ABSTRACT
The major photopigment of the cyanobacterium Acaryochloris marina is chlorophyll d, while its direct biosynthetic precursor, chlorophyll a, is also present in the cell. These pigments, along with the majority of chlorophylls utilized by oxygenic phototrophs, carry an ethyl group at the C-8 position of the molecule, having undergone reduction of a vinyl group during biosynthesis. Two unrelated classes of 8-vinyl reductase involved in the biosynthesis of chlorophylls are known to exist, BciA and BciB. The genome of Acaryochloris marina contains open reading frames (ORFs) encoding proteins displaying high sequence similarity to BciA or BciB, although they are annotated as genes involved in transcriptional control (nmrA) and methanogenesis (frhB), respectively. These genes were introduced into an 8-vinyl chlorophyll a-producing ΔbciB strain of Synechocystis sp. strain PCC 6803, and both were shown to restore synthesis of the pigment with an ethyl group at C-8, demonstrating their activities as 8-vinyl reductases. We propose that nmrA and frhB be reassigned as bciA and bciB, respectively; transcript and proteomic analysis of Acaryochloris marina reveal that both bciA and bciB are expressed and their encoded proteins are present in the cell, possibly in order to ensure that all synthesized chlorophyll pigment carries an ethyl group at C-8. Potential reasons for the presence of two 8-vinyl reductases in this strain, which is unique for cyanobacteria, are discussed.
IMPORTANCE The cyanobacterium Acaryochloris marina is the best-studied phototrophic organism that uses chlorophyll d for photosynthesis. Unique among cyanobacteria sequenced to date, its genome contains ORFs encoding two unrelated enzymes that catalyze the reduction of the C-8 vinyl group of a precursor molecule to an ethyl group. Carrying a reduced C-8 group may be of particular importance to organisms containing chlorophyll d. Plant genomes also contain orthologs of both of these genes; thus, the bacterial progenitor of the chloroplast may also have contained both bciA and bciB.
INTRODUCTION
The process of photosynthesis, in which solar energy is converted into chemical potential energy, is reliant upon light-absorbing chlorophyll (Chl) pigments that are incorporated into the antenna complexes of phototrophic organisms. Structural modifications to the tetrapyrrole macrocycle of these Chls, which influence the pigment-pigment and pigment-protein interactions within light-harvesting antenna complexes, are responsible for the specific absorption and energy transfer features of the photosystem (1–3).
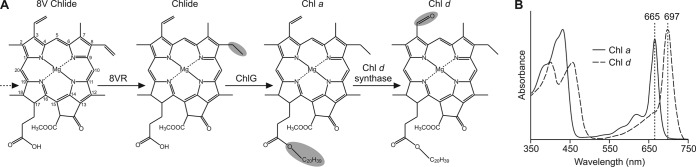
With the exception of the marine cyanobacterial Prochlorococcus spp. (4), the majority of Chls used by oxygenic phototrophs carry an ethyl group at the C-8 position (8E), the product of an 8-vinyl reductase (8VR) acting on a biosynthetic precursor, 8-vinyl (8V) chlorophyllide (Chlide) (5) (Fig. 1A). Two unrelated classes of 8VR are known to exist in oxygenic phototrophs, BciA and BciB.
FIG 1.
The terminal steps in the biosynthesis of Chls a and d. (A) The precursor 8V-Chlide (IUPAC numbered) is reduced to Chlide by an 8VR prior to the addition of phytol to the C-17 propionate side chain by Chl synthase (ChlG). In A. marina, the currently unidentified Chl d synthase oxidizes the C-3 vinyl group of Chl a to a characteristic formyl group. (B) The Chl d synthase-catalyzed oxidation results in a red shift in the Qy absorption maximum of the pigment from 665 nm to 697 nm (in methanol).
BciA was first identified through screening mutants of Arabidopsis thaliana; mutations in the AT5G18660 locus led to the accumulation of 8V- rather than 8E-Chls (6, 7), and recombinant protein produced in Escherichia coli was shown to reduce 8V-Chlide to 8E-Chlide (6). Subsequently, BciA activities were demonstrated for proteins from rice (8), maize and cucumber (9), the green sulfur bacterium Chlorobaculum tepidum (10), and the purple phototrophic bacterium Rhodobacter sphaeroides (11). In vitro assays performed with BciA-type 8VRs from various species showed that NADPH is a reductant for this enzyme (8–10, 12).
Although also utilizing 8E-Chls, the genomes of the majority of cyanobacteria do not contain orthologs of bciA, indicating the existence of a second, unrelated 8VR. Two studies on the model cyanobacterium Synechocystis sp. strain PCC 6803 (Synechocystis) demonstrated that mutants with mutations in open reading frame (ORF) slr1923 were unable to grow under high light intensities and accumulated 8V-Chl a (13, 14). Subsequently, an ortholog of slr1923 from the green sulfur bacterium Chloroherpeton thalassium was shown to complement the Chlorobaculum tepidum bciA mutant, recovering synthesis of 8E-bacteriochlorophyll (BChl) and Chl in this strain, confirming the activity of the second, BciB, class of 8VRs (15). A study on the in vitro activity of the BciB-type 8VR from Chloroherpeton thalassium showed that the enzyme is an flavin adenine dinucleotide (FAD)-containing Fe-S protein, deriving electrons from reduced ferredoxin (16).
Acaryochloris marina is the most widely studied organism utilizing Chl d for photosynthesis (17–19). Chl d differs from Chl a in that it carries a formyl group at C-3 rather than a vinyl group (17) (Fig. 1A), and oxygen labeling experiments confirmed that Chl a is the direct biosynthetic precursor of Chl d (20) (Fig. 1A). The presence of the formyl group red-shifts the Qy absorption band of the unbound pigment by approximately 30 nm compared to that of Chl a (Fig. 1B), and Chl d was found to account for 92% of the total Chl content of the cell (18). It has also been determined that Chl d is used not only for light harvesting as an antenna pigment but also as photochemically active special-pair Chls in both photosystem II (PSII) (21) and PSI (22, 23). The pigment composition of A. marina allows it to efficiently harvest far-red light to drive photosynthesis, an adaptation that permits survival in colonial ascidians (24) and microbial mats (25), where the photosynthetically active radiation is absorbed by the Chl a (with or without Chl b)-containing phototrophs but far-red light is enriched (26).
While most cyanobacteria utilize BciB to provide reduced Chls for photosynthesis, a small number instead use BciA. Uniquely for cyanobacteria sequenced to date, bioinformatic analysis revealed that the two sequenced genomes of Acaryochloris spp. (A. marina MBIC11017 and Acaryochloris sp. strain CCMEE 5410) contain homologs of both bciA and bciB. Here we expressed the A. marina genes in a mutant of Synechocystis unable to synthesize 8E-Chl a in an attempt to determine whether both ORFs encoded functional 8VRs. Heterologous expression of both genes restored the ability of the strain to grow under high-light conditions and to synthesize reduced Chl a. RNA and protein level analyses of A. marina cells demonstrated that both BciA and BciB are present in vivo. We hypothesize that two 8VRs are employed to ensure that only Chls carrying 8E groups are synthesized in these strains; possible penalties for the presence of 8V-Chl d are discussed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strain JM109 (27) transformed with pPD-FLAG (28) plasmids was grown in a rotary shaker at 37°C in LB medium supplemented with 30 μg · ml−1 kanamycin. Synechocystis strains were grown photoautotrophically in a rotary shaker under moderate (50 μmol photons · m−2 · s−1)- or high (250 μmol photons · m−2 · s−1)-light conditions at 30°C in liquid BG-11 medium (29) supplemented with 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid], pH 8.2. A. marina was grown photoautotrophically in a rotary shaker under moderate-light conditions (50 μmol photons · m−2 · s−1) at 28°C in liquid MBG-11 medium (25, 30) supplemented with 10 mM TES, pH 8.2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| E. coli JM109 | Cloning strain for pPD constructs | Promega |
| A. marina MBIC11017 | WT | R. Blankenshipa |
| Synechocystis strains | ||
| PCC 6803 | WT | R. Sobotkab |
| ΔbciB mutant | Emr replacement of central portion of slr1923 in WT | 11 |
| ΔbciB::nmrA(Am) mutant | AM1_2394 and Kmr replacement of psbAII in ΔbciB mutant | This study |
| ΔbciB::frhB(Am) mutant | AM1_2849 and Kmr replacement of psbAII in ΔbciB mutant | This study |
| Plasmids | ||
| pPD-FLAG | Cloning site and Kmr flanked by psbAII up- and downstream regions; Ampr | 28 |
| pPD[nmrA] | AM1_2394 with encoded His6 tag cloned into pPD-FLAG (NdeI/BglII) | This study |
| pPD[frhB] | AM1_2849 with encoded His6 tag cloned into pPD-FLAG (NdeI/BglII) | This study |
Departments of Biology and Chemistry, Washington University, St. Louis, MO.
Institute of Microbiology, Department of Phototrophic Microorganisms, Trebon, Czech Republic.
Construction of Synechocystis mutants containing A. marina genes.
The PCR primers used in this study are listed in Table S1 in the supplemental material. The frhB gene was amplified from A. marina MBIC11017 genomic DNA using primers frhBF and frhBR, with the reverse primer encoding a C-terminal hexahistidine tag. The PCR product was digested and cloned into the NdeI/BglII sites of pPD-FLAG vector, and the resulting plasmid was named pPD[frhB]. The construction of pPD[nmrA] was similar to that described for pPD[frhB] except that overlap extension PCR was used to generate full-length nmrA containing a silent mutation removing an internal NdeI site found in the native gene. The regions up- and downstream of this restriction site were amplified using the primer pairs nmrA1F/nmrA1R and nmrA2F/nmrA2R, respectively. Primers nmrA1R and nmrA2F were designed to be inversely complementary to each other and did not contain the NdeI site. These amplicons were used as the template for overlap extension PCR with primers nmrA1F and nmrA2R, generating the full-length nmrA. The sequenced plasmids were introduced into the Synechocystis ΔbciB strain (11). Transformants were selected on solid BG-11 medium containing 10 μg · ml−1 kanamycin and fully segregated by incrementally doubling the concentration of antibiotic to 80 μg · ml−1. Fully segregated Synechocystis strains were confirmed by colony PCR using primers pPDCheckF and pPDCheckR.
Extraction and analysis of pigments.
Chls were extracted from Synechocystis cell pellets after washing in 20 mM HEPES (pH 7.2) by adding 9 pellet volumes of 0.2% (vol/vol) ammonia in methanol, vortex mixing for 30 s, and incubating on ice for 20 min. The extracts were clarified by centrifugation (15,000 × g for 5 min at 4°C), and the supernatants were immediately analyzed on an Agilent 1200 high-pressure liquid chromatography (HPLC) system. Chl a species were separated on a Phenomenex Aqua C18 reverse-phase column (5-μm particle size, 125-Å pore size, 250 mm by 4.6 mm) using a method modified from that of van Heukelem et al. (31). Solvents A and B were 80:20 (vol/vol) methanol-500 mM ammonium acetate and 80:20 (vol/vol) methanol-acetone, respectively. Pigments were eluted at 1 ml · min−1 at 40°C on a linear gradient of 92 to 94% solvent B over 25 min, increasing to 100% to wash the column. Elution of Chl a species was monitored by checking absorbance at 665 nm.
RNA isolation and RT-PCR analysis.
Total A. marina RNA was isolated from a culture at mid-exponential growth phase using the hot TRIzol method (32) and subsequently treated with the Turbo DNA-free kit (Ambion). The cDNA synthesis and PCR amplification were performed in a single reaction with gene-specific primers using the MyTaq one-step reverse transcription-PCR (RT-PCR) kit (Bioline) according to the manufacturer's instructions. Primer pairs RTnmrAF/RTnmrAR, RTfrhBF/RTfrhBR, and RTrnpBF/RTrnpBR (see Table S1 in the supplemental material) were used to detect transcripts of nmrA, frhB, and the reference gene rnpB, respectively. One hundred nanograms of purified RNA was used in a 50-μl reaction mixture, and the thermal cycling conditions were as follows: 30 min at 45°C, 2 min at 95°C, and then 30 cycles of 10 s at 95°C, 10 s at 60°C, and 30 s at 72°C. No-RT controls were included for each sample by omitting the reverse transcriptase from the reaction mixture. Ten microliters of each PCR product was separated on a 2% agarose gel and visualized by staining with ethidium bromide.
Proteomic analysis of A. marina.
A cell pellet from 1 ml of culture (A750 = 0.62) was resuspended in 50 μl 2% SDS–60 mM dithiothreitol (DTT) and mixed with an equal volume of silicon carbide beads (1 mm; BioSpec). The cells were incubated at 95°C for 90 s and then vortexed for 30 s. This treatment was repeated 3 more times before centrifugation at 12,000 × g for2 min. The supernatant was transferred to a new vial, the beads were washed with 50 μl 2% SDS–60 mM DTT, and the supernatants were pooled. Total cell lysate protein was precipitated, dissolved in 8 M urea, reduced, S-alkylated, and digested with a combination of endoproteinase LysC and trypsin according to the method of Zhang et al. (33). Two micrograms of each proteolytic enzyme was used based on a 1:25 (wt/wt) enzyme/substrate ratio, as estimated by the method of Kalb and Bernlohr (34). After digestion, trifluoroacetic acid (TFA) was added to a final concentration of 0.5%, the peptide fragments desalted using a C18 spin column (Thermo Scientific), and the eluate dried by vacuum centrifugation. The sample was redissolved in 0.1% TFA–3% acetonitrile, and 450 ng was analyzed in duplicate by nanoflow liquid chromatography (Dionex Ultimate 3000 RSLCnano; Thermo Scientific) coupled to a mass spectrometer (MS) (Q Exactive HF Orbitrap; Thermo Scientific) operating with data-dependent acquisition of 10 tandem MS (MS/MS) scans per MS scan, all in centroid mode. Resolution settings for MS and MS/MS scans were 120,000 and 30,000, respectively, with automatic gain control targets of 1e6 and 1e5 to a 60-ms maximum. Peptides were separated using a 75-min gradient from 97% solvent A (0.1% formic acid) to 50% solvent B (0.08% formic acid, 80% acetonitrile). Mass spectra were converted to Mascot generic files using MSConvert (www.proteowizard.sourceforge.net). Protein identification was carried out by searching against the A. marina proteome database (strain MBIC 11017; release date, 3 October 2015; 8,172 entries) (www.uniprot.org/proteomes/UP000000268) using Mascot Daemon v. 2.5.1 running with Mascot Server v. 2.5 (Matrix Science), merging both duplicates into a single search, specifying trypsin as the enzyme in the search parameters and allowing for one missed cleavage. S-Carbamidomethyl-cysteine and methionine oxidation were selected as fixed and variable modifications, respectively. MS and MS/MS tolerances were set to 0.01 Da and false-discovery rates determined by searching a decoy database composed of reversed protein sequences.
Phylogenetic analysis of BciA and BciB.
To investigate the evolutionary context of A. marina BciA and BciB, homologs were identified in the species listed in Table S2 in the supplemental material through the use of the A. marina protein sequences as the query in blastp searches of the predicted proteomes or in tblastn searches of the genome assemblies where gene annotations were not available. Amino acid alignments were generated for BciA and BciB using MUSCLE (35) with default settings, and phylogenies were obtained with RAxML (36) version 8.2.4, using the automated protein model assignment algorithm and a gamma model of rate heterogeneity (-m PROTGAMMAAUTO). For comparative purposes, the 16S rRNA sequences of the same organisms were obtained from the SILVA database, and a nucleotide alignment was constructed using MUSCLE with default settings. 16S rRNA phylogenies were obtained with RAxML using the general time-reversible model of nucleotide substitution with the CAT approximation of rate heterogeneity (-m GTRCAT).
Data set accession numbers.
The complete data set has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://proteomecentral.proteomexchange.org) with the identifiers PXD003139 and 10.6019/PXD003139.
RESULTS
Expression of candidate 8VR-encoding genes from A. marina in a Synechocystis ΔbciB mutant.
ORFs encoding proteins with high sequence similarity to both BciA and BciB found in the genome sequence of A. marina (26) are annotated as genes encoding a transcriptional regulator (nmrA) and a subunit of a NiFe hydrogenase, responsible for coenzyme F420 reduction in archaeal methanogenesis (frhB), respectively. In order to test the possible 8VR activities of the encoded proteins, nmrA and frhB were expressed in a ΔbciB mutant of Synechocystis (11) that is unable to synthesize 8E-Chl a and, as a consequence, is unable to grow under high-light conditions (13, 14). The A. marina genes were integrated into the genome of Synechocystis ΔbciB in place of the psbAII gene (one of three ORFs encoding the D1 protein of photosystem II; deletion of a single copy of the gene does not affect photosynthetic capability [37]) (Fig. 2A). To determine whether the recombinant proteins were produced in Synechocystis, samples from cultures of wild-type (WT), ΔbciB, ΔbciB::nmrA(Am) and ΔbciB::frhB(Am) strains, grown under moderate light intensity, were disrupted by bead beating, and the soluble and membrane fractions were separated by centrifugation. The membrane fractions were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane, which was probed with an antibody raised against Synechocystis BciB and, in the absence of an antibody raised against BciA, a commercial anti-His6 antibody (Bethyl Laboratories, Inc.) (Fig. 2B). The blot indicates that the recombinant proteins are present, confirming the effective expression of the A. marina genes when under the control of the psbAII promoter.
FIG 2.
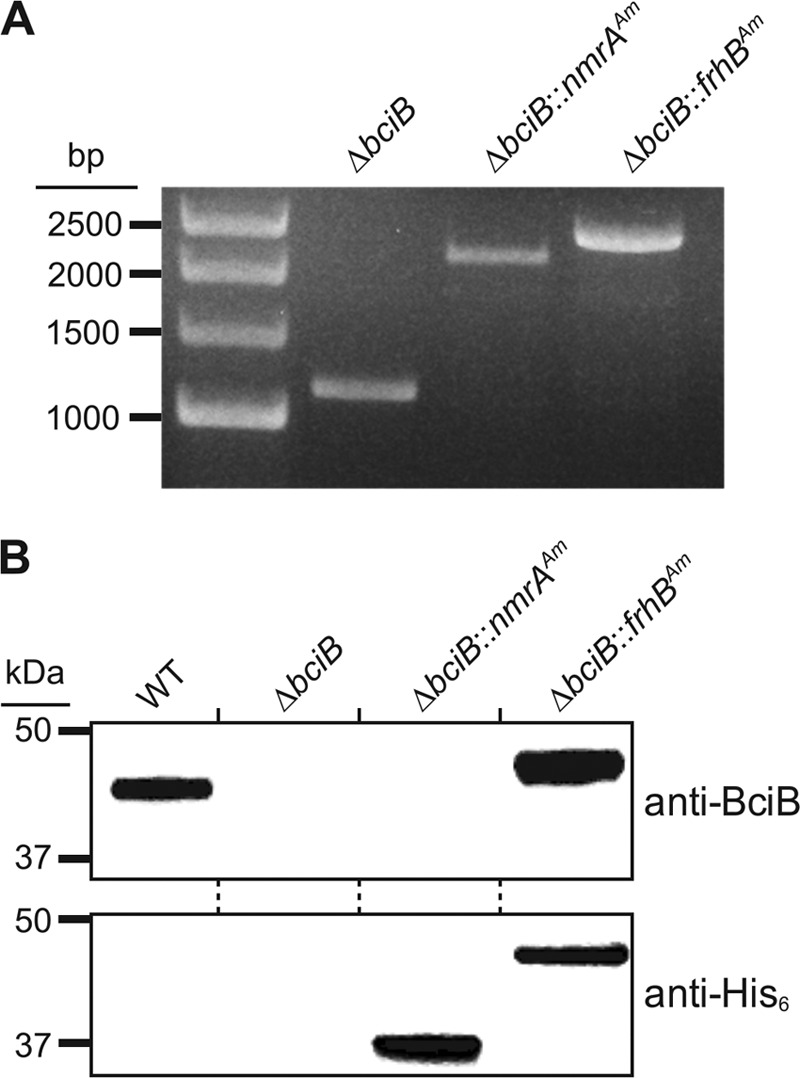
Construction of Synechocystis strains designed to express putative A. marina 8VR-encoding genes. (A) Isolation of fully segregated Synechocystis ΔbciB strains containing genes from A. marina, confirmed by colony PCR amplifying the psbAII locus. (B) Expression of recombinant proteins was confirmed by resolving membrane fractions from the described strains by SDS-PAGE, transferring to a membrane, and probing with anti-BciB and anti-His6 antibodies.
Functional testing of recombinant proteins.
The strains expressing A. marina genes, along with the WT and ΔbciB strains, were tested for their ability to grow under high light. Patches of cells were incubated under constant illumination on solid medium. As expected, the ΔbciB strain was unable to grow under high light, consistent with previously published results (11, 13, 14), while complementation with both nmrA and frhB restored the ability of the ΔbciB strain to grow under these conditions, comparable to the growth of the WT (Fig. 3A).
FIG 3.
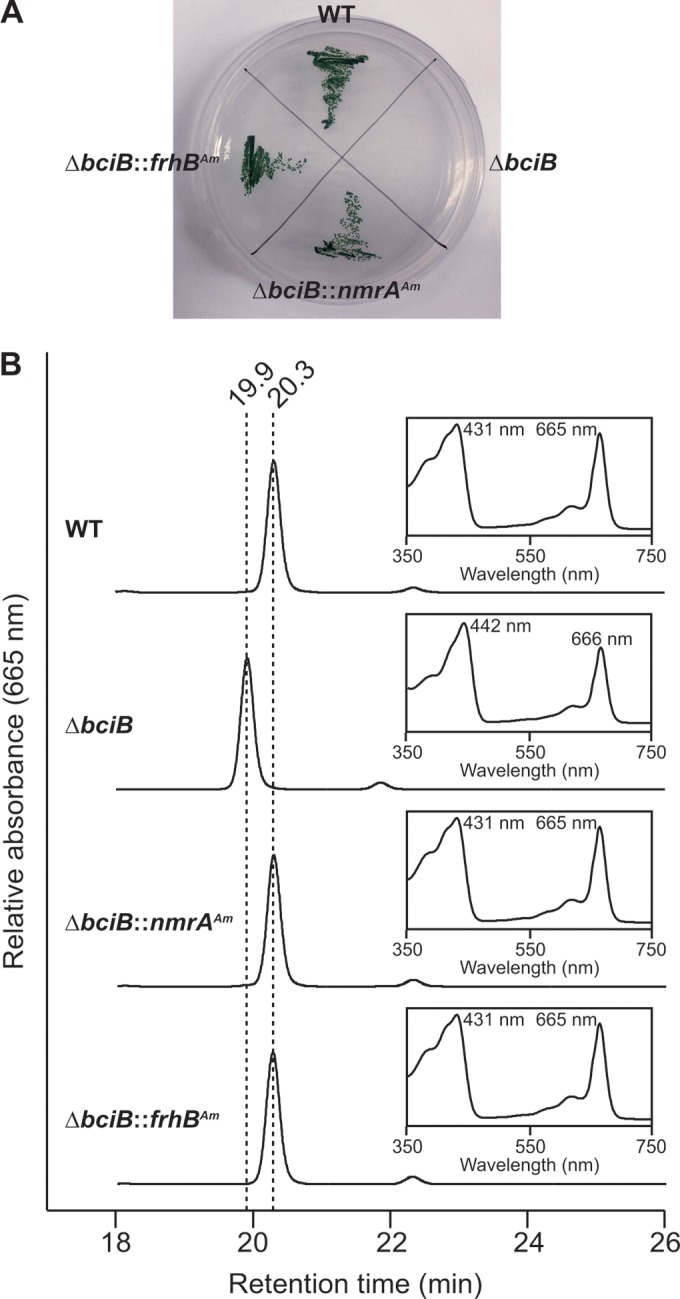
Growth and pigment analysis of described strains of Synechocystis. (A) Strains tested for growth under high light intensity on solid medium. (B) HPLC elution profiles and absorption spectra (inset) of Chls extracted from strains grown under moderate light intensity. Retention times of 20.3 and 19.9 min and Soret absorption maxima at 431 and 442 nm are indicative of 8E-Chl a and 8V-Chl a, respectively, in the HPLC solvents.
Chls from these strains grown in liquid medium under normal light were extracted and analyzed by HPLC (Fig. 3B). The Chl extracted from the ΔbciB mutant had a retention time 0.4 min shorter than that of Chl from the WT (Fig. 3B). Analysis of the absorbance profiles of these peaks demonstrated that the Soret band maximum from the ΔbciB spectrum is red shifted by 11 nm relative to that from the WT spectrum, indicating that it is the 8V form of the pigment (11, 13, 14). The retention times and absorbance profiles of the Chl peaks from both the ΔbciB::nmrA(Am) and ΔbciB::frhB(Am) strains were identical to those of the Chl peaks from the WT (Fig. 3B). Therefore, expression of either nmrA or frhB successfully recovers the WT status, with respect to 8E-Chl a synthesis, and thus we propose that they be reassigned as bciA and bciB, respectively.
Identification of 8VR utilized by A. marina.
In order to determine which of the 8VRs A. marina utilizes for Chl biosynthesis or whether both proteins are employed, transcription of bciA and bciB was checked by RT-PCR, and the presence of the cognate proteins was determined by mass spectrometry. Total A. marina RNA was isolated from a culture at mid-exponential growth phase, treated with DNase to remove any genomic DNA, and used as the template for one-step RT-PCR in which cDNA synthesis and PCR amplification were performed in a single reaction. The housekeeping gene rnpB, encoding the RNA subunit of RNase P, was included as positive control. The amplicon generated by RT-PCR displayed a single band with the expected sizes for all the three genes when analyzed by agarose gel electrophoresis (Fig. 4): 140 bp for bciA, 142 bp for bciB, and 106 bp for rnpB. The absence of bands in the no-RT controls eliminated the possibility of genomic DNA contamination. Therefore, we can conclude that both nmrA and frhB are actively transcribed under the conditions tested in A. marina. Mass spectrometry analysis was performed to verify the presence of NmrA and FrhB proteins in A. marina. Proteins extracted from an A. marina whole-cell lysate were treated with a combination of endoproteinase LysC and trypsin to generate peptide fragments which were then analyzed by nano-liquid chromatography (LC)-MS/MS. Mass spectra, consisting of both peptide ion masses and their product ion profiles, were used as inputs for searching against the A. marina reference proteome database. In total, 1,470 proteins were identified, including both NmrA and FrhB, as shown in Table 2.
FIG 4.
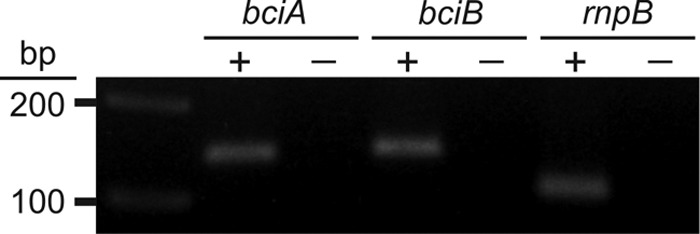
Detection of transcripts of bciA and bciB in A. marina by RT-PCR. Reactions for bciA and bciB, along with the rnpB housekeeping control, were performed with the inclusion (+) and omission (−) of reverse transcriptase to ensure that samples were not contaminated by genomic DNA.
TABLE 2.
Identification of BciA and BciB by proteomic analysis
| Protein | Mass (Da) | MOWSE scorea | Sequence coverage (%) | Peptidesb |
|---|---|---|---|---|
| BciA | 36,780 | 216 | 27 | R.ILVLGGTGTIGR.A, R.ATVAELVK.R, K.FLAEQVFK.N, R.QFYGVVSCLASR.T, R.ESGLIYSIVRPTAYFK.S, K.SVPPGFLNAIATVLGGIAK.I, R.LVDGSEEAERGDFAVF.- |
| BciB | 45,492 | 58 | 7 | R.TPEEVLAAR.V, R.SVQDSLGLEK.L, R.AGLQTFLETTSR.S |
The two 8VRs were identified by database searching with a P value of <0.05 indicating significance, with MOWSE scores representing the inverse of the probability that a match is a random event. The false-discovery rate for this search was 0.75%.
Tryptic peptides are shown with flanking amino acid residues separated by periods.
Phylogenetic analysis of BciA and BciB.
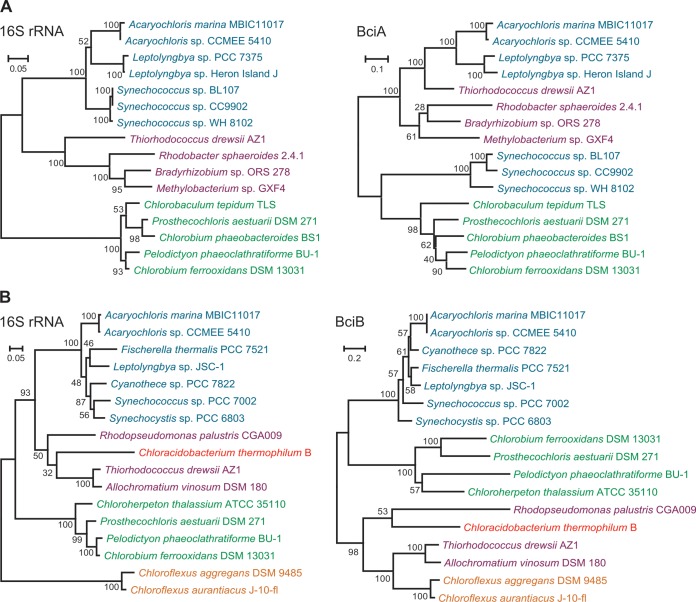
Comparisons of the phylogenies obtained by maximum-likelihood analysis of BciA and BciB amino acid alignments with those obtained by analysis of 16S rRNA alignments from the same species are shown in Fig. 5A and B, respectively. The phylogenetic positions of A. marina BciA and BciB are both broadly consistent with those shown for A. marina in the 16S rRNA trees, suggesting that the bciA and bciB genes have not been acquired by horizontal transfer. However, the positions of Synechococcus spp. in the BciA tree and the clade containing the green sulfur bacteria in the BciB tree are inconsistent with the 16S rRNA phylogeny, indicating that there may have been lateral transfer events during the evolution of both bciA and bciB.
FIG 5.
Phylogenetic relationships among 8VR protein sequences compared with parent organism 16S rRNA phylogenies. Maximum-likelihood phylogenies of BciA (A) and BciB (B) homologs, compared with 16S rRNA phylogenies of the same organisms, are shown. The BciA and BciB trees were constructed from amino acid alignments using the PROTGAMMAAUTO model in RAxML version 8.2.4. The rRNA trees were constructed from nucleotide alignments using the GTRCAT model. The numbers on branches indicate the percent bootstrap support from 100 replicates, and the scale bars indicate the specified number of amino acid or nucleotide substitutions per site. Example organisms from cyanobacteria (cyan), purple nonsulfur bacteria (purple), green sulfur bacteria (green), green filamentous bacteria (amber), and Acidobacteria (red) are included.
DISCUSSION
With the current absence of a genetic system for targeted mutagenesis of A. marina, we were unable to determine if the loss of a single 8VR-encoding gene, or loss of 8VR function via disruption of both nmrA and frhB, would have a negative effect on viability of the cells. Recently, Watabe and coworkers have described the first successful mutagenesis of A. marina cells using a transposon-based system (38); they reported the isolation of a mutant with a transposon insertion mutation in a gene involved in molybdenum cofactor biosynthesis. This mutant could be functionally complemented via introduction of the WT copy of the disrupted gene in trans. It is hoped that further development of this method may yield a system for routine targeted mutagenesis in A. marina and other cyanobacteria of interest, allowing the determination of factors involved in far-red-light utilization, including the biosynthesis of Chl d. Further, identifying the genes involved in such a process is of significant interest with the recent discovery that some strains of terrestrial cyanobacteria utilizing Chl a when grown in white light possess the ability to initiate synthesis of Chls d and f when cultured in far-red light, coupled with the extensive remodeling of their photosynthetic complexes, a response termed “far-red-light photoacclimation” (FaRLiP) (39).
Loss of 8VR activity in A. marina would result in the production of 8V-Chl a and 8V-Chl d. 8V-Chl a dominates in Prochlorococcus spp. (4) and in a recently isolated strain of the marine eukaryotic protist Alexandrium ostenfeldii (40), and the unreduced form of the pigment is tolerated in plant and cyanobacterial mutants with lesions in 8VR-encoding genes (6, 13, 14). 8V-Chl d, however, has yet to be detected in nature. Chl d was first reported as a minor pigment in various species of red microalgae (41), although it was later determined that Acaryochloris spp. attached to the surface of the alga were the true source of the pigment (42). Chl a can also be readily oxidized to Chl d during pigment extraction (43–45). Further, a study by Loughlin et al. determined that vinyl groups of naturally occurring Chls can spontaneously oxidize at C-3, yielding Chl d-like pigments, and/or at C-8, yielding novel 8-formyl versions of these Chls (46). The authors measured the Soret/Qy ratios of the substrate and product for each oxidation, comparing the absorption intensity of the high-energy, blue-most-absorbing band of the pigment to that of the lower-energy, red-most-absorbing band. Interestingly, Chls a, d and f, the latter carrying a formyl group at C-2, have Soret/Qy ratios of <1.0, and the ratios of the 8V forms of Chls a and d are 1.15 and 0.99, respectively. However, the Soret/Qy ratios of both 8-formyl Chl a and 8-formyl Chl d are 2.34. If the oxidation of the vinyl group at C-3 to yield Chl d occurs spontaneously in vivo or if the enzyme catalyzing the oxidation is not specific for the C-3 vinyl group, these 8-formyl pigments would be utilized for light harvesting and photochemistry and may result in impaired red-light absorption, thus negating the advantage A. marina holds in its ecological niche conferred by using the far-red-absorbing Chl d. This may explain why the two sequenced species in this genus employ two unrelated 8VRs: reduced Chls can be synthesized due to the presence of an alternative enzyme under conditions in which one of the reductants is limiting; e.g., 8V reduction by BciA may dominate when cellular levels of ferredoxin are depleted under iron-limiting conditions. Interestingly, the genomes of the cyanobacterial strains using the FaRLiP response sequenced thus far do not contain multiple copies of 8VR-encoding genes. However, unlike in Acaryochloris spp., Chl d is not a dominant pigment, making up only 1 to 2% of the total Chls in the cell (47). We intend to explore the consequences of accumulation of 8V-Chl d, and possibly 8-formyl Chl d, once the targeted genetic manipulation of A. marina is possible.
The utilization of unrelated enzymes to catalyze a single reaction would not be uncommon in phototrophic organisms. The magnesium protoporphyrin monomethyl ester cyclase and Pchlide oxidoreductase enzymes exist in two distinct classes in oxygenic phototrophs, each employing different reaction mechanisms (48). As with A. marina, many strains of green sulfur bacteria appear to employ multiple 8VRs for (B)Chl biosynthesis, containing either genes encoding enzymes of both classes or more than one copy of bciB (15). However, the activities of different conventional 8VRs from the same organism had not been demonstrated until this study. Interestingly, the enzyme catalyzing the first committed step in true BChl biosynthesis in organisms using BChl a, Chlide oxidoreductase (COR), is able to use both 8V- and 8E-Chlide substrates, but in each case the product pigment carries an 8E group, demonstrating a surprising additional 8VR activity (49). All known BChl a-utilizing phototrophs other than Roseiflexus spp. also contain a bciA gene (50). Removal of 8VR function in Rhodobacter sphaeroides, which naturally produces BChl a, resulted in the switch to the biosynthesis of BChl b, the pigment with the lowest energy-absorbing property of any naturally occurring photopigment (51), leading to the proposal that multiple 8VR activities ensure against the formation of BChl b in these organisms. The presence of multiple 8VRs in green sulfur bacteria may also ensure that methylation of the C-8 group is possible; deletion of bciA in Chlorobaculum tepidum prevented this methylation and resulted in aberrant assembly of the chlorosome, the specialized light-harvesting antenna in these organisms (52). Similarly, we propose here that Acaryochloris spp. employ two 8VRs to prevent the synthesis of pigments deficient in red/far-red absorption.
The genomes of many plant species, including A. thaliana and rice, which rely on BciA for 8V group reduction, contain orthologs of bciB which appeared to have become redundant in these species. However, Meguro et al. demonstrated that the bciB ortholog in A. thaliana encodes an enzyme involved in the conversion of Chl b back to Chl a (53), a process important for greening, acclimation to light intensity, and senescence in higher plants. This enzyme is proposed to have evolved from a diatom BciB and now catalyzes a new step in pigment biosynthesis (53). Of the sequenced cyanobacteria and prochlorophytes, only Acaryochloris spp. appear to contain both bciA and bciB, and our phylogenetic analysis indicates that neither of the genes was acquired by lateral gene transfer. These observations may provide insights when considering the cyanobacterial progenitor of the chloroplast.
Supplementary Material
ACKNOWLEDGMENTS
We thank Min Chen (University of Sydney, Australia) for kindly supplying A. marina genomic DNA at the beginning of the project and Robert Blankenship (Washington University, St. Louis, MO) for providing viable A. marina cells.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00925-15.
REFERENCES
- 1.Green BR, Anderson JM, Parson WW. 2004. Photosynthetic membranes and their light-harvesting antennas, p 1–28. In Green BR, Parson WW (ed), Light-harvesting antennas in photosynthesis. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 2.Scheer H. 2006. An overview of chlorophylls and bacteriochlorophylls, p 1–26. In Grimm B, Porra RJ, R̈udiger W, Scheer H (ed), Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 3.Melkozernov AN, Blankenship RE. 2006. Photosynthetic functions of chlorophylls, p 397–412. In Grimm B, Porra RJ, R̈udiger W, Scheer H (ed), Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 4.Chisholm SW, Olson RJ, Zettler ER, Goericke R, Waterbury JB, Welschmeyer NA. 1988. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334:340–343. doi: 10.1038/334340a0. [DOI] [Google Scholar]
- 5.Canniffe DP, Chidgey JW, Hunter CN. 2014. Elucidation of the preferred routes of C8-vinyl reduction in chlorophyll and bacteriochlorophyll biosynthesis. Biochem J 462:433–440. doi: 10.1042/BJ20140163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagata N, Tanaka R, Satoh S, Tanaka A. 2005. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17:233–240. doi: 10.1105/tpc.104.027276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakanishi H, Nozue H, Suzuki K, Kaneko Y, Taguchi G, Hayashida N. 2005. Characterization of the Arabidopsis thaliana mutant pcb2 which accumulates divinyl chlorophylls. Plant Cell Physiol 46:467–473. doi: 10.1093/pcp/pci053. [DOI] [PubMed] [Google Scholar]
- 8.Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X. 2010. Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiol 153:994–1003. doi: 10.1104/pp.110.158477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P, Wan C, Xu Z, Wang P, Wang W, Sun C, Ma X, Xiao Y, Zhu J, Gao X, Deng X. 2013. One divinyl reductase reduces the 8-vinyl groups in various intermediates of chlorophyll biosynthesis in a given higher plant species, but the isozyme differs between species. Plant Physiol 161:521–534. doi: 10.1104/pp.112.208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez Maqueo Chew A, Bryant DA. 2007. Characterization of a plant-like protochlorophyllide a divinyl reductase in green sulfur bacteria. J Biol Chem 282:2967–2975. doi: 10.1074/jbc.M609730200. [DOI] [PubMed] [Google Scholar]
- 11.Canniffe DP, Jackson PJ, Hollingshead S, Dickman MJ, Hunter CN. 2013. Identification of an 8-vinyl reductase involved in bacteriochlorophyll biosynthesis in Rhodobacter sphaeroides and evidence for the existence of a third distinct class of the enzyme. Biochem J 450:397–405. doi: 10.1042/BJ20121723. [DOI] [PubMed] [Google Scholar]
- 12.Nagata N, Tanaka R, Tanaka A. 2007. The major route for chlorophyll synthesis includes [3,8-divinyl]-chlorophyllide a reduction in Arabidopsis thaliana. Plant Cell Physiol 48:1803–1808. doi: 10.1093/pcp/pcm153. [DOI] [PubMed] [Google Scholar]
- 13.Ito H, Yokono M, Tanaka R, Tanaka A. 2008. Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J Biol Chem 283:9002–9011. doi: 10.1074/jbc.M708369200. [DOI] [PubMed] [Google Scholar]
- 14.Islam MR, Aikawa S, Midorikawa T, Kashino Y, Satoh K, Koike H. 2008. slr1923 of Synechocystis sp. PCC6803 is essential for conversion of 3,8-divinyl(proto)chlorophyll(ide) to 3-monovinyl(proto)chlorophyll(ide). Plant Physiol 148:1068–1081. doi: 10.1104/pp.108.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Bryant DA. 2011. Multiple types of 8-vinyl reductases for (bacterio)chlorophyll biosynthesis occur in many green sulfur bacteria. J Bacteriol 193:4996–4998. doi: 10.1128/JB.05520-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders AH, Golbeck JH, Bryant DA. 2013. Characterization of BciB: a ferredoxin-dependent 8-vinyl-protochlorophyllide reductase from the green sulfur bacterium Chloroherpeton thalassium. Biochemistry 52:8442–8451. doi: 10.1021/bi401172b. [DOI] [PubMed] [Google Scholar]
- 17.Miyashita H, Adachi K, Kurano N, Ikemoto H, Chihara M, Miyachi S. 1996. Chlorophyll d as a major pigment. Nature 338:402. [Google Scholar]
- 18.Miyashita H, Adachi K, Kurano N, Ikemot H, Chihara M, Miyach S. 1997. Pigment composition of a novel oxygenic photosynthetic prokaryote containing chlorophyll d as the major chlorophyll. Plant Cell Physiol 38:274–281. doi: 10.1093/oxfordjournals.pcp.a029163. [DOI] [Google Scholar]
- 19.Miyashita H, Ikemoto H, Kurano N, Miyachi S, Chihara M. 2003. Acaryochloris marina gen. et sp. nov. (cyanobacteria), an oxygenic photosynthetic prokaryote containing Chl d as a major pigment. J Phycol 39:1247–1253. doi: 10.1111/j.0022-3646.2003.03-158.x. [DOI] [Google Scholar]
- 20.Schliep M, Crossett B, Willows RD, Chen M. 2010. 18O labeling of chlorophyll d in Acaryochloris marina reveals that chlorophyll a and molecular oxygen are precursors. J Biol Chem 285:28450–28456. doi: 10.1074/jbc.M110.146753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomo T, Okubo T, Akimoto S, Yokono M, Miyashita H, Tsuchiya T, Noguchi T, Mimuro M. 2007. Identification of the special pair of photosystem II in a chlorophyll d-dominated cyanobacterium. Proc Natl Acad Sci U S A 104:7283–7288. doi: 10.1073/pnas.0701847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Q, Miyashita H, Iwasaki I, Kurano N, Miyachi S, Iwaki M, Itoh S. 1998. A photosystem I reaction center driven by chlorophyll d in oxygenic photosynthesis. Proc Natl Acad Sci U S A 95:13319–13323. doi: 10.1073/pnas.95.22.13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomo T, Kato Y, Suzuki T, Akimoto S, Okubo T, Noguchi T, Hasegawa K, Tsuchiya T, Tanaka K, Fukuya M, Dohmae N, Watanabe T, Mimuro M. 2008. Characterization of highly purified photosystem I complexes from the chlorophyll d-dominated cyanobacterium Acaryochloris marina MBIC 11017. J Biol Chem 283:18198–18209. doi: 10.1074/jbc.M801805200. [DOI] [PubMed] [Google Scholar]
- 24.Kühl M, Chen M, Ralph PJ, Schreiber U, Larkum AWD. 2005. A niche for cyanobacteria containing chlorophyll d. Nature 433:820. doi: 10.1038/433820a. [DOI] [PubMed] [Google Scholar]
- 25.Miller SR, Augustine S, Olson TL, Blankenship RE, Selker J, Wood AM. 2005. Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proc Natl Acad Sci U S A 102:850–855. doi: 10.1073/pnas.0405667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao J, Honchak BM, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Miyashita H, Page L, Ramakrishna P, Satoh S, Sattley WM, Shimada Y, Taylor HL, Tomo T, Tsuchiya T, Wang ZT, Raymond J, Mimuro M, Blankenship RE, Touchman JW. 2008. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci U S A 105:2005–2010. doi: 10.1073/pnas.0709772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 28.Hollingshead S, Kopecná J, Jackson PJ, Canniffe DP, Davison PA, Dickman MJ, Sobotka R, Hunter CN. 2012. Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium-protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J Biol Chem 287:27823–27833. doi: 10.1074/jbc.M112.352526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61. [Google Scholar]
- 30.Swingley WD, Hohmann-Marriott MF, Le Olson T, Blankenship RE. 2005. Effect of iron on growth and ultrastructure of Acaryochloris marina. Appl Environ Microbiol 71:8606–8610. doi: 10.1128/AEM.71.12.8606-8610.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Heukelem L, Lewitus AJ, Kana TM, Craft NE. 1994. Improved separations of phytoplankton pigments using temperature-controlled high performance liquid chromatography. Mar Ecol Prog Ser 114:303–313. doi: 10.3354/meps114303. [DOI] [Google Scholar]
- 32.Pinto FL, Thapper A, Sontheim W, Lindblad P. 2009. Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol Biol 10:79. doi: 10.1186/1471-2199-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Liu H, Blankenship RE, Gross ML. 2015. Isotope-encoded carboxyl group footprinting for mass spectrometry-based protein conformational studies. J Am Soc Mass Spectrom doi: 10.1007/s13361-015-1260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalb VF, Bernlohr RW. 1977. A new spectrophotometric assay for protein in cell extracts. Anal Biochem 82:362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- 35.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansson C, Debus RJ, Osiewacz HD, Gurevitz M, McIntosh L. 1987. Construction of an obligate photoheterotrophic mutant of the cyanobacterium Synechocystis 6803. Plant Physiol 85:1021–1025. doi: 10.1104/pp.85.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watabe K, Mimuro M, Tsuchiya T. 2015. Establishment of the forward genetic analysis of the chlorophyll d-dominated cyanobacterium Acaryochloris marina MBIC 11017 by applying in vivo transposon mutagenesis system. Photosynth Res 125:255–265. doi: 10.1007/s11120-015-0082-4. [DOI] [PubMed] [Google Scholar]
- 39.Gan F, Zhang S, Rockwell NC, Martin SS, Lagarias JC, Bryant DA. 2014. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345:1312–1317. doi: 10.1126/science.1256963. [DOI] [PubMed] [Google Scholar]
- 40.Rodríguez F, Kremp A, Garrido JL, Sobrino C, Johnsen G, Riobó P, Franco J, Aamot I, Ramilo I, Sanz N. 2015. Divinyl chlorophyll a in the marine eukaryotic protist Alexandrium ostenfeldii (Dinophyceae). Environ Microbiol doi: 10.1111/1462-2920.13042. [DOI] [PubMed] [Google Scholar]
- 41.Manning WM, Strain HH. 1943. Chlorophyll d, a green pigment of red algae. J Biol Chem 151:1–19. [Google Scholar]
- 42.Murakami A, Miyashita H, Iseki M, Adachi K, Mimuro M. 2004. Chlorophyll d in an epiphytic cyanobacterium of red algae. Science 303:1633. doi: 10.1126/science.1095459. [DOI] [PubMed] [Google Scholar]
- 43.Holt AS, Morley HV. 1959. A proposed structure for chlorophyll d. Can J Chem 37:507–514. doi: 10.1139/v59-069. [DOI] [Google Scholar]
- 44.Holt AS. 1961. Further evidence of the relation between 2-desvinyl-2-formyl-chlorophyll a and chlorophyll d. Can J Bot 39:327–331. doi: 10.1139/b61-026. [DOI] [Google Scholar]
- 45.Fukusumi T, Matsuda K, Mizoguchi T, Miyatake T, Ito S, Ikeda T, Tamiaki H, Oba T. 2012. Non-enzymatic conversion of chlorophyll-a into chlorophyll-d in vitro: a model oxidation pathway for chlorophyll-d biosynthesis. FEBS Lett 586:2338–2341. doi: 10.1016/j.febslet.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 46.Loughlin PC, Willows RD, Chen M. 2014. In vitro conversion of vinyl to formyl groups in naturally occurring chlorophylls. Sci Rep 4:6069. doi: 10.1038/srep06069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gan F, Shen G, Bryant DA. 2014. Occurrence of far-red light photoacclimation (FaRLiP) in diverse cyanobacteria. Life 5:4–24. doi: 10.3390/life5010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galperin MY, Walker DR, Koonin EV. 1998. Analogous enzymes: independent inventions in enzyme evolution. Genome Res 8:779–790. [DOI] [PubMed] [Google Scholar]
- 49.Tsukatani Y, Yamamoto H, Harada J, Yoshitomi T, Nomata J, Kasahara M, Mizoguchi T, Fujita Y, Tamiaki H. 2013. An unexpectedly branched biosynthetic pathway for bacteriochlorophyll b capable of absorbing near-infrared light. Sci Rep 3:1217–1223. doi: 10.1038/srep01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanada S, Takaichi S, Matsuura K, Nakamura K. 2002. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium which lacks chlorosomes. Int J Syst Evol Microbiol 52:187–193. doi: 10.1099/00207713-52-1-187. [DOI] [PubMed] [Google Scholar]
- 51.Canniffe DP, Hunter CN. 2014. Engineered biosynthesis of bacteriochlorophyll b in Rhodobacter sphaeroides. Biochim Biophys Acta 1837:1611–1616. doi: 10.1016/j.bbabio.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez Maqueo Chew A, Frigaard N-U, Bryant DA. 2007. Bacteriochlorophyllide c C-82 and C-121 methyltransferases are essential for adaptation to low light in Chlorobaculum tepidum. J Bacteriol 189:6176–6184. doi: 10.1128/JB.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A. 2011. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 23:3442–2453. doi: 10.1105/tpc.111.089714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.