Abstract
Hepatitis B virus (HBV) enters hepatocytes via its receptor, human sodium taurocholate cotransporting polypeptide (hNTCP). So far, HBV infection has been achieved only in human hepatic cells reconstituted with hNTCP and not in cells of mouse origin. Here, the first mouse liver cell line (AML12) which gains susceptibility to HBV upon hNTCP expression is described. Thus, HBV infection of receptor-expressing mouse hepatocytes does not principally require a human cofactor but can be triggered by endogenous murine determinants.
TEXT
Hepatitis B virus (HBV) is a hepatotropic, enveloped DNA virus replicating via an RNA intermediate (1). Infections with HBV and coinfections with hepatitis delta virus (HDV), which uses the HBV envelope for dissemination, increase the risk for liver cirrhosis and carcinoma (2). Both viruses exploit human sodium taurocholate cotransporting polypeptide (hNTCP), a hepatic bile salt transporter, as an essential entry receptor (3, 4). Hitherto, only hepatic human liver cells expressing hNTCP have been shown to become susceptible for HBV (3, 5, 6). In contrast, HDV infections can be established through hNTCP supplementation in nonhuman and even nonhepatic cells.
Several hNTCP-expressing mouse liver cell lines are resistant to HBV infection (3, 6–8). Likewise, HBV transgenic mice cannot form covalently closed circular DNA (cccDNA), the transcriptional viral template (9). Such previous results have led to interpretations that mouse cell lines might lack a factor needed for HBV replication—a hypothesis strongly supported by recent work by Lempp et al. (8)—or, alternatively, might express a restriction factor that prevents cccDNA formation. However, Cui et al. recently described a mouse liver cell line inducibly expressing HBV from an integrate (AML12HBV10) capable of forming cccDNA (10). Here, we provide evidence that AML12 cells complemented with hNTCP gain susceptibility to HBV, including cccDNA formation and antigen (Ag) expression.
Generation of hNTCP-expressing cell lines.
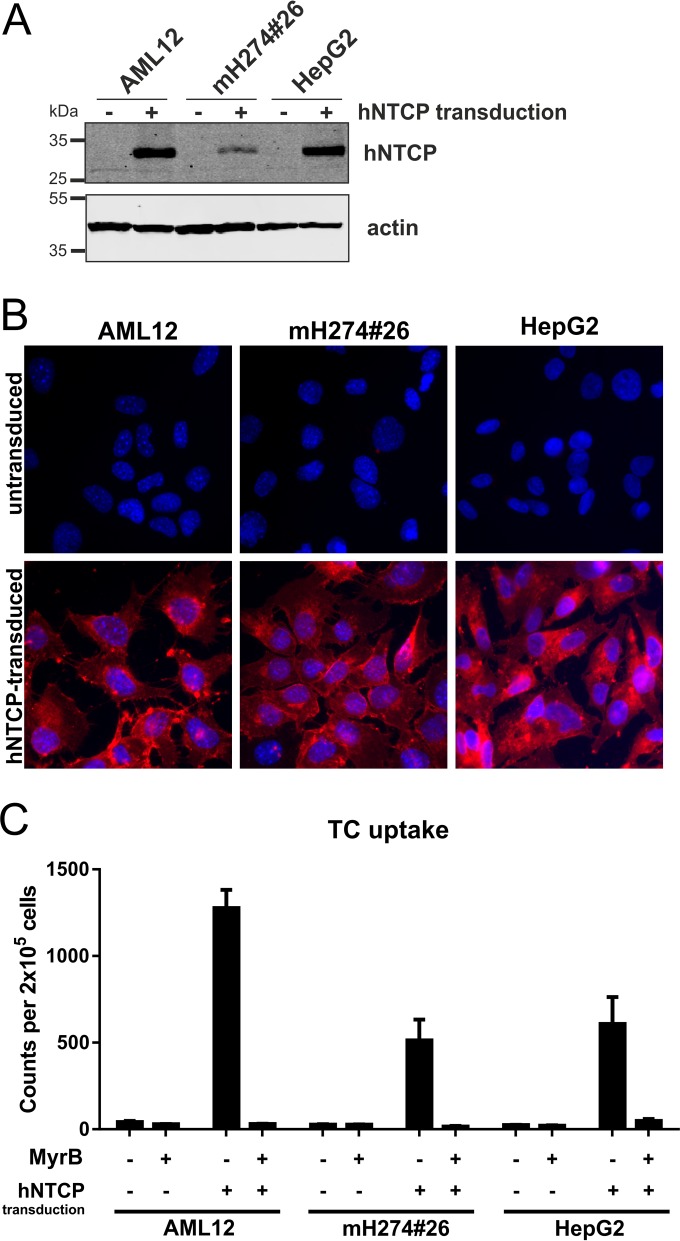
Mouse AML12 cells were tested for their ability to support HBV infection after stable hNTCP transduction. For controls, we implemented the HepG2 cell line, which becomes susceptible to HBV upon hNTCP transduction, and the mouse liver cell line mH274#26, which remains refractory (3, 4, 8). Stable transduction was achieved using lentiviruses encoding hNTCP. Western blot analysis (Fig. 1A) confirmed expression of hNTCP exclusively in the transduced lines. The hNTCP-expressing cells bound Myrcludex B-atto (myristoyl-GTNLSVPNPLGFFPDHQLDPAFGANSNNPDWDFNPNKDHWPEANKVG-atto [MyrB-atto]), a fluorescently labeled entry inhibitor of HBV and HDV that specifically binds to NTCP (Fig. 1B) (11). This indicates correct folding and localization to the plasma membrane. Functionality of hNTCP was further confirmed by its ability to transport 3H-labeled taurocholate (Fig. 1C) (3, 12–14).
FIG 1.
Characterization of mouse and human cell lines stably expressing hNTCP. AML12 (obtained from ATCC CRL-2254), mH274#26 (8), and HepG2 cells were transduced with lentiviruses coexpressing hNTCP and a puromycin resistance gene and selected to establish a stable cell pool. (A) Total cell lysates of the transduced (+) or parental (−) cells were treated with PNGase F (New England BioLabs) and analyzed by Western blotting using a hNTCP-specific antibody (Sigma-Aldrich). (B) The respective transduced (lower row) and parental (upper row) cell lines were incubated for 30 min with 200 µM atto565-labeled Myrcludex B, washed, and analyzed by fluorescence microscopy. (C) Cell lines were incubated with 3H-TC (Hartmann Analytic) for 15 min at 37°C, washed, and lysed, and scintillation counts of the total lysates were measured and the results normalized to the cell number. TC uptake was inhibited by treatment of the cells with 2 μM MyrB (13).
hNTCP expression confers susceptibility to HDV infection.
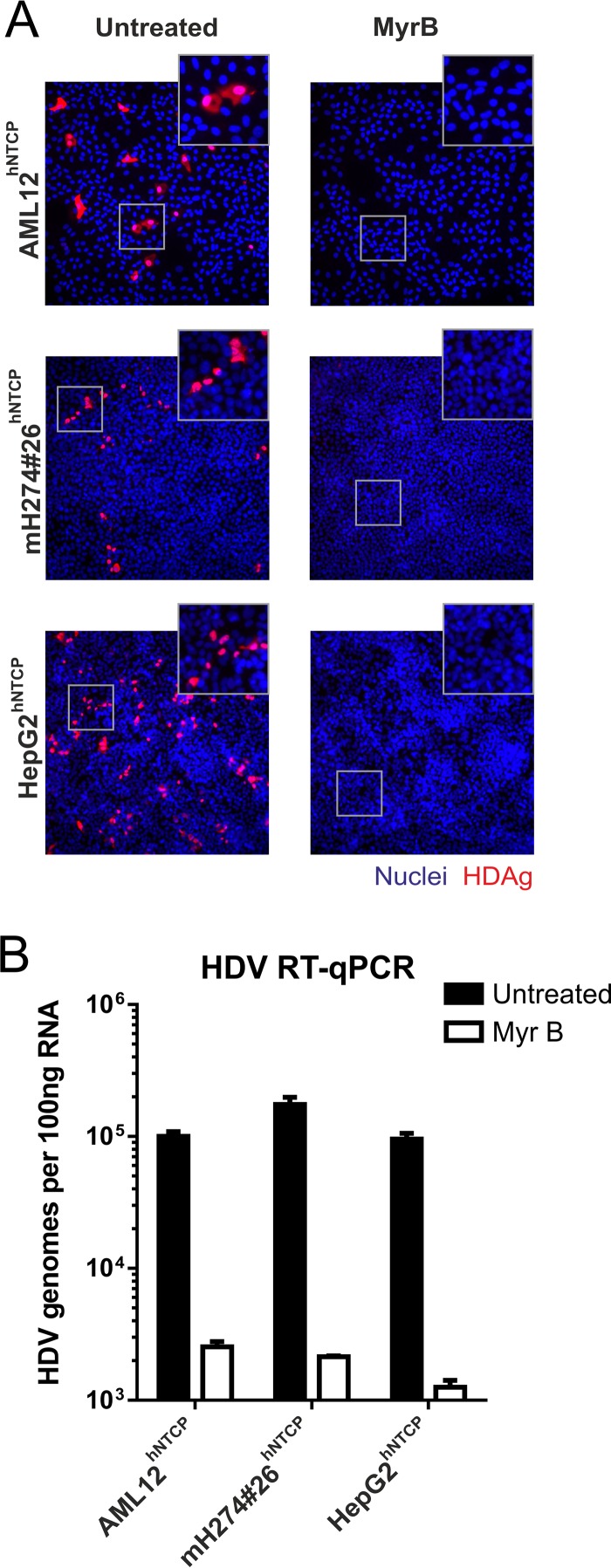
Cell lines were infected with HDV, and replication was analyzed by immunofluorescent staining of hepatitis delta virus antigen (HDAg) (Fig. 2A) and quantification of intracellular HDV RNA (Fig. 2B). All infected cell lines showed comparable infection levels, indicating that hNTCP expression is sufficient to overcome the restriction in the parental cells. Inhibition by MyrB verified that entry proceeds via the NTCP-mediated pathway. The observed susceptibilities to HDV rule out a possible limitation by a lack of heparan sulfate proteoglycans, which are required prior to NTCP engagement (15, 16)
FIG 2.
HDV infection of stable hNTCP-expressing mouse and human liver cell lines. AML12hNTCP, mH274#26hNTCP, and HepG2hNTCP cells were seeded in 24-well plates and infected 2 days later with HDV by inoculation of the cells with 4 IU/cell HDV (8) in the presence of 4% polyethylene glycol (PEG) in medium supplemented with 2% dimethyl sulfoxide (DMSO) for 16 h. Entry inhibition was achieved with 500 nM Myrcludex B 30 min prior and during virus inoculation. (A) At day 5 postinfection, cells were fixed with 4% paraformaldehyde (PFA) and permeabilized and HDAg was stained using patient serum (Vuda) and a fluorescently labeled goat-anti-human secondary antibody (Thermo Fisher Scientific). Images (×200 magnification) were acquired on an inverted fluorescence microscope (Leica). (B) Total RNA was extracted from the cells at day 5 postinfection, reverse transcribed, and analyzed by quantitative PCR using primers and probes specific for HDV as previously described (8, 18).
AML12hNTCP cells are susceptible to HBV infection.
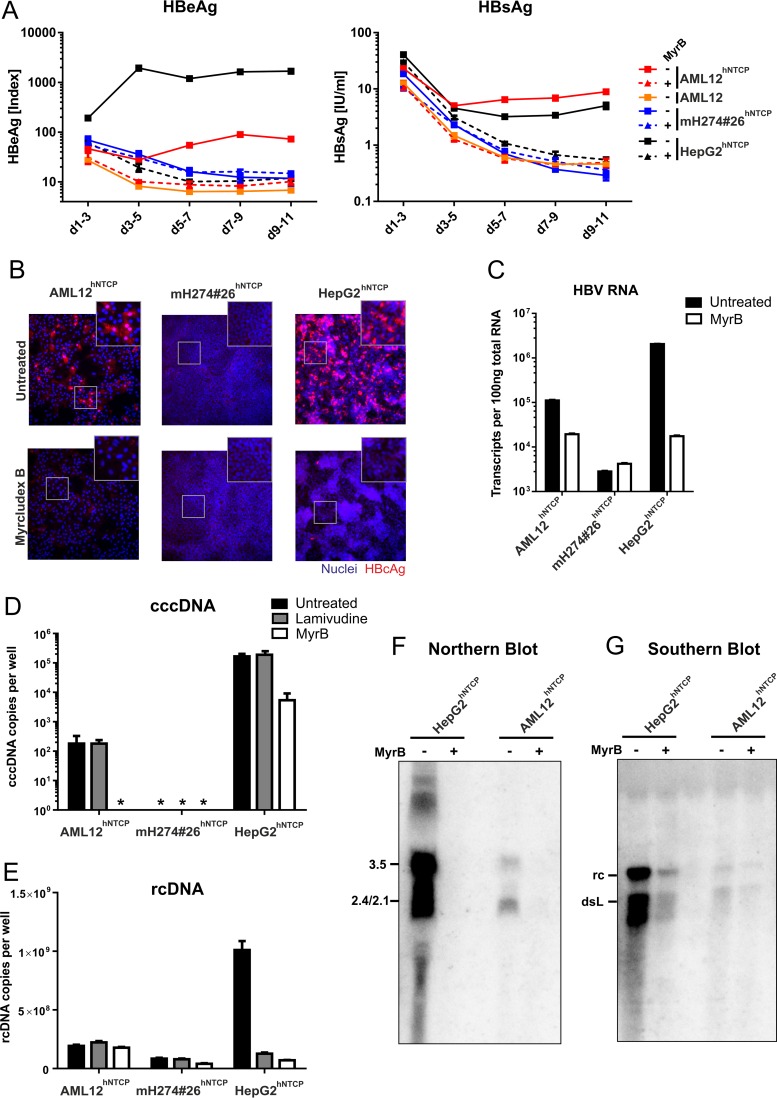
Cells were inoculated with HBV and intracellular nucleic acids, and secreted viral proteins were analyzed. HBeAg and HBsAg were detectable in the supernatant of infected HepG2hNTCP and AML12hNTCP cells but not in mH274#26hNTCP or parental AML12 cells (Fig. 3A). MyrB treatment abrogated antigen secretion. HBeAg levels of infected AML12hNTCP cells were ∼10× lower than HepG2hNTCP cell levels; however, secreted HBsAg levels were comparable. Furthermore, intracellular HBcAg was readily detected in infected AML12hNTCP and HepG2hNTCP cells but not in mH274#26hNTCP cells (Fig. 3B). The levels of HBV RNA (Fig. 3C) corresponded to the HBeAg measurement, with an ∼10× difference between AML12hNTCP and HepG2hNTCP cells. This difference was also observed in a corresponding Northern blot analysis (Fig. 3F). For quantification of HBV cccDNA and relaxed-circular DNA (rcDNA), an additional treatment was performed with the reverse transcriptase inhibitor lamivudine to inhibit rcDNA formation while cccDNA remained unaffected. In accordance with HBcAg staining results, cccDNA was detected in infected AML12hNTCP and HepG2hNTCP cells but not in mH274#26hNTCP cells (Fig. 3D). As expected, the levels of cccDNA remained unchained upon lamivudine treatment; however, absolute levels differed significantly between infected AML12hNTCP and HepG2hNTCP cells. In the absence of lamivudine, rcDNA was readily detected in infected HepG2hNTCP cells by quantitative PCR (qPCR) (Fig. 3E) and Southern blot analysis (Fig. 3G). However, in infected AML12hNTCP cells, rcDNA was barely detectable. Residual signals in the MyrB- or lamivudine-treated samples probably reflects remaining viral input.
FIG 3.
HBV infection of stable hNTCP-expressing cell lines. AML12, AML12hNTCP, mH274#26hNTCP, and HepG2hNTCP cells were seeded in 24-well plates and infected 2 days later by inoculation with HBV at a multiplicity of genome equivalents (MGE) of 1,500 (genotype D, cell culture derived) in the presence of 4% PEG and 2% DMSO for 16 h as described previously (8). For entry inhibition, cells were treated with 500 nM Myrcludex B 30 min prior and during virus inoculation. (A) Supernatants of the cells were collected every second day, and secreted viral markers HBeAg (left) and HBsAg (right) were quantified by ELISA as previously described (8). Each data point represents the mean of the results of three biological replicates. d, day. (B) At day 11 postinfection, cells were fixed and immunostained with an antibody (Dako) recognizing viral HBcAg (red, ×200 magnification). (C) RNA from infected cells was extracted, reverse transcribed, and quantified using primers binding to the HBV core open reading frame (ORF), thereby detecting pregenomic, precore, and possibly spliced HBV RNA (4). (D and E) Total DNA was extracted from cells at day 10 postinfection. Lamivudine (10 μM) was added at day 3 postinfection. rcDNA was quantified with HBV-specific primers (E). Following digestion of rcDNA with T5 exonuclease (New England BioLabs), cccDNA (D) was specifically quantified using primers spanning the gap region (Bingqian Qu, unpublished data). Values below the limit of quantitation are marked by asterisks (*). (F and G) AML12hNTCP and HepG2hNTCP cells were seeded in 10 cm-diameter dishes and infected with HBV at an MGE of 1,500 in the presence or absence of 1 μM MyrB as described above. (F) Eight days after infection, total cellular RNA was extracted, separated on a 1.2% agarose gel, blotted on a nylon membrane, and analyzed with a 32P-labeled HBV-specific probe of genotype D (Northern blotting). (G) Ten days after infection, cytosolic DNA was extracted by treatment of the cells with NP-40 lysis buffer, centrifugation, proteinase K treatment of the supernatant, and subsequent phenol-chloroform DNA extraction. The DNA was separated on a 1.2% agarose gel and analyzed by Southern blotting as described above.
In order to exclude the possibility of contamination of the mouse AML12hNTCP cells with a human cell line, we performed a multiplex cell contamination test by the use of Multiplexion (Heidelberg, Germany) (17). The results confirmed that the AML12hNTCP cell line contained only cells of mouse origin (data not shown). We further selected single-cell clones. All clones retained their TC transporter function (Fig. 4A) and showed comparable levels of susceptibility to HBV with variations in infection rates ranging up to 2-fold (Fig. 4B to D).
FIG 4.
HBV infection of single-cell clones derived from AML12hNTCP. Single-cell clones of AML12hNTCP were picked, expanded, seeded in 24-well plates and analyzed for their ability to transport 3H-TC (see Fig. 1 for details) (A) or infected by inoculation with HBV at an MGE of 1,500 in the presence or absence of MyrB as described in the Fig. 3 legend (B, C, and D). The supernatant of the infected cells was collected from day 7 to day 9 postinfection and analyzed for secreted HBeAg (B) and HBsAg (C). Cells were fixed and stained with an antibody against viral HBcAg (D).
We describe for the first time a mouse liver cell line that, solely upon expression of the hNTCP receptor, becomes susceptible to HBV. In contrast to other murine cell lines (5), AML12hNTCP cells support HBV entry, including the formation of cccDNA, cccDNA-driven transcription of viral RNAs, expression of HBcAg, and secretion of HBeAg and HBsAg. Notably, the infected cells showed only barely detectable levels of rcDNA, which might hint at a defect in nucleocapsid stability and, consequently, in rcDNA synthesis. However, this needs further investigation. The successful infection with HBV contradicts the assumption that a highly species-specific factor is principally lacking in cells of mouse origin, supporting and extending a report from Cui et al., who provided evidence that AML12 cells are capable of forming cccDNA (10). Taking into account that nonsusceptible hNTCP-expressing mouse cells can be rescued by fusion with HepG2 cells, we assume that AML12 cell lines express a cofactor which is present in human but missing in many mouse hepatic cells (8). This cofactor might affect the stability of the HBV nucleocapsids (10) and may be involved in the release and/or the transport of the viral genome to the nucleus. AML12 cells are derived from a transgenic mouse overexpressing human transforming growth factor α (TGFα). In order to examine if TGFα was the limiting factor, we performed rescue experiments by overexpression or addition of recombinant TGFα to previously described HBV-restricted mouse cell lines (8). However, TGFα could not rescue HBV susceptibility (data not shown). Identification of the limiting factor(s), e.g., by differential transcriptome analysis, may help to generate immunocompetent mouse models that fully support HBV infection.
ACKNOWLEDGMENTS
F.A.L. is member of the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (HBIGS), Heidelberg University. We thank Steven Dooley for providing an additional batch of AML12 cells for preliminary experiments and Michael Nassal for his generous support in performing additional experiments in his laboratory. We thank Franziska Schlund and Christa Kuhn for cell culture experiments, Yi Ni and Volker Lohmann for lentiviral constructs, Christina Kaufman for MyrB-atto labeling, and Ralf Bartenschlager for his continuous support.
We have no conflicts of interest to declare.
REFERENCES
- 1.Trépo C, Chan HL, Lok A. 2014. Hepatitis B virus infection. Lancet 384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 2.Alfaiate D, Deny P, Durantel D. 2015. Hepatitis delta virus: from biological and medical aspects to current and investigational therapeutic options. Antiviral Res 122:112–129. doi: 10.1016/j.antiviral.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S. 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winer BY, Ploss A. 2015. Determinants of hepatitis B and delta virus host tropism. Curr Opin Virol 13:109–116. doi: 10.1016/j.coviro.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Peng B, He W, Zhong G, Qi Y, Ren B, Gao Z, Jing Z, Song M, Xu G, Sui J, Li W. 2013. Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide. J Virol 87:7977–7991. doi: 10.1128/JVI.03540-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Zhuang Q, Wang Y, Zhang T, Zhao J, Zhang Y, Zhang J, Lin Y, Yuan Q, Xia N, Han J. 2014. HBV life cycle is restricted in mouse hepatocytes expressing human NTCP. Cell Mol Immunol 11:175–183. doi: 10.1038/cmi.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lempp FA, Mutz P, Lipps C, Wirth D, Bartenschlager R, Urban S. 11 November 2015. Evidence that hepatitis B virus replication in mouse cells is limited by the lack of a host cell dependency factor. J Hepatol doi: 10.1016/j.jhep.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 9.Guidotti LG, Matzke B, Schaller H, Chisari FV. 1995. High-level hepatitis B virus replication in transgenic mice. J Virol 69:6158–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui X, Guo JT, Hu J. 2015. Hepatitis B virus covalently closed circular DNA formation in immortalized mouse hepatocytes associated with nucleocapsid destabilization. J Virol 89:9021–9028. doi: 10.1128/JVI.01261-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lempp FA, Urban S. 2014. Inhibitors of hepatitis B virus attachment and entry. Intervirology 57:151–157. doi: 10.1159/000360948. [DOI] [PubMed] [Google Scholar]
- 12.König A, Döring B, Mohr C, Geipel A, Geyer J, Glebe D. 2014. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J Hepatol 61:867–875. doi: 10.1016/j.jhep.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Nkongolo S, Ni Y, Lempp FA, Kaufman C, Lindner T, Esser-Nobis K, Lohmann V, Mier W, Mehrle S, Urban S. 2014. Cyclosporin A inhibits hepatitis B and hepatitis D virus entry by cyclophilin-independent interference with the NTCP receptor. J Hepatol 60:723–731. doi: 10.1016/j.jhep.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 14.Yan H, Peng B, Liu Y, Xu G, He W, Ren B, Jing Z, Sui J, Li W. 2014. Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J Virol 88:3273–3284. doi: 10.1128/JVI.03478-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamas Longarela O, Schmidt TT, Schoneweis K, Romeo R, Wedemeyer H, Urban S, Schulze A. 2013. Proteoglycans act as cellular hepatitis delta virus attachment receptors. PLoS One 8:e58340. doi: 10.1371/journal.pone.0058340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urban S. 2015. Entry and entry inhibition of hepatitis B (HBV) and hepatitis delta virus (HDV) into hepatocytes. Hepatology doi: 10.1002/hep.28308. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt M, Pawlita M. 2009. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Res 37:e119. doi: 10.1093/nar/gkp581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferns RB, Nastouli E, Garson JA. 2012. Quantitation of hepatitis delta virus using a single-step internally controlled real-time RT-qPCR and a full-length genomic RNA calibration standard. J Virol Methods 179:189–194. doi: 10.1016/j.jviromet.2011.11.001. [DOI] [PubMed] [Google Scholar]