ABSTRACT
The alarmone (p)ppGpp regulates transcription, translation, replication, virulence, lipid synthesis, antibiotic sensitivity, biofilm formation, and other functions in bacteria. Signaling nucleotide cyclic di-GMP (c-di-GMP) regulates biofilm formation, motility, virulence, the cell cycle, and other functions. In Mycobacterium smegmatis, both (p)ppGpp and c-di-GMP are synthesized and degraded by bifunctional proteins RelMsm and DcpA, encoded by relMsm and dcpA genes, respectively. We have previously shown that the ΔrelMsm and ΔdcpA knockout strains are antibiotic resistant and defective in biofilm formation, show altered cell surface properties, and have reduced levels of glycopeptidolipids and polar lipids in their cell wall (K. R. Gupta, S. Kasetty, and D. Chatterji, Appl Environ Microbiol 81:2571–2578, 2015, http://dx.doi.org/10.1128/AEM.03999-14). In this work, we have explored the phenotypes that are affected by both (p)ppGpp and c-di-GMP in mycobacteria. We have shown that both (p)ppGpp and c-di-GMP are needed to maintain the proper growth rate under stress conditions such as carbon deprivation and cold shock. Scanning electron microscopy showed that low levels of these second messengers result in elongated cells, while high levels reduce the cell length and embed the cells in a biofilm-like matrix. Fluorescence microscopy revealed that the elongated ΔrelMsm and ΔdcpA cells are multinucleate, while transmission electron microscopy showed that the elongated cells are multiseptate. Gene expression analysis also showed that genes belonging to functional categories such as virulence, detoxification, lipid metabolism, and cell-wall-related processes were differentially expressed. Our results suggests that both (p)ppGpp and c-di-GMP affect some common phenotypes in M. smegmatis, thus raising a possibility of cross talk between these two second messengers in mycobacteria.
IMPORTANCE Our work has expanded the horizon of (p)ppGpp and c-di-GMP signaling in Gram-positive bacteria. We have come across a novel observation that M. smegmatis needs (p)ppGpp and c-di-GMP for cold tolerance. We had previously shown that the ΔrelMsm and ΔdcpA strains are defective in biofilm formation. In this work, the overproduction of (p)ppGpp and c-di-GMP encased M. smegmatis in a biofilm-like matrix, which shows that both (p)ppGpp and c-di-GMP are needed for biofilm formation. The regulation of cell length and cell division by (p)ppGpp was known in mycobacteria, but our work shows that c-di-GMP also affects the cell size and cell division in mycobacteria. This is perhaps the first report of c-di-GMP regulating cell division in mycobacteria.
INTRODUCTION
Nucleotide second messengers are utilized across all domains of life (1, 2). To counter innumerable threats and to regulate physiological functions, bacteria exploit a repertoire of signaling nucleotides, (p)ppGpp, cyclic di-GMP (c-di-GMP), c-di-AMP, cGMP, and cAMP (1–3). Each second messenger signaling module controls the response to a specific set of cues and brings about changes in gene expression pattern accordingly. For example, the alarmone (p)ppGpp is generally synthesized when bacteria are stressed by lack of amino acids or other carbon sources (4). Similarly, the signaling nucleotide c-di-GMP is synthesized when bacteria switch from a motile to a sessile lifestyle (5).
Monofunctional RelA and bifunctional Rel proteins synthesize (p)ppGpp, while bifunctional SpoT and Rel proteins degrade it (4, 6, 7). The alarmone (p)ppGpp regulates processes such as transcription, translation, and replication (8). It also regulates secondary metabolite production (9, 10), biofilm formation (11–13), lipid synthesis (14–16), and toxin-antitoxin production and persistence in various bacteria (17, 18). Enzymes called diguanylate cyclases (DGCs), containing a GGDEF sequence motif in the active site, synthesize c-di-GMP from two molecules of GTP. Phosphodiesterases (PDEs) containing either an EAL or an HD-GYP motif degrade c-di-GMP (5, 19, 20). Once synthesized, c-di-GMP principally regulates biofilm formation in various bacteria. Apart from biofilm formation, c-di-GMP regulates flagellar biosynthesis, virulence, cell morphology, and the cell cycle (5, 20–22).
In mycobacteria, (p)ppGpp is synthesized and broken down by the dual-function enzyme Rel, encoded by the rel gene (23). The alarmone (p)ppGpp is required for long-term survival of Mycobacterium smegmatis during nutrient starvation (24). In the same way, Mycobacterium tuberculosis lacking the RelMtb protein shows reduced long-term survival in the lungs of mice and in a mouse hypoxic granuloma model and cannot form tubercle lesions in a guinea pig model of infection (25–28). In M. smegmatis, c-di-GMP is synthesized and broken down by a bifunctional protein, DcpA, encoded by the dcpA gene (29, 30). The long-term survival of the ΔdcpA mutant is compromised during nutritional stress (29). c-di-GMP controls the expression of lipid transport and metabolism genes through a transcription factor, LtmA, in M. smegmatis (31). It also regulates pathogenicity and dormancy in M. tuberculosis (32).
In our previous work, we have shown that both ΔrelMsm and ΔdcpA strains are relatively more antibiotic tolerant, show altered cell surface properties, and have reduced amounts of glycopeptidolipids (GPLs) and polar lipids in their cell walls compared to the wild-type (WT) M. smegmatis (33). We found that impairing (p)ppGpp and c-di-GMP signaling in M. smegmatis caused similar effects on “macroscopic” surface-related properties such as colony morphology, sliding motility, and biofilm formation, which gave us the hint that there could be more phenotypes that could be commonly regulated by these second messengers. Hence, in the current work, we have explored the effects of impairment of (p)ppGpp and c-di-GMP signaling on stress tolerance, cellular morphology, and gene expression patterns and showed that both of the second messengers do influence some common phenotypes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All the strains used in this study are listed in Table 1. M. smegmatis mc2 155 (the wild type [WT]), the knockout strains (the ΔrelMsm and ΔdcpA strains), the complemented strains (relComp and dcpAComp strains), and the overexpression strains (relOE and dcpAOE strains) were cultivated in MB7H9 (Difco) broth containing 0.2% (vol/vol) glycerol as the carbon source and 0.05% Tween 80 or on MB7H9 medium solidified with 1.5% (wt/vol) agar. The overexpression was achieved by shifting the primary cultures of the relOE or dcpAOE strain to 42°C. The antibiotics hygromycin and kanamycin were used at a concentration of 40 μg/ml.
TABLE 1.
List of strains used in the study
| M. smegmatis strain | Characteristic(s) | Reference or source |
|---|---|---|
| mc2 155 (WT) | Wild-type strain | 49 |
| ΔrelMsm strain | Strain in which the rel gene has been replaced with Hygr cassette | Lab strain (24) |
| ΔdcpA strain | Strain in which the dcpA gene has been replaced with Kanr cassette | Lab strain (29) |
| relComp strain | ΔrelMsm strain complemented with rel gene; Kanr | Gift from Christina Stallings (50) |
| dcpAComp strain | ΔdcpA complemented with dcpA gene; Kanr Hygr | Lab strain (29) |
| relOE strain | WT strain transformed with pMV261 plasmid containing rel gene under heat shock promoter | Lab strain |
| dcpAOE strain | WT strain transformed with pMV261 plasmid containing dcpA gene under heat shock promoter | Lab strain |
The growth curve experiments were performed in MB7H9 broth containing either 0.02% (vol/vol) or 0.2% (vol/vol) glycerol and 0.05% Tween 80. Growth of three biological replicates for each strain was measured. The initial optical density at 600 nm (OD600) of all the cultures was set at 0.02. The cultures were incubated at 37°C, and aliquots were regularly withdrawn to measure OD600.
Confocal microscopy, scanning electron microscopy, and transmission electron microscopy (TEM) analyses.
The protocol for fluorescence microscopy was adapted from the work of Ghosh et al. (34). M. smegmatis cells were fixed overnight with 1× phosphate-buffered saline (PBS) containing 1% toluene and 2% Triton X-100. Postfixation, nuclei were stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 5 min. The cells were imaged using a Zeiss Axio Imager fluorescence microscope under an 100× objective.
The procedure for scanning electron microscopy was adapted as previously described (35). Briefly, M. smegmatis cells were placed on poly-l-lysine-coated coverslips for 30 min. Unbound cells were washed off using 1× PBS. The bound cells were fixed with 0.1 M sodium cacodylate buffer containing 2% paraformaldehyde and 2.5% glutaraldehyde at 4°C overnight. The fixing solution was decanted, and the fixed cells were washed with 1× PBS. This was followed by fixing the cells with 2% OsO4 in 0.1 M sodium cacodylate buffer for 2 h at room temperature. Cells were then sequentially dehydrated using ethanol at different concentrations for 10 min each (50%, 70%, 90%, 95%, and 100%). The dehydrated samples were dried into a CO2 critical point drier, followed by gold sputter coating, and imaged in an FEI Quanta 200 scanning electron microscope.
Transmission electron microscopy was carried out by adapting the protocol from the work of Ghosh et al. (34). M. smegmatis cells were fixed with 0.1 M sodium cacodylate buffer containing 2% paraformaldehyde and 2% glutaraldehyde at 4°C overnight followed by postfixing with 0.1 M sodium cacodylate containing 0.8% K3Fe(CN)6 and 2% OsO4 for 2 h at room temperature. Postfixation, cells were embedded in 1.5% molten agarose. The agar blocks were cut into very fine pieces which were subjected to sequential ethanol dehydration at increasing concentrations for 10 min each (30%, 50%, 70%, 90%, and 100%). Samples were infiltrated overnight with a mixture containing equal volumes of Spurr's resin and acetone at room temperature. The infiltrated samples were mixed with Spurr's resin in an embedding mold and allowed to set at 68°C for 48 h. Ultrathin sections were cut using a Boeckeler powerTome X microtome and picked up on a copper grid. The grids were stained with 1% methanolic uranyl acetate, followed by Reynolds' lead citrate, and imaged in an FEI Tecnai G2 Spirit Biotwin 120-kV transmission electron microscope.
Microarray analysis and quantitative real-time PCR (qRT-PCR) experiments.
WT, ΔrelMsm, and ΔdcpA strains were grown in 100-ml cultures in MB7H9 medium containing 0.2% glycerol for 48 h. Twenty-five milliliters of this culture was centrifuged, and the cell pellet was resuspended in RNAlater solution for 4 h and centrifuged. The cell pellet was suspended in 1× PBS, snap-frozen in liquid nitrogen, and stored at −80°C until RNA extraction. RNA extraction, labeling, and hybridization were carried out at Genotypic Technology, Bangalore, India. RNA was isolated using Qiagen's RNeasy minikit, and its concentration and purity were determined by measuring the A260/280 ratio. The integrity of total RNA was checked by an Agilent 2100 bioanalyzer with the RNA 6000 Nano Labchip. RNA samples having an A260/280 ratio between 1.8 and 2.2 and an A260/230 ratio between 0.5 and 2.4 were chosen for further analysis. The RNAs were then labeled using Agilent's Quick-Amp labeling kit. Labeled cRNAs were generated using the random hexamer method which was followed by T7 promoter-based linear amplification. Agilent's in situ kit was used to perform hybridization. Gene expression analysis was carried out using Genespring GX 11 software using percentile normalization. We independently normalized microarray data using the variance stabilization normalization method (36).
Genes that showed an absolute fold change of 2 or more in the microarray were considered for qRT-PCR analysis for the validation of microarray data. RNA was isolated using the TRIzol method for qRT-PCR analysis. The cDNA synthesis was carried out using the Applied Biosystems cDNA synthesis kit according to the protocol provided by the manufacturer. The cDNA generated with random primers was used for qRT-PCRs with SYBR green (Applied Biosystems) as the indicator dye. The RNA levels were normalized with respect to the rpoC gene, which encodes the β′ subunit of RNA polymerase (37). The experiments were performed for at least two biological replicates. Each replicate was performed in triplicate.
Microarray data accession number.
The microarray data have been uploaded on NCBI's Gene Expression Omnibus (GEO) with accession number GSE69681.
RESULTS
Second messengers (p)ppGpp and c-di-GMP are required for the maintenance of proper growth under stress conditions.
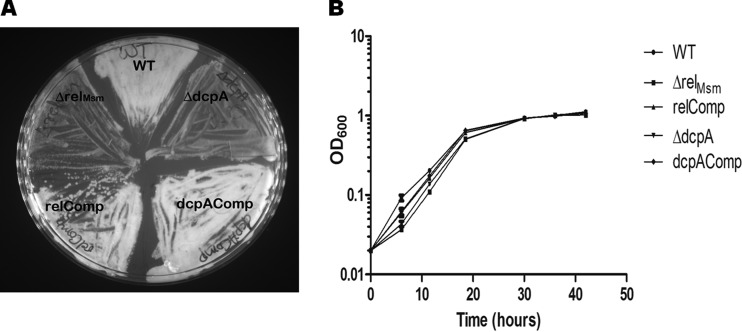
It was observed that whenever glycerol stocks of the ΔrelMsm and ΔdcpA strains were plated onto MB7H9 agar, the growth of these knockout strains was slower than those of the WT and the respective complemented strains (Fig. 1A). Whenever the cultures of the ΔrelMsm and ΔdcpA strains were subjected to overnight cold shock at 4°C and a loopful of bacteria was plated from these cultures, the growth of the knockout strains was comparatively slow (data not shown). These observations implied that both (p)ppGpp and c-di-GMP influence the growth of M. smegmatis. To confirm this observation, we compared the growth of the WT, the knockout strains, and the complemented strains in MB7H9 medium containing 0.2% glycerol as a carbon source and also under nutrient deprivation conditions under which MB7H9 medium contained 10-fold-less carbon source, i.e., 0.02% glycerol. The growth of the knockout strains was slow compared with those of the WT and respective complemented strains during lag and log phase under nutrient deprivation conditions. However, the knockout strains attained growth equal to that of the WT by the time that the cultures had reached stationary phase (Fig. 1B). We did not find any significant growth difference between the knockout strains, the WT, and the respective complemented strains when cells were grown in nutrient-rich medium (see Fig. S1 in the supplemental material).
FIG 1.
Comparison of growth profiles of the WT, knockout, and complemented strains under stress conditions. (A) The knockout strains grow slowly when subjected to cold shock before being plated on MB7H9 medium containing 0.2% glycerol as a carbon source. The plate was incubated for 48 h at 37°C. (B) The initial growth of the knockout strains is slower in the carbon-starved MB7H9 medium containing only 0.02% glycerol as a carbon source. The growth of the knockout strains becomes equal to that of the WT and respective complemented strains when culture reaches mid-log phase.
Signaling nucleotides (p)ppGpp and c-di-GMP control cell morphology and cell division.
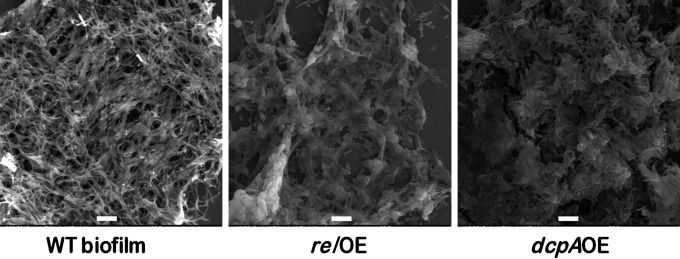
Escherichia coli strains completely devoid of (p)ppGpp [(p)ppGpp0 strains] have been shown to be elongated (16, 38, 39). It had previously been shown in our laboratory that carbon starvation or the overexpression of E. coli relA in M. smegmatis shortened the cell lengths and that some cells assumed coccoid morphology (40). Similarly, the ΔrelMsm strain of M. smegmatis was also elongated. Hence, we wondered if c-di-GMP also regulates the cell length in M. smegmatis in the same manner as (p)ppGpp. High levels of (p)ppGpp and c-di-GMP were achieved by overexpressing the RelMsm and DcpA proteins. Surprisingly, we found that both of the overexpression strains were embedded in a biofilm-like matrix and that it was very difficult to observe the individual cells of relOE and dcpAOE strains (Fig. 2). Some of the cells in the scanning electron micrographs did show coccoid morphology (see Fig. S2 in the supplemental material). We also quantified the biofilm-like matrix using the standard crystal violet method and found that the amount of the matrix was larger in the relOE and dcpAOE strains than in the WT and the vector control strain (see Fig. S3). Thus, high levels of c-di-GMP can also reduce the cell length of M. smegmatis (see Fig. S2).
FIG 2.
Scanning electron micrographs of planktonic cultures of relOE and dcpAOE strains and biofilm culture of WT strain of M. smegmatis. High levels of (p)ppGpp and c-di-GMP in the planktonic cultures of the strains which were shaken at 180 rpm caused the relOE and dcpAOE cells to become embedded in a biofilm-like matrix. The biofilm culture of WT M. smegmatis has been used as a control.
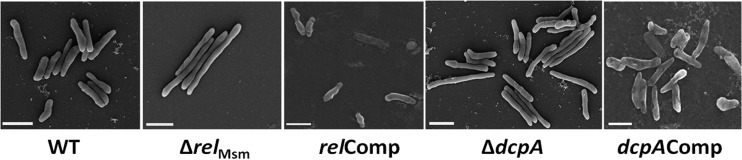
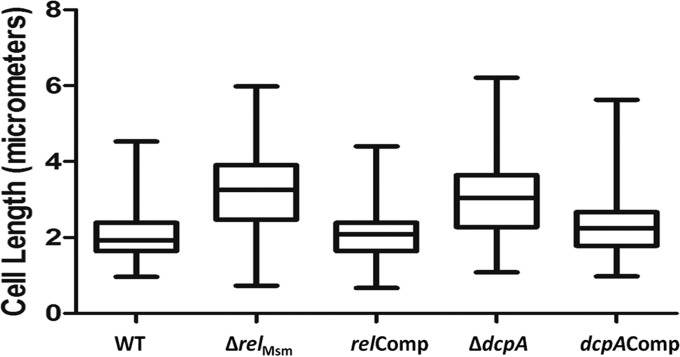
Since high levels of (p)ppGpp and c-di-GMP reduced the cell lengths of M. smegmatis, we next posed the question of whether the ΔrelMsm and ΔdcpA strains would be longer than the WT strains. The scanning electron micrographs revealed that the cells of the ΔrelMsm and ΔdcpA strains were longer than those of the WT strain (Fig. 3). We measured lengths of at least 200 cells of each strain from different micrographs. The median cell lengths of the ΔrelMsm and ΔdcpA strains were 3.24 μm and 3.04 μm, respectively, compared with 1.92 μm for the WT, 2.08 μm for the relComp strain, and 2.24 μm for the dcpAComp strain (Fig. 4). This experiment hinted that apart from (p)ppGpp, the cyclic nucleotide c-di-GMP also influences the cell size of M. smegmatis. Taken together, we would like to conclude that high levels of (p)ppGpp and c-di-GMP reduce the cell length and that absence or low levels increase the cell length.
FIG 3.
Scanning electron micrographs of the WT, knockout, and complemented strains. It is evident that the knockout strains are more elongated than the WT and respective complemented strains. Bar, 2 μm.
FIG 4.
Cell length distribution analysis using a box plot. Lengths of at least 200 cells of each strain from different electron micrographs were measured and plotted using the box plot analysis function from GraphPad Prism 5. The whiskers in the plot represent minimum and maximum values. The median lengths were found to be 1.92 μm for the WT, 3.24 μm for the ΔrelMsm strain, 2.08 μm for the ΔdcpA strain, 3.04 μm for the relComp strain, and 2.24 μm for the dcpAComp strain.
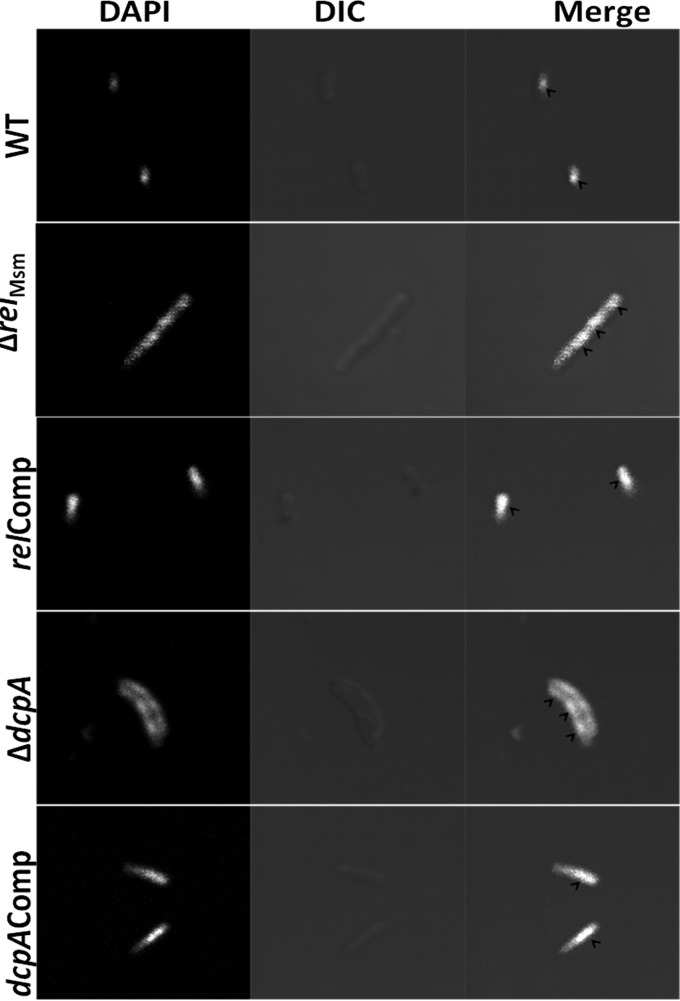
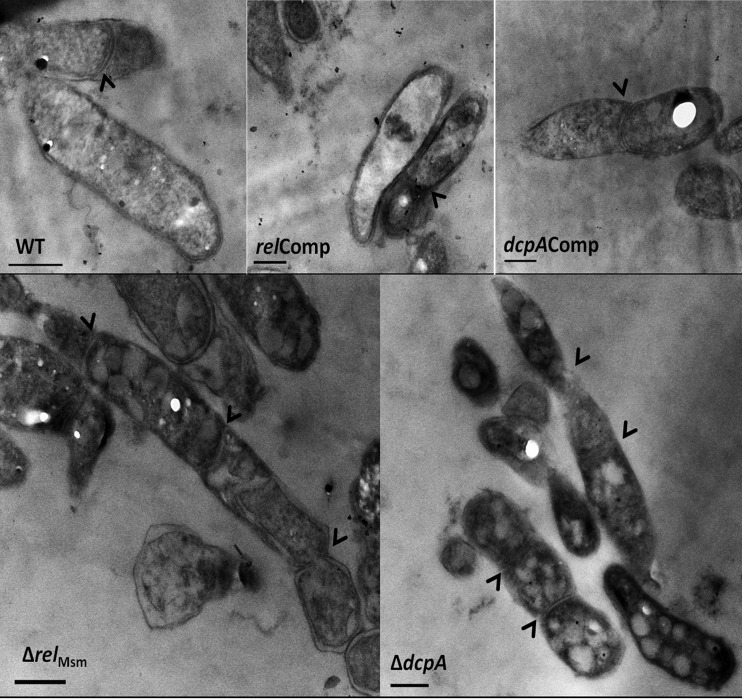
We next asked the question of whether the elongated ΔrelMsm and ΔdcpA cells are multinucleate. Confocal microscopy of DAPI-stained cells revealed that the elongated knockout cells appeared filamentous and had at least two or more nuclei (Fig. 5; see also Fig. S4 to S8 in the supplemental material). This left us with the question of whether the cytoplasm in the case of multinucleate cells is continuous or multiseptate. Ultrastructural studies unveiled that the elongated knockout strains were multiseptate (Fig. 6), indicating that the signaling nucleotide (p)ppGpp or c-di-GMP also regulates the cell division process in M. smegmatis. Thus, transmission electron microscopy (TEM) studies revealed the novel role of c-di-GMP in regulating the cell division process in the Gram-positive soil saprophyte M. smegmatis.
FIG 5.
Confocal micrographs of DAPI-stained cells. Images were acquired at an ×100 magnification. It is evident that the elongated ΔrelMsm ΔdcpA cells have multiple nuclei, as shown by arrowheads in the merged section of the image. Bar, 1 μm. DIC, differential interference contrast.
FIG 6.
Ultrastructural analysis of the WT, knockout, and complemented strains using transmission electron microscopy. It can be noted from the transmission electron micrographs that the elongated ΔrelMsm and ΔdcpA cells are multiseptate, unlike the WT and the respective complemented strain. Arrowheads represent the positions of septa. Bars, 500 nm.
Signaling nucleotides (p)ppGpp and c-di-GMP regulate the expression of a large number of genes.
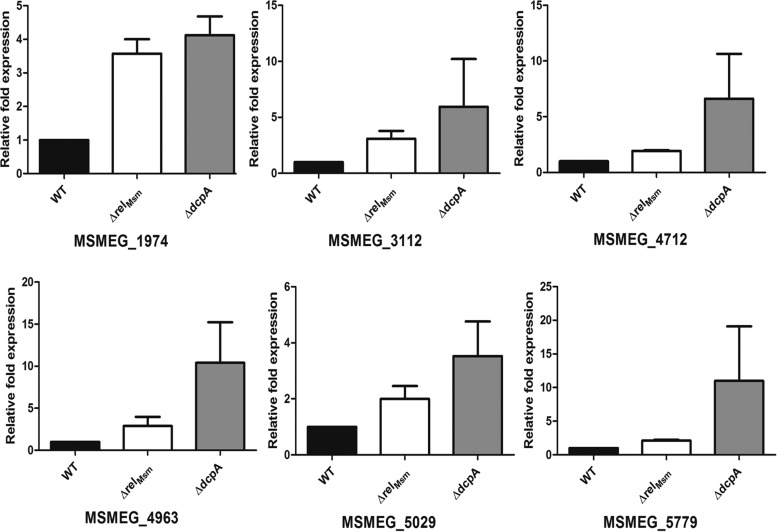
To explain the phenotypes displayed by the ΔrelMsm and ΔdcpA strains, gene expression analysis was carried out using microarrays. The microarray data showed that the gene expression profiles of both of the knockouts were markedly different from that of the WT. Genes with absolute fold changes of 2 or more were considered to be significantly upregulated. This cutoff value revealed that 143 genes in the ΔrelMsm strain and 43 genes in the ΔdcpA strain were upregulated by 2-fold or more (see Data set S1 in the supplemental material). Most of these genes belonged to functional categories such as intermediary metabolism and respiration, virulence, detoxification and adaptation, cell wall and cell processes, and lipid metabolism. Six representative genes in these categories which were upregulated in both of the knockout strains were selected for qRT-PCR analysis to validate the microarray data (Table 2). The upregulation of genes MSMEG_1974 and MSMEG_5029, which are involved in processes of virulence, detoxification, and adaptation, was confirmed by qRT-PCR (Fig. 7). Similarly, qRT-PCR analysis of MSMEG_4712 and MSMEG_5779, genes involved in cell-wall-related processes, and MSMEG_3112 and MSMEG_4963, genes involved in intermediary metabolism and respiration, correlated well with microarray data. We also selected the principal bacterial cell division gene, ftsZ, for qRT-PCR analysis, as downregulation of this gene is known to make bacterial cells elongated, even though microarray data showed no significant differences in its expression level (41). The qRT-PCR analysis showed that in the knockout strains, the levels of ftsZ were significantly downregulated (see Fig. S9 in the supplemental material).
TABLE 2.
List of genes selected for qRT-PCR analysisa
| Gene | Gene product | Functional classification | Fold upregulation in: |
|
|---|---|---|---|---|
| ΔrelMsm strain | ΔdcpA strain | |||
| MSMEG_1974 | Propane monooxygenase coupling protein | Virulence, detoxification, and adaptation | 7.84 | 3.53 |
| MSMEG_3112 | Estradiol 17-beta-dehydrogenase 8 | Intermediary metabolism and respiration | 2.34 | 2.14 |
| MSMEG_4712 | TetR family protein transcriptional regulator | Cell wall and cell processes | 2.96 | 2.12 |
| MSMEG_4963 | Pyruvate dehydrogenase E1 component, alpha subunit | Intermediary metabolism and respiration | 11.64 | 3.06 |
| MSMEG_5029 | Peptide synthetase ScpsB, putative | Virulence, detoxification, and adaptation | 3.47 | 2.5 |
| MSMEG_5779 | Phosphate import ATP-binding protein PstB | Cell wall and cell processes | 4.51 | 3.4 |
Representative genes that were upregulated by 2-fold or more in both ΔrelMsm and ΔdcpA strains were selected for qRT-PCR analysis to validate the microarray data.
FIG 7.
Quantitative real-time PCR analysis. Six genes, upregulated more than 2-fold in the microarray, were chosen for qRT-PCR analysis. Almost all genes were upregulated in both strains by at least 2-fold, thus validating the microarray data. The experiment was performed in at least two biological replicates, and each replicate was performed in triplicate. The figure shows one of the replicates. The graphs were plotted using GraphPad Prism 5.
In order to gain more information from the microarray data, we performed gene enrichment analysis in which the upregulated genes were assigned to various gene ontologies, i.e., functional classes or categories (see Fig. S10 and S11 in the supplemental material). In the ΔrelMsm strain, ontologies related to cell division, cell cycle, cell wall organization, reactive oxygen species metabolism, peptidoglycan synthesis, lipid metabolism, superoxide metabolism, response to oxidative stress, and various metabolic pathways were enriched (see Fig. S10). Similarly, enrichment of ontologies such as response to oxidative stress, one-carbon metabolism, and various other metabolism-related ontologies suggests that these processes are altered in the ΔdcpA strain (see Fig. S11). Thus, gene expression studies revealed some common and some different processes controlled by the second messengers (p)ppGpp and c-di-GMP.
DISCUSSION
Our results show that (p)ppGpp and c-di-GMP regulate not only common phenotypes such as growth under stress, cell size, and cell division but also global gene expression in M. smegmatis. The initial observation that glycerol stocks of the ΔrelMsm and ΔdcpA strains grew slowly on the solid medium was confirmed by giving an overnight cold shock at 4°C and plating the strains on MB7H9 medium. We found that the knockout strains did grow slowly post-cold shock. We next checked if the nutrient stress would produce the similar effect. This was, indeed, the case. The initial growth of the knockout strains in carbon-starved cultures was slow, but the growth became equal to that of the WT and the complemented strains by the time that cultures reached stationary phase. The Δrel mutant of Bacillus subtilis grows slowly in exponential phase; however, its growth becomes equal to that of the WT strain when the culture reaches stationary phase (42). Our results are further supported by the findings that the ppGpp0 strain of E. coli has slower growth than the WT strain (16, 38). Our result is also in agreement with the findings in M. tuberculosis where deletion of relMtb results in a significantly lower aerobic growth rate (25). It has also been reported previously that a stringent response does not affect the growth rate of M. smegmatis (26). Our data show that aerobic growth rates of both the ΔrelMsm and ΔdcpA strains are lower than that of the WT strain under stressed conditions but not under normal growth conditions. Thus, the results presented here show that both (p)ppGpp and c-di-GMP are needed for the maintenance of proper growth under stress conditions. So far, we have not come across any report on the role of c-di-GMP in regulating bacterial growth. Hence, we report a novel role for c-di-GMP in mycobacterial growth.
We have reported earlier that both ΔrelMsm and ΔdcpA strains of M. smegmatis are defective in biofilm formation (33). It is well known that high levels of (p)ppGpp or c-di-GMP are required for biofilm formation (1, 4, 5, 43). Hence, we reasoned that increasing the levels of (p)ppGpp or c-di-GMP in M. smegmatis through overexpression of Rel or DcpA should confer a biofilm-like state. As revealed by the scanning electron micrographs, the relOE and dcpAOE strains were found to be encased in a biofilm-like matrix (Fig. 2). This matrix was also visible in phase-contrast images (data not shown). The point worth mentioning here is that both the relOE and dcpAOE strains were grown under shaking conditions, and not under static conditions as required for biofilm formation. We carried out crystal violet quantification of the biofilm-like matrix formed by the overexpression strains. The results showed that the amount of biofilm formed by the relOE or dcpAOE strain is relatively larger (see Fig. S2 in the supplemental material). Consequently, it can be said that the high levels of second messengers (p)ppGpp and c-di-GMP in M. smegmatis not only reduce its cell length but also produce a biofilm-like matrix.
The (p)ppGpp0 mutant of E. coli is longer than the WT cells (38). The ΔrelMsm mutant of M. smegmatis is also elongated and multiseptate (26). Plant pathogens Erwinia amylovora and Pseudomonas syringae lacking (p)ppGpp are also elongated compared with the respective WT strains (44, 45). We found that both ΔrelMsm and ΔdcpA mutants are elongated, multinucleate, and multiseptate. Thus, our work is in agreement with the previously reported role of (p)ppGpp in controlling cell size and cell division. c-di-GMP regulates the cell cycle transition in Gram-negative Caulobacter crescentus (21, 22). It has been shown that high levels of c-di-GMP inhibit sporulation septation and low levels induce precocious hypersporulation without formation of aerial hyphae in Gram-positive Streptomyces venezuelae (46). Our work shows that c-di-GMP is also involved in cell division in Gram-positive mycobacteria. Thus, the data show that both (p)ppGpp and c-di-GMP are also involved in the regulation of the cell cycle in M. smegmatis.
Gene expression analysis of the ΔrelMsm and ΔdcpA strains brought to light the fact that many genes were differentially expressed in the knockout strains. To validate the microarray data, a few genes whose expression was significantly altered were selected for qRT-PCR analysis. Several genes related to cell wall metabolism showed differential expression in both of the knockout strains (see Data set S1 in the supplemental material). Upregulation of genes such as MSMEG_4712 and MSMEG_5779, involved in cell wall processes, confirms that cell wall metabolism is affected in the knockout strains (Fig. 7). Thus, differential expression of genes involved in cell-wall-related processes in ΔrelMsm and ΔdcpA strains might also explain our previous work where we have shown that the ΔrelMsm and ΔdcpA strains are defective in synthesis of cell wall glycopeptidolipids and polar lipid synthesis (33).
Both (p)ppGpp and c-di-GMP not only affect the metabolism but also are needed for adaptation to stress conditions. Thus, the lack or low levels of (p)ppGpp or c-di-GMP may affect the ability of bacteria to adjust their metabolism under stress conditions. Differential expression of genes such as MSMEG_1974 and MSMEG_5029, belonging to the “virulence, detoxification, and adaptation” functional class, as well as MSMEG_3112 and MSMEG_4963, belonging to the “intermediary metabolism and respiration” category, might explain the initial slow growth of the ΔrelMsm and ΔdcpA strains during cold shock and nutrient stress. This is also corroborated by the enrichment of gene ontologies related to various metabolic processes such as macromolecular biosynthesis, nucleic acid metabolism, metabolism of purine-containing compounds, and catabolism in both ΔrelMsm and ΔdcpA strains (see Fig. S10 and S11 in the supplemental material).
Since depletion of ftsZ has been linked to a filamentous phenotype in bacteria (41, 47), and the ΔrelMsm and ΔdcpA strains exhibited the filamentous phenotype, we checked expression of the principal cell division control gene ftsZ. The qRT-PCR analysis showed that it was downregulated in both the knockout strains, thus explaining the filamentous phenotype shown by ΔrelMsm and ΔdcpA strains (see Fig. S9 in the supplemental material). Enrichment of ontologies related to cell division and the cell cycle further corroborates the finding that the cell division process is affected in the ΔrelMsm strain (see Fig. S10). We did not find enrichment of these ontologies in the ΔdcpA strain. This is because only 43 genes could be considered, according to the 2-fold cutoff for the gene enrichment analysis for the ΔdcpA strain. However, when the gene enrichment analysis was performed with a reduced cutoff, 1.5-fold, the ontologies related to cell wall organization, cell division, and cell cycle also became enriched (data not shown). Altogether, microscopy analyses, qRT-PCR studies, and gene enrichment analyses prove that (p)ppGpp and c-di-GMP regulate the cell division process in M. smegmatis.
We have previously reported that both ΔrelMsm and ΔdcpA strains show resistance not only to multiple classes of antibiotics but also to various antifolates (33). Gene enrichment analysis showed the enrichment of ontologies related to metabolism of reactive oxygen species and superoxide in the ΔrelMsm strain and showed that the response to oxidative stress ontology was enriched in the ΔdcpA strain (see Fig. S10 and S11 in the supplemental material). The enrichment of one-carbon metabolism ontology in both ΔrelMsm and ΔdcpA strains may also explain the resistance of these strains to various antifolates. It is well known that irrespective of the mode of action, the bactericidal antibiotics kill the bacteria by mainly increasing the concentration of free radicals, which increases the oxidative stress within the bacterial cell (48). Hence, the enrichment of the ontologies related to reactive oxygen species metabolism, superoxide metabolism, and oxidative stress in the knockout strains also explains the previously reported multidrug resistance of the ΔrelMsm and ΔdcpA strains of M. smegmatis.
Taken together, our work shows that the hitherto-unreported phenotypes of stress tolerance, cell size, and cell division are being influenced by c-di-GMP in M. smegmatis. Our results suggest that both (p)ppGpp and c-di-GMP regulate the common phenotypes of growth, cell size, and cell division. The striking similarity between the phenotypes regulated by (p)ppGpp and those regulated by c-di-GMP may also suggest a potential cross talk between these second messengers in mycobacteria.
Supplementary Material
ACKNOWLEDGMENTS
K.R.G. and D.C. thank the Department of Biotechnology, Government of India, for funding this work. K.R.G. and P.B. thank the Council for Scientific and Industrial Research for a senior research fellowship.
K.R.G. acknowledges P. Anushya for proofreading the manuscript, Grace Chongloi for her extensive help in preparing the figures for the manuscript, Preeti Garai for her extensive help during confocal microscopy analysis, and Geetha Melangath for her help in analyzing microarray data. K.R.G. and D.C. acknowledge Aswin Sai Narain Seshasayee for normalizing the microarray data using the variance stabilization method.
K.R.G. and D.C. designed the experiments and wrote the paper. K.R.G. performed the experiments. K.R.G. and P.B. analyzed microarray data. P.B. carried out gene enrichment analysis. K.R.G. and S.S.I. performed transmission electron microscopy experiments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00126-16.
REFERENCES
- 1.Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. 2013. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev 42:305–341. doi: 10.1039/C2CS35206K. [DOI] [PubMed] [Google Scholar]
- 2.Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonough KA, Rodriguez A. 2012. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol 10:27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 5.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 6.Chatterji D, Ojha A. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol 4:160–165. doi: 10.1016/S1369-5274(00)00182-X. [DOI] [PubMed] [Google Scholar]
- 7.Jain V, Kumar M, Chatterji D. 2006. ppGpp: stringent response and survival. J Microbiol 44:1–10. [PubMed] [Google Scholar]
- 8.Srivatsan A, Wang JD. 2008. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol 11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Ochi K. 1987. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J Bacteriol 169:3608–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M. 2007. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol 8:R161. doi: 10.1186/gb-2007-8-8-r161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor CM, Beresford M, Epton HA, Sigee DC, Shama G, Andrew PW, Roberts IS. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J Bacteriol 184:621–628. doi: 10.1128/JB.184.3.621-628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemos JA, Brown TA Jr, Burne RA. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect Immun 72:1431–1440. doi: 10.1128/IAI.72.3.1431-1440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pizer LI, Merlie JP. 1973. Effect of serine hydroxamate on phospholipid synthesis in Escherichia coli. J Bacteriol 114:980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seyfzadeh M, Keener J, Nomura M. 1993. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci U S A 90:11004–11008. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 18.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 19.Pesavento C, Hengge R. 2009. Bacterial nucleotide-based second messengers. Curr Opin Microbiol 12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lori C, Ozaki S, Steiner S, Bohm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 23.Avarbock D, Salem J, Li LS, Wang ZM, Rubin H. 1999. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233:261–269. doi: 10.1016/S0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- 24.Mathew R, Ojha AK, Karande AA, Chatterji D. 2004. Deletion of the rel gene in Mycobacterium smegmatis reduces its stationary phase survival without altering the cell-surface associated properties. Curr Sci 86:149–153. [Google Scholar]
- 25.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol 182:4889–4898. doi: 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahl JL, Arora K, Boshoff HI, Whiteford DC, Pacheco SA, Walsh OJ, Lau-Bonilla D, Davis WB, Garza AG. 2005. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J Bacteriol 187:2439–2447. doi: 10.1128/JB.187.7.2439-2447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med 200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC. 2010. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202:1397–1404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bharati BK, Sharma IM, Kasetty S, Kumar M, Mukherjee R, Chatterji D. 2012. A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 158:1415–1427. doi: 10.1099/mic.0.053892-0. [DOI] [PubMed] [Google Scholar]
- 30.Sharma IM, Prakash S, Dhanaraman T, Chatterji D. 2014. Characterization of a dual-active enzyme, DcpA, involved in cyclic diguanosine monophosphate turnover in Mycobacterium smegmatis. Microbiology 160:2304–2318. doi: 10.1099/mic.0.080200-0. [DOI] [PubMed] [Google Scholar]
- 31.Li W, He ZG. 2012. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res 40:11292–11307. doi: 10.1093/nar/gks923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong Y, Zhou X, Fang H, Yu D, Li C, Sun B. 2013. Cyclic di-GMP mediates Mycobacterium tuberculosis dormancy and pathogenicity. Tuberculosis (Edinb) 93:625–634. doi: 10.1016/j.tube.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Gupta KR, Kasetty S, Chatterji D. 2015. Novel functions of (p)ppGpp and cyclic di-GMP in mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of Mycobacterium smegmatis. Appl Environ Microbiol 81:2571–2578. doi: 10.1128/AEM.03999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh S, Indi SS, Nagaraja V. 2013. Regulation of lipid biosynthesis, sliding motility, and biofilm formation by a membrane-anchored nucleoid-associated protein of Mycobacterium tuberculosis. J Bacteriol 195:1769–1778. doi: 10.1128/JB.02081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahl JL. 2004. Electron microscopy analysis of Mycobacterium tuberculosis cell division. FEMS Microbiol Lett 240:15–20. doi: 10.1016/j.femsle.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.S96. [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee R, Chatterji D. 2005. Evaluation of the role of sigma B in Mycobacterium smegmatis. Biochem Biophys Res Commun 338:964–972. doi: 10.1016/j.bbrc.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 38.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 39.Vinella D, D'Ari R, Jaffe A, Bouloc P. 1992. Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J 11:1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ojha AK, Mukherjee TK, Chatterji D. 2000. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect Immun 68:4084–4091. doi: 10.1128/IAI.68.7.4084-4091.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Hajj ZW, Newman EB. 2015. An Escherichia coli mutant that makes exceptionally long cells. J Bacteriol 197:1507–1514. doi: 10.1128/JB.00046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ababneh QO, Herman JK. 2015. RelA inhibits Bacillus subtilis motility and chaining. J Bacteriol 197:128–137. doi: 10.1128/JB.02063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He H, Cooper JN, Mishra A, Raskin DM. 2012. Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol 194:2962–2972. doi: 10.1128/JB.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ancona V, Lee JH, Chatnaparat T, Oh J, Hong JI, Zhao Y. 2015. The bacterial alarmone (p)ppGpp activates the type III secretion system in Erwinia amylovora. J Bacteriol 197:1433–1443. doi: 10.1128/JB.02551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatnaparat T, Li Z, Korban SS, Zhao Y. 2015. The bacterial alarmone (p)ppGpp is required for virulence and controls cell size and survival of Pseudomonas syringae on plants. Environ Microbiol doi: 10.1111/1462-2920.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ. 2014. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lutkenhaus JF, Wolf-Watz H, Donachie WD. 1980. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol 142:615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 49.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. [DOI] [PubMed] [Google Scholar]
- 50.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. 2009. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.