Abstract
Feline leukemia virus (FeLV) subgroups have emerged in infected cats via the mutation or recombination of the env gene of subgroup A FeLV (FeLV-A), the primary virus. We report the isolation and characterization of a novel env gene, TG35-2, and report that the TG35-2 pseudotype can be categorized as a novel FeLV subgroup. The TG35-2 envelope protein displays strong sequence identity to FeLV-A Env, suggesting that selection pressure in cats causes novel FeLV subgroups to emerge.
TEXT
Feline leukemia viruses (FeLVs) are pathogenic retroviruses of domestic cats (1, 2), which are classified into subgroups A (the parent virus), B, C, D, and T based on their interference and in vitro host range properties (3, 4, 5, 6, 7, 8). Subgroups B and D arose from the recombination of FeLV-A env and the env genes of endogenous FeLV or endogenous retroviruses in the genomes of domestic cats (ERV-DCs) (7, 9, 10). Subgroups C and T possibly arose from mutations in FeLV-A env (11, 12). The recombination or mutation of env often alters the interference and host ranges of FeLVs by affecting their receptor usage (5, 6, 13, 14, 15, 16).
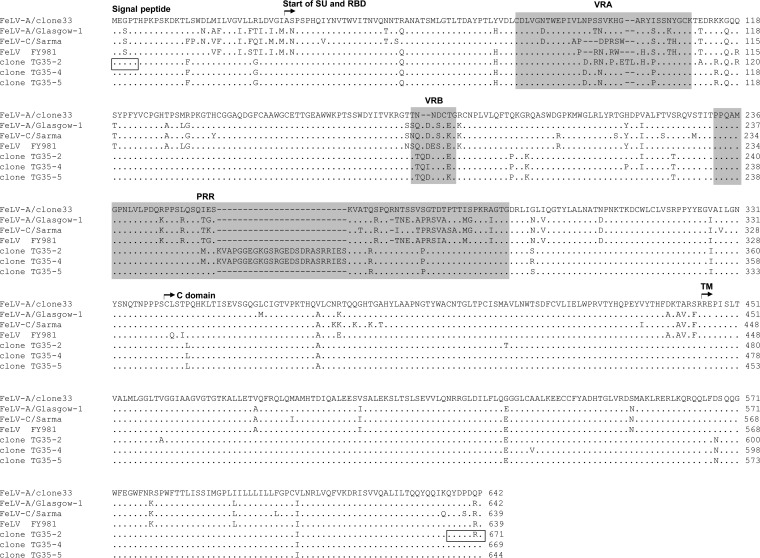
FeLV env genes were isolated by PCR from the blood DNA of a 1-year-old castrated male cat, TG35, with a bite injury, stomatitis, loss of appetite, and FeLV infection, although he had been vaccinated with inactivated FeLV (genotype III) (16). Five clones (TG35-1 to -5) were isolated, and we focused on TG35-2, TG35-4, and TG35-5. The env sequences of these clones showed strong similarity (Fig. 1), and the viruses clustered phylogenetically with those of genotype I/clade I FeLV, found mainly in Japan (16). The encompassing variable region A (VRA) of TG35-2 Env differs at eight amino acids from those of the TG35-4 and TG35-5 Env proteins. The proline-rich regions of TG35-2 and TG35-4, but not TG35-5, contain an inserted sequence of 25 amino acids (Fig. 1) not found in the cat genome database and of unknown origin.
FIG 1.
Receptor-binding domains (RBDs), proline-rich regions (PRRs), C domains, and TM regions of the Env protein are shown for FeLV-A clone 33 (17), FeLV-A Glasgow-1 (9), FeLV-C Sarma (19), FeLV FY981 (21), FeLV TG35-2 (16), TG35-4 (accession number LC029807), and TG35-5 (accession number LC029808). The variable regions, VRA and VRB, are also shown. Dots indicate identical residues, and dashes indicate spaces that were introduced for the amino acid alignment. Boxes indicate the positions of the PCR primers (16). The Env sequences were aligned with the Genetyx program (Genetyx Corporation, Tokyo, Japan).
To identify the FeLV subgroup to which this viral strain belongs, we used an interference assay (16) and generated β-galactosidase (LacZ)-encoding pseudotype viruses expressing TG35-2, TG35-4, or TG35-5 envelope (Env) proteins in GPLac cells (7). Pseudotype viruses TG35-2, -4, and -5 infected uninfected HEK293T cells (Table 1). However, HEK293T cells preinfected with FeLV-A/clone 33 (293T/clone 33 cells) (17) or FeLV-A/Glasgow-1 (293T/Glasgow-1 cells) (9) were infected by pseudotype virus TG35-2, but not by TG35-4 or TG35-5. Neither cell type was infected by FeLV-A/clone 33 or FeLV-A/Glasgow-1. Therefore, only the TG35-4 and TG35-5 viruses interfered with FeLV-A. Neither the TG35-2, TG35-4, nor TG35-5 pseudotype interfered with other subgroups of FeLV, or with retroviruses such as ERV-DC10, a replication-competent feline ERV (7) (Table 1). Therefore, FeLV TG35-4 and TG35-5 belong to the FeLV-A subgroup. However, TG35-2 could not be categorized.
TABLE 1.
Receptor interference assay using various pseudotyped FeLVs in HEK293T cells
| Preinfecting virus | Titer of pseudotyped virusa: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FeLV-A |
FeLV-B (GA) | FeLV-C (Sarma) | FeLV (FY981) | FeLV-D (ON-T) | Clone |
ERV-DC10 | Ampho-MLV | ||||
| Clone 33 | Glasgow-1 | TG35-2 | TG35-4 | TG35-5 | |||||||
| None | (4.1 ± 0.4) × 103 | (4.9 ± 0.3) × 103 | (2.1 ± 0.0) × 105 | (7.1 ± 0.6) × 103 | (1.3 ± 1.0) × 103 | (4.1 ± 0.2) × 104 | (1.1 ± 0.1) × 104 | (1.1 ± 0.6) × 103 | (2.3 ± 1.3) × 103 | (1.9 ± 0.0) × 103 | (8.2 ± 0.4) × 104 |
| FeLV-A (clone 33) | 0 | 0 | (1.8 ± 0.3) × 105 | (5.3 ± 0.3) × 103 | (1.6 ± 1.0) × 103 | (4.6 ± 0.5) × 104 | (7.1 ± 0.3) × 103 | 0 | 0 | (2.4 ± 0.2) × 103 | (1.6 ± 0.1) × 105 |
| FeLV-A (Glasgow-1) | 0 | 0 | (1.6 ± 0.2) × 105 | (3.0 ± 0.4) × 103 | (1.4 ± 0.9) × 103 | (4.8 ± 0.5) × 104 | (9.0 ± 1.0) × 103 | 0 | 0 | (1.9 ± 0.1) × 103 | (1.2 ± 0.0) × 105 |
| FeLV-B (GA) | (1.6 ± 0.0) × 103 | (4.1 ± 1.1) × 103 | 0 | (4.3 ± 0.6) × 103 | (2.9 ± 0.8) × 103 | (4.2 ± 0.2) × 104 | (6.6 ± 0.3) × 103 | (1.1 ± 0.8) × 103 | (1.6 ± 1.1) × 103 | (1.1 ± 0.0) × 103 | (9.8 ± 1.5) × 104 |
| FeLV-C (Sarma) | (2.1 ± 0.0) × 103 | (1.0 ± 0.0) × 103 | (1.6 ± 0.1) × 105 | 0 | 0 | (4.5 ± 0.4) × 104 | (1.0 ± 0.1) × 104 | (8.4 ± 6.3) × 102 | (1.1 ± 0.8) × 103 | (1.2 ± 0.1) × 103 | (1.4 ± 0.1) × 105 |
| FeLV-D (33/ON-T) | (2.2 ± 0.2) × 103 | (5.1 ± 0.7) × 103 | (1.6 ± 0.2) × 105 | (3.3 ± 0.4) × 103 | (1.9 ± 1.3) × 103 | 0 | (6.0 ± 0.2) × 103 | (1.0 ± 0.7) × 103 | (1.3 ± 0.5) × 103 | (2.6 ± 0.1) × 103 | (1.2 ± 0.1) × 105 |
| 33TGE2 | (4.3 ± 0.4) × 103 | (8.3 ± 1.1) × 103 | (1.5 ± 0.0) × 105 | (1.1 ± 0.0) × 104 | (2.2 ± 1.5) × 103 | (5.8 ± 0.7) × 104 | 0 | (1.0 ± 0.6) × 103 | (1.4 ± 0.6) × 103 | (2.4 ± 0.4) × 103 | (1.2 ± 0.1) × 105 |
| ERV-DC10 | (2.8 ± 0.1) × 103 | (6.4 ± 0.8) × 103 | (1.9 ± 0.1) × 105 | (6.9 ± 0.7) × 103 | (2.0 ± 1.2) × 103 | (3.5 ± 0.2) × 104 | (1.0 ± 0.1) × 104 | (1.0 ± 0.5) × 103 | (1.4 ± 0.5) × 103 | 0 | (1.1 ± 0.0) × 105 |
| Ampho-MLV | ND | ND | ND | ND | ND | ND | (1.1 ± 0.1) × 104 | ND | ND | ND | 0 |
The indicated FeLV env genes inserted into the pFUΔss expression vector were used to prepare the LacZ pseudotype viruses. GPLac cells, an env-negative packaging cell line containing a LacZ-encoding retroviral vector (7), were transfected (with ScreenFect Reagent; Wako, Osaka, Japan) with each env expression vector. Supernatants were passed through 0.45-μm filters and used for infection assays. pFUΔss clone 33 (FeLV-A/clone 33 env), pFUΔss A5 [FeLV-A/Glasgow-1(pFGA5) env], pFUΔss GB (FeLV-B/Gardner-Arnstein env), pFUΔss SC (FeLV-C/Sarma env), pFUΔss ON-T (FeLV-D/ON-T env), pFUΔss DC10 (ERV-DC10 env), and pFUΔss 4070A (amphotropic MLV/4070A env) have been described previously (7). pFUΔss TG35-2 (TG35-2 env), pFUΔss TG35-4 (TG35-4 env), pFUΔss TG35-5 (TG35-5 env), and pFUΔss FY33 (the SU was derived from FeLV FY981, and the transmembrane [TM] protein was derived from FeLV-A clone 33) were newly constructed in this study. The target cells used were FeLV-A-, FeLV-B-, FeLV-C-, FeLV-D-, FeLV/33TGE2-, ERV-DC10-, and Ampho-MLV (4070A)-infected HEK293T cells. Infectious clone 33TGE2 was constructed from FeLV clone 33 (but contained the TG35-2 env gene) and was used to transfect HEK293T cells, thus establishing persistently infected cells. Viral titers were determined as IU per milliliter with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining, as previously described (7). The data are the means from 3 to 4 independent experiments, with their standard errors. Ampho-MLV, amphotropic MLV; ND, not done.
We next constructed a replication-competent virus (33TGE2) containing the TG35-2 env gene and the LTR, gag, and pol genes of FeLV-A clone 33 (GeneArt; Thermo Fisher Scientific, Waltham, MA). HEK293T cells were transfected with virus p33TGE2, and productive replication was confirmed by detecting the FeLV p27 antigen. 33TGE2-infected HEK293T cells (293T/33TGE2) were successfully infected with FeLV-A (clone 33), FeLV-A (Glasgow-1), FeLV-B (Gardner-Arnstein) (18), FeLV-C (Sarma) (19), FeLV-D (ON-T) (7), TG35-4, TG35-5, ERV-DC10, and amphotropic murine leukemia virus (MLV) 4070A (16), but not with the TG35-2 pseudotype. However, the pseudotype virus infected 293T/FeLV-A, 293T/FeLV-B, 293T/FeLV-C, 293T/FeLV-D, 293T/ERV-DC10, and 293T/4070A cells (Table 1). FeLV 33TGE2 did not interfere with xenotropic MLV (X-MLV) from 22RV.1 cells (20) or vice versa (the viral infectious titers were 8 × 104 and 2 × 105 infectious units [IU]/ml, respectively) in HEK293T cells, but neither virus infected HEK293T cells already infected with itself. Thus, FeLV 33TGE2 displayed the same interference behavior as the TG35-2 pseudotype.
When we examined AH927 feline cells, the TG35-2 pseudotype virus infected AH927 cells infected with FeLV-A, FeLV-B, or FeLV-C, but not 33TGE2-infected AH927 cells (Table 2), as seen with human HEK293T cells (Table 1). To determine whether TG35-2 interferes with FeLV FY981, which uses the THTR1, FLVCR1, and FLVCR2 receptors (21), we constructed a plasmid expressing a chimeric gene-synthesized FY981 env gene (FY33) encoding the surface glycoprotein SU of FeLV FY981 and the transmembrane (TM) protein of FeLV clone 33. Pseudotype virus FY981 infected 293T/33TGE2 and AH927/33TGE2 cells, but not FeLV-C-infected cells, so FY981 was newly categorized as FeLV-C (Tables 1 and 2). We then tested whether TG35-2 uses known FeLV receptors, such as THTR1 (15), PIT1 (5), PIT2 (22), FLVCR1 (13, 14), or FLVCR2 (21), for cell entry. The TG35-2 virus infected neither MDTF cells nor MDTF cells expressing any of these receptors (Table 3).
TABLE 2.
Receptor interference assay using various pseudotype FeLV Env proteins in feline AH927 cells
| Cell line | Titer of pseudotyped virusa: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| FeLV-A |
FeLV-B (GA) | FeLV-C (Sarma) | FeLV (FY981) | Clone |
Ampho-MLV | ||||
| Clone 33 | Glasgow-1 | TG35-2 | TG35-4 | TG35-5 | |||||
| AH927 | (1.7 ± 0.2) × 104 | (4.2 ± 0.4) × 104 | (3.5 ± 0.1) × 104 | (5.1 ± 0.3) × 103 | (1.1 ± 0.2) × 104 | (4.0 ± 0.4) × 103 | (3.4 ± 0.5) × 103 | (4.7 ± 0.2) × 103 | (7.9 ± 0.8) × 104 |
| AH927/clone 33 | 0 | 0 | (8.7 ± 1.6) × 104 | (6.3 ± 1.6) × 103 | (1.6 ± 0.1) × 104 | (5.7 ± 0.7) × 103 | 0 | 0 | (7.7 ± 1.7) × 104 |
| AH927/Glasgow-1 | 0 | 0 | (9.8 ± 0.6) × 104 | (5.3 ± 0.4) × 103 | (1.9 ± 0.0) × 104 | (7.5 ± 0.5) × 103 | 0 | 0 | (6.2 ± 0.7) × 104 |
| AH927/GA | (4.5 ± 0.7) × 104 | (1.4 ± 0.4) × 105 | 0 | (9.7 ± 0.9) × 103 | (2.1 ± 0.2) × 104 | (9.2 ± 1.3) × 103 | (1.2 ± 0.1) × 104 | (1.8 ± 0.2) × 104 | (1.3 ± 0.1) × 105 |
| AH927/Sarma | (3.1 ± 0.2) × 104 | (1.1 ± 0.1) × 105 | (8.4 ± 0.9) × 104 | 0 | 0 | (1.1 ± 0.2) × 104 | (9.3 ± 0.5) × 103 | (1.2 ± 0.1) × 104 | (9.0 ± 0.7) × 104 |
| AH927/33TGE2 | (3.7 ± 0.4) × 104 | (7.5 ± 0.4) × 104 | (1.8 ± 1.6) × 104 | (9.5 ± 1.4) × 103 | (1.3 ± 0.1) × 104 | 0 | (4.8 ± 1.0) × 103 | (9.8 ± 0.6) × 103 | (3.1 ± 0.2) × 104 |
| 104C1 | 0 | 0 | 0 | (1.6 ± 0.9) × 103 | (2.3 ± 1.2) × 103 | 0 | ND | ND | (5.5 ± 0.2) × 104 |
| CRFK | (1.5 ± 0.1) × 104 | (4.7 ± 0.6) × 104 | (9.1 ± 0.4) × 104 | (2.8 ± 0.4) × 103 | (3.7 ± 0.4) × 104 | (1.4 ± 1.2) × 103 | ND | ND | (9.1 ± 0.5) × 104 |
The target cells used for the viral interference assay were uninfected feline AH927 cells or FeLV-A/clone33-, FeLV-A/Glasgow-1-, FeLV-B/GA-, FeLV-C/Sarma-, or FeLV/33TGE2-infected feline AH927 cells, 104C1 cells (guinea pig), or CRFK cells (feline). Viral titers were determined as IU per milliliter with X-Gal staining as previously described (7). The data are the means from three independent experiments, with their standard errors. ND, not done. Ampho-MLV, amphotropic MLV.
TABLE 3.
Receptor usage by FeLVsc
| Cell line | FeLV-A (clone 33)a | FeLV-B (GA)b | FeLV (FY981)a | Clone TG35-2a | 33TGE2b | Ampho-MLVa | Mock |
|---|---|---|---|---|---|---|---|
| MDTF | − | − | + | − | − | ++ | − |
| MDTF/feTHTR-1 | ++ | − | ND | − | − | ND | − |
| MDTF/fePit-1 | − | ++ | ND | − | − | ND | − |
| MDTF/fePit-2 | − | + | ND | − | − | ND | − |
| MDTF/hFLVCR1 | − | ND | ++ | − | ND | ++ | − |
| MDTF/hFLVCR2 | − | ND | ++ | − | ND | ++ | − |
| MDTF/feFLVCR1 | − | ND | ++ | − | ND | ++ | − |
The indicated env genes were used to prepare the LacZ pseudotyped viruses.
Replication-competent viruses carrying the LacZ-encoding retroviral reporter were used for infection.
Each retroviral expression plasmid encoding feline THTR1 (feTHTR1), feline PIT1 (fePit-1), feline PIT2 (fePit-2), feline FLVCR1 (feFLVCR1), human FLVCR1 (hFLVCR1), or human FLVCR2 (hFLVCR2) was expressed in MDTF cells under G418 selection. The pooled G418-resistant cells were tested in the viral infection assay using the indicated viruses. Titers were determined from three experiments with X-Gal staining. −, infection titer of 0; +, 1 to 103 IU/ml; ++, 103 to 105 IU/ml. ND, not done. Ampho-MLV, amphotropic MLV.
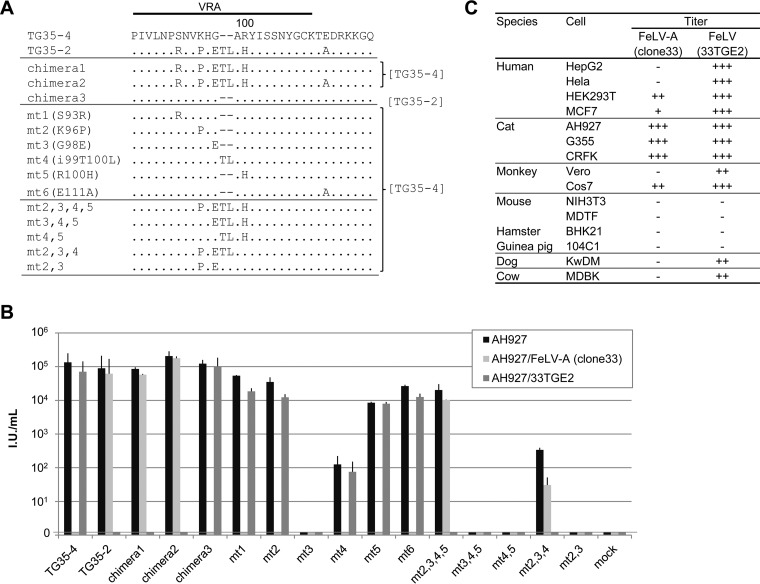
We then determined the minimal changes in the Env protein required to produce the TG35-2 phenotype. When the VRA of TG35-2 was replaced with that of TG35-4, the pseudotyped viruses (chimeras 1 and 2) showed the phenotype of TG35-2, and conversely, when the VRA of TG35-4 was replaced with that of TG35-2, the pseudotyped virus (chimera 3) showed the phenotype of FeLV-A/TG35-4. Furthermore, pseudotyped viruses (mt2,3,4,5 and mt2,3,4) generated by the site-directed mutagenesis of the VRA showed that a substitution of 2 to 3 amino acids in addition to the insertion of threonine and leucine in the VRA of TG35-4 conferred the TG35-2 phenotype (Fig. 2A and B). Therefore, specific amino acids within the VRA are responsible for the TG35-2 and FeLV-A phenotypes.
FIG 2.
Determination of the amino acids in the Env protein that are required for the TG35-2 phenotype and the host range of FeLV 33TGE2. (A) The indicated mutant FeLV env genes, constructed in either the TG35-2 or TG35-4 env gene, were generated with site-directed mutagenesis or recombination of the VRA in the pFUΔss vector (7). The env sequences other than the VRA, derived from TG35-2 or TG35-4 env, are described at right. (B) GPLac cells were transfected with the indicated FeLV env genes inserted into the pFUΔss expression vector. Supernatants were passed through 0.45-μm filters and used for infection assays in AH927 cells, AH927/FeLV-A (clone 33) cells, and AH927/33TGE2 cells. Titers were determined with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining (7). The data shown are the averages from several independent experiments, with their standard errors. (C) Replication-competent virus or its LacZ-encoding pseudovirus, produced by the transfection of 293Lac cells (HEK293T cells stably containing a LacZ-encoding retroviral vector) with either FeLV-A clone 33 or the FeLV 33TGE2 plasmid, was used for the infection assay. The indicated target cells were used to determine the host ranges of the viruses. Titers were determined with X-Gal staining. −, zero infection titer; +, 101 to 103 IU/ml; ++, 103 to 105 IU/ml; +++, >105 IU/ml. The data shown are the averages from three independent experiments.
The host range of TG35-2 was determined using infectious clone 33TGE2 (Fig. 2C). 33TGE2 infected a broad range of cell lines yielding high titers, whereas FeLV-A displayed restricted infection. Neither virus infected mouse, hamster, or guinea pig cells. Therefore, the host range properties of FeLV 33TGE2 and FeLV-A differ.
We have characterized a novel FeLV that does not interfere with known feline retroviruses, including FeLV-D and ERV-DC10. A pseudotype virus expressing TG35-2 Env infected FeLV-A-, FeLV-B-, and FeLV-C-infected cells, and all cells that express the receptors THTR1, PIT1/2, and FLVCR1 (data not shown). TG35-2 did not infect guinea pig cells (104C1), which are permissive for FeLV-C and FY981 (FeLV FY33) (Table 2) (4, 21, 23), suggesting that TG35-2 differs from FeLV-C and FY981. MLV 4070A was used as a positive control because it and some FeLV-B viruses use PIT2 for viral entry (24, 25, 26). The wide host range of TG35-2 is not attributable to its xenotropic nature (27). A pseudotype virus expressing TG35-2 Env infected FeLV-D- or ERV-DC10-infected cells, but the receptors of neither virus have been identified. FeLV-T requires FeLIX for infection (6), but TG35-2 infected cells without FeLIX. Artificial mutation of the FeLV-A env gene changed its receptor to that of porcine endogenous retrovirus A (PERV-A) (28), but the host range of PERV-A differs from that of TG35-2 (28, 29), and PERV-A isolated from PK15 cells (30) did not interfere with FeLV 33TGE2 in HEK293T cells (data not shown). A mutagenesis analysis confirmed that subtle changes in the VRA altered the interference patterns of the TG35-2 and FeLV-A phenotypes, but some of these Env mutants did not infect AH927 cells (Fig. 2B), although they expressed the Env protein, as demonstrated with Western blotting (data not shown). Artificial mutation of the FeLV env gene may not alter its receptor. However, subtle mutation of the env gene may predispose the virus to enhanced replication in vivo and subsequent conversion to different FeLV subgroups (31, 32, 33). We also found an intermediary between the env genes of FeLV-A and FeLV-B (16). Therefore, the emergence of a novel FeLV subgroup may be mediated by intermediaries arising through several steps, possibly under selection pressure from cats (e.g., by vaccination).
We propose that FeLV-E is a novel interference subgroup of FeLV.
Nucleotide sequence accession numbers.
The sequences reported here have been deposited in DDBJ/EMBL/GenBank under accession numbers LC029807 and LC029808.
ACKNOWLEDGMENTS
We are grateful to Kenji Baba (Yamaguchi University) for providing the MDBK cells, to Yoshinao Kubo (Nagasaki University) for the MDTF cells, and to the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University, for the COS-7 cells. We are grateful to Julie Overbaugh (Fred Hutchinson Cancer Research Center, Seattle, WA) for providing the feline THTR1, feline PIT1, and PIT2 expression plasmids and to Janis L. Abkowitz (University of Washington, Seattle, WA) for providing the feline and human FLVCR1 and human FLVCR2 expression plasmids.
This study was partly supported by JSPS KAKENHI, grant number 15H04602.
REFERENCES
- 1.Hisasue M, Nagashima N, Nishigaki K, Fukuzawa I, Ura S, Katae H, Tsuchiya R, Yamada T, Hasegawa A, Tsujimoto H. 2009. Myelodysplastic syndromes and acute myeloid leukemia in cats infected with feline leukemia virus clone33 containing a unique long terminal repeat. Int J Cancer 124:1133–1141. doi: 10.1002/ijc.24050. [DOI] [PubMed] [Google Scholar]
- 2.Hartmann K. 2011. Clinical aspects of feline immunodeficiency and feline leukemia virus infection. Vet Immunol Immunopathol 143:190–201. doi: 10.1016/j.vetimm.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarma PS, Log T. 1973. Subgroup classification of feline leukemia and sarcoma viruses by viral interference and neutralization tests. Virology 54:160–169. doi: 10.1016/0042-6822(73)90125-6. [DOI] [PubMed] [Google Scholar]
- 4.Jarrett O, Laird HM, Hay D. 1973. Determinants of the host range of feline leukaemia viruses. J Gen Virol 20:169–175. doi: 10.1099/0022-1317-20-2-169. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi Y, Vile RG, Simpson G, O'Hara B, Collins MK, Weiss RA. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol 66:1219–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson MM, Lauring AS, Burns CC, Overbaugh J. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 7.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J Virol 86:8634–8644. doi: 10.1128/JVI.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito J, Watanabe S, Hiratsuka T, Kuse K, Odahara Y, Ochi H, Kawamura M, Nishigaki K. 2013. Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J Virol 87:12029–12040. doi: 10.1128/JVI.01267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart MA, Warnock M, Wheeler A, Wilkie N, Mullins JI, Onions DE, Neil JC. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol 58:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overbaugh J, Riedel N, Hoover EA, Mullins JI. 1988. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature 332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 11.Donahue PR, Quackenbush SL, Gallo MV, deNoronha CM, Overbaugh J, Hoover EA, Mullins JI. 1991. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol 65:4461–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigby MA, Rojko JL, Stewart MA, Kociba GJ, Cheney CM, Rezanka LJ, Mathes LE, Hartke JR, Jarrett O, Neil JC. 1992. Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes. J Gen Virol 73:2839–2847. doi: 10.1099/0022-1317-73-11-2839. [DOI] [PubMed] [Google Scholar]
- 13.Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL. 2000. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093–1099. [PubMed] [Google Scholar]
- 14.Tailor CS, Willett BJ, Kabat D. 1999. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J Virol 73:6500–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe S, Kawamura M, Odahara Y, Anai Y, Ochi H, Nakagawa S, Endo Y, Tsujimoto H, Nishigaki K. 2013. Phylogenetic and structural diversity in the feline leukemia virus env gene. PLoS One 8:e61009. doi: 10.1371/journal.pone.0061009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishigaki K, Hanson C, Thompson D, Yugawa T, Hisasue M, Tsujimoto H, Ruscetti S. 2002. Analysis of the disease potential of a recombinant retrovirus containing Friend murine leukemia virus sequences and a unique long terminal repeat from feline leukemia virus. J Virol 76:1527–1532. doi: 10.1128/JVI.76.3.1527-1532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins JI, Casey JW, Nicolson MO, Burck KB, Davidson N. 1981. Sequence arrangement and biological activity of cloned feline leukemia virus proviruses from a virus-productive human cell line. J Virol 38:688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riedel N, Hoover EA, Gasper PW, Nicolson MO, Mullins JI. 1986. Molecular analysis and pathogenesis of the feline aplastic anemia retrovirus, feline leukemia virus C-Sarma. J Virol 60:242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knouf EC, Metzger MJ, Mitchell PS, Arroyo JD, Chevillet JR, Tewari M, Miller AD. 2009. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J Virol 83:7353–7356. doi: 10.1128/JVI.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalev Z, Duffy SP, Adema KW, Prasad R, Hussain N, Willett BJ, Tailor CS. 2009. Identification of a feline leukemia virus variant that can use THTR1, FLVCR1, and FLVCR2 for infection. J Virol 83:6706–6716. doi: 10.1128/JVI.02317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson MM, Lauring AS, Robertson S, Dirks C, Overbaugh J. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J Virol 75:10563–10572. doi: 10.1128/JVI.75.22.10563-10572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarma PS, Log T, Jain D, Hill PR, Huebner RJ. 1975. Differential host range of viruses of feline leukemia-sarcoma complex. Virology 64:438–446. doi: 10.1016/0042-6822(75)90121-X. [DOI] [PubMed] [Google Scholar]
- 24.Miller DG, Edwards RH, Miller AD. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci U S A 91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Zeijl M, Johann SV, Closs E, Cunningham J, Eddy R, Shows TB, O'Hara B. 1994. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci U S A 91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boomer S, Eiden M, Burns CC, Overbaugh J. 1997. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol 71:8116–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stieler K, Schulz C, Lavanya M, Aepfelbacher M, Stocking C, Fischer N. 2010. Host range and cellular tropism of the human exogenous gammaretrovirus XMRV. Virology 399:23–30. doi: 10.1016/j.virol.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 28.Mazari PM, Linder-Basso D, Sarangi A, Chang Y, Roth MJ. 2009. Single-round selection yields a unique retroviral envelope utilizing GPR172A as its host receptor. Proc Natl Acad Sci U S A 106:5848–5853. doi: 10.1073/pnas.0809741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J Virol 72:9986–9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patience C, Takeuchi Y, Weiss RA. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat Med 3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 31.Stewart H, Adema KW, McMonagle EL, Hosie MJ, Willett BJ. 2012. Identification of novel subgroup A variants with enhanced receptor binding and replicative capacity in primary isolates of anaemogenic strains of feline leukaemia virus. Retrovirology 9:48. doi: 10.1186/1742-4690-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohn JL, Linenberger ML, Hoover EA, Overbaugh J. 1994. Evolution of feline leukemia virus variant genomes with insertions, deletions, and defective envelope genes in infected cats with tumors. J Virol 68:2458–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohn JL, Moser MS, Gwynn SR, Baldwin DN, Overbaugh J. 1998. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J Virol 72:2686–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]