ABSTRACT
In vertebrates, TFEB (transcription factor EB) and MITF (microphthalmia-associated transcription factor) family of basic Helix-Loop-Helix (bHLH) transcription factors regulates both lysosomal function and organ development. However, it is not clear whether these 2 processes are interconnected. Here, we show that Mitf, the single TFEB and MITF ortholog in Drosophila, controls expression of vacuolar-type H+-ATPase pump (V-ATPase) subunits. Remarkably, we also find that expression of Vha16-1 and Vha13, encoding 2 key components of V-ATPase, is patterned in the wing imaginal disc. In particular, Vha16-1 expression follows differentiation of proneural regions of the disc. These regions, which will form sensory organs in the adult, appear to possess a distinctive endolysosomal compartment and Notch (N) localization. Modulation of Mitf activity in the disc in vivo alters endolysosomal function and disrupts proneural patterning. Similar to our findings in Drosophila, in human breast epithelial cells we observe that impairment of the Vha16-1 human ortholog ATP6V0C changes the size and function of the endolysosomal compartment and that depletion of TFEB reduces ligand-independent N signaling activity. Our data suggest that lysosomal-associated functions regulated by the TFEB-V-ATPase axis might play a conserved role in shaping cell fate.
KEYWORDS: autophagy, lysosome, mitf, Notch signaling, patterning, SOP, TFEB, V-ATPase
Introduction
The V-ATPase is a conserved 15-subunit enzyme that pumps H+ across cell membranes.1 V-ATPase ensures acidification of the endolysosomal compartment, an essential process for cargo trafficking, sorting and degradation during endocytosis and autophagy. In addition, V-ATPase-mediated acidification of secretory vesicles is necessary for secretion and, in neurons, for neurotransmitter release. In gastric, renal, and osteoclast cells, plasma membrane V-ATPase acidifies the extracellular milieu, supporting specialized function in the respective organs.2 Recent evidence indicates that V-ATPase activity is central for the regulation of signaling pathways that control cell proliferation, patterning and survival. Indeed, we and others have demonstrated that V-ATPase is required for productive N and wingless (wg) signaling in Drosophila and mammals.3–10 However, it is unclear how V-ATPase activity might assist major signaling pathways that shape cell fate.
In vertebrates, TFEB, a member of the TFEB-MITF bHLH family of transcription factors, functions as a regulator of lysosomal biogenesis and autophagy in an axis with V-ATPase and MTOR that senses the nutritional status of the cell. 11–13 TFEB transcriptionally controls more than 400 lysosomal- and autophagy-related genes, including subunits of the V-ATPase by binding to specific E-box sequences (termed CLEAR sites) of target genes. 14,15 In mammals, the TFEB-MITF family encodes 4 members: TFEB, TFE3, TFEC and MITF. Interestingly, MITF has been shown to be essential for eye development and for development of specialized cell types, including osteoclasts, melanocytes and mast cells.16–18 Similar to TFEB, MITF and TFE3 transcriptionally regulate endolysosomal genes suggesting that the TFEB-MITF family might control organ development by regulating signaling in the endolysosomal system.19,20 Both MITF and V-ATPase have been implicated in a wide range of cancers but the functions that, when altered, contribute to tumorigenesis are currently obscure.21,22
A single ortholog of vertebrate TFEB-MITF transcription factors is encoded by the Drosophila genome.23 Overexpression of Drosophila Mitf in eye imaginal discs perturbs eye development, suggesting that the functions of the TFEB-MITF family in tissue patterning are evolutionarily conserved.24 Despite this, it is unknown whether Drosophila Mitf controls transcription of orthologs of TFEB target genes, including those encoding V-ATPase subunits, whether it controls endolysosomal biogenesis and autophagy and finally how it functions in regulation of tissue patterning.
Here, we show that Drosophila Mitf regulates lysosomal biogenesis and expression of multiple V-ATPase genes in vivo, indicating that Mitf is the Drosophila ortholog of vertebrate TFEB. Interestingly, we find that expression of Vha16-1 and Vha13, encoding 2 key subunits of V-ATPase follows N-dependent patterning, and that both are perturbed by Mitf misexpression in vivo. Experiments in human cells suggest that TFEB, V-ATPase and lysosomes are deeply connected with N signaling regulation. Our data indicate that the TFEB-V-ATPase axis might operate at the endolysosome as a conserved unit that supports N signaling during cell fate determination.
Results
Drosophila Mitf is the functional ortholog of vertebrate TFEB
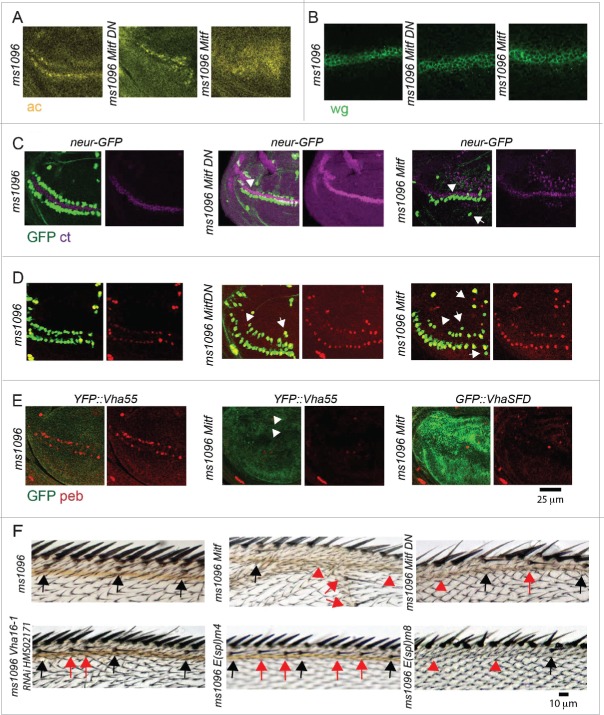
To explore whether Drosophila Mitf possesses functions of mammalian TFEB in vivo, we first characterized expression and function of endogenous and overexpressed Mitf in the wing imaginal disc of Drosophila melanogaster, an epithelial organ that will give rise to the adult thorax and wing (Fig. 1). By in situ hybridization, we observed that endogenous Mitf mRNA is expressed at low uniform level in wing disc tissue (Fig. 1A). This finding was consistent with expression of endogenous Mitf protein (Fig. 1B), using a specific antibody that we have generated (Fig. S1A; Material and Methods). Upon overexpression of both a functional Mitf and a dominant negative form that cannot bind DNA (Mitf DN)24 in the wing pouch with Bxms1096-Gal4 (ms1096; Fig. 1B, Fig. S1B and C), large cytoplasmic puncta and nuclear localization in a subset of cells were detected (Fig. 1C and D). Approximately half of the puncta were positive for YFP-Lamp1, which localizes to lysosomes,25 with a slight increase when Mitf is overexpressed (Fig. 1D, quantification in E). These data are consistent with the reported TFEB localization in mammalian cells.26,27
Figure 1.
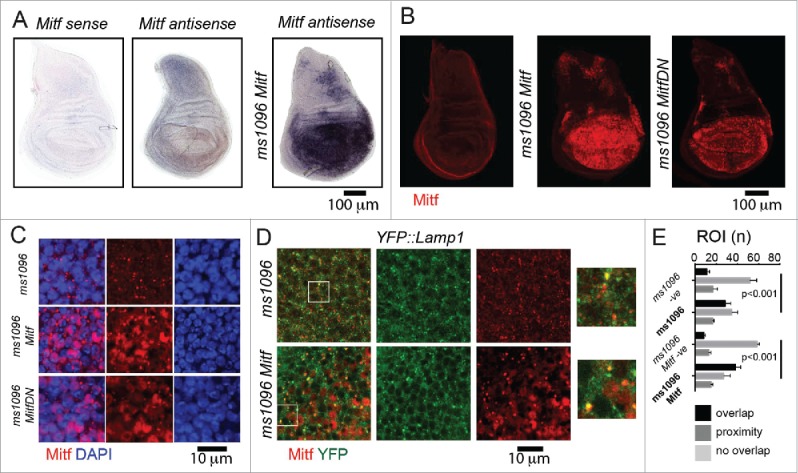
Mitf localization in wing imaginal discs. (A) In situ hybridization using labeled sense and antisense RNA probe for Mitf transcripts in wing discs from yellow white (control) animals and from animals overexpressing Mitf in wing disc (ms1096 Mitf). The sense probe has been used as a negative control. Dorsal is up, anterior to the left. All wing discs shown in figures are oriented as such. (B) Control wing disc and wing disc overexpressing Mitf or Mitf DN stained with anti- Drosophila Mitf antibody. (C and D) High magnifications of wing pouch tissue of the indicated genotype stained as indicated. Note that Mitf is present in the nucleus when overexpressed and in some lysosomes (close-up in D). (E) Quantification of colocalization of YFP:Lamp1 and Mitf. Colocalization across 80 regions of interest (ROI) per discs is shown as overlap. Proximity denotes nonoverlapping signal that falls within the ROI. Averages of 3 discs per genotype are graphed. -ve controls are obtained by rotating one of the 2 channels by 90 degrees.
To test whether Drosophila Mitf promotes activation of catabolic pathways, we labeled acidified lysosomes in wild-type and Mitf-overexpressing discs with the acidophilic dye LysoTracker Red (LTR). Compared to the control, Mitf overexpression increased the size of LTR-positive puncta, indicating that Mitf might control lysosomal biogenesis (Fig. 2A, quantification in B). To determine whether Mitf regulates autophagy, we labeled discs to detect ref(2)P (human SQSTM1/p62), and Atg8a (human MAP1LC3/LC3). Overexpression of Mitf led to a mild increase in the ref(2)P and Atg8a signal (Fig. 2C and D), relative to the basal low levels observed in control discs, suggesting that Mitf might affect autophagy. Finally, we find that overexpression of Mitf in the wing discs led to formation of a low number of apoptotic cells, as shown by expression of activated product of the gene Decay/caspase 3, which are normally not present in control discs (Fig. S1D).
Figure 2.
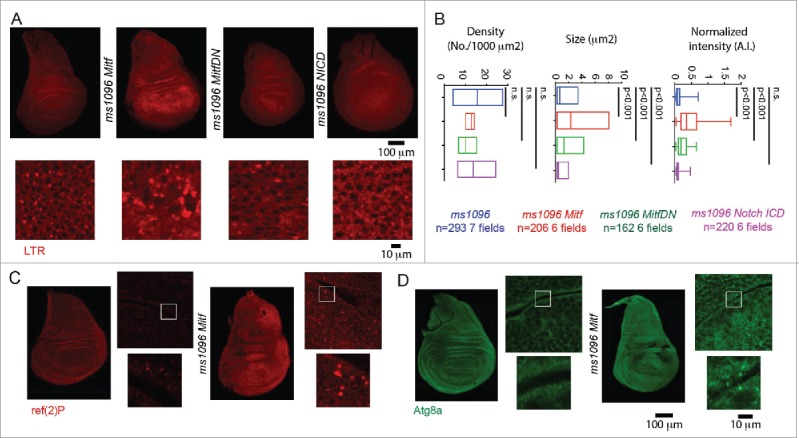
Mitf regulates lysosomal biogenesis. (A) Wing discs of the indicated genotypes subjected to incorporation of LTR. High magnifications of portions of the overexpressing tissue are shown below the discs. (B) Quantification of LTR puncta density, size and normalized intensity are graphed for the indicated samples. Overexpression of Mitf or Mitf DN increases LTR incorporation in wing disc cells, while overexpression of activated NICD slightly reduces it. (C and D) Immunolocalization of ref(2)P and Atg8a in discs of the indicated genotype. Side panels are higher magnifications of the overexpressing tissue with insets shown below them.
To assess whether Mitf acts as a master regulator of lysosomal gene expression in Drosophila, we took advantage of available GFP and YFP knock-in lines in the Drosophila orthologs of a subset of TFEB target genes (Fig. 3A). We used 3 lines with insertions in genes encoding components of the cytoplasmic V1 sector of V-ATPase: YFP::Vha55/V1B, GFP::VhaSFD/V1H, and GFP::Vha13/V1G. Two lines with insertions in genes encoding components of the membrane-embedded V0 sector: GFP::Vha16-1/V0c and GFP::VhaAC45/AC45 (see Fig. 3B for a schematic of the V-ATPase). Finally, we used YFP::Lamp1, tagging the single Drosophila gene whose product is the ortholog of mammalian Lysosomal-associated membrane protein 1 (LAMP1).25,28,29 Complementation analysis with existing mutants and deficiencies reveals that most knock-in lines in V-ATPase genes behaved as loss-of-function mutants (Table S1), but that all were viable and fertile in heterozygosis. The absence of dominant effects indicates that the exon traps can be used in heterozygosity to study regulation of expression in vivo.
Figure 3.
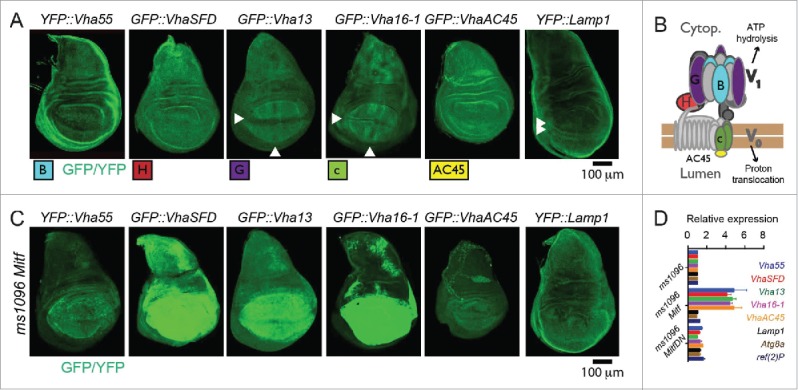
Mitf controls transcription of V-ATPase genes. (A) Pattern of expression in wing discs of the tagged lines for the indicated subset of Drosophila orthologs of TFEB target genes. Arrowheads point to areas with distinctive pattern of expression. (B) Schematics of the structure and function of V-ATPase with positioning of the subunits analyzed. (C) Pattern of expression in wing discs of the tagged lines for the indicated subset of Drosophila orthologs of TFEB target genes upon overexpression of Mitf. Note that V-ATPase subunit expression but not Lamp1 expression is controlled by Mitf. (D) Transcript levels of the indicated putative Mitf targets in wing discs by Q-PCR. RpL32 has been used as housekeeping control. The values represents means ± s.d of 2 independent experiments.
To investigate whether Drosophila Mitf regulates expression of putative target genes, we overexpressed Mitf in wing discs and assayed expression of the tagged lines. All displayed variable upregulation upon Mitf overexpression, with the exception of YFP::Lamp1 (Fig. 3C). Consistent with this, expression of the endogenous mRNA of the V-ATPase subunits analyzed was increased 4- to 5-fold upon Mitf overexpression, compared to the control or to expression of Mitf DN, when analyzed by Q-PCR. In contrast, expression of Lamp1, ref(2)P, or Atg8a, which are TFEB targets in mammalian cells14,15 was not upregulated (Fig. 3D). Upregulation of expression of V-ATPase subunits but not of ref(2)P, Atg8a or Lamp1 was consistent with the higher abundance of E-Boxes in conserved regions of the V-ATPase subunit genes analyzed, than in corresponding regions of ref(2)P, Atg8a or Lamp1 (Table S2). These data indicate that, similar to vertebrate orthologs, Drosophila Mitf regulates transcription of V-ATPase subunit genes.
Overall, in Drosophila epithelial tissue, Mitf localizes and acts similarly to vertebrate TFEB.
Expression of Vha16-1 in SOPs depends on Mitf and N signaling
Surprisingly, while expression of most knock-in lines appeared uniform in wing imaginal discs, expression of GFP::Vha16-1, GFP::Vha13 and YFP::Lamp1 appeared patterned (Fig. 3A), suggesting that function of the lysosome-associated proteins might be developmentally regulated in epithelial tissue. One exclusive feature of GFP::Vha16-1 expression, that is not displayed by GFP::Vha13, is elevated expression in proneural clusters (PNCs), as revealed by colocalization of GFP::Vha16-1 with the PNC marker ac (achaete) (Fig. 4A; Fig. S1E).30 The GFP::Vha16-1 signal is specific to Vha16-1 as the GFP-exon insertion tags Vha16-1 protein and did not alter significantly mRNA expression (Fig. S1F to H). Also, the GFP::Vha16-1 signal in PNCs is unlikely due to morphology differences between SOPs and surrounding cells or to the transmembrane nature of the protein. In fact, no pattern was observed in wing discs of GFP::CG8668 larvae (Fig. S1I), in which GFP is tagging the gene coding for the plasma membrane transmembrane glycosyl transferase CG8668/Resille.31 Finally, elevated expression of endogenous Vha16-1 mRNA could be observed by in situ hybridization (Fig. 4B, arrow).
Figure 4.
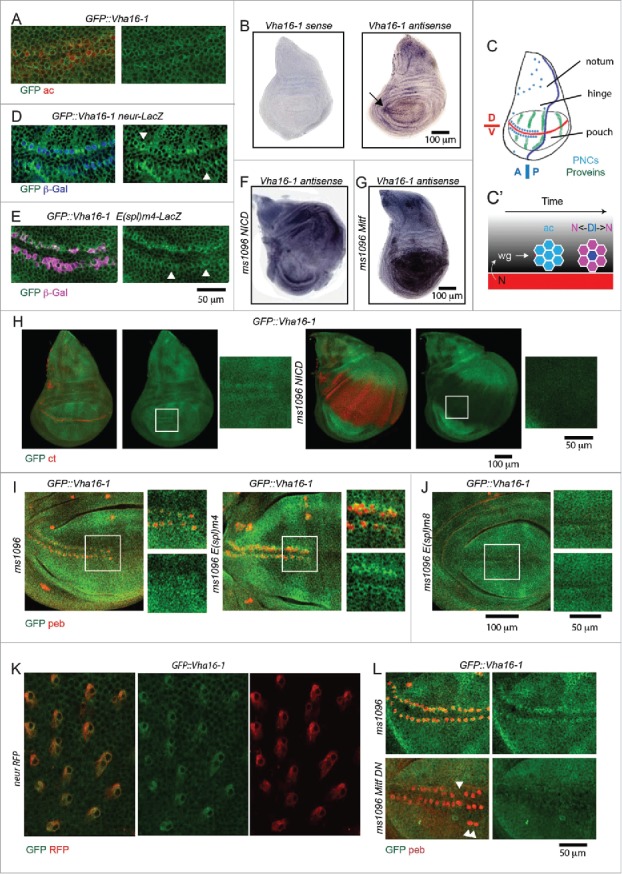
Vha16-1 expression is elevated in SOPs. (A) A high magnification of the anterior part of the wing pouch of wing discs of the indicated genotypes stained as indicated. Note that GFP::Vha16-1 expression is elevated in ac-expressing tissue. (B) In situ hybridization using labeled sense and antisense RNA probes for Vha16-1 transcripts in control wing discs. Note the high expression in 2 stripes of tissue abutting the anterior D/V boundary (arrow) when using the antisense probe. (C) Schematic representation of the patterning of a larval wing disc. The patterning features relevant for this study are indicated. Dl, N ligand Delta. (D and E) A high magnification of the anterior part of the wing pouch of wing discs of the indicated genotypes stained as indicated. Note that GFP::Vha16-1 expression is elevated in neur-LacZ-positive cell (D, arrowheads) and low in E(spl)m4 -positive cells (E, arrowheads). Arrowheads in E indicate examples of SOP cells that are not E(spl)m4-positive and have high GFP::Vha16-1 expression. (F and G) In situ hybridization using an antisense RNA probe for Vha16-1 transcripts of discs overexpressing NICD or Mitf. Note the very different transcriptional modulation of Vha16-1 expression upon overexpression. (H) Discs of the indicated genotype stained to detect the N target cut (ct). Note that GFP::Vha16-1 expression is very low and not patterned in ct -positive tissue. Insets enlarging corresponding areas of the pouch are show on the sides of each disc. (I and J) GFP::Vha16-1 wing disc overexpressing the N target E(Spl) genes m8, and m4 under ms1096-Gal4. Note that overexpression of E(Spl)m8 results in loss of sensory organs and of GFP::Vha16-1 expression at the anterior margin, while overexpression of E(Spl)m4 leads to formation of ectopic SOPs expressing GFP::Vha16-1. Insets enlarging corresponding areas of the pouch are show on the sides of each disc. (K) Pupal nota of the indicated genotype dissected 20 h after puparium formation. Note that elevated GFP::Vha16-1 expression is maintained along the SOP lineage. (L) High magnifications of the anterior part of the wing pouch of wing discs of the indicated genotypes stained as indicated. The image is a maximal projection of several sections. Note that overexpression of Mitf DN disrupts the pattern of GFP::Vha16-1 expression in SOPs and leads to missing and ectopic SOPs (arrowheads).
In larval wing discs, PNCs straddling the anterior dorsoventral (D/V) boundary are set by a N-instructed wg gradient and gradually develop into SOPs by the N-dependent lateral inhibition process (see Fig. 4C for a schematic of relevant disc regions, PNC patterning and SOP development at the D/V boundary).32-34 We found that when the SOPs are formed, elevation of GFP::Vha16-1 expression appeared restricted to cells positive for the SOPs marker neur-LacZ (Fig. 4D; Fig. S1E). Remarkably, expression of CD8-GFP, a membrane-tagged form of GFP, under the control of 2 independent Gal4 elements inserted in the 5′UTR of Vha16-1 (Vha16-1::Gal4; Fig. S1F) could be detected in variable subsets of SOPs (Fig. S1J and K). Despite the fact that expression of CD8-GFP only partially recapitulated the pattern of GFP::Vha16-1 expression, these data indicate that expression in SOPs is likely to be controlled by the Vha16-1 promoter. This, together with the fact that not all ac or neur-LacZ-positive cells are GFP::Vha16-1-positive and vice-versa (Fig. S1E), suggests that Vha16-1 expression in SOPs might be tightly temporarily regulated during SOP development.
Interestingly, expression of GFP::Vha16-1 was low in cells positive for E(spl)m4-lacZ (Fig. 4E), a N target that identifies non-SOP cells, in which N signaling is active, within differentiated PNC cluster.35 Consistent with this, at the dorso-ventral (D/V) wing margin and in intervein regions, which display high expression of the N signaling reporter E(spl)mβ-lacZ,35 GFP::Vha16-1 and GFP::Vha13 expression was at its lowest (Fig. 3A; S2A). Considering such inverse correlation, we assayed whether Vha16-1 expression was downregulated by activation of N signaling. To this end, we overexpressed N intracellular domain (NICD), an active form of N, in the wing pouch with Bxms1096-Gal4. By in situ hybridization, we observed that endogenous Vha16-1 expression in the wing pouch was reduced (Fig. 4F), compared to control discs (Fig. 4B), or discs overexpressing Mitf (Fig 4G). Similar results were obtained in GFP::Vha16-1 and GFP::Vha13-expressing discs, but not in YFP::Vha55, GFP::VhaSFD, GFP::VhaAC45 or GFP::CG8668-expressing discs, which display uniform expression (Fig. 4H, Fig. S2B to D), indicating that N could regulate expression of these 2 V-ATPase subunits. To test whether such regulation applies to N-mediated lateral inhibition, we generated ectopic wing margin SOPs by over-expression of the N target E(Spl)m4, which antagonizes N signaling activation, and we found expression of GFP::Vha16-1 in ectopic clusters marked with an antibody against the SOP marker pebbled (peb; also called hindsight or hnt) (Fig. 4I). In contrast, ectopic expression of E(Spl)m8, a N target that is known to enforce lateral inhibition, led to disappearance of SOPs differentiation and associated GFP::Vha16-1 expression (Fig. 4J). This evidence indicates that GFP::Vha16-1 expression follows N-dependent PNC differentiation. During pupal life, SOPs undergo N-dependent asymmetric cell divisions to generate the differentiated cells that compose the adult mechano-sensory organ.34 Interestingly, elevated GFP::Vha16-1 expression was present in the SOP lineage also during pupal development (Fig. 4K). Together with the evidence presented above, these data suggest that high levels of Vha16-1 might be crucial for correct SOP establishment and also for subsequent development of mechano-sensory organs.
Changes in V-ATPase subunit expression induced by N activation might correlate with lysosomal functionality. In fact, we have found that in NICD-overexpressing discs LTR puncta were slightly smaller, compared to control (Fig. 2A and B). However, this is unlikely due to control of Mitf by N signaling. Indeed, when we monitored expression of endogenous Mift upon overexpression of NICD by QPCR, we found no change (Fig. S2E). However, elevation of Vha16-1 expression in SOPs depends on Mitf. In fact, when we overexpressed Mitf DN, we found strongly reduced GFP::Vha16-1 expression in existing anterior boundary SOPs (Fig. 4L). We also found alteration in the stereotypic pattern of SOPs (Fig. 4L arrowheads), prompting us to evaluate the importance of Mitf in SOP differentiation.
Proneural development is supported by the TFEB-V-ATPase axis
To determine whether Mitf might influence the proneural differentiation cascade that leads to SOP formation, we overexpressed Mitf and Mitf DN in the wing pouch and assessed for possible alteration of patterning of the PNCs and SOPs straddling the anterior D/V boundary that will give rise to the mechano-sensory bristles of the adult wing margin. 34 We found perturbation of PNC patterning as revealed by broadening of expression of the PNC marker Ac, compared to control discs (Fig. 5A, Fig. S2F). This is unlikely to be due to changes in wg or N signaling because overexpression of Mitf did not change expression of wg and of the N target cut (ct) at the D/V margin, compared to control wing discs (Fig. 5B and C, Fig. S2F). To assess SOP differentiation, we analyzed discs expressing neur-GFP or stained for peb or ct, which marks sense organs and non-neuronal cells in the hinge and notum.36 Interestingly, misexpression of Mitf or Mitf-DN led to loss and ectopic peb-, neur- or ct-positive cells (Fig. 5C and D, Fig. S2F). Strikingly, in YFP::Vha55 or GFP::VhaSFD or GFP::Vha16-1 discs overexpressing Mitf, ectopic and normal peb-positive cells were not present. This is not the case in YFP::Vha55 discs, which showed a normal pattern (Fig. 5E). Thus, when high amounts of mutant tagged forms of V-ATPase components are present, the ability of Mitf to generate normal and ectopic SOPs observed in overexpressing discs is prevented, suggesting that Mitf requires V-ATPase to support SOP development. Consistent with this, the wing margin of adult animals displayed missing or ectopic mechano-sensory bristles, similar to animals in which the activity of N target genes of the E(Spl) cluster was modulated (Fig. 5F). To test whether this is the case also upon impairment of Vha16-1, we used in vivo RNAi (see Fig. S1F for details). Expression of a Vha16-1 RNAi hairpin in the whole wing pouch led to specific reduction of endogenous Vha16-1 mRNA expression and of GFP expression in GFP::Vha16-1 wing disc, indicating that the RNAi line is on target (Fig. S2G and H). Expression of Vha16-1 RNAi also led to the formation of an adult wing margin with ectopic SOPs (Fig. 5F), indicating that the Vha16-1 is involved in correct SOP development.
Figure 5.
Misexpression of Mitf perturbs SOP development. (A and D) High magnification of the anterior part of the wing disc pouch of the indicated genotype stained as indicated. Note that Mitf and Mitf DN overexpression results in perturbation of the expression of ac protein (A), no perturbation of wg or ct expression at the D/V boundary (B and C), formation of misplaced or ectopic neur-GFP- ct -, (C) and neur-GFP- peb - (D) positive cells. Some of the ectopic ct and peb -positive cells are negative for neur-GFP and could represent incomplete SOP commitment. (E) Presence of normal and ectopic peb -positive cells is reduced to different extent in YFP::Vha55 discs overexpressing Mitf, when compared to YFP::GFP discs not overexpressing Mitf. A similar lack of peb -positive cells is observed in GFP::VhaSFD discs overexpressing Mitf. Note that the genetic null tagged forms of these genes are highly expressed, due to induction by Mitf. (F) High magnification of the antero-distal dorsal area of the margin of adult wings of the indicated genotypes. The stereotypic position of sensory margin bristles is shown by black arrows. Expression of the indicated constructs in wing discs results in loss (red arrowheads) or misplacement of sensory bristles (red arrows).
Interestingly, expression of Vha16-1 RNAi in the pupal notum with pannier-Gal4 (pnr) led to a decrease in size of the adult thorax, which is formed by the fusion of the left and right nota. This phenotype is coupled with depigmentation and misorientation of bristles, a known effect in Drosophila of reduced V-ATPase and lysosomal activity6,37 (Fig. 6A). Importantly, the density of microchaeta, which derive from pupal SOPs, was also increased independent of thorax size (Fig. 6B), suggesting that reduction of Vha16-1 might weaken N -mediated lateral inhibition during pupal life. These effects are specific to depletion of Vha16-1, as they were rescued by concomitant overexpression of RNAi-resistant Vha16-1 tagged with HA (Vha16-1-HA, see Fig. S1F for details; Fig. 6A and B). Similar results were obtained by downregulating VhaPPA1-1, the gene encoding the component of the membrane-embedded V0 sector c” (Fig. S2I), as previously reported.38 However, Vha16-1 is not sufficient to promote ectopic PNC formation. In fact, compared to controls, overexpression of Vha16-1-HA per se in the wing pouch or notum did not perturb microcheta formation (Fig. 6B), suggesting that the patterning activity of Vha16-1 requires additional factors. Overall, these data indicate that Mitf and components of the V-ATPase might be functional elements of the proneural patterning machinery in wing discs epithelia.
Figure 6.
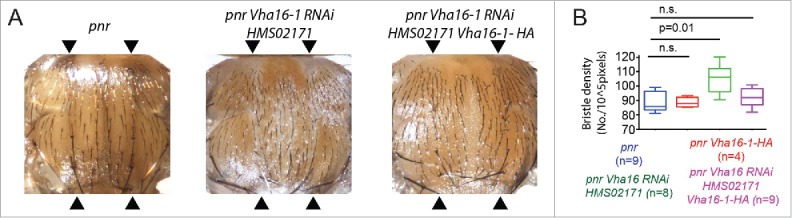
Vha16-1 is required for in pupal SOPs differentiation. (A) Phenotypic defects associated with RNAi-mediated knockdown of Vha16-1 expression with the indicated RNAi line using the driver pannier-Gal4 (pnr) compared to control. Defects are rescued by concomitant expression of the RNAi lines and a RNAi-resistant Vha16-1-HA construct. The domain of pannier-Gal4 expression is delimited by arrowheads. (B) Quantification of the number of bristle/area (bristle density) relative to the experiment as in A. Statistical analysis is based on Kruskal Wallis Test with Dunn multiple comparison relative to control.
PNCs possess a distinctive lysosomal compartment
Is the function of V-ATPase and Mitf in proneural development linked to regulation of the endolysosomal system? To assess this, we tested whether PNCs possess an endolysosomal compartment that is different to that of surrounding cells. Consistent with observations in Figs. 1 and 2, expression of YFP::Lamp1 was mildly upregulated in PNC regions abutting the D/V margin, whereas we did not detect significant differences in endogenous expression or localization of Mitf across the wing pouch (Fig. 7A). To further assess lysosome abundance, we labeled acidified compartments in the disc with LTR. We found that LTR incorporation was high in PNCs compared to other epithelial cells of the disc, suggesting that PNCs might possess more lysosomes than surrounding cells (Fig. 7B). Upon ubiquitous expression of GFP-hLamp1 in the disc with actin-GAL4, we found that PNCs were more GFP-positive than surrounding cells (Fig. 7C). GFP-hLamp1 is a lysosome-anchored GFP form that has been developed as a sensor for lysosomal degradation, because the GFP is exposed to the lysosomal lumen, where it is degraded by resident hydrolases. In conditions of impaired lysosome function, GFP-hLamp1 is less degraded leading to a high GFP signal.9 Thus, PNC cells possess lysosomes with less degradative capacity than surrounding cells. In contrast, localization of Syx7 (Syntaxin7), a marker of early endosomes,40 was uniform across the disc tissue (Fig. 7D). Consistent with previous evidence indicating transcriptional down-regulation of N in PNCs,41,42 we found that overall N protein levels in the endolysosomal system of PNCs were lower than in the rest of the disc (Fig. 7D). We next determined N stability in the endolysosomal system of PNCs and pro-vein cells. To this end, we analyzed expression of Ni-GFP4-Cherry5, a functional N form tagged with fast-maturing, pH-sensitive GFP and a slow-maturing, pH-insensitive mCherry. It has been recently reported that the GFP signal of such N form indicates the newly synthetized N found at the plasma membrane, while the mCherry signal highlights old N molecules that reach the endolysosomal compartment on their way to degradation.43 Using this sensor, we found that the amount of mCherry-positive N in the endolysosomal compartment was higher in the PNCs than in surrounding cells (Fig. 7E). Overall, these data indicate that PNCs might possess an expanded, less degradative and more N -rich lysosomal system than surrounding cells.
Figure 7.
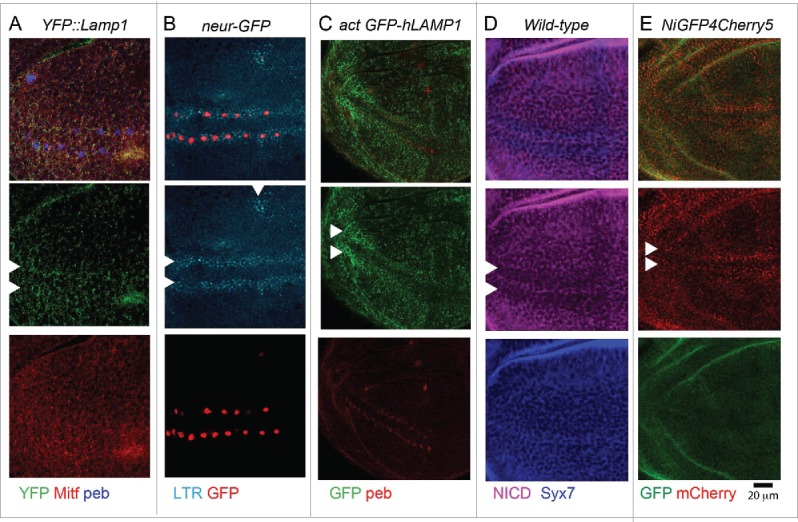
The endolysosomal system at sites of PNC development is distinctive. (A and E) High magnification of the anterior part of the wing pouch of wing discs of the indicated genotypes, stained as indicated. Arrowheads point to the approximate location of PNC. Note that compared to surrounding epithelial cells, PNC cells show a slightly higher amount of YFP::Lamp1-positive lysosomes (A), a higher number of acidified organelles (B) and of GFP-hLAMP1 puncta (C), overall less N (D) and more endolysosomal N (E).
Endolysosomal regulation by TFEB-V-ATPase in human epithelial cells
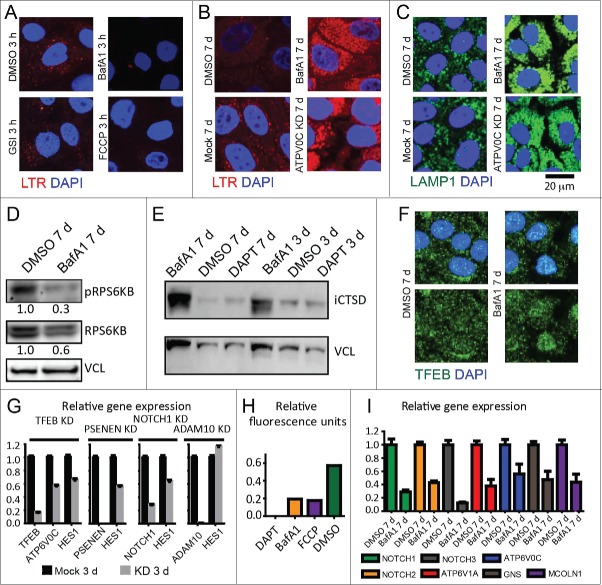
We and others previously reported that V-ATPase sustains N signaling activity in Drosophila epithelial tissue.9,10 Recently, we have shown that mild reduction of V-ATPase activity affects N signaling also in human MCF10A breast epithelial cells in culture, which endogenously express N receptors,7 suggesting that MCF10A cells could be a relevant model to study the impact of the TFEB-V-ATPase axis on functionality of the endolysosomal system and on N signaling activation. Acute treatment of MCF10A cells with the V-ATPase inhibitor bafilomycin A1 (BafA1) or with FCCP, which dissipates lysosomal acidification without impairing V-ATPase activity,44 led to strong reduction in LTR incorporation when compared to mock-treated controls or cells treated with the γ-secretase inhibitor (GSI) N-[2S-(3,5-difluorophenyl)acetyl]-L-alanyl-2-phenyl-1,1-dimethylethyl ester (DAPT), that blocks N signaling activation (Fig. 8A). In sheer contrast, chronic reduction of V-ATPase activity by treatment with a very low dose of BafA1 (3 nM) over 7 d, a regimen that did not prevent MCF10A growth (Fig. S2J),7 caused expansion of the lysosomal compartment, as revealed by LTR incorporation or LAMP1 staining. Similar effects were found 7 d after siRNA-mediated downregulation of ATP6V0C/V-ATPase subunit c expression (Fig. 8B and C). In these conditions, phosporylation of RPS6KB/S6K and maturation of CTSD (cathepsin D), which are measures of MTOR signaling and lysosomal activity, respectively,13,45 were impaired (Fig. 8D and E). Consistent with the described compensatory feedback loop between TFEB and V-ATPase activity,27 the effects described above correlated with increased localization of TFEB in the nucleus (Fig. 8F). These data indicate that in MCF10A cells lysosomal compartment size and function are inversely correlated by the activity of the TFEB-V-ATPase axis.
Figure 8.
The TFEB-V-ATPase axis regulates lysosomal function and basal N signaling in human cells. (A) LTR assay indicates that acute treatment with both BafA1 and FCCP, which dissipates lysosomal pH independently of V-ATPase, impairs acidification in MCF10A cells. (B and C) Chronic inhibition of V-ATPase or KD of ATP6V0C in contrast causes an expansion of the acidified (B) and LAMP1-positive lysosomal organelles (C). (D) Relative to the control, western blot analysis of MTOR signaling of lysate from MCF10A cells treated as indicated reveals a reduction in the level of phosphorylated RPS6KB (pS6K), a measure of active MTOR signaling. (E) Western blot analysis of MCF10A lysates treated as indicated shows that chronic inhibition of V-ATPase leads to accumulation of the 52 and 44 KDa immature CTSD forms (iCTSD), a sign of impaired lysosomal function. (F) Chronic V-ATPase inhibition results in high levels of nuclear TFEB compared to control. (G) siRNA against TFEB reduce the level of TFEB, ATP6V0C and HES1 mRNA expression compared to control. A similar reduction in the level of N signaling is observed after knocking down the expression of the γ-secretase component, PSENEN or of the NOTCH1 receptor. In contrast, efficient knockdown of ADAM10, which is dispensable for ligand-independent signaling, does not reduce HES1 levels. (H) Chronic treatment with both BafA1 and FCCP compromises γ-secretase substrate cleavage. (I) Chronic inhibition of the V-ATPase reduces the levels of expression of N receptors among other genes.
To test whether in MCF10A cells V-ATPase might sustain N signaling via TFEB, we reduced TFEB expression by RNAi. We found that TFEB knockdown led to reduced expression of ATP6V0C, the human ortholog of Vha16-1 (Fig. 8G). In addition, TFEB knockdown decreased basal N signaling activity by 50%, a reduction that is similar to knockdown of the γ-secretase component PSENEN or of NOTCH1, the main N receptor expressed by MCF10A7 (Fig. 8G). The observed reduction in N activity was most likely ligand-independent, as no reduction was observed by efficient knockdown of ADAM10, which is required for ligand-dependent N activity (Fig. 8G).46 These data indicate that in MCF10A cells, TFEB is required to sustain basal levels of ligand-independent N signaling activity, which has been proposed to originate in the endolysosomal system in Drosophila.47
To assess how N signaling activation in MCF10A cells could be regulated by changes in the endolysosomal system, we assayed activity of γ-secretase, the enzyme that catalyzes the final cleavage of the N receptor during proteolytic activation. To this end, we treated cells with the BafA1 or with the proton ionophore carbonyl cyanide-p-trifluoreomethoxyphenylhydrazone (FCCP). We then measured γ-secretase activity on a cellular fraction containing endosomes and lysosomes and observed that upon BafA1 or FCCP treatment, γ-secretase activity was reduced to an intermediate level, when compared to the mock-treated control, or to cells treated with γ-secretase inhibitor DAPT (Fig. 8H). In addition, we analyzed expression of N receptors. Compared to mock-treated cells, we observed a reduction in the expression levels of N receptors and of lysosomal genes, but not reductions of GADPH, a housekeeping gene (Fig. 8I; S2K). These data indicate that reduced N signaling activity in cells with low V-ATPase activity might be due to reduced γ-secretase cleavage or to N expression or both. Overall, these data suggest that regulation of lysosomal activity by the TFEB-V-ATPase axis might sustain N signaling.
Discussion
In this study, we report that Drosophila Mitf regulates lysosomal biogenesis and expression of subunits of the V-ATPase pump. In epithelial tissue, we find that Mitf resides in lysosomes and, when overexpressed, in the nucleus, where it is transcriptionally active. These observations are in accordance with findings in mice and C. elegans in which TFEB (HLH-30 in C. elegans) moves to the nucleus to induce lysosomal biogenesis and autophagy 11,20,48,49 and reveal for the first time that Drosophila Mitf is the functional ortholog of mammalian TFEB. Interestingly, we show that, differently from mammalian cells, Drosophila Lamp1, Atg8a and ref(2)P are not modulated by Mitf in wing discs and that their promoters do not contain as many and as conserved E-Boxes as V-ATPase subunits. In addition, we find only slight changes in the protein level of Atg8a and ref(2)P in the wing tissue in overexpressing discs. This evidence suggests that the set of Mitf-TFEB target genes in Drosophila might be limited compared to other metazoans and mostly restricted to V-ATPase subunit genes.
Interestingly, in wing discs we observed a diverse expression pattern of Lamp1 and of Vha16-1 and Vha13 encoding 2 subunits of the V-ATPase, which are transcriptionally controlled by Mitf. This indicates that key components of lysosomes are developmentally regulated and follow patterning of disc during development. Mis-expression experiments in vivo also revealed that Mitf might act downstream of developmental signaling to regulate expression of V-ATPase subunits and proneural differentiation. Mitf is unlikely to crosstalk to or interfere with bHLH factors involved in PNC development, such as E(Spl) because patterning perturbations are observed also by overexpression of Mitf DN, which is unable to bind DNA.24 Thus, our findings strongly suggest that the developmental and lysosomal biogenesis function of the TFEB-MITF bHLH family of transcription factors coexist in a single gene in Drosophila and might in large part coincide. Further experiments will be needed to address the molecular nature of the interplay between Mitf and V-ATPase subunit genes with factors involved in proneural development.
A complication to the scenario proposed above is that the expression pattern of Vha55, VhaSFD and VhaAC45 is uniform in the discs and unchanged by modulation of developmental signaling. Vha16-1 encodes subunit V0c that, together with the product of VhaPPA1-1, constitute the membrane-embedded proteo-lipidic ring, the rotor of V-ATPase, while VhaAC45 encodes a subunit that has been recently suggested to cap the proteo-lipidic ring on the lumenal side.50,51 Vha13 encodes subunit V1G that forms with V0a and V1E the 3 stalks that make up the stator of V-ATPase. Finally, Vha55/V1B encodes part of the ATPase motor (peripheral membrane V1domain), while VhaSFD/V1H, that in yeast prevents ATPase activity of the V1 catalytic domain when not assembled on the V0 proteolipid-containing proton channel.50–52 Indeed, the motor is thought to be removed upon low nutrition states in both yeast and insects,53,54 although recent in vivo yeast experiments and in vitro experiments in mammals suggest a more subtle rearrangement rather than complete separation, with the only release of Vha44/V1C, a known regulator of nutrient-mediated coupling of the V1 and V0 sector.55 Thus, it is not clear whether the differences in expression that we observe reflect a particular part of the pump, nutrients coupling or moonlighting functions of single subunits. Interestingly, we have recently shown that forced expression of Drosophila Vha44, encoding V1C, results in a sharp increase of GFP::Vha16-1 and decrease in GFP::VhaSFD in wing discs,56 indicating that pump functionality in vivo involves complex and currently unclear regulation of subunit expression and/or turnover. Despite this, the common aspect of Vha16-1 and Vha13 patterned expression, and the similar phenotypes of downregulation of Vha16-1 and VhaPPA1-1 and the ability of mutant Vha55 and VhaSFD to interfere with the effect of Mitf overexpression on SOP development, all suggest that pump activity, rather than moonlighting functions of the single subunits, might be the developmentally important function. In contrast, the strong elevation of expression in SOPs, which is exclusively observed for Vha16-1 and which is maintained during pupal life, could hint to additional moonlighting functions of Vha16-1 that might not involve the V-ATPase pump. Concerning this, the proteo-lipid ring is thought to assist membrane-fusion processes,57,58 while Vha16-1 has been reported to be part of the gap junctions.59,60
A possible reason for modulated V-ATPase subunit expression might be the necessity to change functionality of endolysosomal compartment of differentiating cells. In mammals, cell differentiation from monocyte to macrophages results in a large expansion of the lysosomal compartment and increased V-ATPase expression.61 in vivo and in human cells, we observed differences in the distribution and functionality of endolysosomal compartments in the PNC regions that in part correlate with changes in expression of V-ATPase components. Differences in lysosomal compartment distribution and activity could be a mode of biasing signaling by altering endocytic trafficking, a major route of signaling regulation.62 We propose that one of the signaling pathways most impacted by regulation of lysosomal function might be N signaling. This evidence is in accordance with previous reports in Drosophila showing that V-ATPase activity is required for N signaling activation and for bristle specification.9,10,38 Several studies have demonstrated that N activation is exquisitely sensitive to endolysosomal events.47,63–66 Thus, differences in V-ATPase expression and Mitf activity could channel N into specific compartments for either N receptor stabilization and signaling activation, or to accelerate receptor degradation, ultimately reducing signaling. Such scenario is likely to apply specifically to ligand-independent N signaling activity that has been shown to enhance N signaling activation in different Drosophila epithelial organs including follicle cells, blood cells and wing imaginal discs.47,64,67 Recent work has also demonstrated the existence of alternative routes of N activation in mammalian cells.68 In line with this, our experiments in human cells indicate that rising of the pH, either by reducing V-ATPase activity or by dissipating pH irrespective of V-ATPase, results in reduced basal N activation correlating with reduced γ-secretase activity. In support of this, evidence suggests that pH might also control γ-secretase efficiency, a process that could boost signaling activation at early steps of PNC development.69 Overexpression in Drosophila wing imaginal discs of Trpml/Mucolipin, a lysosomal calcium channel that is a target of TFEB,14 strongly enhances ligand-independent activation of Notch, which is calcium-sensitive.47,70 Interestingly, lysosomal calcium regulation has been recently implicated in regulation of TFEB activity in mammalian cells.71 Thus, it is possible that ligand-independent basal activation of N might be an integral part of TFEB regulatory loops. Together with modulation of N signaling, we also observe changes in expression of NOTCH and other genes that might indirectly result from the TFEB-mediated transcriptional regulation associated with rearrangement of the endolysosomal compartment. Whether N phenotypes are dependent on MTOR, TORC1 and nutrient metabolism, which in mammalian cells operate with TFEB at the endolysosome,13,72 remains to be assessed.
In summary, we propose a model (Fig. 9) for early step of PNCs development in which Mitf and components of V-ATPase might be important to set the correct level of N signaling activity. Although it requires further testing, such model integrates the developmental and lysosomal functions of the TFEB-Mift family of bHLH transcription factors and might provide a framework for our understanding of misregulation of the TFEB-V-ATPase axis in cancer.
Figure 9.
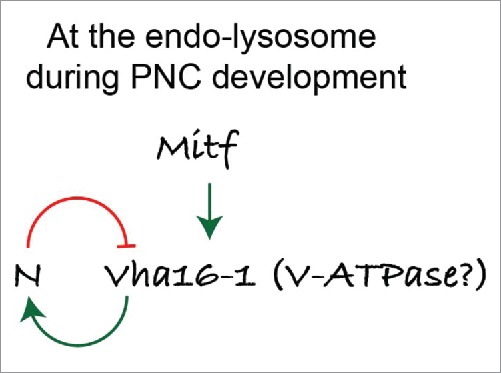
Proposed role of lysosomes, and of the TFEB-V-ATPase axis in patterning. A model for the activity of Mitf and V-ATPase in PNC regions during Drosophila wing disc development. [Change to “N” or “N/Notch.”]
Materials and methods
Genetics
The GFP trap lines GFP::Vha16-1 (G00007), GFP::VhaAC45 (ZCL0366), GFP::Vha13(CA07644), YFP::Vha55 (CPTI100063), GFP::VhaSFD (G00259) were obtained from large-scale random transposon insertion projects.28,29 The YFP::Lamp1 insertion line CPTI001775 is from the Drosophila Genomics and Genetic Resources (DGGR, Kyoto, Japan). 25 GFP::CG8668 (117-2) was a gift of J. Zallen (Memorial Sloan Kettering Cancer Center, New York, NY USA). The G/YFP cassette, which is inserted within the gene of interest, behaves as an extra exon and undergoes splicing and translation to generate a chimeric protein. Lines obtained from the Bloomington Drosophila Stock Center (BDSC) are: UAS N RNAi 14E (7078); UAS Vha16-1 RNAi (40923); neur-LacZ (4369); UASmCD8GFP (5137); pannier-Gal4 (3039); ms1096-Gal4 (8860); neur-Gal4 A101(6393); Df(2R)BSC326 (24351); Df(2R)ED1791 (9063); Df(3R)ED6025 (8964) P{lacW}Vha55j2E9 (12128). P{EPgy2}VhaSFD EY04644 (15758). The Vha16-1-Gal4 lines NP5271 and NP3437 were obtained from from DGGR. UAS VhaPPA1-1 RNAi (v47188) is from the Vienna Drosophila Rnai Center (VDRC, Vienna, Austria). UAS NICD was a gift of M. Fortini (Thomas Jefferson University, Philadelphia, PA USA). E(spl)mβLacZ, and E(spl)m4-LacZ were gifts from E. Lai (Memorial Sloan Kettering Cancer Center, New York, NY USA). UAS Mitf and UAS MitfEA (DN) were gifts of F. Pignoni (SUNY Upstate Medical University, Syracuse, NY USA). neur-GFP and NiGFP4mCherry543 were gifts of Francois Schweisguth, ActGal4, GFP-hLamp1 was a gift from Helmut Kramer (UT Southwestern, Dallas, TX USA). UAS E(spl)m8, and m4 were gifts of C. Delidakis (IMBB, Heraklion, Greece). Misexpression in either larval or adult tissues was achieved using the Gal4-UAS system.73 Genotypes of the experiments presented in the figures are listed in Table S3.
Generation of transgenic Drosophila lines
For the generation of the UAS Vha16-1-HA fly strain, we cloned the Vha16-1 gene using the following primers: forward 5′ GATCGAATTCATGTCTTCTGAAGTGAGCAG 3′ and reverse 5′ GATCTCTAGATTAGGCGTAGTCAGGCACGTCGTAAGGATATTTCGTGTACAGGTAAATGGC 3′. The HA tag was inserted at the C terminus of the protein. The amplicon of Vha16-1-HA was inserted into the pUAST expression vector and injected into the ZH-86fb landing site (Basler lab). Transgenesis was performed by Genetic Services, Inc.
Genomic DNA extraction
Files were homogenized with a pestle in lysis buffer (100 mM Tris-HCl, pH 7.5, 100 mM EDTA, 100 mM NaCl, 0.5% SDS [Sigma, T3253, E9884, S9888, L4522, respectively]) and proteinase K (Sigma, P2308). After incubation for 30 min at 70°C, a solution composed of 1 part 5 M KAc and 2.5 parts 6 M LiCL was added to the mix (Sigma, P1190, L0505). Isopropanol and 70% ethanol (Sigma, 190764, 459844) were then added to allow DNA precipitation. DNA was resuspended in autoclaved water and 1 μl of genomic DNA was used as a template for PCR reactions.
Production of a polyclonal antibody against the first 4 exons of the Drosophila Mitf
A polypeptide that lacks the basic helix-loop-helix leucine zipper domains was selected as immunogen to prevent cross-reactions with other bHLH-Zip proteins. A fragment of the whole Mitf cDNA was amplified by PCR using the Taq Polymerase (Promega, M3175), using a template genomic DNA of flies carrying the construct UAS Mitf24 and the following primers: BamHI-1st exon Mitf: 5′ GATCGGATCCATGACGGAATCTGGAATCG 3′; SalI-4th exon Mitf: 5′ GATCGTCGACTTACGAATGATGGTAGCTCAGAGAC 3′. The PCR product was inserted into the prokaryotic expression vector pGEX, containing the GST sequence (pGEX-GST), using BamHI and SALI sites. Expression was carried out in the E. coli rosetta (Millipore, 70956) adding IPTG 0.5 mM overnight at 18°C. Purification was performed by the IFOM antibody-service facility according to standard protocols. Purified peptides were consigned to Eurogentech for rabbit immunizations. Sera affinity purification was performed by the IFOM antibody-service facility, using AminoLink® Kit (Pierce Biotechnology, 44890).
Immunohistochemistry
Drosophila immunostainings were performed as described previously.74 For immunostaining experiments in human cells, MCF10A cells were fixed in 4% PFA at room temperature (RT) and permeabilized in 0.1% Triton X-100 (Sigma, T8787) diluted in phosphate-buffered saline (PBS 1X; Sigma, P5493). They were then incubated in 3% BSA (Sigma, A9418) dissolved in PBS 1X blocking solution for 30 min and incubated at RT in primary antibody diluted in blocking solution for 1 h followed by incubation at RT with secondary antibody diluted in PBS 1X for 1 h. The following Drosophila primary antibodies for the following antigens were used: Mitf (Rabbit 1:200); GFP (chicken 1:1000; Abcam ab13970); ref(2)P (rabbit 1:100; kind gift of Tor Erik Rusten, Oslo University, Oslo, Norway), cleaved Decay/caspase 3 (rabbit 1:200; Cell Signaling Technology, 9661), Atg8a (rat 1:300; a kind gift of G. Juhasz, Eotvos Lorand University, Budapest, Hungary), rabbit Syx7 (rabbit 1:500).40 N (mouse 1:100; Developmental Studies Hybridoma Bank [DSHB], C17.9C6), peb (mouse, 1:40; DSHB, 1G9-s), β–Gal (mouse, 1:25; DSHB, 401a-c), ct (mouse, 1:100; DSHB, 2B10), wg (mouse, 1:100; DSHB, 4D4-s), ac (mouse, 1:50; DSHB, anti-achaete). The following antibodies for human cells were used: anti-LAMP1 (rabbit, 1:300; Sigma, L1418) and anti-human TFEB (mouse, 1:250; Bethyl Laboratories, A303-673A). Alexa Fluor 488- or Alexa Fluor 647- (Life Technologies, A-21202 [mouse], A-21206 [rabbit], A-21203 [rat], A-31571 [mouse], A-11039 [chicken]) and Cy3-conjugated secondary antibodies (Jackson Immunoresearch Laboratories, 715-165-150 [mouse], 711-165-154 [rabbit], 712-165-150 [rat]) were used. Phalloidin-TRITC (Sigma, P1951) has been used to mark F-actin. All images shown are confocal sections taken with TCS microscope (Leica, Heidelberg, Germany) using 20x/NA 0.5, 40x/NA 1.25 or 63x/NA 1.4 oil-immersion lenses. Digital images were processed and assembled using ImageJ, Photoshop and Illustrator with minimal manipulations. All images are single confocal sections unless otherwise stated.
In situ experiments
Single strand sense and antisense RNA probes for Vha16-1 and Mitf were generated using the following primers:
T7-Vha16-1_fwd: 5′ TAATACGACTCACTATAGGGAGAGTGTCAAGCCAATGAGCAAC 3′
T3_Vha16-1_rev: 5′ AATTAACCCTCACTAAAGGGAGAAGTTGGTTTCCGCAGTTGAC 3′
T7_Mitf_fwd: 5′ TAATACGACTCACTATAGGGAGAGTGCTGCAGGTCAGTACAGTG 3′
T3_Mitf_rev: 5′AATTAACCCTCACTAAAGGGAGAGG-CGAAATAGGAGCTGAGG 3′
As templates we used a plasmid carrying the Vha16-1 cDNA (pFLC-I RH30178) or the genomic DNA from UAS Mitf flies.
PCR product has been used as a template for in vitro transcription with the T3/T7 polymerase (Promega, P207B/P208C) according to manufacturer's instructions.
In situ experiments were performed in wing imaginal discs. One probe lacking the most conserved basic HLH-Zip (bHLH-Zip) domain was used against Mitf mRNAs. This probe has been designed to prevent cross-reactions with other mRNAs encoding bHLH-Zip proteins. One probe used for Vha16-1 was designed in the 4th exon. The probes were labeled with digoxigenin-UTP (Roche 11209256910) and expression patterns revealed using anti-digoxigenin antibodies conjugated to alkaline phosphatase (Roche, 11093274910). Alkaline phosphatase activity was revealed using the NBT and BCIP substrates (Roche, 11383213001 and 11383221001).
LTR assay
The LTR assay was performed by adding LTR (DND-99; Life Technologies, L-7528) directly into the culture medium of cells or to M3 medium (Shields and Sang M3 insect medium (Sigma, S3652) of ex vivo wing discs after dissection. Cells or wing discs were incubated for 30 and 5 min respectively in medium containing 1 μM LTR. They were then rinsed twice with PBS1X and mounted immediately in antifade-glycerol 1:1 solution for confocal examination. Where indicated, MCF10A cells were pretreated for with 3 nM BafA1 (Sigma, B1793), 3 μM DAPT (Sigma, D5942), 1 μM FCCP (Sigma, C2920) or as DMSO (Euroclone, EMR385250) for the indicated periods or the indicated genes had been knocked down for 7 d prior to the LTR assay.
Quantifications
To quantify colocalization of YFP::Lamp1 with Mitf, wing imaginal discs were immunostained using anti-GFP and anti-Mitf antibodies. Images were recorded as optical z-sections. Three sets of z-stack of 20 sections each with step size of 0.24 μm were analyzed using imageJ for each sample. Clear and distinct YFP::Lamp1-positive compartments were selected blindly with no other channel visible using a region of interest (ROI) with a surface area of 4 μm2. Each YFP::Lamp1 compartment was subsequently manually assessed for colocalization with Mitf. 240 ROIs were analyzed for each sample in total and score for “overlap,” “no overlap” or “proximity” colocalization of the 2 signals.
For quantification of LTR analysis, optical sections corresponding to 6 fields of 3 independent wing discs for each sample were analyzed. Particles with a discrete dimension size that ranged from 5 to infinity pixels were then considered. For each particle the following measurements were performed: number of puncta, area of puncta, integrated density density of puncta (sum of the pixel intensities divided by the number of pixels).
To count bristles, adult torax microchaete of female's nota were considered. The area of each notum was measured and the density of bristles calculated by dividing number of bristles by the area.
Cell culture and siRNA knockdown of human cells
MCF10A cells were cultured in DMEM/F12 (1:1; Invitrogen, 31331-093) supplemented with 5% horse serum (Invitrogen, 16050-122), 10 μg/ml insulin (Roche, 11376497001), 0.5 μg/ml hydrocortisone (Sigma, H0888-1G), 100 ng/ml cholera toxin (Sigma, C8052-2mg) and freshly added 20 ng/ml EGF (Vinci Biochem, BPS-90201-3).
To knock down genes, siRNA against the desired genes were transfected into MCF10A cells using lipofectamine RNAi-Max transfection reagent (Invitrogen, 13778-150) remove following the manufacturer's instructions. After transfection, the cells were placed under normal cell culture conditions for the indicated time. Then, samples were processed for RNA or protein extraction or immunofluorescence. siRNA duplexes against all genes were purchased from Dharmacon (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Q-PCR assays
Wing imaginal discs (40 discs per sample) were dissected in Shields and Sang M3 insect medium (Sigma, S3652) and total RNA was extracted using TRIZOL Reagent (invitrogen, 15596-026) and RNAase Mini kit (Qiagen, 74104). In human cells, 72 h after siRNA transfection, the cells were harvested for the Q-PCR assay by scraping them in 1 ml of ice cold DPBS (Thermo Fisher Scientific). They were then pelleted by centrifugation at maximum speed for 5 min. The RNA was extracted from cells using the RNAeasy mini kit following the manufacturer's instruction. cDNA from both wing discs and human cells was generated from 1 μg of RNA using the SuperScript VILO cDNA Synthesis kit (Invitrogen, 11754050), according to manufacturer's protocol. 5 ng of cDNA was amplified (in triplicate). RT-PCR was carried out on the ABI/Prism 7900 HT Sequence Detector System (Applied Biosystems, Carlsbad, CA, USA), using a pre-PCR step of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C.
The following primers (5′-3′) were designed from Universal Probe Library Roche (UPL):
Vha16-1 fwd 5′cacaacaacaacagatagacaaacg 3′ Vha16-1 rev 5′gaagctgctgctgatgttgat 3′
Vha55 fwd 5′atcgctgtcgcgtttgat 3′ Vha55 rev 5′agagtggtccttacgggtcat 3′
VhaSFD fwd 5′aggtgctgaagcagctatcc 3′ VhaSFD rev 5′ctctacgtcggcggtaatgt 3′
Vha13 fwd 5′ aggagttcgaggccaagc 3′ Vha13 rev 5′ccaggatgaacgggtcct 3′
VhaAC45 fwd 5′ccctgtttgtgaccttcgag 3′ VhaAC45 rev 5′cactcgaactgcttgctgat 3′
Lamp1 fwd 5′ gctttcctttatgcaaattcatc 3′ Lamp1 rev 5′gctgaaccgtttgattttcc 3′
Ref(2)P fwd 5′ agacagagcccctgaatcct 3′ Ref(2)P rev 5′ggcgtctttcctgctctgt 3′
ATG8a fwd 5′ catgggctccctgtacca 3′ Atg8a rev 5′ctcatcggagtaggcaatgt 3′
RPL32_fwd: 5′cggatcgatatgctaagctgt 3′ RPL32 rev 5′ cgacgcactctgttgtcg 3′
To assay Mift expression, the following Applied Biosystem (Applied Biosystems, Carlsbad, CA, USA) probes were used: Mitf: Dm02749950_m1 RPL32: Dm02151827_g1. Amplicon expression in each sample was normalized to the mRNA content of the housekeeping gene RpL32-RA. Note that ms1096-Gal4 is expressed in approximately 50% of the disc tissue. The following Applied Biosystem probes for human genes were used: HES1: Hs00172878_m1, NOTCH1: Hs00413187-m1, NOTCH2: Hs00225747-m1, NOTCH3: Hs00166432_m1, GADPH: Hs99999905_m1, ATP6V0C: Hs00798308_sH, ATP6V1A: Hs01097169_m1, GNS: hs00157741_m1, MCOLN1: hs00220937_m1, PSENEN: Hs00708570_s1, TFEB: hs01065085_m1 and ADAM10: Hs00153853_m1. The reactions were performed by the IFOM-Q-PCR service facility.
Western blots
Wing imaginal discs (50 discs per sample) were dissected in M3 medium, then lysed in RIPA buffer freshly supplemented with protease inhibitors (Calbiochem, 539134). MCF10A cells were treated by adding 3 nM BafA1 or DMSO as negative control directly into the medium. The cells were placed in normal culture conditions for the indicated time before harvesting for western blot. Cell pellets were resuspended in appropriate volumes of RIPA buffer freshly supplemented with protease inhibitor cocktail set III (Calbiochem, 539134). The cell lysate was then cleared by centrifugation at full speed at 4°C for 30 min. The supernatants were recovered and quantified by standard methods. Samples from both wing discs or cells were denatured by adding β-mercaptoethanol containing loading dye and by heating for 5 min at 98°C. They were then resolved on 8 – 12% polyacrylamide gels. After blotting onto nitrocellulose membranes, the membranes were stained with rabbit anti-Mitf diluted at 1:1000; anti-CTSD/cathepsin D (goat, 1:500; Santa Cruz Biotechnology, sc-6486); anti-phospho-RPS6KB/pS6K (Thr389; rabbit, 1:1000; Cell Signaling Technology, 9205); anti-RPS6KB/p70 S6 Kinase (rabbit, 1:1000; Cell Signaling Technology, 2708). Normalization of cell extracts was performed with mouse anti-αTub84B (1:10,000; Sigma, T6074) anti-LAMP1 (rabbit, 1:300; Sigma, L1418) or mouse-anti VCL/Vinculin (1:5000; Sigma, V4139). Goat anti-rabbit, goat anti-mouse (1:8000) HRP-conjugated secondary antibodies (GE Healthcare, NA934 and NXA931) were used. Signal was detected using Pierce ECL Western Blotting Substrate (Thermo Scientific, 34080–34095), and imaged using a Chemidoc molecular imager (Bio-Rad, Hercules CA USA).
Gamma-secretase assay
To perform the γ-secretase assay, 80% confluent MCF10A cells were pretreated for 3 h with 3 nM BafA1, 1 μM FCCP, DMSO as negative control or with 3 μM DAPT as a positive control (Sigma B1793, C2920, D2650, D5942, respectively). The treated cells were then scraped in 1 ml of ice cold fractionation buffer (1 mM EGTA, 50 mM sucrose, 20 mM HEPES, pH 7.4) supplemented with freshly added 5 mM glucose, protease inhibitor cocktail at 1:200, and the respective compounds used for pretreatment (All from Sigma, St. Louis, MI USA). Cells were then homogenized by passing through a 23 G needle 5 times. The homogenate was then centrifuged at 2000 g at 4°C to remove the nuclei, unbroken cells and large cellular debris. The post-nuclear supernatant was collected and centrifuged at maximum speed at 4°C for 20 min to pellet the light endomembranes which were then resuspended in 1 ml of fractionation buffer containing the respective compounds and prewarmed at 37°C. 8 μM γ-secretase fluorogenic substrate (Calbiochem, 565764) was then added to each light endomembrane suspension and mixed well. The γ-secretase fluorogenic substrate is internally quenched and only fluoresces when cleaved by γ-secretase. The light endomembrane suspensions were then transferred into black 96 multi-well plates (Corning, New York, NY USA) in triplicate (200 μL per well) and incubated at 37°C for 12 h before reading fluorescence. Fluorescence measurements were taken using a Wallac 1420 VICTOR plate reader (Perkin Elmer, Waltham, MA USA).
Sequence analysis
Genome alignments (maf files) for the genomes of D. melanogaster (dm6), D. simulans (droSim1), D. yakuba (droYak1), D. erecta (droEre2), D. pseudoobscura (droPse3), D. miranda (droMir2), D. ananassae (droAna3), D. virilis (droVir3), D. grimshawi (droGri2) and D. mojavensis (droMoj3) were obtained from the UCSC genome database. Maf files were processed and realigned according to the phylCRM algorithm's genome preparation75 and aligned regions in the range −2000 to +2000 from the transcription start site were extracted for analysis.
Extracted genomic regions were searched for occurrences of potential Mitf sites according to the rules presented for the core CANNTG binding76 and for the base specificities in the first flaking sites 5′ and 3′ of the E-boxes.77
Statistical analysis
Mitf colocalization data were subjected to a Chi-square test, with Bonferroni correction for multiple comparison analyses.
Bristle data and LTR data were subjected a Kruskal Wallis Test with the Dunn multiple comparison relative to control. All analyses were performed with imageJ and statistically analyzed and graphed with GraphPad Prism.
Supplementary Material
Abbreviations
- ac
Achaete
- Atg8a
autophagy-related 8a
- BafA1
bafilomycin A1
- bHLH
basic helix-loop-helix
- DN
dominant negative
- E(spl)
enhancer of split
- GFP
green fluorescent protein
- Lamp1
lysosomal-associated membrane protein 1
- LTR
LysoTracker Red
- MITF
microphthalmia-associated transcription factor
- N
Notch
- NICD
N intracellular domain
- peb
pebbled/hindsight
- PNC
proneural cluster
- ref(2)P
refractory to sigma P
- S2
Schneider-2
- RNAi
RNA interference
- ROI
region of interest
- RT
room temperature
- SOP
sensory organ precursor
- TFEB
transcription factor EB
- V-ATPase
vacuolar-type H+- ATPase
- WT
wild-type
- wg
wingless
- YFP
yellow fluorescent protein
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank G. Ossolengo for technical help.
Funding
Work in the Vaccari lab is supported by the AIRC Investigator Grant #15954, by the Telethon Grant GPP13225 and by the Cariplo Foundation.
References
- [1].Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 2007; 8:917-29; PMID:17912264; http://dx.doi.org/ 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- [2].Hinton A, Bond S, Forgac M. V-ATPase functions in normal and disease processes. Pflugers Arch 2009; 457:589-98; PMID:18026982; http://dx.doi.org/ 10.1007/s00424-007-0382-4 [DOI] [PubMed] [Google Scholar]
- [3].Buechling T, Bartscherer K, Ohkawara B, Chaudhary V, Spirohn K, Niehrs C, Boutros M. Wnt/Frizzled signaling requires dPRR, the Drosophila homolog of the prorenin receptor. Curr Biol 2010; 20:1263-8; PMID:20579883; http://dx.doi.org/ 10.1016/j.cub.2010.05.028 [DOI] [PubMed] [Google Scholar]
- [4].Cruciat C-M, Ohkawara B, Acebron SP, Karaulanov E, Reinhard C, Ingelfinger D, Boutros M, Niehrs C. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science 2010; 327:459-63; PMID:20093472; http://dx.doi.org/ 10.1126/science.1179802 [DOI] [PubMed] [Google Scholar]
- [5].Gleixner EM, Canaud G, Hermle T, Guida MC, Kretz O, Helmstädter M, Huber TB, Eimer S, Terzi F, Simons M. V-ATPase/mTOR signaling regulates megalin-mediated apical endocytosis. Cell Rep 2014; 8:10-9; PMID:24953654; http://dx.doi.org/ 10.1016/j.celrep.2014.05.035 [DOI] [PubMed] [Google Scholar]
- [6].Hermle T, Saltukoglu D, Grünewald J, Walz G, Simons M, Gru J, Division R. Regulation of Frizzled-dependent planar polarity signaling by a V-ATPase subunit. Curr Biol 2010; 20:1269-76; PMID:20579879; http://dx.doi.org/ 10.1016/j.cub.2010.05.057 [DOI] [PubMed] [Google Scholar]
- [7].Kobia F, Duchi S, Deflorian G, Vaccari T. Pharmacologic inhibition of vacuolar H+ ATPase reduces physiologic and oncogenic Notch signaling. Mol Oncol 2014; 8:207-20; PMID:24309677; http://dx.doi.org/ 10.1016/j.molonc.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lange C, Prenninger S, Knuckles P, Taylor V, Levin M, Calegari F. The H(+) vacuolar ATPase maintains neural stem cells in the developing mouse cortex. Stem Cells Dev 2011; 20:843-50; PMID:21126173; http://dx.doi.org/ 10.1089/scd.2010.0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vaccari T, Duchi S, Cortese K, Tacchetti C, Bilder D. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development 2010; 137:1825-32; PMID:20460366; http://dx.doi.org/ 10.1242/dev.045484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yan Y, Denef N, Schüpbach T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev Cell 2009; 17:387-402; PMID:19758563; http://dx.doi.org/ 10.1016/j.devcel.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; PMID:21617040; http://dx.doi.org/ 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423; http://dx.doi.org/ 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334:678-83; PMID:22053050; http://dx.doi.org/ 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al.. A gene network regulating lysosomal biogenesis and function. Science 2009; 325:473-7; PMID:19556463 [DOI] [PubMed] [Google Scholar]
- [15].Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 2011; 20:3852-66; PMID:21752829; http://dx.doi.org/ 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- [16].Hemesath TJ, Steingrímsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994; 8:2770-80; PMID:7958932; http://dx.doi.org/ 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- [17].Hodgkinson CA, Moore KJ, Nakayama A, Steingrímsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 1993; 74:395-404; PMID:8343963; http://dx.doi.org/ 10.1016/0092-8674(93)90429-T [DOI] [PubMed] [Google Scholar]
- [18].Hughes MJ, Lingrel JB, Krakowsky JM, Anderson KP. A helix-loop-helix transcription factor-like gene is located at the mi locus. J Biol Chem 1993; 268:20687-90; PMID:8407885 [PubMed] [Google Scholar]
- [19].Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen H-W, Graeber TG, von Euw E, Ribas A, De Robertis EM. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci U S A 2015; 112:E420-9; PMID:25605940; http://dx.doi.org/ 10.1073/pnas.1424576112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Martina JA, Diab HI, Li H, Puertollano R. Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cell Mol Life Sci 2014; 71:2483-97; PMID:24477476; http://dx.doi.org/ 10.1007/s00018-014-1565-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martinez-Zaguilan R, Lynch RM, Martinez GM, Gillies RJ. Vacuolar-type H(+)-ATPases are functionally expressed in plasma membranes of human tumor cells. Am J Physiol Cell Physiol 1993; 265:C1015-29 [DOI] [PubMed] [Google Scholar]
- [22].Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res 2010; 23:27-40; PMID:19995375; http://dx.doi.org/ 10.1111/j.1755-148X.2009.00653.x [DOI] [PubMed] [Google Scholar]
- [23].Hallsson JH, Haflidadóttir BS, Schepsky A, Arnheiter H, Steingrímsson E. Evolutionary sequence comparison of the Mitf gene reveals novel conserved domains. Pigment Cell Res 2007; 20:185-200; PMID:17516926; http://dx.doi.org/ 10.1111/j.1600-0749.2007.00373.x [DOI] [PubMed] [Google Scholar]
- [24].Hallsson JH, Haflidadóttir BS, Stivers C, Odenwald W, Arnheiter H, Pignoni F, Steingrímsson E. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 2004; 167:233-41; PMID:15166150; http://dx.doi.org/ 10.1534/genetics.167.1.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Takáts S, Nagy P, Varga Á, Pircs K, Kárpáti M, Varga K, Kovács AL, Hegedűs K, Juhász G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol 2013; 201:531-9; PMID:Can't; http://dx.doi.org/ 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012; 8:903-14; PMID:22576015; http://dx.doi.org/ 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al.. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095-108; PMID:22343943; http://dx.doi.org/ 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al.. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 2007; 175:1505-31; PMID:17194782; http://dx.doi.org/ 10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A 2001; 98:15050-5; PMID:11742088; http://dx.doi.org/ 10.1073/pnas.261408198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cubas P, de Celis JF, Campuzano S, Modolell J. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev 1991; 5:996-1008; PMID:2044965; http://dx.doi.org/ 10.1101/gad.5.6.996 [DOI] [PubMed] [Google Scholar]
- [31].Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell 2006; 11:459-70; PMID:17011486; http://dx.doi.org/ 10.1016/j.devcel.2006.09.007 [DOI] [PubMed] [Google Scholar]
- [32].Diaz-Benjumea FJ, Cohen SM. Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development 1995; 121:4215-25; PMID:8575321 [DOI] [PubMed] [Google Scholar]
- [33].Rulifson EJ, Blair SS. Notch regulates wingless expression and is not required for reception of the paracrine wingless signal during wing margin neurogenesis in Drosophila. Development 1995; 121:2813-24; PMID:7555709 [DOI] [PubMed] [Google Scholar]
- [34].Hartenstein V, Posakony JW. A dual function of the Notch gene in Drosophila sensillum development. Dev Biol 1990; 142:13-30; PMID:2227090; http://dx.doi.org/ 10.1016/0012-1606(90)90147-B [DOI] [PubMed] [Google Scholar]
- [35].Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev 1995; 9:2609-22; PMID:7590239; http://dx.doi.org/ 10.1101/gad.9.21.2609 [DOI] [PubMed] [Google Scholar]
- [36].Blochlinger K, Jan LY, Jan YN. Transformation of sensory organ identity by ectopic expression of Cut in Drosophila. Genes Dev 1991; 5:1124-35; PMID:1676691; http://dx.doi.org/ 10.1101/gad.5.7.1124 [DOI] [PubMed] [Google Scholar]
- [37].Akbar MA, Ray S, Kramer H. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol Biol Cell 2009; 20:1705-14; PMID:19158398; http://dx.doi.org/ 10.1091/mbc.E08-03-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 2009; 458:987-92; PMID:19363474; http://dx.doi.org/ 10.1038/nature07936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pulipparacharuvil S, Akbar MA, Ray S, Sevrioukov EA, Haberman AS, Rohrer J, Krämer H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci 2005; 118:3663-73; PMID:16046475; http://dx.doi.org/ 10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- [40].Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol 2005; 7:1132-9; http://dx.doi.org/ 10.1038/ncb1324 [DOI] [PubMed] [Google Scholar]
- [41].Bray SJ. Expression and function of Enhancer of split bHLH proteins during Drosophila neurogenesis. Perspect Dev Neurobiol 1997; 4:313-23; PMID:9171445 [PubMed] [Google Scholar]
- [42].De Celis JF, Garcia-Bellido A, Bray SJ. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development 1996; 122:359-69; PMID:8565848 [DOI] [PubMed] [Google Scholar]
- [43].Couturier L, Trylinski M, Mazouni K, Darnet L, Schweisguth F. A fluorescent tagging approach in Drosophila reveals late endosomal trafficking of Notch and Sanpodo. J Cell Biol 2014; 207:351-63; PMID:25365996; http://dx.doi.org/ 10.1083/jcb.201407071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Steinberg BE, Huynh KK, Brodovitch A, Jabs S, Stauber T, Jentsch TJ, Grinstein S. A cation counterflux supports lysosomal acidification. J Cell Biol 2010; 189:1171-86; PMID:20566682; http://dx.doi.org/ 10.1083/jcb.200911083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gieselmann V, Pohlmann R, Hasilik A, Von Figura K. Biosynthesis and transport of cathepsin D in cultured human fibroblasts. J Cell Biol 1983; 97:1-5; PMID:6863385; http://dx.doi.org/ 10.1083/jcb.97.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Groot AJ, Cobzaru C, Weber S, Saftig P, Blobel CP, Kopan R, Vooijs M, Franzke C-W. Epidermal ADAM17 is dispensable for notch activation. J Invest Dermatol 2013; 133:2286-8; PMID:23657465; http://dx.doi.org/ 10.1038/jid.2013.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shimizu H, Woodcock SA, Wilkin MB, Trubenová B, Monk NAM, Baron M. Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell 2014; 157:1160-74; PMID:24855951; http://dx.doi.org/ 10.1016/j.cell.2014.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu C-C, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al.. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 2013; 4:2267; PMID:23925298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 2013; 15:668-76; PMID:Can't; http://dx.doi.org/ 10.1038/ncb2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Allan AK, Du J, Davies SA, Dow JAT. Genome-wide survey of V-ATPase genes in Drosophila reveals a conserved renal phenotype for lethal alleles. Physiol Genomics 2005; 22:128-38; PMID:15855386; http://dx.doi.org/ 10.1152/physiolgenomics.00233.2004 [DOI] [PubMed] [Google Scholar]
- [51].Rawson S, Phillips C, Huss M, Tiburcy F, Wieczorek H, Trinick J, Harrison MA, Muench SP. Structure of the Vacuolar H(+)-ATPase Rotary Motor Reveals New Mechanistic Insights. Structure 2015; 23:461-71; PMID:25661654; http://dx.doi.org/ 10.1016/j.str.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Parra KJ, Keenan KL, Kane PM. The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J Biol Chem 2000; 275:21761-7; PMID:10781598; http://dx.doi.org/ 10.1074/jbc.M002305200 [DOI] [PubMed] [Google Scholar]
- [53].Kane PM. Disassembly and Reassembly of the Yeast Vacuolar H+-ATPase in Vivo. J Biol Chem 1995; 270:17025-32; PMID:7622524 [PubMed] [Google Scholar]
- [54].Sumner JP, Dow JA, Earley FG, Klein U, Jäger D, Wieczorek H. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J Biol Chem 1995; 270:5649-53; PMID:7890686; http://dx.doi.org/ 10.1074/jbc.270.10.5649 [DOI] [PubMed] [Google Scholar]
- [55].Tabke K, Albertmelcher A, Vitavska O, Huss M, Schmitz H-P, Wieczorek H. Reversible disassembly of the yeast V-ATPase revisited under in vivo conditions. Biochem J 2014; 462:185-97; PMID:24805887; http://dx.doi.org/ 10.1042/BJ20131293 [DOI] [PubMed] [Google Scholar]
- [56].Petzoldt AG, Gleixner EM, Fumagalli A, Vaccari T, Simons M. Elevated expression of the V-ATPase C subunit triggers JNK-dependent cell invasion and overgrowth in a Drosophila epithelium. Dis Model Mech 2013; 6:689-700; PMID:23335205; http://dx.doi.org/ 10.1242/dmm.010660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Strasser B, Iwaszkiewicz J, Michielin O, Mayer A. The V-ATPase proteolipid cylinder promotes the lipid-mixing stage of SNARE-dependent fusion of yeast vacuoles. EMBO J 2011; 30:4126-41; PMID:21934648; http://dx.doi.org/ 10.1038/emboj.2011.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liégeois S, Benedetto A, Garnier J-M, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol 2006; 173:949-61; PMID:16785323; http://dx.doi.org/7983150 10.1083/jcb.200511072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Finbow ME, Goodwin SF, Meagher L, Lane NJ, Keen J, Findlay JB, Kaiser K. Evidence that the 16 kDa proteolipid (subunit c) of the vacuolar H(+)-ATPase and ductin from gap junctions are the same polypeptide in Drosophila and Manduca: molecular cloning of the Vha16k gene from Drosophila. J Cell Sci 1994; 107 (Pt 7:1817-24; PMID:7983150 [DOI] [PubMed] [Google Scholar]
- [60].Dunlop J, Jones PC, Finbow ME. Membrane insertion and assembly of ductin: a polytopic channel with dual orientations. EMBO J 1995; 14:3609-16; PMID:7641680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lee BS, Underhill DN, Crane MK, Gluck SL. Transcriptional Regulation of the Vacuolar H[+]-ATPase B2 Subunit Gene in Differentiating THP-1 Cells. J Biol Chem 1995; 270:7320-9; PMID:7706273; http://dx.doi.org/ 10.1074/jbc.270.13.7320 [DOI] [PubMed] [Google Scholar]
- [62].Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, Di Fiore PP. Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol Rev 2012; 92:273-366; PMID:22298658; http://dx.doi.org/ 10.1152/physrev.00005.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, Baron M, Matsuno K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 2004; 131:5527-37; PMID:15496440; http://dx.doi.org/ 10.1242/dev.01448 [DOI] [PubMed] [Google Scholar]
- [64].Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual beta-arrestin. Nat Cell Biol 2005; 7:1191-201; PMID:16284625; http://dx.doi.org/ 10.1038/ncb1327 [DOI] [PubMed] [Google Scholar]
- [65].Thompson BJ, Mathieu J, Sung H-H, Loeser E, Rørth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell 2005; 9:711-20; PMID:16256745; http://dx.doi.org/ 10.1016/j.devcel.2005.09.020 [DOI] [PubMed] [Google Scholar]
- [66].Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell 2005; 9:687-98; PMID:16256743; http://dx.doi.org/ 10.1016/j.devcel.2005.09.019 [DOI] [PubMed] [Google Scholar]
- [67].Palmer WH, Jia D, Deng W-M. Cis-interactions between Notch and its ligands block ligand-independent Notch activity. Elife 2014 8:3. PMID:25486593; http://dx.doi.org/23171551 10.7554/eLife.04415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zheng L, Saunders CA, Sorensen EB, Waxmonsky NC, Conner SD. Notch signaling from the endosome requires a conserved dileucine motif. Mol Biol Cell 2013; 24:297-307; PMID:23171551; http://dx.doi.org/ 10.1091/mbc.E12-02-0081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Pasternak SH, Bagshaw RD, Guiral M, Zhang S, Ackerley CA, Pak BJ, Callahan JW, Mahuran DJ. Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J Biol Chem 2003; 278:26687-94; PMID:12736250; http://dx.doi.org/ 10.1074/jbc.M304009200 [DOI] [PubMed] [Google Scholar]
- [70].Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, Sklar J, Aster JC. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol 2000; 20:1825-35; PMID:10669757; http://dx.doi.org/ 10.1128/MCB.20.5.1825-1835.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al.. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 2015; 17:288-99; PMID:25720963; http://dx.doi.org/ 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci 2013; 126:1713-9; PMID:23641065; http://dx.doi.org/ 10.1242/jcs.125773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 2002; 34:1-15; PMID:12324939; http://dx.doi.org/ 10.1002/gene.10150 [DOI] [PubMed] [Google Scholar]
- [74].Tognon E, Vaccari T. Notch Signaling - Methods Mol Biol 2014; 1187:63-78. PMID:25053481; http://dx.doi.org/ 10.1007/978-1-4939-1139-4_5 [DOI] [PubMed] [Google Scholar]
- [75].Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML. Systematic identification of mammalian regulatory motifs' target genes and functions. Nat Methods 2008; 5:347-53; PMID:18311145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].De Masi F, Grove CA, Vedenko A, Alibés A, Gisselbrecht SS, Serrano L, Bulyk ML, Walhout AJM. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res 2011; 39:4553-63; PMID:21335608; http://dx.doi.org/ 10.1093/nar/gkr070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJM. A Multiparameter Network Reveals Extensive Divergence between C. elegans bHLH Transcription Factors. Cell 2009; 138:314-27; PMID:19632181; http://dx.doi.org/ 10.1016/j.cell.2009.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.