Abstract
Regulating responses to stress is critical for all bacteria, whether they are environmental, commensal, or pathogenic species. For pathogenic bacteria, successful colonization and survival in the host are dependent on adaptation to diverse conditions imposed by the host tissue architecture and the immune response. Once the bacterium senses a hostile environment, it must enact a change in physiology that contributes to the organism's survival strategy. Inappropriate responses have consequences; hence, the execution of the appropriate response is essential for survival of the bacterium in its niche. Stress responses are most often regulated at the level of gene expression and, more specifically, transcription. This minireview focuses on mechanisms of regulating transcription initiation that are required by Mycobacterium tuberculosis to respond to the arsenal of defenses imposed by the host during infection. In particular, we highlight how certain features of M. tuberculosis physiology allow this pathogen to respond swiftly and effectively to host defenses. By enacting highly integrated and coordinated gene expression changes in response to stress, M. tuberculosis is prepared for battle against the host defense and able to persist within the human population.
INTRODUCTION
The survival of any organism relies on its ability to sense and respond to changes in its environment. For bacteria, stress responses are primarily mediated through the regulation of gene expression. By integrating multiple molecular approaches to gene regulation, pathogenic bacteria are able to orchestrate condition-specific patterns that promote survival and pathogenesis in the face of a strong immune response. This minireview focuses on mechanisms of transcription regulation required for stress responses in one of the most successful and deadly pathogens in the world, Mycobacterium tuberculosis. M. tuberculosis has coexisted with humans for >50,000 years (1) and continues to cause more than 1.5 million deaths a year (2). The coevolution of M. tuberculosis with the human host response to infection has resulted in a pathogen that is specialized for long-term infection in people. Tuberculosis is a complex disease that requires the bacteria to multiply within phagocytes, survive extracellularly in hypoxic and necrotic granulomas, and endure a robust immune response to persist in the host. During infection, the host immune response restrains M. tuberculosis from proliferating by imposing a battery of defenses, including reactive oxygen and nitrogen stress, hypoxia, acid stress, genotoxic stress, cell surface stress, and starvation (3). Despite this onslaught of attacks, M. tuberculosis is able to persist for the lifetime of the host, indicating that this pathogen has highly effective molecular mechanisms to resist host-inflicted damage. In order to enact these defenses and facilitate this specialized lifestyle, M. tuberculosis executes a complex, interconnected web of stress responses that rely on changes in gene expression. In fact, M. tuberculosis is well suited to respond quickly to diverse stresses in a coordinated fashion. For instance, the RNA polymerase (RNAP) bears kinetic properties that allow it to be easily modulated by accessory factors. Compared to other obligate human pathogens, M. tuberculosis encodes the highest ratio of σ factors to genome size (4), which allows the bacterium to tailor its expression profile in response to a given environment. Even during exponential growth in culture, traditionally thought of as a relatively stress-free environment, M. tuberculosis expresses its entire complement of σ factors (5–7), indicating that M. tuberculosis is poised to quickly respond to stress. M. tuberculosis also integrates stress responses into basic cellular processes; as a result, some stress-associated transcriptional regulators are essential in M. tuberculosis. In this minireview, we discuss features of the mycobacterial transcription apparatus that position M. tuberculosis to be ready to respond to host attacks, the networks of factors that contribute to these responses, and how this culminates in a successful pathogenic strategy. The general strategies to be discussed are illustrated in Fig. 1, and individual factors touched on in this minireview are summarized in Fig. 2.
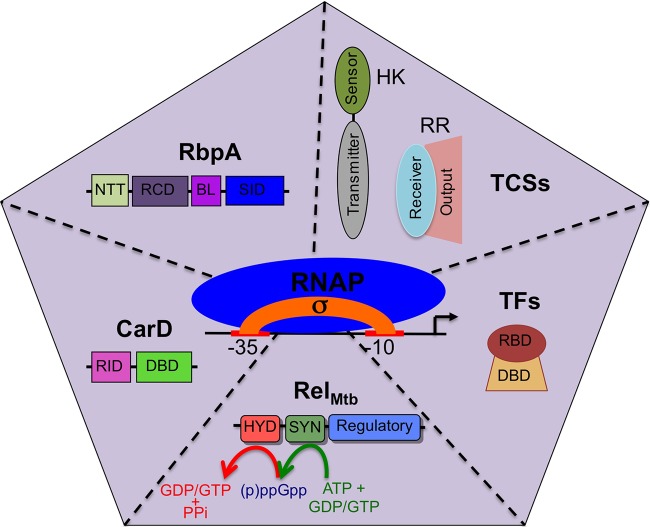
FIG 1.
Summary of the branches of transcriptional regulation that are discussed in this minireview. The illustration shows 6 types of factors (σ factors, CarD, RbpA, TCSs, TFs, and RelMtb) that modulate RNAP activity at promoters to mediate reprogramming of the expression profile in M. tuberculosis in response to different environments. A σ factor associates with the core RNAP to form the RNAP holoenzyme, which is then modified by the other factors shown in the sections of the pentagon. Domains of each protein are shown. For CarD, RID is the RNAP interaction domain and DBD is the DNA binding domain. For RbpA, NTT is the N-terminal tail, RCD is the RbpA core domain, BL is the basic linker, and SID is the sigma interaction domain. For RelMtb, HYD is the (p)ppGpp hydrolase domain and SYN is the (p)ppGpp synthetase domain. For TFs, RBD is the RNAP binding domain and DBD is the DNA binding domain. In the presence of a given stress, these factors coordinate their responses to effectively respond to host attacks.
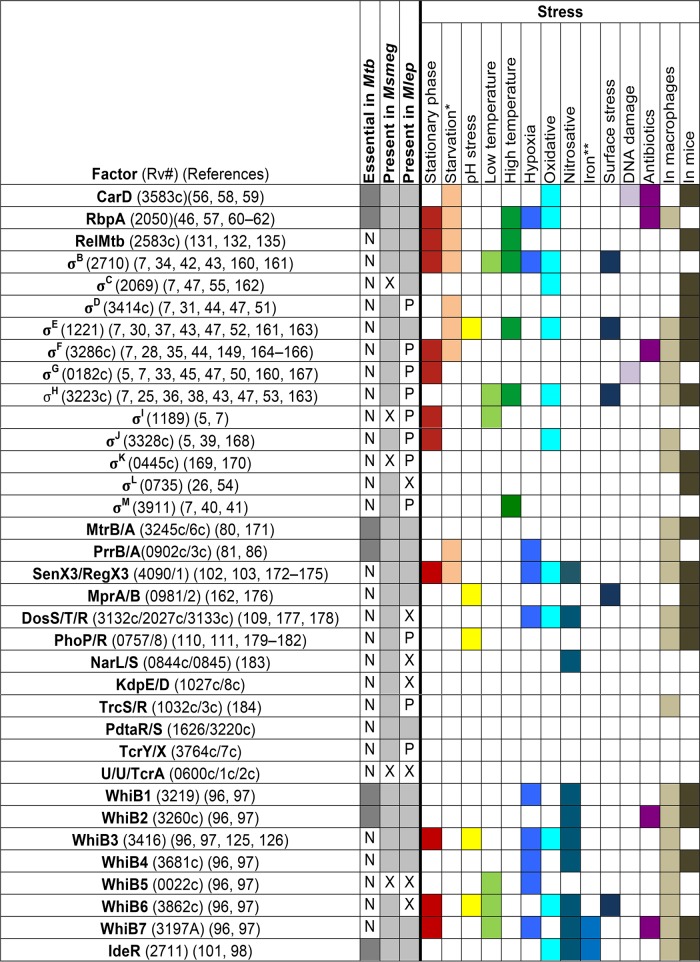
FIG 2.
Conservation of M. tuberculosis regulatory factors and the stresses that the factors are associated with in M. tuberculosis (160–184). The left side of the table designates whether the gene for a transcriptional regulator is essential (shaded) or not essential (N) in M. tuberculosis (Mtb) and whether that gene is conserved (shaded) or not conserved (X) or exists as a pseudogene (P) in the environmental saprophytic M. smegmatis (Msmeg) or the obligate pathogen M. leprae (Mlep). The right side of the table indicates whether a particular stress condition has been associated with a given transcriptional regulator. Involvement in the response to a particular stress is designated by shading of the box and may represent expression profiling data or phenotypic analysis of mutants. An unshaded square indicates that the factor is not induced, is not important for survival, or has not been studied under that particular condition. U, unnamed factor; *, starvation (including nutrient, phosphate, and nitrogen starvation); **, iron-depleted or iron-replete conditions. See specific references for more information.
THE MYCOBACTERIAL RNA POLYMERASE—READY TO RESPOND
Transcription is achieved in all bacteria by a single core RNAP enzyme, consisting of the essential subunits β and β′ and 2 α subunits along with the nonessential ω subunit (8, 9). To recognize and bind promoter sequences upstream from genes, the core RNAP associates with a σ subunit to form an RNAP holoenzyme. Most transcriptional regulation occurs at the level of initiation (10), and transcription factors (TFs) can mediate this regulation by directly affecting the polymerase-promoter interaction, manipulating the equilibrium between closed and open RNAP-promoter complexes (RPc and RPo, respectively), or affecting rates of promoter escape (11, 12). The majority of studies on the mechanisms of transcription initiation and its regulation have used Escherichia coli as a model system. However, multiple groups have recently shown that Mycobacterium bovis RNAP, which differs from the M. tuberculosis RNAP by only one amino acid (aa), exhibits an inherently unstable RPo complex compared to E. coli RNAP on the same promoter (13, 14). In these reports, saturating concentrations of M. bovis RNAP σA holoenzyme were found to be incapable of opening a large percentage of the promoters, leaving the majority of bound complexes in the closed state. It has been proposed (14) that the presence or absence of lineage-specific insertions within RNAP could contribute to the inherent differences in stability of the promoter complexes formed by M. bovis versus E. coli RNAP. Notably, RNAPs from Bacillus subtilis, Thermus aquaticus, and Thermus thermophilus have also been found to generate relatively unstable open promoter complexes (15–17). Based on these observations, it is worth considering that the properties of E. coli RNAP may not be representative of most bacterial RNAPs and that there may be significant lineage-specific variation in enzyme kinetics. The inherent instability of RNAP-promoter complexes would allow the mycobacterial RNAP to be poised to respond to changes in the environment by being easily modified in activity by additional factors.
σ FACTORS: THE GENERALS OF STRESS RESPONSES
The first determinant of gene expression in response to different conditions is the activity of the σ factor repertoire. Each σ factor binds a specific promoter sequence, thus determining what promoters are targeted by the RNAP holoenzyme for transcription. Changes in σ factor activity in response to different stresses and conditions are able to shift a bacterium's expression profile. The σ factor network of M. tuberculosis includes one essential housekeeping group 1 σ factor (σA), one stress-responsive group 2 σ factor (σB), and 11 group 3 and 4 alternative σ factors that also function as environmentally responsive regulators (σC to σM) (4, 6, 18). This broad panel of σ factors allows M. tuberculosis to tune its transcriptional response for a large and diverse set of conditions. All of the σ factors in M. tuberculosis belong to the σ70 family, whose members in E. coli recognize two sequences in the promoter DNA, the −10 element (recognized by sigma region 2.4) and the −35 element (recognized by sigma region 4.2) (19). M. tuberculosis promoters contain a conserved −10 sequence that is essential and sometimes sufficient for transcription, while the −35 sequences are less conserved (19–21). The spacer region between the −10 and −35 elements in M. tuberculosis also varies dramatically compared to E. coli promoters (19, 22, 23). These differences in promoter elements may reflect the sigma diversity in M. tuberculosis (19, 23).
The activity of σ factors in M. tuberculosis is most often regulated by anti-σ factors that inactivate their cognate σ factors until a signal is received to liberate the σ factor for action. Specifically, σB, σD, σE, σF, σH, σK, σL, and σM are all regulated by a cognate anti-σ factor (24–32). A putative anti-σ factor has also been proposed for σG (33). To investigate under which conditions a particular σ factor is active, the expression levels of σ factors have been studied in vitro under many physiologically relevant conditions, but transcriptional upregulation of a given σ factor does not necessarily equate to σ factor activity. Therefore, σ factor gene deletion or overexpression strains have been used to determine the functional role of individual σ factors in response to stress. These data are summarized here and together paint a picture of an intricate circuitry of transcriptional regulation that integrates multiple σ factor regulons under many conditions (Fig. 3 and 4), allowing M. tuberculosis to respond to the arsenal of attacks from the host.
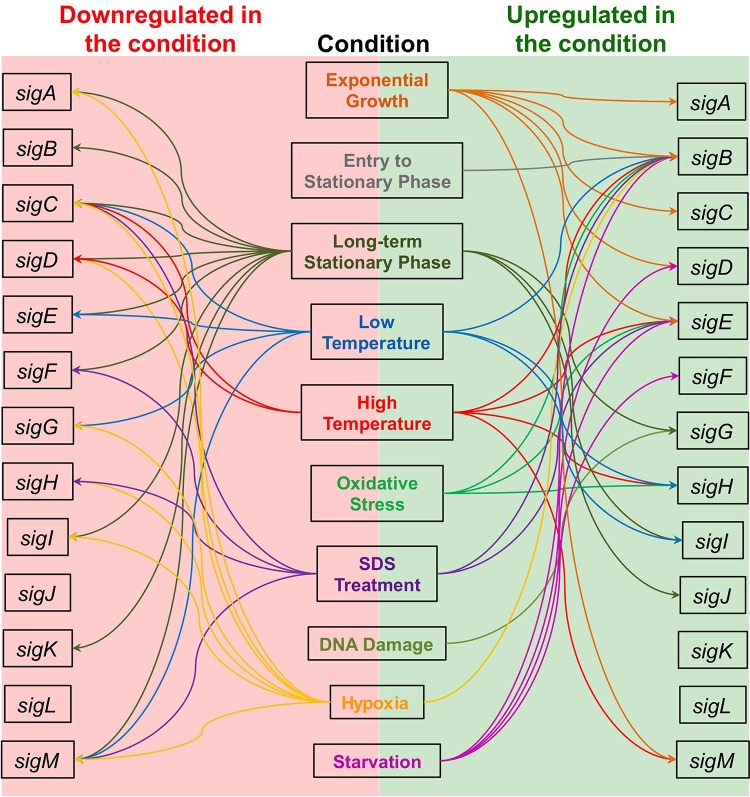
FIG 3.
Transcriptional regulation of M. tuberculosis σ factor genes in response to various stresses. Transcriptional responses of σ factor genes of M. tuberculosis include responses to entry to stationary phase, the long-term stationary phase, mild cold shock (room temperature), heat shock (45°C), oxidative stress, exposure to SDS, DNA damage, hypoxia, and starvation. The σ factor genes that are transcriptionally upregulated in response to a stress are diagramed with arrows to the right, and the σ factor genes that are transcriptionally downregulated are shown with arrows to the left. The σ factor genes that are highly expressed during exponential growth in culture are shown as being upregulated under this condition. Where no arrow is present to connect a σ factor gene to a particular stress, this indicates that expression of the σ factor gene is not significantly changed during exposure to that stress or has not been studied under that particular condition. References are available in the text.
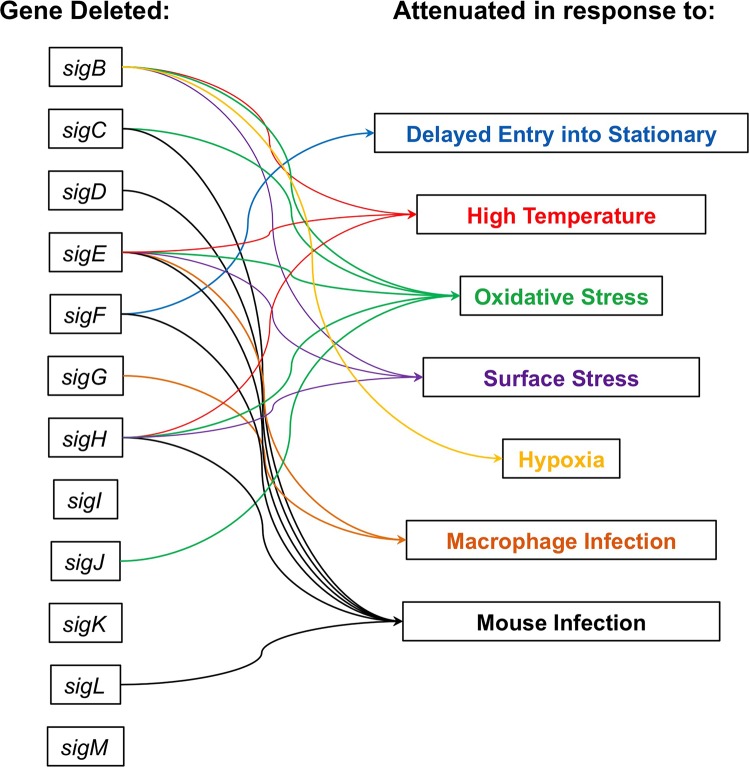
FIG 4.
Effects of σ factor gene deletions on stress responses in M. tuberculosis. Arrows indicate whether deletion of a σ factor gene causes delayed entry into stationary phase, decreased survival during heat shock, decreased survival during oxidative stress, decreased survival during surface stress or changes in cell permeability, decreased survival during hypoxia, decreased survival in macrophages, or decreased immunopathology during mouse infection. Where no arrow is present to connect a σ factor gene to a particular stress, this indicates that deletion of that σ factor gene did not significantly change survival during exposure to that stress or has not been studied under that particular condition. References are available in the text.
During exponential growth of M. tuberculosis in culture, sigA, sigB, sigC, sigD, sigE, and sigM are the most highly expressed σ factor genes (7). Upon entry into stationary phase, levels of sigB transcripts increase (34). Strains with a disrupted sigF gene grow to a density three times greater than that seen with wild-type cultures in stationary phase, suggesting that σF may have a key role in regulating this transition (35). Later in stationary phase, there is a global change in regulation of σ factors resulting in downregulation of most of the σ factor genes, with the exception of sigG, sigI, and sigJ, which are upregulated in long-term stationary cultures (5, 7). σH is a central regulator of the response of M. tuberculosis to both heat and oxidative stress through regulation of sigE, sigB, heat shock proteins, thioredoxin reductase/thioredoxin, and synthesis of mycothiol precursors (36). In addition to σB, σE, and σH, survival during oxidative stress is also dependent on σC and σJ (6, 36–39). sigM is also induced during exposure to heat in the M. tuberculosis CDC1551 strain but not in M. tuberculosis H37Rv, indicating strain-specific regulation of σ factor expression (24, 40, 41). Cold temperatures induce expression of sigB, sigH, and sigI while repressing transcription of sigC, sigE, sigG, and sigM (7). σI is the most highly induced σ factor during cold shock and has been proposed to be important for the bacterium's survival in aerosol particles between hosts (7). Deletion of sigB, sigE, or sigH has been shown to increase M. tuberculosis's sensitivity to cell surface stress (6, 37, 42, 43). Expression of sigB is also upregulated under hypoxic conditions (7) and σB is the only σ factor shown to impact the sensitivity of M. tuberculosis to hypoxia (42). Deletion of sigF induces permeability changes in the cell envelope, although this does not affect sensitivity to tested surface stresses (35, 44). In vitro studies have shown that sigG is induced upon DNA damage but that deletion of sigG does not sensitize strains to DNA damage (45). sigB, sigD, sigE, and sigF have all been shown to be upregulated during prolonged nutrient starvation (46).
Evidence that alternative σ factors are important in M. tuberculosis during infection has come from cell culture and animal infection models. sigE, sigF, sigG, sigH, and sigJ are upregulated during infection of macrophages (47, 48), and both sigE and sigG are necessary for survival within macrophages (37, 49, 50). Deletion of sigB, sigG, sigJ, or sigM has no effect in animal models (6, 39, 40, 50). Deletion of sigD, sigE, sigH, or sigL results in a delayed time to death without affecting bacterial burden (51–54), while deletion of sigC or sigF results in a delayed time to death and a decrease in bacterial burden during acute (sigC) or chronic (sigF) infection (35, 44, 55). The importance of individual σ factors during infection and for survival under stressful conditions highlights both their central role in guiding M. tuberculosis's stress response and the diverse adverse conditions encountered by M. tuberculosis during infection.
CarD AND RbpA—MAINTAINING THE PEACE, BUT READY TO DEFEND
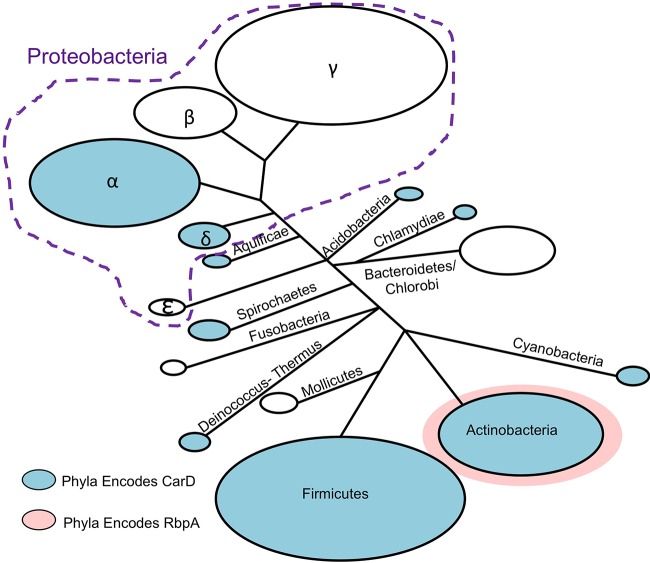
The next branch of transcriptional regulation during stress responses involves RNAP-binding proteins that further modify gene expression from a given holoenzyme. CarD and RbpA are RNAP-binding proteins in M. tuberculosis that were each originally identified in experiments looking for genes upregulated in response to stress (56, 57). carD expression is upregulated in response to oxidative stress, starvation, and a broad panel of antibiotics. CarD activity is required for survival under the same conditions as well as for virulence in a mouse model of infection (56, 58, 59). rbpA is upregulated during oxidative stress, stationary phase, starvation, hypoxia, high temperatures, and treatment with antibiotics and during infection in macrophages (46, 57, 60–62). Overexpression of rbpA in mycobacteria also improves resistance to the antibiotic rifampin (63). CarD and RbpA both act by stabilizing the inherently unstable mycobacterial RNAP-promoter complexes, albeit by different mechanisms. While the presence of RbpA is limited to actinobacteria, CarD is present in members of numerous other bacterial phyla (56, 64, 65), including Bacillus and Thermus, where purified RNAPs also generate relatively unstable open promoter complexes (15–17), but not in E. coli, where RNAP generally forms stable open complexes (13, 14) (Fig. 5). carD and rbpA are essential in M. tuberculosis even during growth in nutrient-rich cultures (56, 66–68), indicating a general role in promoting efficient gene expression that also allows the RNAP to optimally respond to stress.
FIG 5.
Phylogenetic distribution of CarD and RbpA. The BLAST database of completed genomes was searched for homologs of M. tuberculosis CarD and RbpA. Homologs of each protein were schematically drawn on a phylogenetic tree using a previously calculated phylogenetic distribution of bacteria based on the sequence conservation of RNAP subunits (185). Blue-shaded phyla have members that encode CarD homologs, members of the pink-shaded phylum (actinobacteria) encode RbpA, and phyla that are not shaded do not encode CarD or RbpA.
CarD interacts with the RNAP β-subunit β1-lobe through an N-terminal RNAP interaction domain (RID) and with DNA via a C-terminal basic patch (56, 58, 59, 65, 69, 70). In mycobacteria cultured under nutrient-rich conditions, CarD associates with RNAP-promoter complexes throughout the genome to enhance RPo stability (14, 58, 59, 65). Using a bulk fluorescence assay to measure the effects of CarD on transcription initiation kinetics, it was shown that CarD associates with RPo with high affinity and slows the rate of DNA closing by preventing bubble collapse and that CarD associates with RPc with lower affinity and increases the rate of DNA opening (13). Importantly, the concentration of CarD in cells is sufficient for both of these activities to be physiologically relevant (13). These two activities of CarD change the kinetics of open complex formation such that the M. bovis RNAP more closely mirrors the E. coli RNAP (13, 14). The interactions between CarD and both DNA and RNAP are required for CarD activity (13). In addition, a conserved tryptophan within the C-terminal basic patch is also important for CarD's effects on RNAP-promoter complex stability and, based on structural studies, has been proposed to serve as a wedge at the upstream edge of the transcription bubble that prevents bubble collapse (59, 64, 65). Taken together, the inherently weak transcription initiation activity of M. bovis RNAP and CarD's global promoter localization suggest that CarD may be a general member of the mycobacterial transcription machinery.
RbpA consists of a central RbpA core domain (RCD) flanked by an unstructured 26-aa N-terminal tail and a C-terminal σ interaction domain (SID) linked to the RCD by a 15-aa basic linker (BL) (68, 71, 72). RbpA forms a stable binary complex with the σ2-domain of group 1 (σA in M. tuberculosis) and certain group 2 (σB in M. tuberculosis) σ factors through its SID (68, 71, 72), with additional contacts made between the N terminus and the σ factor (68). Based on structural modeling, the RbpA BL domain and adjacent residues interact with the DNA phosphate backbone of the nontemplate strand upstream of the −10 promoter element in the RPo conformation (68). Additional contacts between RbpA and RNAP β have been proposed based on cross-linking experiments (63, 73, 74), but the recent structural modeling of RbpA onto an RNAP-promoter open complex would be incompatible with these interactions (71), suggesting that further analysis will be needed to resolve these inconsistencies. RbpA has been shown to increase the affinity of the σ factor to the core RNAP, increase the affinity of RNAP holoenzyme to promoter DNA, and facilitate the formation of RPo (71, 75, 76), all of which could contribute to the ability of RbpA to promote RNAP-promoter complex formation and stability. The housekeeping σ factor σA has been reported to have an affinity for M. tuberculosis RNAP core enzyme similar to that of the alternative σ factor σF (74), in which case RbpA may be necessary to improve σA affinity and competitiveness for RNAP under conditions that require the activity of σA. In E. coli, in contrast, σ70 has a very high affinity to the RNAP core enzyme and thus can outcompete other σ factors under conditions where it is required without accessary factors such as RbpA. The RbpA SID and BL are important and sufficient to partially activate transcription in vitro (71, 72), but full activation of transcription requires the full-length protein, although the function of the N terminus of RbpA remains elusive.
Based on structural modeling performed with the information currently available, association of CarD and RbpA with the same RNAP holoenzyme is feasible (71), but why M. tuberculosis requires both CarD and RbpA activities is unknown. CarD and RbpA transcriptional regulatory activities have thus far been analyzed only on limited promoters under limited conditions. However, their roles and effects at individual promoters likely depend on the kinetic properties of individual RNAP-promoter complexes and the presence of additional transcriptional regulators. The roles for CarD and RbpA during stress responses indicate that their effects on RPo stability also provide a mechanism for adjusting gene expression during the switch between different physiological states in response to stress. Indeed, RPo formation and stability comprise a commonly regulated step of transcription initiation during stress responses in bacteria, including during the stringent response (77, 78). It is possible that CarD and RbpA are important in stabilizing transcription complexes activated by stress-responsive transcription factors or alternative σ factors. While the functions of CarD and RbpA in stress responses remain unclear, the diversity of the stresses that they respond to suggests that they are acting at a common point shared among numerous stress responses.
ESSENTIAL TCSs AND TFs—ALWAYS ON THE LOOKOUT FOR HOSTILITY
M. tuberculosis encodes 12 complete two-component systems (TCSs), which are classically recognized as bacterial systems that sense and respond to stress and changes in the environment (79). Each TCS consists of at least one sensor histidine kinase (HK) that responds to specific environmental conditions by autophosphorylation and phosphotransfer to its cognate response regulator (RR), which then binds DNA and activates transcription of a specific regulon (79). Two TCSs in M. tuberculosis, MtrAB (80) and PrrAB (81), are essential for growth under unstressed culture conditions and have been integrated into the basic physiology of the bacteria. The HK MtrB colocalizes with cell division machinery at the bacterial septa and poles (82). Upon stimulation by an unknown signal, MtrB phosphorylates its cognate RR MtrA, which then binds DNA and activates transcription of a regulon that includes essential replication and cell division genes dnaA and ripA as well as the fbpB and rpfB genes that encode proteins with roles during infection (82–84). Integration of a TCS with the cell division machinery could allow these slowly replicating bacteria to sense environmental stress and abort cell division if unfavorable conditions surface. The second essential TCS in M. tuberculosis is the PrrAB system. The RR PrrA can bind DNA in the unphosphorylated state, but its binding affinity increases once phosphorylated by HK PrrB (85). The stimulus that results in activation of the PrrAB TCS has not been characterized, but expression of the prrAB operon is induced by nitrogen limitation and growth inside macrophages (81, 86), suggesting a possible role for this TCS under these conditions.
M. tuberculosis also encodes a series of essential iron-binding transcription factors (TF). M. tuberculosis does not contain functional homologues of the common redox-sensing TFs, FNR, SoxR, and OxyR, that allow other bacteria to sense and respond to redox state and reactive nitrogen and oxygen species (87–91). Instead, M. tuberculosis encodes a 7-member family of WhiB iron-sulfur (Fe-S) cluster TFs that sense the redox state in the cell and regulate gene expression accordingly (92). Of these, whiB1 and whiB2 are predicted to be essential, although their regulons have yet to be defined (93–95). whiB1 is also upregulated during hypoxia and within infected mouse lungs (96, 97). WhiB2 may play a role in cell cycle progression, as a conditional whiB2 mutant in Mycobacterium smegmatis was filamentous during depletion (95). The iron-binding TF IdeR is also essential for M. tuberculosis viability (98). IdeR dimerizes when bound to iron (99) and binds DNA as a dimer to inhibit transcription of genes involved in iron uptake and storage in order to promote adaptation to changing levels of iron (100, 101). By reducing levels of intracellular iron that can catalyze formation of reactive oxygen species, IdeR protects M. tuberculosis from oxidative and nitrosative stress and is important for survival in macrophages and mice (98, 100, 101). The essentiality of whiB1, whiB2, and ideR indicates a particular need for M. tuberculosis to couple redox sensing and iron availability with basic cellular processes to maintain homeostasis.
NONESSENTIAL TCSs AND TFs: SPECIAL FORCES OF THE STRESS RESPONSE TEAM
In addition to the essential TCSs and TFs mentioned above, M. tuberculosis maintains 10 nonessential TCSs and a number of nonessential TFs that are not required for bacterial growth in vitro but respond to particular stresses.
The SenX3/RegX3 TCS is activated under low-phosphate conditions to regulate expression of genes encoding proteins involved in phosphate uptake, translation, lipid metabolism, DNA replication, and DNA repair (102, 103). The SenX3/RegX3 TCS is important for optimal M. tuberculosis growth during phosphate starvation and for survival in macrophages and mice where the bacteria encounter low phosphate levels (102).
The DosRST system responds to nitric oxide and hypoxia to activate the “dormancy regulon” in M. tuberculosis (104). This TCS contains 2 separate HKs, DosS and DosT, that are both capable of activating the DosR RR. DosS acts as a redox sensor and DosT as a hypoxia sensor, illustrating the integration and differentiation of M. tuberculosis stress responses (105). Genetic disruption of the dosRST TCS results in reduced bacterial survival under low-oxygen conditions, in mouse models that develop hypoxic lesions, and in a nonhuman primate macaque model of infection (106–109).
The PhoPR TCS is stimulated by low pH (110). The PhoP regulon includes multiple genes involved in cellular lipid synthesis, dosR, dosS, and genes involved in the ESX1 secretion system (111, 112). M. tuberculosis strains deficient in PhoPR activity display defects in replication in mice and macrophages (111–113). Supporting the idea of a role in M. tuberculosis virulence, mutations in phoPR in M. bovis and Mycobacterium africanum are associated with reduced mycobacterial virulence (114). In addition, M. tuberculosis phoPR mutants have defects in cell morphology and lipid production in the absence of stress, suggesting that PhoPR is required to maintain normal cell physiology under all growth conditions (113).
The MprAB TCS regulates expression of a subset of genes in the DosR regulon, the stress-responsive chaperone pepD, and the espA operon, which encodes ESX-1 substrates (115–118). The MprAB TCS also activates expression of sigB and sigE in response to envelope stress and indirectly regulates the stringent response mediator M. tuberculosis rel gene (relMtb) through σE activity (119, 120). Deletion of this TCS compromises M. tuberculosis viability during a persistent infection in mice but renders M. tuberculosis hypervirulent in macrophages, suggesting a role for this TCS in allowing the bacteria to appropriately respond to their specific in vivo niche (80, 121).
Genes encoding six additional TCSs, KdpDE, TrcRS, TcrXY, NarLS, PtdaRS, and Rv0600c/Rv0601c/TcrA, have been identified in the M. tuberculosis genome but have yet to be investigated in detail (79).
M. tuberculosis encodes five nonessential Fe-S cluster WhiB TF family members that have been implicated in a variety of cellular responses (96, 97). In particular, WhiB3, WhiB4, and WhiB5 impact M. tuberculosis virulence (122–124). Of these, WhiB3 has been studied in the most detail. WhiB3 promotes mycobacterial lipid regulation, and whiB3 mutants demonstrate altered macrophage cytokine release and reduced pathology in vivo, without directly impacting bacterial titers (125, 126). A model has been proposed in which WhiB3 senses the intracellular redox state and redirects lipid synthesis pathways to cope with reductive stress generated by host lipid catabolism during infection (125).
M. tuberculosis encodes a number of other known and predicted TFs not highlighted in this review. Recently, researchers overexpressed 200 predicted TFs in M. tuberculosis and performed chromatin immunoprecipitation sequencing experiments and microarray analyses to catalogue a genome-wide characterization of TF binding events and target gene expression (127–129). These reports describe 16,000 binding sites for 154 TFs and identify regulatory routes for ∼70% of the genome. The complex regulatory circuits that were uncovered highlight how much remains to be investigated regarding how M. tuberculosis regulates transcription to integrate precise stress responses.
THE STRINGENT RESPONSE: WHEN RATIONS RUN LOW
The stringent response is a conserved global stress response in bacteria that provides an additional layer of gene regulation in harsh environments. The stringent response is best characterized during amino acid starvation, when the RelMtb enzyme senses uncharged tRNAs in ribosomes and responds by transferring the pyrophosphate (PPi) group from ATP to GDP and GTP to synthesize hyperphosphorylated guanine nucleotides ppGpp and pppGpp [collectively called (p)ppGpp] (130). (p)ppGpp then coordinates downstream regulation of bacterial physiology and mediates changes in the transcriptional profile to support survival during stress. Deletion of relMtb led to differential expression of 159 genes during starvation, including genes involved in coordinating metabolic rate reduction, production of mycobacterial cell wall and lipids, secreted proteins, and cell division machinery (131). (p)ppGpp synthesis by RelMtb is required for survival under low-nutrient conditions, in long-term culture, and during infection in animal models, all indicative of a strict requirement for RelMtb during exposure to stress (131–135). In E. coli, (p)ppGpp directly affects transcription initiation by binding the RNAP (136, 137). In contrast, in a number of Gram-positive bacteria, (p)ppGpp inhibits GTP biosynthesis by directly interacting with GTP synthesis enzymes, which impacts gene expression by altering initiating nucleotide levels (137–140). Although (p)ppGpp has not been demonstrated to directly bind M. tuberculosis RNAP or GTP synthesis enzymes, (p)ppGpp has been reported to influence mycobacterial RNAP activity in vitro, suggesting that the mechanism of (p)ppGpp action in M. tuberculosis transcriptional modulation requires further investigation (136, 138, 141).
RelMtb also encodes a second distinct catalytic domain that hydrolyzes (p)ppGpp into PPi and GDP or GTP (142). It was recently shown that (p)ppGpp hydrolysis by RelMtb is important for growth and normal physiology in culture and during infection (135). These observations suggest that RelMtb constitutively produces (p)ppGpp independently of activation during nutrient limitation and may act continuously to maintain M. tuberculosis homeostasis under all growth conditions in addition to its role in survival during stress.
FINAL THOUGHTS
In order to respond to host-derived stresses, M. tuberculosis has evolved a complex network of strategies to modify gene expression and promote survival. The responses to different stresses are integrated and coordinated, often resulting in overlapping regulons and stress responders (Fig. 2, 3, and 4). Not only do these highly effective stress response strategies protect M. tuberculosis from host immunity, but the resulting changes in physiology also contribute to antibiotic tolerance, which precludes eradication of the infection (143–148). The recalcitrance of M. tuberculosis in response to antibiotic therapy has led to an increase in drug-resistant M. tuberculosis infections to the point that we are not equipped to successfully battle the M. tuberculosis epidemic (2). Therefore, new therapeutic strategies that target M. tuberculosis stress responses could increase the susceptibility of the bacteria to both the immune system and antibiotic treatment.
As an obligate pathogen, M. tuberculosis is specialized for survival in a mammalian host. Analysis of the conservation of transcriptional regulators across different mycobacterial species reveals some interesting patterns that reflect their respective lifestyles (Fig. 2). Mycobacterium leprae is an even more specialized pathogen than M. tuberculosis and has undergone a drastic reduction in genetic material to the point that this degenerate genome has retained only 4 functional σ factor genes (sigA, sigB, sigC, and sigE) and 5 TCSs. On the other end of the spectrum, environmental mycobacteria such as Mycobacterium smegmatis must adapt to a larger diversity of conditions within a larger range of environments. As such, M. smegmatis encodes 28 σ factors to facilitate a more versatile lifestyle. In addition, even when a transcriptional regulator is conserved across mycobacterial species, it can be coopted to perform a function specific for a particular species. For example, σF homologs are differentially regulated and activated in M. tuberculosis, M. smegmatis, and M. bovis (7, 29, 149).
Finally, this minireview is in no way exhaustive in terms of all of the mechanisms of transcriptional regulation that M. tuberculosis employs to respond to stress. In particular, there is a growing area of research into the roles of nucleoid-associated proteins and small RNAs (150–155). M. tuberculosis also contains 11 serine/threonine protein kinases (STPKs) that, like TCSs, are involved in signal transduction pathways that aid M. tuberculosis in adaptation to its environment (156). However, unlike TCSs that consist of HKs that activate RRs to directly modulate M. tuberculosis transcription, STPKs are single proteins that phosphorylate numerous downstream targets (156). Although STPKs do not directly affect M. tuberculosis transcription, they do influence gene expression by modifying the activity of other M. tuberculosis proteins with more-direct roles in transcription, such as σ factors, nucleoid-associated proteins, anti-anti-σ factors, and TCSs (24, 154, 157–159). These and other aspects of gene regulation further add to the complexity of stress responses in M. tuberculosis.
ACKNOWLEDGMENTS
We thank Michael Caparon, Katherine Mann, and Jerome Prusa for careful reading of the manuscript and providing their insightful comments.
C.L.S. is supported by a Beckman Young Investigator Award from the Arnold and Mabel Beckman Foundation, an Interdisciplinary Research Initiative grant from the Children's Discovery Institute of Washington University and St. Louis Children's Hospital, and grants GM107544 and AI111696-01 from the National Institutes of Health. K.F. is supported by a pilot award from the Center for Women's Infectious Disease Research at Washington University School of Medicine. A.L.G. is supported by a National Institute of General Medical Sciences (NIGMS) Cell and Molecular Biology Training Grant (grant GM007067) and the Stephen I. Morse Graduate Fellowship.
Biographies

Kelly Flentie is currently a postdoctoral associate in the laboratory of Christina Stallings at Washington University in St. Louis. She obtained her B.S. in microbiology from the University of Kansas in 2004. As an undergraduate, she completed her honors research studying the pathogenesis of Shigella flexneri under the guidance of William Picking. She completed her Ph.D. at Washington University in St. Louis in 2011 in the laboratory of David Piwnica-Worms. In her doctoral thesis research, she investigated interactions between Salmonella and cancer cells, with an emphasis on characterizing bacterial behaviors that could be coopted for novel cancer treatment or diagnostic strategies. Her current research focuses on interrogating mechanisms of stress tolerance in Mycobacterium tuberculosis and identifying new ways to target this pathogen.

Ashley L. Garner is a Ph.D. student at Washington University in St. Louis. In 2008, she received a B.S. in microbiology at the University of California, Davis, where she interned in the laboratory of Michele Igo, studying an autotransporter in the plant pathogen Xylella fastidiosa. After graduating, she worked for the R&D Department of Novozymes, studying cellulosic ethanol production. She joined the laboratory of Christina Stallings in 2010, where she currently researches the mycobacterial transcription regulator CarD.

Christina L. Stallings is an assistant professor in the Department of Molecular Microbiology at Washington University in St. Louis. She received her Ph.D. with distinction from Columbia University College of Physicians and Surgeons, where she performed her thesis work on alphaherpesviruses in the laboratory of Saul Silverstein. She then transitioned to another fascinating and chronic pathogen, Mycobacterium tuberculosis, for her postdoctoral research in Michael Glickman's laboratory at the Sloan-Kettering Institute. She started in her faculty position at Washington University in St. Louis in 2010, and research in her laboratory seeks to dissect the molecular mechanisms involved in M. tuberculosis pathogenesis and stress responses.
REFERENCES
- 1.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, Gagneux S. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol 6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2015. Global tuberculosis report 2015. WHO, Geneva, Switzerland. [Google Scholar]
- 3.Stallings CL, Glickman MS. 2010. Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infect 12:1091–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigue S, Provvedi R, Jacques PÉ, Gaudreau L, Manganelli R. 2006. The σ factors of Mycobacterium tuberculosis. FEMS Microbiol Rev 30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Coates ARM. 2001. Increased levels of sigJ mRNA in late stationary phase cultures of Mycobacterium tuberculosis detected by DNA array hybridisation. FEMS Microbiol Lett 202:59–65. doi: 10.1111/j.1574-6968.2001.tb10780.x. [DOI] [PubMed] [Google Scholar]
- 6.Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J Bacteriol 186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol 31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 8.Murakami KS, Darst SA. 2003. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13:31–39. doi: 10.1016/S0959-440X(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 9.Saecker RM, Record MT, Dehaseth PL. 2011. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat Rev Microbiol 2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 11.Rojo F. 2001. Mechanisms of transcriptional repression. Curr Opin Microbiol 4:145–151. doi: 10.1016/S1369-5274(00)00180-6. [DOI] [PubMed] [Google Scholar]
- 12.Lee DJ, Minchin SD, Busby SJW. 2012. Activating transcription in bacteria. Annu Rev Microbiol 66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 13.Rammohan J, Ruiz Manzano A, Garner AL, Stallings CL, Galburt EA. 2015. CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res 43:3272–3285. doi: 10.1093/nar/gkv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis E, Chen J, Leon K, Darst SA, Campbell EA. 2015. Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res 43:433–445. doi: 10.1093/nar/gku1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whipple FW, Sonenshein AL. 1992. Mechanism of initiation of transcription by Bacillus subtilis RNA polymerase at several promoters. J Mol Biol 223:399–414. doi: 10.1016/0022-2836(92)90660-C. [DOI] [PubMed] [Google Scholar]
- 16.Xue Y, Hogan BP, Erie DA. 2000. Purification and initial characterization of RNA polymerase from Thermus thermophilus strain HB8. Biochemistry 39:14356–14362. doi: 10.1021/bi0012538. [DOI] [PubMed] [Google Scholar]
- 17.Miropolskaya N, Ignatov A, Bass I, Zhilina E, Pupov D, Kulbachinskiy A. 2012. Distinct functions of regions 1.1 and 1.2 of RNA polymerase subunits from Escherichia coli and Thermus aquaticus in transcription initiation. J Biol Chem 287:23779–23789. doi: 10.1074/jbc.M112.363242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wösten M. 1998. Eubacterial sigma-factors. FEMS Microbiol Rev 22:127–150. [DOI] [PubMed] [Google Scholar]
- 19.Newton-Foot M, Gey van Pittius NC. 2013. The complex architecture of mycobacterial promoters. Tuberculosis 93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Bashyam MD, Kaushal D, Dasgupta SK, Tyagi AK. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol 178:4847–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal N, Tyagi AK. 2006. Mycobacterial transcriptional signals: requirements for recognition by RNA polymerase and optimal transcriptional activity. Nucleic Acids Res 34:4245–4257. doi: 10.1093/nar/gkl521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kremer L, Baulard A, Estaquier J, Content J, Capron A, Locht C. 1995. Analysis of the Mycobacterium tuberculosis 85A antigen promoter region. J Bacteriol 177:642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bashyam MD, Tyagi AK. 1998. Identification and analysis of “extended −10” promoters from mycobacteria. J Bacteriol 180:2568–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sachdeva P, Misra R, Tyagi AK, Singh Y. 2010. The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J 277:605–626. doi: 10.1111/j.1742-4658.2009.07479.x. [DOI] [PubMed] [Google Scholar]
- 25.Song T, Dove SL, Lee KH, Husson RN. 2003. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol Microbiol 50:949–959. doi: 10.1046/j.1365-2958.2003.03739.x. [DOI] [PubMed] [Google Scholar]
- 26.Hahn M, Raman S, Anaya M, Husson RN. 2005. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J Bacteriol 187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saïd-Salim B, Mostowy S, Kristof AS, Behr MA. 2006. Mutations in Mycobacterium tuberculosis Rv0444c, the gene encoding anti-SigK, explain high level expression of MPB70 and MPB83 in Mycobacterium bovis. Mol Microbiol 62:1251–1263. doi: 10.1111/j.1365-2958.2006.05455.x. [DOI] [PubMed] [Google Scholar]
- 28.DeMaio J, Zhang Y, Ko C, Bishai WR. 1997. Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tuber Lung Dis 78:3–12. doi: 10.1016/S0962-8479(97)90010-1. [DOI] [PubMed] [Google Scholar]
- 29.Singh AK, Singh BN. 2008. Conservation of sigma F in mycobacteria and its expression in Mycobacterium smegmatis. Curr Microbiol 56:574–580. doi: 10.1007/s00284-008-9126-8. [DOI] [PubMed] [Google Scholar]
- 30.Donà V, Rodrigue S, Dainese E, Palù G, Gaudreau L, Manganelli R, Provvedi R. 2008. Evidence of complex transcriptional, translational, and posttranslational regulation of the extracytoplasmic function sigma factor σE in Mycobacterium tuberculosis. J Bacteriol 190:5963–5971. doi: 10.1128/JB.00622-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider JS, Sklar JG, Glickman MS. 2014. The Rip1 protease of mycobacterium tuberculosis controls the SigD regulon. J Bacteriol 196:2638–2645. doi: 10.1128/JB.01537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sklar JG, Makinoshima H, Schneider JS, Glickman MS. 2010. M. tuberculosis intramembrane protease Rip1 controls transcription through three anti-sigma factor substrates. Mol Microbiol 77:605–617. doi: 10.1111/j.1365-2958.2010.07232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaudion AE. 2011. The role of the ECF sigma factor SigG in Mycobacterium tuberculosis. Ph.D. thesis. MRC National Institute for Medical Research, London, United Kingdom. [Google Scholar]
- 34.Hu Y, Coates ARM. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol 181:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P, Ruiz RE, Li Q, Silver RF, Bishai WR. 2000. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect Immun 68:5575–5580. doi: 10.1128/IAI.68.10.5575-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raman S, Song T, Puyang X, Jacobs WRJ, Husson RN. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J Bacteriol 183:6119–6125. doi: 10.1128/JB.183.20.6119-6125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol 41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 38.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. 2002. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol Microbiol 45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, Kendall S, Stoker NG, Coates ARM. 2004. The Mycobacterium tuberculosis sigJ gene controls sensitivity of the bacterium to hydrogen peroxide. FEMS Microbiol Lett 237:415–423. doi: 10.1111/j.1574-6968.2004.tb09725.x. [DOI] [PubMed] [Google Scholar]
- 40.Raman S, Puyang X, Cheng TY, Young DC, Moody DB, Husson RN. 2006. Mycobacterium tuberculosis SigM positively regulates Esx secreted protein and nonribosomal peptide synthetase genes and down regulates virulence-associated surface lipid synthesis. J Bacteriol 188:8460–8468. doi: 10.1128/JB.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal N, Woolwine SC, Tyagi S, Bishai WR. 2007. Characterization of the Mycobacterium tuberculosis sigma factor SigM by assessment of virulence and identification of SigM-dependent genes. Infect Immun 75:452–461. doi: 10.1128/IAI.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fontán PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, Fattorini L, Schoolnik GK, Smith I. 2009. The Mycobacterium tuberculosis sigma factor B is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J Bacteriol 191:5628–5633. doi: 10.1128/JB.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dutta NK, Mehra S, Kaushal D. 2010. A Mycobacterium tuberculosis sigma factor network responds to cell-envelope damage by the promising anti-mycobacterial thioridazine. PLoS One 5:e10069. doi: 10.1371/journal.pone.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geiman D, Kaushal D, Ko C. 2004. Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect Immun 72:1733–1745. doi: 10.1128/IAI.72.3.1733-1745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smollett KL, Dawson LF, Davis EO. 2011. SigG does not control gene expression in response to DNA damage in Mycobacterium tuberculosis H37Rv. J Bacteriol 193:1007–1011. doi: 10.1128/JB.01241-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 47.Graham JE, Clark-Curtiss JE. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc Natl Acad Sci U S A 96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cappelli G, Volpe E, Grassi M, Liseo B, Colizzi V, Mariani F. 2006. Profiling of Mycobacterium tuberculosis gene expression during human macrophage infection: upregulation of the alternative sigma factor G, a group of transcriptional regulators, and proteins with unknown function. Res Microbiol 157:445–455. doi: 10.1016/j.resmic.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Lee J-H, Geiman DE, Bishai WR. 2008. Role of stress response sigma factor SigG in Mycobacterium tuberculosis. J Bacteriol 190:1128–1133. doi: 10.1128/JB.00511-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaudion A, Dawson L, Davis E, Smollett K. 2013. Characterisation of the mycobacterium tuberculosis alternative sigma factor SigG: its operon and regulon. Tuberculosis 93:482–491. doi: 10.1016/j.tube.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calamita H, Ko C, Tyagi S, Yoshimatsu T, Morrison NE, Bishai WR. 2005. The Mycobacterium tuberculosis SigD sigma factor controls the expression of ribosome-associated gene products in stationary phase and is required for full virulence. Cell Microbiol 7:233–244. [DOI] [PubMed] [Google Scholar]
- 52.Ando M, Yoshimatsu T, Ko C, Converse PJ, Bishai WR. 2003. Deletion of Mycobacterium tuberculosis sigma factor E results in delayed time to death with bacterial persistence in the lungs of aerosol-infected mice. Infect Immun 71:7170–7172. doi: 10.1128/IAI.71.12.7170-7172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaushal D, Schroeder BG, Tyagi S, Yoshimatsu T, Scott C, Ko C, Carpenter L, Mehrotra J, Manabe YC, Fleischmann RD, Bishai WR. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci U S A 99:8330–8335. doi: 10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dainese E, Rodrigue S, Delogu G, Provvedi R, Laflamme L, Brzezinski R, Fadda G, Smith I, Gaudreau L, Palù G, Manganelli R. 2006. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor sigma L and roles in virulence and in global regulation of gene expression. Infect Immun 74:2457–2461. doi: 10.1128/IAI.74.4.2457-2461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun R, Converse PJ, Ko C, Tyagi S, Morrison NE, Bishai WR. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol Microbiol 52:25–38. doi: 10.1111/j.1365-2958.2003.03958.x. [DOI] [PubMed] [Google Scholar]
- 56.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. 2009. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol Microbiol 42:1007–1020. doi: 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- 58.Weiss LA, Harrison PG, Nickels BE, Glickman MS, Campbell EA, Darst SA, Stallings CL. 2012. Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J Bacteriol 194:5621–5631. doi: 10.1128/JB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garner AL, Weiss LA, Ruiz Manzano A, Galburt EA, Stallings CL. 2014. CarD integrates three functional modules to promote efficient transcription, antibiotic tolerance, and pathogenesis in mycobacteria. Mol Microbiol 93:682–697. doi: 10.1111/mmi.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing KG, Young DB, Butcher PD. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 61.Provvedi R, Boldrin F, Falciani F, Palù G, Manganelli R. 2009. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155:1093–1102. doi: 10.1099/mic.0.024802-0. [DOI] [PubMed] [Google Scholar]
- 62.Murphy DJ, Brown JR. 2007. Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis 7:84. doi: 10.1186/1471-2334-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dey A, Verma AK, Chatterji D. 2010. Role of an RNA polymerase interacting protein, MsRbpA, from Mycobacterium smegmatis in phenotypic tolerance to rifampicin. Microbiology 156:873–883. doi: 10.1099/mic.0.033670-0. [DOI] [PubMed] [Google Scholar]
- 64.Bae B, Chen J, Davis E, Leon K, Darst SA, Campbell EA. 2015. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. Elife 4:e08505. doi: 10.7554/eLife.08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srivastava DB, Leon K, Osmundson J, Garner AL, Weiss LA, Westblade LF, Glickman MS, Landick R, Darst SA, Stallings CL, Campbell EA. 2013. Structure and function of CarD, an essential mycobacterial transcription factor. Proc Natl Acad Sci U S A 110:12619–12624. doi: 10.1073/pnas.1308270110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forti F, Mauri V, Dehò G, Ghisotti D. 2011. Isolation of conditional expression mutants in Mycobacterium tuberculosis by transposon mutagenesis. Tuberculosis (Edinb) 91:569–578. doi: 10.1016/j.tube.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 67.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 68.Bortoluzzi A, Muskett FW, Waters LC, Addis PW, Rieck B, Munder T, Schleier S, Forti F, Ghisotti D, Carr MD, O'Hare HM. 2013. Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors. J Biol Chem 288:14438–14450. doi: 10.1074/jbc.M113.459883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.García-Moreno D, Abellón-Ruiz J, García-Heras F, Murillo FJ, Padmanabhan S, Elías-Arnanz M. 2010. CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucleic Acids Res 38:4586–4598. doi: 10.1093/nar/gkq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gulten G, Sacchettini JC. 2013. Structure of the Mtb CarD/RNAP β-lobes complex reveals the molecular basis of interaction and presents a distinct DNA-binding domain for Mtb CarD. Structure 21:1859–1869. doi: 10.1016/j.str.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hubin EA, Tabib-Salazar A, Humphrey LJ, Flack JE, Olinares PDB, Darst SA, Campbell EA, Paget MS. 2015. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc Natl Acad Sci U S A 112:7171–7176. doi: 10.1073/pnas.1504942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tabib-Salazar A, Liu B, Doughty P, Lewis RA, Ghosh S, Parsy M-L, Simpson PJ, O'Dwyer K, Matthews SJ, Paget MS. 2013. The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase. Nucleic Acids Res 41:5679–5691. doi: 10.1093/nar/gkt277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dey A, Verma AK, Chatterji D. 2011. Molecular insights into the mechanism of phenotypic tolerance to rifampicin conferred on mycobacterial RNA polymerase by MsRbpA. Microbiology 157:2056–2071. doi: 10.1099/mic.0.047480-0. [DOI] [PubMed] [Google Scholar]
- 74.Hu Y, Morichaud Z, Chen S, Leonetti J-P, Brodolin K. 2012. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the σ A-containing RNA polymerase holoenzyme. Nucleic Acids Res 40:6547–6557. doi: 10.1093/nar/gks346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu Y, Morichaud Z, Perumal AS, Roquet-Baneres F, Brodolin K. 2014. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res 42:10399–10408. doi: 10.1093/nar/gku742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verma AK, Chatterji D. 2014. Dual role of MsRbpA: transcription activation and rescue of transcription from the inhibitory effect of rifampicin. Microbiology 160:2018–2029. doi: 10.1099/mic.0.079186-0. [DOI] [PubMed] [Google Scholar]
- 77.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 78.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parish T. 2014. Two-component regulatory systems of Mycobacteria, p 209–223. In Hatfull GF, Jacobs WR (ed), Molecular genetics of Mycobacteria, 2nd ed ASM Press, Washington, DC. doi: 10.1128/microbiolspec.MGM2-0010-2013. [DOI] [Google Scholar]
- 80.Zahrt TC, Deretic V. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J Bacteriol 182:3832–3838. doi: 10.1128/JB.182.13.3832-3838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haydel SE, Malhotra V, Cornelison GL, Clark-Curtiss JE. 2012. The prrAB two-component system is essential for Mycobacterium tuberculosis viability and is induced under nitrogen-limiting conditions. J Bacteriol 194:354–361. doi: 10.1128/JB.06258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Plocinska R, Purushotham G, Sarva K, Vadrevu IS, Pandeeti EVP, Arora N, Plocinski P, Madiraju MV, Rajagopalan M. 2012. Septal localization of the Mycobacterium tuberculosis MtrB sensor kinase promotes MtrA regulon expression. J Biol Chem 287:23887–23899. doi: 10.1074/jbc.M112.346544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajagopalan M, Dziedzic R, Al Zayer M, Stankowska D, Ouimet M-C, Bastedo DP, Marczynski GT, Madiraju MV. 2010. Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J Biol Chem 285:15816–15827. doi: 10.1074/jbc.M109.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma AK, Chatterjee A, Gupta S, Banerjee R, Mandal S, Mukhopadhyay J, Basu J, Kundu M. 2015. MtrA, an essential response regulator of the MtrAB two-component system, regulates the transcription of resuscitation-promoting factor B of Mycobacterium tuberculosis. Microbiology 161:1271–1281. doi: 10.1099/mic.0.000087. [DOI] [PubMed] [Google Scholar]
- 85.Ewann F, Locht C, Supply P. 2004. Intracellular autoregulation of the Mycobacterium tuberculosis PrrA response regulator. Microbiology 150:241–246. doi: 10.1099/mic.0.26516-0. [DOI] [PubMed] [Google Scholar]
- 86.Ewann F. 2002. Transient requirement of the PrrA-PrrB two-component system for early intracellular multiplication of Mycobacterium tuberculosis. Infect Immun 70:2256–2263. doi: 10.1128/IAI.70.5.2256-2263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Crack JC, Green J, Cheesman MR, Le Brun NE, Thomson AJ. 2007. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc Natl Acad Sci U S A 104:2092–2097. doi: 10.1073/pnas.0609514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu M, Imlay JA. 2011. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol 79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS. 1996. Nitrosative stress: activation of the transcription factor OxyR. Cell 86:719–729. doi: 10.1016/S0092-8674(00)80147-6. [DOI] [PubMed] [Google Scholar]
- 90.Poole RK, Anjum MF, Membrillo-Hernández J, Kim SO, Hughes MN, Stewart V. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J Bacteriol 178:5487–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu J, Weiss B. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J Bacteriol 174:3915–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saini V, Farhana A, Glasgow JN, Steyn AJC. 2012. Iron sulfur cluster proteins and microbial regulation: implications for understanding tuberculosis. Curr Opin Chem Biol 16:45–53. doi: 10.1016/j.cbpa.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith LJ, Stapleton MR, Fullstone GJM, Crack JC, Thomson AJ, Le Brun NE, Hunt DM, Harvey E, Adinolfi S, Buxton RS, Green J. 2010. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem J 432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Raghunand TR, Bishai WR. 2006. Mycobacterium smegmatis whmD and its homologue Mycobacterium tuberculosis whiB2 are functionally equivalent. Microbiology 152:2735–2747. doi: 10.1099/mic.0.28911-0. [DOI] [PubMed] [Google Scholar]
- 95.Gomez JE, Bishai WR. 2000. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc Natl Acad Sci U S A 97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Larsson C, Luna B, Ammerman NC, Maiga M, Agarwal N, Bishai WR. 2012. Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1–7 in redox environments. PLoS One 7:e37516. doi: 10.1371/journal.pone.0037516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Geiman DE, Raghunand TR, Agarwal N, Bishai WR. 2006. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob Agents Chemother 50:2836–2841. doi: 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun 70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wisedchaisri G, Holmes RK, Hol WGJ. 2004. Crystal structure of an IdeR-DNA complex reveals a conformational change in activated IdeR for base-specific interactions. J Mol Biol 342:1155–1169. doi: 10.1016/j.jmb.2004.07.083. [DOI] [PubMed] [Google Scholar]
- 100.Gold B, Rodriguez GM, Marras SA, Pentecost M, Smith I. 2001. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol Microbiol 42:851–865. [DOI] [PubMed] [Google Scholar]
- 101.Pandey R, Rodriguez GM. 2014. IdeR is required for iron homeostasis and virulence in Mycobacterium tuberculosis. Mol Microbiol 91:98–109. doi: 10.1111/mmi.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parish T, Smith DA, Roberts G, Betts J, Stoker NG. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149:1423–1435. doi: 10.1099/mic.0.26245-0. [DOI] [PubMed] [Google Scholar]
- 103.Rifat D, Bishai WR, Karakousis PC. 2009. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J Infect Dis 200:1126–1135. doi: 10.1086/605700. [DOI] [PubMed] [Google Scholar]
- 104.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med 198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kumar A, Toledo JC, Patel RP, Lancaster JR, Steyn AJC. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A 104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mehra S, Foreman TW, Didier PJ, Ahsan MH, Hudock TA, Kissee R, Golden NA, Gautam US, Johnson A-M, Alvarez X, Russell-Lodrigue KE, Doyle LA, Roy CJ, Niu T, Blanchard JL, Khader SA, Lackner AA, Sherman DR, Kaushal D. 2015. The DosR regulon modulates adaptive immunity and is essential for Mycobacterium tuberculosis persistence. Am J Respir Crit Care Med 191:1185–1196. doi: 10.1164/rccm.201408-1502OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Voskuil MI, Schlesinger LS. 2015. Toward resolving the paradox of the critical role of the DosR regulon in Mycobacterium tuberculosis persistence and active disease. Am J Respir Crit Care Med 191:1103–1105. doi: 10.1164/rccm.201503-0424ED. [DOI] [PubMed] [Google Scholar]
- 108.Gautam US, Sikri K, Vashist A, Singh V, Tyagi JS. 2014. Essentiality of DevR/DosR interaction with SigA for the dormancy survival program in Mycobacterium tuberculosis. J Bacteriol 196:790–799. doi: 10.1128/JB.01270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gautam US, McGillivray A, Mehra S, Didier PJ, Midkiff CC, Kissee RS, Golden NA, Alvarez X, Niu T, Rengarajan J, Sherman DR, Kaushal D. 2015. DosS Is required for the complete virulence of mycobacterium tuberculosis in mice with classical granulomatous lesions. Am J Respir Cell Mol Biol 52:708–716. doi: 10.1165/rcmb.2014-0230OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abramovitch RB, Rohde KH, Hsu F-F, Russell DG. 2011. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol 80:678–694. doi: 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, Brodin P, Garnier T, Gicquel B, Martin C, Leclerc C, Cole ST, Brosch R. 2008. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog 4:e33. doi: 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gonzalo-Asensio J, Mostowy S, Harders-Westerveen J, Huygen K, Hernández-Pando R, Thole J, Behr M, Gicquel B, Martín C. 2008. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One 3:e3496. doi: 10.1371/journal.pone.0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. 2006. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol 60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 114.Gonzalo-Asensio J, Malaga W, Pawlik A, Astarie-Dequeker C, Passemar C, Moreau F, Laval F, Daffe M, Martin C, Brosch R, Guilhot C. 2014. Evolutionary history of tuberculosis shaped by conserved mutations in the PhoPR virulence regulator. Proc Natl Acad Sci U S A 111:11491–11496. doi: 10.1073/pnas.1406693111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.White MJ, He H, Penoske RM, Twining SS, Zahrt TC. 2010. PepD participates in the mycobacterial stress response mediated through MprAB and SigE. J Bacteriol 192:1498–1510. doi: 10.1128/JB.01167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bretl DJ, He H, Demetriadou C, White MJ, Penoske RM, Salzman NH, Zahrt TC. 2012. MprA and DosR coregulate a Mycobacterium tuberculosis virulence operon encoding Rv1813c and Rv1812c. Infect Immun 80:3018–3033. doi: 10.1128/IAI.00520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.He H, Bretl DJ, Penoske RM, Anderson DM, Zahrt TC. 2011. Components of the Rv0081-Rv0088 locus, which encodes a predicted formate hydrogenlyase complex, are coregulated by Rv0081, MprA, and DosR in Mycobacterium tuberculosis. J Bacteriol 193:5105–5118. doi: 10.1128/JB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pang X, Samten B, Cao G, Wang X, Tvinnereim A, Chen RX-L, Howard ST. 2013. MprAB regulates the espA operon in Mycobacterium tuberculosis and modulates ESX-1 function and host cytokine response. J Bacteriol 195:66–75. doi: 10.1128/JB.01067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pang X, Vu P, Byrd TF, Ghanny S, Soteropoulos P, Mukamolova GV, Wu S, Samten B, Howard ST. 2007. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153:1229–1242. doi: 10.1099/mic.0.29281-0. [DOI] [PubMed] [Google Scholar]
- 120.Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodrigue S, Basu J, Kundu M. 2007. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol 65:261–276. doi: 10.1111/j.1365-2958.2007.05814.x. [DOI] [PubMed] [Google Scholar]
- 121.Zahrt TC, Wozniak C, Jones D, Trevett A. 2003. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect Immun 71:6962–6970. doi: 10.1128/IAI.71.12.6962-6970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Burian J, Ramon-Garcia S, Howes CG, Thompson CJ. 2012. WhiB7, a transcriptional activator that coordinates physiology with intrinsic drug resistance in Mycobacterium tuberculosis. Expert Rev Anti Infect Ther 10:1037–1047. doi: 10.1586/eri.12.90. [DOI] [PubMed] [Google Scholar]
- 123.Casonato S, Cervantes Sánchez A, Haruki H, Rengifo González M, Provvedi R, Dainese E, Jaouen T, Gola S, Bini E, Vicente M, Johnsson K, Ghisotti D, Palù G, Hernández-Pando R, Manganelli R. 2012. WhiB5, a transcriptional regulator that contributes to Mycobacterium tuberculosis virulence and reactivation. Infect Immun 80:3132–3144. doi: 10.1128/IAI.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chawla M, Parikh P, Saxena A, Munshi M, Mehta M, Mai D, Srivastava AK, Narasimhulu KV, Redding KE, Vashi N, Kumar D, Steyn AJC, Singh A. 2012. Mycobacterium tuberculosis WhiB4 regulates oxidative stress response to modulate survival and dissemination in vivo. Mol Microbiol 85:1148–1165. doi: 10.1111/j.1365-2958.2012.08165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJC. 2009. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog 5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Steyn AJC, Collins DM, Hondalus MK, Jacobs WR, Kawakami RP, Bloom BR. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci U S A 99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Turkarslan S, Peterson EJR, Rustad TR, Minch KJ, Reiss DJ, Morrison R, Ma S, Price ND, Sherman DR, Baliga NS. 2015. A comprehensive map of genome-wide gene regulation in Mycobacterium tuberculosis. Sci Data 2:150010. doi: 10.1038/sdata.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Minch KJ, Rustad TR, Peterson EJR, Winkler J, Reiss DJ, Ma S, Hickey M, Brabant W, Morrison B, Turkarslan S, Mawhinney C, Galagan JE, Price ND, Baliga NS, Sherman DR. 2015. The DNA-binding network of Mycobacterium tuberculosis. Nat Commun 6:5829. doi: 10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baloni P, Chandra N. 2015. Architectural plan of transcriptional regulation in Mycobacterium tuberculosis. Trends Microbiol 23:123–125. doi: 10.1016/j.tim.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 130.Avarbock D, Avarbock A, Rubin H. 2000. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry 39:11640–11648. [DOI] [PubMed] [Google Scholar]
- 131.Dahl JL, Kraus CN, Boshoff HIM, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A 100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol 182:4889–4898. doi: 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klinkenberg LG, Lee J-H, Bishai WR, Karakousis PC. 2010. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202:1397–1404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med 200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weiss LA, Stallings CL. 2013. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J Bacteriol 195:5629–5638. doi: 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. 2013. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Vassylyev DG, Ross W, Gourse RL. 2008. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol 377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu K, Myers AR, Pisithkul T, Claas KR, Satyshur KA, Amador-Noguez D, Keck JL, Wang JD. 2015. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell 57:735–749. doi: 10.1016/j.molcel.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Krásný L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tare P, Mallick B, Nagaraja V. 2013. Co-evolution of specific amino acid in sigma 1.2 region and nucleotide base in the discriminator to act as sensors of small molecule effectors of transcription initiation in mycobacteria. Mol Microbiol 90:569–583. doi: 10.1111/mmi.12384. [DOI] [PubMed] [Google Scholar]
- 142.Avarbock A, Avarbock D, Teh J-S, Buckstein M, Wang Z, Rubin H. 2005. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44:9913–9923. doi: 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- 143.Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 64:2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Deb C, Lee C-M, Dubey VS, Daniel J, Abomoelak B, Sirakova TD, Pawar S, Rogers L, Kolattukudy PE. 2009. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sarathy J, Dartois V, Dick T, Gengenbacher M. 2013. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:1648–1653. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baek S-H, Li AH, Sassetti CM. 2011. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol 9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cunningham-Bussel A, Bange FC, Nathan CF. 2013. Nitrite impacts the survival of Mycobacterium tuberculosis in response to isoniazid and hydrogen peroxide. Microbiologyopen 2:901–911. doi: 10.1002/mbo3.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Franzblau SG, DeGroote MA, Cho SH, Andries K, Nuermberger E, Orme IM, Mdluli K, Angulo-Barturen I, Dick T, Dartois V, Lenaerts AJ. 2012. Comprehensive analysis of methods used for the evaluation of compounds against Mycobacterium tuberculosis. Tuberculosis (Edinb) 92:453–488. doi: 10.1016/j.tube.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 149.Michele TM, Ko C, Bishai WR. 1999. Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob Agents Chemother 43:218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, Perkins TT, Parkhill J, Dougan G, Young DB. 2011. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog 7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haning K, Cho SH, Contreras LM. 2014. Small RNAs in mycobacteria: an unfolding story. Front Cell Infect Microbiol 4:96. doi: 10.3389/fcimb.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pelly S, Bishai WR, Lamichhane G. 2012. A screen for non-coding RNA in Mycobacterium tuberculosis reveals a cAMP-responsive RNA that is expressed during infection. Gene 500:85–92. doi: 10.1016/j.gene.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.DiChiara JM, Contreras-Martinez LM, Livny J, Smith D, McDonough KA, Belfort M. 2010. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic Acids Res 38:4067–4078. doi: 10.1093/nar/gkq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gupta M, Sajid A, Sharma K, Ghosh S, Arora G, Singh R, Nagaraja V, Tandon V, Singh Y. 2014. HupB, a nucleoid-associated protein of Mycobacterium tuberculosis, is modified by serine/threonine protein kinases in vivo. J Bacteriol 196:2646–2657. doi: 10.1128/JB.01625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gordon BRG, Li Y, Wang L, Sintsova A, Van Bakel H, Tian S, Navarre WW, Xia B, Liu J. 2010. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Prisic S, Husson RN. 2014. Mycobacterium tuberculosis serine/threonine protein kinases, p 681–708. In Hatfull GF, Jacobs WR (ed), Molecular genetics of Mycobacteria, 2nd ed ASM Press, Washington, DC. doi: 10.1128/microbiolspec.MGM2-0006-2013. [DOI] [Google Scholar]
- 157.Park ST, Kang C-M, Husson RN. 2008. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 105:13105–13110. doi: 10.1073/pnas.0801143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Greenstein AE, MacGurn JA, Baer CE, Falick AM, Cox JS, Alber T. 2007. M. tuberculosis Ser/Thr protein kinase D phosphorylates an anti-anti-sigma factor homolog. PLoS Pathog 3:e49. doi: 10.1371/journal.ppat.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chao JD, Papavinasasundaram KG, Zheng X, Chávez-Steenbock A, Wang X, Lee GQ, Av-Gay Y. 2010. Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in Mycobacterium tuberculosis. J Biol Chem 285:29239–29246. doi: 10.1074/jbc.M110.132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee JH, Karakousis PC, Bishai WR. 2008. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J Bacteriol 190:699–707. doi: 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.He H, Hovey R, Kane J, Singh V, Zahrt TC. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J Bacteriol 188:2134–2143. doi: 10.1128/JB.188.6.2134-2143.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Abdul-Majid K-B, Ly LH, Converse PJ, Geiman DE, McMurray DN, Bishai WR. 2008. Altered cellular infiltration and cytokine levels during early Mycobacterium tuberculosis sigC mutant infection are associated with late-stage disease attenuation and milder immunopathology in mice. BMC Microbiol 8:151. doi: 10.1186/1471-2180-8-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dubnau E, Fontán P, Manganelli R, Soares-Appel S, Smith I. 2002. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect Immun 70:2787–2795. doi: 10.1128/IAI.70.6.2787-2795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.DeMaio J, Zhang Y, Ko C, Young DB, Bishai WR. 1996. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hartkoorn RC, Sala C, Uplekar S, Busso P, Rougemont J, Cole ST. 2012. Genome-wide definition of the SigF regulon in Mycobacterium tuberculosis. J Bacteriol 194:2001–2009. doi: 10.1128/JB.06692-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Williams EP, Lee J-H, Bishai WR, Colantuoni C, Karakousis PC. 2007. Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J Bacteriol 189:4234–4242. doi: 10.1128/JB.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rand L, Hinds J, Springer B, Sander P, Buxton RS, Davis EO. 2003. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol Microbiol 50:1031–1042. doi: 10.1046/j.1365-2958.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- 168.Dutta NK, Mehra S, Didier PJ, Roy CJ, Doyle LA, Alvarez X, Ratterree M, Be NA, Lamichhane G, Jain SK, Lacey MR, Lackner AA, Kaushal D. 2010. Genetic requirements for the survival of tubercle bacilli in primates. J Infect Dis 201:1743–1752. doi: 10.1086/652497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Veyrier F, Saïd-Salim B, Behr MA. 2008. Evolution of the mycobacterial SigK regulon. J Bacteriol 190:1891–1899. doi: 10.1128/JB.01452-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Talaat AM, Lyons R, Howard ST, Johnston SA. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci U S A 101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, Jagannath C, Madiraju MVVS, Rajagopalan M. 2006. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol 60:643–657. doi: 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 172.Rickman L, Saldanha JW, Hunt DM, Hoar DN, Colston MJ, Millar JBA, Buxton RS. 2004. A two-component signal transduction system with a PAS domain-containing sensor is required for virulence of Mycobacterium tuberculosis in mice. Biochem Biophys Res Commun 314:259–267. doi: 10.1016/j.bbrc.2003.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.James JN, Hasan Z, Ioerger TR, Brown AC, Personne Y, Carroll P, Ikeh M, Tilston-Lunel NL, Palavecino C, Sacchettini JC, Parish T. 2012. Deletion of SenX3-RegX3, a key two-component regulatory system of Mycobacterium smegmatis, results in growth defects under phosphate-limiting conditions. Microbiology 158:2724–2731. doi: 10.1099/mic.0.060319-0. [DOI] [PubMed] [Google Scholar]
- 174.Singh N, Kumar A. 2015. Virulence factor SenX3 is the oxygen-controlled replication switch of Mycobacterium tuberculosis. Antioxid Redox Signal 22:603–613. doi: 10.1089/ars.2014.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rifat D, Belchis DA, Karakousis PC. 2014. senX3-independent contribution of regX3 to Mycobacterium tuberculosis virulence. BMC Microbiol 14:265. doi: 10.1186/s12866-014-0265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zahrt TC, Deretic V. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc Natl Acad Sci U S A 98:12706–12711. doi: 10.1073/pnas.221272198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kendall SL, Movahedzadeh F, Rison SCG, Wernisch L, Parish T, Duncan K, Betts JC, Stoker NG. 2004. The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis (Edinb) 84:247–255. doi: 10.1016/j.tube.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 178.Converse PJ, Karakousis PC, Klinkenberg LG, Kesavan AK, Ly LH, Allen SS, Grosset JH, Jain SK, Lamichhane G, Manabe YC, McMurray DN, Nuermberger EL, Bishai WR. 2009. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect Immun 77:1230–1237. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ferrer NL, Gomez AB, Neyrolles O, Gicquel B, Martin C. 2010. Interactions of attenuated Mycobacterium tuberculosis phoP mutant with human macrophages. PLoS One 5:e12978. doi: 10.1371/journal.pone.0012978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Baker JJ, Johnson BK, Abramovitch RB. 2014. Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources. Mol Microbiol 94:56–69. doi: 10.1111/mmi.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pérez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martín C. 2001. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol Microbiol 41:179–187. doi: 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- 182.Tan S, Sukumar N, Abramovitch RB, Parish T, Russell DG. 2013. Mycobacterium tuberculosis responds to chloride and pH as synergistic cues to the immune status of its host cell. PLoS Pathog 9:e1003282. doi: 10.1371/journal.ppat.1003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Malhotra V, Agrawal R, Duncan TR, Saini DK, Clark-Curtiss JE. 2015. Mycobacterium tuberculosis response regulators, DevR and NarL, interact in vivo and co-regulate gene expression during aerobic nitrate metabolism. J Biol Chem 290:8294–8309. doi: 10.1074/jbc.M114.591800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Haydel SE, Benjamin WH, Dunlap NE, Clark-Curtiss JE. 2002. Expression, autoregulation, and DNA binding properties of the Mycobacterium tuberculosis TrcR response regulator. J Bacteriol 184:2192–2203. doi: 10.1128/JB.184.8.2192-2203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lane WJ, Darst SA. 2010. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J Mol Biol 395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]