Abstract
Tristetraprolin (TTP) acts by binding to AU-rich elements in certain mRNAs, such as tumor necrosis factor (TNF) mRNA, and increasing their decay rates. TTP knockout mice exhibit a profound inflammatory syndrome that is largely due to increased TNF levels. Although TTP's effects on gene expression have been well studied in cultured cells, little is known about its functions in intact tissues. We performed deep RNA sequencing on spleens from TTP knockout mice that were also deficient in both TNF receptors (“triple knockout” mice) to remove the secondary effects of excess TNF activity. To help identify posttranscriptionally regulated transcripts, we also compared changes in mature mRNA levels to levels of transiently expressed pre-mRNA. In the triple knockout spleens, levels of 3,014 transcripts were significantly affected by 1.5-fold or more, but only a small fraction exhibited differential mRNA/pre-mRNA changes suggestive of increased mRNA stability. Transferrin receptor mRNA, which contains two highly conserved potential TTP binding sites, was significantly upregulated relative to its pre-mRNA. This was reflected in increased transferrin receptor expression and increased splenic iron/hemosiderin deposition. Our results suggest that TTP deficiency has profound effects on the splenic transcriptome, even in the absence of secondary increases in TNF activity.
INTRODUCTION
Members of the tristetraprolin (TTP) family of CCCH tandem zinc finger proteins can bind to AU-rich elements (AREs) in the 3′ untranslated regions (3′UTR) of certain mRNAs and promote their turnover. The ideal binding site appears to be UUAUUUAUU, although variations of this sequence can be tolerated with relatively minor changes in affinity (1–5). TTP and its other family members can bind with high affinity to these AREs and then promote the removal of the poly(A) tail and the ultimate destruction of the transcript (6).
The protein members of the TTP family in mice are as follows: TTP itself, encoded by Zfp36; ZFP36L1, also known as BRF1, ERF1, and CMG1, encoded by Zfp36l1; ZFP36L2, also known as BRF2 and ERF2, encoded by Zfp36l2; and ZFP36L3, encoded by Zfp36l3. The identification of TTP, the best-studied member of this family, as an mRNA-destabilizing protein was first recognized by analyzing the phenotype of TTP knockout (KO) mice, which exhibited a severe systemic inflammatory phenotype with polyarticular erosive arthritis, weight loss, dermatitis, autoimmunity, and myeloid hyperplasia (7). The nature of this syndrome suggested excess tumor necrosis factor alpha (TNF) as a causative agent. This was confirmed by the observations that treatment with anti-TNF antibodies or interbreeding the TTP KO mice with mice lacking both TNF receptors prevented the development of the inflammatory phenotype (7, 8). Subsequent studies on bone marrow-derived macrophages from these animals demonstrated that the TNF mRNA is a direct target of TTP and that the binding of TTP to binding sites in the 3′UTR of TNF mRNA could promote its decay (9, 10).
Since the early demonstration of TNF mRNA as an important physiological target of TTP, many other mRNAs have been identified as real or potential TTP targets (11–14). Many of these putative target transcripts have been identified and studied in primary cells derived from the TTP KO mice or in cultured cells in which TTP was knocked down by other techniques. However, little is known about the effect of TTP deficiency on gene expression in intact tissues of any organism, although TTP is expressed in all vertebrate lineages examined to date except birds (15).
We have begun to identify the effects of TTP on the transcriptomes of tissues in intact organisms by exploring the effect of TTP deficiency on mRNA levels in spleens of adult mice. We chose the spleen for this initial analysis because TTP mRNA and protein expression in this tissue are the highest seen in normal mouse tissues (16; D. J. Stumpo and P. J. Blackshear, unpublished data) and because of its key role in immune responses. Our hypothesis was that TTP deficiency in intact animals would lead to stabilization of a set of mRNA “targets,” leading to steady-state increases in their concentrations. However, the severe systemic inflammatory phenotype seen in the TTP KO animals, largely due to the chronic TNF excess, makes it potentially difficult to identify physiological targets of TTP in intact tissues because of the profound effects of TNF itself on gene expression (17). In particular, one of the hallmarks of TTP deficiency is myeloid hyperplasia, resulting in increases in myeloid cells in many tissues, including spleen (7), making comparisons between TTP KO and control spleens somewhat problematic.
In order to circumvent this problem, we performed deep mRNA sequencing analysis (RNA-Seq) on 10- to 14-week-old “triple KO” mice, which are deficient in both TNF receptors 1 and 2, in addition to TTP. The triple KO mice are externally normal phenotypically until much later in life, when they develop a still-unexplained form of myeloid hyperplasia (8). Both wild-type (WT) and TNF receptor 1- and 2-deficient “double KO” mice were used as controls. We found that a large fraction of the splenic transcriptome was altered in the triple KO spleens, with many fewer changes seen in the double KO mice. However, most of these alterations could be accounted for by increases in pre-mRNA, reflecting changes in transcription. Among the few whose mRNA increased out of proportion to its pre-mRNA was the type 1 transferrin receptor (TFRC) mRNA. Its increased mRNA and protein expression was associated with an increase in iron deposition in the triple KO spleens that was not seen in the controls.
These studies describe a “molecular phenotype” for the splenic transcriptome of TTP-deficient mice in the setting of TNF receptor deficiency and highlight the complexity of the systemic syndrome resulting from the genetic ablation of TTP expression.
MATERIALS AND METHODS
Animals.
All animal procedures were approved by the NIEHS Animal Care and Use Committee. Tnfr1−/−/Tnfr2−/− (double KO) and Zfp36−/−/Tnfr1−/−/Tnfr2−/− (triple KO) mice were generated as described previously (8). Briefly, the triple heterozygous mice were generated on a mixed background (75% C57BL/6NTac and 25% 129/SVEV) and were crossbred to generate three separate lines (WT, Tnfr1−/−/Tnfr2−/−, and Zfp36−/−/Tnfr1−/−/Tnfr2−/− mice). These three mouse lines were maintained in a homozygous state through brother-sister matings. The experimental animals were derived from the homozygous mouse lines and were matched for age (10 to 14 weeks) and sex (male).
Tissue collection.
Spleens were removed from 10- to 14-week-old WT, double KO, and triple KO male mice after euthanasia with CO2 and were stored in RNA Later (Ambion Life Technologies, Carlsbad, CA) or in 10% neutral buffered formalin for histopathology.
RNA extraction.
Total RNA was extracted from each spleen using the GE Healthcare Illustra RNAspin MiniRNA isolation kit according to the manufacturer's instructions (GE Healthcare, Little Chalfont, United Kingdom). RNA concentration and purity were determined by measuring absorbance at 260 and 260/280 nm, respectively, on Tecan's Infinite 200 PRO NanoQuant, using i-control software (Tecan, Mannedorf, Switzerland). The quality of RNA was determined using the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA).
Library preparation and RNA deep sequencing.
Library preparation and sequencing were performed at the NIH Intramural Sequencing Center (http://www.nisc.nih.gov), using 100-bp paired-end reads on the Illumina HiSeq2000 platform (Illumina, Inc., San Diego, CA). Detailed methods have been described previously (15). Analysis of RNA-Seq data was performed beginning with fastq sequence files of 100-base paired-end reads and subjecting them to read quality and length trimming using sickle version 1.200 in paired-end mode (http://github.com/najoshi/sickle). One million reads per sample were aligned to the mm10 transcriptome using bowtie version 0.12.8 (18), from which the average distance of properly aligned paired-end reads was calculated. The length of both reads (200) was subtracted from this average distance to determine the inner mate distance.
Trimmed reads were mapped to the mm10 mouse genome using TopHat2 version 2.0.4 (19) with the empirically determined inner mate distance described above, guided by a transcript GTF file for mouse genome assembly mm10. Where applicable, alignments of technical replicates were merged using samtools version 0.1.9-2 (20), from which the sequence read coverage was produced using BEDtools (21) and converted to bigwig format using BigWig and BigBed tools (22) for the UCSC genome browser. TopHat2 splice junctions were combined per sample group using only the splice boundary regions, to produce one consolidated set of junctions per sample group, and converted to bigBed format for the UCSC genome browser. Differential expression testing was performed using Cuffdiff version 2.1.1 (23), using the mm10 iGenome transcripts (https://support.illumina.com/sequencing/sequencing_software/igenome.html) prepared by Illumina for the mean values from each respective WT control group versus its matched double KO and triple KO groups. Each sample group contained four separate biological samples from four individual mice. Significant changes were required to have “OK” status from Cuffdiff and were filtered to have a P value of 0.05 or less after Benjamini-Hochberg correction for multiple testing (24), an absolute fold change of at least 1.5, and where one sample group had at least 1.5 fragments per kilobase per million mapped reads (FPKM).
Ingenuity pathway analysis.
The differentially expressed transcripts from the triple KO spleens were subjected to Ingenuity pathway analysis (IPA) to investigate the biological networks and pathways that were enriched in the spleen under these conditions (Qiagen, Redwood City, CA). IPA generates canonical pathways and biological networks that are overrepresented in the queried data set.
Identification of TTP binding sites.
We used a custom TTP binding site search application to scan the 3′UTR of potential target transcripts for the presence of possible high-affinity TTP binding sites: UAUUUAU (7-mer), UUAUUUAU and UAUUUAUU (8-mers), and UUAUUUAUU (9-mer) (available on request).
Histochemistry, iron staining, IHC, and digital quantitation.
Slides were stained with hematoxylin and eosin (H&E) for routine histology and with Perl's Prussian blue stain (Mallory's method) to test for the presence of iron in the tissue. For immunohistochemistry (IHC), formalin-fixed, paraffin-embedded tissues were first deparaffinized and rehydrated. Antigen retrieval was performed in a steamer using 10 mM citrate buffer (pH 6.0) retrieval solution (Biocare Medical, Concord, CA). Endogenous peroxidase was blocked with 3% hydrogen peroxide. Nonspecific sites were blocked using 10% normal goat, rabbit, or donkey serum (Jackson ImmunoResearch, West Grove, PA) for 20 min at room temperature. An avidin/biotin blocking solution (Vector Laboratories, Burlingame, CA) was applied to the sections to block any endogenous biotin in the tissues. For Ly6G IHC, the sections were incubated with a rat anti-mouse monoclonal Ly6G antibody (BioLegend, San Diego, CA) at a dilution of 1:500 for 30 min at room temperature. Negative-control sections were exposed to rat IgG2a at the same dilution. Sections were then incubated with a rabbit anti-rat biotin-conjugated secondary antibody for 30 min at a 1:500 dilution, followed by Vectastain Elite ABC reagent RTU (Vector Laboratories, Burlingame, CA). For myeloperoxidase (Mpo) IHC, spleen sections were incubated with rabbit anti-human polyclonal Mpo (Dako Corp., Carpinteria, CA) antibody, or with rabbit immunoglobulin as a negative control, at dilutions of 1:1,500 for 30 min at room temperature. Sections were incubated with biotinylated goat anti-rabbit (Vector Laboratories, Burlingame, CA) secondary antibody at a dilution of 1:500 for 30 min, followed by incubation with Vectastain Elite ABC reagent as described above for Ly6G staining. For Ter119 IHC, the sections were incubated with rat anti-mouse TER-119 monoclonal antibody (BD Pharmingen, San Diego, CA) at a 1:10 dilution for 1 h at room temperature. Rat IgG2b (BioLegend, San Diego, CA) was used as a negative control. Sections were then incubated with a biotinylated rabbit anti-rat IgG (Vector Laboratories, Burlingame, CA) secondary antibody at a dilution of 1:500 for 30 min, followed by the Vectastain Elite ABC reagent as described above. For F4/80 IHC, sections were incubated with a rat anti-mouse F4/80 antibody (BioLegend, San Diego, CA) at a dilution of 1:500 for 60 min at room temperature. Purified rat IgG2a was used as a negative control. Sections were then incubated with a biotinylated rabbit anti-rat secondary antibody (Vector Laboratories, Burlingame, CA) for 30 min at a 1:500 dilution, followed by the Vectastain Elite ABC reagent, as described above. The antigen-antibody complexes were visualized using 3-diaminobenzidine (DAB) chromogen (Dakocytomation, Carpenteria, CA) for 6 min at room temperature. Finally, the sections were counterstained with hematoxylin, dehydrated through graded ethanol, cleared in xylene, and coverslipped.
Immunostained sections were graded by a pathologist and given a score ranging between 0 and 4. For Ly6G staining, grading was based on the density of positively stained cells within the red pulp and on the presence of clusters of contiguous positive-staining cells, usually beneath the capsule but sometimes within the outer third of the red pulp. For Ter119 staining, grading was performed as follows: 1, occasional clusters of nucleated erythroid precursors beneath the capsule; 2, small clusters of nucleated erythroid cells lining much of the subcapsular area or scattered beneath the capsule and within the red pulp; 3, clusters of erythroid precursors not only beneath the capsule but in clusters within the red pulp, with the clusters also being larger than those in grade 2.
For Prussian blue positive iron quantitation, slides were scanned with an Aperio Scanscope XT digital slide scanner (Aperio Technologies, Vista, CA). The resulting images were viewed using Aperio ImageScope v. 12.0.1.5027, and image analysis was performed using the Aperio colocalization algorithm. This algorithm calculates the contribution of multiple stains at every pixel location in the image, which in this case involved identification of Prussian blue staining. The data were expressed as the percentage of the Prussian blue-stained particles in the tissue over the total tissue area.
RNA probe preparation for in situ hybridization histochemistry.
A plasmid (BB.A19/BS+) for preparing the mouse RNA probes for in situ hybridization was constructed as follows. Briefly, a 185-bp fragment of a mouse TTP cDNA (bp 843 to 1027, NM_011756.4) was excised from a mouse TTP cDNA clone (25) using the restriction enzymes BamHI and BglII and ligated into the BamHI cloning site of vector Bluescribe (BS+; Stratagene). The orientation of the construct was determined by sequencing. To obtain the antisense RNA probe, the construct was linearized using the restriction enzyme XbaI, followed by in vitro transcription using T7 RNA polymerase in the presence of α-[35S]UTP (New England Nuclear, Boston, MA; >1,000 Ci/mmol). Similarly, to obtain a sense probe, the construct was linearized using the restriction enzyme EcoRI, followed by in vitro transcription using T3 RNA polymerase. For in situ hybridization, spleen sections were prepared as described previously (26). Briefly, adult WT mice were euthanized, and spleens were removed, frozen on dry ice with Tissue-Tek embedding compound (Miles Scientific, Naperville IL), and stored at −80°C until being sectioned. Twelve-micrometer frozen sections were prepared on a cryotome, thaw-mounted on twice-gelatin-coated slides, and fixed with 4% paraformaldehyde in 0.12 M phosphate-buffered saline (pH 7.4) for 5 min at room temperature. Hybridization was done as described before (26, 27). Sections were developed initially using Kodak XAR-2 X-ray film and thereafter by nuclear emulsion autoradiography (Kodak NTB-3; 1:1 with water). The exposure times were 1 to 14 days with film and 1 to 6 weeks with nuclear emulsion. The sections were subsequently stained with cresyl violet in order to counterstain the cell bodies.
Exon-intron statistical analysis.
In order to compare combined exon expression for a given transcript with combined “intron” expression, the latter representing pre-mRNA, we downloaded UCSC RefSeq transcript models and combined them to produce one set of exons and introns per gene. In order to remove the potential effects of read alignments near the intron-exon boundaries, five bases from each end of each intron were removed. Reads were assigned to exons and introns per gene using featureCounts version 1.4.6. Total exon reads and total intron reads per gene were used for statistical analysis in R (version 3.1.1), using Bioconductor (28) package limma version 3.22.1 (29). The counts were normalized using the voom method (30), which also modeled the mean variance to assign observational-level weights for downstream linear modeling. Data were configured to test for interactions between exon and intron differential changes for each genotype versus its respective control group. Statistical hits were filtered to have Benjamini-Hochberg adjusted P values of <0.05 and absolute fold changes of >1.5.
DCQ.
To supplement the immunohistochemical staining for specific cells types, we applied the digital cell quantitation (DCQ) method for digital quantitation of cell types in the various samples (31) (http://www.dcq.tau.ac.il). This method attempts to analyze the relative enrichment of certain immune cell subsets, based on the RNA-Seq data comparisons and known cell-type-specific markers. We applied this method to the RNA-Seq data from spleens of all three genotypes. Mean enrichment values were compared between the two sets of WT versus mutant samples using Student's unpaired, two-tailed t tests.
Nucleotide sequence accession number.
All of the RNA-Seq data have been deposited in NCBI GEO under accession number GSE60243.
RESULTS
Splenic phenotype and TTP mRNA expression.
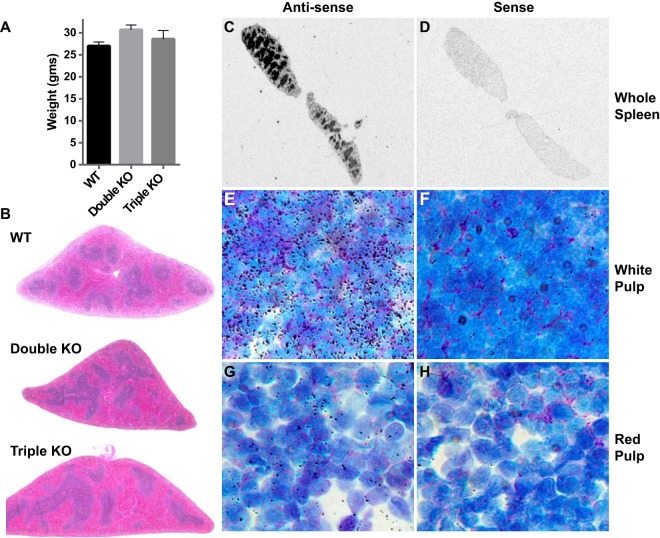
Mice were used at the age of 10 to 14 weeks, when body weights and external phenotype are essentially normal in the triple KO mice (8). The conventional TTP KO mice already exhibit failure of weight gain and other manifestations of the TTP deficiency syndrome by this age (7). The lack of effect of genotype on body weights in the present set of triple KO mice was confirmed (Fig. 1A). Routine necropsy of other tissues did not show evident abnormalities in any of the triple or double KO animals. Histological examination revealed mild expansion of the red pulp with mature erythrocytes and an increase in the amount of extramedullary hematopoiesis (EMH) in the triple KO mouse spleens. No other major histopathological changes were observed upon low-power microscopic examination of the spleens after hematoxylin and eosin (H&E) staining (Fig. 1B).
FIG 1.
Body weights, spleen TTP mRNA expression, and spleen histopathology. (A) Average body weights ± standard error of the mean (SEM) of the male mice used in the study at 10 to 14 weeks of age. These weights were not significantly different from each other. (B) H&E-stained spleen sections from WT, double KO, and triple KO mice. Original magnification, ×2. (C and D) Film autoradiograms of spleen sections from a normal adult mouse after in situ hybridization using an 35S-labeled TTP antisense riboprobe (C) and a corresponding sense probe (D), developed and imaged in parallel. The antisense probe revealed intense signals in the white pulp, whereas little or no signal was detected using the sense probe. (E and G) Emulsion autoradiograms of regions of white pulp (E) and red pulp (G) from a WT mouse spleen, showing intense TTP mRNA signals in the lymphocytes of the white pulp (E) but much lower signals in the cells of the red pulp (G). (F and H) No hybridization signal was detected with the sense probe under these conditions for panels E and G, respectively. Original magnification for panels E to H, ×600.
The expression pattern of TTP mRNA in normal adult mouse spleen was characterized using in situ hybridization histochemistry (ISHH). We used a radiolabeled antisense TTP probe to detect TTP mRNA and used the corresponding sense probe as a negative control. The film autoradiograph with the antisense probe revealed a strong signal in the white pulp of the spleen, with a lower signal in the red pulp (Fig. 1C). The sense probe produced little signal at the same exposures (Fig. 1D). Emulsion autoradiographs of cells in the white pulp (Fig. 1E) and the red pulp (Fig. 1G) showed intense coverage of silver grains over the lymphocytes of the white pulp but much less signal over the mixed cell populations of the red pulp, suggesting that the lymphocytes of the white pulp express particularly high levels of TTP mRNA. The sense probe produced little signal at the same exposure for both the white pulp and the red pulp (Fig. 1F and H). This was the most intense hybridization signal seen in a survey of tissues from a normal adult mouse performed in the same experiment.
RNA-Seq data quality control.
We performed RNA-Seq on 16 individual spleen samples, comparing 4 WT to 4 double KO samples in the first experiment and 4 WT to 4 triple KO samples in the second experiment. The total read numbers per sample ranged from 55,982,176 to 139,541,704, and after quality control, 81.1% to 87.9% of the original reads from all samples were mapped to the mouse transcriptome (Table 1).
TABLE 1.
Summary of RNA-Seq reads obtained and mapped to the UCSC mouse genome (mm10) by TopHat2 version 2.0.4a
| Sample | Sample group | Total reads | Total filtered reads | % filtered reads | Tophat2 total reads used | Tophat2 mapped reads | % Tophat2 mapped | % original reads mapped |
|---|---|---|---|---|---|---|---|---|
| ES594 | WT | 59,739,172 | 54,207,483 | 90.7% | 51,675,521 | 48,601,337 | 94.1 | 81.4 |
| ES595 | WT | 57,152,858 | 52,875,700 | 92.5 | 50,730,261 | 47,964,101 | 94.5 | 83.9 |
| ES596 | WT | 58,833,186 | 54,250,597 | 92.2 | 51,983,177 | 48,554,589 | 93.4 | 82.5 |
| ES597 | WT | 61,523,552 | 56,214,437 | 91.4 | 53,791,001 | 50,569,820 | 94.0 | 82.2 |
| ES598 | Double KO | 57,775,842 | 53,563,505 | 92.7 | 51,429,249 | 48,165,044 | 93.7 | 83.4 |
| ES600 | Double KO | 57,768,736 | 53,587,435 | 92.8 | 51,405,002 | 46,872,085 | 91.2 | 81.1 |
| ES628 | Double KO | 58,727,920 | 54,463,663 | 92.7 | 52,325,308 | 49,201,172 | 94.0 | 83.8 |
| ES629 | Double KO | 55,982,176 | 51,588,651 | 92.2 | 49,477,295 | 46,444,930 | 93.9 | 83.0 |
| ES333 | WT | 122,435,492 | 117,028,201 | 95.6 | 113,414,331 | 105,959,643 | 93.4 | 86.5 |
| ES334 | WT | 92,048,820 | 88,045,246 | 95.7 | 85,442,873 | 80,418,792 | 94.1 | 87.4 |
| ES335 | WT | 111,930,898 | 106,605,448 | 95.2 | 103,046,163 | 97,020,593 | 94.2 | 86.7 |
| ES336 | WT | 139,541,704 | 133,974,689 | 96.0 | 130,360,240 | 122,726,693 | 94.1 | 87.9 |
| ES337 | Triple KO | 113,175,782 | 103,321,345 | 91.3 | 99,264,043 | 94,070,208 | 94.8 | 83.1 |
| ES338 | Triple KO | 136,755,774 | 124,372,707 | 90.9 | 119,283,030 | 112,261,751 | 94.1 | 82.1 |
| ES339 | Triple KO | 110,055,756 | 99,835,607 | 90.7 | 95,783,561 | 90,098,345 | 94.1 | 81.9 |
| ES340 | Triple KO | 117,766,678 | 106,854,914 | 90.7 | 102,476,075 | 95,920,466 | 93.6 | 81.4 |
Total reads as well as the percentage of reads removed for quality control are shown. In addition, the percentages of good-quality reads that were aligned to the reference genome are shown. Double KO group, four WT and four double KO spleen RNA samples sequenced on a lane of Illumina HiSeq2000 generated between 55 and 61 million raw reads per biological replicate. Triple KO group, four WT and four triple KO spleen RNA samples sequenced on a lane of Illumina HiSeq2000 generated between 92 and 139 million reads per biological replicate.
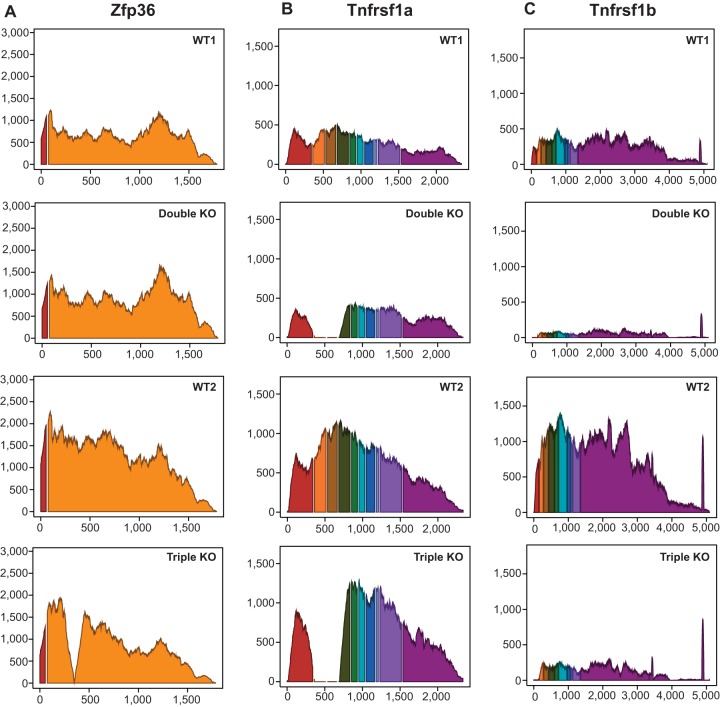
The expression patterns for the three genetically modified transcripts (Zfp36, Tnfrsf1a, and Tnfrsf1b) are shown as histograms for the reads mapped to the annotated exons of the three genes (Fig. 2). These data confirm the absence of critical exons in the transcripts expressed from the knocked out genes. Although the TTP mRNA is expressed as a fusion mRNA with Neo sequences in the middle, repeated studies have demonstrated no expression of TTP protein in cells and tissues derived from these mice (7, 16, 32).
FIG 2.
Average read coverage showing the annotated exons of the three genetically modified transcripts (Zfp36, Tnfrsf1a, and Tnfrsf1b mRNAs) used in the two comparisons. The y axis represents the average sequence read depth at each position for the four samples, and the x axis represents the nucleotide position along the transcript exons, including a fixed 10-nucleotide gap at the site of each intron. Each exon in each transcript is in a different color. (A) Interruption of the reads mapped to the 5′ end of exon 2 of Zfp36, caused by the insertion of the neomycin resistance element, in the triple KO samples. (B) Exons 2 and 3 of Tnfrsf1a were disrupted, as is evident from the absence of reads for those two exons in both the double KO and triple KO samples. (C) Reads were absent from exon 1 in the Tnfrsf1b transcript in both the double and triple KO samples.
We compared the relative expression of TTP mRNA and its family members, the Zfp36l1, Zfp36l2, and Zfp36l3 mRNAs, in the two WT sample groups. Compared to the other family members, Zfp36l1 mRNA was expressed at the highest level in this tissue (mean fragments per kilobase of transcript per million mapped reads [FPKM] for the two WT sample groups, 140, followed by Zfp36l2 mRNA [73] and Zfp36/TTP [52]). Zfp36l3 is a rodent-specific, placenta-specific TTP family member (33) and was not detected in the spleen. We found no significant differences among the various mutant groups in the expression of Zfp36l2 mRNA. Zfp36l1 mRNA, the most highly expressed TTP family member in the WT samples, was found to be decreased in the triple KO spleens by approximately 1.4-fold (P < 0.005), although it was not significantly affected in the double KO spleens. These findings indicate that the other TTP family members expressed in spleen did not increase in the setting of TTP deficiency in a compensatory manner and that TTP mRNA was the most poorly expressed of the three major TTP family members expressed in this tissue, representing only about 20% of the total “TTP transcript equivalents” in the whole tissue.
Differentially expressed transcripts.
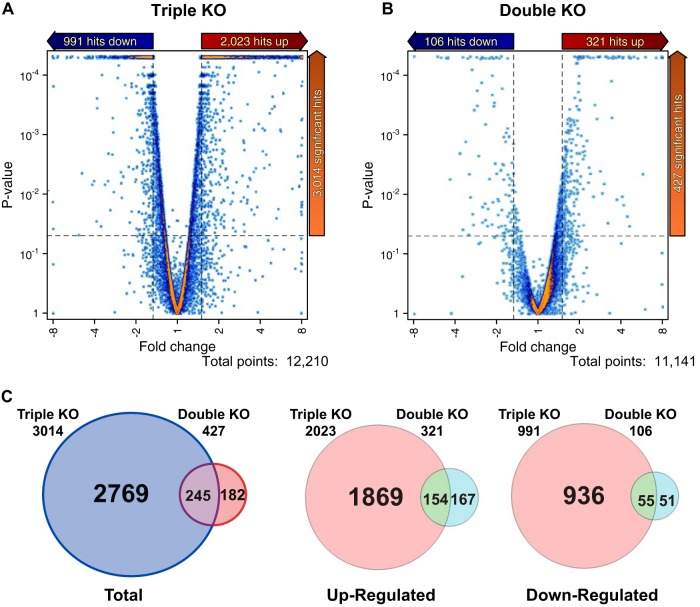
Sequence reads were mapped to the mm10 RefSeq transcriptome using TopHat 2, and relative expression levels were calculated using cufflinks and CuffDiff 2 and expressed as FPKM. We discarded transcripts with mean expression levels of less than 1.5 FPKM in any sample group. Of the remaining expressed transcripts, representing approximately 12,210 gene models in the triple KO spleens, 3,014 (24.6%) were differentially expressed compared to the WT, using the following criteria: P value of ≤0.05, fold change of ≥1.5, and expression level of ≥1.5 FPKM. In contrast, only 427 (3.8%) of expressed transcripts, representing approximately 11,141 gene models, were differentially expressed in the double KO spleens compared to the WT, using the same criteria (Fig. 3A to C). Of the 3,014 transcripts differentially expressed in the triple KO spleens, 2,023 (67.1%) were upregulated and 991 (32.8%) were downregulated, and of the 427 differentially expressed transcripts in the double KO spleens, 321 (75.1%) were upregulated and 106 (24.8%) were downregulated (Fig. 3A to C).
FIG 3.
(A and B) Differential transcript expression in the triple KO versus WT (A) and double KO versus WT (B) comparisons. Volcano plots of transcript expression in the two comparisons are shown. The dashed lines indicate significance cutoffs of P < 0.05 and fold change cutoffs of 1.5. Only transcripts that were expressed above the abundance cutoff of 1.5 FPKM are shown. (C) Venn diagrams of unique and common transcripts between the two RNA-Seq data sets that met these criteria.
The transcripts that were significantly upregulated or downregulated by more than 1.5-fold in the triple KO spleens compared to the WT spleens are listed in Tables S1 and S2 in the supplemental material, respectively.
These data demonstrate that many transcripts were differentially expressed in the triple KO spleens, even when the animals exhibited no external signs of the TTP deficiency phenotype. Most of these transcripts were not similarly affected in the double KO mice, supporting the concept that most of the observed changes in gene expression were either primary or secondary consequences of the TTP deficiency, even in the setting of absent TNF activity.
Evaluation of previously identified or suggested TTP targets.
Next we evaluated the expression levels of known previously identified or suggested TTP target transcripts, as summarized in reference 11 (Table 2). Previously identified or suggested targets that were upregulated in the triple KO spleens but not in the double KO spleens were mRNAs encoded by Il1b, Il12, Ier3, Ccl3, Cxcl1, Il10, Ifng, Il1a, Tlr4, Plau, Pim1, E2f1, and Cd36. Three genes encoding previously identified or suggested targets, Clmp, Lats2, and Thbd, showed decreased expression in the triple KO spleens. Most of the other previously identified or suggested targets listed in reference 11 were not significantly affected in the triple KO spleens.
TABLE 2.
Comparison of previously identified TTP targets from the literature to spleen RNA-Seq data
| Gene symbol | Significant double KO | Significant triple KO | FPKMc |
P value |
||||
|---|---|---|---|---|---|---|---|---|
| WT controls for double KO | Double KO | WT controls for triple KO | Triple KO | Double KO | Triple KO | |||
| Tnf | No | No | 3.31 | 4.44 | 2.99 | 3.58 | 2.43E−01 | 1.74E−01 |
| Csf2 | No | No | 0.13 | 0.14 | 0.10 | 0.17 | 9.22E−01 | 1.00E+00 |
| Il3 | No | No | 0.00 | 0.00 | 0.00 | 0.00 | 1.00E+00 | 1.00E+00 |
| Ptgs2 | No | No | 0.09 | 0.07 | 0.11 | 0.18 | 7.31E−01 | 1.00E+00 |
| Pitx2 | No | No | 0.00 | 0.00 | 0.00 | 0.00 | 6.13E−01 | 1.00E+00 |
| Serpinb2 | No | No | 1.72 | 1.08 | 2.21 | 2.94 | 2.38E−01 | 4.49E−02 |
| Zfp36 | No | No | 41.34 | 59.50 | 62.91 | 54.72 | 1.20E−02 | 1.75E−01 |
| Nos2 | No | No | 0.41 | 1.06 | 1.04 | 1.04 | 4.26E−03 | 9.82E−01 |
| Il2 | No | No | 0.05 | 0.09 | 0.02 | 0.12 | 6.19E−01 | 1.00E+00 |
| Il1b | No | Yes | 11.08 | 13.77 | 13.23 | 41.91 | 2.53E−01 | 5.00E−05 |
| Il12a | No | Yes | 4.60 | 5.88 | 4.57 | 3.06 | 3.23E−01 | 3.91E−03 |
| Il12b | No | Yes | 0.74 | 0.51 | 1.27 | 2.12 | 3.63E−01 | 6.00E−04 |
| Cxcl2 | No | No | 0.04 | 0.04 | 0.05 | 0.10 | 9.12E−01 | 1.00E+00 |
| Ccl20 | No | No | 0.00 | 0.01 | 0.00 | 0.01 | 4.48E−01 | 1.00E+00 |
| Il6 | No | No | 0.11 | 0.14 | 0.08 | 0.15 | 7.67E−01 | 1.00E+00 |
| Ier3 | No | Yes | 0.84 | 1.05 | 1.07 | 2.06 | 6.14E−01 | 4.50E−04 |
| Lif | No | No | 0.44 | 0.44 | 0.60 | 0.30 | 9.80E−01 | 8.07E−02 |
| Plk3 | No | No | 2.70 | 3.07 | 3.98 | 4.12 | 6.04E−01 | 7.79E−01 |
| Ccnd1 | No | No | 14.73 | 15.24 | 26.48 | 31.46 | 8.18E−01 | 1.03E−01 |
| Myc | No | No | 10.83 | 13.22 | 22.93 | 28.92 | 3.37E−01 | 8.61E−02 |
| Ccl2 | No | No | 0.42 | 0.13 | 0.40 | 0.66 | 1.47E−01 | 4.86E−02 |
| Ccl3 | No | Yes | 0.63 | 0.54 | 0.44 | 0.90 | 7.85E−01 | 3.85E−03 |
| Vegfa | No | No | 5.54 | 6.92 | 8.59 | 8.27 | 5.27E−01 | 7.44E−01 |
| Tcf3 | No | No | 8.46 | 11.19 | 15.80 | 15.36 | 4.03E−01 | 5.60E−01 |
| Cxcl1 | No | Yes | 0.29 | 0.15 | 0.13 | 0.34 | 3.60E−01 | 1.19E−02 |
| Il10 | No | Yes | 0.24 | 0.23 | 0.20 | 0.76 | 9.64E−01 | 5.00E−05 |
| Clmp | No | Yes | 1.36 | 1.16 | 2.95 | 1.14 | 5.97E−01 | 5.00E−05 |
| Ifng | No | Yes | 0.40 | 0.64 | 0.36 | 0.73 | 3.59E−01 | 2.95E−03 |
| Ube3a | No | No | 16.84 | 13.16 | 17.12 | 17.74 | 1.18E−01 | 8.10E−01 |
| Il1a | No | Yes | 3.34 | 3.16 | 3.85 | 6.80 | 8.26E−01 | 5.00E−05 |
| Tlr4 | No | Yes | 8.71 | 7.78 | 14.91 | 20.81 | 4.89E−01 | 1.35E−03 |
| Plau | No | Yes | 1.74 | 2.22 | 2.93 | 4.53 | 3.82E−01 | 6.00E−04 |
| Plaur | No | No | 17.51 | 22.61 | 23.75 | 23.60 | 1.20E−01 | 9.51E−01 |
| Mmp1a | No | No | 0.00 | 0.00 | 0.00 | 0.00 | 1.00E+00 | 1.00E+00 |
| Ecscr | No | No | 3.38 | 3.68 | 3.02 | 3.76 | 7.67E−01 | 1.21E−01 |
| Hif1a | No | No | 28.20 | 27.88 | 48.21 | 39.36 | 9.37E−01 | 5.66E−02 |
| Lats2 | No | Yes | 15.55 | 18.80 | 36.59 | 25.36 | 3.22E−01 | 3.00E−04 |
| Dusp6 | No | No | 18.06 | 22.22 | 35.62 | 28.49 | 1.65E−01 | 3.07E−02 |
| Slc10a2 | No | No | 0.02 | 0.01 | 0.01 | 0.01 | 8.55E−01 | 1.00E+00 |
| Thbd | No | Yes | 16.96 | 20.47 | 35.93 | 27.00 | 2.00E−01 | 5.25E−03 |
| Il23a | No | No | 0.01 | 0.01 | 0.01 | 0.03 | 9.89E−01 | 1.00E+00 |
| Cdkn1a | No | No | 2.04 | 1.60 | 2.84 | 3.16 | 4.78E−01 | 4.40E−01 |
| Il17a | No | No | 0.00 | 0.00 | 0.00 | 0.01 | 1.00E+00 | 1.00E+00 |
| Pim1 | No | Yes | 18.19 | 27.38 | 32.43 | 75.61 | 8.49E−03 | 5.00E−05 |
| Serpinh1 | No | No | 10.27 | 13.77 | 19.01 | 17.47 | 2.67E−01 | 4.66E−01 |
| Ahrr | No | No | 0.31 | 0.36 | 0.53 | 0.55 | 6.72E−01 | 7.68E−01 |
| E2f1 | No | Yes | 2.48 | 2.89 | 4.42 | 15.42 | 5.38E−01 | 5.00E−05 |
| Cd36 | No | Yes | 27.86 | 27.08 | 69.50 | 107.73 | 8.68E−01 | 3.19E−03 |
| Cxcr4 | No | No | 50.97 | 59.70 | 69.03 | 61.98 | 2.99E−01 | 3.19E−01 |
a FPKM, fragments per kilobase of exon per million fragments mapped.
Strikingly, the TNF mRNA, the first-discovered and best-studied physiological target of TTP (7, 9, 10, 34), was not significantly affected in the triple KO spleens, despite being expressed at low but readily detectable levels in these samples (mean FPKM values: triple KO, 3.5; WT controls for triple KO, 2.9; double KO, 4.4; WT controls for double KO, 3.3). The differences between experimental and control means were not significant for this transcript in either of the two-way comparisons. These results highlight the potential differences between previous studies in single cell types, such as lipopolysaccharide (LPS)-stimulated macrophages or serum-stimulated fibroblasts, and the current study in whole tissues from intact animals studied under normal laboratory conditions.
Identification of transcripts with optimum TTP binding sites.
The 2,023 transcripts that were increased in the triple KO versus WT comparison included 154 of the 245 transcripts that were upregulated in both the triple and the double KO data sets but excluded 147 that were upregulated only in the double KO data set. Binding site analysis found that 395 of the 2,023 upregulated transcripts contained at least one potential ideal TTP binding site (7-mer, 8-mer, or 9-mer) (see Table S3 in the supplemental material), and 335 of the 991 downregulated transcripts contained at least one potential ideal TTP binding site in their 3′UTR. The most highly upregulated transcript in the triple KO mice that also contained one or more potential TTP binding sites encoded apolipoprotein L 11b (APOL11B). Apol11b mRNA was upregulated by 367-fold in the triple KO spleens and 3.5-fold in the double KO spleens. This transcript contained a single 8-mer potential TTP binding site. We should note that minor variations in the optimum consensus binding site sequence can be tolerated with fairly minor changes in binding affinity, at least to a synthetic human TZF domain peptide (1). It is therefore possible that “true” TTP target transcripts that were excluded from our list because they did not contain ideal binding sites could still use these slightly suboptimal sequences in a physiological context.
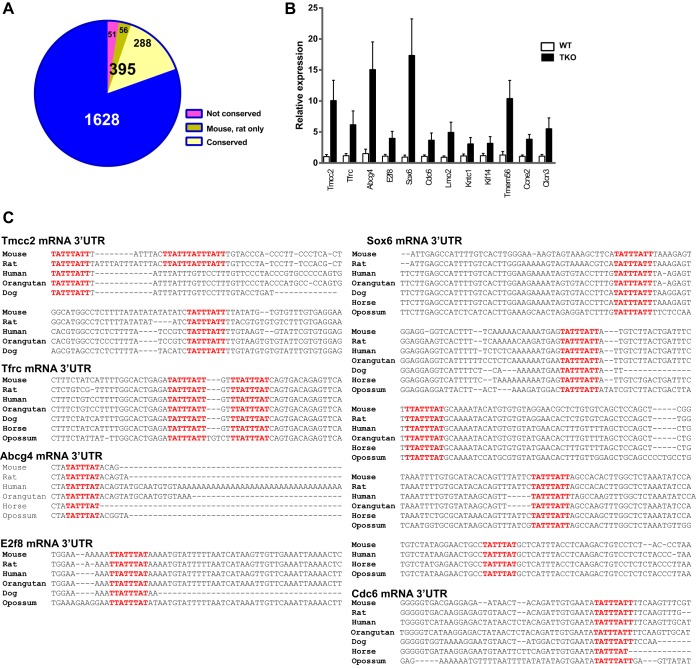
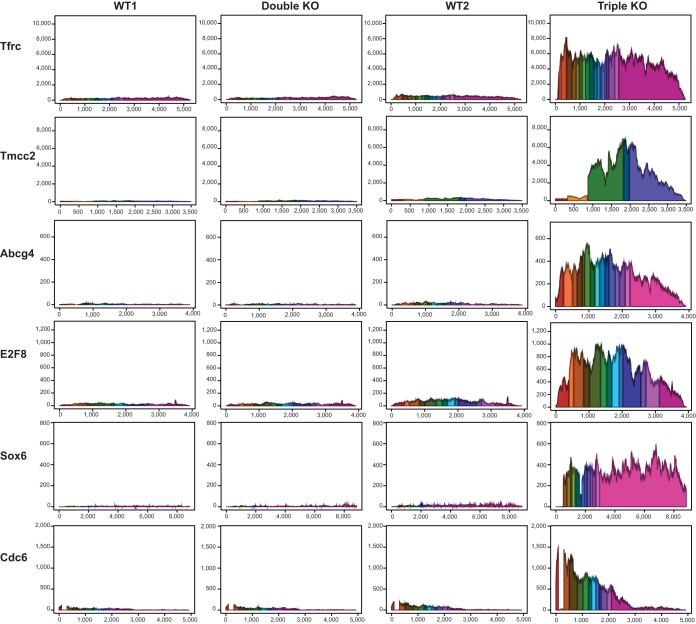
The 395 potential target transcripts listed in Table S3 in the supplemental material were then examined for conservation of TTP binding sites, determined by examination of sequence tracks on the University of California at Santa Cruz (UCSC) genome browser. Of the 395, 56 transcripts contained sites that were conserved in mouse and rat only, 288 transcripts contained sites that were conserved in rat as well as multiple other mammals, including humans, and 51 transcripts contained sites that exhibited no obvious conservation in other mammals (Fig. 4A; see Table S3 in the supplemental material). Quantitative real-time PCR analysis confirmed the significantly increased expression of the top 12 transcripts in the triple KO spleens relative to WT levels (Fig. 4B). The top six transcripts that were upregulated in the triple KO but not in the double KO spleens and contained highly conserved potential TTP binding sites (Fig. 4C), were as follows: transmembrane and coiled coil domain family 2 (Tmcc2) mRNA, 23.2-fold; type I transferrin receptor (Tfrc), 18.5-fold; ATP binding cassette, subfamily G, member 4 (Abcg4), 17.0-fold; E2f transcription factor 8 (E2f8), 13-fold; SRY (sex-determining region Y)-box6 (Sox 6), 11.9-fold; and cell division cycle 6 (Cdc6), 11.7-fold. Figure 5 shows the reads mapped to these six transcripts in the four genotype comparisons, Table 3 shows the fold change of expression, and Fig. 4C shows in each case the conserved binding sites in mouse and several other mammalian species.
FIG 4.
Transcripts with potential TTP binding sites in the 3′UTR. (A) Pie chart depicting 395 of the upregulated 2,023 transcripts from the triple KO versus WT comparison that contained at least one potential ideal TTP binding site (7-mer) in their 3′UTR. (B) Quantitative real-time PCR data showing the relative expression of the top 12 potential TTP target transcripts in triple KO spleen versus WT spleen. (C) Alignments of potential TTP binding sites in the 3′UTR of six possible TTP targets in various mammalian species. The GenBank accession numbers for the mRNA sequences used in these alignments are NM_178874 (Tmcc2), NM_011638 (Tfrc), NM_138955 (Abcg4), NM_001013368 (E2f8), NM_001025559 (Sox6), and NM_001025779 (Cdc6). The highlighted sequences represent potential 7-mer (TATTTAT), 8-mer (TTATTTAT and TATTTATT), and 9-mer (TTATTTATT) TTP binding sites that were conserved in the species shown.
FIG 5.
Average read coverage of the annotated exons of six top potential TTP target mRNAs. The y axis represents the average sequence read depth at each position, and the x axis represents the nucleotide position along the transcript exons, including a fixed 10-nucleotide gap at each intron. Each exon in each transcript is a different color. As shown, Tfrc, Tmcc2, Abcg4, E2f8, Sox6, and Cdc6 mRNAs were by far the most abundant in the triple KO group.
TABLE 3.
Top six potential TTP targets with highly conserved TTP binding sites in the 3′UTRa
| Gene symbol | Product | Fold change |
Significant |
TTP binding site | ||
|---|---|---|---|---|---|---|
| DKO | TKO | DKO | TTP KO | |||
| Tmcc2 | Transmembrane and coiled coil domain family 2 | 1.5 | 23.2 | No | Yes | One 8-mer–9-mer overlap (mouse, rat), one 9-mer highly conserved |
| Tfrc | Transferrin receptor (p90, CD71) | 1.1 | 18.5 | No | Yes | Two 8-mers highly conserved |
| Abcg4 | ATP binding cassette, subfamily G, member 4 | 1.0 | 17.0 | No | Yes | One 7-mer highly conserved |
| E2f8 | E2F transcription factor 8 | 1.4 | 13.0 | No | Yes | One 8-mer highly conserved |
| Sox6 | SRY (sex-determining region Y)-box 6 | 1.0 | 11.9 | No | Yes | Five 8-mers, one 7-mer, all highly conserved |
| Cdc6 | Cell division cycle 6 | 1.4 | 11.7 | No | Yes | One 8-mer highly conserved |
The top 6 potential TTP target mRNAs were upregulated in the triple KO spleens but not in the double KO spleens and contain highly conserved potential TTP binding sites in their 3′UTR. DKO, double knockout; TKO, triple knockout.
Pathway analysis.
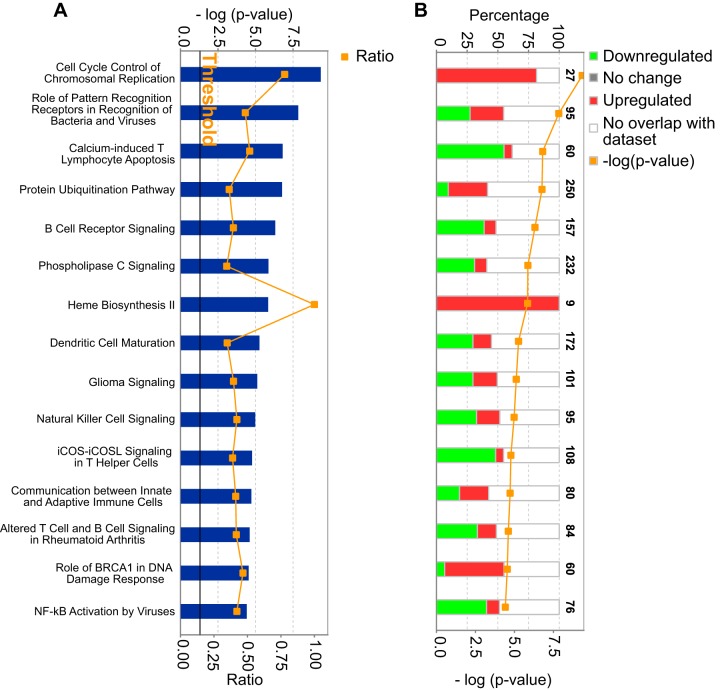
To explore physiological systems potentially regulated in the spleen by the TTP deficiency, we subjected the differentially expressed transcripts in the triple KO-WT comparison to pathway analysis using the Ingenuity pathway analysis (IPA) application (Qiagen, Redwood City, CA). The “core analysis” function was used to investigate the differentially expressed genes in the triple KO spleens in the context of biological processes, pathways, and networks. In this analysis, 10 of the 15 top-scoring networks were immune response pathways, either innate or adaptive (Fig. 6A). This is consistent with known functions of TTP to regulate levels of transcripts encoding immune and inflammatory response proteins. The other five significant pathways were as follows: cell cycle control of chromosomal replication, heme biosynthesis II, glioma signaling, role of BRCA1 in DNA damage response, and the protein ubiquitination pathway. Interestingly, all the mRNAs that were enriched in the topmost canonical pathway, i.e., cell cycle control of chromosomal replication, were found to be upregulated in the triple KO mice versus controls (Fig. 6B). Several mRNAs in these pathways also contain highly conserved potential TTP binding sites in their 3′UTR and thus might serve as direct TTP targets. Of the four remaining canonical pathways, the heme biosynthesis II pathway was particularly interesting, since mRNAs encoding all nine enzymes in this pathway were upregulated in the triple KO mice (Fig. 6B). However, none of these mRNAs contained obvious TTP binding sites in the 3′UTR of their mRNAs, making it unlikely that they were direct targets of TTP. This suggests that the apparent dysregulation of the heme biosynthesis pathway might be an indirect or secondary effect of the TTP deficiency.
FIG 6.
Ingenuity pathway analysis (IPA) of differentially expressed transcripts in the triple KO spleen. Shown are IPA data indicating major pathways that were significantly modulated by the transcripts whose expression differed in the triple KO spleens compared to the WT spleens. Only the top 15 pathways with the most significant changes are shown in each case. (A) The P values for each pathway are indicated by the bars and are expressed as −1 times the log of the P value. The orange line represents the ratio of the number of transcripts in a given pathway that met the cutoff criteria divided by the total number of transcripts that make up that pathway. The black threshold line corresponds to a P value of 0.05. (B) Each of the stacked bars indicates the percentage of significantly upregulated transcripts in that pathway, significantly downregulated transcripts, and transcripts in the pathway that did not overlap our data set. The number at the top of each bar represents the total number of genes in that particular pathway.
Effect of TTP deficiency on TFRC mRNA levels.
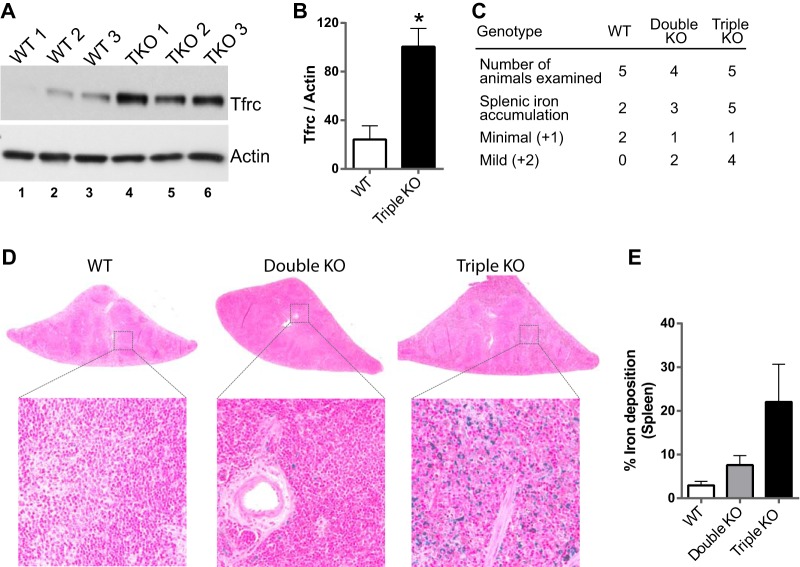
The type I transferrin receptor (TFRC) mRNA was found to be upregulated by 18.5-fold only in the triple KO spleens compared to controls and was not affected in the double KO spleens. This mRNA contains two highly conserved 8-mers (UAUUUAUU and UUAUUUAU) in its 3′UTR (Fig. 4C). In addition, we found that the expression of the TFRC protein was significantly upregulated in the triple KO spleens (Fig. 7A and B). To determine whether the triple KO mice exhibited enhanced iron uptake in the spleen, as might be expected from the marked increase in TFRC mRNA and protein expression, we evaluated the deposition of iron/hemosiderin pigment. The WT and double KO mice had minimal deposits, but there were major deposits in the triple KO spleens (Fig. 7C). Figure 7D and E show the ferric iron staining (Prussian blue reaction; Mallory method) of the spleens and quantitation of the ferric iron staining, respectively.
FIG 7.
Transferrin receptor expression and iron accumulation. (A) Western blot demonstrating increased TFRC protein expression in three triple KO spleens compared to three WT spleens, with immunoreactive actin used as a loading control. (B) Quantitation of the immunoblots shown in panel A, using ImageJ. Quantitation is represented as the mean ratio of TFRC staining to actin staining ± SEM. *, P ≤ 0.05; n = 3. (C) Prevalence and severity of Prussian blue-positive iron accumulation in the splenic red pulp in five WT, four double KO, and five triple KO animals. Minimal, normal to 10% increase; mild, 10 to 25% increase. (D) Slides of spleen after staining with Perl's Prussian blue, depicting accumulation of iron/hemosiderin. The presence of iron is indicated by the dark blue staining in the triple KO spleen, whereas almost no staining was seen in the WT and double KO spleens. (E) Quantitation of staining shown in panel D, for which images from five WT, four double KO, and five triple KO animals were used for quantitation. Quantitation is represented as the mean ± SEM of the percentage of the amount of iron particles in the tissue over the total tissue area.
Analysis of exonic versus intronic RNA-Seq reads.
Because we considered that the large number of upregulated transcripts in triple KO spleens, even those with conserved TTP family member binding sites, were unlikely to all be direct targets of TTP in this tissue, we attempted to measure both mature transcript levels, represented by conventional mRNA-Seq, and immature pre-mRNA levels in the same samples. Although total cellular RNA was used to prepare the sequence libraries, which then included a poly(A) selection step, we would expect a certain low concentration of the transient intermediates representing pre-mRNA to be present in each sample. We developed a single statistic for each pre-mRNA, based on total intron reads per gene included in the mRNA-Seq data. We reasoned that the transcript levels would represent the steady-state results of transcription versus decay, whereas the pre-mRNA levels would be an indicator of transcription alone. For transcripts that were upregulated in the triple KO spleens, this method should allow us to conclude either that the increases in mature transcript levels were accompanied by proportionate changes in pre-mRNA levels, indicating a transcriptional or other secondary response to the triple KO genotype (such as changes in cellular composition), or that the increases in mRNA levels were disproportionately higher than the pre-mRNA increases, suggesting a primary change in mRNA stability.
For this comparison, we analyzed spleen RNA-Seq data from the two independent experimental comparisons (double KO versus WT and triple KO versus WT) and compared total exon and intron read counts for each annotated mouse transcript. We focused on the 395 transcripts that were significantly upregulated in the triple KO spleens compared to control and double KO spleens and contained at least one minimal 7-mer TTP family member binding site.
We first calculated the ratios of the average double KO and triple KO exon reads to the average WT exon reads for each group of samples and excluded 10 of the original 395 transcripts because of low read counts in some of the samples, leaving 385 evaluable transcripts that were significantly upregulated in the triple KO versus control but not the double KO versus control groups. We then ranked these transcripts according to the ratio of average triple KO exon reads to average WT exon reads. These data are shown in Table S4 in the supplemental material. For most members of this set of transcripts that were upregulated in the triple KO spleens, there was remarkable agreement between the upregulation of exon reads seen in the absence of TTP and the upregulation of intron reads in the same samples. A Pearson's correlation comparing the two triple KO data sets yielded an r value of 0.97, indicating a very strong positive correlation.
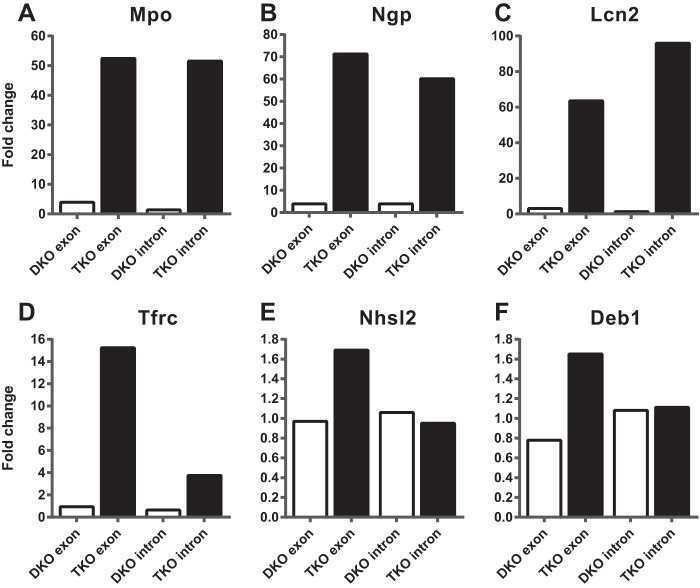
Data from some of the most highly upregulated transcripts are shown in Fig. 8; note that these are illustrative examples and do not contain obvious potential TTP binding sites. Figure 8A to C show the ratios of average exon and intron reads for the double KO over WT and triple KO over WT for three transcripts that are highly expressed in neutrophils: Mpo mRNA, encoding myeloperoxidase (GenBank accession number NP_034954); Ngp mRNA, encoding neutrophilic granule protein precursor (NP_032720); and Lcn2 mRNA, encoding neutrophil gelatinase-associated lipocalin precursor (NP_032517). These values show large increases in the levels of these three transcripts in the triple KO exons compared to their respective controls and also show the minimal increases that occurred in the double KO spleens. Remarkably, the patterns of intron expression were almost identical to the patterns of exon expression for these three transcripts (Fig. 8A to C), strongly suggesting that mature mRNA accumulation was mirrored by pre-mRNA accumulation in these cases. In other words, there was little evidence that the increased exon reads were due to TTP-induced changes in posttranscriptional processes such as mRNA stability. These data strongly suggest that the dramatic changes in accumulation of these neutrophil-enriched transcripts that were seen in the triple KO spleens were due to secondary effects of the systemic TTP deficiency, possibly reflecting an increase in neutrophils in the spleen (see below).
FIG 8.
Exonic versus intronic RNA-Seq reads. (A to C) Ratios of average exon and intron reads for the double KO/WT and the triple KO/WT for three transcripts highly expressed in neutrophils (Mpo, Ngp, and Lcn2 mRNAs). Note that the patterns of intron and exon expression are almost identical in the triple KO spleens. (D to F) Ratios of average exon and intron reads for the double KO/WT and the triple KO/WT for the three transcripts, Tfrc, Nhsl2, and Deb1 mRNAs. Note that the increased exon expression in the triple KO spleen was not closely matched by the patterns of intron expression for these transcripts.
At the other end of the spectrum was a small group of transcripts for which increases in exon expression in the triple KO mice were not clearly matched by commensurate increases in intron expression. These were identified by first comparing “ratios of ratios” for exon and intron reads. For this comparison, we identified upregulated transcripts whose ratios [i.e., (triple KO/WT exons)/(triple KO/WT introns)] were greater than 1.5. There were only 52 transcripts of this type (indicated in green in Table S4 in the supplemental material). For those transcripts whose ratios of ratios were less than 1.5, we also subjectively evaluated the histograms of the relative expression levels and identified five additional transcripts of this type (indicated in red in Table S4 in the supplemental material). Three examples of transcripts selected by these methods are shown in Fig. 8D to F; these are Tfrc mRNA, discussed above, along with Nhsl2 and Deb1 mRNAs. Nhsl2 encodes NHS (Nance-Horan syndrome)-like protein 2 (NP_001157082), and Deb1 encodes SS-18 like protein 2 (NP_081070), thought to be a protein phosphatase 2A regulatory subunit. The Nhsl2 transcript contains two potential TTP binding site 7-mers, only one of which is conserved in humans; the Deb1 transcript contains one potential TTP binding site 8-mer, which is conserved in humans and many other mammals; and the Tfrc transcript's two potential binding sites are discussed above.
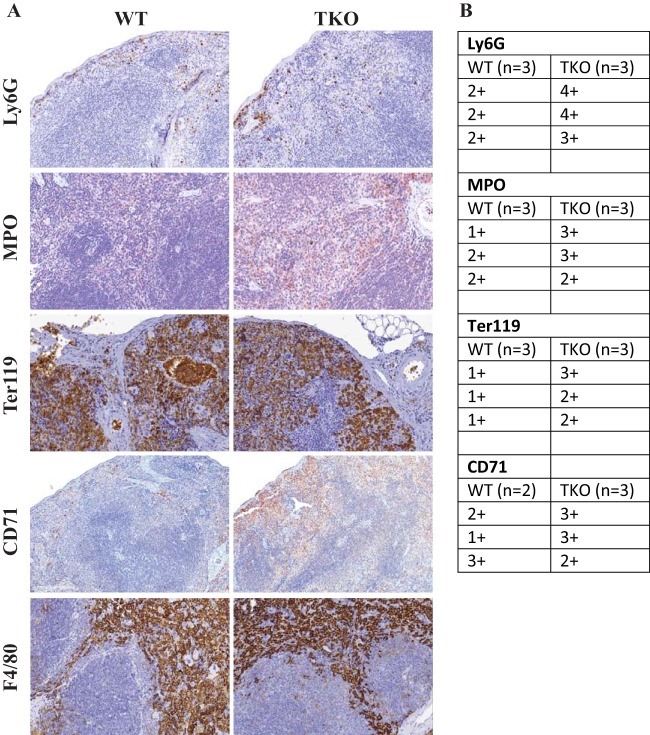
Immunohistochemical staining in triple KO spleen.
Since a large number of the transcripts that were upregulated in the triple KO spleens seemed likely to be secondary responses to TTP deficiency and not direct TTP targets, we sought to determine if genotype-specific changes in splenic cellular composition could be at least partly responsible for such effects. We stained the spleen sections for neutrophils (Ly6G), myeloid cells (myeloperoxidase [Mpo]), macrophages (F4/80), cells of erythroid lineage (Ter119), and erythroid precursors (CD71) (Fig. 9). Immunohistochemically stained slides were graded as described in Materials and Methods. Although not readily apparent in the examples shown in Fig. 9, there were modestly increased numbers of neutrophils and overall cells of myeloid lineage, as demonstrated by consistently higher scores for both Ly6G and Mpo staining, in the triple KO spleens. This could represent either extramedullary myelopoiesis or neutrophil infiltration of the tissue or both. Staining for cells of erythroid lineage using Ter119 antibody demonstrated the presence of increased numbers of cells of erythroid lineage in the triple KO spleens. Staining with the CD71 antibody for transferrin receptors, which are highly expressed on the surface of cells of erythroid lineage, also demonstrated increased erythroid precursors in the triple KO spleens, suggesting possible increased extramedullary hematopoiesis. F4/80 staining for macrophages was not different when comparing the WT and triple KO spleens.
FIG 9.
Immunohistochemical staining of splenic cell populations. (A) Immunohistochemical staining for neutrophils (Ly6G), myeloid cells (Mpo), cells of erythroid lineage (Ter119), erythroid precursors (CD71), and macrophages (F4/80). (B) Grades for positive staining for Ly6G, Mpo, Ter119, and CD71. Ter119 stains both nucleated red cells and mature erythrocytes. For grading, only nucleated red cells were counted.
DCQ of immune cells in double and triple KO spleens.
To further address the question of possible changes in cellular composition of the triple KO spleens, we applied the method of digital cell quantitation (DCQ) (31). This method uses the genome-wide transcriptional profiles of a tissue to identify changes in the quantities of at least 213 immune cell subpopulations. We first compared average enrichment scores (relative cell quantity) of immune cell subtypes between the four WT and four triple KO spleens. Eleven cell subtypes were significantly enriched using this method; these included all six subsets of granulocytes evaluated, one subset (of eight evaluated) of monocytes, one subset (of 18) of macrophages, one subset (of 36) of dendritic cells, one subset (of 12) of NK cells, and one subset (of 14) of stem cells. Thirteen cell subtypes exhibited decreased abundance using this method, including 11 subsets (of 22) of αβ T cells and one subset each of B cells (of 30) and dendritic cells (of 36). In contrast, the spleens from the double KO mice exhibited significant changes in only five cell subtypes, which did not overlap those seen in the triple KO analysis. The upregulated cell subtypes included one each of stromal cells and monocytes, and the downregulated cell subtypes included one each of macrophages, dendritic cells, and γδ T cells. These data support the notion that the cellular composition of the triple KO spleen was somewhat perturbed as a result of the whole-body TTP deficiency, even in the setting of TNF receptor deficiency. The significantly affected cell types are listed in Table S5 in the supplemental material.
DISCUSSION
Most studies of the effects of members of the TTP family of RNA binding and destabilizing proteins on transcript levels have relied largely on the use of cultured cells, in which mRNA decay rates can be monitored readily after knocking out or down the relevant TTP family member. Given TTP's extreme transcriptional inducibility in cultured cells, such experiments have most often utilized acute stimuli, such as LPS stimulation of macrophages, or serum stimulation of fibroblasts, to induce TTP protein and its potential target mRNAs (10, 32). These approaches have yielded valuable information on target mRNAs in specific cell types and have also informed studies on mechanisms of action of this family of proteins. However, little is known about the role of TTP in various tissues and cells of intact animals under normal laboratory conditions, even though TTP mRNA can be detected readily in essentially all tissues in mice under these conditions.
TTP has long been viewed as being of particular relevance to myeloid cells such as macrophages, since they are the primary source of TNF, the cytokine whose excess is largely responsible for the severe, systemic inflammatory syndrome characteristic of TTP deficiency (7, 8, 10). However, two recent reports have found that myeloid cell-specific TTP KO mice do not exhibit the same inflammatory phenotype as the TTP KO mice under normal laboratory conditions; it is not until they are stimulated with low doses of an innate immune system activator like LPS that they exhibit profound TNF excess and endotoxin shock (35, 36). The roles of other cell types, such as fibroblasts, dendritic cells, and Th17 lymphocytes, in the complete TTP deficiency syndrome have also been highlighted recently (37, 38). We therefore have begun to examine the effects of TTP deficiency on transcript levels in the tissues of the adult mouse, in hopes of beginning to define the physiological roles of TTP in whole tissues under unstimulated conditions.
An important consideration is the selection of mouse genotype for studies of this type. Because the severe inflammatory phenotype characteristic of conventional “whole-body” TTP KO mice, largely the consequence of TNF excess, may make interpretation of the results difficult because of the major alterations in tissue morphology and gene expression caused by the chronic TNF excess, we evaluated the effects of TTP deficiency in the setting of TNF receptor deficiency, given that these mice exhibit essentially no abnormal external phenotypes for the first several months of life under laboratory conditions (8). A drawback of this approach is the necessity to evaluate two sets of controls, i.e., WT mice and mice with normal TTP expression but lacking both TNF receptors.
We chose spleen as our initial test organ because of its relatively high level of TTP expression, among the highest in mouse tissues surveyed (16), as well as its importance in immune surveillance. A major drawback of this tissue is its extreme cellular heterogeneity, illustrated by the high-level expression of TTP transcripts in the white pulp of normal spleens, compared to much lower expression levels in the red pulp. However, for the evaluation of the effects of TTP deficiency on transcript levels in an intact tissue, it was reassuring that the phenotype of the triple KO mice at 3 months of age included normal body weight, normal to only slightly enhanced spleen weight, and apparently normal spleen morphology except for possible expansion of the red pulp, at least by staining with hematoxylin and eosin. We could therefore focus on the effects of TTP deficiency on gene expression patterns without too much concern about gross anatomical abnormalities.
Our RNA-Seq analysis appeared to be of adequate depth to determine quantitative differences in gene expression among the three groups of animals, with high percentages of reads being mappable to the reference transcriptome. Using the criteria of FPKM of ≥1.5, at least a 1.5-fold change in mean levels, and a P value of ≤0.05 for the mean comparisons, we found that transcripts from 3,014 genes, or 24.6% of the total number that were expressed at greater than 1.5 FPKM, were significantly increased or decreased in the triple KO spleens compared to the WT controls. This result indicates that a remarkably high fraction of the splenic transcriptome is altered under basal conditions. Therefore, despite the normal external appearance and body weights of the mice and the apparently normal spleen morphology, it is clear that these genetic manipulations had a profound effect on the transcriptome of this tissue. We anticipated that significant increases in transcript levels in the triple KO mice would result from a combination of stabilization of direct mRNA targets of TTP within one or more spleen cell populations and changes in gene expression that were secondary results of TTP deficiency in the spleen as well as in the rest of the body. Similarly, the downregulated transcripts would be expected to be entirely indirect or secondary products of the TTP deficiency but would nonetheless be part of the “molecular phenotype” that characterizes the effects of whole-body TTP deficiency in this tissue.
For the upregulated transcripts, we attempted to gain some insight into the relative numbers of potential direct TTP targets versus indirect or secondary transcript changes by comparing the increases in exon reads, representing mature mRNA, to intron reads, representing nascent pre-mRNA. In general, we found striking positive correlations between the exon and intron reads, suggesting that mRNA and pre-mRNA were affected in parallel in most cases. This was substantiated by comparisons between the exon and intron reads for the evaluable transcripts that were both upregulated and had potential TTP binding sites. Remarkably, we found that approximately 85% of the 385 evaluable transcripts upregulated in triple KO spleens exhibited closely parallel changes in exon and intron reads, suggesting that their changes in steady-state levels in the tissue were largely if not entirely due to changes in transcription that were secondary to the systemic TTP deficiency. This left only 57 transcripts, or 14.8%, that exhibited increased exon reads that were not closely matched by the intron reads, representing possible direct TTP targets in this tissue. There are several potential problems with this approach, including the fact that we did not attempt to isolate nuclear pre-mRNA but relied instead on total cellular RNA from the tissue. Nonetheless, this method for comparing mRNA to pre-mRNA expression can be applied to conventional mRNA-Seq data sets, such as those used in this study, and may be useful in the identification of transcripts that are modified posttranscriptionally.
This initial exon-intron analysis also suggested that some of the transcript changes observed in the triple KO spleens could be due to changes in the cellular composition of the triple KO tissues. In order to test this, we took two different approaches: first, we immunostained the spleen sections for the presence of various cell types, and second, we utilized the digital cell quantitation (DCQ) method (31) to investigate the relative quantities of various immune cell subpopulations in the triple KO spleens. Consistent with the impression of increased EMH observed in the H&E-stained sections of the triple KO mouse spleens, there were increases in staining for neutrophils and overall myeloid cells in the triple KO spleens, suggesting either extramedullary myelopoiesis or neutrophil infiltration of the tissue or both. These results were supported by the DCQ analysis, which suggested enrichment of all six subsets of neutrophils in the triple KO spleens. These results suggest that the major increases in mRNAs encoding neutrophil granule proteins in our data set are the result of increased numbers of neutrophils in the triple KO spleens. Along the same lines, CD71 and Ter119 immunostaining showed evidence of increased extramedullary erythropoiesis, which may be one of the reasons for the enrichment of mRNAs encoding erythrocyte lineage proteins, such as hemoglobins. We suspect that many other changes in transcript levels could be due to other types of secondary effects, such as responses to changes in circulating cytokine levels other than TNF.
We also compared the list of transcripts upregulated in triple KO mice to those known or suspected to be TTP targets from previous literature (11). Only a few of these were also elevated in the present study. Most surprisingly, given that it was the first-discovered and best-studied physiological target of TTP, the TNF mRNA itself was not increased in triple KO spleens (7, 9, 10, 34). This example highlights the differences between previous studies in single cell types, such as LPS-stimulated macrophages, and studies in tissues with heterogeneous mixtures of cells, in which macrophages are both unstimulated and represent a relatively minor fraction of cells. It also may reflect that, at the level of the whole tissue in unstimulated mice, TTP transcripts represented only a minor component of total “TTP equivalents,” with Zfp36l1 mRNA levels being by far the greatest, as indicated by FPKM values. The relative predominance of Zfp36l1 mRNA is seen in most other tissues of the mouse, as determined by mRNA-Seq (see, e.g., reference 39), and it may be that this transcript and its encoded protein play a larger role than TTP in regulating ARE-containing transcript levels in tissues of unstressed organisms.
Nonetheless, we observed several transcripts for which the data support a direct role of the TTP deficiency in mediating their abnormal accumulation in the triple KO spleens. One example of a highly upregulated mRNA with conserved binding sites is the transcript encoding the type 1 transferrin receptor, TFRC. This is a widely expressed type II transmembrane glycoprotein that plays a critical role in transmembrane iron transport (40). It exhibited increased numbers of exon reads in the triple KO spleens that appeared to be out of proportion to the changes in intron reads. Its mRNA contains two ideal potential TTP binding sites that are conserved in many mammalian species. Its protein expression was also significantly increased. These changes were accompanied by greatly increased iron deposition in the spleens of the triple KO mice but not in the WT or double KO mice, raising the possibility that the increased TFRC expression plays a role in this pathology. TFRC mRNA has recently been suggested as a TTP target in experiments involving cultured mouse embryonic fibroblasts (MEFs) from TTP KO mice (41). A link between the two TTP family members expressed in Saccharomyces cerevisiae, CTH1p and CTH2p, and the regulation of cellular iron homeostasis also has been demonstrated previously (42, 43). Although the TFRC mRNA may well be a direct target of TTP in one or more splenic cell types, we cannot exclude a primary or contributory secondary effect of TTP deficiency on Tfrc gene transcription.
A possible alternative mechanism for the enhanced iron deposition is an increase in heme biosynthesis. This pathway comprises nine enzymes, with the first and the rate-limiting step represented by delta-aminolevulinate synthase (ALAS). This activity is expressed as two distinct proteins: a housekeeping form (ALAS1) and an erythroid cell-specific form (ALAS2). Previous studies have shown a strong correlation between the expression levels of Alas2 mRNA and most other mRNAs of heme pathway enzymes (44), which could explain the elevated expression of mRNAs encoding all nine enzymes in the triple KO spleens, even though none of these mRNAs contains an optimal TTP binding site in its 3′UTR. Physiologically, chronic inflammation is known to cause iron accumulation in macrophages of the reticuloendothelial system (45, 46). This can lead to anemia resulting from functional iron deficiency, also known as “anemia of chronic disease” or anemia of inflammation. Our results may represent a similar condition, in which TTP deficiency leads to chronic inflammation and increased expression of the transferrin receptor, resulting in iron accumulation in the spleen.
In conclusion, TTP deficiency, even in the setting of TNF receptor deficiency, can lead to many gene expression changes in the spleen, resulting in a molecular phenotype for this syndrome in this tissue. Our data suggest that relatively few of the transcripts changed under these conditions are likely to be direct targets for TTP, indicating that most of the changes in mRNA accumulation in this setting either are indirect responses to systemic changes in, for example, circulating cytokine levels, or are due to changes in cellular infiltration. Other types of experiments will be necessary to sort out the specific splenic cell types in which TTP deficiency leads to accumulation of direct target transcripts, including potential overexpression studies, studies with mice stimulated with, e.g., LPS, thiouracil labeling (47), cell-specific KOs, and mRNA immunoprecipitation studies, possibly combined in each case with sorting of relevant cell populations. These data also suggest that parallel studies in other tissues might reveal other regulatory possibilities in tissues and cells not previously evaluated for roles of TTP in normal physiology.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to the NIH Intramural Sequencing Center for the RNA-Seq data. We thank Norris Flagler for digital quantitation of slides and the Histology and the Immunohistochemistry cores for technical support. We also thank Mike Fessler and Don Cook for critical comments on the manuscript.
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences and National Institute of Mental Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01068-15.
REFERENCES
- 1.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. 2004. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J Biol Chem 279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 2.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. 2002. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J Biol Chem 277:48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- 3.Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X, Zhou P. 2003. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J Biol Chem 278:19947–19955. doi: 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- 4.Lai WS, Carrick DM, Blackshear PJ. 2005. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J Biol Chem 280:34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- 5.Lai WS, Perera L, Hicks SN, Blackshear PJ. 2014. Mutational and structural analysis of the tandem zinc finger domain of tristetraprolin. J Biol Chem 289:565–580. doi: 10.1074/jbc.M113.466326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai WS, Kennington EA, Blackshear PJ. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol 23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. 1996. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445–454. doi: 10.1016/S1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 8.Carballo E, Blackshear PJ. 2001. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 98:2389–2395. doi: 10.1182/blood.V98.8.2389. [DOI] [PubMed] [Google Scholar]
- 9.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. 1999. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 19:4311–4323. doi: 10.1128/MCB.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 11.Brooks SA, Blackshear PJ. 2013. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta 1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross CR, Brennan-Laun SE, Wilson GM. 2012. Tristetraprolin: roles in cancer and senescence. Ageing Res Rev 11:473–484. doi: 10.1016/j.arr.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanduja S, Blanco FF, Young LE, Kaza V, Dixon DA. 2012. The role of tristetraprolin in cancer and inflammation. Front Biosci (Landmark Ed) 17:174–188. doi: 10.2741/3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciais D, Cherradi N, Feige JJ. 2013. Multiple functions of tristetraprolin/TIS11 RNA-binding proteins in the regulation of mRNA biogenesis and degradation. Cell Mol Life Sci 70:2031–2044. doi: 10.1007/s00018-012-1150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai WS, Stumpo DJ, Kennington EA, Burkholder AB, Ward JM, Fargo DL, Blackshear PJ. 2013. Life without TTP: apparent absence of an important anti-inflammatory protein in birds. Am J Physiol Regul Integr Comp Physiol 305:R689–R700. doi: 10.1152/ajpregu.00310.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao H, Tuttle JS, Blackshear PJ. 2004. Immunological characterization of tristetraprolin as a low abundance, inducible, stable cytosolic protein. J Biol Chem 279:21489–21499. doi: 10.1074/jbc.M400900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Goeddel DV. 2002. TNF-R1 signaling: a beautiful pathway. Science 296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 18.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B Met 57:289–300. [Google Scholar]
- 25.Lai WS, Stumpo DJ, Blackshear PJ. 1990. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem 265:16556–16563. [PubMed] [Google Scholar]
- 26.Blackshear PJ, Manzella JM, Stumpo DJ, Wen L, Huang JK, Oyen O, Young WS III. 1989. High level, cell-specific expression of ornithine decarboxylase transcripts in rat genitourinary tissues. Mol Endocrinol 3:68–78. doi: 10.1210/mend-3-1-68. [DOI] [PubMed] [Google Scholar]
- 27.Young WS III, Mezey E, Siegel RE. 1986. Quantitative in situ hybridization histochemistry reveals increased levels of corticotropin-releasing factor mRNA after adrenalectomy in rats. Neurosci Lett 70:198–203. doi: 10.1016/0304-3940(86)90463-5. [DOI] [PubMed] [Google Scholar]
- 28.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth G. 2005. Limma: linear models for microarray data, p 397–420. In Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY. [Google Scholar]
- 30.Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altboum Z, Steuerman Y, David E, Barnett-Itzhaki Z, Valadarsky L, Keren-Shaul H, Meningher T, Mendelson E, Mandelboim M, Gat-Viks I, Amit I. 2014. Digital cell quantification identifies global immune cell dynamics during influenza infection. Mol Syst Biol 10:720. doi: 10.1002/msb.134947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. 2006. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol 26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blackshear PJ, Phillips RS, Ghosh S, Ramos SB, Richfield EK, Lai WS. 2005. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol Reprod 73:297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 34.Carballo E, Gilkeson GS, Blackshear PJ. 1997. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (−/−) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFalpha overproduction. J Clin Invest 100:986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu LQ, Stumpo DJ, Blackshear PJ. 2012. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J Immunol 188:5150–5159. doi: 10.4049/jimmunol.1103700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kratochvill F, Machacek C, Vogl C, Ebner F, Sedlyarov V, Gruber AR, Hartweger H, Vielnascher R, Karaghiosoff M, Rulicke T, Muller M, Hofacker I, Lang R, Kovarik P. 2011. Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Mol Syst Biol 7:560. doi: 10.1038/msb.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molle C, Zhang T, Ysebrant de Lendonck L, Gueydan C, Andrianne M, Sherer F, Van Simaeys G, Blackshear PJ, Leo O, Goriely S. 2013. Tristetraprolin regulation of interleukin 23 mRNA stability prevents a spontaneous inflammatory disease. J Exp Med 210:1675–1684. doi: 10.1084/jem.20120707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu LQ, Lai WS, Bradbury A, Zeldin DC, Blackshear PJ. 2015. Tristetraprolin (TTP) coordinately regulates primary and secondary cellular responses to proinflammatory stimuli. J Leukoc Biol 97:723–736. doi: 10.1189/jlb.3A0214-106R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darshan D, Frazer DM, Anderson GJ. 2010. Molecular basis of iron-loading disorders. Expert Rev Mol Med 12:e36. doi: 10.1017/S1462399410001687. [DOI] [PubMed] [Google Scholar]
- 41.Bayeva M, Khechaduri A, Puig S, Chang HC, Patial S, Blackshear PJ, Ardehali H. 2012. mTOR regulates cellular iron homeostasis through tristetraprolin. Cell Metab 16:645–657. doi: 10.1016/j.cmet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puig S, Askeland E, Thiele DJ. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 43.Puig S, Vergara SV, Thiele DJ. 2008. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab 7:555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meguro K, Igarashi K, Yamamoto M, Fujita H, Sassa S. 1995. The role of the erythroid-specific delta-aminolevulinate synthase gene expression in erythroid heme synthesis. Blood 86:940–948. [PubMed] [Google Scholar]
- 45.Theurl I, Aigner E, Theurl M, Nairz M, Seifert M, Schroll A, Sonnweber T, Eberwein L, Witcher DR, Murphy AT, Wroblewski VJ, Wurz E, Datz C, Weiss G. 2009. Regulation of iron homeostasis in anemia of chronic disease and iron deficiency anemia: diagnostic and therapeutic implications. Blood 113:5277–5286. doi: 10.1182/blood-2008-12-195651. [DOI] [PubMed] [Google Scholar]
- 46.Weiss G, Goodnough LT. 2005. Anemia of chronic disease. N Engl J Med 352:1011–1023. doi: 10.1056/NEJMra041809. [DOI] [PubMed] [Google Scholar]
- 47.Gay L, Karfilis KV, Miller MR, Doe CQ, Stankunas K. 2014. Applying thiouracil tagging to mouse transcriptome analysis. Nat Protoc 9:410–420. doi: 10.1038/nprot.2014.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.