PubMed] [Google Scholar]
- 660.Zhang J, Wang J, Ng S, Lin Q, Shen HM. Development of a novel method for quantification of autophagic protein degradation by AHA labeling. Autophagy 2014; 10:901-12; http://dx.doi.org/ 10.4161/auto.28267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 2008; 283:22847-57; http://dx.doi.org/ 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 662.Kabuta T, Furuta A, Aoki S, Furuta K, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem 2008; 283:23731-8; http://dx.doi.org/ 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663.Saitoh Y, Fujikake N, Okamoto Y, Popiel HA, Hatanaka Y, Ueyama M, Suzuki M, Gaumer S, Murata M, Wada K, et al.. p62 plays a protective role in the autophagic degradation of polyglutamine protein oligomers in polyglutamine disease model flies. J Biol Chem 2015; 290:1442-53; http://dx.doi.org/ 10.1074/jbc.M114.590281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 2007; 171:513-24; http://dx.doi.org/ 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 2005; 280:40282-92; http://dx.doi.org/ 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 666.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al.. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007; 447:859-63; http://dx.doi.org/ 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 667.Tomek K, Wagner R, Varga F, Singer CF, Karlic H, Grunt TW. Blockade of fatty acid synthase induces ubiquitination and degradation of phosphoinositide-3-kinase signaling proteins in ovarian cancer. Mol Cancer Res 2011:1767-79; http://dx.doi.org/ 10.1158/1541-7786.MCR-10-0467. [DOI] [PubMed] [Google Scholar]
- 668.Zimmermann AC, Zarei M, Eiselein S, Dengjel J. Quantitative proteomics for the analysis of spatio-temporal protein dynamics during autophagy. Autophagy 2010; 6:1009-16; http://dx.doi.org/ 10.4161/auto.6.8.12786. [DOI] [PubMed] [Google Scholar]
- 669.Kristensen AR, Schandorff S, Hoyer-Hansen M, Nielsen MO, Jaattela M, Dengjel J, Andersen JS. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics: MCP 2008; 7:2419-28; http://dx.doi.org/ 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- 670.Furuya N, Kanazawa T, Fujimura S, Ueno T, Kominami E, Kadowaki M. Leupeptin-induced appearance of partial fragment of betaine homocysteine methyltransferase during autophagic maturation in rat hepatocytes. J Biochem (Tokyo) 2001; 129:313-20; http://dx.doi.org/ 10.1093/oxfordjournals.jbchem.a002859. [DOI] [PubMed] [Google Scholar]
- 671.Ueno T, Ishidoh K, Mineki R, Tanida I, Murayama K, Kadowaki M, Kominami E. Autolysosomal membrane-associated betaine homocysteine methyltransferase. Limited degradation fragment of a sequestered cytosolic enzyme monitoring autophagy. J Biol Chem 1999; 274:15222-9; http://dx.doi.org/ 10.1074/jbc.274.21.15222. [DOI] [PubMed] [Google Scholar]
- 672.Overbye A, Saetre F, Hagen LK, Johansen HT, Seglen PO. Autophagic activity measured in whole rat hepatocytes as the accumulation of a novel BHMT fragment (p10), generated in amphisomes by the asparaginyl proteinase, legumain. Autophagy 2011; 7:1011-27; http://dx.doi.org/ 10.4161/auto.7.9.16436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Seglen PO, Overbye A, Saetre F. Sequestration assays for mammalian autophagy. Methods Enzymol 2009; 452:63-83; http://dx.doi.org/ 10.1016/S0076-6879(08)03605-7. [DOI] [PubMed] [Google Scholar]
- 674.Mercer CA, Kaliappan A, Dennis PB. Macroautophagy-dependent, intralysosomal cleavage of a betaine homocysteine methyltransferase fusion protein requires stable multimerization. Autophagy 2008; 4:185-94; http://dx.doi.org/ 10.4161/auto.5275. [DOI] [PubMed] [Google Scholar]
- 675.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol 2003; 33:1250-9; http://dx.doi.org/ 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 676.Taylor GS, Long HM, Haigh TA, Larsen M, Brooks J, Rickinson AB. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J Immunol 2006; 177:3746-56; http://dx.doi.org/ 10.4049/jimmunol.177.6.3746. [DOI] [PubMed] [Google Scholar]
- 677.Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J 1989; 8:2241-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Venerando R, Miotto G, Kadowaki M, Siliprandi N, Mortimore GE. Multiphasic control of proteolysis by leucine and alanine in the isolated rat hepatocyte. Am J Physiol 1994; 266:C455-61. [DOI] [PubMed] [Google Scholar]
- 679.Häussinger D, Hallbrucker C, vom Dahl S, Lang F, Gerok W. Cell swelling inhibits proteolysis in perfused rat liver. Biochem J 1990; 272:239-42; http://dx.doi.org/ 10.1042/bj2720239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680.vom Dahl S, Häussinger D. Cell hydration and proteolysis control in liver. Biochem J 1995; 312:988-9; http://dx.doi.org/ 10.1042/bj3120988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci USA 2013; 110:6400-5; http://dx.doi.org/ 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell 2005; 16:5843-56; http://dx.doi.org/ 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Manjithaya R, Jain S, Farre JC, Subramani S. A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J Cell Biol 2010; 189:303-10; http://dx.doi.org/ 10.1083/jcb.200909154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Journo D, Mor A, Abeliovich H. Aup1-mediated regulation of Rtg3 during mitophagy. J Biol Chem 2009; 284:35885-95; http://dx.doi.org/ 10.1074/jbc.M109.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem 2008; 283:32386-93; http://dx.doi.org/ 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, et al.. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell 2009; 20:4730-8; http://dx.doi.org/ 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 2009; 17:98-109; http://dx.doi.org/ 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 2009; 17:87-97; http://dx.doi.org/ 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 689.Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol 1998; 141:625-36; http://dx.doi.org/ 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 690.Nazarko TY, Nicaud JM, Sibirny AA. Observation of the Yarrowia lipolytica peroxisome-vacuole dynamics by fluorescence microscopy with a single filter set. Cell Biol Int 2005; 29:65-70; http://dx.doi.org/ 10.1016/j.cellbi.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 691.Roetzer A, Gratz N, Kovarik P, Schuller C. Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol 2010; 12:199-216; http://dx.doi.org/ 10.1111/j.1462-5822.2009.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Bormann C, Sahm H. Degradation of microbodies in relation to activities of alcohol oxidase and catalase in Candida boidinii. Arch Microbiol 1978; 117:67-72; http://dx.doi.org/ 10.1007/BF00689353. [DOI] [PubMed] [Google Scholar]
- 693.Clare DA, Duong MN, Darr D, Archibald F, Fridovich I. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal Biochem 1984; 140:532-7; http://dx.doi.org/ 10.1016/0003-2697(84)90204-5. [DOI] [PubMed] [Google Scholar]
- 694.Vachova L, Kucerova H, Devaux F, Ulehlova M, Palkova Z. Metabolic diversification of cells during the development of yeast colonies. Environ Microbiol 2009; 11:494-504; http://dx.doi.org/ 10.1111/j.1462-2920.2008.01789.x. [DOI] [PubMed] [Google Scholar]
- 695.Stasyk OV, Nazarko TY, Sibirny AA. Methods of plate pexophagy monitoring and positive selection for ATG gene cloning in yeasts. Methods Enzymol 2008; 451:229-39; http://dx.doi.org/ 10.1016/S0076-6879(08)03216-3. [DOI] [PubMed] [Google Scholar]
- 696.Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci 1999; 112:4079-87. [DOI] [PubMed] [Google Scholar]
- 697.Mukaiyama H, Oku M, Baba M, Samizo T, Hammond AT, Glick BS, Kato N, Sakai Y. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells 2002; 7:75-90; http://dx.doi.org/ 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- 698.Tuttle DL, Dunn WA Jr.. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci 1995; 108 (Pt 1):25-35. [DOI] [PubMed] [Google Scholar]
- 699.Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy 2005; 1:37-45; http://dx.doi.org/ 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Veenhuis M, Douma A, Harder W, Osumi M. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch Microbiol 1983; 134:193-203; http://dx.doi.org/ 10.1007/BF00407757. [DOI] [PubMed] [Google Scholar]
- 701.Monosov EZ, Wenzel TJ, Luers GH, Heyman JA, Subramani S. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J Histochem Cytochem 1996; 44:581-9; http://dx.doi.org/ 10.1177/44.6.8666743. [DOI] [PubMed] [Google Scholar]
- 702.Wiemer EA, Wenzel T, Deerinck TJ, Ellisman MH, Subramani S. Visualization of the peroxisomal compartment in living mammalian cells: dynamic behavior and association with microtubules. J Cell Biol 1997; 136:71-80; http://dx.doi.org/ 10.1083/jcb.136.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Monastyrska I, van der Heide M, Krikken AM, JAKW Kiel, van der Klei IJ, Veenhuis M. Atg8 is essential for macropexophagy in Hansenula polymorpha. Traffic 2005; 6:66-74; http://dx.doi.org/ 10.1111/j.1600-0854.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 704.Devenish RJ, Prescott M, Turcic K, Mijaljica D. Monitoring organelle turnover in yeast using fluorescent protein tags. Methods Enzymol 2008; 451:109-31; http://dx.doi.org/ 10.1016/S0076-6879(08)03209-6. [DOI] [PubMed] [Google Scholar]
- 705.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell 2013; 26:9-18; http://dx.doi.org/ 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 706.TK Kerppola. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nature Protocols 2006; 1:1278-86; http://dx.doi.org/ 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 707.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 2006; 40:61-6; http://dx.doi.org/ 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 708.Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell 2008; 14:365-76; http://dx.doi.org/ 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709.He Y, Deng YZ, Naqvi NI. Atg24-assisted mitophagy in the foot cells is necessary for proper asexual differentiation in Magnaporthe oryzae. Autophagy 2013; 9:1818-27; http://dx.doi.org/ 10.4161/auto.26057. [DOI] [PubMed] [Google Scholar]
- 710.Kanki T, Klionsky DJ. The molecular mechanism of mitochondria autophagy in yeast. Mol Microbiol 2010; 75:795-800; http://dx.doi.org/ 10.1111/j.1365-2958.2009.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem 2007; 282:5617-24; http://dx.doi.org/ 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- 712.H Abeliovich. Stationary-phase mitophagy in respiring Saccharomyces cerevisiae. Antioxid Redox Sign 2011; 14:2003-11; http://dx.doi.org/ 10.1089/ars.2010.3807. [DOI] [PubMed] [Google Scholar]
- 713.East DA, Fagiani F, Crosby J, Georgakopoulos ND, Bertrand H, Schaap M, Fowkes A, Wells G, Campanella M. PMI: a DeltaPsim independent pharmacological regulator of mitophagy. Chem Biol 2014; 21:1585-96; http://dx.doi.org/ 10.1016/j.chembiol.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714.Aksam EB, Koek A, JAKW Kiel, Jourdan S, Veenhuis M, van der Klei IJ. A peroxisomal lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy 2007; 3:96-105; http://dx.doi.org/ 10.4161/auto.3534. [DOI] [PubMed] [Google Scholar]
- 715.Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell 2003; 14:129-41; http://dx.doi.org/ 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen EL, Millen J, Goldfarb DS, Thumm M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell 2008; 19:4492-505; http://dx.doi.org/ 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 717.Farre JC, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol 2009; 21:522-30; http://dx.doi.org/ 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 718.Kvam E, Goldfarb DS. Structure and function of nucleus-vacuole junctions: outer-nuclear-membrane targeting of Nvj1p and a role in tryptophan uptake. J Cell Sci 2006; 119:3622-33; http://dx.doi.org/ 10.1242/jcs.03093. [DOI] [PubMed] [Google Scholar]
- 719.Millen JI, Krick R, Prick T, Thumm M, Goldfarb DS. Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Autophagy 2009; 5:75-81; http://dx.doi.org/ 10.4161/auto.5.1.7181. [DOI] [PubMed] [Google Scholar]
- 720.Mijaljica D, Prescott M, Devenish RJ. A late form of nucleophagy in Saccharomyces cerevisiae. PloS One 2012; 7:e40013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 721.Shoji JY, Kikuma T, Arioka M, Kitamoto K. Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PloS One 2010; 5:e15650; http://dx.doi.org/ 10.1371/journal.pone.0015650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Shoji J-Y, Kikuma T, Arioka M, Kitamoto K. Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PloS One 2010; 5:e15650; http://dx.doi.org/ 10.1371/journal.pone.0015650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723.He M, Kershaw MJ, Soanes DM, Xia Y, Talbot NJ. Infection-associated nuclear degeneration in the rice blast fungus Magnaporthe oryzae requires non-selective macro-autophagy. PloS One 2012; 7:e33270; http://dx.doi.org/ 10.1371/journal.pone.0033270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724.Maheshwari R. Nuclear behavior in fungal hyphae. FEMS Microbiol Lett 2005; 249:7-14; http://dx.doi.org/ 10.1016/j.femsle.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 725.Shoji J-Y, Craven KD. Autophagy in basal hyphal compartments: A green strategy of great recyclers. Fungal Biol Rev 2011; 25:79-83; http://dx.doi.org/ 10.1016/j.fbr.2011.04.001. [DOI] [Google Scholar]
- 726.Voigt O, Poggeler S. Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora. Autophagy 2013; 9:33-49; http://dx.doi.org/ 10.4161/auto.22398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 727.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell 2005; 16:1593-605; http://dx.doi.org/ 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 2002; 3:825-37; http://dx.doi.org/ 10.1016/S1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Abeliovich H, Darsow T, Emr SD. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J 1999; 18:6005-16; http://dx.doi.org/ 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Abeliovich H, Zarei M, Rigbolt KT, Youle RJ, Dengjel J. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat Commun 2013; 4:2789; http://dx.doi.org/ 10.1038/ncomms3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Overbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy 2007; 3:300-22; http://dx.doi.org/ 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- 732.Petroi D, Popova B, Taheri-Talesh N, Irniger S, Shahpasandzadeh H, Zweckstetter M, Outeiro TF, Braus GH. Aggregate clearance of alpha-synuclein in Saccharomyces cerevisiae depends more on autophagosome and vacuole function than on the proteasome. J Biol Chem 2012; 287:27567-79; http://dx.doi.org/ 10.1074/jbc.M112.361865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Shahpasandzadeh H, Popova B, Kleinknecht A, Fraser PE, Outeiro TF, Braus GH. Interplay between sumoylation and phosphorylation for protection against alpha-synuclein inclusions. J Biol Chem 2014; 289:31224-40; http://dx.doi.org/ 10.1074/jbc.M114.559237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 734.Wafa K, MacLean J, Zhang F, Pasumarthi KB. Characterization of growth suppressive functions of a splice variant of cyclin D2. PloS One 2013; 8:e53503; http://dx.doi.org/ 10.1371/journal.pone.0053503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 735.Ju JS, Miller SE, Jackson E, Cadwell K, Piwnica-Worms D, Weihl CC. Quantitation of selective autophagic protein aggregate degradation in vitro and in vivo using luciferase reporters. Autophagy 2009; 5:511-9; http://dx.doi.org/ 10.4161/auto.5.4.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Hohn A, Sittig A, Jung T, Grimm S, Grune T. Lipofuscin is formed independently of macroautophagy and lysosomal activity in stress-induced prematurely senescent human fibroblasts. Free Radical Bio Med 2012; 53:1760-9; http://dx.doi.org/ 10.1016/j.freeradbiomed.2012.08.591. [DOI] [PubMed] [Google Scholar]
- 737.Jung T, Hohn A, Catalgol B, Grune T. Age-related differences in oxidative protein-damage in young and senescent fibroblasts. Arch Biochem Biophys 2009; 483:127-35; http://dx.doi.org/ 10.1016/j.abb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 738.Fuentealba RA, Marasa J, Diamond MI, Piwnica-Worms D, Weihl CC. An aggregation sensing reporter identifies leflunomide and teriflunomide as polyglutamine aggregate inhibitors. Hum Mol Genet 2012; 21:664-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Allophagy: A macroautophagic process degrading spermatozoid-inherited organelles. Autophagy 2012; 8:421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Sato M, Sato K. Maternal inheritance of mitochondrial DNA: Degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy 2012; 8:424-5. [DOI] [PubMed] [Google Scholar]
- 741.Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 2011; 334:1144-7. [DOI] [PubMed] [Google Scholar]
- 742.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 2011; 334:1141-4; http://dx.doi.org/ 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 743.Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem 2012; 393:547-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 744.Fiesel FC, Ando M, Hudec R, Hill AR, Castanedes-Casey M, Caulfield TR, Moussaud-Lamodiere EL, Stankowski JN, Bauer PO, Lorenzo-Betancor O, et al.. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 746.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al.. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014; 510:162-6. [DOI] [PubMed] [Google Scholar]
- 747.Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 2010; 139:1740-52; http://dx.doi.org/ 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 2007; 462:245-53; http://dx.doi.org/ 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Dong H, Cheung SH, Liang Y, Wang B, Ramalingam R, Wang P, Sun H, Cheng SH, Lam YW. “Stainomics”: identification of mitotracker labeled proteins in mammalian cells. Electrophoresis 2013; 34:1957-64; http://dx.doi.org/ 10.1002/elps.201200557. [DOI] [PubMed] [Google Scholar]
- 750.Mauro-Lizcano M, Esteban-Martinez L, Seco E, Serrano-Puebla A, Garcia-Ledo L, Figueiredo-Pereira C, Vieira HL, Boya P. New method to assess mitophagy flux by flow cytometry. Autophagy 2015; 11:833-43; http://dx.doi.org/ 10.1080/15548627.2015.1034403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Presley AD, Fuller KM, Arriaga EA. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr B, Analytical technologies in the biomedical and life sciences 2003; 793:141-50; http://dx.doi.org/ 10.1016/S1570-0232(03)00371-4. [DOI] [PubMed] [Google Scholar]
- 752.Keij JF, Bell-Prince C, Steinkamp JA. Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs. Cytometry 2000; 39:203-10; http://dx.doi.org/. [DOI] [PubMed] [Google Scholar]
- 753.Poot M, Zhang YZ, Kramer JA, Wells KS, Jones LJ, Hanzel DK, Lugade AG, Singer VL, Haugland RP. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J Histochem Cytochem 1996; 44:1363-72; http://dx.doi.org/ 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- 754.Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, Fiesel FC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy 2010; 6:871-8; http://dx.doi.org/ 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 755.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010; 12:119-31; http://dx.doi.org/ 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 756.Diot A, Hinks-Roberts A, Lodge T, Liao C, Dombi E, Morten K, Brady S, Fratter C, Carver J, Muir R, et al.. A novel quantitative assay of mitophagy: Combining high content fluorescence microscopy and mitochondrial DNA load to quantify mitophagy and identify novel pharmacological tools against pathogenic heteroplasmic mtDNA. Pharmacol Res 2015; 100:24-35; http://dx.doi.org/ 10.1016/j.phrs.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 757.Dagda RK, Cherra SJ III, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 2009; 284:13843-55; http://dx.doi.org/ 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy 2008; 4:770-82; http://dx.doi.org/ 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al.. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 2005; 25:1025-40; http://dx.doi.org/ 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 760.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol 2011; 18:1042-52; http://dx.doi.org/ 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 761.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol 2010; 191:1367-80; http://dx.doi.org/ 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762.Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem 2011; 286:19630-40; http://dx.doi.org/ 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 763.Amadoro G, Corsetti V, Florenzano F, Atlante A, Ciotti MT, Mongiardi MP, Bussani R, Nicolin V, Nori SL, Campanella M, et al.. AD-linked, toxic NH2 human tau affects the quality control of mitochondria in neurons. Neuobiol Dis 2014; 62:489-507; http://dx.doi.org/ 10.1016/j.nbd.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 764.Chang TK, Shravage BV, Hayes SD, Powers CM, Simin RT, WaDe Harper J, Baehrecke EH. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol 2013; 15:1067-78; http://dx.doi.org/ 10.1038/ncb2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 765.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron 2015; 85:257-73; http://dx.doi.org/ 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 766.Yang JY, Yang WY. Spatiotemporally controlled initiation of Parkin-mediated mitophagy within single cells. Autophagy 2011; 7:1230-8; http://dx.doi.org/ 10.4161/auto.7.10.16626. [DOI] [PubMed] [Google Scholar]
- 767.Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy 2012; 8:1462-76; http://dx.doi.org/ 10.4161/auto.21211. [DOI] [PubMed] [Google Scholar]
- 768.Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Nardacci R, Piacentini M, Campanella M, et al.. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.JJ Lemasters. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3). Redox Biol 2014; 2:749-54; http://dx.doi.org/ 10.1016/j.redox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Manjithaya R, Nazarko TY, Farre JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett 2010; 584:1367-73; http://dx.doi.org/ 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Till A, Lakhani R, Burnett SF, Subramani S. Pexophagy: the selective degradation of peroxisomes. Int J Cell Biol 2012; 2012:512721; http://dx.doi.org/ 10.1155/2012/512721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O'Rahilly S, et al.. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev 2006; 58:726-41; http://dx.doi.org/ 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- 773.Walter KM, Schonenberger MJ, Trotzmuller M, Horn M, Elsasser HP, Moser AB, Lucas MS, Schwarz T, Gerber PA, Faust PL, et al.. Hif-2alpha promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab 2014; 20:882-97; http://dx.doi.org/ 10.1016/j.cmet.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 774.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, Maclean KH, Inoki K, Guan K-L, Shen J, Person MD, et al.. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA 2010; 107:4153-8; http://dx.doi.org/ 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 775.Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, Wogan GN. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of mTORC1. Proc Natl Acad Sci USA 2013; 110:E2950-7; http://dx.doi.org/ 10.1073/pnas.1307736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 776.Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, et al.. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol 2013; 15:1186-96; http://dx.doi.org/ 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, et al.. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Luiken JJ, van den Berg M, Heikoop JC, Meijer AJ. Autophagic degradation of peroxisomes in isolated rat hepatocytes. FEBS Lett 1992; 304:93-7; http://dx.doi.org/ 10.1016/0014-5793(92)80596-9. [DOI] [PubMed] [Google Scholar]
- 779.Yokota S. Formation of autophagosomes during degradation of excess peroxisomes induced by administration of dioctyl phthalate. Eur J Cell Biol 1993; 61:67-80. [PubMed] [Google Scholar]
- 780.D'Eletto M, Farrace MG, Rossin F, Strappazzon F, Giacomo GD, Cecconi F, Melino G, Sepe S, Moreno S, Fimia GM, et al.. Type 2 transglutaminase is involved in the autophagy-dependent clearance of ubiquitinated proteins. Cell Death Differ 2012; 19:1228-38; http://dx.doi.org/ 10.1038/cdd.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 781.Nardacci R, Sartori C, Stefanini S. Selective autophagy of clofibrate-induced rat liver peroxisomes. Cytochemistry and immunocytochemistry on tissue specimens and on fractions obtained by Nycodenz density gradient centrifugation. Cell Mol Biol 2000; 46:1277-90. [PubMed] [Google Scholar]
- 782.Huybrechts SJ, Van Veldhoven PP, Brees C, Mannaerts GP, Los GV, Fransen M. Peroxisome dynamics in cultured mammalian cells. Traffic 2009; 10:1722-33; http://dx.doi.org/ 10.1111/j.1600-0854.2009.00970.x. [DOI] [PubMed] [Google Scholar]
- 783.Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, et al.. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 2013; 126:939-52; http://dx.doi.org/ 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 784.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol 2010; 189:671-9; http://dx.doi.org/ 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 785.Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al.. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2012; 2:120080; http://dx.doi.org/ 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 786.Yang KC, Ma X, Liu H, Murphy J, Barger PM, Mann DL, Diwan A. TNF-Receptor Associated Factor-2 Mediates Mitochondrial Autophagy. Circ Heart Fail 2014; 8:175-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet 2011; 20:1726-37; http://dx.doi.org/ 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 788.Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, et al.. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 2010; 15:887-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 789.Mauro-Lizcano . New method to assess mitophagy flux by flow cytometry. Autophagy 2015; 11:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 790.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J 2014; 33:282-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 791.Ivatt RM, Sanchez-Martinez A, Godena VK, Brown S, Ziviani E, Whitworth AJ. Genome-wide RNAi screen identifies the Parkinson disease GWAS risk locus SREBF1 as a regulator of mitophagy. Proc Natl Acad Sci USA 2014; 111:8494-9; http://dx.doi.org/ 10.1073/pnas.1321207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, Noguchi A, Springer D, Bocharov AV, Eggerman TL, et al.. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest 2011; 121:3701-12; http://dx.doi.org/ 10.1172/JCI44736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 793.Klinkenberg M, Gispert S, Dominguez-Bautista JA, Braun I, Auburger G, Jendrach M. Restriction of trophic factors and nutrients induces PARKIN expression. Neurogenetics 2012; 13:9-21; http://dx.doi.org/ 10.1007/s10048-011-0303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 794.Parganlija D, Klinkenberg M, Dominguez-Bautista J, Hetzel M, Gispert S, Chimi MA, Drose S, Mai S, Brandt U, Auburger G, et al.. Loss of PINK1 Impairs Stress-Induced Autophagy and Cell Survival. PloS One 2014; 9:e95288; http://dx.doi.org/ 10.1371/journal.pone.0095288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 795.Lyamzaev KG, Nepryakhina OK, Saprunova VB, Bakeeva LE, Pletjushkina OY, Chernyak BV, Skulachev VP. Novel mechanism of elimination of malfunctioning mitochondria (mitoptosis): formation of mitoptotic bodies and extrusion of mitochondrial material from the cell. Biochim Biophys Acta 2008; 1777:817-25; http://dx.doi.org/ 10.1016/j.bbabio.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 796.Davis CH, Kim KY, Bushong EA, Mills EA, Boassa D, Shih T, Kinebuchi M, Phan S, Zhou Y, Bihlmeyer NA, et al.. Transcellular degradation of axonal mitochondria. Proc Natl Acad Sci USA 2014; 111:9633-8; http://dx.doi.org/ 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 797.Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp Cell Res 2008; 314:3531-41; http://dx.doi.org/ 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 798.Ezaki J, Kominami E, Ueno T. Peroxisome degradation in mammals. IUBMB Life 2011; 63:1001-8; http://dx.doi.org/ 10.1002/iub.537. [DOI] [PubMed] [Google Scholar]
- 799.Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Phys 2008; 148:142-55; http://dx.doi.org/ 10.1104/pp.108.122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Phys 2009; 149:885-93; http://dx.doi.org/ 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 801.Michaeli S, Honig A, Levanony H, Peled-Zehavi H, Galili G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 2014; 26:4084-101; http://dx.doi.org/ 10.1105/tpc.114.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 802.Spitzer C, Li F, Buono R, Roschzttardtz H, Chung T, Zhang M, Osteryoung KW, Vierstra RD, Otegui MS. The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 2015; 27:391-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 803.Changou CA, Chen YR, Xing L, Yen Y, Chuang FY, Cheng RH, Bold RJ, Ann DK, Kung HJ. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc Natl Acad Sci USA 2014; 111:14147-52; http://dx.doi.org/ 10.1073/pnas.1404171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 804.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 805.Kurz T, Eaton JW, Brunk UT. The role of lysosomes in iron metabolism and recycling. Int J Biochem Cell Biol 2011; 43:1686-97; http://dx.doi.org/ 10.1016/j.biocel.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 806.Terman A, Kurz T. Lysosomal iron, iron chelation, and cell death. Antioxid Redox Sign 2013; 18:888-98; http://dx.doi.org/ 10.1089/ars.2012.4885. [DOI] [PubMed] [Google Scholar]
- 807.Asano T, Komatsu M, Yamaguchi-Iwai Y, Ishikawa F, Mizushima N, Iwai K. Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol Cell Biol 2011; 31:2040-52; http://dx.doi.org/ 10.1128/MCB.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Bauckman KA, Haller E, Flores I, Nanjundan M. Iron modulates cell survival in a Ras- and MAPK-dependent manner in ovarian cells. Cell Death Dis 2013; 4:e592; http://dx.doi.org/ 10.1038/cddis.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 809.De Domenico I, Ward DM, Kaplan J. Autophagy, ferritin and iron chelation. Autophagy 2010; 6:157; http://dx.doi.org/ 10.4161/auto.6.1.10587. [DOI] [PubMed] [Google Scholar]
- 810.Sturm B, Goldenberg H, Scheiber-Mojdehkar B. Transient increase of the labile iron pool in HepG2 cells by intravenous iron preparations. Eur J Biochem 2003; 270:3731-8; http://dx.doi.org/ 10.1046/j.1432-1033.2003.03759.x. [DOI] [PubMed] [Google Scholar]
- 811.Nagl W. ‘'Plastolysomes' - Plastids involved in the autolysis of the embryo-suspensor in Phaseolus. Zeitschrift Pflanzenphysiol 1977; 85:45-51; http://dx.doi.org/ 10.1016/S0044-328X(77)80263-8. [DOI] [Google Scholar]
- 812.Gartner PJ, Nagl W. Acid phosphatase activity in plastids (plastolysomes) of senescing embryo-suspensor cells. Planta 1980; 149:341-9; http://dx.doi.org/ 10.1007/BF00571168. [DOI] [PubMed] [Google Scholar]
- 813.van Doorn WG, Kirasak K, Sonong A, Srihiran Y, van Lent J, Ketsa S. Do plastids in Dendrobium cv. Lucky Duan petals function similar to autophagosomes and autolysosomes? Autophagy 2011; 7:584-97; http://dx.doi.org/ 10.4161/auto.7.6.15099. [DOI] [PubMed] [Google Scholar]
- 814.Parra-Vega V, Corral-Martínez P, Rivas-Sendra A, Segui-Simarro JM. Formation and excretion of autophagic plastids (plastolysomes) in Brassica napus embryogenic microspores. Front Plant Sci 2015; 6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 815.Gonzalez-Melendi P, Uyttewaal M, Morcillo CN, Hernandez Mora JR, Fajardo S, Budar F, Lucas MM. A light and electron microscopy analysis of the events leading to male sterility in Ogu-INRA CMS of rapeseed (Brassica napus). J Exp Bot 2008; 59:827-38; http://dx.doi.org/ 10.1093/jxb/erm365. [DOI] [PubMed] [Google Scholar]
- 816.Newcomb EH. Fine structure of protein-storing plastids in bean root tips. J Cell Biol 1967; 33:143-63; http://dx.doi.org/ 10.1083/jcb.33.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009; 458:1131-5; http://dx.doi.org/ 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 818.Koenig U, Fobker M, Lengauer B, Brandstetter M, Resch GP, Groger M, Plenz G, Pammer J, Barresi C, Hartmann C, et al.. Autophagy facilitates secretion and protects against degeneration of the Harderian gland. Autophagy 2015; 11:298-313; http://dx.doi.org/ 10.4161/15548627.2014.978221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 819.Shi Y, Han JJ, Tennakoon JB, Mehta FF, Merchant FA, Burns AR, Howe MK, McDonnell DP, Frigo DE. Androgens promote prostate cancer cell growth through induction of autophagy. Mol Endocrinol 2013; 27:280-95; http://dx.doi.org/ 10.1210/me.2012-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 820.O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab 2009; 10:430-5; http://dx.doi.org/ 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821.Inokuchi-Shimizu S, Park EJ, Roh YS, Yang L, Zhang B, Song J, Liang S, Pimienta M, Taniguchi K, Wu X, et al.. TAK1-mediated autophagy and fatty acid oxidation prevent hepatosteatosis and tumorigenesis. J Clin Invest 2014; 124:3566-78; http://dx.doi.org/ 10.1172/JCI74068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822.Lee JH, Budanov AV, Talukdar S, Park EJ, Park HL, Park HW, Bandyopadhyay G, Li N, Aghajan M, Jang I, et al.. Maintenance of metabolic homeostasis by Sestrin2 and Sestrin3. Cell Metab 2012; 16:311-21; http://dx.doi.org/ 10.1016/j.cmet.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 823.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al.. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 2013; 15:647-58; http://dx.doi.org/ 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 824.AM Cuervo. Preventing lysosomal fat indigestion. Nat Cell Biol 2013; 15:565-7; http://dx.doi.org/ 10.1038/ncb2778. [DOI] [PubMed] [Google Scholar]
- 825.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nature Rev Mol Cell Biol 2013; 14:283-96; http://dx.doi.org/ 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 826.Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci USA 2010; 107:16320-4; http://dx.doi.org/ 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 827.Heck MV, Azizov M, Stehning T, Walter M, Kedersha N, Auburger G. Dysregulated expression of lipid storage and membrane dynamics factors in Tia1 knockout mouse nervous tissue. Neurogenetics 2014; 15:135-44; http://dx.doi.org/ 10.1007/s10048-014-0397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 828.Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol 2012; 32:1733-44; http://dx.doi.org/ 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 829.Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 2013; 4:2111; http://dx.doi.org/ 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- 830.Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al.. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 2013; 32:2336-47; http://dx.doi.org/ 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 831.De Meyer GR, Grootaert MO, Michiels CF, Kurdi A, Schrijvers DM, Martinet W. Autophagy in vascular disease. Circ Res 2015; 116:468-79; http://dx.doi.org/ 10.1161/CIRCRESAHA.116.303804. [DOI] [PubMed] [Google Scholar]
- 832.Brown AJ, Jessup W. Oxysterols and atherosclerosis. Atherosclerosis 1999; 142:1-28; http://dx.doi.org/ 10.1016/S0021-9150(98)00196-8. [DOI] [PubMed] [Google Scholar]
- 833.He C, Zhu H, Zhang W, Okon I, Wang Q, Li H, Le YZ, Xie Z. 7-Ketocholesterol induces autophagy in vascular smooth muscle cells through Nox4 and Atg4B. Am J Pathol 2013; 183:626-37; http://dx.doi.org/ 10.1016/j.ajpath.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 834.Martinet W, Schrijvers DM, Timmermans JP, Bult H. Interactions between cell death induced by statins and 7-ketocholesterol in rabbit aorta smooth muscle cells. Br J Pharmacol 2008; 154:1236-46; http://dx.doi.org/ 10.1038/bjp.2008.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 835.Zarrouk A, Vejux A, Mackrill J, O'Callaghan Y, Hammami M, O'Brien N, Lizard G. Involvement of oxysterols in age-related diseases and ageing processes. Ageing Res Rev 2014; 18:148-62; http://dx.doi.org/ 10.1016/j.arr.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 836.Monier S, Samadi M, Prunet C, Denance M, Laubriet A, Athias A, Berthier A, Steinmetz E, Jurgens G, Negre-Salvayre A, et al.. Impairment of the cytotoxic and oxidative activities of 7 beta-hydroxycholesterol and 7-ketocholesterol by esterification with oleate. Biochem Biophys Res Commun 2003; 303:814-24; http://dx.doi.org/ 10.1016/S0006-291X(03)00412-1. [DOI] [PubMed] [Google Scholar]
- 837.Nury T, Zarrouk A, Mackrill JJ, Samadi M, Durand P, Riedinger JM, Doria M, Vejux A, Limagne E, Delmas D, et al.. Induction of oxiapoptophagy on 158N murine oligodendrocytes treated by 7-ketocholesterol-, 7beta-hydroxycholesterol-, or 24(S)-hydroxycholesterol: Protective effects of alpha-tocopherol and docosahexaenoic acid (DHA; C22:6 n-3). Steroids 2015; 99:194-203; http://dx.doi.org/ 10.1016/j.steroids.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 838.Nury T, Zarrouk A, Vejux A, Doria M, Riedinger JM, Delage-Mourroux R, Lizard G. Induction of oxiapoptophagy, a mixed mode of cell death associated with oxidative stress, apoptosis and autophagy, on 7-ketocholesterol-treated 158N murine oligodendrocytes: impairment by alpha-tocopherol. Biochem Biophys Res Commun 2014; 446:714-9; http://dx.doi.org/ 10.1016/j.bbrc.2013.11.081. [DOI] [PubMed] [Google Scholar]
- 839.Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015; 522:359-62; http://dx.doi.org/ 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 840.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem 2006; 281:30299-304; http://dx.doi.org/ 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 841.Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci 2014; 127:4078-88; http://dx.doi.org/ 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy 2007; 3:285-7; http://dx.doi.org/ 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 843.Klionsky DJ, Cuervo AM, Dunn WA Jr., Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy 2007; 3:413-6; http://dx.doi.org/ 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- 844.Bolender RP, Weibel ER. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of threatment. J Cell Biol 1973; 56:746-61; http://dx.doi.org/ 10.1083/jcb.56.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 845.Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al.. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015; 522:354-8; http://dx.doi.org/ 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 846.Lipatova Z, Segev N. A Role for Macro-ER-Phagy in ER Quality Control. PLoS Genet 2015; 11:e1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 847.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 2008; 10:602-10; http://dx.doi.org/ 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 848.Ossareh-Nazari B, Nino CA, Bengtson MH, Lee JW, Joazeiro CA, Dargemont C. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J Cell Biol 2014; 204:909-17; http://dx.doi.org/ 10.1083/jcb.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 849.Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA 2012; 109:15942-6; http://dx.doi.org/ 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 850.Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol 2012; 14:1314-21; http://dx.doi.org/ 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 851.Zhang P, Zhang H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep 2013; 14:568-76; http://dx.doi.org/ 10.1038/embor.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 852.Brown CR, Chiang H-L. A selective autophagy pathway that degrades gluconeogenic enzymes during catabolite inactivation. Commun Integr Biol 2009; 2:177-83; http://dx.doi.org/ 10.4161/cib.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 853.Schule T, Rose M, Entian KD, Thumm M, Wolf DH. Ubc8p functions in catabolite degradation of fructose-1, 6-bisphosphatase in yeast. EMBO J 2000; 19:2161-7; http://dx.doi.org/ 10.1093/emboj/19.10.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 854.Schork SM, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem 1995; 270:26446-50; http://dx.doi.org/ 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- 855.Regelmann J, Schule T, Josupeit FS, Horak J, Rose M, Entian KD, Thumm M, Wolf DH. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol Biol Cell 2003; 14:1652-63; http://dx.doi.org/ 10.1091/mbc.E02-08-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 856.Hung GC, Brown CR, Wolfe AB, Liu J, Chiang HL. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem 2004; 279:49138-50; http://dx.doi.org/ 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- 857.Chiang H-L, Schekman R, Hamamoto S. Selective uptake of cytosolic, peroxisomal, and plasma membrane proteins into the yeast lysosome for degradation. J Biol Chem 1996; 271:9934-41; http://dx.doi.org/ 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 858.Huang PH, Chiang H-L. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol 1997; 136:803-10; http://dx.doi.org/ 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 859.Alibhoy AA, Giardina BJ, Dunton DD, Chiang H-L. Vid30 is required for the association of Vid vesicles and actin patches in the vacuole import and degradation pathway. Autophagy 2012; 8:29-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 860.Brown CR, Wolfe AB, Cui D, Chiang H-L. The vacuolar import and degradation pathway merges with the endocytic pathway to deliver fructose-1,6-bisphosphatase to the vacuole for degradation. J Biol Chem 2008; 283:26116-27; http://dx.doi.org/ 10.1074/jbc.M709922200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 861.Chiang MC, Chiang H-L. Vid24p, a novel protein localized to the fructose-1, 6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J Cell Biol 1998; 140:1347-56; http://dx.doi.org/ 10.1083/jcb.140.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 862.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 1995; 128:779-92; http://dx.doi.org/ 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 863.Brown CR, Hung GC, Dunton D, Chiang H-L. The TOR complex 1 is distributed in endosomes and in retrograde vesicles that form from the vacuole membrane and plays an important role in the vacuole import and degradation pathway. J Biol Chem 2010; 285:23359-70; http://dx.doi.org/ 10.1074/jbc.M109.075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 864.Brown CR, Dunton D, Chiang H-L. The vacuole import and degradation pathway utilizes early steps of endocytosis and actin polymerization to deliver cargo proteins to the vacuole for degradation. J Biol Chem 2010; 285:1516-28; http://dx.doi.org/ 10.1074/jbc.M109.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 865.Webster P. Cytoplasmic bacteria and the autophagic pathway. Autophagy 2006; 2:159-61; http://dx.doi.org/ 10.4161/auto.2826. [DOI] [PubMed] [Google Scholar]
- 866.Dubuisson JF, Swanson MS. Mouse infection by Legionella, a model to analyze autophagy. Autophagy 2006; 2:179-82; http://dx.doi.org/ 10.4161/auto.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 867.Jordan TX, Randall G. Manipulation or capitulation: virus interactions with autophagy. Microbes Infect 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 868.Knodler LA, Celli J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol 2011; 13:1319-27; http://dx.doi.org/ 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 869.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323-35; http://dx.doi.org/ 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 870.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev 2011; 240:92-104; http://dx.doi.org/ 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 871.Dong X, Levine B. Autophagy and viruses: adversaries or allies? J Innate Immun 2013; 5:480-93; http://dx.doi.org/ 10.1159/000346388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 872.Wang C, Symington JW, Mysorekar IU. ATG16L1 and pathogenesis of urinary tract infections. Autophagy 2012; 8:1693-4; http://dx.doi.org/ 10.4161/auto.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 873.Choy A, Roy CR. Autophagy and bacterial infection: an evolving arms race. Trends Microbiol 2013; 21:451-6; http://dx.doi.org/ 10.1016/j.tim.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 874.Mostowy S, Cossart P. Bacterial autophagy: restriction or promotion of bacterial replication? Trends Cell Biol 2012; 22:283-91; http://dx.doi.org/ 10.1016/j.tcb.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 875.Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S, Imamoto F, Noda T, Yoshimori T. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell 2011; 22:2290-300; http://dx.doi.org/ 10.1091/mbc.E10-11-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 876.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012; 482:414-8; http://dx.doi.org/ 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 877.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 2009; 183:5909-16; http://dx.doi.org/ 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 878.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10:1215-21; http://dx.doi.org/ 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 879.Tumbarello DA, Manna PT, Allen M, Bycroft M, Arden SD, Kendrick-Jones J, Buss F. The autophagy receptor TAX1BP1 and the molecular motor myosin VI are required for clearance of Salmonella Typhimurium by autophagy. PLoS Pathog 2015;11:e1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 880.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al.. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011; 333:228-33; http://dx.doi.org/ 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 881.Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci USA 2009; 106:6226-31; http://dx.doi.org/ 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 882.Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol 2003; 5:455-68; http://dx.doi.org/ 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 883.Shahnazari S, Brumell JH. Mechanisms and consequences of bacterial targeting by the autophagy pathway. Current opinion in microbiology 2011; 14:68-75; http://dx.doi.org/ 10.1016/j.mib.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 884.Klionsky DJ, Eskelinen EL, Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I'm confused. Autophagy 2014; 10:549-51; http://dx.doi.org/ 10.4161/auto.28448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 885.Li X, Ye Y, Zhou X, Huang C, Wu M. Atg7 enhances host defense against infection via downregulation of superoxide but upregulation of nitric oxide. J Immunol 2015; 194:1112-21; http://dx.doi.org/ 10.4049/jimmunol.1401958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 886.Ye Y, Tan S, Zhou X, Li X, Jundt MC, Lichter N, Hidebrand A, Dhasarathy A, Wu M. Inhibition of p-IkappaBalpha Ubiquitylation by Autophagy-Related Gene 7 to Regulate Inflammatory Responses to Bacterial Infection. J Infect Dis 2015; 212:1816-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 887.Yuan K, Huang C, Fox J, Laturnus D, Carlson E, Zhang B, Yin Q, Gao H, Wu M. Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. J Cell Sci 2012; 125:507-15; http://dx.doi.org/ 10.1242/jcs.094573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 888.Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, Thomas BJ, Malosse C, Gantier MP, Casillas LN, et al.. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 2014; 15:623-35; http://dx.doi.org/ 10.1016/j.chom.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 889.Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nature reviews Immunology 2015; 15:375-87; http://dx.doi.org/ 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 890.McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z. Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. J Biol Chem 2011; 286:22147-59; http://dx.doi.org/ 10.1074/jbc.M110.192500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 891.Mao Y, Da L, Tang H, Yang J, Lei Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem Biophys Res Commun 2011; 415:68-74; http://dx.doi.org/ 10.1016/j.bbrc.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 892.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 2007; 1:23-35; http://dx.doi.org/ 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 893.Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol 2007; 81:12128-34; http://dx.doi.org/ 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 894.Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol 2009; 83:12164-71; http://dx.doi.org/ 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 895.Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe 2012; 12:334-45; http://dx.doi.org/ 10.1016/j.chom.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 896.Liang CEX, Jung JU. Downregulation of autophagy by herpesvirus Bcl-2 homologs. Autophagy 2008; 4:268-72; http://dx.doi.org/ 10.4161/auto.5210. [DOI] [PubMed] [Google Scholar]
- 897.Hernaez B, Cabezas M, Munoz-Moreno R, Galindo I, Cuesta-Geijo MA, Alonso C. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr Mol Med 2013; 13:305-16; http://dx.doi.org/ 10.2174/156652413804810736. [DOI] [PubMed] [Google Scholar]
- 898.Alonso C, Galindo I, Cuesta-Geijo MA, Cabezas M, Hernaez B, Munoz-Moreno R. African swine fever virus-cell interactions: from virus entry to cell survival. Virus Res 2013; 173:42-57; http://dx.doi.org/ 10.1016/j.virusres.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 899.Galindo I, Hernaez B, Diaz-Gil G, Escribano JM, Alonso C. A179L, a viral Bcl-2 homologue, targets the core Bcl-2 apoptotic machinery and its upstream BH3 activators with selective binding restrictions for Bid and Noxa. Virology 2008; 375:561-72; http://dx.doi.org/ 10.1016/j.virol.2008.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 900.Gannage M, Ramer PC, Munz C. Targeting Beclin 1 for viral subversion of macroautophagy. Autophagy 2010; 6:166-7; http://dx.doi.org/ 10.4161/auto.6.1.10624. [DOI] [PubMed] [Google Scholar]
- 901.MS Killian. Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Res Ther 2012; 9:16; http://dx.doi.org/ 10.1186/1742-6405-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 902.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, et al.. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 2009; 186:255-68; http://dx.doi.org/ 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 903.Nardacci R, Amendola A, Ciccosanti F, Corazzari M, Esposito V, Vlassi C, Taibi C, Fimia GM, Del Nonno F, Ippolito G, et al.. Autophagy plays an important role in the containment of HIV-1 in nonprogressor-infected patients. Autophagy 2014; 10:1167-78; http://dx.doi.org/ 10.4161/auto.28678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 904.Zhang H, Monken CE, Zhang Y, Lenard J, Mizushima N, Lattime EC, Jin S. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy 2006; 2:91-5; http://dx.doi.org/ 10.4161/auto.2.2.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 905.Heaton NS, Randall G. Dengue virus and autophagy. Viruses 2011; 3:1332-41; http://dx.doi.org/ 10.3390/v3081332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 906.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA 2009; 106:14046-51; http://dx.doi.org/ 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 907.Collins CA, De Maziere A, van Dijk S, Carlsson F, Klumperman J, Brown EJ. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog 2009; 5:e1000430; http://dx.doi.org/ 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 908.Moreau K, Lacas-Gervais S, Fujita N, Sebbane F, Yoshimori T, Simonet M, Lafont F. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell Microbiol 2010; 12:1108-23; http://dx.doi.org/ 10.1111/j.1462-5822.2010.01456.x. [DOI] [PubMed] [Google Scholar]
- 909.Grasso D, Ropolo A, Lo Re A, Boggio V, Molejon MI, Iovanna JL, Gonzalez CD, Urrutia R, Vaccaro MI. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem 2011; 286:8308-24; http://dx.doi.org/ 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 910.Gorbunov NV, Kiang JG. Autophagy-Mediated Innate Defense Mechanism in Crypt Paneth Cells Responding to Impairment of Small Intestine Barrier after Total-Body Gamma-Photon Irradiation In: Gorbunov NV, ed. Autophagy: Principles, Regulation and Roles in Disease. Hauppauge, NY: NOVA SCIENCE PUBLISHERS, INC., 2011:61-84. [Google Scholar]
- 911.Seglen PO, Gordon PB, Tolleshaug H, Høyvik H. Use of [3H]raffinose as a specific probe of autophagic sequestration. Exp Cell Res 1986; 162:273-7. [DOI] [PubMed] [Google Scholar]
- 912.Kopitz J, Kisen GO, Gordon PB, Bohley P, Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol 1990; 111:941-53; http://dx.doi.org/ 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 913.Gordon PB, Seglen PO. Autophagic sequestration of [14C]sucrose, introduced into rat hepatocytes by reversible electro-permeabilization. Exp Cell Res 1982; 142:1-14; http://dx.doi.org/ 10.1016/0014-4827(82)90402-5. [DOI] [PubMed] [Google Scholar]
- 914.Seglen PO, Luhr M, Mills IG, Saetre F, Szalai P, Engedal N. Macroautophagic cargo sequestration assays. Methods 2015; 75:25-36; http://dx.doi.org/ 10.1016/j.ymeth.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 915.Boland B, Smith DA, Mooney D, Jung SS, Walsh DM, Platt FM. Macroautophagy is not directly involved in the metabolism of amyloid precursor protein. J Biol Chem 2010; 285:37415-26; http://dx.doi.org/ 10.1074/jbc.M110.186411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 916.Nair U, Thumm M, Klionsky DJ, Krick R. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy 2011; 7:1546-50; http://dx.doi.org/ 10.4161/auto.7.12.18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 917.Plomp PJ, Gordon PB, Meijer AJ, Høyvik H, Seglen PO. Energy dependence of different steps in the autophagic-lysosomal pathway. J Biol Chem 1989; 264:6699-704. [PubMed] [Google Scholar]
- 918.Høyvik H, Gordon PB, Berg TO, Strømhaug PE, Seglen PO. Inhibition of autophagic-lysosomal delivery and autophagic lactolysis by asparagine. J Cell Biol 1991; 113:1305-12; http://dx.doi.org/ 10.1083/jcb.113.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 919.Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy 2006; 2:39-46; http://dx.doi.org/ 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 920.Lorenz H, Hailey DW, Lippincott-Schwartz J. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat Methods 2006; 3:205-10; http://dx.doi.org/ 10.1038/nmeth857. [DOI] [PubMed] [Google Scholar]
- 921.McNeil PL, Murphy RF, Lanni F, Taylor DL. A method for incorporating macromolecules into adherent cells. J Cell Biol 1984; 98:1556-64; http://dx.doi.org/ 10.1083/jcb.98.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 922.Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem 2002; 277:763-73; http://dx.doi.org/ 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 923.Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 2012; 23:896-909; http://dx.doi.org/ 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 924.Kovács AL, Laszlo L, Kovács J. Effect of amino acids and cycloheximide on changes caused by vinblastine, leupeptin and methylamine in the autophagic/lysosomal system of mouse hepatocytes in vivo. Exp Cell Res 1985; 157:83-94; http://dx.doi.org/ 10.1016/0014-4827(85)90154-5. [DOI] [PubMed] [Google Scholar]
- 925.Swanson MS, Byrne BG, Dubuisson JF. Kinetic analysis of autophagosome formation and turnover in primary mouse macrophages. Methods Enzymol 2009; 452:383-402; http://dx.doi.org/ 10.1016/S0076-6879(08)03623-9. [DOI] [PubMed] [Google Scholar]
- 926.Beugnet A, Tee AR, Taylor PM, Proud CG. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J 2003; 372:555-66; http://dx.doi.org/ 10.1042/bj20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 927.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al.. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 2007; 26:663-74; http://dx.doi.org/ 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 928.Jomain-Baum M, Garber AJ, Farber E, Hanson RW. The effect of cycloheximide on the interaction between mitochondrial respiration and gluconeogenesis in guinea pig and rat liver. J Biol Chem 1973; 248:1536-43. [PubMed] [Google Scholar]
- 929.Garber AJ, Jomain-Baum M, Salganicoff L, Farber E, Hanson RW. The effects of cycloheximide on energy transfer in rat and guinea pig liver mitochondria. J Biol Chem 1973; 248:1530-5. [PubMed] [Google Scholar]
- 930.Mora R, Dokic I, Kees T, Huber CM, Keitel D, Geibig R, Brugge B, Zentgraf H, Brady NR, Regnier-Vigouroux A. Sphingolipid rheostat alterations related to transformation can be exploited for specific induction of lysosomal cell death in murine and human glioma. Glia 2010; 58:1364-83. [DOI] [PubMed] [Google Scholar]
- 931.Bright NA, Lindsay MR, Stewart A, Luzio JP. The relationship between lumenal and limiting membranes in swollen late endocytic compartments formed after wortmannin treatment or sucrose accumulation. Traffic 2001; 2:631-42; http://dx.doi.org/ 10.1034/j.1600-0854.2001.20906.x. [DOI] [PubMed] [Google Scholar]
- 932.Deter RL. Quantitative characterization of dense body, autophagic vacuole, and acid phosphatase-bearing particle populations during the early phases of glucagon-induced autophagy in rat liver. J Cell Biol 1971; 48:473-89; http://dx.doi.org/ 10.1083/jcb.48.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 933.Deter RL. Analog modeling of glucagon-induced autophagy in rat liver. I. Conceptual and mathematical model of telolysosome-autophagosome-autolysosome interaction. Exp Cell Res 1975; 94:122-6; http://dx.doi.org/ 10.1016/0014-4827(75)90538-8. [DOI] [PubMed] [Google Scholar]
- 934.Deter RL. Analog modeling of glucagon-induced autophagy in rat liver. II. Evaluation of iron labeling as a means for identifying telolysosome, autophagosome and autolysosome populations. Exp Cell Res 1975; 94:127-39; http://dx.doi.org/ 10.1016/0014-4827(75)90539-X. [DOI] [PubMed] [Google Scholar]
- 935.Deter RL, Baudhuin P, de Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol 1967; 35:C11-6; http://dx.doi.org/ 10.1083/jcb.35.2.C11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 936.Deter RL, de Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol 1967; 33:437-49; http://dx.doi.org/ 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 937.Stromhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J 1998; 335 (Pt 2):217-24; http://dx.doi.org/ 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 938.Deter RL. Electron microscopic evaluation of subcellular fractions obtained by ultracentrifugation In: Hayat MA, ed. Principles and Techniques of Electron Microscopy. New York: Van Nostrand Reinhold Co, 1973:199-235. [Google Scholar]
- 939.Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol 1982; 93:144-54; http://dx.doi.org/ 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 940.Wattiaux R, Wattiaux-De Coninck S, Ronveaux-Dupal M-F, Dubois F. Isolation of rat liver lysosomes by isopycnic centrifugation in a metrizamide gradient. J Cell Biol 1978; 78:349-68; http://dx.doi.org/ 10.1083/jcb.78.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 941.Rodriguez-Navarro JA, Rodriguez L, Casarejos MJ, Solano RM, Gomez A, Perucho J, Cuervo AM, Garcia de Yebenes J, Mena MA. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neuobiol Dis 2010; 39:423-38; http://dx.doi.org/ 10.1016/j.nbd.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 942.Weibel ER, Bolender RP. Stereological techniques for electron microscopic morphometry In: Hayat MA, ed. Principles and Techniques of Electron Microscopy. New York: Van Nostrand Reinhold Co, 1973:237-96. [Google Scholar]
- 943.Baudhuin P, Evrard P, Berthet J. Electron microscopic examination of subcellular fractions. I. The preparation of representative samples from suspensions of particles. J Cell Biol 1967; 32:181-91; http://dx.doi.org/ 10.1083/jcb.32.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 944.Baudhuin P, Berthet J. Electron microscopic examination of subcellular fractions. II. Quantitative analysis of the mitochondrial population isolated from rat liver. J Cell Biol 1967; 35:631-48; http://dx.doi.org/ 10.1083/jcb.35.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 945.Storrie B, Madden EA. Isolation of subcellular organelles. Methods Enzymol 1990; 182:203-25; http://dx.doi.org/ 10.1016/0076-6879(90)82018-W. [DOI] [PubMed] [Google Scholar]
- 946.Balch WE, Rothman JE. Characterization of protein transport between successive compartments of the Golgi apparatus: asymmetric properties of donor and acceptor activities in a cell-free system. Arch Biochem Biophys 1985; 240:413-25; http://dx.doi.org/ 10.1016/0003-9861(85)90046-3. [DOI] [PubMed] [Google Scholar]
- 947.Graham JM. Isolation of lysosomes from tissues and cells by differential and density gradient centrifugation In: Bonifacino JS, Dasso M, Harfod JB, Lippincott-Schwartz J and Yamada KM, eds. Current Protocols in Cell Biology: John Wiley & Sons, Inc, 2000:Unit 3.6. [DOI] [PubMed] [Google Scholar]
- 948.Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem 1997; 272:5606-15; http://dx.doi.org/ 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 949.He C, Sumpter R Jr., Levine B. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy 2012; 8:1548-51; http://dx.doi.org/ 10.4161/auto.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 950.Sarkar C, Zhao Z, Aungst S, Sabirzhanov B, Faden AI, Lipinski MM. Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy 2014:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 951.Iwai-Kanai E, Yuan H, Huang C, Sayen MR, Perry-Garza CN, Kim L, Gottlieb RA. A method to measure cardiac autophagic flux in vivo. Autophagy 2008; 4:322-9; http://dx.doi.org/ 10.4161/auto.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 952.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest 2007; 117:1782-93; http://dx.doi.org/ 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 953.Castillo K, Valenzuela V, Matus S, Nassif M, Onate M, Fuentealba Y, Encina G, Irrazabal T, Parsons G, Court FA, et al.. Measurement of autophagy flux in the nervous system in vivo. Cell Death Dis 2013; 4:e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 954.Matus S, Valenzuela V, Hetz C. A new method to measure autophagy flux in the nervous system. Autophagy 2014; 10:710-4; http://dx.doi.org/ 10.4161/auto.28434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 955.Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G, Court FA, van Zundert B, Hetz C. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 2013; 9:1308-20; http://dx.doi.org/ 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 956.Chiarelli R, Agnello M, Roccheri MC. Sea urchin embryos as a model system for studying autophagy induced by cadmium stress. Autophagy 2011; 7:1028-34; http://dx.doi.org/ 10.4161/auto.7.9.16450. [DOI] [PubMed] [Google Scholar]
- 957.Morici G, Agnello M, Spagnolo F, Roccheri MC, Di Liegro CM, Rinaldi AM. Confocal microscopy study of the distribution, content and activity of mitochondria during Paracentrotus lividus development. J Microsc 2007; 228:165-73; http://dx.doi.org/ 10.1111/j.1365-2818.2007.01860.x. [DOI] [PubMed] [Google Scholar]
- 958.Martinet W, De Meyer GR, Andries L, Herman AG, Kockx MM. Detection of autophagy in tissue by standard immunohistochemistry: possibilities and limitations. Autophagy 2006; 2:55-7; http://dx.doi.org/ 10.4161/auto.2217. [DOI] [PubMed] [Google Scholar]
- 959.Holt SV, Wyspianska B, Randall KJ, James D, Foster JR, Wilkinson RW. The development of an immunohistochemical method to detect the autophagy-associated protein LC3-II in human tumor xenografts. Toxicol Pathol 2011; 39:516-23; http://dx.doi.org/ 10.1177/0192623310396903. [DOI] [PubMed] [Google Scholar]
- 960.Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol 2009; 452:1-12; http://dx.doi.org/ 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 961.Dehay B, Bove J, Rodriguez-Muela N, Perier C, Recasens A, Boya P, Vila M. Pathogenic lysosomal depletion in Parkinson's disease. J Neurosci 2010; 30:12535-44; http://dx.doi.org/ 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 962.Daniels BH, McComb RD, Mobley BC, Gultekin SH, Lee HS, Margeta M. LC3 and p62 as diagnostic markers of drug-induced autophagic vacuolar cardiomyopathy: a study of 3 cases. Am J Surg Pathol 2013; 37:1014-21; http://dx.doi.org/ 10.1097/PAS.0b013e3182863fa8. [DOI] [PubMed] [Google Scholar]
- 963.Hiniker A, Daniels BH, Lee HS, Margeta M. Comparative utility of LC3, p62 and TDP-43 immunohistochemistry in differentiation of inclusion body myositis from polymyositis and related inflammatory myopathies. Acta Neuropathol Commun 2013; 1:29; http://dx.doi.org/ 10.1186/2051-5960-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 964.Lee HS, Daniels BH, Salas E, Bollen AW, Debnath J, Margeta M. Clinical utility of LC3 and p62 immunohistochemistry in diagnosis of drug-induced autophagic vacuolar myopathies: a case-control study. PloS One 2012; 7:e36221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 965.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab 2011; 13:655-67; http://dx.doi.org/ 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 966.Hamada K, Terauchi A, Nakamura K, Higo T, Nukina N, Matsumoto N, Hisatsune C, Nakamura T, Mikoshiba K. Aberrant calcium signaling by transglutaminase-mediated posttranslational modification of inositol 1,4,5-trisphosphate receptors. Proc Natl Acad Sci USA 2014; 111:E3966-75; http://dx.doi.org/ 10.1073/pnas.1409730111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 967.Rodriguez-Muela N, Koga H, Garcia-Ledo L, de la Villa P, de la Rosa EJ, Cuervo AM, Boya P. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell 2013; 12:478-88; http://dx.doi.org/ 10.1111/acel.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 968.Esteban-Martinez L, Boya P. Autophagic flux determination in vivo and ex vivo. Methods 2015; 75:79-86; http://dx.doi.org/ 10.1016/j.ymeth.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 969.McMahon J, Huang X, Yang J, Komatsu M, Yue Z, Qian J, Zhu X, Huang Y. Impaired autophagy in neurons after disinhibition of mammalian target of rapamycin and its contribution to epileptogenesis. J Neurosci 2012; 32:15704-14; http://dx.doi.org/ 10.1523/JNEUROSCI.2392-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 970.Herrando-Grabulosa M, Casas C, Aguilera J. The C-terminal domain of tetanus toxin protects motoneurons against acute excitotoxic damage on spinal cord organotypic cultures. J Neurochem 2013; 124:36-44; http://dx.doi.org/ 10.1111/jnc.12062. [DOI] [PubMed] [Google Scholar]
- 971.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 2011; 13:589-98; http://dx.doi.org/ 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 972.Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, Blaauw B, Urciuolo A, Tiepolo T, Merlini L, et al.. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med 2010; 16:1313-20; http://dx.doi.org/ 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 973.Bloemberg D, McDonald E, Dulay D, Quadrilatero J. Autophagy is altered in skeletal and cardiac muscle of spontaneously hypertensive rats. Acta Physiol (Oxf) 2014; 210:381-91; http://dx.doi.org/ 10.1111/apha.12178. [DOI] [PubMed] [Google Scholar]
- 974.Ogata T, Oishi Y, Higuchi M, Muraoka I. Fasting-related autophagic response in slow- and fast-twitch skeletal muscle. Biochem Biophys Res Commun 2010; 394:136-40; http://dx.doi.org/ 10.1016/j.bbrc.2010.02.130. [DOI] [PubMed] [Google Scholar]
- 975.Yamada E, Bastie CC, Koga H, Wang Y, Cuervo AM, Pessin JE. Mouse skeletal muscle fiber-type-specific macroautophagy and muscle wasting are regulated by a Fyn/STAT3/Vps34 signaling pathway. Cell Rep 2012; 1:557-69; http://dx.doi.org/ 10.1016/j.celrep.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 976.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, et al.. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012; 481:511-5; http://dx.doi.org/ 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 977.Haspel J, Shaik RS, Ifedigbo E, Nakahira K, Dolinay T, Englert JA, Choi AM. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy 2011; 7:629-42; http://dx.doi.org/ 10.4161/auto.7.6.15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 978.Bell RM, Mocanu MM, Yellon DM. Retrograde heart perfusion: the Langendorff technique of isolated heart perfusion. J Mol Cell Cardiol 2011; 50:940-50; http://dx.doi.org/ 10.1016/j.yjmcc.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 979.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PloS One 2011; 6:e20975; http://dx.doi.org/ 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 980.Gottlieb RA, Finley KD, Mentzer RM Jr.. Cardioprotection requires taking out the trash. Basic Res Cardiol 2009; 104:169-80; http://dx.doi.org/ 10.1007/s00395-009-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 981.Avagliano L, Virgili E, Garo C, Quadrelli F, Doi P, Samaja M, Bulfamante GP, Marconi AM. Autophagy and human parturition: evaluation of LC3 expression in placenta from spontaneous or medically induced onset of labor. BioMed Res Intl 2013; 2013:689768; http://dx.doi.org/ 10.1155/2013/689768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 982.Hung TH, Hsieh TT, Chen SF, Li MJ, Yeh YL. Autophagy in the human placenta throughout gestation. PloS One 2013; 8:e83475; http://dx.doi.org/ 10.1371/journal.pone.0083475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 983.Signorelli P, Avagliano L, Virgili E, Gagliostro V, Doi P, Braidotti P, Bulfamante GP, Ghidoni R, Marconi AM. Autophagy in term normal human placentas. Placenta 2011; 32:482-5; http://dx.doi.org/ 10.1016/j.placenta.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 984.Hung TH, Chen SF, Lo LM, Li MJ, Yeh YL, Hsieh TT. Increased autophagy in placentas of intrauterine growth-restricted pregnancies. PloS One 2012; 7:e40957; http://dx.doi.org/ 10.1371/journal.pone.0040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 985.Chang YL, Wang TH, Chang SD, Chao AS, Hsieh PC, Wang CN. Increased autophagy in the placental territory of selective intrauterine growth-restricted monochorionic twins. Prenatal Diag 2013; 33:187-90; http://dx.doi.org/ 10.1002/pd.4040. [DOI] [PubMed] [Google Scholar]
- 986.Oh SY, Choi SJ, Kim KH, Cho EY, Kim JH, Roh CR. Autophagy-related proteins, LC3 and Beclin-1, in placentas from pregnancies complicated by preeclampsia. Reprod Sci 2008; 15:912-20; http://dx.doi.org/ 10.1177/1933719108319159. [DOI] [PubMed] [Google Scholar]
- 987.Avagliano L, Danti L, Doi P, Felis S, Guala M, Locatelli A, Maffeo I, Mecacci F, Plevani C, Simeone S, et al.. Autophagy in placentas from acidotic newborns: an immunohistochemical study of LC3 expression. Placenta 2013; 34:1091-4; http://dx.doi.org/ 10.1016/j.placenta.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 988.Perry CN, Kyoi S, Hariharan N, Takagi H, Sadoshima J, Gottlieb RA. Novel methods for measuring cardiac autophagy in vivo. Methods Enzymol 2009; 453:325-42; http://dx.doi.org/ 10.1016/S0076-6879(08)04016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 989.Munafo DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci 2001; 114:3619-29. [DOI] [PubMed] [Google Scholar]
- 990.Carloni S, Buonocore G, Balduini W. Protective role of autophagy in neonatal hypoxia-ischemia induced brain injury. Neuobiol Dis 2008; 32:329-39; http://dx.doi.org/ 10.1016/j.nbd.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 991.Carloni S, Girelli S, Scopa C, Buonocore G, Longini M, Balduini W. Activation of autophagy and Akt/CREB signaling play an equivalent role in the neuroprotective effect of rapamycin in neonatal hypoxia-ischemia. Autophagy 2010; 6:366-77; http://dx.doi.org/ 10.4161/auto.6.3.11261. [DOI] [PubMed] [Google Scholar]
- 992.Carloni S, Albertini MC, Galluzzi L, Buonocore G, Proietti F, Balduini W. Increased autophagy reduces endoplasmic reticulum stress after neonatal hypoxia-ischemia: Role of protein synthesis and autophagic pathways. Exp Neurol 2014; 57:192-9. [DOI] [PubMed] [Google Scholar]
- 993.Ginet V, Puyal J, Clarke PG, Truttmann AC. Enhancement of autophagic flux after neonatal cerebral hypoxia-ischemia and its region-specific relationship to apoptotic mechanisms. Am J Pathol 2009; 175:1962-74; http://dx.doi.org/ 10.2353/ajpath.2009.090463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 994.Puyal J, Vaslin A, Mottier V, Clarke PG. Postischemic treatment of neonatal cerebral ischemia should target autophagy. Annals Neurol 2009; 66:378-89; http://dx.doi.org/ 10.1002/ana.21714. [DOI] [PubMed] [Google Scholar]
- 995.Penas C, Font-Nieves M, Fores J, Petegnief V, Planas A, Navarro X, Casas C. Autophagy, and BiP level decrease are early key events in retrograde degeneration of motoneurons. Cell Death Differ 2011; 18:1617-27; http://dx.doi.org/ 10.1038/cdd.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 996.Y Uchiyama. Autophagic cell death and its execution by lysosomal cathepsins. Arch Histol Cytol 2001; 64:233-46; http://dx.doi.org/ 10.1679/aohc.64.233. [DOI] [PubMed] [Google Scholar]
- 997.Udelnow A, Kreyes A, Ellinger S, Landfester K, Walther P, Klapperstueck T, Wohlrab J, Henne-Bruns D, Knippschild U, Wurl P. Omeprazole inhibits proliferation and modulates autophagy in pancreatic cancer cells. PloS One 2011; 6:e20143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 998.Weber SM, Levitz SM. Chloroquine interferes with lipopolysaccharide-induced TNF-alpha gene expression by a nonlysosomotropic mechanism. J Immunol 2000; 165:1534-40; http://dx.doi.org/ 10.4049/jimmunol.165.3.1534. [DOI] [PubMed] [Google Scholar]
- 999.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005; 307:593-6; http://dx.doi.org/ 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 1000.Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J 2011; 30: 4642-51; http://dx.doi.org/ 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1001.Akagi Y, Isaka Y, Akagi A, Ikawa M, Takenaka M, Moriyama T, Yamauchi A, Horio M, Ueda N, Okabe M, et al.. Transcriptional activation of a hybrid promoter composed of cytomegalovirus enhancer and beta-actin/beta-globin gene in glomerular epithelial cells in vivo. Kidney Int 1997; 51:1265-9; http://dx.doi.org/ 10.1038/ki.1997.172. [DOI] [PubMed] [Google Scholar]
- 1002.Kimura T, Takabatake Y, Takahashi A, Kaimori JY, Matsui I, Namba T, Kitamura H, Niimura F, Matsusaka T, Soga T, et al.. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol 2011; 22:902-13; http://dx.doi.org/ 10.1681/ASN.2010070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1003.Hartleben B, Godel M, Meyer-Schwesinger C, Liu S, Ulrich T, Kobler S, Wiech T, Grahammer F, Arnold SJ, Lindenmeyer MT, et al.. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest 2010; 120:1084-96; http://dx.doi.org/ 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1004.Vandrovcova J, Anaya F, Kay V, Lees A, Hardy J, de Silva R. Disentangling the role of the tau gene locus in sporadic tauopathies. Curr Alzheimer Res 2010; 7:726-34; http://dx.doi.org/ 10.2174/156720510793611619. [DOI] [PubMed] [Google Scholar]
- 1005.Chen YS, Chen SD, Wu CL, Huang SS, Yang DI. Induction of sestrin2 as an endogenous protective mechanism against amyloid beta-peptide neurotoxicity in primary cortical culture. Exp Neurol 2014; 253:63-71; http://dx.doi.org/ 10.1016/j.expneurol.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 1006.Tofaris GK, Spillantini MG. Physiological and pathological properties of alpha-synuclein. Cell Mol Life Sci 2007; 64:2194-201; http://dx.doi.org/ 10.1007/s00018-007-7217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1007.EE Wanker. Protein aggregation and pathogenesis of Huntington's disease: mechanisms and correlations. Biol Chem 2000; 381:937-42. [DOI] [PubMed] [Google Scholar]
- 1008.Sandri M, Coletto L, Grumati P, Bonaldo P. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J Cell Sci 2013; 126:5325-33; http://dx.doi.org/ 10.1242/jcs.114041. [DOI] [PubMed] [Google Scholar]
- 1009.Bentmann E, Haass C, Dormann D. Stress granules in neurodegeneration–lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J 2013; 280:4348-70; http://dx.doi.org/ 10.1111/febs.12287. [DOI] [PubMed] [Google Scholar]
- 1010.Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: much more than mitophagy. Trends in Neurosci 2014; 37:315-24; http://dx.doi.org/ 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1011.Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, Dargemont C. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep 2010; 11:548-54; http://dx.doi.org/ 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1012.Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 2014; 17:17-23; http://dx.doi.org/ 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1013.Ebrahimi-Fakhari D, Wahlster L, Hoffmann GF, Kolker S. Emerging role of autophagy in pediatric neurodegenerative and neurometabolic diseases. Pediatr Res 2014; 75:217-26; http://dx.doi.org/ 10.1038/pr.2013.185. [DOI] [PubMed] [Google Scholar]
- 1014.Lee KM, Hwang SK, Lee JA. Neuronal autophagy and neurodevelopmental disorders. Exp Neurobiol 2013; 22:133-42; http://dx.doi.org/ 10.5607/en.2013.22.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1015.Yasin SA, Ali AM, Tata M, Picker SR, Anderson GW, Latimer-Bowman E, Nicholson SL, Harkness W, Cross JH, Paine SM, et al.. mTOR-dependent abnormalities in autophagy characterize human malformations of cortical development: evidence from focal cortical dysplasia and tuberous sclerosis. Acta Neuropathol 2013; 126:207-18; http://dx.doi.org/ 10.1007/s00401-013-1135-4. [DOI] [PubMed] [Google Scholar]
- 1016.Salminen A, Kaarniranta K, Haapasalo A, Hiltunen M, Soininen H, Alafuzoff I. Emerging role of p62/sequestosome-1 in the pathogenesis of Alzheimer's disease. Prog Neurobiol 2012; 96:87-95; http://dx.doi.org/ 10.1016/j.pneurobio.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 1017.Seidel K, Brunt ER, de Vos RA, Dijk F, van der Want HJ, Rub U, den Dunnen WF. The p62 antibody reveals various cytoplasmic protein aggregates in spinocerebellar ataxia type 6. Clin Neuropathol 2009; 28:344-9; http://dx.doi.org/ 10.5414/NPP28344. [DOI] [PubMed] [Google Scholar]
- 1018.Harada H, Warabi E, Matsuki T, Yanagawa T, Okada K, Uwayama J, Ikeda A, Nakaso K, Kirii K, Noguchi N, et al.. Deficiency of p62/Sequestosome 1 causes hyperphagia due to leptin resistance in the brain. J Neurosci 2013; 33:14767-77; http://dx.doi.org/ 10.1523/JNEUROSCI.2954-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1019.Merenlender-Wagner A, Malishkevich A, Shemer Z, Udawela M, Gibbons A, Scarr E, Dean B, Levine J, Agam G, Gozes I. Autophagy has a key role in the pathophysiology of schizophrenia. Mol Psychiatr 2015; 20:126-32; http://dx.doi.org/ 10.1038/mp.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1020.Dresner E, Agam G, Gozes I. Activity-dependent neuroprotective protein (ADNP) expression level is correlated with the expression of the sister protein ADNP2: deregulation in schizophrenia. Eur Neuropsychopharm 2011; 21:355-61; http://dx.doi.org/ 10.1016/j.euroneuro.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 1021.Nishino I. Autophagic vacuolar myopathy. Semin Pediatr Neurol 2006; 13:90-5; http://dx.doi.org/ 10.1016/j.spen.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 1022.Girolamo F, Lia A, Amati A, Strippoli M, Coppola C, Virgintino D, Roncali L, Toscano A, Serlenga L, Trojano M. Overexpression of autophagic proteins in the skeletal muscle of sporadic inclusion body myositis. Neuropath Appl Neuro 2013; 39:736-49; http://dx.doi.org/ 10.1111/nan.12040. [DOI] [PubMed] [Google Scholar]
- 1023.Temiz P, Weihl CC, Pestronk A. Inflammatory myopathies with mitochondrial pathology and protein aggregates. J Neurol Sci 2009; 278:25-9; http://dx.doi.org/ 10.1016/j.jns.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 1024.Maugeri N, Campana L, Gavina M, Covino C, De Metrio M, Panciroli C, Maiuri L, Maseri A, D'Angelo A, Bianchi ME, et al.. Activated platelets present high mobility group box 1 to neutrophils, inducing autophagy and promoting the extrusion of neutrophil extracellular traps. J Thromb Haemost 2014; 12:2074-88; http://dx.doi.org/ 10.1111/jth.12710. [DOI] [PubMed] [Google Scholar]
- 1025.Screen M, Raheem O, Holmlund-Hampf J, Jonson PH, Huovinen S, Hackman P, Udd B. Gene expression profiling in tibial muscular dystrophy reveals unfolded protein response and altered autophagy. PloS One 2014; 9:e90819; http://dx.doi.org/ 10.1371/journal.pone.0090819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1026.Brady S, Squier W, Sewry C, Hanna M, Hilton-Jones D, Holton JL. A retrospective cohort study identifying the principal pathological features useful in the diagnosis of inclusion body myositis. BMJ Open 2014; 4:e004552; http://dx.doi.org/ 10.1136/bmjopen-2013-004552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1027.Lin NY, Beyer C, Giessl A, Kireva T, Scholtysek C, Uderhardt S, Munoz LE, Dees C, Distler A, Wirtz S, et al.. Autophagy regulates TNFalpha-mediated joint destruction in experimental arthritis. Ann Rheum Dis 2013; 72:761-8; http://dx.doi.org/ 10.1136/annrheumdis-2012-201671. [DOI] [PubMed] [Google Scholar]
- 1028.Lin NY, Stefanica A, Distler JH. Autophagy: a key pathway of TNF-induced inflammatory bone loss. Autophagy 2013; 9:1253-5; http://dx.doi.org/ 10.4161/auto.25467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1029.Tchetina EV, Poole AR, Zaitseva EM, Sharapova EP, Kashevarova NG, Taskina EA, Alekseeva LI, Semyonova LA, Glukhova SI, Kuzin AN, et al.. Differences in Mammalian target of rapamycin gene expression in the peripheral blood and articular cartilages of osteoarthritic patients and disease activity. Arthritis 2013; 2013:461486; http://dx.doi.org/ 10.1155/2013/461486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1030.Mitroulis I, Kourtzelis I, Kambas K, Chrysanthopoulou A, Ritis K. Evidence for the involvement of mTOR inhibition and basal autophagy in familial Mediterranean fever phenotype. Hum Immunol 2011; 72:135-8; http://dx.doi.org/ 10.1016/j.humimm.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 1031.Bachetti T, Chiesa S, Castagnola P, Bani D, Di Zanni E, Omenetti A, D'Osualdo A, Fraldi A, Ballabio A, Ravazzolo R, et al.. Autophagy contributes to inflammation in patients with TNFR-associated periodic syndrome (TRAPS). Ann Rheum Dis 2013; 72:1044-52; http://dx.doi.org/ 10.1136/annrheumdis-2012-201952. [DOI] [PubMed] [Google Scholar]
- 1032.Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, De Rycke R, Noppen S, Delforge M, Willems J, Vandenabeele P. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res 2011; 21:290-304; http://dx.doi.org/ 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1033.Mitroulis I, Kambas K, Chrysanthopoulou A, Skendros P, Apostolidou E, Kourtzelis I, Drosos GI, Boumpas DT, Ritis K. 2011. Neutrophil extracellular trap formation is associated with IL-1beta and autophagy-related signaling in gout. PLoS One. 6, e29318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1034.Kambas K, Mitroulis I, Apostolidou E, Girod A, Chrysanthopoulou A, Pneumatikos I, Skendros P, Kourtzelis I, Koffa M, Kotsianidis I, et al.. Autophagy mediates the delivery of thrombogenic tissue factor to neutrophil extracellular traps in human sepsis. PloS One 2012; 7:e45427; http://dx.doi.org/ 10.1371/journal.pone.0045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1035.Chrysanthopoulou A, Mitroulis I, Apostolidou E, Arelaki S, Mikroulis D, Konstantinidis T, Sivridis E, Koffa M, Giatromanolaki A, Boumpas DT, et al.. Neutrophil extracellular traps promote differentiation and function of fibroblasts. J Pathol 2014; 233:294-307; http://dx.doi.org/ 10.1002/path.4359. [DOI] [PubMed] [Google Scholar]
- 1036.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J 2011; 30:4701-11; http://dx.doi.org/ 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1037.Kambas K, Chrysanthopoulou A, Vassilopoulos D, Apostolidou E, Skendros P, Girod A, Arelaki S, Froudarakis M, Nakopoulou L, Giatromanolaki A, et al.. Tissue factor expression in neutrophil extracellular traps and neutrophil derived microparticles in antineutrophil cytoplasmic antibody associated vasculitis may promote thromboinflammation and the thrombophilic state associated with the disease. Ann Rheum Dis 2013. [DOI] [PubMed] [Google Scholar]
- 1038.Masini M, Bugliani M, Lupi R, del Guerra S, Boggi U, Filipponi F, Marselli L, Masiello P, Marchetti P. Autophagy in human type 2 diabetes pancreatic beta cells. Diabetologia 2009; 52:1083-6; http://dx.doi.org/ 10.1007/s00125-009-1347-2. [DOI] [PubMed] [Google Scholar]
- 1039.Mizukami H, Takahashi K, Inaba W, Tsuboi K, Osonoi S, Yoshida T, Yagihashi S. Involvement of oxidative stress-induced DNA damage, endoplasmic reticulum stress, and autophagy deficits in the decline of beta-cell mass in Japanese type 2 diabetic patients. Diabetes Care 2014; 37:1966-74; http://dx.doi.org/ 10.2337/dc13-2018. [DOI] [PubMed] [Google Scholar]
- 1040.Ost A, Svensson K, Ruishalme I, Brannmark C, Franck N, Krook H, Sandstrom P, Kjolhede P, Stralfors P. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med 2010; 16:235-46; http://dx.doi.org/ 10.2119/molmed.2010.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1041.Kosacka J, Kern M, Kloting N, Paeschke S, Rudich A, Haim Y, Gericke M, Serke H, Stumvoll M, Bechmann I, et al.. Autophagy in adipose tissue of patients with obesity and type 2 diabetes. Mol Cell Endocrinol 2015; 409:21-32; http://dx.doi.org/ 10.1016/j.mce.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 1042.Stienstra R, Haim Y, Riahi Y, Netea M, Rudich A, Leibowitz G. Autophagy in adipose tissue and the beta cell: implications for obesity and diabetes. Diabetologia 2014; 57:1505-16; http://dx.doi.org/ 10.1007/s00125-014-3255-3. [DOI] [PubMed] [Google Scholar]
- 1043.Berton G. Editorial: Gigantism: a new way to prolong neutrophil life. J Leukocyte Biol 2014; 96:505-6; http://dx.doi.org/ 10.1189/jlb.3CE0214-107R. [DOI] [PubMed] [Google Scholar]
- 1044.Dyugovskaya L, Berger S, Polyakov A, Lavie L. The development of giant phagocytes in long-term neutrophil cultures. J Leukocyte Biol 2014; 96:511-21; http://dx.doi.org/ 10.1189/jlb.0813437. [DOI] [PubMed] [Google Scholar]
- 1045.Galluzzi L, Kepp O, Kroemer G. Enlightening the impact of immunogenic cell death in photodynamic cancer therapy. EMBO J 2012; 31:1055-7; http://dx.doi.org/ 10.1038/emboj.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1046.Panzarini E, Inguscio V, Fimia GM, Dini L. Rose Bengal Acetate PhotoDynamic Therapy (RBAc-PDT) induces exposure and release of damage-associated molecular patterns (DAMPs) in human HeLa cells. PloS One 2014; 9:e105778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1047.Santin G, Bottone MG, Malatesta M, Scovassi AI, Bottiroli G, Pellicciari C, Croce AC. Regulated forms of cell death are induced by the photodynamic action of the fluorogenic substrate, Hypocrellin B-acetate. J Photochem Photobiol B 2013; 125:90-7; http://dx.doi.org/ 10.1016/j.jphotobiol.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 1048.Maes H, Rubio N, Garg AD, Agostinis P. Autophagy: shaping the tumor microenvironment and therapeutic response. Trends Mol Med 2013; 19:428-46; http://dx.doi.org/ 10.1016/j.molmed.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 1049.Garg AD, Krysko DV, Vandenabeele P, Agostinis P. The emergence of phox-ER stress induced immunogenic apoptosis. Oncoimmunology 2012; 1:786-8; http://dx.doi.org/ 10.4161/onci.19750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1050.Garg AD, Martin S, Golab J, Agostinis P. Danger signalling during cancer cell death: origins, plasticity and regulation. Cell Death Differ 2014; 21:26-38; http://dx.doi.org/ 10.1038/cdd.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1051.Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013; 31:51-72; http://dx.doi.org/ 10.1146/annurev-immunol-032712-100008. [DOI] [PubMed] [Google Scholar]
- 1052.Dudek AM, Garg AD, Krysko DV, De Ruysscher D, Agostinis P. Inducers of immunogenic cancer cell death. Cytokine Growth Fact Rev 2013; 24:319-33; http://dx.doi.org/ 10.1016/j.cytogfr.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 1053.Garg AD, Dudek AM, Ferreira GB, Verfaillie T, Vandenabeele P, Krysko DV, Mathieu C, Agostinis P. ROS-induced autophagy in cancer cells assists in evasion from determinants of immunogenic cell death. Autophagy 2013; 9:1292-307; http://dx.doi.org/ 10.4161/auto.25399. [DOI] [PubMed] [Google Scholar]
- 1054.Garg AD, Krysko DV, Verfaillie T, Kaczmarek A, Ferreira GB, Marysael T, Rubio N, Firczuk M, Mathieu C, Roebroek AJ, et al.. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J 2012; 31:1062-79; http://dx.doi.org/ 10.1038/emboj.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1055.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, et al.. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 2011; 334:1573-7; http://dx.doi.org/ 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 1056.Bian S, Sun X, Bai A, Zhang C, Li L, Enjyoji K, Junger WG, Robson SC, Wu Y. P2X7 integrates PI3K/AKT and AMPK-PRAS40-mTOR signaling pathways to mediate tumor cell death. PloS One 2013; 8:e60184; http://dx.doi.org/ 10.1371/journal.pone.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1057.Di Virgilio F. Liaisons dangereuses: P2X(7) and the inflammasome. Trends Pharmacol Sci 2007; 28:465-72; http://dx.doi.org/ 10.1016/j.tips.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 1058.Garg AD, Dudek AM, Agostinis P. Calreticulin surface exposure is abrogated in cells lacking, chaperone-mediated autophagy-essential gene, LAMP2A. Cell Death Dis 2013; 4:e826; http://dx.doi.org/ 10.1038/cddis.2013.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1059.Garg AD, Dudek AM, Agostinis P. Autophagy-dependent suppression of cancer immunogenicity and effector mechanisms of innate and adaptive immunity. Oncoimmunology 2013; 2:e26260; http://dx.doi.org/ 10.4161/onci.26260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1060.Hermans G, Casaer MP, Clerckx B, Guiza F, Vanhullebusch T, Derde S, Meersseman P, Derese I, Mesotten D, Wouters PJ, et al.. Effect of tolerating macronutrient deficit on the development of intensive-care unit acquired weakness: a subanalysis of the EPaNIC trial. Lancet Resp Med 2013; 1:621-9; http://dx.doi.org/ 10.1016/S2213-2600(13)70183-8. [DOI] [PubMed] [Google Scholar]
- 1061.Vanhorebeek I, Gunst J, Derde S, Derese I, Boussemaere M, Guiza F, Martinet W, Timmermans JP, D'Hoore A, Wouters PJ, et al.. Insufficient activation of autophagy allows cellular damage to accumulate in critically ill patients. J Clin Endocrinol Metab 2011; 96:E633-45; http://dx.doi.org/ 10.1210/jc.2010-2563. [DOI] [PubMed] [Google Scholar]
- 1062.Czaja MJ, Ding WX, Donohue TM Jr., Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH, et al.. Functions of autophagy in normal and diseased liver. Autophagy 2013; 9:1131-58; http://dx.doi.org/ 10.4161/auto.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1063.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 2012; 142:938-46; http://dx.doi.org/ 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1064.Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy 2012; 8:719-30; http://dx.doi.org/ 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1065.Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cortes E, Honig LS, Dauer W, Consiglio A, et al.. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci 2013; 16:394-406; http://dx.doi.org/ 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1066.Napolitano G, Johnson JL, He J, Rocca CJ, Monfregola J, Pestonjamasp K, Cherqui S, Catz SD. Impairment of chaperone-mediated autophagy leads to selective lysosomal degradation defects in the lysosomal storage disease cystinosis. EMBO Mol Med 2015; 7:158-74; http://dx.doi.org/ 10.15252/emmm.201404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1067.Venugopal B, Mesires NT, Kennedy JC, Curcio-Morelli C, Laplante JM, Dice JF, Slaugenhaupt SA. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol 2009; 219:344-53; http://dx.doi.org/ 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- 1068.Franch HA. Pathways of proteolysis affecting renal cell growth. Curr Opin Nephrol Hypertens 2002; 11:445-50; http://dx.doi.org/ 10.1097/00041552-200207000-00012. [DOI] [PubMed] [Google Scholar]
- 1069.Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int 2004; 65:2135-44; http://dx.doi.org/ 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 1070.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, et al.. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PloS One 2008; 3:e3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1071.Wasko MC, Hubert HB, Lingala VB, Elliott JR, Luggen ME, Fries JF, Ward MM. Hydroxychloroquine and risk of diabetes in patients with rheumatoid arthritis. JAMA 2007; 298:187-93; http://dx.doi.org/ 10.1001/jama.298.2.187. [DOI] [PubMed] [Google Scholar]
- 1072.Merlini L, Nishino I, Consortium for Autophagy in Muscular D. 201st ENMC International Workshop: Autophagy in muscular dystrophies–translational approach, 1-3 November 2013, Bussum, The Netherlands. Neuromuscular Disord 2014; 24:546-61; http://dx.doi.org/ 10.1016/j.nmd.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 1073.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 2007; 131:1137-48; http://dx.doi.org/ 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1074.Aits S, Gustafsson L, Hallgren O, Brest P, Gustafsson M, Trulsson M, Mossberg AK, Simon HU, Mograbi B, Svanborg C. HAMLET (human alpha-lactalbumin made lethal to tumor cells) triggers autophagic tumor cell death. Int J Cancer 2009; 124:1008-19; http://dx.doi.org/ 10.1002/ijc.24076. [DOI] [PubMed] [Google Scholar]
- 1075.Koike M, Shibata M, Tadakoshi M, Gotoh K, Komatsu M, Waguri S, Kawahara N, Kuida K, Nagata S, Kominami E, et al.. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am J Pathol 2008; 172:454-69; http://dx.doi.org/ 10.2353/ajpath.2008.070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1076.Hou YC, Hannigan AM, Gorski SM. An executioner caspase regulates autophagy. Autophagy 2009; 5:530-3; http://dx.doi.org/ 10.4161/auto.5.4.8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1077.Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjorkoy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol 2010; 190:523-31; http://dx.doi.org/ 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1078.Piras A, Gianetto D, Conte D, Bosone A, Vercelli A. Activation of autophagy in a rat model of retinal ischemia following high intraocular pressure. PloS One 2011; 6:e22514; http://dx.doi.org/ 10.1371/journal.pone.0022514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1079.Schwarze PE, Seglen PO. Reduced autophagic activity, improved protein balance and enhanced in vitro survival of hepatocytes isolated from carcinogen-treated rats. Exp Cell Res 1985; 157:15-28; http://dx.doi.org/ 10.1016/0014-4827(85)90148-X. [DOI] [PubMed] [Google Scholar]
- 1080.Liu Y, Shoji-Kawata S, Sumpter RM Jr., Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, et al.. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci USA 2013; 110:20364-71; http://dx.doi.org/ 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1081.Santoni M, Amantini C, Morelli MB, Liberati S, Farfariello V, Nabissi M, Bonfili L, Eleuteri AM, Mozzicafreddo M, Burattini L, et al.. Pazopanib and sunitinib trigger autophagic and non-autophagic death of bladder tumour cells. Brit J Cancer 2013; 109:1040-50; http://dx.doi.org/ 10.1038/bjc.2013.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1082.Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C, Rotiroti D, Morrone LA, Bagetta G, Corasaniti MT. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis 2011; 2:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1083.Denton D, Nicolson S, Kumar S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ 2012; 19:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1084.Beaulaton J, Lockshin RA. Ultrastructural study of the normal degeneration of the intersegmental muscles of Anthereae polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J Morphol 1977; 154:39-57; http://dx.doi.org/ 10.1002/jmor.1051540104. [DOI] [PubMed] [Google Scholar]
- 1085.PG Clarke. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol 1990; 181:195-213. [DOI] [PubMed] [Google Scholar]
- 1086.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al.. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1087.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nature Rev Mol Cell Biol 2008; 9:1004-10; http://dx.doi.org/ 10.1038/nrm2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1088.Richard VR, Beach A, Piano A, Leonov A, Feldman R, Burstein MT, Kyryakov P, Gomez-Perez A, Arlia-Ciommo A, Baptista S, et al.. Mechanism of liponecrosis, a distinct mode of programmed cell death. Cell Cycle 2014; 13:3707-26; http://dx.doi.org/ 10.4161/15384101.2014.965003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1089.Sheibani S, Richard VR, Beach A, Leonov A, Feldman R, Mattie S, Khelghatybana L, Piano A, Greenwood M, Vali H, et al.. Macromitophagy, neutral lipids synthesis, and peroxisomal fatty acid oxidation protect yeast from “liponecrosis”, a previously unknown form of programmed cell death. Cell Cycle 2014; 13:138-47; http://dx.doi.org/ 10.4161/cc.26885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1090.Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, Bazan NG, Blagosklonny MV, Blomgren K, Borner C, et al.. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ 2009; 16:1093-107; http://dx.doi.org/ 10.1038/cdd.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1091.Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM, Adam D, Alnemri ES, Altucci L, Andrews D, Annicchiarico-Petruzzelli M, et al.. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ 2015; 22:58-73; http://dx.doi.org/ 10.1038/cdd.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1092.Minina EA, Bozhkov PV, Hofius D. Autophagy as initiator or executioner of cell death. Trends Plant Sci 2014; 19:692-7. [DOI] [PubMed] [Google Scholar]
- 1093.van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, et al.. Morphological classification of plant cell deaths. Cell Death Differ 2011; 18:1241-6; http://dx.doi.org/ 10.1038/cdd.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1094.Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J 2010; 64:151-64. [DOI] [PubMed] [Google Scholar]
- 1095.Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D, Sanchez-Vera V, Suarez MF, Zhivotovsky B, Daniel G, et al.. Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol 2013; 203:917-27; http://dx.doi.org/ 10.1083/jcb.201307082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1096.Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jorgensen LB, Jones JD, Mundy J, Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 2009; 137:773-83; http://dx.doi.org/ 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 1097.Giusti C, Tresse E, Luciani MF, Golstein P. Autophagic cell death: analysis in Dictyostelium. Biochim Biophys Acta 2009; 1793:1422-31; http://dx.doi.org/ 10.1016/j.bbamcr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 1098.Luciani MF, Giusti C, Harms B, Oshima Y, Kikuchi H, Kubohara Y, Golstein P. Atg1 allows second-signaled autophagic cell death in Dictyostelium. Autophagy 2011; 7:501-8; http://dx.doi.org/ 10.4161/auto.7.5.14957. [DOI] [PubMed] [Google Scholar]
- 1099.Uchikawa T, Yamamoto A, Inouye K. Origin and function of the stalk-cell vacuole in Dictyostelium. Dev Biol 2011; 352:48-57; http://dx.doi.org/ 10.1016/j.ydbio.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 1100.Guimar[a]es CA, Benchimol M, Amarante-Mendes GP, Linden R. Alternative programs of cell death in developing retinal tissue. J Biol Chem 2003; 278:41938-46; http://dx.doi.org/ 10.1074/jbc.M306547200. [DOI] [PubMed] [Google Scholar]
- 1101.Lossi L, Gambino G, Mioletti S, Merighi A. In vivo analysis reveals different apoptotic pathways in pre- and postmigratory cerebellar granule cells of rabbit. J Neurobiol 2004; 60:437-52; http://dx.doi.org/ 10.1002/neu.20032. [DOI] [PubMed] [Google Scholar]
- 1102.Lossi L, Alasia S, Salio C, Merighi A. Cell death and proliferation in acute slices and organotypic cultures of mammalian CNS. Prog Neurobiol 2009; 88:221-45; http://dx.doi.org/ 10.1016/j.pneurobio.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 1103.Thorburn A. I think autophagy controls the death of my cells: what do I do to get my paper published? Autophagy 2011; 7:455-6; http://dx.doi.org/ 10.4161/auto.7.5.14797. [DOI] [PubMed] [Google Scholar]
- 1104.Kaushik S, Bandyopadhyay U, Sridhar S, Kiffin R, Martinez-Vicente M, Kon M, Orenstein SJ, Wong E, Cuervo AM. Chaperone-mediated autophagy at a glance. J Cell Sci 2011; 124:495-9; http://dx.doi.org/ 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1105.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol 2010; 23:184-9; http://dx.doi.org/ 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1106.Kaushik S, Cuervo AM. Methods to monitor chaperone-mediated autophagy. Methods Enzymol 2009; 452:297-324; http://dx.doi.org/ 10.1016/S0076-6879(08)03619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1107.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci 1990; 15:305-9; http://dx.doi.org/ 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 1108.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 1996; 273:501-3; http://dx.doi.org/ 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 1109.Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci 2000; 113:4441-50. [DOI] [PubMed] [Google Scholar]
- 1110.Finn PF, Mesires NT, Vine M, Dice JF. Effects of small molecules on chaperone-mediated autophagy. Autophagy 2005; 1:141-5; http://dx.doi.org/ 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 1111.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol 2008; 28:5747-63; http://dx.doi.org/ 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1112.Aniento F, Emans N, Griffiths G, Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol 1993; 123:1373-87; http://dx.doi.org/ 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1113.Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem 2000; 275:27447-56. [DOI] [PubMed] [Google Scholar]
- 1114.Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun 2011; 2:386; http://dx.doi.org/ 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1115.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 2011; 20:131-9; http://dx.doi.org/ 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1116.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P, Saint R, Fleischmann BK, et al.. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol 2010; 20:143-8; http://dx.doi.org/ 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 1117.Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lullmann-Rauch R, Hartmann D, Heeren J, et al.. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell 2004; 15: 3132-45; http://dx.doi.org/ 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1118.Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell 2002; 13:3355-68; http://dx.doi.org/ 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1119.Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J 2007; 26:313-24; http://dx.doi.org/ 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1120.Fujiwara Y, Furuta A, Kikuchi H, Aizawa S, Hatanaka Y, Konya C, Uchida K, Yoshimura A, Tamai Y, Wada K, et al.. Discovery of a novel type of autophagy targeting RNA. Autophagy 2013; 9:403-9; http://dx.doi.org/ 10.4161/auto.23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1121.Fujiwara Y, Hase K, Wada K, Kabuta T. An RNautophagy/DNautophagy receptor, LAMP2C, possesses an arginine-rich motif that mediates RNA/DNA-binding. Biochem Biophys Res Commun 2015; 460:281-6; http://dx.doi.org/ 10.1016/j.bbrc.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 1122.Fujiwara Y, Kikuchi H, Aizawa S, Furuta A, Hatanaka Y, Konya C, Uchida K, Wada K, Kabuta T. Direct uptake and degradation of DNA by lysosomes. Autophagy 2013; 9:1167-71; http://dx.doi.org/ 10.4161/auto.24880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1123.Hase K, Fujiwara Y, Kikuchi H, Aizawa S, Hakuno F, Takahashi S, Wada K, Kabuta T. RNautophagy/DNautophagy possesses selectivity for RNA/DNA substrates. Nucleic Acids Res 2015; 43:6439-49; http://dx.doi.org/ 10.1093/nar/gkv579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1124.Furuta A, Kikuchi H, Fujita H, Yamada D, Fujiwara Y, Kabuta T, Nishino I, Wada K, Uchiyama Y. Property of lysosomal storage disease associated with midbrain pathology in the central nervous system of lamp-2-deficient mice. Am J Pathol 2015; 185:1713-23; http://dx.doi.org/ 10.1016/j.ajpath.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 1125.Rothaug M, Stroobants S, Schweizer M, Peters J, Zunke F, Allerding M, D'Hooge R, Saftig P, Blanz J. LAMP-2 deficiency leads to hippocampal dysfunction but normal clearance of neuronal substrates of chaperone-mediated autophagy in a mouse model for Danon disease. Acta Neuropathol Commun 2015; 3:6; http://dx.doi.org/ 10.1186/s40478-014-0182-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1126.Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, Vakeel P, Stadel D, Haas A, Saftig P, et al.. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol 2013; 23:430-5; http://dx.doi.org/ 10.1016/j.cub.2013.01.064. [DOI] [PubMed] [Google Scholar]
- 1127.Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem 2008; 283:1437-44; http://dx.doi.org/ 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 1128.Carra S, Seguin SJ, Landry J. HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy 2008; 4:237-9; http://dx.doi.org/ 10.4161/auto.5407. [DOI] [PubMed] [Google Scholar]
- 1129.Niemann A, Baltes J, Elsasser HP. Fluorescence properties and staining behavior of monodansylpentane, a structural homologue of the lysosomotropic agent monodansylcadaverine. J Histochem Cytochem 2001; 49:177-85; http://dx.doi.org/ 10.1177/002215540104900205. [DOI] [PubMed] [Google Scholar]
- 1130.Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, Domingo D, Yahalom J. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res 2001; 61:439-44. [PubMed] [Google Scholar]
- 1131.Florez-McClure ML, Linseman DA, Chu CT, Barker PA, Bouchard RJ, Le SS, Laessig TA, Heidenreich KA. The p75 neurotrophin receptor can induce autophagy and death of cerebellar Purkinje neurons. J Neurosci 2004; 24:4498-509; http://dx.doi.org/ 10.1523/JNEUROSCI.5744-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1132.Moriyasu Y, Hattori M, Jauh G-Y, Rogers JC. Alpha tonoplast intrinsic protein is specifically associated with vacuole membrane involved in an autophagic process. Plant Cell Physiol 2003; 44:795-802; http://dx.doi.org/ 10.1093/pcp/pcg100. [DOI] [PubMed] [Google Scholar]
- 1133.Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, Nixon RA. Autophagy failure in Alzheimer's disease and the role of defective lysosomal acidification. Eur J Neurosci 2013; 37:1949-61; http://dx.doi.org/ 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1134.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol 1995; 66:3-14. [PubMed] [Google Scholar]
- 1135.Hoyer-Hansen M, Bastholm L, Mathiasen IS, Elling F, Jaattela M. Vitamin D analog EB1089 triggers dramatic lysosomal changes and Beclin 1-mediated autophagic cell death. Cell Death Differ 2005; 12:1297-309; http://dx.doi.org/ 10.1038/sj.cdd.4401651. [DOI] [PubMed] [Google Scholar]
- 1136.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 2004; 117:2687-97; http://dx.doi.org/ 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 1137.Fogel JL, Thein TZ, Mariani FV. Use of LysoTracker to detect programmed cell death in embryos and differentiating embryonic stem cells. J Vis Exp 2012; 68; doi: 10.3791/4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1138.Freundt EC, Czapiga M, Lenardo MJ. Photoconversion of Lysotracker Red to a green fluorescent molecule. Cell Res 2007; 17:956-8; http://dx.doi.org/ 10.1038/cr.2007.80. [DOI] [PubMed] [Google Scholar]
- 1139.Oeste CL, Seco E, Patton WF, Boya P, Perez-Sala D. Interactions between autophagic and endo-lysosomal markers in endothelial cells. Histochem Cell Biol 2013; 139:659-70; http://dx.doi.org/ 10.1007/s00418-012-1057-6. [DOI] [PubMed] [Google Scholar]
- 1140.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov 2007; 6:304-12; http://dx.doi.org/ 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 1141.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20:355-62; http://dx.doi.org/ 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1142.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 2008; 4:600-6; http://dx.doi.org/ 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1143.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 2009; 186:773-82; http://dx.doi.org/ 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1144.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, et al.. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem 2005; 280:20722-9; http://dx.doi.org/ 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 1145.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2000; 275:992-8; http://dx.doi.org/ 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 1146.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, et al.. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem 2011; 286:9587-97; http://dx.doi.org/ 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1147.Crisan TO, Plantinga TS, van de Veerdonk FL, Farcas MF, Stoffels M, Kullberg BJ, van der Meer JW, Joosten LA, Netea MG. Inflammasome-independent modulation of cytokine response by autophagy in human cells. PloS One 2011; 6:e18666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1148.Kleinnijenhuis J, Oosting M, Plantinga TS, van der Meer JW, Joosten LA, Crevel RV, Netea MG. Autophagy modulates the Mycobacterium tuberculosis-induced cytokine response. Immunology 2011; 134:341-8; http://dx.doi.org/ 10.1111/j.1365-2567.2011.03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1149.Peral de Castro C, Jones SA, Ni Cheallaigh C, Hearnden CA, Williams L, Winter J, Lavelle EC, Mills KH, Harris J. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol 2012; 189:4144-53; http://dx.doi.org/ 10.4049/jimmunol.1201946. [DOI] [PubMed] [Google Scholar]
- 1150.Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, et al.. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 2014; 16:1069-79; http://dx.doi.org/ 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 1151.Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, et al.. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol 2014; 10:1013-9; http://dx.doi.org/ 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 1152.Chen J, Chen MX, Fogo AB, Harris RC, Chen JK. mVps34 deletion in podocytes causes glomerulosclerosis by disrupting intracellular vesicle trafficking. J Am Soc Nephrol 2013; 24:198-207; http://dx.doi.org/ 10.1681/ASN.2012010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1153.Cantino D, Mosso R, Baccino FM. Changes induced by fasting and cycloheximide in the vacuolar apparatus of rat hepatocytes. A morphometric investigation. Boll Soc Ital Biol Sper 1979; 55:1884-9. [PubMed] [Google Scholar]
- 1154.Kovács J. Morphometric study of the effect of leupeptin, vinblastine, estron acetate and cycloheximide on the autophagic vacuole-lysosomal compartments in mouse seminal vesicle cells. Virchows Arch B Cell Pathol Incl Mol Pathol 1983; 42:83-93; http://dx.doi.org/ 10.1007/BF02890372. [DOI] [PubMed] [Google Scholar]
- 1155.Papadopoulos T, Pfeifer U. Regression of rat liver autophagic vacuoles by locally applied cycloheximide. Lab Investig 1986; 54:100-7. [PubMed] [Google Scholar]
- 1156.Rumpelt HJ, Albring M, Thoenes W. Prevention of D-galactosamine-induced hepatocellular autophagocytosis by cycloheximide. Virchows Arch B Cell Pathol 1974; 16:195-203; http://dx.doi.org/ 10.1007/BF02894074. [DOI] [PubMed] [Google Scholar]
- 1157.Rumpelt HJ, Weisbach T. Effect of cycloheximide on glucagon-induced autophagy. Quantitative examinations on hepatocytes in the rat. Am J Pathol 1978; 91:49-55. [PMC free article] [PubMed] [Google Scholar]
- 1158.Kovács AL, Kovács J. Autophagocytosis in mouse seminal vesicle cells in vitro. Temperature dependence and effects of vinblastine and inhibitors of protein synthesis. Virchows Arch B Cell Pathol Incl Mol Pathol 1980; 32:97-104; http://dx.doi.org/ 10.1007/BF02889018. [DOI] [PubMed] [Google Scholar]
- 1159.Rodemann HP, Dittmann K, Toulany M. Radiation-induced EGFR-signaling and control of DNA-damage repair. Int J Radiat Biol 2007; 83:781-91; http://dx.doi.org/ 10.1080/09553000701769970. [DOI] [PubMed] [Google Scholar]
- 1160.Chaachouay H, Ohneseit P, Toulany M, Kehlbach R, Multhoff G, Rodemann HP. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother Oncol 2011; 99:287-92; http://dx.doi.org/ 10.1016/j.radonc.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 1161.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res 2008; 68:1485-94; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 1162.Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal 2010; 3:ra31. [DOI] [PubMed] [Google Scholar]
- 1163.Seglen PO, Gordon PB. Effects of lysosomotropic monoamines, diamines, amino alcohols, and other amino compounds on protein degradation and protein synthesis in isolated rat hepatocytes. Mol Pharmacol 1980; 18:468-75. [PubMed] [Google Scholar]
- 1164.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA 2011; 108:11121-6; http://dx.doi.org/ 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1165.Pellegrini P, Strambi A, Zipoli C, Hagg-Olofsson M, Buoncervello M, Linder S, De Milito A. Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: implications for cancer therapies. Autophagy 2014; 10:562-71; http://dx.doi.org/ 10.4161/auto.27901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1166.Fischer S, Ronellenfitsch MW, Thiepold AL, Harter PN, Reichert S, Kogel D, Paschke R, Mittelbronn M, Weller M, Steinbach JP, et al.. Hypoxia enhances the antiglioma cytotoxicity of B10, a glycosylated derivative of betulinic acid. PloS One 2014; 9:e94921; http://dx.doi.org/ 10.1371/journal.pone.0094921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1167.Gonzalez P, Mader I, Tchoghandjian A, Enzenmuller S, Cristofanon S, Basit F, Debatin KM, Fulda S. Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ 2012; 19:1337-46; http://dx.doi.org/ 10.1038/cdd.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1168.Potze L, Mullauer FB, Colak S, Kessler JH, Medema JP. Betulinic acid-induced mitochondria-dependent cell death is counterbalanced by an autophagic salvage response. Cell Death Dis 2014; 5:e1169; http://dx.doi.org/ 10.1038/cddis.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1169.Broniatowski M, Flasinski M, Wydro P. Investigation of the interactions of lupane type pentacyclic triterpenes with outer leaflet membrane phospholipids–Langmuir monolayer and synchrotron X-ray scattering study. J Colloid Interface Sci 2012; 381:116-24; http://dx.doi.org/ 10.1016/j.jcis.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 1170.Chen Y, Sun R, Wang B. Monolayer behavior of binary systems of betulinic acid and cardiolipin: thermodynamic analyses of Langmuir monolayers and AFM study of Langmuir-Blodgett monolayers. J Colloid Interface Sci 2011; 353:294-300; http://dx.doi.org/ 10.1016/j.jcis.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 1171.Gao M, Lau PM, Kong SK. Mitochondrial toxin betulinic acid induces in vitro eryptosis in human red blood cells through membrane permeabilization. Arch Toxicol 2014; 88:755-68. [DOI] [PubMed] [Google Scholar]
- 1172.Wei P, Zhang L, Lu Y, Man N, Wen L. C60(Nd) nanoparticles enhance chemotherapeutic susceptibility of cancer cells by modulation of autophagy. Nanotechnology 2010; 21:495101; http://dx.doi.org/ 10.1088/0957-4484/21/49/495101. [DOI] [PubMed] [Google Scholar]
- 1173.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 1998; 8:397-403; http://dx.doi.org/ 10.1016/S0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 1174.S Mehdi. Cell-penetrating inhibitors of calpain. Trends Biochem Sci 1991; 16:150-3; http://dx.doi.org/ 10.1016/0968-0004(91)90058-4. [DOI] [PubMed] [Google Scholar]
- 1175.Holen I, Gordon PB, Seglen PO. Inhibition of hepatocytic autophagy by okadaic acid and other protein phosphatase inhibitors. Eur J Biochem 1993; 215:113-22; http://dx.doi.org/ 10.1111/j.1432-1033.1993.tb18013.x. [DOI] [PubMed] [Google Scholar]
- 1176.Sasaki K, Murata M, Yasumoto T, Mieskes G, Takai A. Affinity of okadaic acid to type-1 and type-2A protein phosphatases is markedly reduced by oxidation of its 27-hydroxyl group. Biochem J 1994; 298:259-62; http://dx.doi.org/ 10.1042/bj2980259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1177.Robinson DG, Albrecht S, Moriyasu Y. The V-ATPase inhibitors concanamycin A and bafilomycin A lead to Golgi swelling in tobacco BY-2 cells. Protoplasma 2004; 224:255-60; http://dx.doi.org/ 10.1007/s00709-004-0070-6. [DOI] [PubMed] [Google Scholar]
- 1178.Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, et al.. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab 2014; 20:526-40; http://dx.doi.org/ 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 1179.Wu YC, Wu WK, Li Y, Yu L, Li ZJ, Wong CC, Li HT, Sung JJ, Cho CH. Inhibition of macroautophagy by bafilomycin A1 lowers proliferation and induces apoptosis in colon cancer cells. Biochem Biophys Res Commun 2009; 382:451-6; http://dx.doi.org/ 10.1016/j.bbrc.2009.03.051. [DOI] [PubMed] [Google Scholar]
- 1180.Ostenfeld MS, Hoyer-Hansen M, Bastholm L, Fehrenbacher N, Olsen OD, Groth-Pedersen L, Puustinen P, Kirkegaard-Sorensen T, Nylandsted J, Farkas T, et al.. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation. Autophagy 2008; 4:487-99; http://dx.doi.org/ 10.4161/auto.5774. [DOI] [PubMed] [Google Scholar]
- 1181.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1182.Garcia-Garcia A, Anandhan A, Burns M, Chen H, Zhou Y, Franco R. Impairment of Atg5-dependent autophagic flux promotes paraquat- and MPP(+)-induced apoptosis but not rotenone or 6-hydroxydopamine toxicity. Toxicol Sci 2013; 136:166-82; http://dx.doi.org/ 10.1093/toxsci/kft188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1183.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest 2008; 118:79-88; http://dx.doi.org/ 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1184.Poole B, Ohkuma S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J Cell Biol 1981; 90:665-9; http://dx.doi.org/ 10.1083/jcb.90.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1185.Matsuoka K, Higuchi T, Maeshima M, Nakamura K. A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 1997; 9:533-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1186.Arstila AU, Nuuja IJ, Trump BF. Studies on cellular autophagocytosis. Vinblastine-induced autophagy in the rat liver. Exp Cell Res 1974; 87:249-52; http://dx.doi.org/ 10.1016/0014-4827(74)90477-7. [DOI] [PubMed] [Google Scholar]
- 1187.Hirsimaki Y, Arstila AU, Trump BF. Autophagocytosis: in vitro induction by microtuble poisons. Exp Cell Res 1975; 92:11-4; http://dx.doi.org/ 10.1016/0014-4827(75)90630-8. [DOI] [PubMed] [Google Scholar]
- 1188.Kominami E, Hashida S, Khairallah EA, Katunuma N. Sequestration of cytoplasmic enzymes in an autophagic vacuole-lysosomal system induced by injection of leupeptin. J Biol Chem 1983; 258:6093-100. [PubMed] [Google Scholar]
- 1189.Réz G, Fellinger E, Reti M, Biczo I, Kovács AL. Time course of quantitative morphological changes of the autophagic-lysosomal compartment of murine seminal vesicle epithelial cells under the influence of vinblastine. J Submicrosc Cytol Pathol 1990; 22:529-34. [PubMed] [Google Scholar]
- 1190.Oliva O, Réz G, Pálfia Z, Fellinger E. Dynamics of vinblastine-induced autophagocytosis in murine pancreatic acinar cells: influence of cycloheximide post-treatments. Exp Mol Pathol 1992; 56:76-86; http://dx.doi.org/ 10.1016/0014-4800(92)90025-7. [DOI] [PubMed] [Google Scholar]
- 1191.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 2009; 7:e38; http://dx.doi.org/ 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1192.Fleming A, Noda T, Yoshimori T, Rubinsztein DC. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol 2011; 7:9-17; http://dx.doi.org/ 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 1193.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009; 284:8023-32; http://dx.doi.org/ 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1194.Yu K, Toral-Barza L, Shi C, Zhang WG, Lucas J, Shor B, Kim J, Verheijen J, Curran K, Malwitz DJ, et al.. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res 2009; 69:6232-40; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- 1195.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, Vincent JP, Ellston R, Jones D, Sini P, et al.. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res 2010; 70:288-98; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 1196.Roscic A, Baldo B, Crochemore C, Marcellin D, Paganetti P. Induction of autophagy with catalytic mTOR inhibitors reduces huntingtin aggregates in a neuronal cell model. J Neurochem 2011; 119:398-407; http://dx.doi.org/ 10.1111/j.1471-4159.2011.07435.x. [DOI] [PubMed] [Google Scholar]
- 1197.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, Haas-Kogan D, James CD, Oakes SA, Debnath J, et al.. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal 2010; 3:ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1198.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 2010; 11:467-78; http://dx.doi.org/ 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1199.Yamamoto A, Yue Z. Autophagy and its normal and pathogenic States in the brain. Annu Rev Neurosci 2014; 37:55-78; http://dx.doi.org/ 10.1146/annurev-neuro-071013-014149. [DOI] [PubMed] [Google Scholar]
- 1200.Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci USA 2010; 107:16982-7; http://dx.doi.org/ 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1201.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, et al.. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol 2008; 4:295-305; http://dx.doi.org/ 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1202.Palomo GM, Cerrato T, Gargini R, Diaz-Nido J. Silencing of frataxin gene expression triggers p53-dependent apoptosis in human neuron-like cells. Hum Mol Genet 2011; 20:2807-22; http://dx.doi.org/ 10.1093/hmg/ddr187. [DOI] [PubMed] [Google Scholar]
- 1203.Bolinches-Amoros A, Molla B, Pla-Martin D, Palau F, Gonzalez-Cabo P. Mitochondrial dysfunction induced by frataxin deficiency is associated with cellular senescence and abnormal calcium metabolism. Front Cell Neurosci 2014; 8:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1204.Sakagami H, Kawase M, Wakabayashi H, Kurihara T. Factors that affect the type of cell death induced by chemicals. Autophagy 2007; 3:493-5; http://dx.doi.org/ 10.4161/auto.4594. [DOI] [PubMed] [Google Scholar]
- 1205.Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 2002; 277:33105-14; http://dx.doi.org/ 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- 1206.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al.. Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447:1121-5. [DOI] [PubMed] [Google Scholar]
- 1207.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032-6; http://dx.doi.org/ 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 1208.Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, et al.. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 2012; 11:397-409; http://dx.doi.org/ 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1209.Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu CG, Yang JM. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 2009; 5:816-23; http://dx.doi.org/ 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1210.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ 2007; 14: 146-57; http://dx.doi.org/ 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 1211.Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, Kirschnek S, Gaffal E, Landsberg J, Hellmuth J, et al.. 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med 2008; 14:1256-63; http://dx.doi.org/ 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 1212.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J 2008; 27:1110-21; http://dx.doi.org/ 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1213.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med 2007; 204:25-31; http://dx.doi.org/ 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1214.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, et al.. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy 2008; 4:309-14; http://dx.doi.org/ 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 1215.Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, et al.. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 2009; 11:1355-62; http://dx.doi.org/ 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1216.Kimball SR, Siegfried BA, Jefferson LS. Glucagon represses signaling through the mammalian target of rapamycin in rat liver by activating AMP-activated protein kinase. J Biol Chem 2004; 279:54103-9; http://dx.doi.org/ 10.1074/jbc.M410755200. [DOI] [PubMed] [Google Scholar]
- 1217.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 1995; 270:2320-6; http://dx.doi.org/ 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 1218.Klionsky DJ, Meijer AJ, Codogno P, Neufeld TP, Scott RC. Autophagy and p70S6 kinase. Autophagy 2005; 1:59-61; http://dx.doi.org/ 10.4161/auto.1.1.1536. [DOI] [PubMed] [Google Scholar]
- 1219.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 1998; 273:3963-6; http://dx.doi.org/ 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 1220.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol 2005; 170:1101-11; http://dx.doi.org/ 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1221.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem 2010; 285:11061-7; http://dx.doi.org/ 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1222.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, et al.. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA 2007; 104:19023-8; http://dx.doi.org/ 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1223.Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al.. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-[b], and Bcl-2. Mol Cell 2007; 25:193-205; http://dx.doi.org/ 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 1224.Decuypere JP, Kindt D, Luyten T, Welkenhuyzen K, Missiaen L, De Smedt H, Bultynck G, Parys JB. mTOR-Controlled Autophagy Requires Intracellular Ca(2+) Signaling. PloS One 2013; 8:e61020; http://dx.doi.org/ 10.1371/journal.pone.0061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1225.Pereira GJ, Hirata H, Fimia GM, do Carmo LG, Bincoletto C, Han SW, Stilhano RS, Ureshino RP, Bloor-Young D, Churchill G, et al.. Nicotinic acid adenine dinucleotide phosphate (NAADP) regulates autophagy in cultured astrocytes. J Biol Chem 2011; 286:27875-81; http://dx.doi.org/ 10.1074/jbc.C110.216580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1226.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al.. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013; 494:201-6; http://dx.doi.org/ 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1227.Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S. Targeting gamma-herpesvirus 68 Bcl-2-mediated down-regulation of autophagy. J Biol Chem 2014; 289:8029-40; http://dx.doi.org/ 10.1074/jbc.M113.515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1228.Winter G, Hazan R, Bakalinsky AT, Abeliovich H. Caffeine induces macroautophagy and confers a cytocidal effect on food spoilage yeast in combination with benzoic acid. Autophagy 2008; 4:28-36; http://dx.doi.org/ 10.4161/auto.5127. [DOI] [PubMed] [Google Scholar]
- 1229.Saiki S, Sasazawa Y, Imamichi Y, Kawajiri S, Fujimaki T, Tanida I, Kobayashi H, Sato F, Sato S, Ishikawa K, et al.. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy 2011; 7:176-87; http://dx.doi.org/ 10.4161/auto.7.2.14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1230.Tsabar M, Eapen VV, Mason JM, Memisoglu G, Waterman DP, Long MJ, Bishop DK, Haber JE. Caffeine impairs resection during DNA break repair by reducing the levels of nucleases Sae2 and Dna2. Nucleic Acids Res 2015; 43:6889-901; http://dx.doi.org/ 10.1093/nar/gkv520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1231.Fu J, Shao CJ, Chen FR, Ng HK, Chen ZP. Autophagy induced by valproic acid is associated with oxidative stress in glioma cell lines. Neuro-oncology 2010; 12:328-40; http://dx.doi.org/ 10.1093/neuonc/nop005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1232.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, et al.. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 2011; 471:74-9; http://dx.doi.org/ 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1233.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al.. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci USA 2012; 109:11206-10; http://dx.doi.org/ 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1234.Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, et al.. Function and molecular mechanism of acetylation in autophagy regulation. Science 2012; 336:474-7; http://dx.doi.org/ 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 1235.Katagiri N, Kuroda T, Kishimoto H, Hayashi Y, Kumazawa T, Kimura K. The nucleolar protein nucleophosmin is essential for autophagy induced by inhibiting Pol I transcription. Sci Rep 2015; 5:8903; http://dx.doi.org/ 10.1038/srep08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1236.Kreiner G, Bierhoff H, Armentano M, Rodriguez-Parkitna J, Sowodniok K, Naranjo JR, Bonfanti L, Liss B, Schutz G, Grummt I, et al.. A neuroprotective phase precedes striatal degeneration upon nucleolar stress. Cell Death Differ 2013; 20:1455-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1237.Furuya N, Liang XH, Levine B. Autophagy and cancer In: Klionsky DJ, ed. Autophagy. Georgetown, TX: Landes Bioscience, 2004:241-55. [Google Scholar]
- 1238.de Medina P, Paillasse MR, Segala G, Khallouki F, Brillouet S, Dalenc F, Courbon F, Record M, Poirot M, Silvente-Poirot S. Importance of cholesterol and oxysterols metabolism in the pharmacology of tamoxifen and other AEBS ligands. Chem Phys Lipids 2011; 164:432-7. [DOI] [PubMed] [Google Scholar]
- 1239.de Medina P, Payre B, Boubekeur N, Bertrand-Michel J, Terce F, Silvente-Poirot S, Poirot M. Ligands of the antiestrogen-binding site induce active cell death and autophagy in human breast cancer cells through the modulation of cholesterol metabolism. Cell Death Differ 2009; 16:1372-84. [DOI] [PubMed] [Google Scholar]
- 1240.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, et al.. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol 2007; 3:331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1241.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and [a]-synuclein. J Biol Chem 2007; 282:5641-52. [DOI] [PubMed] [Google Scholar]
- 1242.Kruger U, Wang Y, Kumar S, Mandelkow EM. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol Aging 2012; 33:2291-305. [DOI] [PubMed] [Google Scholar]
- 1243.Koshkina NV, Briggs K, Palalon F, Curley SA. Autophagy and enhanced chemosensitivity in experimental pancreatic cancers induced by noninvasive radiofrequency field treatment. Cancer 2014; 120:480-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1244.Cardenas C, Miller RA, Smith I, Bui T, Molgo J, Muller M, Vais H, Cheung KH, Yang J, Parker I, et al.. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 2010; 142:270-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1245.Decuypere JP, Bultynck G, Parys JB. A dual role for Ca2+ in autophagy regulation. Cell Calcium 2011; 50:242-50. [DOI] [PubMed] [Google Scholar]
- 1246.Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, et al.. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ 2009; 16:1006-17. [DOI] [PubMed] [Google Scholar]
- 1247.Dayan F, Bilton RL, Laferriere J, Trottier E, Roux D, Pouyssegur J, Mazure NM. Activation of HIF-1alpha in exponentially growing cells via hypoxic stimulation is independent of the Akt/mTOR pathway. J Cell Physiol 2009; 218:167-74. [DOI] [PubMed] [Google Scholar]
- 1248.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 2009; 29:2570-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1249.Yamashita S, Yurimoto H, Murakami D, Yoshikawa M, Oku M, Sakai Y. Lag-phase autophagy in the methylotrophic yeast Pichia pastoris. Genes Cells 2009; 14:861-70. [DOI] [PubMed] [Google Scholar]
- 1250.van Zutphen T, Baerends RJ, Susanna KA, de Jong A, Kuipers OP, Veenhuis M, van der Klei IJ. Adaptation of Hansenula polymorpha to methanol: a transcriptome analysis. BMC Genomics 2010; 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1251.Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol 2006; 47:1641-52. [DOI] [PubMed] [Google Scholar]
- 1252.Yano K, Suzuki T, Moriyasu Y. Constitutive autophagy in plant root cells. Autophagy 2007; 3:360-2. [DOI] [PubMed] [Google Scholar]
- 1253.Gordon PB, Kisen GO, Kovacs AL, Seglen PO. Experimental characterization of the autophagic-lysosomal pathway in isolated rat hepatocytes. Biochem Soc Symp 1989; 55:129-43. [PubMed] [Google Scholar]
- 1254.Poli A, Gordon PB, Schwarze PE, Grinde B, Seglen PO. Effects of insulin and anchorage on hepatocytic protein metabolism and amino acid transport. J Cell Sci 1981; 48:1-18. [DOI] [PubMed] [Google Scholar]
- 1255.Schliess F, Reissmann R, Reinehr R, vom Dahl S, Häussinger D. Involvement of integrins and Src in insulin signaling toward autophagic proteolysis in rat liver. J Biol Chem 2004; 279:21294-301. [DOI] [PubMed] [Google Scholar]
- 1256.vom Dahl S, Dombrowski F, Schmitt M, Schliess F, Pfeifer U, Haussinger D. Cell hydration controls autophagosome formation in rat liver in a microtubule-dependent way downstream from p38MAPK activation. Biochem J 2001; 354:31-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1257.vom Dahl S, Stoll B, Gerok W, Häussinger D. Inhibition of proteolysis by cell swelling in the liver requires intact microtubular structures. Biochem J 1995; 308 (Pt 2):529-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1258.Klionsky DJ, Bruford EA, Cherry JM, Hodgkin J, Laulederkind SJ, Singer AG. In the beginning there was babble. Autophagy 2012; 8:1165-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1259.Kovacs AL, Zhang H. Role of autophagy in Caenorhabditis elegans. FEBS Lett 2010; 584:1335-41. [DOI] [PubMed] [Google Scholar]
- 1260.Wu F, Li Y, Wang F, Noda NN, Zhang H. Differential function of the two Atg4 homologues in the aggrephagy pathway in Caenorhabditis elegans. J Biol Chem 2012; 287:29457-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1261.Zhang H, Wu F, Wang X, Du H, Wang X, Zhang H. The two C. elegans ATG-16 homologs have partially redundant functions in the basal autophagy pathway. Autophagy 2013; 9:1965-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1262.Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al.. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 2009; 136:308-21. [DOI] [PubMed] [Google Scholar]
- 1263.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, et al.. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis 2010; 1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1264.Samara C, Syntichaki P, Tavernarakis N. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ 2008; 15:105-12. [DOI] [PubMed] [Google Scholar]
- 1265.Alberti A, Michelet X, Djeddi A, Legouis R. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 2010; 6:622-33. [DOI] [PubMed] [Google Scholar]
- 1266.Manil-Segalen M, Lefebvre C, Jenzer C, Trichet M, Boulogne C, Satiat-Jeunemaitre B, Legouis R. The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev Cell 2014; 28:43-55; http://dx.doi.org/ 10.1016/j.devcel.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 1267.Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev 2007; 21:2161-71; http://dx.doi.org/ 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1268.Liang Q, Yang P, Tian E, Han J, Zhang H. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Autophagy 2012; 8:1426-33; http://dx.doi.org/ 10.4161/auto.21163. [DOI] [PubMed] [Google Scholar]
- 1269.Yang P, Zhang H. The coiled-coil domain protein EPG-8 plays an essential role in the autophagy pathway in C. elegans. Autophagy 2011; 7:159-65; http://dx.doi.org/ 10.4161/auto.7.2.14223. [DOI] [PubMed] [Google Scholar]
- 1270.SenGupta T, Torgersen ML, Kassahun H, Vellai T, Simonsen A, Nilsen H. Base excision repair AP endonucleases and mismatch repair act together to induce checkpoint-mediated autophagy. Nat Commun 2013; 4:2674; http://dx.doi.org/ 10.1038/ncomms3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1271.Schiavi A, Maglioni S, Palikaras K, Shaik A, Strappazzon F, Brinkmann V, Torgovnick A, Castelein N, De Henau S, Braeckman BP, et al.. Iron-starvation-induced mitophagy mediates lifespan extension upon mitochondrial stress in C. elegans. Curr Biol 2015; 25:1810-22; http://dx.doi.org/ 10.1016/j.cub.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 1272.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al.. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008; 10:676-87; http://dx.doi.org/ 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1273.Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy 2008; 4:870-3; http://dx.doi.org/ 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- 1274.Schiavi A, Torgovnick A, Kell A, Megalou E, Castelein N, Guccini I, Marzocchella L, Gelino S, Hansen M, Malisan F, et al.. Autophagy induction extends lifespan and reduces lipid content in response to frataxin silencing in C. elegans. Exp Gerontol 2013; 48:191-201; http://dx.doi.org/ 10.1016/j.exger.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1275.Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 2015; 521:525-8; http://dx.doi.org/ 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- 1276.Zhang H, Chang JT, Guo B, Hansen M, Jia K, Kovacs AL, Kumsta C, Lapierre LR, Legouis R, Lin L, et al.. Guidelines for monitoring autophagy in Caenorhabditis elegans. Autophagy 2015; 11:9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1277.Alers S, L{o}ffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K, Campbell DG, Fehrenbacher B, Schaller M, Wesselborg S, et al.. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy 2011; 7:1424-33; http://dx.doi.org/ 10.4161/auto.7.12.18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1278.Brown WR, Hubbard SJ, Tickle C, Wilson SA. The chicken as a model for large-scale analysis of vertebrate gene function. Nature reviews Genetics 2003; 4:87-98; http://dx.doi.org/ 10.1038/nrg998. [DOI] [PubMed] [Google Scholar]
- 1279.Wang L, Rodrigues NA, Wu Y, Maslikowski BM, Singh N, Lacroix S, Bedard PA. Pleiotropic action of AP-1 in v-Src-transformed cells. J Virol 2011; 85:6725-35; http://dx.doi.org/ 10.1128/JVI.01013-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1280.Baba TW, Giroir BP, Humphries EH. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology 1985; 144:139-51; http://dx.doi.org/ 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 1281.Perez-Martin M, Perez-Perez ME, Lemaire SD, Crespo JL. Oxidative Stress Contributes to Autophagy Induction in Response to Endoplasmic Reticulum Stress in Chlamydomonas reinhardtii. Plant Physiol 2014; 166:997-1008; http://dx.doi.org/ 10.1104/pp.114.243659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1282.Perez-Perez ME, Couso I, Crespo JL. Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 2012; 8:376-88; http://dx.doi.org/ 10.4161/auto.18864. [DOI] [PubMed] [Google Scholar]
- 1283.Mauvezin C, Ayala C, Braden CR, Kim J, Neufeld TP. Assays to monitor autophagy in Drosophila. Methods 2014; 68:134-9; http://dx.doi.org/ 10.1016/j.ymeth.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1284.Kim M, Semple I, Kim B, Kiers A, Nam S, Park HW, Park H, Ro SH, Kim JS, Juhasz G, et al.. Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy 2015; 11:1358-72; http://dx.doi.org/ 10.1080/15548627.2015.1063766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1285.Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol 2008; 181:655-66; http://dx.doi.org/ 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1286.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 2009; 30:588-98; http://dx.doi.org/ 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1287.Anding AL, Baehrecke EH. Vps15 is required for stress induced and developmentally triggered autophagy and salivary gland protein secretion in Drosophila. Cell Death Differ 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1288.Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol 2008; 182:1127-39; http://dx.doi.org/ 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1289.Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PloS One 2012; 7:e44214; http://dx.doi.org/ 10.1371/journal.pone.0044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1290.Hindle SJ, Elliott CJ. Spread of neuronal degeneration in a dopaminergic, Lrrk-G2019S model of Parkinson disease. Autophagy 2013; 9:936-8; http://dx.doi.org/ 10.4161/auto.24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1291.Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development 2013; 140:1321-9; http://dx.doi.org/ 10.1242/dev.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1292.Marinkovic D, Zhang X, Yalcin S, Luciano JP, Brugnara C, Huber T, Ghaffari S. Foxo3 is required for the regulation of oxidative stress in erythropoiesis. J Clin Invest 2007; 117:2133-44; http://dx.doi.org/ 10.1172/JCI31807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1293.McIver SC, Kang YA, DeVilbiss AW, O'Driscoll CA, Ouellette JN, Pope NJ, Camprecios G, Chang CJ, Yang D, Bouhassira EE, et al.. The exosome complex establishes a barricade to erythroid maturation. Blood 2014; 124:2285-97; http://dx.doi.org/ 10.1182/blood-2014-04-571083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1294.Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell 2009; 36:667-81; http://dx.doi.org/ 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1295.Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 2004; 104:3136-47; http://dx.doi.org/ 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 1296.Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al.. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 2009; 36:682-95; http://dx.doi.org/ 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1297.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 2008; 112:1493-502; http://dx.doi.org/ 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1298.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA 2010; 107:832-7; http://dx.doi.org/ 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1299.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature 2008; 454:232-5; http://dx.doi.org/ 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1300.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, et al.. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA 2007; 104:19500-5; http://dx.doi.org/ 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1301.Josefsen L, Droce A, Sondergaard TE, Sørensen JL, Bormann J, Schäfer W, Giese H, Olsson S. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium gaminearum. Autophagy 2012; 8:326-37. [DOI] [PubMed] [Google Scholar]
- 1302.Nadal M, Gold SE. The autophagy genes ATG8 and ATG1 affect morphogenesis and pathogenicity in Ustilago maydis. Mol Plant Pathol 2010; 11:463-78; http://dx.doi.org/ 10.1111/j.1364-3703.2010.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1303.Pollack JK, Harris SD, Marten MR. Autophagy in filamentous fungi. Fungal Genet Biol 2009; 46:1-8; http://dx.doi.org/ 10.1016/j.fgb.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 1304.Richie DL, Fuller KK, Fortwendel J, Miley MD, McCarthy JW, Feldmesser M, Rhodes JC, Askew DS. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot Cell 2007; 6:2437-47; http://dx.doi.org/ 10.1128/EC.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1305.Voigt O, Poggeler S. Self-eating to grow and kill: autophagy in filamentous ascomycetes. Appl Microbiol Biot 2013; 97:9277-90; http://dx.doi.org/ 10.1007/s00253-013-5221-2. [DOI] [PubMed] [Google Scholar]
- 1306.Kim Y, Islam N, Moss BJ, Nandakumar MP, Marten MR. Autophagy induced by rapamycin and carbon-starvation have distinct proteome profiles in Aspergillus nidulans. Biotechnol Bioeng 2011; 108:2705-15; http://dx.doi.org/ 10.1002/bit.23223. [DOI] [PubMed] [Google Scholar]
- 1307.Pinan-Lucarre B, Balguerie A, Clave C. Accelerated cell death in Podospora autophagy mutants. Eukaryot Cell 2005; 4:1765-74; http://dx.doi.org/ 10.1128/EC.4.11.1765-1774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1308.Deng YZ, Naqvi NI. A vacuolar glucoamylase, Sga1, participates in glycogen autophagy for proper asexual differentiation in Magnaporthe oryzae. Autophagy 2010; 6:455-61; http://dx.doi.org/ 10.4161/auto.6.4.11736. [DOI] [PubMed] [Google Scholar]
- 1309.Deng YZ, Ramos-Pamplona M, Naqvi NI. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy 2009; 5:33-43; http://dx.doi.org/ 10.4161/auto.5.1.7175. [DOI] [PubMed] [Google Scholar]
- 1310.Knuppertz L, Hamann A, Pampaloni F, Stelzer E, Osiewacz HD. Identification of autophagy as a longevity-assurance mechanism in the aging model Podospora anserina. Autophagy 2014; 10:822-34; http://dx.doi.org/ 10.4161/auto.28148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1311.Asakura M, Ninomiya S, Sugimoto M, Oku M, Yamashita S, Okuno T, Sakai Y, Takano Y. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 2009; 21:1291-304; http://dx.doi.org/ 10.1105/tpc.108.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1312.Liu XH, Lu JP, Zhang L, Dong B, Min H, Lin FC. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell 2007; 6:997-1005; http://dx.doi.org/ 10.1128/EC.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1313.Nguyen LN, Bormann J, Le GT, Starkel C, Olsson S, Nosanchuk JD, Giese H, Schafer W. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection. Fungal Genet Biol 2011; 48:217-24; http://dx.doi.org/ 10.1016/j.fgb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 1314.Duan Z, Chen Y, Huang W, Shang Y, Chen P, Wang C. Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii. Autophagy 2013; 9:538-49; http://dx.doi.org/ 10.4161/auto.23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1315.Deng YZ, Ramos-Pamplona M, Naqvi NI. Methods for functional analysis of macroautophagy in filamentous fungi. Methods Enzymol 2008; 451:295-310; http://dx.doi.org/ 10.1016/S0076-6879(08)03220-5. [DOI] [PubMed] [Google Scholar]
- 1316.Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci USA 2009; 106:15967-72; http://dx.doi.org/ 10.1073/pnas.0901477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1317.Liu TB, Liu XH, Lu JP, Zhang L, Min H, Lin FC. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy 2010; 6:74-85; http://dx.doi.org/ 10.4161/auto.6.1.10438. [DOI] [PubMed] [Google Scholar]
- 1318.Penalva MA, Galindo A, Abenza JF, Pinar M, Calcagno-Pizarelli AM, Arst HN, Pantazopoulou A. Searching for gold beyond mitosis: Mining intracellular membrane traffic in Aspergillus nidulans. Cell Log 2012; 2:2-14; http://dx.doi.org/ 10.4161/cl.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1319.Pinar M, Pantazopoulou A, Penalva MA. Live-cell imaging of Aspergillus nidulans autophagy: RAB1 dependence, Golgi independence and ER involvement. Autophagy 2013; 9:1024-43; http://dx.doi.org/ 10.4161/auto.24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1320.Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci USA 2012; 109:6981-6; http://dx.doi.org/ 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1321.Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA 2010; 107:7811-6; http://dx.doi.org/ 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1322.Deng Y, Qu Z, Naqvi NI. The role of snx41-based pexophagy in magnaporthe development. PloS One 2013; 8:e79128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1323.Piggott N, Cook MA, Tyers M, Measday V. Genome-wide fitness profiles reveal a requirement for autophagy during yeast fermentation. Genes Genomes Genetics 2011; 1:353-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1324.Cebollero E, Gonzalez R. Induction of autophagy by second-fermentation yeasts during elaboration of sparkling wines. Appl Environ Microbiol 2006; 72:4121-7; http://dx.doi.org/ 10.1128/AEM.02920-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1325.Marks VD, Ho Sui SJ, Erasmus D, van der Merwe GK, Brumm J, Wasserman WW, Bryan J, van Vuuren HJ. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res 2008; 8:35-52; http://dx.doi.org/ 10.1111/j.1567-1364.2007.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1326.Mendes-Ferreira A, Sampaio-Marques B, Barbosa C, Rodrigues F, Costa V, Mendes-Faia A, Ludovico P, Leao C. Accumulation of non-superoxide anion reactive oxygen species mediates nitrogen-limited alcoholic fermentation by Saccharomyces cerevisiae. Appl Environ Microbiol 2010; 76:7918-24; http://dx.doi.org/ 10.1128/AEM.01535-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1327.Rossignol T, Dulau L, Julien A, Blondin B. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 2003; 20:1369-85; http://dx.doi.org/ 10.1002/yea.1046. [DOI] [PubMed] [Google Scholar]
- 1328.Teixeira MC, Raposo LR, Mira NP, Lourenco AB, Sa-Correia I. Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol. Appl Environ Microbiol 2009; 75:5761-72; http://dx.doi.org/ 10.1128/AEM.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1329.Yoshikawa K, Tanaka T, Furusawa C, Nagahisa K, Hirasawa T, Shimizu H. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res 2009; 9:32-44; http://dx.doi.org/ 10.1111/j.1567-1364.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- 1330.Hazan R, Levine A, Abeliovich H. Benzoic acid, a weak organic acid food preservative, exerts specific effects on intracellular membrane trafficking pathways in Saccharomyces cerevisiae. Appl Environ Microbiol 2004; 70:4449-57; http://dx.doi.org/ 10.1128/AEM.70.8.4449-4457.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1331.Singletary K, Milner J. Diet, autophagy, and cancer: a review. Cancer Epidemiol Biomark Prev 2008; 17:1596-610; http://dx.doi.org/ 10.1158/1055-9965.EPI-07-2917. [DOI] [PubMed] [Google Scholar]
- 1332.Su CL, Chen FN, Won SJ. Involvement of apoptosis and autophagy in reducing mouse hepatoma ML-1 cell growth in inbred BALB/c mice by bacterial fermented soybean products. Food Chem Toxicol 2011; 49:17-24; http://dx.doi.org/ 10.1016/j.fct.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 1333.Abeliovich H, Gonzalez R. Autophagy in food biotechnology. Autophagy 2009; 5:925-9; http://dx.doi.org/ 10.4161/auto.5.7.9213. [DOI] [PubMed] [Google Scholar]
- 1334.Berger B, Abdalla FC, Cruz-Landim C. Effect of narcosis with CO2 on the ovarian development in queens of Apis mellifera (Hymenoptera, Apini). Sociobiology 2005; 45:261-70. [Google Scholar]
- 1335.Silva-Zacarin ECM, Tomaino GA, Brocheto-Braga MR, Taboga SR, Silva de Moraes RLM. Programmed cell death in the larval salivary glands of Apis mellifera (Hymenoptera, Apidae). J Biosci 2007; 32:309-28; http://dx.doi.org/ 10.1007/s12038-007-0031-2. [DOI] [PubMed] [Google Scholar]
- 1336.Gregorc A, Bowen ID. Programmed cell death in the honey-bee (Apis mellifera L.) larvae midgut. Cell Biol Int 1997; 21:151-8; http://dx.doi.org/ 10.1006/cbir.1997.0127. [DOI] [PubMed] [Google Scholar]
- 1337.Navajas M, Migeon A, Alaux C, Martin-Magniette M, Robinson G, Evans J, Cros-Arteil S, Crauser D, Le Conte Y. Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC Genomics 2008; 9:301; http://dx.doi.org/ 10.1186/1471-2164-9-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1338.Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res 2013; 73:3-7; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 1339.Takahashi A, Kimura T, Takabatake Y, Namba T, Kaimori J, Kitamura H, Matsui I, Niimura F, Matsusaka T, Fujita N, et al.. Autophagy guards against cisplatin-induced acute kidney injury. Am J Pathol 2012; 180:517-25; http://dx.doi.org/ 10.1016/j.ajpath.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 1340.Colasanti T, Vomero M, Alessandri C, Barbati C, Maselli A, Camperio C, Conti F, Tinari A, Carlo-Stella C, Tuosto L, et al.. Role of alpha-synuclein in autophagy modulation of primary human T lymphocytes. Cell Death Dis 2014; 5:e1265; http://dx.doi.org/ 10.1038/cddis.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1341.Spruessel A, Steimann G, Jung M, Lee SA, Carr T, Fentz AK, Spangenberg J, Zornig C, Juhl HH, David KA. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. BioTechniques 2004; 36:1030-7. [DOI] [PubMed] [Google Scholar]
- 1342.Espina V, Edmiston KH, Heiby M, Pierobon M, Sciro M, Merritt B, Banks S, Deng J, VanMeter AJ, Geho DH, et al.. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics: MCP 2008; 7:1998-2018; http://dx.doi.org/ 10.1074/mcp.M700596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1343.Barth S, Glick D, Macleod KF. Autophagy: assays and artifacts. J Pathol 2010; 221:117-24; http://dx.doi.org/ 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1344.Domart MC, Esposti DD, Sebagh M, Olaya N, Harper F, Pierron G, Franc B, Tanabe KK, Debuire B, Azoulay D, et al.. Concurrent induction of necrosis, apoptosis, and autophagy in ischemic preconditioned human livers formerly treated by chemotherapy. J Hepatol 2009; 51:881-9; http://dx.doi.org/ 10.1016/j.jhep.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 1345.Jahania SM, Sengstock D, Vaitkevicius P, Andres A, Ito BR, Gottlieb RA, Mentzer RM Jr. Activation of the homeostatic intracellular repair response during cardiac surgery. J Am Coll Surgeons 2013; 216:719-26; discussion 26-9; http://dx.doi.org/ 10.1016/j.jamcollsurg.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1346.Singh KK, Yanagawa B, Quan A, Wang R, Garg A, Khan R, Pan Y, Wheatcroft MD, Lovren F, Teoh H, et al.. Autophagy gene fingerprint in human ischemia and reperfusion. J Thor Cardio Surg 2014; 147:1065-72 e1; http://dx.doi.org/ 10.1016/j.jtcvs.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 1347.Nyman E, Brannmark C, Palmer R, Brugard J, Nystrom FH, Stralfors P, Cedersund G. A hierarchical whole-body modeling approach elucidates the link between in Vitro insulin signaling and in Vivo glucose homeostasis. J Biol Chem 2011; 286:26028-41; http://dx.doi.org/ 10.1074/jbc.M110.188987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1348.Adkins Y, Schie IW, Fedor D, Reddy A, Nguyen S, Zhou P, Kelley DS, Wu J. A novel mouse model of nonalcoholic steatohepatitis with significant insulin resistance. Lab Investig 2013; 93:1313-22; http://dx.doi.org/ 10.1038/labinvest.2013.123. [DOI] [PubMed] [Google Scholar]
- 1349.Lake AD, Novak P, Hardwick RN, Flores-Keown B, Zhao F, Klimecki WT, Cherrington NJ. The adaptive endoplasmic reticulum stress response to lipotoxicity in progressive human nonalcoholic fatty liver disease. Toxicol Sci 2014; 137:26-35; http://dx.doi.org/ 10.1093/toxsci/kft230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1350.Sinha RA, Farah BL, Singh BK, Siddique MM, Li Y, Wu Y, Ilkayeva OR, Gooding J, Ching J, Zhou J, et al.. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology 2014; 59:1366-80; http://dx.doi.org/ 10.1002/hep.26667. [DOI] [PubMed] [Google Scholar]
- 1351.Gonzalez-Rodriguez A, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, Vargas-Castrillon J, Lo Iacono O, Corazzari M, Fimia GM, et al.. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis 2014; 5:e1179; http://dx.doi.org/ 10.1038/cddis.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1352.Buzgariu W, Chera S, Galliot B. Methods to investigate autophagy during starvation and regeneration in hydra. Methods Enzymol 2008; 451:409-37; http://dx.doi.org/ 10.1016/S0076-6879(08)03226-6. [DOI] [PubMed] [Google Scholar]
- 1353.Chera S, Buzgariu W, Ghila L, Galliot B. Autophagy in Hydra: a response to starvation and stress in early animal evolution. Biochim Biophys Acta 2009; 1793:1432-43; http://dx.doi.org/ 10.1016/j.bbamcr.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 1354.Chera S, de Rosa R, Miljkovic-Licina M, Dobretz K, Ghila L, Kaloulis K, Galliot B. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. J Cell Sci 2006; 119:846-57; http://dx.doi.org/ 10.1242/jcs.02807. [DOI] [PubMed] [Google Scholar]
- 1355.B Galliot. Autophagy and self-preservation: a step ahead from cell plasticity? Autophagy 2006; 2:231-3; http://dx.doi.org/ 10.4161/auto.2706. [DOI] [PubMed] [Google Scholar]
- 1356.Galliot B, Miljkovic-Licina M, de Rosa R, Chera S. Hydra, a niche for cell and developmental plasticity. Seminars Cell Dev Biol 2006; 17:492-502; http://dx.doi.org/ 10.1016/j.semcdb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 1357.Sala-Mercado JA, Wider J, Undyala VV, Jahania S, Yoo W, Mentzer RM Jr., Gottlieb RA, Przyklenk K. Profound cardioprotection with chloramphenicol succinate in the swine model of myocardial ischemia-reperfusion injury. Circulation 2010; 122:S179-84; http://dx.doi.org/ 10.1161/CIRCULATIONAHA.109.928242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1358.Botting KJ, McMillen IC, Forbes H, Nyengaard JR, Morrison JL. Chronic hypoxemia in late gestation decreases cardiomyocyte number but does not change expression of hypoxia-responsive genes. J Am Heart Assoc 2014; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1359.Wang KC, Brooks DA, Summers-Pearce B, Bobrovskaya L, Tosh DN, Duffield JA, Botting KJ, Zhang S, Caroline McMillen I, Morrison JL. Low birth weight activates the renin-angiotensin system, but limits cardiac angiogenesis in early postnatal life. Physiol Rep 2015; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1360.Zhang S, Regnault TR, Barker PL, Botting KJ, McMillen IC, McMillan CM, Roberts CT, Morrison JL. Placental adaptations in growth restriction. Nutrients 2015; 7:360-89; http://dx.doi.org/ 10.3390/nu7010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1361.Derde S, Vanhorebeek I, Guiza F, Derese I, Gunst J, Fahrenkrog B, Martinet W, Vervenne H, Ververs EJ, Larsson L, et al.. Early parenteral nutrition evokes a phenotype of autophagy deficiency in liver and skeletal muscle of critically ill rabbits. Endocrinology 2012; 153:2267-76; http://dx.doi.org/ 10.1210/en.2011-2068. [DOI] [PubMed] [Google Scholar]
- 1362.Gunst J, Derese I, Aertgeerts A, Ververs EJ, Wauters A, Van den Berghe G, Vanhorebeek I. Insufficient autophagy contributes to mitochondrial dysfunction, organ failure, and adverse outcome in an animal model of critical illness. Crit Care Med 2013; 41:182-94; http://dx.doi.org/ 10.1097/CCM.0b013e3182676657. [DOI] [PubMed] [Google Scholar]
- 1363.Lopez-Alonso I, Aguirre A, Gonzalez-Lopez A, Fernandez AF, Amado-Rodriguez L, Astudillo A, Batalla-Solis E, Albaiceta GM. Impairment of autophagy decreases ventilator-induced lung injury by blockade of the NF-kappaB pathway. Am J Physiol Lung Cell Mol Physiol 2013; 304:L844-52; http://dx.doi.org/ 10.1152/ajplung.00422.2012. [DOI] [PubMed] [Google Scholar]
- 1364.Sun Y, Li C, Shu Y, Ju X, Zou Z, Wang H, Rao S, Guo F, Liu H, Nan W, et al.. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci Signal 2012; 5:ra16. [DOI] [PubMed] [Google Scholar]
- 1365.Sobolewska A, Motyl T, Gajewska M. Role and regulation of autophagy in the development of acinar structures formed by bovine BME-UV1 mammary epithelial cells. Eur J Cell Biol 2011; 90:854-64; http://dx.doi.org/ 10.1016/j.ejcb.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 1366.Motyl T, Gajewska M, Zarzynska J, Sobolewska A, Gajkowska B. Regulation of autophagy in bovine mammary epithelial cells. Autophagy 2007; 3:484-6; http://dx.doi.org/ 10.4161/auto.4491. [DOI] [PubMed] [Google Scholar]
- 1367.Sobolewska A, Gajewska M, Zarzynska J, Gajkowska B, Motyl T. IGF-I, EGF, and sex steroids regulate autophagy in bovine mammary epithelial cells via the mTOR pathway. Eur J Cell Biol 2009; 88:117-30; http://dx.doi.org/ 10.1016/j.ejcb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 1368.Facey CO, Lockshin RA. The execution phase of autophagy associated PCD during insect metamorphosis. Apoptosis 2010; 15:639-52; http://dx.doi.org/ 10.1007/s10495-010-0499-3. [DOI] [PubMed] [Google Scholar]
- 1369.Malagoli D, Abdalla FC, Cao Y, Feng Q, Fujisaki K, Gregorc A, Matsuo T, Nezis IP, Papassideri IS, Sass M, et al.. Autophagy and its physiological relevance in arthropods: Current knowledge and perspectives. Autophagy 2010; 6:575-88; http://dx.doi.org/ 10.4161/auto.6.5.11962. [DOI] [PubMed] [Google Scholar]
- 1370.Mpakou VE, Nezis IP, Stravopodis DJ, Margaritis LH, Papassideri IS. Programmed cell death of the ovarian nurse cells during oogenesis of the silkmoth Bombyx mori. Dev Growth Differ 2006; 48:419-28; http://dx.doi.org/ 10.1111/j.1440-169X.2006.00878.x. [DOI] [PubMed] [Google Scholar]
- 1371.Mpakou VE, Nezis IP, Stravopodis DJ, Margaritis LH, Papassideri IS. Different modes of programmed cell death during oogenesis of the silkmoth Bombyx mori. Autophagy 2008; 4:97-100; http://dx.doi.org/ 10.4161/auto.5205. [DOI] [PubMed] [Google Scholar]
- 1372.Sumithra P, Britto CP, Krishnan M. Modes of cell death in the pupal perivisceral fat body tissue of the silkworm Bombyx mori L. Cell Tissue Res 2010; 339:349-58; http://dx.doi.org/ 10.1007/s00441-009-0898-3. [DOI] [PubMed] [Google Scholar]
- 1373.Tettamanti G, Grimaldi A, Casartelli M, Ambrosetti E, Ponti B, Congiu T, Ferrarese R, Rivas-Pena ML, Pennacchio F, Eguileor M. Programmed cell death and stem cell differentiation are responsible for midgut replacement in Heliothis virescens during prepupal instar. Cell Tissue Res 2007; 330:345-59; http://dx.doi.org/ 10.1007/s00441-007-0449-8. [DOI] [PubMed] [Google Scholar]
- 1374.Khoa DB, Takeda M. Expression of autophagy 8 (Atg8) and its role in the midgut and other organs of the greater wax moth, Galleria mellonella, during metamorphic remodelling and under starvation. Insect Mol Biol 2012; 21:473-87; http://dx.doi.org/ 10.1111/j.1365-2583.2012.01152.x. [DOI] [PubMed] [Google Scholar]
- 1375.Gai Z, Zhang X, Islam M, Wang X, Li A, Yang Y, Li Y, Peng J, Hong H, Liu K. Characterization of Atg8 in lepidopteran insect cells. Arch Insect Biochem 2013; 84:57-77. [DOI] [PubMed] [Google Scholar]
- 1376.Goncu E, Parlak O. Some autophagic and apoptotic features of programmed cell death in the anterior silk glands of the silkworm, Bombyx mori. Autophagy 2008; 4:1069-72; http://dx.doi.org/ 10.4161/auto.6953. [DOI] [PubMed] [Google Scholar]
- 1377.Zhou S, Zhou Q, Liu Y, Wang S, Wen D, He Q, Wang W, Bendena WG, Li S. Two Tor genes in the silkworm Bombyx mori. Insect Mol Biol 2010; 19:727-35; http://dx.doi.org/ 10.1111/j.1365-2583.2010.01026.x. [DOI] [PubMed] [Google Scholar]
- 1378.Zhang X, Hu ZY, Li WF, Li QR, Deng XJ, Yang WY, Cao Y, Zhou CZ. Systematic cloning and analysis of autophagy-related genes from the silkworm Bombyx mori. BMC Mol Biol 2009; 10:50; http://dx.doi.org/ 10.1186/1471-2199-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1379.Romanelli D, Casati B, Franzetti E, Tettamanti G. A molecular view of autophagy in Lepidoptera. Biomed Res Int 2014; 2014:902315; http://dx.doi.org/ 10.1155/2014/902315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1380.Li Q, Deng X, Huang Z, Zheng S, Tettamanti G, Cao Y, Feng Q. Expression of autophagy-related genes in the anterior silk gland of the silkworm (Bombyx mori) during metamorphosis. Can J Zool 2011; 89:1019-26; http://dx.doi.org/ 10.1139/z11-075. [DOI] [Google Scholar]
- 1381.Casati B, Terova G, Cattaneo AG, Rimoldi S, Franzetti E, de Eguileor M, Tettamanti G. Molecular cloning, characterization and expression analysis of ATG1 in the silkworm, Bombyx mori. Gene 2012; 511:326-37; http://dx.doi.org/ 10.1016/j.gene.2012.09.086. [DOI] [PubMed] [Google Scholar]
- 1382.Godefroy N, Hoa C, Tsokanos F, Le Goff E, Douzery EJ, Baghdiguian S, Martinand-Mari C. Identification of autophagy genes in Ciona intestinalis: a new experimental model to study autophagy mechanism. Autophagy 2009; 5:805-15; http://dx.doi.org/ 10.4161/auto.8995. [DOI] [PubMed] [Google Scholar]
- 1383.Martinand-Mari C, Vacelet J, Nickel M, Worheide G, Mangeat P, Baghdiguian S. Cell death and renewal during prey capture and digestion in the carnivorous sponge Asbestopluma hypogea (Porifera: Poecilosclerida). J Exp Biol 2012; 215:3937-43; http://dx.doi.org/ 10.1242/jeb.072371. [DOI] [PubMed] [Google Scholar]
- 1384.Thomé RG, Santos HB, Arantes FP, Domingos FF, Bazzoli N, Rizzo E. Dual roles for autophagy during follicular atresia in fish ovary. Autophagy 2009; 5:117-9; http://dx.doi.org/ 10.4161/auto.5.1.7302. [DOI] [PubMed] [Google Scholar]
- 1385.Santos HB, Thome RG, Arantes FP, Sato Y, Bazzoli N, Rizzo E. Ovarian follicular atresia is mediated by heterophagy, autophagy, and apoptosis in Prochilodus argenteus and Leporinus taeniatus (Teleostei: Characiformes). Theriogenology 2008; 70:1449-60; http://dx.doi.org/ 10.1016/j.theriogenology.2008.06.091. [DOI] [PubMed] [Google Scholar]
- 1386.Santos HB, Sato Y, Moro L, Bazzoli N, Rizzo E. Relationship among follicular apoptosis, integrin beta1 and collagen type IV during early ovarian regression in the teleost Prochilodus argenteus after induced spawning. Cell Tissue Res 2008; 332:159-70; http://dx.doi.org/ 10.1007/s00441-007-0540-1. [DOI] [PubMed] [Google Scholar]
- 1387.Santos HB, Rizzo E, Bazzoli N, Sato Y, Moro L. Ovarian regression and apoptosis in the South American teleost Leporinus taeniatus Lutken (Characiformes, Anostomidae) from the São Francisco Basin. 2005; 67:1446-59. [Google Scholar]
- 1388.Couve E, Schmachtenberg O. Autophagic activity and aging in human odontoblasts. J Dent Res 2011; 90:523-8; http://dx.doi.org/ 10.1177/0022034510393347. [DOI] [PubMed] [Google Scholar]
- 1389.Gonzalez-Estevez C. Autophagy in freshwater planarians. Methods Enzymol 2008; 451:439-65; http://dx.doi.org/ 10.1016/S0076-6879(08)03227-8. [DOI] [PubMed] [Google Scholar]
- 1390.Gonzalez-Estevez C, Felix DA, Aboobaker AA, Salo E. Gtdap-1 promotes autophagy and is required for planarian remodeling during regeneration and starvation. Proc Natl Acad Sci USA 2007; 104:13373-8; http://dx.doi.org/ 10.1073/pnas.0703588104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1391.Toyooka K, Moriyasu Y, Goto Y, Takeuchi M, Fukuda H, Matsuoka K. Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2006; 2:96-106; http://dx.doi.org/ 10.4161/auto.2.2.2366. [DOI] [PubMed] [Google Scholar]
- 1392.Corral-Martinez P, Parra-Vega V, Segui-Simarro JM. Novel features of Brassica napus embryogenic microspores revealed by high pressure freezing and freeze substitution: evidence for massive autophagy and excretion-based cytoplasmic cleaning. J Exp Bot 2013; 64:3061-75. [DOI] [PubMed] [Google Scholar]
- 1393.Le Bars R, Marion J, Le Borgne R, Satiat-Jeunemaitre B, Bianchi MW. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun 2014; 5:4121; http://dx.doi.org/ 10.1038/ncomms5121. [DOI] [PubMed] [Google Scholar]
- 1394.Shin KD, Lee HN, Chung T. A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells 2014; 37:399-405; http://dx.doi.org/ 10.14348/molcells.2014.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1395.Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011; 7:993-1010; http://dx.doi.org/ 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1396.Zientara-Rytter K, Lukomska J, Moniuszko G, Gwozdecki R, Surowiecki P, Lewandowska M, Liszewska F, Wawrzynska A, Sirko A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011; 7:1145-58; http://dx.doi.org/ 10.4161/auto.7.10.16617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1397.Minina EA, Sanchez-Vera V, Moschou PN, Suarez MF, Sundberg E, Weih M, Bozhkov PV. Autophagy mediates caloric restriction-induced lifespan extension in Arabidopsis. Aging Cell 2013; 12:327-9; http://dx.doi.org/ 10.1111/acel.12048. [DOI] [PubMed] [Google Scholar]
- 1398.van Doorn WG, Papini A. Ultrastructure of autophagy in plant cells: a review. Autophagy 2013; 9:1922-36; http://dx.doi.org/ 10.4161/auto.26275. [DOI] [PubMed] [Google Scholar]
- 1399.Moriyasu Y, Inoue Y. Use of protease inhibitors for detecting autophagy in plants. Methods Enzymol 2008; 451:557-80; http://dx.doi.org/ 10.1016/S0076-6879(08)03232-1. [DOI] [PubMed] [Google Scholar]
- 1400.Moriyasu Y, Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Phys 1996; 111:1233-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1401.Inoue Y, Moriyasu Y. Autophagy is not a main contributor to the degradation of phospholipids in tobacco cells cultured under sucrose starvation conditions. Plant Cell Physiol 2006; 47:471-80; http://dx.doi.org/ 10.1093/pcp/pcj013. [DOI] [PubMed] [Google Scholar]
- 1402.Takatsuka C, Inoue Y, Matsuoka K, Moriyasu Y. 3-methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol 2004; 45:265-74; http://dx.doi.org/ 10.1093/pcp/pch031. [DOI] [PubMed] [Google Scholar]
- 1403.Besteiro S, Brooks CF, Striepen B, Dubremetz J-F. Autophagy protein Atg3 is essential for maintaining mitochondrial integrity and for normal intracellular development of Toxoplasma gondii tachyzoites. PLoS Pathog 2011; 7:e1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1404.Calvo-Garrido J, Carilla-Latorre S, Kubohara Y, Santos-Rodrigo N, Mesquita A, Soldati T, Golstein P, Escalante R. Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy 2010; 6:686-701; http://dx.doi.org/ 10.4161/auto.6.6.12513. [DOI] [PubMed] [Google Scholar]
- 1405.Tung SM, Unal C, Ley A, Pena C, Tunggal B, Noegel AA, Krut O, Steinert M, Eichinger L. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell Microbiol 2010; 12:765-80; http://dx.doi.org/ 10.1111/j.1462-5822.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- 1406.Bozzaro S, Eichinger L. The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets 2011; 12:942-54; http://dx.doi.org/ 10.2174/138945011795677782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1407.Schlegel M, Hülsmann N. Protists – A textbook example for a paraphyletic taxon. Org Divers Evol 2007; 7:166-72; http://dx.doi.org/ 10.1016/j.ode.2006.11.001. [DOI] [Google Scholar]
- 1408.Kitamura K, Kishi-Itakura C, Tsuboi T, Sato S, Kita K, Ohta N, Mizushima N. Autophagy-related Atg8 localizes to the apicoplast of the human malaria parasite Plasmodium falciparum. PloS One 2012; 7:e42977; http://dx.doi.org/ 10.1371/journal.pone.0042977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1409.Barquilla A, Crespo JL, Navarro M. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci USA 2008; 105:14579-84; http://dx.doi.org/ 10.1073/pnas.0802668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1410.Hain AU, Bartee D, Sanders NG, Miller AS, Sullivan DJ, Levitskaya J, Meyers CF, Bosch J. Identification of an Atg8-Atg3 protein-protein interaction inhibitor from the medicines for Malaria Venture Malaria Box active in blood and liver stage Plasmodium falciparum parasites. J Med Chem 2014; 57:4521-31; http://dx.doi.org/ 10.1021/jm401675a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1411.Hain AU, Weltzer RR, Hammond H, Jayabalasingham B, Dinglasan RR, Graham DR, Colquhoun DR, Coppens I, Bosch J. Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J Struct Biol 2012; 180:551-62; http://dx.doi.org/ 10.1016/j.jsb.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1412.Navale R, Atul , Allanki AD, Sijwali PS. Characterization of the autophagy marker protein Atg8 reveals atypical features of autophagy in Plasmodium falciparum. PloS One 2014; 9:e113220; http://dx.doi.org/ 10.1371/journal.pone.0113220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1413.Morais P, Lamas J, Sanmartin ML, Orallo F, Leiro J. Resveratrol induces mitochondrial alterations, autophagy and a cryptobiosis-like state in scuticociliates. Protist 2009; 160:552-64; http://dx.doi.org/ 10.1016/j.protis.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 1414.Yakisich JS, Kapler GM. The effect of phosphoinositide 3-kinase inhibitors on programmed nuclear degradation in Tetrahymena and fate of surviving nuclei. Cell Death Differ 2004; 11:1146-9; http://dx.doi.org/ 10.1038/sj.cdd.4401473. [DOI] [PubMed] [Google Scholar]
- 1415.Akematsu T, Pearlman RE, Endoh H. Gigantic macroautophagy in programmed nuclear death of Tetrahymena thermophila. Autophagy 2010; 6:901-11; http://dx.doi.org/ 10.4161/auto.6.7.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1416.Akematsu T, Fukuda Y, Attiq R, Pearlman RE. Role of class III phosphatidylinositol 3-kinase during programmed nuclear death of Tetrahymena thermophila. Autophagy 2014; 10:209-25; http://dx.doi.org/ 10.4161/auto.26929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1417.Liu ML, Yao MC. Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila. Eukaryot Cell 2012; 11:494-506; http://dx.doi.org/ 10.1128/EC.05296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1418.Thorgaard GH, Bailey GS, Williams D, Buhler DR, Kaattari SL, Ristow SS, Hansen JD, Winton JR, Bartholomew JL, Nagler JJ, et al.. Status and opportunities for genomics research with rainbow trout. Comp Biochem Phys B 2002; 133:609-46; http://dx.doi.org/ 10.1016/S1096-4959(02)00167-7. [DOI] [PubMed] [Google Scholar]
- 1419.Govoroun M, Le Gac F, Guiguen Y. Generation of a large scale repertoire of Expressed Sequence Tags (ESTs) from normalised rainbow trout cDNA libraries. BMC Genomics 2006; 7:196; http://dx.doi.org/ 10.1186/1471-2164-7-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1420.Rexroad CE III, Lee Y, Keele JW, Karamycheva S, Brown G, Koop B, Gahr SA, Palti Y, Quackenbush J. Sequence analysis of a rainbow trout cDNA library and creation of a gene index. Cytogenetic Genome Res 2003; 102:347-54; http://dx.doi.org/ 10.1159/000075773. [DOI] [PubMed] [Google Scholar]
- 1421.Rise ML, von Schalburg KR, Brown GD, Mawer MA, Devlin RH, Kuipers N, Busby M, Beetz-Sargent M, Alberto R, Gibbs AR, et al.. Development and application of a salmonid EST database and cDNA microarray: data mining and interspecific hybridization characteristics. Genome Res 2004; 14:478-90; http://dx.doi.org/ 10.1101/gr.1687304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1422.Salem M, Rexroad CE III, Wang J, Thorgaard GH, Yao J. Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics 2010; 11:564; http://dx.doi.org/ 10.1186/1471-2164-11-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1423.Polakof S, Panserat S, Craig PM, Martyres DJ, Plagnes-Juan E, Savari S, Aris-Brosou S, Moon TW. The metabolic consequences of hepatic AMP-kinase phosphorylation in rainbow trout. PloS One 2011; 6:e20228; http://dx.doi.org/ 10.1371/journal.pone.0020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1424.Seiliez I, Gabillard JC, Skiba-Cassy S, Garcia-Serrana D, Gutierrez J, Kaushik S, Panserat S, Tesseraud S. An in vivo and in vitro assessment of TOR signaling cascade in rainbow trout (Oncorhynchus mykiss). Am J Physiol Reg Integ Comp Physiol 2008; 295:R329-35; http://dx.doi.org/ 10.1152/ajpregu.00146.2008. [DOI] [PubMed] [Google Scholar]
- 1425.Seiliez I, Gabillard J-C, Riflade M, Sadoul B, Dias K, Avérous J, Tesseraud S, Skiba S, Panserat S. Amino acids downregulate the expression of several autophagy-related genes in rainbow trout myoblasts. Autophagy 2012; 8:364-75. [DOI] [PubMed] [Google Scholar]
- 1426.Chiarelli R, Agnello M, Bosco L, Roccheri MC. Sea urchin embryos exposed to cadmium as an experimental model for studying the relationship between autophagy and apoptosis. Mar Environ Res 2014; 93:47-55; http://dx.doi.org/ 10.1016/j.marenvres.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 1427.Umemiya R, Matsuo T, Hatta T, Sakakibara S, Boldbaatar D, Fujisaki K. Cloning and characterization of an autophagy-related gene, ATG12, from the three-host tick Haemaphysalis longicornis. Insect Biochem Molec 2007; 37:975-84; http://dx.doi.org/ 10.1016/j.ibmb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 1428.Kawano S, Umemiya-Shirafuji R, Boldbaatar D, Matsuoka K, Tanaka T, Fujisaki K. Cloning and characterization of the autophagy-related gene 6 from the hard tick, Haemaphysalis longicornis. Parasitol Res 2011; 109:1341-9; http://dx.doi.org/ 10.1007/s00436-011-2429-x. [DOI] [PubMed] [Google Scholar]
- 1429.Umemiya-Shirafuji R, Matsuo T, Liao M, Boldbaatar D, Battur B, Suzuki HI, Fujisaki K. Increased expression of ATG genes during nonfeeding periods in the tick Haemaphysalis longicornis. Autophagy 2010; 6:473-81; http://dx.doi.org/ 10.4161/auto.6.4.11668. [DOI] [PubMed] [Google Scholar]
- 1430.Umemiya-Shirafuji R, Galay RL, Maeda H, Kawano S, Tanaka T, Fukumoto S, Suzuki H, Tsuji N, Fujisaki K. Expression analysis of autophagy-related genes in the hard tick Haemaphysalis longicornis. Vet Parasitol 2014; 201:169-75; http://dx.doi.org/ 10.1016/j.vetpar.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 1431.de la Fuente J, Kocan KM, Almazan C, Blouin EF. RNA interference for the study and genetic manipulation of ticks. Trends Parasitol 2007; 23:427-33; http://dx.doi.org/ 10.1016/j.pt.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 1432.Ayllón N, Villar V, Galindo RC, Kocan KM, Šíma R, López JA, Vázquez J, Alberdi P, Cabezas-Cruz A, Kopáček P, et al.. Systems biology of tissue-specific response to Anaplasma phagocytophilum reveals differentiated apoptosis in the tick vector Ixodes scapularis. PLoS Genet 2015; 11:e1005120; http://dx.doi.org/ 10.1371/journal.pgen.1005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1433.Genomic Resources Development C, Contreras M, de la Fuente J, Estrada-Pena A, Grubhoffer L, Tobes R. Genomic resources notes accepted 1 April 2014 - 31 May 2014. Mol Ecol Resour 2014; 14:1095. [DOI] [PubMed] [Google Scholar]
- 1434.Lee E, Koo Y, Ng A, Wei Y, Luby-Phelps K, Juraszek A, Xavier RJ, Cleaver O, Levine B, Amatruda JF. Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy 2014; 10:572-87; http://dx.doi.org/ 10.4161/auto.27649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1435.Sasaki T, Lian S, Qi J, Bayliss PE, Carr CE, Johnson JL, Guha S, Kobler P, Catz SD, Gill M, et al.. Aberrant autolysosomal regulation is linked to the induction of embryonic senescence: differential roles of Beclin 1 and p53 in vertebrate Spns1 deficiency. PLoS Genet 2014; 10:e1004409; http://dx.doi.org/ 10.1371/journal.pgen.1004409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1436.He C, Bartholomew CR, Zhou W, Klionsky DJ. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy 2009; 5:520-6; http://dx.doi.org/ 10.4161/auto.5.4.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1437.Komoike Y, Shimojima K, Liang JS, Fujii H, Maegaki Y, Osawa M, Fujii S, Higashinakagawa T, Yamamoto T. A functional analysis of GABARAP on 17p13.1 by knockdown zebrafish. J Hum Genet 2010; 55:155-62; http://dx.doi.org/ 10.1038/jhg.2010.1. [DOI] [PubMed] [Google Scholar]
- 1438.Dowling JJ, Low SE, Busta AS, Feldman EL. Zebrafish MTMR14 is required for excitation-contraction coupling, developmental motor function and the regulation of autophagy. Hum Mol Genet 2010; 19:2668-81; http://dx.doi.org/ 10.1093/hmg/ddq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1439.Makky K, Tekiela J, Mayer AN. Target of rapamycin (TOR) signaling controls epithelial morphogenesis in the vertebrate intestine. Dev Biol 2007; 303:501-13; http://dx.doi.org/ 10.1016/j.ydbio.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1440.Moreau K, Fleming A, Imarisio S, Lopez Ramirez A, Mercer JL, Jimenez-Sanchez M, Bento CF, Puri C, Zavodszky E, Siddiqi F, et al.. PICALM modulates autophagy activity and tau accumulation. Nat Commun 2014; 5:4998; http://dx.doi.org/ 10.1038/ncomms5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1441.Hishiya A, Salman MN, Carra S, Kampinga HH, Takayama S. BAG3 directly interacts with mutated alphaB-crystallin to suppress its aggregation and toxicity. PloS One 2011; 6:e16828; http://dx.doi.org/ 10.1371/journal.pone.0016828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1442.Ruparelia AA, Oorschot V, Vaz R, Ramm G, Bryson-Richardson RJ. Zebrafish models of BAG3 myofibrillar myopathy suggest a toxic gain of function leading to BAG3 insufficiency. Acta Neuropathol 2014; 128:821-33; http://dx.doi.org/ 10.1007/s00401-014-1344-5. [DOI] [PubMed] [Google Scholar]
- 1443.Mostowy S, Boucontet L, Mazon Moya MJ, Sirianni A, Boudinot P, Hollinshead M, Cossart P, Herbomel P, Levraud JP, Colucci-Guyon E. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog 2013; 9:e1003588; http://dx.doi.org/ 10.1371/journal.ppat.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1444.van der Vaart M, Korbee CJ, Lamers GE, Tengeler AC, Hosseini R, Haks MC, Ottenhoff TH, Spaink HP, Meijer AH. The DNA Damage-Regulated Autophagy Modulator DRAM1 Links Mycobacterial Recognition via TLP-MYD88 to Authophagic Defense. Cell Host Microbe 2014; 15:753-67; http://dx.doi.org/ 10.1016/j.chom.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 1445.Varga M, Sass M, Papp D, Takacs-Vellai K, Kobolak J, Dinnyes A, Klionsky DJ, Vellai T. Autophagy is required for zebrafish caudal fin regeneration. Cell Death Differ 2014; 21:547-56; http://dx.doi.org/ 10.1038/cdd.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1446.Benato F, Skobo T, Gioacchini G, Moro I, Ciccosanti F, Piacentini M, Fimia GM, Carnevali O, Dalla Valle L. Ambra1 knockdown in zebrafish leads to incomplete development due to severe defects in organogenesis. Autophagy 2013; 9:476-95; http://dx.doi.org/ 10.4161/auto.23278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1447.Skobo T, Benato F, Grumati P, Meneghetti G, Cianfanelli V, Castagnaro S, Chrisam M, Di Bartolomeo S, Bonaldo P, Cecconi F, et al.. Zebrafish ambra1a and ambra1b knockdown impairs skeletal muscle development. PloS One 2014; 9:e99210; http://dx.doi.org/ 10.1371/journal.pone.0099210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1448.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol 2009; 452:13-23; http://dx.doi.org/ 10.1016/S0076-6879(08)03602-1. [DOI] [PubMed] [Google Scholar]
- 1449.Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al.. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 2012; 37:986-97; http://dx.doi.org/ 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1450.Varma H, Gangadhar NM, Letso RR, Wolpaw AJ, Sriramaratnam R, Stockwell BR. Identification of a small molecule that induces ATG5-and-cathepsin-l-dependent cell death and modulates polyglutamine toxicity. Exp Cell Res 2013; 319:1759-73; http://dx.doi.org/ 10.1016/j.yexcr.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1451.Kong-Hap MA, Mouammine A, Daher W, Berry L, Lebrun M, Dubremetz JF, Besteiro S. Regulation of ATG8 membrane association by ATG4 in the parasitic protist Toxoplasma gondii. Autophagy 2013; 9:1334-48; http://dx.doi.org/ 10.4161/auto.25189. [DOI] [PubMed] [Google Scholar]
- 1452.Jayabalasingham B, Voss C, Ehrenman K, Romano JD, Smith ME, Fidock DA, Bosch J, Coppens I. Characterization of the ATG8-conjugation system in 2 Plasmodium species with special focus on the liver stage: possible linkage between the apicoplastic and autophagic systems? Autophagy 2014; 10:269-84; http://dx.doi.org/ 10.4161/auto.27166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1453.Tomlins AM, Ben-Rached F, Williams RA, Proto WR, Coppens I, Ruch U, Gilberger TW, Coombs GH, Mottram JC, Muller S, et al.. Plasmodium falciparum ATG8 implicated in both autophagy and apicoplast formation. Autophagy 2013; 9:1540-52; http://dx.doi.org/ 10.4161/auto.25832. [DOI] [PubMed] [Google Scholar]
- 1454.Mizushima N, Sahani MH. ATG8 localization in apicomplexan parasites: apicoplast and more? Autophagy 2014; 10:1487-94; http://dx.doi.org/ 10.4161/auto.32183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1455.Haldar AK, Piro AS, Pilla DM, Yamamoto M, Coers J. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PloS One 2014; 9:e86684; http://dx.doi.org/ 10.1371/journal.pone.0086684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1456.Ohshima J, Lee Y, Sasai M, Saitoh T, Su Ma J, Kamiyama N, Matsuura Y, Pann-Ghill S, Hayashi M, Ebisu S, et al.. Role of mouse and human autophagy proteins in IFN-gamma-induced cell-autonomous responses against Toxoplasma gondii. J Immunol 2014; 192:3328-35; http://dx.doi.org/ 10.4049/jimmunol.1302822. [DOI] [PubMed] [Google Scholar]
- 1457.Zhao YO, Khaminets A, Hunn JP, Howard JC. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog 2009; 5:e1000288; http://dx.doi.org/ 10.1371/journal.ppat.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1458.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, et al.. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature 2014; 509:366-70; http://dx.doi.org/ 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 1459.Taguchi Y, Imaoka K, Kataoka M, Uda A, Nakatsu D, Horii-Okazaki S, Kunishige R, Kano F, Murata M. Yip1A, a novel host factor for the activation of the IRE1 pathway of the unfolded protein response during Brucella infection. PLoS Pathog 2015; 11:e1004747; http://dx.doi.org/ 10.1371/journal.ppat.1004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1460.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, Virgin HW, Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 2012; 11:33-45; http://dx.doi.org/ 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1461.Ferguson TA, Green DR. Autophagy and phagocytosis converge for better vision. Autophagy 2014; 10:165-7; http://dx.doi.org/ 10.4161/auto.26735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1462.Mehta P, Henault J, Kolbeck R, Sanjuan MA. Noncanonical autophagy: one small step for LC3, one giant leap for immunity. Curr Opin Immunol 2014; 26:69-75; http://dx.doi.org/ 10.1016/j.coi.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 1463.Scarlatti F, Maffei R, Beau I, Ghidoni R, Codogno P. Non-canonical autophagy: an exception or an underestimated form of autophagy? Autophagy 2008; 4:1083-5; http://dx.doi.org/ 10.4161/auto.7068. [DOI] [PubMed] [Google Scholar]
- 1464.Takeshita F, Kobiyama K, Miyawaki A, Jounai N, Okuda K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy 2008; 4:67-9; http://dx.doi.org/ 10.4161/auto.5055. [DOI] [PubMed] [Google Scholar]
- 1465.Deretic V, Jiang S, Dupont N. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol 2012; 22:397-406; http://dx.doi.org/ 10.1016/j.tcb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1466.Cleyrat C, Darehshouri A, Steinkamp MP, Vilaine M, Boassa D, Ellisman MH, Hermouet S, Wilson BS. Mpl traffics to the cell surface through conventional and unconventional routes. Traffic 2014; 15:961-82; http://dx.doi.org/ 10.1111/tra.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1467.Hughes T, Rusten TE. Origin and evolution of self-consumption: autophagy. Adv Exp Med Biol 2007; 607:111-8; http://dx.doi.org/ 10.1007/978-0-387-74021-8. [DOI] [PubMed] [Google Scholar]
- 1468.JA Kiel. Autophagy in unicellular eukaryotes. Philos Trans R Soc B 2010; 365:819-30; http://dx.doi.org/ 10.1098/rstb.2009.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1469.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389-402; http://dx.doi.org/ 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1470.Pertsemlidis A, Fondon JW III. Having a BLAST with bioinformatics (and avoiding BLASTphemy). Genome Biol 2001; 2:REVIEWS2002; http://dx.doi.org/ 10.1186/gb-2001-2-10-reviews2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1471.B Rost. Twilight zone of protein sequence alignments. Protein engineering 1999; 12:85-94; http://dx.doi.org/ 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 1472.Duszenko M, Ginger ML, Brennand A, Gualdron-Lopez M, Colombo MI, Coombs GH, Coppens I, Jayabalasingham B, Langsley G, de Castro SL, et al.. Autophagy in protists. Autophagy 2011; 7:127-58; http://dx.doi.org/ 10.4161/auto.7.2.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1473.Rigden DJ, Michels PA, Ginger ML. Autophagy in protists: Examples of secondary loss, lineage-specific innovations, and the conundrum of remodeling a single mitochondrion. Autophagy 2009; 5:784-94; http://dx.doi.org/ 10.4161/auto.8838. [DOI] [PubMed] [Google Scholar]
- 1474.Katsani KR, Irimia M, Karapiperis C, Scouras ZG, Blencowe BJ, Promponas VJ, Ouzounis CA. Functional genomics evidence unearths new moonlighting roles of outer ring coat nucleoporins. Sci Rep 2014; 4:4655; http://dx.doi.org/ 10.1038/srep04655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1475.Mei Y, Su M, Soni G, Salem S, Colbert CL, Sinha SC. Intrinsically disordered regions in autophagy proteins. Proteins 2014; 82:565-78; http://dx.doi.org/ 10.1002/prot.24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1476.Promponas VJ, Ouzounis CA, Iliopoulos I. Experimental evidence validating the computational inference of functional associations from gene fusion events: a critical survey. Brief Bioinform 2014; 15:443-54; http://dx.doi.org/ 10.1093/bib/bbs072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1477.Homma K, Suzuki K, Sugawara H. The Autophagy Database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res 2011; 39:D986-90; http://dx.doi.org/ 10.1093/nar/gkq995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1478.Turei D, Foldvari-Nagy L, Fazekas D, Modos D, Kubisch J, Kadlecsik T, Demeter A, Lenti K, Csermely P, Vellai T, et al.. Autophagy Regulatory Network - a systems-level bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy 2015; 11:155-65; http://dx.doi.org/ 10.4161/15548627.2014.994346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1479.Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci 2013; 126:3237-47. [DOI] [PubMed] [Google Scholar]
- 1480.Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J Cell Sci 2014; 127:3-9; http://dx.doi.org/ 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- 1481.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 2010; 584:1379-85; http://dx.doi.org/ 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 1482.Kalvari I, Tsompanis S, Mulakkal NC, Osgood R, Johansen T, Nezis IP, Promponas VJ. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy 2014; 10:913-25; http://dx.doi.org/ 10.4161/auto.28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1483.Dosztanyi Z, Meszaros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009; 25:2745-6; http://dx.doi.org/ 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1484.Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Kruger D, Grebnev G, Kuban M, et al.. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res 2014; 42:D259-66; http://dx.doi.org/ 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1485.Wu D, Huang Y, Kang JJ, Li KN, Bi XM, Zhang T, Jin NN, Hu YF, Tan PW, Zhang L, et al.. ncRDeathDB: a comprehensive bioinformatics resource for deciphering network organization of the ncRNA-mediated cell death system. Autophagy 2015; 11:1917-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1486.Li Y, Zhuang L, Wang Y, Hu Y, Wu Y, Wang D, Xu J. Connect the dots: a systems level approach for analyzing the miRNA-mediated cell death network. Autophagy 2013; 9:436-9; http://dx.doi.org/ 10.4161/auto.23096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1487.Xu J, Li YH. miRDeathDB: a database bridging microRNAs and the programmed cell death. Cell Death Differ 2012; 19:1571; http://dx.doi.org/ 10.1038/cdd.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1488.Xu J, Wang Y, Tan X, Jing H. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 2012; 8:873-82; http://dx.doi.org/ 10.4161/auto.19629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1489.Tavassoly I, Parmar J, Shajahan-Haq AN, Clarke R, Baumann WT, Tyson JJ. Dynamic Modeling of the Interaction Between Autophagy and Apoptosis in Mammalian Cells. CPT Pharmacometrics Syst Pharmacol 2015; 4:263-72; http://dx.doi.org/ 10.1002/psp4.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1490.Tavassoly I. Dynamics of Cell Fate Decision Mediated by the Interplay of Autophagy and Apoptosis in Cancer Cells: Mathematical Modeling and Experimental Observations. Springer, 2015. [Google Scholar]
- 1491.Borlin CS, Lang V, Hamacher-Brady A, Brady NR. Agent-based modeling of autophagy reveals emergent regulatory behavior of spatio-temporal autophagy dynamics. Cell Commun Signal 2014; 12:56; http://dx.doi.org/ 10.1186/s12964-014-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1492.Martin KR, Barua D, Kauffman AL, Westrate LM, Posner RG, Hlavacek WS, Mackeigan JP. Computational model for autophagic vesicle dynamics in single cells. Autophagy 2013; 9:74-92; http://dx.doi.org/ 10.4161/auto.22532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1493.Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL, et al.. A comprehensive glossary of autophagy-related molecules and processes (2nd) edition). Autophagy 2011; 7:1273-94; http://dx.doi.org/ 10.4161/auto.7.11.17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1494.Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, Gewirtz DA, Kroemer G, Levine B, Mizushima N, et al.. A comprehensive glossary of autophagy-related molecules and processes. Autophagy 2010; 6:438-48; http://dx.doi.org/ 10.4161/auto.6.4.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1495.Rosich L, Xargay-Torrent S, Lopez-Guerra M, Campo E, Colomer D, Roue G. Counteracting autophagy overcomes resistance to everolimus in mantle cell lymphoma. Clin Cancer Res 2012; 18:5278-89; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-0351. [DOI] [PubMed] [Google Scholar]
- 1496.Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, Cuervo AM. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol 2013; 9:374-82; http://dx.doi.org/ 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1497.De Mei C, Ercolani L, Parodi C, Veronesi M, Vecchio CL, Bottegoni G, Torrente E, Scarpelli R, Marotta R, Ruffili R, et al.. Dual inhibition of REV-ERBbeta and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene 2015; 34:2597-608; http://dx.doi.org/ 10.1038/onc.2014.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1498.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 2008; 19:4651-9; http://dx.doi.org/ 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1499.Vanrell MC, Cueto JA, Barclay JJ, Carrillo C, Colombo MI, Gottlieb RA, Romano PS. Polyamine depletion inhibits the autophagic response modulating Trypanosoma cruzi infectivity. Autophagy 2013; 9:1080-93; http://dx.doi.org/ 10.4161/auto.24709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1500.Song W, Zukor H, Liberman A, Kaduri S, Arvanitakis Z, Bennett DA, Schipper HM. Astroglial heme oxygenase-1 and the origin of corpora amylacea in aging and degenerating neural tissues. Exp Neurol 2014; 254:78-89; http://dx.doi.org/ 10.1016/j.expneurol.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1501.Song W, Zukor H, Lin SH, Liberman A, Tavitian A, Mui J, Vali H, Fillebeen C, Pantopoulos K, Wu TD, et al.. Unregulated brain iron deposition in transgenic mice over-expressing HMOX1 in the astrocytic compartment. J Neurochem 2012; 123:325-36; http://dx.doi.org/ 10.1111/j.1471-4159.2012.07914.x. [DOI] [PubMed] [Google Scholar]
- 1502.Zukor H, Song W, Liberman A, Mui J, Vali H, Fillebeen C, Pantopoulos K, Wu TD, Guerquin-Kern JL, Schipper HM. HO-1-mediated macroautophagy: a mechanism for unregulated iron deposition in aging and degenerating neural tissues. J Neurochem 2009; 109:776-91; http://dx.doi.org/ 10.1111/j.1471-4159.2009.06007.x. [DOI] [PubMed] [Google Scholar]
- 1503.Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem J 2009; 421:29-42; http://dx.doi.org/ 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1504.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al.. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009; 458:732-6; http://dx.doi.org/ 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 1505.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, et al.. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res 2012; 72:3360-71; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 1506.Yang D, Zhao Y, Liu J, Sun Y, Jia L. Protective autophagy induced by RBX1/ROC1 knockdown or CRL inactivation via modulating the DEPTOR-MTOR axis. Autophagy 2012; 8:1856-8; http://dx.doi.org/ 10.4161/auto.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1507.Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis 2012; 3:e386; http://dx.doi.org/ 10.1038/cddis.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1508.Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S, et al.. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res 2008; 68:8022-30; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 1509.Liu TJ, Koul D, LaFortune T, Tiao N, Shen RJ, Maira SM, Garcia-Echevrria C, Yung WK. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther 2009; 8:2204-10; http://dx.doi.org/ 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1510.Pirola L, Frojdo S. Resveratrol: one molecule, many targets. IUBMB Life 2008; 60:323-32; http://dx.doi.org/ 10.1002/iub.47. [DOI] [PubMed] [Google Scholar]
- 1511.Vingtdeux V, Giliberto L, Zhao H, Chandakkar P, Wu Q, Simon JE, Janle EM, Lobo J, Ferruzzi MG, Davies P, et al.. AMP-activated protein kinase signaling activation by resveratrol modulates amyloid-beta peptide metabolism. J Biol Chem 2010; 285:9100-13; http://dx.doi.org/ 10.1074/jbc.M109.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1512.Puissant A, Auberger P. AMPK- and p62/SQSTM1-dependent autophagy mediate Resveratrol-induced cell death in chronic myelogenous leukemia. Autophagy 2010; 6:655-7; http://dx.doi.org/ 10.4161/auto.6.5.12126. [DOI] [PubMed] [Google Scholar]
- 1513.Vingtdeux V, Chandakkar P, Zhao H, d'Abramo C, Davies P, Marambaud P. Novel synthetic small-molecule activators of AMPK as enhancers of autophagy and amyloid-[b] peptide degradation. The FASEB J 2011; 25:219-31; http://dx.doi.org/ 10.1096/fj.10-167361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1514.Wong VK, Li T, Law BY, Ma ED, Yip NC, Michelangeli F, Law CK, Zhang MM, Lam KY, Chan PL, et al.. Saikosaponin-d, a novel SERCA inhibitor, induces autophagic cell death in apoptosis-defective cells. Cell Death Dis 2013; 4:e720; http://dx.doi.org/ 10.1038/cddis.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1515.Gordon PB, Holen I, Fosse M, Rotnes JS, Seglen PO. Dependence of hepatocytic autophagy on intracellularly sequestered calcium. J Biol Chem 1993; 268:26107-12. [PubMed] [Google Scholar]
- 1516.Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 2011; 42:731-43; http://dx.doi.org/ 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1517.Zhang L, Dai F, Cui L, Jing H, Fan P, Tan X, Guo Y, Zhou G. Novel role for TRPC4 in regulation of macroautophagy by a small molecule in vascular endothelial cells. Biochim Biophys Acta 2015; 1853:377-87; http://dx.doi.org/ 10.1016/j.bbamcr.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 1518.Casarejos MJ, Solano RM, Gomez A, Perucho J, de Yebenes JG, Mena MA. The accumulation of neurotoxic proteins, induced by proteasome inhibition, is reverted by trehalose, an enhancer of autophagy, in human neuroblastoma cells. Neurochem Int 2011; 58:512-20; http://dx.doi.org/ 10.1016/j.neuint.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 1519.Fernandez-Estevez MA, Casarejos MJ, Lopez Sendon J, Garcia Caldentey J, Ruiz C, Gomez A, Perucho J, de Yebenes JG, Mena MA. Trehalose reverses cell malfunction in fibroblasts from normal and Huntington's disease patients caused by proteosome inhibition. PloS One 2014; 9:e90202; http://dx.doi.org/ 10.1371/journal.pone.0090202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1520.Carpenter JE, Jackson W, Benetti L, Grose C. Autophagosome formation during varicella-zoster virus Infection following endoplasmic reticulum stress and the unfolded protein response. J Virol 2011; 85:9414-24; http://dx.doi.org/ 10.1128/JVI.00281-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1521.Lu Y, Dong S, Hao B, Li C, Zhu K, Guo W, Wang Q, Cheung KH, Wong CW, Wu WT, et al.. Vacuolin-1 potently and reversibly inhibits autophagosome-lysosome fusion by activating RAB5A. Autophagy 2014; 10:1895-905; http://dx.doi.org/ 10.4161/auto.32200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1522.Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G, Kraft C, Peter M. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy 2010; 6:1168-78; http://dx.doi.org/ 10.4161/auto.6.8.13849. [DOI] [PubMed] [Google Scholar]
- 1523.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 2010; 30:1049-58; http://dx.doi.org/ 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1524.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA 2009; 106:17049-54; http://dx.doi.org/ 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1525.Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, Sinha S, Gaestel M, Levine B. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. eLife 2015; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1526.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013; 152:290-303; http://dx.doi.org/ 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1527.Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J 1999; 344 Pt 2:427-31; http://dx.doi.org/ 10.1042/bj3440427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1528.Peterson RT, Beal PA, Comb MJ, Schreiber SL. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem 2000; 275:7416-23; http://dx.doi.org/ 10.1074/jbc.275.10.7416. [DOI] [PubMed] [Google Scholar]
- 1529.Nicot AS, Lo Verso F, Ratti F, Pilot-Storck F, Streichenberger N, Sandri M, Schaeffer L, Goillot E. Phosphorylation of NBR1 by GSK3 modulates protein aggregation. Autophagy 2014; 10:1036-53; http://dx.doi.org/ 10.4161/auto.28479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1530.Rosner M, Fuchs C, Siegel N, Valli A, Hengstschlager M. Functional interaction of mammalian target of rapamycin complexes in regulating mammalian cell size and cell cycle. Hum Mol Genet 2009; 18:3298-310; http://dx.doi.org/ 10.1093/hmg/ddp271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1531.Shin S, Wolgamott L, Yu Y, Blenis J, Yoon SO. Glycogen synthase kinase (GSK)-3 promotes p70 ribosomal protein S6 kinase (p70S6K) activity and cell proliferation. Proc Natl Acad Sci USA 2011; 108:E1204-13; http://dx.doi.org/ 10.1073/pnas.1110195108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1532.Ro SH, Semple IA, Park H, Park H, Park HW, Kim M, Kim JS, Lee JH. Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1. FEBS J 2014; 281:3816-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1533.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan J-L, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008; 181:497-510; http://dx.doi.org/ 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1534.Xue L, Fletcher GC, Tolkovsky AM. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol Cell Neurosci 1999; 14:180-98; http://dx.doi.org/ 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- 1535.Zhang N, Chen Y, Jiang R, Li E, Chen X, Xi Z, Guo Y, Liu X, Zhou Y, Che Y, et al.. PARP and RIP 1 are required for autophagy induced by 11'-deoxyverticillin A, which precedes caspase-dependent apoptosis. Autophagy 2011; 7:598-612; http://dx.doi.org/ 10.4161/auto.7.6.15103. [DOI] [PubMed] [Google Scholar]
- 1536.Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 2010; 142:590-600; http://dx.doi.org/ 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1537.Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy 2007; 3: 374-6; http://dx.doi.org/ 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 1538.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al.. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005; 435:677-81; http://dx.doi.org/ 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 1539.Nazarko TY. Atg37 regulates the assembly of the pexophagic receptor protein complex. Autophagy 2014; 10:1348-9; http://dx.doi.org/ 10.4161/auto.29073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1540.Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Kuttner V, Bhukel A, Marino G, Pietrocola F, Harger A, Zimmermann A, et al.. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab 2014; 19:431-44; http://dx.doi.org/ 10.1016/j.cmet.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1541.Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al.. Regulation of autophagy by cytosolic acetyl-coenzyme a. Mol Cell 2014; 53:710-25; http://dx.doi.org/ 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 1542.Nandi N, Tyra LK, Stenesen D, Kramer H. Acinus integrates AKT1 and subapoptotic caspase activities to regulate basal autophagy. J Cell Biol 2014; 207:253-68; http://dx.doi.org/ 10.1083/jcb.201404028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1543.Haberman AS, Akbar MA, Ray S, Kramer H. Drosophila acinus encodes a novel regulator of endocytic and autophagic trafficking. Development 2010; 137:2157-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1544.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al.. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol 2009; 11:1233-40; http://dx.doi.org/ 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 1545.Till A, Lipinski S, Ellinghaus D, Mayr G, Subramani S, Rosenstiel P, Franke A. Autophagy receptor CALCOCO2/NDP52 takes center stage in Crohn disease. Autophagy 2013; 9:1256-7; http://dx.doi.org/ 10.4161/auto.25483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1546.Eby KG, Rosenbluth JM, Mays DJ, Marshall CB, Barton CE, Sinha S, Johnson KN, Tang L, Pietenpol JA. ISG20L1 is a p53 family target gene that modulates genotoxic stress-induced autophagy. Mol Cancer 2010; 9:95; http://dx.doi.org/ 10.1186/1476-4598-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1547.Kang R, Tang D, Livesey KM, Schapiro NE, Lotze MT, Zeh HJ 3rd. The Receptor for Advanced Glycation End-products (RAGE) protects pancreatic tumor cells against oxidative injury. Antioxid Redox Sign 2011; 15:2175-84; http://dx.doi.org/ 10.1089/ars.2010.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1548.Kang R, Tang D, Livesey KM, Schapiro NE, Lotze MT, Zeh HJ. The receptor for advanced glycation end-products (RAGE) protects pancreatic tumor cells against oxidative injury. Antioxid Redox Sign 2011; 15:2175-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1549.Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep 2011; 12:149-56; http://dx.doi.org/ 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1550.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol 1998; 143:1883-98; http://dx.doi.org/ 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1551.Viana R, Aguado C, Esteban I, Moreno D, Viollet B, Knecht E, Sanz P. Role of AMP-activated protein kinase in autophagy and proteasome function. Biochem Biophys Res Commun 2008; 369:964-8; http://dx.doi.org/ 10.1016/j.bbrc.2008.02.126. [DOI] [PubMed] [Google Scholar]
- 1552.Hadano S, Otomo A, Kunita R, Suzuki-Utsunomiya K, Akatsuka A, Koike M, Aoki M, Uchiyama Y, Itoyama Y, Ikeda JE. Loss of ALS2/Alsin exacerbates motor dysfunction in a SOD1-expressing mouse ALS model by disturbing endolysosomal trafficking. PloS One 2010; 5:e9805; http://dx.doi.org/ 10.1371/journal.pone.0009805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1553.Otomo A, Kunita R, Suzuki-Utsunomiya K, Ikeda JE, Hadano S. Defective relocalization of ALS2/alsin missense mutants to Rac1-induced macropinosomes accounts for loss of their cellular function and leads to disturbed amphisome formation. FEBS Lett 2011; 585:730-6; http://dx.doi.org/ 10.1016/j.febslet.2011.01.045. [DOI] [PubMed] [Google Scholar]
- 1554.Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, Marsella C, Piselli P, Gretzmeier C, Dengjel J, et al.. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell 2014; 31:734-46; http://dx.doi.org/ 10.1016/j.devcel.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 1555.Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF, De Zio D, Nazio F, Antonioli M, D'Orazio M, et al.. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol 2015; 17:20-30; http://dx.doi.org/ 10.1038/ncb3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1556.Fu M, St-Pierre P, Shankar J, Wang PT, Joshi B, Nabi IR. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell 2013; 24:1153-62; http://dx.doi.org/ 10.1091/mbc.E12-08-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1557.Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology 2014; 59:505-17; http://dx.doi.org/ 10.1002/hep.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1558.Lee KY, Oh S, Choi YJ, Oh SH, Yang YS, Yang MJ, Lee K, Lee BH. Activation of autophagy rescues amiodarone-induced apoptosis of lung epithelial cells and pulmonary toxicity in rats. Toxicol Sci 2013; 136:193-204; http://dx.doi.org/ 10.1093/toxsci/kft168. [DOI] [PubMed] [Google Scholar]
- 1559.Seglen PO, Berg TO, Blankson H, Fengsrud M, Holen I, Stromhaug PE. Structural aspects of autophagy. Adv Exp Med Biol 1996; 389:103-11; http://dx.doi.org/ 10.1007/978-1-4613-0335-0. [DOI] [PubMed] [Google Scholar]
- 1560.Meijer AJ, Codogno P. AMP-activated protein kinase and autophagy. Autophagy 2007; 3:238-40; http://dx.doi.org/ 10.4161/auto.3710. [DOI] [PubMed] [Google Scholar]
- 1561.Katsiarimpa A, Anzenberger F, Schlager N, Neubert S, Hauser MT, Schwechheimer C, Isono E. The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell 2011; 23:3026-40; http://dx.doi.org/ 10.1105/tpc.111.087254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1562.Katsiarimpa A, Kalinowska K, Anzenberger F, Weis C, Ostertag M, Tsutsumi C, Schwechheimer C, Brunner F, Huckelhoven R, Isono E. The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell 2013; 25:2236-52; http://dx.doi.org/ 10.1105/tpc.113.113399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1563.Costa R, Morrison A, Wang J, Manithody C, Li J, Rezaie AR. Activated protein C modulates cardiac metabolism and augments autophagy in the ischemic heart. J Thromb Haemost 2012; 10:1736-44; http://dx.doi.org/ 10.1111/j.1538-7836.2012.04833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1564.Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem 2011; 286:13704-13; http://dx.doi.org/ 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1565.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 2009; 5:527-49; http://dx.doi.org/ 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1566.Wang P, Xu TY, Wei K, Guan YF, Wang X, Xu H, Su DF, Pei G, Miao CY. ARRB1/beta-arrestin-1 mediates neuroprotection through coordination of BECN1-dependent autophagy in cerebral ischemia. Autophagy 2014; 10:1535-48; http://dx.doi.org/ 10.4161/auto.29203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1567.Keller KE, Yang YF, Sun YY, Sykes R, Acott TS, Wirtz MK. Ankyrin repeat and suppressor of cytokine signaling box containing protein-10 is associated with ubiquitin-mediated degradation pathways in trabecular meshwork cells. Mol Vis 2013; 19:1639-55. [PMC free article] [PubMed] [Google Scholar]
- 1568.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 2010; 29:4424-35; http://dx.doi.org/ 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 1569.Sheng Z, Ma L, Sun JE, Zhu LJ, Green MR. BCR-ABL suppresses autophagy through ATF5-mediated regulation of mTOR transcription. Blood 2011; 118:2840-8; http://dx.doi.org/ 10.1182/blood-2010-12-322537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1570.Klionsky DJ, Cregg JM, Dunn WA Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al.. A unified nomenclature for yeast autophagy-related genes. Dev Cell 2003; 5:539-45; http://dx.doi.org/ 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 1571.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 1997; 192:245-50; http://dx.doi.org/ 10.1016/S0378-1119(97)00084-X. [DOI] [PubMed] [Google Scholar]
- 1572.Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y. Apg2p functions in autophagosome formation on the perivacuolar structure. J Biol Chem 2001; 276:30452-60; http://dx.doi.org/ 10.1074/jbc.M102346200. [DOI] [PubMed] [Google Scholar]
- 1573.Wang C-W, Kim J, Huang W-P, Abeliovich H, Stromhaug PE, Dunn WA Jr., Klionsky DJ. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem 2001; 276:30442-51; http://dx.doi.org/ 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1574.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al.. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488-92; http://dx.doi.org/ 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 1575.Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf DH, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol 1997; 179:1068-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1576.Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J 2006; 273:2553-62; http://dx.doi.org/ 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 1577.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature 1998; 395:395-8; http://dx.doi.org/ 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 1578.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell 1999; 10:1337-51; http://dx.doi.org/ 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1579.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell 1999; 10:1367-79; http://dx.doi.org/ 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1580.Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 2000; 148:465-80; http://dx.doi.org/ 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1581.Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 2005; 280:18283-90; http://dx.doi.org/ 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- 1582.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J 1999; 18:5234-41; http://dx.doi.org/ 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1583.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA Jr., Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 2001; 153:381-96; http://dx.doi.org/ 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1584.Kamber RA, Shoemaker CJ, Denic V. receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol Cell 2015; 59:372-81; http://dx.doi.org/ 10.1016/j.molcel.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1585.Lin L, Yang P, Huang X, Zhang H, Lu Q, Zhang H. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J Cell Biol 2013; 201:113-29; http://dx.doi.org/ 10.1083/jcb.201209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1586.Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1587.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 1997; 192:207-13; http://dx.doi.org/ 10.1016/S0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 1588.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem 1998; 273:22284-91; http://dx.doi.org/ 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 1589.Epple UD, Suriapranata I, Eskelinen E-L, Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol 2001; 183: 5942-55; http://dx.doi.org/ 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1590.Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem 2001; 276:2083-7; http://dx.doi.org/ 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1591.van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell 2014; 25:290-301; http://dx.doi.org/ 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1592.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J 1999; 18:3888-96; http://dx.doi.org/ 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1593.Massey DC, Parkes M. Genome-wide association scanning highlights two autophagy genes, ATG16L1 and IRGM, as being significantly associated with Crohn's disease. Autophagy 2007; 3:649-51; http://dx.doi.org/ 10.4161/auto.5075. [DOI] [PubMed] [Google Scholar]
- 1594.Yang SK, Hong M, Zhao W, Jung Y, Baek J, Tayebi N, Kim KM, Ye BD, Kim KJ, Park SH, et al.. Genome-wide association study of Crohn's disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 2014; 63:80-7; http://dx.doi.org/ 10.1136/gutjnl-2013-305193. [DOI] [PubMed] [Google Scholar]
- 1595.Chew LH, Setiaputra D, Klionsky DJ, Yip CK. Structural characterization of the Saccharomyces cerevisiae autophagy regulatory complex Atg17-Atg31-Atg29. Autophagy 2013; 9:1467-74; http://dx.doi.org/ 10.4161/auto.25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1596.Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, Yip CK, Klionsky DJ. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci USA 2013; 110:E2875-84; http://dx.doi.org/ 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1597.Mao K, Chew LH, Yip CK, Klionsky DJ. The role of Atg29 phosphorylation in PAS assembly. Autophagy 2013; 9:2178-9; http://dx.doi.org/ 10.4161/auto.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1598.Leber R, Silles E, Sandoval IV, Mazon MJ. Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole. J Biol Chem 2001; 276:29210-7; http://dx.doi.org/ 10.1074/jbc.M101438200. [DOI] [PubMed] [Google Scholar]
- 1599.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell 2001; 7:1131-41; http://dx.doi.org/ 10.1016/S1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1600.Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem 2002; 277:30198-207; http://dx.doi.org/ 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1601.Deng YZ, Qu Z, He Y, Naqvi NI. Sorting nexin Snx41 is essential for conidiation and mediates glutathione-based antioxidant defense during invasive growth in Magnaporthe oryzae. Autophagy 2012; 8:1058-70; http://dx.doi.org/ 10.4161/auto.20217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1602.Suriapranata I, Epple UD, Bernreuther D, Bredschneider M, Sovarasteanu K, Thumm M. The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J Cell Sci 2000; 113:4025-33. [DOI] [PubMed] [Google Scholar]
- 1603.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell 2006; 17:5094-104; http://dx.doi.org/ 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1604.Legakis JE, Yen W-L, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy 2007; 3:422-32; http://dx.doi.org/ 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- 1605.Tucker KA, Reggiori F, Dunn WA Jr., Klionsky DJ. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J Biol Chem 2003; 278:48445-52; http://dx.doi.org/ 10.1074/jbc.M309238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1606.Monastyrska I, Kiel JAKW, Krikken AM, Komduur JA, Veenhuis M, van der Klei IJ. The Hansenula polymorpha ATG25 gene encodes a novel coiled-coil protein that is required for macropexophagy. Autophagy 2005; 1:92-100; http://dx.doi.org/ 10.4161/auto.1.2.1832. [DOI] [PubMed] [Google Scholar]
- 1607.Cao Y, Klionsky DJ. Atg26 is not involved in autophagy-related pathways in Saccharomyces cerevisiae. Autophagy 2007; 3:17-20; http://dx.doi.org/ 10.4161/auto.3371. [DOI] [PubMed] [Google Scholar]
- 1608.Yamashita S, Oku M, Wasada Y, Ano Y, Sakai Y. PI4P-signaling pathway for the synthesis of a nascent membrane structure in selective autophagy. J Cell Biol 2006; 173:709-17; http://dx.doi.org/ 10.1083/jcb.200512142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1609.Yen W-L, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell 2007; 18:581-93; http://dx.doi.org/ 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1610.Stasyk OV, Stasyk OG, Mathewson RD, Farre JC, Nazarko VY, Krasovska OS, Subramani S, Cregg JM, Sibirny AA. Atg28, a novel coiled-coil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris. Autophagy 2006; 2:30-8; http://dx.doi.org/ 10.4161/auto.2226. [DOI] [PubMed] [Google Scholar]
- 1611.Kawamata T, Kamada Y, Suzuki K, Kuboshima N, Akimatsu H, Ota S, Ohsumi M, Ohsumi Y. Characterization of a novel autophagy-specific gene, ATG29. Biochem Biophys Res Commun 2005; 338:1884-9; http://dx.doi.org/ 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- 1612.Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun 2007; 356:405-10; http://dx.doi.org/ 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- 1613.Watanabe Y, Noda NN, Kumeta H, Suzuki K, Ohsumi Y, Inagaki F. Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J Biol Chem 2010; 285:30026-33; http://dx.doi.org/ 10.1074/jbc.M110.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1614.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JAKW. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 2007; 3:106-16. [DOI] [PubMed] [Google Scholar]
- 1615.Nazarko VY, Nazarko TY, Farre JC, Stasyk OV, Warnecke D, Ulaszewski S, Cregg JM, Sibirny AA, Subramani S. Atg35, a micropexophagy-specific protein that regulates micropexophagic apparatus formation in Pichia pastoris. Autophagy 2011; 7:375-85; http://dx.doi.org/ 10.4161/auto.7.4.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1616.Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J 2012; 31:2852-68; http://dx.doi.org/ 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1617.Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol 2013; 203:299-313; http://dx.doi.org/ 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1618.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009; 5:973-9; http://dx.doi.org/ 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 1619.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009; 5:649-62; http://dx.doi.org/ 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 1620.Honig A, Avin-Wittenberg T, Ufaz S, Galili G. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 2012; 24:288-303; http://dx.doi.org/ 10.1105/tpc.111.093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1621.Dehay B, Ramirez A, Martinez-Vicente M, Perier C, Canron MH, Doudnikoff E, Vital A, Vila M, Klein C, Bezard E. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc Natl Acad Sci USA 2012; 109:9611-6; http://dx.doi.org/ 10.1073/pnas.1112368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1622.Gusdon AM, Zhu J, Van Houten B, Chu CT. ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neuobiol Dis 2012; 45:962-72; http://dx.doi.org/ 10.1016/j.nbd.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1623.Niu H, Rikihisa Y. Ats-1: a novel bacterial molecule that links autophagy to bacterial nutrition. Autophagy 2013; 9:787-8; http://dx.doi.org/ 10.4161/auto.23693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1624.Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci USA 2012; 109:20800-7; http://dx.doi.org/ 10.1073/pnas.1218674109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1625.Isakson P, Bjoras M, Boe SO, Simonsen A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood 2010; 116:2324-31; http://dx.doi.org/ 10.1182/blood-2010-01-261040. [DOI] [PubMed] [Google Scholar]
- 1626.Orfali N, McKenna SL, Cahill MR, Gudas LJ, Mongan NP. Retinoid receptor signaling and autophagy in acute promyelocytic leukemia. Exp Cell Res 2014; 324:1-12; http://dx.doi.org/ 10.1016/j.yexcr.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1627.Vanhee C, Zapotoczny G, Masquelier D, Ghislain M, Batoko H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 2011; 23:785-805; http://dx.doi.org/ 10.1105/tpc.110.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1628.Papp D, Kovacs T, Billes V, Varga M, Tarnoci A, Hackler L Jr, et al. AUTEN-67, an autophagy-enhancing drug candidate with potent antiaging and neuroprotective effects. Autophagy 2015; 11:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1629.Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol 1990; 110:1923-33; http://dx.doi.org/ 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1630.Schulze RJ, Weller SG, Schroeder B, Krueger EW, Chi S, Casey CA, McNiven MA. Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol 2013; 203:315-26; http://dx.doi.org/ 10.1083/jcb.201306140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1631.Gundara JS, Robinson BG, Sidhu SB. Evolution of the “autophagamiR”. Autophagy 2011; 7:1553-4; http://dx.doi.org/ 10.4161/auto.7.12.17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1632.Mijaljica D, Nazarko TY, Brumell JH, Huang WP, Komatsu M, Prescott M, Simonsen A, Yamamoto A, Zhang H, Klionsky DJ, et al.. Receptor protein complexes are in control of autophagy. Autophagy 2012; 8:1701-5; http://dx.doi.org/ 10.4161/auto.21332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1633.Shpilka T, Welter E, Borovsky N, Amar N, Mari M, Reggiori F, Elazar Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J 2015; 34:2117-31; http://dx.doi.org/ 10.15252/embj.201490315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1634.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J 2009; 28:889-901; http://dx.doi.org/ 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1635.Sebti S, Prebois C, Perez-Gracia E, Bauvy C, Desmots F, Pirot N, Gongora C, Bach AS, Hubberstey AV, Palissot V, et al.. BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc Natl Acad Sci USA 2014; 111:4115-20; http://dx.doi.org/ 10.1073/pnas.1313618111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1636.Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J Biol Chem 2012; 287:16256-66; http://dx.doi.org/ 10.1074/jbc.M112.348250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1637.Lindqvist LM, Heinlein M, Huang DC, Vaux DL. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc Natl Acad Sci USA 2014; 111:8512-7; http://dx.doi.org/ 10.1073/pnas.1406425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1638.Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, Yasui H, Ueda H, Akazawa Y, Nakayama H, et al.. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun 2015; 6:7527; http://dx.doi.org/ 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1639.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity 2012; 36:947-58; http://dx.doi.org/ 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1640.Liang X, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672-6; http://dx.doi.org/ 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 1641.Hurley JH, Schulman BA. Atomistic autophagy: the structures of cellular self-digestion. Cell 2014; 157:300-11; http://dx.doi.org/ 10.1016/j.cell.2014.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1642.Cheng B, Xu A, Qiao M, Wu Q, Wang W, Mei Y, Wu M. BECN1s, a short splice variant of BECN1, functions in mitophagy. Autophagy 2015; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1643.He C, Wei Y, Sun K, Li B, Dong X, Zou Z, Liu Y, Kinch LN, Khan S, Sinha S, et al.. Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell 2013; 154:1085-99; http://dx.doi.org/ 10.1016/j.cell.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1644.Yang LJ, Chen Y, He J, Yi S, Wen L, Zhao J, Zhang BP, Cui GH. Betulinic acid inhibits autophagic flux and induces apoptosis in human multiple myeloma cells in vitro. Acta Pharmacol Sin 2012; 33:1542-8; http://dx.doi.org/ 10.1038/aps.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1645.Minoia M, Boncoraglio A, Vinet J, Morelli FF, Brunsting JF, Poletti A, Krom S, Reits E, Kampinga HH, Carra S. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy 2014; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1646.Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell 1994; 79:341-51; http://dx.doi.org/ 10.1016/0092-8674(94)90202-X. [DOI] [PubMed] [Google Scholar]
- 1647.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem 2012; 287:19094-104; http://dx.doi.org/ 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1648.Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab 2015; 3:4; http://dx.doi.org/ 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1649.Landes T, Emorine LJ, Courilleau D, Rojo M, Belenguer P, Arnaune-Pelloquin L. The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Rep 2010; 11:459-65; http://dx.doi.org/ 10.1038/embor.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1650.Kasper LH, Boussouar F, Boyd K, Xu W, Biesen M, Rehg J, Baudino TA, Cleveland JL, Brindle PK. Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J 2005; 24:3846-58; http://dx.doi.org/ 10.1038/sj.emboj.7600846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1651.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol 2007; 27:6229-42; http://dx.doi.org/ 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1652.Feng X, Liu X, Zhang W, Xiao W. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J 2011; 30:3397-415; http://dx.doi.org/ 10.1038/emboj.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1653.Shaw J, Yurkova N, Zhang T, Gang H, Aguilar F, Weidman D, Scramstad C, Weisman H, Kirshenbaum LA. Antagonism of E2F-1 regulated Bnip3 transcription by NF-kappaB is essential for basal cell survival. Proc Natl Acad Sci USA 2008; 105:20734-9; http://dx.doi.org/ 10.1073/pnas.0807735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1654.Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, et al.. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest 2007; 117:2825-33; http://dx.doi.org/ 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1655.Glick D, Zhang W, Beaton M, Marsboom G, Gruber M, Simon MC, Hart J, Dorn GW 2nd, Brady MJ, Macleod KF. BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol 2012; 32:2570-84; http://dx.doi.org/ 10.1128/MCB.00167-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1656.Melser S, Chatelain EH, Lavie J, Mahfouf W, Jose C, Obre E, Goorden S, Priault M, Elgersma Y, Rezvani HR, et al.. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab 2013; 17:719-30; http://dx.doi.org/ 10.1016/j.cmet.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 1657.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, et al.. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 2014; 23:3579-95; http://dx.doi.org/ 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1658.O'Farrell F, Wang S, Katheder N, Rusten TE, Samakovlis C. Two-tiered control of epithelial growth and autophagy by the insulin receptor and the ret-like receptor, stitcher. PLoS Biol 2013; 11:e1001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1659.Ikeda H, Hideshima T, Fulciniti M, Perrone G, Mimura N, Yasui H, Okawa Y, Kiziltepe T, Santo L, Vallet S, et al.. PI3K/p110{delta} is a novel therapeutic target in multiple myeloma. Blood 2010; 116:1460-8; http://dx.doi.org/ 10.1182/blood-2009-06-222943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1660.Xia HG, Zhang L, Chen G, Zhang T, Liu J, Jin M, Ma X, Ma D, Yuan J. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy 2010; 6:61-6; http://dx.doi.org/ 10.4161/auto.6.1.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1661.Zitvogel L, Kepp O, Senovilla L, Menger L, Chaput N, Kroemer G. Immunogenic tumor cell death for optimal anticancer therapy: the calreticulin exposure pathway. Clin Cancer Res 2010; 16:3100-4; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-2891. [DOI] [PubMed] [Google Scholar]
- 1662.Obeid M, Tesniere A, Ghiringhelli F, Fimia GM, Apetoh L, Perfettini JL, Castedo M, Mignot G, Panaretakis T, Casares N, et al.. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med 2007; 13:54-61; http://dx.doi.org/ 10.1038/nm1523. [DOI] [PubMed] [Google Scholar]
- 1663.Garg AD, Agostinis P. ER stress, autophagy and immunogenic cell death in photodynamic therapy-induced anti-cancer immune responses. Photoch Photobio Sci 2014; 13:474-87; http://dx.doi.org/ 10.1039/c3pp50333j. [DOI] [PubMed] [Google Scholar]
- 1664.Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 2005; 280:29060-6; http://dx.doi.org/ 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 1665.Demarchi F, Bertoli C, Copetti T, Tanida I, Brancolini C, Eskelinen E-L, Schneider C. Calpain is required for macroautophagy in mammalian cells. J Cell Biol 2006; 175:595-605; http://dx.doi.org/ 10.1083/jcb.200601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1666.Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang X, Jin H, Xu H, Chen Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 2010; 1:468-77; http://dx.doi.org/ 10.1007/s13238-010-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1667.Li H, Wang P, Sun Q, Ding WX, Yin XM, Sobol RW, Stolz DB, Yu J, Zhang L. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res 2011; 71:3625-34; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1668.Garcia-Marcos M, Ear J, Farquhar MG, Ghosh P. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol Biol Cell 2011; 22:673-86; http://dx.doi.org/ 10.1091/mbc.E10-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1669.Latterich M, Frohlich KU, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 1995; 82:885-93; http://dx.doi.org/ 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- 1670.Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen E-L, Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol 2010; 190:965-73; http://dx.doi.org/ 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1671.Joubert PE, Meiffren G, Gregoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain PO, Vidal M, Lotteau V, et al.. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 2009; 6:354-66; http://dx.doi.org/ 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 1672.Orlotti NI, Cimino-Reale G, Borghini E, Pennati M, Sissi C, Perrone F, Palumbo M, Daidone MG, Folini M, Zaffaroni N. Autophagy acts as a safeguard mechanism against G-quadruplex ligand-mediated DNA damage. Autophagy 2012; 8:1185-96; http://dx.doi.org/ 10.4161/auto.20519. [DOI] [PubMed] [Google Scholar]
- 1673.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al.. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol 2007; 9:218-24; http://dx.doi.org/ 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 1674.Budina-Kolomets A, Hontz RD, Pimkina J, Murphy ME. A conserved domain in exon 2 coding for the human and murine ARF tumor suppressor protein is required for autophagy induction. Autophagy 2013; 9:1553-65; http://dx.doi.org/ 10.4161/auto.25831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1675.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab 2010; 21:142-50; http://dx.doi.org/ 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1676.Dice J. Chaperone-mediated autophagy. Autophagy 2007; 3:295-9; http://dx.doi.org/ 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 1677.Agarraberes F, Terlecky S, Dice J. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol 1997; 137:825-34; http://dx.doi.org/ 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1678.Cuervo A, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 1996; 273:501-3; http://dx.doi.org/ 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 1679.Mitsuhashi S, Hatakeyama H, Karahashi M, Koumura T, Nonaka I, Hayashi YK, Noguchi S, Sher RB, Nakagawa Y, Manfredi G, et al.. Muscle choline kinase beta defect causes mitochondrial dysfunction and increased mitophagy. Hum Mol Genet 2011; 20:3841-51; http://dx.doi.org/ 10.1093/hmg/ddr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1680.Fedorko M. Effect of chloroquine on morphology of cytoplasmic granules in maturing human leukocytes–an ultrastructural study. J Clin Invest 1967; 46:1932-42; http://dx.doi.org/ 10.1172/JCI105683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1681.Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J 2010; 29:606-18; http://dx.doi.org/ 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1682.Chen YF, Kao CH, Chen YT, Wang CH, Wu CY, Tsai CY, Liu FC, Yang CW, Wei YH, Hsu MT, et al.. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev 2009; 23:1183-94; http://dx.doi.org/ 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1683.Yang Z, Geng J, Yen W-L, Wang K, Klionsky DJ. Positive or negative regulatory roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae Mol Cell 2010; 38:250-64; http://dx.doi.org/ 10.1016/j.molcel.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1684.Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, Cotman SL. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem 2006; 281:20483-93; http://dx.doi.org/ 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- 1685.Chandrachud U, Walker MW, Simas AM, Heetveld S, Petcherski A, Klein M, Oh H, Wolf P, Zhao WN, Norton S, et al.. Unbiased Cell-based Screening in a Neuronal Cell Model of Batten Disease Highlights an Interaction between Ca2+ Homeostasis, Autophagy, and CLN3 Protein Function. J Biol Chem 2015; 290:14361-80; http://dx.doi.org/ 10.1074/jbc.M114.621706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1686.Cortese A, Tucci A, Piccolo G, Galimberti CA, Fratta P, Marchioni E, Grampa G, Cereda C, Grieco G, Ricca I, et al.. Novel CLN3 mutation causing autophagic vacuolar myopathy. Neurology 2014; 82:2072-6; http://dx.doi.org/ 10.1212/WNL.0000000000000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1687.Wang F, Wang H, Tuan HF, Nguyen DH, Sun V, Keser V, Bowne SJ, Sullivan LS, Luo H, Zhao L, et al.. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet 2014; 133:331-45; http://dx.doi.org/ 10.1007/s00439-013-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1688.Yen W-L, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol 2010; 188:101-14; http://dx.doi.org/ 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1689.Lancel S, Montaigne D, Marechal X, Marciniak C, Hassoun SM, Decoster B, Ballot C, Blazejewski C, Corseaux D, Lescure B, et al.. Carbon monoxide improves cardiac function and mitochondrial population quality in a mouse model of metabolic syndrome. PloS One 2012; 7:e41836; http://dx.doi.org/ 10.1371/journal.pone.0041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1690.Chen LL, Song JX, Lu JH, Yuan ZW, Liu LF, Durairajan SS, Li M. Corynoxine, a Natural Autophagy Enhancer, Promotes the Clearance of Alpha-Synuclein via Akt/mTOR Pathway. J Neuroimmune Pharm 2014:380-7; http://dx.doi.org/ 10.1007/s11481-014-9528-2. [DOI] [PubMed] [Google Scholar]
- 1691.Lu JH, Tan JQ, Durairajan SS, Liu LF, Zhang ZH, Ma L, Shen HM, Chan HY, Li M. Isorhynchophylline, a natural alkaloid, promotes the degradation of alpha-synuclein in neuronal cells via inducing autophagy. Autophagy 2012; 8:98-108 (see also the erratum in Autophagy 2012; 8:864-6); 10.4161/auto.8.1.18313. [DOI] [PubMed] [Google Scholar]
- 1692.Smith RE, Farquhar MG. Lysosome function in the regulation of the secretory process in cells of the anterior pituitary gland. J Cell Biol 1966; 31:319-47; http://dx.doi.org/ 10.1083/jcb.31.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1693.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HWI, Kyei GB, Johansen T, Vergne I, et al.. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 2010; 32:329-41; http://dx.doi.org/ 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1694.Song H, Pu J, Wang L, Wu L, Xiao J, Liu Q, Chen J, Zhang M, Liu Y, Ni M, et al.. ATG16L1 phosphorylation is oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein phosphatase 1 which determines the fate of cardiomyocytes during hypoxia/reoxygenation. Autophagy 2015:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1695.Sun LL, Li M, Suo F, Liu XM, Shen EZ, Yang B, Dong MQ, He WZ, Du LL. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet 2013; 9:e1003715; http://dx.doi.org/ 10.1371/journal.pgen.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1696.Campbell EM, Fares H. Roles of CUP-5, the Caenorhabditis elegans orthologue of human TRPML1, in lysosome and gut granule biogenesis. BMC Cell Biol 2010; 11:40; http://dx.doi.org/ 10.1186/1471-2121-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1697.Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet 2001; 28:64-8. [DOI] [PubMed] [Google Scholar]
- 1698.Hersh BM, Hartwieg E, Horvitz HR. The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc Natl Acad Sci USA 2002; 99:4355-60; http://dx.doi.org/ 10.1073/pnas.062065399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1699.Sun T, Wang X, Lu Q, Ren H, Zhang H. CUP-5, the C. elegans ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy 2011; 7:1308-15; http://dx.doi.org/ 10.4161/auto.7.11.17759. [DOI] [PubMed] [Google Scholar]
- 1700.Bruns C, McCaffery JM, Curwin AJ, Duran JM, Malhotra V. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol 2011; 195:979-92; http://dx.doi.org/ 10.1083/jcb.201106098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1701.Wang M, Tan W, Zhou J, Leow J, Go M, Lee HS, Casey PJ. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem 2008; 283:18678-84; http://dx.doi.org/ 10.1074/jbc.M801855200. [DOI] [PubMed] [Google Scholar]
- 1702.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 1995; 131:591-602; http://dx.doi.org/ 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1703.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 2008; 4:e24; http://dx.doi.org/ 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1704.Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol 2011; 21:1507-14; http://dx.doi.org/ 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1705.Netea-Maier RT, Plantinga TS, Van De Veerdonk FL, Smit JW, Netea MG. Modulation of inflammation by autophagy: consequences for human disease. Autophagy 2015:0; http://dx.doi.org/ 10.1080/15548627.2015.1071759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1706.Koren I, Reem E, Kimchi A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr Biol 2010; 20:1093-8; http://dx.doi.org/ 10.1016/j.cub.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 1707.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol 2002; 157:455-68; http://dx.doi.org/ 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1708.Buraschi S, Neill T, Goyal A, Poluzzi C, Smythies J, Owens RT, Schaefer L, Torres A, Iozzo RV. Decorin causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci USA 2013; 110:E2582-91; http://dx.doi.org/ 10.1073/pnas.1305732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1709.DeVorkin L, Go NE, Hou Y-CC, Moradian A, Morin GB, Gorski SM. The Drosophila effector caspase Dcp-1 regulates mitochondrial dynamics and autophagic flux via SesB. J Cell Biol 2014; 205:477-92; http://dx.doi.org/ 10.1083/jcb.201303144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1710.Hu G, McQuiston T, Bernard A, Park YD, Qiu J, Vural A, Zhang N, Waterman SR, Blewett NH, Myers TG, et al.. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol 2015; 17:930-42; http://dx.doi.org/ 10.1038/ncb3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1711.Molitoris JK, McColl KS, Swerdlow S, Matsuyama M, Lam M, Finkel TH, Matsuyama S, Distelhorst CW. Glucocorticoid elevation of dexamethasone-induced gene 2 (Dig2/RTP801/REDD1) protein mediates autophagy in lymphocytes. J Biol Chem 2011; 286:30181-9; http://dx.doi.org/ 10.1074/jbc.M111.245423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1712.Slavov N, Botstein D. Decoupling nutrient signaling from growth rate causes aerobic glycolysis and deregulation of cell size and gene expression. Mol Biol Cell 2013; 24:157-68; http://dx.doi.org/ 10.1091/mbc.E12-09-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1713.Kohler K, Brunner E, Guan XL, Boucke K, Greber UF, Mohanty S, Barth JM, Wenk MR, Hafen E. A combined proteomic and genetic analysis identifies a role for the lipid desaturase Desat1 in starvation-induced autophagy in Drosophila. Autophagy 2009; 5:980-90; http://dx.doi.org/ 10.4161/auto.5.7.9325. [DOI] [PubMed] [Google Scholar]
- 1714.Shahnazari S, Yen W-L, Birmingham CL, Shiu J, Namolovan A, Zheng YT, Nakayama K, Klionsky DJ, Brumell JH. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe 2010; 8:137-46; http://dx.doi.org/ 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1715.Lu Z, Baquero MT, Yang H, Yang M, Reger AS, Kim C, Levine DA, Clarke CH, Liao WS, Bast RC Jr. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy 2014; 10:1071-92; http://dx.doi.org/ 10.4161/auto.28577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1716.Mao K, Liu X, Feng Y, Klionsky DJ. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 2014; 10:652-61; http://dx.doi.org/ 10.4161/auto.27852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1717.Dagda RK, Gusdon AM, Pien I, Strack S, Green S, Li C, Van Houten B, Cherra SJ 3rd, Chu CT. Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson's disease. Cell Death Differ 2011; 18:1914-23; http://dx.doi.org/ 10.1038/cdd.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1718.Kwon MH, Callaway H, Zhong J, Yedvobnick B. A targeted genetic modifier screen links the SWI2/SNF2 protein domino to growth and autophagy genes in Drosophila melanogaster. G3 (Bethesda) 2013; 3:815-25; http://dx.doi.org/ 10.1534/g3.112.005496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1719.Gomez-Santos C, Ferrer I, Santidrian AF, Barrachina M, Gil J, Ambrosio S. Dopamine induces autophagic cell death and alpha-synuclein increase in human neuroblastoma SH-SY5Y cells. J Neurosci Res 2003; 73:341-50; http://dx.doi.org/ 10.1002/jnr.10663. [DOI] [PubMed] [Google Scholar]
- 1720.McPhee CK, Logan MA, Freeman MR, Baehrecke EH. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature 2010; 465:1093-6; http://dx.doi.org/ 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1721.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell 2012; 151:1501-12; http://dx.doi.org/ 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1722.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 2007; 3:597-9; http://dx.doi.org/ 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 1723.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, et al.. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 2008; 4:330-8; http://dx.doi.org/ 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 1724.Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM. Identification of regulators of chaperone-mediated autophagy. Mol Cell 2010; 39:535-47; http://dx.doi.org/ 10.1016/j.molcel.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1725.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 2005; 19:15-26; http://dx.doi.org/ 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 1726.Talloczy Z, Jiang W, Virgin HWT, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA 2002; 99:190-5; http://dx.doi.org/ 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1727.Zhao X, Fang Y, Yang Y, Qin Y, Wu P, Wang T, Lai H, Meng L, Wang D, Zheng Z, et al.. Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy 2015:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1728.Kim S, Naylor SA, DiAntonio A. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc Natl Acad Sci USA 2012; 109:E1072-81; http://dx.doi.org/ 10.1073/pnas.1120320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1729.Berge T, Leikfoss IS, Harbo HF. From Identification to Characterization of the Multiple Sclerosis Susceptibility Gene CLEC16A. Int J Mol Sci 2013; 14:4476-97; http://dx.doi.org/ 10.3390/ijms14034476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1730.Soleimanpour SA, Gupta A, Bakay M, Ferrari AM, Groff DN, Fadista J, Spruce LA, Kushner JA, Groop L, Seeholzer SH, et al.. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell 2014; 157:1577-90; http://dx.doi.org/ 10.1016/j.cell.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1731.Li Y, Zhao Y, Hu J, Xiao J, Qu L, Wang Z, Ma D, Chen Y. A novel ER-localized transmembrane protein, EMC6, interacts with RAB5A and regulates cell autophagy. Autophagy 2013; 9:150-63; http://dx.doi.org/ 10.4161/auto.22742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1732.Poluzzi C, Casulli J, Goyal A, Mercer TJ, Neill T, Iozzo RV. Endorepellin evokes autophagy in endothelial cells. J Biol Chem 2014; 289:16114-28; http://dx.doi.org/ 10.1074/jbc.M114.556530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1733.Tian E, Wang F, Han J, Zhang H. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy 2009; 5:608-15; http://dx.doi.org/ 10.4161/auto.5.5.8624. [DOI] [PubMed] [Google Scholar]
- 1734.Cullup T, Kho AL, Dionisi-Vici C, Brandmeier B, Smith F, Urry Z, Simpson MA, Yau S, Bertini E, McClelland V, et al.. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet 2013; 45:83-7; http://dx.doi.org/ 10.1038/ng.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1735.Li S, Yang P, Tian E, Zhang H. Arginine methylation modulates autophagic degradation of PGL granules in C. elegans. Mol Cell 2013; 52:421-33; http://dx.doi.org/ 10.1016/j.molcel.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 1736.Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR, Knecht E, Rubinsztein DC. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet 2010; 19:2867-76; http://dx.doi.org/ 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1737.Bockler S, Westermann B. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev Cell 2014; 28:450-8; http://dx.doi.org/ 10.1016/j.devcel.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 1738.Sinha S, Roy S, Reddy BS, Pal K, Sudhakar G, Iyer S, Dutta S, Wang E, Vohra PK, Roy KR, et al.. A lipid-modified estrogen derivative that treats breast cancer independent of estrogen receptor expression through simultaneous induction of autophagy and apoptosis. Mol Cancer Res 2011; 9:364-74; http://dx.doi.org/ 10.1158/1541-7786.MCR-10-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1739.Wang L, Yu C, Lu Y, He P, Guo J, Zhang C, Song Q, Ma D, Shi T, Chen Y. TMEM166, a novel transmembrane protein, regulates cell autophagy and apoptosis. Apoptosis 2007; 12:1489-502; http://dx.doi.org/ 10.1007/s10495-007-0073-9. [DOI] [PubMed] [Google Scholar]
- 1740.Yu C, Wang L, Lv B, Lu Y, Zeng L, Chen Y, Ma D, Shi T. TMEM74, a lysosome and autophagosome protein, regulates autophagy. Biochem Biophys Res Commun 2008; 369:622-9; http://dx.doi.org/ 10.1016/j.bbrc.2008.02.055. [DOI] [PubMed] [Google Scholar]
- 1741.Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, et al.. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 2011; 144:253-67; http://dx.doi.org/ 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1742.Abrahamsen H, Stenmark H. Protein secretion: unconventional exit by exophagy. Curr Biol 2010; 20:R415-8; http://dx.doi.org/ 10.1016/j.cub.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 1743.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 2010; 188:527-36; http://dx.doi.org/ 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1744.Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol 2010; 188:537-46; http://dx.doi.org/ 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1745.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, Murino L, Tagliaferri R, Brunetti-Pierri N, Isacchi A, et al.. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci USA 2010; 107:14621-6; http://dx.doi.org/ 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1746.Lisa-Santamaria P, Jimenez A, Revuelta JL. The protein factor-arrest 11 (Far11) is essential for the toxicity of human caspase-10 in yeast and participates in the regulation of autophagy and the DNA damage signaling. J Biol Chem 2012; 287:29636-47; http://dx.doi.org/ 10.1074/jbc.M112.344192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1747.McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T, Tooze SA. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J 2012; 31:1931-46; http://dx.doi.org/ 10.1038/emboj.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1748.Vaccari I, Carbone A, Previtali SC, Mironova YA, Alberizzi V, Noseda R, Rivellini C, Bianchi F, Del Carro U, D'Antonio M, et al.. Loss of Fig4 in both Schwann cells and motor neurons contributes to CMT4J neuropathy. Hum Mol Genet 2015; 24:383-96; http://dx.doi.org/ 10.1093/hmg/ddu451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1749.Romano S, D'Angelillo A, Pacelli R, Staibano S, De Luna E, Bisogni R, Eskelinen EL, Mascolo M, Cali G, Arra C, et al.. Role of FK506-binding protein 51 in the control of apoptosis of irradiated melanoma cells. Cell Death Differ 2010; 17:145-57; http://dx.doi.org/ 10.1038/cdd.2009.115. [DOI] [PubMed] [Google Scholar]
- 1750.Gassen NC, Hartmann J, Zschocke J, Stepan J, Hafner K, Zellner A, Kirmeier T, Kollmannsberger L, Wagner KV, Dedic N, et al.. Association of FKBP51 with priming of autophagy pathways and mediation of antidepressant treatment response: evidence in cells, mice, and humans. PLoS Med 2014; 11:e1001755; http://dx.doi.org/ 10.1371/journal.pmed.1001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1751.Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, et al.. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell 2002; 2:157-64; http://dx.doi.org/ 10.1016/S1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 1752.Dunlop EA, Seifan S, Claessens T, Behrends C, Kamps MA, Rozycka E, Kemp AJ, Nookala RK, Blenis J, Coull BJ, et al.. FLCN, a novel autophagy component, interacts with GABARAP and is regulated by ULK1 phosphorylation. Autophagy 2014; 10:1749-60; http://dx.doi.org/ 10.4161/auto.29640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1753.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 2013; 202:1107-22; http://dx.doi.org/ 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1754.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 2013; 52: 495-505; http://dx.doi.org/ 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1755.Huett A, Ng A, Cao Z, Kuballa P, Komatsu M, Daly MJ, Podolsky DK, Xavier RJ. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J Immunol 2009; 182:4917-30; http://dx.doi.org/ 10.4049/jimmunol.0803050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1756.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol 2010; 12:665-75; http://dx.doi.org/ 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 1757.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circulation Research 2010; 107:1470-82; http://dx.doi.org/ 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1758.Attaix D, Bechet D. FoxO3 controls dangerous proteolytic liaisons. Cell Metab 2007; 6:425-7; http://dx.doi.org/ 10.1016/j.cmet.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 1759.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al.. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 2012; 14:177-85; http://dx.doi.org/ 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 1760.Ryu HH, Jun MH, Min KJ, Jang DJ, Lee YS, Kim HK, Lee JA. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging 2014; 35:2822-31; http://dx.doi.org/ 10.1016/j.neurobiolaging.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 1761.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, {O}vervatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 2010; 188:253-69; http://dx.doi.org/ 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1762.Lakhani R, Vogel KR, Till A, Liu J, Burnett SF, Gibson KM, Subramani S. Defects in GABA metabolism affect selective autophagy pathways and are alleviated by mTOR inhibition. EMBO Mol Med 2014; 6:551-66; http://dx.doi.org/ 10.1002/emmm.201303356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1763.Ogier-Denis E, Couvineau A, Maoret JJ, Houri JJ, Bauvy C, De Stefanis D, Isidoro C, Laburthe M, Codogno P. A heterotrimeric Gi3-protein controls autophagic sequestration in the human colon cancer cell line HT-29. J Biol Chem 1995; 270:13-6; http://dx.doi.org/ 10.1074/jbc.270.1.13. [DOI] [PubMed] [Google Scholar]
- 1764.Ogier-Denis E, Houri JJ, Bauvy C, Codogno P. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J Biol Chem 1996; 271:28593-600; http://dx.doi.org/ 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- 1765.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem 2001; 276:1701-6; http://dx.doi.org/ 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 1766.Mata IF, Samii A, Schneer SH, Roberts JW, Griffith A, Leis BC, Schellenberg GD, Sidransky E, Bird TD, Leverenz JB, et al.. Glucocerebrosidase gene mutations: a risk factor for Lewy body disorders. Arch Neurol 2008; 65:379-82; http://dx.doi.org/ 10.1001/archneurol.2007.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1767.Mitsui J, Mizuta I, Toyoda A, Ashida R, Takahashi Y, Goto J, Fukuda Y, Date H, Iwata A, Yamamoto M, et al.. Mutations for Gaucher disease confer high susceptibility to Parkinson disease. Arch Neurol 2009; 66:571-6; http://dx.doi.org/ 10.1001/archneurol.2009.72. [DOI] [PubMed] [Google Scholar]
- 1768.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, et al.. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. New Engl J Med 2009; 361:1651-61; http://dx.doi.org/ 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1769.Osellame LD, Rahim AA, Hargreaves IP, Gegg ME, Richard-Londt A, Brandner S, Waddington SN, Schapira AH, Duchen MR. Mitochondria and quality control defects in a mouse model of Gaucher disease–links to Parkinson's disease. Cell Metab 2013; 17:941-53; http://dx.doi.org/ 10.1016/j.cmet.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1770.Webster BR, Scott I, Han K, Li JH, Lu Z, Stevens MV, Malide D, Chen Y, Samsel L, Connelly PS, et al.. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci 2013; 126:4843-9; http://dx.doi.org/ 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1771.Moreau K, Rubinsztein DC. The plasma membrane as a control center for autophagy. Autophagy 2012; 8:861-3; http://dx.doi.org/ 10.4161/auto.20060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1772.Todd LR, Damin MN, Gomathinayagam R, Horn SR, Means AR, Sankar U. Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol Biol Cell 2010; 21:1225-36; http://dx.doi.org/ 10.1091/mbc.E09-11-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1773.Kalamidas SA, Kotoulas OB. Glycogen autophagy in newborn rat hepatocytes. Histol Histopathol 2000; 15:1011-8. [DOI] [PubMed] [Google Scholar]
- 1774.Delbridge LM, Mellor KM, Taylor DJ, Gottlieb RA. Myocardial autophagic energy stress responses–macroautophagy, mitophagy, and glycophagy. Am J Physiol Heart Circ Physiol 2015; 308:H1194-204; http://dx.doi.org/ 10.1152/ajpheart.00002.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1775.Mellor KM, Varma U, Stapleton DI, Delbridge LM. Cardiomyocyte glycophagy is regulated by insulin and exposure to high extracellular glucose. Am J Physiol Heart Circ Physiol 2014; 306:H1240-5; http://dx.doi.org/ 10.1152/ajpheart.00059.2014. [DOI] [PubMed] [Google Scholar]
- 1776.Li B, Castano AP, Hudson TE, Nowlin BT, Lin S-L, Bonventre JV, Swanson KD, Duffield JS. The melanoma-associated transmembrane glycoprotein Gpnmb controls trafficking of cellular debris for degradation and is essential for tissue repair. FASEB J 2010; 24:4767-81; http://dx.doi.org/ 10.1096/fj.10-154757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1777.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 2013; 153: 1461-74; http://dx.doi.org/ 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1778.Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Liu Q, Ruan K, et al.. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 2012; 336:477-81; http://dx.doi.org/ 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 1779.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al.. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J 2010; 29:969-80; http://dx.doi.org/ 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1780.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 2003; 115:727-38; http://dx.doi.org/ 10.1016/S0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 1781.Bohensky J, Shapiro IM, Leshinsky S, Terkhorn SP, Adams CS, Srinivas V. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy 2007; 3:207-14; http://dx.doi.org/ 10.4161/auto.3708. [DOI] [PubMed] [Google Scholar]
- 1782.Mellor HR, Harris AL. The role of the hypoxia-inducible BH3-only proteins BNIP3 and BNIP3L in cancer. Cancer Metastasis Rev 2007; 26:553-66; http://dx.doi.org/ 10.1007/s10555-007-9080-0. [DOI] [PubMed] [Google Scholar]
- 1783.Mimouna S, Bazin M, Mograbi B, Darfeuille-Michaud A, Brest P, Hofman P, Vouret-Craviari V. HIF1A regulates xenophagic degradation of adherent and invasive Escherichia coli (AIEC). Autophagy 2014; 10:2333-45; http://dx.doi.org/ 10.4161/15548627.2014.984275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1784.Roberts DJ, Miyamoto S. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ 2015; 22:248-57; http://dx.doi.org/ 10.1038/cdd.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1785.Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, et al.. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene 2010; 29:5299-310; http://dx.doi.org/ 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1786.Thorburn J, Horita H, Redzic J, Hansen K, Frankel AE, Thorburn A. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ 2009; 16:175-83; http://dx.doi.org/ 10.1038/cdd.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1787.Mao K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol 2011; 193:755-67; http://dx.doi.org/ 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1788.Pfaffenwimmer T, Reiter W, Brach T, Nogellova V, Papinski D, Schuschnig M, Abert C, Ammerer G, Martens S, Kraft C. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep 2014; 15:862-70; http://dx.doi.org/ 10.15252/embr.201438932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1789.Tanaka C, Tan LJ, Mochida K, Kirisako H, Koizumi M, Asai E, Sakoh-Nakatogawa M, Ohsumi Y, Nakatogawa H. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J Cell Biol 2014; 207:91-105; http://dx.doi.org/ 10.1083/jcb.201402128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1790.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell 2009; 36:15-27; http://dx.doi.org/ 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1791.Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ 2008; 15:1460-71; http://dx.doi.org/ 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1792.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 1989; 246:382-5; http://dx.doi.org/ 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 1793.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J 2006; 25:3921-33; http://dx.doi.org/ 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1794.Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic 2002; 3:388-96; http://dx.doi.org/ 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 1795.Xu C, Liu J, Hsu LC, Luo Y, Xiang R, Chuang TH. Functional interaction of heat shock protein 90 and Beclin 1 modulates Toll-like receptor-mediated autophagy. FASEB J 2011; 25:2700-10; http://dx.doi.org/ 10.1096/fj.10-167676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1796.Bandhyopadhyay U, Kaushik S, Vartikovsky L, Cuervo AM. Dynamic organization of the receptor for chaperone-mediated autophagy at the lysosomal membrane. Mol Cell Biol 2008; 28:5747-63; http://dx.doi.org/ 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1797.Li B, Hu Q, Wang H, Man N, Ren H, Wen L, Nukina N, Fei E, Wang G. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ 2010; 17:1773-84; http://dx.doi.org/ 10.1038/cdd.2010.55. [DOI] [PubMed] [Google Scholar]
- 1798.Cilenti L, Ambivero CT, Ward N, Alnemri ES, Germain D, Zervos AS. Inactivation of Omi/HtrA2 protease leads to the deregulation of mitochondrial Mulan E3 ubiquitin ligase and increased mitophagy. Biochim Biophys Acta 2014; 1843:1295-307; http://dx.doi.org/ 10.1016/j.bbamcr.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 1799.Kang S, Fernandes-Alnemri T, Alnemri ES. A novel role for the mitochondrial HTRA2/OMI protease in aging. Autophagy 2013; 9:420-1; http://dx.doi.org/ 10.4161/auto.22920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1800.Kang S, Louboutin JP, Datta P, Landel CP, Martinez D, Zervos AS, Strayer DS, Fernandes-Alnemri T, Alnemri ES. Loss of HtrA2/Omi activity in non-neuronal tissues of adult mice causes premature aging. Cell Death Differ 2013; 20: 259-69; http://dx.doi.org/ 10.1038/cdd.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1801.Coll NS, Smidler A, Puigvert M, Popa C, Valls M, Dangl JL. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ 2014; 21:1399-408; http://dx.doi.org/ 10.1038/cdd.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1802.Kim J, Cheon H, Jeong YT, Quan W, Kim KH, Cho JM, Lim YM, Oh SH, Jin SM, Kim JH, et al.. Amyloidogenic peptide oligomer accumulation in autophagy-deficient beta cells induces diabetes. J Clin Invest 2014; 124:3311-24; http://dx.doi.org/ 10.1172/JCI69625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1803.Rivera JF, Costes S, Gurlo T, Glabe CG, Butler PC. Autophagy defends pancreatic beta cells from human islet amyloid polypeptide-induced toxicity. J Clin Invest 2014; 124:3489-500; http://dx.doi.org/ 10.1172/JCI71981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1804.Shigihara N, Fukunaka A, Hara A, Komiya K, Honda A, Uchida T, Abe H, Toyofuku Y, Tamaki M, Ogihara T, et al.. Human IAPP-induced pancreatic beta cell toxicity and its regulation by autophagy. J Clin Invest 2014; 124:3634-44; http://dx.doi.org/ 10.1172/JCI69866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1805.Lotze MT, Buchser WJ, Liang X. Blocking the interleukin 2 (IL2)-induced systemic autophagic syndrome promotes profound antitumor effects and limits toxicity. Autophagy 2012; 8:1264-6; http://dx.doi.org/ 10.4161/auto.20752. [DOI] [PubMed] [Google Scholar]
- 1806.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al.. Intrinsically disordered protein. J Mol Graph Model 2001; 19:26-59; http://dx.doi.org/ 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 1807.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci 2002; 27:527-33; http://dx.doi.org/ 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 1808.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000; 41:415-27; http://dx.doi.org/. [DOI] [PubMed] [Google Scholar]
- 1809.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 1999; 293:321-31; http://dx.doi.org/ 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 1810.Peng Z, Yan J, Fan X, Mizianty MJ, Xue B, Wang K, Hu G, Uversky VN, Kurgan L. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci 2015; 72:137-51; http://dx.doi.org/ 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1811.De Guzman RN, Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding. Biochemistry 2005; 44:490-7; http://dx.doi.org/ 10.1021/bi048161t. [DOI] [PubMed] [Google Scholar]
- 1812.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry 2002; 41:6573-82; http://dx.doi.org/ 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 1813.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 2008; 18:756-64; http://dx.doi.org/ 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 1814.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett 2005; 579:3346-54; http://dx.doi.org/ 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 1815.Peng Z, Xue B, Kurgan L, Uversky VN. Resilience of death: intrinsic disorder in proteins involved in the programmed cell death. Cell Death Differ 2013; 20:1257-67; http://dx.doi.org/ 10.1038/cdd.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1816.Popelka H, Uversky VN, Klionsky DJ. Identification of Atg3 as an intrinsically disordered polypeptide yields insights into the molecular dynamics of autophagy-related proteins in yeast. Autophagy 2014; 10:1093-104; http://dx.doi.org/ 10.4161/auto.28616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1817.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al.. Classification of intrinsically disordered regions and proteins. Chem Rev 2014; 114:6589-631; http://dx.doi.org/ 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1818.Uversky VN. Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Des 2013; 19:4191-213; http://dx.doi.org/ 10.2174/1381612811319230005. [DOI] [PubMed] [Google Scholar]
- 1819.Pejaver V, Hsu WL, Xin F, Dunker AK, Uversky VN, Radivojac P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci 2014; 23:1077-93; http://dx.doi.org/ 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1820.Chiang HS, Maric M. Lysosomal thiol reductase negatively regulates autophagy by altering glutathione synthesis and oxidation. Free Radical Bio Med 2011; 51:688-99; http://dx.doi.org/ 10.1016/j.freeradbiomed.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 1821.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, et al.. The IKK complex contributes to the induction of autophagy. EMBO J 2010; 29:619-31; http://dx.doi.org/ 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1822.Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell 2011; 22:4124-33; http://dx.doi.org/ 10.1091/mbc.E11-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1823.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, Garcia E, Dinkins C, Leuba F, Wu L, et al.. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 2010; 32:654-69; http://dx.doi.org/ 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1824.Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol 2005; 26:523-8; http://dx.doi.org/ 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 1825.Dortet L, Mostowy S, Samba-Louaka A, Gouin E, Nahori MA, Wiemer EA, Dussurget O, Cossart P. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog 2011; 7:e1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1826.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006; 313:1438-41; http://dx.doi.org/ 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 1827.Bugnicourt A, Mari M, Reggiori F, Haguenauer-Tsapis R, Galan JM. Irs4p and Tax4p: two redundant EH domain proteins involved in autophagy. Traffic 2008; 9:755-69; http://dx.doi.org/ 10.1111/j.1600-0854.2008.00715.x. [DOI] [PubMed] [Google Scholar]
- 1828.Namkoong S, Lee KI, Lee JI, Park R, Lee EJ, Jang IS, Park J. The integral membrane protein ITM2A, a transcriptional target of PKA-CREB, regulates autophagic flux via interaction with the vacuolar ATPase. Autophagy 2015; 11:756-68; http://dx.doi.org/ 10.1080/15548627.2015.1034412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1829.Yogev O, Goldberg R, Anzi S, Yogev O, Shaulian E. Jun proteins are starvation-regulated inhibitors of autophagy. Cancer Res 2010; 70:2318-27; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-3408. [DOI] [PubMed] [Google Scholar]
- 1830.Taylor R Jr., Chen PH, Chou CC, Patel J, Jin SV. KCS1 deletion in Saccharomyces cerevisiae leads to a defect in translocation of autophagic proteins and reduces autophagosome formation. Autophagy 2012; 8:1300-11; http://dx.doi.org/ 10.4161/auto.20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1831.Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, et al.. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell 2009; 36:131-40; http://dx.doi.org/ 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1832.Stepkowski TM, Kruszewski MK. Molecular cross-talk between the NRF2/KEAP1 signaling pathway, autophagy, and apoptosis. Free Radical Bio Med 2011; 50:1186-95; http://dx.doi.org/ 10.1016/j.freeradbiomed.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 1833.Puustinen P, Rytter A, Mortensen M, Kohonen P, Moreira JM, Jaattela M. CIP2A oncoprotein controls cell growth and autophagy through mTORC1 activation. J Cell Biol 2014; 204:713-27; http://dx.doi.org/ 10.1083/jcb.201304012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1834.Feng MM, Baryla J, Liu H, Laurie GW, McKown RL, Ashki N, Bhayana D, Hutnik CM. Cytoprotective effect of lacritin on human corneal epithelial cells exposed to benzalkonium chloride in vitro. Curr Eye Res 2014; 39:604-10; http://dx.doi.org/ 10.3109/02713683.2013.859275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1835.Ma P, Beck SL, Raab RW, McKown RL, Coffman GL, Utani A, Chirico WJ, Rapraeger AC, Laurie GW. Heparanase deglycanation of syndecan-1 is required for binding of the epithelial-restricted prosecretory mitogen lacritin. J Cell Biol 2006; 174:1097-106; http://dx.doi.org/ 10.1083/jcb.200511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1836.Wang N, Zimmerman K, Raab RW, McKown RL, Hutnik CM, Talla V, Tyler MFT, Lee JK, Laurie GW. Lacritin rescues stressed epithelia via rapid forkhead box O3 (FOXO3)-associated autophagy that restores metabolism. J Biol Chem 2013; 288:18146-61; http://dx.doi.org/ 10.1074/jbc.M112.436584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1837.Eskelinen E-L, Illert A, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell 2002; 13:3355-68; http://dx.doi.org/ 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1838.Eskelinen E-L, Schmidt C, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lullmann-Rauch R, Hartmann D, Heeren J, et al.. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell 2004; 15: 3132-45; http://dx.doi.org/ 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1839.Tanaka Y, Guhde G, Suter A, Eskelinen E-L, Hartmann D, Lullmann-Rauch R, Janssen P, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in Lamp-2-deficient mice. Nature 2000; 406:902-6; http://dx.doi.org/ 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 1840.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, et al.. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 2000; 406:906-10; http://dx.doi.org/ 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 1841.Bertolo C, Roa S, Sagardoy A, Mena-Varas M, Robles EF, Martinez-Ferrandis JI, Sagaert X, Tousseyn T, Orta A, Lossos IS, et al.. LITAF, a BCL6 target gene, regulates autophagy in mature B-cell lymphomas. Br J Haematol 2013; 162:621-30; http://dx.doi.org/ 10.1111/bjh.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1842.Boya P. Lysosomal function and dysfunction: mechanism and disease. Antioxid Redox Sign 2012; 17:766-74; http://dx.doi.org/ 10.1089/ars.2011.4405. [DOI] [PubMed] [Google Scholar]
- 1843.Gabande-Rodriguez E, Boya P, Labrador V, Dotti CG, Ledesma MD. High sphingomyelin levels induce lysosomal damage and autophagy dysfunction in Niemann Pick disease type A. Cell Death Differ 2014; 21:864-75; http://dx.doi.org/ 10.1038/cdd.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1844.Rodriguez-Muela N, Hernandez-Pinto AM, Serrano-Puebla A, Garcia-Ledo L, Latorre SH, de la Rosa EJ, Boya P. Lysosomal membrane permeabilization and autophagy blockade contribute to photoreceptor cell death in a mouse model of retinitis pigmentosa. Cell Death Differ 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1845.Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B. Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 2013; 25:4085-100; http://dx.doi.org/ 10.1105/tpc.113.113407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1846.Carew JS, Espitia CM, Esquivel JA II, Mahalingam D, Kelly KR, Reddy G, Giles FJ, Nawrocki ST. Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem 2011; 286:6602-13; http://dx.doi.org/ 10.1074/jbc.M110.151324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1847.Zou J, Yue F, Jiang X, Li W, Yi J, Liu L. Mitochondrion-associated protein LRPPRC suppresses the initiation of basal levels of autophagy via enhancing Bcl-2 stability. Biochem J 2013; 454:447-57; http://dx.doi.org/ 10.1042/BJ20130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1848.Zou J, Yue F, Li W, Song K, Jiang X, Yi J, Liu L. Autophagy inhibitor LRPPRC suppresses mitophagy through interaction with mitophagy initiator Parkin. PloS One 2014; 9:e94903; http://dx.doi.org/ 10.1371/journal.pone.0094903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1849.Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet 2009; 18:4022-34; http://dx.doi.org/ 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1850.Ng ACY, Eisenberg JM, Heath RJW, Huett A, Robinson CM, Nau GJ, Xavier RJ. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci USA 2011; 108:4631-8; http://dx.doi.org/ 10.1073/pnas.1000093107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1851.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 1997; 243:240-6; http://dx.doi.org/ 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 1852.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, Lynch JP, Uehara T, Sepulveda AR, Davis LE, et al.. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci USA 2012; 109:8253-8; http://dx.doi.org/ 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1853.Amaravadi RK, Winkler JD. Lys05: a new lysosomal autophagy inhibitor. Autophagy 2012; 8:1383-4; http://dx.doi.org/ 10.4161/auto.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1854.Pineda CT, Ramanathan S, Fon Tacer K, Weon JL, Potts MB, Ou YH, White MA, Potts PR. Degradation of AMPK by a Cancer-Specific Ubiquitin Ligase. Cell 2015; 160:715-28; http://dx.doi.org/ 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1855.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem 1994; 269:11492-7. [PubMed] [Google Scholar]
- 1856.Xie R, Nguyen S, McKeehan K, Wang F, McKeehan WL, Liu L. Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. J Biol Chem 2011; 286:10367-77; http://dx.doi.org/ 10.1074/jbc.M110.206532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1857.Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jaattela M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J 2009; 28:677-85; http://dx.doi.org/ 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1858.Ogier-Denis E, Pattingre S, El Benna J, Codogno P. Erk1/2-dependent phosphorylation of Galpha-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J Biol Chem 2000; 275: 39090-5; http://dx.doi.org/ 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- 1859.Fu MM, Nirschl JJ, Holzbaur EL. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev Cell 2014; 29:577-90; http://dx.doi.org/ 10.1016/j.devcel.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1860.Raciti M, Lotti LV, Valia S, Pulcinelli FM, Di Renzo L. JNK2 is activated during ER stress and promotes cell survival. Cell Death Dis 2012; 3:e429; http://dx.doi.org/ 10.1038/cddis.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1861.Keil E, Hocker R, Schuster M, Essmann F, Ueffing N, Hoffman B, Liebermann DA, Pfeffer K, Schulze-Osthoff K, Schmitz I. Phosphorylation of Atg5 by the Gadd45beta-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ 2013; 20:321-32; http://dx.doi.org/ 10.1038/cdd.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1862.Menon MB, Dhamija S, Kotlyarov A, Gaestel M. The problem of pyridinyl imidazole class inhibitors of MAPK14/p38alpha and MAPK11/p38beta in autophagy research. Autophagy 2015; 11:1425-7; http://dx.doi.org/ 10.1080/15548627.2015.1059562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1863.Menon MB, Kotlyarov A, Gaestel M. SB202190-induced cell type-specific vacuole formation and defective autophagy do not depend on p38 MAP kinase inhibition. PloS One 2011; 6:e23054; http://dx.doi.org/ 10.1371/journal.pone.0023054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1864.Colecchia D, Strambi A, Sanzone S, Iavarone C, Rossi M, Dall'Armi C, Piccioni F, Verrotti Di Pianella A, Chiariello M. MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy 2012; 8:1724-40; http://dx.doi.org/ 10.4161/auto.21857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1865.Wang Z, Zhang J, Wang Y, Xing R, Yi C, Zhu H, Chen X, Guo J, Guo W, Li W, et al.. Matrine, a novel autophagy inhibitor, blocks trafficking and the proteolytic activation of lysosomal proteases. Carcinogenesis 2013; 34:128-38; http://dx.doi.org/ 10.1093/carcin/bgs295. [DOI] [PubMed] [Google Scholar]
- 1866.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, et al.. Crosstalk between the cGAS DNA Sensor and Beclin-1 Autophagy Protein Shapes Innate Antimicrobial Immune Responses. Cell Host Microbe 2014; 15:228-38; http://dx.doi.org/ 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1867.Lorente M, Torres S, Salazar M, Carracedo A, Hernandez-Tiedra S, Rodriguez-Fornes F, Garcia-Taboada E, Melendez B, Mollejo M, Campos-Martin Y, et al.. Stimulation of ALK by the growth factor midkine renders glioma cells resistant to autophagy-mediated cell death. Autophagy 2011; 7:1071-3; http://dx.doi.org/ 10.4161/auto.7.9.15866. [DOI] [PubMed] [Google Scholar]
- 1868.Lorente M, Torres S, Salazar M, Carracedo A, Hernandez-Tiedra S, Rodriguez-Fornes F, Garcia-Taboada E, Melendez B, Mollejo M, Campos-Martin Y, et al.. Stimulation of the midkine/ALK axis renders glioma cells resistant to cannabinoid antitumoral action. Cell Death Differ 2011; 18:959-73; http://dx.doi.org/ 10.1038/cdd.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1869.Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, Deretic V. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol 2015; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1870.Bagniewska-Zadworna A, Byczyk J, Eissenstat DM, Oleksyn J, Zadworny M. Avoiding transport bottlenecks in an expanding root system: xylem vessel development in fibrous and pioneer roots under field conditions. Am J Bot 2012; 99:1417-26; http://dx.doi.org/ 10.3732/ajb.1100552. [DOI] [PubMed] [Google Scholar]
- 1871.van Doorn WG, Woltering EJ. Many ways to exit? Cell death categories in plants. Trends Plant Sci 2005; 10:117-22; http://dx.doi.org/ 10.1016/j.tplants.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 1872.Eastwood MD, Cheung SW, Lee KY, Moffat J, Meneghini MD. Developmentally programmed nuclear destruction during yeast gametogenesis. Dev Cell 2012; 23:35-44; http://dx.doi.org/ 10.1016/j.devcel.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 1873.Wang P, Lazarus BD, Forsythe ME, Love DC, Krause MW, Hanover JA. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc Natl Acad Sci USA 2012; 109:17669-74; http://dx.doi.org/ 10.1073/pnas.1205748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1874.Oku M, Warnecke D, Noda T, Muller F, Heinz E, Mukaiyama H, Kato N, Sakai Y. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J 2003; 22:3231-41; http://dx.doi.org/ 10.1093/emboj/cdg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1875.Ding WX, Guo F, Ni HM, Bockus A, Manley S, Stolz DB, Eskelinen EL, Jaeschke H, Yin XM. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J Biol Chem 2012; 287:42379-88; http://dx.doi.org/ 10.1074/jbc.M112.413682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1876.Ding WX, Li M, Biazik JM, Morgan DG, Guo F, Ni HM, Goheen M, Eskelinen EL, Yin XM. Electron microscopic analysis of a spherical mitochondrial structure. J Biol Chem 2012; 287:42373-8; http://dx.doi.org/ 10.1074/jbc.M112.413674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1877.Seca H, Lima RT, Lopes-Rodrigues V, Guimaraes JE, Almeida GM, Vasconcelos MH. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr Drug Targets 2013; 14:1135-43; http://dx.doi.org/ 10.2174/13894501113149990185. [DOI] [PubMed] [Google Scholar]
- 1878.Pennati M, Lopergolo A, Profumo V, De Cesare M, Sbarra S, Valdagni R, Zaffaroni N, Gandellini P, Folini M. miR-205 impairs the autophagic flux and enhances cisplatin cytotoxicity in castration-resistant prostate cancer cells. Biochem Pharmacol 2014; 87:579-97; http://dx.doi.org/ 10.1016/j.bcp.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 1879.Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014; 7:ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1880.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 2005; 8:3-5; http://dx.doi.org/ 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 1881.Choi YJ, Hwang KC, Park JY, Park KK, Kim JH, Park SB, Hwang S, Park H, Park C, Kim JH. Identification and characterization of a novel mouse and human MOPT gene containing MORN-motif protein in testis. Theriogenology 2010; 73:273-81; http://dx.doi.org/ 10.1016/j.theriogenology.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 1882.Frost LS, Lopes VS, Bragin A, Reyes-Reveles J, Brancato J, Cohen A, Mitchell CH, Williams DS, Boesze-Battaglia K. The Contribution of Melanoregulin to Microtubule-Associated Protein 1 Light Chain 3 (LC3) Associated Phagocytosis in Retinal Pigment Epithelium. Mol Neurobiol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1883.Frost LS, Mitchell CH, Boesze-Battaglia K. Autophagy in the eye: implications for ocular cell health. Exp Eye Res 2014; 124:56-66; http://dx.doi.org/ 10.1016/j.exer.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1884.Bhutia SK, Kegelman TP, Das SK, Azab B, Su ZZ, Lee SG, Sarkar D, Fisher PB. Astrocyte elevated gene-1 induces protective autophagy. Proc Natl Acad Sci USA 2010; 107:22243-8; http://dx.doi.org/ 10.1073/pnas.1009479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1885.Wu Y, Cheng S, Zhao H, Zou W, Yoshina S, Mitani S, Zhang H, Wang X. PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep 2014; 15:973-81; http://dx.doi.org/ 10.15252/embr.201438618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1886.Al-Qusairi L, Prokic I, Amoasii L, Kretz C, Messaddeq N, Mandel JL, Laporte J. Lack of myotubularin (MTM1) leads to muscle hypotrophy through unbalanced regulation of the autophagy and ubiquitin-proteasome pathways. FASEB J 2013; 27:3384-94; http://dx.doi.org/ 10.1096/fj.12-220947. [DOI] [PubMed] [Google Scholar]
- 1887.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic 2010; 11:468-78; http://dx.doi.org/ 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 1888.Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J 2009; 28:2244-58; http://dx.doi.org/ 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1889.Zou J, Zhang C, Marjanovic J, Kisseleva MV, Majerus PW, Wilson MP. Myotubularin-related protein (MTMR) 9 determines the enzymatic activity, substrate specificity, and role in autophagy of MTMR8. Proc Natl Acad Sci USA 2012; 109:9539-44; http://dx.doi.org/ 10.1073/pnas.1207021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1890.Hnia K, Kretz C, Amoasii L, Bohm J, Liu X, Messaddeq N, Qu CK, Laporte J. Primary T-tubule and autophagy defects in the phosphoinositide phosphatase Jumpy/MTMR14 knockout mice muscle. Adv Biol Reg 2012; 52:98-107; http://dx.doi.org/ 10.1016/j.advenzreg.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 1891.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, et al.. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol 2007; 17:1817-25; http://dx.doi.org/ 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 1892.Brandstaetter H, Kishi-Itakura C, Tumbarello DA, Manstein DJ, Buss F. Loss of functional MYO1C/myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome-lysosome fusion. Autophagy 2014; 10:2310-23; http://dx.doi.org/ 10.4161/15548627.2014.984272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1893.Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol 2012; 14:1024-35; http://dx.doi.org/ 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1894.Kuo HP, Lee DF, Chen CT, Liu M, Chou CK, Lee HJ, Du Y, Xie X, Wei Y, Xia W, et al.. ARD1 stabilization of TSC2 suppresses tumorigenesis through the mTOR signaling pathway. Sci Signal 2010; 3:ra9; http://dx.doi.org/ 10.1126/scisignal.2000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1895.Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H, Patel R, Liu D, Qin ZH, Shih IM, et al.. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene 2012; 31:1055-64; http://dx.doi.org/ 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1896.Wang P, Guan YF, Du H, Zhai QW, Su DF, Miao CY. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy 2012; 8:77-87; http://dx.doi.org/ 10.4161/auto.8.1.18274. [DOI] [PubMed] [Google Scholar]
- 1897.Naydenov NG, Harris G, Morales V, Ivanov AI. Loss of a membrane trafficking protein alphaSNAP induces non-canonical autophagy in human epithelia. Cell Cycle 2012; 11:4613-25; http://dx.doi.org/ 10.4161/cc.22885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1898.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 2008; 135:1311-23; http://dx.doi.org/ 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1899.Bonapace L, Bornhauser BC, Schmitz M, Cario G, Ziegler U, Niggli FK, Schafer BW, Schrappe M, Stanulla M, Bourquin JP. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest 2010; 120:1310-23; http://dx.doi.org/ 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1900.Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Codogno P. Regulation of autophagy by NF{kappa}B transcription factor and reactives oxygen species. Autophagy 2007; 3:390-2; http://dx.doi.org/ 10.4161/auto.4248. [DOI] [PubMed] [Google Scholar]
- 1901.Criado O, Aguado C, Gayarre J, Duran-Trio L, Garcia-Cabrero AM, Vernia S, San Millan B, Heredia M, Roma-Mateo C, Mouron S, et al.. Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum Mol Genet 2012; 21:1521-33; http://dx.doi.org/ 10.1093/hmg/ddr590. [DOI] [PubMed] [Google Scholar]
- 1902.Cervia D, Perrotta C, Moscheni C, De Palma C, Clementi E. Nitric oxide and sphingolipids control apoptosis and autophagy with a significant impact on Alzheimer's disease. J Biol Reg Homeos Ag 2013; 27:11-22. [PubMed] [Google Scholar]
- 1903.Rabkin SW. Nitric oxide-induced cell death in the heart: the role of autophagy. Autophagy 2007; 3:347-9; http://dx.doi.org/ 10.4161/auto.4054. [DOI] [PubMed] [Google Scholar]
- 1904.Zang L, He H, Ye Y, Liu W, Fan S, Tashiro S, Onodera S, Ikejima T. Nitric oxide augments oridonin-induced efferocytosis by human histocytic lymphoma U937 cells via autophagy and the NF-kappaB-COX-2-IL-1beta pathway. Free Rad Res 2012; 46:1207-19; http://dx.doi.org/ 10.3109/10715762.2012.700515. [DOI] [PubMed] [Google Scholar]
- 1905.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al.. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010; 11:55-62; http://dx.doi.org/ 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 1906.Aveleira CA, Botelho M, Carmo-Silva S, Pascoal JF, Ferreira-Marques M, Nobrega C, Cortes L, Valero J, Sousa-Ferreira L, Alvaro AR, et al.. Neuropeptide Y stimulates autophagy in hypothalamic neurons. Proc Natl Acad Sci USA 2015; 112:E1642-51; http://dx.doi.org/ 10.1073/pnas.1416609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1907.Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM. NRBF2 regulates macroautophagy as a component of Vps34 Complex I. Biochem J 2014; 461:315-22; http://dx.doi.org/ 10.1042/BJ20140515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1908.Lu J, He L, Behrends C, Araki M, Araki K, Jun Wang Q, Catanzaro JM, Friedman SL, Zong WX, Fiel MI, et al.. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun 2014; 5:3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1909.Judith D, Mostowy S, Bourai M, Gangneux N, Lelek M, Lucas-Hourani M, Cayet N, Jacob Y, Prevost MC, Pierre P, et al.. Species-specific impact of the autophagy machinery on Chikungunya virus infection. EMBO Rep 2013; 14:534-44; http://dx.doi.org/ 10.1038/embor.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1910.Kong DK, Georgescu SP, Cano C, Aronovitz MJ, Iovanna JL, Patten RD, Kyriakis JM, Goruppi S. Deficiency of the transcriptional regulator p8 results in increased autophagy and apoptosis, and causes impaired heart function. Mol Biol Cell 2010; 21:1335-49; http://dx.doi.org/ 10.1091/mbc.E09-09-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1911.Chang KY, Tsai SY, Wu CM, Yen CJ, Chuang BF, Chang JY. Novel phosphoinositide 3-kinase/mTOR dual inhibitor, NVP-BGT226, displays potent growth-inhibitory activity against human head and neck cancer cells in vitro and in vivo. Clin Cancer Res 2011; 17:7116-26; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-0796. [DOI] [PubMed] [Google Scholar]
- 1912.Liu XM, Sun LL, Hu W, Ding YH, Dong MQ, Du LL. ESCRTs cooperate with a selective autophagy receptor to mediate vacuolar targeting of soluble cargos. Mol Cell 2015; 59:1035-42; http://dx.doi.org/ 10.1016/j.molcel.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 1913.Gundara JS, Zhao J, Robinson BG, Sidhu SB. Oncophagy: harnessing regulation of autophagy in cancer therapy. Endocr Relat Cancer 2012; 19:R281-95; http://dx.doi.org/ 10.1530/ERC-12-0325. [DOI] [PubMed] [Google Scholar]
- 1914.Mijaljica D. Autophagy in 2020 and beyond: eating our way into a healthy future. Autophagy 2010; 6:194-6; http://dx.doi.org/ 10.4161/auto.6.1.10992. [DOI] [PubMed] [Google Scholar]
- 1915.Zhang CF, Gruber F, Ni C, Mildner M, Koenig U, Karner S, Barresi C, Rossiter H, Narzt MS, Nagelreiter IM, et al.. Suppression of autophagy dysregulates the antioxidant response and causes premature senescence of melanocytes. J Invest Dermatol 2015; 135:1348-57; http://dx.doi.org/ 10.1038/jid.2014.439. [DOI] [PubMed] [Google Scholar]
- 1916.Zhao Y, Zhang CF, Rossiter H, Eckhart L, Konig U, Karner S, Mildner M, Bochkov VN, Tschachler E, Gruber F. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. J Invest Dermatol 2013; 133:1629-37; http://dx.doi.org/ 10.1038/jid.2013.26. [DOI] [PubMed] [Google Scholar]
- 1917.Bertolin G, Ferrando-Miguel R, Jacoupy M, Traver S, Grenier K, Greene AW, Dauphin A, Waharte F, Bayot A, Salamero J, et al.. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. Autophagy 2013; 9:1801-17; http://dx.doi.org/ 10.4161/auto.25884. [DOI] [PubMed] [Google Scholar]
- 1918.Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, McBride HM, Park DS, Fon EA. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep 2012; 13:378-85; http://dx.doi.org/ 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1919.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy 2013; 9:1750-7; http://dx.doi.org/ 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1920.Meissner C, Lorenz H, Hehn B, Lemberg MK. Intramembrane protease PARL defines a negative regulator of PINK1- and PARK2/Parkin-dependent mitophagy. Autophagy 2015:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1921.Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep 2012; 2:1002; http://dx.doi.org/ 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1922.Durcan TM, Tang MY, Perusse JR, Dashti EA, Aguileta MA, McLelland GL, Gros P, Shaler TA, Faubert D, Coulombe B, et al.. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1923.Ren H, Fu K, Mu C, Li B, Wang D, Wang G. DJ-1, a cancer and Parkinson's disease associated protein, regulates autophagy through JNK pathway in cancer cells. Cancer Lett 2010; 297:101-8; http://dx.doi.org/ 10.1016/j.canlet.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 1924.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 2010; 191:933-42; http://dx.doi.org/ 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1925.Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem 2011; 117:856-67; http://dx.doi.org/ 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- 1926.Shi G, Lee JR, Grimes DA, Racacho L, Ye D, Yang H, Ross OA, Farrer M, McQuibban GA, Bulman DE. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson's disease. Hum Mol Genet 2011; 20:1966-74; http://dx.doi.org/ 10.1093/hmg/ddr077. [DOI] [PubMed] [Google Scholar]
- 1927.Munoz-Gamez JA, Rodriguez-Vargas JM, Quiles-Perez R, Aguilar-Quesada R, Martin-Oliva D, de Murcia G, Menissier de Murcia J, Almendros A, Ruiz de Almodovar M, Oliver FJ. PARP-1 is involved in autophagy induced by DNA damage. Autophagy 2009; 5:61-74; http://dx.doi.org/ 10.4161/auto.5.1.7272. [DOI] [PubMed] [Google Scholar]
- 1928.Huang Q, Shen HM. To die or to live: the dual role of poly(ADP-ribose) polymerase-1 in autophagy and necrosis under oxidative stress and DNA damage. Autophagy 2009; 5:273-6; http://dx.doi.org/ 10.4161/auto.5.2.7640. [DOI] [PubMed] [Google Scholar]
- 1929.Thayyullathil F, Rahman A, Pallichankandy S, Patel M, Galadari S. ROS-dependent prostate apoptosis response-4 (Par-4) up-regulation and ceramide generation are the prime signaling events associated with curcumin-induced autophagic cell death in human malignant glioma. FEBS Open Bio 2014; 4:763-76; http://dx.doi.org/ 10.1016/j.fob.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1930.Wang LJ, Chen PR, Hsu LP, Hsu WL, Liu DW, Chang CH, Hsu YC, Lee JW. Concomitant induction of apoptosis and autophagy by prostate apoptosis response-4 in hypopharyngeal carcinoma cells. Am J Pathol 2014; 184:418-30; http://dx.doi.org/ 10.1016/j.ajpath.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 1931.Silvente-Poirot S, Poirot M. Cholesterol metabolism and cancer: the good, the bad and the ugly. Current Opin Pharmacol 2012; 12:673-6; http://dx.doi.org/ 10.1016/j.coph.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 1932.Bock BC, Tagscherer KE, Fassl A, Kramer A, Oehme I, Zentgraf HW, Keith M, Roth W. The PEA-15 protein regulates autophagy via activation of JNK. J Biol Chem 2010; 285:21644-54; http://dx.doi.org/ 10.1074/jbc.M109.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1933.Murrow L, Malhotra R, Debnath J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol 2015; 17:300-10; http://dx.doi.org/ 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1934.Leu JI-J, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell 2009; 36:15-27; http://dx.doi.org/ 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1935.Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 2013; 25:4967-83; http://dx.doi.org/ 10.1105/tpc.113.116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1936.Ano Y, Hattori T, Oku M, Mukaiyama H, Baba M, Ohsumi Y, Kato N, Sakai Y. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. Mol Biol Cell 2005; 16:446-57; http://dx.doi.org/ 10.1091/mbc.E04-09-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1937.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, et al.. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol 2008; 9:908-16; http://dx.doi.org/ 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1938.Seglen PO, Gordon PB, Holen I. Non-selective autophagy. Semin Cell Biol 1990; 1:441-8. [PubMed] [Google Scholar]
- 1939.He C, Klionsky DJ. Atg9 trafficking in autophagy-related pathways. Autophagy 2007; 3:271-4; http://dx.doi.org/ 10.4161/auto.3912. [DOI] [PubMed] [Google Scholar]
- 1940.Huang H, Kawamata T, Horie T, Tsugawa H, Nakayama Y, Ohsumi Y, Fukusaki E. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J 2015; 34:154-68; http://dx.doi.org/ 10.15252/embj.201489083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1941.Meijer AJ, Klionsky DJ. Vps34 is a phosphatidylinositol 3-kinase, not a phosphoinositide 3-kinase. Autophagy 2011; 7:563-4; http://dx.doi.org/ 10.4161/auto.7.6.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1942.Devereaux K, Dall'Armi C, Alcazar-Roman A, Ogasawara Y, Zhou X, Wang F, Yamamoto A, De Camilli P, Di Paolo G. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PloS One 2013; 8:e76405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1943.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem 2005; 280:33076-82; http://dx.doi.org/ 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 1944.Roppenser B, Grinstein S, Brumell JH. Modulation of host phosphoinositide metabolism during Salmonella invasion by the type III secreted effector SopB. Methods Cell Biol 2012; 108:173-86; http://dx.doi.org/ 10.1016/B978-0-12-386487-1.00009-2. [DOI] [PubMed] [Google Scholar]
- 1945.Cuesta-Geijo MA, Galindo I, Hernaez B, Quetglas JI, Dalmau-Mena I, Alonso C. Endosomal maturation, Rab7 GTPase and phosphoinositides in African swine fever virus entry. PloS One 2012; 7:e48853; http://dx.doi.org/ 10.1371/journal.pone.0048853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1946.Jin N, Mao K, Jin Y, Tevzadze G, Kauffman EJ, Park S, Bridges D, Loewith R, Saltiel AR, Klionsky DJ, et al.. Roles for PI(3,5)P2 in nutrient sensing through TORC1. Mol Biol Cell 2014; 25:1171-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1947.Wang H, Sun HQ, Zhu X, Zhang L, Albanesi J, Levine B, Yin H. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc Natl Acad Sci USA 2015; 112:7015-20; http://dx.doi.org/ 10.1073/pnas.1507263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1948.Dou Z, Chattopadhyay M, Pan JA, Guerriero JL, Jiang YP, Ballou LM, Yue Z, Lin RZ, Zong WX. The class IA phosphatidylinositol 3-kinase p110-β subunit is a positive regulator of autophagy. J Cell Biol 2010; 191:827-43; http://dx.doi.org/ 10.1083/jcb.201006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1949.Lindmo K, Brech A, Finley KD, Gaumer S, Contamine D, Rusten TE, Stenmark H. The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy 2008; 4:500-6; http://dx.doi.org/ 10.4161/auto.5829. [DOI] [PubMed] [Google Scholar]
- 1950.Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic 2002; 3:416-27; http://dx.doi.org/ 10.1034/j.1600-0854.2002.30605.x. [DOI] [PubMed] [Google Scholar]
- 1951.Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Hum Mol Genet 2010; 19:R28-37; http://dx.doi.org/ 10.1093/hmg/ddq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1952.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al.. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA 2010; 107:378-83; http://dx.doi.org/ 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1953.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem 2004; 279:20663-71; http://dx.doi.org/ 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1954.Shahab S, Namolovan A, Mogridge J, Kim PK, Brumell JH. Bacterial toxins can inhibit host cell autophagy through cAMP generation. Autophagy 2011; 7:957-65. [DOI] [PubMed] [Google Scholar]
- 1955.Yao Z, Delorme-Axford E, Backues SK, Klionsky DJ. Atg41/Icy2 regulates autophagosome formation. Autophagy 2015; 11:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1956.McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, et al.. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 2015; 57: 39-54; http://dx.doi.org/ 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 1957.Broadley K, Larsen L, Herst PM, Smith RA, Berridge MV, McConnell MJ. The novel phloroglucinol PMT7 kills glycolytic cancer cells by blocking autophagy and sensitizing to nutrient stress. J Cell Biochem 2011; 112:1869-79; http://dx.doi.org/ 10.1002/jcb.23107. [DOI] [PubMed] [Google Scholar]
- 1958.Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R, Robenek H, Proikas-Cezanne T, Deretic V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol 2014; 24:609-20; http://dx.doi.org/ 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1959.Bhullar KS, Rupasinghe HP. Polyphenols: multipotent therapeutic agents in neurodegenerative diseases. Oxid Med Cell Longev 2013; 2013:891748; http://dx.doi.org/ 10.1155/2013/891748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1960.Macedo D, Tavares L, McDougall GJ, Vicente Miranda H, Stewart D, Ferreira RB, Tenreiro S, Outeiro TF, Santos CN. (Poly)phenols protect from alpha-synuclein toxicity by reducing oxidative stress and promoting autophagy. Hum Mol Genet 2015; 24:1717-32; http://dx.doi.org/ 10.1093/hmg/ddu585. [DOI] [PubMed] [Google Scholar]
- 1961.Hasima N, Ozpolat B. Regulation of autophagy by polyphenolic compounds as a potential therapeutic strategy for cancer. Cell Death Dis 2014; 5:e1509; http://dx.doi.org/ 10.1038/cddis.2014.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1962.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci 2013; 126:1713-9; http://dx.doi.org/ 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1963.Palomer X, Capdevila-Busquets E, Botteri G, Salvado L, Barroso E, Davidson MM, Michalik L, Wahli W, Vazquez-Carrera M. PPARbeta/delta attenuates palmitate-induced endoplasmic reticulum stress and induces autophagic markers in human cardiac cells. Int J Cardiol 2014; 174:110-8; http://dx.doi.org/ 10.1016/j.ijcard.2014.03.176. [DOI] [PubMed] [Google Scholar]
- 1964.Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev 2000; 14:1027-47. [PubMed] [Google Scholar]
- 1965.Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev 1995; 59:94-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1966.Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, Goliaei B, Peyvandi AA. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench 2014; 7:17-31. [PMC free article] [PubMed] [Google Scholar]
- 1967.Le Guezennec X, Brichkina A, Huang YF, Kostromina E, Han W, Bulavin DV. Wip1-dependent regulation of autophagy, obesity, and atherosclerosis. Cell Metab 2012; 16:68-80; http://dx.doi.org/ 10.1016/j.cmet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 1968.Uddin MN, Ito S, Nishio N, Suganya T, Isobe KI. Gadd34 induces autophagy through the suppression of the mTOR pathway during starvation. Biochem Biophys Res Comm 2011; 407:692-8. [DOI] [PubMed] [Google Scholar]
- 1969.Peti W, Nairn AC, Page R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J 2013; 280:596-611; http://dx.doi.org/ 10.1111/j.1742-4658.2012.08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1970.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al.. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 2015; 17:288-99; http://dx.doi.org/ 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1971.Eisenberg-Lerner A, Kimchi A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ 2012; 19:788-97; http://dx.doi.org/ 10.1038/cdd.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1972.Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure 2012; 20:15-27; http://dx.doi.org/ 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1973.Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell 2012; 47:339-48; http://dx.doi.org/ 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1974.Krick R, Busse RA, Scacioc A, Stephan M, Janshoff A, Thumm M, Kuhnel K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc Natl Acad Sci USA 2012; 109:E2042-9; http://dx.doi.org/ 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1975.Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem 2012; 287:31681-90; http://dx.doi.org/ 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1976.Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol Cell 2015; 58:1053-66; http://dx.doi.org/ 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1977.Starokadomskyy P, Dmytruk KV. A bird's-eye view of autophagy. Autophagy 2013; 9:1121-6; http://dx.doi.org/ 10.4161/auto.24544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1978.Neely KM, Green KN, Laferla FM. Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a {gamma}-secretase-independent manner. J Neurosci 2011; 31:2781-91; http://dx.doi.org/ 10.1523/JNEUROSCI.5156-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1979.Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr.. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl 2005; 44:7342-72; http://dx.doi.org/ 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 1980.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods 2007; 4:798-806; http://dx.doi.org/ 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 1981.Popelka H, Klionsky DJ. Posttranslationally-modified structures in the autophagy machinery: an integrative perspective. FEBS J 2015; 282:3474-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1982.Huang YH, Al-Aidaroos AQ, Yuen HF, Zhang SD, Shen HM, Rozycka E, McCrudden CM, Tergaonkar V, Gupta A, Lin YB, et al.. A role of autophagy in PTP4A3-driven cancer progression. Autophagy 2014; 10:1787-800; http://dx.doi.org/ 10.4161/auto.29989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1983.Martin KR, Xu Y, Looyenga BD, Davis RJ, Wu CL, Tremblay ML, Xu HE, MacKeigan JP. Identification of PTPsigma as an autophagic phosphatase. J Cell Sci 2011; 124:812-9; http://dx.doi.org/ 10.1242/jcs.080341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1984.Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Munch J, Kirchhoff F, Simonsen A, et al.. TRIM Proteins Regulate Autophagy and Can Target Autophagic Substrates by Direct Recognition. Dev Cell 2014; 30:394-409; http://dx.doi.org/ 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1985.Nagy G, Ward J, Mosser DD, Koncz A, Gergely P Jr., Stancato C, Qian Y, Fernandez D, Niland B, Grossman CE, et al.. Regulation of CD4 expression via recycling by HRES-1/RAB4 controls susceptibility to HIV infection. J Biol Chem 2006; 281:34574-91; http://dx.doi.org/ 10.1074/jbc.M606301200. [DOI] [PubMed] [Google Scholar]
- 1986.Fernandez DR, Telarico T, Bonilla E, Li Q, Banerjee S, Middleton FA, Phillips PE, Crow MK, Oess S, Muller-Esterl W, et al.. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol 2009; 182:2063-73; http://dx.doi.org/ 10.4049/jimmunol.0803600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1987.Caza TN, Fernandez DR, Talaber G, Oaks Z, Haas M, Madaio MP, Lai ZW, Miklossy G, Singh RR, Chudakov DM, et al.. HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann Rheum Dis 2014; 73:1888-97; http://dx.doi.org/ 10.1136/annrheumdis-2013-203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1988.Talaber G, Miklossy G, Oaks Z, Liu Y, Tooze SA, Chudakov DM, Banki K, Perl A. HRES-1/Rab4 promotes the formation of LC3(+) autophagosomes and the accumulation of mitochondria during autophagy. PloS One 2014; 9:e84392; http://dx.doi.org/ 10.1371/journal.pone.0084392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1989.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011; 80:125-56; http://dx.doi.org/ 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 1990.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nature Rev Mol Cell Biol 2009; 10:513-25; http://dx.doi.org/ 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 1991.Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 2004; 117:4837-48; http://dx.doi.org/ 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 1992.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, et al.. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012; 37:223-34; http://dx.doi.org/ 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1993.Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 2012; 197:659-75; http://dx.doi.org/ 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1994.Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep 2013; 14:450-7; http://dx.doi.org/ 10.1038/embor.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1995.Jean S, Cox S, Nassari S, Kiger AA. Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. EMBO Rep 2015; 16:297-311; http://dx.doi.org/ 10.15252/embr.201439464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1996.Munafo DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic 2002; 3:472-82; http://dx.doi.org/ 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- 1997.Ylä-Anttila P, Mikkonen E, Happonen KE, Holland P, Ueno T, Simonsen A, Eskelinen E-L: RAB24 facilitates clearance of autophagic compartments during basal conditions. Autophagy 2015, 10:1833-48 DOI: 10.1080/15548627.2015.108652210.1080/15548627.2015.1086522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1998.Hirota Y, Tanaka Y. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell Mol Life Sci 2009; 66:2913-32; http://dx.doi.org/ 10.1007/s00018-009-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1999.Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell 2008; 19:2916-25; http://dx.doi.org/ 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2000.Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol 2011; 192:839-53; http://dx.doi.org/ 10.1083/jcb.201008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2001.Chen XW, Leto D, Xiong T, Yu G, Cheng A, Decker S, Saltiel AR. A Ral GAP complex links PI 3-kinase/Akt signaling to RalA activation in insulin action. Mol Biol Cell 2011; 22:141-52; http://dx.doi.org/ 10.1091/mbc.E10-08-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2002.Gentry LR, Martin TD, Reiner DJ, Der CJ. Ral small GTPase signaling and oncogenesis: More than just 15minutes of fame. Biochim Biophys Acta 2014; 1843:2976-88; http://dx.doi.org/ 10.1016/j.bbamcr.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2003.Martin TD, Chen XW, Kaplan RE, Saltiel AR, Walker CL, Reiner DJ, Der CJ. Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol Cell 2014; 53:209-20; http://dx.doi.org/ 10.1016/j.molcel.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2004.Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2010; 21:2257-69; http://dx.doi.org/ 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2005.Shirakawa R, Fukai S, Kawato M, Higashi T, Kondo H, Ikeda T, Nakayama E, Okawa K, Nureki O, Kimura T, et al.. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem 2009; 284:21580-8; http://dx.doi.org/ 10.1074/jbc.M109.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2006.Oeckinghaus A, Postler TS, Rao P, Schmitt H, Schmitt V, Grinberg-Bleyer Y, Kuhn LI, Gruber CW, Lienhard GE, Ghosh S. kappaB-Ras proteins regulate both NF-kappaB-dependent inflammation and Ral-dependent proliferation. Cell Rep 2014; 8:1793-807; http://dx.doi.org/ 10.1016/j.celrep.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2007.Punnonen EL, Reunanen H, Hirsimaki P, Lounatmaa K. Filipin labelling and intramembrane particles on the membranes of early and later autophagic vacuoles in Ehrlich ascites cells. Virchows Archiv B, Cell Pathol 1988; 54:317-26; http://dx.doi.org/ 10.1007/BF02899229. [DOI] [PubMed] [Google Scholar]
- 2008.Opipari AJ, Tan L, Boitano AE, Sorenson DR, Aurora A, Liu JR. Resveratrol-induced autophagocytosis in ovarian cancer cells. Cancer Res 2004; 15:696-703; http://dx.doi.org/ 10.1158/0008-5472.CAN-03-2404. [DOI] [PubMed] [Google Scholar]
- 2009.Ogier-Denis E, Petiot A, Bauvy C, Codogno P. Control of the expression and activity of the Galpha-interacting protein (GAIP) in human intestinal cells. J Biol Chem 1997; 272:24599-603; http://dx.doi.org/ 10.1074/jbc.272.39.24599. [DOI] [PubMed] [Google Scholar]
- 2010.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2007; 18:4180-9; http://dx.doi.org/ 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2011.Yonekawa T, Gamez G, Kim J, Tan AC, Thorburn J, Gump J, Thorburn A, Morgan MJ. RIP1 negatively regulates basal autophagic flux through TFEB to control sensitivity to apoptosis. EMBO Rep 2015; 16:700-8; http://dx.doi.org/ 10.15252/embr.201439496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2012.Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, Macintosh GC. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci USA 2011; 108:1093-8; http://dx.doi.org/ 10.1073/pnas.1009809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2013.Haud N, Kara F, Diekmann S, Henneke M, Willer JR, Hillwig MS, Gregg RG, Macintosh GC, Gartner J, Alia A, et al.. rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci USA 2011; 108:1099-103; http://dx.doi.org/ 10.1073/pnas.1009811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2014.Xu C, Feng K, Zhao X, Huang S, Cheng Y, Qian L, Wang Y, Sun H, Jin M, Chuang TH, et al.. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy 2014; 10:2239-50.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2015.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009; 284:12297-305; http://dx.doi.org/ 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2016.Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 2011; 7:737-47; http://dx.doi.org/ 10.4161/auto.7.7.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2017.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan K-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008; 10:935-45; http://dx.doi.org/ 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2018.White E. Exploiting the bad eating habits of Ras-driven cancers. Genes Dev 2013; 27:2065-71; http://dx.doi.org/ 10.1101/gad.228122.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2019.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA 2004; 101:18030-5; http://dx.doi.org/ 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2020.Stankov MV, El Khatib M, Kumar Thakur B, Heitmann K, Panayotova-Dimitrova D, Schoening J, Bourquin JP, Schweitzer N, Leverkus M, Welte K, et al.. Histone deacetylase inhibitors induce apoptosis in myeloid leukemia by suppressing autophagy. Leukemia 2014; 28:577-88; http://dx.doi.org/ 10.1038/leu.2013.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2021.Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A, et al.. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics 2011; 10:M110 006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2022.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen W-L, Griffith J, Nag S, Wang K, Moss T, et al.. SNARE proteins are required for macroautophagy. Cell 2011; 146:290-302; http://dx.doi.org/ 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2023.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, et al.. SNARE proteins are required for macroautophagy. Cell 2011; 146:290-302; http://dx.doi.org/ 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2024.Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell 2001; 12:3690-702; http://dx.doi.org/ 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2025.Jiang S, Dupont N, Castillo EF, Deretic V. Secretory versus degradative autophagy: unconventional secretion of inflammatory mediators. J Innate Immun 2013; 5:471-9; http://dx.doi.org/ 10.1159/000346707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2026.Mostowy S, Bonazzi M, Hamon MA, Tham TN, Mallet A, Lelek M, Gouin E, Demangel C, Brosch R, Zimmer C, et al.. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe 2010; 8:433-44; http://dx.doi.org/ 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 2027.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al.. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science 2010; 329:229-32; http://dx.doi.org/ 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 2028.Lee JH, Budanov AV, Karin M. Sestrins orchestrate cellular metabolism to attenuate aging. Cell Metab 2013; 18:792-801; http://dx.doi.org/ 10.1016/j.cmet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2029.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, Lee HE, Kang D, Rhee SG. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab 2013; 17:73-84; http://dx.doi.org/ 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 2030.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008; 134:451-60; http://dx.doi.org/ 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2031.Park HW, Park H, Ro SH, Jang I, Semple IA, Kim DN, Kim M, Nam M, Zhang D, Yin L, et al.. Hepatoprotective role of Sestrin2 against chronic ER stress. Nat Commun 2014; 5:4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2032.Ben-Sahra I, Dirat B, Laurent K, Puissant A, Auberger P, Budanov A, Tanti JF, Bost F. Sestrin2 integrates Akt and mTOR signaling to protect cells against energetic stress-induced death. Cell Death Differ 2013; 20:611-9; http://dx.doi.org/ 10.1038/cdd.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2033.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al.. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 2007; 9:1142-51; http://dx.doi.org/ 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2034.Wong AS, Lee RH, Cheung AY, Yeung PK, Chung SK, Cheung ZH, Ip NY. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson's disease. Nat Cell Biol 2011; 13:568-79; http://dx.doi.org/ 10.1038/ncb2217. [DOI] [PubMed] [Google Scholar]
- 2035.Zhang C, Li A, Zhang X, Xiao H. A novel TIP30 protein complex regulates EGF receptor signaling and endocytic degradation. J Biol Chem 2011; 286:9373-81; http://dx.doi.org/ 10.1074/jbc.M110.207720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2036.Khan MM, Strack S, Wild F, Hanashima A, Gasch A, Brohm K, Reischl M, Carnio S, Labeit D, Sandri M, et al.. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy 2014; 10:123-36; http://dx.doi.org/ 10.4161/auto.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2037.Belaid A, Ndiaye PD, Klionsky DJ, Hofman P, Mograbi B. Signalphagy: Scheduled signal termination by macroautophagy. Autophagy 2013; 9:1629-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2038.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA 2008; 105:3374-9; http://dx.doi.org/ 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2039.Webster BR, Scott I, Traba J, Han K, Sack MN. Regulation of autophagy and mitophagy by nutrient availability and acetylation. Biochim Biophys Acta 2014; 1841:525-34; http://dx.doi.org/ 10.1016/j.bbalip.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2040.Pi H, Xu S, Reiter RJ, Guo P, Zhang L, Li Y, Li M, Cao Z, Tian L, Xie J, et al.. SIRT3-SOD2-mROS-dependent autophagy in cadmium-induced hepatotoxicity and salvage by melatonin. Autophagy 2015; 11:1037-51; http://dx.doi.org/ 10.1080/15548627.2015.1052208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2041.Polletta L, Vernucci E, Carnevale I, Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T, Schutkowski M, Pellegrini L, et al.. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy 2015; 11:253-70; http://dx.doi.org/ 10.1080/15548627.2015.1009778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2042.Takasaka N, Araya J, Hara H, Ito S, Kobayashi K, Kurita Y, Wakui H, Yoshii Y, Yumino Y, Fujii S, et al.. Autophagy induction by SIRT6 through attenuation of insulin-like growth factor signaling is involved in the regulation of human bronchial epithelial cell senescence. J Immunol 2014; 192:958-68; http://dx.doi.org/ 10.4049/jimmunol.1302341. [DOI] [PubMed] [Google Scholar]
- 2043.Araki S, Izumiya Y, Rokutanda T, Ianni A, Hanatani S, Kimura Y, Onoue Y, Senokuchi T, Yoshizawa T, Yasuda O, et al.. Sirt7 Contributes to Myocardial Tissue Repair by Maintaining TGF-beta Signaling Pathway. Circulation 2015. [DOI] [PubMed] [Google Scholar]
- 2044.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature 2008; 451:350-4; http://dx.doi.org/ 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 2045.Bhardwaj V, Kanagawa O, Swanson PE, Unanue ER. Chronic Listeria infection in SCID mice: requirements for the carrier state and the dual role of T cells in transferring protection or suppression. J Immunol 1998; 160:376-84. [PubMed] [Google Scholar]
- 2046.Liu H, Ma Y, He HW, Wang JP, Jiang JD, Shao RG. SLC9A3R1 stimulates autophagy via BECN1 stabilization in breast cancer cells. Autophagy 2015:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2047.Catalina-Rodriguez O, Kolukula VK, Tomita Y, Preet A, Palmieri F, Wellstein A, Byers S, Giaccia AJ, Glasgow E, Albanese C, et al.. The mitochondrial citrate transporter, CIC, is essential for mitochondrial homeostasis. Oncotarget 2012; 3:1220-35; http://dx.doi.org/ 10.18632/oncotarget.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2048.Jung J, Genau HM, Behrends C. Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol Cell Biol 2015; 35:2479-94; http://dx.doi.org/ 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2049.Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, et al.. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015; 519:477-81; http://dx.doi.org/ 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2050.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al.. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015; 347:188-94; http://dx.doi.org/ 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2051.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013; 13:722-37; http://dx.doi.org/ 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2052.Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, Oren M, Kimchi A. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell 2006; 22:463-75; http://dx.doi.org/ 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 2053.Morelli E, Ginefra P, Mastrodonato V, Beznoussenko GV, Rusten TE, Bilder D, Stenmark H, Mironov AA, Vaccari T. Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila. Autophagy 2014; 10:2251-68; http://dx.doi.org/ 10.4161/15548627.2014.981913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2054.Batelli S, Peverelli E, Rodilossi S, Forloni G, Albani D. Macroautophagy and the proteasome are differently involved in the degradation of alpha-synuclein wild type and mutated A30P in an in vitro inducible model (PC12/TetOn). Neuroscience 2011; 195:128-37; http://dx.doi.org/ 10.1016/j.neuroscience.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2055.Song JX, Lu JH, Liu LF, Chen LL, Durairajan SS, Yue Z, Zhang HQ, Li M. HMGB1 is involved in autophagy inhibition caused by SNCA/alpha-synuclein overexpression: a process modulated by the natural autophagy inducer corynoxine B. Autophagy 2014; 10:144-54; http://dx.doi.org/ 10.4161/auto.26751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2056.Knaevelsrud H, Soreng K, Raiborg C, Haberg K, Rasmuson F, Brech A, Liestol K, Rusten TE, Stenmark H, Neufeld TP, et al.. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol 2013; 202:331-49; http://dx.doi.org/ 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2057.Barnett TC, Liebl D, Seymour LM, Gillen CM, Lim JY, Larock CN, Davies MR, Schulz BL, Nizet V, Teasdale RD, et al.. The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe 2013; 14:675-82; http://dx.doi.org/ 10.1016/j.chom.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2058.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al.. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011; 147:223-34; http://dx.doi.org/ 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2059.Pietrocola F, Lachkar S, Enot DP, Niso-Santano M, Bravo-San Pedro JM, Sica V, Izzo V, Maiuri MC, Madeo F, Marino G, et al.. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ 2015; 22:509-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2060.Ghidoni R, Houri JJ, Giuliani A, Ogier-Denis E, Parolari E, Botti S, Bauvy C, Codogno P. The metabolism of sphingo(glyco)lipids is correlated with the differentiation-dependent autophagic pathway in HT-29 cells. Eur J Biochem 1996; 237:454-9; http://dx.doi.org/ 10.1111/j.1432-1033.1996.0454k.x. [DOI] [PubMed] [Google Scholar]
- 2061.Lavieu G, Scarlatti F, Sala G, Levade T, Ghidoni R, Botti J, Codogno P. Is autophagy the key mechanism by which the sphingolipid rheostat controls the cell fate decision? Autophagy 2007; 3:45-7; http://dx.doi.org/ 10.4161/auto.3416. [DOI] [PubMed] [Google Scholar]
- 2062.Rong Y, McPhee C, Deng S, Huang L, Chen L, Liu M, Tracy K, Baehreck EH, Yu L, Lenardo MJ. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci USA 2011; 108:7826-31; http://dx.doi.org/ 10.1073/pnas.1013800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2063.Chen Q, Yue F, Li W, Zou J, Xu T, Huang C, Zhang Y, Song K, Huang G, Xu G, et al.. Potassium Bisperoxo (1,10-phenanthroline) Oxovanadate (bpV(phen)) Induces Apoptosis and Pyroptosis and Disrupts the P62-HDAC6 Interaction to Suppress the Acetylated Microtubule-dependent Degradation of Autophagosomes. J Biol Chem 2015; 290:26051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2064.Tambe Y, Yamamoto A, Isono T, Chano T, Fukuda M, Inoue H. The drs tumor suppressor is involved in the maturation process of autophagy induced by low serum. Cancer Lett 2009; 283:74-83; http://dx.doi.org/ 10.1016/j.canlet.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 2065.Mesquita FS, Thomas M, Sachse M, Santos AJ, Figueira R, Holden DW. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog 2012; 8:e1002743; http://dx.doi.org/ 10.1371/journal.ppat.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2066.Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, Souquere S, Marino G, Lachkar S, Senovilla L, et al.. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell 2012; 48:667-80; http://dx.doi.org/ 10.1016/j.molcel.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 2067.Wang CW. Stationary phase lipophagy as a cellular mechanism to recycle sterols during quiescence. Autophagy 2014; 10:2075-6; http://dx.doi.org/ 10.4161/auto.36137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2068.Wang CW, Miao YH, Chang YS. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol 2014; 206:357-66; http://dx.doi.org/ 10.1083/jcb.201404115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2069.Wilkinson DS, Jariwala JS, Anderson E, Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V, Dillin A, et al.. Phosphorylation of LC3 by the Hippo kinases STK3/STK4 is essential for autophagy. Mol Cell 2015; 57:55-68; http://dx.doi.org/ 10.1016/j.molcel.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2070.Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu CP, et al.. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med 2013; 19:1478-88; http://dx.doi.org/ 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2071.Renna M, Schaffner C, Winslow AR, Menzies FM, Peden AA, Floto RA, Rubinsztein DC. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J Cell Sci 2011; 124:469-82; http://dx.doi.org/ 10.1242/jcs.076489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2072.Lu Y, Zhang Z, Sun D, Sweeney ST, Gao FB. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol Cell 2013; 52:264-71; http://dx.doi.org/ 10.1016/j.molcel.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2073.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al.. Autophagosomes form at ER-mitochondria contact sites. Nature 2013; 495:389-93; http://dx.doi.org/ 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 2074.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38{alpha} MAPK through mAtg9 and p38IP. EMBO J 2010; 29:27-40; http://dx.doi.org/ 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2075.Lopergolo A, Nicolini V, Favini E, Dal Bo L, Tortoreto M, Cominetti D, Folini M, Perego P, Castiglioni V, Scanziani E, et al.. Synergistic cooperation between sunitinib and cisplatin promotes apoptotic cell death in human medullary thyroid cancer. J Clin Endocrinol Metab 2014; 99:498-509; http://dx.doi.org/ 10.1210/jc.2013-2574. [DOI] [PubMed] [Google Scholar]
- 2076.Jackson DJ, Worheide G. Symbiophagy and biomineralization in the “living fossil” Astrosclera willeyana. Autophagy 2014; 10:408-15; http://dx.doi.org/ 10.4161/auto.27319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2077.Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, et al.. Inhibition of autophagy by TAB2 and TAB3. EMBO J 2011; 30:4908-20; http://dx.doi.org/ 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2078.Takaesu G, Kobayashi T, Yoshimura A. TGFbeta-activated kinase 1 (TAK1)-binding proteins (TAB) 2 and 3 negatively regulate autophagy. J Biochem 2012; 151:157-66; http://dx.doi.org/ 10.1093/jb/mvr123. [DOI] [PubMed] [Google Scholar]
- 2079.Nagahara Y, Takeyoshi M, Sakemoto S, Shiina I, Nakata K, Fujimori K, Wang Y, Umeda E, Watanabe C, Uetake S, et al.. Novel tamoxifen derivative Ridaifen-B induces Bcl-2 independent autophagy without estrogen receptor involvement. Biochem Biophys Res Comm 2013; 435:657-63; http://dx.doi.org/ 10.1016/j.bbrc.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 2080.Bose JK, Huang CC, Shen CK. Regulation of autophagy by neuropathological protein TDP-43. J Biol Chem 2011; 286:44441-8; http://dx.doi.org/ 10.1074/jbc.M111.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2081.Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, et al.. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 2011; 332:966-70; http://dx.doi.org/ 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2082.Newman AC, Scholefield CL, Kemp AJ, Newman M, McIver EG, Kamal A, Wilkinson S. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-kappaB signalling. PloS One 2012; 7:e50672; http://dx.doi.org/ 10.1371/journal.pone.0050672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2083.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell 2012; 47:535-46; http://dx.doi.org/ 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2084.Alfaiz AA, Micale L, Mandriani B, Augello B, Pellico MT, Chrast J, Xenarios I, Zelante L, Merla G, Reymond A. TBC1D7 mutations are associated with intellectual disability, macrocrania, patellar dislocation, and celiac disease. Hum Mutat 2014; 35:447-51; http://dx.doi.org/ 10.1002/humu.22529. [DOI] [PubMed] [Google Scholar]
- 2085.Capo-Chichi JM, Tcherkezian J, Hamdan FF, Decarie JC, Dobrzeniecka S, Patry L, Nadon MA, Mucha BE, Major P, Shevell M, et al.. Disruption of TBC1D7, a subunit of the TSC1-TSC2 protein complex, in intellectual disability and megalencephaly. J Med Genet 2013; 50:740-4; http://dx.doi.org/ 10.1136/jmedgenet-2013-101680. [DOI] [PubMed] [Google Scholar]
- 2086.Pomerantz JL, Baltimore D. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J 1999; 18:6694-704; http://dx.doi.org/ 10.1093/emboj/18.23.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2087.Neill T, Torres A, Buraschi S, Owens RT, Hoek JB, Baffa R, Iozzo RV. Decorin induces mitophagy in breast carcinoma cells via peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha) and mitostatin. J Biol Chem 2014; 289:4952-68; http://dx.doi.org/ 10.1074/jbc.M113.512566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2088.Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, Piao Z, Ashida H, Yoshida M, Kakuta S, et al.. A tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe 2011; 9:376-89; http://dx.doi.org/ 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 2089.Li L, Khatibi NH, Hu Q, Yan J, Chen C, Han J, Ma D, Chen Y, Zhou C. Transmembrane protein 166 regulates autophagic and apoptotic activities following focal cerebral ischemic injury in rats. Exp Neurol 2012; 234:181-90; http://dx.doi.org/ 10.1016/j.expneurol.2011.12.038. [DOI] [PubMed] [Google Scholar]
- 2090.Oz-Levi D, Ben-Zeev B, Ruzzo EK, Hitomi Y, Gelman A, Pelak K, Anikster Y, Reznik-Wolf H, Bar-Joseph I, Olender T, et al.. Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am J Hum Genet 2012; 91:1065-72; http://dx.doi.org/ 10.1016/j.ajhg.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2091.Oz-Levi D, Gelman A, Elazar Z, Lancet D. TECPR2: a new autophagy link for neurodegeneration. Autophagy 2013; 9:801-2; http://dx.doi.org/ 10.4161/auto.23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2092.D'Eletto M, Farrace MG, Falasca L, Reali V, Oliverio S, Melino G, Griffin M, Fimia GM, Piacentini M. Transglutaminase 2 is involved in autophagosome maturation. Autophagy 2009; 5:1145-54; http://dx.doi.org/ 10.4161/auto.5.8.10040. [DOI] [PubMed] [Google Scholar]
- 2093.Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, et al.. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest 2009; 119:1359-72; http://dx.doi.org/ 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2094.Salazar M, Lorente M, Garcia-Taboada E, Hernandez-Tiedra S, Davila D, Francis SE, Guzman M, Kiss-Toth E, Velasco G. The pseudokinase tribbles homologue-3 plays a crucial role in cannabinoid anticancer action. Biochim Biophys Acta 2013; 1831:1573-8; http://dx.doi.org/ 10.1016/j.bbalip.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 2095.Velasco G, Sanchez C, Guzman M. Towards the use of cannabinoids as antitumour agents. Nat Rev Cancer 2012; 12:436-44; http://dx.doi.org/ 10.1038/nrc3247. [DOI] [PubMed] [Google Scholar]
- 2096.Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J 2009; 28:3015-26; http://dx.doi.org/ 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2097.Lok CN, Sy LK, Liu F, Che CM. Activation of autophagy of aggregation-prone ubiquitinated proteins by timosaponin A-III. J Biol Chem 2011; 286:31684-96; http://dx.doi.org/ 10.1074/jbc.M110.202531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2098.He P, Peng Z, Luo Y, Wang L, Yu P, Deng W, An Y, Shi T, Ma D. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy 2009; 5:52-60; http://dx.doi.org/ 10.4161/auto.5.1.7247. [DOI] [PubMed] [Google Scholar]
- 2099.Boada-Romero E, Letek M, Fleischer A, Pallauf K, Ramon-Barros C, Pimentel-Muinos FX. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J 2013; 32:566-82; http://dx.doi.org/ 10.1038/emboj.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2100.Shi CS, Kehrl JH. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy 2010; 6:986-7; http://dx.doi.org/ 10.4161/auto.6.7.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2101.Matsuzawa Y, Oshima S, Takahara M, Maeyashiki C, Nemoto Y, Kobayashi M, Nibe Y, Nozaki K, Nagaishi T, Okamoto R, et al.. TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy. Autophagy 2015; 11:1052-62; http://dx.doi.org/ 10.1080/15548627.2015.1055439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2102.Jacinto E. What controls TOR? IUBMB Life 2008; 60:483-96; http://dx.doi.org/ 10.1002/iub.56. [DOI] [PubMed] [Google Scholar]
- 2103.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009; 137:873-86; http://dx.doi.org/ 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2104.Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J 2007; 405:513-22; http://dx.doi.org/ 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2105.Vlahakis A, Graef M, Nunnari J, Powers T. TOR complex 2-Ypk1 signaling is an essential positive regulator of the general amino acid control response and autophagy. Proc Natl Acad Sci USA 2014; 111:10586-91; http://dx.doi.org/ 10.1073/pnas.1406305111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2106.Renna M, Bento CF, Fleming A, Menzies FM, Siddiqi FH, Ravikumar B, Puri C, Garcia-Arencibia M, Sadiq O, Corrochano S, et al.. IGF-1 receptor antagonism inhibits autophagy. Hum Mol Genet 2013; 22:4528-44; http://dx.doi.org/ 10.1093/hmg/ddt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2107.Arias E, Koga H, Diaz A, Mocholi E, Patel B, Cuervo AM. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol Cell 2015; 59:270-84; http://dx.doi.org/ 10.1016/j.molcel.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2108.N'Guessan P, Pouyet L, Gosset G, Hamlaoui S, Seillier M, Cano CE, Seux M, Stocker P, Culcasi M, Iovanna JL, et al.. Absence of Tumor Suppressor Tumor Protein 53-Induced Nuclear Protein 1 (TP53INP1) Sensitizes Mouse Thymocytes and Embryonic Fibroblasts to Redox-Driven Apoptosis. Antioxid Redox Sign 2011; 15:1639-53; http://dx.doi.org/ 10.1089/ars.2010.3553. [DOI] [PubMed] [Google Scholar]
- 2109.Sancho A, Duran J, Garcia-Espana A, Mauvezin C, Alemu EA, Lamark T, Macias MJ, DeSalle R, Royo M, Sala D, et al.. Absence of Tumor Suppressor Tumor Protein 53-Induced Nuclear Protein 1 (TP53INP1) Sensitizes Mouse Thymocytes and Embryonic Fibroblasts to Redox-Driven Apoptosis. PloS One 2012; 7:e34034; http://dx.doi.org/ 10.1371/journal.pone.0034034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2110.Seillier M, Peuget S, Gayet O, Gauthier C, N'Guessan P, Monte M, Carrier A, Iovanna JL, Dusetti NJ. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell Death Differ 2012; 19:1525-35; http://dx.doi.org/ 10.1038/cdd.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2111.Seillier M, Pouyet L, N'Guessan P, Nollet M, Capo F, Guillaumond F, Peyta L, Dumas JF, Varrault A, Bertrand G, et al.. Defects in mitophagy promote redox-driven metabolic syndrome in the absence of TP53INP1. EMBO Mol Med 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2112.Mauvezin C, Orpinell M, Francis VA, Mansilla F, Duran J, Ribas V, Palac{i}n M, Boya P, Teleman AA, Zorzano A. The nuclear cofactor DOR regulates autophagy in mammalian and Drosophila cells. EMBO Rep 2010; 11:37-44; http://dx.doi.org/ 10.1038/embor.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2113.Nowak J, Archange C, Tardivel-Lacombe J, Pontarotti P, Pebusque MJ, Vaccaro MI, Velasco G, Dagorn JC, Iovanna JL. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell 2009; 20:870-81; http://dx.doi.org/ 10.1091/mbc.E08-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2114.Sala D, Ivanova S, Plana N, Ribas V, Duran J, Bach D, Turkseven S, Laville M, Vidal H, Karczewska-Kupczewska M, et al.. Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes. J Clin Invest 2014; 124:1914-27; http://dx.doi.org/ 10.1172/JCI72327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2115.Cang C, Zhou Y, Navarro B, Seo YJ, Aranda K, Shi L, Battaglia-Hsu S, Nissim I, Clapham DE, Ren D. mTOR regulates lysosomal ATP-sensitive two-pore Na+ channels to adapt to metabolic state. Cell 2013; 152:778-90; http://dx.doi.org/ 10.1016/j.cell.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2116.Lin PH, Duann P, Komazaki S, Park KH, Li H, Sun M, Sermersheim M, Gumpper K, Parrington J, Galione A, et al.. Lysosomal two-pore channel subtype 2 (TPC2) regulates skeletal muscle autophagic signaling. J Biol Chem 2015; 290:3377-89; http://dx.doi.org/ 10.1074/jbc.M114.608471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2117.Funasaka T, Tsuka E, Wong RW. Regulation of autophagy by nucleoporin Tpr. Sci Rep 2012; 2:878; http://dx.doi.org/ 10.1038/srep00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2118.Zou S, Chen Y, Liu Y, Segev N, Yu S, Liu Y, Min G, Ye M, Zeng Y, Zhu X, et al.. Trs130 participates in autophagy through GTPases Ypt31/32 in Saccharomyces cerevisiae. Traffic 2013; 14: 233-46; http://dx.doi.org/ 10.1111/tra.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2119.Hua F, Li K, Yu JJ, Lv XX, Yan J, Zhang XW, Sun W, Lin H, Shang S, Wang F, et al.. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun 2015; 6:7951; http://dx.doi.org/ 10.1038/ncomms8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2120.Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Egia A, Lorente M, Vazquez P, Torres S, Iovanna JL, Guzman M, et al.. TRB3 links ER stress to autophagy in cannabinoid anti-tumoral action. Autophagy 2009; 5:1048-9; http://dx.doi.org/ 10.4161/auto.5.7.9508. [DOI] [PubMed] [Google Scholar]
- 2121.Francisco R, Perez-Perarnau A, Cortes C, Gil J, Tauler A, Ambrosio S. Histone deacetylase inhibition induces apoptosis and autophagy in human neuroblastoma cells. Cancer Lett 2012; 318:42-52; http://dx.doi.org/ 10.1016/j.canlet.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 2122.Micale L, Fusco C, Augello B, Napolitano LM, Dermitzakis ET, Meroni G, Merla G, Reymond A. Williams-Beuren syndrome TRIM50 encodes an E3 ubiquitin ligase. Eur J Hum Genet 2008; 16:1038-49; http://dx.doi.org/ 10.1038/ejhg.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2123.Fusco C, Micale L, Augello B, Mandriani B, Pellico MT, De Nittis P, Calcagni A, Monti M, Cozzolino F, Pucci P, et al.. HDAC6 mediates the acetylation of TRIM50. Cell Signal 2014; 26:363-9; http://dx.doi.org/ 10.1016/j.cellsig.2013.11.036. [DOI] [PubMed] [Google Scholar]
- 2124.Fusco C, Micale L, Egorov M, Monti M, D'Addetta EV, Augello B, Cozzolino F, Calcagni A, Fontana A, Polishchuk RS, et al.. The E3-ubiquitin ligase TRIM50 interacts with HDAC6 and p62, and promotes the sequestration and clearance of ubiquitinated proteins into the aggresome. PloS One 2012; 7:e40440; http://dx.doi.org/ 10.1371/journal.pone.0040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2125.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al.. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001; 294:1704-8; http://dx.doi.org/ 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 2126.Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, et al.. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol 2001; 306:717-26; http://dx.doi.org/ 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- 2127.Gatliff J, East D, Crosby J, Abeti R, Harvey R, Craigen W, Parker P, Campanella M. TSPO interacts with VDAC1 and triggers a ROS-mediated inhibition of mitochondrial quality control. Autophagy 2014; 10:2279-96; http://dx.doi.org/ 10.4161/15548627.2014.991665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2128.Geisler S, Vollmer S, Golombek S, Kahle PJ. UBE2N, UBE2L3 and UBE2D2/3 ubiquitin-conjugating enzymes are essential for parkin-dependent mitophagy. J Cell Sci 2014; 127:3280-93; http://dx.doi.org/ 10.1242/jcs.146035. [DOI] [PubMed] [Google Scholar]
- 2129.Fiesel FC, Moussaud-Lamodiere EL, Ando M, Springer W. A specific subset of E2 ubiquitin-conjugating enzymes regulate Parkin activation and mitophagy differently. J Cell Sci 2014; 127:3488-504; http://dx.doi.org/ 10.1242/jcs.147520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2130.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al.. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 2008; 134:668-78; http://dx.doi.org/ 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 2131.Muller M, Kotter P, Behrendt C, Walter E, Scheckhuber CQ, Entian KD, Reichert AS. Synthetic quantitative array technology identifies the Ubp3-Bre5 deubiquitinase complex as a negative regulator of mitophagy. Cell Rep 2015; 10:1215-25; http://dx.doi.org/ 10.1016/j.celrep.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 2132.N'Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 2009; 10:173-9; http://dx.doi.org/ 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2133.Chan EYW, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 2007; 282:25464-74; http://dx.doi.org/ 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 2134.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 2010; 22:132-9; http://dx.doi.org/ 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 2135.Dorsey FC, Rose KL, Coenen S, Prater SM, Cavett V, Cleveland JL, Caldwell-Busby J. Mapping the phosphorylation sites of Ulk1. J Proteome Res 2009; 8:5253-63; http://dx.doi.org/ 10.1021/pr900583m. [DOI] [PubMed] [Google Scholar]
- 2136.Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, Sue C, Gevaert K, De Strooper B, Verstreken P, et al.. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet 2014; 23:5227-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2137.Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 2014; 510:370-5. [DOI] [PubMed] [Google Scholar]
- 2138.Taillebourg E, Gregoire I, Viargues P, Jacomin AC, Thevenon D, Faure M, Fauvarque MO. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy 2012; 8:767-79; http://dx.doi.org/ 10.4161/auto.19381. [DOI] [PubMed] [Google Scholar]
- 2139.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 2006; 8:688-99; http://dx.doi.org/ 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 2140.Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, Kim DH. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell 2015; 57:207-18; http://dx.doi.org/ 10.1016/j.molcel.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2141.Munson MJ, Allen GF, Toth R, Campbell DG, Lucocq JM, Ganley IG. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2142.Pirooz SD, He S, Zhang T, Zhang X, Zhao Z, Oh S, O'Connell D, Khalilzadeh P, Amini-Bavil-Olyaee S, Farzan M, et al.. UVRAG is required for virus entry through combinatorial interaction with the class C-Vps complex and SNAREs. Proc Natl Acad Sci USA 2014; 111:2716-21; http://dx.doi.org/ 10.1073/pnas.1320629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2143.Kosta A, Roisin-Bouffay C, Luciani MF, Otto GP, Kessin RH, Golstein P. Autophagy gene disruption reveals a non-vacuolar cell death pathway in Dictyostelium. J Biol Chem 2004; 279:48404-9; http://dx.doi.org/ 10.1074/jbc.M408924200. [DOI] [PubMed] [Google Scholar]
- 2144.Oku M, Nishimura T, Hattori T, Ano Y, Yamashita S, Sakai Y. Role of Vac8 in formation of the vacuolar sequestering membrane during micropexophagy. Autophagy 2006; 2:272-9; http://dx.doi.org/ 10.4161/auto.3135. [DOI] [PubMed] [Google Scholar]
- 2145.Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev 1990; 54:266-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2146.Hoffman M, Chiang H-L. Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics 1996; 143:1555-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2147.Zhang C, Lee S, Peng Y, Bunker E, Giaime E, Shen J, Zhou Z, Liu X. PINK1 triggers autocatalytic activation of Parkin to specify cell fate decisions. Curr Biol 2014; 24:1854-65; http://dx.doi.org/ 10.1016/j.cub.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2148.Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol 1997; 138:517-29; http://dx.doi.org/ 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2149.Fader CM, Sanchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta 2009; 1793:1901-16; http://dx.doi.org/ 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 2150.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell 2011; 146:303-17; http://dx.doi.org/ 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2151.Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell 2010; 21:1001-10; http://dx.doi.org/ 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2152.Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol 2009; 187:875-88; http://dx.doi.org/ 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2153.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 2010; 6:217-27; http://dx.doi.org/ 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2154.Donohue E, Tovey A, Vogl AW, Arns S, Sternberg E, Young RN, Roberge M. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J Biol Chem 2011; 286:7290-300; http://dx.doi.org/ 10.1074/jbc.M110.139915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2155.Kaelin WG., Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 2008; 8:865-73; http://dx.doi.org/ 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 2156.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science 2005; 307:727-31; http://dx.doi.org/ 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 2157.Vaccaro MI, Ropolo A, Grasso D, Iovanna JL. A novel mammalian trans-membrane protein reveals an alternative initiation pathway for autophagy. Autophagy 2008; 4:388-90; http://dx.doi.org/ 10.4161/auto.5656. [DOI] [PubMed] [Google Scholar]
- 2158.Calvo-Garrido J, King JS, Munoz-Braceras S, Escalante R. Vmp1 regulates PtdIns3P signaling during autophagosome formation in Dictyostelium discoideum. Traffic 2014; 15:1235-46; http://dx.doi.org/ 10.1111/tra.12210. [DOI] [PubMed] [Google Scholar]
- 2159.Molejon MI, Ropolo A, Re AL, Boggio V, Vaccaro MI. The VMP1-Beclin 1 interaction regulates autophagy induction. Sci Rep 2013; 3:1055; http://dx.doi.org/ 10.1038/srep01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2160.Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J Cell Sci 2013; 126:1307-16; http://dx.doi.org/ 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 2161.Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol 2009; 21:543-51; http://dx.doi.org/ 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2162.Clancey LF, Beirl AJ, Linbo TH, Cooper CD. Maintenance of melanophore morphology and survival is cathepsin and vps11 dependent in zebrafish. PloS One 2013; 8:e65096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2163.Uttenweiler A, Schwarz H, Neumann H, Mayer A. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol Biol Cell 2007; 18:166-75; http://dx.doi.org/ 10.1091/mbc.E06-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2164.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci 2004; 117:4239-51; http://dx.doi.org/ 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 2165.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al.. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell 2010; 38:265-79; http://dx.doi.org/ 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2166.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, et al.. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 2010; 6:330-44; http://dx.doi.org/ 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 2167.Kast DJ, Zajac AL, Holzbaur EL, Ostap EM, Dominguez R. WHAMM Directs the Arp2/3 Complex to the ER for Autophagosome Biogenesis through an Actin Comet Tail Mechanism. Curr Biol 2015; 25:1791-7; http://dx.doi.org/ 10.1016/j.cub.2015.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2168.Haack TB, Hogarth P, Kruer MC, Gregory A, Wieland T, Schwarzmayr T, Graf E, Sanford L, Meyer E, Kara E, et al.. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am J Hum Genet 2012; 91:1144-9; http://dx.doi.org/ 10.1016/j.ajhg.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2169.Abidi A, Mignon-Ravix C, Cacciagli P, Girard N, Milh M, Villard L. Early-onset epileptic encephalopathy as the initial clinical presentation of WDR45 deletion in a male patient. Eur J Hum Genet 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2170.Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, Kasai-Yoshida E, Sawaura N, Nishida H, Hoshino A, et al.. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 2013; 45:445-9, 9e1; http://dx.doi.org/ 10.1038/ng.2562. [DOI] [PubMed] [Google Scholar]
- 2171.Biagosch CA, Hensler S, Kühn R, Meitinger T, Prokisch HT. ALEN-mediated mutagenesis as a tool to generate disease models for diseases caused by dominant de novo mutations. Eur J Hum Genet 2014; 22:153. [Google Scholar]
- 2172.Maiese K, Chong ZZ, Shang YC, Wang S. Targeting disease through novel pathways of apoptosis and autophagy. Expert Opin Ther Tar 2012; 16:1203-14; http://dx.doi.org/ 10.1517/14728222.2012.719499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2173.Petherick KJ, Williams AC, Lane JD, Ordonez-Moran P, Huelsken J, Collard TJ, Smartt HJ, Batson J, Malik K, Paraskeva C, et al.. Autolysosomal beta-catenin degradation regulates Wnt-autophagy-p62 crosstalk. EMBO J 2013; 32:1903-16; http://dx.doi.org/ 10.1038/emboj.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2174.Kaser A, Blumberg RS. Endoplasmic reticulum stress in the intestinal epithelium and inflammatory bowel disease. Semin Immunol 2009; 21:156-63; http://dx.doi.org/ 10.1016/j.smim.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2175.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005; 120:159-62. [DOI] [PubMed] [Google Scholar]
- 2176.Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D, Molgo J, Diaz J, Lavandero S, Harper F, et al.. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ 2007; 14:1029-39. [DOI] [PubMed] [Google Scholar]
- 2177.Kweon Y, Rothe A, Conibear E, Stevens TH. Ykt6p is a multifunctional yeast R-SNARE that is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell 2003; 14:1868-81; http://dx.doi.org/ 10.1091/mbc.E02-10-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2178.Cebollero E, van der Vaart A, Zhao M, Rieter E, Klionsky DJ, Helms JB, Reggiori F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol 2012; 22:1545-53; http://dx.doi.org/ 10.1016/j.cub.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2179.Cheng J, Fujita A, Yamamoto H, Tatematsu T, Kakuta S, Obara K, Ohsumi Y, Fujimoto T. Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat Commun 2014; 5:3207. [DOI] [PubMed] [Google Scholar]
- 2180.Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, Campellone KG, Heo WD, Gruenheid S, Meyer T, et al.. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 2011; 7:17-26; http://dx.doi.org/ 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2181.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 2010; 11:1246-61; http://dx.doi.org/ 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 2182.Pozuelo-Rubio M. Regulation of autophagic activity by 14-3-3zeta proteins associated with class III phosphatidylinositol-3-kinase. Cell Death Differ 2011; 18:479-92; http://dx.doi.org/ 10.1038/cdd.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2183.Vantaggiato C, Crimella C, Airoldi G, Polishchuk R, Bonato S, Brighina E, Scarlato M, Musumeci O, Toscano A, Martinuzzi A, et al.. Defective autophagy in spastizin mutated patients with hereditary spastic paraparesis type 15. Brain 2013; 136:3119-39; http://dx.doi.org/ 10.1093/brain/awt227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2184.Lin JF, Lin YC, Lin YH, Tsai TF, Chou KY, Chen HE, Hwang TI. Zoledronic acid induces autophagic cell death in human prostate cancer cells. J Urol 2011; 185:1490-6; http://dx.doi.org/ 10.1016/j.juro.2010.11.045. [DOI] [PubMed] [Google Scholar]
- 2185.Schneider EM, Lorezn M, Walther P. Autophagy as a hallmark of hemophagocytic diseases In: Gorbunov N, ed. Autophagy: Principles, Regulation and Roles in Disease: Nova Science Publishers, 2012. [Google Scholar]
- 2186.Ryhanen T, Hyttinen JM, Kopitz J, Rilla K, Kuusisto E, Mannermaa E, Viiri J, Holmberg CI, Immonen I, Meri S, et al.. Crosstalk between Hsp70 molecular chaperone, lysosomes and proteasomes in autophagy-mediated proteolysis in human retinal pigment epithelial cells. J Cell Mol Med 2009; 13:3616-31; http://dx.doi.org/ 10.1111/j.1582-4934.2008.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2187.Amadoro G, Corsetti V, Florenzano F, Atlante A, Bobba A, Nicolin V, Nori SL, Calissano P. Morphological and bioenergetic demands underlying the mitophagy in post-mitotic neurons: the pink-parkin pathway. Front Aging Neurosci 2014; 6:18; http://dx.doi.org/ 10.3389/fnagi.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]