ABSTRACT
The stringent response, mediated by the (p)ppGpp synthetase RelA and the RNA polymerase-binding protein DksA, is triggered by limiting nutrient conditions. For some bacteria, it is involved in regulation of virulence. We investigated the role of two DksA-like proteins from the Gram-negative nitrogen-fixing symbiont Sinorhizobium meliloti in free-living culture and in interaction with its host plant Medicago sativa. The two paralogs, encoded by the genes SMc00469 and SMc00049, differ in the constitution of two major domains required for function in canonical DksA: the DXXDXA motif at the tip of a coiled-coil domain and a zinc finger domain. Using mutant analyses of single, double, and triple deletions for SMc00469 (designated dksA), SMc00049, and relA, we found that the ΔdksA mutant but not the ΔSMc00049 mutant showed impaired growth on minimal medium, reduced nodulation on the host plant, and lower nitrogen fixation activity in early nodules, while its nod gene expression was normal. The ΔrelA mutant showed severe pleiotropic phenotypes under all conditions tested. Only S. meliloti dksA complemented the metabolic defects of an Escherichia coli dksA mutant. Modifications of the DXXDXA motif in SMc00049 failed to establish DksA function. Our results imply a role for transcriptional regulator DksA in the S. meliloti-M. sativa symbiosis.
IMPORTANCE The stringent response is a bacterial transcription regulation process triggered upon nutritional stress. Sinorhizobium meliloti, a soil bacterium establishing agriculturally important root nodule symbioses with legume plants, undergoes constant molecular adjustment during host interaction. Analyzing the components of the stringent response in this alphaproteobacterium helps understand molecular control regarding the development of plant interaction. Using mutant analyses, we describe how the lack of DksA influences symbiosis with Medicago sativa and show that a second paralogous S. meliloti protein cannot substitute for this missing function. This work contributes to the field by showing the similarities and differences of S. meliloti DksA-like proteins to orthologs from other species, adding information to the diversity of the stringent response regulatory system.
INTRODUCTION
Bacteria employ the stringent response as a mechanism to adjust global gene expression to adverse nutrient conditions. For example, when Escherichia coli encounters amino acid starvation, the ribosome-bound protein RelA (1, 2) recognizes uncharged tRNA molecules and synthesizes guanosine tetraphosphate and guanosine pentaphosphate (referred to here as ppGpp) from GTP and ATP (3). The alarmone ppGpp and the regulatory protein DksA subsequently bind RNA polymerase (RNAP) and alter the kinetic properties of promoter/RNAP complexes (4). In particular, ppGpp and DksA reduce the lifetime of promoter/RNAP complexes by inhibiting the transition from closed to intermediate complex formation on promoters depending on the primary sigma factor RpoD (σ70) (4, 5). In addition, RNAP dissociation can allow alternative sigma factors to bind RNAP, leading to the regulation of distinct promoter sets important for adjustment to new environmental conditions (3, 6). In some bacteria, ppGpp executes the stringent response in a different way, not involving RNAP binding (7, 8).
The overall processes of the stringent response are similar in a wide variety of bacteria, but structural and functional variations occur. Some bacterial genomes carry a single spoT/relA gene that encodes a bifunctional protein comprising a ppGpp synthetase and a hydrolase domain (9–11). Others possess more than one DksA-like protein; in most cases, it is unknown which protein participates in the stringent response. The structures, functions, and interactions of RelA, its homolog SpoT, and the DksA protein have been studied most extensively in E. coli. SpoT is a ppGpp-hydrolyzing enzyme but also has some synthetase activity (12). DksA is a small protein of 151 amino acids and consists of three major domains: a globular domain, a coiled-coil (CC) domain, and a C-terminal α-helix (13, 14). The CC domain, which protrudes into the secondary channel of RNAP (14), consists of two α-helices and contains a conserved DXXDXA motif at its tip, the linker region between the two helices. The DksA globular domain has been shown to bind to the rim helices located in the β′ subunit of RNAP (15). It is formed by the C and N termini of DksA and harbors a CXXCX17CXXC zinc finger motif (13). In addition, recent work in E. coli identified the RNAP β subunit sequence insertion 1 as a binding site for the DksA C-terminal helix (16). Structural as well as amino acid substitution analyses of DksA proteins from Pseudomonas aeruginosa, Rhodobacter sphaeroides, and E. coli indicate that the conserved amino acid motif DXXDXA at the tip of the CC domain is critical for DksA function as a transcriptional regulator (17–19). The globular domain with conserved cysteines seems to play an important role in P. aeruginosa DksA2 (17) and might be involved in proper folding of E. coli DksA (18).
DksA proteins appear in diverse regulatory schemes. In some bacterial pathogens, DksA proteins are critical for bacterial virulence (20–24). It was recently proposed that Salmonella DksA may act as a sensor for reactive oxygen and nitrogen species due to its redox-active thiols (25). Of the two DksA-like proteins present in Rhodobacter sphaeroides, only one was shown to regulate the stringent response and to be necessary for photosynthetic growth and utilization of exogenous amino acids (19). The function of the other DksA-like protein was not identified (19). Of the five DksA paralogs from P. aeruginosa, two (DksAPa and DksA2Pa) were investigated, and both were shown to be involved in stringent response processes (26, 27).
The genome of Sinorhizobium meliloti, a Gram-negative alphaproteobacterium, also encodes two DksA paralogs (SMc00469 and SMc00049) (28). S. meliloti is a model organism for studying microbe-plant interactions. Upon plant signal recognition, it establishes a symbiotic interaction with its host Medicago sativa (alfalfa) by invading the roots and converting molecular nitrogen into plant-usable ammonia. Nitrogen fixation takes place in specialized root nodules; there, the bacteria differentiate into bacteroids in a distinct symbiosome compartment within host cells and fix nitrogen usable by the plant (29, 30). In the transition from free-living growth to symbiotic life within the plant cells, bacteria face changing environmental inputs, and all stages of symbiosis require adaptations of gene expression. RelA is essential for nodule formation (31), and global transcription profiling analyses performed on Medicago truncatula and S. meliloti nodules suggest the involvement of the stringent response in bacteroid differentiation as well (32). Expression studies with a relA and a dksA (SMc00469) mutant in cultures exposed to a nitrogen or carbon downshift showed that most of the relA-dependent transcriptional changes are also dksA dependent (33). Functional studies of S. meliloti dksA or the paralog SMc00049 are lacking.
We took a genetic approach to investigate the role of stringent response-related genes in S. meliloti free-living culture and in symbiosis with M. sativa. This work describes the function of dksA in a symbiotic Gram-negative alphaproteobacterium and shows for the first time its involvement in effective symbiosis with its host plant.
MATERIALS AND METHODS
Bacterial growth conditions and assays.
S. meliloti and E. coli strains were grown in Luria broth (LB) (34) medium at 30°C and 37°C, respectively. Solid medium contained 1.5% agar. Appropriate antibiotics were added at the following final concentrations: 500 μg/ml streptomycin, 10 μg/ml tetracycline, 25 μg/ml gentamicin (5 μg/ml for E. coli), and 50 μg/ml ampicillin. Minimal medium (M9) for S. meliloti contained M9 salts (35), 0.5 mg/liter biotin, and 1 mM magnesium sulfate; M9 for E. coli contained M9 salts, 2 mM magnesium sulfate, and 0.1 mM calcium chloride. Where indicated, filter-sterilized solutions of carbon sources were added to sterile medium. For spotting assays, bacterial cultures were grown in LB, harvested in mid-exponential phase, washed in 10 mM magnesium sulfate, and adjusted to an optical density at 600 nm (OD600) of 0.1. Serial dilutions of 10−1, 10−2, and 10−3 were made, and 3 μl of each was spotted on agar plates from left to right. Colony growth was monitored after 1 to 3 days of incubation. For growth curves, precultures of S. meliloti strains were grown in liquid LB medium containing appropriate antibiotics. Two sets of cells were harvested from each culture; one set was resuspended in LB and the other set in M9 medium with antibiotics. Cultures were adjusted to an OD600 of 0.05, and 200 μl of each was transferred to a 96-well plate. The plate was sealed with a sealing tape (Breathe-Easy; Diversified Biotech) to decrease evaporation. Growth was monitored using a PerkinElmer Victor ×3 plate reader. A600 values were measured every 10 min at 30°C, and continuous double orbital shaking at normal speed was applied between reads.
Plant growth conditions and assays.
Medicago sativa seed sterilization and germination as well as preparation of bacterial strains for inoculation were done as described previously (36). For nodulation assays, root tips of plants grown on buffered nodulation medium (BNM) agar plates (37) were spot inoculated 2 to 4 days after germination with 1 μl of bacterial suspension. The nodules were usually counted (number per plant) at 7, 10, 14, and 21 days postinoculation. Nitrogen fixation in root nodules was determined by acetylene reduction assay (38). Plants grown on BNM agar plates were transferred to 30-ml glass tubes on strips of wet Whatman paper 10 or 21 days postinoculation with S. meliloti strains. Each tube contained two plants. After injection of 1 ml acetylene and 2 to 5 h of incubation, production of ethylene as a measure of nitrogenase activity was monitored on a Shimadzu GC-8A1F gas chromatograph with a Porapak N column and flame ionization detector.
Deletion strain construction.
Strains and plasmids used for this study are listed in Table S1 in the supplemental material. Primer sequences are listed in Table S2 in the supplemental material. We used S. meliloti strain CL150 (39) as wild-type (WT) background for all experiments, because a point mutation in an anti-sigma factor is repaired in this strain compared to Rm1021 (28), avoiding sigma factor competition. Markerless single-deletion strains for SMc00469 (dksA), SMc00049, and SMc02659 (relA) were made by homologous recombination. An upstream sequence including the gene's start ATG and a downstream sequence including the stop codon were amplified, fused, and ligated into a modified version of the suicide vector pJQ200SK (40). The lengths of the upstream and downstream sequences were for dksA 657 bp and 831 bp, for SMc00049 501 bp and 576 bp, and for relA 703 bp and 528 bp. The resulting plasmids pKW201, pKW202, and pKW203 were transferred into CL150 by conjugation with the helper strain B001 (41), and single crossover events were identified by selection for plasmid-derived gentamicin resistance. Double crossover events that either restored the WT situation or generated the genomic deletion were found by selection for sucrose resistance and screening for gentamicin sensitivity. Deletion strains were identified by PCR and streaked for single colonies three times on LB medium supplemented with streptomycin. Double and triple mutants were constructed by transduction of the individual mutants with N3 phage lysates (42) of the strains carrying single crossovers of the other mutants. Selection for double crossovers and confirmation of the deletions were carried out as just described.
Construction of complementation plasmids and strains.
For the complementation of the E. coli dksA mutant, the open reading frames (ORFs) including the stop codon plus the predicted ribosome-binding site of dksASm or SMc00049 were cloned into pINIIIA1 (43) via XbaI and HindIII. The E. coli dksA mutant strain RLG6348 (5) and the corresponding WT strain RLG4996 (5) were transformed with the resulting plasmids pKW222 and pKW223, as well as with the empty vector and the positive control, pINIIIA1 harboring E. coli dksA (pRLG6333, recovered from strain RLG6333 [5]). For the complementation of the S. meliloti dksA mutant, the ORFs of dksASm, SMc00049, and dksAEc including the stop codon and the ribosome-binding site were cloned into pRF771 (31) via XbaI and BamHI, resulting in plasmids pKW224, pKW225, and pKW226, respectively. dksASm and SMc00049 were amplified by PCR; dksAEc was excised from pRLG6333. Genes in those constructs are expressed from the strong Salmonella enterica serovar Typhimurium tryptophan promoter (trpp). To generate plasmids for gene expression driven by their native promoters, ORFs plus promoter sequences were cloned into promoterless pDW76 (31) via XbaI and BamHI. The gene-specific promoter sequences were derived from published transcription start site (TSS) data (39, 44). For dksA, a 195-bp region upstream of the dksA translation start was chosen. For SMc00049, no TSS directly upstream could be determined. However, there is a TSS located 197 bp upstream of the start ATG of the upstream gene SMc00048 (44). SMc00049 and SMc00048 are only 61 bp apart, making it possible that the two genes are transcribed as an operon from the same promoter. Therefore, two different putative promoter regions were chosen. One is 235 bp upstream of the SMc00049 start ATG; the second is a 794-bp sequence that includes the complete SMc00048 ORF, plus another 297 bp upstream of it, so that it includes the putative SMc00048 TSS. The corresponding plasmids pKW227, pKW228, and pKW229 were transferred into S. meliloti ΔdksA via conjugation, and transconjugants were identified by selection for tetracycline resistance. Amino acid substitutions and chimeras of DksA and SMc00049 were generated by site-directed mutagenesis, using suitable primers and overlap extension PCR (45), and cloned into pRF771 via XbaI and BamHI and into pINIIIA1 via XbaI and HindIII.
RNA isolation and qPCR.
S. meliloti WT strain CL150 was grown in liquid LB or M9 sucrose (15 mM) medium supplemented with streptomycin and harvested in mid-exponential phase. CL150-induced nodules on M. sativa plants were harvested at 10 and 21 days postinoculation. Sample handling, RNA isolation, and cDNA synthesis procedures were carried out as described previously (32). Quantitative PCR (qPCR) was performed using the DyNAmo Flash SYBR green qPCR kit (Thermo Scientific, Rockford, IL) and a CFX Connect real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). Data were analyzed using the ΔΔCT method (where CT is threshold cycle) (46). Expression was normalized to pdhB, and LB medium was chosen as the calibrator condition. The reference gene pdhB has been used previously for similar comparisons (32).
Luteolin treatment and β-galactosidase assay.
Bacterial cells were grown overnight in liquid medium and diluted to an OD of 0.2 in the same medium. At an OD of 0.4 to 0.5, cultures of each bacterial strain were split. Luteolin (3 μM; Calbiochem, La Jolla, CA) was added to one batch, an equal volume of dimethylformamide, the luteolin solvent, was added to the other batch, and cultures were incubated with shaking at 30°C for 6 h. Cells were harvested and permeabilized as described previously (47). The β-galactosidase substrate 4-methylumbelliferryl β-d-galactopyranoside (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 1.25 mg/ml; after 2 h of incubation at 37°C in the dark, fluorescent product was measured with a plate reader (excitation, 365 nm; emission, 455 nm).
RESULTS
S. meliloti SMc00469 (DksA) and SMc00049 possess distinct protein motifs.
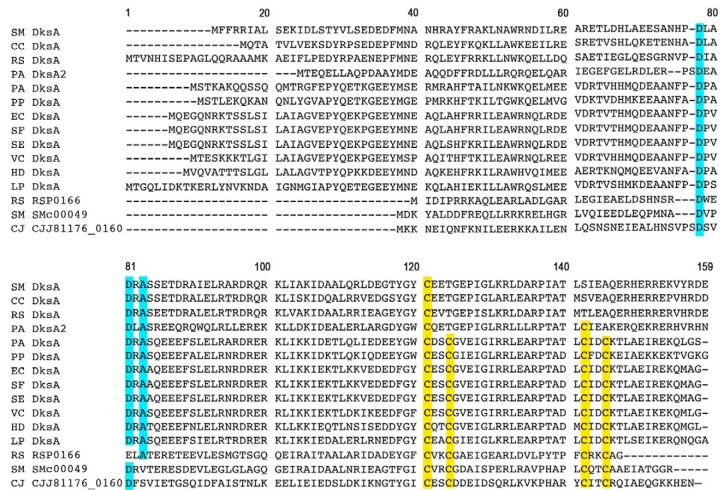
The amino acid sequences of described DksA homologs from different bacteria were aligned to compare structural/functional motifs (Fig. 1). The two S. meliloti paralogs, designated DksA and SMc00049, are 23.3% identical to each other. S. meliloti DksA (DksASm) possesses the complete DXXDXA motif and only one conserved cysteine within the zinc finger, whereas SMc00049 contains a DXXDXV sequence and all four conserved zinc finger cysteines. In addition, SMc00049 is missing the first 24 amino acids found in DksASm. The sole E. coli DksA (DksAEc) homolog has both the complete DXXDXA motif and the four conserved zinc finger cysteines and shares higher overall sequence identity with DksASm (38.6%) than with SMc00049 (24.1%). The two DksA-like proteins in the alphaproteobacterium R. sphaeroides (DksARs and RSP0166) are very similar to the S. meliloti orthologs. DksARs shares 58.2% identity with DksASm, and SMc00049 and RSP0166 are 42.9% identical. As in S. meliloti, the two paralogs of R. sphaeroides share very low sequence identity (21.5%).
FIG 1.
Alignment of various DksA homologs. Different bacterial genomes harbor one or more DksA homologs, which differ in the presence or absence of a complete DXXDXA motif (highlighted in blue), or in the presence of one, two, or four conserved cysteines within the zinc finger motif (yellow). Amino acid sequences (from NCBI, confirmed by comparison to original papers) were aligned using Geneious version 8.0.3 (52). The order of protein sequences represents overall protein similarity. CC, Caulobacter crescentus; CJ, Campylobacter jejuni; EC, Escherichia coli; HD, Haemophilus ducreyi; LP, Legionella pneumophila; PA, Pseudomonas aeruginosa; PP, Pseudomonas putida; RS, Rhodobacter sphaeroides; SE, Salmonella enterica; SF, Shigella flexneri; SM, Sinorhizobium meliloti; VC, Vibrio cholerae.
A second alignment includes more DksA orthologs from other rhizobia (see Fig. S1 in the supplemental material). While most of them possess one homolog, Rhizobium sp. NGR234, Rhizobium etli, and Rhizobium leguminosarum genomes each encode two proteins that are much like the ones from S. meliloti: one of them is highly similar to DksASm (91.8 to 98.6%), while the other one is more similar to SMc00049 (38.2 to 83.3%) than to DksASm. Mesorhizobium loti and Bradyrhizobium japonicum, the microsymbionts of Lotus japonicus and soybean (Glycine max), respectively, possess only one DksA protein, carrying a complete DXXDXA motif and one conserved cysteine. A phylogenetic tree of these DksA-like and SMc00049-like proteins that we constructed (see Fig. S2 in the supplemental material) shows that the latter form a separate branch and thus are most likely distinct from DksAs. The two S. meliloti proteins differ both in sequence length and in the makeup of their two main structural motifs; we aimed to discover if they both possess DksA activity and to investigate their respective roles in S. meliloti.
S. meliloti dksA complements an E. coli dksA mutant.
To assess whether DksASm and SMc00049 are functional DksA proteins, we tested for complementation of an E. coli dksA mutant that is growth deficient on minimal medium without amino acids (48) and is a convenient host strain to test the activity of heterologous DksA proteins (19, 26). The metabolic defect was complemented when dksAEc or dksASm was expressed from a plasmid, controlled by the strong promoter of the outer membrane protein gene (lpp) (Fig. 2). In contrast, SMc00049 was unable to restore growth on minimal medium. All strains grew equally well on rich medium (Fig. 2). To ensure that expression of the S. meliloti genes, in particular SMc00049, had no negative effects on E. coli growth on minimal medium in general, the same plasmids were expressed in the wild-type (WT) background. All the corresponding strains grew equally well on rich and minimal media (Fig. 2). Our complementation assays show that S. meliloti DksA likely functions as a transcriptional regulator, similar to the native protein in E. coli.
FIG 2.
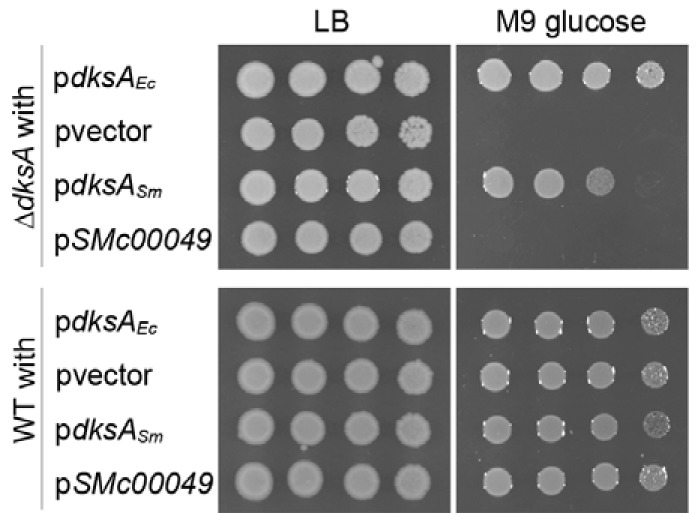
S. meliloti dksA, but not SMc00049, complements the metabolic defect of an E. coli dksA mutant. Growth of an E. coli ΔdksA strain and the corresponding WT strain carrying either an empty vector (pvector) or a plasmid expressing E. coli dksA (pdksAEc), S. meliloti dksA (pdksASm), or SMc00049 (pSMc00049) on rich (LB) and on minimal medium with 0.4% glucose (M9).
Growth of the S. meliloti dksA mutant is impaired on minimal medium.
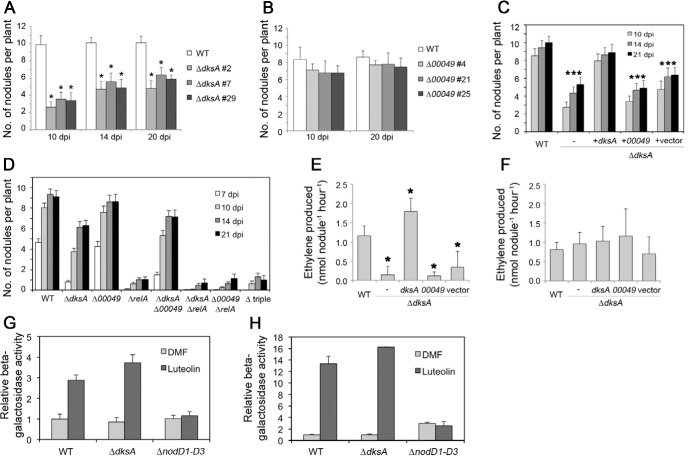
To investigate the importance of the dksA-like genes and the stringent response in general in S. meliloti, markerless deletion mutants for dksA, SMc00049, and relA were constructed in the CL150 WT background (39). The effect of the mutations on bacterial behavior in culture was tested by monitoring growth on solid M9 minimal medium containing ammonium chloride as the sole nitrogen source, supplemented with 15 mM sucrose. ΔdksA, ΔSMc00049, and ΔrelA strains showed growth similar to that of the WT on the control medium (LB), with marginally lower cell numbers for ΔdksA and ΔrelA mutants (Fig. 3A). Growth of the relA mutant was negligible on minimal medium, similar to the previously described behavior of a partial deletion mutant (31). Growth of the ΔSMc00049 strain was indistinguishable from that of the WT under the conditions tested, whereas growth of the ΔdksA mutant was impaired on minimal medium (Fig. 3A). We observed similar results on minimal medium with glucose, arabinose, succinate, or glutamate (data not shown), with the impairment most obvious on sucrose.
FIG 3.
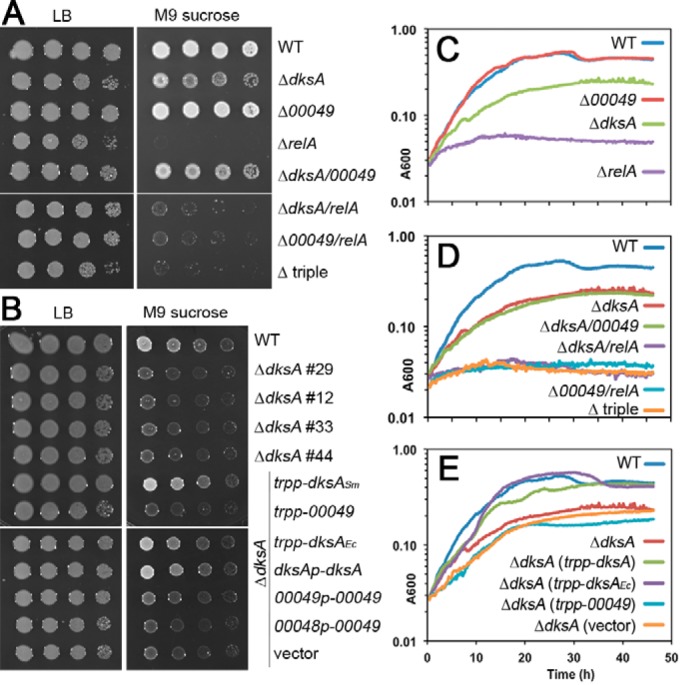
Metabolic phenotypes of S. meliloti dksA, SMc00049, and relA deletion mutants. (A) Growth of ΔdksA, ΔSMc00049, and ΔrelA mutants and corresponding double and triple mutants on rich medium (LB) and on solid M9 minimal medium supplemented with 15 mM sucrose; images were taken after 3 days of incubation. (B) Growth of four independent dksA mutant strains and ΔdksA strains carrying complementation plasmids expressing dksASm, SMc00049, or dksAEc either from the strong tryptophan promoter (trpp) or their native promoters (see Materials and Methods). (C, D, and E) Growth curves of WT, dksA, SMc00049 and relA single, double, and triple mutants and ΔdksA strains with complementation plasmids in M9 sucrose medium. For each strain, a representative graph of three independent biological experiments is shown.
Growth kinetics in liquid minimal medium were consistent with behaviors on solid medium: growth of the ΔrelA mutant leveled off quickly, barely displaying any exponential growth; the ΔdksA mutant growth was slower than WT growth and never reached the same saturation level; the ΔSMc00049 mutant growth was indistinguishable from that of the WT. The doubling times were 2.81 ± 0.07 h for the WT, 3.02 ± 0.13 for the ΔSMc00049 mutant, and 4.43 ± 0.51 h for the ΔdksA mutant in minimal medium with sucrose (Fig. 3C). To analyze possible additive or compensatory effects of the dksA, SMc00049, and relA mutations, we generated corresponding double and triple mutants. In growth tests on solid and in liquid minimal medium, the mutants behaved like the corresponding single mutant with the strongest phenotype (Fig. 3A and D).
We used complementation tests to confirm that the ΔdksA phenotypes are in fact caused by the missing DksA activity in S. meliloti. dksASm, SMc00049, or dksAEc was expressed from plasmids either under the control of a strong promoter (trpp; comprising the promoter and leader sequence of the Salmonella Typhimurium tryptophan operon) or driven by the genes' own promoters (except for dksAEc; see Materials and Methods for details on promoter sequence choice). Both dksASm and dksAEc restored colony growth in the ΔdksA background to WT levels on minimal medium, proving that the mutant phenotype was indeed a consequence of loss of DksA activity (Fig. 3B and E). In contrast, introducing SMc00049 into the mutant did not result in better growth on minimal medium, no matter which promoter was used. In addition to the complementation strains, four independent isolates of the dksA mutant derived from four different single crossover events were analyzed; they all showed the same degree of growth impairment on minimal medium but grew like the WT on rich medium (Fig. 3B). The similar behaviors of these independent strains suggest that the detected phenotypes were due to the dksA deletion and not a secondary mutation. These results show that in amino acid-free minimal medium, dksA, but not SMc00049, is important for maximum growth efficiency when sucrose is the sole carbon source. relA is indispensable for proper bacterial growth in this medium.
It is noteworthy that in the context of free-living behavior we also examined motility and high-temperature tolerance and found that both ΔdksA and ΔSMc00049 mutants showed WT-like motility (data not shown), but the ΔdksA mutant showed reduced growth in liquid medium at 37°C, whereas the ΔSMc00049 mutant grew like WT (see Fig. S4A in the supplemental material). Consistent with its apparent requirement under free-living conditions under nutrient stress, we found that dksA expression was induced 3-fold in minimal medium compared to expression in rich medium (see Fig. S5 in the supplemental material).
Nodulation efficiency and nitrogenase activity are decreased in plants infected with the ΔdksA mutant.
We assessed the performance of the dksA and SMc00049 mutants in symbiosis by observing their ability to form nodules on M. sativa host plants. The nodulation rate was significantly reduced on plants infected with ΔdksA strains (Fig. 4A) compared to WT S. meliloti-inoculated plants. Over 3 weeks, the number of ΔdksA nodules per plant increased gradually but remained significantly lower than nodulation with WT bacteria. In general, the nodule number does not increase further after 21 days postinoculation (dpi) in these assays. Several independent assays (data not shown) revealed that the nodulation rate induced by the ΔdksA mutant compared to the WT strain could be as low as 3.4% at 7 dpi and up to 70.3% at 21 dpi. dksA transcript levels in WT nodules at either 10 or 21 dpi were not induced compared to levels in rich medium (see Fig. S5 in the supplemental material). While mature nodules formed by ΔdksA resembled WT nodules, their development appeared to be delayed. At 10 dpi, for example, WT nodules had already turned pink, suggesting active nitrogen fixation, whereas all ΔdksA nodules were still white or only pale pink. To determine whether those nodules fix nitrogen efficiently, the acetylene reduction assay (ARA; conversion of acetylene to ethylene) was used as a proxy for nitrogenase activity. At 11 dpi, low levels of ethylene were formed by ΔdksA-infected plants, suggesting very low nitrogenase activity (Fig. 4E), whereas WT nodules already showed much higher levels of nitrogen fixation. This phenotype could be complemented by expression of trpp-dksA from a plasmid but not by introducing trpp-SMc00049 or the vector control. In contrast to what was seen at 11 dpi, nitrogenase activity per nodule did not significantly differ between WT, ΔdksA, ΔdksAp(trpp-dksA), or ΔdksAp(trpp-SMc00049) nodules at 21 dpi (Fig. 4F).
FIG 4.
Symbiotic phenotypes of S. meliloti dksA, SMc00049, and relA deletion mutants and nodC expression in ΔdksA. (A to D) Nodulation on M. sativa plants inoculated with the indicated S. meliloti strains: three independent ΔdksA (A) and ΔSMc00049 strains (B); ΔdksA strains expressing the indicated genes from a plasmid (C); single, double, and triple mutants of dksA, SMc00049, and relA (D). Error bars indicate standard errors (n = 10 to 20) from one representative experiment each (A to C) or from a pool of 2 to 4 experiments (D); dpi, days postinoculation. (E and F) Nitrogenase activity, assessed by the rate of acetylene reduction. Activity per nodule in plants inoculated with S. meliloti WT, ΔdksA, or ΔdksA expressing SMc00049 or the empty vector at 11 dpi (E) and at 21 dpi (F). Error bars indicate standard deviations (n = 10). (G and H) Relative β-galactosidase activity of a translational nodC-lacZ fusion in WT, ΔdksA, and ΔnodD1-D3 (deletion of nodD1, nodD2, and nodD3) backgrounds in LB medium (G) and M9 sucrose medium (H). nodC expression was induced in the cells by a 6-hour incubation with luteolin. Asterisks in panels A, C, and E indicate a significant difference from the WT according to Student's t test (P ≤ 0.05).
No significant difference in symbiotic behavior was observed between the ΔSMc00049 mutant and WT strains (Fig. 4B). The relA single mutant induced very few nodules: ΔrelA nodulation rates were 3.9 to 7.4% between 10 dpi and 21 dpi compared to the WT strain (Fig. 4D). The few nodules formed by ΔrelA strains were white, i.e., nonfixing, as also reported previously for a partial deletion mutant of S. meliloti relA (31).
Expressing dksASm or dksAEc from trpp in the dksA mutant restored WT-like nodulation (Fig. 4C; see also Fig. S3 in the supplemental material), whereas trpp-SMc00049 or the empty vector induced few nodules compared to WT, similar to the ΔdksA strain (Fig. 4C). We also tested if dksA is able to complement the relA mutant by constitutive expression from a plasmid. Indeed, dksA mostly restored growth on minimal medium (Fig. 5A) and increased nodulation to WT levels (Fig. 5B), indicating that DksA acts in the same pathway as RelA. In this context, we also investigated if the ΔdksA mutant displays succinoglycan overproduction as was described for a relA mutant (31), but a qualitative analysis of fluorescence on calcofluor white plates suggested no elevated succinoglycan levels in the dksA mutant (see Fig. S4 in the supplemental material).
FIG 5.
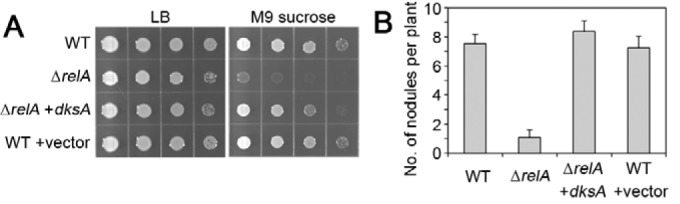
Constitutive expression of dksA recovers metabolic and symbiotic phenotypes in the ΔrelA strain. (A) Growth of WT and ΔrelA strains, the ΔrelA strain expressing dksA from the tryptophan promoter, and the WT strain carrying the empty vector on rich LB and M9 sucrose medium. (B) Nodulation on M. sativa plants at 21 days after inoculation with the indicated strains. Error bars indicate standard errors (n = 20; one experiment).
The double and triple mutants typically displayed nodulation rates of the corresponding single mutant with the strongest phenotype (Fig. 4D). Thus, the ΔdksA ΔSMc00049 mutant induced a nodule number per plant similar to that seen with the ΔdksA strain (Fig. 4D). However, we noted that the nodule number was intermediate between those of ΔdksA and ΔSMc00049 mutants with a significant difference from both at 10 dpi and a nonsignificant difference at 14 and 21 dpi.
Because the ΔdksA mutant displayed a developmental delay during early symbiosis, we asked whether Nod factor biosynthesis might be altered. As a proxy, we measured nod gene expression via a nodABC-lacZ reporter in strains treated with the plant flavonoid luteolin, which induces nod gene expression. The effect of a dksA mutation was not significant: expression increased 2.9 times and 4.4 times in WT and S. meliloti ΔdksA cells, respectively, in rich medium (Fig. 4G) and 13.4 times and 16.1 times, respectively, in minimal medium (Fig. 4H). The ΔnodD1D2D3 strain was used as a negative control, since it is lacking all three paralogs of the transcription factor NodD responsible for flavonoid-induced nod expression. The symbiosis effect of a dksA mutation is not due to a lack of nod gene induction.
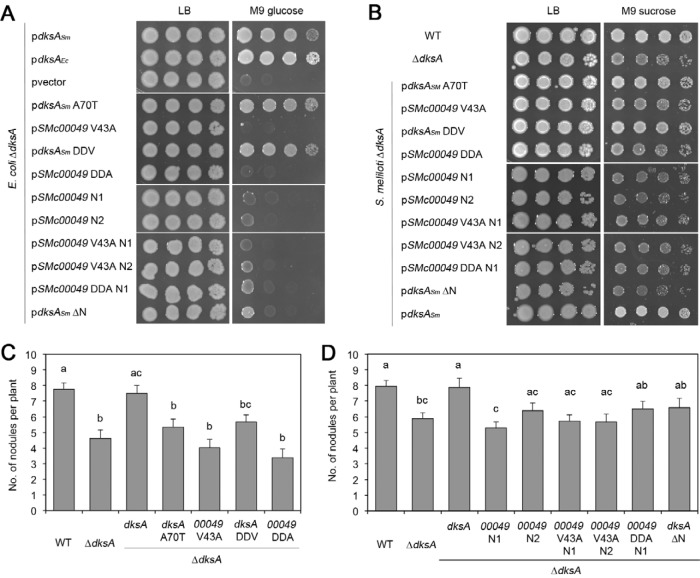
Modifications of the DXXDXA motif and DksA/SMc00049 chimeras fail to establish function in SMc00049.
Previous work showed that the aspartates as well as the alanine in the DXXDXA motif are crucial for activity (14, 15, 17, 18). We constructed variants of DksA and SMc00049, modifying the motif, to investigate if we could abolish or establish DksA activity. In addition, since the two proteins differ in the lengths of their N termini (Fig. 1), we made chimeras by fusing the DksA N terminus to SMc00049 and a DksAΔN variant with the first 25 amino acids deleted (protein starts at M26 of native sequence). The native DksA motif is DLADRA (Fig. 1); the mutants are A70T (DLADRT) and DDV (DVPDRV). The latter variant mimics the native SMc00049 sequence. The SMc00049 mutants are V43A (DVPDRA) and DDA (DLADRA), with the latter one mimicking the native DksA sequence. For the chimeras, two different N-terminal DksA sequences were chosen to be attached to SMc00049: (i) amino acids M1-F25 (N1) were fused to SMc00049 D2, basically adding the first 24 amino acids that are not present in SMc00049; (ii) M1-Y33 (N2) were fused to SMc00049 F9, to add a slightly longer sequence of DksA, covering part of the N-terminal coiled-coil helix 1 (see results from Lennon et al. [19]); SMc00049 F9 is a conserved residue in this helix. These N termini were also attached in the same manner to the SMc00049 amino acid substitution mutants as described above.
None of the amino acid substitutions or chimeras greatly affected the functionality of DksA or rendered SMc00049 functional either in E. coli (Fig. 6A) or in S. meliloti grown on minimal medium (Fig. 6B). However, deleting the N terminus of DksA led to loss of function (Fig. 6A and B). The results for symbiotic interaction were overall similar: during nodulation of alfalfa plants, the SMc00049 amino acid substitution mutants likewise did not rescue the ΔdksA phenotype; the DksA variants A70T and DDV induced higher nodule numbers than ΔdksA alone; however, this difference was not statistically significant (Fig. 6C); chimera SMc00049+N1 did not restore WT nodulation, and the other chimeras and DksAΔN showed nodulation rates between WT and ΔdksA mutant strains (Fig. 6D), suggesting incomplete complementation.
FIG 6.
Modifications of conserved amino acids and chimeras fail to render SMc00049 functional. (A) The growth of E. coli ΔdksA expressing the indicated genes from a plasmid was monitored on rich (LB) or minimal medium with 0.4% glucose (M9). For plasmid construction, see Materials and Methods. (B) Complementation of S. meliloti ΔdksA. Growth of the indicated spotted strains on rich (LB) or minimal medium (M9 supplemented with 15 mM sucrose). (C and D) Nodulation induced by DksA and SMc00049 amino acid substitution mutants (C) and chimeras (D). Bars show the average numbers of nodules per M. sativa plant 21 days after inoculation with S. meliloti WT, ΔdksA, or ΔdksA expressing the indicated genes from a plasmid. Error bars indicate standard errors (n = 37 to 144, from pools of 2 to 4 experiments). The same letters above bars indicate means not significantly different; different letters indicate significant difference with a P value of ≤0.05 according to one-way analysis of variance (ANOVA) and Tukey-Kramer's test. DDV, SMc00049's DVPDRV motif; DDA, DksA's DLADRA motif; N1 and N2, different DksA N termini (see the text for details).
DISCUSSION
We investigated the involvement of central players of the stringent response under free-living and symbiotic conditions in the alphaproteobacterium Sinorhizobium meliloti. A ΔrelA strain, which cannot produce ppGpp, was severely defective in growth on medium without amino acids (Fig. 3A). This mutant likely is unable to activate biosynthetic pathways and the stress response effectively, so that growth is impaired in the absence of complex nutrients. Deleting ΔSMc00049 did not affect S. meliloti growth, and the gene was not able to substitute for the lack of dksA in E. coli, suggesting a distinct function or lack of function. In line with our observation of poor growth of ΔdksA in minimal medium, an insertion mutant of P. aeruginosa dksA also showed reduced growth on M9 glucose medium (49), as does a dksA mutant from Vibrio cholerae (21), while an E. coli dksA mutant completely lost the ability to grow on minimal medium (48). However, similar deletion mutants of S. meliloti dksA and relA have been described as growing well on minimal medium (33). The contrasting results of those experiments may arise from the use of different S. meliloti genetic backgrounds, different minimal media, or both.
S. meliloti dksA mutants have not previously been described in a symbiosis context. We found that while the ΔSMc00049 mutant shows WT-like behavior (Fig. 4B), the nodulation rate of ΔdksA strains on M. sativa is significantly reduced compared to that of the WT (Fig. 4A) and that nodule development is delayed. This delay might start at an early stage of nodulation or during the infection process. It is possible that the growth impairment seen under free-living conditions is also the cause of the symbiotic defect: reduced growth in minimal medium without amino acids might resemble a stress that is similar to one encountered in the rhizosphere, limiting bacterial growth and reducing root colonization and leading to a less efficient invasion process. Our assay, using pure cultures (no competition) and a sufficient inoculum, might obscure such effects from being observed. Alternatively, DksA may have a direct role in the regulation of certain gene sets that are directly involved in early invasion processes, e.g., genes relevant for plant signal reception, bacterial signal (Nod factor) production, root hair attachment, or infection thread formation. Our reporter gene assay showed that in the ΔdksA mutant at least nod gene induction by luteolin is normal (Fig. 4G and H), both under rich and minimal medium conditions, implying that Nod factor is produced. It remains possible that other aspects of early symbiosis are affected. In any case, the dksA mutation would only delay these processes rather than completely block them, since the ΔdksA mutant eventually forms nodules on the host plant. Perhaps another regulatory protein partially compensates for the loss of DksA, or perhaps the DksA-dependent genes affected in the mutant are necessary for maximum symbiotic efficiency only but not essential in general. These hypotheses provide a basis for further experimentation. To date, there are no reports on characterizations of DksA proteins from other rhizobia, most of which possess either one or two orthologs (see Fig. S1 and S2 in the supplemental material).
Given the behavior of ΔrelA, ΔdksA, and ΔSMc00049 double and triple mutants, it is likely that the already strong defects of the ΔrelA mutant mask any further impairments resulting from the additional deletion of dksA. Transcriptional profiling in E. coli suggests that DksA can work synergistically with, independently from, or antagonistically to ppGpp (50). While the phenotypes of relA and dksA single mutants in E. coli show considerable overlap, overexpression of dksA in the ΔrelA mutant can compensate for some of the relA defects, including amino acid auxotrophy, which indicates that DksA might act independently from ppGpp (51). For S. meliloti, global transcription analyses of free-living bacterial cultures of WT, ΔrelA, and ΔdksA mutants exposed to nitrogen or carbon starvation have shown that most RelA-dependent gene expression changes are also DksA dependent (33). Our complementation findings (Fig. 5) suggest that at least parts of the gene regulation that requires relA can be accomplished by elevated dksA expression.
It appears that SMc00049 either has no function or has a yet-unknown function independent of dksA or relA, since its deletion had no additional effect on their individual deletions. SMc00049 expression was induced in minimal medium and additionally in 21-day-old nodules (see Fig. S5 in the supplemental material), despite the fact that we found no phenotypes different from WT behavior under these conditions or stages in strains lacking SMc00049. These transcript level data imply, however, that there may be environments, such as late symbiosis stages and nitrogen-free medium, where SMc00049 plays some important role, perhaps in combination with an as-yet-unidentified stressor. Alternatively, SMc00049 might be cotranscribed with the immediately upstream gene SMc00048, encoding a conserved hypothetical protein, which might be involved directly or indirectly in SMc00049 expression or protein function.
The reason for the functional differences between DksASm and SMc00049 may lie in their primary protein structures. This and previous studies have shown via complementation or transcription experiments that when DksA has a DXXDXA motif at the tip of the CC domain, it functions as a transcriptional regulator (5, 15, 17, 18). Accordingly, DksA activity is lost when the second D in the motif is mutated (14, 17–19). DksA also becomes inactive when the A in the motif is replaced by threonine in E. coli and R. sphaeroides (18, 19). In contrast, the corresponding A70 appears not to be critical for the function of DksASm, because substitution mutants were still able to complement ΔdksA (Fig. 6B and D). However, the N terminus likely is important to provide at least structural stability, as its deletion led to loss of function under free-living conditions (Fig. 6A and B), which has not been described before for a DksA protein.
Constructing a DXXDXA motif via either a V43A substitution or introduction of the complete DLADRA motif of DksA did not render SMc00049 functional (Fig. 6A, B, and C). Thus, the nature of this motif alone is not sufficient to explain SMc00049's lack of activity. In addition, mimicking DksA even more by fusing its N terminus to SMc00049 failed to assign activity to SMc00049 (Fig. 6A, B, and D), indicating that the structures of the two proteins are still distinct. SMc00049, RSP0166 from R. sphaeroides, and a DksA-like protein from Campylobacter jejuni (Fig. 1) all possess an incomplete DXXDXA motif, four conserved cysteines in the zinc finger, and a shorter N terminus than that found in other DksA homologs. Since SMc00049 and RSP0166 are nonfunctional for complementing dksA mutants (this work and reference 19, respectively), it would be interesting to see if the C. jejuni protein has DksA activity, which has not yet been tested (24).
The role of the zinc finger motif for DksA function is less clear. The zinc finger itself may stabilize DksA's internal structure by connecting the C-terminal helix with the globular domain (17), which binds to the rim helices of RNAP (15, 17) and is important for DksA/RNAP stability. In addition, mutating single or multiple conserved cysteine residues strongly reduced or abolished P. aeruginosa DksA2's activity on transcription inhibition (17). The fact that the number of cysteines in the predicted zinc finger motif of DksA-like proteins (one, two, or four cysteines) varies strongly among different bacteria might suggest other, perhaps species-specific, requirements for this motif than transcriptional regulation during the stringent response.
This paper lays the foundation for further investigation of DksA-dependent developmental and transcriptional processes in the plant-microbe symbiosis, which will shed light on the role of DksA in mediating the bacterial response to the symbiosis environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wilma Ross and Richard L. Gourse, University of Wisconsin, Madison, for providing the E. coli dksA mutant strain, Markus W. Covert, Stanford University, for use of the plate reader, Virginia Walbot for use of the qPCR detection system, Hector Trujillo for assistance with spotting assays, and the members of the Long lab for comments and discussions on the manuscript. We declare that we have no conflict of interest.
K.W. was supported by the German Research Foundation postdoctoral fellowship.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00013-16.
REFERENCES
- 1.Cashel M, Gentry DR, Hernandes VJ, Vinella D. 1996. The stringent response, p 1458–1496. In Neidhardt FC. (ed), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 2.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. 2002. Dissection of the mechanism for the stringent factor RelA. Mol Cell 10:779–788. doi: 10.1016/S1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 3.Potrykus K, Cashel M. 2008. (p) ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 4.Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. 2009. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev 23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Jishage M, Kvint K, Shingler V, Nystrom T. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev 16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krasny L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J 23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu K, Bittner AN, Wang JD. 2015. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol 24:72–79. doi: 10.1016/j.mib.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, Rubin H. 2005. Functional regulation of the opposing (p) ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44:9913–9923. doi: 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- 10.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117:57–68. doi: 10.1016/S0092-8674(04)00260-0 [Erratum, 117:415, doi:.] [DOI] [PubMed] [Google Scholar]
- 11.Mittenhuber G. 2001. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J Mol Microbiol Biotechnol 3:585–600. [PubMed] [Google Scholar]
- 12.Gentry DR, Cashel M. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol 19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 13.Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. 2013. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J Mol Biol 425:82–93. doi: 10.1016/j.jmb.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. 2004. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, Lee JH, Butcher SE, Gourse RL. 2012. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev 26:2634–2646. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parshin A, Shiver AL, Lee J, Ozerova M, Schneidman-Duhovny D, Gross CA, Borukhov S. 2015. DksA regulates RNA polymerase in Escherichia coli through a network of interactions in the secondary channel that includes sequence insertion 1. Proc Natl Acad Sci U S A 112:E6862–E6871. doi: 10.1073/pnas.1521365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman R, Biswas T, Danhart EM, Foster MP, Tsodikov OV, Artsimovitch I. 2013. DksA2, a zinc-independent structural analog of the transcription factor DksA. FEBS Lett 587:614–619. doi: 10.1016/j.febslet.2013.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Lennon CW, Ross W, Gourse RL. 2012. Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J Mol Biol 416:503–517. doi: 10.1016/j.jmb.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lennon CW, Lemmer KC, Irons JL, Sellman MI, Donohue TJ, Gourse RL, Ross W. 2014. A Rhodobacter sphaeroides protein mechanistically similar to Escherichia coli DksA regulates photosynthetic growth. mBio 5:e01105-14. doi: 10.1128/mBio.01105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. 2010. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol Microbiol 76:200–219. doi: 10.1111/j.1365-2958.2010.07094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal RR, Bag S, Dasgupta S, Das B, Bhadra RK. 2012. Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J Bacteriol 194:5638–5648. doi: 10.1128/JB.00518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma AK, Payne SM. 2006. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol Microbiol 62:469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- 23.Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss R III, Foster JW. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol Microbiol 34:112–123. doi: 10.1046/j.1365-2958.1999.01581.x. [DOI] [PubMed] [Google Scholar]
- 24.Yun J, Jeon B, Barton YW, Plummer P, Zhang Q, Ryu S. 2008. Role of the DksA-like protein in the pathogenesis and diverse metabolic activity of Campylobacter jejuni. J Bacteriol 190:4512–4520. doi: 10.1128/JB.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henard CA, Tapscott T, Crawford MA, Husain M, Doulias PT, Porwollik S, Liu L, McClelland M, Ischiropoulos H, Vazquez-Torres A. 2014. The 4-cysteine zinc-finger motif of the RNA polymerase regulator DksA serves as a thiol switch for sensing oxidative and nitrosative stress. Mol Microbiol 91:790–804. doi: 10.1111/mmi.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crecy-Lagard V. 2011. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol Microbiol 79:700–715. doi: 10.1111/j.1365-2958.2010.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perron K, Comte R, van Delden C. 2005. DksA represses ribosomal gene transcription in Pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol Microbiol 56:1087–1102. doi: 10.1111/j.1365-2958.2005.04597.x. [DOI] [PubMed] [Google Scholar]
- 28.Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thebault P, Vandenbol M, Vorholter FJ, Weidner S, Wells DH, Wong K, Yeh KC, Batut J. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 29.Gage DJ. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells DH, Long SR. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol Microbiol 43:1115–1127. doi: 10.1046/j.1365-2958.2002.02826.x. [DOI] [PubMed] [Google Scholar]
- 32.Barnett MJ, Toman CJ, Fisher RF, Long SR. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc Natl Acad Sci U S A 101:16636–16641. doi: 10.1073/pnas.0407269101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krol E, Becker A. 2011. ppGpp in Sinorhizobium meliloti: biosynthesis in response to sudden nutritional downshifts and modulation of the transcriptome. Mol Microbiol 81:1233–1254. doi: 10.1111/j.1365-2958.2011.07752.x. [DOI] [PubMed] [Google Scholar]
- 34.Glazebrook J, Walker GC. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol 204:398–418. doi: 10.1016/0076-6879(91)04021-F. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook S, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed, vol 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 36.Oke V, Long SR. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol Microbiol 32:837–849. doi: 10.1046/j.1365-2958.1999.01402.x. [DOI] [PubMed] [Google Scholar]
- 37.Ehrhardt DW, Atkinson EM, Long SR. 1992. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- 38.Turner GL, Gibson AH. 1980. Measurement of nitrogen fixation by indirect means, p 111–138. In Bergersen FJ. (ed), Methods for evaluating biological nitrogen fixation. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 39.Schlüter JP, Reinkensmeier J, Barnett MJ, Lang C, Krol E, Giegerich R, Long SR, Becker A. 2013. Global mapping of transcription start sites and promoter motifs in the symbiotic alpha-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics 14:156. doi: 10.1186/1471-2164-14-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 41.Griffitts JS, Carlyon RE, Erickson JH, Moulton JL, Barnett MJ, Toman CJ, Long SR. 2008. A Sinorhizobium meliloti osmosensory two-component system required for cyclic glucan export and symbiosis. Mol Microbiol 69:479–490. doi: 10.1111/j.1365-2958.2008.06304.x. [DOI] [PubMed] [Google Scholar]
- 42.Martin MO, Long SR. 1984. Generalized transduction in Rhizobium meliloti. J Bacteriol 159:125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masui Y, Mizuno T, Inouye M. 1984. Novel high-level expression cloning vehicles: 104-fold amplification of Escherichia coli minor protein. Nat Biotechnol 2:81–85. doi: 10.1038/nbt0184-81. [DOI] [Google Scholar]
- 44.Barnett MJ, Bittner AN, Toman CJ, Oke V, Long SR. 2012. Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J Bacteriol 194:4983–4994. doi: 10.1128/JB.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Griffith KL, Wolf RE Jr. 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun 290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- 48.Brown L, Gentry D, Elliott T, Cashel M. 2002. DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol 184:4455–4465. doi: 10.1128/JB.184.16.4455-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jude F, Kohler T, Branny P, Perron K, Mayer MP, Comte R, van Delden C. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J Bacteriol 185:3558–3566. doi: 10.1128/JB.185.12.3558-3566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aberg A, Fernandez-Vazquez J, Cabrer-Panes JD, Sanchez A, Balsalobre C. 2009. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol 191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol 189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.