Abstract
Histo-blood group antigens (HBGAs) are important binding factors for norovirus infections. We show that two human milk oligosaccharides, 2′-fucosyllactose (2′FL) and 3-fucosyllactose (3FL), could block norovirus from binding to surrogate HBGA samples. We found that 2′FL and 3FL bound at the equivalent HBGA pockets on the norovirus capsid using X-ray crystallography. Our data revealed that 2′FL and 3FL structurally mimic HBGAs. These results suggest that 2′FL and 3FL might act as naturally occurring decoys in humans.
TEXT
Mothers' milk has long been seen as a great source of infant nutrition and protection against a large number of pathogens. Human milk oligosaccharides (HMOs), the third-most-abundant (10 to 15 g/liter) components of human milk, are thought to be in part accountable for these health benefits (1). HMOs are unconjugated complex glycans, and more than 200 isomers are known. HMOs consist of combinations of different monosaccharide building blocks, including fucose, glucose, galactose, N-acetylglucosamine, and the sialic acid derivative N-acetyl-neuraminic acid. HMOs have been demonstrated to protect against human noroviruses, rotavirus, and certain bacteria (reviewed in reference 2).
Human noroviruses are also known to interact with histo-blood group antigens (HBGAs), and the interaction is thought to be important for infection (3–6). HBGAs can be found as soluble antigens in saliva and are expressed on epithelial cells. HBGAs consist of monosaccharide building blocks similar to those of HMOs, and at least nine different HBGA types have been found to interact with human norovirus (7–12). HMOs are thought to act as a “receptor decoy” for certain pathogens, since HMOs and HBGAs mimic each other structurally. However, little is known about how HMOs block norovirus infections. One study found that human milk was able to block genogroup I genotype 1 (GI.1) and GII.4 norovirus strains from binding to saliva samples (13). A follow-up study suggested that certain HMOs might compete with the HBGA binding sites on the GI.1 and GII.4 norovirus capsid (14). Despite the fact that human noroviruses are the dominant cause of acute gastroenteritis, there are still no suitable antivirals or vaccines commercially available.
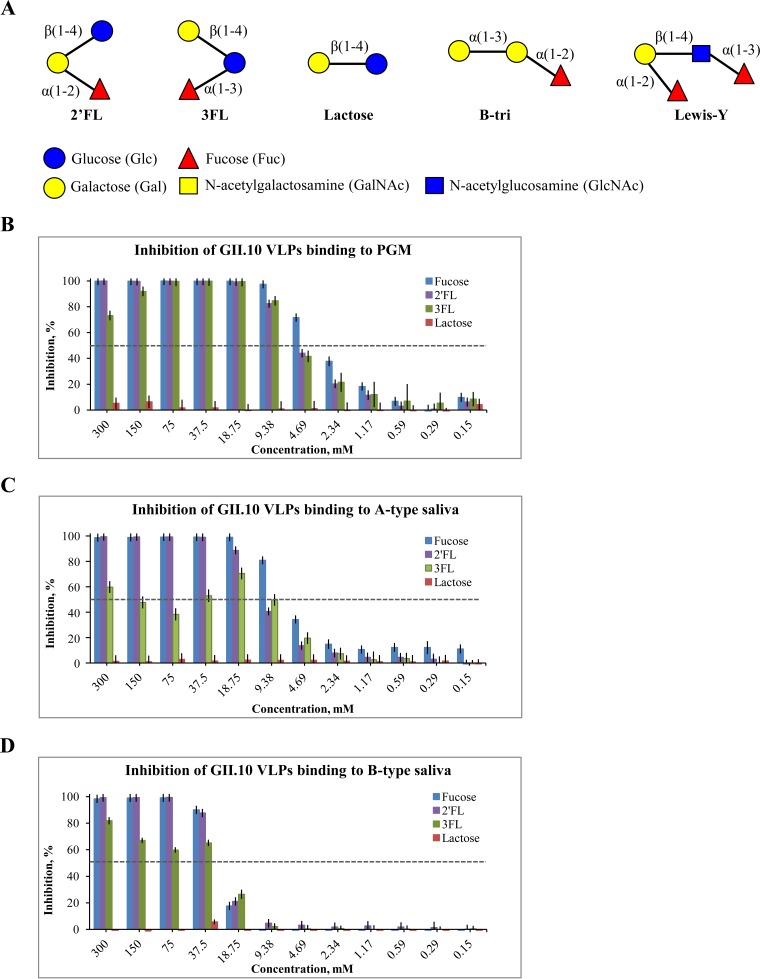
In this study, we analyzed the ability of two HMOs, i.e., 2′-fucosyllactose (2′FL) and 3-fucosyllactose (3FL), to block GII.10 norovirus virus-like particles (VLPs) from binding to HBGAs (Fig. 1A). A slightly modified blocking enzyme-linked immunosorbent assay (ELISA) was developed using both porcine gastric mucin type III (PGM) and human saliva (A and B types) (3, 15). The PGM sample was confirmed to contain a mixture of A and H types using specific anti-HBGA monoclonal antibodies (data not shown).
FIG 1.
HMOs and blocking of binding of norovirus VLPs to HBGAs. (A) Schematic representation of HMOs and HBGAs. The 2′FL is an α-l-fucose-(1-2)-β-d-galactose-(1-4)-α-d-glucose; 3FL is an α-l-fucose-(1-3)-[β-d-galactose-(1-4)]-β-d-glucose; lactose is a β-d-galactose-(1-4)-α-d-glucose; B-trisaccharide (B-tri) is an α-l-fucose-(1-2)-α-d-galactose-(3-)-N-acetyl-α-d-galactosamine; and Lewis Y-tetrasaccharide (Lewis-Y) is an α-l-fucose-(1-2)-β-d-galactose-(1-4)-N-acetyl-β-d-glucosamine-(3-1)-α-l-fucose. (B) Inhibition of binding of GII.10 VLPs to PGM. (C) Inhibition of binding of GII.10 VLPs to A-type saliva. (D) Inhibition of binding of GII.10 VLPs to B-type saliva. All experiments were performed in triplicate (standard deviations are shown). The half-maximal inhibitory concentration (IC50) cutoff is shown as a dashed line.
The GII.10 VLPs were expressed and purified as previously described (16). The untreated VLPs were first examined for binding to PGM and saliva samples using a direct ELISA. Maxisorp 96-well plates were coated with 100 μl per well of 10 μg/ml PGM for 4 h at room temperature. The saliva samples were processed in a similar way, except that the saliva was heated at 95°C for 10 min and briefly centrifuged and then the supernatant was diluted 1:100 in phosphate-buffered saline (PBS) and coated on plates overnight at 4°C. Plates were washed three times with PBS containing 0.1% Tween 20 (PBS-T) and blocked with 5% skim milk (SM)–PBS overnight at 4°C. Serial dilutions of the VLPs were added to the wells and incubated for 2 h at room temperature (RT). The plates were washed, and 100 μl per well of a GII.10 rabbit polyclonal antibody (1:20,000) diluted in PBS-T–SM was added. Plates were incubated for 1.5 h at 18°C and washed as before. Then, 100 μl per well of horseradish peroxidase (HRP)-conjugated goat α-rabbit antibody (1:40,000) diluted in PBS-T–SM was added and the mixture was incubated overnight at 4°C. Plates were developed with 100 μl per well of o-phenylenediamine and H2O2 in the dark for 30 min at RT. The reaction was stopped with 50 μl per well of 3 N hydrochloric acid and the absorbance measured at an optical density of 490 nm (OD490). We found that 2.5 μg/ml VLPs produced OD490 values of 2.9, 2.2, and 2.9 for PGM, A-type saliva, and B-type saliva, respectively (data not shown). Therefore, we proceeded to test the HMO inhibition using 2.5 μg/ml VLPs with PGM and saliva samples.
The inhibition study was performed with an identical ELISA format, except that the VLPs were first mixed with serial diluted HMOs. The 2′FL and 3FL oligosaccharides were synthesized by whole-cell biocatalysis (17) and diluted to 1 M in distilled water. 2′FL, 3FL, lactose (negative control), and fucose (positive control) were each serially diluted in PBS containing 2.5 μg/ml VLPs and incubated for 2 h at RT. The plates were prepared as described above, and then 100 μl per well of the treated-VLP dilution series was added in triplicate wells. The OD490 value of the untreated VLPs was set as the reference value corresponding to 100% binding, and the percentage of inhibition was calculated as follows: [1 − (treated VLP mean OD490/mean reference OD490)] × 100.
We found that 2′FL and 3FL were able to block GII.10 VLPs from binding to PGM, A-type saliva, and B-type saliva in a mostly dose-dependent manner. The half-maximal inhibitory concentration (IC50) in these assays was determined using Prism software (version 6.0). The IC50s in the PGM assay for 2′FL and 3FL were 5.5 mM and 5.6 mM, respectively (Fig. 1B). For A-type saliva assay, the IC50s for 2′FL and 3FL were 11.2 mM and 9.7 mM, respectively (Fig. 1C). However, the inhibition by 3FL appeared to have a biphasic pattern in the A-type saliva; i.e., the inhibition decreased slightly at higher concentrations. This behavior could be due to the multiple fucose binding sites on the P dimer (18), which was shown to have multistep cooperative binding (19). For B-type saliva assays, the IC50s for 2′FL and 3FL were 26.9 mM and 30.2 mM, respectively (Fig. 1D). The IC50s of fucose were 3.2 mM, 6.3 mM, and 27.1 mM in PGM, A-type saliva, and B-type saliva, respectively. Lactose showed no inhibition at any concentration, indicating a lack of binding to the VLPs. Our previous results showed that fucose and HBGA H2 trisaccharide had relatively weak (∼0.5 mM) affinities to the GII.10 P domain (20). Therefore, considering that 2′FL and 3FL might also have similar affinities and since mothers' milk contains ∼1 to 5 mM 2′FL/3FL, these HMOs might compete against HBGA binding. Importantly, one study found that human milk containing high levels of 2-linked fucosyl oligosaccharides reduced the incidence of calicivirus diarrhea in infants (21).
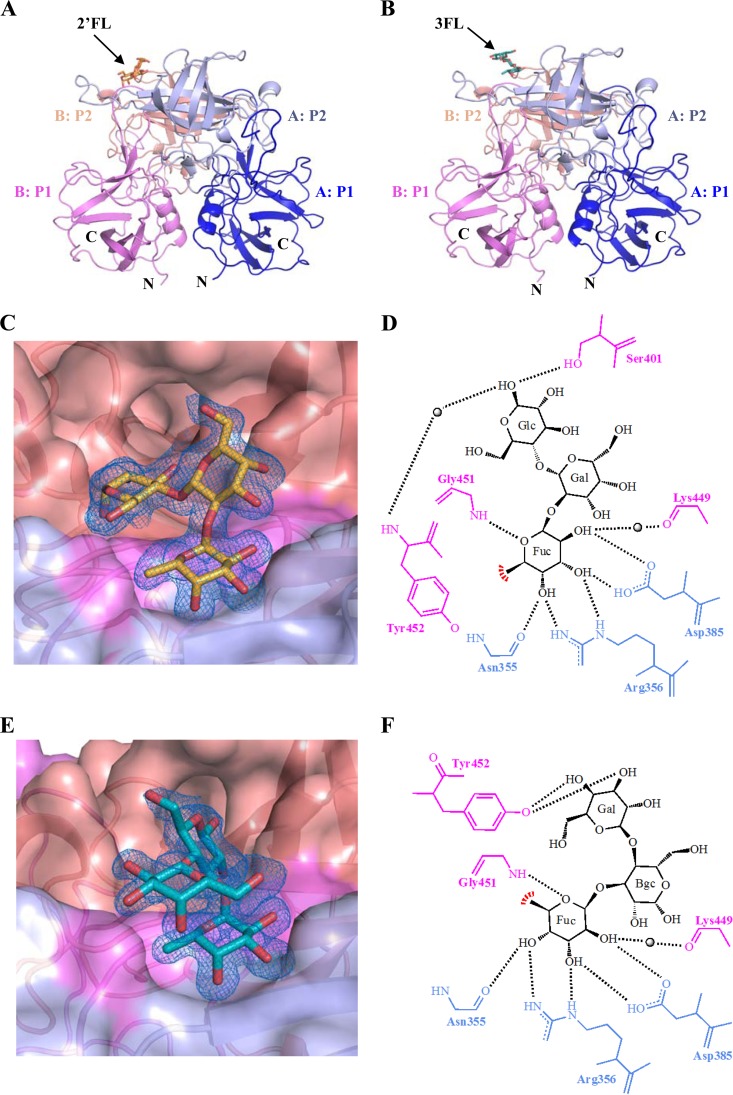
In order to better understand the 2′FL and 3FL binding interactions from a structural perspective, we solved the X-ray crystal structures of the GII.10 P domains in complex with 2′FL and 3FL. The GII.10 P domain was prepared as described earlier (22). The P domain and HMOs were cocrystalized in a 1:1:1 mixture of protein sample (∼2 mg/ml), mother liquor (0.2 M sodium nitrate, 0.1 M bis-tris propane [pH 7.5], 20% [wt/vol] polyethylene glycol [PEG] 3350), and a 30 to 60 molar excess of HMOs for 2 to 6 days at 18°C. Prior to flash freezing, crystals were transferred to a cryoprotectant containing mother liquor, a 30 molar excess of HMO, and 30% ethylene glycol. X-ray diffraction data were processed as previously described (16). Briefly, the structures were solved using molecular replacement with subsequent refinement with PHENIX (23). The HMOs were added to the models at the final stages of structural refinement in order to reduce bias during refinement. Structures were validated with Molprobity (24) and Procheck (25). HMO interactions were analyzed using Accelrys Discovery Studio (Version 4.1). Atomic coordinates and structure factors were deposited in the Protein Data Bank.
Data collection and refinement statistics for P domain HMO complex structures are provided in Table 1. In both structures, only one HMO bound per dimer, which was similar to results of our previous GII.10 HBGA study (22) (Fig. 2). The electron density was well defined for all saccharide units, indicating that the HMOs were firmly held by the P domain. The 2′FL was held in place by a network of hydrophilic and hydrophobic interactions at the dimeric interface (Fig. 2C and D). The fucose of 2′FL was held by six direct hydrogen bonds, two from the side chain of Asp385, two from the side chain of Arg356, one from the main chain of Asn355, and one from the main chain of Gly451. A hydrophobic interaction was provided from Tyr452. The central galactose of 2′FL was not held with any residues, while the terminal glucose was held with one hydrogen bond from the side chain of Ser401 and a water-mediated interaction from the main chain of Tyr452.
TABLE 1.
Data collection and refinement statistics of GII.10 P domain in complex with 2′FL and 3FLa
| Characteristic | GII.10 P domain result(s)b |
|
|---|---|---|
| 2′FL 5HZB | 3FL 5HZA | |
| Data collection | ||
| Space group | P1211 | P1211 |
| Cell dimensions | ||
| a, b, c (Å) | 65.68, 78.90, 71.07 | 65.71, 78.17, 71.84 |
| α, β, γ (°) | 90, 102.3, 90 | 90, 102.3, 90 |
| Resolution range (Å) | 42.81–1.55 (1.61–1.55) | 49.61–1.35 (1.39–1.35) |
| Rmerge | 3.38 (30.71) | 4.99 (37.67) |
| I/σI | 15.47 (2.39) | 9.84 (2.02) |
| Completeness (%) | 92.30 (93.18) | 91.72 (88.38) |
| Redundancy | 2.2 (2.1) | 2.3 (2.3) |
| CC1/2 | 0.99 (0.83) | 0.99 (0.74) |
| Refinement | ||
| Resolution range (Å) | 42.81–1.55 | 49.61–1.35 |
| No. of reflections | 94,095 | 142,739 |
| Rwork/Rfree | 16.36/19.53 | 16.08/17.89 |
| No. of atoms | 5,577 | 5,751 |
| Protein | 4,809 | 4,871 |
| Ligand/ion | 49 | 81 |
| Water | 719 | 799 |
| Average B factors (Å2) | ||
| Protein | 21.00 | 16.70 |
| Ligand/ion | 29.40 | 21.00 |
| Water | 32.70 | 29.10 |
| RMSD | ||
| Bond length (Å) | 0.007 | 0.006 |
| Bond angle (°) | 1.09 | 1.06 |
The data set was collected from a single crystal. RMSD, root mean square deviation.
Values in parentheses are for the highest-resolution shell.
FIG 2.
GII.10 P dimer binding interaction with HMOs. (A) The X-ray crystal structure of the GII.10 P domain dimer and 2′FL complex determined to 1.55 Å resolution and colored according to monomers (chain A and chain B) and P1 and P2 subdomains, i.e., chain A P1 (blue), chain A P2 (light blue), chain B P1 (violet), and chain B P2 (salmon). (B) The X-ray crystal structure of the GII.10 P domain dimer and 3FL complex determined to 1.35-Å resolution and colored as described for panel A. (C) A closeup surface and ribbon representation of the GII.10 and 2′FL (orange sticks) complex structure, showing a simulated annealing difference omit map (blue mesh) of 2′FL contoured at 2.0 σ. (D) The GII.10 P domain binding interaction with 2′FL showing α-fucose (Fuc), α-galactose (Gal), and β-glucose (Glc). The black lines represent the hydrogen bonds, the red line represents the hydrophobic interaction with the aromatic ring of Tyr452, and the black spheres represent water. Hydrogen bond distances were less than 3.3 Å. (E) A closeup surface and ribbon representation of the GII.10 and 3FL (deep teal sticks) complex structure, showing a simulated annealing difference omit map of 3FL contoured at 2.0 σ. (F) GII.10 P domain binding interaction with 3FL showing α-fucose (Fuc), β-galactose (Bgc), and β-glucose (Glc).
The 3FL was also held at the dimeric interface (Fig. 2E and F). The fucose of 3FL was held by the same set of residues as 2′FL (Asp385, Arg356, Asn355, Gly451, and Tyr452). One additional water-mediated interaction with the fucose was provided from the main chain of Lys449. The central glucose of 3FL was not held with any residues, while the terminal galactose was held with two hydrogen bonds from the side chain of Tyr452.
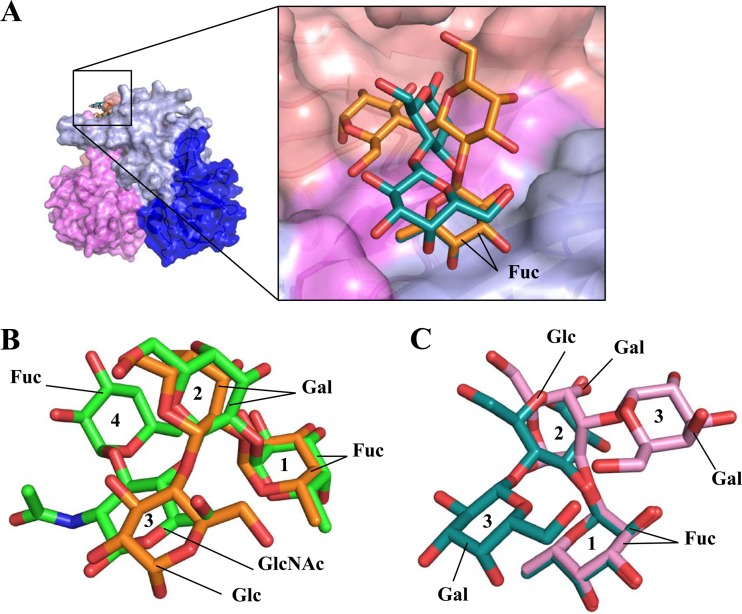
A number of similarities with and differences from the HMO and HBGA complex structures were observed. The five residues that held the fucose of HMOs also interacted with the fucose of HBGAs, and the fucoses of HMOs and HBGAs were identically positioned on the P domain (Fig. 3) (22). The central saccharides of both HMOs and HBGAs were poorly held by the P domain, while the terminal saccharides interacted with various residues (22). Interestingly, the 2′FL essentially mimicked the first three saccharides of the Lewis-Y tetrasaccharide, where fucoses were identically positioned, the second saccharides kinked up, and the third saccharides were lowered on the protein (Fig. 2C and 3B). Moreover, the third saccharides in both 2′FL and Lewis-Y made direct hydrogen bonds with the side chain of Ser401 (22). The 3FL was positioned on the P domain in a manner quite unlike that seen with other HBGAs (Fig. 3C). The glucose twists upward, and the terminal galactose turns and partially overlaps the fucose (Fig. 2E). Altogether, these data showed that the P domain is capable of binding HMOs and HBGAs in copious orientations, despite the fact that the fucoses were always held in identical positions.
FIG 3.
HMO and HBGA binding to GII.10 norovirus. (A) Surface representation of the GII.10 P domain in complex with 2′FL (orange sticks) and 3FL (deep-teal sticks). The fucose units of the HMO were positioned similarly on the P domain, whereas the other saccharide units were differently oriented. (B) Superposition of 2′FL and Lewis-Y tetrasaccharide (green sticks) showed that the 2′FL saccharide units essentially mimicked the orientations of first three saccharides of Lewis-Y. The saccharides are numbered (1, 2, 3, and 4) for viewing. (C) Superposition of 3FL and B-trisaccharide (pink sticks) indicated that the fucose units were similarly positioned, whereas the other saccharide units were orientated differently.
Our results provide the structural basis for understanding the HMO binding to the human norovirus GII.10 P domain. We found that both 2′FL and 3FL were capable of blocking binding of GII.10 norovirus VLPs to PGM and saliva samples. Another study also showed that GII.4 VLPs bound HMOs, which indicates a possible common binding event among genetically distinct GII noroviruses (14). Indeed, our previous studies showed that the GII.4 and GII.10 HBGA binding pockets were structurally comparable, having a common set of HBGA binding residues (22, 26). Superposition of 2′FL and 3FL onto GII.4 P domain structures indicated that the GII.4 strains might also bind HMOs at an equivalent site on the P domain (data not shown).
Currently, there is no treatment or vaccine available for human norovirus infections, which cause a massive burden of disease worldwide. The data from this study indicated that 2′FL and 3FL might function as norovirus antivirals by blocking the HBGA binding site. Importantly, 2′FL has already been shown to be a safe food supplement for infant formula (27). Interestingly, fucose alone was also capable of blocking GII.10 VLP binding to PGM and saliva samples. We previously showed that fucose could bind to four sites on the P dimer in a dose-dependent manner (18). Considering that mothers' milk contains 20 to 30 mg/liter of fucose, fucose alone may also work as a norovirus antiviral. Further clinical studies performed with 2′FL, 3FL, and fucose, but also with more-complex HMO structures, are highly anticipated.
Protein structure accession numbers.
Atomic coordinates and structure factors determined in this work were deposited in the Protein Data Bank (GII.10 P domain-2′FL, PDB code 5HZB; GII.10 P domain-3FL, PDB code 5HZA).
ACKNOWLEDGMENTS
We are grateful to Christoph Schall and Thilo Stehle for the initial use of their X-ray home source. We acknowledge the European Synchrotron Radiation Facility (ID23-1, ID30-A1, and ID30-B) for provision of synchrotron radiation facilities.
Jennewein Biotechnologie GmbH holds patents for the synthesis of 2′FL and 3FL (no. WO2010070104 and WO2010142305).
Funding Statement
Funding for this study was also provided to Anna Koromyslova and Grant Hansman by the CHS Foundation, the Helmholtz-Chinese Academy of Sciences (HCJRG-202).
REFERENCES
- 1.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 2.Etzold S, Bode L. 2014. Glycan-dependent viral infection in infants and the role of human milk oligosaccharides. Curr Opin Virol 7:101–107. doi: 10.1016/j.coviro.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Huang P, Farkas T, Marionneau S, Zhong W, Ruvoen-Clouet N, Morrow AL, Altaye M, Pickering LK, Newburg DS, LePendu J, Jiang X. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J Infect Dis 188:19–31. doi: 10.1086/375742. [DOI] [PubMed] [Google Scholar]
- 4.Huang P, Farkas T, Zhong W, Tan M, Thornton S, Morrow AL, Jiang X. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol 79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrington PR, Lindesmith L, Yount B, Moe CL, Baric RS. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J Virol 76:12335–12343. doi: 10.1128/JVI.76.23.12335-12343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rockx BH, Vennema H, Hoebe CJ, Duizer E, Koopmans MP. 2005. Association of histo-blood group antigens and susceptibility to norovirus infections. J Infect Dis 191:749–754. doi: 10.1086/427779. [DOI] [PubMed] [Google Scholar]
- 7.Tan M, Jiang X. 2005. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol 13:285–293. doi: 10.1016/j.tim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Tan M, Jiang X. 2011. Norovirus-host interaction: multi-selections by human histo-blood group antigens. Trends Microbiol 19:382–388. doi: 10.1016/j.tim.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol 81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bu W, Mamedova A, Tan M, Xia M, Jiang X, Hegde RS. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J Virol 82:5340–5347. doi: 10.1128/JVI.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubota T, Kumagai A, Ito H, Furukawa S, Someya Y, Takeda N, Ishii K, Wakita T, Narimatsu H, Shirato H. 2012. Structural basis for the recognition of Lewis antigens by genogroup I norovirus. J Virol 86:11138–11150. doi: 10.1128/JVI.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. 2011. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J Virol 85:8635–8645. doi: 10.1128/JVI.00848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X, Huang P, Zhong W, Tan M, Farkas T, Morrow AL, Newburg DS, Ruiz-Palacios GM, Pickering LK. 2004. Human milk contains elements that block binding of noroviruses to human histo-blood group antigens in saliva. J Infect Dis 190:1850–1859. doi: 10.1086/425159. [DOI] [PubMed] [Google Scholar]
- 14.Shang J, Piskarev VE, Xia M, Huang P, Jiang X, Likhosherstov LM, Novikova OS, Newburg DS, Ratner DM. 10 September 2013. Identifying human milk glycans that inhibit norovirus binding using surface plasmon resonance. Glycobiology doi: 10.1093/glycob/cwt077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindesmith LC, Debbink K, Swanstrom J, Vinje J, Costantini V, Baric RS, Donaldson EF. 2012. Monoclonal antibody-based antigenic mapping of norovirus GII.4-2002. J Virol 86:873–883. doi: 10.1128/JVI.06200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koromyslova AD, Hansman GS. 2015. Nanobody binding to a conserved epitope promotes norovirus particle disassembly. J Virol 89:2718–2730. doi: 10.1128/JVI.03176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weichert S, Jennewein S, Hufner E, Weiss C, Borkowski J, Putze J, Schroten H. 2013. Bioengineered 2′-fucosyllactose and 3-fucosyllactose inhibit the adhesion of Pseudomonas aeruginosa and enteric pathogens to human intestinal and respiratory cell lines. Nutr Res 33:831–838. doi: 10.1016/j.nutres.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Koromyslova AD, Leuthold MM, Bowler MW, Hansman GS. 2015. The sweet quartet: binding of fucose to the norovirus capsid. Virology 483:203–208. doi: 10.1016/j.virol.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Mallagaray A, Lockhauserbaumer J, Hansman G, Uetrecht C, Peters T. 2015. Attachment of norovirus to histo blood group antigens: a cooperative multistep process. Angew Chem Int Ed Engl 54:12014–12019. doi: 10.1002/anie.201505672. [DOI] [PubMed] [Google Scholar]
- 20.Hansman GS, Shahzad-Ul-Hussan S, McLellan JS, Chuang GY, Georgiev I, Shimoike T, Katayama K, Bewley CA, Kwong PD. 2012. Structural basis for norovirus inhibition and fucose mimicry by citrate. J Virol 86:284–292. doi: 10.1128/JVI.05909-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, Farkas T, Chaturvedi P, Pickering LK, Newburg DS. 2004. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr 145:297–303. doi: 10.1016/j.jpeds.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 22.Hansman GS, Biertumpfel C, Georgiev I, McLellan JS, Chen L, Zhou T, Katayama K, Kwong PD. 2011. Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J Virol 85:6687–6701. doi: 10.1128/JVI.00246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. 1992. Stereochemical quality of protein structure coordinates. Proteins 12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 26.Singh BK, Leuthold MM, Hansman GS. 2015. Human noroviruses' fondness for histo-blood group antigens. J Virol 89:2024–2040. doi: 10.1128/JVI.02968-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coulet M, Phothirath P, Allais L, Schilter B. 2014. Pre-clinical safety evaluation of the synthetic human milk, nature-identical, oligosaccharide 2′-O-fucosyllactose (2′FL). Regul Toxicol Pharmacol 68:59–69. doi: 10.1016/j.yrtph.2013.11.005. [DOI] [PubMed] [Google Scholar]