Abstract
RNA-dependent RNA polymerase (RdRP) plays key roles in RNA silencing to generate double-stranded RNAs. In model organisms, such as Caenorhabditis elegans and Neurospora crassa, two types of small interfering RNAs (siRNAs), primary siRNAs and secondary siRNAs, are expressed; RdRP produces secondary siRNAs de novo, without using either Dicer or primers, while primary siRNAs are processed by Dicer. We reported that human telomerase reverse transcriptase (TERT) has RdRP activity and produces endogenous siRNAs in a Dicer-dependent manner. However, de novo synthesis of siRNAs by human TERT has not been elucidated. Here we show that the TERT RdRP generates short RNAs that are complementary to template RNAs and have 5′-triphosphorylated ends, which indicates de novo synthesis of the RNAs. In addition, we confirmed short RNA synthesis by TERT in several human carcinoma cell lines and found that TERT protein levels are positively correlated with RdRP activity.
INTRODUCTION
Telomerase reverse transcriptase (TERT) is known as the catalytic subunit of telomerase and is expressed at high levels in cancer cells but only at low levels in normal human somatic cells. TERT elongates telomeres by its RNA-dependent DNA polymerase (RdDP) activity, using the telomerase RNA component (TERC) as the template. TERT and TERC assemble and form telomerase; however, there is a population of TERT proteins that are not assembled into the telomerase complex (1). Several lines of evidence indicate that TERT plays roles independent of telomere maintenance; therefore, nonassembled TERT may be involved in complexes other than telomerase.
RNA silencing is a sequence-specific gene regulatory mechanism mediated by double-stranded RNAs (dsRNAs). RNA-dependent RNA polymerase (RdRP) is a key player in RNA silencing, and the polymerase is found in a variety of organisms, including fungi, plants, and worms (2). Although insects and mammals lack sequence homologues of cell-encoded RdRPs, phylogenetic and structural analyses of TERT revealed that TERT is closely related to RdRPs of RNA viruses as well as to retroviral RdDPs (3). In fact, we found that TERT generates dsRNA in a primer-dependent manner and works as an RdRP by a mechanism similar to that for cell-encoded RdRPs (4, 5). Both viral RdRPs and cell-encoded RdRPs transcribe single-stranded RNA (ssRNA) from template RNA, not only in a primer-dependent manner but also in a primer-independent manner. However, primer-independent initiation of RNA synthesis by TERT, a human RdRP, remains to be elucidated.
To analyze the characteristics of the RdRP activity of human TERT, we established an in vitro RdRP assay in which we analyzed the RdRP activity of TERT immune complexes immunoprecipitated from cell lysates by use of an anti-human TERT monoclonal antibody (MAb) (IP-RdRP assay) (5). Here we investigated the detailed characteristics of RNAs processed through the IP-RdRP assay. The results indicate that TERT RdRP produces short RNAs in a primer-independent manner. The relationship between TERT protein levels and the RdRP activity of TERT was further confirmed in various carcinoma cell lines.
MATERIALS AND METHODS
Reagents.
The following reagents were used for the IP-RdRP assay: cOmplete EDTA-free protease inhibitor cocktail (Roche), 3′-dATP (TriLink BioTechnologies), 3′-dCTP (TriLink BioTechnologies), 3′-dGTP (TriLink BioTechnologies), 3′-dUTP (TriLink BioTechnologies), β-rubromycin (Enzo Life Sciences), VX-222 (Selleckchem), and α-amanitin (Nacalai Tesque). Pefabloc SC [4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride] (AEBSF; Roche) was used for IP followed by the telomeric repeat amplification protocol (IP-TRAP assay).
Antibodies.
Anti-human TERT MAbs (clones 10E9-2 and 2E4-2) were generated as reported previously (5). Briefly, sense and antisense oligonucleotides corresponding to 304 to 460 amino acids of human TERT were cloned into the plasmid pET-30a(+) (Novagen). A recombinant carboxyl-terminally His-tagged TERT protein containing 157 amino acids (positions 304 to 460) was overexpressed in Escherichia coli and purified with a nickel-agarose column. Recombinant purified TERT was used as an immunogen to stimulate production of anti-human TERT MAbs in mice by using standard methodologies (5). A sequential screening strategy was used to identify hybridomas producing anti-human TERT MAbs.
Primary antibodies used for immunoblotting were as follows: an anti-phospho-histone H3 (Ser10) polyclonal antibody (06-570; Millipore), an anti-SNAIL polyclonal antibody (ab17732; abcam), an anti-human TWIST mouse MAb (clone Twist2C1a; Bio Matrix Research), and an anti-β-actin mouse MAb (clone AC-15; Sigma-Aldrich). The following antibodies were used for immunofluorescence analysis: an anti-human TERT MAb (clone 2E4-2), an anti-TRF2 polyclonal antibody (IMG-148A; Imgenex), an anti-human Ki-67 antigen mouse MAb (clone MIB-1; Dako), Alexa Fluor 488-conjugated donkey anti-mouse IgG(H+L) (Life Technologies), and Alexa Fluor 568-conjugated donkey anti-goat IgG(H+L) (Life Technologies).
Peptide array.
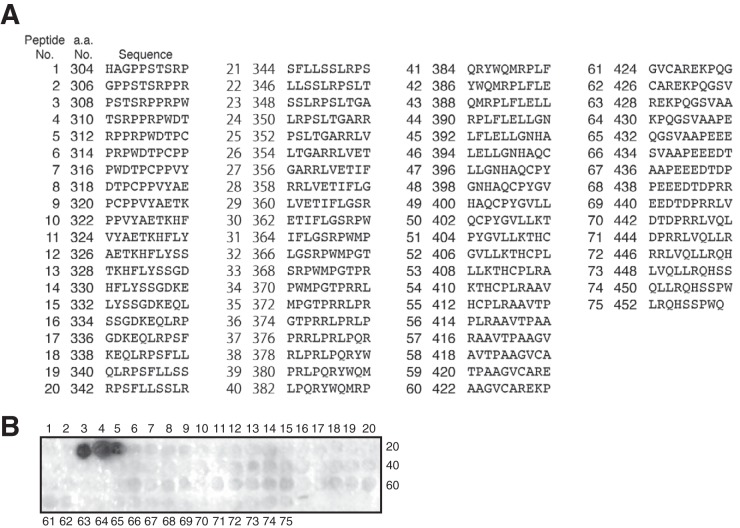
A peptide array was created as described previously (5). Seventy-five peptides derived from a truncated version of human TERT (304 to 460 amino acids) were covalently bound to a continuous cellulose membrane. The panel of peptides was then probed with an anti-human TERT MAb (clone 2E4-2), and bound antibody was detected using a Pep spot assay (Funakoshi) according to the manufacturer's protocol.
Cell culture, mitotic cell synchronization, and transfection of small interfering RNAs (siRNAs).
The human cervical carcinoma cell line HeLa, the simian virus 40 (SV40)-transformed human embryonic kidney cell line 293T, and the human hepatocellular carcinoma (HCC) cell lines HepG2, HLE, and HLF were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (IFS). The human ovarian carcinoma (OC) cell lines were cultured as follows: RMG-I cells were cultured in Ham's F-12 medium supplemented with 10% IFS, TOV-21G cells were cultured in MCDB105-medium 199 (1:1) supplemented with 10% IFS, and PEO1 and PEO14 cells were cultured in RPMI 1640 medium supplemented with 10% IFS and 2 mM sodium pyruvate (Gibco).
Mitotic cell synchronization was performed as described previously (5). Briefly, cells were switched to medium containing 2.5 mM thymidine (Nacalai Tesque) and incubated for 24 h. Six hours after release, cells were incubated in medium containing 0.1 μg/ml nocodazole (Invitrogen) for 14 h. After shake-off, mitotic cells were retrieved. For suppression of TERT expression (see Fig. 2C, D, and G), HeLa cells were transfected with siRNAs by use of Lipofectamine 2000 (Invitrogen). After 48 h of incubation, cells were treated with 0.1 μg/ml nocodazole (Invitrogen) for 16 h. 293T cells were transfected with siRNAs by use of Lipofectamine 2000 (Invitrogen) twice, with the transfections separated by 48 h, and cells were harvested 24 h after the second transfection. The sequences used for the indicated siRNAs were as follows (6): 5′-GUGUCUGUGCCCGGGAGAATT-3′ and 5′-UUCUCCCGGGCACAGACACTT-3′ for TERT siRNA 1 and 5′-GCAUUGGAAUCAGACAGCATT-3′ and 5′-UGCUGUCUGAUUCCAAUGCTT-3′ for TERT siRNA 2. Mission siRNA universal negative control 1 (Sigma-Aldrich Japan) was used as a negative control (NC).
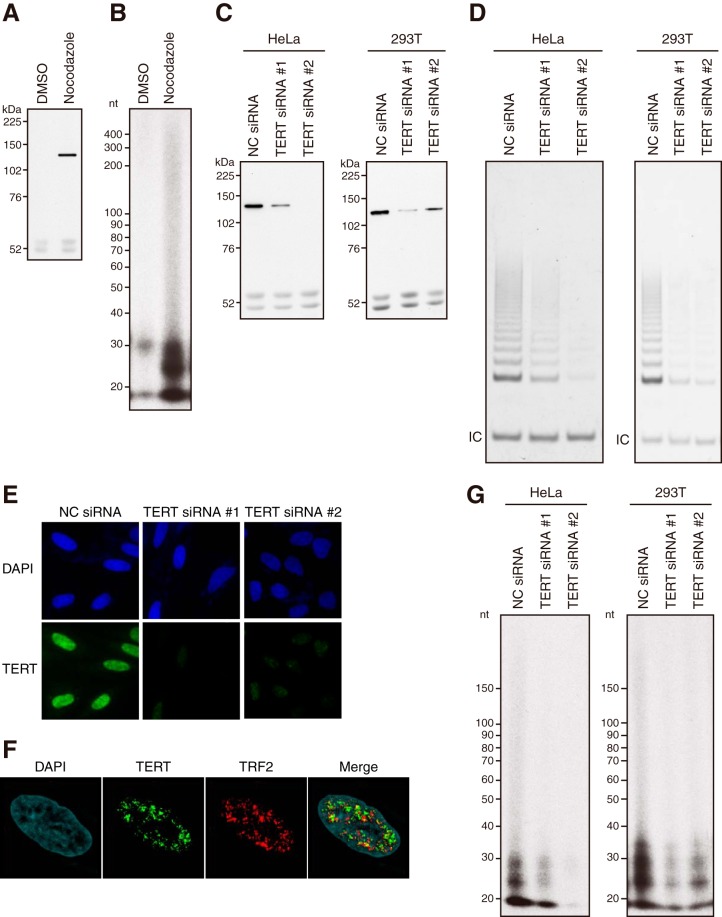
FIG 2.
Enzymatic activities of endogenous TERT. Endogenous TERT was immunoprecipitated from HeLa and 293T cells by use of an anti-human TERT MAb (clone 10E9-2). An anti-human TERT MAb (clone 2E4-2) was used for immunoblotting. (A) IP-IB of endogenous TERT. HeLa cells were treated with nocodazole (manipulated) or DMSO (unmanipulated). The signals at ≈52 kDa indicate heavy chains of IgG. (B) IP-RdRP assay using HeLa cells treated with nocodazole or DMSO. [α-32P]UTP and a synthetic RNA (RNA 1) were used. (C and D) HeLa cells treated with nocodazole or 293T cells were transfected with TERT-specific (TERT siRNA 1 and TERT siRNA 2) or NC siRNA. (C) IP-IB of endogenous TERT. The signals at ≈52 kDa indicate heavy chains of IgG. (D) IP-TRAP assay. IC, internal control. (E) Immunofluorescence of HeLa cells transfected with TERT-specific (TERT siRNA 1 and TERT siRNA 2) or NC siRNA. TERT was immunostained with an anti-human TERT MAb (clone 2E4-2). (F) Colocalization of endogenous TERT with TRF2 in HeLa cells. An anti-human TERT MAb (clone 2E4-2) and an anti-TRF2 antibody were used for staining. DAPI, 4′,6-diamidino-2-phenylindole. (G) IP-RdRP assay using [α-32P]UTP and synthetic RNA (RNA 1). HeLa and 293T cells were prepared in the same way as that for panels C and D.
MTT assay.
Cells were seeded in 96-well dishes at 2.5 × 103 cells/well. After 96 h of incubation, the amount of viable cells was quantified using a cell proliferation kit I [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay] (Roche) according to the manufacturer's protocol.
Immunofluorescence.
HeLa cells were seeded in 8-well CultureSlides (BD Falcon) at 1.5 × 104 cells/well 1 day before transfection. TERT-specific siRNAs (TERT siRNA 1 and TERT siRNA 2) or Mission siRNA universal negative control 1 (Sigma-Aldrich Japan) was transfected twice, with the transfections separated by 48 h, by use of Lipofectamine 2000 (Invitrogen). Cells were fixed for immunofluorescence staining 48 h after the second transfection.
To synchronize HeLa cells in S phase, 1.4 × 104 HeLa cells/well were seeded in 8-well CultureSlides (BD Falcon) and treated with 2 mM thymidine (Nacalai Tesque) for 14 h. Eleven hours after release, cells were incubated with 2 mM thymidine (Nacalai Tesque) again for 14 h. Four hours after release, cells were fixed for immunofluorescence analysis.
Immunofluorescence staining was performed as described previously (5). The cells were observed by fluorescence microscopy (IX-81 microscope without DSU disk; Olympus), spinning disk confocal microscopy (IX-81 microscope with DSU disk; Olympus), or confocal microscopy (Fluoview FV10i microscope; Olympus).
Silkworm pupae overexpressing recombinant human TERT (rTERT).
Human TERT was expressed in the silkworm-baculovirus system produced by ProCube (Sysmex Corp.). The TERT gene was inserted into a transfer vector (Sysmex Corp.), based on the pUC19 vector, for Bombyx mori nucleopolyhedrovirus (BmNPV). This transfer vector was cotransfected with baculovirus DNA (BmNPV CPd strain) (7) into a Bombyx mori-derived cell line (BmN) (8). After 7 days of incubation, the recombinant baculovirus was injected into the bodies of silkworm pupae, which were harvested 6 days after infection. In total, 5 ml of homogenization buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], and phenylthiourea) was added per pupa, and pupae were homogenized using an SH-IIM homogenizer (Elmex). The homogenates were immediately centrifuged at 100,000 × g at 4°C for 1 h. The supernatants, containing TERT proteins, were collected and used for the IP-immunoblot (IP-IB) and IP-RdRP assays.
Partial purification of rTERT.
Twenty milligrams of silkworm lysate overexpressing rTERT was incubated with 40 μl of anti-FLAG M2–agarose (Sigma) overnight at 4°C. The beads were washed three times with lysis buffer A (0.5% NP-40, 20 mM Tris-HCl [pH 7.4], and 150 mM NaCl) and eluted with FLAG peptides (Sigma). The eluate was then incubated with 10 μg of an anti-human TERT MAb (clone 10E9-2) and 40 μl of Pierce protein A plus agarose (Thermo Scientific) overnight at 4°C to obtain partially purified rTERT.
IP-IB assay of TERT.
For human cell lines, 1 × 107 cells were lysed in 1 ml of lysis buffer A. After sonication, lysates were cleared of insoluble material by centrifugation at 21,000 × g at 4°C for 15 min. One milliliter of lysate was preabsorbed with 40 μl of Pierce protein A plus agarose for 30 min at 4°C. Preabsorbed lysate was mixed with 10 μg of an anti-human TERT MAb (clone 10E9-2) and 40 μl of Pierce protein A plus agarose and incubated overnight at 4°C. Immune complexes were washed three times with lysis buffer A, eluted in 2× SDS loading buffer (2% β-mercaptoethanol, 20% glycerol, 4% SDS, and 100 mM Tris-HCl [pH 6.8]), and then subjected to SDS-PAGE on 8% polyacrylamide gels. An anti-human TERT MAb (clone 2E4-2) and MouseTrueBlot Ultra (eBioscience) were used for immunoblotting.
The amount of immunoprecipitated TERT proteins was estimated by SDS-PAGE after Coomassie brilliant blue staining against bovine serum albumin (BSA). With 10 μg of an anti-human TERT MAb (clone 10E9-2), 66 ng of rTERT was obtained from a lysate containing 20 mg of protein. ImageJ software was used to quantify the amounts of endogenous TERT proteins on the immunoblots.
Synthetic RNAs.
Synthetic RNAs used as templates for the IP-RdRP assay were as follows: RNA 1, 5′-GGGAUCAUGUGGGUCCUAUUACAUUUUAAACCCA-3′, and RNA 2, 5′-GGGUUUAAAAUGUAAUAGGACCCACAUGAUCCCA-3′. The synthetic RNAs had hydroxyl groups at both the 5′ and 3′ ends. There are no sequences identical to that of either RNA 1 or RNA 2 in the human or Bombyx mori genome. A 3′-fold-back structure was not predicted for RNA 1 or RNA 2 (9). Synthetic RNAs were reported as templates for RNA polymerization by Qβ replicase, a virus-encoded RdRP (9).
IP-RdRP assay.
The IP-RdRP assay was performed as described previously (5). For IP-RdRP assay of human cell lines, 1 × 107 cells were lysed in 1 ml of lysis buffer A. After sonication, lysates were cleared of insoluble material by centrifugation at 21,000 × g at 4°C for 15 min. One milliliter of lysate, prepared from either cell cultures or silkworm pupae, was preabsorbed with 40 μl of Pierce protein A plus agarose for 30 min at 4°C. The preabsorbed lysate was mixed with 10 μg of an anti-human TERT MAb (clone 10E9-2) and 40 μl of Pierce protein A plus agarose and incubated overnight at 4°C. Immune complexes were washed four times with 1× acetate buffer (10 mM HEPES-KOH [pH 7.8], 100 mM potassium acetate, and 4 mM MgCl2) containing 10% glycerol, 0.1% Triton-X, and 0.06× cOmplete EDTA-free protease inhibitor cocktail and once with AGC solution {1× acetate buffer containing 10% glycerol and 0.02% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)} containing 2 mM CaCl2. The bead suspension was treated with 0.25 U/μl micrococcal nuclease (MNase) at 25°C for 15 min. Immunoprecipitates were subsequently washed twice with AGC solution containing 3 mM EGTA and once with 1× acetate buffer containing 0.02% CHAPS. Finally, 40 μl of reaction mixture was prepared by combining 20 μl of the bead suspension, 6 μl of [α-32P]UTP (3,000 Ci/mmol) or [γ-32P]ATP (3,000 Ci/mmol), 25 ng/μl (final concentration) of RNA template, and supplements and was incubated at 32°C for 2 h. The final concentrations of ribonucleotides were 1 mM ATP, 0.2 mM GTP, 10.5 μM UTP, and 0.2 mM CTP. The resulting products were treated with proteinase K to stop the reaction, purified several times with phenol-chloroform until the white interface disappeared, and precipitated using ethanol. For the UTP incorporation assay, RdRP products were treated with RNase I (2 U; Promega) at 37°C for 2 h to digest ssRNAs, followed by proteinase K treatment, phenol-chloroform purification, and ethanol precipitation. The IP-RdRP products were electrophoresed in a 10% polyacrylamide gel containing 7 M urea and detected by autoradiography. The amounts of products were compared by densitometry (ImageJ). The amount of product generated with nocodazole-treated HeLa cells was used for normalization.
IP-TRAP assay.
For the TRAP assay using immunoprecipitated TERT, the TERT protein was immunoprecipitated as described for the IP-RdRP assay, without sonication. Immune complexes were washed three times with lysis buffer A and then suspended in 35 μl of TRAP lysis buffer (pH 7.5) (10 mM Tris-HCl, 1 mM MgCl2, 1 mM EGTA, 0.5% CHAPS, 10% glycerol, 100 μM Pefabloc SC, and 0.035% 2-mercaptoethanol). The TRAP assay was performed with 2 μl of the suspension as described previously (5).
RT-qPCR.
A cell lysate of nocodazole-treated HeLa cells was prepared as described for the IP-RdRP assay, and total RNA was extracted from the lysate by use of acid phenol-CHCl3 (Ambion) followed by ethanol precipitation. The TERT immune complex was immunoprecipitated from the cell lysate, treated with MNase, and washed as described for the IP-RdRP assay. Total RNA was then extracted from the TERT immune complex by use of TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription was performed using PrimeScript reverse transcriptase (RT; TaKaRa) with pd(N)6 random hexamers (GE Healthcare). A TaqMan gene expression assay (Hs03454202_s1; Applied Biosystems) was used for quantitative PCR (qPCR) analysis of TERC.
Next-generation sequencing.
Ten batches of IP-RdRP products were pooled into one library for efficiency. The library for deep sequencing of IP-RdRP products was constructed using a NEBNext multiplex small RNA library prep set for Illumina sequencing (New England BioLabs) essentially according to the manufacturer's instructions. Libraries were sequenced using a HiSeq 2000 sequencer (Illumina).
Ten batches of IP-RdRP products and the 3′ SR adaptor for Illumina sequencing were mixed and denatured at 98°C for 1 min. Thereafter, 3′ adaptor ligation was performed using the denatured mixture, 3′ ligation reaction buffer, and 3′ ligation enzyme mix at 25°C for 1 h. Half the amount (0.5 μl) of SR RT primer for Illumina sequencing was added to the mixture, and then the mixture was denatured at 98°C for 1 min. To remove pyrophosphate from the 5′ ends of triphosphorylated RNA, the RNA mixture was incubated with RNA 5′ pyrophosphohydrolase (RppH) (New England BioLabs) and simultaneously incubated with 5′ SR adaptor for Illumina sequencing, 5′ ligation reaction buffer, and 5′ ligase enzyme mix for 5′ adaptor ligation at 25°C for 1 h. After the rest of the SR RT primer for Illumina sequencing was added, the mixture was denatured at 70°C for 2 min, and then reverse transcription and 12 PCR cycles were performed according to the manufacturer's protocol. The PCR products were purified by PAGE extraction, and six additional PCR cycles were performed to obtain sufficient amounts of products for sequencing with a HiSeq 2000 instrument.
Adapter sequences were identified and removed from sequencing reads for all libraries. Sequences of 6 nucleotides (nt) or longer were aligned to the RNA templates (RNA 1 and RNA 2) used for the IP-RdRP assay by using the blastn program of NCBI BLAST (version 2.2.28+).
Immunoblotting.
Immunoblot analysis was performed as described previously (5).
Statistics.
Simple regression analyses were performed using Statcel3 software (OMS Publishing). P values of <0.05 were considered significant.
RESULTS
Endogenous TERT is related to RdRP activity as well as telomerase activity.
We reported previously that TERT protein expression is enriched in the mitotic phase in HeLa cells (5). To immunoprecipitate and detect endogenous TERT, we generated a series of anti-human TERT MAbs (clones 10E9-2 [5] and 2E4-2) (Fig. 1) (5). We performed immunoprecipitation (IP) with clone 10E9-2 followed by immunoblotting with clone 2E4-2 and confirmed that TERT protein expression was enriched in HeLa cells treated with nocodazole, in concordance with our previous report (5) (Fig. 2A). To investigate the detailed mode of action of RdRP activity by TERT, we modified the original IP-RdRP assay (5) by using a chemically synthesized RNA of 34 nt (RNA 1), which is more uniform than RNAs transcribed by T7 or SP6 RNA polymerases in vitro, as the template. Production of radioactive products was observed specifically in HeLa cells treated with nocodazole (Fig. 2B). This result suggested that RNA 1 would be utilized as a template for TERT RdRP activity and that endogenous TERT detected by immunoblotting would be responsible for the product synthesis. To further validate the specificity of the antibodies, expression of endogenous TERT in HeLa and 293T cells was suppressed by use of TERT-specific siRNAs, and IP (clone 10E9-2) followed by immunoblotting (clone 2E4-2) (Fig. 2C) and the telomerase assay (Fig. 2D) was performed. Suppression of TERT expression by TERT-specific siRNAs eliminated the TERT protein signal in immunoblots (Fig. 2C) and the telomerase activities (Fig. 2D) for both cell lines. The results demonstrate that endogenous TERT is immunoprecipitated by clone 10E9-2 and detected by clone 2E4-2. The specificity of clone 2E4-2 was further confirmed by immunocytochemistry. Distinct nuclear staining, which was diminished by use of TERT-specific siRNAs, was observed in HeLa cells stained with clone 2E4-2 (Fig. 2E). In addition, the nuclear staining was colocalized with TRF2 in HeLa cells synchronized at S phase (Fig. 2F). The immunofluorescence results again demonstrated specific detection of endogenous TERT by clone 2E4-2. Furthermore, TERT-specific siRNAs eliminated the IP-RdRP products (Fig. 2G), indicating that the radioactive products were produced by TERT.
FIG 1.
Confirmation of the specificity of anti-human TERT MAbs. (A) Peptides used to map the TERT epitope (5). The truncated version of human TERT (amino acids [a.a.] 304 to 460) used to generate MAbs was divided into 75 peptides. (B) Epitope mapping for clone 2E4-2 by use of the peptide array. Numbers indicate specific peptides.
De novo RNA synthesis by TERT RdRP.
To determine whether the products found in the IP-RdRP assay were indeed RNA strands produced by polymerase activity, we investigated the requirement for ribonucleotides in this assay. Exclusion of ribonucleotides abolished almost all the products found in the IP-RdRP assay for HeLa cells treated with nocodazole (Fig. 3A). Production was also inhibited by 3′-deoxyribonucleotide 5′-triphosphates (3′-dNTPs), a type of chain-terminating ribonucleotides, in a dose-dependent manner (Fig. 3B). These results indicate that all four types of ribonucleotide triphosphates are required to synthesize the IP-RdRP products and that the products are not generated by the terminal transferase activity (10) but by the polymerase activity.
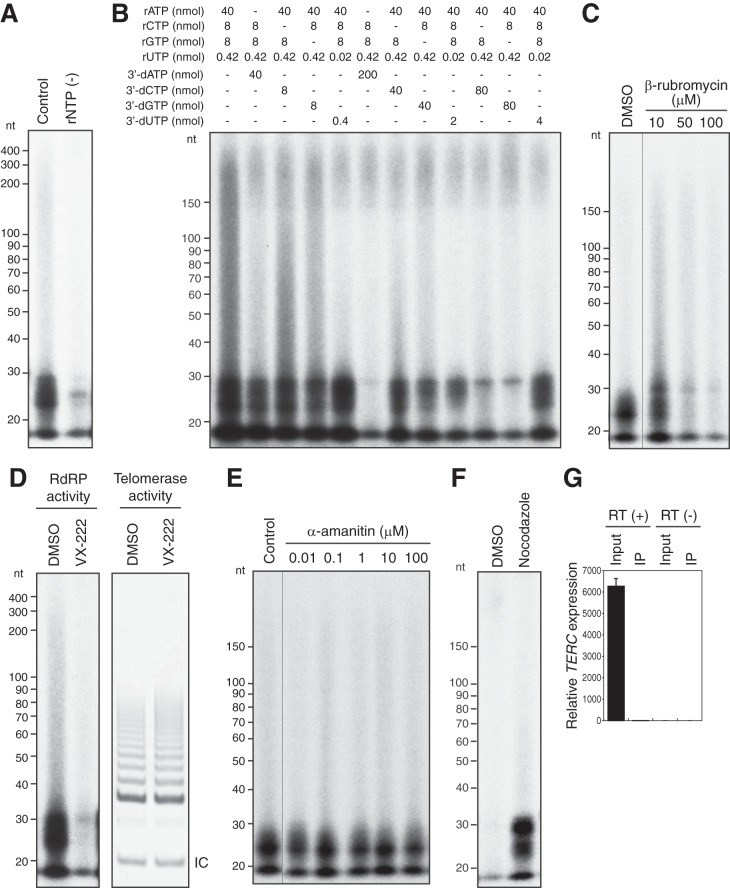
FIG 3.
TERT RdRP generates short RNA strands de novo. The IP-RdRP assay was performed with HeLa cells treated with nocodazole, using [α-32P]UTP (A to E) or [γ-32P]ATP (F). Synthetic RNA (RNA 1) was used as a template. (A) IP-RdRP assay. All four types of ribonucleotides (control) or rUTP alone [rNTP (−)] was added. (B) Supplemented ribonucleotides in the IP-RdRP assay were replaced by 3′-dNTPs at the indicated concentrations. (C) Suppression of RNA synthesis by a telomerase inhibitor. β-Rubromycin was used as an inhibitor in the IP-RdRP assay. (D) Suppression of RNA synthesis but not telomerase activity by an RdRP inhibitor. HeLa cells treated with nocodazole were used for the IP-RdRP and IP-TRAP assays. VX-222 (100 μM) was used as an inhibitor. IC, internal control. (E) No effects of α-amanitin on RNA synthesis. α-Amanitin at the indicated concentrations was used for the IP-RdRP assay. (F) IP-RdRP assay using [γ-32P]ATP. HeLa cells treated with nocodazole or DMSO were used. (G) RT-qPCR of TERC in HeLa cells treated with nocodazole. Total RNA was extracted from cell lysate (input; 0.13%) or TERT immune complexes (IP; 2%) prepared for the RdRP assay. RT (−), the reverse transcriptase reaction was performed without reverse transcriptase.
We next investigated whether RdRP activity of TERT is responsible for RNA synthesis. First, we performed the IP-RdRP assay with either a telomerase inhibitor (β-rubromycin) (11) or dimethyl sulfoxide (DMSO; control). We previously confirmed that β-rubromycin inhibits telomerase activity (5), and as shown in Fig. 3C, RNA synthesis in this assay was remarkably inhibited by β-rubromycin. We next used VX-222, an RdRP inhibitor for NS5B of hepatitis C virus (HCV) (12). VX-222 fully inhibited RNA synthesis, while the same amount of VX-222 did not affect telomerase activity (Fig. 3D). These results strongly suggest that the TERT RdRP produces RNAs in the IP-RdRP assay. RNA polymerase II (Pol II) is reportedly responsible for RdRP activity in Saccharomyces cerevisiae (13) and mice (14); therefore, we excluded the possibility that Pol II is responsible for RNA production in this assay. For this purpose, we performed the IP-RdRP assay with α-amanitin, a well-characterized Pol II inhibitor. We found no effects of α-amanitin on RNA synthesis (Fig. 3E), which suggests that Pol II is not responsible for RNA synthesis in this assay. Taken together, our results confirmed that human TERT synthesizes short RNAs through its RdRP activity.
We next investigated how short RNAs are synthesized by TERT RdRP. Since no primers were provided as supplements in our IP-RdRP assay and the 3′-fold-back structure was not predicted for the template RNA 1 (9), short RNA synthesis in the IP-RdRP assay was assumed to be primer independent (de novo). RNA species synthesized de novo carry characteristic 5′-triphosphate termini, while siRNAs cleaved by Dicer bear monophosphorylated 5′ ends (15–17). To determine whether the short RNA products have a 5′-triphosphate structure, we performed the IP-RdRP assay with [γ-32P]ATP, which specifically labels products with 5′-triphosphate termini (17). We found radioactive IP-RdRP products specifically in HeLa cells treated with nocodazole (Fig. 3F). The lengths of the products were identical to those observed in the IP-RdRP assay using [α-32P]UTP (Fig. 2B). This result indicates the de novo synthesis of RNA species by TERT RdRP in the assay.
As described above, there are two populations of TERT proteins: those that assemble with TERC and those that do not (1). To examine whether TERC plays any roles in de novo RNA synthesis, we monitored the TERC level in immune complexes prepared for the RdRP assay. As shown in Fig. 3G, we were unable to detect TERC in the TERT immune complexes used for the RdRP assay, suggesting that TERT synthesized the RNAs independently of TERC.
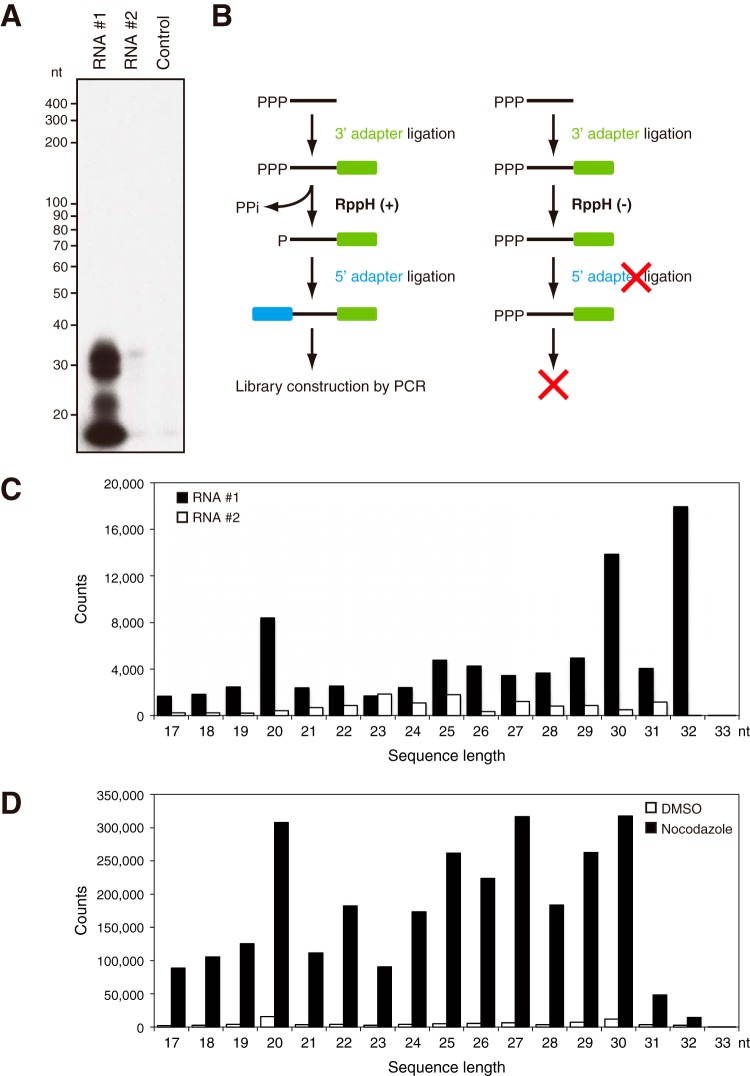
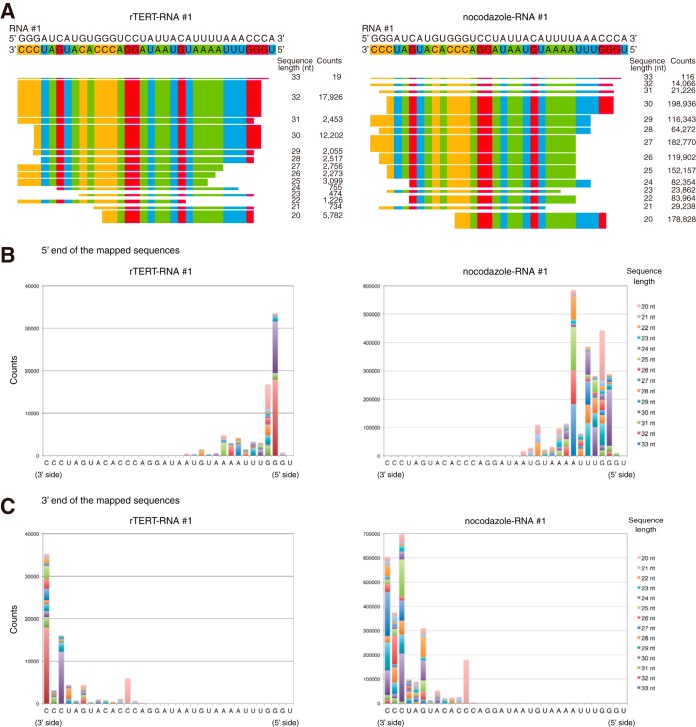
Some types of small silencing RNAs demonstrate a characteristic bias in the arrangement of residues as well as in their length. To clarify the characteristics of the primary structure of RNAs synthesized by TERT RdRP, we investigated these RNAs by deep sequencing. We first analyzed IP-RdRP products of rTERT produced by silkworm pupae (Bombyx mori) to elucidate the fundamental characteristics of TERT RdRP products. We partially purified rTERT from silkworm pupae by IP with clone 10E9-2 and then performed the RdRP reaction (Fig. 4A). Intriguingly, rTERT demonstrated a remarkable template RNA preference in the RdRP reaction. Specifically, rTERT effectively produced RNAs from template RNA 1 but not from template RNA 2 (Fig. 4A). rTERT and template RNA 2 did not synthesize any RNA products; therefore, we used this combination as a negative control (NC) and prepared two sets of IP-RdRP products for deep sequencing: the products from RNA 1 (designated rTERT-RNA 1) and those from RNA 2 (designated rTERT-RNA 2) (Table 1, library 1). For library construction, we treated the IP-RdRP products with RppH (18) to enable the ligation of 5′ adapters to de novo-synthesized RNAs, which have 5′-triphosphate ends (Fig. 4B). In total, 182 or 104 million sequences were obtained from each IP-RdRP product (Table 1). Among them, sequences of 6 nt or longer were mapped onto the template RNAs used in the IP-RdRP assay. The sequences used for mapping were classified into five categories (type A to D and “other”) (Table 1) according to their sequence identity or similarity to either template RNA or its complementary strand. We specifically focused on type D sequences, which are completely complementary to a part of the template RNA sequence. Because the silkworm genome does not contain any sequences that are nearly identical to RNA 1 or RNA 2, type D sequences found in the libraries should be the antisense products of synthetic template RNAs. Consistent with the IP-RdRP assay results (Fig. 4A), the number of type D sequences was 14-fold higher in the rTERT-RNA 1 set than in the rTERT-RNA 2 set (251,672 versus 17,945) (Table 1). The length distribution of type D sequences traced the features of the IR-RdRP assay (Fig. 4A and C); there was a predominance of rTERT-RNA 1 over rTERT-RNA 2, with the highest counts for ≈30-nt products, indicating successful library construction for the IP-RdRP products. We next investigated if there were any biases in residues at the 5′ or 3′ ends of type D sequences (>20 nt) of the rTERT-RNA 1 set. In total, 88.3% of type D sequences in rTERT-RNA 1 possessed purine residues at their 5′ ends (A, 17.7%; G, 70.7%) (Fig. 5A and B). As for 3′ ends, 84.6% of type D sequences terminated with C, and 60.0% of them were placed at the 5′ end of template RNA 1 (Fig. 5A and C).
FIG 4.
Properties of short RNAs synthesized by TERT RdRP. (A) IP-RdRP assay using a lysate of silkworm pupae overexpressing rTERT, an anti-human TERT MAb (clone 10E9-2), and [α-32P]UTP. Synthetic RNAs (RNA 1 and RNA 2) were used as templates. The IP-RdRP assay without template RNA was performed as the NC (control). (B) Scheme for the preparation of a small RNA library for deep sequencing analysis of IP-RdRP products. RppH was used to convert 5′ ends of triphosphate to monophosphate. The library was successfully constructed from 5′-triphosphorylated RNAs with RppH treatment. (C) Length distributions of type D sequences in rTERT-RNA 1 and rTERT-RNA 2. IP-RdRP products of rTERT were analyzed by deep sequencing. (D) Length distributions of type D sequences in DMSO-RNA 1 and nocodazole-RNA 1. Short RNA products in the IP-RdRP assay using HeLa cells were analyzed by deep sequencing.
TABLE 1.
Classification of sequences obtained by deep sequencinga
| Sequence typeb | No. of reads |
|||
|---|---|---|---|---|
| Library 1 |
Library 2 |
|||
| rTERT-RNA 1 | rTERT-RNA 2 | DMSO-RNA 1 | Nocodazole-RNA 1 | |
| A | 1,159,555 | 14,776 | 31,355 | 8,804 |
| B | 39,340,949 | 43,335,025 | 93,651,643 | 54,932,629 |
| C | 0 | 0 | 0 | 0 |
| D | 251,672 | 17,945 | 118,320 | 4,287,182 |
| Other | 141,370,214 | 60,833,729 | 68,867,660 | 102,515,449 |
| Total reads | 182,122,390 | 104,201,475 | 162,668,978 | 161,744,064 |
The RNA products of the IP-RdRP assay were analyzed by deep sequencing. The obtained sequences (>6 nt) were mapped to the template sequence and classified into five categories (type A to D and “other”), as indicated. The number of total reads indicates the number of sequences used for mapping.
Type A, completely identical to template RNA; type B, completely identical to a part of the sequence of template RNA; type C, completely complementary to template RNA; type D, completely complementary to a part of the sequence of template RNA.
FIG 5.
Start and end positions of short RNAs synthesized by TERT RdRP. The results of deep sequencing analysis of short RNA products in the IP-RdRP assay are shown. (A) Mapping of type D sequences in rTERT-RNA 1 (left) and nocodazole-RNA 1 (right). The most frequent sequences of each sequence length are presented. Counts of each sequence are listed. (B and C) 5′ ends (B) and 3′ ends (C) of type D sequences obtained from rTERT-RNA 1 and nocodazole-RNA 1.
We next sequenced IP-RdRP products prepared from HeLa cells treated with DMSO or nocodazole, using RNA 1 as the template (DMSO-RNA 1 and nocodazole-RNA 1 of library 2) (Table 1). The sequencing library was constructed with RppH treatment as indicated in Fig. 4B. It is noteworthy that nocodazole-RNA 1 demonstrated a remarkably larger number of type D sequences than DMSO-RNA 1 (4,287,182 versus 118,320) (Table 1). The results were consistent with the IP-RdRP assay using radioactivity (Fig. 2B and 3F). The size distribution of type D sequences indicated the specific production of 20- to 30-nt RNAs by TERT, which was enriched in cells in the mitotic phase (Fig. 4D). The human genome does not contain consecutive sequences of 19 nt or longer that are identical or complementary to template RNA 1. Therefore, we concluded that type D sequences of 19 nt or longer were not due to contamination with cellular RNAs but were the products of the IP-RdRP assay. Specifically, the products were RNA strands newly synthesized by TERT RdRP, using RNA 1 as the template. We further investigated the 5′ and 3′ ends of type D sequences (>20 nt) of nocodazole-RNA 1. Nocodazole-RNA 1 demonstrated an apparently different distribution of 5′-terminal residues from that of rTERT-RNA 1: 68.0% of sequences started with a purine residue (A, 33.9%; G, 34.1%), while 32.0% possessed a U at the 5′ end (Fig. 5A and B). In addition to 5′ ends, the 3′ ends of type D sequences were more varied in nocodazole-RNA 1 than in rTERT-RNA 1 (Fig. 5A and C).
TERT protein levels correlate with RdRP activity.
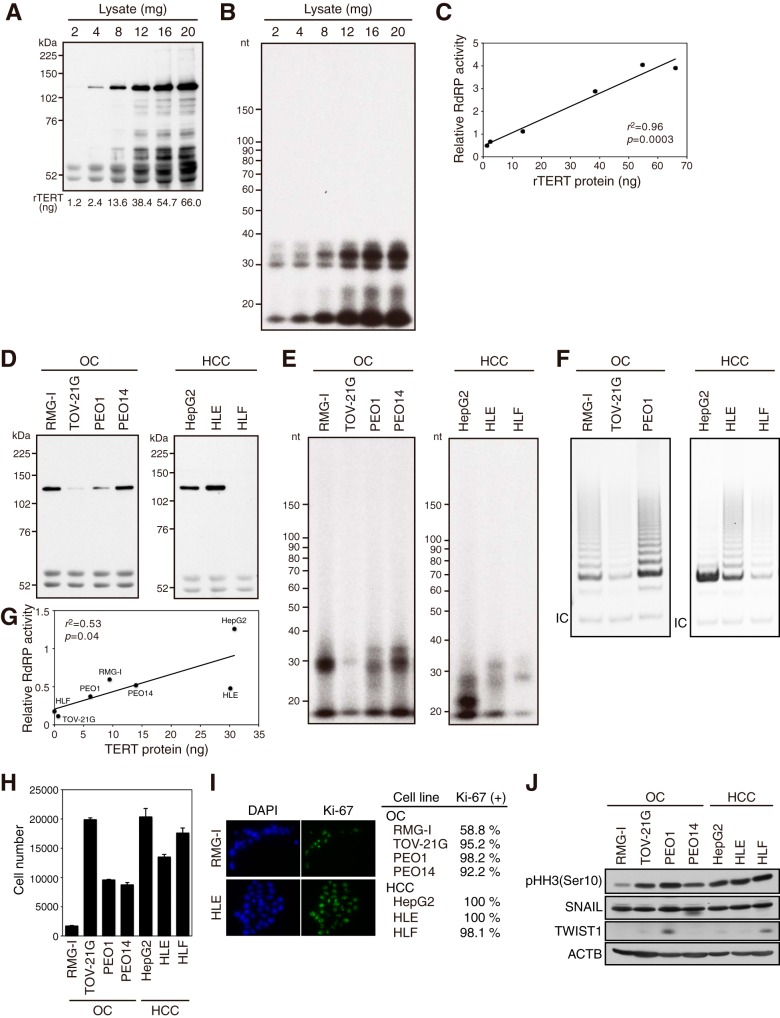
We found an association between TERT protein levels and RdRP activity, as well as telomerase activity, in both HeLa and 293T cells (Fig. 2). To clearly demonstrate the relationship between TERT protein levels and RdRP activity, the levels of rTERT protein (Fig. 6A) and its RdRP activity (Fig. 6B) were analyzed in various amounts of silkworm lysate. The results indicate a significant positive correlation between the level of rTERT and RdRP activity (Fig. 6C).
FIG 6.
TERT protein levels are correlated with RdRP activity. (A to C) Positive correlation between the level of rTERT and RdRP activity. IP-IB (A) and IP-RdRP (B) assays were performed with the indicated amounts of silkworm lysate overexpressing rTERT. The amount of rTERT immunoprecipitated from each lysate was calculated by comparing the signals obtained by IP-IB. (C) Protein levels versus relative RdRP activities for rTERT. Relative RdRP activities were plotted in arbitrary units. (D to G) Expression levels and enzymatic activities of TERT protein in OC and HCC cell lines. (D) IP-IB of OC and HCC cell lines. The signals at ≈52 kDa indicate heavy chains of IgG. (E) IP-RdRP assays of OC and HCC cell lines. (F) IP-TRAP assays of OC and HCC cell lines. IC, internal control. (G) TERT protein level versus relative RdRP activity for each of the OC and HCC cell lines. Mean values for two independent experiments were plotted. (H) Cell growth of OC and HCC cell lines as analyzed by MTT assay. (I) Ki-67-positive ratios were calculated based on immunofluorescence analysis. Representative images are shown. (J) Immunoblot analysis of OC and HCC cell lines.
Since we were able to calculate the rTERT level and RdRP activity in vitro, we monitored the TERT levels and RdRP activities in human cancer cell lines. We first calculated that up to 1,500 molecules of TERT were expressed per HeLa cell, consistent with a previous report (1). TERT protein levels and RdRP activities were then analyzed in human ovarian carcinoma (OC) and hepatocellular carcinoma (HCC) cell lines. TERT expression levels varied among the cell lines (Fig. 6D and G); TERT protein levels obtained from each IP were high (≈30 ng) for HepG2 and HLE cells but low (<1 ng) for TOV-21G and HLF cells. Short RNA products of the IP-RdRP assay were found in OC and HCC cell lines with moderate to high levels of TERT proteins (RMG-I, PEO1, PEO14, HepG2, and HLE), while these products were almost absent in cell lines with low levels of TERT (TOV-21G and HLF) (Fig. 6E). Telomerase activity results were comparable to the RdRP product results: moderate to high levels of telomerase activity were detected in the cell lines with RdRP products, and telomerase activity was very weak in the cell lines with little product (Fig. 6F). The association between the levels of TERT proteins and RdRP products was in agreement with the results obtained using HeLa cells treated with or without nocodazole (Fig. 2A and B), as well as with the results for TERT suppression in both HeLa and 293T cells (Fig. 2C, D, and G). Simple regression analysis revealed a positive correlation between levels of endogenous TERT protein and RdRP activity (Fig. 6G). Taken together, these data suggest that TERT protein levels are the rate-limiting factor for the RdRP activity.
We next examined characteristics of OC and HCC cell lines and TERT protein/RdRP activity. We analyzed cell proliferation as gauged by cell growth (Fig. 6H), Ki-67 expression (Fig. 6I), and phospho-histone H3 (Ser10) levels (Fig. 6J) and metastatic potentialities as gauged by the expression levels of epithelial-mesenchymal transition (EMT)-related genes, such as SNAIL and TWIST1 (Fig. 6J). In these analyses, however, we were unable to find any conclusive relationships between TERT protein/RdRP activity and cell proliferation or EMT-related gene expression.
DISCUSSION
RNA polymerization by telomerase was first described for Tetrahymena (19). Using the telomerase RNA components as the template, Tetrahymena telomerase incorporated rGTP as well as dTTP to synthesize the chimeric product [d(TT)r(GGGG)]n, which was complementary to the template RNA. DNA elongation from the RNA primer by Tetrahymena telomerase was also described in the same study (19), which indicated the competency of the enzyme to bind dsRNAs. We previously reported an RNA primer requirement for synthesis of dsRNA by human TERT through its RdRP activity (4). A fundamental difference between DNA and RNA polymerases is that RNA polymerases do not necessarily require primers to initiate nucleotide polymerization, while DNA polymerases do. Indeed, both viral RdRPs and cell-encoded RdRPs initiate cRNA strand synthesis in either a primer-dependent or -independent manner (15, 16, 20, 21). The biological significance of primer-independent RNA polymerization by these RdRPs has been demonstrated. For example, RdRPs of RNA viruses use a primer-independent (de novo) initiation mechanism for complete replication of their genomes (22, 23). RdRPs in Caenorhabditis elegans, specifically RRF-1 and EGO-1, perform de novo synthesis of 22-nt RNAs, known as secondary siRNAs, which contribute to amplification of RNA silencing signals triggered by primary siRNAs (15, 16). In this study, we confirmed the occurrence of de novo RNA synthesis by the RdRP activity of TERT, suggesting that TERT shares common features with viral and cell-encoded RdRPs in the initiation step of RNA polymerization and that there may be unknown biological processes mediated by the de novo-synthesized RNAs in human cells expressing TERT.
Functional small RNAs are often classified based on their length and nucleotide bias. For instance, C. elegans expresses three different classes of small RNAs: 26G-, 22G-, and 21U-RNAs (24, 25). The names of these RNAs reflect the size of each RNA and the preference for a G or U residue at the 5′ end. Among these RNA classes, 22G-RNAs are secondary siRNAs synthesized de novo by RdRP (25, 26). In addition to 22G-RNA in C. elegans, viral RdRPs usually start de novo RNA synthesis with purine residues (22, 23). Specifically, G is the most universal initiation nucleotide, and a complementary C at or near the 3′ terminus of the RNA template is recognized as the initiation site by viral RdRPs (21). The RNA templates used in this study have a 5′-CCCA-3′ sequence at the 3′ end, and we found that 70.7% of cRNA strand synthesis in the rTERT-RNA 1 set was initiated with G residues, most of which were mapped on the 5′-CCCA-3′ sequence. This result indicates that TERT preserves the common property of RdRPs in priming de novo RNA synthesis.
The preference for 5′ purine residues in nocodazole-RNA 1, however, was lower than that reported for RdRP of C. elegans (RRF-1). In a sequencing analysis of the RNA products of RRF-1, more than 90% of the products possessed a purine residue at the 5′ end (17). The proportion of 5′ purines in nocodazole-RNA 1 (68.0%) was also lower than that in rTERT-RNA 1 (88.3%). Conversely, stringency in the initiation nucleotide would be relaxed in endogenous TERT (nocodazole-RNA 1) compared to rTERT (rTERT-RNA 1). These results imply that there is a mechanism(s) that enhances the flexibility of de novo RNA synthesis by endogenous TERT and thereby increases the diversity of RNA species produced in human cells.
Increased TERT expression and subsequent telomerase activation are common features of human cancers. A recent study by Borah et al. demonstrated that both TERT mRNA and protein levels correlate strongly with telomerase activity in urothelial carcinoma cell lines (27). As a counterpart to research focusing on telomerase activity, we analyzed the protein levels and RdRP activity of endogenous TERT in OC and HCC cells in this study, and we found that TERT protein levels were positively correlated with RdRP activity. It will be important to revisit why most human cancers express TERT based on the understanding that TERT is a multifunctional protein; not only telomere maintenance by telomerase activity but also RNA regulation by RdRP activity would be crucial for human carcinogenesis.
In the past decade, many different types of functional small RNAs have been reported, and we now know that these RNA-related pathways regulate diverse physiological and pathological processes. As found in plants and worms, de novo synthesis of RNAs by TERT RdRP might shed further light on the mechanism underlying RNA silencing in human cells, especially malignant cells. Given that the TERT expression level is critical for the carcinogenic process of various cancers (27–30), in addition to its authentic telomerase activity, the RdRP activity of TERT might be a novel promising molecular target for cancer treatment.
ACKNOWLEDGMENTS
This work was supported in part by the Project for Development of Innovative Research on Cancer Therapeutics (P-DIRECT), Japan Agency for Medical Research and Development (Kenkichi Masutomi), by the Takeda Science Foundation (Yoshiko Maida), and by a research grant of the Princess Takamatsu Cancer Research Fund (grant 13-24520; Yoshiko Maida).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Xi L, Cech TR. 2014. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res 42:8565–8577. doi: 10.1093/nar/gku560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siomi H, Siomi MC. 2009. On the road to reading the RNA-interference code. Nature 457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 4.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. 2009. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maida Y, Yasukawa M, Okamoto N, Ohka S, Kinoshita K, Totoki Y, Ito TK, Minamino T, Nakamura H, Yamaguchi S, Shibata T, Masutomi K. 2014. Involvement of telomerase reverse transcriptase in heterochromatin maintenance. Mol Cell Biol 34:1576–1593. doi: 10.1128/MCB.00093-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto N, Yasukawa M, Nguyen C, Kasim V, Maida Y, Possemato R, Shibata T, Ligon KL, Fukami K, Hahn WC, Masutomi K. 2011. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc Natl Acad Sci U S A 108:20388–20393. doi: 10.1073/pnas.1015171108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T, Kanaya T, Okazaki H, Ogawa K, Usami A, Watanabe H, Kadono-Okuda K, Yamakawa M, Sato H, Mori H, Takahashi S, Oda K. 1997. Efficient protein production using a Bombyx mori nuclear polyhedrosis virus lacking the cysteine proteinase gene. J Gen Virol 78:3073–3080. doi: 10.1099/0022-1317-78-12-3073. [DOI] [PubMed] [Google Scholar]
- 8.Maeda S, Mitsuhashi J. 1989. Gene transfer vectors of a baculovirus Bombyx mori and their use for expression of foreign genes in insect cells, p 167–181. In Invertebrate cell system applications. CRC Press, Boca Raton, FL. [Google Scholar]
- 9.Zamora H, Luce R, Biebricher CK. 1995. Design of artificial short-chained RNA species that are replicated by Q beta replicase. Biochemistry 34:1261–1266. doi: 10.1021/bi00004a020. [DOI] [PubMed] [Google Scholar]
- 10.Lue NF, Bosoy D, Moriarty TJ, Autexier C, Altman B, Leng S. 2005. Telomerase can act as a template- and RNA-independent terminal transferase. Proc Natl Acad Sci U S A 102:9778–9783. doi: 10.1073/pnas.0502252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushina Y, Takeuchi T, Sugawara F, Yoshida H. 2012. Anti-cancer targeting telomerase inhibitors: beta-rubromycin and oleic acid. Mini Rev Med Chem 12:1135–1143. doi: 10.2174/138955712802762220. [DOI] [PubMed] [Google Scholar]
- 12.Yi G, Deval J, Fan B, Cai H, Soulard C, Ranjith-Kumar CT, Smith DB, Blatt L, Beigelman L, Kao CC. 2012. Biochemical study of the comparative inhibition of hepatitis C virus RNA polymerase by VX-222 and filibuvir. Antimicrob Agents Chemother 56:830–837. doi: 10.1128/AAC.05438-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehmann E, Brueckner F, Cramer P. 2007. Molecular basis of RNA-dependent RNA polymerase II activity. Nature 450:445–449. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]
- 14.Wagner SD, Yakovchuk P, Gilman B, Ponicsan SL, Drullinger LF, Kugel JF, Goodrich JA. 2013. RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J 32:781–790. doi: 10.1038/emboj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 16.Pak J, Fire A. 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 17.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J 26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deana A, Celesnik H, Belasco JG. 2008. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 19.Collins K, Greider CW. 1995. Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J 14:5422–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makeyev EV, Bamford DH. 2002. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell 10:1417–1427. doi: 10.1016/S1097-2765(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 21.van Dijk AA, Makeyev EV, Bamford DH. 2004. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol 85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 22.Kao CC, Singh P, Ecker DJ. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 23.Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol 74:851–863. doi: 10.1128/JVI.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. 2012. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet 8:e1002702. doi: 10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D Jr, Mello CC. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, Chen CC, Chaves DA, Duan S, Kasschau KD, Fahlgren N, Yates JR III, Mitani S, Carrington JC, Mello CC. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, Costello JC, Theodorescu D, Cech TR. 2015. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347:1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. 2013. Highly recurrent TERT promoter mutations in human melanoma. Science 339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, Tsuji S, Donehower LA, Slagle BL, Nakamura H, Yamamoto S, Shinbrot E, Hama N, Lehmkuhl M, Hosoda F, Arai Y, Walker K, Dahdouli M, Gotoh K, Nagae G, Gingras MC, Muzny DM, Ojima H, Shimada K, Midorikawa Y, Goss JA, Cotton R, Hayashi A, Shibahara J, Ishikawa S, Guiteau J, Tanaka M, Urushidate T, Ohashi S, Okada N, Doddapaneni H, Wang M, Zhu Y, Dinh H, Okusaka T, Kokudo N, Kosuge T, Takayama T, Fukayama M, Gibbs RA, Wheeler DA, Aburatani H, Shibata T. 2014. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 30.Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Kramer A, Roncaioli JL, Sand F, Heuckmann JM, Ikram F, Schmidt R, Ackermann S, Engesser A, Kahlert Y, Vogel W, Altmuller J, Nurnberg P, Thierry-Mieg J, Thierry-Mieg D, Mariappan A, Heynck S, Mariotti E, Henrich KO, Gloeckner C, Bosco G, Leuschner I, Schweiger MR, Savelyeva L, Watkins SC, Shao C, Bell E, Hofer T, Achter V, Lang U, Theissen J, Volland R, Saadati M, Eggert A, de Wilde B, Berthold F, Peng Z, Zhao C, Shi L, Ortmann M, Buttner R, Perner S, Hero B, Schramm A, Schulte JH, Herrmann C, O'Sullivan RJ, Westermann F, Thomas RK, Fischer M. 2015. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]