ABSTRACT
One of the challenges of developing influenza A vaccines is the diversity of antigenically distinct isolates. Previously, a novel hemagglutinin (HA) for H5N1 influenza was derived from a methodology termed computationally optimized broadly reactive antigen (COBRA). This COBRA HA elicited a broad antibody response against H5N1 isolates from different clades. We now report the development and characterization of a COBRA-based vaccine for both seasonal and pandemic H1N1 influenza virus isolates. Nine prototype H1N1 COBRA HA proteins were developed and tested in mice using a virus-like particle (VLP) format for the elicitation of broadly reactive, functional antibody responses and protection against viral challenge. These candidates were designed to recognize H1N1 viruses isolated within the last 30 years. In addition, several COBRA candidates were designed based on sequences of H1N1 viruses spanning the past 100 years, including modern pandemic H1N1 isolates. Four of the 9 H1N1 COBRA HA proteins (X1, X3, X6, and P1) had the broadest hemagglutination inhibition (HAI) activity against a panel of 17 H1N1 viruses. These vaccines were used in cocktails or prime-boost combinations. The most effective regimens that both elicited the broadest HAI response and protected mice against a pandemic H1N1 challenge were vaccines that contained the P1 COBRA VLP and either the X3 or X6 COBRA VLP vaccine. These mice had little or no detectable viral replication, comparable to that observed with a matched licensed vaccine. This is the first report describing a COBRA-based HA vaccine strategy that elicits a universal, broadly reactive, protective response against seasonal and pandemic H1N1 isolates.
IMPORTANCE Universal influenza vaccine approaches have the potential to be paradigm shifting for the influenza vaccine field, with the goal of replacing the current standard of care with broadly cross-protective vaccines. We have used COBRA technology to develop an HA head-based strategy that elicits antibodies against many H1 strains that have undergone genetic drift and has potential as a “subtype universal” vaccine. Nine HA COBRA candidates were developed, and these vaccines were used alone, in cocktails or in prime-boost combinations. The most effective regimens elicited the broadest hemagglutination inhibition (HAI) response against a panel of H1N1 viruses isolated over the past 100 years. This is the first report describing a COBRA-based HA vaccine strategy that elicits a broadly reactive response against seasonal and pandemic H1N1 isolates.
INTRODUCTION
Influenza vaccine efficacy is constantly undermined by antigenic variation in the circulating viral strains, particularly in the hemagglutinin (HA) and neuraminidase (NA) proteins. Current influenza vaccination strategies rely on changing the HA and NA components of the annual human influenza vaccine to ensure that they antigenically match circulating influenza strains (1, 2). Developing an influenza vaccine that is capable of providing broad and long-lasting protective antibody responses remains the central challenge for influenza virus research. Protection against influenza infection is mediated primarily by neutralizing antibodies (NAbs) (3–5). However, the virus undergoes genetic variation to avoid the host immune response generating unique epitopes not present in the parental strain while maintaining other shared epitopes. This genetic drift allows the virus to escape from NAbs. Ideally, upon infection with a variant virus strain, memory B cells, which recognize the shared epitopes between the parental and variant virus strains, should become activated. Simultaneously, naive B cells should respond to the presence of novel epitopes in variant strains. In this scenario, antibodies should be generated to both shared and unique epitopes resulting in neutralization of the new variant virus strain. However, in reality, the activation of naive B cells and generation of NAbs to novel epitopes are not effectively induced during infection, particularly in those populations undergoing immunosenescence, such as in the elderly (6, 7).
Several approaches are under way to develop a more effective influenza vaccine that will recognize most, if not all, circulating influenza strains. Immune responses directed at more conserved influenza proteins such as matrix (M2e) or nucleoprotein (NP) are in development, but these vaccines function through disease modulation and appear not to be as protective as needed to prevent morbidity and mortality following a high-dose influenza virus challenge (8). While these approaches and many other approaches have merit, several research groups (9), including ours, continue efforts toward the generation of an influenza vaccine capable of eliciting a potent, broadly reactive HA-specific antibody responses that protects against both seasonal and novel pandemic influenza strains that have undergone genetic drift. These vaccines induce immunity targeting both the globular head and stem regions of HA.
In order to overcome antigenic drift and mismatch of current influenza vaccines for pandemic and seasonal viruses, we reported a new methodology of antigen design using multiple rounds of consensus building to generate candidates termed computationally optimized broadly reactive antigens (COBRAs). This method was initially designed to address the diversity of H5N1 highly pathogenic avian influenza (10–13), and COBRA H5 designs were shown to elicit increased breadth of antibody responses to 25 H5N1 strains that were isolated over a 12-year span. In this report, a similar approach was used to generate COBRA HA antigens to elicit antibody responses to a panel of 17 H1N1 viruses isolated over almost a 100-year span of time. Each of the COBRA HA antigens were expressed on virus-like particles (VLPs) to be used as proof-of-concept antigens for immunogenicity and efficacy testing in the murine model. These candidates were used individually, in cocktails or in prime-boost regimens to evaluate the effect on elicited antibodies and protective efficacy compared to wild-type H1N1 HA antigens.
MATERIALS AND METHODS
Antigen construction and synthesis.
Influenza A hemagglutinin (HA) nucleotide sequences isolated from human H1N1 infections were downloaded from the NCBI Influenza Virus Resource database (13). Nucleotide sequences were translated into protein sequences using the standard genetic code. Full-length sequences from H1N1 viral infections isolated from swine and human sources between 1918 and 2012 were used in this analysis. For each round of consensus generation, multiple alignment analysis was applied, and the consensus sequence was generated using AlignX (Vector NTI). The final amino acid sequence, termed computationally optimized broadly reactive antigen (COBRA), was reverse translated and optimized for expression in mammalian cells, including codon usage and RNA optimization (Genewiz, Washington, DC, USA). Nine H1N1 HA constructs were synthesized and inserted into the pTR600 expression vector, as previously described (14). We designed COBRA HA antigens to represent different antigenic spaces. For example, HA COBRA designated as A1, was generated with HA sequences from human viruses isolated between 1918 and 2005, whereas A5 was generated using HA sequences from human viruses isolated between 1918 and 2011, including the pandemic 2009 strains (Table 1). Additionally, X1 HA COBRA sequence was generated using sequences from 1918 to 2012 but layered in a different way than A5. X2 HA COBRA was based upon sequences from viruses isolated between 1933 and 1947 which represents the post-1918 time period but early viral evolution in humans. X3 HA COBRA was designed using sequences from viruses isolated over a recent 30-year period (1978 to 2008), X5 HA COBRA uses sequences from the last 20 years (1982 to 2012), and X6 HA COBRA was based upon sequences representing the last ∼10 years (1999 to 2012). The HA amino acid sequences were aligned, and the most common amino acid at each position was determined resulting in primary consensus sequences representing each antigenic group within each subclade (Fig. 1). Each COBRA HA structure was generated using the 3D-JIGSAW algorithm (15, 16), and renderings were performed using MacPyMol (Fig. 2). A phylogenetic tree was inferred from hemagglutinin amino acid sequences using the maximum likelihood method, and clade/subclade groupings were identified using Jalview (Dundee, United Kingdom) (Fig. 3A).
TABLE 1.
H1N1 COBRA designations and descriptions
| H1N1 COBRA designation | Descriptiona |
|---|---|
| A1 | H1 antigenic space from viruses isolated from 1918 to 2005 |
| A5 | H1 antigenic space from viruses isolated from 1918 to 2011 |
| X1 | H1 antigenic space from viruses isolated from 1918 to 2012 |
| X2 | H1 antigenic space from viruses isolated from 1933 to 1947 |
| X3 | H1 antigenic space from viruses isolated from 1978 to 2008 |
| X4 | H1 antigenic space of A1, deglycosylated at aa142 and aa172 |
| X5 | H1 antigenic space from viruses isolated from 1982 to 2012 |
| X6 | H1 antigenic space from viruses isolated from 1999 to 2012 |
| P1 | H1 antigenic space covering viruses isolated from 1933 to 1957 and 2009 to 2011 and swine sequences isolated from 1931 to 1998 |
aa142, amino acid at position 142; aa172, amino acid at position 172.
FIG 1.
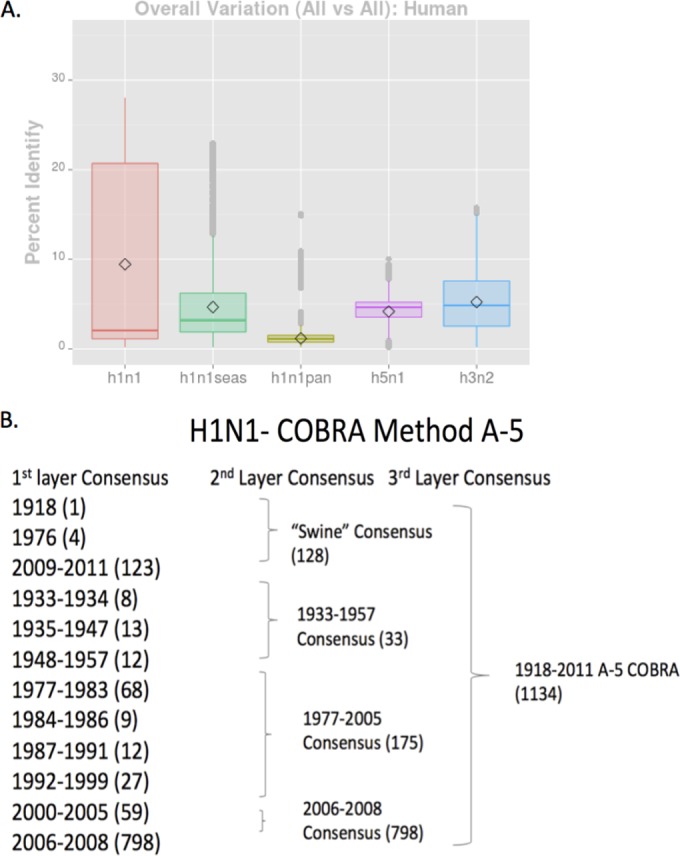
Influenza HA diversity and example of COBRA design. (A) Diversity of HA per subtype. The percent identity values in HA amino acid sequences from three subtypes (H1N1, H3N2, and H5N1) are displayed. For H1N1, seasonal H1N1 (green bar), pandemic H1N1 (yellow bar), and all H1N1 (red bar) are displayed along with H5N1 (purple bar) and H3N2 (blue bar). For each subtype, the diamond symbol indicates the mean value and the horizontal line indicates the median value for each subtype. (B) Schematic example of the layered consensus used for H1N1 COBRA design. Primary consensus sequences are designed to incorporate the most common amino acid at each location based upon different eras. The number of sequences per time period is listed in parentheses. These primary consensus sequences are then grouped into second-layer and third-layer COBRA sequences. The final COBRA HA sequence represents the layered set of sequence from a particular period of time.
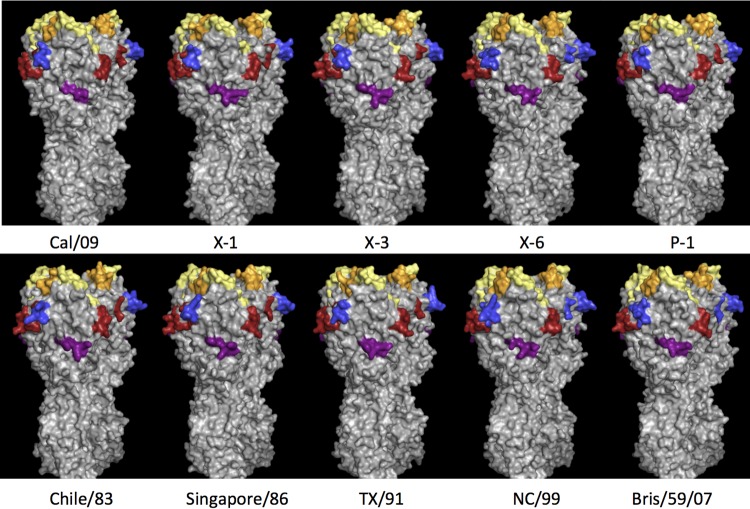
FIG 2.
Schematic of four COBRA HA proteins and six wild-type H1N1 HA trimerized proteins are depicted. Each COBRA HA structure presented is generated using the 3D-JIGSAW algorithm based on reference 13, and renderings were performed using MacPyMol. The Sa site is shown in yellow, the Sb site in orange, the Ca1 site in blue, the Ca2 site in red, and the Cb site in purple.
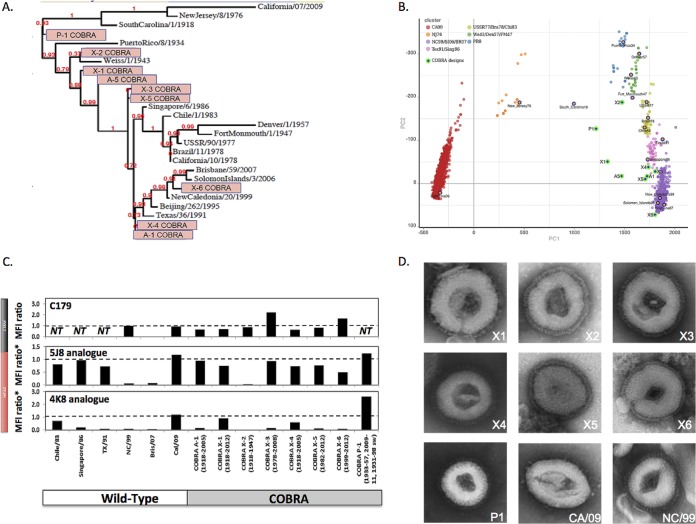
FIG 3.
Characterization of the H1N1 influenza COBRA HA vaccines. (A) The unrooted phylogenetic tree was inferred from HA amino acid sequences derived from 17 representative H1N1 isolates and also the COBRA HA using the maximum likelihood method. Sequences were aligned with MUSCLE 3.7 software, and the alignment was refined by Gblocks 0.91b software. Phylogeny was determined using the maximum likelihood method with PhyML software. Trees were rendered using TreeDyn 198.3 software (45). The NCBI accession numbers for the HA sequences used in phylogeny inference were obtained through the Influenza Virus Resource (46). (B) Genetic map of the HA of H1N1 viruses isolated between 1918 and 2013 that was generated from the numbers of amino acid substitutions between strains in the antigenic map. The two principal components, principal component 1 (PC1) and principal component 2 (PC2), are shown on the x and y axes. The clusters are colored according to the era of isolation. The spheres that represent each COBRA HA are shown in green. (C) Binding of neutralizing anti-HA monoclonal antibodies to mammalian expressed COBRA HA proteins. Wild-type and COBRA HAs expressed on the surfaces of HEK 293FT cells were labeled with stem-specific C179 monoclonal antibody or analogues to head-specific 5J8 and 4K8 monoclonal antibodies before analysis by flow cytometry. Mean fluorescence intensity (MFI) values were corrected by subtracting background fluorescence in mock-transfected cells and are shown as a ratio compared to MFI obtained with wild-type HA reference (A/New Caledonia/20/1999 HA for C179 monoclonal antibody or A/California/07/2009 for 4K8 and 5J8 monoclonal antibody analogues). The presence of an asterisk (MFI ratio*) indicates that the ratios have been normalized for HA protein expression levels based on binding of control antibody to conserved HA stem epitope, which is necessary to correct for differential levels of expression. NT, not tested. (D) Electron micrographs are presented for seven COBRA HA VLP vaccines (X1, X2, X3, X4, X5, X6, and P1) and two wild-type HA VLP vaccines (CA/09 and NC/99).
In vitro expression.
Human embryonic kidney (HEK) 293T cells (1 × 106) were transiently transfected with 3 μg DNA expressing each COBRA or wild-type HA gene cassette. Cells were incubated for 72 h at 37°C and then lysed with 1% Triton X-100 and clarified supernatant harvested following centrifugation. Cell lysates were then electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAG) and transferred to a polyvinylidene difluoride (PVDF) membrane. The blot was probed with pooled mouse antisera from infections with A/Brisbane/57/2007 and A/California/07/2009 viruses. HA-antibody complexes were then detected using goat anti-mouse IgG labeled with horseradish peroxidase (HRP) (Southern Biotech, Birmingham, AL, USA). HRP activity was detected using chemiluminescent substrate (Pierce Biotechnology, Rockford, IL, USA) and exposed to X-ray film (Thermo Fisher, Pittsburgh, PA, USA).
Functional characterization.
To determine receptor-binding characteristics, virus-like particles (VLPs) containing COBRA HA proteins were purified from the supernatants of mammalian cell lines as previously described (13). HEK 293T cells were transiently transfected with plasmids expressing HIV Gag, COBRA HA, and neuraminidase NA (A/mallard/Alberta/24/01; H7N3) and incubated for 72 h at 37°C. Supernatants were collected, and VLPs were purified via ultracentrifugation (100,000 × g through 20% glycerol, weight per volume) for 4 h at 4°C. The pellets were subsequently resuspended in phosphate-buffered saline (PBS), pH 7.2, and stored at −80°C until use. Protein concentration was determined by Micro BCA Protein Assay Reagent kit (Pierce Biotechnology, Rockford, IL, USA). COBRA HA VLPs were prepared in various amounts as measured by total HA protein, and each individual preparation was 2-fold serially diluted in v-bottom microtiter plates. An equal volume of 1% horse erythrocytes (RBCs) (Lampire Biological Products, Pipersville, PA, USA) in PBS was added to the diluted VLPs and incubated for 60 min at room temperature. The HA titer was determined by the reciprocal dilution of the last well that contained agglutinated RBCs.
Cartography methods.
Human, influenza A (subtype H1N1) hemagglutinin protein sequences were obtained from the NCBI Influenza Virus Resource database, trimmed to remove signal peptides, transmembrane regions, and cytoplasmic tails, and the resulting ectodomain sequences were aligned using MAFFT (17). The pairwise dissimilarity matrix was calculated from the multiple-sequence alignment based on the Hamming distance between pairs of sequences with no prior assumptions regarding the function or structure of the sequences. Principal component analysis (PCA) was applied to the dissimilarity matrix for the purpose of dimension reduction and to facilitate visualization of the relative distances between HA proteins. The first two or three principal components were retained for visualizing protein relationships in sequence space and represent a reasonable approximation of the general structure of the phylogenetic tree. Calculations were performed using custom scripts written in python and R.
Antigenicity determination by monoclonal antibody binding.
To confirm antigenicity of cell surface-expressed COBRA HAs, HEK 293FT cells (1 × 106) were transiently transfected with 2 μg DNA encoding each COBRA HA or wild-type HA using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. Transfected cells were harvested 24 h posttransfection by gentle dissociation with TrypLE Express (Life Technologies) and labeled by using a LIVE/DEAD Fixable Far Red Dead Cell Stain kit (Life Technologies) to determine cell viability. Labeled cells were blocked in PBS containing 0.1% bovine serum albumin (BSA) prior to staining with 0.4 μg of anti-HA stem monoclonal antibody (MAb) C179 (TaKaRa) or anti-HA MAb analogues to human neutralizing antibodies directed against the head regions (5J8 [18]), and 4K8 [12]), followed by staining with Alexa Fluor 488-labeled anti-mouse or anti-human IgG secondary antibody (Life Technologies). Stained cells were analyzed on a Becton Dickinson FACSCalibur flow cytometer, and the results were analyzed using FlowJo software. All mean fluorescence intensity (MFI) values were corrected by subtracting background fluorescence in mock-transfected cells. Corrected MFI values were expressed as a ratio using A/New Caledonia/20/99 HA as a reference for C179 and A/California/07/09 as a reference for the 5J8 and 4K8 MAb analogues. Anti-HA head MAb binding values were further standardized for expression levels by correcting MFI values based on binding of a MAb directed against a conserved epitope in the HA stem region.
Vaccine preparation.
HEK 293T cells were transiently transfected with plasmids expressing HIV-1 Gag (optimized for expression in mammalian cells), NA (A/mallard/Alberta/24/01; H7N3) (optimized for expression in mammalian cells), and COBRA HA or HA from wild-type H1N1 strains and incubated for 72 h at 37°C (Medigen Inc., Rockville, MD, USA). Supernatants were collected, and cell debris was removed by low-speed centrifugation followed by vacuum filtration through a 0.22-μm sterile filter. VLPs were purified via ultracentrifugation (100,000 × g through 20% glycerol, weight per volume) for 4 h at 4°C. The pellets were subsequently resuspended in PBS (pH 7.2) and stored in single-use aliquots at −80°C until use. Total protein concentration was determined by using a Micro BCA Protein Assay Reagent kit (Pierce Biotechnology, Rockford, IL, USA).
HA-specific content was determined by densitometry of reducing SDS-PAGE with the HA band identified by Western blotting. Purified VLPs or other HA-containing samples and appropriate standards were electrophoresed on a 4 to 12% Bis-Tris SDS-PAG in morpholinepropanesulfonic acid (MOPS) buffer and transferred to a PVDF membrane (Life Technologies, Grand Island, NY). The blot was probed with mouse monoclonal antibody 15B7 (Immune Technology Corporation, New York, NY), and the HA-antibody complexes were detected by transilluminating fluorescence from a biotinylated goat anti-mouse IgG and the WesternDot 625 Western blot kit (Life Technologies). A gel run under identical conditions was stained with Sypro Ruby stain (Life Technologies). Gels and blots were imaged, and the migration distances of all bands were recorded using a Chemi-Doc XRS+ camera system with QuantityOne software (Bio-Rad, Hercules, CA). Molecular weights were calculated for all bands relative to MagicMarkXP standards (Life Technologies). The HA band was identified on the Sypro Ruby-stained gel as the one migrating at the same molecular weight for HA as determined from the Western blot. The densities of HA bands from recombinant A/Puerto Rico/8/1934 HA standards (Immune Technology Corp.) were used to generate standard curves. The amounts of HA in the purified VLP lanes were calculated by interpolation from the data of the standard curve. Experiments were performed in duplicate or triplicate, and multiple exposure times were analyzed for all iterations. Inactivated influenza virus (IIV) split vaccines were provided by Sanofi Pasteur.
Electron microscopy.
VLP samples were adsorbed onto freshly glow-discharged carbon-stabilized Parlodion-coated 400-mesh copper grids (Polysciences, Inc., Warrington, PA). The grids were then rinsed with buffer containing 20 mM Tris (pH 7.4) and 120 mM KCl, negatively stained with 1% phosphotungstic acid (pH 7.2), and then dried by blotting onto filter paper (Whatman no. 1). VLPs were visualized on a Hitachi H-7600 transmission electron microscope (Hitachi High Technologies America, Schaumburg, IL) operating at 80 kV and digitally captured with a charge-coupled-device (CCD) camera at 5-megapixel resolution (Advanced Microscopy Techniques Corp., Danvers, MA).
Mouse studies.
BALB/c mice (Mus musculus, females, 6 to 8 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME, USA) and housed in microisolator units and allowed free access to food and water and were cared for under USDA guidelines for laboratory animals. All procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC). Mice (16 mice per group) were vaccinated with purified VLPs (3.0 μg) based upon HA content from the densitometry assay or they were vaccinated with the monovalent inactivated split influenza vaccine, via intramuscular injection at week 0 and then boosted with the same vaccine at the same dose at weeks 4 and 8. In other cases, mice were primed with one VLP vaccine and boosted with a different vaccine. In addition, some mice were administered a cocktail of H1N1 COBRA VLP vaccines (1.5-μg dose of each of the two vaccines) or phosphate-buffered saline alone as a mock vaccination. Vaccines at each dose were formulated with an emulsified squalene-in-water adjuvant (Sanofi Pasteur, Lyon, France). The final concentration after mixing 1:1 with VLPs is 2.5% squalene. Twenty-eight days after each vaccination, blood samples were collected from anesthetized mice via the retro-orbital plexus and transferred to a microcentrifuge tube. The tubes were centrifuged, and serum samples were removed and frozen at −20°C ± 5°C.
Four weeks after final vaccination, mice were challenged intranasally with 1 × 106 PFU (10 50% lethal doses [LD50]) of the A/California/07/2009 (H1N1) in a volume of 50 μl. After infection, mice were monitored daily for weight loss, disease signs, and death for 14 days after infection. At days 2 and 3 postinfection, five mice were sacrificed per time point for determination of viral lung titers. Individual body weights and death were recorded for each group on each day after inoculation. The experimental endpoint was defined as >20% weight loss. All procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (19), Animal Welfare Act (20), and Biosafety in Microbiological and Biomedical Laboratories (21).
HAI assay.
The hemagglutination inhibition (HAI) assay was used to assess functional antibodies to the HA able to inhibit agglutination of turkey erythrocytes. The protocol was adapted from the CDC laboratory-based influenza surveillance manual (22). To inactivate nonspecific inhibitors, sera were treated with receptor-destroying enzyme (RDE) (Denka Seiken, Co., Japan) prior to being tested (14, 23–26). Briefly, three parts of RDE was added to one part of sera and incubated overnight at 37°C. RDE was inactivated by incubation at 56°C for ∼30 min. RDE-treated sera were diluted in a series of twofold serial dilutions in in v-bottom microtiter plates. An equal volume of each H1N1 virus, adjusted to approximately 8 hemagglutination units (HAU)/50 μl, was added to each well. The plates were covered and incubated at room temperature for 20 min, and then 1% turkey erythrocytes (Lampire Biologicals, Pipersville, PA, USA) in PBS were added. Red blood cells were stored at 4°C and used within 72 h of preparation. The plates were mixed by agitation and covered, and the RBCs were allowed to settle for 1 h at room temperature (18). The HAI titer was determined by the reciprocal dilution of the last well that contained nonagglutinated RBCs. Positive and negative serum controls were included for each plate. All mice were negative (HAI ≤ 1:10) for preexisting antibodies to currently circulating human influenza viruses prior to vaccination.
Plaque assay.
Madin-Darby canine kidney (MDCK) cells were plated (5 × 105) in each well of a six-well plate. Samples were diluted (final dilution factors of 100 to 10−6) and overlaid onto the cells in 100 μl of Dulbecco modified Eagle medium (DMEM) supplemented with penicillin-streptomycin and incubated for 1 h. Samples were removed, cells were washed twice, and medium was replaced with 2 ml of L15 medium plus 0.8% agarose (Cambrex, East Rutherford, NJ, USA) and incubated for 72 h at 37°C with 5% CO2. Agarose was removed and discarded. The cells were fixed with 10% buffered formalin and then stained with 1% crystal violet for 15 min. Following thorough washing in distilled water (dH2O) to remove excess crystal violet, the plates were allowed to dry, the numbers of plaques were counted, and the numbers of PFU per milliliter were calculated.
Statistical analysis.
Statistical significance of the antibody data was determined using a two-way analysis of variance (ANOVA) with Bonferroni's posttest to analyze differences between each vaccine group for the different test antigens (multiparametric). Differences in weight loss, sickness score, and viral titers were analyzed by two-way ANOVA, followed by Bonferroni's posttest for each vaccine group at multiple time points. Statistical significance was defined as P value of <0.05. Statistical analyses were done using GraphPad Prism software.
Nucleotide sequences.
The sequences for all nine COBRA HAs have been reported in U.S. patent filings patent US 9,212,207 B2, 14/388,726, and filing patent US 14/126,550.
RESULTS
Design of H1N1 COBRA HA genes.
Previous studies from our laboratory used the COBRA methodology to design H5N1 HA sequences to account for various clades and subclades of H5N1, as well as outbreaks using sequences obtained from viruses collected over an approximately 5-year span of time (13). A similar method was designed for the generation of COBRA HA genes for H1N1; however, these designs took into account the evolution of H1N1 influenza viruses isolated in humans over a ca. 100-year span of time, and therefore, the diversity of the HAs from H1N1 influenza viruses is quite large (Fig. 1A). Viruses isolated represent different antigenic eras of H1N1 HA sequences that result in a higher diversity of sequence variation. The COBRA design utilized these chronologically different eras of H1N1 HA sequences to account for the unique antigenic types of HA domains. HA sequences were designed both chronologically, as well as by species from which influenza virus was isolated. An example of the COBRA sequence alignments is shown in Fig. 1B. Primary consensus sequences were then aligned, and the most common amino acid was chosen resulting in secondary consensus sequences. The secondary and tertiary consensus sequences were aligned, and the most common amino acid at each position was selected resulting in the final consensus sequence referred to by one of 9 H1N1 COBRA HA designations (Table 1). Three of the H1N1 COBRA designs, A1, A5, and X1, encompassed HA sequences that represented the last ∼100 years of H1N1 history (Table 1). A1 contains only seasonal H1N1 HA sequences. The X2 sequence represented a historical era of H1N1 from 1933 to 1947, and three COBRA sequences represented the modern era of H1N1 with X3 (1978 to 2008), X5 (1982 to 2012), and X6 (1999 to 2012). X3 does not contain pandemic H1N1 HA sequences, but X5 and X6 do contain sequences representing the post-2009 pandemic H1N1 era. X4 is the same sequence as A1; however, the glycosylation sites at amino acid 142 and 177 have been mutated to prevent glycosylation. Last, P1 represents a unique set of H1N1 HA sequences isolated from both human and swine species. The sequences used in the methodology included HA sequences isolated from humans between 1933 and 1957 and between 2009 and 2011, as well as swine HA sequences from viruses isolated between 1933 and 1998. Using a predictive structural modeling of COBRA H1N1 HA sequences, three-dimensional trimerized HA proteins were designed for five COBRA HA sequences and five wild-type HA sequences (Fig. 2).
VLP characterization of the H1N1 COBRA HA VLP vaccines.
The design and characterization of the COBRA HA proteins have been described previously (10–13). Despite nearly identical predicted structures, the COBRA HA proteins did have subtle differences in the major antigenic binding and receptor-binding sites. Phylogenetic analysis of the COBRA HA proteins indicated that each molecule clustered with different representative wild-type vaccine strains (Fig. 3A). The P1 COBRA HA located on the tree with the swine-like viruses (A/California/07/2009 [CA/09], New Jersey/8/1976 [NJ/76], and South Carolina/1/1918 [SC/1918]) and the X1, X2, and A5 COBRA HAs clustered with historical H1N1 viruses. The X3, X4, X5, X6, and A1 H1N1 COBRA HA proteins clustered with the more modern seasonal H1N1 viruses. Similar to the phylogenetic analysis, each COBRA HA locates on a two-dimensional PCA plot with different clusters of seasonal H1N1 influenza viruses (Fig. 3B). Furthermore, a BLAST search using each of the COBRA HA sequences revealed that each sequence was a unique sequence that has not been isolated from the environment (data not shown).
Binding of monoclonal antibodies to the influenza virus HA head and stem domains.
COBRA or wild-type HA proteins were expressed on the surfaces of HEK 293FT cells and used to test the binding of three previously characterized monoclonal antibodies using flow cytometry (17, 27–29). The monoclonal antibody C179 binds to conformational epitopes in the conserved stem domain of H1N1 HA proteins (30). As expected, C179 monoclonal antibody bound all tested wild-type and COBRA HA proteins although with varying efficiencies indicating exposure of different known epitopes on the surfaces of the various HA proteins. In contrast, the two HA head-specific monoclonal antibodies bound to some, but not all, COBRA and wild-type HA proteins (Fig. 3C). The 5J8 monoclonal antibody bound to the HA from modern seasonal isolates A/Chile/1/1983 (Chile/83), A/Texas/36/91 (TX/91), A/Singapore/6/86 (Singapore/86), and A/California/07/2009 (CA/09). Significantly, 5J8 and 4K8 antibodies failed to bind the HA from A/New Caledonia/20/1999 (NC/99) or A/Brisbane/59/2007 (Bris/07), consistent with the poor neutralizing activity exhibited by these antibodies against both influenza H1N1 strains (28, 29). In the case of COBRA antigens, 5J8 bound to the majority of designs except for X2, which was not recognized by any of the head-specific antibodies tested. In contrast, the 4K8 antibody efficiently bound to X1 and P1 with similar antibody binding to pandemic CA/09 HA. Interestingly, 4K8 also bound to X4, but not to A1, suggesting that removal of glycosylation sites on A1 may have contributed to unmasking the 4K8 epitope. Overall, the pattern of monoclonal antibody binding indicates that each of the COBRA HA proteins have differently exposed epitopes and therefore may have different antigenic properties.
Characterization of virus-like particle vaccines expressing COBRA HA.
COBRA and wild-type HA VLP vaccines were adsorbed and visualized by electron microscopy (Fig. 3D). Each VLP had a classic viral core structure with visible spike proteins representing HA and NA gene products. With the exception of CA/09 VLPs, the surface glycoproteins were uniform in size and distribution. CA/09 VLPs had fewer and more irregular surface glycoproteins (Fig. 3D). All wild-type and COBRA VLPs were designed to express a mismatched NA from the N3 subtype (taxonomy identifier [ID] 333366, http://www.ncbi.nlm.nih.gov).
Vaccinated mice challenged with pandemic H1N1 influenza virus.
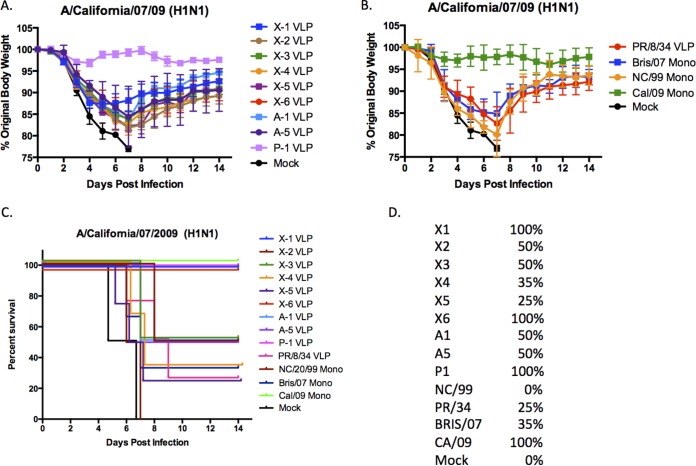
To determine the protective efficacy of each H1N1 COBRA vaccine, mice were vaccinated using different regimens and then challenged with CA/09 influenza virus. BALB/c mice (n = 11) were vaccinated twice at 4-week intervals via intramuscular injection with purified COBRA HA VLPs (3 μg based upon HA content) with one of the nine H1N1 COBRA HA VLP vaccines plus an oil-in-water-based adjuvant (proprietary to Sanofi Pasteur). Mice were then challenged with CA/09 influenza virus (1 × 106 PFU) on day 84. Mice vaccinated with P1 VLPs or the CA/09 influenza monovalent inactivated influenza vaccine (IIV) were protected from weight loss and death, while mock-vaccinated animals rapidly lost weight and reached experimental endpoints between 6 and 7 days postinfection (dpi) (Fig. 4). All other COBRA VLP-vaccinated mice lost between 10 and 20% of body weight by day 6 or 7 postinfection and then slowly began to recover weight (Fig. 4A). Even though mice vaccinated with the X6 VLP lost weight, all the mice survived the influenza virus challenge (Fig. 4C). In contrast, other mouse groups demonstrating weight loss ultimately had between 25% (X5) and 50% (X2, X3, A1, and A5) survival. Similar results were observed using seasonal H1N1 IIV (Fig. 4B). Mock-vaccinated mice all died by day 7 postinfection.
FIG 4.
H1N1 influenza virus challenge of mice. (A and B) BALB/c mice (11 mice/group) were vaccinated on days 0, 28, and 56 with each vaccine plus the AF03 adjuvant and infected on day 84 with 1 × 10e+6 PFU of the H1N1 isolate A/California/07/2009 (CA/09). Mice were monitored daily for weight loss over a 14-day observation period. Three mice were sacrificed on day 3 postinfection to assess lung titers; therefore, the weights for 5 mice were recorded for the entire 14 days. Some mice were vaccinated with one virus (Mono). (C) Kaplan-Meier survival curves for vaccinated mice challenged with CA/09 influenza virus. (D) Percent survival per vaccine group is listed to the right of the graph.
H1N1 COBRA HA VLP vaccines elicit broadly reactive antibody responses.
In order to determine the breadth of the vaccine-elicited antibodies, serum was collected from vaccinated mice on day 84, following the third vaccination (Fig. 5). Mice vaccinated with each of the three monovalent IIV vaccines elicited antibodies that had HAI activity against different sets of H1N1 influenza viruses (Fig. 5K to M). Mice vaccinated with the Bris/07 or NC/99 IIV vaccine had HAI activity exceeding 1:40 against four seasonal viruses isolated from 1995 to 2007 with little or no activity against the remaining 15-member H1N1 viral panel. Vaccination with the CA/09 IIV vaccine elicited high HAI antibodies against the two pandemic H1N1 strains (CA/09 and Philadelphia/1/2013 [Phil/13]) and SC/18 (Fig. 5M). All vaccinated mice had high-titer anti-HA antibodies that bound to recombinant HA derived from the CA/09 and Bris/07 viruses as detected by enzyme-linked immunosorbent assay (ELISA) (data not shown).
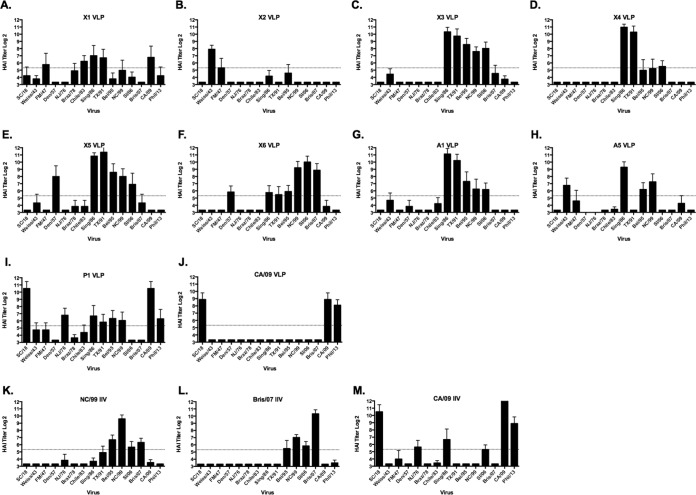
FIG 5.
Hemagglutination inhibition (HAI) serum antibody titers induced by vaccination of mice with COBRA VLP vaccines. HAI titers were determined for each group of mice (n = 11) vaccinated three times (days 0, 28, and 56) with 1 of the 9 COBRA VLP vaccines or with 3 of the inactivated influenza virus (IIV) vaccines against a panel of 15 H1N1 influenza viruses. The CA/09 VLP was used as a control wild-type HA VLP. Values are the geometric mean titers plus standard errors of the means (SEM) (error bars) from antisera collected on day 84. Dotted lines indicate the 1:40 HAI titer.
Although all COBRA HA VLP vaccines elicited similar IgG titers, COBRA-vaccinated animals had HAI antibodies that recognized different sets of viruses from the H1N1 panel (Fig. 5). Mice vaccinated with the X3, X5, X6, and A1 VLP vaccine candidates had HAI titers against a set of seasonal influenza viruses from 1986 to 2006. Only mice vaccinated with the X6 VLP candidates detected the Bris/07 virus and the viruses representing the viruses circulating in 1943 to 1983 were occasionally detected. The X4 and A1 VLP vaccine candidates had high HAI activity against Sing/86 and TX/91 but little or low HAI activity against other viruses in the panel (Fig. 5D and E). Mice vaccinated with the X1 VLPs had low-titer antibodies that recognized viruses from 1978 to 1991 (Fig. 5A). The X2 VLP candidate had robust HAI activity against the oldest viruses in the panel (1934 to 1947) and little activity against any of the other H1N1 strains (Fig. 5B). The only VLP candidate that elicited HAI-specific antibodies against the pandemic H1N1 strains were P1 and X1. All of the mice vaccinated with P1 had HAI activity against both CA/09 and Phil/13 viruses. In addition to the two pandemic H1N1 viruses, mice vaccinated with P1 also had low HAI activity against four other seasonal H1N1 viruses, as well as NJ/76 and SC/18 for a total of eight viruses recognized by the vaccine-elicited sera (Fig. 5I). This was the broadest HAI activity elicited by any of the H1N1 COBRA HA VLP vaccine candidates.
In order to enhance the breadth of elicited HAI activity by H1N1 COBRA HA VLP vaccine candidates, mice were vaccinated with different combinations of the COBRA VLPs in a prime-boost strategy, and sera were tested for HAI activity on day 56 postvaccination. Mice were vaccinated with one of four H1N1 COBRA VLP candidates (X1, X3, X6, and P1) in multiple prime-boost combinations. Mice vaccinated with prime-boost combinations of the X1 and X6 COBRA VLPs had little HAI activity against any of the 15 viruses in the panel (data not shown), whereas mice primed with X3 and boosted with X6 had HAI activity against Sing/86, Beijing/1995 (Bei/95), NC/99, and Solomon Island/2006 (SI/06) (data not shown). Other vaccine combinations elicited little or low HAI activity. There was no activity against pandemic H1N1 (2009 to 2013) or historical H1N1 strains (1934 to 1957) by any of these combinations of vaccine candidates. In contrast, mice vaccinated with combinations containing the P1 COBRA VLP candidate elicited antibodies that recognized different sets of H1N1 viruses. Mice primed with P1 VLPs and then boosted with either X3 or X6 VLPs had high HAI activity against four seasonal (A/Weiss/43 [Weiss/43], Sing/86, Bei/95, and NC/99) and the two pandemic H1N1 viruses. When X3 VLPs were used to prime mice and those mice were boosted with P1, HAI activity against the same four seasonal viruses were detected, but there was no HAI detected against the two pandemic viruses. Interestingly, priming with P1 COBRA VLPs elicits higher HAI titers than boosting with P1 COBRA VLPs. P1 and X1 combinations were quite poor. Priming with P1 and boosting with X1 elicited HAI activity against only the pandemic strains, and there was no detectable HAI activity when mice were primed with X1 VLPs and boosted with any other COBRA VLP candidate. Interestingly, the HAI activity of mice primed with P1 and boosted with X3 VLPs was similar to the HAI activities of mice treated with combinations of P1 and X6 VLPs.
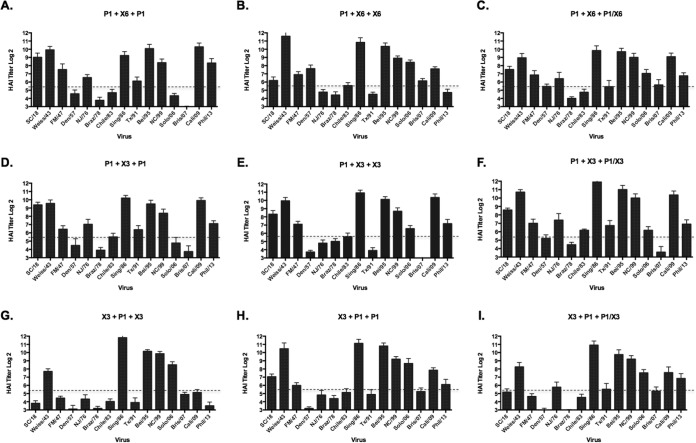
To test additional potential immunization regimens, some groups of mice received a third vaccination on day 56 with one of the vaccine candidates administered in the two prior immunizations (Fig. 6). Mice primed with P1 VLPs and then boosted with either X6 or X3 COBRA VLP vaccines and then boosted again with P1 VLPs had serum HAI activity against the same 10 viruses (out of 15) in the panel (Fig. 6A and D). When the same prime-boost regimens were instead followed by a third immunization with X3 or X6 COBRA VLPs, the animals had HAI activity against the same seasonal viruses in the panel but reduced HAI activity against the SC/18, NJ/76, CA/09, and Phil/13 (Fig. 6B and E). Priming with X3 COBRA VLPs and boosting with P1 VLPs followed by another boost of P1 VLPs elicited HAI activity in 9 of the 15 viruses (Fig. 6H), whereas administering X3 as the third VLP vaccination elicited antibodies with HAI activity against only 5 seasonal viruses (Fig. 6G).
FIG 6.
Hemagglutination inhibition serum antibody titers induced by vaccination of mice with prime-boost regimens using COBRA VLP vaccines. HAI titers were determined for each group of mice vaccinated three times (on days 0, 28, and 56) with combinations of the 3 COBRA VLP vaccines against a panel of 15 H1N1 influenza viruses. Panels C, F, and I were vaccinated with a cocktail of the prime-boost vaccines as the third vaccination. Values are the geometric mean titers plus standard errors of the means (SEM) (error bars) from antisera collected on day 84. Dotted lines indicate the 1:40 HAI titer.
Heterologous cocktail third vaccination.
On day 56, some groups of mice that were given a heterologous prime-boost vaccination regimen were divided into subgroups and given a cocktail of their first two vaccinations as their third vaccination (Fig. 6C, F, and I). Mice primed with P1 COBRA VLPs, boosted with X6 COBRA VLPs, followed by a third cocktail vaccination of P1/X6, had HAI activity against 10 viruses in the H1N1 panel on day 84 (Fig. 6C). Similar results were observed using a similar regimen with P1 and X3 VLP combinations (Fig. 6F). Both sets of mice had HAI activity against SC/18, Weiss/43, Sing/86, Bei/95, NC/99, and CA/09. To a lesser extent, A/Fort Monmouth/1/47 (FM/47), Chile/83, A/Texas/36/91 (TX/91), SI/06, Bris/07, and Phil/13 were also detected. Mice administered X3 as the prime, followed by P1 VLP boost and boosted again with the P1/X3 VLP cocktail had similar activities but lower titers to SC/18, CA/09, and Phil/13 (Fig. 6I).
HAI activity against H1N1 strains by cocktail mixtures of H1N1 COBRA HA VLP vaccines.
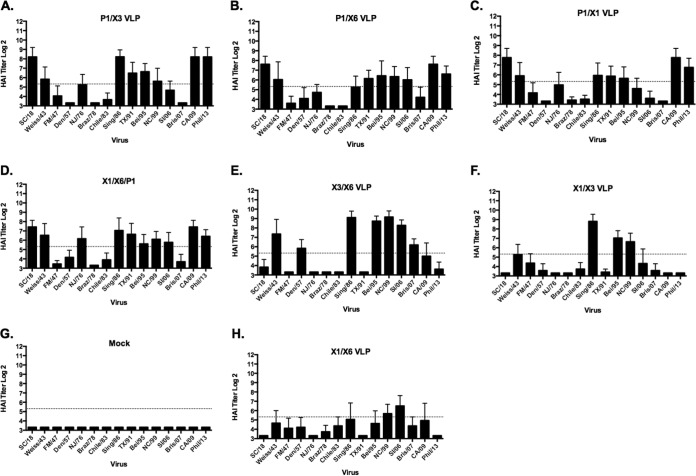
Since each of the H1N1 COBRA HA proteins elicit HAI activity to different sets of H1N1 viral strains, we decided to mix cocktails of COBRA VLP candidates together with the goal of stimulating the broadest breadth of HAI activity against the panel of 15 H1N1 isolates (Fig. 7). The main rationale for using cocktails was to increase the breadth of HAI responses over and above what can be achieved using a monovalent approach, using an immunization regimen more compatible with a final vaccine product. Mice were vaccinated with different combinations of two H1N1 COBRA VLP vaccine candidates simultaneously (1.5-μg dose of each VLP vaccine). On day 84, after three vaccinations, mice vaccinated with a cocktail of X1 and X3 VLPs (Fig. 7F) had HAI activity against three viruses in the panel (Sing/86, Bei/95, and NC/99), and mice vaccinated with a cocktail of X1 and X6 VLPs (Fig. 7H) recognized only NC/99 and SI/06. However, mice vaccinated with X3 and X6 VLPs simultaneously had HAI activity against seven seasonal H1N1 influenza viruses (Weiss/43, A/Denver/1/1957 [Den/57], Sing/86, Bei/95, NC/99, SI/06, and Bris/07) (Fig. 7E). The X3/X6 cocktail combination was the only COBRA vaccine to effectively elicit antibodies that detected Bris/07 and Den/57. When mice were vaccinated with cocktails containing the P1 VLPs, all mice had HAI activity against SC/18, CA/09, and Phil 13 (Fig. 7A to D). Cocktails of P1 and X6 elicited antibodies with HAI activity against viruses from 1986 to 2006 but little or no activity against earlier H1N1 isolates or Bris/07 (Fig. 7B). Cocktails of P1 and X3 elicited antibodies with HAI activity against viruses from 1986 to 2006, as well as NJ/76, Weiss/43, but low HAI activity against CA/09 and Phil/13 (Fig. 7A). Mice vaccinated with a triple cocktail of X1, X6, and P1 (1 μg of each VLP vaccine) had HAI activity against 10 H1N1 viruses in the panel, albeit generally at low levels (Fig. 7D).
FIG 7.
Hemagglutination inhibition serum antibody titers induced by vaccination of mice with cocktails of COBRA VLP vaccines. HAI titers were determined for each group of mice vaccinated three times (days 0, 28, and 56) with cocktails of two or three COBRA VLP vaccines against a panel of 15 H1N1 influenza viruses. Values are the geometric mean titers plus SEM from antisera collected on day 84. Dotted lines indicate the 1:40 HAI titer.
HAI activity against H1N1 strains by cocktail mixtures of wild-type H1N1 HA VLP vaccines.
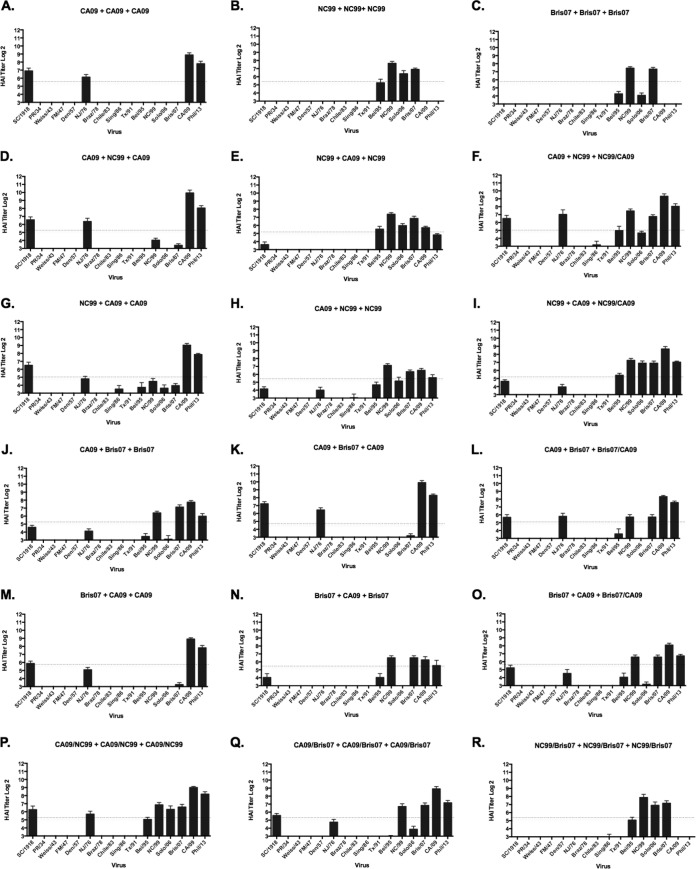
In order to determine the breadth of HAI activity against the panel of H1N1 strains that was unique to the HA COBRA vaccines, three wild-type HA genes were selected from the NC/99, Bris/07, and CA/09 influenza strains and used to generate VLPs with N3 NA in a similar manner as the COBRA VLPs. These VLP vaccines were used to vaccinate mice in a prime-boost-boost regimen with homologous VLPs or in various combinations of heterologous vaccination (Fig. 8). At both week 8 after two vaccinations (data not shown) and week 12 after the third vaccination, mice vaccinated with NC/99 and Bris/07 VLPs or CA/09 VLPs (three times) elicited antibodies that had HAI activity against each homologous virus and 1 to 3 of the other 17 viruses in the panel (Fig. 8A to C). When these wild-type HA VLPs were used in a cocktail or prime-boost regimen, there was no increase in the breadth of HAI activity against the panel of viruses, but rather a blending of the two responses (Fig. 8). This is in contrast with the same regimens using COBRA HA VLPs, where the elicitation of antibodies expanded the number of viruses in the H1N1 panel (Fig. 6 and 7). Therefore, the COBRA HA VLPs, when used in combination, had more HAI activity against a broader number of viruses not included in the sequences used to design the COBRA HA sequence. The wild-type HA VLP elicited antibodies with a narrow breadth of HAI activity than the COBRA HA proteins did.
FIG 8.
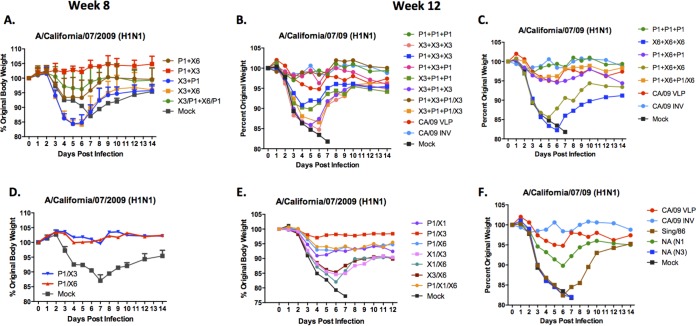
Hemagglutination inhibition serum antibody titers induced by vaccination of mice with prime-boost and cocktail regimens using wild-type VLP vaccines. HAI titers were determined for each group of mice vaccinated three times (days 0, 28, and 56) with combinations of the three COBRA VLP vaccines against a panel of 15 H1N1 influenza viruses. (A to C) Wild-type HA VLP administered three times; (D to I) prime-boost of CA09 and NC99; (J to O) prime-boost of CA09 and Bris07; (P) cocktails of CA09/NC99 administered three times; (Q) cocktails of CA09/Bris07 administered three times; (R) cocktails of NC99/Bris07 administered three times. Values are the geometric mean titers plus SEM from antisera collected on day 84. Titers below the limit of detection were assigned a value of 5 for the purpose of calculating the geometric mean titer and are not visible on this scale. Dotted lines indicate the 1:40 HAI titer.
Vaccinated mice challenged with CA/09 virus.
Groups of mice were challenged with CA/09 influenza virus (1 × 106 PFU) on day 56 and day 84. Mice vaccinated with any prime-boost combination including the P1 VLPs had an average weight loss of less than 7%, except for mice primed with X1 and boosted with P1, which lost 13% of their original weight by day 6 postinfection following infection at day 56 (Fig. 9A) and at day 84 (Fig. 9B and C). All other prime-boost vaccinated mice lost between 10 and 20% of body weight by day 6 or 7 postinfection. Mice that did not reach 20% weight loss by day 7 typically recovered. All mock-vaccinated mice lost greater than 20% body weight, and all were humanely euthanized by day 7 postinfection.
FIG 9.
H1N1 influenza virus challenge of mice vaccinated with cocktails or prime-boost regimens. BALB/c mice (11 mice/group) vaccinated three times (days 0, 28, and 56) with each vaccine plus AF03 adjuvant were infected with 1 × 10e+6 PFU of the H1N1 isolate A/California/07/2009 (CA/09). Mice were monitored daily for weight loss over a 14-day observation period. Three mice were sacrificed on day 3 postinfection to assess lung titers; therefore, the weights for 5 mice were recorded for the entire 14 days. Percent original weight for mice vaccinated by two different VLP vaccines (or cocktails) by prime-boost-boost and challenged on day 56 (A), vaccinated by two different VLP vaccines (or cocktails for the second boost) by prime-boost and challenged on day 84 (B and C), vaccinated with cocktails of two COBRA VLP vaccines and challenged on day 56 (D), vaccinated with cocktails of two COBRA VLP vaccines and challenged on day 84 (E), and vaccinated with control vaccines and challenged on day 84 (F).
For cocktail-vaccinated mice, again vaccine regimens containing P1 VLPs were better protected against CA/09 challenge than mice vaccinated with other COBRA VLP vaccine candidate cocktails (Fig. 9D and E). Mice vaccinated with a cocktail of P1 and X3 VLPs had no signs of morbidity and weight loss, whereas mice vaccinated with other cocktails containing P1 VLPs lost on average 5 to 7% body weight. Mice vaccinated with cocktail candidates not containing P1 VLPs lost between 15 and 20% body weight, with few mice surviving beyond day 7 in any group. Mice vaccinated with P1 COBRA VLPs were protected against CA/09 virus challenge as well as mice vaccinated with control CA/09 VLPs or CA/09 IIV monovalent vaccines were (Fig. 9F). Interestingly, vaccination of mice with N1 neuraminidase (CA/09), but not N3, protected mice against severe weight loss and disease.
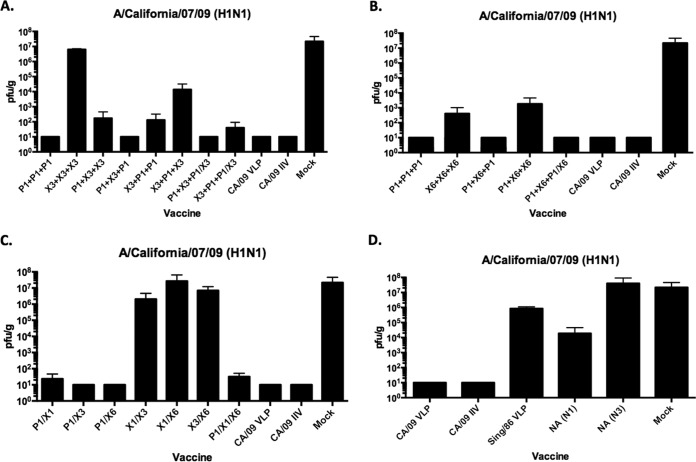
On day 3 postinfection, all mice primed with P1 VLPs had little or no detectable viral titers in their lungs (Fig. 10A and B), which was similar to mice vaccinated with either of the CA/09 vaccines. Mice primed with X3 or X6 COBRA VLPs and boosted with P1 VLPs had moderate viral lung titers. Mice vaccinated with any other prime-boost regimens of COBRA VLPs, except X6, had viral lung titers greater than 10e+6 PFU/g, which was similar to mock-vaccinated mice. Mice vaccinated with any cocktail containing P1 VLPs had virtually no detectable viral titers in their lungs (Fig. 10C and D). Viral lung titers were greater than 10e+6 PFU/g in mice vaccinated with any other cocktail regimen.
FIG 10.
Viral lung titers in vaccinated mice. Vaccinated BALB/c mice challenged on day 84 postvaccination had lungs collected (3 mice/group/time point) on day 3 postinfection from COBRA VLP-vaccinated mice plus controls. Viral lung titers are listed as PFU per gram of lung tissue. (A and B) Prime-boost-boost. (C) Cocktails. (D) Control immunizations.
DISCUSSION
Current licensed influenza vaccines induce hemagglutination inhibition (HAI) antibodies that do not cross-react with influenza strains that have undergone genetic drift. There is an urgent unmet need for more broadly protective seasonal influenza vaccines with greater breadth, enhanced potency, and durability of HAI responses. It is currently impossible to predict which antigenic variants may evolve, and therefore, novel vaccine candidates are needed that will elicit immunity to potential variants. Recently, we designed a new technology termed COBRA as a universal/more broadly reactive vaccine approach for both emerging/pandemic influenza viruses and currently circulating influenza subtypes. We have published a series of studies demonstrating the effectiveness of the COBRA HA antigens for H5N1 (11–13).
This method was applied to address the diversity within the H1N1 HA and yields a vaccine that elicits increased breadth of antibody response within the H1 influenza subtype. The method uses multiple rounds of consensus building based upon not only the phylogenetic sequence of each isolate but also the outbreak and specific time that each isolate was collected, thereby eliminating the bias in the number of sequences uploaded to online databases. Often similar or virtually identical HA sequences are listed in NCBI or GISAID databases that represent individual strains. This is primarily due to the fact that numerous isolates are collected and sequenced from a single outbreak from a human or zoonotic source in a single location at one specific period of time. Therefore, in reality, often these sequences actually represent one “antigenic type” or “sequence” of HA collected from thousands of viral isolates and not thousands of individual viral isolates. This repetition of HA sequences in the database accounts for the limitation inherent in using these sequences for identification of highly immunogenic, cross-reactive sets of epitopes for vaccine design. Using multiple rounds of consensus building and weighting each of these isolates for sequence, outbreak group, and time of isolation eliminates this bias and allows COBRA designs to retain highly immunogenic and cross-reactive epitopes.
In this study, nine independent COBRA HA genes were designed to elicit anti-HA antibodies directed at the head domain of the H1N1 HA. Some of the COBRA HA proteins were designed using H1N1 HA sequences that represent specific recent periods of time (X3, X5, and X6), whereas other HA COBRA proteins were designed using HA sequences from viruses isolated from 1918 to present (A1, A5, and X1) to cover almost the entire known antigenic history of H1N1 influenza virus evolution in humans (Table 1). Other HA COBRA proteins were designed to address specific hypotheses (X2, X4, and P1).
Four of the H1N1 COBRA HA proteins (X1, X3, X6, and P1) were further characterized. Of all the COBRA vaccine candidates, the P1 COBRA VLPs elicited antibodies that had HAI activity against both pandemic and seasonal influenza strains (Fig. 5). Even though X1, X3, and X6 did not prevent weight loss following CA/09 virus challenge, these vaccines did elicit HAI antibodies against most seasonal H1N1 strains isolated in the last 30 years (Fig. 5), which would indicate that these vaccines would elicit broadly reactive immune response against seasonal influenza strains. Interestingly, X1 and X6 did protect all mice against CA/09 challenge (Fig. 4). In fact, X6 had little virus detected in the lungs of challenge mice, even though there were no HAI titers. While this may seem unusual, in a previous study, ferrets exposed to FM47 virus had low HAI titers, but it did protect animals against direct infection and virus was not transmitted to other ferrets (31). Future vaccine studies using these COBRA VLPs in ferrets will allow for multiple challenges using both seasonal and pandemic H1N1 viruses to demonstrate true protective efficacy against a diverse panel of H1N1 viruses.
While there was not a single H1 COBRA design that could elicit HAI antibodies to all viruses in the panel, collectively these vaccines elicited a polyclonal antibody response that did recognize all viruses in the panel. We speculated that presenting COBRA HA VLP vaccines to the immune system in a prime-boost-boost regimen or mixed together in a cocktail would elicit antibody responses able to recognize most, if not all, the H1N1 viruses in the panel. We recently demonstrated that a cocktail of three H5N1 COBRA VLPs elicited antibodies that recognized H5N1 viruses from 11 H5N1 clades/subclades isolated over a 12-year span (10). There are structural changes between HA proteins from different eras that may expose different epitopes on the HA molecule. Individually, each of these H1N1 COBRA HA VLP vaccines most likely elicited antibodies that neutralized antigenically related H1N1 viruses in the panel based upon similarities in the structures of their HA molecules. The four leading candidates were used in various prime-boost-boost combinations. Any combination using X1, X3, or X6 COBRA VLPs resulted in broad HAI activity against the seasonal H1N1 viruses in the panel but little or no activity against the pandemic/swine strains (SC/18, NJ/76, CA/09, and Phil/13). When these three COBRA VLPs were used in combination with P1 VLPs, the breadth of HAI activity extended to pandemic strains. The breadth of HAI activity was more significant than the HAI activity elicited by combinations of VLP vaccines with wild-type HA protein.
It appears that the priming vaccination dose is a significant factor in determining the viruses in the panel that will be recognized by the vaccine-induced antibodies. The priming dose drives the immune response and influences how the host responds to subsequent booster vaccinations. If the prime vaccination was X3 COBRA VLPs, for example, it takes two booster vaccinations with P1 VLPs to elicit antibodies with HAI activity against pandemic strains (Fig. 6H). Priming with P1 COBRA sets the B cell repertoire to elicit antibodies that recognize a specific set of H1N1 viruses that include pandemic H1N1 strains. Interestingly, using a cocktail of the first two COBRA vaccines as the third vaccination resulted in the greatest breadth of HAI activity, activity greater than using a single COBRA VLP alone (Fig. 6). Using the P1/X6 combination as an example, a third immunization with the P1/X6 cocktail resulted in seroprotection against 13 strains, whereas P1 or X6 used individually resulted in seroprotection against fewer strains. Similar trends were observed for other COBRA vaccine combinations. All studies were performed in immunologically naive mice that had no preexisting immunity to influenza, and the observed vaccine-induced immune responses were always consistently broad. Most humans have a varied and extensive repertoire of preexisting memory B cells with circulating antibodies that recognize previous H1N1 strains (32–34). The current trivalent influenza virus (IIV) has a single H1N1 strain that can recall some, but not all, B cell memory responses. However, a COBRA HA vaccine has the potential to recall a broader repertoire of memory responses and therefore protect against more antigenic H1N1 variants.
While both P1 COBRA VLPs and CA/09 VLPs elicit similar HAI antibody titers against CA/09, CA/09 VLPs elicited higher HAI titers against Phil/13, a pandemic H1N1 lineage virus, but not a true genetic drift variant (Fig. 5 to 8). However, we expect that once true genetic drift variants emerge, by definition antibodies elicited by CA/09 virus will not recognize these new variants; however, P1 COBRA-elicited antibodies may still have the potential to recognize these future genetic drift variants. In addition, only P1 can elicit antibodies that neutralize seasonal H1N1 strains, whereas CA/09 VLPs do not elicit functional antibodies to any seasonal strains. Therefore, P1 COBRA HA is a unique antigen that has different epitopes for induction of more broadly reactive antibodies at lower titers than HA proteins that elicit a more narrow breadth of high HAI activity.
Developing a universal vaccine strategy for seasonal influenza virus subtypes has the potential to greatly impact human health. A vaccine that protects against influenza variants within and across subtypes will improve our understanding of the basic biology, evolution, and transmission of influenza A virus. There are more than 40 universal influenza vaccine candidates in development (for reviews, see references 9, 35, and 36), but almost all of them elicit immune responses that only modulate influenza-induced disease and are not likely to offer the same protective capability as a well-matched HA-based vaccine. The COBRA HA vaccines are one of the few universal influenza vaccine candidates that elicit immune responses that offer protection against influenza disease. The COBRA approach refocuses the HA immunogen so as to elicit functional antibodies that are broad in coverage within a subtype. COBRA HA-elicited antibodies had potent HAI activity against a broad panel of H1N1 strains isolated over several decades that was superior to the antibodies elicited by wild-type HA vaccines (Fig. 8). In contrast to head-based universal strategies, anti-HA stem-based approaches elicit antibodies that recognize the stem portion of HA in isolates across several strains within a subtype (37) and multiple subtypes within a group (38). Stem-induced antibodies, just as head-based elicited antibodies, may function through disease modulation. However, there is no correlate of protection for stem-induced antibodies for human vaccine studies. Some groups have demonstrated that stem-induced antibodies may have suboptimal neutralization potency compared to antibodies directed against HA head domain (39, 40), whereas other studies have demonstrated high binding potency of these antibodies (41). These stem epitopes may be less accessible than HA head epitopes, and therefore, newer presentation and delivery strategies have been developed to present HA stem regions to the immune system more effectively (42, 43). COBRA HA-elicited antibodies have HAI activity against the receptor binding site, which has benefits over other universal vaccine candidates. Induction of HAI antibodies is accepted as a surrogate/correlate for protective efficacy in humans (44). Therefore, COBRA-based vaccine candidates have a clear path forward to clinical proof of concept, unlike many other universal vaccine candidates.
Universal influenza vaccine approaches have the potential to be paradigm shifting for influenza vaccine landscape, with the goal of replacement of seasonal vaccines with universal ones specifying enhanced efficacy through their breadth and the potential for increased durability through the use of adjuvants. The H1N1 COBRA HA VLP vaccines described in this report elicit anti-head antibody responses against a broad number of H1N1 isolates that cover the past 100 years. The ability of COBRA HA designs to elicit HAI responses against future antigenically distinct H1 viruses for which the current H1 component is a mismatch will provide a critical assessment for their feasibility as “subtype universal” vaccine candidates.
ACKNOWLEDGMENTS
This study was supported by a Sponsored Research Agreement from Sanofi Pasteur.
We thank Joshua DiNapoli, Greg Kirchenbaum, and Terianne Wong for helpful discussions and reviews. We also thank James Allen for technical assistance and Terianne Wong for inputting sequences and technical work on pymol structures of H1N1 HA molecules. We thank Scott Hensley for providing the A/Philadelphia/1/2013 influenza viruses. Other H1N1 viruses were obtained from the Influenza Reagent Resource.
REFERENCES
- 1.Ellebedy AH, Webby RJ. 2009. Influenza vaccines. Vaccine 27(Suppl 4):D65–D68. doi: 10.1016/j.vaccine.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monto AS. 2010. Seasonal influenza and vaccination coverage. Vaccine 28(Suppl 4):D33–D44. doi: 10.1016/j.vaccine.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 3.Simmons CP, Bernasconi NL, Suguitan AL, Mills K, Ward JM, Chau NV, Hien TT, Sallusto F, Ha DQ, Farrar J, de Jong MD, Lanzavecchia A, Subbarao K. 2007. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med 4:e178. doi: 10.1371/journal.pmed.0040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puck JM, Glezen WP, Frank AL, Six HR. 1980. Protection of infants from infection with influenza A virus by transplacentally acquired antibody. J Infect Dis 142:844–849. doi: 10.1093/infdis/142.6.844. [DOI] [PubMed] [Google Scholar]
- 5.Murphy BR, Clements ML. 1989. The systemic and mucosal immune response of humans to influenza A virus. Curr Top Microbiol Immunol 146:107–116. [DOI] [PubMed] [Google Scholar]
- 6.Frasca D, Diaz A, Romero M, Landin AM, Phillips M, Lechner SC, Ryan JG, Blomberg BB. 2010. Intrinsic defects in B cell response to seasonal influenza vaccination in elderly humans. Vaccine 28:8077–8084. doi: 10.1016/j.vaccine.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goronzy JJ, Weyand CM. 2013. Understanding immunosenescence to improve responses to vaccines. Nat Immunol 14:428–436. doi: 10.1038/ni.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao S, Kong W-P, Wei C-J, Van Hoeven N, Gorres J, Nason M, Andersen H, Tumpey T, Nabel G. 2010. Comparative efficacy of hemagglutinin, nucleoprotein, and matrix 2 protein gene-based vaccination against H5N1 influenza in mouse and ferret. PLoS One 5:e9812. doi: 10.1371/journal.pone.0009812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchenbaum GA, Ross TM. 2014. Eliciting broadly protective antibody responses against influenza. Curr Opin Immunol 28:71–76. doi: 10.1016/j.coi.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Crevar CJ, Carter DM, Lee KY, Ross TM. 2015. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum Vaccin Immunother 11:572–583. doi: 10.1080/21645515.2015.1012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giles BM, Bissel SJ, Dealmeida DR, Wiley CA, Ross TM. 2012. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin Vaccine Immunol 19:128–139. doi: 10.1128/CVI.05533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, Ross TM. 2012. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis 205:1562–1570. doi: 10.1093/infdis/jis232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giles BM, Ross TM. 2011. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29:3043–3054. doi: 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross TM, Xu Y, Bright RA, Robinson HL. 2000. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat Immunol 1:127–131. doi: 10.1038/77802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 16.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Askonas B, McMichael A, Webster RG. 1982. The immune response to influenza viruses and the problem of protection against infection, p 159–188. In Beare AS. (ed), Basic and applied influenza research. CRC Press, Boca Raton, FL. [Google Scholar]
- 19.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 20.US Congress. 1966. Animal Welfare Act of 1966 intended to regulate the transport, sale and handling of dogs, cats, guinea pigs, nonhuman primates, hamsters and rabbits intended to use for research or other purposes. Public Law 89-544. United States Code, Title 7. Sections 2131-2156. [Google Scholar]
- 21.US Department of Health and Human Services. 2009. Biosafety in microbiological and biomedical laboratories, 5th ed HHS publication no. (CDC) 21-1112 US Department of Health and Human Services, Washington, DC. [Google Scholar]
- 22.Gillim-Ross L, Subbarao K. 2006. Emerging respiratory viruses: challenges and vaccine strategies. Clin Microbiol Rev 19:614–636. doi: 10.1128/CMR.00005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, Povinelli L, Cox NJ, Klimov AI. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 24.Bright RA, Ross TM, Subbarao K, Robinson HL, Katz JM. 2003. Impact of glycosylation on the immunogenicity of a DNA-based influenza H5 HA vaccine. Virology 308:270–278. doi: 10.1016/S0042-6822(03)00008-4. [DOI] [PubMed] [Google Scholar]
- 25.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell JA, Green TD, Bright RA, Ross TM. 2003. Induction of heterosubtypic immunity to influenza A virus using a DNA vaccine expressing hemagglutinin-C3d fusion proteins. Vaccine 21:902–914. doi: 10.1016/S0264-410X(02)00539-X. [DOI] [PubMed] [Google Scholar]
- 27.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Briney BS, Smith SA, Basler CF, Crowe JE Jr. 2011. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol 187:3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE Jr. 2011. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol 85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter DM, Bloom CE, Nascimento EJ, Marques ET, Craigo JK, Cherry JL, Lipman DJ, Ross TM. 2013. Sequential seasonal H1N1 influenza virus infections protect ferrets against novel 2009 H1N1 influenza virus. J Virol 87:1400–1410. doi: 10.1128/JVI.02257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter DM, Lu HR, Bloom CE, Crevar CJ, Cherry JL, Lipman DJ, Ross TM. 2012. Complex patterns of human antisera reactivity to novel 2009 H1N1 and historical H1N1 influenza strains. PLoS One 7:e39435. doi: 10.1371/journal.pone.0039435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, Ellebedy AH, Carter DM, Andrews SF, Zheng NY, Huang M, Huang Y, Strauss D, Shaz BH, Hodinka RL, Reyes-Teran G, Ross TM, Wilson PC, Ahmed R, Bloom JD, Hensley SE. 2014. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci U S A 111:15798–15803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross TM, Lin CJ, Nowalk MP, Huang HH, Spencer SM, Shay DK, Sambhara S, Sundaram ME, Friedrich T, Sauereisen S, Bloom CE, Zimmerman RK. 2014. Influence of pre-existing hemagglutination inhibition titers against historical influenza strains on antibody response to inactivated trivalent influenza vaccine in adults 50-80 years of age. Hum Vaccin Immunother 10:1195–1203. doi: 10.4161/hv.28313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Wang L, Compans RW, Wang BZ. 2014. Universal influenza vaccines, a dream to be realized soon. Viruses 6:1974–1991. doi: 10.3390/v6051974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang YH, Seong BL. 2014. Options and obstacles for designing a universal influenza vaccine. Viruses 6:3159–3180. doi: 10.3390/v6083159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirchenbaum GA, Carter DM, Ross TM. 2015. Sequential infection in ferrets with antigenically distinct seasonal H1N1 influenza viruses boosts hemagglutinin stalk-specific antibodies. J Virol 90:1116–1128. doi: 10.1128/JVI.02372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krammer F, Palese P. 2013. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3:521–530. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellebedy AH, Ahmed R. 2012. Re-engaging cross-reactive memory B cells: the influenza puzzle. Front Immunol 3:53. doi: 10.3389/fimmu.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, Mulligan M, Das SR, Yewdell JW, Mehta AK, Wilson PC, Ahmed R. 2012. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Radosevic K. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 43.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, Graham BS. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 44.Tsang TK, Cauchemez S, Perera RA, Freeman G, Fang VJ, Ip DK, Leung GM, Malik Peiris JS, Cowling BJ. 2014. Association between antibody titers and protection against influenza virus infection within households. J Infect Dis 210:684–692. doi: 10.1093/infdis/jiu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J Virol 82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]