ABSTRACT
Oxidative phosphorylation using multiple-component, membrane-associated protein complexes is the most effective way for a cell to generate energy. Here, we systematically investigated the multiple protein-protein interactions of the denitrification apparatus of the pathogenic bacterium Pseudomonas aeruginosa. During denitrification, nitrate (Nar), nitrite (Nir), nitric oxide (Nor), and nitrous oxide (Nos) reductases catalyze the reaction cascade of NO3− → NO2− → NO → N2O → N2. Genetic experiments suggested that the nitric oxide reductase NorBC and the regulatory protein NosR are the nucleus of the denitrification protein network. We utilized membrane interactomics in combination with electron microscopy colocalization studies to elucidate the corresponding protein-protein interactions. The integral membrane proteins NorC, NorB, and NosR form the core assembly platform that binds the nitrate reductase NarGHI and the periplasmic nitrite reductase NirS via its maturation factor NirF. The periplasmic nitrous oxide reductase NosZ is linked via NosR. The nitrate transporter NarK2, the nitrate regulatory system NarXL, various nitrite reductase maturation proteins, NirEJMNQ, and the Nos assembly lipoproteins NosFL were also found to be attached. A number of proteins associated with energy generation, including electron-donating dehydrogenases, the complete ATP synthase, almost all enzymes of the tricarboxylic acid (TCA) cycle, and the Sec system of protein transport, among many other proteins, were found to interact with the denitrification proteins. This deduced nitrate respirasome is presumably only one part of an extensive cytoplasmic membrane-anchored protein network connecting cytoplasmic, inner membrane, and periplasmic proteins to mediate key activities occurring at the barrier/interface between the cytoplasm and the external environment.
IMPORTANCE The processes of cellular energy generation are catalyzed by large multiprotein enzyme complexes. The molecular basis for the interaction of these complexes is poorly understood. We employed membrane interactomics and electron microscopy to determine the protein-protein interactions involved. The well-investigated enzyme complexes of denitrification of the pathogenic bacterium Pseudomonas aeruginosa served as a model. Denitrification is one essential step of the universal N cycle and provides the bacterium with an effective alternative to oxygen respiration. This process allows the bacterium to form biofilms, which create low-oxygen habitats and which are a key in the infection mechanism. Our results provide new insights into the molecular basis of respiration, as well as opening a new window into the infection strategies of this pathogen.
INTRODUCTION
The most effective mode of biological energy generation is oxygen respiration. During this process, electrons are transported from donor molecules like NADH via membrane-localized large multiprotein complexes along a redox cascade to the electron acceptor oxygen. The free energy generated is recovered via the formation of a H+/Na+ gradient at the membrane, ultimately driving ATP synthesis by ATP synthases (1). Under conditions of oxygen depletion, many microorganisms are capable of replacing oxygen by an alternative electron acceptor, including nitrate, sulfate, fumarate, or dimethyl sulfoxide (DMSO) (2). Furthermore, the electron acceptors are combined with a whole variety of electron donors by these organisms. In this context, every electron donor and acceptor pair requires its own complex enzymatic system, either soluble or membrane associated and often accompanied by additional intermediate transfer complexes, like the bc1 complex. The structure and biochemistry of these different enzyme complexes and enzymatic systems are well understood (1). In contrast, much less is known about the dynamic interactions of these protein complexes in the respiratory chains. For humans (3), cattle (4), mice (5), spinach (6), potatoes (7), and yeast species (8, 9), the existence of supracomplexes has been proposed and proven. Similarly, membrane-localized multienzyme complexes were investigated in great detail for bacterial and plant photosynthesis (10–12). For prokaryotes, only a few investigations on supramolecular formation are available for the sulfide oxidase-oxygen reductase supercomplex of the bacterium Aquifex aeolicus (13) and the respirasome of Paracoccus denitrificans (14). However, none of the investigations reached the level of protein-protein interaction elucidation.
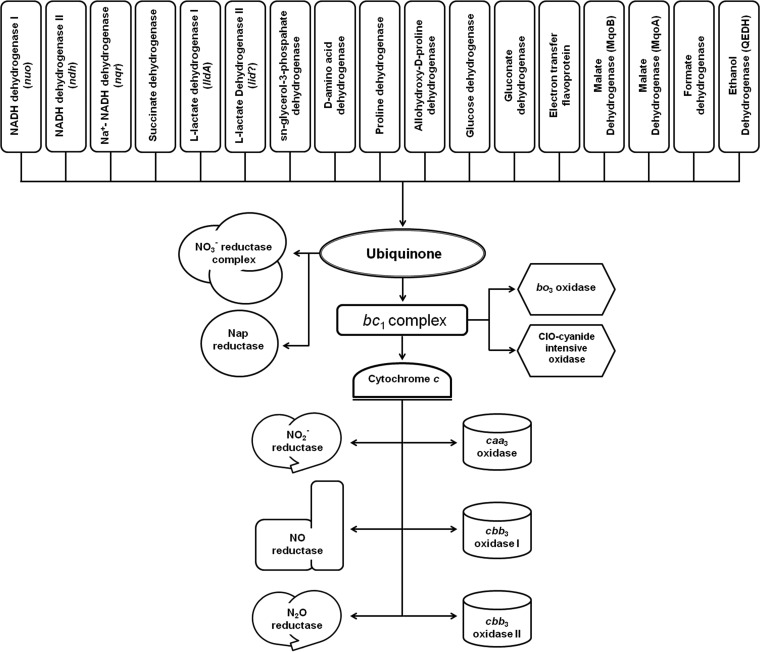
Here, we studied the supermolecular organization of the denitrification machinery of Pseudomonas aeruginosa. In the environment and during infection, P. aeruginosa forms biofilms, which during anaerobic growth are dependent on anaerobic respiration via denitrification (15). Four different multiprotein complexes receive electrons from various electron donors (Fig. 1) via ubiquinone and intermediate complexes to catalyze the following final reduction steps: NO3− → NO2− → NO → N2O → N2. During the first step of denitrification in various proteobacteria, respiratory nitrate reductase (NarGHI) catalyzes the following reaction: NO3− + 2e− + 2H+ → NO2− + H2O. Multiple biochemical and structural data revealed that these quinol-nitrate oxidoreductases receive electrons from ubiquinol via the two heme groups of the membrane-spanning small subunit NarI. Electrons are transferred through a linear arrangement of Fe-S clusters of the NarI-attached subunit NarH to the active site of the molybdenum cofactor (MoCo) containing subunit NarG. NarH and NarG are directed into the cytoplasm (16–18). The protein NarJ and cardiolipins are involved in the complex assembly of the various protein subunits with their corresponding cofactors (19, 20). P. aeruginosa additionally contains the periplasmic nitrate reductase NapAB. Electrons are supplied by the membrane-anchored tetraheme cytochrome c NapC. Interestingly, NapABC can compensate for NarGHI deficiency (21). The periplasmic nitrite reductase NirS catalyzes the next reaction as follows: NO2− + 2H+ + e− → NO + H2O. The P. aeruginosa enzyme is a cytochrome cd1 carrying an electron-receiving heme c and a catalytic site with heme d1 (3,8-dioxo-17-acrylate-porphyrindione) (22–24). The two c-type cytochromes NirM and NirC act as electron donors (25). NirN and NirF form a stable complex with the nitrite reductase NirS during enzyme maturation (26). NirF is involved in heme d1 insertion (27). The final step of heme d1 biosynthesis, localized mainly in the periplasm, involves the uroporphyrin III c-methyltransferase NirE (28), the siroheme decarboxylase NirDL/NirGH (29), and a novel electron-bifurcating dehydrogenase NirN (30). Nitrate from the environment gets transported into the cytoplasm by the nitrate/nitrite antiporter NarK2 and converted by the activity of NarGHI into nitrite, which then returns via NarK2 into the periplasm to serve as the substrate for NirS (31). Then, the integral cytoplasmic membrane protein nitric oxide reductase NorBC catalyzes the following reaction: 2NO + 2H+ + 2e− → N2O + H2O. The heterodimeric P. aeruginosa enzyme utilizes heme c and heme b for electron delivery to a heme b and nonheme iron in the active site that coordinates the two NO substrates (32, 33). Interestingly, the small subunit C covers most of the surface of the enzyme toward the periplasm (32, 33). The ATP-binding protein NirQ seems to be involved in NorCB maturation (34). For the final step, the periplasmic enzyme nitrous-oxide reductase NosZ catalyzes the following reaction: N2O + 2e− + 2H+ → N2 + H2O. Catalysis of the head-to-tail homodimeric enzyme requires two copper centers (35, 36). The periplasmic domain of the transmembrane iron-sulfur flavoprotein NosR is essential for NosZ function (37). The periplasmic lipoprotein NosL anchored to the outer membrane is a copper binding protein involved in NosZ maturation (38, 39).
FIG 1.
Proposed respiratory chains of P. aeruginosa based on a recent review (41).
However, the denitrification further relies on electron-donating primary dehydrogenases and on multiple other components of the electron transport chain (Fig. 1) (40–43). Recently, we described the existence of a stable NirS-DnaK-FliC complex in the periplasm of P. aeruginosa that was involved in flagellum assembly, using a combined membrane and periplasmic proteomics and electron microscopy approach (44). Here, we employed this experimental approach (45–47) to determine the nature of the protein-protein interactions of the denitrification machinery.
MATERIALS AND METHODS
Construction of the bait expression vectors.
Escherichia coli DH10b (Invitrogen, Germany) served as the cloning host. E. coli was grown in Luria-Bertani (LB) medium, at 37°C with shaking at 200 rpm. Ampicillin and carbenicillin were used at 100 μg/ml. P. aeruginosa PAO1 (DSMZ, Braunschweig, Germany) was used as the wild-type strain (48). The mini-Tn5-luxCDABE transposon mutants of P. aeruginosa (nosR, norC, norB, and nosZ mutants) served as the hosts for the physiological characterization of the denitrification process (49). The mutants were obtained from an in-house mutant collection. The P. aeruginosa strains were grown at 37°C in LB medium with shaking at 200 rpm. Carbenicillin was supplemented at 250 μg/ml. The anaerobic cultures were supplemented with 50 mM KNO3 as described before (50). The E. coli vector pJET1.2 (Thermo Scientific, Germany) was employed as the initial cloning vector. The E. coli and P. aeruginosa shuttle vector pUCP20t (51) was modified by the removal of the BsaI restriction site at position 3259 of the plasmid via site-directed mutagenesis (Table 1). The resulting plasmid was called pAS40. In this study, multiple plasmids were constructed and multiple E. coli and P. aeruginosa strains were used; all primers for amplification, plasmids, and strains are listed in Tables 1 and 2.
TABLE 1.
List of bacterial strains and plasmids used in this study
| Plasmid or straina | Relevant characteristic(s)b | Source or reference |
|---|---|---|
| Plasmids | ||
| pJET1.2 | Apr; carrier vector; toxin expressed upon self-ligation | Thermo Scientific, Darmstadt, Germany |
| pAS40 | Cbr; expression vector for P. aeruginosa; based on pUCP20t without BsaI (GGTCTC→GCTCTC) | Laboratory collection |
| pJET1.2-nosR | Apr; pJET1.2 carrying nosR | This study |
| pJET1.2-norC | Apr; pJET1.2 carrying norC | This study |
| pJET1.2-norCB | Apr; pJET1.2 carrying norCB | This study |
| pJET1.2-nosPR | Apr; pJET1.2 carrying nos 500-bp upstream region (includes promoter) | This study |
| pJET1.2-nosZ | Apr; pJET1.2 carrying nosZ | This study |
| pAS40-Strep-tag II (Z) | Cbr; pAS40 carrying the Strep-tag II tag; suitable for nosZ cloning | This study |
| pAS40-His6× (R, C, B) | Cbr; pAS40 carrying the His6× tag; suitable for nosR, norC, and norB cloning | This study |
| pAS40-nosR-His6× | Cbr; pAS40 carrying His6×-tagged nosR | This study |
| pAS40-norC-His6× | Cbr; pAS40 carrying His6×-tagged norC | This study |
| pAS40-norCB-His6× | Cbr; pAS40 carrying norCB with His6×-tagged norB | This study |
| pAS40-nosZ-Strep-tag II | Cbr; pAS40 carrying Strep-tag II-tagged nosZ | This study |
| Strains | ||
| E. coli | ||
| DH10b | Parental strain | Invitrogen, Darmstadt, Germany |
| DH10b/pJET1.2-nosZ | DH10b carrying P. aeruginosa nosZ | This study |
| DH10b/pJET1.2-nosZPR | DH10b carrying nosZ promoter | This study |
| DH10b/pJET1.2-nosR | DH10b carrying P. aeruginosa nosR | This study |
| DH10b/pJET1.2-norC | DH10b carrying P. aeruginosa norC | This study |
| DH10b/pJET1.2-norCB | DH10b carrying P. aeruginosa norCB | This study |
| DH10b/pAS40-nosZ | DH10b harboring expression vector pAS40-nosZ | This study |
| DH10b/pAS40-nosR | DH10b harboring expression vector pAS40-nosR | This study |
| DH10b/pAS40-norC | DH10b harboring expression vector pAS40-norC | This study |
| DH10b/pAS40-norCB | DH10b harboring expression vector pAS40-norCB | This study |
| P. aeruginosa | ||
| PAO1 wild type | Parental strain | DSMZ, Braunschweig, Germany |
| PAO1/pAS40 | PAO1 wild type carrying pAS40 | This study |
| PAO1 nosR | PAO1 nosR::mini-Tn5-luxCDABE | 49 |
| PAO1 norC | PAO1 norC::mini-Tn5-luxCDABE | 49 |
| PAO1 norB | PAO1 norB::mini-Tn5-luxCDABE | 49 |
| PAO1 nosZ | PAO1 nosZ::mini-Tn5-luxCDABE | 49 |
| PAO1 nosR-pnosR-His6× | PAO1 nosR harboring nosR under the control of the nos promoter and encoding a His6× tag | This study |
| PAO1 norC-pnorC-His6× | PAO1 norC harboring norC under the control of the nor promoter and encoding a His6× tag | This study |
| PAO1 norB-pnorB-His6× | PAO1 norB harboring norCB under the control of the nor promoter and encoding a His6× tag at the norB gene | This study |
| PAO1 nosZ-pnosZ-Strep-tag II | PAO1 nosZ harboring nosZ under the control of the nos promoter and encoding a Strep-tag II tag | This study |
R, C, and B indicate that the plasmid was used for the cloning of nosR, norC, and norB His6× fusion genes, respectively. Z indicates that the plasmid is suitable for nosZ fusion gene cloning.
Antibiotic resistance: Apr, ampicillin resistance; Cbr, carbenicillin resistance.
TABLE 2.
List of primers used in this studya
| Primer | Sequence (5′ to 3′) |
|---|---|
| nosRFw | CACCGGTCTCGGGTGTATCTCACCTGCGGCAA |
| nosRRv | CCTCGGTCTCTGAGGTTCTCCGCGGGTATCCG |
| nosRHis6×Fw | AATTGGTGTGAGACCAGATCTGATATCGGTCTCACCTCCATCATCATCATCATCATTGA |
| nosRHis6×Rv | AGCTTCAATGATGATGATGATGATGGAGGTGAGACCGATATCAGATCTGGTCTCACACC |
| norCFw | GCCAGGTCTCGCTGATGCTCTACTGGCACATGGTC |
| norCRv | CAAGGGTCTCTACCCTCCTTGTTCGGCGGCCA |
| norCHis6×Fw | AATTCTGATGAGACCAGATCTGATATCGGTCTCAGGGTCATCATCATCATCATCATTGA |
| norCHis6×Rv | AGCTTCAATGATGATGATGATGATGACCCTGAGACCGATATCAGATCTGGTCTCATCAG |
| norCBFw | GCCAGGTCTCGCTGATGCTCTACTGGCACATGGTC |
| norCBRv | CAAGGGTCTCTGGCGGCCGCCTTGCCGCGCCG |
| norCBHis6×Fw | AATTCTGATGAGACCAGATCTGATATCGGTCTCACGCCCATCATCATCATCATCATTGA |
| norCBHis6×Rv | AGCTTCAATGATGATGATGATGATGGGCGTGAGACCGATATCAGATCTGGTCTCATCAG |
| nosZFw | TACCGGTCTCTGGTAACCTCTGACCTGGCTCC |
| nosZRv | GGCTGGTCTCTAGCCTTTTCCACCAGCATCCG |
| nosPRFw | GATAGGTCTCGTATCTCACCTGCGGCAACGAC |
| nosPRRv | GGTAGGTCTCGTACCGGCGAAAAAAGGGACGC |
| nosZStrep-tag IIFw | AATTTATCCGAGACCAGATCTGATATCGGTCTCCGGCTTGGAGCCACCCGCAGTTCGAAAAATGA |
| nosZStrep-tag IIRv | AGCTTCATTTTTCGAACTGCGGGTGGCTCCAAGCCGGAGACCGATATCAGATCTGGTCTCGGATA |
All primers were purchased from Invitrogen, Darmstadt, Germany. Within the primer sequences, the BsaI restriction site is underlined, and the first 4 and the last 4 nucleotides of the amplicon sequence are in bold. The BglII-EcoRV restriction sequence is in italics.
Nitrite and nitrate consumption rate determination.
In order to determine the nitrite reduction activity of the diverse P. aeruginosa strains that were tested, a nitrate/nitrite colorimetric test was used according to the manufacturer's guidelines (Roche, Germany).
P. aeruginosa interactomic studies using NosR, NorC, and NorB as bait proteins.
For our interaction studies, NorB, NorC, and NosR were produced as tagged proteins in an appropriate defined genetic background. For this purpose, a plasmid toolbox based on the Golden Gate cloning strategy was developed (52). The respective genes were fused at their 3′ ends to the corresponding tags in order to avoid interference with signal peptide recognition and protein insertion into the membrane. To mimic natural protein production, the native promoters were used. The nosR, norC, and norCB genes, including a region between 400 and 500 bp upstream of their start codon, were PCR amplified using the primers nosRFw/nosRRv, norCFw/norCRv, and norCBFw/norCBRv, respectively. The nosZ gene, including the promoter sequence, was PCR amplified from the nosRZDFYL operon using the nosZFw/nosZRv and nosPRFw/nosPRRv primers (Table 2). The respective PCR products were cloned into the BsaI restriction sites of the carrier vector pJET1.2 (Table 1). The second acceptor plasmid encoded the His6× tag. The plasmids pJET1.2-nosR, pJET1.2-norC, and pJET1.2-norB were united with the plasmid pAS40 and subjected to 50 cycles of amplification via PCR using the individually chosen primers (nosRHis6×Fw/nosRHis6×Rv, norCHis6×Fw/norCHis6×Rv, norCBHis6×Fw/norCBHis6×Rv, and nosZStrep-tagIIFw/nosZStrep-tagIIRv), followed by a 5-min restriction with BsaI and a 5-min ligation using T4 DNA ligase. Depending on the combination of gene constructs, the following final products were generated: pAS40-nosR-His6×, pAS40-norCB-His6×, pAS40-norC-His6×, and pAS40-nosZ-Strep-tag II, respectively (Table 1). The corresponding P. aeruginosa nosR, norC, norB, and nosZ transposon mutant strains were transformed with the complementing plasmid, respectively.
The resulting strains were PAO1 nosR-pnosR-His6×, PAO1 norC-pnorC-His6×, PAO1 norB-pnorB-His6×, and PAO1 nosZ-pnosZ-Strep-tag II. The expression vector pAS40 was introduced into the P. aeruginosa PAO1 wild type and employed as the negative control for the interactomic studies (Table 1). The P. aeruginosa strains were grown to the exponential growth phase (optical density at 578 nm [OD578] of 0.8). The protein complexes were cross-linked in vivo as previously described (44). Formaldehyde, used as the cross-linking agent, was injected into the anaerobic denitrifying culture in the late stationary phase to a final concentration of 0.125% (vol/vol). The cultures were further incubated by shaking at 150 rpm to facilitate cross-linking penetration at 37°C for 20 additional min. The free aldehyde groups were quenched by adding 135 mM glycine, and the cultures were further incubated at 37°C for 5 min with shaking at 150 rpm. Then the cells were harvested at 3,000 × g for 20 min at 4°C. The resulting cell pellet was washed twice with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4 [pH 7.3]). Because of interest in the interactome of the outer membrane, the periplasm, the inner membrane, and the cytoplasm, the corresponding fractions were separated and isolated at 4°C. For this purpose, washed cells were resuspended in 0.65 pellet volume of lysis buffer (50 mM NaH2PO4, 300 mM NaCl [pH 8.0]) with 1 tablet of protease inhibitor cocktail (Roche, Germany) per 10 ml of buffer. The cells were lysed by using a French press at 1,100 lb/in2 and 4°C. The cell debris was removed by centrifugation at 20,000 × g for 30 min at 4°C. In order to isolate the membrane-bound proteins, lysates were ultracentrifuged at 100,000 × g for 1 h at 4°C. Subsequently, 200 μg of the membrane proteins of the pellet fraction was dissolved for 1 h at 4°C in 500 μl of 20 mM potassium buffer (pH 8.0) supplemented with 2% Triton X-100. The cell debris and insolubilized membranes were removed by centrifugation at 20,000 × g for 30 min at 4°C. The solubilized membrane proteins present in the supernatant were mixed for 1 h at 4°C with nickle-nitrilotriacetic acid (Ni-NTA) agarose previously equilibrated with NPI-10 buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [Protino]; Macherey-Nagel GmbH, Düren, Germany). The column was washed with imidazole at increasing concentrations (20, 30, 40, and 50 mM in NPI-20, NPI-30, NPI-40, and NPI-50 buffers consecutively). Recombinantly tagged NosR, NorC, and NorB proteins, along with their potential interaction partners, were eluted from the column by addition of 300 mM imidazole in NPI-300 buffer. The protein fractions of interest were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein bands were stained with Coomassie brilliant blue according to the manufacturer's instructions. Primary antibodies against His6× tags were reacted with glutathione S-transferase (GST)–alkaline phosphatase-conjugated secondary antibodies. Stained SDS-PAGE gels and Western blots were visualized and quantified on a GS-800 calibrated densitometer (Bio-Rad).
In order to assess whether the detected bound protein interaction partners constituted specific interaction partners or were artifacts derived from potential high protein abundances within the membrane fractions, P. aeruginosa PAO1 membranes were isolated, and the residing proteins were solubilized as described above. The solubilized membrane and membrane-associated proteins were extracted (53). Protein samples of 0.1 to 0.5 mg/ml were analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Experiments were carried out in three independent biological replicates.
LC-MS/MS analyses of protein complexes and data interpretation.
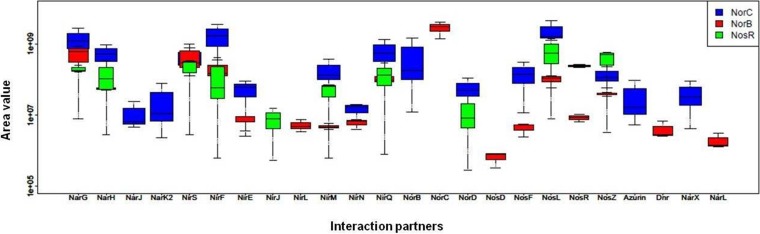
As described above, the PAO1 membrane proteins were solubilized, and the remaining detergent was removed by methanol-chloroform protein extraction. Protein samples were loaded onto the LC-MS/MS. Thereby, it was feasible to evaluate the interaction partners' enrichment within the copurifications in comparison to the natural abundances of these proteins in the membrane fraction. The proteins that were significantly enriched in the copurification in contrast to the controls were defined as specific interactors. Several cytoplasmic and periplasmic proteins were present in the cross-linked samples but did not appear in the control samples, indicating their overall low abundance and enrichment via bait binding. The protein abundance index (PAI), as previously introduced (47), was used to attribute a value to each protein of the sample mixture, and “area values” were deduced. Here, we used the “peak area calculation quantification” method of Proteome Discoverer to determine the PAI values. The method integrates the MS signal intensities of each peptide eluting from the chromatographic column over time, resulting in corresponding peptide peak areas. The average of the peak areas from the three most abundant distinct peptides defines the individual PAI value (47, 54). The results were plotted in a box plot (see Fig. 5).
FIG 5.
Potential interaction partners for NorB, NorC, and NosR involved in denitrification. The box plot represents interaction partners found for NorC, NorB, and NosR plotted against the area value. Only the interaction partners found in all replicates for each protein are depicted here. The heights of the boxes enclose the difference between the first and the third quartiles of the values comprising 50% of the data. The lines extending vertically (whiskers) denote the maximal and minimal values encountered. The median (second quartile) is presented as a black band within the boxes. The area value (y axis) is given in logarithmic scale. The R program was used to generate this plot.
Ni-NTA agarose eluates from triplicate experiments for the isolation of the cross-linked complexes to the respective NosR-His6×, NorC-His6×, and NorB-His6× bait proteins were subsequently evaluated by LC-MS/MS as previously described (55). The eluted proteins with their attached copurified interaction partners were identified by quantitative mass spectrometry (AP-QMS) (56). The LC-MS/MS analyses were performed on a Dionex UltiMate 3000 RSLCnano system connected to an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, Germany). The protein complexes eluted first were digested by trypsin protease treatment with a protein/protease ratio of about 50:1 at 37°C overnight. Subsequently, peptides were loaded onto a C18 precolumn (3-μm RP18 beads, Acclaim, 75 μm by 20 mm; Dionex, Thermo Scientific, Germany) and washed for 3 min at a flow rate of 6 μl/min. The peptides were separated on a C18 analytical column (3-μm, Acclaim PepMap RSLC, 75 μm by 25 cm; Dionex) at a flow rate of 350 μl/min via a linear 120-min gradient from 100% buffer A (0.1% formic acid in water) to 25% buffer B (99.9% acetonitrile with 0.1% formic acid), followed by a 50-min gradient from 25% buffer A to 80% buffer B. The LC system was operated with Chromeleon software (version 6.8; Dionex), which was embedded in the Xcalibur software suite (version 2.1; Thermo Scientific, Germany). The effluent from the column was electrosprayed (PicoTip emitter needles; Thermo Fischer Omnilab, Germany) into the mass spectrometer (Orbitrap Velos Pro). The mass spectrometer was controlled and operated in the data-dependent mode of the Xcalibur software, allowing the automatic selection of a maximum of 10 double- and triple-charged peptides and their subsequent fragmentation. Peptide fragmentation was carried out using LTQ settings (minimum signal, 2,000; isolation width, 4; normalized collision energy, 35; default charge state, 4; and activation time, 10 ms). The MS/MS raw data files were processed via the Proteome Discoverer program (version 1.4; Thermo Scientific) on a Mascot server (version 2.3.02; Matrix Science) using a P. aeruginosa PAO1 database extracted from Swiss-Prot. The following search parameters were used: enzyme, trypsin; maximum missed cleavages, 1; fixed modification, MMTS(C); variable modifications, oxidation (M); peptide tolerance, 10 ppm; and MS/MS tolerance, 0.4 Da. The results were evaluated and quantitated using Proteome Discoverer 1.4 (53, 55, 57).
Antibody generation.
All polyclonal antibodies were raised in rabbits against purified proteins or synthetic peptides representing soluble loops of the membrane proteins of interest. For NosZ antibodies, the protein was recombinantly produced in P. aeruginosa and purified to apparent homogeneity via its Strep-tag II using affinity chromatography (IBA, Germany). The protein eluates were concentrated using Vivaspin 15 (10-kDa cutoff) centrifugal concentrators (Sartorius AG, Germany). For the generation of anti-NosR, anti-NarH, anti-NirS, and anti-NorC polyclonal antibodies, synthetic peptides were used. The amino acid residues 50 to 61, 102 to 116, 118 to 132, and 106 to 130 for NorC and 190 to 203 and 404 to 418 for NosR were selected for this purpose. These amino acid residues corresponded to medium-length periplasmic loops of these proteins, resulting in soluble peptides. For the generation of anti-NarH polyclonal antibodies, a mixture of three peptides corresponding to amino acid residues 1 to 276, 277 to 392, and 418 to 513 was selected. Anti-NirS polyclonal antibodies were raised in rabbits with a three-peptide mixture corresponding to amino acid residues 379 to 392, 526 to 540, and 541 to 555, respectively.
Double immunogold labeling-based colocalization studies using electron microscopy.
The P. aeruginosa PAO1 wild-type strain was grown anaerobically in LB broth supplemented with 50 mM nitrate for 8 h. The bacteria were fixed with 1% formaldehyde in the growth medium overnight at 5°C. After centrifugation, the cells were resuspended in Tris-EDTA (TE) buffer (10 mM Tris-Cl, 1 mM EDTA [pH 7.5]) with the addition of 10 mM glycine to quench free aldehyde groups. Afterward, the bacteria were dehydrated and subsequently embedded in Lowicryl K4M resin. After polymerization by ultraviolet light treatment, ultrathin sections were cut with a diamond knife, collected onto Butvar-coated nickel grids, and incubated with specific antibodies (44). First, sections were incubated with the first round of purified IgG antibodies overnight at 5°C. After a washing with PBS, the bound antibodies were rendered visible by adding protein A/G gold nanoparticles (15 nm in size; 1:75 dilution of the stock solution) for 30 min at room temperature. Sections were washed with PBS containing 0.1% Tween 100 and further incubated with a protein A solution (0.1 mg/ml) for 15 min at room temperature. After an additional washing in PBS, the sections were incubated for 3 h at room temperature with the second round of a different purified IgG. The antibody dilutions were established at 1:25 for the first IgG and at 1:25 for the second charge. After PBS washing, the sections were incubated with protein A/G gold nanoparticles (10 nm in size; 1:200 dilution of the stock solution) for 30 min at room temperature. Subsequently, the sections were washed with PBS containing 0.1% Tween 100, TE buffer, and distilled water. The sections were counterstained with 4% aqueous uranyl acetate for 1 min. The samples were then examined in a TEM910 transmission electron microscope (Carl Zeiss, Germany) at an acceleration voltage of 80 kV. The images were recorded digitally at calibrated magnifications with a slow-scan charge-coupled-device (CCD) camera (ProScan, 1,024 by 1,024 pixels) with ITEM software (Olympus Soft Imaging Solutions, Germany). The contrast and brightness were adjusted with Adobe Photoshop CS4.
RESULTS AND DISCUSSION
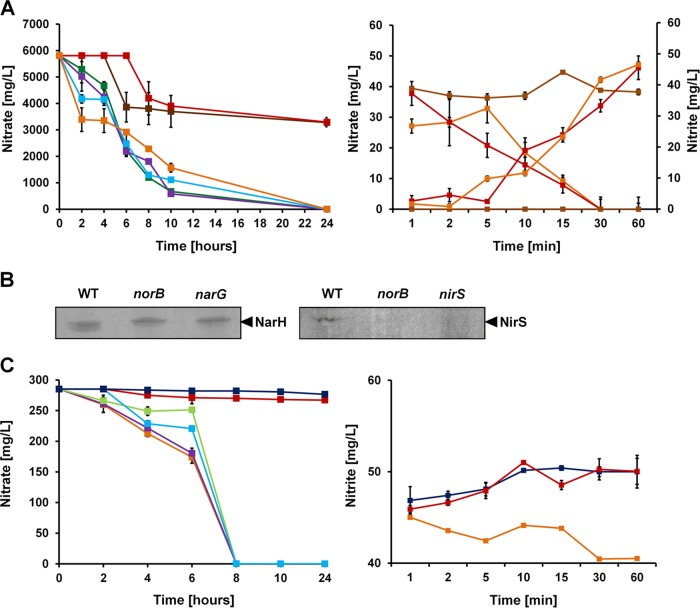
Mutants of the P. aeruginosa nitric oxide oxidoreductase genes norBC and of the regulatory gene nosR are defective in nitrate and nitrite reduction activity.
The selection of the initial bait proteins for our interactomic approach was based on the assumption that the protein network of denitrification should be stably localized, most likely anchored to the membrane. Only NarI, NorB, NorC, and NosR were confirmed as integral membrane proteins. In order to investigate the membrane proteins NorBC and NosR for additional structural functions in the overall denitrifying process, the nitrate and nitrite consumption of the corresponding P. aeruginosa mutants was determined. The values obtained were compared to those for mutants of the cytoplasmic nitrate reductase gene (narG) and periplasmic nitrite reductase gene (nirS) (Fig. 2A). The norB mutant showed a reduced level of conversion of nitrate to nitrite similar to that of the narG nitrate reductase mutant (Fig. 2A). Anti-NarH antibodies detected intact nitrate reductase in the norB mutant (Fig. 2B), and a cell extract of the norB mutant exhibited normal nitrate reductase activity. The norB mutant was also defective in nitrite reduction, with activities similar to those of the nirS nitrite reductase mutant (Fig. 2C). The norC and nosR mutants also showed reduced nitrite reduction activity. The overall nitrite conversion of this mutant was somewhat delayed, indicating an initial defect and a following compensatory role of other components of the system. It was known from previous experiments that NorBC influences NirS formation and stability (58), although the underlying mechanism was unclear. Consistent with this observation, our anti-NirS antibodies failed to detect significant amounts of nitrite reductase in the norB mutant (Fig. 2B), and only residual nitrite reductase activity was found in the cell extracts of the norB mutant. Since NorBC and NosR are not enzymatically involved in nitrate and nitrite reduction and indications for their involvement in narGHIJ and nirS gene expression have not been observed, an essential alternative contribution to the overall denitrification process must be deduced. Therefore, we tested NorBC and NosR for their function in denitrification protein network formation.
FIG 2.
In vivo and in vitro nitrate and nitrite conversion of P. aeruginosa denitrification mutants. (A) The in vivo nitrate consumption by the wild type (WT) (orange) and narG (brown), norB (red), norC (green), nosZ (lilac), and nosR (blue) mutant strains is shown in the left panel. The in vitro nitrate utilization and nitrite production of the wild type (red) and norB (orange) and narG (brown) mutant strains were determined with cell extracts from the corresponding strains. (B) The cell extracts of the indicated strains were tested for the presence of the nitrate reductase subunit NarH (left) and the nitrite reductase NirS (right) using Western blot analyses. (C) The in vivo nitrite conversion of the wild type (orange) and nosZ (lilac), norC (green), norB (red), nosR (blue), and nirS (black) mutant strains is shown in the left panel. The in vitro nitrite utilization by the wild type (orange) and the norB (red) and nirS (black) mutant strains was determined using cell extracts from the corresponding strains (right panel).
Interactomic studies using P. aeruginosa NosR, NorC, and NorB.
Double immunolabeling of ultrathin sections, combined with transmission electron microscopy, was employed to verify some of the observed protein-protein interactions.
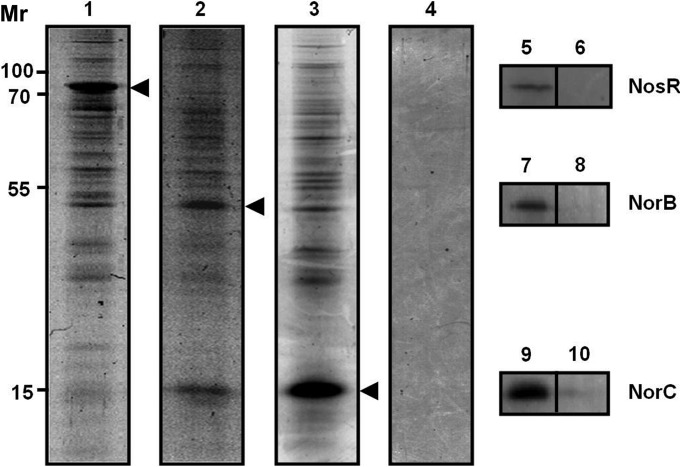
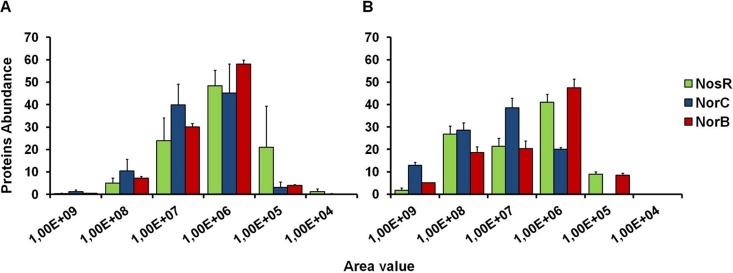
For the interactomic approach, protein eluates from the affinity material were trichloroacetic acid (TCA) precipitated and analyzed by SDS-PAGE (Fig. 3). The NosR-His6× eluates contained a protein with a relative molecular weight of approximately 80,000 ± 5,000 as a major component, which was identified as NosR-His6× by immunoblot detection (Fig. 3 lane 5). The SDS-PAGE showed a number of additional protein bands visible above and below NosR-His6×, which represented potential interaction partners (Fig. 3, lane 1). Likewise, the NorC-His6× and NorB-His6× eluates contained prominent proteins, which were identified as NorC-His6× and NorB-His6×, respectively (Fig. 3, lanes 7 and 9). Again, several potential interaction partners were observed (Fig. 3, lanes 2 and 3). As expected, the control reactions with PAO1 carrying the control vector did not result in copurification of additional proteins (Fig. 3, lanes 4, 6, 8, and 10). Similarly, production of a His-tagged cytosolic control protein (porphobilinogen synthase [HemB]) under identical conditions did not yield the observed bound proteins (not shown). The abundance distributions of the potential interaction partners of NosR, NorC, and NorB related to denitrification are illustrated in graphs (Fig. 4) representing the concentration distributions of the potential interaction partners for the various bait proteins with NosR, NorC, and NorB determined after copurification. The percentage of peptides found for certain proteins (y axis) was plotted against the corresponding area value (x axis). The potential interaction partners involved in denitrification are shown in Fig. 4A, and all the other proteins are depicted in the Fig. 4B.
FIG 3.
Interactomics of the NosR, NorC, and NorB membrane proteins with multiple potential interaction partners. Tagged bait proteins were produced as fusion proteins in P. aeruginosa, chemically cross-linked to interaction partners, and affinity purified. The fractions were resolved on an SDS-PAGE gel and visualized by Coomassie brilliant blue staining. The copurification fractions for NosR (lane 1), NorB (lane 2), NorC (lane 3), and the control reaction (PAO1/pAS40) (lanes 6, 8, and 10) are shown as indicated. Western blot analyses showed the presence of the NosR-His6× fusion protein (lane 5), NorB-His6× fusion protein (lane 7), and NorC-His6× fusion protein (lane 9). Mr represents the relative molecular weight (×1,000). The recombinant strains PAO1 nosR-pnosR-His6×, PAO1 norC-pnorC-His6×, and PAO1 norB-pnorB-His6× were subjected to these interactomic studies.
FIG 4.
Abundance distribution of potential interaction partners of NosR, NorC, and NorB nonrelated or related to denitrification. Graphs representing the concentration distributions of potential interaction partners for the various bait proteins with NosR, NorC, and NorB were determined after copurification. The percentages of peptides found for certain proteins were plotted against the corresponding area value. The potential interaction partners involved in denitrification are shown in panel A, while all the other proteins are depicted in panel B. Data for this plot were obtained from triplicate experiments with corresponding averages and standard deviations.
The various affinity chromatographic eluates were then analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). A protein abundance index (PAI) was attributed to each protein of the sample mixture, and an “area value” was calculated, as previously described (47). The percentages of the overall potential interaction partners were plotted against the corresponding area value ranges. The standard deviations were calculated from biological triplicates and integrated into a box plot (Fig. 5). Most of the potential interaction partners were detected at the area value range of 106 to 107, and high-affinity interaction partners were in the area value range of 108 to 109. Abundant proteins in an average bacterial cell, such as chaperones and translation elongation factors, were found within an area value of approximately 106. Therefore, proteins within higher area values were considered specific interaction partners, and those below a 106 area value were not further considered in this study. The candidate proteins were separated into the categories of “denitrification-related proteins” and “other proteins,” as shown in Table 3 (see also Table S1 in the supplemental data). This approach does not distinguish between proteins directly bound to the prey proteins and indirectly bound proteins cross-linked and attached to the whole protein complex via already-associated proteins.
TABLE 3.
Determination of NorB/C and NosR interaction partners from the denitrification process via binding, cross-linking, affinity copurification, and proteomic identificationa
| PA no. | Interaction partner | Result for: |
||||
|---|---|---|---|---|---|---|
| NorB | NorC | NosR | NarH | NosZ | ||
| PA4292 | Azurin | ND | 0.6 | ND | ||
| PA0527 | Transcriptional regulator Dnr | ∞ | ND | ND | ||
| PA3875 | Respiratory nitrate reductase alpha chain NarG | 0.98 | 7.08 | 2.46 | ||
| PA3874 | Respiratory nitrate reductase beta chain NarH | 1.26 | 12.06++ | 3.41++ | − | ++ |
| PA3873 | Respiratory nitrate reductase delta chain NarJ | ND | ∞ | ND | ||
| PA3876 | Nitrite extrusion protein NarK2 | ND | 3.11 | ND | ||
| PA3879 | Two-component response regulator NarL | ∞ | ND | ND | ||
| PA3878 | Two-component sensor NarX | ND | ∞ | ND | ||
| PA0510 | Uroporphyrin-III c-methyltransferase NirE | ∞ | ∞ | ND | ||
| PA0516 | Heme d1 biosynthesis protein NirF | 18.89 | 163.94 | 14.42 | ||
| PA0511 | Heme d1 biosynthesis protein NirJ | ND | ND | ∞ | ||
| PA0514 | Heme d1 biosynthesis protein NirL | ∞ | ND | ND | ||
| PA0518 | Cytochrome c551 NirM | 0.11 | 4.17 | 1.00 | ||
| PA0509 | Putative c-type cytochrome NirN | 0.96 | 2.27 | ND | ||
| PA0520 | Regulatory protein NirQ | 1.75 | 10.54 | 2.26 | ||
| PA0519 | Nitrite reductase NirS | 2.17 | 1.97+++ | 0.98++ | ++ | +++ |
| PA0524 | Nitric oxide reductase subunit B NorB | 435.73 | ND | |||
| PA0523 | Nitric oxide reductase subunit C NorC | 10.61 | ND++ | ++ | +++ | |
| PA0525 | Probable denitrification protein NorD | ND | 15.73 | 3.81 | ||
| PA3393 | NosD protein | ∞ | ND | ND | ||
| PA3394 | NosF protein | ∞ | ∞ | ND | ||
| PA3396 | NosL protein | 2.82 | 68.91 | 18.65 | ||
| PA3391 | Regulatory protein NosR | 1.91 | 52.88++ | ++ | +++ | |
| PA3392 | Nitrous oxide reductase NosZ | 0.77 | 2.61+++ | 7.11+++ | ++ | − |
The LC-MS/MS data reveal the enrichment of these proteins during the copurification, given as area values (fold enrichment) in comparison to the negative control. Each number represents the average area of the protein in copurifications divided by the average area of the protein in the control fraction. −, protein present in the control fraction but not in the copurification; ∞, protein found present in the cross-linked, copurified fraction but not in the control fraction; ++, specific interaction with the tagged bait protein in the immunogold labeling experiments; +++, highly specific interaction with the tagged bait protein in the immunogold labeling experiments. Values in bold correspond to the proteins with a strong interaction in the copurification experiments. The proteins in the “Interaction partner” column are ordered alphabetically. ND, not determined.
The nitric oxide reductase NorBC represents one major assembly platform protein for the denitrification protein network.
Crystal structures of the heterodimeric nitric oxide reductase have revealed that subunit B forms a tight complex with its second subunit C. In agreement, our interactomic results showed the strongest interaction of NorB with NorC and vice versa, which provided a proof of principle of the approach (Table 3). Overall, the interactions of the majority of the proteins detected with NorC were significantly stronger than those with NorB with only a few exceptions. Most likely, this difference is due to the NorBC structural arrangement where most of the NorC subunit is exposed toward the periplasm. Alternatively, differences in the solubilities of NorB and NorC in the experiments might explain these results. The differences in the affinity of NorB (bait) for NorC (prey) of 10 and of NorC (bait) for NorB (prey) of 435 support this assumption (Table 3).
Nitrate reductase NarGHI is interacting with the NorBC platform.
The cytoplasmic nitrate reductase subunits NarG and NarH were found to be strongly bound to NorB and NorC (Table 3). No interaction was detected for the membrane subunit NarI. Moreover, the nitrate/nitrite antiporter NarK2 interacts with NorC (Table 3). In nitrate respiration, nitrate has to be transported into the cytoplasm, and cytotoxic nitrite has to be promptly returned to the periplasm to minimize damage to the cytoplasmic components, so integration of NarK2 into the protein network is not unexpected (31). Interestingly, no detectable interaction of the various proteins tested with the periplasmic nitrate reductase NapAB was observed.
Integration of the nitrite reductase NirS, its maturation apparatus, and azurin to NorBC.
The strongest interaction of NorBC (bait) for an isolated prey protein was detected for the periplasmic, membrane-attached lipoprotein NirF, the last enzyme in heme d1 synthesis and a cofactor of nitrite reductases (26). For the final insertion of heme d1 into NirS, the system uses NirN, a structural homologue of NirS without nitrite reductase activity. NirS, NirF, and NirN form a stable complex during nitrite reductase maturation (26). In agreement, protein interactions of NorB and NorC with NirN were detected in our study (Table 3). Furthermore, strong interactions between NorC and the c-type cytochrome NirM, which serves as an electron donor for the nitrite reductase, were measured (Table 3) (25). Additionally, an interaction of NorC with the ATP-binding protein NirQ was observed (Table 3). This is consistent with the observation that deletion of the nirQ gene resulted in the simultaneous loss of nitrite and NO reduction in P. aeruginosa (34). Posttranslational enhancement of nitrite and nitric oxide reductase activity was observed for P. aeruginosa NirQ activity, similar to the posttranslational activation of 1,5-bisphosphate carboxylase/oxygenase by the NirQ homologue (34). By analogy, NirQ might act as a nitrite reductase- and nitric oxide reductase-specific assembly factor.
Azurin was also found to attach to the protein network via NorC. Azurin, like cytochrome c, mediates the transfer of electrons from the cytochrome bc1 complex to various terminal reductases (41). It is known that cytochrome c551 NirM, the electron donor to nitrite reductase NirS, forms a tight complex with azurin (59).
Surprisingly, the nitrite reductase NirS itself showed a stronger interaction with NorB than with NorC (Table 3), especially when the relative observed interaction strengths of the two proteins were taken into account. Nevertheless, our immunoelectron microscopy approach confirmed the NirS-NorC interaction (Fig. 6). Thus, the nitrite reductase NirS is bound to the denitrification supercomplex via NorB, while the electron donor system NirM and the enzyme maturation machinery NirN-NirF-NirQ, interacting with NirS, are bound via NorC.
FIG 6.
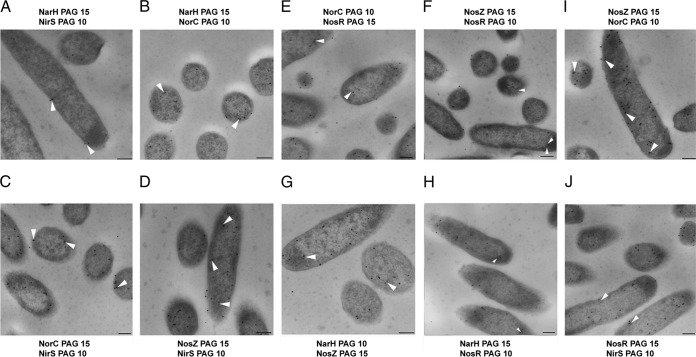
Double immunogold labeling of NosR, NarH, NirS, NorC, and NosZ to determine their in vivo colocalization in the P. aeruginosa cell. The names of the proteins that were immunogold labeled and tested for colocalization are shown above and below each photograph. The numbers for each protein represent the gold particle size employed for each labeling. Bars, 200 nm. The arrows indicate proteins as colocalized only if the distance between them was <25 nm.
Nitrous oxide reductase NosZ and assembly protein NosL are integrated into the protein network via NosR.
Originally, NosR was discovered as a component necessary for the expression of nitrous oxide reductase (60). NosR from Pseudomonas stutzeri is a polytopic membrane protein required for the formation of nitrous oxide reductase NosZ. However, NosZ purified from a nosR-deficient background revealed nitrous oxide reductase activity in vitro (61).
NosR is an iron-sulfur flavoprotein with its flavin cofactor directed toward the periplasm and redox centers toward the cytoplasm. The exact biochemical function of the protein is currently unknown. NosR was found to be strongly attached to the NorBC platform via NorC, which, in turn, had strong interactions with the nitrate and nitrite reductase systems. In agreement with these results, tagged NosR bait indeed “captured” NorBC as prey (Table 3). Moreover, strong interactions of NosR with nitrate reductase subunits NarG and NarH, as well as with NirE, NirQ, nitrite reductase NirS, and NirM, were observed, indicating a tightly bound complex at the NorBC-NosR platform. Most important, further interactions were detected with the nitrous oxide reductase NosZ (Table 3), whereas NosZ exhibited only weak interactions with the NorBC proteins. Consequently, NosZ is bound to the denitrification protein network via NosR. The nitrous oxide reductase NosZ is a homodimeric metalloprotein containing two unusual copper centers (39). These centers require additional proteins for maturation. Multiple proteins encoded by the nosDFYL tatE operon were proposed to be involved in this process (61). We detected strong protein interactions of NosL with NorC and NosR. Achromobacter cycloclastes NosL was identified as a copper binding protein (39). The protein contains a putative lipobox, a signal peptide that targets the protein for translocation through the inner membrane, followed by cleavage of the signal leader sequence and aminoacylation of the N-terminal cysteine to diacylglycerol. NosL is most likely shuttled through the periplasm where the attached diacylglycerol anchor tethers the protein to the outer membrane.
Immunogold labeling for in vivo colocalization of NosR and the four reductases of the denitrification pathway in P. aeruginosa.
Antibodies specific for NarH, NirS, NorC, NosZ, and NosR were raised and used to determine whether these proteins colocalize in P. aeruginosa. Double immunogold labeling was performed with all possible combinations of the two proteins, one protein of the pair being labeled with nanogold particles of 15 nm in size and the other protein of the pair with particles of 10 nm in size. As shown in Fig. 6, all proteins seem to colocalize to a certain extent. As can be deduced from the quantitative data from the double labeling studies (Table 4), all of the pairs involving proteins localized within the cytoplasmic membrane and/or in the periplasm showed a higher percentage of colocalization (well above 1) compared with those of pairs of proteins separated by the cytoplasmic membrane (with an average of colocalization of less than 1). Thus, colocalization of the pairs NosZ-NirS, NorC-NirS, NosZ-NorC, and NosZ-NosR was more frequent than that of the pairs NarH-NirS, NarH-NosZ, NarH-NorC, NosR-NirS, NarH-NosR, and NorC-NosR.
TABLE 4.
Quantification of in vivo protein colocalizations using double immunogold labeling studies and electron microscopya
| Fig. 6 panel | Interaction | Protein immunolabeled (avg no. of proteins that colocalized in 20 cells) with: |
Total no. of colocalizations in 20 cells | Avg no. of colocalizations in 20 cells | |
|---|---|---|---|---|---|
| PAG15 | PAG10 | ||||
| A | NarH-NirS | NarH (3) | NirS (1.95) | 3 | 0.15 |
| B | NarH-NorC | NarH (7.5) | NorC (4.8) | 14 | 0.7 |
| C | NorC-NirS | NorC (8.35) | NirS (3.65) | 25 | 1.25 |
| D | NosZ-NirS | NosZ (8.6) | NirS (4.6) | 25 | 1.25 |
| E | NosR-NorC | NosR (9.95) | NorC (2.7) | 17 | 0.85 |
| F | NosZ-NosR | NosZ (7.45) | NosR (4.4) | 20 | 1.7 |
| G | NosZ-NarH | NosZ (8.25) | NarH (3) | 12 | 0.6 |
| H | NarH-NosR | NarH (6.05) | NosR (3.15) | 12 | 0.6 |
| I | NosZ-NorC | NosZ (10.5) | NorC (7.2) | 36 | 1.8 |
| J | NosR-NirS | NosR (8.05) | NirS (3.55) | 22 | 1.1 |
For 20 longitudinal sections with approximately the same area, large and small gold particles and the colocalization events in those cells were counted. The average colocalization value was then calculated for the 20 cells. PAG15 and PAG10, protein A-gold particles with diameters of 15 and 10 nm, respectively.
Thus, the results of the immunogold colocalization experiments are consistent with the conclusions of the interactomic experiments. One possibility we considered was that protein pairs more functionally distant in the denitrification process, i.e., regarding the reduction steps that they perform, such as nitrate reductase and nitrous oxide reductase, might also be more physically separated from one another than from their functional neighbor enzymes. Similarly, proteins involved in intermediate steps, such as NirS and NorCB, might be separated from proteins mediating reductions at the beginning or end of the catalytic chain by an intermediate distance. With this notion in mind, the colocalization experiments suggest that the first two enzymes of the pathway, Nar and Nir, are attached to the third, Nor, while the fourth protein, Nos, is mainly attached to NosR. Interestingly, the distance between the gold particles of NarH and NosZ correspond approximately to the sum of the sizes (in nanometers) of the NarH-NorC and NosZ-NorC pairs.
Electron-donating primary dehydrogenases, ATPase, TCA cycle enzyme, and proteins of protein-translocating Sec are part of the protein network, among others.
In addition to the proteins involved in the denitrification pathway, other proteins of the electron transport chain were identified as being bound to the NorBC-NosR platform (see Table SA1 in the supplemental material). These included electron-donating NADH dehydrogenase Nuo (PA2638 to -2644), proline dehydrogenase PutA (PA0782), l-lactate dehydrogenase (PA4771), succinate dehydrogenase (PA1582 to -1584), d-amino acid dehydratase (PA3357), and malate:quinone oxidoreductase (PA3452). Two of these enzymes participate in the TCA cycle. Similarly, succinyl-coenzyme A (succinyl-CoA) synthetase (PA1588), isocitrate dehydrogenase (PA2624), citrate synthase (PA1580), 2-oxoglutarate dehydrogenase (PA1585), and enzymes of the pyruvate knot, including various subunits of pyruvate dehydrogenases (PA5015), pyruvate kinase (PA4329), phosphopyruvate hydratase (PA3035), acetyl-CoA carboxylase (PA3112, PA3639, and PA4848), and phosphoenolpyruvate synthase (PA1770) were also found to be bound. The cofactor-synthesizing enzymes of heme, cytochrome c, the iron-sulfur cluster, the molybdenum cofactor, and ubiquinone were also part of the supercomplex (see Table SA1). Additionally, multiple subunits of the FoF1 ATP synthase (PA5553 to -5560) were part of the megacomplex. Furthermore, various subunits of the chemotactic complexes CheA (PA1458), CheZ (PA1457), ChpA (PA0413), and PctC (PA4307) and the aerotaxis receptor AER (PA1561) were found to be bound to NorBC-NosR. Surprisingly, multiple subunits of the nucleotide-metabolizing ribonucleotide reductases NrdJ (PA5496/97) and NrdAB (PA1156 and PA1155) and of the iron-storage protein bacterioferritin (PA3531 and PA4235) were also found to be strongly attached to the denitrification platform. FtsH (PA4751), FtsA (PA4408), FtsZ (PA4407), ZipA (PA1528), MniD (PA3244), and MreB (PA4481), which are all involved in cell division, were mainly found attached to NorC.
Model for a nitrate respirasome of P. aeruginosa.
Currently, respirasome research—the investigation of megacomplexes involved in respiration—focuses almost exclusively on the mitochondrial electron transport chain of eukaryotes (3–10). Here, we describe a highly structured, well-organized membrane-attached protein-protein network in bacteria involved in anaerobic energy generation (Fig. 7). From the data obtained, one can deduce the presence of a dynamic megacomplex mediating diverse membrane-associated and periplasmic functions of the cell envelope, including, inter alia, electron donor systems, cofactor-synthesizing complexes, ATPase activity, flagellum assembly and motility, cell wall and outer membrane formation and integrity, cell division, chemotaxis, iron-sulfur protein formation, stress responses and cellular defenses against noxious agents and biotic attacks, DNA and RNA metabolism, and signal transduction and transport processes, many of which are central to the pathogenic lifestyle of P. aeruginosa (Fig. 8). Overall, a complex network of interacting proteins, functionally connecting the central physiological functions of the bacterial cytoplasmic cell membrane, was detected. Similar complex and dynamic protein structures might provide the basis for the coordination of membrane biochemistry in other bacteria.
FIG 7.
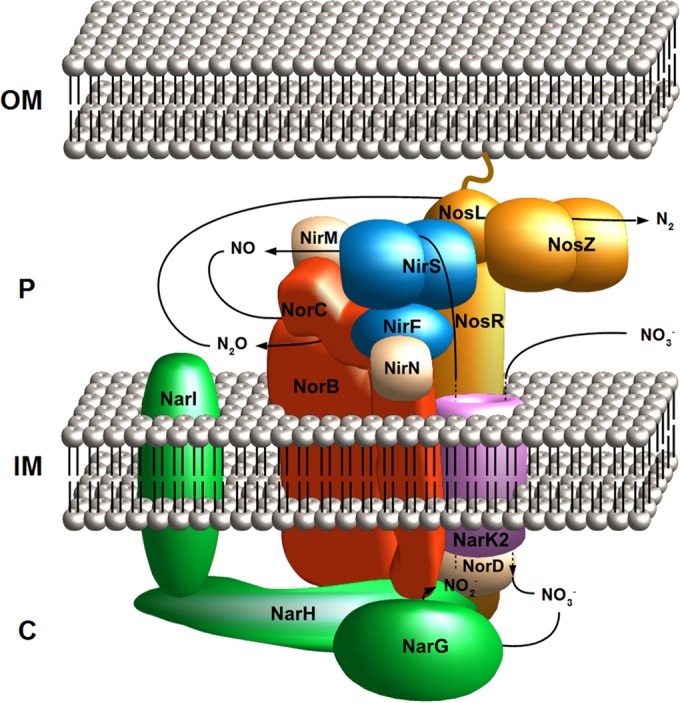
Denitrification supercomplex model proposed in this study. The figure is based on the interactomic and immunogold labeling data. A tight complex of the nitrate (Nar), nitrite (Nir), NO (Nor), and N2O (Nos) reductase enzymes with their corresponding assembly and cofactor proteins was observed. OM, outer membrane; P, periplasm; IM, inner membrane; C, cytoplasm.
FIG 8.
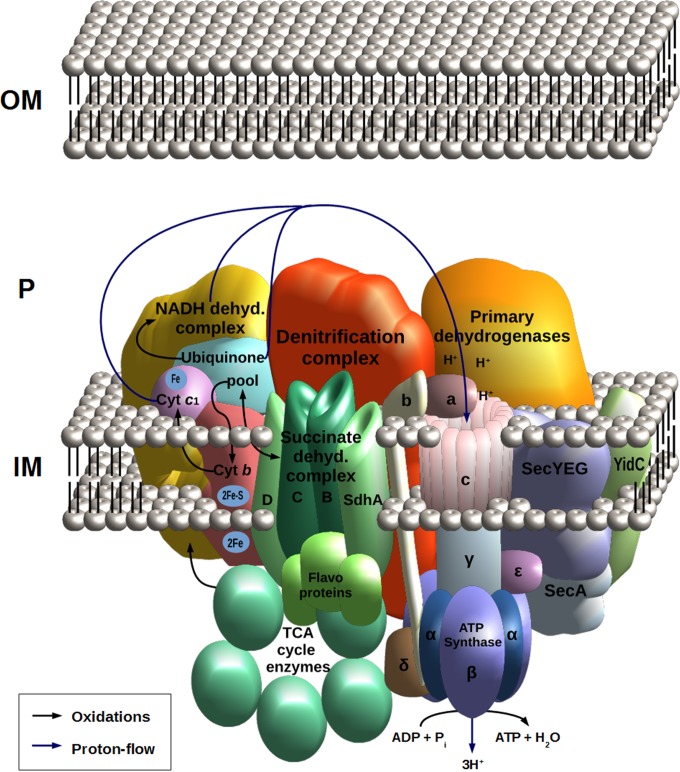
Schematic model of the respirasome relying on the NorCB-NosR supracomplex. OM, outer membrane; P periplasm; IM, inner membrane.
Supplementary Material
ACKNOWLEDGMENTS
This work was generously supported by ERC grant IPBSL, awarded to K.N.T. and Ricardo Amils, and funding by the Deutsche Forschungsgemeinschaft (Forschergruppe PROTRAIN) to M.J. and D.J.
We thank BioMed Proofreading LLC for their support. We also thank Simone Virus for her outstanding technical support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00055-16.
REFERENCES
- 1.Jahn M, Jahn D. 2012. Electron transfer reactions and oxidative phosphorylation. In Michal G, Schomburg D (ed), Biochemical pathways: an atlas of biochemistry and molecular biology, 2nd ed Wiley, New York, NY. [Google Scholar]
- 2.Tielen P, Schobert M, Härtig E, Jahn D. 2012. Anaerobic regulatory networks in bacteria. In Fillaux AAM. (ed), Bacterial regulatory networks. Horizon Academic Press, Norwich, United Kingdom. [Google Scholar]
- 3.Moreno-Lastres D, Fontanesi F, Garcia-Consuegra I, Martin MA, Arenas J, Barrientos A, Ugalde C. 2012. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab 15:324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. 2011. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci U S A 108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, Navas P, Cruz R, Carracedo A, Lopez-Otin C, Perez-Martos A, Fernandez-Silva P, Fernandez-Vizarra E, Enriquez JA. 2013. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 6.Krause F, Reifschneider NH, Vocke D, Seelert H, Rexroth S, Dencher NA. 2004. “Respirasome”-like supercomplexes in green leaf mitochondria of spinach. J Biol Chem 279:48369–48375. doi: 10.1074/jbc.M406085200. [DOI] [PubMed] [Google Scholar]
- 7.Bultema JB, Braun HP, Boekema EJ, Kouril R. 2009. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta 1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Marques I, Dencher NA, Videira A, Krause F. 2007. Supramolecular organization of the respiratory chain in Neurospora crassa mitochondria. Eukaryot Cell 6:2391–2405. doi: 10.1128/EC.00149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schägger H, Pfeiffer K. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudkina NV, Folea IM, Boekema EJ. 2015. Towards structural and functional characterization of photosynthetic and mitochondrial supercomplexes. Micron 72:39–51. doi: 10.1016/j.micron.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Liu LN. 2016. Distribution and dynamics of electron transport complexes in cyanobacterial thylakoid membranes. Biochim Biophys Acta 1857:256−265. doi: 10.1016/j.bbabio.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niederman RA. 2016. Development and dynamics of the photosynthetic apparatus in purple phototrophic bacteria. Biochim Biophys Acta 1857:232−246. doi: 10.1016/j.bbabio.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Prunetti L, Infossi P, Brugna M, Ebel C, Giudici-Orticoni MT, Guiral M. 2010. New functional sulfide oxidase-oxygen reductase supercomplex in the membrane of the hyperthermophilic bacterium Aquifex aeolicus. J Biol Chem 285:41815–41826. doi: 10.1074/jbc.M110.167841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stroh A, Anderka O, Pfeiffer K, Yagi T, Finel M, Ludwig B, Schagger H. 2004. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J Biol Chem 279:5000–5007. [DOI] [PubMed] [Google Scholar]
- 15.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI0213870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Einsle O, Kroneck PM. 2004. Structural basis of denitrification. Biol Chem 385:875–883. [DOI] [PubMed] [Google Scholar]
- 17.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NC. 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol 10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 18.Dias JM, Than ME, Humm A, Huber R, Bourenkov GP, Bartunik HD, Bursakov S, Calvete J, Caldeira J, Carneiro C, Moura JJ, Moura I, Romao MJ. 1999. Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods. Structure 7:65–79. doi: 10.1016/S0969-2126(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 19.Lanciano P, Vergnes A, Grimaldi S, Guigliarelli B, Magalon A. 2007. Biogenesis of a respiratory complex is orchestrated by a single accessory protein. J Biol Chem 282:17468–17474. doi: 10.1074/jbc.M700994200. [DOI] [PubMed] [Google Scholar]
- 20.Arias-Cartin R, Grimaldi S, Arnoux P, Guigliarelli B, Magalon A. 2012. Cardiolipin binding in bacterial respiratory complexes: structural and functional implications. Biochim Biophys Acta 1817:1937–1949. doi: 10.1016/j.bbabio.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Van Alst NE, Sherrill LA, Iglewski BH, Haidaris CG. 2009. Compensatory periplasmic nitrate reductase activity supports anaerobic growth of Pseudomonas aeruginosa PAO1 in the absence of membrane nitrate reductase. Can J Microbiol 55:1133–1144. doi: 10.1139/W09-065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nurizzo D, Silvestrini MC, Mathieu M, Cutruzzola F, Bourgeois D, Fulop V, Hajdu J, Brunori M, Tegoni M, Cambillau C. 1997. N-terminal arm exchange is observed in the 2.15 A crystal structure of oxidized nitrite reductase from Pseudomonas aeruginosa. Structure 5:1157–1171. doi: 10.1016/S0969-2126(97)00267-0. [DOI] [PubMed] [Google Scholar]
- 23.Rinaldo S, Giardina G, Castiglione N, Stelitano V, Cutruzzola F. 2011. The catalytic mechanism of Pseudomonas aeruginosa cd1 nitrite reductase. Biochem Soc Trans 39:195–200. doi: 10.1042/BST0390195. [DOI] [PubMed] [Google Scholar]
- 24.Farver O, Brunori M, Cutruzzola F, Rinaldo S, Wherland S, Pecht I. 2009. Intramolecular electron transfer in Pseudomonas aeruginosa cd1 nitrite reductase: thermodynamics and kinetics. Biophys J 96:2849–2856. doi: 10.1016/j.bpj.2008.12.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa N, Arai H, Igarashi Y. 2001. Two c-type cytochromes, NirM and NirC, encoded in the nir gene cluster of Pseudomonas aeruginosa act as electron donors for nitrite reductase. Biochem Biophys Res Commun 288:1223–1230. doi: 10.1006/bbrc.2001.5919. [DOI] [PubMed] [Google Scholar]
- 26.Nicke T, Schnitzer T, Munch K, Adamczack J, Haufschildt K, Buchmeier S, Kucklick M, Felgentrager U, Jansch L, Riedel K, Layer G. 2013. Maturation of the cytochrome cd1 nitrite reductase NirS from Pseudomonas aeruginosa requires transient interactions between the three proteins NirS, NirN and NirF. Biosci Rep 33:e00048. doi: 10.1042/BSR20130043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bali S, Warren MJ, Ferguson SJ. 2010. NirF is a periplasmic protein that binds d1 heme as part of its essential role in d1 heme biogenesis. FEBS J 277:4944–4955. doi: 10.1111/j.1742-4658.2010.07899.x. [DOI] [PubMed] [Google Scholar]
- 28.Storbeck S, Saha S, Krausze J, Klink BU, Heinz DW, Layer G. 2011. Crystal structure of the heme d1 biosynthesis enzyme NirE in complex with its substrate reveals new insights into the catalytic mechanism of S-adenosyl-l-methionine-dependent uroporphyrinogen III methyltransferases. J Biol Chem 286:26754–26767. doi: 10.1074/jbc.M111.239855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haufschildt K, Schmelz S, Kriegler TM, Neumann A, Streif J, Arai H, Heinz DW, Layer G. 2014. The crystal structure of siroheme decarboxylase in complex with iron-uroporphyrin III reveals two essential histidine residues. J Mol Biol 426:3272–3286. doi: 10.1016/j.jmb.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Adamczack J, Hoffmann M, Papke U, Haufschildt K, Nicke T, Broring M, Sezer M, Weimar R, Kuhlmann U, Hildebrandt P, Layer G. 2014. NirN protein from Pseudomonas aeruginosa is a novel electron-bifurcating dehydrogenase catalyzing the last step of heme d1 biosynthesis. J Biol Chem 289:30753–30762. doi: 10.1074/jbc.M114.603886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma V, Noriega CE, Rowe JJ. 2006. Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 72:695–701. doi: 10.1128/AEM.72.1.695-701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y. 2010. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 33.Shiro Y. 2012. Structure and function of bacterial nitric oxide reductases: nitric oxide reductase, anaerobic enzymes. Biochim Biophys Acta 1817:1907–1913. doi: 10.1016/j.bbabio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi NR, Arai H, Kodama T, Igarashi Y. 1998. The nirQ gene, which is required for denitrification of Pseudomonas aeruginosa, can activate the RubisCO from Pseudomonas hydrogenothermophila. Biochim Biophys Acta 1381:347–350. doi: 10.1016/S0304-4165(98)00045-2. [DOI] [PubMed] [Google Scholar]
- 35.Wüst A, Schneider L, Pomowski A, Zumft WG, Kroneck PM, Einsle O. 2012. Nature's way of handling a greenhouse gas: the copper-sulfur cluster of purple nitrous oxide reductase. Biol Chem 393:1067–1077. doi: 10.1515/hsz-2012-0177. [DOI] [PubMed] [Google Scholar]
- 36.Pomowski A, Zumft WG, Kroneck PM, Einsle O. 2011. N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase. Nature 477:234–237. doi: 10.1038/nature10332. [DOI] [PubMed] [Google Scholar]
- 37.Wunsch P, Zumft WG. 2005. Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration. J Bacteriol 187:1992–2001. doi: 10.1128/JB.187.6.1992-2001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taubner LM, McGuirl MA, Dooley DM, Copie V. 2004. 1H, 13C, 15N backbone and sidechain resonance assignments of apo-NosL, a novel copper(I) binding protein from the nitrous oxide reductase gene cluster of Achromobacter cycloclastes. J Biomol NMR 29:211–212. doi: 10.1023/B:JNMR.0000019240.29152.ac. [DOI] [PubMed] [Google Scholar]
- 39.McGuirl MA, Bollinger JA, Cosper N, Scott RA, Dooley DM. 2001. Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. J Biol Inorg Chem 6:189–195. doi: 10.1007/s007750000190. [DOI] [PubMed] [Google Scholar]
- 40.Dell'acqua S, Moura I, Moura JJ, Pauleta SR. 2011. The electron transfer complex between nitrous oxide reductase and its electron donors. J Biol Inorg Chem 16:1241–1254. doi: 10.1007/s00775-011-0812-9. [DOI] [PubMed] [Google Scholar]
- 41.Williams HD, Zlosnik JE, Ryall B. 2007. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv Microb Physiol 52:1–71. doi: 10.1016/S0065-2911(06)52001-6. [DOI] [PubMed] [Google Scholar]
- 42.Ugidos A, Morales G, Rial E, Williams HD, Rojo F. 2008. The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ Microbiol 10:1690–1702. doi: 10.1111/j.1462-2920.2008.01586.x. [DOI] [PubMed] [Google Scholar]
- 43.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. 2010. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol 12:1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 44.Borrero-de Acuña JM, Molinari G, Rohde M, Dammeyer T, Wissing J, Jänsch L, Arias S, Jahn M, Schobert M, Timmis KN, Jahn D. 2015. A periplasmic complex of the nitrite reductase NirS, the chaperone DnaK, and the flagellum protein FliC is essential for flagellum assembly and motility in Pseudomonas aeruginosa. J Bacteriol 197:3066–3075. doi: 10.1128/JB.00415-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee PY, Bae KH, Kho CW, Kang S, Lee do H, Cho S, Lee SC, Park BC, Park SG. 2008. Interactome analysis of yeast glutathione peroxidase 3. J Microbiol Biotechnol 18:1364–1367. [PubMed] [Google Scholar]
- 46.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. 2010. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics 9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ong SE, Mann M. 2005. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol 1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 48.Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schock U, Pohl TM, Wiehlmann L, Tummler B. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol 192:1113–1121. doi: 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FS, Hancock RE. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res 15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benkert B, Quack N, Schreiber K, Jaensch L, Jahn D, Schobert M. 2008. Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiology 154:3053–3060. doi: 10.1099/mic.0.2008/018929-0. [DOI] [PubMed] [Google Scholar]
- 51.Schweizer HP, Klassen T, Hoang T. 1996. Improved methods for gene analysis and expression in Pseudomonas spp., p 229–237. In Nakazawa T, Furukawa K, Haas D, Silver S (ed), Molecular biology of pseudomonads. ASM Press, Washington, DC. [Google Scholar]
- 52.Engler C, Gruetzner R, Kandzia R, Marillonnet S. 2009. Golden Gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wessel D, Flugge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 54.Lambert JP, Ivosev G, Couzens AL, Larsen B, Taipale M, Lin ZY, Zhong Q, Lindquist S, Vidal M, Aebersold R, Pawson T, Bonner R, Tate S, Gingras AC. 2013. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat Methods 10:1239–1245. doi: 10.1038/nmeth.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Düvel J, Bertinetti D, Moller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser HG, Jansch L, Herberg FW, Haussler S. 2012. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods 88:229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 56.Howden AJ, Geoghegan V, Katsch K, Efstathiou G, Bhushan B, Boutureira O, Thomas B, Trudgian DC, Kessler BM, Dieterich DC. 2013. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods 10:343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaake RM, Wang X, Huang L. 2010. Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry. Mol Cell Proteomics 9:1650–1665. doi: 10.1074/mcp.R110.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zumft WG, Braun C, Cuypers H. 1994. Nitric oxide reductase from Pseudomonas stutzeri. Primary structure and gene organization of a novel bacterial cytochrome bc complex. Eur J Biochem 219:481–490. [DOI] [PubMed] [Google Scholar]
- 59.Santini S, Bizzarri AR, Yamada T, Beattie CW, Cannistraro S. 2014. Binding of azurin to cytochrome c 551 as investigated by surface plasmon resonance and fluorescence. J Mol Recognit 27:124–130. doi: 10.1002/jmr.2346. [DOI] [PubMed] [Google Scholar]
- 60.Cuypers H, Viebrock-Sambale A, Zumft WG. 1992. NosR, a membrane-bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri. J Bacteriol 174:5332–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Honisch U, Zumft WG. 2003. Operon structure and regulation of the nos gene region of Pseudomonas stutzeri, encoding an ABC-type ATPase for maturation of nitrous oxide reductase. J Bacteriol 185:1895–1902. doi: 10.1128/JB.185.6.1895-1902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.