ABSTRACT
Three waves of human infection with H7N9 influenza viruses have concluded to date, but only viruses within the first wave (isolated between March and September 2013) have been extensively studied in mammalian models. While second- and third-wave viruses remain closely linked phylogenetically and antigenically, even subtle molecular changes can impart critical shifts in mammalian virulence. To determine if H7N9 viruses isolated from humans during 2013 to 2015 have maintained the phenotype first identified among 2013 isolates, we assessed the ability of first-, second-, and third-wave H7N9 viruses isolated from humans to cause disease in mice and ferrets and to transmit among ferrets. Similar to first-wave viruses, H7N9 viruses from 2013 to 2015 were highly infectious in mice, with lethality comparable to that of the well-studied A/Anhui/1/2013 virus. Second- and third-wave viruses caused moderate disease in ferrets, transmitted efficiently to cohoused, naive contact animals, and demonstrated limited transmissibility by respiratory droplets. All H7N9 viruses replicated efficiently in human bronchial epithelial cells, with subtle changes in pH fusion threshold identified between H7N9 viruses examined. Our results indicate that despite increased genetic diversity and geographical distribution since their initial detection in 2013, H7N9 viruses have maintained a pathogenic phenotype in mammals and continue to represent an immediate threat to public health.
IMPORTANCE H7N9 influenza viruses, first isolated in 2013, continue to cause human infection and represent an ongoing public health threat. Now entering the fourth wave of human infection, H7N9 viruses continue to exhibit genetic diversity in avian hosts, necessitating continuous efforts to monitor their pandemic potential. However, viruses isolated post-2013 have not been extensively studied, limiting our understanding of potential changes in virus-host adaptation. In order to ensure that current research with first-wave H7N9 viruses still pertains to more recently isolated strains, we compared the relative virulence and transmissibility of H7N9 viruses isolated during the second and third waves, through 2015, in the mouse and ferret models. Our finding that second- and third-wave viruses generally exhibit disease in mammals comparable to that of first-wave viruses strengthens our ability to extrapolate research from the 2013 viruses to current public health efforts. These data further contribute to our understanding of molecular determinants of pathogenicity, transmissibility, and tropism.
INTRODUCTION
Since their initial detection in humans in the spring of 2013, low-pathogenicity avian influenza (LPAI) H7N9 viruses have represented a continued public health threat in Southeast Asia, with over 700 laboratory-confirmed cases of human infection to date (1). Human cases have been limited to China or among travelers who visited China before returning to their home country (2). Asymptomatic H7N9 virus-infected chickens appear to be central to the persistence and expansion of this outbreak (3); accordingly, poultry contact and visitation of live poultry markets has been linked with H7N9 virus infection (4, 5), and the closure of live poultry markets has been associated with a decline of new human infections in 2013 and 2014 (6, 7). Limited family clusters of H7N9 virus infection have been reported (8, 9), but human-to-human transmission has remained a rarely documented and unsustainable event (10), while human infections continue to occur following exposure to H7N9 viruses circulating in avian reservoirs (11).
To date, three completed epidemiologic waves of human cases with H7N9 viruses have taken place in Southeast Asia (9). The first wave of human infection (30 March to 30 September 2013) resulted in over 130 confirmed cases with a >30% fatality rate, with human infection detected in mainland China and Thailand. The second wave (1 October 2013 to 30 September 2014) yielded over 260 additional cases, with the majority of cases detected in Guangdong and Zhejiang provinces of China (9) and additional cases detected in Hong Kong, Thailand, and Malaysia. The third wave (1 October 2014 to 30 September 2015) resulted in over 220 reported cases in China, Hong Kong, and Canada. Disease symptoms, severity, and mortality generally have held constant between all three waves, and no pronounced antigenic drift has been detected from 2013 to 2015 (2). However, increased case-fatality rates reported in one province in China between cases from the first and second waves suggested that despite high homology between viruses isolated from different waves, it was possible that H7N9 viruses developed an enhanced ability to cause disease in humans (12). Previous studies with H7 subtype avian influenza viruses have demonstrated that even subtle changes can indicate enhanced adaptation to mammals, including increased transmissibility among mammalian hosts (6, 13–15), underscoring the need for ongoing surveillance and characterization of emerging H7N9 viruses as they continue to cause human infections.
The majority of published studies examining H7N9 viruses have focused on first-wave viruses only. However, as H7N9 viruses continue to cause human infection, the increased temporal, geographic, and genetic distribution of these viruses necessitates continued investigation (11). Notably, second- and third-wave viruses have not been extensively examined in vitro or in vivo, and the contributing role of precursor viruses to H7N9 pathogenicity is still understudied (16). Here, we characterized the mammalian pathogenicity and transmissibility of first-, second-, and third-wave H7N9 viruses. We found that H7N9 viruses possess comparable virulence in the mouse and ferret over three waves of human infection. Similar to viruses from the first wave, H7N9 viruses isolated from the second and third waves maintain transmissibility in the ferret model in the presence of direct contact and exhibit limited transmissibility by respiratory droplets. Generally comparable replication kinetics in human respiratory tract cells and the pH of fusion between viruses from all waves indicates that H7N9 viruses continue to remain a pandemic threat.
MATERIALS AND METHODS
Viruses.
H7N9 influenza A viruses used in this study are shown in Table 1. Virus stocks were propagated in the allantoic cavity of 10-day-old embryonated hens' eggs at 37°C for 42 h. Allantoic fluid from multiple eggs was pooled, clarified by centrifugation, and frozen in aliquots until use at −80°C. Virus stocks were tested by standard plaque assay in Madin-Darby canine kidney (MDCK) cells (ATCC, Manassas, VA) for determination of PFU titer (17). All experiments were conducted under biosafety level 3 containment, including enhancements as required by the U.S. Department of Agriculture and the National Select Agent Program (18).
TABLE 1.
LPAI H7N9 viruses used in this study
| Virus name | Abbreviation | Wave | Patient outcome | Reference or source |
|---|---|---|---|---|
| A/shoveler/Egypt/00215-NAMRU3/2007 | Shv/Egypt/07 | NAa | NA | 61 |
| A/Anhui/1/2013 | Anhui/1/13 | First | Fatal | 45 |
| A/Taiwan/1/2013 | Taiwan/1/13 | First | Survived | 62 |
| A/Hong Kong/5942/2013 | HK/5942/13 | Second | Survived | This study |
| A/Taiwan/1/2014 | Taiwan/1/14 | Second | Survived | 38 |
| A/Taiwan/2/2014 | Taiwan/2/14 | Second | Survived | 38 |
| A/Hong Kong/734/2014 | HK/734/14 | Second | Fatal | This study |
| A/Hong Kong/56/2015 | HK/56/15 | Third | Survived | This study |
| A/British Columbia/1/2015 | BC/1/15 | Third | Survived | 63 |
NA, not applicable.
Ethics statement.
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the Centers for Disease Control and Prevention and were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited facility.
Mouse experiments.
Female BALB/c mice (Jackson Laboratories), 6 to 8 weeks of age, were anesthetized with 2,2,2-tribromoethanol in tert-amyl alcohol (Avertin; Sigma-Aldrich, St. Louis, MO). Mice were inoculated with 106 PFU of virus by either the intranasal (i.n.) route in a 50-μl volume diluted in phosphate-buffered saline (PBS) or the ocular (i.o.) route, with 5 μl of virus diluted in PBS dropped onto the corneal surface of the right eye and massaged in by using the eyelids following corneal scarification as described previously (19). The 50% mouse infectious dose (MID50) and 50% lethal dose (LD50) were determined by inoculating mice i.n. with 10-fold serial dilutions of virus. Five mice per group were monitored daily for 14 days for morbidity and to obtain the LD50, and 3 mice were euthanized day 3 postinoculation (p.i.) for the collection of lungs for titration to obtain the MID50 for each virus. Any mouse that lost >25% of preinfection body weight was euthanized. The replication and systemic spread of each virus was determined by harvesting the eyes, nose, lung, and brain of mice (n = 3) on days 3 and 6 p.i., with tissues titrated by standard plaque assay (limit of detection, 10 PFU).
Ferret experiments.
Male Fitch ferrets (Triple F Farms), 6 to 8 months of age and serologically negative by standard hemagglutination inhibition for currently circulating influenza viruses, were used in this study. Ferrets were housed in a Duo-Flo Bioclean mobile environmental enclosure (Lab Products, Seaford, DE) for the duration of each experiment. Ferrets were inoculated i.n. with 106 PFU of each indicated virus in a 1-ml volume diluted in PBS and observed daily for clinical signs and symptoms of infection, with nasal washes (NW), conjunctival washes (CW), and rectal swabs (RS) collected on alternate days p.i. to measure virus shedding as previously described (20). Three additional ferrets inoculated with 106 PFU of each virus were euthanized day 3 p.i. for the assessment of virus replication and systemic spread as previously described (21). All samples were titrated by standard plaque assay. For histopathological analyses, respiratory tissues from animals euthanized day 3 p.i. were fixed in 10% neutral buffered formalin and embedded in paraffin. Four-micrometer sections from formalin-fixed, paraffin-embedded specimens were stained with hematoxylin and eosin (H&E) for histopathologic evaluation.
Virus transmissibility was assessed by placing a naive ferret in the same cage as an inoculated ferret (to assess transmission in the presence of direct contact [DC]) or in an adjacent cage with modified side walls to allow air exchange in the absence of direct or indirect contact between animals (to assess transmission by respiratory droplets [RD]) as previously described (22). Serum was collected on days 17 to 23 p.i./postcontact (p.c.) to measure seroconversion to homologous virus.
Cell culture and viral replication.
The human bronchial epithelial cell line Calu-3 (ATCC) was grown on membrane inserts and cultured as previously described (17). Cells were grown to confluence on 6-well inserts for 1 week to achieve a transepithelial resistance of >1,000 Ω. To measure replication kinetics, virus was added apically in serum-free medium to cells at a multiplicity of infection (MOI) of 0.01 and incubated 1 h before washing and culturing for the duration of the experiment at 37°C or 33°C. Aliquots of culture supernatant at the indicated times p.i. were immediately frozen at −80°C until titration for the presence of infectious virus by standard plaque assay. The statistical significance of replication kinetics in Calu-3 cells was determined by 2-way analysis of variance (ANOVA) with a Bonferroni posttest.
pH threshold for influenza virus fusion activation.
The influenza virus-induced hemolysis assay was carried out as a surrogate assay to measure the pH threshold for H7 influenza virus fusion activation as described previously (23). Briefly, 50 μl of H7 viruses in triplicate containing 128 hemagglutinin (HA) units or PBS mock-treated control were incubated with 50 μl of 1% (vol/vol) turkey red blood cells (TRBC) in 96-well plates at 4°C for 1 h. The virus and TRBC mixture was pelleted and resuspended in 150 μl of citric fusion buffer (20 mM HEPES, 2 mM CaCl2, 150 mM NaCl, 20 mM citric acid monohydrate/sodium citrate tribasic dehydrate) at various pH values at 37°C for 1 h to induce hemolysis. The released hemoglobin resulting from virus fusion with TRBC was measured by the optical density (OD) at 405 nm after removing nonlysed TRBC pellets at 2,000 rpm for 5 min and subtracting the background OD value from the PBS mock control at each pH value. The maximal hemolysis at a given pH was normalized to 100% and the hemolysis at other pH values was expressed as a percentage of the maximal hemolysis; results were plotted as averages from three independent experiments.
RESULTS
H7N9 virus information and genetic diversity.
To study the mammalian pathogenicity and transmissibility of H7N9 viruses from 2013 to 2015, we first identified representative viruses associated with human infection from each of the first three waves (Table 1). All human isolates tested in this study were likely the result of independent avian exposures to the virus and originated from different geographical areas. All patients infected with the indicated viruses exhibited respiratory symptoms, ranging from mild (i.e., influenza-like illness) to severe (i.e., acute respiratory distress syndrome); two isolates were from fatal cases. A representative precursor avian H7N9 virus (shv/Egypt/07) was included in the study to better contextualize phenotypic changes associated with mammalian adaptation.
Since their first detection in 2013, the spread of H7N9 viruses throughout China has led to the establishment of multiple lineages and virus genotypes (3). The spread of H7N9 viruses throughout China and potential ongoing reassortment with circulating H9N2 viruses had led to the establishment of a diverse array of viral genotypes (24, 25). Sequence alignments of the viruses isolated from the second and third waves revealed several differences from the well-studied first-wave Anhui/1 virus (3, 24). While most of these differences were present only in selected isolates and were not consistent across viruses from each epidemic wave, selected mutations (most notably in the polymerase genes) did appear to become more common in the second and/or third waves of the epidemic. Nonetheless, the internal genes of all viruses isolated from humans described here maintained greater than 99% amino acid identity with Anhui/1/13 virus (data not shown). Table 2 highlights these residues as well as those in the hemagglutinin that are believed to influence antigenicity and receptor binding. As is typical of most H7N9 human isolates, seven of the eight human-derived H7N9 viruses included in this study possess a lysine at PB2 position 627. Taiwan/2/14 virus bears a glutamic acid at position 627 but features a 701 asparagine residue, which, like 627, is associated with mammalian adaptation in this subtype (26). Similarly, with the exception of one first-wave isolate, all viruses in this study maintain a leucine at position 226 (H3 numbering) of the hemagglutinin, which is also associated with mammalian host adaptation (27). Mutations in the HA and polymerase genes between viruses isolated from the first, second, and third waves prompted us to choose representative viruses from each wave for further characterization in in vitro and in vivo models.
TABLE 2.
Amino acid substitutions in the proteins of three waves of H7N9 viruses
| Virus | Wave | Substitution in: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HAa |
PB2 |
PA | ||||||||||
| 57 | 132 | 186 | 226 | 276 | 191 | 559 | 570 | 627 | 701 | 394 | ||
| Anhui/1/13 | First | R | T | V | L | N | K | N | M | K | D | N |
| Taiwan/1/13 | First | R | T | V | P | N | K | N | M | K | D | N |
| HK/5942/13 | Second | R | A | V | L | N | E | T | I | K | D | N |
| Taiwan/1/14 | Second | R | T | V | L | N | E | T | I | K | D | D |
| Taiwan/2/14 | Second | R | T | V | L | N | E | T | I | E | N | N |
| HK/734/14 | Second | K | A | V | L | N | E | T | I | K | D | D |
| HK/56/15 | Third | K | T | A | L | D | E | T | M | K | D | D |
| BC/1/15 | Third | R | T | V | L | N | E | T | M | K | D | D |
H3 numbering.
Pathogenesis of first-, second-, and third-wave H7N9 viruses in mice.
H7N9 viruses isolated from the beginning of the H7N9 outbreak in 2013 were found to cause moderate to severe disease in mice (28–32), but viruses isolated from 2014 to 2015 have not been examined as extensively. We studied the pathogenicity of selected first (Taiwan/1/13)-, second (HK/5942/13)-, and third (BC/1/15)-wave viruses in the BALB/c mouse as they compared to the well-characterized, first-wave Anhui/1 virus (Table 3). Regardless of the wave of isolation, all H7N9 viruses tested exhibited a high degree of infectivity and comparable virulence in this species, leading to severe (≥20%) weight loss and pronounced mortality. Viral titers on days 3 and 6 p.i. were high in both the lung (≥log10 5.5 PFU/ml) and nose (≥log10 2.2 PFU/ml), comparable to H5N1 viruses, which cause similar morbidity and mortality in mice (21). However, viral titers in the brain were not detected for any virus on day 6 p.i. (data not shown).
TABLE 3.
Mouse infectivity and tissue viral titers of H7N9 viruses
| Virus | % wt lossa (day p.i.) | Infectivityb |
Viral titer by day p.i.c |
||||
|---|---|---|---|---|---|---|---|
| MID50 | LD50 | 3 |
6 |
||||
| Lung | Nose | Lung | Nose | ||||
| Anhui/1d | 22.2 (7) | 0.25 | 3.4 | 7.2 ± 0.6 | 3.2 ± 0.4 | 5.5 ± 0.5 | 2.2 ± 0.7 |
| Taiwan/1/13 | 27.4 (5) | 0.25 | 3.5 | 7.4 ± 0.1 | 5.0 (1/3)e | 6.9 ± 0.6 | 2.4 ± 0.2 |
| HK/5942/13 | 19.6 (5) | 0.10 | 3.2 | 7.7 ± 0.1 | 4.2 ± 0.3 | 7.0 ± 0.3 | 3.1 ± 0.5 |
| BC/1/15 | 22.4 (5) | 0.25 | 2.8 | 7.3 ± 0.6 | 3.6 ± 0.2 | 7.3 ± 0.1f | 3.2 ± 0.2f |
Mean maximum percent weight loss (5 mice per group) following inoculation with 106 PFU.
Fifty percent mouse infectious dose (MID50) and 50% lethal dose (LD50) are expressed as the log10 PFU required to give one MID50 or one LD50, respectively.
Virus endpoint titers are expressed as the mean log10 PFU/ml ± SD from 3 mice per tissue. The limit of virus detection was 10 PFU.
Includes data published previously in reference 28.
Virus was detected in only 1 of 3 inoculated mice.
Due to mice not surviving to day 6 p.i. with BC/1/15 virus, titers reflect two mice inoculated with 106 PFU and one mouse inoculated with 105 PFU.
Unlike most H7 subtype viruses, H7N9 viruses have not been associated with ocular disease in humans. However, as the eye represents a secondary route of virus entry to mount a respiratory infection, we examined the ability of H7N9 viruses from three waves to infect mice following ocular inoculation (Table 4). As reported previously and confirmed here, mice inoculated i.o. with Anhui/1 virus possessed detectable virus in the nose, but it was detected only sporadically in the eye or lung day 3 or 6 p.i. (16, 28). However, Taiwan/1/13, HK/5942/13, and BC/1/15 viruses exhibited the capacity to replicate to detectable titers in all three tissues following i.o. inoculation. These findings indicate that while H7N9 viruses from three waves do not maintain the ocular tropism observed with H7 viruses, they nonetheless possess the ability to use the eye as a portal of entry in mammals.
TABLE 4.
Replication of H7N9 influenza viruses following ocular inoculation in mice
| Tissue | Day p.i. | Virus titera |
|||
|---|---|---|---|---|---|
| Anhui/1/13b | Taiwan/1/13 | HK/5942/13 | BC/1/15 | ||
| Eye | 3 | 1.5 (1/7) | 2.1 ± 0.1 | 2.9 ± 0.3 | 1.9 ± 0.5 (2/4) |
| Nose | 3 | 2.7 ± 0.9 (4/7) | 3.9 ± 0.8 | 2.3 ± 0.5 | 2.5 ± 0.4 (3/4) |
| Lung | 3 | 0/7 | 2.1 (1/3) | 1.4 ± 0.1 | 2.2 ± 0.7 (3/4) |
| Eye | 6 | 2.1 ± 1.2 (2/7) | 2.1 ± 0.5 | 2.2 ± 0.1 | 1.4 ± 0.2 (3/4) |
| Nose | 6 | 2.8 ± 0.8 (6/7) | 2.8 ± 0.6 | 2.7 ± 1.2 | 3.4 ± 0.2 |
| Lung | 6 | 1.6 (1/7) | 1.5 (2/3) | 5.1 ± 0.5 (2/3) | 2.0 ± 0.6 (3/4) |
Mean virus titers in mice inoculated with 106 PFU/5 μl of virus following corneal scarification. Virus titers are expressed as the mean log10 PFU/ml ± SD among mice with positive virus detection (data are representative of all mice examined unless denoted in parentheses, in which the number of mice with positive virus detection/total number of mice in the group is indicated). The limit of virus detection was 10 PFU.
Includes data published previously in reference 28.
Pathogenesis of first-, second-, and third-wave H7N9 viruses in ferrets.
The presence of amino acid changes among H7N9 viruses indicative of mammalian adaptation warrants the examination of recently isolated H7N9 viruses in multiple mammalian models (33). We first examined the precursor LPAI H7N9 virus shv/Egypt/07 in ferrets, which shares high surface glycoprotein amino acid identity with Anhui/1/13 virus and was previously shown to exhibit reduced virulence in the mouse model compared with human isolates from 2013 (28). Ferrets inoculated with this avian virus exhibited mild weight loss (<3%), and virus was detected only in respiratory tract tissues, with no extrapulmonary spread to ocular, intestinal, or brain tissues (Table 5), comparable to other LPAI viruses isolated from avian species in this model (30).
TABLE 5.
Replication of H7N9 influenza viruses in ferrets
| Characteristic | Value(s) for: |
||||
|---|---|---|---|---|---|
| Shv/Egypt/07 | Anhui/1/13f | Taiwan/1/13 | HK/5942/13 | BC/1/15 | |
| Clinicala | |||||
| Feverb | 1.6 | 1.6 | 1.9 | 1.8 | 1.8 |
| wt lossc | 2.2 | 8.8 | 4.4 | 15.1 | 11.3 |
| Virus sample source and titerd | |||||
| CWe | 0/3 | 6/11 | 1/3 | 3/3 | 6/6 |
| RSe | 0/3 | 7/11 | 1/3 | 0/3 | 1/6 |
| Conj | 0/3 | 1.8 (1/3) | 2.2 ± 0.4 (2/3) | 0/3 | 1.8 ± 0.4 |
| Int | 0/3 | 0/3 | 1.7 ± 0.5 (2/3) | 1.6 (1/3) | 2.4 (1/3) |
| Nas Tur | 5.8 ± 0.4 | 6.2 ± 0.2 | 5.6 ± 0.6 | 5.4 ± 0.4 | 6.1 ± 0.6 |
| Trachea | 3.5 ± 0.6 (2/3) | 5.5 ± 0.6 | 5.3 ± 0.8 | 3.3 (1/3) | 5.1 ± 0.4 |
| Lung | 3.5 ± 2.2 (2/3) | 5.6 ± 0.3 | 5.0 ± 0.9 | 5.0 ± 1.1 | 6.5 ± 0.1 |
| OB | 0/3 | 4.9 (1/3) | 0/3 | 4.0 (1/3) | 4.1 ± 2.1 (2/3) |
| Bn Ant | 0/3 | 2.7 (1/3) | 0/3 | 1.8 ± 0.7 | 3.8 ± 0.1 (2/3) |
Clinical signs, symptoms, and observations detected through 14 days p.i.
Mean maximum rise in body temperature in degrees centigrade (baseline body temperature, 37.7 to 39.3°C).
Percent mean maximum weight loss in ferrets.
Unless specified otherwise, virus titers are expressed as the mean log10 PFU/g or ml ± SD among ferrets with positive virus detection on day 3 p.i. (data are representative of 3 ferrets examined, unless denoted in parentheses). The limit of virus detection was 10 PFU. Conj, surrounding conjunctival tissue; Int, pooled (duodenum, jejunum, and ileum) intestinal tissue; Nas Tur, nasal turbinates; OB, olfactory bulb; Bn Ant, anterior brain.
Number of ferrets with detectable virus in conjunctival washes (CW) or rectal swabs (RS) collected on days 1, 3, and 5 p.i./total number of ferrets tested.
Includes data published previously in reference 28.
The mild to moderate disease in ferrets caused by infection with first-wave H7N9 viruses has been established (28, 30, 31, 34, 35). We next examined if H7N9 viruses isolated from the second and third waves exhibited virulence comparable to that of first-wave viruses in the ferret model. Infection with all H7N9 viruses examined caused a transient rise in body temperature in ferrets, ranging from 1.6 to 1.9°C above baseline (Table 5). Second- and third-wave viruses (HK/5942/13 and BC/1/15) induced somewhat greater weight loss in ferrets compared to first-wave H7N9 viruses (Anhui/1/13 and Taiwan/1/13). All H7N9 viruses isolated from humans replicated efficiently throughout the ferret respiratory tract, reaching mean peak titers ranging from log10 5.4 to 6.2 PFU/ml in nasal turbinates and log10 5.0 to 6.5 PFU/g in lung tissues. Virus was detected sporadically and at low titer (≤2.4 log10 PFU/ml or g) in ocular and intestinal samples. First-wave H7N9 viruses were detected in the olfactory bulb or brain of 1 out of 6 ferrets (Table 5) (28), while second- and third-wave viruses were detected in these tissues more frequently but at titers comparable to those of the first-wave viruses.
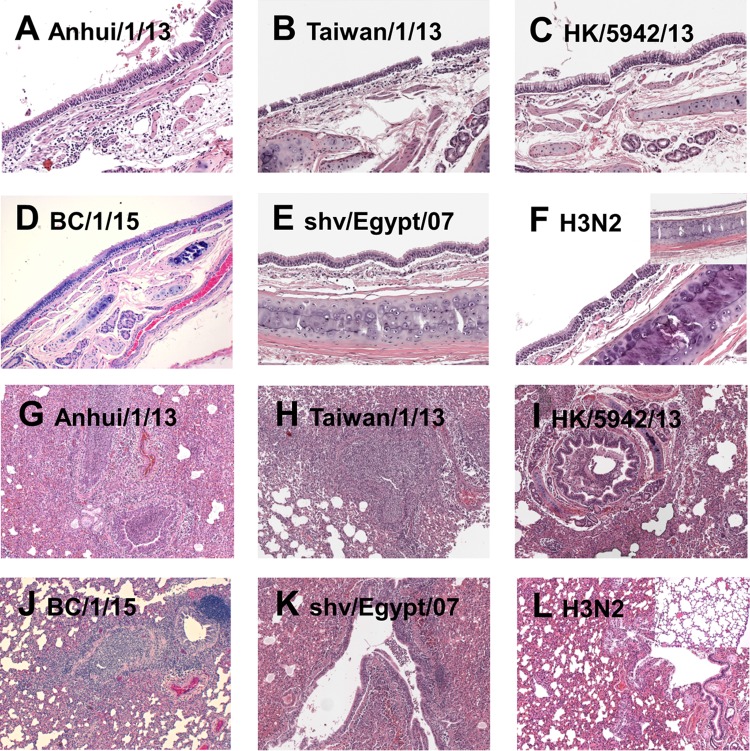
Histopathologic evaluation of tissues collected day 3 p.i. from H7N9 virus-infected ferrets revealed a spectrum of inflammation depending on the virus used to infect, although striking differences between H7N9 isolates were not observed (Fig. 1). Airway tissues showed minimal to moderate tracheobronchitis (Fig. 1A to E), while the lung tissues show mild to severe bronchopneumonia (Fig. 1G to K) in different groups of infected animals. The most severe inflammation was observed in Anhui/1 virus-infected ferrets (Fig. 1A and G), whereas infection with the seasonal H3N2 virus A/Texas/50/12 (28) resulted in reduced inflammation in these tissues (Fig. 1F and L). Together, these data suggest that second- and third-wave H7N9 viruses generally have maintained the moderate virulence observed with the first-wave H7N9 virus.
FIG 1.
Representative images of histopathologic evaluation of ferrets infected with H7N9 and seasonal influenza viruses. Histopathologic evaluation using H&E staining shows a spectrum of inflammation in the respiratory tissue following infection with Anhui/1/13 (first wave) (A and G), Taiwan/1/13 (first wave) (B and H), HK/5942/13 (second wave) (C and I), BC/1/15 (third wave) (D and J), shv/Egypt/07 (precursor H7N9) (E and K), and A/Texas/50/12 (seasonal H3N2) (F and L) viruses. The airway tissues show minimal to moderate tracheobronchitis (A to F), while the lung tissues show mild to severe bronchopneumonia (G to L) in different groups of infected animals. The normal histology of ferret airway and lung tissues are shown as inserts in panels F and L. Original magnifications: airway tissue, ×200; lung tissue, ×50.
Transmissibility of first-, second-, and third-wave H7N9 viruses in ferrets.
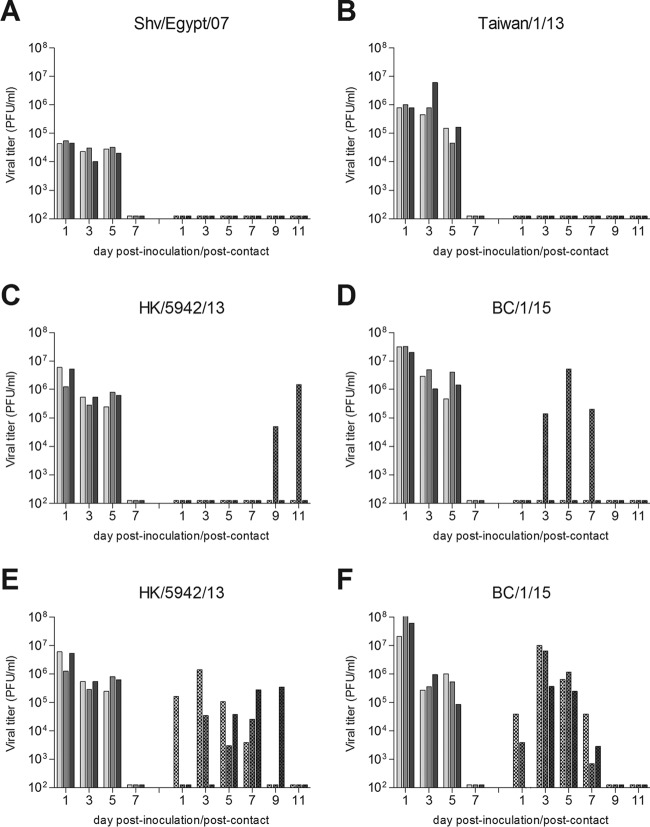
Avian H7N9 viruses isolated before or concurrent with the first detection of human cases have demonstrated no or only limited transmission in the ferret model by either respiratory droplets or in the presence of direct contact (30, 31, 36). In contrast, H7N9 viruses isolated in 2013, during the first wave of the outbreak, demonstrated the capacity for efficient transmission between cohoused ferrets and limited transmission in the more stringent respiratory droplet model (28, 30, 31, 34, 35, 37). To determine if H7N9 viruses isolated since the first wave have maintained this transmission profile, ferrets were inoculated with selected H7N9 viruses from each of the first three waves. Twenty-four hours later, a naive ferret was placed in an adjacent cage that permits air exchange between ferret pairs but without direct or indirect contact. Consistent with previous findings, an avian precursor virus, shv/Egypt/07, did not transmit via respiratory droplets: no virus was detected in nasal washes, and no seroconversion to homologous virus was noted (Fig. 2A). Taiwan/1/13 virus differs in the M gene from Anhui/1/13 virus and does not possess a lysine at position 226 of the HA gene (38). While Anhui/1/13 virus previously was shown to transmit between members of 2 out of 6 ferret pairs (28), Taiwan/1/13 virus did not transmit to any ferrets by this route (Fig. 2B), highlighting strain-specific differences in virus transmissibility between first-wave H7N9 viruses. The second- and third-wave H7N9 viruses (HK/5942/13 and BC/1/15) demonstrated comparable transmissibility with Anhui/1/13 virus, with virus detected in nasal wash specimens of 1/3 ferrets for both viruses and seroconversion of 2/3 ferrets for HK/5942/13 virus and 1/3 ferrets for BC/1/15 virus by the end of the experiment (Fig. 2C and D). The efficient transmission of first-wave H7N9 viruses among cohoused animals (8/8 ferrets [28]) was maintained with both of the second- and third-wave H7N9 viruses tested here (Fig. 2E and F). Therefore, we can conclude that no enhancement in transmission capability occurred among the second- and third-wave H7N9 viruses evaluated here.
FIG 2.
Transmissibility of H7N9 viruses in ferrets. Three ferrets were inoculated with 106 PFU of virus, and nasal washes were collected from each ferret on the indicated days p.i. (solid bars). A naive ferret was placed in a cage with perforated side walls adjacent to each inoculated ferret 24 h p.i. (A to D) or in the same cage as an inoculated ferret (E and F), and nasal washes were collected from each contact ferret on the indicated days p.c. (hatched bars) to assess virus transmission. Avian precursor (Shv/Egypt/07) (A), first-wave (Taiwan/1/13) (B), second-wave (HK/5942/13) (C and E), and third-wave (BC/1/15) (D and F) H7N9 influenza viruses were tested. The limit of virus detection was 100 PFU.
Replication of H7N9 viruses in human bronchial epithelial cells.
First-wave H7N9 viruses isolated from the first human cases in 2013 demonstrated the ability to replicate efficiently in the human bronchial epithelial cell line Calu-3 at 37°C but less efficiently at a lower temperature (33°C) that is representative of the upper airways (28). To determine if H7N9 viruses from the second and third waves have maintained this property, we evaluated the replication kinetics of H7N9 viruses from all three waves in Calu-3 cells at both temperatures (Fig. 3). Titers of all eight H7N9 viruses examined were significantly lower at 33°C than at 37°C at both 12 and 24 h p.i. (P < 0.001) before reaching comparable mean titers at 48 h p.i. Despite the strain-specific differences observed between viruses isolated within each wave, Anhui/1/13, Taiwan/1/14, Taiwan/2/14, and BC/1/15 replicated most efficiently at 37°C in Calu-3 cells, with mean titers of >108 PFU/ml by 24 h p.i. Ultimately, all H7N9 viruses tested reached peak mean titers of >107 PFU/ml by 48 h p.i., demonstrating the efficient replication capability of H7N9 viruses from all three waves in this cell type. These results indicate that while H7N9 viruses isolated over three waves replicate to high titer in human bronchial epithelial cells, they nonetheless have not acquired the ability to replicate efficiently at the temperature of the upper respiratory tract, a characteristic shared by other avian influenza viruses (39).
FIG 3.
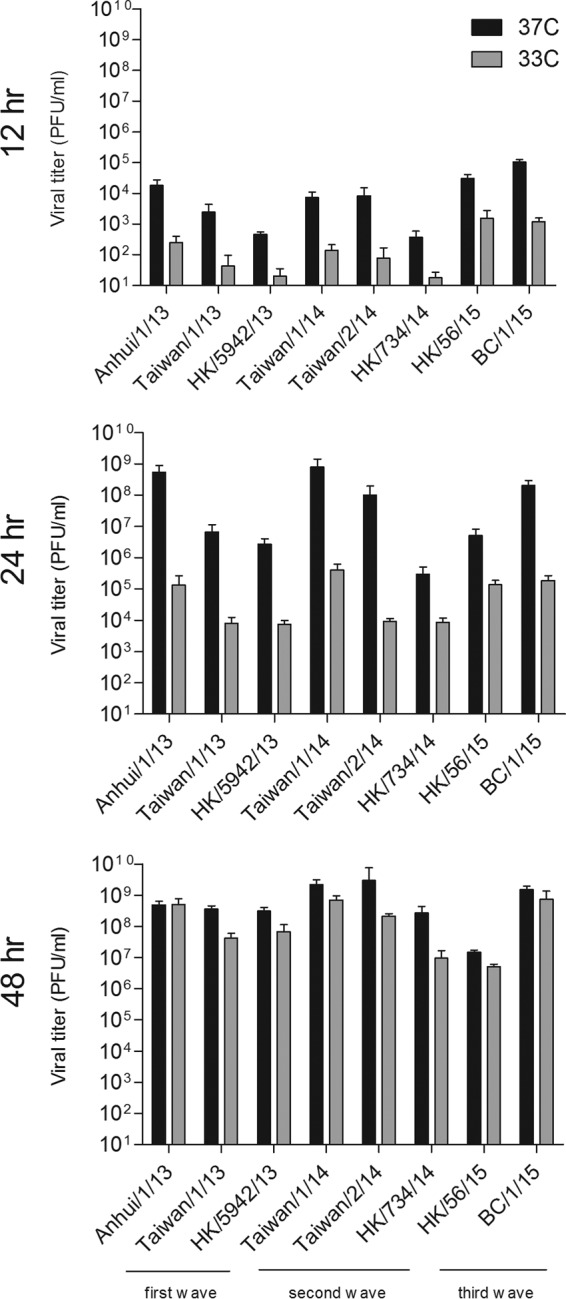
Replication kinetics of H7N9 viruses in polarized human airway epithelial cells. Calu-3 cells were inoculated with influenza viruses at an MOI of 0.01 and cultured at either 37°C or 33°C. Culture supernatants were collected at the times indicated, and virus titers were determined by plaque assay. Titer values represent the means plus standard deviations. The limit of detection was 10 PFU.
pH fusion threshold of first-, second-, and third-wave H7N9 viruses.
It has been shown recently that the H7N9 virus Anhui/1/13 has a relatively elevated pH threshold (pH 5.8) for fusion (as determined by a virus-mediated cell-cell fusion assay) compared to that of a seasonal human H3N2 virus (pH 5.4) (40). Here, we used a virus-induced hemolysis assay to evaluate the pH fusion of H7N9 viruses from all three waves of H7N9 infection. Consistent with previous findings, the first-wave Anhui/1/13 virus maintained greater than 30% of its maximal hemolysis ability at pH 5.8 (Fig. 4). All other first- and second-wave H7N9 viruses tested shared pH profiles for fusion similar to that of Anhui/1/13 virus, showing approximately 10% or more of their maximal fusion activities at a fusion threshold of pH 5.8 (Fig. 4 and data not shown). However, H7N9 viruses from the third wave (BC/1/15 and HK/56/15) showed a slightly reduced pH threshold for fusion compared to that of the first- and second-wave H7N9 viruses. The highest pH value for the fusion of third-wave H7N9 viruses was 5.6, at which 20% of maximal fusion activity was remaining, indicating that the pH threshold for third-wave viruses has lowered 0.2 pH units, from pH 5.8 to pH 5.6. A reduction in the pH fusion threshold has been associated with avian influenza virus adaptation to mammalian hosts (41). Although highly transmissible influenza viruses typically resist fusion until reaching the acidic environment of the mammalian upper respiratory tract, the reduced pH fusion threshold of third-wave H7N9 viruses indicates a sign of adaptation toward humans, although further study is required due to the limited number of isolates from each wave tested here.
FIG 4.
pH fusion threshold of H7N9 viruses. The hemolysis of turkey red blood cells following incubation with influenza virus or PBS mock-treated control at the indicated pH values was measured by optical density (OD) at 405 nm. The percentage of maximal hemolysis at each pH value was expressed as the average from three independent repeats. Solid shapes, first-wave H7N9 viruses; open shapes, second-wave H7N9 viruses; cross-hatch marks, third-wave H7N9 viruses.
DISCUSSION
Since their initial detection in 2013, H7N9 viruses have exhibited a great expansion in both genetic diversity and geographical distribution (3, 25), representing an ongoing pandemic threat. Well-characterized mammalian H7N9 challenge models have been used widely to evaluate a broad range of parameters critical to public health, including the induction of inflammatory responses, the generation of effective vaccine approaches, and antiviral efficacy (42–44). However, these studies largely rely on the use of the very first human isolates from Anhui and Shanghai from spring 2013 (45). As H7N9 viruses continue to evolve (3), it is critical to determine if the pathogenesis and potential for transmission among mammals of currently circulating isolates remain similar to that of first-wave H7N9 viruses; while our study is not inclusive of all potential isolates from each wave, the representative viruses from each wave tested here provide a first attempt to analyze this virus subtype over three calendar years as it continues to cause human infection.
The generally comparable pathogenesis observed in both mouse and ferret models following intranasal inoculation with H7N9 viruses (Tables 3 and 5 and Fig. 2) is consistent with studies which have demonstrated that minor HA sequence variation between H7N9 viruses does not substantially influence virus attachment to the mammalian respiratory tract (46). Infectivity (MID50), lethality (LD50), and morbidity (weight loss) of H7N9 viruses in BALB/c mice were similar to those of highly pathogenic influenza viruses H5N1 and H7N7 in this model (13, 21), highlighting the capacity of this LPAI virus to cause severe mammalian disease. However, infection with the first-wave virus Taiwan/1/13, while maintaining a virulent phenotype in mice, resulted in reduced morbidity and systemic spread of virus in ferrets compared with other H7N9 viruses associated with human infection (Table 5), in agreement with a prior study which also reported enhanced virulence in mice but not ferrets with this virus (47). While Taiwan/1/13 was the only virus in this study not bearing a lysine at position 226 in the HA, it is unlikely that this position alone contributed to the difference in mammalian virulence observed here, as the first-wave virus A/Shanghai/1/13, which exhibited virulence in mice and ferrets comparable to that of Anhui/1/13 virus, also lacks a lysine at this position (Table 2) (48). However, several NA variants of A/Taiwan/1/13 virus were detected in the original clinical isolate of this virus, with selected variants exhibiting reduced susceptibility to oseltamivir and attenuated phenotypes in vitro (47), suggesting that this isolate is unique compared with other first-wave isolates tested in previous studies.
H7N9 subtype viruses continue to distinguish themselves from other Eurasian and North American lineage H7 subtype viruses in their absence of an ocular tropism, which is typically observed with this subtype (49). Despite maintaining a strong tropism for the respiratory tract, the frequency of H7N9 virus detection in murine tissues following i.o. inoculation is comparable to that of other H7 subtype viruses, exceeding that observed with H5N1, seasonal, or 2009 H1N1 pandemic viruses (19, 50). These findings are in agreement with previous work in the ferret model, demonstrating the ability of a first-wave H7N9 virus to cause infection in ferrets following ocular exposure (51). While potential correlations between ocular tropism detected in the murine model and human susceptibility to influenza viruses following ocular exposure still are poorly understood, these data underscore the necessity of evaluating influenza viruses for the capacity to cause mammalian infection following alternate inoculation routes, even in the absence of a demonstrated tropism outside the respiratory tract.
As observed in this study with shv/Egypt/07 virus, H7N9 viruses isolated from poultry from multiple waves have demonstrated reduced transmissibility in the ferret model compared with viruses isolated from humans (30, 31, 52). While this can be explained in part by sequence differences often present between poultry- and human-isolated viruses (notably at PB2 residue 627), further examination of how these viruses diverge will allow for a more precise understanding of the adaptation these viruses have undergone to cause human disease. Mutations in the polymerase genes highlighted in Table 2 may have increased in frequency as a result of selection during circulation in poultry, but they appear to have minimal impact on virus replication and transmission in mammals. The limited transmissibility by respiratory droplets of second- and third-wave H7N9 viruses in ferrets tested here aligns with other studies which have reported transmission primarily among cohoused animals (52). Importantly, the similar transmission profile of H7N9 viruses between first-wave isolates and viruses isolated during the second and third waves suggest that vaccine and antiviral candidates which limit the transmissibility of the commonly employed Anhui/1/13 virus would maintain a comparable efficacy against more recently detected H7N9 viruses (53).
A previous study suggested that a high viral load (and associated induction of host responses) following the infection of bronchial epithelial cells with first-wave H7N9 viruses results in decreased integrity of the epithelial monolayer, leading to the infection of underlying endothelial cells, which ultimately may contribute to the severe pneumonia associated with H7N9 virus infection in humans (54). The high capacity for H7N9 viruses to replicate in a human respiratory tract cell type shown in this study (Fig. 3) indicates that H7N9 viruses isolated during the second and third waves have maintained this property. The induction of host innate immune responses represents a critical component of the virulent phenotype of H5N1 viruses in mammals (55). Interestingly, first-wave H7N9 viruses have been shown to be poor inducers of proinflammatory cytokines and chemokines compared with other avian viruses (such as H5N1) associated with severe human respiratory disease (54, 56, 57). While not examined here, these data suggest that H7N9 viruses from 2013 to 2015 would function similarly to first-wave viruses in this regard.
The study of viral fusion pH has attracted increased attention due to recent evidence that this property influences virus stability, transmission, and host adaptation (58, 59). In general, human influenza viruses tend to fuse at relatively lower pH values in order to avoid being inactivated prior to reaching the acidic nasal passageways, whereas most avian influenza viruses preferentially fuse at higher pH values, leading to the potential advantage of more rapid fusion at early endosomes during viral entry (60). As demonstrated in the ferret transmission model, airborne-transmissible H5N1 viruses acquired not only receptor binding switching mutations but also mutations lowering fusion pH, emphasizing that having a reduced pH for fusion might be necessary for avian influenza virus to initiate airborne transmission in mammalian hosts (58, 59). The relatively high fusion threshold (pH 5.8) of novel H7N9 viruses shown in our study and by other investigators (34, 40) may partially account for its limited transmission in humans. Interestingly, both third-wave H7N9 viruses examined here possessed slightly shifted pH fusion thresholds (pH 5.6) compared with those of first- and second-wave viruses, which indicates improved viral survival prior to reaching the acidic upper airways. However, based on the available H7N9 virus sequences from different waves, we cannot identify any mutations in the HA, NA, or M genes that are shared by all H7N9 viruses from specific waves, suggesting that the substitutions modulating viral fusion pH are strain specific. Continued studies individually examining fusion thresholds of newly emerging H7N9 viruses, and the impact these findings may have on transmission potential, are warranted.
It is likely that H7N9 viruses will continue to cause human infection in China, leading to persistent reassortment, diversity, and geographic spread. Our findings that viruses from the first three waves have maintained a pathogenic phenotype in mammals and upheld a robust transmissibility in the presence of direct contact with a capacity for transmission by respiratory droplets suggest that these viruses remain as immediate a threat to public health as when they were first detected in humans in 2013. However, given the phylogenetic diversity among H7N9 viruses and the relatively small number of viruses tested in vivo from each wave here, further studies with additional isolates from these waves are warranted to confirm the absence of strain-specific effects. As human infection with H7N9 viruses has reached a fourth wave of human cases, a greater understanding of how these viruses continue to evolve, and how this may influence mammalian pathogenicity and transmissibility, will support the continued generation of preventative measures required to limit and prevent the spread of H7N9 viruses to humans.
ACKNOWLEDGMENTS
We thank the China CDC, Taiwan CDC, and Hong Kong Department of Health, as part of the WHO Global Influenza Surveillance and Response System (GISRS), for facilitating access to viruses.
H.M.C. and X.S. were supported by the Oak Ridge Institute for Science and Education.
The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency.
REFERENCES
- 1.WHO. 2013-2015. WHO risk assessment of human infections with avian influenza A(H7N9) virus. WHO, Geneva, Switzerland: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/RiskAssessment_H7N9_23Feb20115.pdf?ua=1 Accessed 9 June 2015. [Google Scholar]
- 2.WHO. 2006-2015. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. WHO, Geneva, Switzerland: http://www.who.int/influenza/vaccines/virus/201502_zoonotic_vaccinevirusupdate.pdf?ua=1 Accessed 9 June 2015. [Google Scholar]
- 3.Lam TT, Zhou B, Wang J, Chai Y, Shen Y, Chen X, Ma C, Hong W, Chen Y, Zhang Y, Duan L, Chen P, Jiang J, Zhang Y, Li L, Poon LL, Webby RJ, Smith DK, Leung GM, Peiris JS, Holmes EC, Guan Y, Zhu H. 2015. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature 522:102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 4.Liu B, Havers F, Chen E, Yuan Z, Yuan H, Ou J, Shang M, Kang K, Liao K, Liu F, Li D, Ding H, Zhou L, Zhu W, Ding F, Zhang P, Wang X, Yao J, Xiang N, Zhou S, Liu X, Song Y, Su H, Wang R, Cai J, Cao Y, Wang X, Bai T, Wang J, Feng Z, Zhang Y, Widdowson MA, Li Q. 2014. Risk factors for influenza A(H7N9) disease–China, 2013. Clin Infect Dis 59:787–794. doi: 10.1093/cid/ciu423. [DOI] [PubMed] [Google Scholar]
- 5.Yuan J, Lau EH, Li K, Leung YH, Yang Z, Xie C, Liu Y, Liu Y, Ma X, Liu J, Li X, Chen K, Luo L, Di B, Cowling BJ, Tang X, Leung GM, Wang M, Peiris M. 2015. Effect of live poultry market closure on avian influenza A(H7N9) virus activity in Guangzhou, China, 2014. Emerg Infect Dis 21:1784–1793. doi: 10.3201/eid2110.150623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Wu JT, Cowling BJ, Liao Q, Fang VJ, Zhou S, Wu P, Zhou H, Lau EH, Guo D, Ni MY, Peng Z, Feng L, Jiang H, Luo H, Li Q, Feng Z, Wang Y, Yang W, Leung GM. 2014. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: an ecological study. Lancet 383:541–548. doi: 10.1016/S0140-6736(13)61904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Yu X, Pu X, Yang X, Kou Y, Zhou Y, Qian X, Xie L, Pan J. 2015. The diversity of avian influenza virus subtypes in live poultry markets before and during the second wave of A(H7N9) infections in Hangzhou, China. Emerg Microbes Infect 4:e14. doi: 10.1038/emi.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin Y, Horby PW, Tsang TK, Chen E, Gao L, Ou J, Nguyen TH, Duong TN, Gasimov V, Feng L, Wu P, Jiang H, Ren X, Peng Z, Li S, Li M, Zheng J, Liu S, Hu S, Hong R, Farrar JJ, Leung GM, Gao GF, Cowling BJ, Yu H. 2015. Differences in the epidemiology of human cases of avian influenza A(H7N9) and A(H5N1) viruses infection. Clin Infect Dis 61:563–571. doi: 10.1093/cid/civ345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Paquette SG, Zhang L, Leon AJ, Liu W, Xiuming W, Huang L, Wu S, Lin P, Chen W, Fang X, Zeng T, Kelvin N, Farooqui A, Kelvin DJ. 2015. The third wave: H7N9 endemic reassortant viruses and patient clusters. J Infect Dev Ctries 9:122–127. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Zhang Y, Fang L, Halloran ME, Ma M, Liang S, Kenah E, Britton T, Chen E, Hu J, Tang F, Cao W, Feng Z, Longini IM Jr. 2015. Household transmissibility of avian influenza A (H7N9) virus, China, February to May 2013 and October 2013 to March 2014. Euro Surveill 20:21056. doi: 10.2807/1560-7917.ES2015.20.10.21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farooqui A, Leon AJ, Huang L, Wu S, Cai Y, Lin P, Chen W, Fang X, Zeng T, Liu Y, Zhang L, Su T, Chen W, Ghedin E, Zhu H, Guan Y, Kelvin DJ. 2015. Genetic diversity of the 2013-14 human isolates of influenza H7N9 in China. BMC Infect Dis 15:109. doi: 10.1186/s12879-015-0829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang XY, Chai CL, Li FD, He F, Yu Z, Wang XX, Shang XP, Liu SL, Lin JF. 2015. Epidemiology of human infections with avian influenza A(H7N9) virus in the two waves before and after October 2013 in Zhejiang province, China. Epidemiol Infect 143:1839–1845. doi: 10.1017/S095026881400257X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belser JA, Lu X, Maines TR, Smith C, Li Y, Donis RO, Katz JM, Tumpey TM. 2007. Pathogenesis of avian influenza (H7) virus infection in mice and ferrets: enhanced virulence of Eurasian H7N7 viruses isolated from humans. J Virol 81:11139–11147. doi: 10.1128/JVI.01235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, Van Hoeven N, Donis R, Busch J, McBride R, Paulson JC, Katz JM, Tumpey TM. 2008. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc Natl Acad Sci U S A 105:7558–7563. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutton TC, Finch C, Shao H, Angel M, Chen H, Capua I, Cattoli G, Monne I, Perez DR. 2014. Airborne transmission of highly pathogenic H7N1 influenza virus in ferrets. J Virol 88:6623–6635. doi: 10.1128/JVI.02765-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao L, Xu L, Zhu H, Deng W, Chen T, Lv Q, Li F, Yuan J, Xu Y, Huang L, Li Y, Liu J, Yao Y, Yu P, Chen H, Qin C. 2014. Transmission of H7N9 influenza virus in mice by different infective routes. Virol J 11:185. doi: 10.1186/1743-422X-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, Tumpey TM, Katz JM. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type I interferon response in polarized human bronchial epithelial cells. J Virol 81:12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chosewood LC, Wilson DE, Centers for Disease Control and Prevention, National Institutes of Health. 2009. Biosafety in microbiological and biomedical laboratories, 5th ed U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institutes of Health, Washington, DC. [Google Scholar]
- 19.Belser JA, Wadford DA, Xu J, Katz JM, Tumpey TM. 2009. Ocular infection of mice with influenza A (H7) viruses: a site of primary replication and spread to the respiratory tract. J Virol 83:7075–7084. doi: 10.1128/JVI.00535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belser JA, Gustin KM, Maines TR, Pantin-Jackwood MJ, Katz JM, Tumpey TM. 2012. Influenza virus respiratory infection and transmission following ocular inoculation in ferrets. PLoS Pathog 8:e1002569. doi: 10.1371/journal.ppat.1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol 79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai Le Q, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM. 2006. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shelton H, Roberts KL, Molesti E, Temperton N, Barclay WS. 2013. Mutations in haemagglutinin that affect receptor binding and pH stability increase replication of a PR8 influenza virus with H5 HA in the upper respiratory tract of ferrets and may contribute to transmissibility. J Gen Virol 94:1220–1229. doi: 10.1099/vir.0.050526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie S, Jia W, Lin Y, Xing K, Ren X, Qi W, Liao M. 2015. Third wave of influenza A(H7N9) virus from poultry, Guangdong Province, China, 2014-2015. Emerg Infect Dis 21:1657–1660. doi: 10.3201/eid2109.150635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui L, Liu D, Shi W, Pan J, Qi X, Li X, Guo X, Zhou M, Li W, Li J, Haywood J, Xiao H, Yu X, Pu X, Wu Y, Yu H, Zhao K, Zhu Y, Wu B, Jin T, Shi Z, Tang F, Zhu F, Sun Q, Wu L, Yang R, Yan J, Lei F, Zhu B, Liu W, Ma J, Wang H, Gao GF. 2014. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5:3142. [DOI] [PubMed] [Google Scholar]
- 26.Yamayoshi S, Fukuyama S, Yamada S, Zhao D, Murakami S, Uraki R, Watanabe T, Tomita Y, Neumann G, Kawaoka Y. 2015. Amino acids substitutions in the PB2 protein of H7N9 influenza A viruses are important for virulence in mammalian hosts. Sci Rep 5:8039. doi: 10.1038/srep08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 28.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mok CK, Lee HH, Chan MC, Sia SF, Lestra M, Nicholls JM, Zhu H, Guan Y, Peiris JM. 2013. Pathogenicity of the novel A/H7N9 influenza virus in mice. mBio 4:e00362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Z, Yang Y, Feng Y, Shi B, Chen L, Zheng Y, Tian D, Song Z, Xu C, Qin B, Zhang X, Guan W, Liu F, Yang T, Yang H, Zeng D, Zhou W, Hu Y, Zhou X. 2013. Infection of inbred BALB/c and C57BL/6 and outbred Institute of Cancer Research mice with the emerging H7N9 avian influenza virus. Emerg Microbes Infect 2:e50. doi: 10.1038/emi.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe T, Watanabe S, Maher EA, Neumann G, Kawaoka Y. 2014. Pandemic potential of avian influenza A (H7N9) viruses. Trends Microbiol 22:623–631. doi: 10.1016/j.tim.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, Smith DJ, van den Brand JM, Burke DF, Kuiken T, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2013. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 501:560–563. doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. 2013. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341:183–186. doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- 36.Belser JA, Davis CT, Balish A, Edwards LE, Zeng H, Maines TR, Gustin KM, Martinez IL, Fasce R, Cox NJ, Katz JM, Tumpey TM. 2013. Pathogenesis, transmissibility, and ocular tropism of a highly pathogenic avian influenza A (H7N3) virus associated with human conjunctivitis. J Virol 87:5746–5754. doi: 10.1128/JVI.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu L, Bao L, Deng W, Dong L, Zhu H, Chen T, Lv Q, Li F, Yuan J, Xiang Z, Gao K, Xu Y, Huang L, Li Y, Liu J, Yao Y, Yu P, Li X, Huang W, Zhao X, Lan Y, Guo J, Yong W, Wei Q, Chen H, Zhang L, Qin C. 2014. Novel avian-origin human influenza A(H7N9) can be transmitted between ferrets via respiratory droplets. J Infect Dis 209:551–556. doi: 10.1093/infdis/jit474. [DOI] [PubMed] [Google Scholar]
- 38.Yang JR, Kuo CY, Huang HY, Wu FT, Huang YL, Cheng CY, Su YT, Wu HS, Liu MT. 2015. Characterization of influenza A (H7N9) viruses isolated from human cases imported into Taiwan. PLoS One 10:e0119792. doi: 10.1371/journal.pone.0119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, Garcia-Sastre A, Sasisekharan R, Katz JM, Tumpey TM. 2009. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A 106:3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabbard JD, Dlugolenski D, Van Riel D, Marshall N, Galloway SE, Howerth EW, Campbell PJ, Jones C, Johnson S, Byrd-Leotis L, Steinhauer DA, Kuiken T, Tompkins SM, Tripp R, Lowen AC, Steel J. 2014. Novel H7N9 influenza virus shows low infectious dose, high growth rate, and efficient contact transmission in the guinea pig model. J Virol 88:1502–1512. doi: 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galloway SE, Reed ML, Russell CJ, Steinhauer DA. 2013. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog 9:e1003151. doi: 10.1371/journal.ppat.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao G, Liu C, Kou Z, Gao T, Pan T, Wu X, Yu H, Guo Y, Zeng Y, Du L, Jiang S, Sun S, Zhou Y. 2014. Differences in the pathogenicity and inflammatory responses induced by avian influenza A/H7N9 virus infection in BALB/c and C57BL/6 mouse models. PLoS One 9:e92987. doi: 10.1371/journal.pone.0092987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marjuki H, Mishin VP, Chesnokov AP, De La Cruz JA, Fry AM, Villanueva J, Gubareva LV. 2014. An investigational antiviral drug, DAS181, effectively inhibits replication of zoonotic influenza A virus subtype H7N9 and protects mice from lethality. J Infect Dis 210:435–440. doi: 10.1093/infdis/jiu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krammer F, Albrecht RA, Tan GS, Margine I, Hai R, Schmolke M, Runstadler J, Andrews SF, Wilson PC, Cox RJ, Treanor JJ, Garcia-Sastre A, Palese P. 2014. Divergent H7 immunogens offer protection from H7N9 virus challenge. J Virol 88:3976–3985. doi: 10.1128/JVI.03095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 46.Siegers JY, Short KR, Leijten LM, de Graaf M, Spronken MI, Schrauwen EJ, Marshall N, Lowen AC, Gabriel G, Osterhaus AD, Kuiken T, van Riel D. 2014. Novel avian-origin influenza A (H7N9) virus attachment to the respiratory tract of five animal models. J Virol 88:4595–4599. doi: 10.1128/JVI.03190-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marjuki H, Mishin VP, Chesnokov AP, Jones J, De La Cruz JA, Sleeman K, Tamura D, Nguyen HT, Wu HS, Chang FY, Liu MT, Fry AM, Cox NJ, Villanueva JM, Davis CT, Gubareva LV. 2015. Characterization of drug-resistant influenza A(H7N9) variants isolated from an oseltamivir-treated patient in Taiwan. J Infect Dis 211:249–257. doi: 10.1093/infdis/jiu447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. 2013. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- 49.Belser JA, Rota PA, Tumpey TM. 2013. Ocular tropism of respiratory viruses. Microbiol Mol Biol Rev 77:144–156. doi: 10.1128/MMBR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belser JA, Wadford DA, Pappas C, Gustin KM, Maines TR, Pearce MB, Zeng H, Swayne DE, Pantin-Jackwood M, Katz JM, Tumpey TM. 2010. Pathogenesis of pandemic influenza A (H1N1) and triple-reassortant swine influenza A (H1) viruses in mice. J Virol 84:4194–4203. doi: 10.1128/JVI.02742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belser JA, Gustin KM, Katz JM, Maines TR, Tumpey TM. 2014. Influenza virus infectivity and virulence following ocular-only aerosol inoculation of ferrets. J Virol 88:9647–9654. doi: 10.1128/JVI.01067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luk GS, Leung CY, Sia SF, Choy KT, Zhou J, Ho CC, Cheung PP, Lee EF, Wai CK, Li PC, Ip SM, Poon LL, Lindsley WG, Peiris M, Yen HL. 2015. Transmission of H7N9 influenza viruses with polymorphism at PB2 residue 627 in chickens and ferrets. J Virol 89:9939–9951. doi: 10.1128/JVI.01444-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong H, Zhang Q, Gu C, Shi J, Deng G, Ma S, Liu J, Chen P, Guan Y, Jiang Y, Chen H. 2015. A live attenuated vaccine prevents replication and transmission of H7N9 virus in mammals. Sci Rep 5:11233. doi: 10.1038/srep11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng H, Belser JA, Goldsmith CS, Gustin KM, Veguilla V, Katz JM, Tumpey TM. 2015. A(H7N9) virus results in early induction of proinflammatory cytokine responses in both human lung epithelial and endothelial cells and shows increased human adaptation compared with avian H5N1 virus. J Virol 89:4655–4667. doi: 10.1128/JVI.03095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maines TR, Szretter KJ, Perrone L, Belser JA, Bright RA, Zeng H, Tumpey TM, Katz JM. 2008. Pathogenesis of emerging avian influenza viruses in mammals and the host innate immune response. Immunol Rev 225:68–84. doi: 10.1111/j.1600-065X.2008.00690.x. [DOI] [PubMed] [Google Scholar]
- 56.Chan MC, Chan RW, Chan LL, Mok CK, Hui KP, Fong JH, Tao KP, Poon LL, Nicholls JM, Guan Y, Peiris JS. 2013. Tropism and innate host responses of a novel avian influenza A H7N9 virus: an analysis of ex-vivo and in-vitro cultures of the human respiratory tract. Lancet Respir Med 1:534–542. doi: 10.1016/S2213-2600(13)70138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Josset L, Zeng H, Kelly SM, Tumpey TM, Katze MG. 2014. Transcriptomic characterization of the novel avian-origin influenza A (H7N9) virus: specific host response and responses intermediate between avian (H5N1 and H7N7) and human (H3N2) viruses and implications for treatment options. mBio 5:e01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long JS, Benfield CT, Barclay WS. 2015. One-way trip: influenza virus' adaptation to gallinaceous poultry may limit its pandemic potential. Bioessays 37:204–212. doi: 10.1002/bies.201400133. [DOI] [PubMed] [Google Scholar]
- 61.Gerloff NA, Jones J, Simpson N, Balish A, Elbadry MA, Baghat V, Rusev I, de Mattos CC, de Mattos CA, Zonkle LE, Kis Z, Davis CT, Yingst S, Cornelius C, Soliman A, Mohareb E, Klimov A, Donis RO. 2013. A high diversity of Eurasian lineage low pathogenicity avian influenza A viruses circulate among wild birds sampled in Egypt. PLoS One 8:e68522. doi: 10.1371/journal.pone.0068522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang SY, Lin PH, Tsai JC, Hung CC, Chang SC. 2013. The first case of H7N9 influenza in Taiwan. Lancet 381:1621. doi: 10.1016/S0140-6736(13)60943-5. [DOI] [PubMed] [Google Scholar]
- 63.WHO. 1 February 2015, posting date. Human infection with avian influenza A(H7N9) virus--Canada. WHO, Geneva, Switzerland: http://www.who.int/csr/don/01-february-2015-avian-influenza/en/ Accessed 9 June 2015. [Google Scholar]