Abstract
Histone chaperones, like nucleosome assembly protein 1 (Nap1), play a critical role in the maintenance of chromatin architecture. Here, we use the GAL locus in Saccharomyces cerevisiae to investigate the influence of Nap1 on chromatin structure and histone dynamics during distinct transcriptional states. When the GAL locus is not expressed, cells lacking Nap1 show an accumulation of histone H2A-H2B but not histone H3-H4 at this locus. Excess H2A-H2B interacts with the linker DNA between nucleosomes, and the interaction is independent of the inherent DNA-binding affinity of H2A-H2B for these particular sequences as measured in vitro. When the GAL locus is transcribed, excess H2A-H2B is reversed, and levels of all chromatin-bound histones are depleted in cells lacking Nap1. We developed an in vivo system to measure histone exchange at the GAL locus and observed considerable variability in the rate of exchange across the locus in wild-type cells. We recapitulate this variability with in vitro nucleosome reconstitutions, which suggests a contribution of DNA sequence to histone dynamics. We also find that Nap1 is required for transcription-dependent H2A-H2B exchange. Altogether, these results indicate that Nap1 is essential for maintaining proper chromatin composition and modulating the exchange of H2A-H2B in vivo.
INTRODUCTION
The basic unit of chromatin is the nucleosome, which forms when DNA is wrapped around two copies each of the four core histones arranged as two histone H2A-H2B dimers and a histone H3-H4 tetramer (1). Nucleosomes are highly dynamic, capable of multiple structural transitions between completely assembled and entirely disassembled structures (2). Indeed, H2A-H2B and H3-H4 are actively exchanged during both DNA replication-dependent and -independent events (3–9). Chromatin transitions have the potential to profoundly affect gene expression, and a diverse spectrum of factors, including histone chaperones, participate in this process (10).
Histone chaperones are histone-binding proteins that facilitate nucleosome assembly and/or disassembly in an ATP-independent fashion (11–13). The histone chaperone nucleosome assembly protein 1 (Nap1) is a highly conserved chaperone that binds H2A-H2B in vitro with nanomolar affinity (12, 14) in a conformation that shields interfaces required for nucleosome assembly (15). Although functional in the assembly of nucleosomes in vitro (16), a number of studies support a role for Nap1 in transcription-dependent processes of disassembly of nucleosomes. Nap1 is critical for the eviction of histones during transcription in a mammalian in vitro system (17), and Nap1 (with the ATP-dependent chromatin remodeler RSC [remodels the structure of chromatin]) can facilitate the elongation of RNA polymerase II (RNAPII) on chromatin templates using yeast in vitro systems (18, 19). Our previous in vivo studies indicated that Nap1 prevents excess H2A-H2B accumulation on chromatin (20), and here, we expand our analysis to investigate the role of Nap1 in histone exchange and occupancy. As a model system, we use the GAL locus in yeast under transcriptionally repressed and activated conditions.
The Saccharomyces cerevisiae GAL gene cluster (GAL7, GAL1, and GAL10) is a powerful system for studying chromatin dynamics in vivo under variable levels of gene expression. Transcription of GAL genes is repressed in the presence of glucose and coordinately activated in the presence of galactose (21, 22). The region is characterized by high levels of nucleosome occupancy in the repressed state (23–25). Upon activation, histones are acetylated (26–33), and a majority are evicted (24, 34–36) with the help of chromatin-remodeling factors (37) and histone chaperones (38).
Although chromatin structure and histone modifications at the GAL locus are well characterized, probing of histone dynamics has been confounded by the fact that typical exchange studies utilize a tagged histone expression system regulated by a galactose-inducible promoter (4–9). We therefore constructed a novel exchange system in which the expression of a hemagglutinin (HA)-tagged histone (HAH2B or H3HA) is regulated by the antibiotic doxycycline. This allowed us to examine histone exchange at these model genes without altering the carbon source. We found that certain nucleosomes are highly dynamic and require Nap1 for high levels of histone H2A-H2B exchange. Nap1 is also needed for maintaining normal histone density and preventing excess H2A-H2B accumulation.
MATERIALS AND METHODS
Yeast strains, plasmids, and culture conditions.
The following S. cerevisiae yeast strains were purchased from Thermo Scientific Open Biosystems: the wild-type (wt) strain (BY4741 MATa his3Δ1 ura3Δ0 leu2Δ0 met15Δ0) (catalog number YSC1048) and the nap1Δ strain (BY4741 nap1Δ::Kanr) (catalog number YSC6273-201936599). The strain containing tandem affinity purification (TAP)-tagged Htz1 was purchased from GE Dharmacon (BY4741 Htz1-TAP::HIS3MX6) (catalog number YSC1178-202233238). TAP-tagged Htz1 in the nap1Δ strain was generated by homologous recombination of a PCR product derived from TAP-tagged Htz1 in the wild-type strain. For replication-independent histone exchange studies, α-factor (catalog number RP01002; GenScript) was added to arrest cells in the G1 stage. To prevent α-factor degradation by Bar1 (39), a BAR1 gene deletion strain (bar1Δ) (catalog number YSC6273-201920294; Thermo Scientific) was used as the wild type, and the nap1Δ strain was generated in this background by using established protocols (40).
Plasmids for doxycycline-regulated HAH2B and H3HA expression were generated by subcloning from the galactose-regulated plasmids pGAL1 HA-H2B and pGAL1 H3-HA (both containing 3 contiguous HA sequences [3×HA]), which were generously provided by Michel Strubin (4). A NotI-BamHI DNA fragment from pGAL1 HA-H2B encoding HAH2B was cloned into the doxycycline-regulated pCM188 plasmid (ATCC 87660) by using standard techniques (41). The H3HA gene was amplified from the pGAL1 H3-HA plasmid via PCR using Pfu Turbo. The primers used for PCR were designed to incorporate a BamHI or EagI site near the ends of the PCR product. The resulting product, the H3HA-encoding DNA, was cleaved with BamHI and EagI and cloned into the doxycycline-regulated pCM188 plasmid.
To analyze histone density and transcription levels of the GAL genes after continuous transcriptional activation, wild-type and nap1Δ strains were grown overnight in either yeast extract-peptone (YP)–glucose (2%) medium or YP-galactose (2%) medium, diluted to an optical density at 600 nm (OD600) of ∼0.2, and then allowed to undergo 2 cell doublings. When cultures reached an OD600 of 0.8 to 1.0, the cells were collected and subjected to chromatin immunoprecipitation (ChIP), immunoblot, or RNA abundance analysis. For the histone exchange experiments, wild-type and nap1Δ strains (both in the bar1Δ background) were cultured in YP glucose or YP galactose medium continuously (>24 h) to early log phase (OD600 = 0.2). α-Factor was then added to the medium to a final concentration of 5 μM, and after 90 to 120 min, cells were arrested in G1 phase. Shmooing was confirmed visually by using a microscope. Doxycycline was added to a final concentration of 1 μg/ml in YP glucose medium or to a final concentration of 3 μg/ml in YP galactose medium to maintain a consistent histone degradation rate. Following doxycycline treatment, cells were collected every hour for 5 h and subjected to chromatin immunoprecipitation.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (42) assays were performed as previously described (20). The cells were cross-linked with 1% formaldehyde and lysed by bead beating. The cross-liked chromatin was then sheared into small fragments of ∼200 bp with sonication. Antibodies and Sepharose beads were added to immunoprecipitate the cross-linked proteins of interest. Cross-linking of the immunoprecipitated material was reversed by incubation overnight at 65°C, and the samples were then treated with proteinase K and RNase A, followed by DNA extraction using the phenol-chloroform method. The isolated DNA was analyzed by quantitative real-time PCR (qPCR). The commercially available antibodies used in this study were anti-H2B (catalog number 39237; Active Motif), anti-H3 (catalog number 1791; Abcam), and anti-HA (catalog number sc-7392; Santa Cruz). ChIP assays were performed in biological triplicates. Unnormalized occupancy was directly calculated from the change in the quantification cycle (ΔCq) values between the immunoprecipitated DNA and the input control DNA. For the exchange assays, the ratio of the unnormalized HA-tagged histone occupancy over the respective unnormalized total (endogenous plus HA-tagged) histone occupancy at each region for each time point of doxycycline treatment was calculated. The average ratio from three biological replicates at the 0-h time point (T0) was set as 100% histone persistence, and the average ratios at the remaining time points were calculated as a percentage of histone persistence relative to that at T0. The histone persistence percentages are shown as means ± standard deviations in Fig. S7 in the supplemental material for comparison between the wt and nap1Δ strains. Heat maps (see Fig. 5) were generated by assigning colors to the blocks representing each region during the time course according to the histone persistence percentages. For all other ChIP assays, the total occupancy of each protein at the GAL region was normalized to its respective occupancy at a telomere-proximal position (chromosome VI positions 269571 to 269488), which served as the internal control.
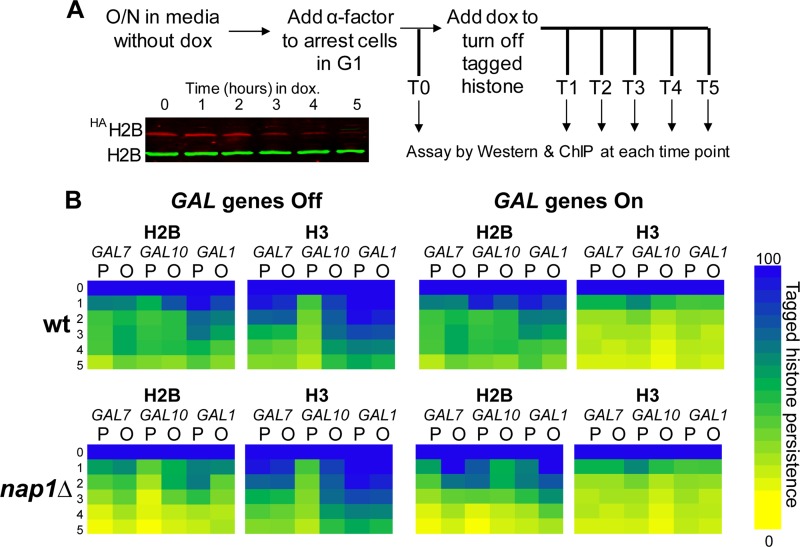
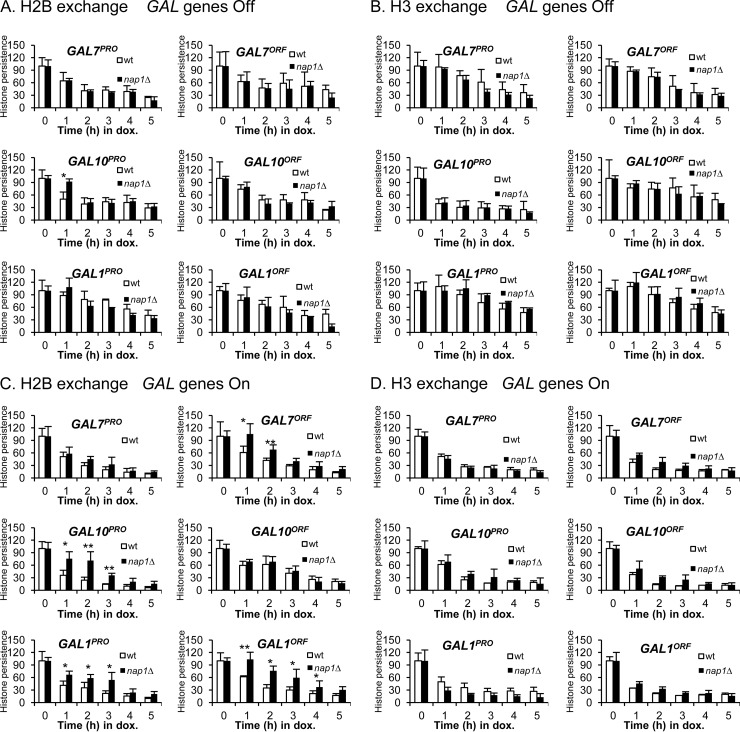
FIG 5.
Histone exchange rates at the GAL locus are distinct, and Nap1 is involved in H2A-H2B exchange. (A) Schematic of histone exchange assays with a representative Western blot showing decreasing HA-tagged H2B levels and unchanged endogenous H2B levels. O/N, overnight; dox, doxycycline. (B) Heat maps showing tagged histone persistence over the 5-h time course. Persistence is defined as the percentage of tagged histone occupancy (detected with anti-HA) over endogenous histone occupancy (detected with anti-H3) under transcriptionally repressed (“Off”) or activated (“On”) conditions in the wt and nap1Δ strains. The tagged histone persistence at T0 was set as 100% (dark blue). Bright yellow indicates that tagged histone was completely exchanged (0% tagged histone persistence). Promoter (P) and ORF (O) regions are indicated.
RNA abundance.
S1 nuclease protection assays to quantify the expression levels of HAH2B, GAL7, GAL10, GAL1, and tRNAw were performed as previously described (43). RNA was extracted with the traditional hot-phenol method. For each reaction, 32P-labeled S1 probes were incubated with 40 μg of RNA at 55°C overnight. The hybridized samples were digested by S1 nuclease at 37°C for 30 min and separated by 8% acrylamide sequencing gels. The gels were exposed to phosphor screens, visualized by using a Typhoon PhosphorImager, and analyzed by using ImageQuant software.
Immunoblotting of total protein levels.
Cells were harvested at log phase, suspended in lysis buffer (120 mM Tris-HCl [pH 6.8], 12% [vol/vol] glycerol, 3.4% [wt/vol] SDS, 200 mM dithiothreitol [DTT], 0.004% [wt/vol] bromophenol blue), and incubated at 95°C and on ice for 5 min each. Insoluble cell debris was removed by centrifugation, and total protein was separated on a 15% SDS-PAGE gel. The same antibodies against HA, H2B, and H3 that were used for ChIP analysis were used for protein detection. For the quantification of total protein levels during histone exchange, the protein level of each strain at T0 was set as 100%; the protein levels at the remaining time points were calculated as a percentage relative to that at T0.
In vitro H2A-H2B dimer-binding competition assay.
Förster resonance energy transfer (FRET)-based competition assays were conducted in a 384-well format using Alexa Fluor 488-labeled Xenopus laevis histone H2A-H2B (with a threonine 118-to-cysteine mutation in H2B for labeling purposes) as described previously (44). The final reaction buffer contained 20 mM Tris-HCl (pH 7.5), 300 mM NaCl, 5% (vol/vol) glycerol, 0.01% (vol/vol) NP-40, 0.01% (vol/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1 mM DTT, and 1 mM EDTA (pH 8.0), and the reaction volume was 40 μl. Each PCR-prepared fragment was titrated against 10 nM histone H2A-H2B in the presence 50 nM 59-bp DNA. Each reaction was performed in duplicate, and control titrations containing only one labeled component were used to correct the raw FRET signal to produce FRETcorrected. Each experiment contained an internal control titration of recombinant 147-bp Widom 601 DNA (data not shown). Reaction mixtures were pipetted into a microplate and scanned by using a Typhoon Trio variable-mode imager (GE). Three scans were performed, using excitation/emission wavelengths of 488/520 nm, 633/670 nm, or 488/670 nm. Fluorescence was quantified by using ImageQuant software. GAL fragment titration produced a sigmoidal curve from which a 50% inhibitory concentration (IC50) was determined by using GraphPad Prism software. Data were normalized and fit to have a minimum of 0 and a maximum of 100%. At least two independent titrations were performed for each fragment, and all IC50 values used in the mean calculation were derived from data with an R2 of >0.98.
In vitro micrococcal nuclease analysis of chromatin.
In vitro micrococcal nuclease (MNase) digestion assays were performed according to established protocols (45). Canonical mononucleosomes and trinucleosomes were reconstituted by salt dilution using a purified recombinant Xenopus laevis octamer on a 207-bp DNA fragment containing the 147-bp Widom 601 nucleosome-positioning sequence with 30 bp of flanking linker DNA (46). Nucleosomal DNA or free DNA (5 μg; 0.17 μM) was used for each MNase digestion. MNase digestion was performed in the absence and presence of excess recombinant Xenopus laevis histone H2A/H2B dimer. A 3-fold molar excess of histone H2A-H2B dimer was added to a final concentration of 0.51 μM, and the mixture was incubated for 5 min, followed by MNase digestion at room temperature for the indicated times. Reactions were quenched with EDTA to a final concentration of 12.5 mM. The DNA was purified by the addition of SDS (0.5%) and proteinase K (0.2 mg/ml), and the mixture was incubated at 50°C for 30 min, phenol-chloroform extracted, and ethanol precipitated. The samples were separated on a 6% native polyacrylamide gel and stained with SYBR gold.
Micrococcal nuclease analysis of chromatin structure in vivo.
Detailed protocols for micrococcal nuclease analysis of the chromatin structure in vivo can be found in the supplemental material. Nuclei were isolated from wild-type (BY4741) and nap1Δ cells essentially as described previously (47). Indirect end-labeling analysis was completed as described previously (24), and primer extension analysis was performed as described previously (48). The membranes and gels were exposed to phosphor storage screens, visualized on a Typhoon PhosphorImager, and quantitated by using ImageQuant TL software.
Reconstitution studies of GAL fragments.
Nucleosome reconstitution was performed by using the salt dilution method (49). Xenopus laevis histone octamers were titrated against each PCR-prepared DNA fragment. Reaction mixtures were separated on 5% native gels, stained with ethidium bromide, and visualized by UV.
RESULTS
Nap1 prevents excess histone H2A-H2B accumulation on linker DNA.
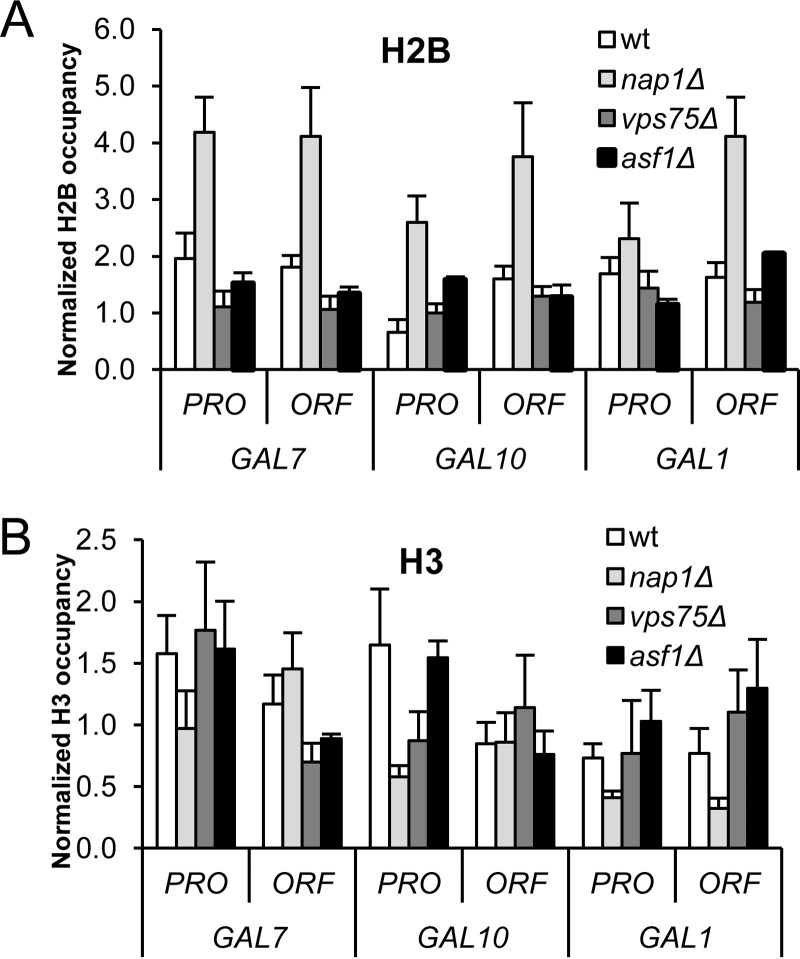
We previously showed that a knockout of the histone chaperone NAP1 in S. cerevisiae results in an atypical chromatin structure with excess H2A-H2B at most positions across the GAL locus (20). We next asked whether this was a general feature of Nap family proteins by assaying a strain deleted for Vps75, a histone chaperone with a structure similar to that of Nap1 (50). We utilized chromatin immunoprecipitation (ChIP) assays to visualize H2A-H2B (via H2B) and H3-H4 (via H3) occupancy at six different regions of the GAL locus. The relative positions of the sites were selected based on extensive studies of nucleosome positioning, histone density, and histone variant H2A.Z occupancy (51–55) and include three promoter (PRO) and three open reading frame (ORF) sites (see Fig. S1 in the supplemental material). The GAL10 and GAL1 genes are divergently transcribed and share one upstream activating sequence (UAS) (23, 56, 57). As shown previously (20), loss of Nap1 (nap1Δ) causes a general increase in the level of H2A-H2B compared to that in the wild-type strain at all sites but one (Fig. 1A). At these sites, the occupancy of H3-H4 remains at or slightly below wild-type levels (Fig. 1B). In contrast, deletion of VPS75 (vps75Δ) does not significantly change H2A-H2B or H3-H4 occupancy across the GAL locus. We also deleted ASF1 (asf1Δ), a histone chaperone structurally distinct from Nap1 and Vps75 (58), and detected no significant changes in H2A-H2B or H3-H4 occupancy. Thus, excess H2A-H2B accumulation is not a shared feature of strains deleted for Nap family proteins or other histone chaperones but is unique to the nap1Δ strain. Together, these data indicate that preventing this atypical chromatin is a unique function of Nap1.
FIG 1.
Excess H2A-H2B accumulation is observed with loss of Nap1 but not Vps75 or Asf1. Shown is the occupancy of histone H2B (A) or histone H3 (B) at the GAL locus under transcriptionally repressed conditions (glucose) in the wild-type (wt), nap1Δ, vps75Δ, and asf1Δ strains as determined by ChIP assays. Data are represented as the means from 3 biological replicates ± standard deviations, normalized to the levels of occupancy at a telomere-proximal location.
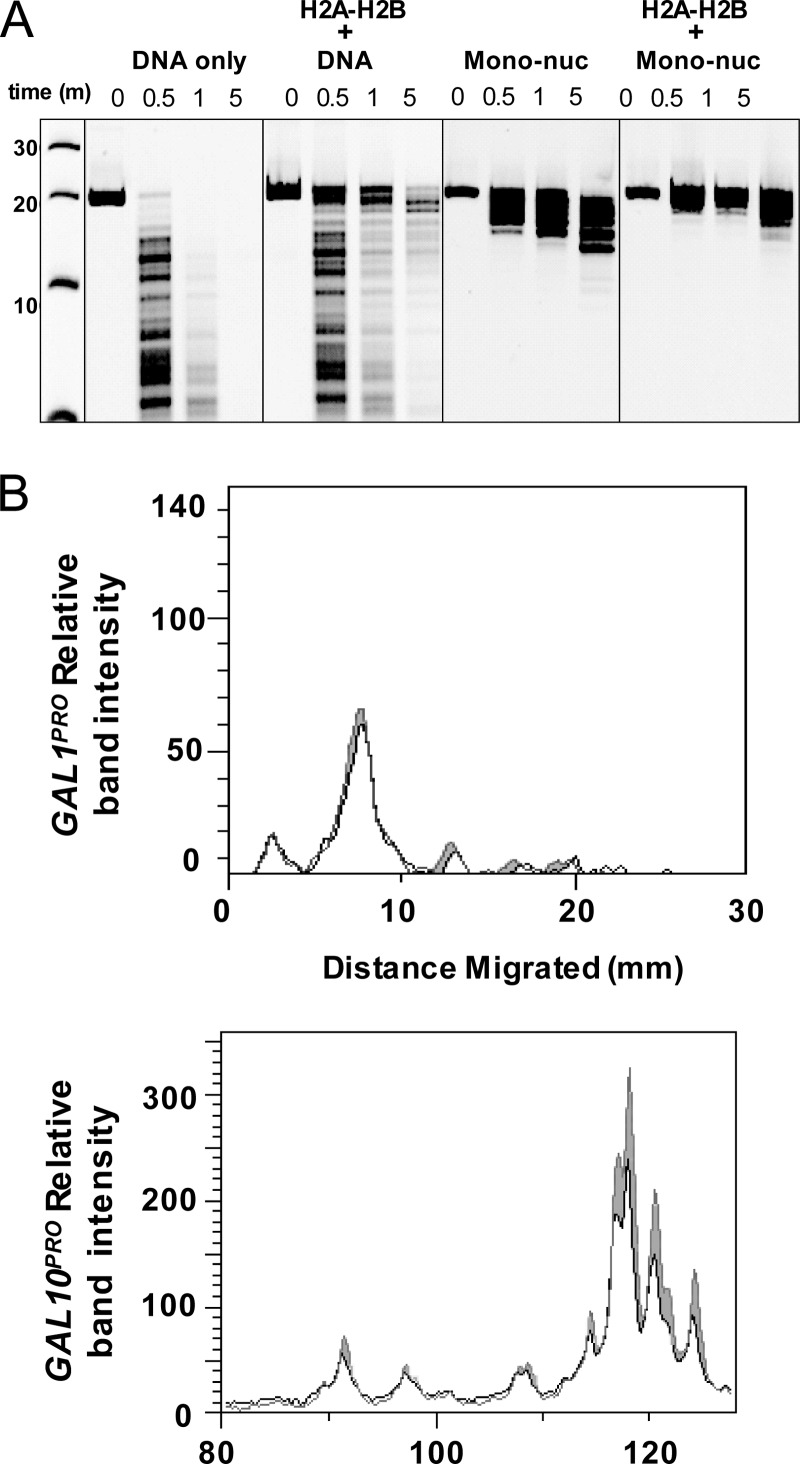
We next characterized chromatin with H2A-H2B enrichment biochemically. To mimic the atypical chromatin observed in vivo in the nap1Δ strain, mononucleosomes were incubated in the presence (and absence) of excess H2A-H2B, and the micrococcal nuclease (MNase) digestion patterns were compared (Fig. 2A). We used a 207-bp DNA fragment carrying the well-characterized 147-bp Widom 601 nucleosome-positioning sequence (59) and 30 bp of flanking linker DNA. As expected, free DNA was rapidly digested by MNase. Incubation of DNA with H2A-H2B resulted in modest protection from MNase. A clearly defined protected region was not observed, consistent with the nonspecific binding of H2A-H2B to DNA. MNase digestion of mononucleosomes revealed a well-defined 140- to 180-bp region (depending on the digestion time) of protection, indicating that the linker DNA is digested more rapidly than nucleosomal DNA (45). Mononucleosomes with excess H2A-H2B displayed significantly increased resistance to MNase digestion and broader DNA protection surrounding the nucleosome site. This suggests that excess H2A-H2B binds to the flanking linker DNA.
FIG 2.
H2A-H2B accumulation protects DNA from MNase digestion. (A) In vitro analysis of chromatin with and without excess H2A-H2B. Shown are data from PAGE analysis of MNase digestion of naked DNA alone, DNA with H2A-H2B, mononucleosomes (Mono-nuc), and mononucleosomes with H2A-H2B, assessed on a 207-bp DNA fragment. (B) Chromatin from wild-type and nap1Δ cells was digested with MNase in vivo, and the cleavage products were analyzed by primer extension assays. Quantitation of the primer extension products from the GAL1 promoter (top) and the GAL10 promoter (bottom) is shown in black for nap1Δ cells and in gray for wild-type cells. The GAL10 promoter, which has excess H2A-H2B as determined by ChIP assays, is more protected from MNase cleavage in nap1Δ cells.
We also performed MNase digestion assays on two other nucleosome templates: a 621-bp DNA fragment capable of forming trinucleosomes with three consecutive 207-bp Widom 601 nucleosome-positioning sequences with linker DNA (see Fig. S2A in the supplemental material) and a 588-bp native promoter DNA fragment without detectable positioning sequences (60) (see Fig. S2B in the supplemental material). Similar results were observed for all DNA fragments: the addition of excess H2A-H2B to reconstituted nucleosomes significantly increased resistance to MNase digestion. These results indicate that excess H2A-H2B binds to linker DNA in the context of both mono- and multinucleosome templates.
We next assayed how excess H2A-H2B affects chromatin architecture in vivo by measuring MNase digestion patterns of chromatin from wild-type and nap1Δ cells. Cleavage patterns within the GAL locus were assessed by high-resolution (single-base-pair) reiterative primer extension analysis. The GAL1 and GAL10 promoters were analyzed, since the GAL1 promoter had normal levels and the GAL10 promoter had excess levels of H2A-H2B in the nap1Δ strain as determined by ChIP analysis (Fig. 1). As expected, at the GAL1 promoter, there were no differences observed in the MNase digestion patterns of chromatin from wild-type and nap1Δ cells (Fig. 2B; see also Fig. S3A in the supplemental material). The GAL10 promoter region is hypersensitive to MNase cleavage, and this is reflected in the amount of product that terminates within the hypersensitive region (Fig. 2B; see also Fig. S3B in the supplemental material). Although subtle, MNase cleavage of the chromatin from nap1Δ cells is reproducibly reduced in the region of GAL10 that exhibits excess H2A-H2B by ChIP analysis compared to the chromatin from wild-type cells. The minor reduction in MNase cleavage is not unexpected. Nucleosomes in the GAL locus evaluated at high resolution in fact occupy multiple positions (36). Thus, the excess dimer is most likely binding at multiple positions, leading to the subtle difference seen in the primer extension products. Taken together, the in vitro and in vivo MNase results suggest that the excess H2A-H2B is binding to the linker DNA.
Occupancy of the histone variant H2A.Z is unchanged in the nap1Δ strain.
Histone H2A.Z (Htz1 in yeast) is a variant of histone H2A that substitutes for the canonical major histone H2A in a wide, but nonrandom, genomic distribution (61). H2A.Z is involved in transcription regulation, gene activation and silencing, DNA repair, and chromosomal stability (62). Since Nap1 also interacts with H2A.Z (63), we investigated whether the occupancy of H2A.Z is altered in the absence of Nap1. Chromatin tandem affinity purification (TAP) was performed by using TAP-tagged Htz1 (Htz1-TAP) in the wild-type or nap1Δ strain with analyses of the GAL locus genes. In wild-type cells, significant occupancy of Htz1 was detected across the region, and this occupancy was unaffected by the loss of Nap1 (see Fig. S4 in the supplemental material). These results indicate that Nap1 is not required for normal levels of Htz1 occupancy at these specific regions, nor does Htz1 accumulate in the same manner as major H2A in this region in the absence of Nap1.
Histone H2A-H2B accumulation in the nap1Δ strain is independent of DNA sequence.
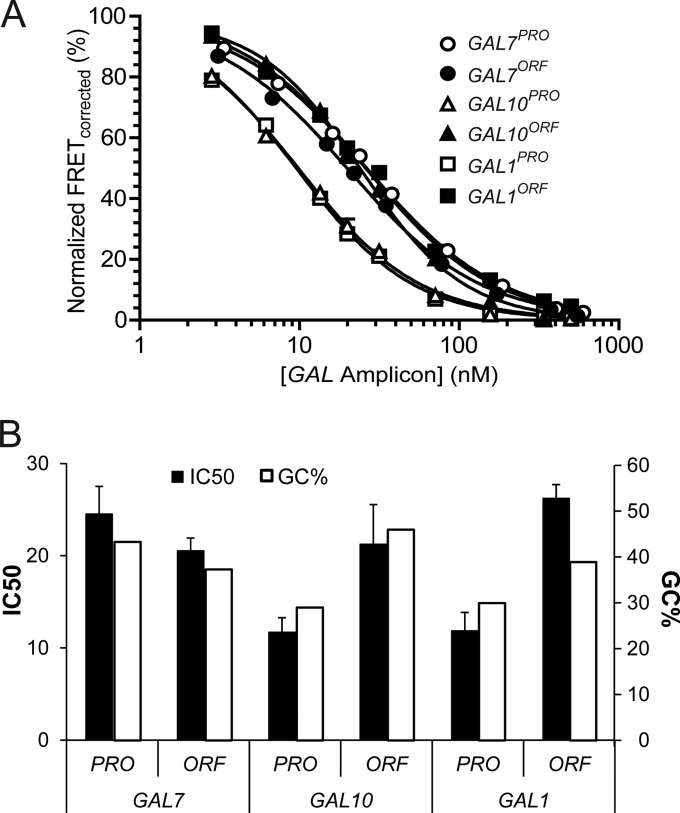
The various degrees of H2A-H2B accumulation across the GAL locus in nap1Δ cells (Fig. 1) prompted us to test the role of DNA sequence. Some studies suggest that DNA sequence is a major factor in nucleosome positioning (64, 65), while others argue that it may be only a partial factor (66–69). To test the role of sequence in vitro, we used a Förster resonance energy transfer (FRET) competition assay (44) to compare H2A-H2B affinities for the six different GAL promoter (PRO) and open reading frame (ORF) sequences, all of ∼150 bp. We first measured FRET of a complex between H2A-H2B and a 59-bp DNA fragment containing the H2A-H2B-binding region of the well-characterized Widom 601 nucleosome-positioning sequence (59). We titrated each of the GAL locus DNAs and measured the half-maximal inhibitory concentration (IC50) values as the FRET signal decreased to zero (Fig. 3A). A lower IC50 reflects a lower dissociation constant (Kd) or a tighter interaction between the GAL locus DNA and H2A-H2B. All GAL locus DNAs were able to fully compete with the 59-bp DNA for H2A-H2B, indicating efficient H2A-H2B-GAL DNA interactions. Slight differences in IC50s were observed among the GAL DNAs (10 to 30 nM), which did not correlate with H2A-H2B accumulation in nap1Δ cells. For example, the lowest IC50 was measured with the GAL10PRO and GAL1PRO DNAs, which in fact possessed the highest and lowest levels, respectively, of H2A-H2B accumulation in vivo. We noticed, however, that the in vitro binding of H2A-H2B correlates with the DNA GC content (Fig. 3B). DNA with a lower CG content is more flexible (70), consistent with the lower Kd for H2A-H2B binding. Overall, these data indicate that the DNA sequences themselves do not account for the varied H2A-H2B accumulation across the GAL locus in the absence of Nap1 in vivo. This implies the involvement of other extrinsic factors.
FIG 3.
All six GAL DNA fragments bind histone H2A-H2B with comparable affinities. H2A-H2B-DNA-binding affinities were measured with a FRET-based competition assay. DNA fragments (∼150 bp) from the six promoter (PRO) and open reading frame (ORF) sites of GAL7, GAL10, and GAL1 were titrated in the presence of 50 nM the 59-bp dimer-binding region of the 601 sequence and analyzed for binding to Alexa Fluor 488-labeled histone H2A-H2B. (A) Representative competition curves. Each data point reflects the mean of results from duplicate measurements ± 1 standard error of the mean. The errors bars are small and in many cases not visible. All curves used for statistics had an R2 value of ≥0.98. (B) Mean IC50 values for at least 2 replicates and percent GC contents for the DNA sequences. Error bars reflect 1 standard error of the mean.
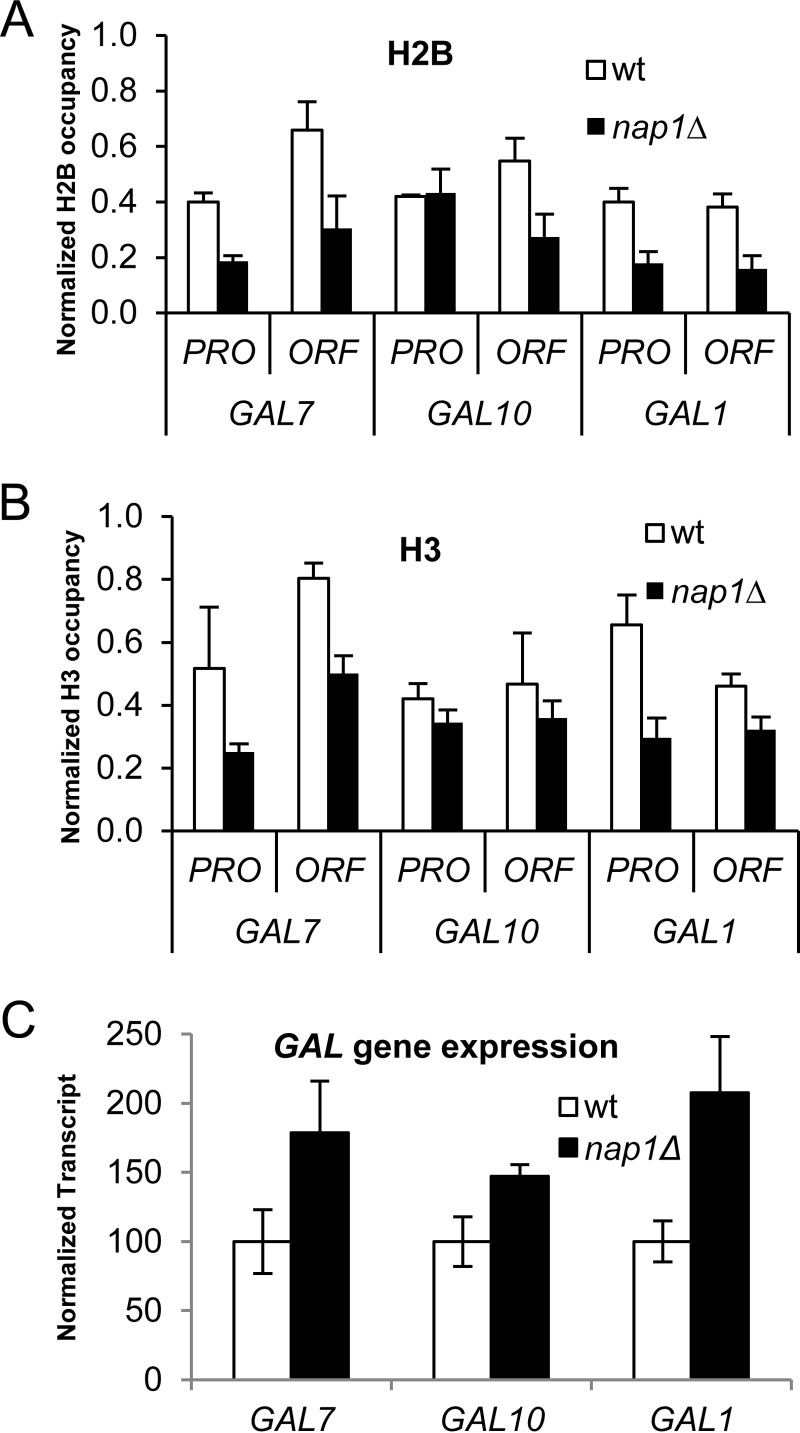
Transcription removes accumulated histone H2A-H2B in the nap1Δ strain.
The above-mentioned accumulation of H2A-H2B in the nap1Δ strain was observed under repressed conditions for GAL gene expression (glucose medium). We next asked if this atypical H2A-H2B distribution remained when the GAL genes were actively transcribed. Histone occupancy was assayed by using ChIP after growth under inducing conditions (galactose medium) for 24 h or ∼10 generations. The ratio of H2A-H2B occupancy (H2B) to H3-H4 occupancy (H3) at each location revealed no significant difference between the wild-type and nap1Δ strains or between different GAL loci (Fig. 4A and B; see also Fig. S5A in the supplemental material). This indicates that excess H2A-H2B does not stably accumulate during active transcription. It is important to note, however, that the occupancies of both H2B and H3 were lower in the nap1Δ strain than in the wild type at every site tested except GAL10PRO, where the occupancy in the nap1Δ strain was similar to that in the wild type. Analysis of extracts confirmed that this decrease in histone occupancy is not due to lower overall levels of histones in the nap1Δ strain (see Fig. S5B in the supplemental material). The lower nucleosome occupancy in galactose in cells lacking Nap1 thus reflects a change in histone deposition, supporting a nucleosome assembly role for Nap1. Since the transcribing GAL locus in the nap1Δ strain has lower histone occupancy, we tested whether this impacted gene expression using S1 nuclease protection assays (Fig. 4C). High expression levels were observed from the GAL genes in nap1Δ cells relative to the wild-type strain. Thus, the reduced histone density at the GAL locus observed in the absence of Nap1 is linked to increased transcription levels, implicating Nap1 in the repression of gene expression via maintenance of histone occupancy, potentially through nucleosome assembly in vivo.
FIG 4.
H2A-H2B accumulation is not maintained in the nap1Δ strain at the GAL locus under induced conditions. (A and B) The occupancies of H2B (A) or H3 (B) in the wt strain and the nap1Δ strain under active transcription were determined by ChIP assays. (C) Transcript levels of GAL genes in the wt and nap1Δ strains were detected by S1 nuclease protection assays with 32P-labeled probes specific to GAL7, GAL10, GAL1, and tRNAw. The transcript levels of tRNAw were used as an internal control for normalization. Bars reflect the means from 3 biological replicates ± standard deviations.
Establishment of an in vivo doxycycline-regulated histone exchange system.
To further investigate the role of Nap1, we developed a method to measure histone exchange at the GAL locus. We created an in vivo system in which the expression of epitope-tagged histones is controlled by using doxycycline (Fig. 5A). Previously reported histone exchange studies utilized a system in which tagged histone expression is GAL promoter regulated and therefore induced by the presence of galactose (4–9). Since we wanted to evaluate histone exchange specifically at the GAL locus, we needed a different means to control histone expression. We subcloned HA-tagged versions of histones H2B (HAH2B) and H3 (H3HA) (4) into a doxycycline-controlled expression vector. In this system, expression of the HA-tagged histones is repressed when doxycycline is present. These plasmids were introduced into both the wild-type and nap1Δ strains, in a bar1Δ background. The bar1Δ background increases α-factor sensitivity, which arrests cells in G1 (39), such that subsequent studies are replication independent. As expected, the addition of doxycycline resulted in transcriptional repression of the HA-tagged histone and did not affect GAL gene expression in either wild-type or nap1Δ cells (see Fig. S6 in the supplemental material). After the addition of doxycycline, the level of total H2B protein (endogenous plus HAH2B, detected by anti-H2B antibody) was not affected, whereas the level of HAH2B protein (detected by anti-HA antibody) gradually diminished (see Fig. S7A and B in the supplemental material). After 5 h of repression by doxycycline, the HAH2B level dropped to <20% of the predoxycycline levels, with no impact on endogenous histone levels.
We next utilized ChIP assays to measure the occupancy of HAH2B and total H2B at a telomere-proximal location as a standard for the assay. H2B exchange was observed at this location (see Fig. S7C and D in the supplemental material). During the 5 h following doxycycline addition, the decrease in HAH2B protein levels correlated directly with the decrease in HAH2B occupancy, whereas the occupancy of total H2B was not affected by doxycycline treatment. Histone exchange data for the telomere-proximal control region with doxycycline-regulated H3HA were similar to those obtained with HAH2B (data not shown), and it is important to note that neither H2B (tagged or untagged) nor H3 (tagged or untagged) levels were impacted by the deletion of Nap1 (see Fig. S8 in the supplemental material).
Histone exchange rates vary across the GAL locus.
We utilized the doxycycline-regulated histone exchange system to determine exchange rates at the GAL locus. We tested both wild-type and nap1Δ strains and compared them under repressed and transcriptionally active conditions (Fig. 5B). To visualize and compare the complex histone exchange patterns at the different GAL loci, the percentage of HA-tagged histone that persists at each site is shown as a heat map. Under transcriptionally repressed conditions, exchange of both H2B and H3 was observed in the wild-type strain at all promoter and open reading frame sites tested (Fig. 6A and B). However, the exchange kinetics differ, depending on the site. Histone H2B exchange is the least dynamic at GAL1PRO and GAL1ORF, where the exchange rate is similar to the exchange rate at the telomere-proximal region. The most dynamic exchange was observed at GAL10PRO, where persistence of the tagged derivative was down to 50% within an hour. In general, H2B exchange is significantly more dynamic than H3 exchange across the GAL locus, a trend that was also shown in a previous study (4). The one exception to this trend is GAL10PRO, where the exchange of H3 is as dynamic as that of H2B. The rapid exchange of both H2A-H2B and H3-H4 at GAL10PRO indicates a highly dynamic chromatin structure at this location, even when transcription is off. Under transcriptionally repressed conditions, there was very little impact of the loss of Nap1 on histone H2B or H3 exchange at the promoter and open reading frame of all GAL sites tested, except for a decrease in the exchange of H2B at the otherwise highly dynamic GAL10PRO site. The absence of Nap1 reduces H2B exchange but does not affect H3 exchange. This indicates that the “old” or tagged version of H2B is being reassembled into chromatin in the absence of Nap1. This suggests that when present, Nap1 facilitates the incorporation of “new” histones and not the reassembly of the old histones, at least at this highly dynamic position. It is important to note that these studies were performed with cell cycle-arrested cells, so this Nap1 function is independent of histone deposition during replication.
FIG 6.
Histone persistence varies depending upon the time of shutoff and the specific GAL region. Histone persistence is defined as the relative percentage of tagged histone occupancy (detected with anti-HA) over endogenous histone occupancy (detected with anti-H2B or anti-H3) under GAL gene transcriptionally repressed (“Off”) or activated (“On”) conditions in the wt and nap1Δ strains. The histone persistence at T0 was set as 100%. (A and B) H2B (A) and H3 (B) persistence at the GAL locus under transcriptionally repressed conditions. (C and D) H2B (C) and H3 (D) persistence under continuous transcriptionally active conditions. P, promoter; O, ORF. *, P value of <0.05; **, P value of <0.01.
When GAL gene transcription was activated, the exchange kinetics of both H2B and H3 in the wild-type strain were highly dynamic across the locus (Fig. 5 and 6C and D), to a level comparable to that of GAL10PRO in the absence of transcription. This rapid transcription-mediated histone exchange is consistent with previously reported observations that nucleosomes are highly dynamic at the GAL locus upon the activation of transcription (20, 25, 30, 31, 71). In the nap1Δ strain, H2B exchange kinetics were less dynamic across the region, whereas histone H3 exchange was similar to that of the wild type (Fig. 6C and D). Therefore, when transcription is active and chromatin is highly dynamic in the wild-type strain, H2A-H2B exchange is consistently slower in the absence of Nap1.
In vitro nucleosome reconstitution recapitulates some of the in vivo dynamics of the GAL locus.
The various regions of the GAL locus exhibit distinctive exchange profiles in the wild-type strain (Fig. 5B). Since DNA sequence can directly affect nucleosome stability and contribute to in vivo nucleosome dynamics (64, 65), we tested the fragments for their intrinsic ability to form nucleosomes. Using the 601 positioning sequence (59) as a control, the GAL fragments were analyzed for the formation of stable, positioned nucleosomes by salt reconstitution with native histones (49) (see Fig. S9 in the supplemental material). This allowed us to group the GAL fragments into three classes, represented by 601 (601 and GAL7ORF), the GAL10 promoter (GAL10PRO, GAL10ORF, and GAL7PRO), and the GAL1 promoter (GAL1PRO and GAL1ORF). In some cases, multiple forms of DNA-histone complexes were evident. To identify the histone composition of the complexes, nucleosome reconstitution was performed with fluorescently labeled histones (see Fig. S10 in the supplemental material). In addition, heat-shifting and lower-salt experiments were used to determine if complexes were well positioned (see Fig. S11 in the supplemental material). The results indicate that the 601 class forms hexasomes and positioned nucleosomes at low concentrations of the octamer, with an increase in the amount of nucleosomes as the octamer is titrated. The GAL1 promoter class formed stable, positioned nucleosomes more efficiently than did 601, in that only a single band was observed even at low octamer levels. Both of the fragments in this class (GAL1 promoter and GAL1 ORF) had the least dynamic nucleosomes in vivo in the exchange assay for both H2B and H3, suggesting that sequence plays an important role in these properties. The GAL10 promoter class formed intermediate complexes and different forms of nucleosomes at every ratio in vitro, suggesting that nucleosomes are not stable and/or that there are multiple nucleosome positions. The GAL10 promoter region was the highly dynamic site in the in vivo exchange assay (Fig. 5), but the other two fragments in this unstable in vitro class (GAL10 ORF and GAL7 promoter) did not share this behavior in vivo.
DISCUSSION
We previously found that Nap1 prevents the formation of atypical chromatin characterized by excess H2A-H2B across the GAL locus under transcriptionally repressed conditions (20). Here, we show that H2A-H2B accumulation does not occur in a strain deleted for the Nap1 family member Vps75 or the H3-H4 histone chaperone Asf1. In addition, this effect is specific for the major histone variants, as H2A.Z does not accumulate in these same regions. Although H2A-H2B accumulation varies at different locations in the absence of Nap1, we found this is not likely due to variable H2A-H2B-DNA binding, as in vitro affinities did not correlate with in vivo accumulation. The excess H2A-H2B appears to bind between nucleosomes in the linker regions, as evidenced by in vitro and in vivo MNase protection assays. Notably, the process of transcription results in a loss of the excess H2A-H2B, and in fact, the absence of Nap1 leads to lower overall histone density on chromatin, indicative of critical nucleosome assembly functions of Nap1.
Nucleosomal histones are readily exchanged into and out of chromatin (4, 5). This histone exchange is a combination of partial nucleosome disassembly and reassembly, and histone chaperones are directly involved in this process (3, 7, 8, 72–74). However, studies on the requirement for histone chaperones in histone exchange within nucleosomes have focused primarily on H3-H4 histone chaperones (7, 9, 75). Also, most histone exchange studies utilize a galactose-inducible promoter for the regulation of tagged histone expression (4–9); therefore, GAL locus gene transcription is affected when the expression of the tagged histones is turned on or off. In this study, we designed a doxycycline-regulated histone exchange system that does not affect GAL gene transcription. Histone exchange is generally observed across the GAL locus under repressed conditions, with the exchange of both H2B and H3 being highly dynamic at the GAL10 promoter (GAL10PRO). Furthermore, the highly dynamic H2B exchange is Nap1 dependent, whereas H3 exchange is not. It is intriguing that even without active transcription, the nucleosome at the GAL10 promoter is highly dynamic. One unique feature of the chromatin at this site is a lower H2B occupancy in general; the GAL10 promoter has only half the H2B occupancy of other GAL sites (20). Moreover, in the absence of Nap1, this decrease in H2B occupancy is reversed (20), and the H2A-H2B exchange rate is severely reduced. Importantly, the GAL10 promoter region has the binding site for the Gal4-Gal80 activator complex and the RSC chromatin-remodeling complex under transcriptionally repressed conditions (55). In vitro studies have shown that RSC promotes histone transfer (76). In the presence of Nap1, RSC releases one H2A-H2B from the nucleosome to form a hexasome, and the dimer is transferred to Nap1 (19). This combination of highly dynamic Nap1-dependent H2A-H2B exchange, a 2-fold reduction in H2B occupancy, and the presence of an RSC-binding site supports a model whereby hexasome formation likely occurs at the GAL10 promoter in vivo.
Taken together, the findings presented here advance our understanding of the diverse roles of the histone chaperone Nap1 in chromatin regulation in vivo (see Fig. S12 in the supplemental material): (i) removal of atypical histone H2A-H2B under transcriptionally repressed conditions, (ii) maintenance of proper histone density on chromatin during activated transcription (i.e., transcription-coupled assembly), and (iii) facilitation of histone H2A-H2B exchange within highly dynamic chromatin regions whether the locus is repressed or activated. The active exchange of H2A-H2B could lead to the destabilization of the H3-H4 tetramer; thereby, Nap1 could also contribute to nucleosome disassembly. This is consistent with the findings that deletion of NAP1 reverses the cryptic transcript phenotype (interpreted as the by-product of underassembled chromatin) observed in mutant strains defective for other chromatin regulators (77) and that Nap1 has an important function in the eviction of histones during transcription in a mammalian in vitro system (17). Importantly, these varied functions of Nap1 are not redundant with other histone chaperones in vivo. Since Nap1 has a high affinity for H3-H4 tetramers as well as H2A-H2B dimers (14), Nap1 has the potential to function independently in these various roles. However, a large body of work indicates collaborative functional activities between Nap1 and other chaperones as well as ATP-dependent chromatin remodelers and histone acetyltransferases (13, 17–19, 60, 77–79). How the combinatorial action of these factors contributes to histone occupancy and dynamics in vivo remains to be elucidated.
Supplementary Material
ACKNOWLEDGMENTS
Plasmids with tagged histones were kind gifts from Michel Strubin. We thank the Protein Expression and Purification (PEP) Facility at Colorado State University for purified histones and Kyle W. Martin for assistance with the FRET assays.
Funding Statement
The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00835-15.
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Dechassa ML, Tremethick DJ. 2012. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linger J, Tyler JK. 2006. Global replication-independent histone H4 exchange in budding yeast. Eukaryot Cell 5:1780–1787. doi: 10.1128/EC.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamai A, Imoberdorf RM, Strubin M. 2007. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell 25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 6.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Kim HJ, Seol JH, Han JW, Youn HD, Cho EJ. 2007. Histone chaperones regulate histone exchange during transcription. EMBO J 26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gat-Viks I, Vingron M. 2009. Evidence for gene-specific rather than transcription rate-dependent histone H3 exchange in yeast coding regions. PLoS Comput Biol 5:e1000282. doi: 10.1371/journal.pcbi.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamai A, Puglisi A, Strubin M. 2009. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol Cell 35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 10.van Bakel H, Tsui K, Gebbia M, Mnaimneh S, Hughes TR, Nislow C. 2013. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet 9:e1003479. doi: 10.1371/journal.pgen.1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loyola A, Almouzni G. 2004. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta 1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Park YJ, Luger K. 2006. Structure and function of nucleosome assembly proteins. Biochem Cell Biol 84:549–558. doi: 10.1139/o06-088. [DOI] [PubMed] [Google Scholar]
- 13.Zlatanova J, Seebart C, Tomschik M. 2007. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J 21:1294–1310. doi: 10.1096/fj.06-7199rev. [DOI] [PubMed] [Google Scholar]
- 14.Andrews AJ, Downing G, Brown K, Park YJ, Luger K. 2008. A thermodynamic model for Nap1-histone interactions. J Biol Chem 283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Arcy S, Martin KW, Panchenko T, Chen X, Bergeron S, Stargell LA, Black BE, Luger K. 2013. Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A-H2B in an unconventional tetrameric form. Mol Cell 51:662–677. doi: 10.1016/j.molcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii-Nakata T, Ishimi Y, Okuda A, Kikuchi A. 1992. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J Biol Chem 267:20980–20986. [PubMed] [Google Scholar]
- 17.Luebben WR, Sharma N, Nyborg JK. 2010. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc Natl Acad Sci U S A 107:19254–19259. doi: 10.1073/pnas.1009650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorch Y, Maier-Davis B, Kornberg RD. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A 103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuryan BG, Kim J, Tran NN, Lombardo SR, Venkatesh S, Workman JL, Carey M. 2012. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc Natl Acad Sci U S A 109:1931–1936. doi: 10.1073/pnas.1109994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. 2010. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell 37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Citron BA, Donelson JE. 1984. Sequence of the Saccharomyces GAL region and its transcription in vivo. J Bacteriol 158:269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohr D, Venkov P, Zlatanova J. 1995. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J 9:777–787. [DOI] [PubMed] [Google Scholar]
- 23.Lohr D, Hopper JE. 1985. The relationship of regulatory proteins and DNaseI hypersensitive sites in the yeast GAL1-10 genes. Nucleic Acids Res 13:8409–8423. doi: 10.1093/nar/13.23.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavalli G, Thoma F. 1993. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J 12:4603–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han M, Grunstein M. 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 26.Burley SK, Roeder RG. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu Rev Biochem 65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 27.Starr DB, Hawley DK. 1991. TFIID binds in the minor groove of the TATA box. Cell 67:1231–1240. doi: 10.1016/0092-8674(91)90299-E. [DOI] [PubMed] [Google Scholar]
- 28.Bhaumik SR, Raha T, Aiello DP, Green MR. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev 18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larschan E, Winston F. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhaumik SR, Green MR. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dudley AM, Rougeulle C, Winston F. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev 13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol 25:3461–3474. doi: 10.1128/MCB.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol 20:634–647. doi: 10.1128/MCB.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 35.Lohr D, Lopez J. 1995. GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1-10 and GAL80 genes. J Biol Chem 270:27671–27678. doi: 10.1074/jbc.270.46.27671. [DOI] [PubMed] [Google Scholar]
- 36.Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. 2008. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol 6:2928–2939. doi: 10.1371/journal.pbio.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemieux K, Gaudreau L. 2004. Targeting of Swi/Snf to the yeast GAL1 UASG requires the mediator, TAF IIs, and RNA polymerase II. EMBO J 23:4040–4050. doi: 10.1038/sj.emboj.7600416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. 2008. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol 28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan RK, Otte CA. 1982. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol 2:21–29. doi: 10.1128/MCB.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longtine M, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. [DOI] [PubMed] [Google Scholar]
- 41.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman GG, Smith JA, Struhl K (ed). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY. [Google Scholar]
- 42.Cavallini B, Faus I, Matthes H, Chipoulet J-M, Winsor B, Egly JM, Chambon P. 1989. Cloning of the gene encoding the yeast protein BTF1, which can substitute for the human TATA box-binding factor. Proc Natl Acad Sci U S A 86:9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Fletcher AG, Cheung V, Winston F, Stargell LA. 2008. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol 28:1393–1403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hieb AR, D'Arcy S, Kramer MA, White AE, Luger K. 2012. Fluorescence strategies for high-throughput quantification of protein interactions. Nucleic Acids Res 40:e33. doi: 10.1093/nar/gkr1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. 2011. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun 2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luger K, Rechsteiner TJ, Richmond TJ. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304:3–19. doi: 10.1016/S0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 47.Reese JC, Zhang H, Zhang Z. 2008. Isolation of highly purified yeast nuclei for nuclease mapping of chromatin structure. Methods Mol Biol 463:43–53. doi: 10.1007/978-1-59745-406-3_3. [DOI] [PubMed] [Google Scholar]
- 48.Ryan MP, Stafford GA, Yu L, Cummings KB, Morse RH. 1999. Assays for nucleosome positioning in yeast. Methods Enzymol 304:376–399. doi: 10.1016/S0076-6879(99)04023-9. [DOI] [PubMed] [Google Scholar]
- 49.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. 2004. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375:23–44. [DOI] [PubMed] [Google Scholar]
- 50.Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. 2008. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol 15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 52.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 53.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang C, Pugh BF. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, Hicks J, Bryant GO, Ptashne M. 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnston M, Davis RW. 1984. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol 4:1440–1448. doi: 10.1128/MCB.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West RW, Yocum RR, Ptashne M. 1984. Yeast GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol Cell Biol 4:2467–2478. doi: 10.1128/MCB.4.11.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daganzo SM, Erzberger JP, Lam WM, Skordalakes E, Zhang R, Franco AA, Brill SJ, Adams PD, Berger JM, Kaufman PD. 2003. Structure and function of the conserved core of histone deposition protein Asf1. Curr Biol 13:2148–2158. doi: 10.1016/j.cub.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 59.Lowary PT, Widom J. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 60.Sharma N, Nyborg JK. 2008. The coactivators CBP/p300 and the histone chaperone NAP1 promote transcription-independent nucleosome eviction at the HTLV-1 promoter. Proc Natl Acad Sci U S A 105:7959–7963. doi: 10.1073/pnas.0800534105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A 102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zlatanova J, Thakar A. 2008. H2A.Z: view from the top. Structure 16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 63.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 64.Deniz O, Flores O, Battistini F, Perez A, Soler-Lopez M, Orozco M. 2011. Physical properties of naked DNA influence nucleosome positioning and correlate with transcription start and termination sites in yeast. BMC Genomics 12:489. doi: 10.1186/1471-2164-12-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hornung G, Oren M, Barkai N. 2012. Nucleosome organization affects the sensitivity of gene expression to promoter mutations. Mol Cell 46:362–368. doi: 10.1016/j.molcel.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bai L, Ondracka A, Cross FR. 2011. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell 42:465–476. doi: 10.1016/j.molcel.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jansen A, van der Zande E, Meert W, Fink GR, Verstrepen KJ. 2012. Distal chromatin structure influences local nucleosome positions and gene expression. Nucleic Acids Res 40:3870–3885. doi: 10.1093/nar/gkr1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jansen A, Verstrepen KJ. 2011. Nucleosome positioning in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 75:301–320. doi: 10.1128/MMBR.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perales R, Zhang L, Bentley D. 2011. Histone occupancy in vivo at the 601 nucleosome binding element is determined by transcriptional history. Mol Cell Biol 31:3485–3496. doi: 10.1128/MCB.05599-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinogradov AE. 2003. DNA helix: the importance of being GC-rich. Nucleic Acids Res 31:1838–1844. doi: 10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhaumik SR, Green MR. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol 22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Byrum S, Mackintosh SG, Edmondson RD, Cheung WL, Taverna SD, Tackett AJ. 2011. Analysis of histone exchange during chromatin purification. J Integr OMICS 1:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Byrum SD, Taverna SD, Tackett AJ. 2011. Quantitative analysis of histone exchange for transcriptionally active chromatin. J Clin Bioinform 1:17. doi: 10.1186/2043-9113-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. 2011. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol 9:e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Das C, Tyler JK. 2013. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta 1819:332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowe CE, Narlikar GJ. 2010. The ATP-dependent remodeler RSC transfers histone dimers and octamers through the rapid formation of an unstable encounter intermediate. Biochemistry 49:9882–9890. doi: 10.1021/bi101491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xue YM, Kowalska AK, Grabowska K, Przybyt K, Cichewicz MA, Del Rosario BC, Pemberton LF. 2013. Histone chaperones Nap1 and Vps75 regulate histone acetylation during transcription elongation. Mol Cell Biol 33:1645–1656. doi: 10.1128/MCB.01121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Del Rosario BC, Pemberton LF. 2008. Nap1 links transcription elongation, chromatin assembly, and messenger RNP complex biogenesis. Mol Cell Biol 28:2113–2124. doi: 10.1128/MCB.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. 2007. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J 26:2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.