ABSTRACT
The HIV envelope binds cellular CD4 and undergoes a range of conformational changes that lead to membrane fusion and delivery of the viral nucleocapsid into the cellular cytoplasm. This binding to CD4 reveals cryptic and highly conserved epitopes, the molecular nature of which is still not fully understood. The atomic structures of CD4 complexed with gp120 core molecules (a form of gp120 in which the V1, V2, and V3 loops and N and C termini have been truncated) have indicated that a hallmark feature of the CD4-bound conformation is the bridging sheet minidomain. Variations in the orientation of the bridging sheet hairpins have been revealed when CD4-liganded gp120 was compared to CD4-unliganded trimeric envelope structures. Hence, there appears to be a number of conformational transitions possible in HIV-1 monomeric gp120 that are affected by CD4 binding. The spectrum of CD4-bound conformations has been interrogated in this study by using a well-characterized panel of conditional, CD4-induced (CD4i) monoclonal antibodies (MAbs) that bind HIV-1 gp120 and its mutations under various conditions. Two distinct CD4i epitopes of the outer domain were studied: the first comprises the bridging sheet, while the second contains elements of the V2 loop. Furthermore, we show that the unliganded extended monomeric core of gp120 (coree) assumes an intermediate CD4i conformation in solution that further undergoes detectable rearrangements upon association with CD4. These discoveries impact both accepted paradigms concerning gp120 structure and the field of HIV immunogen design.
IMPORTANCE Elucidation of the conformational transitions that the HIV-1 envelope protein undergoes during the course of entry into CD4+ cells is fundamental to our understanding of HIV biology. The binding of CD4 triggers a range of gp120 structural rearrangements that could present targets for future drug design and development of preventive vaccines. Here we have systematically interrogated and scrutinized these conformational transitions using a panel of antibody probes that share a specific preference for the CD4i conformations. These have been employed to study a collection of gp120 mutations and truncations. Through these analyses, we propose 4 distinct sequential steps in CD4i transitions of gp120 conformations, each defined by antibody specificities and structural requirements of the HIV envelope monomer. As a result, we not only provide new insights into this dynamic process but also define probes to further investigate HIV infection.
INTRODUCTION
Viral tropism is mediated by the specific binding of the viral spike protein to its corresponding cell surface receptor. Evolution has driven human immunodeficiency virus (HIV) to elaborate on this canonical paradigm, introducing a series of orchestrated sequential events involving two receptors: CD4 as a primary receptor (1–3) and a chemokine receptor (CXCR4 or CCR5) as a subsequent coreceptor (4–10). However, many critical details of the molecular mechanisms by which CD4 triggers a number of conformational rearrangements within gp120 to assemble a coreceptor binding site and how this ultimately leads to gp41-mediated membrane fusion are still missing. Obviously, it would be extremely beneficial to have high-resolution atomic structures for the viral spike before it encounters CD4 and serial snapshots of the structural transitions that the gp120 subunits undergo until gp41 steps in to drive membrane fusion. However, this has proven extremely challenging, in part due to the fact that the HIV-1 envelope exists in dynamic equilibrium among an ensemble of conformations (11–17).
In 1998, the first structure of the monomeric HIV-1 gp120 subunit was solved (18) but only when its N and C termini; variable loops V1, V2, and V3; and sugar moieties were removed and the remaining “core” was further stabilized via binding to CD4 along with a Fab of a gp120-specific monoclonal antibody (MAb) (MAb 17b). Nonetheless, this tripartite crystal proved extremely informative and provided the first glimpse of the gp120 structure in a CD4-bound state. Compared to the atomic structure of an unliganded simian immunodeficiency virus (SIV) envelope (19), it was proposed that the four-β-stranded “bridging sheet,” consistently found in a variety of HIV-1 gp120/CD4/Fab cocrystal structures (16, 18, 20–23) yet absent from the SIV structure, was a defining structural hallmark of the CD4-bound conformation. Subsequently, Kwon and collaborators discovered that by extension of the N terminus of monomeric gp120 core and retention of the base of the V3 loop (yet still V1 to V3 depleted), one could generate high-quality crystals in the absence of both CD4 and a stabilizing Fab, thus providing atomic structures for an extended core version (coree) of unliganded HIV-1 gp120 (24). Unexpectedly, the fully assembled four-stranded bridging sheet, previously taken as the epitome of the CD4-bound conformation, persisted in all the analyzed coree structures. This led Kwon et al. to propose that the default structure of monomeric gp120, depleted of its variable loops (V1 to V3), assumes an energetically favorable ground-state “CD4-bound conformation” characterized by a fully formed bridging sheet. It was postulated that within the trimer, interactions between the gp120 protomers and the variable loops lock the envelope into a higher-energy state (24). Binding of the trimer to CD4 triggers a series of conformational changes, shifting the variable loops to allow each monomer to snap into a preferred “ground state,” thus opening up the trimer and enabling the binding of the second coreceptor followed by gp41 transitions and, ultimately, membrane fusion (14, 15, 17, 24–28) (also see Fig. 5 in reference 24). Cryo-electron microscopy (EM) tomography analyses integrated with X-ray structures of stabilized, fully cleaved, soluble Env trimers have been extremely effective in generating a detailed model for the HIV spike (clade A BG505 SOSIP.664 gp140 [17, 29–31], the clade B B41 SOSIP.664 trimer [22], and the clade C ZM197M and DU422 SOSIP.664 trimers [32]). A surprising feature of the CD4-unliganded gp120 molecule is the presence of a four-stranded bridging sheet, in contrast to the SIV structure discussed above (29–31). However, this structure in the trimer is distinct from bridging sheets seen in the monomeric gp120/CD4 cocrystals (16, 18, 20–23) and the unliganded gp120 coree structures (24). Whereas the two versions of the bridging sheet have the same four-β-strand composition, they differ with regard to the juxtapositions and relative orientations of these β-strands. Hence, CD4 still plays a role in inducing conformational reorientations within the bridging sheet. A better definition of what actually occurs within the gp120 protomer as a result of CD4 binding is essential to our understanding of the biology of the mechanisms of HIV infection of CD4 cells.
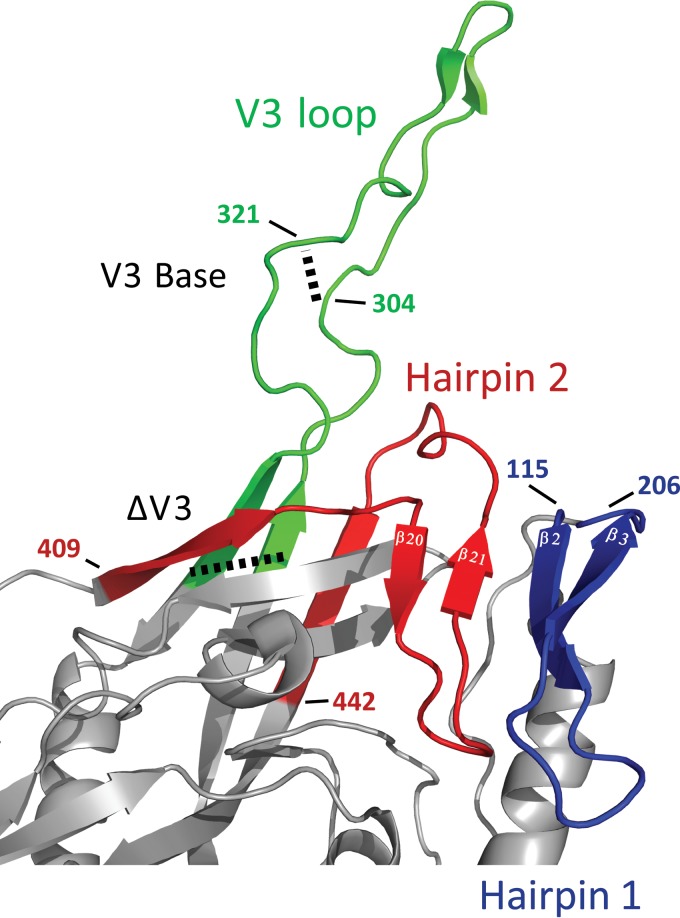
FIG 5.
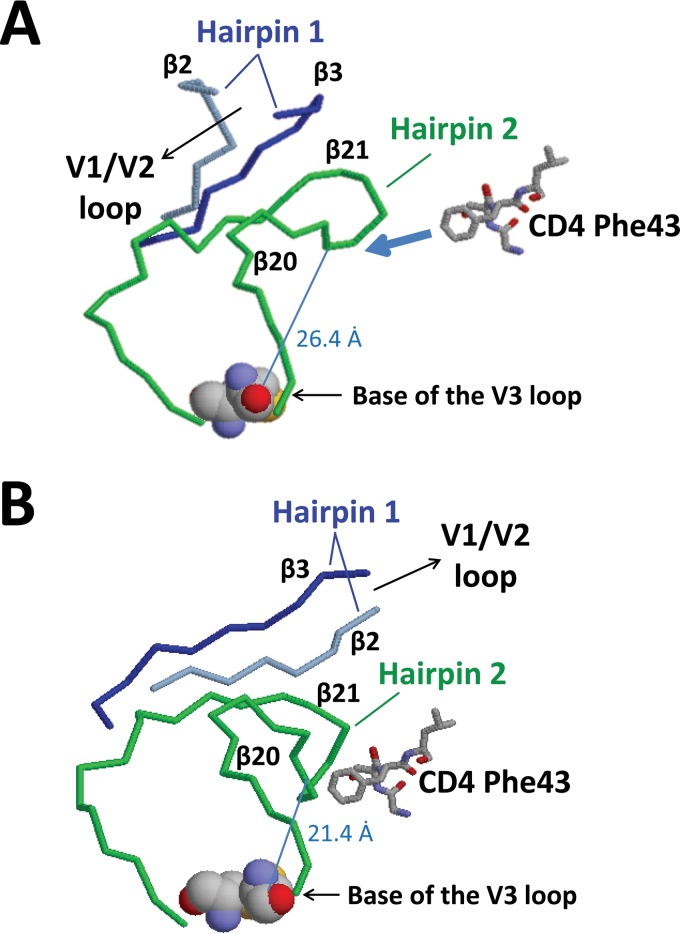
Orientation of the bridging sheet hairpins in relation to the V3 loop. The hairpins that constitute the bridging sheet (hairpins 1 and 2, shown in blue and red, respectively) and the V3 loop (green) are shown in the CD4-liganded conformation (PDB accession number 2B4C). Note that hairpin 1 (β2-β3) is the base for the V1/V2 loops (the V1/V2 loops are missing in the crystal structure) and that the V3 loop protrudes between the proximal and distal aspects of hairpin 2 (β20-β21). The locations of the different V3 loop truncations (V3 base and ΔV3) are marked with dashed lines. For the sake of consistency with our mutants, the numbering is based on BaL gp120 (see Materials and Methods).
In a desire to reveal structural elements that discriminate and delineate the essence of the CD4-induced (CD4i) conformational rearrangements in the HIV envelope, we and others have investigated gp120 structures using “conditional” probes, MAbs, that specifically define epitopes associated with CD4i transitions of gp120 (33–41). Here we describe the analysis of gp120 with a select panel of five well-established CD4i MAbs that target the core outer domain. Our results illustrate that gp120 undergoes a range of CD4i conformations that go beyond that of the previously reported structure of monomeric coree.
MATERIALS AND METHODS
Antibodies and reagents.
The outer domain-specific CD4i MAbs used in this study are described in Table 1.
TABLE 1.
List of antibodies used in this study
| Antibody | Biological source | Source | Description | References |
|---|---|---|---|---|
| 17b | Human MAb | J. E. Robinson, Tulane University | Binds the gp120 bridging sheet, as determined by X-ray structure determination | 18, 38 |
| 21c | Human MAb | J. E. Robinson | Binds a hybrid epitope comprised of gp120 and CD4, as determined by X-ray structure determination | 23, 39 |
| 19e | Human MAb | J. E. Robinson | Specific for the gp120-CD4 complex | 42, 43 |
| N12-i15 | Human MAb | Institute of Human Virology | Specific for the gp120-CD4 complex | 41, 65 |
| CG10 | Murine MAb | Tel Aviv University | Specific for the gp120-CD4 complex; a chimeric human version of this MAb was produced and used as part of this study | 44–46 |
MAbs 17b (18, 38) and 19e (42, 43) were kindly provided by J. Robinson (Tulane University Medical Center, USA). MAb N12-i15 was isolated by Y. Guan and colleagues, including one of us (G.K.L.), at the Institute of Human Virology, as described previously (41). MAb 21c was kindly provided by R. Diskin (Division of Biology, California Institute of Technology, USA) (23, 39). MAbs CG10, CG9, 1B6, and LG4 were isolated at Tel Aviv University. The chimeric human IgG version of the murine CG10 CD4i MAb (44–46) was produced by cloning the heavy and light chain variable-domain sequences of this MAb into the pMAZ-IgH and pMAZ-IgL vectors designed for the production of IgG1 antibodies in mammalian cells, as described previously (47). The vectors were then used to transiently transfect HEK 293T cells as described below. MAbs LG4 and 1B6 are both used as capture reagents that bind gp120 (MAb LG4 targets a conserved epitope at the carboxy terminus of gp120, “SGGPLGVAPTKAKRRVVQREKRAD,” while 1B6 binds gp120 without interfering with binding by CD4i MAbs or CD4 [48]). MAb CG9 binds to CD4 (46). Soluble CD4 (sCD4) (domains D1 to D4) was a kind gift from GlaxoSmithKline. HIVIg is a pool of purified IgGs from HIV-infected individuals, kindly provided by Nabi, Inc. The m2 peptide is a phage-displayed, 14-amino-acid-long, cysteine-constrained peptide (CDRRDLPDWAIRAC). Binding of gp120 by the m2 peptide allosterically induces CD4i epitopes (48).
HIV envelopes.
Except for gp120 from the CDC451 strain (which was purchased as soluble monomeric gp120 from Advanced BioScience Labs, USA) and the monomeric and SOSIP forms of BG505 (kindly provided by J. Moore, Weill Cornell Medical College, USA), all of the other gp120s used in this study were produced by transient transfection of HEK 293T cells (see below). For R2, a vector containing a codon-optimized R2 gp120/gp160 gene was kindly provided by G. V. Quinnan and C. C. Broder (Uniformed Services University of the Health Sciences, USA). For JR-FL, a vector containing a codon-optimized JR-FL gp120 gene was kindly provided by R. Pantophlet (Scripps Institute, USA). For BaL, a codon-optimized BaL gp120 gene from the pNGVL-FLSC-RT vector, provided by A. L. DeVico (Institute of Human Virology, USA) was recloned into the pCDNA3 expression vector (Invitrogen, USA). For YU2, a vector encoding the YU2 gp160 gene was kindly provided by Joseph G. Sodroski (Dana Farber Cancer Institute, USA). For extended gp120 cores, vectors encoding extended core gp120 genes were kindly provided by Y. D. Kwon and P. D. Kwong (National Institutes of Health, Bethesda, MD, USA). The BaL gp120 truncation mutants were constructed based on the codon-optimized BaL gene by using overlap PCR. The resulting mutated env genes were verified by sequencing and cloned into the pCDNA3 expression vector.
Amino acid numbering system.
In view of the fact that all the genetic manipulations, truncations, and mutations of gp120 reported here are derived from BaL, the numbering system used throughout this study is based on BaL gp120. In order to assist in comparisons with other HIV-1 envelope molecules, we have indicated landmark cysteine residues in the figures and text.
Production of gp120 and antibodies by transient transfection.
HIV gp120s and chimeric human CG10 were produced by transient transfection of HEK 293T cells with 10 μg of the appropriate gp120-encoding vector or 10 μg each of the chimeric CG10 heavy chain- and light chain-encoding vectors per 10-cm plate by the calcium phosphate method. HEK 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) complemented with 10% fetal calf serum (Biological Industries, Beit Haemek, Israel). Spent medium was collected at 48 h posttransfection. The chimeric CG10 MAb was purified from spent medium by using protein G affinity chromatography, dialyzed against phosphate-buffered saline (PBS), and quantified by UV spectrometry. The gp120 proteins were filtered and concentrated by using Amicon Ultracel Centricon with a 50,000-molecular-weight (MW) cutoff (Millipore) according to the manufacturer's instructions. The gp120s were then quantified by binding to HIVIg against gp120s of a known concentration and tested for functional folding in a quality control enzyme-linked immunosorbent assay (ELISA) (ligand overlay of Western blots was also used as described previously [46, 49]). In the quality control ELISA, mutated envelopes were tested for binding to the stringent CG10 MAb in the presence of sCD4, showing that the produced envelopes were able to bind sCD4 and undergo CD4i conformational changes. Both of these traits were taken as an indication of proper protein folding. Further confirmation of the functional configuration of gp120 and its mutants was demonstrated by binding of conformation-demanding MAbs, such as b12 (50, 51) and b6 (52, 53), as well as the glycomoiety-specific MAb 2G12. SDS-PAGE analyses of the gp120 preparations indicated the presence of both monomeric and oligomeric forms, as previously reported (54–63). The aberrant disulfide-linked oligomers continued to bind CD4 as expected. Moreover, Coutu and Finzi recently compared the binding kinetics of non-fast protein liquid chromatography (FLPC)-purified gp120 with FPLC-purified monomeric gp120 by surface plasmon resonance and confirmed that the affinity constants for CD4 in both preparations are not statistically different (some differences in antibody on-rates were measured) (55). It is noteworthy that all the conclusions reported here are based exclusively on positive results, i.e., the acquisition of robust binding as a result of CD4 association with the gp120s tested. Nonetheless, a number of the gp120 truncations were FPLC purified according to methods described previously by Coutu and Finzi (55), and the monomers (>95% purity) were tested, compared to FPLC-purified wild-type (wt) BaL monomeric gp120 where indicated, and found in all cases to respond the same as non-FPLC-purified gp120. Furthermore, Western blot analyses confirmed that both monomeric gp120 and the β-mercaptoethanol-sensitive oligomers (54, 56–63) continued to bind CD4, which further induced both relaxed (e.g., 17b) as well as stringent (e.g., CG10) MAb binding, as previously described (46, 49).
Envelope ELISA.
ELISA plates were coated overnight at 4°C with 100 μl of 7.5 μg/ml of capture MAb (either LG4 or 1B6) or 5 μg/ml gp120 diluted in PBS. The wells were blocked for 1 h at 37°C by using 5% nonfat dry milk and 20% horse serum in PBS. Capture of gp120 in MAb-coated wells was carried out by adding 5 μg/ml gp120 to the wells for 1 h at room temperature (RT). Detecting MAbs were added at 2.5 μg/ml for 1 h at RT. Horseradish peroxidase (HRP)-conjugated secondary antibodies (e.g., anti-human in the case of the human-derived CD4i MAbs) were then diluted 1:5,000 and added to the wells for 1 h at RT. All antibodies and proteins were added in blocking solution. Between incubations, the wells were washed three times with 0.05% Tween 20 in PBS (PBST). Finally, the wells were reacted with 100 μl of the 3,3',5,5'-tetramethybenzidine (TMB/E) ELISA substrate (Chemicon International, USA). Absorbance was measured at 650 nm. Unless stated otherwise, gp120 from the BaL strain was used in the reported assays. For BG505 SOSIP.664 gp140/BG505 gp120 ELISAs and extended core ELISAs, CD4i MAbs were used as capture reagents. BG505 gp120 or BG505 SOSIP.664 gp140 or extended envelope cores were added to the wells in the presence or absence of sCD4 (4 μg/ml). Bound extended envelope cores were detected with biotin-conjugated HIVIg (2.5 μg/ml) followed by HRP-conjugated streptavidin. For competition ELISAs, the murine version of the CG10 MAb was added at 10 to 15 μg/ml to wells containing captured gp120 and sCD4 (gp120 and sCD4 concentrations were the same as those for the regular envelope ELISA) and incubated for 1 h at RT. The competing human CD4i MAbs were then added (2.5 μg/ml), and the ELISA was continued as described above. In ELISAs utilizing biotinylated antibodies, the blocking buffer used was 3% bovine serum albumin (BSA) in PBST. Detection of bound biotinylated antibody was done with HRP-conjugated streptavidin at a dilution of 1:2,500 in blocking buffer. Concentrations of reagents used in ELISAs are as follows: coating antibodies were used at 7.5 μg/ml, detection antibodies were used at 2.5 to 3.5 μg/ml, HIVIg was used at 10 μg/ml, CD4 was used at 5 μg/ml, and gp120s were used at 5 μg/ml. Graphs were generated and statistical significance of ELISA results was calculated (two-tailed Student's t test) by using GraphPad Prism version 6.01 for Windows (GraphPad Software, La Jolla, CA, USA). The results from three independent experiments are shown. Results from different experiments were normalized according to envelope binding by HIVIg. Bars represent standard deviations.
RESULTS
Here we focus on the primary and direct effects of CD4 binding to HIV-1 envelope, namely, the conformational transitions of gp120, using MAbs that can discriminate between different facets of this process. Such conformational changes have been reported for both the outer and inner domains of gp120 (35–37). Here we have specifically analyzed the changes associated with the outer domain harboring the CD4 binding site itself.
CD4i MAbs can be divided into distinct CD4-relaxed and -stringent subsets.
Numerous CD4i MAbs have been described, whose dependence on CD4 for gp120 binding ranges from marginal to absolute (18, 21, 23, 42, 62, 64–66). These MAbs are generally designated CD4i MAbs. We focused on and tested a collection of outer domain-specific CD4i MAbs against envelopes from several HIV-1 strains: gp120 from clade B strains BaL, CDC451, YU2, JR-FL, and R2 in addition to trimeric BG505 SOSIP.664 gp140 and the BG505 gp120 monomer (clade A). The BG505 SOSIP.664 trimer was used to ascertain the effects of trimerization on CD4i MAb binding, while the R2 strain can infect CD4-negative cells (67), constitutively exposes several CD4i epitopes (68), and therefore could theoretically bind CD4i MAbs in the absence of CD4.
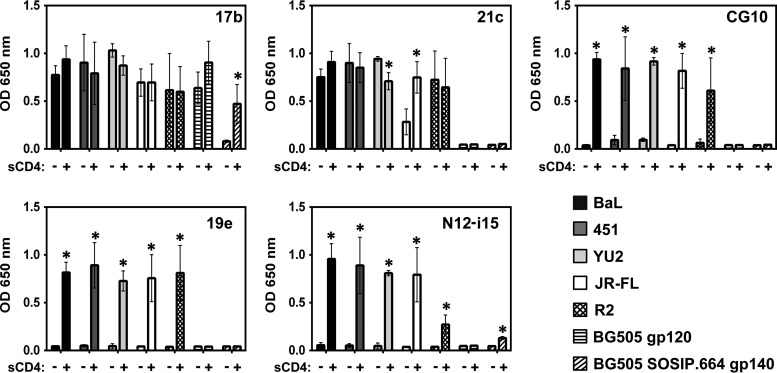
As shown in Fig. 1, the dependence on CD4 for gp120 binding of the selected, representative panel of five CD4i MAbs to gp120 is variable (the MAbs of the panel are described in Table 1). One of these, MAb 17b, has been used extensively as a “gold standard” CD4i MAb (11, 24, 39, 62, 69–76). The prominent CD4i status of MAb 17b stems from the fact that cell surface trimeric envelope requires CD4 for 17b binding (38), as is also the case for the soluble BG505 SOSIP.664 trimer (Fig. 1). In contrast, MAb 17b shows little requirement for CD4 when binding to full-length monomeric gp120, consistent with the conclusion of Kwon et al. that monomeric gp120 assumes a default CD4-bound conformation (24). MAb 21c also shows a similar CD4-relaxed pattern of binding to the clade B gp120s while not binding at all to the clade A BG505 monomer or the BG505 SOSIP trimer. In contrast, the other three CD4i MAbs, CG10, 19e, and N12-i15, bound all clade B gp120s in a totally CD4-stringent manner yet did not bind the clade A BG505 monomer and the BG505 SOSIP trimer. Thus, although some variation in binding can be seen on a subtype-specific level, CD4i MAbs can be divided into two distinct classes: relaxed binders, which can bind monomeric gp120 in the absence of CD4 (e.g., 17b and 21c), and stringent binders, whose binding to gp120 is completely dependent on CD4 (CG10, 19e, and N12-i15).
FIG 1.
Stringent CD4i MAbs do not bind gp120 in the absence of sCD4. An ELISA was used to analyze CD4i MAb binding to gp120s from different strains and to a gp140 trimer. MAbs 17b and 21c show binding to gp120 independent of sCD4 (relaxed binding). MAbs CG10, 19e, and N12-i15 show no binding to gp120 in the absence of sCD4 (stringent binding). Only MAb 17b was cross-reactive with the clade A BG505 SOSIP trimer and its monomer, showing relaxed binding to the monomer and stringent binding to the trimer. Statistically significant differences (P < 0.05) between the “−sCD4” and “+sCD4” columns are marked with an asterisk. OD, optical density.
Mapping distinct CD4i regions on gp120.
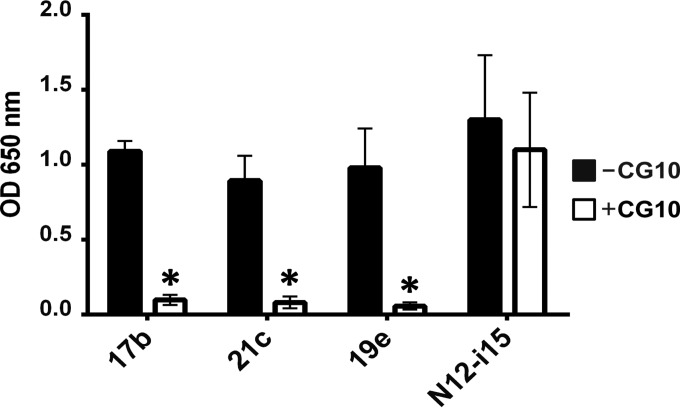
Does the CD4-bound state constitute a single epitope or region, or are there a number of different epitopes in the HIV-1 gp120 outer domain that are affected by CD4 binding? For this, we conducted a competitive ELISA, thereby testing the ability of the bridging sheet binding MAb CG10 (44–46) to compete against the other four MAbs of the panel (Fig. 2). It should be noted that the region of MAb 17b and 21c binding within gp120 is the bridging sheet minidomain, as ascertained by cocrystallization of these MAbs in complex with gp120 and CD4 (18, 23), and that MAb 19e binding has also been mapped to this region (42). Therefore, the corresponding epitopes of four of the CD4i MAbs overlap aspects of the bridging sheet, as also indicated by the ability of MAb CG10 to totally obstruct binding by MAbs 17b, 21c, and 19e (Fig. 2). Binding of the stringent MAb N12-i15, however, shows almost no competition with MAb CG10 and therefore defines an independent epitope that has no immediate overlap on the bridging sheet. Thus, we can conclude that conformational rearrangements in gp120 due to CD4 binding include at least two outer domain epitopes of gp120, which are spatially separated enough to preclude competition by antibodies.
FIG 2.
MAb N12-i15 defines a CD4i region upon gp120 distinct from the bridging sheet. Competition ELISAs were carried out in order to discern whether all the detected CD4i epitopes cluster to one area on gp120 or not. Binding of CD4i MAbs to gp120BaL-sCD4 was measured in either the absence or the presence of prebound MAb CG10. MAbs 17b, 21c, and 19e compete strongly with MAb CG10. MAb N12-i15 defines a CD4i epitope sufficiently separated from the bridging sheet so as to show no competition with MAb CG10. Statistically significant differences (P < 0.05) between the “−CG10” and “+CG10” columns are marked with an asterisk.
Stringent CD4i MAbs bind conformationally induced gp120 in the absence of CD4.
The stringent binding nature of MAbs CG10, 19e, and N12-i15 could hypothetically be the result of the CD4 protein directly contributing pivotal contact residues necessary for MAb recognition. We previously assessed the contribution of such hypothetical critical contact residues within CD4 using a molecule other than CD4. A short 14-amino-acid peptide (designated the m2 peptide [CDRRDLPDWAIRAC]), shown to bind to gp120 and allosterically induce the CD4i conformation, i.e., without occluding the CD4 binding site itself, was used in place of CD4 (48). We showed that the m2 peptide induces the epitopes of the CD4i conformation recognized by the three stringent MAbs CG10, 19e, and N12-i15 and allows binding to gp120 in the absence of CD4. m2 peptide induction of a CD4i conformation occurs without occluding the CD4 binding site, thus illustrating an alternative route toward achieving a conformation resembling a CD4-bound state. Together, this experiment (48) and previous studies of CG10 (62, 77) demonstrate that MAbs CG10, 19e, and N12-i15 can bind in the total absence of CD4, provided that gp120 is induced to display the stringent CD4i epitopes. This illustrates that even if some residues of CD4 contact these MAbs, they are not a prerequisite for stable binding.
Contributions of the V1, V2, and V3 loops to the CD4i conformation.
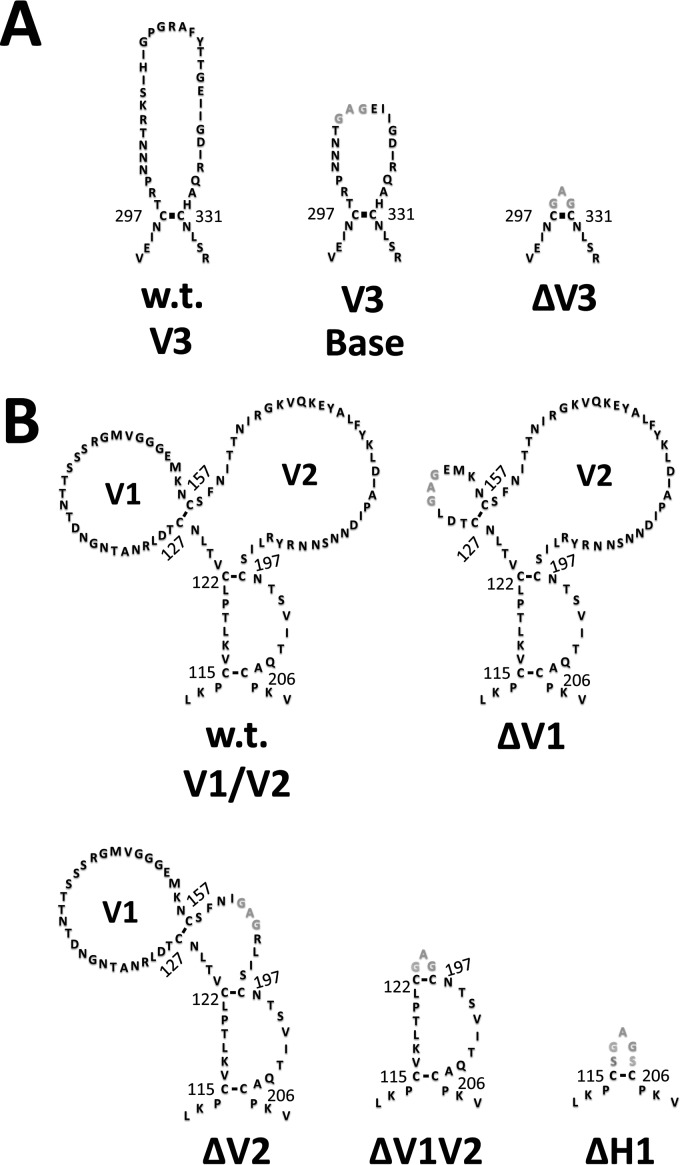
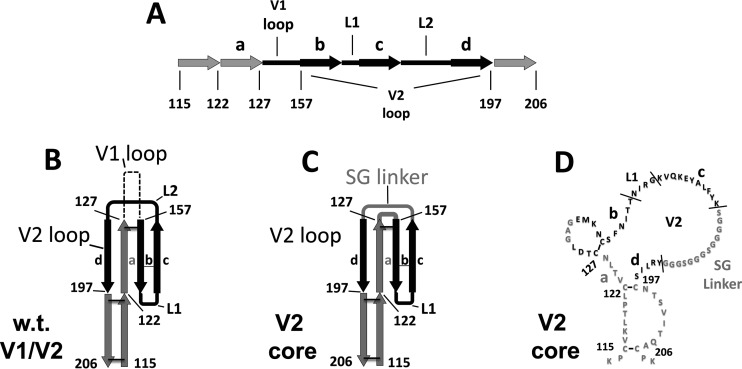
Next, we analyzed the effects of the V1/V2/V3 loops and bridging sheet hairpin 1 (the base of the V1/V2 loops) on the CD4-bound conformation as interpreted by CD4i MAb binding. Using BaL gp120 as the prototype, a number of mutants were generated. For the sake of clarity, Fig. 3 to 5 provide structural schematics of the collection of gp120 mutations and truncations analyzed. The V3 loop was either partially or fully deleted (the V3 base and ΔV3 mutants, respectively) (Fig. 3A), and the V1 and V2 loops were either both truncated (ΔV1V2 mutant) or truncated separately (the ΔV1 and ΔV2 mutants) (Fig. 3B). Additionally, the V2 core mutant (described in detail in the legend of Fig. 4) was constructed based on the crystal structure of the V1/V2 loops bound to a neutralizing antibody (78). This crystal structure showed that the V2 loop assumes a “Greek key” fold. The V2 core mutant was generated by truncating the V1 loop and replacing the parts of the V2 loop that do not directly participate in the Greek key fold (the loop designated L2) with a Gly-Ser linker, allowing us to test the importance of the Greek key V2 loop residues for CD4i MAb binding. Finally, the bridging sheet hairpin 1 (the base of the V1/V2 loops) was truncated (along with the V1/V2 loops) (Fig. 3B) in order to test if the CD4i MAbs could bind in the absence of a complete four-stranded bridging sheet. Figure 5 depicts the spatial relationship between the V3 loop and the bridging sheet minidomain comprised of hairpin 1 (the β2-β3 excursion from the gp120 inner domain) and hairpin 2 (the β20-β21 excursion from the gp120 outer domain).
FIG 3.
Schematic of the wt and modified forms of the gp120 V1/V2 and V3 loops. The sequences of wt and modified V3 (A) and V1/V2 (B) loops are shown with disulfide bonds depicted as black lines. Amino acid numbering corresponds to BaL gp120. Landmark cysteine residues are numbered for convenience. wt sequences are shown in black, and altered sequences are shown in gray. (A) V3 loop mutants, including (i) the wt V3 loop, (ii) V3 base (gp120 with a partially truncated V3 loop), and (iii) ΔV3 (gp120 with a truncated V3 loop). (B) V1/V2 loop mutants, including (i) wt V1/V2 loops, (ii) ΔV1 (gp120 with a truncated V1 loop), (iii) ΔV2 (gp120 with a truncated V2 loop), (iv) ΔV1V2 (gp120 with truncated V1/V2 loops), and (v) ΔH1 (gp120 with truncated V1/V2 loops and stem).
FIG 4.
Comparison of wt V1/V2 loops and the V2 core mutant. The crystal structure of the V1/V2 loops from gp120 of strain ZM109 reveals that the V2 loop assumes a Greek key fold (PDB accession number 3U2S) (78). The Greek key fold is comprised of four antiparallel β-strands (designated a to d) connected by the V1 loop (dashed) and two small loops (designated L1 and L2), which are part of the V2 loop (black). Based upon this crystal structure, the V2 core mutant was constructed by using gp120 from the BaL strain (the same strain upon which all of the envelope mutants in this study are based). The V2 core mutant was constructed by removing most of the V1 loop and replacing the parts of the V2 loop that do not participate in the Greek fold (loop L2) with a Ser-Gly linker. Numbering refers to adjacent cysteine residues and is based on the BaL strain, with disulfide bonds shown as black bars. Shown are the V1 loop (dashed), the V2 loop (black), and disulfide bonds (black bars between cysteine residues). (A) Linearized schematic of the wt V1/V2 loop fold. (B) Schematic of the fold of the wt V1/V2 loops. (C) Schematic of the fold of the V2 core mutant. (D) Sequence of the V2 core mutant.
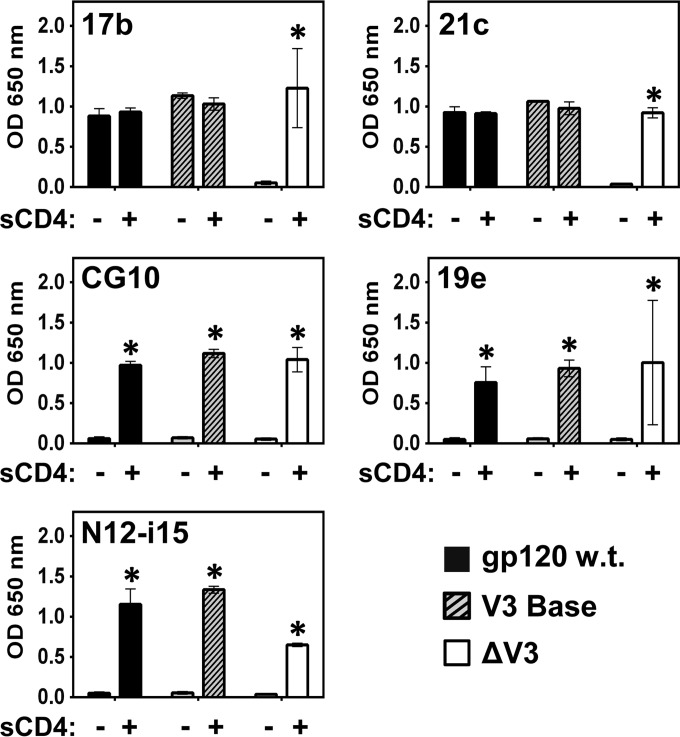
The three variations of V3 loop structures (depicted in Fig. 3A) were examined with regard to their impact on the binding of the CD4i MAb panel. As is shown in Fig. 6, total removal of the V3 loop, including its base (ΔV3), rendered MAbs 17b and 21c absolutely stringent for CD4 (an effect previously demonstrated [23, 75]). Relaxed binding (no requirement for CD4) could be regained provided that the base of V3 was kept in place. Thus, removal of the V3 loop in its entirety has a profound negative effect on 17b and 21c binding that can be overcome by maintaining even a few residues of the base of the V3 loop. MAbs CG10, 19e, and N12-i15 continued to demonstrate stringent binding to all the V3 loop-truncated gp120 constructs.
FIG 6.
V3 loop modifications affect binding by relaxed CD4i MAbs. A binding ELISA was carried out to ascertain the effects of V3 loop truncations on binding by the CD4i MAb panel to gp120BaL. Full truncation of the V3 loop (ΔV3) results in stringent binding by MAbs 17b and 21c, an effect not seen for the V3 base mutant. Binding by the stringent CD4i MAbs CG10, 19e, and N12-i15 was not affected by V3 loop truncations. Statistically significant differences (P < 0.05) between the “−sCD4” and “+sCD4” columns are marked with an asterisk.
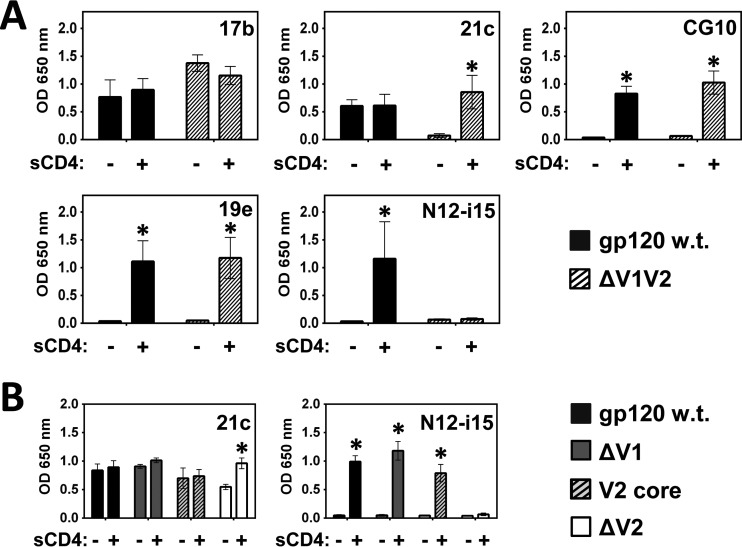
Next, we tested the effects of systematic truncations in the region of the V1 and V2 loops and the base of these loops, hairpin 1, on CD4i MAb binding (constructs depicted in Fig. 3B and 4). Removal of the V1 and V2 loops (ΔV1V2 mutant), while maintaining an intact V3 loop, did not affect binding by MAbs 17b, CG10, and 19e, while MAb 21c was rendered stringent for CD4, and binding of N12-i15 was irreversibly lost (Fig. 7A). Hence, it is clear that at least one of these variable loops impacts the binding of MAbs 21c and N12-i15.
FIG 7.
CD4i MAbs 21c and N12-i15 are sensitive to V1/V2 loop modifications. A binding ELISA was performed to test whether binding by the CD4i MAbs to gp120BaL is affected by the complete truncation of the V1/V2 loops (ΔV1V2) (A) or selective modification of the V1 or V2 loop (B). (A) MAb 21c becomes stringent and MAb N12-i15 loses all binding when the V1/V2 loops are fully truncated. (B) Selective modifications within the V1/V2 loops indicate that the V1 loop is not necessary for binding by MAbs 21c and N12-i15, but the core of the V2 loop (the V2 loop residues which fold into a Greek key conformation) (Fig. 4) is required. Statistically significant differences (P < 0.05) between the “−sCD4” and “+sCD4” columns are marked with an asterisk.
Selective removal of the V1 versus the V2 loop reveals that the V1 loop plays no or little role in MAb 21c or N12-i15 binding, in contrast to the requirement for a V2 loop (Fig. 7B) (23). This conclusion can be refined further by showing that a gp120 mutant that retains at least the V2 core residues (V2 core mutant described in the legend of Fig. 4 and ELISA results shown in Fig. 7B) binds MAbs 21c and N12-i15 at wt levels. These core V2 loop residues were found to adopt a Greek key structure when the V1 and V2 loops were expressed on a protein scaffold and cocrystallized with a broadly cross-neutralizing MAb (78) and again when trimeric BG505 SOSIP.664 gp140 was crystallized in the absence of sCD4 (29–31) and are sufficient to allow wt-level binding.
In addition, by comparing the V2 loop sequences of strains BaL, 451, YU2, and JR-FL, which are stringently bound by MAb N12-i15, with the V2 loop sequence of strain R2, which is barely recognized by MAb N12-i15, we gain some indication as to which residues within the Greek key may be critical for N12-i15 binding. Figure 8 shows the sequence alignment of the different V2 loops, with the four residues that differ in strain R2 within the Greek key structure marked. These differences may explain the reduced binding of N12-i15 to R2 compared to the other clade B isolates (Fig. 1).
FIG 8.
Sequence alignment of the gp120 V2 loop from different HIV strains. The gp120 V2 loop sequences from the different HIV strains tested against MAb N12-i15 for binding were aligned by using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/) and compared. The numbering system used is based on the BaL strain for consistency. Strand c of the Greek key fold based on the crystal structure of V1/V2 loops from the ZM109 strain (78) is shown. As MAb N12-i15 was shown to bind to elements within the Greek key fold of the V2 loop, the four residues different in the R2 V2 loop located within the Greek key fold are highlighted in dark gray. The V2 loop of the BG505 strain (both the monomer and the BG505 SOSIP trimer) is shown for comparison only, as MAb N12-i15 does not bind this strain (Fig. 1).
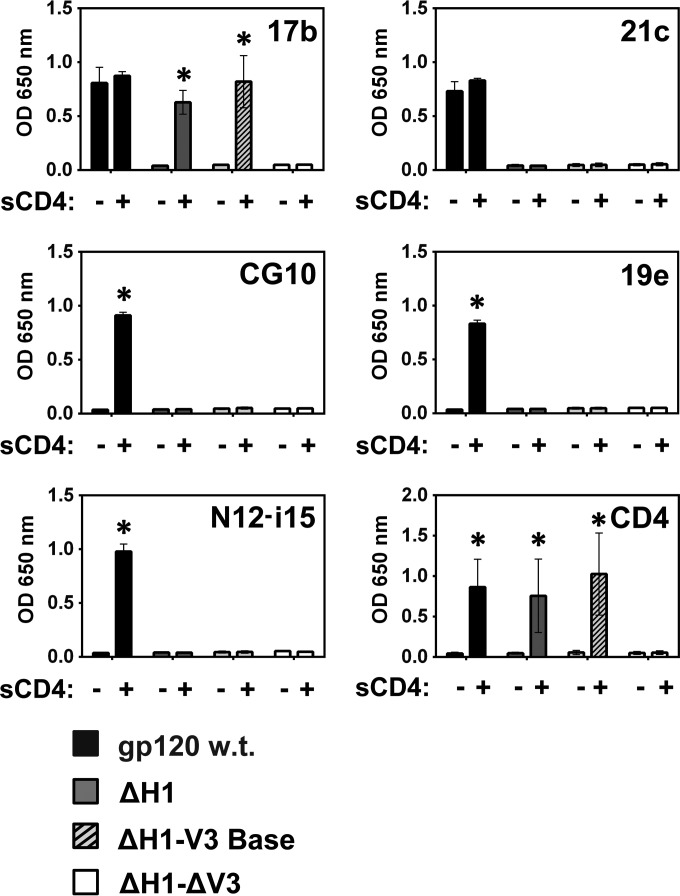
Finally, we asked whether the CD4i MAbs actually require an intact four-stranded bridging sheet. For this, we removed hairpin 1 (the stem of the V1/V2 loops) altogether, thus producing the ΔH1 mutant. The ΔH1 mutant was combined with the above-described V3 loop base and ΔV3 truncations (both the ΔH1 and ΔV3 truncations are shown in Fig. 3). MAbs 21c, CG10, 19e, and N12-i15 showed no binding to the ΔH1 constructs, even in the presence of CD4 (Fig. 9). Surprisingly, however, MAb 17b bound the ΔH1 construct although with a strict requirement for CD4. This contrasts with a previous report using a mutant very similar to ΔH1 in which MAb 17b failed to bind in a pulldown experiment (75). The binding of MAb 17b to the ΔH1 mutants persisted, provided that at least the base of the V3 loop was maintained. No recovery of binding could be found for 17b in the ΔH1-ΔV3 construct, probably as a result of the lack of binding to CD4. Thus, MAb 17b is indeed a CD4i MAb. However, its association with gp120 does not indicate a functional bridging sheet but rather hairpin 2 in a conformation that allows CD4 binding and MAb recognition. A summary of the binding activities of the CD4i MAbs tested is given in Table 2.
FIG 9.
MAb 17b binds gp120 in the absence of bridging sheet hairpin 1 in a CD4-dependent manner. A binding ELISA was performed to test whether the CD4i MAbs and sCD4 bind wt gp120BaL or the following bridging sheet hairpin 1 gp120BaL mutants (truncation sequences shown in Fig. 3): ΔH1 (gp120 with truncated hairpin 1), ΔH1-V3 base (gp120 with truncated hairpin 1 and a partially truncated V3 loop), and ΔH1-ΔV3 (gp120 with truncated hairpin 1 and a fully truncated V3 loop). Only MAb 17b showed binding to the ΔH1 mutant and only in the presence of sCD4, provided that at least the base of the V3 loop was retained. Note that as sCD4 levels were detected by using a biotinylated anti-CD4 MAb and HRP-conjugated streptavidin (see Materials and Methods), a slightly different OD at 650 nm scale (y axis) is used. Although ELISAs are semiquantitative, the strict requirement for CD4 binding is considerable and statistically significant (P < 0.05), as indicated by an asterisk.
TABLE 2.
Summary of CD4i MAb binding to different gp120 mutantsa
| Protein | Binding |
||||
|---|---|---|---|---|---|
| 17b | 21c | CG10 | 19e | N12-i15 | |
| wt | |||||
| Absence of sCD4 | + | + | − | − | − |
| Presence of sCD4 | + | + | + | + | + |
| V3 base | |||||
| Absence of sCD4 | + | + | − | − | − |
| Presence of sCD4 | + | + | + | + | + |
| ΔV3 | |||||
| Absence of sCD4 | − | − | − | − | − |
| Presence of sCD4 | + | + | + | + | + |
| ΔV12 | |||||
| Absence of sCD4 | + | − | − | − | − |
| Presence of sCD4 | + | + | + | + | − |
| ΔV1 | |||||
| Absence of sCD4 | + | + | − | − | − |
| Presence of sCD4 | + | + | + | + | + |
| V2 core | |||||
| Absence of sCD4 | + | + | − | − | − |
| Presence of sCD4 | + | + | + | + | + |
| ΔV2 | |||||
| Absence of sCD4 | + | +* | − | − | − |
| Presence of sCD4 | + | + | + | + | − |
| ΔH1 | |||||
| Absence of sCD4 | − | − | − | − | − |
| Presence of sCD4 | + | − | − | − | − |
| ΔH1-V3 base | |||||
| Absence of sCD4 | − | − | − | − | − |
| Presence of sCD4 | + | − | − | − | − |
| ΔH1-ΔV3 | |||||
| Absence of sCD4 | − | − | − | − | − |
| Presence of sCD4 | − | − | − | − | − |
| YU2 coree | |||||
| Absence of sCD4 | + | − | − | − | − |
| Presence of sCD4 | + | − | + | − | − |
Results for binding in the absence or presence of sCD4 are shown. The asterisk denotes binding at ∼50% of binding to wt gp120.
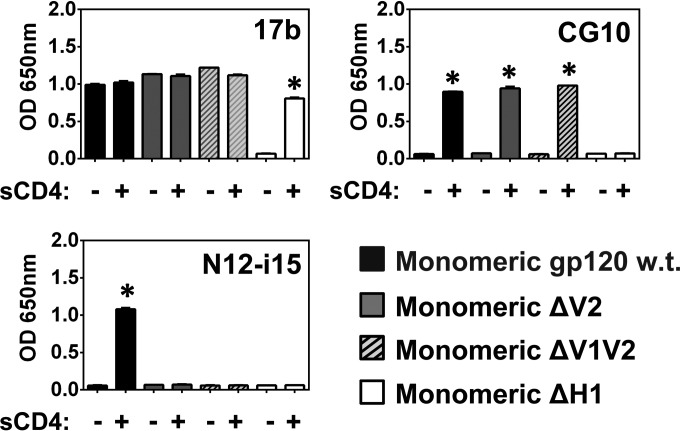
Binding activities of purified monomeric gp120 and its mutations.
The formation of oligomeric forms of gp120 in transfected HEK 293T cells due to aberrant disulfide bridges is well documented (55, 58, 79–81). Oligomerization can result specifically from rearranged disulfides in the V2 loop region. Hence, we tested the CD4 dependence of binding of MAbs 17b, CG10, and N12-i15 that define CD4i epitopes associated with hairpin 2, the bridging sheet, and the V2 loop, respectively, using FPLC-purified monomeric gp120 in addition to 4 relevant mutations. As shown in Fig. 10, MAb 17b binds to wt BaL purified monomeric gp120 in the absence of CD4 as do the ΔV2 and ΔV1V2 truncations, with little improvement in binding with the addition of CD4. However, as illustrated in Fig. 9, the purified monomer of ΔH1 binds 17b in a stringent CD4-dependent manner. The bridging sheet defining stringent MAb CG10 is totally dependent on the presence of CD4 for binding purified monomeric wt gp120 or the ΔV1V2 and ΔV2 truncations. The addition of CD4 to the ΔH1 monomer does not support CG10 recognition. Finally MAb N12-i15 is also stringently dependent on CD4 binding for recognition of WT monomeric gp120; however, no binding is demonstrated for any of the truncations missing the V2 loop, as illustrated in Fig. 7 and 9.
FIG 10.
Binding of MAbs 17b, CG10, and N12-i15 to FPLC-purified monomeric gp120. The gp120 monomers of the truncations described in the legends of Fig. 3B, 7, and 9 were purified by FPLC and tested by an ELISA for MAb binding in the presence and absence of CD4, as indicated. Binding to purified monomeric full-length BaL gp120 is given for comparison. Patterns of binding to monomeric gp120 were identical to the patterns of binding to mixed monomeric and oligomeric preparations. Statistically significant differences (P < 0.05) between the “−sCD4” and “+sCD4” columns are marked with an asterisk.
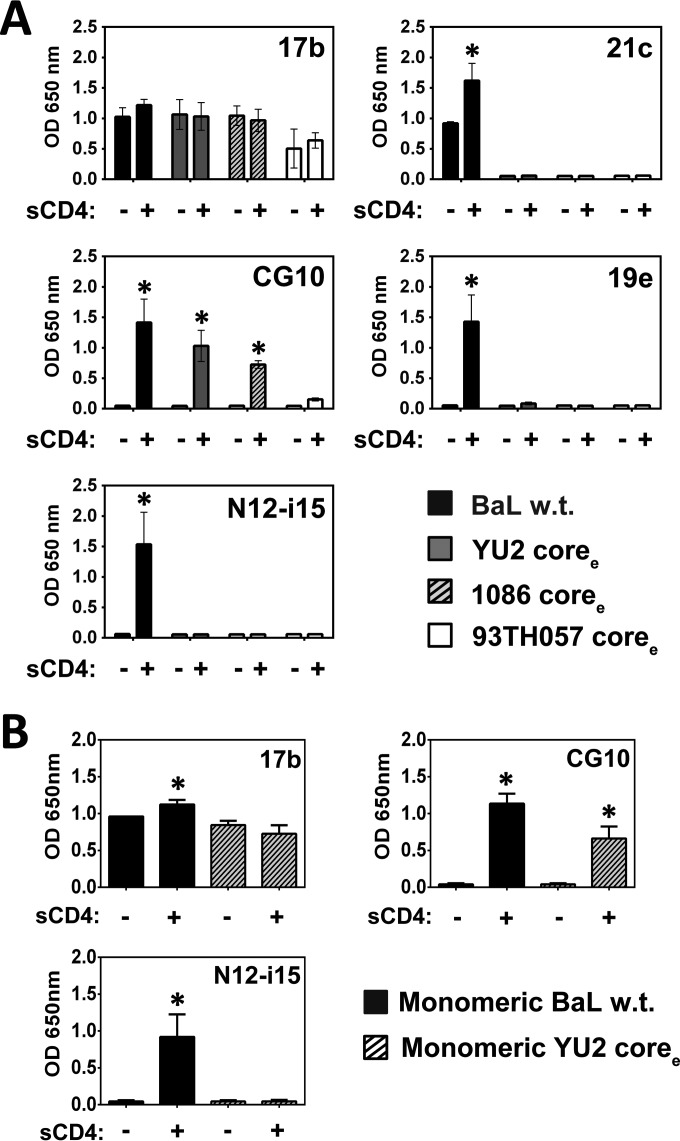
Extended core gp120s do not present stringent CD4i epitopes.
The coree constructs produced and characterized previously by Kwon et al. (24) have been proposed to represent the CD4-bound conformation. These constructs contain most of the N and C termini and only the V3 loop base, and hairpin 1 is truncated just below the cysteine disulfide (C122-C197), thus replacing the V1 and V2 loops with a short Gly-Gly linker. We expressed three representative coree structures, clade B YU2 coree, clade C C1086 coree, and clade E 93TH057 coree (kindly provided by Y. D. Kwon and P. D. Kwong), and tested their binding to the five CD4i MAbs of the panel, in the presence and absence of CD4, the logic being that if the coree gp120s indeed represent the ultimate CD4-bound conformation, CD4i MAbs CG10 and 19e should bind them in the absence of sCD4. As shown in Fig. 11A, MAb 17b binds wt gp120 and the coree proteins equally well in the presence and absence of CD4 (with slightly lower binding to 93TH057 coree). CD4i MAbs 21c and 19e showed no binding, even in the presence of CD4, to any of the coree constructs, possibly as a result of the truncations introduced into the tip of hairpin 1 of the bridging sheet in the construction of the coree proteins (24). As expected, no binding was detected for MAb N12-i15, as the V1-V2 loops are missing. However, MAb CG10 bound to the coree constructs in a strictly CD4-dependent manner. This assay was repeated with the monomeric, FPLC-purified YU2 coree protein, with similar results (Fig. 11B).
FIG 11.
MAb CG10 retains stringent binding to coree gp120s. A binding ELISA was performed to test whether the CD4i MAbs bind the different coree gp120s. (A) MAb 17b binds all of the coree gp120s with or without sCD4, while MAbs 21c and 19e do not bind coree gp120s at all. MAb CG10 retained stringent binding to the coree gp120s. (B) A binding ELISA was performed to test whether monomeric, FPLC-purified YU2 coree protein bound the CD4i MAbs similarly to the mixed monomeric and oligomeric preparations shown in panel A. Binding to purified monomeric full-length BaL gp120 is given for comparison. Patterns of binding to the CD4i MAbs tested persisted. Statistically significant differences (P < 0.05) between the “−sCD4” and “+sCD4” columns are marked with an asterisk.
DISCUSSION
HIV mediates target cell infection through binding of 2 to 3 spikes to cell surface CD4 (82). This constitutes the first step in the dynamic process in which structural elements of the envelope shift and rearrange to gain function, ultimately leading to gp41-mediated membrane fusion and the introduction of the viral nucleocapsid into the cytoplasm of target cells (83). A key player in this process is gp120, whose outer domain directly binds CD4 and undergoes a range of conformational transitions. One way to investigate such dynamic processes is to use conditional probes, probes that can discriminate between the various transition states of a given target. The range of CD4i conformational transitions within gp120 was therefore systematically interrogated by using a panel of well-characterized conditional probes, the CD4i MAbs, and led to a number of conclusions, as discussed below.
MAb 17b has often been taken as a gold standard indicator for the CD4-bound conformation (11, 24, 39, 62, 69–76). This MAb was originally described by Thali et al., who recognized that native cell surface HIV-1 spike was absolutely dependent on CD4 for 17b binding, in contrast, however, to the unrestricted ability of this MAb to bind monomeric gp120 in the absence of CD4 (38).
Here we report that MAb 17b binds the ΔH1 mutant complexed with CD4. This is surprising, as hairpin 1 constitutes half of the bridging sheet and contributes 8 out of 18 contact residues for 17b (with the other 10 contact residues residing at the base of hairpin 2 [18]). Nonetheless, gp120 devoid of hairpin 1 binds MAb 17b robustly albeit in a strictly CD4-dependent manner. These results point to the unique role and conformation of hairpin 2 (discussed below). This structural feature assumes the required conformation recognized by MAb 17b independent of CD4 binding when the gp120 monomer dissociates from the trimeric spike. Furthermore, two conditions under which 17b binding to monomeric gp120 becomes stringently dependent on CD4 binding have been found: (i) truncation of the V3 loop base and (ii) removal of hairpin 1. In gp120, the β2 strand of hairpin 1 aligns with the descending flank of hairpin 2 (β21), forming a series of hydrogen bonds. When this stabilizing interaction with hairpin 1 is maintained, CD4 binding can compensate for the absence of a V3 loop base. Removal of hairpin 1 can be compensated for by CD4 binding so long as at least the V3 loop base is in place.
What makes the V3 loop base so critical for 17b binding? This might be explained by the fact that the V3 loop naturally splays the two strands at the base of hairpin 2. Without this splaying effect, the overall orientation of hairpin 2 becomes distorted and unrecognized by 17b. In the absence of the V3 loop base, CD4 still binds gp120 and restores the required conformation of hairpin 2 (18), thus gaining 17b recognition. This hypothesis is supported by Zhou and collaborators, who report that tethering of the bridging sheet hairpin 2 to the inner domain, in approximation of the CD4i conformation, reduced the entropy of interactions with CD4 by 60% and increased the on-rate and affinity of MAb 17b in the absence of CD4 (84). In conclusion, a critical aspect for a CD4-bound conformation is a functional hairpin 2, a feature that is measured by 17b recognition.
The physical juxtaposition of hairpins 1 and 2, via the alignment of β2 with β21, is the essence of a bridging sheet, a feature that can be detected by MAb 21c. In contrast to 17b, MAb 21c requires intimate contacts with the extended aspects of hairpin 1 in addition to residues of hairpin 2. The ΔH1 mutation lacks these essential elements of the 21c epitope, which obviously cannot be restored by CD4; hence, no 21c binding is measured.
Critical evaluation of the essential elements of the V2 loop and how they are involved in CD4i transitions of gp120 was accomplished by studying MAb N12-i15. The stringent dependence of N12-i15 binding on CD4 and the V2 loop delineates a second measureable aspect of the CD4i conformation of gp120. The lack of N12-i15 competition against the other MAbs of the described panel emphasizes the uniqueness of this structural feature of the CD4-bound conformation.
Finally, whereas it has been proposed that the coree structures reported by Kwon et al. (24) represent the default CD4-bound conformation of monomeric gp120, our results clearly indicate that this is not the full picture. The stringent MAb CG10 binds to coree structures but only in complex with CD4. This clearly illustrates that there are still structural features lacking in coree that can be further induced upon association with CD4. This assertion is further strengthened by a recent single-molecule fluorescence resonance energy transfer (smFRET) study which showed that the viral spike can undergo transitions between three distinct conformations (14). The authors of that study assigned the first of these to the “closed” conformation and the second to the “CD4-bound” conformation seen in the tripartite gp120-CD4-17b crystal structures, while the third conformation could not be assigned a defined structure but was also favored after the addition of CD4. Interestingly, the viral spikes could sample all three conformations even when unliganded and continued to fluctuate between the two CD4-associated conformations after the addition of CD4, with conformational dynamics being strain specific. This multiplicity of conformations associated with binding by CD4 clearly shows that more work is needed to clarify their exact structural nature. Here, our analyses of the CD4i stringent MAbs may provide specific probes to better understand and scrutinize various gp120 transitional states.
Taking these results together, along with the crystal structures of CD4-unliganded trimeric gp140 and the CD4-bound gp120 core, we postulate the following sequence of conformational transitions that are triggered upon CD4 binding (illustrated in Fig. 12):
Insertion of CD4 into its binding pocket in gp120 drives residue Phe43 of CD4 forward, pushing the tip of hairpin 2 inwards and downwards (this transition brings the tip of hairpin 2 closer to the base of the V3 loop by 5 Å, as shown in Fig. 12).
The shift and reorientation of strand β21 disrupt hydrogen bonding with strand β3.
This allows hairpin 1 to fully extend, flipping the V1/V2 loops outward.
Repositioning of the V1/V2 loops involves twisting of hairpin 1, thereby displacing β3 in favor of β2, which can then form hydrogen bonds with β21, thus stabilizing a complete bridging sheet.
The newly formed bridging sheet with the central β2-β21 antiparallel orientation becomes exposed and accessible for coreceptor binding.
FIG 12.
Comparison of the gp120 bridging sheet in the CD4-unliganded and CD4-bound conformations. The structures of the bridging sheet elements in the crystal of CD4-unliganded trimer (PDB accession number 4NCO) (A) and the crystal of CD4-bound gp120 core (PDB accession number 1G9M) (B) were compared. Hairpin 1 (β2 [blue]-β3 [gray]), hairpin 2 (β20-β21 [green]), and the orientation of the V1/V2 loops are indicated (V1/V2 loops are not shown in the unliganded crystal and are truncated in the CD4-bound crystal). Residue Phe43 of CD4 is also shown (schematically in the unliganded crystal). The distance between carbon α of Trp422 in the tip of hairpin 2 and carbon α of Cys297 at the base of the V3 loop is also indicated (the cysteine residues at the base of the V3 loop are shown in space-fill for orientation). (A) In the unliganded crystal, β3 of hairpin 1 forms hydrogen bonds with β21 of hairpin 2, and the V1/V2 loops are oriented toward the trimer apex. (B) After binding to CD4, hairpin 1 twists and flips. Now, β2 of hairpin 1 forms hydrogen bonds with β21 of hairpin 2, and the V1/V2 loops are oriented away from the trimer apex. In addition, the distance between the tip of hairpin 2 and the base of the V3 loop is decreased by 5 Å.
Obviously, more defining MAbs combined with structural studies will further contribute to our understanding of these events and possibly provide insights in the quest for an AIDS vaccine. Despite many years of intense research, we are still far from achieving this goal. There seems to be increasing support for the idea that immunogens that are as close as possible to the natural trimeric conformation of HIV-1 envelope will provide a more relevant and hopefully potent vaccine (85, 86). Understanding the atomic details of envelope structures in the bound and free states is definitely important and extremely useful. Simpler epitope-based vaccines, accentuating neutralizing epitopes, may prove to be an efficient alternative for vaccine design. A better understanding of the dynamics of HIV-1 envelope interactions with its receptors, and in particular those of monomeric gp120, might contribute to the ultimate task of generating such an epitope-based vaccine for AIDS (87).
ACKNOWLEDGMENTS
This research was supported by grants from the Israel Science Foundation (J.M.G.), the Bill and Melinda Gates Foundation (grant OPP1033109) (G.K.L.), the National Institutes of Health (grant R01 AI087181) (G.K.L), and the Frankel Foundation and a donation by Peter Kraus. J.M.G. is the incumbent of the David Furman Chair of Immunobiology in Cancer. G.K. is the recipient of the Jakov, Miriana, and Jorge Saia doctoral fellowship. A.R.B. received the Dan David doctoral fellowship.
We acknowledge Yongjun Guan, James Robinson, and Ron Diskin for provision of antibodies; John Moore for providing the BG505 monomer and the BG505 SOSIP trimer; Young Do Kwon and Peter D. Kwong for providing the gp120 cores; and Itai Benhar and Limor Nahary for their assistance.
REFERENCES
- 1.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 2.McDougal JS, Mawle A, Cort SP, Nicholson JK, Cross GD, Scheppler-Campbell JA, Hicks D, Sligh J. 1985. Cellular tropism of the human retrovirus HTLV-III/LAV. I. Role of T cell activation and expression of the T4 antigen. J Immunol 135:3151–3162. [PubMed] [Google Scholar]
- 3.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 5.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 6.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. doi: 10.1016/S0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 7.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 8.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158. doi: 10.1016/S0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 9.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 10.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 11.Dey B, Pancera M, Svehla K, Shu Y, Xiang SH, Vainshtein J, Li Y, Sodroski J, Kwong PD, Mascola JR, Wyatt R. 2007. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J Virol 81:5579–5593. doi: 10.1128/JVI.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. 2009. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog 5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong L, Huang CC, Coales SJ, Molnar KS, Skinner J, Hamuro Y, Kwong PD. 2010. Local conformational stability of HIV-1 gp120 in unliganded and CD4-bound states as defined by amide hydrogen/deuterium exchange. J Virol 84:10311–10321. doi: 10.1128/JVI.00688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munro JB, Mothes W. 2015. Structure and dynamics of the native HIV-1 Env trimer. J Virol 89:5752–5755. doi: 10.1128/JVI.03187-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward AB, Wilson IA. 2015. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci 40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 20.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329–1339. doi: 10.1016/S0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 21.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, Julien JP, Burton DR, Wilson IA, Sanders RW, Klasse PJ, Ward AB, Moore JP. 2015. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol 89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diskin R, Marcovecchio PM, Bjorkman PJ. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol 17:608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. 2013. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger EA. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl A):S3–S16. [PubMed] [Google Scholar]
- 27.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci U S A 108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog 8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Julien JP, Lee JH, Ozorowski G, Hua Y, Torrents de la Pena A, de Taeye SW, Nieusma T, Cupo A, Yasmeen A, Golabek M, Pugach P, Klasse PJ, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci U S A 112:11947–11952. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeVico AL, Rahman R, Welch J, Crowley R, Lusso P, Sarngadharan MG, Pal R. 1995. Monoclonal antibodies raised against covalently crosslinked complexes of human immunodeficiency virus type 1 gp120 and CD4 receptor identify a novel complex-dependent epitope on gp 120. Virology 211:583–588. doi: 10.1006/viro.1995.1441. [DOI] [PubMed] [Google Scholar]
- 34.Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J Virol 85:7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gohain N, Tolbert WD, Acharya P, Yu L, Liu T, Zhao P, Orlandi C, Visciano ML, Kamin-Lewis R, Sajadi MM, Martin L, Robinson JE, Kwong PD, DeVico AL, Ray K, Lewis GK, Pazgier M. 2015. Cocrystal structures of antibody N60-i3 and antibody JR4 in complex with gp120 define more cluster A epitopes involved in effective antibody-dependent effector function against HIV-1. J Virol 89:8840–8854. doi: 10.1128/JVI.01232-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis GK, Finzi A, DeVico AL, Pazgier M. 2015. Conformational masking and receptor-dependent unmasking of highly conserved Env epitopes recognized by non-neutralizing antibodies that mediate potent ADCC against HIV-1. Viruses 7:5115–5132. doi: 10.3390/v7092856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, Pazgier M, Schon A, Freire E, Routy JP, Smith AB III, Park J, Jones DM, Courter JR, Melillo BN, Kaufmann DE, Hahn BH, Permar SR, Haynes BF, Madani N, Sodroski JG, Finzi A. 2015. CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc Natl Acad Sci U S A 112:E2687–E2694. doi: 10.1073/pnas.1506755112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses 18:1207–1217. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- 40.Zhang PF, Cham F, Dong M, Choudhary A, Bouma P, Zhang Z, Shao Y, Feng YR, Wang L, Mathy N, Voss G, Broder CC, Quinnan GV Jr. 2007. Extensively cross-reactive anti-HIV-1 neutralizing antibodies induced by gp140 immunization. Proc Natl Acad Sci U S A 104:10193–10198. doi: 10.1073/pnas.0608635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan Y, Pazgier M, Sajadi MM, Kamin-Lewis R, Al-Darmarki S, Flinko R, Lovo E, Wu X, Robinson JE, Seaman MS, Fouts TR, Gallo RC, Devico AL, Lewis GK. 2013. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc Natl Acad Sci U S A 110:E69–E78. doi: 10.1073/pnas.1217609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis GK, Fouts TR, Ibrahim S, Taylor BM, Salkar R, Guan Y, Kamin-Lewis R, Robinson JE, Devico AL. 2011. Identification and characterization of an immunogenic hybrid epitope formed by both HIV gp120 and human CD4 proteins. J Virol 85:13097–13104. doi: 10.1128/JVI.05072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denisova G, Stern B, Raviv D, Zwickel J, Smorodinsky NI, Gershoni JM. 1996. Humoral immune response to immunocomplexed HIV envelope glycoprotein 120. AIDS Res Hum Retroviruses 12:901–909. doi: 10.1089/aid.1996.12.901. [DOI] [PubMed] [Google Scholar]
- 45.Enshell-Seijffers D, Denisov D, Groisman B, Smelyanski L, Meyuhas R, Gross G, Denisova G, Gershoni JM. 2003. The mapping and reconstitution of a conformational discontinuous B-cell epitope of HIV-1. J Mol Biol 334:87–101. doi: 10.1016/j.jmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Gershoni JM, Denisova G, Raviv D, Smorodinsky NI, Buyaner D. 1993. HIV binding to its receptor creates specific epitopes for the CD4/gp120 complex. FASEB J 7:1185–1187. [DOI] [PubMed] [Google Scholar]
- 47.Mazor Y, Barnea I, Keydar I, Benhar I. 2007. Antibody internalization studied using a novel IgG binding toxin fusion. J Immunol Methods 321:41–59. doi: 10.1016/j.jim.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Roitburd-Berman A, Dela G, Kaplan G, Lewis GK, Gershoni JM. 2013. Allosteric induction of the CD4-bound conformation of HIV-1 Gp120. Retrovirology 10:147. doi: 10.1186/1742-4690-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gershoni JM, Lapidot M, Zakai N, Loyter A. 1986. Protein blot analysis of virus receptors: identification and characterization of the Sendai virus receptor. Biochim Biophys Acta 856:19–26. doi: 10.1016/0005-2736(86)90004-0. [DOI] [PubMed] [Google Scholar]
- 50.Burton DR, Barbas CF III, Persson MA, Koenig S, Chanock RM, Lerner RA. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A 88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Stiehm ES, Bryson YJ, Cao Y, Moore JP, Ho DD, Barbas CF. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 52.Zhang MY, Xiao X, Sidorov IA, Choudhry V, Cham F, Zhang PF, Bouma P, Zwick M, Choudhary A, Montefiori DC, Broder CC, Burton DR, Quinnan GV Jr, Dimitrov DS. 2004. Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody. J Virol 78:9233–9242. doi: 10.1128/JVI.78.17.9233-9242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roben P, Moore JP, Thali M, Sodroski J, Barbas CF III, Burton DR. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol 68:4821–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Center RJ, Earl PL, Lebowitz J, Schuck P, Moss B. 2000. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J Virol 74:4448–4455. doi: 10.1128/JVI.74.10.4448-4455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coutu M, Finzi A. 2015. HIV-1 gp120 dimers decrease the overall affinity of gp120 preparations for CD4-induced ligands. J Virol Methods 215–216:37–44. doi: 10.1016/j.jviromet.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 56.Doms RW, Earl PL, Moss B. 1991. The assembly of the HIV-1 env glycoprotein into dimers and tetramers. Adv Exp Med Biol 300:203–219; discussion 220–201. doi: 10.1007/978-1-4684-5976-0_13. [DOI] [PubMed] [Google Scholar]
- 57.Earl PL, Doms RW, Moss B. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci U S A 87:648–652. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finzi A, Pacheco B, Zeng X, Kwon YD, Kwong PD, Sodroski J. 2010. Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J Virol Methods 168:155–161. doi: 10.1016/j.jviromet.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hallenberger S, Tucker SP, Owens RJ, Bernstein HB, Compans RW. 1993. Secretion of a truncated form of the human immunodeficiency virus type 1 envelope glycoprotein. Virology 193:510–514. doi: 10.1006/viro.1993.1156. [DOI] [PubMed] [Google Scholar]
- 60.Owens RJ, Compans RW. 1990. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology 179:827–833. doi: 10.1016/0042-6822(90)90151-G. [DOI] [PubMed] [Google Scholar]
- 61.Schawaller M, Smith GE, Skehel JJ, Wiley DC. 1989. Studies with crosslinking reagents on the oligomeric structure of the env glycoprotein of HIV. Virology 172:367–369. doi: 10.1016/0042-6822(89)90142-6. [DOI] [PubMed] [Google Scholar]
- 62.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol 72:4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas DJ, Wall JS, Hainfeld JF, Kaczorek M, Booy FP, Trus BL, Eiserling FA, Steven AC. 1991. gp160, the envelope glycoprotein of human immunodeficiency virus type 1, is a dimer of 125-kilodalton subunits stabilized through interactions between their gp41 domains. J Virol 65:3797–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guan Y, Sajadi MM, Kamin-Lewis R, Fouts TR, Dimitrov A, Zhang Z, Redfield RR, DeVico AL, Gallo RC, Lewis GK. 2009. Discordant memory B cell and circulating anti-Env antibody responses in HIV-1 infection. Proc Natl Acad Sci U S A 106:3952–3957. doi: 10.1073/pnas.0813392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acharya P, Tolbert WD, Gohain N, Wu X, Yu L, Liu T, Huang W, Huang CC, Kwon YD, Louder RK, Luongo TS, McLellan JS, Pancera M, Yang Y, Zhang B, Flinko R, Foulke JS Jr, Sajadi MM, Kamin-Lewis R, Robinson JE, Martin L, Kwong PD, Guan Y, DeVico AL, Lewis GK, Pazgier M. 2014. Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J Virol 88:12895–12906. doi: 10.1128/JVI.02194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV Jr. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol 76:644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Young KR, Teal BE, Brooks Y, Green TD, Bower JF, Ross TM. 2004. Unique V3 loop sequence derived from the R2 strain of HIV-type 1 elicits broad neutralizing antibodies. AIDS Res Hum Retroviruses 20:1259–1268. doi: 10.1089/aid.2004.20.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol 76:7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB III, Sodroski J, Freire E. 2006. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan W, Bazick J, Sodroski J. 2006. Characterization of the multiple conformational states of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J Virol 80:6725–6737. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. 1999. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry 38:9405–9416. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- 73.Martin G, Burke B, Thai R, Dey AK, Combes O, Ramos OH, Heyd B, Geonnotti AR, Montefiori DC, Kan E, Lian Y, Sun Y, Abache T, Ulmer JB, Madaoui H, Guerois R, Barnett SW, Srivastava IK, Kessler P, Martin L. 2011. Stabilization of HIV-1 envelope in the CD4-bound conformation through specific cross-linking of a CD4 mimetic. J Biol Chem 286:21706–21716. doi: 10.1074/jbc.M111.232272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol 76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol 69:5723–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fouts TR, Tuskan R, Godfrey K, Reitz M, Hone D, Lewis GK, DeVico AL. 2000. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J Virol 74:11427–11436. doi: 10.1128/JVI.74.24.11427-11436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee S, Peden K, Dimitrov DS, Broder CC, Manischewitz J, Denisova G, Gershoni JM, Golding H. 1997. Enhancement of human immunodeficiency virus type 1 envelope-mediated fusion by a CD4-gp120 complex-specific monoclonal antibody. J Virol 71:6037–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Go EP, Cupo A, Ringe R, Pugach P, Moore JP, Desaire H. 30 December 2015. Native conformation and canonical disulfide bond formation are interlinked properties of HIV-1 Env glycoproteins. J Virol doi: 10.1128/JVI.01953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Go EP, Zhang Y, Menon S, Desaire H. 2011. Analysis of the disulfide bond arrangement of the HIV-1 envelope protein CON-S gp140 deltaCFI shows variability in the V1 and V2 regions. J Proteome Res 10:578–591. doi: 10.1021/pr100764a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kassa A, Dey AK, Sarkar P, Labranche C, Go EP, Clark DF, Sun Y, Nandi A, Hartog K, Desaire H, Montefiori D, Carfi A, Srivastava IK, Barnett SW. 2013. Stabilizing exposure of conserved epitopes by structure guided insertion of disulfide bond in HIV-1 envelope glycoprotein. PLoS One 8:e76139. doi: 10.1371/journal.pone.0076139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brandenberg OF, Magnus C, Rusert P, Regoes RR, Trkola A. 2015. Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry. PLoS Pathog 11:e1004595. doi: 10.1371/journal.ppat.1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prabakaran P, Dimitrov AS, Fouts TR, Dimitrov DS. 2007. Structure and function of the HIV envelope glycoprotein as entry mediator, vaccine immunogen, and target for inhibitors. Adv Pharmacol 55:33–97. doi: 10.1016/S1054-3589(07)55002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O'Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. 2015. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog 11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gershoni JM, Roitburd-Berman A, Siman-Tov DD, Tarnovitski Freund N, Weiss Y. 2007. Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs 21:145–156. doi: 10.2165/00063030-200721030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]