Abstract
The yeast spindle pole body (SPB) is the functional equivalent of the mammalian centrosome. Centrosomes and SPBs duplicate exactly once per cell cycle by mechanisms that use the mother structure as a platform for the assembly of the daughter. The conserved Sfi1 and centrin proteins are essential components of the SPB duplication process. Sfi1 is an elongated molecule that has, in its center, 20 to 23 binding sites for the Ca2+-binding protein centrin. In the yeast Saccharomyces cerevisiae, all Sfi1 N termini are in contact with the mother SPB whereas the free C termini are distal to it. During S phase and early mitosis, cyclin-dependent kinase 1 (Cdk1) phosphorylation of mainly serine residues in the Sfi1 C termini blocks the initiation of SPB duplication (“off” state). Upon anaphase onset, the phosphatase Cdc14 dephosphorylates Sfi1 (“on” state) to promote antiparallel and shifted incorporation of cytoplasmic Sfi1 molecules into the half-bridge layer, which thereby elongates into the bridge. The Sfi1 C termini of the two Sfi1 layers localize in the bridge center, whereas the N termini of the newly assembled Sfi1 molecules are distal to the mother SPB. These free Sfi1 N termini then assemble the new SPB in G1 phase. Recruitment of Sfi1 molecules into the anaphase SPB and bridge formation were also observed in Schizosaccharomyces pombe, suggesting that the Sfi1 bridge cycle is conserved between the two organisms. Thus, restricting SPB duplication to one event per cell cycle requires only an oscillation between Cdk1 kinase and Cdc14 phosphatase activities. This clockwork regulates the “on”/“off” state of the Sfi1-centrin receiver.
INTRODUCTION
The mammalian centrosome and the yeast spindle pole body (SPB) are both able to nucleate microtubules (MTs) from tubulin subunits and are therefore collectively named MT organizing centers (MTOCs). A second common feature is the duplication of the centrosome and SPB just once during each cell division cycle. In each case, it is the mother centrosome or SPB that provides the platform for the assembly of the daughter (1–3).
In yeast, the SPB is absolutely critical for mitotic spindle formation and chromosome segregation. This is because alternative MT assembly pathways, such as the RCC1/Ran-GTP- or augmin-dependent MT nucleation pathways, are absent from yeast (4, 5). In contrast, human cells can assemble a mitotic spindle even in the absence of centrosomes because of centrosome-independent MT formation and the presence of spindle assembly pathways. Recent studies have shown that the ability of human cells to tolerate loss of centrosomes or centrosome overduplication is reliant upon the inactivation of the p53 tumor suppressor gene that otherwise arrests these abnormal cells in G1 phase (6, 7). How cells sense centrosome number defects is currently not understood. Yet it is becoming clearer why such a surveillance mechanism is important. Centrosome overamplification may contribute to cell transformation or enhance the aggressive nature of already transformed cells (8, 9). It is therefore not surprising that overamplification of centrosomes is a common feature of cancer cells. Thus, understanding how cells control centrosome and SPB duplication is an important mission objective.
The mammalian centrosome consists of two or four centrioles, depending on the cell cycle phase. Centrioles are barrels of nine triplet MTs and carry the duplication capacity of centrosomes. They also provide a scaffold upon which the pericentriolar material (PCM) can assemble. The PCM is a network of proteins that surrounds the centrioles to anchor MT nucleation activity in the form of γ-tubulin complexes to centrosomes (1). In contrast, the SPB does not contain centrioles. Instead, SPBs are multilayered or amorphous structures consisting of multiple copies of a small subset of proteins. In budding yeast, only 18 proteins constitute the SPB, fewer than the complement of the mammalian centrosome, which consists of >50 proteins (2, 10). Because the yeast nuclear envelope (NE) remains intact in mitosis (“closed mitosis”), SPBs become embedded in the NE in a way similar to that seen with nuclear pore complexes. The Saccharomyces cerevisiae SPB is embedded in the NE throughout the cell cycle. In Schizosaccharomyces pombe, the SPB sits on the cytoplasmic side of the NE during interphase and becomes inserted into the NE prior to mitosis. This process is reversed with mitotic exit (11, 12). NE insertion enables the SPB to simultaneously organize two spatially separated sets of MT arrays, the nuclear and the cytoplasmic MTs, with functions in chromosome segregation in mitosis and nuclear positioning, respectively. Here we discuss recent developments in analysis of the molecular mechanism of SPB duplication in the model organisms S. cerevisiae (budding yeast) and S. pombe (fission yeast). We compare these findings with human centriole duplication data.
SPB DUPLICATION IN BUDDING YEAST
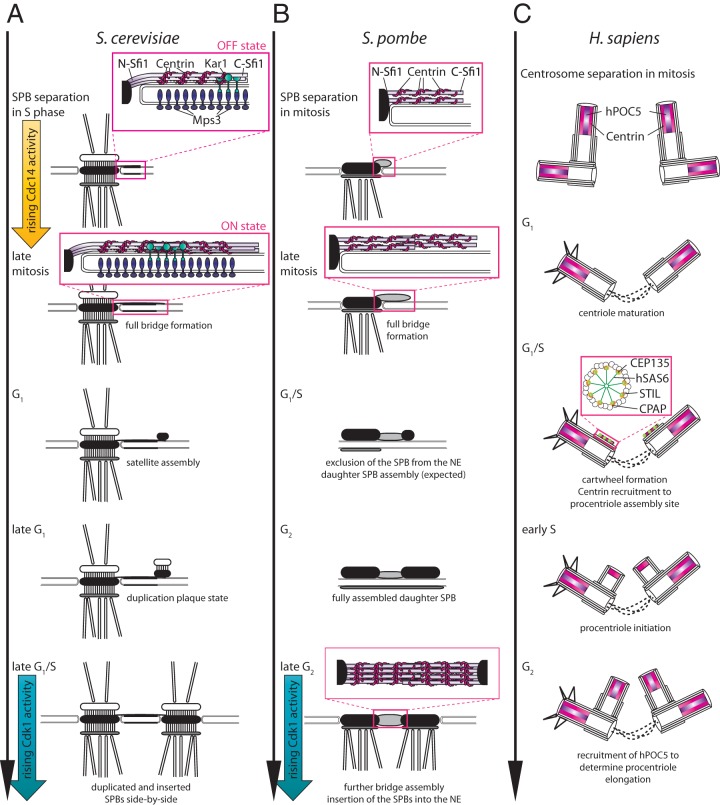
The budding yeast SPB is a multilayered structure. The central plaque is the SPB substructure that is close to the NE. The outer and inner plaques on the cytoplasmic and nuclear sides of the SPB organize the nuclear and cytoplasmic MTs, respectively. The half-bridge is an additional SPB substructure that is important for SPB duplication. The half-bridge is attached to one side of the central plaque and sits on top of the nuclear and cytoplasmic sides of the NE. In late mitosis, the half-bridge doubles its length to develop into the bridge structure (13–15). It is the distal end of the bridge that, in G1 phase, assembles the daughter SPB precursor, the satellite (16, 17). With rising cyclin-dependent kinase 1 (Cdk1) activity, the satellite enlarges and becomes embedded in the NE. Upon NE insertion, the nuclear half of the SPB assembles from within the nucleus (18–20). The two side-by-side SPBs are at first still connected by the bridge (17). In order to facilitate bipolar spindle formation, the two SPBs separate through fission of the SPB bridge center in a process that is largely driven by Cdk1 activity in S phase (21–23) (Fig. 1A).
FIG 1.
Comparison of SPB and centrosome duplication results. (A) The SPB duplication cycle in S. cerevisiae. In S phase, each of the two separated SPBs carries a half-bridge (exemplarily, only one SPB is shown), which becomes duplicated into a full bridge in late mitosis upon the activation of the phosphatase Cdc14 in anaphase (“on” state). The distal tip of the bridge serves as an assembly platform for the satellite, the SPB precursor, which grows after start into the duplication plaque and ultimately becomes inserted into the nuclear envelope. This results in two side-by-side SPBs. For spindle assembly in mitosis, the bridge has to become severed in its center. This process is promoted by Cdk1 activity (“off” state). (B) The duplicated S. pombe SPBs become embedded in the nuclear envelope in G2/M phase at the same time as the bridge is severed into two half-bridges promoted by mitotic Cdk1 activity. We propose that the orientation of SpSfi1 molecules within the half-bridge is the same as in budding yeast: the N terminus of all SPB-associated SpSfi1 molecules is next to the mother SPB (mSPB); C-SpSfi1 is distal to the mSPB. Shortly after mitotic entry, while the core SPB is still embedded in the NE, the half-bridge starts its first SpSfi1 recruitment phase, which probably results in the formation of the full bridge. After exclusion of the SPB from the NE in G1 phase, there is most probably, in analogy to the S. cerevisiae SPB, the formation of a daughter S. pombe SPB precursor, which grows further during S/G2 phase. (C) Centrosome duplication in human cells. The localization of human centrin and the centrin-binding protein hPOC5 is shown. Since little is known about the exact localization of hSFI1 or its function, it was not included in this cartoon. In G2 phase/mitosis, the centrosome linker resolves and two centrosomes (a pair of two centrioles) are instrumental to organize the mitotic spindle. After completion of mitosis, G1-phase cells have two centrioles (a mother and a daughter), which are connected by the centrosome linker (dotted line). Centriole duplication is initiated in G1/S phase with the recruitment of cartwheel proteins (hSAS6, CEP135, STIL, and CPAP) and centrin at the procentriole assembly site. In early S phase, the procentrioles form and recruit the centrin-binding protein hPOC5 to their distal end in G2 phase. hPOC5 drives procentriole elongation.
The steps that ultimately lead to SPB duplication have been known since the analysis of the SPB duplication cycle by electron microscopy (EM) in the 1970s (16). However, a molecular understanding of budding yeast SPB duplication was achieved only with the discovery of half-bridge/bridge components. The yeast centrin CDC31 gene was identified in the famous Hartwell screen for conditional lethal cell cycle mutants (24, 25). cdc31-1 cells fail to duplicate the SPB at the restrictive temperature and arrest cell cycle progression in mitosis due to the stimulation of the spindle assembly checkpoint (SAC) (26–28). Cdc31, like calmodulin, consists of 4 EF hands that have the potential to bind Ca2+. KAR1 was originally discovered as a gene involved in karyogamy (29). It was subsequently found that it has a second and essential function in SPB duplication (30). Cdc31 binds directly to Kar1, and elegant genetic analysis performed in the Rose laboratory identified a complex genetic relationship between the CDC31 and KAR1 genes (31–33). A screen for Cdc31 interacting proteins identified Sfi1, which, like centrin, is a conserved component of MTOCs (34). The gene encoding the SUN domain protein Mps3 (monopolar spindle 3), an integral membrane protein, was identified by screening a temperature-sensitive collection of yeast cells for defects in SPB duplication (35, 36). Kar1, Cdc31, and Sfi1 are components of the cytoplasmic side of the half-bridge (27, 31, 34). In contrast, the integral membrane protein Mps3 sits on the nuclear side (37). Because relatively little is known about the molecular functions of Mps3 that promote and support SPB duplication, we do not discuss Mps3 further here.
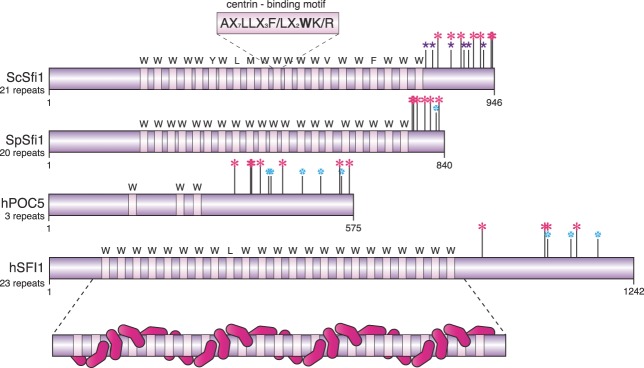
Understanding the molecular mechanisms that underlie the initial steps of SPB duplication has relied upon structural data, superresolution microscopy, the SPB duplication phenotype of SFI1 phosphomutants, mathematical modeling, and biochemistry. One key observation was the finding by Kilmartin and colleagues that Sfi1 is a long, mainly α-helical protein that spans the entire length of the cytoplasmic side of the half-bridge (15, 34). Immuno-EM and superresolution data suggest that all Sfi1 molecules in the half-bridge have the same orientation: the Sfi1 N termini are associated with the central plaque of the mother SPB, while the C terminus is placed in a position distal from the central plaque (13–15, 34). This suggests that the long α-helical central part that contains the 21 Cdc31 binding sites (each 23 to 35 amino acids in length) is aligned along the cytoplasmic side of the NE and is the principal determinant of the length of the half-bridge (Fig. 2). An additional important observation was that the first step in SPB duplication, the conversion from the half-bridge to the bridge, is driven by the antiparallel and staggered incorporation of cytoplasmic Sfi1 molecules into the SPB-bound Sfi1 (13–15). This means that, within the bridge, the C termini of the two antiparallel Sfi1 layers sit at the center (14, 34). The N termini of the newly incorporated Sfi1 molecules are located in a position distal from the mother SPB (and ∼120 nm away). It has been proposed that these free N-Sfi1 molecules provide a platform upon which the satellite can assemble on the cytoplasmic side of the NE (15, 34). However, how N-Sfi1 regulates and initiates satellite assembly is not yet understood.
FIG 2.
Domain alignment of yeast and human centrin-binding proteins. The centrin-binding motifs (marked with W, Y, L, M, V, or F) of ScSfi1, SpSfi1, hPOC5 (isoform 1), and hSFI1 (isoform a) are shown schematically. The predicted centrin binding consensus motif of SFI1 molecules is shown in the box above ScSfi1. In addition, centrin (pink) binding to hSFI1 is indicated. Structural data suggest that each centrin binds to binding sites at SFI1 at a 60° angle with respect to the precursor centrin. Cyclin-dependent kinase and polo-like kinase phosphorylation sites within the C terminus of ScSfi1, SpSfi1, hPOC5, and hSFI1 were either published (ScSfi1) or predicted by the GPS software. Purple asterisk, confirmed Cdk1 site; blue asterisk, predicted cyclin-dependent kinase site; pink asterisk, predicted polo-like kinase site.
Cdk1 has both a positive impact and a negative impact on promoting and inhibiting SPB duplication at different cell cycle phases (38). Thus, it is likely that Cdk1 directly regulates one or several of the proteins that together assemble the half-bridge and bridge. In this respect, the observation that six Cdk1 sites in the C terminus of Sfi1 regulate bridge formation was a breakthrough (21–23) (Fig. 2). Analysis of phosphoinhibitory and phosphomimetic SFI1 mutants revealed a key role for phosphorylation of C-Sfi1 by Cdk1 in both the fission of the bridge in S phase and the prevention of SPB reduplication in mitosis (“off” state). This analysis also explains why SPB duplication is restricted to G1 phase, when Cdk1 activity is low. The conserved phosphatase Cdc14 dephosphorylates Sfi1 (22, 23, 39) to promote the conversion of the half-bridge to the full bridge (“on” state). Because budding yeast Cdc14 becomes active at anaphase onset (40), this relationship indicates that Sfi1 is already incorporated into the half-bridge during anaphase. This timing was confirmed through the reappraisal of bridge assembly by fluorescence quantification that revealed incorporation of Sfi1 into the SPB already during mitosis (14). Structured illumination microscopy (SIM) showed that the step of conversion of half-bridge to bridge accompanies this anaphase incorporation (13, 14). This revised timing now places the conversion of half-bridge to bridge earlier in the cell cycle than the time at which it had originally been concluded to occur as a result of analyses by electron microscopy (13, 16). Strikingly, premature activation of Cdc14 in early mitosis can induce SPB overduplication. Breakage of the Cdk1/Cdc14 cycle alone is sufficient to induce SPB duplication (23). Thus, the basic regulatory machinery of the SPB of the budding yeast can be accounted for by oscillations of alternating activities of Cdk1 kinase and Cdc14 phosphatase. The C terminus of Sfi1 seems to be the main target of this clockwork relationship.
The simple elegance of the interplay between Sfi1 and Cdk1/Cdc14 oscillations raises the issue of the underlying molecular basis of the complex genetic interactions between CDC31 and KAR1 that was revealed 20 years ago by Rose and coworkers (33, 41). Since Cdc31 interacts with a region of Kar1 that is important for SPB duplication (31), genetic interactions between the two genes should be anticipated. Mutations in KAR1 that affect Cdc31 binding may be compensated for by adaptive mutations in CDC31. However, mutations in CDC31 that change certain acidic amino acids to hydrophobic residues (e.g., in cdc31-16; D131N) suppress the deletion of the normally essential KAR1 gene (32, 33). This finding is inconsistent with such a simple KAR1-CDC31 reciprocal amino acid exchange suppression model and so is suggestive of a more complex functional relationship between KAR1 and CDC31.
Electron microscopic analysis of the viable kar1Δ cdc31-16 cells revealed that the Sfi1 layer was no longer aligned parallel to the nuclear envelope, as seen in KAR1 wild-type and KAR1 cdc31-16 cells (14). This bridge arching phenotype is consistent with a role for Kar1 in anchoring the central portion of the Sfi1-Sfi1 layer on the NE via the embedding of its hydrophobic C-terminal tail in the outer NE membrane (42). This model is further supported by the observation that in vivo cross-linking between Sfi1-green fluorescent protein (Sfi1-GFP) and the NE-tethered GFP-binding protein (GBP) bypassed the normally essential requirement for the region of Kar1 that interacts with Sfi1-Cdc31 (14). Furthermore, in vitro binding assays revealed an interaction between Kar1 and the C terminus of Sfi1 and C-terminal Cdc31 binding sites but not with Sfi1-N and Cdc31 binding sites close to the Sfi1 N terminus. These data are indicative of a preferential interaction between Kar1 and Sfi1 in the center of the bridge that harbors the C termini from the two opposing Sfi1 layers. The placement of Kar1 in the bridge center by PALM and dSTORM superresolution microscopy using two different labeling approaches (mMaple and GFP nanobodies) further supports this model (14). A single-particle averaging SIM approach detected Kar1 along the bridge. However, fluorescence intensity measurements revealed a localization of yellow fluorescent protein-Kar1 (YFP-Kar1) along the mother-satellite axis that was strikingly reminiscent of that of the C terminus of Sfi1 (15), albeit with a slight shift in the full-width half-maximum value by 20 nm (see Fig. 3D in reference 13). These data are fully consistent with binding of Kar1 to Sfi1 C termini and the adjacent Cdc31 binding sites.
The arched bridge phenotype raises the issue of how the bridge is stabilized in kar1Δ cdc31-16 cells. At the heart of this issue is the finding that Cdc31 can decorate the center of Sfi1 through recruitment to around 21 conserved centrin-binding motifs (15, 34). In vitro studies by Li et al. (15) indicated that predicted Cdc31 binding sites in several Sfi1 subfragments are fully occupied by Cdc31. Currently, it is unclear how many of these sites in Sfi1 are occupied by Cdc31 in vivo. Based on SIM averaging, it was suggested that an overexpressed YFP-Cdc31 product associates asymmetrically with the bridge (13). However, this result is surprising in light of the uniform distribution of the Cdc31 binding sites through the central region of Sfi1. An explanation may lie in the fact that tagging of Cdc31 impairs its function such that the localization of the tagged molecule used in this study may not reflect the distribution of wild-type Cdc31 (13, 43). In this respect, it is interesting that localization of wild-type Cdc31 by immuno-EM with antibodies did not give this type of asymmetric distribution; rather, the label was uniformly detected along the entire length of the half-bridge and bridge (31).
Li and colleagues further analyzed the structure of several Sfi1-Cdc31 crystals (15). Because of longitudinal interactions between Cdc31 molecules that were bound to neighboring binding sites of Sfi1, we suggest that the Cdc31 scaffold stabilizes the long α-helix of Sfi1 (15). One Sfi1-Cdc31 crystal showed the structure of Sfi1-Cdc31 filaments with antiparallel orientation. This structure revealed lateral interactions between Cdc31 molecules of neighboring strands (15). Most notably, D107, D131, and E148 sat at the interface between two trans-interacting Cdc31 molecules. Interestingly, these residues are the mutated ones in the kar1Δ suppressing alleles: CDC31-12 (E148A), CDC31-14 (D107Y), CDC31-16 (D131N), and CDC31-17 (E148Q) (33). Free energy calculations of the Cdc31 interactions indicated a gain in free energy upon mutation of E148, D107, or E148 to hydrophobic or less polar amino acids. Although the gain in free energy for each individual interaction site would be small, the stabilizing impact of each mutation would be additive due to the putative overlap of ∼50 antiparallel Sfi1 molecules in the bridge center (14).
This model predicts overlaps between Cdc31 binding sites from the two antiparallel Sfi1 layers at the center of the bridge. The finding that the C terminus of Sfi1 (i.e., the entire C terminus, including the 6 Cdk1 sites [sfi1ΔC, 151 codons—or 54 codons in the case of sfi1-229]) can be deleted without impairing viability or bridge stability supports this model (14, 21). However, this is only the case at lower temperatures since the bridge of G1/S-phase-arrested sfi1ΔC cells is unstable and breaks at 37°C (14).
On the basis of these findings, we propose that the C-terminal region of Sfi1 is the receiver of cell cycle signals because Cdk1 phosphorylation sites in this region regulate the formation of the antiparallel Sfi1 arrays. Phosphoinhibitory mutations in these sites impair bridge severing, and the two SPBs remain joined. These cells therefore arrest cell cycle progression in mitosis with a monopolar spindle. However, deleting the C terminus of Sfi1 supports growth at 23°C. Because the bridge-stabilizing forces in sfi1ΔC cells are modest, as they rely solely upon the overlapping antiparallel Sfi1-Cdc31 sites (with probably only approximately 3 to 4 sites per antiparallel Sfi1 dimer), bridge severing can now be executed by the kinesin-5 Cin8 motor that pushes the two S-phase SPBs apart to induce a stress that physically breaks the bridge independently of the normal control at Sfi1 C termini (14, 21). In sfi1ΔC cells, cell cycle-dependent regulation of bridge fission arises from the cell cycle-dependent accumulation of Cin8 in S phase (44). The Cdh1 anaphase-promoting complex (APCCdh1) that is itself, in turn, controlled by Cdk1 and Cdc14 regulates Cin8 stability during the cell cycle (45, 46).
SPB DUPLICATION IN FISSION YEAST
The fission yeast SPB is an amorphous structure that is not constantly embedded in the nuclear envelope. The mother and daughter SPBs reside on the cytoplasmic side of the NE and insert into an opening within the NE during G2/M phase before the two SPBs separate (11, 12, 47). In G1 phase, the SPBs move back to sit, once more, on the cytoplasmic side of the NE (Fig. 1B).
The SPB duplication process in fission yeast is much less well characterized than in budding yeast because SPB substructures and duplication intermediates are not resolved as well by electron microscopy as they are in budding yeast. However, immunoelectron microscopy localized fission yeast Cdc31 to the bridge of side-by-side SPBs or to a site next to SPBs in an anaphase cell (48). S. pombe Sfi1 (SpSfi1) perfectly colocalizes with Cdc31 at the SPB half-bridge/bridge in high-resolution fluorescence microscopy, and both proteins are essential for SPB duplication (48–50). The interdependency of the SpSfi1-Cdc31 SPB localization is consistent with an interaction of the two proteins (49). Interestingly, electron micrographs reveal a much thicker bridge in fission yeast than in its budding yeast counterpart (12, 51). One could speculate that the thickness of the budding yeast bridge may be accounted for by the presence of a single S. cerevisiae Sfi1 (ScSfi1)-Cdc31 molecule layer, whereas in fission yeast SpSfi1-Cdc31 may represent a number of layers that are piled into a broader assembly. Perhaps this arrangement in S. pombe is stabilized by a SpSfi1-SpCdc31 cross-linking protein.
Electron microscopy studies have so far failed to pinpoint the time at which half-bridge elongation takes place in fission yeast. However, light microscopy revealed an interesting biphasic nature with respect to the incorporation of SpSfi1 into the mother SPB after SPB separation with mitotic entry (49, 50). The first phase, during which SpSfi1 levels at the SPB increase 1.5-fold, stretches from early anaphase until mid-septation. The second phase, exhibiting slightly slower incorporation, persists from septation to the end of the G2 phase. While these experiments document incorporation of SpSfi1 into the SPB, they do not resolve the issue of when in the cell cycle the conversion from half-bridge to bridge takes place. However, elegant execution point experiments showed that it is SpSfi1-Cdc31 that becomes incorporated during early anaphase that is critical for SPB duplication in the next cell cycle (49). Therefore, most likely, the half-bridge expands into the bridge during this cell cycle phase in a manner similar to that seen with budding yeast SPBs (13). Interestingly, one-third of SpSfi1 molecules dissociate from the SPBs at mitotic onset as SPBs separate. Such a large-scale departure suggests the presence of a pool of loosely attached SpSfi1 proteins (49). The function of this SpSfi1 pool is unclear. This phase of SpSfi1 loss contrasts with bridge fission in budding yeast, which is accompanied by an even distribution of ScSfi1 between the two SPBs without the obvious loss of any ScSfi1 molecules (14). Superresolution microscopy not only should reveal SpSfi1 orientation in the half-bridge/bridge but also should provide a better insight into the nature and early interphase function of the dissociating SpSfi1 pool.
As outlined above, the behavior of S. pombe Sfi1 is somehow more complex than that of its homologue in budding yeast. The regulation of the SpSfi1-centrin complex most likely also differs between the two organisms. As mentioned before, phosphoregulation of the ScSfi1 C terminus by Cdk1 and Cdc14 controls ScSfi1 in budding yeast (22, 23). Puzzlingly, the C terminus of fission yeast Sfi1 is much shorter than that of its budding yeast counterpart and contains only one Cdk1 consensus site (Fig. 2). Whether this site is used to regulate SpSfi1 at the SPB has not yet been addressed. Interestingly, SpSfi1 harbors 6 putative sites for phosphorylation by the polo-like kinase (named Plo1 for fission yeast). This raises the possibility that fission yeast Plo1 has assumed the function executed by Cdk1 in budding yeast. In this respect, it is interesting that the budding yeast polo-like kinase (named Cdc5) also participates, to some extent, in ScSfi1 regulation (22) (Fig. 2). While Plo1 phosphorylation of SpSfi1 is an untested possibility, it is now quite clear that phosphorylation of fission yeast Cdc31 at serine 15 by Cdk1 promotes bridge separation into the two half-bridges (49). However, as Cdc31S15A mutant cells are only delayed in SPB separation, additional mechanisms must be in place to promote bridge severing in fission yeast. This additional mechanism could be mechanical, as promoted by the kinesin Cin8 in budding yeast (14, 44), or regulatory (SpSfi1-Cdc31 phosphorylation by Cdk1 or Plo1) or a combination of the two. Interestingly, S. cerevisiae Cdc31 does not contain Cdk1 phosphorylation sites in the N-terminal extension. Thus, it remains to be determined whether ScCdc31 is also regulated to promote either bridge assembly or bridge severing.
Taking the data together, we propose that antiparallel duplication of the Sfi1 layer is the licensing event for SPB duplication in budding yeast and, most likely, also in fission yeast. Phosphorylation of ScSfi1 and SpCdc31 by Cdk1 promotes bridge severing after SPB duplication, depending on the organism. Currently, it is unclear whether the fission yeast genome encodes a KAR1 homologue. Thus, we do not understand how the half-bridge/bridge is anchored to the nuclear envelope in S. pombe.
CENTRIN AND CENTRIN-BINDING PROTEINS AT HUMAN CENTROSOMES
Centrin is essential for MTOC duplication in yeast, Tetrahymena thermophile, and Chlamydomonas reinhardtii (32, 33, 48, 52, 53). The literature pertaining to centrin function in mammalian cells is much more controversial. Early small interfering RNA (siRNA) depletion experiments investigating human centrin 2 (hCetn-2) (one of three human centrin genes) in HeLa cells revealed an essential role in centriole duplication (54). In contrast, hCetn-2 deletion in the human telomerase reverse transcriptase (hTERT)-immortalized retinal pigment epithelial cell line RPE1 affected not centrioles but instead cilogenesis (55). Moreover, centrosome duplication was not impaired by gene knockout of all three vertebrate centrins in chicken DT40 cells (56). Although altogether striking and puzzling, the latter finding is far from conclusive. DT40 cells are p53−/− (57), and it would be interesting to see whether this result would also be true for p53-positive human cells. Adaptation is another possibility to account for this result. Budding yeast calmodulin Cmd1 is able to interact with the Cdc31 binding sites, and, vice versa, CDC31 overexpression suppresses the growth defect of conditional lethal cmd1-1 mutant cells (58). Thus, it is entirely plausible that other EF hand proteins may be suppressing the essential requirement of centrins in centrosome duplication in DT40 centrin-null cells. Purification of the known vertebrate centrin-binding proteins POC5 and SFI1 from centrin-deficient cells and determination of whether other EF hand proteins can compensate for the role of centrin would be highly revealing (34, 59).
In human cells, a module of proteins that initiate centriole duplication in G1/S phase comprises CEP152 (named “asterless” in Drosophila), CEP63, and polo-like kinase 4 (PLK4) (60). This module assembles SAS6, CEP135, STIL, and CPAP into a platform, the so-called cartwheel (Fig. 1C), which initiates formation of the MTs forming the wall of the centrioles. Thus, it fulfils the initiating function in centrosome duplication that Sfi1 and centrin have in yeast. Although centrin is already recruited to the procentriole early in the assembly process (52, 61) (Fig. 1C), the functions of the centrin-binding proteins in the initial steps of centriole duplication remain elusive. This is in part because very little information is available on the role of hSFI1. It is known to interact only with centrin in vitro via 23 well-conserved binding repeats (Fig. 2) and to localize to centrioles (34, 62, 63). The human POC5 (hPOC5) protein is a centrin-binding protein that has only three binding repeats (Fig. 2). It is recruited to the procentrioles in G2 phase (Fig. 1C), where it plays a crucial role in initiation of the procentriole elongation (59), and it has been shown that mutations in this gene are associated with idiopathic scoliosis (64). The location of its function in the centriole duplication pathway therefore sits downstream of the PLK4 and SAS6 module.
The lack of centrioles in yeast probably made PLK4-SAS6 regulation dispensable. More downstream regulators of centriole duplication may have taken over SPB duplication control upon centriole loss in yeast. To test this notion, it will be important to understand the cell cycle regulation of hSFI1, hPOC5, and centrin by kinases and phosphatases. Both centrin-binding proteins show a number of predicted human CDK1 sites and polo-like kinase sites (human cells have four polo-like kinases) in their C-terminal regions (Fig. 2) that might constitute targets for cell cycle regulation. In addition, it will be interesting to determine whether hSFI1 can also form the antiparallel assemblies formed by ScSfi1 during bridge formation. Also, the exact localization of hSFI1 at the centrosome has yet to be determined and it remains unclear whether it plays any role in centriole formation or elongation.
BROADER PERSPECTIVE
Multidisciplinary approaches have disclosed the molecular mechanisms of the initial steps of SPB duplication in budding yeast. The simplicity of the system and of its regulation is surprising, since a small set of proteins can initiate SPB duplication in the G1 phase of the cell cycle. Nevertheless, many open issues remain regarding the embedding of Sfi1 in the SPB, how the N terminus of Sfi1 assembles the satellite, and the role of the SUN domain protein Mps3 in the duplication process. Whether all principles of bridge formation and regulation are conserved in S. pombe remains to be clarified. Analysis of S. pombe SPB duplication by superresolution microscopy, a technique that has been performed recently in budding yeast (13, 14), will shed further light on the duplication process. Remarkably little is known about the molecular function of hSFI1 and centrin in human cells. Applying approaches based on the lessons learnt from studies in S. cerevisiae will be helpful here.
ACKNOWLEDGMENT
We thank the Deutsche Forschungsgemeinschaft (DFG) for funding.
Funding Statement
This work, including the efforts of Diana Rüthnick and Elmar Schiebel, was funded by DFG (SFB638 and Schi295/5-2)
REFERENCES
- 1.Nigg EA, Stearns T. 2011. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seybold C, Schiebel E. 2013. Spindle pole bodies. Curr Biol 23:R858–R860. doi: 10.1016/j.cub.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Kilmartin JV. 5 September 2014. Lessons from yeast: the spindle pole body and the centrosome. Philos Trans R Soc Lond B Biol Sci doi: 10.1098/rstb.2013.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104:83–93. doi: 10.1016/S0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 6.Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, Mitchell BJ, Desai A, Gahman TC, Shiau AK, Oegema K. 30 April 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4; Science doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambrus BG, Uetake Y, Clutario KM, Daggubati V, Snyder M, Sluder G, Holland AJ. 2015. p53 protects against genome instability following centriole duplication failure. J Cell Biol 210:63–77. doi: 10.1083/jcb.201502089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serçin O, Larsimont JC, Karambelas AE, Marthiens V, Moers V, Boeckx B, Le Mercier M, Lambrechts D, Basto R, Blanpain C. 2016. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat Cell Biol 18:100–110. doi: 10.1038/ncb3270. [DOI] [PubMed] [Google Scholar]
- 9.Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Thery M, Pellman D. 2014. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 510:167–171. doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 11.McCully EK, Robinow CF. 1971. Mitosis in the fission yeast Schizosaccharomyces pombe: a comparative study with light and electron microscopy. J Cell Sci 9:475–507. [DOI] [PubMed] [Google Scholar]
- 12.Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. 1997. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell 8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns S, Avena JS, Unruh JR, Yu Z, Smith SE, Slaughter BD, Winey M, Jaspersen SL. 15 September 2015. Structured illumination with particle averaging reveals novel roles for yeast centrosome components during duplication. Elife doi: 10.7554/eLife.08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seybold C, Elserafy M, Ruthnick D, Ozboyaci M, Neuner A, Flottmann B, Heilemann M, Wade RC, Schiebel E. 2015. Kar1 binding to Sfi1 C-terminal regions anchors the SPB bridge to the nuclear envelope. J Cell Biol 209:843–861. doi: 10.1083/jcb.201412050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Sandercock AM, Conduit P, Robinson CV, Williams RL, Kilmartin JV. 2006. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J Cell Biol 173:867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byers B, Goetsch L. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol 124:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams IR, Kilmartin JV. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol 145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilmartin JV, Goh PY. 1996. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J 15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- 19.Sundberg HA, Goetsch L, Byers B, Davis TN. 1996. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J Cell Biol 133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott S, Knop M, Schlenstedt G, Schiebel E. 1999. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc Natl Acad Sci U S A 96:6205–6210. doi: 10.1073/pnas.96.11.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson VE, Prudden J, Prochnik S, Giddings TH Jr, Hardwick KG. 2007. Novel sfi1 alleles uncover additional functions for Sfi1p in bipolar spindle assembly and function. Mol Biol Cell 18:2047–2056. doi: 10.1091/mbc.E06-10-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elserafy M, Saric M, Neuner A, Lin TC, Zhang W, Seybold C, Sivashanmugam L, Schiebel E. 2014. Molecular mechanisms that restrict yeast centrosome duplication to one event per cell cycle. Curr Biol 24:1456–1466. doi: 10.1016/j.cub.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 23.Avena JS, Burns S, Yu Z, Ebmeier CC, Old WM, Jaspersen SL, Winey M. 2014. Licensing of yeast centrosome duplication requires phosphoregulation of Sfi1. PLoS Genet 10:e1004666. doi: 10.1371/journal.pgen.1004666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baum P, Furlong C, Byers B. 1986. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci U S A 83:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartwell LH, Culotti J, Pringle JR, Reid BJ. 1974. Genetic control of the cell division cycle in yeast. Science 183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 26.Schild D, Ananthaswamy HN, Mortimer RK. 1981. An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics 97:551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. 1993. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol 123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss E, Winey M. 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol 132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conde J, Fink GR. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A 73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose MD, Fink GR. 1987. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell 48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- 31.Spang A, Courtney I, Grein K, Matzner M, Schiebel E. 1995. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J Cell Biol 128:863–877. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biggins S, Rose MD. 1994. Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J Cell Biol 125:843–852. doi: 10.1083/jcb.125.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallen EA, Ho W, Winey M, Rose MD. 1994. Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae. Genetics 137:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilmartin JV. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol 162:1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaspersen SL, Ghosh S. 2012. Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes. Nucleus 3:226–236. doi: 10.4161/nucl.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaspersen SL, Giddings THJ, Winey M. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol 159:945–956. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner JM, Smoyer CJ, Stensrud ES, Alexander R, Gogol M, Wiegraebe W, Jaspersen SL. 2011. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J Cell Biol 193:489–507. doi: 10.1083/jcb.201011017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haase SB, Winey M, Reed SI. 2001. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat Cell Biol 3:38–42. doi: 10.1038/35050543. [DOI] [PubMed] [Google Scholar]
- 39.Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR. 2011. Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem 286:5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 2:709–718. doi: 10.1016/S1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 41.Biggins S, Ivanovska I, Rose MD. 1996. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol 133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vallen EA, Hiller MA, Scherson TY, Rose MD. 1992a. Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J Cell Biol 117:1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer T, Rodriguez-Navarro S, Pereira G, Racz A, Schiebel E, Hurt E. 2004. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol 6:840-U844. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 44.Crasta K, Huang P, Morgan G, Winey M, Surana U. 2006. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J 25:2551–2563. doi: 10.1038/sj.emboj.7601136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visintin R, Prinz S, Amon A. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 46.Zachariae W, Schwab M, Nasmyth K, Seufert W. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka K, Kanbe T. 1986. Mitosis in the fission yeast Schizosaccharomyces pombe as revealed by freeze-substitution electron microscopy. J Cell Sci 80:253–268. [DOI] [PubMed] [Google Scholar]
- 48.Paoletti A, Bordes N, Haddad R, Schwartz CL, Chang F, Bornens M. 2003. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol Biol Cell 14:2793–2808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouhlel IB, Ohta M, Mayeux A, Bordes N, Dingli F, Boulanger J, Velve Casquillas G, Loew D, Tran PT, Sato M, Paoletti A. 2015. Cell cycle control of spindle pole body duplication and splitting by Sfi1 and Cdc31 in fission yeast. J Cell Sci 128:1481–1493. doi: 10.1242/jcs.159657. [DOI] [PubMed] [Google Scholar]
- 50.Lee IJ, Wang N, Hu W, Schott K, Bahler J, Giddings TH Jr, Pringle JR, Du LL, Wu JQ. 2014. Regulation of spindle pole body assembly and cytokinesis by the centrin-binding protein Sfi1 in fission yeast. Mol Biol Cell 25:2735–2749. doi: 10.1091/mbc.E13-11-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uzawa S, Li F, Jin Y, McDonald KL, Braunfeld MB, Agard DA, Cande WZ. 2004. Spindle pole body duplication in fission yeast occurs at the G1/S boundary but maturation is blocked until exit from S by an event downstream of cdc10+. Mol Biol Cell 15:5219–5230. doi: 10.1091/mbc.E04-03-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stemm-Wolf AJ, Morgan G, Giddings TH Jr, White EA, Marchione R, McDonald HB, Winey M. 2005. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell 16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koblenz B, Schoppmeier J, Grunow A, Lechtreck KF. 2003. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J Cell Sci 116:2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- 54.Salisbury JL, Suino KM, Busby R, Springett M. 2002. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol 12:1287–1292. doi: 10.1016/S0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 55.Prosser SL, Morrison CG. 2015. Centrin2 regulates CP110 removal in primary cilium formation. J Cell Biol 208:693–701. doi: 10.1083/jcb.201411070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dantas TJ, Wang Y, Lalor P, Dockery P, Morrison CG. 2011. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J Cell Biol 193:307–318. doi: 10.1083/jcb.201012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ulrich E, Boehmelt G, Bird A, Beug H. 1992. Immortalization of conditionally transformed chicken cells: loss of normal p53 expression is an early step that is independent of cell transformation. Genes Dev 6:876–887. doi: 10.1101/gad.6.5.876. [DOI] [PubMed] [Google Scholar]
- 58.Geier BM, Wiech H, Schiebel E. 1996. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding-sites in the spindle pole body components Kar1p and Spc110p. J Biol Chem 271:28366–28374. doi: 10.1074/jbc.271.45.28366. [DOI] [PubMed] [Google Scholar]
- 59.Azimzadeh J, Hergert P, Delouvee A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M. 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol 185:101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu J, Hagan IM, Glover DM. 2015. The centrosome and its duplication cycle. Cold Spring Harb Perspect Biol 7:a015800. doi: 10.1101/cshperspect.a015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Middendorp S, Paoletti A, Schiebel E, Bornens M. 1997. Identification of a new mammalian centrin gene, more closely related to Sachharomyces cerevisiae CDC31 gene. Proc Natl Acad Sci U S A 94:9141–9146. doi: 10.1073/pnas.94.17.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinez-Sanz J, Kateb F, Assairi L, Blouquit Y, Bodenhausen G, Abergel D, Mouawad L, Craescu CT. 2010. Structure, dynamics and thermodynamics of the human centrin 2/hSfi1 complex. J Mol Biol 395:191–204. doi: 10.1016/j.jmb.2009.10.041. [DOI] [PubMed] [Google Scholar]
- 63.Martinez-Sanz J, Yang A, Blouquit Y, Duchambon P, Assairi L, Craescu CT. 2006. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J 273:4504–4515. doi: 10.1111/j.1742-4658.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- 64.Patten SA, Margaritte-Jeannin P, Bernard JC, Alix E, Labalme A, Besson A, Girard SL, Fendri K, Fraisse N, Biot B, Poizat C, Campan-Fournier A, Abelin-Genevois K, Cunin V, Zaouter C, Liao M, Lamy R, Lesca G, Menassa R, Marcaillou C, Letexier M, Sanlaville D, Berard J, Rouleau GA, Clerget-Darpoux F, Drapeau P, Moldovan F, Edery P. 2015. Functional variants of POC5 identified in patients with idiopathic scoliosis. J Clin Invest 125:1124–1128. doi: 10.1172/JCI77262. [DOI] [PMC free article] [PubMed] [Google Scholar]