Abstract
Background:
We investigated possible biomarkers for endometriosis (EM) using the ClinProt technique and proteomics methods.
Methods:
We enrolled 50 patients with EM, 34 with benign ovarian neoplasms and 40 healthy volunteers in this study. Serum proteomic spectra were generated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MS) combined with weak cationic exchange (WCX) magnetic beads. Possible biomarkers were analyzed by a random and repeat pattern model-validation method that we designed, and ClinProtools software, results were refined using online liquid chromatography-tandem MS.
Results:
We found a cluster of 5 peptides (4210, 5264, 2660, 5635, and 5904 Da), using 3 peptides (4210, 5904, 2660 Da) to discriminate EM patients from healthy volunteers, with 96.67% sensitivity and 100% specificity. We selected 4210 and 5904 m/z, which differed most between patients with EM and controls, and identified them as fragments of ATP1B4, and the fibrinogen alpha (FGA) isoform 1/2 of the FGA chain precursor, respectively.
Conclusions:
ClinProt can identify EM biomarkers, which – most notably – distinguish even early-stage or minimal disease. We found 5 stable peaks at 4210, 5264, 2660, 5635, and 5904 Da as potential EM biomarkers, the strongest of which were associated with ATP1B4 (4210 Da) and FGA (5904 Da); this indicates that ATP1B4 and FGA are associated with EM pathogenesis.
Keywords: ClinProt, Endometriosis; Liquid Chromatography-tandem Mass Spectrometry, Magnetic Bead; Matrix-assisted Laser Desorption/Ionization Time-of-flight Mass Spectrometry; Proteomics
INTRODUCTION
Endometriosis (EM) is the presence of endometrial-like tissue outside the uterus and is associated with chronic intrapelvic inflammation. It is most common in the ovaries but may appear in the pelvic cavity, peritoneum, other pelvic organs, and even distant organs such as lung, pleura or brain.[1] It occurs in roughly 5–10% of women during their childbearing years. Although the only way to confirm an EM diagnosis is by laparoscopy or other surgery, the positive rate for laparoscopic diagnosis is only 18–60%.[2]
Mean delay between symptom onset and the surgical EM diagnosis is an estimated 6.7 years. Each affected woman loses on average 10.8 h of work weekly, mainly from reduced effectiveness while working.[3] Despite many treatments used to reduce symptoms, EM has a 67% 5-year recurrence rate.[4] Thus, EM both reduces patients’ quality-of-life and generates a huge economic burden.[5,6] Timely diagnosis is critical.
We previously investigated the applicability of 2-DE gels and peptide mass mapping to identify candidate endometrial proteins by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) protein chip array technology, to detect proteomic patterns in serum of women with EM.[7,8] However, SELDI technology has very low reproducibility, and cannot detect low-abundance proteins. The ClinProt system facilitates preparation and acquisition of complex proteomic profile patterns and their comparative analyses.[9] It can optimize searches for biomarkers. In the study, we used ClinProt technology to explore and identify biomarkers from serum of EM patients.
METHODS
Serum samples
In this study, we enrolled 50 patients who had been clinically diagnosed with EM and confirmed postsurgery by 2 senior pathologists. Of these patients, 28 had ovarian EM, 11 had adenomyosis, 8 had adenomyosis complicated with ovarian EM, and 3 had peritoneal and abdominal wall EM. Their average age was 37 ± 8 years (range: 18–51 years). Their disease was staged according to the revised American Fertility Society (AFS) classification. As controls, we also enrolled 34 patients with benign ovarian neoplasms, including 19 with mature ovary teratoma, 8 with epithelial neoplasms, 2 with inflammatory masses, and 5 with other benign tumor types; their average age was 34 ± 8 years (range: 19–49 years). Finally, we enrolled 40 healthy women controls who were undergoing health check-ups during the enrollment period, for whom serology, X-ray, and B-ultrasound of their pelvic and abdominal cavities excluded gynecologic diseases and other complications of primary hypertension and renal disorders; the average age of these healthy controls was 38 ± 9 years (range: 23–54 years). Blood samples from all patients were collected preoperatively. Subjects were of reproductive age and had normal menstrual cycles. They had no other diseases on physical examination or by biochemical tests. None had received any hormonal treatment within 3 months before this study. All subjects signed a consent form before participating in this research protocol which was approved by the Institutional Review Board of Peking University People's Hospital.
All blood samples were obtained by venipuncture and centrifuged at 3000 r/min for 5 minutes at room temperature (25°C). The serum was aliquoted and stored at −80°C. Repeated freeze/thaw cycles during further proceedings were avoided to protect protein integrity.
Serum pretreatment with magnetic beads
We used 10 samples from each group to select appropriate magnetic particles; finally WCX was chosen to fractionate samples, following manufacturers’ instructions through a standard protocol (ClinProt™, Bruker Daltonics, USA). For each sample, 5 μl of plasma was mixed with 5 μl of beads. Samples were purified through 3 steps – binding, washing, and elution, during which binding incubation took 1 minute. We eluted 5 μl of each sample; the purified plasma samples were further diluted 4-, 8-, and 16-fold.
Data acquisition by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
We prepared diluted solutions for matrix-assisted laser desorption/ionization TOF MS (MALDI-TOF MS) by mixing 1 μl each of diluted samples with 0.5 μl of matrix solution containing 2 g/L α-cyano-4-hydroxycinnamic acid, 10 nmol/L angiotensin II, 10 nmol/L adrenocorticotropic hormone 18–39 (Bruker Daltonics), and 10 ml/L formic acid in 500 ml/L acetonitrile and allowing each droplet to dry on the MALDI sample plate (Bruker Daltonics). We used an Auto Flex II MALDI-TOF mass spectrometer (Bruker Daltonics), operated in positive ion linear or reflectron mode. Briefly, all spectra were processed by automatic baseline subtraction, peak detection, recalibration, and peak area calculation, according to the predefined settings.
Statistical analysis
When we used ClinProtools Software (Bruker Daltonics) to search for differential proteins and design models,[10] we found that not only the number, mass of candidate proteins, but also the potential importance of these proteins; that is, candidate proteins with lower peak intensities appeared to be less meaningful in this context. We, therefore, tried to improve the analytic process.
(A) First, we used ClinProtools to identify the proteins with the greatest differential expression among the three groups. (B) Second, we processed more than 100 stochastic analyses of the EM group versus the healthy controls (total 90 samples – 50 EM and 40 healthy controls), which were then randomly sorted into either the model group or the test group. Of the proteins found by ClinProtools, we identified those with expressions that varied >30% between the two groups. (C) Third, we used ClinProtools to process Supervised Neural Network (SNN) arithmetic (signal/noise threshold: >5; threshold basic peak intensity: 0.1–0.25) and used leave-1-out arithmetic to find the most plausible candidate proteins. (D) Fourth, we combined the 10 initial proteins that showed the highest sensitivity in our first step with other proteins to evaluate the diagnosis veracity in combination. (E) Fifth, we regarded proteins found in the above steps to correlate with differences in disease states to be likely biomarkers; and (F) Sixth, we tried to identify those candidate proteins.
Identification of endometriosis-specific serum peptides
We used online liquid chromatography-tandem MS (LC-MS/MS) (LTQ Orbitrap XL, Thermo Fisher Ltd., USA) to further identify proteins. Our MS analysis used the following source parameters: Nano ion source; spray voltage 2.0 kV; 60 m scanning time; MS mode of data-dependent and dynamic exclusion; mass range: 300–2000 Da; Orbitrap for MS1 at 100,000 resolution; and LTQ for collision-induced dissociation and MS2. The 6 most intense ions were chosen as parent ions. Obtained chromatograms were analyzed with Bioworks Browser 3.3.1 SP1; resulting mass lists were used for database searching using Sequest™ (IPI Human 3.45). Parameters for generating peak lists were as: Parent ion: 50 μg/g; fragment mass relative accuracy: 1 Da.
RESULTS
Quality control for the ClinProt System
Stabilization of the mass spectrum using different sample runs was the means of verifying reproducibility of the ClinProt System, so in this study we tested the system stabilization first. We also used commercial standard sample as a reference to compare different chips and experimental runs in which six typical proteins were detected; the results show that the mean coefficient of variation (CV) was < 10%, which indicates good system reproducibility [Figure 1] and indicates that our results are credible.
Figure 1.
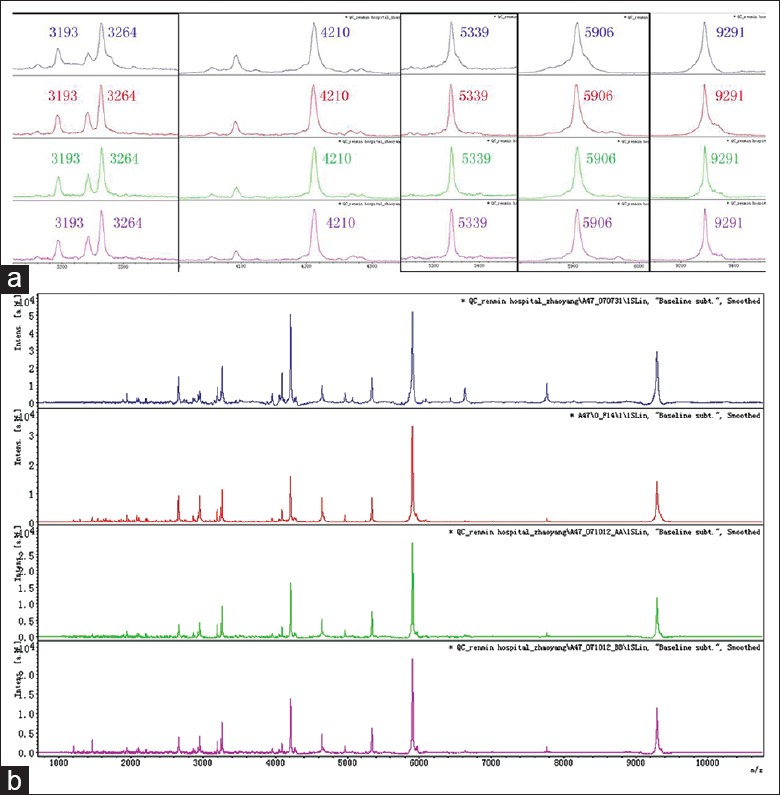
Protein standard fingerprints in different experimental runs. (a) Six proteins in different experimental runs; (b) Entire standard fingerprint in different experimental runs. Blue: 1st run; red: 2nd run; green: 3rd run; purple: 4th run.
The preliminary results of biomarker exploring
First, we constructed models from ClinProtools analyses of 30 EM patients and 20 controls, and then we used another 20 patients and 20 controls to test the model. We found many differentially expressed proteins [Figure 2], which scored high in the cross test, which indicates a very stable model [Table 1]. From the four table analysis, sensitivity and specificity were 92.0% and 97.5%, respectively. Positive and negative prediction rates were 97.8% and 90.70%, respectively. However, as we found no differentially expressed proteins when we used 30 EM patients and 20 benign controls to construct models [Table 2], we tried to establish whether patients with early-stage EM and healthy controls showed differentially expressed proteins.
Figure 2.
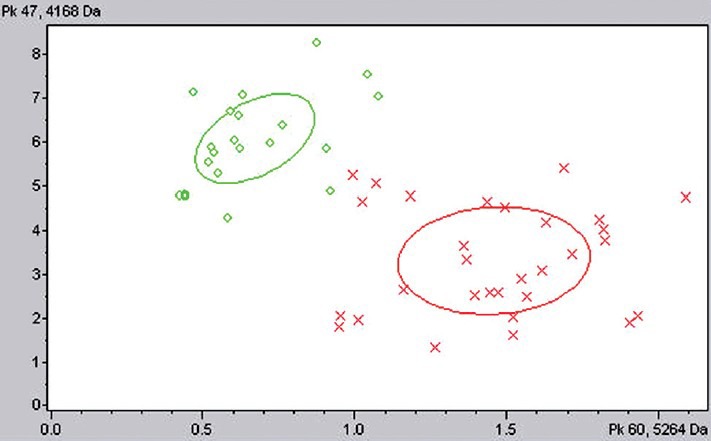
Distribution of endometriosis (EM) and control. X: EM samples; O: Control samples.
Table 1.
Model and test results for patients with EM and healthy controls
| Groups | Serum sample | n | Correct | False | Accuracy (%) | Cross-validation (%) |
|---|---|---|---|---|---|---|
| Model | Control | 20 | 20 | 0 | 100.00 | 100 |
| EM patients | 30 | 29 | 1 | 96.67 | 90 | |
| Test | Control | 20 | 19 | 1 | 95.00 | |
| EM patients | 20 | 17 | 3 | 85.00 |
EM: Endometriosis.
Table 2.
Model and test results for patients with EM and those with benign gynecological tumors (controls)
| Groups | Serum sample | n | Correct | False | Accuracy (%) | Cross-validation (%) |
|---|---|---|---|---|---|---|
| Model | Benign controls | 30 | 2 | 28 | 6.67 | 26.67 |
| EM patients | 20 | 20 | 0 | 100.00 | 100.00 | |
| Test | Benign controls | 20 | 0 | 20 | 0 | - |
| EM patients | 14 | 14 | 0 | 100.00 | - |
EM: Endometriosis.
The AFS system classifies EM into 4 stages, in which patients with Stage I-II EM normally have only pelvic disease and no ovarian neoplasms; we considered them to have early-stage disease for this study. Notably, we found these early-stage patients to have two groups of differential proteins with average cross-validation of 86.25%. These proteins merit wider attention; our next work will focus on them.
Exploring peaks of differentially expressed proteins
The ClinProt system analyzes proteins by their protein fingerprint, and constructs an optimized model, in which numbers and mass of candidate proteins may vary when target samples changed, and its calculations might not seem biologically plausible for all the protein peaks. We, therefore, tried to find the most stable model and appropriate differential proteins. We independently constructed a reiterative model test method and used several statistical paths, after many random testing runs and associative steps to get the most stable and accurate proteins.
Differential proteins between endometriosis and control groups
We then randomly distributed the EM and healthy control samples into the modeling and testing groups, on which we carried out 100 random analyses of these candidate proteins, which showed a 99.75% mean recognition accuracy rate, and mean validation accuracy rates of 85% for EM and 99% for healthy controls. Table 3 shows proteins with > 30% occurrence frequency.
Table 3.
Proteins with expression that varies by ≥30% between healthy controls and patients with EM
| Mass (Da) | Frequency (%) | Change in EM |
|---|---|---|
| 4209.64 | 100 | Down |
| 5263.87 | 89 | Up |
| 4247.51 | 87 | Down |
| 4963.78 | 84 | Up |
| 2659.26 | 69 | Up |
| 4168.75 | 63 | Down |
| 8106.89 | 60 | Down |
| 5805.74 | 59 | Up |
| 8142.45 | 53 | Down |
| 3240.52 | 52 | Up |
| 5634.22 | 40 | Up |
| 5864.94 | 39 | Up |
| 7009.98 | 37 | Down |
| 7398.71 | 37 | Down |
| 4266.52 | 32 | Down |
| 5754.13 | 31 | Up |
Da: Daltons; EM: Endometriosis.
Stable differential proteins among groups
Combining the sensitivity and specificity of each protein with reiterative random tests, we used ClinProtools SNN (signal-to-noise: >5; correlation threshold base peak: 0.1–0.25), and the leave-1-out method to cross test, which gave the differential proteins combination for each group [Table 4].
Table 4.
Differential proteins selected by ClinProtools software
| Protein combinations (Da) | Sensitivity (%) | Specificity (%) | Average cross verification (%) |
|---|---|---|---|
| EM patients vs. healthy controls | |||
| 4209.26 | 96.67 | 100 | 97.77 |
| 5904.97 | |||
| 2658.79 | |||
| Other benign tumors vs. healthy controls | |||
| 5264.28 | 100.00 | 100 | 100.00 |
| Stage I–II EM vs. healthy controls | |||
| 5905.37 | 100.00 | 100 | 97.73 |
| 4209.68 | |||
| Stage III–IV EM vs. healthy controls | |||
| 2659.98 | 100.00 | 100 | 96.30 |
| 4210.09 | |||
| 3191.88 |
Da: Daltons; EM: Endometriosis.
Confirmation of stable differential proteins
After testing with the random repeat-validation self-determination system, the stable differential protein peaks were for 4210, 5264, 2660, 5635, and 5904 Da [Figure 3]. The 5635 Da peak distinguished patients with Stage I–II EM from healthy controls very well. Peak intensity of some proteins was transformed with greater disease stage: 2660 Da, 5264 Da, 5635 Da, 5904 Da gradually increased and 4210Da decreased compared with normal. This pattern implies that those proteins affect disease development.
Figure 3.
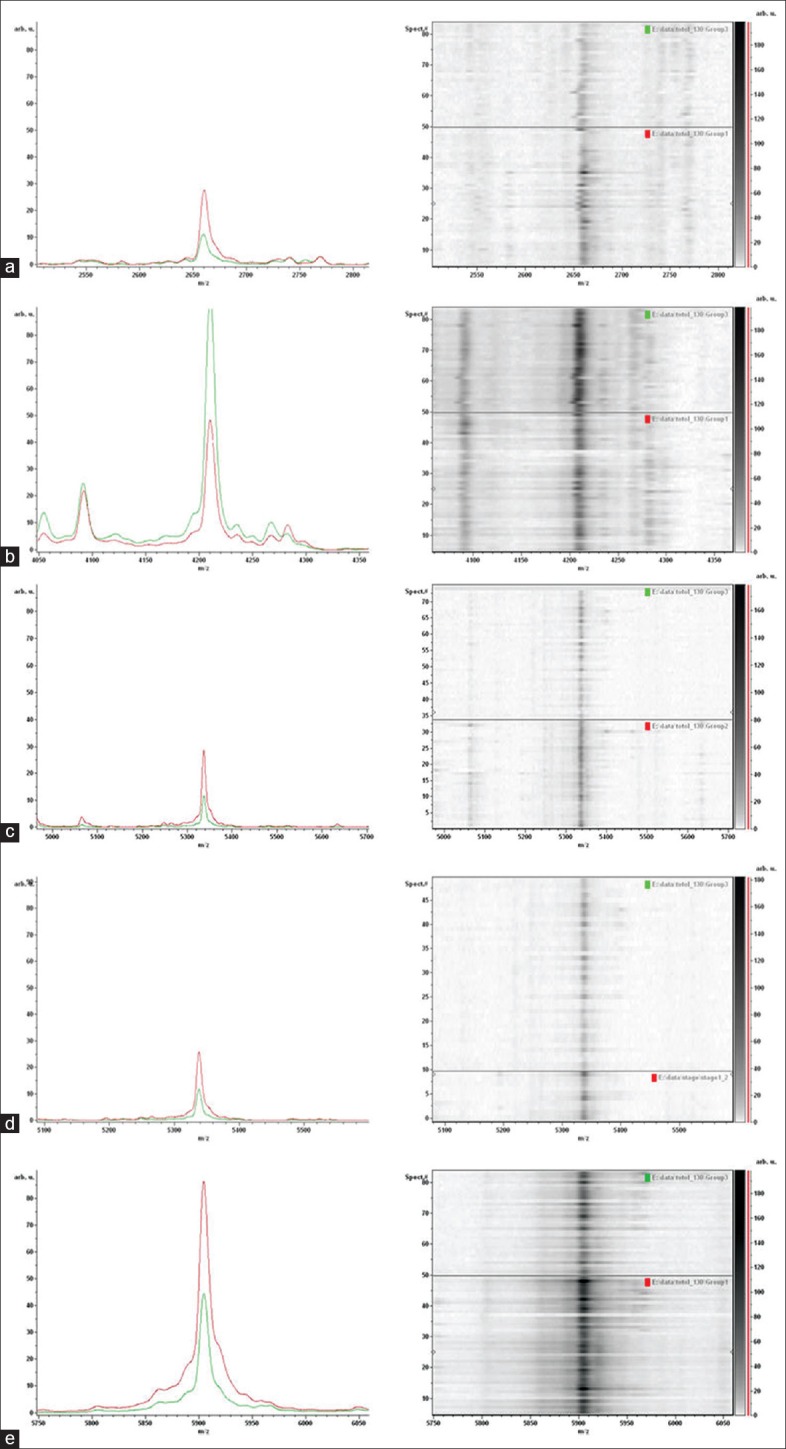
Differential protein peaks in typical samples, and in electrophoresis gel bands. Left: Differential protein peaks; red: Patient samples; green: Healthy controls; Right: Electrophoresis gel bands; Upper: Healthy controls; lower: Patients. (a) 2660 Da; (b) 4210 Da; (c) 5264 Da; (d) 5635 Da; and (e) 5904 Da.
Identification of endometriosis-specific serum peptides by liquid chromatography-tandem mass spectrometry
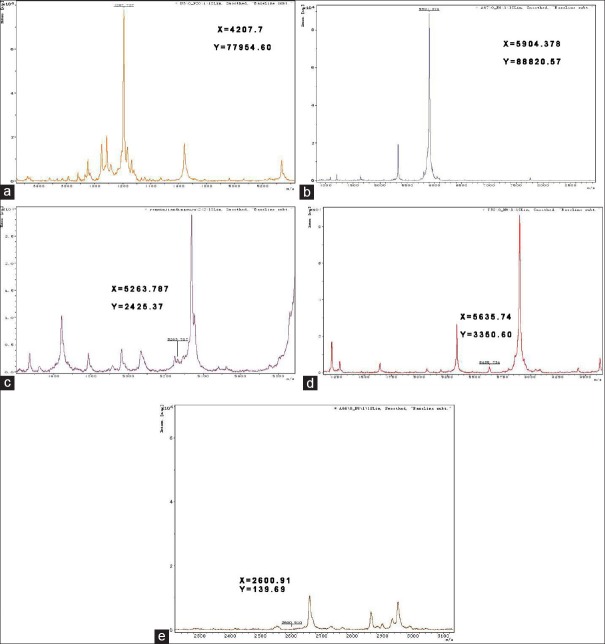
We collected data for nine samples using an Auto Flex II MALDI-TOF/TOF operated in linear mode for 400 shots to draw peptide mass fingerprints for of 4210, 5904, 5264, 5635, and 2600 m/z [Figure 4]. Of these five peaks, 4210 and 5904 m/z had high discrepancy peaks without interference peaks, which facilitate follow-up peptide identification; whereas peak intensities of 5263, 5635, and 2600 m/z were too low or had intensive interference peaks, which impede follow-up identification. The two selected peaks (4210 m/z, 5904 m/z) also showed the biggest differences between EM patients and controls [Figure 5]. LC-MS/MS showed the amino acid sequence of the 4210 m/z peptide as IMSEYLWDPERRMFLARTGQSWSLILLIYFFFY, a fragment of ATP1B4; and the 5904 m/z peptide sequence as SSSYSKQFTSSTSYNRGDSTFESKSYKMADEAGSE ADHEGTHSTKRGHAKSRPV, which is a fragment of the fibrinogen alpha (FGA) isoform 1/2 of the FGA chain precursor.
Figure 4.
Peptide mass fingerprinting of (a) 4210 m/z; (b) 5904 m/z; (c) 5264 m/z; (d) 5635 m/z; and (e) 2600 m/z.
Figure 5.
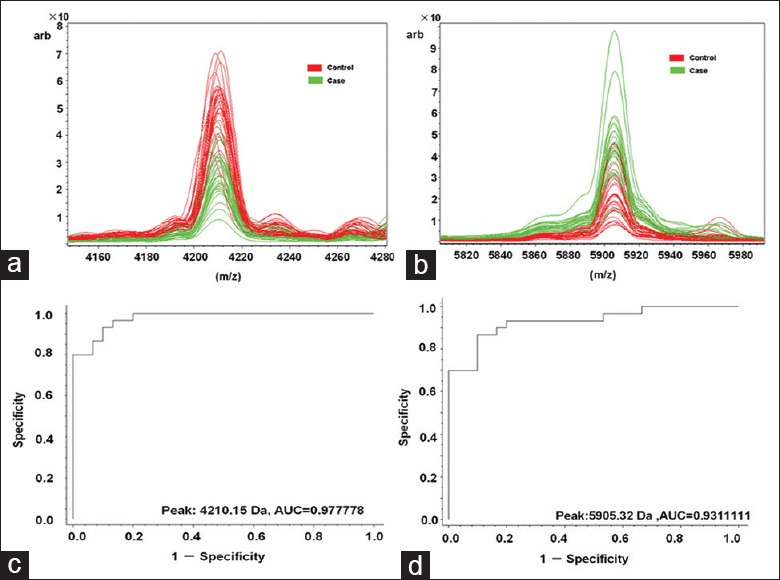
Distribution and performance of serum peptide biomarker for (a) 4210 m/z and (b) 5904 m/z. Receiver operator characteristic of (c) 4210 m/z and (d) 5905 m/z distinguishes patients with endometriosis (EM) from healthy controls. Red: Controls; green: Patients with EM.
DISCUSSION
Although many EM patients develop dysmenorrhea, infertility or chronic pelvic pain long before forming detectable ovarian neoplasms, it is difficult to diagnose in its early stages, even in women who develop EM in puberty.[11]
For the last decade, researchers have used SELDI technology to explore biomarkers for EM, and have found differential peaks at 8141, 6096, 5894, 3269, 3956.83, 11710.70, 6986.45, 4974, 5813, and 4290 Da.[12,13] These results did not accord well, partly because of variations in samples, and possibly because their data were simply imported to the analysis software with little further statistical analysis for in-depth validation. Some investigators got different proteins using the same type of sample in different sites or times, which made identifying these proteins much harder.
Matrix-assisted laser desorption/ionization TOF MS is a sensitive and effective approach for identifying potential biomarkers of health and disease.[14] In our previous study, WCX was a superior means of finding differential proteins and establishing steady models.[15] Here, we enriched low-abundant serum peptides/proteins by WCX magnetic beads and performed serum protein profiling through MALDI-TOF-MS, which offers a wide selection. It is also noninvasive and might allow dispensing with tissue biopsies.
In this study, we used the ClinProt System to explore serum biomarkers of EM, and found many differential proteins between EM patients and healthy controls from which we constructed a model with high sensitivity and specificity. Although we also used serum from patients with other benign ovarian neoplasms as a control group, we found no proteins that significantly differed between them and the EM patients. Further studies showed differences in protein fingerprints between patients with minimal and early-stage EM and healthy controls, and also significant differential proteins that can stably distinguish these two groups, which may aid in early diagnosis of EM.
Other technologies use different serum EM markers, possibly because different systems focus on different mass area of target proteins. The advantage of our system is that it facilitates identification of those proteins which has been otherwise so time-consuming.
As the weakest aspect of SELDI is reproducibility, we focused on this point. First, we designed a strictly standardized experimental operation: Sample collection and storage, transport, magnetic beads selection, equilibrium for matrix formula, data collection. We then used the CV value for several typical peaks in different samples to evaluate the stability of the system. We randomly chose a commercial sample as the standard serum; all CV values of these selected peaks were <10%, which indicates high stability. Inter- and intra-group consistency was also satisfactory. To assess stability of differential proteins in different groups, we first found cross-validation results (average diagnosis values from different samples) to construct model and validation groups; in our study all results were ≥85%, which indicted a stable model. We then used randomly selected samples to construct our models, and found the differential proteins remained when groups changed. Thereafter, the biggest challenge was to confirm the differential proteins.
We also found results to change when using a different analysis system, which we thought was because of variations in bioinformatics. Because complicated algorithms are based on provided sampling data, and may modify parameters to make analyses more suitable for the data at hand, they may change with different sample sets. If the protein fingerprint differed dramatically between two groups, random combinations for each 2-protein set could show strong sensitivity and specificity for the model, but would change with the variables.
After employing these methods, we found differential proteins at 4210, 5264, 2660, 5635, 5904 Da to distinguish between patients with EM and healthy controls. These proteins have high diagnostic accuracy, either singly or combined. We also found that the 5635 Da peak is associated with early-stage or minimal EM and could help monitor early disease. LC-MS/MS sequencing showed the 4210 Da peptide to correspond uniquely to ATP1B4, and the 5904 Da protein to FGA isoform 1/2 subunit.
ATP1B4 is crucial in forming functionally active X, K-ATPases.[16] In a previous study, we discovered that the ATP synthase β subunit is expressed differently in patients with EM than in normal controls. We considered that there is a close relationship between ATP function and EM generation and development. EM results from a menstrual bleeding countercurrent, the formation of which depends on functions that use ATP enzymes (ATP synthesis, signal transduction, metabolic regulation, etc.).[7,17]
Fibrinogen alpha fibrinogen is a glycoprotein secreted by hepatocytes,[18] which mainly affects blood coagulation and hemostasis.[19] Fibrinogen is considered to be an acute-phase reactant protein; it increases during tumor progression, apparently as an ongoing inflammatory response to tumor. Some studies suggest that tumor cells could promote coagulation by interacting with endothelial cells and platelets, and then releasing active biologics that activate platelets, thus increasing the fibrinogen in the blood of cancer patients. FGA, isoform 1 of FGA chain precursor, has also been found (using ClinProt) to significantly differ between adult patients with acute lymphocytic leukemia and healthy controls, and is a potential biomarkers for forecasting relapse, and monitoring residual disease and therapeutic response in these patients.[20] Although EM is not cancer, it displays malignant behavior. Nothnick et al. utilized a novel mouse model in which eutopic endometrial fragments used to induce EM were deficient in miR-451 expression, which resulted in fewer ectopic lesions in vivo and was associated with differential expression of alpha polypeptide isoform 2 precursor.[21] As this polypeptide contained arginyl-glycyl-aspartic acid (RGD) cell adhesion motifs that could modify early-stage lesion development, RGD peptides might help prevent recurrent EM development.
Our results using ClinProt to identify biomarkers for EM screening indicate that it is effective even for early-stage or minimal EM. We found five stable peaks with molecular weights of 4210, 5264, 2660, 5635, and 5904 Da as potential EM biomarkers. Furthermore, our results indicate that ATP1B4 and FGA are associated with EM pathogenesis; these associations merit further study.
Footnotes
Edited by: Li-Min Chen
Source of Support: This work was supported by grants from the National Natural Science Foundation of China (No. 30772319 and 81170544) and supported by Peking University People's Hospital Research and Development Funds (RDB2007-10).
Conflict of Interest: None declared.
REFERENCES
- 1.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–99. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 2.Walter AJ, Hentz JG, Magtibay PM, Cornella JL, Magrina JF. Endometriosis: Correlation between histologic and visual findings at laparoscopy. Am J Obstet Gynecol. 2001;184:1407–11. doi: 10.1067/mob.2001.115747. [DOI] [PubMed] [Google Scholar]
- 3.Rogers PA, D’Hooghe TM, Fazleabas A, Giudice LC, Montgomery GW, Petraglia F, et al. Defining future directions for endometriosis research: Workshop report from the 2011 World Congress of Endometriosis in Montpellier, France. Reprod Sci. 2013;20:483–99. doi: 10.1177/1933719113477495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selçuk I, Bozdað G. Recurrence of endometriosis; risk factors, mechanisms and biomarkers; review of the literature. J Turk Ger Gynecol Assoc. 2013;14:98–103. doi: 10.5152/jtgga.2013.52385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simoens S, Dunselman G, Dirksen C, Hummelshoj L, Bokor A, Brandes I, et al. The burden of endometriosis: Costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod. 2012;27:1292–9. doi: 10.1093/humrep/des073. [DOI] [PubMed] [Google Scholar]
- 6.Klein S, D’Hooghe T, Meuleman C, Dirksen C, Dunselman G, Simoens S. What is the societal burden of endometriosis-associated symptoms? A prospective Belgian study. Reprod Biomed Online. 2014;28:116–24. doi: 10.1016/j.rbmo.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Niu Y, Feng J, Guo H, Ye X, Cui H. Use of proteomic analysis of endometriosis to identify different protein expression in patients with endometriosis versus normal controls. Fertil Steril. 2006;86:274–82. doi: 10.1016/j.fertnstert.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Feng J, Chang XH, Li ZX, Wu XY, Cui H. Effect of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry on identifing biomarkers of endometriosis. Chin Med J. 2009;122:373–6. [PubMed] [Google Scholar]
- 9.Xiang Y, Xu Q, Tan W, He S, Shi X, Zhang W, et al. Serum biomarkers of Keshan disease assessed using a protein profiling approach based on ClinProt technique. Protein J. 2014;33:344–53. doi: 10.1007/s10930-014-9567-9. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Yang J, Gao Y, Zhao L, Liu L, Qin Y, et al. Identification of potential serum proteomic biomarkers for clear cell renal cell carcinoma. PLoS One. 2014;9:e111364. doi: 10.1371/journal.pone.0111364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laufer MR, Sanfilippo J, Rose G. Adolescent endometriosis: Diagnosis and treatment approaches. J Pediatr Adolesc Gynecol. 2003;16:S3–11. doi: 10.1016/s1083-3188(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Zheng W, Yu JK, Jiang WZ, Mu L, Zhang SZ. Artificial neural networks combined with surface-enhanced laser desorption/ionization mass spectra distinguish endometriosis from healthy population. Fertil Steril. 2007;88:1700–2. doi: 10.1016/j.fertnstert.2007.01.055. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Lang J, Zhou Q, Shan D, Li Q. Detection of endometriosis with the use of plasma protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Fertil Steril. 2007;87:988–90. doi: 10.1016/j.fertnstert.2006.08.095. [DOI] [PubMed] [Google Scholar]
- 14.Aldred S, Grant MM, Griffiths HR. The use of proteomics for the assessment of clinical samples in research. Clin Biochem. 2004;37:943–52. doi: 10.1016/j.clinbiochem.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Yi Li, Chang XH, Tian Li, Hu XH, Ye X, Cui H. Selection of magnetic bead for identification serum biomarker s in endometr iosis with Clinprot: A preliminary study (in Chinese) BME Clin Med. 2008;12:189–92. [Google Scholar]
- 16.Pestov NB, Ahmad N, Korneenko TV, Zhao H, Radkov R, Schaer D, et al. Evolution of Na, K-ATPase beta m-subunit into a coregulator of transcription in placental mammals. Proc Natl Acad Sci U S A. 2007;104:11215–20. doi: 10.1073/pnas.0704809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehasseb MK, Taylor AH, Pringle JH, Bell SC, Habiba M. Enhanced invasion of stromal cells from adenomyosis in a three-dimensional coculture model is augmented by the presence of myocytes from affected uteri. Fertil Steril. 2010;94:2547–51. doi: 10.1016/j.fertnstert.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 18.Weisel JW, Stauffacher CV, Bullitt E, Cohen C. A model for fibrinogen: Domains and sequence. Science. 1985;230:1388–91. doi: 10.1126/science.4071058. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa N, Sasaki S. Location of the binding site “b” for lateral polymerization of fibrin. Thromb Res. 1990;57:183–95. doi: 10.1016/0049-3848(90)90318-7. [DOI] [PubMed] [Google Scholar]
- 20.Bai J, He A, Huang C, Yang J, Zhang W, Wang J, et al. Serum peptidome based biomarkers searching for monitoring minimal residual disease in adult acute lymphocytic leukemia. Proteome Sci. 2014;12:49. doi: 10.1186/s12953-014-0049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nothnick WB, Graham A, Holbert J, Weiss MJ. miR-451 deficiency is associated with altered endometrial fibrinogen alpha chain expression and reduced endometriotic implant establishment in an experimental mouse model. PLoS One. 2014;9:e100336. doi: 10.1371/journal.pone.0100336. [DOI] [PMC free article] [PubMed] [Google Scholar]