ABSTRACT
Chikungunya virus (CHIKV) is a rapidly emerging mosquito-borne human pathogen causing major outbreaks in Africa, Asia, and the Americas. The cell entry pathway hijacked by CHIKV to infect a cell has been studied previously using inhibitory compounds. There has been some debate on the mechanism by which CHIKV enters the cell: several studies suggest that CHIKV enters via clathrin-mediated endocytosis, while others show that it enters independently of clathrin. Here we applied live-cell microscopy and monitored the cell entry behavior of single CHIKV particles in living cells transfected with fluorescent marker proteins. This approach allowed us to obtain detailed insight into the dynamic events that occur during CHIKV entry. We observed that almost all particles fused within 20 min after addition to the cells. Of the particles that fused, the vast majority first colocalized with clathrin. The average time from initial colocalization with clathrin to the moment of membrane fusion was 1.7 min, highlighting the rapidity of the cell entry process of CHIKV. Furthermore, these results show that the virus spends a relatively long time searching for a receptor. Membrane fusion was observed predominantly from within Rab5-positive endosomes and often occurred within 40 s after delivery to endosomes. Furthermore, we confirmed that a valine at position 226 of the E1 protein enhances the cholesterol-dependent membrane fusion properties of CHIKV. To conclude, our work confirms that CHIKV enters cells via clathrin-mediated endocytosis and shows that fusion occurs from within acidic early endosomes.
IMPORTANCE Since its reemergence in 2004, chikungunya virus (CHIKV) has spread rapidly around the world, leading to millions of infections. CHIKV often causes chikungunya fever, a self-limiting febrile illness with severe arthralgia. Currently, no vaccine or specific antiviral treatment against CHIKV is available. A potential antiviral strategy is to interfere with the cell entry process of the virus. However, conflicting results with regard to the cell entry pathway used by CHIKV have been published. Here we applied a novel technology to visualize the entry behavior of single CHIKV particles in living cells. Our results show that CHIKV cell entry is extremely rapid and occurs via clathrin-mediated endocytosis. Membrane fusion from within acidic early endosomes is observed. Furthermore, the membrane fusion capacity of CHIKV is strongly promoted by cholesterol in the target membrane. Taking these findings together, this study provides detailed insight into the cell entry process of CHIKV.
INTRODUCTION
Chikungunya virus (CHIKV) is a human arboviral pathogen that was first isolated from a febrile patient in East Africa in 1952 (1). Since then, numerous small CHIKV outbreaks have been reported in Africa and Asia at irregular intervals. In 2004, the virus reemerged and spread rapidly around the world (1, 2). At the end of 2013, the first autochthonous case of CHIKV was reported in the Americas (3). Within 1.5 year, the virus has spread over 45 countries within Central and South America and caused more than 1.6 million infections (3). CHIKV often leads to chikungunya fever, which is characterized by high fever, headache, overall weakness, and joint pain (4). Chikungunya fever is mostly self-limiting, yet symptoms can be severe and disabling; as many as 80% of patients experience recurrent joint pains for months to years after infection (5–7). No vaccine or specific antiviral treatment is available to prevent or treat CHIKV infection (2, 4).
CHIKV is an alphavirus belonging to the Togaviridae family, which also includes Semliki Forest virus (SFV), Sindbis virus (SINV), Ross River virus (RRV), and Venezuelan equine encephalitis virus (VEEV). Alphavirus cell entry and membrane fusion are facilitated by the viral glycoproteins E1 and E2. Of these proteins, E2 is responsible for receptor binding and E1 facilitates the low-pH-dependent membrane fusion process (8, 9). Multiple receptors that facilitate SFV, SINV, RRV, and VEEV cell entry have been identified, but none of these receptors appear to be crucial (10–16). The receptors identified act predominantly as attachment factors to capture the virus. Upon virus-receptor interaction, the virus is internalized via clathrin-mediated endocytosis (CME) (9, 17, 18). Then the virus is transported to Rab5-positive early endosomes, where membrane fusion predominantly occurs (9, 19, 20). For VEEV, however, infection of mosquito cells has been reported to depend on Rab7-positive late endosomes as well (18, 21). In addition, liposomal membrane fusion studies have shown that besides low pH, target membrane cholesterol and sphingomyelin (SPM) are also required for SFV and SINV fusion (22–25).
Whereas the cell entry pathway of SFV, SINV, and VEEV is well studied, relatively few data have been published on CHIKV cell entry. To date, prohibitin, TIM-1, and glycosaminoglycans have been reported to function as receptors for CHIKV, but infection can also occur in the absence of these molecules (26). Thus, CHIKV receptors also appear to act mainly by facilitating the initial virus-cell contact. Contradictory reports have been published on the route of cell entry. Initially, dynamin was found to be important for CHIKV cell entry. Dynamin is involved in numerous cell entry pathways, such as clathrin-mediated endocytosis, caveolar endocytosis (20), and phagocytosis (27). CHIKV infection was also found to depend on Eps15 (28), a mediator of both clathrin-dependent (29) and clathrin-independent (30) cell entry pathways. One study showed that small interfering RNAs (siRNAs) against the clathrin heavy chain did not interfere with CHIKV infection in HEK239T cells (28), suggesting that CHIKV infects cells via a clathrin-independent pathway. However, siRNAs against clathrin did inhibit CHIKV infection of U-2 OS cells, primary human umbilical vein endothelial cells (31), and the mosquito cell line C6/36 (32). The majority of the results thus suggest that CHIKV cell entry occurs mainly via CME.
The intracellular trafficking behavior of CHIKV is poorly understood, although a few reports describe the function of early and late endosomes within infection. For example, Bernard et al. found that integrity of Rab7-positive endosomes is not required for CHIKV infection of HEK293T cells (28), suggesting that CHIKV fuses from within early endosomes. In the mosquito cell line C6/36, however, CHIKV infection was dependent on both Rab5- and Rab7-positive endosomes (32), indicating that fusion might also occur from Rab5/Rab7-positive maturing endosomes and Rab7-positive late endosomes. Furthermore, and in line with the findings for other alphaviruses, low-pH-dependent CHIKV fusion is strongly promoted by target membrane cholesterol and sphingomyelin (33, 34).
Here we dissected the cell entry pathway of CHIKV and elucidated the dynamics involved in virus-cell binding, internalization, trafficking to endosomes, and membrane fusion. In addition, the site of membrane fusion is unraveled. Cell entry by CHIKV was visualized by single-particle tracking of fluorescently labeled virions in living cells expressing fluorescent marker proteins. This approach allowed us to obtain accurate insight into the dynamic virus-host interactions that occur until the moment of membrane fusion.
MATERIALS AND METHODS
Cells.
Green monkey kidney BS-C-1 cells (ATCC CCL-26) were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Lonza), 25 mM HEPES, penicillin (100 U/ml), and streptomycin (100 U/ml). Green monkey kidney Vero-WHO cells (ATCC CCL-81) were cultured in DMEM (Gibco) supplemented with 5% FBS (Lonza), penicillin (100 U/ml), and streptomycin (100 U/ml). Baby hamster kidney cells (BHK-21 cells; ATCC CCL-10) were cultured in RPMI medium (Gibco) supplemented with 10% FBS (Lonza), penicillin (100 U/ml), and streptomycin (100 U/ml). Finally, human adenocarcinoma HeLa cells (ATCC CCL-2) were cultured in DMEM (Gibco) supplemented with 10% FBS (Lonza), penicillin (100 U/ml), and streptomycin (100 U/ml). All cells were maintained at 37°C under 5% CO2.
Virus production, purification, and labeling.
The infectious clone-derived CHIKV strains LS3 and LS3-GFP have been described previously (35). CHIKV strain LS3-226A is identical to LS3 except that it has an alanine at position 226 of the E1 protein instead of a valine. The LS3-226A clone was generated by quick-change mutagenesis and standard cloning techniques (details available upon request). Infectious virus was produced essentially as described by Scholte et al. (35) except that BHK-21 cells were transfected with in vitro-transcribed RNA transcripts by electroporation with a Gene Pulser Xcell system (Bio-Rad) set at 1.5 kV, 25 μF, and 200 Ω. At 24 h posttransfection, the medium was harvested and was used to inoculate Vero-WHO cultures at a multiplicity of infection (MOI) of 0.01 to produce large CHIKV working stocks.
For the production of purified CHIKV preparations, monolayers of BHK-21 cells were inoculated with CHIKV LS3 at an MOI of 4. At 24 h postinfection (p.i.), the supernatant was harvested and was cleared of cell debris by low-speed centrifugation. CHIKV particles were subsequently pelleted by ultracentrifugation in a Beckman type 19 rotor at 54,000 × g for 2.5 h at 4°C. The virus pellet was resuspended overnight in HNE buffer (5 mM HEPES, 150 mM NaCl, 0.1 mM EDTA [pH 7.4]) and was purified by ultracentrifugation on a sucrose density gradient (20 to 50% [wt/vol] in HNE) in a Beckman SW41 rotor at 50,000 × g for 18 h at 4°C. The 40%-to-45% section containing the virus was harvested, aliquoted, and stored at −80°C.
For microscopy studies, CHIKV was labeled with the lipophilic fluorescent probe 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD; Life Technologies) essentially as described previously for dengue virus (DENV) (36). For this purpose, 2 × 1011 genome-containing particles (GCPs) of purified CHIKV were mixed with 2 nmol DiD in dimethyl sulfoxide (DMSO; final concentration, 2%) in a final volume of 50 to 60 μl. The mixture was incubated for 30 min at room temperature in the dark. Next, unincorporated dye was removed by size exclusion chromatography on Sephadex G-50 Fine (Pharmacia) columns. DiD-labeled CHIKV was stored at 4°C in the dark and was used within 2 days.
The number of individual DiD-labeled virus particles was estimated by fluorescence microscopy as described previously by van der Schaar et al. (36). DiD-labeled viruses were detected by epifluorescence microscopy in a Leica Biosystems 6000B instrument using a 635-nm helium-neon laser. Analysis was carried out using the ParticleAnalyzer plugin of ImageJ.
As a control, CHIKV was treated with diethylpyrocarbonate (DEPC; Sigma-Aldrich). DEPC was freshly dissolved in cold ethanol to obtain a 1 M stock and was diluted in phosphate-buffered saline (PBS) to a final concentration of 2 mM prior to use. Then 5 × 108 GCPs of DiD-labeled CHIKV LS3 were diluted in 2 mM DEPC and were treated for 30 min at room temperature in the dark.
Virus for the bulk fusion assay was labeled biosynthetically with pyrene, as described previously (34). Briefly, BHK-21 cells were cultured in the presence of 15 μg/ml of 1-pyrenehexadecanoic acid (Invitrogen) 48 h prior to infection. BHK-21 cells were infected at an MOI of 4, and at 24 h p.i., virus was harvested and purified as described above.
Virus quantification.
The infectious virus titer was determined by a standard plaque assay. The number of PFU was determined on Vero-WHO cells at 37°C. Plaques were counted 2 days after infection. Furthermore, reverse transcriptase quantitative PCR (RT-qPCR) was used to determine the number of GCPs, as described previously (34).
Pharmacological inhibitors, siRNAs, and plasmids.
Chlorpromazine, methyl-β-cyclodextrin, and cholesterol (water soluble) were all purchased from Sigma-Aldrich. Ammonium chloride (NH4Cl) was obtained from Merck and Pitstop2 from Abcam. All chemicals were dissolved and stored according to the manufacturer's instructions. The cytotoxicity of the compounds was tested using a standard MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay. MTT was purchased from Sigma and was used at a final concentration of 0.45 mg/ml. Concentrations at which compounds reduced the cellular metabolic activity to <75% were considered cytotoxic and were excluded from further analysis.
ON-TARGETplus SMARTpool siRNAs against the human CTLC gene (encoding clathrin heavy chain [CHC]; L-004001-01), the ON-TARGETplus Non-targeting Pool (D-001810-10), and the transfection agent DharmaFECT 2 (T-2002-01) were purchased from Dharmacon. The target sequences of 3 of the 4 siRNAs in the ON-TARGETplus SMARTpool against human CTLC were identical (100%) in the human and green monkey CTLC genes.
The clathrin-LCa-eYFP plasmid was a kind gift from Xiaowei Zhuang (Harvard University, Cambridge, MA, USA). The Rab5-wt-GFP plasmid and its dominant negative mutant Rab5-S34N-GFP were generously provided by P. van der Sluijs (University Medical Center, Utrecht, The Netherlands). The pGL-wt-Rab7 plasmid, containing a green fluorescent protein (GFP) reporter gene, was obtained from Gary R. Whittaker (Cornell University, College of Veterinary Medicine, Ithaca, NY, USA).
The inhibitory effects of the chemical inhibitors, siRNAs, and dominant negative mutant were checked using the following controls. Alexa Fluor 633-conjugated transferrin (3 μg/ml; 15-min incubations; Life Technologies) was used as a control for chlorpromazine, Pitstop2, and the siRNAs. LysoTracker Green (5 μM; incubation for 30 min; Life Technologies) staining was used as a control for NH4Cl. Dextran-Texas Red (25 μg/ml; incubation for 30 min; Life Technologies) was used as a control for the Rab5 constructs.
Microscopic fusion assay.
The membrane fusion capacity of CHIKV was determined by a microscopy-based fusion assay, as described previously (37). Briefly, BS-C-1 cells were seeded into 8-well Lab-Tek II chambered coverglass slides (Nunc) to obtain a subconfluent monolayer the next day. Cells were washed three times with serum-free, phenol red-free MEM (Gibco), after which phenol red-free MEM supplemented with 1% glucose was added. DiD-labeled CHIKV was added to the cells at an MOI of 20. Cells were subsequently incubated at 37°C for 30 min to allow viral fusion. Next, unbound virus was removed by washing three times with serum-free, phenol red-free MEM, and fresh phenol red-free MEM was added. Microscopic analysis was carried out using the Leica Biosystems 6000B instrument by randomly selecting fields using differential interference contrast (DIC) settings. A total of 20 random snapshots were taken per experiment in both the DIC and DiD channels. Snapshots were analyzed using the ParticleAnalyzer plugin of ImageJ. The total area of fluorescent spots was quantified in arbitrary units (AU) for each snapshot, and values were averaged for each experiment.
To study the route of entry, several endocytic inhibitors were used. The microscopy-based fusion assay was performed as described above except that the cells were pretreated with the inhibitor of interest. NH4Cl (50 mM) was added 1 h prior to the start of the experiments and Pitstop2 (25 μM) 15 min in advance. CHIKV was added to the cells in the presence of the inhibitor. Both inhibitors were diluted in phenol red-free MEM supplemented with 1% glucose.
Single-particle tracking of DiD-labeled CHIKV.
Single-particle tracking experiments were performed as described previously for DENV (37). Unless indicated otherwise, 1.25 × 106 BS-C-1 cells were transfected with 5 μg of plasmid DNA by electroporation using a Gene Pulser Xcell system (Bio-Rad) and a square wave pulse (100 V, 25 ms). Subsequently, BS-C-1 cells were seeded into 8-well Lab-Tek II chambered coverglass slides (Nunc) to obtain 50 to 70% confluence on the day of tracking.
Directly before the experiment, cells were washed three times with phenol red-free MEM, and phenol red-free MEM supplemented with 1% glucose was added to the cells. GLOX, a glucose oxidase solution, was added to prevent phototoxicity (37). Cells were mounted on the Leica Biosystems 6000B microscope and were kept at 37°C throughout the whole experiment. DiD-labeled CHIKV was added in situ, and image series were recorded at 1 frame per s for 25 to 30 min. To localize the nucleus and plasma membrane of the cell, DIC snapshots were taken before and after the imaging. Image analysis and processing were carried out by ImageJ and Imaris x64, release 7.6.1. Particles smaller than 40 AU were considered individual particles and were selected for further analysis. To avoid the chance of misinterpretation, particles that fused in close proximity (<3 μm) to the nucleus were excluded from tracking behavior analysis, since cells are thicker within this region, and movement in the z axis cannot be detected with our microscope (see Fig. S1 in the supplemental material).
Flow cytometry analysis to determine the number of infected cells.
Flow cytometry analysis was used to assess the effects of the pharmacological inhibitor Pitstop2, siRNAs against CTLC, and the Rab5 dominant negative mutant on CHIKV infection. For the inhibitor studies, Pitstop2 (25, 12.5, or 6.25 μM) was diluted in BS-C-1 medium containing 2% FBS. Cells were preincubated with Pitstop2 for 15 min, after which CHIKV LS3-GFP was added to the cells at an MOI of 1, and infection was allowed for 1.5 h at 37°C. Then BS-C-1 medium containing 10% FBS was added, and incubation was continued overnight in the presence of the inhibitor. At 18 h p.i., cells were fixed with 4% paraformaldehyde (PFA) and were analyzed by flow cytometry.
For the siRNA experiments, siRNA was transfected into cells according to the manufacturer's protocol. At 72 h posttransfection, HeLa cells were infected with CHIKV LS3-GFP at an MOI of 5. At 18 h p.i., cells were fixed with 4% PFA and were analyzed by flow cytometry.
For infection studies with the Rab5 dominant negative mutant, BS-C-1 and HeLa cells were transfected with either the Rab5-wt-GFP plasmid or its dominant negative mutant Rab5-S34N-GFP using Lipofectamine 3000 according to the manufacturer's instructions. At 24 h posttransfection, HeLa cells were infected with CHIKV LS3 at an MOI of 5. BS-C-1 cells were infected at 48 h posttransfection with CHIKV LS3 at an MOI of 1. At 18 h p.i., cells were fixed with 4% PFA, stained with a rabbit anti-E2-stem antibody (1:1,000) obtained from G. Pijlman (Wageningen University, Wageningen, The Netherlands) and an Alexa Fluor 647-conjugated chicken anti-rabbit antibody (1:300; Life Technologies), and analyzed by flow cytometry.
Liposomal bulk fusion assay.
Liposomes (large unilamellar vesicles) of 200 nm were prepared by freeze-thaw extrusion as described previously (24, 34). Unless otherwise specified, liposomes consisted of phosphatidylcholine (PC) from egg yolk, phosphatidylethanolamine (PE) prepared from transphosphatidylation of egg PC, sphingomyelin (SPM) from porcine brain, and cholesterol from ovine wool (all from Avanti Polar Lipids, Alabaster, AL) in a molar ratio of 1:1:1:1.5. Fusion of pyrene-labeled CHIKV with liposomes was monitored in a Fluorolog 3-22 fluorometer (BFi OPTiLAS, Alphen aan den Rijn, The Netherlands) as described previously (24, 34, 38). Briefly, 4 × 1010 pyrene-labeled CHIKV particles were mixed with 6 × 1010 liposomes in a total volume of 665 μl HNE buffer at 37°C with continuous stirring. Fusion was triggered by the addition of 35 μl of 0.1 M morpholineethanesulfonic acid (MES) with 0.2 M acetic acid pretitrated with NaOH to achieve the desired pH. Excitation and emission wavelengths were 345 and 480 nm, respectively. The fusion scale was set such that 0% fusion corresponded to the initial excimer fluorescence and 100% fusion to the signal obtained after the addition of 35 μl of 0.2 M octaethylene glycol monododecyl ether (C12E8; Sigma-Aldrich, Steinheim, Germany), which causes an infinite dilution of the probe. The extent of fusion was determined using the average fluorescent signal between 50 and 60 s after the pH drop.
Cholesterol depletion assay.
BS-C-1 cells were depleted of cholesterol by using various concentrations (7.5, 5, or 2.5 mM) of methyl-β-cyclodextrin. Cells were plated in 8-well Lab-Tek II chambered coverglass slides as described above for the microscopic CHIKV fusion assay. Next, methyl-β-cyclodextrin in phenol red-free MEM was added to the cells, and incubation was continued for 1 h at 37°C. Then the cells were washed three times with phenol red-free MEM, and the experiment was continued as a standard fusion assay. Cholesterol depletion was reversed by the addition of a medium containing water-soluble cholesterol (200 μg/ml) to the cells. In this case, after the treatment with methyl-β-cyclodextrin, cells were washed three times and were incubated for 30 min at 37°C in the presence of cholesterol. Subsequently, the cells were again washed three times to remove free cholesterol, and the experiment was continued as a standard fusion assay.
Statistics.
Statistical analysis was carried out using the two-tailed Student t test in GraphPad Prism software (version 5). A P value of <0.05 was considered significant.
RESULTS
Characteristics of DiD-labeled chikungunya virus.
To visualize the dynamics of CHIKV cell entry and membrane fusion, we labeled the virus with the lipophilic fluorescent probe DiD. This probe was chosen because of its self-quenching properties at a high surface density, as described previously (36). DiD was incorporated into CHIKV particles such that its fluorescence was largely quenched but still allowed the detection of single virus particles. Membrane fusion is measured as a sudden increase in fluorescence intensity due to dilution of the probe in the target cell membrane.
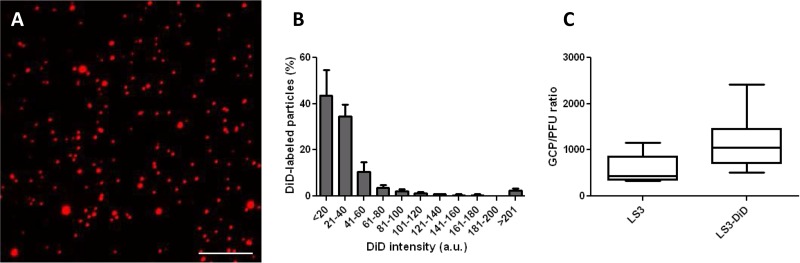
For this study, the well-characterized synthetic CHIKV strain LS3 was used (35). The amino acid sequences of LS3 E1 and E2 are identical to those of the clinical isolate LR2006-OPY1, and therefore, it is expected that these viruses will exhibit the same cell entry behavior. The probe DiD was added at a concentration of 2 nmol per 2 × 1011 virus particles. At this ratio, a uniformly labeled virus preparation was seen (Fig. 1A). Figure 1B shows the total fluorescence intensities of individual CHIKV particles derived from three distinct labeling procedures. Approximately 80% of the spots had a fluorescence intensity below 40 AU. Next, the labeling efficiency was assessed. For this purpose, the number of DiD-labeled particles was counted in 25 random image areas. In parallel, qPCR was used to determine the GCP titer. In line with our previous work on DENV, approximately 2% of the total number of particles present in solution are visualized under the conditions of the experiment (based on three individual experiments) (36). Thereafter, viral infectivity was calculated by dividing the number of GCPs by the number of PFU in five independent experiments. The GCP/PFU ratio was 579 (95% confidence interval [95% CI], 237, 920) for unlabeled virus and 1,156 (95% CI, 467, 1,845) for DiD-labeled virus (Fig. 1C). Although the GCP/PFU ratio increased 2-fold upon DiD labeling, the increase was not statistically significant, suggesting that the overall infectivities of the virus preparations are comparable. Taking these results together, under these labeling conditions, a uniform labeling is achieved that visualizes 2% of the total number of CHIKV particles present in solution. For further experiments, only particles with a fluorescence intensity lower than 40 AU were used, because these likely represent single virus particles.
FIG 1.
Characteristics of DiD-labeled CHIKV. (A) Representative images showing DiD-labeled CHIKV particles. Bar, 10 μm. (B) Histogram of the DiD intensity of CHIKV. DiD intensity was determined by fluorescence microscopy using a 635-nm laser. More than 80,000 particles (from 3 individual experiments) were used for this analysis. Error bars represent standard deviations. a.u., arbitrary units. (C) Boxplot showing the GCP/PFU ratios of unlabeled CHIKV and DiD-labeled CHIKV. Boxes show the 25th and 75th percentiles, while whiskers show the 5th and 95th percentiles. Data represent the results of 5 individual experiments carried out in duplicate.
Rapid chikungunya virus cell entry and fusion in BS-C-1 cells.
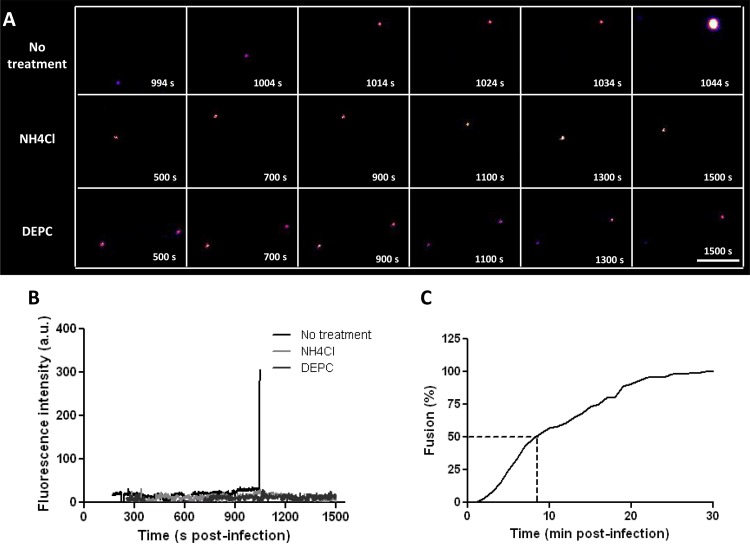
The cell entry behavior of CHIKV was studied in the kidney epithelial cell line BS-C-1, since epithelial cells are thought to contribute to viremia during natural infection (1, 39). Indeed, CHIKV infection of BS-C-1 cells at an MOI of 1 resulted in 2.6 × 106 PFU/ml progeny virions at 24 h p.i., demonstrating that these cells are permissive to CHIKV. Moreover, BS-C-1 cells are relatively flat, which allows us to capture complete viral trajectories from virus-cell binding until the moment of membrane fusion. Figure 2A (no treatment) shows a cell entry trajectory of a single CHIKV particle. Initially, the DiD signal is low but constant over time. At 1,044 s p.i., a sudden increase in fluorescence is seen, which is indicative of the moment of membrane fusion (Fig. 2A and B, no treatment). After fusion, the intensity of the DiD signal remains high for a long time (up to 30 min); the DiD-labeled compartment is highly dynamic, and DiD-labeled structures frequently pinch off to be transported elsewhere, reflecting the highly dynamic nature of endocytic vesicles. A movie showing the DiD signal over time until the moment of membrane fusion is available in the supplemental material (see Movie S1). Under the conditions of this experiment, on average, 1.2 fusion events are recorded per experiment.
FIG 2.
CHIKV cell entry and membrane fusion in BS-C-1 cells. (A) Filmstrips showing the fluorescence intensities of DiD-labeled CHIKV particles over time. In the top filmstrip, an increase in fluorescence intensity, indicative of membrane fusion, is seen at 1,044 s postinfection. The center and bottom strips show the DiD signals in NH4Cl-treated cells and for DEPC-inactivated CHIKV, respectively. Virus particles were tracked until 1,500 s postinfection. Images are artificially colored in order to show differences in fluorescence intensity clearly, with purple and yellow representing low and high intensities, respectively. Bar, 5 μm. (B) Fluorescence intensities of the particles shown in panel A, plotted against time. (C) Percentage of fused CHIKV particles as a function of time. In total, 113 trajectories were analyzed. The time of fusion was defined as the moment when the fluorescence intensity increased >2-fold within 1 to 2 s.
To ensure that the increase in fluorescence indeed reflects membrane fusion, we next performed tracking experiments in BS-C-1 cells treated with NH4Cl. NH4Cl is known to neutralize the endosomal pH, thereby inhibiting the membrane fusion activity of CHIKV (28, 40, 41). Indeed, in the presence of NH4Cl, the infectivity of CHIKV was reduced >4 log units from that for the positive control (5.1 × 102 PFU/ml for NH4Cl-treated cells versus 1.2 × 107 PFU/ml for nontreated cells at an MOI of ∼5). Under these conditions, NH4Cl-treated cells showed no toxicity as measured by the MTT assay (see Fig. S2A in the supplemental material). In 12 independent tracking experiments, we were able to analyze 102 single virus trajectories. Only one particle fused, indicating that NH4Cl indeed severely hampers the membrane fusion activity of CHIKV. Again, under these conditions, no cytotoxicity was seen (see Fig. S2B). Furthermore, endosomal acidification was efficiently blocked (see Fig. S3A in the supplemental material). A representative nonfusogenic DiD-CHIKV trajectory is shown in Fig. 2A, and the fluorescence intensity is plotted in Fig. 2B (NH4Cl).
As a second control, we inactivated the membrane fusion properties of CHIKV by DEPC treatment. DEPC treatment of viral particles is known to covalently modify histidines on viral glycoproteins, thereby abolishing viral fusion without changing the protein structure (42, 43). DiD-labeled CHIKV was treated with 2 mM DEPC for 30 min at room temperature. Direct titration of DEPC-treated CHIKV by a plaque assay revealed that the virus was completely noninfectious. Next, 11 independent tracking experiments were performed, and the fluorescence intensities of all particles remained constant for the duration of the experiment. An example is shown in Fig. 2A and B (DEPC). Together, these results indicate that a sudden dramatic increase in fluorescence indeed reflects the moment of membrane fusion.
Next, we analyzed how fast CHIKV particles can fuse upon addition to cells. For this analysis, 133 fusion events recorded in 93 independent experiments were used. We found that CHIKV cell entry is a very rapid process, with the first fusion events occurring within 2 min p.i. Half of all fusion events investigated occurred within the first 9 min after infection, and more than 95% of all fusion events occurred within 22 min p.i. (Fig. 2C).
Chikungunya virus entry occurs via clathrin-mediated endocytosis.
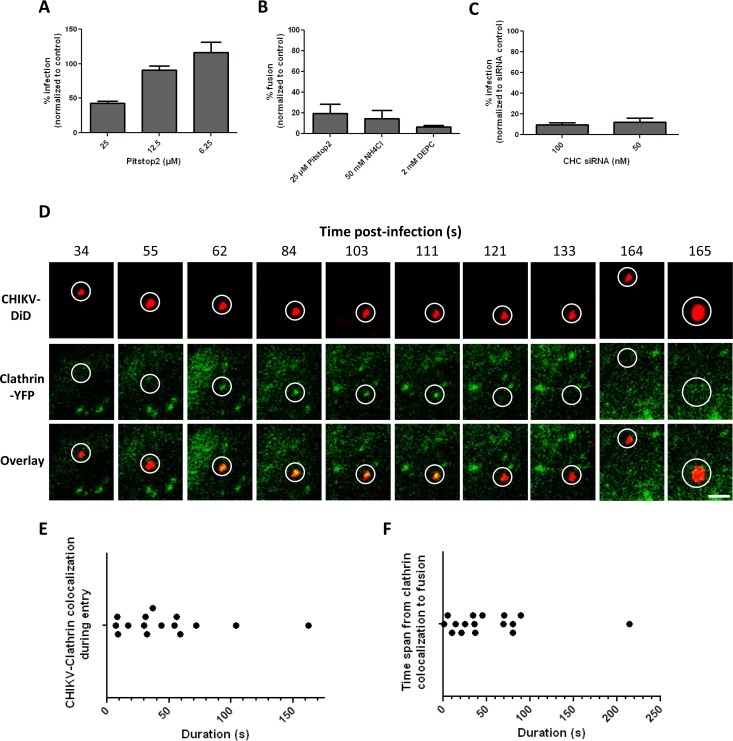
We next aimed to obtain more-detailed insight into the dynamic virus-host interactions that occur during virus entry. Alphaviruses are generally considered to enter cells via CME (9, 17, 18). For CHIKV, however, conflicting reports have been published, and therefore, we first investigated whether CHIKV cell entry is indeed mediated by clathrin. For this purpose, CME was perturbed with two widely used small-compound inhibitors: chlorpromazine and Pitstop2. For both compounds, cytotoxicity was tested using the MTT assay, and transferrin was used to determine whether the compound was biologically active (44, 45). Unfortunately, chlorpromazine was found to be toxic at concentrations higher than 30 μM (see Fig. S2C in the supplemental material), while at this concentration, only a mere 30% reduction in transferrin uptake was seen (see Fig. S3B in the supplemental material). Therefore, this compound was unsuitable for study of the role of CME in our experimental setup. In contrast, transferrin uptake was almost completely abolished (>95%) at a concentration of 25 μM Pitstop2 (see Fig. S3C), which did not cause any measurable cytotoxicity (see Fig. S2D and E).
BS-C-1 cells were pretreated with Pitstop2, and infection was allowed for 18 h in the presence of the inhibitor. Indeed, a substantial drop in infection was found upon treatment of the cells with 25 μM Pitstop2 (Fig. 3A). But because residual infectivity was seen, we next employed a more direct microscopic fusion assay using DiD-labeled CHIKV. If viral entry is affected by Pitstop2, a decrease in membrane fusion activity should be detected. DiD-labeled CHIKV was allowed to enter and fuse within cells for 30 min, after which unbound particles were washed away. Next, random microscopic images were taken, and the fluorescence intensity was quantified. First, the fusion assay was validated using NH4Cl-treated cells and DEPC-treated virus as described above. Figure S4 in the supplemental material shows representative images of both treatment conditions and the nontreated virus control. The quantification of the total fluorescence intensity is shown in Fig. 3B. Indeed, NH4Cl treatment severely hampered the membrane fusion capacity of the virus. Furthermore, almost no fusion was observed using DEPC-treated virus. Importantly, a dramatic inhibition of CHIKV fusion was seen in Pitstop2-treated cells (Fig. 3B), suggesting that CHIKV indeed enters BS-C-1 cells via CME. To further confirm CHIKV cell entry via CME, we attempted to knock down clathrin heavy chain (CHC) expression using siRNAs. BS-C-1 cells were difficult to transfect at high efficiency, however, and we failed to inhibit transferrin uptake efficiently in BS-C-1 cells using these siRNAs, despite testing a variety of transfection conditions and agents (an example is given in Fig. S3D in the supplemental material). We then used HeLa cells, which can be transfected efficiently and are permissive for CHIKV (39). Indeed, transferrin uptake was efficiently inhibited in anti-CHC siRNA-transfected HeLa cells (see Fig. S3E). Next, CHIKV infectivity was assessed in CHC siRNA-transfected HeLa cells by flow cytometry. The number of infected cells was as much as 90% lower than that for cells transfected with a nontargeting siRNA control (Fig. 3C). We also attempted to use HEK293T cells, but despite multiple efforts, these cells were detached from the plate upon transfection, thereby preventing further experiments. Taken together, the results presented above show that CHIKV enters BS-C-1 and HeLa cells via CME.
FIG 3.
CHIKV infects cells via clathrin-mediated endocytosis. (A) Flow cytometry analysis of CHIKV infection in the presence of Pitstop2. BS-C-1 cells were preincubated with Pitstop2 for 15 min, after which cells were infected with CHIKV LS3-GFP (MOI, 1) and were incubated for an additional 18 h. Data represent results of two individual experiments performed in triplicate. Error bars represent standard deviations. (B) CHIKV cell entry and membrane fusion in the presence of inhibitors. The percentage of fusion is normalized to that for a nontreated positive control. Data are from at least three individual experiments. (C) Flow cytometry analysis of CHIKV infection in anti-CHC siRNA-transfected HeLa cells. Cells were transfected with anti-CHC siRNAs, incubated for 72 h, and infected with CHIKV LS3-GFP (MOI, 5). The percentage of infection is relative to that of HeLa cells transfected with a nontargeting siRNA control. Data represent results of two individual experiments performed in triplicate. (D) Time series of a cell expressing clathrin-YFP (green) upon infection with DiD-labeled CHIKV particles (red). White circles indicate the position of the virus in each panel. All images are equally enhanced for visual purposes. Bar, 1 μm. (E) Dot plot showing the duration of clathrin colocalization of 17 CHIKV particles. Only particles that fused were used in the analysis. Each dot represents 1 particle. (F) Dot plot showing the time of membrane fusion minus the last time point of clathrin colocalization. The graph shows data for 16 individual virus particles; each dot represents 1 particle.
To reveal the dynamics of CHIKV entry via CME, we next transfected BS-C-1 cells with yellow fluorescent protein (YFP)-labeled clathrin and simultaneously tracked the clathrin and CHIKV-DiD signals in living cells. The vast majority of particles (17 of 19 fusion-positive CHIKV particles) colocalized with clathrin prior to fusion. An example of CME of CHIKV is shown in the filmstrips in Fig. 3D. Furthermore, two movies that focus on virus-clathrin colocalization (see Movies S2C and E) and three movies demonstrating that clathrin colocalization precedes membrane fusion (see Movies S2A, B, and D) are included in the supplemental material. The average duration of colocalization between CHIKV and clathrin was 48.1 s (standard error of the mean [SEM], ±10.6; n = 15 [2 particles were excluded because the duration of colocalization could not be estimated]) (Fig. 3E), which is in line with the kinetics of clathrin-coated pit formation and internalization as described by Schelhaas et al. (46). We also determined the time between the disappearance of the clathrin signal and the moment of fusion. On average, fusion occurred 51.9 s after clathrin colocalization (SEM, ±13.0; n = 16 [1 particle was excluded because the exact time of clathrin disappearance could not be determined]). Moreover, approximately 95% of the fusion events occurred within 90 s after colocalization, indicating that CHIKV fuses rapidly after entry (Fig. 3F).
Based on the kinetics described above, it can be estimated that CHIKV entry (from initial clathrin colocalization to membrane fusion) is completed within roughly 1 min 40 s. When the fusion kinetics alone were assessed, it was found that approximately 50% of all fusion events occurred within the first 9 min p.i. (Fig. 2C). The difference between these time points indicates that most CHIKV particles spend a relatively long time searching for a cellular attachment factor.
Chikungunya fusion occurs mainly from within early endosomes.
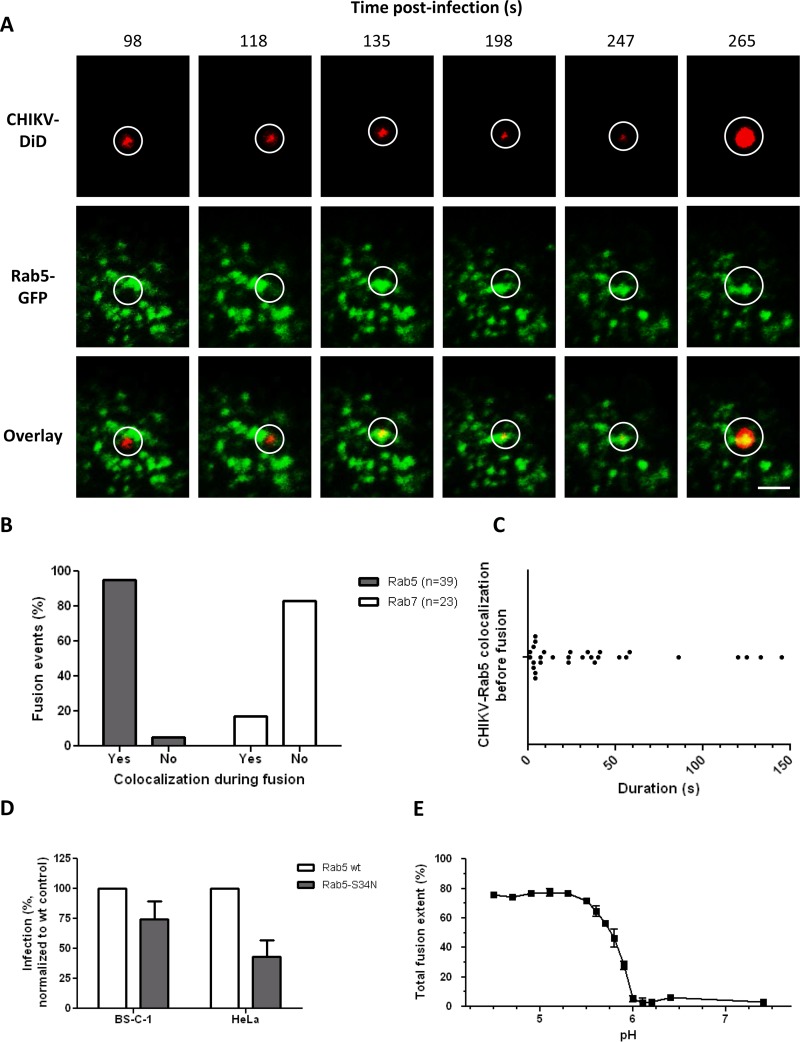
We next addressed whether CHIKV fuses from within early or late endosomal compartments. First, we tracked DiD-labeled CHIKV particles in BS-C-1 cells that had first been transfected with Rab5-GFP, a marker for early endosomes. Figure 4A shows a filmstrip of a single CHIKV particle fusing from within a Rab5-positive compartment. At 98 and 118 s p.i., the virus is seen in close proximity to Rab5-positive structures; however, no colocalization is seen. Colocalization is visible at 135 s p.i. and continues until the moment of membrane fusion at 265 s p.i. Three movies showing a CHIKV fusion event in which CHIKV is colocalized with Rab5 are available in the supplemental material (see Movies S3A, B, and C).
FIG 4.
CHIKV interacts with Rab5-positive endosomes at the time of fusion. (A) Time series of a cell expressing Rab5-GFP (green) upon infection with DiD-labeled CHIKV particles (red). Circles indicate the position of the virus in each panel. All images are equally enhanced for visual purposes. Bar, 2 μm. (B) Bar graph showing the percentages of fusion events in Rab5-positive and Rab7-positive structures. In total, 39 fusion events were analyzed in Rab5-GFP-expressing cells, and 23 fusion events in Rab7-GFP-expressing cells. (C) Dot plot demonstrating how long CHIKV resides in Rab5-positive organelles prior to fusion. A total of 30 individual fusion events were analyzed. Each dot represents 1 fusion event. (D) CHIKV infectivity in cells expressing wt-Rab5-GFP or dominant negative Rab5-GFP. Infection was quantified and normalized to the level for the wt-Rab5-GFP control. For both cell lines, two individual experiments were carried out in triplicate. Error bars represent standard deviations. (E) pH-dependent membrane fusion properties of LS3 as determined by a bulk fusion assay. At least three measurements were performed per pH value.
In total, 39 fusion events were recorded in Rab5-transfected cells. Of these fusion events, 37 (95%) occurred while CHIKV was colocalized with Rab5 (Fig. 4B). Next, tracking was performed in cells transfected with Rab7-GFP, a specific marker for late endosomes. A total of 23 fusion events were recorded, only 4 of which (17%) occurred while CHIKV was colocalized with a Rab7-positive structure (Fig. 4B). Together, these results show that CHIKV fuses predominantly from within Rab5-positive early endosomes. The remaining particles fuse from within Rab5/Rab7-positive maturing endosomes or Rab7-positive late endosomes.
Further analysis of the CHIKV trajectories in Rab5-transfected cells revealed that CHIKV resided, on average, 37.6 s (SEM, ±7.8; n = 30) in early endosomes before fusing. Interestingly, though, approximately 40% of all particles fused almost immediately (within 10 s) after colocalization (Fig. 4C). This finding might reflect the maturation state of the endosome at the time the virus is delivered, since the pH gradually drops during endosomal maturation (47, 48).
To further confirm that fusion occurs from within early endosomes, we assayed the number of infected cells upon transfection with the dominant negative mutant Rab5-S34N. Rab5-S34N caused a 25% reduction in infection from that with the wild-type (wt) control (Fig. 4D), but this reduction was not as pronounced as expected on the basis of our single-particle tracking results. This might, again, be related to the overall low transfection efficiency, and thus low expression levels, of the Rab5 constructs in BS-C-1 cells. As a control, we next analyzed the uptake of dextran-Texas Red in Rab5-transfected cells (49, 50). In line with the results for CHIKV, we only found a mild (25%) reduction in dextran-Texas Red uptake by Rab5-S34N-transfected cells from that with the Rab5 wild-type control (see Fig. S3F in the supplemental material). Furthermore, if we arbitrarily change the gating to include only cells with high GFP expression (see Fig. S5A and B in the supplemental material), the inhibition of infection is more pronounced (up to 49% [see Fig. S5C]). Collectively, the data indeed suggest that the expression level of Rab5-S34N in BS-C-1 cells is too low to actively block endocytosis. For additional proof that fusion occurs from within Rab5-positive early endosomes, we also assessed the effect of Rab5-S34N in HeLa cells. As expected, a more pronounced inhibition of infection was found in HeLa cells than in BS-C-1 cells (Fig. 4D). Again, the inhibition of infection was comparable to the inhibition of dextran uptake (see Fig. S3F).
The pH values within early endosomes typically range from 6.8 to 5.5 (48, 51). Earlier reports showed that the pH threshold for fusion is strain specific (34), yet all strains reported thus far fused at early endosomal pH values. To confirm that CHIKV-LS3 is also able to fuse at an early endosomal pH, we assessed the pH-dependent membrane fusion properties of CHIKV-LS3 by use of a bulk fusion assay (24, 25, 34). Here, pyrene-labeled CHIKV is mixed with liposomes consisting of PC, PE, SPM, and cholesterol, and fusion is triggered by the addition of a low-pH buffer. Upon fusion, the pyrene phospholipids are diluted in the liposomes, resulting in a decrease in fluorescence intensity, which can be monitored continuously. Figure 4E shows the total fusion extent as a function of pH. No fusion was seen at a neutral pH (7.4). CHIKV fusion was first observed at pH 5.9, and maximal fusion was seen at pH values lower than 5.5. Thus, the threshold of CHIKV fusion was pH 5.9, which indicates that CHIKV-LS3 fusion can indeed occur from within early endosomes.
A valine at position 226 in the E1 protein increases the cholesterol dependency of CHIKV fusion.
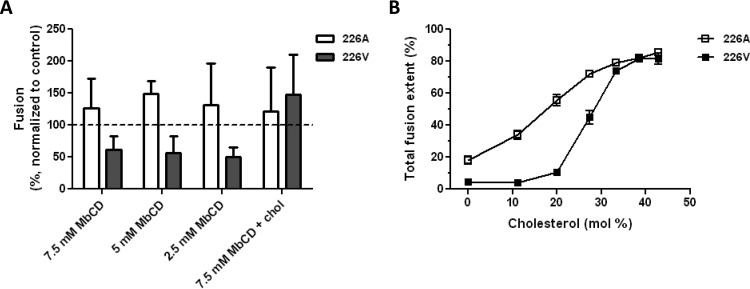
Since cholesterol has been found to be important in alphavirus fusion and infection (24, 25, 52, 53), we subsequently determined the effect of cholesterol on CHIKV cell entry and fusion. Residue 226 in E1 is an important determinant of the cholesterol dependency of alphavirus infection (54, 55). For CHIKV, increased cholesterol dependency of infection was seen when E1 had a valine residue instead of an alanine residue at position 226 (28, 56). To study the effect of the E1-A226V mutation on membrane fusion, we mutated the infectious clone of LS3 (E1-226V) to create a virus with an alanine at this position (LS3-226A). The two strains exhibited similar growth kinetics in Vero cells (data not shown). The membrane fusion activities of both strains were first evaluated by the microscopic fusion assay with cells that were depleted of cholesterol by use of various concentrations of methyl-β-cyclodextrin (7.5, 5, or 2.5 mM). Under these conditions, no cytotoxicity was seen in the MTT assay (see Fig. S2F in the supplemental material). Under identical infection conditions, the fluorescence intensities measured for LS3-226A were 1.6 times higher than those measured for LS3-226V, suggesting that LS3-226A fuses more efficiently with the host cell membrane than LS3-226V. Hence, all data were normalized to the data for the positive control (no inhibitor) of the corresponding virus strain. There was a clear difference in cholesterol dependency between the two LS3 strains. Whereas cholesterol depletion had no effect on LS3-226A entry and/or fusion, it strongly inhibited the entry and/or fusion of LS3-226V (Fig. 5A). Importantly, upon replenishment of membrane cholesterol, the extent of LS3-226V fusion was restored to control levels (Fig. 5A).
FIG 5.
CHIKV infection and membrane fusion are promoted by target membrane cholesterol. (A) Effect of cholesterol (chol) depletion on the cell entry and fusion of LS3-226A and LS3-226V. BS-C-1 cells were depleted of cholesterol using methyl-β-cyclodextrin (MbCD) at 7.5, 5, or 2.5 mM for 1 h. Then the cells were infected with either LS3-226A or LS3-226V (MOI, 20), and the extent of membrane fusion was measured at 30 min postinfection. As a control, cells were replenished with water-soluble cholesterol (at 200 μg/ml for 30 min) and were infected. Data are from at least three individual experiments. Error bars represent standard deviations. (B) Bulk fusion assay data showing the cholesterol-dependent membrane fusion properties of LS3-226A and LS3-226V. Open squares represent LS3-226A, whereas filled squares represent LS3-226V. At least three measurements per data point were performed.
To investigate more specifically whether cholesterol has an effect on membrane fusion, we next employed the bulk fusion assay. First, we determined the pH threshold for LS3-226A fusion. Compared to the pH threshold of 5.9 for LS3-226V fusion (Fig. 4E), the pH threshold for LS3-226A fusion was slightly elevated, to pH 6.1. Next, viral fusion was measured using liposomes with increasing concentrations of cholesterol. The bulk fusion assay revealed that the presence of cholesterol in the target membrane promotes the membrane fusion capacities of both LS3-226V and LS3-226A (Fig. 5B). Yet LS3-226A fusion was also observed in the absence of cholesterol in the target membrane, indicating that cholesterol is not strictly required for fusion. In contrast, LS3-226V did not fuse with liposomes when the cholesterol concentrations were ≤11 mol%. Together, these results confirm that the E1-A226V mutation indeed increases the cholesterol dependency of CHIKV fusion.
DISCUSSION
This paper describes the mechanistic and kinetic events that occur during CHIKV entry into BS-C-1 cells. We show that almost all particles that fused first colocalized with clathrin structures. The time of colocalization is consistent with the reported overall life span of clathrin-coated structures (46). CHIKV particles fuse predominantly from within Rab5-positive early endosomes. The process of CHIKV entry into cells is extremely fast: 50% of the particles fused within 1.7 min after colocalization with clathrin. Furthermore, 40% of the particles fused instantly upon delivery to acidic endosomes. Finally, we showed that a valine at position 226 in E1 increased the cholesterol dependency of CHIKV fusion over that for a virus with an alanine at this position.
The observation that CHIKV enters cells via CME is in line with two earlier reports on CHIKV cell entry (31, 32). In these studies, significant inhibition of viral infectivity was seen in cells treated with drugs or siRNAs that have been proposed to interfere with CME. In contrast, another study showed that siRNAs against the clathrin heavy chain did not interfere with infection, and a clathrin-independent but Eps15-dependent CHIKV entry pathway was proposed (28). This suggests that CHIKV entry is strain and/or target cell dependent. The receptor utilized by the virus to infect cells may be an important factor here, especially since CHIKV has been proposed to interact with multiple receptors (57–60). Another possibility is that CHIKV has the capacity to hijack multiple entry pathways. In that case, inhibitor studies may not identify the dominant cell entry pathway of the virus, since under inhibiting conditions, the virus will enter through another pathway. In BS-C-1 cells, residual infectivity was seen in cells treated with the CME inhibitor Pitstop2, while transferrin uptake was abolished. Residual infectivity was also found in CHC siRNA-transfected HeLa cells, albeit to a lesser extent. In fact, in all of the studies discussed above, residual infectivity was seen, indicating that CHIKV indeed has the capacity to infect cells via multiple pathways. An advantage of the approach used in this study is that we were able to monitor CHIKV cell entry in the absence of inhibitors. The results presented in this paper clearly show that 89% of the particles that fused first colocalized with clathrin. Taking the data together, and in line with the findings for other alphaviruses, we conclude that CME is the major cell entry pathway of CHIKV.
Almost all viruses that are internalized by the cell via endocytosis are first delivered to Rab5-positive early endosomes before being targeted to Rab5/Rab7-positive maturing endosomes, Rab7-positive late endosomes, and lysosomes (20). Previous studies on alphaviruses showed that SFV fused from within early endosomes, and VEEV was found to fuse from maturing and/or late endosomes (18, 21, 61, 62). Our single-particle tracking data show that CHIKV fuses predominantly from within early endosomes. Approximately 40% of the particles fused within 10 s after colocalization with Rab5, demonstrating that CHIKV almost instantly fuses with the endosomal membrane. The time during which CHIKV resides within endosomes is probably controlled by the pH of the endosomal lumen. Instant fusion may occur when the particles are delivered to endosomes where the pH is lower than the pH threshold for fusion. We show here that the pH threshold for CHIKV-LS3 fusion is 5.9, although strain-specific pH-dependent properties exist (34). The short time during which CHIKV resides in endosomes is in sharp contrast with the behavior of dengue virus particles, which spend minutes in late endosomal compartments prior to fusion. For dengue virus, negatively charged lipids are required to complete the fusion process (63), and this dependency likely prevents fusion from within early endosomes. Our results are in line with an earlier study of Bernard et al., who showed that CHIKV infection of HEK-293T cells is dependent on the integrity of early endosomes, but not on late endosomes (28). In mosquito cells, both early and late endosomes are required for CHIKV infection, suggesting that in these cells, CHIKV is trafficked to maturing/late endosomes before fusion (32). This discrepancy may be related to the different virus strains used but could also be related to potential differences in the endosomal pH between cells (64).
To our knowledge, this is the first study that describes the kinetics of CHIKV entry at the single-particle level. The first fusion events were detected 2 min p.i., and 50% of all fusion events observed occurred within 9 min p.i. This corresponds nicely with data described for SFV (65). In the latter study, the authors added NH4Cl at different time points p.i. and assessed the number of infected cells by flow cytometry. The addition of NH4Cl at 2 to 3 min p.i. did not prevent SFV infection, and its addition at 6 min p.i. led only to a 50% reduction in the number of infected cells from that for the untreated control. The earlier SFV study and our results indicate that both SFV and CHIKV have very rapid entry kinetics. Besides assessing the overall time to fusion, we also determined here the kinetics of the major steps in CHIKV entry. Half of the particles fused within 1.7 min after colocalization with clathrin, demonstrating that virus particles spend a relatively long time searching for a receptor. This also shows that once the virus colocalizes with clathrin, the entry process is extremely fast.
Last, we confirmed that position 226 in the CHIKV E1 protein is an important determinant of the cholesterol-dependent membrane fusion properties of the virus. The E1-A226V mutation was first reported in a clinical CHIKV strain during the 2005-to-2006 outbreak on La Réunion and was associated with CHIKV vector specificity (56, 66), the pH threshold of fusion in cell fusion assays (67, 68), and the host immune response to the virus (69, 70). Furthermore, the E1-A226V mutation was linked to increased cholesterol dependency of CHIKV infection (28, 56, 68). We confirmed that the E1-A226V mutation influences the pH threshold for fusion by use of a direct bulk fusion assay and demonstrated that higher concentrations of cholesterol are needed within the target membrane to facilitate efficient fusion. Our results are in agreement with reports published on other alphaviruses (25, 52, 55).
Studying the cell entry pathway at the single-particle level is important, because it provides detailed and quantitative information on the dynamic events involved in CHIKV cell entry. Furthermore, by measuring fusion instead of infection as a readout of CHIKV cell entry, we avoid artifacts of inhibitors that exert effects not only during entry but also at later stages of infection. Therefore, this study enhances our understanding of the cell entry process of CHIKV and alphaviruses in general.
Supplementary Material
ACKNOWLEDGMENTS
T.E.H. was supported by an ASPASIA grant from the Dutch Scientific Organization (NWO) to J.M.S. I.C.A. was supported by the Marie Curie Initial Training Network EUVIRNA (grant agreement 264286).
Funding Statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03184-15.
REFERENCES
- 1.Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol 8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 2.WHO. 2015. Chikungunya. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs327/en/. [Google Scholar]
- 3.CDC. 2015. Chikungunya virus: geographic distribution. CDC, Atlanta, GA: http://www.cdc.gov/chikungunya/geo/index.html. [Google Scholar]
- 4.CDC. 2015. Chikungunya virus: symptoms. CDC, Atlanta, GA: http://www.cdc.gov/chikungunya/symptoms/index.html. [Google Scholar]
- 5.Essackjee K, Goorah S, Ramchurn SK, Cheeneebash J, Walker-Bone K. 2013. Prevalence of and risk factors for chronic arthralgia and rheumatoid-like polyarthritis more than 2 years after infection with chikungunya virus. Postgrad Med J 89:440–447. doi: 10.1136/postgradmedj-2012-131477. [DOI] [PubMed] [Google Scholar]
- 6.Schilte C, Staikovsky F, Couderc T, Madec Y, Carpentier F, Kassab S, Albert ML, Lecuit M, Michault A. 2013. Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl Trop Dis 7:e2137. doi: 10.1371/journal.pntd.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, Pierre V. 2009. Post-epidemic Chikungunya disease on Reunion Island: course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl Trop Dis 3:e389. doi: 10.1371/journal.pntd.0000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voss JE, Vaney M-C, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 9.Solignat M, Gay B, Higgs S, Briant L, Devaux C. 2009. Replication cycle of chikungunya: a re-emerging arbovirus. Virology 393:183–197. doi: 10.1016/j.virol.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang KS, Schmaljohn AL, Kuhn RJ, Strauss JH. 1991. Antiidiotypic antibodies as probes for the Sindbis virus receptor. Virology 181:694–702. doi: 10.1016/0042-6822(91)90903-O. [DOI] [PubMed] [Google Scholar]
- 11.Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. 1992. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol 66:4992–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klimstra WB, Nangle EM, Smith MS, Yurochko AD, Ryman KD. 2003. DC-SIGN and L-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J Virol 77:12022–12032. doi: 10.1128/JVI.77.22.12022-12032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smit JM, Waarts B-L, Kimata K, Klimstra WB, Bittman R, Wilschut J. 2002. Adaptation of alphaviruses to heparan sulfate: interaction of Sindbis and Semliki forest viruses with liposomes containing lipid-conjugated heparin. J Virol 76:10128–10137. doi: 10.1128/JVI.76.20.10128-10137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Linn M, Eble JA, Lübken C, Slade RW, Heino J, Davies J, Suhrbier A. 2005. An arthritogenic alphavirus uses the α1β1 integrin collagen receptor. Virology 336:229–239. doi: 10.1016/j.virol.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Malygin AA, Bondarenko EI, Ivanisenko VA, Protopopova EV, Karpova GG, Loktev VB. 2009. C-terminal fragment of human laminin-binding protein contains a receptor domain for Venezuelan equine encephalitis and tick-borne encephalitis viruses. Biokhimiia 74:1328–1336. doi: 10.1134/S0006297909120050. [DOI] [PubMed] [Google Scholar]
- 16.Kielian M, Chanel-Vos C, Liao M. 2010. Alphavirus entry and membrane fusion. Viruses 2:796–825. doi: 10.3390/v2040796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helenius A, Kartenbeck J, Simons K, Fries E. 1980. On the entry of Semliki Forest virus into BHK-21 cells. J Cell Biol 84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolokoltsov AA, Fleming EH, Davey RA. 2006. Venezuelan equine encephalitis virus entry mechanism requires late endosome formation and resists cell membrane cholesterol depletion. Virology 347:333–342. doi: 10.1016/j.virol.2005.11.051. [DOI] [PubMed] [Google Scholar]
- 19.Marsh M, Bolzau E, Helenius A. 1983. Penetration of Semliki Forest virus from acidic prelysosomal vacuoles. Cell 32:931–940. doi: 10.1016/0092-8674(83)90078-8. [DOI] [PubMed] [Google Scholar]
- 20.Yamauchi Y, Helenius A. 2013. Virus entry at a glance. J Cell Sci 126:1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- 21.Colpitts TM, Moore AC, Kolokoltsov AA, Davey RA. 2007. Venezuelan equine encephalitis virus infection of mosquito cells requires acidification as well as mosquito homologs of the endocytic proteins Rab5 and Rab7. Virology 369:78–91. doi: 10.1016/j.virol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White J, Helenius A. 1980. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc Natl Acad Sci U S A 77:3273–3277. doi: 10.1073/pnas.77.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieva JL, Bron R, Corver J, Wilschut J. 1994. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J 13:2797–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smit JM, Bittman R, Wilschut J. 1999. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J Virol 73:8476–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waarts B-L, Bittman R, Wilschut J. 2002. Sphingolipid and cholesterol dependence of alphavirus membrane fusion. Lack of correlation with lipid raft formation in target liposomes. J Biol Chem 277:38141–38147. doi: 10.1074/jbc.M206998200. [DOI] [PubMed] [Google Scholar]
- 26.van Duijl-Richter MKS, Hoornweg TE, Rodenhuis-Zybert IA, Smit JM. 2015. Early events in chikungunya virus infection—from virus cell binding to membrane fusion. Viruses 7:3647–3674. doi: 10.3390/v7072792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gold ES, Underhill DM, Morrissette NS, Guo J, McNiven MA, Aderem A. 1999. Dynamin 2 is required for phagocytosis in macrophages. J Exp Med 190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernard E, Solignat M, Gay B, Chazal N, Higgs S, Devaux C, Briant L. 2010. Endocytosis of chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PLoS One 5:e11479. doi: 10.1371/journal.pone.0011479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirchhausen T, Owen D, Harrison SC. 2014. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb Perspect Biol 6:a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. 2005. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci U S A 102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M. 2013. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog 9:e1003835. doi: 10.1371/journal.ppat.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee RCH, Hapuarachchi HC, Chen KC, Hussain KM, Chen H, Low SL, Ng LC, Lin R, Ng MM-L, Chu JJH. 2013. Mosquito cellular factors and functions in mediating the infectious entry of chikungunya virus. PLoS Negl Trop Dis 7:e2050. doi: 10.1371/journal.pntd.0002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo S-C, Chen Y-J, Wang Y-M, Tsui P-Y, Kuo M-D, Wu T-Y, Lo SJ. 2012. Cell-based analysis of Chikungunya virus E1 protein in membrane fusion. J Biomed Sci 19:44. doi: 10.1186/1423-0127-19-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Duijl-Richter M, Blijleven J, van Oijen A, Smit J. 2015. Chikungunya virus fusion properties elucidated by single-particle and bulk approaches. J Gen Virol 96:2122–2132. doi: 10.1099/vir.0.000144. [DOI] [PubMed] [Google Scholar]
- 35.Scholte FEM, Tas A, Martina BEE, Cordioli P, Narayanan K, Makino S, Snijder EJ, van Hemert MJ. 2013. Characterization of synthetic Chikungunya viruses based on the consensus sequence of recent E1-226V isolates. PLoS One 8:e71047. doi: 10.1371/journal.pone.0071047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Schaar HM, Rust MJ, Waarts B-L, van der Ende-Metselaar H, Kuhn RJ, Wilschut J, Zhuang X, Smit JM. 2007. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. J Virol 81:12019–12028. doi: 10.1128/JVI.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayala-Nuñez NV, Wilschut J, Smit JM. 2011. Monitoring virus entry into living cells using DiD-labeled dengue virus particles. Methods 55:137–143. doi: 10.1016/j.ymeth.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Thompson BS, Moesker B, Smit JM, Wilschut J, Diamond MS, Fremont DH. 2009. A therapeutic antibody against West Nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog 5:e1000453. doi: 10.1371/journal.ppat.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Le Roux K, Prevost MC, Fsihi H, Frenkiel MP, Blanchet F, Afonso PV, Ceccaldi PE, Ozden S, Gessain A, Schuffenecker I, Verhasselt B, Zamborlini A, Saïb A, Rey FA, Arenzana-Seisdedos F, Desprès P, Michault A, Albert ML, Schwartz O. 2007. Characterization of reemerging chikungunya virus. PLoS Pathog 3:e89. doi: 10.1371/journal.ppat.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dean RT, Jessup W, Roberts CR. 1984. Effects of exogenous amines on mammalian cells, with particular reference to membrane flow. Biochem J 217:27–40. doi: 10.1042/bj2170027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Axelsson MAB, Karlsson NG, Steel DM, Ouwendijk J, Nilsson T, Hansson GC. 2001. Neutralization of pH in the Golgi apparatus causes redistribution of glycosyltransferases and changes in the O-glycosylation of mucins. Glycobiology 11:633–644. doi: 10.1093/glycob/11.8.633. [DOI] [PubMed] [Google Scholar]
- 42.Stauffer F, De Miranda J, Schechter MC, Carneiro FA, Salgado LT, Machado GF, Da Poian AT. 2007. Inactivation of vesicular stomatitis virus through inhibition of membrane fusion by chemical modification of the viral glycoprotein. Antiviral Res 73:31–39. doi: 10.1016/j.antiviral.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Stauffer F, De Miranda J, Schechter MC, Queiroz FA, Santos NO, Alves AMB, Da Poian AT. 2007. New chemical method of viral inactivation for vaccine development based on membrane fusion inhibition. Vaccine 25:7885–7892. doi: 10.1016/j.vaccine.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 44.Le Roy C, Wrana JL. 2005. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol 6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 45.Liu AP, Aguet F, Danuser G, Schmid SL. 2010. Local clustering of transferrin receptors promotes clathrin-coated pit initiation. J Cell Biol 191:1381–1393. doi: 10.1083/jcb.201008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schelhaas M, Shah B, Holzer M, Blattmann P, Kühling L, Day PM, Schiller JT, Helenius A. 2012. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog 8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuesta-Geijo MA, Galindo I, Hernáez B, Quetglas JI, Dalmau-Mena I, Alonso C. 2012. Endosomal maturation, Rab7 GTPase and phosphoinositides in African swine fever virus entry. PLoS One 7:e48853. doi: 10.1371/journal.pone.0048853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huotari J, Helenius A. 2011. Endosome maturation. EMBO J 30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schnatwinkel C, Christoforidis S, Lindsay MR, Uttenweiler-Joseph S, Wilm M, Parton RG, Zerial M. 2004. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol 2:E261. doi: 10.1371/journal.pbio.0020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coyne CB, Shen L, Turner JR, Bergelson JM. 2007. Coxsackievirus entry across epithelial tight junctions requires occludin and the small GTPases Rab34 and Rab5. Cell Host Microbe 2:181–192. doi: 10.1016/j.chom.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mellman I. 1992. The importance of being acid: the role of acidification in intracellular membrane traffic. J Exp Biol 172:39–45. [DOI] [PubMed] [Google Scholar]
- 52.Kielian MC, Helenius A. 1984. Role of cholesterol in fusion of Semliki Forest virus with membranes. J Virol 52:281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phalen T, Kielian M. 1991. Cholesterol is required for infection by Semliki Forest virus. J Cell Biol 112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatterjee PK, Vashishtha M, Kielian M. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J Virol 74:1623–1631. doi: 10.1128/JVI.74.4.1623-1631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu YE, Cassese T, Kielian M. 1999. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J Virol 73:4272–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wintachai P, Wikan N, Kuadkitkan A, Jaimipuk T, Ubol S, Pulmanausahakul R, Auewarakul P, Kasinrerk W, Weng W-Y, Panyasrivanit M, Paemanee A, Kittisenachai S, Roytrakul S, Smith DR. 2012. Identification of prohibitin as a Chikungunya virus receptor protein. J Med Virol 84:1757–1770. doi: 10.1002/jmv.23403. [DOI] [PubMed] [Google Scholar]
- 58.Moller-Tank S, Kondratowicz AS, Davey RA, Rennert PD, Maury W. 2013. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J Virol 87:8327–8341. doi: 10.1128/JVI.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silva LA, Khomandiak S, Ashbrook AW, Weller R, Heise MT, Morrison TE, Dermody TS. 2014. A single-amino-acid polymorphism in Chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J Virol 88:2385–2397. doi: 10.1128/JVI.03116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fongsaran C, Jirakanwisal K, Kuadkitkan A, Wikan N, Wintachai P, Thepparit C, Ubol S, Phaonakrop N, Roytrakul S, Smith DR. 2014. Involvement of ATP synthase β subunit in chikungunya virus entry into insect cells. Arch Virol 159:3353–3364. doi: 10.1007/s00705-014-2210-4. [DOI] [PubMed] [Google Scholar]
- 61.Sieczkarski SB, Whittaker GR. 2003. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4:333–343. doi: 10.1034/j.1600-0854.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 62.Leung JY-S, Ng MM-L, Chu JJH. 2011. Replication of alphaviruses: a review on the entry process of alphaviruses into cells. Adv Virol 2011:249640. doi: 10.1155/2011/249640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaitseva E, Yang S-T, Melikov K, Pourmal S, Chernomordik LV. 2010. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog 6:e1001131. doi: 10.1371/journal.ppat.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sipe DM, Jesurum A, Murphy RF. 1991. Absence of Na+,K+-ATPase regulation of endosomal acidification in K562 erythroleukemia cells. Analysis via inhibition of transferrin recycling by low temperatures. J Biol Chem 266:3469–3474. [PubMed] [Google Scholar]
- 65.Vonderheit A, Helenius A. 2005. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol 3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vazeille M, Moutailler S, Coudrier D, Rousseaux C, Khun H, Huerre M, Thiria J, Dehecq J-S, Fontenille D, Schuffenecker I, Despres P, Failloux A-B. 2007. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One 2:e1168. doi: 10.1371/journal.pone.0001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsetsarkin KA, McGee CE, Higgs S. 2011. Chikungunya virus adaptation to Aedes albopictus mosquitoes does not correlate with acquisition of cholesterol dependence or decreased pH threshold for fusion reaction. Virol J 8:376. doi: 10.1186/1743-422X-8-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gay B, Bernard E, Solignat M, Chazal N, Devaux C, Briant L. 2012. pH-dependent entry of chikungunya virus into Aedes albopictus cells. Infect Genet Evol 12:1275–1281. doi: 10.1016/j.meegid.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 69.Bordi L, Meschi S, Selleri M, Lalle E, Castilletti C, Carletti F, Chiappini R, Di Caro A, Capobianchi MR. 2011. Chikungunya virus isolates with/without A226V mutation show different sensitivity to IFN-α, but similar replication kinetics in non human primate cells. New Microbiol 34:87–91. [PubMed] [Google Scholar]
- 70.Priya R, Dhanwani R, Patro IK, Rao PVL, Parida MM. 2013. Differential regulation of TLR mediated innate immune response of mouse neuronal cells following infection with novel ECSA genotype of Chikungunya virus with and without E1:A226V mutation. Infect Genet Evol 20:396–406. doi: 10.1016/j.meegid.2013.09.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.